Abstract
Obesity is associated with cognitive decline. Recent observations in mice propose an adipose tissue (AT)–brain axis. We identified 188 genes from RNA sequencing of AT in three cohorts that were associated with performance in different cognitive domains. These genes were mostly involved in synaptic function, phosphatidylinositol metabolism, the complement cascade, anti-inflammatory signaling, and vitamin metabolism. These findings were translated into the plasma metabolome. The circulating blood expression levels of most of these genes were also associated with several cognitive domains in a cohort of 816 participants. Targeted misexpression of candidate gene ortholog in the Drosophila fat body significantly altered flies memory and learning. Among them, down-regulation of the neurotransmitter release cycle–associated gene SLC18A2 improved cognitive abilities in Drosophila and in mice. Up-regulation of RIMS1 in Drosophila fat body enhanced cognitive abilities. Current results show previously unidentified connections between AT transcriptome and brain function in humans, providing unprecedented diagnostic/therapeutic targets in AT.
The adipose tissue-brain axis regulates cognitive performance in humans, mice, and flies.
INTRODUCTION
There is an increased awareness that peripheral tissues modulate brain function shaping different cognitive domains. Recent studies in rodents revealed that restoring glucose metabolism of myeloid cells reversed cognitive decline in aging (1), whereas selective gastrointestinal vagal afferent ablation impaired hippocampus-dependent episodic and spatial memory in rats (2). The liver has also been proposed to play a major role in regulating feeding behavior in mice (3).
Obesity is associated with increased risk of cognitive impairment (4, 5). An increased fatness in middle-aged adults (<65 years) predicts decreased cognitive abilities in late life, mainly memory, executive functioning, and learning (6–8). However, the precise mechanisms involved remain largely unknown.
The brain regulates adipose tissue performance. For instance, sympathetic neuroadipose connections mediate lipolysis (9) and cytokine production (10), while neuroimmune interactions are involved in thermoregulation (11). Recent observations in rodents show that adipose tissue also regulates brain function. Genetically modified mice engineered to release the peptide NaKtide in adipocytes led to improved hippocampal memory through the inhibition of Na- and K-dependent adenosine triphosphatase (Na,K-ATPase) signaling in adipocytes (12). Visceral adipose NLR family Pyrin Domain Containing 3 (NLRP3) was linked to impaired memory in obese mice via interleukin-1R1 (IL-1R1) on CX3CR1+ cells, leading to microglial activation (13). An increased abundance of beige adipocytes in subcutaneous fat restored hippocampal synaptic plasticity in mice through induction of anti-inflammatory cytokine IL-4, protecting from obesity-induced cognitive impairment (14).
We have investigated in humans and animal models the mechanisms involved in the bidirectional interactions between adipose tissue gene expression and cognitive abilities. An RNA sequencing (RNA-seq) analysis of adipose tissue identified genes associated with different cognitive domains in discovery and different validation human cohorts. The gene expression of those genes most consistently associated with a battery of neuropsychological tests was then targeted in the fat body of Drosophila and in adipose tissue of mice to evaluate the possible functional implications. Last, we studied the expression of several of these genes in peripheral blood mononuclear cells (PBMCs) in association with cognitive traits in humans.
RESULTS
Adipose tissue gene expression is linked to several cognitive domains
To gain insights into the fat-brain axis, we performed a transcriptomics analysis (RNA-seq) of visceral adipose tissue (VAT) from 17 subjects with severe obesity [body mass index (BMI) > 35 mg/kg2] aged 28 to 60 years (table S1). Ten different cognitive tests assessed the functional performance of three cognitive domains: memory, executive function, and attention. The tests and their interpretation are shown in Materials and Methods.
Following normalization by trimmed mean of M value and calculating weights to account for variations in precision between observations, we fitted robust linear regression models with empirical Bayes moderation of the standard errors to identify gene transcripts associated with each cognitive test score controlling for age, sex, BMI, and years of education, all confounding factors know to affect cognitive function. There are other lifestyle-related factors known to cause or contribute to cognitive function such as physical activity, diet, or alcohol use (15). However, we found no significant associations of any of the 10 cognitive tests with physical activity (table S2), macronutrients, alcohol intake, glycemia, or hemoglobin A1c (fig. S1). Note that all subjects had morbid obesity (table S1).
We observed significant associations with gene transcripts in all cognitive tests at Benjamini-Hochberg procedure for false discovery rate (pFDR) of <0.05 (Fig. 1; fig. S2, A and B; and tables S3 to S12). Among these genes, NUDT2, AMPH, UNC5B, and OAT were those with most significant associations across the different cognitive domains (significant in seven, six, six, and eight cognitive tests at pFDR of <0.05, respectively). The attributed action of the proteins encoded by these genes was concordant with their function in the central nervous system (CNS), but their expression in adipose tissue in association with cognition is unprecedented. Variants in the NUDT2 gene have been recently associated with intellectual disability (16, 17). AMPH encodes for a protein highly concentrated in presynaptic terminals (18) and is one of the six proteins altered in the cerebrospinal fluid of patients with mild cognitive impairment and Alzheimer’s disease (19). Amph in Drosophila is an important postsynaptic protein that regulates the location of key components of glutamatergic signaling and dendrite dynamics, maturation, and synaptogenesis (20, 21). OAT is the gene that encodes for the enzyme ornithine aminotransferase (OAT), which plays a key role in transforming ornithine and arginine into glutamate and γ-aminobutyric acid as well as major excitatory and inhibitory neurotransmitters, respectively (22). In addition, genes involved in tryptophan metabolism such as KYNU, QPRT, and INMT were among the most significant genes associated with inhibitory control (Fig. 1A), attention (fig. S2A), and executive function (Fig. 1B). Consistently, subjects with obesity had impaired memory and inhibitory control through alterations in tryptophan metabolism (23, 24).
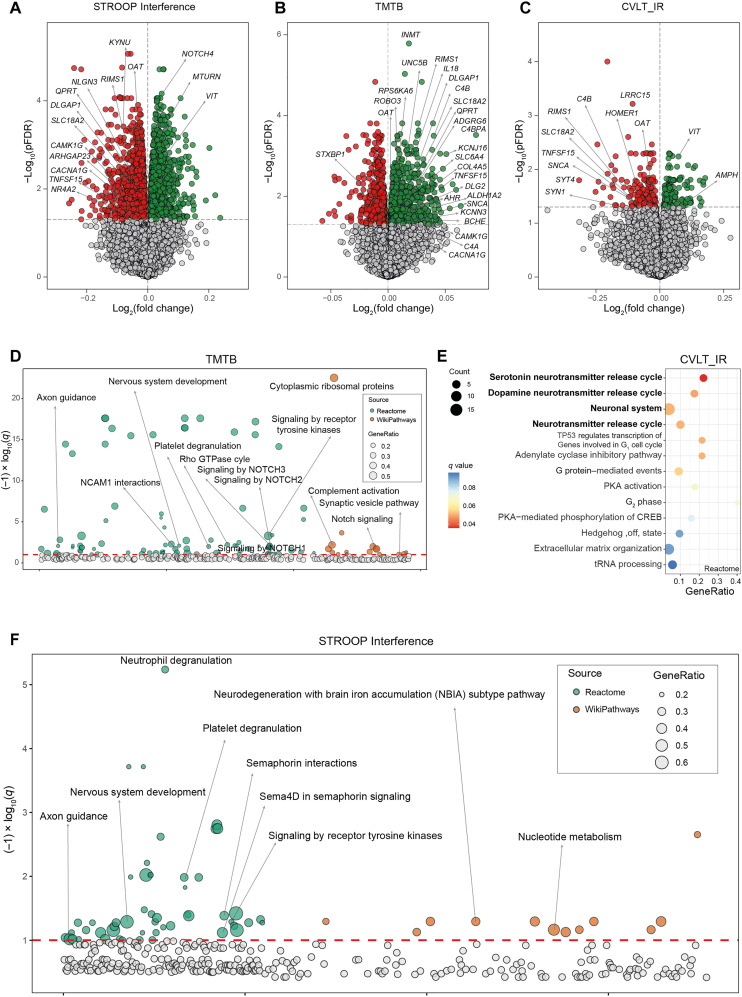
Fig. 1. Associations of VAT gene expression and cognitive domains in the discovery cohort.
Volcano plots of differentially expressed genes in the VAT associated with (A) the STROOP Interference tests, (B) the trail making test part B (TMTB), and (C) California verbal learning test immediate recall (CVLT_IR) scores in discovery cohort (IRONMET, n = 17) identified by limma-voom analysis controlling for age, BMI, sex, and education years. The log2 fold change associated with a unit change in the cognitive test score and the log10 P values adjusted for multiple testing (pFDR) are plotted for each gene. Differentially expressed genes (pFDR < 0.05) are colored in red and green indicating down-regulation and up-regulation, respectively. (D) Manhattan-like plot of pathways significantly associated (q < 0.1) with the TMTB in the VAT identified from a pathway overrepresentation analysis mapping significant genes to the Reactome and WikiPathways databases. (E) Dot plot of pathways significantly associated (q < 0.1) with the CVLT_IR in the VAT identified from a pathway overrepresentation analysis mapping significant genes to the Reactome database. Dots are colored by the q value. (F) Manhattan-like plot of pathways significantly associated (q < 0.1) with the STROOP Interference in the VAT identified from a pathway overrepresentation analysis mapping significant genes to the Reactome and WikiPathways databases. In the Manhattan-like plots, the bubble size represents the ratio of input genes that are annotated in a pathway (GeneRatio). CREB, adenosine 3′,5′-monophosphate response element–binding protein; tRNA, transfer RNA; NCAM1, Neural Cell Adhesion Molecule 1; TP53, Tumor Protein 53; PKA, Protein Kinase CAMP-Activated Catalytic Subunit Alpha.
To facilitate interpretation and identify relevant adipose tissue pathways associated with cognitive function, we next performed overrepresentation analyses mapping significant genes to the Reactome and WikiPathways databases included in the ConsensusPathDB. We found an overrepresentation of pathways with important role in the CNS associated with measurements of inhibitory control, working memory, immediate memory, and attention (Fig. 1, D to F; fig. S2, C to F; and tables S13 to S17). Executive function was associated with axon guidance, nervous system development, synaptic vesicle pathway, and signaling by receptor tyrosine kinases (Fig. 1D). The neuronal system and the serotonin and dopamine neurotransmitter release cycles were the most significant pathways associated with immediate memory (Fig. 1E) and attention. The axon guidance and the nervous system development were also overrepresented in the inhibitory control (Fig. 1F) and attention domains (fig. S2C). Semaphorin signaling and interactions (Fig. 1F), which play an important role in adult neuronal plasticity (25), were associated with inhibitory control, while alterations in complement activation were linked to executive function. We also found an overrepresentation of pathway involved in inflammation associated with the different cognitive domains. Hence, alterations in both notch signaling and complement activation were significantly associated with working memory (Fig. 1D).
Adipose tissue gene expression was associated with several cognitive functions later in life
We next sought to validate these results performing an RNA-seq of both VAT and subcutaneous adipose tissue (SAT) in an independent longitudinal cohort of 22 subjects with severe obesity aged 23 to 57 years that underwent the same battery of cognitive tests (table S1). Patients from this cohort underwent the same battery of cognitive tests that patients in the discovery cohort. However, in this case, the cognitive testing was performed 2 to 3 years after the collection of adipose tissue samples. Once more, we did not find any significant association of physical activity, diet, alcohol intake, glucose, or hemoglobin A1c levels (fig. S1). Thus, models were adjusted for age, sex, BMI, and education years. We again found significant genes associated with all cognitive test at a pFDR of <0.05 both in VAT (Fig. 2, A to C, and tables S18 to S28) and in SAT (fig. S3 and tables S29 to S37). Consistent with the previous results, enrichment analyses highlighted an overrepresentation of pathways with important roles in the CNS (tables S38 to S40). Axon guidance, nervous system development, neuronal system, and tryptophan (Fig. 2, D and E) were replicated as the pathways most significantly associated with attention (Fig. 2, D and E), while semaphorin signaling was associated with inhibitory control (Fig. 2F). The activation of the complement system in VAT was again strongly associated with the performance in several cognitive domains (Fig. 2, D to F). In SAT (tables S41 to S43), pathways related to synaptic transmission, neuronal system, and one‑carbon metabolism were associated with both long-term (fig. S3, D and E) and short-term memory (fig. S3, F and G). Complement activation in SAT was again among the pathways most significantly linked to attention (fig. S3, H and I).
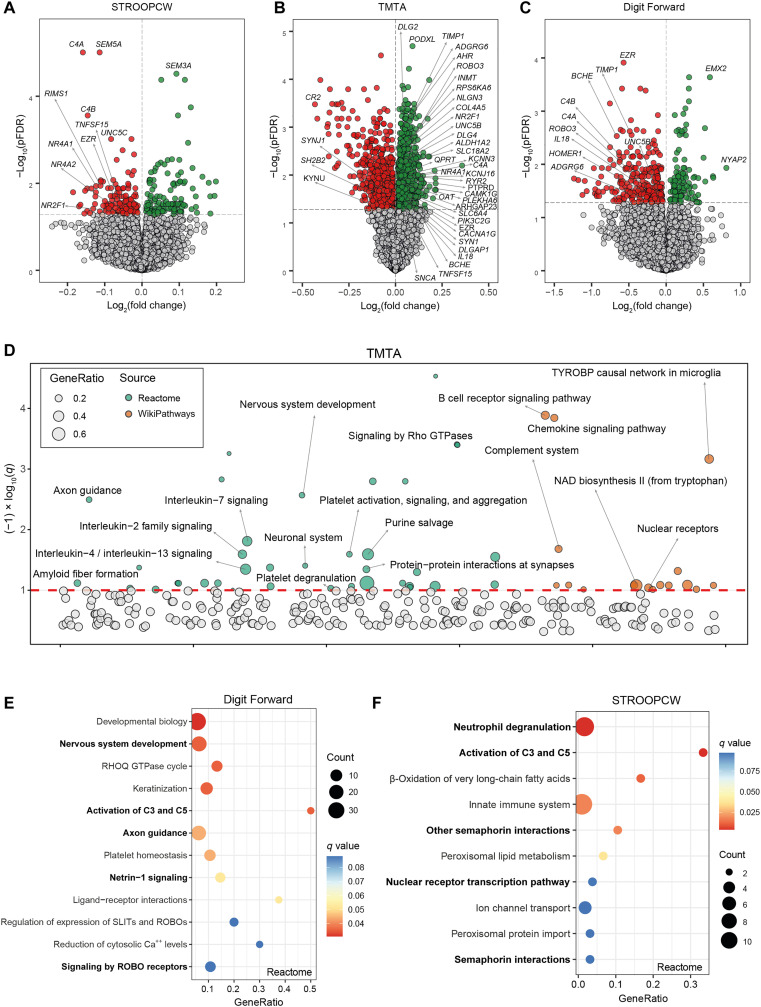
Fig. 2. Longitudinal associations of VAT gene expression at baseline and the scores in different cognitive domains later in life.
Volcano plots of differentially expressed genes in the VAT at baseline associated with (A) the STROOP color word tests (STROOPCW), (B) the TMTA, and (C) the Forward Digit Span scores 2 to 3 years later in the validation cohort (INTESTINE, n = 22) identified by limma-voom analysis controlling for age, BMI, sex, and education years. The log2 fold change associated with a unit change in the cognitive test score and the log10 P values adjusted for multiple testing (pFDR) are plotted for each gene. Differentially expressed genes (pFDR < 0.05) are colored in red and green indicating down-regulation and up-regulation, respectively. (D) Manhattan-like plot of pathways significantly associated (q < 0.1) with the TMTA in the VAT identified from a pathway overrepresentation analysis mapping significant genes to the Reactome and WikiPathways databases. (E) Dot plot of pathways significantly associated (q < 0.1) with the STROOPCW and (F) the Forward Digit Span in the VAT identified from a pathway overrepresentation analysis mapping significant genes to the Reactome database. The x axis in the dot plots and the bubble size in the Manhattan-like plots represent the ratio of input genes that are annotated in a pathway (GeneRatio). Dots are colored by the q value. NAD, nicotinamide adenine dinucleotide; TYROBP, Transmembrane Immune Signaling Adaptor TYROBP; SLIT, Slit Guidance Ligand; ROBO, Roundabout Guidance Receptor.
The gene encoding for Ezrin (EZR) was one with most associations across the different cognitive domains in VAT (significant in four tests at a pFDR of <0.05), being the most significantly associated gene with the digit span forward score (Fig. 2C and table S22). In line with previous findings, complement-related genes were particularly and strongly associated with inhibitory control (Fig. 2A). This cognitive function was also associated with early immediate genes from the orphan nuclear receptor 4A family (NR4A1 and NR4A2). In particular, NR4A2 was the most significant gene associated with the STROOPI test (which evaluates inhibitory control), while NR4A1 was among the top genes associated with trail making test part B (TMTB) (executive function). In SAT, NR4A1 and NR4A2 were negatively associated with long-term memory (fig. S3A), while the complement genes were particularly associated with attention (fig. S3C). A gene from the NR4A (NR4A3) had again the strongest negative association with inhibitory control (table S35). Last, AMPH was the gene most significantly associated with digit span forward test (table S31) and the total digit span test (table S33).
Adipose tissue genes linked to cognitive function are involved in neuronal system and synaptic formation
To gain further insights into the coincident associations between both cohorts, we performed an enrichment analysis considering only those genes significantly associated with at least one cognitive test in all three tissues (VAT from discovery and VAT and SAT from validation cohorts), and a total of 188 coincident genes were found (Fig. 3A and table S44). The most overrepresented pathways from the subset of genes included mostly pathways with key roles in synaptic function (Fig. 3, B and C; fig. S4, A and B; and table S45) and inflammation (tryptophan metabolism and complement activation). Mapping these genes to the Search Tool for the Retrieval of Interacting Proteins/Genes (STRING) database to construct a gene-gene interaction network, a cluster of highly interconnected genes mainly involved in neuronal system and synapse formation and maintenance (26) (DLG4, DLGAP1, DLG2, ATP1B1, HOMER1, NLGN3, SNCA, BCHE, RIMS1, SLC18A2, ADRB2, EZR, PODXL, and NCAM2) was disclosed (Fig. 3D).
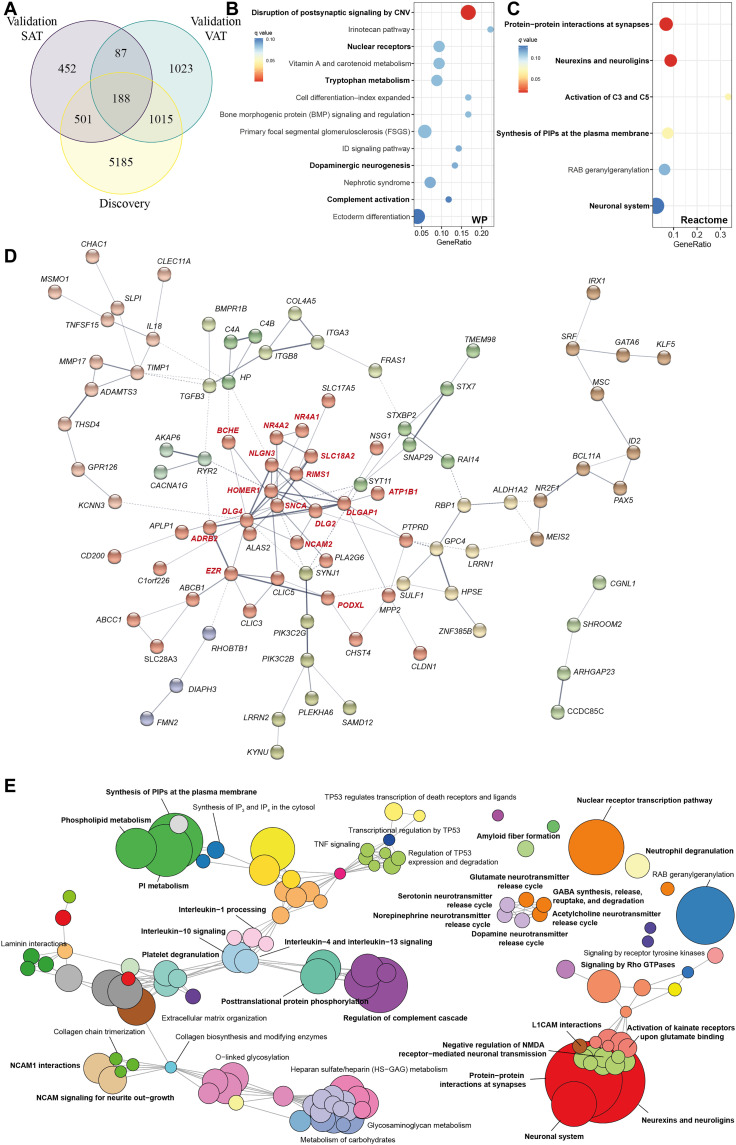
Fig. 3. Pathway analysis of differentially expressed genes in common among the VAT and SAT in the discovery and validation cohorts (n = 188).
(A) Venn diagram representing the overlap of significant genes associated with at least one cognitive test in VAT of the discovery cohort, the VAT of the validation cohort, and the SAT of the validation cohort. (B) Dot plot of significantly overrepresented pathways (q < 0.1) mapping common differentially expressed genes (n = 188) to the WikiPathways and (C) Reactome databases. The x axis in the dot plots represents the ratio of input genes that are annotated in a pathway (GeneRatio). Dots are colored by the q value. (D) Gene-gene interaction network constructed using common differentially expressed genes via the STRING database. The network nodes are genes, and the edges represent the predicted functional interactions. The thickness indicates the degree of confidence prediction of the interaction. Functional gene clusters are colored on the basis of the Markov cluster algorithm (MCL) with an inflation parameter of 1.4. Only connected nodes are shown. A highly connected functional cluster (in red) was detected comprising genes with important roles in the CNS. (E) Enrichment map of the interrelation of significant pathways identified using an active subnetwork oriented approach. Each color displays a cluster of related pathways using a threshold for kappa statistics = 0.35. The size of the nodes corresponds to its −log10(pFDR). The thickness of the edges between nodes corresponds to the kappa statistic between the two nodes. IP3, inositol 1,4,5-trisphosphate; IP4, inositol 1,4,5,6-tetrakisphosphate; PI, phosphatidylinositol; TNF, tumor necrosis factor; NMDA, N-methyl-d-aspartate; CNV, copy number variations; L1CAM, L1 Cell Adhesion Molecule.
To obtain a better picture of the associated pathways, we leveraged the interaction information from the gene-gene interaction network to identify distinct active subnetwork and then performed an active subnetwork–oriented enrichment analysis on these subnetworks. Using this approach, we identified a total of 120 overrepresented Reactome pathways (pFDR < 0.05). To reduce complexity derived from this large number of pathways, we performed a hierarchical clustering to collapse redundant pathways and ease the identification of mechanisms relevant to cognition (table S46). Figure 3E shows the functional network of the clustered pathways. Of 53 clusters, the most significant one included pathways related to the neuronal system and synaptic function (RIMS1, SLC18A2, DLG2, DLG4, DLGAP1, HOMER1, NLGN3, PTPRD, BCHE, and KCNN3) (Fig. 4A). This cluster was strongly connected with three other cluster involved in the synthesis and metabolism of phospholipids, particularly phosphatidylinositols (PIPs) (PIK3C2B, PIK3C2G, SYNJ1, and PLEKHA6), the signaling by Rho guanosine triphosphatase (GTPases) (ARHGAP23, DIAPH3, DLG4, RHOBTB1, SGO2, and SRF), and the release of neurotransmitters (RIMS1 and SLC18A2) (Fig. 4B).
Fig. 4. Selected clusters from active subnetwork oriented pathway analysis of common significant genes among the VAT and SAT in the discovery and validation cohorts (n = 188) and integration with circulating metabolites.
(A) Dot plot of enrichment analysis results performed on active subnetworks grouped by selected clusters. The x axis represents the fold enrichment defined as the ratio of the frequency of input genes annotated in a pathway to the frequency of all genes annotated to that pathway. The dot size indicates the number of differentially expressed genes in a given pathway. Dots are colored by the –log10(pFDR), with red indicating high significance. (B) Gene-concept network depicting significant genes involved in enriched pathways from selected clusters. The dot size of the pathways represents the −log10(pFDR). Pathways with the same color correspond to the same cluster. (C) Correlation circle plot for the integration of the adipose tissue genes, metabolites (NMR, HPLC-ESI-MS/MS in positive and negative mode), and cytokines and neurotoxic proteins using a multiblock PLS model in canonical mode. Strongly positively associated variables or groups of variables are projected close to one another on the correlation circle (<0° angle). The variables or groups of variables strongly negatively associated are projected diametrically opposite (<180° angle) on the correlation circle. Variables not correlated are situated <90° from one another.
To gain better insights into potential mechanisms linking adipose tissue to cognition, we next performed a multiblock Partial Least Squares (PLS) analysis to integrate these 188 adipose tissue genes with the plasma levels of metabolites measured by nuclear magnetic resonance (NMR), high-performance liquid chromatography–electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) in positive and negative modes and the circulating levels of cytokines and neurotoxic proteins [IL-6, IL-4, Tau, α-synuclein, and amyloid-β42 (Aβ42)]. The expression levels of adipose tissue genes involved in phospholipid metabolism, such as PIK3C2B, PLA2G6, or SYNJ1, had a strong association with several circulating phospholipids and lysophospholipids (Fig. 4C). Although lipids are essential to brain function and, in particular, phospholipids are crucial for synaptic plasticity, neurotransmission, and memory (27), the brain can synthesize a limited number of lipids. However, plasma fatty acids can cross the Blood-Brain Barrier (BBB) as nonesterified free fatty acids or esterified in lysophospholipids such as lysophosphatidylcholine (LPC) (28, 29). Notably, some of the genes from the phospholipid metabolism cluster codified for phospholipase A2 (PLA2G6), which catalyzes the hydrolysis of the sn-2 position of glycerophospholipids to yield nonesterified fatty acids and lysophospholipids. Hence, these phospholipids could be mediating the effects of adipose tissue on cognition.
We also identified three highly interconnected clusters of genes related to inflammatory response that included the complement cascade (C4A and C4B) and the IL-4, IL-10, IL-13 interleukin signaling (IL18 and TIMP1). Chronic low-grade inflammation is a hallmark of obesity, and the association between obesity and cognitive decline has recently been shown to be mediated by inflammation (30, 31). In addition, a complement-dependent synapse elimination has been recently identified as a mechanism for memory loss (32). In line with our findings, NLRP3 from the VAT has recently shown to impair cognition in mice by activating microglial IL-1 receptor (7), whereas IL-4 has shown to mediate the protective effects of beige fat on obesity-induced cognitive impairment by restoring synaptic plasticity in the hippocampus (14). Consistent with recent findings that inhibition of Na,K-ATPase signaling resulted in improved cognitive function (12), we also identified a cluster containing the Na+/K+ ATPase transporting subunit beta 1 gene (ATP1B1). Notably, the Na,K-ATPase signaling pathway requires the activation of PIP 3-kinase, and we identified a cluster of pathways involved in PIP metabolism strongly associated with cognition (Fig. 4B). Last, the genes involved in the inflammatory cluster (C4A, C4B, IL18, and TIMP1) had a strong positive association with the circulating levels of IL-6 and a strong negative association with the IL-4 levels in plasma (Fig. 4C). Therefore, these adipokines could be released by the adipose tissue and mediate its effects on cognition. Accumulating evidence indicates that peripheral inflammation contributes to increased neuroinflammation (33) and could be a potential causal mechanisms involved in the obesity-related cognitive impairment (34, 35).
Other significant clusters included the nuclear receptor transcription (NR4A1, NR4A2, and NR2F1) and the metabolism of vitamins (ABCC1, TCN2), particularly vitamin A (RBP1 and GPC4). Recent studies have shown that NR4A2 activation enhances long-term memory in young mice and ameliorates age-related memory impairments in old mice (36). Several studies have also suggested that water-soluble vitamins (C, E, and Bs) may affect cognitive performance by reducing reactive oxygen species generation and proinflammatory mediators such as NLRP3 inflammasome (37). Retinoic acid, the active form of vitamin A, is essential for regulating synaptic plasticity in regions of the brain involved in memory and learning (38). Notably, we found a strong positive association between the expression levels of genes involved in the metabolism of vitamins in the adipose tissue, particularly TCN2 and ABCC1, and the circulating levels of retinoic acid (Fig. 4C). Notably, RBP1, which encodes for cellular retinol binding protein 1 involved in the transport of retinol, had a strong negative association with the plasma levels of retinoic acid. Together, these results suggest that the adipose tissue could modulate cognitive performance through the metabolism of vitamin A.
From the identified cluster of genes in Fig. 4B, SLC18A2 and RIMS1 (regulating synaptic exocytosis 1) were those with the largest number of significant associations across the different cognitive domains in the three tissues analyzed. SLC18A2 encodes for VMAT2 (vesicle monoamine transporter member 2), a protein that transports cytoplasmic monoamine neurotransmitters (dopamine, norepinephrine, histamine, and serotonin) into presynaptic vesicles and has been linked to the pathophysiology of different neuropsychiatric and neurological disorders (39). The Drosophila and mouse RIMS1 orthologues have shown to be a central component in the regulation of synaptic vesicle exocytosis and the neurotransmitter release (40). Mice lacking Rim1α show severely impaired learning and memory (41), and RIMS1 has recently been identified as the highest-ranked candidate gene in autism spectrum disorder (42).
Morbid obesity modulates the direction of the associations between adipose tissue genes and cognition
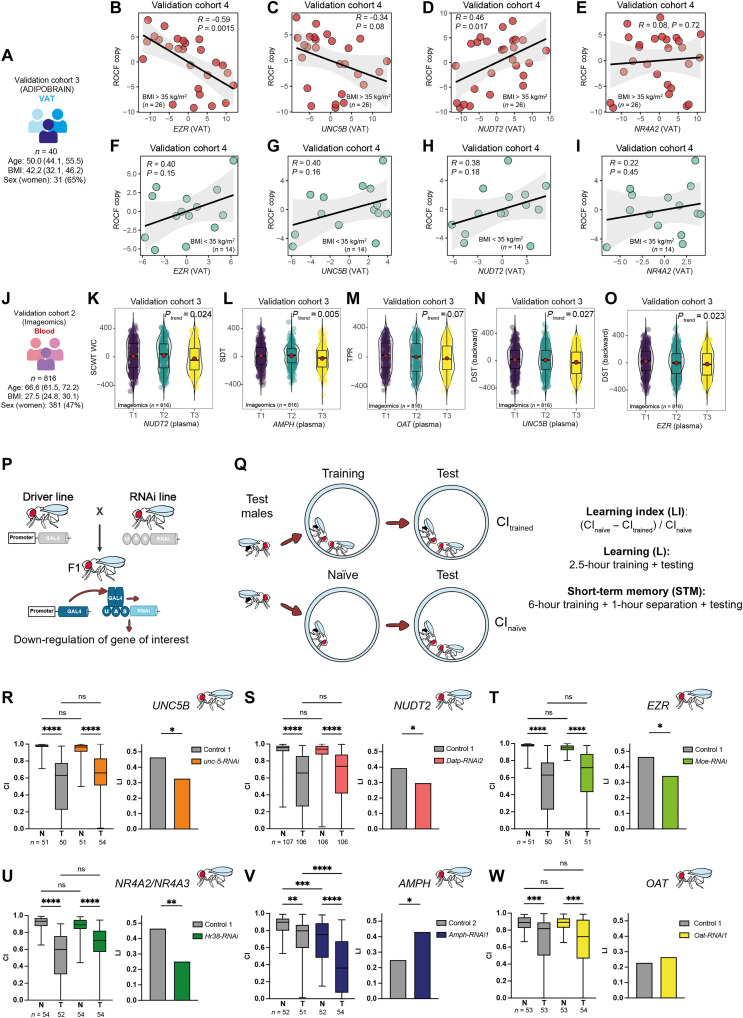
We sought to further validate these results using an additional cohort. Hence, we measured the expression levels of EZR, UNC5B, NUDT2, and NR4A2 in the VAT from a third independent validation cohort (ADIPOBRAIN cohort) consisting of n = 40 patients with and without obesity aged 40 to 64 years old (Fig. 5A and table S1) who underwent the Rey-Osterrieth complex figure (ROCF) test 4 to 10 years later. The ROCF is one of the top 10 tests used by neuropsychologists for the evaluation of visuospatial constructional ability, visual memory, and executive function (43). Consistent with our findings in the discovery cohort (including only patients with morbid obesity), the expression of EZR (Fig. 5B) and UNC5B (Fig. 5C) at baseline was strongly negatively associated with the ROCF copy scores in subjects with morbid obesity (BMI > 35 kg/m2). Conversely, the expression of these genes was positively associated with executive function in subjects with a BMI of <35 kg/m2 (Fig. 5, F and G). The expression of NUDT2 was positively associated with the ROCF copy performance in subjects with morbid obesity (Fig. 5D) but not in subjects with a BMI of <35 kg/m2 (Fig. 5H), whereas we did not find any significant association with the expression of NR4A2 (Fig. 5, E and I).
Fig. 5. Associations of expression levels of selected genes with cognition in a validation cohort, the Aging Imageomics cohort, and D. melanogaster.
(A) Main baseline characteristics of the validation cohort 3 (ADIPOBRAIN). Scatter plots of the partial Spearman’s rank correlations (adjusted for age, BMI, sex, and education years) between ROCF copy scores and the VAT expression levels of (B) EZR, (C) UNC5B, (D) NUDT2, and (E) NR4A2 in patients with a BMI of >35 kg/m2 from the ADIPOBRAIN cohort. (F to I) The same associations but in patients with a BMI of <35 kg/m2. (J) Main baseline characteristics of the validation cohort 2 (Imageomics). Violin plots of the score in several cognitive tests and the tertiles (T1, T2, and T3) of the circulating expression levels of selected genes: (K) normalized STROOP color word test–color word (SCWT CW) versus NUDT2; (L) symbol digit test (SDT) versus AMPH; (M) total paired recall (TFP) versus OAT; (N) normalized digit span test (DST) backward versus UNC5B; and (O) normalized DST backward versus EZR. The ranked residuals after controlling for age, BMI, gender, and education level are plotted. Overall significance was assessed using a Mann-Kendall trend test. (P) Scheme of the RNA interference via the UAS-GAL4 system. (Q) Courtship conditioning paradigm. (R to W) The graphs display courtship conditioning paradigm results of fat body promoter line w; C7-GAL4; UAS-Dcr-2 crossed with RNAi lines targeting (R) unc-5, (S) Datp, (T) Moe, (U) Hr38, (V) Amph, (W) Oat, and their corresponding genetic background controls (controls 1 and 2). Boxplots represent the CI of naïve (N) and trained (T) males. Significance of courtship suppression upon training was assessed with Kruskal-Wallis nonparametrical test and post hoc Dunn’s multiple comparison test. LI statistical significance was determined with the nonparametrical bootstrap analysis with 10,000 iterations. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. ns, not significant.
Blood expression levels of key genes linked to cognitive performance
To further validate these findings and provide noninvasive biomarkers of cognitive function as an alternative to adipose tissue biopsies, we measured the expression of some of the most promising genes in the circulation of n = 816 individuals from the general population aged ≥50 years taking part in the Aging Imageomics study (Fig. 5J and table S1). Initially, we validated those genes with the largest number of associations across the different neuropsychological tests in the discovery cohort (NUDT2, AMPH, UNC5B, and OAT) and validation cohort 1 (EZR and NR4A2). In agreement with our findings, the circulating levels of NUDT2 were negatively associated with executive function (Fig. 5K), while the circulating expression of AMPH was negatively associated with processing speed (Fig. 5L) and memory (assessed by the total free recall; Ptrend = 0.02). Increased levels of OAT and UNC5B tended to be associated with delayed memory (Fig. 5M) and executive function (Fig. 5N). Last, we also found consistent negative associations between the circulating expression of EZR and executive functioning (Fig. 5O) and the quintiles of NR4A2 with memory (Ptrend = 0.027).
Down-regulation of candidate genes in the fat body of Drosophila melanogaster improves cognition
We next assessed the causal effect of the adipose tissue expression of these candidate genes (AMPH, UNC5B, NUDT2, OAT, NR4A2, NR4A3, and EZR) on cognitive function using the model organism D. melanogaster (fruit flies). A main advantage of Drosophila is the possibility of manipulating gene expression in a tissue-specific manner by means of the UAS-GAL4 system (Fig. 5P) (44). To evaluate Drosophila cognition, we used the courtship conditioning paradigm to measure associative learning and memory capabilities (45) . Male courtship behavior is conditioned by the exposure to nonreceptive premated females resulting in a suppression of courtship after a learning process. Male courtship rates can be assessed at different times after training periods with premated females. Learning is assessed immediately after the training period, and short-term memory is typically assessed 1 hour after training. The courtship index (CI; the percentage of time spent on courtship during an 8-min interval) of trained and of socially naïve male is used to calculate the learning index (LI). A higher LI is indicative of a better learning or short-term memory (Fig. 5Q).
We down-regulated Amph, unc-5, Datp, Oat, Hr38, and Moe (the well-conserved orthologues of AMPH, UNC5B, NUDT2, OAT, NR4A2/NR4A3, and EZR, respectively) specifically in the fat body of Drosophila, which is the equivalent of the adipose tissue in humans. The fat body promoter line w; C7-GAL4; UAS-Dcr-2 was crossed with at least one RNA interference (RNAi) line targeting each gene of interest (table S47). Learning capabilities were assessed immediately after training using the courtship conditioning assay. Down-regulation of unc-5, Datp, Hr38, and Moe lead to a significant decrease in learning capabilities compared to their corresponding genetic background controls in at least one of the RNAi lines tested (Fig. 5, R to U). Conversely, down-regulation of Amph lead to an improvement of learning capabilities for one RNAi line (Fig. 5V). Down-regulation of Oat in the fat body was lethal for one RNAi line or did not affect learning captivities when using a second RNAi line (Fig. 5W). Our findings demonstrate that specific down-regulation of some of these genes in the fat body of Drosophila modulates learning, thereby suggesting a direct effect on cognition uniquely depending on its expression and/or function in fat body.
Neurotransmitter release cycle–associated genes SLC18A2 and RIMS1 modulate cognition in mice adipose tissue or Drosophila fat body
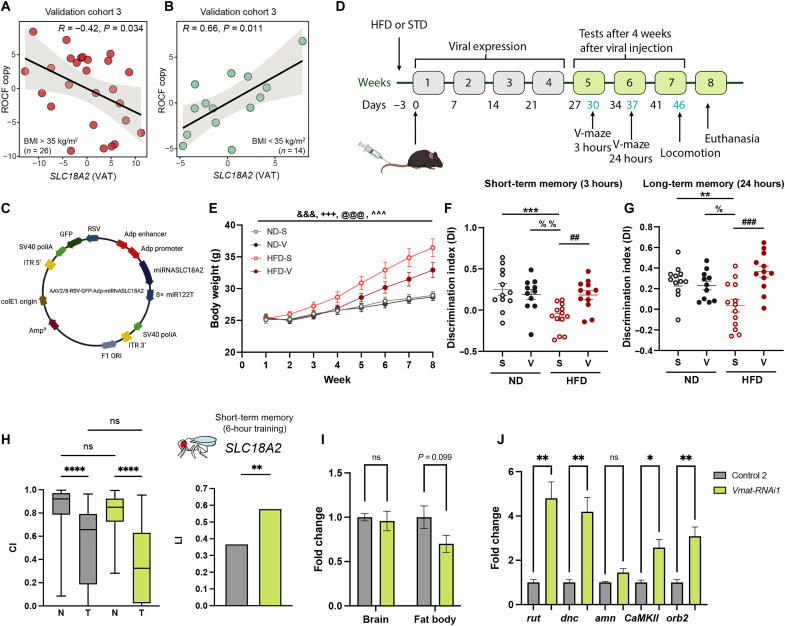
Among the genes associated with cognition in both VAT and SAT from the discovery and validation cohort 1, the cluster of genes involved in the release of neurotransmitters (RIMS1 and SLC18A2) (Fig. 4B) had the largest number of significant associations across different cognitive domains. Therefore, we also validated these associations in the third validation cohort (ADIPOBRAIN). Again, we found that in subjects with morbid obesity (Fig. 6A), the expression of SLC18A2 at baseline was strongly negatively associated with the ROCF copy performance 4 to 10 years later, whereas it was positively associated in subjects with a BMI of <35 kg/m2 (Fig. 6B).
Fig. 6. Associations of SLC18A2 with cognition in the validation cohort 3 and altered expression in preclinical models.
Scatter plots of the partial Spearman’s correlations (adjusted for age, BMI, sex, and education years) between the SLC18A2 VAT expression and the ROCF copy scores in (A) morbid obese patients (BMI > 35 kg/m2) and (B) patients with a BMI of <35 kg/m2 (ADIPOBRAIN cohort, n = 40). (C) Schematic representation of the AAV packaging plasmid used to down-regulate SLC18A2 gene expression. (D) Timeline of events and mice cognitive testing. Arrows indicate punctual events such as virus injection, tests, and euthanasia. (E) Weekly body weight (in grams). Means ± SEM; n = 12 normal diet + saline (ND-S), n = 11 normal diet + virus (ND-V), n = 13 high fat diet + saline (HFD-S), n = 12 high fat diet + virus (HFD-V). &&&P < 0.001 week effect; +++P < 0.001 diet effect, @@@P < 0.001 week × diet interaction, and ^^^P < 0.001 week × treatment interaction (three-way ANOVA). (F and G) Short-term (3 hours) and long-term (24 hours) memory using the novel object recognition test. Dots with the means ± SEM. **P < 0.01, ***P < 0.001 ND-S versus HFD-S, %P < 0.05, %%P < 0.01 ND-V versus HFD-S, and ##P < 0.01, ###P < 0.001 HFD-S vs HFD-V (two-way ANOVA). (H) Male short-term memory in the courtship-conditioning paradigm. Control 2 (w; C7-GAL4/+; UAS-Dcr-2/+) and Vmat-RNAi1 fat body–specific knockdown flies (w; C7-GAL4/+; UAS-Dcr-2/Vmat-RNAi1). (I) Relative gene expression of Vmat in fly heads and fat body and (J) rutabaga (rut), dunce (dnc), amnesiac (amn), homer, CAMKll, and orb2 in fly heads of Vmat-RNAi1 fat body–specific knockdown flies and its corresponding genetic background control. Means ± SEM. (t test: *P < 0.05, **P < 0.01, and ****P < 0.0001). Data are based on a minimum of five biological and two technical replicates. (C) and (D) were created with BioRender.com.
To demonstrate a possible causal effect of adipose tissue SLC18A2 on cognition, we studied two preclinical models. In a mice model, we used an AAV2/8-RSV-GFP-Adp-miRNASLC18A2 vector that expresses a miRNA to selectively down-regulate slc18a2 gene expression in adipose tissue (Fig. 6C). Eight-week-old mice were fed a normal chow diet (ND; n = 23) or a high-fat diet (HFD; n = 25) for 8 weeks (Fig. 6D). Three days after the start of the diet, each dietary group was divided into two groups and injected intravenously via tail vein either with the AAV2/8-RSV-GFP-Adp-miRNASLC18A2 vector or with saline. The expression levels of green fluorescent protein (GFP) contained in the vector as a reporter gene was significantly increased in both inguinal and mesenteric white adipose tissue (iWAT and mWAT) of mice from the vector-injected groups compared to the saline groups (fig. S5, A and B), thereby indicating that the vector was able to reach the adipose tissue. We also found a significant decrease in the solute carrier family 18 member A2 (slc18a2) protein levels in the mWAT of virus-injected mice fed an HFD compared to their respective controls (fig. S5, C and D). Four weeks after injection, we assessed short- and long-term memory and locomotor activity. As expected, mice fed an HFD gained more weight in parallel to food intake compared with those fed an ND (Fig. 6E and fig. S6, A and B). There is good evidence that obesity affects cognition in rodents (35). Consistently, mice fed an HFD had lower short- and long-term memory than mice fed an ND (Fig. 6, F and G). Down-regulation of slc18a2 in the mice adipose tissue reversed the memory impairment induced by an HFD (Fig. 6, F and G). We observed no differences in the exploration time between groups (fig. S6, C and D). Although we found a diet effect on locomotion, down-regulation of slc18a2 had no substantial effect (fig. S6, E to H).
To further assess the role of adipose tissue SLC18A mRNA on cognition, we performed RNAi-mediated knockdown of Vmat (the orthologue in Drosophila) using the w; C7-GAL4; UAS-Dcr-2 promoter line for specific knockdown in the fat body (the equivalent of adipose tissue in humans). The courtship-conditioning paradigm was performed, and short-term memory was assessed 1 hour after training. Males of Vmat-RNAi1were able to suppress courtship significantly better than the corresponding genetic background control flies (Fig. 6H). Consequently, the LI was significantly increased (Vmat-RNAi1: LI = 0.577, control 2: LI = 0.366, P = 0.0064), indicating that Vmat-RNAi1 presented more short-term memory (Fig. 6H). Next, we performed real-time polymerase chain reaction (RT-PCR) to prove that Vmat was specifically knocked down in the fat body but not in the fly head (Fig. 6I), indicating that when decreasing Vmat expression exclusively in the fat body flies are able to learn better. To unravel a potential mechanism through which Vmat expression in the fat body could influence cognition, we performed RT-PCR to evaluated the expression of rutabaga (rut), dunce (dnc), amnesiac (amn), homer, CAMKll, and orb2 in Drosophila heads. These genes are part of the cyclic adenosine 5′-monophosphate pathway, which have been demonstrated to influence learning and short-term memory in the courtship-conditioning assay (46). RNAi-mediated knockdown of Vmat in the fat body lead to a significant increase in memory related genes (rut, dnc, caMKll, and orb) in the fly head (Fig. 6J). To further validate our results, we used an independent RNAi line against Vmat (Vmat-RNAi2). When down-regulating Vmat in the fat body with RNAi2, we observed a significant increase in short-term memory (Vmat-RNAi2: LI = 0.650, control 1: LI = 0.437, P = 0.002) (fig. S7A) and a significant overexpression of dnc, CAMKll, and orb2 in the fly head (fig. S7B), confirming our previous data.
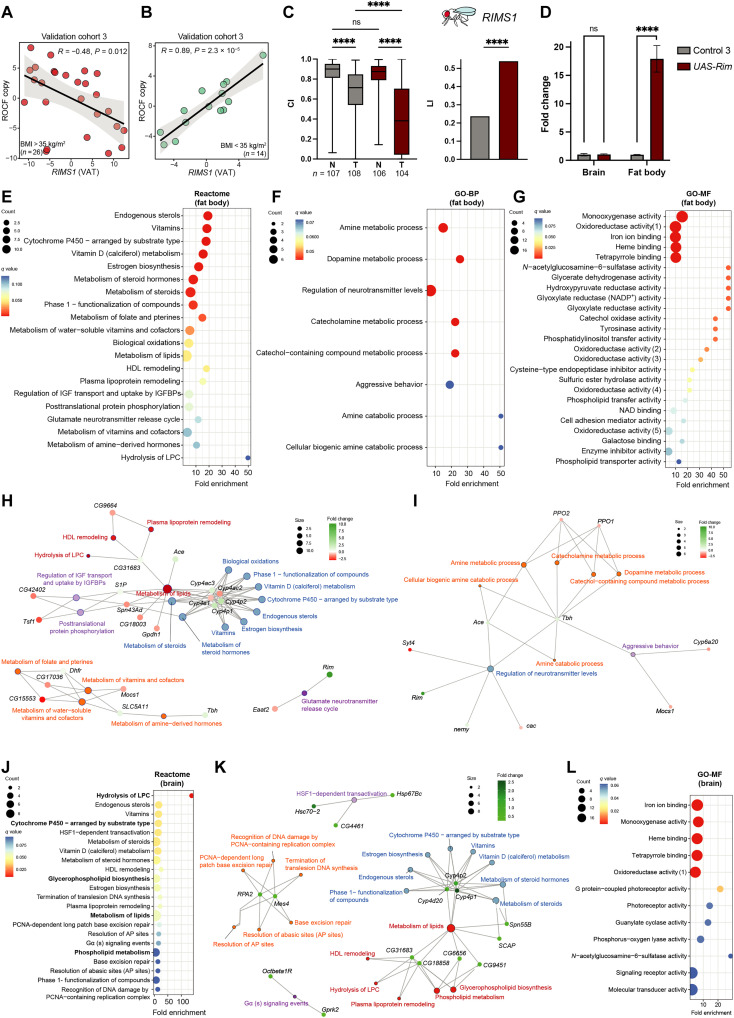
In the third validation cohort, VAT RIMS1 mRNA was also evaluated. In line with the results obtained for SLC18A2, the expression of RIMS1 was negatively associated with ROCF in subjects with severe obesity (Fig. 7A) but positively in subjects with a BMI of <35 kg/m2 (Fig. 7B). To prove a possible causal effect, we assessed learning in Drosophila after down-regulation or overexpression of Rim (the well-conserved orthologue of RIMS1) in the fat body. A significant decrease in CI after learning was observed in two independent RNAi lines and corresponding genetic background controls, but we did not observe significant differences in the LI (fig. S7, C and D). However, when Rim was overexpressed (UAS-Rim) in the fat body, males were able to suppress courtship significantly better than the corresponding genetic background control flies (Fig. 7C), resulting in a significant increase in the LI (UAS-Rim: LI = 0.539, control 3: LI = 0.237, P = 0.0001) (Fig. 7D). Our results indicate that flies that overexpress Rim exclusively in the fat body are able to learn better. Next, we used RT-PCR to prove that Rim was uniquely targeted in the fat body but not in the brain (Fig. 7D). Again, learning improvement was exclusively promoted by Rim altered expression in the fat body and not due to a “leakage” of the RNAi in the brain.
Fig. 7. Associations of RIMS1 with cognition in the validation cohort 3 and Rim overexpression in Drosophila fat body effects in learning and gene expression profiles.
(A and B) Scatter plots of the partial Spearman’s rank correlations (adjusted for age, BMI, sex, and education years) between the VAT expression levels of RIMS1 and the ROCF copy scores in (A) morbid obese patients (BMI > 35 kg/m2) and (B) patients with a BMI of <35 kg/m2 from the ADIPOBRAIN cohort (n = 40). (C) Rim overexpression in the Drosophila fat body and associations with learning. Differences in CI between naïve and trained males were assessed with Kruskal-Wallis nonparametrical test and post hoc Dunn’s multiple comparison test. LIs to assess either short-term memory (for Vmat) or learning (for Rim) were calculated from CIs as specified in Materials and Methods. Statistical significance was determined with the nonparametrical bootstrap analysis with 10,000 replicates. ****P < 0.0001. (D) Rim relative expression assessed by quantitative RT-PCR (qRT-PCR) in fly brain or fat body in UAS-Rim fat body–specific overexpression flies and its corresponding genetic background control. Data are derived from a minimum of five biological and two technical replicates. (E) Dot plot of significantly (q < 0.1) overrepresented Reactome pathways, (F) GO biological processes (GO-BP), and (G) molecular functions (GO-MF) associated with the miss expressed genes in the fat body of UAS-Rim flies. (H and I) Gene-concept network depicting significant genes involved in enriched Reactome pathways [from (E)] and GO-GO-BP [from (F)]. (J) Overrepresentation analysis of pathways (q > 0.1) associated with differentially expressed gene transcript in UAS-Rim fly heads based on the Reactome database. (K) Gene-concept network associated with significant genes involved in Reactome pathways of (J). (L) Significantly overrepresented GO molecular functions of significant gene transcripts in fly heads (q > 0.1). HDL, high-density lipoprotein; HSF1, heat shock factor 1; IGF, insulin-like growth factor; IGFBP, insulin-like growth factor binding protein; NADP+, nicotinamide adenine dinucleotide phosphate; PCNA, proliferating cell nuclear antigen.
To elucidate the potential mechanisms underlying the overexpression of Rim in learning, we performed an mRNA-seq in the fat body and fly heads of UAS-Rim flies that overexpressed Rim specifically in the adipocytes. We identified the differentially expressed genes between the flies overexpressing Rim and the corresponding genetic backgrounds in the fat body (table S48) and fly heads (table S49). In the fat body, consistent with our results in humans, an enrichment analysis based on Reactome identified an overrepresentation of pathways involved in vitamin metabolism and lipid metabolism, including the hydrolysis of LPC and the glutamate neurotransmitter release cycle (Fig. 7, E and H). It also identified genes involved in other pathways involved in biological oxidations and cytochrome P450 (Cyp4e1, Cyp4ac2, Cyp4ac3, Cyp4p1, and Cyp4p2). Notably, a gene ontology (GO) enrichment analysis identified mainly biological processes participating in neurotransmitter regulation, in particular, the metabolism of biogenic amines (dopamine, serotonin, and norepinephrine), which is strongly consistent with our findings in humans (Fig. 7, F and I). At the molecular function level, we identified several functions involved in monooxygenase and oxidoreductase activity (Fig. 7G). Notably, we also identified several functions involved in the PIP transfer and phospholipid transfer and transport. These results were replicated in the fly heads. We found again an overrepresentation of pathways involved in the metabolism of lipids (particularly phospholipids and the hydrolysis of LPC), vitamins, and biological oxidations (table S50). An analysis based only on up-regulated genes also identified alterations in DNA metabolism (Fig. 7, J and K), while a GO analysis identified once more molecular functions related to monooxygenase and oxidoreductase activity (Fig. 7L).
Oxidative damage and reduction of antioxidants are associated with a decline in cognitive function due to neurodeterioration (47). Vitamins can act as potent antioxidant molecules protecting neurons from oxidative stress and improving overall neuronal functioning (48). Reducing oxidative stress in Drosophila has been associated with an increase in lifespan (49) and improves age-related memory impairment (47). Similarly, monooxygenase and oxidoreductase enzymes are contributing to the cellular levels of oxygen reactive species, and cytochrome P450 family members have been found to be up-regulated in the presence of oxidative stress having a detoxification and antioxidant function in Drosophila (50). Notably, low-grade inflammation is also associated with cellular oxidative imbalance and one of the physiopathological mechanisms behind obesity-associated syndromes (51). Phospholipids are also abundant molecules in Drosophila brain, where they play essential roles in cognition, by both supplying energy for brain activity and particip4ating in membrane trafficking and neurotransmission (52). Similar to mammals, main source of phospholipids in Drosophila are lipid droplets inside fat body cells (53). In Drosophila, phospholipid homeostasis has been found to be important for the regulation of dendrite morphogenesis (54), and lipid imbalance is used as a target to protect against Aβ42-induced cytotoxicity in Alzheimer's disease (AD) fly model (55).
Overall, we have demonstrated that overexpression of Rim uniquely in Drosophila fat body is sufficient to promote an increment in learning and that the effect of Rim overexpression in the fat body can mediate changes at the transcriptional level in the adipose tissue itself and in peripheral tissues as the brain modulating pathways associated with neuronal functioning, cellular oxidative balance and detoxification, and genes from the phospholipid metabolism. Together, these results suggest that altering Vmat or Rim expression in the fat body mediates changes in gene expression in the head (brain) throughout a putative fat body-to-brain axis.
DISCUSSION
While the brain regulates adipose tissue performance, here, we show that the reverse is also plausible. We found that the expression of genes linked to axon guidance, nervous system development, neuronal system, tryptophan, and inflammation activity in human adipose tissue were all linked to cognition in three different cohorts of subjects.
SLC18A2 expression in the brain has been linked to the pathophysiology of different neuropsychiatric and neurological disorders (39). Similarly, decreased expression of Rim1α in the brain of mice led to impaired learning and memory (41) . For this reason, and given the associations of both SLC18A2 and RIMS mRNA in the adipose tissue with cognitive function, we selected these neurotransmitter release cycle–associated genes to test their possible functional relevance. Knockdown of slc18A2 exclusively in adipose tissue reversed the memory impairment induced by an HFD in mice (Fig. 6, F and G). Down-regulation of Vmat in the fat body of two different lines of Drosophila also led to improved short-term memory. Overexpressing Rim [the orthologue of regulating synaptic membrane exocytosis 1 (Rims1)] exclusively in the fat body also led to an increment in learning abilities in Drosophila. The altered expression of other genes linked to cognition in humans (AMPH, UNC5B, NUDT2, NR4A2/NR4A3, and EZR) in the fat body of Drosophila again modified cognition of the latter.
In the search of more accessible biomarkers, we evaluated the expression of most of these genes in PBMCs. PBMC mRNA levels of adipogenic genes decreased after weight loss in subjects with obesity (56), while an HFD-induced cognitive disruption led to alterations in the PBMC transcriptome in rats (57). Here, we show that the expression of genes in PBMCs that were most associated with cognition in adipose tissue (NUDT2, AMPH, UNC5B, OAT, EZR, and NR4A2) were also linked to cognitive traits in a cohort of 816 subjects.
The main limitation of the current study is that it is only focused on morbid obese subjects (BMI > 35 mg/kg2); hence, our results may not be generalizable to other populations. Findings should be confirmed in longitudinal, larger samples of subjects to generalize these results to the general population.
Together, these findings suggest a systemic developmental program that changes in parallel in different tissues and cells, being reflected in neuronal networks. Current findings hint at potential therapeutic targets: Peripheral administration of miRNAs targeting the adipose tissue and influencing cognition seems far safer than targeting the brain. Current observations also provide information about useful biomarkers in predicting cognitive decline and in monitoring response to therapy using accessible sources.
MATERIALS AND METHODS
Clinical cohorts
In discovery cohort (IRONMET, n = 17) (58, 59), from January 2016 to October 2017, a cross-sectional case-control study was undertaken in the Endocrinology Department of Dr. Josep Trueta University Hospital (Girona, Spain). In 17 morbidly obese patients (BMI > 35 mg/kg2) aged 28 to 60 years old, adipose tissue–stranded RNA-seq was analyzed. In validation cohort 1 (INTESTINE, n = 22) (58), consecutively, 22 morbidly obese (BMI > 35 kg/m2) subjects with different degrees of insulin action (measured using hyperinsulinemic-euglycaemic clamp) were recruited at the Endocrinology Service of the Dr. Josep Trueta University Hospital to validate adipose tissue RNA-seq analysis.
In both cohorts, all subjects were of Caucasian origin and reported a body weight stable for at least 3 months before the study. Subjects were studied in the postabsorptive state. Exclusion criteria were previous type 2 diabetes mellitus, chronic inflammatory systemic diseases, and acute or chronic infections in the previous month; severe disorders of eating behavior or major psychiatric antecedents; neurological diseases, history of trauma or injured brain, language disorders; and excessive alcohol intake (≥40 g of OH/day in women or 80 g of OH/day in men). Body fat composition was estimated using bioelectrical impedance analysis (BC-418, Tanita Corporation of America, Illinois, USA).
This protocol was revised, validated, and approved by the ethics committee of the Dr. Josep Trueta University Hospital. The purpose of the study was explained to participants, and they signed written informed consent before being enrolled in the study.
In validation cohort 2 (Imageomics, n = 816) (58, 59), the Aging Imageomics study is an observational study including participants from two independent cohort studies (MESGI50 and MARK). Detailed description of the cohorts can be found elsewhere (60). Briefly, the MESGI50 cohort included a population aged ≥50 years old, while the MARK cohort included a random sample of patients aged 35 to 74 years with intermediate cardiovascular risk. Eligibility criteria included ages of ≥50 years, dwelling in the community, no history of infection during the past 15 days, no contraindications for magnetic resonance imaging, and consent to be informed of potential incidental findings. The Aging Imageomics study protocol was approved by the ethics committee of the Dr. Josep Trueta University Hospital.
In validation cohort 3 (ADIPOBRAIN, n = 40), all the samples and data of the participants included in this study were provided by the FATBANK (platform promoted by the CIBEROBN and coordinated by the IDIBGI Biobank) and integrated in the Spanish National Biobank Network. A total of 40 patients, ages 40 to 64 years, were included. From these, 26 subjects had morbid obesity (BMI > 35 mg/kg2), aged between 40 and 63. Patients had no systemic disease other than obesity, and all were free of any infection in the month before the study. Liver disease [specifically tumor disease and Hepatitis C Virus (HCV) infection] and thyroid dysfunction were specifically excluded. Adipose tissue samples were obtained from VAT depots during elective surgical procedures.
Neuropsychological assessment
The primary end point in all these cohorts was the study of cognitive function. In these subjects, the following tests were collected (58, 59).
Digit span tests
The digit span is a subtest of the Wechsler Adult Intelligence Scale-III (61). It is based on numbers and includes the forward and the backward digit span tests. In the forward digit span test, the examinee repeats a number sequence in the same order as presented. This constitutes a measure not only of working memory but also of attention. In the backward digit span task, the examinee repeats in reverse order series of digits that became gradually longer. This is an executive task particularly dependent on working memory. A higher score reflects a better working memory. In a standardization sample of 394 participants (aged 16 to 89 years), the reliability coefficient was very high, ranging from 0.94 to 0.97 (62).
Trail making test
The TMT is composed by two parts: TMTA and TMTB. The TMTA (greater focus on attention) consisted of a standardized page in which numbers 1 to 25 are scattered within the circles, and participants were asked to connect the numbers in order as quickly as possible. Before starting the test, a six-item practice test was administered to the participants to make sure they understood both tasks. A maximum time of 300 s was allowed before suspending the test. The direct scores of TMTA were the time in seconds taken to complete each task. In the same way, the TMTB (greater focus on executive function) consisted of an alternating sequence of numbered circles and letters (63, 64). In both tests, shorter times to completion indicate better performance.
California verbal learning test-II
The California verbal learning test-II (CVLT) is used to assess verbal learning and memory (65). It consists of five learning tests in which a list of words (list A) is presented and the subject is asked, immediately after each presentation, to recall as much words as possible. Then, an interference list (list B) is presented, and the subject is asked to repeat the same task. CVLT immediate recall score is a result of the first five tests and provides information about the learning process. In the short delay test, the patient is asked to recall list A, free (CVLT short-delayed free recall) or with semantic facilitation (CVLT short-delayed cued recall). After 20 min, in which nonverbal tasks are carried on, the subjects are asked to repeat a list of words that the examinee has previously presented without semantic facilitation (the CVLT long-delayed free recall) to assess long-term memory. A higher score reflects a better memory function. About 50 min are necessary to administrate this test, and its reliability ranges from 0.78 to 0.94 (66).
STROOP test (Golden’s version)
The STROOP test was used to assess cognitive flexibility, selective attention, inhibition, and information-processing speed. This version consists of three different parts: (i) 100 words (color names) are printed in black ink, and the subject is asked to read them as fast as possible; (ii) 100 “XXX” are printed in color ink (green, blue, and red), and the subject is asked to name as fast as possible the ink color; and (iii) 100 color names (from the first page) were printed in color ink (from the second page), the color name and the ink color do not match, and the subject is asked to name the ink color (and not to read the color name). The subject is given 45 s for each task. After the 45 s, the last item completed is noted, obtaining three scores: one for each part of the test (“W,” “C,” and “CW”). The interference (“I”) index was also obtained from the subtraction CW – CW′, where CW′ = (C × W)/(C + W). Standard administration procedures were followed as indicated in the test manual (67).
Memory binding test
The memory binding test consists of 16 category cues two word lists containing 16 word items in each list. Both lists are learned and recalled by controlled learning and cued and paired recall. The memory binding test produces four principal measures, the total paired recall, total free recall, total delayed free recall, and total delayed paired recall.
Symbol digit test
The symbol digit test is a subtest of the Wechsler Adult Intelligence Scale-III. It includes a coding key that shows nine abstract symbols, each paired with a number, and below the key, series of symbols were presented. The participants were asked to write down the corresponding numbers associated with the abstract symbols as quickly as possible. This task is a measure of information processing speed. The number of correct substitutions during a 90-s interval was scored, and higher numbers indicate better performance.
General cognition
A composite cognitive score was computed as the sum of the standardized scores of each individual for each test included in the Aging Imageomics cohort.
Adipose tissue collection and handling
Adipose tissue samples were obtained from SAT and VAT during elective surgical procedures (cholecystectomy, surgery of abdominal hernia, and gastric bypass surgery) (68). Both SAT and VAT samples were collected from the abdomen, following standard procedures. Samples of adipose tissue were immediately transported to the laboratory (5 to 10 min). The handling of tissue was carried out under strictly aseptic conditions. Adipose tissue samples were washed in phosphate-buffered saline, cut off with forceps and scalpel into small pieces (100 mg), and immediately flash-frozen in liquid nitrogen before stored at −80°C.
VAT- and SAT-stranded RNA-seq (discovery IRONMET; validation cohort 1 INTESTINE)
VAT and SAT RNA purification was performed using RNeasy-Tissue Mini-Kit (QIAGEN). Total RNA was quantified by Qubit RNA BR Assay kit (Thermo Fisher Scientific), and the integrity was checked by using the RNA Kit (15NT) on 5300 Fragment Analyzer System (Agilent Technologies) (68).
The RNA-seq libraries were prepared with Illumina TruSeq Stranded Total RNA Sample Preparation kit following the manufacturer’s recommendations with some modifications. Briefly, in function of availability, 100 to 500 ng of total RNA was ribosomal RNA depleted using the RiboZero Magnetic Gold Kit and fragmented by divalent cations. The strand specificity was achieved during the second-strand synthesis performed in the presence of deoxyuridine triphosphate. The cDNA was adenylated and ligated to Illumina platform compatible IDT adaptors with unique dual indexes with unique molecular identifiers (Integrated DNA Technologies) for paired-end sequencing. The ligation products were enriched with 15 PCR cycles, and the final library was validated on an Agilent 2100 Bioanalyzer with the DNA 7500 assay (Agilent Technologies). The libraries were sequenced on NovaSeq 6000 (Illumina) in a fraction of sequencing flow cell with a read length of 2 × 101 base pairs following the manufacturer’s protocol for dual indexing. Image analysis, base calling, and quality scoring of the run were processed using the manufacturer’s software Real Time Analysis (RTA v3.4.4) and followed by generation of FASTQ sequence files.
RNA-seq reads were mapped against human reference genome (GRCh38) using STAR software version 2.5.3a (69) with ENCODE parameters. Genes were quantified using RSEM version 1.3.0 (70) with default parameters and using the annotation file from GENCODE version 29. Only protein-coding genes that were expressed >1 count per million (cpm) in at least 10 samples were considered for the association of gene expression with clinical variables.
Circulating gene expression analysis (validation cohort 2, Aging Imageomics)
Total RNA was purified from blood using PAXgene Blood RNA Kit (QIAGEN, Gaithersburg, MD). RNA concentrations were assessed with Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Total RNA was reverse-transcribed to cDNA using High Capacity cDNA Archive Kit (Applied Biosystems, Darmstadt, Germany). Gene expression was assessed by RT-PCR using the LightCycler 480 Real-Time PCR System (Roche Diagnostics SL, Barcelona, Spain), using SYBR green technology suitable for relative genetic expression quantification. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as endogenous control.
The commercially predesigned KiCqStart primers used were as follows: NR4A2, 5′-GACTATCAAATGAGTGGAGATG-3′ (forward) and 5′-GACCTGTATGCTAATCGAAG-3′ (reverse); OAT, 5′-CAGACCTGATATAGTCCTCC-3′ (forward) and 5′-CTTCTTCTA AAACCTCAAGGG-3′ (reverse); amphiphysin (AMPH), 5′-CCTGACATAAAGAATCGCATC-3′ (forward) and 5′-CAAACACTTTCTGTGCTTTC-3′ (reverse); nudix hydrolase 2 (NUDT2), 5′-TTCTGCTTCGGTCCTTAG-3′ (forward) and 5′-AACTCAATTGCATTGTTGTC-3′ (reverse); Unc-5 netrin receptor B (UNC5B), 5′-AGGACAGTTACCACAACC-3′ (forward) and 5′-GGGGATCTCCTGGTATTTG-3′ (reverse); EZR, 5′-TGACTTTGTGTTTTATGCCC-3′ (forward) and 5′-CTCCTTTTCTT CTCTGTTTCC-3′ (reverse); GAPDH, 5′-ACAGTTGCCATGTAGACC-3′ (forward) and 5′-TTGAGCACAGGGTACTTTA-3′ (reverse).
VAT expression of candidate genes by RT-PCR (validation cohort 3 ADIPOBRAIN)
Total RNA was purified from VAT using RNeasy Lipid Tissue Mini Kit (QIAGEN, Izasa, SA). RNA concentrations were assessed with Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE), and the integrity was checked by Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA). Total RNA was reverse-transcribed to cDNA using High Capacity cDNA Archive Kit (Applied Biosystems, Darmstadt, Germany). Gene expression was assessed by RT-PCR using a LightCycler 480 Real-Time PCR System (Roche Diagnostics SL, Barcelona, Spain), using TaqMan technology suitable for relative genetic expression quantification. Peptidylprolyl isomerase A (PPIA) and GAPDH were used as endogenous control. The commercially available and prevalidated TaqMan primer/probe sets used were as follows: SLC18A2 (Hs00996835_m1); RIMS1 (Hs01112189_m1), PPIA (Hs99999904_m1), and GAPDH (4352934). The commercially predesigned KiCqStart primers used were as follows: NUDT2, EZR, UNC5B, NR4A2, and PPIA.
Plasma HPLC-ESI-MS/MS metabolomics (discovery IRONMET)
Metabolites were extracted from plasma samples with methanol (containing phenylalanine-C13 as an internal standard) according to previously described methods (58, 59, 71). Briefly, samples (30 μl) of cold methanol were added to 10 μl of each sample, vortexed for 1 min, and incubated for one hour at −20°C. Samples were homogenized using FastPrep-24 (MP Biomedicals) and were incubated overnight in a rocker at 4°C. Then, all samples were centrifuged for 3 min at 12,000g, and the supernatant was recovered and filtered with a 0.2 μm Eppendorf filter. Two microliters of the extracted sample were applied onto a reversed-phase column (Zorbax SB-Aq 1.8 μm, 2.1 × 50 mm; Agilent Technologies) equipped with a precolumn (Zorbax-SB-C8 Rapid Resolution Cartridge 2.1 × 30 mm, 3.5 μm; Agilent Technologies) with a column temperature of 60°C. The flow rate was 0.6 Ml/min. Solvent A was composed of water containing 0.2% acetic acid, and solvent B was composed of methanol containing 0.2% acetic acid. The gradient started at 2% B, increased to 98% B in 13 min, and held at 98% B for 6 min. Posttime was established in 5 min.
Data were collected in positive and negative electrospray modes time of flight operated in full-scan mode at 50 to 3000 mass/charge ratio in an extended dynamic range (2 GHz), using N2 as the nebulizer gas (5 liters/min at 350°C). The capillary voltage was 3500 V with a scan rate of 1 scan/s. The ESI source used a separate nebulizer for the continuous, low-level (10 liters/min) introduction of reference mass compounds 121.050873 and 922.009798, which were used for continuous, online mass calibration. MassHunter Data Analysis software (Agilent Technologies, Barcelona, Spain) was used to collect the results, and MassHunter Qualitative Analysis software (Agilent Technologies, Barcelona, Spain) was used to obtain the molecular features of the samples, representing different, comigrating ionic species of a given molecular entity using the Molecular Feature Extractor algorithm (Agilent Technologies, Barcelona, Spain). We selected samples with a minimum of two ions. Multiple charge states were forbidden. Compounds from different samples were aligned using a retention time window of 0.1% ± 0.25 min and a mass window of 20.0 parts per million (ppm) ± 2.0 mDa. We selected only those present in at least 50% of the samples of one group and corrected for individual bias.
Plasma 1H NMR metabolomics (discovery IRONMET)
Plasma samples were thawed at room temperature. For each sample, 400 μl of plasma were combined with 200 μl of phosphate buffer [9% (w/v) NaCl and 100% D2O] that contained 10 mM of 3-trimethylsilyl-1-[2,2,3,3-2H4] (TSP) (58, 59). Samples were mixed with the use of a vortex and centrifuged (10,000g) for 10 min. Then, a 550 μl aliquot was transferred into a 5 mm NMR tube before NMR analysis. 1H spectra of low–molecular weight metabolites were performed using a Carr-Purcell-Meiboom-Gill (CPMG) sequence (RD–90°–[t–180°– t]n–ACQ-FID) with spin-echo delay of 400 μs (for a total T2 filter of 210 ms) allowing an efficient attenuation of the lipid NMR signals. The CPMG sequence generates spectra edited by T2 relaxation times, reducing broad resonances from high–molecular weight compounds facilitating the observation of low–molecular weight metabolites. The total acquisition time was 2.73 s with a Recycling Delay (RD) of 2 s, and the 90° pulse length was automatically calibrated for each sample at around 11.1 μs. For each sample, eight dummy scans were followed by 256 scans and collected in 64,000 points over a spectral width of 20 ppm. TSP was used a general reference for NMR samples because it does not introduce any additional signals apart from the sharp methylsilyl resonance at 0 ppm. In addition, a high concentration of TSP was used to release low–weight metabolites with high affinity for serum proteins by binding competition with TSP.
All 1H NMR spectra were recorded at 300 K on an Avance III 600 spectrometer (Bruker, Germany) operating at a proton frequency of 600.20 MHz using a 5 mm Broad Band Multinuclear (PABBO) gradient probe and automatic sample changer with a cooling rack at 4°C.
Plasma cytokines and neurotoxic proteins
Commercially available enzyme-linked immunosorbent assay (ELISA) kits were used to measure the plasma of IL-6 (RND-HS600C, human IL-6 Quantikine HS ELISA kit, R&D Systems), IL-4 (RND-HS400, human IL-4 Quantikine HS ELISA kit, R&D Systems), α-synuclein (RND-DY1338-05, human α-synuclein DuoSet ELISA, R&D Systems), Aβ42 [RND-DAB142, human Aβ (amino acids 1 to 42) Quantikine ELISA kit, R&D Systems], and tau (NBP2-62749, human tau ELISA kit, Novus Biologicals) following the manufacturer’s protocol.
D. melanogaster experiments
D. melanogaster stocks and maintenance
Flies were raised on standard medium (cornmeal, sugar, and yeast) and cultured according to standard procedures at 28°C in a 12:12-hour light-dark cycle.
Conditional RNAi lines targeting the genes of interest are summarized in table S47. v60000 (control 1) and v60100 (control 2) were obtained from Vienna Drosophila RNAi Centre (http://stockcenter.vdrc.at/control/main) and used as the corresponding genetic background controls for KK and GD RNAi line collections, respectively. The Rim overexpression line (UAS-Rim), stock BL78050, referred to as UAS-Rim and the corresponding genetic background control (BL36304; control 3) were obtained from the Bloomington Drosophila Stock Centre (http://flystocks.bio.indiana.edu/). UAS-Rim was subsequently isomerized for six generations into the (BL36304; control 3) genetic background.
RNAi expression in the fat body was induced by w; C7-GAL4; UAS-Dcr-2 driver line, which carries a fat tissue–specific promoter, driving the expression of GAL4 specifically on Drosophila adiposities, was provided by M. Jindra (72).
Drosophila courtship conditioning
Drosophila courtship conditioning was used to assess Drosophila learning and memory capabilities. Courtship conditioning was performed as described previously (45). RNAi and overexpression lines together with their corresponding genetic background controls (table S47) were crossed with w; C7-GAL4; UAS-Dcr-2 driver line and raised at 28°C to down-regulate or overexpress the corresponding targeted genes in the adipose tissue. Males were collected at eclosion.
and kept in isolation until 7 days of age at 25°C. Males were randomly assigned to either trained or naïve groups. Learning capabilities were assessed by pairing individual males with a single premated wild-type female for 2.5 hours during the training period. Immediately after training, the males were tested to assess learning. Short-term memory was assessed by pairing individual males with a single premated wild-type female for 6 hours during the training period, and, subsequently, the male was individually isolated for 1 hour before testing. Testing was performed by transferring male to a 1-cm-diameter chamber with in the presence of a premated female and filmed for 8 min. The courtship behavior of male flies toward females was quantified manually from videos. The mean CIs (defined as the percentage of time spent on courtship during an 8-min interval) of trained males and naïve males were used to calculate the LI, which is defined as the per cent reduction in mean courtship activity in trained males compared with naïve males; LI = (CInaïve − CItrained)/CInaïve. For all conditions, male-female pairs were analyzed on a minimum of three different test days, and the data were pooled.
Nonparametrical statistical bootstrapping analysis was preformed to assess differences in LI (learning and short-term memory) between RNAi knockdown or overexpression lines and their corresponding genetic background controls. The R script provided by Koemans et al. (45) was used to perform the bootstrapping analysis. Briefly, CI values were randomly sampled with replacement to generate 10,000 hypothetical LIs, which were used to determine the 95% confidence interval of the difference between the index of the control and the index of the knockdown and the probability that this difference is >0. t test or one-way analysis of variance (ANOVA) was performed to assess statistical differences in CI of the compared conditions.
mRNA extraction and cDNA synthesis
Seven-day-old male were selected for quantitative RT-PCR (qRT-PCR) or RNA-seq. A total of 10 adult heads or 10 fat bodies were collected per sample and transferred to RNAlater solution (Sigma-Aldrich). Total RNA was extracted using the Arcturus PicoPure RNA Purification kit (Thermo Fisher Scientific). To avoid amplification from genomic DNA, deoxyribonuclease treatment was performed using the DNA-free Ambion kit, and RNA was reverse-transcribed into cDNA using the High-Capacity cDNA reverse transcription (Applied Biosystems) according to the manufacturers’ procedures.
Quantitative real-time polymerase chain reaction
Quantitative PCRs were performed using the LightCycler 480 SYBR Green Master (Roche) in a LightCycler 480 II machine (Roche). Primer sequences and the reference genes RNApol2 and 1tub23cf are provided in table S51. For each condition, a minimum of five biological replicates and two technical replicates were analyzed. Differential gene expression was calculated using the 2ΔΔCt method. The average threshold cycle (Ct) value for each sample was calculated and subtracted from the geometric mean Ct value of the reference genes RNApol2 and 1tub23cf to calculate the ΔCt value. t test (GraphPad Prism version 5.00 for Windows) was used for calculations of P values.
mRNA sequencing of fat body and fly heads
The starting material for sequencing library preparation was total RNA. The samples were quantified using the Qubit RNA BR Assay kit (Thermo Fisher Scientific), and RNA integrity was estimated with the RNA 6000 Nano Bioanalyzer 2100 Assay (Agilent Technologies). The RNA-seq libraries were constructed using the KAPA Stranded mRNA-Seq Illumina Platforms Kit (Roche), following the manufacturer’s recommendations. The size and quality of the libraries were assessed using a High Sensitivity DNA Bioanalyzer assay (Agilent Technologies). The libraries were sequenced on a NovaSeq 6000 (Illumina) in paired-end mode, with a read length of 2 × 51 base pairs, following the manufacturer’s protocol for dual indexing. Image analysis, base calling, and quality scoring of the run were processed using the manufacturer’s software, Real Time Analysis (RTA 3.4.4). RNA-seq reads were mapped D. melanogaste reference genome (BDGP6.32) with STAR/2.7.8a (ref1) with ENCODE parameters (69) with ENCODE parameters. Gene quantification was performed with RSEM/1.3.0 (70) with default parameters and using ensembl109 annotation. Only protein-coding genes that were expressed >1 cpm in at least 10 samples were considered.
Mice experiment
Animals
Male wild-type C57BL/6J (Charles River, France) 8-week-old mice were housed individually in controlled laboratory conditions (21° ± 1°C, 55 ± 10%) with illumination at 12-hour cycles (lights on at 7:30 a.m. and off at 7:30 p.m.). Animals were fed ad libitum for 8 weeks either with an ND (~3.514 kcal/kg: 10% fat, 66% carbohydrate, and 24% protein; n = 23) or an HFD (~5.228 kcal/kg: 60% fat, 16% carbohydrate, and 24% protein; n = 25). ND and HFD were obtained from Altromin Spezialfutter GmbH & Co. (KG, Lage, Germany), references C1090-10, and C1090-60, respectively. Three days after the start of the diet, weight-matched mice of each group (ND or HFD) were divided into two groups and injected intravenously via tail vein with 200 μl of either the AAV8-RSV-GFP-Adp-miRNASLC18A2 vector containing 1 × 1012 Genome Copies (GC) (Fig. 6C) in phosphate-buffered saline or with saline. Adeno-associated virus (AAV) vector was produced by the Viral Production Unit, UAB (www.viralvector.eu/). The AAV2/8-RSV-GFP-Adp-miRNASLC18A2 vector expresses the GFP and an miRNA designed to down-regulate SLC18A2 gene expression. It has a recombinant AAV2/8 serotype that has been demonstrated to be effective at targeting adipose tissue (73). Adipose selectivity was enhanced by incorporating a proximal promoter and distal enhancer of the adiponectin gene into the AAV vector. To decrease vector expression in the liver, this AAV incorporates eight copies of the liver-specific miR-122T into the 3′ untranslated region of the expression cassette (Fig. 6C) (73). Bodyweight and food intake were monitored weekly for the entire protocol. All experiments were performed during the light phase. Mice were euthanized by decapitation, and mWAT was collected and frozen with dry ice. All procedures were conducted in strict accordance with the guidelines of the European Communities Directive 86/609/EEC regulating animal experimentation and were approved by the local ethical committee (Comitè Ètic d’Experimentació Animal-Parc de Recerca Biomèdica de Barcelona, CEEA-PRBB, agreement no. 9687).
Experimental procedure
Behavioral tasks aimed to assess short- and long-term memory (novel object recognition in V-maze), and locomotor activity (horizontal and vertical activity assessment) was performed chronologically as indicated in (Fig. 6D) using standardized tests.
Novel object recognition test (V-MAZE)
Object-recognition memory was assayed in a V-shaped maze (30 cm long × 4.5 cm wide × 15 3 cm height each corridor) (74, 75). On day 1 of the test, mice were habituated for 9 min to the maze. Then, 24 hours later, mice were put back in the maze for 9 min, two identical objects were presented, and the time that the mice spent exploring each object was recorded. The mice were again placed in the maze 3 hours (30 days after the AAV injection to assess short-term memory) or 24 hours (37 days after the AAV injection to assess long-term memory) later for 9 min, with one of the familiar objects replaced with a novel object. The total time spent exploring each of the two objects (novel and familiar) was computed. A discrimination index was calculated as the difference between the times spent exploring the novel object divided by the total time exploring both objects. A higher discrimination index is considered to reflect greater memory retention for the familiar object (76, 77).
Locomotion
On day 46 after AAV injection, mice underwent a 1-hour locomotion test in individual locomotor activity boxes (10.8 × 20.3 × 18.6 cm; Imetronic, Pessac, France) equipped with infrared sensors to detect locomotor activity and an infrared plane to detect rearing (76, 77).
Statistical analysis
The number of animals (n) in each experimental condition is indicated in figure legends. All graphs and statistical comparisons were performed using GraphPad Prism software version 8.0 for Mac (GraphPad Software, San Diego, CA, USA). To test whether the data were normally distributed, the Kolmogorov-Smirnov test was applied. Comparison of means between different groups was performed using two-way ANOVA, followed by Newman-Keuls multiple comparisons test as post hoc test. Three-way ANOVA was performed when required to test the evolution over time. Results are expressed as individual values with the means ± SEM, specified in the figure legend. A P < 0.05 was used to determine statistical significance.
RNA expression
RNA purification was performed using RNeasy Lipid Tissue Mini Kit (QIAGEN, Hilden, Germany), and concentrations were measured using a Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Integrity was checked by Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA). Total RNA was reversed transcribed to cDNA using High Capacity cDNA Archive Kit (Applied Biosystems, Darmstadt, Germany). Gene expression was assessed by RT-PCR using a LightCycler 480 Real-Time PCR System (Roche Diagnostics SL, Barcelona, Spain), using SYBR Green technology suitable for relative genetic expression quantification. The RT-PCR reaction was performed in a final volume of 10 μl. The cycle program consisted of an initial denaturing of 10 min at 95°C and then 40 cycles of 15 s denaturizing phase at 95°C and 1 min annealing and extension phase at 60°C. A Ct value was obtained for each amplification curve and then a ΔCt was first calculated by subtracting the Ct value for endogenous control from the Ct value for each sample. Fold changes compared with the endogenous control were then determined by calculating 2 − ΔCt. β-Actin (Actb) was used as housekeeping. Actb and GFP were analyzed by SYBR green technology using the following primer sets: Actb, 5′-GATGTATGAAGGCTTTGGTC-3′ (forward) and 5′-TGTGCACTTTTATTGGTCTC-3′ (reverse); GFP, 5′-CAACAGCCACAACGTCTATATCATG-3′ (forward) and 5′-ATGTTGTGGCGGATCTTGAAG-3′ (reverse).
Protein analysis
Adipose tissue samples (100 mg) were homogenized using radioimmunoprecipitation lysis buffer system (sc-24948, Santa Cruz Biotechnology, CA) at 4°C for 30 min. Cellular debris were eliminated by centrifugation at 13,000g for 15 min (4°C). Protein concentration was determined using the RC/DC Protein Assay (Bio-Rad Laboratories, Hercules, CA). For Western blot analysis, samples were resolved by SDS–polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, CA). Membrane was exposed overnight at 4°C to primary antibody VMAT2/SLC18A2 (EB06558) at 1:250 dilution (VWR International EuroLab, SL) diluted in 2% bovine serum albumin – 1× tris-buffered saline containing 0.1% Tween 20, following the recommendations of the manufacturer. After secondary antibody incubation (anti-goat horseradish peroxidase), signal was detected using enhanced chemiluminescence horseradish peroxidase substrate (Millipore) and analyzed with a luminescent image analyzer ChemiDoc MP Imaging System (Bio-Rad Laboratories, Hercules, CA). TGX Stain-Free gels (#4568096, Bio-Rad Laboratories, Hercules, CA) were used as protein loading control.
Statistical analysis
Differential expression gene analyses were performed on gene counts using the “limma” R package (78). First, low expressed genes were filtered, so that only genes with more than 10 reads in at least two samples were selected. RNA-seq data were then normalized for RNA composition using the trimmed mean of M value as implemented in edgeR package (79). Normalized counts were then converted to log2 count per million with associated precision weights to account for variations in precision between different observations using the “voom” function with donor’s age, BMI, sex, and education years as covariates. A robust linear regression model adjusted for the previous covariates was then fitted to the data using the “lmFit” function with the option method = “robust,” to limit the influence of outlying samples. Last, an empirical Bayes method was applied to borrow information between genes with the “eBayes” function. P values were adjusted for multiple comparisons using the pFDR. A cutoff for the pFDR of <0.05 was used a threshold for statistical significance. The functional roles of differentially expressed genes were characterized using overrepresentation analyses based on the Reactome and WikiPathways databases using ConsensusPathDB (80). Pathway significance was assessed using a hypergeometric test, and a Storey procedure (q value) was applied for multiple testing corrections. Statistical significance was set at q < 0.1. For differentially expressed genes simultaneously in the VAT and SAT of the discovery and validation cohorts, data were further analyzed using functional gene-gene interaction networks mapping genes to the STRING database (which integrates known and predicted protein/gene interactions) (81). Then, functional local clusters in the interaction network were determined using a Markov cluster algorithm (MCL) with an inflation parameter = 1.4. Active interacting sources including text mining, experiments, databases, coexpression, and cooccurrence and an interaction score of >0.4 were used to construct the interaction networks. In addition, we integrated the information provided from differential expression analysis, gene-gene interaction networks, and pathway overrepresentation analysis using the R package “pathfinder” with default parameters (82). First, significant genes were mapped onto a STRING gene-gene interaction network. Then, active subnetworks of interconnected genes (including genes that are not significant themselves but connect significant genes) in this gene-gene interaction network were identified. Last, separate pathway overrepresentation analyses based on the Reactome databases were performed for each active subnetwork using the significant genes in each of the active subnetworks and genes in the Protein-protein Interaction Network (PIN) as the background genes. Pathway statistical significance was set at the pFDR of <0.05. Significantly enriched pathways were clustered via hierarchical clustering. Genes in each pathway were used to calculate the pairwise kappa statistics, a chance-corrected measure of cooccurrence between pathways. The distance matrix 1-kappa statistic was used for agglomerative hierarchical clustering and a threshold of 0.35 for the kappa statistics was used to define strong relation. Partial Spearman’s correlation analysis controlling for age, BMI, sex, and education level was used to determine the correlation between circulating levels of genes and the neurocognitive tests scores. As it works by obtaining the residuals of the ranked variables after removing the effect of the ranked covariates, scatter and violin plots were generated with the ranked residuals of the model adjusting for selected covariates. Nonparametric monotonic trends according to the gene tertiles or quintiles for the selected cognitive tests were assessed by the Mann-Kendall trend test.
Acknowledgments
Funding: This study was partially funded by Instituto de Salud Carlos III (ISCIII, Madrid, Spain) through the project PI18/01022 and PI21/01361 to J.M.F.-R. and the projects PI20/01090 (cofunded by the European Union under the European Regional Development Fund “A way to make Europe”) and CP18/00009 (cofunded by the European Union under the European Social Fund “Investing in your future”) to J.M.-P. and Generalitat de Catalunya 2021 SGR 01263. Albert Einstein College of Medicine owns the copyright for the Memory Binding Test and makes it available as a service to the research community but charges for commercial use (for permission requests contact the AECOM at biotech@einstein.yu.edu); the Spanish and Catalan adaptations used were provided by the Barcelona Beta Brain Research Center and the Pasqual Maragall Foundation with the AECOM’s permission. For further information about these versions, contact Nina Gramunt at ngramunt@pmaragall.org. M.A.-R. is funded by Instituto de Salud Carlos III, Río Hortega (CM19/00190). N.O.-C. is funded by Instituto de Salud Carlos lll, PFIS (FI19/00293). A.C.-N. is funded by Instituto de Salud Carlos lll, Sara Borrell (CD20/00051). IDIBGI is a CERCA center from the “CERCA Programme/Generalitat de Catalunya.”
Author contributions: J.M.-P. researched the data, performed the statistical analysis, and wrote the manuscript. N.O.-C., M.A.-R., and J.L.-L. researched the data. A.C.-N. and L.d.l.V.-C. performed and wrote the Drosophila experiments. V.C.-A., E.M.-G., and R.M. performed and wrote the mouse experiment. A.M.-A. performed the neuropsychological examination. J.M.M.-N., J.P., J.G.-O., R.R., and F.V. contributed to the discussion and reviewed the manuscript. J.M.F.-R. performed the statistical analysis and wrote the manuscript. J.M.-P. and J.M.F.-R. carried out the conception and coordination of the study. All authors participated in final approval of the version to be published. J.M.F.-R. is the guarantor of this work and, hence, had full access to all the data in the study and takes responsibility for the integrity of the data.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Legends for tables S1 to S51
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S51
REFERENCES AND NOTES
- 1.P. S. Minhas, A. Latif-Hernandez, M. R. McReynolds, A. S. Durairaj, Q. Wang, A. Rubin, A. U. Joshi, J. Q. He, E. Gauba, L. Liu, C. Wang, M. Linde, Y. Sugiura, P. K. Moon, R. Majeti, M. Suematsu, D. Mochly-Rosen, I. L. Weissman, F. M. Longo, J. D. Rabinowitz, K. I. Andreasson, Restoring metabolism of myeloid cells reverses cognitive decline in ageing. Nature 590, 122–128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A. N. Suarez, T. M. Hsu, C. M. Liu, E. E. Noble, A. M. Cortella, E. M. Nakamoto, J. D. Hahn, G. De Lartigue, S. E. Kanoski, Gut vagal sensory signaling regulates hippocampus function through multi-order pathways. Nat. Commun. 9, 2181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.H. Endle, G. Horta, B. Stutz, M. Muthuraman, I. Tegeder, Y. Schreiber, I. F. Snodgrass, R. Gurke, Z.-W. Liu, M. Sestan-Pesa, K. Radyushkin, N. Streu, W. Fan, J. Baumgart, Y. Li, F. Kloss, S. Groppa, N. Opel, U. Dannlowski, H. J. Grabe, F. Zipp, B. Rácz, T. L. Horvath, R. Nitsch, J. Vogt, AgRP neurons control feeding behaviour at cortical synapses via peripherally derived lysophospholipids. Nat. Metab. 4, 683–692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Y. Qu, H. Y. Hu, Y. N. Ou, X. N. Shen, W. Xu, Z. T. Wang, Q. Dong, L. Tan, J. T. Yu, Association of body mass index with risk of cognitive impairment and dementia: A systematic review and meta-analysis of prospective studies. Neurosci. Biobehav. Rev. 115, 189–198 (2020). [DOI] [PubMed] [Google Scholar]
- 5.X. Tang, W. Zhao, M. Lu, X. Zhang, P. Zhang, Z. Xin, R. Sun, W. Tian, M. A. Cardoso, J. Yang, R. Simó, J. B. Zhou, C. D. A. Stehouwer, Relationship between central obesity and the incidence of cognitive impairment and dementia from cohort studies involving 5,060,687 participants. Neurosci. Biobehav. Rev. 130, 301–313 (2021). [DOI] [PubMed] [Google Scholar]
- 6.A. K. Dahl, L. B. Hassing, E. I. Fransson, M. Gatz, C. A. Reynolds, N. L. Pedersen, Body mass index across midlife and cognitive change in late life. Int. J. Obes. (Lond) 37, 296–302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.H. Gardener, M. Caunca, C. Dong, Y. K. Cheung, T. Rundek, M. S. V. Elkind, C. B. Wright, R. L. Sacco, Obesity measures in relation to cognition in the Northern Manhattan study. J. Alzheimers Dis. 78, 1653–1660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. C. Farruggia, D. M. Small, Effects of adiposity and metabolic dysfunction on cognition: A review. Physiol. Behav. 208, 112578 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.W. Zeng, R. M. Pirzgalska, M. M. A. Pereira, N. Kubasova, A. Barateiro, E. Seixas, Y. H. Lu, A. Kozlova, H. Voss, G. G. Martins, J. M. Friedman, A. I. Domingos, Sympathetic neuro-adipose connections mediate leptin-driven lipolysis. Cell 163, 84–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.F. Cardoso, R. G. J. Klein Wolterink, C. Godinho-Silva, R. G. Domingues, H. Ribeiro, J. A. da Silva, I. Mahú, A. I. Domingos, H. Veiga-Fernandes, Neuro-mesenchymal units control ILC2 and obesity via a brain–adipose circuit. Nature 597, 410–414 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S. F. Morrison, K. Nakamura, Central mechanisms for thermoregulation. Annu. Rev. Physiol. 81, 285–308 (2019). [DOI] [PubMed] [Google Scholar]
- 12.K. Sodhi, R. Pratt, X. Wang, H. V. Lakhani, S. S. Pillai, M. Zehra, J. Wang, L. Grover, B. Henderson, J. Denvir, J. Liu, S. Pierre, T. Nelson, J. I. Shapiro, Role of adipocyte Na,K-ATPase oxidant amplification loop in cognitive decline and neurodegeneration. iScience 24, 103262 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D. H. Guo, M. Yamamoto, C. M. Hernandez, H. Khodadadi, B. Baban, A. M. Stranahan, Visceral adipose NLRP3 impairs cognition in obesity via IL-1R1 on CX3CR1+ cells. J. Clin. Investig. 130, 1961–1976 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D. H. Guo, M. Yamamoto, C. M. Hernandez, H. Khodadadi, B. Baban, A. M. Stranahan, Beige adipocytes mediate the neuroprotective and anti-inflammatory effects of subcutaneous fat in obese mice. Nat. Commun. 12, 4623 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. Kivipelto, F. Mangialasche, T. Ngandu, Lifestyle interventions to prevent cognitive impairment, dementia and Alzheimer disease. Nat. Rev. Neurol. 14, 653–666 (2018). [DOI] [PubMed] [Google Scholar]
- 16.S. Anazi, S. Maddirevula, E. Faqeih, H. Alsedairy, F. Alzahrani, H. E. Shamseldin, N. Patel, M. Hashem, N. Ibrahim, F. Abdulwahab, N. Ewida, H. S. Alsaif, H. Al Sharif, W. Alamoudi, A. Kentab, F. A. Bashiri, M. Alnaser, A. H. Alwadei, M. Alfadhel, W. Eyaid, A. Hashem, A. Al Asmari, M. M. Saleh, A. Alsaman, K. A. Alhasan, M. Alsughayir, M. Al Shammari, A. Mahmoud, Z. N. Al-Hassnan, M. Al-Husain, R. Osama Khalil, N. Abd Elmeguid, A. Masri, R. Ali, T. Ben-Omran, P. Elfishway, A. Hashish, A. Ercan Sencicek, M. State, A. M. Alazami, M. A. Salih, N. Altassan, S. T. Arold, M. Abouelhoda, S. M. Wakil, D. Monies, R. Shaheen, F. S. Alkuraya, Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol. Psychiatry 22, 615–624 (2017). [DOI] [PubMed] [Google Scholar]
- 17.F. Diaz, S. Khosa, D. Niyazov, H. Lee, R. Person, M. M. Morrow, R. Signer, N. Dorrani, A. Zheng, M. Herzog, R. Freundlich, J. B. Birath, Y. Cervantes-Manzo, J. A. Martinez-Agosto, C. Palmer, S. F. Nelson, B. L. Fogel, S. K. Mishra, Novel NUDT2 variant causes intellectual disability and polyneuropathy. Ann. Clin. Transl. Neurol. 7, 2320–2325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.K. Takei, V. I. Slepnev, V. Haucke, P. De Camilli, Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat. Cell Biol. 1, 33–39 (1999). [DOI] [PubMed] [Google Scholar]
- 19.S. Bergström, J. Remnestål, J. Yousef, J. Olofsson, I. Markaki, S. Carvalho, J. C. Corvol, K. Kultima, L. Kilander, M. Löwenmark, M. Ingelsson, K. Blennow, H. Zetterberg, B. Nellgård, F. Brosseron, M. T. Heneka, B. Bosch, R. Sanchez-Valle, A. Månberg, P. Svenningsson, P. Nilsson, Multi-cohort profiling reveals elevated CSF levels of brain-enriched proteins in Alzheimer’s disease. Ann. Clin. Transl. Neurol. 8, 1456–1470 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Y. Yu, T. Niccoli, Z. Ren, N. S. Woodling, B. Aleyakpo, G. Szabadkai, L. Partridge, PICALM rescues glutamatergic neurotransmission, behavioural function and survival in a Drosophila model of Aβ42 toxicity. Hum. Mol. Genet. 29, 2420–2434 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.C. Sheng, U. Javed, M. Gibbs, C. Long, J. Yin, B. Qin, Q. Yuan, Experience-dependent structural plasticity targets dynamic filopodia in regulating dendrite maturation and synaptogenesis. Nat. Commun. 9, 3362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A. Ginguay, L. Cynober, E. Curis, I. Nicolis, Ornithine aminotransferase, an important glutamate-metabolizing enzyme at the crossroads of multiple metabolic pathways. Biology (Basel) 6, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.M. Arnoriaga-Rodríguez, J. Mayneris-Perxachs, O. Contreras-Rodríguez, A. Burokas, J. A. Ortega-Sanchez, G. Blasco, C. Coll, C. Biarnés, A. Castells-Nobau, J. Puig, J. Garre-Olmo, R. Ramos, S. Pedraza, R. Brugada, J. C. Vilanova, J. Serena, J. Barretina, J. Gich, V. Pérez-Brocal, A. Moya, X. Fernández-Real, L. Ramio-Torrentà, R. Pamplona, J. Sol, M. Jové, W. Ricart, M. Portero-Otin, R. Maldonado, J. M. Fernández-Real, Obesity-associated deficits in inhibitory control are phenocopied to mice through gut microbiota changes in one-carbon and aromatic amino acids metabolic pathways. Gut 70, 2283–2296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. Arnoriaga-Rodríguez, J. Mayneris-Perxachs, A. Burokas, O. Contreras-Rodríguez, G. Blasco, C. Coll, C. Biarnés, R. Miranda-Olivos, J. Latorre, J. M. Moreno-Navarrete, A. Castells-Nobau, M. Sabater, M. E. Palomo-Buitrago, J. Puig, S. Pedraza, J. Gich, V. Pérez-Brocal, W. Ricart, A. Moya, X. Fernández-Real, L. Ramió-Torrentà, R. Pamplona, J. Sol, M. Jové, M. Portero-Otin, R. Maldonado, J. M. Fernández-Real, Obesity impairs short-term and working memory through gut microbial metabolism of aromatic amino acids. Cell Metab. 32, 548–560.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 25.D. Carulli, F. de Winter, J. Verhaagen, Semaphorins in adult nervous system plasticity and disease. Front. Synaptic Neurosci. 13, 672891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.T. C. Südhof, Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.V. García-Morales, F. Montero, D. González-Forero, G. Rodríguez-Bey, L. Gómez-Pérez, M. J. Medialdea-Wandossell, G. Domínguez-Vías, J. M. García-Verdugo, B. Moreno-López, Membrane-derived phospholipids control synaptic neurotransmission and plasticity. PLOS Biol. 13, e1002153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R. W. Mitchell, G. M. Hatch, Fatty acid transport into the brain: Of fatty acid fables and lipid tails. Prostaglandins Leukot. Essent. Fatty Acids 85, 293–302 (2011). [DOI] [PubMed] [Google Scholar]
- 29.L. N. Nguyen, D. Ma, G. Shui, P. Wong, A. Cazenave-Gassiot, X. Zhang, M. R. Wenk, E. L. K. Goh, D. L. Silver, Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature 509, 503–506 (2014). [DOI] [PubMed] [Google Scholar]
- 30.K. Bourassa, D. A. Sbarra, Body mass and cognitive decline are indirectly associated via inflammation among aging adults. Brain Behav. Immun. 60, 63–70 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Y. Yang, G. S. Shields, Q. Wu, Y. Liu, H. Chen, C. Guo, The association between obesity and lower working memory is mediated by inflammation: Findings from a nationally representative dataset of U.S. adults. Brain Behav. Immun. 84, 173–179 (2020). [DOI] [PubMed] [Google Scholar]
- 32.C. Wang, H. Yue, Z. Hu, Y. Shen, J. Ma, J. Li, X. D. Wang, L. Wang, B. Sun, P. Shi, L. Wang, Y. Gu, Microglia mediate forgetting via complement-dependent synaptic elimination. Science 367, 688–694 (2020). [DOI] [PubMed] [Google Scholar]
- 33.R. Dantzer, J. C. O’Connor, G. G. Freund, R. W. Johnson, K. W. Kelley, From inflammation to sickness and depression: When the immune system subjugates the brain. Nat. Rev. Neurosci. 9, 46–56 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A. A. Miller, S. J. Spencer, Obesity and neuroinflammation: A pathway to cognitive impairment. Brain Behav. Immun. 42, 10–21 (2014). [DOI] [PubMed] [Google Scholar]
- 35.O. Guillemot-Legris, G. G. Muccioli, Obesity-induced neuroinflammation: Beyond the hypothalamus. Trends Neurosci. 40, 237–253 (2017). [DOI] [PubMed] [Google Scholar]
- 36.S. Chatterjee, E. N. Walsh, A. L. Yan, K. P. Giese, S. Safe, T. Abel, Pharmacological activation of Nr4a rescues age-associated memory decline. Neurobiol. Aging 85, 140–144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.H. Shah, F. Dehghani, M. Ramezan, R. B. Gannaban, Z. F. Haque, F. Rahimi, S. Abbasi, A. C. Shin, Revisiting the role of vitamins and minerals in Alzheimer’s disease. Antioxidants (Basel) 12, 415 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. U. Wołoszynowska-Fraser, A. Kouchmeshky, P. McCaffery, Vitamin A and retinoic acid in cognition and cognitive disease. Annu. Rev. Nutr. 40, 247–272 (2020). [DOI] [PubMed] [Google Scholar]
- 39.J. Ng, A. Papandreou, S. J. Heales, M. A. Kurian, Monoamine neurotransmitter disorders - Clinical advances and future perspectives. Nat. Rev. Neurol. 11, 567–584 (2015). [DOI] [PubMed] [Google Scholar]
- 40.P. S. Kaeser, L. Deng, M. Fan, T. C. Südhof, RIM genes differentially contribute to organizing presynaptic release sites. Proc. Natl. Acad. Sci. U.S.A. 109, 11830–11835 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.C. M. Powell, S. Schoch, L. Monteggia, M. Barrot, M. F. Matos, N. Feldmann, T. C. Südhof, E. J. Nestler, The presynaptic active zone protein RIM1α is critical for normal learning and memory. Neuron 42, 143–153 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.N. Krumm, T. N. Turner, C. Baker, L. Vives, K. Mohajeri, K. Witherspoon, A. Raja, B. P. Coe, H. A. Stessman, Z. X. He, S. M. Leal, R. Bernier, E. E. Eichler, Excess of rare, inherited truncating mutations in autism. Nat. Genet. 47, 582–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M. S. Shin, S. Y. Park, S. R. Park, S. H. Seol, J. S. Kwon, Clinical and empirical applications of the Rey-Osterrieth complex figure test. Nat. Protoc. 1, 892–899 (2006). [DOI] [PubMed] [Google Scholar]
- 44.A. H. Brand, N. Perrimon, Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 45.T. S. Koemans, C. Oppitz, R. A. T. Donders, H. Van Bokhoven, A. Schenck, K. Keleman, J. M. Kramer, Drosophila courtship conditioning as a measure of learning and memory. J. Vis. Exp. 2017, 55808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.N. Raun, S. Jones, J. M. Kramer, Conditioned courtship suppression in Drosophila melanogaster. J. Neurogenet. 35, 154–167 (2021). [DOI] [PubMed] [Google Scholar]
- 47.M. Haddadi, S. R. Jahromi, B. K. C. Sagar, R. K. Patil, T. Shivanandappa, S. R. Ramesh, Brain aging, memory impairment and oxidative stress: A study in Drosophila melanogaster. Behav. Brain Res. 259, 60–69 (2014). [DOI] [PubMed] [Google Scholar]
- 48.D. Wang, L. Qian, H. Xiong, J. Liu, W. S. Neckameyer, S. Oldham, K. Xia, J. Wang, R. Bodmer, Z. Zhang, Antioxidants protect PINK1-dependent dopaminergic neurons in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 103, 13520–13525 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.S. Bahadorani, P. Bahadorani, J. P. Phillips, A. J. Hilliker, The effects of vitamin supplementation on Drosophila life span under normoxia and under oxidative stress. J. Gerontol. A Biol. Sci. Med. Sci. 63, 35–42 (2008). [DOI] [PubMed] [Google Scholar]
- 50.W. G. Roeterdink, K. E. Strecker, C. C. Hayden, M. H. M. Janssen, D. W. Chandler, Imaging the rotationally state-selected NO(A,n) product from the predissociation of the A state of the NO-Ar van der Waals cluster. J. Chem. Phys. 130, 134305 (2009). [DOI] [PubMed] [Google Scholar]
- 51.T. Kawai, M. V. Autieri, R. Scalia, Adipose tissue inflammation and metabolic dysfunction in obesity. Am. J. Physiol. Cell Physiol. 320, C375–C391 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Z. Liu, X. Huang, Lipid metabolism in Drosophila: Development and disease. Acta Biochim. Biophys. Sin (Shanghai) 45, 44–50 (2013). [DOI] [PubMed] [Google Scholar]
- 53.R. P. Kühnlein, The contribution of the Drosophila model to lipid droplet research. Prog. Lipid Res. 50, 348–356 (2011). [DOI] [PubMed] [Google Scholar]
- 54.S. Meltzer, J. A. Bagley, G. L. Perez, C. E. O’Brien, L. DeVault, Y. Guo, L. Y. Jan, Y. N. Jan, Phospholipid homeostasis regulates dendrite morphogenesis in Drosophila sensory neurons. Cell Rep. 21, 859–866 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.J. Muffat, D. W. Walker, S. Benzer, Human ApoD, an apolipoprotein up-regulated in neurodegenerative diseases, extends lifespan and increases stress resistance in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 7088–7093 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.A. Costa, B. Reynés, J. Konieczna, M. Martín, M. Fiol, A. Palou, D. Romaguera, P. Oliver, Use of human PBMC to analyse the impact of obesity on lipid metabolism and metabolic status: A proof-of-concept pilot study. Sci. Rep. 11, 18329 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.M. Cifre, A. Palou, P. Oliver, Cognitive impairment in metabolically-obese, normal-weight rats: Identification of early biomarkers in peripheral blood mononuclear cells. Mol. Neurodegener. 13, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.J. Mayneris-Perxachs, A. Castells-Nobau, M. Arnoriaga-Rodríguez, M. Martin, L. de la Vega-Correa, C. Zapata, A. Burokas, G. Blasco, C. Coll, A. Escrichs, C. Biarnés, J. M. Moreno-Navarrete, J. Puig, J. Garre-Olmo, R. Ramos, S. Pedraza, R. Brugada, J. C. Vilanova, J. Serena, J. Gich, L. Ramió-Torrentà, V. Pérez-Brocal, A. Moya, R. Pamplona, J. Sol, M. Jové, W. Ricart, M. Portero-Otin, G. Deco, R. Maldonado, J. M. Fernández-Real, Microbiota alterations in proline metabolism impact depression. Cell Metab. 34, 681–701.e10 (2022). [DOI] [PubMed] [Google Scholar]
- 59.J. Mayneris-Perxachs, A. Castells-Nobau, M. Arnoriaga-Rodríguez, J. Garre-Olmo, J. Puig, R. Ramos, F. Martínez-Hernández, A. Burokas, C. Coll, J. M. Moreno-Navarrete, C. Zapata-Tona, S. Pedraza, V. Pérez-Brocal, L. Ramió-Torrentà, W. Ricart, A. Moya, M. Martínez-García, R. Maldonado, J. M. Fernández-Real, Caudovirales bacteriophages are associated with improved executive function and memory in flies, mice, and humans. Cell Host Microbe 30, 340–356.e8 (2022). [DOI] [PubMed] [Google Scholar]
- 60.J. Puig, C. Biarnes, S. Pedraza, J. C. Vilanova, R. Pamplona, J. M. Fernández-Real, R. Brugada, R. Ramos, G. Coll-de-Tuero, L. Calvo-Perxas, J. Serena, L. Ramió-Torrentà, J. Gich, L. Gallart, M. Portero-Otin, A. Alberich-Bayarri, A. Jimenez-Pastor, E. Camacho-Ramos, J. Mayneris-Perxachs, V. Pineda, R. Font, A. Prats-Puig, M.-L. Gacto, G. Deco, A. Escrichs, B. Clotet, R. Paredes, E. Negredo, B. Triaire, M. Rodríguez, A. Heredia-Escámez, R. Coronado, W. de Graaf, V. Prevost, A. Mitulescu, P. Daunis-I-Estadella, S. Thió-Henestrosa, F. Miralles, V. Ribas-Ripoll, M. Puig-Domingo, M. Essig, C. R. Figley, T. D. Figley, B. Albensi, A. Ashraf, J. H. C. Reiber, G. Schifitto, U. Md Nasir, C. Leiva-Salinas, M. Wintermark, K. Nael, J. Vilalta-Franch, J. Barretina, J. Garre-Olmo, The aging imageomics study: Rationale, design and baseline characteristics of the study population. Mech. Ageing Dev. 189, 111257 (2020). [DOI] [PubMed] [Google Scholar]
- 61.D. Wechsler, WAIS-IV. Escala de inteligencia de Wechsler para adultos-IV, in Manual técnico y de interpretación (NCS Pearson, Madrid, Inc. Edici., 2012). [Google Scholar]
- 62.E. Strauss, E. M. S. Sherman, O. Spreen, A Compendium of Neuropsychological Tests: Administration, Norms, and Commentary (Oxford Univ. Press, 3rd Oxfo., 2006). [Google Scholar]
- 63.J. D. Corrigan, N. S. Hinkeldey, Relationships between parts A and B of the trail making test. J. Clin. Psychol. 43, 402–409 (1987). [DOI] [PubMed] [Google Scholar]
- 64.M. D. Lezak, Neuropsychological assessment in behavioral toxicology - Developing techniques and interpretative issues. Scand. J. Work Environ. Health 10 (suppl. 1), 25–29 (1984). [PubMed] [Google Scholar]
- 65.D. C. Delis, J. H. Kramer, E. Kaplan, B. A. Ober, Manual for the California Verbal Learning Test, (CVLT-II) (The Psychological Corporation, 2000).
- 66.A. M. Paolo, A. I. Tröster, J. J. Ryan, Test-retest stability of the California verbal learning test in older persons. Neuropsychology 11, 613–616 (1997). [DOI] [PubMed] [Google Scholar]
- 67.C. Golden, A manual for the clinical and experimental use of the Stroop color and word test, in Faculty Books and Book Chapters. Stoelting Company, 751–758 (1978).
- 68.J. Latorre, J. Mayneris-Perxachs, N. Oliveras-Cañellas, F. Ortega, F. Comas, J. M. Fernández-Real, J. M. Moreno-Navarrete, Adipose tissue cysteine dioxygenase type 1 is associated with an anti-inflammatory profile, impacting on systemic metabolic traits. EBioMedicine 85, 104302 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.B. Li, C. N. Dewey, RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.W. R. Wikoff, G. Pendyala, G. Siuzdak, H. S. Fox, Metabolomic analysis of the cerebrospinal fluid reveals changes in phospholipase expression in the CNS of SIV-infected macaques. J. Clin. Investig. 118, 2661–2669 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.J. Rynes, C. D. Donohoe, P. Frommolt, S. Brodesser, M. Jindra, M. Uhlirova, Activating transcription factor 3 regulates immune and metabolic homeostasis. Mol. Cell. Biol. 32, 3949–3962 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.S. M. O’neill, C. Hinkle, S. J. Chen, A. Sandhu, R. Hovhannisyan, S. Stephan, W. R. Lagor, R. S. Ahima, J. C. Johnston, M. P. Reilly, Targeting adipose tissue via systemic gene therapy. Gene Ther. 21, 653–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.E. Puighermanal, G. Marsicano, A. Busquets-Garcia, B. Lutz, R. Maldonado, A. Ozaita, Cannabinoid modulation of hippocampal long-term memory is mediated by mTOR signaling. Nat. Neurosci. 12, 1152–1158 (2009). [DOI] [PubMed] [Google Scholar]
- 75.A. Busquets-Garcia, E. Puighermanal, A. Pastor, R. De La Torre, R. Maldonado, A. Ozaita, Differential role of anandamide and 2-arachidonoylglycerol in memory and anxiety-like responses. Biol. Psychiatry 70, 479–486 (2011). [DOI] [PubMed] [Google Scholar]
- 76.M. Gomis-González, L. Galera-López, M. Ten-Blanco, A. Busquets-Garcia, T. Cox, R. Maldonado, A. Ozaita, Protein kinase C-gamma knockout mice show impaired hippocampal short-term memory while preserved long-term memory. Mol. Neurobiol. 58, 617–630 (2021). [DOI] [PubMed] [Google Scholar]
- 77.A. Escudero-Lara, D. Cabañero, R. Maldonado, Kappa opioid receptor modulation of endometriosis pain in mice. Neuropharmacology 195, 108677 (2021). [DOI] [PubMed] [Google Scholar]
- 78.M. E. Ritchie, B. Phipson, D. Wu, Y. Hu, C. W. Law, W. Shi, G. K. Smyth, Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.M. D. Robinson, D. J. McCarthy, G. K. Smyth, edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.A. Kamburov, U. Stelzl, H. Lehrach, R. Herwig, The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 41, D793–D800 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.D. Szklarczyk, A. L. Gable, D. Lyon, A. Junge, S. Wyder, J. Huerta-Cepas, M. Simonovic, N. T. Doncheva, J. H. Morris, P. Bork, L. J. Jensen, C. von Mering, STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.E. Ulgen, O. Ozisik, O. U. Sezerman, PathfindR: An R package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front. Genet. 10, 858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for tables S1 to S51
Tables S1 to S51