Abstract
TAR DNA-binding protein-43 (TDP-43) proteinopathies are accompanied by the pathological hallmark of cytoplasmic inclusions in the neurodegenerative diseases, including frontal temporal lobar degeneration-TDP and amyotrophic lateral sclerosis. We found that transthyretin accumulates with TDP-43 cytoplasmic inclusions in frontal temporal lobar degeneration-TDP human patients and transgenic mice, in which transthyretin exhibits dramatic expression decline in elderly mice. The upregulation of transthyretin expression was demonstrated to facilitate the clearance of cytoplasmic TDP-43 inclusions through autophagy, in which transthyretin induces autophagy upregulation via ATF4. Of interest, transthyretin upregulated ATF4 expression and promoted ATF4 nuclear import, presenting physical interaction. Neuronal expression of transthyretin in frontal temporal lobar degeneration-TDP mice restored autophagy function and facilitated early soluble TDP-43 aggregates for autophagosome targeting, ameliorating neuropathology and behavioural deficits. Thus, transthyretin conducted two-way regulations by either inducing autophagy activation or escorting TDP-43 aggregates targeted autophagosomes, suggesting that transthyretin is a potential modulator therapy for neurological disorders caused by TDP-43 proteinopathy.
Keywords: TTR, TDP-43, proteinopathy, ATF4, FTLD
Chu et al. report that transthyretin (TTR) co-aggregates with TDP-43 in the brains of FTLD-TDP patients and transgenic mice. TTR potentiates autophagy activity via ATF4 and facilitates the targeting of soluble TDP-43 aggregates into autophagosomes for degradation.
Introduction
The pathological inclusions of trans-activation response (TAR) DNA-binding protein 43 (TDP-43) are neuro-pathosignatures that form cytoplasmic/nuclear accumulation, accounting for 45% in frontal temporal lobar degeneration (FTLD) and 97% in amyotrophic lateral sclerosis (ALS).1 Patients with FTLD present with neuronal loss and gliosis in the frontal and temporal lobes of the brain that cause progressive dysfunction in cognition, behaviour, language and motor function.2,3 ALS is characterized by severe loss of motor neurons leading to gradual paralysis and respiratory failure within 2–5 years.4,5 Physiologically, TDP-43 shuttles between the nuclear and cytoplasmic compartments, which are predominantly located in the nucleus. It binds to both DNA and RNA and has multiple functions to regulate RNA splicing, processing, trafficking and translation, which involves various biological processes, including apoptosis, cell division and axonal transport.1,6,7 Histopathologically, TDP-43 has been found to accumulate in the nucleus and cytoplasm and form ubiquitination/hyperphosphorylation-enriched inclusions leading to neurotoxicity, which is accompanied by TDP-43 depletion in the nucleus and sequestered in the cytoplasmic inclusions, resulting in the abolition of TDP-43 function, known as TDP-43 proteinopathy.8,9 In addition, the proteolytic cleavages of TDP-43 by caspases typically generate truncated carboxyl-terminal fragments of 25 and 35 kDa, which enhances TDP-43 aggregation and deteriorates neuronal dysfunction.10–12 Finding a way to prevent or reduce TDP-43 aggregation caused neurotoxicity may protect neurons from damage.
The unfolded protein response (UPR) activation has been shown to respond to protein aggregation caused by endoplasmic reticulum (ER) stress in several neurodegenerative disorders, including FTLD and ALS.13 Among the three signal branches in UPR, activation of protein-kinase-like ER kinase (PERK) leads to repression of general translation via eukaryotic initiation factor 2α (eIF2α) phosphorylation; reportedly, the overloaded TDP-43 burden accordingly triggers eIF2α activation.14,15 Attenuation of the general protein translation allows neuronal cells to deal with the excessive TDP-43 protein load. However, prolonged UPR is harmful given to failing to restore cellular homeostasis. Activated PERK selectively stimulates ATF4 activation triggering CHOP and BIM downstream death signal cascade, which induces neuronal apoptosis, eventually leading to severe disease progression.16 Notably, autophagy is a lysosomal degradative process that is one of the major protein quality control systems, which also can be triggered by the UPR signalling.17 It acts to recycle obsolete cellular constituents and eliminate damaged organelles and aggregated proteins to maintain cellular homeostasis.18 Functionally, autophagy plays an indispensable role as a vital biosensor to disturbed homeostasis, which undergoes autophagic induction, autophagosome biogenesis, substrate recognition and eventually lysosomal degradation.19 Dysfunction of each step in the process of autophagy may cause an imbalance in homeostasis leading to pathogenesis, in which disturbance of proteostasis by autophagy impairment manifests as one of the key causes of neurodegeneration.
Transthyretin (TTR), a 55 kDa homotetrameric protein, is primarily synthesized by the liver and the choroid plexus, functioning as a transporter responsible for the trafficking of the thyroid hormone thyroxine (T4) and retinol by binding to retinol-binding protein in serum and CSF.20 The mutant forms of TTR are prone to amyloid TTR aggregates, which deposit in the peripheral and autonomic nerves and heart causing distinct amyloidogenic diseases, such as familial amyloidotic polyneuropathy and familial amyloidotic cardiomyopathy.21 TTR has been found a significant reduction in CSF of Alzheimer’s disease.22 Reportedly, native TTR has a neuroprotective role in Alzheimer’s disease on several aspects as follows: (i) TTR is involved in Aβ peptide binding to prevent amyloid fibril formation and Aβ deposition; (ii) TTR has neprilysin-like proteolytic activity and is able to cleave Aβ and (iii) overexpression of human TTR in the Alzheimer’s disease mouse model is capable of normalizing its cognitive function.23–25 Despite the elevation of TTR levels have been reported in plasma and CSF of several neurodegenerative diseases, including FTLD and ALS.25 Given that TTR involves the clearance of Aβ aggregates to reach neuroprotection, the underlying mechanism of how TTR manages to shelter the CNS remains unclear. It is proposed that TTR may have a function involving cellular proteostasis in neuroprotection. Furthermore, perturbation of TDP-43 expression is sufficient to elicit pathogenesis. Therefore, TDP-43 proteinopathy is warranted to elucidate the neuroprotective function of TTR in neurons.
Previously, we established an FTLD-TDP mouse model with transgenic neuron-specific overexpression of TDP-43 in the forebrain using the Ca2+/calmodulin-dependent kinase II (CamKII) promoter (TDP-43 Tg mice).26 The common TDP-43 pathology has been verified in an age-dependent manner in TDP-43 Tg mice, which includes ubiquitin-positive TDP-43 cytoplasmic inclusions, nuclear depletion and increased active caspase 3. Consequently, the accompanying phenotypic characteristics of learning/memory deficits, motor dysfunction, hippocampal atrophy and upregulated gliosis mimic FTLD-TDP clinico-neuropathological features. Therefore, neuronal overexpression of TDP-43 successfully imitated TDP-43 proteinopathy-induced FTLD in TDP-43 Tg mice, which serves as a fit model for therapeutic studies.
To address the neuroprotective function of TTR, TDP-43 Tg mice mimic FTLD-TDP and TDP-43 proteinopathy mimetic cells, both in vivo and in vitro models were used to examine the function of TTR in modulating proteostasis.26–29 Considering the proteostatistic perturbations triggering ER stress, we further validated the potential engagement of TTR in the two essential systems of the UPR and autophagy in maintaining proteostasis. We found that TTR potentiates a novel role in manipulating autophagic activation to achieve neuroprotection, which presents therapeutic feasibility for ER-deranged neurological diseases.
Materials and methods
Plasmid constructs
The TDP-43 expression plasmid was created as previously described.26 Mouse TTR cDNA cut from pGEM-T vector (Promega Corporation, Madison, WI, USA) was subcloned into the EcoRV site of pNN265 vector (provided by E. Kandel, Columbia University, New York, NY, USA), named 265-TTR. Next, 265-TTR cut from pNN265 vector was again subcloned into the NotI site of pMM403_CaMKII-promoter vector (provided by E. Kandel).30 And then CaMKII-promoter-265-TTR was cut using SfiI for microinjection to generate TTR transgenic mice. For ATF4 constructs, mouse ATF4 cDNA was amplified by reverse transcription-PCR and cloned into mammalian expression vector FUGW (Addgene, Inc., Watertown, MA, USA). Three different truncated forms of mouse ATF4 cDNA, named ATF41-270 (containing amino acids 1–270), ATF41-303 (containing amino acids 1–303) and ATF4126-349 (containing amino acids 126–349), were PCR-amplified, flanked with cloning sites BamHI and AgeI, and inserted into mammalian expression vector FUGW that contains a green fluorescent protein (GFP) sequence. The integrity of all cDNA and production of the fusion proteins were confirmed by DNA sequencing and western blot, respectively.
Animal models
The procedures for the mice handling in animal experiments were followed the guidelines of the Institutional Animal Care and Use Committee of National Cheng Kung University (NCKU). All the experiments were under review and approved by the Institutional Animal Care and Use Committee at NCKU, Taiwan. Mice were all bred at NCKU laboratory animal centre in a pathogen-free room, housed no more than 5 per cage under a 12-h light-dark cycle, provided with standard rodent chow and sterilized water ad libitum. The TDP-43 Tg mouse model carried full-length mouse TDP-43 cDNA under the transcription control of an 8.5-kb CaMKII promoter and overexpressed TDP-43 in the forebrain. Genotyping by PCR was used to identify whether mice were transgene-positive. At 6-month-old, cytoplasmic TDP-43 aggregation accompanied with nuclear TDP-43 depletion appears in the brain of TDP-43 Tg mice, which mimics the hallmark of TDP-43 proteinopathy in the brains of human FTLD-TDP patients.26 Furthermore, the establishment of TTR Tg mouse model carried with full-length mouse TTR cDNA under the transcription control of an 8.5-kb CaMKII promoter and overexpressed TTR in the forebrain. Genotyping by PCR was used to identify whether mice were transgene-positive. Moreover, the TDP-43/TTR Tg mouse model was generated by crossbreeding TDP-43 Tg mice with TTR Tg mice to overexpress TDP-43 and TTR in the forebrain. Genotyping by PCR was used to identify whether mice were double-transgene-positive. For protein extraction, mice were sacrificed by rapid cervical dislocation and the forebrain regions were isolated. For IF staining, mice were anaesthetized by Isoflurane prior to transcardial perfusion.
Urea-soluble fraction preparation
Assay for the insoluble proteins, tissues were dissected and extracted with serial buffers following with increased reducing agents present as previously described.10 Briefly, the forebrain tissues were sequentially extracted at 5 ml/g (volume/weight) with low-salt (LS) buffer (10 mM Tris, pH 7.5, 5 mM EDTA, 1 mM DTT, 10% sucrose containing protease inhibitors), high-salt Triton X-100 (TX) buffer (LS buffer + 1% Triton X-100 + 0.5 M NaCl), myelin flotation buffer (TX buffer containing 30% sucrose) and sarkosyl (SARK) buffer (LS buffer + 1% N-lauroyl-sarcosine + 0.5 M NaCl). Finally, the SARK-insoluble fractions were further extracted with 0.25 ml/g in urea buffer (7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulphonate, 30 mM Tris, pH 8.5). The urea-soluble proteins were ready for the analysis in western and dot blotting.
dSTORM super-resolution microscopy
The super-resolution microscopy was performed as described previously.7,31 Briefly, the automated inverted Olympus IX-83 microscope was constructed with a ×60, 1.49 numerical aperture, total internal reflection fluorescence, oil-immersion objective (APON; Olympus Corporation), a multiline laser source (405, 488, 561 and 640 nm; Andor Technology, Ltd.) and other appropriate equipment including single-molecular localization signals separating filters (Andor Technology), electron-multiplying charge-coupled device camera (iXon Ultra 897; Andor Technology). An oxygen-scavenging buffer system and fluorophore-conjugated secondary antibodies CFTM568-conjugated Fab fragment (Biotium) and Alexa Fluor® 647-conjugated Fab fragment (Jackson ImmunoResearch) labelled cellular samples was prepared for dSTORM microscopy. The dSTORM imaging buffers used were Buffer A (10 mM TRIS, pH 8.0, 50 mM NaCl), Buffer B (50 mM TRIS, pH 8.0, 10 mM NaCl and 10% glucose), GLOX solution [56 mg glucose oxidase, 200 μl of catalase (17 mg/ml), 800 μl of Buffer A] and MEA solution (77 mg MEA, 1 ml of 0.25 M HCl). Each chamber with 20-mm coverslip was added with 500 μl of imaging buffer, which mixed with 5 μl of GLOX solution, 50 μl of MEA solution and 445 μl of Buffer B on ice. Before dSTORM acquisition, conventional fluorescence microscopy was acquired to define the region of interest. In dSTORM imaging, 50 ms exposure time (20 frames/second) were applied to record 10 000 frames of the sequential activated photoswitchable fluorophores. The acquired image series were further analysed by MetaMorph® Super-Resolution System (Molecular Devices) to reconstruct dSTORM images. For TDP-43 puncta size analysis, 10 images for control cells and 12 images for TTR-expressed cells were applied to analysis by MetaMorph software, of which 321 puncta for control cells and 369 puncta for TTR-expressed cells were quantified. For distance analysis of TDP-43 and LC3 puncta, 53 images were applied to analysis by MetaMorph software, of which 33 cells for control and 20 cells for TTR overexpression were applied for distance analysis between TDP-43 and LC3 puncta. Overall, 142 TDP-43-LC3 puncta distances were calculated for controls and 140 TDP-43-LC3 puncta distances were calculated for TTR-expressed cells.
Fluorescence recovery after photobleaching assay
ATF4-GFP alone or ATF4-GFP/mCherry-TTR-overexpressed human embryonic kidney 293 (HEK293) cells were respectively cultured on glass-bottom dishes. Fluorescence recovery after photobleaching (FRAP) experiments were performed on a confocal laser scanning microscope equipped with a 50-mW, 405-nm laser for photostimulation, in which cells were maintained under 37°C and 5% CO2 conditions (FV1000, Olympus). The power of illumination was set at 100% (452 μW) to photobleach the fluorescent proteins located at the nuclear region for 6 ms. The fluorescence images of GFP- and/or mCherry-labelled proteins were recorded at 512 × 512 pixels and monitored for 30-min time periods. Recovery curves have been normalized with background subtraction of the photobleaching region and analysed using Olympus Fluoview software.
Morris water maze task
For spatial learning test, the Morris water maze task was performed as described previously.26,32 Mice were subjected to training four trials per session and one session a day. For a complete test, a total of six sessions in 6 days were performed. The time spent by individual mice swimming in the water to reach the hidden platform was recorded as the escape latency.
Rotarod test
For the motor function test, the rotarod performance assessment was performed followed by the description that follows.26 The mice were placed on a rotating rod at the speed of 20 rpm, and the integral time spent on the rod was measured. When a mouse stayed on the rod until the end of 2 min trial, this was recorded as 120 s.
Human samples
Post-mortem brain samples of normal individuals and patients with FTLD were kindly provided by Dr Lee-Way Jin by the Alzheimer’s Disease Center at University of California Davis, Sacramento, CA, USA. Informed consented autopsies to share research tissue after death were obtained from all patients with Institutional Review Board approval (Supplementary Table 1). The study was approved by the Institutional Review Board of NCKU Hospital (B-ER-103-180) on the basis of the ethical standards prescribed by the World Medical Association Declaration of Helsinki and the Department of Health and Human Services Belmont Report.
Statistical analysis
All data were statistically analysed and represented using GraphPad Prism (GraphPad Software, San Diego, CA, USA). All experiments were repeated at least three times. For immunoreactive cells (Supplementary material), the images pictured by confocal or widefield microscopy from each section were acquired with at least five random views of the cortex or hippocampus, of which 8–10 sections within the dorsal to ventral cortex or hippocampus were analysed per mouse. The TissueFAXS system has the capability to conduct whole slide imaging automatically, acquiring multiple samples analysis with identical setting parameters such as intensity. Therefore, for the immunoreactive cells analysed by the TissueFAXS system (TissueGnostics GmbH), eight sections from each mouse for every group [wild-type (WT), TDP-43 Tg and TDP-43/TTR Tg mice] were acquired using the TissueFAXS image system together for each analysis, and a total of five to six mice per group were analysed using TissueQuest software. Two group comparisons were analysed by a two-tailed Student’s t-test. For multiple groups, the difference comparisons of statistical significances in each independent experiment among groups were evaluated using one-way analysis of variance (ANOVA) followed by post hoc Tukey’s test. For the Morris water maze task, multiple comparisons were analysed by repeated measures ANOVA with the Bonferroni test. Differences are considered statistically significant at *P < 0.05, **P ≤ 0.01 and ***P ≤ 0.001 as indicated by the asterisks.
Data availability
The original contributions presented in the study are included in the article and Supplementary material. Further inquiries can be directed to the corresponding author on reasonable request.
Results
TTR is involved in TDP-43 proteinopathy in FTLD-TDP human patients
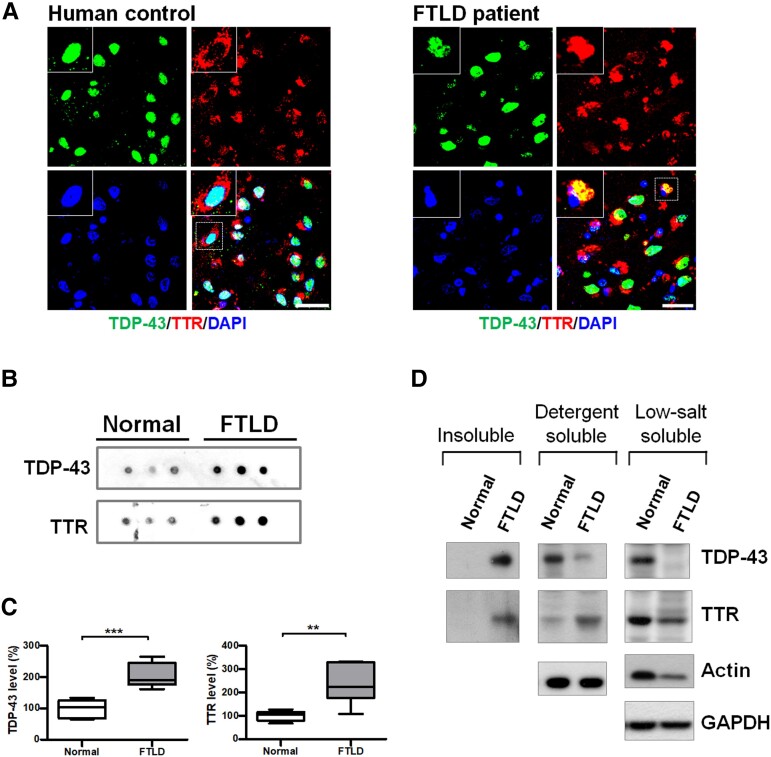
In view of TTR’s multifaceted functions, such as preventing Aβ aggregation/deposition and cleaving Aβ peptide,25 we determined whether TTR played a role in maintaining cellular proteostasis such as with pathological TDP-43 removal. To ascertain the involvement of TTR in FTLD-TDP, we first validated whether TTR was associated with TDP-43 proteinopathy. Post-mortem brain specimens were used to survey the distribution of TTR and TDP-43 using immunofluorescence (IF) staining. Data from human brain slides revealed that TDP-43 mainly localized in the nucleus and TTR predominantly distributed in the cytoplasm in healthy human control (Fig. 1A). However, TDP-43 shuttled from the nucleus to cytoplasmic accumulation as a characteristic of the disease phenotype and showed spatial colocalization with TTR in FTLD-TDP human patient sections (Fig. 1A). Furthermore, to understand the role of TTR in TDP-43 aggregation, we characterized the levels of TTR and TDP-43 in the insoluble fractions from normal and FTLD-TDP human frontal cortices using the dot-blot technique (Fig. 1B). The quantitative results from the dot-blot analysis showed that the insoluble TTR level was significantly increased (∼2-fold) in FTLD-TDP patients compared to normal control, along with an abundance of TDP-43 (∼2-fold increase) in the insoluble fraction of FTLD-TDP (Fig. 1C). Western blot analysis of sequential fractionation extracts revealed that TTR specifically co-precipitated with TDP-43 in the insoluble fraction of FTLD-TDP but not in the normal control (Fig. 1D). Subsequently, membrane-bound organelles/proteins extracted from detergent-soluble fractions showed a heightened level of TTR in FTLD-TDP relative to the normal control, and comparatively, high-level soluble TDP-43 was obtained only from the normal human brain extracts (Fig. 1D). LS fractions of normal and FTLD human samples revealed that TDP-43 and TTR exhibited greater levels in the normal control than in FTLD-TDP (Fig. 1D). Together, analyses of the human brain samples suggest that TTR is potentially involved in the pathogenesis of FTLD-TDP and possibly has a function related to the formation or clearance of TDP-43 cytoplasmic inclusions.
Figure 1.
TTR atypically mislocalizes to TDP-43 inclusions in human patients with FTLD-TDP. (A) The specimens from the frontal cortices were immunostained with TDP-43 (green, upper left) and TTR (red, upper right) for normal and FTLD-TDP human patients. Nuclei were counterstained with DAPI (blue, bottom left). The magnified view shown in the upper left is indicated with a white square line. Scale bar = 30 μm. (B) Dot blotting of TDP-43 and TTR in the insoluble fractions from human brains of normal and FTLD-TDP individuals. (C) Quantification of insoluble TDP-43 and TTR in the insoluble fractions from human brains of normal and FTLD-TDP individuals. Data are shown as box-and-whisker plots (min to max). The central horizontal line within the box indicates the median value. (TDP-43 level: ***P = 0.0004; TTR level: **P = 0.0036, t-test) n = 6 human brain tissue extracts. (D) Comparative analyses of brain extracts were fractionated from insoluble (urea-soluble), detergent-soluble and LS soluble fractions of normal and FTLD humans by western blotting.
TTR co-occurrence in TDP-43 inclusions is correlated to the pathogenesis of TDP-43 proteinopathy
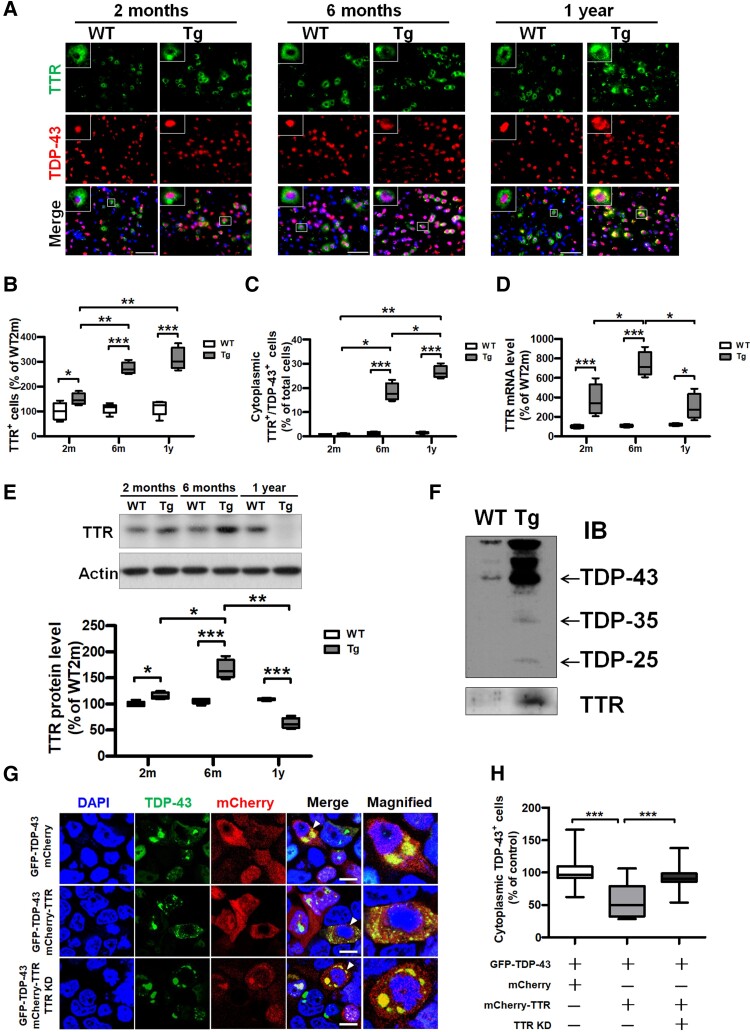
To better understand the correlation between TDP-43 proteinopathy and TTR, we further measured the cytoplasmic co-occurrence between TTR and pathogenic TDP-43 aggregates changing with age. Double IF staining for TDP-43 and TTR was conducted to detect their cellular distribution in WT and TDP-43 Tg mice at 2, 6 and 12 months old, where the cytoplasmic localization of TDP-43 increased with age and displayed evident co-occurrence with TTR (Fig. 2A). Quantification of fluorescence images, TTR-positive cells displayed a significant increase in TDP-43 Tg mice than in WT mice at all 2, 6 and 12-month-old; another aspect, TTR and TDP-43 double-positive cells also had a substantial increase in TDP-43 Tg mice than in WT mice at 6- and 12-month-old (Fig. 2B and C). In addition, TTR-positive cells, as well as cells with cytoplasmic TDP-43 inclusions,26 showed increased following along with age in TDP-43 Tg mice, indicating disease progression (Fig. 2B and C). Of which cytoplasmic co-occurrence of TTR and TDP-43 double-positive cell number, as well as TTR-positive cells, presented a dramatic increase beginning at the age of 6 months in TDP-43 Tg mice (Fig. 2B and C). Furthermore, expression of TTR mRNA significantly increased in TDP-43 Tg mice relative to WT mice at all ages of 2, 6 and 12 months (Fig. 2D). On the other hand, expression of TTR protein exhibited significant elevation before 6 months in TDP-43 Tg mice compared to WT mice but showed a decrease at 1 year (Fig. 2E). In TDP-43 Tg mice, TTR mRNA and protein levels elevated with age before 6 months, whereas TTR levels in both mRNA and protein were dramatically decreased at 1 year (Fig. 2D and E). These data indicate that the expression of TTR is highly associated with TDP-43 pathogenesis and reveal that TTR potentially affects the cytoplasmic aggregation of TDP-43 after the age of 6 months later in TDP-43 Tg mice. Analysis of the brain insoluble fraction revealed an apparent increase in insoluble TDP-43 accumulation in TDP-43 Tg mice compared with WT mice (Fig. 2F). Moreover, TTR was clearly detected in the insoluble fraction from TDP-43 Tg mice along with a heightened level of insoluble TDP-43 (Fig. 2F). Together, these data indicate that TTR potentially co-aggregates with TDP-43 and has a regulatory role in TDP-43 pathogenesis.
Figure 2.
Dysregulated TTR mislocalizes in TDP-43 inclusions in vivo and in vitro, wherein functional TTR expression facilitates the removal of cytoplasmic TDP-43 aggregates in vitro. (A) IF staining of TDP-43 (red) and TTR (green) in the brain tissue specimens from WT mice as well as TDP-43 Tg mice with age. Nuclei were counterstained with DAPI (blue). The enlarged images shown in the upper left are indicated with a white square line representing the co-occurrence of TDP-43 and TTR. Scale bar = 100 μm. (B) The number of TTR-immunoreactive cells and (C) TDP-43- and TTR double immunoreactive cells was quantified from the brain tissue specimens of WT mice and TDP-43 Tg mice at 2 and 6 months and 1 year old. (TTR+ cells: P < 0.0001, TDP-43+/TTR+ cells: P < 0.0001; repeated measures ANOVA with Bonferroni post hoc test) n = 8 sections per mouse; n = 5 mice per group. (D) Total RNA extracted from the forebrain tissues of WT mice and TDP-43 Tg mice at the indicated age were applied to calculate the mRNA level of TTR (P < 0.0001; repeated measures ANOVA with Bonferroni post hoc test). (E) The brain tissue lysates from WT and TDP-43 Tg mice extracted from detergent-soluble fractions at the indicated age were used to analyse the protein levels of TTR (P < 0.0001; repeated measures ANOVA with Bonferroni post hoc test). (F) Western blot analysis of TDP-43 and TTR in insoluble fractions from WT and TDP-43 Tg mice at 6 months. (G) HEK293 cells were overexpressed with TDP-43-GFP and administered with MG132, respectively combined with mCherry only, mCherry-TTR or mCherry-TTR overexpression along with TTR knockdown (TTR KD), which represent with confocal images. The magnified view is shown in the rightmost indicated by a white arrow. Nuclei were counterstained with DAPI (blue). Scale bar = 12 μm. (H) The numbers of cells with cytoplasmic TDP-43 inclusions were quantified from each view of microscopic images (P < 0.0001; one-way ANOVA followed by post hoc Tukey test). Data are shown as box-and-whisker plots (min to max). The central horizontal line within the box indicates the median value. Statistically significant differences are denoted by asterisks (*P < 0.05, **P ≤ 0.01 and ***P ≤ 0.001).
After confirming the involvement of TTR in TDP-43 pathogenesis, we further examined its effect on TDP-43 to promote or prevent cytoplasmic aggregation. On the basis of previous reports, MG132 treatment triggers the nuclear depletion of TDP-43 and forms large cytoplasmic inclusions closely mimicking TDP-43 pathology in the disease.28,29,33 An in vitro cellular model was established to investigate TDP-43 proteinopathy, in which GFP-TDP-43 plasmid and MG-132 were introduced into HEK293 cells to mimic TDP-43 proteinopathy. Furthermore, TDP-43 proteinopathy mimetic cells, combined with the expression of mCherry control or mCherry-TTR and with specific TTR mRNA knockdown, were used to investigate TTR influence on TDP-43 cytoplasmic inclusions (Fig. 2G). Confocal images revealed that TDP-43 aggregates were apparently mislocalized to the cytoplasm in control GFP-TDP-43/mCherry-overexpressed cells (Fig. 2G and H). Interestingly, the cytoplasmic accumulation of TDP-43 inclusions was significantly reduced following mCherry-TTR overexpression compared to that in the mCherry control, in which the number of cells with TDP-43 puncta was virtually diminished, and the size of puncta was visually decreased in TTR-overexpressed cells (∼0.5-fold decrease) (Fig. 2G and H). However, TDP-43 aggregates eliminated by TTR overexpression were reverted by a specific knockdown of TTR mRNA (∼1.6-fold increase) (Fig. 2G and H). Taken together, these results indicate that overexpression of TTR contributes to the clearance of TDP-43 aggregates represented as cellular protection in TDP-43 proteinopathy.
TTR induces activation of autophagy independent of the UPR pathway
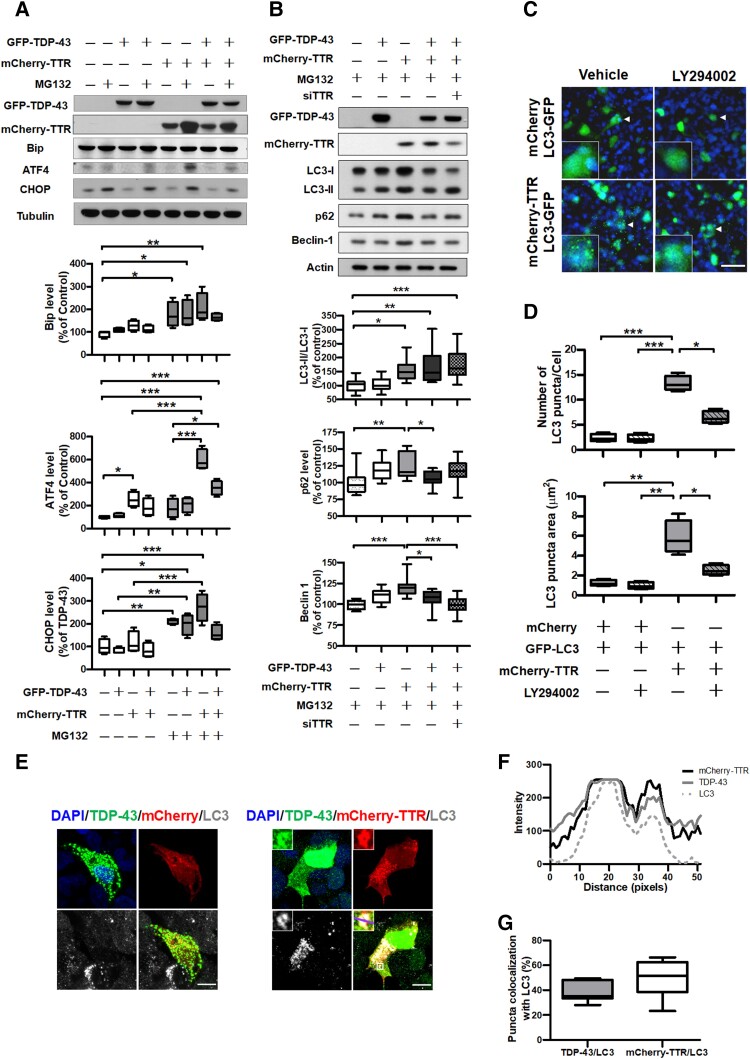
Protein aggregation disturbs ER homeostasis, further triggering UPR signalling. Overexpression of TDP-43 has been reported to activate eIF2α to reduce the burden of overload proteins and, additionally, eIF2α drives ATF4 activation. A cascade of the UPR downstream target genes, which control protein folding, autophagy and CHOP-mediated apoptosis in response to a failure of ER homeostasis recovery.14,34,35 To elucidate the effect of TTR on the cytoplasmic TDP-43 aggregates, we initially evaluated the UPR cascade for TDP-43 proteinopathy affected by overexpression of TTR through validation of the UPR onset indicator, Bip and its downstream regulators, ATF4 and CHOP, by immunoblotting (Fig. 3A). Quantitative analysis of the immunoblot data revealed that the three UPR-associated factors, Bip, ATF4 and CHOP, were strongly upregulated with MG132 treatment (Fig. 3A). Nevertheless, the increased level reactive to TTR overexpression was only observed in ATF4 (∼2.6-fold), and the expression levels of Bip and CHOP remained unaffected (Fig. 3A). These results indicate that the UPR signalling is upregulated on TDP-43 proteinopathy. In contrast, TTR expression shows no additive effect on TDP-43 proteinopathy-stimulated UPR activation due to Bip and CHOP being absent from significant expression changes. Interestingly, TTR mediated the elevation of ATF4 levels in the presence or absence of MG132 treatment (Fig. 3A). ATF4 plays a crucial role in autophagy activation that is regulated dependent or independent of the PERK pathway.36 Therefore, we further investigated the effect of TTR on autophagy, in which the autophagy markers, LC3, p62/SQSTM1 and Beclin-1, involved in autophagy activity, were applied to evaluate the regulation of autophagy by TTR using immunoblotting (Fig. 3B). Quantification of the immunoblotting results revealed an increased conversion of LC3-I to LC3-II and upregulated Beclin-1 and p62 on TTR overexpression (LC3-II/I ∼1.5-fold, Beclin-1 ∼1.2-fold, p62 ∼1.3-fold increases) (Fig. 3B). Notably, co-expression of TDP-43 and TTR showed promotion of autophagic flux with substantial decreases in p62 and Beclin-1, reflecting TDP-43 degradation (∼0.5-fold decrease) (Fig. 3B). Moreover, TTR expression-contingent knockdown of TTR mRNA displayed increases of LC-II/LC3-I and p62 levels (LC3-II/I ∼1.3-fold; p62 ∼1.2-fold), accompanying a ∼1.5-fold increase in TDP-43 compared to TTR overexpression (Fig. 3B). These results underline that TTR facilitates the induction of autophagy progression independent of the UPR signalling.
Figure 3.
TTR induces autophagy activation that enhances autophagosome formation and spatial colocalization and promotes ATF4 upregulation to be independent of the UPR signalling. The cell lysates of TDP-43 proteinopathy mimicked in HEK293 cells with or without TTR expression were subjected to determine intracellular pathway activation. Western blot analyses of (A) the key factors related to the UPR signalling and (B) autophagy-associated markers responded to TTR. Densitometric quantification of each factor was normalized by Tubulin and Actin, respectively (UPR: Bip P = 0.0025, ATF4 P < 0.0001, CHOP P < 0.0001; Autophagy: LC3-II/I P < 0.0001, p62 P = 0.0028, Beclin-1 P < 0.0001). (C) Confocal images of LC3-GFP transfected HEK293 cells were co-expressed with mCherry or mCherry-TTR applied to evaluate LC3-GFP puncta formation with or without 10 μM LY294002 administration. The magnified view is shown in the lower left indicated with a white arrow. Nuclei were counterstained with DAPI (blue). Scale bar = 50 μm. (D) Quantification of LC3-GFP puncta number per cell and puncta area were used to assess the change of LC3-GFP puncta affected by mCherry-TTR expression. Administration of 10 μM LY294002 inhibited LC3-GFP puncta formation (Puncta number P < 0.0001, puncta area P < 0.0001). (E) Representative IF images of LC3 staining in TDP-43 proteinopathy mimetic cells with respective mCherry control and mCherry- =TTR expression. Nuclei were counterstained with DAPI (blue). Scale bar = 10 μm. The magnified view shown in the upper left is indicated with a white square line, and a purple transversal line shown on the magnified merge image was selected to present RGB plot in F. (F) The pixel intensities (y-axis) of mCherry-TTR (black line), TDP-43 (grey line) and LC3 (grey dashed line) display the distribution of fluorescence across the selected purple line in E, in which the consistent intensity change profiles of mCherry-TTR, TDP-43 and LC3 indicate their colocalization feature. (G) The percentages of TDP-43 and TTR puncta that, respectively, colocalize with LC3 were quantified. Data are shown as box-and-whisker plots (min to max). The central horizontal line within the box indicates the median value. Statistical analysis with one-way ANOVA followed by a post hoc Tukey test. Statistically significant differences are denoted by asterisks (*P < 0.05, **P ≤ 0.01 and ***P ≤ 0.001).
To verify the effect of TTR overexpression on the induction of autophagy, we investigated autophagosome formation under TTR overexpression in HEK293 cells. Using confocal microscopy, we observed that TTR overexpression facilitated an increase in the number of LC3-positive puncta, indicating the upregulation of autophagosome formation compared to the control (Fig. 3C). However, this effect was blocked when cells were treated with the PI3K inhibitor, LY294002, to inhibit the autophagic process (Fig. 3C). Quantification of the number and size of LC3 puncta in each cell showed that LC3 puncta number (∼5.5-fold increase) and area (∼4.7-fold increase) had robust increases in TTR expression, while administration of LY294002 substantially inhibited this effect (Fig. 3D). These data indicate that the overexpression of TTR facilitates autophagy activation and progression.
Furthermore, the triple IF staining technique was applied to inspect the spatial correlation among TTR, TDP-43 and LC3 in TDP-43 proteinopathy mimetic cellular model. On the basis of confocal imaging, a comparison between cells with or without TTR overexpression under prolonged MG132 treatment showed that cells without TTR expression followed by depletion of nuclear TDP-43 pool and a lesser extent of LC3-positive puncta formation (Fig. 3E left). In contrast, cells with TTR expression were capable of preserving a majority of TDP-43 in the nucleus and upregulating autophagosome formation, showing triple puncta colocalization among LC3, TTR and TDP-43 (Fig. 3E right and Fig. 3F). Further quantification of TDP-43 and TTR’s respective intracellular colocalization with LC3 showed that TDP-43 had ∼39.6% colocalization with LC3, and TTR revealed 49.5% colocalization with LC3 (Fig. 3G). These data indicate that TTR overexpression involves the upregulation of autophagosome formation and potentially facilitates the elimination of TDP-43 aggregates through autophagosome targeting.
TTR promotes TDP-43 aggregates targeted to autophagosomes for degradation
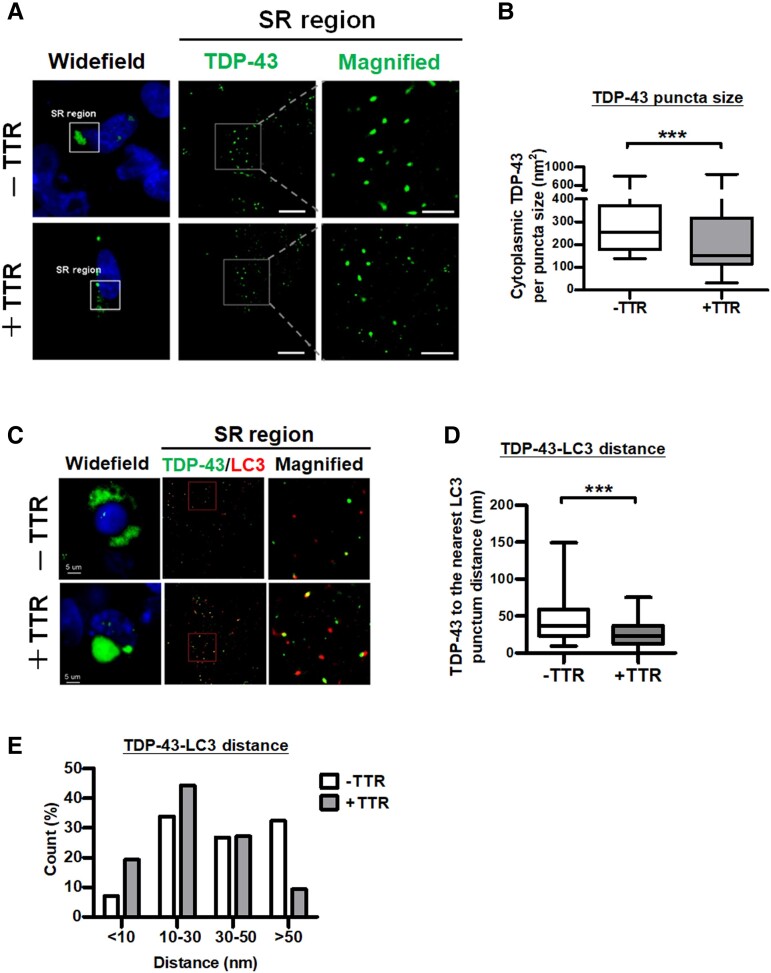
As previously reported, TDP-43 aggregates have an identified size distribution of 40–400 nm and, typically, the diameter of an autophagosome is 500–1500 nm in mammals.37,38 Supposedly, TTR targets smaller TDP-43 aggregates for autophagic degradation. Considering that the resolution limit of confocal optical microscopy defined by Abbe’s diffraction limit is ∼200 nm, super-resolution imaging (∼20 nm resolution) enables better detection of endogenous molecules inside subcellular structures such as autophagosomes and synapses.7,31,39,40 To characterize the potential role of TTR in the elimination of TDP-43 aggregates, we further performed an advanced technique of super-resolution microscopy to clarify the variation of TDP-43 puncta and its relationship with TTR and LC3. Super-resolution imaging of cytoplasmic TDP-43 accumulation demonstrated that cellular TDP-43 proteinopathy in conjunction with TTR expression reduced the size of TDP-43 puncta compared to the control (Fig. 4A). Quantification of TDP-43 puncta size revealed that cellular TDP-43 proteinopathy lacking TTR expression significantly accumulated a larger TDP-43 puncta size (average 288 nm2) than TTR conjunct expressed cells (average TDP-43 puncta size 214 nm2) (Fig. 4B). These results demonstrate that overexpression of TTR facilitates the elimination of cytoplasmic TDP-43 aggregates resulting in a reduced TDP-43 puncta size under TDP-43 proteinopathy.
Figure 4.
TTR facilitates TDP-43 aggregates targeting autophagosomes for degradation. TDP-43 proteinopathy mimicked in HEK293 cells was co-expressed with mCherry control or mCherry-TTR for super-resolution dSTORM microscopy. (A) Left: Widefield fluorescence images selected higher-order TDP-43 assembly (green) region in the cytoplasm for further (middle) super-resolution imaging is denoted by a white square line. Right: A white square line with two extended dash lines indicates the magnification of assembled TDP-43 puncta from a super-resolution image. Nuclei were counterstained with DAPI (blue). Scale bar = 20 μm. (B) Quantifications of TDP-43 puncta size with or without TTR expression were resolved from dSTORM images (***P < 0.0001, t-test). (C) Left: The region acquired for super-resolution microscopy was selected from the widefield fluorescence image. Middle: Super-resolution imaging of the condition cells was immunostained with TDP-43 and LC3. Right: The magnified view displaying TDP-43 and LC3 distribution is indicated by the red square line. Scale bar = 5 μm. Quantifications of adjacent TDP-43 and LC3 puncta distributions resolved from dSTORM images are shown as (D) the mean distances (P < 0.0001, t-test) and (E) the distribution proportion of the distances. The distance proportions were calculated according to the number of TDP-43-LC3 puncta distances distributed in the indicated ranges of the grand total counts shown as a bar chart in E. The rest of the data are shown as box-and-whisker plots (min to max). The central horizontal line within the box indicates the median value. Statistically significant differences are denoted by asterisks (***P ≤ 0.001).
Furthermore, to understand the relationship between TTR-induced LC3 and TDP-43 aggregates, super-resolution microscopy was performed to measure the distance between the cytoplasmic TDP-43 punctum and its nearest LC3 punctum under cellular TDP-43 proteinopathy with or without TTR overexpression. By comparison, super-resolution images from the conjunct mCherry-TTR-overexpressed cells presented the most TDP-43 puncta colocalized with LC3 compared to mCherry control (Fig. 4C). Measurement of the minimal distance between TDP-43 and LC3 punctum resulted in a mean distance of 41.3 nm for cells without TTR and 25.9 nm for cells with TTR expression, in which TDP-43 to LC3 distance showed significant difference with or without conjunct TTR expression (Fig. 4D). Proportional discrimination of the measured distances between TDP-43 and LC3 punctum revealed that 32.4% were located at a wide-range distance larger than 50 nm (Fig. 4E). Among them, 6.3% with a 90–150 nm broader distance in cells without TTR conjunct expression. In contrast, cells with TTR conjunct expression had only 9.3% with a distance range larger than 50 nm, showing no punctum distance situated at 90–150 nm (Fig. 4E). Based on proportional counts, the short-range punctum distance between TDP-43 and LC3 was mainly identified in cells with TTR co-expression, which held 19.3% less than 10 nm and overall 63.6% beneath the 30 nm range (Fig. 4E). Regarding cells without TTR conjunct expression, the short-range distance <10 nm was only 7.0% and, overall, 40.8% had a distance beneath the 30 nm range (Fig. 4E). In general, the punctum distance between TDP-43 and LC3 was 40.8% less than 30 nm and 59.2% larger than 30 nm, showing nearly hypodispersion in mCherry control, indicating a random distribution of LC3 near the TDP-43 aggregates. Comparatively, cellular TDP-43 proteinopathy with TTR expression remarkably narrowed the distance between TDP-43 and LC3 puncta, showing a major punctum distance beneath the 30-nm range. Together, these data prove that overexpression of TTR contributes to TDP-43 aggregates targeting autophagosomes and facilitates their removal.
TTR involves direct interaction with ATF4 to facilitate its nuclear translocation and autophagy induction
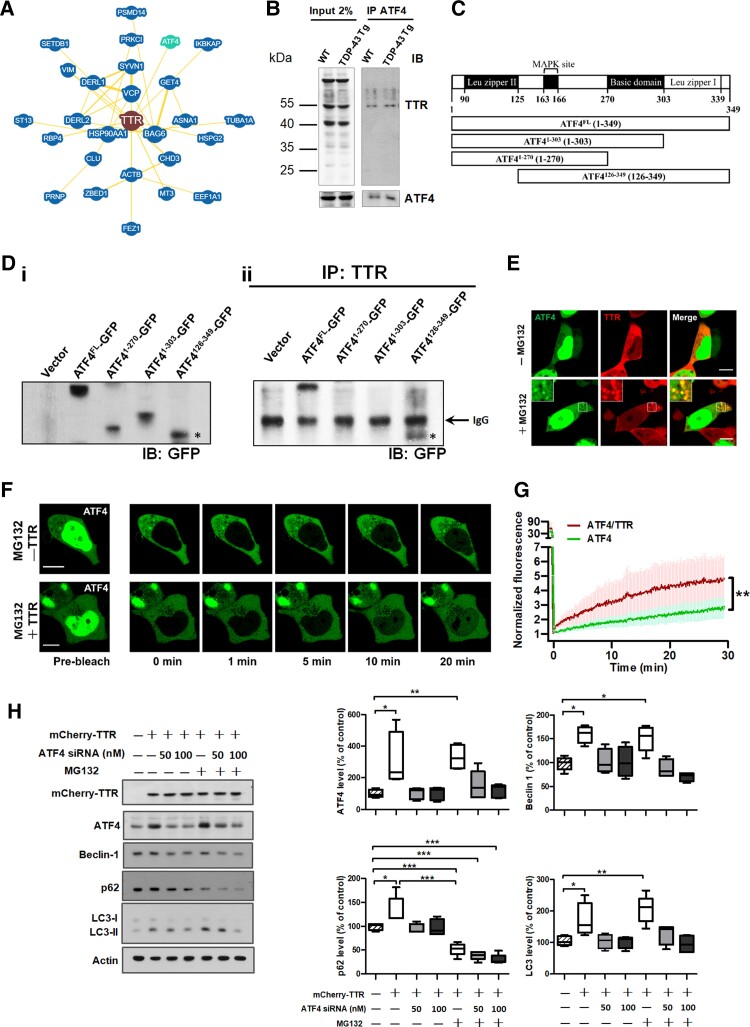
The next question that arose was how autophagy was induced by TTR. To answer this question, we further analysed the protein–protein interaction network of TTR using the BioGRID database (https://thebiogrid.org/) to obtain TTR protein interactome41 (Fig. 5A). The skeleton of TTR interactome has drawn our attention to ATF4 due to its significant upregulation on TTR expression, as evaluated previously (Fig. 3A). In addition, ATF4 activation has been reported to induce autophagy in response to ER stress.36,42 Thus, we further verified the interaction between ATF4 and TTR in the brain tissue lysate of WT and FTLD-TDP Tg mice by co-immunoprecipitation (Co-IP), in which targeting to capture ATF4 enabled Co-IP of the endogenous TTR (Fig. 5B). Furthermore, an in vitro validation was conducted to gain insight into which domain region of ATF4 involved in the interaction with TTR. To the best of our knowledge, ATF4 contains three well-defined domains: leucine zipper I, basic domain and leucine zipper II.43 Thus, the full-length DNA construct encompassing the full-length ATF4-fused GFP (ATF4FL-GFP) was first generated, and accordingly, the designated truncations of ATF4 (ATF41-270-GFP, ATF41-303-GFP and ATF4126-349-GFP) were created to confirm the interaction between ATF4 and TTR by Co-IP assay (Fig. 5C). Primarily, the designed ATF4 plasmid constructs were transfected into HEK293 cells to assess their protein expression capability and size accuracy [Fig. 5D(i)]. The Co-IP assay was performed on protein lysates from HEK293 cells, which were transiently transfected with the full-length and other domain-based truncation of ATF4-fused with GFP, using an anti-TTR antibody for immunoprecipitation followed by immunoblotting with anti-GFP antibody in western blot analysis [Fig. 5D(ii)]. The analysis results demonstrated that TTR complexed with the full-length ATF4 and also immunoprecipitated with the ATF4126-349 truncated protein, which displayed multiple bands in immunoblot indicating post-translational modifications (PTMs) on the C-terminal basic leucine zipper (bZIP) domain of ATF4 [Fig. 5D(ii)]. Given that ATF4 function is extensively regulated by PTMs, modifications to its C-terminal bZIP domain are potentially required for TTR binding. Together, ATF4 physically interacts with TTR via its C-terminal bZIP domain, potentially contingent on PTMs.
Figure 5.
A direct physical interaction between TTR and ATF4 promotes the nuclear import of ATF4, mediating autophagy upregulation. (A) BioGRID database showing the protein interaction network of TTR highlights a bona fide interaction between TTR (brown) and ATF4 (green). Minimum evidence set to two for BioGRID database searching. (B) The forebrain tissue lysates from the age of 6-month-old WT and TDP-43 Tg mice extracted by RIPA buffer were subjected to immunoprecipitation of ATF4 and further detected the Co-IP of TTR by western blot analysis. (C) Schematic diagram of the domain structures of full-length and truncations of ATF4 designated to clarify its region involved in TTR binding. [D(i)] Validation of the full-length and truncated forms of ATF4 fused with GFP was expressed in HEK293 cells by western blotting. [D(ii)] Immunoprecipitation of TTR from HEK293 cells with full-length and truncations of ATF4 fused with GFP expression was subjected to western blot analysis for their interaction by immunoblotting with anti-GFP antibody. * indicates the relative size of ATF4126-349,-GFP which multiple bands from IP result reflect PTMs on this region responsible for TTR recognition. The black arrow indicates the IgG band. (E) HEK293 cells were transfected with ATF4-GFP and mCherry-TTR under conditions with or without prolonged MG132 treatment (24 h). Colocalization of higher-order assemblies of ATF4-GFP and mCherry-TTR was shown under prolonged MG132 treatment (bottom) but absent colocalization in cells without MG132 treatment (top). Scale bar = 10 μm. (F) Fluorescence recovery after nuclear photobleaching (nuclear FRAP) was performed in ATF4-GFP transfected HEK293 cells with or without mCherry-TTR co-expression under prolonged MG132 treatment. Selected cells were imaged before nuclear FRAP by confocal microscopy (pre-bleach), and subsequently, the region of nuclear fluorescence was photobleached by irradiating with the full power of the 405-nm laser (452 μW). Right panels: Nuclear fluorescence recovery was recorded by time-lapse imaging, in which the selected images were remade by their corresponding acquisition time. Scale bar = 10 μm. (G) The time course curves of nuclear fluorescence recovery display exponential increases for cells with single ATF4 or ATF4/TTR co-expression. (P = 0.0018, t-test) Both FRAP curves were normalized by pre-bleaching values, in which the co-expression of ATF4 and TTR showed a significantly higher intensity of ATF4 nuclear import than single ATF4 expression (n = 5). (H) Western blot assay of TTR overexpression effect on ATF4 and autophagic key factors, including Beclin-1, p62/SQSTM1, LC3-I/LC3-II. Comprehensively, ATF4 knockdown and MG132 administration effects on TTR-induced autophagy activation were also validated. Densitometric quantification of each factor was normalized by Actin (ATF4 P = 0.0003, Beclin-1 P < 0.0001, p62 P < 0.0001, LC3 P < 0.0001). Data are shown as box-and-whisker plots (min to max). The central horizontal line within the box indicates the median value. Statistical analysis with one-way ANOVA followed by post hoc Tukey test. Statistically significant differences are denoted by asterisks (*P < 0.05, **P ≤ 0.01 and ***P ≤ 0.001).
To perceive the spatial localization of TTR and ATF4, confocal microscopy was conducted to inspect the distributions of ATF4-GFP and mCherry-TTR inside HEK293 cells with or without prolonged MG132 treatment. Confocal imaging of the cellular distribution of ATF4 and TTR revealed that most cells showed a common localization of ATF4 in the nucleus, and partial cells had a proportion of ATF4 shuttled to the cytoplasm, while TTR predominantly localized to the cytoplasm. After prolonged MG132 treatment (>24 h) of ATF4 and TTR co-expressed cells, selective imaging of the cells with ATF4 located in both the nucleus and cytoplasm showed that ATF4 generated cytoplasmic puncta and colocalized with the puncta of TTR (Fig. 5E bottom). However, a lack of MG132 treatment resulted in the absence of ATF4 puncta co-occurring with TTR puncta (Fig. 5E top). This indicates a higher-order assembly of ATF4 and TTR on prolonged stress.
Moreover, to discern the dynamic behaviour the effect of TTR on ATF4 function, FRAP analysis was used to study whether TTR regulated the nucleocytoplasmic translocation of ATF4. ATF4-GFP alone or with mCherry-TTR, the fluorescent labelling proteins were transiently expressed in cells with prolonged MG132 treatment (>24 h). In cells with ATF4 alone or in conjunction with TTR expression, ATF4 localized to both the nucleus and cytoplasm was selected to carry out FRAP, in which the nuclear fluorescent proteins were photobleached by irradiation with high laser power and, subsequently, the nuclear fluorescence recovery was recorded by acquiring time-lapse imaging (Fig. 5F). The recovered fluorescence was attributed to the influx of cytoplasmic unbleached ATF4-GFP. Furthermore, a comparison of cells with ATF4 alone or in conjunction with TTR expression showed that a faster and heightened nuclear fluorescence recovery of ATF4 was obtained in ATF4/TTR co-expressed cells over the imaging time period (Fig. 5F). Analysis of time-lapse photography revealed that ATF4 and TTR co-expressed cells showed a faster rate of recovery and greater ATF4 fluorescence intensity exponentially, with an ∼2-fold increased fluorescence plateau than single ATF4-expressed cells according to the recorded time period (Fig. 5G). This demonstrates that TTR facilitates the nuclear translocation of ATF4 in response to cellular stress.
To elucidate the essential role of ATF4 in TTR-mediated activation of autophagy, we made use of specific siRNA to knock down ATF4 mRNA in TTR-overexpressed cells. Primarily, TTR overexpression induced ATF4 upregulation (∼3-fold increase) followed by an increase in the expression levels of autophagic marker proteins, including Beclin 1, p62/SQSTM1 and LC3, compared to the control (Beclin-1 ∼1.6-fold; p62 ∼1.4-fold; LC3 ∼1.6-fold) (Fig. 5H). Furthermore, TTR-induced increases in autophagic marker proteins were reduced by knockdown of ATF4 with specific siRNA in a dose-dependent manner, in which the elevated concentration of ATF4 antisense oligomer levelled down the expression of autophagic markers counteracting the TTR effect (Fig. 5H). On the other hand, knockdown of overexpressed TTR also contributed to the downregulation of ATF4 level (Supplementary Fig. 1). As described before, TDP-43 proteinopathy mimetic cellular model was administered with MG132. Additionally, MG132 is regarded as a proteasome inhibitor while the ubiquitin-proteasome system and autophagy are functionally coupled in that inhibition of proteasome activity with MG132 is compensated with autophagy activation.44 Therefore, cells expressing TTR in combination with MG132 treatment were included in the comparison, which presented significantly reduced levels of p62/SQSTM1 and Beclin 1, and showed no statistically significant difference in ATF4 and LC3 levels, compared to TTR-expressed control cells, indicating the elicitation of autophagic flux on MG132 treatment (Fig. 5H). Subsequently, all the autophagic markers were downregulated on ATF4 knockdown in a siRNA dose-dependent manner under MG132 treatment as well as TTR-expressed control cells, indicating that the effect of TTR-induced autophagy upregulation was counteracted by ATF4 knockdown (Fig. 5H). These results support the hypothesis that TTR engages with ATF4 to trigger autophagy activation.
Engineering of neuronal TTR expression for the establishment of TTR-overexpressed TDP-43 Tg mice
Considering that the expression levels of TTR mRNA and protein significantly decreased during disease progression in TDP-43 Tg mice after 6 months old (Fig. 2D and E). Therefore, we examined whether TTR overexpression could have the same effect in vivo to facilitate the removal of cytoplasmic TDP-43 inclusions attenuating TDP-43 pathology. Primarily, TTR transgenic (TTR Tg) mice were generated by pronuclear microinjection of CaMKII promoter-driven TTR plasmid into fertilized embryos (Supplementary Fig. 2A), in which TTR was selectively overexpressed in the forebrain of the transgenic mice. Founders carrying TTR transgene were identified by genotyping PCR (Supplementary Fig. 2B). The protein levels of TTR in the brains of all selected TTR Tg mice and non-Tg mice were determined by immunoblotting. Quantification of the immunoblotting results showed that the protein level of TTR was increased in the brains of all selected transgenic lines of mice harbouring the TTR transgene (Supplementary Fig. 2C). Furthermore, brains derived from non-Tg and TTR Tg mice were dissected into the cortex, hippocampus and cerebellum, which were subjected to western blot analysis of TTR protein level. Comparatively, TTR expression was elevated in the cortex and hippocampus of TTR Tg mice compared to non-Tg mice, while no significant difference was observed in the cerebellum (Supplementary Fig. 2D). Additionally, the distribution of TTR in the brain areas was determined by IF staining of brain tissue sections. TTR signals were apparently increased in the cortex and hippocampus of TTR Tg mice compared to non-Tg mice (Supplementary Fig. 3). These data support that TTR Tg mice were successfully generated with neuronal TTR expression in the brain areas of the cortex and hippocampus. Next, we crossbred TTR Tg mice with TDP-43 Tg mice to generate double transgenic mice (TDP-43/TTR Tg) harbouring TTR and TDP-43 transgenes. A series of validations, including genotyping PCR, western blotting and fluorescence microscopy, were used to screen the crossbred TDP-43/TTR Tg mice and were subsequently subjected to confirmation of the effect of TTR on TDP-43 proteinopathy.
Neuronal TTR expression ameliorates TDP-43 proteinopathy-caused pathology and behavioural deficits
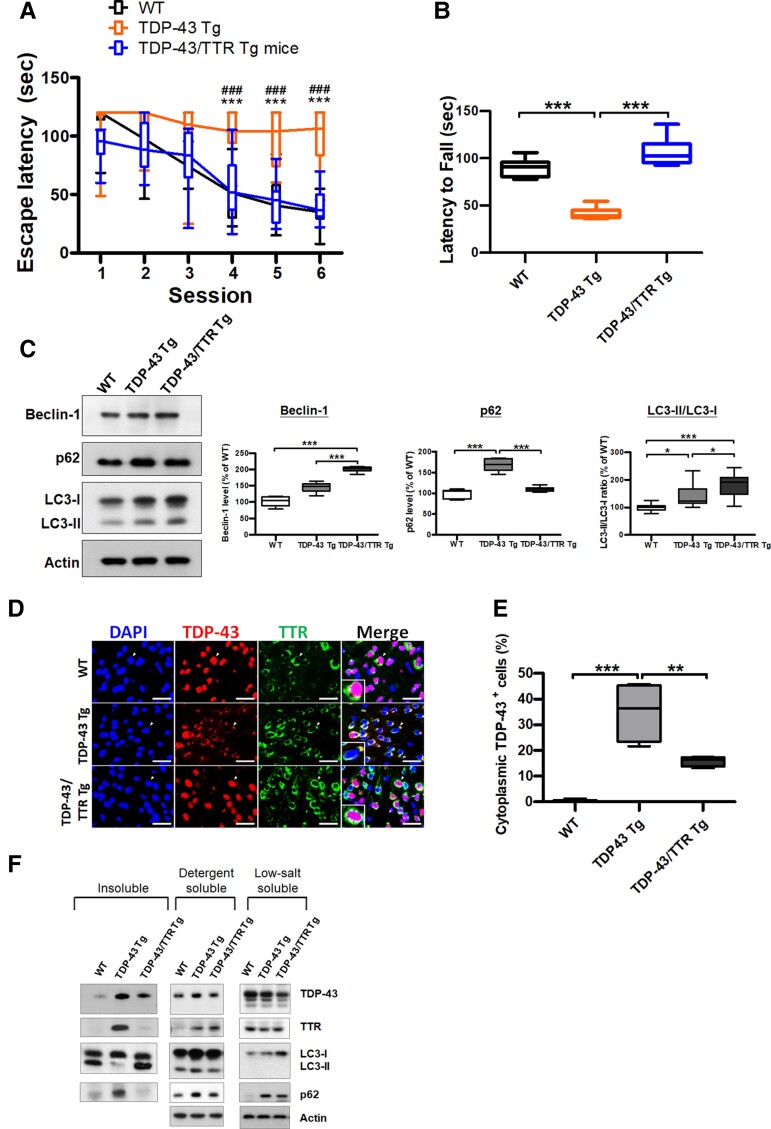
Primarily, we attempted to evaluate whether the neuron-specific expression of TTR was accompanied by an improvement in behavioural performance. Behavioural tests were performed to assess the cognition and motor function in WT, TDP-43 Tg and TDP-43/TTR Tg mice at 6 months. The Morris water maze test was used to survey the variation in spatial memory, in which the escape latency to the platform was monitored and analysed. Plotting of the daily trials showed that WT mice had a reduced latency in every trial; relatively, the continuous long latencies until the last trial were measured in TDP-43 Tg mice (Fig. 6A). Furthermore, neuronal TTR expression significantly shortened the time to the platform in TDP-43/TTR Tg mice, showing a comparable latency curve with WT mice (Fig. 6A). The rotarod task was conducted to determine their variation in motor coordination, during which the time of latency to fall was recorded and analysed. Compared to the longer-lasting time on the rotarod of WT mice, a quick fall was observed in TDP-43 Tg mice, indicating feeble motor activity (Fig. 6B). Strikingly, neuronal expression of TTR restored TDP-43 proteinopathy-induced motor defects in TDP-43/TTR Tg mice, characterized by a latency comparable to that of WT mice (Fig. 6B). Collectively, neuronal expressed TTR has a neuroprotective role in ameliorating TDP-43 proteinopathy-caused neuronal toxicity in TDP-43/TTR Tg mice, which progress towards better behavioural outcomes than TDP-43 Tg mice.
Figure 6.
Neuron-specific expression of native TTR attenuates TDP-43 cytoplasmic accumulation via autophagy, ameliorating behaviour deficits caused by TDP-43 proteinopathy. (A) Morris water maze task was performed at 6 months in WT, TDP-43 Tg and TDP-43/TTR Tg mice. (P < 0.0001, repeated measures ANOVA, Bonferroni post hoc test) Neuronal TTR expression significantly improves cognitive function, shown by better escape latency in TDP-43/TTR Tg mice than TDP-43 Tg mice. (B) The rotarod test was performed to evaluate motor function of WT, TDP-43 Tg and TDP-43/TTR Tg mice at 6 months (P < 0.0001). Neuronal TTR expression alleviates motor defects shown longer latency to fall in TDP-43/TTR Tg mice compared to TDP-43 Tg mice. n = 10 mice per group. (C) Left panel: Western blot analysis of autophagy markers Beclin-1, p62/SQSTM1 and LC3 in the forebrain tissue lysates from WT, TDP-43 Tg and TDP-43/TTR Tg mice at 6 months. Right panels: The densitometries from western blot results were quantified and normalized by Actin (Beclin-1 P < 0.0001, p62 P < 0.0001, LC3 P < 0.0001, one-way ANOVA with Tukey test). (D) Double IF staining for TDP-43 (red) and TTR (green) in the forebrain tissue specimens of WT, TDP-43 Tg and TDP-43/TTR Tg mice. The magnified view presents the colocalization of TDP-43 and TTR in the cytoplasm shown in the lower left. Nuclei were counterstained with DAPI (blue). Scale bar = 50 μm. (E) Quantitative analysis of the cytoplasmic TDP-43 inclusions in the forebrain tissue among WT, TDP-43 Tg and TDP-43/TTR Tg mice (P < 0.0001, one-way ANOVA with Tukey test). Neuronal expression of TTR reduced the cytoplasmic accumulation of TDP-43 in TDP-43/TTR Tg mice compared to TDP-43 Tg mice. (F) Immunoblots were probed with TDP-43, TTR and autophagy markers, p62/SQSTM1 and LC3, in the insoluble (urea-soluble), detergent-soluble and LS soluble fractions from the brain tissue extractions of WT, TDP-43 Tg and TDP-43/TTR Tg mice. Escape latency data are shown as overlaying box-and-whisker plots (min to max) and line plots. The rest of the data are shown as box-and-whisker plots (min to max). The central horizontal line within the box indicates the median value. Statistically significant differences are denoted by asterisks (*P < 0.05, **P ≤ 0.01 and ***P ≤ 0.001). n = 10 sections per mouse; n = 5 mice per group.
Next, both TDP-43 Tg and TDP-43/TTR Tg mice, as well as WT mice, were subjected to verification of TTR effects in vivo, as observed in vitro, showing that TTR manipulates autophagy activation against TDP-43 proteinopathy. On the basis of our previous studies, autophagy failure was identified in TDP-43 Tg mice, characterized by downregulation of LC3 and accumulation of p62/SQSTM1.26,45 Additionally, results from our in vitro observations demonstrated that TTR overexpression facilitates the elimination of TDP-43 aggregates via autophagy. As expected, restoration of autophagy function by neuronal expression of TTR in TDP-43 Tg mice might alleviate TDP-43 pathology. Initially, we attempted to understand whether neuronal TTR expression would revive autophagy activity in vivo. The brain tissue lysates from 6-month-old WT, TDP-43 Tg and TDP-43/TTR Tg mice were subjected to western blotting to analyse the changes in the expression levels of the autophagic markers, Beclin-1, p62/SQSTM1 and LC3 (Fig. 6C). Compared to WT mice, the heightened p62/SQSTM1 level implicated autophagy failure in TDP-43 Tg mice (∼1.7-fold increase) (Fig. 6C). Compared to both WT and TDP-43 Tg mice, the quantitative data indicated that neuronal expression of TTR gave rise to significant upregulation of Beclin 1 and conversion of LC3-I to LC3-II, as observed in TDP-43/TTR Tg mice (Beclin-1 ∼2-fold; LC3-II/I ∼1.8-fold) (Fig. 6C). Additionally, compared to the significant accumulation of p62/SQSTM1 in TDP-43 Tg mice (∼1.7-fold increase), the level of p62 in TDP-43/TTR Tg mice was similar to that in WT mice (Fig. 6C). Together, these results demonstrate that neuronal expression of TTR facilitates the activation of autophagy in the brains of TDP-43/TTR Tg mice. Furthermore, to determine whether neuronal TTR-induced autophagy enabled to ease TDP-43 pathology, the forebrain tissue specimens from 6-month-old WT, TDP-43 Tg and TDP-43/TTR Tg mice were subjected to double IF staining of TDP-43 and TTR to validate the variance of cytoplasmic TDP-43 mislocalization by confocal microscopy (Fig. 6D). Quantification of confocal images showed that compared to WT mice, the significantly increased cytoplasmic TDP-43 inclusions in TDP-43 Tg mice (∼142-fold) comparatively decreased in TDP-43/TTR Tg mice (∼0.4-fold reduction) (Fig. 6E). These results demonstrate that neuronal TTR-induced autophagy promotes the removal of cytoplasmic TDP-43 inclusions in the forebrain of TDP-43/TTR Tg mice. Moreover, to comprehend whether neuronal TTR expression substantially reduced the insoluble TDP-43 aggregates in vivo, the insoluble fractions derived from the forebrain tissues of 6-month-old WT, TDP-43 Tg and TDP-43/TTR Tg mice were used to analyse the insoluble protein levels of TDP-43, TTR, p62/SQSTM1 and LC3 by immunoblotting (Fig. 6F). Analysis of the insoluble fractions from brain extracts confirmed that the heightened levels of TDP-43 were simply traced in TDP-43 Tg mice, and neuronal TTR expression levelled down insoluble TDP-43 aggregates (Fig. 6F). Notably, an elevated level of insoluble TTR observed in TDP-43 Tg mice has retrieved its solubility in TDP-43/TTR Tg mice (Fig. 6F). In addition, a diminished conversion of LC3-I to LC3-II and an increase of p62/SQSTM1 accumulation in the insoluble fraction of TDP-43 Tg mice indicated autophagy impairment, and neuronal TTR expression restored autophagy function accompanying the increased LC3-I to LC3-II conversion and decreased p62/SQSTM1 accumulation (Fig. 6F). Furthermore, detergent-soluble fractions for the membrane-bound proteins presented a higher level of TDP-43 in TDP-43 Tg mice than in WT and TDP-43/TTR Tg mice and, comparatively, a higher level of TTR was apparently detected in TDP-43/TTR Tg mice (Fig. 6F). Accordingly, autophagy impairment corresponding to LC3 and p62 accumulations were also identified in the detergent-soluble fraction of TDP-43 Tg mice (Fig. 6F). Comparatively, higher-level TDP-43 within LS fractions was detected to be dominant in WT mice, and lower-level TTR within LS soluble fractions was obtained in TDP-43 Tg mice (Fig. 6F). Apparently, a higher level of LS soluble LC3-I form was identified in TDP-43/TTR Tg mice; accordingly, a higher level of LS soluble p62 was observed in TDP-43 Tg and TDP-43/TTR Tg mice. Altogether, in agreement with the in vitro findings, neuronal expression of TTR mediates autophagy activation in vivo, further facilitating the elimination of cytoplasmic TDP-43 inclusions and the reduction of TDP-43 truncations.
TTR facilitates TDP-43 aggregates targeting autophagosomes and nuclear translocation of ATF4
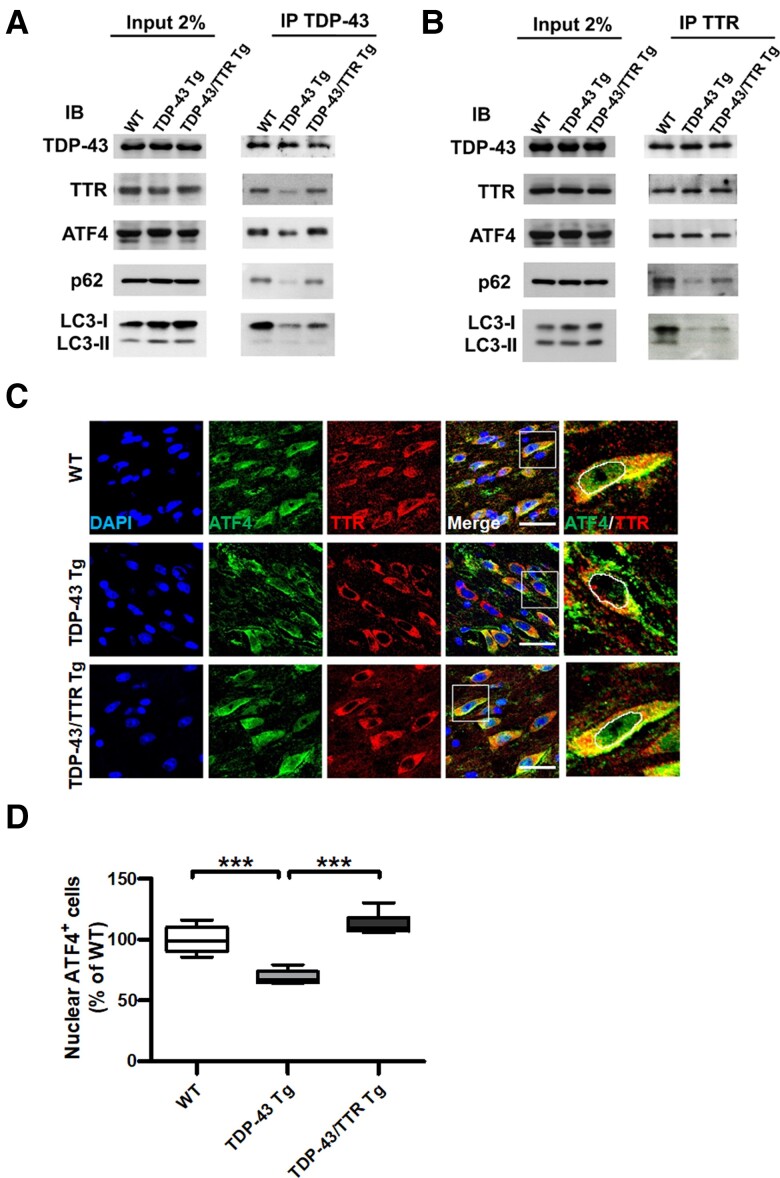
To clarify TTR-mediated clearance of TDP-43 aggregates through autophagy and its functional interaction with ATF4 in vivo, we further determined the TTR-associated protein complex from the brain tissue lysates of 6-month-old WT, TDP-43 Tg and TDP-43/TTR Tg mice using Co-IP and immunoblotting. Primarily, immunoprecipitation of TDP-43 was performed to verify the functional complex of TDP-43 interacting with TTR, p62/SQSTM1 and LC3, along with the additional participation of ATF4 (Fig. 7A). Analysis of the Co-IP results indicated that TDP-43 reduced its binding ability to TTR, p62/SQSTM1, LC3 and the additional ATF4 association in TDP-43 Tg mice compared to WT mice, and that neuronal expression of TTR recovered the association of TDP-43 with interacting proteins in TDP-43/TTR Tg mice (Fig. 7A). These results suggest that, compared to WT and TDP-43/TTR Tg mice, the immunoprecipitated TDP-43 from the lysate of TDP-43 Tg mice only partially retained the correct conformation and binding ability to interact with TTR, which is in complex with less p62 for targeting LC3-conjugated autophagosomes, indicating autophagy impairment. Interestingly, probing of TDP-43 interacting proteins comprised ATF4, and the decreased solubility of TDP-43 in TDP-43 Tg mice was accompanied by reduced interaction with ATF4 (Fig. 7A). Alternatively, reverse confirmation was conducted by immunoprecipitation of TTR to examine its dynamic properties of interacting with TDP-43, p62, LC3 and ATF4 (Fig. 7B). Inspection of the Co-IP results showed that TTR made the most use of its binding ability to associate with TDP-43 and ATF4, characterized by comparable binding intensities among WT, TDP-43 Tg and TDP-43/TTR Tg mice (Fig. 7B). However, compared to WT mice, the equivalent binding level of TTR to TDP-43 in TDP-43 Tg mice could not completely achieve the recognition of p62 for further targeting to LC3-conjugated autophagosomes, which showed decreased binding levels of p62 and LC3 in the soluble fraction of brain tissue lysate (Fig. 7B). Furthermore, neuronal TTR expression notably restored the capability of interacting with p62 and LC3 in TDP-43/TTR Tg mice (Fig. 7B). These data support that TTR associates with early soluble aggregated TDP-43 facilitating its recognition by p62 for targeted LC3-conjugated autophagosomes. Furthermore, ATF4 is associated with TTR, which is possibly involved in TDP-43 interactome. Accordingly, to elucidate whether TTR facilitates the nuclear import of ATF4 in vivo, brain tissue specimens from the age of 6-month-old WT, TDP-43 Tg and TDP-43/TTR Tg mice were used to examine the cellular distribution of ATF4 and TTR by double IF staining. Confocal imaging of the brain tissue specimens revealed that instead of the common nuclear localization, ATF4 was more evenly distributed in the nucleus and cytoplasm of neurons in WT and TDP-43/TTR mice. Comparatively, most ATF4 was localized in the cytoplasm in TDP-43 Tg mice, of which TTR was predominantly retained in the cytoplasm of neurons in all three types of mouse (Fig. 7C). Imaging-based quantification showed that compared to WT mice, a significantly reduced level of nuclear ATF4 was observed in TDP-43 Tg mice, and the neuronal expression of TTR substantially repaired the nuclear import of ATF4 in TDP-43/TTR Tg mice (Fig. 7D). Taken together, TTR has a dual role in the removal of TDP-43 aggregates, which either facilitates the nuclear entry of ATF4 or escorts TDP-43 aggregates to autophagosomes.
Figure 7.
TTR forms a complex with TDP-43 and ATF4 mediating two-way regulations for either targeting TDP-43 aggregates to autophagosomes or promoting nuclear translocation of ATF4. (A) Immunoprecipitation of TDP-43 or (B) TTR from the brain tissue lysates of WT, TDP-43 Tg and TDP-43/TTR Tg mice at 6 months followed by western blot analysis of the indicated interacting proteins. Representative western blots of proteins detected from (Left panel) crude tissue lysates and (Right panel) Co-IP elutes. IB, immunoblotting; IP, immunoprecipitation. (C) Confocal images of double IF staining ATF4 (green) and TTR (red) in the forebrain tissue specimens from 6 months old of WT, TDP-43 Tg and TDP-43/TTR Tg mice. The rightmost panel displays the magnified view of ATF4 and TTR merged image circling out the nuclear region, as indicated by a white square line in the previous image. The nuclei were counterstained with DAPI (Blue). Scale bar = 30 μm. (D) Quantification of the nuclear localization of ATF4 was processed by TissueQuest software in the TissueFAXS system (TissueGnostics GmbH) (P < 0.0001). The basal nuclear fluorescence intensity was set up based on ATF4 intensity in WT and gathered the number of cells with ATF4 intensity equal or above baseline intensity. Data are shown as box-and-whisker plots (min to max). The central horizontal line within the box indicates the median value. Statistical analysis with one-way ANOVA followed by post hoc Tukey test. Statistically significant differences are denoted by asterisks (***P ≤ 0.001). n = 8 sections per mouse; n = 6 mice per group.
Neuronal TTR expression strengthens neuronal survival and alleviates microgliosis and astrogliosis
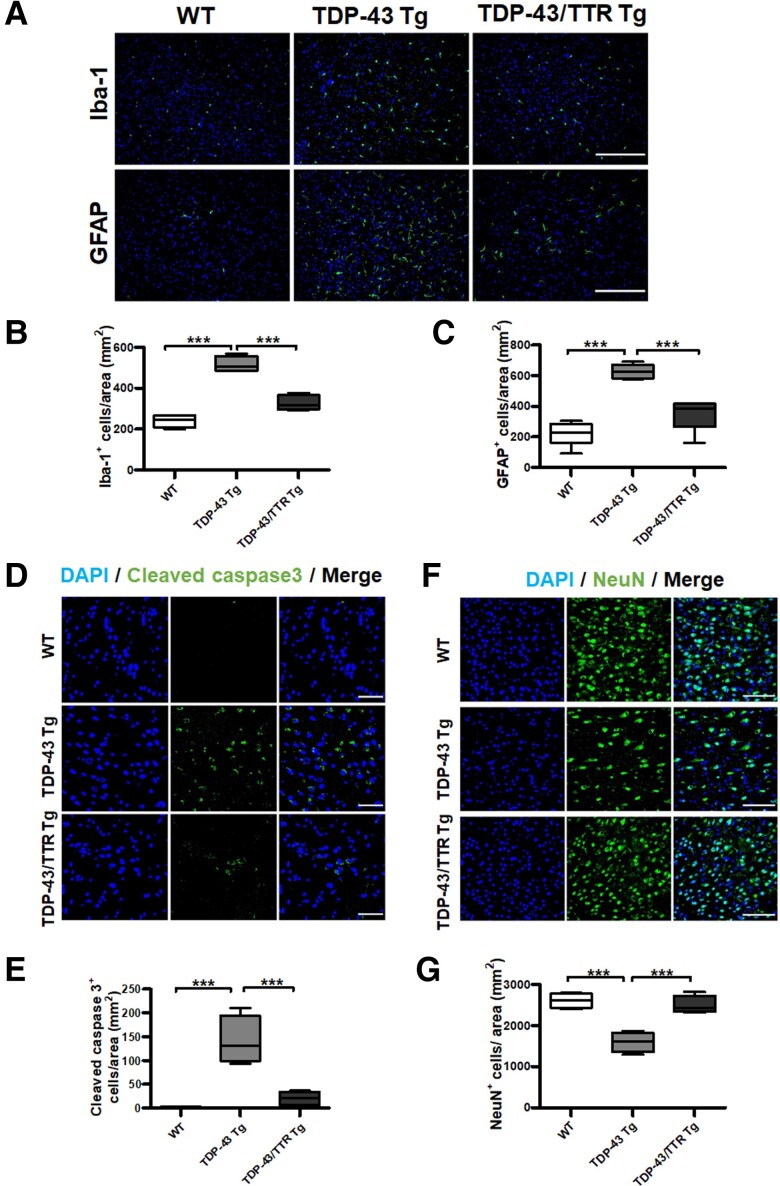
Based on our previous findings, reactive gliosis and apoptosis are the neuropathological characteristics of TDP-43 Tg mice.26,45 Primarily, the activation of microglia or astrocytes was validated by the immunoreactivity of their respective markers, Iba-1 and GFAP, in the brain tissue specimens from the age of 1-year-old WT, TDP-43 Tg and TDP-43/TTR Tg mice using fluorescence microscopy. Widefield fluorescence imaging of the brain tissue specimens showed that activated microglia and astrocytes were observed in TDP-43 Tg mice with increased immunoreactivity of Iba-1 and GFAP compared to WT mice. Furthermore, reduced Iba-1 and GFAP immunoreactivity was observed in TDP-43/TTR Tg mice (Fig. 8A). Quantitative data evinced that the levels of Iba-1- and GFAP-positive cells presented an ∼3-fold increase in TDP-43 Tg mice compared to WT mice, and neuronal TTR expression substantially downregulated the levels of Iba-1- and GFAP-positive cells in TDP-43/TTR Tg mice (Fig. 8B and C). These results indicate that activated microglia and astrocytes are significantly upregulated in TDP-43 Tg mice compared to WT mice, and neuronal expression of TTR alleviated microgliosis and astrogliosis in TDP-43/TTR Tg mice compared to TDP-43 Tg mice. Furthermore, to verify the cellular apoptosis affected by TTR, the active caspase-3 in brain tissue sections from the age of 1-year-old WT, TDP-43 Tg and TDP-43/TTR Tg mice were analysed by immunostaining. Active caspase-3 expression was greatly increased in TDP-43 Tg mice (∼82.4-fold) compared to WT mice, and neuronal expression of TTR fundamentally mitigated TDP-43 proteinopathy-triggered cellular apoptotic death in TDP-43/TTR Tg mice (Fig. 8D and E). Correspondingly, neuronal survival was further examined to ascertain the neuroprotective role of TTR. The neuronal marker NeuN was used to inspect neuronal survival in the age of 1-year-old WT, TDP-43 Tg and TDP-43/TTR Tg mice by immunostaining. Analysis of the imaging results showed that significant neuronal loss was confirmed in TDP-43 Tg mice (∼39% decrease) compared to WT mice, and neuronal expression of TTR markedly rescued TDP-43 proteinopathy-caused neuronal damage in TDP-43/TTR Tg mice (∼86% restoration) (Fig. 8F and G). Altogether, neuronal expression of TTR fundamentally ameliorates neurotoxicity caused by TDP-43 proteinopathy to diminish apoptotic neuronal death and reduce microgliosis and astrogliosis. These results further confirmed that the neuronal expressed TTR engages autophagy regulation to mitigate neuropathology, proving the improvement of spatial learning/memory abilities and the restoration of motor function as demonstrated in Fig. 6.
Figure 8.
Neuronal expression of TTR alleviates TDP-43 proteinopathy-caused neuropathology, presenting reduced microgliosis and astrogliosis, diminished neuronal apoptosis and increased neuronal survival. (A) Representative images of (top) Iba-1 and (bottom) GFAP were immunostained in the forebrain tissue specimens of 1-year-old WT, TDP-43 Tg, TDP-43/TTR Tg mice. Nuclei were counterstained with DAPI (blue). Scale bar = 200 μm. The number of cells with (B) Iba-1- and (C) GFAP immunoreactivity were quantified by TissueQuest software. The numbers of microglial and astrocytic cells were significantly decreased upon neuronal TTR expression in TDP-43/TTR Tg mice compared to TDP-43 Tg mice (Iba-1 P = 0.0002, GFAP P < 0.0001). (D) Representative images from the brain tissue specimens from the age of 1-year-old WT, TDP-43 Tg, TDP-43/TTR Tg mice were immunostained with cleaved caspase 3. Scale bar = 50 μm. (E) The number of cleaved caspase 3-positive cells represented apoptotic death cells was quantified by TissueQuest software. Cellular death by apoptosis was substantially reduced on neuronal TTR expression in TDP-43/TTR Tg mice compared to TDP-43 Tg mice (P = 0.0002). (F) Representative images of the brain tissue specimens were immunostained with NeuN in WT, TDP-43 Tg and TDP-43/TTR Tg mice at the age of 1 year. Scale bar = 100 μm. (G) Quantification of NeuN-positive neurons evaluated neuronal survival using TissueQuest software (P = 0.0002). Nuclei were counterstained with DAPI (blue). Data are shown as box-and-whisker plots (min to max). The central horizontal line within the box indicates the median value. Statistical analysis with one-way ANOVA followed by post hoc Tukey test. Statistically significant differences are denoted by asterisks (***P < 0.001). n = 8 sections per mouse; n = 6 mice per group.
Discussion
The currently known TTR plays an essential role in enhancing neurite outgrowth and maintaining the learning/memory competence of the brain.25 Moreover, uncovering of TTR acts against Aβ aggregation and promotes Aβ degradation due to its neuroprotective role in AD. In this study, TDP-43 proteinopathy-induced cytotoxicity was used to investigate the general neuroprotective function of TTR in vitro and in vivo. The proportion of insoluble TTR was increased in FTLD-TDP of human patients and transgenic mice (TDP-43 Tg mice). Accordingly, the cytoplasmic TTR aggregates showed colocalization with TDP-43 inclusions in fluorescent images. Although the upregulation of TTR has been detected in FTLD and ALS,25 increased TTR levels were coherently observed before 6 months old. However, drastically decreased TTR was identified in one-year-old TDP-43 Tg mice. Regarding the reduced TTR levels affected by downregulation of mRNA and protein expression in old age, it is plausible that the decreased expression level of TTR is due to large-scale neuronal death caused by disease progression response to ageing in TDP-43 Tg mice.46 Supposedly, TTR coaggregated with TDP-43 in the cytoplasmic inclusions and the reduced expression level of TTR on aging accelerates disease deterioration in FTLD-TDP.
The most notable functions of TTR are its proteolytic activity to cleave Aβ peptide and prevent amyloid fibril formation of Aβ in a chaperone-like manner, which contributes to proteostasis of the CNS.47–49 However, lowered TTR levels in CSF have been characterized in Alzheimer’s disease patients, considering that TTR sequesters Aβ to protect the brain from disease progression50 and, eventually, the amyloidogenic process causes the functional defect of TTR concomitant with neuronal death. However, several neurological diseases have shown increased levels of TTR, including FTLD and ALS.25 Regarding the other aspects, the elevation or reduction of TTR levels in ALS remains debatable,51,52 which is potentially related to the variance of the disease process and severity in ALS. Reportedly, upregulation of TTR in ALS has been found in the rapid progression of the disease when compared with slow progression.53 Supposedly, the variation of TTR levels in FTLD/ALS in accordance with our findings reveals that the elevated TTR occurs in early disease onset, in response to the loss of proteostasis, and the reduced TTR level reflects neuronal damage in the late stage with severe cellular function loss.
Considering that two primary pathways, the UPR and autophagy, are responsible for maintaining cellular proteostasis,54 we found that TTR mediates the clearance of cytoplasmic TDP-43 inclusions via autophagy under TDP-43 proteinopathy in vitro and in vivo. Several neurodegenerative diseases caused by proteostatic perturbations commonly trigger the UPR activation, such as TDP-43 proteinopathy in FTLD and ALS.55 In line with previous studies, the general upregulation of the UPR was also identified in TDP-43 proteinopathy mimetic cellular model with a significant increase in the UPR onset indicator, Bip. Furthermore, Bip level, which was not affected by TTR expression, indicated that TTR-governed elimination of TDP-43 aggregates was not mediated by the UPR signalling. In particular, overexpression of TTR followed by ATF4 upregulation was ineffective in activating its downstream pro-apoptotic factor, CHOP, suggesting that TTR-stimulated ATF4 elevation against death signalling. Alternatively, autophagy was substantially upregulated in response to TTR expression, accompanied by significant increases in the expression of autophagy-related markers, Beclin-1, p62/SQSTM1 and LC3-I/II conversion in vitro and in vivo. Surprisingly, ATF4 proven in TTR interactome mapping links the connection of TTR-mediated autophagy56,57 (Fig. 5). ATF4 plays a crucial role in regulating the activation of autophagy-related genes, which can be dependent or independent of PERK activation in the UPR signalling.36,58 In addition, TTR has been reported to contribute to transcriptional and translational upregulation of insulin-like growth factor receptor I (IGF-IR) in vitro, as a transcription inducer.59 Therefore, the upregulation of ATF4 reactive to TTR signified that ATF4 is the downstream regulator of TTR. In our observations, the elevation of ATF4 level responsive to TTR expression was accompanied by activation of autophagic markers and, sequentially, knockdown of ATF4 abrogated TTR-induced autophagy activation (Fig. 5H). Supporting this, TTR endows a novel function mediating autophagy activation via ATF4 independent of the UPR signalling to facilitate the abolition of cytoplasmic TDP-43 aggregates.
In our findings, TTR expression induced the upregulation of autophagosome formation and reflected its association with TDP-43 and LC3 by their fluorescence colocalization. Super-resolution imaging further demonstrated that TTR effectively mediated the reduced TDP-43 puncta size and narrowed down the distance between TDP-43 and LC3 (Fig. 4). Ex vivo assays of brain tissue protein fractionations revealed that abundant TDP-43 as well as accumulated p62/SQSTM1 showed simultaneously elevated levels in both insoluble and soluble fractions in TDP-43 Tg mice; in contrast, WT and TDP-43/TTR Tg mice showed dominantly soluble TDP-43 (Fig. 6F). These indicated that neuronal TTR expression facilitates the balance of TDP-43 levels and prevents the formation of insoluble TDP-43. Accordingly, the heightened TTR level was predominantly obtained in the detergent-soluble fraction in TDP-43/TTR Tg mice, suggesting that overexpressed neuronal TTR is mainly a membrane-bound form working in concert with TTR-facilitated autophagosome targeting of aggregated TDP-43. Basically, fractionation extracts derived from normal and FTLD humans displayed the same trends of TDP-43 and TTR levels in both insoluble and soluble fractions as mouse models (Fig. 1D). Alternatively, brain lysates extracted by strong detergent [radioimmunoprecipitation assay (RIPA)] lysis buffer of WT, TDP-43 Tg and TDP-43/TTR Tg mice showed unremarkable discrimination of TDP-43 and TTR levels in the input protein analysis (Fig. 7A). Speculatively, both membrane-bound and LS soluble forms of TDP-43 and TTR were extracted together by RIPA buffer, and overloaded TDP-43 was left in the detergent-insoluble pellet. Nevertheless, overexpressed TTR accompanied aggregated TDP-43 for degradation. Furthermore, ex vivo analysis of Co-IP results showed that a large proportion of soluble TDP-43 lost interaction with TTR as well as p62/SQSTM1 and LC3 in TDP-43 proteinopathy indicating autophagy impairment, and neuronal TTR expression restored autophagosome targeting complex formation for soluble TDP-43 aggregates (Fig. 7A). Importantly, TTR bound to TDP-43 under normal circumstances, suggesting that TTR chaperons early soluble forms of TDP-43 aggregates for autophagosome targeting. However, the pathological TDP-43 aggregates either improperly assembled with TTR and factors related to degradation such as p62/SQSTM1 or failed to be recognized by those degradation factors in TDP-43 proteinopathy, which turned into insoluble aggregates and eventually grew into cytoplasmic inclusions. These pathological TDP-43 inclusions were characterized by the increased colocalization of cytoplasmic TDP-43 and TTR with accumulation ex vivo and in vitro, which appeared intensified insoluble TTR levels and autophagy markers in the insoluble fractions of TDP-43 proteinopathy ex vivo. Furthermore, neuronal TTR expression rescued autophagosome targeting of TDP-43 aggregates, in which neuronal TTR recovered chaperoning with early soluble TDP-43 aggregates and facilitated their correct engulfment into autophagosomes. Proposedly, TTR coordinates with p62/SQSTM1 targeting early soluble TDP-43 aggregates to autophagosomes for degradation.
Synergistically, TTR is characterized as an ATF4 binding partner, which is potentially regulated by PTMs on the C-terminal bZIP domain of ATF4. Several PTMs have been identified in the C-terminal bZIP of ATF4, including phosphorylation, acetylation and ubiquitination, which presumably control heterodimerization to regulate the binding affinity and specificity of ATF4 to its interacting partners.60 Furthermore, we found that TTR interacted with ATF4 under normal circumstances and generated higher-order assembly on stress, disclosing the colocalization of their cytoplasmic puncta (Fig. 5E). ATF4 ordinarily displays widespread distribution throughout the hippocampal neuron, showing direct interaction with GABAB receptor.61,62 Notably, ER stress-induced upregulation of histone deacetylase 4 (HDAC4) causes cytoplasmic retention of ATF4, in which HDAC4 constraint ATF4 transcription activity to prevent its downstream activation of apoptosis-related factors such as CHOP and TRB3.43 Normally, HDAC4 is retained in the cytoplasm of neurons, suggesting that there is regular modulation between HDAC4 and ATF4 in the cytoplasm of neurons.63 In light of TTR contact with the C-terminal bZIP domain of ATF4, HDAC4 interacts with the N terminus of ATF4, in which the N-terminal leucine zipper domain is required for ATF4 binding to HDAC4.43 It is proposed that TTR functions counter to HDAC4 in neurons, where HDAC4 prefers to retain ATF4 in the cytoplasm. Conversely, TTR prefers to promote the nuclear translocation of ATF4. Our findings confirmed that TTR efficiently and abundantly boosted ATF4 import into the nucleus on stress on the basis of a FRAP assay (Fig. 5F and G). Previous studies have reported that TTR has a synergistic effect with IGF-IR, characterized by direct interaction of TTR with IGF-IR and triggering IGF-IR nuclear translocation to activate downstream signalling.59,64 Accordingly, our data demonstrated that impaired nuclear import of ATF4 in TDP-43 proteinopathy was retrieved by neuronal TTR expression in vivo. Importantly, TTR-induced ATF4 nuclear translocation, which facilitated autophagy upregulation instead of CHOP-mediated apoptosis. Here, we showed that TTR directly binds to the C-terminal bZIP domain of ATF4, potentially in a PTM-dependent manner, and promotes the nuclear import of ATF4 on stress.
Taken as a whole, TTR orchestrated the degradation of TDP-43 aggregates in two ways: autophagosome targeting and autophagy activation. We disclosed that TTR was associated with TDP-43 and ATF4 and was in complex with p62/SQSTM1 and LC3 ex vivo (Fig. 7A and B). However, a proportion of soluble TDP-43 failed to engage with ATF4 and TTR together with p62/SQSTM1 and LC3 in TDP-43 proteinopathy. Comparatively, the successful binding of TTR, ATF4 and TDP-43 could not promise the following p62/SQSTM1 and LC3 recruitment. It is worth noting the TTR joint with ATF4 as part of the TDP-43 complex, suggesting that TTR routinely binds to ATF4 and is associated with early soluble TDP-43 aggregates, as indicated by immunoprecipitation of TTR, showing equivalent binding levels with ATF4 and TDP-43. Instead, immunoprecipitation of TDP-43 showed a portion disengaged with TTR and ATF4 binding in TDP-43 proteinopathy, which was potentially due to low soluble pathological TDP-43 aggregates that could not be properly recognized. On the other hand, the interaction of TTR and TDP-43 deficiently docks to p62/SQSTM1 for autophagosome targeting in TDP-43 proteinopathy, possibly because of gradual pathological aggregation. Subsequently, the pathological aggregates become insoluble detergent-resistant inclusions with TTR, p62 and LC3 presented in an insoluble fraction. Thus, TTR conducts a symphony of signals to regulate the clearance of cytoplasmic TDP-43 aggregates through autophagy. Neuronal TTR expression recovered defective autophagy in maintaining TDP-43 proteostasis, which normalized the nuclear import of ATF4, the complex formation of early soluble TDP-43 aggregates with TTR, p62/SQSTM1 and LC3 and the reduction of cytoplasmic TDP-43 inclusions. Neuropathologically, neuronal TTR expression remarkably counterbalances TDP-43 proteinopathy-caused pathological effects, which presented with increased neuronal survival and decreased apoptotic cells and gliosis, further ameliorating behavioural deficits.
Here we demonstrate that TTR potentiates the dual function of achieving neuroprotection in TDP-43 proteinopathy, in which TTR either escorts TDP-43 aggregates targeting autophagosomes for degradation or ATF4 targeting the nucleus for autophagy upregulation. Importantly, neuronal TTR expression retrieves autophagy function via ATF4 and facilitates TDP-43 aggregates docking to autophagosomes, which attenuate TDP-43 proteinopathy-caused neurotoxicity, ensuring better behavioural outcomes. Herein, we uncovered TTR endowed with two-way regulation to maintain cellular proteostasis via autophagy, which endorses TTR as a modulator therapy for neurodegenerative diseases caused by TDP-43 proteinopathy.
Supplementary Material
Acknowledgements
The authors appreciate the University of California Davis Alzheimer’s Disease Center for gathering and offering the human brain samples funded by the National Institute on Aging (NIA, grant no. P30AG10129).
Contributor Information
Yuan-Ping Chu, Institute of Clinical Medicine, College of Medicine, National Cheng Kung University, Tainan, Taiwan.
Lee-Way Jin, Department of Pathology and Laboratory Medicine, UC Davis Medical Center, CA, USA.
Liang-Chao Wang, Division of Neurosurgery, Department of Surgery, National Cheng Kung University Hospital, College of Medicine, National Cheng Kung University, Tainan, Taiwan.
Pei-Chuan Ho, Institute of Clinical Medicine, College of Medicine, National Cheng Kung University, Tainan, Taiwan.
Wei-Yen Wei, Institute of Clinical Medicine, College of Medicine, National Cheng Kung University, Tainan, Taiwan.
Kuen-Jer Tsai, Institute of Clinical Medicine, College of Medicine, National Cheng Kung University, Tainan, Taiwan; Research Center of Clinical Medicine, National Cheng Kung University Hospital, College of Medicine, National Cheng Kung University, Tainan, Taiwan.
Funding
This study was performed, in part, with support from Ministry of Science and Technology, Taiwan (MOST-106-2628-B-006-001-MY4, MOST-111-2320-B-006-038-MY3) and National Cheng Kung University Hospital.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neary D, Snowden JS, Gustafson L, et al. Frontotemporal lobar degeneration: A consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. [DOI] [PubMed] [Google Scholar]
- 3. Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010;6:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown RH, Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;377:162–172. [DOI] [PubMed] [Google Scholar]
- 5. Neudert C, Oliver D, Wasner M, Borasio GD. The course of the terminal phase in patients with amyotrophic lateral sclerosis. J Neurol. 2001;248:612–616. [DOI] [PubMed] [Google Scholar]
- 6. Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008;105:797–806. [DOI] [PubMed] [Google Scholar]
- 7. Wong CE, Jin LW, Chu YP, Wei WY, Ho PC, Tsai KJ. TDP-43 proteinopathy impairs mRNP granule mediated postsynaptic translation and mRNA metabolism. Theranostics. 2021;11:330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geser F, Martinez-Lage M, Robinson J, et al. Clinical and pathological continuum of multisystem TDP-43 proteinopathies. Arch Neurol. 2009;66:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem. 2008;283:13302–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. [DOI] [PubMed] [Google Scholar]
- 11. Nonaka T, Kametani F, Arai T, Akiyama H, Hasegawa M. Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum Mol Genet. 2009;18:3353–3364. [DOI] [PubMed] [Google Scholar]
- 12. Zhang YJ, Xu YF, Cook C, et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2009;106:7607–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scheper W, Hoozemans JJ. The unfolded protein response in neurodegenerative diseases: A neuropathological perspective. Acta Neuropathol. 2015;130:315–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. [DOI] [PubMed] [Google Scholar]
- 15. Kim HJ, Raphael AR, LaDow ES, et al. Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet. 2014;46:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matus S, Lopez E, Valenzuela V, Nassif M, Hetz C. Functional contribution of the transcription factor ATF4 to the pathogenesis of amyotrophic lateral sclerosis. PLoS ONE. 2013;8:e66672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Senft D, Ronai ZA. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci. 2015;40:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scheper W, Nijholt DAT, Hoozemans JJM. The unfolded protein response and proteostasis in Alzheimer disease: Preferential activation of autophagy by endoplasmic reticulum stress. Autophagy. 2011;7:910–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glick D, Barth S, Macleod KF. Autophagy: Cellular and molecular mechanisms. J Pathol. 2010;221:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gião T, Saavedra J, Cotrina E, et al. Undiscovered roles for transthyretin: from a transporter protein to a new therapeutic target for Alzheimer's disease. Int J Mol Sci. 2020;21:2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buxbaum JN. Transthyretin and the transthyretin amyloidoses. In: Uversky VN and Fink AL, editors. Protein misfolding, aggregation, and conformational diseases: part B: Molecular mechanisms of conformational diseases. Springer, USA; 2007. p. 259–283. [Google Scholar]
- 22. Saponaro F, Kim JH, Chiellini G. Transthyretin stabilization: an emerging strategy for the treatment of Alzheimer's disease? Int J Mol Sci. 2020;21:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buxbaum JN, Ye Z, Reixach N, et al. Transthyretin protects Alzheimer's mice from the behavioral and biochemical effects of Abeta toxicity. Proc Natl Acad Sci U S A. 2008;105:2681–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kerridge C, Belyaev ND, Nalivaeva NN, Turner AJ. The Aβ-clearance protein transthyretin, like neprilysin, is epigenetically regulated by the amyloid precursor protein intracellular domain. J Neurochem. 2014;130:419–431. [DOI] [PubMed] [Google Scholar]
- 25. Vieira M, Saraiva MJ. Transthyretin: A multifaceted protein. Biomol Concepts. 2014;5:45–54. [DOI] [PubMed] [Google Scholar]
- 26. Tsai KJ, Yang CH, Fang YH, et al. Elevated expression of TDP-43 in the forebrain of mice is sufficient to cause neurological and pathological phenotypes mimicking FTLD-U. J Exp Med. 2010;207:1661–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scotter EL, Vance C, Nishimura AL, et al. Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J Cell Sci. 2014;127:1263–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Homma S, Beermann ML, Boyce FM, Miller JB. Expression of FSHD-related DUX4-FL alters proteostasis and induces TDP-43 aggregation. Ann Clin Transl Neurol. 2015;2:151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun X, Song J, Huang H, Chen H, Qian K. Modeling hallmark pathology using motor neurons derived from the family and sporadic amyotrophic lateral sclerosis patient-specific iPS cells. Stem Cell Res Ther. 2018;9:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. [DOI] [PubMed] [Google Scholar]
- 31. Wong CE, Chu YP, Tsai KJ. Structure-based analysis of protein cluster size for super-resolution microscopy in the nervous system. Biomedicines. 2022;10:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsai KJ, Tsai YC, Shen CK. G-CSF rescues the memory impairment of animal models of Alzheimer's disease. J Exp Med. 2007;204:1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shahheydari H, Ragagnin A, Walker AK, et al. Protein quality control and the amyotrophic lateral sclerosis/frontotemporal dementia continuum. Front Mol Neurosci. 2017;10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ito Y, Yamada M, Tanaka H, et al. Involvement of CHOP, an ER-stress apoptotic mediator, in both human sporadic ALS and ALS model mice. Neurobiol Dis. 2009;36:470–476. [DOI] [PubMed] [Google Scholar]
- 36. Luhr M, Torgersen ML, Szalai P, et al. The kinase PERK and the transcription factor ATF4 play distinct and essential roles in autophagy resulting from tunicamycin-induced ER stress. J Biol Chem. 2019;294:8197–8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prasad A, Bharathi V, Sivalingam V, Girdhar A, Patel BK. Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front Mol Neurosci. 2019;12:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shibutani ST, Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res. 2014;24:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lauterbach MA. Finding, defining and breaking the diffraction barrier in microscopy—A historical perspective. Optical Nanoscopy. 2012;1:8. [Google Scholar]
- 40. Kedia S, Ramakrishna P, Netrakanti PR, et al. Alteration in synaptic nanoscale organization dictates amyloidogenic processing in Alzheimer's disease. iScience. 2021;24:101924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rzymski T, Milani M, Pike L, et al. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 2010;29:4424–4435. [DOI] [PubMed] [Google Scholar]
- 43. Zhang P, Sun Q, Zhao C, et al. HDAC4 protects cells from ER stress induced apoptosis through interaction with ATF4. Cell Signal. 2014;26:556–563. [DOI] [PubMed] [Google Scholar]
- 44. Ding WX, Ni HM, Gao W, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang IF, Guo BS, Liu YC, et al. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci U S A. 2012;109:15024–15029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mattson MP, Magnus T. Ageing and neuronal vulnerability. Nat Rev Neurosci. 2006;7:278–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Costa R, Ferreira-da-Silva F, Saraiva MJ, Cardoso I. Transthyretin protects against A-beta peptide toxicity by proteolytic cleavage of the peptide: a mechanism sensitive to the Kunitz protease inhibitor. PLoS ONE. 2008;3:e2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwarzman AL, Gregori L, Vitek MP, et al. Transthyretin sequesters amyloid beta protein and prevents amyloid formation. Proc Natl Acad Sci U S A. 1994;91:8368–8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu L, Murphy RM. Kinetics of inhibition of beta-amyloid aggregation by transthyretin. Biochemistry. 2006;45:15702–15709. [DOI] [PubMed] [Google Scholar]
- 50. Li X, Buxbaum JN. Transthyretin and the brain re-visited: Is neuronal synthesis of transthyretin protective in Alzheimer's disease? Mol Neurodegener. 2011;6:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Iridoy MO, Zubiri I, Zelaya MV, et al. Neuroanatomical quantitative proteomics reveals common pathogenic biological routes between amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Int J Mol Sci. 2018;20:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ryberg H, An J, Darko S, et al. Discovery and verification of amyotrophic lateral sclerosis biomarkers by proteomics. Muscle Nerve. 2010;42:104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brettschneider J, Lehmensiek V, Mogel H, et al. Proteome analysis reveals candidate markers of disease progression in amyotrophic lateral sclerosis (ALS). Neurosci Lett. 2010;468:23–27. [DOI] [PubMed] [Google Scholar]
- 54. Ren H, Zhai W, Lu X, Wang G. The cross-links of endoplasmic reticulum stress, autophagy, and neurodegeneration in Parkinson's disease. Front Aging Neurosci. 2021;13:691881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hetz C, Saxena S. ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol. 2017;13:477–491. [DOI] [PubMed] [Google Scholar]
- 56. Chen S, Fragoza R, Klei L, et al. An interactome perturbation framework prioritizes damaging missense mutations for developmental disorders. Nat Genet. 2018;50:1032–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rual JF, Venkatesan K, Hao T, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. [DOI] [PubMed] [Google Scholar]
- 58. B'Chir W, Maurin AC, Carraro V, et al. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vieira M, Gomes JR, Saraiva MJ. Transthyretin induces insulin-like growth factor I nuclear translocation regulating its levels in the hippocampus. Mol Neurobiol. 2015;51:1468–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wortel IMN, van der Meer LT, Kilberg MS, van Leeuwen FN. Surviving stress: modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol Metab. 2017;28:794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vernon E, Meyer G, Pickard L, et al. GABA(B) receptors couple directly to the transcription factor ATF4. Mol Cell Neurosci. 2001;17:637–645. [DOI] [PubMed] [Google Scholar]
- 62. Nehring RB, Horikawa HPM, El Far O, et al. The metabotropic GABAB receptor directly interacts with the activating transcription factor 4. J Biol Chem. 2000;275:35185–35191. [DOI] [PubMed] [Google Scholar]
- 63. Bolger TA, Yao TP. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J Neurosci. 2005;25:9544–9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vieira M, Leal SS, Gomes CM, Saraiva MJ. Evidence for synergistic action of transthyretin and IGF-I over the IGF-I receptor. Biochim Biophys Acta. 2016;1862:797–804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article and Supplementary material. Further inquiries can be directed to the corresponding author on reasonable request.