Abstract
Brome mosaic virus (BMV), a positive-strand RNA virus in the alphavirus-like superfamily, encodes two RNA replication proteins. The 1a protein has putative helicase and RNA-capping domains, whereas 2a contains a polymerase-like domain. Saccharomyces cerevisiae expressing 1a and 2a is capable of replicating a BMV RNA3 template produced by in vivo transcription of a DNA copy of RNA3. Although insufficient for RNA3 replication, the expression of 1a protein alone results in a dramatic and specific stabilization of the RNA3 template in yeast. As one step toward understanding 1a-induced stabilization of RNA3, the interactions involved, and its possible relation to RNA replication, we have identified the cis-acting sequences required for this effect. We find that 1a-induced stabilization is mediated by a 150- to 190-base segment of the RNA3 intergenic region corresponding to a previously identified enhancer of RNA3 replication. Moreover, this segment is sufficient to confer 1a-induced stability on a heterologous β-globin RNA. Within this intergenic segment, partial deletions that inhibited 1a-induced stabilization in yeast expressing 1a alone resulted in parallel decreases in the levels of negative- and positive-strand RNA3 replication products in yeast expressing 1a and 2a. In particular, a small deletion encompassing a motif corresponding to the box B element of RNA polymerase III promoters dramatically reduced the ability of RNAs to respond to 1a or 1a and 2a. These and other findings suggest that 1a-induced stabilization likely reflects an early template selection step in BMV RNA replication.
The alphavirus-like superfamily is a group of positive-strand RNA viruses that includes both animal and plant viruses (15). Although the members of this superfamily have different virion structures, hosts, and genome organizations, their nonstructural proteins have extensive similarity, suggesting common replication mechanisms. Brome mosaic virus (BMV), a member of the alphavirus-like superfamily, has a genome consisting of 5′-capped RNA1, RNA2, and RNA3 (for reviews, see references 1, 27 and 42). RNA1 and RNA2 encode the 1a and 2a proteins, respectively, which are required for RNA replication (12, 20, 22). The 109-kDa 1a protein has two major domains, one having sequence similarity to DEAD box helicases (16) and the other having similarity to alphavirus nsP1 proteins, which have been shown to have m7G methyltransferase and guanylyltransferase activities presumed to function in viral RNA capping (3, 28, 40). The 94-kDa 2a protein has sequence similarity to RNA-dependent RNA polymerases (5). In both plant cells (36) and yeast cells (35a) 1a and 2a colocalize in an endoplasmic reticulum-associated replication complex that is the site of BMV-specific RNA synthesis. RNA3 is a dicistronic RNA whose protein products are dispensable for RNA replication but required for productive infection (4). The 5′ gene encodes the 32-kDa 3a protein, which is essential for cell-to-cell movement of bromovirus infection (4, 30). The 3′ gene encodes the coat protein, which is required for encapsidation and long-range vascular spread of the virus (4, 29a, 38). The coat protein gene is expressed by synthesis of a subgenomic mRNA, RNA4, from the negative-strand RNA3 replication intermediate.
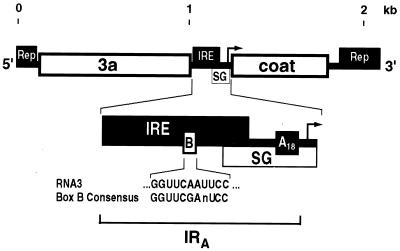
In vitro and in vivo studies have identified 5′, 3′, and intergenic sequences within RNA3 that are required for efficient RNA replication and subgenomic RNA synthesis (Fig. 1) (10, 11, 25, 29). The 250-base intergenic region plays an important role in both processes, containing the promoter for subgenomic mRNA synthesis and sequences involved in RNA3 replication (Fig. 1). While the conserved BMV tRNA-like 3′ end functions as a minimal negative-strand promoter in vitro (29), experiments with Saccharomyces cerevisiae expressing 1a, 2a, and RNA3 templates demonstrated that sequences within the intergenic region of RNA3 stimulate negative-strand synthesis in vivo approximately 100-fold (35). The same study showed that intergenic sequences play a role in in vivo assembly of a functional, isolatable RNA replication complex. A 150- to 200-base subset of the intergenic region, the intergenic replication enhancer (IRE), was implicated in RNA replication in plant protoplasts (11). A striking feature of this segment is a motif matching the box B sequence of RNA polymerase III promoters and thus also the conserved TΨC loop of tRNAs (11, 26). Deletion of the box B element impairs RNA3 replication (34, 41). The same box B motif is also found in the 5′ noncoding regions of BMV RNA1 and RNA2 (11).
FIG. 1.
Schematic diagram of BMV RNA3 showing cis-acting signals involved in RNA replication and subgenomic RNA4 synthesis. The 3a and coat protein ORFs are shown as open boxes. cis-acting signals required for RNA3 replication in vivo, including the IRE, are represented by solid boxes above RNA3 (Rep and IRE). cis-acting signals required for subgenomic mRNA synthesis (SG) in vivo are represented by an open box below RNA3. An expansion of the approximately 250-base RNA3 intergenic region is shown, indicating the position of the oligo(A) tract and box B motif, which is shown compared to the cellular RNA polymerase III promoter box B consensus sequence. IRA (indicated with a bracket) is a 210-base segment of the intergenic region from the 3a coding region up to and including the oligo(A) tract that was used in several experiments.
Recently, it has been shown that in yeast, in the absence of 2a and hence also of RNA replication, 1a dramatically and selectively increases the stability and accumulation of a DNA-derived RNA3 transcript (20). However, increased levels of RNA3 did not lead to increased RNA3 translation. These and other observations suggested that the 1a-RNA3 interaction leading to 1a-induced stabilization might be involved in directing RNA3 to replication rather than the potentially competing fates of translation, encapsidation, or degradation.
As one step toward understanding 1a-induced stabilization of RNA3, the interactions involved, and its possible relation to RNA replication, we have identified the cis-acting sequences required for this effect. We show here that 1a-induced stabilization is mediated by a 150- to 190-base segment of the RNA3 5′-proximal intergenic region corresponding to the IRE and that these sequences are sufficient to direct 1a-induced stabilization of heterologous RNAs. Partial intergenic deletions that inhibited 1a-induced stabilization in cis in yeast expressing 1a alone similarly inhibited in cis the accumulation of negative- and positive-strand RNA3 replication products in yeast expressing 1a and 2a, further supporting a close link between 1a-induced RNA3 stabilization and RNA3 replication.
MATERIALS AND METHODS
Yeast strain, cell growth, and transformation.
Yeast strain YPH500 (MATα ura3-52 lys2-801 ade2-101 tyr1-Δ63 his3-Δ200 leu2-Δ1) was used throughout. Yeast cultures were grown at 30°C in defined synthetic medium containing 2% glucose or 2% galactose (6). Relevant amino acids were omitted to maintain selection for any plasmids present. Plasmid transformation was by the LiOAc-polyethylene glycol method (19).
Plasmids and plasmid constructions.
Standard procedures were used for all DNA manipulations (39). The sequences of PCR-generated DNA fragments were confirmed by automated DNA sequencing, and the overall structures of all plasmids were confirmed by restriction analysis.
The 1a expression cassette of pB1CT19 (21) (ADH1 promoter-1a open reading frame [ORF]-ADH1 polyadenylation signal) was inserted into the EcoRV site of pRS423, a yeast 2μm plasmid that contains a HIS3 selectable marker (8) to generate pB1MS6. This allowed the use of a matching control plasmid lacking the 1a cassette (pRS423) in control yeast strains without 1a. BMV 2a protein was expressed from pB2CT15 (21), a yeast 2μm plasmid that contains a LEU2 selectable marker.
All plasmids expressing RNAs tested for 1a-induced stabilization were based on YCplac22, a yeast CEN4 centromeric plasmid containing a TRP1 selectable marker and the multiple-cloning site from pUC19 (14). RNA names used in the text and figures in this paper are indicated in parentheses following plasmid names. Transcripts were expressed by using a GAL1 promoter fragment derived from pMII93 (18), which has a SnaBI site at the transcription start site. Wild-type RNA3 plasmid pB3 (laboratory designation, pB3RQ39) (18) and a number of additional plasmids described below also include a hepatitis delta ribozyme for 3′ end formation.
(i) Intermediate plasmids.
Several intermediate plasmids facilitated construction of plasmids expressing RNA3 derivatives. pPG948, a pBluescript II SK(+) (Stratagene) derivative whose polylinker has been modified to SacI-BglII-XbaI-EcoRV-BamHI-ClaI, and pPG977, which contains a human β-globin ORF (31), were the generous gifts of Pam Green. pB3TP10 is a BMV RNA3 clone with BamHI and BglII sites introduced by site-directed mutagenesis immediately preceding and following the 3a ORF, respectively (32). pB3MJ23 (20) is a pB3 derivative with the pB3TP10 BamHI site. pADH1 was made by inserting a T4 DNA polymerase-treated 0.5-kb HindIII-BamHI fragment from pB1CT19 (21) containing the ADH1 polyadenylation signal into the EcoRV site of pPG948. pADH1 had the orientation BglII-5′ ADH1 3′-BamHI. pIR was made by inserting a mung bean nuclease-treated 0.2-kb BglII fragment from pB3TP10, corresponding to a 210-base segment of the RNA3 intergenic region, into the EcoRV site of pPG948. pIR had the orientation BglII-5′ intergenic region segment 3′-BamHI. pMS97 is pBluescript II SK(−) containing a 0.8-kb EcoRI-PstI GAL1 promoter and leader fragment from pB2YT2 (the generous gift of Y. Tomita and M. Ishikawa). A PstI site was introduced immediately upstream of the β-globin ORF of pPG977 by PCR with the primers d(GCGGCTGCAGTTATAATGGTGCACCTGACTCC) and d(GCGGGATCCGATTCGAGGTC); the resulting 0.5-kb fragment was digested with PstI and BamHI and inserted into pMS97 digested with PstI and BamHI to generate pMS98. pMS130, a yeast shuttle vector containing the GAL1 promoter, leader, and β-globin ORF, was made by sequentially inserting the 0.1-kb EcoRI-BamHI and 1.2-kb EcoRI fragments of pMS98 into YCplac22.
(ii) RNA3 deletion plasmids.
pB3Δ3′ (in vivo transcription plasmid for ShRNA3; laboratory designation, pB3MS13) is a 1-kb BglII-BamHI deletion derivative of pB3 that removes all RNA3 sequences 3′ of the intergenic oligo(A) tract through the hepatitis delta ribozyme. pB3MS63 (3′RNA3) has a 1.0-kb BglII (T4 DNA polymerase-treated)-BamHI fragment from pB3 containing the RNA3 coat protein ORF, RNA3 3′ untranslated region (UTR), and hepatitis delta ribozyme replacing the 2.2-kb SnaBI-BamHI fragment of pB3. pB3MS89 (fs-3′RNA3) is similar to pB3MS63 except that (i) the second in-frame ATG has been replaced by ATC by replacing the SalI-BssHI fragment in the coat ORF with the complementary synthetic oligonucleotides d(TCGACTTCAGGAACTGGTAAGATCACTCG) and d(CGCGCGAGTGATCTTACCAGTTCCTGAAG) and (ii) a frameshift mutation was introduced by digestion with SalI, treatment with T4 DNA polymerase, and religation. pB3MS60 (5′RNA3) was made by replacing the 1.0-kb BglII-BamHI fragment of pB3 with the 0.5-kb BglII-BamHI fragment of pADH1. pB3MS46 (RNA3ΔIRA) was made by replacing the 0.6-kb ClaI-BglII fragment of pB3 with the 0.4-kb ClaI-BglII fragment from pB3TP10, removing most of the intergenic region from RNA3. pB3MS45 (ΔIRA) was generated by replacing the 1.0-kb BglII-BamHI fragment of pB3MS46 with the 0.5-kb BglII-BamHI fragment of pADH1. pB3MS95 (Δ3a) was generated by replacing the 2.2-kb BamHI fragment of pB3MJ23 with the 0.2-kb BglII-BamHI fragment of pIR followed by insertion of the 0.5-kb BglII-BamHI fragment of pADH1 into the BamHI site of the resulting plasmid. pB3MS107 (Δ5′UTR) was generated by replacing the 0.6-kb SnaBI-ClaI fragment of pB3MS60 with the 0.5-kb BamHI (T4 DNA polymerase-treated)-ClaI fragment from pB3TP10. pB3MS108 (Δ5′UTRΔ3a) was generated by replacing the 2.2-kb SnaBI-BamHI fragment of pB3 with the 0.2-kb BglII-BamHI fragment from pIR followed by insertion of the 0.5-kb BglII-BamHI fragment of pADH1 into the BamHI site of the resulting plasmid.
(iii) β-Globin plasmids.
pMS72 (RNA3−IRA) and pMS73 (RNA3+IRA) were made by replacing the 2.2-kb BamHI fragment of pB3MJ23 with a 0.5-kb BglII-BamHI fragment from pPG977 containing the β-globin ORF and subsequent insertion of the 0.5-kb BglII-BamHI fragment from pADH1 (to generate pMS72) or sequential insertion of the 0.2- and 0.5-kb BglII-BamHI fragments from pIR and pADH1, respectively (to generate pMS73), into the BamHI site of the resulting plasmid. pMS99 (GAL−IRA) and pMS100 (GAL+IRA) were made by replacing the 1.2-kb EcoRI fragment (containing the GAL1 promoter, the RNA3 leader, and a portion of the β-globin ORF) from pMS72 and pMS73 with the analogous EcoRI fragment from pMS98. pMS110 (PGK−IRA) and pMS111 (PGK+IRA) were generated by replacing the 2.2-kb SnaBI-PstI fragment of pB3 with the complementary oligonucleotides d(AGTAATTATCTACTTTTTACAACAAATATAAAAACACTGCA) and (GTGTTTTTATATTTGTTGTAAAAAGTAGATAATTACT), corresponding to the PGK leader sequence, followed by insertion into the PstI site of the resulting plasmid of the 1.0-kb PstI fragment from pMS99 (to generate pMS110) or the 1.2-kb PstI fragment from pMS100 (to generate pMS111). pMS176 (None−IRA) and pMS177 (None+IRA) were generated by replacing the 2.2-kb SnaBI-BamHI fragment of pB3 with the 1.0- and 1.2-kb PstI (T4 DNA polymerase-treated)-BamHI fragments from pMS99 and pMS100, respectively.
(iv) Partial intergenic region deletions.
Deletions within the intergenic region were generated by PCR with pIR as a template. PCR products containing the various intergenic partial deletions were then cloned into pPG948 as XbaI-BamHI fragments. The 5′ deletions were generated with the T7 primer [d(GTAATACGACTCACTATAGGGC)] and either d(GGCCGGTCTAGAGATTAAGCAAGCTGGGGAGAC) (5′Δ32), d(GCTCTAGAGATGAGACCCCCGACAGCCG) (5′Δ45), or d(GGCCGGTCTAGAGATCTGCTCGTTTGGGTTCAATTC) (5′Δ79). The 3′ deletions were generated with the reverse primer [d(GGAAACAGCTATGACCATG)] and either d(CGGGATCCGATAATAATAACTCAGACACACAAC) (3′ΔA), d(CGGGATCCGATCACAAACATGGATAACCTCCCCCG) (3′Δ52), or d(CGGGATCCGATCCGAGGACCTATCTCAAC) (3′Δ73). A 14-base deletion encompassing box B (RNA3 bases 1101 to 1114) was generated by a two-step PCR procedure. In the first step, the pIR template was amplified with either the T7 primer (see above) and d(ACCTTACAACGGCGTGTTGAG) or the reverse primer (see above) and d(AACACGCCGTTGTAAGGTACGAGACGCGAGCGCTGATCC). The resulting fragments were then combined and reamplified with the T7 and reverse primers.
The intergenic region partial deletions described above were introduced into the β-globin reporter plasmids by inserting them as BglII-BamHI fragments into the BamHI site of pMS130, followed by insertion of the 0.5-kb BglII-BamHI fragment from pADH1 into the BamHI sites of the resulting plasmids. The same intergenic partial deletions were introduced into the RNA3 context by inserting them as BglII-BamHI fragments into the BglII site of pMS46.
RNA analysis.
Yeast strains expressing selected RNAs and 1a or 1a plus 2a proteins as indicated were first grown to saturation in synthetic medium containing galactose. To insure full induction, the yeast strains were then passaged twice more in fresh galactose medium, with the cells harvested each time in mid-logarithmic phase (optical density at 600 nm [OD600], 0.4 to 0.7). To analyze RNA accumulation, approximately 2 OD600 units of final mid-logarithmic culture were harvested by centrifugation and frozen on dry ice prior to extraction of total RNA as described previously (21). Polyadenylated RNA was isolated from total RNA with the PolyAT Tract System (Promega). To analyze the decay of RNAs transcribed from the galactose-inducible, glucose-repressible GAL1 promoter, the mid-logarithmic culture was harvested by centrifugation and resuspended at 1.0 OD600 unit/ml in fresh glucose-containing medium. At selected times following glucose addition, 2-ml samples of culture were removed, harvested by centrifugation, and frozen on dry ice prior to extraction of total RNA.
RNA blotting was performed as described previously (31), with formaldehyde-agarose gels and Nytran nylon membranes (Schleicher & Schuell). For RNA detection, 32P-labeled hybridization probes were generated by random priming from plasmid restriction fragment templates (9). Alternatively, strand-specific 32P-labeled RNA probes for positive- and negative-strand RNA3 were generated as described previously (20). Radioactive signals were detected and measured with a Molecular Dynamics PhosphorImager model 425 imaging system.
RESULTS
Coat gene and 3′ UTR are dispensable for 1a-induced RNA3 stabilization.
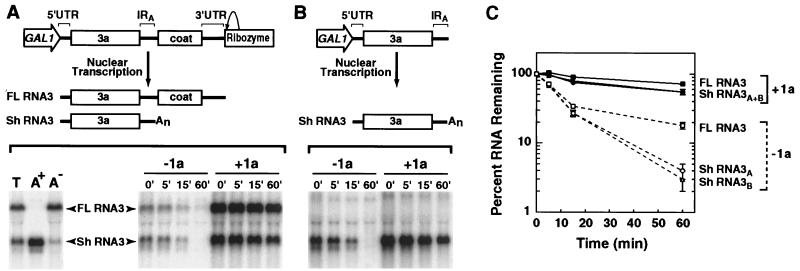
The 240-base RNA3 intergenic region (Fig. 1) contains the IRE, an 18-base oligo(A) tract followed by a BglII site, and the core promoter for subgenomic mRNA synthesis. Since many of the experiments described below use a large 5′ subset (210 bases) of the intergenic region extending from the 3′ end of the 3a gene to the BglII site immediately following the oligo(A) tract, this region is hereafter referred to as IRA (Fig. 1). pB3 is a plasmid using the galactose-inducible, glucose-repressible GAL1 promoter and a 3′ ribozyme to produce a replicatable RNA3 transcript in vivo (18). Mapping the cis-acting sequences required for 1a-induced RNA3 stabilization was serendipitously advanced by the discovery that pB3 produced not only full-length RNA3 but also a shorter transcript that, as shown below, is truncated at a fortuitous polyadenylation site in IRA. This 1.2-kb transcript was not detected previously with a probe complementary to the 3′ 200 bases of RNA3 (18, 20) but was detected together with full-length, 2.1-kb RNA3 by using a probe complementary to all of RNA3 (Fig. 2A, lane T).
FIG. 2.
1a-induced stabilization of RNA3 and a 3′-truncated RNA3 derivative. Plasmids expressing cDNA corresponding to full-length RNA3 or a truncated derivative were cotransformed into yeast with either a 1a-expressing plasmid or the corresponding empty vector, pRS423. The resulting yeast strains were analyzed as described in Materials and Methods. (A) The schematic at the top depicts the RNA3 expression cassette and its resulting full-length (FL) and short (Sh) transcripts. Relevant features of the RNA3 cDNA and the flanking GAL1 promoter and ribozyme sequences are indicated. After galactose induction of the GAL1 promoter, total RNA from yeast containing this plasmid was fractionated on oligo(dT) beads. Total (5 mg) (T), poly(A)-enriched (0.5 mg) (A+), and poly(A)-depleted (5 mg) (A−) RNA fractions were electrophoresed on an agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized to a 32P-labeled DNA probe complementary to the entire RNA3 cDNA (lower left). RNA stability in the absence (−1a) or presence (+1a) of 1a was assessed by glucose repression of RNA3 transcription. Equal amounts of RNA prepared from yeast harvested at the indicated times following glucose repression were analyzed by Northern blotting and hybridization as described above. A representative Northern blot is shown on the lower right. (B) The schematic at the top shows an RNA3 expression cassette, lacking sequences 3′ of the RNA3 IRA, and the transcript resulting from this cassette. The stability of this transcript in the absence (−1a) and presence (+1a) of 1a was assessed as described above. A representative Northern blot is shown below. (C) Radioactive signals from three to seven stability analyses of the RNAs depicted in panels A and B were measured with a PhosphorImager, normalized to starting RNA levels (100%), averaged, and plotted with error bars on a logarithmic scale versus time. Stability in the presence (+1a) or absence (−1a) of 1a is plotted with solid symbols and solid lines or open symbols and dashed lines, respectively. The error bars represent standard errors of the mean and are included for all points, but in some cases they are obscured by the symbols used to plot the average values.
Because the existence of a shortened form of RNA3 had potentially significant implications for this and many other studies utilizing pB3 derivatives, we further characterized this RNA. We found that the short RNA depended on galactose-induced, DNA-dependent transcription of pB3 but not on exression of the BMV 1a or 2a protein. Also, the short RNA was absent when a selectable RNA3 replicon (18, 21) was maintained by RNA replication in the absence of ongoing pB3 transcription. Thus, the short RNA was not a product of BMV RNA replication but of in vivo transcription of RNA3 cDNA. Moreover, probes complementary to the 3a ORF or IRA detected the short transcript whereas a probe complementary to the coat protein ORF did not.
From these results and studies showing that a variety of AU-rich sequences function as mRNA polyadenylation signals in yeast (17), we suspected that the short transcript was initiated by the GAL1 promoter but was terminated and polyadenylated in the AU-rich IRA. This was tested in two ways. First, total RNA isolated from galactose-induced, pB3-containing yeast was fractionated with oligo(dT) paramagnetic beads into poly(A)-enriched and -depleted fractions (Fig. 2A, bottom left panel). Despite an 18-base oligo(A) tract in the intergenic subgenomic RNA promoter (Fig. 1) (10), full-length RNA3 was not recovered in the poly(A)-enriched fraction, remaining instead in the poly(A)-depleted fraction. In contrast, the short transcript was greatly enriched in the poly(A) fraction, indicating that it possessed a poly(A) tail long enough to interact efficiently with the oligo(dT) beads and longer or more accessible than that in the intergenic region of full-length RNA3. Second, RNA3 sequences 3′ of the intergenic oligo(A) tract and the 3′ flanking ribozyme were deleted from pB3. As expected, the resulting plasmid, pB3Δ3′, produced a defined transcript that comigrated with the pB3 short transcript, despite the absence of an explicit 3′ polyadenylation signal or ribozyme (Fig. 2B).
Next, the in vivo stability of the short transcript was determined in the presence and absence of 1a. The in vivo stability of RNA3 transcribed from pB3 and its derivatives can be measured by rapidly repressing the GAL1 promoter with glucose and monitoring surviving RNA levels by Northern blotting. The RNA half-life (t1/2) can be determined from the slope of a plot of the surviving RNA level on a logarithmic scale against time after transcription repression. The results of such half-life analyses were highly reproducible. For each RNA stability test described in this paper, three to seven independent experiments were performed and averaged to generate the illustrated decay curves with standard error bars.
As shown in Fig. 2, the short RNAs derived from pB3 and pB3Δ3′ were indistinguishable in stability analyses and showed 1a-responsive behavior similar to that of full-length RNA3. In the absence of 1a, all three RNAs decayed rapidly, with an initial t1/2 of 8 to 9 min. For the short transcripts, this decay rate remained nearly constant through the period examined, so that only a few percent of the starting RNA survived after 60 min. For full-length RNA3, decay slowed after 80 to 90% of the initial full-length RNA3 had decayed. Similar prolonged survival of 10 to 20% of the starting RNA after 60 min has been seen for all BMV and hybrid RNAs containing the BMV tRNA-like 3′ end, which apparently stabilizes a small subpopulation of RNA even in the absence of 1a (Fig. 3 and 4) (7a).
FIG. 3.
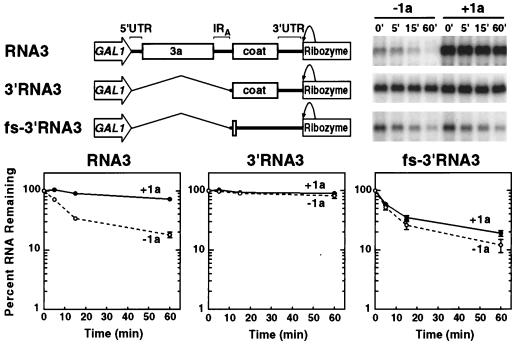
RNA3 sequences 3′ of the intergenic oligo(A) tract do not direct 1a-induced RNA stabilization. DNA expression cassettes for 3′RNA3, which lacks RNA3 sequences 5′ of and including the intergenic oligo(A) tract, and fs-3′RNA3, a frameshifted version of 3′RNA3 that does not produce coat protein, are diagrammed at top left. The stabilities of these RNAs in the absence (−1a) and presence (+1a) of 1a were analyzed by glucose repression as described in the legend to Fig. 2, and representative Northern blots are shown at the right. The results for full-length RNA3 are shown for comparison. The results of three independent stability analyses of each RNA were averaged and plotted with error bars, as in Fig. 2.
FIG. 4.
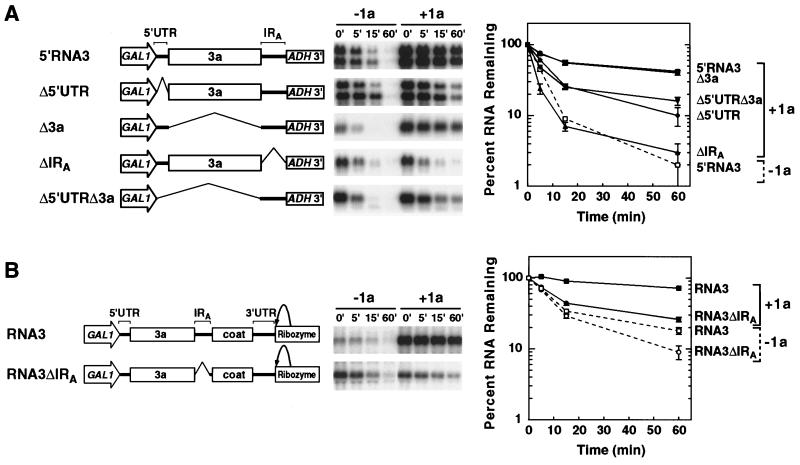
IRA requirement for 1a-induced stabilization of RNA3. The stabilities of the indicated RNA3 deletion derivatives in the absence (−1a) and presence (+1a) of 1a were analyzed by glucose repression as described in the legend to Fig. 2. DNA expression cassettes used to generate the RNAs are diagrammed to the left of representative Northern blots. GAL1 promoter, RNA3 sequences, ADH1 polyadenylation signal, and ribozyme sequences are indicated. Deletions are represented as bent lines. For each RNA, the results of three independent stability analyses were averaged and are plotted to the right of the Northern blots, as in Fig. 2. (A) Stability assays of 5′RNA3, Δ5′UTR, Δ3a, ΔIRA, and Δ5′UTRΔ3a. For 5′RNA3 and Δ5′UTR, for which the long and short transcripts behaved similarly, results are plotted only for the longer transcripts derived from the ADH1 polyadenylation site. Because decay curves in the absence of 1a were similar for all the tested RNAs, for clarity only that of 5′RNA3 is shown (open symbol and dashed line). (B) Stability assays of RNA3ΔIRA. The results for full-length RNA3 are shown for comparison.
In the presence of 1a, full-length RNA3 and both short transcripts were highly stabilized, with half-lives of >60 min (i.e., with 55 to 70% of the starting RNA level surviving after 60 min [Fig. 2C]). It should be noted that for these extremely long-lived RNAs, this assay actually underestimates stability, as continued cell growth during the course of the assay (20 to 25% increase in cell numbers) causes a corresponding decrease in RNA levels independent of decay. Consistent with their enhanced stability, these RNAs accumulated to three- to sixfold-higher levels in the presence of 1a (Fig. 2A and B). Thus, the short transcript contained cis-acting sequences sufficient for full 1a-induced stabilization.
RNA3 coat gene and 3′ UTR do not support 1a-induced RNA stabilization.
To test whether the 3′ portion of RNA3 contained similar 1a-responsive signals, a plasmid expressing RNA3 sequences 3′ of the intergenic oligo(A) tract was made (Fig. 3). In yeast, the resulting transcript, 3′RNA3, was highly stable both in the presence and absence of 1a, with virtually no decay detected 60 min following transcription repression (Fig. 3). The high, 1a-independent stability of 3′RNA3 appeared likely to be due to interaction with coat protein, since expression of BMV coat protein stabilizes RNA4 and a number of other BMV-derived RNAs in yeast (23), and immunoblot analysis confirmed that 3′RNA3 expresses coat protein in yeast (data not shown). To block coat protein synthesis, a four-base frameshifting insertion was introduced immediately after the coat protein initiation codon (Fig. 3, fs-3′RNA3). In the same RNA, the second in-frame AUG (at coat protein codon 9) was mutated to AUC to preclude reinitiation of translation, which could lead to production of a functional, N-terminally truncated coat protein (38). These mutations prevent coat protein production while minimizing changes in the RNA itself. As expected, yeast transcribing fs-3′RNA3 failed to produce coat protein, as assessed by immunoblotting (data not shown).
Unlike 3′RNA3 or wild-type RNA3, fs-3′RNA3 decayed rapidly in either the absence or presence of 1a (Fig. 3). In the absence of 1a, its decay was nearly identical to that of full-length RNA3: i.e., most of the RNA decayed with an initial t1/2 of ∼5 min, followed by slower decay of the remaining 10 to 20%. In the presence of 1a, fs-3′RNA3 showed no detectable increase in steady-state accumulation, one of the hallmarks of 1a’s effect on full-length RNA3 and Sh RNA3 (Fig. 2, compare 0-min time points with those in Fig. 3 [Northern blots]). Careful measurement revealed a slight elevation of approximately 5% in the fs-3′RNA3 decay curve in the presence of 1a versus that in the absence of 1a (Fig. 3, right-hand graph; note that the logarithmic scale exaggerates the magnitude of effects in the lower part of the graph). However, a similar, low-level response to 1a has also been observed for all nonviral transcripts analyzed to date (Fig. 5) (see Discussion). Thus, in contrast to the selective high-level stabilization of wild-type RNA3 by 1a, the slight reduction in the fs-3′RNA3 decay rate appears to represent a low-level, sequence-nonspecific effect of 1a on RNAs in general.
FIG. 5.
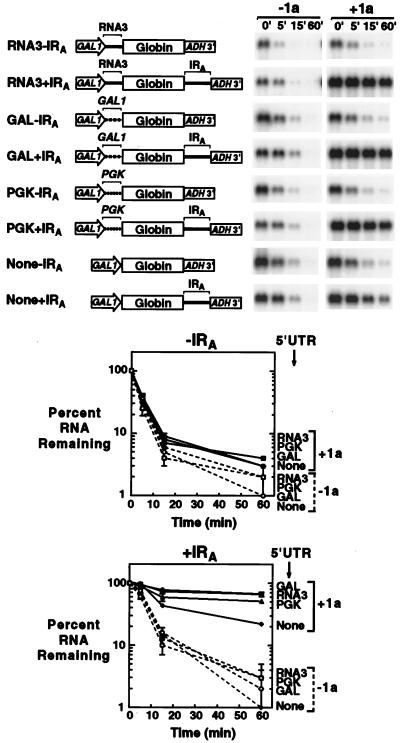
RNA3 IRA confers 1a-induced stability on a heterologous RNA. The stabilities of β-globin reporter RNAs in the absence (−1a) and presence (+1a) of 1a were analyzed by glucose repression as described in the legend to Fig. 2, except that hybridization utilized a 32P-labeled DNA probe complementary to the β-globin ORF. DNA expression cassettes used to generate the RNAs are diagrammed to the left of representative Northern blots. The GAL1 promoter, various 5′UTRs (see Results), the β-globin ORF, RNA3 IRA, and ADH1 polyadenylation signal were included or not as indicated. Each RNA is designated by its 5′ UTR and whether the RNA3 IRA is absent (−IRA) or present (+IRA). The results of three independent stability analyses were averaged and plotted as in Fig. 2 for RNAs lacking (upper plot) or possessing (lower plot) the RNA3 IRA. Stability in the presence (+1a) or absence (−1a) of 1a is plotted with solid symbols and solid lines or open symbols and dashed lines, respectively.
An intergenic segment is the major determinant of 1a-induced stabilization of RNA3.
The results described above showed that the short transcript shown in Fig. 2, consisting of the 5′ half of RNA3, contained signals that were necessary and sufficient for 1a-induced stabilization. Accordingly, we made and tested deletions of the three major segments of this RNA, i.e., the 5′ UTR, 3a ORF, and IRA (Fig. 4A). To provide a starting point for these deletions, the plasmid shown in Fig. 2B was modified by adding the yeast ADH1 gene polyadenylation signal immediately following IRA (Fig. 4A, 5′RNA3). The explicit ADH1 polyadenylation signal was added to provide a common 3′ end to all deletion variants in this series, since our further experiments had shown that self-directed 3′ end formation from the fortuitous polyadenylation site in IRA depended on sequences in IRA and flanking portions of the 3a ORF. As expected, the starting plasmid and its derivative lacking the 5′ UTR each produced two major transcripts in yeast: a shorter transcript resulting from the fortuitous polyadenylation signal in IRA and a longer transcript resulting from the ADH1 3′ polyadenylation site. No significant differences in stability were seen between the longer ADH1-terminated and shorter self-terminated transcripts (Fig. 4A and data not shown). Consequently, the longer, ADH1-derived RNAs from these 5′RNA3 and Δ5′UTR plasmids were used for comparison to the single major ADH1-derived RNAs of Δ3a, ΔIRA, and Δ5′UTRΔ3a.
In the absence of 1a, all of the RNAs shown in Fig. 4A decayed rapidly, with near-first-order kinetics and half-lives of 3 to 4 min. In the presence of 1a, 5′RNA3 accumulated to approximately 2.5-fold-higher levels than in its absence and was markedly stabilized, with a half-life of approximately 50 min.
Deleting the 3a ORF (Δ3a) resulted in an RNA whose 1a-responsive behavior was indistinguishable from that of 5′RNA3 (Fig. 4A). Deleting IRA (ΔIRA) resulted in an RNA that neither accumulated to higher levels nor showed significant stabilization in the presence of 1a (t1/2 ≈ 3 min). Thus, in the context of the 5′ half of RNA3, IRA plays a vital role in 1a-dependent stabilization. Deleting the 5′ UTR, alone or in combination with the 3a ORF, resulted in an intermediate level of 1a responsiveness: the resulting RNAs (Δ5′UTR and Δ5′UTRΔ3a) decayed much more slowly in the presence of 1a than in its absence but more rapidly than 5′RNA3 in the presence of 1a. While this shows that 5′ untranslated sequences can facilitate 1a-induced stabilization, experiments described below show that this effect does not depend on specific viral sequences.
The role of IRA in 1a stimulation of RNA3 stability and accumulation was further tested by IRA deletion from full-length RNA3 (RNA3ΔIRA). As shown by the Northern blots in Fig. 4B, IRA deletion largely abolished RNA3 responsiveness to 1a. While 1a increased wild-type RNA3 accumulation up to sixfold, the data in Fig. 4B and independent repetitions of this experiment showed that the accumulation of RNA3ΔIRA in galactose-induced yeast was unchanged in the presence and absence of 1a. Moreover, after glucose inhibition of GAL1-promoted transcription, RNA3ΔIRA decay in either the presence or absence of 1a was rapid relative to that of wild-type RNA3 plus 1a. As illustrated by the graphs in Fig. 4B, the average residual amount of RNA3ΔIRA persisting 60 min after transcription repression increased slightly in the presence of 1a. However, this increase was similar in magnitude to the nonspecific effects of 1a on nonviral RNAs (Fig. 5, Northern blots) (see Discussion), the amount of RNA3ΔIRA surviving was extremely small compared to the amount of RNA3 stabilized by 1a (Fig. 4B, compare 60-min time points in Northern blots), and the decay curve of RNA3ΔIRA even in the presence of 1a was similar to that of wild-type RNA3 minus 1a.
An RNA3 intergenic segment confers 1a-induced stabilization on a heterologous RNA.
To further assess which RNA3 sequences were required for 1a-induced stabilization, we tested several chimeric RNAs containing a human β-globin ORF (31) followed by IRA and the yeast ADH1 polyadenylation signal (Fig. 5). A parallel set of chimeric RNAs lacking IRA was also tested. Because the deletions shown in Fig. 4A showed that loss of the 5′ UTR partially inhibited 1a-induced stabilization, a possible sequence-specific role of the RNA3 5′ UTR was tested by comparing four pairs of β-globin hybrids. Three pairs of hybrids had 5′ UTRs from RNA3 or two yeast genes, GAL1 and PGK. The fourth pair of hybrids had only six nucleotides preceding the β-globin initiator AUG and thus lacked a functional 5′ leader for this ORF (see Discussion). The stability of all of these RNAs was assessed in vivo in the absence and presence of 1a.
In the absence of 1a, all of the hybrid RNAs decayed rapidly, with half-lives of 3 to 6 min (Fig. 5). In the presence of 1a, all of the hybrid RNAs lacking IRA sequences also decayed rapidly, with half-lives of 3 to 4 min (Fig. 5, Northern blots and upper plot). However, 1a did result in a slight but consistent increase in the residual amount of RNA remaining 60 min following transcription repression. This was true not only for the β-globin RNA containing the RNA3 5′ UTR but also for those containing the GAL1 or PGK 5′ UTRs, or no explicit 5′ UTR, and thus lacking viral sequences entirely. We have detected similar low-level responses to 1a by every RNA tested in this manner, including hybrid GUS (β-glucuronidase) RNAs lacking viral sequences (data not shown). Thus, 1a exerts a low-level, sequence-nonspecific effect on RNAs in general.
In contrast to this low-level, nonspecific response, all of the hybrid RNAs containing IRA sequences and explicit 5′ UTRs were dramatically stabilized in the presence of 1a, with >50% of the RNA persisting 1 h following transcriptional repression (Fig. 5, Northern blots and lower plot). Also, prior to repression of transcription, these RNAs accumulated to greater than twofold-higher levels in the presence of 1a than in its absence. No significant differences were observable between 1a-induced stabilizations of hybrid RNAs with RNA3 or GAL1 5′ UTRs, and only a minor reduction in stability was seen with the PGK 5′ UTR. A hybrid RNA lacking an explicit 5′ UTR exhibited 1a-responsive behavior comparable to that of the Δ5′UTR and Δ5′UTRΔ3a RNAs (Fig. 4), i.e., 1a-induced stabilization was evident but reduced compared to that of RNAs containing explicit 5′ UTRs. Similar results were obtained with a parallel set of hybrid RNAs containing the GUS ORF in place of the β-globin ORF (data not shown). Thus, the RNA3 IRA is sufficient to confer 1a-dependent stabilization on a heterologous, nonviral RNA. Moreover, although the presence of an explicit 5′ UTR enhances stabilization, there is no requirement for specific RNA3 sequences in the 5′ UTR.
Identification of sequences within the RNA3 intergenic region required for 1a-induced stabilization.
As shown above, a β-globin mRNA with the GAL1 5′ UTR and BMV IRA (Fig. 5, GAL+IRA) was as responsive to stabilization by 1a as wild-type RNA3. Accordingly, we used this context to further explore the boundaries of contributing IRA sequences in the absence of any other RNA3 sequences, which conceivably could confuse the analysis by providing redundant functions or other effects.
The stability of β-globin-IRA reporter RNAs containing partial IRA deletions was assessed in yeast lacking or expressing 1a (Fig. 6). In the absence of 1a, all of these RNAs were short lived, with half-lives of 3 to 5 min. In the presence of 1a, partial deletions from the 5′ side of IRA showed graded effects. An RNA lacking the 5′ 32 bases of IRA was stabilized by 1a at a significant level, but lower than that of the hybrid bearing the full IRA; i.e., 24 rather than 67% of the transcript persisted 1 h after transcriptional repression (Fig. 6, upper decay plot). Deletion of 45 and 79 bases from the 5′ end of IRA resulted in RNAs which showed only slight or no stabilization, respectively, by 1a.
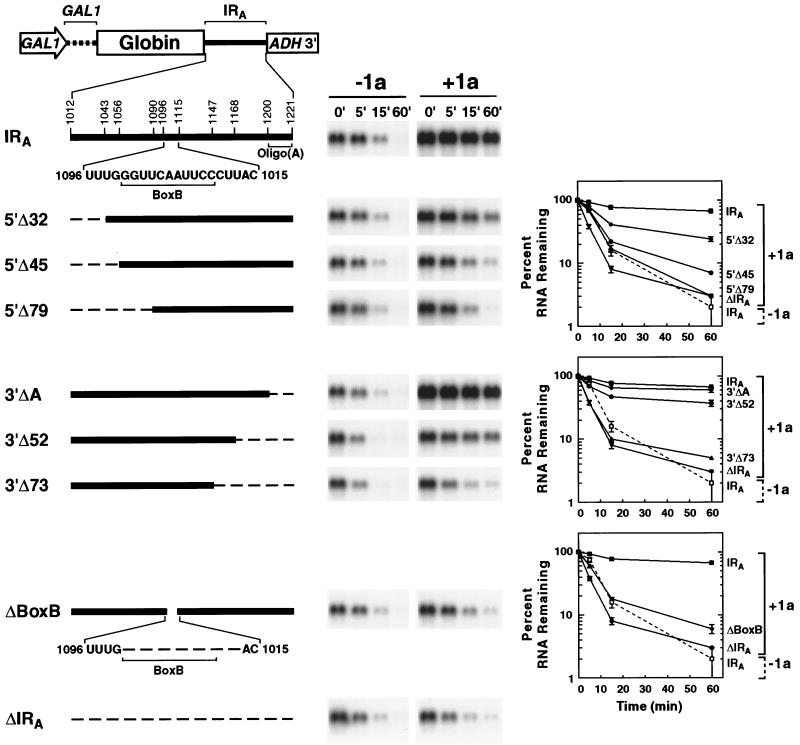
FIG. 6.
Deletion analysis of the RNA3 IRA segment. A series of partial IRA deletions was tested for 1a-induced stabilization of β-globin RNA. Stability in the absence (−1a) and presence (+1a) of 1a was analyzed for each RNA by glucose repression as described in the legend to Fig. 5. A general diagram of the DNA expression cassettes used to generate the RNAs is shown at the top, with the GAL1 promoter and 5′ UTR, β-globin ORF, IRA, and ADH1 polyadenylation signal indicated. A detailed expansion of IRA, including the box B motif and oligo(A) tract, is shown below, with numbering corresponding to the base position in RNA3. For each derivative, the extent of IRA deletion is shown by a dashed line on an IRA schematic to the left of a representative Northern blot. Results for β-globin RNAs containing the intact IRA (IRA; at top) or lacking IRA (ΔIRA; at bottom) are shown for comparison. The results of three independent stability analyses were averaged and plotted as in Fig. 2 for 5′ (upper graph), 3′ (center graph), and box B (lower graph) deletions. Because decay curves in the absence of 1a were similar for all of the tested RNAs, for clarity only that of the β-globin RNA containing the intact IRA is shown on each graph (open symbol and dashed line).
At the 3′ end of IRA, deletion of the oligo(A) tract had no effect on 1a-induced stabilization (Fig. 6, middle decay plot). Extending this deletion to 52 bases from the IRA 3′ end had a modest but discernible inhibitory effect on the ability of the RNA to be stabilized by 1a (37% of the RNA persisted 1 h after transcriptional repression). Deletion of 73 bases from the IRA 3′ end resulted in an RNA that showed little if any stabilization by 1a. Thus, sequences contributing minimal 1a responsiveness are encompassed within a 125- to 150-base region of IRA (from bases 1043 to 1168 of RNA3) while full 1a responsiveness requires additional flanking sequences (within bases 1012 to 1200 of RNA3).
The IRA segment required for 1a responsiveness contains an 11-base motif corresponding to the box B element of RNA polymerase III promoters and thus also to the conserved TΨC loop of tRNAs. Deleting this motif severely inhibits RNA3 replication (34, 41). To test whether this motif is required for 1a-induced stabilization, a 14-base region containing the box B motif and flanking 3′ pyrimidine residues conserved with the 5′ box B regions of BMV RNA1 and RNA2 (11) was deleted from the hybrid β-globin-IRA RNA (ΔBox B). In the absence of 1a, the ΔBoxB RNA decayed comparably to the other tested β-globin RNAs (t1/2 ≈ 5 min). In the presence of 1a, the ΔBoxB RNA was only slightly more stable than in its absence and strikingly less stable than a reporter RNA containing the intact IRA (Fig. 6, lower decay plot). Thus, the box B element appears to play a major role in 1a-induced stabilization.
Sequences required for 1a-induced stabilization are also required for RNA3 replication.
The approximately 150-base IRA segment defined above as required for 1a-induced stabilization of β-globin corresponds well to an intergenic segment previously defined as important for RNA3 replication in plant protoplasts, the IRE (11). To determine whether the IRA sequences responsible for 1a-induced stabilization are similarly required for RNA3 replication in yeast, all partial IRA deletions (Fig. 6) were engineered into full-length RNA3 (Fig. 7). Since this introduced some polylinker sequences flanking IRA, a reconstructed RNA3 containing the same extra bases was used as a positive control (rWT). The 1a-stimulated increase in steady-state RNA3 accumulation under continuous galactose induction of GAL1-promoted RNA3 transcription, an easily measured indicator of 1a-induced stabilization, was determined for the modified RNAs in order to assess whether their 1a-responsive behavior paralleled that of the analogous β-globin RNAs. Partial IRA deletions that caused severe reductions in 1a-induced stabilization of chimeric β-globin RNAs (5′Δ45, 5′Δ79, 3′Δ73, and ΔBoxB) also prevented significant 1a-induced accumulation of RNA3 (Fig. 7). However, in the context of RNA3, two partial IRA deletions that modestly reduced 1a-induced stabilization of chimeric β-globin RNAs (5′Δ32 and 3′Δ52) increased 1a-stimulated RNA3 accumulation over that seen for RNA3 containing the full IRA. Careful stability measurements of these RNAs in the presence and absence of 1a indicated that they were stabilized by 1a to a level similar to or greater than that of wild-type RNA3 (71 to 81% of starting RNA surviving 60 min after transcription repression). Possible reasons for the unusually high 1a responsiveness of these RNAs are considered further in the Discussion.
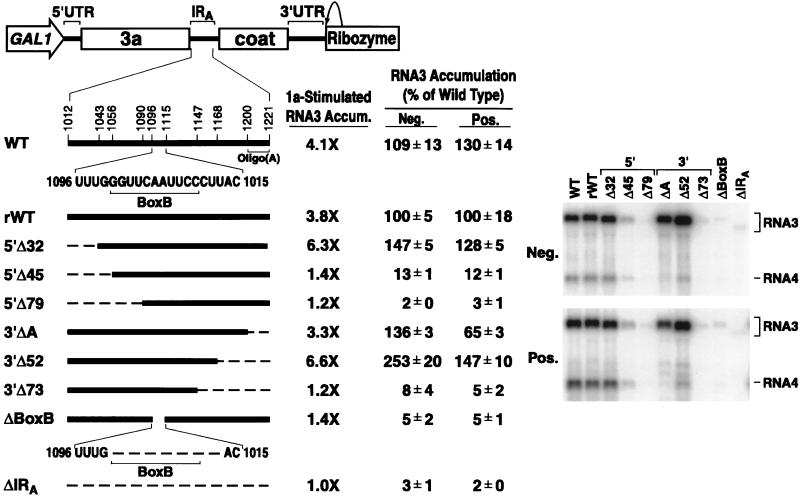
FIG. 7.
Effect of IRA deletions on RNA3 replication in yeast. The partial IRA deletions shown in Fig. 6 were engineered into RNA3. A general diagram of the DNA expression cassettes used to generate the RNAs is shown, with GAL1 promoter, RNA3, and ribozyme sequences indicated. A detailed expansion of IRA, including the box B motif and oligo(A) tract, is shown below, with numbering corresponding to the base position in RNA3. For each RNA3 derivative, the extent of IRA deletion is shown by a dashed line on an IRA schematic at the left. rWT is a reconstructed RNA3 that contains extra polylinker sequences flanking IRA that are present in all the deletion derivatives. To measure 1a-stimulated RNA3 accumulation, plasmids expressing the indicated RNA3 derivatives were cotransformed into yeast with either a 1a-expressing plasmid or the corresponding empty vector, pRS423. Equal amounts of total RNA prepared from the resulting galactose-induced yeast were analyzed by Northern blotting with a 32P-labeled DNA probe complementary to the entire RNA3 cDNA. Radioactive signals corresponding to each RNA3 derivative in the absence or presence of 1a were measured with a PhosphorImager. Two to three independent transformants were analyzed for each RNA3 derivative in the presence and absence of 1a. 1a-stimulated RNA3 accumulation is shown as the fold increase in accumulation in the presence of 1a versus that in its absence. To assay RNA replication, plasmids expressing the indicated RNA3 derivatives were cotransformed with 1a- and 2a-expressing plasmids. Equal amounts of total RNA prepared from the resulting galactose-induced yeast were analyzed by Northern blotting with a single-stranded, 32P-labeled RNA probe complementary to either negative- or positive-strand RNA3, as indicated. Representative Northern blots are shown, with the migration positions of RNA3 and RNA4 indicated. Positive- and negative-strand RNA3 accumulation was quantified for three independent transformants of each RNA3 derivative and averaged and is presented as a percentage of rWT accumulation ± standard error of the mean.
Each of these RNA3 derivatives was also introduced into yeast expressing both the 1a and 2a BMV replication proteins, and the accumulation of RNA3 replication products was analyzed. Negative-strand RNA3 and the subgenomic coat protein mRNA, RNA4, are produced only by 1a- and 2a-directed RNA-dependent RNA synthesis (12, 20, 22). Positive-strand RNA3 replication products can also be analyzed because BMV RNA replication increases positive-strand RNA3 accumulation in yeast more than 50-fold over the accumulation of GAL1-promoted, DNA-derived RNA3 transcripts in the absence of RNA replication (20).
As shown in Fig. 7, IRA deletions that supported significant 1a-induced stabilization of β-globin RNA or RNA3 supported wild-type or higher levels of negative- and positive-strand RNA3 accumulation while deletions that supported weak or no 1a-induced stabilization of these RNAs showed corresponding inhibition of RNA3 replication. Deletion of the 5′ 32 bases or the 3′ 52 bases of the IRA segment resulted in negative-strand accumulation to 147 and 253% of rWT levels, respectively, paralleling the increased 1a-induced stabilization of these RNA3 derivatives. Conversely, deleting the 5′ 45 and 79 bases reduced negative-strand RNA3 accumulation to 13 and 2% of wild-type levels and deleting the 3′ 73 bases reduced negative-strand RNA3 accumulation to 8% of wild-type levels. A small deletion encompassing box B also reduced negative-strand RNA3 accumulation to 5% of wild-type levels.
The effects of partial IRA deletions on positive-strand RNA3 accumulation generally paralleled those for negative strands, except for deletion of the intergenic oligo(A), which modestly increased negative-strand RNA3 (136% of wild type) while decreasing positive-strand RNA3 (65% of wild type). As expected from prior studies of the subgenomic mRNA promoter (10) (Fig. 1), this same oligo(A) deletion and deletions of sequences further-upstream severely inhibited production of subgenomic mRNA relative to that of positive-strand RNA3.
DISCUSSION
A subset of RNA3 intergenic sequences directs 1a-induced stabilization.
Using a combination of deletion and gain-of-function analyses, we have shown here that a 150- to 190-base, 5′-proximal portion of the RNA3 intergenic region is necessary and sufficient for the dramatic increase in RNA3 stability induced by the BMV 1a RNA replication protein. All RNA3 derivatives containing this region were strongly stimulated in stability and accumulation by 1a expression, while any RNA3 derivatives lacking this region showed only low-level, nonspecific responses to 1a. Moreover, transfer of the same intergenic sequences to foreign RNAs containing the β-globin or GUS ORFs rendered these RNAs similarly responsive to 1a.
Our findings with chimeric β-globin RNAs were generally consistent with those obtained with RNA3s carrying comparable IRA deletions. However, two partial IRA deletions, 5′Δ32 and 3′Δ52, reduced 1a-induced stabilization of IRA-containing β-globin RNAs, whereas in RNA3 these deletions resulted in stabilization to a level comparable to or slightly higher than that of wild-type RNA3 as well as parallel increases in accumulation of RNA3 replication products in yeast expressing both 1a and 2a. The difference in stabilization of β-globin and RNA3 derivatives could be due to some portion of RNA3 outside of IRA being able to compensate for these particular deletions from the 5′ and 3′ ends of the IRA. It is perhaps not surprising that the ends of the minimal sequence required for 1a-induced stabilization are slightly different in the two contexts tested, especially if secondary structure, which could be influenced by flanking sequences, is important (see below). It should also be noted that in vivo transcription of RNA3 containing the 3′Δ52 deletion failed to generate the truncated, 1a-stabilizable transcript (Sh RNA3 [Fig. 2]) that wild-type RNA3 sequences produce in yeast. If, as discussed below, 1a or some other factor is limiting for stabilization, then it would be expected that the absence of this potential competitor RNA would result in stabilization higher than that seen for wild-type RNA3. 3′Δ53 also greatly reduced subgenomic mRNA synthesis, thus making more negative-strand RNA3 available for replication.
Relation of 1a-induced stabilization to RNA replication.
Multiple results suggest that IRA-mediated, 1a-induced stabilization of RNA3 is related to RNA3 replication. In addition to the well-established role of 1a as an essential RNA replication factor (12, 20, 22, 24), the intergenic element found here to direct 1a-induced RNA stabilization corresponds to the IRE, a crucial, 50- to 100-fold in vivo enhancer of RNA3 replication in plant cells (11) and in yeast (Fig. 7). The inhibitory and stimulatory effects of partial IRA deletions on 1a-dependent RNA3 stabilization correlated well with their effects on (1a plus 2a)-dependent RNA3 replication (Fig. 7). Even the two mutants that increased 1a-stimulated RNA3 accumulation above that of wild type (5′Δ32 and 3′Δ52) resulted in parallel increases in accumulation of positive- and negative-strand RNA3 replication products.
The role of the IRE as an enhancer of overall RNA3 replication might be solely the result of enhancing negative-strand synthesis, since sequences within IRA stimulate negative-strand RNA3 synthesis in vivo by 100-fold (35). Furthermore, sequence exchanges between BMV and a closely related bromovirus, cowpea chlorotic mottle virus, showed that the preference of BMV 1a and 2a for replicating BMV RNA3 over that of cowpea chlorotic mottle virus is controlled in trans by 1a (43) and in cis by the RNA3 intergenic region (32). Thus, independent results connect 1a with the RNA3 intergenic region in RNA replication and show that these elements are jointly involved in selecting RNA3 templates for replication. Consistent with a role in template selection prior to initiating negative-strand RNA synthesis, the 1a-IRE interaction underlying 1a-induced stabilization requires neither BMV 2a protein, which is essential for negative-strand RNA synthesis (20), nor, as shown here, the 3′-terminal RNA3 sequences that function as the negative-strand initiation site.
1a-induced RNA3 stabilization also inhibits RNA3 translation (20). As first suggested for bacteriophage Qβ (44), inhibiting the translation of positive-strand viral RNAs could be a prerequisite for initiating negative-strand synthesis, to prevent ribosomes from blocking the opposing passage of viral polymerase. Thus, a combination of independent results suggest that direct or indirect interaction of 1a with the IRE may be involved in recruiting RNA3 templates into the replication complex for negative-strand synthesis while diverting them from the competing process of translation. Translation inhibition by 1a could be a significant factor in 1a-mediated RNA3 stabilization, since the degradation of many RNAs is also linked to their translation (33).
The role of the IRE in BMV RNA-dependent RNA synthesis may be formally similar to that of an enhancer element in DNA-dependent RNA synthesis: from a position distal to the initiation site, the IRE directs interaction of the template with one or more components of the RNA synthesis complex, thus facilitating subsequent recognition and initiation at the linked start site. Similar elements distal to the site of negative-strand initiation have been identified for bacteriophage Qβ (7) and recently for poliovirus (13). In parallel with the BMV RNA3 IRE, interaction of these distal sites with Qβ replicase and poliovirus 3CD protein, respectively, inhibit translation and promote negative-strand RNA synthesis.
Prior protoplast (11) and yeast (35) studies showed low (approximately 1 to 2% of wild-type) levels of negative-strand synthesis and RNA replication for RNA3 derivatives lacking intergenic region sequences. Our RNA replication results (Fig. 7) agree with these findings. The low template activity of RNA3 derivatives lacking the IRE might reflect a residual level of RNA replication in the absence of the 1a function(s) underlying 1a-induced RNA stabilization. However, we also find that 1a has a low-level, nonspecific effect on all RNAs tested, including nonviral RNAs (Fig. 5). Such low-level nonspecific effects might be expected if 1a or associated factors must interact with and scan the pool of cytoplasmic RNAs in search of appropriate templates. Thus, even inefficient IRE-independent RNA replication might proceed by a parallel, albeit nonspecific, interaction with 1a.
Role of box B element in 1a-dependent RNA3 stabilization and replication.
1a-induced RNA stabilization was severely impaired by a small deletion encompassing the central box B element of the IRE (Fig. 6). Mutation of the box B motif in the RNA3 IRE similarly reduces RNA3 replication in both yeast (Fig. 7) and plant cells (34, 41). This motif, which is also present in the 5′ UTRs of RNA1 and RNA2 (2), corresponds to box B of RNA polymerase III promoters and thus to the conserved TΨC loop of tRNAs (26). While our results do not show whether 1a interacts directly or indirectly with the IRE, or whether 1a-induced RNA stabilization depends on host factors that interact independently with the IRE, the box B element could represent a site of interaction with host factors.
Despite its importance, the box B motif alone was unable to support 1a-induced RNA stabilization or RNA3 replication without 5′ and 3′ flanking sequences (Fig. 6 and 7). These flanking sequences might contribute independent recognition sites, higher-order structure, or both. Since the IRE was capable of functioning in multiple non-RNA3 contexts (e.g., β-globin and GUS), any essential higher-order RNA structure must be self-contained within the IRE.
Additional effects on 1a-dependent RNA stabilization.
When expressed alone, IRA showed significant 1a-induced stabilization (Fig. 4A) and thus contains the only sequences essential in cis for this effect. However, RNAs containing a functional intergenic region segment but lacking natural 5′ UTRs (Δ5′UTR and Δ5′UTRΔ3a [Fig. 4A] and None+IRA [Fig. 5]) were less efficiently stabilized by 1a than their counterparts having 5′ UTRs. The ability of nonviral GAL1 and PGK 5′ UTRs to substitute for the RNA3 leader (Fig. 5) indicates that any requirement for a 5′ UTR does not involve specific viral sequences and thus may be linked to 5′ UTR binding of translation-associated factors or to translation itself. For both the 3a and β-globin RNAs lacking natural 5′ UTRs, the first AUG (corresponding to the start of the 3a and β-globin ORFs) is only six bases from the 5′ end of the mRNA and thus is too close for efficient translation initiation in yeast (45), resulting in translation of relatively short, out-of-frame ORFs from the second AUG. Consequently, the absence of a natural 5′ UTR may result in these RNAs being degraded by the efficient nonsense-mediated mRNA decay pathway (37) before 1a or associated factors can interact with them. Alternatively, efficient translation of an ORF immediately upstream of the IRE sequences might be required for efficient 1a-induced stability.
Another factor influencing the extent of RNA stabilization appeared to be the amount of 1a present. During the course of these studies, we observed that expressing 1a from the plasmid used here (pB1MS6) (see Materials and Methods) increased RNA3 accumulation only 3- to 6-fold, or less than the 7- to 10-fold increase seen with a different plasmid (20). The higher efficiency of 1a stabilization in the latter case correlated with a higher level of 1a expression, as determined by RNA blotting. Still higher 1a expression from the strong GAL1 promoter led to even higher levels of 1a-induced stabilization. Thus, under the conditions used here, 1a protein was limiting for 1a-induced stabilization.
Further experiments are in progress to determine whether 1a interacts directly or indirectly with the IRE, whether the resulting interaction is transient or stable, and how 1a action leads to increased RNA3 stability. The results of these studies should provide additional insights into the mechanisms involved in positive-strand RNA virus RNA replication.
ACKNOWLEDGMENTS
We thank Sanjeev Shah for excellent technical assistance and members of our laboratory for helpful discussions throughout the course of this work.
This work was supported by the National Institutes of Health through grant GM35072. P. Ahlquist is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 2.Ahlquist P, Dasgupta R, Kaesberg P. Nucleotide sequence of the brome mosaic virus genome and its implications for viral replication. J Mol Biol. 1984;172:369–383. doi: 10.1016/s0022-2836(84)80012-1. [DOI] [PubMed] [Google Scholar]
- 3.Ahola T, Laakkonen P, Vihinen H, Kaariainen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison R, Thompson C, Ahlquist P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc Natl Acad Sci USA. 1990;87:1820–1824. doi: 10.1073/pnas.87.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988;16:9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 7.Barrera I, Schuppli D, Sogo J M, Weber H. Different mechanisms of recognition of bacteriophage Q beta plus and minus strand RNAs by Q beta replicase. J Mol Biol. 1993;232:512–521. doi: 10.1006/jmbi.1993.1407. [DOI] [PubMed] [Google Scholar]
- 7a.Chen, J., and P. Ahlquist. Unpublished results.
- 8.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Genes. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 10.French R, Ahlquist P. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J Virol. 1988;62:2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French R, Ahlquist P. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J Virol. 1987;61:1457–1465. doi: 10.1128/jvi.61.5.1457-1465.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.French R, Janda M, Ahlquist P. Bacterial gene inserted in an engineered RNA virus: efficient expression in monocotyledonous plant cells. Science. 1986;231:1294–1297. doi: 10.1126/science.231.4743.1294. [DOI] [PubMed] [Google Scholar]
- 13.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 15.Goldbach R. Genome similarities between plant and animal RNA viruses. Microbiol Sci. 1987;4:197–202. [PubMed] [Google Scholar]
- 16.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. A novel superfamily of nucleotide triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988;235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Z, Sherman F. 3′-end-forming signals of yeast mRNA. Trends Biochem Sci. 1996;21:477–481. doi: 10.1016/s0968-0004(96)10057-8. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa M, Janda M, Krol M A, Ahlquist P. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J Virol. 1997;71:7781–7790. doi: 10.1128/jvi.71.10.7781-7790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janda M, Ahlquist P. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc Natl Acad Sci USA. 1998;95:2227–2232. doi: 10.1073/pnas.95.5.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 22.Kiberstis P A, Loesch-Fries L S, Hall T C. Viral protein synthesis in barley protoplasts inoculated with native and fractionated brome mosaic virus RNA. Virology. 1981;112:804–808. doi: 10.1016/0042-6822(81)90331-7. [DOI] [PubMed] [Google Scholar]
- 23.Krol, M., and P. Ahlquist. Unpublished results.
- 24.Kroner P A, Young B M, Ahlquist P. Analysis of the role of brome mosaic virus 1a protein domains in RNA replication, using linker insertion mutagenesis. J Virol. 1990;64:6110–6120. doi: 10.1128/jvi.64.12.6110-6120.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh L E, Dreher T W, Hall T C. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 1988;16:981–995. doi: 10.1093/nar/16.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsh L E, Hall T C. Evidence implicating a tRNA heritage for the promoters of positive-strand RNA synthesis in brome mosaic and related viruses. Cold Spring Harbor Symp Quant Biol. 1987;52:331–341. doi: 10.1101/sqb.1987.052.01.038. [DOI] [PubMed] [Google Scholar]
- 27.Marsh L E, Pogue G P, Huntley C C, Hall T C. Insight to replication strategies and evolution of (+) strand RNA viruses provided by brome mosaic virus. Oxf Surv Plant Mol Cell Biol. 1991;7:297–334. [Google Scholar]
- 28.Mi S, Stollar V. Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli. Virology. 1991;184:423–427. doi: 10.1016/0042-6822(91)90862-6. [DOI] [PubMed] [Google Scholar]
- 29.Miller W A, Bujarski J J, Dreher T W, Hall T C. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986;187:537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- 29a.Mise, K., and P. Ahlquist. Unpublished results.
- 30.Mise K, Ahlquist P. Host-specificity restriction by bromovirus cell-to-cell movement protein occurs after initial cell-to-cell spread of infection in nonhost plants. Virology. 1995;206:276–286. doi: 10.1016/s0042-6822(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 31.Newman T C, Ohme-Takagi M, Taylor C B, Green P J. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell. 1993;5:701–714. doi: 10.1105/tpc.5.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pacha R F, Ahlquist P. Use of bromovirus RNA3 hybrids to study template specificity in viral RNA amplification. J Virol. 1991;65:3693–3703. doi: 10.1128/jvi.65.7.3693-3703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peltz S W, Jacobson A. mRNA turnover in Saccharomyces cerevisiae. In: Belasco J, Brawerman G, editors. Control of messenger RNA stability. San Diego, Calif: Academic Press, Inc.; 1993. pp. 291–328. [Google Scholar]
- 34.Pogue G P, Marsh L E, Connell J P, Hall T C. Requirements for ICR-like sequences in the replication of brome mosaic virus genome RNA. Virology. 1992;188:742–753. doi: 10.1016/0042-6822(92)90529-x. [DOI] [PubMed] [Google Scholar]
- 35.Quadt R, Ishikawa M, Janda M, Ahlquist P. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc Natl Acad Sci USA. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Restrepo-Hartwig, M., and P. Ahlquist. Unpublished results.
- 36.Restrepo-Hartwig M, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Echevarria M J, Czaplinski K, Peltz S. Making sense of nonsense in yeast. Trends Biochem Sci. 1996;21:433–438. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- 38.Sacher R, Ahlquist P. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J Virol. 1989;63:4545–4552. doi: 10.1128/jvi.63.11.4545-4552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Scheidel L M, Stollar V. Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1. Virology. 1991;181:490–499. doi: 10.1016/0042-6822(91)90881-b. [DOI] [PubMed] [Google Scholar]
- 41.Smirnyagina E, Hsu Y H, Chua N, Ahlquist P. Second-site mutations in the brome mosaic virus RNA3 intercistronic region partially suppress a defect in coat protein mRNA transcription. Virology. 1994;198:427–436. doi: 10.1006/viro.1994.1054. [DOI] [PubMed] [Google Scholar]
- 42.Sullivan M, Ahlquist P. cis-acting signals in Bromovirus RNA replication and gene expression: networking with viral proteins and host factors. Semin Virol. 1997;8:221–230. [Google Scholar]
- 43.Traynor P, Ahlquist P. Use of bromovirus RNA2 hybrids to map cis- and trans-acting functions in a conserved RNA replication gene. J Virol. 1990;64:69–77. doi: 10.1128/jvi.64.1.69-77.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber H, Billeter M A, Kahane S, Weissmann C, Hindley J, Porter A. Molecular basis for repressor activity of Qβ replicase. Nat New Biol. 1972;237:166–169. doi: 10.1038/newbio237166a0. [DOI] [PubMed] [Google Scholar]
- 45.Yoon H, Donahue T. Control of translation initiation in Saccharomyces cerevisiae. Mol Microbiol. 1992;6:1413–1419. doi: 10.1111/j.1365-2958.1992.tb00861.x. [DOI] [PubMed] [Google Scholar]