Abstract
Introduction:
Most patients with pancreatic cancer present with advanced stage, incurable disease. However, patients with high-grade precancerous lesions and many patients with low-stage disease can be cured with surgery, suggesting that early detection has the potential to improve survival. While serum CA19.9 has been a long-standing biomarker used for pancreatic cancer disease monitoring, its low sensitivity and poor specificity have driven investigators to hunt for better diagnostic markers.
Areas covered:
This review will cover recent advances in genetics, proteomics, imaging, and artificial intelligence, which offer opportunities for the early detection of curable pancreatic neoplasms.
Expert opinion:
From exosomes, to circulating tumor DNA, to subtle changes on imaging, we know much more now about the biology and clinical manifestations of early pancreatic neoplasia than we did just five years ago. The overriding challenge, however, remains the development of a practical approach to screen for a relatively rare, but deadly, disease that is often treated with complex surgery. It is our hope that future advances will bring us closer to an effective and financially sound approach for the early detection of pancreatic cancer and its precursors.
Keywords: Pancreas, pancreatic cancer, early detection, screening, ctDNA, liquid biopsy
1. Introduction
NORC, a research institute at the University of Chicago, recently estimated that only 14% of all cancers diagnosed in the United States are detected by screening [1]. Even in this background of low overall screening rates, pancreatic cancer stands out as a deadly malignancy without a widely-accepted screening program. Most patients with pancreatic cancer are not diagnosed until after the cancer has locally progressed or metastasized, and the prognosis for patients with advanced pancreatic cancer is dismal [2]. By contrast, patients diagnosed with non-invasive precancerous lesions can be cured, and patients diagnosed with low-stage invasive pancreatic cancers have promising survival rates [3–8]. These and other findings suggest that the earlier detection of pancreatic cancer and its precursors offers hope to improve survival for this disease. Indeed, Goggins and Canto have recently reported, in an uncontrolled trial, that the median survival for patients with a screen-detected pancreatic cancer (9.8 years) is more than six times longer than the median survival for patients with pancreatic cancer diagnosed outside of surveillance (1.5 years) [9].
An enormous challenge for the early detection of pancreatic cancer and its precursors is the relative rarity of the disease. While it has been estimated that 62,210 Americans were diagnosed with pancreatic cancer in 2022, the population of the country by the end of 2022 was about 334 million [2]. This means that only 0.0186 percent of the population developed pancreatic cancer that year. Screening tests with sensitivities and specificities near 100% would be needed to effectively detect most of these cancers in the asymptomatic general population without unnecessarily alarming too many people with false positives or overtreating lesions that would never have progressed even without treatment [10]. Another essential characteristic of an effective screening test is its ability to detect curable disease. For pancreatic cancer, that’s primarily Stage I disease, but few patients are diagnosed with Stage I pancreatic cancer outside of pancreatic surveillance programs. Most biomarker studies involve patients with advanced-stage disease whose biomarker alterations often reflect derangements associated with advanced disease.
The pressing question, then, is how will we get to a sensitive, specific, and cost-effective approach to the early diagnosis of pancreatic cancer and its precursors? To help answer this question, we first describe available screening approaches, as well as some potential advances on the horizon. We then review the performance characteristics needed for a pancreatic cancer screening program, and we describe ways to improve screening performance by prioritizing high-risk populations. Finally, we look forward to emerging technologies and opportunities.
2. CA19–9
Cancer antigen 19–9 (CA19–9, or sialylated Lewis (a) antigen) is one of the best-known markers of pancreatic cancer [11]. CA19–9, a monosialoganglioside, is produced by most pancreatic cancers, and CA19–9 levels are easily measured in the blood. Serum CA19–9 levels have proven to be useful in predicting resectability and in monitoring treatment response in patients known to have pancreatic cancer [12–16]. Warshaw and colleagues, for example, found that a postoperative decline in CA19–9 levels is a strong predictor of survival after surgical resection [17]. However, several other malignancies, and even a number of benign conditions, including obstructive jaundice, lung disease, liver failure, and acute and chronic pancreatitis, are also associated with elevated serum CA19–9 levels [18]. Furthermore, germline variants in the CA19–9 synthetic pathway influence serum levels, including those in FUT3, the enzyme responsible for CA19–9 synthesis [19]. Depending on race, as much as 10% of the population lack any functional FUT3 [20,21]. Pancreatic cancers that arise in these “Lewis antigen negative” individuals will not make significant amounts CA19–9, and their cancers are, therefore, generally non-detectable using this marker [19,22].
3. Other serum protein and glycoprotein markers
Other potential serum markers of pancreatic cancer have been identified using a variety of technologies, including serial analysis of gene expression (SAGE), gene expression arrays, mRNA sequencing, and mass spectrometry (Table 1) [23–29]. For example, one of genes first found to be overexpressed in pancreatic cancer by SAGE was tissue inhibitor of metalloproteinase type 1 (TIMP-1). Hanash and colleagues have found that the combination of serum TIMP1 and CA19–9 levels is more sensitive in detecting presymptomatic pancreatic cancers than CA19–9 levels alone [23,29].
Table 1:
Selected circulating protein markers of pancreatic cancer
| Marker | Full name(s) | Function | References |
|---|---|---|---|
| ApoA2 isoforms | Apolipoprotein A2 | Component of high-density lipoprotein particles | [30,31] |
| CA125 | Carcinoma antigen 125, Mucin 16 | O-glycosylated protein that functions in forming a protective mucous barrier | [32,33] |
| CA19–9 | Cancer antigen 19–9, carbohydrate antigen sialyl Lewis a | A glycosphingolipid that functions in embryogenesis that is secondarily absorbed to red blood cells | [31,34–36] |
| CEA | Carcinoembryonic antigen, carcinoembryonic Antigen-Related Cell Adhesion Molecule 5 | Cell surface glycoprotein that functions in cell adhesion | [19,33,35,37,38] |
| CPA1 | Carboxypeptidase A1 | A zinc metalloprotease that cleaves select dietary proteins | [34] |
| DUPAN-2 | Duke pancreatic monoclonal antigen type 2 | A precursor of CA 19–9 | [39] |
| GDF15/MIC-1 | Growth differentiation factor 15 | A secreted ligand in the transforming growth factor-beta superfamily of proteins | [40,41] |
| LRG1 | Leucine rich alpha-2-glycoprotein 1 | Functions in protein-protein interactions, cell adhesion, and signal transduction | [42,43] |
| MUC5AC | Mucin 5AC | Protects mucosal surfaces, and functions in phosphatidylinositol-mediated signaling | [44] |
| SPP1 | Osteopontin | Attachment of osteoclasts to mineralized bone matrix (hydroxyapatite) | [33,38,45,46] |
| THBS2 | Thrombospondin-2 | A glycoprotein that mediates cell-cell and cell-matrix interactions | [47–49] |
| TIMP1 | Tissue inhibitor of metalloproteinases 1 | As the name suggests, inhibits matrix metalloproteinases | [23,50] |
Several technologies have been employed to identify other potential new protein and glycoprotein blood markers since the discovery of TIMP1 [24,51–54]. For example, Cao and colleagues, as part of the Clinical Proteomic Tumor Analysis Consortium (CPTAC), conducted a comprehensive proteomic, phosphoproteomic, and glycoproteomic analysis of 140 pancreatic cancers. They identified over 200 proteins expressed at higher levels in tumors compared to normal pancreas and macrodissected normal pancreatic ducts [24]. Importantly, many of these proteins were similarly abundant in low-stage cancers, and a number of them were predicted to be secreted proteins, making them attractive targets for early detection [24]. Glycoproteomic analyses in this study identified 75 N-linked glycoproteins upregulated more than two-fold in the cancers [24]. As expected, mucin-type O-linked glycoproteins associated with CA19–9 were found to be upregulated in pancreatic malignancy [24]. As with the proteomic analysis, many of the glycoproteins identified were also upregulated in low-stage disease, suggesting that they may be good markers for early detection [24]. Thus, a host of potential new markers of early pancreatic cancer have been identified, and several, such as thrombospondin 2, have been shown to improve upon the diagnostic accuracy of CA19–9 alone [48].
The added value of these newly discovered blood-based proteins, when used individually as markers for pancreatic cancer, is unclear. Boyd and colleagues conducted a meta-analysis of 28 primary studies and found that while several novel protein biomarkers have moderate diagnostic accuracy, they do not outperform CA19–9 in distinguishing between patients with and without pancreatic cancer [11]. When they are used in combination, however, these new markers may, however, be useful in Lewis antigen non-secretors. Kane and colleagues conducted a similar analysis using PRISMA standards and found that pooling biomarkers, particularly TIMP-1, CEA, CA125, and CA19–9, can improve the diagnostic accuracy over CA19–9 alone [55].
4. Using genotyping to improve blood-based protein and glycoprotein markers
One challenge to using the protein and glycoprotein markers identified to date is that the expression of these markers can be influenced by inherited gene variants, creating significant person-to-person variability in the normal ranges of the blood levels of these markers [19,34,56]. Abe and colleagues proposed to overcome this challenge by using germline genotyping to create germline variant-specific tumor marker reference ranges for CA19–9, CA125 and carcinoembryonic antigen (CEA)[19,34]. Tanaka and colleagues performed a similar study evaluating serum carboxypeptidase A (CPA) and CA19–9 levels, coupled with determinations of germline CPA1 and germline Lewis and secretor gene genotypes, in 345 controls and 190 patients with pancreatic cancer and found that the combination of germline variant-stratified CPA and CA19–9 levels achieved specificity levels >98% for pancreatic cancer [34]. Thus, “personalized” biomarker marker reference ranges offer one potential route for improving the early detection of pancreatic cancer.
5. Branch chain amino acids
Mayers and colleagues reported that elevated plasma levels of branched chain amino acids are a predictive marker for developing pancreatic cancer [57]. Using blood samples from four large prospective cohort studies, the team found that the branched chain amino acids isoleucine, leucine and valine, were significantly elevated in some individuals years before they developed pancreatic cancer [57]. The authors hypothesized that early pancreatic cancers are associated with an increased whole-body breakdown of proteins and that the resultant increase in branched chain amino acids could be used as a marker for the early detection of pancreatic cancer [57].
In addition, it appears that dietary intake high in branched chain amino acids is associated with pancreatic cancer risk and that branched chain amino acid metabolism can contribute to the growth of pancreatic cancer [58–62]. Subsequent studies have validated these findings and have confirmed that branched chain amino acids can be elevated years, and sometimes even a decade, before patients become symptomatic and are diagnosed with pancreatic cancer [62].
6. Metabolites
Alterations in the blood levels of a number of metabolites have been reported in patients with pancreatic cancer [43,63]. Mahajan and colleagues reported that a test incorporating the serum levels of multiple metabolites with CA19–9 could achieve an area under the curve (AUC) of >90% in distinguishing patients with pancreatic cancer from controls [63]. Similarly, Fahrmann and colleagues reported that a plasma metabolite panel, which included acetylspermidine, diacetylspermine, an indole-derivative, and two lysophosphatidylcholines, improved the performance of CA19–9, LRG1 and TIMP1 in detecting low-stage pancreatic cancer [43].
One of the challenges of discovering metabolites specific for pancreatic cancer, and indeed of any blood-based protein or glycoprotein marker of pancreatic cancer, is that patients with pancreatic cancer often have a number of comorbidities in addition to their cancer that can dramatically impact the analytes measured. For example, virtually all cancers that arise in the head of the pancreas obstruct the distal common bile duct as it runs through the pancreas, and, as noted with CA19–9, this biliary obstruction will alter the metabolism of some analytes [64]. Thus, markers identified by comparing healthy controls to patients with pancreatic cancer may be detecting non-specific physiologic changes, such as biliary obstruction, rather than the development of pancreatic cancer itself. As a result, alterations in analytes may not be specific for pancreatic cancer if applied to real-world populations [64].
7. Exosomes
Extracellular vesicles function in cell-cell communication [65]. Exosomes, a type of extracellular vesicle, have an average diameter of 100 nanometers and can contain a variety of materials including nucleic acids and proteins [65]. Cancer cells may secrete exosomes, which have been shown to increase the susceptibility of distant organs to metastatic seeding and growth [66]. Since exosomes are shed into extracellular spaces, including blood, exosomal “cargo” is an attractive target for early detection. Indeed, Nakamura and others reported that exosomal miRNAs in the blood could serve as markers for early pancreatic cancer [67–70]. As is true for the blood protein markers, the combination of exosomal markers with serum CA19–9 levels may prove more sensitive and specific than any one marker in isolation [71].
8. Circulating tumor DNA
One of the most active targets for the early detection of pancreatic and other cancers is circulating tumor DNA (ctDNA). Pancreatic cancer, like almost all other cancers, is driven by the accumulation of somatic mutations in a set of well-defined genes [52,72–74]. DNA harboring these cancer-specific somatic mutations is released in small quantities into the blood, and this ctDNA can be detected using modern sequencing methods [33,75–79]. Here we briefly describe some of the many different cancer-specific changes that can be detected in ctDNA.
The most common alterations used in ctDNA tests are somatic intragenic mutations [33,77]. The KRAS gene is somatically mutated in ~95% of pancreatic cancers, and extremely sensitive tests have been developed to detected rare mutant KRAS alleles admixed with thousands of wild-type KRAS alleles [75,80–82]. For example, MacGregor-Das and colleagues determined the plasma levels of mutant KRAS and GNAS in 67 patients with pancreatic cancer and in 73 healthy controls using digital next generation sequencing. They found that a third of the patients with low-stage cancer had detectable mutations in their blood, while mutant alleles were rarely detected in healthy individuals [80].
Aberrantly methylated DNA is another somatic cancer-specific change that can be detected in blood [78,83–86]. Ying and colleagues reported that a four gene methylation panel, which included the ADAMTS1, BNC1, LRFN5, and PXDN genes, was accurate in detecting pancreatic cancer, and Kandimalla and colleagues reported a separate methylation panel that produced areas under the curve (AUCs) of 0.85 in distinguishing patients with low-stage pancreatic cancer from controls [87,88].
Velculescu and others have described novel approaches to cancer detection based on the patterns of DNA fragmentation detected in the blood [76,79]. They reported that cell free DNA in blood samples from healthy individuals reflected the nucleosomal patterns of normal white blood cells, while patients with cancer had abnormal circulating DNA fragmentation patterns. These circulating DNA fragmentation patterns could be used to accurately identify the presence of a cancer and, remarkably, often predict the organ of origin of that cancer [79]. Adding to these observations, Mouliere and colleagues found that integrating the analyses of DNA fragment size improved the detection of cancer-specific mutations in ctDNA [76]. This fragmentation-based approach is being commercialized through the biotech company Delfi.
Jamshidi and colleagues recently compared some of the many approaches to detecting cancer-specific DNA in blood [89]. Using data from the Circulating Cell-free Genome Atlas study, they were able to compare whole-genome methylation, single nucleotide variants, somatic copy number alterations, and fragmentation pattern approaches. They found that the whole genome methylation approach was among the most sensitive of the methods and best predicted cancer origin. Some of the authors of the Jamshidi study are affiliated with the cancer screening company Grail, which is pursuing a methylation-based approach to multi-cancer detection.
Combining ctDNA markers with other markers will likely improve the accuracy of early detection tests. For example, Cohen and colleagues reported high sensitivity and specificity with a blood test that combines ctDNA testing for somatic mutations with a panel of serum protein biomarkers [38]. In this study, the combinatorial approach achieved a specificity of 99.5% with sensitivity of 64% [38].
Unfortunately, ctDNA levels are significantly lower in patients with low-stage cancers than they are in patients with high-stage disease, meaning that approaches based on ctDNA are more likely to detect advanced cancers than they are to detect early curable cancers [89]. For example, the company GRAIL recently reported the findings for a trial screening 6,662 individuals over the age of 50 using their multi-cancer early detection test [90]. Only 36 cancers were detected, and about half of them were advanced cancers (stage III or IV). The United Kingdom is currently performing a large, randomized trial to evaluate whether the GRAIL test (Galleri) can reduce the incidence of advanced cancers (stage III and IV) 3–4 years after randomization [91].
Another commercialized cancer screening test from the company Thrive Earlier Detection has clinical data available to evaluate. In a large non-randomized study of Thrive’s multi-analyte blood-based screening test performed by some of the authors of this review, 26 cancers were detectable among 9,911 apparently healthy subjects using a combined ctDNA and protein approach [92]. Of the 26 cancers identified, 17 were advanced (stage III or stage IV), one was stage unknown, and only eight were low-stage cancers (stage I or II) [92]. In addition, some of the 26 patients with screen detectable cancers already showed symptoms. The challenges of ctDNA-based screening are significantly greater if the goal is to detect curable precancerous lesions, because non-invasive lesions, even large precancers with high-grade dysplasia, appear not to release significant amounts of mutant DNA into the blood [93].
Although mutant DNA from precancerous lesions in the pancreas may not be shed into the blood in significant quantities, it is frequently shed into the pancreatic duct system (since precursor lesions arise in the ducts), and this DNA will eventually pass into the stool [94–97]. Yu and colleagues tested secretin-stimulated pancreatic juice samples collected at the time of endoscopy and found that mutant DNA could be detected in patients with invasive pancreatic cancer as well as patients with an intraductal papillary mucinous neoplasm [94]. Of note, the study included at-risk patients who were not known to have pancreatic cancer, and four of these at-risk study subjects later developed pancreatic cancer. Remarkably, two of these four had SMAD4/TP53 gene mutations detected in their pancreatic juice over a year before they were clinically diagnosed with cancer. These results highlight the potential value of biosamples obtained from sources closer to the organ being screened.
Nucleic acid-based screening assays have already appeared in clinics. There are at least six companies with tests for the early detection of cancer testing in various phases of development (Table 2). Three are developing tests based on methylation of ctDNA (EarlyDiagnostics, GRAIL and Singlera), one a test based on hydoxy-methylation of ctDNA (Bluestar/ClearNote Health), one uses ctDNA fragment length (DELFI), and finally one is developing a test based on the combination of ctDNA mutations and tumor protein markers (Thrive/Exact Biosciences).
Table 2:
Nucleic Acid-based screening assays for early detection1
| Company/PI | Test Name | Sample | Analyte/Assay | Status | Clinical Trials (NCT number) | References2 |
|---|---|---|---|---|---|---|
| Bluestar Genomics/ ClearNote Health | Avantect pancreatic cancer test | PB | 5-OH-methyl-cytosine | Development/ Live |
NCT03869814
NCT05188586 NCT05188573 |
[98,99] |
| Delfi | NA | PB | ctDNA, molecule length |
Development, anticipated 2023 |
NCT04825834
NCT05306288 |
[79,100–102] |
| EarlyDiagnostics | cfMethyl-Seq | PB | ctDNA methylation | Development | ND, Trials planned for liver and lung cancer | [103–105] |
| Exact Biosciences/ Thrive Early Detection |
CancerSEEK/ Detect-A |
PB | ctDNA and protein markers | Development |
NCT04213326
NCT04213326 |
[92] |
| Grail | Galleri | PB | ctDNA methylation | Live |
NCT05481697
NCT05611632 NCT05205967 |
[106–108] |
| Singlera Genomics | PanSeer, PDACatch | PB | ctDNA methylation |
Development |
NCT05336058
NCT05159544 NCT05431621 NCT05485077 NCT05444491 NCT05432128 NCT05536089 NCT05336539 NCT03828396 NCT05626985 NCT03685669 |
[109–113] |
Assays for earlier detection of cancer are also referred to as multi-cancer early detection (MCED), in addition to liquid biopsy.
As determined by an author or funding from the company, or use of the test in the study. NA: not available. ND: none detected in clinicaltrails.gov. PB= peripheral blood.
9. Autoantibodies
The somatic mutations that accumulate in neoplastic cells as they progress to invasive pancreatic cancer may create neoantigens when the genes altered are translated into proteins, and some people will develop antibodies to these neoantigens [114–117]. As reviewed by Dumstrei and colleagues, these novel autoantibodies have the potential to serve as markers for the detection of pancreatic cancer [114]. Their review concluded that single autoantibodies make poor markers, while multiple serum autoantibodies, when combined with other blood markers, such as CA19–9, can have better diagnostic performance. Nonetheless, most reports on the value of autoantibodies have not been validated across multiple studies, and the overall value of this approach has not been established [114].
10. Imaging
A variety of imaging modalities have been used to detect pancreatic neoplasia, as well as the changes that can occur in the body secondary to pancreatic cancer (Table 3) [118–124]. Endoscopic ultrasound (EUS) is probably the most sensitive of the pancreatic imaging modalities, and it has been used to detect asymptomatic precursor lesions, including intraductal papillary mucinous neoplasms, small invasive pancreatic cancers, and subtle secondary changes in the pancreas suggestive of multiple microscopic precursor lesions (pancreatic intraepithelial neoplasia (PanIN)) [120–126].
Table 3:
Comparison of imaging modalities
| Modality | Region of Interest | Resolution | Ionizing Radiation | Advantages | Disadvantages |
|---|---|---|---|---|---|
| Pancreatic protocol CT | Whole abdomen | Millimeter | Yes | • Easily accessible • Relatively cheap • High spatial resolution for detection of small detections and local staging • Detection of distant metastatic disease |
• Small masses can be occult on CT • Concerns with radiation safety limit its potential use as screening modality • Contraindicated in patients with renal insufficiency or contrast allergy |
| MRI/MRCP | Whole abdomen | Millimeter | No | • Accurate detection of pancreatic cysts and suspicious features • Accurate detection of pancreatic duct abnormality • Can detect pancreatic masses that are occult on CT • Can be performed without contrast for patients with renal insufficiency or contrast allergy |
• Less accessible and more expensive than CT • More operator dependent than CT |
| Endoscopic ultrasound | Pancreas and adjacent organs only | Millimeter | No | • Accurate characterization of pancreatic mass and pancreatic duct abnormality • Obtain fine needle aspiration for tissue diagnosis |
• Invasive procedure • Rare procedural complications • Limited assessment of disease extent outside pancreas |
CT= Computed tomography, MRCP=magnetic resonance cholangiopancreatography, MRI= magnetic resonance imaging, US= ultrasound
Computed tomography and magnetic resonance imaging can also provide detailed images of the pancreas, and a number of subtle changes, including dilatation of the main pancreatic duct, focal pancreatic atrophy and an abrupt cut-off of the duct, may be present years before patients are diagnosed with pancreatic cancer [127–129]. Image analysis with 3D rendering techniques like Cinematic Rendering have the potential for earlier lesion detection by looking at numerous features, such as pancreatic texture, rather than simply looking for a mass [130]. Textural changes may prove to be among the earliest finding and have often been the critical finding in review of pre-diagnostic CT scans (Figure 1).
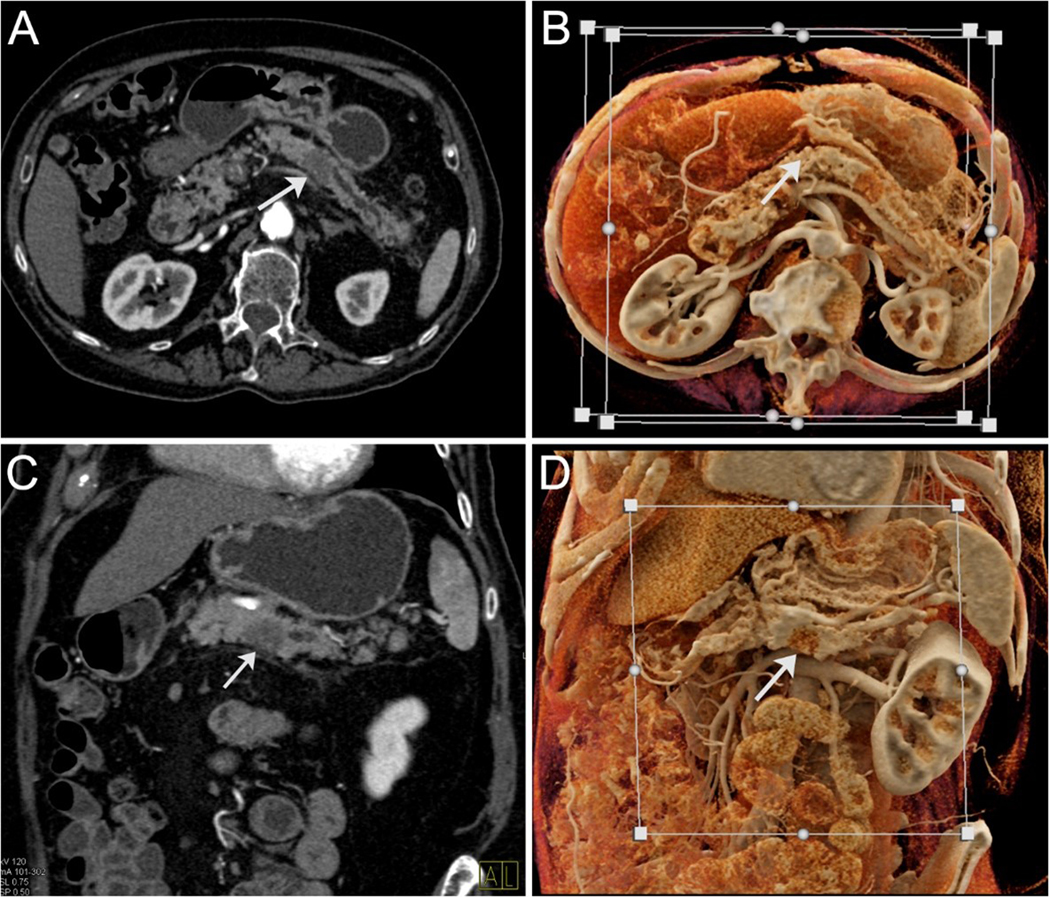
Figure 1:
Computed tomography of a patient with pancreatic cancer (arrows). Standard axial (A) and coronal (B) views demonstrating a hypointense mass in the body of the pancreas with upstream dilatation of the main pancreatic duct. Cinematic rendering (C and D), in similar planes, highlights the mass.
Harinck and colleagues compared endoscopic ultrasound to magnetic resonance imaging in a screening study of 139 asymptomatic high-risk individuals [131]. They found that the two imaging techniques are complementary, each with their own strengths and weaknesses.
Improvements in both image resolution and image analysis are on the horizon [132]. For example, photon counting CT has the potential to deliver 5 times higher resolution than standard CT [132,133]. In addition, several recent technological advances in image analysis have allowed investigators to detect features in images that the human eye might not perceive, and these have the potential to improve the sensitivity and specificity of imaging as a screening tool. Chu and colleagues, for example, applied radiomics, the mathematical integration of features such as shape, size, volume and texture, to CT of the pancreas and showed that radiomics features helped differentiate scans from patients with pancreatic cancer from scans from healthy controls, with sensitivities of 100% and specificities of 98.5% [134]. As discussed later in the section on artificial intelligence, these and other new technologies have the potential to improve the performance of imaging techniques.
The general challenge with using imaging for screening is that these tests tend to be expensive, some involve radiation (CT scans), and some (EUS) are invasive. Therefore, imaging-based screening programs cannot easily be applied to the general population.
11. New-onset diabetes
Pancreatic cancers cause a number of profound changes in the body. Some of these changes occur before patients develop symptoms, and these changes may therefore serve as early detection markers. Perhaps the most notable of these are alterations in blood glucose levels [135–140]. Chari and others have shown that many or even most patients with pancreatic cancer develop either hyperglycemia or overt diabetes, sometimes years before their cancer is diagnosed [141]. However, there are challenges with detecting early pancreatic cancer based on a diagnosis of new-onset diabetes (NOD) alone; first, only one in several hundred individuals with NOD will have pancreatic cancer, and second, diabetes associated with pancreatic cancer becomes more likely with increasing tumor burden [136,139,142,143]. Nonetheless, several groups have suggested a potential simple approach to screening for pancreatic cancer: all elderly patients with new onset diabetes could be prioritized for pancreatic imaging [135–140].
12. Other clinical features of pancreatic cancer
Muscle wasting is also commonly found in patients with pancreatic cancer and, like new onset diabetes in the elderly, muscle wasting could serve as an early indicator of pancreatic cancer [144–146]. For example, in the IMPACT study out of Italy, 73% of individuals with pancreatic cancer had sarcopenia at presentation [145]. Similarly, many patients with pancreatic cancer develop depression, and some even develop depression before they are diagnosed with cancer [147,148].
The problem with indirect clinical markers of pancreatic cancer is that they are common, non-specific, and all too often the health care professionals who care for these medical problems are unaware of their association with pancreatic cancer. Individuals who develop diabetes are treated by primary care physicians or endocrinologists, not oncologists. Even if these specialists are increasingly becoming aware of the association between physiological changes and pancreatic cancer, they have not been provided with well-established diagnostic algorithms to pursue this possibility.
13. Screening without intent
Artificial intelligence (AI) is one possible approach to overcoming many of the challenges noted above. For example, AI-based algorithms have been used to identify patterns in health care records that might indicate an increased likelihood of having pancreatic cancer [149–151]. Such algorithms are only as good as the information available in the medical record, but could eventually be used to automatically notify the patient’s health care provider that their patient has an increased risk of developing pancreatic cancer, and these algorithms could even suggest the best next steps to evaluate the patient. Such an approach, however, would require careful clinical validation to ensure it is providing actionable results without causing undue alarm.
AI can also be applied to images, and AI-based algorithms have the potential to be trained to detect early curable pancreatic cancers in CT scans [152–155]. These AI-based algorithms can achieve similar diagnostic performance to subspecialized academic radiologists and can potentially elevate the performance of an average community radiologist to the level of an expert. AI-based algorithms may have a role in early detection as reviews of pre-diagnostic CT scans from patients who subsequently developed pancreatic cancer have identified small, curable pancreatic cancers that went undetected by radiologists [118,119]. These small cancers produced subtle changes, such as focal atrophy of the gland, faint enhancement changes in the gland, dilatation of the main pancreatic duct, or an abrupt cut-off of the duct [118,119]. AI-based algorithms can be trained to recognize these subtle changes on pre-diagnostic (median time to diagnosis 386 days) CT scans with AUC up to 0.98 in a retrospective study, which can lead to significantly earlier diagnosis [153]. Since so many people in the United States have CT scans for reasons unrelated to their pancreas, AI-based algorithms could run in the background with the potential to detect early, curable pancreatic cancers [151,156]. Importantly, the application of AI-based algorithms to routine abdominal imaging would not incur the expense, added radiation exposure, and inconvenience of a population-wide screening program. Instead, AI-based algorithms can take advantage of what is already being done for other clinical indications and serve as a peer review mechanism to identify potentially clinically significant findings that are overlooked by radiologists [156,157]. On the other hand, relying solely on ad-hoc imaging will limit the potential reach of this approach since most people do not undergo abdominal imaging.
14. Low incidence as a barrier to screening
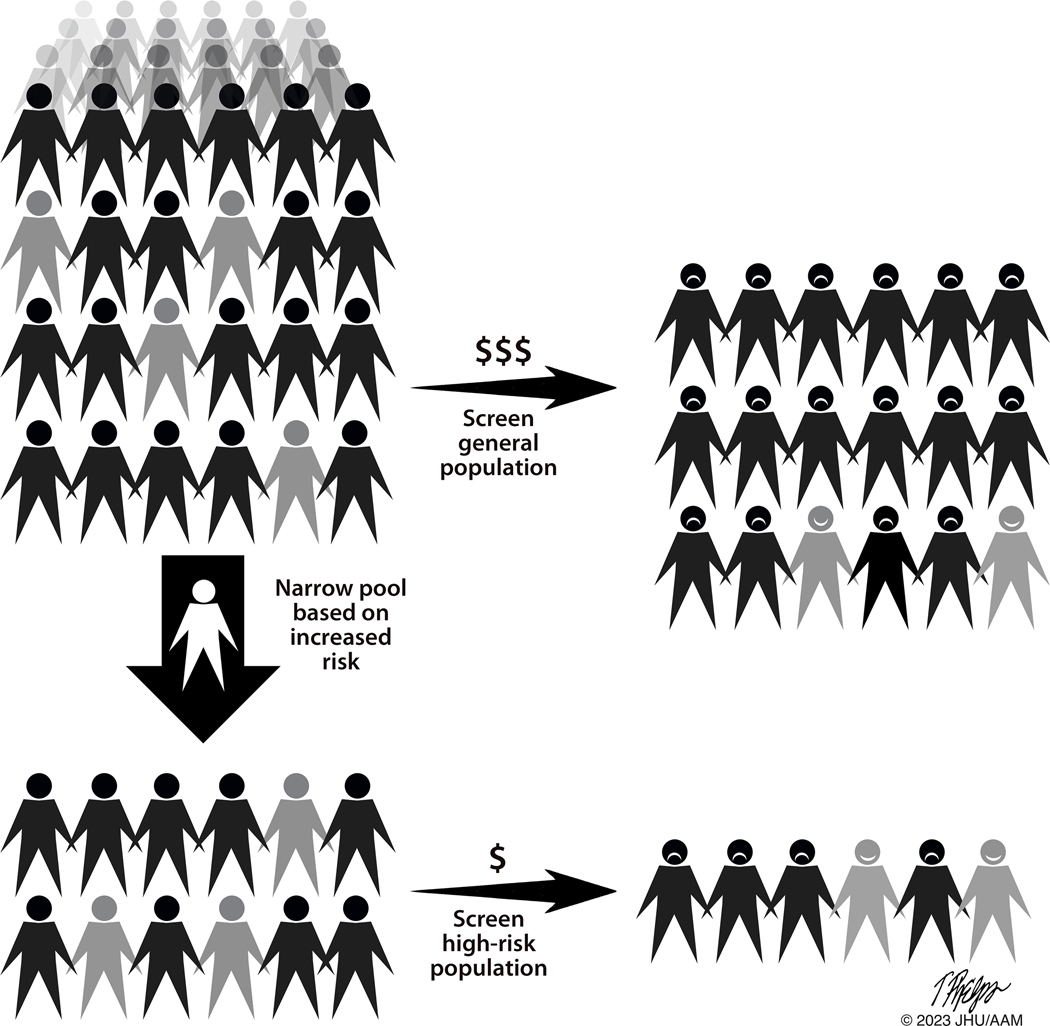
As noted earlier, although pancreatic cancer is deadly when it strikes, it is a relatively rare disease. Data from the Surveillance, Epidemiology and End Results (SEER) program of the National Cancer Institute have been used to estimate that, in the United States, the annual rate of new cases of pancreatic cancer is 13.3 per 100,000 for men and 11.1 per 100,000 for women [158]. Screening for a disease that strikes such a small fraction of the population is problematic [10,89]. One could improve the odds by focusing on the older adults, as 90% of pancreatic cancers occur in individuals 55 and older. However, tests with anything short of near perfect specificity will lead to numerous false positive results, needlessly alarming healthy people and potentially leading to unnecessary, costly, and potentially harmful interventions (Figure 2) [10,158–160]. Conversely, tests with low sensitivity will lower the number of true positive tests, thereby increasing the cost of the test per cancer detected and potentially offer false reassurance to participants. For example, we previously estimated that if 100,000 Americans over the age of 55 were screened using a test with a specificity of 98% and a sensitivity of 100%, the test would produce 1,999 false positive results for every 68 true positive results [10]. Screening tests for diseases such as pancreatic cancer that have a low annual incidence of disease must therefore have extraordinarily high specificity to avoid the negative consequences of large numbers of false positive results relative to true positives [89,159]. As Gray and colleagues stated in their classic 2008 paper on screening, “All screening programs do harm; some do good as well, and, of these, some do more good than harm at reasonable cost“ [159]. The question then becomes: how can one improve both pre-test probability and test performance to reduce the number of false positives?
Figure 2:
The rarity of pancreatic cancer in the general population (top left) remains a significant challenge to screening for the disease. Screening the general population, even with a highly sensitive and specific test, will result in numerous false positives (frowning faces) relative to true positives (smiling faces). Efforts to define populations with the greatest risk (lower left), will improve the ratio of true positives to false positives. (Grey indicates individuals affected with a high-grade precancer or early invasive pancreatic cancer. In the interest of space, the proportions of individuals affected in each group are not accurate).
15. Defining individuals with the greatest risk
Any successful screening program for pancreatic cancer must include efforts to focus the program on those with a risk of developing the disease that is sufficiently increased to overcome the adverse risks of screening. Age, family history of pancreatic cancer, germline gene status, and new onset diabetes mellitus are all associated with an increased risk, and the magnitude of this risk has been quantified for each, making them plausible approaches to identifying high-risk groups for screening (Table 4).
Table 4:
Risk factors for pancreatic cancer
| Risk Factor | Prevalence and Associated Risk | References |
|---|---|---|
| Non-genetic | ||
| Cigarette Smoking | 1.7 fold increased risk vs to never smokers. | [161] |
| Family history of pancreatic cancer | 2.5 fold increased risk for individuals with a first degree relative with pancreatic cancer, risk increases with additional affected relatives and/or family members who developed cancer before age 50. | [162] |
| Long Standing Diabetes | 1.5 – 2-fold increased risk of PDAC for individuals with diabetes >3 years in duration. | [163–166] |
| New onset Diabetes | <0.3% to 0.8% of new diabetes develop pancreatic ductal adenocarcinoma primarily within one year of diabetes diagnosis. | [143,167,168] |
| Obesity | 1.6 fold increased risk in obese vs normal weight. | [169] |
| Genes | ||
| ATM pathogenic variant | Occurs in 1–3% of PDAC cases, 6.5 fold increased risk. | [170–172] |
| BRCA1 | Occurs in 1% of PDAC, 2–4 fold increased risk. | [173–176] |
| BRCA2 pathogenic variant | Occurs in 2–7% of PDAC cases, 3.5–10 fold increased risk. |
[174–180] |
| CDKN2A pathogenic variant | Occurs <0.5% of PDAC cases, 34 -fold increased risk. | [175,181] |
| CPA1/CPB1 variants | <1%; lifetime risk not defined | [182,183] |
| GWAS Variants & Polygenic Risk Scores | European ancestry populations: 1q32.1 (NR5A2), 1p36.33 (NOC2L), 2p13.3 (ETAA1), 3q29 (TP63), 5p15.33 (CLPTM1L, TERT), 7p14.1 (INHBA), 8q21.11 (HNF4G) 8q24.21 (MYC), 9q34.2 (ABO), 13q12.2 (PDX1), 13q22.1 (KLF5), 16q23.1 (BCAR1), 17q12 (HNF1B) 17q25.1 (LINC00673), 18q21.32 (GRP), and 22q12.1 (ZNRF). China: 21q21.3, 5p13.1, 21q22.3, 22q13.32 and 10q26.11: Japan 6p25.3, 12p11.21 and 7q36.2. In Europeans ~4 fold higher risk for those with the top 10% of polygenic risk (combined weighted average of associated loci). | [184–189] |
| Mismatch Repair Gene | Occurs in up to 1% of PDAC, 8 fold increased risk. | [190] |
| PALB2 pathogenic variant | Occurs <0.5% of PDAC cases, up to 10-fold risk. | [191] |
| PRSS1 | Very Rare; >40 fold. | [192–194] |
| STK11 | Very Rare: 10–40 fold. | [195–197] |
Having a first-degree family member with pancreatic cancer increases one’s risk of developing precancerous and invasive pancreatic cancer, and having multiple family members increases the risk further [162,198–202]. The impact of family cancer history on pancreatic cancer risk is illustrated in a prospective study of 21,141 individuals in 4,433 families enrolled in the National Familial Pancreas Tumor Registry, conducted by Porter and colleagues [162]. They found that individuals in familial pancreatic cancer kindreds, defined as families in which at-least two first-degree family members had been diagnosed with pancreatic cancer, had a 4.86-fold increased risk of developing pancreatic cancer themselves [162]. Furthermore, the risk in the familial pancreatic cancer kindreds increased as the number of first-degree relatives (FDRs) with pancreatic cancer increased. Individuals with one FDR with pancreatic cancer had a 3.46-fold increased risk, those with two FDRs with pancreatic cancer had a 5.44-fold increased risk, and those with three or more FDRs with pancreatic cancer had a 10.78-fold increased risk [162]. The risk was even higher in families in which one of the family members developed pancreatic cancer before the age of 50 [162,203].
The risk associated with a family history of cancer can be refined if the pathogenic germline variant driving that risk can be identified (Table 4) [122,170–172,182,194,204–214]. For example, individuals with a pathogenic germline ATM variant have a 6.5-fold increased risk of developing pancreatic cancer [172]. Looking at other genes, carriers of pathogenic germline BRCA2 variants have a 3.5–10-fold increased risk of developing pancreatic cancer, carriers of a pathogenic germline CDKN2A variants have a 34-fold increased risk, and carriers of a pathogenic germline STK11 variant have a 10–40-fold increases risk [122,170–172,182,194,204–214]. Establishing age-specific risk estimates for all pancreatic cancer susceptibility genes will be important to optimize clinical surveillance protocols and early detection initiatives.
New onset diabetes, as discussed earlier, can be the first symptom of pancreatic cancer. While early studies suggested that elderly individuals with new-onset diabetes have up to an eight-fold increased risk of developing pancreatic cancer compared to healthy individuals, population-based studies in the Veterans Administration health system have shown that this risk is lower [141,167].
Other risk factors can be added to the mix (Table 4). For example, cigarette smokers have double the risk, obesity increases risk, and individuals of non-O blood groups are almost twice as likely to develop pancreatic cancer than are individuals of blood type O [215–217]. Unfortunately, even when one combines most of the leading risk factors together (including cigarette smoking, obesity, diabetes, family history and non-O ABO blood group), relatively few people have a combination of risk factors suggesting a risk sufficient to warrant screening with currently available tests [218]. As a result, it is challenging to identify a significant number of individuals who have a very high risk of developing pancreatic cancer and who would most benefit from screening for pancreatic cancer [218]. Furthermore, those with the greatest individual risk for pancreatic cancer may not contribute most to the total incidence of pancreatic cancer. As a result, a highly-targeted pancreatic cancer screening program with good performance is not guaranteed to markedly reduce the total population-level mortality. Most pancreatic cancers, for instance, are not associated with a known high-risk germline genetic predisposition [215]. To achieve the greatest population-level impact, moderate or even low-risk individuals might need to be included in a screening program, exposing many of them to unnecessary testing. This has been referred to as the “prevention paradox [219].”
16. Costs
The last words of J. Gray’s statement on screening, “All screening programmes do harm; some do good as well, and, of these, some do more good than harm at reasonable cost,” remind us that costs need to be considered when instituting a screening program [159]. One estimate of the financial costs, using Medicare and national average pricing, suggested that a screening program based on magnetic resonance imaging/magnetic resonance cholangiopancreatography could be affordable if applied to a high-risk population [220]. In particular, using Medicare data, the cost per “year of life added” was $638.62 for screening individuals with the Peutz-Jeghers syndrome and $945.33 for individuals with hereditary pancreatitis [220]. The costs were even lower, only $356.42, for obese smokers over the age of 50 with new-onset diabetes [220]. Corral and colleagues came to a similar conclusion, that screening can be financially reasonable for individuals with a very high (>20-fold) increased risk of pancreatic cancer [221].
The direct financial costs are, however, not the only costs of screening. One also has to consider the anxiety generated and the real risk of harm caused by false positive results. False positives can lead to additional testing and sometimes even to unnecessary invasive procedures. Unneeded biopsies may cause bleeding or pancreatitis in a minority of cases, while surgical overtreatment of low-risk precancerous lesions can lead to substantial recovery times, post-operative complications, and lifelong insulin dependence.
17. Actual results from screening
We have described the basis for screening and theories on how to improve the odds of screening, but what about real-life efforts to screen for pancreatic cancer? As shown in Table 5, there have been many studies targeting at-risk populations using different technologies [120–126]. Most studies employed combinations of imaging modalities, including endoscopic ultrasound, magnetic resonance imaging, and computed tomography. Imaging modalities are often alternated during sequential screening rounds, as each modality has its own advantages and disadvantages. For example, endoscopic ultrasound is a more sensitive and specific than CT, particularly for early disease, but requires an invasive approach [222]. CT, on the other hand, exposes the individual being screened to radiation.
Table 5:
Screening studies that included at least 100 study participants
| First author | Year | Total number of participants | Primary mode of screening | Number of PDACs | Surgically resected PDACS | Surgeries for high-grade dysplasia | Surgeries for benign/low-risk lesions | Reference |
|---|---|---|---|---|---|---|---|---|
| Al-Sukhni | 2012 | 262 | MRI | 3 | 1 | 0 | 2 | [123] |
| Canto | 2018 | 354 | EUS, CT, MRI | 14 | 10 | 10 | 23 | [120] |
| Dbouk | 2022 | 1461 | EUS, CT, MRI | 10 | 8 | 3 | 5 | [9] Overlap with Canto (2018) |
| Harinck | 2016 | 139 | EUS, MRI | 1 | 1 | 0 | 1 | [131] |
| Klatte | 2022 | 347 | MRI, EUS | 36 | 27 | 0 | 8 | [122] Overlap with Vasen (2016) |
| Konings | 2017 | 186 | MRI, EUS | 2 | 1 | 0 | 2 | [223] |
| Lachter | 2018 | 123 | EUS | 1 | 1 | 0 | 0 | [224] |
| Ludwig | 2011 | 109 | MRI, EUS | 1 | 1 | 1 | 4 | [225] |
| Overbeek | 2022 | 366 | MRI, EUS | 10 | 6 | 0 | 8 | [121] |
| Paiella | 2019 | 187 | MRI, EUS | 5 | 2 | 0 | 2 | [226] |
| Vasen | 2016 | 411 | MRI, EUS | 15 | 11 | 4 | 14 | [125] |
| Zubarik | 2011 | 546 | CA19–9, EUS | 1 | 1 | 0 | 1 | [124] |
CT=computed tomography, EUS=endoscopic ultrasound, MRI= magnetic resonance imaging, PDAC= pancreatic ductal adenocarcinoma
Table 5 describes selected studies of pancreatic cancer screening that included at least 100 subjects. In these studies, surgically resectable pancreatic cancers were detected by screening in a minority of participants. Many, though not all, of these screen-detected cancers were low stage [120–126,223,225,226]. For example, Dbouk and colleagues enrolled 1,461 high-risk individuals in the Cancer of the Pancreas Screening (CAPS) program. Seven of the 10 patients who were diagnosed with pancreatic cancer in this study had stage I disease, and three individuals with imaging abnormalities resulting from high-grade dysplasia were also successfully detected [9]. Similarly, Klatte and colleagues, detected 36 pancreatic cancers when they screened 347 individuals with a germline pathogenic variant in the CDKN2A gene [122]. In their study, 83% of the cancers were considered resectable at the time of imaging, and one-third were Stage I [122]. These and similar studies have shown that potentially curable cancers and high-risk precancerous lesions can be identified in highly-selected cohorts.
These screening programs, however, came at the cost of resecting a number of benign or low-risk lesions, thereby exposing those patients to an unnecessary major surgical procedure. Unnecessary surgery represents the most substantial documented harm of pancreatic cancer screening, and to date this harm has not been entirely preventable in practice. The Dbouk et al study, for example, reported that five patients underwent surgery for concerning imaging findings that turned out to be related to low-grade PanIN and intraductal papillary mucinous neoplasms. Although none of these patients had a significant surgical complication, the risk of low-grade dysplasia progressing to pancreatic cancer is quite low, so it’s debatable whether these five patients benefitted from having their pancreas resection. In the Klatte et al study, seven patients underwent pancreaticoduodenectomy or distal pancreatectomy for benign low-risk lesions. All of these patients were alive at follow up. Studies of low-risk pancreatic cyst surveillance have also reported significant rates of overtreatment. Tamburrino and colleagues found that in a cohort of 961 patients receiving surveillance for a cyst, 40 were surgically overtreated while only 16 were classified as correctly treated [227]. Although immediate surgical complications have been rare, these procedures can be life-altering for patients. Many develop diabetes, some develop difficult to control diabetes, and many require substantial recovery time [228]. Any widely-adopted screening program would need to minimize such adverse events.
As noted earlier, individuals with screen-detected pancreatic cancers live much longer than individuals whose cancers are detected because they developed symptoms [9]. However, because pancreatic cancer screening studies have not used randomized controlled methodology, positive results are subject to the possibility of lead-time and length-time biases potentially inflating the apparent survival benefit [229]. But the accumulating results of surveillance using imaging (EUS/MRI) techniques demonstrate that, in the right populations, the screening of asymptomatic high-risk individuals is possible and will detect some surgically resectable early stage cancers, likely leading to improved survival for those individuals.
18. Looking forward
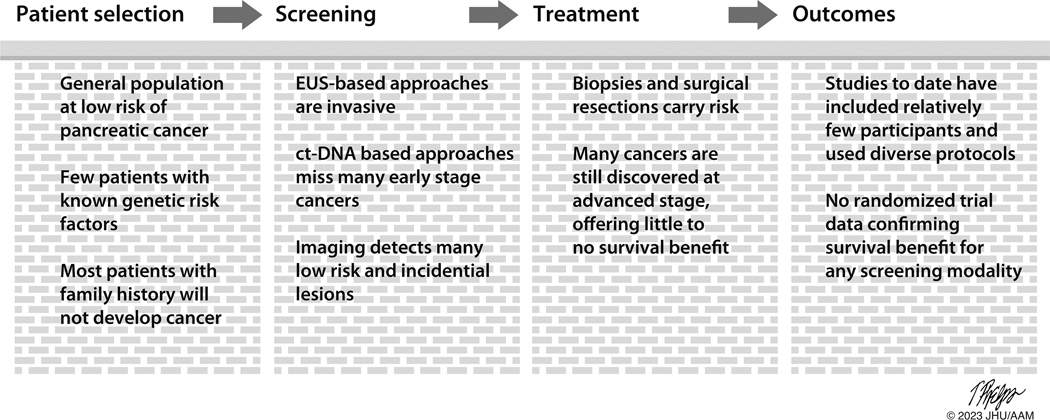
Screening for pancreatic cancer presents enormous opportunities, and yet it also poses enormous challenges. Advanced pancreatic cancers are deadly, with little hope for cure. Yet high-grade precancerous lesions and low-stage invasive cancers can be cured. The practical challenges to finding and treating these curable lesions are substantial (Figure 3). Pancreatic cancer is a rare disease in the overall population, and the organ lies deep in the abdomen. False positive results from screening the general population will be too common and can produce real patient harm. The challenge, then, is to develop a screening program that minimizes patient harm, maximizes patient benefit, and does so at a reasonable cost to society [159].
Figure 3:
Barriers to screening for pancreatic cancer.
One can easily envision that in the not too distant future, AI-based algorithms will be integrated into electronic patient medical records, looking for family history, germline gene status, smoking, obesity, and new onset diabetes [230]. These systems could alert health care providers when a patient appears to be at risk for pancreatic cancer. In parallel, AI-based algorithms will be quietly running in the background of radiology practices, analyzing the many abdominal CT scans that are performed every day. These algorithms will look for subtle changes in the pancreas, unseen by the radiologist, which could alert practitioners of the patient’s cancer risk. In addition, advances in non-invasive DNA-based screening tests will improve on the sensitivity and specificity of existing ctDNA approaches, increasing the yield of low-stage cancers and decreasing false positives. New protein and glycoprotein-based markers of pancreatic cancer will also continue to be discovered, enhancing the arsenal of biomarkers available for screening programs. No single marker will be sufficient; instead, a combination of markers, used in the right setting, will provide the sensitivity and specificity needed to screen for pancreatic cancer in a cost-effective manner.
As we move forward, our zeal to help those at-risk for developing pancreatic cancer must always be balanced with an understanding of the costs of screening. These costs not only include financial burdens to the medical system and patients, but also the psychological and medical harms of false positive results. Surgical overtreatment of low-risk precursor lesions will especially need to be minimized. Exciting advances should also, whenever possible, be subjected to the rigor of randomized controlled trials to prove their benefit in real-world populations.
AI-based applications will present their own challenges. These include potential legal and privacy issues when using data obtained for other clinical indications. Patients should be given the opportunity to provide informed consent before being subjected to any screening program, especially one using a novel modality such as artificial intelligence. Patients and clinicians alike may feel distrust or discomfort when it comes to applying an opaque third-party algorithm to intimate personal health data. The use of ancillary AI testing could also increase patient costs in a fee-for-service payment system.
19. Conclusion
At first glance, pancreatic cancer seems to be an ideal target for screening and early detection. It is a highly lethal cancer, and most patients do not develop signs and symptoms until late in the course of their illness. A variety of methods have been developed to screen for early disease. From blood CA19–9 levels to circulating tumor DNA, each method has advantages and disadvantages, and it is likely that the best approach will incorporate multiple complementary modalities. It is also likely that artificial intelligence, whether applied to electronic patient medical records or to image analysis, will play a role in identifying the highest risk individuals. Screening for pancreatic cancer, however, must take into account the rarity of the disease in the general population, the costs of false positive test results, and the real potential for overtreatment of low-risk lesions.
20. Expert Opinion
If we wait until patients develop symptoms from pancreatic cancer, most will die within months of their diagnosis. There are, however, a number of potential approaches to the earlier diagnosis of pancreatic cancer and its precursors in asymptomatic individuals. The unresolved challenge is that none of the existing approaches have the sensitivity and specificity needed to effectively screen the general population. While combining tests can improve sensitivities, even these combinations fall short. Existing approaches to screening, therefore, have to be focused on populations at greatest risk of the disease. Towards this end, screening efforts to date have targeted individuals with a strong family history of pancreatic cancer or a known pathogenic germline variant that predisposes to pancreatic cancer. Other groups with a heightened cancer risk that have not yet been clinically targeted include cigarette smokers, obese individuals, and adults with new onset diabetes mellitus. Screening programs that target high-risk groups and utilize imaging modalities like magnetic resonance imaging and endoscopic ultrasound have generated promising results. These programs, however, have thus far been relatively expensive and limited in scope.
Artificial intelligence (AI) has the potential to be a significant advance. AI-based algorithms can be applied to medical records and to imaging. AI-based algorithms can be trained to scour electronic patient medical records and imaging scans for subtle patterns, which may not be recognizable to the human eye, yet that indicate an increased risk of pancreatic cancer. Clinicians could then be alerted and even guided on the best ways to evaluate their patients further. AI-based algorithms have already been built that can evaluate computed tomography (CT) and other scans for subtle changes caused by low-stage, curable, pancreatic neoplasms. These algorithms could quietly run in the background, evaluate the millions of CT and other scans that are already being performed for other indications, and have the potential to identify changes in the pancreas that radiologists could easily miss in their day to day practice. Again, the program could alert the radiologist to look more closely at the images of the pancreas, and even suggest the best next steps to evaluate the subtle AI-detected changes. This approach has the advantage of not requiring additional testing until an abnormality is discovered. But enthusiasm about AI technologies must be tempered by the yet unproven real-world benefit and privacy concerns of these tools.
The growing emphasis on early detection of pancreatic cancer and its precursors should be balanced with a consideration of the costs and harms of screening. Costs are incurred during acquisition of the screening cohort, initial testing, confirmatory imaging and biopsies, surgery and other treatments, and post-treatment follow up and care. Screening can also cause serious patient harm. This harm includes the psychological stresses that individuals will feel when they are told that they are at increased risk, the worries from an initial result, and the medical complications that can occur from unnecessary biopsies and surgeries. The pancreas lies deep in the abdomen, and surgical resection of the head of the gland (the pancreatoduodenectomy) is associated with a 1–2% operative mortality rate and significant morbidity. If this surgery is done for a false positive screening test or low-risk precursor lesion, we have done significant harm to the individual.
As we look forward to the next five to ten years, we hope that new technical advances will improve the sensitivity and specificity of screening for pancreatic cancer. We also anticipate a better understanding of who is at risk, and of which groups will benefit most from screening. The development of new AI-based tools might improve screening approaches and help refine the identification of at-risk populations. The challenge, then, will be to fulfill these goals in a cost-effective manner that minimizes patient harm and to prove, prospectively and rigorously, the real-world value of these interventions before widespread implementation.
Article highlights:
Most patients with pancreatic cancer present with advanced, incurable disease.
Early detection and treatment offer the best hope for cure.
A variety of approaches, from novel blood tests to artificial intelligence driven algorithms to interpret CAT scans, offer hope for improved early detection.
All early detection approaches, however, come with added costs and potential harms from false positives.
New technical advances are needed to improve the sensitivity and specificity of screening for pancreatic cancer.
Funding
This article was supported by (U01210170, R01CA176828 CA62924), Susan Wojcicki and Dennis Troper, and by a Stand Up To Cancer-Lustgarten Foundation Pancreatic Cancer Interception Translational Cancer Research Grant (Grant Number: SU2C-AACR-DT25-17). Stand Up To Cancer is a program of the Entertainment Industry Foundation. SU2C research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. Sol Goldman Pancreatic Cancer Research Center, Rolfe Pancreatic Cancer Foundation.
Footnotes
Declaration of interest
Under a license agreement between Thrive Earlier Detection Corp., a subsidiary of Exact Sciences Corp. and the Johns Hopkins University, Dr. Lennon and the University are entitled to royalty distributions related to a technology discussed in this publication. This agreement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict-of-interest policies. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewers Disclosure
Peer reviewers on this manuscript have no relevant financial relationships or otherwise to disclose.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.NORC. Only 14% of cancers are detected through a preventive screening test. https://cancerdetection.norc.org/, (2022).
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin, 72(1), 7–33 (2022). [DOI] [PubMed] [Google Scholar]
- 3. Blackford AL, Canto MI, Klein AP, Hruban RH, Goggins M. Recent trends in the incidence and survival of Stage 1A Pancreatic Cancer: A Surveillance, Epidemiology, and End Results analysis. J Natl Cancer Inst, 112(11), 1162–1169(2020). *This study suggests that low-stage pancreatic cancers are being detected.
- 4.Assarzadegan N, Thompson E, Salimian K et al. Pathology of intraductal papillary mucinous neoplasms. Langenbecks Arch Surg, 406(8); 2643–2655 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hur C, Tramontano AC, Dowling EC et al. Early Pancreatic Ductal Adenocarcinoma Survival Is Dependent on Size: Positive Implications for Future Targeted Screening. Pancreas, 45(7), 1062–1066 (2016). *Patients with low-stage cancers have better survival.
- 6.Marchegiani G, Andrianello S, Malleo G et al. Does Size Matter in Pancreatic Cancer?: Reappraisal of Tumour Dimension as a Predictor of Outcome Beyond the TNM. Ann Surg, 266(1), 142–148 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Leonhardt CS, Kinny-Koster B, Hank T et al. Resected Early-Onset Pancreatic Cancer: Practices and Outcomes in an International Dual-Center Study. Ann Surg Oncol, 30(4):2433–2443 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda Y, Saiura A, Takahashi Y et al. Asymptomatic Pancreatic Cancer: Does Incidental Detection Impact Long-Term Outcomes? J Gastrointest Surg, 21(8), 1287–1295 (2017). [DOI] [PubMed] [Google Scholar]
- 9. Dbouk M, Katona BW, Brand RE et al. The Multicenter Cancer of Pancreas Screening Study: Impact on Stage and Survival. J Clin Oncol, 40(28), 3257–3266 (2022). **Patients with scree-detected pancreatic cancers live longer.
- 10.Lennon AM, Wolfgang CL, Canto MI et al. The early detection of pancreatic cancer: what will it take to diagnose and treat curable pancreatic neoplasia? Cancer Res, 74(13), 3381–3389 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd LNC, Ali M, Leeflang MMG et al. Diagnostic accuracy and added value of blood-based protein biomarkers for pancreatic cancer: A meta-analysis of aggregate and individual participant data. EClinicalMedicine, 55, 101747 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santucci N, Facy O, Ortega-Deballon P, Lequeu JB, Rat P, Rat P. CA 19–9 predicts resectability of pancreatic cancer even in jaundiced patients. Pancreatology, 18(6), 666–670 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Colloca GA, Venturino A, Guarneri D. Tumor growth kinetics by CA 19–9 in patients with unresectable pancreatic cancer receiving chemotherapy: A retrospective analysis. Pancreatology, 20(6), 1189–1194 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Berger AC, Garcia M Jr., Hoffman JP et al. Postresection CA 19–9 predicts overall survival in patients with pancreatic cancer treated with adjuvant chemoradiation: a prospective validation by RTOG 9704. J Clin Oncol, 26(36), 5918–5922 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugiura T, Uesaka K, Kanemoto H et al. Serum CA19–9 is a significant predictor among preoperative parameters for early recurrence after resection of pancreatic adenocarcinoma. J Gastrointest Surg, 16(5), 977–985 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Tzeng CW, Balachandran A, Ahmad M et al. Serum carbohydrate antigen 19–9 represents a marker of response to neoadjuvant therapy in patients with borderline resectable pancreatic cancer. HPB (Oxford), 16(5), 430–438 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrone CR, Finkelstein DM, Thayer SP, Muzikansky A, Fernandez-delCastillo C, Warshaw AL. Perioperative CA19–9 levels can predict stage and survival in patients with resectable pancreatic adenocarcinoma. J Clin Oncol, 24(18), 2897–2902 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Park BK, Seo JH et al. Carbohydrate antigen 19–9 elevation without evidence of malignant or pancreatobiliary diseases. Sci Rep, 10(1), 8820 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abe T, Koi C, Kohi S et al. Gene Variants That Affect Levels of Circulating Tumor Markers Increase Identification of Patients With Pancreatic Cancer. Clin Gastroenterol Hepatol, 18(5), 1161–1169 e1165 (2020). *Germline genetics can be used to inform normal ranges of blood markers.
- 20.Guo M, Luo G, Lu R et al. Distribution of Lewis and Secretor polymorphisms and corresponding CA19–9 antigen expression in a Chinese population. FEBS Open Bio, 7(11), 1660–1671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mark K.Fung BJG, Hillyer Christopher D., Westhoff Connie M.. Technical Manual, 18th edition. American Association of Blood Banks, Bethesda, MD: (2014). [Google Scholar]
- 22.Luo G, Fan Z, Cheng H et al. New observations on the utility of CA19–9 as a biomarker in Lewis negative patients with pancreatic cancer. Pancreatology, 18(8), 971–976 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Zhou W, Sokoll LJ, Bruzek DJ et al. Identifying markers for pancreatic cancer by gene expression analysis. Cancer Epidemiol Biomarkers Prev, 7(2), 109–112 (1998). [PubMed] [Google Scholar]
- 24. Cao L, Huang C, Cui Zhou D et al. Proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell, 184(19), 5031–5052 e5026 (2021). The largest study of proteins expressed in pancreatic cancer.
- 25.Argani P, Rosty C, Reiter RE et al. Discovery of new markers of cancer through serial analysis of gene expression: prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res, 61(11), 4320–4324 (2001). [PubMed] [Google Scholar]
- 26.Argani P, Iacobuzio-Donahue C, Ryu B et al. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE). Clin Cancer Res, 7(12), 3862–3868 (2001). [PubMed] [Google Scholar]
- 27.Jahan R, Ganguly K, Smith LM et al. Trefoil factor(s) and CA19.9: A promising panel for early detection of pancreatic cancer. EBioMedicine, 42, 375–385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkinson C, Elliott V, Menon U et al. Evaluation in pre-diagnosis samples discounts ICAM-1 and TIMP-1 as biomarkers for earlier diagnosis of pancreatic cancer. J Proteomics, 113, 400–402 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Capello M, Bantis LE, Scelo G et al. Sequential Validation of Blood-Based Protein Biomarker Candidates for Early-Stage Pancreatic Cancer. J Natl Cancer Inst, 109(4) djw266, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato Y, Kobayashi T, Nishiumi S et al. Prospective Study Using Plasma Apolipoprotein A2-Isoforms to Screen for High-Risk Status of Pancreatic Cancer. Cancers (Basel), 12(9): 2625(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honda K, Katzke VA, Husing A et al. CA19–9 and apolipoprotein-A2 isoforms as detection markers for pancreatic cancer: a prospective evaluation. Int J Cancer, 144(8), 1877–1887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng Q, Shi S, Liang C et al. Diagnostic Accuracy of a CA125-Based Biomarker Panel in Patients with Pancreatic Cancer: A Systematic Review and Meta-Analysis. J Cancer, 8(17), 3615–3622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cohen JD, Li L, Wang Y et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science, 359(6378), 926–930 (2018). *Study from the Vogelstein group highlighting the usefulness of combining ctDNA and other blood based biomarkers for the detrcetion of cancer.
- 34. Tanaka H, Tamura K, Abe T et al. Serum Carboxypeptidase Activity and Genotype-Stratified CA19–9 to Detect Early-Stage Pancreatic Cancer. Clin Gastroenterol Hepatol, 20(10), 2267–2275 e2262 (2022). *Germline genetics can be used to inform normal ranges of blood markers.
- 35.Ge L, Pan B, Song F et al. Comparing the diagnostic accuracy of five common tumour biomarkers and CA19–9 for pancreatic cancer: a protocol for a network meta-analysis of diagnostic test accuracy. BMJ Open, 7(12), e018175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fahrmann JF, Schmidt CM, Mao X et al. Lead-Time Trajectory of CA19–9 as an Anchor Marker for Pancreatic Cancer Early Detection. Gastroenterology, 160(4), 1373–1383 e1376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Manen L, Groen JV, Putter H et al. Elevated CEA and CA19–9 serum levels independently predict advanced pancreatic cancer at diagnosis. Biomarkers, 25(2), 186–193 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Cohen JD, Javed AA, Thoburn C et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci U S A, 114(38), 10202–10207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omiya K, Oba A, Inoue Y et al. Serum DUPAN-2 could be an Alternative Biological Marker for CA19–9 Non-secretors with Pancreatic Cancer. Ann Surg, Jan 25 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Song J, Sokoll LJ, Pasay JJ et al. Identification of Serum Biomarker Panels for the Early Detection of Pancreatic Cancer. Cancer Epidemiol Biomarkers Prev, 28(1), 174–182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koopmann J, Buckhaults P, Brown DA et al. Serum macrophage inhibitory cytokine 1 as a marker of pancreatic and other periampullary cancers. Clin Cancer Res, 10(7), 2386–2392 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Park J, Choi Y, Namkung J et al. Diagnostic performance enhancement of pancreatic cancer using proteomic multimarker panel. Oncotarget, 8(54), 93117–93130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fahrmann JF, Bantis LE, Capello M et al. A Plasma-Derived Protein-Metabolite Multiplexed Panel for Early-Stage Pancreatic Cancer. J Natl Cancer Inst, 111(4), 372–379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaur S, Smith LM, Patel A et al. A Combination of MUC5AC and CA19–9 Improves the Diagnosis of Pancreatic Cancer: A Multicenter Study. Am J Gastroenterol, 112(1), 172–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koopmann J, Fedarko NS, Jain A et al. Evaluation of osteopontin as biomarker for pancreatic adenocarcinoma. Cancer Epidemiol Biomarkers Prev, 13(3), 487–491 (2004). [PubMed] [Google Scholar]
- 46.Rychlikova J, Vecka M, Jachymova M et al. Osteopontin as a discriminating marker for pancreatic cancer and chronic pancreatitis. Cancer Biomark, 17(1), 55–65 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Peng HY, Chang MC, Hu CM, Yang HI, Lee WH, Chang YT. Thrombospondin-2 is a Highly Specific Diagnostic Marker and is Associated with Prognosis in Pancreatic Cancer. Ann Surg Oncol, 26(3), 807–814 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Kim J, Bamlet WR, Oberg AL et al. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19–9 blood markers. Sci Transl Med, 9(398) eaah5583(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Udgata S, Takenaka N, Bamlet WR et al. THBS2/CA19–9 Detecting Pancreatic Ductal Adenocarcinoma at Diagnosis Underperforms in Prediagnostic Detection: Implications for Biomarker Advancement. Cancer Prev Res (Phila), 14(2), 223–232 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slater EP, Fendrich V, Strauch K et al. LCN2 and TIMP1 as Potential Serum Markers for the Early Detection of Familial Pancreatic Cancer. Transl Oncol, 6(2), 99–103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL et al. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol, 160(4), 1239–1249 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Network. CGAR. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell, 32(2), 185–203 e113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas JK, Kim MS, Balakrishnan L et al. Pancreatic Cancer Database: an integrative resource for pancreatic cancer. Cancer Biol Ther, 15(8), 963–967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gronborg M, Kristiansen TZ, Iwahori A et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics, 5(1), 157–171 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Laura E.Kane GSM, Mylod Eimear, O’Brien Rebecca M., O’Connell Fiona, , Buckley Croí E., Arlow Jennifer, Nguyen Khanh, Mockler David, Meade Aidan D., Ryan Barbara M., and Maher Stephen G. Diagnostic Accuracy of Blood-based updates Biomarkers for Pancreatic Cancer: A Systematic Review and Meta-analysis. Cancer Research Communications, 2(10), 1229–1243 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawai S, Oba-Shinjo SM, Ito LS, Uno M, Marie SKN, Hamajima N. Factors associated with serum CA19–9 levels among healthy children: a cross-sectional study. BMC Clin Pathol, 12, 23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mayers JR, Wu C, Clish CB et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med, 20(10), 1193–1198 (2014). *One of the first reports of the utility of branched-chain amino acids.
- 58.Rossi M, Turati F, Strikoudi P et al. Dietary intake of branched-chain amino acids and pancreatic cancer risk in a case-control study from Italy. Br J Nutr, 1–19 (2022). [DOI] [PubMed] [Google Scholar]
- 59.Li JY, Sun F, Yang CL et al. GEO data mining and TCGA analysis reveal altered branched chain amino acid metabolism in pancreatic cancer patients. Aging (Albany NY), 13(8), 11907–11918 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JH, Cho YR, Kim JH et al. Branched-chain amino acids sustain pancreatic cancer growth by regulating lipid metabolism. Exp Mol Med, 51(11), 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li JT, Yin M, Wang D et al. BCAT2-mediated BCAA catabolism is critical for development of pancreatic ductal adenocarcinoma. Nat Cell Biol, 22(2), 167–174 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Katagiri R, Goto A, Nakagawa T et al. Increased Levels of Branched-Chain Amino Acid Associated With Increased Risk of Pancreatic Cancer in a Prospective Case-Control Study of a Large Cohort. Gastroenterology, 155(5), 1474–1482 e1471 (2018). [DOI] [PubMed] [Google Scholar]
- 63. Mahajan UM, Oehrle B, Sirtl S et al. Independent Validation and Assay Standardization of Improved Metabolic Biomarker Signature to Differentiate Pancreatic Ductal Adenocarcinoma From Chronic Pancreatitis. Gastroenterology, 163(5), 1407–1422 (2022). *Metabolites may also be useful markers.
- 64.Kern SE. Why your new cancer biomarker may never work: recurrent patterns and remarkable diversity in biomarker failures. Cancer Res, 72(23), 6097–6101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science, 367(6478) eaau6977(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao Y, Bado I, Wang H, Zhang W, Rosen JM, Zhang XH. Metastasis Organotropism: Redefining the Congenial Soil. Dev Cell, 49(3), 375–391 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nakamura K, Zhu Z, Roy S et al. An Exosome-based Transcriptomic Signature for Noninvasive, Early Detection of Patients With Pancreatic Ductal Adenocarcinoma: A Multicenter Cohort Study. Gastroenterology, 163(5), 1252–1266 e1252 (2022). *Exosomes are a hot area for early detection.
- 68.Xu YF, Hannafon BN, Zhao YD, Postier RG, Ding WQ. Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget, 8(44), 77028–77040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romano R, Picca A, Eusebi LHU et al. Extracellular Vesicles and Pancreatic Cancer: Insights on the Roles of miRNA, lncRNA, and Protein Cargos in Cancer Progression. Cells, 10(6)1361 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang S, Che SP, Kurywchak P et al. Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol Ther, 18(3), 158–165 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiao D, Dong Z, Zhen L et al. Combined Exosomal GPC1, CD82, and Serum CA19–9 as Multiplex Targets: A Specific, Sensitive, and Reproducible Detection Panel for the Diagnosis of Pancreatic Cancer. Mol Cancer Res, 18(2), 300–310 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Jones S, Zhang X, Parsons DW et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science, 321(5897), 1801–1806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Biankin AV, Waddell N, Kassahn KS et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature, 491(7424), 399–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waddell N, Pajic M, Patch AM et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature, 518(7540), 495–501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bettegowda C, Sausen M, Leary RJ et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med, 6(224), 224ra224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mouliere F, Chandrananda D, Piskorz AM et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med, 10(466) eaat4921(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Phallen J, Sausen M, Adleff V et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med, 9(403): eaan2415 (2017). **Highlights the potential of DNA fragmentation patterns.
- 78.Moss J, Magenheim J, Neiman D et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun, 9(1), 5068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cristiano S, Leal A, Phallen J et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature, 570(7761), 385–389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Macgregor-Das A, Yu J, Tamura K et al. Detection of Circulating Tumor DNA in Patients with Pancreatic Cancer Using Digital Next-Generation Sequencing. J Mol Diagn, 22(6), 748–756 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Le Calvez-Kelm F, Foll M, Wozniak MB et al. KRAS mutations in blood circulating cell-free DNA: a pancreatic cancer case-control. Oncotarget, 7(48), 78827–78840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allenson K, Castillo J, San Lucas FA et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol, 28(4), 741–747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Omura N, Li CP, Li A et al. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol Ther, 7(7), 1146–1156 (2008). *Abnormal patterns of methylation as a target for early detection.
- 84.Tan AC, Jimeno A, Lin SH et al. Characterizing DNA methylation patterns in pancreatic cancer genome. Mol Oncol, 3(5–6), 425–438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vincent A, Omura N, Hong SM, Jaffe A, Eshleman J, Goggins M. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res, 17(13), 4341–4354 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yi JM, Guzzetta AA, Bailey VJ et al. Novel methylation biomarker panel for the early detection of pancreatic cancer. Clin Cancer Res, 19(23), 6544–6555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ying L, Sharma A, Chhoda A et al. Methylation-based Cell-free DNA Signature for Early Detection of Pancreatic Cancer. Pancreas, 50(9), 1267–1273 (2021). [DOI] [PubMed] [Google Scholar]
- 88.Kandimalla R, Xu J, Link A et al. EpiPanGI Dx: A Cell-free DNA Methylation Fingerprint for the Early Detection of Gastrointestinal Cancers. Clin Cancer Res, 27(22), 6135–6144 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jamshidi A, Liu MC, Klein EA et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell, 40(12), 1537–1549 e1512 (2022). **A comparison of different DNA-based blood tests.
- 90.https://www.businesswire.com/news/home/20220911005035/en/. CORRECTING and REPLACING GRAIL Announces Final Results From the PATHFINDER Multi-Cancer Early Detection Screening Study at ESMO Congress 2022. (2022).
- 91.Neal RD, Johnson P, Clarke CA et al. Cell-Free DNA-Based Multi-Cancer Early Detection Test in an Asymptomatic Screening Population (NHS-Galleri): Design of a Pragmatic, Prospective Randomised Controlled Trial. Cancers (Basel), 14(19):4818 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lennon AM, Buchanan AH, Kinde I et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science, 369(6499):EABB9601 (2020). *The Thrive Earlier Detection company’s study using a combination of markers.
- 93.Nassar FJ, Msheik ZS, Nasr RR, Temraz SN. Methylated circulating tumor DNA as a biomarker for colorectal cancer diagnosis, prognosis, and prediction. Clin Epigenetics, 13(1), 111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yu J, Sadakari Y, Shindo K et al. Digital next-generation sequencing identifies low-abundance mutations in pancreatic juice samples collected from the duodenum of patients with pancreatic cancer and intraductal papillary mucinous neoplasms. Gut, 66(9), 1677–1687 (2017). **Obtaining biosamples closer to the source may improve markers.
- 95.Suenaga M, Yu J, Shindo K et al. Pancreatic Juice Mutation Concentrations Can Help Predict the Grade of Dysplasia in Patients Undergoing Pancreatic Surveillance. Clin Cancer Res, 24(12), 2963–2974 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Majumder S, Raimondo M, Taylor WR et al. Methylated DNA in Pancreatic Juice Distinguishes Patients With Pancreatic Cancer From Controls. Clin Gastroenterol Hepatol, 18(3), 676–683 e673 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kisiel JB, Yab TC, Taylor WR et al. Stool DNA testing for the detection of pancreatic cancer: assessment of methylation marker candidates. Cancer, 118(10), 2623–2631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sjostrom M, Zhao SG, Levy S et al. The 5-Hydroxymethylcytosine Landscape of Prostate Cancer. Cancer Res, 82(21), 3888–3902 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guler GD, Ning Y, Ku CJ et al. Detection of early stage pancreatic cancer using 5-hydroxymethylcytosine signatures in circulating cell free DNA. Nat Commun, 11(1), 5270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mathios D, Johansen JS, Cristiano S et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat Commun, 12(1), 5060 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Foda ZH, Annapragada AV, Boyapati K et al. Detecting liver cancer using cell-free DNA fragmentomes. Cancer Discov, 13(3):616–631 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van ‘t Erve I, Medina JE, Leal A et al. Metastatic colorectal cancer treatment response evaluation by ultra-deep sequencing of cell-free DNA and matched white blood cells. Clin Cancer Res, 29(5):899–909(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stackpole ML, Zeng W, Li S et al. Cost-effective methylome sequencing of cell-free DNA for accurately detecting and locating cancer. Nat Commun, 13(1), 5566 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li W, Li Q, Kang S et al. CancerDetector: ultrasensitive and non-invasive cancer detection at the resolution of individual reads using cell-free DNA methylation sequencing data. Nucleic Acids Res, 46(15), e89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kang S, Li Q, Chen Q et al. CancerLocator: non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biol, 18(1), 53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li BT, Janku F, Jung B et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol, 30(4), 597–603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Razavi P, Li BT, Brown DN et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med, 25(12), 1928–1937 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, Consortium C. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol, 31(6), 745–759 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu H, Guo S, Liu X et al. Noninvasive detection of pancreatic ductal adenocarcinoma using the methylation signature of circulating tumour DNA. BMC Med, 20(1), 458 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen X, Gole J, Gore A et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat Commun, 11(1), 3475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mo S, Dai W, Wang H et al. Early detection and prognosis prediction for colorectal cancer by circulating tumour DNA methylation haplotypes: A multicentre cohort study. EClinicalMedicine, 55, 101717 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cai G, Cai M, Feng Z et al. A Multilocus Blood-Based Assay Targeting Circulating Tumor DNA Methylation Enables Early Detection and Early Relapse Prediction of Colorectal Cancer. Gastroenterology, 161(6), 2053–2056 e2052 (2021). [DOI] [PubMed] [Google Scholar]
- 113.Jia Z, Wang Y, Xue J et al. DNA methylation patterns at and beyond the histological margin of early-stage invasive lung adenocarcinoma radiologically manifested as pure ground-glass opacity. Clin Epigenetics, 13(1), 153 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Dumstrei K, Chen H, Brenner H. A systematic review of serum autoantibodies as biomarkers for pancreatic cancer detection. Oncotarget, 7(10), 11151–11164 (2016). *Autoantibodies have not proven particularly useful.
- 115.Nagayoshi Y, Nakamura M, Matsuoka K et al. Profiling of autoantibodies in sera of pancreatic cancer patients. Ann Surg Oncol, 21 Suppl 3, S459–465 (2014). [DOI] [PubMed] [Google Scholar]
- 116.Bracci PM, Zhou M, Young S, Wiemels J. Serum autoantibodies to pancreatic cancer antigens as biomarkers of pancreatic cancer in a San Francisco Bay Area case-control study. Cancer, 118(21), 5384–5394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li J, Wang LJ, Ying X et al. Immunodiagnostic value of combined detection of autoantibodies to tumor-associated antigens as biomarkers in pancreatic cancer. Scand J Immunol, 75(3), 342–349 (2012). [DOI] [PubMed] [Google Scholar]
- 118.Toshima F, Watanabe R, Inoue D et al. CT Abnormalities of the Pancreas Associated With the Subsequent Diagnosis of Clinical Stage I Pancreatic Ductal Adenocarcinoma More Than 1 Year Later: A Case-Control Study. AJR Am J Roentgenol, 217(6), 1353–1364 (2021). [DOI] [PubMed] [Google Scholar]
- 119.Nakahodo J, Kikuyama M, Fukumura Y et al. Focal pancreatic parenchyma atrophy is a harbinger of pancreatic cancer and a clue to the intraductal spreading subtype. Pancreatology, 22(8), 1148–1158 (2022). [DOI] [PubMed] [Google Scholar]
- 120.Canto MI, Almario JA, Schulick RD et al. Risk of Neoplastic Progression in Individuals at High Risk for Pancreatic Cancer Undergoing Long-term Surveillance. Gastroenterology, 155(3), 740–751 e742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Overbeek KA, Levink IJM, Koopmann BDM et al. Long-term yield of pancreatic cancer surveillance in high-risk individuals. Gut, 71(6), 1152–1160 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Klatte DCF, Boekestijn B, Wasser M et al. Pancreatic Cancer Surveillance in Carriers of a Germline CDKN2A Pathogenic Variant: Yield and Outcomes of a 20-Year Prospective Follow-Up. J Clin Oncol, 40(28), 3267–3277 (2022). **Highlights the value of screening high-risk groups.
- 123. Al-Sukhni W, Borgida A, Rothenmund H et al. Screening for pancreatic cancer in a high-risk cohort: an eight-year experience. J Gastrointest Surg, 16(4), 771–783 (2012). **Highlights the value of screening high-risk groups.
- 124.Zubarik R, Gordon SR, Lidofsky SD et al. Screening for pancreatic cancer in a high-risk population with serum CA 19–9 and targeted EUS: a feasibility study. Gastrointest Endosc, 74(1), 87–95 (2011). [DOI] [PubMed] [Google Scholar]
- 125. Vasen H, Ibrahim I, Ponce CG et al. Benefit of Surveillance for Pancreatic Cancer in High-Risk Individuals: Outcome of Long-Term Prospective Follow-Up Studies From Three European Expert Centers. J Clin Oncol, 34(17), 2010–2019 (2016). **Highlights the value of screening high-risk groups.
- 126.Brune K, Abe T, Canto M et al. Multifocal neoplastic precursor lesions associated with lobular atrophy of the pancreas in patients having a strong family history of pancreatic cancer. Am J Surg Pathol, 30(9), 1067–1076 (2006). [PMC free article] [PubMed] [Google Scholar]
- 127.Singh DP, Sheedy S, Goenka AH et al. Computerized tomography scan in pre-diagnostic pancreatic ductal adenocarcinoma: Stages of progression and potential benefits of early intervention: A retrospective study. Pancreatology, 20(7), 1495–1501 (2020). [DOI] [PubMed] [Google Scholar]
- 128.Chen W, Butler RK, Zhou Y, Parker RA, Jeon CY, Wu BU. Prediction of Pancreatic Cancer Based on Imaging Features in Patients With Duct Abnormalities. Pancreas, 49(3), 413–419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gangi S, Fletcher JG, Nathan MA et al. Time interval between abnormalities seen on CT and the clinical diagnosis of pancreatic cancer: retrospective review of CT scans obtained before diagnosis. AJR Am J Roentgenol, 182(4), 897–903 (2004). [DOI] [PubMed] [Google Scholar]
- 130.Javed AA, Young RWC, Habib JR et al. Cinematic Rendering: Novel Tool for Improving Pancreatic Cancer Surgical Planning. Curr Probl Diagn Radiol, 51(6), 878–883 (2022). [DOI] [PubMed] [Google Scholar]
- 131. Harinck F, Konings IC, Kluijt I et al. A multicentre comparative prospective blinded analysis of EUS and MRI for screening of pancreatic cancer in high-risk individuals. Gut, 65(9), 1505–1513 (2016). **Comparison of EUS vs. MRI for screening.
- 132.Thomsen FSL, Horstmeier S, Niehoff JH, Pena JA, Borggrefe J. Effective Spatial Resolution of Photon Counting CT for Imaging of Trabecular Structures is Superior to Conventional Clinical CT and Similar to High Resolution Peripheral CT. Invest Radiol, 57(9), 620–626 (2022). [DOI] [PubMed] [Google Scholar]
- 133.Grunz JP, Heidenreich JF, Lennartz S et al. Spectral Shaping Via Tin Prefiltration in Ultra-High-Resolution Photon-Counting and Energy-Integrating Detector CT of the Temporal Bone. Invest Radiol, 57(12), 819–825 (2022). [DOI] [PubMed] [Google Scholar]
- 134. Chu LC, Park S, Kawamoto S et al. Utility of CT Radiomics Features in Differentiation of Pancreatic Ductal Adenocarcinoma From Normal Pancreatic Tissue. AJR Am J Roentgenol, 213(2), 349–357 (2019). **Comparison of CT vs. MRI for screening.
- 135.Ben Q, Xu M, Ning X et al. Diabetes mellitus and risk of pancreatic cancer: A meta-analysis of cohort studies. Eur J Cancer, 47(13), 1928–1937 (2011). [DOI] [PubMed] [Google Scholar]
- 136.Sharma A, Kandlakunta H, Nagpal SJS et al. Model to Determine Risk of Pancreatic Cancer in Patients With New-Onset Diabetes. Gastroenterology, 155(3), 730–739 e733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nagpal SJS, Kandlakunta H, Her T et al. Pancreatic ductal adenocarcinoma is associated with a unique endocrinopathy distinct from type 2 diabetes mellitus. Pancreatology, 20(5), 929–935 (2020). [DOI] [PubMed] [Google Scholar]
- 138.Chari ST, Leibson CL, Rabe KG, Ransom J, de Andrade M, Petersen GM. Probability of pancreatic cancer following diabetes: a population-based study. Gastroenterology, 129(2), 504–511 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Setiawan VW, Stram DO, Porcel J et al. Pancreatic Cancer Following Incident Diabetes in African Americans and Latinos: The Multiethnic Cohort. J Natl Cancer Inst, 111(1), 27–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hart PA, Andersen DK, Mather KJ et al. Evaluation of a Mixed Meal Test for Diagnosis and Characterization of PancrEaTogEniC DiabeTes Secondary to Pancreatic Cancer and Chronic Pancreatitis: Rationale and Methodology for the DETECT Study From the Consortium for the Study of Chronic Pancreatitis, Diabetes, and Pancreatic Cancer. Pancreas, 47(10), 1239–1243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pannala R, Basu A, Petersen GM, Chari ST. New-onset diabetes: a potential clue to the early diagnosis of pancreatic cancer. Lancet Oncol, 10(1), 88–95 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huang BZ, Pandol SJ, Jeon CY et al. New-Onset Diabetes, Longitudinal Trends in Metabolic Markers, and Risk of Pancreatic Cancer in a Heterogeneous Population. Clin Gastroenterol Hepatol, 18(8), 1812–1821 e1817 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Chari ST, Leibson CL, Rabe KG, Ransom J, de AM, Petersen GM. Probability of pancreatic cancer following diabetes: a population-based study. Gastroenterology, 129(2), 504–511 (2005). **Classic paper highlighting new onset diabetes as an indicator of pancreatic cancer in the elderly.
- 144.Sah RP, Sharma A, Nagpal S et al. Phases of Metabolic and Soft Tissue Changes in Months Preceding a Diagnosis of Pancreatic Ductal Adenocarcinoma. Gastroenterology, 156(6), 1742–1752 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Basile D, Parnofiello A, Vitale MG et al. The IMPACT study: early loss of skeletal muscle mass in advanced pancreatic cancer patients. J Cachexia Sarcopenia Muscle, 10(2), 368–377 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sugimoto M, Farnell MB, Nagorney DM et al. Decreased Skeletal Muscle Volume Is a Predictive Factor for Poorer Survival in Patients Undergoing Surgical Resection for Pancreatic Ductal Adenocarcinoma. J Gastrointest Surg, 22(5), 831–839 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kenner BJ. Early Detection of Pancreatic Cancer: The Role of Depression and Anxiety as a Precursor for Disease. Pancreas, 47(4), 363–367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Seoud T, Syed A, Carleton N et al. Depression Before and After a Diagnosis of Pancreatic Cancer: Results From a National, Population-Based Study. Pancreas, 49(8), 1117–1122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kenner BJ, Abrams ND, Chari ST et al. Early Detection of Pancreatic Cancer: Applying Artificial Intelligence to Electronic Health Records. Pancreas, 50(7), 916–922 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Baecker A, Kim S, Risch HA et al. Do changes in health reveal the possibility of undiagnosed pancreatic cancer? Development of a risk-prediction model based on healthcare claims data. PLoS One, 14(6), e0218580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Chu LC, Park S, Kawamoto S et al. Application of Deep Learning to Pancreatic Cancer Detection: Lessons Learned From Our Initial Experience. J Am Coll Radiol, 16(9 Pt B), 1338–1342 (2019). *Radiomics and artificial intelligence have great potential.
- 152.Park HJ, Shin K, You MW et al. Deep Learning-based Detection of Solid and Cystic Pancreatic Neoplasms at Contrast-enhanced CT. Radiology, 306(1), 140–149 (2023). [DOI] [PubMed] [Google Scholar]
- 153.Mukherjee S, Patra A, Khasawneh H et al. Radiomics-based Machine-learning Models Can Detect Pancreatic Cancer on Prediagnostic Computed Tomography Scans at a Substantial Lead Time Before Clinical Diagnosis. Gastroenterology, 163(5), 1435–1446 e1433 (2022). [DOI] [PubMed] [Google Scholar]
- 154.Chen PT, Wu T, Wang P et al. Pancreatic Cancer Detection on CT Scans with Deep Learning: A Nationwide Population-based Study. Radiology, 306(1), 172–182 (2023). [DOI] [PubMed] [Google Scholar]
- 155.Suman G, Panda A, Korfiatis P, Goenka AH. Convolutional neural network for the detection of pancreatic cancer on CT scans. Lancet Digit Health, 2(9), e453 (2020). [DOI] [PubMed] [Google Scholar]
- 156.Mettler FA Jr., Mahesh M, Bhargavan-Chatfield M et al. Patient Exposure from Radiologic and Nuclear Medicine Procedures in the United States: Procedure Volume and Effective Dose for the Period 2006–2016. Radiology, 295(2), 418–427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Thomas SP, Fraum TJ, Ngo L et al. Leveraging Artificial Intelligence to Enhance Peer Review: Missed Liver Lesions on Computed Tomographic Pulmonary Angiography. J Am Coll Radiol, 19(11), 1286–1294 (2022). [DOI] [PubMed] [Google Scholar]
- 158.SEER. Cancer stat facts: Pancreatic cancer. https://seer.cancer.gov/statfacts/html/pancreas.html, (2023).
- 159. Gray JA, Patnick J, Blanks RG. Maximising benefit and minimising harm of screening. BMJ, 336(7642), 480–483 (2008). **A wonderful paper that sets the bar for screening tests.
- 160.Singhi AD, Koay EJ, Chari ST, Maitra A. Early Detection of Pancreatic Cancer: Opportunities and Challenges. Gastroenterology, 156(7), 2024–2040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bosetti C, Lucenteforte E, Silverman DT et al. Cigarette smoking and pancreatic cancer: an analysis from the International Pancreatic Cancer Case-Control Consortium (Panc4). Ann Oncol, 23(7), 1880–1888 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Porter N, Laheru D, Lau B et al. Risk of Pancreatic Cancer in the Long-Term Prospective Follow-Up of Familial Pancreatic Cancer Kindreds. J Natl Cancer Inst, 114(12), 1681–1688 (2022). **Family history can be used to quantify the risk of pancreatic cancer.
- 163.Everhart J, Wright D. Diabetes mellitus as a risk factor for pancreatic cancer. A meta-analysis. JAMA, 273(20), 1605–1609 (1995). [PubMed] [Google Scholar]
- 164.Huxley R, Ansary-Moghaddam A, Berrington de Gonzalez A, Barzi F, Woodward M. Type-II diabetes and pancreatic cancer: a meta-analysis of 36 studies. Br J Cancer, 92(11), 2076–2083 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bosetti C, Rosato V, Li D et al. Diabetes, antidiabetic medications, and pancreatic cancer risk: an analysis from the International Pancreatic Cancer Case-Control Consortium. Ann Oncol, 25(10), 2065–2072 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Elena JW, Steplowski E, Yu K et al. Diabetes and risk of pancreatic cancer: a pooled analysis from the pancreatic cancer cohort consortium. Cancer Causes Control, 24(1), 13–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gupta S, Vittinghoff E, Bertenthal D et al. New-onset diabetes and pancreatic cancer. Clin Gastroenterol Hepatol, 4(11), 1366–1372(2006). [DOI] [PubMed] [Google Scholar]
- 168.Munigala S, Singh A, Gelrud A, Agarwal B. Predictors for Pancreatic Cancer Diagnosis Following New-Onset Diabetes Mellitus. Clin Transl Gastroenterol, 6(10), e118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Arslan AA, Helzlsouer KJ, Kooperberg C et al. Anthropometric measures, body mass index, and pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan). Arch Intern Med, 170(9), 791–802 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Roberts NJ, Norris AL, Petersen GM et al. Whole Genome Sequencing Defines the Genetic Heterogeneity of Familial Pancreatic Cancer. Cancer Discov, 6(2), 166–175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Roberts NJ, Jiao Y, Yu J et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov, 2(1), 41–46 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hsu FC, Roberts NJ, Childs E et al. Risk of Pancreatic Cancer Among Individuals With Pathogenic Variants in the ATM Gene. JAMA Oncol, 7(11), 1664–1668 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Thompson D, Easton DF. Cancer Incidence in BRCA1 mutation carriers. J.Natl.Cancer Inst, 94(18), 1358–1365 (2002). [DOI] [PubMed] [Google Scholar]
- 174.Mocci E, Milne RL, Mendez-Villamil EY et al. Risk of pancreatic cancer in breast cancer families from the breast cancer family registry. Cancer Epidemiol Biomarkers Prev, 22(5), 803–811 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hu C, Hart SN, Polley EC et al. Association Between Inherited Germline Mutations in Cancer Predisposition Genes and Risk of Pancreatic Cancer. JAMA, 319(23), 2401–2409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Li S, Silvestri V, Leslie G et al. Cancer Risks Associated With BRCA1 and BRCA2 Pathogenic Variants. J Clin Oncol, 40(14), 1529–1541 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Goggins M, Schutte M, Lu J et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res, 56(23), 5360–5364 (1996). [PubMed] [Google Scholar]
- 178.Holter S, Borgida A, Dodd A et al. Germline BRCA Mutations in a Large Clinic-Based Cohort of Patients With Pancreatic Adenocarcinoma. J Clin Oncol, 33(28), 3124–3129 (2015). [DOI] [PubMed] [Google Scholar]
- 179.Yurgelun MB, Chittenden AB, Morales-Oyarvide V et al. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet Med, 21(1), 213–223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shindo K, Yu J, Suenaga M et al. Deleterious Germline Mutations in Patients With Apparently Sporadic Pancreatic Adenocarcinoma. J Clin Oncol, 35(30), 3382–3390 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Goldstein AM, Struewing JP, Fraser MC, Smith MW, Tucker MA. Prospective risk of cancer in CDKN2A germline mutation carriers. J Med Genet, 41(6), 421–424 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tamura K, Yu J, Hata T et al. Mutations in the pancreatic secretory enzymes CPA1 and CPB1 are associated with pancreatic cancer. Proc Natl Acad Sci U S A, 115(18), 4767–4772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kawamoto M, Kohi S, Abe T et al. Endoplasmic stress-inducing variants in CPB1 and CPA1 and risk of pancreatic cancer: A case-control study and meta-analysis. Int J Cancer, 150(7), 1123–1133 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Childs EJ, Mocci E, Campa D et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet, 47(8), 911–916 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhong J, Jermusyk A, Wu L et al. A Transcriptome-Wide Association Study Identifies Novel Candidate Susceptibility Genes for Pancreatic Cancer. J Natl Cancer Inst, 112(10), 1003–1012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang M, Wang Z, Obazee O et al. Three new pancreatic cancer susceptibility signals identified on chromosomes 1q32.1, 5p15.33 and 8q24.21. Oncotarget, 7(41), 66328–66343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Klein AP, Wolpin BM, Risch HA et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat Commun, 9(1), 556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wu C, Miao X, Huang L et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet, 44(1), 62–66 (2011). [DOI] [PubMed] [Google Scholar]
- 189.Low SK, Kuchiba A, Zembutsu H et al. Genome-wide association study of pancreatic cancer in Japanese population. PLoS One, 5(7), e11824 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kastrinos F, Mukherjee B, Tayob N et al. Risk of pancreatic cancer in families with Lynch syndrome. JAMA, 302(16), 1790–1795 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Jones S, Hruban RH, Kamiyama M et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science, 324(5924), 217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lowenfels AB, Maisonneuve P, DiMagno EP et al. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J Natl Cancer Inst, 89(6), 442–446 (1997). [DOI] [PubMed] [Google Scholar]
- 193.Lowenfels AB, Maisonneuve P, Whitcomb DC, Lerch MM, DiMagno EP. Cigarette smoking as a risk factor for pancreatic cancer in patients with hereditary pancreatitis. JAMA, 286(2), 169–170 (2001). [DOI] [PubMed] [Google Scholar]
- 194. Shelton CA, Umapathy C, Stello K, Yadav D, Whitcomb DC. Hereditary Pancreatitis in the United States: Survival and Rates of Pancreatic Cancer. Am J Gastroenterol, 113(9), 1376 (2018). **Individuals with hereditary pancreatitis have an elevated risk of developing pancreatic cancer.
- 195.van Lier MG, Wagner A, Mathus-Vliegen EM, Kuipers EJ, Steyerberg EW, van Leerdam ME. High cancer risk in Peutz-Jeghers syndrome: a systematic review and surveillance recommendations. Am J Gastroenterol, 105(6), 1258–1264(2010). [DOI] [PubMed] [Google Scholar]
- 196.Giardiello FM, Brensinger JD, Tersmette AC et al. Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology, 119(6), 1447–1453 (2000). [DOI] [PubMed] [Google Scholar]
- 197.Giardiello FM, Welsh SB, Hamilton SR et al. Increased risk of cancer in the Peutz-Jeghers syndrome. N Engl J Med, 316(24), 1511–1514 (1987). [DOI] [PubMed] [Google Scholar]
- 198.Shi C, Klein AP, Goggins M et al. Increased Prevalence of Precursor Lesions in Familial Pancreatic Cancer Patients. Clin Cancer Res, 15(24), 7737–7743 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang L, Brune KA, Visvanathan K et al. Elevated cancer mortality in the relatives of patients with pancreatic cancer. Cancer Epidemiol Biomarkers Prev, 18(11), 2829–2834 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang W, Chen S, Brune KA, Hruban RH, Parmigiani G, Klein AP. PancPRO: risk assessment for individuals with a family history of pancreatic cancer. J Clin Oncol, 25(11), 1417–1422 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Tersmette AC, Petersen GM, Offerhaus GJ et al. Increased risk of incident pancreatic cancer among first-degree relatives of patients with familial pancreatic cancer. Clin Cancer Res, 7(3), 738–744 (2001). **One of the first papers to use family history to quantify the risk of pancreatic cancer.
- 202.Klein AP, Brune KA, Petersen GM et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res, 64(7), 2634–2638 (2004). [DOI] [PubMed] [Google Scholar]
- 203.Brune KA, Lau B, Palmisano E et al. Importance of age of onset in pancreatic cancer kindreds. J Natl Cancer Inst, 102(2), 119–126 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Al-Sukhni W, Rothenmund H, Borgida AE et al. Germline BRCA1 mutations predispose to pancreatic adenocarcinoma. Hum Genet, 124(3), 271–278 (2008). [DOI] [PubMed] [Google Scholar]
- 205.Cho JH, Bang S, Park SW, Chung JB, Song SY. BRCA2 mutations as a universal risk factor for pancreatic cancer has a limited role in Korean ethnic group. Pancreas, 36(4), 337–340 (2008). [DOI] [PubMed] [Google Scholar]
- 206.de Snoo FA, Bishop DT, Bergman W et al. Increased risk of cancer other than melanoma in CDKN2A founder mutation (p16-Leiden)-positive melanoma families. Clin Cancer Res, 14(21), 7151–7157 (2008). [DOI] [PubMed] [Google Scholar]
- 207.Goldstein AM, Fraser MC, Struewing JP et al. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med, 333(15), 970–974 (1995). [DOI] [PubMed] [Google Scholar]
- 208.Ibrahim I, Sibinga Mulder BG, Bonsing B et al. Risk of multiple pancreatic cancers in CDKN2A-p16-Leiden mutation carriers. Eur J Hum Genet, 26(8), 1227–1229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Iqbal J, Ragone A, Lubinski J et al. The incidence of pancreatic cancer in BRCA1 and BRCA2 mutation carriers. Br J Cancer, 107(12), 2005–2009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Zhen DB, Rabe KG, Gallinger S et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet Med, 17(7), 569–577 (2015). *Germline variants can be used to quantify the risk of pancreatic cancer.
- 211.Abe T, Blackford AL, Tamura K et al. Deleterious Germline Mutations Are a Risk Factor for Neoplastic Progression Among High-Risk Individuals Undergoing Pancreatic Surveillance. J Clin Oncol, 37(13), 1070–1080 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lim W, Olschwang S, Keller JJ et al. Relative frequency and morphology of cancers in STK11 mutation carriers. Gastroenterology, 126(7), 1788–1794 (2004). [DOI] [PubMed] [Google Scholar]
- 213.Kimura H, Klein AP, Hruban RH, Roberts NJ. The Role of Inherited Pathogenic CDKN2A Variants in Susceptibility to Pancreatic Cancer. Pancreas, 50(8), 1123–1130 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Maisonneuve P, Lowenfels AB. Chronic pancreatitis and pancreatic cancer. Dig Dis, 20(1), 32–37 (2002). [DOI] [PubMed] [Google Scholar]
- 215.Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol, 18(7), 493–502 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gong Y, Yang YS, Zhang XM et al. ABO blood type, diabetes and risk of gastrointestinal cancer in northern China. World J Gastroenterol, 18(6), 563–569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wolpin BM, Chan AT, Hartge P et al. ABO blood group and the risk of pancreatic cancer. J Natl Cancer Inst, 101(6), 424–431 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Klein AP, Lindstrom S, Mendelsohn JB et al. An absolute risk model to identify individuals at elevated risk for pancreatic cancer in the general population. PLoS One, 8(9), e72311 (2013). *Relatively few very-high risk individuals are identified when risk factors are combined.
- 219.Rose G.Sick individuals and sick populations. Int J Epidemiol, 30(3), 427–432; discussion 433–424 (2001). [DOI] [PubMed] [Google Scholar]
- 220.Bruenderman E, Martin RC 2nd. A cost analysis of a pancreatic cancer screening protocol in high-risk populations. Am J Surg, 210(3), 409–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Corral JE, Das A, Bruno MJ, Wallace MB. Cost-effectiveness of Pancreatic Cancer Surveillance in High-Risk Individuals: An Economic Analysis. Pancreas, 48(4), 526–536 (2019). [DOI] [PubMed] [Google Scholar]
- 222.Kitano M, Yoshida T, Itonaga M, Tamura T, Hatamaru K, Yamashita Y. Impact of endoscopic ultrasonography on diagnosis of pancreatic cancer. J Gastroenterol, 54(1), 19–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Konings IC, Harinck F, Poley JW et al. Prevalence and Progression of Pancreatic Cystic Precursor Lesions Differ Between Groups at High Risk of Developing Pancreatic Cancer. Pancreas, 46(1), 28–34 (2017). [DOI] [PubMed] [Google Scholar]
- 224.Lachter J, Rosenberg C, Hananiya T et al. Screening to Detect Precursor Lesions of Pancreatic Adenocarcinoma in High-risk Individuals: A Single-center Experience. Rambam Maimonides Med J, 9(4):e0029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ludwig E, Olson SH, Bayuga S et al. Feasibility and yield of screening in relatives from familial pancreatic cancer families. Am J Gastroenterol, 106(5), 946–954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Paiella S, Capurso G, Cavestro GM et al. Results of First-Round of Surveillance in Individuals at High-Risk of Pancreatic Cancer from the AISP (Italian Association for the Study of the Pancreas) Registry. Am J Gastroenterol, 114(4), 665–670 (2019). [DOI] [PubMed] [Google Scholar]
- 227. Tamburrino D, Cortesi P, Facchetti R et al. Real-world costs and dynamics of surveillance in patients who underwent surgery for low-risk branch duct intraductal papillary mucinous neoplasms. Eur J Surg Oncol, 49(1), 137–141 (2023). *Highlights the costs of overtreatment.
- 228.Brentnall TA, Bronner MP, Byrd DR, Haggitt RC, Kimmey MB. Early diagnosis and treatment of pancreatic dysplasia in patients with a family history of pancreatic cancer. Ann Intern Med, 131(4), 247–255 (1999). [DOI] [PubMed] [Google Scholar]
- 229.Black WC, Welch HG. Advances in diagnostic imaging and overestimations of disease prevalence and the benefits of therapy. N Engl J Med, 328(17), 1237–1243 (1993). [DOI] [PubMed] [Google Scholar]
- 230.Placido D, Yuan B, Hjaltelin JX et al. A deep learning algorithm to predict risk of pancreatic cancer from disease trajectories. Nat Med, 29(5):1113–1122 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]