Abstract
A precore-deficient mutant of duck hepatitis B virus (DHBV) produced by site-directed mutagenesis was tested for its ability to compete with wild-type virus in a mixed infection of 3-day-old ducklings. The mutation was shown to produce a cis-acting defect, resulting in a replication rate that was about one-half that of wild-type virus. Accordingly, wild-type virus was rapidly selected during the spread of infection. During the chronic phase of the infection, however, two selection patterns were seen. In 4 of 10 ducks, the wild-type virus slowly replaced the precore mutant. In another four ducks, the precore mutant virus slowly replaced the wild-type virus. In the remaining two ducklings, ratios of wild-type and precore mutant virus fluctuated, with wild-type virus slowly predominating. The replacement of wild-type virus was not due to the emergence of a rapidly replicating variant of the precore mutant, since genomes cloned from the infected ducks retained their original replication defect. Replacement of wild-type virus, however, correlated with elevated anti-core antibody titers, which continued to increase with time. The selection of a precore-negative strain of DHBV may be analogous to the selection for precore mutants of HBV during chronic hepatitis in humans.
Hepadnaviruses are a small group of DNA viruses that replicate their genomes through reverse transcription of an RNA intermediate (9, 23, 35, 38). These viruses have been found in humans (6, 32, 39), woodchucks and ground squirrels (22, 41, 42), and several species of waterfowl, including ducks (24, 36). All hepadnaviruses share similar genetic structures consisting of the three genetic regions essential for replication, i.e., the core, P protein, and envelope regions (reviewed in references 7, 29). In addition to these three genetic regions, the mammalian hepadnaviruses contain a fourth gene, commonly called the X gene, whose function in replication has not been clearly defined (45). The core regions of all known hepadnaviruses can be divided into two functional units that encode two overlapping protein products in the same reading frame (21). One product, the capsid protein, is a viral structural protein essential for RNA packaging, reverse transcription, and virus assembly. The second product, the precore protein, is a nonstructural protein translated from an mRNA that contains an upstream in-frame AUG followed by a small number of codons that encode a type I signal recognition sequence. The precore protein is translocated into the endoplasmic reticulum as it is translated, where the signal recognition sequence is cleaved and the basic C terminus of the protein is proteolytically removed. Subsequent to this processing, the precore protein is transported through the Golgi apparatus and secreted from the cell (10, 37). Processed precore protein is found in the blood of animals with chronic hepadnavirus infections and is called “e antigen” (20). The e antigen has long been a convenient serological marker associated with high levels of viremia in hepatitis B virus (HBV)-infected humans (44).
Stop codons engineered into the precore open reading frame of duck HBV (DHBV) or woodchuck hepatitis virus (WHV) do not inhibit virus replication or prevent infection (34), although precore-negative WHV mutants have been reported to produce only transient infection of woodchucks (4). Moreover, spontaneous precore-negative mutants arise commonly in chronic HBV or WHV infections and can emerge as the predominant genotype (2, 3, 19, 31, 43). The basis for selection of precore mutants in chronic hepadnavirus infection is not known. Published evidence implicates the precore protein in HBV as a regulator of replication (1, 28, 30, 33), but in DHBV, no increase in replication rate has been observed in engineered precore mutants (44a).
It has been proposed that the precore protein functions through the production of extracellular e antigen to modulate the T-cell response to core antigen. This effect was demonstrated for one core antigen-e antigen T-cell epitope in e antigen- and core antigen-producing transgenic mice (25–27). By several assays, the presence of e antigen caused a reduction or elimination of the Th1-dependent immunoglobulin G2 (IgG2) anti-core antibody response in e antigen-transgenic mice in favor of a Th2-dependent IgG1 antibody response. Since Th1 response is thought to be important in virus clearance, it was proposed that the suppression of this response might favor chronic infections and produce an overall benefit for the virus in the efficiency of viral transmission. However, e antigen may also target cells for T-cell killing in the presence of an effective T-cell response, resulting in the selection of e antigen-negative virus mutants.
The experiments we report here were undertaken to examine selection of DHBV variants during a chronic infection. We carried out a mixed infection of ducklings with wild-type virus and a precore-defective mutant DHBV with a weak replication defect. We found that after an initial expected wild-type enrichment during the spread of infection, the wild-type virus was unexpectedly eliminated in favor of the precore mutant in some birds. This results suggests that the phenotype of a precore DHBV mutant may confer a selective advantage under some conditions during chronic infection, resulting in the selection of precore-minus mutants, as has been observed with HBV.
MATERIALS AND METHODS
Animals.
One-day-old ducklings were obtained from Metzer Farms (Redlands, Calif.), and congenitally infected birds were identified by dot hybridization. Only virus-free birds were used in the experiments.
Plasmids and mutants.
Viral genomes were cloned as head-to-tail dimers in plasmid pSP65. The wild-type DHBV used in this study was DHBV 16 (19). The mutant DHBV 16, called 2619A, was kindly provided by Wengang Yang. This mutant contained a single-nucleotide substitution of A for T at nucleotide position 2619, which created a stop codon, TGA, at position 34 in the precore open reading frame. The nucleotide substitution also caused a cis-acting replication defect that reduced the replication rate of this mutant by about 40% (44a) (see below).
Transfections and production of virus inocula.
Production and concentration of infectious virus after transfection of LMH cells was performed as previously described (15, 40). Enveloped virus concentrations were determined by selective extraction of viral DNA from virus preparations, by agarose gel electrophoresis, and by Southern blot hybridization. Virus was concentrated by polyethylene glycol precipitation as previously described (16). Viral DNA was determined by comparison of the hybridization signal with that obtained from known amounts of cloned viral DNA run in the same gel. Procedures used in the analysis of viral DNA replicative intermediates, agarose gel electrophoresis, and blot hybridization were previously published (40).
Analysis of viral DNA in the serum.
The level of viremia was determined by quantitation of viral DNA in the serum by dot hybridization and phosphorimage analysis. Serum (2 μl) was applied directly to nylon membranes, denatured by brief treatment with alkali, and neutralized with 0.2 M Trizma HCl. DNA was detected on the filter by hybridization with a riboprobe specific for the minus strand, as previously described. For analysis of the viral genotype, serum (10 μl) was mixed with 10 μl of 0.2 N NaOH and incubated for 1 h at 37°C to disrupt the virus and denature the viral DNA. The samples were neutralized by the addition of 10 μl of 0.2 N HCl, cleared by brief microcentrifugation, and diluted with the addition of 30 μl of TE (10 mM Tris-HCl, pH 7.5, 1 mM EDTA). Five microliters of the sample was then used in a 50-μl PCR mixture.
PCR and sequencing.
Amplification of the serum viral DNA was carried out with a primer set corresponding to nucleotides 2548 to 2571 (biotinylated plus strand) and 2840 to 2818 (minus strand). The standard PCR buffer contained DNA template; 200 μM (each) dATP, dGTP, dCTP, and TTP; 50 mM KCl; 10 mM Tris-HCl, pH 8.3; 1.5 mM MgCl2; 0.02% gelatin; and 38 pmol of each primer in a final volume of 50 μl, with 2.5 U of Taq DNA polymerase (Sigma). Amplification was carried out for 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 45 s. The biotinylated PCR products (40 μl) were adsorbed with 20 μl of strepavidin-coated M-280 Dynabeads (Dynal Corp.) suspended in a solution of 20 mM Tris-HCl (pH 8.0)–2 M NaCl–1 mM EDTA, and washed two times with 50 μl of TE with the help of a magnetic particle concentrator (catalog no. 120.04; Dynal). The nonbiotinylated strand was released from the beads by denaturation in 0.1 N NaOH (50 μl), the denaturing solution was removed, and the beads were washed two times with 50 μl of TE. Washed beads with specifically bound biotinylated plus-strand products were used directly in sequencing reactions with a minus-strand primer (nucleotides 2747 to 2729).
Quantitation of viral genotypes.
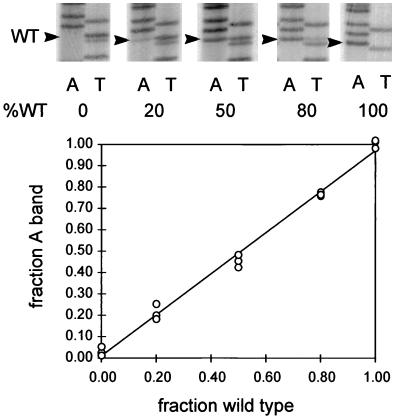
Sequencing gels were used to determine the ratio of the two genotypes in the samples of amplified DNAs. In order to evaluate this assay, we performed an experiment using known ratios of plasmids containing the two genomes, 2619A and wild type (2619T). Plasmid mixtures (50 pg) containing 0, 20, 50, 80, and 100% wild type were amplified, and the plus strand was sequenced with a minus-strand primer as described above. The A bands (wild-type minus strand) and the T bands (mutant minus strand) at position 2619 were quantitated by phosphorimaging and normalized to the intensity of the corresponding immediately preceding A or T band in the respective lanes of the sequencing gel. This normalization corrected for variations in the amount of radioactivity loaded in each lane. Each corrected value was then normalized to the sum of the two corrected values to determine the fraction of the ladder that was due to either mutant or wild-type sequences, assuming that at position 2619 only two genotypes existed. Three examples of these experimentally determined values were plotted against the standard ratios in the templates and are shown in Fig. 1. Standardization curves thus obtained showed a direct relationship between the fraction of a genotype in the template and the experimentally determined ratio of the wild-type and mutant residues at position 2619. For all the data presented in this paper, standardization curves were used to determine the ratio of wild-type genomes to mutant genomes in the PCR templates.
FIG. 1.
PCR sequencing assay for serum virus genotype. DNAs (50 pg total) consisting of the indicated ratios of wild type (WT) to mutant 2619A dimer plasmids were linearized by digestion with the single-cut enzyme SalI and subjected to PCR amplification. The plus strand, containing the biotinylated primer, was isolated, and two sequencing reactions were performed with dideoxyadenosine triphosphate (lanes A) for detection of the wild-type 2619T residue in the plus strand and with dideoxythymidine triphosphate (lanes T) for detection of the 2619A residue. The signals in the A lanes and the T lanes at position 2619 were corrected for loading and normalized, and the fraction of the wild-type signal was plotted against the fraction of wild-type plasmid DNA in the template. The results of separate loadings of a single pair of reactions are shown. The curve was calculated by linear regression analysis.
Enzyme-linked immunosorbent assay for antibody to core antigen.
Assays for antibody to core antigen were performed in 96-well microtiter plates coated with recombinant DHBV core protein produced in Escherichia coli. The wells were incubated with 100 ng of core protein, blocked, and incubated with samples diluted serially 1:5, starting with a 1:100 dilution, for 1 h at 37°C. Bound duck Ig was detected by incubation with horseradish peroxidase-conjugated rabbit anti-duck IgG (heavy plus light chains) (Nordic Immunology, Tilberg, The Netherlands) diluted 1:10,000 and by reaction with the color reagent o-phenylenediamine. One unit was defined as the reciprocal of the dilution that produced an optical density at 495 nm of 0.5. The titers (see Fig. 4B) were normalized to a standard serum sample run with every set of assays. The standard serum value was between 2,600 and 19,000 U per ml in different assays.
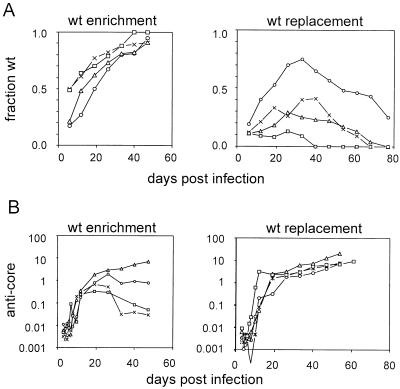
FIG. 4.
Analysis of serum genotype and anti-core antibody titers during chronic mixed infection. The separate graphs show the results for four birds in which continuous enrichment of wild-type virus was observed (wt enrichment) and four birds in which wild-type virus was replaced by the 2619A virus (wt replacement). (A) The fraction of wild-type virus in the serum, determined by PCR sequencing assay, was plotted against the time postinfection. (B) The anti-core antibody titers for the two birds shown in panel A, normalized to a standard anti-core duck serum, were plotted against the time postinfection. Symbols are used to represent results from the same individual birds in panels A and B as follows. Left-hand graphs: x, bird 11; ○, bird 30; ▵, bird 40; □, bird 41. Right-hand graphs: x, bird 8; ○, bird 10; ▵, bird 14; □, bird 34.
PCR amplification, cloning, and analysis of serum virus.
Serum viral DNA was amplified by PCR before cloning, according to the strategy previously described (8). Total viral DNA was purified from serum by protease digestion and phenol extraction. Serum (50 μl) was added to 200 μl of digestion buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.2% sodium dodecyl sulfate) containing 500 mg of pronase per ml. After 1 h at 37°C, the digested sample was extracted with an equal volume of phenol and the nucleic acids were recovered by ethanol precipitation. The pellet was dissolved in water and used for PCR amplification, using a primer pair that primed DNA synthesis at positions corresponding to either end of the complete minus-strand DNA. The primers used contained a SapI recognition sequence (underlined) positioned such that cleavage of the primers from the ends of the linear DNA would generate a full-length linear copy of the DHBV genome that could be ligated at the SapI “sticky” ends. The sequences of the primers used were as follows: 5′ CCC GCT CTT CA/G AAT TAC ACC CCT CTC 3′ (plus strand) and 5′ CCC GCT CTT CA/T TCT TAA GTT CCA CAT AGC CTA 3′ (minus strand). Amplification was carried out in a volume of 50 μl of standard PCR buffer, using 2.5 U of Taq I DNA polymerase (Sigma) and 0.5 U of Pwo DNA polymerase (Boehringer-Mannheim). The amplification reaction was carried out for 35 cycles of 94°C for 5 s and 68°C for 4 min. In subsequent reactions the denaturation time was extended to 15 s and annealing was extended to 45 s at 58°C, with elongation for 4 min at 72°C to increase sensitivity.
The amplified linear DNA was purified by low-melting-temperature agarose gel electrophoresis to remove excess primers and then cleaved by SapI. SapI recognition was provided by the underlined sequence, resulting in cleavage at 1 and 4 nucleotides downstream to leave a 3-nucleotide protruding 5′ end. The SapI-cleaved DNA was repurified through a second low-melting-temperature agarose gel and then religated to produce head-to-tail multimers, which were cleaved by EcoRI to produce monomers. EcoRI-linearized DNA was cloned initially in pSP65 as monomers, which were then isolated and recloned as dimers in pSP65.
Plasmids containing DHBV dimer inserts were purified through CsCl gradients containing ethidium bromide and used to transfect LMH cells (14), as previously described (5). Enveloped virus in the supernatant fluid was isolated by the DNase I-pronase method (15), and the virus yield was determined by blot hybridization, or the genotype was determined by PCR and direct sequencing.
Calculation of the relative growth rate of mutant 2619A during spread of infection.
The rate of increase in the enrichment (E) of wild-type virus (WT) relative to 2619A (Mut) was determined experimentally to be 0.30 log10E per 24 h (Fig. 2). Enrichment of wild-type virus between two points in time, t = t1 and t = t2, is defined as follows: E = [WT(t2)/Mut(t2)]/[WT(t1)/Mut(t1)] or E = [WT(t2)/WT(t1)/[Mut(t2)/Mut(t1)]. If the replication rates of WT and Mut viruses are determined by the first-order rate constants, kWT and kMut, respectively, then E = exp(kWT · Δt)/exp(kMut · Δt). The following equation gives the relative growth rate of the 2619A virus: kMut/kWT = 1 − (1/kWT) · (1nE)/Δt = 1 − (1/kWT) · 0.69, where (log10E)/Δt = 0.30 day−1 or (1nE)/Δt = 0.69 day−1. For a doubling time of 9.4 h (0.39 days) during the spread of wild-type DHBV in ducklings (12), kWT is equal to (1n2)/0.39, or 1.77 day−1. Similarly, for a doubling time of 16 h (13), kWT is equal to 1.03 day−1. Using these two estimates of kWT, we calculated the relative growth rate of 2619A virus to be between 61 and 37% of that of the wild type.
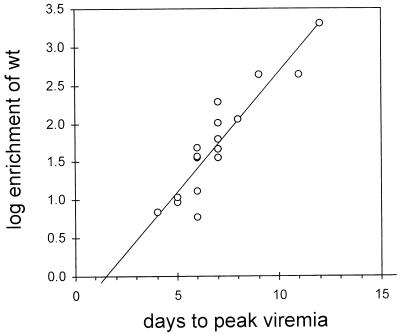
FIG. 2.
Enrichment of wild-type (wt) virus during spread of infection. The ratio of wild-type virus to 2619A virus at peak viremia was divided by the ratio of wild type to 2619A in the inoculum to obtain the enrichment, E. The logarithm of E (log10E) was plotted against the time at which peak viremia occurred, and a linear regression curve was calculated. The slope of the curve was determined to be 0.30 log10E per day, i.e., wild-type virus was enriched over the 2619A virus approximately twofold each day during the spread of infection until peak viremia.
RESULTS
A series of mutant DHBV genomes defective in the production of the precore protein were constructed and tested for the ability to replicate after transfection into LMH cells, infection of primary duck hepatocytes, or inoculation into newly hatched ducklings (44a). One mutant, 2619A, was partially defective in replication yet able to establish a chronic infection in vivo. This mutant was selected for the present study. The purpose of this study was to determine how the relative replication rates of two viruses influence their selection in a chronic infection. For this purpose we established a mixed infection with two competing virus strains and measured the rate of replacement of the slower-replicating strain. As the faster-replicating virus, we used the parent DHBV 16, which was wild type for precore production.
Competition of wild type and 2619A during spread of infection and measurement of replication defect.
Thirty ducklings were inoculated at 3 days of age with a dose of either 106, 107, or 108 viral genomes containing a mixture of wild-type and 2619A virus in ratios of 1:5, 1:50, or 1:500. Infected ducklings were bled daily after infection until peak viremia was reached and weekly thereafter. The viral DNA titers and the genotype of virus in the blood were determined. Twenty-four birds developed a viremia which peaked within 4 to 12 days postinoculation, and a mixed infection was detected in 19 of these (the lower detection limit for a genotype was about 5% of the total). All of the birds showed enrichment of the wild-type virus compared with the inoculum. The virological data for the group of 19 ducks with mixed infection are presented in Table 1.
TABLE 1.
Enrichment of wild-type virus during spread of infection
| Bird no. | Day of peak viremia | Inoculum | Fraction wta in inoculum | Fraction wta at peak viremia | Enrichment |
|---|---|---|---|---|---|
| 8b | 4 | 108 | 0.02 | 0.12 | 7 |
| 5 | 5 | 108 | 0.2 | 0.7 | 9 |
| 36 | 5 | 107 | 0.2 | 0.7 | 9 |
| 10b | 5 | 108 | 0.02 | 0.19 | 11 |
| 4 | 6 | 108 | 0.2 | 0.6 | 6 |
| 21 | 6 | 106 | 0.2 | 0.9 | 36 |
| 11b | 6 | 108 | 0.02 | 0.5 | 49 |
| 40b | 6 | 107 | 0.02 | 0.21 | 13 |
| 16b | 6 | 108 | 0.002 | 0.07 | 38 |
| 28 | 7 | 106 | 0.02 | 0.8 | 196 |
| 37 | 7 | 107 | 0.2 | 0.9 | 36 |
| 14b | 7 | 108 | 0.002 | 0.11 | 62 |
| 41b | 7 | 107 | 0.02 | 0.49 | 47 |
| 34b | 7 | 106 | 0.002 | 0.11 | 62 |
| 30b | 7 | 106 | 0.002 | 0.17 | 102 |
| 23 | 8 | 106 | 0.02 | 0.7 | 114 |
| 25 | 9 | 106 | 0.02 | 0.9 | 441 |
| 39 | 11 | 106 | 0.02 | 0.9 | 441 |
| 33 | 12 | 106 | 0.002 | 0.8 | 1,996 |
wt, wild type.
Selected for follow-up study.
Enrichment of the wild-type virus indicated that the wild-type virus replicated more rapidly than the competing mutant virus, 2619A. Enrichment of the wild-type virus would be expected to increase according to the amount of growth of the two viruses required to achieve peak viremia. Therefore, more enrichment should occur with smaller inocula; however, we found that the time that elapsed between inoculation and peak viremia was a better predictor of enrichment than the size of the inoculum. As can be seen in Fig. 2, the logarithm of the enrichment, E (expressed as the ratio of wild type to mutant at peak viremia divided by the ratio in the inoculum), was linearly proportional to the time required to achieve peak viremia, suggesting that the effective inoculum size, as opposed to the amount of virus injected, varied among the individual birds. The slope of the regression curve for this plot was determined to be 0.30 log E/day. Using the data of Jilbert et al. (12, 13) showing that the doubling time of DHBV in ducklings inoculated under similar conditions was 9.4 to 16 h, it was calculated that growth rates for 2619A of 61 and 37% that of wild-type virus, respectively, would produce the observed amount of wild-type enrichment (see Materials and Methods). These data confirm the presence of a replication defect in mutant virus 2619A.
Changes in the ratio of genotypes during follow-up.
A group of 10 ducklings with mixed, predominantly 2619A, infections were selected for follow-up studies to determine the rate of replacement of the mutant by the wild-type virus. Assays for viral DNA in the blood, the results of which are presented in Table 2, showed that this group of ducks remained persistently infected during the course of the experiment. Viremia was characterized by an initial peak followed by a rapid decrease in viral-DNA-containing particles in the blood, which eventually stabilized at levels 1 to 3 orders of magnitude below the initial peak. This pattern has been previously reported in experimental infections of ducklings (11, 17, 18).
TABLE 2.
Virus titers in 10 birds with mixed infections
| Days p.i.a | Virus titer (107 genomes) in bird no.:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 10 | 11 | 13 | 14 | 16 | 30 | 34 | 40 | 41 | |
| 3 | 156 | 5 | 4 | 17 | 3 | 5 | 2 | 1 | 3 | 1 |
| 4 | 689 | 71 | 6 | 877 | 6 | 13 | 4 | 9 | 15 | 4 |
| 5 | 408 | 714 | 50 | 1,127 | 6 | 245 | 3 | 17 | 525 | 6 |
| 6 | 283 | 611 | 369 | 244 | 223 | 563 | 34 | 342 | 858 | 197 |
| 7 | 3 | 460 | 196 | 57 | 1,133 | 120 | 590 | 897 | 3 | 478 |
| 8 | 75 | 237 | 61 | 58 | 83 | 2 | 409 | 3 | 75 | 481 |
| 10 | 1 | 11 | 10 | 15 | 19 | 2 | 127 | 2 | 62 | 38 |
| 12 | 6 | 49 | 5 | 126 | 5 | 1 | 51 | 5 | 51 | 69 |
| 19 | 1 | 82 | 4 | 69 | 14 | 1 | 4 | 1 | 6 | 3 |
| 26 | 1 | 31 | 4 | 33 | 25 | 3 | 1 | 4 | 9 | 13 |
| 33 | 1 | 33 | 12 | 59 | 22 | 4 | 1 | 2 | 17 | 34 |
| 40 | <1 | 11 | 10 | 43 | 9 | 2 | 1 | 2 | 11 | 28 |
| 47 | <1 | 11 | 1 | 60 | 3 | 2 | <1 | 1 | 2 | 3 |
| 54 | <1 | 7 | *b | 29 | 2 | 5 | * | 2 | * | * |
| 61 | <1 | 9 | * | 47 | 1 | 9 | * | 9 | * | * |
| 68 | <1 | 2 | * | 14 | <1 | 14 | * | 5 | * | * |
| 77 | <1 | 1 | * | 35 | <1 | 17 | * | 4 | * | * |
p.i., postinoculation.
*, birds sacrificed at day 47.
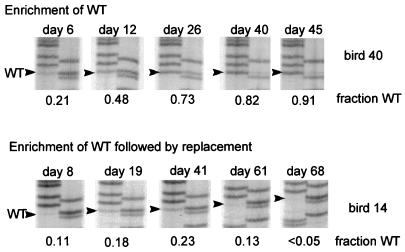
The region of viral DNA containing the mutation was amplified from each serum sample by PCR and subjected to direct sequencing to determine the ratio of wild type to 2619A in the serum virus. Examples of the data for two birds showing different enrichment patterns are shown in Fig. 3. Serum samples from bird 40 showed a steady enrichment of the wild-type genotype during the course of the experiment, until the precore mutant virus was no longer detected. This result was expected, since the wild-type virus replicated faster than the precore mutant. In contrast, the genotype of serum virus for bird 14 was initially enriched for the wild type (days 8, 19, and 41), but the pattern of enrichment was reversed at later times (days 61 and 68), and the precore mutant eventually replaced the wild-type virus.
FIG. 3.
Examples of two different patterns of selection during chronic infection. The results of PCR sequencing assays for serum samples obtained at various times postinfection are shown for two ducklings. In the upper panels (bird 40), the wild type (WT)-specific band is seen to increase continuously during chronic infection, while in the lower set of panels (bird 14) the initial enrichment of wild type is followed by replacement with the 2619A-specific band.
The data for 8 of the 10 birds followed are shown in Fig. 4A. Four ducklings with 17 to 50% wild-type virus in the blood at peak viremia, the start of the follow-up samples, showed a continuous, gradual enrichment in wild-type virus until day 47 postinfection, at which time the wild-type genotype made up 91 to 100% of the viral DNA (Fig. 4A, left graph). Because further increases in the ratios of wild-type virus to mutant virus could not be accurately calculated beyond this point, the follow-up study on these four birds was terminated. In a second group of four ducklings the proportion of wild-type virus showed a sustained decrease or disappearance following an initial increase (Fig. 4A, right graph). In two ducklings, birds 13 and 16, the wild-type genotype increased slowly, with intermittent periods of decrease (not shown).
Rate of wild-type virus enrichment during chronic infection.
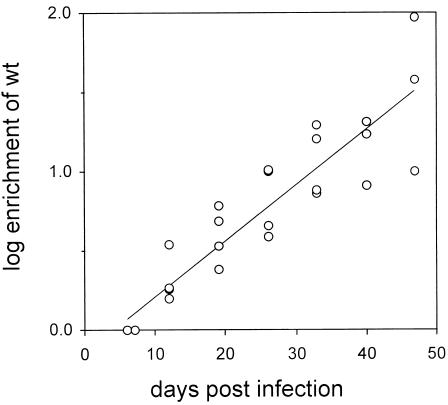
In birds in which wild-type virus eventually became the dominant genotype, the rate of enrichment was much lower after the liver was fully infected than during the spread of infection. In order to calculate an average enrichment rate (E/Δt) during this phase of the infection, we used the serum genotype data obtained for birds 11, 30, 40, and 41 from the time of peak viremia through 47 days postinfection to calculate the enrichment (E) for wild-type virus (see Materials and Methods). The log10E values were plotted against time postinfection to obtain the graph shown in Fig. 5. As with the enrichment during the spread of infection (Fig. 2), the increase in logE was roughly linear over time, and a linear regression curve was calculated. The slope of the regression, log10E/Δt, was determined to be 0.035 log10E/Δt, or approximately 0.12 times that calculated for the enrichment during the spread of infection.
FIG. 5.
Enrichment of wild-type virus during chronic infection. The ratios of wild-type virus to 2619A virus in the serum of ducks 11, 30, 40, and 41 on days 6 through 47 were determined and used to calculate the enrichment, E, over the earliest time point (day 6 for birds 11 and 40 and day 7 for birds 30 and 41). The linear regression was calculated for the combined data for all four birds.
Anti-core antibody response in birds with predominant wild-type or 2619A virus.
The selection against wild-type virus in four birds (birds 8, 10, 14, and 34) did not obviously correlate with the any virological parameters of the individual birds shown in Tables 1 and 2, including viremia, body weight (not shown), dose of infection, or size of the inoculum. All birds showing selection against wild-type virus were infected with inocula containing ratios of wild type to 2619A of 1:50 or less, but the importance of this correlation is uncertain. Since the wild-type virus expressed precore protein and the 2619A virus did not, it was possible that precore production formed the basis for a selection against wild-type virus that differed among individual birds. Such a selection could be mediated by the individual immunological response of each infected bird. As an initial indicator of the immunological response against precore and core epitopes we measured the titers of total anti-core antibody of the eight individual birds represented in Fig. 4A at various times postinfection. The results of these assays are shown in the corresponding graphs of Fig. 4B.
Differences between the two groups of birds showing different patterns of strain selection could be seen. Selection against wild-type virus was correlated with elevated levels of anticore antibody that increased throughout the follow-up period. Anti-core antibody titers in the group of birds showing no selection against wild-type virus were more variable, differing by more than 2 orders of magnitude within the group. In three of four birds, anti-core antibody titers were stable or decreased 10-fold or more from an early peak value. These results suggest that the immunological response to precore and core epitopes differed substantially among individual birds in a manner that could be related to the relative enrichment of the two strains.
Analysis of the replication properties of the predominant 2619A strain in birds 8, 12, 14, and 34.
It was possible that chronic infection selected for a virus that replicated more rapidly than either the wild-type or the 2619A virus in the original inoculum and which eventually replaced both of the infecting strains. This replacement would appear in our assay as an enrichment of either wild-type or 2619A virus, depending on which genetic marker was carried by the variant at position 2619. If this were the case, viruses carrying the 2619A mutation predominating at late times postinfection in birds 8, 10, 14, and 34 would show a replication rate that was enhanced over that of wild-type virus. In order to measure the growth properties of these viruses, viral genomes from birds showing the presence of only the 2619A mutant, i.e., birds 8, 14, and 34, were amplified from serum obtained at 77 days postinfection, using the strategy devised for high-fidelity PCR of complete hepadnavirus genomes (8). In addition, serum DNA obtained at 77 days from bird 12, which never showed detectable wild-type virus (not shown), was also amplified. Two viral genomes from each amplified sample were cloned as head-to-tail dimers in plasmid pSP65. The cloned viral genomes were then compared with wild-type genomes for their relative rates of replication in LMH cells and in ducklings.
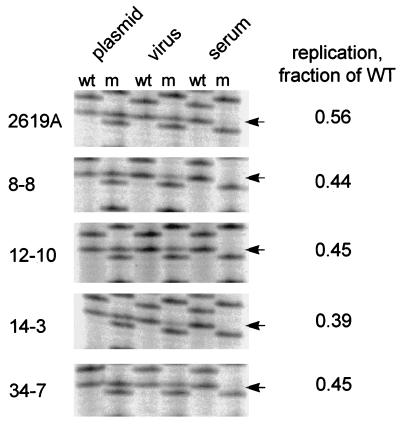
Of the 10 genomes that were cloned and tested, 9 clones were able to produce enveloped virus after transfection into LMH cells (data not shown). If these 2619A genomes were selected in vivo because they had acquired the ability to replicate faster than wild-type virus, we should observe their enrichment during competition with wild-type virus either in vitro or in vivo. To test this prediction, eight 2619A dimer clones were cotransfected with a wild-type dimer clone into LMH cells at a ratio of 1:1, and the virus produced in the medium was injected into 3-day-old ducklings. The ratios of wild-type genomes to 2619A genomes in the plasmid mixture, in virus from the culture medium, and in virus from the serum of the infected ducklings at peak viremia were compared by PCR and sequencing. Examples of these assays are presented in Fig. 6, and the combined results of all the assays on these clones are shown in Table 3. We observed that all 2619A clones retained a replication defect in LMH cells that was comparable to that of the 2619A parent in the original inoculum. Neither the authentic 2619A mutant nor any of the cloned genomes were detected in the serum after passage in ducklings, consistent with the high level of enrichment of wild-type virus shown in Fig. 2. This result indicates that the wild-type virus was not replaced by a faster-replicating variant of the precore mutant.
FIG. 6.
PCR sequencing assay for the relative replication of 2619A clones obtained from serum. Whole DHBV genomes were amplified and cloned from the sera of birds 8, 12, 14, and 34 obtained at day 77 postinfection. The genomes were subcloned as dimers in pSP65 and cotransfected with a wild-type (wt) DHBV plasmid into LMH cells, and the supernatants were used to infect ducklings (107 viral genomes per duckling). PCR sequencing was performed on the original plasmid mixture (plasmid), on DNA from enveloped virus from the supernatants of the transfected cells (virus), and on viral DNA from serum of the infected ducklings (serum). The yield of mutant viruses (m) in the culture supernatants was used to calculate the replication rate of the mutant relative to that of the wild type (WT) (right). Mutant virus was not detected in serum from any of the mixed infections of ducklings.
TABLE 3.
Growth properties of 2619A clones isolated from ducks
| Clone | Relative replication rate of 2619A in vitroa | 2619A in serumb |
|---|---|---|
| 2619A | 0.56, 0.58, 0.54 | ND |
| 8-4 | 0.27 | NDc |
| 8-8 | 0.44, 0.28 | ND |
| 12-9 | 0.30 | ND |
| 12-10 | 0.78, 0.45 | ND |
| 14-2 | 0.66 | ND |
| 14-3 | 0.39, 0.38 | ND |
| 34-7 | 0.56, 0.45 | ND |
| 34-8 | 0.53 | ND |
Expressed as the fraction of wild type in LMH supernatants. Each value is the result from an independent transfection.
ND, not detected.
This mutant failed to make enveloped virus in single transfections.
DISCUSSION
The results of our experiments suggest that selection between two competing strains of DHBV during the chronic phase of an infection is not necessarily determined by the relative replication rates of the two strains. We have examined the behavior of two strains of DHBV with different replication rates during mixed infections lasting up to 12 weeks postinfection. During the initial stage, in which infection spread throughout the liver, strain selection was always determined by the relative growth rates of the two viruses. The evidence for this conclusion was the fact that the faster-replicating wild-type virus was always highly enriched, and the extent of enrichment was proportional to the amount of growth that preceded peak viremia (Fig. 2). The replication rate of the precore mutant, 2619A, was estimated to be 37 and 61% of that of the wild-type virus based on the data from this study combined with two previously estimated replication rates for wild-type virus in ducklings (12, 13). This range was inclusive of the relative replication rates of the parental 2619A genome calculated in transfected LMH cells, i.e., 54 to 58% of that of the wild type (Table 3), and differences in the estimates can probably be attributed to a combination of various experimental errors and assumptions. In any case, the replication rate appeared to be the overriding factor in strain selection of DHBV when replication was not limited by the number of susceptible cells, i.e., during the spread of infection.
Strain selection during the chronic phase of infection, when all hepatocytes were infected, differed in two respects from that observed during the spread of infection. In 4 of 10 birds, wild-type virus continued to be enriched, but at a greatly reduced rate. While enrichment occurred at the rate of 0.3 log10E day−1 (Fig. 2) during the spread of infection, the rate of enrichment during chronic infection was around 0.035 ln10E day−1 (Fig. 5). This result may be a reflection of the dynamic state of the infection after the liver is fully infected. That is, continued competition between the two virus strains may be limited by the rate at which newly susceptible cells appear in the liver or by the rate at which covalently closed circular DNA molecules in cells that are already infected are replaced by newly synthesized molecules. In these circumstances, the rate of enrichment can be used to calculate the dynamic state of the infection in vivo (44b).
In a second group of four birds, strain selection was determined by factors not directly related to replication rate. In these birds the faster-replicating wild-type virus was replaced by the precore mutant despite its slower replication rate. This result indicates that a selective advantage of the mutant virus was expressed that was sufficient to overcome its replication disadvantage. Replacement of the wild-type virus by the 2619A virus in these birds was correlated with elevated titers of anti-core antibody. The reason for differences in the anti-core antibody titers is not known, but we can suggest three possibilities that are not mutually exclusive. First, the anti-core antibody titers in birds producing higher levels of e antigen might be reduced correspondingly by titration with cross-reacting soluble e antigen in the blood. Thus, birds with high production of e antigen from wild-type virus would have lower levels of anti-core and anti-e antibody. The natural responses of DHBV-infected ducks to core and e antigens and the cross-reactivity of these antigens have not been characterized, and therefore it is difficult to evaluate this explanation. Alternatively, the anti-core antibody response might be a reflection of the level of antigen stimulation caused by release of viral cores from injured hepatocytes. Hepatocyte injury could be part of the mechanism of selection against wild-type virus in favor of the precore mutant. Thus, higher anti-core antibody titers would be found in birds in which wild-type virus was being replaced by the precore mutant. Finally, the anti-core antibody-specific B-cell response could reflect the strength or quality of the Th response, which in turn would influence the T-cell-mediated immune pressure on infected cells in the liver. In this scenario, the wild-type virus would be more sensitive to the cellular immune response in the liver than the precore-minus mutant.
The mechanism for a putative immunological selection against wild-type virus in favor of our precore mutant is not known. Presumably, such a selection would operate at the level of the infected cell, since the precore protein is not incorporated into virus particles. The core and precore proteins of HBV are generally considered to be antigenically identical at the T-cell level, but it is possible that (i) the precore region may encode one or more unique T-cell epitopes or (ii) part of the precore protein is proteolytically processed to produce peptides that are recognized by unique precore-specific T-cell receptors on lymphocytes.
Alternatively, immunological selection might depend on the growth properties of the 2619A mutant. Precore variants have been widely observed to emerge during chronic HBV infections, leading to the speculation that such variants have been selected on the basis of an enhanced rate of replication. This explanation does not appear to apply in our experiments, since the precore mutant selected in vivo replicated more slowly than the wild type when subjected to a second passage in vitro and in vivo. In fact, it is possible that strain selection in our experiment might have depended on the reduced replication rate of the 2619A mutant if, for example, the cellular immune response were able to distinguish relatively small differences in replication rate as the basis for immune pressure. In humans, the emergence of precore variants of HBV is often associated with exacerbations of liver disease, consistent with an immunological selection against wild-type virus-infected cells. This hypothesis would imply that the emergence of precore variants may be determined by selective pressures that are associated with the disease and that the disease itself is not an inherent property of precore-minus variants.
Finally, it is possible that precore expression or wild-type replication is toxic in some animals for reasons unrelated to the immune response. This hypothesis does not depend on an immunological selection occurring, but toxicity of wild-type replication may produce higher levels of anti-core antibody from greater antigenic stimulation by cores released from dying hepatocytes. Individual differences among birds could be related to differences in their genetic backgrounds, for example.
It is commonly assumed that expression of the precore protein results in some benefit to DHBV during some stage of its life cycle. These studies do not contradict this view; they indicate, however, that precore protein may be disadvantageous to the virus under some conditions. In fact, if the precore protein or e antigens act at a very early phase of infection to influence the course of the immune response, this influence may not have been exerted in our experiments because the inoculum contained a large excess of precore-minus virus. Thus, it is possible that the positive function of the precore protein in hepadnaviruses may be expressed only during a limited window in the viral life cycle.
ACKNOWLEDGMENTS
We express appreciation to Andrew Kuhn, Josh Ramey, and Bai-Hua Zhang for technical assistance and to Wengang Yang, Raymond Lenhoff, and Carolyn Luscombe for helpful suggestions and advice during the course of these experiments. We thank W. S. Mason for helpful advice on the manuscript.
This work was supported by HSS grant CA-42542.
REFERENCES
- 1.Baumert T F, Marrone A, Vergalla J, Liang T J. Naturally occurring mutations define a novel function of the hepatitis B virus core promoter in core protein expression. J Virol. 1998;72:6785–6795. doi: 10.1128/jvi.72.8.6785-6795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonino F, Rosina F, Rizzetto M, Rizzi R, Chiaberge E, Tardanico R, Callea F, Verme G. Chronic hepatitis in HBsAg carriers with serum HBV-DNA and anti-HBe. Gastroenterology. 1986;90:1268–1273. doi: 10.1016/0016-5085(86)90395-1. [DOI] [PubMed] [Google Scholar]
- 3.Carman W, Jacyna M R, Hadziyannis S, Karayiannis P, McGarvey M J, Makris A, Thomas H C. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;ii:588–591. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 4.Chen H S, Kew M C, Hornbuckle W E, Tennant B C, Cote P J, Gerin J L, Purcell R H, Miller R H. The precore gene of the woodchuck hepatitis virus genome is not essential for viral replication in the natural host. J Virol. 1992;66:5682–5684. doi: 10.1128/jvi.66.9.5682-5684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condreay L, Aldrich C, Coates L, Mason W S, Wu T-T. Efficient duck hepatitis B virus production by an avian tumor cell line. J Virol. 1990;64:3249–3258. doi: 10.1128/jvi.64.7.3249-3258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dane D S, Cameron C H, Briggs M. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet. 1970;i:695–698. doi: 10.1016/s0140-6736(70)90926-8. [DOI] [PubMed] [Google Scholar]
- 7.Ganem D. Hepadnaviridae and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2703–2737. [Google Scholar]
- 8.Gunther S, Li B C, Miska S, Kruger D H, Meisel H, Will H A. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol. 1995;69:5437–5444. doi: 10.1128/jvi.69.9.5437-5444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang M, Summers J. Infection initiated by the RNA pregenome of a DNA virus. J Virol. 1991;65:5435–5439. doi: 10.1128/jvi.65.10.5435-5439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jean-Jean O, Levrero M, Will H, Perricaudet M, Rossignol J M. Expression mechanism of the hepatitis B virus (HBV) C gene and biosynthesis of HBe antigen. Virology. 1989;170:99–106. doi: 10.1016/0042-6822(89)90356-5. [DOI] [PubMed] [Google Scholar]
- 11.Jilbert A R, Botten J A, Miller D S, Bertram E M, Hall P M, Kotlarski J, Burrell C J. Characterization of age- and dose-related outcomes of duck hepatitis B virus infection. Virology. 1998;244:273–282. doi: 10.1006/viro.1998.9095. [DOI] [PubMed] [Google Scholar]
- 12.Jilbert A R, Freiman J S, Burrell C J, Holmes M, Gowans E J, Rowland R, Hall P, Cossart Y E. Virus-liver cell interactions in duck hepatitis B virus infection. A study of virus dissemination within the liver. Gastroenterology. 1988;95:1375–1382. doi: 10.1016/0016-5085(88)90375-7. [DOI] [PubMed] [Google Scholar]
- 13.Jilbert A R, Miller D S, Scougall C A, Turnbull H, Burrell C J. Kinetics of duck hepatitis B virus infection following low dose virus inoculation: one virus DNA genome is infectious in neonatal ducks. Virology. 1996;226:338–345. doi: 10.1006/viro.1996.0661. [DOI] [PubMed] [Google Scholar]
- 14.Kawaguchi T, Nomura K, Hirayama Y, Kitagawa T. Establishment and characterization of a chicken hepatocellular carcinoma cell line LMH. Cancer Res. 1987;47:4460–4464. [PubMed] [Google Scholar]
- 15.Lenhoff R, Summers J. Construction of avian hepadnavirus variants with enhanced replication and cytopathicity in primary hepatocytes. J Virol. 1994;68:5706–5713. doi: 10.1128/jvi.68.9.5706-5713.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenhoff R, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenhoff, R., C. A. Luscombe, and J. Summers. Acute liver injury following infection with a cytopathic strain of duck hepatitis B virus. Hepatology, in press. [DOI] [PubMed]
- 18.Lenhoff, R. L., C. A. Luscombe, and J. Summers. Competition in vivo between a cytopathic variant and a wild type duck hepatitis B virus. Virology, in press. [DOI] [PubMed]
- 19.Li D H, Newbold J E, Cullen J M. Natural populations of woodchuck hepatitis virus contain variant precore and core sequences including a premature stop codon in the epsilon motif. Virology. 1996;220:256–262. doi: 10.1006/viro.1996.0311. [DOI] [PubMed] [Google Scholar]
- 20.Magnius L O, Espmark J A. New specificities in Australia antigen positive sera distinct from the Le Bouvier determinants. J Immunol. 1972;109:1017–1021. [PubMed] [Google Scholar]
- 21.Mandart E, Kay A, Galibert F. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J Virol. 1984;49:782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marion P L, Oshiro L S, Regnery D C, Scullard G H, Robinson W S. A virus in Beechey ground squirrels that is related to hepatitis B virus of humans. Proc Natl Acad Sci USA. 1980;77:2941–2945. doi: 10.1073/pnas.77.5.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason W S, Aldrich C, Summers J, Taylor J M. Asymmetric replication of duck hepatitis B virus DNA in liver cells (free minus strand DNA) Proc Natl Acad Sci USA. 1982;79:3997–4001. doi: 10.1073/pnas.79.13.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mason W S, Seal G, Summers J. A virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol. 1980;36:829–836. doi: 10.1128/jvi.36.3.829-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milich D R. Influence of T-helper cell subsets and crossregulation in hepatitis B virus infection. J Viral Hepat. 1997;4(Suppl. 2):48–59. doi: 10.1111/j.1365-2893.1997.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 26.Milich D R, Chen M K, Hughes J L, Jones T E. The secreted hepatitis B precore antigen can modulate the immune response to the nucleocapsid: a mechanism for persistence. J Immunol. 1998;160:2013–2021. [PubMed] [Google Scholar]
- 27.Milich D R, Schodel F, Hughes J L, Jones J E, Peterson D L. The hepatitis B virus core and e antigens elicit different Th cell subsets: antigen structure can affect Th cell phenotype. J Virol. 1997;71:2192–2201. doi: 10.1128/jvi.71.3.2192-2201.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moriyama K, Okamoto H, Tsuda F, Mayumi M. Reduced precore transcription and enhanced core-pregenome transcription of hepatitis B virus DNA after replacement of the precore-core promoter with sequences associated with e antigen-seronegative persistent infections. Virology. 1996;226:269–280. doi: 10.1006/viro.1996.0655. [DOI] [PubMed] [Google Scholar]
- 29.Nassal M, Schaller H. Hepatitis B virus replication—an update. J Viral Hepat. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 30.Pult I, Chouard T, Wieland S, Klemenz R, Yaniv M, Blum H E. A hepatitis B virus mutant with a new hepatocyte nuclear factor 1 binding site emerging in transplant-transmitted fulminant hepatitis B. Hepatology. 1997;25:1507–1515. doi: 10.1002/hep.510250633. [DOI] [PubMed] [Google Scholar]
- 31.Raimondo G, Stemler M, Schneider R, Wildner G, Squadrito G, Will H. Latency and reactivation of a precore mutant hepatitis B virus in a chronically infected patient. J Hepatol. 1990;11:374–380. doi: 10.1016/0168-8278(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 32.Robinson W S, Clayton D A, Greenman R L. DNA of a human hepatitis B virus candidate. J Virol. 1974;14:384–391. doi: 10.1128/jvi.14.2.384-391.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scaglioni P P, Melegari, Wands J R. Biologic properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame. Virology. 1997;233:374–381. doi: 10.1006/viro.1997.8594. [DOI] [PubMed] [Google Scholar]
- 34.Schlicht H J, Salfeld J, Schaller H. The duck hepatitis B virus pre-C region encodes a signal sequence which is essential for synthesis and secretion of processed core proteins but not for virus formation. J Virol. 1987;61:3701–3709. doi: 10.1128/jvi.61.12.3701-3709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seeger C, Ganem D, Varmus H E. Biochemical and genetic evidence for the hepatitis B virus replication strategy. Science. 1986;232:477–484. doi: 10.1126/science.3961490. [DOI] [PubMed] [Google Scholar]
- 36.Sprengel R, Kaleta E F, Will H. Isolation and characterization of a hepatitis B virus endemic in herons. J Virol. 1988;62:3832–3839. doi: 10.1128/jvi.62.10.3832-3839.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Standring D N, Ou J H, Masiarz F R, Rutter W J. A signal peptide encoded within the precore region of hepatitis B virus directs the secretion of a heterogeneous population of e antigens in Xenopus oocytes. Proc Natl Acad Sci USA. 1988;85:8405–8409. doi: 10.1073/pnas.85.22.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 39.Summers J, O’Connell A P, Millman I. Genome of hepatitis B virus: restriction enzyme cleavage and structure of the DNA extracted from Dane particles. Proc Natl Acad Sci USA. 1975;72:4597–4601. doi: 10.1073/pnas.72.11.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Summers J, Smith P, Huang M, Yu M. Regulatory and morphogenetic effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Summers J, Smolec J M, Snyder R L. A virus similar to hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci USA. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Testut P, Renard C A, Terradillos O, Vitvitski-Trepo L, Tekaia F, Degott C, Blake J, Boyer B, Buendia M A. A new hepadnavirus endemic in arctic ground squirrels in Alaska. J Virol. 1996;70:4210–4219. doi: 10.1128/jvi.70.7.4210-4219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas H C. The emergence of envelope and precore/core variants of hepatitis B virus: the potential role of antibody selection. J Hepatol. 1995;22:1–8. [PubMed] [Google Scholar]
- 44.Trepo C, Zoulim F, Alonso C, Petit M A, Pichoud C, Vitvitski L. Diagnostic markers of viral hepatitis B and C. Gut. 1993;34:S20–S25. doi: 10.1136/gut.34.2_suppl.s20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Yang, W., and J. Summers. Unpublished data.
- 44b.Zhang, Y.-Y., and J. Summers. Unpublished data.
- 45.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]