Abstract
Thermodynamically predicted secondary structure analysis of the 3′-terminal 305 nucleotides (nt) of the rubella virus (RUB) genome, a region conserved in all RUB defective interfering RNAs, revealed four stem-loop (SL) structures; SL1 and SL2 are both located in the E1 coding region, while SL3 and SL4 are within the 59-nt 3′ untranslated region (UTR) preceding the poly(A) tract. SL2 is a structure shown to interact with human calreticulin (CAL), an autoantigen potentially involved in RUB RNA replication and pathogenesis. RNase mapping indicated that SL2 and SL3 are in equilibrium between two conformations, in the second of which the previously proposed CAL binding site in SL2, a U-U bulge, is not formed. Site-directed mutagenesis of the 3′ UTR with a RUB infectious clone, Robo302, revealed that most of the 3′ UTR is required for viral viability except for the 3′-terminal 5 nt and the poly(A) tract, although poly(A) was rapidly regenerated during subsequent replication. Maintenance of the overall SL3 structure, the 11-nt single-stranded sequence between SL3 and SL4, and the sequences forming SL4 were all important for viral viability. Studies on the interaction between host factors and the 3′ UTR showed the formation of three RNA-protein complexes by gel mobility shift assay, and UV-induced cross-linking detected six host protein species, with molecular masses of 120, 80, 66, 55, 48, and 36 kDa, interacting with the 3′ UTR. Site-directed mutagenesis of SL2 by nucleotide substitutions showed that maintenance of SL2 stem rather than the U-U bulge was critical in CAL binding since mutants having the U-U bulge base paired had a similar binding activity for CAL as the native structure whereas mutants having the SL2 stem destabilized had much lower binding activity. However, all of these mutations gave rise to viable viruses when introduced into Robo302, indicating that binding of CAL to SL2 is independent of viral viability.
Rubella virus (RUB) is a significant human pathogen which causes rubella, or German measles. RUB is the sole member of the Rubivirus genus within the Togaviridae family. Like genomes of the members of Alphavirus, the other genus in the togavirus family, the genome of RUB consists of a single-stranded, plus-sense RNA genome (9,762 nucleotides [nt] in length) which is 5′ capped and 3′ polyadenylated and contains two open reading frames (ORFs): the 5′-proximal ORF encodes nonstructural proteins including the RNA-dependent RNA polymerase (RdRp), which functions in viral RNA replication, and the 3′-proximal ORF encodes the viral structural proteins, the capsid protein (C), and two envelope glycoproteins, E1 and E2. RUB RNA replication initiates with the synthesis of a genomic RNA complement of minus polarity by the viral RdRp. This minus-strand RNA is then used as a template for the synthesis of both genomic RNA and the subgenomic RNA from which the structural proteins are translated (reviewed in reference 14).
The 3′ terminus of the genomes of plus-strand RNA viruses is postulated to contain the cis-acting elements required for the synthesis of the minus-strand RNA. In alphaviruses, a conserved 19-nt stretch preceding the poly(A) tract was found critical for viral viability (26); further analysis showed that some of the nucleotides in this stretch were more important than others (24). However, there is no sequence homology between RUB and alphaviruses at the 3′ end of the genome (reviewed in reference 14). A prominent stem-and-loop (SL) structure 59 nt from the 3′-terminal poly(A) tract of the RUB genome has been noticed by several groups (13, 36). This SL consists of a UAUA loop and a 13-nt GC-rich stem which contains a U-U bulge. Analysis of the interaction between host factors and the 3′ end of the RUB genome demonstrated that three cellular proteins with molecular masses of 68, 63, and 61 kDa, respectively, were able to specifically bind to this SL (37); the 61-kDa protein was later identified as calreticulin (CAL) (38, 50), a ubiquitous calcium binding protein found in most eukaryotic cells. CAL is located primarily in the endoplasmic reticulum and has been found to function as a molecular chaperone in the maturation of the cytomegalovirus glycoprotein B (60), the influenza virus hemagglutinin (17, 18, 41), hepatitis C virus E2 and E1 (9), and the human immunodeficiency virus gp160 envelope glycoprotein (39). In all three cases, CAL binds to the nascent polypeptide. Thus, the finding that CAL binds with the RUB RNA was surprising. Although the CAL binding site was not mapped, the interaction of CAL with the U-U bulge appears to be important since deletion of the loop dramatically reduced the binding of all three proteins in cell lysates (37). The phosphorylation state of CAL was found to be altered in RUB-infected cells, and this change correlated with increased binding of CAL in cell lysates to the SL (38, 50). Using a bacterially expressed maltose binding protein (MBP)-CAL fusion, it was later found that MBP-CAL can be autophosphorylated and both the autophosphorylation sites and RNA-binding activities of MBP-CAL were localized to the N terminus of the CAL sequences (2), which is the most conserved domain among CALs from different species (reviewed in reference 23). The binding of CAL to the RUB SL implied a role in regulation of RUB RNA replication. A number of studies have shown specific interactions between viral genomic RNAs and cellular proteins and have indicated a role of such binding in viral RNA replication or translation (1, 5, 6, 10, 15, 16, 21, 25, 27, 35, 37, 38, 40, 42, 43, 50, 52, 53).
The interaction between the RUB genome and CAL has also been proposed to be related to viral pathogenesis, particularly arthritis, which is a frequent complication of rubella in adults (up to 25% of female vaccines suffer from transient arthalgia or arthritis). CAL was classified as an autoantigen as a result of several studies that have shown that the antisera from patients suffering from autoimmune disease, such as onchocerciasis (47, 48), can cross-react with CAL, and several autoantigens, such as DR2Dw4/DR3 (33, 56) and Ro/SS-A (29, 30), have been reported to show sequence homology with CAL. It was later found that CAL can interact with hY RNA and might function as a molecular chaperone in the formation of a Ro/SS-A RNP complex (8). Interestingly, CAL must be dephosphorylated to bind with this RNA species. Two other host factors with molecular masses of 52 and 59 kDa were found to interact with an SL at the 5′ end of the RUB genome (42), and one of these was later identified as the La autoantigen (43). Significantly, rubella patients were found to have increased levels of anti-La antibodies (43). A mutagenic study of the 5′ SL element using Robo302, a RUB infectious clone, showed that this structure functions in translation rather than in RNA replication (45).
Previous studies of RUB defective interfering (DI) RNAs generated from serial undiluted passages found that the 3′-terminal 305 nt were retained in all such DI RNAs (7, 11). Therefore, this region must contain the 3′ cis-acting elements required for RUB genome replication. In this study, we used Robo302 to begin analysis of this region to map the 3′ cis-acting elements required for RUB RNA replication, including the SL structure that binds CAL.
MATERIALS AND METHODS
RNA secondary structure predictions.
Optimal RNA secondary structures were predicted by the method of Zuker and Steigler (61), using the RNAFOLD program in the Wisconsin Package (Genetics Computer Group, Madison, Wis.).
Viruses, cell lines, and transfection.
The F-Therien (fTH; originally obtained from J. Chantler) and HPV77 (obtained from Viral Antigens Inc., Memphis, Tenn.) strains of RUB were propagated and titered by plaque assay described previously (57).
Vero cells (American Type Culture Collection) were maintained in Dulbecco’s modified Eagle medium (DMEM; Gibco/BRL, Gaithersburg, Md.) containing 5% fetal bovine serum and gentamicin (10 μg/ml) at 35°C under 5% CO2. For transfection, duplicate 60-mm2 plates of 80% confluent Vero cells were washed with phosphate-buffered saline twice and Opti-MEM (Gibco/BRL) once before transfection. RNA transcripts were mixed with Lipofectamine and Opti-MEM (1:40) and inoculated onto the Vero cells. After a 4-h incubation with occasional rocking, one plate was overlaid with plaque assay agar whereas DMEM containing 2% fetal bovine serum was added to the other. Generally in cells transfected with Robo302 transcripts, cytopathic effect (CPE) appeared 3 to 4 days posttransfection in the plates maintained in DMEM. Single plaques (if present) from each transfection were picked, and virus was eluted in DMEM and amplified once in Vero cells to produce stocks for sequencing to confirm the mutation and for the subsequent analysis of the mutant phenotype.
Molecular cloning, in vitro transcription, and reverse transcription.
Standard recombinant DNA techniques were used, with minor modifications. Restriction enzymes were purchased from New England BioLabs (Beverly, Mass.) or Boehringer Mannheim Biochemicals (Indianapolis, Ind.), while T4 DNA ligase was obtained from New England BioLabs.
For transfection, 5′-capped RNA transcripts were runoff transcribed (transcription terminates at the end of the restricted template) from the wild-type (wt) Robo302 virus or one of the mutagenized derivatives that had been linearized with EcoRI. The reaction contained reaction buffer (40 mM Tris-HCl [pH 7.5], 6 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol [DTT]) (New England BioLabs), 1 mM ATP, CTP, GTP, and UTP (Pharmacia, Piscataway, N.J.), 2 mM RNA cap analog [m7G(5′)pppG] (New England BioLabs), RNasin (1 U/μl; Boehringer Mannheim Biochemicals), and SP6 RNA polymerase (1 to 2 U/μl; Epicentre, Madison, Wis.). The plasmid DNA used as a template in these reactions was prepared by standard miniprep procedures.
RNA transcripts for RNase probing or competition assays were synthesized by using T7 RNA polymerase (New England BioLabs) under the same conditions described above for SP6 transcription with no RNA cap analog added to the reaction (these plasmids contained the T7 promoter rather than the SP6 promoter used in Robo302 constructs). The plasmids used in these reactions were prepared from large cultures and CsCl purified. 32P-labeled pUCRUB3′110-fTH, pUCRUB3′110-HPV, or 498-ΔA T7 RNA polymerase transcripts used in gel mobility shift assays were transcribed from EcoRI-restricted plasmid templates by incorporating 10 μCi of [32P]CTP (3,000 Ci/mmol; Amersham) per μl into transcription buffer which contained unlabeled CTP diluted 1:40. 32P-labeled probes containing SL2 only were transcribed from pUC18-SL2 constructs restricted with Bsp120I, using the same conditions. The 32P-labeled probes were electrophoresed in 8% polyacrylamide–8 M urea gels, visualized by autoradiography, and eluted in 0.5 M ammonium acetate–0.1% sodium dodecyl sulfate (SDS)–1 mM EDTA at 37°C overnight (5). The eluted probe was harvested by ethanol precipitation in the presence of 1 μg of glycogen and dissolved in water. The probe concentration was calculated by determining the percentage of radiolabel incorporated into probe by liquid scintillation spectrophotometry and multiplying by the amount of CTP in the reaction mixture.
Reverse transcription and primer extension reactions were carried out with Superscript reverse transcriptase (Gibco/BRL). Primers were first annealed with template RNA in Superscript reverse transcription buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2) by denaturing at 80°C for 5 min and slowly cooling to room temperature followed by the addition of DTT to 10 mM, each deoxynucleoside triphosphate (Pharmacia) to 1 mM, Superscript reverse transcriptase (10 U/μl), and RNasin (1 U/μl). The reaction was incubated at 45°C for 1 h and then stopped by boiling for 5 min. In primer extension reactions, actinomycin D (20 μg/ml) was added to the reaction buffer. To prepare 5′-end-labeled primers for primer extension, [γ-32P]ATP (5 μCi/μl; 3,000 Ci/mmol; Amersham) was used to label 200 ng of primer in 20 μl of reaction buffer (70 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 5 mM DTT) (New England BioLabs) containing 1 U of T4 polynucleotide kinase per μl.
Enzymatic probing of RNA secondary structure.
Two plasmids containing the T7 RNA polymerase promoter followed by the 3′ 110 nt of either the fTH (pUCRUB3′110-fTH) or HPV77 (pUCRUB3′110-HPV) strain, a 20-nt poly(A) tract, and an EcoRI site for linearization were constructed. Intracellular RNA from fTH- or HPV77-infected Vero cells was extracted at 48 h postinfection by using Tri-reagent (Molecular Research Center, Cincinnati, Ohio) according to the manufacturer’s protocols. First-strand cDNA was primed with primer 105 (Table 1) and then amplified by PCR using primer 105 as the reverse primer and forward primer 337 (Table 1). PCR amplification using Ex-taq DNA polymerase (Panvera, Madison, Wis.) was done as described previously (44). The HindIII-EcoRI-restricted PCR fragment was ligated with HindIII-EcoRI-restricted pUC18 (Gibco/BRL).
TABLE 1.
DNA primers used in this study
| Manipulation | Primer | Sequence (5′-3′)a | Useb | Polarityc |
|---|---|---|---|---|
| Site-directed mutagenesis of the 3′ UTR | 52 | CGGGATCCACGCCCAC | Cloning (9169 to 9182) | + |
| 313 | GTGGGCCCCCGCGGGAAACC | ∗S 9711 (C to G) | + | |
| 315 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTCCACTAGCGCGGCGCTATAG | Δ 9705 to 9762 | − | |
| 316 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTGTGGCCTAGTGCGGGTTTCG | Δ 9728 to 9762 | − | |
| 322 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTCGGGGCTCTAGTGGCCTAGT | Δ 9745 to 9762 | − | |
| 323 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTGCAACAGGTGCGGGGATCTA | Δ 9755 to 9762 | − | |
| 324 | TGGGCCCTAGCTCCCCGCACCTGTTGC | ∗Δ 9708 to 9733 | + | |
| 327B | CGCGCTAGTGGGCCCGCGGGCGAAACCCGCACTA | ∗S 9706 (C to G); 9709 (C to G) | + | |
| 328 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTTATACAGCAACAGGTGCGGGG | Δ 9762 | − | |
| 334 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTATACAGCAACAGGTGCGGGG | Δ 9761 to 9762 | − | |
| 335 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTTACAGCAACAGGTGCGGGGA | Δ 9760 to 9762 | − | |
| 340 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTAGCAACAGGTGCGGGGATCT | Δ 9757 to 9762 | − | |
| 341 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTCAGCAACAGGTGCGGGGATC | Δ 9758 to 9762 | − | |
| 359 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTCAGCAACAGGTGCGGGGAT | Δ 9759 to 9762 | − | |
| 360 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTxxAGCAACAGGTGCGGGGAT | Δ 9759 to 9762; S 9757 and 9758 (GU to xx) | − | |
| 368 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTxxCAGCAACAGGTGCGGGGA | Δ 9760 to 9762; S 9758 and 9759 (UA to xx) | − | |
| 390 | CACTAGATCCCCGCATGCTGTATAGAAAAA | ∗Δ 9745 to 9749 | + | |
| 391 | AGTGGGCCCCCGCGCCCGCACTAGGCCACT | ∗Δ 9714 to 9718 | + | |
| 392 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTCTATAGCAACAGGTGCGGGGATCTA | Δ 5956 to 9757 | − | |
| 393 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTCTATAACAGGTGCGGGGATCTAGTG | Δ 9753 to 9757 | − | |
| 524 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTCTATAACAACAGGTGCGGGAATC | S 9655 (U to A) | + | |
| 447 | GTGGGCCCCCGCGCGCCCGCACTAGGCCAC | ∗Δ 9718 to 9720 | + | |
| 448 | GCACTAGGCCACTAGGCACCTGTTGCTGTA | ∗Δ 9739 to 9744 | + | |
| 449 | CTAGATCCCCGCACTTGCTGTATAGAAA | ∗Δ 9749 to 9751 | + | |
| 450 | CGTGAATTCTTTTTTTTTTTTTTTTTTTTCTATATCTAGACAGGTGCGGGGATC | S 9753 to 9757 | − | |
| 545 | CGTGAATTC(G/A)ACAACAGGTGCGGGGATC | Δ 9758 to 9762, poly(A) and 9755; S 9757 (G to C or U) | + | |
| 523 | GCTAGTGGGCCCCCGCCGAAACCGCGACTAGGCCACTAG | ∗Δ 9709; SW 9710 to 9712 with 9720 to 9722 | + | |
| 523B | GCTAGTGGGCCCCCCGCCGAAACCGCGACTAGGCCACTAG | ∗Sw 9710 to 9712 with 9720 to 9722 | + | |
| 573 | CGTGAATTCTTTTTTTTTTTCTATTGTGCAACAGGTGCGGGA | S 9756 to 9758 (UGU to ACA) | − | |
| 574 | CGTGAATTCTTTTTTTTTTTCTATACACGTACAGGTGCGGGAATC | S 9753 to 9755 (UGC to ACG) | − | |
| 575 | CGTGAATTCTTTTTTTTTTTCTATACAGCATGTGGTGCGGGAATCTAG | S 9750 to 9752 (UGU to ACA) | − | |
| 576 | CGTGAATTCTTTTTTTTTTTCTATCACGACCACGGTGCGGGAATCTAG | S 9750 to 9758 (UGUUGCUGU to GUGGUCGUG | − | |
| 344B | AGCGCCGCGCTAGTGGATCCCCGCACCTGT | ∗Δ 9702 to 9747 | + | |
| 344C | AACCCGCACTAGGCCGATCCCCGCACCTGT | ∗Δ 9744 to 9747 | + | |
| 345 | GTGCGGGGATCTAGTCACTAGCGCGGCGCA | ∗Δ 9703 to 9733 | + | |
| 507 | CGTGAATTCTATACAGCAACAGG | Δ poly(A) | − | |
| 508A | CGTGAATTCTTTTTTTTTTTCTATACAGCAACAGG | Δ 10 A’s from poly(A) | − | |
| 508T | CGTGAATTCAAAAAAAAAACTATACAGCAACAGG | Δ poly(A); S 10 T’s with A | − | |
| 509 | CGTGAATTCAGCAACAGGTGCGGGGATC | Δ 9758 to 9762 and poly(A) | − | |
| Site-directed mutagenesis of SL2 | 430 | GCTATAGCGCCGCGC(A/G)AGTGGGCCCCCGCG | ∗S 9658 (U to A or G) | + |
| 419 | GCTGTGCCAAATGCATGTACTACTTGCGCG | ∗S 9655 (U to A) | + | |
| 435 | CCAATGCATGTACTACCTTCGCGGCGCTATAGCG | S 9674 (U to C); 9676 (G to U) | + | |
| 436 | CCAATGCATGTACTACTTGCGCGGGGCTATAGCGCCGCGC | S 9680 (C to G) | + | |
| 444 | CCAATGCATGTACTGCTGGCGCGGCGCTATAG | S 9675 (U to G) | + | |
| 461 | CCAATGCATGTACTACTTGCGGGGCGCTATAGCGCC | S 9677 (C to A); 9679 (C to G) | + | |
| 330 | ATAGCGCCGCGCTAGCGGGCCCCCGCGCGAA | ∗S 9701 (U to G) | − | |
| Construction of plasmids for RNase probing and RNA probes | 337 | TCGAAGCTTAATACGACTCACTATACGTTGTACTACTTGCGCG | Cloning (9663 to 9680) | + |
| 420 | TCGAAGCTTAATACGACTCACTATACGATGCATGTACTACTTGCGCG | Cloning (9659 to 9680) | + | |
| 498 | TCGAAGCTTAATACGACTCACTATAGGCCCCCGCGCGAAA | Cloning (9703 to 9718) | + | |
| 105 | ACGTGAATTCTTTTTTTTTTTTTTTTTTTT | Cloning) [EcoRI plus poly(A)] | − |
Restriction sites used for cloning are underlined, and the T7 RNA polymerase promoter is in italics. x, any nucleotide other than the WT one; (X/X), either of the two nucleotides was included in the primer.
b With primers used for cloning, the nucleotide numbers in the RUB genome to which the primer corresponds are given in parentheses. Mutagenic primers were used to delete (Δ), make substitutions (S) at, or switch (Sw) the indicated nucleotides. Primers used in the asymmetric mutagenesis strategy are marked with asterisks.
+, primers colinear with the RUB genome; −, primers complementary to the RUB genome.
RNA transcripts from EcoRI-linearized pUCRUB3′110 were resuspended in RNase-free water after removal of the DNA template by digestion with RNase-free DNase (Promega, Madison, Wis.). The RNA transcripts were denatured in RNA renaturing buffer (10 mM Tris-HCl [pH 7.0], 10 mM MgCl2, 100 mM KCl) (22) at 80°C for 5 min, slowly cooled to room temperature, and placed in an ice slurry bath. For digestion with mung bean nuclease, the transcripts were renatured in mung bean nuclease reaction buffer (50 mM sodium acetate [pH 5.0], 30 mM NaCl, 1 mM ZnSO4). In each reaction, approximately 500-ng aliquots of RNA transcripts were subjected to digestion with various amounts (0.001 to 0.1 U) of RNase A (Sigma, St. Louis, Mo.), RNase T1 (0.005 to 0.1 U; Pharmacia), RNase T2 (0.05 to 0.5 U); (Pharmacia), or mung bean nuclease (1 to 10 U; New England BioLabs), or the double-stranded RNase, RNase V1 (0.001 to 0.1 U; Pharmacia), in a total volume of 20 μl at room temperature or in an ice slurry bath for 30 min followed by the addition of equal volume of stop solution (0.5 M sodium acetate) and two phenol-chloroform extractions (51). The digested RNA fragments were adjusted to a concentration of 0.3 M sodium acetate, precipitated with 3 volumes of 100% ethanol and 1 μg of glycogen (Boehringer Mannheim Biochemicals), and resuspended in water.
5′ 32P-labeled primer 358 [5′-TTTTTTCTATACAGCAACAG-3′; complementary to 3′ the 14 nt of the RUB genome plus 6 A’s of the poly(A) tract] was purified by electrophoresis in an 8% polyacrylamide–8 M urea gel. The labeled primer was visualized by autoradiography and eluted in buffer containing 0.5 M ammonium acetate, 10 mM MgCl2, 1 mM EDTA, and 0.1% SDS at 37°C for overnight followed by ethanol precipitation. The eluted probe was resuspended in 50 μl of water. For each extension reaction, RNA transcripts in Superscript reverse transcriptase reaction buffer were annealed with approximately 10,000 cpm of 32P-labeled primer 358. After primer extension, an equal volume of loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF) was added to each reaction followed by denaturation at 95°C for 2 min and electrophoresis in an 8% polyacrylamide–8 M urea sequencing gel.
Site-directed mutagenesis.
Mutations were created by PCR using primers containing the desired mutations, Ex-taq polymerase, and EcoRI-linearized Robo302 plasmid as a template under conditions optimized for amplification of the high-G+C RUB sequences (39). Primers for constructing mutants using PCR amplification are listed in Table 1. Generally, the PCR products were purified by phenol-chloroform extraction and ethanol precipitation with sodium acetate at a final concentration of 0.3 M. The DNA was then pelleted by centrifugation, dissolved in water, and digested with appropriate restriction enzymes for cloning. Following cloning, mutations were confirmed by DNA sequencing using a dideoxy-chain termination sequencing kit from United States Biochemicals (Cleveland, Ohio).
To construct the deletion mutants near the 3′ terminus (315, 316, 322, 323, 328, 334, 335, 340, 341, 359, 360, 368, 392, 393, 524, 507, 508A, 508T, and 509), the mutagenic primers contained an EcoRI site (corresponding to the unique EcoRI site in Robo302 used for runoff transcription), a tract of 20 T’s, and sequence complementary to the RUB 3′ end extending 15 nt beyond the deletion. PCR amplification was done with the mutagenic primer and primer 52 (Table 1; colinear with nt 9167 to 9186 of the RUB genome; a unique BamHI site at nt 9170 of the RUB genome within this sequence used for cloning is underlined). The BamHI-EcoRI-restricted PCR fragments were used to replace the corresponding fragment in Robo302.
To construct point mutations in SL2 (419, 430-AAG/GAG, and 330) and mutants in SL3 (313, 327, 324, 344B, 344C, 345, 391, 447, 523, and 523B), the hinge region (448 and 344C), or SL4 (390, 449, and 450), an asymmetric PCR amplification starting from single-stranded oligonucleotides was devised. Briefly, a single-stranded DNA was amplified by asymmetric PCR using a colinear mutagenic primer (Table 1; labeled with asterisks) and EcoRI-linearized Robo302 template; this single-stranded DNA product, which extended from the mutation through the poly(A) tract, was then used as a template for the second round of asymmetric PCR using primer 105 [oligod(T) plus an EcoRI site] to amplify a complementary strand from the first-round PCR product. The third-round PCR BamHI-EcoRI fragment and second-round PCR product were amplified by using primer 52 and an EcoRI-linearized Robo302, respectively as templates. The BamHI-EcoRI-restricted PCR fragment was introduced into Robo302. The yield of mutations using this strategy was between 25 and 80%.
One of the mutations from the asymmetric PCR introduced an NsiI site (419) upstream from SL2 (nt 9665) and was used to introduce point mutation in SL2 (435, 436, 444, and 461).. The infectivity of 419 transcripts was tested and gave the same titer as Robo302. An NsiI-EcoRI PCR fragment was amplified with primer 105 and a mutagenic primer consisting of RUB sequences containing the NsiI site and the desired point mutation in SL2. The NsiI-EcoRI-restricted fragment was used to replace the corresponding region in clone 419.
Confirmation of mutation by RT-PCR and automated sequencing.
Single plaques were picked from transfection plates and amplified by one passage in Vero cells (in the case of occasional mutants that produced no plaques, medium from the DMEM plate was amplified). Once the CPE became obvious, intracellular RNA was extracted with Tri-reagent and used as a template for reverse transcription-PCR (RT-PCR). First-strand cDNA was synthesized with primer 105 and amplified by PCR with primers 105 and 52 (Table 1). The PCR fragments were examined on a 1% agarose gel and purified with a Wizard PCR Prep kit (Promega). The 3′-terminal 188 nt from these PCR fragments were amplified asymmetrically by using primer 451 (5′-GGGCTGCTGCCCATTGG-3′; colinear to RUB nt 9577 to 9599) and an ABI PRISM Dye Cycle Sequencing Ready Reaction kit (PE Applied Biosystems) and purified on Centrisep columns (Princeton Separations, Adelphia, N.J.), and the nucleotide sequences were determined by cycle sequencing using an ABI 373 sequencer (The Perkin-Elmer Corporation, Foster City, Calif.).
Purification of MBP-CAL, in vitro kinase reaction, and dephosphorylation reaction.
A pMAL-c2 vector construct encoding an MBP-CAL fusion protein was provided by C. D. Atreya (Center for Biologies Evaluation and Research, Food and Drug Administration, Bethesda, Md.). The fusion protein was expressed in Escherichia coli DH5α and purified by maltose chromatography according to the manufacturer’s protocol followed by dialysis against 10 mM Tris-HCl (pH 7.4)–10 mM KCl–5% glycerol buffer at 4°C overnight and storage at −20°C. The amount of protein was determined with a Bio-Rad (Hercules, Calif.) protein assay. The in vitro kinase reaction was carried out by incubating 2 μg of purified MBP-CAL in kinase reaction buffer at 30°C for 30 min as described previously (50).
Dephosphorylation of MBP-CAL was done with calf intestine alkaline phosphatase (CIAP; Promega). One microgram of MBP-CAL was incubated with 1 U of CIAP per ml in the presence of 0.1 mM Zn2+, a cation essential for CIAP activity, with cytolysis buffer (40 mM KCl, 25 mM Tris-HCl [pH 7.5], 1% Triton X-100, 10 μg of leupeptin per ml, and 1× Complete EDTA-free protease inhibitor cocktail [Boehringer Mannheim]) at 37°C for 30 min.
Preparation of cell lysates.
Cell lysates were prepared as previously described (37). Briefly, confluent monolayers of Vero cells in 75-cm2 T flasks were washed with ice-cold phosphate-buffered saline twice, scraped with a rubber policeman, and pelleted by centrifugation at 400 × g for 10 min. Pelleted cells were resuspended in 200 μl of cytolysis buffer as described above and allowed to incubate on ice for 30 min. The lysate was clarified by centrifugation at 11,000 × g for 15 min, and the supernatant was collected. The amount of protein was determined by a Bio-Rad protein assay.
In vitro synthesis of RNA probe.
Construction of plasmids containing the T7 RNA polymerase promoter followed by the 3′ 110 nt of each SL2 mutant, a 20-nt poly(A) tract, and an EcoRI site was done by PCR amplification using primers 420 and 105 (Table 1), with EcoRI-linearized SL2 mutant DNA (435, 436, 444, 461, and 430-GAG) as a template. The HindIII-EcoRI-restricted PCR fragments were cloned into pUC18. Construction of plasmids containing the T7 RNA polymerase promoter followed by the 3′ untranslated region (UTR) was also done by PCR using primers 498 (Table 1) and 105 [with poly(A) tract and an EcoRI site] or 509 [Table 1; complementary to RUB genome nt 9739 to 9757 to produce a 3′ UTR without the poly(A) and the 3′ terminal 5 nt but with an EcoRI site], and the HindIII-EcoRI-restricted fragment was cloned into pUC18 (resulting in 498 and 498-ΔA, respectively).
32P-labeled 498-ΔA T7 RNA polymerase transcripts were transcribed from EcoRI-restricted 498-ΔA plasmid. Probes containing SL2 only were transcribed from each pUC18-SL2 construct that had been restricted with Bsp120I. In both cases, the 32P-labeled probes were electrophoresed in 8% polyacrylamide–8 M urea gels, visualized by autoradiography, and eluted in 0.5 M ammonium acetate–0.1% SDS–1 mM EDTA at 37°C overnight (5). The eluted probe was harvested by ethanol precipitation in the presence of 1 μg of glycogen. The probe was dissolved in water, and the radioactivity was determined by liquid scintillation spectrophometry.
Gel mobility shift assay.
Various amounts of cell lysate or purified MBP-CAL were incubated with 10,000 cpm of the different 32P-labeled probes (∼0.6 to 0.9 ng) in cytolysis buffer containing 100 ng of poly(I)-poly(C) (P-L Biochemicals, Inc., Milwaukee, Wis.) and 10 U of RNasin in a final volume of 10 μl for 15 min on ice or 30 min at room temperature. In competition assays, various amounts of unlabeled competitor RNAs were preincubated with the cell lysates prior to addition of the probe. RNA-protein complexes were electrophoresed in nondenaturing 4% polyacrylamide gels (50:1, polyacrylamide-to-bisacrylamide ratio) in Tris-borate-EDTA buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA) at room temperature. The gel was then dried, and the RNA-protein complexes were visualized by autoradiography.
UV-induced cross-linking.
Binding reactions were set up as described above with 32P-labeled RNA probe (∼70,000 cpm) and 20 to 40 μg of cell lysate in each reaction. The binding reactions were then transferred to an ice-water bath and irradiated with a 254-nm UV lamp (UV Crosslinker; Fisher Scientific, Pittsburgh, Pa.) placed 3 cm from the reaction for 30 min. After irradiation, 1 U of RNase A was added to the reaction mixture and incubated for another 15 min at room temperature to remove unprotected RNA probe. The UV cross-linked products were boiled in Laemmli sample buffer for 2 min and resolved on a discontinuous SDS–10% acrylamide gel. The complexes were then visualized by autoradiography (5, 37).
Nucleotide sequence accession number.
The sequence shown in Fig. 1A has been assigned GenBank accession no. M15240.
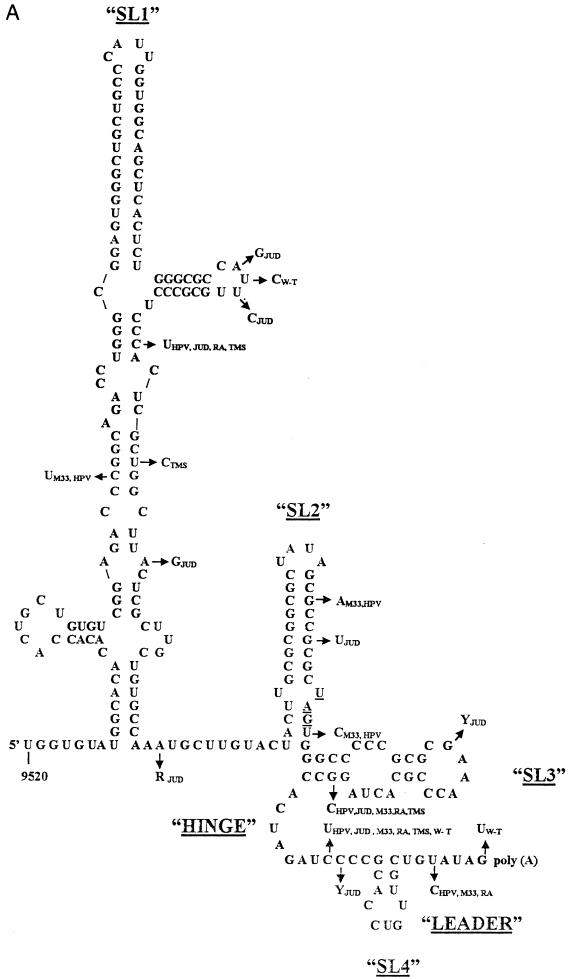
FIG. 1.
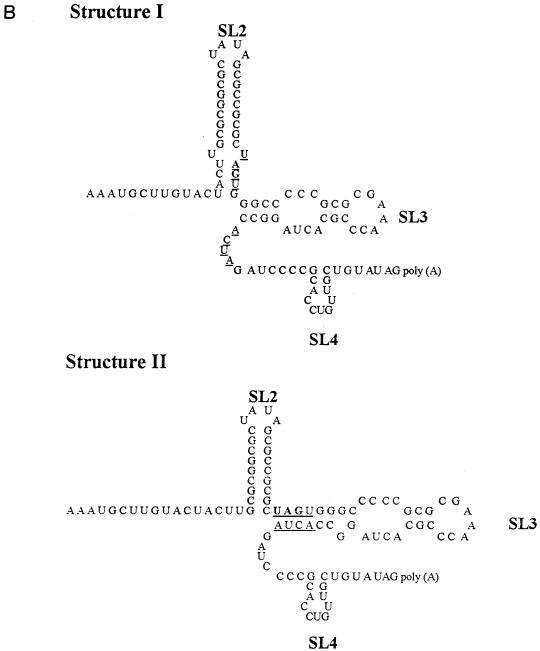
(A) Thermodynamically predicted secondary structure of the 3′-terminal 240 nt of the RUB genome (W-Therien strain). A structure with four prominent SLs (SL1 to SL4) and a ΔG of ∼−99.8 kcal/mol was predicted; SL2 is the structure shown to interact with CAL. Both SL1 and SL2 are within the E1 coding region; the UAG stop codon, located at the base of SL2, is underlined. SL3 and SL4 are within the 3′ UTR and are connected by a single-stranded hinge region. SL4 is followed by a 7-nt leader and the poly(A) tract. Nucleotide variations found in other RUB strains are indicated by arrows. fTH, F-Therien (clear-plaque variant derived from W-Therien) (accession no. D00156); M33, M33 (X05259); HPV, HPV77 (vaccine strain derived from M33) (M30776); JUD, Judith (L19420); THM, Thomas (L19421); RA, RA27/3 (vaccine strain currently used in the United States) (L78917). (B) Alternative structures of SL2 and SL3. Structure I (top) was predicted for the W- and F-Therien strains (ΔG ∼−30 kcal/mol); structure II (bottom) was predicted for the M33/HPV77, Judith, RA27/3, and Thomas strains (both are drawn by reference to the W-Therien sequence) (ΔG ∼−25 Kcal/mol in the RA27/3 strain). The complementary UAGU and ACUA stretches which pair to form the longer SL3 in the second structure are underlined.
RESULTS
Thermodynamically predicted secondary structure of the 3′ cis-acting elements.
The secondary structure of the 3′-terminal 305 nt of the RUB genome (the region conserved in DI RNAs) was analyzed by using the sequence of our standard lab strain, fTH, and four SLs were predicted (Fig. 1A). SL1 (ΔG ∼ −63.2 kcal/mol), the largest, was between nt 9527 and 9647. SL2 (ΔG ∼ −20.3 kcal/mol), which has been shown to interact with CAL, is in the coding sequences at the exact C terminus of the E1 protein coding sequence (nt 9671 through 9702). In the 3′ UTR were SL3 (nt 9703 through 9730; ΔG ∼ −7.9 kcal/mol) and a very short SL, SL4 (nt 9742 through 9753; ΔG ∼ −0.8 kcal/mol). SL4 is connected with SL3 by an 11-nt single-stranded hinge region and followed by a 7-nt single-stranded leader sequence preceding the poly(A) tract.
Nucleotide variations within this sequence among six other RUB strains are marked in Fig. 1A by arrows. Most nucleotide variations in SL1 occurred within bulges or loop regions, and those that occurred within stems did not affect base pairing, which explained why SL1 is well conserved among these strains by thermodynamic prediction. Interestingly, in some of these strains an alternative structure of SL2 and SL3 consisting of a shorter SL2, which retains the GC-rich stem but not the U-U bulge, and a longer SL3 stem which utilizes an ACUA stretch from the single-stranded hinge region to base pair with UAGU from SL2 was predicted (Fig. 1B). Nucleotide variations in SL2 were noticed in both the M33/HPV77 and Judith strains within the GC-rich stem at nt 9689 and 9692, respectively, but not at the U-U bulge. In both strains, these variations led to the formation of the alternative structure because of a lower ΔG of SL2. However, this alternative structure was also formed by the RA27/3 sequences which lacked these variations in SL2. Most of the other nucleotide variations occurred in the predicted hinge region and the 7-nt leader; only one significant nucleotide variation was found in SL3.
Analysis of the secondary structure of SL2 and SL3 by RNase probing.
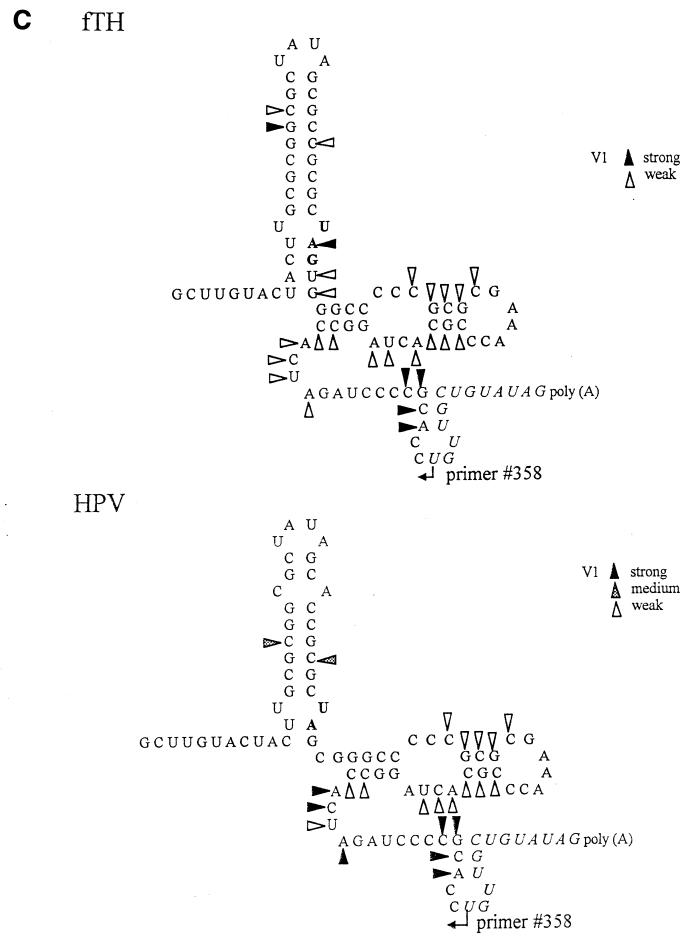
Considering the apparent importance of the unpaired U loop in CAL binding (37), it was important to confirm the secondary structure of SL2 and SL3. Therefore, we performed RNase probing using RNA transcripts from pUCRUB3′110-fTH and -HPV, plasmids containing the 3′ 90 nt plus a 20-nt poly(A) tract derived from the fTh and HPV77 strains, which are predicted to have the first and the second SL2-SL3 conformations, respectively. RNA transcripts from these two constructs were subjected to digestion by several RNases, and the results of RNase digestion were resolved by primer extension. A representative gel is shown in Fig. 2A, and the digestion observed is summarized in Fig. 2B (single-stranded RNases) and C (double-stranded RNase V1).
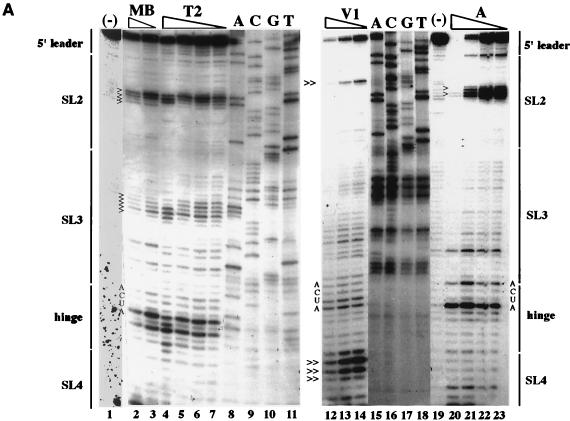
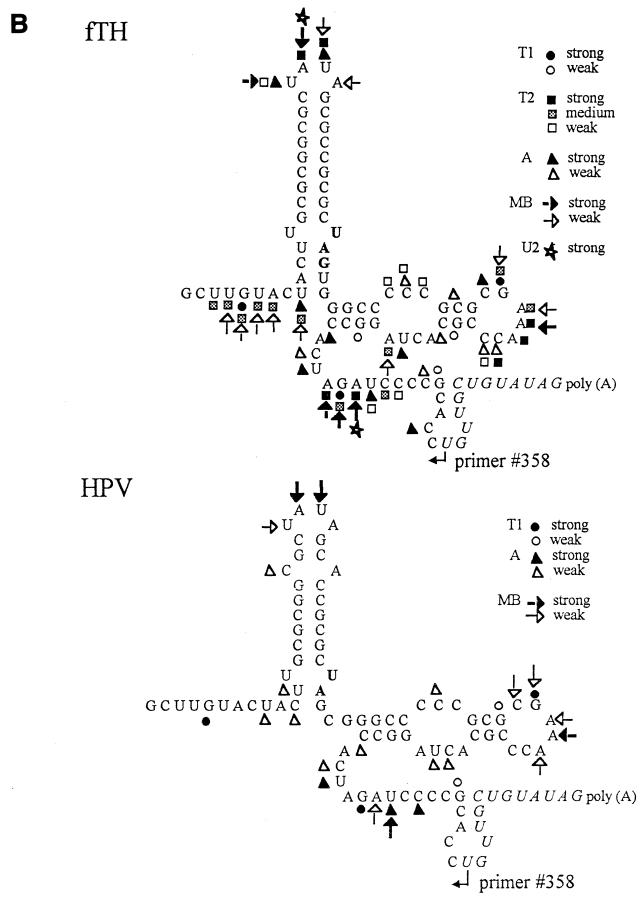
FIG. 2.
Analysis of SL2/SL3 conformations by RNase probing. RNA probes consisting of the 3′-terminal 90 nt plus a poly(A) tract from the fTH or HPV77 (HPV) strain were digested with a battery of single- or double-stranded RNases, and the digestion pattern was resolved by primer extension. (A) Results of primer extension from pUC3′RUB110-fTH transcripts digested in a 20-μl reaction with no RNase (−; lanes 1 and 19); mung bean nuclease (MB; cleaves single-stranded RNA with no nucleotide specificity), 10 (lane 2) and 5 (lane 3) U; RNase T2 (cleaves single-stranded RNA with no nucleotide specificity), 0.5 (lane 4), 0.25 (lane 5), 0.1 (lane 6), and 0.05 (lane 7) U; RNase V1 (cleaves double-stranded RNA with no nucleotide specificity), 0.1 (lane 12), 0.05 (lane 13), and 0.025 (lane 14) U; and RNase A (cleaves single-stranded RNA with preference for C and U), 0.1 (lane 20), 0.05 (lane 21), 0.025 (lane 22), and 0.01 (lane 23) U. In lanes 8 to 11 and 15 to 18 are the sequencing ladders for orientation produced by using the primer extension primer and plasmid pUCRUB3′110-fTH as a template. Structural regions within the probe are shown on both margins. Digestion landmarks highlighted include the loops of SL2 and SL3 (>), both of which are sensitive to the single-stranded RNases but not RNase V1 (the SL3 loop is not sensitive to RNase A because it contains a GAAA sequence), and the single efficient RNase V1 digestion site in SL2 and the digestion of the 5′ side of the SL4 stem by RNase V1 (≫). The overall results of RNase probing on both probes are summarized in panels B (single-stranded RNases) and C (double-stranded RNase V1). Because the primer was complementary to the 11 nt preceding the poly(A) tract (italics), digestion within these nucleotides could not be resolved. Single-stranded RNases: T1, RNase T1 (G residue); T2, RNase T2 (no specificity); A, RNase A (C and U residues); MB, mung bean nuclease (no specificity); U2, RNase U2 (A residue).
Probing with several single-stranded RNases (RNases A, T1, T2, and U2 and mung bean nuclease) confirmed that structure I exists in pUCRUB3′110-fTH transcripts since the ACUU at the 5′ base of SL2 was protected whereas the ACUA in the hinge region (critical in formation of the long SL3 of structure II) was sensitive (these nucleotides are indicated in the gel in Fig. 2A), as predicted in structure I but not in structure II. However, inefficient digestion by double-stranded RNase V1 was also observed in ACUA in the hinge region (Fig. 2A), in comparison to the other nucleotides in the hinge region, which were digested only by single-stranded RNases. This suggests that an equilibrium between these structures may exist. Digestion of pUCRUB3′110-HPV transcripts revealed that the CUA sequence in the hinge region (base pairing in both SL2 of structure I and SL3 of structure II is shortened by the presence of a C at nt 9699 in this strain) was susceptible to both single- and double-stranded nucleases. These data are also consistent with equilibrium but indicate that structure II is more prevalent in HPV77 than in fTH.
In both strains, single-stranded RNase probing confirmed the loop regions of both SL2 and SL3 (highlighted in Fig. 2A) and the bulge of SL3; however, no digestion by these nucleases occurred at the U-U bulge in SL2. Interestingly, although the stem of SL2 and lower stem of SL3 were resistant to single-stranded RNase digestion, the double-stranded RNase, RNase V1, digested only one site in the SL2 GC-rich stem (Fig. 2A) and digested only inefficiently at the base of SL2 and within SL3; digestion within SL3 by this RNase included both the upper stem and some nucleotides on either side of this stem predicted to be unpaired. Interestingly, three strong cleavages by RNase V1 occurred in the 5′ stem of SL4 (Fig. 2A), indicating that SL4 exists under the experimental conditions even though it is not predicted to be as stable as SL2 or SL3.
Site-directed mutagenesis of 3′-terminal 90 nt.
To analyze the significance of the sequences within the 3′-terminal 90 nt of the RUB genome on replication, site-directed mutagenesis was performed with Robo302, a RUB infectious clone. For each mutant construct, RNA transcripts from at least four individual plasmids were used for transfection. For those mutants which did not produce CPE within three passages, mutagenesis, cloning, and transfection were repeated one more time to confirm the observation, while for those mutations which yielded viable virus, the nucleotide sequence was confirmed after one or two passages of the mutant virus in Vero cells by RT-PCR amplification of the 3′-terminal 600 nt and sequencing of the amplification product.
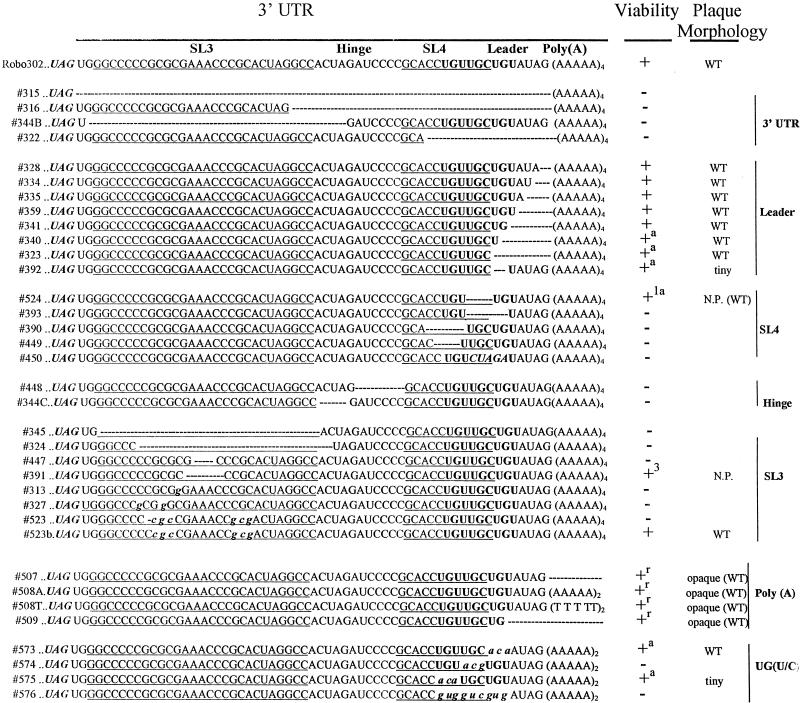
(i) Site-directed mutagenesis of the 3′ UTR.
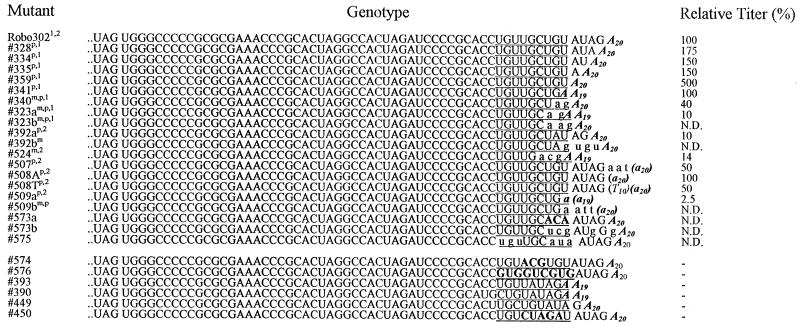
Deletion mutagenesis was used to probe the 3′ UTR; the mutants are listed in Fig. 3. Large deletions, including one encompassing the complete UTR (315) as well as most of a number of smaller deletions, were nonviable, indicating that most of the 3′ UTR is critical for RUB replication. An exception was the 3′-terminal 5 nt as deletions of 1 (328), 2 (334), 3 (335), 4 (359) or all 5 (341) of these nt were viable. These mutated transcripts had transfection efficiencies similar to those of Robo302 transcripts; the plaque morphology and titer produced by each of these mutant viruses were also similar to those for Robo302 virus (∼106 to 107 PFU/ml). Viruses were also recovered from mutated transcripts with the 3′-terminal 6 (340) and 7 (323) nt deleted; however, the transfection efficiency of these mutated transcripts was lower (indicative of reversions occurring), and sequence analysis revealed that the viruses recovered had an AG added between the deletion and the poly(A) (Fig. 4). The titer produced by these viruses was about 10- to 100-fold less than that of the WT Robo302 virus (∼105 PFU/ml). Deletion of nt 6 to 10 (393) abolished infectivity completely, indicating that the specific sequence is critical for viability rather than the length of this region. Further mutagenesis demonstrated the importance of nt 6 and 7, since a dramatic reduction in transfection efficiency was observed with mutant transcripts lacking these two nucleotides (392). This mutant virus also formed tiny plaques and grew to a 10-fold-lower titer than did Robo302. Virus recovered from this mutant in one transfection preserved the mutated sequence, while virus recovered from a second independent transfection had an insertion and a rearrangement of the downstream 5 nt (UAGUGU).
FIG. 3.
Site-directed mutagenesis of the 3′ UTR. The Robo302 sequence of the 3′ UTR beginning with the UAG stop codon of the E1 gene is given on the top line, and the structural features of the UTR are delineated [in addition to those shown in Fig. 1, a UG(U/C) triplet is marked in bold]. Among the mutations created, deletions are indicated by dashes and substitutions are indicated by lowercase letters. Viability is indicated as (nonviable), + (viable with plaques or CPE apparent in transfected cultures), +1 (CPE apparent after one passage), and +3 (CPE after three passages). Sequences were confirmed on all viable mutants after one amplification in Vero cells; those designated +a contained alterations in the mutant sequence (Fig. 4), while those designated +r contained regenerated poly(A) tracts and formed opaque plaques in transfected cells but clear plaques in the subsequent passages. Mutants which formed plaques similar to those of Robo302 on transfection plaques are inicated as “WT,” while those that formed tiny plaques on transfection plaques or no plaques are indicated as “tiny” and “N.P.,” respectively. Plaque morphologies indicated in parentheses are those formed after subsequent passaging for amplification.
FIG. 4.
Characterization of viruses with mutations in SL4, the leader, and the poly(A) tract. Sequences determined from 15 viable mutants are given (a and b indicate results of independent transfections); altered nucleotides not present in the original transcripts are in lowercase. For comparison, sequences of six nonviable mutants are also given at the bottom. To determine replication ability, mutant viruses isolated from transfection plaques (p) or medium (m) were amplified by one passage in Vero cells, and the titers of the amplified stocks were determined. These stocks were then used for infection of Vero cells (in the first experiment [superscript “1”], which compared the replication of Robo302 and eight mutants, the multiplicity of infection [MOI] was 5; in the second experiment [superscript “2”], which compared Robo302 and six additional mutants, the MOI was 0.01). The infected culture media were harvested at 2 (MOI = 5) or 3 (MOI = 0.01) days postinfection, and the titer (PFU per milliliter) was determined. The relative titer produced by each mutant is shown in comparison with the Robo302 titer (set at 100%) produced in the same experiment (N.D., not determined). The proposed critical stretch of UG(U/C) triplets is underlined.
Although SL4 is not a particularly stable structure, mutagenesis demonstrated the importance of its sequences: deletion of the entire or part of SL4 loop (390 and 449) or destabilization of SL4 by replacing the stem sequences with an XbaI site (TCTAGA) (450) all abolished viral viability. A mutation with a deletion of the 3′ stem of SL4 (524) was viable with a reduced transfection efficiency; sequence confirmation showed that the viruses recovered had the downstream leader rearranged.
Interestingly, a UG(U/C) triplet occurs between nt 5 and 13 from the 3′ terminus. Mutants with deletions or point mutations in the leader or SL4 which resulted in disruption of the integrity within this sequence were lethal, while viable mutants, including mutants that exhibited rearrangements following transfection, either maintained this sequence or encroached on only the 3′ member of the triplet (Fig. 4). Since most nucleotides in these UG(U/C) triplets share a keto group, it is possible that the moiety in this sequence is important for RUB RdRp recognition, similar to the finding that functional groups are most important in subgenomic promoter recognition by the brome mosaic virus RdRp (49). To test this possibility, we constructed four mutations with changes in this sequence: 573, a transversion mutation of the first UG(U/C) (nt 5 to 7 to ACA); 574, a transversion mutation of the second UG(U/C) (nt 8 to 10 to ACG); 575, a transversion mutation of the third UG(U/C) (nt 11 to 13 to ACA); and 576, a mutant maintaining the keto group by changing the entire sequence (UGUUGCUGU) to GUGGUCGUG. 573 transcripts had a transfection efficiency of approximately 10% of the efficiency of Robo302 transcripts and 574 was lethal, while only two plaques were recovered from the 575-transfected plate (575 plaques were tiny even after two passages in Vero cells). Sequence analysis showed that the virus recovered from both 573 and 575 had sequence changes; significantly, the ACA inserted in 575 reverted to UGU. 576 was lethal, ruling out the necessity of a simple string of keto groups. However, the maintenance of the first two triplets (UGUUGC) may still be significant in terms of functional groups being important in RdRp promoter activity. It is to be noted, however, that the mutation in 574 was located in the stem of SL4, while 573 and 575 were located in either side of the SL4 3′ stem; thus, these results again stressed the importance of SL4. Overall, these mutations reconfirmed our original finding that nt 5 to 14 are critical in viral viability.
Two deletions within the hinge region, 448 and 344C, were also lethal. The latter of these mutants deleted the ACUA stretch (nt 3729 to 3732) (344C) by which the alternative SL2-SL3 conformation could be formed. Deletion of all (345) or the upper part (324) of SL3 was not tolerated, although viruses were recovered from a mutation that deleted the SL3 loop (391). However, this mutant did not produce CPE until after three passages. Point mutations within SL3, C9706 (to G) and C9708 (to G) (327) or C9711 (to G) (313) (both constructed to create a SacII site) were not viable as was mutant 523, which had the sequences of the upper stem switched with a C inadvertently deleted from the bulge. All three of these mutations introduced subtle alterations into the SL3 structure and changed the calculated thermodynamic stability (Fig. 5). Interestingly, a mutant (523B) which replaced the missing C in 523 was viable; 523B transcripts had a transfection efficiency similar to that of Robo302 and produced WT plaques. Taken together, these data indicate that maintenance of the overall structure of SL3 is critical to viral replication; however, the sequence in the upper stem is not important.
FIG. 5.
Thermodynamically predicted secondary structure of SL3 in virus transcripts with mutations in this region. The native SL3 structure (WT) is at the top. Point mutations are underlined. The entropy of each mutant is also given.
The role of the poly(A) tract was also investigated (Fig. 4). Transcripts without poly(A) (507) and with U10 replacing the poly(A) tract (508T) had specific infectivities equivalent to those of Robo302 transcripts, although these mutants produced opaque plaques instead of clear plaques in transfected cells. After one passage for amplification, these viruses produced clear WT plaques, and sequence analysis showed that these mutants had a poly(A) tract regenerated at the 3′ end. In the case of poly(A) deletion (507), the poly(A) was added to the EcoRI linearization site whereas the U10 mutants had poly(A) added after a stretch of 10 U’s (508T). Viable viruses were also recovered from mutants had both the poly(A) tract and the 3′-terminal 5 nt deleted (509); the poly(A) tract was also regenerated without the 3′-terminal 5 nt in these viruses.
(ii) Mutagenesis in SL2.
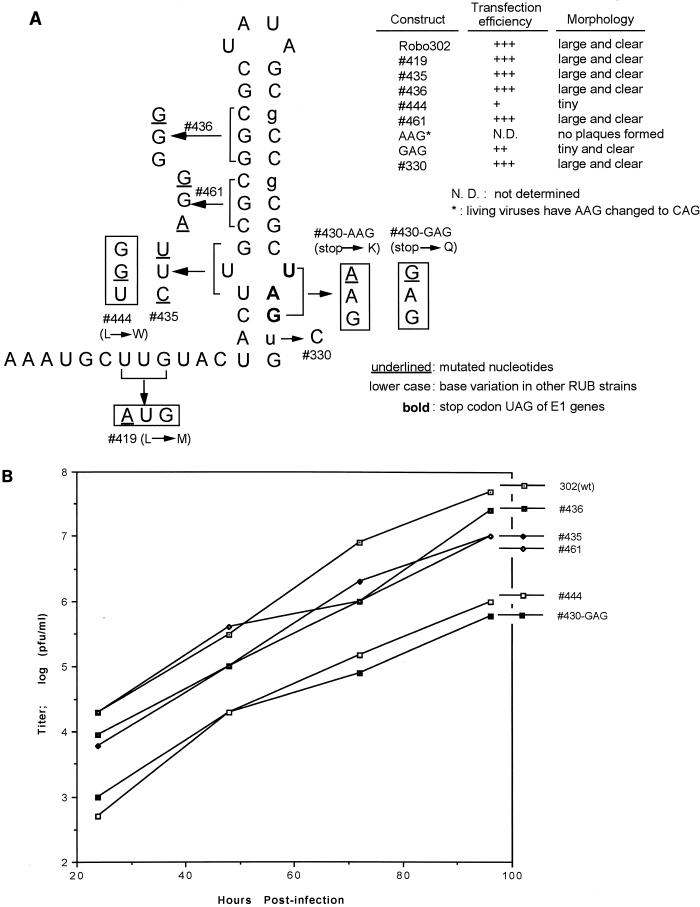
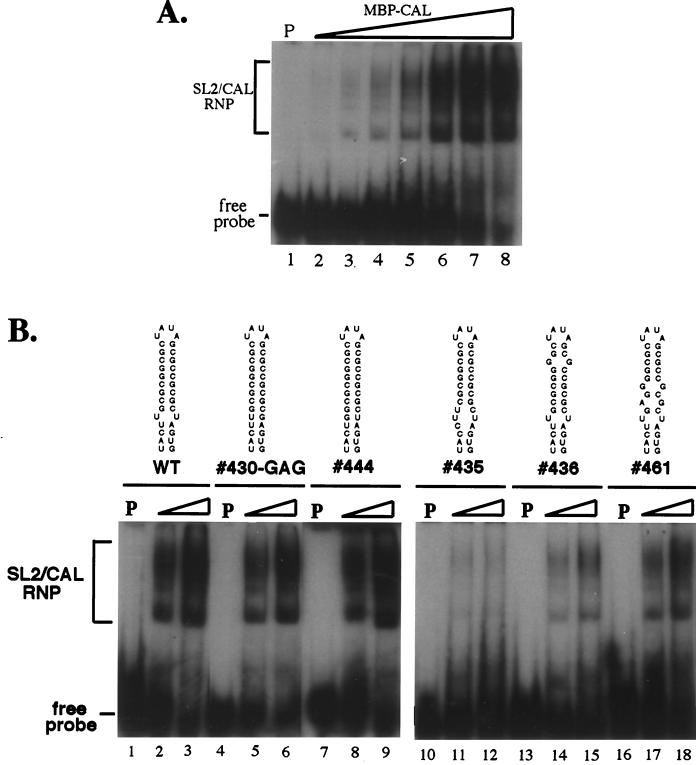
Because SL2 is within the coding region of E1, mutagenesis was done by creation of point mutations. The point mutations constructed in SL2 included mutations which destabilized the GC-rich stem (436 and 461) or disrupted the U-U bulge by either opening it (435) or base pairing it (444 and 430) (Fig. 6A). Two control constructs were also made: 330, which had the same sequence as the HPV strain at nt 9699; and 419, which had a mutation upstream from SL2 which introduced an NsiI site for cloning. Most of these mutations in SL2 using Robo302 were made at third codons to conserve the coding of E1. The exceptions were 444, in which a leucine (UUG) was replaced with a tryptophan (UGG), 430, in which the stop codon (UAG) at the end of the E1 was changed to lysine (AAG) or glutamic acid (GAG), and 419, in which a leucine (UUG) was changed to a methionine (AUG) (Fig. 6A). All of the mutant transcripts which retained the amino acid sequence (435, 436, 461, and 330) had specific infectivities similar to those of Robo302 transcripts and produced similar plaques; however, two mutants with changes in amino acid sequences (444 and 430-GAG) had lower specific infectivities and formed tiny plaques (Fig. 6A, insert). 419 had a specific infectivity similar to that of Robo302. Additionally, the 430-AAG (Lys) mutant did not form plaques, and the sequence of virus recovered from transfection fluid was found to have changed to CAG (glutamine). These results indicated that the SL2 structure could accommodate substantial changes in structure without affecting viability. The growth curves of the stable SL2 mutants were also characterized (Fig. 6B). Mutants 435, 436, and 461 (mutations which destabilized the SL2 structure) had growth curves similar to that of Robo302, while 444 and 430-GAG (mutants which base paired the entire structure) produced approximately 10-fold-lower titers after two passages.
FIG. 6.
(A) Mutagenesis of SL2. Because SL2 is located in the E1 coding region, analysis was done by creating point mutations that resulted in silent changes (436) or synonymous codons (435 and 461). However, to eliminate the U-U bulge, we created two missense mutations (444 and 430-AAG/GAG) which led to changes in coding as indicated. Mutation 419 was made to create an NsiI site for cloning, and 330 was made to conform to HPV77 sequence. The bold characters are the stop codon ending the E1 gene; the nucleotide variations found in other RUB strains are in lowercase. Transfection efficiencies of mutant transcripts are shown as +++ (equivalent to that of Robo302); ++ (about 30 to 50% of that of Robo302) +, (about 5 to 10% of that of Robo302), and N.D., not determinable, but transfected cells inoculated with medium showed CPE. The plaque morphology described is that produced following transfection. Following amplification of 430-AAG from transfection medium, the sequence was found to have changed to CAG. (B) Growth curves of SL2 mutants. Vero cells were infected with viruses amplified from transfection plaques at an MOI of 0.01 PFU/cell. At indicated times postinfection, aliquots of the infected culture media were harvested and titered on Vero cells.
Interaction between CAL and mutated SL2 RNA.
To investigate the significance of the SL2 mutations on the interaction between CAL and SL2, binding between a purified MBP-CAL fusion protein and gel-purified 32P-labeled SL2 RNA probe from each of the individual SL2 mutants was analyzed by gel mobility shift assay. The probes contained additional 10 nt (430-GAG, 435, 436, 444, and 461) or 8 nt (pUCRUB3′110-fTH and -HPV) at their 5′ ends but had the exact 3′ end of SL2 (due to the presence of a Bsp120I site immediately following SL2) and thus were similar to those used by Nakhasi et al. in their binding assays (2, 37). Binding of MBP-CAL to SL2 RNA from Robo302 is shown in Fig. 7A; as can be seen, increasing amounts of MBP-CAL–SL2 RNA complexes were formed with increasing input concentrations of protein. It was reported that MBP-CAL is able to be autophosphorylated in vitro and that in vitro phosphorylation was required for the interaction between MBP-CAL and SL2 RNA (2). However, in our hands, in vitro phosphorylation was not required for binding, as shown in Fig. 7A. Interestingly, we found that in vitro phosphorylation was extremely difficult to detect and efficient phosphorylation of MBP-CAL in vitro required addition of a Vero cell lysate (incubation of MBP-CAL in a phosphorylation reaction, either with or without cell lysate, made no difference on its ability to bind SL2). In one experiment, autophosphorylation was detectable; however, this reaction used a preparation MBP-CAL which had been stored frozen for a long period of time (∼6 months), and prolonged autoradiography was required for detection (in comparison with the companion reaction containing a Vero cell lysate, the intensity of the MBP-CAL band produced by autophosphorylation was only about 3.5% of the band produced in the presence of cell lysate) (data not shown). To test the possibility that MBP-CAL was phosphorylated in the bacteria, either by autophosphorylation or by bacterial kinases, MBP-CAL was dephosphorylated with CIAP. The efficiency of CIAP dephosphorylation of MBP-CAL phosphorylated in vitro in the presence of [32P]ATP and Vero cell lysate was about 85% (data not shown). To examine whether CIAP treatment would affect the binding activity of MBP-CAL, untreated and CIAP-treated MBP-CAL fusion proteins were incubated with SL2 probe and analyzed by gel mobility shift assay; however, no significant difference was observed between treated and untreated proteins (data not shown). Thus, although SL2 binding by MBP-CAL appeared to be largely independent of phosphorylation, because dephosphorylation may not have been complete, we cannot rule out that phosphorylation of key sites is required for SL2 binding.
FIG. 7.
Binding of MBP-CAL to SL2 probes. (A) Robo302 SL2 probe (<1 ng) was incubated in a binding reaction with various amounts of MBP-CAL prior to resolution of binding by electrophoresis in a nondenaturing polyacrylamide gel. Lane 1, control (0 μM MBP-CAL); lanes 2 to 7, 0.125, 0.25, 0.5, 1, 2, and 4 μM MBP-CAL. (B) SL2 probes containing sequences of WT Robo302 (lane 1 to 3) and mutations 430-GAG (lanes 4 to 6), 444 (lanes 7 to 9), 435 (lanes 10 to 12), 436 (lanes 13 to 15), and 461 (lanes 16 to 18) were incubated with 0 μM (lanes 1, 4, 7, 10, 13, and 16), 0.25 μM (lanes 2, 5, 8, 11, 14, and 17), or 1 μM (lane 3, 6, 9, 12, 15, and 18) MBP-CAL. After binding, formation of RNP complexes was determined by electrophoresis in polyacrylamide gels under nondenaturing conditions. P, probe only.
Interaction of MBP-CAL with SL2 is dependent on SL2 structure.
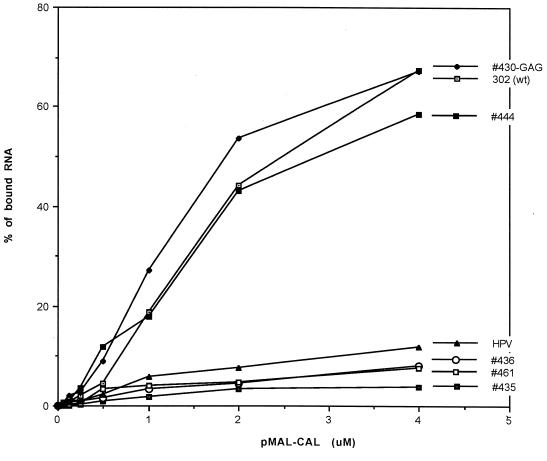
To compare the binding activities of MBP-CAL to the SL2 mutants, 32P-labeled SL2 probe containing each of the mutations was incubated in a binding reaction with increasing amounts of MBP-CAL (Fig. 7B). Interestingly, mutations in which the U-U bulge was base paired (444 and 430-GAG) had a binding activity similar to that of Robo302 SL2 (Fig. 7B, left panel) while mutations with destabilization in the U-U bulge (435) or the GC-rich stem (436 and 461) had a lower binding activity (Fig. 7B, right panel), indicating that maintenance of the entire SL2 stem structure is critical in CAL binding. The interaction between MBP-CAL and mutant SL2s was also quantified as the percentage of total radioactivity in the MBP-CAL–SL2 RNP over an extensive range of input MBP-CAL concentrations (Fig. 8); an SL2 probe with the HPV77 sequence was also included in these assays. 444 and 430-GAG had high activity similar to that of Robo302, reaching 50% binding between 2 and 3 μM MBP-CAL, while 435, 436, 461, and HPV77 never attained over 5 to 10% binding. Considering that viruses containing these mutations were viable and most replicated to similar titers as did Robo302 virus, the interaction between SL2 and CAL does not appear to be critical for RUB replication.
FIG. 8.
Comparison of binding activities of Robo302 and mutated SL2s to MBP-CAL. Robo302 and mutant SL2 probes were incubated with increasing amounts of MBP-CAL, and formation of RNP complexes was determined by gel mobility shift (an example of binding of MBP-CAL and Robo302 SL2 is shown in Fig. 6A). The binding was quantitated as the percentage of total radioactivity (bound plus unbound) in RNP complexes.
Investigation of interaction between cellular factors and 3′ UTR.
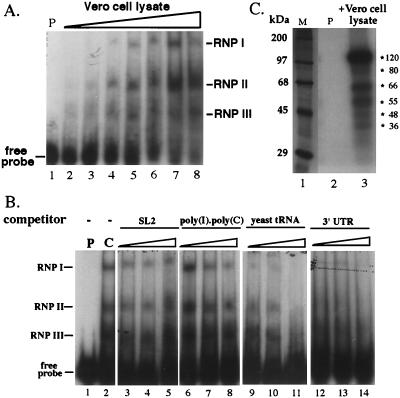
We wished to know whether the finding that the 3′ UTR is critical in RUB replication is related to its interaction with host factors. As shown in Fig. 9A, three RNP complexes were detected by gel mobility shift assay when a 32P-labeled 3′ UTR probe was incubated with a Vero cell lysate, indicated as RNP I, RNP II, and RNP III. RNP II was detectable when incubated with a lower amount of cell lysate (∼0.75 μg), while detection of RNP I and III required higher amounts of cell lysate, indicating that the host factors composing RNP II either had a higher activity for the RUB 3′ UTR or were more abundant in the cell lysate preparations. The specificities of these three RNPs were examined by competition assay using several competitors. The specific competitor, 3′ UTR, was able to outcompete the probe for binding at the lowest concentration used (20× molar excess), while nonspecific competitors SL2 RNA and poly(I)-poly(C) did not outcompete for binding even at the highest concentration used. However, tRNA was able to outcompete the 3′ UTR probe at the 20× molar excess for RNP I and 150× excess for RNPs II and III. Six proteins with molecular masses of 120, 80, 66, 55, 48, and 36 kDa were detected after UV-induced cross-linking; of these, the 120-, 66-, and 55-kDa proteins were the three major species (Fig. 9C).
FIG. 9.
Binding of cellular factors to the 3′ UTR. (A) 32P-labeled UTR probe (lacking both the poly(A) tract and the 3′-terminal 5 nt was incubated with 0 μg (lane 1), 0.2 μg (lane 2), 0.375 μg (lane 3), 0.75 μg (lane 4), 1.5 μg (lane 5), 3 μg (lane 6), 6 μg (lane 7), and 12 μg (lane 8) of cytoplasmic lysate from uninfected Vero cells, and formation of complexes was detected by electrophoresis in nondenaturing polyacrylamide gels. The three RNP complexes detected are indicated on the right as RNP I, II, and III. (B) Increasing amounts of different competitor RNAs, including both nonspecific [SL2, poly(I)-poly(C), and yeast tRNA] and specific (3′ UTR RNA transcripts) competitors, were incubated with 4 μg of Vero cell lysate prior to the addition of the 32P-labeled probe. Lane 5: 1 (P), probe alone; 2 (C), positive control without any competitor; 3 to 5, SL2 at 20× (lane 3), 50× (lane 4), or 150× (lane 5) molar excess; 6 to 8, poly(I)-poly(C) at 50 ng (lane 6), 100 ng (lane 7), or 200 ng (lane 8) [the standard binding assay buffer containing contains 100 ng of poly(I)-poly(C)]; 9 to 11, yeast tRNA at 20× (lane 9), 50× (lane 10), or 150× (lane 11) molar excess; 12 to 14, 3′ UTR RNA at 20× (lane 12), 50× (lane 13), or 150× (lane 14) molar excess. (C) 3′ UTR probe in the absence (lane 2, P [probe only]) or presence (lane 3) of 30 μg of Vero cell lysates was exposed to UV light and digested with RNase. Cross-linked proteins were revealed by SDS-polyacrylamide gel electrophoresis. Approximate positions of size markers (lane M) are given on the right.
DISCUSSION
The elements in the 3′-terminal 110 nt in the RUB genome, including the poly(A) tract, critical in virus replication were analyzed in this study by site-directed mutagenesis using a RUB infectious clone, Robo302. Previous studies on RUB DI RNAs found that the 3′-terminal 305 nt were retained in all DI RNAs generated during serial undiluted passage, and it was concluded that these sequences contained the 3′ cis-acting elements required for replication (7, 11). Thermodynamically predicted secondary structures of this 305-nt region revealed four prominent SL structures, including two SLs (SL1 and SL2) in the E1 coding region and two in the 59-nt 3′ UTR (SL3 and SL4). Because of its size and location in the E1 coding region, SL1 was not analyzed in this study.
Two alternative structures containing SL2 and SL3 were predicted, depending on the virus strain. Because previous studies indicated that the interaction of SL2 with CAL was dependent on the presence of the U-U bulge (37), which did not exist in the second structure, RNase mapping was done to analyze the SL2/SL3 structure in both the fTh and HPV77 strains, in which the first and second structures, respectively, were predicted. The RNase mapping data were consistent with the formation of the first structure in both strains. It is likely, however, that the two structures achieve equilibrium in both strains since both single- and double-stranded RNase digestion of the ACUA in the hinge region instrumental in forming the second structure was observed in both strains. However, this could also be due to nonspecific digestion by RNase V1, since it has been noticed that RNase V1 may cleave within a single-stranded region if base stacking occurs (28). RNase mapping also confirmed the loops of SL2 and SL3 and the predominantly single-stranded nature of the hinge region between SL3 and SL4. The bulge and upper stem of SL3 were digested by single- and double-stranded RNases in a pattern that was generally, but not completely, consistent with the predicted structure. Single-stranded RNases failed to cleave the unpaired U loop in SL2, and the double-stranded RNase did not cut the lower stem of SL3 and cleaved at only one major site in the GC-rich SL2 stem. These data possibly indicate the existence of a tertiary structure between SL2 and SL3 that could explain the inaccessibility of SL2 and unexpected aspects of the cleavage pattern of SL3. The existence of SL4 is tentative, as it is a thermodynamically weak structure and varies to some extent among strains according to the thermodynamic prediction. However, RNase mapping confirmed its existence.
Our mutagenesis data revealed that most of the 3′ UTR is required for replication, in contrast to the case for both alphaviruses (24) and picornaviruses (54), in which large deletions within the 3′ UTR can be accommodated. Recently, mutagenesis resulting in complete removal of the 3′ UTR from poliovirus type 1 and human rhinovirus type 14 infectious clones with recovery of viable virus was reported (54). However, these mutant transcripts had much lower transfection efficiency than WT transcripts with the 3′ UTR, and the 3′ UTR minus-strand virus was impaired in replication (onset of CPE was observed at 18 to 24 h posttransfection with the latter mutants, compared with 8 h posttransfection with WT virus). The only region in the RUB 3′ UTR that could be deleted is the single-stranded leader region in which nt 1 to 5 could be deleted, and the resulting virus was phenotypically WT. This is also in contrast to alphaviruses, in which the nucleotide preceding the poly(A) tract was necessary for viability, although the insertion of nucleotides between this nucleotide (a C) and the poly(A) could be tolerated (24). Addition of nonviral sequence in the RUB single-stranded leader region was tolerated since mutants having poly(A) replaced with 10 U’s (508T) or with an EcoRI restriction site (507 and 509) were viable and retained these sequences along with a regenerated poly(A) tract; these viruses were also phenotypically WT.
The only two nucleotides in the single-stranded leader sequence required for viability were nt 6 and 7. Deletions of these nucleotides either by themselves or in the context of the entire leader resulted in a dramatic reduction in transfection efficiency, and addition of nucleotides downstream prior to the poly(A) tract occurred in the viruses which were recovered; the replication of these viruses was impaired to some extent. Addition of nonviral sequences at a similar location by viral RdRp has been described for Sindbis virus (SIN) (46), although an AU-rich motif was added instead of the G residue usually added in RUB 3′ terminus [although nonviral AAT or AATT also exists in some poly(A)− mutants, these sequences are actually copied from an EcoRI protruding end used for runoff transcription]. Despite SL4 not being a particularly stable structure according to thermodynamic prediction, RNase mapping data indicated that SL4 existed in the native sequence and that the nucleotides within this small SL are critical in viral replication. A number of deletions within the SL4 sequence were lethal (only clone 524, in which nt 8 to 10 were deleted, was viable) as were most of the mutations which changed the sequence (only clone 575, in which the UGU in the loop was changed to ACA, was viable). However, the virus recovered from 575 restored the SL4 sequence, while the virus recovered from 524 restored all but the last nucleotide. From available data, we cannot conclude whether the primary sequence or the SL4 structure is the important feature. While the SL4 sequences were maintained in all viable mutants and revertants, the SL4 structure is not predicted thermodynamically in some of them when the complete 3′ 110-nt segment is folded.
Deletions in the hinge region were also lethal. Interestingly, one of these deletions of the ACUA, which formed part of the second SL2/SL3 structure (344C), was lethal, which implied that formation of this alternative structure might be important in viral replication. While the sequences in the upper stem of SL3 could be switched, mutations which changed the structure were lethal. An exception was mutant 391, in which the SL3 GAAAC loop was deleted. However, the mutant was highly attenuated. Given these findings, it is tempting to speculate that the significance of SL3 is in specific interactions with the RUB RdRp and/or cellular factors since bulges or loops in an RNA secondary structure have been implicated as specific recognition sites for RNA binding proteins because they expose RNA backbones and bases to interaction while the regular helical RNA stems are less useful (reviewed in reference 32). The requirement of a secondary structure is in contrast to alphaviruses, which share a consensus 19-nt stretch immediately preceding the poly(A) tract which contains no significant secondary structure.
Like SIN replication (19), RUB replication can apparently occur independent of the poly(A) tract; however, also as for SIN virus, the poly(A) tract was rapidly regenerated. The role of poly(A) in eukaryotic mRNAs is proposed to be in mRNA stability and translational efficiency (reviewed in reference 59). Opaque plaques were formed after transfection by mutants with defects in poly(A) tract; after one passage when poly(A) regeneration was detected, these viruses produced large, clear plaques and grew to titers similar to Robo302 (Fig. 4). It is not clear how the nontemplated polyadenylation occurs. Unlike the case for coronavirus (20) or vesicular stomatitis virus (3), which might utilize a short poly(U) as a template for polyadenylation, polyadenylation of RUB genomic RNA is not due to polymerase reiterating at a poly(U) stretch since there are no such elements found in the 3′ UTR or its minus-strand complement in the RUB genome. The cellular cytoplasmic polyadenylation machinery, cellular terminal transcriptase-like enzymes, and the viral RdRp were implicated in the regeneration of the poly(A) tract in SIN (46).
Because of its location within the E1 coding sequences, substitution mutagenesis was used to investigate the interaction of SL2 with CAL and its effect on virus replication. The CAL binding site on SL2 has never been precisely mapped, although it has been shown that deletion of the U-U bulge abolished binding of cellular proteins (37), including the one later identified as CAL, and the binding of MBP-CAL to SL2 was abolished when the U-U bulge was replaced with A-A, C-C, or G-G bulges (1a). Because of the constraints imposed by E1 coding, we were unable to make these mutations. Therefore, we constructed mutations that either base paired the U-U bulge or created other bulges in this SL structure. We found that the mutations with the U-U bulge base paired had as great an affinity for MBP-CAL as did the native SL2 with the U-U bulge. This finding does not necessarily contradict the findings of Nakhasi et al., since all mutants maintained at least one of the two U residues, and it has been shown that the members of a U-U bulge in an SL structure can be hydrogen bonded (58), which would also explain why no cleavage was observed at the U-U bulge with any single-stranded RNase. In contrast, a mutant in which the SL in the region of the U-U pair was destabilized by the introduction of noncomplementary nucleotides had the lowest activity for MBP-CAL. In addition to the U-U bulge, maintenance of the GC-rich stem in SL2 was also critical in CAL binding since reduction in binding activity was also observed in mutants which destabilized the GC-rich SL2 stem, including 436, which was based on the sequence of the M33/HPV77 strain. (Although binding with CAL was first performed with an HPV77 probe, the unpaired CA bulge in the upper stem of SL2 of HPV77 [Fig. 2B] was changed to a CG base pair in subsequent studies [37]). All of these findings indicate that maintenance of a long stem is the primary requirement for MBP-CAL binding. Interestingly, CAL was also found to bind with hY RNA (8), whose structure also exhibits a long stem which was confirmed by enzymatic structure probing (55). Therefore, it is possible that SL2 mimics the structure feature of hY RNA which is recognized by CAL.
Although many attempts have been made to determine the significance of the host factor-viral RNA interaction on virus replication, most of the studies were done with a reporter gene system. Ours is the first direct study of the effects of a host factor’s interaction with a viral cis-acting element in virus replication using a virus infectious clone. Interestingly, despite the differences observed in the binding activity between SL2 mutants and MBP-CAL, maintenance of the SL2 structure required for CAL binding was not required for viral replication. Mutants having lower binding affinities (435, 436, and 461) without changes in amino acid sequence had similar transfection efficiencies, formed similar plaques, and grew to titers that were similar to or somewhat lower than those for Robo302. Mutants having U-U bulge base paired exhibited lower transfection efficiencies, tiny plaques, and lower titers, and some of these mutants (444 and 430-AAG) reverted to the WT Robo302 sequence within a few passages. This is likely due to the changes in these mutants of the amino acid sequence in the C terminus of E1, which is proposed to function as a cytoplasmic tail of the E1 proteins that interacts with the capsid in virions. In one case, a CAG revertant was recovered from the 430-AAG mutant, which, interestingly, resulted in glutamine residue; 430-GAG which was stable encoded a glutamic acid at that position.
Altogether, these data indicate that binding of CAL to SL2 is independent of RUB replication. However, despite the decreased MBP-CAL binding affinities of some of the viable SL2 mutants, some binding was still observed. Thus, it is still possible that CAL and SL2 in the RUB genome interact in infected cells, particularly if local CAL concentrations are high. In this regard, RUB RNA replication was recently confirmed to occur within subcellular vesicles of lysosomal origin (31). Since CAL is found primarily associated with endoplasmic reticulum membranes, it would have to be relocalized to interact with the RUB RNA during replication. Relocalization of cellular proteins that bind viral RNA has been found upon viral infection (10, 27; reviewed in reference 4) although relocalization of La was not observed during SIN replication (40). It would be of interest to determine if CAL relocalizes in RUB-infected cells.
In contrast to SL2, our mutagenesis data clearly demonstrated the importance of the 3′ UTR in RUB replication; however, the function could be in translational efficiency as well as serving as a promoter for minus-strand RNA synthesis. Nakhasi et al. showed that the RUB 3′ sequences enhanced translation of a reporter gene expressed in context with the 5′ SL structure (38, 42). However, binding of cellular factors has been shown to a number of putative cis-acting replicational elements, and thus binding of cellular proteins to the 3′ UTR was investigated. Three RNA complexes were resolved by gel mobility shift assay using a 3′ UTR probe lacking the 3′-terminal 5 nt, and poly(A) and UV-induced cross-linking revealed that six protein species bound to the 3′ UTR probe. The interaction was not outcompeted by nonspecific competitors such as SL2 RNA or poly(I)-poly(C) but was inhibited by 150× molar excess amounts of yeast tRNA. However, we also found that the interaction between SL2 and MBP-CAL can be outcompeted by a 30× molar excess of yeast tRNA (data not shown). Thus, the interaction between these cellular factors and the 3′ UTR appears to be specific. However, further characterization of these proteins and these interactions with the 3′ UTR is necessary to define their role(s) in viral replication.
ACKNOWLEDGMENTS
We thank C. D. Atreya for his generous gift of the MBP-CAL recombinant clone and suggestions on its use, Jerry Blackwell and Kostia Pugachev for technical and critical advice, and Ping Chiang for synthesizing oligonucleotides and assisting in automated sequencing.
This research was funded by grant AI 21789 from NIH. M.-H.C. was supported in part by Georgia State University Research Program Enhancement.
REFERENCES
- 1.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes and RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Atreya, C. D. Personal communication.
- 2.Atreya C D, Singh N K, Nakhasi H L. The rubella virus RNA binding activity of human calreticulin is localized to the N-terminal domain. J Virol. 1995;69:3848–3851. doi: 10.1128/jvi.69.6.3848-3851.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr J N, Whelan S P J, Wertz G W. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belsham G J, Sonenberg N. RNA protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell J L, Brinton M A. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J Virol. 1995;69:5650–5658. doi: 10.1128/jvi.69.9.5650-5658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blyn L B, Chen R, Semler B L, Ehrenfeld E. Host cell proteins binding to domain IV of the 5′ noncoding region of poliovirus RNA. J Virol. 1995;69:4381–4389. doi: 10.1128/jvi.69.7.4381-4389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, M.-H., and T. K. Frey. Unpublished data.
- 8.Cheng S-T, Nguyen T-Q, Yang Y-S, Capra D, Sontheimer R D. Calreticulin binds hYRNA and the 52-kDa polypeptide component of the Ro/SS-A ribonucleoprotein autoantigen. J Immunol. 1996;156:4484–4491. [PubMed] [Google Scholar]
- 9.Choukhi A, Ung S, Wychowski C, Dubuisson J. Involvement of endoplasmic reticulum chaperons in the folding of hepatitis C virus glycoproteins. J Virol. 1998;72:3851–3858. doi: 10.1128/jvi.72.5.3851-3858.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De B P, Gupta S, Zhao H, Drazba J A, Banerjee A K. Specific interaction in vitro and in vivo of glyceraldehyde-3-phosphate dehydrogenase and La protein with cis-acting RNAs of parainfluenza virus type 3. J Biol Chem. 1996;271:24728–24735. doi: 10.1074/jbc.271.40.24728. [DOI] [PubMed] [Google Scholar]
- 11.Derdeyn C A, Frey T K. Characterization of defective-interfering RNAs of rubella virus generated during serial undiluted passage. Virology. 1995;206:216–226. doi: 10.1016/S0042-6822(95)80036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel J-P, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frey T K, Marr L D, Hemphill M L, Dominguez G. Molecular cloning and sequencing of the region of the rubella virus genome cloning for glycoprotein E1. Virology. 1986;53:228–232. doi: 10.1016/0042-6822(86)90446-0. [DOI] [PubMed] [Google Scholar]
- 14.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukushi S, Kurihara C, Ishiyama N, Hoshino F B, Oya A, Katayama K. The sequence element of the internal ribosomal entry site and a 25-kilodalton of translation of hepatitis C virus RNA. J Virol. 1997;71:1662–1666. doi: 10.1128/jvi.71.2.1662-1666.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez A L, Denova-Ocampo M, Racaniello V R. Attenuation mutations in the poliovirus 5′ untranslated region alter its interaction with polypyrimidine tract-binding protein. J Virol. 1997;71:3826–3833. doi: 10.1128/jvi.71.5.3826-3833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hebert D N, Foellmer B, Helenius A. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza virus hemagglutinin in microsomes. EMBO J. 1996;15:2961–2968. [PMC free article] [PubMed] [Google Scholar]
- 18.Hebert D N, Zhang J X, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill K R, Hajjou M, Hu J Y, Raju R. RNA-RNA recombination in Sindbis virus: roles of the 3′ conserved motif, poly(A) tail, and nonviral sequences of template RNAs in polymerase recognition and template switching. J Virol. 1997;71:2693–2704. doi: 10.1128/jvi.71.4.2693-2704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann M A, Brain D A. The 5′ end of coronavirus minus-strand RNAs contain a short poly(U) tract. J Virol. 1991;65:6331–6333. doi: 10.1128/jvi.65.11.6331-6333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito Y, Lai M M C. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knapp G. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- 23.Krause K-H, Michalak M. Calreticulin. Cell. 1997;88:439–443. doi: 10.1016/s0092-8674(00)81884-x. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn R J, Hong Z, Strauss J H. Mutagenesis of the 3′ nontranslated region of Sindbis virus RNA. J Virol. 1990;64:1465–1476. doi: 10.1128/jvi.64.4.1465-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leopardi R, Hukkanen V, Vainionpaa R, Salmi A A. Cell proteins bind to sites within the 3′ noncoding region and the positive-strand leader sequence of measles virus RNA. J Virol. 1993;67:785–790. doi: 10.1128/jvi.67.2.785-790.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levis R, Weiss B G, Huang H V, Schlesinger S. Deletion mapping of Sindbis virus DI RNAs derived from cDNA defines the sequences essential for replication and packaging. Cell. 1986;44:137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- 27.Li H P, Zhang X, Duncan R, Comai L, Lai M M. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowman H B, Draper D E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986;261:5396–5403. [PubMed] [Google Scholar]
- 29.Lu J, Willis A C, Sim R B. A calreticulin-like protein co-purifies with a ’60 kD’ component of Ro/SSA, but is not recognized by antibodies in Sjoegren’s syndrome sera. Clin Exp Immunol. 1993;94:429–434. doi: 10.1111/j.1365-2249.1993.tb08213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lux F A, McCauliffe D P, Buettner D W, Lucius R, Capra J D, Sontheimer R D, Lieu T-S. Serological cross-reactivity between a human Ro/SS-A autoantigen (calreticulin) and the lambda RAL-1 antigen of Onchocerca volvulus. J Clin Investig. 1992;89:1945–1951. doi: 10.1172/JCI115801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magliano D, Marshall J A, Bowden D S, Vardaxis N, Meanger J, Lee J-Y. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- 32.Mattaj I W. RNA recognition: a family matter? Cell. 1993;73:837–840. doi: 10.1016/0092-8674(93)90265-r. [DOI] [PubMed] [Google Scholar]
- 33.Max H, Halder T, Kalbus M, Gnau V, Jung G, Kalbacher H. A 16 mer peptide of the human autoantigen calreticulin is a most prominent HLA-DR4Dw4-associated self-peptide. Hum Immunol. 1994;41:39–45. doi: 10.1016/0198-8859(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 34.McCauliffe D P, Zappi E, Lieu T-S, Michalak M, Sontheimer R D, Capra J D. A human Ro/SS-A autoantigen is the homologue of calreticulin and is highly homologous with onchocercal RAL-1 antigen and an Aplysia “memory molecule.”. J Clin Investig. 1990;86:332–335. doi: 10.1172/JCI114704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakhasi H L, Thomas D, Zheng D, Liu T-Y. Nucleotide sequence of capsid, E2 and E1 protein genes of rubella virus vaccine strain RA27/3. Nucleic Acids Res. 1989;17:4393–4394. doi: 10.1093/nar/17.11.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakhasi H L, Rouault T A, Haile D J, Liu T-Y, Klausner R D. Specific high-affinity binding of host cell proteins to the 3′ region of rubella virus RNA. New Biol. 1990;2:255–264. [PubMed] [Google Scholar]
- 38.Nakhasi H L, Singh N K, Pogue G P, Cao X-Q. Identification and characterization of host factor interactions with cis-acting elements of rubella virus. Arch Virol. 1994;9:255–267. doi: 10.1007/978-3-7091-9326-6_26. [DOI] [PubMed] [Google Scholar]
- 39.Otteken A, Moss B. Calreticulin interacts with newly synthesized human immunodeficiency virus type 1 envelope glycoprotein, suggesting a chaperon function similar to that of calnexin. J Biol Chem. 1996;271:97–103. doi: 10.1074/jbc.271.1.97. [DOI] [PubMed] [Google Scholar]
- 40.Pardigon N E, Strauss J H. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J Virol. 1996;70:1173–1181. doi: 10.1128/jvi.70.2.1173-1181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson J R, Ora A, Van P N, Helenius A. Transient, lectin-like association of calreticulin with folding intermediates of cellular and viral glycoproteins. Mol Biol Cell. 1995;6:1173–1184. doi: 10.1091/mbc.6.9.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pogue G P, Cao X-Q, Singh N K, Nakhasi H L. 5′ sequences of rubella virus RNA stimulate translation of chimeric RNAs and specifically interact with two host-encoded proteins. J Virol. 1993;67:7106–7117. doi: 10.1128/jvi.67.12.7106-7117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pogue G, Hofmann P, Duncan R, Best J M, Etherington J, Sontheimer R D, Nakhasi H L. Autoantigens interact with cis-acting elements of rubella virus RNA. J Virol. 1996;70:6269–6277. doi: 10.1128/jvi.70.9.6269-6277.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pugachev K V, Abernathy E S, Frey T K. Improvement of the specific infectivity of the rubella virus (RUB) infectious clone: determinants of cytopathogenicity induced by RUB map to the nonstructural proteins. J Virol. 1997;71:562–568. doi: 10.1128/jvi.71.1.562-568.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pugachev K V, Frey T K. Effects of defined mutations in the 5′ nontranslated region of rubella virus genomic RNA on virus viability and macromolecule synthesis. J Virol. 1998;72:641–650. doi: 10.1128/jvi.72.1.641-650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raju R, Hajjou M, Hill K R, Botta V, Botta S. In vivo addition of poly(A) tail and AU-rich sequences to the 3′ terminus of Sindbis virus RNA genome: a novel 3′ end repair pathway. J Virol. 1999;73:2410–2419. doi: 10.1128/jvi.73.3.2410-2419.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rokeach L A, Haselby J A, Meilof J F, Smeenk R J T, Unnasch T R, Greene B M, Hoch S O. Characterization of the autoantigen calreticulin. J Immunol. 1991;147:3031–3039. [PubMed] [Google Scholar]
- 48.Rokeach L A, Zimmerman P A, Unnasch T R. Epitopes of the Onchocerca volvulus RAL1 antigen, a member of calreticulin family of proteins, recognized by sera from patients with onchocerciasis. Infect Immun. 1994;62:3696–3704. doi: 10.1128/iai.62.9.3696-3704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siegel R W, Bellon L, Beigelman L, Kao C C. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1998;95:11613–11618. doi: 10.1073/pnas.95.20.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh N K, Atreya C D, Nakhasi H L. Identification of calreticulin as a rubella virus RNA binding protein. Proc Natl Acad Sci USA. 1994;91:12770–12774. doi: 10.1073/pnas.91.26.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skinner M A, Racaniello V R, Dunn G, Cooper J, Minor P D, Almond J W. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J Mol Biol. 1989;207:379–392. doi: 10.1016/0022-2836(89)90261-1. [DOI] [PubMed] [Google Scholar]
- 52.Sriskanda V S, Pruss G, Ge X, Vance V B. An eight-nucleotide sequence in the potato virus X 3′ untranslated region is required for both host proteins binding and viral multiplication. J Virol. 1996;70:5266–5271. doi: 10.1128/jvi.70.8.5266-5271.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Svitkin Y V, Meerovitch K, Lee H S, Dholakia J N, Kenan D J, Agol V I, Sonenberg N. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J Virol. 1994;68:1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Todd S, Towner J S, Brown D M, Semler B L. Replication-competent picornavirus with complete genomic RNA 3′ noncoding region deletions. J Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Gelder C W G, Thijssen J P H M, Klaassen E C J, Sturchler C, Krol A, van Venrooij W J, Pruijn G J M. Common structural features of the Ro RNP associated hY1 and hY5 RNAs. Nucleic Acids Res. 1994;22:2498–2506. doi: 10.1093/nar/22.13.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verreck F A W, Elferink D, Vermeulen C J, Amons R, Breedveld F, De-Vries R R P, Koning F. DR2Dw4/DR53 molecules contain a peptide from the autoantigen calreticulin. Tissue Antigens. 1995;45:270–275. doi: 10.1111/j.1399-0039.1995.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 57.Wang C-Y, Dominguez G, Frey T K. Construction of rubella virus genome-length cDNA clones and synthesis of infectious RNA transcripts. J Virol. 1994;68:3550–3557. doi: 10.1128/jvi.68.6.3550-3557.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y-X, Huang S, Draper D E. Structure of a U-U pair within a conserved ribosomal RNA hairpin. Nucleic Acids Res. 1996;24:2662–2672. doi: 10.1093/nar/24.14.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990;15:277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- 60.Zheng Z, Maidji E, Tugizov S, Pereira L. Mutations in the carboxyl-terminal hydrophobic sequence of human cytomegalovirus glycoprotein B alter transport and protein chaperone binding. J Virol. 1996;70:8029–8040. doi: 10.1128/jvi.70.11.8029-8040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuker M, Stiegler P. Optimal computer folding large RNA sequence using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]