Abstract
Although species can arise through hybridization, compelling evidence for hybrid speciation has been reported only rarely in animals. Here, we present phylogenomic analyses on genomes from 12 macaque species and show that the fascicularis group originated from an ancient hybridization between the sinica and silenus groups ~3.45 to 3.56 million years ago. The X chromosomes and low-recombination regions exhibited equal contributions from each parental lineage, suggesting that they were less affected by subsequent backcrossing and hence could have played an important role in maintaining hybrid integrity. We identified many reproduction-associated genes that could have contributed to the development of the mixed sexual phenotypes characteristic of the fascicularis group. The phylogeny within the silenus group was also resolved, and functional experimentation confirmed that all extant Western silenus species are susceptible to HIV-1 infection. Our study provides novel insights into macaque evolution and reveals a hybrid speciation event that has occurred only very rarely in primates.
Comparative phylogenomic analysis of major groups of macaque uncovered an ancient hybrid origin of a macaque lineage.
INTRODUCTION
Interspecific hybridization can facilitate species adaptation by introducing new genetic material and novel allelic combinations (1). In some circumstances, hybridization may result in the near-instantaneous formation of new species, i.e., hybrid speciation, thereby promoting biodiversity (2). Hybrid speciation has been an important mode of speciation during plant evolution (3). However, hybrid speciation has been considered to occur only rarely in animals owing to the fact that the derived species from interspecific hybridization typically tend to be evolutionarily less fit (e.g., Dobzhansky-Muller incompatibility) or are only weakly reproductively isolated from their parental species and hence could be swamped by one of their parents (4).
Both theoretical and empirical studies suggest that those species groups that have experienced rapid adaptive radiation are prone to hybridization because the incompatibilities between these species are weak (1). Hybrid lineages are more readily established when hybrids mate assortatively with each other (5) and/or when they became adapted to a new environment (6). An increasing number of empirical studies have documented the establishment of hybrid lineages in rapidly radiating groups, including butterflies, canids, baboon, birds, and bears (7–11), suggesting that hybrid speciation may be more common and hence more important than previously thought. Nevertheless, concrete evidence of hybrid speciation in animals remains scarce and putative examples are often highly contentious (12, 13), impeding our understanding of the genomic mechanisms underlying hybrid speciation. In particular, it is often challenging to reliably differentiate between hybrid speciation and post-speciation genetic introgression (4), which may be further complicated by the stochastic sorting of ancestral polymorphisms among descendant lineages, a process known as incomplete lineage sorting (ILS) (14). Thus, a comparative phylogenomic framework to unequivocally demonstrate hybrid speciation has yet to be established.
The genus of macaques (Macaca) represents an excellent model in which to study the interplay between interspecific hybridization and speciation. As one of the most successful primate lineages, macaques now include 23 species that are widely distributed across South, East, and Southeast Asia, the only exception being M. sylvanus, which is confined to the Atlas mountains in North Africa (15). Paleontological and molecular data suggest that macaques originated in North Africa ~7 million years (Ma) ago and then experienced a burst of speciation in Asia during the past 5 Ma (16, 17). Some macaque species are widely used as laboratory models for studying human disease and for vaccine development (18). Although members of this genus are morphologically and behaviorally distinct, interspecific hybridization is potentially possible between any geographically overlapping pair of species because their reproductive isolation is likely to be incomplete. This notion is supported by both field observations (19, 20) and molecular studies (21, 22). While some macaque species (e.g., the fascicularis group of macaques and the stump-tailed macaque) exhibit distinctive mixed phenotypes, which could, in principle, have arisen by genomic admixture (23, 24), the broader role of hybridization and its specific outcomes remain largely unknown. Here, we perform multiple genome analyses to ascertain species phylogeny in the macaque genus, assess the role of interspecific hybridization in speciation, and explore the genetic basis of mixed phenotypes in macaque species.
RESULTS
We generated 10 high-quality macaque genome assemblies (tables S1 to S3) using the long-read sequencing strategy of Nanopore and long fragment read technologies (stLFR and 10X Genomics), respectively. Scaffold N50 sizes of these newly assembled genomes ranged from 17.7 to 33.2 Mb, while the Benchmarking Universal Single-Copy Orthologs (BUSCO) completeness scores ranged from ~93.3 to 94.5%. The final annotated gene numbers ranged from 20,662 to 21,811 between different species using a combination of ab initio and homology-based gene prediction approaches. We also included two previously published genomes of M. mulatta (25) and M. nemestrina, and one outgroup species Papio hamadryas (26). Thus, our genome assemblies cover all known macaque species groups (Fig. 1A) (17).
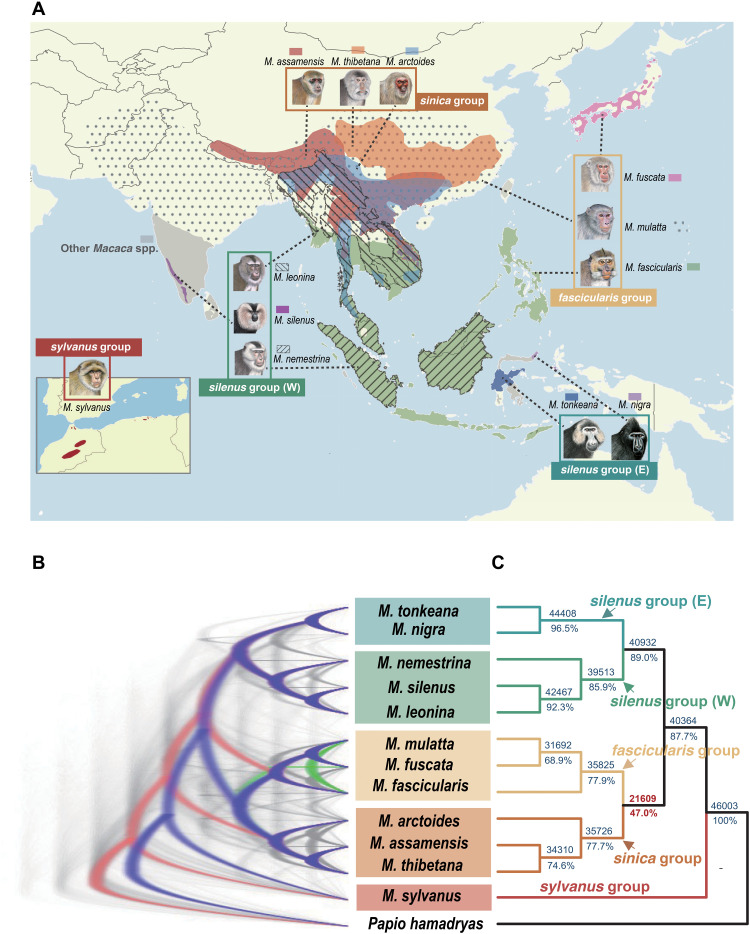
Fig. 1. Distribution map and the discordance of phylogeny.
(A) Distribution map of macaque species used in this study. Nomenclature follows Delson (16) in defining the species group. We further split the silenus group into Western (W) and Eastern (E) forms on the basis of their similar divergence to other species groups. Macaque drawings are copyright, 2013, Stephen D. Nash, International Union for Conservation of Nature Species Survival Commission Primate Specialist Group and are used with permission. All species except M. mulatta and M. nemestrina were newly sequenced in this study. (B) DensiTree plot for 50-kb window trees. Blue, red, and green colors represent the first, second, and third most common topologies, respectively, whereas gray represents other topologies. (C) Majority-rule consensus tree of all 50-kb window trees (n = 46,003). The numbers above the branches indicate the absolute number of topologies supporting the splits, whereas the numbers below represent the percentage values.
We first performed pairwise whole-genome alignments against the Chinese rhesus macaque (M. mulatta, rheMacS) genome using the LASTZ program (27) and then merged all of them into multiple genome alignments with MULTIZ (28). These genomes showed high collinearity, with the aligned base pairs spanning more than 2.7 Gb or 90% of the reference assembly (fig. S1 and table S5). To deduce the correct branching pattern of the sequenced macaque species and to characterize the heterogeneous phylogenetic signals across genomes, we partitioned the genome alignments into nonoverlapping windows of 50 kb and then performed maximum likelihood (ML) analyses for each window sequence. We initially obtained the multispecies coalescent-based species tree (ASTRAL and STAR; fig. S3) from windows with high bootstrap values (e.g., mean value of >80%). Although the topologies obtained from these two methods were mutually consistent and largely in accord with recent studies (17, 21), we observed substantial genealogical discordance within and between species groups among all window trees (n = 7392 alternative topologies; Fig. 1, B and C, and fig. S5), mostly pertaining to the phylogenetic position of the fascicularis group with respect to the sinica and silenus groups. The discordance of this position was not a trivial consequence of the choice of window size because analyses using either smaller (20 kb) or larger (100 kb) window sizes yielded broadly similar results (figs. S6 to S9). To better understand the interrelationship of macaque groups with incongruent phylogenies, we pruned the trees so as to include five ingroup species, each representing a particular lineage proposed by previous studies (16, 17). Two topologies were found to dominate: One supported a sister relationship between the fascicularis and sinica groups (T1), whereas the other (T2) favored the fascicularis group as a sister lineage to the silenus group. These two tree topologies were supported by a total of 78% of windows interleavedly distributed across the genome, with 47 and 31% referring to T1 and T2, respectively (Fig. 2, A and B). Specifically, we found that the X chromosome, where genomic incompatibilities normally first develop during speciation (29), exhibited a nearly equal proportion of T1 and T2 topologies versus the autosomes irrespective of window size (fig. S10). Such widespread mixed ancestry of the autosomes and the X chromosome led us to speculate that the fascicularis group may have originated from an ancient hybridization between the progenitors of the sinica group and those of the silenus group.
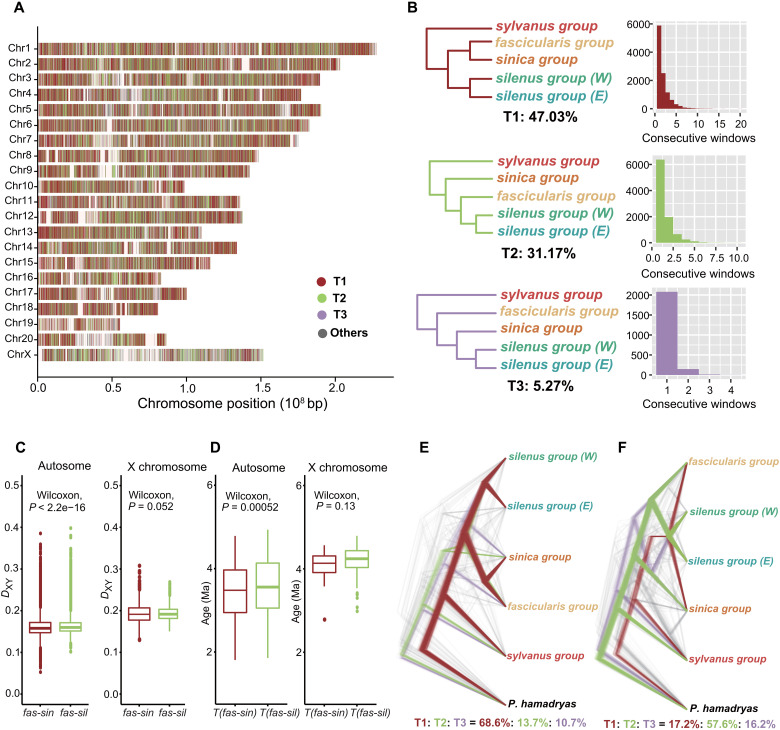
Fig. 2. Phylogenetic relationship of the five major macaque lineages across the genome.
(A) Distribution of the three most common topologies by reference to the Chinese rhesus macaque (M. mulatta) genome. Colored bands represent tree topologies of each 50-kb window. White interval regions denote missing data. (B) Three most common trees (T1 to T3) recovered by ML analysis and their frequency in consecutive 50-kb windows. Values below the tree refer to the percentage of windows recovering that topology. The outgroup P. hamadryas is not shown. (C) Genetic divergence (DXY) and (D) estimated divergence times from MCMCTREE between the fascicularis group and its two putative parental lineages (sinica and silenus) based on autosomal 50-kb window sequences (left) and the X chromosome (right), respectively. P values were estimated by the Wilcoxon rank sum test. The substitution rates estimated from MCMCTREE for the autosomes and X chromosome were 1.2 × 10−9 per site per year and 0.9 × 10−9 per site per year, respectively. (E) DensiTree plot of ML trees derived from the 1% windows with the lowest absolute D-statistic values using T1, and (F) T2, as the tested relationship. The values below show the percentage of windows that recovered the topology.
If the hybrid origination hypothesis is correct, we would expect to observe an equal level of sequence divergence between the hybrid species and its two parental lineages. To test this postulate, we compared the sequence divergence (DXY) and relative divergence time between fascicularis versus sinica and fascicularis versus silenus for the autosomes and X chromosome separately to allow for their different rates of evolution (30). We found no obvious difference in these two statistics calculated using only the sex chromosome data (both P values > 0.05, Wilcoxon’s test; Fig. 2, C and D). With the autosomes, although both parameters were statistically significant (P < 0.001, Wilcoxon’s test; Fig. 2, C and D), the mean DXY value (0.1616 versus 0.1631) and the mean estimated age (3.45 Ma versus 3.56 Ma) between the fascicularis group and its two progenitors were only slightly different. The observed pattern therefore concurs with that expected under the hybrid speciation scenario.
However, extensive post-speciation gene flow may also have produced a similar signature (31). To test this possibility, we performed D-statistic analyses in a sliding window (32). If either T1 or T2 represent the true species phylogeny, the windows least affected by gene flow (absolute D-statistic values close to zero) would overwhelmingly support one of the tested trees, whereas hybridization would support both (33). Our results supported the latter prediction showing that the 1% of windows with the lowest absolute D-statistic values supported T1 and T2, respectively, when using T1 and T2 as the tested topologies (Fig. 2, E and F). Furthermore, on the basis that regions of lower recombination are “cold spots” for introgression (29), we classified the trees by recombination rate and found that T1 and T2 were still dominant in the low recombination rate regions of 0 to 0.1 cM/Mb (fig. S11). This observation was even more evident with a smaller window size (20 kb), where the ratio of T1 and T2 was almost identical (T1:T2 = 32%:31%; fig. S13), probably due to the averaging effect with the larger window size. These findings corroborate the view that hybrid speciation, rather than post-speciation gene flow, was responsible for the pattern of genomic mosaicism.
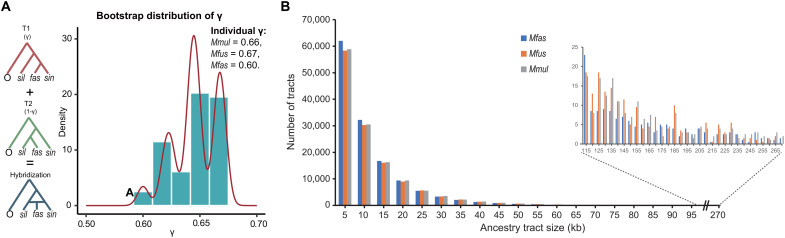
Another underlying assumption of hybrid speciation is that all the individuals from the hybrid species are uniformly admixed. However, if ancient directional gene flow and ILS were to have occurred, it is likely that not all individuals in the hybrid group would have contained the same amount and length distribution of introgressed alleles because subsequent recombination and negative selection would have acted so as to purge the deleterious intruder alleles from the acceptor genome during a long period of divergence (34). To test this prediction, we used HyDe (35) and LOTER (36) to quantify the genomic contributions of the two parental lineages (sinica and silenus) to each of the hybrid species in the fascicularis group. The results were consistent with our expectation under the hybridization hypothesis, showing that all three species from the fascicularis group were uniformly mixed and exhibited a general exponential decay of the tract size (Fig. 3, A and B, and table S7), irrespective of whether the species are geographically isolated (M. fuscata) or widely distributed (M. mulatta and M. fascicularis). This result was further corroborated by PhyloNet-MPL analyses, an alternative method that allows for ILS and hybridization simultaneously, based on the maximum pseudo-likelihood method (37), where the section of the fascicularis group was invariably identified as a reticulate node in the scenarios allowing one and two past hybridization events (Fig. 4A and fig. S17).
Fig. 3. Genomic ancestry inherent in two parental lineages.
(A) Density plot of estimated γ values across 500 bootstrap replicates in HyDe analyses. γ represents the estimated probability of inheritance from the ancestor of the sinica group, whereas 1-γ represents the probability of inheritance from the silenus group. The mean γ values of the three hybrid species are given top right. Mmul, M. mulatta; Mfas, M. fascicularis; Mfus, M. fuscata. (B) Distribution of tract sizes (in 5-kb bins) of sinica group ancestry in three hybrid species.
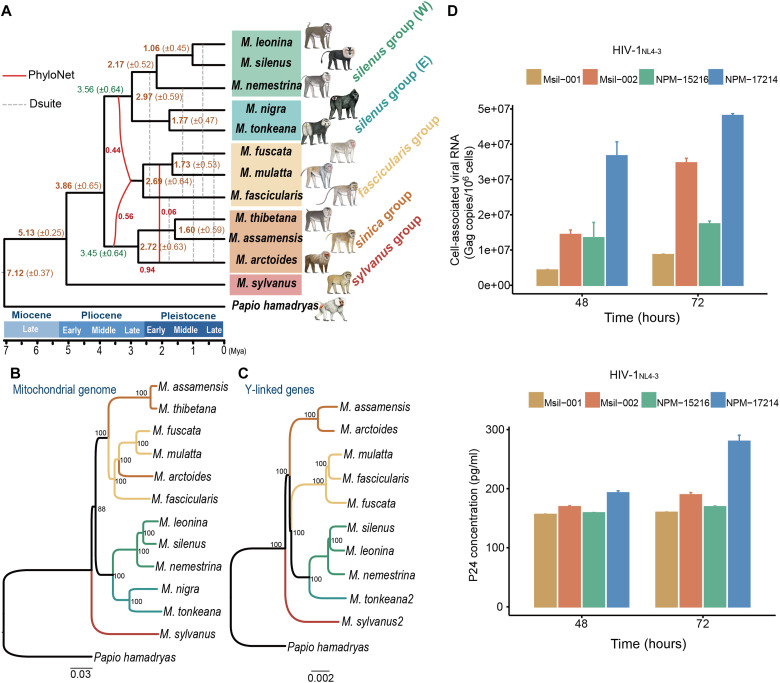
Fig. 4. Hybrid origin of the fascicularis group.
(A) Schema of the reticulated evolutionary relationships between macaque species, illustrating the hybridization and major admixture events and approximate divergence times (Ma ± 1 SD) inferred from autosomal windows. The solid red line denotes the interspecific gene flow obtained from PhyloNet analyses, whereas the gray dashed line denotes the gene flow inferred from Dsuite. For the Dsuite results, we show the intersection gene flow events modeled from the two most common trees, i.e., one supporting the sister relationship between the fascicularis and sinica groups (fig. S5A), the other supporting the sister relationship between the fascicularis and silenus groups (fig. S5B). (B) Mitochondrial and (C) Y chromosomal trees. Numbers at nodes refer to bootstrap values. M. tonkeana2 and M. sylvanus2 are two additional sequenced male samples. Detailed sample information is shown in table S1. (D) Plasma viral load in PBMC aliquots from two lion-tailed macaques (Msil-001 and Msil-002) after 48 and 72 hours of infection with HIV-1NL4-3 virus. Two northern pig-tailed macaques (NPM-15216 and NPM-17214) were used as positive controls.
Together, these phylogenomic analyses concur in terms of providing consistent support for the hybrid origin hypothesis of the fascicularis group but were not informative regarding its direction in the initial hybridization. Because the mitochondrial genome and the Y chromosome are representative of the maternal and paternal lineages, respectively, we next performed phylogenetic analyses based on mitochondrial and Y chromosome sequences (~480 kb in length) to determine the hybridization pattern. We found a clear topology discrepancy between the mitochondrial and the Y chromosomal data, with the fascicularis group having a mitochondrial genome similar to that of the sinica group and a Y chromosome similar to that of the silenus group (Fig. 4, B and C), suggesting that the ancient hybridization occurred predominantly or exclusively between proto-sinica group females and proto-silenus group males.
With this resolved phylogeny, we further inferred directional gene flow events between macaque groups after the hybrid origin of the fascicularis group. The results obtained revealed a complex network of ancestral admixture in Macaca (Fig. 4A and fig. S20). It is interesting to note that after the hybrid origin, various species of the fascicularis group maintained gene flow with the descendant species of the two parental groups. Among them, the most notable direction of gene flow occurred between the stump-tailed macaque (M. arctoides) and the common ancestor of mulatta/fuscata. On the basis of discrepancies between mitochondrial and Y chromosomal topologies, it has been previously proposed that the stump-tailed macaque originated from hybridization between sinica and mulatta/fuscata macaques (22). However, our analyses are consistent with another phylogenomic study (38) that indicated the stump-tailed macaque to be a member of the sinica group with a low level of introgression from mulatta/fuscata (0.05 ≤ γ ≤ 0.09; table S7). In the phylogenetic trees based on low recombination rate regions and the X chromosome (figs. S14 and S15), the stump-tailed macaque also clustered with the sinica group, suggesting that mitochondrial introgression from mulatta/fuscata macaques occurred after the initial speciation.
Our phylogeny and introgression network also revealed a branching pattern within the silenus group that contradicts the previous morphological hypothesis, which suggested that the northern-tailed macaque (M. leonina) and southern-tailed macaque (M. nemestrina) are sister species (39). Although these two species are close ecologically and geographically and are also similar in phenotypic appearance (Fig. 1A), all our phylogenomic analyses with different genomic data types support the postulate that the lion-tailed macaque (M. silenus) from the South Indian Western Ghats and M. leonina are sister species (Fig. 4, A to C, and figs. S14 and S15). Our previous understanding of the phylogeny of the silenus group was also confused by the notion that M. leonina and M. nemestrina were the only two Old World primates known to be susceptible to HIV-1 (human immunodeficiency virus type 1) infection due to the highly unusual retrotranspositional insertion of a cyclophilin A2 (CypA2) gene into the 3′ untranslated region of the TRIM5 locus (40, 41). On the basis of the orthologous sequence, we identified the same TRIM5-CypA2 fusion gene in M. silenus (fig. S25), suggesting that this retrotranspositional event evolved in the common ancestor of the Western silenus group (W) at least 2.17 Ma (Fig. 4A). In vitro infection experiments using peripheral blood mononuclear cells (PBMCs) confirmed that M. silenus can be infected with HIV-1 (Fig. 4D).
The success or otherwise of a given hybrid speciation depends upon whether the hybridization events give rise to an established, persistent, morphologically and ecologically distinct hybrid lineage (42). Today, the species of the fascicularis group are well segregated from their two parental groups by either ecogeographic barriers or behavioral differences (43). The extant members of the fascicularis group, however, display a distinctive mixture of their parental species’ characteristics as a result of the ancient hybridization. For example, the unique bluntly bilobed and narrow penile morphology in males, and the bright red sexual skin with little or no evident swelling in females, are approximately intermediate between the sinica and silenus groups (Fig. 5D) (23, 43). As these morphological traits are mostly associated with the reproductive system and often have a polygenic basis, it is reasonable to expect that genes related to these traits should also exhibit a mosaic pattern with respect to the putative parental lineages.
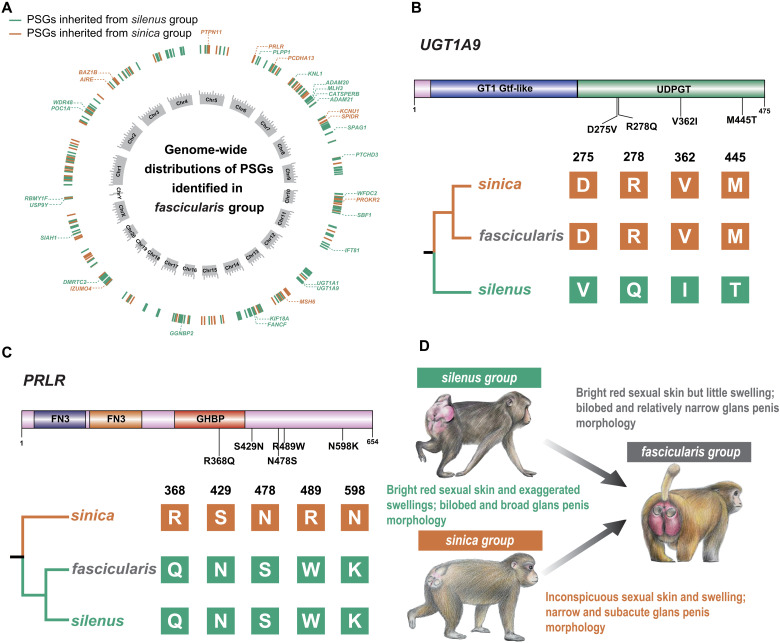
Fig. 5. The genetic basis underlying the mixture of phenotypes in the fascicularis group macaques.
(A) Genomic distribution of PSGs identified in the fascicularis group. Only PSGs associated with reproductive functions are depicted. (B and C) are two examples of reproduction-related PSGs with the largest number of fixed nonsynonymous mutations that were inherited from the sinica and silenus groups, respectively. The position of the mutation and predicted functional domain are both shown. (D) Diagram showing the three-pole morphocline in macaque (23, 24). Species of the fascicularis group generally exhibit a mixture of glans penis and sexual skin morphology from their parental species groups (silenus and sinica), although there is variation within the group. Examples of species are M. nemestrina (silenus group), M. mulatta (fascicularis group), and M. thibetana (sinica group). Macaque drawings by J. Shi.
To identify the genetic mechanisms underlying these mixed phenotypes, we used a recently developed method (44) to detect genes that have been subject to positive selection in the fascicularis group and each of its parental species. In addition, we applied three criteria to filter those positively selected genes (PSGs) for the purpose of minimizing the potential bias of the small sample size in each group (see Materials and Methods for details). We identified 216 PSGs in the fascicularis group that were inherited from the sinica lineage (table S14). Functional annotation identified 22 genes as being overrepresented in several gene regulatory networks related to reproductive function, such as male gamete generation [Gene Ontology (GO):0048232] and sexual reproduction (GO:0019953) (P < 0.001; table S15). Of these genes, seven (UGT1A9, ADAM20, WFDC2, FANCF, DMRTC2, SIAH1, and POC1A) exhibit sinica-derived nonsynonymous substitutions that have become fixed in known functional domain regions (Fig. 5B and fig. S23), whereas another two (WDR48 and PLPP1) have sinica-derived mutations in the 1-kb upstream region (table S14). Of special interest is a gene involved in flavonoid glucuronidation (UGT1A9); all four sinica-derived nonsynonymous mutations were found to be located in the UDPGT domain region (Fig. 5B). The UGT1A9 gene encodes UDP glucuronosyltransferase, which catalyzes the glucuronidation of endogenous estrogen hormones into water-soluble excretable metabolites (45). Because estrogen is crucial for the development of sexual swellings (46), it is certainly conceivable that the shared substitutions in this gene between fascicularis and sinica may have contributed to the reduced sexual swelling in females of both groups. In similar vein, the comparison involving fascicularis-silenus versus the sinica group identified 195 PSGs in the fascicularis group that were inherited from the silenus lineage (table S16). Functional annotation revealed 10 to be related to human reproductive phenotypes (HP0000134: female hypogonadism; HP:0000137: abnormality of the ovaries, ovarian disease, P < 0.005; table S17). Four of them (PRLR, PTPN11, KCNU1, and MSH6) harbor silenus-derived nonsynonymous mutations, fixed in functional domain regions (Fig. 5C and fig. S24), whereas three (IZUMO4, BAZ1B, and SPIDR) have silenus-derived mutations all occurring in 1-kb upstream regions (table S16). Together, these results provide additional genetic evidence to support the mosaic model of reproductive morphologies observed in the fascicularis group.
DISCUSSION
The origin of a new species from hybridization between two preexisting species is one of the most spectacular modes of speciation and has consequently attracted the attention of evolutionary biologists for decades (47, 48). Although traditionally considered to be more prevalent in plants, hybrid speciation has in recent years been recognized as being more common in animals than previously thought, including examples from both invertebrates and vertebrates (49). However, convincing evidence of hybrid speciation in animals remains scarce, and our understanding of the underlying genomic mechanisms is still quite limited. Here, our phylogenomic analyses using different data types and methods as well as morphological evidence, have provided consistent support for the hypothesis that the fascicularis group of macaques originated from an ancient hybridization between the sinica and silenus groups, thereby providing us with an unparalleled opportunity to investigate the factors driving the origin and maintenance of hybrid species in higher animals.
On the basis of the age estimates derived from the putatively neutral sequences of autosomal windows (Fig. 4A), our results suggest that the ancient hybrid formation of the fascicularis group occurred ~3.45 to 3.56 Ma, soon after the initial separation of the two parental lineages (proto-sinica and proto-silenus) ~3.86 Ma. The divergence time of the two parental lineages and subsequent hybridization coincided with the rapid glacio-eustatic fluctuations in the early-middle Pliocene (50). As a very dynamic geographical region in Asia, the land bridge (Isthmus of Kra) between the Malay peninsula and the Sunda region was repeatedly affected by glacio-eustatic fluctuations during the early-middle Pliocene. We therefore speculate that the early sea level highstand (when the sea level was above the edge of the continental shelf) may have led to the initial separation of proto-sinica and proto-silenus, while the subsequent lowering of the sea level facilitated the secondary contact required for hybridization. However, because no obvious postzygotic isolation mechanisms were observed among macaque species, the most insurmountable difficulties would be how hybrid integrity was maintained and reinforced after the initial hybridization. Prezygotic barriers, such as ecological divergence or/and geographic isolation, are thus likely to have been of greater importance in the establishment of hybrid taxa. We propose that geographic isolation may not be a necessary requirement for hybrid speciation because of the continual overlapping distribution between members of the fascicularis group of macaques (e.g., M. mulatta and M. fascicularis) and their parental group species (Fig. 1A). Nevertheless, the fascicularis group macaques are now well segregated from their two parental species by ecological barriers and/or behavioral differences (43, 51). In particular, the fascicularis group macaques show distinctive mixed sexual phenotypes, and the footprints of natural selection have been detected in these characteristics, suggesting that assortative mating behavior may have played an important role in the origin and maintenance of the fascicularis group as a hybrid species. In primates, primary and secondary sexual characteristics are often the target of male-female mate choice/sexual selection (52, 53). Such novel combinations of sexually selected traits in the fascicularis group may have led to the emergence of novel mate preferences, which could have promoted the establishment of a distinct lineage.
We further examine the genomic makeups from the origination of a new species as the consequence of hybridization and how the subsequent genomic changes in the hybrid lineage facilitated the establishment of reproductive barriers toward both parental species. We found that the sex chromosomes (X) and regions of low recombination display more conspicuous patterns of mosaicism. Generally, these regions are considered to be less permeable to introgression as they are less permissive of foreign genes due to lower hybrid fitness (54). However, species that have experienced rapid adaptive radiation provide just such an opportunity because these incompatible regions in the descendant lineages are likely to be less genetically divergent from each other, allowing them to hybridize and produce viable offspring (29). In similar vein, these less permeable regions could also serve as a strong barrier against further parental introgression in hybrids and hence would be expected to exhibit more mosaicism than other autosomal regions once hybridization occurred (55, 56).
Last, we have resolved several long-standing evolutionary conundrums during the rapid speciation of macaques, involving controversies surrounding phylogeny and complex ancestral hybridizations. For example, our analyses confirm that the stump-tailed macaque is a member of the sinica group with a low level of introgression from mulatta/fuscata, and that the lion-tailed macaque (M. silenus) and M. leonina are sister species, both of which can be infected by HIV. Considering the high species diversity of macaques and the strong lineage structure within some species, genome sequencing data from all macaque species and different subspecies will undoubtedly help to further elucidate the complex evolutionary history and biogeography of this genus. We are, however, confident that our conclusions regarding the hybrid speciation of the fascicularis group will not change with the acquisition of new sequence data.
In summary, our study reports the occurrence of an unusual ancient hybridization event in primates and illustrates how speciation through natural hybridization can arise via the reshuffling of standing genetic variation and how hybrid species may maintain their genetic integrity through the action of selection. Our study provides both a strategy and a pipeline of genome analyses to identify hybrid speciation, which should pave the way for the identification and exploration of further such events in the future.
MATERIALS AND METHODS
Sample information and ethics statement
A total of 12 macaque blood or DNA samples were obtained for this project. Ten were used for full-genome assembly, whereas samples from the other two males, from M. tonkeana and M. sylvanus, respectively, were used for whole-genome shotgun resequencing. These samples were collected from multiple sources in China and Germany. Detailed information is given in table S1. All samples were collected legally and in accordance with the policy of the Animal Care and Use Ethics of Kunming Institute of Zoology (approval ID: SMKX-20180701-01 and SMKX-2021-01-002), which conforms to the regulatory standards for the human care and treatment of animals in research.
Genome sequencing, assembly, and annotation
The 10 newly assembled macaque genomes were sequenced on different platforms due to the protracted process of collecting tissue samples (see table S1 for details). We used Supernova (version 2.0.0, 10X Genomics Inc., Pleasanton, CA, USA) with default parameters to assemble the reads generated from the stLFR and 10X Genomics Chromium reads platforms. For the Nanopore long reads, we first performed self-error correction for all Nanopore long reads using NextDenovo software (v2.4.0) and then assembled them into contigs using wtdbg-1.2.8 (57). The raw assemblies were further polished by Illumina short reads in Pilon v 1.22 (58) three times under the default settings. The completeness of the new macaque genomes was assessed by BUSCO (v3.0.2) (59) based on the mammal-specific set of 4104 single-copy orthologs (mammalia_odb9).
Repeat elements were predicted by RepeatMasker v4.0.6 (60). For protein-coding gene annotation, we first obtained protein sequences from five well-annotated mammalian species, namely, human (Homo sapiens, GCA_000001405.28), chimpanzee (Pan troglodytes, GCA_002880755.3), gorilla (Gorilla gorilla, GCA_900006655.3), orangutan (Pongo abelii, GCA_002880775.3), and mouse (Mus musculus, GCA_000001635.8). These protein sequences were mapped to each de novo genome using TBLASTN v2.2.26 (61) with an E value cutoff of 1 × 10−5. Proteins with multiple adjacent hits were connected to each other using genBlastA v1.0.4 (62). We filtered out those candidate loci with homologous block lengths shorter than 30% of the length of the query protein. For the ab initio prediction, we used Augustus v3.0.3 (63) with optimized parameters trained from 1000 randomly selected homologous genes. Last, we integrated all gene sets to form a comprehensive and nonredundant gene set using in-house Perl scripts.
Whole-genome alignments
We first performed pairwise whole-genome alignments against Chinese rhesus macaque (M. mulatta, rheMacS) using the LASTZ program (27) (v 1.04.03) under the following parameters: K = 4500, l = 3000, Y = 15,000, E = 150, H = 2000, O = 600, T = 2. The original alignments were then processed by the chainNet package (64) to generate the reciprocal best net alignment (default parameters, except for axtChain where we used “-minScore = 5,000 -linearGap = medium”). Subsequently, we used the “maf-swap” (65) command (LAST software) to sort the alignment results and obtain the optimal pairwise synteny blocks between genomes. Last, we used MULTIZ (v 11.2) (28) to merge all pairwise alignments into multiple genome alignments using rheMacS as the reference.
Phylogenetic analysis
Sliding window tree and signal of conflict
On the basis of the multiple genome alignment, we used scripts provided in (66) to partition the alignment (20 autosomes and one X chromosome) into nonoverlapping windows of 50 kb without considering the protein-coding content. Meanwhile, we excluded windows that contained more than 10% gaps or hard-masked repeat sequences. For each window of the alignment, an ML tree was constructed using RAxML v8.1.15 (67) with the GTRGAMMA model and rapid bootstrapping for 100 replicates and specifying P. hamadryas as the outgroup. The number of alternative tree topologies and their relative frequency were categorized by PhyBin v0.3 program (68). Conflicts between the gene trees and their frequencies were summarized using the CONSENSE program in PHYLIP v3.697 package (69). The coalescent species-tree was estimated from the aforementioned sliding window trees by two summary coalescent-based methods, ASTRAL (70) and STAR (71). Because the major disagreement among all topologies concerned the relative placement of the fascicularis group (Fig. 1, B and C), for the sake of simplicity, we pruned the trees so as to include only five ingroup species: M. sylvanus, M. thibetana, M. silenus, M. nigra, and M. mulatta. These five species represented the sylvanus group, sinica group, silenus group (E), silenus group (W), and fascicularis group, respectively, and were selected on the basis of their longest scaffold N50 size. Similarly, we used PhyBin v0.3 (68) to estimate the categories of alternative topologies and their relative frequencies. We also evaluated smaller (20 kb) and larger (100 kb) window sizes, but this did not change our results materially. We therefore used the 50-kb block size for all subsequent analyses.
Mitochondrial genome tree
The mitochondrial genome sequence of each species was obtained from Illumina short reads using NOVOplasty 2.4 (72). K-mer was set to 33, and the mitogenome of M. sylvanus (AJ309865) downloaded from the National Center for Biotechnology Information (NCBI) database was used as a starting reference. The reliability of mitochondrial contig assemblies was further validated via BLAST searches against the reference mitogenome (AJ309865). The D-loop and all transfer RNA (tRNA) genes were not used owing to their high mutation rate and high rate of loss. Protein-coding genes were translated into amino acid sequences to ensure open reading frames and to avoid NUMTs (nuclear mitochondrial DNA segments). An additional 18 macaque mitochondrial genome sequences from NCBI were added to expand our datasets and to further validate our sample identification. The aligned genomes were partitioned into protein-coding genes and noncoding fragments, and the protein-coding genes were further partitioned into first, second, and third codon positions. PartitionFinder v2.1.6 (73) was used to evaluate the best partitioning scheme under the Bayesian information criterion. The ML analyses were carried out using RAxML v8.1.15 with 1000 bootstrap replications under the best partition scheme and the GTRGAMMA model.
Y chromosome phylogeny
We confined our analyses to five Y-linked genes (TSPY10, SRY, ZFY, USP9Y, and RPS4Y1) owing to the substantial technical challenges presented by the sequence alignment of repetitive sequence regions of the Y chromosome. Seven of our genome assemblies were from males. To expand our dataset, we generated whole-genome shotgun sequences of two male samples from M. tonkeana and M. sylvanus, and further downloaded one male sample from M. nemestrina (SRR5947292). The Illumina short reads of each species were mapped to the Chinese rhesus macaque genome (rheMacS) using BWA-MEM v0.7.12 (74) with the default settings. After obtaining the bam files, we used ANGSD (75) to obtain the consensus sequence of these genes with the following filtering parameters: -b bamfiles.txt -minQ 20 -minMapQ 20 -remove_bads -uniqueOnly -rf region.txt -dohaplocall 1 -doCounts 1. The concatenation of the abovementioned five genes comprised a total of 480,333 base pairs (bp). Again, we performed the phylogenetic analyses using RAxML v.8.1.15 under the GTRGAMMA model with 1000 bootstrap replications.
Phylogenetic signal relative to the local recombination rate
No recombination map is currently available for the reference genome of the Chinese rhesus macaque (rheMacS). To construct this, we used the liftover tool (76) to convert the latest fine-scale linkage map of Indian rhesus macaque (rheMac8) (77). First, the genome of rheMac8 was aligned to that of rheMacS using the LASTZ program v1.04.03 (27) with the same parameters mentioned for the genome alignments. After obtaining the chain file, which records the links of the reciprocal best orthologous regions of the genome, we transferred the coordinates of the recombination map of rheMac8 to rheMacS coordinates using the program liftover (76). Only the successfully derived liftover positions (~96.3%) were used in downstream analyses.
Hybridization and introgression analyses
Although ILS certainly underpins some aspects of phylogenetic discordance within and between clades, it is unlikely to be the only explanation for the notable differences we observed across all window trees. Therefore, we used several different methods to test for hybridization and introgression events in the presence of ILS. We first used the InferNetwork_MPL program in PhyloNet (37) to reconstruct optimal phylogenetic networks between all macaque species. Owing to the extensive computational requirements, we performed this analysis on 2000 topologies that were randomly selected and had a mean bootstrap support threshold of more than 80%. Three independent analyses were performed assuming reticulation scenarios ranging from 1 to 3, and each analysis executed 100 runs. The branch lengths and inheritance probabilities of the returned species networks were optimized under full-likelihood by specifying the “-po” option [full parameters: InferNetwork_MPL (all) h –b 75 -n 5 -di -po -x 100 -pl 10].
HyDe is another powerful and computationally efficient method to detect the level of interspecies hybridization based on site pattern frequencies of single-nucleotide polymorphism (SNP) data (35). To acquire high-quality SNPs for our analyses, we used the “BWA-GATK-SAMtools” pipeline (78) and further filtered the SNPs using the following criteria: (i) removal of high probability miscalls around 6 bp from predicted indels, (ii) removal of sites with consensus quality of <40, (iii) removal of sites with triallelic alleles and indels, and (iv) removal of sites present in <90% of individuals. We first assessed whether ancient hybridization events had occurred between species groups (sylvanus group, silenus group, sinica group, and fascicularis group) using the “run_hyde.py” script. P. hamadryas was used as an outgroup, and sites with missing/ambiguous bases were ignored (--ignore_amb_sites). Next, an analysis at the individual level was performed using the “individual_hyde.py” script to detect hybridization in individuals within species groups that had significant levels of admixture. Last, bootstrap resampling (500 replicates in “bootstrap_hyde.py” script) was performed on individuals within the hybrid group to obtain a distribution of gamma values to assess heterogeneity in levels of gene flow. The tract size of two parental ancestry was inferred by LOTER (36). The genomic distributions of each ancestry were calculated using custom R and Perl scripts.
Because HyDe is a powerful method with which to identify hybrids when the parental contribution is symmetrical, we further applied D-statistic analysis (also known as the ABBA-BABA test) (79) to explore broader introgression patterns between lineages and species. We performed these analyses in Dsuite package (80), which can estimate the D statistics across all possible triplets at one time based on the user-specified topology and can also assign gene flow to specific, possibly internal, branches. Our former steps suggested that the fascicularis group was of hybrid origin; thus, two bifurcating topologies could be used: one supported the sister relationship between the fascicularis and sinica groups (fig. S5A), whereas the other supported the sister relationship between the fascicularis and silenus groups (fig. S5B). We used the program Dtrios in Dsuite to calculate the D statistics of all possible combinations of species trios as well as species group trios. P. hamadryas was used as the outgroup (O) in all analyses. The resulting P values were further adjusted by Benjamini and Hochberg (BH) correction for multiple testing bias via the p.adjust function in R. Using a significance threshold (α = 0.05) that was BH-corrected for a total of 221 comparisons (the maximum number of trios involving a given pair), z scores of >2.1 were considered significant. We conservatively display only z scores of >3 as a significant signal of introgression. Results from Dtrios were further processed using the Fbranch function to generate a matrix of z scores, and a heatmap of the matrix was visualized using the dtools.py script, which is provided within the Dsuite package.
Following the logic of Zhang et al. (33), we computed the D statistics in sliding windows to test phylogenetic hypotheses, as the genomic region least affected by introgression should reflect the true relationship. For example, if the phylogenetic relationship of T1 were true, then the genomic region least affected by gene flow should be dominated by T1 topology. In this scenario, we set P1 as the sinica group, P2 as the fascicularis group, and P3 as the silenus group. The sylvanus group was treated as the outgroup (O) because it was always located outside the other three Asian species groups (Fig. 1B). The same was true for T2. We used the Python script ABBABABAwindows.py from (32) to compute the D statistics with window size set to 50 kb, and at least 100 biallelic SNPs were allowed per window (-w 50,000 -m 100). Windows with absolute D-statistic values close to zero (e.g., low 1% windows) were regarded as the genomic regions least affected by gene flow, whereas the highest D-statistic values (e.g., top 1% windows) were regarded as those most affected by gene flow.
Hybridization and divergence time estimates
To minimize the negative impact of post-speciation gene flow upon divergence time estimation, we used the genomic window sequences that supported the most common (fig. S5A) and second most common (fig. S5B) trees to infer the timing of species divergence and hybridization because they only differed in the placement of the fascicularis group. According to the annotation of the rheMacS assembly, we filtered any genomic window that contained exonic sequences, and the 10-kb regions flanking them on either side, to reduce the potential bias introduced by selection at linked sites (e.g., background selection and hitchhiking) into our analysis (81). Furthermore, to ensure that each genomic window was phylogenetically independent, we only retained the windows that had a distance of at least 100 kb from each other. After applying these filtering strategies, a total of 1663 windows were obtained on the autosomes and 103 on the X chromosome, respectively. For each window sequence, we used the MCMCTREE program in PAML v.4.9e package (82) to estimate the relative divergence time for the tree imputed from that window. Two well-justified and widely used fossil record–based calibration points were used to calibrate evolutionary time (16, 17): (i) the split between African (M. sylvanus) and Asian macaques: 4.5 to 6.5 Ma and (ii) the split between Macacina and Papionina: 5.8 to 8.0 Ma. The prior of the overall substitution rate was estimated using the BASEML program in PAML (82), assuming a mean split of the most recent common ancestor of Macacina and Papionina at 7 Ma, based on the GTR + Γ substitution model (rgene gamma = 1, 7.27, 1). The gamma-Dirichlet prior for the rate-drift parameter (sigma2 gamma) was set to G (1, 4.5, 1). Fossil record constraints were scaled to units of 100 Ma, and the constraints of minimum and maximum bounds were soft, with the default 2.5% probability that allowed bounds to be violated. A Markov chain Monte Carlo (MCMC) chain was first run for 1,000,000 generations as burn-in, then sampled every 500 generations, until 10,000 samples had been collected. Node ages for each topology were obtained using ape v5.1 package (83) in R.
Identification of PSGs under hybridization
To identify candidate genes that may have fulfilled key functions in hybrids, we used a recently developed method (44) to identify PSGs in the fascicularis group and each of its parental groups (sinica and silenus groups). Under the null hypothesis of a neutral model, polymorphism within a lineage and divergence observed between two lineages at homologous loci would be highly correlated, whereas positive selection would reduce levels of polymorphism relative to that associated with divergence. We performed two independent analyses. First, we grouped fascicularis and sinica together (group 1) and compared them to the silenus group (group 2) to identify the PSGs that originated from the sinica group. Similarly, to identify PSGs originating from the silenus group, we grouped fascicularis and silenus together (group 1) and compared them to the sinica group (group 2). Both coding regions and 1-kb upstream sequences were considered in our analyses because these mutations were deemed more likely to have a major effect on gene function. For each gene, we counted the number of polymorphic sites (SNPs) in group 1 (supposition A) and the number of fixed differences [the SNPs with population genetic diffierentiation (FST) value > 0.95] between group 1 and group 2 (supposition B) based on the filtered SNP file for all species. Then, a test was performed by comparing the ratio of A/B to the genome-wide average A/B, which was calculated as the sum of A and B values across all genes analyzed. A Pearson’s chi-square test on the 2 × 2 contingency table was used to obtain the significance score that rejected the null hypothesis A(gene)/B(gene) = A(genome-wide)/B(genome-wide) (44). We annotated all SNP variants using SnpEff software (version 5.0c) (84) and recorded the number of fixed nonsynonymous mutations and fixed 1-kb upstream mutations in each gene using in-house Perl scripts. PSGs were further filtered using the following criteria: (i) the Yates’ corrected P value should be significant (<0.01) in the hybrid group and one of the parental groups, but not significant (>0.05) in another parental group; (ii) the number of fixed nonsynonymous mutations together with 1-kb upstream mutations was within the top 2.5% of gene; and (iii) the topology of the gene tree should be consistent with the relationship being tested (built with the following parameters: iqtree -s SNP_data.fasta -st DNA -nt AUTO -ntmax 5 -mem 5G -bb 1000 -bnni -o Pham -m MFP+ASC -quiet -redo). Functional enrichment analyses among the PSGs were performed using the web-based toolkit WebGestalt (85). The top 40 most significant function categories from the overrepresentation analysis were reported (tables S15 and S17).
HIV infection experiment
Blood samples from two lion-tailed macaques were provided by G. Zoo (female Msil-001, male Msil-002), China, and used to perform the HIV infection experiment. As a positive control, two northern pig-tailed macaques (NPM-15216 and NPM-17216) from Kunming Primate Research Center, Kunming Institute of Zoology, Chinese Academy of Science were used.
Whole blood was collected into EDTA vacutainer tubes by venipuncture, and PBMCs were separated by Ficoll (GE Healthcare) gradient centrifugation. PBMCs were activated with concanavalin A (Sigma-Aldrich, USA) and interleukin-2 (IL-2) (5 U/ml). After 72 hours of activation, the PBMCs were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and IL-2 (10 U/ml). Freshly activated PBMCs were infected with HIV-1NL4-3 particles at a multiplicity of infection (MOI) of 0.05. At 4 hours post-infection (hpi), cells were washed four times with phosphate-buffered saline and resuspended in fresh RPMI–10% FBS supplemented with IL-2. Supernatants were collected after 48 and 72 hpi. The levels of viral RNA in supernatant were quantified by a real-time polymerase chain reaction (PCR) method based on amplification of an HIV-1NL4-3–derived Gag coding sequence described previously with slight modification (41). For each sample, total cell-associated RNA was extracted by RNAiso Plus (TaKaRa) reagent and dissolved in 50 μl of diethyl pyrocarbonate H2O. Then, the cell-associated viral RNA was determined using the RNA-Direct Real-time PCR Master Mix Kit (Toyobo). The primers were 6F (5′-CATGTTTTCAGCATTATCAGAAGGA-3′) and 84R (5′-TGCTTGATGTCCCCCCACT-3′), and the probe was 5′-FAM-CCACCCCACAAGA-TTTAAACACCATGCTAA-TAMRA-3′. PCRs were performed on the ABI Vii7 Sequence Detection System under conditions of 1 cycle of 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. HIV-1–specific p24 antigen was measured by enzyme-linked immunosorbent assay (ELISA) using the HIV-1 p24 ELISA Kit (ZeptoMetrix, USA).
The M. silenus TRIM5 and TRIMCyp genes were amplified by TRIM5/Cyp-F: 5′-ATGGCTTCTGGAATCCTGCTTAATGTA-3′, TRIM5-R: 5′-TCAAGAGCTTGGTGAGCACAGAGTCA-3′, and TRIMCyp-R: 5′-TTATTCGAGTTGTCCACAGTCAGCA-3′. The PCR conditions were 94°C for a 3-min hot start, followed by 30 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 120 s, and a final extension of 10 min at 72°C. The PCR products were analyzed on a 1% agarose gel with ethidium bromide, purified using a DNA gel extraction kit (Generay Biotech, Shanghai, China), cloned into pMD19-T simple vector (Takara, Dalian, China), and finally sequenced.
Acknowledgments
We thank W. Wang, X. Ren, and X. Li for their early assistance with this project.
Funding: This work was funded by the National Key Research and Development Program of China (nos. 2021YFF0702700 and 2022YFF0710901), the National Natural Science Foundation of China (no. 31822048 to D.-D.W. and no. 32270500 to B.-L.Z.), Yunnan Applied Basic Research Projects (no. 2019FI010 to D.-D.W.), CAS Light of West China Program (xbzg-zdsys-202213) and the Animal Branch of the Germplasm Bank of Wild Species of Chinese Academy of Science (the Large Research Infrastructure Funding). This work was also partially supported by a Villum Investigator Grant to G.Z. (no. 25900) and the National Natural Science Foundation of China to Y.-T.Z. (U1802284). T.M.-B. was supported by funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (no. 864203), BFU2017-86471-P (MINECO/FEDER, UE), and Howard Hughes International Early Career.
Author contributions: Conceptualization and project administration: D.-D.W., G.Z., and Y.-T.Z. Data analysis and visualization: B.-L.Z., Z.W., S.W., Y.S., Y.D., L.Z., J.C., and L.W. HIV infection experiments: W.P., M.-T.L., and W.-Q.H. Sample collection and provider: W.C., M.-M.Y., Y.W., H.F.-B., S.M., H.M., F.W., L.K., T.M.-B., and C.R. Writing—original draft: B.-L.Z. and D.-D.W. Writing—review and editing: B.-L.Z., D.-D.W., G.Z., Y.-T.Z., J.L., D.N.C., M.H.S., Z.L., M.L., X.-G.Q., C.R., T.M.-B., H.M., and Z.W.
Competing interests: L.K. is an employee of Illumina Inc. The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw sequencing data from this project were deposited in the Genome Warehouse (GWH) database under project accession number PRJCA007326.
Supplementary Materials
This PDF file includes:
Figs. S1 to S27
Tables S1 to S17
REFERENCES AND NOTES
- 1.O. Seehausen, Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207 (2004). [DOI] [PubMed] [Google Scholar]
- 2.R. Abbott, D. Albach, S. Ansell, J. W. Arntzen, S. J. E. Baird, N. Bierne, J. Boughman, A. Brelsford, C. A. Buerkle, R. Buggs, R. K. Butlin, U. Dieckmann, F. Eroukhmanoff, A. Grill, S. H. Cahan, J. S. Hermansen, G. Hewitt, A. G. Hudson, C. Jiggins, J. Jones, B. Keller, T. Marczewski, J. Mallet, P. Martinez-Rodriguez, M. Most, S. Mullen, R. Nichols, A. W. Nolte, C. Parisod, K. Pfennig, A. M. Rice, M. G. Ritchie, B. Seifert, C. M. Smadja, R. Stelkens, J. M. Szymura, R. Vainola, J. B. W. Wolf, D. Zinner, Hybridization and speciation. J. Evol. Biol. 26, 229–246 (2013). [DOI] [PubMed] [Google Scholar]
- 3.L. H. Rieseberg, J. H. Willis, Plant speciation. Science 317, 910–914 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M. Schumer, G. G. Rosenthal, P. Andolfatto, How common is homoploid hybrid speciation? Evolution 68, 1553–1560 (2014). [DOI] [PubMed] [Google Scholar]
- 5.M. C. Melo, C. Salazar, C. D. Jiggins, M. Linares, Assortative mating preferences among hybrids offers a route to hybrid speciation. Evolution 63, 1660–1665 (2009). [DOI] [PubMed] [Google Scholar]
- 6.B. L. Gross, L. H. Rieseberg, The ecological genetics of homoploid hybrid speciation. J. Hered. 96, 241–252 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J. Mavarez, C. A. Salazar, E. Bermingham, C. Salcedo, C. D. Jiggins, M. Linares, Speciation by hybridization in Heliconius butterflies. Nature 441, 868–871 (2006). [DOI] [PubMed] [Google Scholar]
- 8.B. M. vonHoldt, J. A. Cahill, Z. Fan, I. Gronau, J. Robinson, J. P. Pollinger, B. Shapiro, J. Wall, R. K. Wayne, Whole-genome sequence analysis shows that two endemic species of North American wolf are admixtures of the coyote and gray wolf. Sci. Adv. 2, e1501714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.S. Lamichhaney, F. Han, M. T. Webster, L. Andersson, B. R. Grant, P. R. Grant, Rapid hybrid speciation in Darwin's finches. Science 359, 224–228 (2018). [DOI] [PubMed] [Google Scholar]
- 10.J. Rogers, M. Raveendran, R. A. Harris, T. Mailund, K. Leppala, G. Athanasiadis, M. H. Schierup, J. Cheng, K. Munch, J. A. Walker, M. K. Konkel, V. Jordan, C. J. Steely, T. O. Beckstrom, C. Bergey, A. Burrell, D. Schrempf, A. Noll, M. Kothe, G. H. Kopp, Y. Liu, S. Murali, K. Billis, F. J. Martin, M. Muffato, L. Cox, J. Else, T. Disotell, D. M. Muzny, J. Phillips-Conroy, B. Aken, E. E. Eichler, T. Marques-Bonet, C. Kosiol, M. A. Batzer, M. W. Hahn, J. Tung, D. Zinner, C. Roos, C. J. Jolly, R. A. Gibbs, K. C. Worley; Baboon Genome Analysis Consortium , The comparative genomics and complex population history of Papio baboons. Sci. Adv. 5, eaau6947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.T. Zou, W. Kuang, T. Yin, L. Frantz, C. Zhang, J. Liu, H. Wu, L. Yu, Uncovering the enigmatic evolution of bears in greater depth: The hybrid origin of the Asiatic black bear. Proc. Natl. Acad. Sci. U.S.A. 119, e2120307119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.P. A. Hohenlohe, L. Y. Rutledge, L. P. Waits, K. R. Andrews, J. R. Adams, J. W. Hinton, R. M. Nowak, B. R. Patterson, A. P. Wydeven, P. A. Wilson, B. N. White, Comment on “Whole-genome sequence analysis shows two endemic species of North American wolf are admixtures of the coyote and gray wolf”. Sci. Adv. 3, e1602250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.G. G. Rosenthal, M. Schumer, P. Andolfatto, How the manakin got its crown: A novel trait that is unlikely to cause speciation. Proc. Natl. Acad. Sci. U.S.A. 115, E4144–E4145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.C. Scornavacca, N. Galtier, Incomplete lineage sorting in mammalian phylogenomics. Syst. Biol. 66, 112–120 (2017). [DOI] [PubMed] [Google Scholar]
- 15.P. Fan, Y. Liu, Z. Zhang, C. Zhao, C. Li, W. Liu, Z. Liu, M. Li, Phylogenetic position of the white-cheeked macaque (Macaca leucogenys), a newly described primate from southeastern Tibet. Mol. Phylogenet. Evol. 107, 80–89 (2017). [DOI] [PubMed] [Google Scholar]
- 16.E. Delson, Fossil macaques, phyletic relationships and a scenario of deployment, in The Macaques: Studies in Ecology, Behavior, and Evolution, D. G. Lindburg, Ed. (van Nostrand–Reinhold, 1980), pp. 10–30. [Google Scholar]
- 17.C. Roos, M. Kothe, D. M. Alba, E. Delson, D. Zinner, The radiation of macaques out of Africa: Evidence from mitogenome divergence times and the fossil record. J. Hum. Evol. 133, 114–132 (2019). [DOI] [PubMed] [Google Scholar]
- 18.M. B. Gardner, P. A. Luciw, Macaque models of human infectious disease. ILAR J. 49, 220–255 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.E. L. Bynum, D. Z. Bynum, J. Supriatna, Confirmation and location of the hybrid zone between wild populations of Macaca tonkeana and Macaca hecki in central Sulawesi, Indonesia. Am. J. Primatol. 43, 181–209 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Y. Hamada, N. Urasopon, I. Hadi, S. Malaivijitnond, Body size and proportions and pelage color of free-ranging Macaca mulatta from a zone of hybridization in Northeastern Thailand. Int. J. Primatol. 27, 497–513 (2006). [Google Scholar]
- 21.Y. Song, C. Jiang, K.-H. Li, J. Li, H. Qiu, M. Price, Z.-X. Fan, J. Li, Genome-wide analysis reveals signatures of complex introgressive gene flow in macaques (genus Macaca). Zool. Res. 42, 433–449 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A. J. Tosi, J. C. Morales, D. J. Melnick, Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution 57, 1419–1435 (2003). [DOI] [PubMed] [Google Scholar]
- 23.J. E. Fa, The genus Macaca: A review of taxonomy and evolution. Mamm. Rev. 19, 45–81 (1989). [Google Scholar]
- 24.J. Fooden, Provisional classification and key to living species of macaques (primates: Macaca). Folia Primatol. (Basel) 25, 225–236 (1976). [DOI] [PubMed] [Google Scholar]
- 25.Y. He, X. Luo, B. Zhou, T. Hu, X. Meng, P. A. Audano, Z. N. Kronenberg, E. E. Eichler, J. Jin, Y. Guo, Y. Yang, X. Qi, B. Su, Long-read assembly of the Chinese rhesus macaque genome and identification of ape-specific structural variants. Nat. Commun. 10, 4233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Y. Shao, L. Zhou, F. Li, L. Zhao, B.-L. Zhang, F. Shao, J.-W. Chen, C.-Y. Chen, X.-P. Bi, X.-L. Zhuang, H.-L. Zhu, J. Hu, Z. Sun, X. Li, D. Wang, I. Rivas-González, S. Wang, Y.-M. Wang, W. Chen, G. Li, H.-M. Lu, Y. Liu, L. Kuderna, K. Farh, P.-F. Fan, L. Yu, M. Li, Z.-J. Liu, G. P. Tiley, A. D. Yoder, C. Roos, T. Hayakawa, T. Marques-Bonet, J. Rogers, D. N. Cooper, H. Li, M. H. Schierup, Y.-G. Yao, Y.-P. Zhang, W. Wang, X.-G. Qi, G. Zhang, D.-D. Wu, Phylogenomic analyses provide insights into primate genomic and phenotypic evolution. Science, (2021). [DOI] [PubMed] [Google Scholar]
- 27.R. S. Harris, “Improved pairwise alignment of genomic DNA,” thesis, The Pennsylvania State University (2007). [Google Scholar]
- 28.M. Blanchette, W. J. Kent, C. Riemer, L. Elnitski, A. F. Smit, K. M. Roskin, R. Baertsch, K. Rosenbloom, H. Clawson, E. D. Green, D. Haussler, W. Miller, Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14, 708–715 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.N. B. Edelman, P. B. Frandsen, M. Miyagi, B. Clavijo, J. Davey, R. B. Dikow, G. Garcia-Accinelli, S. M. Van Belleghem, N. Patterson, D. E. Neafsey, R. Challis, S. Kumar, G. R. P. Moreira, C. Salazar, M. Chouteau, B. A. Counterman, R. Papa, M. Blaxter, R. D. Reed, K. K. Dasmahapatra, M. Kronforst, M. Joron, C. D. Jiggins, W. O. McMillan, F. Di Palma, A. J. Blumberg, J. Wakeley, D. Jaffe, J. Mallet, Genomic architecture and introgression shape a butterfly radiation. Science 366, 594–599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.B. Charlesworth, J. A. Coyne, N. H. Barton, The relative rates of evolution of Sex chromosomes and autosomes. Am. Nat. 130, 113–146 (1987). [Google Scholar]
- 31.M. C. Fontaine, J. B. Pease, A. Steele, R. M. Waterhouse, D. E. Neafsey, I. V. Sharakhov, X. F. Jiang, A. B. Hall, F. Catteruccia, E. Kakani, S. N. Mitchell, Y. C. Wu, H. A. Smith, R. R. Love, M. K. Lawniczak, M. A. Slotman, S. J. Emrich, M. W. Hahn, N. J. Besansky, Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347, 1258524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. H. Martin, J. W. Davey, C. D. Jiggins, Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 32, 244–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D. Zhang, F. E. Rheindt, H. She, Y. Cheng, G. Song, C. Jia, Y. Qu, P. Alstrom, F. Lei, Most genomic loci misrepresent the phylogeny of an avian radiation because of ancient gene flow. Syst. Biol. 70, 961–975 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. W. Nachman, B. A. Payseur, Recombination rate variation and speciation: Theoretical predictions and empirical results from rabbits and mice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 409–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.P. D. Blischak, J. Chifman, A. D. Wolfe, L. S. Kubatko, HyDe: A Python package for genome-scale hybridization detection. Syst. Biol. 67, 821–829 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.T. Dias-Alves, J. Mairal, M. G. B. Blum, Loter: A software package to infer local ancestry for a wide range of species. Mol. Biol. Evol. 35, 2318–2326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Y. Yu, L. Nakhleh, A maximum pseudo-likelihood approach for phylogenetic networks. BMC Genomics 16 (suppl. 10), S10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Z. Fan, A. Zhou, N. Osada, J. Yu, J. Jiang, P. Li, L. Du, L. Niu, J. Deng, H. Xu, J. Xing, B. Yue, J. Li, Ancient hybridization and admixture in macaques (genus Macaca) inferred from whole genome sequences. Mol. Phylogenet. Evol. 127, 376–386 (2018). [DOI] [PubMed] [Google Scholar]
- 39.C. Abegg, B. Thierry, Macaque evolution and dispersal in insular South-East Asia. Biol. J. Linn. Soc. 75, 555–576 (2002). [Google Scholar]
- 40.C. H. Liao, Y. Q. Kuang, H. L. Liu, Y. T. Zheng, B. Su, A novel fusion gene, TRIM5-Cyclophilin A in the pig-tailed macaque determines its susceptibility to HIV-1 infection. AIDS 21 (Suppl 8), S19–S26 (2007). [DOI] [PubMed] [Google Scholar]
- 41.W. Pang, G.-H. Zhang, J. Jiang, H.-Y. Zheng, L.-T. Zhang, X.-L. Zhang, J.-H. Song, M.-X. Zhang, J.-W. Zhu, A.-H. Lei, R.-R. Tian, X.-M. Liu, L. Zhang, G. Gao, L. Su, Y.-T. Zheng, HIV-1 can infect northern pig-tailed macaques (Macaca leonina) and form viral reservoirs in vivo. Sci. Bull. (Beijing) 62, 1315–1324 (2017). [DOI] [PubMed] [Google Scholar]
- 42.G. N. Feliner, I. Alvarez, J. Fuertes-Aguilar, M. Heuertz, I. Marques, F. Moharrek, R. Pineiro, R. Riina, J. A. Rossello, P. S. Soltis, I. Villa-Machio, Is homoploid hybrid speciation that rare? An empiricist’s view. Heredity (Edinb.) 118, 513–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.J. Fooden, Comparative review of fascicularis-group species of macaques (primates: Macaca). Fieldiana Zool. 2006, 1–43 (2006). [Google Scholar]
- 44.Z. Wang, Y. Jiang, H. Bi, Z. Lu, Y. Ma, X. Yang, N. Chen, B. Tian, B. Liu, X. Mao, T. Ma, S. P. DiFazio, Q. Hu, R. J. Abbott, J. Q. Liu, Hybrid speciation via inheritance of alternate alleles of parental isolating genes. Mol. Plant 14, 208–222 (2021). [DOI] [PubMed] [Google Scholar]
- 45.J. Lepine, O. Bernard, M. Plante, B. Tetu, G. Pelletier, F. Labrie, A. Belanger, C. Guillemette, Specificity and regioselectivity of the conjugation of estradiol, estrone, and their catecholestrogen and methoxyestrogen metabolites by human uridine diphospho-glucuronosyltransferases expressed in endometrium. J. Clin. Endocrinol. Metab. 89, 5222–5232 (2004). [DOI] [PubMed] [Google Scholar]
- 46.L. R. Gesquiere, E. O. Wango, S. C. Alberts, J. Altmann, Mechanisms of sexual selection: Sexual swellings and estrogen concentrations as fertility indicators and cues for male consort decisions in wild baboons. Horm. Behav. 51, 114–125 (2007). [DOI] [PubMed] [Google Scholar]
- 47.R. J. Abbott, M. J. Hegarty, S. J. Hiscock, A. C. Brennan, Homoploid hybrid speciation in action. Taxon 59, 1375–1386 (2010). [Google Scholar]
- 48.J. Mallet, Hybrid speciation. Nature 446, 279–283 (2007). [DOI] [PubMed] [Google Scholar]
- 49.J. Mavarez, M. Linares, Homoploid hybrid speciation in animals. Mol. Ecol. 17, 4181–4185 (2008). [DOI] [PubMed] [Google Scholar]
- 50.D. S. Woodruff, Neogene marine transgressions, palaeogeography and biogeographic transitions on the Thai–Malay Peninsula. J. Biogeogr. 30, 551–567 (2003). [Google Scholar]
- 51.J. Fooden, Ecogeographic segregation of macaque species. Primates 23, 574–579 (1982). [Google Scholar]
- 52.T. Caro, K. Brockelsby, A. Ferrari, M. Koneru, K. Ono, E. Touche, T. Stankowich, The evolution of primate coloration revisited. Behav. Ecol. 32, 555–567 (2021). [Google Scholar]
- 53.C. Dubuc, L. J. N. Brent, A. K. Accamando, M. S. Gerald, A. MacLarnon, S. Semple, M. Heistermann, A. Engelhardt, Sexual skin color contains information about the timing of the fertile phase in free-ranging Macaca mulatta. Int. J. Primatol. 30, 777–789 (2009). [Google Scholar]
- 54.O. Seehausen, R. K. Butlin, I. Keller, C. E. Wagner, J. W. Boughman, P. A. Hohenlohe, C. L. Peichel, G. P. Saetre; C. Bank, A. Brannstrom, A. Brelsford, C. S. Clarkson, F. Eroukhmanoff, J. L. Feder, M. C. Fischer, A. D. Foote, P. Franchini, C. D. Jiggins, F. C. Jones, A. K. Lindholm, K. Lucek, M. E. Maan, D. A. Marques, S. H. Martin, B. Matthews, J. I. Meier, M. Most, M. W. Nachman, E. Nonaka, D. J. Rennison, J. Schwarzer, E. T. Watson, A. M. Westram, A. Widmer, Genomics and the origin of species. Nat. Rev. Genet. 15, 176–192 (2014). [DOI] [PubMed] [Google Scholar]
- 55.T. O. Elgvin, C. N. Trier, O. K. Torresen, I. J. Hagen, S. Lien, A. J. Nederbragt, M. Ravinet, H. Jensen, G. P. Saetre, The genomic mosaicism of hybrid speciation. Sci. Adv. 3, e1602996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.K. Kunte, C. Shea, M. L. Aardema, J. M. Scriber, T. E. Juenger, L. E. Gilbert, M. R. Kronforst, Sex chromosome mosaicism and hybrid speciation among tiger swallowtail butterflies. PLOS Genet. 7, e1002274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. Ruan, H. Li, Fast and accurate long-read assembly with wtdbg2. Nat. Methods 17, 155–158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.B. J. Walker, T. Abeel, T. Shea, M. Priest, A. Abouelliel, S. Sakthikumar, C. A. Cuomo, Q. Zeng, J. Wortman, S. K. Young, A. M. Earl, Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS ONE 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.F. A. Simao, R. M. Waterhouse, P. Ioannidis, E. V. Kriventseva, E. M. Zdobnov, BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 60.G. Benson, Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.S. F. Altschul, W. Gish, W. Miller, E. W. Myers, D. J. Lipman, Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 62.R. She, J. S.-C. Chu, K. Wang, J. Pei, N. Chen, genBlastA: Enabling BLAST to identify homologous gene sequences. Genome Res. 19, 143–149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O. Keller, M. Kollmar, M. Stanke, S. Waack, A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27, 757–763 (2011). [DOI] [PubMed] [Google Scholar]
- 64.W. J. Kent, R. Baertsch, A. Hinrichs, W. Miller, D. Haussler, Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. U.S.A. 100, 11484–11489 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.S. M. Kielbasa, R. Wan, K. Sato, P. Horton, M. C. Frith, Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.L. Chen, Q. Qin, Y. Jiang, K. Wang, Z. Lin, Z. Li, F. Bibi, Y. Yang, J. Wang, W. Nie, W. Su, G. Liu, Q. Li, W. Fu, X. Pan, C. Liu, J. Yang, C. Zhang, Y. Yin, Y. Wang, Y. Zhao, C. Zhang, Z. Wang, Y. Qin, W. Liu, B. Wang, Y. Ren, R. Zhang, Y. Zeng, R. R. da Fonseca, B. Wei, R. Li, W. Wan, R. Zhao, W. Zhu, Y. Wang, S. Duan, Y. Gao, Y. Zhang, C. Chen, C. Hvilsom, C. W. Epps, L. G. Chemnick, Y. Doug, S. Mirarab, H. R. Siegismund, O. A. Ryder, M. T. P. Gilbert, H. A. Lewin, G. Zhang, R. Heller, W. Wang, Large-scale ruminant genome sequencing provides insights into their evolution and distinct traits. Science 364, eaav6202 (2019). [DOI] [PubMed] [Google Scholar]
- 67.A. Stamatakis, RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.R. R. Newton, I. L. Newton, PhyBin: Binning trees by topology. PeerJ 1, e187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.J. Felsenstein, PHYLIP—Phylogeny inference package (version 3.2). Cladistics 5, 164–166 (1989). [Google Scholar]
- 70.S. Mirarab, R. Reaz, M. S. Bayzid, T. Zimmermann, M. S. Swenson, T. Warnow, ASTRAL: Genome-scale coalescent-based species tree estimation. Bioinformatics 30, i541–i548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.L. Liu, L. Yu, D. K. Pearl, S. V. Edwards, Estimating species phylogenies using coalescence times among sequences. Syst. Biol. 58, 468–477 (2009). [DOI] [PubMed] [Google Scholar]
- 72.N. Dierckxsens, P. Mardulyn, G. Smits, NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.R. Lanfear, B. Calcott, S. Y. W. Ho, S. Guindon, Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701 (2012). [DOI] [PubMed] [Google Scholar]
- 74.H. Li, Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN] (16 March 2013).
- 75.T. S. Korneliussen, A. Albrechtsen, R. Nielsen, ANGSD: Analysis of next generation sequencing data. BMC Bioinform. 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.R. M. Kuhn, D. Haussler, W. J. Kent, The UCSC Genome Browser and associated tools. Brief. Bioinform. 14, 144–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.C. Xue, N. Rustagi, X. Liu, M. Raveendran, R. A. Harris, M. G. Venkata, J. Rogers, F. Yu, Reduced meiotic recombination in rhesus macaques and the origin of the human recombination landscape. PLOS ONE 15, e0236285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.A. O. Ayoola, B.-L. Zhang, R. P. Meisel, L. M. Nneji, Y. Shao, O. B. Morenikeji, A. C. Adeola, S. I. Ng'ang'a, B. G. Ogunjemite, A. O. Okeyoyin, C. Roos, D.-D. Wu, Population genomics reveals incipient speciation, introgression, and adaptation in the African mona monkey (Cercopithecus mona). Mol. Biol. Evol. 38, 876–890 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.N. Patterson, P. Moorjani, Y. Luo, S. Mallick, N. Rohland, Y. Zhan, T. Genschoreck, T. Webster, D. Reich, Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.M. Malinsky, M. Matschiner, H. Svardal, Dsuite—Fast D-statistics and related admixture evidence from VCF files. Mol. Ecol. Resour. 21, 584–595 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.A. H. Freedman, I. Gronau, R. M. Schweizer, D. Ortega-Del Vecchyo, E. Han, P. M. Silva, M. Galaverni, Z. Fan, P. Marx, B. Lorente-Galdos, H. Beale, O. Ramirez, F. Hormozdiari, C. Alkan, C. Vila, K. Squire, E. Geffen, J. Kusak, A. R. Boyko, H. G. Parker, C. Lee, V. Tadigotla, A. Wilton, A. Siepel, C. D. Bustamante, T. T. Harkins, S. F. Nelson, E. A. Ostrander, T. Marques-Bonet, R. K. Wayne, J. Novembre, Genome sequencing highlights the dynamic early history of dogs. PLOS Genet. 10, e1004016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Z. Yang, PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 83.E. Paradis, K. Schliep, ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 84.P. Cingolani, A. Platts, L. L. Wang, M. Coon, T. Nguyen, L. Wang, S. J. Land, X. Lu, D. M. Ruden, A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Y. Liao, J. Wang, E. J. Jaehnig, Z. Shi, B. Zhang, WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S27
Tables S1 to S17