Abstract
Background
Cytoplasmic polyadenylation element binding (CPEB) proteins are sequence-specific RNA-binding proteins that control translation via cytoplasmic polyadenylation. We previously reported that CPEB1 or CPEB4 knockdown suppresses TAK1 and SMAD signaling in an in vitro study.
Objective
This study aimed to investigate whether suppression of CPEB1 or CPEB4 expression inhibits scar formation in a mice model of acute dermal wound healing.
Methods
CPEB1 and CPEB4 expression levels were suppressed by siRNA treatment. Skin wounds were created by pressure-induced ulcers in mice. Images of the wound healing were obtained using a digital camera and contraction was measured by ImageJ. mRNA and protein expression was analyzed using quantitative real time polymerase chain reaction and western blotting, respectively.
Results
Wound contraction was significantly decreased by pre-treatment with CPEB1 or CPEB4 siRNA compared to the control. Suppression of CPEB1 or CPEB4 expression decreased TAK1 signaling by reducing the levels of TLR4 and TNF-α, phosphorylated TAK1, p38, ERK, JNK, and NF-κB-p65. Decreased levels of phosphorylated SMAD2 and SMAD3 indicated a reduction in SMAD signaling as well. Consequently, the expression of α-SMA, fibronectin, and type I collagen decreased.
Conclusion
CPEB1 siRNA or CPEB4 siRNA inhibit scar formation by modulating the TAK1 and SMAD signaling pathways. Our study highlights CPEB1 and CPEB4 as potential therapeutic targets for the treatment of scar formation.
Keywords: Cicatrix, CPEB1, CPEB4, MAP3K7 signaling, SMAD signaling, Wound healing
INTRODUCTION
Wound healing is a complex, well-orchestrated, multicellular process usually characterized by three phases: inflammation, proliferation, and remodeling1. These processes are involved in the communication and interplay between several cell types, including keratinocytes, fibroblasts, endothelial cells, macrophages, and platelets1. Moreover, multiple signaling pathways participate in the healing process and are regulated by numerous growth factors, cytokines and chemokines2. Deregulation of any of these steps can ultimately delay healing or lead to excessive scar formation such as hypertrophic scars (HTS)1.
Upon skin injury, when toll-like receptors (TLRs) are activated by danger signals, a series of intracellular signaling pathways involving the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and mitogen-activated protein kinases (MAPKs), which induce the expression of many TLR target genes, are activated3. Transforming growth factor-β1 activated kinase 1 (TAK1), also known as mitogen-activated protein kinase kinase kinase (MAP3K7), is a downstream molecule of the TLR signaling pathway. It is important for the activation of the transcription factor, NF-κB, and for that of p38 MAPK, c-Jun terminal kinase (JNK), and extracellular signal-regulated kinase 1/2 (ERK1/2) signaling3,4. Transforming growth factor-β1 (TGF-β1) is a potent inducer of fibroblast proliferation and/or differentiation into myofibroblasts that produce extracellular matrix (ECM), composed of collagen, fibronectin, connective tissue growth factor, etc., to form the clot in the wound5. However, excessive TGF-β1 produced from an unbalanced healing process leads to over-scarring, called as HTS, a serious burn sequela in clinical practice5. TGF-β1 acts through SMAD-dependent pathways, including phosphorylation and activation of SMAD 2 and SMAD 3 by TGF-β receptor 1 (TGFR1)6. However, TGF-β1 can also activate SMAD-independent pathways to modify cellular functions, including the TAK1, phosphatidylinositol 3-kinase/AKT, and Rho-like GTPase signaling pathways6.
The mammalian cytoplasmic polyadenylation element binding (CPEB) protein is a sequence-specific RNA-binding protein that controls poly(A) tail length and polyadenylation-induced translation of mRNAs. It is important for cell proliferation and differentiation, cellular senescence, neuronal synaptic plasticity, and immune response7. In vertebrates, CPEB-like proteins are composed of four paralogs (CPEB1~4), and many studies have elucidated the biological functions of CPEB1 and CPEB4 in health and disease7,8.
Our previous work showed that CPEB1 or CPEB4 knockdown inhibits TAK1 and SMAD signaling in lipopolysaccharide (LPS)-induced human macrophages and TGF-β1-induced fibroblasts that are derived from human skin, respectively9. Therefore, in the present study, we investigated the effects of suppression of CPEB1 or CPEB4 expression on wound healing in mice models of pressure ulcers. Our findings indicate that suppression of CPEB1 or CPEB4 expression inhibited TAK1 and SMAD signaling led to decreased scar formation. These results, combined with previous in vitro studies, indicate that CPEB1 and CPEB4 could be potential targets for the treatment of dermal fibrotic scarring.
MATERIALS AND METHODS
siRNA transfection and pressure ulcer mouse model
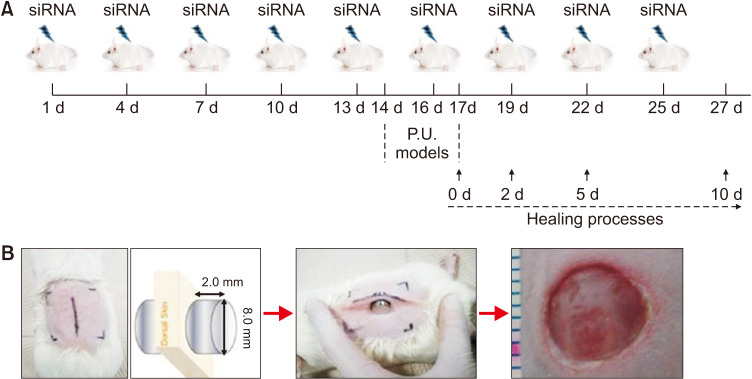
SMARTpool siRNAs for the negative control, and knockdown of CPEB1 and CPEB4, were purchased from Dharmacon (Lafayette). ICR mice (8-week-old) were purchased from Korean Animal Technology (Koatech). All animal experiments were conducted in the animal laboratory of Hangang Sacred Heart Hospital, Hallym University, in accordance with the Guidelines for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal protocols were approved by the Animal Research Ethics Board of Hallym University (HMC2017-2-1121-30). Mice were randomly classified into four groups: (1) without any intervention (NC), (2) treated with the negative control siRNA (si-Ct)/atelocollagen mixture, (3) treated with CPEB1 siRNA (si-CP1)/atelocollagen mixture, and (4) treated with CPEB4 siRNA (si-CP4)/atelocollagen mixture. Each siRNA was mixed with atelocollagen, according to the manufacturer’s instructions (Koken)10. Mice were injected with 100 µl of atelocollagen containing 3 µg11 of CPEB1 or CPEB4 siRNA, or negative control siRNA intravenously (i.v.) via the tail vein, once every three days for nine consecutive times (Fig. 1A). After the fifth injection (day 14), mice were anesthetized and maintained using 100% oxygen with 2.5% isoflurane (Hana Pharm). The dorsal skins of mice were shaved and sterilized with 70% alcohol. Then it was gently pulled up and loaded with two circular ceramic magnetic plates (1.2 g, 1500.0 Gauss, 8.0 mm diameter, and 2.0 mm thickness) as Fig. 1B, to creating a 5.0 mm of skin thick bridge between the two magnets. The magnets were placed for 12 hours, and the removal time was 12 hours as a single ischemia-reperfusion (I/R) cycle. The mice were provided food and water ad libitum. After three cycles, a complete pressure ulcer (P. U.) was created12. Wounds were allowed to be exposed to air, and dry naturally forming a scab.
Fig. 1. Design of siRNA-treatment and wound model in mice. (A) Flowchart of atelocollagen-mediated siRNA transfer procedure, modeling of P.U. wound, and tissue preparation times. siRNA/atelocollagen mixture was injected via the tail vein once in three days, nine times in total. On day 14, P.U. was modelled in mice using circular magnets on the dorsal skin for 3 days. Wound tissues were collected on 0, 2, 5, and 10 days after wound creation. (B) Illustration showing the application of magnets for wound creation in mice. P.U.: pressure ulcer.
Measurement of wound contraction
To analyze wound contraction, images were captured using a digital camera (Nikon) on days 0, 2, 5, and 10 post-wounding. Quantification of wound closure was carried out by Image J software (National Institutes of Health) and normalized with the wound size at day 0 being taken as 100% (https://imagej.nih.gov/ij).
Quantitative polymerase chain reaction (qPCR)
Wound tissues were obtained 2, 5, and 10 days after wound formation. Total RNA was extracted, concentration was measured and RNA was reverse-transcribed to cDNA, for qPCR as previously described9. The following primers were used: CPEB1, 5′-TTTCAAGCCTTCG CATTTCCC-3′ (forward) and 5′-GGACCCAACGCCCATCTTTA-3′ (reverse); CPEB4, 5′-CCTTCTTCCTCCACTATA-3′ (forward) and 5′-GAGAGCACCATTAT TAGC-3′. The relative fold mRNA expression of each target gene was evaluated by the 2-ΔΔCt method, using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the normalization control13.
Western blot
Wound tissue samples were obtained 2 and 5 days after wounding. Total protein sample preparation, electrophoresis, development, and imaging analyses were performed as previously described9. The primary antibodies used were rabbit polyclonal anti-CPEB1 and anti-CPEB4 (Thermo Fisher Scientific); rabbit polyclonal anti-TLR4 (Cusabio Biotech); rabbit polyclonal anti-phospho-TAK1, anti-TAK1, anti-p38, anti-phospho-JNK, anti-JNK, anti-phospho-ERK, anti-phospho-NF-κB-p65, anti-β-actin, and anti-phospho-SMAD2; rabbit polyclonal anti-phospho-SMAD3 and anti-SMAD3 (ST John’s laboratory); rabbit polyclonal anti-SMAD2, mouse polyclonal anti-phospho-p38, and anti-ERK (Cell Signaling Technology); rabbit polyclonal anti-NF-κB-p65, anti-type I collagen, mouse polyclonal anti-α-SMA, and monoclonal anti-fibronectin (Abcam); and mouse monoclonal anti-phospho-IκBα, anti-IκBα, and anti-GAPDH (Santa Cruz Biotechnology). The secondary antibodies used were peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG (Merck Millipore).
Statistical analysis
All results are presented as the mean±standard error of the mean (SEM). The Mann–Whitney U test was used for comparisons between two groups. Statistical analyses were performed using PASW Statistics 24 (IBM Corp.), and significance was set to p<0.05.
RESULTS
Atelocollagen-mediated siRNA transfer suppression of CPEB1 or CPEB4 expression in wound tissues of mice
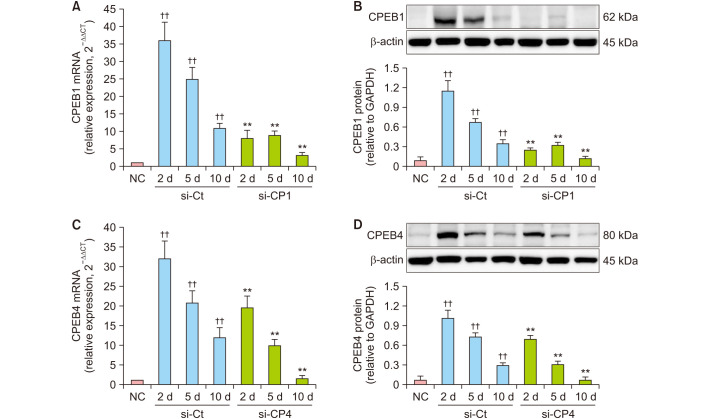
To confirm efficiency of atelocollagen-mediated siRNA transfer via tail vein on mouse, we examined mRNA and protein expression of CPEB1 and CPEB4 by qRT-PCR and western blotting respectively, in wound tissues obtained on day 2 (inflammation phase), 5 (proliferation phase), and 10 (remodeling phase) after wound creation. The mRNA and protein expression of CPEB1 or CPEB4 was significantly higher in the wound tissue of control mice (si-Ct) than in the normal skin tissue (NC) in each healing phase (p<0.01) (Fig. 2). However, treatment with si-CP1 or si-CP4 significantly suppressed the mRNA and protein expression of CPEB1 or CPEB4 in wound tissues compared to si-Ct corresponding to each healing phase (p<0.01) (Fig. 2).
Fig. 2. Treatment with si-CP1 or si-CP4 decreased mRNA and protein expression of CPEB1 or CPEB4 in wound tissues of mice. RT-PCR and western blotting analysis for the expression of CPEB1 or CPEB4 on day 2, 5, and 10 in wound tissues undergoing the healing process. (A) RT-PCR result showing si-CP1 decreased the increased mRNA expression of CPEB1 in wound tissues compared to si-Ct corresponding to each healing phase. (B) Western blot result showing si-CP1 decreased the increased protein expression of CPEB1 in wound tissues compared to si-Ct corresponding to each healing phase. (C) RT-PCR result showing si-CP4 decreased the increased mRNA expression of CPEB4 in wound tissues compared to si-Ct corresponding to each healing phase. (D) Western blot result showing si-CP4 decreased the increased protein expression of CPEB4 in wound tissues compared to si-Ct corresponding to each healing phase. Data are expressed as mean±standard error (n=10). RT-PCR: quantitative real-time PCR, NC: normal control, si-Ct: negative control siRNA, si-CP1: CPEB1 siRNA, si-CP4: CPEB4 siRNA, P.U.: pressure ulcer. ††p<0.01 for comparison with NC, and **p<0.01 for comparison with si-Ct.
Suppression of CPEB1 and CPEB4 decreased wound contraction
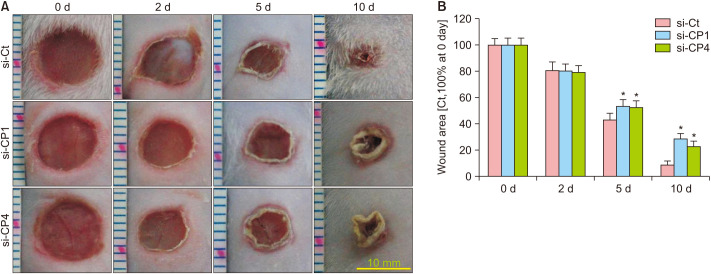
Wound contraction was not significantly different on day 2 for the comparison of si-Ct with si-CP1 and si-Ct with si-CP4 groups (Fig. 3). Moreover, on days 5 and 10, contraction was remarkably decreased in the si-CP1 and si-CP4 groups compared to that in the si-Ct group (p<0.05) (Fig. 3). These results suggested that the rate of wound healing was delayed by treatment with si-CP1 or si-CP4.
Fig. 3. Treatment with si-CP1 or si-CP4 decreased wound contraction in mice. (A) Representative images of wound contraction on day 2, 5, and 10 undergoing the healing process. Scale bar=10 mm. (B) Quantification of wound contraction. Wound contraction was expressed as a percentage relative to the initial wound surface area, which was considered to be 100%. Data are expressed as mean±standard error (n=10). si-Ct: negative control siRNA, si-CP1: CPEB1 siRNA, si-CP4: CPEB4 siRNA, P.U.: pressure ulcer. *p<0.05 for the comparison with si-Ct.
Suppression of CPEB1 and CPEB4 decreased inflammation related-TAK1 signaling
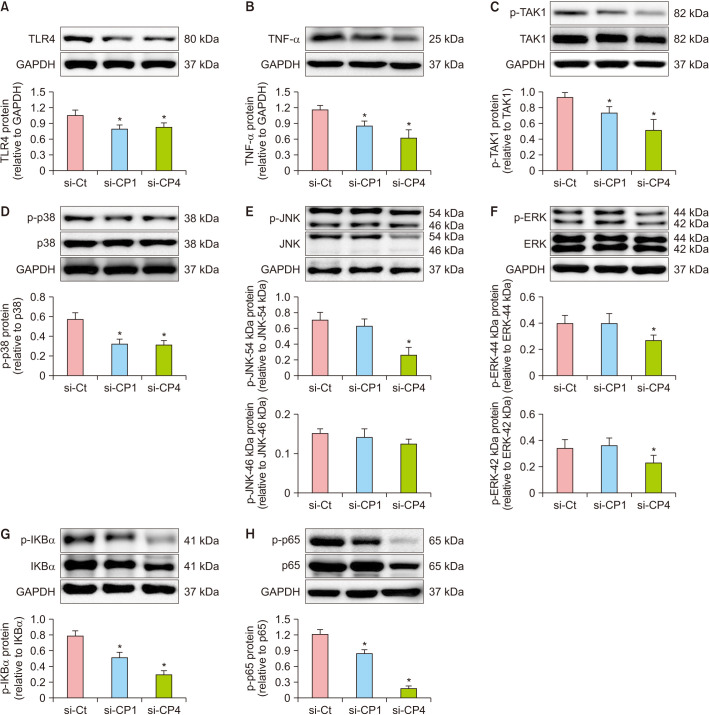
To explore the functional mechanism of CPEB1 and CPEB4 in inflammation, we measured changes in TLR4-TAK1 signaling on day 2 after the wound creation. TLR4 and TNF-α protein expression levels were significantly decreased in the si-CP1 and si-CP4 groups compared to the si-Ct group (p<0.05) (Fig. 4A, B). The phosphorylation levels of TAK1 and p38 were decreased in the si-CP1 and si-CP4 groups compared to those in the si-Ct group (p<0.05) (Fig. 4C, D). Although phosphorylation of the JNK isoform at 54 kDa was decreased (p<0.05), the levels of the JNK isoform at 46 kDa unaltered in the si-CP4 group (Fig. 4E). Moreover, phosphorylation of isoforms 54 and 46 kDa of JNK was not affected in the si-CP1 group (Fig. 4E). Phosphorylation of ERK was significantly decreased in the si-CP4 group (p<0.05) but was unchanged in the si-CP1 group (Fig. 4F). Phosphorylation of both IKBα and NF-κB-p65 was decreased in the si-CP1 and si-CP4 groups compared to the si-Ct group (p<0.05) (Fig. 4G, H). These results indicated that si-CP1 and si-CP4 decreased the inflammatory response of wound healing by suppressing TLR-TAK1 signaling.
Fig. 4. Treatment with si-CP1 or si-CP4 decreased inflammatory signaling in the mouse model of P.U. Protein expression was measured by western blot analysis on day 2 after wound creation. (A) CPEB1 or CPEB4 siRNA decreased protein expression of TRL4 in wound tissues compared to si-Ct. (B) CPEB1 or CPEB4 siRNA decreased protein expression of TNF-α in wound tissues compared to si-Ct. (C) CPEB1 or CPEB4 siRNA decreased phosphorylation of TAK1 in wound tissues compared to si-Ct. (D) CPEB1 or CPEB4 siRNA decreased phosphorylation of p38 in wound tissues compared to si-Ct. (E) si-CP1 not changed phosphorylation of JNK in wound tissues compared to si-Ct. si-CP4 decreased phosphorylation of JNK isoform at 54 kDa, and not changed phosphorylation of isoforms at 46 kDa of JNK in wound tissues compared to si-Ct. (F) si-CP1 not changed phosphorylation of ERK in wound tissues compared to si-Ct. si-CP4 decreased phosphorylation of ERK in wound tissues compared to si-Ct. (G) CPEB1 or CPEB4 siRNA decreased phosphorylation of IKBα in wound tissues compared to si-Ct. (H) CPEB1 or CPEB4 siRNA decreased phosphorylation of NF-κB-p65 in wound tissues compared to si-Ct. Data are expressed as mean±standard error (n=10). si-Ct: negative control siRNA, si-CP1: CPEB1 siRNA, si-CP4: CPEB4 siRNA, P.U.: pressure ulcer. *p<0.05 for the comparison with si-Ct.
Suppression of CPEB1 and CPEB4 decreased SMAD and TAK1 signaling
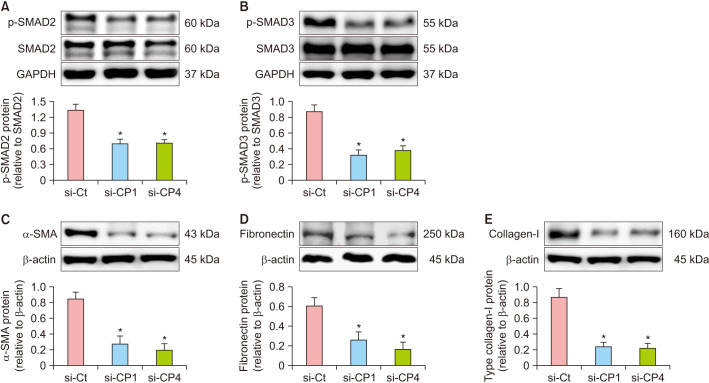
We determined the changes in SMAD signaling and the expression of its related scarring markers in tissues obtained on day 5 and 10 after wound creation. The phosphorylation of SMAD2 and SMAD3 was significantly decreased in the si-CP1 and si-CP4 groups compared to that in the si-Ct group (p<0.05) (Fig. 5A, B; Supplementary Fig. 1A, B). Moreover, the expression of the myofibroblast marker, α-SMA, was significantly decreased in the si-CP1 and si-CP4 groups compared to that in the si-Ct group (p<0.05) (Fig. 5C; Supplementary Fig. 1C). Furthermore, the levels of fibronectin and collagen-I, both novel marker of scars, were dramatically decreased in the si-CP1 and si-CP4 groups on day 5 (p<0.05) (Fig. 5D, E). In addition, the phosphorylation TAK1 was significantly decreased in the si-CP1 and si-CP4 groups compared to that in the si-Ct group on day 10 (p<0.05) (Supplementary Fig. 1D). The pro-inflammatory cytokines, TNF-α and IL-6 were decreased in the si-CP1 and si-CP4 groups on day 10 (p<0.05) (Supplementary Fig. 1E, F). These results suggest that si-CP1 or si-CP4 decreased the scarring response of wound healing by suppressing SMAD and TAK1 signaling.
Fig. 5. Treatment with si-CP1 or si-CP4 decreased SMAD signaling and expression of scar markers in the mouse model of P.U. Protein expression was measured by western blot analysis on day 5 after wound creation. (A) CPEB1 or CPEB4 siRNA decreased phosphorylation of SMAD2 in wound tissues compared to si-Ct. (B) CPEB1 or CPEB4 siRNA decreased phosphorylation of SMAD3 in wound tissues compared to si-Ct. (C) CPEB1 or CPEB4 siRNA decreased protein expression of α-SMA in wound tissues compared to si-Ct. (D) CPEB1 or CPEB4 siRNA decreased protein expression of fibronectin in wound tissues compared to si-Ct. (E) CPEB1 or CPEB4 siRNA decreased protein expression of collagen-I in wound tissues compared to si-Ct. Data are expressed as mean±standard error (n=10). si-Ct: negative control siRNA, si-CP1: CPEB1 siRNA, si-CP4: CPEB4 siRNA, P.U.: pressure ulcer. *p<0.05 for comparison with si-Ct.
DISCUSSION
In this study, we found that CPEB1 and CPEB4 were significantly increased in the wound tissues of mice compared to normal tissues suggesting that CPEB1 and CPEB4 are involved in wound healing. However, RNAi-mediated suppression of CPEB1 or CPEB4 expression decreased wound contraction during TLR4-TAK1 signaling in inflammatory phases and SMAD signaling of proliferative phases during wound healing. These results indicate that CPEB1 and CPEB4 play important roles in scar formation during wound healing.
In our previous study, we reported that levels of both CPEB1 and CPEB4 were increased in LPS-treated macrophages; however, knockdown of their expression suppressed TAK1 signaling by reducing the phosphorylation levels of TAK1, p38, ERK, JNK, IkBα, as well as NF-κB-p65 in LPS-treated macrophages9. Accumulating evidence supports the pathological roles of TAK1 signaling in the inflammatory response, which is becoming a potential therapeutic target for inflammatory disorders6. Suppression or deletion of TAK1 prevents neuronal death in cerebral ischemia or inhibits renal and pulmonary inflammation14,15,16. Inflammatory response is the first essential stage and plays an important role in the normal wound healing process. An acute inflammatory reaction generates an antimicrobial host defense, resulting in the elimination of infectious pathogens and wound debris, followed by resolution, which is mediated mainly by tissue-resident and recruited macrophages, both originating from embryonic and circulating monocytes, respectively4. M1 macrophages accumulate and are activated in the inflammatory phase of wound healing and produce pro-inflammatory cytokines, including IL-1, IL-6, and TNF-α7. These pro-inflammatory cytokines have a positive therapeutic effect on wound healing, particularly TNF-α, which exerts a growth-promoting activity on fibroblasts and enhances collagen deposition17. Clinical research has indicated that prolonged inflammation is a risk factor for the pathological development of HTS; therefore, they are defined as weakly inflamed pathological scars18. It has been observed that most of the intervention modalities for the clinical treatment of HTS are accompanied by reducing inflammation18.
The wound healing proliferative phase consists of granulation tissue formation, angiogenesis, and re-epithelialization4,5. Granulation tissue formation is characterized by a high density of fibroblasts, macrophages, capillaries, and ECM4,5. Scarring is a fibrous tissue that is physiologically essential for dermal wound healing in adults. However, excessive fibrotic scarring often leads to HTS, which has a high incidence of up to 91% in burn patients with deep dermal injuries7. It is well accepted in HTS formation that M2 macrophage-produced TGF-β1 is a theoretical basis, which acts through SMAD-dependent or independent pathways and stimulates secretion and deposition of excessive ECM protein by fibroblasts (myofibroblasts), that raises the normal skin surface area7. In addition, HTS-derived fibroblasts overexpress TGF-β1, α-SMA, type I and III collagen, and fibronectin19. A previous in vitro study reported that protein expression of both CPEB1 and CPEB4 increased in TGF-β1–induced human dermal fibroblasts; knockdown of their expression by siRNA decreased phosphorylation levels of SMAD2 and inhibited the expression of α-SMA, type I collagen, and fibronectin in fibroblasts treated with TGF-β19.
In the present study, we observed that CPEB1 and CPEB4 were upregulated on day 2 (inflammatory stage) and day 5 (proliferative stage) in the process of wound healing. Our previous work indicated that mRNA levels of CPEB1 and CPEB4 in HTS tissues are 1.8 and 2.2 times higher than those in normal skin tissues, respectively9. Other studies have also reported that the levels of both CPEB1 and CPEB4 were increased in human cirrhotic liver tissues compared with those in control tissues20. These evidences indicate that both CPEB1 and CPEB4 are involved in tissue repair or fibrotic scarring. CPEB1 depletion inhibits up-regulation of iNOS, NO, and ROS production that are induced by LPS stimulation21. CPEB1 activation induces angiogenesis by promoting the expression of vascular endothelial growth factor (VEGF); conversely, knockdown of CPEB1 downregulates angiogenic capacity20. A recent study showed that in an LPS-induced sepsis model, depletion of CPEB4 in mouse macrophages impaired resolution of inflammation22. However, CPEB4 is necessary for IL-22 production in CD4+T and innate lymphoid (ILC3) cells23. Moreover, CPEB4 promotes tissue regeneration in acute transient intestinal inflammation and is conducive to the development of colorectal cancer23. In addition, knockdown of all the subtypes of CPEB proteins (CPEB1, 2, 3, and 4) increases the phosphorylation levels of TAK1 and p38, activates NF-κB, and augments the production of IL-6 in mouse RAW 264.7 macrophages24. It is well known that CPEBs bind to CPEs located in the 3′ untranslated region (UTR) of specific mRNAs and control their translation by favoring their poly(A) tail elongation. Furthermore, they modulate the formation of alternative 3′-UTRs, which contain AU-rich elements, and mediate mRNA stability of several pro-inflammatory or anti-inflammatory cytokines25,26. However, to date, the mechanism by which CPEB regulates TAK1 and SMAD signaling is unclear and is a task for future studies. In addition, the CPEB2 regulate hippocampus-dependent synaptic plasticity and long-term memory27, controls translation of hypoxia-inducible factor 1α (hif-1α) mRNA in response to oxidative stress28, and suppress tumorigenesis in breast epithelial cells by inhibit the EMT, migration, invasion, and proliferation29. The CPEB3 also involved in synaptic plasticity and spatial memory27. Circular RNA, mmu-Cpeb3_0007 was related to cardiac hypertrophy and cardiac fibrosis based on gene set enrichment analysis in mouse model of cardiac hypertrophy30. In addition, CPEB3 suppresses EMT by blocking the interaction between colorectal cancer cells and tumor-associated macrophages via decreasing the IL-6/STAT3 signaling31.
In present study, suppression of CPEB1 or CPEB4 expression by RNAi inhibited wound scar formation, which seems to delay the wound healing rather than inhibits HTS formation. In vitro study, we demonstrated that the CPEB1 or CPEB4 knockdown suppressed TGF-β1–induced fibrosis9. Moreover, mRNA expression of CPEB1 and CPEB4 are significantly higher in HTS of patients compared to normal skin, they matched with HTS9. The results of both in vitro and vivo study suggest CPEB1 or CPEB4 is related to HTS formation. Wound healing of mice is mediated by the panniculus carnosus muscle, which, importantly, the skin of mice is structurally and functionally different with humans. Therefore, the wound of mice cannot form HTS32. Nevertheless, mice models can help to understand the skin diseases and predict treatment efficacy. In the future, it is worthwhile to evaluate the inhibition of CPEB1 or CPEB4 on the formation of HTS in HTS models using medium or large mammals.
In conclusion, this study reported that CPEB1 or CPEB4 were upregulated in the process of wound healing, and siRNA-mediated suppression of CPEB1 or CPEB4 inhibited inflammation and scar formation through TAK1 and SMAD signaling in a mice model of pressure-induced wounds (Supplementary Fig. 2). Our findings indicate that CPEB1 and CPEB4 are involved in the scarring response, providing new insights into the pathogenesis of dermal fibrosis.
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
FUNDING SOURCE: This research was supported by the Hallym University Research Fund and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1D1A1A02018478, 2020R1I1A3074338, and 2020R1I1A1A01074923).
DATA SHARING STATEMENT
Data supporting the results of this study are available from corresponding authors upon reasonable request.
SUPPLEMENTARY MATERIALS
Supplementary data can be found via http://anndermatol.org/src/sm/ad-22-210-s001.pdf.
Treatment with si-CP1 or si-CP4 decreased inflammation-related signaling and fibrosis-related signaling in the mouse model of P.U. Protein expression was measured by western blot analysis on day 10 after wound creation. (A) CPEB1 or CPEB4 siRNA decreased phosphorylation of SMAD2 in wound tissues compared to si-Ct. (B) CPEB1 or CPEB4 siRNA decreased phosphorylation of SMAD3 in wound tissues compared to si-Ct. (C) CPEB1 or CPEB4 siRNA decreased expression of α-SMA in wound tissues compared to si-Ct. (D) CPEB1 or CPEB4 siRNA decreased phosphorylation of TAK1 in wound tissues compared to si-Ct. (E) CPEB1 or CPEB4 siRNA decreased expression of TNF-α in wound tissues compared to si-Ct. (F) CPEB1 or CPEB4 siRNA decreased expression of IL-6 in wound tissues compared to si-Ct. Data are expressed as mean±standard error (n=10). si-Ct: negative control siRNA, si-CP1: CPEB1 siRNA, si-CP4: CPEB4 siRNA, P.U.: pressure ulcer. *p<0.05 for comparison with si-Ct.
Overview the effects of treatment with si-CPEB1 or si-CPEB4 on wound healing in an mice model of pressure ulcers. si-CPEB: siRNA targeted to cytoplasmic polyadenylation element binding protein.
References
- 1.Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev. 2003;83:835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- 2.Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- 3.Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. 2012;4:a006049. doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakurai H. Targeting of TAK1 in inflammatory disorders and cancer. Trends Pharmacol Sci. 2012;33:522–530. doi: 10.1016/j.tips.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Zhu Z, Ding J, Tredget EE. The molecular basis of hypertrophic scars. Burns Trauma. 2016;4:2. doi: 10.1186/s41038-015-0026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-β: the master regulator of fibrosis. Nat Rev Nephrol. 2016;12:325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 7.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Fernández-Miranda G, Méndez R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res Rev. 2012;11:460–472. doi: 10.1016/j.arr.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Cui HS, Joo SY, Cho YS, Kim JB, Seo CH. CPEB1 or CPEB4 knockdown suppresses the TAK1 and Smad signalings in THP-1 macrophage-like cells and dermal fibroblasts. Arch Biochem Biophys. 2020;683:108322. doi: 10.1016/j.abb.2020.108322. [DOI] [PubMed] [Google Scholar]
- 10.Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci U S A. 2005;102:12177–12182. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JC, Lai S, Guo X, Zhang X, de Crombrugghe B, Sonnylal S, et al. Attenuation of fibrosis in vitro and in vivo with SPARC siRNA. Arthritis Res Ther. 2010;12:R60. doi: 10.1186/ar2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanzafame RJ, Stadler I, Cunningham R, Muhlbauer A, Griggs J, Soltz R, et al. Preliminary assessment of photoactivated antimicrobial collagen on bioburden in a murine pressure ulcer model. Photomed Laser Surg. 2013;31:539–546. doi: 10.1089/pho.2012.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Neubert M, Ridder DA, Bargiotas P, Akira S, Schwaninger M. Acute inhibition of TAK1 protects against neuronal death in cerebral ischemia. Cell Death Differ. 2011;18:1521–1530. doi: 10.1038/cdd.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma FY, Tesch GH, Ozols E, Xie M, Schneider MD, Nikolic-Paterson DJ. TGF-β1-activated kinase-1 regulates inflammation and fibrosis in the obstructed kidney. Am J Physiol Renal Physiol. 2011;300:F1410–F1421. doi: 10.1152/ajprenal.00018.2011. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Liang C, Zhang ZK, Pan X, Peng S, Lee WS, et al. TAK1 inhibition attenuates both inflammation and fibrosis in experimental pneumoconiosis. Cell Discov. 2017;3:17023. doi: 10.1038/celldisc.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piguet PF, Grau GE, Vassalli P. Subcutaneous perfusion of tumor necrosis factor induces local proliferation of fibroblasts, capillaries, and epidermal cells, or massive tissue necrosis. Am J Pathol. 1990;136:103–110. [PMC free article] [PubMed] [Google Scholar]
- 18.Ogawa R. Keloid and hypertrophic scars are the result of chronic inflammation in the reticular dermis. Int J Mol Sci. 2017;18:606. doi: 10.3390/ijms18030606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo CH, Cui HS, Kim JB. Altered KCa3.1 expression following burn injury and the therapeutic potential of TRAM-34 in post-burn hypertrophic scar formation. Transl Res. 2021;236:133–146. doi: 10.1016/j.trsl.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Calderone V, Gallego J, Fernandez-Miranda G, Garcia-Pras E, Maillo C, Berzigotti A, et al. Sequential functions of CPEB1 and CPEB4 regulate pathologic expression of vascular endothelial growth factor and angiogenesis in chronic liver disease. Gastroenterology. 2016;150:982–997.e930. doi: 10.1053/j.gastro.2015.11.038. [DOI] [PubMed] [Google Scholar]
- 21.Kim KC, Hyun Joo S, Shin CY. CPEB1 modulates lipopolysaccharide-mediated iNOS induction in rat primary astrocytes. Biochem Biophys Res Commun. 2011;409:687–692. doi: 10.1016/j.bbrc.2011.05.065. [DOI] [PubMed] [Google Scholar]
- 22.Suñer C, Sibilio A, Martín J, Castellazzi CL, Reina O, Dotu I, et al. Macrophage inflammation resolution requires CPEB4-directed offsetting of mRNA degradation. Elife. 2022;11:e75873. doi: 10.7554/eLife.75873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sibilio A, Suñer C, Fernández-Alfara M, Martín J, Berenguer A, Calon A, et al. Immune translational control by CPEB4 regulates intestinal inflammation resolution and colorectal cancer development. iScience. 2022;25:103790. doi: 10.1016/j.isci.2022.103790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivshina M, Alexandrov IM, Vertii A, Doxsey S, Richter JD. CPEB regulation of TAK1 synthesis mediates cytokine production and the inflammatory immune response. Mol Cell Biol. 2015;35:610–618. doi: 10.1128/MCB.00800-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bava FA, Eliscovich C, Ferreira PG, Miñana B, Ben-Dov C, Guigó R, et al. CPEB1 coordinates alternative 3'-UTR formation with transla tional regulation. Nature. 2013;495:121–125. doi: 10.1038/nature11901. [DOI] [PubMed] [Google Scholar]
- 26.Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol. 2014;14:361–376. doi: 10.1038/nri3682. [DOI] [PubMed] [Google Scholar]
- 27.Lu WH, Yeh NH, Huang YS. CPEB2 activates GRASP1 mRNA translation and promotes AMPA receptor surface expression, long-term potentiation, and memory. Cell Rep. 2017;21:1783–1794. doi: 10.1016/j.celrep.2017.10.073. [DOI] [PubMed] [Google Scholar]
- 28.Chen PJ, Weng JY, Hsu PH, Shew JY, Huang YS, Lee WH. NPGPx modulates CPEB2-controlled HIF-1α RNA translation in response to oxidative stress. Nucleic Acids Res. 2015;43:9393–9404. doi: 10.1093/nar/gkv1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tordjman J, Majumder M, Amiri M, Hasan A, Hess D, Lala PK. Tumor suppressor role of cytoplasmic polyadenylation element binding protein 2 (CPEB2) in human mammary epithelial cells. BMC Cancer. 2019;19:561. doi: 10.1186/s12885-019-5771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Zhou J, Wei Z, Cheng Y, Tian G, Quan Y, et al. Identification of circular RNAs in cardiac hypertrophy and cardiac fibrosis. Front Pharmacol. 2022;13:940768. doi: 10.3389/fphar.2022.940768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong Q, Fang Y, Lai Q, Wang S, He C, Li A, et al. CPEB3 inhibits epithelial-mesenchymal transition by disrupting the crosstalk between colorectal cancer cells and tumor-associated macrophages via IL-6R/STAT3 signaling. J Exp Clin Cancer Res. 2020;39:132. doi: 10.1186/s13046-020-01637-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong VW, Sorkin M, Glotzbach JP, Longaker MT, Gurtner GC. Surgical approaches to create murine models of human wound healing. J Biomed Biotechnol. 2011;2011:969618. doi: 10.1155/2011/969618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Treatment with si-CP1 or si-CP4 decreased inflammation-related signaling and fibrosis-related signaling in the mouse model of P.U. Protein expression was measured by western blot analysis on day 10 after wound creation. (A) CPEB1 or CPEB4 siRNA decreased phosphorylation of SMAD2 in wound tissues compared to si-Ct. (B) CPEB1 or CPEB4 siRNA decreased phosphorylation of SMAD3 in wound tissues compared to si-Ct. (C) CPEB1 or CPEB4 siRNA decreased expression of α-SMA in wound tissues compared to si-Ct. (D) CPEB1 or CPEB4 siRNA decreased phosphorylation of TAK1 in wound tissues compared to si-Ct. (E) CPEB1 or CPEB4 siRNA decreased expression of TNF-α in wound tissues compared to si-Ct. (F) CPEB1 or CPEB4 siRNA decreased expression of IL-6 in wound tissues compared to si-Ct. Data are expressed as mean±standard error (n=10). si-Ct: negative control siRNA, si-CP1: CPEB1 siRNA, si-CP4: CPEB4 siRNA, P.U.: pressure ulcer. *p<0.05 for comparison with si-Ct.
Overview the effects of treatment with si-CPEB1 or si-CPEB4 on wound healing in an mice model of pressure ulcers. si-CPEB: siRNA targeted to cytoplasmic polyadenylation element binding protein.
Data Availability Statement
Data supporting the results of this study are available from corresponding authors upon reasonable request.