Abstract
Over the past century, studies of human pigmentary disorders along with mouse and zebrafish models have shed light on the many cellular functions associated with visible pigment phenotypes. This has led to numerous genes annotated with the ontology term “pigmentation” in independent human, mouse, and zebrafish databases. Comparisons among these datasets revealed that each is individually incomplete in documenting all genes involved in integument-based pigmentation phenotypes. Additionally, each database contained inherent species-specific biases in data annotation, and the term “pigmentation” did not solely reflect integument pigmentation phenotypes. This review presents a comprehensive, cross-species list of 650 genes involved in pigmentation phenotypes that was compiled with extensive manual curation of genes annotated in OMIM, MGI, ZFIN, and GO. The resulting cross-species list of genes both intrinsic and extrinsic to integument pigment cells provides a valuable tool that can be used to expand our knowledge of complex, pigmentation-associated pathways.
Keywords: melanin, melanocyte, pigmentation, skin
1 |. INTRODUCTION
Studies investigating both the complex natural variation in pigmentation along with abnormal pigmentation phenotypes throughout the animal kingdom have built a solid foundation for understanding pigment cell biology. Pigment cells are defined by their ability to perform the distinctive cellular process of producing colored pigments within specialized organelles (Schartl et al., 2016). Mouse and zebrafish models have been crucial for the identification and functional characterization of genes centrally important to pigment cell biology (Bennett & Lamoreux, 2003; Kelsh, Harris, Colanesi, & Erickson, 2009; Mort, Jackson, & Patton, 2015). This is exemplified by the list of 171 cloned murine coat color genes, previously annotated by ESPCR members in 2011 as a pigment cell community resource (https://www.espcr.org/micemut/), and by the ever-growing lists of pigmentation term-annotated genes at the species-specific resources Mouse Genome Informatics (MGI) and the Zebrafish Information Network (ZFIN). In this review, we have systematically compiled a list of genes associated with integument pigmentation phenotypes in human, mouse, or zebrafish. This creation of a multispecies gene list reveals an integrated view of pigmentation-related processes that can be used to better understand the evolution of pigmentation and the underlying functional processes associated with human pigmentary disorders.
For the purposes of this review, only genes altering pigmentation phenotypes related to three neural crest-derived pigment cell lineages—melanocytes, iridophores, and xanthophores—are highlighted given their relevance to human, mouse, and zebrafish (Schartl et al., 2016). Mammalian melanocytes are highly specialized cells that produce eumelanin (black) and pheomelanin (yellow to red) pigment in organelles called melanosomes. In both humans and mice, melanosomes can be transferred to the surrounding keratinocytes of skin and hair in a precisely organized manner. In keratinocytes of the basal epidermal layer of the skin, the melanosomes harboring this transferred pigment form a cap over the cell nucleus and provide protection from UV exposure to the skin (Byers, Maheshwary, Amodeo, & Dykstra, 2003). Deposition of pigment to hair keratinocytes occurs in the hair follicle, and hair pigmentation reflects differences in the type and amount of melanin transferred as well as the pattern of melanosome deposition (Slominski, Tobin, Shibahara, & Wortsman, 2004; Wu & Hammer, 2014). Zebrafish differ from humans and mice, as the pigmented melanosomes produced by melanocytes are retained intracellularly and are not transferred to other cells (Bagnara & Matsumoto, 2006). Another difference in zebrafish is the presence of multiple pigment cell types; in addition to the melanocyte cell lineage, zebrafish have xanthophores, which are responsible for yellow pigment production that is pteridine- or carotenoid-derived, and iridophores, characterized by their production of a light-reflective, purine/pteridine crystal-based pigment (Bagnara & Matsumoto, 2006). In zebrafish, these three lineages form organized, heritable patterns in the integument throughout larval stages that are responsible for the fully developed stripe patterns in adults, allowing investigation of cell migration, patterning, and homotypic and heterotypic pigment cell interactions (Kelsh, 2004; Kelsh et al., 2009; Mahalwar, Singh, Fadeev, Nüsslein-Volhard, & Irion, 2016; Parichy & Spiewak, 2015).
Research utilizing mouse and zebrafish models of pigmentation variation has led to novel pigmentation gene identification and indepth analyses of associated gene functions. Much of what is known about the developmental processes underlying pigmentation has been discovered in zebrafish and murine mutant gene models (Kelsh et al., 2009; Mort et al., 2015). These species are readily amenable to genetic manipulation, permitting evaluation and validation of genes associated with human pigmentary disorders (Cooper, 2017; Mort et al., 2015; Yamaguchi & Hearing, 2014). It should be noted that while humans and mice frequently share similar hair/coat color phenotypic presentations when orthologous genes are altered, notable differences in skin morphology exist between the two species. Human melanocytes are present in hair follicles and throughout the basal epidermis at the epidermal–dermal junction, while in mice the melanocytes reside primarily within epidermal hair follicles, and only occur in the epidermis and dermis of tail, ears, and feet. Zebrafish models also present limitations in the evaluation of the full range of human phenotypes, because of different integument anatomy and because pigment organelle transfer between cells does not occur. However, zebrafish models give the tremendous advantages of rapid organism development and the ability to monitor in vivo melanocyte processes during development at a single cell level, including cell patterning and melanosome intracellular granule dispersal rates.
Pigmentation is a complex process, resulting from many gene interactions across diverse cellular functions. These functions include regulation of pigment cell specification, proliferation, survival, migration, patterning, and positioning of pigment cells within the integument. They also include processes regulating pigment production, pigment organelle cellular localization, and the transfer and retention of pigment-containing vesicles. Genes involved in pigmentation can perform pigment cell-intrinsic functions, or can be associated with pigment cell-extrinsic functions within neighboring keratinocytes or fibroblasts (or neighboring pigment cells in zebrafish), or can be related to broad systemic effects (Irion, Singh, & Nüsslein-Volhard, 2016; Wang et al., 2017; Yamaguchi & Hearing, 2009). Given the complexity of diverse cellular functions associated with pigmentation and both intrinsic and extrinsic modes of action, it is not surprising that alterations in pigmentation often provide a visible phenotypic hallmark associated with pleiotropy, where one gene mutation affects multiple organs and cell types. Therefore, studies of normal skin and hair color variation and disease-associated pigmentation changes can reveal gene functions that are not solely associated with pigmentation, but also with a wide spectrum of human diseases.
This review provides an updated list of human, mouse, and zebrafish pigmentation genes, increasing our understanding of the genes and cellular functions governing integument-based pigmentary phenotypes. Using a pigment phenotype-centric curation approach, we generated a single integrated inventory of genes responsible for pigmentation phenotypes across human, mouse, and zebrafish species. In the future, this unified pigmentation gene list can be a resource for a variety of studies, including cross-species candidate gene analysis, pathway-based investigations, and genome-wide comprehensive studies, aiding researchers in their advancement of the pigment cell biology field.
2 |. ASSEMBLY AND MANUAL CURATION OF AN EXPANDED PIGMENTATION GENE LIST
2.1 |. Gene retrieval
We set out to assemble an updated catalog of genes that affect melanocyte-, xanthophore-, and iridophore-derived integument, hair and skin pigmentation, using the following four publicly available databases: Online Mendelian Inheritance in Man (OMIM, https://www.omim.org), MGI (https://www.informatics.jax.org), ZFIN (https://zfin.org), and the Gene Ontology Consortium (GO, https://www.geneontology.org) (Amberger & Hamosh, 2017; Ashburner et al., 2000; Blake et al., 2017; Howe et al., 2013; Ruzicka et al., 2015; The Gene Ontology Consortium, 2017). First, OMIM, MGI, ZFIN, and GO were each queried using four common ontology terms: pigmentation, melanocyte, melanosome, and melanin. Next, organism-specific visual pigmentation descriptors were used to query OMIM (pigmentary abnormalities, pigmented, hyperpigmented, hyperpigmentation, hypopigmented, hypopigmentation, albinism, café au lait, lentigines, and nevi), MGI (melanoblast, pigment, pigmented, and the mammalian phenotype browser ID MP:0001186), and ZFIN (pigment cell, retinal pigment epithelium, colorless/colorless, melanophore, iridophore, xanthophore, and melanoblast). Finally, GO was queried using all of the annotation terms used for OMIM, MGI, and ZFIN. Ultimately, these iterative database queries resulted in the retrieval of 730, 854, 729, and 587 genes from OMIM, MGI, ZFIN, and GO, respectively (Figure 1a “Retrieved”).
FIGURE 1.
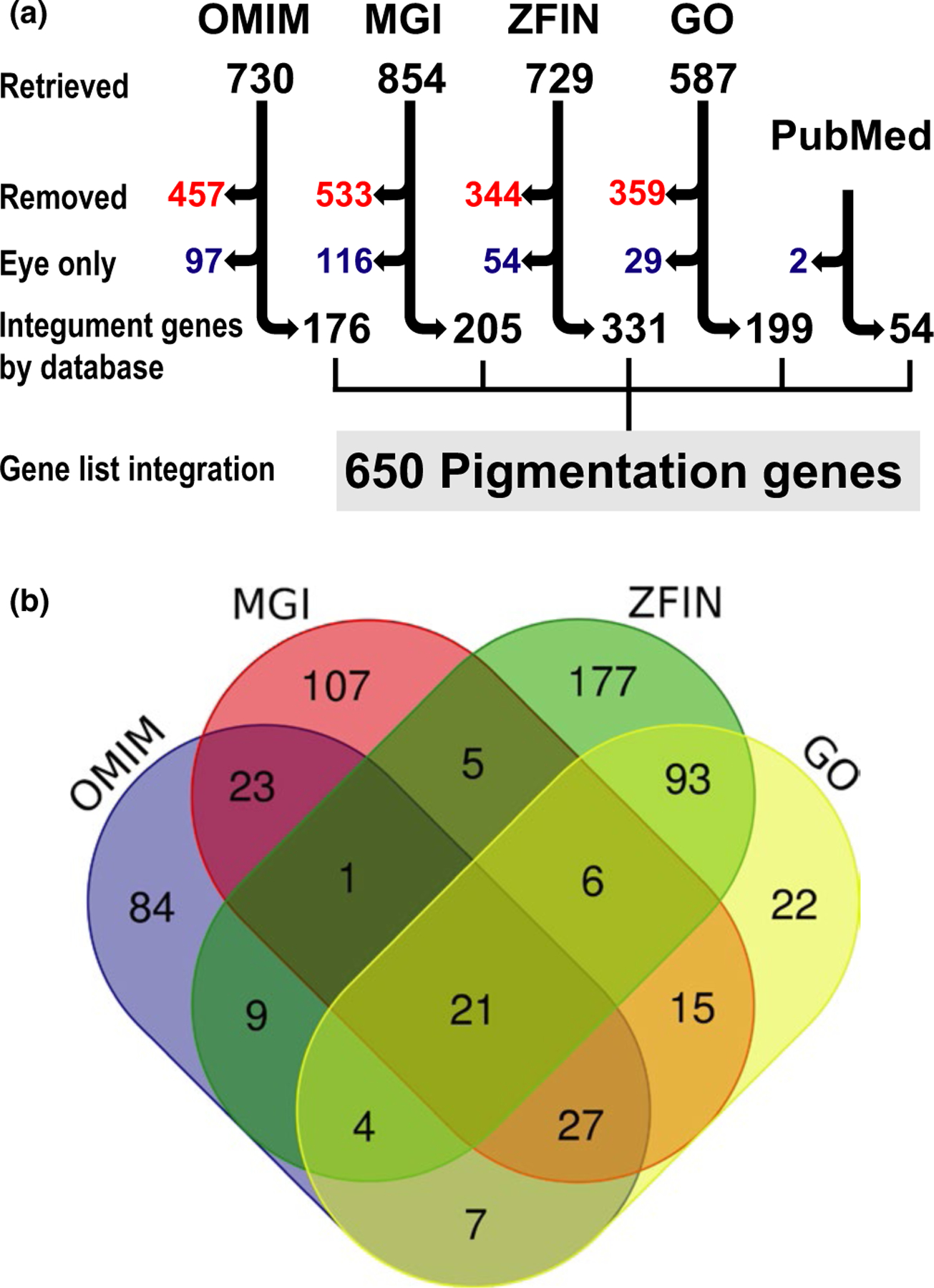
The pigmentation gene list reveals differences among annotation at large-scale databases. (a) Flow chart illustrating the process used for evaluation of genes retrieved from OMIM, MGI, ZFIN, GO, and PubMed that were included on the final list of 650 pigmentation genes. The numbers of genes are indicated at each step: “Retrieved” indicates all unique genes initially retrieved by all search terms; “Removed” indicates genes that did not meet the criteria for retention, “Eye only” indicates genes with pigmentation phenotypes exclusively in the eye that were moved to an eye-specific list (Supporting Information Table S6), and “Integument genes by database” indicates the genes with verified pigmentation phenotypes retrieved from each database (Supporting Information Tables S1–S5). Finally, “Gene list integration” indicates the final list of 650 pigmentation phenotype genes, generated by merging the lists from each database (Supporting Information Table S7). The 56 PubMed genes were not identified with systematic query terms, but were manually discovered and added during literature searches used to validate genes retrieved from the four databases. (b) Venn diagram depicting the overlap of pigment gene annotation in OMIM, MGI, ZFIN, and GO. Note the minimal overlap among databases, especially OMIM, MGI, and ZFIN. Venn diagram was generated with the Venn diagram tool from the Bioinformatics and Evolutionary Genomics/Van de Peer Lab website (http://bioinformatics.psb.ugent.be/webtools/Venn)
Differences in each of these databases impacted both the terms used and the manner in which searches were performed. For example, the OMIM search utility is solely text-based, without overlaid ontology annotation, and also restricts the total number of query line outputs that are available for batch download. Thus, OMIM required more specific keywords for phenotype/gene retrieval. In contrast, MGI data are contained within a clearly defined ontology structure, without line output limitations. MGI queries were performed using “smart search”-related parameters under the “Phenotypes, Alleles & Disease model” search (https://www.informatics.jax.org/allele). ZFIN data were obtained using searches restricted to gene and phenotype terms, and then limited further using queries directed toward “affected anatomy,” “affected biological process,” and “phenotype statement.” GO is a structured ontology database, comprised of annotations derived from both experimental findings and annotations inferred across all species. Thus, GO queries retrieved genes from many species, of which we retained only human, mouse, and zebrafish.
2.2 |. Manual curation
The gene lists retrieved by the multiple queries described above were subsequently subjected to a series of manual analyses to verify their association with a pigmentation phenotype. Each gene-associated phenotype description was manually evaluated using the database description, the primary literature references cited by each database, and additional primary literature if needed. Only protein-coding genes with a published pigmentation phenotype were retained. These included genes affecting pigmentation processes and/or cells related to melanin, melanosomes, pigment production, pigment transport, melanocytes, iridophores, or xanthophores at either the whole organism or cellular level. Since this was a list defined by pigment phenotypes, genes did not have to be expressed in melanocytes to be retained on the gene list, but could be expressed in neighboring cells such as keratinocytes. During validation of the genes retrieved from OMIM, MGI, ZFIN, and GO, 56 additional genes with published pigmentation phenotypes were identified and confirmed by PubMed literature searches (Figure 1a, https://www.ncbi.nlm.nih.gov/pubmed).
Many of the genes retrieved by automated text-based searches did not meet our criteria for inclusion as pigmentation genes. Genes were excluded from the lists obtained at each individual database if the text annotation fit one or more of the following categories: (a) uncloned or non-protein-coding genes; (b) description of normal pigmentation; (c) non-melanin pigment-related phenotype description (e.g., heme, lipofuscin, liver, and other pigments); (d) irrelevant text (e.g., query term matched text description of a different gene, or text in linked references/abstracts that was related to a different gene/phenotype); (e) suggested gene-phenotype linkage only, or misattribution of pigment phenotypes to a gene residing in close proximity; (f) relation to retinal pigment epithelium in a descriptive manner (e.g., description of gross developmental delay or position from which a measurement was performed); (g) insufficient data (e.g., data not shown, no published data, or expression of mRNA or protein described in a pigmented tissue, but without any functional or phenotypic data); (h) annotation derived solely from computationally made inferences (e.g., GO-inferred cellular localizations and inferred annotation by Phylogenetic trees, UniProt Keywords with GO Terms, and Biological Aspect of Ancestor); (i) fewer than three unrelated human patients that have been identified for a gene-correlated phenotype; (j) human skin lesions with pigment changes appearing secondary to keratin and epidermal structural abnormalities; (k) neuromelanin-related annotation; (l) annotations associated with a transgenic mouse line if only one insertion event was reported, and thus we could not verify if a pigmentation phenotype was associated with transgene expression rather than disruption of a different gene at the insertion site; and (m) extracutaneous melanocyte pigmentation phenotypes of the mouse harderian gland or inner ear strial cells. Ultimately, 47%–63% of the genes retrieved from each database were excluded using these 13 criteria. Exact numbers of genes excluded from each database are shown (Figure 1a “Removed”) and a full list of excluded genes can be obtained upon request to the authors.
2.3 |. A cross‐species, integrated, integument gene list
Following manual curation to confirm pigment cell-associated phenotypes, each of the individual database lists were subdivided to distinguish genes associated with integument phenotypes from those associated only with pigmentation phenotypes in the eye. The genes which exhibited visible hair and skin/integument pigmentation phenotypes from OMIM, MGI, ZFIN, GO, and PubMed were retained in individual, database-specific lists of genes with integument-associated phenotypes, found in Supporting Information Tables S1–S5. These five tables include brief phenotype descriptions for each gene, which can be used to classify subgroups of genes based upon phenotypes or cellular processes. Of note, some of these genes had both eye- and integument-associated pigment phenotypes; if these were present in the same organism, they were retained in the integument-associated lists. However, genes with pigmentation phenotype descriptions limited only to the retina or melanin-producing eye cells (iris, choroid and RPE) in human and mouse, or to melanophores and iridophores of the eye in zebrafish were separated into a species-integrated list and were not analyzed further (Figure 1a “Eye only”; Supporting Information Table S6). This is a diverse and complex list, in which many genes reflect eye pigmentation changes arising secondary to defects in non-pigmented cells of the eye. For example, many of these genes are associated with Retinitis pigmentosa. The characteristic retinal pigmentary abnormalities of Retinitis pigmentosa are, in most cases, initiated by mutations in genes which cause photoreceptor cell degeneration. Subsequently, the closely associated RPE cells are affected by the photoreceptor abnormalities, undergoing abnormal migration and pigment deposition in perivascular regions of the retina (Dias et al., 2018; Verbakel et al.., 2018).
As a final step, we generated a cross-species list of pigmentation genes. The integument pigmentation gene lists from each database (Figure 1a “Integument genes by database”; Supporting Information Tables S1–S5) were integrated into a single list of 650 genes (Figure 1a “Gene list integration”; Supporting Information Table S7). Species-specific orthologs for each gene were obtained from Ensembl (Biomart tool; www.ensembl.org); when Ensembl-predicted orthology was unclear, individual genes were examined at ZFIN and/or NCBI to confirm conserved genome location (synteny), amino acid homology, and/or functional complementation. If multiple orthologs were confirmed by these criteria, they were included in the list, and notations were made to indicate if one or both of the genes had a known pigmentation phenotype in zebrafish (Supporting Information Table S7). The cross-species pigmentation list is organized to facilitate the identification of subcategories of interest: for example, the 605 genes with unambiguous orthologs in all three species can be identified (Supporting Information Table S7, “Orthologs across species” column), or the 552 genes with phenotypes only reported in the body/integument can be identified (Supporting Information Table S7, “Pigment phenotype location” column). Of note, eight genes on the gene list have pigmentation phenotypes in species other than human, mouse, and zebrafish: Alx3 (Rhabdomys pumilio), LVRN (Felis catus), shroom2 (Xenopus laevis), sox5 (Oryzias latipes), and NR4A3, TRPM1, SLC36A1, and STX17 (Equus caballus). These eight genes were identified with PubMed searches or from OMIM annotation (which is not exclusive to humans and thus includes descriptions of pigmentation genes in other organisms) and were subsequently retained because of their relevance to the pigmentation gene list.
As illustrated in Figure 1b, no single database provided a comprehensive annotation of the cross-species pigmentation gene list. In particular, there was minimal overlap among genes annotated for pigmentation in OMIM, MGI, and ZFIN, suggesting that for numerous genes, pigmentation phenotypes are not yet fully characterized across human, mouse, and zebrafish. Importantly, GO pigment annotation did not fully overlap with the other three databases, indicating that pigmentation- and melanocyte-related annotations for these genes are not completely captured across species in GO. Overall, this combination of genes from multiple databases across human, mouse, and zebrafish species provided a tremendous increase in pigmentation gene number compared to any single database.
3 |. MOUSE AND ZEBRAFISH MODELS COMPRISE THE MAJORITY OF THE INTEGRATED INTEGUMENT GENE LIST
The pigmentation gene list contains 128 genes with human phenotypes, 243 genes with mouse phenotypes, and 325 genes with zebrafish phenotypes (these sublists are indicated in the “Species with phenotype” column of Supporting Information Table S7). To better visualize the distribution of genes with pigmentation phenotypes across the three species, the genes were subdivided based upon the organism(s) in which the phenotype was reported (Figure 2a). This highlighted the moderate overlap among these three species: the vast majority of the genes exhibited phenotypes in only mouse or zebrafish models (480, 74%), indicating the utility of these two models in revealing pigmentation genes, and also suggesting these genes as candidates for future analyses in human pigmentation. Of note, one Hermansky–Pudlak syndrome-associated gene, AP3D1, is only listed as showing a mouse phenotype because the published report of a single patient did not meet our criteria of three unrelated patients for inclusion on the human phenotype list (Ammann et al., 2016; Montoliu & Marks, 2017). However, the presence of a well-characterized mouse model harboring a mutation in this gene lends strong support that this gene is a bona fide human pigmentation mutant. Fifty-one of the genes have both a human pigmentation phenotype and an animal model that recapitulates the phenotype. Seventy-seven genes have only been identified to date with human pigmentation phenotypes; these genes will be excellent candidates for mouse and zebrafish studies, to test for cross-species conservation of pigmentation functions.
FIGURE 2.
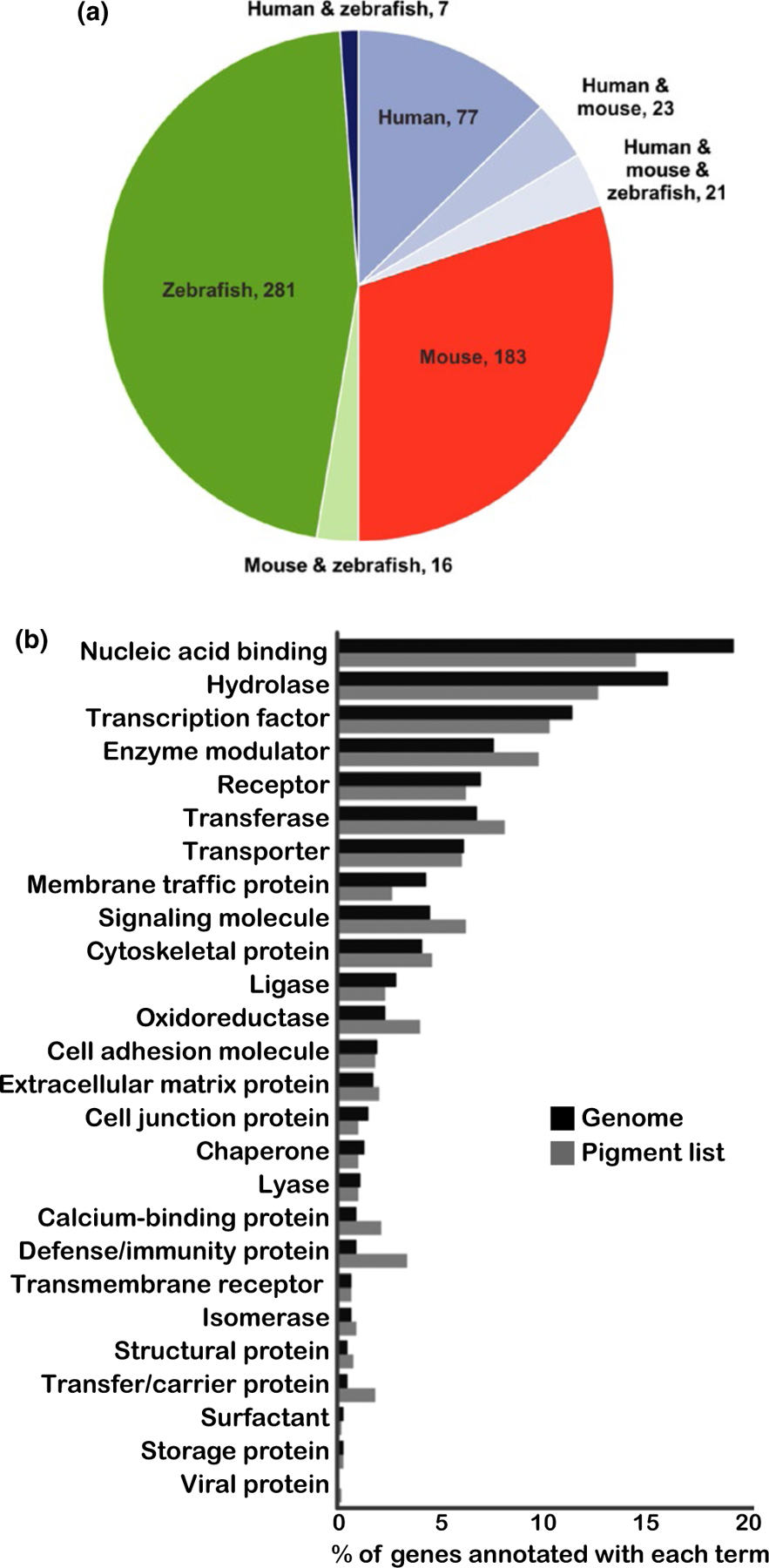
Animal models comprise the majority of the pigmentation gene list, and the pigmentation gene list spans numerous protein classes. (a) Pie chart illustrating the distribution of human, mouse, and zebrafish pigmentation phenotypes of the pigmentation gene list. For simplicity, cell-specific phenotypes and other animal models were excluded from this figure. A total of 531 genes (82% of the pigmentation gene list) were annotated with pigmentation phenotypes in mouse and/or zebrafish; of these, 480 were reported in mouse or zebrafish, but not human. The importance of animal models to a broader understanding of pigmentation is indicated by the large number of pigmentation genes that are recognized because of mouse and zebrafish phenotypes. (b) PANTHER statistical overrepresentation analysis for protein class in the pigmentation gene list. Of the 650 pigmentation genes, 638 human genes were annotated at PANTHER and thus were used for protein class analysis. The pigmentation genes show 26 distinct protein class designations (light gray bars), revealing complexity of protein classes that affect pigmentation. The distribution of the pigmentation genes in these protein classes is similar to that of the total genome (dark gray bars)
Importantly, 98 genes (15%) have a documented pigmentation phenotype in both cutaneous melanocytes and one or more of the three pigmented cell types in the eye: iris, choroid, or RPE (see Supporting Information Table S7 for complete list). These include 28 well-characterized pigmentation genes that display phenotypes in both humans and animal models. Examples include the Hermansky–Pudlak syndrome genes AP3B1, BLOC1S3, BLOC1S6, HPS1, HPS3, HPS4, HPS5, HPS6, and DTNBP1, which function in melanosome transport; the Waardenburg syndrome-related genes MITF, SOX10, PAX3, EDN3, EDNRB, and SNAI2, which are involved in neural crest development and survival; and albinism-related genes TYR, TYRP1, GPR143, OCA2, SLC24A5, SLC45A2, and LRMDA, which regulate pigment synthesis or melanosome function. Interestingly, for 44 of these 98 genes with both integument and eye pigment phenotypes, zebrafish is the only species with documented phenotypes in both cell types. Some of these have eye-related phenotypes but no documented dermal component in humans, such as BBS1 and BBS2; these genes exhibit Bardet–Biedl syndrome-associated retinitis pigmentosa in humans, while in zebrafish, both retinal phenotypes and integument melanosome transport defects have been observed. Examples such as BBS1 and BBS2 suggest underappreciated integumentary functions in human and mouse for additional genes currently reported with eye and integument phenotypes only in zebrafish.
4 |. DIVERSITY OF PIGMENTATION GENE FUNCTION
This expanded, cross-species pigmentation gene list provides the opportunity to identify a broad spectrum of protein complexes, cellular pathways, and functions that contribute to pigmentation variation. To assess the diversity of protein functions captured within the pigmentation gene list, we utilized PANTHER database software tools (Protein ANnotation THrough Evolutionary Relationship; https://pantherdb.org, [Mi et al., 2017]). Of the 650 pigment genes, 638 human genes were annotated at PANTHER and were found within 26 distinct protein class designations (Figure 2b, light gray bars). This number of protein classes reflects distribution across a wide array of protein types, demonstrating a diverse array of proteins and their associated cellular processes that influence pigmentation.
Two of the three largest functional classes of proteins represented on the pigment gene list are “nucleic acid binding” and “transcription factor,” reflecting the over 90 genes that are associated with chromatin binding or DNA transcription. The second most frequently represented protein class, “hydrolase,” is a broad category of enzymes with hydrolysis catalytic activity. This class encompasses numerous genes with diverse molecular functions, including ATP synthases, metalloproteases, phosphatases, and cysteine proteases. Interestingly, the pigmentation gene list shows no enrichment for any given protein class, as the distribution of protein classes within the pigmentation genes is similar in distribution to that of the entire genome (Figure 2b, dark gray bars). This suggests that all protein classes contribute to maintenance of normal pigment cell function and that gene disruption across a broad range of functions seems equally likely to result in a pigmentation phenotype.
5 |. SPECIES‐INTEGRATED VIEW OF BIOLOGICAL PROCESSES REGULATING PIGMENTATION
The gene list was further analyzed using a PANTHER enrichment analysis tool to identify biological processes that were statistically overrepresented in the species-integrated list of genes, or in the species-specific sublists of genes with phenotypes reported in human, mouse, or zebrafish (Figure 3). The integration of pigmentation phenotype-related data from all three species (Figure 3a, black bars) highlights the utility of a cross-species approach in revealing a more comprehensive view of the genes that regulate pigmentation, as 21 additional processes emerge that are not enriched on the species-specific gene lists. These additional processes reflect cellular functions that have been studied broadly across all three species. They include proteins with small molecule transporter and channel function, proteins that mediate vesicular transport, endocytosis and exocytosis, and numerous proteins involved in cell communication and signal transduction. It is striking that these categories most closely align to fundamental functions of pigment cells; the production of pigment in a membrane-bound organelle, as well as regulation of that organelle’s localization in response to extracellular signals.
FIGURE 3.
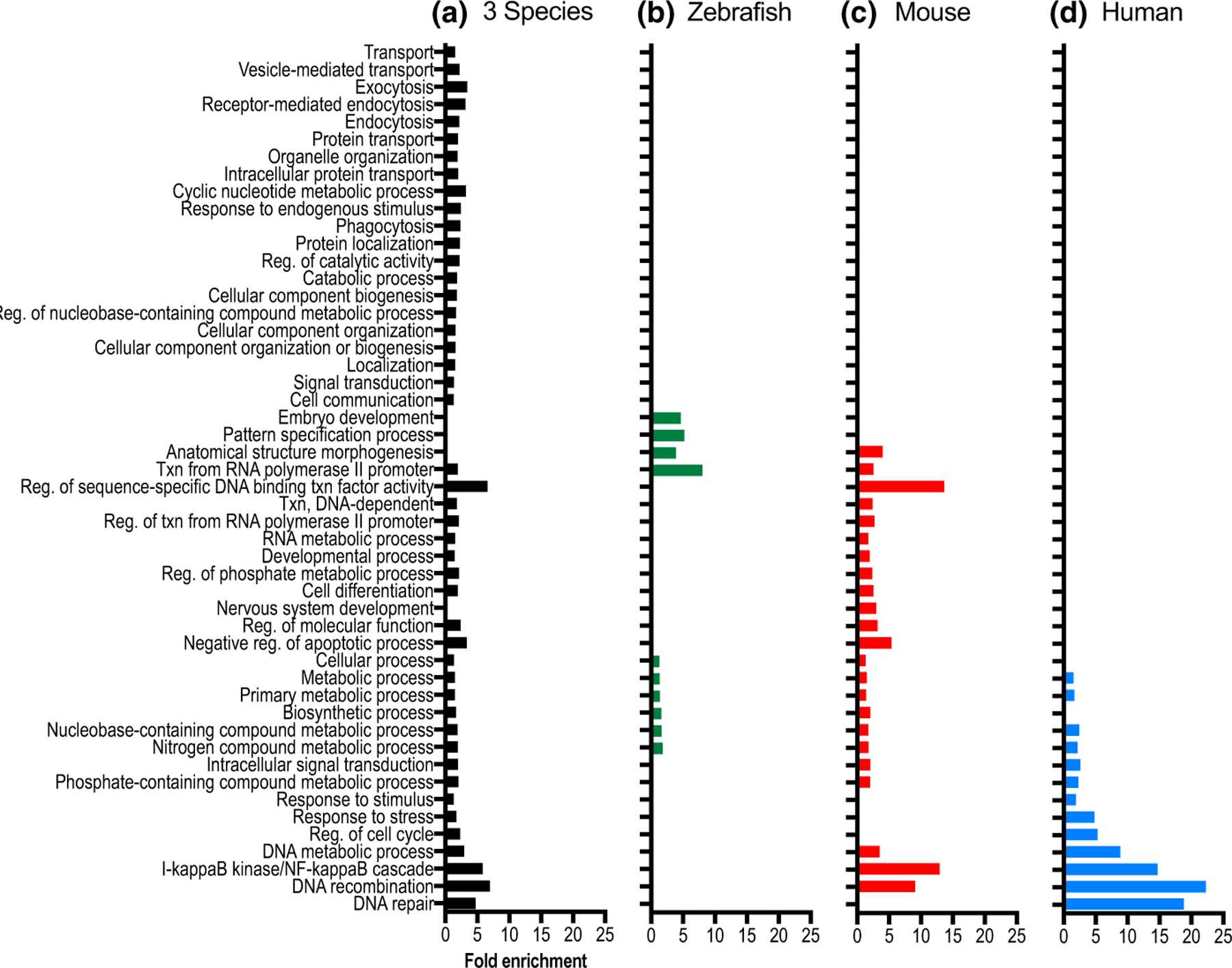
PANTHER statistical overrepresentation analysis identifies biological processes that are significantly overrepresented within the pigmentation gene list. The significantly overrepresented biological processes identified using PANTHER GO BioSlim analyses are shown for four different lists of genes, containing pigmentation phenotypes in (a) any of the three species (black); (b) zebrafish (green); (c) mouse (red); and (d) human (blue). The three species list (a) shows 21 additional processes that are not present in the analysis from species-specific data. Note that several overrepresented species-specific processes are also seen in human, mouse, and zebrafish that are not present in the other species-specific analyses (b–d). The total number of Panther gene annotations available for analysis in each species list was as follows: three species list, 638 genes; human, 127 genes; mouse, 240 genes; and zebrafish, 337 genes. Abbreviations for BioSlim terms include Txn (Transcription) and Reg (Regulation). Fold enrichment (FDR) values ranged from 1.11e–2 to 1.23e–15
On the species-integrated pigmentation gene list, there are over 50 pigmentation genes associated with pigment cell vesicle biogenesis and movement within the cell, which are broadly annotated in PANTHER under “transport,” and more specifically under “vesicle mediated transport,” “exocytosis,” and “endocytosis.” The proteins encoded by these genes span multiple components of the vesicular trafficking pathway and include vacuolar sorting proteins, RAB GTPases, and members of the AP-3, BLOC-1, BLOC-2, and BLOC-3 complexes (Crawford et al., 2017; Lloyd-Jones et al., 2017; Marks, Heijnen, & Raposo, 2013; Sitaram & Marks, 2012). Also categorized under the term “transport” are proteins that function as channels and molecular transporters, which are known to directly influence melanin synthesis by affecting melanosome pH as well as calcium, cysteine, and copper levels in melanosomes (Bellono & Oancea, 2014). The crucial importance of proper regulation of these melanosomal properties is supported by the identification of transporters with pigmentation phenotypes in all three species, such as ATP7A, OCA2, SLC45A2, and SLC24A5 (Bellono & Oancea, 2014; Bellono, Escobar, Lefkovith, Marks, & Oancea, 2014; Bin et al., 2015; Dooley et al., 2013; Lamason et al., 2005; Newton et al., 2001; Setty et al., 2008; Vogel et al., 2008). Adding to the complexity of melanosomal regulation are many additional transporters and channel proteins that to date have been discovered with pigmentation phenotypes in a single species, including CTNS, SLC17A5, and SLC29A3 (human); Slc7a11, Slc30a4, Slc31a1, and Clcn7, along with its interacting partner Ostm1 (mouse); and slc12a2, slc16a2, slc17a6b, slc22a7b.1, slc2a1a, slc2a1b, slc2a11b, and slc40a1 (zebrafish).
Additionally, the integrated species list of pigmentation genes contains nearly 100 proteins associated with signal transduction pathways. While the signaling pathway members EDN3, EDNRB, KIT, KITLG, MC1R, NF1, POMC, and PTPN11 have pigmentation phenotypes in all three species, over half of the genes involved in signal transduction processes only have phenotypes characterized in zebrafish or mouse models. This reiterates that most of the pigmentation genes have only been analyzed in animal models (Figure 2a), thus providing numerous candidate genes with potential to also be responsible for pigmentation-related phenotypes in humans.
The zebrafish and mouse model gene lists (Figure 3b,c, green and red bars) show overrepresentation of 16 biological processes that are not overrepresented in the human-specific list (Figure 3d, blue bars). Additionally, four biological processes are overrepresented in the human list, but not in the two animal models. These interesting differences may be present because the overrepresentation analyses revealed research focus biases of investigators, species-specific attributes that may facilitate research of selected cellular processes, or the ability of animal models to assess gene function in the context of embryonic lethality. For example, the ease with which biological function can be assayed in each organism appears to correlate with differential species-specific distributions. The zebrafish-exclusive over-represented biological processes “embryo development” and “pattern specification process” (Figure 3b) may reflect the strength of zebrafish as a model system for early developmental analysis. The genes annotated in these categories include multiple transcription factors (hoxa13a, hoxa13b, zic2a, zic2b, meox1, mespaa, mespab, mespba, mespbb, pax7a, and pax7b), which are involved in regulating developmental pigment cell patterning and function.
The human-specific overrepresentation of pigmentation genes regulating DNA repair (Figure 3d) may reflect species-specific attributes. One of the primary functions of melanin production by dermal melanocytes in humans is to protect the skin from the damaging effects to DNA caused by ultraviolet radiation (UVR, (D’Orazio, Jarrett, Amaro-Ortiz, & Scott, 2013)). Melanocytes respond to UVR by a variety of signaling mechanisms, resulting in the upregulation of pigmentation and activation of the Nucleotide Excision Repair (NER) pathway to correct abnormal DNA photo-products (Abdel-Malek, Kadekaro, & Swope, 2010). Research focused on the human disorder Xeroderma pigmentosum (XP), a condition of extreme UV sensitivity caused by underlying defects in DNA repair that are reflected in pigmentary abnormalities and cancer susceptibility, has linked XP to mutations in eight NER genes (DDB2, ERCC2, ERCC3, ERCC4, ERCC5, POLH, XPA, and XPC (Koch, Simon, Ebert, & Carell, 2016)).
Also represented within the DNA repair and DNA recombination categories are genes that function in replication and chromatin/telomere stability. Genes regulating these cellular processes in human and mouse melanocytes frequently share a hair graying/melanocyte loss phenotype, which may arise from the loss of melanocyte stem cell progenitors (Carrero, Soria-Valles, & López-Otín, 2016; Sahin & DePinho, 2010). Human disorders and the corresponding mutated genes on the pigmentation gene list that can present phenotypically with hair graying include Rothmund–Thomson syndrome (RECQL4), Werner syndrome (WRN), Bloom syndrome (BLM), ataxia telangiectasia (ATM), and some cases of dyskeratosis congenita (e.g., TERT and CTC1 (Arora et al., 2014; Ballew & Savage, 2013; Lebel & Monnat, 2017; Lu, Jin, & Wang, 2017; Teive et al., 2015)). Along with mouse models for two of the human disorders described above (Recql4 and Atm (Hoki et al., 2003; Inomata et al., 2009; Mann et al., 2005)), hair graying with age also occurs in mice with mutations in Mcm2 and Ercc2, which function in DNA replication and DNA damage repair, respectively (de Boer et al., 2002; Pruitt, Bailey, & Freeland, 2007).
6 |. PLEIOTROPY OF PIGMENTATION GENES
While this list focused on pigmentation phenotypes, it is important to note that disruption of pigment-associated genes can have pleiotropic effects. Numerous human disorders present with both pigmentation abnormalities and distinct phenotypes in other tissues, thus illustrating that pigmentation mutations can be a visible sentinel for anomalies in a variety of cell types. The tissues and phenotypes frequently affected beyond melanocytes include neural crest-derived tissues, such as glial cells of the central and peripheral nervous systems, bleeding disorders, immunodeficiencies, and neurological abnormalities (Bondurand & Sham, 2013; Huizing, Helip-Wooley, Westbroek, Gunay-Aygun, & Gahl, 2008; Marks et al., 2013; Reissmann & Ludwig, 2013; Weider & Wegner, 2017). In this pigmentation gene list, over 75% of the 128 human genes with pigmentation phenotypes have additional phenotypes affecting other tissues. This demonstrates that the genes governing pigmentation are linked to a wide variety of cellular pathways with significant clinical implications.
To further illustrate the extensive pleiotropy of pigmentation-associated genes, a subset of 296 mouse genes from the pigmentation gene list that are specifically annotated in MGI with the mammalian phenotype (MP) term “pigmentation” were analyzed for additional MP terms (Figure 4). Strikingly, 271 of these pigmentation genes (92%) are annotated with at least five additional high-level MP terms. The most commonly occurring phenotype annotations were integument, growth/size/body region, mortality/aging, nervous system, and vision/eye. The high frequency of pleiotropic effects on a wide range of organ systems provides strong evidence for the clinical relevance of pigmentation genes across a broad range of human diseases.
FIGURE 4.

Extensive phenotypic pleiotropy is present in mouse models for pigmentation genes. The 296 genes annotated in MGI with the mammalian phenotype (MP) term “pigmentation phenotype” were evaluated for additional MP terms, and the graph illustrates the percentage of these 296 genes with additional annotations. Clinical relevance of these pigmentation genes is indicated by the most commonly occurring high-level phenotype annotations: integument, growth/size/body region, mortality/aging, nervous system, and vision/eye
7 |. THE NEED FOR IMPROVED PIGMENTATION ONTOLOGY
The extensive manual curation that was required to assemble this pigmentation gene list highlights problems with the wide array of ontology terms that are used across research platforms to describe the spectrum of visible pigmentation phenotypes. Historically, researchers have used similar phenotypes, shared by multiple genes, to suggest common pathways and/or functions and to direct research hypotheses. Accurate and consistent descriptions of phenotypes are important to facilitate this work; however, phenotypic annotation is complex and can be difficult to standardize across species, in part because not all annotation terms have unique meanings. For example, “hypopigmentation” can be used in mice interchangeably with “white spotting” or “belly spot” to refer to a localized region of skin and hair that completely lacks pigment cells (e.g., Tfap2a, Sox10, Mitf, Zic2). Alternatively, “hypopigmentation” may refer to a “dilution of color” resulting from an overall reduction in pigment across the entirety of the body (e.g., Rab27a, Tyrp1, Vps33a). Similarly, the phrase “gray hair” may refer to at least two phenotypes: a reduction in transport of eumelanin- or pheomelanin-containing melanosomes (e.g., Rab27a, Myo5a); or premature differentiation or apoptosis of melanocyte stem cell function associated with aging (e.g., Polg, Atr, Bcl2). Thus, in many cases the same annotation term can be used to describe pigmentation phenotypes that result from very distinct mechanisms. Further complications result from species-specific differences in ontology. For example, numerous qualifiers describe lower pigment levels in zebrafish, such as “reduced amount,” “decreased amount,” “colorless,” and “absent;” in sum, these types of qualifiers add phenotypic specificity to over 150 distinct melanocyte-associated phenotype statements in zebrafish. While this provides greater clarity for zebrafish, it also complicates automated data integration and phenotype-based queries. These examples serve as a reminder that we, as pigment cell researchers, need to accurately and completely describe these diverse pigment phenotypes, apply our descriptions uniformly, and actively engage to improve database annotation within our areas of expertise.
8 |. ASSESSING PIGMENTATION BIOLOGY IN THE FUTURE
Pigment biology researchers today have vast, publicly available data collections at their disposal, but utilization of these datasets to their full capability can be difficult, and each list may have gaps in information. For these reasons, we generated this hand-curated pigmentation gene list, producing a comprehensive resource across human, mouse, and zebrafish that can be used in a variety of ways to promote further discoveries in pigment cell biology. For example, this list contains numerous understudied genes that fall within the same protein family or share functions with well-characterized genes at the center of pigment biology. Notably, the pigmentation gene list includes over 90 transcriptional regulatory factors. While in the past, studies on individual transcription factors have been a focus of pigmentation research, now future analyses can broaden to include these new genes, thus determining how these numerous factors and multi-component complexes interact to coordinately regulate transcription. In another example of the utility of a well-curated pigment gene list, growing gene expression and protein interaction databases will certainly provide novel candidate genes and pathways which can be rapidly implicated in pigment cell biology based on their association with genes on the pigmentation gene list.
Additionally, this comprehensive list of pigmentation-related genes and associated pathways will have great utility in prioritizing, assessing, and validating genes in close proximity to new loci identified in Genome Wide Association Studies (GWAS). Pigmentation is clearly a multigenic trait, and the catalog of human GWAS loci associated with normal pigmentation variation is expanding through new studies of ethnically diverse populations (Crawford et al., 2017; Hernandez-Pacheco et al., 2017; Lloyd-Jones et al., 2017; Martin et al., 2017; Rawofi et al., 2017). For example, genomic variants in close proximity to the DDB1 genomic locus have been identified in two separate GWAS analyses that studied pigmentation variation in individuals of African descent (Crawford et al., 2017; Lloyd-Jones et al., 2017). The replication of this locus in two studies suggests that DDB1, or neighboring genes within the region of linkage disequilibrium, contributes to human pigment variation. While DDB1 is not currently associated with pigmentation phenotypes, it forms a complex with the protein encoded by the NER/XP gene DDB2, and this DDB1/DDB2 complex recruits NER pathway members to facilitate DNA damage repair (Chu & Chang, 1988; Palomera-Sanchez & Zurita, 2011). Since the pigmentation gene list contains eight NER pathway members that impact pigmentation, DDB1 is an excellent candidate gene at this locus to subject to future in-depth analysis.
Although this cross-species pigmentation gene list is quite extensive, much still remains to be discovered regarding the genes and pathways that govern pigmentation. More than 180 genes/loci at MGI that are indexed with mouse pigmentation phenotypes remain uncloned, and there are more than 50 reported human conditions with abnormal pigmentation phenotypes for which the molecular basis is unknown (https://www.omim.org). Large-scale phenotyping screens such as those performed by the International Mouse Phenotyping Consortium (Meehan et al., 2017) will also continue to identify unexpected genes affecting pigmentation. Furthermore, data exist that suggest the relevance of particular pathways and biochemical functions for pigment cell biology, yet the details have not been elucidated, such as systemic hormonal effects on pigmentation, or immune system interactions with melanocytes (Harris et al., 2018; Yamaguchi & Hearing, 2009). In the future, we suggest that a broad view that incorporates the larger number of genes and pathways that affect pigmentation across multiple species will continue to expand the horizons of pigmentation biology, leading to new discoveries and a deeper understanding of the complexities of pigmentation.
Supplementary Material
Funding information
National Institutes of Health, National Human Genome Research Institute, Grant/Award Number: 1ZIAHG000136-18
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- Abdel-Malek ZA, Kadekaro AL, & Swope VB (2010). Stepping up melanocytes to the challenge of UV exposure. Pigment Cell & Melanoma Research, 23(2), 171–186. 10.1111/j.1755-148X.2010.00679.x. [DOI] [PubMed] [Google Scholar]
- Amberger JS, & Hamosh A (2017). Searching online mendelian inheritance in man (OMIM): A knowledgebase of human genes and genetic phenotypes. Current Protocols in Bioinformatics, 58, 1.2.1–1.2.12. 10.1002/cpbi.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammann S, Schulz A, Krägeloh-Mann I, Dieckmann NMG, Niethammer K, Fuchs S, … Ehl S (2016). Mutations in AP3D1 associated with immunodeficiency and seizures define a new type of Hermansky-Pudlak syndrome. Blood, 127(8), 997–1006. 10.1182/blood-2015-09-671636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora H, Chacon AH, Choudhary S, McLeod MP, Meshkov L, Nouri K, & Izakovic J (2014). Bloom syndrome. International Journal of Dermatology, 53(7), 798–802. 10.1111/ijd.12408. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, … Sherlock G (2000). Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics, 25(1), 25–29. 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnara JT, & Matsumoto J (2006). Comparative anatomy and physiology of pigment cells in nonmammalian tissues. In Nordlund JJ, Boissy RE, Hearing VJ, King RA & Ortonne J-P (Eds.), The pigmentary system: Physiology and pathophysiology (2nd ed., pp. 11–59). Oxford, UK: Wiley-Blackwell. [Google Scholar]
- Ballew BJ, & Savage SA (2013). Updates on the biology and management of dyskeratosis congenita and related telomere biology disorders. Expert Review of Hematology, 6(3), 327–337. 10.1586/ehm.13.23. [DOI] [PubMed] [Google Scholar]
- Bellono NW, Escobar IE, Lefkovith AJ, Marks MS, & Oancea E (2014). An intracellular anion channel critical for pigmentation. eLife, 3, e04543. 10.7554/eLife.04543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellono NW, & Oancea EV (2014). Ion transport in pigmentation. Archives of Biochemistry and Biophysics, 563, 35–41. 10.1016/j.abb.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DC, & Lamoreux ML (2003). The color loci of mice–a genetic century. Pigment Cell Research, 16(4), 333–344. 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- Bin B-H, Bhin J, Yang SH, Shin M, Nam Y-J, Choi D-H, … Lee TR (2015). Membrane-associated transporter protein (MATP) regulates melanosomal ph and influences tyrosinase activity. PLoS ONE, 10(6), e0129273. 10.1371/journal.pone.0129273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Eppig JT, Kadin JA, Richardson JE, Smith CL, Bult CJ,& the Mouse Genome Database Group (2017). Mouse Genome Database (MGD)-2017: Community knowledge resource for the laboratory mouse. Nucleic Acids Research, 45(D1), D723–D729. 10.1093/nar/gkw1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondurand N, & Sham MH (2013). The role of SOX10 during enteric nervous system development. Developmental Biology, 382(1), 330–343. 10.1016/j.ydbio.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Byers HR, Maheshwary S, Amodeo DM, & Dykstra SG (2003). Role of cytoplasmic dynein in perinuclear aggregation of phagocytosed melanosomes and supranuclear melanin cap formation in human keratinocytes. The Journal of Investigative Dermatology, 121(4), 813–820. 10.1046/j.1523-1747.2003.12481.x. [DOI] [PubMed] [Google Scholar]
- Carrero D, Soria-Valles C, & López-Otín C (2016). Hallmarks of progeroid syndromes: Lessons from mice and reprogrammed cells. Disease Models & Mechanisms, 9(7), 719–735. 10.1242/dmm.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G, & Chang E (1988). Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science, 242(4878), 564–567. [DOI] [PubMed] [Google Scholar]
- Cooper CD (2017). Insights from zebrafish on human pigment cell disease and treatment. Developmental Dynamics, 246(11), 889–896. 10.1002/dvdy.24550. [DOI] [PubMed] [Google Scholar]
- Crawford NG, Kelly DE, Hansen MEB, Beltrame MH, Fan S, Bowman SL, … Tishkoff SA (2017). Loci associated with skin pigmentation identified in African populations. Science, 358(6365), eaan8433. 10.1126/science.aan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, … … J. H. (2002). Premature aging in mice deficient in DNA repair and transcription. Science, 296(5571), 1276–1279. 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- Dias MF, Joo K, Kemp JA, Fialho SL, da Silva Cunha A, Woo SJ, & Kwon YJ (2018). Molecular genetics and emerging therapies for retinitis pigmentosa: Basic research and clinical perspectives. Progress in Retinal and Eye Research, 63, 107–131. 10.1016/j.preteyeres.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Dooley CM, Schwarz H, Mueller KP, Mongera A, Konantz M, Neuhauss SCF, … Geisler R (2013). Slc45a2 and V-ATPase are regulators of melanosomal pH homeostasis in zebrafish, providing a mechanism for human pigment evolution and disease. Pigment Cell & Melanoma Research, 26(2), 205–217. 10.1111/pcmr.12053. [DOI] [PubMed] [Google Scholar]
- D’Orazio J, Jarrett S, Amaro-Ortiz A, & Scott T (2013). UV radiation and the skin. International Journal of Molecular Sciences, 14(6), 12222–12248. 10.3390/ijms140612222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ML, Fufa TD, Palmer JW, Joshi SS, Larson DM, Incao A, … Pavan WJ (2018). A direct link between MITF, innate immunity, and hair graying. PLoS Biology, 16(5), e2003648. 10.1371/journal.pbio.2003648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Pacheco N, Flores C, Alonso S, Eng C, Mak ACY, Hunstman S, … Pino-Yanes M (2017). Identification of a novel locus associated with skin colour in African-admixed populations. Scientific Reports, 7, 44548. 10.1038/srep44548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoki Y, Araki R, Fujimori A, Ohhata T, Koseki H, Fukumura R, … … M. (2003). Growth retardation and skin abnormalities of the Recql4-deficient mouse. Human Molecular Genetics, 12(18), 2293–2299. 10.1093/hmg/ddg254. [DOI] [PubMed] [Google Scholar]
- Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, Frazer K, … Westerfield M (2013). ZFIN, the Zebrafish model organism database: Increased support for mutants and transgenics. Nucleic Acids Research, 41(D1), D854–D860. 10.1093/nar/gks938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Helip-Wooley A, Westbroek W, Gunay-Aygun M, & Gahl WA (2008). Disorders of lysosome-related organelle biogenesis: Clinical and molecular genetics. Annual Review of Genomics and Human Genetics, 9, 359–386. 10.1146/annurev.genom.9.081307.164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata K, Aoto T, Binh NT, Okamoto N, Tanimura S, Wakayama T, … Nishimura EK (2009). Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell, 137(6), 1088–1099. 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- Irion U, Singh AP, & Nüsslein-Volhard C (2016). The developmental genetics of vertebrate color pattern formation: Lessons from zebrafish. Current Topics in Developmental Biology, 117, 141–169. 10.1016/bs.ctdb.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Kelsh RN (2004). Genetics and evolution of pigment patterns in fish. Pigment Cell Research, 17(4), 326–336. 10.1111/j.1600-0749.2004.00174.x. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Harris ML, Colanesi S, & Erickson CA (2009). Stripes and belly-spots – A review of pigment cell morphogenesis in vertebrates. Seminars in Cell & Developmental Biology, 20(1), 90–104. 10.1016/j.semcdb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch SC, Simon N, Ebert C, & Carell T (2016). Molecular mechanisms of xeroderma pigmentosum (XP) proteins. Quarterly Reviews of Biophysics, 49, e5. 10.1017/S0033583515000268. [DOI] [PubMed] [Google Scholar]
- Lamason RL, Mohideen M-A-P-K, Mest JR, Wong AC, Norton HL, Aros MC, … Cheng KC (2005). SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science, 310(5755), 1782–1786. 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- Lebel M, & Monnat RJ (2017). Werner syndrome (WRN) gene variants and their association with altered function and age-associated diseases. Ageing Research Reviews, 41, 82–97. 10.1016/j.arr.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Lloyd-Jones LR, Robinson MR, Moser G, Zeng J, Beleza S, Barsh GS, … Visscher PM (2017). Inference on the genetic basis of eye and skin color in an admixed population via bayesian linear mixed models. Genetics, 206(2), 1113–1126. 10.1534/genetics.116.193383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Jin W, & Wang LL (2017). Aging in Rothmund-Thomson syndrome and related RECQL4 genetic disorders. Ageing Research Reviews, 33, 30–35. 10.1016/j.arr.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Mahalwar P, Singh AP, Fadeev A, Nüsslein-Volhard C, & Irion U (2016). Heterotypic interactions regulate cell shape and density during color pattern formation in zebrafish. Biology Open, 5(11), 1680–1690. 10.1242/bio.022251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MB, Hodges CA, Barnes E, Vogel H, Hassold TJ, & Luo G (2005). Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Human Molecular Genetics, 14(6), 813–825. 10.1093/hmg/ddi075. [DOI] [PubMed] [Google Scholar]
- Marks MS, Heijnen HFG, & Raposo G (2013). Lysosome-related organelles: Unusual compartments become mainstream. Current Opinion in Cell Biology, 25(4), 495–505. 10.1016/j.ceb.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Lin M, Granka JM, Myrick JW, Liu X, Sockell A, … Henn BM (2017). An unexpectedly complex architecture for skin pigmentation in Africans. Cell, 171(6), 1340–1353.e14. 10.1016/j.cell.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan TF, Conte N, West DB, Jacobsen JO, Mason J, Warren J, … Smedley D (2017). Disease model discovery from 3,328 gene knockouts by The International Mouse Phenotyping Consortium. Nature Genetics, 49(8), 1231–1238. 10.1038/ng.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, & Thomas PD (2017). PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Research, 45(D1), D183–D189. 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoliu L, & Marks MS (2017). A new type of syndromic albinism associated with mutations in AP3D1. Pigment Cell & Melanoma Research, 30(1), 5–7. 10.1111/pcmr.12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort RL, Jackson IJ, & Patton EE (2015). The melanocyte lineage in development and disease. Development, 142(4), 620–632. 10.1242/dev.106567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton JM, Cohen-Barak O, Hagiwara N, Gardner JM, Davisson MT, King RA, & Brilliant MH (2001). Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. American Journal of Human Genetics, 69(5), 981–988. 10.1086/324340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomera-Sanchez Z, & Zurita M (2011). Open, repair and close again: Chromatin dynamics and the response to UV-induced DNA damage. DNA Repair, 10(2), 119–125. 10.1016/j.dnarep.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Parichy DM, & Spiewak JE (2015). Origins of adult pigmentation: Diversity in pigment stem cell lineages and implications for pattern evolution. Pigment Cell & Melanoma Research, 28(1), 31–50. 10.1111/pcmr.12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt SC, Bailey KJ, & Freeland A (2007). Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells, 25(12), 3121–3132. 10.1634/stemcells.2007-0483. [DOI] [PubMed] [Google Scholar]
- Rawofi L, Edwards M, Krithika S, Le P, Cha D, Yang Z, … Parra EJ (2017). Genome-wide association study of pigmentary traits (skin and iris color) in individuals of East Asian ancestry. PeerJ, 5, e3951. 10.7717/peerj.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann M, & Ludwig A (2013). Pleiotropic effects of coat colour-associated mutations in humans, mice and other mammals. Seminars in Cell & Developmental Biology, 24(6–7), 576–586. 10.1016/j.semcdb.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Ruzicka L, Bradford YM, Frazer K, Howe DG, Paddock H, Ramachandran S, … Westerfield M (2015). ZFIN, The zebrafish model organism database: Updates and new directions. Genesis, 53(8), 498–509. 10.1002/dvg.22868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, & DePinho RA (2010). Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature, 464(7288), 520–528. 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M, Larue L, Goda M, Bosenberg MW, Hashimoto H, & Kelsh RN (2016). What is a vertebrate pigment cell? Pigment Cell & Melanoma Research, 29(1), 8–14. 10.1111/pcmr.12409. [DOI] [PubMed] [Google Scholar]
- Setty SRG, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, & Marks MS (2008). Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature, 454(7208), 1142–1146. 10.1038/nature07163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram A, & Marks MS (2012). Mechanisms of protein delivery to melanosomes in pigment cells. Physiology, 27(2), 85–99. 10.1152/physiol.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski A, Tobin DJ, Shibahara S, & Wortsman J (2004). Melanin pigmentation in mammalian skin and its hormonal regulation. Physiological Reviews, 84(4), 1155–1228. 10.1152/physrev.00044.2003. [DOI] [PubMed] [Google Scholar]
- Teive HAG, Moro A, Moscovich M, Arruda WO, Munhoz RP, Raskin S, & Ashizawa T (2015). Ataxia-telangiectasia - A historical review and a proposal for a new designation: ATM syndrome. Journal of the Neurological Sciences, 355(1–2), 3–6. 10.1016/j.jns.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2017). Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Research, 45(D1), D331–D338. 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbakel SK, van Huet RAC, Boon CJF, den Hollander AI, Collin RWJ, Klaver CCW, … Klevering BJ (2018). Non-syndromic retinitis pigmentosa. Progress in Retinal and Eye Research, 10.1016/j.preteyeres.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Vogel P, Read RW, Vance RB, Platt KA, Troughton K, & Rice DS (2008). Ocular albinism and hypopigmentation defects in Slc24a5−/− mice. Veterinary Pathology, 45(2), 264–279. 10.1354/vp.45-2-264. [DOI] [PubMed] [Google Scholar]
- Wang Y, Viennet C, Robin S, Berthon J-Y, He L, & Humbert P (2017). Precise role of dermal fibroblasts on melanocyte pigmentation. Journal of Dermatological Science, 88(2), 159–166. 10.1016/j.jdermsci.2017.06.018. [DOI] [PubMed] [Google Scholar]
- Weider M, & Wegner M (2017). SoxE factors: Transcriptional regulators of neural differentiation and nervous system development. Seminars in Cell & Developmental Biology, 63, 35–42. 10.1016/j.semcdb.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Wu X, & Hammer JA (2014). Melanosome transfer: It is best to give and receive. Current Opinion in Cell Biology, 29, 1–7. 10.1016/j.ceb.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, & Hearing VJ (2009). Physiological factors that regulate skin pigmentation. BioFactors (Oxford, England), 35(2), 193–199. 10.1002/biof.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, & Hearing VJ (2014). Melanocytes and their diseases. Cold Spring Harbor Perspectives in Medicine, 4(5), a017046. 10.1101/cshperspect.a017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


