Abstract
Expression of EBNA-1 protein is required for the establishment and maintenance of the Epstein-Barr virus (EBV) genome during latent infection. During type I latency, the BamHI Q promoter (Qp) gives rise to EBNA-1 expression. The dominant regulatory mechanism for Qp appears to be mediated through the Q locus, located immediately downstream of the transcription start site. Binding of EBNA-1 to the Q locus represses Qp constitutive activity, and repression has been reported to be overcome by an E2F family member that binds to the Q locus and displaces EBNA-1 (N. S. Sung, J. Wilson, M. Davenport, N. D. Sista, and J. S. Pagano, Mol. Cell. Biol. 14:7144–7152, 1994). These data suggest that the final outcome of Qp activity is reciprocally controlled by EBNA-1 and E2F. Since E2F activity is cell cycle regulated, Qp activity and EBNA-1 expression are predicted to be regulated in a cell cycle-dependent manner. Proliferation of the type I latently infected cell line, Akata, was synchronized with the use of the G2/M blocking agent nocodazole. From 65 to 75% of cells could be made to peak in S phase without evidence of viral reactivation. Following release from G2/M block, EBNA-1 mRNA levels declined as the synchronized cells entered the G1 phase of the cell cycle. As cells proceeded into S phase, EBNA-1 mRNA levels increased parallel to the peak in cell numbers in S phase. However, EBNA-1 protein levels showed no detectable change during the cell cycle, most likely due to the protein’s long half-life as estimated by inhibition of protein synthesis by cycloheximide. Finally, in Qp luciferase reporter assays, the activity of Qp was shown to be regulated by cell cycle and to be dependent on the E2F sites within the Q locus. These findings demonstrate that transcriptional activity of Qp is cell cycle regulated and indicated that E2F serves as the stimulus for this regulation.
Following primary cytolytic infection of epithelial cells in the oropharynx, Epstein-Barr virus (EBV) infects B lymphocytes, in which it establishes latent infection. EBV can establish three types of latency (I, II, and III) associated with malignancy, each of which is characterized by the differential expression of a group of latency proteins. One of these proteins, Epstein-Barr nuclear antigen 1 (EBNA-1), is expressed during all three types of latent infection and is the only protein absolutely required for maintenance of latency (reviewed in reference 24). During latency, the EBV genome is maintained as an episome which requires EBNA-1 for replication (39, 69). EBNA-1’s function is to act in trans by binding to 24 sites clustered within the viral genome, called the origin of latent replication (oriP), and regulate episomal replication (4, 23, 46).
Although EBNA-1 expression is common to the three types of latency, the promoter used for its expression differs. During type III latency, EBNA-1 is expressed from the BamHI C and W promoters (Cp/Wp) (reviewed in reference 24). In type I and II latency, Cp/Wp are inactive, and EBNA-1 is expressed from the BamHI Q promoter (Qp) (38, 53, 63). In addition to latent expression, EBNA-1 mRNA is apparently transcribed during the viral lytic cycle from a fourth promoter called the BamHI F promoter (Fp) (25, 38, 52). Fp was originally misidentified as the promoter used for EBNA-1 expression during type I latency, but this conclusion was based on experiments in which a portion of the cells were undergoing spontaneous cytolytic replication (49, 54).
The control of Qp and EBNA-1 expression during type I latency has received intensive study. Qp appears to be controlled by two important regulatory elements. The first is an interferon-stimulated response element (ISRE) which lies upstream of the Qp transcriptional start site and is bound by members of the interferon regulatory factor (IRF) family (37, 51, 71). These factors include positive and negative regulatory proteins which appear to be important for the constitutive activity of Qp. Immediately downstream of the start site lie two binding sites for the EBNA-1 protein (4, 46). These sites, called the Q locus, are the only other EBNA-1 binding sites outside of oriP. They are lower-affinity binding sites relative to oriP, and binding of EBNA-1 to these sites represses Qp activity (4, 23, 50, 56, 57, 63). Furthermore, this repression appears to override the effects of positive regulatory elements that lie upstream of the Qp start site.
These data raise an important question: how is Qp activated during type I latency when EBNA-1 is present and able to repress Qp? We have reported that a member of the E2F family of cellular transcription factors upregulates Qp in the presence of EBNA-1 (56). Additionally, E2F-1 binds in vitro to sequences within the Q locus that partially overlap the two EBNA-1 binding sites. These data suggest that EBNA-1 and E2F control the final outcome of Qp activity. When EBNA-1 is bound to the Q locus, Qp is repressed. E2F can overcome repression by binding to the Q locus, displacing or competing with EBNA-1 binding and thereby activating Qp.
E2F constitutes a family of cellular transcription factors which regulate cellular promoters for proteins important in cell cycle progression. E2F transcriptional activity is cell cycle regulated (reviewed in references 2, 20, 32, and 34). In early G1, E2F exists in a complex with the underphosphorylated form of the retinoblastoma (Rb) tumor suppressor protein. As G1 proceeds, Rb becomes hyperphosphorylated, releasing E2F which is now able to transactivate its responsive genes (reviewed in references 2, 11, 20, 32, 34, and 65). Many of these genes, including those encoding DNA polymerase α, ribonucleotide reductase (RR), cyclins A and E, and HsOrc1, are responsible for synthesis of cellular DNA and/or progression of the cell cycle (12, 17, 41, 42, 48, 55). Presumably, one or more of these genes is involved in replication of EBV episomes which are replicated by host enzymes during latency (67, 69).
Since E2F appears to activate Qp, we hypothesized that Qp activity and expression of EBNA-1 are cell cycle regulated (56). To test this prediction, we first established a system to synchronize growth of a type I latently infected cell line. EBNA-1 message and protein levels were then measured following synchronization to determine if their expression is cell cycle regulated. We show that expression of EBNA-1 mRNA is clearly linked to cell cycle but that the level of EBNA-1 protein does not vary detectably, perhaps due to its long half-life. We also demonstrate that mutation of the E2F sites in Qp luciferase reporter constructs abolished cell cycle regulation of the promoter.
MATERIALS AND METHODS
Cell culture, synchronization, and fluorescence-activated cell sorting (FACS) analysis.
Akata (59), Raji (45), DG75 (5), X50-7 (66), and BL41 958 cells (19) were maintained in RPMI 1640–10% fetal bovine serum (FBS)–penicillin-streptomycin (pen-strep). Human embryonic lung fibroblasts (HELs; gift of S. Bachenheimer) and NIH 3T3 mouse fibroblasts were maintained in Dulbecco modified Eagle medium-high glucose (DMEM-H)–10% FBS–pen-strep.
Akata cells were synchronized in G2/M by treatment with nocodazole (0.04 μg/ml) for 14 h. Following block, cells were washed and replated with fresh RPMI 1640–10% FBS–pen-strep; samples were taken every 3 h. For each time point, 4 × 106 cells were fixed with 80% ethanol for FACS analysis. The remaining cells were harvested, and either total RNA or protein lysates were prepared.
HELs were synchronized by serum starvation in DMEM-H–1% FBS–pen-strep for 48 h and then replated in DMEM-H–10% FBS–pen-strep. At 0 and 18 h following replating, 4 × 106 cells were fixed with 80% ethanol for FACS analysis, and the remaining cells were harvested for protein lysates.
To determine DNA content and cell cycle status of each time point, 2 × 106 fixed cells were spun down, washed once with 1× phosphate-buffered saline (PBS)–1% bovine serum albumin, and resuspended in 1.12% sodium citrate–0.1% Triton X-100–2 μg of RNase A per ml–50 μg of propidium iodine per ml. Data were acquired on a Becton Dickinson FACSStarPlus and analyzed by using ModFit.
RNA preparation and EBNA-1 mRNA analysis.
Total RNA was prepared from time point samples and from control cell lines by using a Qiagen RNeasy midi kit. For Northern analysis of EBNA-1 mRNA, 50-μg aliquots of total RNA from synchronized Akata cells at each time point or controls were electrophoresed on 1% agarose-formaldehyde gels and transferred to nitrocellulose (BA85; Schleicher & Schuell). The nitrocellulose was cut in two, and EBNA-1 mRNA expression was detected in the top half with a riboprobe made from the EBNA-1 open reading frame (ORF) (minus Gly-Ala repeat). β-Actin expression was determined by probing the bottom half with a random-primed labeled cDNA probe (gift of W. E. Miller).
Use of CHX to estimate EBNA-1 protein half-life.
Akata and X50-7 cells were treated with cycloheximide (CHX; 50 μg/ml; Sigma) for 0, 1, 3, 6, 12, 24, 36, and 48 h. At each time point, cells were harvested, and protein lysates were prepared.
Protein lysates and immunoblotting for viral and cellular proteins.
Total cellular protein was prepared from control cell lines and time point samples. Cells were washed once with 1× PBS and then resuspended in lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 5 mM dithiothreitol, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, Complete [GIBCO-BRL]). After freezing and thawing three times, debris was spun down at 4°C for 15 min, and the supernatant fluid was transferred to new tubes. Protein concentration was determined by the Bradford protein assay.
For EBNA-1 Western analysis of synchronized cells, 75-μg aliquots of lysates from each time point were separated by polyacrylamide gel electrophoresis (PAGE) on a 8% sodium dodecyl sulfate (SDS)–polyacrylamide gel. For controls, 75 and 25 μg of DG75 and Raji lysates, respectively, were run. For CHX experiments, 100 μg of each CHX-treated Akata time point sample and 25 μg of each CHX-treated X50-7 time point sample were used. Additionally, 100 μg of DG75 and BL41 958 were run as controls. Proteins were transferred to either Immobilon-P (Millipore) or NitroPlus (MSI), and EBNA-1 protein was detected with monoclonal antibody (MAb) EBNA.OT1x (10) (gift of J. Middeldorp).
IRF-1 expression was determined by separating 25 μg of each CHX-treated X50-7 time point sample and 100 μg of each CHX-treated Akata time point sample on an SDS–8% polyacrylamide gel. As controls, 100 μg of DG75 and BL41 958 lysates were run. Following transfer to NitroPlus (MSI), IRF-1 protein was detected with polyclonal antibody (PAb) C-20 (Santa Cruz Biotechnology).
Expression of the EBV immediate-early protein Z was determined by separating 50 μg of total protein from each time point on an SDS–12% polyacrylamide gel, transferring the material to Immobilon-P (Millipore), and detecting Z protein with a BZLF-1 MAb.
For E2F-1 Western analysis, 50 μg of extract from each time point was electrophoresed on an SDS–10% polyacrylamide gel. As controls, 150 μg of serum-starved and released HEL extracts were run. Following transfer to Immobilon-P (Millipore), E2F-1 protein was detected with MAb KH95 (Santa Cruz).
For all Western analyses, proteins were visualized with either anti-mouse immunoglobulin (Ig)-horseradish peroxidase (Amersham) for MAbs EBNA.OT1x, BZLF-1, and KH95 or anti-rabbit Ig-horseradish peroxidase for PAb C-20 (Amersham) and enhanced chemiluminescence (Amersham).
Luciferase reporter constructs.
pQLUC was cloned by digesting the previously described pF2 chloramphenicol acetyltransferase (CAT) reporter construct (56) with BamHI and XbaI. The resulting Qp sequence corresponding to −173 to +115 relative to the Qp transcription start site was blunt-end cloned into the SmaI site of pGL-2 Basic (Promega). pQEcoCAT (56) was digested with BamHI and XbaI to release Qp sequence −173 to +115, which was then blunt-end cloned into the SmaI site of pGL-2 Basic. This construct, pQEcoLUC, is identical to pQLUC except for a 3-bp mutation in the downstream E2F site (QpE2Fb) within the Q locus. pQ2LUC was constructed by digesting the construct pQ2CAT, which contains Qp sequence −173 to +5, with HindIII and XbaI. The resulting Qp fragment was then blunt-end cloned into the SmaI site of pGL-2 Basic. The pHsOrc1-Luc(−1053) construct (41) was kindly provided by J. Nevins.
Transfection and synchronization of NIH 3T3 mouse fibroblasts and luciferase assays.
The day before transfection, NIH 3T3 cells were plated in two 100-mm-diameter dishes per time point at 5 × 106 cells per dish with normal growth medium (DMEM-H, 10% FBS, pen-strep). The following day each plate was cotransfected with 5 μg of one luciferase reporter construct and 1 μg of pCMV-β-gal (Clontech), using the SuperFect transfection reagent as specified by the manufacturer (Qiagen). The cells were allowed to recover for 18 to 20 h and were then placed in starvation medium (DMEM-H, 0.5% FBS, pen-strep) for 48 h. Following starvation, the cells were replated in growth medium and time points were taken every 6 h. For each time point, two 100-mm-diameter dishes were harvested, combined, and washed with 1× PBS. Half of the sample was used for FACS analysis as described for the Akata synchronizations, and the other half was used for protein extracts for β-galactosidase (β-Gal) and luciferase assays.
Luciferase assays were performed as previously described (41). Each sample was done in duplicate, and the amount of protein lysate used in each assay was normalized by β-Gal activity.
RESULTS
Synchronization of type I latently infected Akata cells.
To determine whether the expression of EBNA-1 mRNA and protein is coordinated with the cell cycle in type I latency, growth of Akata cells was synchronized with nocodazole. The Akata cell line was selected because it expresses the type I latency phenotype in which EBNA-1 mRNA arises exclusively from Qp (Fig. 1A and B). In our experience, this cell line is also tightly latent with virtually no cells undergoing spontaneous lytic replication, as indicated by the absence of Z-protein expression. Therefore, the EBNA-1 detected is not contaminated by lytic EBNA-1 expression thought to arise from Fp (Fig. 1A and B) (25, 38, 52).
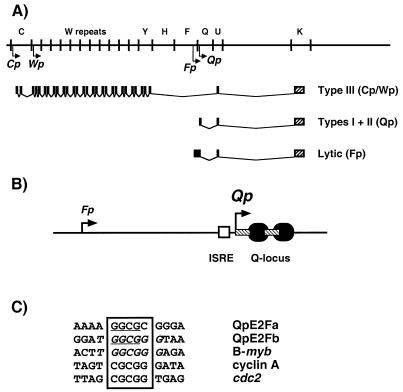
FIG. 1.
Promoters used for EBNA-1 expression. (A) The top line represents part of the EBV genome, some of the BamHI restriction fragments, and the promoters used for EBNA-1 expression (small arrows). The lower portion shows the splice structure of the EBNA-1 mRNAs and their latency expression patterns. Black rectangles represent exons, and the hatched rectangles designate the EBNA-1 ORF. (B) Structure of Qp. The large and small arrows designate the Qp (+1) and Fp (−193) start sites, respectively. EBNA-1 binding sites are represented as black ovals (+11 to +54); hatched rectangles represent the E2F-1 DNase I footprints (+1 to +14 and +24 to +43; QpE2Fa and QpE2Fb, respectively) (56); the ISRE is depicted as the open rectangle (−18 to −5) (37, 51, 71). (C) Comparison of the E2F sites from the B-myb (6), cyclin A (55), and cdc2 (61) promoters with the Qp E2F sites, QpE2Fa and QpE2Fb (56). The box indicates the core element identified in the B-myb and cdc2 E2F sites. The italicized nucleotides show the matches between B-myb and QpE2Fb. The underlined sequence in QpE2Fa and QpE2Fb indicates the mutations of Nonkwelo et al. (36).
For all experiments described, cells were blocked with nocodazole and then allowed to progress through the cell cycle by replating in fresh drug-free medium (see Materials and Methods). Following drug removal, samples were taken every 3 h. For each time point, DNA content and cell cycle status were determined by staining with propidium iodide followed by FACS analysis. The remaining cells were harvested to prepare either total RNA or protein.
Nocodazole effectively synchronized Akata cells in the G2/M phase of the cell cycle. Consistently, 65 to 85% of Akata cells were blocked in G2/M, as determined by FACS analysis (Fig. 2A and see Fig. 4A, 0 h). Following drug removal, the cells proceeded through the cell cycle, with a majority of cells in G1 at 6 h and in S phase at 15 h (Fig. 2A and see Fig. 4A). Drug treatment had no effect on cell viability (data not shown).
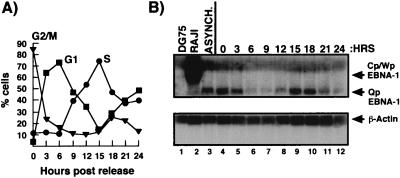
FIG. 2.
Expression of EBNA-1 mRNA is cell cycle regulated. (A) Following release from nocodazole block, cells from each 3-h time point were stained with propidium iodide and analyzed by FACS to determine cell cycle status. (B) Expression of EBNA-1 and β-actin was analyzed by Northern hybridization using 50 μg of total Akata RNA from each time point (lanes 4 to 12), asynchronous Akata cells (lane 3), Raji cells (lane 2), and DG75 cells (lane 1). The top panel demonstrates EBNA-1 mRNA expression. Arrows mark EBNA-1 mRNA initiating from Cp/Wp and Qp. The lower panel shows β-actin expression (marked with an arrow). This is a representative result from four separate time course experiments.
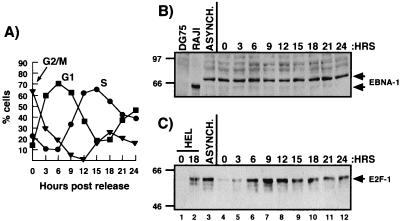
FIG. 4.
Level of EBNA-1 protein does not vary detectably with cell cycle in type I latency. (A) Cell cycle status of each time point following release of Akata cells from nocodazole block was determined by FACS analysis. (B) EBNA-1 expression was determined by Western analysis; 75-μg aliquots of total cellular protein from each time point (lanes 4 to 12), from asynchronous akata cells (lane 3), and from DG75 cells (lane 1) and 25 μg from Raji cells (lane 2) were used. (C) E2F-1 levels were determined by Western analysis; 50 μg of total protein from each time point along with 150 μg of 0-h HELs (serum starved; lane 1) and 18-h HELs (late G1/early S; lane 2) were electrophoresed. Sizes in panels B and C are indicated in kilodaltons.
EBNA-1 mRNA is expressed in a cell cycle-dependent manner during type I latency.
To determine the expression of EBNA-1 mRNA during the cell cycle, total RNA was prepared from synchronized cells at each 3-h time point. Equivalent amounts of RNA from each time point were electrophoresed on formaldehyde gels, and EBNA-1 expression was determined by Northern analysis with a riboprobe of the EBNA-1 ORF (Fig. 2B). Northern analysis demonstrated that EBNA-1 mRNA expression varies during the cell cycle in Akata cells. As cells enter G1, EBNA-1 expression decreases, with the least expression at the G1 peak or 6 h following release from nocodazole block (lane 6). Expression then increases and peaks, with a corresponding increase and peak in the percentage of S-phase cells at 15 h (lane 9). As controls for EBNA-1 expression, DG75, an EBV-negative Burkitt lymphoma line, lacks EBNA-1 expression, as expected, in contrast to the EBV-positive Raji cell line, in which EBNA-1 mRNA is expressed. Raji cells exhibit the type III latency program in which EBNA-1 expression arises from Cp/Wp (Fig. 1A). This EBNA-1 mRNA has a size of approximately 3.5 kb, in contrast to Akata EBNA-1 mRNA, which is approximately 2.5 kb, indicative of type I latency (49).
These data demonstrate that expression of EBNA-1 transcripts is regulated in a cell cycle-dependent manner during type I latency. Because Qp expresses EBNA-1 mRNA during type I latency, these data also suggest that Qp activity is cell cycle regulated. The decrease in EBNA-1 mRNA expression during G1 progression and its subsequent increase during S phase suggest that Qp is inactive at the G1 peak and becomes active as cells progress through S phase.
Nocadozole treatment does not induce the viral cytolytic cycle.
It is important to establish that synchronization of Akata cells with nocodazole does not induce the viral cytolytic cycle. Among the many agents that can induce latently EBV-infected cell lines to enter the lytic cycle is the cell cycle-blocking agent hydroxyurea, which induces Akata cells (data not shown). EBNA-1 mRNA is thought to be expressed during the lytic cycle from another promoter, Fp, which lies 193 bp upstream of the latent Qp start site (Fig. 1B). Both latent and lytic messages would have the same splice structure, with the Fp message containing an additional 193 nucleotides at its 5′ end (Fig. 1A) (25, 38, 49, 52–54). Therefore, Northern analysis does not permit the resolution of EBNA-1 messages arising from Qp and Fp.
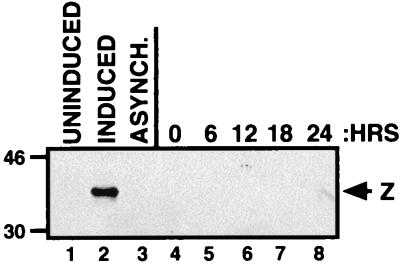
To determine if nocodazole induces the cytolytic cycle, Akata cells were synchronized by nocodazole treatment as described above. The cell cycle status at each time point is shown in Fig. 4A. From each 6-h time point, equivalent amounts of total cellular protein were separated by SDS-PAGE, and expression of the EBV immediate-early protein Z was determined (Fig. 3). Treatment of Akata cells with IgG induces the viral lytic cycle and the expression of Z (lane 2) (58). Figure 3 shows that Z is detected in neither the asynchronous Akata cells nor at any time point following release from nocodazole (lanes 3 to 8). Therefore, nocodazole does not induce the viral lytic cycle. It follows that the EBNA-1 mRNA detected in Fig. 2 is due to latent cell cycle-regulated expression from Qp, not from lytic Fp expression.
FIG. 3.
Treatment of Akata cells with nocodazole does not induce the viral lytic cycle. Expression of the immediate-early protein Z was determined by Western blotting as a marker for lytic induction. Fifty micrograms of total cellular protein was used for each time point following release from nocodazole block (lanes 4 to 8). Controls include 50 μg of total protein from uninduced Akata (lane 1), induced Akata (treated for 24 h with IgG; lane 2), and asynchronous Akata (lane 3) cells. Sizes are indicated in kilodaltons.
The steady-state level of EBNA-1 protein does not vary detectably during the cell cycle in type I latency.
To examine EBNA-1 protein expression throughout the cell cycle, Akata cells were again synchronized with nocodazole. From each 3-h time point, total protein was prepared, and expression of EBNA-1 protein was determined by Western analysis. FACS analysis demonstrates that at 0 h following release, a majority of the cells are in G2/M (Fig. 4A). The cells then progress into G1, with a G1 peak at 6 h, and then into S phase, with an S-phase peak at 15 h. Western analysis shows that there is no detectable change in the steady-state level of EBNA-1 protein during the cell cycle (Fig. 4B, lanes 3 to 12). The size difference of the EBNA-1 protein in Raji compared with Akata cells is due to the variable internal Gly-Ala repeat unit within the EBNA-1 protein (compare lanes 2 and 3) (21).
As a control for correct cell cycle expression, E2F-1 protein levels at each time point were analyzed by Western blotting (Fig. 4C). In fibroblasts, E2F-1 regulates its own promoter (22). Furthermore, its protein expression increases during G1 with a late G1/early S-phase peak. As a control for E2F-1 expression, HELs were synchronized in G0 by serum starvation. Serum-starved HELs do not express E2F-1 (lane 1). At 18 h following release from starvation, HELs enter late G1/early S phase and display E2F-1 expression (lane 2). Following release of Akata cells from nocodazole block, E2F-1 expression increases, with an increase in G1 cells, and peaks at 9 h, which corresponds to late G1/early S phase (lanes 4 to 7). These data demonstrate that nocodazole treatment does not affect the proper expression of a cell cycle-regulated protein. Furthermore, the apparent lack of cell cycle-dependent expression of EBNA-1 protein is not an artifact due to nocodazole treatment.
EBNA-1 has a long half-life.
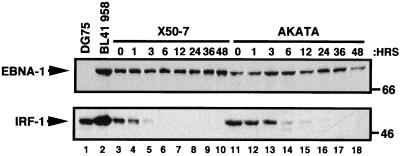
The lack of any detectable change in the expression of the EBNA-1 protein during the cell cycle may be due to its stability. To determine the half-life of the EBNA-1 protein, both Akata and X50-7 cells were treated with CHX to block protein synthesis. This approach allows the estimation of the half-life of endogenous EBNA-1 in both type I and type III latency. Following CHX treatment, total protein was prepared from cells at various time points, and the half-life of the EBNA-1 protein was estimated by Western analysis (Fig. 5, upper panel). In both Akata (type I) and X50-7 (type III) cells, the EBNA-1 protein appears to have a half-life in excess of 36 to 48 h. The data presented here extend those in a previous report which indicated that EBNA-1 is stable up to 20 h (26).
FIG. 5.
EBNA-1 protein has a half-life in excess of 36 to 48 h. In the upper panel, EBNA-1 expression levels were determined by Western analysis. For X50-7 (lanes 3 to 10) and Akata (lanes 11 to 18) cells, 25 and 100 μg of total cellular protein, respectively, were used from each time point. For controls, 100 μg of total cellular protein for both DG75 (lane 1) and BL41 958 (lane 2) cells were used. In the lower panel, the bottom half of the membrane used for the EBNA-1 analysis in the upper panel was probed for IRF-1 expression by Western analysis. Sizes are indicated in kilodaltons.
As an internal control, these same lysates were examined for the IRF-1 protein, which is reported to have a half-life of 30 min (Fig. 5, lower panel) (64). In X50-7 and Akata cells, the IRF-1 protein has apparent half-lives of 30 min and 2 h, respectively. The degradation of IRF-1 and stability of EBNA-1 demonstrate that the estimated half-life of EBNA-1 is not an artifact of CHX treatment.
Taken together, these data show that EBNA-1 has an extended half-life. The lack of any significant degradation of EBNA-1 up to 24 h covers the time span of the synchronization experiment in Fig. 4. This result would explain why there is an apparent lack of cell cycle-dependent expression of the EBNA-1 protein. Interestingly, this same result has been obtained for other E2F- and cell cycle-dependent genes, as discussed below.
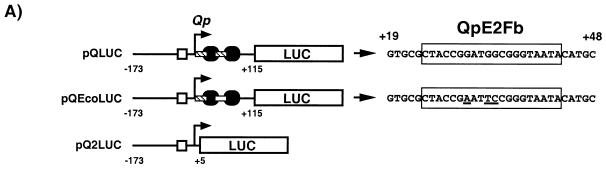
Mutation of the E2F sites within the Q locus abolishes cell cycle regulation of Qp activity.
To determine if Qp activity is dependent on the cell cycle, we made use of a luciferase reporter system which has demonstrated the cell cycle-dependent transcriptional activity of other E2F-responsive promoters (17, 41, 42, 55). Qp sequence from −173 to +115 relative to the Qp transcriptional start site was cloned into the luciferase reporter construct pGL-2 Basic (Fig. 6A, pQLUC). This construct contains the ISRE, the Qp start site, and the entire Q locus but lacks the Fp start site, which might obscure Qp activity. In addition, two other reporter plasmids were constructed as controls. The first, pQEcoLUC, is identical in sequence to pQLUC, with a 3-bp mutation of the downstream E2F site in the Q locus (QpE2Fb) to an EcoRI site (Fig. 6A). A CAT reporter construct containing this mutation has been shown to be repressed by EBNA-1. However, this EBNA-1-mediated repression could not be overcome by the overexpression of E2F-1 (56). The second construct, pQ2LUC, contains Qp sequence −173 to +5 and lacks both E2F binding sites as well as the two EBNA-1 binding sites of the Q locus. These constructs were cotransfected with a β-Gal expression vector into NIH 3T3 mouse fibroblasts, and the transfectants were serum starved in 0.5% serum for 48 h to synchronize the cells in G0/G1. Following starvation, the cells were released by the addition of normal growth medium and time points were taken every 6 h. From each time point, cells were sampled for FACS analysis as well as for β-Gal and luciferase assays.
FIG. 6.
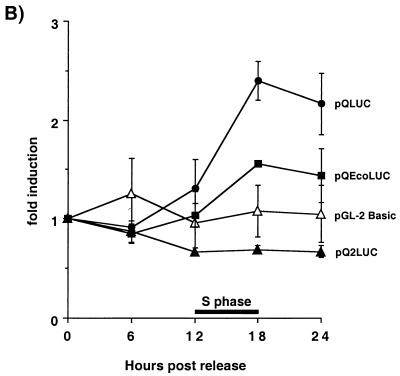
Qp activity is induced during S phase and is dependent on the E2F sites within the Q locus. (A) Structure of the Qp luciferase (LUC) reporter constructs. The bent arrow marks the Qp start site; the open square is the ISRE; black ovals and hatched rectangles are the EBNA-1 and E2F binding sites, respectively, within the Q locus. The upstream and downstream E2F sites are designated QpE2Fa and QpE2Fb, respectively, in Fig. 1 and the text. The nuleotide sequence of the QpE2Fb site (boxed sequence) in pQLUC and pQEcoLUC is shown at the right of each reporter construct. The three mutated base pairs within pQEcoLUC are underlined. (B) Fold induction of the Qp luciferase reporter constructs and the vector control, pGL-2 Basic. β-Gal assays were performed on each time point to normalize the amount of protein lysate used in the luciferase assays. The time of S phase is shown as a black bar spanning from 12 to 18 h after release from serum starvation.
Serum starvation arrested approximately 80 to 85% of the cells in G0/G1, as determined by FACS analysis (data not shown). The cells entered S phase 12 h following the addition of growth medium and traversed S between 12 and 18 h (data not shown). Concurrent with entry into and progression through S phase, luciferase activity of pQLUC was induced an average of 2.4-fold (Fig. 6B). Results shown summarize three experiments. In another experiment where the cells had been synchronized by serum starvation with 0.1% serum, 2.5-fold activation of pQLUC was observed (data not shown). In contrast to pQLUC, there was no induction of luciferase activity of pQ2LUC, which lacks both E2F sites. Additionally, there was diminished activity of the pQEcoLUC construct, which retains one E2F site. Both results are the average of two independent experiments (Fig. 6B). Identical results were obtained for both promoter constructs in an experiment where the transfectants were synchronized with 0.1% serum: 1.7-fold induction of pQEcoLUC and no induction of pQ2LUC (data not shown). These data indicate that Qp transcriptional activity is linked to the cell cycle through the two E2F sites which lie within the Q locus. Furthermore, they suggest that the cell cycle-regulated expression of EBNA-1 mRNA during type I latency is at the level of transcription and not due to mRNA stability.
As a positive control, the E2F-responsive pHsOrc1-Luc(−1053) construct was used to demonstrate the efficiency of the transfection and synchronization method. HsOrc1 is the human homolog of the yeast ORC1 protein, which is involved in DNA replication. The promoter for the HsOrc1 gene contains E2F sites which link HsOrc1 expression to the cell cycle (41). As previously reported, pHsOrc1-Luc(−1053) demonstrated cell cycle activity following release from serum starvation (41). As the cells proceeded into and through S phase, luciferase activity increased approximately 7.6- and 10-fold at 18 and 24 h, respectively, validating the system (data not shown).
DISCUSSION
We have reported that a member of the E2F family activates Qp and have predicted that Qp activity and EBNA-1 expression might therefore be cell cycle regulated (56). This report demonstrates that expression of EBNA-1 mRNA is clearly regulated by the cell cycle during type I latency, with mRNA levels lowest during G1 and subsequently increasing and peaking during S phase. This result suggests that Qp activity is also cell cycle regulated, with corresponding activity during G1 and S phases. Consistent with this hypothesis, Qp activity was induced during S phase in synchronized NIH 3T3 cells. In addition, the induction of Qp was diminished by mutation of the QpE2Fb site and abolished by deletion of both E2F binding sites within the Q locus. Taken with our earlier report (56), these results indicate that an E2F protein probably plays a key role in Qp activity and are consistent with the idea that E2F and EBNA-1 act reciprocally to regulate the promoter.
The E2F family of transcription factors includes six E2F proteins (E2F-1, -2, -3, -4, -5, and -6) as well as two E2F-related proteins, DP-1 and DP-2, which heterodimerize with the E2Fs (reviewed in references 2, 8, 16, 20, 34, and 62). Although the exact function of each E2F remains unclear, it is known that E2F-1, -2, and -3 become activated during the G1 phase of the cell cycle upon their release from hyperphosphorylated Rb (reviewed in references 2, 20, and 33). Availability of these E2Fs leads to the transactivation of many genes required for cellular DNA replication and progression of the cell cycle (12, 17, 22, 41, 42, 48, 55). It is interesting that expression of the E2F-1 protein, which was used as a control for proper cell cycle expression in synchronized Akata cells, occurs during G1, with a peak in late G1 (Fig. 4), whereas the induction of EBNA-1 mRNA expression begins and the levels peak in S phase, 6 h later (Fig. 2). This apparent discrepancy can be explained. The induction and peak of expression of many E2F-dependent genes occur at different points in the cell cycle (reviewed in reference 32). For example, promoter activity and expression of both cyclin E and cyclin A are dependent on E2F (12, 17, 42, 55). However, their expression patterns differ. Cyclin E mRNA expression is induced during G1 and peaks at the G1/S boundary (13, 27). In contrast, cyclin A expression is induced at the G1/S boundary and does not peak until late S to G2 phase, similar to EBNA-1 mRNA levels (13, 18, 44). In another system, the mRNA expression of both cyclins can be induced by E2F-1 expression, although cyclin A is expressed much later than cyclin E (12). These data demonstrate the complexities of E2F regulation. Qp activity and EBNA-1 mRNA expression could be regulated by any member of the E2F family. Furthermore, expression of EBNA-1 mRNA may be dependent on E2F-1 but not necessarily mirror E2F-1 expression.
Although EBNA-1 mRNA levels clearly change, the steady-state level of the protein does not appear to vary with cell cycle due to the stability of EBNA-1. This is not unprecedented. Cellular genes whose mRNA expression is cell cycle dependent, but without corresponding changes in protein levels, have been described. For example, the mRNA expression for both subunits of ribonucleotide reductase (RR) is induced by E2F and regulated in a cell cycle-dependent manner (7, 12, 29). In proliferating cells, both mRNAs are low in G1 and increase during S phase. However, protein levels of the R1/M1 subunit of RR do not appear to change (29). The authors conclude that this lack of cell cycle-dependent expression is due to the long half-life of the R1/M1 protein (15 to 24 h) (7, 14, 29). In contrast to R1/M1, the R2/M2 subunit of RR has a half-life of 3 h, and protein levels vary with cell cycle in proliferating cells (7, 15). Similar to R1/M1, EBNA-1 is cell cycle regulated at the mRNA level (Fig. 2) but not detectably at the protein level (Fig. 4). The EBNA-1 protein has an extended half-life in excess of 36 to 48 h (Fig. 5). Another possibility is that there are small differences in EBNA-1 protein levels which can not be distinguished by the available assay method.
As part of this study, pulse-labeling experiments to examine EBNA-1 protein synthesis during the cell cycle were attempted without success in synchronized Akata cells. Cells were labeled with [35S]methionine for 3 h at the G2/M, G1, and S-phase time points (0, 6, and 15 h, respectively). EBNA-1 was then immunoprecipitated with either a MAb or one of several PAbs (generous gifts of J. Middeldorp). In contrast to type III cells, levels of EBNA-1 protein in Akata cells are typically 20- to 50-fold lower (data not shown). The large amount of Akata extract needed to immunoprecipitate EBNA-1 efficiently, as detected by Western analysis, resulted in heavy 35S background so that labeled EBNA-1 could not be distinguished.
Cell cycle regulation of EBNA-1 during type I latency may serve several functions in episomal maintenance and viral gene regulation. EBNA-1 functions as the origin-binding protein of EBV and is required for replication and maintenance of the viral episome during latency (39, 69). The episome is known to replicate once per cell cycle, concurrently with cellular DNA (1, 68). Therefore, cell cycle-dependent expression of EBNA-1 mRNA may ensure that a certain level of EBNA-1 is maintained to populate the 24 binding sites at oriP at the time when the episome is replicated.
The exact function of EBNA-1 at oriP is unknown. Possible functions include the recruitment of the host DNA polymerase during S phase or segregation of episomes to daughter cells during mitosis (40, 43, 47, 70). The cell cycle regulation of viral DNA replication in cycling B lymphocytes may not be mediated by the level of EBNA-1 protein because EBNA-1 is present throughout the cell cycle (Fig. 4). Regulation of replication by EBNA-1 must be at some other level. For example, EBNA-1 may interact with a component of the cellular DNA replication machinery in a cell cycle-dependent manner. A recent report has shown the interaction of EBNA-1 with RPA, the single-strand DNA-binding protein (70). Additionally, EBNA-1 may bind to oriP at precise times during the cell cycle. Interestingly, none of the sequences in the spaces between the 24 EBNA-1 binding sites in oriP contain E2F sites or bind E2F-1 in vitro (56). Although not detected in Fig. 4, cell cycle-dependent phosphorylation of EBNA-1 may regulate either of these or other unknown functions of EBNA-1.
A recent report claims that Qp is not cell cycle regulated (51). This conclusion was reached by methods similar to those used in this study but did not reveal variation in luciferase activity with cell cycle. However, Schaefer et al. (51) chose a construct which contains 1,730 nucleotides of sequence upstream of Qp which includes the Fp transcriptional start site plus sequence of unknown function. It has been demonstrated that Fp and Qp reporter constructs have multiple sites of transcriptional initiation (35). Therefore, it is not clear that Qp is the site of transcriptional initiation in this construct when transfected into NIH 3T3 cells. For the luciferase experiments presented in this report, Qp promoter sequences were cloned such that Fp and other extraneous viral sequences which might obscure Qp activity were not included. This construct, pQLUC, demonstrated a 2.4-fold induction in luciferase activity coincident with the progression of the synchronized NIH 3T3 cells through S phase. Interestingly, this is the same level of induction observed for the cyclin E promoter, which is also E2F responsive (17).
Schaefer et al. proposed a model for the alleviation of EBNA-1 repression and activation of Qp whereby an HMG-I(Y)-containing complex might bind directly downstream of the ISRE and overlap the transcriptional start site (51). This complex would also overlap the 5′ end of the upstream EBNA-1 binding site in the Q locus. Therefore, the putative HMG-I(Y)-containing complex could displace or inhibit the binding of EBNA-1 to the 5′ site of the Q locus. However, this model leaves important questions unanswered. First, it does not sufficiently explain how EBNA-1 is dislocated from the 3′ EBNA-1 binding site in the Q locus. Second, and more importantly, precise mutation of the sequences supposedly bound by HMG-I(Y) in vitro have absolutely no effect on promoter activity (36).
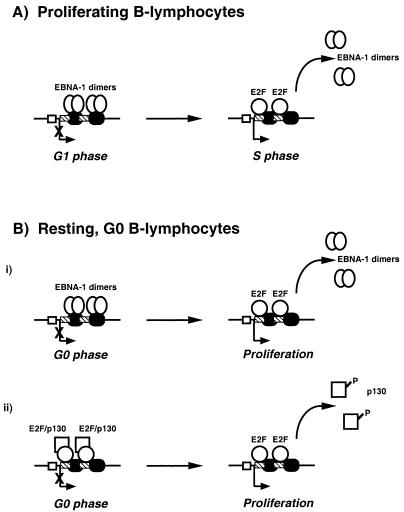
Other data support the idea that the E2F sites in Qp are required for full promoter activity (36). Nonkwelo et al. showed that in proliferating lymphocytes, mutation of the core sequences of either E2F-binding element (Fig. 1C, QpE2Fa and QpE2Fb) reduced promoter activity by 50 to 70% in the absence of EBNA-1 (36). These data raise the possibility that QpE2Fa and QpE2Fb have different roles in cycling versus noncycling cells (Fig. 7). In cycling B lymphocytes, E2F would overcome EBNA-1 repression and activate Qp as the cells enter S phase, similar to other E2F-responsive promoters (Fig. 7A). In resting, G0 B lymphocytes, E2F-responsive promoters are repressed by a complex of E2F (i.e., E2F-4 or -5) and a member of the pocket protein family (i.e., p107 or p130) (reviewed in references 2, 20, 33, and 34). Therefore, these sites in Qp may be required for repression of Qp during G0. Interestingly, QpE2Fa and QpE2Fb resemble the E2F sites in the cyclin A, B-myb, and cdc2 promoters (Fig. 1C) (6, 28, 55). In particular, none of these E2F elements has the characteristic thymidine residues flanking the 5′ end of the consensus sequence (reviewed in references 2 and 33). Additionally, QpE2Fb matches the B-myb E2F site over seven consecutive base pairs. Furthermore, the E2F sites in both the B-myb and cdc2 promoters appear to be important for repression, as both are occupied during G0 (61, 72). As cells move through G1, these sites are vacated, and expression of both genes is induced. Taken together with the data of Nonkwelo et al. (36) and Sung et al. (56), the data presented in this study suggest that the E2F sites in Qp could be important for the repression of Qp in a nonproliferating, G0 cell (Fig. 7B). This may be the situation in type 0 latency.
FIG. 7.
Model for Qp regulation. (A) Qp regulation in proliferating (cycling), type I B lymphocytes. (B) Regulation of Qp in resting (noncycling), type 0 latency. The bent arrow represents Qp, and the “X” indicates repression of transcription. The hatched rectangles represent the E2F binding sites, and the black ovals indicate the EBNA-1 binding sites in the Q locus. The white rectangle shows the ISRE. Either resting, G0 B-lymphocyte model (i or ii) could then return to the situation in proliferating B lymphocytes (A).
Synthesis of EBNA-1 dependent on the cell cycle may be pertinent in the normal carrier state for EBV in the human host. Others have reported that in the immunocompetent host, EBV episomes reside in a population of small resting G0 B lymphocytes that express little or no EBNA-1 message as detected by reverse transcription-PCR (9, 30, 31, 60). Taken together, these data suggest that in the normal carrier state, EBV episomes exist in nonproliferating B lymphocytes that do not express EBNA-1 message, termed type 0 latency. While EBNA-1 mRNA is not transcribed, detection of EBNA-1 protein is not possible in these cells. However, due to the protein’s stability, preexisting subdetectable amounts of EBNA-1 protein that bind to the Q locus might be present. In these G0 cells, E2F would be transcriptionally inactive, and Qp would also be inactive due to EBNA-1 protein bound at the Q locus (Fig. 7B, model i). As these cells begin to proliferate due to some physiological stimulus, E2F would become active, bind to the Q locus, compete with or displace EBNA-1, and activate Qp. Alternatively, Qp may be inactive in G0 due to a repressive E2F complex bound to the Q locus (Fig. 7B, model ii). As in the first model, when the cells enter the cell cycle, p130 would be hyperphosphorylated, and E2F either would become transcriptionally active or would dissociate from the Q locus, similar to the B-myb and cdc2 promoters, leading to activation of Qp. In either model, EBNA-1 expression is tied to proliferation of the latently infected cell, the cell cycle, and DNA replication. This linkage would ensure that EBNA-1 expression occurs concurrently with cellular and episomal replication so that oriP can be populated by EBNA-1 and the episome can be maintained.
ACKNOWLEDGMENTS
We thank Steve Bachenheimer, Nancy Raab-Traub, Lishan Su, Eng- Shang Huang, and Shannon Kenney for reading the manuscript and helpful discussion of the data. We also thank Valerie Zacny and Luwen Zhang for careful review of the data. We are grateful to Jaap Middeldorp for his gift of MAb EBNA.OT1x and several anti-EBNA-1 PAbs. We thank Joe Nevins for kindly providing the pHsOrc1-Luc(−1053) reporter construct. FACS analysis was performed by Larry Arnold at the Flow Cytometry Core Facility of the University of North Carolina Lineberger Comprehensive Cancer Center and by Erlina Siragusa at the Hanes Lab Flow Cytometry Facility at Duke University.
This work was supported by grants CA19014 and AI42372-01 from the NIH.
REFERENCES
- 1.Adams A. Replication of Epstein-Barr virus genomes in Raji cells. J Virol. 1987;61:1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams P D, Kaelin W G. Transcriptional control by E2F. Semin Cancer Biol. 1995;6:99–108. doi: 10.1006/scbi.1995.0013. [DOI] [PubMed] [Google Scholar]
- 3.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- 4.Ambinder R F, Shah W A, Rawlins D A, Hayward G S, Hayward S D. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J Virol. 1990;64:2369–2379. doi: 10.1128/jvi.64.5.2369-2379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey J M, Cohen M M, Bentwitch Z, Ramot B, Klein E, Klein G. Establishment in culture of a new type of lymphocyte from a “Burkitt-like” lymphoma (line D.G.-75) Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 6.Bennett J D, Farlie P, Watson R J. E2F binding is required but not sufficient for repression of B-myb transcription in quiescent fibroblasts. Oncogene. 1996;13:1073–1082. [PubMed] [Google Scholar]
- 7.Bjorklund S, Skog S, Tribukait B, Thelander L. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry. 1990;29:5452–5458. doi: 10.1021/bi00475a007. [DOI] [PubMed] [Google Scholar]
- 8.Cartwright P, Muller H, Wagener C, Holm K, Helin K. E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene. 1998;17:611–623. doi: 10.1038/sj.onc.1201975. [DOI] [PubMed] [Google Scholar]
- 9.Chen F, Zou J-Z, DiRenzo L, Winberg G, Hu L-F, Klein E, Klein G, Ernberg I. A subpopulation of normal B cells latently infected with Epstein-Barr virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP-1. J Virol. 1995;69:3752–3758. doi: 10.1128/jvi.69.6.3752-3758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen M-R, Middeldorp J M, Hayward S D. Separation of the complex DNA binding domain of EBNA-1 into DNA recognition and dimerization subdomains of novel structure. J Virol. 1993;67:4875–4885. doi: 10.1128/jvi.67.8.4875-4885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobrinik D. Regulatory interaction among E2Fs and cell cycle control proteins. Curr Top Microbiol Immunol. 1996;208:31–61. doi: 10.1007/978-3-642-79910-5_2. [DOI] [PubMed] [Google Scholar]
- 12.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulic V, Lees E, Reed S I. Association of human cyclin A with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 14.Engstrom Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. J Biol Chem. 1985;260:9114–9116. [PubMed] [Google Scholar]
- 15.Eriksson S, Grasland A, Skog S, Thelander L, Tribukait B. Cell cycle dependent expression of mammalian ribonucleotide reductase. The S phase correlated increase in subunit M2 is regulated by de novo protein synthesis. J Biol Chem. 1984;259:11695–11700. [PubMed] [Google Scholar]
- 16.Gaubatz S, Wood J G, Livingston D M. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc Natl Acad Sci USA. 1998;95:9190–9195. doi: 10.1073/pnas.95.16.9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng Y, Eaton E N, Picon M, Roberts J M, Lundberg A S, Gifford A, Sardet C, Weinberg R A. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- 18.Girard F, Strausfeld U, Fernandez A, Lamb N J C. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 19.Gulley M L, Raphael M, Lutz C T, Ross D W, Raab-Traub N. Epstein-Barr virus integration in human lymphomas and lymphoid cell lines. Cancer. 1992;70:185–191. doi: 10.1002/1097-0142(19920701)70:1<185::aid-cncr2820700129>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 20.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 21.Hennessy K, Kieff E. One of two Epstein-Barr virus nuclear antigens contains a glycine-alanine copolymer domain. Pro Natl Acad Sci USA. 1983;80:5665–5669. doi: 10.1073/pnas.80.18.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson D J, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 23.Jones C H, Hayward S D, Rawlins D R. Interaction of lymphocyte-derived Epstein-Barr virus nuclear antigen (EBNA-1) with its DNA-binding sites. J Virol. 1989;63:101–110. doi: 10.1128/jvi.63.1.101-110.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieff E. Epstein-Barr Virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 25.Lear A L, Rowe M, Kurilla M G, Lee S, Henderson S, Kieff E, Rickinson A B. The Epstein-Barr virus (EBV) nuclear antigen 1 BamHI F promoter is activated on entry of EBV-transformed B cells into the lytic cycle. J Virol. 1992;66:7461–7468. doi: 10.1128/jvi.66.12.7461-7468.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levitskaya J, Shapiro A, Leonchiks A, Ciechanover A, Masucci M A. Inhibition of ubiquitin/proteosome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc Natl Acad Sci USA. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lew D J, Dulic V, Reed S I. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 28.Liu N, Lucibello F C, Zwicker J, Engeland K, Muller R. Cell cycle-regulated repression of B-myb transcription: cooperation of an E2F site with a contiguous corepressor element. Nucleic Acids Res. 1996;24:2905–2910. doi: 10.1093/nar/24.15.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann G J, Musgrove E A, Fox R M, Thelander L. Ribonucleotide reductase M1 subunit in cellular proliferation, quiescence, and differentiation. Cancer Res. 1988;48:5151–5156. [PubMed] [Google Scholar]
- 30.Miyashita E M, Yang B, Babcock G J, Thorley-Lawson D A. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyashita E M, Yang B, Lam K M C, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 32.Muller R. Transcriptional regulation during the mammalian cell cycle. Trends Genet. 1995;11:173–178. doi: 10.1016/S0168-9525(00)89039-3. [DOI] [PubMed] [Google Scholar]
- 33.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 34.Nevins J R, Leone G, DeGregori J, Jakoi L. Role of the Rb/E2F pathway in cell growth control. J Cell Physiol. 1997;173:233–236. doi: 10.1002/(SICI)1097-4652(199711)173:2<233::AID-JCP27>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 35.Nonkwelo C, Daniel Henson E B, Sample J. Characterization of the Epstein-Barr virus Fp promoter. Virology. 1995;206:183–195. doi: 10.1016/s0042-6822(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 36.Nonkwelo C, Rif I K, Sample J. The Epstein-Barr virus EBNA-1 promoter Qp requires an initiator-like element. J Virol. 1997;71:354–361. doi: 10.1128/jvi.71.1.354-361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nonkwelo C, Ruf I K, Sample J. Interferon-independent and -induced regulation of Epstein-Barr virus EBNA-1 gene transcription in Burkitt lymphoma. J Virol. 1997;71:6887–6897. doi: 10.1128/jvi.71.9.6887-6897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nonkwelo C, Skinner J, Bell A, Rickinson A, Sample J. Transcription start sites downstream of the Epstein-Barr (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J Virol. 1996;70:623–627. doi: 10.1128/jvi.70.1.623-627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nonoyama M, Pagano J S. Separation of Epstein-Barr virus DNA from large chromosomal DNA in non-virus-producing cells. Nature. 1972;238:169–171. doi: 10.1038/newbio238169a0. [DOI] [PubMed] [Google Scholar]
- 40.Ohno S, Luka J, Lindahl T, Klein G. Identification of a purified complement-fixing antigen as the Epstein-Barr-virus-determined nuclear antigen (EBNA) by its binding to metaphase chromosomes. Proc Natl Acad Sci USA. 1977;74:1605–1609. doi: 10.1073/pnas.74.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohtani K, DeGregori J, Leone G, Herendeen D R, Kelly T J, Nevins J R. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petti L, Sample C, Kieff E. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology. 1990;176:563–574. doi: 10.1016/0042-6822(90)90027-o. [DOI] [PubMed] [Google Scholar]
- 44.Pines J, Hunter T. Human cyclin A is adenovirus E1A-associated protein p60 and behaves differently from cyclin B. Nature. 1990;346:760–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- 45.Pulvertaft R J V. A study of malignant tumors in Nigeria by short-term tissue culture. J Clin Pathol. 1965;18:261–271. doi: 10.1136/jcp.18.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 47.Reedman B M, Klein G. Cellular localization of an Epstein-Barr virus (EBV)-associated complement-fixing antigen in producer and non-producer lymphoblastoid cell lines. Int J Cancer. 1973;11:499–520. doi: 10.1002/ijc.2910110302. [DOI] [PubMed] [Google Scholar]
- 48.Sala A, Nicolaides N C, Englehard A, Bellon T, Lawe D C, Arnold A, Grana X, Giordana A, Calabretta B. Correlation between E2F-1 requirement in the S phase and E2F-1 transactivation of cell cycle-related genes in human cells. Cancer Res. 1994;54:1402–1406. [PubMed] [Google Scholar]
- 49.Sample J, Brooks L, Sample C, Young L, Rowe M, Gregory C, Rickinson A, Kieff E. Restricted Epstein-Barr virus protein expression in Burkitt lymphoma is due to a different Epstein-Barr nuclear antigen 1 transcriptional initiation site. Proc Natl Acad Sci USA. 1991;88:6343–6347. doi: 10.1073/pnas.88.14.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sample J, Daniel Henson E B, Sample C. The Epstein-Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J Virol. 1992;66:4654–4661. doi: 10.1128/jvi.66.8.4654-4661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaefer B C, Paulson E, Strominger J L, Speck S H. Constitutive activation of Epstein-Barr virus (EBV) nuclear antigen 1 gene transcription by IRF1 and IRF2 during restricted EBV latency. Mol Cell Biol. 1997;17:873–886. doi: 10.1128/mcb.17.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaefer B C, Strominger J L, Speck S H. The Epstein-Barr virus BamHI F promoter is an early lytic promoter: lack of correlation with EBNA-1 gene transcription in group I Burkitt’s lymphoma cell lines. J Virol. 1995;69:5039–5047. doi: 10.1128/jvi.69.8.5039-5047.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaefer B C, Strominger J L, Speck S H. Redefining the Epstein-Barr virus-encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc Natl Acad Sci USA. 1995;92:10565–10569. doi: 10.1073/pnas.92.23.10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schaefer B C, Woisetschlaeger M, Strominger J L, Speck S H. Exclusive expression of Epstein-Barr virus nuclear antigen 1 in Burkitt lymphoma arises from a third promoter, distinct from the promoters used in latently infected lymphocytes. Proc Natl Acad Sci USA. 1991;88:6550–6554. doi: 10.1073/pnas.88.15.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sung N S, Wilson J, Davenport M, Sista N D, Pagano J S. Reciprocal regulation of the Epstein-Barr virus BamHI-F promoter by EBNA-1 and an E2F transcription factor. Mol Cell Biol. 1994;14:7144–7152. doi: 10.1128/mcb.14.11.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sung N S, Wilson J, Pagano J S. Characterization of cis-acting regulatory elements of the BamHI-F promoter of EBV. In: Tursz T, Pagano J S, Ablashi D V, de The G, Lenior G, Pearson G R, editors. The Epstein-Barr virus and associated diseases. London, United Kingdom: INSERM/John Libbey Eurotext Limited; 1993. pp. 239–242. [Google Scholar]
- 58.Takada K. Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt’s lymphoma lines. Int J Cancer. 1984;33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- 59.Takada K, Horinouchi K, Ono Y, Aya T, Osato M, Takahashi M, Hayasaka S. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes. 1991;5:147–156. doi: 10.1007/BF00571929. [DOI] [PubMed] [Google Scholar]
- 60.Tierney R J, Steven N, Young L S, Rickinson A B. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tommasi S, Pfeifer G P. In vivo structure of the human cdc2 promoter: release of a p130-E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol Cell Biol. 1995;15:6901–6913. doi: 10.1128/mcb.15.12.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trimarchi J M, Fairchild B, Verona R, Moberg K, Andon N, Lees J A. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc Natl Acad Sci USA. 1998;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsai C-H, Liu S-T, Chang Y-S. Identification of a novel promoter located within the BamHI Q region of the Epstein-Barr virus genome for the EBNA-1 gene. DNA Cell Biol. 1995;14:767–776. doi: 10.1089/dna.1995.14.767. [DOI] [PubMed] [Google Scholar]
- 64.Watanabe N, Sakakibara J, Hovanessian A G, Taniguchi T, Fujita T. Activation of IFN-β element by IRF-1 requires a post-translational event in addition to IRF-1 synthesis. Nucleic Acids Res. 1991;19:4421–4428. doi: 10.1093/nar/19.16.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 66.Wilson G, Miller G. Recovery of Epstein-Barr virus from non-producer neonatal human lymphoid cell transformants. Virology. 1979;95:351–358. doi: 10.1016/0042-6822(79)90490-2. [DOI] [PubMed] [Google Scholar]
- 67.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr virus genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:821–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 70.Zhang D, Frappier L, Gibbs E, Hurwitz J, O’Donnel M. Human RPA (hSSB) interacts with EBNA1, the latent origin binding protein of Epstein-Barr virus. Nucleic Acids Res. 1998;26:631–637. doi: 10.1093/nar/26.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zwicker J, Liu N, Engeland K, Lucibello F C, Muller R. Cell cycle regulation of E2F site occupation in vivo. Science. 1996;271:1595–1597. doi: 10.1126/science.271.5255.1595. [DOI] [PubMed] [Google Scholar]