Abstract
The human immunodeficiency virus (HIV) integrase protein (IN) catalyzes two reactions required to integrate HIV DNA into the human genome: 3′ processing of the viral DNA ends and integration. IN has three domains, the N-terminal zinc-binding domain, the catalytic core, and the C-terminal SH3 domain. Previously, it was shown that IN proteins mutated in different domains could complement each other. We now report that this does not require any overlap between the two complementing proteins; an N-terminal domain, provided in trans, can restore IN activity of a mutant lacking this domain. Only the zinc-coordinating form of the N-terminal domain can efficiently restore IN activity of an N-terminal deletion mutant. This suggests that interaction between different domains of IN is needed for functional multimerization. We find that the N-terminal domain of feline immunodeficiency virus IN can support IN activity of an N-terminal deletion mutant of HIV type 2 IN. These cross-complementation experiments indicate that the N-terminal domain contributes to the recognition of specific viral DNA ends.
Integration of the viral DNA into a chromosome of an infected cell is essential for replication of the human immunodeficiency virus (HIV). The integration process is catalyzed by the integrase protein (IN) and consists of two steps. In the first step, usually two nucleotides are removed from the 3′ ends of the viral DNA (cleavage reaction). In the second step, the newly generated 3′ hydroxyl groups of the viral DNA are coupled to phosphate groups on opposite strands of the target DNA (integration or strand transfer reaction). The single-stranded gaps flanking the integrated viral DNA are repaired, probably by cellular enzymes. The resulting provirus is flanked by a 5-bp direct duplication of target DNA. (For recent reviews on retroviral integration, see references 1, 34, 50, and 69.)
Purified, recombinant IN mediates cleavage and integration of oligonucleotides that mimic the viral DNA ends (6, 10, 33). In the cleavage reaction, IN makes a specific phosphodiester bond near the viral DNA ends accessible for nucleophilic attack. In vitro, several hydroxyl group-containing compounds can serve as the nucleophile, such as water, glycerol, or the 3′ ends of the viral DNA (25, 73). The choice of nucleophile varies between different INs and can be affected by substitutions of residues around the active site (14, 21, 23, 46, 61, 64, 71). Besides cleavage and integration, IN can also mediate a phosphoryl transfer reaction, called disintegration (9). The substrate of the disintegration reaction is similar to the product of the integration of viral DNA ends into target DNA. IN releases the viral DNA in the disintegration reaction and restores the target DNA. In contrast to the integration reaction, disintegration activity does not require specific viral DNA ends; IN can still catalyze this reaction when the viral DNA ends are replaced by a single A nucleotide (9) or by random sequences (60).
Two amino acid sequence motifs are conserved among retroviral and retrotransposon IN proteins (37, 38). The N-terminal domain contains a phylogenetically conserved motif, His(X3–7)His(X23–32)Cys(X2)Cys (13, 32, 37) that coordinates a zinc ion (5, 7). The precise role of the N-terminal domain in the integration process is unknown. Some studies suggested that it is involved in protein-protein interactions, whereas other experiments indicate that it contributes to substrate specificity. Structure elucidation of the N terminus showed that it consists of three-helix bundles that are stabilized by Zn2+ (8, 18). Although its fold is similar to that of DNA-binding proteins, such as the Trp repressor, no direct DNA binding by the N-terminal domain has been observed (37, 47, 56, 74). Zinc induces tetramerization in the full-length protein and enhances Mg2+-dependent IN activity (42, 43, 77). This suggests that the N-terminal domain either contributes to intersubunit contacts within the tetramer or has an allosteric effect on IN that favors tetramerization of the protein. Deletion of the N-terminal domain abolishes Mn2+-dependent aggregation of HIV type 1 (HIV-1) IN (20), again suggesting a possible role of the N-terminal domain in protein-protein interaction. Mutagenesis of the zinc-coordinating residues reduces cleavage up to 90%, whereas integration activity is less affected (23, 63). The HHCC motif may therefore be involved in correct positioning of the viral DNA ends for the 3′ processing reaction. Disintegration activity of mutants in the N-terminal domain on various substrates have indicated that the viral DNA sequences internal to the conserved CA are recognized by the N-terminal domain (67).
The second conserved motif of IN is located in the core of the protein (amino acids 50 to 212). This motif, the acidic triad DD(35)E, is found in all retroviruses, retrotransposons, and some transposases (23, 26, 38, 49, 54). Mutational analyses have shown that replacement of these acidic residues impairs all IN activities (7, 15, 23, 38, 41, 63). This indicates that the DD(35)E motif is part of a single active site of IN. Elucidation of the three-dimensional structure of the core of HIV-1 IN (16) and of avian sarcoma virus IN (3) revealed that IN is a member of a group of structurally related polynucleotidyl transferases (53, 76). It has been shown for avian sarcoma virus IN that two Asp residues of the active site coordinate one metal ion (Mn2+ or Mg2+) (4). The core domain of IN determines target site selection and is involved in binding of the viral DNA termini (27–29, 31, 35, 36, 57).
The C terminus of IN (amino acids 220 to 270) is the least conserved domain. It binds DNA nonspecifically (24, 37, 52, 68, 74), and its structure is similar to that of Src homology 3 (SH3) domains (17, 44). Mutations in the dimer interface of the C-terminal domain can affect oligomerization of the full-length protein (51).
Previously, it has been demonstrated that certain combinations of inactive proteins restore IN activity (22, 65). Mutations in different domains can rescue each other, indicating that each domain belongs to a separate complementation group. This suggests that IN is active as a dimer or higher-order oligomer. All three domains form dimers themselves (8, 16–18, 44). If the intraprotein complementation were to depend on this, one would predict that no functional IN can be formed when an N-terminal domain is mixed with an IN from which the complete N-terminal domain has been removed, since no oligomerization can happen between the two mutant proteins. To investigate whether the N-terminal domain can function in trans, we did complementation experiments using an N-terminal domain and an N-terminal deletion mutant. We show that a structural N-terminal domain restores IN activity of an N-terminal deletion mutant. This suggests that for IN activity, interaction between the N-terminal domain and another IN domain is required. In cross-complementation experiments between feline immunodeficiency virus (FIV) and HIV IN mutants, we demonstrate that the N terminus of FIV IN can restore IN activity of an N-terminal deletion mutant of HIV-2 IN. By using different substrates, we found that the N-terminal domain contributes to specific recognition of the viral DNA ends.
MATERIALS AND METHODS
Construction of protein expression vectors.
The HIV-2 IN gene of pRP279 (62) was cloned into pET-15b vector (Novagen, Madison, Wis.). In the resulting plasmid (pRP1013), the HIV-2 IN gene is fused at its 5′ end to the 3′ end of a His tag and a thrombin cleavage site. The IN gene of FIV was amplified by PCR from pRP817 (71) by using oligonucleotides 93M313 (5′-GGACGCATATGTCCTCTTGGGTTGACAGAATTG-3′) and 93M312 (5′-CCTGCGGATCCTCACTCATCCCCTTCAGGAAG-3′). The PCR product was purified, digested with NdeI and BamHI, and ligated into the NdeI/BamHI-digested vector pET3c, resulting in plasmid pRP824. A double-stranded oligonucleotide (4030/4031) containing codons for six His residues was cloned into NdeI-digested pRP824, resulting in plasmid pRP825. The sequence of oligonucleotide 4030 is 5′-TATGAGAGGATCGCATCACCATCACCATCACAGATC-3′; that of oligonucleotide 4031 is 5′-TAGATCTGTGATGGTGATGGTGATGCGATCCTCTCA-3′. The N-terminal deletion mutant of HIV-2 IN (amino acids 50 to 293) was cloned by PCR using 95A2537 (5′-CGGGGAACATATGCATGGGCAAGTAAATGCAGAAC-3′) as the forward primer and a reverse primer identical to the T7 terminator sequence (5′-GCTAGTTATTGCTCAGCGGTGGATCCATC-3′). The PCR product was cloned into pET-15b, resulting in plasmid pRP1707. The N terminus of FIV IN was amplified by PCR and fused in frame at its 5′ end to the 3′ end of the glutathione S-transferase gene in vector pGEX-2T, resulting in plasmid pRP1713. Cloning of the N-terminal domain of HIV-2 IN has been published previously (18). Mutagenesis of the N-terminal domain of HIV-2 IN was done by the same procedure as that described for site-directed mutagenesis (61). Mutant clones were verified by sequencing.
Protein expression and purification.
Full-length INs from HIV-2 and FIV were expressed and purified as described previously (61). Purification of the N-terminal domain of FIV IN was done as described for the N-terminal domain of HIV-2 IN (18). The N-terminal deletion mutant derived from HIV-2 IN was expressed and purified as follows: plasmid pRP1707 was introduced into E. coli BL21(DE3), containing the plasmid pLysS (55, 58). An overnight culture of this strain was diluted 1:200 in 2 liters of TB medium (55) containing ampicillin and chloramphenicol. The culture was grown at 37°C to an optical density at 615 nm of 0.8. Protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 0.4 mM. The bacteria were harvested after 3 h by centrifugation and resuspended in 30 ml of buffer A (50 mM HEPES [pH 7.5], 1 mM EDTA, 3 mM β-mercaptoethanol). The cell suspension was sonicated and centrifuged for 30 min at 15,000 × g (SS34 rotor; Beckman). The pellet was Dounce homogenized in 30 ml of buffer B (20 mM Tris-HCl [pH 6.7], 1 M NaCl, 0.1 mM EDTA, 3 mM β-mercaptoethanol) and rotated for 15 min at 4°C in the presence of 0.1% Tween 20–5 mM imidazole (pH 6.7). The supernatant was cleared by a 30-min spin at 15,000 × g (SS34 rotor; Beckman) and bound to 2 ml of Ni2+-nitrilotriacetic acid agarose beads (Qiagen) for 2 h. The beads were packed into a column, washed with 20 ml of buffer B, which had been supplemented with 0.1% Tween 20 and 20 mM imidazole (pH 6.7), and subsequently washed with 20 ml of buffer B containing 20 mM imidazole (pH 6.7). The protein was eluted with buffer B, which had been supplemented with 10% glycerol and 200 mM imidazole (pH 6.7). Fractions containing the N-terminal deletion mutant were dialyzed against buffer C (0.5 M NaCl, 20 mM Tris-HCl [pH 6.7], 3 mM β-mercaptoethanol), and the His tag was removed by thrombin digestion overnight (approximately 0.5 NIH units/mg of protein). The protein fraction that still contained a His tag was removed by the addition of 50 μl of Ni2+-nitrilotriacetic acid agarose beads (Qiagen). Thrombin was removed by incubating the protein with benzamidine-Sepharose 6B (Pharmacia). The purified protein was dialyzed to 1 M NaCl–20 mM Tris-HCl (pH 7.6)–1 mM EDTA and coupled to thiopropyl-Sepharose (Pharmacia). After 2.5 h of incubation, the beads were washed with buffer D (10 mM morpholinepropanesulfonic acid [MOPS; pH 7.2], 0.5 mM dithithreitol, 10% glycerol) and stored as a 50% slurry in buffer D at −80°C.
Size exclusion chromatography.
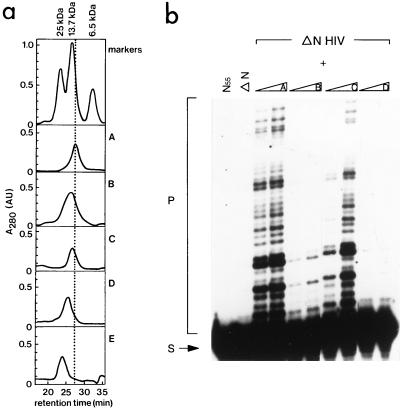
The oligomeric state of the N-terminal domains of the FIV and HIV-2 IN mutants was analyzed on a Superdex 75 HR 10/30 column (Pharmacia) in a buffer containing 150 mM NaCl, 50 mM Tris-HCl (pH 6.7), 3 mM β-mercaptoethanol, and 4 μM ZnCl2. After centrifugation of the sample at 15,000 rpm (in an Eppendorf Microfuge), 100 μl of 0.4 mM protein was injected. The size exclusion column was calibrated with the following globular proteins, unless stated otherwise: 100 μg of aprotinin (6.5 kDa), 125 μg of RNase A (13.7 kDa), and 55 μg of chymotrypsinogen (25 kDa). Protein elution was done at a flow rate of 0.5 ml/min and monitored by UV absorption at 280 nm. Several runs were performed for each protein, in parallel to a wild-type run. A representative profile is shown in Fig. 3a and 5a.
FIG. 3.
(a) Size exclusion chromatography of mutant proteins of the N-terminal domain of HIV-2 IN (amino acids 1 to 55). Proteins were injected on a Superdex 75 HR 10/30 column (Pharmacia), which had been equilibrated in buffer G (150 mM NaCl, 50 mM Tris-HCl [pH 6.7], 3 mM β-mercaptoethanol, 4 μM ZnCl2). The following markers were used: aprotinin (6.5 kDa, 100 μg), RNase A (13.7 kDa, 125 μg), and chymotrypsinogen (25 kDa, 55 μg). Elution profiles are of the following proteins: wild-type IN55 (profile A), mutant F1E;L2D;I5A (profile B), mutant F1E (profile C), and mutant H12L (profile D). Profile E is of the mutant F1E in IN1–55 in the presence of 4 mM EDTA. All proteins were injected at a concentration of 0.4 mM. The retention time in minutes is indicated on the X axis, and the absorption at 280 nm (A280, in arbitrary units) is shown in the Y axis. (b) Complementation between the N-terminal deletion mutant of HIV-2 IN (ΔN HIV) and several mutants of the N-terminal domain of HIV-2 IN (IN55). In the first and second lanes, only IN55 or ΔN is present in the reaction. The N-terminal deletion mutant was incubated with the mutants of profiles A to D of IN55 (see description for panel a), prior to the integration reaction. In each two-lane panel, increasing amounts of IN55 protein were added (20 and 200 pmol, respectively). The migration positions of the substrate (S) and strand transfer products (P) are indicated on the left.
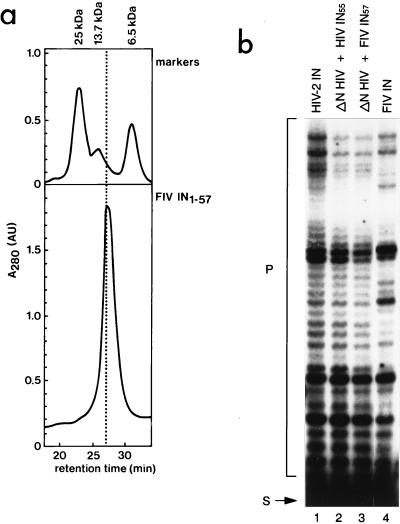
FIG. 5.
(a) Size exclusion chromatography of the N-terminal domain of FIV IN (IN57). FIV IN1–57 was injected at a concentration of 0.4 mM on a Superdex 75 HR 10/30 column (Pharmacia), which had been equilibrated in buffer G (see legend to Fig. 3a). Protein markers were injected prior to each experiment (see legend to Fig. 3a). The retention time (minutes is indicated on the X axis, and the absorption at 280 nm (A280, in arbitrary units) is depicted on the y axis. (b) Cross-complementation between FIV and HIV-2 IN mutants. Integration activities of full-length HIV-2 IN and FIV IN are depicted in lanes 1 and 4, respectively. Prior to the integration assay, an N-terminal deletion mutant of HIV-2 IN (ΔN HIV) was mixed with the N-terminal domain of HIV-2 (HIV IN55) or FIV IN (FIV IN57) (lanes 2 and 3, respectively). The substrate (S) mimics the U5 end of FIV; integration products (P) are indicated.
Cleavage, integration, and disintegration assays.
Cleavage and integration assays were done with oligonucleotides that mimic the viral DNA ends of HIV-2 (63), FIV (71), and Moloney murine leukemia virus (MoMLV) (71). Oligonucleotides were labeled at the 5′ end as described previously (72). The disintegration substrate represents an integration intermediate in which two oligonucleotides representing the U5 and U3 ends of the HIV long terminal repeats are integrated into a target DNA oligonucleotide (60). Prior to each complementation reaction, the proteins were mixed and incubated on ice for 30 min. The reaction volume was 10 μl and contained 0.02 μM oligonucleotide substrate, 20 mM MOPS (pH 7.2), 3 mM dithiothreitol, 1 mM MnCl2, 0.1 μg of bovine serum albumin/μl, and approximately 3 pmol of HIV-2 IN or FIV IN, 7 pmol of an N-terminal deletion mutant coupled to thiopropyl-Sepharose, and 20 pmol to 2 nmol of the N-terminal domain of HIV-2 IN or FIV IN. Cleavage and integration reaction mixtures were incubated at 37°C, and disintegration was carried out at 30°C. Reactions were stopped after 1 h by the addition of formamide loading dye (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol), and the mixtures were incubated at 80°C for 5 min. The samples were analyzed by denaturing polyacrylamide gel electrophoresis followed by autoradiography.
RESULTS
Complementation between the N terminus and an N-terminal deletion mutant.
Structural studies have shown that the N-terminal domain of IN (IN1–55) exists as a dimer in solution and that zinc binding is required for complete folding of this domain (8, 18). However, whether the structured N-terminal domain has a conformation that is active is not known. To address this question, we overexpressed and purified the 55-amino-acid N-terminal domain of HIV-2 IN and mixed it with a deletion mutant lacking the N-terminal region (ΔN mutant; amino acids 50 to 293) (Fig. 1a). As shown in Fig. 1b, IN1–55 restores integration activity of a ΔN mutant, which by itself is inactive. Integration activity increases with increasing concentrations of IN1–55. These results show that IN1–55 has an active conformation.
FIG. 1.
(a) Schematic representation of the three domains of HIV-2 IN. The N-terminal domain contains a helix-turn-helix motif (HTH) and the zinc-coordinating residues HHCC. The catalytic core (spanning amino acids to 50 to 212) has the three active site residues DD(35)E. The C-terminal nonspecific DNA binding domain has an SH3-like fold. For complementation experiments, the N-terminal domain (IN1–55, spanning amino acids 1 to 55) of HIV-2 IN is mixed with the N-terminal deletion mutant of HIV-2 IN (ΔN, comprising amino acids 50 to 293). (b) Complementation between an N-terminal deletion mutant of HIV-2 IN (ΔN HIV) and the N-terminal domain of HIV-2 IN (IN55). In the left lane, IN55 is omitted from the reaction; in the middle and right lanes, increasing amounts of IN55 (20 and 200 pmol) are mixed with the N-terminal deletion mutant (7 pmol) prior to the reaction (see Materials and Methods). The substrate (S) represents the precleaved HIV-2 U5 end; strand transfer products are indicated on the left (P).
Thus far, complementation assays were done with mutant proteins that have an extensive region in common (12, 22, 65). It was unknown whether the common region is needed for the interaction between the mutant proteins or that multimerization occurred between different domains of IN. Here, we show that two mutant proteins without an extensive overlap (only six amino acids) are active when mixed together in an activity assay. This implies that interaction between different domains is essential for formation of a functional multimeric complex, either directly or via DNA.
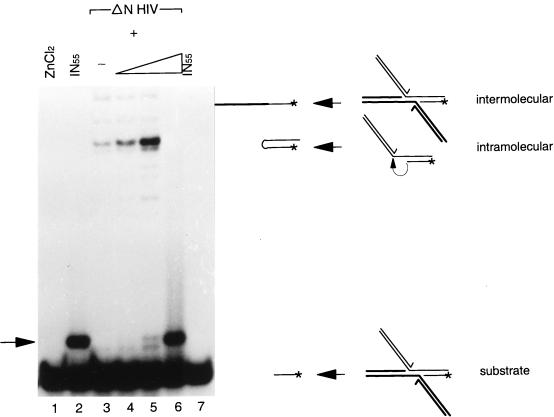
Previously, it has been shown that IN can release viral DNA ends from an integration intermediate (9). This can be done in two ways (Fig. 2). In the intermolecular disintegration reaction, the 3′ hydroxyl group of the target DNA attacks the phosphodiester bond of the viral DNA-target DNA junction of the opposite half molecule. In the intramolecular disintegration reaction, this phosphodiester bond is attacked by the 3′ hydroxyl group of the target DNA in the same half molecule (9, 45). For catalysis of the intramolecular disintegration reaction, specific viral DNA sequences are important (11, 60). The intermolecular disintegration reaction is independent of specific viral DNA sequences (9, 11, 60) and can be catalyzed by the core domain of IN alone (7, 68). Here, we show that IN1–55 enhances intramolecular disintegration activity of a ΔN mutant (Fig. 2, lanes 3 to 5). The intermolecular disintegration activity remains the same. When the concentration of IN1–55 is 200 μM, disintegration activity is lost (lane 6).
FIG. 2.
Disintegration activity in a complementation assay between the N-terminal deletion mutant of HIV-2 (ΔN HIV) and the N-terminal domain of HIV-2 IN (IN55). The disintegration substrate was incubated with 2 nmol of ZnCl2 (lane 1), 2 nmol of IN55 (lane 2), and the N-terminal deletion mutant without IN55 (7 pmol) (lane 3) or with increasing amounts of IN55 (20 pmol, 200 pmol, and 2 nmol [lanes 4 to 6, respectively]). In lane 7, part of the reaction mixture from lane 2 was extracted with phenol and chloroform. The substrate is shown on the right with an arrow indicating the product of the intermolecular disintegration reaction (labeled intermolecular) or to the product of the intramolecular disintegration reaction (intramolecular). The substrate itself runs as a 10-mer on a denaturing 20% polyacrylamide gel. The arrow on the left indicates the migration position of the IN1–55-dependent band.
Under these conditions, a new band appears; this band is dependent on the presence of IN1–55 and appears at high concentrations (Fig. 2, lane 2). Incubation of the disintegration substrate with the ΔN mutant (lane 3) or with zinc (lane 1) does not result in the formation of this band. The interaction between IN1–55 and the disintegration substrate is disrupted by extraction with phenol and chloroform (lane 7). The IN1–55-dependent band and the substrate were eluted from the gel and precipitated with ethanol. Mung bean nuclease digestion showed that this band is not the result of a newly formed covalent bond in the DNA backbone (data not shown). The nature of this band remains mysterious (see Discussion).
Only a well-structured N-terminal domain is active in a complementation assay.
Coordination of zinc by two His and two Cys residues is required for a completely structured IN1–55 (8, 18). Both HIV-1 IN1–55 and HIV-2 IN1–55 elute at the dimer position from a gel filtration column (data not shown and reference 18). The dimer interface of HIV-1 IN1–55 contains residues of the first and third helix (8). We mutated residues in HIV-2 IN1–55 that are either involved in zinc coordination or part of the putative dimer interface. Mutant proteins were subjected to gel filtration experiments and tested for activity in the complementation assay. Gel filtration experiments show that replacement of one of the His residues (H12L) results in a partially disordered IN1–55 (Fig. 3a, compare profiles A and D). The mutant protein containing substitutions of residues in the putative dimer interface elutes earlier from the gel filtration column than wild-type IN1–55, indicating that this protein no longer has a globular shape like wild-type IN1–55 (profile B). Replacement of the first residue (F1E) results in a shift of the elution profile towards the unfolded protein, but not as dramatic a shift as that of mutant B, which contains additional substitutions. Mutant proteins containing substitutions in the dimer interface do not have the expected profile of a globular monomer of 6.3 kDa. Although we replaced all residues of the dimer interface with nonconservative amino acids and introduced them alone or in different combinations into IN1–55, a globular monomer was never observed. This could indicate that a globular form of IN1–55 can only be dimeric. Mutant proteins were tested for complementation activity. As shown in Fig. 3b, replacement of one of the His residues of the zinc binding unit abolishes complementation activity (Fig. 3b, compare two-lane panels A and D). Also, substituting residues in the putative dimer interface of the N terminus of HIV-2 IN impairs complementation activity (Fig. 3b, two-lane panel B). Taken together, these results indicate that only the structured form of IN1–55 is able to complement the ΔN mutant.
Although complementation experiments were performed with the N-terminal deletion mutant coupled to thiopropyl-Sepharose, activity was also observed with a soluble ΔN mutant mixed with IN1–55 (data not shown).
Stable complex formation requires the N-terminal domain in cis.
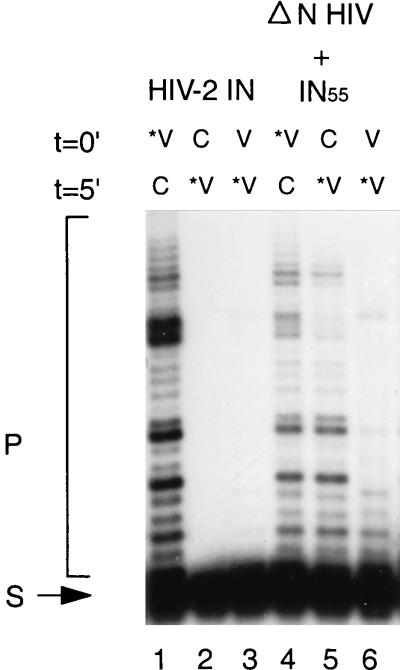
Previously, it has been shown that IN can form a stable complex with the viral DNA substrate (19, 70). This stable interaction between IN and viral DNA is dependent on metal ions and is not competed by an excess of nonspecific DNA or other polyanions, like poly(Asp50, Glu50). Deletion of the HHCC motif or chemical modification of Cys-65 impairs assembly of the stable complex (20). We investigated whether a stable complex can be formed when IN1–55 complements the ΔN mutant in trans. As shown in Fig. 4, full-length IN binds stably to viral DNA after incubation of the protein with Mn2+ and viral DNA ends. Competition by a 25 M excess of nonspecific DNA does not destroy this specific interaction. When IN is first incubated with nonspecific DNA or nonlabeled viral DNA ends in the presence of Mn2+, addition of viral DNA ends does not result in integration activity (Fig. 4, lanes 1 to 3). Under these conditions, full-length IN binds stably to nonlabeled DNA, which is consistent with previous reports (19, 70). When we do the same experiment in a complementation assay, we do not observe a difference in the order of addition of viral or nonspecific DNA (lanes 4 and 5). This indicates that the N terminus is required in cis to the remainder of the protein in order to form a stable complex. Furthermore, these order-of-addition experiments show that in a complementation assay, integration occurs even in the presence of an excess of nonspecific DNA, implying a high on-off rate of initial DNA binding. Assuming that the on-off rate of DNA binding in the complementation assay is independent of the DNA sequence, one would expect the same integration activity whether competition is done with nonlabeled viral or nonspecific DNA. To investigate this, we added a 25 M excess of nonlabeled viral DNA substrate to the reaction mixture, prior to the addition of radiolabeled substrate. As shown in Fig. 4, lane 6, the integration activity was reduced. This is in contrast to the competition with nonspecific DNA and suggests that the IN1–55-ΔN mutant complex binds more tightly to specific DNA than to nonspecific DNA.
FIG. 4.
Stable complex formation between IN mutants and the integration substrate. The proteins were incubated with the reaction mixture containing 1 mM MnCl2 for 5 min at t = 0′, prior to the addition of 0.2 pmol of labeled integration substrate (*V; lanes 1 and 4), 5 pmol of nonspecific DNA (C; lanes 2 and 5), or 5 pmol of nonlabeled integration substrate (V; lanes 3 and 6). After 5 min (t = 5′), 5 pmol of nonspecific DNA was added to the reaction mixtures in lanes 1 and 4 and 0.2 pmol of labeled integration substrate was added to the reaction mixtures in lanes 2, 3, 5, and 6. Incubations were done at room temperature. The integration substrate (S) is radioactively labeled; integration products (P) are indicated. Stable complex formation of full-length HIV-2 IN is shown in lanes 1 to 3. The same experiment was done with an N-terminal deletion mutant of HIV-2 IN (ΔN HIV), which was mixed with the N-terminal domain of HIV-2 IN 30 min prior to the reaction (lanes 4 to 6).
Cross-complementation between HIV and FIV IN.
The N-terminal domains of FIV and HIV-2 INs are 36% identical at the amino acid level. FIV IN contains two additional Ser residues at the N terminus. The N terminus of FIV IN (IN57) was overexpressed and purified in the same way as has been done for IN1–55 of HIV-2. As shown in Fig. 5a, the N-terminal domain of FIV IN elutes at the dimer position from a size exclusion column.
We tested whether the N terminus of FIV IN can complement integration activity of a ΔN mutant of HIV-2 IN. As shown in Fig. 5b, FIV IN1–55 can complement a ΔN mutant of HIV-2 IN at a similar efficiency as in the complementation between HIV-2 IN1–55 and the ΔN mutant of HIV-2 IN (lanes 2 and 3).
Full-length IN proteins from HIV and FIV have different preferences for the sites of integration (Fig. 5b, lanes 1 and 4) (57, 71). Cross-complementation experiments show the same target site preference for an N-terminal deletion mutant of HIV-2 IN that is complemented by either FIV IN1–57 or HIV-2 IN1–55 (Fig. 5b, lanes 2 and 3). This indicates that the N-terminal domain does not control target site preference, which is in agreement with results obtained with chimeric proteins between HIV-1 IN and FIV IN (57).
It has been shown that the choice of nucleophile in the cleavage reaction is different for HIV IN and FIV IN (71). In a cross-complementation assay, the choice of nucleophile is independent of the source of the N-terminal domain (data not shown). This is consistent with results described elsewhere indicating that residues near the active site determine the choice of nucleophile in the cleavage reaction (23, 61, 64).
Cross-complementation activity on different substrates.
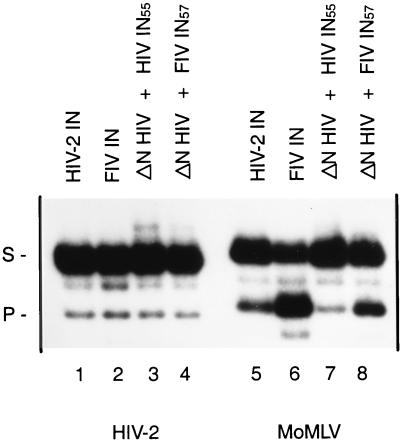
Previous reports have indicated that the efficiency of 3′ processing of different viral DNA substrates varies between HIV-1 IN and FIV IN (71). The most pronounced difference was found for the MoMLV substrate; FIV IN cleaves this substrate much better than HIV IN (Fig. 6, lanes 5 and 6) (71). To address the question of whether the N-terminal domain is involved in viral DNA recognition, a cross-complementation experiment was done using HIV-2 and MoMLV substrates. Protein concentrations were such that the efficiency of 3′ processing of HIV-2 substrate was similar in all reactions (Fig. 6, lanes 1 to 4). The same protein concentrations were used in the cleavage reaction of MoMLV substrate (lanes 5 to 8). For both substrates, HIV IN1–55 and FIV IN1–57 can complement a ΔN mutant in the 3′ processing reaction. Under these conditions the HIV-2 substrate is cleaved at a similar rate in all reactions, whereas the MoMLV substrate is processed more efficiently by FIV IN than by HIV-2 IN (Fig. 6, lanes 1 and 2 and lanes 5 and 6, respectively). Moreover, cleavage of MoMLV substrate is more efficient with FIV IN1–57 than with HIV IN1–57 in the complementation assay (lanes 7 and 8). This indicates that the N-terminal domain of IN is involved in recognition of the specific viral DNA ends.
FIG. 6.
Cross-complementation assay between FIV and HIV-2 mutants on different substrates. Cleavage activity of full-length HIV-2 is shown in lanes 1 and 5; that of FIV IN is shown in lanes 2 and 6. The N-terminal deletion mutant of HIV-2 IN (ΔN HIV; 7 pmol) is complemented by the N-terminal domain of HIV-2 IN (HIV IN55; 0.2 nmol) (lanes 3 and 7) or the N-terminal domain of FIV IN (FIV IN57; 0.1 nmol) (lanes 4 and 8). HIV-2 substrate is used in lanes 1 to 4, and MoMLV substrate is used in lanes 5 to 8. The migration positions of the 28-mer substrate (S) and the 26-mer product (P) are indicated.
DISCUSSION
Structure elucidation of the N-terminal domains of HIV-1 IN and HIV-2 IN revealed a dimeric three-helix bundle, stabilized by a zinc-binding unit (8, 18). The topology of the N-terminal domain is very similar to that of DNA binding proteins, containing a helix-turn-helix motif such as the Trp repressor (48), the Prd paired domain (75), and the N terminus of Tc3 transposase (66). In contrast to the DNA binding proteins, which use the second helix of the helix-turn-helix motif to bind DNA, HIV-1 IN1–55 uses this helix for dimerization (8). Although no direct DNA binding of IN1–55 has been observed, several studies have indicated that the N-terminal domain may be involved in recognition of the viral DNA ends. Since mutations in the HHCC motif affect cleavage more than integration, the N-terminal domain could participate in correct positioning of the viral DNA ends (7, 63, 67). Disintegration activity of HHCC mutants suggested that the N-terminal domain may interact with viral DNA sequences internal to the conserved CA (67). Alternatively, the N-terminal domain could be involved indirectly in viral DNA recognition through the interaction with other domains of IN. Interaction between the HHCC motif and the core has been suggested by chemical modification of specific Cys residues by N-ethylmaleimide, which impairs metal-induced aggregation of IN (20). In addition, it has been shown that preincubation of IN with a C-terminal specific monoclonal antibody interferes with binding of an N-terminal specific monoclonal antibody, indicating that the N- and C-terminal domains are close to each other (2). To dissect the role of the N-terminal domain, we developed an in trans complementation assay. The N-terminal domain (IN1–55) is provided on a separate molecule, unlinked to the catalytic core and C-terminal DNA binding domain. Although high levels of IN activity require an excess of IN1–55, significant levels of reaction products were detected when the molar ratio of IN1–55 to the N-terminal deletion mutant was around 2.
The N-terminal domain complements an N-terminal deletion mutant in trans.
We have shown that the N-terminal domain of IN can restore IN activity of an N-terminal deletion mutant when provided in trans to the active site. Previously, it has been shown that IS10 transposase retains transposition activity after two of its domains that are not covalently attached are mixed (40). In contrast to IN, both domains of IS10 contribute residues to the active site. In trans complementation has been reported for the enhancer region of bacteriophage Mu; when the enhancer is provided on an unlinked DNA molecule, it still can promote correct synapsis of the left and right Mu DNA ends (59). In contrast to the in trans complementation described here, the Mu transposable enhancer is only required in the initial steps of the Mu reaction and is dispensable for cleavage and strand transfer. To restore 3′ processing and strand transfer activity of an N-terminal deletion mutant, IN1–55 needs to be properly folded. The active conformation of IN1–55 is disturbed by nonconservative substitutions in the putative dimer interface, as well as by the loss of zinc coordination (either by replacement of one of the zinc-binding residues or by chelation of zinc by EDTA). Although the introduction of negatively charged residues in the dimer interface would be expected to result in a monomeric protein, no mutant proteins that elute at a monomer position from a size exclusion column were found. Also, not a trace of monomer was observed in zinc-containing IN1–55. This could indicate that IN1–55 cannot exist as a globular folded monomer.
Complementation of disintegration activity.
Intermolecular disintegration of an N-terminal deletion mutant is independent of the presence of IN1–55. This is consistent with the finding that intermolecular disintegration activity is indifferent to zinc (77) and can be carried out in the absence of the N-terminal domain (7, 39, 68). The intramolecular disintegration activity of an N-terminal deletion mutant is enhanced in the presence of increasing amounts of IN1–55. Based on sequence requirements of the intramolecular disintegration reaction, it has been postulated that the intramolecular disintegration activity resembles the 3′ processing reaction (45, 60). The enhanced intramolecular disintegration activity of the N-terminal deletion mutant upon addition of IN1–55 supports this hypothesis. At high concentrations of IN1–55 (0.2 mM), no disintegration activity takes place and a new band appears. This band probably results from a noncovalent interaction between the substrate and IN1–55. It is surprising that the complex survives heating in formamide and denaturing gel electrophoresis. We propose that the N-terminal domain impairs disintegration activity by sequestering the substrate. Support for the interaction between the N terminus of IN and the disintegration substrate comes from photo-cross-linking studies, which showed that the N terminus of IN interacts with the target DNA portion of a disintegration substrate (29).
Stable complex formation.
Stable complex formation between IN and viral DNA is established by incubation of IN with Mn2+, followed by addition of viral DNA (19, 70). Once the viral DNA is bound, nonspecific DNA no longer competes for viral DNA binding but functions as a target for the strand transfer reaction. When nonlabeled DNA is added prior to the viral DNA ends, no integration takes place. Here, we show that integration occurs in a complementation assay even in the presence of an excess of nonspecific DNA. When nonlabeled viral DNA sequences are added prior to the labeled viral DNA substrate, integration activity is reduced. These results suggest that DNA binding by the IN1–55 ΔN complex has a higher off rate than that of full-length IN. In addition, the IN1–55–ΔN complex binds nonspecific DNA less tightly than specific viral DNA ends.
Cross-complementation activity between FIV and HIV-2 IN mutants.
Previously, it has been shown that HIV IN and FIV IN have different preferences for viral DNA substrates, as well as different target site choices. To address which domains contribute to substrate specificity and target site preference, cross-complementation experiments were done by using FIV and HIV-2 IN mutants. We have established that an N-terminal domain of FIV IN can function in trans to an N-terminal deletion mutant of HIV-2 IN. The integration pattern corresponds to that of HIV-2 IN, which is consistent with results obtained with chimeric proteins that have an integration pattern dependent on the source of the core domain of IN (57).
Cross-complementation experiments show that the N-terminal domains of HIV IN and FIV IN contribute to substrate specificity. This is in contrast to the result obtained with a chimeric protein that contains the N-terminal domain of HIV-1 IN fused to the core and C terminus of visna virus IN (35, 36). This chimeric protein has the same substrate specificity as full-length visna virus IN, suggesting that the core domain of visna virus IN alone determines substrate specificity. To extend this conclusion to IN proteins from HIV and other viruses, additional swaps should be tested. Also, chimeric proteins of HIV and FIV IN tested on MoMLV substrate could demonstrate the role of the N-terminal domain in substrate recognition. We cannot fully exclude the possibility that the conformation of the active complex in the complementation assay may be different from that of chimeric proteins and can therefore affect the results. However, since the choice of nucleophile in the cleavage reaction and the target choice in the strand transfer reaction in the complementation between FIV IN1–55 and the ΔN mutant of HIV-2 IN is similar to that of full-length HIV-2 IN, it is not very likely that gross changes in topology have taken place. As mentioned by Katzman and Sudol, the assignment of viral DNA recognition to the central domain of IN does not exclude that other parts of IN can facilitate the formation of a viral DNA complex (36). Recently, cross-linking studies have indeed revealed that binding to the viral DNA ends is not limited to a single domain (27–29, 31). The N-terminal domain may function as a module within the multimeric complex of IN and the viral DNA ends. It is likely that functional multimerization requires interactions between nonequivalent domains, but we cannot exclude the possibility that complementation activity is achieved via the interaction of the N-terminal domain with DNA.
Previously, Heuer and Brown (30) proposed a role of the N-terminal domain in the formation of a bridge between two IN protomers. The N-terminal domain of one protomer would interact with the core of another protomer, thereby approaching target and viral DNA sequences. Binding of the catalytic core to the viral DNA end would be stabilized by its interaction with the N-terminal domain. The results presented here are in agreement with the proposed model in the sense that the N-terminal domain can act in trans to the active site. They suggest that by interacting with the core, the N-terminal domain influences the binding of the complex to the viral DNA ends, which is in accordance with the location of the N-terminal domain in the model of Heuer and Brown.
ACKNOWLEDGMENTS
This work was supported by a grant from The Netherlands AIDS foundation.
We thank Karin van der Linden for construction of pRP825 and appreciate the assistance of Henk Hilkman with the high-performance liquid chromatography. We thank Chris Vos, Henri van Luenen, Piet Borst, and Ramon Puras Lutzke for critically reading the manuscript.
REFERENCES
- 1.Asante-Appiah E, Skalka A M. Molecular mechanisms in retrovirus DNA integration. Antiviral Res. 1997;36:139–156. doi: 10.1016/s0166-3542(97)00046-6. [DOI] [PubMed] [Google Scholar]
- 2.Bizub-Bender D, Kulkosky J, Skalka A M. Monoclonal antibodies against HIV type 1 integrase: clues to molecular structure. AIDS Res Hum Retroviruses. 1994;10:1105–1115. doi: 10.1089/aid.1994.10.1105. [DOI] [PubMed] [Google Scholar]
- 3.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J Mol Biol. 1995;253:333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- 4.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure. 1996;4:89–96. doi: 10.1016/s0969-2126(96)00012-3. [DOI] [PubMed] [Google Scholar]
- 5.Burke C J, Sanyal G, Bruner M W, Ryan J A, LaFemina R L, Robbins H L, Zeft A S, Middaugh C R, Cordingley M G. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J Biol Chem. 1992;267:9639–9644. [PubMed] [Google Scholar]
- 6.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman F D, Engleman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai M, Zheng R, Caffrey M, Craigie R, Clore G M, Gronenborn A M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 9.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 10.Craigie R, Fujiwara T, Bushman F D. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 11.Donzella G A, Jonsson C B, Roth M J. Coordinated disintegration reactions mediated by Moloney murine leukemia virus integrase. J Virol. 1996;70:3909–3921. doi: 10.1128/jvi.70.6.3909-3921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donzella G A, León 0, Roth M J. Implication of a central residue and the HHCC domain of Moloney murine leukemia virus integrase protein in functional multimerization. J Virol. 1998;72:1691–1698. doi: 10.1128/jvi.72.2.1691-1698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doolittle R F, Feng D F, Johnson M S, McClure M A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 14.Dotan I, Scottoline B P, Heuer T S, Brown P O. Characterization of recombinant murine leukemia virus integrase. J Virol. 1995;69:456–468. doi: 10.1128/jvi.69.1.456-468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 16.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 17.Eijkelenboom A P A M, Puras Lutzke R A, Boelens R, Plasterk R H A, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 18.Eijkelenboom A P A M, van den Ent F M I, Vos A, Doreleijers J F, Hard K, Tullius T D, Plasterk R H A, Kaptein R, Boelens R. The solution structure of the amino-terminal HHCC domain of HIV-2 integrase: a three-helix bundle stabilized by zinc. Curr Biol. 1997;7:739–746. doi: 10.1016/s0960-9822(06)00332-0. [DOI] [PubMed] [Google Scholar]
- 19.Ellison V, Brown P O. A stable complex between integrase and viral DNA ends mediates human immunodeficiency virus integration in vitro. Proc Natl Acad Sci USA. 1994;91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellison V, Gerton J, Vincent K A, Brown P O. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J Biol Chem. 1995;270:3320–3326. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- 21.Engelman A. Biochemical characterization of recombinant equine infectious anemia virus integrase. Protein Expr Purif. 1996;8:299–304. doi: 10.1006/prep.1996.0104. [DOI] [PubMed] [Google Scholar]
- 22.Engelman A, Bushman F D, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelman A, Hickman A B, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 26.Fayet O, Ramond P, Polard P, Prere M F, Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol Microbiol. 1990;4:1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- 27.Gerton J L, Brown P O. The core domain of HIV-1 integrase recognizes key features of its DNA substrates. J Biol Chem. 1998;272:25809–25815. doi: 10.1074/jbc.272.41.25809. [DOI] [PubMed] [Google Scholar]
- 28.Gerton J L, Ohgi S, Olsen M, Derisi J, Brown P O. Effects of mutations in residues near the active site of human immunodeficiency virus type 1 integrase on specific enzyme-substrate interactions. J Virol. 1998;72:5046–5055. doi: 10.1128/jvi.72.6.5046-5055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heuer T S, Brown P O. Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-cross-linking. Biochemistry. 1997;36:10655–10665. doi: 10.1021/bi970782h. [DOI] [PubMed] [Google Scholar]
- 30.Heuer T S, Brown P O. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37:6667–6678. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins T M, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson M S, McClure M A, Feng D F, Gray J, Doolittle R F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci USA. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 34.Katz R A, Skalka A M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 35.Katzman M, Sudol M. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J Virol. 1995;69:5687–5696. doi: 10.1128/jvi.69.9.5687-5696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katzman M, Sudol M. Mapping viral DNA specificity to the central region of integrase by using functional human immunodeficiency virus type 1/visna virus chimeric proteins. J Virol. 1998;72:1744–1753. doi: 10.1128/jvi.72.3.1744-1753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan E, Mack J P, Katz R A, Kulkosky J, Skalka A M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulkosky J, Jones K S, Katz R A, Mack J P, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulkosky J, Katz R A, Merkel G, Skalka A M. Activities and substrate specificity of the evolutionarily conserved central domain of retroviral integrase. Virology. 1995;206:448–456. doi: 10.1016/s0042-6822(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 40.Kwon D, Chalmers R M, Kleckner N. Structural domains of IS10 transposase and reconstitution of transposition activity from proteolytic fragments lacking an interdomain linker. Proc Natl Acad Sci USA. 1995;92:8234–8238. doi: 10.1073/pnas.92.18.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leavitt A D, Shiue L, Varmus H E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- 42.Lee S P, Han M K. Zinc stimulates Mg2+-dependent 3′-processing activity of human immunodeficiency virus type 1 integrase in vitro. Biochemistry. 1996;35:3837–3844. doi: 10.1021/bi952056p. [DOI] [PubMed] [Google Scholar]
- 43.Lee S P, Xiao J, Knutson J R, Lewis M S, Han M K. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry. 1997;36:173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- 44.Lodi P J, Ernst J A, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Gronenborn A M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 45.Mazumder A, Engelman A, Craigie R, Fesen M, Pommier Y. Intermolecular disintegration and intramolecular strand transfer activities of wild-type and mutant HIV-1 integrase. Nucleic Acids Res. 1994;22:1037–1043. doi: 10.1093/nar/22.6.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller B, Jones K S, Merkel G W, Skalka A M. Rapid solution assays for retroviral integration reactions and their use in kinetic analyses of wild-type and mutant Rous sarcoma virus integrases. Proc Natl Acad Sci USA. 1993;90:11633–11637. doi: 10.1073/pnas.90.24.11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mumm S R, Grandgenett D P. Defining nucleic acid-binding properties of avian retrovirus integrase by deletion analysis. J Virol. 1991;65:1160–1167. doi: 10.1128/jvi.65.3.1160-1167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Schevitz R W, Zhang R G, Lawson C L, Joachimiak A, Marmorstein R Q, Luisi B F, Sigler P B. Crystal structure of Trp repressor/operator complex at atomic resolution. Nature. 1988;335:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 49.Polard P, Chandler M. Bacterial transposases and retroviral integrases. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 50.Puras Lutzke R A, Plasterk R H A. HIV integrase: a target for drug discovery. Genes Function. 1998;1:289–307. doi: 10.1046/j.1365-4624.1997.00026.x. [DOI] [PubMed] [Google Scholar]
- 51.Puras Lutzke R A, Plasterk R H A. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J Virol. 1998;72:4841–4848. doi: 10.1128/jvi.72.6.4841-4848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puras Lutzke R A, Vink C, Plasterk R H A. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994;22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rice P, Craigie R, Davies D R. Retroviral integrases and their cousins. Curr Opin Struct Biol. 1996;6:76–83. doi: 10.1016/s0959-440x(96)80098-4. [DOI] [PubMed] [Google Scholar]
- 54.Rowland S J, Dyke K G. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990;4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 56.Schauer M, Billich A. The N-terminal region of HIV-1 integrase is required for integration activity, but not for DNA-binding. Biochem Biophys Res Commun. 1992;185:874–880. doi: 10.1016/0006-291x(92)91708-x. [DOI] [PubMed] [Google Scholar]
- 57.Shibagaki Y, Chow S A. Central core domain of retroviral integrase is responsible for target site selection. J Biol Chem. 1997;272:8361–8369. doi: 10.1074/jbc.272.13.8361. [DOI] [PubMed] [Google Scholar]
- 58.Studier F W, Moffat B A. Use of bacteriophage T7 polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 59.Surette M G, Chaconas G. The Mu transpositional enhancer can function in trans: requirement of the enhancer for synapsis but not strand cleavage. Cell. 1992;68:1101–1108. doi: 10.1016/0092-8674(92)90081-m. [DOI] [PubMed] [Google Scholar]
- 60.van den Ent F M I, Vink C, Plasterk R H A. DNA substrate requirements for different activities of the human immunodeficiency virus type 1 integrase protein. J Virol. 1994;68:7825–7832. doi: 10.1128/jvi.68.12.7825-7832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van den Ent F M I, Vos A, Plasterk R H A. Mutational scan of the human immunodeficiency virus type 2 integrase protein. J Virol. 1998;72:3916–3924. doi: 10.1128/jvi.72.5.3916-3924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Gent D C, Elgersma Y, Bolk M W, Vink C, Plasterk R H A. DNA binding properties of the integrase proteins of human immunodeficiency viruses types 1 and 2. Nucleic Acids Res. 1991;19:3821–3827. doi: 10.1093/nar/19.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Gent D C, Oude Groeneger A A M, Plasterk R H A. Mutational analysis of the integrase protein of human immunodeficiencey virus type 2. Proc Natl Acad Sci USA. 1992;89:9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Gent D C, Oude Groeneger A A M, Plasterk R H A. Identification of amino acids in HIV-2 integrase involved in site-specific hydrolysis and alcoholysis of viral DNA termini. Nucleic Acids Res. 1993;21:3373–3377. doi: 10.1093/nar/21.15.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Gent D C, Vink C, Oude Groeneger A A M, Plasterk R H A. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Pouderoyen G, Ketting R F, Perrakis A, Plasterk R H A, Sixma T K. Crystal structure of the specific DNA-binding domain of Tc3 transposase of C. elegans in complex with transposon DNA. EMBO J. 1997;16:6044–6054. doi: 10.1093/emboj/16.19.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vink C, Oude Groeneger A A M, Plasterk R H A. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vink C, Plasterk R H A. The human immunodeficiency virus integrase protein. Trends Genet. 1993;9:433–438. doi: 10.1016/0168-9525(93)90107-s. [DOI] [PubMed] [Google Scholar]
- 70.Vink C, Puras Lutzke R A, Plasterk R H A. Formation of a stable complex between the human immunodeficiency virus integrase protein and viral DNA. Nucleic Acids Res. 1994;22:4103–4110. doi: 10.1093/nar/22.20.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vink C, van der Linden K H, Plasterk R H A. Activities of the feline immunodeficiency virus integrase protein produced in Escherichia coli. J Virol. 1994;68:1468–1474. doi: 10.1128/jvi.68.3.1468-1474.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vink C, van Gent D C, Elgersma Y, Plasterk R H A. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vink C, Yeheskiely E, van der Marel G A, van Boom J H, Plasterk R H A. Site-specific hydrolysis and alcoholysis of human immunodeficiency virus DNA termini mediated by the viral integrase protein. Nucleic Acids Res. 1991;19:6691–6698. doi: 10.1093/nar/19.24.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woerner A M, Marcus-Sekura C J. Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res. 1993;21:3507–3511. doi: 10.1093/nar/21.15.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu W, Rould M A, Jun S, Desplan C, Pabo C O. Crystal structure of a paired domain-DNA complex at 2.5 Å resolution reveals structural basis for Pax developmental mutations. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- 76.Yang W, Steitz T A. Recombining the structures of HIV integrase, RuvC and RNase H. Structure. 1995;3:131–134. doi: 10.1016/s0969-2126(01)00142-3. [DOI] [PubMed] [Google Scholar]
- 77.Zheng R, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]