Abstract
The cytoplasmic aggregation of TAR DNA binding protein-43 (TDP-43), also known as TDP-43 pathology, is the pathological hallmark of amyotrophic lateral sclerosis (ALS). However, the mechanism underlying TDP-43 cytoplasmic mislocalization and subsequent aggregation remains unclear. Here, we show that TDP-43 dimerization/multimerization is impaired in the postmortem brains and spinal cords of patients with sporadic ALS and that N-terminal dimerization–deficient TDP-43 consists of pathological inclusion bodies in ALS motor neurons. Expression of N-terminal dimerization–deficient mutant TDP-43 in Neuro2a cells and induced pluripotent stem cell–derived motor neurons recapitulates TDP-43 pathology, such as Nxf1-dependent cytoplasmic mislocalization and aggregate formation, which induces seeding effects. Furthermore, TDP-DiLuc, a bimolecular luminescence complementation reporter assay, could detect decreased N-terminal dimerization of TDP-43 before TDP-43 pathological changes caused by the transcription inhibition linked to aberrant RNA metabolism in ALS. These findings identified TDP-43 monomerization as a critical determinant inducing TDP-43 pathology in ALS.
TDP-43 monomerization associated with spliceosomal dysfunction induces pathological changes in ALS.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the progressive loss of motor neurons, resulting in skeletal muscle weakness and death within 3 to 5 years after onset (1–3). Common pathological hallmarks in patients with ALS are the cytoplasmic mislocalization and aggregation of hyperphosphorylated TAR DNA binding protein-43 (TDP-43) (4, 5), which constitute the so-called TDP-43 pathology. TDP-43 pathology is observed in almost all sporadic ALS cases, which accounts for 90 to 95% of patients with ALS, and in a subtype of frontotemporal lobar degeneration (FTLD) (2, 3). The significance of TDP-43 in ALS pathogenesis is also supported by genetic evidence that mutations in the TARDBP gene, which encodes TDP-43, are causative for inherited ALS (6). Although several mechanisms have been proposed for TDP-43–mediated neurodegeneration, the exact molecular mechanism remains unclear.
TDP-43 is a ubiquitously expressed and predominantly nuclear RNA binding protein shuttling between the nucleus and the cytoplasm and plays a critical role in multiple aspects of RNA metabolism, such as mRNA splicing, stability, transport, and translation (1–3). TDP-43 consists of four domains: an N-terminal domain (NTD), two RNA recognition motifs (RRMs), and a C-terminal domain (CTD). The CTD, which includes a glycine-rich low-complexity region, is a critical component of TDP-43 pathology in ALS. Multivalent interactions of low-complexity domains are essential for RNA binding proteins to undergo liquid-liquid phase separation (LLPS). Through LLPS, cells can sequester biomolecules into membrane-less organelles that are associated with RNA processing. Almost all ALS-linked mutations are located in the CTD, and cleaved C-terminal fragments of TDP-43 accumulate in the brain and the spinal cord of patients with ALS (2).
Conversely, recent studies have highlighted the functional significance of TDP-43 NTD. While TDP-43 has been shown to exist in a monomer-dimer equilibrium under normal physiological conditions (7, 8), at high concentrations, head-to-tail interactions between the NTDs of TDP-43 monomers could generate higher-order multimers (9, 10). The dimerization/multimerization of the TDP-43 NTD is essential for the physiological splicing function (8, 10–12) and formation of nuclear subdomains via LLPS (9, 13). Meanwhile, TDP-43 interaction via the NTD is reportedly involved in pathological aggregate formation and neurotoxicity (8, 10, 11, 14). Despite the hypothetical mechanistic link between impaired TDP-43 dimerization/multimerization and ALS pathogenesis, no studies have examined the TDP-43 multimerization status in the central nervous system (CNS) tissues of patients with sporadic ALS. Here, we aimed to determine the TDP-43 multimerization status in patients with sporadic ALS and to characterize the downstream events and upstream mechanisms involved in TDP-43 monomer-dimer/multimer abnormalities.
RESULTS
TDP-43 dimerization/multimerization is impaired in the CNS tissue of patients with sporadic ALS
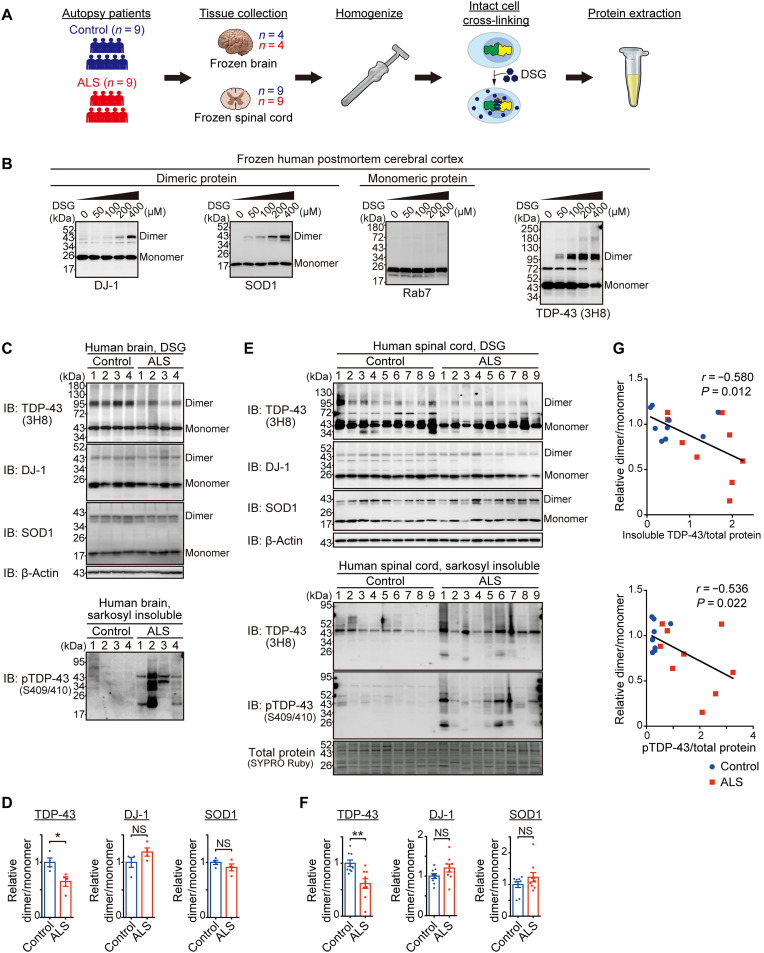
To evaluate the multimerization status of TDP-43 in human postmortem CNS samples, we performed a cross-linking assay using disuccinimidyl glutarate (DSG) (Fig. 1A) (8, 10). DSG is a membrane-permeable homo-bifunctional cross-linker that enables the quantification of the protein dimerization state through immunoblotting in the intracellular physiological environment before cell lysis. DSG cross-linking of human postmortem brain samples allowed us to identify the TDP-43 dimeric and multimeric bands by immunoblotting with an anti–TDP-43 antibody as well as dimeric bands of known dimeric protein controls such as DJ-1 and SOD1, while Rab7, a monomeric protein control, did not show dimeric bands (Fig. 1B and figs. S1 and S2A). These results indicate that the DSG cross-linking assay is suitable for examining the multimerization status of TDP-43 in postmortem CNS tissues. Therefore, we performed a DSG cross-linking assay on postmortem brains (frontal/temporal cortex; table S1) of patients with sporadic ALS with TDP-43 pathology confirmed in the brains and spinal cords by neuropathologists. It was found that the dimer/monomer ratio of TDP-43 was reduced in the affected brain tissues of ALS cases, whereas that of the other dimeric proteins was not (Fig. 1, C and D, and fig. S2, B and C). In contrast, the TDP-43 dimer/monomer ratio was relatively preserved in the cerebellum (fig. S2, D and E), which is consistent with the region specificity of ALS pathology.
Fig. 1. TDP-43 dimerization/multimerization is impaired in the cerebral cortices and spinal cords of patients with sporadic ALS.
(A) Schematic representation of the experimental design of the DSG cross-linking assay using frozen postmortem brain or spinal cord tissues. (B) Representative immunoblots showing a dose-dependent increase in TDP-43 dimer/multimer levels and DJ-1 and SOD1 dimer levels after DSG treatment, with unaffected Rab7 levels. Postmortem frozen cerebral cortex was DSG–cross-linked at the indicated concentrations. The same extracted protein samples were analyzed by immunoblotting for TDP-43; dimeric protein controls, DJ-1 and SOD1; and monomeric protein control, Rab7. The 72 kDa additional band in the TDP-43 immunoblotting is a nonspecific band (see fig. S1). (C) Representative immunoblots (IB) of DSG–cross-linked or sarkosyl-insoluble postmortem brain samples visualized with the indicated antibodies. n = 4, biologically independent samples. (D) Quantification of the dimer/monomer ratio (relative to the mean level of the control samples) of TDP-43, DJ-1, and SOD1 in (C). n = 4, biologically independent samples. (E) Representative immunoblots of DSG–cross-linked or sarkosyl-insoluble samples from postmortem spinal cord tissues visualized with the indicated antibodies. The SYPRO Ruby stain detected the total protein levels. n = 9, biologically independent samples. (F) Quantification of the dimer/monomer ratio (relative to the mean level of the control samples) of TDP-43, DJ-1, and SOD1 in (E). n = 9, biologically independent samples. (G) Correlation dot plots of the dimer/monomer ratio (relative to the mean level of the control samples) of TDP-43 and relative levels of sarkosyl-insoluble TDP-43 (top) or phosphorylated TDP-43 (pTDP-43) (bottom) normalized to the total protein levels (relative to the mean level of all samples) in (E) (Pearson’s correlation method). Data are expressed as means ± SEM (D and F). NS, not significant. *P < 0.05 and **P < 0.01 [unpaired two-sided t test in (D) and (F)].
We examined the TDP-43 multimerization status in the spinal cord tissue of nine patients with ALS and nine control participants to confirm our findings in the representative affected ALS lesions. The TDP-43 dimer/monomer ratio was also lower in the spinal cord of patients with ALS than in controls (Fig. 1, E and F, and fig. S2F). Moreover, the TDP-43 dimer/monomer ratio was negatively correlated with the insoluble and phosphorylated TDP-43 levels in the same spinal cord samples as they were subjected to DSG cross-linking (Fig. 1G). These data indicate that TDP-43 dimerization/multimerization is more impaired in lesions with more severe TDP-43 pathology.
N-terminal dimerization–deficient TDP-43 constitutes pathological inclusion bodies in ALS motor neurons
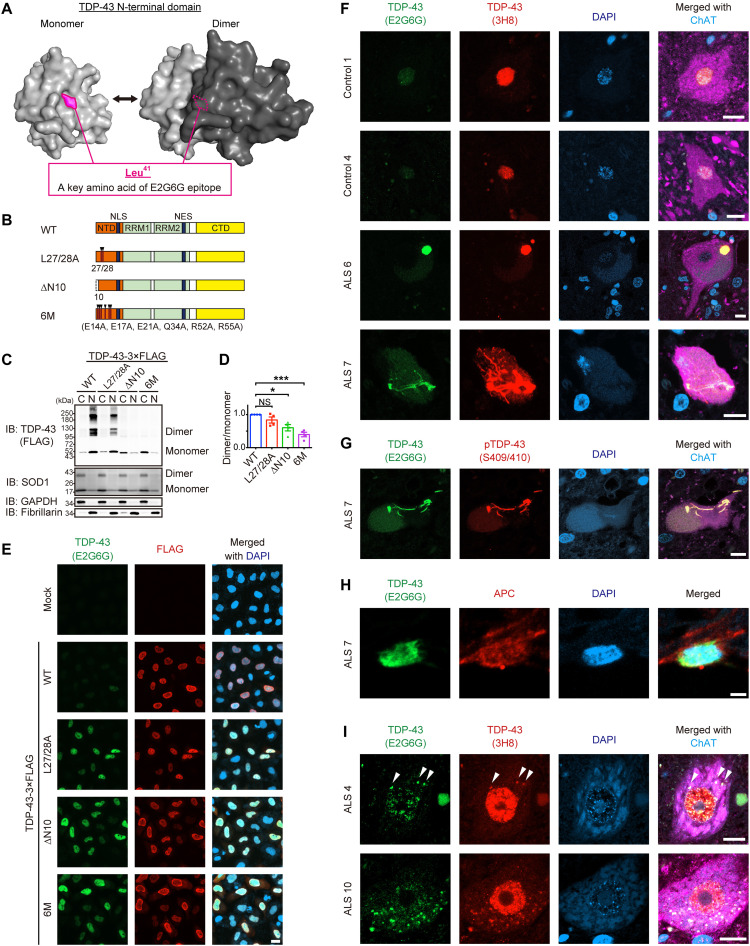
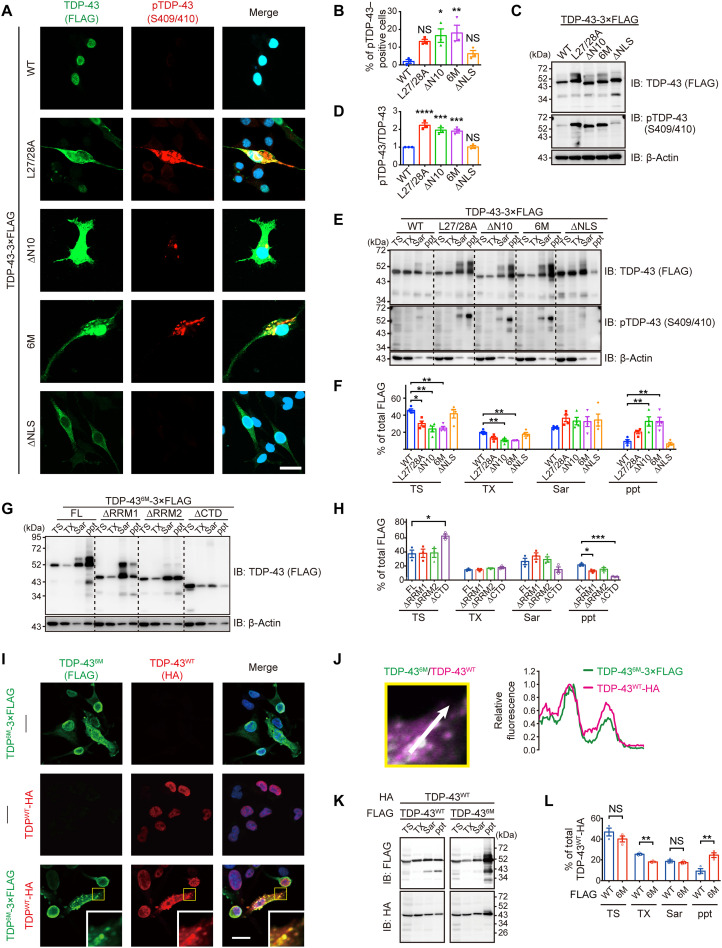
We sought an antibody that recognizes TDP-43 monomers to validate the TDP-43 monomerization status histologically. E2G6G, a monoclonal TDP-43 antibody, recognizes the epitope including Leu41, which is located at the dimer interface of the NTD (Fig. 2A and fig. S3A). To test the ability of this antibody to discriminate N-terminal dimerization–deficient TDP-43 (NDD-TDP-43), we prepared NDD-TDP-43 variants harboring a deletion or mutations in the NTD, which was previously reported to compromise the homodimerization (Fig. 2B) (8, 10, 12). The DSG cross-linking assay revealed that all of these mutants exhibited a propensity for reduced dimerization/multimerization capacity, of which the TDP-436M mutant, carrying six missense mutations in the NTD (E14/17/21A, Q34A, and R52/55A), had the lowest dimerization/multimerization capacity (Fig. 2, C and D). Immunocytochemistry revealed that the E2G6G scarcely reacted with the endogenous and wild-type (WT) TDP-43 (TDP-43WT) but recognized the NDD-TDP-43 mutants (Fig. 2E). The antibody was applied to stain the postmortem spinal cord sections and was found to barely recognize the nuclear physiological TDP-43 in the motor neurons from control cases, but it strongly stained the pathological aggregates with phosphorylated TDP-43 in the motor neurons from ALS cases (Fig. 2, F and G). E2G6G also recognized the glial cytoplasmic inclusions in oligodendrocytes from patients with ALS (Fig. 2H). We further observed the motor neurons from patients with ALS, in which TDP-43 remained in the nucleus. E2G6G stained the TDP-43–positive granular structures in the cytoplasm of some motor neurons stronger than the nuclear TDP-43 (Fig. 2I). These data indicate that E2G6G antibody recognizes NDD-TDP-43, which constitutes the pathological inclusions in ALS motor neurons.
Fig. 2. NDD-TDP-43 constitutes pathological inclusion bodies in ALS.
(A) Three-dimensional models of the monomeric and dimeric NTD of TDP-43 as predicted by AlphaFold2. The critical amino acid of the anti–TDP-43 monoclonal antibody (E2G6G) epitope, Leu41, is indicated. (B) Schematic illustration of the TDP-43WT and three N-terminal dimerization–deficient mutants (NDD mutants). NLS, nuclear localization signal; NES, nuclear export signal. (C) Representative immunoblots of DSG–cross-linked subcellular fractions obtained from Neuro2a cells transiently expressing TDP-43-3×FLAG WT or NDD mutants. Monomeric, dimeric, and multimeric TDP-43 in the cytoplasmic (C) and nuclear (N) fractions were detected using an anti-FLAG antibody. SOD1, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and fibrillarin were used as markers for equal cross-linking, cytoplasmic fractions, and nuclear fractions, respectively. (D) Quantification of the dimer/monomer ratio of TDP-43 (relative to TDP-43WT) in (C). n = 4, biologically independent experiments. Data are expressed as means ± SEM. *P < 0.05 and ***P < 0.001 [analysis of variance (ANOVA) with Tukey’s test]. (E) Representative images of HeLa cells transiently expressing TDP-43 WT or NDD mutants immunostained with E2G6G and anti-FLAG antibody. DAPI, 4′,6-diamidino-2-phenylindole. (F) Representative images of spinal motor neurons from two controls and four patients with sporadic ALS (sALS), with similar results within the groups, immunostained with E2G6G, anti–panTDP-43 (3H8), and anti–choline acetyltransferase (ChAT). (G) Representative images of a spinal motor neuron from a patient with sALS immunostained with E2G6G, anti–pTDP-43 (Ser409/Ser410), and anti-ChAT. (H) Representative images of an oligodendrocyte in the spinal cord from a patient with sALS immunostained with E2G6G and anti-APC. (I) Representative images of spinal motor neurons from a patient with sALS with remaining nuclear TDP-43, immunostained with E2G6G, 3H8, and anti-ChAT. Arrowheads indicate E2G6G-positive cytoplasmic granules. Scale bars, 20 μm (E), 10 μm (F, G, and I), and 2 μm (H).
Monomerization of TDP-43 promotes its mislocalization to the cytoplasm
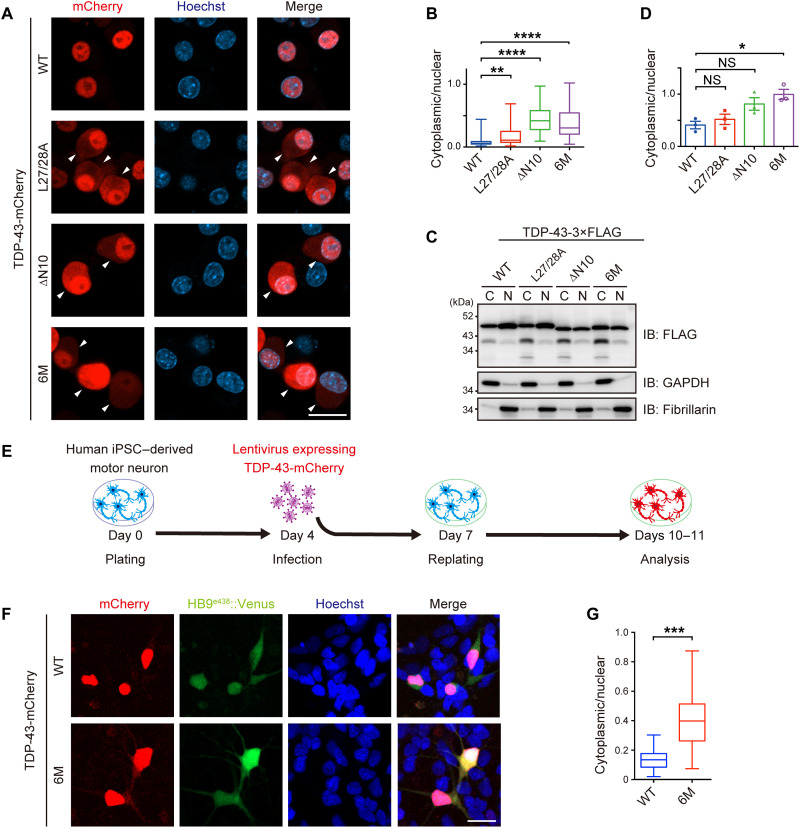
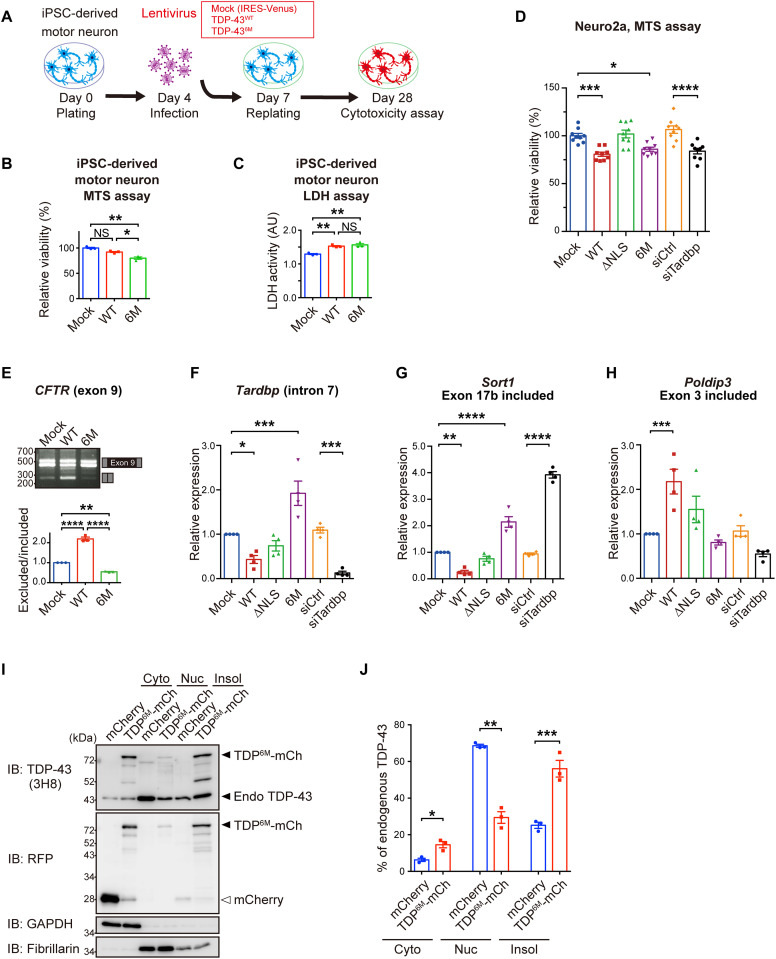
We examined whether a defect in TDP-43 dimerization/multimerization induced TDP-43 pathology in neuronal cell lines. Because the cytoplasmic mislocalization of TDP-43 is one of the characteristics of TDP-43 pathology, we examined the subcellular localization of the WT and the NDD-TDP-43 mutants. The cytoplasmic mislocalization of all the mutants was greater than that of the TDP-43WT in Neuro2a cells, as observed by fluorescent microscopy (Fig. 3, A and B, and fig. S4, A and C) and subcellular fractionation (Fig. 3, C and D, and fig. S4, D and E). Furthermore, the mislocalization of the TDP-436M mutant into the cytoplasm was confirmed in human induced pluripotent stem cell (iPSC)–derived motor neurons, indicating that the monomerization of TDP-43 promotes its cytoplasmic mislocalization in human motor neurons (Fig. 3, E to G, and fig. S5, A and B).
Fig. 3. NDD-TDP-43 mutations induce protein mislocalization into the cytoplasm.
(A) Representative images of Neuro2a cells transiently expressing TDP-43-mCherry WT or NDD mutants. Arrowheads indicate cells with cytoplasmic TDP-43. Scale bar, 10 μm. (B) Box and whisker plots of the cytoplasmic/nuclear ratios of TDP-43-mCherry fluorescence quantified in 100 cells from (A) from three biologically independent experiments. (C) Representative immunoblots of the fractions obtained from Neuro2a cells transiently expressing TDP-43-3×FLAG WT or NDD mutants. (D) Quantification of the cytoplasmic/nuclear ratios of TDP-43-3×FLAG in (C). n = 3, biologically independent experiments. (E and F) Experimental protocol (E) and representative images of human iPSC–derived motor neurons infected with lentivirus expressing HB9e438::TDP-43WT- or HB9e438::TDP-436M-mCherry-IRES-Venus (F). (G) Box and whisker plots of the cytoplasmic/nuclear ratios of TDP-43-mCherry fluorescence quantified in 100 cells from (F) from eight biologically independent experiments. Data are expressed as means ± SEM (D). Box and whisker plots represent the quartiles (boxes), 50th percentiles (center lines), and ranges between the maximum and minimum values (whiskers) in (B) and (G). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 [ANOVA with Tukey’s test in (B) and (D) and unpaired two-sided t test in (G)].
Nuclear RNA export factor-1 mediates the cytoplasmic transport of TDP-43 monomers
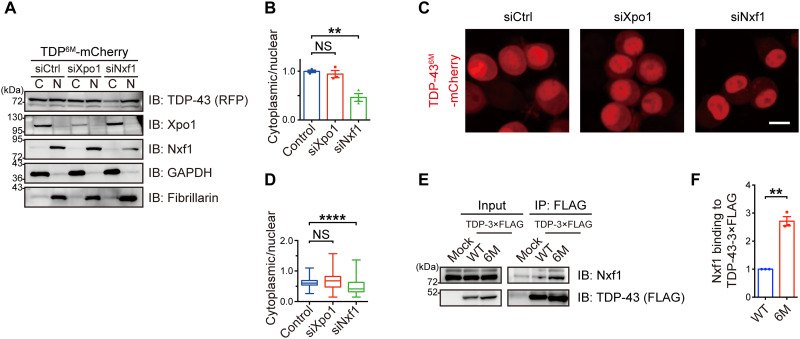
To elucidate the mechanism responsible for TDP-436M cytoplasmic mislocalization, we investigated the involvement of exportin-1 (Xpo1), a protein exporter, and nuclear RNA export factor-1 (Nxf1), a major nuclear RNA exporter component. As previously reported (15, 16), the small interfering RNA (siRNA)–mediated knockdown of Xpo1 did not affect the localization of TDP-43WT and TDP-436M. On the other hand, we found that the suppression of Nxf1 substantially prevented the mislocalization of TDP-436M, while the localization of TDP-43WT was unaffected (Fig. 4, A to D, and figs. S6 and S7). In addition, co-immunoprecipitation experiments revealed that the affinity of TDP-436M for Nxf1 was higher than that of TDP-43WT (Fig. 4, E and F) and was predominantly dependent on the RRM2 domain of TDP-43 (fig. S8). Our findings suggest that Nxf1 recognizes TDP-43 monomers rather than dimers and mediates their nuclear export.
Fig. 4. Nxf1 facilitates cytoplasmic translocation of TDP-43 monomers.
(A) Neuro2a cells transiently expressing TDP-436M-mCherry were treated with control siRNA or siRNAs targeting Xpo1 or Nxf1, followed by nucleocytoplasmic fractionation. Representative immunoblots showing the cytoplasmic (C) and nuclear (N) levels of TDP-43 (RFP), Xpo1, and Nxf1 in the Neuro2a cells expressing TDP-436M-mCherry. GAPDH and fibrillarin were used as cytoplasmic and nuclear markers, respectively. (B) Relative cytoplasmic/nuclear ratios of TDP-43-mCherry in (A) plotted as means ± SEM. n = 3, biologically independent experiments. (C) Representative images of Neuro2a cells transiently expressing TDP-436M-mCherry treated with control siRNA or siRNAs targeting Xpo1 or Nxf1. Scale bar, 10 μm. (D) Box and whisker plots of the cytoplasmic/nuclear ratios of TDP-43-mCherry fluorescence quantified in 100 cells from three independent experiments in (C). The graph shows the quartiles (boxes), 50th percentiles (center lines), and ranges between the maximum and minimum values (whiskers). (E) TDP-43-3×FLAG WT or 6M mutant was transiently transfected in the Neuro2a cells. An anti-FLAG antibody was used for immunoprecipitation of the lysates. Bound endogenous Nxf1 was detected by immunoblotting with an anti-Nxf1 antibody. (F) Relative Nxf1 levels bound to TDP-43-3×FLAG was normalized to the immunoprecipitated FLAG levels (relative to TDP-43WT) in (E) plotted as means ± SEM. n = 3, biologically independent experiments. **P < 0.01 and ****P < 0.0001 [ANOVA with Tukey’s test in (B) and (D) and unpaired two-sided t test in (F)].
TDP-436M mutant recapitulates sarkosyl-insoluble, phosphorylated aggregates as observed in ALS
Because phosphorylated TDP-43 aggregation is another hallmark of TDP-43 pathology in ALS, we investigated whether NDD-TDP-43 variants induce phosphorylated aggregation in Neuro2a cells. All the NDD-TDP-43 variants exhibited high aggregation propensity; the aggregates were immunopositive for phosphorylated TDP-43 and ubiquitin, and partially immunopositive for p62 (Fig. 5, A to D, and figs. S4B and S9). The TDP-43 mutant devoid of nuclear localization signal (TDP-43ΔNLS) localized in the cytoplasm but did not form as many phosphorylated pathological aggregates as NDD-TDP-43 variants. Similarly, in the iPSC-derived motor neurons, TDP-436M expression induced the formation of phosphorylated TDP-43–positive cytoplasmic aggregates (fig. S5, C to E). Pathological aggregation often decreases protein solubility in detergents, leading to the formation of inclusion bodies. Hence, we investigated the detergent solubility of the NDD-TDP-43 variants. Sequential detergent extraction revealed that all NDD-TDP-43 variants except TDP-43ΔNLS were significantly more phosphorylated and enriched in the sarkosyl-insoluble fraction than TDP-43WT (Fig. 5, E and F, and fig. S4, F and G). The TDP-43 nuclear export inhibition by Nxf1 knockdown enhanced TDP-436M phosphorylation and aggregate formation (fig. S10). Moreover, domain analysis using deletion mutants of TDP-436M revealed that the CTD was the key domain for aggregation (Fig. 5, G and H). These findings indicate that NDD-TDP-43 variants recapitulate the TDP-43 pathology observed in ALS.
Fig. 5. NDD-TDP-43 mutations induce phosphorylation, insolubilization, and coaggregation with WT TDP-43.
(A) Representative images of Neuro2a cells transiently expressing TDP-43 WT, NDD mutants, or NLS mutant immunostained with anti-FLAG and anti–pTDP-43. (B) Quantification of the percentage of the pTDP-43–positive cells per FLAG-positive cells in (A). Over 100 cells were quantified per experiment. (C) Representative immunoblots of Neuro2a cells transiently transfected with TDP-43-3×FLAG WT and mutants. (D) Quantification of pTDP-43/FLAG ratios (relative to TDP-43WT) in (C). (E) Representative immunoblots of Neuro2a cells transiently expressing TDP-43-3×FLAG WT and mutants. Proteins sequentially extracted with tris-HCl (TS), 1% Triton X-100 (TX), and 1% sarkosyl (Sar), as well as a sarkosyl-insoluble fraction (ppt), were analyzed by immunoblotting. (F) Quantification of the percentage of each fraction relative to the total amount of TDP-43-3×FLAG in (E). (G) Representative immunoblots of the sequentially extracted lysates from Neuro2a cells transiently expressing TDP-436M-3×FLAG full-length (FL) and deletion mutants. (H) Quantification of the percentage of each fraction to the total amount of TDP-43-3×FLAG in (G). (I) Representative immunofluorescence images of Neuro2a cells transiently expressing TDP-436M-3×FLAG (top), TDP-43WT-HA (middle), or both (bottom). Insets show magnified images of the outlined square area. (J) Line scan analysis of coaggregation of TDP-436M-3×FLAG and TDP-43WT-HA in the inset from (I). (K) Representative immunoblots of the sequentially extracted lysates from Neuro2a cells transiently coexpressing TDP-43WT-3×FLAG or TDP-436M-3×FLAG and TDP-43WT-HA. (L) Quantification of the percentage of TDP-43-3×FLAG in each fraction relative to the total amount of TDP-43WT-HA in (K). Scale bars, 10 μm (A and I). n = 3, biologically independent experiments (B, D, F, H, and L). Data are expressed as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 [ANOVA with Tukey’s test in (B), (D), (F), and (H) and unpaired two-sided t test in (L)].
Recent studies have shown that TDP-43 forms aggregates in a seed-dependent, prion-like manner (17–19). However, it remains unknown whether TDP-43 monomers function as a seed to induce coaggregation with TDP-43WT. Immunocytochemistry analyses revealed that the 3×FLAG-tagged TDP-436M coaggregated with hemagglutinin (HA)–tagged TDP-43WT (Fig. 5, I and J). Sequential detergent extraction also demonstrated that the solubility of the HA-tagged TDP-43WT was reduced in the presence of 3×FLAG-tagged TDP-436M (Fig. 5, K and L). The RRM2 domain of the TDP-43WT was required for interaction with TDP-436M, suggesting the role of the RRM2 domain in recruiting TDP-43WT to these aggregates (fig. S11). These findings suggest that aggregates derived from the TDP-43 monomers function as seeds and trigger coaggregation and sequestration of TDP-43WT.
TDP-436M inhibits the physiological functions of endogenous TDP-43 by sequestrating the endogenous TDP-43 into aggregates
To examine the cytotoxicity of TDP-436M, we performed 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay and lactate dehydrogenase (LDH) assay in iPSC-derived motor neurons and found that TDP-436M exhibited cytotoxicity in human motor neurons (Fig. 6, A to C, and fig. S5, F to H). An MTS assay in Neuro2a cells revealed that TDP-436M induced cytotoxicity at a similar level to that observed with TDP-43WT overexpression or with siRNA-mediated TDP-43 knockdown, whereas TDP-43∆NLS had no cytotoxic effects (Fig. 6D). It has been reported that the overexpression of TDP-43WT induces cytotoxicity despite the absence of aggregate formation and that the toxicities of TDP-43WT and aggregate-forming TDP-43 are induced via different pathways (20). To investigate the differences in the mechanisms of TDP-43WT– and TDP-436M–mediated toxicities, we assessed the splicing function of TDP-436M using the cystic fibrosis transmembrane conductance regulator (CFTR) minigene and quantitative reverse transcription polymerase chain reaction (qRT-PCR) to quantify the known splicing targets of TDP-43 (21–23). TDP-436M lost its ability to skip exon 9 of the CFTR gene and to splice Tardbp, Sort1, and Poldip3 (Fig. 6, E to H). Notably, TDP-436M showed a phenotype similar to that obtained after siRNA-mediated TDP-43 knockdown, and this phenotype was rescued by the additional expression of TDP-43WT (fig. S12), suggesting that TDP-436M induced cytotoxicity by inhibiting the endogenous TDP-43 functions. We further investigated the mechanism by which TDP-436M disrupts the endogenous TDP-43 function. The overexpression of TDP-436M reduced the amount of endogenous TDP-43 in the nucleus and induced its accumulation in the insoluble fraction (Fig. 6, I and J). These results, together with those presented in Fig. 5 (I to L), suggest that TDP-436M inhibits the TDP-43 intrinsic functions by sequestrating the endogenous TDP-43 into the aggregates.
Fig. 6. NDD-TDP-43 mutant impairs endogenous TDP-43 function by sequestrating the endogenous TDP-43 into aggregates.
(A to C) Cell viability and cytotoxicity of iPSC-derived motor neurons infected with lentivirus expressing TDP-43WT or TDP-436M. Experimental protocol (A), MTS assay (B), and LDH assay (C) (relative to mock-transfected cells). (D) Relative viability of Neuro2a cells transiently transfected with TDP-43, control siRNAs, or siRNA targeting TDP-43 (relative to mock-transfected cells) was determined by MTS assay. Triplicate samples were analyzed in three biologically independent experiments. (E) Exon skipping assay for CFTR exon 9. Neuro2a cells were transiently cotransfected with TDP-43 and CFTR minigene reporter plasmids. Splicing of CFTR exon 9 was assessed by RT-PCR (top). Quantification of the exclusion/inclusion ratio of CFTR exon 9 (relative to mock-transfected cells) was performed from the band intensities (bottom). n = 3, biologically independent experiments. (F to H) Quantification of RNA levels of Tardbp intron 7 (F), Sort1 (G), and Poldip3 (H) with indicated exon inclusion. Neuro2a cells were transiently transfected with TDP-43, control siRNAs, or siRNAs targeting TDP-43, and mRNA levels (relative to mock-transfected cells) were analyzed by quantitative RT-PCR. n = 4 biologically independent experiments. (I) Representative immunoblots showing endogenous TDP-43 and mCherry levels in cytoplasmic (Cyto), nuclear (Nuc), and 1% Triton X-100 insoluble fractions (Insol) of Neuro2a cells transiently expressing mCherry or TDP-436M-mCherry. GAPDH and fibrillarin were used as the cytoplasmic and nuclear markers, respectively. (J) Quantification of the percentage of endogenous TDP-43 in each fraction to the total endogenous TDP-43 in (I). n = 3, biologically independent experiments. Data are expressed as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 [ANOVA with Tukey’s test in (B) to (H) and unpaired two-sided t test in (J)].
TDP-DiLuc, a bimolecular luminescence complementation reporter to evaluate TDP-43 dimerization, revealed that spliceosomal integrity is essential for TDP-43 dimerization/multimerization
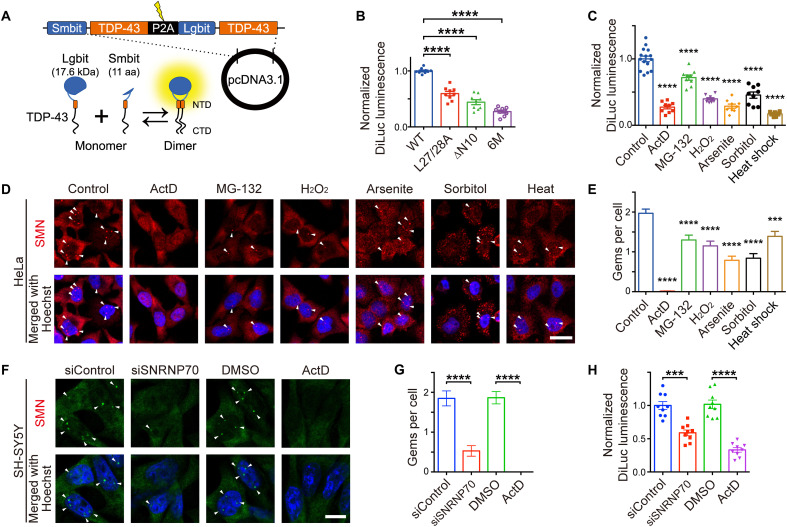
We developed a split luminescent-based reporter, TDP-DiLuc, for high-throughput evaluation of TDP-43 dimerization/multimerization in living cells. TDP-DiLuc is based on the NanoBiT complementation system. NanoBit components, Lgbit and Smbit, are fused to the N-terminal end of TDP-43, allowing for the reconstitution of the active luciferase when TDP-43 dimerizes (Fig. 7A). We first confirmed the decreased luminescence of TDP-DiLuc incorporating NDD-TDP-43 mutants (Fig. 7B and fig. S13, A to C).
Fig. 7. A bimolecular fluorescence complementation assay, TDP-DiLuc, demonstrates that transcription inhibition impairs TDP-43 dimerization/multimerization through spliceosomal defects.
(A) Schematic illustration of TDP-DiLuc, a NanoBiT-based complementation assay to monitor the TDP-43 dimerization/multimerization. aa, amino acids. (B) Relative luminescence in the Neuro2a cells transiently expressing TDP-DiLuc WT or NDD mutants (normalized to mean TDP-43WT levels). Triplicate samples were analyzed in three biologically independent experiments. (C) Quantification of the TDP-43 dimerization under stress conditions. Relative DiLuc luminescence of Neuro2a cells stably expressing TDP-DiLuc WT treated with actinomycin D (ActD), MG-132, H2O2, sodium arsenite, sorbitol, or heat shock (relative to nontreated control cells). Triplicate samples were analyzed in three biologically independent experiments. (D) Representative confocal images of HeLa cells treated with the indicated reagents followed by staining with anti-SMN. Arrowheads indicate SMN-positive Gems. (E) Quantification of Gems per cell in (D). The number of Gems was quantified in 100 cells from three independent experiments. (F) Representative confocal images of SH-SY5Y cells transiently transfected with control siRNA or siRNA targeting SNRNP70. Cells treated with ActD (5 μg/ml; 3 hours) served as the positive control. Arrowheads indicate SMN-positive Gems. DMSO, dimethyl sulfoxide. (G) Quantification of Gems per cell in (F). The number of Gems was quantified in 100 cells from three biologically independent experiments. (H) Quantification of the relative DiLuc luminescence in SH-SY5Y cells transiently transfected with TDP-DiLuc and control siRNA or siRNA targeting SNRNP70. Cells treated with ActD (5 μg/ml; 3 hours) served as the positive control. Triplicate samples were analyzed in three biologically independent experiments. Scale bars, 20 μm (D and F). Data are expressed as means ± SEM (B, C, E, G, and H). ***P < 0.001 and ****P < 0.0001 [ANOVA with Tukey’s test in (B), (C), (E), (G), and (H)].
Various stresses induce the cytoplasmic mislocalization and aggregation of TDP-43 (24–28). To examine whether these stresses cause the pathological changes in TDP-43 by impairing its dimerization/multimerization, we performed a screening using Neuro2a cells stably expressing TDP-DiLuc. Dimerization/multimerization of TDP-43 was impaired to different degrees by all of the examined stresses, including transcription inhibition by actinomycin D (ActD) and energy deprivation (Fig. 7C and figs. S13D and S14). In the ActD-treated cells, DiLuc luminescence was negatively correlated with the E2G6G antibody staining fluorescence (fig. S3, B to E). We also found that RNA is crucial for TDP-43 dimerization/multimerization and for preventing TDP-43 from binding to Nxf1 as both transcription inhibition by ActD and RNA degradation by ribonuclease reduced TDP-43 dimerization/multimerization (fig. S15). TDP-43 is known to localize to nuclear Gems, which are ribonucleoprotein (RNP) granules involved in spliceosomal component maturation (29), but here, TDP-436M lost its localization to the Gems (fig. S16). Hence, we hypothesized that abnormalities in the Gems under these stress conditions might be responsible for TDP-43 monomerization. We found that all stresses decreased the number of Gems represented by the intranuclear survival motor neuron (SMN)-positive puncta (Fig. 7, D and E). In addition, disrupting the spliceosome by the siRNA-mediated knockdown of SNRNP70, a component of the spliceosomal U1 small nuclear RNP, decreased the number of Gems and TDP-43 dimerization/multimerization measured via the TDP-DiLuc assay in the SH-SY5Y neuronal cell line (Fig. 7, F to H). These findings suggest that various stresses, including transcription inhibition, impair TDP-43 dimerization/multimerization, likely through spliceosomal defects.
Decreased TDP-43 dimerization/multimerization induced by transcription inhibition precedes TDP-43 pathological changes
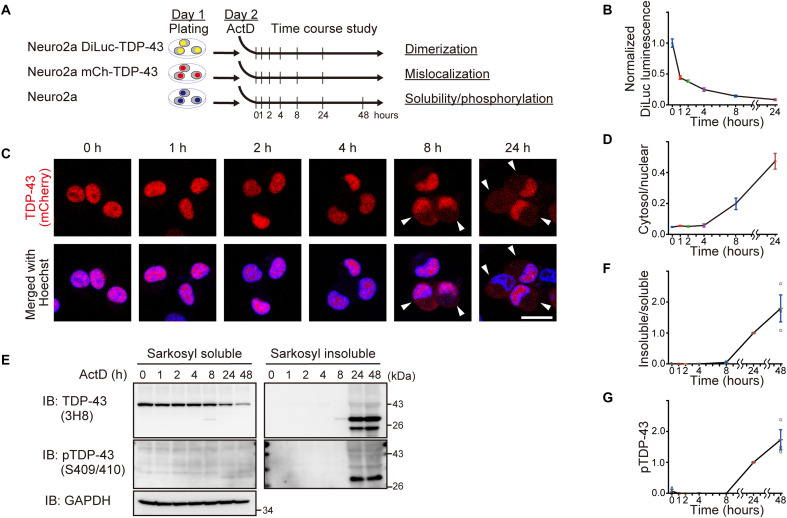
On the basis of the observation of cytoplasmic granules composed of NDD-TDP-43 in motor neurons with the remaining nuclear TDP-43 by immunohistochemistry of ALS spinal cord tissue using E2G6G antibody (Fig. 2I), we hypothesized that the monomerization of TDP-43 NTD might be an early change in ALS and precedes the loss of TDP-43 from the nucleus and the formation of pathological inclusions. Because the administration of ActD eliminated the nuclear Gems, we focused on the effects of ActD-induced transcription inhibition on the temporal sequence of TDP-43 monomerization and pathology. Time-course analyses of TDP-43 alterations in the ActD-treated cells revealed that TDP-43 monomerization, as detected using TDP-DiLuc, preceded TDP-43 pathological changes, such as cytoplasmic mislocalization, reduced solubility, and phosphorylation, despite the sensitivity of each measurement and limitations of the assay (Fig. 8). These results suggest that aberrant RNA metabolism triggers spliceosomal defects, promotes TDP-43 monomerization, and induces ALS-linked TDP-43 pathology.
Fig. 8. Time-course analyses of TDP-43 alterations in ActD-treated cells indicate that TDP-43 pathological changes are preceded by its monomerization.
(A) Experimental protocol of time course analysis of Neuro2a cells treated with ActD (1 μg/ml) for the indicated time. (B) DiLuc luminescence of the Neuro2a cells stably expressing TDP-DiLuc treated with ActD (1 μg/ml) for the indicated time (relative to mean levels at time zero). Triplicate samples were analyzed in three biologically independent experiments. (C) Representative images of Neuro2a cells stably expressing mCherry-TDP-43WT and treated with ActD (1 μg/ml) for the indicated time. Arrowheads indicate cells with cytoplasmic TDP-43. Scale bar, 20 μm. (D) Time course analysis of the cytoplasmic/nuclear ratios of mCherry-TDP-43WT fluorescence quantified in 50 cells from (C) from two biologically independent experiments. (E) Representative immunoblots of sarkosyl-soluble and sarkosyl-insoluble fractions from Neuro2a cells treated with ActD (1 μg/ml) for the indicated time. (F and G) Time course analysis of the insoluble/soluble TDP-43 ratios (F) and levels of pTDP-43 at Ser409/Ser410 in the sarkosyl-insoluble fractions (G) (relative to levels at 24 hours). n = 3, biologically independent experiments. Data are expressed as means ± SEM (B, D, F, and G).
DISCUSSION
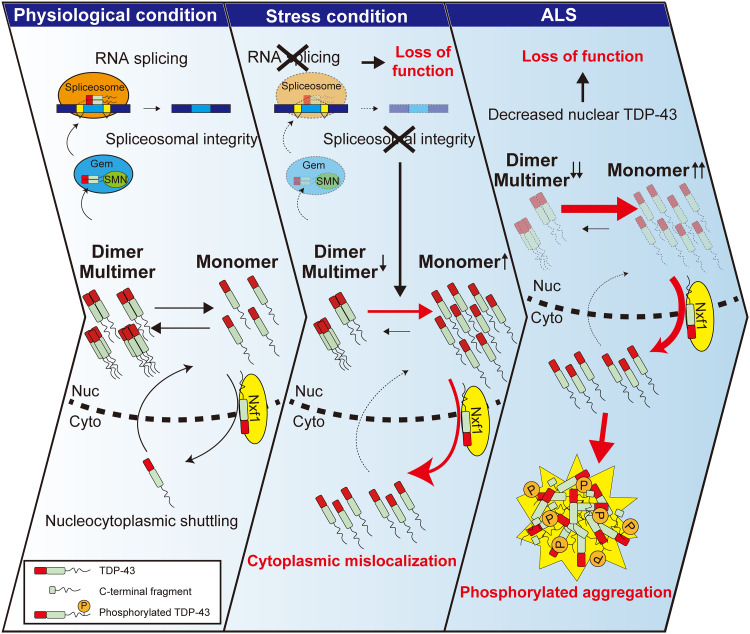
In this study, we found that TDP-43 dimerization/multimerization is impaired in the CNS lesions of patients with ALS that have TDP-43 pathology and pathological inclusion bodies in the ALS motor neurons consist of NDD-TDP-43. TDP-43 monomerization induced its cytoplasmic mislocalization in an Nxf1-dependent manner, along with phosphorylated aggregation of TDP-43, which is a pathological hallmark of ALS. We also demonstrated that monomeric TDP-43 inhibits the splicing and self-regulatory functions of endogenous TDP-43 by sequestering endogenous TDP-43 into aggregates. Furthermore, our bimolecular luminescence complementation assay TDP-DiLuc revealed that various cellular stresses, including transcription inhibition, impaired TDP-43 dimerization/multimerization, which resulted in TDP-43 pathology (Fig. 9).
Fig. 9. Schematic illustration of the role of the TDP-43 monomer-dimer/multimer equilibrium in the development of TDP-43 pathology in ALS.
TDP-43 exists in a monomer-dimer/multimer equilibrium in cells. The NTD of TDP-43 mediates the formation of physiological homodimers, which are essential for its physiological splicing function. The RNA export factor Nxf1 facilitates the nuclear export of monomeric TDP-43 (left). Various types of stresses, including transcription inhibition, impair TDP-43 dimerization/multimerization, likely by inducing spliceosomal defects and disrupting the monomer-dimer/multimer equilibrium, resulting in increased amounts of monomeric nonfunctional TDP-43 (middle). This excess of monomeric TDP-43 is exported from the nucleus in an Nxf1-dependent manner, leading to the cytoplasmic mislocalization of TDP-43 and its subsequent phosphorylation and aggregation, resulting in irreversible TDP-43 pathology in ALS (right).
It is notable that functional dimerization/multimerization of TDP-43 NTD is impaired in the CNS lesions of patients with sporadic ALS and that we identified NDD-TDP-43 as a component of pathological inclusion bodies in ALS motor neurons using E2G6G, an NDD-TDP-43–specific antibody. Furthermore, we found that the TDP-43 dimer/monomer ratio negatively correlated with insoluble and phosphorylated TDP-43 levels in the spinal cord tissue from sporadic ALS, implying that lesions with more monomeric TDP-43 may exhibit more severe TDP-43 pathology. These biochemical results may reflect TDP-43 alterations in neurons and glial cells, especially in oligodendrocytes in which glial cytoplasmic inclusions were immunopositive for E2G6G antibody. Although posttranslational modifications of TDP-43, such as phosphorylation, ubiquitination, C-terminal fragmentation, and acetylation, were observed in postmortem ALS/FTLD brain tissues (4, 5, 30–32), most of these changes are recognized as probably late events associated with TDP-43 pathology. Therefore, the early events and mechanisms preceding TDP-43 pathological changes are poorly understood. We found that the expression of various NDD-TDP-43 mutants in cultured cells comprehensively recapitulated TDP-43 pathological changes such as cytoplasmic mislocalization, phosphorylation, and sarkosyl-insoluble aggregation. It is of great interest that the E2G6G antibody detected cytoplasmic granules even in the morphologically unaffected motor neurons with nuclear TDP-43 of patients with ALS. Moreover, we analyzed the sequential phenotypic changes using a transcription-inhibited cell model and found that decreased TDP-43 dimerization/multimerization preceded TDP-43 pathological changes. Although the detection sensitivity of each phenotypic change may vary, focusing on the functional dimerization/multimerization of TDP-43 may allow the detection of aberrant conditions earlier than the observable pathological changes evaluated in previous studies. Hence, our findings may be important in future research on early pathogenic events related to TDP-43 and in the search for early diagnostic markers of TDP-43 proteinopathy.
Although previous studies have focused on the consequence of defective TDP-43 dimerization/multimerization, such as aberrant splicing functions (8, 10–12), the mechanisms regulating TDP-43 dimerization/multimerization are unknown. A bimolecular green fluorescent protein (GFP) fluorescence complementation (i.e., split-GFP) assay to assess TDP-43 dimerization has been reported (10, 33), but the split-GFP–based complementation is irreversible and probably affects physiological TDP-43 dimerization and functions (34). Therefore, we developed a high-throughput and quantitative assay, TDP-DiLuc, to screen for the upstream mechanisms of TDP-43 dimerization/multimerization; this assay is reversible and has little effect on physiological TDP-43 dimerization/multimerization (35). This assay indicated that the presence of RNA was important for TDP-43 dimerization/multimerization (fig. S15), which suggests that RNA and its associated molecules may play a crucial role in ALS pathogenesis, considering that the RRM2 domain is predominantly involved in the recognition of monomeric TDP-43 by Nxf1 and interaction between TDP-43. It also revealed reduced TDP-43 dimerization/multimerization under various stress conditions involved in ALS pathogenesis. Furthermore, we found that Gem formation was impaired under these stress conditions and that knockdown of SNRNP70, a Gem component, reduced TDP-43 dimerization/multimerization, suggesting that spliceosomal integrity is essential for TDP-43 dimerization/multimerization. Conversely, the NDD-TDP-43 mutant did not localize to Gems, indicating that TDP-43 dimerization/multimerization is required for the proper localization of TDP-43 to the nuclear organelle. We and others previously showed that TDP-43 localizes to nuclear Gems through its association with SMN, playing an essential role in Gem formation and that nuclear Gems collapse in ALS motor neurons (29, 36). These findings suggest that physiological TDP-43 dimerization/multimerization and spliceosomal integrity are closely interrelated and that Gem disruption in ALS may impair TDP-43 dimerization/multimerization and, thus, induce TDP-43 pathology.
A previous study reported that an NDD-TDP-43 mutant (TDP-436M) lost its normal splicing function and formed phosphorylated aggregates, but without significant differences in subcellular localization between the TDP-43WT and the NDD-TDP-43 mutant (10). In addition, other previous studies reporting the localization of NDD-TDP-43 mutants (TDP-43L26/27A and TDP-43ΔN10) did not quantify them (8, 12). Moreover, most of these studies examined the phenotype using a single NDD-TDP-43 mutant or evaluated limited aspects of the pathological changes; therefore, whether a diverse range of NDD-TDP-43 mutants commonly lose splicing function and induce subcellular mislocalization and aggregate formation remains unknown. Compared with previous reports, our study has comprehensively demonstrated that the multimerization status of TDP-43 is essential for determining its nucleocytoplasmic distribution and aggregation propensity; we found that various NDD-TDP-43 mutants uniformly recapitulated TDP-43 pathology.
Meanwhile, there have been several controversial reports regarding NTD-mediated TDP-43 dimerization/multimerization and aggregate formation. One study reported that an NDD-TDP-43 mutant formed phosphorylated aggregates (10), while other studies showed that the NTD was essential for incorporation of TDP-43 into aggregates (8, 14, 37). The latter report artificially induced TDP-43 aggregation through the expression of a TDP-43 mutant tandemly fused with multiple low-complexity domains at the C terminus or the overexpression of an NLS mutant but it did not examine whether the NDD-TDP-43 mutants themselves induced the formation of aggregates. In contrast, our study revealed that the aggregates formed by the NDD-TDP-43 mutants in a CTD-dependent manner served as seeds to sequester the WT or endogenous TDP-43. These findings present the possibility that the monomerization of TDP-43 NTD is critical for the initial induction of aggregates and that interactions through the NTD, as well as the CTD, may be important for the subsequent incorporation of TDP-43 into aggregates. Despite its high aggregation propensity, the NDD-TDP-43 mutant confers toxicity at a similar level to that of TDP-43WT. TDP-43 phosphorylation and aggregate formation have been suggested to be protective cellular reactions (38, 39); therefore, the formation of phosphorylated aggregates may reduce the toxicity of monomeric TDP-43.
TDP-43 is predominantly localized in the nucleus and is constantly shuttling between the nucleus and the cytoplasm (24, 40). Although TDP-43 carries both a classical NLS and a putative nuclear export signal (NES), its nuclear import is mediated by importin-α (24, 41), and several studies have shown that its nuclear export is not dependent on NES-recognizing Xpo1 (15, 16, 42). While these studies have shown that TDP-43 nuclear egress is mediated by passive diffusion and neither Nxf1 nor Aly/REF knockdown affects the nuclear export of TDP-43WT (15), other studies have reported that Nxf1 and other exportins redundantly promote the nuclear egress of TDP-43 (43) and that Nxf1 may interact with TDP-43 through a proteomic approach (44). Recently, another study concluded that TDP-43 nuclear export is independent of Nxf1 based on the observation that the acute depletion of Nxf1 caused a modest delayed inhibition of TDP-43 nuclear export induced by the inhibition of RNA polymerase II (42). Although the association between Nxf1 and TDP-43 nuclear export from these studies seems inconsistent, there are various possible interpretations. First, these studies mainly analyzed the TDP-43WT that was primarily localized in the nucleus. Second, they did not focus on the multimerization status of TDP-43. By focusing on TDP-43 monomerization, we found that ActD-mediated transcriptional inhibition enhances TDP-43 nuclear export, possibly by inducing TDP-43 monomerization, and Nxf1 predominantly recognizes monomeric TDP-43. In addition, Nxf1 knockdown promoted TDP-43 aggregate formation and phosphorylation, suggesting that the active export of monomeric, nonfunctional TDP-43 from the nucleus to the cytoplasm is a defense mechanism in response to cellular stress. Moreover, this result is consistent with previous findings that show that many RNA-binding proteins couple transcription to splicing and mRNA export (45, 46). Furthermore, TDP-43 is involved in mRNA trafficking and facilitates the axonal delivery of target mRNAs (47, 48). While further investigation is needed to determine the extent to which nuclear import and export of TDP-43 contribute to TDP-43 pathology, our findings indicate that the enhanced nuclear export induced by disrupted TDP-43 dimerization/multimerization is as crucial for TDP-43 pathology as the interference of nucleocytoplasmic transporters or the impairment of the nuclear membrane pore complex induced by TDP-43 aggregation (41, 49–52).
It remains debatable whether gain-of-toxicity or loss-of-function mechanisms are involved in TDP-43–mediated neurodegeneration. Overexpression of TDP-43WT is toxic in both in vitro and in vivo models (20, 53), as is increased TDP-43 mRNA expression in the spinal cord or iPSC-derived motor neurons of sporadic ALS and TARDBP mutation carriers (54, 55), which supports the gain-of-toxicity mechanism. Furthermore, the up-regulation of TDP-43 function has been reported in knock-in mice with a TARDBP mutation (23, 56). Conversely, the nuclear clearance of TDP-43 in ALS motor neurons suggests the loss of physiological TDP-43 function. Motor neuron–specific ablation of TDP-43 in mice results in ALS-like phenotypes, including muscle atrophy, weight loss, and motor neuron loss (57, 58). Moreover, TDP-43 knockdown affects numerous RNA splicing events by suppressing the inclusion of cryptic exons (59), and STMN2 and UNC13A, which are critical genes for motor neuron maintenance, are misregulated in patients with sporadic ALS owing to TDP-43 dysfunction (60–63). Thus, despite the suggested gain of function in ALS, accumulating evidence suggests the involvement of loss of function in ALS pathomechanism. Similarly, several studies, including ours, have demonstrated that the splicing function of TDP-43 is diminished by destabilizing its dimerization/multimerization (8, 10–12). Our study revealed that monomeric TDP-43 not only lost its function but also formed aggregates, sequestering endogenous TDP-43 into aggregates by changing it to an aberrant conformation in a prion-like manner, thereby inhibiting its function, which further exacerbated the TDP-43 loss of function. In addition, while TDP-43 expression is strictly regulated by an autoregulatory mechanism (54), monomeric TDP-43 appears to disrupt this autoregulatory mechanism, thereby increasing TDP-43 mRNA levels. Increased cytoplasmic TDP-43 levels and oxidative stress induce endogenous TDP-43 to demix into the cytoplasm, depleting nuclear TDP-43 and causing cell death in the long term (64). Our study showed that the disruption of TDP-43 monomer-dimer/multimer equilibrium may be a relatively early change, followed by cytoplasmic mislocalization of monomeric TDP-43, leading to demixing and sequestering of normal TDP-43 into aggregates. We speculate that this vicious cycle may exacerbate TDP-43 pathology, including nuclear TDP-43 depletion in the long term.
Using two independent assays, i.e., DSG cross-linking and immunohistochemistry with E2G6G antibody, we have shown that the NTD-mediated dimerization/multimerization of TDP-43 is disrupted in ALS-affected lesions. However, the limitations of this study are that neither DSG cross-linking nor E2G6G antibody is sufficiently sensitive for quantifying the absolute total amounts of dimers/multimers or monomers, respectively. Future development of in vivo techniques to measure the absolute amounts of monomeric and dimeric/multimeric forms of TDP-43 will hopefully lead to the detection of early changes and the development of ALS biomarkers.
In conclusion, we provide evidence that TDP-43 monomerization plays an essential role in inducing TDP-43 pathology in ALS. Our findings are similar to those obtained with SOD1 or α-synuclein (65–69), suggesting that the physiological multimerization of aggregate-prone proteins is crucial for maintaining their proper folding and preventing pathological aggregation in various neurodegenerative diseases. Maintaining the monomer-dimer/multimer equilibrium of these pathogenic proteins may represent a common therapeutic target for neurodegenerative diseases.
MATERIALS AND METHODS
Postmortem human tissues
Frozen samples from the brains and spinal cords of patients with sporadic ALS/FTLD or patients with other neurological diseases as controls were obtained by autopsy with informed consent (table S1). ALS was diagnosed according to El Escorial clinical criteria for ALS and the presence of TDP-43 inclusions in lower motor neurons (70). FTLD was diagnosed according to the diagnostic criteria of FTLD and the presence of TDP-43 inclusions in the frontotemporal cortices (71). The neuropathological diagnosis was made by more than two neuropathologists based on immunohistochemical observations, including TDP-43 pathology, and all patients, regardless of the clinical diagnosis of ALS, were screened immunohistochemically for TDP-43 inclusions. The patients were confirmed as sporadic cases through the detailed investigation of their family history, but genetic analyses for ALS/FTLD-linked mutations were not performed. The collection of tissues and their use in this study was approved by the ethics committee of Nagoya University (approval number 328). For immunoblotting analyses, tissues were immediately frozen in liquid nitrogen and stored at −80°C until use. For immunofluorescence analyses, 3 μm sections were prepared from paraffin-embedded tissues, deparaffinized, and incubated at 90°C for 20 min in HistoVT One antigen retrieval solution (Nacalai Tesque Inc., Kyoto, Japan).
Cell culture
Neuro2a and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing glucose (4.5 g/liter) supplemented with 10% (v/v) fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml; all from Thermo Fisher Scientific, MA, USA) at 37°C in a humidified chamber containing 5% (v/v) CO2. SH-SY5Y cells were maintained in Advanced DMEM/F12 (Gibco) supplemented with 10% (v/v) FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml). The transfection procedures of the plasmids and siRNAs were performed using Lipofectamine 2000 and Lipofectamine RNAiMAX (both from Thermo Fisher Scientific), respectively, according to the manufacturer’s instructions.
Neuro2a cells stably expressing TDP-DiLuc or mCherry-TDP-43 were established through the transfection of linearized pTDP-DiLuc or pmCherry-TDP-43, followed by positive selection with G-418 (500 μg/ml; Nacalai Tesque Inc.). The Neuro2a cells stably expressing TDP-DiLuc were exposed to various cellular stresses using the following reagents: ActD (FUJIFILM Wako Pure Chemical Corp., Osaka, Japan; 5 μg/ml for 3 hours unless otherwise stated), Z-Leu-Leu-Leu-H aldehyde (MG-132, Peptide Institute Inc., Osaka, Japan; 10 μM for 4 hours), sorbitol (FUJIFILM Wako; 0.5 M for 30 min), sodium arsenite (Sigma-Aldrich Co. LLC, St. Louis, MO,USA; 0.3 mM for 30 min), or hydrogen peroxide (FUJIFILM Wako; 1 mM for 1 hour). Heat shock was induced by incubating the cells at 42°C for 30 min.
Plasmids
The plasmids encoding TDP-43-3×FLAG and its mutants (ΔNLS, ΔRRM1, ΔRRM2, and ΔCTD), as well as TDP-43-mCherry, were used as previously described (29, 72). For L27/28A, ΔN10, and 6M mutants of TDP-43, mutations were introduced using the PrimeSTAR Mutagenesis Basal Kit (Takara Bio, Shiga, Japan). For the construction of TDP-DiLuc, SmBit-tagged human TDP-43 complementary DNA (cDNA) fused to a 2A self-cleaving peptide sequence from porcine teschovirus-1 (P2A) was amplified using KOD Plus Neo DNA polymerase (TOYOBO, Osaka, Japan) and cloned into the pcDNA3.1(+) vector (Thermo Fisher Scientific). The DNA fragment of LgBit-fused human TDP-43 cDNA was amplified using overlap PCR and was inserted into downstream of the P2A cleavage site described above. The resulting construct, pTDP-DiLuc, was, therefore, arranged on pcDNA3.1(+) in the order of SmBit-TDP-43, P2A, and LgBit-TDP-43.
Antibodies
Antibodies used in this study are as follows: anti–TDP-43 (1:1000; clone 3H8, #MABN45, EMD Millipore, Billerica, MA), anti–TDP-43 (1:1000 for immunoblotting and 1:50 for immunofluorescence; clone E2G6G, #89718, RRID:AB_2920572, Cell Signaling Technology, Danvers, MA, USA), anti–phosphorylated TDP-43 (1:1000 for immunoblotting and 1:5000 for immunofluorescence; pS409/410; clone 11-9, #TIP-PTD-M01, RRID: AB_1961900, Cosmo Bio, Tokyo, Japan), anti–DJ-1 (1:500; #sc-55572, RRID: AB_831639, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-SOD1 (1:1000; #sc-11407, RRID: AB_2193779, Santa Cruz Biotechnology), anti-fibrillarin (1:1000; #2639, RRID: AB_2278087, Cell Signaling Technology), anti–glyceraldehyde-3-phosphate dehydrogenase (1:1000; clone H-12, #sc-166574, RRID: AB_2107296, Santa Cruz Biotechnology), anti–β-actin (1:5000; #5441, RRID: AB_476744, Sigma-Aldrich), anti–choline acetyltransferase (1:50; #AB144P, RRID:AB_2079751, EMD Millipore), anti-adenomatous polyposis coli (APC) (1:100; clone CC-1, #OP80, RRID:AB_2057371, EMD Millipore), anti-FLAG M2 (1:5000; #F1804, RRID: AB_262044, Sigma-Aldrich), anti–DDDDK-tag [1:1000 for immunofluorescence; 5 μg per sample for immunoprecipitation; #PM020, RRID: AB_591224, Medical & Biological Laboratories (MBL) Co LTD, Nagoya, Japan], anti–red fluorescence protein (RFP) (1:1000; #PM005, RRID: AB_591279, MBL), anti-Nxf1 (1:1000; clone D5X4G, #12735, RRID:AB_2798011, Cell Signaling Technology), anti-SMN (1:1000; clone 8/SMN, #610646, RRID:AB_397973, BD Biosciences, Franklin Lakes, NJ, USA), anti-Xpo1 (1:500; clone C-1, #sc-74454, RRID:AB_1122704, Santa Cruz Biotechnology), and anti–HA-tag (1:3000 for immunoblotting and 1:1000 for immunofluorescence; clone TANA2, #M180-3, RRID: AB_10951811, MBL).
Intact-cell cross-linking with DSG
DSG (FUJIFILM Wako) was dissolved in dimethyl sulphoxide (Nacalai Tesque Inc.) to a final concentration of 20 mM immediately before use. Neuro2a cells were harvested using 0.25% (w/v) trypsin-EDTA (1 mM) (FUJIFILM Wako), washed with phosphate-buffered saline (PBS), and resuspended in PBS supplemented with cOmplete EDTA-free protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany) (PBS/PI), followed by incubation with DSG (400 μM unless otherwise stated) for 30 min at room temperature with gentle rotation. The cross-linking reaction was quenched by adding 20 mM tris base (pH 7.4) and by incubating for 15 min at room temperature.
For brain and spinal cord tissue samples, approximately 100 to 200 mg of the cortex or spinal cord was minced and then homogenized using a Dounce tissue grinder (loose, DWK Life Sciences, LLC, Milville, NJ, USA; 10 gentle strokes) in 500 μl of ice-cold PBS/PI. The homogenized tissues were transferred to 15 ml tubes containing 4.5 ml of PBS/PI and centrifuged at 1000 rpm for 5 min. The supernatants were discarded, and the pellets were resuspended in 100 μl of PBS/PI per 100 mg of tissue. The resuspended pellets of approximately 200 μl in volume were incubated with DSG (400 μM unless otherwise stated) at 37°C for 30 min with rotation, followed by quenching with 20 mM tris base (pH 7.4) for 15 min.
Sequential extraction of proteins from postmortem brain and spinal cord or Neuro2a cells
Frozen postmortem brain or spinal cord samples were homogenized in HS buffer [10 mM tris-HCl (pH 7.5) containing 150 mM NaCl, 0.1 mM EDTA, and 1 mM dithiothreitol (DTT)] supplemented with cOmplete protease inhibitor and PhosSTOP phosphatase inhibitors (Roche). An equal volume of HS buffer containing 2% (w/v) sarkosyl was added to each homogenate. The mixtures were incubated for 30 min at 37°C and centrifuged at 100,000g for 30 min at 25°C. The supernatant was used as the sarkosyl-soluble fraction. The pellets were sonicated in SDS sample buffer, boiled for 5 min, and centrifuged at 15,000g for 5 min to remove insoluble debris. The resultant supernatant was used as the sarkosyl-insoluble fraction.
Neuro2a cells were lysed in TS buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 5 mM ethylene glycol tetraacetic acid, cOmplete protease inhibitor, and PhosSTOP phosphatase inhibitors]. The lysates were centrifuged at 20,000g for 30 min at 4°C, and the supernatant was collected as the TS-soluble fraction. The TS-insoluble pellets were resuspended in TX buffer [TS buffer containing 1% (v/v) Triton X-100] and centrifuged at 20,000g for 30 min at 4°C. The supernatant was collected as the TX-soluble fraction. The TX-insoluble pellets were further sonicated in sarkosyl buffer [TS buffer containing 1% (w/v) sarkosyl] and centrifuged at 20,000g for 30 min at 4°C. The supernatant was collected as the sarkosyl-soluble fraction. The remaining pellets were lysed in the SDS sample buffer with sonication, boiled for 5 min, and used as the sarkosyl-insoluble fraction.
Subcellular fractionation
Neuro2a cells were harvested, washed in PBS, and lysed in ice-cold hypotonic buffer [10 mM Hepes, 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 0.5% (v/v) Tween 20, and cOmplete protease inhibitor (pH 7.4)] for 10 min. After centrifugation at 6000g for 1 min, the supernatant was collected as the cytoplasmic fraction. The pellet was resuspended in ice-cold hypertonic buffer [10 mM Hepes, 400 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% (v/v) glycerol, 1 mM DTT, 0.5% (v/v) Tween 20, and cOmplete protease inhibitor (pH 7.4)] and incubated for 30 min at 4°C. After centrifugation at 14,000g at 4°C for 30 min, the resultant supernatant was collected as a nuclear fraction.
Immunoblotting
Protein samples were separated by SDS–polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Immobilon-P, EMD Millipore). After blocking with 2% (w/v) bovine serum albumin in TBS-T [50 mM tris-HCl, 150 mM NaCl, and 0.5% (v/v) Tween 20 (pH 7.4)], the membrane was incubated with the primary antibodies diluted in tris-buffered saline [TBS; 50 mM tris-HCl and 150 mM NaCl (pH 7.4)], followed by incubation with horseradish peroxidase (HRP)–conjugated secondary antibodies. The bound primary antibodies were detected using an Immobilon Crescendo Western HRP substrate (EMD Millipore) according to the manufacturer’s instructions. The total protein contents were detected using SYPRO Ruby protein gel stain (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Live-cell imaging
For the confocal imaging of living cells, cells were seeded and cultured on a four-well cover-slide chamber (AGC Techno Glass Inc., Shizuoka, Japan) coated with poly-d-lysine (Sigma-Aldrich). The cells expressing mCherry-fused TDP-43 and its mutants were stained with Hoechst 33342 (0.5 μg/ml; Thermo Fisher Scientific) for 10 min before observation. Confocal images were obtained using a confocal laser scanning microscope (LSM-700 and LSM-900; Carl Zeiss AG, Oberkochen, Germany) and the equipped software (ZEN; Carl Zeiss AG). Fluorescent intensities of nuclear and cytoplasmic mCherry in the cells were analyzed and quantified using ImageJ software.
iPSC culture and differentiation
The human iPSCs (201B7) were provided by Dr. S. Yamanaka at Kyoto University (73). The maintenance and differentiation of human iPSCs into spinal motor neurons were performed as previously described (74). Briefly, the dissociated human embryoid bodies were plated onto growth factor–reduced Matrigel (33× dilution, thin coated; Corning Incorporated, Corning, NY, USA)–coated 24-well plates at a density of 2 × 105 cells per well and cultured in motor neuron medium (MNM) consisting of media hormone mix or KBM Neural Stem Cell medium (Kohjin Bio, Saitama, Japan) (75) supplemented with 2% B27 supplement (Thermo Fisher Scientific), 1% non-essential amino acids (NEAA), 50 nM retinoic acid (RA), 500 nM purmorphamine, 10 μM cyclic adenosine monophosphate (Sigma-Aldrich), recombinant brain-derived neurotrophic factor (10 ng/ml; R&D Systems, Minneapolis, MN, USA), recombinant glial cell line–derived neurotrophic factor (10 ng/ml; R&D Systems), recombinant human insulin-like growth factor 1 (10 ng/ml; R&D Systems), and l-ascorbic acid (200 ng/ml; Sigma-Aldrich) for 1 week. On day 3 of the adherent differentiation, the iPSC-derived motor neurons were infected with lentivirus expressing TDP-43WT or TDP-436M tagged with mCherry or 3×FLAG tag at the C terminus and Venus fluorescent protein under the control of HB9e438 enhancer and β-globin minimal promoter (74). On day 7, the motor neurons were redissociated using neural tissue dissociation kits (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions and replated onto a 96-well imaging plate (Greiner, Kremsmünster, Austria) at a density of 2 × 104 cells per well in a modified MNM for 2 weeks. For live cell imaging, the cells were processed for the imaging analysis at day 9 of adherent differentiation (2 days after replating) using confocal laser scanning microscope (LSM-900; Carl Zeiss AG). The nuclei were stained using Hoechst33342 (50 ng/ml; Thermo Fisher Scientific). The cells were fixed at 4 weeks of adherent differentiation (3 weeks after replating) and processed for immunocytochemistry and imaging analysis. All experimental procedures using human iPSCs were approved by the ethics committee of Aichi Medical University School of Medicine (approval numbers: 14-004, 2020-213, and 2022-058).
Generation of the HB9e438::TDP-43(WT or 6M)-mCherry-IRES-Venus lentivirus
Lenti-X 293T cells cultured in DMEM supplemented with 10% FBS in 150 mm dishes were transfected with 16 μg of pSIN2-HB9e438-βglo-TDP-43(WT or 6M)-mCherry/3×FLAG-IRES-Venus (a variant of yellow fluorescent protein with fast and efficient maturation) (76) or pSIN2-HB9e438-βglo-TDP-43 (WT or 6M)-mCherry-IRES-Venus and 10 μg of each of the two packaging vectors (pCMV-VSV-G-RSV-Rev and pCAG-HIV-gp; provided by H. Miyoshi) with 200 μl of polyethyleneimine (Polysciences Inc., Warrington, PA, USA), and the medium was changed to the Freestyle 293 expression medium (Thermo Fisher Scientific) the following day. Three days later, the culture supernatant was collected, aliquoted, and stored at −80°C. For the lentiviral infection, lentivirus was added to a motor neuron culture, followed by incubation for 2 hours, after which the total medium was changed to MNM.
Immunofluorescence
Neuro2a, HeLa, or SH-SY5Y cells were fixed with 4% (w/v) paraformaldehyde in a phosphate buffer for 15 min at room temperature and permeabilized in PBS supplemented with 0.5% (v/v) Triton X-100 for 30 min at room temperature. After incubation with the blocking solution [PBS, 5% (v/v) normal serum, and 0.3% (v/v) Triton X-100] for 1 hour at room temperature, the cells were incubated with primary antibodies diluted in the blocking solution overnight at 4°C. The bound primary antibodies were detected using Alexa Fluor–conjugated secondary antibodies (Thermo Fisher Scientific) diluted in the blocking solution. Images were acquired using a confocal laser scanning microscope (LSM-700 and LSM-900; Carl Zeiss AG) and the equipped software (ZEN; Carl Zeiss AG).
Immunoprecipitation
Neuro2a cells were harvested, washed in PBS, and lysed in ice-cold lysis buffer [50 mM tris-HCl, 150 mM NaCl, 2 mM MgCl2, 0.5% (v/v) NP-40, 1 mM DTT, and cOmplete protease inhibitor (pH 7.4]. The lysates were centrifuged at 15,000g for 10 min at 4°C. The supernatant was incubated with an anti-FLAG antibody (MBL) for 1.5 hours at 4°C with gentle agitation, followed by incubation with Protein G sepharose (Cytiva, Tokyo, Japan) for further 1.5 hours. The beads were washed three times with PBS. The immunoprecipitated proteins were eluted with SDS sample buffer at 95°C for 3 min and subjected to immunoblotting.
Cell viability assay
CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS assay, Promega, Madison, WI, USA) and Cytotoxicity LDH Assay Kit-WST (Dojindo, Kumamoto, Japan) were used according to the manufacturer’s instructions. Briefly, iPSC-derived motor neurons were cultured in 96-well plates, and after each treatment, they were incubated with the MTS reagent for 3 hours and then spectrophotometrically measured at 490 and 655 nm for the background using the iMark microplate absorbance reader (Bio-Rad, Hercules, CA, USA). For the cytotoxicity LDH assay, 50 to 100 μl of culture supernatant was used, which was incubated with the substrate for 30 min and spectrophotometrically measured at 490 nm using a plate reader. Neuro2a cells were seeded on poly-d-lysine–coated 96-well plates in DMEM containing glucose (4.5 g/liter) supplemented with 10% (v/v) FBS. After transfection, the cells were differentiated for 48 hours in DMEM containing glucose (1.0 g/liter) supplemented with 2% (v/v) FBS and 2 mM N6,2′-O-dibutyryladenosine-3′,5′-cyclic monophosphate (Nacalai Tesque Inc.). The cell viability was evaluated using the MTS assay.
CFTR minigene assay
The CFTR minigene assay was performed as previously described (77). Briefly, Neuro2a cells were cotransfected with 2 μg of pF5K-TDP-43-3×FLAG and 0.5 μg of pF5K-CFTR exon 9 plasmids (78, 79). Total RNA was extracted from the transfected cells using an RNeasy micro kit (QIAGEN, Venlo, Netherlands). The cDNA was synthesized from 1 μg of total RNA using the PrimeScript II first-strand synthesis kit (Takara Bio). CFTR exon 9 fragments were amplified by PCR using the cDNA and primers, specifically recognizing the minigene construct by PCR, and the PCR products were separated by 2% agarose gel.
RNA isolation and qRT-PCR
Total RNA was isolated using an RNeasy Micro Kit (QIAGEN). The cDNA was synthesized from 1 μg of total RNA using the PrimeScript II first-strand synthesis kit (Takara Bio). qRT-PCR was performed using SYBR Premix Ex Taq II in Thermal Cycler Dice Real Time System II (both from Takara Bio). The mouse Actb gene was used as the internal control for normalization. The primers used for qRT-PCR in this study are listed in table S2.
NanoLuc luciferase assay
Neuro2a cells stably expressing TDP-DiLuc were seeded on 96-well white cell culture plates (Greiner) coated with poly-d-lysine. NanoLuc luciferase assay was performed using Nano-Glo Live Cell Assay System (Promega) according to the manufacturer’s instructions. Luminescence was measured using the SpectraMax Paradigm Multi-Mode Detection Platform (Molecular Devices LLC, San Jose, CA, USA). The relative luminescent intensity was calculated by normalizing the relative luminescence unit using the relative cell viability measured by the MTS assay.
Structural analysis using AlphaFold2
The structures of the monomeric and dimeric NTDs of TDP-43 were predicted using ColabFold (80), a version of the deep neural network AlphaFold2 coupled with Google Colaboratory, using default parameters. The structures were visualized in PyMOL 2.5.5 (Schrödinger Inc., New York, NY, USA).
Statistical analysis
All data obtained from the immunofluorescence, semiquantitative immunoblotting, and qRT-PCR analyses were analyzed using unpaired t tests for the comparison between two groups or one-way analysis of variance (ANOVA) followed by post hoc Tukey’s multiple comparison t test for the comparison among three or more groups. Pearson’s correlation method was used to calculate the correlation coefficient between variables. All statistical analyses were performed using GraphPad Prism software (GraphPad Software, La Jolla, CA, USA).
Acknowledgments
Funding: This work was supported by Grants-in-Aid for Scientific Research JP18H02740, JP19KK0214, JP22H02988, and JP22H00467 from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan/Japan Society for the Promotion of Science (JSPS), as well as AMED under grant numbers JP22ek0109426 and JP23wm0425014 (to K.Y.) and JP22bm0804020 and JP23bm1423003 (to Y.O.).
Author contributions: K.Oi., S.W., and K.Y. designed the study. K.Oi., S.W., and Y.K. performed the experiments and analyzed the data with support from O.K. and A.S. under the supervision of K.Y. K.On. and Y.O. conducted experiments in iPSC-derived motor neurons. Y.I. and M.K. obtained the patients’ autopsy samples and performed neuropathological and clinical diagnoses. K.Oi., S.W., and K.Y. wrote the manuscript. All authors approved the final manuscript.
Competing interests: Y.O. is a scientific advisor of Kohjin Bio Co. Ltd. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Human iPSC and lentiviral vectors can be provided by the RIKEN Bioresource Research Center (BRC)’s pending scientific review and a completed material transfer agreement. Requests for the hiPSC (201B7) should be submitted to RIKEN BRC at https://cell.brc.riken.jp/en/hps/search_hps_en. Requests for the lentivirus vector (pCSII-EF-MCS-IRES-Venus) and the packaging plasmid (pCAG-HIVgp, pCMV-VSV-G-RSV-Rev) should be submitted to RIKEN BRC at https://dna.brc.riken.jp/en/.
Supplementary Materials
This PDF file includes:
Figs. S1 to S16
Tables S1 and S2
REFERENCES AND NOTES
- 1.J. P. Taylor, R. H. Brown Jr., D. W. Cleveland, Decoding ALS: From genes to mechanism. Nature 539, 197–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.P. Tziortzouda, L. Van Den Bosch, F. Hirth, Triad of TDP43 control in neurodegeneration: Autoregulation, localization and aggregation. Nat. Rev. Neurosci. 22, 197–208 (2021). [DOI] [PubMed] [Google Scholar]
- 3.G. Kim, O. Gautier, E. Tassoni-Tsuchida, X. R. Ma, A. D. Gitler, ALS genetics: Gains, losses, and implications for future therapies. Neuron 108, 822–842 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.T. Arai, M. Hasegawa, H. Akiyama, K. Ikeda, T. Nonaka, H. Mori, D. Mann, K. Tsuchiya, M. Yoshida, Y. Hashizume, T. Oda, TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 (2006). [DOI] [PubMed] [Google Scholar]
- 5.M. Neumann, D. M. Sampathu, L. K. Kwong, A. C. Truax, M. C. Micsenyi, T. T. Chou, J. Bruce, T. Schuck, M. Grossman, C. M. Clark, L. F. McCluskey, B. L. Miller, E. Masliah, I. R. Mackenzie, H. Feldman, W. Feiden, H. A. Kretzschmar, J. Q. Trojanowski, V. M. Lee, Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 6.J. Sreedharan, I. P. Blair, V. B. Tripathi, X. Hu, C. Vance, B. Rogelj, S. Ackerley, J. C. Durnall, K. L. Williams, E. Buratti, F. Baralle, J. de Belleroche, J. D. Mitchell, P. N. Leigh, A. Al-Chalabi, C. C. Miller, G. Nicholson, C. E. Shaw, TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Y. Shiina, K. Arima, H. Tabunoki, J. Satoh, TDP-43 dimerizes in human cells in culture. Cell. Mol. Neurobiol. 30, 641–652 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Y. J. Zhang, T. Caulfield, Y. F. Xu, T. F. Gendron, J. Hubbard, C. Stetler, H. Sasaguri, E. C. Whitelaw, S. Cai, W. C. Lee, L. Petrucelli, The dual functions of the extreme N-terminus of TDP-43 in regulating its biological activity and inclusion formation. Hum. Mol. Genet. 22, 3112–3122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A. Wang, A. E. Conicella, H. B. Schmidt, E. W. Martin, S. N. Rhoads, A. N. Reeb, A. Nourse, D. Ramirez Montero, V. H. Ryan, R. Rohatgi, F. Shewmaker, M. T. Naik, T. Mittag, Y. M. Ayala, N. L. Fawzi, A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37, e97452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.T. Afroz, E. M. Hock, P. Ernst, C. Foglieni, M. Jambeau, L. A. B. Gilhespy, F. Laferriere, Z. Maniecka, A. Pluckthun, P. Mittl, P. Paganetti, F. H. T. Allain, M. Polymenidou, Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8, 45 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.L. L. Jiang, W. Xue, J. Y. Hong, J. T. Zhang, M. J. Li, S. N. Yu, J. H. He, H. Y. Hu, The N-terminal dimerization is required for TDP-43 splicing activity. Sci. Rep. 7, 6196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. Mompean, V. Romano, D. Pantoja-Uceda, C. Stuani, F. E. Baralle, E. Buratti, D. V. Laurents, Point mutations in the N-terminal domain of transactive response DNA-binding protein 43 kDa (TDP-43) compromise its stability, dimerization, and functions. J. Biol. Chem. 292, 11992–12006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.H. Yu, S. Lu, K. Gasior, D. Singh, S. Vazquez-Sanchez, O. Tapia, D. Toprani, M. S. Beccari, J. R. Yates 3rd, S. Da Cruz, J. M. Newby, M. Lafarga, A. S. Gladfelter, E. Villa, D. W. Cleveland, HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science 371, eabb4309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.H. Sasaguri, J. Chew, Y. F. Xu, T. F. Gendron, A. Garrett, C. W. Lee, K. Jansen-West, P. O. Bauer, E. A. Perkerson, J. Tong, C. Stetler, Y. J. Zhang, The extreme N-terminus of TDP-43 mediates the cytoplasmic aggregation of TDP-43 and associated toxicity in vivo. Brain Res. 1647, 57–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.H. Ederle, C. Funk, C. Abou-Ajram, S. Hutten, E. B. E. Funk, R. H. Kehlenbach, S. M. Bailer, D. Dormann, Nuclear egress of TDP-43 and FUS occurs independently of Exportin-1/CRM1. Sci. Rep. 8, 7084 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.E. S. Pinarbasi, T. Cagatay, H. Y. J. Fung, Y. C. Li, Y. M. Chook, P. J. Thomas, Active nuclear import and passive nuclear export are the primary determinants of TDP-43 localization. Sci. Rep. 8, 7083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.T. Nonaka, M. Masuda-Suzukake, T. Arai, Y. Hasegawa, H. Akatsu, T. Obi, M. Yoshida, S. Murayama, D. M. Mann, H. Akiyama, M. Hasegawa, Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep. 4, 124–134 (2013). [DOI] [PubMed] [Google Scholar]
- 18.S. Shimonaka, T. Nonaka, G. Suzuki, S. Hisanaga, M. Hasegawa, Templated aggregation of TAR DNA-binding protein of 43 kDa (TDP-43) by seeding with TDP-43 peptide fibrils. J. Biol. Chem. 291, 8896–8907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. Porta, Y. Xu, C. R. Restrepo, L. K. Kwong, B. Zhang, H. J. Brown, E. B. Lee, J. Q. Trojanowski, V. M. Lee, Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat. Commun. 9, 4220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. Yamashita, T. Nonaka, S. Hirai, A. Miwa, H. Okado, T. Arai, M. Hosokawa, H. Akiyama, M. Hasegawa, Distinct pathways leading to TDP-43-induced cellular dysfunctions. Hum. Mol. Genet. 23, 4345–4356 (2014). [DOI] [PubMed] [Google Scholar]
- 21.E. Buratti, T. Dork, E. Zuccato, F. Pagani, M. Romano, F. E. Baralle, Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 20, 1774–1784 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M. Polymenidou, C. Lagier-Tourenne, K. R. Hutt, S. C. Huelga, J. Moran, T. Y. Liang, S. C. Ling, E. Sun, E. Wancewicz, C. Mazur, H. Kordasiewicz, Y. Sedaghat, J. P. Donohue, L. Shiue, C. F. Bennett, G. W. Yeo, D. W. Cleveland, Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. Watanabe, K. Oiwa, Y. Murata, O. Komine, A. Sobue, F. Endo, E. Takahashi, K. Yamanaka, ALS-linked TDP-43(M337V) knock-in mice exhibit splicing deregulation without neurodegeneration. Mol. Brain 13, 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Y. M. Ayala, P. Zago, A. D'Ambrogio, Y. F. Xu, L. Petrucelli, E. Buratti, F. E. Baralle, Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 121, 3778–3785 (2008). [DOI] [PubMed] [Google Scholar]
- 25.F. Hans, H. Glasebach, P. J. Kahle, Multiple distinct pathways lead to hyperubiquitylated insoluble TDP-43 protein independent of its translocation into stress granules. J. Biol. Chem. 295, 673–689 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Y. R. Li, O. D. King, J. Shorter, A. D. Gitler, Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.J. van Eersel, Y. D. Ke, A. Gladbach, M. Bi, J. Gotz, J. J. Kril, L. M. Ittner, Cytoplasmic accumulation and aggregation of TDP-43 upon proteasome inhibition in cultured neurons. PLOS ONE 6, e22850 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.X. Zuo, J. Zhou, Y. Li, K. Wu, Z. Chen, Z. Luo, X. Zhang, Y. Liang, M. A. Esteban, Y. Zhou, X. D. Fu, TDP-43 aggregation induced by oxidative stress causes global mitochondrial imbalance in ALS. Nat. Struct. Mol. Biol. 28, 132–142 (2021). [DOI] [PubMed] [Google Scholar]
- 29.H. Tsuiji, Y. Iguchi, A. Furuya, A. Kataoka, H. Hatsuta, N. Atsuta, F. Tanaka, Y. Hashizume, H. Akatsu, S. Murayama, G. Sobue, K. Yamanaka, Spliceosome integrity is defective in the motor neuron diseases ALS and SMA. EMBO Mol. Med. 5, 221–234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. Neumann, L. K. Kwong, E. B. Lee, E. Kremmer, A. Flatley, Y. Xu, M. S. Forman, D. Troost, H. A. Kretzschmar, J. Q. Trojanowski, V. M. Lee, Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 117, 137–149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.M. Hasegawa, T. Arai, T. Nonaka, F. Kametani, M. Yoshida, Y. Hashizume, T. G. Beach, E. Buratti, F. Baralle, M. Morita, I. Nakano, T. Oda, K. Tsuchiya, H. Akiyama, Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.T. J. Cohen, A. W. Hwang, C. R. Restrepo, C. X. Yuan, J. Q. Trojanowski, V. M. Lee, An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun. 6, 5845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.C. Foglieni, S. Papin, A. Salvade, T. Afroz, S. Pinton, G. Pedrioli, G. Ulrich, M. Polymenidou, P. Paganetti, Split GFP technologies to structurally characterize and quantify functional biomolecular interactions of FTD-related proteins. Sci. Rep. 7, 14013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S. Sakai, S. Watanabe, O. Komine, A. Sobue, K. Yamanaka, Novel reporters of mitochondria-associated membranes (MAM), MAMtrackers, demonstrate MAM disruption as a common pathological feature in amyotrophic lateral sclerosis. FASEB J. 35, e21688 (2021). [DOI] [PubMed] [Google Scholar]
- 35.A. S. Dixon, M. K. Schwinn, M. P. Hall, K. Zimmerman, P. Otto, T. H. Lubben, B. L. Butler, B. F. Binkowski, T. Machleidt, T. A. Kirkland, M. G. Wood, C. T. Eggers, L. P. Encell, K. V. Wood, NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–408 (2016). [DOI] [PubMed] [Google Scholar]
- 36.T. Ishihara, Y. Ariizumi, A. Shiga, T. Kato, C. F. Tan, T. Sato, Y. Miki, M. Yokoo, T. Fujino, A. Koyama, A. Yokoseki, M. Nishizawa, A. Kakita, H. Takahashi, O. Onodera, Decreased number of Gemini of coiled bodies and U12 snRNA level in amyotrophic lateral sclerosis. Hum. Mol. Genet. 22, 4136–4147 (2013). [DOI] [PubMed] [Google Scholar]
- 37.V. Romano, Z. Quadri, F. E. Baralle, E. Buratti, The structural integrity of TDP-43 N-terminus is required for efficient aggregate entrapment and consequent loss of protein function. Prion 9, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.B. Bolognesi, A. J. Faure, M. Seuma, J. M. Schmiedel, G. G. Tartaglia, B. Lehner, The mutational landscape of a prion-like domain. Nat. Commun. 10, 4162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.L. A. Gruijs da Silva, F. Simonetti, S. Hutten, H. Riemenschneider, E. L. Sternburg, L. M. Pietrek, J. Gebel, V. Dotsch, D. Edbauer, G. Hummer, L. S. Stelzl, D. Dormann, Disease-linked TDP-43 hyperphosphorylation suppresses TDP-43 condensation and aggregation. EMBO J. 41, e108443 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. J. Winton, L. M. Igaz, M. M. Wong, L. K. Kwong, J. Q. Trojanowski, V. M. Lee, Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 283, 13302–13309 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A. L. Nishimura, V. Zupunski, C. Troakes, C. Kathe, P. Fratta, M. Howell, J. M. Gallo, T. Hortobagyi, C. E. Shaw, B. Rogelj, Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain 133, 1763–1771 (2010). [DOI] [PubMed] [Google Scholar]
- 42.L. Duan, B. L. Zaepfel, V. Aksenova, M. Dasso, J. D. Rothstein, P. Kalab, L. R. Hayes, Nuclear RNA binding regulates TDP-43 nuclear localization and passive nuclear export. Cell Rep. 40, 111106 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.H. C. Archbold, K. L. Jackson, A. Arora, K. Weskamp, E. M. Tank, X. Li, R. Miguez, R. D. Dayton, S. Tamir, R. L. Klein, S. J. Barmada, TDP43 nuclear export and neurodegeneration in models of amyotrophic lateral sclerosis and frontotemporal dementia. Sci. Rep. 8, 4606 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.B. D. Freibaum, R. K. Chitta, A. A. High, J. P. Taylor, Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 9, 1104–1120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.X. Y. Zhong, P. Wang, J. Han, M. G. Rosenfeld, X. D. Fu, SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell 35, 1–10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.T. Maniatis, R. Reed, An extensive network of coupling among gene expression machines. Nature 416, 499–506 (2002). [DOI] [PubMed] [Google Scholar]
- 47.C. Fallini, G. J. Bassell, W. Rossoll, The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum. Mol. Genet. 21, 3703–3718 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.N. H. Alami, R. B. Smith, M. A. Carrasco, L. A. Williams, C. S. Winborn, S. S. W. Han, E. Kiskinis, B. Winborn, B. D. Freibaum, A. Kanagaraj, A. J. Clare, N. M. Badders, B. Bilican, E. Chaum, S. Chandran, C. E. Shaw, K. C. Eggan, T. Maniatis, J. P. Taylor, Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron 81, 536–543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D. A. Solomon, A. Stepto, W. H. Au, Y. Adachi, D. C. Diaper, R. Hall, A. Rekhi, A. Boudi, P. Tziortzouda, Y. B. Lee, B. Smith, J. C. Bridi, G. Spinelli, J. Dearlove, D. M. Humphrey, J. M. Gallo, C. Troakes, M. Fanto, M. Soller, B. Rogelj, R. B. Parsons, C. E. Shaw, T. Hortobagyi, F. Hirth, A feedback loop between dipeptide-repeat protein, TDP-43 and karyopherin-α mediates C9orf72-related neurodegeneration. Brain 141, 2908–2924 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.L. Guo, H. J. Kim, H. Wang, J. Monaghan, F. Freyermuth, J. C. Sung, K. O'Donovan, C. M. Fare, Z. Diaz, N. Singh, Z. C. Zhang, M. Coughlin, E. A. Sweeny, M. E. DeSantis, M. E. Jackrel, C. B. Rodell, J. A. Burdick, O. D. King, A. D. Gitler, C. Lagier-Tourenne, U. B. Pandey, Y. M. Chook, J. P. Taylor, J. Shorter, Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.C. C. Chou, Y. Zhang, M. E. Umoh, S. W. Vaughan, I. Lorenzini, F. Liu, M. Sayegh, P. G. Donlin-Asp, Y. H. Chen, D. M. Duong, N. T. Seyfried, M. A. Powers, T. Kukar, C. M. Hales, M. Gearing, N. J. Cairns, K. B. Boylan, D. W. Dickson, R. Rademakers, Y. J. Zhang, L. Petrucelli, R. Sattler, D. C. Zarnescu, J. D. Glass, W. Rossoll, TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 21, 228–239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.A. C. Woerner, F. Frottin, D. Hornburg, L. R. Feng, F. Meissner, M. Patra, J. Tatzelt, M. Mann, K. F. Winklhofer, F. U. Hartl, M. S. Hipp, Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 351, 173–176 (2016). [DOI] [PubMed] [Google Scholar]
- 53.T. Philips, J. D. Rothstein, Rodent models of amyotrophic lateral sclerosis. Curr. Protoc. Pharmacol. 69, 5.67.1–5.67.21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.A. Koyama, A. Sugai, T. Kato, T. Ishihara, A. Shiga, Y. Toyoshima, M. Koyama, T. Konno, S. Hirokawa, A. Yokoseki, M. Nishizawa, A. Kakita, H. Takahashi, O. Onodera, Increased cytoplasmic TARDBP mRNA in affected spinal motor neurons in ALS caused by abnormal autoregulation of TDP-43. Nucleic Acids Res. 44, 5820–5836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.B. Bilican, A. Serio, S. J. Barmada, A. L. Nishimura, G. J. Sullivan, M. Carrasco, H. P. Phatnani, C. A. Puddifoot, D. Story, J. Fletcher, I. H. Park, B. A. Friedman, G. Q. Daley, D. J. Wyllie, G. E. Hardingham, I. Wilmut, S. Finkbeiner, T. Maniatis, C. E. Shaw, S. Chandran, Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc. Natl. Acad. Sci. U.S.A. 109, 5803–5808 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.M. A. White, E. Kim, A. Duffy, R. Adalbert, B. U. Phillips, O. M. Peters, J. Stephenson, S. Yang, F. Massenzio, Z. Lin, S. Andrews, A. Segonds-Pichon, J. Metterville, L. M. Saksida, R. Mead, R. R. Ribchester, Y. Barhomi, T. Serre, M. P. Coleman, J. R. Fallon, T. J. Bussey, R. H. Brown Jr., J. Sreedharan, TDP-43 gains function due to perturbed autoregulation in a Tardbp knock-in mouse model of ALS-FTD. Nat. Neurosci. 21, 552–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Y. Iguchi, M. Katsuno, J. Niwa, S. Takagi, S. Ishigaki, K. Ikenaka, K. Kawai, H. Watanabe, K. Yamanaka, R. Takahashi, H. Misawa, S. Sasaki, F. Tanaka, G. Sobue, Loss of TDP-43 causes age-dependent progressive motor neuron degeneration. Brain 136, 1371–1382 (2013). [DOI] [PubMed] [Google Scholar]
- 58.L. S. Wu, W. C. Cheng, C. K. Shen, Targeted depletion of TDP-43 expression in the spinal cord motor neurons leads to the development of amyotrophic lateral sclerosis-like phenotypes in mice. J. Biol. Chem. 287, 27335–27344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. P. Ling, O. Pletnikova, J. C. Troncoso, P. C. Wong, TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science 349, 650–655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.J. R. Klim, L. A. Williams, F. Limone, I. G. S. Juan, B. N. Davis-Dusenbery, D. A. Mordes, A. Burberry, M. J. Steinbaugh, K. K. Gamage, R. Kirchner, R. Moccia, S. H. Cassel, K. Chen, B. J. Wainger, C. J. Woolf, K. Eggan, ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat. Neurosci. 22, 167–179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Z. Melamed, J. Lopez-Erauskin, M. W. Baughn, O. Zhang, K. Drenner, Y. Sun, F. Freyermuth, M. A. McMahon, M. S. Beccari, J. W. Artates, T. Ohkubo, M. Rodriguez, N. Lin, D. Wu, C. F. Bennett, F. Rigo, S. Da Cruz, J. Ravits, C. Lagier-Tourenne, D. W. Cleveland, Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat. Neurosci. 22, 180–190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.A. L. Brown, O. G. Wilkins, M. J. Keuss, S. E. Hill, M. Zanovello, W. C. Lee, A. Bampton, F. C. Y. Lee, L. Masino, Y. A. Qi, S. Bryce-Smith, A. Gatt, M. Hallegger, D. Fagegaltier, H. Phatnani, N. A. Consortium, J. Newcombe, E. K. Gustavsson, S. Seddighi, J. F. Reyes, S. L. Coon, D. Ramos, G. Schiavo, E. M. C. Fisher, T. Raj, M. Secrier, T. Lashley, J. Ule, E. Buratti, J. Humphrey, M. E. Ward, P. Fratta, TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature 603, 131–137 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.X. R. Ma, M. Prudencio, Y. Koike, S. C. Vatsavayai, G. Kim, F. Harbinski, A. Briner, C. M. Rodriguez, C. Guo, T. Akiyama, H. B. Schmidt, B. B. Cummings, D. W. Wyatt, K. Kurylo, G. Miller, S. Mekhoubad, N. Sallee, G. Mekonnen, L. Ganser, J. D. Rubien, K. Jansen-West, C. N. Cook, S. Pickles, B. Oskarsson, N. R. Graff-Radford, B. F. Boeve, D. S. Knopman, R. C. Petersen, D. W. Dickson, J. Shorter, S. Myong, E. M. Green, W. W. Seeley, L. Petrucelli, A. D. Gitler, TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature 603, 124–130 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.F. Gasset-Rosa, S. Lu, H. Yu, C. Chen, Z. Melamed, L. Guo, J. Shorter, S. Da Cruz, D. W. Cleveland, Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339–357.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.R. Rakhit, J. P. Crow, J. R. Lepock, L. H. Kondejewski, N. R. Cashman, A. Chakrabartty, Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J. Biol. Chem. 279, 15499–15504 (2004). [DOI] [PubMed] [Google Scholar]
- 66.M. J. Lindberg, R. Bystrom, N. Boknas, P. M. Andersen, M. Oliveberg, Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants. Proc. Natl. Acad. Sci. U.S.A. 102, 9754–9759 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.U. Dettmer, A. J. Newman, F. Soldner, E. S. Luth, N. C. Kim, V. E. von Saucken, J. B. Sanderson, R. Jaenisch, T. Bartels, D. Selkoe, Parkinson-causing α-synuclein missense mutations shift native tetramers to monomers as a mechanism for disease initiation. Nat. Commun. 6, 7314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.T. Bartels, J. G. Choi, D. J. Selkoe, α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477, 107–110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.S. Nuber, M. Rajsombath, G. Minakaki, J. Winkler, C. P. Muller, M. Ericsson, B. Caldarone, U. Dettmer, D. J. Selkoe, Abrogating native α-synuclein tetramers in mice causes a L-DOPA-responsive motor syndrome closely resembling Parkinson's disease. Neuron 100, 75–90.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.B. R. Brooks, R. G. Miller, M. Swash, T. L. Munsat, D., El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 1, 293–299 (2000). [DOI] [PubMed] [Google Scholar]
- 71.D. Neary, J. S. Snowden, L. Gustafson, U. Passant, D. Stuss, S. Black, M. Freedman, A. Kertesz, P. H. Robert, M. Albert, K. Boone, B. L. Miller, J. Cummings, D. F. Benson, Frontotemporal lobar degeneration: A consensus on clinical diagnostic criteria. Neurology 51, 1546–1554 (1998). [DOI] [PubMed] [Google Scholar]
- 72.S. Watanabe, H. Inami, K. Oiwa, Y. Murata, S. Sakai, O. Komine, A. Sobue, Y. Iguchi, M. Katsuno, K. Yamanaka, Aggresome formation and liquid-liquid phase separation independently induce cytoplasmic aggregation of TAR DNA-binding protein 43. Cell Death Dis. 11, 909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.K. Takahashi, K. Tanabe, M. Ohnuki, M. Narita, T. Ichisaka, K. Tomoda, S. Yamanaka, Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- 74.D. Shimojo, K. Onodera, Y. Doi-Torii, Y. Ishihara, C. Hattori, Y. Miwa, S. Tanaka, R. Okada, M. Ohyama, M. Shoji, A. Nakanishi, M. Doyu, H. Okano, Y. Okada, Rapid, efficient, and simple motor neuron differentiation from human pluripotent stem cells. Mol. Brain 8, 79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Y. Okada, A. Matsumoto, T. Shimazaki, R. Enoki, A. Koizumi, S. Ishii, Y. Itoyama, G. Sobue, H. Okano, Spatiotemporal recapitulation of central nervous system development by murine embryonic stem cell-derived neural stem/progenitor cells. Stem Cells 26, 3086–3098 (2008). [DOI] [PubMed] [Google Scholar]
- 76.T. Nagai, K. Ibata, E. S. Park, M. Kubota, K. Mikoshiba, A. Miyawaki, A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002). [DOI] [PubMed] [Google Scholar]
- 77.S. Watanabe, K. Kaneko, K. Yamanaka, Accelerated disease onset with stabilized familial amyotrophic lateral sclerosis (ALS)-linked mutant TDP-43 proteins. J. Biol. Chem. 288, 3641–3654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.A. D'Ambrogio, E. Buratti, C. Stuani, C. Guarnaccia, M. Romano, Y. M. Ayala, F. E. Baralle, Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 37, 4116–4126 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.F. Pagani, E. Buratti, C. Stuani, M. Romano, E. Zuccato, M. Niksic, L. Giglio, D. Faraguna, F. E. Baralle, Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 275, 21041–21047 (2000). [DOI] [PubMed] [Google Scholar]
- 80.M. Mirdita, K. Schutze, Y. Moriwaki, L. Heo, S. Ovchinnikov, M. Steinegger, ColabFold: Making protein folding accessible to all. Nat. Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S16
Tables S1 and S2