Highlights
-
•
KIAA1429 up-regulated in the OSCC samples and cells, and the high-expression of KIAA1429 promoted OSCC proliferation.
-
•
KIAA1429 positively accelerated the aerobic glycolysis of OSCC, including glucose uptake, lactate production, ATP level and ECAR.
-
•
Mechanistically, m6A reader YTHDF1 recognized the m6A modification site of PGK1 mRNA and enhanced its mRNA stability.
-
•
Our findings demonstrated a novel insight for KIAA1429 on OSCC tumorigenesis via m6A-dependent manner.
Keywords: Oral squamous cell carcinoma, N6-methyladenosine, KIAA1429, Ferroptosis, Aerobic glycolysis
Abstract
N6-methyladenosine (m6A) modification acts as the most prevalent modification on eukaryotic RNA, and its function on oral squamous cell carcinoma (OSCC) is still unclear. Here, the present research aimed to explore the novel function of m6A methyltransferase KIAA1429 in OSCC. Results illustrated that KIAA1429 up-regulated in the OSCC samples and cells. Gain/loss functional assays demonstrated that KIAA1429 repressed the ferroptosis of OSCC. Moreover, KIAA1429 positively accelerated the aerobic glycolysis of OSCC, including glucose uptake, lactate production, ATP level and ECAR. Mechanistically, KIAA1429 could install the m6A modification on the PGK1 mRNA, thereby up-regulating the methylated m6A level. Moreover, m6A reader YTHDF1 recognized the m6A modification site of PGK1 mRNA and enhanced its mRNA stability. Thus, KIAA1429 promoted the OSCC aerobic glycolysis and inhibited the ferroptosis of OSCC through YTHDF1-mediated PGK1 mRNA stability. Taken together, these findings reveal a novel insight for KIAA1429 on OSCC via m6A-dependent manner.
Background
Oral squamous cell carcinoma (OSCC) is considered as major type of head and neck cancer, which induces estimated 200 thousands deaths worldwide [1,2]. Unfortunately, the morbidity and incidence of OSCC have increased recently, especially getting younger patients [3]. Despite great achievement has made in recent decades, the survival of OSCC patients still significantly deteriorates. The negative clinical outcomes encourage clinicians and doctors to look for promising molecular factors to treat the disease.
N6-Methyladenosine (m6A) has been diffusely considered as the most abundant modifications in eukaryotic messenger RNAs [4,5]. The m6A modification is dynamic and reversible in eukaryocytes. Landscape analysis revealed that there are thousands of m6A peaks being identified on over 25% of genomic transcripts. Averagely, there are 1–3 m6A peaks modifications per transcript. The m6A modifications are installed by methyltransferases (writers), e.g. METTL3 (methyltransferase-like 3), METTL14, KIAA1429 and WTAP (Wilms tumor 1-associated protein), while uninstalled by demethyses (erasers), e.g. ALKBH5 and FTO. In OSCC tumorigenesis, more and more literature indicated that m6A modifications wildly regulate the progression [6]. For instance, the stable FTO knockdown inhibits OSCC cells’ viability, tumor growth, colony formation, and increases YAP1 m6A modification at YAP1 mRNA 3′-UTR [7]. Thus, the potential roles of m6A modification are abundant and undiscovered.
Energy metabolism abnormality is commonly observed in the development of human cancers, including OSCC [8,9]. In the tumor energy metabolism, aerobic glycolysis (as known as Warburg effect) provides the main source of energy for cancers. Although glycolysis could be anaerobic or aerobic, in tumor microenvironment, cancer cells tend to up-regulate the glycolysis level to generate ATP. In OSCC, aerobic glycolysis had been illustrated to regulate its pathological development. For instance, in OSCC, TMEM207/WWOX bind to HIF-1α to increase HIF-1α and GLUT-1 expression under normoxic conditions, and promoted tumor growth [10]. These data suggest the critical role of aerobic glycolysis in OSCC.
In this research, our work showed that the novel m6A writer KIAA1429 up-regulated in the OSCC, and KIAA1429 overexpression promoted OSCC proliferation and aerobic glycolysis, and repressed the ferroptosis of OSCC. These findings inspired us that KIAA1429 may act as an oncogene in OSCC. Mechanistically, KIAA1429 could install the m6A modification on the PGK1 mRNA, and cooperated with m6A reader YTHDF1 to enhance PGK1 mRNA stability. These findings could reveal a novel insight for KIAA1429 on OSCC tumorigenesis via m6A-dependent manner.
Methods
Cells and culture
Normal oral epithelium keratinocytes (HOK) and oral tumor cells (SCC-9, CAL-27) were provided from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM added with 10% fetal bovine serum (FBS) and penicillin-streptomycin (1% streptomycin, 1% penicillin, GE Healthcare, Chicago, IL, USA).
Transfection
The vector expressing KIAA1429 full-length sequences or stable silenced sequences was obtained from GenePharma (GenePharma Tech, Shanghai, China). CAL-27 cells were stably transfected with KIAA1429 over-expressing vectors (oe-NC, oe-KIAA1429). SCC-9 cells were stably transfected with KIAA1429 silencing vectors (sh-NC, sh-KIAA1429–1, sh-KIAA1429–2). The down/upregulated expression of KIAA1429 was confirmed by qRT-PCR or western blotting
Real-time quantitative PCR (RT-qPCR)
Total RNAs were isolated/extracted from OSCC cells using Trizol (Sigma, USA) and cDNA reverse transcription was conducted using PrimeScript RT reagent kit (Takara, Dalian, China). The qPCR assay was carried out using SYBR-Green (Vazyme, China). The relative expression was normalized to beta-actin expression and calculated by 2−ΔΔCt, where ΔCt = Ct (gene of interest) − Ct (housekeeping gene). The primer information was shown as follows: KIAA1429, forward ATACTGATGGTCTGGTGCTAAGA, reverse TGGAGGGCTTCCATTAAACTGAT; beata-actin, forward, 5′-CTCCATCCTGGCCTCGCTGT-3′, reverse, 5′- GCTGTCACCTTCACCGTTCC-3′.
Western blot
Cells with the indicated treatment were subjected to protein isolation by RIPA reagents supplemented with 1% proteinase inhibitor (Sigma, USA) and quantified by BCA Protein Assay Kit (Thermo Fisher Scientific). Proteins (∼30 μg) were electrophoresed to 12% SDS-PAGE gels and then transferred to PVDF membranes. Membranes were blocked in BSA (5%) for 2 h and then incubated with primary antibodies (anti-KIAA1429, Cell Signaling Technology, #88,358, 1:1000; anti-PGK1, Cell Signaling Technology, #63,536, 1:1000) overnight at 4 °C. Then, proteins were carefully incubated with secondary antibody for 1 h. Membranes were washed with TBST solution three times. Lastly, signals were detected by ECL detection system (Bio-Rad, California, USA).
Proliferation assay
Colony formation assay was performed to reflect the proliferation of OSCC cells. In short, the both non-transfected and transfected were seeded at the density of 1000 cells/well. After 14 days, the cells were fixed with paraformaldehyde (4%, 30 min) and then stained with 0.1% crystal violet (Beyotime, Shanghai, China) for 30 min. Next, the plates were washed mildly with PBS before being air-dried, and the stained colonies were photographed using a high-resolution camera. The experiments were performed at least in triplicate.
Glucose uptake, lactate production and ATP analysis assay
The transfected OSCC cells were cultured in glucose-free culture medium and then replaced by high-glucose culture medium under normoxic condition. The medium supernatants of OSCC cells were collected for following analysis. The glucose uptake was measured by glucose assay kit (BioVision, Milpitas, California, USA). The lactate production level was measured by lactate assay kit (BioVision) according to the manufacturer's instructions. ATP level was determined by ATP Determination Kit (Thermo Fisher Scientific, Cat. A22066) according to the manufacturer protocol.
Measurement of extracellular acidification rate (ECAR) and oxygen consumption rate (OCR)
The ECAR was detected using a Seahorse XF Glycolysis Stress Test Kit, respectively on Seahorse XF-96 metabolic flux analyzer (Seahorse Bioscience, North Billerica, MA, USA). The OCR was detected using a Seahorse XF Cell Mito Stress Test Kit on Seahorse XF-96 metabolic flux analyzer. In short, cells were seeded in the XF-96 cell culture microplates at 2 × 104 cells/well. For ECAR, cells were added with glucose (10 mM), oligomycin (1 mM) and 2-DG (80 mM). For OCR, wells were added with oligomycin (1 mM), FCCP (1 mM), antimycin A (2 mM) and Rotenone (2 mM). The data was evaluated using the Seahorse XF-96 Wave software.
Measurement of Fe2+ and lipid ROS
The intracellular iron assay for Fe2+ was detected using iron colorimetric assay kit (Applygen, Beijing, China, E1042) according to the product instructions. The lipid ROS level was detected using Dihydroethidium (Beyotime, #S0063) ROS red dye and FerroOrange (DOJINDO, #F374) on flow cytometer and observed by fluorescence microscope.
m6A quantification
The RNA was extracted from cells using TRIzol reagent (Invitrogen). The m6A quantification in total RNA was detected using m6A RNA methylation detection kit (ab185912, Abcam). In short, 200 ng sample RNA was added to the 96-well plates, as well as 50 mL diluted capture antibody and diluted enhancer solution. After the solution, the termination solution was supplemented to cells and read by microplate reader at 450 nm.
Methylated rna immunoprecipitation (MeRIP)-qPCR (MeRIP-PCR)
The MeRIP-PCR was performed to detect the m6A modification on genes or mRNAs. OSCC cells were harvested by ice-cold PBS twice and subsequently centrifugated at 4 °C for 5 min at 1500 rpm. Removing the supernatant, OSCC cells were mixed with 100 μL RIP lysis buffer and incubated with the lysate on ice. m6A antibody (Abcam, ab208577) was added to coat magnetic beads. Then, the m6A modification on PGK1 genes was tested using Magna MeRIP Kit (Millipore, Massachusetts, USA, #CR203146) according to the manufacturer's instructions. After rotation overnight at 4 °C, beads were washed by buffer and RNA was eluted with RIP wash buffer. The RNA enrichment was analyzed by qRT-PCR.
RNA immunoprecipitation (RIP)
The interaction within KIAA1429/YTHDF1 and PGK1 was identified using EZ-Magna RIP Kit (Millipore) according to the manufacturer's protocol. OSCC cells were lysed in complete RIP lysis buffer, and cellular extraction was incubated with protein A/G agarose beads. The beads were conjugated with specific antibodies (anti-YTHDF1, anti-KIAA1429) or control IgG at 4 °C for 2 h. Magnetic beads were bound with antibody in RIP Immunoprecipitation Buffer (Magna RIP Kit) with the extracted and fragmented RNAs. After immunoprecipitation, RNA was extracted subjected to qRT-PCR using primers for normalizing to input.
Fluorescence in situ hybridization (FISH)
The subcellular location was performed using the FISH assay. In short, the slide was deparaffinized with 100% xylene and rehydrated in series alcohol, following by 30 min of 4% (v/v) paraformaldehyde at room temperature. In situ hybridization was carried out using the Hybridization Kit (GenePharma Tech, Shanghai, China) according to the manufacturer's instructions. The slide was counterstained with 20 mL 4,6-diamidino-2-phenylindole (DAPI) to show the cell nucleus.
Stability analysis
Actinomycin D assay was performed to detect the stability of PGK1 mRNA. OSCC cells were seeded in 6-well plates (5 × 105 cells/well). Cells were exposed to 2 μg/ml Actinomycin D (Sigma) for 24 h, and RNA was collected at indicated time points (0, 5, 10 h, or 0, 4, 8 h). The RNA remaining level was analyzed using qRT-PCR and normalized to 0 h values.
Statistical analysis
All these data were expressed as mean value ± standard error (SD) and analyzed by GraphPad Prism 9.0 (GraphPad Software Inc., La Jolla, CA, USA) SPSS 23.0 (SPSS, IBM Corporation, Chicago, IL, USA). The differences between groups were calculated by one-way analysis of variance (ANOVA) and the Student t-test. P<0.05 was considered as statistical significance.
Results
KIAA1429 highly expressed in OSCC and associated with poor prognosis
To test the expression and function of KIAA1429 in OSCC, we search the dataset (HNSC, Head and Neck squamous cell carcinoma, http://gepia.cancer-pku.cn/detail.php?gene=KIAA1429) and found that KIAA1429 level increased in the tumor cohort group comparing to normal group (Fig. 1A). In OSCC cells, KIAA1429 level up-regulated, including mRNA and protein levels (Fig. 1B, 1C). For the clinical analysis, diseases free survival (Fig. 1D) and overall survival (Fig. 1E). In summary, these findings suggested that KIAA1429 highly expressed in OSCC and associated with poor prognosis.
Fig. 1.
KIAA1429 highly expressed in OSCC and associated with poor prognosis. (A) Public dataset (http://gepia.cancer-pku.cn/detail.php?gene=KIAA1429) displayed the expression level of KIAA1429 level in the tumor cohort group (HNSC, Head and Neck squamous cell carcinoma) comparing to normal group. (B) RT-PCR showed the KIAA1429 mRNA level in OSCC cells (SCC-9, CAL-27) and normal cells (HOK). (C) Western blot analysis showed the KIAA1429 protein in OSCC cells (SCC-9, CAL-27) and normal cells (HOK). (E) Diseases free survival and (F) overall survival illustrated the survival rate of OSCC patients. **p<0.01.
KIAA1429 promoted the aerobic glycolysis of OSCC cells
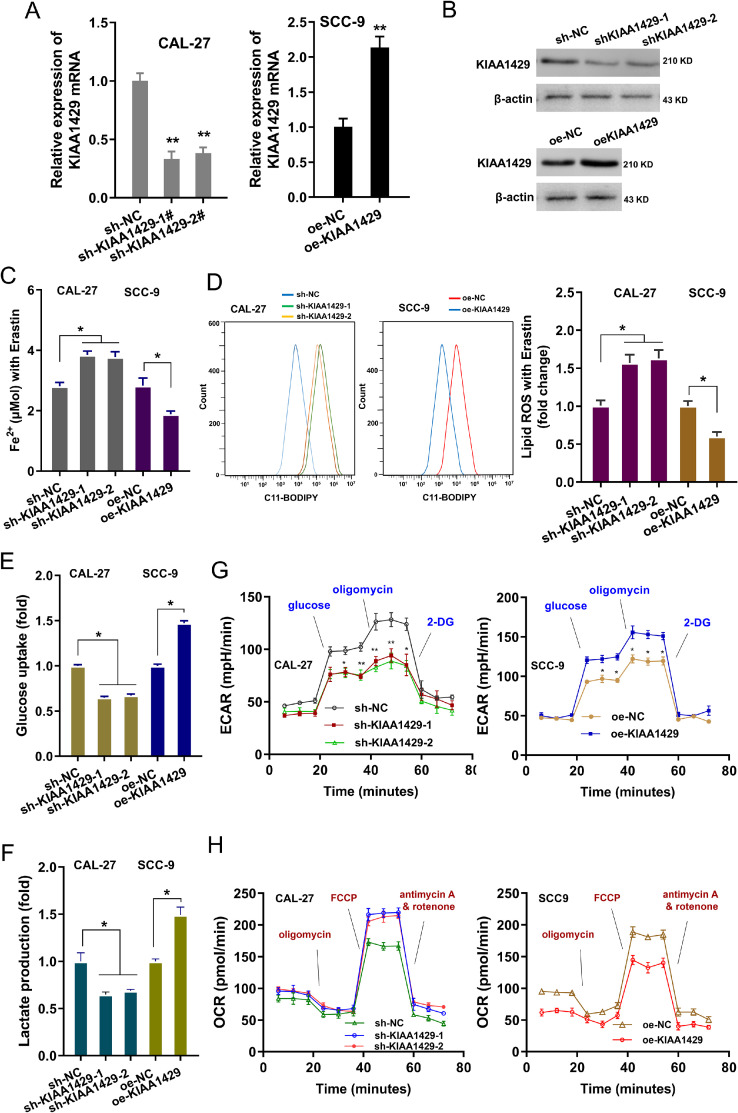
The functions of KIAA1429 were investigated in the OSCC cells (SCC-9, CAL-27). The silencing or overexpression of KIAA1429 was respectively constructed in SCC-9 or CAL-27 (Fig. 2A and B). The intracellular iron assay for Fe2+ indicated that KIAA1429 silencing up-regulated the Fe2+ concentration, and KIAA1429 overexpression repressed the Fe2+ concentration (Fig. 2C). Lipid ROS analysis revealed that KIAA1429 silencing up-regulated the lipid ROS level and KIAA1429 overexpression inhibited the lipid ROS level (Fig. 2D). The aerobic glycolysis of OSCC cells was detected using glucose uptake, lactate production, extracellular acidification rate (ECAR) and oxygen consumption rate (OCR). Results showed that KIAA1429 silencing repressed glucose uptake (Fig. 2E), lactate production (Fig. 2F), and ECAR (Fig. 2G). Besides, KIAA1429 overexpression up-regulated the glucose uptake (Fig. 2E), lactate production (Fig. 2F), and ECAR (Fig. 2G). Moreover, KIAA1429 silencing promoted the OCR, and KIAA1429 overexpression reduced the OCR (Fig. 2H). Taken together, these findings suggested that KIAA1429 promoted the aerobic glycolysis of OSCC cells.
Fig. 2.
KIAA1429 promoted the aerobic glycolysis of OSCC cells. (A) The silencing or overexpression of KIAA1429 was respectively constructed in SCC-9 or CAL-27. The KIAA1429 mRNA was detected using RT-PCR after transfection. (B) Western blot analysis revealed the KIAA1429 protein. (C) The intracellular iron assay for Fe2+ illustrated the Fe2+ level of SCC9 cells with KIAA1429 silencing (sh-KIAA1429–1, sh-KIAA1429–2), and CAL-27 cells with KIAA1429 overexpression (oe-KIAA1429). (D) Lipid ROS analysis was performed to reveal the lipid ROS level of SCC9 cells with KIAA1429 silencing (sh-KIAA1429–1, sh-KIAA1429–2), and CAL-27 cells with KIAA1429 overexpression (oe-KIAA1429). (E) Glucose uptake (F) lactate production (G) extracellular acidification rate (ECAR) and (H) oxygen consumption rate (OCR) were detected in SCC9 cells with KIAA1429 silencing (sh-KIAA1429–1, sh-KIAA1429–2), and CAL-27 cells with KIAA1429 overexpression (oe-KIAA1429). **p<0.01; *p<0.05.
PGK1 acted as the target of KIAA1429 in OSCC
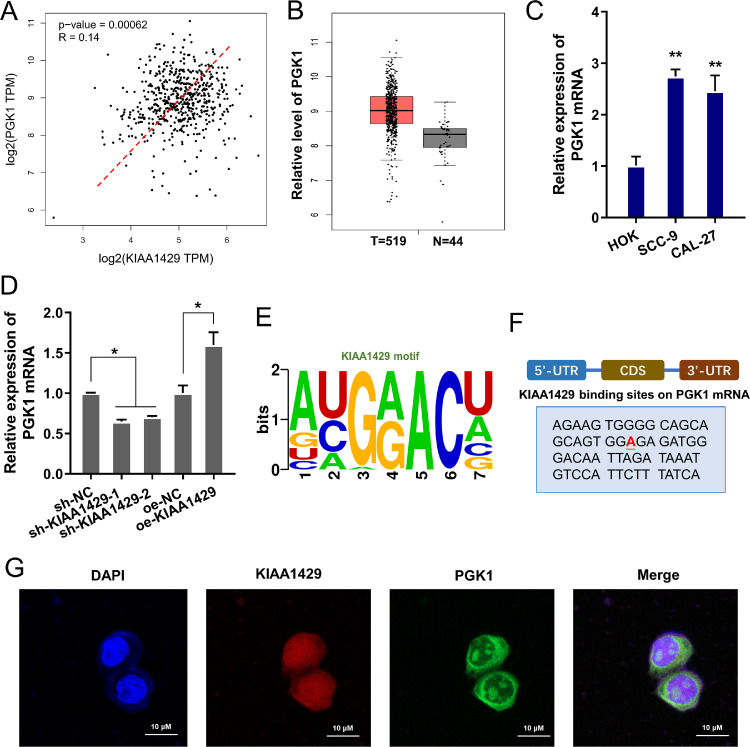
Given that KIAA1429 regulated the aerobic glycolysis of OSCC cells, further research focused on the potential targets for KIAA1429. Phosphoglycerate kinase 1 expression (PGK1) was a glycolytic enzyme regulating tumor cells’ energy metabolism [11,12]. Here, we found that PGK1 positively correlated to the KIAA1429 in tumor cohort group (HNSC) (Fig. 3A). Besides, the PGK1 level up-regulated in the tumor group (Fig. 3B). In OSCC cells, the PGK1 level also up-regulated (Fig. 3C). The level of PGK1 mRNA was reduced upon KIAA1429 silencing and up-regulated upon KIAA1429 overexpression (Fig. 3D). The m6A motif site of PGK1 towards KIAA1429 was GGAC (Fig. 3E) and the binding site of KIAA1429 on PGK1 mRNA was identified (Fig. 3F). The subcellular location of KIAA1429 and PGK1 was detected using FISH, showing the subcellular interaction within KIAA1429 and PGK1 (Fig. 3G). Taken together, these findings suggested that PGK1 acted as the target of KIAA1429 in OSCC.
Fig. 3.
PGK1 acted as the target of KIAA1429 in OSCC. (A) The trends relation of KIAA1429 and PGK1 was analyzed in tumor cohort group (HNSC, Head and Neck squamous cell carcinoma). The data was from (http://gepia.cancer-pku.cn/index.html). (B) The expression level of PGK1 in the tumor cohort group (HNSC) and normal group. (C) RT-PCR showed the PGK1 level in OSCC cells. (D) The level of PGK1 mRNA in OSCC cells with KIAA1429 silencing and KIAA1429 overexpression. (E) The m6A motif site on PGK1 towards KIAA1429. (F) The binding site of KIAA1429 on PGK1 mRNA was identified. (G) The subcellular location of KIAA1429 and PGK1 was detected using FISH. DAPI indicated the cell nucleus. **p<0.01; *p<0.05.
KIAA1429/YTHDF1 enhanced the mRNA stability of PGK1
Given that KIAA1429 positively regulated the fate of PGK1, we further investigated whether m6A reader participate in the course. We noticed that the canonical m6A reader YTHDF1 up-regulated in the OSCC tumor samples (Fig. 4A) and positively correlated to PGK1 (Fig. 4B). In the OSCC cells, YTHDF1 level increased (Fig. 4C). Emerging literature suggested that YTHDF1 could regulate the mRNA stability via m6A-dependent manner, therefore, we tried to investigate whether YTHDF1 mediated the PGK1 mRNA stability. Results indicated that KIAA1429 overexpression promoted the PGK1 mRNA remaining level, suggesting that PGK1 mRNA enhanced the stability of PGK1 mRNA (Fig. 4D). Moreover, the silencing of YTHDF1 reduced the level of PGK1 mRNA, while KIAA1429 overexpression up-regulated the PGK1 mRNA level (Fig. 4E). RIP-PCR analysis revealed that the PGK1 mRNA was significantly enriched by anti-YTHDF1 in OSCC cells, suggesting the molecular interaction within PGK1 and YTHDF1 (Fig. 4F). Moreover, the RIP-PCR analysis showed that KIAA1429 silencing reduced the interaction within PGK1 and YTHDF1, while KIAA1429 overexpression increased the interaction within PGK1 and YTHDF1 (Fig. 4G). Taken together, these findings suggested that KIAA1429/YTHDF1 enhanced the mRNA stability of PGK1.
Fig. 4.
KIAA1429/YTHDF1 enhanced the mRNA stability of PGK1. (A) Public dataset showed the up-regulation of m6A reader YTHDF1 in the OSCC tumor samples. (B) The trends relation of YTHDF1 and PGK1 was analyzed in tumor cohort group (HNSC, Head and Neck squamous cell carcinoma). The data was from (http://gepia.cancer-pku.cn/index.html). (C) RT-PCR showed the YTHDF1 mRNA level in the OSCC cells. (D) RNA stability analysis revealed the PGK1 mRNA remaining level in OSCC cells with Act D treatment of KIAA1429 silencing (sh-KIAA1429–1, sh-KIAA1429–2), and CAL-27 cells with KIAA1429 overexpression (oe-KIAA1429). (E) RNA stability analysis revealed the PGK1 mRNA remaining level in CAL-27 cells with YTHDF1 knockdown (si-YTHDF1) or/and KIAA1429 overexpression (si-YTHDF1 + oe-KIAA1429). (F) RIP-PCR analysis revealed the PGK1 mRNA enrichment by anti-YTHDF1 in OSCC cells normalized to Input. (G) RIP-PCR analysis showed the PGK1 mRNA enrichment by anti-YTHDF1 in OSCC cells with KIAA1429 silencing (sh-KIAA1429–1, sh-KIAA1429–2), and CAL-27 cells with KIAA1429 overexpression (oe-KIAA1429) normalized to Input. **p<0.01; *p<0.05.
KIAA1429 targeted PGK1 to regulate OSCC aerobic glycolysis via YTHDF1/m6A-dependent manner
To identify the function of KIAA1429/YTHDF1/PGK1 on OSCC phenotype, rescue assay was performed in OSCC cells. Firstly, the PGK1 protein level was detected in the co-transfection, and results illustrated that PGK1 protein level increased upon PGK1 overexpression, which was reduced by YTHDF1 silencing or KIAA1429 silencing (Fig. 5A). The intracellular iron assay for Fe2+found that PGK1 overexpression reduced the Fe2+ level, and YTHDF1 silencing or KIAA1429 silencing up-regulated the Fe2+ level (Fig. 5B). Lipid ROS analysis illustrated that PGK1 overexpression reduced the lipid ROS level, and YTHDF1 silencing or KIAA1429 silencing up-regulated the lipid ROS level (Fig. 5C). Glucose analysis illustrated that PGK1 overexpression up-regulated the glucose uptake, while YTHDF1 silencing or KIAA1429 silencing reduced the role of PGK1 overexpression (Fig. 5D). Lactate analysis revealed that PGK1 overexpression increased the lactate production, and YTHDF1 silencing or KIAA1429 silencing inhibited the function of PGK1 overexpression (Fig. 5E). Moreover, extracellular acidification rate (ECAR) analysis showed that PGK1 overexpression enhanced the glycolysis rate and glycolytic capacity, while YTHDF1 silencing or KIAA1429 silencing reversed it (Fig. 5F). Then, oxygen consumption rate (OCR) analysis illustrated that PGK1 overexpression decreased the respiratory rate, while YTHDF1 silencing or KIAA1429 silencing rescued it (Fig. 5G). Taken together, these findings suggested that KIAA1429 targeted PGK1 to regulate OSCC aerobic glycolysis via YTHDF1/m6A-dependent manner.
Fig. 5.
KIAA1429 targeted PGK1 to regulate OSCC aerobic glycolysis via YTHDF1/m6A-dependent manner. (A) Western blot showed the PGK1 protein level in the CAL27 cells with transfection of PGK1 overexpression plasmids (PGK1), YTHDF1 knockdown (si-YTHDF1) and KIAA1429 knockdown (sh-KIAA1429). (B) The intracellular iron assay for Fe2+ illustrated the Fe2+ level of CAL27 cells with transfection of PGK1 overexpression plasmids (PGK1), YTHDF1 knockdown (si-YTHDF1) and KIAA1429 knockdown (sh-KIAA1429). (C) Lipid ROS analysis was performed to reveal the lipid ROS level of CAL27 cells with transfection or co-transfection. (D) Glucose analysis, (E) Lactate analysis, and (F) extracellular acidification rate (ECAR) were performed to detect the glucose uptake, lactate production, ATP level in CAL27 cells with transfection of PGK1 overexpression plasmids (PGK1), YTHDF1 knockdown (si-YTHDF1) and KIAA1429 knockdown (sh-KIAA1429). (G) Oxygen consumption rate (OCR) analysis indicated the basal respiratory rate and maximum respiratory rate in CAL27 cells. **p<0.01; *p<0.05.
Discussion
The pathologic basis of oral squamous cell carcinoma (OSCC) is a mechanism that is difficult to determine very well [13,14]. More and more researches begin to explore the basic molecular mechanism level [15]. With the rise of epigenetics research, we begin to explore the pathological mechanism of oral cancer in this new field.
Here, present research found that KIAA1429 up-regulated in the OSCC, acting as an oncogene in OSCC's tumorigenesis. The gain/loss functional assays revealed that KIAA1429 positively accelerated the proliferation. Importantly, our data found that KIAA1429 also promoted the aerobic glycolysis of OSCC cells, including glucose uptake, lactate production, ATP level, extracellular acidification rate (ECAR) and oxygen consumption rate (OCR). According to these findings, we can draw a conclusion that KIAA1429 aggravated the OSCC tumorigenesis through targeting energy metabolism [16].
The abnormal of energy metabolism acts as a critical hallmark for malignant tumors [17,18]. Aerobic glycolysis, as known as Warburg effect, is a way to obtain energy from sugar degradation metabolism in the presence of oxygen [19,20]. In multiple cancers, aerobic glycolysis serves as the major energy source no matter aerobic or hypoxic [21]. In OSCC, emerging evidence has indicated that aerobic glycolysis is a very important feature of tumorigenesis [22,23]. Thus, the research on the mechanism of glycolysis may bring a new breakthrough for OSCC. Therefore, our findings that KIAA1429 regulates OSCC aerobic glycolysis may bring about breakthroughs in the field.
N6-methyladenosine (m6A) is an abundant nucleotide modification on mRNA, however there is still few studies on its role in OSCC energy metabolism [24,25]. In OSCC, the roles of m6A on its pathophysiological process, especially aerobic glycolysis, are more and more important [26,27]. For instance, m6A reader IGF2BP2 up-regulates in OSCC and acts as a predictor of poor prognosis, and IGF2BP2 promotes the proliferation, migration and Warburg effect of OSCC cell by regulating HK2 mRNA stability [28]. Thus, the novel function of m6A on OSCC energy metabolism is with new attraction and research value.
PGK1 (Phosphoglycerate kinase 1) is a glycolytic enzyme to show a relevant impact of increased phosphoglycerate kinase 1 expression [29]. As one of key metabolic enzymes in the glycolytic, PGK1 catalyzes the reduction of pyruvate to lactate and the reduction of pyruvate to lactate, which correlated to tumorigenesis and malignant progression. Under OSCC's hypoxic conditions, phosphoglycerate PGK1 level is found to be upregulated, resulting in the potentiation of EMT enhancement and stem cell‑like properties.
Our findings showed that the KIAA1429 (m6A ‘writer’) installed the m6A modified level in PGK1 mRNA, and YTHDF1 (m6A ‘reader’) recognized this m6A site on PGK1 mRNA to up-regulate its stability. YTHDF1 is a canonical m6A ‘reader’, which could increase its target mRNAs’ stability [30]. Here, we found that KIAA1429 targeted PGK1 to regulate OSCC aerobic glycolysis and ferroptosis via YTHDF1/m6A-dependent manner.
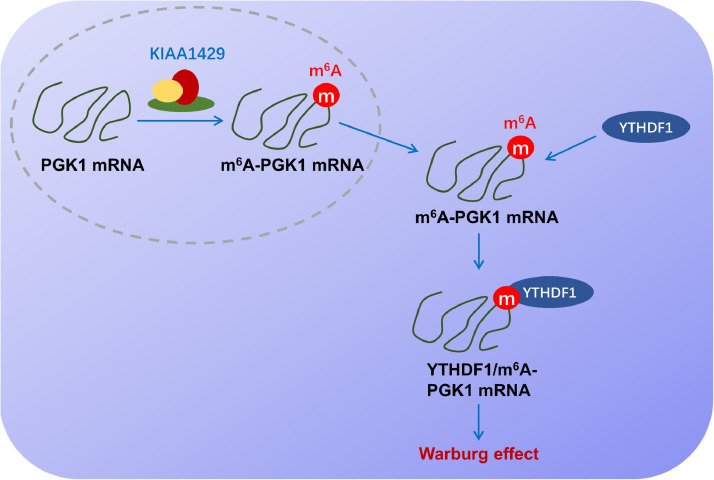
In conclusion, our work revealed a novel insight for KIAA1429 regulating OSCC energy metabolism and ferroptosis, suggesting the essential function of KIAA1429 on OSCC aerobic glycolysis. For the molecular mechanism, KIAA1429 installed the m6A modification on PGK1 mRNA, and YTHDF1 enhanced its mRNAs’ stability (Fig. 6). Overall, these findings might provide a therapeutic strategy for OSCC via m6A-dependent manner.
Fig. 6.
KIAA1429 promotes the aerobic glycolysis of oral squamous cell carcinoma via YTHDF1/m6A/PGK1 manner.
Funding
No funding was received.
Data availability
Not applicable.
Compliance with ethical standards
Ethics approval
The present study was approved by the ethical review committee of the Cangzhou Central Hospital. Patient consent for publications Not applicable.
CRediT authorship contribution statement
Ke Xu: Funding acquisition. Xiaojuan Dai: Supervision. Jincheng Yue: Supervision.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101745.
Appendix. Supplementary materials
References
- 1.Botha H., Farah C.S., Koo K., Cirillo N., McCullough M., Paolini R., Celentano A. The role of glucose transporters in oral squamous cell carcinoma. Biomolecules. 2021;11 doi: 10.3390/biom11081070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dolens E.D.S., Dourado M.R., Almangush A., Salo T.A., Gurgel Rocha C.A., da Silva S.D., Brennan P.A., Coletta R.D. The impact of histopathological features on the prognosis of oral squamous cell carcinoma: a comprehensive review and meta-analysis. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.784924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrari E., Pezzi M.E., Cassi D., Pertinhez T.A., Spisni A., Meleti M. Salivary cytokines as biomarkers for oral squamous cell carcinoma: a systematic review. Int. J. Mol. Sci. 2021;22(13):6795. doi: 10.3390/ijms22136795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen M., Wong C.M. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol. Cancer. 2020;19:44. doi: 10.1186/s12943-020-01172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L., Li H., Wu A., Peng Y., Shu G., Yin G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer. 2019;18:176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang N., Ding C., Zuo Y., Peng Y., Zuo L. N6-methyladenosine and neurological diseases. Mol. Neurobiol. 2022;59:1925–1937. doi: 10.1007/s12035-022-02739-0. [DOI] [PubMed] [Google Scholar]
- 7.Li D.Q., Huang C.C., Zhang G., Zhou L.L. FTO demethylates YAP mRNA promoting oral squamous cell carcinoma tumorigenesis. Neoplasma. 2022;69:71–79. doi: 10.4149/neo_2021_210716N967. [DOI] [PubMed] [Google Scholar]
- 8.Ko S.H., Jung Y. Energy metabolism changes and dysregulated lipid metabolism in postmenopausal women. Nutrients. 2021;13(12):4556. doi: 10.3390/nu13124556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D., Xu X., Ye Q. Metabolism and immunity in breast cancer. Front Med. 2021;15:178–207. doi: 10.1007/s11684-020-0793-6. [DOI] [PubMed] [Google Scholar]
- 10.Bunai K., Okubo H., Hano K., Inoue K., Kito Y., Saigo C., Shibata T., Takeuchi T. TMEM207 hinders the tumour suppressor function of WWOX in oral squamous cell carcinoma. J. Cell Mol. Med. 2018;22:1026–1033. doi: 10.1111/jcmm.13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Q., Yu Z. Phosphoglycerate kinase 1 (PGK1) in cancer: a promising target for diagnosis and therapy. Life Sci. 2020;256 doi: 10.1016/j.lfs.2020.117863. [DOI] [PubMed] [Google Scholar]
- 12.He Y., Luo Y., Zhang D., Wang X., Zhang P., Li H., Ejaz S., Liang S. PGK1-mediated cancer progression and drug resistance. Am. J. Cancer Res. 2019;9:2280–2302. [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson D.E., Burtness B., Leemans C.R., Lui V.W.Y., Bauman J.E., Grandis J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers. 2020;6:92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melo B.A.C., Vilar L.G., Oliveira N.R., Lima P.O., Pinheiro M.B., Domingueti C.P., Pereira M.C. Human papillomavirus infection and oral squamous cell carcinoma - a systematic review. Braz. J. Otorhinolaryngol. 2021;87:346–352. doi: 10.1016/j.bjorl.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danzi F., Pacchiana R., Mafficini A., Scupoli M.T., Scarpa A., Donadelli M., Fiore A. To metabolomics and beyond: a technological portfolio to investigate cancer metabolism. Signal Transduct. Target. Ther. 2023;8:137. doi: 10.1038/s41392-023-01380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finley L.W.S. What is cancer metabolism? Cell. 2023;186:1670–1688. doi: 10.1016/j.cell.2023.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aki T., Funakoshi T., Unuma K., Uemura K. Inverse regulation of GSDMD and GSDME gene expression during LPS-induced pyroptosis in RAW264.7 macrophage cells. Apoptosis Int. J. Program Cell Death. 2022;27:14–21. doi: 10.1007/s10495-022-01708-1. [DOI] [PubMed] [Google Scholar]
- 18.Gupta V.K., Kumar A. Targeting lysophosphatidic acid receptor with Ki16425 impedes T cell lymphoma progression through apoptosis induction, glycolysis inhibition, and activation of antitumor immune response. Apoptosis Int. J. Program Cell Death. 2022;27:382–400. doi: 10.1007/s10495-022-01723-2. [DOI] [PubMed] [Google Scholar]
- 19.Sharma A., Sinha S., Shrivastava N. Therapeutic targeting hypoxia-inducible factor (HIF-1) in cancer: cutting gordian knot of cancer cell metabolism. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.849040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y., Guo Y., Tam K.Y. Targeting glucose metabolism to develop anticancer treatments and therapeutic patents. Expert Opin. Ther. Pat. 2022;32:441–453. doi: 10.1080/13543776.2022.2027912. [DOI] [PubMed] [Google Scholar]
- 21.You Q., Wang J., Yu Y., Li F., Meng L., Chen M., Yang Q., Xu Z., Sun J., Zhuo W., Chen Z. The histone deacetylase SIRT6 promotes glycolysis through the HIF-1α/HK2 signaling axis and induces erlotinib resistance in non-small cell lung cancer. Apoptosis Int. J. Program Cell Death. 2022;27:883–898. doi: 10.1007/s10495-022-01751-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elzakra N., Kim Y. HIF-1α metabolic pathways in human cancer. Adv. Exp. Med. Biol. 2021;1280:243–260. doi: 10.1007/978-3-030-51652-9_17. [DOI] [PubMed] [Google Scholar]
- 23.Kato Y., Maeda T., Suzuki A., Baba Y. Cancer metabolism: new insights into classic characteristics. Jpn. Dent. Sci. Rev. 2018;54:8–21. doi: 10.1016/j.jdsr.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui Y., Liu J., Liu L., Ma X., Gui Y., Liu H., Zhao W. m(6)A-modified circFOXK2 targets GLUT1 to accelerate oral squamous cell carcinoma aerobic glycolysis. Cancer Gene Ther. 2023;30:163–171. doi: 10.1038/s41417-022-00526-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhao W., Cui Y., Liu L., Qi X., Liu J., Ma S., Hu X., Zhang Z., Wang Y., Li H., Wang Z., Liu Z., Wu J. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 2020;27:919–933. doi: 10.1038/s41418-019-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui Y., Liu J., Liu L., Ma X., Gui Y., Liu H., Zhao W. m(6)A-modified circFOXK2 targets GLUT1 to accelerate oral squamous cell carcinoma aerobic glycolysis. Cancer Gene Ther. 2022;30(1):163–171. doi: 10.1038/s41417-022-00526-6. [DOI] [PubMed] [Google Scholar]
- 27.Zhao W., Liu J., Wu J., Ma X., Wang X., Zhang L., Han Z., Yang J., Cui Y., Hu X., Deng J. High-throughput microarray reveals the epitranscriptome-wide landscape of m(6)A-modified circRNA in oral squamous cell carcinoma. BMC Genom. 2022;23:611. doi: 10.1186/s12864-022-08806-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu K., Dai X., Wu J., Wen K. N(6)-methyladenosine (m(6)A) reader IGF2BP2 stabilizes HK2 stability to accelerate the Warburg effect of oral squamous cell carcinoma progression. J. Cancer Res. Clin. Oncol. 2022;148:3375–3384. doi: 10.1007/s00432-022-04093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J., Shin D., Roh J.L. Lipid metabolism alterations and ferroptosis in cancer: paving the way for solving cancer resistance. Eur. J. Pharmacol. 2023;941 doi: 10.1016/j.ejphar.2023.175497. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z., Zhong X., Xia M., Zhong J. The roles and mechanisms of the m6A reader protein YTHDF1 in tumor biology and human diseases. Mol. Ther. Nucleic Acids. 2021;26:1270–1279. doi: 10.1016/j.omtn.2021.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.
Compliance with ethical standards