Abstract
Objectives:
To determine whether ComBat harmonization improves multi-class radiomics-based tissue classification in technically heterogeneous MRI datasets, and to compare the performances to two ComBat variants.
Materials and Methods:
One-hundred patients who had undergone T1-weighted 3D GRE Dixon MRI (two scanners/vendors; 50 patients each) were retrospectively included. Volumes of interest (2.5 cm3) were placed in three disease-free tissues with visually similar appearance on T1 Dixon water images: liver, spleen, and paraspinal muscle. Gray-level histogram (GLH), co-occurrence matrix (GLCM), run-length matrix (GLRLM), and size-zone matrix (GLSZM) radiomic features were extracted. Tissue classification was performed on pooled data from the two centers (1) without harmonization; (2) after ComBat harmonization with empirical Bayes estimation (ComBat-B); and (3) after ComBat harmonization without empirical Bayes estimation (ComBat-NB). Linear discriminant analysis (LDA) with leave-one-out cross-validation was used to distinguish between the three tissue types, using all available radiomic features as input. In addition, a multi-layer perceptron neural network with a random 70:30% split into training and test datasets was used for the same task, but separately for each radiomic feature category.
Results:
LDA-based mean tissue classification accuracies were 52.3% for unharmonized, 66.3% for ComBat-B harmonized, and 92.7% for ComBat-NB harmonized data. For MLP-NN, mean classification accuracies for unharmonized, ComBat-B-harmonized, and ComBat-NB-harmonized test data were: 46.8, 55.1, and 57.5% for GLH; 42.0, 65.3, and 71.0% for GLCM; 45.3, 78.3, and 78.0% for GLRLM; and 48.1, 81.1, and 89.4% for GLSZM. Accuracies were significantly higher for both ComBat-B- and ComBat-NB-harmonized data than for unharmonized data for all feature categories (at P=0.005, respectively). For GLCM (P=0.001) and GLSZM (P=0.005), ComBat-NB harmonization provided slightly higher accuracies than ComBat-B harmonization.
Conclusions:
ComBat harmonization may be useful for multi-center MRI radiomics studies with non-binary classification tasks. The degree of improvement by ComBat may vary between radiomic feature categories, between classifiers, and between ComBat variants.
Keywords: MRI, radiomics, machine learning, harmonization
INTRODUCTION
Radiomics, an analysis technique that extracts quantitative features from medical images, has gathered considerable attention within the last decade.1,2 Radiomics has been applied to the entire spectrum of diagnostic imaging techniques, but most prominently, MRI, CT, and PET, with a focus on disease characterization as well as outcome prediction and prognostication.3–8
A major obstacle for the translation of radiomics to clinical practice is the dependence of radiomic feature values on image acquisition protocols as well as image processing, and associated repeatability.9–13 One strategy to at least partially address this issue is the prospective standardization of image acquisition and reconstruction parameters. However, the latter is mainly feasible for prospective research projects that include a limited numbers of collaborating centers, but less so in a real-world clinical setting where vendor-supplied standard protocols are used. In addition, for MRI, which is also widely used in private practices, differences in magnetic field strength and radiofrequency receiver coils affect signal intensity, adding further complexity.
Therefore, as a more practical and easy to apply approach, mathematical harmonization techniques, which work directly on the calculated radiomic feature values rather than on images, have been proposed. ComBat, which is the most widely used harmonization technique at present, has been shown to remove, or at least decrease, the “batch effect” –e.g., the influence of acquisition protocol differences between centers and scanners– from radiomic features, while preserving their underlying biological and pathophysiological associations.14,15 For clinical MRI data, ComBat has so far predominantly been used for binary classification tasks, i.e., separation of just two tissue, lesion, or outcome classes.3,5,6,15–21
In the present study, our aim was therefore to determine the value of ComBat harmonization of clinical MRI radiomic data from two centers for non-binary tissue classification by machine learning. Further, we aimed to compare the performances of two ComBat variants, as well as the effects of harmonization on radiomic features of different categories.
MATERIALS AND METHODS
Patients and design
A clinical database search was performed at two tertiary care centers (A and B) to identify and retrospectively include 100 patients who had undergone whole-body MRI for routine clinical purposes between 01/2019 and 12/2021. The study was approved by the Institutional Review Boards of the two centers; informed consent was waived. Inclusion criteria were: MRI performed on two specified 3.0 Tesla scanners from different vendors; MRI protocol including a high-resolution T1-weighted 3D gradient-echo (GRE) Dixon sequence (see protocol below); no evidence of disease (malignant or other) in the liver, spleen, or paraspinal musculature, according to routine clinical PET/MRI reports as well as additional evaluation by a board-certified radiologist specializing in hybrid imaging. The single exclusion criterion was the presence of severe MRI artifacts, e.g. due to motion or metal implants, obscuring tissues of interest, as verified by a board-certified radiologist.
MRI protocols
At center A, whole-body MRI from the vertex to the upper thighs was performed on a Signa PET/MR (GE Healthcare, Waukesha, USA) as part of routine PET/MRI examinations. The MRI protocol included an axial 2-point Dixon 3D T1-weighted LAVA-Flex (liver acquisition with volumetric acceleration-flexible) sequence, which was obtained with breath-holding and covered the entire scanned anatomy. Acquisition parameters were: repetition time (TR), 4.06 ms; echo times (TE), 1.67 and 2.23 ms; one acquisition; a 12° flip angle; and a voxel size of 0.98 × 0.98 × 3.8 mm3.
At center B, whole-body MRI from the vertex to the upper thighs was performed on a Biograph mMR (Siemens, Erlangen, Germany) as part of routine PET/MRI examinations. The MRI protocol included an axial 2-point Dixon 3D T1-weighted VIBE (volume interpolated breath-hold) sequence, which covered the entire scanned anatomy. Acquisition parameters were: repetition time (TR), 4.02 ms; echo times (TE) 1.23 and 2.46 ms; one acquisition; 10° flip angle; and a voxel size of 1.34 × 1.34 × 3.0 mm3.
At both centers, Dixon water images were reconstructed from the T1-weighted in- and out-of-phase images, and used for further analysis. These images show relatively low contrast between the three tissues of interest (liver, spleen, muscle), making them visually more difficult to separate (see Fig. 1), and thereby providing a scenario where radiomics is typically applied.
Figure 1.
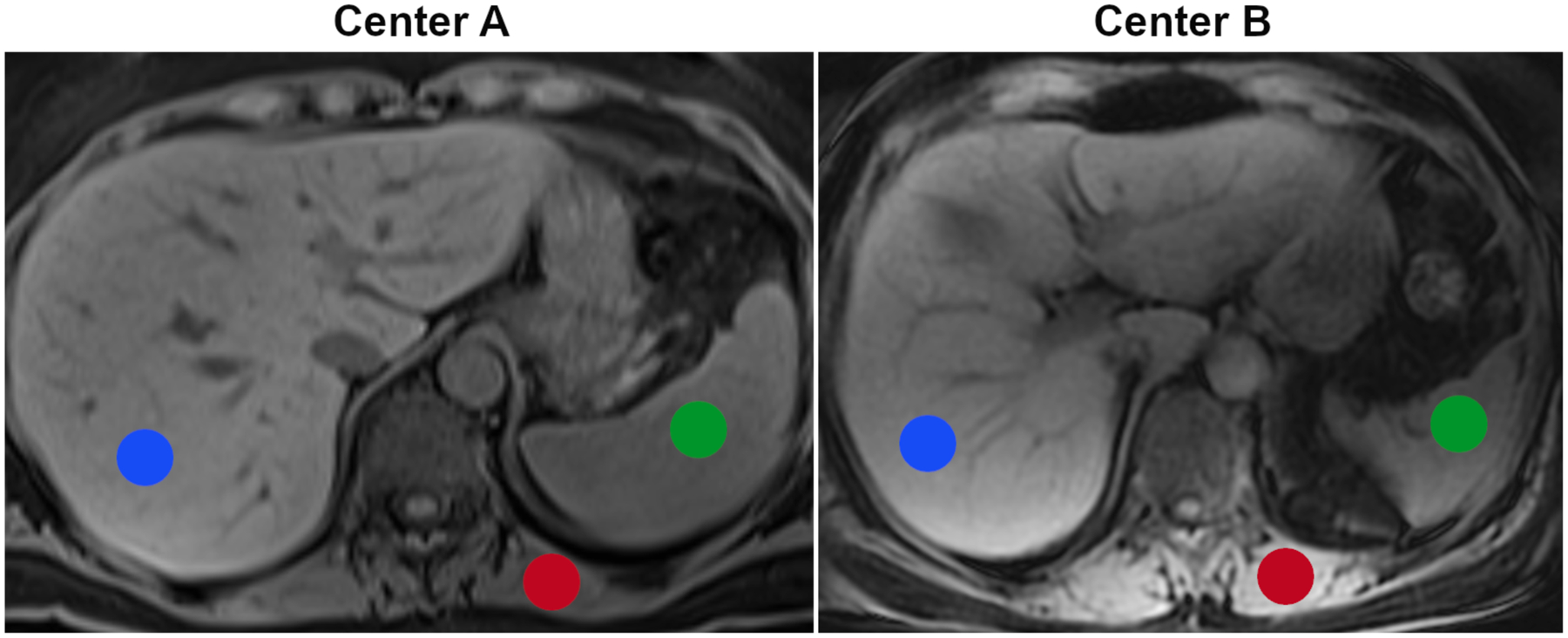
Examples for T1-weighted Dixon MR images from the two centers and volumes of interest for the three tissues of interest (liver, red; spleen, green; and paraspinal muscle, red).
Image analysis and radiomic feature harmonization
Radiomic feature extraction was performed by a board-certified radiologist, using the International Biomarker Standardization Initiative (IBSI)-compliant22 open-source software LIFEx version 7.3.0 (https://lifexsoft.org).23 Manually defined 2.5-cm3 spherical volumes of interest (VOI) were placed in the liver, spleen, and paraspinal musculature, avoiding large vessels and other macrostructures (Fig. 1). The three tissues were chosen because they are relatively homogeneous, meaning that variations in VOI placement are not expected to have a relevant impact on feature values, and because they are sufficiently large to allow placement of VOIs of identical size and shape. Prior to feature extraction, intensity discretization using a fixed bin size, and spatial resampling to 2.0 × 2.0 × 2.0 mm3 voxels were performed. Thirty-three radiomic features from four frequently used feature categories were calculated: gray-level histogram (GLH, n=5); gray-level co-occurrence matrix (GLCM, n=6); gray-level run-length matrix (GLRLM, n=11), and gray-level size-zone matrix (GLSZM, n=11; named gray-level zone-length matrix in LIFEx). For heterogeneity features derived from the GLCM, GLRLM, and GLSZM, individual feature values were automatically calculated by LIFEx as arithmetic means for different 3D orientations and intervoxel distances. For a list of computed features and corresponding equations, see the LIFEx documentation at https://lifexsoft.org/index.php/resources/texture/radiomic-features.
To eliminate or reduce the impact of systematic differences between the MRI datasets of centers A and B while at the same time improving separation of the three tissues, two variants of ComBat harmonization –with and without empirical Bayes assumption (ComBat-B and ComBat-NB)– were applied to the pooled unharmonized radiomic data, separately for the individual tissues analyzed. In brief, ComBat is a data-driven technique originally developed to remove the batch effect (i.e., effects caused by the measurement technique, device, or the sample itself) from genome microarray expression data.24 ComBat works on numerical radiomic feature values without taking actual images, image acquisition parameters, or phantom measurements into account.14 R scripts for the two ComBat variants are available at https://github.com/Jfortin1/neuroCombat_Rpackage/.
Tissue classification and statistics
Linear discriminant analysis (LDA), which reduces feature dimensionality, was performed with leave-on-out cross-validation (LOOCV), taking pooled cases from the two centers and features from all radiomic categories as input. Mean accuracies for separation of the three tissue classes were calculated, independently for unhamonized, ComBat-B-harmonized, and ComBat-NB-harmonized data, and scatterplots based on the LDA functions were used to visualize effects of ComBat harmonization.
In addition, cases were randomly assigned to a training dataset (70%) and a test dataset (30%); case assignments were identical for unharmonized and harmonized datasets. Separately for unharmonized, ComBat-B- and ComBat-NB-harmonized data, and independently for the different feature categories (GLH, GLCM, GLRLM, GLSZM), a multi-layer perceptron neural network (MLP-NN)25 with one hidden layer and a minimum of three neurons was trained to distinguish between liver, spleen, and muscle. These additional experiments were performed because, contrary to LDA, MLP-NN can solve non-linear classification tasks, and because the data split into training and test datasets is the generally recommended strategy to assess model generalizability identify overfitting. Because MLP-NN classification starts with an initial guess at the network parameters, mean accuracies and accuracy ranges based on ten-fold iteration of MLP-NN classification were calculated, separately for training and test datasets, and Wilcoxon signed rank tests were used to assess significant differences between unharmonized and harmonized datasets. Areas under the receiver operating characteristic (ROC) curves (AUC) were calculated for test data using a pair-wise (i.e., 1-versus-2 tissues) approach. All tests were performed using SPSS 28.0.1 (IBM, Armonk, USA). The specified level of significance was P<0.05.
RESULTS
The cohort comprised 100 patients: 50 from center A (23 women and 27 men; mean age 44.4 ± 15.7 years), and 50 from center B (24 women and 26 men; mean age 48.6 ± 16.2 years); 300 VOIs (100 per tissue type) were analyzed.
For pooled unharmonized radiomic data from the two centers, tissue classification based on LDA, which reduced the initially 33 dimensions to two, was overall unsatisfactory, with a mean accuracy of 52.3%. Clearly higher LDA-based mean accuracies were observed for ComBat-B and, even more so, ComBat-NB, at 66.3% and 92.7%, respectively. Scatterplots based on LDA scores also demonstrate markedly superior clustering of data points to the three tissues post harmonization (see Fig. 2).
Figure 2.
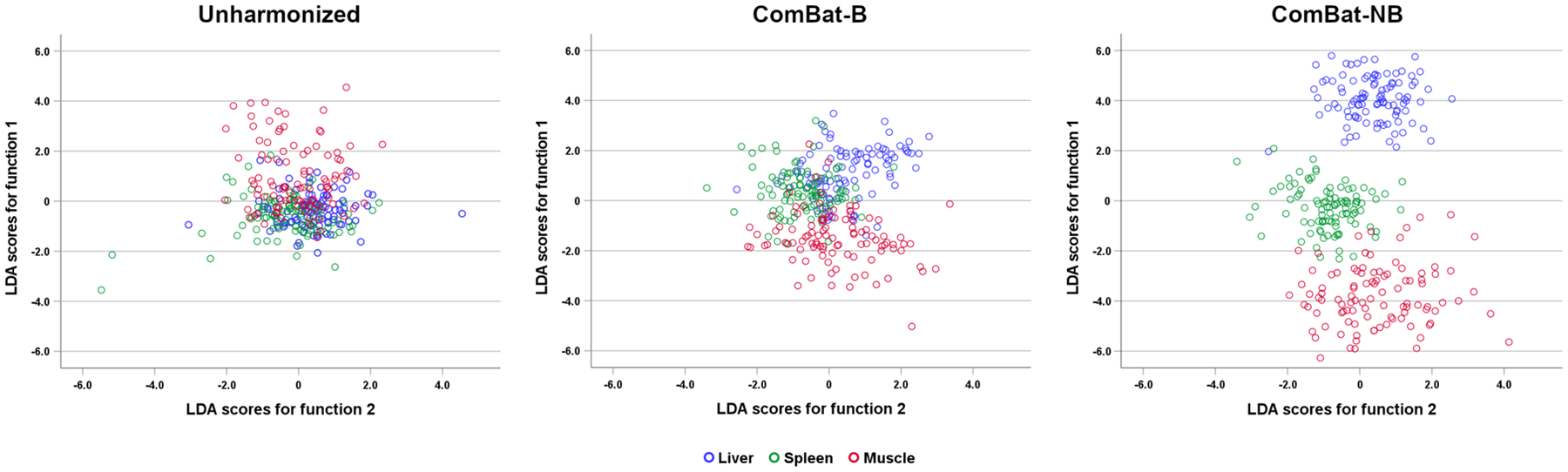
Scatterplots based on feature space dimensionality reduction by linear discriminant analysis. ComBat harmonization, and ComBat-NB (without empirical Bayes assumption) in particular, clearly improve separation of the three tissues of interest.
Tissue classification based on unharmonized data remained unsatisfactory when MLP-NN was applied, with mean accuracies ranging from 42.0–50.1% for the different feature categories (Table 1). This was also confirmed by ROC curves for 1-versus-2 tissue discrimination (Fig. 3). Again, ComBat harmonization markedly improved results: MLP-NN-based mean accuracies differed significantly between unharmonized and ComBat-B, and between unharmonized and ComBat-NB data, at P=0.005 for all features categories, respectively. Improvement was, however, less pronounced for GLH, with a test dataset mean accuracy difference of +8.3 percentage points (p.p.) for ComBat-B, and +10.7 p.p. for ComBat-NB, relative to unharmonized data. The greatest improvement was observed for GLSZM features, with a test dataset mean accuracy difference of +33.0 p.p. for ComBat-B, and +41.3 p.p. for ComBat-NB. These trends were confirmed by 1-versus-2 tissue discrimination experiments (Fig. 3), where almost perfect discrimination based on GLSZM features was achieved following harmonization.
TABLE 1.
Tissue classification accuracies (%) with and without harmonization for four radiomic feature categories, based on MLP-NN
| Unharmonized | ComBat-B | ComBat-NB | ||||
|---|---|---|---|---|---|---|
| Mean | Range | Mean | Range | Mean | Range | |
| GLH: | ||||||
| Accuracy–training | 48.9 | 45.6–51.1 | 69.3 | 61.1–78.9 | 69.8 | 64.4–77.8 |
| Accuracy–test | 46.8 | 43.3–50.5 | 55.1 | 51.4–61.0 | 57.5 | 53.8–63.8 |
| GLCM: | ||||||
| Accuracy–training | 46.0 | 42.2–52.2 | 79.6 | 71.1–87.8 | 83.3 | 81.1–85.6 |
| Accuracy–test | 42.0 | 38.0–47-1 | 65.3 | 62.4–68.1 | 71.0 | 66.7–75.2 |
| GLRLM: | ||||||
| Accuracy–training | 48.5 | 41.1–56.7 | 89.7 | 81.1–97.8 | 89.7 | 81.1–97.8 |
| Accuracy–test | 45.3 | 39.5–49.0 | 78.3 | 64.8–82.4 | 78.0 | 72.9–82.4 |
| GLSZM: | ||||||
| Accuracy–training | 50.1 | 41.1–57.8 | 95.7 | 88.9–100 | 98.6 | 93.3–100 |
| Accuracy–test | 48.1 | 46.2–50.5 | 81.1 | 74.8–87.1 | 89.4 | 86.2–91.9 |
GLH, gray-level histogram; GLCM, gray-level co-occurrence matrix;
GLRLM, gray-level run-length matrix; GLSZM, gray-level size-zone matrix
Figure 3.
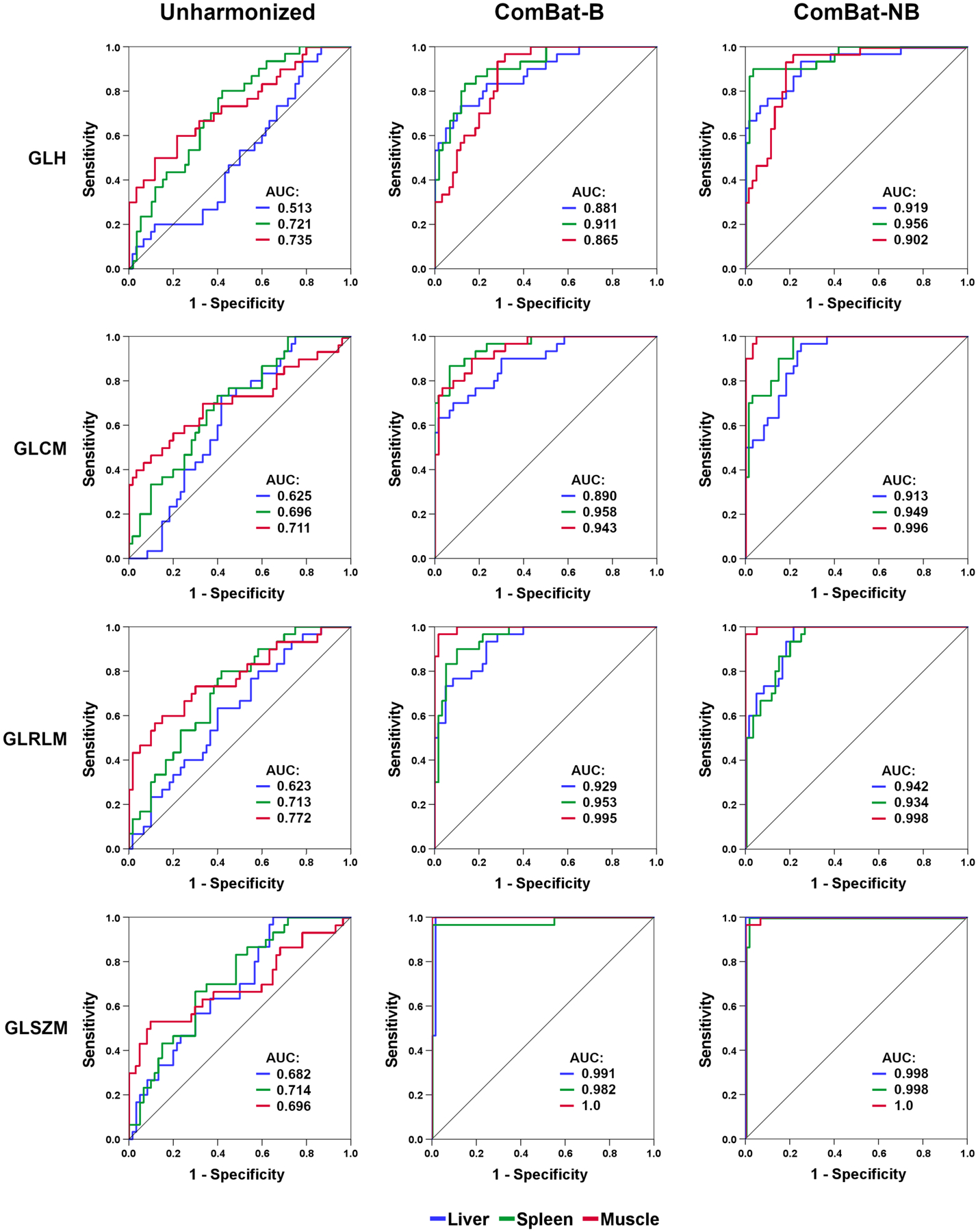
Receiver operating characteristic (ROC) curves for 1-versus-2 tissue classifications, with respective areas under the curve (AUC). Harmonization markedly improves tissue classification for all feature categories, although not to the same degree.
A direct comparison between the two ComBat variants revealed no significant differences in MLP-NN-based accuracy between ComBat-B and ComBat-NB for GLH (P=0.21) and GLRLM features (P=0.66). However, ComBat-NB was superior to ComBat-B when using GLCM (tests dataset mean accuracy difference, +5.7 p.p; P=0.008) or GLSZM (+8.3 p.p.; P=0.005) features.
DISCUSSION
Our results demonstrate that ComBat harmonization can markedly improve MRI radiomics-based tissue classification in technically heterogeneous datasets, even in a multi-class setting, which is becoming increasingly common in radiomics research.2 Contrary to unharmonized radiomic data, for which, with accuracies close to 50%, tissue classification essentially failed, classification based on ComBat harmonized data was clearly more successful (Table 1, Figs. 2 and 3). However, feature harmonization did not improve classification results to the same degree for the different radiomic feature categories included. For first-order, histogram-based features (GLH), which capture relatively basic gray-level statistics such as the mean gray-level value and gray-level percentiles, ComBat led to slightly improved, but still unsatisfactory levels of classification accuracy. On the other hand, for second-order features that reflect the spatial distribution of voxel pairs with pre-defined gray-level values (GLCM), ComBat led to classification accuracies of up to 71% in the test dataset. For higher-order radiomic features (GLRLM, GLSZM) that capture the distribution of runs and areas (zones) of voxels with pre-defined gray-level values, tissue classification after ComBat harmonization was satisfactory, with accuracies up to 89% in the test dataset.
Different variants of ComBat have been proposed for radiomic feature harmonization,14,15,26–28 of which two were evaluated in our study: the “standard” variant with empirical Bayes estimation (ComBat-B) that has been used in several MRI radiomics studies;29–33 and a variant without empirical Bayes estimation (ComBat-NB),32,34 which has been used less frequently,34 but which may be preferable for instance if the number of features is substantially smaller than the number of participants, or if standard ComBat does not fit the data well (see https://github.com/Jfortin1/neuroCombat_Rpackage/). While both variants improved tissue classification, regardless of the classifier used, ComBat-NB was overall superior in our dataset, as evidenced by LDA results that reflecting linear data separability, as well as MLP-NN results for two radiomic feature categories (Table 1), reflecting more sophisticated, non-linear data separability. LDA in particular showed an accuracy difference of +26.4 p.p. in favor of ComBat-NB (Fig. 3). These findings highlight the dependency of perceived harmonization performance on the choice of classifier, and may in part explain why some studies did not report improved classification performance following ComBat harmonization.32
Contrary to binary classification, i.e., separation of only two classes, such as benign and malignant lesions,5,33 or prediction of locoregional spread or control,17,18 treatment response or relapse at a given time-point,19–21,30 for which ComBat has been successfully used, our use of three tissue types with visually similar signal intensity and homogeneity on MRI makes the classification task more complex. Classification was further made difficult by our choice of T1-weighted Dixon images for radiomic feature extraction, where signal intensities showed only minor visible differences between tissues of interest. This is probably also the reason for the poor classification results based on histogram features (GLH), which remained unsatisfactory despite the use of ComBat. Since we extracted radiomic features from disease-free organs, our VOIs were of identical size, thereby eliminating the effect of VOI size differences on radiomic feature values that has previously been observed, and that led to misinterpretation of the impact of radiomics in the past.35 Also, the number of measurements (VOIs) was the same for each tissue class (100 per tissue), eliminating class imbalance as a factor that may impact model performance.36
Our study has several limitations. Our sample size was moderate, and therefore, we included three, rather than more tissue types. Also, images from only two centers were included, which, however, used scanners from different vendors, and slightly different pulse sequence designs and acquisition parameters, resulting in visible differences between MR images (Fig. 1). We limited our evaluation to a T1-weighted Dixon sequence, since it provides high resolution, which impacts radiomic feature values and improves classification.9,37 Notably, spatial resampling to an isotropic voxel size of 2 × 2 × 2 mm3 was used to partly compensate for differences in slice thickness (4 mm in center A and 3 mm in center B); interpolation by factor 2 in this direction was chosen because the results of a previous phantom study suggested that higher interpolation factors may not improve radiomics-based classification further, and may possibly even have negative effects.37 In addition, this pulse sequence is frequently used in clinical practice, for example for whole-body assessment of cancers such as myeloma,38 or for PET/MRI, where it is used for both diagnostic purposes and for PET attenuation correction. Finally, we exclusively evaluated ComBat, but not other harmonization techniques, such as z-scores, which have previously been utilized for a similar task.39 However, a direct comparison between the two harmonization techniques suggested several advantages (such as preservation of the original range of values) of ComBat over z-scores,14 and therefore, we did not explore the use of z-scores further in our study.
In conclusion, the results of our study confirm the benefit of applying ComBat harmonization to MRI radiomics, and show that it performs well even for more challenging, non-binary classification tasks. However, our data suggest that the degree of improvement may vary, in part substantially, between different radiomic feature categories, between classifiers, and between ComBat variants. Therefore, different combinations of these factors should be evaluated at the training stage of multi-center MRI radiomics studies dealing with multi-class data, to determine the optimal approach.
Conflicts of Interest and Source of Funding:
M.E.M. has received honoraria from GE for lectures. For the remaining authors, no conflicts of interest were declared. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748.
REFERENCES
- 1.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fritz B, Yi PH, Kijowski R, et al. Radiomics and Deep Learning for Disease Detection in Musculoskeletal Radiology: An Overview of Novel MRI- and CT-Based Approaches. Invest Radiol. 2023;58:3–13. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Zhang HL, Yin HK, Zhang HK, et al. Comparison of MRI and CT-Based Radiomics and Their Combination for Early Identification of Pathological Response to Neoadjuvant Chemotherapy in Locally Advanced Gastric Cancer. J Magn Reson Imaging. 2022. Dec 17. [DOI] [PubMed] [Google Scholar]
- 4.Makowski MR, Bressem KK, Franz L, et al. De Novo Radiomics Approach Using Image Augmentation and Features From T1 Mapping to Predict Gleason Scores in Prostate Cancer. Invest Radiol. 2021;56:661–668. [DOI] [PubMed] [Google Scholar]
- 5.Duron L, Heraud A, Charbonneau F, et al. A Magnetic Resonance Imaging Radiomics Signature to Distinguish Benign From Malignant Orbital Lesions. Invest Radiol. 2021;56:173–180. [DOI] [PubMed] [Google Scholar]
- 6.Sexauer R, Yang S, Weikert T, et al. Automated Detection, Segmentation, and Classification of Pleural Effusion From Computed Tomography Scans Using Machine Learning. Invest Radiol. 2022;57:552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanduleanu S, Jochems A, Upadhaya T, et al. Non-invasive imaging prediction of tumor hypoxia: A novel developed and externally validated CT and FDG-PET-based radiomic signatures. Radiother Oncol. 2020;153:97–105. [DOI] [PubMed] [Google Scholar]
- 8.Mayerhoefer ME, Riedl CC, Kumar A, et al. Radiomic features of glucose metabolism enable prediction of outcome in mantle cell lymphoma. Eur J Nucl Med Mol Imaging. 2019;46:2760–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayerhoefer ME, Szomolanyi P, Jirak D, et al. Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: an application-oriented study. Med Phys. 2009;36:1236–43. [DOI] [PubMed] [Google Scholar]
- 10.Shafiq-Ul-Hassan M, Zhang GG, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44:1050–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwanenburg A Radiomics in nuclear medicine: robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur J Nucl Med Mol Imaging. 2019;46:2638–2655. [DOI] [PubMed] [Google Scholar]
- 12.Wennmann M, Bauer F, Klein A, Chmelik J, et al. In Vivo Repeatability and Multiscanner Reproducibility of MRI Radiomics Features in Patients With Monoclonal Plasma Cell Disorders: A Prospective Bi-institutional Study. Invest Radiol. 2022; doi: 10.1097/RLI.0000000000000927 [DOI] [PubMed] [Google Scholar]
- 13.Wichtmann BD, Harder FN, Weiss K, et al. Influence of Image Processing on Radiomic Features From Magnetic Resonance Imaging. Invest Radiol. 2022; doi: 10.1097/RLI.0000000000000921 [DOI] [PubMed] [Google Scholar]
- 14.Orlhac F, Eertink JJ, Cottereau AS, et al. A Guide to ComBat Harmonization of Imaging Biomarkers in Multicenter Studies. J Nucl Med. 2022;63:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orlhac F, Lecler A, Savatovski J, et al. How can we combat multicenter variability in MR radiomics? Validation of a correction procedure. Eur Radiol. 2021;31:2272–2280. [DOI] [PubMed] [Google Scholar]
- 16.Tafuri B, Lombardi A, Nigro S, et al. The impact of harmonization on radiomic features in Parkinson’s disease and healthy controls: A multicenter study. Front Neurosci. 2022;16:1012287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourbonne V, Jaouen V, Nguyen TA, et al. Development of a Radiomic-Based Model Predicting Lymph Node Involvement in Prostate Cancer Patients. Cancers (Basel). 2021;13:5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bos P, Martens RM, de Graaf P, et al. External validation of an MR-based radiomic model predictive of locoregional control in oropharyngeal cancer. Eur Radiol. 2022. Dec 3. [DOI] [PubMed] [Google Scholar]
- 19.Bouhamama A, Leporq B, Khaled W, et al. Prediction of Histologic Neoadjuvant Chemotherapy Response in Osteosarcoma Using Pretherapeutic MRI Radiomics. Radiol Imaging Cancer. 2022;4:e210107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bordron A, Rio E, Badic B, et al. External Validation of a Radiomics Model for the Prediction of Complete Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer. Cancers (Basel). 2022;14:1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crombé A, Kind M, Fadli D, et al. Intensity harmonization techniques influence radiomics features and radiomics-based predictions in sarcoma patients. Sci Rep. 2020;10:15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zwanenburg A, Vallières M, Abdalah MA, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295:328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nioche C, Orlhac F, Boughdad S, et al. LIFEx: A Freeware for Radiomic Feature Calculation in Multimodality Imaging to Accelerate Advances in the Characterization of Tumor Heterogeneity. Cancer Res. 2018;78:4786–4789. [DOI] [PubMed] [Google Scholar]
- 24.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118‐127. [DOI] [PubMed] [Google Scholar]
- 25.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44. [DOI] [PubMed] [Google Scholar]
- 26.Carré A, Battistella E, Niyoteka S, et al. AutoComBat: a generic method for harmonizing MRI-based radiomic features. Sci Rep. 2022;12:12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Da-Ano R, Lucia F, Masson I, et al. A transfer learning approach to facilitate ComBat-based harmonization of multicentre radiomic features in new datasets. PLoS One. 2021;16:e0253653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Da-Ano R, Masson I, Lucia F, et al. Performance comparison of modified ComBat for harmonization of radiomic features for multicenter studies. Sci Rep. 2020;10:10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campello VM, Martín-Isla C, Izquierdo C, et al. Minimising multi-centre radiomics variability through image normalisation: a pilot study. Sci Rep. 2022;12:12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Chen J, Zhou R, et al. Machine learning-based multiparametric MRI radiomics for predicting poor responders after neoadjuvant chemoradiotherapy in rectal Cancer patients. BMC Cancer. 2022;22:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acquitter C, Piram L, Sabatini U, et al. Radiomics-Based Detection of Radionecrosis Using Harmonized Multiparametric MRI. Cancers (Basel). 2022;14:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Ammari S, Lawrance L, et al. Radiomics-Based Method for Predicting the Glioma Subtype as Defined by Tumor Grade, IDH Mutation, and 1p/19q Codeletion. Cancers (Basel). 2022;14:1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitney HM, Li H, Ji Y, et al. Multi-Stage Harmonization for Robust AI across Breast MR Databases. Cancers (Basel). 2021;13:4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leithner D, Schöder H, Haug A, et al. Impact of ComBat Harmonization on PET Radiomics-Based Tissue Classification: A Dual-Center PET/MRI and PET/CT Study. J Nucl Med. 2022;63:1611–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welch ML, McIntosh C, Haibe-Kains B, et al. Vulnerabilities of radiomic signature development: The need for safeguards. Radiother Oncol. 2019;130:2–9. [DOI] [PubMed] [Google Scholar]
- 36.Mayerhoefer ME, Materka A, Langs G, et al. Introduction to Radiomics. J Nucl Med. 2020;61:488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayerhoefer ME, Szomolanyi P, Jirak D, et al. Effects of magnetic resonance image interpolation on the results of texture-based pattern classification: a phantom study. Invest Radiol. 2009;44:405–11. [DOI] [PubMed] [Google Scholar]
- 38.Messiou C, Hillengass J, Delorme S, et al. Guidelines for Acquisition, Interpretation, and Reporting of Whole-Body MRI in Myeloma: Myeloma Response Assessment and Diagnosis System (MY-RADS). Radiology. 2019;291:5–13. [DOI] [PubMed] [Google Scholar]
- 39.Chatterjee A, Vallieres M, Dohan A, et al. Creating robust predictive radiomic models for data from independent institutions using normalization. IEEE Trans Radiat Plasma Med Sci. 2019;3:210–215. [Google Scholar]


