Abstract
Histone H2A lysine 119 (H2AK119Ub) is monoubiquitinated by Polycomb repressive complex 1 and deubiquitinated by Polycomb repressive deubiquitinase complex (PR-DUB). PR-DUB cleaves H2AK119Ub to restrict focal H2AK119Ub at Polycomb target sites and to protect active genes from aberrant silencing. The PR-DUB subunits (BAP1 and ASXL1) are among the most frequently mutated epigenetic factors in human cancers. How PR-DUB establishes specificity for H2AK119Ub over other nucleosomal ubiquitination sites and how disease-associated mutations of the enzyme affect activity are unclear. Here, we determine a cryo-EM structure of human BAP1 and the ASXL1 DEUBAD in complex with a H2AK119Ub nucleosome. Our structural, biochemical, and cellular data reveal the molecular interactions of BAP1 and ASXL1 with histones and DNA that are critical for restructuring the nucleosome and thus establishing specificity for H2AK119Ub. These results further provide a molecular explanation for how >50 mutations in BAP1 and ASXL1 found in cancer can dysregulate H2AK119Ub deubiquitination, providing insight into understanding cancer etiology.
The structure of PR-DUB on H2AK119Ub nucleosome sheds light on specificity, activity and several cancer-associated mutations.
INTRODUCTION
Breast cancer type 1 susceptibility protein (BRCA1)–associated protein 1 (BAP1) is a conserved ubiquitin (Ub) C-terminal hydrolase responsible for deubiquitinating monoubiquitinated H2AK119 (H2AK119Ub), a histone posttranslational modification (PTM) associated with gene silencing (1, 2). BAP1 deubiquitinase (DUB) activity requires the deubiquitinase adaptor domain (DEUBAD) of additional sex comb–like 1, 2, or 3 (ASXL1, ASXL2, or ASXL3) (3, 4). In mammals, Polycomb repressive deubiquitinase complex (PR-DUB) activates gene expression by deubiquitinating H2AK119Ub and safeguards Polycomb-mediated repression by removing misplaced H2AK119Ub (5–8), and, in Drosophila, the activity of PR-DUB was shown to regulate Polycomb repressive complex 1 (PRC1)–mediated chromatin compaction and gene silencing (9). Inactivating the catalytic activity of BAP1 abrogates most of its nuclear function, suggesting deubiquitination is central to its role (5–8). The Drosophila homolog Calypso was identified by genetic screening and characterized as PR-DUB because it opposes H2A ubiquitination by PRC1 (2, 10). Besides regulating Polycomb repression, PR-DUB activates enhancers and promotes DNA damage repair (DDR) at double-strand breaks (11–13). Establishing its importance to human health, BAP1/ASXL1 is frequently implicated in various cancers (e.g., uveal melanoma and malignant mesothelioma) and developmental disorders (e.g., Bohring-Opitz syndrome and myelodysplasia) (14–19). Furthermore, mutations in BAP1 are categorized as “BAP1 tumor predisposition syndrome,” characterized by an increased risk of early onset fast-growing and metastatic cancers (1, 11).
Despite its central importance in regulating Polycomb repression, gene activation, and DDR, the primary mechanism by which the PR-DUB specifically deubiquitinates chromatin remains unanswered. While biochemical studies showed that PR-DUB needs nucleosome interaction(s) to be specific for H2AK119Ub and recent crystal structures characterized the architecture and intramolecular interactions in the apo Drosophila Calypso/ASX complex, they did not provide a detailed view of how the complex engages chromatin (3, 20, 21). Furthermore, these studies have not revealed what determines PR-DUB specificity for H2AK119Ub chromatin and how cancer-associated mutations in BAP1 and ASXL1 affect its catalytic activity, thereby providing any insight to disease etiology.
Here, we used a combination of structural, biochemical, and functional approaches to determine the mechanism of nucleosome deubiquitination by human BAP1/ASXL1, one of the most mutated epigenetic complexes among human cancers. We have solved a cryo–electron microscopy (cryo-EM) structure of human BAP1 bound to the ASXL1 DEUBAD in complex with a H2AK119Ub nucleosome. BAP1/ASXL1 forms several anchor points on a nucleosome surface and binds H2AK119Ub in a conformation resembling the Ubiquitin carboxyl-terminal hydrolase 5 (UCH-L5)/ Proteasomal regulatory particle base subunit 13 (RPN13) deubiquitinase (22). Specifically, BAP1 interacts with the nucleosome acidic patch (using regions absent in UCH-L5), while ASXL1 interacts with DNA near the nucleosome DNA exit, and both PR-DUB subunits form a DNA clamp that engages near the nucleosome dyad. All BAP1/ASXL1 anchor points are mediated through conserved lysine/arginine tracts, which, when mutated, compromise PR-DUB activity in vitro and in mouse embryonic stem cells. The position of BAP1/ASXL1 near the dyad interferes with a canonical path of the nucleosomal H2A docking domain, explaining why this portion of histone H2A is disordered in our structure. On the basis of on our results, we propose how the interactions of the PR-DUB complex determine its specificity for H2AK119Ub nucleosomes. Last, our structural, biochemical, and functional data explain the mechanism of >50 distinct mutations found in cancer, spanning BAP1 and ASXL1 surfaces.
RESULTS
The architecture of the BAP1/ASXL1 complex bound to H2AK119Ub nucleosome
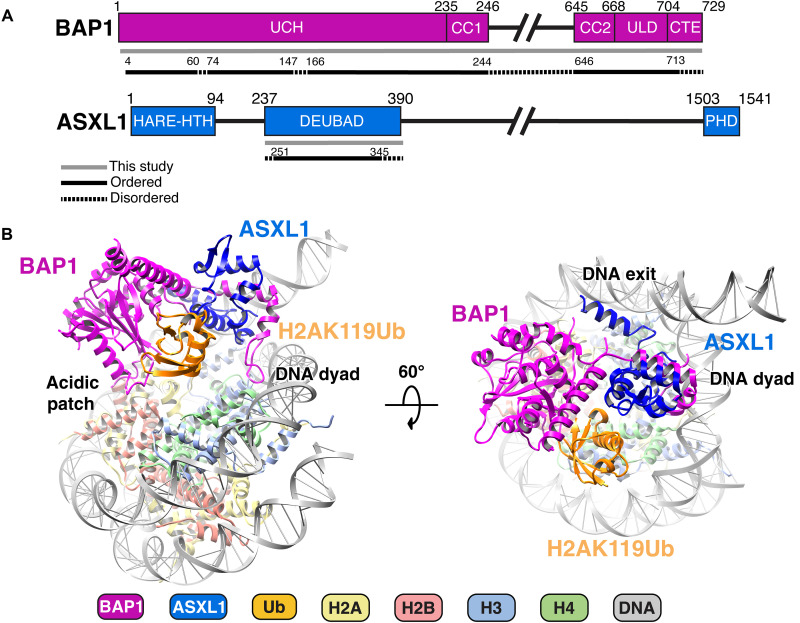
To gain insights into the BAP1/ASXL1 deubiquitination mechanism, we reconstituted full-length human BAP1 (residues 1 to 729) in complex with the DEUBAD of ASXL1 (residues 237 to 390) (Fig. 1A and fig. S1) bound to a nucleosome in which Ub residue G76C was conjugated to histone H2AK119C via a nonhydrolyzable dichloroacetate linkage (hereafter H2AK119Ub) (23). We cross-linked the resulting complex with glutaraldehyde by gradient fixation (GraFix) (fig. S2) (24), froze grids, and collected cryo-EM data on a Titan Krios (300 kV) (table S1). Using these data, we obtained a 3.6-Å-resolution cryo-EM map (figs. S3 and S4 and table S2), which we used to unambiguously model the structures of BAP1 (residues 4 to 60, 74 to 147, 166 to 244, and 646 to 713), ASXL1 (residues 251 to 345), and Ub on the nucleosome (Fig. 1B and fig. S5). The cryo-EM map revealed BAP1 bound to the nucleosome in a catalytic conformation, with clearly resolved BAP1-nucleosome, ASXL1-nucleosome, BAP1-Ub, and ASXL1-Ub interfaces (Fig. 1B and fig. S5). We used a combination of AlphaFold-Multimer prediction (25) and homologous modeling with Drosophila Calypso/ASX crystal structures [Protein Data Bank (PDB) IDs 6hgc and 6cga] (20, 21) to build the model (Fig. 1B).
Fig. 1. Overview of the structure of BAP1/ASXL1 bound to H2AK119Ub nucleosome.
(A) Bar diagram representation of BAP1 and ASXL1 domains. Protein sequences included in this study are in gray line; those resolved in the structure are in black line; and disordered regions are in dashed line. (B) Two different views of the model for the BAP1/ASXL1-H2AK119Ub nucleosome complex with key anchor points highlighted. The figure is color-coded, depicting BAP1 (purple), ASXL1 (dark blue), Ub (orange), H2A (yellow), H2B (salmon), H3 (light blue), H4 (green), and DNA (gray).
Our structure revealed one molecule each of BAP1 and ASXL1 bound on one side of the H2AK119Ub nucleosome, with four main points of contact: BAP1 with the acidic patch, DNA, and Ub, while ASXL1 interfaces with DNA and Ub (Fig. 1B and fig. S5). These multiple contacts between BAP1/ASXL1 and nucleosome provide a rationale for PR-DUB complex specificity toward H2AK119Ub over other H2A or H2B ubiquitinations (e.g., H2AK15Ub or H2BK120Ub) (fig. S6).
The interface of BAP1/ASXL1 with Ub is conserved with other deubiquitinases
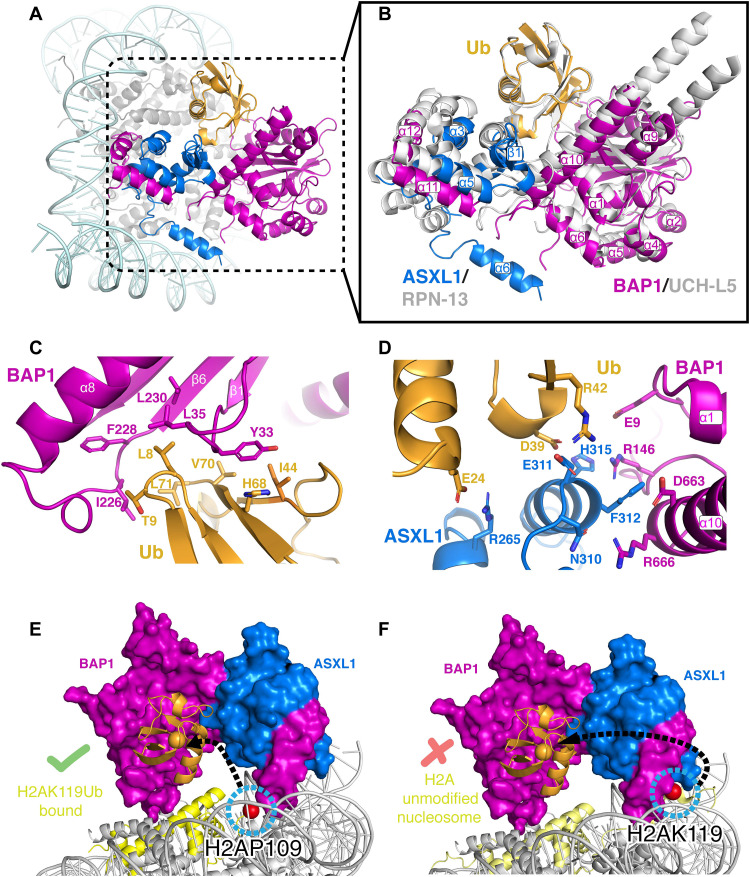
Our structure captures Ub in complex with BAP1/ASXL1 on the nucleosome. We used our cryo-EM map and AlphaFold-Multimer prediction to fit the Ub structure (Fig. 2A). Notably, the interfaces between BAP1/ASXL1 and Ub are conserved with UCH-L5/RPN13 (PDB ID 4uel) (Fig. 2, A and B, and figs. S1 and S7) (22). In our structure, Ub is sandwiched between the BAP1 UCH and ASXL1 DEUBADs (Fig. 2B), where BAP1 uses loops Lβ1-β2 and Lα8-β6 to interact with the Ub canonical “I44 patch” (Fig. 2C) (26). From our structure and comparative analyses, we can propose side-chain interactions at the Ub interface (Fig. 2B). BAP1 residues Y33, L35, L230, F228, and I226 are within distance to establish van der Waals interactions with Ub residues I44, H68, V70, L8, L71, and T9 (Fig. 2C). These residues at the BAP1/ASXL1:Ub interface correspond to those contacting Ub in UCH-L5 and RPN13 (Fig. 2, C and D, and figs. S1 and S7) (22) and also predicted to do so in Drosophila PR-DUB (20, 21). BAP1/ASXL1 also stabilizes Ub by electrostatic interactions on the other face of the I44 patch. In this interface, ASXL1 residues R265, H315, and E311 and BAP1 residue E9 are within the distance to establish interactions with Ub residues E24, D39, and R42 (Fig. 2D). This contrasts with other chromatin modifiers that interact primarily with Ub via one of its hydrophobic patches (I44 patch and “I36 patch”) (26–28). The BAP1/ASXL1-Ub electrostatic interface is stabilized by potential interactions between BAP1 residues R146 (loop Lα6-α7), R663, and D666 (helix α10) and ASXL1 “NEF motif” residues H315, F312, and N310 (Fig. 2D and figs. S1 and S7). These observations explain why mutations to the NEF motif disrupt catalytic activity by destabilizing Ub binding (fig. S1) (3, 21, 29).
Fig. 2. Insights into the mechanism of catalysis by BAP1/ASXL1 and conservation with other deubiquitinases.
(A) Position of Ub engaged with BAP1 and ASXL1 on the nucleosome. (B) BAP1/ASXL1/Ub aligned and superimposed with UCH-L5/RPN13/Ub (PDB ID 4uel; in gray) (22). (C) Hydrophobic interactions between BAP1 and Ub. (D) Electrostatic interactions between BAP1/ASXL1 and Ub. (E) Space-filling representation of our cryo-EM structure with the last observed residue of H2A (H2AP109; blue-circled red sphere). (F) A model of BAP1/ASXL1 from our cryo-EM structure superposed with wild-type (WT; unmodified) nucleosome (PDB ID 1kx5) (30), where the structured H2A docking domain with the unmodified H2AK119 is shown as blue-circled red sphere.
Remodeling of the H2A docking domain allows Ub to reach the catalytic site of BAP1
We next asked how H2AK119Ub reaches the catalytic site of BAP1/ASXL1 complex. In our cryo-EM structure, the last ordered residue in the H2A C-terminal tail is P109 (Fig. 2E). The last visible residue of covalently installed Ub, L71, is 37 Å away from P109 and sandwiched below BAP1/ASXL1, which forms an arc-like structure on the nucleosome surface (Fig. 2E). The remainder of the H2A C-terminal tail, up to residue H2AK119 (connected to G76 of Ub via an isopeptide bond and, in our structure, via a dichloroacetone cross-link between C119 and C76), must be directed to the BAP1 active site for catalysis. This conformation of H2A contrasts with the usual presence of a folded canonical docking domain of H2A in the unoccupied nucleosome (PDB IDs 1kx5 and 6wkr) (30, 31) bound near the DNA dyad and ordered up to residue K128 (Fig. 2F). This suggests that the H2A docking domain must unfold for H2AK119Ub to reach the catalytic site of BAP1/ASXL1. The distance Ub must relocate to reach the BAP1/ASXL1 catalytic site may explain specificity for H2AK119Ub versus H2AK13/15Ub or H2BK120Ub (fig. S6), since the H2A/H2B histone fold would need to be unwrapped for these to reach the deubiquitinase active site (fig. S6). This also reveals why previous studies using a minimal or peptide substrates were unable to explain the specificity for nucleosomal H2AK119Ub (3).
BAP1/ASXL1 clamps the dyad of the nucleosome
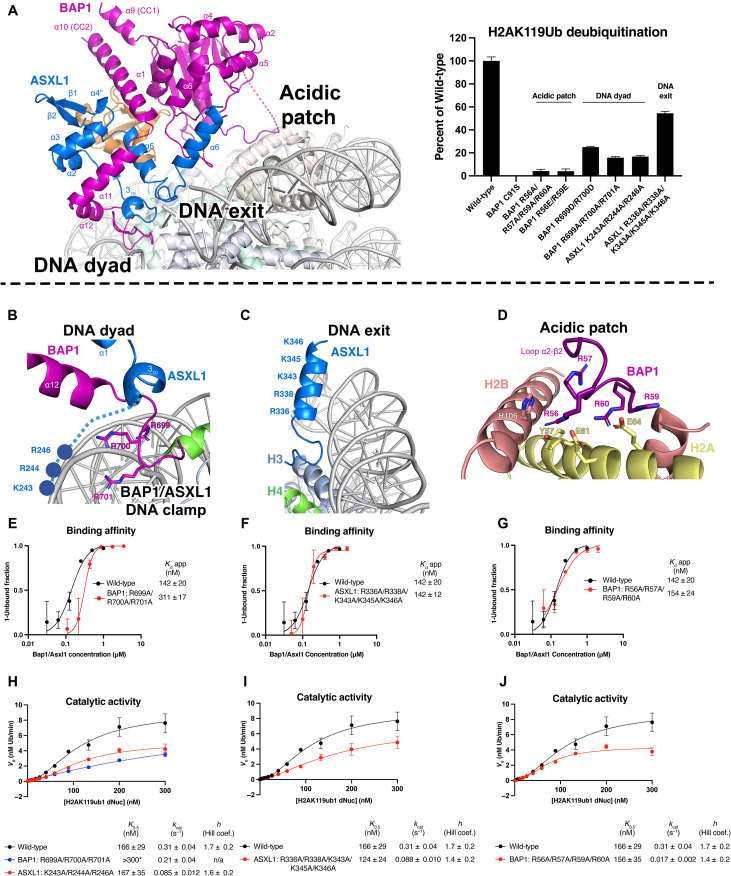
In our structure, BAP1/ASXL1 complex anchors onto the nucleosome surface at three contact points: near the dyad DNA, exit DNA, and the acidic patch. BAP1 residues 672 to 697 and ASXL1 residues 258 to 287/311 to 321 form a helix bundle (Fig. 3A and fig. S1), with ASXL1 helixes αN-310, α1, α2, α3, and α5, folding and wrapping around BAP1 helixes α11 and α12 (Fig. 3A). This helix bundle not only participates in forming the Ub interface but also interacts with nucleosomal DNA near the dyad (Figs. 1B and 3A). The folding of ASXL1 around this part of the BAP1 Ubiquitin C-terminal hydrolase like domain (ULD) region explains previous reports showing that deletion of residues after BAP1 670 impairs complex formation with ASXL1 (3, 22). Adjacent to the α12 helix of BAP1, we were able to trace the backbone of BAP1 up to residue 713, where R699, R700, and R701 approach the minor groove near the DNA dyad (Fig. 3B). These three positively charged, well-ordered residues are within the distance to form electrostatic interactions with the DNA phosphates (fig. S8) and are followed by a C-terminal extension (CTE) region of BAP1 that is needed to recruit PR-DUB to the nucleosomal DNA (3, 20, 21). We further explored the roles of R699, R700, and R701 in BAP1 by mutating them to alanines or reversing their charge (fig. S9). We assembled this mutated BAP1 with ASXL1 and tested the enzymatic activity of the resulting complex by DUB assays with a H2AK119Ub designer nucleosome (dNuc) substrate, observing a substantial reduction relative to wild type (WT) (~84%; Fig. 3A, right, and fig. S9). When we tested nucleosome binding by electromobility shift assay (EMSA), these BAP1 mutations also decreased the apparent affinity relative to WT (Fig. 3E and figs. S10 and S11).
Fig. 3. BAP1/ASXL1 interacts with DNA and acidic patch on the nucleosome.
(A) Left: Overall architecture of the BAP1/ASXL1-H2AK119Ub nucleosome complex with BAP1/ASXL1 anchor points marked. Right: Catalytic activity assays on H2AK119Ub nucleosomes (containing a DUB-cleavable native gamma-lysine isopeptide linkage) and various forms of BAP1/ASXL1 enzyme: WT, catalytically inactive (BAP1 C91S), or mutated nucleosomal anchor points (as labeled). (B) Close-up view of BAP1/ASXL1 DNA clamp contacting the nucleosome near the DNA dyad. (C) Close-up view of the ASXL1 DEUBAD α6 helix, projecting a stretch of basic residues toward a nucleosome DNA exit. (D) Close-up view of the BAP1 R-finger interacting with the acidic patch. (E to G) Quantified electromobility shift assays (EMSAs) of WT heterodimer complex versus mutants of the BAP1 DNA clamp (E), ASXL1 DNA exit (F), and BAP1 interaction with the acidic patch (G). Each data point and error bar indicate the means ± SD from three independent experiments. The SEs of dissociation constant (Kd) are indicated. (H to J) Representative kinetic curves of WT/mutant BAP1/ASXL1 enzymes with H2AK119Ub nucleosomes. Compared relative to WT are mutants of the BAP1 DNA clamp (H), ASXL1 DNA exit (I), and BAP1 interaction with the acidic patch (J). tUI-free Ub sensor was used to monitor the activity of WT BAP1/ASXL1 and its mutants (H to J). A free Ub standard curve was used to calculate the amount of enzymatically generated Ub, with initial velocities (V0) relative to H2AK119ub1 concentration used to determine H2AK119ub1 dNuc K0.5, kcat, and Hill coefficient values by applying an allosteric sigmoidal model. Kinetic constants are represented as ± SD from three independent experiments. Asterisk (*) corresponds to Michaelis-Menten fit instead of allosteric regulation fit. n/a, not applicable.
Immediately before the αN-310 helix, the ASXL1 N-terminal tail also contains a patch of basic residues (K243, R244, and R246) that we hypothesized can form a clamp together with the basic residues of BAP1 near the helix bundle (Fig. 3, A and B, and fig. S1). We mutated these three residues in ASXL1 to alanines, assembled a complex with BAP1, and again observed a substantial loss of DUB activity (~83%) toward H2AK119Ub nucleosomes (Fig. 3A, right). We termed this region the “DNA clamp” because the basic residues of BAP1 (R699, R700, and R701) and ASXL1 (K243, R244, and R246) provide extensive interactions with the double helix near the DNA dyad (Fig. 3B). To further explore enzymatic mechanism, we performed kinetic assays using WT BAP1/ASXL1 and DNA clamp mutants (BAP1 R699A, R700A, and R701A; ASXL1 K243A, R244A, and R246A). The BAP1 mutant complex had compromised catalytic activity where K0.5 (substrate concentration at half-maximal velocity) was increased (>300 nM versus 166 nM WT) and kcat was slightly reduced (0.21 versus 0.31 s−1 WT) (Fig. 3H and table S3). In contrast, the ASXL1 mutant complex showed no effect on binding affinity but reduced kcat (turnover number) (0.085 s−1 versus 0.31 s−1 WT) (Fig. 3H and figs. S10 and S11). The observation that mutations of BAP1 or ASXL1 residues in the DNA clamp have a substantial impact on catalytic activity supports the importance of this region in the H2AK119Ub deubiquitination mechanism (Fig. 3A).
ASXL1 interacts with the DNA at the exit of the nucleosome
Our cryo-EM map showed an extra density emerging from the helix bundle and DNA clamp and approaching the DNA exit. During cryo-EM data processing, this point of contact was only seen when BAP1/ASXL1 was in the catalytic conformation and Ub was stabilized (figs. S4 and S12). The rigid body fit of this region allowed us to assign it to the ASXL1 helix α6 (Fig. 3C), corresponding to Drosophila ASX α6 (PDB ID 6hgc) (20). Even though the map in this region does not allow to assign side-chain contacts, this region of ASXL1 contains a stretch of basic residues (R336, R338, K343, K345, and K346), which we hypothesized may interact with the DNA backbone (Fig. 3C). We mutated these basic residues in ASXL1 (R336, R338, K343, K345, and K346) to alanines, assembled with WT BAP1, and tested DUB activity of the resulting complex. Relative to WT, we observed a moderate reduction of catalytic activity (~46%) (Fig. 3A, right) with no change in the binding affinity (Fig. 3F). Examination of kinetic constants demonstrates the catalytic importance of the ASXL1 DNA exit interface, where swapping the ASXL1 basic residues to alanines had no impact on K0.5 but reduced kcat (0.088 s−1 versus 0.31 s−1 WT) (Fig. 3I).
BAP1 binds the acidic patch with an arginine finger
Another stable anchor between BAP1/ASXL1 and the nucleosome in our cryo-EM map is with the acidic patch, as evidenced by better resolution of this interface (figs. S8 and S13). Here, we were able to assign the side chains of the BAP1 loop Lβ2-α2 residues (R56, R57, R59, and R60) (Fig. 3D and figs. S1 and S8), which formed electrostatic interactions with the acidic patch residues E61, E64, and Y57 of H2A and H106 of H2B (Fig. 3D). This R-finger is the classical interface between chromatin modifiers and the nucleosomal H2A-H2B dimer (27, 32–37). These four arginines are conserved between mammalian BAP1 and Drosophila Calypso, but not in UCH-L5, another DUB that does not deubiquitinate a nucleosome substrate (fig. S1). We tested the functional significance of these arginines (R56, R57, R59, and R60) by mutating them to alanines or glutamic acid, assembling with ASXL1, and performing DUB or EMSA assays with H2AK119Ub nucleosomes. Relative to WT, these BAP1/ASXL1 mutants showed almost a complete loss (~96%) of catalytic activity (Fig. 3A, right) but no change to binding affinity (154 nM versus 142 nM) (Fig. 3G). Determination of the kinetic constants of the BAP1 loop Lβ2-α2 mutant complex revealed a substantial reduction in turnover number (kcat = 0.017 versus 0.31 s−1 WT) and no change of K0.5 (Fig. 3J and table S3). These results show that the binding affinity is not perturbed when disrupting the interaction with the acidic patch, in agreement with a previous report (3). However, we demonstrated that this interface is critical for PR-DUB catalytic activity, closely mirroring the role of arginine finger–acidic patch interactions in the activity of other chromatin enzymes (27, 32, 33, 38).
In conclusion, all anchor points contributed to H2AK119-directed deubiquitination activity, and mutating each interface disrupted catalysis (Fig. 3A, right). We further validated the importance of PR-DUB–nucleosome interfaces by comparing deubiquitination activity on H2AK119Ub nucleosome and peptide substrates. While a catalytic site mutation (BAP1 C91S) abolished DUB activity on both substrates, mutation of PR-DUB—nucleosome interfaces selectively compromised nucleosome deubiquitination (fig. S6C), demonstrating the insight that only a physiological target can provide. Our cryo-EM structure suggests a mechanistic model where BAP1/ASXL1 deubiquitinates chromatin by anchoring to the nucleosomal acidic patch and DNA at the dyad and exit. Complementary in vitro assays (binding affinity, catalytic activity, and kinetics) indicate that these anchor points contribute to enzyme:substrate engagement and catalysis.
BAP1/ASXL1-nucleosome interactions are required for H2AK119Ub deubiquitination in mESCs
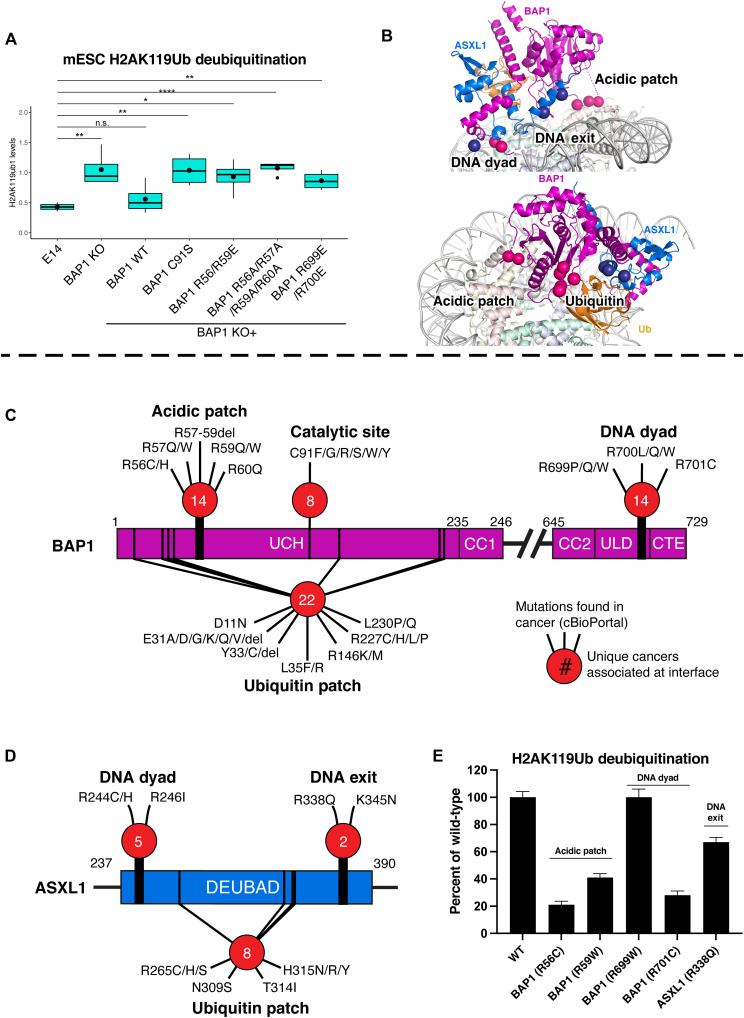
We next sought to examine the in vivo functional importance of the molecular contacts identified in our structural and biochemical analyses. To this end, we stably reexpressed BAP1 R699E/R700E (DNA dyad), BAP1 R56E/R59E, and BAP1 R56A/R57A/R59A/R60A (acidic patch) in BAP1 knockout (KO) embryonic stem cell (ESC) in parallel to a WT or BAP1 catalytic inactive form (C91S). All BAP1 mutants expressed at comparable levels to the BAP1 WT counterpart (Fig. 4A). As we previously reported, the reexpression of WT BAP1 in KO ESC efficiently restored physiological H2AK119Ub levels (6). However, consistent with our in vitro data, the expression of tested BAP1 mutations failed to restore physiological H2AK119Ub levels, similar to catalytically inactive BAP1 (Fig. 4A). Together, these results confirm that the critical residues involved in BAP1/ASXL1 H2AK119Ub nucleosome recognition (Fig. 4B) are required for efficient deubiquitination in vivo.
Fig. 4. BAP1/ASXL1-nucleosome contacts are extensively mutated in cancers.
(A) Western blot analysis of H2AK119Ub levels in total protein extracts from BAP1 WT (E14) or KO mESCs, or KO after reintroduction/stable expression of BAP1 WT and various mutations (C91S, R699E/R700E; R56E/R59E; R56A/R57A/R59A/R60A). t tests were performed as indicated [non significant (n.s.), P > 0.05; *P < 0.05; **P < 0.01; ****P < 0.0001]. (B) Structure of the BAP1/ASXL1-H2AK119Ub nucleosome complex highlighting the three key nucleosome anchors (top) and Ub interaction region (bottom), and relative position of cancer mutations (colored spheres) clustered in these areas. (C and D) Bar diagrams showing cancer-associated point mutations/deletions of BAP1 (C) and ASXL1 (D) at substrate interaction surfaces (B) that can be mechanistically explained by this study. The number of unique cancer types at each interface is inside the red circles. (E) Catalytic activity assays on H2AK119Ub nucleosomes with various forms of BAP1/ASXL1 enzyme: WT or with point mutations found in cancer (from cBioPortal).
Cancer mutations decorate BAP1/ASXL1-nucleosome interfaces
BAP1/ASXL1 is one of the most frequently mutated chromatin-modifying enzymes in cancer, with its disease relevance further highlighted by BAP1 mutations being classified as BAP1 tumor predisposition syndrome (39). BAP1 and ASXL1 cancer-associated mutations are distributed across the entire polypeptide chains, with most driver mutations being truncations and >99% of variants of uncertain significance (VUS) being missense mutations. Most truncation mutations that disrupt function are easily explained by the loss of entire domains or nuclear localization signals (1). In contrast, gain-of-function truncations of the ASXL1 central region may stabilize BAP1 on chromatin and reduce its degradation (40, 41). However, the abundant VUS missense mutations often remain mechanistically unexplained, especially when a protein structure is unavailable (figs. S14 and S15).
The Drosophila PR-DUB crystal structures gave context for those cancer-associated mutations in BAP1-ASXL1 that affect intramolecular contacts within the heterodimer and with Ub but gave no insight to interactions of the deubiquitinase with a H2AK119Ub nucleosome substrate (20, 21). However, our structure provides context for a range of VUS missense mutations that might define deubiquitination activity on nucleosomes (tables S4 and S5). We searched cBioPortal (www.cbioportal.org) and used a curated set of nonredundant studies to both tabulate a list of all cancer mutations across BAP1 and ASXL1 (tables S4 and S5) and annotate the subset of frequent mutations at the interfaces with Ub and the DNA dyad, DNA exit, and acidic patch (Fig. 4B). On BAP1, deletion or nonconservative mutations of the acidic patch–interacting residues R56 to R60 or the DNA dyad interface residues R699 to R701 are each found in 14 different cancers (Fig. 4C). For ASXL1, mutations of the DNA dyad residues R244 and R246 are found in five different cancers (Fig. 4D), while the DNA exit residues R338 and K345 are found in colorectal adenocarcinoma and cholangiocarcinoma (Fig. 4D).
We selected a range of single-point cancer-associated mutations at the PR-DUB–nucleosome interfaces (Fig. 4, C and D) and tested their impact on deubiquitination activity. Mutations to the acidic patch interaction, BAP1 R56C or R59W, reduce deubiquitination activity to 23 and 43% of WT PR-DUB, respectively (Fig. 4E). At the DNA dyad interface, mutating BAP1 R699W had no observable effect, but mutating BAP1 R701C reduced deubiquitination activity to 30% of WT (Fig. 4E), further demonstrating the importance of BAP1 R701 at the DNA minor groove (Fig. 3, B, E, and H, and fig. S8). At the DNA exit interface, mutating ASXL1 R338Q (Fig. 4E) reduced deubiquitination activity to 72% of WT (Fig. 4E).
Together, our structure explains 5% of BAP1 and 1% of ASXL1 VUS missense mutations at key nucleosome interfaces. As such, we can provide key insights into the potential deregulation of BAP1/ASXL1 in 35 cancers ranging from adenoid cystic carcinoma to uveal melanoma and suggest those that may be due, at least in part, to the aberrant accumulation of H2AK119Ub.
DISCUSSION
Our data support a model where BAP1/ASXL1 deubiquitinates H2AK119Ub chromatin by anchoring to the nucleosomal acidic patch, DNA dyad, and the DNA exit regions using conserved arginine and lysine tracts. Fitting the prevailing model for chromatin-modifying proteins (33, 42), the ability of BAP1 to fine-tune deubiquitination activity requires interactions with the nucleosome acidic patch. Previous reports showed that ASXL1 increases the affinity of PR-DUB for Ub and revealed the role of BAP1 CTE region in enabling PR-DUB binding to nucleosomal DNA (3, 20). Here, we show the molecular interactions of BAP1/ASXL1 that direct the PR-DUB complex to the nucleosome and enzymatically characterize these interfaces using a defined H2AK119Ub nucleosome substrate. We describe a previously undefined functional element, the BAP1/ASXL1 DNA clamp, that contains a region of the helix bundle of ASXL1 folded around BAP1 and connecting to its CTE. We show that BAP1 residues that form the clamp are critical for efficient deubiquitination of nucleosome substrate by directing the PR-DUB to its catalytic position on the DNA dyad and clarify the importance of ASXL1 for DUB activity (3). Our cryo-EM structure also provides a rationale as to how PR-DUB mediates substrate specificity for nucleosomal H2AK119Ub over H2AK13/K15Ub and H2BK120Ub. In the latter cases the ubiquitylated residue cannot reach the BAP1/ASXL1 active site without extensive restructuring of the H2A/H2B dimer (Fig. 2, E and F, and fig. S6). We note a functional redundancy in deubiquitination activity between ASXL1, ASXL2, and ASXL3 (3). Our structure reveals that the residues in ASXL1 involved in nucleosome binding are conserved in ASXL2 and ASXL3, explaining their functional redundancy in the PR-DUB (fig. S1). Last, our structure explains VUS cancer mutations that may disrupt deubiquitination activity by abrogating the interaction between BAP1/ASXL1 and substrate nucleosomes.
We showed that residues R699 to R701 of the BAP1 DNA clamp form important interactions with the DNA dyad and lead to the CTE, which extends toward the DNA exit. The deletion of residues of the CTE of BAP1 after H710 has been reported to reduce the recruitment of PR-DUB to endogenous nucleosomes (3, 20, 21). We hypothesize that nonspecific interactions of the positively charged CTE with negatively charged DNA have two important roles. (i) They participate in the proper folding/positioning of the DNA clamp near the DNA dyad to allow PR-DUB to adopt an active conformation. (ii) In a similar fashion as the disordered basic residues of the N-terminal portion of ASXL1, the CTE of BAP1 would help recruit the BAP1/ASXL1 clamp to the DNA. The strong recruitment to nucleosomal DNA via a tract of basic residues and establishing nonspecific interactions may be a common feature of chromatin modifiers (28, 32, 38, 43).
In this study, we primarily discuss a cryo-EM map characterized by a stable binding of the three anchors of BAP1/ASXL1 with the nucleosome acidic patch, DNA clamp, and DNA exit regions. The particles used to calculate this map also contain the Ub stabilized in its catalytic conformation (figs. S4 and S12). This map showed “the best” defined interfaces, which we interpret as the most stable conformation of BAP1/ASXL1 on the H2AK119Ub nucleosome. In addition, during data processing, we observed three-dimensional (3D) classes with alternative conformations of BAP1/ASXL1 on nucleosomes (figs. S4 and S12). In the 3D classification analysis, some of the classes presented higher flexibility in the domains of BAP1/ASXL1 (as evidenced by lower resolution maps) and did not show Ub stabilization in its catalytic conformation (figs. S4 and S12). Thirty-five percent of the particles showed the BAP1/ASXL1 DNA clamp/CTE binding to the DNA exit instead of the dyad (alternative conformation). On the basis of our 3D classification analyses, we propose that, when the PR-DUB is recruited to chromatin, it samples these alternative conformations to find the catalytic position, which is established by pivoting on the nucleosome acidic patch to position the clamp on the dyad.
The BAP1/ASXL1 DNA clamp on the dyad, H2A docking domain in the canonical nucleosome conformation, and histone H1.4 all occupy almost the same location (44, 45). Thus, the BAP1/ASXL1 DNA clamp could be expected to clash with H1.4 bound at the same side of the DNA linker (fig. S12). Notably, BAP1/ASXL1 activity is slightly lower on H1.4 chromatosomes than on nucleosomes (fig. S12), consistent with results published during revision of this study (46). Linker histone H1.4 binds asymmetrically to the linker DNA depending on nucleosome stacking in chromatin (PDB ID 7pfv) (44). We hypothesize that H1.4 binding in this mode may require BAP1/ASXL1 to engage in the alternative conformation at the DNA exit instead of the DNA dyad. In the corollary, this could explain the H1 enrichment observed on chromatin after BAP1/ASXL1 deletion or H2AK119 deubiquitination (6, 47).
Similar to other chromatin complexes, the conformational flexibility of BAP1/ASXL1 on the nucleosome could be subjected to regulation. Both Dot1 and COMPASS are stabilized in their catalytic conformation on a nucleosome by interactions with PTMs such as ubiquitination or acetylation (27, 28). Histone modifications, variants, linker histones, and cofactors could play stabilizing/regulatory roles toward BAP1/ASXL1 and should be the focus of subsequent studies.
MATERIALS AND METHODS
Cloning and mutation of BAP1 and ASXL1
Full-length human BAP1 and the DEUBAD of ASXL1 (amino acids 237 to 390) were cloned into a pFastBac Dual vector by GenScript. ASXL1 was subcloned into pET24a vector for Escherichia coli expression. BAP1 was modified to express alone in SF9 from the pFastBac Dual vector. Mutants were generated using the New England Biolabs (NEB) Q5 Site-Directed Mutagenesis Kit.
Protein expression and purification of BAP1 and ASXL1
BAP1/ASXL1 and BAP1 were expressed in SF9 insect cells (clonal isolate from PLB-Sf-21-AE1) following the Bac-To-Bac Baculovirus Expression System–established protocol (Invitrogen). ASXL1 was expressed in E. coli BL21-CodonPlus (DE3)-RIL by Isopropyl β-D-1-thiogalactopyranoside (IPTG) induction following the established protocols (20).
BAP1/ASXL1 complex and BAP1 alone were purified as previously (29) with minor modifications. First, cells were suspended in resuspension buffer A [20 mM Hepes (pH 8), 150 mM NaCl, 5% glycerol, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP), and Roche EDTA-free protease inhibitor] and lysed using an emulsifier (AvestinEmulsiflexC3). Lysate was precleared by spinning at 7500 relative centrifugal force (RCF) for 20 min. BAP1 was purified by Strep-Tactin XT affinity chromatography resin (IBA Lifesciences). The lysate was incubated with Strep-Tactin XT resin, washed with resuspension buffer A, and sequentially washed with buffer A supplemented with 1 M NaCl and 2 M NaCl. BAP1 was cleaved from resin by incubating overnight with 600 μg of 3C protease. Elution fractions were dialyzed overnight in buffer A [20 mM Hepes (pH 8), 150 mM NaCl, 5% glycerol, and 0.5 mM TCEP], then passed through a HiTrap Q HP 5-ml column, and eluted by an increasing gradient of buffer B [20 mM Hepes (pH 8), 1 M NaCl, 5% glycerol, and 0.5 mM TCEP]. Last, BAP1 or BAP1/ASXL1 was further purified on gel filtration column [HiLoad 16/60 Superdex 200 or Superose 6 Increase 10/300 GL (Cytiva)] using gel filtration buffer C [10 mM Hepes (pH 7.5), 150 mM NaCl, 10% glycerol, and 0.5 mM TCEP]. BAP1 or BAP1/ASXL1 fractions were pooled, concentrated, and snap-frozen.
ASXL1 was purified as previously (20) with some modifications. ASXL1 was expressed in E. coli BL21 (DE3) and collected by centrifugation at 7500 RCF for 15 min and then suspended in lysis buffer D [20 mM Hepes (pH 8), 200 mM NaCl, 5 mM imidazole, 0.5 mM imidazole, and 1× Roche EDTA-free protease inhibitor]. Cells were lysed using an emulsifier (AvestinEmulsiflexC3), and the lysate was cleared by spinning at 7500 RCF for 20 min. ASXL1 was purified by using Ni–nitrilotriacetic acid (NTA) resin (Qiagen). The resin was washed with lysis buffer D and sequentially washed with buffer D supplemented with 1 M NaCl and returned to buffer D. ASXL1 was eluted by cleaving from resin with 3C protease. Elution fractions were concentrated using a 10-KDa cutoff Centricon concentrator and passed over a HiLoad Superdex 75 16/60 pg (Cytiva) using gel filtration buffer C [10 mM Hepes (pH 7.5), 150 mM NaCl, 10% glycerol, and 0.5 mM TCEP]. ASXL1 containing fractions were pooled, concentrated, and snap-frozen.
BAP1/ASXL1 complex assembly
Purified BAP1 was coincubated with an excess of ASXL1 for 1 hour at 4°C before injecting into the Superose 6 Increase 10/300 GL (Cytiva) using gel filtration buffer C [10 mM Hepes (pH 7.5), 150 mM NaCl, 10% glycerol, and 0.5 mM TCEP]. Peak fractions containing BAP1/ASXL1 were pooled, concentrated, and snap-frozen.
Expression and purification of Ub
pET-His-Ub G76C was a generous gift from T. Yao (23). Ub plasmid DNA was transformed into E. coli SoluBL21 (Amsbio, catalog no. C700200) competent cells and grown in 2xYT-Amp medium. Ub G76C (Ub G76C) was expressed as soluble protein by inducing with 0.4 mM IPTG for 4 hours at 37°C upon the culture reaching optical density at 600 nm = 0.4 to 0.6. Bacteria cells were harvested and lysed (AvestinEmulsiflexC3). Protein was purified through Ni-NTA agarose beads (Qiagen) [lysis buffer: 300 mM NaCl, 50 mM tris (pH 8.0), 10 mM imidazole, 5 mM 2-mercaptoethanol (BME), 1x protease inhibitor; elution buffer: 300 mM NaCl, 50 mM tris (pH 8.0), 300 mM imidazole, and 5 mM BME] followed by HiTrap Q HP (GE Healthcare) liquid chromatography column [buffer A: 50 mM NaCl, 20 mM tris (pH 8.0), 0.2 mM EDTA, and 10 mM BME; buffer B: 1 M NaCl, 20 mM tris (pH 8.0), 0.2 mM EDTA, and 10 mM BME]. Purified Ub was then dialyzed against water supplemented with 1 mM acetic acid followed by flash freezing in liquid nitrogen and lyophilized using a Sentry lyophilizer (VirTis).
Purification of Widom 601 DNA 187 bp
A plasmid containing the Widom 601 nucleosome positioning sequence extended to 187 base pairs (bp) and flanked by Eco RV restriction sites (37) was transformed into E. coli DH5α (Thermo Fisher Scientific) and grown in 2xYT-Amp medium overnight at 37°C. The 601 DNA fragment was excised using Eco RV and purified as previously (48).
Expression and purification of WT Xenopus histones and H2AK119C
Plasmids containing WT Xenopus histones were a generous gift from K. Luger, and mutant histone H2AK119C was generated using the Q5 mutagenesis kit (NEB). Briefly, each histone was expressed in E. coli Rosetta (DE3) cells (Novagen), extracted from inclusion bodies, and purified sequentially by size exclusion and anion chromatography as previously (48). Purified histones were freeze dried using a Sentry lyophilizer (VirTis).
Ubiquitination of histone H2AK119C (nonlabile for cryo-EM)
In brief [and as previously; (23)], lyophilized Ub G76C and histone H2AK119C were resuspended (resuspension buffer: 10 mM acetic acid and 7 M urea-deionized) and mixed in the ratio of 2:1. Sodium tetraborate, urea, and TCEP were added to achieve final concentrations of 50 mM, 6 M, and 5 mM, respectively. The mixture was incubated at room temperature for 30 min. Then, an amount of cross-linker [1,3-dichloroacetone (Sigma-Aldrich) diluted in N,N′-dimethylformamide (Sigma-Aldrich)] equal to one-half molar ratio of total sulfhydryl groups was added to the solution and incubated on ice for additional 30 min. The reaction was stopped by addition of BME to a final concentration of 5 mM. Then, the solution was diluted 10 times with denaturing binding buffer [denaturing binding buffer: 50 mM sodium phosphate (NaPi), 50 mM tris (pH 8.0), 300 mM NaCl, 6 M urea, 10 mM imidazole, and 5 mM BME] and purified through Ni-NTA agarose beads (Qiagen) [denaturing elution buffer: 50 mM NaPi, 50 mM tris (pH 8.0), 300 mM NaCl, 6 M urea, 250 mM imidazole, and 5 mM BME]. Purified ubiquitinated H2AK119C (H2AK119Ub) was dialyzed into water supplemented with 1 mM BME and lyophilized using a Sentry lyophilizer (VirTis).
Reconstitution of nucleosomes
Nucleosome substrates were assembled as described (27, 32). Equimolar amounts of each of the four lyophilized histones were dissolved in unfolding buffer [6 M guanidinium hydrochloride, 20 mM tris (pH 7.5), and 5 mM dithiothreitol (DTT)], mixed, and dialyzed into refolding buffer. Octamers were purified through size exclusion chromatography Superdex 200 column (GE Healthcare) in refolding buffer [10 mM tris (pH 7.5), 2 M NaCl, 1 mM EDTA, and 5 mM BME]. Nucleosomes were assembled by combining equimolar ratios of purified Widom 601 DNA and histone octamers and dialyzing the mix overnight with gradient salt dialysis using a peristaltic Rapid Pump (Gilson). Assembled nucleosomes were purified through a Resource Q ion-exchange column (GE Healthcare). Last, purified nucleosomes were dialyzed into dialysis buffer [20 mM tris-HCl (pH 7.5), 1 mM EDTA, and 1 mM DTT], concentrated, and stored at 4°C until use.
Generating native H2AK119Ub nucleosomes and chromatosomes
Nucleosomes were enzymatically modified using Ub, E1, E2, and Polycomb repressive complex 1 as previously (21). H2AK119Ub nucleosomes were purified by precipitation with magnesium chloride as previously (49). Chromatosomes were assembled by adding equimolar amount of H1.4 to nucleosome in assembly buffer [10 mM KCl, 10 mM Hepes (pH 7.5), 0.7 mM EDTA (pH 8), and 1 mM DTT] at 4°C for 1 hour.
PTM-defined nucleosomes (dNucs) and peptides
PTM-defined nucleosomes [all from EpiCypher: rNuc (unmodified), no. 16-0006 (https://www.epicypher.com/products/nucleosomes/mononucleosomes-recombinant-human-biotinylated); H2AK15ub1, no. 16-0399 (https://www.epicypher.com/products/nucleosomes/modified-designer-nucleosomes-dnucs/nucleosome-recombinant-human-h2ak15ub1-dnuc-biotinylated); H2AK119ub1, no. 16-0395 (https://www.epicypher.com/products/nucleosomes/nucleosome-recombinant-human-h2ak119ub1-dnuc-biotinylated); and H2BK120ub1, no. 16-0396 (https://www.epicypher.com/products/nucleosomes/nucleosome-recombinant-human-h2bk120ub1-dnuc-biotinylated)] were assembled through salt dialysis of semisynthetic histones with 5′ biotinylated DNA (147 bp of 601 nucleosome positioning sequence) as previously (50, 51) and confirmed by SDS–polyacrylamide gel electrophoresis (PAGE), immunoblotting, and mass spectrometry (as appropriate). All Kub1 histones for enzymatic assays contain a native gamma-lysine isopeptide linkage cleavable by deubiquitinating (DUB) enzymes (52). H2AK119ub peptide (amino acids 110 to 129) also contains a native gamma-lysine isopeptide linkage.
Free Ub system of detection
Turn-on UQ1UBA-IsoTZnF fusion (tUI)–free Ub sensor (53) was provided by R. Cohen and T. Yao (Colorado State University). Briefly, the sensor is a fusion protein containing Ub binding domains from ubiquilin-1 (amino acids 541 to 586) and isotranspeptidase T (amino acids 163 to 191) and relies on the fluorescence change of a single covalently attached Atto-532 when it engages free Ub (as released by DUB activity on a Kub1 dNuc).
GraFix of BAP1/ASXL1-H2AK119Ub nucleosome
H2AK119Ub nucleosome on 187 bp DNA was saturated with BAP1/ASXL1 and dialyzed in 5% glycerol buffer A2 [20 mM Hepes (pH 7.5), 50 mM KCl, 1 mM DTT, and 5% glycerol]. BAP1/ASXL1-nucleosome complex was cross-linked using GraFix as previously (24). Gradients were produced (Gradient Master, Biocomp Instruments) using a gradient range of 10% glycerol buffer B2 [20 mM Hepes (pH 7.5), 50 mM KCl, 1 mM DTT, and 10% glycerol] to 40% glycerol buffer C2 [20 mM Hepes (pH 7.5), 50 mM KCl, 1 mM DTT, 40% glycerol, and 0.1% glutaraldehyde]. Gradients were spun at 30,000 rpm for 16 hours at 4°C, fractionated, and remnant glutaraldehyde–quenched with 100 mM tris-Cl (pH 7.5) (final). Fractions were analyzed by 4.5% native PAGE [0.2× tris-borate EDTA (TBE)], and those of interest were pooled and dialyzed in cryo-EM buffer [20 mM tris (pH 7.5), 50 mM KCl, 1 mM DTT, and 1% glycerol]. Final samples were concentrated and used for grid preparation.
Grid preparation for cryo-EM
Cryo-EM grids of the BAP1/ASXL1-H2AK119Ub nucleosome complex were prepared with the following established protocol (54). Briefly, Quantifoil R 1.2-μm–hole size Au 300 mesh grids were glow-discharged for 25 s at 15 mA using a PELCO easiGlow. The sample (3 μl) at concentration of 0.1 mg/ml was applied to grids using a FEI Vitrobot Mark IV (FEI Company), at 4°C and 100% humidity. Grids were blotted using filter paper (55/20 mm diameter, TED PELLA) for 3 s with a blot force of 3 and plunge-frozen in liquid ethane cooled to liquid nitrogen temperatures.
Cryo-EM data processing
Data were collected using a Titan Krios G3i using Leginon software (55). All images were recorded using a K3 Summit Gatan direct electron detector camera at a nominal magnification of ×105,000, calibrated physical pixel size of 0.825 Å/pixel (0.4215 in superresolution Å/pixel) using an energy filter width of 20 eV. The images were collected with an exposure of 0.04 s per frame; the total exposure time was 2 s for the BAP1/ASXL1-H2AK119Ub complex, for a total of 50 frames. Per-frame exposure rate was 1.1424 e (charge on the electron)/Å2 leading to a total accumulated electron exposure of 57.12 e/Å2 on the specimen. All the sample images were recorded with a calibrated defocus in the range from −0.9 to −1.9 μm. Movie stacks, acquired in superresolution mode, were corrected for global and local motions (in 11 × 9 patches) using UCSF MotionCor2 v1.2.1 (56) ran through Relion v4.0-beta context (57), resulting in the dose-weighted sums. We imported the motion-corrected dose-weighted images to CryoSPARC v4.1.1 (58) and performed Patch CTF Estimation. We used “blob-based” automated particle picking to obtain 11,876,334 particles and selected 5,510,142 particles for subsequent processing. Particles were then extracted and subjected to reference-free 2D classification into 50 classes in CryoSPARC. A small number of particles (~265 K) were selected from the seven best-looking reference-free 2D classes. We then used CryoSPARC’s “Ab initio” option to generate a 3D initial model from the selected small dataset. This yielded a reconstruction with a clearly defined nucleosome density, which was then used as a template in 3D refinement. For data “in silico” homogenization of our data, we used CryoSPARC’s Ab initio and “Heterogeneous refinement.” After 3D refinement, we used focused classification to improve the density of BAP1/ASXLl in the maps, and the data were subjected to focused classifications. In the BAP1/ASXL1-H2AK119Ub complex, we centered a spherical mask with a 60 Å pixel radius on the BAP1/ASXL1 moiety, which resulted in a clearly resolved BAP1/ASXL1 on the nucleosome at 5.06 Å overall resolution. These particles were subjected to a variability analysis, using a similar mask centered on the BAP1/ASXL1 moiety, using principal component analysis with five modes and an output of 10 clusters, finding one cluster that contained a better-resolved BAP1/ASXL1 clamp-DNA and BAP1/ASXL1-Ub interfaces at 4.35 Å after extraction of the particles. Using the best-scoring particles, we ran Non-Uniform Refinement, which yielded the BAP1/ASXL1-H2AK119Ub complex at 3.6-Å map (map 1). Two additional maps were generated by further refinement and Bayesian Polishing in Relion (59): a map with an improved BAP1:acidic patch interface (map 2) and another map with an improved BAP1:DNA clamp interface (map 3). The final resolutions were established using Fourier shell correlation (FSC) at 0.143 cutoff (60) following a gold-standard refinement. All conversions between Relion and CryoSPARC were performed using Daniel Asarnow’s pyem script (https://github.com/asarnow/pyem). The processing details and summaries are shown in figs. S3 and S4 and table S1.
Model building and refinement
Our cryo-EM reconstructions reveal BAP1/ASXL1 bound to nucleosome, with clearly resolved interfaces. The highest-resolution element in the reconstruction is the nucleosome core particle, akin to other published nucleosome-associated complexes (20, 21). BAP1/ASXL1 is very flexible; however, the quality of our maps allowed unambiguous rigid-body fitting and modeling of the proteins. To model the complex, we used a number of maps; the final BAP1/ASXL1-H2AK119Ub reconstruction from Titan Krios (figs. S3 and S5) at 3.6 Å was used to describe the global architecture of the complex (map 1). The different maps used to build each region are summarized in table S1. We submitted the sequence of our constructs of BAP1, ASXL1, and Ub to AlphaFold-Multimer (25) and evaluated how the predicted models fit into our maps by performing a manual local optimization of the fitting of the best AlphaFold model using UCSF Chimera’s and the “Fit in map” function (61). For the BAP1/ASXL1-H2AK119Ub complex, available x-ray crystal structures or predictions based in homologous modeling were aligned with the AlphaFold-Multimer prediction (25). For the nucleosome, we used PDB IDs 3tu4 (37) and 7k5y (45), and 4uel was used for Ub. We used templates of the UCH-L5 structure 4uel (22) and the homologous Calypso in complex with DEUBAD of ASX 6hgc (20) for modeling on the Swiss-Prot server for BAP1 and ASXL1, respectively. We then used Coot (62) for local adjustments of the secondary elements and side chains into densities, using a combination of crystallographic and AlphaFold-based models. The resulting complete model was refined in PHENIX (63) using a secondary structure, Atomic Displacement Parameters (ADPs), rotamer, and Ramachandran restraints in 100 iterations. The model was then visually inspected, and Ramachandran outliers and problematic regions were fixed manually in Coot (final refinement statistics are summarized in table S2). For assigning the residues interacting with the acidic patch and the DNA dyad of the nucleosome, we used maps with improved resolution at these regions (map 2 and map 3, respectively). An extra density could be observed near the N-terminal tail of BAP1 and the C-terminal tail of ASXL1; however, this region was too disordered in the cryo-EM reconstruction to reliably assign and model.
To validate our structures, we first subjected the atoms to 0.1-Å displacement and then refined it in phenix.real_space_refine against one of the half-maps. This refined model was then converted to a 3D density map and compared against two half-maps and the summed map. We calculated FSC curves with half-map 1 (used for refinement, “work,” shown in our figures with red), half-map 2 (not participating in refinement, hence “free,” green) and the summed map (blue). We then tested the whole model against the 3.6-Å reconstruction (fig. S12). We only observed small differences between the work and the free FSC curves, which indicate lack of overfitting. Figures of the model and cryo-EM densities were prepared using Chimera, ChimeraX, Coot, and PyMOL (61, 62, 64, 65).
Deubiquitinase catalytic activity assay
Deubiquitinase activity of WT BAP1/ASXL1 and indicated mutants toward nucleosome and peptide substrates was assessed by monitoring fluorescence increase for up to 11 min in 1-min (nucleosomes) or 30-s (peptides) intervals at ambient temperature in the presence of Atto-532 labeled tUI-free Ub sensor. A total of 3.33 μl of 3× DUB, 3.33 μl of 3× nucleosome or peptide substrate, and 3.33 μl of 3× tUI-free Ub sensor in DUB buffer [20 mM Hepes (pH 7.5), 20 mM NaCl, 2 mM MgCl2, 0.01% v/v Tween 20, 0.01% w/v bovine serum albumin, and 10 mM DTT] were combined in a 384-well plate (PerkinElmer ProxiPlate-384 Plus F black, no. 6008260), and data were gathered on a Envision 2150 plate reader (PerkinElmer) (excitation, 531 nm; emission, 570 nm; 555 mirror). Nucleosome or peptide (10 nM) and tUI-free Ub sensor (10 nM) were used (final). DUB concentration varied by experiment as follows: activity assays focusing on H2AK119ub1 dNuc, 5 nM; H2AUb and H2AUb-H1.4 nucleosomes, 0.3 nM; multiple unmodified and ubiquinated nucleosomes, 2.5 nM; and H2AK119ub1 peptide, 16 pM. The initial, linear portion of each duplicate (Fig. 3A and fig. S6, B and C) or triplicate (Fig. 4E and fig. S12E) DUB reaction was compared, and data were presented as relative fluorescence units (RFU) per minute. In Figs. 3 and 4, catalytic activity was also expressed as percent of activity of each mutant relative to the WT. The loss of catalytic activity was calculated as percent of activity of each mutant relative to the WT.
Deubiquitination kinetic assays
Kinetic parameters were determined by monitoring BAP1/ASXL1 activity toward H2AK119ub1 dNuc substrate over the course of 11 min in 30-s or 1-min intervals at ambient temperature. DUB and tUI-free Ub sensor concentration were fixed, while H2AK119ub1 dNuc concentration was varied in a 1.5-fold-dilution series. Final DUB concentrations were as follows: wtBAP1/wtASXL1 at 0.5 nM, BAP1 R699A/R700A/R701A/wtASXL1 at 1 nM, wtBAP1/ASXl1 K243A/R244A/R246A at 1 nM, wtBAP1/ASXL1 R336A/R338A/K343A/K345A/K346A at 1 nM, and BAP1 R56/R57/R59/R60A/wtASXL1 at 5 nM. For DUBs tested at 0.5 nM, the substrate was varied from 300 to 3.5 nM; for DUBs tested at 1 nM, from 300 to 5.2 nM; and for DUBs tested at 5 nM, from 300 to 12 nM. In all cases, a no-substrate sample was included for background correction. To calculate the amount of Ub generated enzymatically, a free-ubiquitin standard curve (0.88 to 10 nM; 1.5-fold dilutions) was included, along with a no-ubiquitin sample for background correction. Three independent runs of triplicates were performed. Reaction volumes, tUI-free Ub sensor concentration, assay plates, and plate reader settings were as in the activity assay. H2AK119ub1 dNuc K0.5, kcat, and Hill coefficient (h) values were determined by applying an allosteric sigmoidal model using GraphPad 9.0 (Prism) to initial, linear velocities compared to H2AK119ub1 dNuc concentration, or, alternatively, Michaelis constant (Km) and kcat were determined by applying a Michaelis-Menten model, as appropriate. Michaelis-Menten and allosteric sigmoidal fits were compared for best fit using an F test, and the Michaelis-Menten model was selected unless the P value was <0.05. To determine initial velocities, first, a background correction was applied to the signal of the DUB samples and the standard curve. Second, a linear regression was applied to the background-corrected standard curve at each time point. Third, the standard curve linear regression parameters were used to convert DUB background-corrected RFUs to the concentration of Ub generated at each time point, and a linear regression was fit to the data to generate initial velocity curves.
Electromobility shift assay
Assembled BAP1/ASXL1 complex was dialyzed into EMSA buffer [10 mM Hepes (pH 7.5), 50 mM NaCl, 0.1 mM DTT, and 5% glycerol]. A twofold serial dilution (starting concentration between 1 and 5 μM) of BAP1/ASXL1 complex was made in EMSA buffer. Nonhydrolyzable H2AK119Ub 187-bp nucleosome was added to each reaction to reach a final concentration of 25 nM. Reactions were incubated on ice for 1 hour. Reaction (3.5 μl) was analyzed on 4.5% native polyacrylamide gels (0.2× TBE). Native acrylamide gels were stained with ethidium bromide and visualized using a Geldoc (Bio-Rad). The band quantification was performed with the program Image Lab (Bio-Rad). The amount of BAP1/ASXL1 bound to nucleosomes was determined by measuring the decrease in free nucleosome in each reaction using BAP1/ASXL1-free samples as background. The free DNA was taken under consideration for the calculation of free nucleosome. The apparent dissociation constant (Kd) and the Hill coefficient for each binding curve were calculated by fitting the specific binding with Hill slope equation using the program Prism 9.0 (GraphPad). The final parameters were calculated using for at least three independent experiments (n ≥ 3 per data point). Data were plotted as means ± SE.
Cell lines and cell culture
All cell lines used were generated in the Pasini laboratory and are strain 129P2/Ola: mES cell line E14, mES KO ES c1 cell line, mES KO ES c2 cell line, mES BAP1 KO + BAP1 (pCAG-BAP1 WT), and mES BAP1 KO + BAP1 C91S (pCAG-BAP1 WT). All mouse embryonic stem cell (mESC) lines were grown on 0.1% gelatin-coated dishes in 2i/leukemia inhibitory factor (LIF)–containing Glasgow's minimum essential medium (GMEM) (Euroclone) supplemented with 16% fetal calf serum (Euroclone), 2 mM glutamine (Gibco), penicillin (100 U/ml), streptomycin (0.1 mg/ml; Gibco), 0.1 mM nonessential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 50 μM β-mercaptoethanol phosphate-buffered saline (PBS; Gibco), LIF (1000 U/ml; produced in-house), and glycogen synthase kinase 3β and mitogen-activated protein kinase kinase 1/2 inhibitors (abcr GmbH) to a final concentration of 3 and 1 μM, respectively.
Stable expressing clones BAP1 KO ESC were generated by transfecting 10 μg of pCAG expression vectors encoding N-terminal 2xFlag-hemagglutinin–tagged BAP1 WT or with C91S; R56E/R59E; R699E/R700E; R699A/R700A/R701A; and R56A/R57A/R59A/R60A mutations using Lipofectamine 2000 (Thermo Fisher Scientific), as per the manufacturer’s instructions. Twenty-four hours after transfection ESCs were selected with neomycin (0.5 μg/ml) for 5 days. Cells were then split to clonal density (∼1:50) onto 15-cm plates. Clones were isolated 12 days later and grown further before screening for rescue allele expression by Western blot.
Western blot
Western blot analyses were performed with total protein lysates. All mESCs lines were lysed and sonicated in ice-cold S300 buffer [20 mM tris-HCl (pH 8.0), 300 mM NaCl, 10% glycerol, and 0.2% NP-40] and supplemented with Benzonase (25 U/μl; MilliporeSigma) and protease inhibitors (Roche). Precipitates were removed by centrifugation. Clear lysates were resuspended in Laemmli sample buffer and boiled for 5 min. Protein lysates were separated on SDS-PAGE gels and transferred to nitrocellulose membranes. After probing with the appropriate primary and secondary antibodies, chemiluminescence signals were captured with the ChemiDoc Imaging System (Bio-Rad). Western blots were performed with anti-BAP1 [D7W70, Cell Signaling Technology (CST)], anti-H2AK119ub1 (8240, CST), anti-H3 (ab1791, Abcam).
Quantification and statistical analysis
Protein quantification was done using an A280 extinction coefficient of 106,660 M−1 cm−1 for BAP1/ASXL1 complex on a Nanodrop spectrophotometer (Thermo Fisher Scientific).
Acknowledgments
We thank colleagues for the supply of materials (see Materials and Methods) and B. Wang for helping with data collection at NYU cryo-EM Shared Resource (RRID number SCR_019202). We thank R.E. Cohen and T. Yao’s laboratories for providing the tUI sensor. We thank M. Costantino, K. Yie, and staff of the HPC Core at NYU Langone Health for computer access and support and P. Hare for critical reading of the manuscript. We thank all members of the Armache laboratory for helpful suggestions and discussions.
Funding: Work in the Armache laboratory is supported by grants from the Mark Foundation for Cancer Research and the National Institutes of Health (NIH) (R01GM115882 and R01CA266978). Work in the Pasini laboratory was supported by World Wide Cancer Research (22-0027), by the Italian Association for Cancer Research (AIRC; IG-2017-20290 and IG 2022-27694), and by the European Research Council (ERC; EC-H2020-ERC-CoG-DissectPcG: 725268). S.T. is supported by grants from AIRC (MFAG 2021-26131) and Fondazione Cariplo (Cariplo Giovani 2020-3576). S.R. is a PhD student within the European School of Molecular Medicine (SEMM). Work in the Lu laboratory is supported by R35GM138181. Work in EpiCypher is supported by NIH grant R44GM119893.
Author contributions: J.F.T., M.I.V.-S., and K.-J.A. conceptualized and designed the study. J.F.T., M.I.V.-S., V.G.-L., P.D.I., R.L., and S.A.-A. performed the structural and biochemical experiments. S.L.G., J.M.B., and A.R.H. performed/analyzed enzyme activity and kinetic assays on PTM-defined peptides and nucleosomes (conceptualized and/or synthesized by L.S., S.G., H.F.T., Z.-W.S., R.J.E., A.V., M.J.M., and M.A.C.) under supervision from M.-C.K., S.T., S.R., and D.P. performed studies in mESCs. W.J.R. contributed to data collection and processing. All authors contributed to data analysis, interpretation, and writing of the manuscript with input from K.G., E.N., and C.L.
Competing interests: EpiCypher is a commercial developer and supplier of reagents [e.g., PTM-defined semisynthetic nucleosomes (dNucs)] used in this study. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The structure model was deposited in the Protein Data Bank (PDB ID 8SVF), and the cryo-EM density maps have been deposited in Electron Microscopy Data Bank (EMDB) with the following accession codes: EMD-40791 for building the overall structure, EMD-40790 for the acidic patch interactions, and EMD-40789 for DNA clamp interaction. Further inquiries and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, K.-J.A. (karim-jean.armache@nyulangone.org).
Supplementary Materials
This PDF file includes:
Figs. S1 to S15
Tables S1 to S5
References
REFERENCES AND NOTES
- 1.A. P. Szczepanski, L. Wang, Emerging multifaceted roles of BAP1 complexes in biological processes. Cell Death Discov. 7, 20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J. C. Scheuermann, A. G. de Ayala Alonso, K. Oktaba, N. Ly-Hartig, R. K. McGinty, S. Fraterman, M. Wilm, T. W. Muir, J. Müller, Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D. D. Sahtoe, W. J. Van Dijk, R. Ekkebus, H. Ovaa, T. K. Sixma, BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat. Commun. 7, 10292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.L. Sanchez-Pulido, L. Kong, C. P. Ponting, A common ancestry for BAP1 and Uch37 regulators. Bioinformatics 28, 1953–1956 (2012). [DOI] [PubMed] [Google Scholar]
- 5.N. A. Fursova, A. H. Turberfield, N. P. Blackledge, E. L. Findlater, A. Lastuvkova, M. K. Huseyin, P. Dobrinić, R. J. Klose, BAP1 constrains pervasive H2AK119ub1 to control the transcriptional potential of the genome. Genes Dev. 35, 749–770 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.E. Conway, F. Rossi, D. Fernandez-Perez, E. Ponzo, K. J. Ferrari, M. Zanotti, D. Manganaro, S. Rodighiero, S. Tamburri, D. Pasini, BAP1 enhances Polycomb repression by counteracting widespread H2AK119ub1 deposition and chromatin condensation. Mol. Cell 81, 3526–3541.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J. N. Kuznetsov, T. H. Aguero, D. A. Owens, S. Kurtenbach, M. G. Field, M. A. Durante, D. A. Rodriguez, M. L. King, J. W. Harbour, BAP1 regulates epigenetic switch from pluripotency to differentiation in developmental lineages giving rise to BAP1-mutant cancers. Sci. Adv. 5, eaax1738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.A. Campagne, M. K. Lee, D. Zielinski, A. Michaud, S. le Corre, F. Dingli, H. Chen, L. Z. Shahidian, I. Vassilev, N. Servant, D. Loew, E. Pasmant, S. Postel-Vinay, M. Wassef, R. Margueron, BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat. Commun. 10, 348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. Bonnet, I. Boichenko, R. Kalb, M. le Jeune, S. Maltseva, M. Pieropan, K. Finkl, B. Fierz, J. Müller, PR-DUB preserves Polycomb repression by preventing excessive accumulation of H2Aub1, an antagonist of chromatin compaction. Genes Dev. 36, 1046–1061 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Y. B. Schwartz, V. Pirrotta, Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8, 9–22 (2007). [DOI] [PubMed] [Google Scholar]
- 11.L.-H. Wang, M. A. E. Aberin, S. Wu, S.-P. Wang, The MLL3/4 H3K4 methyltransferase complex in establishing an active enhancer landscape. Biochem. Soc. Trans. 49, 1041–1054 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.L. Wang, Z. Zhao, P. A. Ozark, D. Fantini, S. A. Marshall, E. J. Rendleman, K. A. Cozzolino, N. Louis, X. He, M. A. Morgan, Y. H. Takahashi, C. K. Collings, E. R. Smith, P. Ntziachristos, J. N. Savas, L. Zou, R. Hashizume, J. J. Meeks, A. Shilatifard, Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat. Med. 24, 758–769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.I. H. Ismail, R. Davidson, J. P. Gagné, Z. Z. Xu, G. G. Poirier, M. J. Hendzel, Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res. 74, 4282–4294 (2014). [DOI] [PubMed] [Google Scholar]
- 14.O. Abdel-Wahab, J. Gao, M. Adli, A. Dey, T. Trimarchi, Y. R. Chung, C. Kuscu, T. Hricik, D. Ndiaye-Lobry, L. M. LaFave, R. Koche, A. H. Shih, O. A. Guryanova, E. Kim, S. Li, S. Pandey, J. Y. Shin, L. Telis, J. Liu, P. K. Bhatt, S. Monette, X. Zhao, C. E. Mason, C. Y. Park, B. E. Bernstein, I. Aifantis, R. L. Levine, Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J. Exp. Med. 210, 2641–2659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. Cigognetti, S. Lonardi, S. Fisogni, P. Balzarini, V. Pellegrini, A. Tironi, L. Bercich, M. Bugatti, G. Rossi, B. Murer, M. Barbareschi, S. Giuliani, A. Cavazza, G. Marchetti, W. Vermi, F. Facchetti, BAP1 (BRCA1-associated protein 1) is a highly specific marker for differentiating mesothelioma from reactive mesothelial proliferations. Mod. Pathol. 28, 1043–1057 (2015). [DOI] [PubMed] [Google Scholar]
- 16.M. Cheung, J. R. Testa, BAP1, a tumor suppressor gene driving malignant mesothelioma. Transl. Lung Cancer Res. 6, 270–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.F. Matheus, E. Rusha, R. Rehimi, L. Molitor, A. Pertek, M. Modic, R. Feederle, A. Flatley, E. Kremmer, A. Geerlof, V. Rishko, A. Rada-Iglesias, M. Drukker, Pathological ASXL1 mutations and protein variants impair neural crest development. Stem Cell Rep. 12, 861–868 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L. Shahriyari, M. Abdel-Rahman, C. Cebulla, BAP1 expression is prognostic in breast and uveal melanoma but not colon cancer and is highly positively correlated with RBM15B and USP19. PLOS ONE 14, e0211507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.G. Stålhammar, T. R. O. See, S. Phillips, S. Seregard, H. E. Grossniklaus, Digital image analysis of BAP-1 accurately predicts uveal melanoma metastasis. Transl. Vis. Sci. Technol. 8, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.I. De, E. C. Chittock, H. Grötsch, T. C. R. Miller, A. A. McCarthy, C. W. Müller, Structural basis for the activation of the deubiquitinase calypso by the polycomb protein ASX. Structure 27, 528–536.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 21.M. Foglizzo, A. J. Middleton, A. E. Burgess, J. M. Crowther, R. C. J. Dobson, J. M. Murphy, C. L. Day, P. D. Mace, A bidentate Polycomb repressive-deubiquitinase complex is required for efficient activity on nucleosomes. Nat. Commun. 9, 3932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D. D. Sahtoe, W. J. van Dijk, F. E. Oualid, R. Ekkebus, H. Ovaa, T. K. Sixma, Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol. Cell 57, 887–900 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.L. Long, M. Furgason, T. Yao, Generation of nonhydrolyzable ubiquitin-histone mimics. Methods 70, 134–138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.H. Stark, GraFix: Stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 481, 109–126 (2010). [DOI] [PubMed] [Google Scholar]
- 25.R. Evans, M. O’Neill, A. Pritzel, N. Antropova, A. Senior, T. Green, A. Žídek, R. Bates, S. Blackwell, J. Yim, O. Ronneberger, S. Bodenstein, M. Zielinski, A. Bridgland, A. Potapenko, A. Cowie, K. Tunyasuvunakool, R. Jain, E. Clancy, P. Kohli, J. Jumper, D. Hassabis, Protein complex prediction with AlphaFold-multimer. bioRxiv 10.1101/2021.10.04.463034 (2022). 10.1101/2021.10.04.463034. [DOI]
- 26.D. Komander, M. Rape, The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012). [DOI] [PubMed] [Google Scholar]
- 27.M. I. Valencia-Sánchez, P. de Ioannes, M. Wang, D. M. Truong, R. Lee, J. P. Armache, J. D. Boeke, K. J. Armache, Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation. Science 371, eabc6663 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.E. J. Worden, X. Zhang, C. Wolberger, Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome. eLife 9, e53199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.H. Peng, J. Prokop, J. Karar, K. Park, L. Cao, J. W. Harbour, A. M. Bowcock, S. B. Malkowicz, M. Cheung, J. R. Testa, F. J. Rauscher III, Familial and somatic BAP1 mutations inactivate ASXL1/2-mediated allosteric regulation of BAP1 deubiquitinase by targeting multiple independent domains. Cancer Res. 78, 1200–1213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.C. A. Davey, D. F. Sargent, K. Luger, A. W. Maeder, T. J. Richmond, Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 319, 1097–1113 (2002). [DOI] [PubMed] [Google Scholar]
- 31.V. Kasinath, C. Beck, P. Sauer, S. Poepsel, J. Kosmatka, M. Faini, D. Toso, R. Aebersold, E. Nogales, JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science 371, eabc3393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.M. I. Valencia-Sánchez, P. De Ioannes, M. Wang, N. Vasilyev, R. Chen, E. Nudler, J.-P. Armache, K.-J. Armache, Structural basis of Dot1L stimulation by histone H2B lysine 120 ubiquitination. Mol. Cell 74, 1010–1019.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R. K. McGinty, S. Tan, Recognition of the nucleosome by chromatin factors and enzymes. Curr. Opin. Struct. Biol. 37, 54–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R. K. Mcginty, R. C. Henrici, S. Tan, Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R. D. Makde, J. R. England, H. P. Yennawar, S. Tan, Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A. J. Barbera, J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, K. M. Kaye, The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311, 856–861 (2006). [DOI] [PubMed] [Google Scholar]
- 37.K. J. Armache, J. D. Garlick, D. Canzio, G. J. Narlikar, R. E. Kingston, Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334, 977–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.E. J. Worden, N. A. Hoffmann, C. W. Hicks, C. Wolberger, Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell 176, 1490–1501.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. Carbone, J. W. Harbour, J. Brugarolas, A. Bononi, I. Pagano, A. Dey, T. Krausz, H. I. Pass, H. Yang, G. Gaudino, Biological mechanisms and clinical significance of BAP1 mutations in human cancer. Cancer Discov. 10, 1103–1120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.H. Yang, S. Kurtenbach, Y. Guo, I. Lohse, M. A. Durante, J. Li, Z. Li, H. al-Ali, L. Li, Z. Chen, M. G. Field, P. Zhang, S. Chen, S. Yamamoto, Z. Li, Y. Zhou, S. D. Nimer, J. W. Harbour, C. Wahlestedt, M. Xu, F. C. Yang, Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood 131, 328–341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.L. Wang, N. W. Birch, Z. Zhao, C. M. Nestler, A. Kazmer, A. Shilati, A. Blake, P. A. Ozark, E. J. Rendleman, D. Zha, C. A. Ryan, M. A. J. Morgan, A. Shilatifard, Epigenetic targeted therapy of stabilized BAP1 in ASXL1 gain-of-function mutated leukemia. Nat. Cancer 2, 515–526 (2021). [DOI] [PubMed] [Google Scholar]
- 42.A. Skrajna, D. Goldfarb, K. M. Kedziora, E. M. Cousins, G. D. Grant, C. J. Spangler, E. H. Barbour, X. Yan, N. A. Hathaway, N. G. Brown, J. G. Cook, M. B. Major, R. K. McGinty, Comprehensive nucleosome interactome screen establishes fundamental principles of nucleosome binding. Nucleic Acids Res. 48, 9415–9432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D. Grau, Y. Zhang, C. H. Lee, M. Valencia-Sánchez, J. Zhang, M. Wang, M. Holder, V. Svetlov, D. Tan, E. Nudler, D. Reinberg, T. Walz, K. J. Armache, Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction. Nat. Commun. 12, 714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M. Dombrowski, M. Engeholm, C. Dienemann, S. Dodonova, P. Cramer, Histone H1 binding to nucleosome arrays depends on linker DNA length and trajectory. Nat. Struct. Mol. Biol. 29, 493–501 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.B.-R. Zhou, H. Feng, S. Kale, T. Fox, H. Khant, N. de Val, R. Ghirlando, A. R. Panchenko, Y. Bai, Distinct structures and dynamics of chromatosomes with different human linker histone isoforms. Mol. Cell 81, 166–182.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.W. Ge, C. Yu, J. Li, Z. Yu, X. Li, Y. Zhang, C. P. Liu, Y. Li, C. Tian, X. Zhang, G. Li, B. Zhu, R. M. Xu, Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase. Nature 616, 176–182 (2023). [DOI] [PubMed] [Google Scholar]
- 47.R. Meas, P. Mao, Histone ubiquitylation and its roles in transcription and DNA damage response. DNA Repair 36, 36–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.P. N. Dyer, R. S. Edayathumangalam, C. L. White, Y. Bao, S. Chakravarthy, U. M. Muthurajan, K. Luger, Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004). [DOI] [PubMed] [Google Scholar]
- 49.H. T. Dao, H. Liu, N. Mashtalir, C. Kadoch, T. W. Muir, Synthesis of oriented hexasomes and asymmetric nucleosomes using a template editing process. J. Am. Chem. Soc. 144, 2284–2291 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.L. F. Schachner, K. Jooß, M. A. Morgan, A. Piunti, M. J. Meiners, J. O. Kafader, A. S. Lee, M. Iwanaszko, M. A. Cheek, J. M. Burg, S. A. Howard, M. C. Keogh, A. Shilatifard, N. L. Kelleher, Decoding the protein composition of whole nucleosomes with Nuc-MS. Nat. Methods 18, 303–308 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.M. R. Marunde, H. A. Fuchs, J. M. Burg, I. K. Popova, A. Vaidya, N. W. Hall, M. J. Meiners, R. Watson, S. A. Howard, K. Novitzky, E. M. Anarney, M. A. Cheek, Z.-W. Sun, B. J. Venters, M.-C. Keogh, C. A. Musselman, Nucleosome conformation dictates the histone code. bioRxiv 10.1101/2022.02.21.481373 (2022). 10.1101/2022.02.21.481373. [DOI]
- 52.X. Bi, R. Yang, X. Feng, D. Rhodes, C. F. Liu, Semisynthetic UbH2A reveals different activities of deubiquitinases and inhibitory effects of H2A K119 ubiquitination on H3K36 methylation in mononucleosomes. Org. Biomol. Chem. 14, 835–839 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Y. S. Choi, S. A. Bollinger, L. F. Prada, F. Scavone, T. Yao, R. E. Cohen, High-affinity free ubiquitin sensors for quantifying ubiquitin homeostasis and deubiquitination. Nat. Methods 16, 771–777 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.X. Li, P. Mooney, S. Zheng, C. R. Booth, M. B. Braunfeld, S. Gubbens, D. A. Agard, Y. Cheng, Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. Cheng, C. Negro, J. F. Bruhn, W. J. Rice, S. Dallakyan, E. T. Eng, D. G. Waterman, C. S. Potter, B. Carragher, Leginon: New features and applications. Protein Sci. 30, 136–150 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Q. Zheng, E. Palovcak, J. P. Armache, K. A. Verba, Y. Cheng, D. A. Agard, MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. Zivanov, T. Nakane, B. O. Forsberg, D. Kimanius, W. J. H. Hagen, E. Lindahl, S. H. W. Scheres, New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.A. Punjani, J. L. Rubinstein, D. J. Fleet, M. A. Brubaker, cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 59.J. Zivanov, T. Nakane, S. H. W. Scheres, A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.K. Zhang, Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin, UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 62.P. Emsley, K. Cowtan, Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 63.P. D. Adams, P. V. Afonine, G. Bunkóczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger, P. H. Zwart, PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.E. F. Pettersen, T. D. Goddard, C. C. Huang, E. C. Meng, G. S. Couch, T. I. Croll, J. H. Morris, T. E. Ferrin, UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schrodinger, LLC. (2015).
- 66.M. D. Wilson, S. Benlekbir, A. Fradet-Turcotte, A. Sherker, J. P. Julien, A. McEwan, S. M. Noordermeer, F. Sicheri, J. L. Rubinstein, D. Durocher, The structural basis of modified nucleosome recognition by 53BP1. Nature 536, 100–103 (2016). [DOI] [PubMed] [Google Scholar]
- 67.J. Gao, B. A. Aksoy, U. Dogrusoz, G. Dresdner, B. Gross, S. Onur Sumer, Y. Sun, A. Jacobsen, R. Sinha, E. Larsson, E. Cerami, C. Sander, N. Schultz, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.E. Cerami, J. Gao, U. Dogrusoz, B. E. Gross, S. Onur Sumer, B. A. Aksoy, A. Jacobsen, C. J. Byrne, M. L. Heuer, E. Larsson, Y. Antipin, B. Reva, A. P. Goldberg, C. Sander, N. Schultz, The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.K. Kim, W. Hu, F. Audenet, N. Almassi, A. J. Hanrahan, K. Murray, A. Bagrodia, N. Wong, T. N. Clinton, S. Dason, V. Mohan, S. Jebiwott, K. Nagar, J. Gao, A. Penson, C. Hughes, B. Gordon, Z. Chen, Y. Dong, P. A. Watson, R. Alvim, A. Elzein, S. P. Gao, E. Cocco, A. D. Santin, I. Ostrovnaya, J. J. Hsieh, I. Sagi, E. J. Pietzak, A. A. Hakimi, J. E. Rosenberg, G. Iyer, H. A. Vargas, M. Scaltriti, H. Al-Ahmadie, D. B. Solit, J. A. Coleman, Modeling biological and genetic diversity in upper tract urothelial carcinoma with patient derived xenografts. Nat. Commun. 11, 1975 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.J. P. Sfakianos, E. K. Cha, G. Iyer, S. N. Scott, E. C. Zabor, R. H. Shah, Q. Ren, A. Bagrodia, P. H. Kim, A. Ari Hakimi, I. Ostrovnaya, R. Ramirez, A. J. Hanrahan, N. B. Desai, A. Sun, P. Pinciroli, J. E. Rosenberg, G. Dalbagni, N. Schultz, D. F. Bajorin, V. E. Reuter, M. F. Berger, B. H. Bochner, H. A. Al-Ahmadie, D. B. Solit, J. A. Coleman, Genomic characterization of upper tract urothelial carcinoma. Eur. Urol. 68, 970–977 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.K. A. Hoadley, C. Yau, T. Hinoue, D. M. Wolf, A. J. Lazar, E. Drill, R. Shen, A. M. Taylor, A. D. Cherniack, V. Thorsson, R. Akbani, R. Bowlby, C. K. Wong, M. Wiznerowicz, F. Sanchez-Vega, A. G. Robertson, B. G. Schneider, M. S. Lawrence, H. Noushmehr, T. M. Malta, Cancer Genome Atlas Network, J. M. Stuart, C. C. Benz, P. W. Laird, Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173, 291–304.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.E. Bernard, H. Tuechler, P. L. Greenberg, R. P. Hasserjian, J. E. Arango Ossa, Y. Nannya, S. M. Devlin, M. Creignou, P. Pinel, L. Monnier, G. Gundem, J. S. Medina-Martinez, D. Domenico, M. Jädersten, U. Germing, G. Sanz, A. A. van de Loosdrecht, O. Kosmider, M. Y. Follo, F. Thol, L. Zamora, R. F. Pinheiro, A. Pellagatti, H. K. Elias, D. Haase, C. Ganster, L. Ades, M. Tobiasson, L. Palomo, M. G. Della Porta, A. Takaori-Kondo, T. Ishikawa, S. Chiba, S. Kasahara, Y. Miyazaki, A. Viale, K. Huberman, P. Fenaux, M. Belickova, M. R. Savona, V. M. Klimek, F. P. S. Santos, J. Boultwood, I. Kotsianidis, V. Santini, F. Solé, U. Platzbecker, M. Heuser, P. Valent, K. Ohyashiki, C. Finelli, M. T. Voso, L.-Y. Shih, M. Fontenay, J. H. Jansen, J. Cervera, N. Gattermann, B. L. Ebert, R. Bejar, L. Malcovati, M. Cazzola, S. Ogawa, E. Hellström-Lindberg, E. Papaemmanuil, Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evidence 1, 10.1056/EVIDoa2200008 (2022). [DOI] [PubMed] [Google Scholar]
- 73.A. Zehir, R. Benayed, R. H. Shah, A. Syed, S. Middha, H. R. Kim, P. Srinivasan, J. Gao, D. Chakravarty, S. M. Devlin, M. D. Hellmann, D. A. Barron, A. M. Schram, M. Hameed, S. Dogan, D. S. Ross, J. F. Hechtman, D. F. De Lair, J. J. Yao, D. L. Mandelker, D. T. Cheng, R. Chandramohan, A. S. Mohanty, R. N. Ptashkin, G. Jayakumaran, M. Prasad, M. H. Syed, A. B. Rema, Z. Y. Liu, K. Nafa, L. Borsu, J. Sadowska, J. Casanova, R. Bacares, I. J. Kiecka, A. Razumova, J. B. Son, L. Stewart, T. Baldi, K. A. Mullaney, H. Al-Ahmadie, E. Vakiani, A. A. Abeshouse, A. V. Penson, P. Jonsson, N. Camacho, M. T. Chang, H. H. Won, B. E. Gross, R. Kundra, Z. J. Heins, H.-W. Chen, S. Phillips, H. Zhang, J. Wang, A. Ochoa, J. Wills, M. Eubank, S. B. Thomas, S. M. Gardos, D. N. Reales, J. Galle, R. Durany, R. Cambria, W. Abida, A. Cercek, D. R. Feldman, M. M. Gounder, A. A. Hakimi, J. J. Harding, G. Iyer, Y. Y. Janjigian, E. J. Jordan, C. M. Kelly, M. A. Lowery, L. G. T. Morris, A. M. Omuro, N. Raj, P. Razavi, A. N. Shoushtari, N. Shukla, T. E. Soumerai, A. M. Varghese, R. Yaeger, J. Coleman, B. Bochner, G. J. Riely, L. B. Saltz, H. I. Scher, P. J. Sabbatini, M. E. Robson, D. S. Klimstra, B. S. Taylor, J. Baselga, N. Schultz, D. M. Hyman, M. E. Arcila, D. B. Solit, M. Ladanyi, M. F. Berger, Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.B. L. Manning-Geist, Y. L. Liu, K. A. Devereaux, A. Da Cruz Paula, Q. C. Zhou, W. Ma, P. Selenica, O. Ceyhan-Birsoy, L. A. Moukarzel, T. Hoang, S. Gordhandas, M. M. Rubinstein, C. F. Friedman, C. Aghajanian, N. R. Abu-Rustum, Z. K. Stadler, J. S. Reis-Filho, A. Iasonos, D. Zamarin, L. H. Ellenson, Y. Lakhman, D. L. Mandelker, B. Weigelt, Microsatellite instability-high endometrial cancers with MLH1 promoter hypermethylation have distinct molecular and clinical profiles. Clin. Cancer Res. 28, 4302–4311 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.L. Wu, H. Yao, H. Chen, A. Wang, K. Guo, W. Gou, Y. Yu, X. Li, M. Yao, S. Yuan, F. Pang, J. Hu, L. Chen, W. Liu, J. Yao, S. Zhang, X. Dong, W. Wang, J. Hu, Q. Ling, S. Ding, Y. Wei, Q. Li, W. Cao, S. Wang, Y. Di, F. Feng, G. Zhao, J. Zhang, L. Huang, J. Xu, W. Yan, Z. Tong, D. Jiang, T. Ji, Q. Li, L. Xu, H. He, L. Shang, J. Liu, K. Wang, D. Wu, J. Shen, Y. Liu, T. Zhang, C. Liang, Y. Wang, Y. Shang, J. Guo, G. Liang, S. Xu, J. Liu, K. Wang, M. Wang, Landscape of somatic alterations in large-scale solid tumors from an Asian population. Nat. Commun. 13, 4264 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.F. P. Barthel, K. C. Johnson, F. S. Varn, A. D. Moskalik, G. Tanner, E. Kocakavuk, K. J. Anderson, O. Abiola, K. Aldape, K. D. Alfaro, D. Alpar, S. B. Amin, D. M. Ashley, P. Bandopadhayay, J. S. Barnholtz-Sloan, R. Beroukhim, C. Bock, P. K. Brastianos, D. J. Brat, A. R. Brodbelt, A. F. Bruns, K. R. Bulsara, A. Chakrabarty, A. Chakravarti, J. H. Chuang, E. B. Claus, E. J. Cochran, J. Connelly, J. F. Costello, G. Finocchiaro, M. N. Fletcher, P. J. French, H. K. Gan, M. R. Gilbert, P. V. Gould, M. R. Grimmer, A. Iavarone, A. Ismail, M. D. Jenkinson, M. Khasraw, H. Kim, M. C. M. Kouwenhoven, P. S. La Violette, M. Li, P. Lichter, K. L. Ligon, A. K. Lowman, T. M. Malta, T. Mazor, K. L. McDonald, A. M. Molinaro, D.-H. Nam, N. Nayyar, H. K. Ng, C. Y. Ngan, S. P. Niclou, J. M. Niers, H. Noushmehr, J. Noorbakhsh, D. R. Ormond, C.-K. Park, L. M. Poisson, R. Rabadan, B. Radlwimmer, G. Rao, G. Reifenberger, J. K. Sa, M. Schuster, B. L. Shaw, S. C. Short, P. A. Sillevis Smitt, A. E. Sloan, M. Smits, H. Suzuki, G. Tabatabai, E. G. Van Meir, C. Watts, M. Weller, P. Wesseling, B. A. Westerman, G. Widhalm, A. Woehrer, W. K. A. Yung, G. Zadeh, J. T. Huse, J. F. De Groot, L. F. Stead, R. G. W. Verhaak, The GLASS Consortium , Longitudinal molecular trajectories of diffuse glioma in adults. Nature 576, 112–120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.A. Jusakul, I. Cutcutache, C. H. Yong, J. Q. Lim, M. N. Huang, N. Padmanabhan, V. Nellore, S. Kongpetch, A. W. T. Ng, L. M. Ng, S. P. Choo, S. S. Myint, R. Thanan, S. Nagarajan, W. K. Lim, C. C. Y. Ng, A. Boot, M. Liu, C. K. Ong, V. Rajasegaran, S. Lie, A. S. T. Lim, T. H. Lim, J. Tan, J. L. Loh, J. R. McPherson, N. Khuntikeo, V. Bhudhisawasdi, P. Yongvanit, S. Wongkham, Y. Totoki, H. Nakamura, Y. Arai, S. Yamasaki, P. K.-H. Chow, A. Y. F. Chung, L. L. P. J. Ooi, K. H. Lim, S. Dima, D. G. Duda, I. Popescu, P. Broet, S.-Y. Hsieh, M.-C. Yu, A. Scarpa, J. Lai, D.-X. Luo, A. L. Carvalho, A. L. Vettore, H. Rhee, Y. N. Park, L. B. Alexandrov, R. Gordân, S. G. Rozen, T. Shibata, C. Pairojkul, B. T. Teh, P. Tan, Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 7, 1116–1135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.A. S. Ho, A. Ochoa, G. Jayakumaran, A. Zehir, C. V. Mayor, J. Tepe, V. Makarov, M. G. Dalin, J. He, M. Bailey, M. Montesion, J. S. Ross, V. A. Miller, L. Chan, I. Ganly, S. Dogan, N. Katabi, P. Tsipouras, P. Ha, N. Agrawal, D. B. Solit, P. A. Futreal, A. K. El Naggar, J. S. Reis-Filho, B. Weigelt, A. L. Ho, N. Schultz, T. A. Chan, L. G. Morris, Genetic hallmarks of recurrent/metastatic adenoid cystic carcinoma. J. Clin. Invest. 129, 4276–4289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.K. Schulze, S. Imbeaud, E. Letouzé, L. B. Alexandrov, J. Calderaro, S. Rebouissou, G. Couchy, C. Meiller, J. Shinde, F. Soysouvanh, A.-L. Calatayud, R. Pinyol, L. Pelletier, C. Balabaud, A. Laurent, J.-F. Blanc, V. Mazzaferro, F. Calvo, A. Villanueva, J.-C. Nault, P. Bioulac-Sage, M. R. Stratton, J. M. Llovet, J. Zucman-Rossi, Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 47, 505–511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.P. H. Kim, E. K. Cha, J. P. Sfakianos, G. Iyer, E. C. Zabor, S. N. Scott, I. Ostrovnaya, R. Ramirez, A. Sun, R. Shah, A. M. Yee, V. E. Reuter, D. F. Bajorin, J. E. Rosenberg, N. Schultz, M. F. Berger, H. A. Al-Ahmadie, D. B. Solit, B. H. Bochner, Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur. Urol. 67, 198–201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.J. S. Damrauer, W. Beckabir, J. Klomp, M. Zhou, E. R. Plimack, M. D. Galsky, P. Grivas, N. M. Hahn, P. H. O’Donnell, G. Iyer, D. I. Quinn, B. G. Vincent, D. Z. Quale, S. E. Wobker, K. A. Hoadley, W. Y. Kim, M. I. Milowsky, Collaborative study from the Bladder Cancer Advocacy Network for the genomic analysis of metastatic urothelial cancer. Nat. Commun. 13, 6658 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Y. Sato, T. Yoshizato, Y. Shiraishi, S. Maekawa, Y. Okuno, T. Kamura, T. Shimamura, A. Sato-Otsubo, G. Nagae, H. Suzuki, Y. Nagata, K. Yoshida, A. Kon, Y. Suzuki, K. Chiba, H. Tanaka, A. Niida, A. Fujimoto, T. Tsunoda, T. Morikawa, D. Maeda, H. Kume, S. Sugano, M. Fukayama, H. Aburatani, M. Sanada, S. Miyano, Y. Homma, S. Ogawa, Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 45, 860–867 (2013). [DOI] [PubMed] [Google Scholar]
- 83.R. Xue, L. Chen, C. Zhang, M. Fujita, R. Li, S.-M. Yan, C. K. Ong, X. Liao, Q. Gao, S. Sasagawa, Y. Li, J. Wang, H. Guo, Q.-T. Huang, Q. Zhong, J. Tan, L. Qi, W. Gong, Z. Hong, M. Li, J. Zhao, T. Peng, Y. Lu, K. H. T. Lim, A. Boot, A. Ono, K. Chayama, Z. Zhang, S. G. Rozen, B. T. Teh, X. W. Wang, H. Nakagawa, M.-S. Zeng, F. Bai, N. Zhang, Genomic and transcriptomic profiling of combined hepatocellular and intrahepatic cholangiocarcinoma reveals distinct molecular subtypes. Cancer Cell 35, 932–947.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.C. Curtis, S. P. Shah, S.-F. Chin, G. Turashvili, O. M. Rueda, M. J. Dunning, D. Speed, A. G. Lynch, S. Samarajiwa, Y. Yuan, S. Gräf, G. Ha, G. Haffari, A. Bashashati, R. Russell, S. M. Kinney, METABRIC Group, A. Langerød, A. Green, E. Provenzano, G. Wishart, S. Pinder, P. Watson, F. Markowetz, L. Murphy, I. Ellis, A. Purushotham, A.-L. Børresen-Dale, J. D. Brenton, S. Tavaré, C. Caldas, S. Aparicio, The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.B. Pereira, S.-F. Chin, O. M. Rueda, H.-K. M. Vollan, E. Provenzano, H. A. Bardwell, M. Pugh, L. Jones, R. Russell, S.-J. Sammut, D. W. Y. Tsui, B. Liu, S.-J. Dawson, J. Abraham, H. Northen, J. F. Peden, A. Mukherjee, G. Turashvili, A. R. Green, S. M. Kinney, A. Oloumi, S. Shah, N. Rosenfeld, L. Murphy, D. R. Bentley, I. O. Ellis, A. Purushotham, S. E. Pinder, A.-L. Børresen-Dale, H. M. Earl, P. D. Pharoah, M. T. Ross, S. Aparicio, C. Caldas, The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Y. Y. Li, G. J. Hanna, A. C. Laga, R. I. Haddad, J. H. Lorch, P. S. Hammerman, Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin. Cancer Res. 21, 1447–1456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.M. Kakiuchi, T. Nishizawa, H. Ueda, K. Gotoh, A. Tanaka, A. Hayashi, S. Yamamoto, K. Tatsuno, H. Katoh, Y. Watanabe, T. Ichimura, T. Ushiku, S. Funahashi, K. Tateishi, I. Wada, N. Shimizu, S. Nomura, K. Koike, Y. Seto, M. Fukayama, H. Aburatani, S. Ishikawa, Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat. Genet. 46, 583–587 (2014). [DOI] [PubMed] [Google Scholar]
- 88.F. Petralia, N. Tignor, B. Reva, M. Koptyra, S. Chowdhury, D. Rykunov, A. Krek, W. Ma, Y. Zhu, J. Ji, A. Calinawan, J. R. Whiteaker, A. Colaprico, V. Stathias, T. Omelchenko, X. Song, P. Raman, Y. Guo, M. A. Brown, R. G. Ivey, J. Szpyt, S. G. Thakurta, M. A. Gritsenko, K. K. Weitz, G. Lopez, S. Kalayci, Z. H. Gümüş, S. Yoo, F. da Veiga Leprevost, H.-Y. Chang, K. Krug, L. Katsnelson, Y. Wang, J. J. Kennedy, U. J. Voytovich, L. Zhao, K. S. Gaonkar, B. M. Ennis, B. Zhang, V. Baubet, L. Tauhid, J. V. Lilly, J. L. Mason, B. Farrow, N. Young, S. Leary, J. Moon, V. A. Petyuk, J. Nazarian, N. D. Adappa, J. N. Palmer, R. M. Lober, S. Rivero-Hinojosa, L.-B. Wang, J. M. Wang, M. Broberg, R. K. Chu, R. J. Moore, M. E. Monroe, R. Zhao, R. D. Smith, J. Zhu, A. I. Robles, M. Mesri, E. Boja, T. Hiltke, H. Rodriguez, B. Zhang, E. E. Schadt, D. R. Mani, L. Ding, A. Iavarone, M. Wiznerowicz, S. Schürer, X. S. Chen, A. P. Heath, J. L. Rokita, A. I. Nesvizhskii, D. Fenyö, K. D. Rodland, T. Liu, S. P. Gygi, A. G. Paulovich, A. C. Resnick, P. B. Storm, B. R. Rood, P. Wang, Children’s Brain Tumor Network, Clinical Proteomic Tumor Analysis Consortium , Integrated proteogenomic characterization across major histological types of pediatric brain cancer. Cell 183, 1962–1985.e31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.M. Krauthammer, Y. Kong, B. H. Ha, P. Evans, A. Bacchiocchi, J. P. McCusker, E. Cheng, M. J. Davis, G. Goh, M. Choi, S. Ariyan, D. Narayan, K. Dutton-Regester, A. Capatana, E. C. Holman, M. Bosenberg, M. Sznol, H. M. Kluger, D. E. Brash, D. F. Stern, M. A. Materin, R. S. Lo, S. Mane, S. Ma, K. K. Kidd, N. K. Hayward, R. P. Lifton, J. Schlessinger, T. J. Boggon, R. Halaban, Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 44, 1006–1014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.S. Ren, G.-H. Wei, D. Liu, L. Wang, Y. Hou, S. Zhu, L. Peng, Q. Zhang, Y. Cheng, H. Su, X. Zhou, J. Zhang, F. Li, H. Zheng, Z. Zhao, C. Yin, Z. He, X. Gao, H. E. Zhau, C.-Y. Chu, J. B. Wu, C. Collins, S. V. Volik, R. Bell, J. Huang, K. Wu, D. Xu, D. Ye, Y. Yu, L. Zhu, M. Qiao, H.-M. Lee, Y. Yang, Y. Zhu, X. Shi, R. Chen, Y. Wang, W. Xu, Y. Cheng, C. Xu, X. Gao, T. Zhou, B. Yang, J. Hou, L. Liu, Z. Zhang, Y. Zhu, C. Qin, P. Shao, J. Pang, L. W. K. Chung, J. Xu, C.-L. Wu, W. Zhong, X. Xu, Y. Li, X. Zhang, J. Wang, H. Yang, J. Wang, H. Huang, Y. Sun, Whole-genome and transcriptome sequencing of prostate cancer identify new genetic alterations driving disease progression. Eur. Urol. 73, 322–339 (2018). [DOI] [PubMed] [Google Scholar]
- 91.E. Pleasance, E. Titmuss, L. Williamson, H. Kwan, L. Culibrk, E. Y. Zhao, K. Dixon, K. Fan, R. Bowlby, M. R. Jones, Y. Shen, J. K. Grewal, J. Ashkani, K. Wee, C. J. Grisdale, M. L. Thibodeau, Z. Bozoky, H. Pearson, E. Majounie, T. Vira, R. Shenwai, K. L. Mungall, E. Chuah, A. Davies, M. Warren, C. Reisle, M. Bonakdar, G. A. Taylor, V. Csizmok, S. K. Chan, Z. Zong, S. Bilobram, A. Muhammadzadeh, D. D’Souza, R. D. Corbett, D. M. Millan, M. Carreira, C. Choo, D. Bleile, S. Sadeghi, W. Zhang, T. Wong, D. Cheng, S. D. Brown, R. A. Holt, R. A. Moore, A. J. Mungall, Y. Zhao, J. Nelson, A. Fok, Y. Ma, M. K. C. Lee, J.-M. Lavoie, S. Mendis, J. M. Karasinska, B. Deol, A. Fisic, D. F. Schaeffer, S. Yip, K. Schrader, D. A. Regier, D. Weymann, S. Chia, K. Gelmon, A. Tinker, S. Sun, H. Lim, D. J. Renouf, J. Laskin, S. J. M. Jones, M. A. Marra, Pan-cancer analysis of advanced patient tumors reveals interactions between therapy and genomic landscapes. Nat. Cancer 1, 452–468 (2020). [DOI] [PubMed] [Google Scholar]
- 92.G. Guo, J. Chmielecki, C. Goparaju, A. Heguy, I. Dolgalev, M. Carbone, S. Seepo, M. Meyerson, H. I. Pass, Whole-exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res. 75, 264–269 (2015). [DOI] [PubMed] [Google Scholar]
- 93.D. M. Hyman, S. A. Piha-Paul, H. Won, J. Rodon, C. Saura, G. I. Shapiro, D. Juric, D. I. Quinn, V. Moreno, B. Doger, I. A. Mayer, V. Boni, E. Calvo, S. Loi, A. C. Lockhart, J. P. Erinjeri, M. Scaltriti, G. A. Ulaner, J. Patel, J. Tang, H. Beer, S. D. Selcuklu, A. J. Hanrahan, N. Bouvier, M. Melcer, R. Murali, A. M. Schram, L. M. Smyth, K. Jhaveri, B. T. Li, A. Drilon, J. J. Harding, G. Iyer, B. S. Taylor, M. F. Berger, R. E. Cutler Jr., F. Xu, A. Butturini, L. D. Eli, G. Mann, C. Farrell, A. S. Lalani, R. P. Bryce, C. L. Arteaga, F. Meric-Bernstam, J. Baselga, D. B. Solit, HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 554, 189–194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium , Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.D. Miao, C. A. Margolis, W. Gao, M. H. Voss, W. Li, D. J. Martini, C. Norton, D. Bossé, S. M. Wankowicz, D. Cullen, C. Horak, M. Wind-Rotolo, A. Tracy, M. Giannakis, F. S. Hodi, C. G. Drake, M. W. Ball, M. E. Allaf, A. Snyder, M. D. Hellmann, T. Ho, R. J. Motzer, S. Signoretti, W. G. Kaelin Jr., T. K. Choueiri, E. M. Van Allen, Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359, 801–806 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.P. Johansson, L. G. Aoude, K. Wadt, W. J. Glasson, S. K. Warrier, A. W. Hewitt, J. F. Kiilgaard, S. Heegaard, T. Isaacs, M. Franchina, C. Ingvar, T. Vermeulen, K. J. Whitehead, C. W. Schmidt, J. M. Palmer, J. Symmons, A.-M. Gerdes, G. Jönsson, N. K. Hayward, Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget 7, 4624–4631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.S. Vasaikar, C. Huang, X. Wang, V. A. Petyuk, S. R. Savage, B. Wen, Y. Dou, Y. Zhang, Z. Shi, O. A. Arshad, M. A. Gritsenko, L. J. Zimmerman, J. E. McDermott, T. R. Clauss, R. J. Moore, R. Zhao, M. E. Monroe, Y.-T. Wang, M. C. Chambers, R. J. C. Slebos, K. S. Lau, Q. Mo, L. Ding, M. Ellis, M. Thiagarajan, C. R. Kinsinger, H. Rodriguez, R. D. Smith, K. D. Rodland, D. C. Liebler, T. Liu, B. Zhang, Clinical Proteomic Tumor Analysis Consortium , Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell 177, 1035–1049.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.M. Jamal-Hanjani, G. A. Wilson, N. M. Granahan, N. J. Birkbak, T. B. K. Watkins, S. Veeriah, S. Shafi, D. H. Johnson, R. Mitter, R. Rosenthal, M. Salm, S. Horswell, M. Escudero, N. Matthews, A. Rowan, T. Chambers, D. A. Moore, S. Turajlic, H. Xu, S.-M. Lee, M. D. Forster, T. Ahmad, C. T. Hiley, C. Abbosh, M. Falzon, E. Borg, T. Marafioti, D. Lawrence, M. Hayward, S. Kolvekar, N. Panagiotopoulos, S. M. Janes, R. Thakrar, A. Ahmed, F. Blackhall, Y. Summers, R. Shah, L. Joseph, A. M. Quinn, P. A. Crosbie, B. Naidu, G. Middleton, G. Langman, S. Trotter, M. Nicolson, H. Remmen, K. Kerr, M. Chetty, L. Gomersall, D. A. Fennell, A. Nakas, S. Rathinam, G. Anand, S. Khan, P. Russell, V. Ezhil, B. Ismail, M. Irvin-Sellers, V. Prakash, J. F. Lester, M. Kornaszewska, R. Attanoos, H. Adams, H. Davies, S. Dentro, P. Taniere, B. O’Sullivan, H. L. Lowe, J. A. Hartley, N. Iles, H. Bell, Y. Ngai, J. A. Shaw, J. Herrero, Z. Szallasi, R. F. Schwarz, A. Stewart, S. A. Quezada, J. L. Quesne, P. Van Loo, C. Dive, A. Hackshaw, C. Swanton, TRACERx Consortium , Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S15
Tables S1 to S5
References