Significance
Grass powdery mildews and their cereal hosts engage in a molecular arms race in which the fungus needs to continuously adapt its effector repertoire to avoid recognition by plant immune receptors and maintain virulence activity. We report the structures of several powdery mildew avirulence effectors which adopt a common RALPH scaffold with variations in structural elements that differentiate subfamilies. All tested avirulence effectors lack nuclease and synthetase activities of the ancestral RNase T1/F1 family, suggesting that virulence is associated with neo-functionalization events. Structure-informed engineering turned a sequence-diverged RALPH into a molecule detected by an immune receptor. Thus, the coupling of sequence and structural polymorphisms in RALPH effectors facilitated escape from immune receptor recognition and potential acquisition of new virulence functions.
Keywords: innate immunity, effector, crystal structure, powdery mildew, immune receptor
Abstract
In plants, host–pathogen coevolution often manifests in reciprocal, adaptive genetic changes through variations in host nucleotide-binding leucine-rich repeat immune receptors (NLRs) and virulence-promoting pathogen effectors. In grass powdery mildew (PM) fungi, an extreme expansion of a RNase-like effector family, termed RALPH, dominates the effector repertoire, with some members recognized as avirulence (AVR) effectors by cereal NLR receptors. We report the structures of the sequence-unrelated barley PM effectors AVRA6, AVRA7, and allelic AVRA10/AVRA22 variants, which are detected by highly sequence-related barley NLRs MLA6, MLA7, MLA10, and MLA22 and of wheat PM AVRPM2 detected by the unrelated wheat NLR PM2. The AVR effectors adopt a common scaffold, which is shared with the RNase T1/F1 family. We found striking variations in the number, position, and length of individual structural elements between RALPH AVRs, which is associated with a differentiation of RALPH effector subfamilies. We show that all RALPH AVRs tested have lost nuclease and synthetase activities of the RNase T1/F1 family and lack significant binding to RNA, implying that their virulence activities are associated with neo-functionalization events. Structure-guided mutagenesis identified six AVRA6 residues that are sufficient to turn a sequence-diverged member of the same RALPH subfamily into an effector specifically detected by MLA6. Similar structure-guided information for AVRA10 and AVRA22 indicates that MLA receptors detect largely distinct effector surface patches. Thus, coupling of sequence and structural polymorphisms within the RALPH scaffold of PMs facilitated escape from NLR recognition and potential acquisition of diverse virulence functions.
Interactions of plants with host-adapted pathogens are often subject to a population-level arms race involving competing sets of coevolving genes encoding plant immune receptors and pathogen effectors, the former being essential components for plant immunity and the latter being required for pathogen virulence (1). Intracellular nucleotide-binding domain leucine-rich repeat-containing receptors (NLRs) in plants are key components of the plant immune system and typically detect strain-specific pathogen effectors, known as avirulence (AVR) effectors, to activate immune responses that terminate pathogen proliferation. Canonical plant NLRs share a tripartite domain organization consisting of a variable N-terminal signaling domain, a central nucleotide-binding oligomerization (NOD) domain, followed by a C-terminal leucine-rich repeat region (LRR) with or without a jelly roll/Ig-like (JID) domain (2). The majority of these NLRs carry either a coiled-coil (CC) domain or a Toll-interleukin 1 receptor (TIR) domain at the N terminus and are termed CNLs or TNLs, respectively (2, 3). Pathogen effector perception by NLRs can occur via diverse mechanisms, including direct effector binding to polymorphic LRR and C-JID domains (4–7). Pathogen recognition can also be indirect, with NLRs detecting pathogen effector-mediated modifications of host proteins (guardees) or mimics of these proteins (decoys), including decoys integrated into NLRs (8, 9). Upon effector-mediated activation, canonical CNLs and TNLs undergo extensive structural interdomain rearrangements and oligomerization to form resistosomes composed of five or four NLR protomers, respectively. CNL resistosomes integrate into host cell membranes to act as calcium-permeable channels, whereas TNL resistosomes produce nucleotide-based second messengers for immune signaling (4–7, 10, 11). Ultimately, these NLR-mediated immune responses often result in regulated local death of host cells at sites of attempted pathogen ingress, the so-called hypersensitive response.
In host-adapted pathogens, coevolution with their hosts occurs in iterative cycles and has resulted in genomic expansion of the plant NLR arsenal as well as the pathogen effector complement (1). NLR genes in plants are often organized in complex clusters of paralogous genes, and several examples of allelic series of NLRs have been reported in host populations, with each receptor variant conferring a different effector recognition specificity (12–16). The repertoire of effector genes of pathogenic fungi is much larger (typically hundreds) compared to pathogenic bacteria (a few dozen) and the effectors are often lineage- or species-specific innovations, suggesting that effectors of different fungal lineages evolve rapidly and independently of each other (17, 18). Sequence-relatedness between individual fungal effector genes is often low or undetectable. However, there is increasing evidence that many of these effectors are structurally related. Thus, it is possible that the effector repertoire of pathogenic ascomycetes consists of a limited number of structural folds (17, 19–29). Yet, it is still unclear whether each effector fold is associated with a common biochemical function or serves as a scaffold for diversified virulence activities.
The powdery mildews Blumeria graminis f. sp. hordei (Bgh) and Blumeria graminis f. sp. tritici (Bgt) infect monocotyledonous barley and wheat, respectively, and are widespread, obligate biotrophic ascomycete fungi. Bgh and Bgt each secrete hundreds of candidate effector proteins (CSEPs) to promote pathogen growth. Numerous allelic CNL variants are encoded at the barley Mla or wheat Pm2 or Pm3 resistance loci, each conferring isolate-specific immunity to Bgh or Bgt strains, respectively, with matching AVR effectors (13, 16, 30–35). Although MLA, PM2, and PM3 are phylogenetically unrelated CNLs, receptors encoded by each of these loci with different resistance specificities share >90% sequence identity. Together, this indicates that these polymorphic CNLs in barley and wheat contribute to coevolution with Bgh and Bgt, respectively. The sequence-unrelated paralogous Bgh avirulence effectors AVRA1, AVRA6, AVRA7, AVRA9, and AVRA13 and the sequence-related allelic variants AVRA10/AVRA22 are likely to be recognized directly by barley MLA1, MLA6, MLA7, MLA9, MLA13, MLA10, and MLA22, respectively, through their polymorphic LRR domains (13, 32, 35). AVRPM2, AVRPM3A2/F2, AVRPM3B2/C2, and AVRPM3D3 were identified in Bgt and were shown to be recognized by wheat PM2a and PM3a/PM3f, PM3b/PM3c, and PM3d, respectively, with PM2a and PM3 LRRs also functioning as recognition specificity determinants (16, 34, 36).
Genome-wide AlphaFold2 (AF2) modeling of fungal effectors complements identified extreme expansion of lineage-specific, sequence-unrelated, structurally similar effector families in B. graminis and the rust fungus Puccinia graminis (26). This modeling predicted that at least 70% of all Bgh effectors adopt the common fold of RNase-like proteins associated with haustoria (RALPHs) (13, 16, 30, 32–37). RALPH effectors in a given Bgh or Bgt strain are typically encoded by >400 paralogous genes organized in at least 15 RALPH subfamilies, with no detectable sequence similarity between subfamilies (26, 38–41). All 14 identified AVR effectors in Bgh and Bgt encode variants of predicted RALPH effectors. The structure of a CSEP with unknown avirulence activity, Bgh CSEP0064, features an RNase-like fold and is thought to act as a pseudoenzyme that binds to host ribosomes, thereby inhibiting the action of toxic plant ribosome-inactivating proteins (37). Other modeled RALPH effectors with unknown avirulence activity interact with different barley proteins in vitro and in vivo (42–44). Bgh AVRA1 and the predicted RALPH effector CSEP0491 interact with the barley endoplasmic reticulum-localized J-domain-containing protein HvERdj3 (45).
We report here the crystal structures of four Bgh AVRA effectors and of Bgt AVRPM2 after heterologous expression and purification from Escherichia coli or insect cells. All five AVR effectors adopt the common RALPH scaffold, but they have striking structural differences associated with differentiation of RALPH effector subfamilies. Using biochemical assays, we confirm that all RALPH AVRs tested have lost catalytic activities of the ribonuclease T1/F1 family. The AVRA6 structural template was used for mutagenesis of a RALPH effector belonging to the same subfamily to construct MLA6 gain-of-recognition hybrid effectors upon expression in barley protoplasts and heterologous N. benthamiana. Six amino acid substitutions were sufficient to turn the sequence-diverged effector CSEP0333 into a variant specifically recognized by MLA6. Our findings suggest that coupling of sequence and structural polymorphisms within the RALPH scaffold facilitated both escape from CNL receptor recognition and potential acquisition of new virulence functions, which might explain the proliferation and overabundance of this effector family in B. graminis.
Results
Blumeria graminis AVR Effectors Adopt a Common Structural Scaffold.
To better understand the coevolution of AVR effectors of Bgh with matching barley MLA receptors, we sought to obtain three-dimensional effector structures using X-ray crystallography. To extend this analysis to AVR effectors belonging to the RALPH effector superfamily in a reproductively isolated B. graminis lineage (36), we included the wheat powdery mildew effector AVRPM2, which is detected by wheat Pm2a (31, 36). All AVR effectors were recombinantly expressed without their predicted signal peptides, and throughout this manuscript, we refer to the residue positions in the mature proteins starting with methionine unless otherwise specified. We obtained well-diffracting crystals for AVRA6, AVRA7, AVRA10, AVRA22 of Bgh, and AVRPM2 of Bgt and solved their structures with molecular replacement (SI Appendix, Fig. S1). The data processing and refinement statistics for the structures are outlined in SI Appendix, Table S1.
The cores of the five B. graminis AVR effector proteins are composed of two β-sheets and a central α-helix (Fig. 1A). The first β-sheet is formed by two or three anti-parallel β-strands, of which two contribute an N-terminal β-hairpin (AVRA7, AVRA10, AVRA22, AVRPM2), whereas another β-strand is at the very C-terminus (AVRA6, AVRA10, AVRA22, and AVRPM2). The second β-sheet is formed by three or four antiparallel β-strands, of which at least two pack against the α-helix to stabilize the conformation of the β-sheet (Fig. 1A). The long loop region following the α-helix is stabilized by polar contacts with the central second β-sheet. Two conserved cysteine residues form an intramolecular disulfide bridge that connects the N- and C-terminal ends of the AVR proteins (Fig. 1B).
Fig. 1.
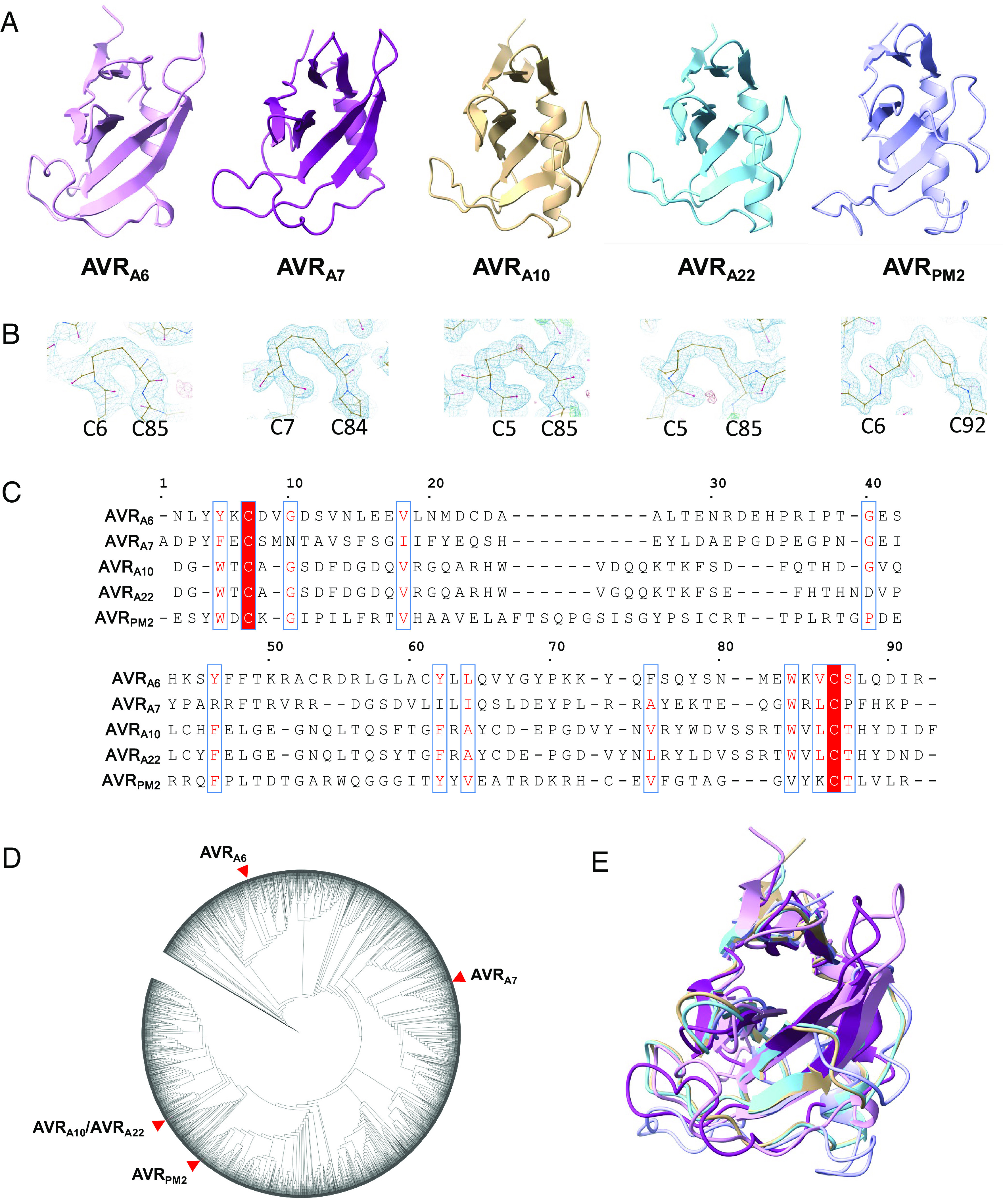
Blumeria graminis AVR effectors adopt a common structural scaffold. (A) Cartoon representation of the crystal structures of AVRA6, AVRA7, AVRA10, AVRA22, and AVRPM2. The effectors exhibit a canonical (α+β) RNase-like fold. (B) Disulfide bonds are conserved in Blumeria AVRs. AVRA6, AVRA7, AVRA10, AVRA22, and AVRPM2 form intramolecular disulfide bridges that connect the N and C termini. The disulfide bridge is indicated in the density map. (C) Amino acid sequences alignment of AVRA6, AVRA7, AVRA10, AVRA22, and AVRPM2 without signal peptides. Red background indicates amino acid similarity. The alignment was generated using ESPript 3.0 (46). (D) Maximum likelihood phylogeny including all predicted CSEPs from B. graminis f. sp. Poae, lolium, avenae, tritici 96224, hordei DH14, secalis S1459, triticale T1-20, and dactylidis. AVRA6, AVRA7, AVRA10, AVRA22, and AVRPM2 are widely separated in the phylogeny. (E) Superposition of AVRA6, AVRA7, AVRA10, AVRA22, and AVRPM2.
Except for allelic AVRA10 and AVRA22, the sequence similarity between the five AVR effectors is extremely low (maximally 40% similarity and 19% identity according to a Needleman–Wunsch BLOSUM62 alignment in SnapGene software; Fig. 1C). Furthermore, based on multiple sequence alignment of the available B. graminis effector complement, they represent distinct effector subfamilies and are widely separated on the corresponding maximum likelihood phylogenetic tree (Fig. 1D). We searched the DALI database of known structures and found that all AVR effectors adopt the common RALPH scaffold (Fig. 1E) (26). Sequence conservation among the AVR effectors is limited to a few residues that are hydrophobic and buried in the cores of the structures. Previous studies have identified the Y/F/WxC-motif as a common feature of powdery mildew effectors, based on sequence similarity analysis (41, 47). The aromatic residue of the Y/F/WxC-motif is buried in the core and forms van-der-Waals contacts with residues in β5 in AVRA6 and AVRA7 or β6 in AVRA10/AVRA22 and AVRPM2. Therefore, the Y/F/WxC-motif contributes to stabilizing the common RALPH fold. A similar role can be assigned to other conserved hydrophobic residues including V18, Y45, Y61, L63, W82, and V84 of AVRA6 (Fig. 1C), suggesting that despite the overall extreme sequence divergence of the RALPH effector family, evolutionary selection has also favored the conservation of less than 20% of residues (Fig. 1C), largely scattered in the primary structure, to maintain a stable common structural scaffold.
Structural Variations of B. graminis AVR Effectors.
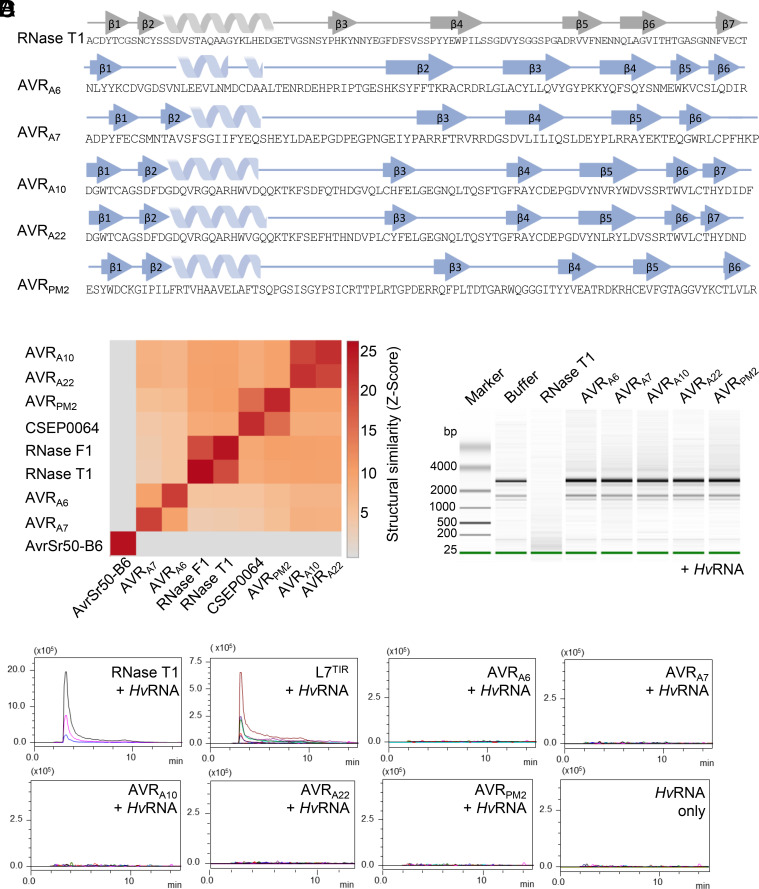
A structure-based similarity search was carried out using the DALI server (48). As anticipated, AVRA6, AVRA7, AVRA10, AVRA22, and AVRPM2 are structurally related to the ribonuclease T1/F1 family, despite the lack of detectable relatedness in their protein sequences (SI Appendix, Fig. S3). The overall structures of AVRA10, AVRA22, and AVRPM2 are more similar to RNase T1 than to AVRA6 and AVRA7, indicating that structural variation among RALPH AVR effectors within a reproductive lineage of B. graminis exceeds the variation relative to the ribonuclease T1/F1 family. Similarly, AVRPM2 in Bgt is structurally more similar to AVRA10 and AVRA22 in Bgh than to AVRA6 and AVRA7 in Bgh, showing that structural dissimilarity between RALPH AVR effectors within a reproductive lineage of B. graminis can be greater than between two powdery mildew lineages (Fig. 2B). Interestingly, the structural relationship among the AVR effectors is consistent with their phylogenetic relationship. Namely, AVRA10/AVRA22 and AVRPM2 are located distant to AVRA6 and AVRA7 in the phylogenetic tree of all Blumeria CSEPs (Fig. 1D).
Fig. 2.
Blumeria graminis AVR effectors are pseudo-RNases with diversified structural features. (A) B. graminis AVR and RNase T1 (9RNT) proteins harboring diversified secondary structural features. β-strands are indicated by arrows, α-helices by spirals. Secondary structures are pictured according to ChimeraX using BioRender. (B) Pairwise comparison between crystal structures using Dali server (48). (C) Recombinant AVR effector proteins lack ribonuclease activity. AVR effectors (1 µM) were coincubated with HvRNA and then analyzed on a Bioanalyzer to evaluate RNA degradation. (D) RNase-like AVR effectors lack 2′, 3′-cNMPs synthetase activity. Samples were subjected to LC-MS/MS for metabolite identification and quantification. Data are depicted as a representative chromatograph of at least three independent experiments.
Although the AVR effector proteins share a general structural similarity with RNase T1, there are striking local structural variations between these proteins. For example, compared with RNase T1, all AVR effectors except AVRA7 possess an additional β-strand (β6 in AVRA6 and AVRPM2, β7 in AVRA10/AVRA22) after the disulfide bridge-forming cysteine in the C-terminus (Fig. 2A and SI Appendix, Fig. S5). Variations in individual structural elements are also evident between the RALPH AVR effectors. The first β-sheet of AVRA10/AVRA22 and AVRPM2 consists of three β-strands, whereas the corresponding β-sheet in AVRA6 and AVRA7 consists of only two β-strands. In AVRA7, the two β-strands of the first β-sheet are located in the N-terminal region, whereas in AVRA6 both N- and C-termini contribute one β-strand each. In AVRA6 and AVRA7, the β-strands of the central β-sheet that face the α-helix are substantially longer than the corresponding β-strands in AVRA10, AVRA22, and AVRPM2. In addition, the loop region connecting β3 and β4 of AVRA7 is much shorter than its equivalent in AVRA6. Finally, the length of the α-helix is also variable among the AVR effectors, except for allelic AVRA10 and AVRA22 (Figs. 1A and 2A). Collectively, this indicates an unexpected structural diversification among RALPH AVR effectors, which is associated with a differentiation of RALPH effector subfamilies despite the maintenance of a common structural scaffold.
Superimposition of RNase T1 (PDB: 9RNT) with AVRA6, AVRA7, AVRA10, AVRA22, and AVRPM2 illustrates structural similarity, but the predicted residues for RNA hydrolysis are not conserved in the AVR effectors (SI Appendix, Fig. S3). We confirmed previous studies that suggested (35, 37, 41) that AVR effectors are pseudo-RNases that cannot hydrolyze total barley RNA (HvRNA) (Fig. 2C and SI Appendix, Fig. S4). RNase T1 has also the capacity to produce 2′, 3′-cyclic nucleotide monophosphates (mainly 2′, 3′ -cGMP), which are putative second messengers in TNL-mediated mediated plant immunity (49). To test whether the B. graminis AVR effectors are catalytically active in producing 2′, 3′ -cNMPs, we coincubated the RNase T1, L7TIR, and powdery mildew AVR effectors with HvRNA as substrate. Liquid chromatography coupled with mass spectrometry (LC-MS) analysis showed that only RNAse T1 and L7TIR could synthesize 2′,3′- cAMP/cGMP under these conditions (Fig. 2D and SI Appendix, Fig. S4B). To test whether the RNase-like effector proteins can bind HvRNA, we performed microscale thermophoresis (MST) experiments. However, the affinity of the AVR effectors for total RNA was not significantly different from the measured affinity of non-RNase-like fold proteins (BSA, GST, and AvrSr50 from P. graminis f. sp. tritici (SI Appendix, Fig. S4C). This strongly suggests that RALPH AVR effectors have lost known enzymatic activities of the RNase T1/F1 family and exhibit variation in the number, position, and length of individual structural elements. This raises the possibility that upon escape from CNL detection, their potential virulence functions were associated with neo-functionalization events involving combined sequence and structural diversification within the common RALPH scaffold.
Structural Determinants of AVRA6 Detection by MLA6.
CSEP0333 is a member of the AVRA6 effector family (family E008) but is not recognized by MLA6 (35, 41). Based on their significant sequence identity (60%), we suspected that this effector had a similar structure to AVRA6 and reasoned that substituting residues in CSEP0333 with those of AVRA6 may convert this family member into an effector recognized by MLA6. Based on the crystal structure, AVRA6 was divided into three units: an N-terminal part comprising the β1 and the α-helix (residues 1–26), a central segment that includes the long loop region and β2 (residues 27–53), and a C-terminal part that includes the β-strands β3–β6 (residues 54–91). Each of these segments was swapped with the corresponding unit in AVRA6 or CSEP0333, resulting in six AVRA6/CSEP0333 effector chimeras (Fig. 3A). We then individually co-expressed the hybrid effectors together with MLA6 in barley protoplasts and quantified cell death by measuring luciferase reporter activity (50). Swapping of either the N-terminal or C-terminal segment of AVRA6 with the corresponding unit in CSEP0333 (constructs A6N and A6C, respectively) did not lead to loss of recognition by MLA6 (Fig. 3 A and B). As anticipated, CSEP0333 with its N-terminal or C-terminal part substituted with the equivalent parts of AVRA6 (constructs B6N and B6C, respectively) did not trigger MLA6-dependent cell death. However, when the central segment was exchanged to that in CSEP0333 (A6M), cell death was completely abolished. Strikingly, swapping the central segment of CSEP0333 with that of AVRA6 was sufficient to activate MLA6-mediated cell death in barley protoplasts (Fig. 3A), i.e., there was a gain in effector recognition. Agrobacterium-mediated individual coexpression of the six effector hybrids with a C-terminal mYFP-tag together with MLA6 in heterologous N. benthamiana produced comparable differential cell death phenotypes (Fig. 3B). All effector proteins were detectable in N. benthamiana leaf extracts (Fig. 3C and SI Appendix, Fig. S7B). This demonstrates that effector detection by the receptor does not require a second barley protein.
Fig. 3.
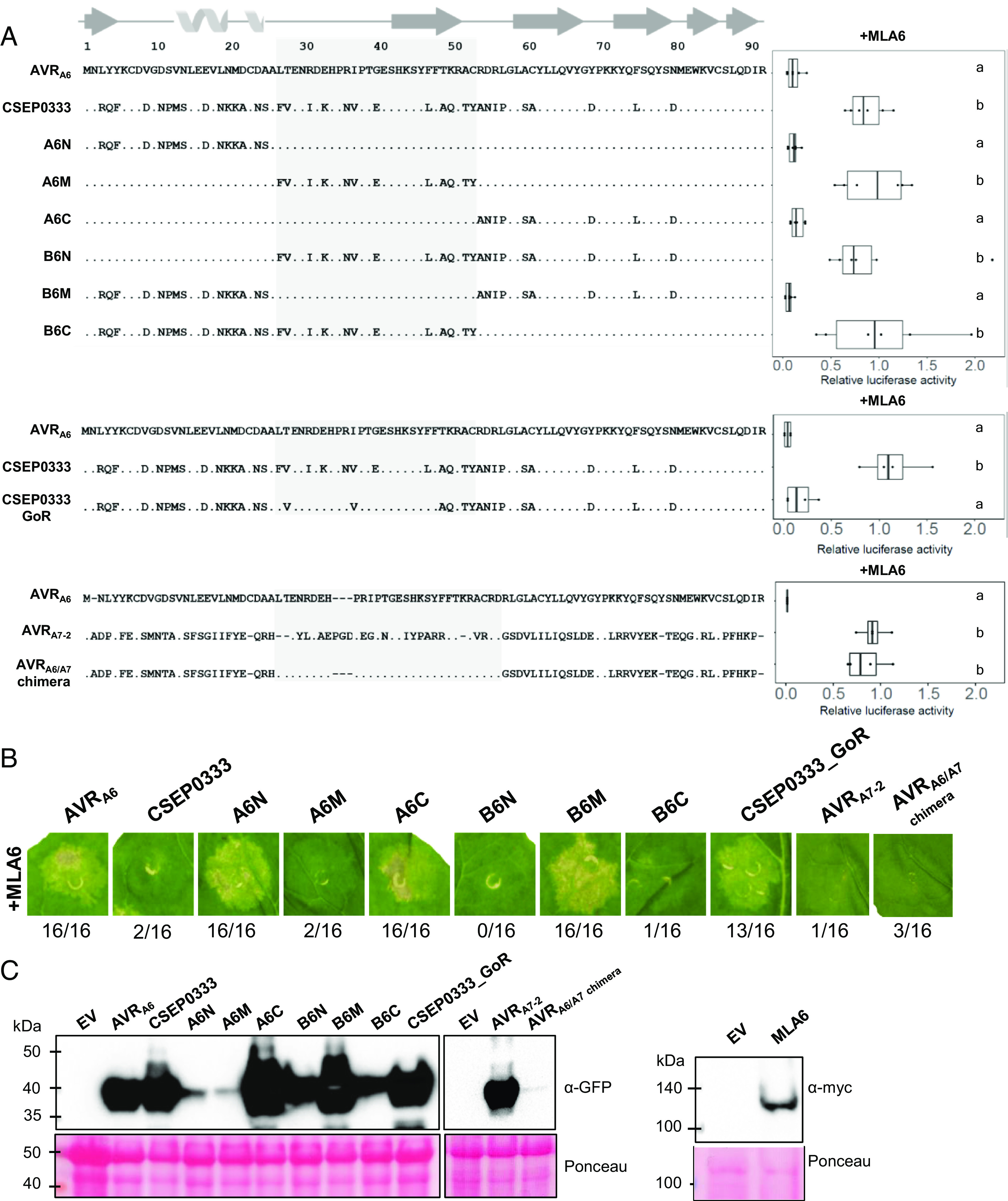
Six amino acids in the central segment of AVRA6 are essential for detection by MLA6. (A) Chimeric effectors were coexpressed with MLA6 in barley protoplasts and cell death was quantified by measuring luciferase reporter activity. Letters indicate results of statistical variance analysis using Kruskal–Wallis test followed by Dunn’s post hoc tests (P < 0.05). Raw relative luciferase measurements and P-values for all protoplast plots are provided in Supplementary Dataset S1. (B) Agrobacterium-mediated coexpression of the six effector chimeras with a C-terminal mYFP-tag together with MLA6 in N. benthamiana produced comparable differential cell death phenotypes. (C) All effector proteins were detectable in N. benthamiana leaf extracts, except for very low amounts of the AVRA6/A7 chimera. A6N and A6M accumulate to lower levels than other CSEP0333 hybrid effectors. Relative protein abundances were consistent across at least three independent experimental replicates.
To define which part of the 12 residues in the central segment of AVRA6 is important for recognition by MLA6, we subdivided the central segment into two parts that encompass the loop region (seven polymorphic residues) or the β2 and loop region connecting β2–β3 (five polymorphic residues). We found that AVRA6 hybrids in which either of these two parts are swapped with the respective unit present in CSEP0333 (constructs M1 and M2) are not recognized by MLA6 (SI Appendix, Figs. S6 and S7). This indicated that residues from both the loop regions as well as β2 are involved in AVRA6 recognition by MLA6. Therefore, we used the constructs M1 and M2 as templates for site-directed mutagenesis to create 19 additional AVRA6 higher-order mutant constructs with different combinations of targeted amino acid substitutions (SI Appendix, Figs. S6 and S7). By comparing cell death induced by different combinations of amino acid substitutions, it was possible to identify residues that are essential for recognition by MLA6. For example, the construct M1F27L/K33E/N36R/E40G did not lead to induction of cell death even though the protein was detectable in N. benthamiana leaf extracts, whereas the construct M1F27L/I31R/K33E/N36R/E40G was able to reduce LUC activity to a level comparable with wild-type AVRA6 (SI Appendix, Figs. S6 and S7). These results indicate that L31R can rescue the phenotype generated by the other four mutations. Cell death induction after cotransfection with MLA6 in protoplasts was only observed for constructs that include the six residues L27, R31, E33, R36, G40, and F47 of AVRA6 (SI Appendix, Figs. S6 and S7). We then introduced the six amino acid substitutions F27L, I31R, K33E, N36R, E40G, and L47F into CSEP0333, and the resulting construct, termed CSEP0333GoR (CSEP0333Gain-of-Recognition), was able to trigger MLA6-dependent cell death both in barley protoplasts and N. benthamiana (Fig. 3 A and B). All constructs that induce cell death when coexpressed with MLA6 were also coexpressed together with MLA7 in barley protoplasts. Only AVRA7 induced MLA7-dependent cell death, confirming recognition specificity of the tested hybrid effectors by MLA6 (SI Appendix, Figs. S6 and S7). In summary, our results show that six amino acid substitutions in the central segment of CSEP0333 are sufficient to turn this sequence-diverged effector into a variant specifically recognized by MLA6. It is therefore possible that CSEP0333 evolved from AVRA6 by immune evasion.
Among the five resolved structures of AVR effectors, AVRA6 is structurally most similar to AVRA7 (Fig. 2B). Nevertheless, based on sequence relatedness, AVRA7 belongs to effector family 29, whereas AVRA6 and CSEP0333 both belong to effector family 8 (41). We constructed a chimeric effector in which the central segment of AVRA7 is swapped for the equivalent part of AVRA6 (AVRA6/A7 chimera). However, coexpression of the AVRA7/AVRA6 hybrid with MLA6 did not induce cell death either in barley protoplasts or N. benthamiana. The AVRA7/AVRA6 chimera also did not cause MLA7-dependent cell death in barley protoplasts, and the hybrid protein was barely detectable in N. benthamiana leaf extracts (Fig. 3 and SI Appendix, Figs. S6 and S7). This shows that the RALPH interfamily AVRA7/AVRA6 hybrid is unstable in planta (Fig. 3C), presumably due to the structural differences between wild-type AVRA6 and AVRA7, which likely hinder the proper folding of the hybrid protein.
MLA6, MLA10, MLA22, and PM2 CNLs Each Recognize Largely Different Surface Patches on the RALPH Scaffold.
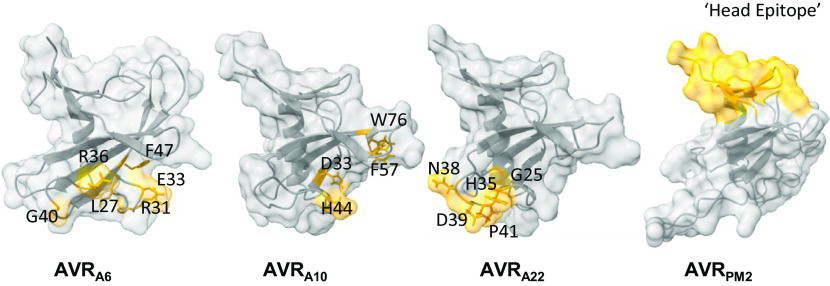
Previously, four amino acids were identified in AVRA10 that cannot be exchanged to the respective residues in AVRA22 without abrogating recognition by MLA10 and five amino acids in AVRA22 that cannot be exchanged to the corresponding residues in AVRA10 without losing MLA22-dependent recognition (35). Furthermore, amino acids that constitute the “head epitope” of AVRPM2 are important for specific recognition by wheat PM2a (31). When mapped onto the structures of these effector proteins, these epitopes are located at different sites (Fig. 4). In AVRA10, one residue is located in the loop region after the α-helix (D33), one residue is in the loop region between β3 and β4 (F57), and two map to the β-strands β3 and β5 (H44 and W76, respectively). By contrast, in AVRA22 the residues important for the recognition by MLA22 are mainly located in the loop region between β2 and β3 (H35, N38, D39, and P41). The residue G25 is located at the end of the α-helix. In AVRA6, the six amino acids that are essential for recognition by MLA6 are located in a surface patch at the loop region between the α-helix and β3. All identified residues are highly surface-exposed (SI Appendix, Fig. S8). Given that MLA receptors likely detect cognate RALPH AVR effectors directly (13), these findings indicate that the interface between an AVRA effector and an MLA receptor is different for each of these matching effector–receptor pairs (see below).
Fig. 4.
MLA6, MLA10, MLA22, and PM2 CNLs each recognize largely distinct surface patches on the common RALPH effector scaffold. Locations of residues in AVRA6, AVRA10, AVRA22, and AVRPM2 that determine the respective MLA6, MLA10, MLA22, and PM2a recognition specificities are highlighted in yellow color. The residues of AVRA10 and AVRA22 required for specific MLA10 and MLA22 recognition as determined in ref. 35. The residues of AVRPM2 important for recognition of PM2 were determined in ref. 31.
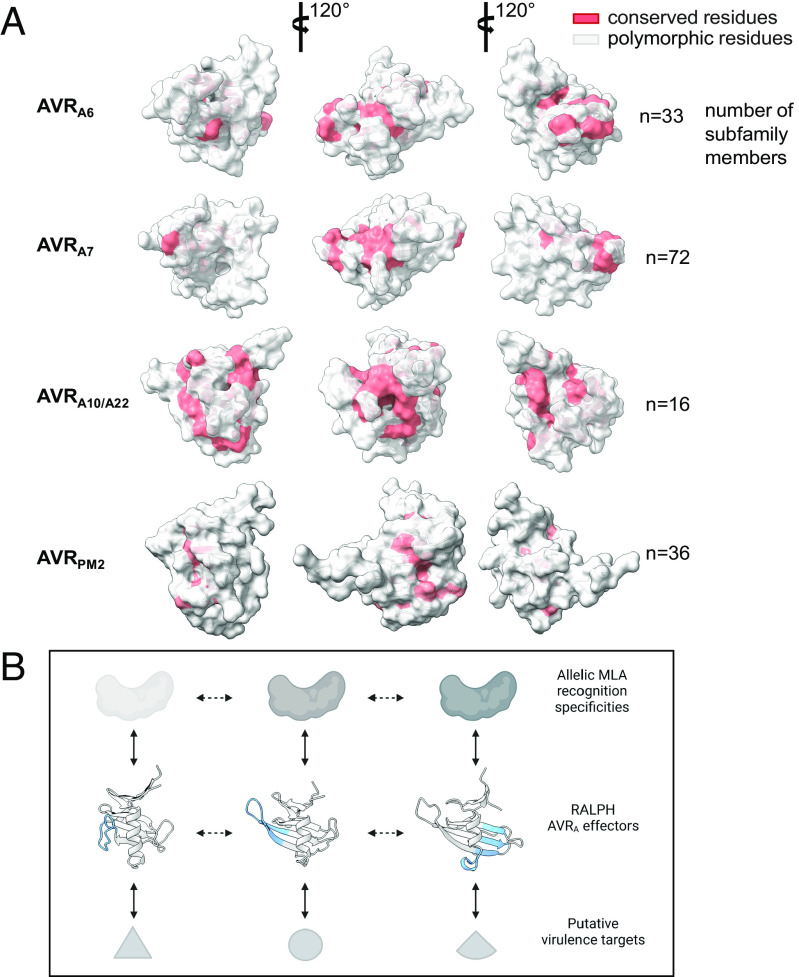
RALPH Effector Subfamilies Harboring Avirulence Effectors Have Different Conserved Surface Arrays.
AVRA6, AVRA7, AVRA10/AVRA22, and AVRPM2 belong to four distinct phylogenetic effector subfamilies (Fig. 1D). We sought to investigate the evolution of these RALPH effectors in their respective subfamilies, by mapping highly polymorphic as well as conserved residues onto the resolved AVR structures. We then highlighted conserved residues (>70%) in the crystal structures (Fig. 5). Most conserved residues are buried in the core of the structure and contribute to maintain the structural scaffold. Similarly, most surface-exposed residues are highly polymorphic. Interestingly, however, some residues are conserved in the respective subfamily, although they have a high relative solvent accessibility (SA). For example, in AVRA6 and AVRA7, in the loop region after the α-helix, three highly exposed residues are conserved: proline at position 38 in AVRA6 (64% SA; 30 out of 33 family members) or position 39 in AVRA7 (72% SA; 71 out of 72 family members); glycine at position 40 in AVRA6 (73% SA; 24 out of 33 family members) or position 41 in AVRA7 (46% SA; 67 out of 72 family members); glutamic acid at position 41 in AVRA6 (42% SA; 26 out of 33 family members) or position 42 in AVRA7 (37% SA; 44 out of 72 family members) (Fig. 5A and Dataset S2). This surface patch appears not to be conserved in AVRA10/AVRA22 and AVRPM2, in which the conserved surface-exposed residues are not confined to a discrete surface patch but rather map to multiple locations.
Fig. 5.
RALPH effector subfamilies harboring avirulence effectors have overlapping or distinct conserved surface arrays. (A) All CSEPs from B. graminis f sp poae, lolium, avenae, tritici 96224, hordei DH14, secalis S1459, triticale T1-20, and dactylidis were subjected to BLASTP. CSEPs that share >30% sequence identity and that are similar in size to the crystallized RALPH AVR effectors were retained for further analysis using Muscle. Red color indicates conserved (70% threshold) residues. (B) Model for MLA receptor and RALPH effector coevolution. Major local structural differences between RALPH AVRA effectors are highlighted in blue. Solid bidirectional arrows indicate selection pressure by coevolving protein pairs, dashed bidirectional arrows represent adaptive genetic changes in RALPH effectors.
Discussion
We have resolved here five crystal structures of AVR effectors from a total of 14 RALPH AVR proteins of B. graminis, each of which was purified to homogeneity and in sufficient quantity (SI Appendix, Figs. S1 and S2). It is possible that the extensive diversity of structural elements found among the crystallized RALPH AVR proteins, combined with the exceptional sequence diversification of surface-exposed residues, makes it challenging to obtain well-diffracting crystals for the remaining 11 AVR effectors (SI Appendix, Fig. S2). This could explain why a wide range of different conditions had to be screened for successful crystallization of the purified proteins, although they share one common structural scaffold (Materials and Methods). Another factor could be that some RALPH AVR effectors adopt a stable conformation only in association with their effector targets inside host cells.
Recently, machine-learning algorithms for the prediction of protein structures, such as AF2, have greatly increased in accuracy (51). However, AF2 is homology-based, and the accuracy of the predictions depends on available experimental structures in the multiple sequence alignment (MSA). The crystal structure of avrSr50-B6 of P. graminis f. sp. tritici revealed that the model predicted by AF2 is largely incorrect (52). This is consistent with the resolved powdery mildew AVR structures. While the topology of predicted AVRA6, AVRA7, and AVRPM2 is close to the real structures, the predictions of AVRA10 and AVRA22 are largely inaccurate. When the crystal structures of AVRA10/AVRA22 are compared to the predicted AF2 model, only 55 atoms (from a total 97 atoms) can be aligned, but with a rmsd of 4.50 Å and 4.90 Å, respectively (SI Appendix, Fig. S10). AVRPM2 of Bgt is in the same family as the previously resolved CSEP0064 of Bgh, which can explain why a subset of effectors including AVRPM2 are predicted more accurately than AVRA10/AVRA22. AVRA10/AVRA22 belong to a relatively small effector subfamily (Fig. 1D and SI Appendix, Fig. S9). Thus, our work on the structural diversification among RALPH AVR effectors illustrates that MSA-based modeling algorithms must be applied with greater caution, because the models captured only a subset of the actual diversity of individual structural elements within a common scaffold.
Despite mounting evidence that fungal effectors with low or undetectable sequence similarity may be structurally related and may share a common scaffold (17, 19, 20, 22, 23, 26), it is still unclear whether this indicates shared scaffold-specific biochemical activity. We have provided evidence that all five RALPH AVR effectors tested have lost both RNase and synthetase catalytic activities of the evolutionarily ancient ribonuclease T1/F1 family. Furthermore, contrary to a previous report on the CSEP0064 RALPH effector from Bgh (37), none of the five RALPH AVR effectors tested in this study has an affinity for RNA that is significantly different to the measured affinity of non-RNase-like fold proteins, including the AvrSr50 effector from P. graminis f. sp. tritici. Our findings rather suggest that virulence activities of RALPH AVR effectors belonging to different subfamilies are the result of neo-functionalization events, which could explain why several tested RALPH effectors were found to interact with different host proteins (37, 42–45). Evolutionary selection of the conserved RALPH framework may indicate the existence of nonvariable functions such as translocation across the fungal and host membranes into host cells and/or stable folding. Similarly, the physiological role of intramolecular disulfide bridges to B. graminis AVR effectors within plant cells requires further investigation.
Sequence-based Bgh genome-wide analysis (41) as well as large-scale structural modeling using AF2 indicate that approximately 70% of candidate secreted effector proteins are RALPHs (26). We have been able to generate, by only six amino acid substitutions, a gain-of-recognition variant of the CSEP0333 effector specifically detected by MLA6 and belonging to the subfamily containing AVRA6, indicating that CSEP0333 may have evolved from AVRA6 by immune evasion. The AVRA effectors AVRA6, AVRA7, AVRA9, AVRA13, and the allelic variants AVRA10/AVRA22 each belong to a different RALPH subfamily with multiple paralogs (13, 32, 35). This suggests that, with the exception of MLA10 and MLA22, each MLA recognition specificity drives sequence diversification within a different RALPH subfamily. Recent structural information on CNL and TNL resistosomes in A. thaliana, N. benthamiana, and wheat, all activated by direct AVR interactions, has shown that multiple surface-exposed effector residues make extensive contacts with their respective neighboring residues in C-terminal LRR or C-JID regions of the corresponding heteromeric receptor complexes (4–7). We identified multiple surface-exposed AVRA6, AVRA10, or AVRA22 residues required for recognition by the corresponding MLA receptors, indicating the existence of a similarly extensive effector–MLA receptor interface region. Together with the variations in the number, position, and length of individual structural elements found between RALPH AVR effectors, natural effector polymorphisms that locally affect individual structural AVR effector features essential for receptor recognition likely facilitate immunoevasion. Thus, our data support an evolutionary model in which changes in individual structural elements of RALPH effectors have contributed to the diversification of RALPH effector subfamilies, whereas nonstructural substitutions of surface-exposed effector residues have mainly driven diversification within a subfamily of RALPH effector paralogs.
The evolutionary model for RALPH effector diversification also provides a plausible framework to explain the molecular coevolutionary arms race between MLA receptors and RALPH AVRA effectors. The coevolutionary dynamics of the barley–Bgh pathosystem must involve iterative cycles of generation and selection of novel MLA recognition specificities, followed by generation and selection of RALPH effectors that evade receptor detection to maintain pathogen virulence. Because i) AVRA6, AVRA7, AVRA9, AVRA13, and the allelic variants AVRA10/AVRA22 each belong to a different RALPH subfamily and are each detected by different MLA recognition specificities, ii) structural effector diversification has driven at least the differentiation of three subfamilies harboring AVRA6 or AVRA7 or AVRA10/AVRA22, and iii) these RALPH effector subfamilies also possess distinct conserved surface patches (Fig. 5A), it is likely that structure-driven RALPH subfamily differentiation is linked to the acquisition of distinct virulence functions. As a corollary, sequence-related RALPH effectors belonging to the same subfamily are probably targeting the same host process to promote virulence. Thus, we propose that proliferation of RALPH effectors in B. graminis genomes and their dominance in the effectorome was driven by both acquisition of novel virulence functions and escape from MLA receptor detection (Fig. 5B). This would also explain why multiple RALPH effector subfamilies and corresponding MLA recognition specificities are simultaneously maintained in host and pathogen populations.
How can allelic MLAs directly recognize effectors with divergent sequences? We identified six residues in AVRA6 that are essential for MLA6-dependent recognition (Fig. 3 and SI Appendix, Figs. S6 and S7). When we introduced these six residues (F27L, I31R, K33E, N36R, E40G, and L47F) into CSEP0333, belonging to the subfamily containing AVRA6, the resulting construct was able to confer MLA6-dependent cell death in barley protoplasts and heterologous N. benthamiana (Fig. 3). However, when we constructed and tested a similar hybrid effector of AVRA7 containing the entire central segment of AVRA6, the resulting hybrid protein was highly unstable and could often not be detected in N. benthamiana leaf extracts, indicating that structure-driven diversification of the two subfamilies containing AVRA6 or AVRA7 is connected to proper folding within subfamily members. Recently, crystal structures of two MAX effector variants APikL2F and APikL2A in complex with effector targets sHMA94 and sHMA25, respectively, revealed that a single polymorphic residue can lead to subtle changes in protein structures that ultimately determine the binding capacity of effectors to their host targets (53). Therefore, it is not surprising that structural elements of AVRA effectors belonging to different subfamilies are not readily interchangeable. Allelic AVRA10 and AVRA22 were shown here to share a highly similar structure, but the residues involved in recognition by MLA10 and MLA22, respectively, locate to different surface patches on the subfamily-specific effector structure. Interestingly, MLA10 and MLA22 are not closely located within the phylogenetic tree of Mla resistance genes (54), which suggests that they evolved to detect AVRA10 and AVRA22, respectively, by convergent evolution. Therefore, MLA receptors have either evolved to detect distinct surface patches on RALPH effectors with highly similar structures or, as seems to be more common, surface patches of structurally diversified RALPHs belonging to different subfamilies. Furthermore, the structure of the wheat powdery mildew AVRPM2 clarified here confirms that the phylogenetically unrelated CNL PM2 has independently evolved the capability to recognize another site on RALPHs, termed the head epitope, in the sister species wheat, although whether this involves direct or indirect effector perception remains to be determined (31). It is likely that regions other than residues of RALPH effectors contacting the LRR contribute to MLA receptor activation. For instance, multiple contact residues in AvrSr35 are needed for binding to the Sr35 LRR domain, leading to a steric clash of other effector surface regions with the Sr35 NOD domain, with both of these processes needed for receptor activation (55).
The common fold adopted by Blumeria effectors raises the question of whether the conserved structural fold evolved convergently (i.e., from independent ancestral proteins) or whether the RALPH AVR effectors share a phylogenetic history which is masked by sequence diversification while the structure is retained (i.e., sequence-diversified from a common ancestor)? Previous work predicted that RALPHs share an intron in the same relative position (35, 41, 56), which points to a common ancestor. Indeed, in the five resolved RALPH AVR effector structures, an intron locates to the same relative position in the loop region between β3 and β4 (which is equivalent to β2 and β3 in AVRA6) (SI Appendix, Fig. S5). Interestingly, this relative intron position is shared with RNase F1 and the catalytic active ribonuclease effectors from hemibiotrophic Zt6 from Zymoseptoria tritici, Fg12 from Fusarium graminearum and SRE1 from Setosphaeria turcica (SI Appendix, Fig. S5) (57–59). We conclude that RALPH AVRAs belonging to different subfamilies have evolved from an ancient ribonuclease that was present in the last common ancestor of Dothideomycetes and Sordariomycetes. Accordingly, maintenance of catalytic ribonuclease activity in the effectorome of Z. tritici, F. graminearum, and S. turcica or its loss in powdery mildew, in tandem with neo-functionalization of multiple virulence functions, is associated with a transition in pathogen lifestyle from hemibiotrophic to obligate biotrophic.
Materials and Methods
Protein Expression and Purification.
Effector sequences (13, 16, 30, 32–35) were put into SignalP-5.0 Server Output - DTU Health Tech to detect signal peptides. The constructs used for effector protein purification have had the signal peptide removed. AVRA1(27–118), AVRA6(25–115), AVRA7(24–112), AVRA9(20–102), AVRA10(21–119), AVRA13(21–122), AVRA22(22–118), AVRPM2(21–119), AVRPM3A2/F2 (24–130), AVRPM3B2/C2(21–130), AVRPM3D3(21–109), AVRPM17(24–110), AVRPM1a (18–155), AVRPM8(17–107), AvrSr50(24–132), and PWL2(21–145) were expressed in E. coli SHuffle cells (NEB) and insect cells as fusion proteins subcloned into pGEX-6P-1(GE Healthcare) or the pFastBac-1 vector (Invitrogen), respectively. The successfully crystallized AVRA6, AVRA10, AVRA22, and AVRPM2 proteins were obtained from E. coli. Although AVRA7 could be purified from E. coli, the successfully crystalized protein was purified from insect cells. These plasmids were used to express effectors with a N-terminal GST-tag followed by a PreScission proteolytic recognition site to remove the GST-tag.
Bacterial cultures were grown at 30 °C to an OD600 of around 0.8 in LB broth and induced with isopropyl-β-D-thiogalactoside (IPTG, Sigma) for 15 to 18 h at 16 °C. The cells were harvested by centrifugation at 6,000 g for 10 min at 4 °C and resuspended in resuspension buffer (25 mM Tris pH 8, 150 mM NaCl, 1 mM PMSF, 1 mM DTT). Bacterial cell suspensions were sonicated for 20 min at 60% power (BANDELIN). Cell debris was removed by centrifugation at 30,000 g for 2 h at 4 °C. The soluble fractions were collected and allowed to flow through GST resin (GE Healthcare). After washing with two column volumes of the same buffer used for resuspension, another 2 mL buffer and 10 μL PreScission protease (GE Healthcare) were added to the column followed by overnight incubation to cleave off the AVR proteins from the GST resin. The cleaved AVR proteins were then eluted and further purified by size-exclusion chromatography using a HiLoad 16/600 Superdex 200 pg gel filtration column (GE Healthcare). Purified proteins were concentrated to 10 to 30 mg mL−1 by using a 10-kDa Amicon centrifugal filter device (Merck), flash-frozen in liquid nitrogen, and stored at -80 °C. Baculoviruses (50 mL) for AVR expression were individually added to 1 L of SF21 insect cells (1.8 × 106 cell mL−1) cultured at 28 °C in Sf-900 II SFM medium. The insect cells were collected 48 h after infection. The purification process is the same as for the E. coli system.
Crystallization, Data Collection, Structure Determination, and Refinement.
The initial crystallization experiments were carried out at 20 °C, using the sitting-drop vapor-diffusion method. For screening, the AVR effector proteins were mixed 1:2, 1:1, and 2:1 with different crystallization buffers using a Mosquito Nanodrop. Out of 16 effector proteins, good diffraction crystals were obtained for only AVRA6, AVRA7, AVRA10, AVRA22, and AVRPM2. After the initial screening, further optimization was performed using a 24-well hanging-drop vapor-diffusion method with an equal volume (1.0 µL) of protein and reservoir solution at 20 °C. Crystals with the best morphology were observed in 20 % w/v polyethylene glycol 3 350, 200 mM sodium fluoride for AVRA6; 1.4 M sodium phosphate monobasic monohydrate/potassium phosphate dibasic pH 8.2 for AVRA7; 0.16 M calcium acetate hydrate, 0.08 M sodium cacodylate trihydrate pH 6.5, 14.4% w/v polyethylene glycol 8,000 for AVRA10; 1.0 M succinic acid pH 7.0, 0.1 M HEPES pH 7.0, 1% w/v polyethylene glycol monomethyl ether 2,000 for AVRA22; 0.1 M BIS-TRIS pH 6.5, 28% w/v polyethylene glycol monomethyl ether 2,000 for the orthorhombic AVRPM2 crystal form; and 20 % w/v polyethylene glycol 3,350, 100 mM BIS-TRIS propane pH 7.5, 200 mM sodium acetate for the monoclinic AVRPM2 crystal form. Crystals were transferred into a cryoprotectant solution containing a reservoir solution with 20% glycerol. The diffraction data were collected at P13 (60) and P14 at the EMBL Hamburg, the X06SA at the SLS and the ID30B (61) at the ESRF as indicated in the SI Appendix, Table S1. The data were processed using XDS or autoProc (62, 63). Data cut-offs were chosen based on statistical significance of CC1/2 values (64). This usually leads to apparently worse statistics in the outer shell but slightly better electron density maps than with more conservative resolution cut-offs (65). For comparison, formal data resolutions determined by an average I/sigI below 2 (mean I/sigI < 2) would be as follows: AVRA6 ~ 2.6 Å; AVRA7 ~ 1.75 Å, AVRA10 ~ 1.6Å; AVRA22 ~1.75 Å; AVARPM2/C2221 ~ 1.5 Å AVRPM2/P21 ~ 1.15 Å. For AVRA10, an overall completeness of only 95% at 1.6 Å could be achieved, with 5 % missing due to the blind region, while the low completeness for the data between 1.6 and 1.38 Å is due to the detector distance. The crystal structures of these five AVR effectors were determined by molecular replacement (MR) with Phenix using structures predicted by AF2 as the initial search model. The models from MR were built automatically by ModelCraft (66) and/or computer-assisted with COOT (67) and subsequently subjected to refinement by Phenix software suite (68). Statistics of diffraction data and refinement of these five effector models are summarized in SI Appendix, Table S1. Structural figures were prepared using the program ChimeraX v1.3 (69). For AVRPM2, the two copies in the monoclinic and the one copy in the orthorhombic space groups are virtually identical (RMSD < 0.35 Å), with the exception of the Arg40–Gly47 loop, which is best defined in chain B of the monoclinic space group and hardly seen in the orthorhombic space group. In all figures, we only show the chain B of the P21 crystal form. Sequence alignments were processed with the ENDscript server (46).
AVRA Effector RNase Activity Assays.
Total RNA was extracted from 9-d-old barley cv. Golden Promise plants using the RNeasy Plant Mini Kit (QIAGEN) and treated with TURBO DNase enzyme (Ambion) to remove genomic DNA. Purified AVRA effectors from E. coli were then incubated with the total barley RNA. The reaction mixture consisted of 1 μg of RNA and 1 μM of protein and was prepared in a buffer containing 15 mM Tris-HCl (pH 8.0), 15 mM NaCl, 50 mM KCl, and 2.5 mM EDTA. The reaction mixture was incubated at 25 °C for 90 min. For analysis using the Bioanalyzer 2100 (Agilent Technologies, USA), 10 μL of the sample were used. RNase T1 (Thermo Scientific) was included as a positive control in the assay.
Production and Detection of 2’, 3’-cNMPs In Vitro.
Barley total RNA (100 ng) was individually incubated with purified AVR effector (1 µM for each), L7TIR (1 µM), and 2.5 µL of RNase T1 (Thermo Scientific) in buffer containing 25 mM Tris-HCl pH 8.0 and 150 mM NaCl at 25 °C for 16 h. The total volume for each reaction was 50 µL. The samples were centrifuged at 12, 000 g for 10 min and the supernatant was applied to LC-MS/MS for metabolite measurement.
Metabolite Measurement by LC-MS/MS.
Chromatography was performed on a Nexera XR 40 series HPLC (Shimadzu) using a Synergi 4 μM Fusion-RP 80 Å 150 × 2 mm column (Phenomenex). The method of determination of 2’, 3’-cNMPs is described in ref. 49.
Transient Gene Expression Assays in Barley Protoplasts.
Entry clones and destination constructs for the expression of AVRA6, AVRA7, MLA6, and MLA7 were previously reported in refs. 13 and 35. Entry clones for CSEP0333 (BLGH_00698) and the chimeric effectors A6N, A6M, A6C, B6N, B6M, B6C, M1, and M2 were generated by gene synthesis based on wild-type codons (GeneArt, Invitrogen). The constructs M1 and M2 were used as templates to generate higher-order mutants by site-directed mutagenesis PCR (NEB, Q5 Site-Directed Mutagenesis Kit) using the primers listed in SI Appendix, Table S2. The integrity of all entry clones was confirmed by Sanger sequencing. For transient expression assays in barley protoplasts and N. benthamiana leaves, the genes were recombined using LR-Clonase II (Thermo Fisher) into the pIPKb002 (SpecR) (70) Gateway-compatible destination vectors. The integrity of all expression vectors was confirmed by Sanger sequencing. The isolation and transfection of barley leaf protoplasts was performed as described in ref. 50. cDNAs of the AVRa effectors chimeras were coexpressed with cDNAs of MLA6 or MLA7 using the pIPKb002 vector with the ubiquitin promoter in barley cv. Golden Promise protoplasts. Protoplast solution (300 μL of 3.5 × 105 cells mL–1) was transfected with 4 μg of LUC reporter construct (pZmUBQ: LUC), 12 μg of Mla plasmid, and 5 μg of the respective AVRa (chimeric) effector or an empty vector.
Transient Gene Expression in N. benthamiana and Protein Detection by Immunoblotting.
N. benthamiana transient gene expression for measuring cell death was previously described in refs. 13 and 35. The detailed description of the experimental procedures is outlined in SI Appendix, SI Materials and Methods.
MST.
The detailed description of the experimental procedures for MST measurements and analysis is outlined in SI Appendix, Materials and Methods.
Phylogenetic Analysis of RALPH Effectors and Detection of Conserved Surface-Exposed Amino Acids.
A maximum likelihood phylogeny was constructed according to ref. 35, including all predicted CSEPs from B. graminis f sp poae, lolium, avenae, tritici 96224, hordei DH14, secalis S1459, triticale T1-20, and dactylidis. The protein sequences of the members of effector subfamilies were aligned using MUSCLE and then displayed by ESPript3 (https://espript.ibcp.fr/ESPript/ESPript/). Conserved residues >70% have been highlighted in the crystal structures.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (PDF)
Acknowledgments
We thank the Alexander von Humboldt Foundation (J.C.), the Max-Planck-Gesellschaft (P.S.-L. and J.C.), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) in the Collaborative Research Centre Grant (SFB-1403 – 414786233 B08 to J.C. and P.S.-L.), and Germany’s Excellence Strategy CEPLAS (EXC-2048/1, Project 390686111; J.C. and P.S.-L.) for funding of this project. Y.C. was funded by a PhD fellowship from the Chinese Scholarship Council (number 201808440401). Crystals were grown in the Cologne Crystallization facility (http://C2f.uni-koeln.de) supported by the DFG (Grant No. INST 216/682-1 FUGG). We thank the staff of the beamlines X06SA (PXI, PSI, Switzerland), EMBL Hamburg at the PETRA III storage ring (DESY, Germany) and ESRF (Grenoble, France) for their help during data collection. AVRA22 and AVRPM2 data were collected at X06SA, AVRA6, and AVRA7 at beamlines P13 (EMBL) and P14 (Proposal MX828) and AVRA7 at ID30B (ESRF) Proposal MX2412). We thank Gleb Bourenkov and Saravanan Panneerselvam for assistance in using these beamlines, Neysan Donnelly for manuscript editing and Petra Köchner for assistance with recombinant DNA work.
Author contributions
Y.C., F.K., J.C., and P.S.-L. designed research; Y.C., F.K., E.L., J.M.G., A.W.L., D.Y., and J.J. performed research; B.K. contributed new reagents/analytic tools; Y.C., F.K., J.M.G., M.U., U.B., and K.T. analyzed data; and Y.C., F.K., J.C., and P.S.-L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Jijie Chai, Email: chai@mpipz.mpg.de.
Paul Schulze-Lefert, Email: schlef@mpipz.mpg.de.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information. Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 8OXH (AVRA6) (71), 8OXL (AVRA7) (72), 8OXK (AVRA10) (73), 8OXJ (AVRA22) (74), 8OXI (AVRPM2#1) (75), and 8PHY (AVRPM2 #2) (76)].
Supporting Information
References
- 1.Saur I. M., Panstruga R., Schulze-Lefert, NOD-like receptor-mediated plant immunity: From structure to cell death. Nat. Rev. Immunol. 21, 305–318 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Hu Z., Chai J., Assembly and architecture of NLR resistosomes and inflammasomes. Annu. Rev. Biophys. 52, 207–228 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Lapin D., Johanndrees O., Wu Z., Li X., Parker J. E., Molecular innovations in plant TIR-based immunity signaling. Plant Cell 34, 1479–1496 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forderer A., et al. , A wheat resistosome defines common principles of immune receptor channels. Nature 610, 532–539 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma S., et al. , Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370, eabe3069 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Martin R., et al. , Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science 370, eabd9993 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y.-B., et al. , Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism. Sci. Adv. 8, eabq5108 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesari S., Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 219, 17–24 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Jones J. D. G., Vance R. E., Dangl J. L., Intracellular innate immune surveillance devices in plants and animals. Science 354, aaf6395 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Wang J., et al. , Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364, eaav5868 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Wang J., et al. , Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Srichumpa P., Brunner S., Keller B., Yahiaoui N., Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 139, 885–895 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saur I. M., et al. , Multiple pairs of allelic MLA immune receptor-powdery mildew AVRA effectors argue for a direct recognition mechanism. Elife 8, e44471 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanzaki H., et al. , Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 72, 894–907 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Ellis J. G., Lawrence G. J., Luck J. E., Dodds P. N., Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourras S., et al. , The AvrPm3-Pm3 effector-NLR interactions control both race-specific resistance and host-specificity of cereal mildews on wheat. Nat. Commun. 10, 2292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Guillen K., et al. , Structure analysis uncovers a highly diverse but structurally conserved effector family in phytopathogenic fungi. PLoS Pathog. 11, e1005228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawksworth D. L., The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycol. Res. 105, 1422–1432 (2001). [Google Scholar]
- 19.Bentham A. R., et al. , A molecular roadmap to the plant immune system. J. Biol. Chem. 295, 14916–14935 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu D. S., et al. , The structural repertoire of Aquarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies. BioRxiv [Preprint] (2022). 10.1101/2021.12.14.472499 (Accessed 12 July 2023). [DOI] [PMC free article] [PubMed]
- 21.De la Concepcion J. C., et al. , Polymorphic residues in rice NLRs expand binding and response to effectors of the blast pathogen. Nat. Plants 4, 576–585 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Di X., et al. , Structure-function analysis of the Fusarium oxysporum Avr2 effector allows uncoupling of its immune-suppressing activity from recognition. New Phytol. 216, 897–914 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazar N., et al. , A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins. PLoS Pathog. 18, e1010664 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derbyshire M. C., Raffaele S., Surface frustration re-patterning underlies the structural landscape and evolvability of fungal orphan candidate effectors. BioRxiv [Preprint] (2023). 10.1101/2023.01.06.522876 (Accessed 12 July 2023). [DOI] [PMC free article] [PubMed]
- 25.Outram M. A., Figueroa M., Sperschneider J., Williams S. J., Dodds P. N., Seeing is believing: Exploiting advances in structural biology to understand and engineer plant immunity. Curr. Opin. Plant Biol. 67, 102210 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Seong K., Krasileva K. V., Prediction of effector protein structures from fungal phytopathogens enables evolutionary analyses. Nat. Microbiol. 8, 174–187 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varden F. A., et al. , Cross-reactivity of a rice NLR immune receptor to distinct effectors from the rice blast pathogen Magnaporthe oryzae provides partial disease resistance. J. Biol. Chem. 294, 13006–13016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., et al. , Crystal structure of the Melampsora lini effector AvrP reveals insights into a possible nuclear function and recognition by the flax disease resistance protein P. Mol. Plant Pathol. 19, 1196–1209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., et al. , A positive-charged patch and stabilized hydrophobic core are essential for avirulence function of AvrPib in the rice blast fungus. Plant J. 96, 133–146 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Müller M. C., et al. , Ancient variation of the AvrPm17 gene in powdery mildew limits the effectiveness of the introgressed rye Pm17 resistance gene in wheat. Proc. Natl. Acad. Sci. USA 119, e2108808119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manser B., et al. , Identification of specificity-defining amino acids of the wheat immune receptor Pm2 and powdery mildew effector AvrPm2. Plant J. 106, 993–1007 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Lu X., et al. , Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc. Natl. Acad. Sci. U.S.A. 113, E6486–E6495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunz L., et al. , The broad use of the Pm8 resistance gene in wheat resulted in hypermutation of the AvrPm8 gene in the powdery mildew pathogen. BMC Biol. 21, 29 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourras S., et al. , Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 27, 2991–3012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauer S., et al. , The leucine-rich repeats in allelic barley MLA immune receptors define specificity towards sequence-unrelated powdery mildew avirulence effectors with a predicted common RNase-like fold. PLoS Pathog. 17, e1009223 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Praz C. R., et al. , AvrPm2 encodes an RNase-like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus. BMC Genomics 213, 1301–1314 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pennington H. G., et al. , The fungal ribonuclease-like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathog. 15, e1007620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frantzeskakis L., et al. , Signatures of host specialization and a recent transposable element burst in the dynamic one-speed genome of the fungal barley powdery mildew pathogen. BMC Genomics 19, 381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kusch S., et al. , Long-term and rapid evolution in powdery mildew fungi. Mol. Ecol. 00, 1–22 (2023). [DOI] [PubMed] [Google Scholar]
- 40.Müller M. C., et al. , A chromosome-scale genome assembly reveals a highly dynamic effector repertoire of wheat powdery mildew. New Phytol. 221, 2176–2189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen C., et al. , Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13, 694 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed A. A., et al. , The barley powdery mildew candidate secreted effector protein CSEP0105 inhibits the chaperone activity of a small heat shock protein. Plant Physiol. 168, 321–333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan H., et al. , The powdery mildew effector CSEP0027 interacts with barley catalase to regulate host immunity. Front. Plant Sci. 12, 733237 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W.-J., et al. , Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Mol. Plant Pathol. 13, 1110–1119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z., et al. , Powdery mildew effectors AVRA1 and BEC1016 target the ER J-domain protein HvERdj3B required for immunity in barley. bioRxiv [Preprint] (2022). 10.1101/2022.04.27.489729 (Accessed 12 July 2023). [DOI] [PMC free article] [PubMed]
- 46.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Godfrey D., et al. , Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics 11, 317 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holm L., Rosenstrom P., Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu D., et al. , TIR domains of plant immune receptors are 2’,3’-cAMP/cGMP synthetases mediating cell death. Cell 185, 2370–2386.e18 (2022). [DOI] [PubMed] [Google Scholar]
- 50.Saur I. M. L., Bauer S., Lu X., Schulze-Lefert P., A cell death assay in barley and wheat protoplasts for identification and validation of matching pathogen AVR effector and plant NLR immune receptors. Plant Methods 15, 118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ortiz D., et al. , The stem rust effector protein AvrSr50 escapes Sr50 recognition by a substitution in a single surface-exposed residue. New Phytol. 234, 592–606 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bentham A. R., et al. , Allelic compatibility in plant immune receptors facilitates engineering of new effector recognition specificities. bioRxiv [Preprint] (2022). 10.1101/2022.10.10.511592 (Accessed 12 July 2023). [DOI] [PMC free article] [PubMed]
- 54.Maekawa T., et al. , Subfamily-specific specialization of RGH1/MLA immune receptors in wild barley. Mol. Plant Microbe Interact. 32, 107–119 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Förderer A., Yu D., Li E., Chai J., Resistosomes at the interface of pathogens and plants. Curr. Opin. Plant Biol. 67, 102212 (2022). [DOI] [PubMed] [Google Scholar]
- 56.Spanu P. D., Cereal immunity against powdery mildews targets RNase-Like Proteins associated with Haustoria (RALPH) effectors evolved from a common ancestral gene. New Phytol. 213, 969–971 (2017). [DOI] [PubMed] [Google Scholar]
- 57.He S., et al. , The secreted ribonuclease SRE1 contributes to Setosphaeria turcica virulence and activates plant immunity. Front. Microbiol. 13, 941991 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kettles G. J., et al. , Characterization of an antimicrobial and phytotoxic ribonuclease secreted by the fungal wheat pathogen Zymoseptoria tritici. New Phytol. 217, 320–331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang B., et al. , Fg12 ribonuclease secretion contributes to Fusarium graminearum virulence and induces plant cell death. J. Integr. Plant Biol. 63, 365–377 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Cianci M., et al. , P13, the EMBL macromolecular crystallography beamline at the low-emittance PETRA III ring for high- and low-energy phasing with variable beam focusing. J. Synchrotron Radiat. 24, 323–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCarthy A. A., et al. , ID30B–A versatile beamline for macromolecular crystallography experiments at the ESRF. J. Synchrotron Radiat. 25, 1249–1260 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kabsch W., Xds. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vonrhein C., et al. , Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karplus P. A., Diederichs K., Linking crystallographic model and data quality. Science 336, 1030–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karplus P. A., Diederichs K., Assessing and maximizing data quality in macromolecular crystallography. Curr. Opin. Stru. Biol. 34, 60–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bond P. S., Cowtan K. D., ModelCraft: An advanced automated model-building pipeline using Buccaneer. Acta Crystallogr. Section D 78, 1090–1098 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Section D 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Himmelbach A., et al. , A set of modular binary vectors for transformation of cereals. Plant Physiol. 145, 1192–1200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao Y., Gebauer J. M., Baumann U., Chai J.. Crystal structure of powdery mildews Blumeria graminis f. sp. hordei AVRA6. Protein Data Bank. https://www.rcsb.org/structure/8OXH. Deposited 2 May 2023. [Google Scholar]
- 72.Cao Y., Gebauer J. M., Baumann U., Chai J.. Crystal structure of powdery mildews Blumeria graminis f. sp. hordei AVRA7. Protein Data Bank. https://www.rcsb.org/structure/8OXL. Deposited 2 May 2023. [Google Scholar]
- 73.Cao Y., Gebauer J. M., Baumann U., Chai J.. Crystal structure of powdery mildews Blumeria graminis f. sp. hordei AVRA10. Protein Data Bank. https://www.rcsb.org/structure/8OXK. Deposited 2 May 2023. [Google Scholar]
- 74.Cao Y., Gebauer J. M., Baumann U., Chai J.. Crystal structure of powdery mildews Blumeria graminis f. sp. hordei AVRA22. Protein Data Bank. https://www.rcsb.org/structure/8OXJ. Deposited 2 May 2023. [Google Scholar]
- 75.Cao Y., Gebauer J. M., Baumann U., Chai J.. Crystal structure of powdery mildews Blumeria graminis f. sp. tritici AVRPM2(#1). Protein Data Bank. https://www.rcsb.org/structure/8OXI. Deposited 2 May 2023. [Google Scholar]
- 76.Cao Y., Gebauer J. M., Baumann U., Chai J.. Crystal structure of powdery mildews Blumeria graminis f. sp. tritici AVRPM2(#2). Protein Data Bank. https://www.rcsb.org/structure/8PHY. Deposited 20 June 2023. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (PDF)
Data Availability Statement
All study data are included in the article and/or supporting information. Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 8OXH (AVRA6) (71), 8OXL (AVRA7) (72), 8OXK (AVRA10) (73), 8OXJ (AVRA22) (74), 8OXI (AVRPM2#1) (75), and 8PHY (AVRPM2 #2) (76)].