Abstract
We have examined the consequences of overexpression of the IκBα and IκBβ inhibitory proteins on the regulation of NF-κB-dependent beta interferon (IFN-β) gene transcription in human cells after Sendai virus infection. In transient coexpression studies or in cell lines engineered to express different forms of IκB under tetracycline-inducible control, the IFN-β promoter (−281 to +19) linked to the chloramphenicol acetyltransferase reporter gene was differentially inhibited in response to virus infection. IκBα exhibited a strong inhibitory effect on virus-induced IFN-β expression, whereas IκBβ exerted an inhibitory effect only at a high concentration. Despite activation of the IκB kinase complex by Sendai virus infection, overexpression of the double-point-mutated (S32A/S36A) dominant repressors of IκBα (TD-IκBα) completely blocked IFN-β gene activation by Sendai virus. Endogenous IFN-β RNA production was also inhibited in Tet-inducible TD-IκBα-expressing cells. Inhibition of IFN-β expression directly correlated with a reduction in the binding of NF-κB (p50-RelA) complex to PRDII after Sendai virus infection in IκBα-expressing cells, whereas IFN-β expression and NF-κB binding were only slightly reduced in IκBβ-expressing cells. These experiments demonstrate a major role for IκBα in the regulation of NF-κB-induced IFN-β gene activation and a minor role for IκBβ in the activation process.
Human interferons (IFNs) are synthesized by leukocytes, macrophages, and epithelial cells in response to virus infection and other pathogenic stimuli. IFNs induce a group of genes encoding proteins with a broad range of antiviral, immunoregulatory, and growth-suppressive activities (reviewed in reference 47). The regulation of alpha/beta IFN (IFN-α and IFN-β) transcription has served as an important model for examining the transcriptional mechanisms controlling virus-inducible gene expression (reviewed in reference 20). IFN-β transcriptional regulation is controlled by the protein-DNA interactions within 110 nucleotides upstream of the intronless structural gene and consisting of multiple overlapping positive and negative regulatory domains. Four positive regulatory domains bind specific members of the NF-κB, IRF, and CREB/ATF transcription factors, as well as the chromatin-associated HMGI(Y) proteins in an induction-specific and cooperative manner; a higher-order structure termed the enhanceosome is formed that, via recruitment of the CBP/p300 coactivator, stimulates IFN-β gene transcription (29). Recently, it has been demonstrated that in cells infected by virus, the newly identified IRF-3 and IRF-7 factors bind to the IFN-β promoter, together with NF-κB and ATF-2/c-Jun. The association of these factors in the IFN-β enhanceosome is thought to create a new protein-protein interface that interacts with the transcriptional coactivator CBP/p300 proteins in response to virus infection, leading to virus-mediated gene activation (16, 24, 26a, 29, 40, 48).
The PRDII domain (−64 to −55) contains the consensus site 5′-GGGAAATTCC-3′ for the binding of NF-κB/Rel transcription factors. Heterodimer or homodimer combinations of NF-κB play an important role in the regulation of a large variety of genes, including cytokines, immune regulatory genes, receptors, and early genes of several viruses. NF-κB binding activity is inducible in most cell types by viruses, double-stranded RNA, cytokines, phorbol esters, and oxygen radicals. NF-κB was initially described as a protein complex composed of two subunits (p50 and p65) retained in the cytoplasm by its association with the inhibitor subunits IκB. Induction resulted in the release of the heterodimer p50-p65 by IκB, translocation to the nucleus, and binding to κB sites. The DNA binding NF-κB and the inhibitory IκB proteins are composed of multiple family members that contribute to the diversity of NF-κB-mediated gene regulation (reviewed in references 2, 3, 27, and 46).
Phosphorylation and degradation of IκB are crucial regulatory events in the activation of NF-κB DNA binding activity. After inducer-mediated stimulation IκBα is phosphorylated within the N-terminal signal response domain at Ser-32 and Ser-36 (6, 7, 43) by the IκB kinase (IKK) (13, 28, 35, 50, 53), ubiquitinated, and subsequently degraded by the 26S proteasome (1, 9, 37). Substitution of Ser-32 and Ser-36 prevents IκBα phosphorylation, ubiquitination, and degradation, thus generating nondegrading, transdominant repressors of IκBα (6, 8, 42). The C-terminal PEST domain of IκBα is involved in the intrinsic stability of the protein, and this region is constitutively phosphorylated by CKII (26, 38).
The inducibility of NF-κB is controlled by different IκB proteins, thus providing an additional level of regulation for NF-κB-dependent gene transcription. Two well-characterized forms, IκBα and IκBβ (44, 49), share several common structural features, including conserved N-terminal signal response, ankyrin repeat, and C-terminal PEST domains. However, IκBα and IκBβ respond differentially to distinct inducers: the level of IκBβ is not affected by tumor necrosis factor alpha (TNF-α) or phorbol myristate acetate and, after lipopolysac- charide or interleukin-1 (IL-1) induction, the degradation and resynthesis of IκBβ occurs more slowly than IκBα (41). IκBβ is also resynthesized in stimulated cells as a hypophosphorylated protein which is able to form stable complexes with NF-κB in the cytosol (32, 49); however, this interaction fails to mask the nuclear localization sequence and DNA binding domain of NF-κB, resulting in NF-κB-IκBβ complexes in the nucleus. This hypophosphorylated form of IκBβ acts as a chaperone, by protecting NF-κB from IκBα and permitting a prolonged activation of gene transcription by NF-κB (39). A model has been proposed for NF-κB activation consisting of two overlapping phases: first, a transient phase mediated mainly through IκBα and, second, a persistent phase of activation mediated by IκBβ (39, 41). Recently, two different isoforms of IκBβ, IκBβ1 (43 kDa) and IκBβ2 (41 kDa), generated as a consequence of RNA processing and differing in their C-terminal PEST domains, have been identified. The relative amounts of these two forms and their degradation in response to stimulation appears to be cell-type specific. Both IκBβ1 and IκBβ2 bind to the same NF-κB subunits and are constitutively phosphorylated (19). Furthermore, IκBα is a stronger inhibitor of NF-κB activity than IκBβ; the inhibitory activity of IκBβ is facilitated on promoters containing HMGI(Y) binding regions (44).
In the present study, the consequences of overexpression of the IκBα and IκBβ inhibitory proteins on the regulation of NF-κB-dependent IFN-β gene transcription after Sendai virus infection was examined. In transient coexpression studies and in stable tetracycline-inducible human 293 cells, wild-type IκBα decreased IFN-β promoter activity, whereas IκB forms with the S32/36A point mutations completely abolished IFN-β gene expression. IκBβ overexpression had minimal effects on IFN-β promoter activity. Analysis of NF-κB protein-DNA complexes in IκB-expressing cells revealed quantitative and temporal alterations in the patterns of NF-κB binding to the PRDII domain after Sendai virus infection. These studies demonstrate differential regulation of IFN-β transcription by IκBα and IκBβ and indicate a major role for IκBα but not IκBβ in IFN-β regulation.
MATERIALS AND METHODS
Generation of plasmids.
For transient transfections, wild-type human IκBα (wtIκBα) or mutated human IκBα cDNA was inserted downstream of the simian virus 40 promoter in the pSVK3 vector (Pharmacia Biotech, Uppsala, Sweden) (5). cDNA encoding IκBβ (a kind gift from Dimitris Thanos) was inserted into SVK3 between the sites EcoRI and XhoI. Mutated human IκBα cDNA was generated as previously described (26). In IκBα (2N), serine 32 and serine 36 are replaced by alanine; in IκBα (3C) serine 283, threonine 291, and threonine 299 were substituted by an alanine residue, and in IκBα-Δ4 22 amino acids were deleted from the terminal end. 2N+3C and 2NΔ4 were combinations of the above plasmids. Constructs for the establishment of stable cell line CMVt-rtTA, CMVt-wtIκBα, and IκBα mutants were generated as previously described (5, 25). cDNA encoding IκBβ (a kind gift of D. Thanos) was inserted into pCMVt-neo vector (4) at the NotI site.
Cell culture and generation of IκB cell lines.
Human 293 cells were cultured in alpha Dulbecco modified Eagle medium (alpha-MEM) supplemented with 10% heat-inactivated fetal bovine serum, glutamine, and antibiotics. CMVt-rtTA 293 cells (26) were cultured in the same medium containing 2.5 ng of puromycin (Sigma) per μl. CMVt-based plasmids (25) expressing wtIκBα, IκBα mutants, or IκBβ were introduced into CMVt-rtTA 293 cells by the calcium phosphate coprecipitation method. Cells were selected at 48 h after transfection in alpha-MEM supplemented with 10% heat-inactivated fetal bovine serum, glutamine, and the antibiotics puromycin (2.5 ng/μl) and G418 (400 μg/ml) (Life Technologies, Inc.). Cell clones resistant to G418 were selected individually after IκB expression levels induced by 1 μg of doxycycline (Dox; Sigma) per ml were examined.
Transfections and CAT reporter gene assays.
Subconfluent 293 cells, CMVt-rtTA 293 cells, or CMVt-rtTA-IκB-expressing 293 cells were transfected with the IFN-β–CAT reporter plasmid, by the calcium phosphate coprecipitation method (17). All of the transfections contained equivalent amounts of DNA standardized with the CMV-B1 vector. In some experiments, cells were infected with Sendai virus (500 hemagglutinating units [HAU]/ml for 90 min). At 24 h after infection (48 h after transfection), cytoplasmic extracts were prepared and the protein concentration was determined by Bradford assay (Bio-Rad). Then, 100 μg of cytoplasmic protein extract was assayed for 2 to 4 h at 37°C as previously described (17). Relative chloramphenicol acetyltransferase (CAT) activity was quantified by scintillation counting of acetylated and nonacetylated chloramphenicol forms.
Western blot analysis.
To characterize IκB expression, wtIκBα and IκBα mutants and IκBβ-expressing cells were cultured in the presence of Dox for various times. Cells were washed twice with phosphate-buffered saline (PBS) and resuspended in lysis buffer containing 10 mM Tris-Cl (pH 8.0), 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 0.5% Nonidet P-40, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and 10 μg of pepstatin, 10 μg of leupeptin, and 10 μg of aprotinin per ml. Whole-cell extracts (20 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 10% gel. Proteins were transferred to Hybond transfer membrane (Amersham, Cleveland, Ohio). Membrane was blocked in a 5% milk-PBS solution for 1 h at room temperature and probed with anti-IκBα (22) or anti-IκBβ G-20 against the N-terminal sequences of IκBβ1 and IκBβ2 (Santa Cruz Biotechnology, Inc., Valencia, Calif.) antibody in 5% milk-PBS at a dilution of 1:1,000 overnight at 4°C. Membranes were washed four times with PBS, incubated with peroxidase-conjugated secondary antibodies (KPL, Gaithersburg, Md.) (goat anti-mouse antibody was used to detect IκBα, and goat anti-rabbit antibody was used to detect IκBβ, each at a dilution of 1:1,000), and visualized with the chemiluminescence detection system as recommended by the manufacturer (NEN-Life Science, Boston, Mass.).
Immunoprecipitation and kinase assay.
For the in vitro kinase assay, cells were left untreated, were treated with TNF-α (10 ng/ml) for 10 min, or were infected with Sendai virus (80 HAU/ml) for different times as indicated. Cells were washed twice in cold PBS and resuspended in lysis buffer containing 20 mM Tris-Cl (pH 7.5), 200 mM NaCl, 0.5% Nonidet P-40, 1 mM sodium orthovanadate, 1 mM NaF, 0.5 mM PMSF, and 5 μg of leupeptin, 5 μg of pepstatin, and 5 μg of aprotinin per ml. Whole-cell extracts (200 μg) were incubated with 500 ng of anti-IKKα antibody H-744 (Santa Cruz) and 30 μl of protein A-Sepharose beads for 2 to 4 h at 4°C. Beads were washed three times with lysis buffer and two times with kinase buffer (50 mM Tris-Cl, pH 8.0; 100 mM NaCl; 2 mM MgCl2; 1 mM sodium orthovanadate; 1 mM NaF; 20 mM β-glycerophosphate; 1 mM DTT; 0.5 mM PMSF; 5 μg [each] of leupeptin, pepstatin, and aprotinin per ml) and then incubated at 30°C for 30 min in kinase buffer containing 5 μCi of [γ-32P]ATP and 2 μg of recombinant GST-IκBα (amino acids [aa] 1–55) or GST-IκBα (aa 1 to 55; S32/36A). Reactions were subjected to SDS-PAGE in a 12% polyacrylamide gel. The gels were dried and subjected to autoradiography.
RNA preparation and RNase protection assay.
Total RNA from 293 cells was extracted with the RNeasy Mini-Kit (Qiagen, Valencia, Calif.). Total RNA (5 μg) was subjected to RNase protection analysis by using a human CK3 cytokine multi-probe template set of the RiboQuant Multi-Probe RPA kit (PharMingen, San Diego, Calif.). Labelled fragments protected from RNase digestion and corresponding to IFN-β mRNA were quantified with the NIH Image 1.60 software package. Values were normalized to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and L32 (housekeeping gene) mRNA levels and then plotted as IFN-β/GAPDH mRNA ratios. Similar results were obtained in three independent experiments.
Electrophoretic mobility shift assay.
Nuclear extracts were prepared as previously described (17). First, 5 μg of nuclear extracts was incubated with 5 μg of poly(dI-dC) (Pharmacia) for 10 min at room temperature in a total volume of nuclear extract buffer containing 0.1% Nonidet P-40. Then, each sample was incubated for 30 min at room temperature in the presence of 100,000 cpm of [γ-32P]ATP-labeled oligonucleotide probe corresponding to the PRDII domain of the IFN-β promoter (5′-GGGAAATTCCGGGAAATTCC-3′). Protein DNA complexes were then separated on a 5% native polyacrylamide gel (60:1 cross-link) in 0.2× TBE. In competition analysis, a 200-fold molar excess of unlabelled oligonucleotide was incubated in the presence of poly(dI-dC) with the nuclear extracts for 30 min before the addition of probe to the extracts. To examine the individual proteins present in the complex, polyclonal subunit-specific antisera against NF-κB were used as previously described (17).
RESULTS
Inhibition of IFN-β promoter activity by IκB overexpression.
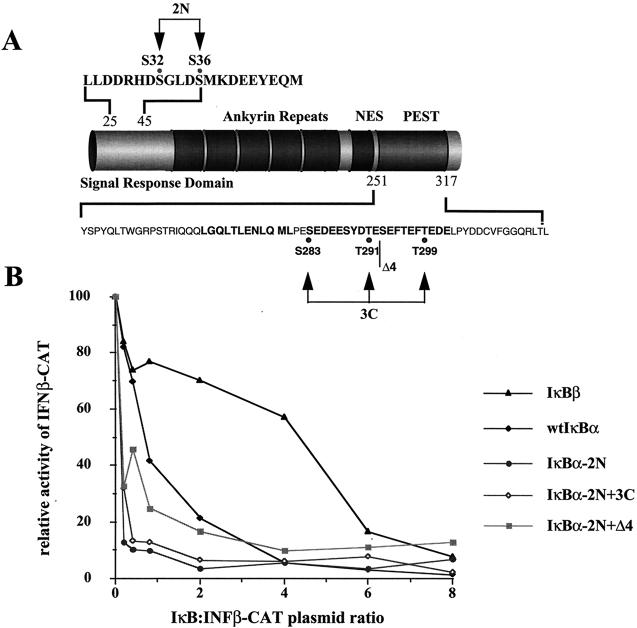
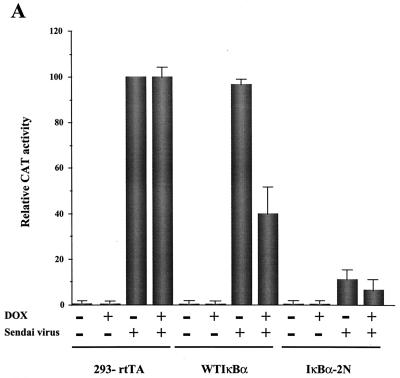
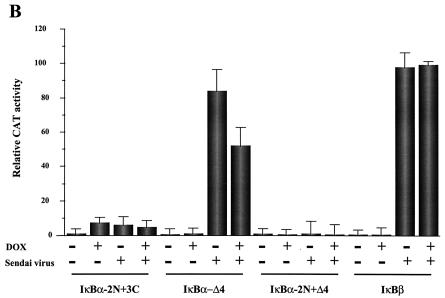
To examine the effect of IκBα and IκBβ on IFN-β promoter activity, 293 cells were cotransfected with a CAT reporter gene driven by the entire IFN-β promoter from −281 to +19 and different expression plasmids encoding wild-type or mutated IκBα and wild-type IκBβ (Fig. 1A). Several mutant forms of IκBα were examined: IκBα-2N, which contains the S32A/S36A double point mutation; IκBα-3C, which contains the S283A/T291A/T299A triple point mutation; IκBα-Δ4, which contains a 22-aa deletion in the PEST C-terminal domain; and the combination mutants IκBα-2N+3C and IκBα-2N+Δ4. At 24 h posttransfection the cells were infected with Sendai virus (500 HAU/ml), and 24 h later the cells were lysed and submitted to CAT reporter gene assay. Full activation of the IFN-β promoter corresponding to a 50- to 100-fold stimulation was observed in Sendai virus-infected cells cotransfected with the IFN-β CAT plasmid (Fig. 1B). Cotransfection of increasing amounts of wtIκBα expressing plasmid resulted in a concentration-dependent decrease of IFN-β promoter activity. IFN-β promoter activity was partially inhibited at low wtIκBα concentrations (IκBα/IFN-β ratio = 0.25) and completely inhibited at higher levels of wtIκBα (IκBα/IFN-β ratio = 4). IκBα mutants containing the S32A/S36A point mutations (IκBα 2N, -2N+Δ4, and -2N+3C) were strongly inhibitory and almost completely inhibited IFN-β promoter activity at the IκBα/IFN-β ratio of 0.25. In contrast, overexpression of IκBβ was a weak inhibitor of IFN-β promoter activity, with a less-than-twofold inhibition of IFN-β activity at a IκBβ/IFN-β ratio of 4.0; complete inhibition of IFN-β promoter activity was observed at the IκBβ/IFN-β ratio of 8. These initial experiments established that IκBα was a stronger inhibitor of IFN-β gene expression than IκBβ; furthermore the nondegrading transdominant forms of IκBα (TD-IκBα) are at least 10-fold more inhibitory than wtIκBα.
FIG. 1.
(A) Schematic of IκBα protein. Mutations of IκBα are as indicated: IκBα-2N, which contains the S32A/S36A double point mutation; IκBα-3C, which contains the S283A/T291A/T299A triple point mutation; IκBα-Δ4, which contains a 22-aa deletion in the PEST C-terminal domain; and the combination mutants IκBα-2N+3C and IκBα-2N+Δ4. (B) IκB expression inhibits IFN-β gene expression. The IFN-β–CAT reporter plasmid (2.5 μg) was cotransfected together with IκB expression plasmids into 293 cells by the calcium phosphate method. After 24 h the cells were infected by Sendai virus (500 HAU/ml), and after a further 24 h the cells were harvested and assayed for CAT activity by using cytoplasmic extracts (50 to 100 μg for 2 h). The ratio of IκB to IFN-β–CAT reporter is indicated on the graph. The relative CAT activity is expressed as a percentage of the activity observed after Sendai virus infection with IFN-β–CAT reporter in the absence of IκB plasmid.
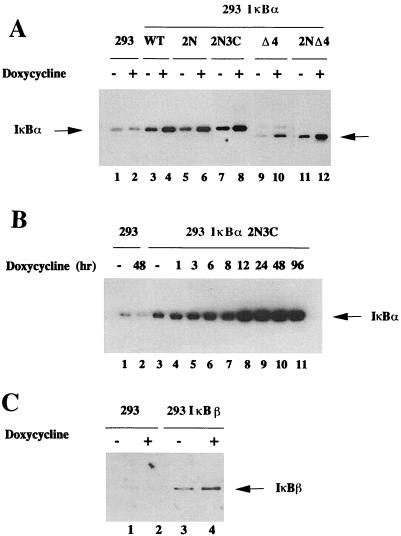
Establishment of human 293 cells inducibly expressing IκBα and IκBβ transgenes.
Human 293 cells constitutively expressing Dox-inducible, reverse-tetracycline-inducible transactivator (rtTA-293) were generated as previously described (25); in a second selection, rtTA-293 cells were established that inducibly expressed wtIκBα, mutant IκBα (-2N, -3C, -2N+3C, -Δ4, and -2NΔ4) and wtIκBβ (Fig. 2). For each construct, 10 to 20 potential clones were expanded individually and screened by immunoblot for protein expression after Dox addition for 48 h. For each IκB construct, at least three inducible clones were selected and utilized for further studies; a representative clone expressing each construct is analyzed for Dox inducibility (Fig. 2A). Clones were also selected for their ability to grow at approximately the same rate as parental 293 cells, since overexpression of different IκBs decreased cell growth and in some clones induced apoptotic cell death (12). Most clones displayed basal IκB expression prior to Dox addition, detected with the MAD3 antibody (Fig. 2A, lanes 3, 5, 7, 9, 11, and 13) and displayed Dox-inducible transgene expression (Fig. 2A, lanes 4, 6, 8, 10, 12, and 14). IκBβ-expressing cells also displayed basal levels of transgene expression (Fig. 2C, lanes 3 and 4).
FIG. 2.
Dox-inducible expression of IκB in rtTA-293 cells. (A) Cells selected for wtIκBα or TD-IκBα expression were induced with Dox for 48 h (+) or not induced (−). (B) Immunoblot analysis of IκBα-2N+3C expression after Dox induction for 0 to 96 h. (C) IκBβ induced expression by Dox for 48 h (+) or not induced (−). Whole-cell extract (20 μg) prepared from induced or uninduced rtTA-293-, wtIκBα-, IκBα-2N-, IκBα-2N+3C-, IκBα-2NΔ4-, and IκBβ-expressing cells were subjected to SDS-PAGE and transferred to nitrocellulose membrane. WtIκBα and IκBα mutants were detected with IκBα-MAD3-specific antibody (33), and wtIκBβ was detected with IκBβ-specific antibody (Santa Cruz Biotechnology).
The kinetics of IκB transgene induction were characterized at various times after Dox induction, and a representative analysis is shown for the IκBα-2N+3C clone (Fig. 2B). This clone possessed basal transgene expression compared to control rtTA-293 cells (compare Fig. 2B, lanes 1 to 3). Between 1 and 3 h after Dox addition, the level of IκBα transgene expression increased (Fig. 2B, lanes 3 to 5) and reached a peak at about 12 h after Dox addition; high-level expression remained constant for 96 h postinduction (Fig. 2B, lanes 8 to 11). Each selected cell line possessed a similar expression pattern; IκBα transgene expression was detectable before Dox induction but increased strongly and progressively following Dox treatment (Fig. 2B). In subsequent experiments, Dox was added 48 h before Sendai virus infection.
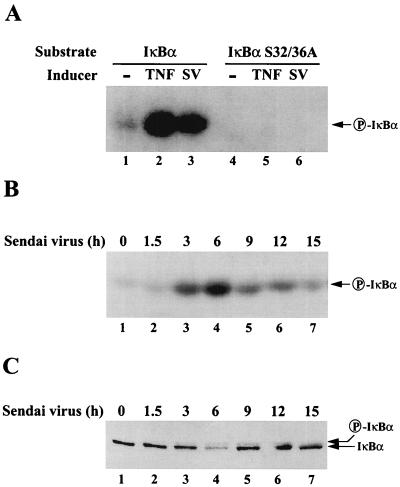
Virus-induced activation of the IKK complex.
To examine the kinetics of Sendai virus-induced activation in 293 cells, the induction of IκBα phosphorylation by the IKK complex was first examined. Sendai virus infection led to activation of the IKK complex as demonstrated by an in vitro kinase assay with immunoprecipitated IKK and the IκBα (aa 1 to 55) protein as substrate; activation of IKK by Sendai virus was similar to the level of activated IKK observed after TNF-α stimulation of 293 cells (Fig. 3A, lanes 1 to 3). No phosphorylation was observed when the IκBα (aa 1 to 55; S32/36A) substrate was used (Fig. 3A, lanes 4 to 6) indicating the specificity of IKK phosphorylation. The kinetics of virus-induced IKK activation demonstrated that IKK activity was maximal at 3 and 6 h after infection (Fig. 3B, lanes 1 to 4) and subsequently decreased between 9 and 15 h (Fig. 3B, lanes 5 to 7). The kinetics of IKK induction also correlated directly with the phosphorylation and degradation of IκBα in virus-infected 293 cells (Fig. 3C). Beginning at 3 h postinfection, a slower-migrating form of phosphorylated IκBα was detected (Fig. 3C, lane 3), while at 6 h the phosphorylated form was detected but the amount of IκBα decreased by fourfold, reflecting phosphorylation-dependent degradation of IκBα (Fig. 3C, lane 4). This kinetic analysis demonstrates that Sendai virus infection of 293 cells leads to activation of the IKK complex and phosphorylation of IκBα.
FIG. 3.
Activation of the IκB kinase complex by Sendai virus infection. (A) Strain 293 cells were either left untreated (lanes 1 and 4), treated with TNF-α (10 ng/ml) for 10 min (lanes 2 and 5), or infected with Sendai virus (80 HAU/ml) for 8 h (lanes 3 and 6). The IκB kinase complex was immunoprecipitated from whole-cell extracts (200 μg) with the anti-IKKα antibody H-744 (Santa Cruz) and assayed for IκBα phosphorylation by using recombinant wild-type GST-IκBα (aa 1 to 55) (lanes 1 to 3) or mutant GST-IκBα (aa 1 to 55; S32/36A) substrate (lanes 4 to 6). (B) Kinetic analysis of IκB kinase activation after Sendai virus infection. Strain 293 cells were infected with Sendai virus for different times as indicated, and the kinase activity was measured as described above. (C) Phosphorylation and degradation of IκBα in response to Sendai virus infection. Whole-cell extracts (20 μg) from 293 cells infected with Sendai virus (80 HAU/ml) for different times as indicated were subjected to SDS-PAGE and transferred to nitrocellulose membrane. IκBα was detected with a monoclonal anti-IκBα antibody. The positions of IκBα and phosphorylated IκBα are indicated by the arrows.
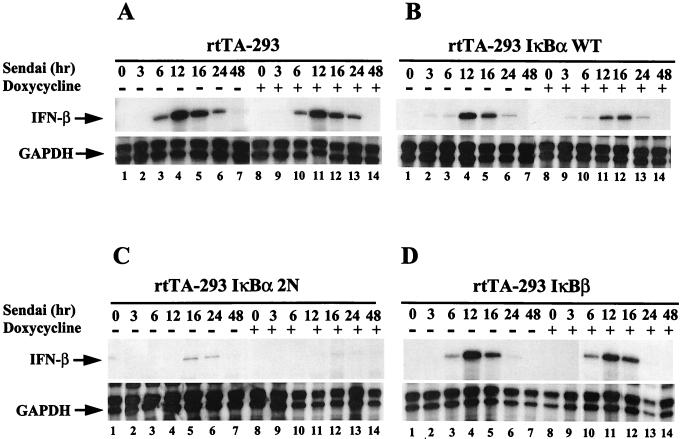
Detection of IFN-β synthesis in IκB-expressing cells.
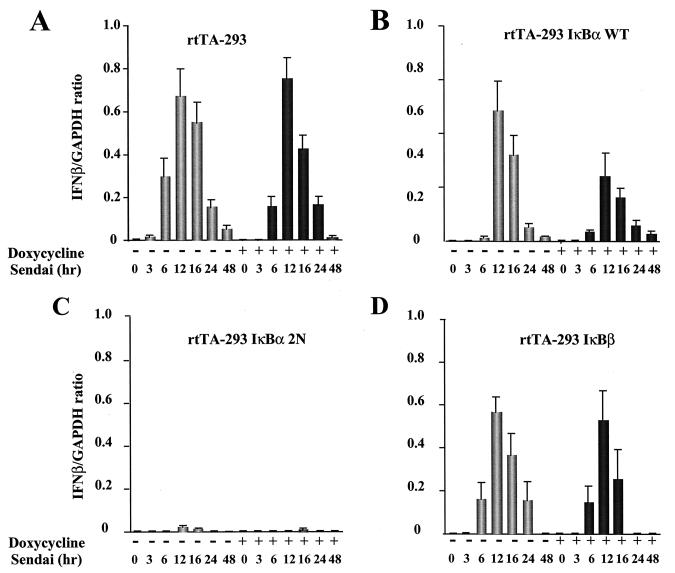
To examine IFN-β inducibility in IκB-expressing cells, total RNA from normal and Sendai virus-infected cells was examined by RNase protection analysis at different times after infection, either with or without Dox addition to increase the level of IκB transgene expression (Fig. 4). In control rtTA-293 cells with or without Dox addition, Sendai virus induced IFN-β mRNA initially at 6 h (Fig. 4A, lanes 1 to 3 and lanes 8 to 10); the amount of mRNA reached a peak at 12 h and thereafter decreased by 24 h (Fig. 4A, lanes 4 to 7 and lanes 11 to 14). In wtIκBα-expressing cells, the induced level of IFN-β was delayed slightly, since only a low level of IFN-β mRNA was detected at 6 h, but again IFN-β mRNA reached a peak of expression at 12 h (Fig. 4B, lanes 1 to 7); the virus-induced level of IFN-β mRNA in wtIκB-expressing cells was not significantly reduced compared to rtTA-expressing cells (compare Fig. 4A and B, lanes 3 to 5, and Fig. 5A and B). Dox induction of the wtIκBα transgene reduced the maximum level of IFN-β mRNA by approximately twofold (Fig. 4B, lanes 8 to 14, and Fig. 5A and B) relative to rtTA-expressing cells, indicating that wtIκB overexpression inhibited but did not completely block IFN-β mRNA expression. However, strikingly, in IκBα2N-expressing cells only low levels of IFN-β mRNA were detected at 12 and 16 h after infection (Fig. 4C, lanes 1 to 7), likely reflecting the leakiness of transgene expression in these cells. Further induction of the IκBα-2N transgene with Dox treatment completely inhibited IFN-β mRNA expression (Fig. 4C, lanes 8 to 14, and Fig. 5C), resulting in an almost 100-fold reduction in IFN-β mRNA levels. Similar low levels of expression were also observed in IκBα-2NΔ4- and -2N+3C-expressing cells with or without Dox addition (data not shown). IFN-β mRNA was also induced in IκBβ-expressing cells at 6 to 16 h after Sendai virus infection (Fig. 4D, lanes 1 to 7); Dox induction of the IκBβ transgene resulted in a partial decrease of this gene expression (Fig. 4D, lanes 8 to 14, and Fig. 5D).
FIG. 4.
mRNA expression of IFN-β in IκBα- and IκBβ-expressing cells after Sendai virus infection. (A) Total RNA (5 μg) prepared from cells 0 to 48 h after Sendai virus infection was used for RNase protection analysis with the human CK3 cytokine template set of the RiboQuant Multi-Probe RPA kit. Cells lines are indicated at the top of each panel. Panels: A, rtTA-293 cells; B, wtIκBα-expressing cells; C, IκBα-2N-expressing cells; D, IκBβ-expressing cells. Where indicated, the cells were pretreated with Dox 48 h prior to Sendai virus infection. As a control, the level of GAPDH is shown.
FIG. 5.
Quantification of IFN-β mRNA expression. Quantification of RNase protection autoradiographs was obtained by normalizing values to the GAPDH and L32 (housekeeping gene) mRNA levels and plotting the values as IFN-β/GAPDH mRNA ratios. Panels: A, rtTA-expressing cells; B, wtIκBα-expressing cells; C, IκBα-2N-expressing cells; D, IκBβ-expressing cells. Lightly shaded columns, no Dox addition; darkly shaded columns, Dox addition (1 μg/ml) for 48 h.
IFN-β transcription is inhibited in IκB-expressing cells.
IκB-expressing 293 cells were transfected with IFN-β–CAT reporter construct containing the IFN-β promoter from −281 to +19; at 24 h after transfection, cells were Sendai virus infected and analyzed at 48 h. Cells, either treated or not treated with Dox, showed the same level of IFN-β-driven CAT activity, indicating that Dox itself had no effect on IFN-β induction. However, IFN-β gene activity was modulated significantly by Dox-induced transgene expression (Fig. 6). wtIκBα- and wtIκBα-Δ4 expressing cells showed a decrease in reporter gene activity after Sendai virus infection, and this activity was reduced by one-half with Dox addition. Also IκBα-2N- and IκBα-2NΔ4-expressing cells displayed very low levels of gene activity, which was completely inhibited after expression of the IκBα transgene. Finally, 293 cells expressing IκBβ did not show a decrease in CAT activity after Dox induction, despite induction of the IκBβ transgene.
FIG. 6.
Inhibition of IFN-β promoter activity in IκB-expressing 293 cells. IFN-β–CAT reporter plasmid (2.5 μg) was transfected into rtTA-293- and IκB-expressing 293 cells by the calcium phosphate method. After 24 h, cells were infected with Sendai virus (500 HAU/ml) for 90 min as indicated. Cultures were harvested at 48 h posttransfection, and 100 μg of the cytoplasmic extracts was prepared from the various cell lines and assayed for CAT activity for 4 h. As indicated, the cells were treated or not treated with Dox 48 h before transfection. The CAT activity observed with extracts of rtTA cells infected by Sendai virus (20 to 30% acetylation) was taken as the 100% value.
Analysis of PRDII DNA binding activity in IκB-expressing cells.
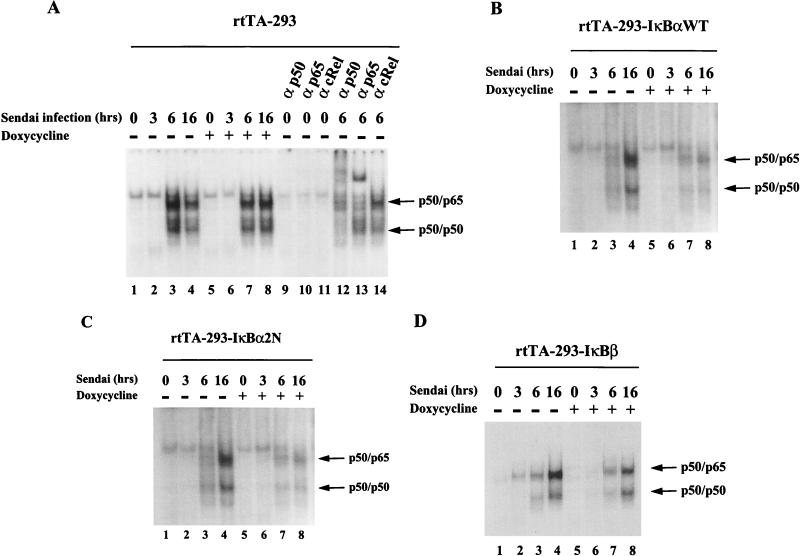
To correlate changes in gene activity with potential changes in NF-κB–PRDII complex formation after virus infection, mobility shift analyses were performed with nuclear extracts from rtTA-293 and IκB-expressing cells (Fig. 7). Subunit composition of the protein-DNA complexes was determined with antibodies specific to p50, p65, and c-Rel, since previous experiments demonstrated that these three NF-κB subunits constituted the majority of NF-κB binding activity on PRDII (17). The specificity of complex formation was controlled by competition with a 200-fold excess of unlabeled PRDII oligonucleotide. For rtTA-293 cells, a specific complex appeared at 6 and at 16 h after Sendai virus infection. Anti-p50 and anti-RelA(p65) antibodies resulted in a prominent shift-up of the inducible complex, demonstrating that the lower complex represented the NF-κB p50-p50 homodimer and the upper complex represented the p50-p65 heterodimer (Fig. 7A). With nuclear extracts from wtIκB-expressing cells infected with Sendai virus, PRDII protein-DNA complex formation corresponding to p50-p50 homodimers and p50-p65 heterodimers were dramatically reduced in intensity and temporally delayed in appearance until 16 h after infection (Fig. 7B). Similarly, in IκBα-2N-expressing 293 cells, NF-κB complex formation was inhibited and detected only at 16 h after Sendai virus infection (Fig. 7C). With IκBβ-expressing cells, formation of NF-κB-PRDII complexes was only slightly reduced at 16 h after infection (Fig. 7D). These results demonstrate that IκBα expression interferes both kinetically and quantitatively with the formation of NF-κB protein-DNA complexes on the PRDII element of IFN-β promoter after virus infection.
FIG. 7.
Analysis of PRDII DNA binding activity in IκB-expressing cells. Nuclear extracts were prepared from rtTA-293-expressing (A), wtIκBα-expressing (B), IκBα-2N-expressing (C), and IκBβ-expressing (D) cells. Cells were infected by Sendai virus (500 HAU/ml) and harvested at the times indicated. Cells were treated with Dox at 48 h prior to Sendai virus infection. Nuclear extracts were incubated in the presence of 5 μg of poly(dI-dC) for 20 min prior to the addition of radiolabelled PRDII probe. For supershift experiments, NF-κB-specific antisera (31) were preincubated in presence of electrophoretic mobility shift assay buffer and poly(dI-dC) prior to the addition of the nuclear extracts.
DISCUSSION
In this study, the potential inhibitory effects of IκBα and IκBβ on IFN-β transcriptional activity were analyzed in transient transfections and in stable 293 cell lines expressing IκB transgenes under Tet-inducible control. In transient-transfection studies, high levels of IFN-β–CAT reporter gene activity were produced after Sendai virus infection, whereas overexpression of wtIκBα inhibited IFN-β transcription in a dose-dependent manner. Overexpression of different mutated forms of IκBα, particularly IκBα-2N(S32A/S36A), completely blocked IFN-β transcription even at low levels of basal expression. IκBα-3C and -Δ4 also inhibited IFN-β transcription more dramatically than wtIκBα. In contrast, IκBβ was a poor inhibitor of IFN-β transcription, indicating a minimal role for IκBβ in the regulation of NF-κB-dependent IFN-β gene expression. Similar results were obtained by measuring IFN-β mRNA accumulation in Tet-inducible rtTA-293 cells expressing the various IκB transgenes. The inhibition of IFN-β transcription in IκBα- and IκBβ-expressing cells correlated directly with the delayed appearance of NF-κB-PRDII complex formation after Sendai virus infection. Overexpression of IκBα or IκBβ impaired NF-κB binding at an early stage of infection, and the later appearance of NF-κB-PRDII complexes at 16 h in IκBα-expressing cells was not sufficient to restore full IFN-β inducibility. Dox-inducible IκBβ expression also resulted in a slightly later appearance of NF-κB binding activity (16 h compared to 6 h in control cells) which decreased IFN-β expression moderately.
The IFN-β promoter contains multiple overlapping positive and negative regulatory domains that bind specific members of the NF-κB, IRF, and ATF transcription factor families in an induction-specific manner, together with the chromatin-associated HMGI(Y) proteins (reviewed in reference 20). Extensive work by the Maniatis and Thanos groups revealed that virus-induced activation of the IFN-β promoter is due to the assembly of a higher-order transcription enhancer complex called an enhanceosome (15, 16, 24, 29, 40, 44). Transcriptional synergy involved in IFN-β activation also requires interaction of all transcription factor activation domains with CBP/p300 (14, 21, 23, 29, 54). A novel domain (aa 322 to 458), termed the synergism domain, was identified in RelA; this domain contains a potential leucine zipper domain present in CBP and CBP-interacting proteins. Through this domain, RelA associates with CBP, and this interaction is essential for transcriptional synergy. The activation domains of the IFN-β transcription factors also interact with CBP in vivo and potentially stabilize the initial association between RelA and CBP. Interestingly, the enhanceosome is able to make contact with components of the basal transcription machinery in vitro (TFIID, TFIIA, TFIIB, and the USA coactivator) (24). Based on these findings, it was proposed that synergistic activation of IFN-β initially involves simultaneous recruitment of RNA polymerase II and the basal transcriptional machinery by the enhanceosome via CBP recruitment by RelA, implicating CBP as a bridge between transcriptional machinery and the IFN-β enhancer. Consistent with the enhanceosome model, we were able to interfere with IFN-β transcription in vivo by preventing the assembly of the complete enhanceosome; continued sequestration of NF-κB in the cytoplasm, particularly by the TD-IκBα forms, prevented formation of the enhanceosome and activation of IFN-β transcription.
The Thanos group demonstrated that the IκBβ inhibitory activity was facilitated by the interaction of NF-κB with HMGI(Y), and a part of their study was based on the analysis of complexes bound on a PRDII probe (44). Our data are complementary with these observations, and in the context of Sendai virus induction in 293 cells, IκBβ does not appear to be involved in the control of IFN-β transcription. A requirement for IκBβ-regulated NF-κB activity may be unnecessary in the context of IFN-β activation because of the rapid and transient nature of IFN-β induction after virus infection. Together with other regulatory protein-DNA interactions, IFN-β induction occurs within the first 6 to 12 h of infection and then is rapidly repressed.
In response to induction by TNF-α or IL-1, the NF-κB-inducing kinase activates the IKK complex that directly phosphorylates IκBα and IκBβ at two amino-terminal serine residues, leading to IκBα and IκBβ ubiquitination and subsequent degradation by the proteasome (36). It has recently been shown that other IKK-associated proteins, such as NEMO/IKKγ and IKAP, regulate the IKK complex and are required for the activation by TNF-α or IL-1 (11, 34, 51). Mitogen-activated protein kinase kinase kinase 1 (MEKK1) has also been identified as an upstream regulator of the IKK complex (30). HTLVI-Tax protein has recently been shown to activate IKKα and IKKβ, leading to NF-κB activation. Furthermore, a dominant negative mutant of NIK blocked Tax induction of NF-κB, thus implicating NIK as a critical upstream regulator (10, 18, 45, 52). Although many viruses induce NF-κB binding activity, this study demonstrates for the first time the activation of the IKK complex by Sendai virus and the subsequent phosphorylation and degradation of IκBα. Strikingly, the kinetics of the IKK activation by Sendai virus temporally reflect not only NF-κB induction but also virus-induced activation of IFN-β mRNA synthesis. At present, the involvement of upstream kinases in the phosphorylation of the IKK complex by Sendai virus remains to be determined.
ACKNOWLEDGMENTS
This study was supported by grants from the Medical Research Council of Canada. M.A. was supported by a FRSQ Santé fellowship, H.N. and C.H. were supported by FCAR studentships, and R.L. was supported by a Fraser, Monat, and McPherson Scholarship from McGill University. J.H. is a Senior Scientist of the Medical Research Council of Canada.
REFERENCES
- 1.Baldi L, Brown K, Franzoso G, Siebenlist U. Critical role for lysines 21 and 22 in signal-induced, ubiquitin-mediated proteolysis of IκBα. J Biol Chem. 1996;271:376–379. doi: 10.1074/jbc.271.1.376. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 3.Beauparlant P, Hiscott J. Biological and biochemical inhibitors of the NF-κB/Rel proteins and cytokine synthesis. Cytokine Growth Factor Rev. 1996;7:175–190. doi: 10.1016/1359-6101(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 4.Beauparlant P, Kwon H, Clarke M, Lin R, Sonenberg N, Wainberg M, Hiscott J. Transdominant mutants of IκBα block Tat-TNF synergistic activation of HIV-1 expression and virus multiplication. J Virol. 1996;70:5777–5785. doi: 10.1128/jvi.70.9.5777-5785.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beauparlant P, Lin R, Hiscott J. The role of the C-terminal domain of IκBα in protein degradation and stability. J Biol Chem. 1996;271:10690–10696. doi: 10.1074/jbc.271.18.10690. [DOI] [PubMed] [Google Scholar]
- 6.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown K, Franzoso G, Baldi L, Carlson L, Mills L, Lin Y-C, Gerstberger S, Siebenlist U. The signal response of IκBα is regulated by transferable N- and C-terminal domains. Mol Cell Biol. 1997;17:3021–3027. doi: 10.1128/mcb.17.6.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of IκB-α proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 10.Chu Z-L, Didonato J, Hawiger J, Ballard D W. The Tax oncoprotein of human T-cell leukemia virus type 1 associates with and persistently activates IκB kinases containing IKKα and IKKβ. J Biol Chem. 1998;273:15891–15894. doi: 10.1074/jbc.273.26.15891. [DOI] [PubMed] [Google Scholar]
- 11.Cohen L, Henzel W J, Baeuerle P A. IKAP is a scaffold protein of the IκB kinase complex. Nature. 1998;395:292–296. doi: 10.1038/26254. [DOI] [PubMed] [Google Scholar]
- 12.DeLuca C, Kwon H J, Pelletier N, Wainberg M A, Hiscott J. NF-κB protects HIV-1 infected myeloid cells from apoptosis. Virology. 1998;244:27–38. doi: 10.1006/viro.1998.9085. [DOI] [PubMed] [Google Scholar]
- 13.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 14.Eckner R. p300 and CBP as transcriptional regulators and targets of oncogenic events. J Biol Chem. 1996;377:685–688. [PubMed] [Google Scholar]
- 15.Escalante C R, Yie J, Thanos D, Aggarwal A K. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 16.Falvo J V, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFNβ gene enhancer by transcription factors and the architectural protein HMG I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 17.Garoufalis E, Kwan I, Lin R, Mustafa A, Pepin N, Roulston A, Lacoste J, Hiscott J. Viral induction of the human interferon beta promoter: modulation of transcription by NF-κB/Rel proteins and interferon regulatory factors. J Virol. 1994;68:4707–4715. doi: 10.1128/jvi.68.8.4707-4715.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geleziunas R, Ferrell S, Lin X, Mu Y, Cunningham E, Grant M, Jr, Connelly M, Hambor J, Marcu K, Greene W. Human T-cell leukemia virus type 1 Tax induction of NF-κB involves activation of the IκB kinase α (IKKα) and IKKβ cellular kinases. Mol Cell Biol. 1998;18:5157–5165. doi: 10.1128/mcb.18.9.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirano F, Chung M, Tanaka H, Maruyama N, Makino I, Moore D, Scheidereit C. Alternative splicing variants of IκBβ establish differential NF-κB signal responsiveness in human cells. Mol Cell Biol. 1998;18:2596–2607. doi: 10.1128/mcb.18.5.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiscott J, Nguyen H, Lin R. Molecular mechanisms of interferon beta gene induction. Sem Virol. 1995;6:161–173. [Google Scholar]
- 21.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T M, Rose D W, Rosenfeld M G, Glass C K. Nuclear integration of JAK/STAT and RAS/AP-1 signalling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaffray E, Wood K M, Hay R T. Domain organization of IκBα and the sites of interaction with NF-κB p65. Mol Cell Biol. 1995;15:2166–2172. doi: 10.1128/mcb.15.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kee B L, Arias J, Montminy M R. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 24.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol Cell. 1998;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 25.Kwon H, Pelletier N, DeLuca C, Genin P, Cisternas S, Lin R, Wainberg M A, Hiscott J. Inducible expression of IκBα repressor mutants interferes with NF-κB activity and HIV-1 replication in Jurkat T cells. J Biol Chem. 1998;273:7431–7440. doi: 10.1074/jbc.273.13.7431. [DOI] [PubMed] [Google Scholar]
- 26.Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol Cell Biol. 1996;16:1401–1409. doi: 10.1128/mcb.16.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.May M J, Ghosh S. Signal transduction through NF-κB. Immunol Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 28.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J W, Young D B, Barbosa M, Mann M. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 29.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 30.Nemoto S, Didonato J, Lin A. Coordinate regulation of IκB kinases by mitogen-activated protein kinase kinase kinase 1 and NF-κB inducing kinases. Mol Cell Biol. 1998;18:7336–7343. doi: 10.1128/mcb.18.12.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pepin N, Roulston A, Lacoste J, Lin R, Hiscott J. Subcellular redistribution of HTLV-1-Tax protein by NF-κB/Rel transcription factors. Virology. 1994;204:706–716. doi: 10.1006/viro.1994.1586. [DOI] [PubMed] [Google Scholar]
- 32.Phillips R J, Ghosh S. Regulation of IκBβ in WEHI 231 mature B cells. Mol Cell Biol. 1997;17:4390–4396. doi: 10.1128/mcb.17.8.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez M S, Michalopoulos I, Arenzana-Seisdedos F, Hay R T. Inducible degradation of IκBα in vitro and in vivo requires the acidic C-terminal domain of the protein. Mol Cell Biol. 1995;15:2413–2419. doi: 10.1128/mcb.15.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothwarf D M, Zandi E, Natoli G, Karin M. IKK-γ is an essential regulatory subunit of the IκB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 35.Régnier C, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 36.Scheidereit C. Docking IκB kinases. Nature. 1998;395:225–226. doi: 10.1038/26121. [DOI] [PubMed] [Google Scholar]
- 37.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz E, Antwerp D, Verma I. Constitutive phosphorylation of IκBα by casein kinase II occurs preferentially at serine 293: requirement for degradation of free IκBα. Mol Cell Biol. 1996;16:3554–3559. doi: 10.1128/mcb.16.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suyang H, Phillips R, Douglas I, Ghosh S. Role of unphosphorylated, newly synthesized IκBβ in persistent activation of NF-κB. Mol Cell Biol. 1996;16:5444–5449. doi: 10.1128/mcb.16.10.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 41.Thompson J E, Phillips R J, Erdjument-Bromage H, Tempst P, Ghosh S. IκB-β regulates the persistent response in a biphasic activation of NF-κB. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 42.Traenckner E B, Phal H L, Henkel T, Schmidt N K, Wilk S, Baeuerle P A. Phosphorylation of human IκBα on serines 32 and 36 controls IκBα proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traenckner E B M, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. Phosphorylation of human IκBα on serines 32 and 36 controls IκBα proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran K, Merika M, Thanos D. Distinct functional properties of IκBα and IκBβ. Mol Cell Biol. 1997;17:5386–5399. doi: 10.1128/mcb.17.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uhlik M, Good L, Xiao G, Harhaj E, Zandi E, Karin M, Sun S-C. NF-κB-inducing kinase and IκB kinase participate in human T-cell leukemia virus 1 Tax-mediated NF-κB activation. J Biol Chem. 1998;273:21132–21136. doi: 10.1074/jbc.273.33.21132. [DOI] [PubMed] [Google Scholar]
- 46.Verma I M, Stevenson J K, Schwarz E M, Antwerp D V, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 47.Vilcek J, Sen G. Interferons and other cytokines. In: Fields B, Knipe D M, Howley P M, editors. Fields virology. New York, N.Y: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- 48.Wathelet M G, Lin C H, Parakh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 49.Weil R, Laurent-Winter C, Israel A. Regulation of IκBβ degradation. Similarities to and differences from IκBα. J Biol Chem. 1997;272:9942–9949. doi: 10.1074/jbc.272.15.9942. [DOI] [PubMed] [Google Scholar]
- 50.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 51.Yamaoka S, Courtois G, Bessia C, Whiteside S T, Weil R, Agou F, Kirk H E, Kay R J, Israel A. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 52.Yin M-J, Christerson L B, Yamamoto Y, Kwak Y-T, Xu S, Mercurio F, Barbosa M, Cobb M H, Gaynor R B. HTLV-I Tax protein binds to MEKK1 to stimulate IκB kinase activity and NF-κB activation. Cell. 1998;93:875–884. doi: 10.1016/s0092-8674(00)81447-6. [DOI] [PubMed] [Google Scholar]
- 53.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E. Two contact regions between STAT1 and CBP/p300 in interferon gamma signalling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]