Abstract
Liquid biopsy is a noninvasive technique that can provide valuable information for disease characterization by using biofluids as a source of biomarkers. Proteins found in biofluids can offer a wealth of information for understanding pathological processes. In this study, we used early-stage clear cell renal cell carcinoma (ccRCC) as a model to explore the proteomic relationships among tissue, plasma, and urine. We analyzed samples of tumor tissue, plasma, and urine from a cohort of 27 ccRCC patients with T1-2 stage and 27 matched healthy controls, using liquid chromatography–mass spectrometry (LC-MS) for proteomic analysis. We integrated the differential proteins found in the three types of samples to explore ccRCC-associated molecular changes. Our results showed that both plasma and urine proteomes could reflect functional changes in tumor tissue. In plasma, cytoskeletal proteins and metabolic enzymes were differentially expressed, while in urine, adhesion molecules and defense proteins showed differential levels. The differential proteins found in plasma and urine both reflect the binding and catalytic activity of tumor tissue. Additionally, proteins only changed in biofluids could reflect body immune response changes, with plasma proteins involved in actin cytoskeleton and oxidative stress, and urine proteins involved in granulocyte adhesion and leukocyte extravasation signaling. Plasma and urine proteins could effectively distinguish RCC from control, with good performances (plasma/urine: 92.6%/92.6% specificity, 96.3%/92.6% sensitivity, and an area under the curve of 0.981/0.97). In conclusion, biofluids could not only reflect functional changes in tumor tissue but also reflect changes in the body's immune response. These findings will benefit the understanding of body biomarkers in tumors and the discovery of potential disease biomarkers.
Keywords: liquid biopsy, plasma, urine, tissue, proteomics, renal cell carcinoma, biomarkers
Graphical Abstract
Highlights
-
•
Tissue, plasma, and urine proteome of ccRCC were integratively profiled.
-
•
Tumor functions reflected by plasma and urine were explored.
-
•
ccRCC plasma and urine biomarkers were tentatively investigated.
In Brief
Liu et al. characterized the proteome of tissue, plasma, and urine from 27 ccRCC patients and 27 healthy controls. All samples were analyzed using data-independent analysis. Both plasma and urine proteome could reflect functional changes of tumor tissue. In plasma, cytoskeletal proteins and metabolic enzymes were differentially expressed. And in urine, adhesion molecular and defense proteins showed differential levels. Plasma and urine proteins could distinct RCC from control with good performances.
Liquid biopsy is a technique that uses human body fluids, including blood, urine, saliva, and even cerebrospinal fluid, as a source of sample to obtain disease-related information. Liquid biopsy is a convenient tool for biomarker discovery or pathological diagnosis. Compared to traditional tissue biopsy, liquid biopsy has advantages of minimally invasive nature and significantly less morbidity. It could be scheduled more frequently to provide a personalized snapshot of disease at successive time points. Additionally, liquid biopsy could reflect tumor heterogeneity present in patients, unlike tissue biopsies which are obtained from only one tumor region (1).
Liquid biopsy, as a powerful supplement to traditional methods, has shown great success in the research and development of new biomarkers in recent years. Circulating tumor cell, circulating tumor DNA, exosome, proteins, and several small moleculars are the mainly testing targets of liquid biopsy (2, 3, 4, 5). However, the clinical application of circulating tumor DNA, circulating tumor cell, and exosomes in tumor is limited by the extraction and separation technology, since their content in biofluids is very low. In contrast, the content of protein in biofluids is relatively high, providing valuable information for disease characterization. Considering that the majority of drug targets are proteins and protein-based analysis is the most common technique utilized in the clinical setting (6), delineating target proteins have direct translational application. A myriad of studies had utilized proteomic technologies to explore the protein profiles of biofluids to identify the differentially expressed proteins associated with tumor.
The network of arteries, veins, and capillaries in contact with organs offers a means for the proteins secreted, shed, or released by tumor tissues. As proteomic technologies continue to be matured, plasma proteome has great potential for biomarker research and clinical applications. Plasma proteins have been used for biomarker discovery for various of tumors, including cancer, autoimmune diseases, viral diseases, and cardiovascular diseases (7, 8). Previous study indicated that plasma E-cadherin performed most favorably from a large panel of plasma proteins in terms of diagnostic and predictive potential in curatively treatable prostate cancer (9). Ting Shu et al. developed a panel of 11 plasma proteins as COVID-19 biomarkers, which could accurately distinguish and predict COVID-19 outcomes (10).
Urine is a proximal biological fluid that may offer a richer source of proteins of interest. Urine has a narrower dynamic range of protein concentration and the reduced abundance of proteins (11). Urine-based proteomics have been widely applied for biomarker discovery and clinical applications. Early in 2015, Tomasz P Radon et al discovered a three-biomarker panel in urine, LYVE-1, REG1A, and TFF1, for pancreatic adenocarcinoma diagnosis (12). Urinary protein panel could distinguish even stage I gastric cancer patients from healthy controls (HC) patients with the area under the curve (AUC) of 0.850 (13). For clear cell renal cell carcinoma (ccRCC) small mass tumor diagnosis, urine proteomics also showed valuable application. Ashley Di Meo et al evaluated 115 urine samples, including 33 renal oncocytoma (≤4 cm), 30 progressive, and 26 nonprogressive ccRCC-small renal mass cases, in addition to 26 healthy controls. A two-protein signature (EPS8L2 and CCT6A) showed significant discriminatory ability (areas under the curve: 0.81) in distinguishing progressive from nonprogressive ccRCC-small renal masses (14).
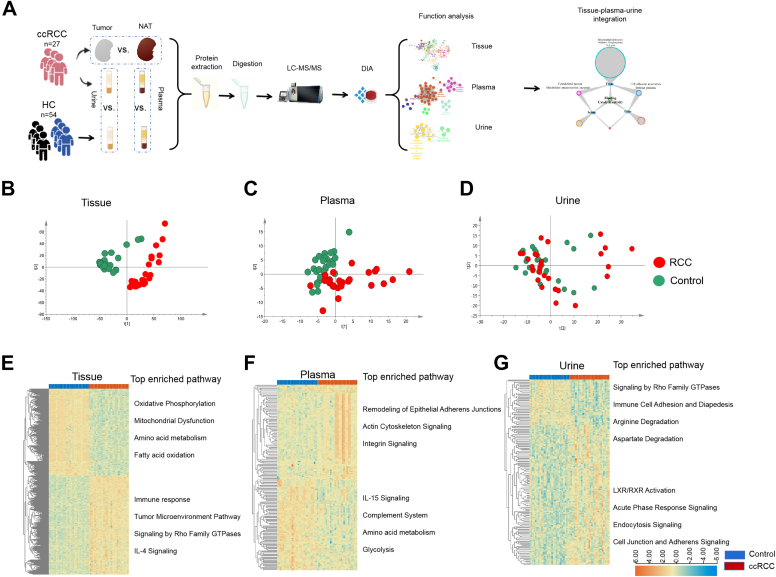
Although most of the published studies have focused on either tumor tissue or a specific biofluid for proteomics analysis, there are limited available data examining the use of a biofluid to serve as a proxy for tumor proteomic changes through simultaneous examination of tissue and biofluids. It is essential to determine how proteomic changes occurring in a tumor are detected in plasma and urine. In present study, early ccRCC (stage I-II) was used as the analysis model. The tumor tissue, plasma, and urine from 27 ccRCC patients (26 with T1 stage and 1 with T2 stage) and 54 matched healthy controls (27 controls for plasma and 27 controls for urine, supplemental Table S1) were analyzed by data-independent acquisition (DIA). In present study, we aimed to explore how proteomic changes occurring in a tumor could be detected in plasma and urine. Thus, we performed integrative analysis of urine/plasma proteome with tissue proteome to discover molecular with high correlation with tumor tissue that may result in the discovery of potential diagnostic targets and biomarkers with high specificity. According to comparisons of the proteomes of tissue, plasma, and urine, we tried to reveal the extent to which plasma and urine could reflect functional changes of tumor in tumor early stage (Fig. 1A).
Fig. 1.
Differentially expressed proteins in tissue, plasma, and urine.A, the workflow of the study. B–D, PCA score plot of proteome of tissue, plasma, and urine. Tissue and plasma proteome showed apparent separation between ccRCC and controls, while urine proteome showed several overlay between ccRCC and controls. E–G, heat map of differentially expressed proteins and the enriched pathways in tissue, plasma, and urine. x-axis t(1) means component 1; y-axis t(2) means component 2. ccRCC, clear cell renal cell carcinoma; PCA, principal component analysis.
Experimental Procedures
Experimental Design and Statistical Rationale
The purpose of this study was to find the proteomic relationships among tissue, plasma, and urine to assistant biofluid biomarker discovery. Overall 81 subjects were enrolled, including 27 ccRCC patients, and 54 healthy controls (27 controls for plasma and 27 controls for urine) were enrolled. Tissue, plasma, and urine spectral libraries were generated using 2D-LC-MS–based data-dependent acquisition (DDA) method, individually. The tumor tissue, adjacent normal tissue, plasma, and urine from ccRCC patients and healthy controls were analyzed by DIA. The variable isolation window of DIA method with different windows was developed for different samples (Detailed data in supplemental Table S2). Samples were injected in a random order for tissue, plasma, and urine, individually. Quality control (QC) sample was injected frequently to monitoring reproducibility of the method. A total of 30 QC samples were injected and the correlation of QC samples was above 0.9, indicating good repeatability. Principal component analysis and heat map virtualization were implemented using the Wu kong data analysis platform (https://www.omicsolution.org/wkomics/main/). Nonparameter Wilcoxon rank-sum test (p < 0.05) and fold change was performed for significance evaluation of proteins between groups for individual omic matrix analysis. For integrative analysis between two omic matrices, only fold change was considered for differential analysis (fold change >1.5) for more pathways and functions coverage. Function and pathway were analyzed using ingenuity pathway analysis (IPA) (http://www.ingenuity.com/products/ipa#/?tab=features) software (QIAGEN), a repository of biologic interactions and functions created from millions of individually modeled relationships that range from the molecular (proteins, genes) to organism (diseases) level.
Sample Collection
This study was approved by the Institutional Review Board of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and abide by the Helsinki Declaration of 1975 (as revised in 2008) concerning Human and Animal Rights and that they followed out the policy concerning Informed Consent as shown on Springer.com. A total of 81 participants, 27 ccRCC patients, 27 healthy controls for plasma control, and 27 healthy controls for urine control were included in this study. All human subjects provided informed consent before participating in this study. Tissue samples were collected from 27 ccRCC patients who were undergoing surgical resection and had received no prior treatment for their disease. Urine and plasma samples were collected from ccRCC patients and matched healthy controls. The urine samples were collected from the first urination in the morning. Samples were centrifuged within 6 h of collection; the supernatants were isolated, aliquoted, and stored at −80 °C until analysis. The plasma samples were collected in the morning from 07:00 AM−09:00 AM after an overnight fast to eliminate dietary disturbances. After collected, all plasma samples were separated following centrifugation at 1000 g for 10 min at 4 °C and were stored at −80 °C.
Sample Preparation
For tissue, approximately 25 to 120 mg of each cryopulverized renal tumor tissues or normal adjacent tissues (NATs) were homogenized separately in an appropriate volume of lysis buffer (2% SDS, 20 mM Tris, Cocktail [1:100 dilution], DNAse [1:100 dilution], RNAse [1:1000 dilution]) by repeated vortexing. Protein concentration was determined by BCA assay (Pierce). Hundred micrograms of protein from each sample were digested with the filter-aided sample preparation (FASP) method. The protein samples were reduced with 20 mM DTT for 5 min at 95 °C and then carboxyamidomethylated with 50 mM iodoacetamide (IAA) at room temperature (RT) in the dark for 45 min. Then, the sample was loaded onto a 30KD ultracentrifugation filter, washed twice with UA buffer (containing 7 M urea and 50 mM Tris), and washed twice with 25 mM NH4HCO3. The treated samples were digested with trypsin (2 μg per 100 g protein) in 25 mM NH4HCO3. The samples were digested at 37 °C overnight. The digested peptides were eluted from the 30 KD filter, and the samples desalted on C18 columns (3 cc, 60 mg, Oasis, Waters Corporation). The desalted peptides were lyophilized by vacuum centrifugation and stored at −80 °C
For urine, 2 ml urine was reduced with 20 mM DTT for 5 min at 95 °C and subsequently alkylated with 50 mM IAA for 45 min at RT in the dark. Then, 6 times volume precooled acetone was added to extract protein. The precipitated protein was redissolved using 200 μl 20 mM Tris buffer. Protein digestion was carried out using FASP method mentioned above.
For plasma, before protein digestion, top 12 high-abundant protein depletion was carried out for 10 μl plasma using spin column (Agilent). Finally, 400 μl high-abundant protein-depleted plasma supernatant was reduced with 20 mM DTT for 5 min at 95 °C and subsequently alkylated with 50 mM IAA for 45 min at RT in the dark. Then, 6 times volume precooled acetone was added to extract protein. The precipitated protein was redissolved using 200 μl 20 mM Tris buffer. Protein digestion was carried out using FASP method mentioned above.
For each type sample, a pooled sample of equal protein amount from each sample was used for spectral library generation and QC analysis.
Offline HPLC Separation for Proteome Library Generation
The pooled peptide sample of each type sample was separated by high-pH RPLC columns (4.6 mm × 250 mm, C18, 3 μm; Waters), respectively. Each pooled sample was loaded onto the column in buffer A1 (99.9% H2O, pH 10). The elution gradient was 5%–30% buffer B1 (90% acetonitrile, pH 10; flow rate, 1 ml/min) for 30 min. The eluted peptides were collected at one fraction per minute. After lyophilization, the 30 fractions were resuspended in 0.1% formic acid and then were concatenated into 15 fractions by combining fractions 1, 16, and so on.
On-Line LC-MS/MS Data Acquisition
The digested peptides were dissolved in 0.1% formic acid and separated on an RP C18 precast integrated capillary LC column (50 μm × 50 cm). The eluted gradient was 5%–30% buffer B2 (0.1% formic acid, 99.9% ACN; flow rate, 0.3 μl/min) for 60 min. A Thermofisher orbitrap fusion lumos mass spectrometer (Thermo Fisher Scientific) was used to analyze the eluted peptides from LC. To generate the spectral library, the fractions from RPLC were analyzed in the DDA mode. The parameters were set as follows: the MS was recorded at 350 to 1500 m/z at a resolution of 60,000 m/z; the maximum injection time was 50 ms, the auto gain control (AGC) was 1E6, and the cycle time was 3 s. MS/MS scans were performed at a resolution of 15,000 with an isolation window of 1.6 Da and a collision energy at 32% (high collision dissociation); the AGC target was 50,000, and the maximum injection time was 30 ms.
For DIA analysis, the variable isolation window DIA method with different windows was developed for different samples (Detailed data in supplemental Table S2). The specific window lists were constructed based on the DDA experiment of the pooled sample. The full scan was set at a resolution of 120,000 over the m/z range of 400 to 900, followed by DIA scans with a resolution of 30,000; the HCD collision energy was 32%, the AGC target was 1E6, and the maximal injection time was 50 ms.
Spectral Library Generation
Tissue, plasma, and urine spectral libraries were generated, individually. To generate a comprehensive spectral library, the pooled sample from each sample was processed. The DDA data were processed using Proteome Discoverer (version 2.3, Thermo Fisher Scientific) software and searched against the human SwissProt database (Homo sapiens, 20,358 SwissProt, 2019_05 version) appended with the indexed retentions time fusion protein sequence (Biognosys). A maximum of two missed cleavages for trypsin was used, and cysteine carbamidomethylation was set as a fixed modification. Methionine oxidation, lysine deamination, and carbamylation (+43) were set as variable modifications. The parent and fragment ion mass tolerances were set to 10 ppm and 0.02 Da, respectively. The applied false discovery rate (FDR) cutoff was 0.01 at the protein level. The results were then imported to Spectronaut Pulsar (version 14, Biognosys) software to generate the spectral library. Additionally, DIA data were also imported into Spectronaut Pulsar software and searched against the human SwissProt database to generate DIA library. The final spectral library was generated by combining DDA and DIA libraries.
Data Analysis
The DIA raw data were loaded to the Spectronaut 14 to calculate peptide retention time based on indexed retentions time data, and Spectronaut provided protein identification and quantitation by matching the retention time, m/z, etc., to peptide library. The retention time prediction type was set to dynamic indexed retentions time, and interference correction at the MS2 level was enabled. The MS1 and MS2 tolerance strategy was set to dynamic. It applied a correction factor to the automatically determined mass tolerance. The correction factor for ms1 and ms2 was all set as 1. The precursor posterior error probability cutoff was set to 1, and precursors that do not satisfy the cutoff will be imputed. The top N (min: 1; max: 3) precursors per peptide were used for quantify calculation. The top N ranking order is determined by a cross-run quality measure. Peptide intensity was calculated by summing the peak areas of their respective fragment ions for MS2. Cross-run normalization was enabled to correct for systematic variance in the LC-MS performance, and a local normalization strategy was used. Normalization was based on the assumption that on average, a similar number of peptides are upregulated and downregulated, and the majority of the peptides within the sample are not regulated across runs and along retention times. Protein inference, which gave rise to the protein groups, was performed on the principle of parsimony using the ID picker algorithm as implemented in Spectronaut Pulsar. All results were filtered by a Q value cutoff of 0.01 (corresponding to an FDR of 1%). Protein intensity was calculated by summing the intensity of their respective peptides (15). Proteins identified in more than 50% of the samples in each group were retained for further analysis. Missing values were imputed based on the k-nearest neighbor method. To make data comparable, the data matrices were auto-scaled as z-score value for subsequent statistical analysis using MetaboAnalyst 5.0 software (https://www.metaboanalyst.ca/). For individual proteomic analysis, differential proteins were defined by p value (nonparameter Wilcoxon rank-sum test) and fold change. For integrative analysis between two proteomic matrices (tissue vs. Plasma, tissue vs. Urine, plasma vs. urine), differential proteins were defined using a cutoff with only fold change for more comprehensive pathways and functions analysis. Differential proteins were submitted to pathway analysis using IPA software (Ingenuity Systems). Gene ontology (GO) terms of molecular function and protein class were used for common proteins enrichment analysis using 'Wu Kong' platform (https://www.omicsolution.com/wkomics/main/). Human swissprot database was used as background. Functional analysis of serum and urine-only changed proteins were performed by ClueGo and CluePedia in cytoscape (version 3.9). The ontology sources we used is “GO-biological process-EBI-uniprot-2022, including 17,400 terms/pathways with 18,085 unique genes”. Network specificity was set as medium. The GO tree internal was set from 3 to 8. For biomarker selection, nonparameter wilcoxon rank-sum test and random forest were performed for significance evaluation of proteins between ccRCC and HC based on all samples. The importance of features was ranked by “p adjusted value” and “mean decrease accuracy”, individually. Features in the top 20 in both models were chosen as potential biomarkers. Then, the potential biomarkers were used to construct prediction model using logistic regression algorithm with 10-fold cross validation. We also used LOOCV method by “pROC” and “caret” package in R to validate the performance of the features.
To make sure the data are comparable with Clinical Proteomic Tumor Analysis Consortium (CPTAC) data, we downloaded the raw DIA data of tissues from ccRCC and control subjects performed by CPTAC. The raw data was re-analyzed using spectronaut software. The analysis parameters and search library were set as the same with our data in present study. Differential proteins were further defined based on the same criteria with our data using p value and fold change.
Results
Spectral Library Generation and Protein Identification in DIA–MS
In present study, to estimate system stability during whole analysis process, the pooled sample was used as a QC to observe the stability of the instrument signal. The QC samples were firstly analyzed by three repeats to ensure the system stability. During the whole analysis process, QC was analyzed before and after all samples and among every 7 to 9 samples. The average Pearson’s correlation coefficient of QC samples was approximately 1 for tissue, plasma, and urine, indicating good system stability (supplemental Fig. S1A). Second, we analyzed protein-wise coefficient of variation (CV) distribution for QC samples and their association with average protein abundance. The medium of protein CV were 0.162, 0.163, and 0.227 for tissue, plasma, and urine, respectively. The dynamic range spanned eight orders of magnitude. As more abundant proteins are generally easier to precisely quantify, protein abundance was negatively correlated with the CVs (supplemental Fig. S1B).
Spectral libraries of ccRCC tissue, plasma, and urine were constructed by DDA and DIA analysis, respectively. Finally, the merged library from the above two analysis strategies was constructed by Spectronaut pulsar. Overall 1372,85/32,673/60,432 precursors, 87,623/15,602/34,560 peptides, and 9643/1563/4514 protein groups were obtained from tissue/plasma/urine spectral library (supplemental Table S3A).
According to above spectral library, 8166 proteins (an average of 6745) in tissue, 1340 proteins (an average of 1017) in plasma, and 4468 proteins (an average of 3497) in urine were detected with a protein FDR <1% (supplemental Table S3, B–D and supplemental Fig. S1C). Proteins with quantitative data in more than 50% of samples in each group were selected for further analysis. Finally, 5927/827/2796 proteins in tissue/plasma/urine were quantified (supplemental Table S3, E–G).
Differential Analysis of ccRCC Proteomes in Tissue, Plasma, and Urine
To systematically investigate relevance of molecular changes between biofluids and tissue, we performed comprehensive analyses of 27 pair of ccRCC tissue and the NAT proteomes, as well as 27 paired of plasma and urine proteomes from ccRCC patients and healthy controls. The integrative analyses were carried out via a tissue-centric manner. Differentially expressed protein and the corresponding biological functions in plasma and urine were compared with those in tissue to investigate the tissue-plasma-urine correlation (Fig. 1A).
First, we analyzed the differential proteomes of the three type samples, respectively. The ccRCC and control samples were distinguishable by principle component analysis of the proteomic results (Fig. 1, B–D). Though urine results showed partial overlay between ccRCC and control samples to some extent, the Orthogonal Partial Least Squares Discrimination Analysis (OPLS-DA) showed statistical significance between two groups (supplemental Fig. S1B).
As a result, the tissue results identified 1480 differential proteins between RCC and NATs, with 691 proteins downregulated and 789 upregulated (Fig. 1E). Functional enrichment analysis revealed significant disturbance of metabolic and energy metabolism, immune response, and cancer mechanism pathway in ccRCC. Decreased OXPHOS and TCA cycle was the response of tumor for low oxygen microenvironment under regulation of transcriptional regulator HIF1α accumulation, which was the fundamental tumor-initiating event during ccRCC (16), while immune response, IL-4 signaling, and tumor microenvironment pathway showed upregulated in tumor (Fig. 1E).
CPTAC group had reported tissue proteomics analysis of 110 treatment-naive ccRCC and 84 paired-matched NAT samples (17). We compared our tissue results with CPTAC data. More than 80% proteins (4859 proteins) were commonly identified, and almost 50% differential proteins (699 proteins) showed significantly differential levels in both studies (supplemental Fig. S2). The top pathways of both studies showed high degree of consistency, with upregulation of immune response pathways (IL-8 signaling, sirtuin signaling pathway, Th1 pathway, etc.) and downregulation of energy metabolism (oxidative phosphorylation, TCA cycle, serotonin degradation, and valine degradation), while, several pathways were significantly enriched only in present study, including clotting-related pathways (extrinsic prothrombin activation pathway, intrinsic prothrombin activation pathway), metabolism pathway (α-tocopherol degradation, aspartate degradation II, aspartate biosynthesis, glutamate degradation II), and immune-related pathways (autophagy, remodeling of epithelial adherens junctions, and Fcγ receptor-mediated phagocytosis in macrophages and monocytes). Metabolism and immune disorders have been reported as the main events occurred in ccRCC (18, 19). Clotting-related pathways showed significantly changed discovered in present study, which have been reported in urological cancers, including renal tumor. Coagulation pathway seems to be activated in urological malignancies. Specific panels of coagulation factors might play a role as screening or prognostic tools in earlier stages of renal cancer (20).
Similarly, plasma proteomic analysis identified 180 differential proteins, with 91 downregulated and 89 upregulated (Fig. 1F). The top three protein classes were cytoskeletal protein (18.2%), defense/immunity protein (10.2%), and metabolite interconversion enzyme (9.1%) (supplemental Fig. S3A). IPA analysis showed that the upregulated proteins were mainly involved in pathways of cellular growth and development, including remodeling of epithelial adhere junction, actin cytoskeleton signaling, and integrin signaling, etc. Plasma downregulated proteins were involved in IL-15 signaling, complement system, amino acid metabolism, and glycolysis (Fig. 1F).
Urine proteomic analysis identified 169 differential proteins, with 41 downregulated and 128 upregulated (Fig. 1G). The top three protein classes were metabolite interconversion enzyme (13.2%), protein modifying enzyme (7.8%), and cell adhesion molecule (6%) (supplemental Fig. S3B). The top enriched pathways of urine differential proteins were immune response (acute phase response signaling, immune cell adhesion, and diapedesis, and endocytosis signaling) and metabolic pathway (arginine degradation and aspartate degradation) (Fig. 1G).
Integrative Analysis of Plasma and Tissue Proteomes
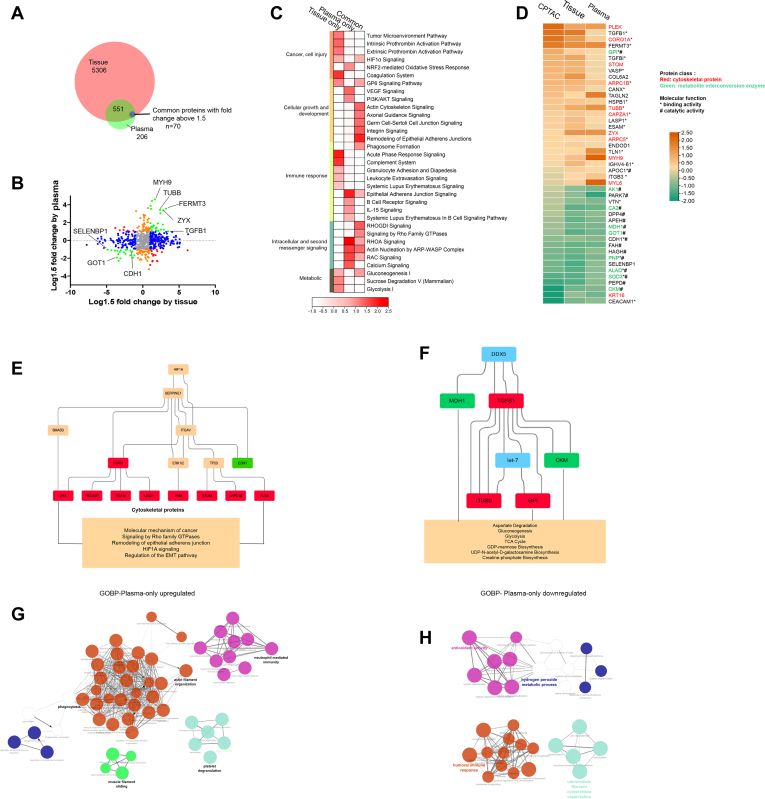
To investigate plasma proteins associated with ccRCC tumor tissue, we analyzed plasma and tissue proteomic results integratively. The tissue proteome covered majority of proteins quantified in the plasma (75%, 621) (Fig. 2A). We applied a cutoff (fold change >1.5) for the common proteins in the two samples, resulting in 70 differential proteins (“common” category) in both tissue and plasma, 262 only in tissue (“tissue only” category), and 87 only in plasma (“plasma-only” category) (Fig. 2B and supplemental Table S4A).
Fig. 2.
Integration of plasma and tissue proteomes discovers consistent changes of cytoskeletal proteins and metabolite interconversion enzymes in early-stage ccRCC.A, Venn diagram of identified proteins in tissue and plasma datasets. Common differential proteins: Proteins with fold change above 1.5 (RCC vs. Control) in both tissue and plasma. B, the distribution of the relative dominance of no change (gray), agree (green), tissue-only (blue), plasma-only (orange), and disagree (red) proteins. Proteins were defined based on the consistency or inconsistency between the tissue and plasma datasets. Agree proteins were those with same direction of change in both tissue and plasma datasets. No change proteins, in both tissue and plasma, were not changed (within 1.5 fold change) between ccRCC and control. Tissue-only proteins varied between ccRCC and control in tissue dataset (larger than 1.5 fold), but not in plasma (within 1.5 fold). Plasma-only proteins varied in plasma (larger than 1.5 fold), but not in tissue (within 1.5 fold). Disagree genes performed opposite direction of change in tissue and plasma. C, pathway enrichment of tissue-plasma agree proteins, tissue-only proteins, and plasma-only proteins. D, proteins varied constantly in CPTAC tissue dataset, present tissue dataset, and plasma dataset. Overall, 54.5% of these “consistent proteins” were cytoskeletal proteins and metabolites interconversion enzymes (marked in red and green). E and F, casual network analysis predicted the upstream regulator, ITGAV, SERPINE1, HIF1, and DDX5, contributing to downstream protein upregulation and pathway disturbance associated with ccRCC pathology. Molecular in red boxes: upregulated proteins in present study. Molecular in green boxes: downregulated proteins in present study; molecular in orange boxes: predicted to be activated status; molecular in blue boxes: predicted to be inhibited status. G and H, GOBP (Go ontology for biological process) terms for plasma-only upregulated proteins(G) and plasma-only downregulated proteins(H). ccRCC, clear cell renal cell carcinoma; CPTAC, Clinical Proteomic Tumor Analysis Consortium.
Comparison analysis of enriched pathway by the three protein categories suggested that common proteome could reflect tumor-associated function disturbance, including cellular growth and development, intracellular and second messenger signaling, and energy supply (gluconeogenesis) (Fig. 2C and supplemental Table S4B).We further investigated differential proteins with the same change trend in tissue and plasma. Overall 45 out of 70 “common” proteins showed consistent change trends. Additionally, these 45 proteins were also quantified in CPTAC study and showed the same change trend in ccRCC with present study (Fig. 2D). Protein class analysis showed 54.5% of these 45 proteins were cytoskeletal proteins and metabolites interconversion enzymes, mainly performed binding and catalytic functions, which indicated that a substantial regulation of cytoskeletal structure and energy metabolism in tumor tissue might be detected in plasma.
These “common” proteins were further functionally analyzed using casual network to explore the upregulator contributing to functional disturbance. Hypoxia factor, HIF1, was found to regulate ITGAV and SERPINE1, which might trigger upregulation of cytoskeletal proteins, then involving in disorder of pathways, including molecular mechanism of cancer, signaling by rho family GTPases, remodeling of epithelial adherens junction, hypoxia inducible factor (HIF)1A signaling, and regulation of the epithelial-mesenchymal transition (EMT) pathway (Fig. 2E). Additionally, a casual network regulating metabolic pathways was found. DDX5 and miRNA let-7 probably contributed to metabolic enzymes changes, which were involved in energy metabolism including aspartate degradation, gluconeogenesis, glycolysis, and TCA cycle (Fig. 2F).
“Plasma-only” proteins were mainly involved in pathways regulating immune response, including epithelial adherens junction signaling, B cell receptor signaling, IL-15 signaling, and systemic lupus erythematosus in B cell signaling pathway (Fig. 2C and supplemental Table S4C). To further identify specific plasma proteomic changes to biological processes, we performed enrichment analyses for GOBP (Go for biological process) terms using “plasma-only” proteins by ClueGo in Cytoscape. The largest cluster of enriched terms for upregulated proteins was related to actin filament organization and phagocytosis (Fig. 2G). Conversely, the downregulated proteins were annotated to antioxidant activity, humoral immune response, and cytoskeleton organization (Fig. 2H).
Integrative Analysis of Urine and Tissue Proteomes
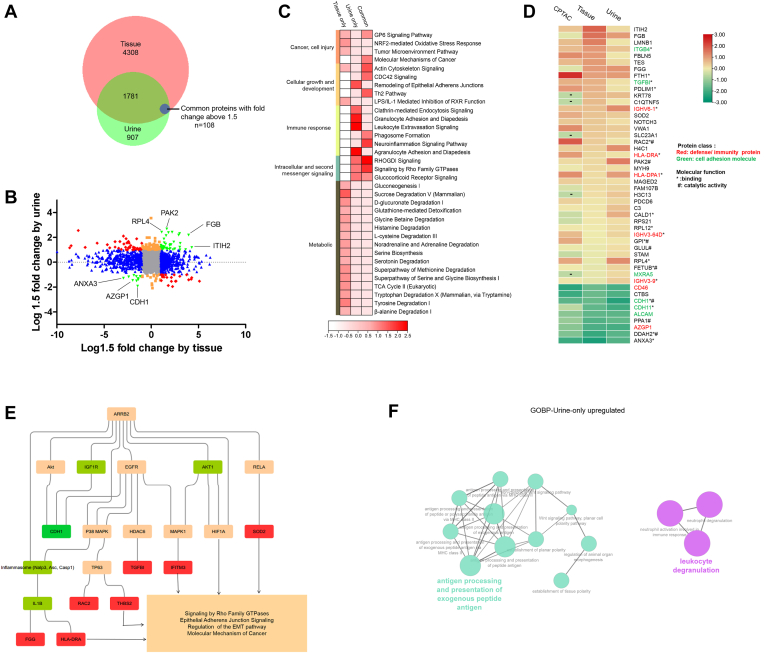
Using similar strategy, we further investigated urine proteome changes associated with ccRCC. The tissue proteome covered 67.6% of proteins (1889) quantified in the urine (Fig. 3A). Comparison analysis resulted in 108 common differential proteins (“common” category), 870 only in tissue (“tissue-only” category), and 74 only in urine (“urine-only” category) (Fig. 3B and supplemental Table S5A).
Fig. 3.
Integration of urine and tissue proteomes discovers consistent changes of cell adhesion molecular and immunity proteins in early-stage ccRCC.A, Venn diagram of identified proteins in tissue and urine datasets. Common differential proteins: Proteins with fold change above 1.5 (RCC vs. Control) in both tissue and urine. B, the distribution of the relative dominance of no change (gray), agree (green), tissue-only (blue), urine-only (orange), and disagree (red) proteins. Proteins were defined based on the consistency or inconsistency between the tissue and urine datasets. Agree proteins were those with the same direction of change in both tissue and urine datasets. No change proteins, in both tissue and urine, were not changed (within 1.5 fold change) between ccRCC and control. Tissue-only proteins varied between ccRCC and control in tissue dataset (larger than 1.5 fold), but not in urine (within 1.5 fold). Urine-only proteins varied in urine (larger than 1.5 fold), but not in tissue (within 1.5 fold). Disagree genes performed opposite direction of change in tissue and urine. C, pathway enrichment of tissue-urine agree proteins, tissue-only proteins, and urine-only proteins. D, proteins varied constantly in CPTAC tissue dataset, present tissue dataset, and urine dataset. Cell adhesion molecular and defense proteins account for the most (marked in green and red). E, casual network analysis predicted ARRB2 as the upregulator of downstream molecular and pathway disturbance. Molecular in red boxes: upregulated proteins in present study. Molecular in green boxes: downregulated proteins in present study; molecular in orange boxes: predicted to be activated status; molecular in grass green boxes: predicted to be inhibited status. F, GOBP (Go ontology for biological process) terms for urine-only upregulated proteins. ccRCC, clear cell renal cell carcinoma; CPTAC, Clinical Proteomic Tumor Analysis Consortium.
IPA enrichment analysis showed that “common” proteins were mainly involved in pathways regulating cell growth/development (actin cytoskeleton signaling, CDC42 signaling, and Th2 pathway) and intracellular and second messenger signaling (RHOGDI signaling, signaling by Rho Family GTPase, and glucocorticoid receptor signaling) (Fig. 3C). “Common” proteins might provide proxy for tumor-associated variations in urine. Overall 48 out of 108 “common” proteins showed consistent change trend in tissue and urine. Additionally, these 48 proteins were also quantified in CPTAC study and showed the same change trend (Fig. 3D). Cell adhesion molecules (ITGB4, TGFBI) and defense proteins (IGHV3-9, HLA-DRA, HLA-DPA1, IGHV6-1, and IGHV3-64D) were upregulated, while cell adhesion molecules (ALCAM, CDH11, CDH1) were downregulated. GO functional enrichment analysis showed that these proteins were mainly about molecular function of binding and catalyzing. Further canonical pathway analysis showed that these proteins were mainly involved in immune response regulation through RHOGDI signaling, pathogen-induced cytokine storm signaling pathway, ILK signaling, etc. Additionally, small molecular pathway, including GDP-mannose biosynthesis, superoxide radicals degradation, vitamin-C transport, and glycolysis, also showed that they are significantly enriched (supplemental Fig. S4). These results suggested that urine proteome changes could reflect immune response and metabolism dysfunction occurred in tissue.
We further performed casual network analysis to explore upregulators of tissue-urine common proteins. Activation of signal transduction inhibitor, ARRB2, was predicted as the upregulator molecule. Activation of cell adhesion and defense molecule could induce dysfunction of epithelial-mesenchymal transition pathway, further regulating tumor-associated variations (21) (Fig. 3E).
“Urine-only” proteins were mainly involved in pathways regulating immune response (Clathrin-mediated endocytosis signaling, granulocyte adhesion and diapedesis, leukocyte extravasation signaling, and agranulocyte adhesion and diapedesis) and intracellular and second messenger signaling. But tumor-associated metabolic pathway did not show significantly enriched in urine proteome. To further identify specific urine proteomic changes to biological processes, we performed enrichment analyses for GOBP terms using “urine-only” proteins. GOBP terms showed that several terms were enriched in the upregulated proteins, including antigen processing and leukocyte degranulation (Fig. 3F).
Integrative Analysis of Plasma and Urine Proteomes
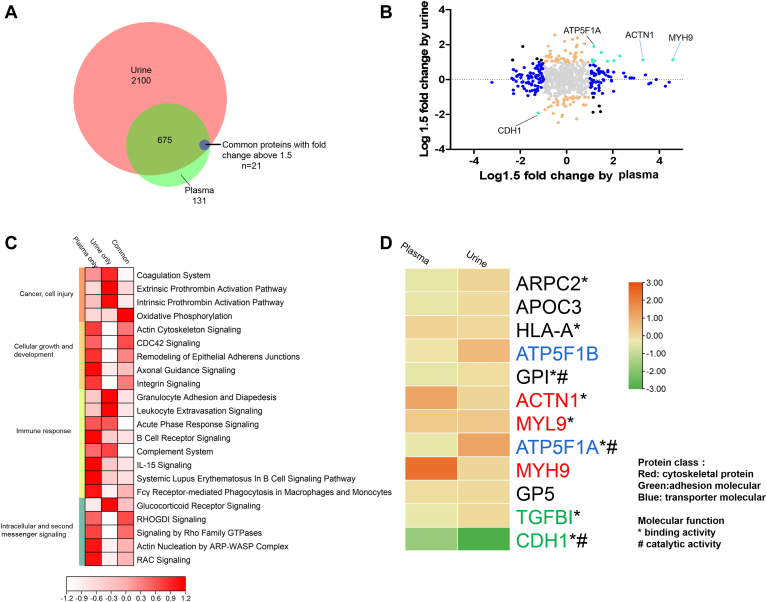
Integration of plasma and urine proteome results showed that 84.2% of plasma proteins (696 ones) were also quantified in urine (Fig. 4A), with 21 common differential proteins (“common” category), 146 only in plasma (“plasma-only” category), and 79 only in urine (“urine-only” category) (Fig. 4B and supplemental Table S6).
Fig. 4.
Integration of plasma and urine proteomes.A, Venn diagram of identified proteins in urine and plasma datasets. Common differential proteins: Proteins with fold change above 1.5 (RCC vs. Control) in both plasma and urine. B, the distribution of the relative dominance of no change (gray), agree (green), plasma-only (blue), urine-only (orange), and disagree (red) proteins. Proteins were defined based on the consistency or inconsistency between the urine and plasma datasets. Agree proteins were those with same direction of change in both urine and plasma datasets. No change proteins, in both urine and plasma, were not changed (within 1.5 fold change) between ccRCC and control. Urine-only proteins varied between ccRCC and control in Urine dataset (larger than 1.5 fold), but not in plasma (within 1.5 fold). Plasma-only proteins varied in plasma (larger than 1.5 fold), but not in urine (within 1.5 fold). Disagree genes performed opposite direction of change in urine and plasma. C, pathway enrichment of plasma-urine agree proteins, plasma-only proteins, and urine-only proteins. D, proteins varied constantly in urine and plasma dataset. These proteins were mainly adhesion molecular and transporter molecular (marked in green and blue). ccRCC, clear cell renal cell carcinoma.
IPA enrichment analysis showed that plasma-urine “common” proteins and “plasma-only” proteins showed similar enriched pathways, including cellular growth/development and intracellular/second messenger signaling. Additionally, immune response was significantly enriched for “plasma-only” and “urine-only” proteins through different pathways. Pathways of B cell receptor signaling, Fcy receptor-mediated phagocytosis in macrophages and monocytes, and IL-15 signaling were enriched in plasma, while granulocyte adhesion and diapedesis and leukocyte extravasation signaling were enriched in urine (Fig. 4C).
Overall 12 out of 21 common proteins showed consistent change trend. The main three protein classes were cytoskeletal proteins (ACTN1, MYH9, MYL9), adhesion molecular (TGFBI, CDH1), and transporter molecules (ATP5F1B, ATP5F1A). GO enrichment showed these proteins were mainly about binding and catalyzing (Fig. 4D).
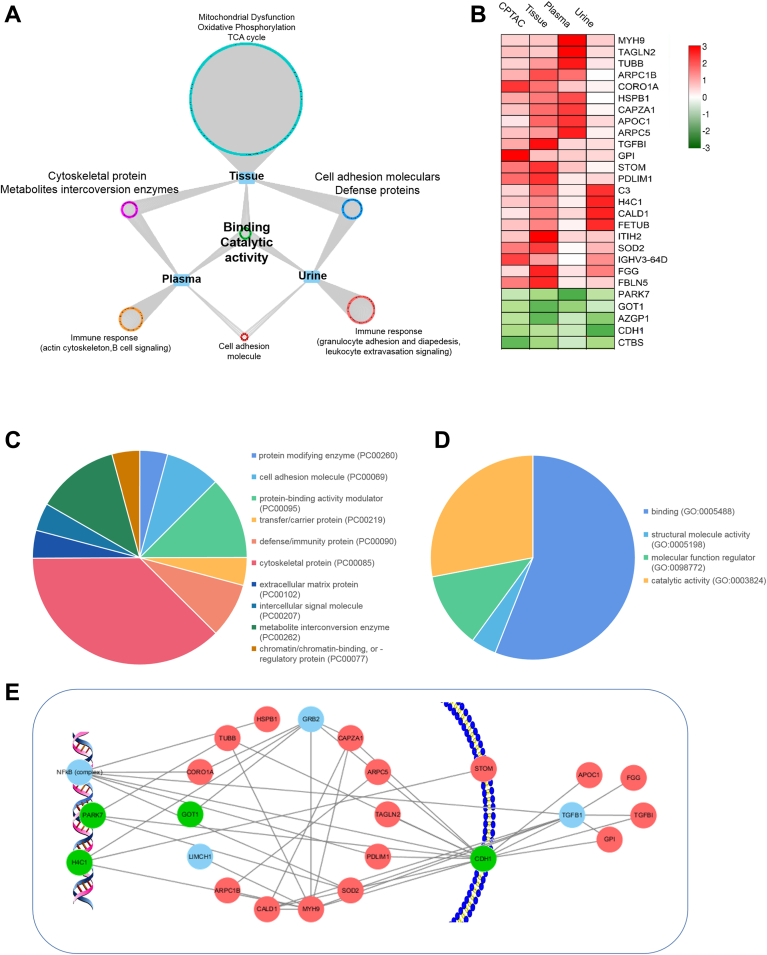
Integrative Analysis of Tissue, Plasma, and Urine Proteomes
According to above integrative analysis, it is evident that both blood and urine are decent proxies of tumoral function changes. In plasma, cytoskeletal proteins and metabolic enzymes expressed differently in early-stage RCC, while in urine, adhesion molecular and defense proteins showed different expression level. Differential proteins in plasma and urine both reflect function of binding and catalytic activity of tumor tissue. Proteins only changed in biofluids could reflect immune response variations triggered by tumor, perhaps through different regulation mechanism. Plasma proteins involved in actin cytoskeleton, B cell signaling and urine proteins involved in granulocyte adhesion, leukocyte extravasation signaling showed specific differential expression levels. (Fig. 5A and Table 1).
Fig. 5.
Integration of tissue, plasma, and urine proteomes.A, summary of molecular and pathway variations transferred from tissue to plasma and urine in early-stage ccRCC. B, proteins varied constantly in CPTAC tissue dataset, present tissue dataset, plasma dataset, and urine dataset. C and D, protein class and molecular function of 27 commonly differential proteins in tissue, plasma, and urine with the same change trend in early-stage ccRCC. E, network of these “tissue-plasma-urine consistent proteins” activate tumor cell invasion and migration, while inhibit tumor cell death, thus promoting ccRCC progression. Red, upregulated proteins; green, downregulated proteins; blue, no detected. ccRCC, clear cell renal cell carcinoma; CPTAC, Clinical Proteomic Tumor Analysis Consortium.
Table 1.
Enriched pathway and function in plasma and urine
| Function | Enriched pathway in plasma | Enriched pathway in urine | ||
|---|---|---|---|---|
| Common with tissue | Plasma only | Common with tissue | Urine only | |
| Cellular growth and development | Actin cytoskeleton signaling | Actin cytoskeleton signaling | ||
| Axonal guidance signaling | CDC42 signaling | |||
| Germ cell-sertoli cell junction signaling | Th2 pathway | |||
| Integrin signaling | ||||
| Remodeling of epithelial adherens junctions | ||||
| Immune response | Phagosome formation | Epithelial adherens junction signaling | Phagosome formation | Clathrin-mediated endocytosis signaling |
| B cell receptor signaling | Neuroinflammation signaling pathway | Granulocyte adhesion and diapedesis | ||
| IL-15 signaling | Leukocyte extravasation signaling | |||
| Systemic lupus erythematosus in B cell signaling pathway | Agranulocyte adhesion and diapedesis | |||
| Intracellular and second messenger signaling | RHOA signaling | RHOGDI signaling | ||
| Actin nucleation by ARP-WASP complex | Signaling by Rho family GTPases | |||
| RAC signaling | Glucocorticoid receptor signaling | |||
| Calcium signaling | ||||
Integration of the three proteomic analyses (tissue, plasma, and urine), we discovered 27 common proteins showing differential levels (fold change >1.5) in at least two omic matrices (Fig. 5B). GO enrichment analysis showed these proteins were mainly cytoskeletal proteins, metabolite interconversion enzymes, and protein-binding modulator. Functions of these proteins were binding and catalyzing (Fig. 5, C and D).
Further protein-protein interaction analysis was performed using these common differential proteins. And a network regulating tumor cell invasion and migration was discovered. Downregulation of nucleoprotein, PARK7 and H1C4, trigger changes of cytoplasmic proteins, including MYH9, SOD2, ARPC1B, CORO1A, TUBB, HSPB1, and GOT1, and membrane protein (CDH1). Downregulation of CDH1 activates extracellular proteins, including APOC1, FGG, TGFBI, and GPI. Interaction of these proteins regulates tumor cell invasion and migration, which plays vital role during ccRCC occurrence and progression (Fig. 5E).
Potential Biofluid Biomarkers for ccRCC Prediction
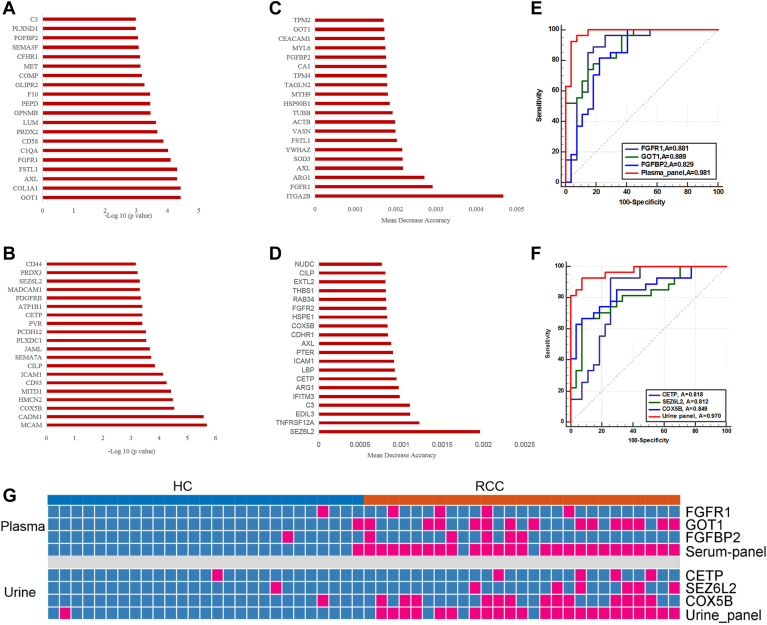
We evaluated the discriminatory performance of differential proteins in plasma and urine. Nonparameter wilcoxon rank-sum test was performed for significance evaluation of proteins between ccRCC and HC. Differential proteins were ranked by the - log 10 (adjusted p value) (Fig. 6, A and B). Next, we evaluated proteins as input variables and identified the most important features for ccRCC distinction using the random forest algorithm. Proteins were ranked by the mean decrease accuracy value (Fig. 6, C and D). The top 20 proteins in the two models were chosen as potential biomarkers. These potential biomarkers were used to construct prediction model using logistic regression algorithm. And proteins showing no significant contribution (p > 0.05) to the model were further excluded.
Fig. 6.
Potential plasma and urine biomarkers for ccRCC.A and B, the top 20 proteins with the lowest FDR values in plasma (A) and urine (B). C and D, the top 20 proteins with the highest mean decrease accuracy value of random forest model in plasma and urine. E and F, ROC curve of plasma panel (E) and urine panel (F) for the diagnostic model for ccRCC. G, discriminant results heat map of plasma FGFR1, GOT1, FGFBP2, the plasma panel, urine CETP, SEZ6L2, COX5B, and the urine panel for ccRCC and HC samples using the cutoff value with a specificity of 95%. Red: positive with a specificity of 95%. ccRCC, clear cell renal cell carcinoma; FDR, false discovery rate; HC, healthy controls.
Finally, three plasma proteins, FGFR1, GOT1, FGFBP2, and three urinary proteins, CETP, SEZ6L2, COX5B, showed good performance for ccRCC prediction. A plasma protein signature consisted of FGFR1, GOT1, and FGFBP2 and a urinary protein signature consisted of CETP, SEZ6L2, and COX5B were built using logistic regression algorithm. The above plasma protein panel achieved 92.6% specificity and 96.3% sensitivity with an AUC of 0.981 (Fig. 6E and supplemental Table S7A). And urinary proteins panel achieved similar results (92.6% specificity, 92.6% sensitivity, an AUC of 0.970) (Fig. 6F and supplemental Table S7B). Further, LOOCV method was used for model validation. For plasma panel, the LOOCV achieved the AUC, sensitivity, and specificity of 0.944, 0.889, and 0.963. For urine panel, the LOOCV achieved the AUC, sensitivity, and specificity of 0.938, 0.926, and 0.852 (supplemental Table S7).
To visualize the protein performance on ccRCC diagnosis prediction, with a specificity of 95%, the cutoff values of each protein and panel were used in the samples. The above plasma panel led to a sensitivity of 92.6% (Fig. 6G and supplemental Table S7C). And the above urine panel led to a sensitivity of 85.2% (Fig. 6G and supplemental Table S7D). The panel could achieve higher sensitivity than a single protein in both plasma and urine.
Discussion
Analysis of tumor tissues is the most direct method to identify dys-regulated protein. Comparing with NATs facilitates comparative proteomic profiling to identify differential proteins, with the end goal of delineating the aberrant cellular processes associated with ccRCC. Biofluid biopsy could offer a minimally invasive strategy for repeat sampling to monitor disease progression and would be more representative of the molecular features associated with tumor genesis (22, 23). In present study, we described a combined analysis of tumor-plasma-urine proteome based on early-stage ccRCC patients and matched controls. This study would not only provide the molecular changes in tumor tissue, plasma, and urine but define to what extent the biofluid proteome might reflect the tissue proteome change in the tumor early stage
Plasma Proteome Reflect Tumor-Associated Changes
The comparison of the plasma and tumor tissue proteome showed that in plasma, cytoskeletal proteins and metabolite conversion enzymes showed consistent change trend with tumor tissue. Cytoskeletal proteins performed vital functions during cancer mechanism (24). In present study, cytoskeletal proteins were involved in ccRCC-associated cell growth and development pathways (remodeling of epithelial adherens junction), immune response pathways (regulation of the EMT pathway, signaling by Rho family GTPases), and cell injury pathways (HIF1A signaling).
Among them, epithelial adherens junction and EMT pathway profoundly impacted prognosis and immunotherapy of ccRCC (25). Several studies showed that EMT could induce the formation of tumor cells during early-stage tumor formation and metastasis, accompanied by specific connective tissue damage and the release of tumor cells into surrounding tissues or blood (26).
Cell injury pathways, HIF1a signaling, was another pathway reflected by plasma proteome. Aberrant accumulation of HIF1a was a consequence event induced by loss of von Hippel-Lindau disease tumor suppressor (VHL) in ccRCC (REF TCGA), which in turn resulted in uncontrolled activation of HIFα-target genes that regulated angiogenesis, glycolysis, and apoptosis. In present plasma proteome, a possible HIF1-serpine1–regulated mechanism was proposed (Fig. 2E). Casual network revealed that HIF-1 act with TP53 through transmembrane receptor, ITGAV. It was reported that HIF-1 facilitated cellular adaptation to mild and moderate hypoxia, while Tp53 was activated to induce apoptosis under severe hypoxia (27).
Additionally, tumor-associated energy metabolic pathways were discovered to be changed in plasma. The landmark TCGA and CPTAC analysis of ccRCC highlighted a key role for metabolic alteration in ccRCC progression (16, 17). Casual network of present plasma proteome suggested that an ATP-dependent RNA helicase DDX5 was probably the upregulator of energy metabolism. Downregulation of malate dehydrogenase 1 and creatine kinase M-type could reduce oxidative phosphorylation process through regulation of mitochondrial NADH supply for oxidative phosphorylation (28). Upregulation of GPI contributed to the glycolytic flux increasing by catalyzing the conversion of glucose-6-phosphate to fructose-6-phosphate (29). These changes indicated that tumor-associated early metabolism changes could be reflected in plasma.
Except for the changes consistent with tumor tissue proteome, plasma proteome also reflected some specific pathway changes. These changes could be the body response to tumor, indirectly associated with tumor occurrence. “Plasma-only” proteins were involved in oxidative stress response, humoral immune response, actin filament organization, and cytoskeleton organization. Actin cytoskeleton plays a central role in regulating immune response, including B cell antigen uptake, polarization and presentation, as well as B cell migration and interaction with T cells (30). Recent studies have suggested a potential role for B cells in the development and progression of ccRCC. It suggested that infiltrated B lymphocytes are present in all stages of RCC, playing a major role in determining tumor formation and advancement, as an essential part in the tumor microenvironment (31). Some evidence proves that B cells can exert a direct or indirect effect on CD8+T cells, acting as a provider of antigen-presenting cells and release cytokines to help T cells perform their duties in tumor sites (32, 33). Moreovver, it was reported that RCC tissues could recruit more B cells than the surrounding normal renal tissues from human clinical RCC samples. Tumor-educated B cells could significantly increase the RCC cell migration and invasion through activation of IL-1β/HIF-2α signals in RCC cells, which could induce the downstream Notch1 signaling pathway (34). However, depth mechanism need to be further investigated. Actin cytoskeleton of immune cells has been recognized as a central mediator of the formation and maturation of the immunological synapse and its signaling and cytolytic activities (35). Additionally, oxidative stress response was enriched for “plasma-only” proteins. Many cytoskeletal proteins are sensitive to reactive oxygen species, and redox regulation has emerged as a pivotal modulator of the actin cytoskeleton and its associated proteins (36). Oxidative stress can effectively induce the occurrence of autophagy in cells because malignant tumor cells need to re-use their organelles to maintain growth (37). ccRCC-induced immune response could be reflected in plasma proteome through actin cytoskeleton and oxidative stress disorders.
Urine Proteome Reflect Tumor-Associated Pathway
In urine, cell adhesion molecules and defense proteins showed consistent change trend with tissue, suggesting tumor-associated pathway disorder might be reflected in urine. Pathways involved in cell growth and development (actin cytoskeleton signaling, CDC42 signaling), immune response (phagosome formation), and intracellular messenger signaling (signaling by Rho family GTPase) showed significantly changed.
Rho family proteins Cdc42 are closely concerned with regulating cell migration, invasion, and cytoskeleton assembling (38). It was suggested that the enhanced migration and invasion of cells in ccRCC were due to Cdc42 and RhoA activity of Rho family, and actin cytoskeletal reorganization alteration may be involved (39). Autophagy is an important regulatory process promoting the disposal of unnecessary or degraded cellular components, tightly linked to almost all cellular processes. It involves the formation of phagosomes that sequester cytoplasmic material and deliver it to lysosomes for degradation (40). During ccRCC development, it tends to disrupt the regulation of the balance between this process and apoptosis, permitting prolonged cell survival and increased replication (41).
In present urine proteome, casual network analysis discovered an upregulator, ARRB2, contributed to urine proteins’ changes through promoting activity of AKT and EGFR. Immunity proteins FGG and HLA-DRA showed up in ccRCC urine, indicating increased inflammation and oxidative stress during ccRCC (42). It was previously reported that FGG is susceptible to oxidation and responsible for coagulation anomalies in renal disease through influencing fibrin formation and properties (43). However, more research at the molecular level is needed to determine the mechanisms.
Proteins only changed in urine were involved in immune response, perhaps through different molecular mechanism with plasma. Pathways of endocytosis signaling, granulocyte adhesion/diapedesis, and leukocyte extravasation signaling showed significantly enriched, reflecting innate and adaptive immune response in urine. Leukocyte extravasation is a multistep process implicated in tumor. This process is essential to get leukocytes to the site of tumor but is also one of the main steps in the metastatic cascade in which cancer cells leave the primary tumor and migrate to target sites through the vascular route (44). It has been reported that leukocytes are much more complex cells having not only effector functions in the innate immune response but also the capacity of modulating the adaptive immune response, via direct interaction with or by producing cytokines that affect dendritic cells and lymphocytes (45). Conclusively, proteins only changed in biofluid could reflect immune response variations from different regulation mechanism.
Integration Analysis of Tissue, Plasma and Urine Proteome
Molecules showing consistent change trend in the three analyses would provide evidences that tumor tissue-associated variations could be reflected in biofluids. Total 27 consistent molecules were involved in the activation of tumor cell invasion and migration. Activation of nucleus regulators, PARK7 and H4C1, contribute to changes of cytoskeletal proteins and metabolite conversion enzymes in cytoplasm and adhesion molecular in plasma membrane. As shown in Figure 5E, adhesion molecular, CDH1, was the hub molecular of the network activating tumor cell invasion and migration and inhibiting tumor cell death. Independent studies have shown a direct correlation between VHL loss and the expression of cadherin in ccRCC. The loss of cadherin was observed in premalignant foci of VHL patients, which concomitantly showed increased CA9 expression (inferring a loss of VHL), suggesting cadherin loss to be an early pathogenic event (46). Evidences showed that cadherin depletion could promote kidney epithelial cells migration and invasion. Knockdown of cadherin in human kidney epithelial cells increased the migration potential in a wound-healing assay and increased the invasive potential of VHL-restored ccRCC cells in a matrigel invasion assay (46). Appreciable changes in the behavior of renal epithelial cells through manipulating a single gene among a myriad of genes implicated the potential value of CDH1 during ccRCC progression.
The clinical use of proteomics in cancer diagnosis has evolved from the direct evaluation of tumor tissue searching for prognostic biomarkers to the use of biofluids as proxies for changes in the cancer of interest. Although it is widely accepted that the latter liquid biopsy approach is more readily translatable to the clinical use, there is no available evidences or studies showing the accuracy of biofluids as a reflection of proteomics changes within tumor tissues. Referring to the concept behind the use of biofluid molecules for cancer diagnosis in clinic, for example, blood prostate specific antigen tests for prostate cancer diagnosis and urine NMP22 test for bladder cancer diagnosis, we assumed that changes in the blood and urine proteomics could accurately reflect alterations in the tumor tissue. In this study, we performed a comprehensive proteomics analysis of tissue, plasma, and urine of early-stage ccRCC simultaneously. In the light of the results, it is evident that both blood and urine are decent proxies of tumoral function changes.
Plasma and Urine Proteins Showed Potential Value for ccRCC Prediction
Utilizing biofluids would offer a minimally invasive strategy to predict and monitor disease and possibly be more representative of the molecular features associated with tumorigenesis. With the goal of identifying candidates for early detection of ccRCC, we applied statistical approach to select potential plasma and urine proteins to construct the optional prediction model. The results revealed that a panel of plasma proteins, FGFR1, GOT1, and FGFBP2, could predict ccRCC with high accuracy (specificity: 92.6%, sensitivity: 96.3%, AUC: 0.981). FGFR1 is a fibroblast growth factor receptor. FGFR1 overexpression has been identified in primary ccRCC (47), and FGFR1 overexpression was shown to be related to a reduced survival outcome in ccRCC patients receiving sorafenib treatment (48). Recent study indicated FGFR1 may be involved in tumor angiogenesis and may represent a promising therapeutic target in metastatic ccRCC (49). GOT1 is a ferroptosis-related gene. It has been reported that GOT1 was associated with overall survival (OS) of ccRCC (50) and inferred to be related to tumor immunity in ccRCC (51).
Urinary protein panel achieved similar performance for ccRCC prediction. A protein panel consisted of CETP, SEZ6L2, and COX5B achieved the AUC of 0.970, sensitivity of 92.6%, and specificity of 92.6%. CETP is one of the most potent endogenous negative regulators of HDL-cholesterol. Cholesterol levels and oxidative stress are key contributors to endothelial damage. It has been indicated that CETP expression negatively impacts endothelial cell function through reducing expression levels of adhesion molecular, intercellular cell adhesion molecule-1, and vascular cell adhesion molecule-1, and diminishing monocyte adhesion (52). Cell adhesion molecular in present study showed decreased expression level in ccRCC, probably resulting from CETP regulation through lipid metabolism pathway. Tissue mitochondrial respiratory chain protein, COX5B has been reported as a potential biomarker for ccRCC (53). ccRCC is characterized by increased glycolysis and reduced activity of the mitochondrial respiratory chain, explained by the VHL–HIF pathway (54). It has recently been shown that COX5B is directly targeted by HIF-1 (55).
Biological fluids such as blood and urine serve as common sources of biomarkers. Blood is often considered to be the ideal source for protein candidates, for that the network of arteries, veins, and capillaries that come in contact with organs offers a means for proteins that are secreted, shed, or otherwise released by tumor tissues to be circulated (56). Changes in blood biomarker levels are unlikely to persist long enough for detection due to homeostasis regulation (57). Therefore, detection of early molecular changes in blood is probably a challenging task. In contrast, urine is the place that most of the wastes in blood are dumped into and thus tolerates changes to a much higher degree. Therefore, early molecular changes are more likely to be magnified and detectable in urine (58, 59). In addition, urine is easily obtained, relatively simple in composition relative to blood. Accumulated changes in urine are not likely to be masked, and some molecular species which are difficult to detect in blood may be detected in urine. In present study, the results showed that plasma and urine molecular showed similar performance for ccRCC discrimination, having potential value for ccRCC early biomarker selection. The potential values for ccRCC diagnosis need further validation using larger cohorts in future work.
Conclusion
In conclusion, this study represents for the first time an integrated proteomics evaluation of tumor, including tissue and biofluids (plasma and urine). Biofluid proteins could reflect mechanism with tumorigenesis. Our study suggested that tumor-associated function changes could be shown up in blood and urine. Differential proteins found in plasma and urine both reflect the binding and catalytic activity of tumor tissue. Plasma and urine cytoskeletal proteins and metabolite conversion enzymes could result in the discovery of novel diagnostic targets of RCC. Integrative analysis of plasma/urine with tissue could discover potential biomarkers of RCC with high specificity. Plasma proteins, FGFR1, GOT1, FGFBP2, and urinary proteins, CETP, SEZ6L2, COX5B, could serve as potential biomarkers for early prediction of RCC. However, several limitations existed. First, considering for the relative small size of sample, the p values were not corrected to obtain more differential proteins to perform functional analysis. Thus, function molecular in plasma and urine should be further validated using cell lines to evaluate the influence on tumor cell biological behavior. Moreover, function experiments on mice model would be performed to validate the potential mechanism of these molecular during tumorigenesis. Above function, validation work were our next work and would be presented in the future. Second, due to the small sample size, the tentatively proposed potential biomarkers need to be further validated using larger cohorts from multiple centers. Third, missing value imputation may lead to inflated p value in differential protein analysis; thus, differential molecular with high degree of missing value need to be further validated using tissue sample.
Data Availability
The datasets generated and/or analyzed during the current study are available in the iProx repository: https://www.iprox.cn/page/PSV023.html;?url=1681310031083RsOl
Password:VFzv
Supplemental data
This article contains supplemental data.
Conflicts of interest
All authors declare no competing interests.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 31901039, 82170524), Beijing Medical Research (No. 2018-7), CAMS Innovation Fund for Medical Sciences (2021-1-I2M-016), and Biologic Medicine Information Center of China, National Scientific Data Sharing Platform for Population and Health.
Author contributions
X. L., M. Z., and C. S. conceptualization; X. L., M. Z., and C. S. methodology; X. L., M. Z., and C. S. data curation; X. L., M. Z., and C. S. writing–original draft; X. L., M. Z., and C. S. writing–review and editing; H. S., J. S., and F. Q. investigation; B. Z. and Z. G. formal analysis; Y. Z., H. N., and W. S. supervision.
Contributor Information
Yushi Zhang, Email: zhangyushi2014@126.com.
Haitao Niu, Email: niuht0532@126.com.
Wei Sun, Email: sunwei@ibms.pumc.edu.cn.
Supplementary Data
References
- 1.Cheng F., Su L., Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget. 2016;7:48832–48841. doi: 10.18632/oncotarget.9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson S.J., Tsui D.W., Murtaza M., Biggs H., Rueda O.M., Chin S.F., et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. New Engl. J. Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 3.Scherer F., Kurtz D.M., Newman A.M., Stehr H., Craig A.F., Esfahani M.S., et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aai8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo W., Sun Y.F., Shen M.N., Ma X.L., Wu J., Zhang C.Y., et al. Circulating tumor cells with stem-like phenotypes for diagnosis, prognosis, and therapeutic response evaluation in hepatocellular carcinoma. Clin. Cancer Res. 2018;24:2203–2213. doi: 10.1158/1078-0432.CCR-17-1753. [DOI] [PubMed] [Google Scholar]
- 5.Raimondo F., Morosi L., Corbetta S., Chinello C., Brambilla P., Della Mina P., et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol. Biosyst. 2013;9:1220–1233. doi: 10.1039/c3mb25582d. [DOI] [PubMed] [Google Scholar]
- 6.Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucl. Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W., Yang Q., Liu B., Zhu Z. Serum proteomics for gastric cancer. Clin. Chim. Acta. 2014;431:179–184. doi: 10.1016/j.cca.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Gemoll T., Roblick U.J., Auer G., Jörnvall H., Habermann J.K. SELDI-TOF serum proteomics and colorectal cancer: a current overview. Arch. Physiol. Biochem. 2010;116:188–196. doi: 10.3109/13813455.2010.495130. [DOI] [PubMed] [Google Scholar]
- 9.Tsaur I., Thurn K., Juengel E., Gust K.M., Borgmann H., Mager R., et al. sE-cadherin serves as a diagnostic and predictive parameter in prostate cancer patients. J. Exp. Clin. Cancer Res. 2015;34:43. doi: 10.1186/s13046-015-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu T., Ning W., Wu D., Xu J., Han Q., Huang M., et al. Plasma proteomics identify biomarkers and pathogenesis of COVID-19. Immunity. 2020;53:1108–11022.e5. doi: 10.1016/j.immuni.2020.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas S., Hao L., Ricke W.A., Li L. Biomarker discovery in mass spectrometry-based urinary proteomics. Proteomics Clin. Appl. 2016;10:358–370. doi: 10.1002/prca.201500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radon T.P., Massat N.J., Jones R., Alrawashdeh W., Dumartin L., Ennis D., et al. Identification of a three-biomarker panel in urine for early detection of pancreatic adenocarcinoma. Clin Cancer Res. 2015;21:3512–3521. doi: 10.1158/1078-0432.CCR-14-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimura T., Dayde D., Wang H., Okuda Y., Iwasaki H., Ebi M., et al. Novel urinary protein biomarker panel for early diagnosis of gastric cancer. Br. J. Cancer. 2020;123:1656–1664. doi: 10.1038/s41416-020-01063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Meo A., Batruch I., Brown M.D., Yang C., Finelli A., Jewett M.A., et al. Searching for prognostic biomarkers for small renal masses in the urinary proteome. Int. J. Cancer. 2020;146:2315–2325. doi: 10.1002/ijc.32650. [DOI] [PubMed] [Google Scholar]
- 15.Zhao M., Liu X., Sun H., Guo Z., Liu X., Sun W. Evaluation of urinary proteome library generation methods on data-independent acquisition MS analysis and its application in normal urinary proteome analysis. Proteomics Clin. Appl. 2019;13 doi: 10.1002/prca.201800152. [DOI] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark D.J., Dhanasekaran S.M., Petralia F., Pan J., Song X., Hu Y., et al. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell. 2019;179:964–983.e31. doi: 10.1016/j.cell.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakimi A.A., Reznik E., Lee C.H., Creighton C.J., Brannon A.R., Luna A., et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29:104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie L., Li H., Zhang L., Ma X., Dang Y., Guo J., et al. Autophagy-related gene P4HB: a novel diagnosis and prognosis marker for kidney renal clear cell carcinoma. Aging. 2020;12:1828–1842. doi: 10.18632/aging.102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alevizopoulos A., Tyritzis S., Leotsakos I., Anastasopoulou I., Pournaras C., Kotsis P., et al. Role of coagulation factors in urological malignancy: a prospective, controlled study on prostate, renal and bladder cancer. Int. J. Urol. 2017;24:130–136. doi: 10.1111/iju.13271. [DOI] [PubMed] [Google Scholar]
- 21.Kaszak I., Witkowska-Piłaszewicz O., Niewiadomska Z., Dworecka-Kaszak B., Ngosa Toka F., Jurka P. Role of cadherins in cancer-A review. Int. J. Mol. Sci. 2020;21:7624. doi: 10.3390/ijms21207624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Meo A., Bartlett J., Cheng Y., Pasic M.D., Yousef G.M. Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol. Cancer. 2017;16:80. doi: 10.1186/s12943-017-0644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato Y., Matoba R., Kato K. Recent advances in liquid biopsy in precision oncology research. Biol. Pharm. Bull. 2019;42:337–342. doi: 10.1248/bpb.b18-00804. [DOI] [PubMed] [Google Scholar]
- 24.Ayanlaja A.A., Hong X., Cheng B., Zhou H., Kanwore K., Alphayo-Kambey P., et al. Susceptibility of cytoskeletal-associated proteins for tumor progression. Cell Mol. Life Sci. 2021;79:13. doi: 10.1007/s00018-021-04101-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Q., Kuai Y., Wang S., Zhu X., Wang H., Liu W., et al. Deep learning-based classification of epithelial-mesenchymal transition for predicting response to therapy in clear cell renal cell carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.782515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pastushenko I., Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Ye X.W., Zhang X.P., Liu F. CSB modulates the competition between HIF-1 and p53 upon hypoxia. Math. Biosci. Eng. 2019;16:5247–5262. doi: 10.3934/mbe.2019262. [DOI] [PubMed] [Google Scholar]
- 28.Kim J.W., Tchernyshyov I., Semenza G.L., Dang C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Mojzikova R., Koralkova P., Holub D., Saxova Z., Pospisilova D., Prochazkova D., et al. Two novel mutations (p.(Ser160Pro) and p.(Arg472Cys)) causing glucose-6-phosphate isomerase deficiency are associated with erythroid dysplasia and inappropriately suppressed hepcidin. Blood Cell Mol. Dis. 2018;69:23–29. doi: 10.1016/j.bcmd.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Burbage M., Keppler S.J. Shaping the humoral immune response: actin regulators modulate antigen presentation and influence B-T interactions. Mol. Immunol. 2018;101:370–376. doi: 10.1016/j.molimm.2018.07.026. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y.Q., Chen W.J., Li W.Y., Pan X.W., Cui X.G. Impact of interaction networks of B cells with other cells on tumorigenesis, progression and response to immunotherapy of renal cell carcinoma: a review. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.995519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi J.Y., Gao Q., Wang Z.C., Zhou J., Wang X.Y., Min Z.H., et al. Margin-infiltrating CD20(+) B cells display an atypical memory phenotype and correlate with favorable prognosis in hepatocellular carcinoma. Clin. Cancer Res. 2013;19:5994–6005. doi: 10.1158/1078-0432.CCR-12-3497. [DOI] [PubMed] [Google Scholar]
- 33.Sharonov G.V., Serebrovskaya E.O., Yuzhakova D.V., Britanova O.V., Chudakov D.M. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat. Rev. Immunol. 2020;20:294–307. doi: 10.1038/s41577-019-0257-x. [DOI] [PubMed] [Google Scholar]
- 34.Li S., Huang C., Hu G., Ma J., Chen Y., Zhang J., et al. Tumor-educated B cells promote renal cancer metastasis via inducing the IL-1β/HIF-2α/Notch1 signals. Cell Death Dis. 2020;11:163. doi: 10.1038/s41419-020-2355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wurzer H., Hoffmann C., Al Absi A., Thomas C. Actin cytoskeleton straddling the immunological synapse between cytotoxic lymphocytes and cancer cells. Cells. 2019;8:463. doi: 10.3390/cells8050463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Q., Huff L.P., Fujii M., Griendling K.K. Redox regulation of the actin cytoskeleton and its role in the vascular system. Free Radic. Biol. Med. 2017;109:84–107. doi: 10.1016/j.freeradbiomed.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y., Jiang M., Chen W., Zhao T., Wei Y. Cancer and ER stress: mutual crosstalk between autophagy, oxidative stress and inflammatory response. Biomed. Pharmacother. 2019;118 doi: 10.1016/j.biopha.2019.109249. [DOI] [PubMed] [Google Scholar]
- 38.Kardash E., Reichman-Fried M., Maître J.L., Boldajipour B., Papusheva E., Messerschmidt E.M., et al. A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat. Cell Biol. 2010;12:47–53. doi: 10.1038/ncb2003. [DOI] [PubMed] [Google Scholar]
- 39.Ni S., Hu J., Duan Y., Shi S., Li R., Wu H., et al. Down expression of LRP1B promotes cell migration via RhoA/Cdc42 pathway and actin cytoskeleton remodeling in renal cell cancer. Cancer Sci. 2013;104:817–825. doi: 10.1111/cas.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang T., Song X., Yang Y., Wan X., Alvarez A.A., Sastry N., et al. Autophagy and hallmarks of cancer. Crit. Rev. Oncog. 2018;23:247–267. doi: 10.1615/CritRevOncog.2018027913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lotze M.T., Maranchie J., Appleman L. Inhibiting autophagy: a novel approach for the treatment of renal cell carcinoma. Cancer J. 2013;19:341–347. doi: 10.1097/PPO.0b013e31829da0d6. [DOI] [PubMed] [Google Scholar]
- 42.Silverstein D.M. Inflammation in chronic kidney disease: role in the progression of renal and cardiovascular disease. Pediatr. Nephrol. 2009;24:1445–1452. doi: 10.1007/s00467-008-1046-0. [DOI] [PubMed] [Google Scholar]
- 43.Baralić M., Robajac D., Penezić A., Miljuš G., Šunderić M., Gligorijević N., et al. Fibrinogen modification and fibrin formation in patients with an end-stage renal disease subjected to peritoneal dialysis. Biochemistry (Mosc) 2020;85:947–954. doi: 10.1134/S0006297920080106. [DOI] [PubMed] [Google Scholar]
- 44.Mondadori C., Crippa M., Moretti M., Candrian C., Lopa S., Arrigoni C. Advanced microfluidic models of cancer and immune cell extravasation: a systematic review of the literature. Front. Bioeng. Biotechnol. 2020;8:907. doi: 10.3389/fbioe.2020.00907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosales C. Neutrophils at the crossroads of innate and adaptive immunity. J. Leukoc. Biol. 2020;108:377–396. doi: 10.1002/JLB.4MIR0220-574RR. [DOI] [PubMed] [Google Scholar]
- 46.Evans A.J., Russell R.C., Roche O., Burry T.N., Fish J.E., Chow V.W., et al. VHL promotes E2 box-dependent E-cadherin transcription by HIF-mediated regulation of SIP1 and snail. Mol. Cell Biol. 2007;27:157–169. doi: 10.1128/MCB.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsimafeyeu I., Demidov L., Stepanova E., Wynn N., Ta H. Overexpression of fibroblast growth factor receptors FGFR1 and FGFR2 in renal cell carcinoma. Scand. J. Urol. Nephrol. 2011;45:190–195. doi: 10.3109/00365599.2011.552436. [DOI] [PubMed] [Google Scholar]
- 48.Ho T.H., Liu X.D., Huang Y., Warneke C.L., Johnson M.M., Hoang A., et al. The impact of FGFR1 and FRS2α expression on sorafenib treatment in metastatic renal cell carcinoma. BMC cancer. 2015;15:304. doi: 10.1186/s12885-015-1302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park J.Y., Kim P.J., Shin S.J., Lee J.L., Cho Y.M., Go H. FGFR1 is associated with c-MYC and proangiogenic molecules in metastatic renal cell carcinoma under anti-angiogenic therapy. Histopathology. 2020;76:838–851. doi: 10.1111/his.14076. [DOI] [PubMed] [Google Scholar]
- 50.Chang K., Yuan C., Liu X. Ferroptosis-related gene signature accurately predicts survival outcomes in patients with clear-cell renal cell carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.649347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong Y., Lin M., Ou D., Huang Z., Shen P. A novel ferroptosis-related 12-gene signature predicts clinical prognosis and reveals immune relevancy in clear cell renal cell carcinoma. BMC cancer. 2021;21:831. doi: 10.1186/s12885-021-08559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wanschel A., Guizoni D.M., Lorza-Gil E., Salerno A.G., Paiva A.A., Dorighello G.G., et al. The presence of Cholesteryl Ester Transfer Protein (CETP) in endothelial cells generates vascular oxidative stress and endothelial dysfunction. Biomolecules. 2021;11:69. doi: 10.3390/biom11010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stein J., Tenbrock J., Kristiansen G., Müller S.C., Ellinger J. Systematic expression analysis of the mitochondrial respiratory chain protein subunits identifies COX5B as a prognostic marker in clear cell renal cell carcinoma. Int. J. Urol. 2019;26:910–916. doi: 10.1111/iju.14040. [DOI] [PubMed] [Google Scholar]
- 54.Hervouet E., Godinot C. Mitochondrial disorders in renal tumors. Mitochondrion. 2006;6:105–117. doi: 10.1016/j.mito.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 55.Soro-Arnaiz I., Li Q.O.Y., Torres-Capelli M., Meléndez-Rodríguez F., Veiga S., Veys K., et al. Role of mitochondrial complex IV in age-dependent obesity. Cell Rep. 2016;16:2991–3002. doi: 10.1016/j.celrep.2016.08.041. [DOI] [PubMed] [Google Scholar]
- 56.Schiess R., Wollscheid B., Aebersold R. Targeted proteomic strategy for clinical biomarker discovery. Mol. Oncol. 2009;3:33–44. doi: 10.1016/j.molonc.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li M., Zhao M., Gao Y. Changes of proteins induced by anticoagulants can be more sensitively detected in urine than in plasma. Sci. China Life Sci. 2014;57:649–656. doi: 10.1007/s11427-014-4661-y. [DOI] [PubMed] [Google Scholar]
- 58.Gao Y. Now is the time to test early urinary biomarkers in large-scale human samples. Sci. China Life Sci. 2019;62:851–853. doi: 10.1007/s11427-019-9562-y. [DOI] [PubMed] [Google Scholar]
- 59.Wei J., Gao Y. Early disease biomarkers can be found using animal models urine proteomics. Expert Rev. Proteomics. 2021;18:363–378. doi: 10.1080/14789450.2021.1937133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the iProx repository: https://www.iprox.cn/page/PSV023.html;?url=1681310031083RsOl
Password:VFzv