Abstract
Rubella virus is a small enveloped positive-strand RNA virus that assembles on intracellular membranes in a variety of cell types. The virus structural proteins contain all of the information necessary to mediate the assembly of virus-like particles in the Golgi complex. We have recently identified intracellular retention signals within the two viral envelope glycoproteins. E2 contains a Golgi retention signal in its transmembrane domain, whereas a signal for retention in the endoplasmic reticulum has been localized to the transmembrane and cytoplasmic domains of E1 (T. C. Hobman, L. Woodward, and M. G. Farquhar, Mol. Biol. Cell 6:7–20, 1995; T. C. Hobman, H. F. Lemon, and K. Jewell, J. Virol. 71:7670–7680, 1997). In the present study, we have analyzed the role of these retention signals in the assembly of rubella virus-like particles. Deletion or replacement of these domains with analogous regions from other type I membrane glycoproteins resulted in failure of rubella virus-like particles to be secreted from transfected cells. The E1 transmembrane and cytoplasmic domains were not required for targeting of the structural proteins to the Golgi complex and, surprisingly, assembly and budding of virus particles into the lumen of this organelle; however, the resultant particles were not secreted. In contrast, replacement or alteration of the E2 transmembrane or cytoplasmic domain, respectively, abrogated the targeting of the structural proteins to the budding site, and consequently, no virion formation was observed. These results indicate that the transmembrane and cytoplasmic domains of E2 and E1 are required for early and late steps respectively in the viral assembly pathway and that rubella virus morphogenesis is very different from that of the structurally similar alphaviruses.
Rubella virus (RV) is the sole member of the genus Rubivirus within the family Togaviridae. Humans are the only natural host for this virus, which causes a mild childhood disease known as German measles (Reviewed in references 8 and 56). The most serious medical consequences of RV infection occur during the first trimester of pregnancy, in which case in utero infection of the fetus often results in a collection of severe malformations known as congenital rubella syndrome. Despite the wealth of clinical and epidemiological information regarding RV, the biology of this virus remains poorly understood. The study of RV has been hampered in large part due to the inherent difficulties associated with growing the virus. Historically, it has been assumed that RV is very similar if not identical with respect to replication and assembly to the better-studied members of the togavirus family, namely, alphaviruses. While it appears that RV and alphaviruses are similar in many respects with regard to entry and replication in host cells (31, 42), it has become clear that the assembly pathways of rubiviruses and alphaviruses are quite different. Some differences of note are that (i) RV generally buds from intracellular membranes, in contrast to alphaviruses, which mature at the plasma membrane; (ii) RV capsid protein does not possess an autoprotease activity (6, 39), whereas alphavirus capsid has a serine protease domain that catalyzes its release from the structural protein precursor (34); and (iii) RV nucleocapsid assembly occurs in association with membranes and is synchronized with virus budding (8). Conversely, alphavirus nucleocapsid assembly occurs independently of membranes and virus budding (reviewed in reference 50).
Rubella virions contain three structural proteins: a capsid protein that complexes with 40S RNA to form a nucleocapsid and two membrane-spanning glycoproteins, E2 and E1, located in the virus envelope (38). The structural proteins are synthesized as a 110-kDa precursor polyprotein which is endoproteolytically cleaved to produce capsid, E2, and E1 (39). E2 and E1 are type I membrane proteins which have amino-terminal, independently functioning signal peptides that facilitate translocation into the endoplasmic reticulum (ER) (17, 20). Capsid protein remains in the cytoplasm in association with membranes and complexes with viral genomic RNA to form nucleocapsids (51). E2 and E1 form a heterodimer in the ER and are transported as a complex to the Golgi apparatus, where they mediate intracellular budding (2, 22).
Our recent studies have focused upon how RV structural proteins and RNA are assembled into virions. We have demonstrated that coordinated expression of capsid, E2, and E1 proteins in mammalian cells results in their assembly into rubella virus-like particles (RLPs) (19). Since alphavirus assembly does not occur with any measurable efficiency in the absence of viral genome (52), the process of genomic RNA-independent viral assembly is seemingly unique to RV among togaviruses. RLPs are very similar to native RV virions in terms of morphology, antigenicity, and immunogenicity (11, 20, 44, 45). In addition, RLPs associate with the plasma membrane of host cells and are found in multivesicular bodies, which indicates that they may be endocytosed in the same manner as infectious virions (reference 19 and our unpublished observations). Thus, RLPs represent a suitable model system to study RV morphogenesis.
In the present study, we have investigated the role of the RV glycoprotein domains in the assembly of RLPs. Our results indicate that the transmembrane (TM) and cytoplasmic (CT) domains of E2 and E1 are required for early and late steps, respectively, in the viral assembly pathway. Furthermore, interaction between the CT domain of E1 and capsid does not appear to be the major interaction which drives the budding reaction as previously hypothesized (19). These results indicate the assembly of RV virions differ significantly from that of alphaviruses and may have implications for the design of recombinant vaccines which use RV as a vector.
MATERIALS AND METHODS
Reagents.
Reagents and supplies were from the following sources. Protein A- and G-Sepharose were purchased from Pharmacia (Alameda, Calif.). Fibronectin, sodium dodecyl sulfate (SDS), dialyzed fetal bovine serum, and bovine serum albumin were purchased from Sigma Chemical Co. (St. Louis, Mo.). Promix [35S]methionine-cysteine (1,000 Ci/mmol) and 14C-labeled protein standards were purchased from Amersham Corp. (Arlington Heights, Ill.). 32Pi (500 mCi/ml) and minimal essential medium (MEM) lacking cysteine and methionine were purchased from ICN Biomedicals (Irvine, Calif.). OptiMEM serum-free medium, fetal bovine serum, and alpha-MEM without nucleosides were obtained from Life Technologies Inc. (Gaithersburg, Md.). DOSPER transfection reagent, Pefabloc, and Pwo polymerase were purchased from Boehringer Mannheim Corporation (Laval, Quebec, Canada). Immobilon-P PVDF (polyvinylidene fluoride) membranes, 0.45-μm pore size, were purchased from Millipore Corporation (Bedford, Mass.). Recombinant endoglycosidase (endo H) was purchased from New England Biolabs (Beverly, Mass.).
Antibodies.
Monoclonal antibodies to RV structural proteins were kindly provided by John Safford, Abbott Laboratories (North Chicago, Ill.), Barbara Pustowoit, University of Leipzig (Leipzig, Germany), and Jerry Wolinski, University of Texas (Houston, Tex.). Human anti-RV was provided by Aubrey Tingle, University of British Columbia (Vancouver, British Columbia, Canada). Rabbit anti-mannosidase II (Man II) was provided by Marilyn G. Farquhar, University of California, San Diego (La Jolla, Calif.). Goat anti-mouse immunoglobulin G (IgG) conjugated to horseradish peroxidase (HRP) was purchased from Bio-Rad Laboratories (Hercules, Calif.). Texas red-conjugated goat anti-mouse IgG and fluorescein isothiocyanate-conjugated donkey anti-rabbit IgG (each double-labeling grade) were purchased from and Jackson ImmunoResearch Laboratories (West Grove, Pa.).
Recombinant plasmids.
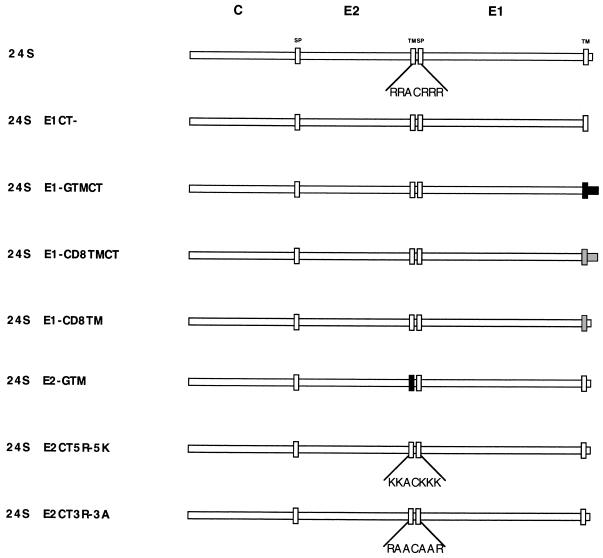
All RV cDNA constructs were subcloned into the expression vector pCMV5 (1) between the EcoRI and HindIII or BamHI sites of the polylinker downstream from the cytomegalovirus promoter. TM and/or CT domains of RV glycoproteins were deleted, altered, or replaced with analogous domains from two other type I membrane glycoproteins, vesicular stomatitis virus (VSV) G protein (46) or CD8 (29), using PCR with Pwo polymerase. Generally, 20 to 30 cycles were used for each reaction to minimize the chances of introducing second-site mutations. All products were verified by DNA sequencing. A schematic diagram of all the constructs is shown in Fig. 1.
FIG. 1.
Schematic of RV 24S expression constructs. The RV sequences are shown as white, whereas VSV G and CD8 sequences are indicated as black and gray, respectively. The signal peptide (SP) and TM domains are indicated by thin rectangles at the beginnings and ends of the E2 and E1 proteins. The 24S cDNA encodes normal capsid (C), E2, and E1. The amino acid sequence of the E2 CT domain located between the E2 TM and E1 signal peptide domains is shown below the construct. Constructs are named to reflect the relevant changes to domains in E2 or E1. For example, 24SE1CT− encodes normal capsid, E2, and an E1 protein which is lacking the CT domain, and 24SE2-GTM encodes normal capsid, E1, and an E2 protein in which the TM domain has been replaced by the analogous region from VSV G protein. Sequences of the mutated E2 CT domains are shown below the 24SE2CT5R-5K and 24SE2CT3R-3A constructs. All cDNA constructs were subcloned between the EcoRI and BamHI or HindIII sites of pCMV5 and stably transfected into CHO cells.
The 24S cDNA encodes the RV structural proteins in the order NH2-C-E2-E1-COOH (5). The 24SE1CT− cDNA encodes capsid-E2 and an E1 gene which lacks the coding region for the 13-amino-acid CT domain (19). In the 24SE1-GTMCT cDNA, the coding regions for the E1 TM and CT domains were replaced by the analogous regions from the VSV G protein (19). Similarly, in 24SE1-CD8TMCT and 24SE1-CD8TM, the coding regions for E1 TM and CT domains or E1 TM domain were replaced with those from CD8. In the 24SE2-GTM cDNA, the E2 TM domain was replaced with the TM domain from VSV G. The coding region of the E2 CT domain in 24SE2CT5R-5K was mutagenized such that five arginine residues were changed to lysines, whereas in 24SE2CT3R-3A three arginines are replaced with three alanines (Fig. 1).
Cell culture and transfection.
CHODG44 cells were cultured and stably transfected exactly as described elsewhere (21). COS cells were transfected by the calcium phosphate method as described elsewhere (48) and were used for metabolic labeling and radioimmunoprecipitation 40 to 48 h posttransfection.
Metabolic labeling and radioimmunoprecipitation.
Biosynthetic labeling with 35S-amino acids, radioimmunoprecipitation, and endo H digestion of RV proteins from transfected cells have been described previously (22). To examine phosphorylation of capsid protein, cells were first cultured for 16 h with 32Pi (500 μCi/ml) in phosphate-free medium. Cell lysis and immunoprecipitations were carried out in the presence of phosphatase inhibitors (10 mM sodium orthovanadate, 50 mM sodium fluoride, 50 mM tetrasodium pyrophosphate).
SDS-PAGE and autoradiography.
Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% polyacrylamide gels and processed for autoradiography or immunoblotting (22) as described below.
Immunofluorescence microscopy.
Cells were grown on fibronectin (10 μg/ml)-coated 12-mm-diameter glass coverslips, fixed with methanol at −20°C, and processed for indirect immunofluorescence as described elsewhere (21).
Immunoblotting and RLP secretion assay.
Secretion of RLPs from transfected cells was assayed by immunoblotting 100,000 × g pellets prepared from clarified conditioned medium by using a monoclonal antibody to capsid protein. Briefly, cells were washed twice with phosphate-buffered saline, then fresh medium was added, and incubation was continued at 37°C for various time periods to allow secretion of RLPs. At specific time periods, the medium was removed and centrifuged at 14,000 × g for 5 min to remove cell-associated material. RLPs were recovered from the precleared medium by centrifugation at 100,000 × g for 60 min at 4°C in a TLS 55 rotor. The 100,000 × g pellets were resuspended and boiled in 2× SDS-gel loading buffer, followed by SDS-PAGE through 10% gels. The proteins were transferred to a PVDF membrane (250 mA for 30 min), using a semidry blotting apparatus (Tyler Research Instruments, Edmonton, Alberta, Canada).
Capsid protein was detected by sequential incubations with a mouse anticapsid monoclonal antibody followed by HRP-conjugated goat anti-mouse antibody and enhanced chemiluminescence (ECL).
Electron microscopy.
Cells grown on fibronectin-coated 12-mm-diameter coverslips were processed for electron microscopy essentially as described elsewhere (15). Briefly, cells were fixed in 1.5% glutaraldehyde–0.1 M cacodylate (pH 7.4) containing 5% sucrose for 1 h at room temperature and then washed three times (5 min each) in cacodylate buffer. Samples were then postfixed with 1% OsO4–0.05 M potassium ferricyanide–0.1 M phosphate buffer (pH 7.4) for 1 h on ice and washed with water three times (5 min each). Cells were dehydrated in a graded series of ethanol and embedded in Epon. Sections were cut parallel to the coverslips, stained with 2% uranyl acetate and lead citrate, and examined in a Philips model 410 electron microscope.
RESULTS
The E1 TM and CT domains are required for secretion of RLPs.
Our previous work indicated that the TM and/or CT domains of RV E1 glycoprotein were required for secretion of RLPs from transfected cells (19). However, in that study it was not determined at which step in the virus assembly pathway that these E1 domains were required. The assembly of RV virions and RLPs can be divided into four parts: (i) synthesis, translocation, and folding of structural proteins in the ER; (ii) targeting of proteins to site of virus assembly (Golgi complex); (iii) assembly of proteins into virus particles; and (iv) secretion of virus particles into the extracellular space. We have recently demonstrated that replacement of the E1 TM and CT domains with the analogous regions from VSV G protein does not affect targeting of E2 and E1 to the Golgi complex (23). Based on our topological model of the RV structural proteins, we predicted that the E1 CT domain interacts with capsid protein to drive the assembly of viral particles into the lumen of the Golgi complex (19). Consequently, replacement or deletion of the E1 CT domain would abrogate the formation of virus particles, possibly by affecting the recruitment of capsid protein to the Golgi complex. Stably transfected CHO cells expressing C, E2, and E1 proteins with deletions or replacements of the TM and CT domains were constructed so that we could test this prediction.
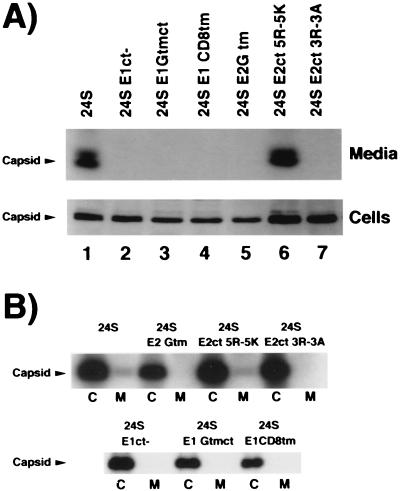
We used a rapid and sensitive immunoblot assay to detect RLP secretion from transfected cells. The RLPs were pelleted from conditioned media by centrifugation at 100,000 × g, and subjected to SDS-PAGE and immunoblotting using a monoclonal antibody to capsid protein as described above. We also monitored the phosphorylation of capsid protein to determine whether mutations in the viral glycoproteins would affect this process. Capsid protein was readily detected in the media of CHO24S cells (Fig. 2A, lane 1). In contrast, deletion of the CT domain or replacement of the TM and/or CT domains of E1 with analogous domains from two other type I membrane glycoproteins, VSV G and CD8, completely abrogated secretion of RLPs into the medium (Fig. 2A, lanes 2 to 4) but did not affect the phosphorylation of capsid protein (Fig. 2B).
FIG. 2.
Mutations in the E2 and E1 TM and CT domains abrogate RLP secretion but not phosphorylation of capsid protein. (A) Medium from CHO cells stably expressing various RV 24S constructs was harvested after 3 h, and RLPs were pelleted by centrifugation at 100,000 × g. The RLPs and cell lysates were subjected to SDS-PAGE on 10% gels and transferred to PVDF membranes. Membranes were probed with mouse anticapsid, followed by probing with goat anti-mouse IgG conjugated to HRP and ECL detection. The gel containing the secreted capsid proteins was run longer to show that capsid protein migrates as a doublet. Identical results were obtained when medium was harvested at 6 h (not shown). (B) CHO cells were labeled for 16 h with 32Pi. Cell lysates and media were subjected to radioimmunoprecipitation with human anti-RV serum and protein A-Sepharose followed by SDS-PAGE and fluorography.
These experiments confirm our previous observations that E1 TM and CT domains are required for secretion of RLPs (19). E2 and E1 were also detected in the cell lysates and media from CHO24S cells by immunoblotting during these time periods but not in the media from cells expressing 24S cDNAs with altered E1 TM and/or CT domains (not shown). All of the transfected CHO cells were subjected to analysis by biosynthetic labeling with [35S]methionine-cysteine and radioimmunoprecipitation using anti-RV sera. These experiments also demonstrated that capsid, E2, and E1 were secreted in a time-dependent manner from CHO24S cells but not CHO24SE1CT−, 24SE1-GTMCT, or 24SE1-CD8TM cells (not shown).
The E1 TM and CT domains are not required for targeting of RV proteins to the Golgi complex.
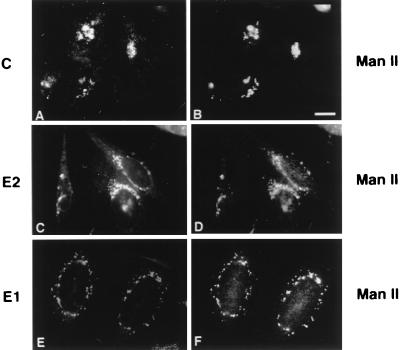
We next sought to determine at which point in the particle assembly pathway the E1 TM and CT domains were required. Biosynthetic labeling and radioimmunoprecipitation of RV antigens from the transfected CHO cells indicated that replacement of the E1 TM and CT domains did not affect the processing and translocation of RV structural proteins (not shown). Therefore, the block in RLP secretion must occur at a later stage in the assembly pathway. The intracellular localization of RV structural proteins was examined by double-label indirect immunofluorescence using monoclonal antibodies to RV structural proteins and a rabbit antibody to the Golgi membrane protein, Man II (53). Consistent with our previous studies, all three of the RV structural proteins were concentrated in the Golgi region of CHO24S cells (Fig. 3).
FIG. 3.
RV structural proteins are concentrated in the Golgi complex. CHO24S cells grown on coverslips were fixed with methanol and processed for double-label indirect immunofluorescence using mouse anticapsid (A), mouse anti-E2 (C), or mouse anti-E1 (E) and rabbit anti-Man II to stain the Golgi complex (B, D, and F). Bar = 10 μm.
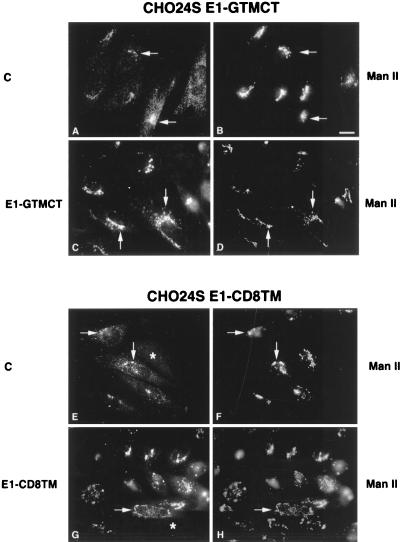
Replacement of the TM domain or both the TM and CT domains of E1 did not affect targeting of E1 to the Golgi complex (Fig. 4C, D, G, and H). Unexpectedly, capsid protein was also concentrated in the Golgi region of these cells (Fig. 4A, B, E, and F), indicating that the TM and/or CT domains of E1 were not required for targeting of capsid to the Golgi complex. In all of the cell lines described above, the intracellular distribution of E2 was indistinguishable from E1 in that it localized to the Golgi complex (not shown). Similarly, stably transfected cells expressing a 24S mutant in which both the E1 TM and CT domains were replaced by the analogous domains from CD8 (24SE1-CD8TMCT) or the CT domain of E1 had been deleted (24SE1CT−) showed characteristics essentially identical to those shown in Fig. 4 with regard to localization of the RV proteins (not shown).
FIG. 4.
The E1 TM and CT domains are not required for targeting of RV structural proteins to the Golgi complex. CHO24SE1-GTMCT (A to D) and 24SE1-CD8TM (E to H) cells were processed for indirect immunofluorescence using mouse antibodies to capsid (A and E) or E1 (C and G) and rabbit anti-Man II (B, D, F, and H). Colocalization between RV structural proteins and the Golgi marker Man II are indicated by arrows. In all cases, E2 showed a distribution similar to that of E1 (not shown). Some of the cells have diminished expression of RV structural proteins (E and G, asterisks). Bar = 10 μm.
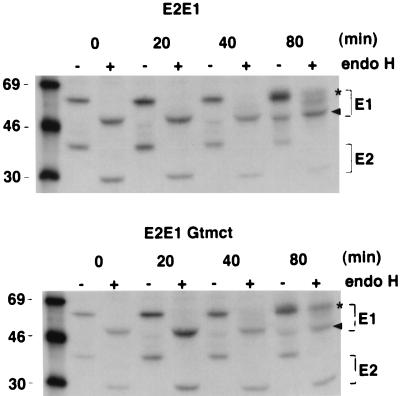
Our recent work demonstrated that the TM and CT domains of E1 comprise an ER retention signal which we believe functions to delay the transport of the E2/E1 heterodimer from the ER to the Golgi complex until the folding of E1 is completed (14). Accordingly, we predicted that replacement of these E1 domains would result in more rapid transport of E2 and E1 from the ER. To test this hypothesis, we conducted biosynthetic labeling experiments with stably transfected CHO cells expressing normal E2 and E1 or E2 and E1-GTMCT (22, 23). The heterogeneous nature of mature E2, which migrates as a 42- to 47-kDa smear, made it difficult to accurately quantitate the rate of ER to Golgi transport for this glycoprotein; therefore, only the rate of ER to Golgi transport of E1 was determined. After 80 min, more than ∼51% of E1-GTMCT was resistant to endo H, whereas at the same time point, <30% of E1 had been converted to the endo H-resistant form (Fig. 5). From these results, we conclude that the E1 TM and CT domains are not required for targeting of E2, E1, or capsid protein to the site of virus budding, the Golgi complex, but they serve to delay transport of the E2/E1 heterodimer from the ER.
FIG. 5.
The E1 TM and CT domains delay transport of E1 to the Golgi complex. Stably transfected CHO cells expressing E2 and E1 or E2 and E1-GTMCT were pulse-labeled with [35S]Met-cys for 10 min and chased for various time periods in the absence of radioactivity before lysis and immunoprecipitation with human anti-RV serum. The radioimmunoprecipitates were incubated with or without endo H, separated on 10% gels, and processed for autoradiography as described in the text. Endo H-resistant (asterisks) and -sensitive (arrowheads) forms of E1 and E1-GTMCT are indicated. Sizes are indicated in kilodaltons.
The E2 TM and CT domains are important for assembly of the E2/E1 heterodimer and transport of the structural proteins to the Golgi complex.
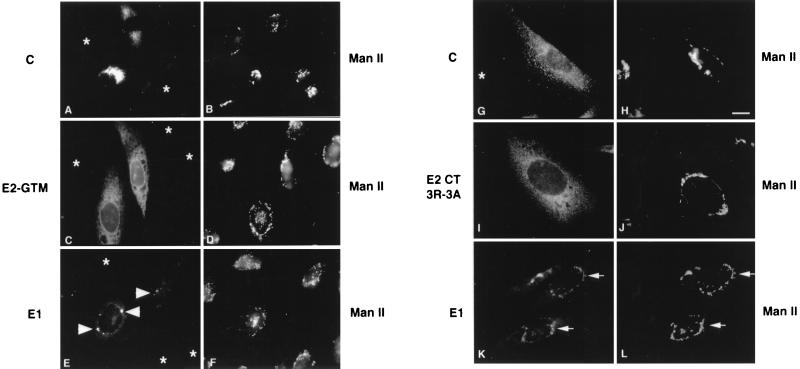
The E2 TM domain contains a Golgi retention signal which is thought to mediate intracellular budding of rubella virions into the lumen of this organelle (23). To assess the importance of this domain for RV particle assembly, the E2 TM was replaced with the TM region from VSV G protein as described elsewhere (23). The cDNA encoding E2-GTM was subcloned into the 24S cDNA for expression in stably transfected CHO cells. As shown in Fig. 2A (lane 5), capsid protein was detected in the cell lysates of CHO24SE2-GTM cells but not the medium, indicating that RLPs were not secreted from these cells. Examination of these cells by indirect immunofluorescence revealed that none of the RV structural proteins were present in the Golgi complex (Fig. 6A, C, and E). Capsid protein was localized throughout the cytoplasm but was not detected in association with the Golgi complex (Fig. 6A and B). E2-GTM was distributed throughout a cytoplasmic reticular network and the nuclear envelope, indicating that this glycoprotein resided in the ER (Fig. 6C and D). Some E1 was also detected in the ER, but the highest concentrations of this protein were found in discrete perinuclear membrane structures (Fig. 6E). Examination of CHO24SE2-GTM cells by electron microscopy revealed the presence of ER-derived smooth tubular membrane nests (see Fig. 10) similar to those found in cells expressing E1 alone (15, 21). Since E2-GTM was not detected in these structures, it is quite likely that E1 and E2-GTM do not associate to form a heterodimer.
FIG. 6.
The E2 TM and CT domains are important for targeting of RV structural proteins to the Golgi complex. CHO24SE2-GTM (A to F) and CHO24SE2CT3R-3A (G to L) cells were processed for indirect immunofluorescence using mouse antibodies to capsid (A and G), E2 (C and I), E1 (E and K), and rabbit anti-Man II (B, D, F, H, J, L). In CHO24SE2-GTM cells, E1 is concentrated in perinuclear membrane structures (E, arrowheads). Colocalization between RV structural proteins and the Golgi marker Man II are indicated by arrows (K and L). Some of the cells have diminished expression of RV structural proteins (A, C, E, and G, asterisks). Bar = 10 μm.
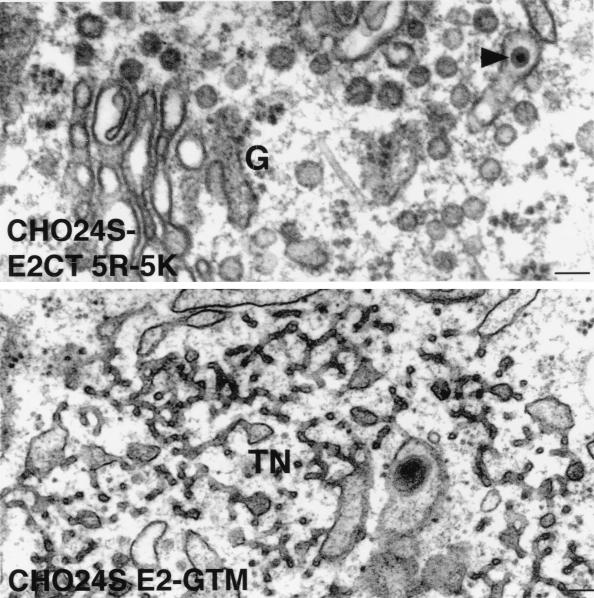
FIG. 10.
The E2 TM domain is required for formation of RLPs. CHO24SE2CT5R-5K and CHO24SE2-GTM cells were prepared for routine morphology by embedding in Epon, and ultrathin sections were examined by electron microscopy. RLPs (arrowheads) can be seen in the lumen of Golgi cisterna (G) of in CHO24SE2CT5R-5K cells but not in CHO24SE2-GTM cells. In the latter cell type, networks of smooth tubular membranes (TN) were also evident in the cytoplasm. Bars = 0.1 μm.
RV capsid has been reported to undergo phosphorylation (32), but it is unknown which enzyme(s) catalyzes this posttranslational modification or where this occurs. Using the Prosite algorithm, we determined that this protein contains two potential protein kinase C phosphorylation sites. Interestingly, the eta and epsilon isoforms of protein kinase C have been localized to the ER and Golgi complex, respectively (4, 28). We took advantage of the fact that two of the 24S mutants resulted in failure of capsid to localize to the Golgi complex to determine whether phosphorylation of capsid protein required proper targeting to the viral budding site. Cell lines were labeled with 32Pi, and RV proteins were immunoprecipitated and subjected to SDS-PAGE and fluorography. Phosphorylation of capsid protein was not dependent on its localization to the Golgi complex or incorporation into virus particles, since cell-associated capsid efficiently incorporated 32Pi in all of the 24S mutants (Fig. 2B).
The predicted CT domain of E2 is a loop of seven amino acids, five of which are arginines (Fig. 1). This domain is likely to be closely associated with the negatively charged phospholipid head groups of cell membranes. To determine if the arginines rather than the overall charge of this domain are important for virus assembly, we changed the five arginines to five lysines by site-directed mutagenesis. The resulting construct, 24SE2CT5R-5K (Fig. 1), was stably expressed in CHO cells and subjected to analysis as described above. Indirect immunofluorescence analysis revealed that in CHO24SE2CT5R-5K cells, all of the RV structural proteins were correctly targeted to the Golgi complex (not shown). Moreover, the capacity for these cells to assemble and secrete RLPs was not noticeably altered (Fig. 2A, lane 6). We also changed the three internal arginine residues to alanines to generate the construct 24SE2CT3R-3A (Fig. 1). The two flanking arginine residues were not changed, as they may be important for the overall topology of the E2 TM and E1 signal peptide domains within the membrane (55). Capsid was not detected in the media of CHO24SE2CT3R-3A cells, indicating that RLP secretion was abrogated by these mutations (Fig. 2A, lane 7). Unexpectedly, these changes to the E2 CT domain also affected the targeting of RV structural proteins in transfected cells. Capsid protein was not associated with the Golgi complex but instead was distributed throughout the cytoplasm in punctate structures (Fig. 6G and H). E1 was present in the Golgi complex, whereas E2CT3R-3A was distributed throughout the ER (Fig. 6I to L). We were puzzled by this result since our previous work indicated that E2 and E1 must form a complex before E1 can be transported efficiently to the Golgi complex (16, 21).
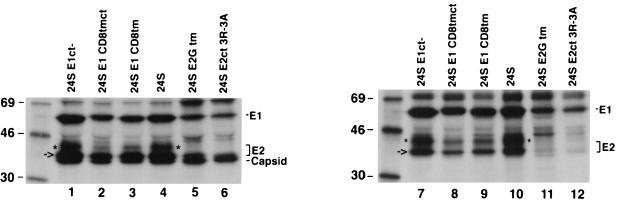
Very little colocalization was observed between E1 and E2-GTM or E2CT3R-3A, suggesting that mutations in the E2 TM and CT domains may affect the assembly of the E2/E1 heterodimer. To address this possibility, we performed coimmunoprecipitation experiments to assay the dimerization of E2 and E1 as described elsewhere (22). Since the expression levels of the stably transfected cell lines varied, the experiments were conducted in transiently transfected COS cells to ensure more uniform expression levels. Transfected cells were pulse-labeled with [35S]Cys-Met and chased for 30 min to allow dimerization of E2 and E1 to occur in the ER. Lysates were divided into two aliquots which were incubated with human anti-RV serum to precipitate all three RV proteins (Fig. 7, lanes 1 to 6) or a monoclonal antibody to E1 (lanes 7 to 12). E2 did not coimmunoprecipitate with E1 in cells expressing 24SE2-GTM or 24SE2CT3R-3A (lanes 11 and 12) even though E2 was clearly present in these cells (lanes 5 and 6). Conversely, alterations in the E1 TM and/or CT domains did not significantly affect the binding of E1 to E2 (lanes 7 to 9). Two species of E2 coprecipitated with E1 under these conditions. The 42-kDa form of E2 is thought to represent a Golgi-processed form of the glycoprotein that contains O-linked carbohydrates (18, 30). Since E2 is not efficiently transported from the ER unless it is bound to E1, only the 39-kDa ER form of E2 is present in cells expressing 24SE2-GTM or 24SE2CT3R-3A (lanes 5 and 6). These results indicate the E2 TM and CT domains are important for dimerization with E1 and its subsequent transport to the Golgi complex.
FIG. 7.
The E2 TM and CT domains are important for assembly of the E2/E1 heterodimer. COS cells were transfected with plasmids encoding wild-type RV 24S cDNA or various 24S mutants encoding altered E1 or E2 glycoproteins. Forty hours posttransfection, cells were pulse-labeled for 15 min with [35S]Met-Cys and chased for 30 min in the absence of radioactivity. Cell lysates were divided into two aliquots which were immunoprecipitated with human anti-RV serum (lanes 1 to 6) or a mouse monoclonal antibody to E1 (lanes 7 to 12). Samples were then subjected to SDS-PAGE on 10% gels followed by fluorography. The positions of capsid, E2, and E1 are indicated. The species of E2 that migrates at 42 kDa (asterisk) represents the Golgi processed form of E2 that contains O-linked sugars. The 39-kDa form of E2 (arrow) represents the ER form of the glycoprotein.
The E1 CT domain is not required for assembly of RLPs.
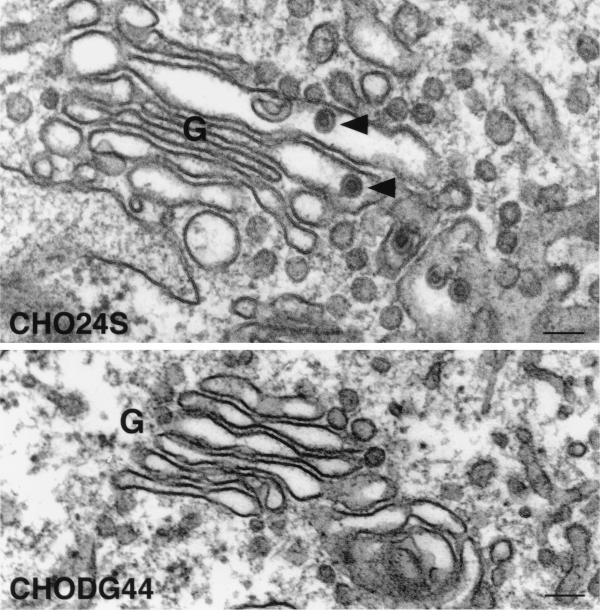
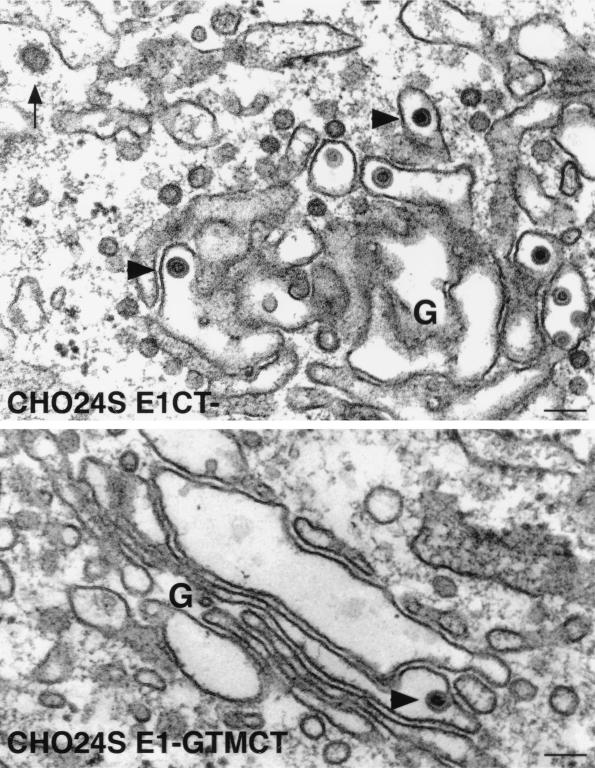
When CHO24S cells were examined by electron microscopy, RLPs could be seen in various stages of assembly and budding into the lumen of the Golgi complex (reference 19 and Fig. 8). RLPs were not detected in the Golgi complex of untransfected CHODG44 cells (Fig. 8). The RLPs are similar in size and appearance to native rubella virions (reviewed in references 8 and 56) in that they are 50 to 60 nm in diameter and have a ∼30-nm electron-dense core (Fig. 8). The CHO24S cells expressing mutant E2 or E1 glycoproteins were subjected to analysis by electron microscopy to determine if RLPs were formed but not secreted. Unexpectedly, RLPs were present in the Golgi complex of both CHO24SE1CT− and CHO24SE1-GTMCT cells (Fig. 9). These particles were indistinguishable from the RLPs found in CHO24S cells (Fig. 8) and were found only in the Golgi complex or associated vacuoles. As expected, RLPs were also found in the Golgi complex of CHO24SE2CT5R-5K cells (Fig. 10). Mutations in E2 which affected targeting of one or more of the RV structural proteins to the Golgi also blocked the formation of intracellular RLPs. Specifically, RLPs were not found in the Golgi complex or any other intracellular membrane structures of CHO24SE2-GTM and CHO24SE2CT-3R-3A cells (not shown). However, in CHO24SE2-GTM cells, but not in cells expressing other 24S mutants, nests of smooth tubular membranes were evident in the cytoplasm (Fig. 10). We assume that these tubular networks are the same E1-containing perinuclear foci shown in Fig. 6E. In previous studies, we determined that these ER-derived structures proliferate when E1 fails to assemble with E2 (15, 21). These results indicate that assembly of RLPs requires targeting of all three RV structural proteins to the Golgi complex.
FIG. 8.
RLPs bud into the Golgi complex of CHO 24S cells. CHO24S and untransfected CHODG44 cells were prepared for routine morphology by embedding in Epon, and ultrathin sections were examined by electron microscopy. RLPs (arrowheads) can be seen in the lumen of stacked Golgi cisterna (G) of CHO24S cells but not in untransfected CHODG44 cells. Bars = 0.1 μm.
FIG. 9.
The E1 CT domain is not required for assembly of RLPs in the Golgi complex. CHO24SE1CT− and CHO24SE1-GTMCT cells were prepared for routine morphology by embedding in Epon, and ultrathin sections were examined by electron microscopy. RLPs (arrowheads) can been seen in the lumen of stacked Golgi cisterna (G) of both cell types. A clathrin-coated vesicle in the vicinity of the Golgi complex is indicated by an arrow. Bars = 0.1 μm.
DISCUSSION
The use of virus-like particles as models has provided a wealth of information about the assembly and replication pathways of many viruses (7, 9, 10, 12, 25–27, 35, 49, 54). Similarly, we reasoned that RLPs could serve as a faithful model system to examine the assembly of rubella virions. In this study we have investigated the role of the RV glycoprotein domains in the assembly of virus-like particles. Our results indicate that the E2 and E1 TM and CT domains function at different stages of the assembly pathway. Ultimately, these findings may have implications for design of recombinant live vaccines using RV as the vector.
The E2 TM and CT domains are required for early events in virus assembly.
We have recently determined that the TM domain of E2 contains a Golgi retention signal which may facilitate the process of intracellular budding (23). The E1 TM and CT domains are not required for retention of E2 and E1 in the Golgi complex, which suggests that the E2 TM domain is the major factor that determines where virus assembly takes place. We reasoned that substituting this domain with the analogous region from a type I plasma membrane protein such as VSV G may shift the site of virus assembly from the Golgi complex to the cell surface. Instead, replacement of the E2 TM domain resulted in failure of the RV structural proteins to be transported to the Golgi complex. Introduction of nonconservative mutations in the CT domain had much the same effect except that a substantial fraction of E1 was transported to the Golgi without E2. The significance of this observation remains to be determined since we have never before observed efficient transport of E1 to the Golgi without E2. It is possible, however, that E1 and E2CT3R-3A dimers are unstable but still able to be transported to the ER-Golgi intermediate compartment (ERGIC), where they dissociate. ERGIC is a sorting station which receives cargo from the ER and directs proteins and lipids forward to the Golgi stack or back to the ER (13). If dissociation does occur in ERGIC, E1 would proceed to the Golgi whereas E2CT3R-3A would be returned to the ER. The fact that E1 is retained in the Golgi complex suggests that this glycoprotein may also contain a retention signal that prevents transport to the plasma membrane.
From this study and our previous work (16), it is now clear that the RV E2 glycoprotein functions early in the viral assembly pathway. We have proposed that E2 acts a scaffold which binds newly synthesized E1 molecules in order to facilitate the maturation of this glycoprotein (22). Accordingly, mutations in the TM and/or CT domain of E2 may affect its ability to bind to E1 or act as a scaffold.
The E1 TM and CT domains are not required for particle assembly but are necessary for secretion.
In contrast to E2 mutants, replacement of the E1 TM and CT domains had no apparent effect on localization of capsid, E2, and E1 to the Golgi complex. Moreover, these domains were not required for the budding of RLPs into this organelle. This unexpected finding is not consistent with our earlier hypothesis that the E1 CT domain mediates the interaction between the glycoprotein spike complex and capsid protein to drive assembly (19). Although RLPs are formed and released into the lumen of the Golgi in the absence of the E1 TM and CT domains, they were not secreted into the medium. Thus, these E1 domains are critical for transport of virus particles between the Golgi and the cell surface. Since the TM and CT domains of E1 are predicted to reside within the RLP envelope and interior, respectively, it is unlikely that they could facilitate this process directly. Rather, we favor the hypothesis that alteration of these domains results in a conformational change in the ectodomain of E1 which affects the quaternary structure of the E2/E1 glycoprotein spike. In turn, the putative alterations in the E1 ectodomain could be due to direct or indirect effects. For example, if specific interactions between the E2 and E1 TM domains occur during heterodimer formation, replacement of the E1 TM domain could ultimately affect the quaternary structure of the E2/E1 complex. An alternative, but not mutually exclusive possibility is that the E1 TM and CT domains are needed to delay the transport of the E2/E1 dimer from the ER until complete maturation of E1 occurs (14). Presumably, delaying E2 and E1 in the ER serves to increase the time required for interaction with lumenal chaperones such as Bip and calnexin, thereby increasing the folding efficiency of these proteins (3). Thus, in the absence of the E1 ER retention signal, the E2/E1 heterodimer is transported to the Golgi at a higher rate, possibly before maturation of E1 and/or E2 is completed.
Although most aberrantly folded proteins are retained in the ER (47), there is mounting evidence that the Golgi complex also has a mechanism to prevent abnormal proteins from reaching the cell surface (36). Moreover, transport of proteins from the Golgi complex to the plasma membrane may require the presence of positive sorting signals that serve to concentrate cargo into exocytic vesicles (37). The presence of altered or denatured E2/E1 glycoprotein spikes on the surface of RLPs could result in failure of the virus particles to be transported to the cell surface by either of these mechanisms.
The mechanism of RV assembly.
An important question that arises from this study is, what are the important interactions that drive RV assembly? It is now clear that interactions between the E1 CT domain and capsid protein are not required for intracellular budding. This leaves a number of possibilities which can ultimately be tested by using the RLP system and/or the RV infectious clone (43). By analogy with alphaviruses (24, 40), binding of the capsid protein to the E2 CT domain could potentially mediate the assembly of virus particles. Our results indicate that it is the net positive charge in the E2 CT domain which is critical for function, rather than the arginine residues per se; therefore, unlike the case for alphaviruses, these interactions would most likely involve electrostatic binding between the arginine residues in the E2 CT domain and acidic residues in capsid protein. Interestingly, there are two closely spaced clusters (positions 143 to 151 and 183 to 189) of aspartic acid and glutamic acid residues in capsid protein that could potentially bind to the arginine residues in the E2 CT domain. Additional interactions between the hydrophobic carboxy terminus of capsid protein and other transmembrane segments of E2 and/or E1 cannot be ruled out at this point. The E2 signal peptide is not cleaved from the carboxy terminus of capsid, and therefore this protein remains membrane associated (51). Consequently, hydrophobic interactions between the E2 signal peptide and the TM domain of E2 within the plane of the Golgi membrane could augment electrostatic interactions between E2 and capsid during particle assembly.
Another possibility is that RV budding is the result of a “push and pull” mechanism which has been proposed for rhabdoviruses (33). This model holds that extrusion of virus components from host cells is effected by the independent but synergistic effects of pushing and pulling forces mediated by cytoplasmic proteins (capsid and/or matrix proteins) and transmembrane glycoproteins, respectively. Retrovirus Gag precursors are able to form virus-like particles in the absence of envelope glycoproteins by pushing the host cell membranes enough to cause budding (7, 9). Although an interaction between the matrix protein and envelope glycoprotein is not absolutely required to drive the budding, it does increase the efficiency of this process 30-fold (33). Presumably, interactions between the CT domain of virus envelope glycoproteins and cytoplasmic viral components are required for fidelity or specificity (41). We are now in the process of testing whether the TM and CT domains of E2 and E1 can direct the incorporation of non-RV membrane proteins into virus particles.
ACKNOWLEDGMENTS
We acknowledge Margaret Hughes for excellent technical assistance. We are grateful to J. Safford, J. Wolinsky, B. Pusowoit, M. Farquhar, and A. Tingle for gifts of antibodies.
This work was supported by grants to T.C.H. from the Alberta Heritage Foundation for Medical Research and the Medical Research Council of Canada. T.C.H. is a scholar of the Alberta Heritage Foundation for Medical Research and the Medical Research Council of Canada.
REFERENCES
- 1.Andersson S, Davis D L, Dahlback H, Jornvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 2.Baron M D, Forsell K. Oligomerisation of the structural proteins of rubella virus. Virology. 1991;185:811–819. doi: 10.1016/0042-6822(91)90552-m. [DOI] [PubMed] [Google Scholar]
- 3.Bergeron J J M, Brenner M B, Thomas D Y, Williams D B. Calnexin: a membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem Sci. 1994;19:124–128. doi: 10.1016/0968-0004(94)90205-4. [DOI] [PubMed] [Google Scholar]
- 4.Chida K, Sagara H, Suzuki Y, Murakami A, Osada S, Ohno S, Hirosawa K, Kuroki T. The eta isoform of protein kinase C is localized on rough endoplasmic reticulum. Mol Cell Biol. 1994;14:3782–3790. doi: 10.1128/mcb.14.6.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke D M, Loo T W, Hui I, Chong P, Gillam S. Nucleotide sequence and in vitro expression of rubella virus 24S subgenomic mRNA encoding the structural proteins E1, E2 and C. Nucleic Acids Res. 1987;15:3041–3057. doi: 10.1093/nar/15.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke D M, Loo T W, McDonald H, Gillam S. Expression of rubella virus cDNA coding for the structural proteins. Gene. 1988;65:23–30. doi: 10.1016/0378-1119(88)90413-1. [DOI] [PubMed] [Google Scholar]
- 7.Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Verdin E, Horth M, Burny A, Bex F. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez S A, Affranchino J L, R. G H, Burny A. Assembly of the matrix protein of simian immunodeficiency virus into virus-like particles. Virology. 1993;194:548–556. doi: 10.1006/viro.1993.1293. [DOI] [PubMed] [Google Scholar]
- 11.Grangeot-Keros L, Pustowoit B, Hobman T C. Evaluation of Cobas Core Rubella IgG E1A recomb, a new enzyme immunoassay based on recombinant rubella-like particles. J Clin Microbiol. 1995;33:2392–2394. doi: 10.1128/jcm.33.9.2392-2394.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haffar O, Garrigues J, Travis B, Moran P, Zarling J, Hu S-L. Human immunodeficiency virus-like, nonreplicating gag-env particles assemble in a recombinant vaccinia virus expression system. J Virol. 1990;64:2653–2659. doi: 10.1128/jvi.64.6.2653-2659.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauri H-P, Schweizer A. The endoplasmic reticulum-Golgi intermediate compartment. Curr Opin Cell Biol. 1992;4:600–608. doi: 10.1016/0955-0674(92)90078-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobman T, Lemon H, Jewell K. Characterization of an endoplasmic reticulum retention signal in the rubella virus E1 glycoprotein. J Virol. 1997;71:7670–7680. doi: 10.1128/jvi.71.10.7670-7680.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobman T, Zhao B, Chan H, Farquhar M. Immunoisolation and characterization of a subdomain of the endoplasmic reticulum that concentrates proteins involved in COPII vesicle biogenesis. Mol Biol Cell. 1998;9:1265–1278. doi: 10.1091/mbc.9.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobman T C. Transport of viral glycoproteins into the Golgi complex. Trends Microbiol. 1993;1:124–130. doi: 10.1016/0966-842X(93)90126-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobman T C, Gillam S. In vitro and in vivo expression of rubella virus E2 glycoprotein: the signal peptide is located in the C-terminal region of capsid protein. Virology. 1989;173:241–250. doi: 10.1016/0042-6822(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 18.Hobman T C, Lundstrom M L, Gillam S. Processing and transport of rubella virus structural proteins in COS cells. Virology. 1990;178:122–133. doi: 10.1016/0042-6822(90)90385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobman T C, Lundstrom M L, Mauracher C A, Woodward L, Gillam S, Farquhar M G. Assembly of rubella virus structural proteins into virus-like particles in transfected cells. Virology. 1994;202:574–585. doi: 10.1006/viro.1994.1379. [DOI] [PubMed] [Google Scholar]
- 20.Hobman T C, Shukin R, Gillam S. Translocation of rubella virus glycoprotein E1 into the endoplasmic reticulum. J Virol. 1988;62:4259–4264. doi: 10.1128/jvi.62.11.4259-4264.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobman T C, Woodward L, Farquhar M G. The rubella virus E1 glycoprotein is arrested in a novel post-ER, pre-Golgi compartment. J Cell Biol. 1992;118:795–811. doi: 10.1083/jcb.118.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobman T C, Woodward L, Farquhar M G. The rubella virus E2 and E1 spike glycoproteins are targeted to the Golgi complex. J Cell Biol. 1993;121:269–281. doi: 10.1083/jcb.121.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobman T C, Woodward L, Farquhar M G. Targeting of a heterodimeric membrane protein complex to the Golgi: rubella virus E2 glycoprotein contains a transmembrane Golgi retention signal. Mol Biol Cell. 1995;6:7–20. doi: 10.1091/mbc.6.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kail M, Hollinshead M, Ansorge W, Pepperkok R, Frank R, Griffiths G, Vaux D. The cytoplasmic domain of alphavirus E2 glycoprotein contains a short linear recognition signal required for viral budding. EMBO J. 1991;10:2343–2351. doi: 10.1002/j.1460-2075.1991.tb07773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karacostas V, Nagashima K, Gonda M A, Moss B. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc Natl Acad Sci USA. 1989;86:8964–8967. doi: 10.1073/pnas.86.22.8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirnbauer R, Taub J, Greenstone H, Roden R, Durst M, Gissmann L, Lowy D R, Schiller J T. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krausslich H-G, Ochsenbauer C, Traenckner A-M, Mergener K, Facke M, Gelderblom H R, Bosch V. Analysis of protein expression and virus-like particle formation in mammalian cell stably expressing HIV-1 gag and env gene products with or without active HIV protease. Virology. 1993;192:605–617. doi: 10.1006/viro.1993.1077. [DOI] [PubMed] [Google Scholar]
- 28.Lehel C, Olah Z, Jakak G, Anderson W B. Protein kinase Cɛ is localized to the Golgi via its zinc-finger domain and modulates Golgi function. Proc Natl Acad Sci. 1995;92:1406–1410. doi: 10.1073/pnas.92.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Littman D R, Thomas Y, Maddon P J, Chess L, Axel R. The isolation and sequence of the gene encoding T8: a molecule defining functional classes of T lymphocytes. Cell. 1985;40:237–246. doi: 10.1016/0092-8674(85)90138-2. [DOI] [PubMed] [Google Scholar]
- 30.Lundstrom M L, Mauracher C A, Tingle A J. Characterization of carbohydrates linked to rubella virus glycoprotein E2. J Gen Virol. 1991;72:843–850. doi: 10.1099/0022-1317-72-4-843. [DOI] [PubMed] [Google Scholar]
- 31.Magliano D, Marshall J, Bowden B D S, Vardaxis N, Meanger J, Lee J-Y. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- 32.Marr L D, Sanchez A, Frey T K. Efficient in vitro translation and processing of the rubella virus structural proteins in the presence of microsomes. Virology. 1991;180:400–405. doi: 10.1016/0042-6822(91)90046-e. [DOI] [PubMed] [Google Scholar]
- 33.Mebatsion T, Konig M, Conzelmann K-K. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 34.Melancon P, Garoff H. Processing of the Semliki Forest virus structural polyprotein: role of the capsid protease. J Virol. 1987;61:1301–1309. doi: 10.1128/jvi.61.5.1301-1309.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mergener K, Facke M, Welker R, Brinkmann V, Gelderblom H R, Krausslich H-G. Analysis of HIV particle formation using transient expression of subviral constructs in mammalian cells. Virology. 1993;186:25–39. doi: 10.1016/0042-6822(92)90058-w. [DOI] [PubMed] [Google Scholar]
- 36.Moolenaar C, Ouwendijk J, Wittpoth M, Wisselaar H, Hauri H-P, Ginsel L, Naim H, Fransen J. A mutation in the highly conserved region in brush-border sucrase-isomaltase and lysosomal α-glucosidase results in Golgi retention. J Cell Sci. 1997;110:557–567. doi: 10.1242/jcs.110.5.557. [DOI] [PubMed] [Google Scholar]
- 37.Munro S. Localization of proteins to the Golgi complex. Trends Cell Biol. 1998;8:11–15. doi: 10.1016/S0962-8924(97)01197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oker-Blom C, Kalkkinen N, Kaariainen L, Pettersson R F. Rubella virus contains one capsid protein and three envelope glycoproteins, E1, E2a, and E2b. J Virol. 1983;46:964–973. doi: 10.1128/jvi.46.3.964-973.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oker-Blom C, Ulmanen I, Kaariainen L, Pettersson R F. Rubella virus 40S genome RNA specifies a 24S subgenomic RNA that codes for a precursor to structural proteins. J Virol. 1984;49:403–408. doi: 10.1128/jvi.49.2.403-408.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owen K E, Kuhn R J. Alphavirus budding is dependent upon the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of E2 envelope glycoprotein. Virology. 1997;230:187–196. doi: 10.1006/viro.1997.8480. [DOI] [PubMed] [Google Scholar]
- 41.Owens R, Rose J K. Cytoplasmic domain requirements for incorporation of a foreign envelope protein into vesicular stomatitis virus. J Virol. 1993;67:360–365. doi: 10.1128/jvi.67.1.360-365.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petruzziello R, Orsi N, Macchia S, Rieti S, Frey T, Mastromarino P. Pathway of rubella virus infectious entry into Vero cells. J Gen Virol. 1996;77:303–308. doi: 10.1099/0022-1317-77-2-303. [DOI] [PubMed] [Google Scholar]
- 43.Pugachev K V, Frey T K. Improvement of the infectivity of the rubella virus infectious clone: mutagenesis of the 5′ noncoding region of the virus. London, Ontario, Canada: American Society for Virology; 1996. p. 99. [Google Scholar]
- 44.Pustowoit B, Grangeot-Keros L, Hobman T C, Hofmann J. Evaluation of recombinant rubella-like particles in a commercial immunoassay for the detection of anti-rubella IgG. Clin Diagn Virol. 1996;5:13–30. doi: 10.1016/0928-0197(95)00150-6. [DOI] [PubMed] [Google Scholar]
- 45.Qiu Z, Ou D, Wu H, Hobman T C, Gillam S. Expression and characterization of virus-like particles containing rubella virus structural proteins. J Virol. 1994;68:4086–4091. doi: 10.1128/jvi.68.6.4086-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose J K, Bergmann J E. Expression from cloned cDNA of cell-surface secreted forms of the glycoprotein of vesicular stomatitis virus in eukaryotic cells. Cell. 1982;30:753–762. doi: 10.1016/0092-8674(82)90280-x. [DOI] [PubMed] [Google Scholar]
- 47.Rose J K, Doms R W. Regulation of protein export from the endoplasmic reticulum. Annu Rev Cell Biol. 1988;4:257–288. doi: 10.1146/annurev.cb.04.110188.001353. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Schalich J, Allison S L, Stiasny K, Mandl C W, Kunz C, Heinz F W. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J Virol. 1996;70:4549–4557. doi: 10.1128/jvi.70.7.4549-4557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suomalainen M, Garoff H, Baron M D. The E2 signal sequence of rubella virus remains part of the capsid protein and confers membrane association in vitro. J Virol. 1990;64:5500–5509. doi: 10.1128/jvi.64.11.5500-5509.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suomalainen M, Liljestrom P, Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velasco A, Hendricks L, Moremen K W, Tulsiani D R P, Touster O, Farquhar M G. Cell type-dependent variations in the subcellular distribution of α-mannosidase I and II. J Cell Biol. 1993;122:39–51. doi: 10.1083/jcb.122.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vennema H, Godeke G-J, Rossen J W A, Voorhout W F, Horzinek M C, Opstelten D-J E, Rottier P J M. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Heijne G. Control of topology and mode of assembly of polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 56.Wolinsky J. Rubella. In: Fields B N, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 899–929. [Google Scholar]