Abstract
Marek’s disease virus, an avian alphaherpesvirus, has been used as an excellent model to study herpesvirus oncogenesis. One of its potential oncogenes, MEQ, has been demonstrated to transform a rodent fibroblast cell line, Rat-2, in vitro by inducing morphological transformation and anchorage- and serum-independent growth and by protecting cells from apoptosis induced by tumor necrosis factor alpha, C2-ceramide, UV irradiation, or serum deprivation. In this report, we show that there is a cell cycle-dependent colocalization of MEQ protein and cyclin-dependent kinase 2 (CDK2) in coiled bodies and the nucleolar periphery during the G1/S boundary and early S phase. To our knowledge, this is the first demonstration that CDK2 is found to localize to coiled bodies. Such an in vivo association and possibly subsequent phosphorylation may result in the cytoplasmic translocation of MEQ protein. Indeed, MEQ is expressed in both the nucleus and the cytoplasm during the G1/S boundary and early S phase. In addition, we were able to show in vitro phosphorylation of MEQ by CDKs. We have mapped the CDK phosphorylation site of MEQ to be serine 42, a residue in the proximity of the bZIP domain. An indirect-immunofluorescence study of the MEQ S42D mutant, in which the CDK phosphorylation site was mutated to a charged residue, reveals more prominent cytoplasmic localization. This lends further support to the notion that the translocation of MEQ is regulated by phosphorylation. Furthermore, phosphorylation of MEQ by CDKs drastically reduces the DNA binding activity of MEQ, which may in part account for the lack of retention of MEQ oncoprotein in the nucleus. Interestingly, the localization of CDK2 in coiled bodies and the nucleolar periphery is observed only in MEQ-transformed Rat-2 cells, implicating MEQ in modifying the subcellular localization of CDK2. Taken together, our data suggest that there is a novel reciprocal modulation between the herpesvirus oncoprotein MEQ and CDK2.
Marek’s disease virus (MDV), an avian alphaherpesvirus, is one of the most potent oncogenic herpesviruses. It elicits the rapid onset of malignant T-cell lymphomas in chickens within several weeks after infection (reviewed in references 11, 35, and 57). The short course of development and polyclonal nature of MDV-induced lymphomas suggest that one or more viral oncogenes are directly involved in the transformation process. Several candidate genes located on the BamHI D, H, I2, L, and Q2 fragments of the MDV genome have been implicated in oncogenesis (8, 61, 63, 67). Among them, MEQ (for “MDV Eco Q”) is most consistently detected in all tumor samples and cell lines (32, 67). MEQ encodes a 339-amino-acid protein with an N-terminal basic region-leucine zipper (bZIP) domain and a C-terminal transactivation domain (32). The bZIP domain has significant homology to that of Jun/Fos family proteins with two stretches of basic residues, termed basic regions 1 and 2 (BR1 and BR2). The transactivation domain is characterized by 2.5 proline-rich repeats. There are at least two sets of DNA response elements to which MEQ binds (59), namely, MEQ response element 1 (MERE1; GAGTGATGA[C/G]TCATC) and MERE2 (PuACACAPy). Heterodimers of MEQ and c-Jun proteins bind the MERE1 site located within the promoter region of the MEQ gene and activate MEQ transcription (58). Consistent with its being a transcription factor, MEQ protein is found in the nucleus (39). The major nuclear localization signal (NLS) has been mapped to BR2. However, MEQ protein can localize to the nucleolus and coiled bodies as well. This novel subnuclear localization suggests that MEQ may be involved in more than transcription. As shown by Xie et al. (75), MEQ expression is required to maintain the transformed phenotype of an MDV tumor cell line, MSB1. In addition, overexpression of MEQ leads to transformation of a rodent fibroblast cell line, Rat-2 (40). MEQ not only induces morphological transformation and anchorage- and serum-independent growth of Rat-2 cells but also protects the transformed cells from apoptosis induced by a variety of means, including tumor necrosis factor alpha, C2-ceramide, UV irradiation, and serum deprivation. At least part of the mechanism seems to be attributed to the induction of bcl-2 expression and the suppression of bax expression by MEQ at the transcriptional level.
In efforts to further understand the transformation mechanism, we examined the cell cycle regulation of MEQ. Here, we report the intriguing observation of a cell cycle-dependent colocalization of MEQ and cyclin-dependent kinase 2 (CDK2) in coiled bodies and the nucleolar periphery during the G1/S boundary and early S phase. We also showed that CDK can phosphorylate MEQ at serine 42, diminishing the DNA binding capacity of MEQ, which may facilitate the nuclear export of MEQ and account for the observed cytoplasmic location of a fraction of MEQ during early S phase. Furthermore, the localization of CDK2 to coiled bodies and the nucleolar periphery is found only in MEQ-transformed Rat-2 cells but not in their untransformed counterparts, implicating MEQ in the alteration of the subcellular localization of CDK2. As discussed below, such a translocation may lead to CDK2 activation, which in turn promotes the G1/S transition and in part accounts for the transforming potential of MEQ.
MATERIALS AND METHODS
Cells.
Rat-2, COS-1, and CV-1 monkey kidney cells (American Type Culture Collection) were maintained in Dulbecco modified Eagle medium (high glucose) supplemented with 10% (vol/vol) calf serum.
Antibodies.
Mouse anti-MEQ monoclonal antibody (MAb) was used at a 1:100 dilution, rabbit anti-MEQ polyclonal antibodies were used at a 1:200 dilution, rabbit anti-p80-coilin polyclonal antibodies was used at a 1:500 dilution, mouse anti-CDK2 (Transduction Lab.) and anti-CDK1 (Oncogene Science) MAbs were used at a 1:100 dilution, and mouse anti-bromodeoxyuridine (BrdU) MAb (Amersham) was used undiluted for immunofluorescent staining. Rabbit anti-MEQ polyclonal antibodies were used at a 1:4,000 dilution for Western blotting.
Cell cycle synchronization and flow cytometry analysis.
Cell cycle synchronization was accomplished by serum deprivation for 3 days to arrest cells at the G0/G1 phase; 1 mM hydroxyurea was used to block cells at the G1/S boundary and/or early S phase; 2 μM etoposide was used to block cells at the G2 phase; and 0.4 μg of nocodazole per ml was used to block cells at the M phase. Cells synchronized at different stages of the cell cycle were then subjected to cell cycle profile analysis by a flow cytometer (Becton Dickinson).
BrdU incorporation assay.
DNA synthesis activity was monitored by incorporation of BrdU (Amersham). Briefly, cells were grown on coverslips inside the six-well plates. Serum deprivation was imposed for 3 days before 50 μM BrdU was added to the medium for 12 h. The cells were fixed with 1% formaldehyde in phosphate-buffered saline (PBS) for 20 min, washed with PBS, and treated with 1 N HCl for 10 min. They were then washed, blocked with 3% bovine serum albumin (BSA)–PBS for 1 h, and stained with anti-BrdU MAb (Amersham) for 1 h at 37°C followed by fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin G (IgG) for 1 h at room temperature.
Indirect immunofluorescence assays and confocal laser scanning microscopy.
Immunofluorescence staining was performed as described previously (39). Briefly, cells were seeded at 5 × 105 cells/well in six-well plates with coverslips inside the plates. The medium was aspirated, and the cells were washed with PBS twice before being fixed with 1% formaldehyde–PBS for 20 min. After another PBS wash, the cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and blocked with 3% BSA–0.1% Tween 20–PBS for 1 h. The cells were then incubated with primary antibodies for 1 h. After the cells were given two washes with PBS containing 0.1% Tween 20, the secondary antibodies conjugated with FITC or Texas red (Vector Labs) were applied for another 1 h, and the cells were analyzed with a Zeiss confocal laser scanning microscope (100× objective).
In vitro mutagenesis.
In vitro mutagenesis was performed by the method described for the Transformer site-directed mutagenesis kit (Clontech). Primers 5′TGGAGGGGGCGTTGGGGA3′ (S42A), 5′TGGAGGGGTCGTTGGGGA3′ (S42D), 5′CATCCCCAACGGGCCCTCCAAAC3′ (S42G), 5′CTTTCTGGGCCCAGACGGAAAAAAAGG3′ (STS29P), and 5′CGCAGGAAGCAGGTCGACTATGTAGAC3′ (T79V) were used to generate point mutations in the MEQ protein.
In vitro kinase assays.
His-tagged MEQ deletion/mutant proteins were first purified with Talon (Clontech). Kinase reactions were carried out in a total volume of 20 μl containing 4 μl of 5× kinase buffer (250 mM Tris [pH 7.4], 125 mM MgCl2, 25 mM MnCl2), 5 μl of CDK1-cyclin B complex (Upstate Biotechnology Inc. [UBI]) or mitogen-activated protein kinase (MAPK; UBI), 1 μg of MEQ protein, and 10 μCi of [γ-32P]ATP (Dupont NEN) at 37°C for 30 min. The reactions were terminated by addition of 10 μl of 50% (vol/vol) acetic acid. Each sample was then resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12.5% polyacrylamide), and the gels were wrapped in Saran Wrap and exposed to X-ray films (Kodak XAR-5) for 3 min (15 min for MAPK). For cyclic AMP-activated protein kinase (PKA; Sigma), the kinase assay was performed in kinase buffer (30 mM potassium phosphate [pH 7.0], 1 mM dithiothreitol, 1 mM EDTA, 150 mM KCl) at room temperature for 30 min. For protein kinase C (PKC; UBI), the kinase reaction was conducted in 50 mM Tris (pH 7.5)–10 mM MgSO4–1 mM dithiothreitol–100 μM CaCl2 in the presence of 40 μg of phosphatidylserine per ml at 37°C for 30 min. The wrapped gels were exposed to X-ray films for 5 min.
In vivo phosphorylation.
CV-1 monkey kidney cells were transfected with meq in the context of an EE (EEEEYMPME) epitope-tagged pTM1 expression vector (cloned into NcoI-EcoRI sites) and infected with recombinant vTF7-3 vaccinia virus as described previously (76). The cells were washed, incubated for 4 h in phosphate-free Dulbecco modified Eagle medium supplemented with 2% (vol/vol) dialyzed calf serum, and then labeled for 2 h with fresh medium containing 250 μCi of [γ-32P]ATP per ml.
Immunoprecipitation.
EE epitope-tagged MEQ and cyclin A, B, and E proteins were expressed in CV-1 cells with the vaccinia virus-T7 polymerase expression system and immunoprecipitated with 20 μl of EE epitope MAb-conjugated Affi-Gel 10 beads (Bio-Rad) as described previously (24).
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays were performed as described previously (59). Briefly, a double-stranded MERE1 (TRE) probe, 5′AATTCAAAAACACATAACATTCGTATATATTTC3′, was labeled with [γ-32P]ATP by Klenow fragment and purified on spin columns. The purified probes (10,000 cpm/reaction) were incubated with MEQ-bZIP protein phosphorylated by a variety of kinases in a buffer containing 25 mM HEPES (pH 7.9), 100 mM KCl, 0.5 mM MgCl2, 1 mg of BSA per ml, 10% glycerol, 5 mM dithiothreitol, and 0.1 mg of poly(dI-dC) in the presence of cold ATP at 37°C for 30 min before being loaded onto a 10% nondenaturing polyacrylamide-Tris-glycine gel.
RESULTS
Cell cycle-dependent cytoplasmic translocation of MEQ proteins.
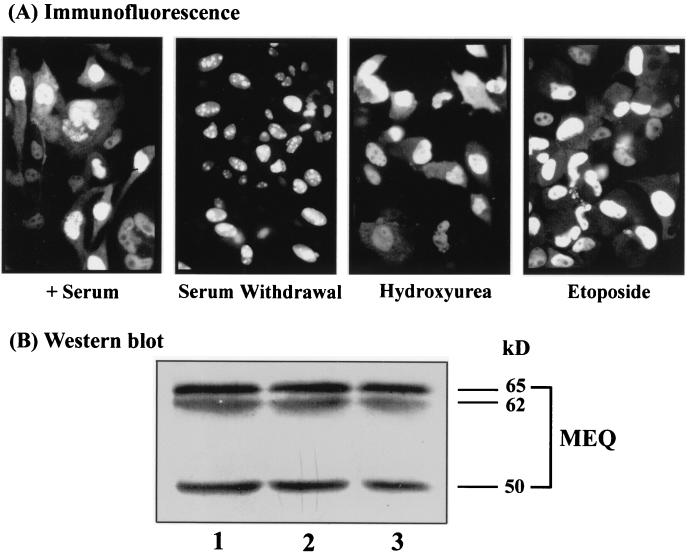
To study the biochemical and transformation properties of MEQ in isolated form, we previously established Rat-2 (MEQ) cells, a pool of clones overexpressing MEQ. These cells exhibited distinct transformed phenotypes, including morphological alterations, anchorage- and serum-independent growth, and resistance to apoptosis (40). We also showed that MEQ is localized primarily in the nucleus, especially in the nucleolus and coiled bodies (39). However, some cytoplasmic staining of MEQ was also observed (Fig. 1A, left panel). One of the hallmarks of transforming proteins is their ability to interact with cellular growth-signaling components and cell cycle regulators. In this study, we set out to examine the subcellular localization of MEQ at different stages of the cell cycle and under different growth conditions. To this end, Rat-2 (MEQ) cells were treated with several cell cycle-blocking agents, which have been routinely used in cell cycle synchronization studies and do not interfere with protein localization per se, and then subjected to indirect immunofluorescence assays to monitor the subcellular localization of MEQ during the course of cell cycle progression. As shown in Fig. 1, the vast majority of MEQ proteins localized to the nucleus and the nucleolus in serum-deprived Rat-2 (MEQ) cells, which are composed of cells primarily arrested in the G0/G1 phase (see below). By contrast, cytoplasmic and nuclear localization of MEQ protein was observed in hydroxyurea-treated cells, which are mostly in early S phase. Likewise, the cytoplasmic localization of MEQ was also detected to a lesser extent in etoposide-treated cells, which are blocked mainly at the S/G2 transition. These preliminary results suggested that subcellular localization of MEQ protein might be cell cycle dependent. During G0/G1 phase, MEQ is strictly localized to the nucleus and nucleolus. As the cell cycle progresses from G1 to early S phase, a significant fraction of MEQ exits into the cytoplasm. We have also analyzed the overall expression levels of MEQ protein during cell cycle progression by Western blotting, as shown in Fig. 1B. Total lysates of Rat-2 (MEQ) cells, which were withdrawn from serum for 3 days (Fig. 1B, lane 1) and released from serum withdrawal for 12 h (lane 2) or 24 h (lane 3) were collected and resolved by SDS-PAGE. As reported previously (40), three bands representing different forms of MEQ were evident, and their levels do not fluctuate as cells progress from G0/G1 to S phase. The cytoplasmic accumulation of MEQ is therefore more probably due to nuclear export than to de novo synthesis. To ensure that our interpretation was accurate, we examined the cell cycle profile of Rat-2 (MEQ) cells compared with that of vector-infected Rat-2 [Rat-2 (Vector)] cells by using cell cycle-blocking agents. As analyzed by flow cytometry (Fig. 2), without any treatment there was a 21% increase in the number of Rat-2 (MEQ) cells in the G2/M phase compared to the number of Rat-2 (Vector) cells, indicating that MEQ transformation perturbs cell cycle control. As also shown in Fig. 2, only 65% of serum-starved Rat-2 (MEQ) cells were arrested at the G0/G1 phase. This observation is consistent with the previously published finding that MEQ is capable of triggering serum-independent growth (40). Meanwhile, 86% of Rat-2 (MEQ) cells treated with 1 mM hydroxyurea, a ribonucleotide reductase inhibitor, were blocked at early S phase. Conversely, when Rat-2 (MEQ) cells were treated with 2 μM etoposide, a topoisomerase II inhibitor, 58% of the cells were blocked in S phase and 12% were blocked at the G2/M phase. More than 50% of the Rat-2 (MEQ) cells were blocked at G2/M phase when treated with an inhibitor of microtubule formation, 0.4 μg of nocodazole per ml. Together with the results of immunofluorescence assays shown in Fig. 1, these data suggest that a fraction of MEQ oncoproteins are localized in the cytoplasm during early S phase.
FIG. 1.
Subcellular localization of MEQ protein. (A) Rat-2 (MEQ) cells were grown in the presence of serum or treated with a variety of cell cycle-blocking vehicles, including serum withdrawal, 1 mM hydroxyurea, 2 μM etoposide, and 0.4 μg of nocodazole per ml. Indirect immunofluorescence assays were performed with rabbit anti-MEQ polyclonal antibodies (1:200 dilution), followed by FITC-conjugated anti-rabbit IgGs (1:600 dilution), and the cells were examined under a fluorescence microscope. Final magnification, ×240. (B) Expression of MEQ protein as determined by Western blotting. Total cell extracts of Rat-2 (MEQ) cells undergoing serum withdrawal for 3 days (lane 1) and released from serum withdrawal for 12 h (lane 2) or 24 h (lane 3) were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The blot was probed with rabbit anti-MEQ polyclonal antibodies (1:4,000 dilution) followed by alkaline phosphatase-conjugated goat anti-rabbit IgGs (1:3,000 dilution).
FIG. 2.
Cell cycle profiles of Rat-2 (MEQ) cells. Rat-2 (MEQ) cells similarly treated with different cell cycle-blocking reagents as described in the legend to Fig. 1 were harvested by trypsinization and incubated with 125 μg of propidium iodide per ml by the method described in the Cycle Test Plus DNA reagent kit (Becton Dickinson), and their cell cycle profiles were analyzed with a flow cytometer (Becton Dickinson).
To substantiate that MEQ proteins are exported into the cytoplasm during the G1/S transition, BrdU incorporation in hydroxyurea-treated Rat-2 (MEQ) cells was measured. Briefly, cells were treated with both 1 mM hydroxyurea and 50 μM BrdU for 12 h after 3 days of serum withdrawal and then doubly labeled with antibodies against MEQ (rabbit polyclonal antibodies) and BrdU (mouse MAb). The results, analyzed by a confocal laser scanning microscopy (Fig. 3), show that MEQ protein is expressed in the nucleus or nucleolus in cells without BrdU incorporation (G0/G1 phase). By contrast, cytoplasmic localization of MEQ protein was found in some cells with BrdU incorporation (early S phase) but not in others (the far-left cell).
FIG. 3.
Cell cycle-dependent cytoplasmic translocation of MEQ oncoprotein during the S phase. Rat-2 (MEQ) cells were serum starved for 3 days before being treated with 1 mM hydroxyurea and 50 μM BrdU. The cells were subsequently doubly labeled with rabbit anti-MEQ polyclonal antibodies (1:200 dilution) and mouse anti-BrdU MAb (undiluted) followed by secondary antibodies and analyzed with a confocal microscope. MEQ protein is expressed in the nucleus or nucleolus in cells without BrdU incorporation (yellow arrows); it is found in the cytoplasm in some cells with BrdU incorporation (early S phase) (blue arrows) but not in others (far-left cell). Final magnification, ×690.
Colocalization of MEQ and CDK2 in coiled bodies.
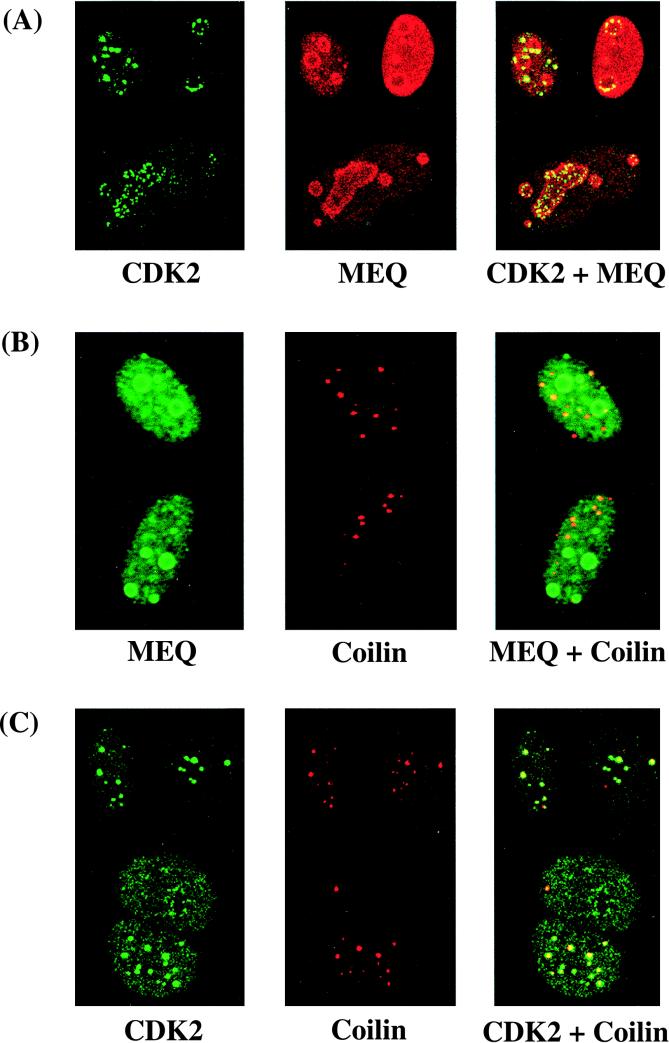
Since the subcellular localization of MEQ protein is cell cycle dependent and the CDK-cyclin complexes are key regulators of cell cycle progression, we sought to explore the possible involvement of CDK-cyclin complexes in the regulation of MEQ protein. Antibodies specific for various CDKs and MEQ were differentially labeled, and the subcellular localization of the respective molecules was examined by indirect immunofluorescent staining assays. Most of the CDKs such as CDK1 were dispersed throughout the nucleoplasm and/or cytoplasm (Fig. 4), which is different from the intense staining of MEQ in the nucleoli and coiled bodies. However, a significant portion of CDK2 colocalizes prominently with MEQ in the nucleolar periphery and nuclear foci resembling coiled bodies (Fig. 5A). In our previous publication (39), we demonstrated the localization of MEQ in coiled bodies by using MAb against fibrillarin as a marker. Since fibrillarin is expressed in both the nucleolus and coiled bodies, we chose to use in the present study rabbit antisera against p80-coilin, a structural protein specific for the coiled body, to confirm that these nuclear foci are coiled bodies in double-labeling assays. As shown in Fig. 5B and C, much of the MEQ protein or CDK2 colocalizes with p80-coilin in coiled bodies. However, some MEQ proteins do not localize to coiled bodies and some coiled bodies in which MEQ is not expressed (Fig. 5B). We interpret this to mean that the localization of MEQ in coiled bodies is not mediated by association with p80-coilin per se but, rather, with another limiting cellular factor(s). We are currently investigating other subnuclear compartments to which MEQ may localize, one of which might be the PML body.
FIG. 4.
CDK1 does not colocalize with the MEQ oncoprotein. Rat-2 (MEQ) cells were doubly labeled with mouse anti-CDK1 MAb (1:100 dilution) and rabbit anti-MEQ polyclonal antibodies (1:200 dilution) followed by secondary antibodies and analyzed with a confocal microscope. Final magnification, ×1,000.
FIG. 5.
Colocalization of MEQ oncoprotein and CDK2 in coiled bodies and the nucleolar periphery. Rat-2 (MEQ) cells were doubly labeled with mouse anti-CDK2 MAb (1:100 dilution) and rabbit anti-MEQ polyclonal antibodies (1:200 dilution) (A), mouse anti-MEQ MAb (1:100 dilution) and rabbit anti-p80 coilin polyclonal antibodies (1:500 dilution) (B), and mouse anti-CDK2 MAb (1:100 dilution) and rabbit anti-p80 coilin polyclonal antibodies (1:500 dilution) (C). FITC- or Texas red-conjugated secondary antibodies (1:600 and 1:300 dilutions, respectively) were then applied, and the cells were analyzed with a confocal microscope. Final magnification, ×1,000.
The localization of CDK2 in coiled bodies is also cell cycle dependent.
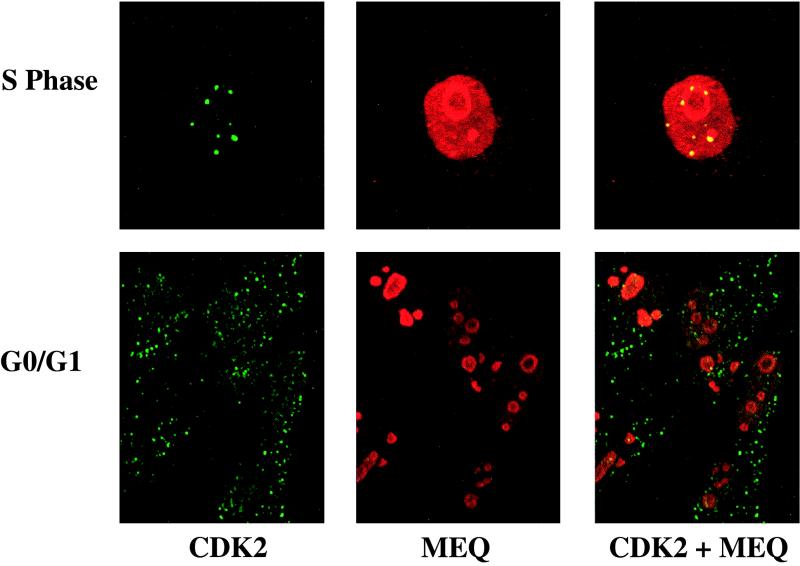
Additional experiments demonstrate that the localization of CDK2 in coiled bodies is cell cycle dependent. Rat-2 (MEQ) cells were blocked at different phases of the cell cycle as described above. As shown in Fig. 6, the colocalization of MEQ and CDK2 in coiled bodies and the nucleolar periphery is observed only during the G1/S boundary and early S phase, whereas CDK2 is dispersed in punctate foci throughout the nucleoplasm and cytoplasm during G0/G1 phase. It is conceivable that colocalization of MEQ and CDK2 in coiled bodies and the nucleolar periphery during the G1/S boundary and early S phase could lead to phosphorylation of MEQ protein and its consequential cytoplasmic retention.
FIG. 6.
Colocalization of MEQ oncoprotein and CDK2 is cell cycle dependent. Rat-2 (MEQ) cells were serum deprived for 3 days to be growth arrested at the G0/G1 phase or treated with 1 mM hydroxyurea to block at the S phase as described in Materials and Methods. These cells were subsequently doubly labeled with mouse anti-CDK2 MAb (1:100 dilution) and rabbit anti-MEQ polyclonal antibodies (1:200 dilution) followed by secondary antibodies and analyzed with a confocal microscope. Final magnification, ×1,000.
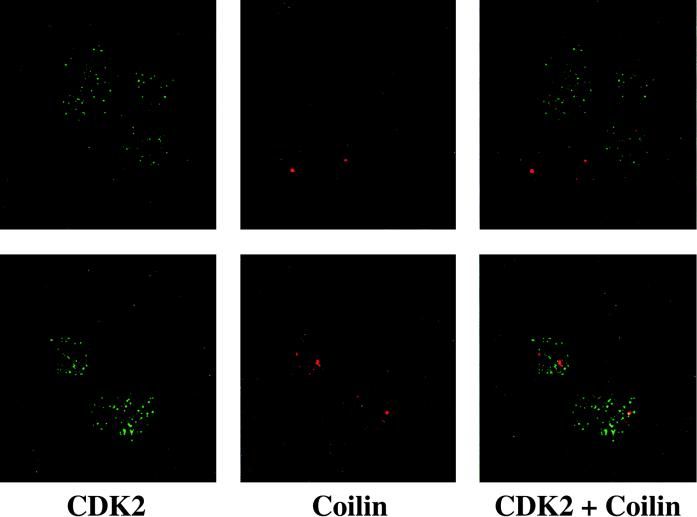
The number and size of coiled bodies usually reflect the proliferation state and metabolic activity of the cells (5). Therefore, it is not surprising to find fewer and smaller coiled bodies in untransformed Rat-2 (Vector) cells than in Rat-2 (MEQ) cells (Fig. 7). Interestingly, in Rat-2 (Vector) cells, CDK2 molecules are not concentrated in coiled bodies with the exception of one (Fig. 7). These data indicate that the localization of CDK2 in coiled bodies may be mediated by MEQ and/or may be transformation associated.
FIG. 7.
CDK2 does not colocalize to coiled bodies in untransformed Rat-2 cells. Untransformed Rat-2 (Vector) cells were doubly labeled with mouse anti-CDK2 MAb (1:100 dilution) and rabbit anti-p80 coilin polyclonal antibodies (1:500 dilution) followed by secondary antibodies and analyzed with a confocal microscope. Two representative fields are shown (top and bottom panels). Final magnification, ×1,000.
Serine 42 of MEQ is the primary site of phosphorylation by CDKs.
Having demonstrated the cell cycle-dependent colocalization of MEQ and CDK2 in vivo, we asked whether there is a direct interaction between CDK2 and MEQ and, specifically, whether MEQ is phosphorylated by CDK2. We showed first that MEQ is phosphorylated in vivo (see Fig. 9D). In vitro immune complex kinase assays showed that MEQ protein is a substrate for phosphorylation by a variety of CDK-cyclin complexes, including CDK1-cyclin B, CDK2-cyclin A, and CDK2-cyclin E, isolated from CV-1 cells with EE epitope MAb-conjugated Affi-Gel 10 beads (Fig. 8). To identify the phosphorylation site(s), we constructed a series of mutants with lesions at serine and threonine residues. We noticed that there is a potential CDK phosphorylation consensus site (S42PSK) located between the BR1 and BR2 regions of MEQ protein. An S42G mutant and two other mutants, STS29P (3 amino acids for one mutation) and T79V, with mutations in the serine or threonine residues were then constructed. In addition, an array of deletion mutants of MEQ carrying various truncations was developed. To ensure that the phosphorylation detected is due to CDK-cyclin and not to contaminating kinases, we used the purified system. Since purified CDK2-cyclin A and CDK2-cyclin E are not available, we used purified CDK1-cyclin B complex (UBI) as the phosphorylating kinase. This is justified because CDK1-cyclin B and CDK2-cyclins have similar substrate specificities in vitro (77) and our data corroborated that the CDK1-cyclin B complex displayed a kinase specificity similar to that of immunoprecipitated CDK-cyclin complexes based on tryptic digestion and two-dimensional gel electrophoresis (data not shown). Figure 9 confirms that MEQ protein is an excellent substrate for CDK phosphorylation. Furthermore, different truncation mutants (as illustrated in Fig. 9A), namely, MEQ-bZIP, MEQ-bZIP (ΔBR1), and MEQ-bZIP (ΔBR2), whose S42PSK sequences are still intact continue to be strongly phosphorylated by CDK1-cyclin B, as does the STS29P mutant (Fig. 9B). By contrast, the phosphorylation is significantly reduced in the S42G mutant, supporting the notion that the S42 residue is the primary CDK phosphorylation site. Based on the amounts of MEQ proteins loaded in individual lanes (Fig. 9B), we estimated there is at least a 10-fold reduction of phosphorylation compared to that in MEQ-bZIP. Intermediate phosphorylation is observed in the T79V mutant, which suggests that RKQT79DY might be a minor CDK phosphorylation site in vitro. Figure 9C showed that the MEQ-bZIP (S42G) mutant protein was phosphorylated by PKA, PKC, and MAPK at a level comparable to that of wild-type MEQ-bZIP protein, suggesting that the S42 residue is a specific site for CDK phosphorylation.
FIG. 9.
The serine 42 residue is the primary site of phosphorylation by CDKs. (A) An array of bacterially expressed truncation or point mutants of MEQ protein was purified with Talon (Clontech). (B) MEQ-bZIP (S42G) is not appreciatively phosphorylated by CDK1-cyclin B. In vitro kinase assays were performed on MEQ bZIP and mutant proteins with CDK1-cyclin B complex in the presence of [γ-32P]ATP at 37°C for 30 min. The samples were resolved by SDS-PAGE and exposed to an X-ray film for 3 min. The same amount of MEQ mutant protein was loaded on a separate SDS-PAGE gel, and the Western blot was detected with rabbit anti-MEQ polyclonal antibodies (1:4,000 dilution). (C) MEQ-bZIP and MEQ-bZIP (S42G) mutant proteins were also phosphorylated by other serine/threonine kinases, including PKA, PKC, and MAPK, in vitro, and the gels were exposed to X-ray films at different intervals (5 min for PKA and PKC and 15 min for MAPK). (D) MEQ is a phosphoprotein in vivo. MEQ, in the context of pTM1 vector with an EE epitope tag, was transfected into CV1 cells with recombinant vaccinia virus. At 24 h later, the cells were labeled with [γ-32P]ATP and MEQ protein was immunoprecipitated with EE epitope MAb-conjugated Affi-Gel 10 beads. Purified protein was resolved by SDS-PAGE, and the blot was probed against MEQ polyclonal antibodies. CV-1 cells were also transfected with PTM1 vector alone as a negative control.
FIG. 8.
MEQ oncoprotein is an excellent substrate for phosphorylation by different CDK-cyclin complexes. In vitro kinase assays were performed on purified MEQ-bZIP protein with EE epitope MAb-immunoprecipitated CDK1-cyclin B, CDK2-cyclin A, and CDK2-cyclin E complexes (24) in a final reaction mixture of 20 μl containing 50 mM Tris (pH 7.4), 1 mM dithiothreitol 10 mM MgCl2, and 10 μCi of [γ-32P]ATP per reaction incubated at 37°C for 30 min. The samples were subjected to SDS-PAGE (12.5% polyacrylamide) and exposed to an X-ray film for 10 min.
Regulation of MEQ by CDK phosphorylation.
Phosphorylation by serine/threonine protein kinases affects the function of transcription factors in many ways, including their DNA-binding property, transactivation potential, and nuclear import and/or export (reviewed in references 7 and 25). Described below are two such properties of MEQ that have been altered by CDK phosphorylation.
(i) Nuclear export of MEQ proteins is facilitated by CDK phosphorylation.
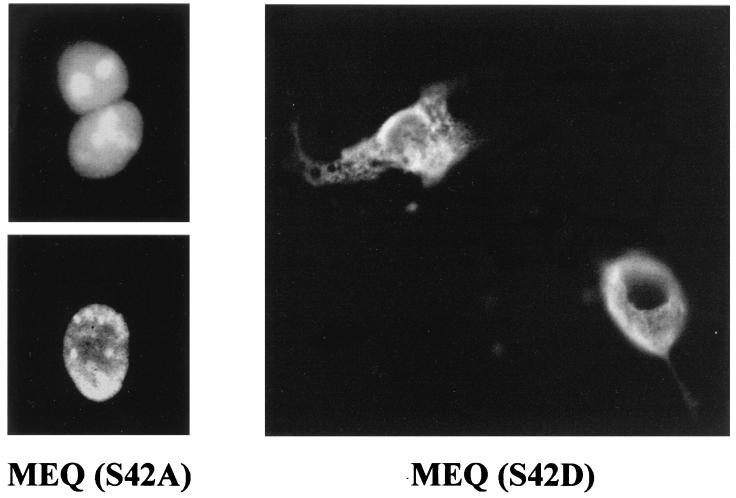
CDK phosphorylation promotes cytoplasmic retention of a number of nuclear proteins in a cell cycle-dependent manner (27, 28, 47, 52, 62). Having demonstrated that MEQ colocalizes with CDK2 in coiled bodies and the nucleolar periphery during S phase and that MEQ is phosphorylated by CDK substantially in vitro at the S42 residue, we wished to explore the possible effects of CDK phosphorylation on subcellular localization of MEQ protein. As shown above, only a fraction of wild-type MEQ is expressed in the cytoplasm of Rat-2 (MEQ) cells during S phase. We hypothesize that the cytoplasmic translocation of MEQ is dependent on its accessibility to CDK phosphorylation and that only MEQ proteins phosphorylated by CDK2 in coiled bodies and the nucleolar periphery would be exported to the cytoplasm. To test our hypothesis, we first mutated the S42 residue into either alanine (A) to render the residue unphosphorylatable or aspartic acid (D) to simulate the constitutively phosphorylated state. MEQ (S42A) and MEQ (S42D) mutants in the context of the pSVL vector were then transiently transfected into COS1 cells. Immunofluorescent staining was performed 2 days after transfection. Figure 10 shows that MEQ (S42A) mutant protein localizes to the nucleus and nucleolus like wild-type MEQ protein. By contrast, the cytoplasmic localization of MEQ (S42D) mutant proteins is clearly enhanced compared to that of wild-type MEQ. Since CDK2 is known to be a kinase that is active during the G1/S transition and S phase and since the cytoplasmic localization of MEQ occurs only during early S phase (see above), these results implicate CDK2 in the regulation of cytoplasmic translocation of MEQ by phosphorylation of its S42 residue.
FIG. 10.
Phosphorylation on the S42 residue is crucial for the cytoplasmic translocation of MEQ oncoprotein. The S42 residue of MEQ protein was mutated into either Asp or Ala in vitro and transfected into COS1 cells in the context of pSVL vector. At 48 h posttransfection, S42A- and S42D-transfected COS1 cells were stained with rabbit anti-MEQ polyclonal antibodies (1:200 dilution) followed by FITC-conjugated secondary antibody (1:600 dilution) and examined under a fluorescence microscope. Final magnification, ×1,000.
(ii) DNA binding activity of MEQ is reduced by CDK phosphorylation.
Another aspect in which transcription factors could be regulated by kinase phosphorylation is in their DNA-binding activity. We previously demonstrated that MEQ protein binds to specific DNA sequences, designated MERE1 and MERE2 (59). A MERE1 (TRE) probe was used in our assays to evaluate the potential regulation of MEQ DNA binding by serine/threonine kinase phosphorylation. MEQ-bZIP protein was first phosphorylated in vitro by CDK1-cyclin B complex, PKA, PKC, or MAPK. The subsequent electrophoretic mobility shift assay shows that MERE1-binding activity of MEQ is diminished by CDK1 phosphorylation compared to the activities of unphosphorylated MEQ or of MEQ phosphorylated by other kinases (Fig. 11). On the other hand, the DNA-binding activity of PKC-phosphorylated MEQ is elevated. These data suggest that the DNA-binding activity of MEQ is negatively regulated by CDK phosphorylation.
FIG. 11.
The DNA-binding activity of the MEQ oncoprotein is alleviated by CDK phosphorylation. MEQ bZIP protein was phosphorylated by a variety of serine/threonine kinases including CDK1, PKA, PKC, and MAPK, as described in the legend to Fig. 9, but in the presence of cold ATP. Electrophoretic mobility shift assays were used to evaluate the MERE1 (TRE)-binding activity of MEQ-bZIP protein as described in Materials and Methods.
DISCUSSION
There is considerable evidence to suggest that MEQ is one of the key components involved in cellular transformation by MDV. We are beginning to understand the mode of action of MEQ during the transformation process (40, 58). Given the strong homology of the bZIP domain of MEQ to the Jun/Fos family of transcription factors and its ability to form dimers with Jun and Fos, it is tempting to speculate that MEQ, like v-jun and v-fos, may transform cells by transcriptional deregulation and subverting the c-jun/c-fos pathways. Indeed, we showed that MEQ is a potent transcriptional regulator which targets TRE and CRE motifs in viral and cellular promoters (59). On the other hand, DNA tumor virus oncoproteins often evolve to converge multiple routes to interfere with cellular pathways, including cell growth and differentiation, apoptosis, and cell cycle regulation of the host cells (see below). In our previous studies, we noticed that MEQ is localized in the nucleolus and coiled bodies and occasionally in the cytoplasm (39) and that it exhibits strong antiapoptotic effects (40). These unique features, which are not shared by c-Jun and c-Fos, suggest that MEQ is engaged in activity other than transcription. This study was aimed at exploring the novel functions of MEQ aside from its being a transcriptional regulator. We found that MEQ colocalizes with CDK2 in coiled bodies and that such colocalization is cell cycle dependent. Although the precise functions of coiled bodies remain elusive, the presence of splicing factors such as small nuclear ribonucleoproteins as well as many nucleolar proteins in coiled bodies has led to the suggestion of its possible participation in mRNA splicing and rRNA synthesis. Coiled bodies may serve as the reservoirs for assembly, maturation, transport, and recycling of splicing factors and nucleolar proteins (reviewed in references 5, 19, and 43). The involvement of coiled bodies during the transformation process is even more obscure. It is known, however, that the formation of coiled bodies, like that of nucleoli, parallels the proliferation state and metabolic activity of cells. Therefore, more and larger coiled bodies are expected to be observed in transformed cells than in untransformed cells (5) (Fig. 7).
To our knowledge, our report is the first to document the localization of CDK2 in coiled bodies. However, CDK2 is not the first kinase to be identified in coiled bodies. PKA (68), DAI/PKR (31), a double-stranded RNA-activated protein kinase, and CDK7-cyclin H-MAT1 complexes (33) have all been detected in coiled bodies. CDK7-cyclin H-MAT1 complexes are especially relevant to our study, since they are not only part of the TFIIH subunit of RNA polymerase II but also include CAK, the CDK-activating kinase (66). CAK is known to activate CDK2 by phosphorylating the T160 residue of CDK2. We report here that CDK2 is localized in coiled bodies during the G1/S boundary and early S phase. Since the antibodies against rat cyclins A and E are not suitable for immunofluorescence, we have studied the subcellular localization of cyclins in human tumor cell lines such as HeLa and MCF-7. Our preliminary data suggests that cyclin E instead of cyclin A is expressed in coiled bodies (data not shown). CDK2-cyclin E is known to trigger G1/S transition, and it also functions in the S phase, including initiation of DNA synthesis (37). We hypothesize that during the course of transformation, MEQ directly or indirectly navigates the translocation of CDK2 to coiled bodies, where they can be activated by CAK. Coincidentally, one of the major substrates of CDK2, the pRb tumor suppressor protein, localizes to PML bodies (65), which are often physically in contact with coiled bodies (15, 26). This close spatial relationship between the cell cycle regulators and gatekeepers could thus confer an advantage to transformed cells in deregulating cell cycle progression. Experiments to determine whether pRb proteins are indeed phosphorylated to a higher degree in Rat-2 (MEQ) cells and whether MEQ is directly involved in the translocation of CDK2 and the regulation of the kinase activity and substrate accessibility are under way.
If MEQ indeed translocates CDK2 into coiled bodies to promote cell cycle progression, it joins a growing list of DNA tumor virus oncoproteins which utilize cell cycle deregulation as a strategy to transform host cells. Several strategies are used by these oncogenic viruses (30, 69). First, some viruses such as adeno-associated virus type 2 (22), human papillomavirus (HPV) type 16 (23), and simian virus 40 (SV40) (70) modulate the expression of cell cycle-regulatory genes. Second, genomes of some viruses encode cyclin homologs on their own, such as the v-cyclin of herpes simplex virus type 1 (HSV-1) (34) and human herpesvirus 8 (38). Third, they sequester tumor suppressor proteins such as (i) Rb, as do SV40 large T antigen (14), adenovirus E1A (74), HPV-16 E7 (16), Epstein-Barr virus EBNA-3C (54), and cytomegalovirus (CMV) IE1 (56) and IE2 (21), or (ii) p53, as do SV40 large T antigen (12), Ad E1B (78), HPV-16 E6 (73), and CMV IE2 (6) and mtrII (48). Fourth, they interact with and stabilize CDK-cyclin complexes, as SV40 large T antigen does with CDK2-cyclin A (1), HSV-1 ICP0 (36) and human T-cell leukemia virus type 1 (HTLV-1) Tax (50) with cyclin D3, human immunodeficiency virus Tat with CDK7 (13, 49) and CDK9-cyclin T (71), HPV-16 E7 with CDK2-cyclin E (44) and CDK2-cyclin A (2), and HTLV-1 Tax (4) and adenovirus E1A and VP16 with CDK8 (20). Fifth, they inactivate CDK inhibitors by association, as p16INK4A is inactivated by HTLV-1 Tax (41, 64), p21WAF1 by HTLV-1 Tax (55) or HPV-16 E7 (18), and p27 KIP1 by adenovirus E1A (42) or HPV-16 E7 (79). Finally, CMV, in a manner similar to that of MDV, induces the nuclear translocation of CDK2 in serum-starved and contact-inhibited cells (9), although translocation into coiled bodies is not reported. It thus seems that deregulation of host cell cycle progression is a common and crucial step during the transformation processes undertaken by DNA tumor viruses. The ability of MEQ to interact with CDK2 in coiled bodies adds yet another clever strategy.
The interaction of MEQ with CDK2 has other important consequences: MEQ becomes phosphorylated, binds weakly to DNA, and, perhaps as a consequence, exits into the cytoplasm. Since only a portion of wild-type MEQ protein in Rat-2 (MEQ) cells interact with CDK2 in coiled bodies and the nucleolar periphery, only a fraction would be phosphorylated by CDK2 and consequently exported to the cytoplasm. By contrast, the MEQ (S42D) mutant was designed to mimic the activation by endogenous CDK2 phosphorylation. As a result, all the MEQ (S42D) mutant proteins are presumably in an active state and their enhanced expression in the cytoplasm is expected. Coincidentally, CDK phosphorylation of nuclear proteins often leads to their cytoplasmic translocation (27, 28, 47, 52, 62), and MEQ is apparently affected in a similar fashion. We have provided evidence that the major CDK2 phosphorylation site in vitro is serine 42, which is located between BR1 and BR2. BR1 and BR2 are the NLS for MEQ protein (39). Phosphorylation within or adjacent to these NLS by kinases could potentially affect their interactions with importin-α and importin-β, the NLS receptor on the nuclear membrane, and hence their entry to the nucleus (reviewed in references 7 and 29). Alternatively, CDK2 phosphorylation of MEQ may enhance the interaction between exportin and a nearby putative nuclear export signal of MEQ (130LTVTLGLL137) (17, 46, 72). Additionally, we have shown that phosphorylation by CDK2 reduces the DNA-binding activity of MEQ (Fig. 11), which may reduce the nuclear retention of this protein. The above mechanisms are not mutually exclusive and must be resolved by further development of MEQ mutants within the specific domains involved in nuclear entry and export. However, one caveat should be borne in mind, i.e., that our hypothesis is founded on the premise that CDK1-cyclin B and CDK2-cyclin E complexes exhibit similar substrate specificities in vitro (reference 77 and our unpublished results). The conclusive proof requires the availability of purified CDK2-cyclin E complex in the future.
A key question, then, is why MEQ needs to assume a cytoplasmic lifestyle during the S phase. We can only speculate that MEQ, with its proline-rich domain which fits the consensus binding site of the SH3 (Src homology) domain, may interact with the cytoplasmic signaling molecule to facilitate transformation. Indeed, we found that MEQ can bind c-Src kinase, although we have no direct evidence that this interaction is functionally relevant to transformation. Alternatively, it is conceivable that enhanced transport of MEQ may be relevant to the maintenance of the latent state of MDV. We noticed that MEQ is expressed early after infections (37a) and that there is a MEQ-binding site at the putative origin of replication. Previous data showed that MEQ binds to this site and thus may modulate the replication potential of the genome (10). Reducing the ability of MEQ to bind DNA and to exit to the cytoplasm may be one way to ensure that the viral genome stays dormant in S phase. This explanation, however, is complicated by the observation that a fraction of MEQ still remains in the nucleus during the S phase. It is noteworthy that shuttling viral proteins between the cytoplasm and nucleus is common for several viruses, including ICP27 (60) and UL11 of HSV-11 (3), human immunodeficiency virus Rev (45), HTLV-1 Rex (53), and influenza virus NS2 (51). In most of these instances, they help transport viral RNAs during replications. MEQ, given its RNA-binding capability, may also be able to function that way. The development of S42D and S42A mutants that have preferences for cytoplasmic or nuclear locations enables us to begin to probe the questions raised above.
In summary, MEQ, like other DNA tumor virus oncoproteins, is versatile and appears to interact with multiple cellular pathways. While their significance is not fully appreciated yet, we demonstrated here the novel interaction of MEQ with CDK2, their colocalization in coiled bodies, and the cell cycle-dependent subcellular localization of MEQ. The information reported and the mutants generated provide a framework for future studies of the role of MEQ in cellular transformation as well as in viral replication.
ACKNOWLEDGMENTS
We thank E. Chan for the anti-p80 coilin antibody and M. Pendergast for assistance with the confocal laser-scanning microscopy. We also thank G. Matera for invaluable discussions and A. W. Grasso for critical reading of the manuscript.
This work was supported by grants from the USDA (93-37204-9340 to L.F.L. and H.-J.K.), the NCI (CA46613 to H.-J.K.), and the council for Tobacco Research (4034 to H.-J.K.). J.-L.L. is the recipient of a USDA fellowship.
REFERENCES
- 1.Adamczewski J P, Gannon J V, Hunt T. Simian virus 40 large T antigen associates with cyclin A and p33cdk2. J Virol. 1993;67:6551–6557. doi: 10.1128/jvi.67.11.6551-6557.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arroyo M, Bagchi S, Raychaudhuri P. Association of the human papillomavirus type 16 E7 protein with the S-phase-specific E2F-cyclin A complex. Mol Cell Biol. 1993;13:6537–6546. doi: 10.1128/mcb.13.10.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines J D, Jacob R J, Simmerman L, Roizman B. The herpesvirus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J Virol. 1995;69:825–833. doi: 10.1128/jvi.69.2.825-833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bex F, McDowall A, Burny A, Gaynor R. The human T-cell leukemia virus type 1 transactivator protein Tax colocalizes in unique nuclear structures with NF-κB proteins. J Virol. 1997;71:3484–3497. doi: 10.1128/jvi.71.5.3484-3497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohmann K, Ferreira J, Santama N, Weis K, Lamond A I. Molecular analysis of the coiled body. J Cell Sci Suppl. 1995;19:107–113. doi: 10.1242/jcs.1995.supplement_19.16. [DOI] [PubMed] [Google Scholar]
- 6.Bonin L R, McDougall J K. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J Virol. 1997;71:5861–5870. doi: 10.1128/jvi.71.8.5861-5870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulikas T. Phosphorylation of transcription factors and control of cell cycle. Crit Rev Eukaryotic Gene Expression. 1995;5:1–77. [PubMed] [Google Scholar]
- 8.Bradley G, Lancz G, Tanaka A, Nonoyama M. Loss of Marek’s disease virus tumorigenicity is associated with truncation of RNAs transcribed within BamHI-H. J Virol. 1989;63:4129–4135. doi: 10.1128/jvi.63.10.4129-4135.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bresnahan W A, Boldogh I, Ma T, Albrecht T, Thompson E A. Cyclin E/Cdk2 activity is controlled by different mechanisms in the G0 and G1 phases of the cell cycle. Cell Growth Differ. 1996;7:1283–1290. [PubMed] [Google Scholar]
- 10.Brunovskis P, Qian Z, Li D, Lee L F, Kung H-J. Functional analysis of the MDV basic-leucine zipper product, MEQ. In: Silva R F, Cheng H H, Coussens P M, Lee L F, Velicer L F, editors. Current research on Marek’s disease. Kennett Square, Pa: American Association of Avian Pathologists; 1996. pp. 265–270. [Google Scholar]
- 11.Calnek B W. Marek’s disease-a model for herpesvirus oncology. Crit Rev Microbiol. 1985;12:293–320. doi: 10.3109/10408418509104432. [DOI] [PubMed] [Google Scholar]
- 12.Carroll R B, Gurney E G. Time-dependent maturation of the simian virus 40 large T antigen-p53 complex studied by using monoclonal antibodies. J Virol. 1982;44:565–573. doi: 10.1128/jvi.44.2.565-573.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O, Peterlin B M. The HIV transactivator TAT binds to the CDK-activating kinase and activities the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 15.de Jong L, Grande M A, Mattern K A, Schul W, van Driel R. Nuclear domains involved in RNA synthesis, RNA processing, and replication. Crit Rev Eukaryotic Gene Expression. 1996;6:215–246. doi: 10.1615/critreveukargeneexpr.v6.i2-3.60. [DOI] [PubMed] [Google Scholar]
- 16.Dyson N, Howley P M, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 17.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 18.Funk J O, Waga S, Harry J B, Espling E, Stillman B, Galloway D A. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gall J G, Tsvetkov A, Wu Z, Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev Genet. 1995;1:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- 20.Gold M O, Tassan J P, Nigg E A, Rice A P, Herrmann C H. Viral transactivators E1A and VP16 interact with a large complex that is associated with CTD kinase activity and contains CDK8. Nucleic Acids Res. 1996;24:3771–3777. doi: 10.1093/nar/24.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermanns J, Schulze A, Jansen-Durr P, Klein-Schmidt J A, Schmidt R, zur Hausen H. Infection of primary cells by adeno-associated virus type 2 results in a modulation of cell cycle-regulatory proteins. J Virol. 1997;71:6020–6027. doi: 10.1128/jvi.71.8.6020-6027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hickman E S, Bates S, Vousden K H. Perturbation of the p53 response by human papillomavirus type 16 E7. J Virol. 1997;71:3710–3718. doi: 10.1128/jvi.71.5.3710-3718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton L E, Templeton D J. The cyclin box and C-terminus of cyclins A and E specify CDK activation and substrate specificity. Oncogene. 1997;14:491–498. doi: 10.1038/sj.onc.1200851. [DOI] [PubMed] [Google Scholar]
- 25.Hunter T, Karin M. The regulation of transcription by phosphorylation. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 26.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jans P A, Ackerman M, Bischoff J R, Beach D H, Peter S. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV-40 T antigen proteins. J Cell Biol. 1991;115:1203–1212. doi: 10.1083/jcb.115.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jans D A, Moll T, Nasmyth K, Jans P. Cyclin-dependent kinase site-regulated signal-dependent nuclear localization of the SW15 yeast transcription factor in mammalian cells. J Biol Chem. 1995;270:17064–17067. doi: 10.1074/jbc.270.29.17064. [DOI] [PubMed] [Google Scholar]
- 29.Jans D A, Hubner S. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol Rev. 1996;76:651–685. doi: 10.1152/physrev.1996.76.3.651. [DOI] [PubMed] [Google Scholar]
- 30.Jansen-Durr P. How viral oncogenes make the cell cycle. Trends Genet. 1996;12:270–275. doi: 10.1016/0168-9525(96)81455-7. [DOI] [PubMed] [Google Scholar]
- 31.Jimenez-Garcia L F, Green S, Mathews M, Spector D L. Organization of the double-stranded RNA-activated protein kinase DAI and virus-associated VA RNA1 in adenovirus-2-infected HeLa cells. J Cell Sci. 1993;106:11–22. doi: 10.1242/jcs.106.1.11. [DOI] [PubMed] [Google Scholar]
- 32.Jones D, Lee L, Liu J-L, Kung H-J, Tillotson J K. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc Natl Acad Sci USA. 1992;89:4042–4046. doi: 10.1073/pnas.89.9.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jordan P, Cunha C, Carmo-Fonseca M. The cdk7-cyclin H-MAT1 complex associated with TFIIH is localized in coiled bodies. Mol Biol Cell. 1997;8:1207–1217. doi: 10.1091/mbc.8.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung J U, Stager M, Desrosiers R C. Virus-encoded cyclin. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato S, Hirai K. Marek’s disease virus. Adv Virus Res. 1985;30:225–277. doi: 10.1016/s0065-3527(08)60452-2. [DOI] [PubMed] [Google Scholar]
- 36.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knudsen E S, Buckmaster C, Chen T T, Feramisco J R, Wang J Y. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 1998;12:2278–2292. doi: 10.1101/gad.12.15.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Lee, L. Unpublished results.
- 38.Li M, Lee H, Yoon D W, Albrecht J C, Fleckenstein B, Neipel F, Jung J U. Kaposi’s sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J-L, Lee L F, Ye Y, Qian Z, Kung H-J. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J Virol. 1997;71:3188–3196. doi: 10.1128/jvi.71.4.3188-3196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J-L, Ye Y, Lee L F, Kung H-J. Transforming potential of the herpesviral oncoprotein MEQ: morphological transformation, serum-independent growth, and inhibition of apoptosis. J Virol. 1998;72:388–395. doi: 10.1128/jvi.72.1.388-395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Low K G, Dorner L F, Fernando D B, Grossman J, Jeang K T, Comb M J. Human T-cell leukemia virus type 1 Tax releases cell cycle arrest induced by p16INK4a. J Virol. 1997;71:1956–1962. doi: 10.1128/jvi.71.3.1956-1962.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mal A, Poon R Y, Nowe P H, Toyoshima H, Hunter T, Harter M L. Inactivation of p27Kip1 by the viral E1A oncoprotein in TGF beta-treated cells. Nature. 1996;380:262–265. doi: 10.1038/380262a0. [DOI] [PubMed] [Google Scholar]
- 43.Matera A G. Of coiled bodies, gems and salmon. J Cell Biochem. 1998;70:181–192. [PubMed] [Google Scholar]
- 44.McIntyre M C, Ruesch M N, Laimin L A. Human papillomavirus E7 oncoproteins bind a single form of cyclin E in a complex with cdk2 and p107. Virology. 1996;215:73–82. doi: 10.1006/viro.1996.0008. [DOI] [PubMed] [Google Scholar]
- 45.Meyer B E, Malim M H. The HIV Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 46.Michael W M, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 47.Moll T, Tebb G, Surana U, Kobitsch H, Nasymth K. The role of phosphorylation and the CDC28 kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- 48.Muralidhar S, Doniger J, Mendelson E, Araujo J C, Kashanchi F, Azumi N, Brady J N, Rosenthal L J. Human cytomegalovirus mtrII oncoprotein binds to p53 and down-regulates p53-activated transcription. J Virol. 1996;70:8691–8700. doi: 10.1128/jvi.70.12.8691-8700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nekhai S, Shukla R R, Kumar A. A human primary T-lymphocyte-derived human immunodeficiency virus type 1 Tat-associated kinase phosphorylates the C-terminal domain of RNA polymerase II and induces CAK activity. J Virol. 1997;71:7436–7441. doi: 10.1128/jvi.71.10.7436-7441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neuveut C, Low K G, Maldarelli F, Schmitt I, Majone F, Grassmann R, Jeang K-T. Human T-cell leukemia virus 1 Tax and cell cycle progression: role of cyclin D-cdk and p110Rb. Mol Cell Biol. 1998;18:3620–3632. doi: 10.1128/mcb.18.6.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Neil R E, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Neill E M, Kaffman A, Jolly E R, O’Shea E K. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 53.Palmeri D, Malim M H. The human T-cell leukemia virus type 1 posttranscriptional trans-activator Rex contains a nuclear export signal. J Virol. 1996;70:6442–6445. doi: 10.1128/jvi.70.9.6442-6445.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parker G A, Crook T, Bain M, Sara E A, Farrell P J, Allday M J. Epstein-Barr virus nuclear antigen (EBNA) 3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene. 1996;13:2541–2549. [PubMed] [Google Scholar]
- 55.Parker S F, Perkins N D, Gitlin S D, Nabel G J. A cooperative interaction of human T-cell leukemia virus type 1 Tax with the p21 cyclin-dependent kinase inhibitor activates the human immunodeficiency virus type 1 enhancer. J Virol. 1996;70:5731–5734. doi: 10.1128/jvi.70.8.5731-5734.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poma E E, Kowalik T F, Zhu L, Sinclair J H, Huang E S. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J Virol. 1996;70:7867–7877. doi: 10.1128/jvi.70.11.7867-7877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Powell P C. Marek’s disease virus in the chicken. Adv Viral Oncol. 1985;5:103–127. [Google Scholar]
- 58.Qian Z, Brunovskis P, Rauscher III F, Lee L, Kung H-J. Transactivation activity of Meq, a Marek’s disease herpesvirus bZIP protein persistently expressed in latently infected transformed T cells. J Virol. 1995;69:4037–4044. doi: 10.1128/jvi.69.7.4037-4044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qian Z, Brunovskis P, Lee L F, Vogt P K, Kung H-J. Novel DNA binding specificities of a putative herpesvirus bZIP oncoprotein. J Virol. 1996;70:7161–7170. doi: 10.1128/jvi.70.10.7161-7170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandri-Gordin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schat K A, Buckmaster A, Ross L J. Partial transcription map of Marek’s disease herpesvirus in lytically infected cells and lymphoblastoid cell lines. Int J Cancer. 1989;44:101–109. doi: 10.1002/ijc.2910440119. [DOI] [PubMed] [Google Scholar]
- 62.Schwab M S, Dreyer C. Protein phosphorylation sites regulate the function of the bipartite NLS of nucleolin. Eur J Cell Biol. 1997;73:287–297. [PubMed] [Google Scholar]
- 63.Sugaya K, Bradley N, Nonoyama M, Tanaka A. Latent transcripts of Marekís disease virus are clustered in the short and long repeat regions. J Virol. 1990;64:5773–5782. doi: 10.1128/jvi.64.12.5773-5782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki T, Kitao S, Matsushime H, Yoshida M. HTLV-1 Tax protein interacts with cyclin-dependent kinase inhibitor p16INK4A and counteracts its inhibitory activity towards CDK4. EMBO J. 1996;15:1607–1614. [PMC free article] [PubMed] [Google Scholar]
- 65.Szekely L, Pokrovskaja K, Jiang W Q, de The H, Ringertz N, Klein G. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J Virol. 1996;70:2562–2568. doi: 10.1128/jvi.70.4.2562-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tassan J-P, Schultz S J, Bartek J, Nigg E A. Cell cycle analysis of the activity, subcellular localization, and subunit composition of human CAK (CDK-activating kinase) J Cell Biol. 1994;127:467–478. doi: 10.1083/jcb.127.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tillotson J K, Kung H-J, Lee L F. The 3rd International Symposium on Marek’s Disease. Osaka, Japan: Japanese Association on Marek’s Disease; 1988. Accumulation of viral transcripts coding for a DNA binding protein in Marek’s disease tumor cells; pp. 128–134. [Google Scholar]
- 68.Trinczek B, Robert-Nicoud M, Schwoch G. In situ localization of cAMP-dependent protein kinases in nuclear and chromosomal structures: relation to transcriptional activity. Eur J Cell Biol. 1993;60:196–202. [PubMed] [Google Scholar]
- 69.Vousden K H. Regulation of the cell cycle by viral oncoproteins. Semin Cancer Biol. 1995;6:109–116. doi: 10.1006/scbi.1995.0014. [DOI] [PubMed] [Google Scholar]
- 70.Watanabe G, Howe A, Lee R J, Albanese C, Shu I W, Karnezis A N, Zon L, Kyriakis J, Rundell K, Pestell R G. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc Natl Acad Sci USA. 1996;93:12861–12866. doi: 10.1073/pnas.93.23.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 72.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 73.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 74.Whyte P, Williamson N M, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 75.Xie Q, Anderson A S, Morgan R W. Marek’s disease virus (MDV) ICP4, pp38, and meq genes are involved in the maintenance of transformation of MDCC-MSB1 MDV-transformed lymphoblastoid cells. J Virol. 1996;70:1125–1131. doi: 10.1128/jvi.70.2.1125-1131.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yan M, Dai T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R, Templeton D J. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 77.Yasuda H, Kamijo M, Ohba Y. The characterization of cdc2 kinase and CDK2. Yakugaku Zasshi. 1993;113:829–846. doi: 10.1248/yakushi1947.113.12_829. [DOI] [PubMed] [Google Scholar]
- 78.Zantema A, Schrier P I, Davis-Olivier A, van Laar T, Vaessen R T, van der Eb A J. Adenovirus serotype determines association and localization of the large E1B tumor antigen with cellular tumor antigen p53 in transformed cells. Mol Cell Biol. 1985;5:3084–3091. doi: 10.1128/mcb.5.11.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zerfass-Thome K, Zwerschke W, Mannhardt B, Tindle R, Botz J W, Jansen-Durr P. Inactivation of the cdk inhibitor p27KIP1 by the human papillomavirus type 16 E7 oncoprotein. Oncogene. 1996;13:2323–2330. [PubMed] [Google Scholar]