Abstract
The mitochondrial genomes of Bilateria are relatively conserved in their protein-coding, rRNA, and tRNA gene complement, but the order of these genes can range from very conserved to very variable depending on the taxon. The supposedly conserved gene order of Annelida has been used to support the placement of some taxa within Annelida. Recently, authors have cast doubts on the conserved nature of the annelid gene order. Various factors may influence gene order variability including, among others, increased substitution rates, base composition differences, structure of noncoding regions, parasitism, living in extreme habitats, short generation times, and biomineralization. However, these analyses were neither done systematically nor based on well-established reference trees. Several focused on only a few of these factors and biological factors were usually explored ad-hoc without rigorous testing or correlation analyses. Herein, we investigated the variability and evolution of the annelid gene order and the factors that potentially influenced its evolution, using a comprehensive and systematic approach. The analyses were based on 170 genomes, including 33 previously unrepresented species. Our analyses included 706 different molecular properties, 20 life-history and ecological traits, and a reference tree corresponding to recent improvements concerning the annelid tree. The results showed that the gene order with and without tRNAs is generally conserved. However, individual taxa exhibit higher degrees of variability. None of the analyzed life-history and ecological traits explained the observed variability across mitochondrial gene orders. In contrast, the combination and interaction of the best-predicting factors for substitution rate and base composition explained up to 30% of the observed variability. Accordingly, correlation analyses of different molecular properties of the mitochondrial genomes showed an intricate network of direct and indirect correlations between the different molecular factors. Hence, gene order evolution seems to be driven by molecular evolutionary aspects rather than by life history or ecology. On the other hand, variability of the gene order does not predict if a taxon is difficult to place in molecular phylogenetic reconstructions using sequence data or not. We also discuss the molecular properties of annelid mitochondrial genomes considering canonical views on gene evolution and potential reasons why the canonical views do not always fit to the observed patterns without making some adjustments. [Annelida; compositional biases; ecology; gene order; life history; macroevolution; mitochondrial genomes; substitution rates.]
Mitochondrial genomes are the remainder of the genome of an α-proteobacteria ancestor, which was incorporated into the eukaryotic cell by endosymbiosis (e.g., Martijn et al. 2018; Zardoya 2020). As a result of gene transfer to the nuclear genome and gene loss due to functional redundancy, mitochondrial genomes contain only a subset of the genes of the original complement: those that are relevant for their function. This is especially true for Bilateria, in which the usually circular mitochondrial genome contains only 13 protein-coding, two rRNA, and 22 tRNA genes and has an average size of 16 kb—though ranging from 11 to 50 kb (Zardoya 2020). Given the small number of genes (37), the gene order of mitochondrial genomes can be determined relatively easily. It has been suggested that gene order might be a powerful tool to reconstruct deep evolutionary histories as gene rearrangements should be unique and are unlikely to have evolved independently (see Boore 1999, 2006). This view appears to be supported by the conservative mitochondrial gene order in different large groups, such as vertebrates and insects (see Bernt et al. 2013; Cameron 2014). However, the gene order is substantially more variable in other bilaterian taxa such as Mollusca, Bryozoa, Tunicata, and Acari (e.g., Boore et al. 2004; Shao et al. 2006; Waeschenbach et al. 2006; Gissi et al. 2010; Stach et al. 2010; Bernt et al. 2013; Sun et al. 2020; Varney et al. 2021).
Besides their potential for phylogenetics, mitochondrial genomes can also inform our understanding of the evolution of gene rearrangement and its drivers (Bernt et al. 2013; Zardoya 2020). For example, increased substitution rates or biases in nucleotide or amino acid frequencies correlate with gene rearrangements in mitochondrial genomes (Shao et al. 2003; Hassanin et al. 2005; Podsiadlowski and Braband 2006; Xu et al. 2006; Min and Hickey 2007; Bernt et al. 2013; Luo et al. 2015). Bernt et al. (2013), investigating the mitochondrial genome evolution across all Metazoa, suggested that the possible molecular mechanisms underlying both increased substitution and rearrangement rates could be relaxed repair mechanisms or high mutational stress in combination with the lesser importance of mitochondrial efficiency. However, it is not certain if these are the causes of the actual rearrangement process or, rather, its signature. Another potential explanation for the increased rate of rearrangements is the presence of larger noncoding regions (Zhang et al. 2021). Others have suggested that tRNAs could act as mobile elements facilitating rearrangements or that recombination among the genomes within individual mitochondria occurred (Saccone et al. 1999; Kajander et al. 2000; Dowton and Campbell 2001; Luo et al. 2015). As possible biological causes, Bernt et al. (2013) also suggested an endoparasitic life style, anoxic conditions, high metabolic rate, or shorter generation time. Others also highlighted the presence of oxidative stress as another factor for rearrangements (Kajander et al. 2000; Dowton and Campbell 2001).
The presence of a conservative mitochondrial gene order has also been observed in Annelida, especially when considering only the protein-coding and rRNA genes (e.g., Boore and Brown 2000; Jennings and Halanych 2005; Vallès and Boore 2006; Zhong et al. 2008, 2011; Richter et al. 2015). Annelida comprises about 17 000 species in all marine, limnic, and terrestrial habitats on Earth (Weigert and Bleidorn 2016). Additionally, in the last decade, tremendous progress has been achieved with respect to the backbone phylogeny of Annelida and within different annelid subgroups using phylogenomic approaches (for review see Weigert and Bleidorn 2016; Struck 2019). On the other hand, in the same period, deviations from the supposed conservative mitochondrial gene order have been reported in increasing numbers (e.g., Mwinyi et al. 2009; Shen et al. 2009; Golombek et al. 2013; Aguado et al. 2016; Weigert et al. 2016; Seixas et al. 2017; Zhang et al. 2018; Alves et al. 2020; Tempestini et al. 2020; Sun et al. 2021). These range from small changes affecting only a few genes, usually tRNAs, to completely different organizations of the mitochondrial gene order in some taxa, especially in early diverging clades of the annelid phylogeny. It has therefore been suggested that the conservative gene order is restricted to Pleistoannelida (sensu Struck 2011) (Weigert et al. 2016). However, even within Pleistoannelida larger rearrangements of the gene order occur (Wu et al. 2009; Aguado et al. 2016; Seixas et al. 2017; Zhang et al. 2018). Recently, it has even been shown that a high variability can also occur within a genus (Tempestini et al. 2020; Sun et al. 2021). Hence, the conservative nature of annelid gene order has been called into question. On the other hand, many mitochondrial genomes of taxa with supposedly conservative gene order have only been published as release notes and as such have been subject to less scrutiny (e.g., Chen et al. 2019; Shekhovtsov and Peltek 2019; Zhou et al. 2020).
Suggested possible reasons for the increased rearrangement rates within Annelida have been similar to those for animals in general: increased substitution rates, oxidative stress, or lifestyles such as inhabiting calcareous tubes or the deep sea (Zhang et al. 2018; Tempestini et al. 2020; Sun et al. 2021). Additionally, increased genome size in general has been associated with an increase in noncoding regions and the presence of introns (Sun et al. 2021). However, for both Annelida and Metazoa these analyses were not done systematically but were either focused on only specific taxa of interest (e.g., only one genus, family, or order) or looked only into a single possible factor such as substitution rate. Biological causes have not been explored systematically and when biological causes are proposed they are usually explored in an ad-hoc manner, without rigorous testing. Fortunately, due to developments in sequencing technology the number of available genomes is steadily increasing. For example, as of 17 November 2021, 211 partial or complete mitochondrial genomes of annelid species with a length of at least 12 kb were deposited in NCBI and publicly accessible via a BLAST search. Annelida comprises both taxa with a conservative gene order and taxa that strongly deviate from this order along a well-established phylogeny. Hence, systematically investigating gene order evolution in Annelida can serve as a case study to understand gene order evolution in Metazoa.
In this study, we complemented the publicly available mitochondrial genomes as of 17 November 2021 with 33 new mitochondrial genomes. We then determined the evolution of the order of mitochondrial protein-coding and rRNA genes given the annelid phylogeny and evaluated conservation in the gene order of Annelida. We did the same for gene orders although accounting for tRNAs and compared both results. The influence of different molecular properties related to genome size, substitution rates, base frequencies, and nucleotide composition biases on the variability of the gene order was systematically assessed. This assessment included a total of 706 variables measuring different molecular properties. We also evaluated whether the degree of gene-rearrangement was related to problems in reconstructing the phylogeny of Annelida using only mitochondrial genome data. Finally, we also systematically assessed the influence of different biological factors on the variability of the gene orders.
Material and Methods
Determination of Mitochondrial Genomes and the Nuclear 18S rRNA for New Species
Mitochondrial genomes and nuclear 18S rRNA were sequenced anew for 36 annelid species of 32 families including 14 families with no mitochondrial genome in NCBI as of 17.11.2021 (Table 1). Specimens were preserved in 70% ethanol, RNAlater, or snap frozen in liquid nitrogen and then stored at −70 °C. Genomic DNA was extracted using the DNeasy Tissue Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocols, with at least two elution steps to increase the amount of DNA. For Apharyngtus punicus, Arenicola marina, Lumbrineris fragilis and Trilobodrilus axi, available EST data were mined for mitochondrial genes and additionally, up to nine genes were amplified using universal primers. Species-specific primers were designed based on these genes with the aid of the Primer3Plus web-interface (Untergasser et al. 2007). The fragments between the genes were amplified and sequenced by primer walking in six or more fragments ranging in size from 1 to 3 kb. The amplification used the QIAGEN® Multiplex PCR Kit (Qiagen, Hilden, Germany) (20 µL reaction: 10 µL multiplex mix, 2 µL Q solution, 1.6 µL 10 pmol/µL forward primer, 1.6 µL 10 pmol/µL reverse primer, 2.5 µL genomic DNA and 2.3 µL water) and a touchdown PCR (initial denaturation: 15ʹ 95 °C; 15 cycles: 35″ 94 °C, 90″ 55 °C or 60 °C (decreasing 1 °C at each cycle), 90″ 72 °C; 25 cycles: 35″ 94 °C, 90″ 50 °C or 55 °C, 90″ 72 °C; final elongation: 10ʹ 72°C). PCR fragments of the expected sizes were excised from TBE agarose gel and purified via NucleoSpin® Gel and PCR Clean-up (Macherey-Nagel, Düren, Germany). Additionally, the nuclear 18S rRNA was obtained as described by Golombek et al. (2013). Purified fragments were sent to Macrogen Europe (Amsterdam, Netherlands) for sequencing. All sequences were assembled using SeqMan II (DNASTAR Inc., Madison, WI, USA).
Table 1.
List of species sequenced in this project as well as additional information concerning the sequences
| Species | Family | mito | 18S | WGAa | HiSeqb | Read length (bp) | #reads | Bpc |
|---|---|---|---|---|---|---|---|---|
| Alitta succinea | Nereididae | Yes | No | Yes | 4000 | 150 | 57 672 716 | 15 758 |
| Apharyngtus punicus | nad,e | Yes | Yes | nad | nad | nad | nad | 13 893 |
| Arenicola marina | Arenicolidaee | Yes | No | nad | nad | nad | nad | 15 730 |
| Aricidea sp. | Paraonidaee | Yes | Yes | Yes | 2000 | 100 | 33 925 182 | 15 621 |
| Chone infundibuliformis | Sabellidae | Yes | Yes | No | 2000 | 100 | 13 376 834 | 15 142 |
| Cirriformia tentaculata | Cirratulidae | Yes | Yes | Yes | 2000 | 100 | 19 933 218 | 15 828 |
| Eunice pennata | Eunicidae | Yes | Yes | No | 2000 | 100 | 15 759 656 | 17 210 |
| Flabelligera mundata | Flabelligeridae | Yes | Yes | Yes | 2000 | 100 | 20 228 430 | 16 047 |
| Goniadides aciculata | Goniadidae | Yes | Yes | Yes | 4000 | 150 | 86 763 ,502 | 15 242 |
| Hyalinoecia tubicola | Onuphidae | Yes | Yes | No | 2000 | 100 | 33 643 220 | 16 563 |
| Laetmonice producta | Aphroditidae | Yes | Yes | No | 2000 | 100 | 42 857 914 | 16 004 |
| Laonice sp. | Spionidae | Yes | Yes | Yes | 2000 | 100 | 16 195 838 | 15 821 |
| Lumbrineris fragilis | Lumbrineridae | Yes | No | nad | nad | nad | nad | 16 445 |
| Marphsa aegypti | Eunicidae | Yes | Yes | Yes | 4000 | 150 | 69 364 192 | 15 157 |
| Mesonerilla intermedia | Nerillidaee | Yes | Yes | Yes | 2000 | 100 | 18 748 494 | 14 843 |
| Microphthalmus listensis | nae | Yes | Yes | Yes | 2000 | 100 | 16 675 624 | 13 656 |
| Naineris cf. setosa | Orbiniidae | Yes | Yes | No | 4000 | 150 | 66 489 860 | 15 670 |
| Neanthes acuminata | Nereididae | Yes | Yes | Yes | 4000 | 150 | 57 191 536 | 15 939 |
| Oenone fulgida | Oenonidaee | Yes | Yes | No | 4000 | 150 | 73 853 716 | 15 390 |
| Ophelina acuminata | Opheliidaee | Yes | Yes | No | 2000 | 100 | 36 213,768 | 15 655 |
| Ophiodromus pugettensis | Hesionidaee | Yes | Yes | No | 2000 | 100 | 33 602 076 | 14 989 |
| Owenia fusiformis | Oweniidae | No | Yes | No | 2000 | 100 | 39 712 492 | 16 204 |
| Perinereis fayedensis | Nereididae | Yes | Yes | No | 4000 | 150 | 58434 094 | 15 955 |
| Phyllochaetopterus sp. | Chaetopteridae | No | Yes | No | 2000 | 100 | 25 904 248 | 16 087 |
| Phyllodoce groenlandica | Phyllodocidaee | Yes | Yes | No | 2000 | 100 | 25 030,704 | 12 391 |
| Pisionidens tchesunovi | Pisionidae | Yes | Yes | Yes | 4000 | 150 | 66 487 630 | 14 969 |
| Polygordius lacteus | Polygordiidaee | Yes | Yes | No | 2000 | 100 | 30 412 116 | 16 672 |
| Protodorvillea kefersteini | Dorvilleidae | Yes | Yes | Yes | 2000 | 100 | 14 289 310 | 16 918 |
| Protodriloides chaetifer | Protodriloidaee | Yes | Yes | Yes | 2000 | 100 | 12 455 902 | 14 726 |
| Protodrilus rubropharyngeus | Protodrilidaee | Yes | Yes | Yes | 2000 | 100 | 34 170 334 | 15 118 |
| Sabellaria alveolata | Sabellariidaee | Yes | Yes | No | 2000 | 100 | 15 392 278 | 15 276 |
| Saccocirrus burchelli | Saccocirridae | Yes | Yes | Yes | 2000 | 100 | 15 379 308 | 15 303 |
| Spirobranchus triqueter | Serpulidae | Yes | Yes | No | 2000 | 100 | 17 694 620 | 17 039 |
| Streptosyllis sp. THS1 | Syllidae | No | Yes | No | 2000 | 100 | 23 024 282 | 14 972 |
| Stygocapitella josemariobrancoi | Parergodrilidaee | Yes | Yes | Yes | 2000 | 100 | 23 604 522 | 14 149 |
| Trilobodrilus axi | Dinophilidae | Yes | No | nad | nad | nad | nad | 12 482 |
aWhole genome amplification used.
bHiSeq machine used.
cSize of the mitochondrial genome after assembly.
dna = not applicable.
eFirst mitochondrial genome for the family.
For all other species, a genome skimming approach was applied. To increase the amount of genomic DNA, the whole genome of some species was amplified using the illustra GenomiPhi HY DNA Amplification Kit according to manufacturer’s instructions (Table 1). A genomic DNA shot-gun library including fragmentation and tagging adaptors for multiplexing was prepared and sequenced as 100 bp or 150 bp paired-end on an Illumina HiSeq2000 (Genterprise Genomics, Mainz, Germany) or HiSeq4000 (Norwegian Sequencing Center, Oslo), respectively. Number of reads per species ranged from 12 455 902 to 86 763 502 and was on average 34 400 631. The reads of each species were assembled into contigs using Spades 3.11 (Bankevich et al. 2012) with kmers set to 21 bp, 33 bp, and 55 bp, an average fragment size of 350 bp, and using otherwise default parameters. Using protein sequence information of the 13 protein-coding mitochondrial genes of Platynereis dumerilii (AF178678) as well as the sequence of the 18S of Stygocapitella subterranea (AF412810) as query sequences in TBLASTN or BLASTN searches, respectively, we searched in the assembled contigs for fragments of the mitochondrial genome as well as the genomic rRNA gene cluster. If possible and required, the mitochondrial genomes were closed using species-specific primers as described above for the four other species.
Compilation and Annotation of Mitochondrial Genome and 18S Data
In addition to our own mitochondrial genomes, we retrieved mitochondrial genomes available from NBCI as of 17 November 2021 (Supplementary online Appendix 1, http://dx.doi.org/10.5061/dryad.8w9ghx3pm). We used tblastx with the COI barcode of Parergodrilus heideri (KY503040), and the records were limited to Annelida (taxid:6340) and sequence length from 12 000 to 200 000 bp. If more than one mitochondrial genome per species was present, only the larger one was kept. Additionally, we retrieved 18S rRNA sequences from NCBI using MegaBlast and AF412810 to match the mitochondrial genomes. The following order was applied for the matching. First, if possible, the 18S was from the same individual. This was the case mostly for our own sequences, but also for some mitochondrial genomes generated from genome skimming data such as Glyceridae and Aphroditiformia. If this was not possible, the sequence was taken from the same species. If this was also not possible and there was no species already representing the genus with both a mitochondrial genome and 18S sequence, an 18S sequence was taken from another species of the same genus. Finally, the mitochondrial genomes of Cirriformia cf. tentaculata, Perinereis aibuhitensis, Ramisyllis multicaudata and Trypanosyllis sp. were excluded as these species had either too short or very divergent 18S sequences, making the alignment problematic and in two cases introduced extremely long branches (LBs) in the tree. In total, 168 mitochondrial genomes and 18S sequences from 69 annelid families were included in the following analyses, of which 33 were new mitochondrial genomes and 32 new 18S sequences generated in this study. Due to redundancy, lack of matching 18S sequence or too divergent 18S sequences, 75 mitochondrial genomes found with the tblastx search were not included. To these datasets, we added two outgroup taxa, Lineus viridis (Nemertea) and Terebratulina retusa (Brachiopoda) as they exhibit the ancestral lophotrochozoan mitochondrial gene order as shown by Bernt et al. (2013).
All 170 mitochondrial genomes were annotated for protein-coding, rRNA, and tRNA genes using MITOS2 (Al Arab et al. 2017; Donath et al. 2019), RefSeq 63 Metazoa as the reference dataset, and the invertebrate mitochondrial genetic code (NCBI code table 5). Finally, all genomes were manually investigated to detect problematic issues and possible genes not found by MITOS2. Specifically, the positions of the mitochondrial rRNAs and tRNAs were checked. The gene order as well as the sequences of the individual mitochondrial genes were compiled using custom-made shell scripts (available at GitHub “CompileDatasets.sh,” and “CheckFileNames.sh”).
Generate Reference Tree for the Macroevolutionary Analyses Using a Constraint Tree and Nuclear 18S Data
We wanted to compare the evolution of the mitochondrial genome in comparison to the nuclear genome including substitution rates along branches derived from the nuclear genome. We did this to avoid tautological reasoning inherent in comparing the evolution of the mitochondrial genome against a tree that would derive branch lengths and topology from the same data source. Hence, we recovered a constraint tree (Supplementary online Appendix 2) combining the results from recent, mostly phylogenomic studies (i.e., Erséus et al. 2020, Struck et al. 2007, 2015; Zhong et al. 2011; Anderson et al. 2017; Zhang et al. 2018; Phillips et al. 2019; Struck 2019; Tilic et al. 2020). The backbone of this constraint tree was based on the large-scale phylogenomic studies shown in Struck (2019), although the relationships within specific groups such as Crassiclitellata or Sabellida were based on studies targeting these groups (e.g., Anderson et al. 2017; Tilic et al. 2020). However, this constraint tree still has some unresolved nodes and does not provide branch length as it is a composite tree derived from multiple different studies. As the available phylogenomic data is very sparse and does not match the representative mitochondrial genome dataset, we used nuclear 18s rRNA sequences as a proxy for the nuclear genome to resolve these nodes and obtain branch lengths. Due to the high availability of the 18S data, we could match nuclear data very well to our mitochondrial genome dataset. The 18S sequences were aligned using MAFFT with automatic selection of the best alignment method, which was FFT-NS-i with the iterative refinement method (max. 2 iterations) (Katoh et al. 2005). The 3ʹ and 5ʹ ends of the alignment were trimmed using AliView (Larsson 2014), such that fewer than 10 sequences had no information on the first and last column of the alignment. Potentially nonhomologous positions were masked using AliScore and AliCut with gaps treated as ambiguous data and default settings (Kück et al. 2010). Next, we used IQtree (Nguyen et al. 2015) using the constraint tree and automatic model selection (-m MFP) for both the unmasked and masked datasets. The model chosen according to the Bayesian information criterion was TIM2e+R10 for the masked dataset and TN+F+I+G4 for the unmasked one. For the tree obtained using the masked or unmasked dataset, a ultrametric tree with relative ages was generated using chronos of the APE package (Paradis and Schliep 2018) in R (R Core Team 2020) with a lambda of 1 and a correlated model. Herein, we will refer to these four trees (masked, unmasked, and ultrametric) based on 18S data as the reference trees (e.g., Supplementary online Appendices 3 and 4).
Determine the Different Structural and Sequence-Based Properties of Each Species for Different Parts of the Mitochondrial Genomes
For the complete mitochondrial genome sequences, we determined genome size, frequencies of nucleotides, AT, and purines, and the AG, AT, CT, and GC skew values using BaCoCa.v1.109 (Kück and Struck 2014) as well as normalized relative composition frequency variability (nRCFV) values adjusted for the number of positions, character states, and taxa using nRCFVReader (Fleming and Struck 2023). Additionally, we added the number of duplicated genes, of introns, and of intergenic regions as well as the average size of the intergenic regions using a custom-made shell script (available at GitHub “RetrieveIntergenicParts.sh”).
For each protein-coding gene, alignments were generated with Mega 11.0.10 (Tamura et al. 2021) using MUSCLE, the invertebrate mitochondrial code (NCBI code table 5) and default settings to obtain both an amino acid alignment and a nucleotide alignment based on the amino acid one. For each rRNA gene, a nucleotide alignment was generated using also Mega 11.0.10 without codons and default settings. Three supermatrices were generated containing nucleotide alignments; one with all genes, one with only the protein-coding genes, and one with only rRNA genes. Additionally, one supermatrix was generated containing all amino-acid alignments of protein-coding genes. FASconCAT-G v1.05 was used to generate the supermatrices (Kück and Longo 2014). For each gene and supermatrix, to obtain tree-based measurements and get pairwise patristic distances, branch lengths were estimated using a constraint tree based on the 18S reference tree (i.e., if species were lacking in a dataset, they were excluded from the tree) and IQtree (Nguyen et al. 2015) with automatic model selection (-m MFP for the single genes and –m MFP+MERGE for the supermatrices). For the supermatrices, we implemented the Partitionfinder option of IQtree. The selected models are shown in Supplementary online Appendix 5. Using BaCoCa.v1.109, we determined the frequencies of nucleotides, gaps, AT, and purines or of amino acids, gaps hydrophobic, polar, positively charged, neutral, and negatively-charged, and the AG, AT, CT, and GC skew values for nucleotides. Normalized RCFV values were obtained using nRCFVReader. We also calculated the evolutionary rates, long branch (LB) scores, and tip-to-root distances using TreSpEx v1.2 using the function e and the IQtree trees (Struck 2014).
For the gene order, we conducted distance analyses using CRex (Bernt et al. 2007) and mapped the gene order evolution on the 18S reference tree using TreeRex (Bernt et al. 2008). Each gene order started with COX1 at position 1 and then the gene order was manually aligned by including not applicable (NA) for genes in the missing region of incomplete mitochondrial genomes and a gap (–) for the most likely position in complete genomes as well as by removing duplicated genes. The aligned gene orders with and without tRNAs (with NA and gaps removed again) were uploaded to CRex and analyses were run using default settings with the following exception: “remove duplicates” was disabled. The distances matrices “common interval,” “breakpoints,” and “reversal distance” (RD) were obtained. For the TreeRex analyses, the species were sorted based on their phylogenetic affiliation and the aligned gene order without tRNAs was modified in the following way to reduce missingness in the dataset. Lost genes (i.e., gaps) and lacking genes (i.e., NA) were included in accordance with the closest relatives. The TreeRex analysis was run using all three consistency methods (strong, weak, and parsimonious weak), the 18S reference tree, and modified aligned gene order file without tRNAs. For the gene orders both with and without tRNAs and without the outgroup gene orders, we also calculated rearrangement frequencies (RF) for the individual genes and rearrangement scores (RS) for the individual gene orders using qMGR (Zhang et al. 2020). To this end, the numbers of the different gene orders were counted using “Count_SequenceOrders.R” and the most common gene order was used as the ground pattern.
Correlation Analyses of Mitochondrial Properties Without Reference Tree
We analyzed the correlation of the three gene order distance matrices with and without tRNA to each other in R. We visualized the correlation using ggscatter with a regular line, confidence interval, and a correlation coefficient based on the Pearson method. We conducted a visual inspection of the data normality using Q–Q plots (quantile-quantile plots) as there were too many data points for the Shapiro–Wilk normality test. As normality could not be assumed, we also determined Spearman’s ρ rank correlation coefficient. Additionally, using only the reversal distances (RD) we compared the datasets with and without tRNAs to one another. We generated boxplots of all pairwise RDs as well as the mean of each species and plotted the mean RD values of the two datasets in comparison to each other. Moreover, we compared the mean RD values to the mean positional differences of both the nucleotide and the amino acid dataset given both the maximum likelihood (ML) and Bayesian trees. The mean positional difference reflects the difference in placement of species in the unconstrained mitochondrial phylogenetic analyses (see below) to the 18S reference tree. To obtain it, the following procedure was used. Firstly, for both the unconstrained and reference tree, the pairwise distances based on equal-spaced cladograms were calculated. This meant that each branch had a length of 1 and, hence, the pairwise distance reflected the number of branches connecting two species in a tree. Secondly, the absolute difference between the two distance matrices of the unconstrained and reference trees was calculated. Thirdly, the mean value of these differences was estimated for each species. This mean value captures not only the position of the species itself but also how other species have changed their position in relation to this species. Accordingly, the higher the value, the more the position of the species has changed in comparison to other species in the tree. For example, a value of 2 means that the paths connecting one species to the others are on average two branches longer in either the reference or the unconstrained tree, although a value of 6 means that it is six branches. The mean RD and mean positional difference are plotted against each other. Additionally, we compared the amino acid and nucleotide datasets to each other for the ML and Bayesian trees. To conduct these analyses we used the libraries “ape,” “dplyr,” “ggplot2,” “ggpubr,” “tidyr,” “TreeDist,” and “TreeTools” (Wickham 2016, 2020; Smith 2019, 2020; Kassambara 2020; Wickham et al. 2020) (see R scripts “ExploreReverseDistance.R” and “CorrelationsGeneOrder.r” on GitHub).
The matrix of compiled sequence properties contained 170 species and 706 variables. Before conducting the correlation analyses, we explored the distribution of the sequence properties across species and genes using the R script “ExplorationDataAnalyses.R” (see GitHub) with the libraries “dplyr,” “ggplot2,” “tidyr,” “ggpubr,” “ggpmisc,” and “data.table” (Wickham 2016, 2020; Dowle and Srinivasan 2020; Kassambara 2020; Wickham et al. 2020; Aphalo 2021). For the correlation analyses, we first calculated the correlation coefficients of each variable to the others. Second, we retrieved all correlated pairs, which were highly correlated (R2 > 0.5 or < −0.5), and reduced them to one variable. The resulting matrix had 80 variables. Third, to reduce the number of variables further, we conducted a hierarchical clustering analysis using the “average” method and the correlation coefficients of the remaining variables. We also generated a correlogram. All groups of clustered variables with cutoff values of 80% of the maximal height were determined and reduced to one variable. The final resulting matrix had 25 variables. Fourth, we repeated the hierarchical clustering analysis combined with a correlogram. For conducting these steps, we used the libraries “corrplot,” “corrr,” “data.table,” “dplyr,” “fastcluster,” “ggpubr,” “graphics,” “Hmisc,” and “tidyverse” (Müllner 2013; Wei and Simko 2017; Wickham et al. 2019; Dowle and Srinivasan 2020; E 2020; Kuhn et al. 2020) (see R script “CorrelationsSeqProperties_Final.R” on GitHub). Finally, we retrieved all groups of correlated or clustered variables using custom-made shell scripts (see GitHub “RetrieveCorrelatedGroups.sh” and “CompileBothGroupsTogether.sh”). Using the libraries “gplots” and “RColorBrewer” (Neuwirth 2014; Warnes et al. 2020), we generated heatmaps with breaks set at 0.001, 0.01, 0.02, 0.03, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, and 0.9 (see GitHub “GenerateHeatmap.R”).
Macroevolutionary Analyses based on Reference Tree
Using the masked, ultrametric 18S reference tree, we assessed if different mitochondrial properties and life history traits predict the gene order rearrangements considering the phylogeny. First, we tested if the gene orders themselves show phylogenetic signal. We tested both the actual gene orders with and without tRNAs as categorical characters and the mean RD values of each species as continuous characters. Then we used phylogenetic least square regression (pgls) where each of the 706 variables was tested against the mean RD values of both gene orders with and without tRNA as response variables. In all cases, Pagel’s λ was determined using a maximum likelihood approach. When this resulted in optimization problems, λ was set to 1. This was the case for 12 variables when the mean RD values of the gene order with tRNAs were the response and for 86 variables when tRNAs were excluded. All variables which were significant predictors at α = 0.001 were kept. For these significant variables, we conducted correlation analyses including hierarchical clustering using the “average” method and generating correlation networks with R2 connection thresholds at 0.7 and correlograms (see R script “pgls_all.R” on GitHub).
We scored several life history traits including those previously suggested as potentially important for genome evolution (see Introduction). Life history data were collected from the literature and further completed based on the authors’ observations (Supplementary online Appendix 33). We also used a pgls approach to test if life history traits were correlated with RD values (both with and without tRNAs). Taxa for which data for certain traits were missing or found to be polymorphic were excluded from the analyses for the corresponding trait. For conducting these steps, we used the R libraries “caper,” “corrplot,” “corrr,” “Hmisc,” and “tidyverse” (Orme et al. 2018) (see R script “pgls_with_tRNA.R” and “pgls_without_tRNA.R” on GitHub).
Phylogenetic Reconstruction
Finally, we reconstructed the phylogeny based on mitochondrial sequence data using the concatenated nucleotide and amino acid sequence data both with and without the constraint tree with Maximum Likelihood; and using the nucleotide and amino acid data without the constraint tree with Bayesian approaches. The alignments of the individual protein-coding and rRNA genes were masked with AliScore and AliCut treating gaps as ambiguous data and concatenated using FASconCAT-G v1.05. For both the nucleotide and the amino acid supermatrix, an ML tree was reconstructed using IQtree v. 1.6.12 with automatic model selection and merging of partitions (MFP+MERGE) to implement a partitioned ML analysis and 1000 rapid bootstrap replicates (Kalyaanamoorthy et al. 2017; Hoang et al. 2018). The selected model schemes are given in Supplementary online Appendix 5. For analyses applying constraints, the constraint tree (Supplementary online Appendix 2) was provided again. For the Bayesian analyses, both the nucleotide and the amino acid datasets were each run with two chains for 40 000 generations each using PhyloBayes-MPI v1.8 (Lartillot et al. 2013). The CAT-GTR model with a discrete gamma distribution with four categories was applied implementing site-heterogeneous substitution model that better captures the complexity of the substitution pattern across sites and genes. Convergence and the burnin of the chains were determined using bpcomp, tracecomp, and Tracer v1.7.1 (Rambaut et al. 2018). The burnin for both was reached after 10 000 generations, but in both cases, the two chains did not converge on the same optimum. Accordingly, we summarized only the trees of the better chain sampling every 10th generation and a burnin of 10 000 generations.
Results
Gene Order in Annelida
Complete mitochondrial genomes were determined for 140 of the species included in this study. The complete complement of protein coding (PC) and rRNA genes were available for an additional nine species. Hence, the gene order for PC and rRNA genes can be established for 149 species. These complete mitochondrial genomes had a median size of 15 356 bp with the first and third quartiles at 14 991 bp to 15 846 bp (Supplementaryonline Appendix 6a). Duplicated genes were found in 14 species and introns in four species (Supplementary online Appendix 6a). The duplicated genes were all tRNAs (i.e., trnA, trnD, trnH, trnK, trnL1, trnM, trnQ, trnS1, trnT, trnY) whilst the introns were found in COX1 or NAD6. The median number of intergenic regions was 13, with the first and third quartiles at 10 and 16. The average size of the intergenic regions per species was large, with a median of 83.5 bp and the first and third quartiles at 55 and 158 bp (Supplementary online Appendix 6a). Meanwhile, the outliers were above 400 bp and as high as around 3000 bp. However, genome size was not found to correlate with the average size of intergenic regions (Supplementary online Appendix 6b). Additionally, the number of intergenic regions, duplicated genes, and introns were also dispersed in their distribution, with no obvious trend correlated to genome size.
Considering gene orders with tRNAs, the 140 species exhibit 73 different orders. One order is clearly dominant and is found 39 times, two orders are found 7 times, two 4 times, two thrice, seven twice and 59 are unique. The dominant gene order is present in species such as the leech Erpobdella japonica, the earthworm Lumbricus terrestris, the siboglinid Osedax rubiplumus, the terebellid Pista cristata, the arenicolid Arenicola marina, and the oenonid Oenone fulgida, and hence is found in both Errantia and Sedentaria, the two major annelid groups. The average RF value per individual gene is 20.78 and the average RS value per gene order is 23.30. The genes most affected by rearrangements are trnG, trnS2, trnY, trnL2, and trnA: all show values above 40. The most affected gene orders are those of Typosyllis antoni, Sabella spallanzanii, Boccardiella hamata, Owenia fusiformis, Phyllochaetopterus sp., Chaetopterus variopedatus and Ophryotrocha labronica with values equal to or above 50 (Supplementary online Appendix 7a and b). However, neither the RF nor the RS values reveal any specific pattern in relation to the position of the genes in the ground pattern, specific gene categories, or phylogenetic position of the species.
When excluding tRNAs, differences in gene order become less apparent. In this case, of the 149 species with the full complement of PC and rRNA genes, 28 different gene orders can be found. One order is clearly dominant and found in 110 species. Three gene orders are found thrice, six twice, and 18 are unique. The average RF value per individual gene is 9.80 and the average RS value per unique gene order is 11.85. The genes most affected by rearrangements are nad1 and nad3 with values above 14, although the gene orders most affected are those of Typosyllis antoni, Sabella spallanzanii, Boccardiella hamata, Phyllochaetopterus sp., and Chaetopterus variopedatus with values above 20 (Supplementary Appendix 7c&d). However, again neither the RF nor the RS values reveal any specific pattern in relation to the ground pattern, specific gene categories, or phylogenetic position of the species.
We then considered the gene orders of all species. The three different distances (common interval, breakpoint, and reversal distance) used are highly correlated with each other independent of whether tRNAs were included or not. The R2 values were above 0.75 or below −0.75 although Spearman’s ρ values were even higher—above 0.9 or below −0.9 (Supplementary Appendix 8). Because Q–Q plots showed that normality could not be assumed (data not shown), the Spearman’s ρ values are more reliable. In the following analyses, we concentrate on reversal distances (RD) only. Not surprisingly, and in agreement with RF and RS values, the RD values for the gene orders including tRNAs are higher than those without tRNAs independent of whether all values are used or only the mean for each species (Supplementary online Appendix 9a and b). In the latter case, the distribution of values is usually more limited. For gene orders without tRNAs, the median reversal distance is 3.0 and 1.8, which is also not surprising as 74% of species show the same gene order. However, for gene orders with tRNAs, which are more variable, the values are low—10.0 and 9.8. This indicates that most different gene orders are relatively similar to each other. Additionally, the mean RD values of gene orders with and without tRNAs are highly correlated with each other (Supplementary online Appendix 9c). Hence, conclusions drawn from the reduced gene order can be used as a proxy for the complete gene order as has been done before.
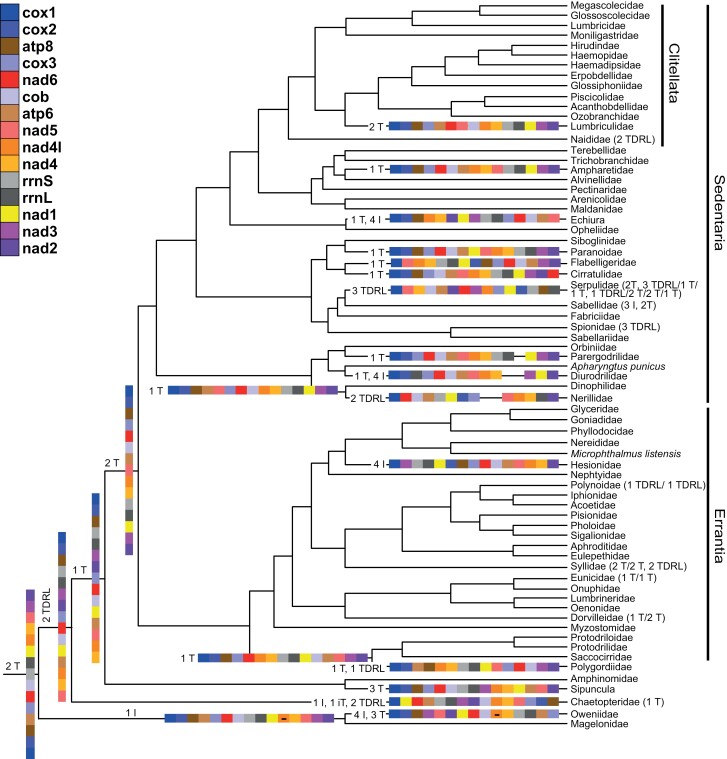
We also reconstructed the evolution of the gene orders without tRNAs given the reference tree using TreeREx. The gene order at the root of the annelid tree of life is only two transpositions (T) different from the supposed lophotrochozoan ground pattern and it is very similar to that of Magelonidae, from which it is different only by an inversion (I) of NAD4L (Fig. 1, Supplementary online Appendix 10). To obtain the pattern of Oweniidae from the pattern of Magelonididae, four inversions, and three transpositions are needed. For the clade comprising all annelid taxa except Oweniidae and Magelonidae, the gene order is very similar to the dominant gene order found in the 110 extant species. Only three transpositions are needed to change one to the other, with the gene order found in Amphinomidae as an intermediate pattern. The dominant pattern is reconstructed as the ground pattern of Pleistoannelida (Sedentaria + Errantia). In total, rearrangements along 38 branches (out of a possible 334) can explain the deviations from the dominant pattern. Hence, rearrangements only occur along around 11% of branches. Moreover, 17 of these 38 branches are within taxonomic families. Along 23 of these 38, the rearrangement did not require more than three transpositions and/or inversions. In contrast, more complex TDRLs rearrangements were required at 12 branches, five in combination with transpositions and/or inversions. Considering only the “family” level shown in Figure 1, half of the complex rearrangements occur in taxa not belonging to either Errantia or Sedentaria, although the other half are found in the two clades of interstitial families (i.e., Dinophilidae, Diurodrilidae, Nerillidae, Parergodrilidae, Polygordiidae, Protodrilidae, Protodriloidae, and Saccocirridae) and the one leading to Serpulidae. Interestingly, most interstitial families show gene orders deviating from the ground pattern, but the degree of deviation is different among them. However, by far the most divergent patterns are found within Serpulidae (Sedentaria), followed by Polynoidae, Syllidae, and Dorvilleidae (all Errantia) (Fig. 1, Supplementary Appendix 10).
Figure 1.
Reconstruction of the gene order evolution without tRNAs of the TRex analysis (Supplementary online Appendix 10) reduced to the family level and excluded the outgroups. Gene orders are given by a code. The legend of the color code is in the upper left corner. Each new gene order is indicated at its node of origin and the most likely evolutionary scenario leading to it from the gene order of its ancestral node is given (I = Inversion; T = Transposition; iT = inverse Transposition; R = Reversal; TDRL = Tandem Duplication Random Loss). If new gene orders originated within a family, this is indicated by its corresponding evolutionary scenario in brackets after the name. If more than one new gene order evolved, they are separated by a slash.
Sequence-Based Properties of Annelid Mitochondrial Genomes
We first compared different sequenced-based properties at the gene level. Evolutionary rates were measured, as the mean pairwise patristic distance of each species showed that the rates were quite similar across the nucleotide datasets, although much more variability was able to be observed across the amino acid datasets (Supplementary online Appendix 11). Overall genes and both types of data, the range of rates is similar. Interestingly, the rates in ATP and most NAD genes are clearly higher in the amino acid dataset than in the corresponding nucleotide dataset. The more canonical view—that the evolutionary rates of amino acid datasets are lower—cannot be observed. With values below 2, the COX1, COX3, and COB genes have the lowest rates at both the nucleotide and amino acid levels.
The proportion of gaps is the lowest in NAD1 and COX1 with values below 20% (Supplementary online Appendix 12a). The highest proportions are observed in ATP6 and rRNAS with values above 40%. AT content is relatively similar across the genes and ranges from 60 to 70% with the lowest values occurring in COX3 and the highest in NAD6 (Supplementary online Appendix 12b). No apparent differences can be observed between structural and protein-coding genes or the different gene families. This is different for the proportion of hydrophobic amino acids, which varies substantially between genes (online Appendix 12c). The lowest proportion occurs in the ATP6, COB, and COX genes with values generally below 55%. The lowest values can be found in COX2, although the highest values (usually above 60%) are present in NAD1 and NAD2 and the largest variation in values can be observed in ATP8. Comparing the different frequencies of amino acid and nucleotide classes against each other does not translate into a strong correlation (i.e., R2 >> 0.5, Supplementary online Appendix 13). Only frequencies of neutral amino acids strongly correlate with negative ones and weakly with positive ones (i.e., R2 of 0.84 and around 0.5), but positive and negative ones do not correlate with each other (Supplementary online Appendix 13a–c).
The AT skew was usually negative, evidencing that T occurs more often than A (Supplementary online Appendix 14a). This trend was reversed only in the structural rRNA genes, although COX2 AT is roughly equally distributed across the species. Similarly, GC is also skewed towards the pyrimidine C across all species (Supplementary online Appendix 14b). The most prominent skew is present in ATP8. Within pyrimidines, there is consistently a skew toward T and within purines towards A in agreement with the higher AT content across genes (Supplementary online Appendix 14c and d). Interestingly, of all skew values, the AG skew exhibits the highest variation across genes. Plotting AT against CG skews as well as CT against AG skews showed that both the whole genome and the protein-coding genes have generally similar patterns (Supplementary online Appendix 15). In the plot of the AT to CG skews, the values in the plot of the protein-coding genes are shifted slightly towards Ts, which is not surprising given the difference in AT skews of protein-coding and structural genes. Most species exhibit negative values in both skew values, and no species has positive values in both (Supplementary online Appendix 15b). A few species possess a negative skew in one and a positive one in the other. On the other hand, most species have a positive AG skew and a negative CT skew and only very few have a positive CT or a negative AG skew, but never a positive CT and a negative AG skew together (Supplementary online Appendix 15c and d).
Assessing the degree of base composition heterogeneity across genes reveals that the nRCFV values for both nucleotides and amino acids are relatively similar across all genes with slightly higher values in NAD2, 4, and 5 (Supplementary online Appendix 16). Finally, nRCFV values from the whole genome and all protein-coding genes datasets for nucleotides as well as the nRCFV values of nucleotides and amino acids of the protein-coding genes were very highly correlated with R2 values above 0.75 (Supplementary online Appendix 17).
General Correlations Among Sequence-Based Properties of Annelid Mitochondrial Genomes
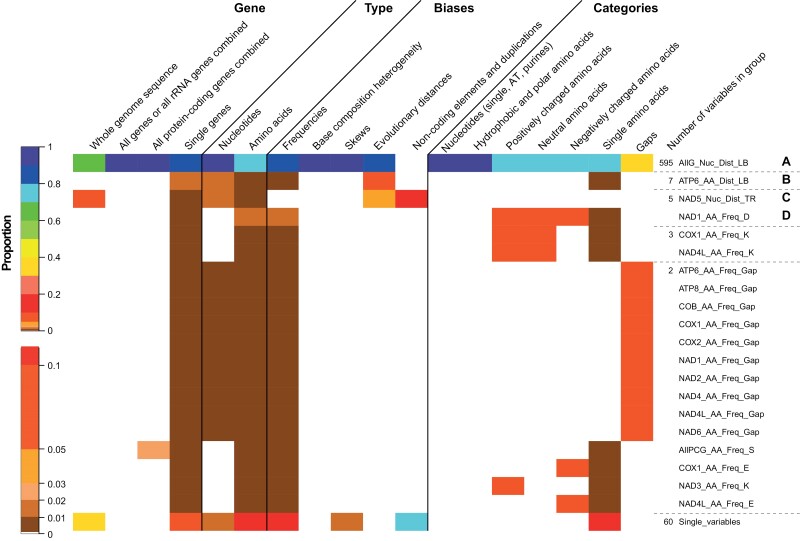
To get a better understanding of the correlation properties, we conducted all-against-all correlation analyses of the 706 variables resulting in 497 730 R2 values. The density plot of the values showed that only a minority of values have R2 above 0.5 or below −0.5 (Supplementary online Appendix 18). Next, we grouped all variables which were highly positively or negatively correlated with each other into one group. This resulted in 80 groups. Interestingly, 595 out of the 706 variables were correlated with each other, at least indirectly (group A in Fig. 2). In the three next largest groups B–D, only seven, five, and five variables were correlated with each other, respectively. On the other hand, 60 variables were not correlated with any other variable. Considering variables of different combinations of genes, more than 95% were in the largest group A (Fig. 2). In both single genes and the whole genome, more than 60% were in group A. However, within the whole genome the remaining variables were in group C (1–2%) or not correlated (30–40%), although in the single genes of the remaining ones 5–10% are not correlated or are distributed along all the other groups. Hence, most variables, which are not part of group A, are variables related to single genes. Although over 90% of the variables relating to nucleotides are in group A, only 70–80% of the amino acid-based variables are present in this group. The nucleotide variables not in group A relate to gap properties (Fig. 2). In addition, all nRCFV and most skew variables were in group A, although this was only the case for 80-90% of the frequency- and evolutionary-distance-based variables. The remaining frequency variables were distributed among the other groups or not correlated, although distance variables were in the groups B and C. The structural elements were in group B (20%) or were not correlated (80%). With respect to the different nucleotide and amino acid categories, all nucleotide categories as well as hydrophobic and polar amino acids were part of group A. A large proportion of all other amino-acid categories related variables are also part of group A (70–80%). The remaining variables of these categories were either correlated with fewer than seven other variables or, in the case of single amino acids, no variables at all (Fig. 2). Hence, the variables that are not part of group A are predominantly related to amino acid properties of single genes and predominantly those related to frequencies. Variables related to gaps are also usually not part of group A, with a low value of 30–40% (Fig. 2). The correlogram of the remaining variables after the reduction of the groups to one of the variables shows that these are not strongly correlated (Supplementary online Appendix 19a).
Figure 2.
Heatmap showing the proportion each group of correlated variables has for different properties shown on top. The color scheme for the heatmap is shown to the left and was done manually to capture the small differences in the low proportion range better. To the right, the name of one variable represents the group. Additionally, the number of variables within the group is indicated. Single variables represent all the variables, which were not correlated with another variable.
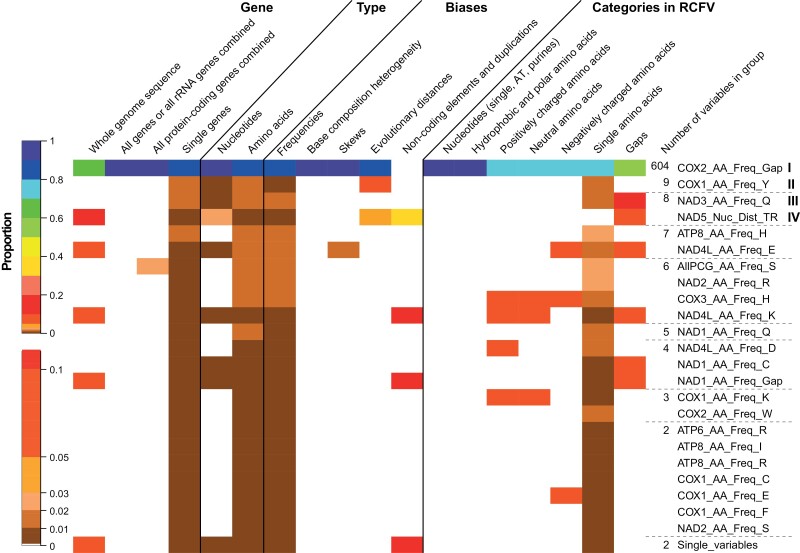
We reduced the number of variables further based on hierarchical clustering using a cutoff value of 80% of the maximal height (Supplementary online Appendix 20a). This resulted in 25 groups. The number of variables in the largest group (group I) increased only to 604, but the number of ungrouped variables reduced substantially to only two (Fig. 3). Moreover, for other groups, the number of included variables also generally increased and the three next largest groups (II-IV) encompassed nine, eight and eight variables, respectively. The patterns described above for the grouping based only on R2 values are maintained, and more gap variables are associated with group I. Comparing heatmaps across the rows reveals no discernable patterns. Several variables with different properties are usually grouped together and the groups do not comprise clusters based on specific properties. For example, often groups (rows in Fig. 3) contain nucleotide- and amino acid-based, frequency-based, and amino acid charge-based variables. Additionally, structural properties such as genome size or number of intergenic regions group with different variables relating to different aspects even though they are not part of group I. Hence, with the exception that most variables are correlated with each other at least in some way, the grouping does not reveal any specific pattern with respect to biological properties such as structural information or frequencies. Finally, the correlogram and the hierarchical cluster show that the remaining 25 variables are not correlated with each other and exhibit deep splits in the hierarchical clustering (Supplementary online Appendices 19b and 20b).
Figure 3.
Heatmap showing the proportion each group of correlated and clustered variables has for different properties shown on top. The color scheme for the heatmap is shown to the left and was done manually to capture the small differences in the low proportion range better. To the right, the name of one variable represents the group. Additionally, the number of variables within the group is indicated.
Macroveolutionary Analyses of Sequence Properties, Life-History and Ecological Traits and Gene Order
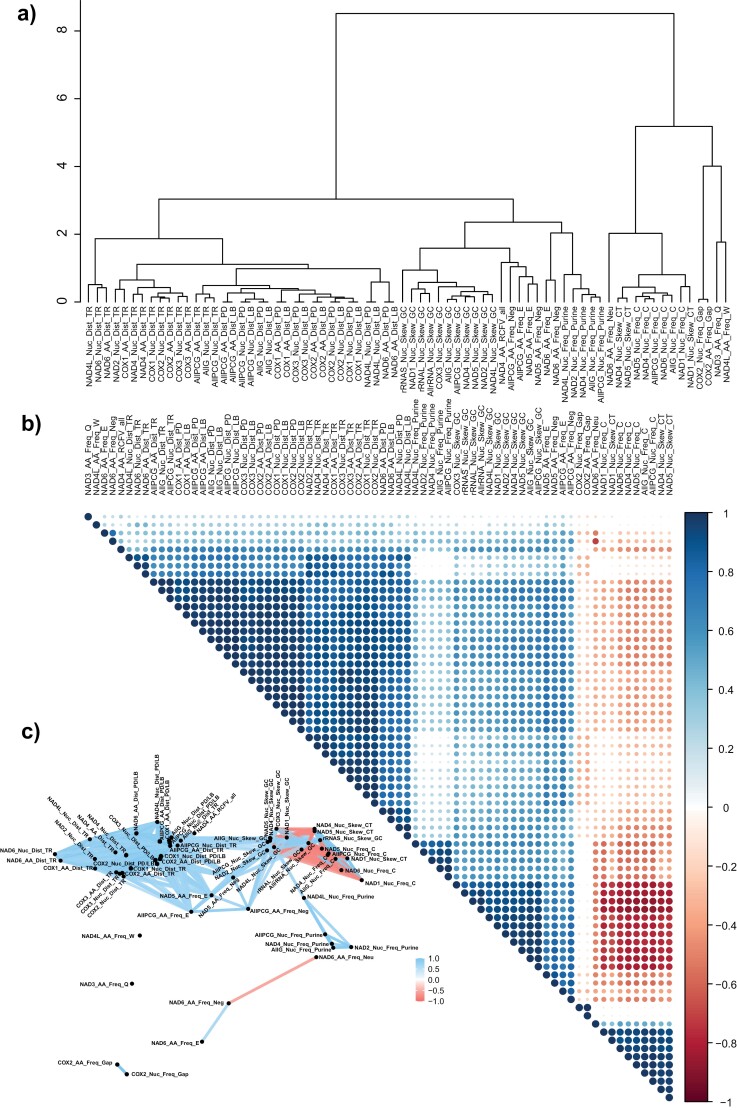
Both mean RD values and actual gene orders with and without tRNAs show strong phylogenetic signals. The λ values are very high, with values of at least 0.982. Of the 706 sequence-based variables, 72 were significantly correlated with gene order without tRNAs (p < 0.001) (Supplementary online Appendix 21). These 72 variables explain at least 5.8% and up to 23.0% of the variance. The correlation analyses showed that most of these variables are highly correlated (Fig. 4). In the clustering analysis, correlogram and correlation network, the frequencies of the amino acids tryptophan (W) and glutamine in NAD4L and NAD4, respectively, and of gaps in COX2, are clearly set apart from the other variables. In the correlation network and correlogram, the frequencies of negative and neutral amino acids and the amino acid glutamic acid (E) of NAD6 are also set apart, although within the clustering analysis, they belong to two different clusters, each containing different frequency variables. All other variables are highly correlated with each other in the correlogram and especially in the correlation network. In the clustering analysis, three major clusters are visible. One contains only variables related to evolutionary rate-related measurements (i.e., _Dist_) for different single genes and combinations of genes. The second largest cluster contains GC skew values, nRCFV of amino acids in NAD4, and different frequencies of amino acids and purines in NAD genes, all protein-coding genes, and all genes. The third cluster comprises predominantly CT skew values and frequencies of cytosine in NAD genes, all protein-coding genes, and all genes. Hence, with respect to the gene order, without tRNA, evolutionary rate is a class with very strong predictors that explains 5.9–12.0% of the variance (Supplementary online Appendix 21). However, the variables relating to composition are stronger predictors. Among the four variables NAD6_AA_Freq_Neg, NAD6_AA_Freq_Neu, NAD5_AA_Freq_E, and NAD6_AA_Freq_E, three are part of a separate group in the correlation network, and explain more of the variance (23.0%, 16.3%, 15.9%, and 12.8%) than the best variable related to evolutionary rate, COX2_AA_Dist_TR (12.0%). On the other hand, three of the four not highly correlated variables (i.e., COX2_Nuc_Freq_Gap, COX2_AA_Freq_Gap, NAD3_AA_Freq_Q) conversely explain only 6.3%, 6.7%, and 6.8%, although the fourth one, NAD4L_AA_Freq_W, explains 11.0%.
Figure 4.
(A) Hierarchical clustering of the variables, which are significant predictors for the gene order without tRNAs. (B) Correlogram of the same predictors. (C) Correlation network of the same predictors.
Considering the gene order with tRNAs, the number of significant variables increases to 105 and explains between 5.7% and 16.6% of the variance (Supplementary online Appendix 21). The number of variables relating to evolutionary rates (i.e., _Dist_) only increased from 35 to 43, but variables relating to base composition (i.e., _RCFV_, _Freq_, and _Skew_) instead increased from 37 to 62. Additionally, with the number of intergenic regions, one variable relating to structural information was also among the significant variables, explaining 7.1% of the variance. The correlation among these variables is less pronounced and only a few groups are prominent (online Appendices 22–24). The three most prominent variables comprise those related to evolutionary rate, GC, and CT skews (online Appendices 23 and 24). When gene order is considered with tRNA, the strongest predictors are related to base composition, but in this case, the base composition of ATP8 and rRNAL genes. Variables related to base composition explain between 5.7% and 13.4% of the variance. However, variables related to evolutionary rate explain more of the variance, even though they are not the strongest predictors. With values between 13.6% and 16.6%, eight variables have better adjusted R2 values than the best base-composition-related variable. Seven of the eight relate to COX genes. In general, evolutionary rate-based variables explain between 6.6% and 16.6% of the variance (Supplementary online Appendix 21). Moreover, the explanatory power is in general much lower in comparison to the gene order without tRNAs (maximum of 16.6% vs. 23.0%). In summary, base-composition measurements are the class that predicts gene order best, with and without tRNAs. In contrast, evolutionary rate variables have a higher explanatory power in the case of gene orders with tRNAs, but composition-based variables explain more without tRNAs. Structural variables such as the number of intergenic regions have relatively little prediction and explanatory power.
Comparing which individual variables are shared among both sets of significant predictors, we find that only 42 are shared among both (Supplementary online Appendix 21). Hence, 30 of the variables with gene order without tRNAs as the response are not found in the analysis of the gene order with tRNAs and vice versa 63 are not found by the other analysis. Of the shared variables, the majority, 26 out of 42, relate to measurements of evolutionary rate, although the remaining 16 reflect base composition. More specifically, the evolutionary-rate variables predominantly relate to measurements of COX genes, and the base-composition variables to GC skew values of NAD or rRNA genes. Hence, although individually base-composition variables occur more often than evolutionary-rate ones, in the shared variables the evolutionary-rate variables clearly dominate. This means that different base-composition variables predict gene order evolution depending on the inclusion of tRNAs, although prediction by evolutionary-rate variables is relatively similar between both gene orders. However, in both analyses, the strongest predictors are the shared variables relating to base-composition (Supplementary online Appendix 21). For the gene order without tRNAs, except for NAD4_Nuc_Skew_CT, all base-composition variables have a lower AIC than the evolutionary-rate variables. For the gene order with tRNAs, the difference is not as clear-cut, but eight still have a lower AIC than any evolutionary-rate variable. In both cases, the strongest predictor of the shared variables is rRNAL_Nuc_Skew_GC. However, considering the explanatory power, evolutionary-rate variables are generally stronger in both analyses. Among the ten variables with the highest adjusted R2 values only three are base-composition variables for the gene orders without tRNAs and only two with tRNAs. The highest explanatory power, with 12.0% and 16.6%, respectively, has COX1_AA_Dist_TR in both analyses (Supplementary online Appendix 21). Hence, across both responses, the GC skews of rRNA and NAD genes are generally strong predictors of gene order evolution, but taking the explanatory power into account, evolutionary rates of especially COX genes explain more variation in gene order.
Finally, we also assessed the combination of best variables from two or three categories (i.e., evolutionary rate, base composition, number of genes, or regions) found among the significant predictors in the analyses. The best variable was chosen for each category, either from the pool of shared variables or from each pool independently (Table 2). For gene order without tRNAs, the shared variables explain less variation than the best single variable, giving higher AIC values and even less explanatory power if the interaction between the shared variables is considered. However, the combination of independent variables results in lower AIC values and an increased explanatory power from 23% to almost 30%, again independent of the interaction. For gene order with tRNAs, the combination of independent variables also resulted in lower AIC values and higher explanatory power. However, in contrast, the combination of shared variables obtained higher AIC values, but higher explanatory powers than the other three models in Table 2 and also a higher explanatory power than the single individual variable with the highest explanatory power: COX1_AA_Dist_TR (16.6%). Hence, considering different factors of sequence evolution and their interaction together resulted in a slight improvement of the model’s predictive abilities and explanatory power for the evolution of both gene orders, but the two models are not necessarily the same. Moreover, the combination of these variables is different from the pattern observed among shared individual characters above.
Table 2.
Combination of best variables from the two or three detected categories predicting the gene order with and without tRNAs
| Predictors | Best of | Interaction | AICa | Adj. R2b |
|---|---|---|---|---|
| Without tRNAs | ||||
| rRNAL_Nuc_Skew_GC + NAD4L_Nuc_Dist_TR | Shared | No | 541.186 | 0.128 |
| rRNAL_Nuc_Skew_GC × NAD4L_Nuc_Dist_TR | Shared | Yes | 542.545 | 0.126 |
| NAD6_AA_Freq_Neg + NAD6_AA_Dist_TR | Independent | No | 515.616 | 0.298 |
| NAD6_AA_Freq_Neg × NAD6_AA_Dist_TR | Independent | Yes | 515.909 | 0.299 |
| NAD6_AA_Freq_Negc | 529.005 | 0.230 | ||
| With tRNAs | ||||
| rRNAL_Nuc_Skew_GC + COX2_AA_Dist_TR | Shared | No | 779.362 | 0.211 |
| rRNAL_Nuc_Skew_GC × COX2_AA_Dist_TR | Shared | Yes | 778.899 | 0.218 |
| ATP8_AA_Freq_P + COB_Nuc_Dist_TR + Whole_Nuc_Num_IG | Independent | No | 729.679 | 0.118 |
| ATP8_AA_Freq_P × COB_Nuc_Dist_TR × Whole_Nuc_Num_IG | Independent | Yes | 722.795 | 0.178 |
| ATP8_AA_Freq_Pc | 738.588 | 0.079 | ||
Note: Best variables were chosen among the shared ones between both gene orders or independently with in each gene order based on the Akaike information criterion (AIC) value.
aAkaike information criterion (AIC) value of the different combinations of predictors.
bAdjusted R2 (adj. R2) of the different combinations of predictors.
cFor comparative reasons, the AIC and adjusted R2 of the best in dependent variable are also provided.
Models using life history traits as predictors of mean RD values (both with and without tRNAs) had no significance at P < 0.001. Considering the gene order without tRNAs, the trait “Type of sexual reproduction” had the best P value of 0.0613 as a predictor. Not surprisingly, in comparison to the sequence-based results, the explanatory power is relatively low at 4.0%. All other traits had lower explanatory powers of 1.2% or less and higher P values of at least 0.07. Considering the gene order with tRNAs, the best P value was 0.0018 for “High altitude,” and the explanatory power was slightly higher at 5.1%. The trait “Habitat” had an even slightly higher explanatory power of 5.5%, but only the second-best P value of 0.0776. All other traits and this gene order have P values higher than 0.16 and the explanatory power is much lower (below 1%). In summary, none of the life history traits measured significantly predict mitochondrial genome evolution.
Mitochondrial Phylogenetic Tree and Gene Order
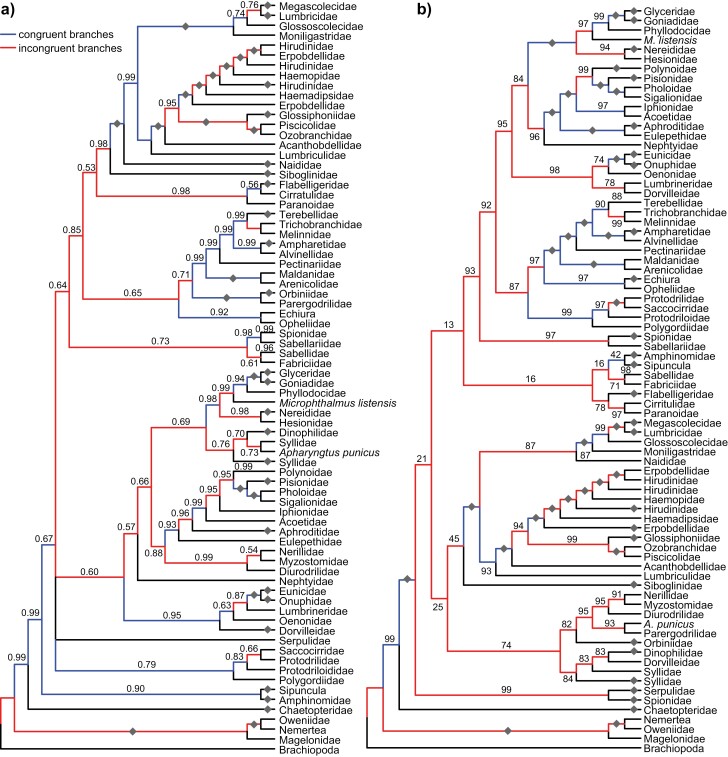
The phylogenies obtained using the mitochondrial data are very incongruent with the nuclear 18S reference tree, independent of Bayesian or ML analyses (Fig. 5 and Supplementary online Appendices 25 and 26). For example, none of the analyses recovered the monophyly of Annelida. The nemertean species are placed within Annelida when the tree is rooted with the brachiopod species. This is especially prominent in the ML analysis of the nucleotide data (Supplementary online Appendix 26b). Most branches are indicated as incongruent rather than congruent. Despite the high degree of incongruence independent of the type of data and analysis, posterior probabilities and bootstrap values are high at several nodes including incongruent nodes with values above or equal to 0.95 or 95 (Fig. 5 and Supplementary online Appendices 27–30).
Figure 5.
Cladograms of the (A) PB and (B) ML tree, respectively, obtained with mitochondrial amino acid data reduced to the family level. Congruence and incongruence of branches is indicated (see legend in the upper left corner). Posterior probabilities and bootstrap values are given above the branches. Gray diamonds indicate a posterior probability of 1.00 or a bootstrap value of 100. Values at terminal branches indicate collapsed branch.
The Robinson-Fould (RFo) distance to the reference tree is lowest for the Bayesian analysis using amino acid data with a value of 154. In contrast to the other analyses, it shows more similarity to the reference tree in the basal branching patterns (Fig. 5a and Supplementary online Appendix 25a). The next best tree is the Bayesian analysis of nucleotide data with a RFo value of 162. This probably stems from a large polytomy in the tree (Supplementary online Appendix 25b). Of the two ML analyses, the analysis based on amino acid data has a lower value (174) compared with the nucleotide dataset (196). In all analyses of the mitochondrial datasets, long-branched taxa such as Nerillidae, Diurodrilidae, Myzostomidae, Dorvilleidae, and Dinophilidae were grouped together, and this effect is more apparent in the ML analyses than in the Bayesian ones (Supplementary online Appendices 27–30). The longest branch by far leads to Serpulidae. Moreover, the families Erpobdellidae, Hirudinidae, and Syllidae are not recovered as monophyletic.
The higher incongruence of the nucleotide dataset in comparison to the AA dataset is also seen in the mean positional differences (Supplementary online Appendix 31a). The median for the nucleotide dataset is at least one mean positional difference higher, independent of the analysis used. This means that the path connecting one species to any other species is on average almost one branch longer in either the reference or the unconstrained tree than in the AA dataset. Interestingly, although the Bayesian analysis of the amino acid data also has the lowest mean positional difference—showing a similar pattern to the RFo values—the second-best dataset is the ML of the amino acid and the worst is the Bayesian analysis of the nucleotide data, which was the second best according to RFo. Hence, polytomies seem to have a lower impact in this case. However, there is a slight correlation between both datasets and analyses (R2 = 0.50–0.65) indicating that some incongruence to the reference tree is shared (Supplementary online Appendix 31b–e). Finally, we also checked if the deviated gene orders correlated with the degree of incongruence in species placements with respect to the reference tree (Supplementary online Appendix 32). For both the AA and the nucleotide dataset independent of the method of tree reconstruction, the mean positional differences do not correlate with the mean RD values with or without tRNAs. In all eight cases, the R2 values are low or negative.
Discussion
Gene Order Evolution of Annelida
In contrast to recent publications (Tempestini et al. 2020; Sun et al. 2021) proposing blows to the conservative gene order, our study showed that the gene order is overall highly conserved across Annelida as suggested previously. That holds even when considering tRNAs, which are considered to be more variable. Furthermore, the gene order is identical in many species (28%) across Pleistoannelida (i.e., Sedentaria and Errantia). In addition, it does not matter if one only looks at gene orders at the family or genus level. About half of the new gene orders are found within taxonomic families and there is no obvious difference in the type of rearrangement processes causing these new orders. Hence, the general result is not due to a biased sampling at the family level. For the gene order without tRNAs, the pattern found in most annelids is the ground pattern of Pleistoannelida as previously suggested by Weigert et al. (2016). The ground pattern of Annelida is different from this, which is very similar to the supposed ground pattern of Lophotrochozoa as well as to the gene order still found today in Magelonidae. At a first glance, the pattern in groups not part of Pleistoannelida appears to be more variable than within Pleistoannelida. However, the corresponding patterns of Amphinomidae and Sipuncula seem to be conserved within the taxon, although two patterns occur within Chaetopteridae, which are different by one transposition. Magelonidae and Oweniidae are thus far only represented by a single species and the question of whether these patterns are conserved or variable within the family remains to be addressed. Given that these groups span the same or deeper evolutionary times than Pleistoannelida, but comprise substantially fewer species, the variability of the pattern within these groups is not more variable than in Pleistoannelida. It appears that the gene order was more flexible early in annelid evolution and stabilized afterward in each different lineage. In Pleistoannelida, which comprises the vast majority of extant annelid biodiversity, the gene order is generally conserved, but in individual groups, families, and genera it appears to be more variable. Serpulidae (see also Sun et al. 2021), Dorvilleidae (see also Tempestini et al. 2020), and interstitial families seem especially variable.
Considering only individual factors, the best predictors for gene order evolution with and without tRNAs are variables related to base composition in less conservative genes such as NAD, ATP, and rRNA genes. The number of intergenic regions is also a significant predictor for gene order with tRNAs but has neither a very strong predictive ability nor a high explanatory power in comparison to variables based on base composition or evolutionary rate. Although base-composition variables also have the highest explanatory power for gene order evolution without tRNAs, it is variables linked to evolutionary rate that have the highest evolutionary power for gene order evolution with tRNAs. Interestingly, considering only the variables shared by both gene orders, GC skew variables of NAD or rRNA genes are the strongest predictors, although measurements of evolutionary rates in COX genes have the highest explanatory power. The best combinations of the different categories together explain about 29% and 22% of the variability observed in both gene orders along a phylogeny, respectively. Hence, our analyses support hypotheses proposing biases in nucleotide or amino acid frequencies in mitochondrial genomes, especially GC skew, increased substitution rates and to a much lower degree the increased presence of noncoding regions affecting gene order evolution (Shao et al. 2003; Hassanin et al. 2005; Podsiadlowski and Braband 2006; Xu et al. 2006; Min and Hickey 2007; Bernt et al. 2013; Luo et al. 2015; Zhang et al. 2021; Lei et al. 2022). The combination of the first two factors seems to facilitate gene order evolution in Annelida. Bernt et al. (2013) suggested that relaxed repair mechanisms could be the reason for this. Relaxed repair mechanisms would result in increased rates and an increase in the likelihood of the fixation of a mutation. This can also lead to stronger biases in base composition as, for example, transversions are less likely to be repaired. Moreover, due to the appearance of new stop codons earlier in the gene, intergenic noncoding regions can appear. High mutational stress, together with a lower importance of mitochondrial efficiency, can also explain the same patterns as relaxed repair mechanisms (Bernt et al. 2013). However, how both relaxed repair mechanisms and high mutational stress with low mitochondrial efficiency can cause high gene order rearrangement is not clear. Another suggestion has been that structural changes such as the switching of the ORI cause a switch of the GC skew and accordingly increase evolutionary rates (Jakovlić et al. 2021; Lei et al. 2022). Considering just the shared variables between both gene orders, GC skews are the strongest predictors of gene order evolution and the evolutionary rate has the highest explanatory power. However, considering the variables independently as well as their combinations, other base composition variables are stronger predictors. Moreover, the prediction of ORI motifs in annelids was not possible with certainty and as such nor were ORI switches (Lei et al. 2022). Hence, although structural changes could drive changes in sequence evolution, evidence for this is still weak considering the mixed signal in our results. Finally, it may well be that there is no causal relationship between sequence-based and structural changes in mitochondrial genomes and that both are affected similarly by the same cause, such as a lower importance of mitochondrial efficiency. For example, both could indicate that the taxon went through a condition where it could accept less effective oxygen consumption and ATP production, allowing more mutations to accumulate with time at both the sequence and the structural level. This might have allowed the passage through an adaptive valley toward a better-adapted solution for this taxon, stabilizing on a new gene order.
Different life-history traits have been suggested, which might allow for a lower mitochondrial efficiency than is usually needed. These traits comprise parasitism, shorter generation time or life styles such as inhabiting calcareous tubes, the deep sea, or anoxic environments (Kajander et al. 2000; Dowton and Campbell 2001; Bernt et al. 2013; Zhang et al. 2018; Tempestini et al. 2020; Sun et al. 2021). However, none of the herein tested life-history traits: including parasitism, generation time, and different specific habitats (interstitium, deep sea, methane seeps, hot vents, extreme environments in general, or biomineralization) were significant at the 0.001-level. Additionally, the trait “High altitude,” the only one significant at the 0.05-level for one of the gene orders, could explain only 5.1% of the variability in the respective gene order. Moreover, closer examination of the trait revealed that it seems to explain the evolution in a single clitellate species (i.e., Haemadipsa crenata), but the majority of the observed variation is not explained. Hence, the biological causes for gene order rearrangement in Annelida are not revealed yet. Living in extreme environments seems insufficient to cause gene order rearrangements, neither when extreme environments were considered individually nor together using both multiple and absence/presence coding. The same is true for short generation times and biomineralization. However, the variation in gene order could be influenced by other biological and ecological factors not suggested so far and hence not considered herein.
Additionally, our results about the evolution of the mitochondrial gene order in Annelida do not support the hypothesis that a rearrangement of the gene order generally triggers a phase of several subsequent rearrangement events due to a destabilized gene order. Analyses of mitochondrial genome evolution in chromodorean nematodes have recently suggested that mitochondrial genome evolution is highly discontinuous. More specifically, a “long period of stasis in gene order and content (is) punctuated by a rearrangement event, (and) such a destabilized mitogenome is much more likely to undergo subsequent rearrangement events, resulting in an exponentially accelerated evolutionary rate of mitogenomic rearrangements” (Zou et al. 2017). At first glance, the pattern found within Pleistoannelida would at least partially fit this hypothesis. From this conserved ground pattern, changes in the ground pattern have occurred along several branches. Moreover, the rearranged gene orders seem to cluster in some regions of the tree. Specifically, interstitial and cirratuliform families (e.g., Dinophilidae, Polygordiidae, Cirratulidae) and Serpulidae show more variability in their gene orders. Recent findings that several species within genera like Hydroides (Serpulidae) and Ophryotrocha (Dorvilleidae) also exhibit different gene orders (Tempestini et al. 2020; Sun et al. 2021) are in line with this hypothesis. However, closer examination at the level of protein-coding and rRNA genes shows that often the new gene order is the same in a clade after the rearrangement. For example, in Protodriliformia the gene order is the same for Protodrilidae, Protodriloidae, and Saccocirridae. The same is true for Ampharetidae, Dinophilidae, Cirratulidae, within Polynoidae, and Marphysa. Additionally, these new gene orders usually evolved independently from the ground pattern and not from another rearranged gene order. On the other hand, the taxon sampling is still limited and many families with deviating gene orders are represented by only one species. Increased taxon sampling might provide more cases like those observed in Hydroides and Ophryotrocha (Tempestini et al. 2020; Sun et al. 2021). However, it is not a rule in Annelida that one rearrangement event necessarily triggers destabilization and several subsequent rearrangements.
Our results also did not support the suggestion that tRNAs could act as facilitators of rearrangements (Saccone et al. 1999; Kajander et al. 2000; Dowton and Campbell 2001; Luo et al. 2015). Although reversal distances (RD) with and without tRNAs are strongly correlated and both RD and RF values with tRNAs are higher than without, a detailed comparison of the RF results with and without tRNAs shows no indication of a facilitating function. Considering the gene order with tRNAs, the six highest values are tRNAs and prior to the first protein-coding gene. Moreover, although the taxa with the highest RS values do not change much according to gene order with and without tRNAs, this is not the case for the protein-coding and rRNA genes. Except for NAD1, the genes with the highest RF values are often among the ones lowest in the other order.
Properties of Annelid Mitochondrial Genomes
Investigating the sequence-based properties of mitochondrial genomes in Annelida also revealed that some traditional views on gene evolution are not entirely applicable to the clade. First, it is generally stated that molecular evolution is faster on the nucleotide than on the amino acid level, as there are only four possible states and more possible synonymous substitutions. However, our data show that this is not the case and, in several genes, the evolutionary rate at the amino acid level is higher than at the nucleotide level. Given that, each substitution at the amino acid must also have at least one substitution at the nucleotide level, although the opposite is not necessary, this result is counterintuitive. One should have in mind that in this study the system captures the molecular evolution of several hundreds of millions of years (e.g., Parry et al. 2014; Chen et al. 2020). At the nucleotide level, there are only four possible character states and hence the evolutionary rate will reach saturation much earlier than amino acid data with 20 possible states (Philippe et al. 2011). Accordingly, given the 20 possible states, amino acid data can reach higher overall rates at these deep time scales than nucleotide data. This can be seen by the fact that the distribution around the median rate is much narrower in the nucleotide data than it is in the amino acid. Therefore, this is another reason to favor amino acid data over nucleotide data in phylogenetic reconstructions in deep time (e.g., Ren et al. 2005; Philippe et al. 2011).
A second canonical view often states that structural genes like rRNAs have more and larger insertion/deletions (indels) than protein-coding genes as the latter require indels dividable by three to avoid frame-shift mutations. This is generally supported by our data, but the median values of several protein-coding genes (i.e., ATP6 and 8, COX2, NAD2, 3, 5, and 6), with values above 30%, are also very high. Other protein-coding genes like COB and COX1 have substantially lower values, below 20%. Hence, although structural genes can sustain high levels of insertions or deletions, so can several protein-coding mitochondrial genes. Interestingly, these protein-coding genes are not necessarily among the fastest-evolving genes at the amino acid level. The reason why some genes sustain more indels than others is an interesting question. Possible reasons could be that indels in these proteins 1) affect the core protein structures less, as they possess more domains with lesser structural importance and 2) have less deleterious effects on elementary biological processes such as catalytic activity, ligase activity, electron transport, or catabolic processes (de la Chaux et al. 2007; Lin et al. 2017).
Other aspects, such as AT and GC skews, AT richness, and the few differences between the whole genome and the protein-coding genes, have been reported before for mitochondrial genomes in both Metazoa and Annelida (e.g., Bernt et al. 2013; Sun et al. 2021). More interestingly, our correlation studies showed that the correlation network between different properties like base composition heterogeneity, skews, evolutionary rate, and structural components is complicated and intricate. Although few correlations were strong, with R2 values above 0.5 or below −0.5, groupings of correlated variables based on specific groups (e.g., specific skew values) occurred only when considering significant predictors. In contrast, the vast majority (84%) of variables were placed in one group. This means that although most are not strongly correlated directly with each other, they are connected indirectly to one another. Hence, it will not be straightforward to predict the change in one variable due to the change in another.
Mitochondrial Genomes and the Phylogeny of Annelida
The phylogenetic least square regression (pgls) analyses showed that both the RD distances and the actual gene orders have very strong phylogenetic signals. However, given that the most dominant gene order is shared among most taxa and that these are not closely related to each other, the actual resolution power of the gene order in phylogenetic reconstructions is low as this would result in a large, basal polytomy. On the other hand, identical gene orders provide additional evidence for the monophyly of groups comprising all taxa with the same gene order. Such groups might also contain taxa with other derived gene orders, but not the typical annelid order. In our analyses, we did not observe reversals in gene orders and mitochondrial gene order in Annelida can, thereby, be considered a Dollo character (Le Quesne 1974). It evolves once and never returns to its previous stage. This property of the annelid mitochondrial gene orders has been mentioned before (e.g., Bleidorn et al. 2007; Golombek et al. 2013).
Considering the phylogenetic reconstructions based on mitochondrial sequence information, we observed strong incongruence to the constrained phylogeny based on phylogenomic data. Strong incongruence between nuclear and mitochondrial data has been observed and has been related to the lower resolution power and faster evolutionary rate of mitochondrial genomes (e.g., Zhong et al. 2011; Bernt et al. 2013). In our analyses, the incongruence was independent of amino acid or nucleotide data used and the method applied. The incongruence is only slightly stronger for the nucleotide data in agreement with a potentially higher degree of saturation in this kind of data. Interestingly, the incongruence observed in the amino acid and nucleotide data as well as between the different methods is weakly correlated. Hence, both types of data and analysis share some incongruencies, but also exhibit substantial differences in some parts of the tree. For example, independent of Bayesian or ML approaches, the amino acid data fails to recover monophyly of Syllidae, although the nuclear data does. Finally, the observed incongruence is not correlated with the degree of gene order changes in a taxon as measured by the RD distance. Hence, even though increased evolutionary rate and base composition heterogeneity are good predictors of gene order rearrangement, this does not translate necessarily into problems concerning the phylogenetic reconstruction of a taxon. This implies that, even if a taxon has a highly derived mitochondrial gene order, its phylogenetic placement may not be incongruent in comparison to phylogenomic datasets. Hence, mitochondrial gene order in Annelida cannot be used to assess if a taxon will be problematic to place in the annelid phylogeny.
Supplementary Material
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.8w9ghx3pm.
Acknowledgment
We thank the Norwegian Sequencing Center and Birgitte Lisbeth Graae Thorbek (DNA lab, NHM, UiO) for their assistance in sequencing using Illumina Hi-Seq 4000. We appreciate the comments by the editors and reviewers on earlier drafts and James Fleming for language editing and providing nRCFVReader before publication. This is NHM Evolutionary Genomics lab contribution nr 31.
Contributor Information
Torsten H Struck, Natural History Museum, University of Oslo, P.O. Box 1172, Blindern, 0318 Oslo, Norway; Centre of Molecular Biodiversity Research, Zoological Research Museum Alexander Koenig Bonn 53113, Germany; FB05 Biology/Chemistry; University of Osnabrück, Osnabrück 49069, Germany.
Anja Golombek, Centre of Molecular Biodiversity Research, Zoological Research Museum Alexander Koenig Bonn 53113, Germany; FB05 Biology/Chemistry; University of Osnabrück, Osnabrück 49069, Germany.
Christoph Hoesel, FB05 Biology/Chemistry; University of Osnabrück, Osnabrück 49069, Germany.
Dimitar Dimitrov, Department of Natural History, University Museum of Bergen, University of Bergen, P.O. Box 7800, 5020 Bergen, Norway.
Asmaa Haris Elgetany, Natural History Museum, University of Oslo, P.O. Box 1172, Blindern, 0318 Oslo, Norway; Zoology Department, Faculty of Science, Damietta University, New Damietta, Central zone, 34517, Egypt.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (grants DFG-STR 683/5-2 and DFG-STR 683/8-1 to T.H.S.), by the Research Council Norway (project number 300587 to T.H.S.) as well as by the Egyptian Government to A.H.E. for a research stay at the NHM of UiO. T.H.S. and D.D. received additional support by the Norwegian Metacenter for Computational Science (NOTUR; project numbers NN9408K, NS9408K, and NN9601K).
Data Availability Statement
Alignments and the associated trees of the reference and supermatrix datasets are available at TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S30262).
The sequence data are deposited in NCBI under the accession numbers OQ702995-OQ703026 and OQ729891-OQ729923 (see also Supplementary online Appendix 1).
The table of the life history traits with the coding scheme and sources provided are available in Supplementary online Appendix 33 (http://dx.doi.org/10.5061/dryad.8w9ghx3pm).
Script used in the analyses is also available from the GitHub Repository: https://github.com/torstenstruck/AnnelidaMitoGenomes.
References
- Aguado M.T., Richter S., Sontowski R., Golombek A., Struck T.H., Bleidorn C.. 2016. Syllidae mitochondrial gene order is unusually variable for Annelida. Gene 594:89–96. [DOI] [PubMed] [Google Scholar]
- Al Arab M., Höner zu Siederdissen C., Tout K., Sahyoun A.H., Stadler P.F., Bernt M.. 2017. Accurate annotation of protein-coding genes in mitochondrial genomes. Mol. Phylogenet. Evol. 106:209–216. [DOI] [PubMed] [Google Scholar]
- Alves P.R., Halanych K.M., Santos C.S.G.. 2020. The phylogeny of Nereididae (Annelida) based on mitochondrial genomes. Zool. Scr. 49:366–378. [Google Scholar]
- Anderson F.E., Williams B.W., Horn K.M., Erséus C., Halanych K.M., Santos S.R., James S.W.. 2017. Phylogenomic analyses of Crassiclitellata support major Northern and Southern Hemisphere clades and a Pangaean origin for earthworms. BMC Evol. Biol. 17:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphalo P.J. 2021. ggpmisc: Miscellaneous extensions to “ggplot2.” R package version 0.4.5. https://CRAN.R-project.org/package=ggpmisc.
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A.. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M., Bleidorn C., Braband A., Dambach J., Donath A., Fritzsch G., Golombek A., Hadrys H., Jühling F., Meusemann K., Middendorf M., Misof B., Perseke M., Podsiadlowski L., von Reumont B., Schierwater B., Schlegel M., Schrödl M., Simon S., Stadler P.F., Stöger I., Struck T.H.. 2013. A comprehensive analysis of bilaterian mitochondrial genomes and phylogeny. Mol. Phylogenet. Evol. 69:352–364. [DOI] [PubMed] [Google Scholar]
- Bernt M., Merkle D., Middendorf M.. 2008. An algorithm for inferring mitogenome rearrangements in a phylogenetic tree. In: Nelson C.E., Vialette S. editors. Comparative genomics. Berlin, Heidelberg: Springer Berlin Heidelberg. p. 143–157. [Google Scholar]
- Bernt M., Merkle D., Ramsch K., Fritzsch G., Perseke M., Bernhard D., Schlegel M., Stadler P., Middendorf M.. 2007. CREx: inferring genomic rearrangements based on common intervals. Bioinformatics 23:2957–2958. [DOI] [PubMed] [Google Scholar]
- Bleidorn C., Eeckhaut I., Podsiadlowski L., Schult N., McHugh D., Halanych K.M., Milinkovitch M.C., Tiedemann R.. 2007. Mitochondrial genome and nuclear sequence data support Myzostomida as part of the annelid radiation. Mol. Biol. Evol. 24:1690–1701. [DOI] [PubMed] [Google Scholar]
- Boore J.L. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore J.L. 2006. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol. Evol. 21:439–446. [DOI] [PubMed] [Google Scholar]
- Boore J.L., Brown W.M.. 2000. Mitochondrial genomes of Galathealinum, Helobdella and Platynereis: sequence and gene arrangements comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol. Biol. Evol. 17:87–106. [DOI] [PubMed] [Google Scholar]
- Boore J.L., Medina M., Rosenberg L.A.. 2004. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod graptacme eborea and the bivalve Mytilus edulis. Mol. Biol. Evol. 21:1492–1503. [DOI] [PubMed] [Google Scholar]
- Cameron S.L. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu. Rev. Entomol. 59:95–117. [DOI] [PubMed] [Google Scholar]
- Chen H., Parry L.A., Vinther J., Zhai D., Hou X., Ma X.. 2020. A Cambrian crown annelid reconciles phylogenomics and the fossil record. Nature 583:249–252. [DOI] [PubMed] [Google Scholar]
- Chen P., Shen X., Cai Y., Ji N., Li Y., Ge T., Liu S.. 2019. The complete mitochondrial genome of Glycera chirori Izuka (Annelida: Polychaeta): an evidence of conservativeness between gene arrangement and phylogenesis in Glycera. Mitochondrial DNA Part B 4:3746–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Chaux N., Messer P.W., Arndt P.F.. 2007. DNA indels in coding regions reveal selective constraints on protein evolution in the human lineage. BMC Evol. Biol. 7:191–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donath A., Jühling F., Al-Arab M., Bernhart S.H., Reinhardt F., Stadler P.F., Middendorf M., Bernt M.. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47:10543–10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowle M., Srinivasan A.. 2020. data.table: extension of `data.frame`. R package version 1.13.4.
- Dowton M., Campbell N.J.H.. 2001. Intramitochondrial recombination – is it why some mitochondrial genes sleep around? Trends Ecol. Evol. 16:269–271. [DOI] [PubMed] [Google Scholar]
- E H.J.F. 2020. Hmisc: harrell miscellaneous. R package version 4.4-2.
- Erséus C., Williams B.W., Horn K.M., Halanych K.M., Santos S.R., James S.W., Creuzé des Châtelliers M., Anderson F.E.. 2020. Phylogenomic analyses reveal a Palaeozoic radiation and support a freshwater origin for clitellate annelids. Zool. Scr. 49:614–640. [Google Scholar]
- Fleming J.F., Struck T.H.. 2023. nRCFV: a new, dataset-size-independent metric to quantify compositional heterogeneity in nucleotide and amino acid datasets. BMC Bioinf. 24:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissi C., Pesole G., Mastrototaro F., Iannelli F., Guida V., Griggio F.. 2010. Hypervariability of Ascidian mitochondrial gene order: exposing the myth of deuterostome organelle genome stability. Mol. Biol. Evol. 27:211–215. [DOI] [PubMed] [Google Scholar]
- Golombek A., Tobergte S., Nesnidal M.P., Purschke G., Struck T.H.. 2013. Mitochondrial genomes to the rescue – Diurodrilidae in the myzostomid trap. Mol. Phylogenet. Evol. 68:312–326. [DOI] [PubMed] [Google Scholar]
- Hassanin A., LéGer N., Deutsch J.. 2005. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst. Biol. 54:277–298. [DOI] [PubMed] [Google Scholar]
- Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovlić I., Zou H., Zhao X.-M., Zhang J., Wang G.-T., Zhang D.. 2021. Evolutionary history of inversions in directional mutational pressures in crustacean mitochondrial genomes: implications for evolutionary studies. Mol. Phylogenet. Evol. 164:107288. [DOI] [PubMed] [Google Scholar]
- Jennings R.M., Halanych K.M.. 2005. Mitochondial genomes of Clymenella torquata (Maldanidae) and Rifta pachyptila (Siboglinidae): evidence for conserved gene order in Annelida. Mol. Biol. Evol. 22:210–222. [DOI] [PubMed] [Google Scholar]
- Kajander O.A., Rovio A.T., Majamaa K., Poulton J., Spelbrink J.N., Holt I.J., Karhunen P.J., Jacobs H.T.. 2000. Human mtDNA sublimons resemble rearranged mitochondrial genoms found in pathological states. Hum. Mol. Genet. 9:2821–2835. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A. 2020. ggpubr: “ggplot2” based publication ready plots. R package version 0.4.0.
- Katoh K., Kuma K.-i, Toh H., Miyata T.. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M., Jackson S., Cimentada J.. 2020. corrr: correlations in R. R package version 0.4.3.
- Kück P., Longo G.. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 11:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück P., Meusemann K., Dambach J., Thormann B., von Reumont B.M., Wägele J.W., Misof B.. 2010. Parametric and non-parametric masking of randomness in sequence alignments can be improved and leads to better resolved trees. Front. Zool. 7:10. doi: 10.1186/s12983-014-0081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück P., Struck T.H.. 2014. BaCoCa—A heuristic software tool for the parallel assessment of sequence biases in hundreds of gene and taxon partitions. Mol. Phylogenet. Evol. 70:94–98. [DOI] [PubMed] [Google Scholar]
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N., Rodrigue N., Stubbs D., Richer J.. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62:611–615. [DOI] [PubMed] [Google Scholar]
- Le Quesne W.J. 1974. The uniquely evolved character concept and its cladistic application. Syst. Zool. 23:513–517. [Google Scholar]
- Lei H.-P., Jakovlić I., Zou H., Zhang D.. 2022. Exploring the chaotic relationships of Annelida produced by mitogenomic data: skew inversions and their effect on evolutionary studies. Research Square. PREPRINT, doi: 10.21203/rs.3.rs-1828162/v1, 13.10.2022, preprint: not peer reviewed. [DOI] [Google Scholar]
- Lin M., Whitmire S., Chen J., Farrel A., Shi X., Guo J.-t.. 2017. Effects of short indels on protein structure and function in human genomes. Sci. Rep. 7:9313. doi: 10.1038/s41598-017-09287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y.-J., Satoh N., Endo K.. 2015. Mitochondrial gene order variation in the brachiopod Lingula anatina and its implications for mitochondrial evolution in lophotrochozoans. Mar. Geonomics 24:31–40. [DOI] [PubMed] [Google Scholar]
- Martijn J., Vosseberg J., Guy L., Offre P., Ettema T.J.G.. 2018. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557:101–105. [DOI] [PubMed] [Google Scholar]
- Min X.J., Hickey D.A.. 2007. DNA asymmetric strand bias affects the amino acid composition of mitochondrial proteins. DNA Res. 14:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwinyi A., Meyer A., Bleidorn C., Lieb B., Bartolomaeus T., Podsiadlowski L.. 2009. Mitochondrial genome sequence and gene order of Sipunculus nudus give additional support for an inclusion of Sipuncula into Annelida. BMC Genomics 10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müllner D. 2013. fastcluster: fast hierarchical, agglomerative clustering routines for R and python. J Stat Softw. 53:1–18. [Google Scholar]
- Neuwirth E. 2014. RColorBrewer: ColorBrewer palettes. R package version 1.1-2.
- Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme D., Freckleton R., Thomas G., Petzoldt T., Fritz S., Isaac N., Pearse W.. 2018. caper: comparative analyses of phylogenetics and evolution in R. R package version 1.0.1.
- Paradis E., Schliep K.. 2018. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35:526–528. [DOI] [PubMed] [Google Scholar]
- Parry L., Tanner A., Vinther J.. 2014. Frontiers in Palaeontology—the origin of annelids. Palaeontolgy 57:1–13. [Google Scholar]
- Philippe H., Brinkmann H., Lavrov D.V., Littlewood D.T.J., Manuel M., Wörheide G., Baurain D.. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9:e10006023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.J., Dornburg A., Zapfe K.L., Anderson F.E., James S.W., Erséus C., Moriarty Lemmon E., Lemmon A.R., Williams B.W.. 2019. Phylogenomic analysis of a putative missing link sparks reinterpretation of leech evolution. Genome Biol. Evol 11:3082–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsiadlowski L., Braband A.. 2006. The complete mitochondrial genome of the sea spider Nymphon gracile (Arthropoda: Pycnogonida). BMC Genomics 7:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A.. 2018. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67:901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Tanaka H., Yang Z.. 2005. An empirical examination of the utility of codon-substitution models in phylogeny reconstruction. Syst. Biol. 54(808):808–818. [DOI] [PubMed] [Google Scholar]
- Richter S., Schwarz F., Hering L., Böggemann M., Bleidorn C.. 2015. The utility of genome skimming for phylogenomic analyses as demonstrated for glycerid relationships (Annelida, Glyceridae). Genome Biol. Evol 7:3443–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone C., De Giorgi C., Gissi C., Pesole G., Reyes A.. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene 238:195–209. [DOI] [PubMed] [Google Scholar]
- Seixas V.C., Russo C.A.M., Paiva P.C.. 2017. Mitochondrial genome of the Christmas tree worm Spirobranchus giganteus (Annelida: Serpulidae) reveals a high substitution rate among annelids. Gene 605:43–53. [DOI] [PubMed] [Google Scholar]
- Shao R., Barker S.C., Mitani H., Takahashi M., Fukunaga M.. 2006. Molecular mechanisms for the variation of mitochondrial gene content and gene arrangement among chigger mites of the genus leptotrombidium (Acari: Acariformes). J. Mol. Evol. 63:251–261. [DOI] [PubMed] [Google Scholar]
- Shao R., Dowton M., Murrell A., Barker S.C.. 2003. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol. Biol. Evol. 20:1612–1619. [DOI] [PubMed] [Google Scholar]
- Shekhovtsov S.V., Peltek S.E.. 2019. The complete mitochondrial genome of Aporrectodea rosea (Annelida: Lumbricidae). Mitochondrial DNA Part B 4:1752–1753. [Google Scholar]
- Shen X., Ma X., Ren J., Zhao F.. 2009. A close phylogenetic relationship between Sipuncula and Annelida evidenced from the complete mitochondrial genome sequence of Phascolosoma esculenta. BMC Genomics 10:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.R. 2019. TreeTools: create, modify and analyse phylogenetic trees.
- Smith M.R. 2020. Information theoretic generalized Robinson–Foulds metrics for comparing phylogenetic trees. Bioinformatics 36:5007–5013. [DOI] [PubMed] [Google Scholar]
- Stach T., Braband A., Podsiadlowski L.. 2010. Erosion of phylogenetic signal in tunicate mitochondrial genomes on different levels of analysis. Mol. Phylogenet. Evol. 55:860–870. [DOI] [PubMed] [Google Scholar]
- Struck T.H. 2011. Direction of evolution within Annelida and the definition of Pleistoannelida. J. Zool. Syst. Evol. Res. 49:340–345. [Google Scholar]
- Struck T.H. 2014. TreSpEx—detection of misleading signal in phylogenetic reconstructions based on tree information. Evol. Bioinform. 10:51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck T.H. 2019. 7.2 Phylogeny. In: Purschke G., Böggemann M., Westheide W. editors. Annelida basal groups and pleistoannelida, Sedentaria I. Berlin/Boston: De Gruyter. p. 37–68. [Google Scholar]
- Struck T.H., Golombek A., Weigert A., Franke Franziska A., Westheide W., Purschke G., Bleidorn C., Halanych K.M.. 2015. The evolution of annelids reveals two adaptive routes to the interstitial realm. Curr. Biol. 25:1993–1999. [DOI] [PubMed] [Google Scholar]
- Struck T.H., Schult N., Kusen T., Hickman E., Bleidorn C., McHugh D., Halanych K.M.. 2007. Annelida phylogeny and the status of Sipuncula and Echiura. BMC Evol. Biol. 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Li Q., Kong L., Yu H.. 2020. Evolution of mitochondrial gene arrangements in Arcidae (Bivalvia: Arcida) and their phylogenetic implications. Mol. Phylogenet. Evol. 150:106879. [DOI] [PubMed] [Google Scholar]
- Sun Y., Daffe G., Zhang Y., Pons J., Qiu J.-W., Kupriyanova E.K.. 2021. Another blow to the conserved gene order in Annelida: evidence from mitochondrial genomes of the calcareous tubeworm genus Hydroides. Mol. Phylogenet. Evol. 160:107124. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Kumar S.. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38:3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempestini A., Massamba-N’Siala G., Vermandele F., Beaudreau N., Mortz M., Dufresne F., Calosi P.. 2020. Extensive gene rearrangements in the mitogenomes of congeneric annelid species and insights on the evolutionary history of the genus Ophryotrocha. BMC Genomics 21:815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilic E., Sayyari E., Stiller J., Mirarab S., Rouse G.W.. 2020. More is needed—Thousands of loci are required to elucidate the relationships of the “flowers of the sea” (Sabellida, Annelida). Mol. Phylogenet. Evol. 151:106892. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A.M.. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallès Y., Boore J.L.. 2006. Lophotrochozoan mitochondrial genomes. Integr. Comp. Biol. 46:544–557. [DOI] [PubMed] [Google Scholar]
- Varney R.M., Brenzinger B., Malaquias M.A.E., Meyer C.P., Schrödl M., Kocot K.M.. 2021. Assessment of mitochondrial genomes for heterobranch gastropod phylogenetics. BMC Ecol Evol. 21:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waeschenbach A., Telford M.J., Porter J.S., Littlewood D.T.J.. 2006. The complete mitochondrial genome of Flustrellidra hispida and the phylogenetic position of Bryozoa among the Metazoa. Mol. Phylogenet. Evol. 40:195–207. [DOI] [PubMed] [Google Scholar]
- Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S., et al. 2020. gplots: various R programming tools for plotting data. R package version 3.1.1.
- Wei T., Simko V.. 2017. R package “corrplot”: visualization of a Correlation Matrix (Version 0.84).
- Weigert A., Bleidorn C.. 2016. Current status of annelid phylogeny. Org. Divers. Evol. 16:345–362. [Google Scholar]
- Weigert A., Golombek A., Gerth M., Schwarz F., Struck T.H., Bleidorn C.. 2016. Evolution of mitochondrial gene order in Annelida. Mol. Phylogenet. Evol. Part A 94:196–206. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- Wickham H. 2020. tidyr: Tidy Messy data. R package version 1.1.2.
- Wickham H., Averick M., Bryan J., Chang W., McGowan L.D.A., François R., Grolemund G., Hayes A., Henry L., Hester J., Kuhn M., Pedersen T., Miller E., Bache S., Müller K., Ooms J., Robinson D., Seidel D., Spinu V., Takahashi K., Vaughan D., Wilke C., Woo K., Yutani H.. 2019. Welcome to the Tidyverse. J Open Source Softw 4:1686. [Google Scholar]
- Wickham H., François R., Henry L., Müller K.. 2020. dplyr: a grammar of data manipulation. R package version 1.0.2.
- Wu Z., Shen X., Sun M., Ren J., Wang Y., Huang Y., Liu B.. 2009. Phylogenetic analyses of complete mitochondrial genome of Urechis unicinctus (Echiura) support that echiurans are derived annelids. Mol. Phylogenet. Evol. 52:558–562. [DOI] [PubMed] [Google Scholar]
- Xu W., Jameson D., Tang B., Higgs P.G.. 2006. The relationship between the rate of molecular evolution and the rate of genome rearrangement in animal mitochondrial genomes. J. Mol. Evol. 63:375–392. [DOI] [PubMed] [Google Scholar]
- Zardoya R. 2020. Recent advances in understanding mitochondrial genome diversity [version 1; peer review: 4 approved]. F1000Res 9:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kan X., Miao G., Hu S., Sun Q., Tian W.. 2020. qMGR: a new approach for quantifying mitochondrial genome rearrangement. Mitochondrion 52:20–23. [DOI] [PubMed] [Google Scholar]
- Zhang J., Miao G., Hu S., Sun Q., Ding H., Ji Z., Guo P., Yan S., Wang C., Kan X., Nie L.. 2021. Quantification and evolution of mitochondrial genome rearrangement in Amphibians. BMC Ecol. Evolut. 21:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sun J., Rouse G.W., Wiklund H., Pleijel F., Watanabe H.K., Chen C., Qian P.-Y., Qiu J.-W.. 2018. Phylogeny, evolution and mitochondrial gene order rearrangement in scale worms (Aphroditiformia, Annelida). Mol. Phylogenet. Evol. 125:220–231. [DOI] [PubMed] [Google Scholar]
- Zhong M., Hansen B., Nesnidal M.P., Golombek A., Halanych K.M., Struck T.H.. 2011. Detecting the symplesiomorphy trap: a multigene phylogenetic analysis for terebelliform annelids. BMC Evol. Biol. 11:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M., Struck T.H., Halanych K.M.. 2008. Phylogenetic information from three mitochondrial genomes of Terebelliformia (Annelida) worms and duplication of the methionine tRNA. Gene 416:11–21. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Li Y., Cheng H., Halanych K.M., Wang C.. 2020. The mitochondrial genome of the bone-eating worm Osedax rubiplumus (Annelida, Siboglinidae). Mitochondrial DNA Part B 5:2267–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Jakovlić I., Chen R., Zhang D., Zhang J., Li W.-X., Wang G.-T.. 2017. The complete mitochondrial genome of parasitic nematode Camallanus cotti: extreme discontinuity in the rate of mitogenomic architecture evolution within the Chromadorea class. BMC Genomics 18:840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Alignments and the associated trees of the reference and supermatrix datasets are available at TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S30262).
The sequence data are deposited in NCBI under the accession numbers OQ702995-OQ703026 and OQ729891-OQ729923 (see also Supplementary online Appendix 1).
The table of the life history traits with the coding scheme and sources provided are available in Supplementary online Appendix 33 (http://dx.doi.org/10.5061/dryad.8w9ghx3pm).
Script used in the analyses is also available from the GitHub Repository: https://github.com/torstenstruck/AnnelidaMitoGenomes.