Abstract
In this report we describe a novel, bipartite vesicular stomatitis virus (VSV) replication system which was used to study the effect of mutations in the transcription start sequence on transcript initiation and 5′-mRNA modifications. The bipartite replication system consisted of two genomic RNAs, one of which (VSVΔG) was a recombinant VSV genome with the G gene deleted and the other (GFC) contained the G gene and two non-VSV reporter genes (green fluorescent protein [GFP] and chloramphenicol acetyltransferase [CAT]). Coinfection of cells with these two components resulted in high-level virus production and gave titers similar to that from wild-type-VSV-infected cells. Mutations were introduced within the first 3 nucleotides of the transcription start sequence of the third gene (CAT) of GFC. The effects of these changes on the synthesis and accumulation of CAT transcripts during in vivo transcription (e.g., in infected cells), and during in vitro transcription were determined. As we had reported previously (E. A. Stillman and M. A. Whitt, J. Virol. 71:2127–2137, 1997), changing the first and third nucleotides (NT-1 and NT-3) reduced CAT transcript levels in vivo to near undetectable levels. Similarly, changing NT-2 to a purine also resulted in the detection of very small amounts of CAT mRNA from infected cells. In contrast to the results in vivo, the NT-1C mutant and all of the second-position mutants produced near-wild-type amounts of CAT mRNA in the in vitro system, indicating that the mutations did not prevent transcript initiation per se but, rather, generated transcripts that were unstable in vivo. Oligo (dT) selection and Northern blot analysis revealed that the transcripts produced from these mutants did not contain a poly(A)+ tail and were truncated, ranging in size from 40 to 200 nucleotides. Immunoprecipitation analysis of cDNA-RNA hybrids with an antibody that recognizes trimethylguanosine revealed that the truncated mutant transcripts were not properly modified at the 5′ end, indicating the transcripts either were not capped or were not methylated. This is the first demonstration that transcript initiation and capping/methylation are separable events during VSV transcription. A model is proposed in which polymerase processivity is linked to proper 5′-end modification. The model suggests that a proofreading mechanism exists for VSV and possibly other nonsegmented minus-strand RNA viruses, whereby if some transcripts do not become capped during transcription in a normal infection, a signal is transduced such that the polymerase undergoes abortive elongation and the defective transcript is terminated prematurely and subsequently degraded.
Vesicular stomatitis virus (VSV) is an enveloped, nonsegmented minus-strand RNA virus that is considered the prototype for the Rhabdoviridae family. Because the genome is in the negative, or noncoding sense, the polymerase must be packaged in the virion during virus assembly and remain associated with the ribonucleocapsid core during virus entry and uncoating. The VSV polymerase is composed of two subunits, the phosphoprotein (P) and the large subunit (L). Once in the cell cytoplasm, the VSV RNA-dependant RNA polymerase is responsible for transcribing each of the five genes. In addition to the initial transcription of viral mRNAs, the polymerase is responsible for replication of the VSV genomic RNA via synthesis of a full-length positive-sense replicative intermediate, which is then used as a template for synthesis of full-length progeny genomes. The genome as well as the replicative intermediate must be encapsidated by the nucleocapsid (N) protein to serve as a template for the VSV polymerase.
One of the characteristic features of VSV transcription is that mRNA synthesis is both sequential and polar (3, 24, 25). Discontinuous transcription is not unique to VSV and occurs with all nonsegmented, minus-strand RNA viruses; however, there are differences that are specific for each virus family. During VSV transcription, the polymerase first transcribes a small (47-nucleotide) RNA called the leader from the extreme 3′ end of the genome, and then each of the five mRNAs encoding the viral proteins are synthesized in the order they appear from the 3′ end of the genome. For the gene junctions that have been studied, the downstream gene is transcribed approximately 30% less than the upstream gene (24), and as a consequence, the abundance of the five mRNAs also follows the order of genes on the genome (N > P > M > G > L).
Since the entire VSV life cycle is carried out in the cytoplasm of the cell, the virus cannot utilize the normal host machinery to carry out posttranscriptional modifications such as capping and polyadenylation. Therefore, these activities must be performed by the VSV polymerase (17, 31, 35, 39, 43). Analysis of the cap structure found on VSV transcripts indicates that the mechanism responsible for capping and methylation must differ from the general eukaryotic capping pathways. Both the α and β phosphates of the 5′-5′ triphosphate linkage of the guanosine cap are contributed by a presumed GDP donor (1, 2). Therefore, the addition of the 5′-5′ guanosine cap cannot be mediated by a covalent nucleotidyl transfer reaction with GMP as occurs during typical eukaryotic capping (7). Available evidence indicates that the P-L polymerase complex mediates the capping reaction, since mRNAs generated during in vitro transcription reactions with purified virions are efficiently capped. While the L protein does have some sequence homology to nucleotide binding proteins, it shows no homology to covalent nucleotidyl transferases (45) and has never been shown to mediate the capping reaction in trans. Recently it was shown that the VSV polymerase must associate with a host protein, translation elongation factor 1 (EF-1), to be transcriptionally active (13). The authors proposed that the GTP-GDP binding properties of the α subunit of EF-1 may play a role in the VSV capping reaction (13).
Another feature of VSV capping-methylation is that these reactions are intimately linked with transcription. Presynthesized VSV mRNAs lacking these modifications are not appropriately modified if added directly to VSV transcription complexes, whereas typical eucaryotic capping and methylation occur posttranscriptionally. Precisely how VSV transcript initiation and capping are coupled remains to be determined. Early studies demonstrated that short oligonucleotides representing the 5′ end of VSV transcripts can be produced in incomplete (minus UTP) in vitro reaction mixtures and that the 5′ termini of these short oligonucleotides contained a triphosphate. These results led the authors to hypothesize that capping occurs following de novo transcript initiation (10). On the other hand, the unique nature of the VSV cap suggests that it may arise through an RNA cleavage event. Recently a model was proposed which suggested that individual transcripts are “initiated” and capped following a GDP-dependent, polymerase-mediated cleavage event and that capping occurs as a result of the GDP-dependent cleavage (44).
In contrast to the uncertain role of the polymerase in the capping of VSV transcripts, the L protein has a methyltransferase activity, which has been shown through the analysis of methylation-defective host range mutants (17–19). During in vitro transcription in the presence of the methyl donor S-adenosyl-l-methionine (SAM), VSV transcripts are doubly methylated on the blocking G at the N-7 position and on the first A at the O-21 position (1) to generate the cap I structure (m7G5′ppp5′AmpApCpApG…). Interestingly, in vivo-generated transcripts exhibit some heterogeneity in their degree of methylation since the VSV mRNAs can be further methylated at the first A (N-6 position) and at the second A (N-6 and O-21 positions) [m7G5′ppp5′(m)Amp(m)AmpCpApG…] (31, 39). The additional methylation is thought to be catalyzed by cytoplasmic host cell enzymes (31) following VSV-specific methylation at the 2′-O-ribose of the first A nucleotide (18).
While the addition of a G cap is thought to be obligatorily coupled to transcription, this does not appear to be the case for methylation. In vitro transcription in the absence of the methyl donor SAM occurs efficiently, producing capped yet unmethylated transcripts (1, 16, 19, 35, 51); therefore methylation of the nascent transcript is not a prerequisite for transcription. Interestingly, during in vitro transcription in the presence of the methylation inhibitor S-adenosylhomocystine (SAH), nonmethylated transcripts that contain large heterogeneous poly(A) tails are generated, indicating that there may be some connection between modification at the 5′ end of the transcript and polyadenylation (20, 21, 41).
The cis-acting signals that direct the polymerase to polyadenylate and terminate the upstream mRNA and then to reinitiate transcription at the downstream gene are contained in a set of 23 conserved nucleotides at each VSV gene junction (42). The exact mechanism by which the polymerase executes its multiple activities as it interacts with these cis-acting sequences has not been elucidated. However, mutational analyses of these conserved cis-acting signals have provided some insight into their functional importance (5, 6, 22, 47, 48). The conserved sequence 3′-AUACU7-5′, which is found at the end of each gene, is critical for both polyadenylation and termination of VSV transcripts (5, 22). When this sequence is encountered, the polymerase reiteratively copies, or stutters, over the seven uridinylate residues to produce a poly(A) tail approximately 150 nucleotides in length (43). Immediately following the polyadenylation signal are two nontranscribed intergenic nucleotides, which are usually 3′-GA-5′ (38). There is some evidence that this dinucleotide contributes to the termination signal of the upstream transcript, because certain nucleotide substitutions resulted in higher levels of readthrough transcription at the mutated gene junction (6, 48). Following the intergenic dinucleotide is the transcription start sequence 3′-UUGUCnnUAC-5′ (with n being any nucleotide). The 5′ ends of all VSV mRNAs contain the complement of this sequence. We have previously shown that the first 3 nucleotides of the VSV 5′ transcription start sequence are the most critical for efficient gene expression because mutations within the first 3 nucleotides severely reduce the levels of mRNA produced from the mutant gene in vivo (48).
Mutational analysis of the cis-acting signals described above were performed with modified and shorter versions of the VSV genome containing a reporter gene or viral genes. One limitation of these minigenome systems is that the N, P, and L proteins are expressed in trans and that replication and transcription are carried out in the presence of vaccinia virus. In addition, mutations that affect transcription are often identified by measuring the reporter gene activity or by directly measuring the levels of transcript present in the minigenome-infected cells. Therefore, mutations that affect transcript stability cannot be differentiated from those that affect transcript initiation.
In this report, we describe a novel, bipartite VSV replication system which was used to further study the functional importance of the transcription start sequence on transcript initiation and 5′-end mRNA modifications. The bipartite replication system consisted of two genomic RNAs, one of which (VSVΔG) was a recombinant VSV genome with the G-protein gene deleted and the other (GFC) contained the G protein gene and two non-VSV reporter genes (green fluorescent protein [GFP] and chloramphenicol acetyltransferase [CAT]). The effects of mutations within the 5′ start sequence of the CAT gene of GFC were determined for in vivo transcription (in GFC/ΔG-infected cells) and following in vitro transcription. Our data indicate that some mutations in the 5′ transcription start sequence can affect transcript initiation while others affect 5′-end modifications. Furthermore, our results suggest that transcript initiation and capping-methylation are separable events and that the processivity of the VSV polymerase can be influenced by the addition of 5′-end modifications.
MATERIALS AND METHODS
Expression plasmids and minigenome constructs.
Plasmids encoding the VSVI N, P, and L proteins have been described elsewhere (14, 46). The plasmid encoding pBS-VSVΔG was obtained from C. Robison (37). To generate the wild-type pBS-GFC minigenome cDNA, an AflII-NheI fragment containing the GFP(S65-T) gene, which was obtained from a minigenome (pBS-GMF) encoding the VSVI M- and G-protein genes upstream of the GFP gene, was subcloned into pBS-GMG (46) which had been digested with the same restriction enzymes. NheI is located in the 3′ untranslated region of the G gene, and AflII lies within the trailer region of pBS-GMG. Sequences corresponding to the P-gene transcriptional start site were used to drive expression of the GFP gene in GMF. In addition, a unique XhoI site was introduced into the GFP 5′ untranslated region and an SphI site was introduced in the 3′ untranslated region. All of these sequence elements were maintained in the construction of the GFC cDNA. Following ligation of the AflII-NheI fragment, the resulting minigenome contained the G-protein gene and the GFP gene flanked by the VSV leader and trailer regions, respectively. Next, the CAT gene was subcloned into this G-GFP minigenome between the trailer sequence and the GFP gene. The GFP-CAT gene junction was derived from the M-G intergenic region (including 107 nucleotides of the M 3′ untranslated region and 29 nucleotides of the G 5′ untranslated region). To position the GFP gene stop codon next to the M gene 3′ untranslated region, we designed an oligonucleotide primer (IG-MUT-2; 5′-CATTCGGCATGCGCTAGTCTAACTTCTAGC-3′) that contained an SphI site (bold) at its 5′ end and sequences that overlapped the M 3′ untranslated region at its 3′ end. To position the G-protein 5′ untranslated region adjacent to the CAT gene start codon, we designed an oligonucleotide primer (IG-MUT-1; 5′-CTAAGTACAAGTCGACTCTAGAAGTGACGCGTAAACAGATC-3′) which contained a SalI site (underlined) and a XbaI site (bold italic) at its 5′ end and sequences that overlapped the G 5′ untranslated region. These primers were used to amplify the M gene-G gene intergenic region from pBS-GMMG (46). The resulting PCR product was digested with SalI and SphI and used in the three-way ligation with an AflII-XhoI fragment, which contained the CAT gene (CAT was previously fused to the trailer in pBS-GMMG-CAT [a gift from J. Rose, Yale University]), and the G-GFP minigenome vector, which had been cut with AflII and SphI to create pBS-GFC-1. SalI and XhoI have compatible overlapping ends. To replace the G 3′ untranslated sequence (present at the 3′ end of the CAT gene and adjacent to the trailer sequence) with the L gene 3′ untranslated region, an oligonucleotide primer was designed (t-L-CAT; 5′CAC AAGAGGGTCTTAAGGATCAAAGTTTTTTTCATACTTAAAGTTTGGA GTCTCCTCATGATTTTTACGCCCCGCCCTGCCACTC-3′) that contained sequences that overlapped the AflII site in the trailer region and extended through the polyadenylation signal and the 3′ untranslated region of the L gene and then overlapped 21 nucleotides into the CAT gene coding sequence (bold italic). This primer and the IG-MUT-2 primer were used to amplify a fragment in which the CAT gene was fused to the L 3′ untranslated region and the trailer. The PCR product was digested with XbaI and AflII and used to replace the CAT gene in pBS-GFC-1 to create pBS-GFC-2, which was the parental minigenome for all of the mutants in this study. PCR-amplified regions, except for the CAT gene coding sequence, were sequenced by using the dideoxynucleotide method and Sequenase.
To generate minigenome mutants with the desired nucleotide changes at the GFP-CAT gene junction, the IG-MUT-1 and IG-MUT-2 oligonucleotide primers were used to amplify the M-G gene junction from GMMG mutants containing nucleotide changes within the 5′ start sequence of the G-protein gene (48). The PCR products were digested with SphI and XbaI, and the resulting 147-bp fragments were used to replace the corresponding regions in the wild-type pBS-GFC-2.
Recovery of GFC/ΔG.
Recovery of VSVΔG was carried out essentially as described previously (49). For recovery of GFC/ΔG, baby hamster kidney (BHK-21) cells in 35-mm-diameter dishes were infected with vTF7-3 and transfected with 10 μg of plasmid DNA containing either wild-type or mutant GFC minigenomes, together with 5, 4, and 1 μg of plasmids encoding the N, P, and L proteins, respectively, by using TransfectACE as described previously (40). At 5 h posttransfection, the medium was removed and the BHK-21 cells were infected with recombinant VSVΔG (approximate multiplicity of infection of 3). At 2 days posttransfection/infection, the medium was collected and vTF7-3 was removed via filtration through a 0.2-μm syringe filter. This P1 supernatant was then passaged onto fresh BHK-21 cells. At 24 h postinfection, dishes containing GFC/ΔG were identified by observing cytopathic effects and GFP expression. To ensure that aberrant viruses were not recovered, the levels of G and GFP mRNAs expressed from these viruses were determined by Northern blot analysis and compared to those of wild-type GFC/ΔG. For all of the experiments, an unselected (non-plaque-purified) population of a low-passage virus was used to reduce the likelihood that a variant with a nonrepresentative transcription phenotype would be picked inadvertently.
Purification of GFC/ΔG virus and in vitro transcription reactions.
Approximately 2.5 × 107 BHK-21 cells in a 15-cm dish were infected with approximately 350 to 2 × 103 PFU of GFC/ΔG. Viruses were harvested at 18 to 22 h postinfection and pelleted from the supernatant. Virus resuspended in TNE (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA) was then gradient purified on a 20 to 45% sucrose gradient in TN (10 mM Tris-HCl [pH 7.5], 150 mM NaCl). All viral bands were collected from the gradient, pelleted, and then resuspended in TNE. The viral protein concentration was determine by a bicinchoninic acid protein assay (Pierce). In vitro transcription reactions were carried out essentially as described previously (34). Gradient-purified virus (15 μg) was incubated at 30°C for 90 min in 50 mM HEPES (pH 8.0)–100 mM NaCl–5 mM MgCl2–4 mM dithiothreitol–1 mM each ATP, CTP, GTP, and UTP–0.1% Triton N-101–40 U of RNasin (Promega)–1 mM SAM (volume, 100 μl).
RNA analysis by primer extension and Northern blot assays.
Following in vitro transcription, the RNA was extracted with phenol-chloroform and precipitated with 95% ethanol. Poly(A) isolation was performed with the Oligotex mRNA isolation system (Qiagen). For in vivo-generated transcripts, BHK-21 cells were infected at an approximate multiplicity of infection of 1 with either wild-type or mutant GFC/ΔG virus. The cells were harvested 6 to 10 h postinfection, and total RNA was isolated by the method of Chomczynski and Sacchi (11). For primer extensions, we used oligonucleotide GFP-5 (5′-GTGCCCATTAACATCACCATC-3′), complementary to a sequence at nucleotides 73 to 94 from the 5′ end of the GFP transcript and oligonucleotide CAT-1 (5′-CAACGGTGGTATATCCAGTG-3′) complementary to a sequence at nucleotides 56 to 75 from the 5′ end of the CAT mRNA. These oligonucleotides were end labeled in a standard kinase reaction as previously described (48). The primer extension reaction was carried out essentially as previously described (48), and the products were analyzed on a 6% sequencing gel. For Northern blot analysis, in vitro-transcribed RNA was extracted with phenol-chloroform, precipitated with 95% ethanol, fractionated on a 2% agarose-formaldehyde gel, and transferred to Nytran (Schleicher & Schuell) essentially as described previously (46). GFP- and CAT-specific RNA species were visualized by using the end-labeled oligonucleotide probes, GFP-5 and CAT-1, under the conditions described in reference 46. Band intensities for both primer extension and Northern blot assays were determined with the Molecular Dynamics Storm 860 PhosphorImager.
Immunoselection with anti-cap antibody.
Total in vitro-generated transcripts were phenol-chloroform extracted, ethanol precipitated, hybridized with at least 5 × 104 cpm of the end-labeled GFP-5 and CAT-1 oligonucleotide primers in 30 μl of hybridization buffer [40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), [pH 6.8], 1 mM EDTA, 0.4 M NaCl, 80% formamide], which was incubated at 85°C for 5 min, and immediately transferred to 30°C for an overnight incubation. The reverse transcription reaction was performed with SuperScript II (GibcoBRL), an RNase H-negative murine reverse transcriptase. Half of the reaction mixture was brought up in volume to 250 μl with NET (150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100), and RNA-cDNA hybrids were immunoselected with an anti-cap monoclonal antibody, H-20 (a generous gift from Reinhard Luhrmann [8]) essentially as described previously (15). A 1-μl volume of the H-20 ascites was incubated with the capped-RNA–cDNA hybrids for 1 h at 4°C and then incubated with a rabbit anti-mouse immunoglobulin G antibody for 30 min at 4°C. Then 40 μl of a 10.5% (wt/vol) solution of Pansorbin cells (Calbiochem) was added, and complexes were formed at 4°C for 4 h. Following three washes with NET, immunoselected as well as nonimmunoselected RNA-cDNA hybrids were phenol extracted, ethanol precipitated, and fractionated on a 6% polyacrylamide sequencing gel.
RESULTS
In this study we have continued our investigation into the role of the VSV gene start sequence during transcription. Our previous data from saturation mutagenesis of a VSV gene start sequence indicated that the first three positions of the start sequence are the most critical to attain near-wild-type levels of transcript in vivo. One conclusion was that the mutations within the first 3 nucleotides affected transcript initiation; however, we could not rule out the possibility that transcripts were synthesized but were unstable and quickly degraded. Therefore, we used an in vitro transcription assay which allowed us to analyze the transcripts generated from the various mutants in an environment in which unstable transcripts would not be subjected to degradation.
Recovery of VSV as a bipartite virus.
For the in vitro transcription assays, we needed to generate relatively large quantities of recombinant VSV particles containing specific mutations in a gene start sequence. To this end, we developed a system in which we could recover a VSV that also contained two non-VSV reporter genes. This recombinant version of VSV was recovered from cDNA as a bipartite virus. One genome (VSVΔG) contained the full-length VSV genome with the G gene deleted, while the other genome (GFC) contained the G-protein gene upstream of two non-VSV reporter genes (GFP and CAT) (Fig. 1). These miniviruses complemented each other; therefore, coinfection of cells in culture resulted in propagation of the bipartite virus (GFC/ΔG). These bipartite viruses were grown to high titers and gradient purified. Northern blot analysis of genomic RNA isolated from purified GFC/ΔG virus confirmed that both genomic RNA species were present in approximately the same amounts (data not shown).
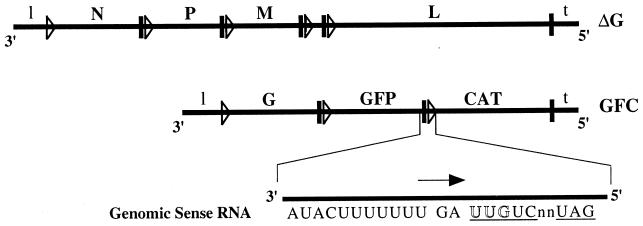
FIG. 1.
Schematic representation of the two genomes of GFC/ΔG. The cDNAs used to recover this bipartite virus were constructed so that the GFP and CAT gene sequences in GFC contained all of the cis-acting sequences necessary for gene expression from a VSV genome. The coding region of the G gene in ΔG was deleted, but the conserved transcription start and polyadenylation-termination signals of G were left intact. The heavy vertical lines represent the polyadenylation/stop sequence and the nontranscribed intergenic dinucleotide, and the triangles represent the 5′ start sequence. The sequence at the GFP-CAT gene junction is enlarged. The CAT gene start sequence is underlined, and the arrow indicates the direction of mRNA transcription. Mutations were introduced at nucleotide positions 1, 2, or 3 (outline font).
Since VSV transcription is sequential and polar, the N, P, M, and L VSV genes in VSVΔG were kept in the same gene order and location as found in wild-type VSV to ensure that these proteins were expressed at the appropriate levels. The GFP and CAT genes were positioned as the second and third cistrons, respectively, on the GFC genome and therefore were expressed at sufficiently high levels for transcript analysis. This bipartite-virus system also allowed us to make mutations that dramatically reduced transcription at the GFP-CAT gene junction without affecting the growth of the bipartite virus, since these genes were not required for viral growth and were not upstream of any other essential VSV genes. A total of 10 recombinant bipartite viruses that contained either the wild-type start sequence or nucleotide changes within the start sequence of the CAT reporter gene were generated.
Analysis of bipartite virus transcripts made in vivo.
Previously, saturation mutagenesis analysis was performed within the G gene 5′ start sequence in the context of an M- and G-expressing minigenome (48). To determine if the same mutations in the CAT gene 5′ start sequence had a similar effect on the level of CAT transcripts during a GFC/ΔG infection, total RNA from wild-type- or mutant-GFC/ΔG-infected cells was analyzed by a primer extension assay to detect GFP and CAT transcripts (Fig. 2, lanes V). The GFP-specific primer was complementary to nucleotides 73 to 94 of the GFP mRNA, and the CAT-specific primer was complementary to nucleotides 56 to 75 of the CAT mRNA. The relative level of CAT mRNA expressed, which is defined as the ratio of CAT to GFP primer extension products, is shown in the bar graph above each respective lane. For in vivo-generated RNA, the major product mapped to the first nucleotide of the 5′ mRNA start site for both CAT mRNA and GFP mRNA. The 5′ start sites were identified by migration of the primer extension products to the appropriate position relative to the sequencing ladder (examples shown in the right-hand-most lane of panels B and C only). The results indicated that the level of CAT mRNA expressed by each of the mutants during in vivo transcription was similar to the level of G mRNA expressed in the previous minigenome system (48) when mutants with the same nucleotide changes were compared. Specifically, nucleotide changes within the first 3 nucleotides of the start sequence severely decreased the amount of the corresponding mRNA, except for the NT-2C mutation, in which there was only a moderate decrease in the level of the corresponding transcript.
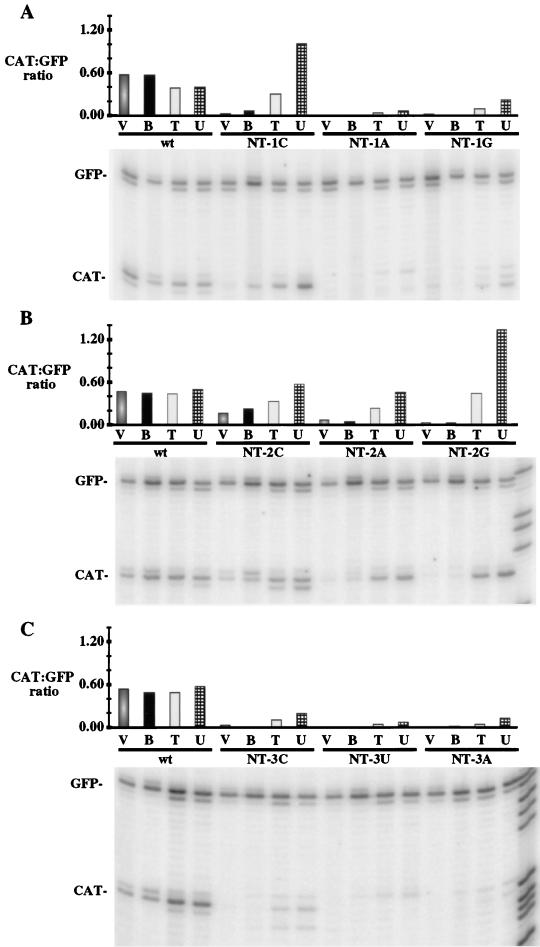
FIG. 2.
Primer extension analysis of RNAs purified from wild-type (wt) and mutant GFC/ΔG-infected cells and from in vitro transcription reactions. Four different types of RNA generated from either wild-type or mutant viruses were analyzed. Lanes V contain total RNA isolated from wild-type or mutant GFC/ΔG-infected cells; lanes B contain oligo (dT)-bound poly(A) RNAs from in vitro transcription reactions; lanes T contain total RNA from in vitro reactions; lanes U contain in vitro-generated RNA that did not bind the oligo(dT) column. Each RNA type was hybridized to end-labeled oligonucleotides complementary to regions in the GFP mRNA and CAT mRNA. Following extension, the products were resolved on a denaturing 6% polyacrylamide gel and quantified with a PhosphorImager. The major primer extension product corresponded to the first nucleotide of each mRNA as identified by comparison with a DNA sequencing ladder (one lane of each sequencing reaction is shown in the right-hand-most lane in panels B and C only). The minor bands may result from incomplete extension and the larger products may result from extension through the 5′ cap or the addition of one nontemplated nucleotide by reverse transcriptase. Band intensities were determined with a STORM PhosphorImager and ImageQuant software. The ratio of CAT to GFP extension products was calculated and is shown in the bar graphs above each lane. (A) NT-1 mutants; (B) NT-2 mutants; (C) NT-3 mutants.
Mutations in the start sequence affect postinitiation events in vitro.
To determine the level of CAT mRNA expressed from wild-type GFC/ΔG and each of the mutants during in vitro transcription, we used primer extension and the same GFP- and CAT-specific primers as described above. The RNA populations examined included total (unfractionated) RNA (Fig. 2, lanes T) as well as both polyadenylated [oligo(dT) bound; lanes B] and nonpolyadenylated (unbound; lanes U) RNAs. Again, the relative level of CAT mRNA expressed and detected via primer extension from the wild-type virus and each of the mutants was determined by calculating the ratio of CAT product to GFP product as shown in the bar graph above each respective lane. For wild-type virus and each of the mutants, the relative amount of poly(A) CAT transcript generated in vitro was similar to the amount of CAT mRNA isolated from cells during in vivo transcription (compare lanes B to lanes V for each of the viruses). Therefore, the amount of polyadenylated RNA expressed by each of the mutants during in vitro transcription corresponded to the amount of stable CAT transcripts isolated from infected cells in vivo.
While the amounts of both poly(A)-containing and unfractionated (total) CAT transcript (relative to the GFP transcript) generated in vitro were similar to those detected in vivo for wild-type virus, large difference were observed for some of the mutants. For example, the amount of total (unfractionated) in vitro CAT transcript produced by the NT-1C virus (Fig. 2A) and all of the second-position mutant viruses (Fig. 2B) was larger than that of transcripts isolated from infected cells (in vivo-generated transcripts) or of in vitro-generated transcripts that contained poly(A) tails (compare lanes T to lanes V and B for each of these mutants). Therefore, most of the CAT transcripts produced from the mutants (with the exception of NT-2C) did not contain a poly(A) tail. Interestingly, the NT-2C virus (Fig. 2B), which expressed moderate levels of stable CAT mRNA in vivo, also produced a moderate amount of poly(A) CAT transcript during in vitro transcription. These data suggest that many of the mutations in the first 3 nucleotides of the start sequence affected a postinitiation event and that their in vivo phenotypes resulted from mRNA degradation.
The NT-1G and NT-1A viruses (Fig. 2A) and all third-position mutants (Fig. 2B) also had similar phenotypes, in that some of the CAT transcripts generated in vitro did not appear to be polyadenylated (compare lanes T to lanes B for each of these mutants). However, these specific mutations also appeared to affect transcript initiation, since the relative levels of CAT transcript detected in the total RNA samples were significantly lower (5 to 25%) than that expressed from wild-type virus (compare lanes T for these mutants to lane T for the wild-type virus). Overall, these data indicate that some mutations in the VSV gene start sequence can affect postinitiation events in vitro such that transcripts are initiated but are not ultimately polyadenylated whereas other mutations reduce transcript initiation.
Mutations in the VSV start sequence affect the processivity of the polymerase.
To determine the length of the nonpolyadenylated transcripts, we used Northern blot analysis to detect the CAT and GFP RNA species produced during an in vitro transcription reaction for wild-type GFC/ΔG and each mutant virus (Fig. 3A). The same oligonucleotide probes that were used in the primer extension assay were used to detect either GFP-specific RNA species (Fig. 3A, top, GFP mRNA only) or CAT specific species (Fig. 3A, bottom). The CAT-specific probe also detected the dicistronic GFP-CAT mRNA as well as the GFC replicative intermediate (Fig. 3A, bottom). The ratio of full-length CAT mRNA to GFP mRNA was determined for each mutant (results not shown). The results indicated that the relative level of full-length CAT mRNA expressed by each mutant corresponded to (i) the amount of polyadenylated RNA produced during in vitro transcription and (ii) the amount of stable CAT transcripts isolated from infected cells in vivo.
FIG. 3.
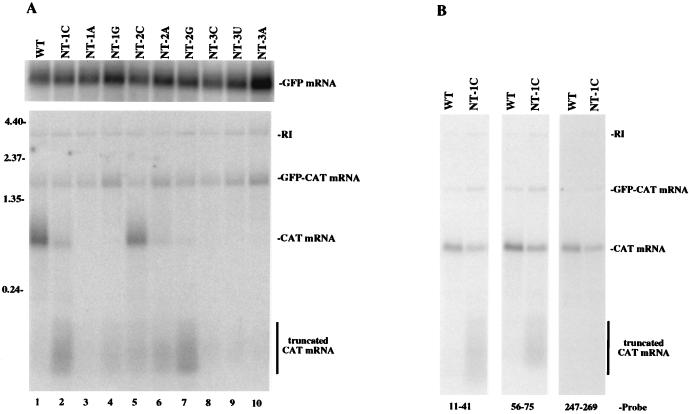
Northern blot analysis of in vitro-generated RNA from wild-type (WT) and mutant GFC/ΔG. Following in vitro transcription, total RNA was isolated, fractionated on a 2% agarose-formaldehyde gel, and transferred to a nylon membrane. (A) Membranes were probed with an end-labeled oligonucleotide complementary to a region in the GFP mRNA (nucleotides 73 to 94 [top]) or with an end-labeled oligonucleotide complementary to a region in the CAT mRNA (nucleotides 56 to 75 [bottom]). (B) A membrane containing wild-type and NT-1C RNA was probed with end-labeled oligonucleotides complementary to different regions in the CAT mRNA (the regions are noted on the figure).
Interestingly, smaller CAT RNA species were also detected for each of the mutants. These truncated products were approximately 75 to 200 nucleotides long. In accordance with the primer extension data, the mutations which affected synthesis of polyadenylated transcripts and not transcript initiation (e.g., NT-1C virus and all of the second-position mutant viruses) synthesized high levels of this truncated CAT mRNA species (Fig. 3A, lanes 2 and 5 to 7). In contrast, the mutations which greatly affected transcript initiation (NT-1A and all third-position mutants) did not generate high levels of this truncated CAT mRNA (Fig. 3A, lanes 3 and 8 to 10). These results indicated that some mutations within the CAT gene start sequence affect the processivity of the polymerase during in vitro transcription.
It is possible that the truncated CAT species resulted from degradation of the full-length mRNA. If so, it might be expected that oligonucleotide probes complementary to different regions of the mRNA could recognize these truncated RNAs equally well. To determine if the heterogeneous populations of truncated CAT transcripts resulted from random degradation, total in vitro-synthesized RNA from wild-type GFC/ΔG and the NT-1C mutant virus was examined by Northern blot analysis with three different CAT-specific oligonucleotide probes that were each complementary to a different region of the CAT gene (Fig. 3B). The probe that was complementary to nucleotides 11 to 41 detected a smear of CAT RNA that contained shorter RNAs than did the probe that was complementary to nucleotides 56 to 75, while the probe that was complementary to nucleotides 247 to 269 did not detect the truncated CAT species. These results indicated that the truncated species did not result from random degradation of the CAT transcript in vitro; however, we cannot exclude the possibility that they represent specific products arising from 3′-end degradation. Second, these results showed that the truncated CAT mRNAs were heterogeneous in size and ranged from approximately 40 to less than 247 nucleotides.
The primer extension data and the Northern blot data both indicated that the major effect of third-position mutations was on transcript initiation. These assays were performed with an oligonucleotide probe that was complementary to nucleotides 56 to 75; therefore, RNA species shorter than 56 to 75 nucleotides would not be detected. To ascertain if CAT mRNA was indeed initiated from the NT3 mutants but was not detected by the probe, total in vitro-generated RNA from the wild type and NT3 mutants was hybridized with a labeled oligonucleotide probe complementary to nucleotides 11 to 41. There was not a significant increase in the level of the truncated CAT mRNA when this oligonucleotide was used as a probe (data not shown). This result suggested that while the NT3 mutants did produce a small amount of the truncated mRNA species, the major effect of the third-position changes was on transcript initiation.
Mutations in the start sequence inhibit capping or methylation by the polymerase.
In addition to transcription, the polymerase is responsible for capping and methylating the 5′ end of nascent transcripts (17, 19, 31, 35, 39). This capping and methylation occurs concomitantly during transcription; therefore the conserved sequences at the start site could potentially serve as a signal for these processes.
To determine if the nucleotide changes in the CAT start sequence affected the ability of the polymerase to cap and/or methylate the nascent CAT transcript, we used a monoclonal antibody specific for trimethyl guanosine (8). This antibody also recognizes the methylated cap structure at the 5′ end of transcripts and can be used to immunoprecipitate capped mRNAs. First, total RNA from in vitro transcription reactions with either wild-type or mutant viruses was used in primer extension assays with the CAT- and GFP-specific primers described above, together with an RNase H-minus reverse transcriptase to prevent degradation of the RNA in the cDNA-RNA complex. The cDNA-RNA hybrid complexes were then immunoprecipitated. The products of both nonimmunoprecipitated and immunoprecipitated primer extension reactions were visualized on a sequencing gel (Fig. 4). The GFP mRNA served as an internal, positive control, since this transcript should be capped and methylated normally. For the wild-type virus (lanes 13 and 14 or lanes 15 and 16), the CAT-to-GFP ratios for both the nonimmunoprecipitated and the immunoprecipitated cDNAs were similar, indicating that both the GFP and CAT mRNAs were capped and methylated to the same extent. However, the relative amounts of CAT transcript that were immunoprecipitated from the mutants were either significantly decreased or undetectable (lanes 2, 4, 6, 10, 12, 18, 20, and 22). These data indicated that the majority of the CAT transcripts generated in vitro from the mutants were not capped and/or methylated. Therefore, mutations within the first 3 nucleotides of the CAT 5′ start sequence affected the ability of the polymerase to cap and/or methylate the nascent transcript.
FIG. 4.
Analysis of the cap structure on wild-type (WT) and mutant CAT transcripts. Following in vitro transcription, total RNA was hybridized to end-labeled oligonucleotides complementary to regions in the GFP mRNA (nucleotides 73 to 94) and CAT mRNA (nucleotides 56 to 75). Following extension with reverse transcriptase (RNase H-minus), RNA-cDNA hybrids were immunoprecipitated with an antibody that recognized trimethylguanosine. Products from both nonimmunoprecipitated (−) and immunoprecipitated (+) samples were resolved on a denaturing 6% polyacrylamide gel.
The only exception to the defect in 5′-end modification was the NT-2C mutant (lanes 7 and 8). Interestingly, in the nonimmunoprecipitated sample, there were two CAT primer extension products. One product mapped to the correct 5′ end of the CAT transcript, and one was 2 nucleotides shorter, suggesting that the polymerase initiated transcription at an alternative site. Furthermore, the product that mapped to the correct 5′ start site was immunoprecipitated (lane 8), while none of the product that mapped to the alternative start site was recognized by the anti-cap antibody. For the NT-1C, NT-2A and NT-2G mutants, which produced low levels of full-length CAT mRNA as detected by primer extension and Northern blot analysis, longer exposures indicated that a very small amount of CAT transcript was also immunoprecipitated. The data from the NT-2C mutant and the other mutants indicated that there was a correlation between the ability of the polymerase to cap and/or methylate a nascent transcript and the processivity of the polymerase during synthesis of that transcript.
Lack of methylation does not induce premature transcript termination.
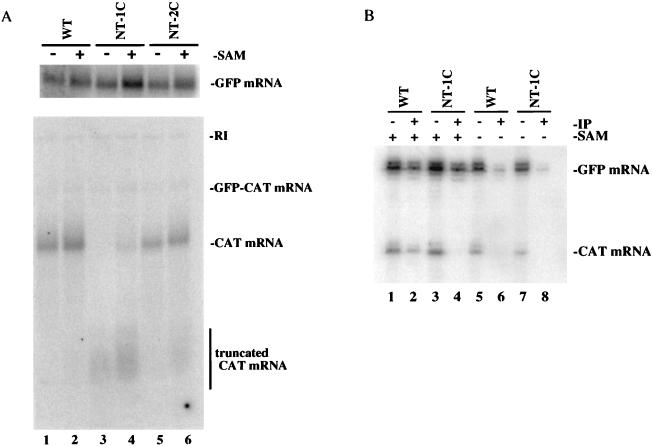
It has been well documented in other in vitro transcription studies that the absence of SAM during VSV in vitro transcription results in full-length transcripts that are capped but not methylated (4, 19, 35), suggesting that transcription is not affected by the lack of methylation. To determine if this result is observed during GFC/ΔG in vitro transcription, transcripts were generated in vitro in either the presence or absence of SAM for the wild-type, NT-1C, and NT-2C viruses. Northern blot analysis with GFP- and CAT-specific probes showed that full-length GFP and CAT mRNAs were generated during wild-type GFC/ΔG transcription in the absence or presence of SAM (Fig. 5A, lanes 1 and 2). Full-length GFP mRNAs were also generated in the absence or presence of SAM during in vitro transcription of the NT-1C and NT-2C mutant viruses (Fig. 5A, top, lanes 3 to 6); however, the profile of the CAT transcripts differed slightly between the transcripts generated in the absence or presence of SAM. For the NT-1C mutant virus, truncated CAT mRNA was synthesized in the absence of SAM but full-length CAT mRNA was undetectable (compare lanes 3 and 4). In contrast, full-length CAT mRNAs were synthesized from the NT-2C mutant virus in both the presence and absence of SAM, but very little truncated CAT mRNA was produced in the absence of SAM (compare lanes 5 and 6). While the implications of these results are not known, it is clear that some mutations in the 5′ start sequence have a greater effect on the polymerase in the absence of a methyl donor during in vitro transcription.
FIG. 5.
Analysis of transcripts generated in the presence or absence of SAM. (A) Northern blot analysis. Total RNA synthesized in the presence or absence of SAM was probed essentially as described in the legend to Fig. 3. (B) Immunoprecipitation analysis. Total RNA synthesized in the presence or absence of SAM was hybridized to end-labeled oligonucleotides complementary to regions in the GFP mRNA (nucleotides 73 to 94) and CAT mRNA (nucleotides 56 to 75). Following extension, the RNA-cDNA hybrids were immunoprecipitated essentially as described in the legend to Fig. 4. WT, wild type.
To examine whether the anti-cap antibody recognized unmethylated transcripts, we used the immunoprecipitation assay to compare transcripts produced in the presence and absence of SAM for the wild-type and NT-1C viruses. Figure 5B shows that transcripts generated from both the wild-type and NT-1C viruses in the presence of SAM were recognized by the antibody (lanes 2 and 4) while transcripts generated in the absence of SAM were inefficiently immunoprecipitated (lanes 6 and 8). As predicted, these data support the conclusion that the H-20 antibody does not bind to unmethylated transcripts but binds only to capped transcripts.
DISCUSSION
To study the role of the 5′ start sequence during VSV transcription, we used a reverse genetics system in which VSV was recovered from cDNA as a bipartite virus and one of the genomes encoded two reporter genes, GFP and CAT. This bipartite-virus system has several advantages over the other existing reverse genetic systems. First, this recombinant virus expresses all of the VSV proteins and does not require helper VSV proteins to be provided in trans for replication and transcription, as is necessary in the minivirus systems that have been used previously (5, 6, 22, 47, 48). Therefore, transcription, replication, and, ultimately, virus propagation are not limited by the efficiency of plasmid transfections. Second, while the vaccinia virus-T7 RNA polymerase system is used initially to recover the virus, the vaccinia virus is removed after the first passage since GFC/ΔG is self-propagating. This eliminates potential problems associated with the presence of cytoplasmic capping enzymes encoded by vaccinia virus, since the activities of these enzymes may mask the effects of mutations that affect VSV capping. Third, this system is especially suited for the analysis of the VSV transcription signals, since mutations can be made at the GFP-CAT gene junction without affecting the expression of essential VSV genes. Because high-titer supernatants and large quantities of the bipartite virus can be produced and purified, the effect of mutations can easily be examined both in vivo and in vitro.
The results of this study confirmed our previous conclusions that the first 3 nucleotides of the 5′ start sequence are critical for gene expression in vivo. Interestingly, mutations within the first 3 nucleotides of the start sequence did not always result in the same phenotype during in vitro transcription. The mutant viruses examined in this study fell into two groups when the amount of CAT mRNA generated in vitro was determined. The primary effect observed for the first group (NT-1G, NT-1A, and all third-position mutants) was on transcription initiation, since the amount of total CAT transcript generated in vitro was severely reduced compared to wild-type levels. In contrast, transcript initiation for the second group (NT-1C and all second-position mutants) ranged from 50% to near-wild-type levels. However, the majority of these mRNAs were not polyadenylated. These data show that the first 3 nucleotides of the start sequence play a role in transcript initiation as well as in postinitiation events during in vitro transcription and that specific nucleotide positions in the 5′ start sequence have differing effects on these events.
Further characterization of the mutant CAT transcripts lacking a poly(A) tail revealed that they did not contain the appropriate 5′ cap structure and were not fully elongated, suggesting that proper capping and/or methylation is important for polymerase processivity. The relative amount of mature CAT mRNA (i.e., capped, methylated, and polyadenylated) that was generated in vitro from each of the mutants correlated with the amount of CAT transcripts produced in vivo. Therefore, the results of the in vitro transcription assays probably parallel the transcriptional events that occur in vivo. For example, we predict that transcripts from the NT-1C and all NT-2 mutants are initiated in vivo but quickly become degraded due to the lack of an appropriate cap structure, the incomplete elongation, and the lack of a poly(A) tail. To our knowledge, this is the first time that the 5′ start sequence of a negative-strand RNA virus has been shown to play a role not only in transcript initiation but also in capping and/or methylation and polymerase processivity.
The data from our study may also explain the results obtained by Luk et al. (30), who used primer extension assays to map the start site of the L protein transcript in VSVNJ-infected cells, as well as after transcription in vitro. The G-L intergenic junction contains an extra 19 nucleotides following the conserved 3′-GA-5′ dinucleotide. Analysis of in vivo-generated transcripts indicated that only the consensus start sequence was used, whereas it appeared that in vitro-generated transcripts were initiated at two distinct sites, one following the intergenic dinucleotide at a nonconsensus start and one at the consensus start sequence. Data obtained with a GFC/ΔG virus that contains this variant intergenic region at the GFP-CAT gene junction indicated that transcripts that were initiated at a nonconsensus start sequence during in vitro transcription were not polyadenylated and were not detected in RNA samples isolated from infected cells (data not shown). Therefore, we speculate that during a wild-type VSVNJ infection, transcripts are initiated at the nonconsensus start site but they do not have the appropriate postinitiation modifications and are quickly degraded.
At this point, we have not been able to definitively determine whether the nucleotide changes in the 5′ start sequence affect the addition of the 5′-GpppA cap or if they affect only methylation. It has been previously documented for wild-type VSV that full-length, nonmethylated transcripts are synthesized during in vitro transcription reactions in the absence of a methyl donor (4, 19, 35). We have also shown here that in vitro transcription of wild-type GFC/ΔG proceeds normally in the absence of a methyl donor and that these transcripts are not efficiently recognized by an antibody specific for trimethyl guanosine. These data suggest that methylation is not required for synthesis of mature mRNAs; therefore, we favor the hypothesis that the major defect of the truncated CAT transcripts is the lack of a 5′-GpppA cap. However, in vitro transcription of two mutant GFC/ΔG viruses indicated that in the absence of a methyl donor, the VSV polymerase synthesizes slightly different amounts of full-length and truncated CAT transcripts. Possibly the nucleotide change in the 5′ start sequence in addition to the lack of a methyl donor further affects polymerase processivity. Experiments to determine if these truncated CAT transcripts are capped but not methylated are under way.
The second defect of these transcripts was that they were not fully elongated, indicating that mutations in the 5′ start sequence also affect the processivity of the polymerase. Whether this is a direct effect on polymerase processivity or a secondary effect resulting from incomplete modification of the 5′ end by capping and/or methylation requires additional research. Interestingly, the majority of the truncated CAT transcripts were heterogeneous in length, from 41 to less than 247 nucleotides, which corresponds to the first quarter of the CAT gene. These results suggest that the abortive elongation event probably occurs within a window and is not a random event occurring at all positions along the CAT gene (i.e., truncated transcripts do not range in size up to the length of the CAT gene). Presumably, transcripts that reach this critical length are completely elongated and subsequently polyadenylated.
The mechanism by which these mutations affect VSV polymerase processivity is not known, but there are various mechanisms by which transcriptional elongation is controlled during prokaryotic, eukaryotic, and viral transcription (52). The occurrence of mutations at the 5′ end of nascent VSV transcripts affecting downstream polymerase processivity is reminiscent of the Tat-TAR (Tat activation response element) interaction during human immunodeficiency virus (HIV) transcription. The interaction of the 5′ end of nascent HIV transcripts containing the TAR sequence with the HIV transactivating protein, Tat, increases polymerase processivity. Mutations in the TAR sequence alter the interaction with Tat and other cellular transcription elongation factors and ultimately affect the ability of the polymerase to efficiently elongate the transcript (26, 28, 29). In another example, the Gre-A and Gre-B proteins have been shown to stimulate a cleavage reaction in which 2 to 10 nucleotides are removed from the 3′ end of the nascent transcript (9). Presumably, this cleavage reaction prevents the formation of arrested transcription complexes and promotes transcript elongation (27). For respiratory syncytial virus, it was recently demonstrated that the M2 (ORF1) protein functions as a transcription elongation factor during transcription (12). While regulation of transcriptional elongation to control gene expression is commonly used, the effects on VSV polymerase processivity observed in this study are not indicative of a mechanism for regulation of transcript levels, since the mutations we have made have not been found in natural VSV isolates. Instead, the mechanism of abortive elongation may have evolved to prevent the synthesis of mRNAs that have not been properly modified at the 5′ end.
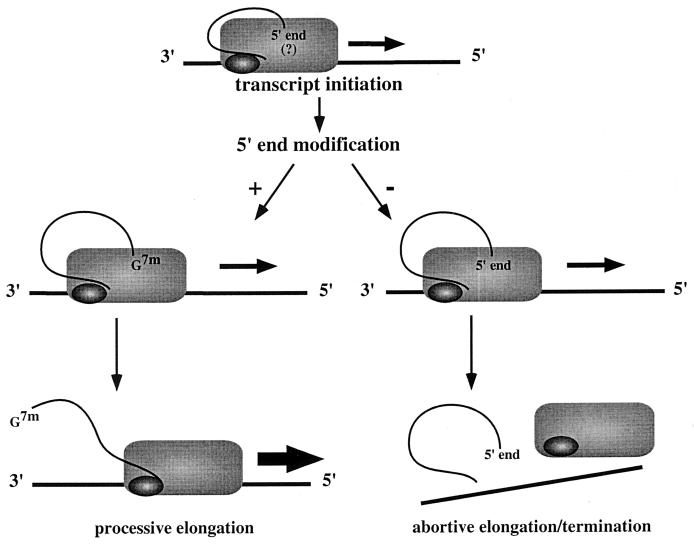
The findings of this study have provided new insights into the events that occur during VSV transcription initiation and elongation. One of the interesting observations is that the inability to correctly modify the 5′ end of the nascent transcript correlates with an increase in abortive elongation. One possible model to account for this observation is that following initiation the 5′ end of the nascent transcript maintains contact with the polymerase (Fig. 6). Modification of the 5′ end of the transcript may occur during transcription of the first 200 nucleotides. Once capping and methylation is completed, the 5′ end of the transcript is released, which may eliminate steric hindrance or induce a conformational change in the polymerase so that it can proceed along the template. In the start site mutants, the polymerase does not properly modify the 5′ end of the nascent transcript, and so the appropriate signals are not sent, which causes the polymerase to stall and terminate transcription prematurely.
FIG. 6.
Processive-elongation and abortive-elongation models. The encapsidated RNA template is shown as a horizontal line. The L and P proteins of the polymerase complex are represented by the rectangle and oval, respectively. Following transcript initiation, the polymerase generates the appropriate cap structure on the 5′ end of the nascent mRNA, thereby allowing processive elongation. Failure to appropriately modify the 5′ end results in abortive elongation, termination, and release of a truncated RNA.
A thorough understanding of the mechanism of VSV transcription requires an understanding of the spatially and temporally connected transcriptional events that occur at the intergenic junctions. Other than the cis-acting signals which direct these events, there are additional considerations that must be taken into account when generating models to describe VSV transcription. For example, how does the polymerase interact with an encapsidated helical template? Recently, it has been demonstrated that certain genetic elements must be positioned along the same face of the helical nucleocapsid at the 3′ end of the paramyxovirus simian virus 5 antigenome to generate a functional antigenomic promoter (32). Similar results have also been demonstrated for another paramyxovirus, Sendai virus, in which a hexamer motif, (GNNNNN)3, found at nucleotides 79 to 96 appears to be critical for both the genome and antigenome promoter function (50). While these results suggest that the helical nature of the encapsidated template affects how the polymerase interacts with the genome, the exact nature of the polymerase interaction with an encapsidated helical template remains very poorly understood. These studies, in addition to earlier experiments examining leader-polymerase interactions (23), indicate that the polymerase may contact a large area of the template and that downstream transcriptional events could therefore affect upstream events. The recent evidence that the cellular elongation factor, EF-1, is required for transcription in vitro (13) and the evidence that phosphorylation and multimerization states of the P protein modulate the transcriptional and replicative activities of the polymerase (33, 36) contribute to the emerging models of transcription for VSV as well as for other minus-strand RNA viruses.
ACKNOWLEDGMENTS
We are indebted to Reinhard Lührmann (Institut für Molekularbiologie und Tumorforschung, Universität Marburg) for the generous gift of the H-20 ascites fluid containing the antitrimethylguanosine specific monoclonal antibody, and we thank Daniel Kolakofsky (University of Geneva) for directing us to him. We also acknowledge Martin Chalfie (Columbia University) for providing the original GFP cDNA and Jack Rose (Yale University School of Medicine) for providing the GMMG-CAT plasmid. The technical assistance of Carolyn Matthews, Felicia Waller, Zorina Bowen, and Gipsy Majumdar is greatly appreciated. Oligonucleotides were synthesized by the Molecular Resource Center at UT—Memphis. The PhosphorImager was made available also through the Molecular Resource Center at UT—Memphis.
This work was supported by NIH grant GM-53726 (to M.A.W.).
REFERENCES
- 1.Abraham G, Rhodes D, Banerjee A K. 5′-terminal structure of methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell. 1975;5:51–58. doi: 10.1016/0092-8674(75)90091-4. [DOI] [PubMed] [Google Scholar]
- 2.Abraham G, Rhodes D, Banerjee A K. Novel initiation of RNA synthesis in vitro by vesicular stomatitis virus. Nature (London) 1975;255:37–40. doi: 10.1038/255037a0. [DOI] [PubMed] [Google Scholar]
- 3.Ball L A, White C N. Order of transcription of genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee A K. 5′-terminal cap structure in eukaryotic messenger ribonucleic acids. Microbiol Rev. 1980;44:175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr J N, Whelan S P, Wertz G W. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barr J N, Whelan S P, Wertz G W. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J Virol. 1997;71:1794–1801. doi: 10.1128/jvi.71.3.1794-1801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisaillon M, Lemay G. Viral and cellular enzymes involved in synthesis of mRNA cap structure. Virology. 1997;236:1–7. doi: 10.1006/viro.1997.8698. [DOI] [PubMed] [Google Scholar]
- 8.Bochnig P, Reuter R, Bringmann P, Luhrmann R. A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. Eur J Biochem. 1987;168:461–467. doi: 10.1111/j.1432-1033.1987.tb13439.x. [DOI] [PubMed] [Google Scholar]
- 9.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 10.Chanda P K, Banerjee A K. Identification of promoter-proximal oligonucleotides and a unique dinucleotide, pppGpC, from in vitro transcription products of vesicular stomatitis virus. J Virol. 1981;39:93–103. doi: 10.1128/jvi.39.1.93-103.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das T, Mathur M, Gupta A K, Janssen G M, Banerjee A K. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 alphabetagamma for its activity. Proc Natl Acad Sci USA. 1998;95:1449–1454. doi: 10.1073/pnas.95.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredericksen B L, Whitt M A. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J Virol. 1995;69:1435–1443. doi: 10.1128/jvi.69.3.1435-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin D, Kolakofsky D. A novel mechanism for the initiation of Tacaribe arenavirus genome replication. J Virol. 1990;64:6196–6203. doi: 10.1128/jvi.64.12.6196-6203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hefti E, Bishop D H L. The 5′ sequence of VSV in vitro transcription product RNA (+/− SAM) Biochem Biophys Res Commun. 1976;68:393–400. doi: 10.1016/0006-291x(76)91158-x. [DOI] [PubMed] [Google Scholar]
- 17.Hercyk N, Horikami S M, Moyer S A. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology. 1988;163:222–225. doi: 10.1016/0042-6822(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 18.Horikami S M, De Ferra F, Moyer S A. Characterization of the infection of permissive and nonpermissive cells by host range mutants of vesicular stomatitis virus defective in RNA methylation. Virology. 1984;138:1–15. doi: 10.1016/0042-6822(84)90142-9. [DOI] [PubMed] [Google Scholar]
- 19.Horikami S M, Moyer S A. Host range mutants of vesicular stomatitis virus defective in in vitro RNA methylation. Proc Natl Acad Sci USA. 1982;79:7694–7698. doi: 10.1073/pnas.79.24.7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt D M. Effect of analogues of S-adenosylmethionine on in vitro polyadenylation by vesicular stomatitis virus. J Gen Virol. 1989;70:535–542. doi: 10.1099/0022-1317-70-3-535. [DOI] [PubMed] [Google Scholar]
- 21.Hunt D M, Mehta R, Hutchinson K L. The L protein of vesicular stomatitis virus modulates the response of the polyadenylic acid polymerase to S-adenosylhomocysteine. J Gen Virol. 1988;69:2555–2561. doi: 10.1099/0022-1317-69-10-2555. [DOI] [PubMed] [Google Scholar]
- 22.Hwang L N, Englund N, Pattnaik A K. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J Virol. 1998;72:1805–1813. doi: 10.1128/jvi.72.3.1805-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isaac C L, Keene J D. RNA polymerase-associated interactions near template promoter sequences of defective interfering particles of vesicular stomatitis virus. J Virol. 1982;43:241–249. doi: 10.1128/jvi.43.1.241-249.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iverson L E, Rose J K. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 25.Iverson L E, Rose J K. Sequential synthesis of 5′-proximal vesicular stomatitis virus mRNA sequences. J Virol. 1982;44:356–365. doi: 10.1128/jvi.44.1.356-365.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao S Y, Calman A F, Luciw P A, Peterlin B M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 27.Kassavetis G A, Geiduschek E P. RNA polymerase marching backward. Science. 1993;259:944–945. doi: 10.1126/science.7679800. [DOI] [PubMed] [Google Scholar]
- 28.Kato H, Sumimoto H, Pognonec P, Chen C H, Rosen C A, Roeder R G. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 1992;6:655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- 29.Laspia M F, Rice A P, Mathews M B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- 30.Luk D, Masters P S, Gill D S, Banerjee A K. Intergenic sequences of the vesicular stomatitis virus genome (New Jersey serotype): evidence for two transcription initiation sites within the L gene. Virology. 1987;160:88–94. doi: 10.1016/0042-6822(87)90048-1. [DOI] [PubMed] [Google Scholar]
- 31.Moyer S A, Banerjee A K. In vivo methylation of vesicular stomatitis virus and its host cell messenger RNA species. Virology. 1976;70:339–351. doi: 10.1016/0042-6822(76)90276-2. [DOI] [PubMed] [Google Scholar]
- 32.Murphy S K, Ito Y, Parks G D. A functional antigenomic promoter for the paramyxovirus simian virus 5 requires proper spacing between an essential internal segment and the 3′ terminus. J Virol. 1998;72:10–19. doi: 10.1128/jvi.72.1.10-19.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pattnaik A K, Hwang L, Li T, Englund N, Mathur M, Das T, Banerjee A K. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J Virol. 1997;71:8167–8175. doi: 10.1128/jvi.71.11.8167-8175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peluso R W, Richardson J C, Talon J, Lock M. Identification of a set of proteins (C′ and C) encoded by the bicistronic P gene of the Indiana serotype of vesicular stomatitis virus and analysis of their effect on transcription by the viral RNA polymerase. Virology. 1996;218:335–342. doi: 10.1006/viro.1996.0202. [DOI] [PubMed] [Google Scholar]
- 35.Rhodes D, Banerjee A K. 5′-terminal sequence of vesicular stomatitis virus genome RNA. J Virol. 1976;17:33–42. doi: 10.1128/jvi.17.1.33-42.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson J C, Peluso R W. Inhibition of VSV genome RNA replication but not transcription by monoclonal antibodies specific for the viral P protein. Virology. 1996;216:26–34. doi: 10.1006/viro.1996.0031. [DOI] [PubMed] [Google Scholar]
- 37.Robison, C. Personal communication.
- 38.Rose J K. Complete intergenic and flanking gene sequences from the genome of vesicular stomatitis virus. Cell. 1980;19:415–421. doi: 10.1016/0092-8674(80)90515-2. [DOI] [PubMed] [Google Scholar]
- 39.Rose J K. Heterogeneous 5′-terminal structures occur on vesicular stomatitis virus mRNAs. J Biol Chem. 1975;250:8098–8104. [PubMed] [Google Scholar]
- 40.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 41.Rose J K, Lodish H F, Brock M L. Giant heterogeneous polyadenylic acid on vesicular stomatitis virus mRNA synthesized in the presence of S-adenosylhomocysteine. J Virol. 1977;21:683–693. doi: 10.1128/jvi.21.2.683-693.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnell M J, Buonocore L, Whitt M A, Rose J K. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J Virol. 1996;70:2318–2323. doi: 10.1128/jvi.70.4.2318-2323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schubert M, Keene J D, Herman R C, Lazzarini R A. Site on the vesicular stomatitis virus genome specifying polyadenylation at the end of the L gene. J Virol. 1980;34:550–559. doi: 10.1128/jvi.34.2.550-559.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shuman S. A proposed mechanism of mRNA synthesis and capping by vesicular stomatitis virus. Virology. 1997;227:1–6. doi: 10.1006/viro.1996.8305. [DOI] [PubMed] [Google Scholar]
- 45.Shuman S, Schwer B. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 46.Stillman E A, Rose J K, Whitt M A. Replication and amplification of novel vesicular stomatitis virus minigenomes encoding viral structural proteins. J Virol. 1995;69:2946–2953. doi: 10.1128/jvi.69.5.2946-2953.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stillman E A, Whitt M A. The length and sequence composition of vesicular stomatitis virus intergenic regions affect mRNA levels and the site of transcript initiation. J Virol. 1998;72:5565–5572. doi: 10.1128/jvi.72.7.5565-5572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stillman E A, Whitt M A. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takada A, Robison C, Goto H, Sanchez A, Murti K G, Whitt M A, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tapparel C, Maurice D, Roux L. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol. 1998;72:3117–3128. doi: 10.1128/jvi.72.4.3117-3128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toneguzzo F, Ghosh H P. Characterization and translation of methylated and unmethylated vesicular stomatitis virus mRNA synthesized in vitro by ribonucleoprotein particles from vesicular stomatitis virus-infected cells. J Virol. 1976;17:477–491. doi: 10.1128/jvi.17.2.477-491.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright S. Regulation of eukaryotic gene expression by transcriptional attenuation. Mol Biol Cell. 1993;4:661–668. doi: 10.1091/mbc.4.7.661. [DOI] [PMC free article] [PubMed] [Google Scholar]