Abstract
We mapped and cloned SKI7, a gene that negatively controls the copy number of L-A and M double-stranded RNA viruses in Saccharomyces cerevisiae. We found that it encodes a nonessential 747-residue protein with similarities to two translation factors, Hbs1p and EF1-α. The ski7 mutant was hypersensitive to hygromycin B, a result also suggesting a role in translation. The SKI7 product repressed the expression of nonpolyadenylated [non-poly(A)] mRNAs, whether capped or uncapped, thus explaining why Ski7p inhibits the propagation of the yeast viruses, whose mRNAs lack poly(A). The dependence of the Ski7p effect on 3′ RNA structures motivated a study of the expression of capped non-poly(A) luciferase mRNAs containing 3′ untranslated regions (3′UTRs) differing in length. In a wild-type strain, increasing the length of the 3′UTR increased luciferase expression due to both increased rates and duration of translation. Overexpression of Ski7p efficiently cured the satellite virus M2 due to a twofold-increased repression of non-poly(A) mRNA expression. Our experiments showed that Ski7p is part of the Ski2p-Ski3p-Ski8p antiviral system because a single ski7 mutation derepresses the expression of non-poly(A) mRNA as much as a quadruple ski2 ski3 ski7 ski8 mutation, and the effect of the overexpression of Ski7p is not obtained unless other SKI genes are functional. ski1/xrn1Δ ski2Δ and ski1/xrn1Δ ski7Δ mutants were viable but temperature sensitive for growth.
The 5′ cap (m7G5′ppp5′Xp) and the 3′ poly(A) structures of eukaryotic mRNAs are essential for efficient translation and mRNA stability. The poly(A) tail acts synergistically with the 5′ cap structure in translation (11, 13; reviewed in reference 37). The first step in mRNA degradation is the shortening of this poly(A) tail, followed by a decapping reaction, producing an uncapped nonpolyadenylated [non-poly(A)] mRNA which is then subject to rapid 5′ → 3′ degradation (9, 29; reviewed in reference 16).
This intermediate of mRNA degradation resembles the mRNAs of the L-A and M double-stranded RNA (dsRNA) viruses of the yeast Saccharomyces cerevisiae, which naturally lack both the 5′ cap and the 3′ poly(A) tail (6, 46). The L-A and M mRNAs are nonetheless expressed, but their expression is sensitive to chromosomal mutations affecting pathways involving these 5′ or 3′ structures. The ski mutants were first isolated on the basis of their superkiller phenotype (35, 47). The SKI2, SKI3, SKI4, SKI6, SKI7, and SKI8 products repress the copy number of L-A and M viruses, and mutations in these genes result in a higher level of expression of the secreted killer toxin encoded by M dsRNA (3, 35, 47, 53; reviewed in reference 50). The SKI1 gene (47), later proved to be XRN1, encodes the 5′ → 3′ exoribonuclease specific for uncapped cellular mRNAs and is responsible for a major pathway of mRNA decay (14, 18, 29, 43, 44). In a ski1 mutant, uncapped viral mRNAs are more stable, causing, for example, increased toxin production (24). The SKI2, SKI3, and SKI8 genes (26, 34, 53) encode a system that specifically blocks the expression of 3′ non-poly(A) mRNAs (24). The nuclear location of Ski3p (34) and the similarity of Ski2p to a nucleolar human homolog (21, 53) suggested that they carry out a function taking place in the nucleus and inducing specific repression of non-poly(A) mRNA translation (24).
Ski6p is homologous to bacterial RNase PH (4), an enzyme responsible for trimming a few nucleotides from the 3′ end of tRNA precursors and 5S rRNA during their maturation (22). Indeed, ski6 mutants produce defective 60S subunits (4), and Ski6p (Rrp41p) was localized in a complex of 3′ → 5′ exoribonucleases involved in 5.8S rRNA processing (28). These abnormalities lead to derepressed expression of non-poly(A) mRNA in ski6 mutants (4), with effects on both the initial rate and the duration of translation from electroporated non-poly(A) mRNAs. Mutations in the SKI2, SKI3, SKI6, or SKI8 gene can also slow the 3′ → 5′ degradation of special mRNA decay intermediates whose degradation by the predominant 5′ → 3′ degradation system is blocked (1). Because the control of translation and mRNA turnover are multiply intertwined, distinguishing the primary effects of the SKI genes is difficult, but the overall effect on viral replication appears to be enhanced expression of the viral mRNA which lacks 3′ poly(A).
Since ski mutations (including, as we show here, ski7) primarily affect the expression of non-poly(A) mRNAs, we examined the role of the 3′ untranslated region (3′UTR) in non-poly(A) mRNA expression. The 3′UTR is known to have specific functions in many systems (49). Furthermore, all cellular mRNAs are believed to undergo deadenylation, and non-poly(A) mRNAs can be present in the polysomal fraction under special circumstances (14, 33). Thus, when the mRNA remains capped and is non-poly(A), its 3′UTR may itself serve as an element influencing mRNA expression (reviewed in references 15 and 49). In CHO (Chinese hamster ovary) cells, increased 3′UTR length substantially improved the efficiency of translation initiation, suggesting a model in which the 3′UTR acts through more efficient ribosome recycling (45). A modest stabilization of mRNAs with longer 3′UTRs was also observed but was interpreted as a secondary phenomenon. There is also evidence for prokaryotes that events occurring during termination can affect the efficiency of ribosome recycling, the fourth step of translation (17, 32).
We show here that the ski7 mutation increases both the initial rate and the duration of the expression of non-poly(A) mRNAs and that this effect is independent of the length of the 3′UTR. The effect of the ski7 mutation on the expression of non-poly(A) mRNAs explains the superkiller phenotype of the mutant because the viral mRNAs lack 5′ poly(A). Overexpression of Ski7p cured the M2 satellite RNA due to increased repression of the expression of non-poly(A) mRNAs. The increased block of the translation of non-poly(A) mRNAs and the resulting elimination of M2 depended on the function of other SKI genes. This and other genetic experiments indicated that SKI7 is part of the SKI2-SKI3-SKI8 antiviral system.
MATERIALS AND METHODS
Genetic mapping of SKI7.
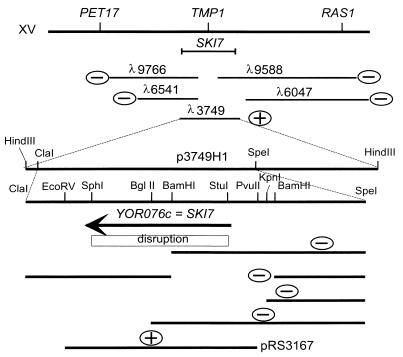
M2 dsRNA is a killer toxin-encoding satellite of the L-A virus (5, 27, 52). Above 32°C, M2 propagation depends on two genes, MKT1 and MKT2 (maintenance of K2), which are not needed by M1 dsRNA (51, 52). ski mutations suppress this requirement of M2 for MKT genes (35). Many laboratory strains carry mkt1 mutations. We crossed mkt1 ski7 M2 strains with a set of multiply marked strains designed for genetic mapping (12), which also proved to be mkt1. We found that ski7 was tightly linked to pet17 (parental ditype [PD], 34; nonparental ditype [NPD], 0; tetratype [T], 8; 9.5 cM) and linked to ade2 (PD, 24; NPD, 0; T, 22; 24 cM) on the right arm of chromosome XV. Analysis of individual tetrads showed that the order was CEN15-PET17-SKI7, placing SKI7 very close to TMP1/CDC21 (Fig. 1).
FIG. 1.
Cloning of SKI7. The ski7-1 mutation was localized by meiotic mapping to a region near TMP1/CDC21. Various clones from the Olson bank (36) were tested for complementation of ski7-1. Subcloning proved that SKI7 was YOR076c; this finding was confirmed by the phenotype of the disruption and complementation of ski7-1 by pRS3167 (see Materials and Methods).
Cloning of SKI7.
Strain RV603 (MATa ura3 leu2 ski7-1 mkt1; L-A-HN M2) is stably K2+ at either 25 or 32°C, but if it becomes SKI+, then it will be K2− after growth at 32°C (Table 1). Several λ clones of yeast DNA (36) located near tmp1 were tested by cotransformation of λ clone DNA and pBM2240, the latter cut with EcoRI and XhoI (10). Transformants were isolated at 25°C, grown at 25 or 32°C, and then tested for killer activity. Only λ3749 (ATCC 70213) gave recombinants which complemented ski7-1 (Fig. 1). The pBM2240 derivative carrying the insert of λ3749 was isolated, and subclones were made by cleavage with HindIII and ligation to pRS316 cut with the same enzyme. One of these, p3749H1, containing an 8.9-kb insert, complemented ski7-1. Subclones of p3749H1 were made by cleavage with ClaI and SpeI and insertion into pRS316 cut with the same enzymes. One of these, p3749L, contained a 5-kb insert and complemented ski7-1. Deletions of p3749L were made by cutting with ClaI-BamHI, BamHI-NotI, BglII-ClaI, KpnI, or BamHI followed by ligation (Fig. 1). None of these deletion plasmids complemented ski7-1. The ends of several of these clones were sequenced and found to match a region of chromosome XV near TMP1/CDC21 (48), as expected. The results indicated that the open reading frame YOR076c was responsible for the complementation of ski7-1. This finding was confirmed by cloning the 3,155-bp EcoRV-PvuII fragment containing YOR076c into pRS316 cut with SmaI and showing that the resulting plasmid, pRS3167, complemented ski7-1 (Fig. 1).
TABLE 1.
Strains of Saccharomyces cerevisiae
| Strain | Description |
|---|---|
| RV603 | MATa ura3 leu2 ski7-1 mkt1; L-A-HN M2 |
| B117 | MATa ura3 his3 trp1 ski7::HIS3; M-o L-A-HN; from 2959 |
| 2959 | MATa ura3 his3 trp1 SKI; M2 L-A-HN |
| B959 | 2959 K− |
| 3973 | MATa ura3 his3 leu2 ski2::HIS3 |
| 3974 | MATa ura3 his3 leu2 ski3::URA3 |
| 3975 | MATα ura3 his3 leu2 ski7::HIS3 |
| 4602-2B | MATα ura3 his3 leu2 trp1 |
| 3963 | MATα ura3 his3 leu2 trp1 ski3::URA3 ski2::HIS3 ski8::URA3 ski7::HIS3 |
| RV493 | MATa ura3 ade3 his leu2 trp1 ski6-2 mkt1; L-A-HN M2 |
Plasmid constructions.
The insert from pRS3167 (see above) was excised as a SpeI-HindIII fragment and inserted into pBluescript SK(+), creating pSKSKI7. To prepare a genomic disruption of SKI7, the StuI-SphI fragment including nearly all of the SKI7 open reading frame was replaced with the HIS3 gene on a SmaI-SphI fragment from pJJ215 (19), creating pSKI7::HIS3. Multicopy YEpSKI7 was created by the insertion of the same pRS3167 SpeI-HindIII fragment containing SKI7 into YEp351 cut with SpeI and HindIII.
Disruption of SKI7.
A diploid strain, 2959 (MATa ura3 his3 trp1; M2 L-A-HN) × 2966 (MATα leu2 his3 trp1 pho3 pho5; L-A-o M-o), was transformed with linearized pSKI7::HIS3. His+ diploids were selected, and the genomic replacement of SKI7 was confirmed by Southern blotting. Positive diploids were sporulated, and the segregation observed was 2 His+ Ski−:2 His− Ski+ (four tetrads). Tetrad analyses were confirmed by Southern blotting. As the SKI7 disruptant was viable, strain B117 was constructed from strain 2959 by the same procedure.
Strain 5X47 was the toxin-sensitive strain used as an indicator in the killer assay (35).
Expression of luciferase mRNAs.
The luciferase mRNA expression plasmids pT7-luc [no poly(A); untranslated region, 22 bases] and pT7-luc50A [50-mer poly(A) tail] (13) were linearized with SmaI and DraI, respectively. Plasmids allowing the transcription of luciferase mRNAs with repeated copies of a 20-base 3′UTR (5′TCTACAGCATATCTGGATCTGGATCC3′) followed (or not) by a 50-mer poly(A) tail (5′CCA25GTTATA25TTTAAA3′) were kindly provided by Daniel Gallie (45). For RNA synthesis, pT7-luc with several copies of the above 3′UTR sequence was linearized with BamHI, and pT7-luc(3′UTR)n+50(A) was linearized with DraI. Transcripts were synthesized with an Ambion MEGAscript transcription kit in the presence or absence of the cap analog 5′m7GpppG in accordance with the manufacturer’s instructions. After DNase I treatment and precipitation with LiCl, RNAs were passed over G-50 columns (5 prime-3 prime SELECT-B) and quantitated by both measurement of the optical density (OD) at 260 nm and comparison with known concentration standards on agarose gels.
RNA electroporation was done as described previously (11) with minor modifications. Two micrograms of RNA was used for electroporation, and cells were assayed for luciferase activity after different times of growth at 30°C. Luciferase activity was assayed as previously described (24).
Drug sensitivity test.
Isogenic wild-type and ski7 cells grown in YPAD to the log phase were diluted to an OD at 600 nm of 0.15; 0.015-, 0.0015, and 5-μl aliquots were spotted on YPAD plates with or without drug (the viabilities of both strains appeared to be identical and proportional to the measured OD on YPAD). The concentration of hygromycin B was 100 μg/ml, and that of cycloheximide was 100 ng/ml.
RESULTS
The SKI7 product is similar to translation factors.
Like other ski (superkiller) mutations, ski7 confers cold sensitivity for growth on cells carrying M dsRNA, indicating the importance in nature of the SKI genes. We genetically mapped ski7 close to CDC21/TMP1 on chromosome XV by using the suppression by ski7 of a defect in propagation of the M2 virus. We obtained λ clones including this small area and cloned the gene from one of them (see Materials and Methods) (Fig. 1). Since the clone complementing the ski7 mutation was obtained from DNA that maps close to CDC21/TMP1, we have cloned SKI7 and not a suppressor. Deletion analysis (Fig. 1) proved that SKI7 is identical to YOR076c, previously identified in the yeast genome project (48). A deletion-substitution ski7 mutation produced the superkiller phenotype but did not produce a growth defect, showing that SKI7 is not essential (see Materials and Methods).
The 747-residue protein Ski7p is similar to yeast Hbs1p (24.4% identity and 58% similarity in 582 amino acids), a factor whose overexpression suppresses the growth defect of an ssb1 ssb2 double mutant. SSB1 and SSB2 products are known to be associated with the ribosome during peptide synthesis (31). However, overexpression of Hbs1p does not suppress ski7-1 (data not shown). Ski7p also resembles the translation elongation factors EF-Tu and EF-1α (45 identities in a 154-residue region of similarity) (Fig. 2) and the yeast translation termination factor Sup35p (data not shown), including a GTPase consensus sequence. These similarities suggest that Ski7p is involved in translation.
FIG. 2.
Ski7p is similar to Hbs1p (31) and yeast EF-1α (Tef1p or Tef2p) (40). Black boxes show residues identical to those in Ski7p; dashes show gaps.
Ski7p specifically blocks the expression of non-poly(A) mRNAs.
Because the ski7 mutation, like ski1, ski2, ski3, ski6, and ski8, increases the expression of viral uncapped non-poly(A) mRNAs, we used electroporation of luciferase mRNAs to study the effect of a SKI7 disruption (Table 2). In a wild-type strain, capped (C+) polyadenylated (A+) mRNA was expressed 38 to 42 times better than C+ non-poly(A) (A−) mRNA (Table 2). In the isogenic SKI7 disruptant, C+ A+ mRNA was expressed only 2 to 5.5 times better than C+ A− mRNA. Thus, the SKI7 disruption increases the expression of C+ A− mRNA 7- to 20-fold. A similar effect on uncapped (C−) A− mRNA expression was also seen, with a seven- to ninefold increase in expression in the ski7::HIS3 disruptant (Table 2). In contrast, no increased expression of C− A+ mRNA in the mutant was seen; a decrease of two- to threefold was observed (Table 2). This modest effect of the absence of a cap in the mutant might also explain why C− A− mRNA expression is not enhanced in the ski7 mutant as strongly as is that of C+ A− mRNA.
TABLE 2.
Ski7p blocks the expression of non-poly(A) mRNAsa
| Expt | Strain | Expression of the following luciferase mRNA:
|
|||
|---|---|---|---|---|---|
| C+ A+ | C− A+ | C+ A− | C− A− | ||
| 1 | Wild type | 33.4 (100) | 2.9 (8.8) | 0.88 (2.6) | 0.19 (0.6) |
| ski7::HIS3 | 26.3 (100) | 1.26 (4.8) | 13.5 (51) | 1.76 (6.7) | |
| Ratio of ski7 to wild type | 0.79 (1.0) | 0.43 (0.54) | 15.3 (20) | 9.3 (11) | |
| 2 | Wild type | 26.9 (100) | 2.25 (8.4) | 0.64 (2.4) | 0.12 (0.4) |
| ski7::HIS3 | 30 (100) | 0.80 (2.7) | 5.4 (18) | 0.81 (2.7) | |
| Ratio of ski7 to wild type | 1.1 (1.0) | 0.35 (0.32) | 8.4 (7.5) | 6.8 (6.7) | |
Wild-type (B959) and isogenic ski7::HIS3 (B117) strains were electroporated with 2 μg of the indicated mRNA made from pT7-luc [for non-poly(A) mRNAs] or pT7-luc50A (for polyadenylated mRNAs). C+ and C− indicate mRNAs with and without the 5′ cap, respectively; A+ and A− indicate mRNAs with and without the 3′ poly(A), respectively. Luciferase was measured 2 h after electroporation as described in Materials and Methods and is reported in light units per microgram of protein. Numbers in parentheses for strain data are percentages of the C+ A+ value. Numbers in parentheses are ratios assuming a ratio of 1.0 for C+ A+ mRNA.
In a ski7 mutant, the lack of a 3′ poly(A) structure appears less critical. The poly(A) structure is well known to be important in both the initial rate and the duration of translation of mRNAs. Since 3′UTRs can also play roles in translation (45) and in mRNA stability (30), it is possible that the ski7 mutation derepresses 3′UTR function itself, compensating for the loss of the poly(A) tail or its natural absence on viral mRNAs. We thus examined the effect of the length of the 3′UTR on mRNA expression in a wild-type strain or an isogenic ski7 mutant.
3′UTR length influences non-poly(A) mRNA expression, and the ski7 mutation has an additive effect.
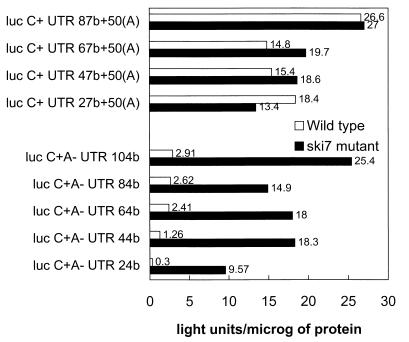
We used constructs allowing the production in vitro of capped luciferase mRNAs with a 3′UTR sequence composed of repeats of a 20-nucleotide sequence (45) (see Materials and Methods). Expression from these mRNAs was measured 2 h after their electroporation into spheroplasts prepared from wild-type and isogenic ski7 strains (Fig. 3). Increasing the length of the 3′UTR from 24 bases to 104 bases resulted in a 9.7-fold increase in expression in the wild-type strain (Fig. 3). With a poly(A) tail, the difference between these mRNA constructs was at most twofold (Fig. 3). We confirmed with yeast cells the observation made with CHO cells (45) that increasing the length of the 3′UTR improves expression when mRNAs are non-poly(A).
FIG. 3.
Luciferase expression in isogenic wild-type (B959) and ski7Δ mutant (B117) strains 2 h after electroporation with 2 μg of the indicated mRNAs. All mRNAs had 5′ cap structures. The lengths of the 3′UTRs are indicated (one to five identical 20-bp units were inserted 3′ of the luc coding region) (45) (see Materials and Methods). microg, microgram.
The ski7 mutation increased expression from all of these non-poly(A) mRNAs, regardless of the length of the 3′UTR (Fig. 3 and Table 3). As in the wild-type strain, the expression of polyadenylated mRNAs in a ski7 strain was not strongly dependent on the length of the 3′UTR (Fig. 3).
TABLE 3.
3′UTR length and SKI7 effects on mRNA translation and stabilitya
| Strain | C+ A− RNAb | Initial rate | Ft1/2 | Maximum activity |
|---|---|---|---|---|
| Wild type | UTR 4b | 0.9 ± 0.1 | 3.7 ± 0.1 | 0.15 ± 0.01 |
| UTR 24b | 2.9 ± 0.1 | 3.2 ± 0.2 | 0.2 ± 0.01 | |
| UTR 64b | 10.3 ± 2.4 | 7.1 ± 0.1 | 1.9 ± 0.6 | |
| UTR 104b | 10.8 ± 0.6 | 8.4 ± 0.4 | 1.7 ± 0.3 | |
| ski7::HIS3 | UTR 4b | 3.9 ± 0.5 | 13.7 ± 0.3 | 1.4 ± 0.1 |
| UTR 24b | 16.3 ± 3.1 | 17.2 ± 2.5 | 4.4 ± 0.1 | |
| UTR 64b | 18.0 ± 3.3 | 31.6 ± 0.3 | 8.6 ± 0.2 | |
| UTR 104b | 21.2 ± 4.8 | 34.4 ± 3.5 | 13.7 ± 0.5 |
Wild-type (B959) and isogenic mutant (B117) strains were electroporated with the indicated C+ A− luciferase mRNAs. From the time course of luciferase synthesis, we determined the initial rate of luciferase accumulation (initial rate) (in light units [×100] per minute per microgram of protein) and functional stability (Ft1/2) (in minutes required to reach one-half maximum luciferase activity). Maximum activity is reported in light units per microgram of protein. The control had 0.005 light unit/μg of protein. Errors indicate the variation of results within one experiment with identical preparations of RNA and cells.
4b, 24b, etc., indicate the number of bases in the 3′UTR.
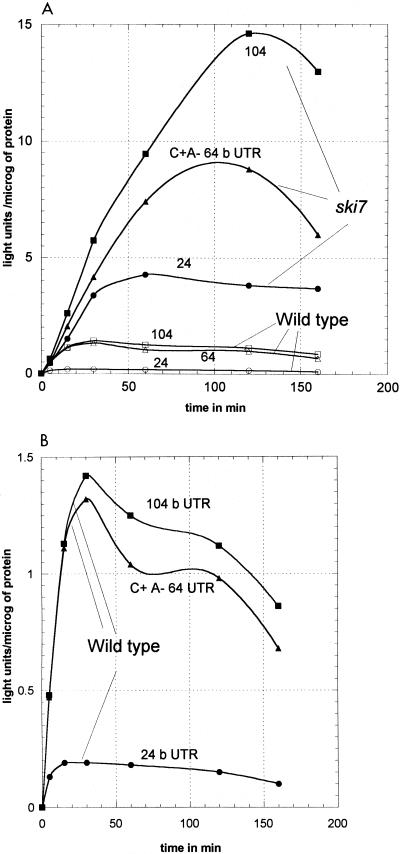
From the time course of luciferase accumulation, the luc-64b-3′UTR or luc-104b-3′UTR mRNAs were found to induce a 3- to 4-fold-higher initial rate and a 2.5-fold-increased duration of expression compared to the luc-4b-3′UTR mRNA (Table 3), leading to a maximum luciferase activity that was 10-fold higher (Table 3).
The initial rate of expression for the luc-24b-3′UTR mRNA was 5.5-fold higher in the mutant than in the wild type (Fig. 4B and Table 3). The functional half-life was increased five times, and overall expression was 24-fold higher. Similarly, the initial rate for the luc-4b-3′UTR mRNA was increased fourfold in the ski7 mutant. Thus, the ski7 mutation clearly increases both the initial rate of expression and the duration of expression of these RNAs.
FIG. 4.
Time course of luciferase accumulation in cells electroporated with 2 μg of C+ A− mRNAs with various lengths of 3′UTRs. Isogenic wild-type (B959) and ski7Δ mutant (B117) strains were used. (B) Wild-type results on an expanded scale. microg, microgram.
One test of whether the ski7 mutation produces its effects by derepressing the activity of the 3′UTR is to examine an mRNA that has almost no 3′UTR. We used a C+ A− mRNA with a 3′UTR only 4 nucleotides long. We still saw a 4-fold increase in the initial expression rate in the ski7 strain and a 3.7-fold increase in the functional half-life (Table 3). These results show that Ski7p does not act by repressing the activity of the 3′UTR. Conversely, a comparison of luc-4b-3′UTR and luc-64b-3′UTR mRNAs in the ski7 strain showed increases in the initial expression rate and in the functional half-life comparable to those seen in the wild-type host (Table 3). These results indicate that the 3′UTR effect is not dependent on Ski7p.
We also noted that while functional half-lives and initial expression rates are undoubtedly connected, there is no one-to-one relationship. For example, the initial expression rates for luc-24b-3′UTR in a wild-type strain and luc-4b-3′UTR in a ski7 mutant were similar, but their duration of expression differed by fourfold. Likewise, the functional half-lives for luc-24b-3′UTR and luc-4b-3′UTR in the ski7 mutant were similar, but their initial expression rates were fourfold different.
Antibiotic sensitivity of the ski7 mutant.
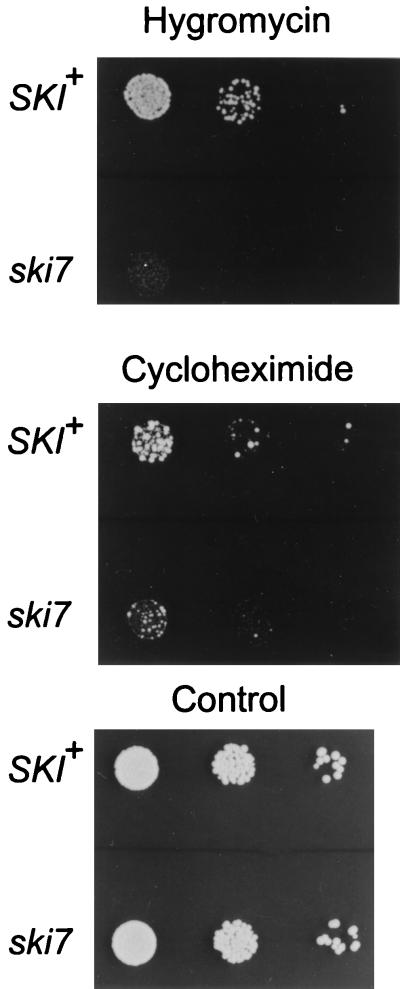
Although the ski7 mutant had an apparently normal polysome profile (data not shown), many mutations affecting components of the translation apparatus are found to be hypersensitive to hygromycin B (7, 38) and other drugs that increase translational errors (23). Figure 5 shows that the ski7 mutant strain was hypersensitive to hygromycin B and slightly hypersensitive to cycloheximide (but not to paromomycin; data not shown), supporting the notion that SKI7 affects ribosome function.
FIG. 5.
Hypersensitivity of the ski7Δ mutant to hygromycin B and cycloheximide. Dilutions of suspended wild-type (B959) and ski7Δ mutant (B117) strains were spotted on rich medium containing hygromycin B, cycloheximide, or neither drug (control).
SKI7 is a regulator of non-poly(A) mRNA expression.
We examined the effect of the overproduction of Ski7p on viral propagation and non-poly(A) mRNA expression. SKI7 was subcloned on a multicopy plasmid and used to transform RV603, a ski7-1 mutant. ski mutants are able to propagate M2 dsRNA at 25 and 32°C and have a superkiller phenotype (Table 4) (see Materials and Methods). When the ski7 mutation was complemented by a single copy of SKI7 (plasmid pRS3167), the strain remained a killer at 25 but lost the virus at 32°C. This result is expected from a wild-type strain containing LA-HN and M2 viruses (35). Interestingly, SKI7 present on a multicopy plasmid (YEpSKI7) induced the loss of killer toxin production and M2 dsRNA at every temperature (Table 4 and data not shown).
TABLE 4.
SKI7 present in a high copy number cures M2 dsRNAa
| Pladmid used with ski7-1 mutant RV603 | Killer toxin secretion at the following temp (°C):
|
|
|---|---|---|
| 25 | 32 | |
| pRS316 (single-copy vector) | ++++ | +++ |
| pRS3167 (+ SKI7) | + | − |
| YEp351 (high-copy vector) | ++++ | +++ |
| YEpSKI7 (+ SKI7) | − | − |
The sci7-1 strain RV603 was transformed at 25°C with the single-copy plasmids pRS316 and pRS3167 and with the high-copy-number plasmids YEp351 and YEpSKI7. Transformants were streaked for single colonies at the indicated temperature, and the colonies were then tested for toxin secretion at 20°C.
We further analyzed the expression of non-poly(A) mRNAs in a ski7 mutant or wild-type strain in the presence or absence of SKI7 on a multicopy plasmid. Figure 3 shows that a ski7 mutant strain expressed luc-24b-3′UTR, luc-64b-3′UTR, and luc-104b-3′UTR 31.9-, 7.5-, and 8.7-fold better than a wild-type strain, respectively. Ski7p overproduction in a wild-type strain (or a ski7 mutant strain) reduced the expression of C+ A− mRNAs, particularly those with luc-64b-3′UTR and luc-104b-3′UTR, to levels twofold below those seen in a wild-type strain with the vector alone (Table 5). We speculate that this stronger repression of expression of non-poly(A) mRNAs was responsible for the observed loss of the virus after several cell divisions.
TABLE 5.
Overproduction of Ski7p blocks the expression of non-poly(A) mRNAsa
| Strain | C+ A− RNA | Luciferase expression with:
|
Ratio (YEp351/YEpSKI7) | |
|---|---|---|---|---|
| YEp351 | YEpSKI7 | |||
| ski7::HIS3 3975 | UTR 24b | 12.7 | 0.40 | 31.9 |
| UTR 64b | 14.9 | 1.33 | 11.2 | |
| UTR 104b | 20.9 | 1.33 | 15.7 | |
| Wild type 4602-2B | UTR 24b | 0.16 | 0.11 | 1.5 |
| UTR 64b | 2.9 | 1.2 | 2.4 | |
| UTR 104b | 2.2 | 1.0 | 2.2 | |
| ski6-2 RV493 | UTR 24b | 10.0 | 7.9 | 1.3 |
| UTR 64b | 10.5 | 11.5 | 0.9 | |
| UTR 104b | 11.2 | 9.2 | 1.2 | |
| ski2::HIS3 3973 | UTR 24b | 19.0 | 17.3 | 1.1 |
| UTR 64b | 30.2 | 31.2 | 1.0 | |
| UTR 104b | 29.9 | 26.6 | 1.1 | |
| ski3::URA3 3974 | UTR 24b | 12.2 | 20.2 | 0.6 |
| UTR 64b | 25.9 | 25.4 | 1.0 | |
| UTR 104b | 24.6 | 33.5 | 0.7 | |
| Quadruple mutant 3963 (ski2 ski3 ski7 ski8) | UTR 24b | 15.7 | 18.4 | 0.8 |
| UTR 64b | 17.1 | 16.3 | 1.1 | |
| UTR 104b | 16.6 | 16.6 | 1.0 | |
The indicated C+ A− luciferase RNAs were electroporated into strains with different SKI genes and in the presence of SKI7 on a multicopy plasmid or the vector alone. Luciferase expression is reported in light units per microgram of protein. The control had less than 0.005 light unit/μg of protein. Values are averages of measurements from two independent experiments. Each measurement was made in duplicate.
Thus, inactivation of Ski7p produces derepression and its overproduction produces repression of C+ A− mRNA expression. We conclude that the level of expression of Ski7p is critical for regulating non-poly(A) mRNA expression and M2 virus propagation.
Ski7p activity in different ski backgrounds.
Single deletions of SKI2, SKI3, SKI7, or SKI8 do not affect cell viability (34, 42, 53; this work). Within this group, multiple ski deletion strains were easily constructed. For example, the cross 4592-1C (MATa leu2 ura3 his3 ski8::URA3 ski7::HIS3) × 4593-6B (MATα trp1 ura3 his3 ski3::URA3 ski2::HIS3) gave 90% spore germination at 30°C. A quadruple disruptant (strain 3963) was confirmed by a subsequent cross. Overexpression of SKI7 in strain 3963 did not modify the expression of luciferase mRNAs (Table 5). Nor did overproduction of Ski7p alter expression in single ski2, ski3, or ski6 strains. Further, adding ski2, ski3, and ski8 mutations to a ski7 mutant did not alter either the initial rates or the duration of translation of different luciferase mRNAs (Table 6). These results suggest that the different Ski proteins are related in function.
TABLE 6.
A quadruple ski mutant is similar to an ski7 mutanta
| Strain | C+ A− RNA | Initial rate | Ft1/2 | Maximum activity |
|---|---|---|---|---|
| Quadruple mutant 3963 (ski2::HIS3 ski3::URA3 ski7::HIS3 ski8::HIS3) | UTR 24b | 18.6 | 18 | 5.0 |
| UTR 64b | 25.7 | 41 | 9.7 | |
| UTR 104b | 26.2 | 32.5 | 11.0 | |
| Single mutant 3975 (ski7::HIS3) | UTR 24b | 17.9 | 18.5 | 4.3 |
| UTR 64b | 24.5 | 32 | 8.8 | |
| UTR 104b | 30.2 | 32.5 | 13.2 |
Strain 3963 or 3975 was electroporated with the indicated C+ A− mRNAs, and the kinetics of luciferase synthesis were measured as described in Materials and Methods. Initial rate, initial rate of luciferase expression in light units (×100) per minute per microgram of protein. Ft1/2, functional stability in minutes. Maximum activity is reported in light units per microgram of protein. The control had 0.005 light unit/μg of protein.
ski1/xrn1 ski7 and ski1/xrn1 ski2 double mutants are viable but temperature sensitive for growth.
The cold-sensitive and temperature-sensitive growth of ski2, ski3, ski4, ski6, ski7, or ski8 mutants depends on M dsRNA (35) and is believed to be due to its high copy number. However, a ski2 mutation is known to also result in increased copy numbers of L-A, L-BC, and 20S RNAs (3, 25). It has been reported that ski1/xrn1 ski2 and ski1/xrn1 ski3 double mutants are lethal even if they lack L-A and M dsRNAs (18). Since ski1/xrn1 mutants were first isolated based on their increased expression of viral mRNAs (47) and should derepress the expression of L-A, L-BC, 20S, and 23S RNAs, all of which apparently lack 5′ caps and are found in nearly all laboratory strains, it is possible that the reported lethality of ski1 ski2 or ski1 ski3 is due to viral effects.
We crossed deletion mutants of ski1/xrn1 marked with URA3 with ski7::HIS3 and ski2::HIS3 strains. The doubly heterozygous diploids were subcloned several times at 39°C in an attempt to cure viruses before sporulation (41), but this procedure cured only L-A and M dsRNAs. Germination at 30°C produced no viable double mutants, as was previously observed for ski1/xrn1 ski2 and ski1/xrn1 ski3, but germination at 20°C produced almost the expected proportions of viable ski1/xrn1 ski7 and ski1/xrn1 ski2 segregants (Table 7). The double-mutant segregants were unable to grow at 30°C, as expected, and showed prominent L-BC dsRNA bands (data not shown). Because there is no known method for curing 20S RNA or 23S RNA and heat curing of L-BC dsRNA is variable and ineffective in these strains, it remains unclear whether the growth defect in these double mutants was due to effects of these RNA replicons or to translation and mRNA stability effects on the host.
TABLE 7.
Viability of xrn1/ski1 ski2 and xrn1/ski1 ski7 mutantsa
| Cross | Parents | Temp (°C) of germination | No. of xrn1 ski spores/total no. of spores |
|---|---|---|---|
| 4605 | xrn1 × ski2 | 20 | 7/48 (12/48 expected) |
| 4596 | xrn1 × ski2 | 30 | 0/70 |
| 4602 | xrn1 × ski7 | 20 | 5/24 (6/24 expected) |
| 4597 | xrn1 × ski7 | 30 | 0/76 |
In each cross, ski1, ski2, and ski7 were detected in the meiotic segregants by auxotrophy. Cross 4596 was D223-2A (MATa ura3 his3 trp1 xrn1::URA3) × 4588-6C (MATα ura3 his3 trp1 ski2::HIS3); crosses 4597 and 4602 were D223-2A × 4587-1A (MATα ura3 his3 trp1 leu2 ski7::HIS3); and cross 4605 was 4602-3C (MATα trp1 his3 ura3 xrn1::HIS3) × 4591-2B (MATa leu2 his3 ura3 ski2::HIS3).
DISCUSSION
Viral mRNAs, 3′UTRs, and SKI7.
Mutation of SKI7 produces an increased copy number of L-A and M viruses and an associated superkiller phenotype. We demonstrate that this mutation specifically induces the expression of non-poly(A) luciferase mRNAs, thus explaining its effect on viruses, since L-A and M viral mRNAs are non-poly(A). Thus, the SKI7 product is a repressor of non-poly(A) mRNA expression. This effect on luciferase or viral mRNAs having only a 3′UTR [but no poly(A)] led us to question its possible involvement in this regulation by SKI genes. As has been shown in other systems (e.g., 45, 49), we find that 3′UTRs are not passive elements in mRNA expression but elevate both the initial rates and the duration of translation of electroporated mRNAs. 3′UTRs are also known to be the locations of certain specific signals affecting the processes of translation and mRNA turnover. We confirm that general 3′UTR actions can be detected only when the mRNA has been deadenylated, because the addition of a poly(A) tail largely overwhelms 3′UTR effects on translation.
The poly(A) structure and the poly(A) binding protein, Pab1p, act synergistically with the factors associated with the 5′ cap in initiation. The poly(A) structure may also play roles in translation termination, in reinitiation, and in protecting mRNA from degradation from both 3′ and 5′ ends. The 3′UTRs could affect any of these functions.
Knowing the 3′UTRs can promote mRNA expression in the absence of poly(A), we considered the possibility that Ski7p is normally a repressor of such a 3′UTR function. ski7 mutants are enhanced for the functions to which the 3′UTR contributes, with increased initial rates and duration of translation. However, mRNAs which have essentially no 3′UTR also show a similar effect of ski7 mutations. This fact indicates that Ski7p and the 3′UTR both affect related events but that Ski7p does not act by repressing 3′UTR function.
Translation efficiency and mRNA stability.
Translation and mRNA turnover are intertwined in complex and poorly understood ways. Translation may either protect an mRNA or lead to its decay. For example, in nonsense-mediated decay, failure of translation of an mRNA leads to mRNA degradation (16). In contrast, there are several cases in which mRNA degradation requires translation of the mRNA to be degraded (2, 39). The 5′ cap and 3′ poly(A) structures each promote translation and protect the mRNA from degradation.
If one assumes that initial rates of translation of electroporated mRNA reflect translation efficiency and that functional half-life reflects mRNA stability, the experiments reported here show effects of ski7 mutations on both. Which is the primary effect? The relationship of translation kinetics to initiation rates and mRNA degradation depends on several assumptions, not directly tested here. Arguing for an effect on translation efficiency are the Ski7p effect on initial rates of translation, the similarity of Ski7p to the known translation components HBS1p and EF1-α, and the fact that Ski7p inactivation produces hypersensitivity to drugs known to affect ribosomal function. Even with longer 3′UTRs, which would be expected to protect an mRNA from 3′ degradation, a substantial effect of a ski7 mutation is seen. However, ski2, ski3, ski7, and ski8 mutations also affect the functional half-life of mRNA (24; this work), effects which could be primary, as suggested by others (1), or secondary to translation effects. It is also possible that ski7 affects both translation efficiency and mRNA turnover. For example, the UPF genes have recently been found to function both in translation elongation (e.g., in maintenance of the reading frame) and in nonsense-mediated decay (8, 16). Further work is necessary to distinguish these possibilities.
Relationships of SKI genes.
Although ski1Δ leads to slowed growth (20) and ski6Δ is lethal (4), ski7Δ does not affect cell growth if M dsRNA is absent. Even a quadruple ski2 ski3 ski7 ski8 mutant is healthy and has kinetics of translation of C+ A− luciferase mRNA similar to those of any of the single mutants. This finding indicates that these four Ski proteins are functionally related, perhaps part of a common complex or part of a common pathway. Ski7p may be the limiting component, since its overexpression can eliminate M2 from a normal strain, but its overexpression does not suppress other ski mutations.
Although xrn1/ski1Δ ski2Δ, and xrn1/ski1Δ ski3Δ double mutants have been reported to be lethal (18), we find that at least xrn1/ski1Δ ski2Δ and xrn1/ski1Δ ski7Δ are viable at 20°C, although they do fail to grow at 30°C, the temperature at which the earlier experiments were performed. Several yeast RNA replicons, because they lack both the 5′ cap and 3′ poly(A), are normally subject to repression by the SKI genes. It is thus possible that this conditional lethal phenotype is due to these other replicons. There is no known method of curing 20S RNA or 23S RNA, and the ability of L-BC to be cured is variable and difficult at best, making testing of this question impractical at present.
ACKNOWLEDGMENTS
We particularly thank Daniel Gallie for providing numerous plasmids. We thank Arlen Johnson for providing the xrn1::URA3 allele. We thank Tom Dever and Alan Hinnebusch for many helpful comments on the manuscript. Christina Pfund and Elizabeth Craig kindly provided the HBS1 plasmid.
REFERENCES
- 1.Anderson J S J, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachurski C J, Theodorakis N G, Coulson R M, Cleveland D W. An amino-terminal tetrapeptide specifies cotranslational degradation of beta-tubulin but not alpha-tubulin mRNAs. Mol Cell Biol. 1994;14:4076–4086. doi: 10.1128/mcb.14.6.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball S G, Tirtiaux C, Wickner R B. Genetic control of L-A and L-BC dsRNA copy number in killer systems of Saccharomyces cerevisiae. Genetics. 1984;107:199–217. doi: 10.1093/genetics/107.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benard L, Carroll K, Valle R C P, Wickner R B. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol Cell Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bostian K A, Sturgeon J A, Tipper D J. Encapsidation of yeast killer double-stranded ribonucleic acids: dependence of M on L. J Bacteriol. 1980;143:463–470. doi: 10.1128/jb.143.1.463-470.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruenn J, Keitz B. The 5′ ends of yeast killer factor RNAs are pppGp. Nucleic Acids Res. 1976;3:2427–2436. doi: 10.1093/nar/3.10.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crouzet M, Izgu F, Grant C M, Tuite M F. The allosuppressor gene SAL4 encodes a protein important for maintaining translational fidelity in Saccharomyces cerevisiae. Curr Genet. 1988;14:537–543. doi: 10.1007/BF00434078. [DOI] [PubMed] [Google Scholar]
- 8.Cui Y, Dinman J D, Peltz S W. Mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 10.Erickson J R, Johnston M. Direct cloning of yeast genes from an ordered set of lambda clones in Saccharomyces cerevisiae by recombination in vivo. Genetics. 1993;134:151–157. doi: 10.1093/genetics/134.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett J G, Gallie D R. RNA delivery in Saccharomyces cerevisiae using electroporation. Yeast. 1992;8:1007–1014. doi: 10.1002/yea.320081203. [DOI] [PubMed] [Google Scholar]
- 12.Gaber R F, Culbertson M R. Frameshift suppression in Saccharomyces cerevisiae. IV. New suppressors among spontaneous co-revertants of the group II his4-206 and leu2-3 frameshift mutations. Genetics. 1982;101:345–367. doi: 10.1093/genetics/101.3-4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 14.Hsu C L, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson R J, Standart N. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell. 1990;62:15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- 16.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 17.Janosi L, Mottagui-Tabar S, Isaksson L A, Sekine Y, Ohtsubo E, Zhang S, Goon S, Nelken S, Shuda M, Kaji A. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 1998;17:1141–1151. doi: 10.1093/emboj/17.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson A W, Kolodner R D. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol. 1995;15:2719–2727. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones J S, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 20.Larimer F W, Stevens A. Disruption of the gene XRN1, coding for a 5′→3′ exoribonuclease, restricts yeast cell growth. Gene. 1990;95:85–90. doi: 10.1016/0378-1119(90)90417-p. [DOI] [PubMed] [Google Scholar]
- 21.Lee S-G, Lee I, Kang C, Song K. Identification and characterization of a human cDNA homologous to yeast SKI2. Genomics. 1994;25:660–666. doi: 10.1016/0888-7543(95)80008-a. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Pandit S, Deutscher M P. 3′ Exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2856–2861. doi: 10.1073/pnas.95.6.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Liebman S W. A translational fidelity mutation in the universally conserved sarcin/ricin domain of 25S yeast ribosomal RNA. RNA. 1996;2:254–263. [PMC free article] [PubMed] [Google Scholar]
- 24.Masison D C, Blanc A, Ribas J C, Carroll K, Sonenberg N, Wickner R B. Decoying the cap− mRNA degradation system by a double-stranded RNA virus and poly(A)− mRNA surveillance by a yeast antiviral system. Mol Cell Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumoto Y, Fishel R, Wickner R B. Circular single-stranded RNA replicon in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:7628–7632. doi: 10.1073/pnas.87.19.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto Y, Sarkar G, Sommer S S, Wickner R B. A yeast antiviral protein, SKI8, shares a repeated amino acid sequence pattern with beta-subunits of G proteins and several other proteins. Yeast. 1993;8:43–51. doi: 10.1002/yea.320090106. [DOI] [PubMed] [Google Scholar]
- 27.Maule A P, Thomas P D. Strains of yeast lethal to brewery yeasts. J Inst Brew (London) 1973;79:137–141. [Google Scholar]
- 28.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome, a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 29.Muhlrad D, Decker C J, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullner E W, Kuhn L C. A stem-loop in the 3′ untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988;53:815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- 31.Nelson R J, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig E A. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 32.Pavlov M Y, Freistroffer D V, MacDougall J, Buckingham R H, Ehrenberg M. Fast recycling of Escherichia coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J. 1997;16:4134–4141. doi: 10.1093/emboj/16.13.4134. . (Erratum 16:5480.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Proweller A, Butler S. Efficient translation of poly(A)-deficient mRNAs in Saccharomyces cerevisiae. Genes Dev. 1994;8:2629–2640. doi: 10.1101/gad.8.21.2629. [DOI] [PubMed] [Google Scholar]
- 34.Rhee S K, Icho T, Wickner R B. Structure and nuclear localization signal of the SKI3 antiviral protein of Saccharomyces cerevisiae. Yeast. 1989;5:149–158. doi: 10.1002/yea.320050304. [DOI] [PubMed] [Google Scholar]
- 35.Ridley S P, Sommer S S, Wickner R B. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol Cell Biol. 1984;4:761–770. doi: 10.1128/mcb.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riles L, Dutchik J E, Baktha A, McCauley B K, Thayer E C, Leckie M P, Braden V V, Depke J E, Olson M V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993;134:81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 38.Sandbaken M G, Lupisella J A, DiDimenico B, Chakraburtty K. Protein synthesis in yeast: structural and functional analysis of the gene encoding elongation factor 3. J Biol Chem. 1990;265:15838–15844. [PubMed] [Google Scholar]
- 39.Savant-Bhonsale S, Cleveland D W. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a > 20S degradation complex. Genes Dev. 1992;6:1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- 40.Schirmaier F, Philippsen P. Identification of two genes coding for the translation elongation factor EF-1. EMBO J. 1984;3:3311–3315. doi: 10.1002/j.1460-2075.1984.tb02295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommer S S, Wickner R B. Co-curing of plasmids affecting killer double-stranded RNAs of Saccharomyces cerevisiae: [HOK], [NEX], and the abundance of L are related and further evidence that M1 requires L. J Bacteriol. 1982;150:545–551. doi: 10.1128/jb.150.2.545-551.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sommer S S, Wickner R B. Gene disruption indicates that the only essential function of the SKI8 chromosomal gene is to protect Saccharomyces cerevisiae from viral cytopathology. Virology. 1987;157:252–256. doi: 10.1016/0042-6822(87)90338-2. [DOI] [PubMed] [Google Scholar]
- 43.Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′ mononucleotides by a 5′ → 3′ mode of hydrolysis. J Biol Chem. 1980;255:3080–3085. [PubMed] [Google Scholar]
- 44.Stevens A, Maupin M K. A 5′ → 3′ exoribonuclease of Saccharomyces cerevisiae: size and novel substrate specificity. Arch Biochem Biophys. 1987;252:339–347. doi: 10.1016/0003-9861(87)90040-3. [DOI] [PubMed] [Google Scholar]
- 45.Tanguay R L, Gallie D R. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol Cell Biol. 1996;16:146–156. doi: 10.1128/mcb.16.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiele D J, Hannig E M, Leibowitz M J. Genome structure and expression of a defective interfering mutant of the killer virus of yeast. Virology. 1984;137:20–31. doi: 10.1016/0042-6822(84)90004-7. [DOI] [PubMed] [Google Scholar]
- 47.Toh-e A, Guerry P, Wickner R B. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J Bacteriol. 1978;136:1002–1007. doi: 10.1128/jb.136.3.1002-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valens M, Bohn C, Daignan-Fornier B, Dang V D, Bolotin-Fukuhara M. The sequence of a 54.7 kb fragment of yeast chromosome XV reveals the presence of two tRNAs and 24 new open reading frames. Yeast. 1997;13:379–390. doi: 10.1002/(SICI)1097-0061(19970330)13:4<379::AID-YEA85>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 49.Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 50.Wickner R B. Double-stranded RNA virus of yeast. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickner R B. Killer systems in Saccharomyces cerevisiae: three distinct modes of exclusion of M2 double-stranded RNA by three species of double-stranded RNA, M1, L-A-E, and L-A-HN. Mol Cell Biol. 1983;3:654–661. doi: 10.1128/mcb.3.4.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wickner R B. Plasmids controlling exclusion of the K2 killer double-stranded RNA plasmid of yeast. Cell. 1980;21:217–226. doi: 10.1016/0092-8674(80)90129-4. [DOI] [PubMed] [Google Scholar]
- 53.Widner W R, Wickner R B. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol Cell Biol. 1993;13:4331–4341. doi: 10.1128/mcb.13.7.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]