Abstract
Background:
Genome-wide association studies have discovered blocks of common variants—likely transcriptional-regulatory—associated with major depressive disorder (MDD), though the functional subset and their biological impacts remain unknown. Likewise, why depression occurs in females more frequently than males is unclear. We therefore tested the hypothesis that risk-associated functional variants interact with sex and produce greater impact in female brains.
Methods:
We developed techniques to directly measure regulatory variant activity and sex interactions using massively parallel reporter assays (MPRAs) in the mouse brain in vivo, in a cell type-specific manner, and applied these approaches to measure activity of >1,000 variants from >30 MDD loci.
Results:
We identified extensive sex-by-allele effects in mature hippocampal neurons, suggesting sex-differentiated impacts of genetic risk may underlie sex bias in disease. Unbiased informatics approaches indicated that functional MDD variants recurrently disrupt a number of transcription factor binding motifs, including those of sex hormone receptors. We confirmed a role for the latter by performing MPRAs in neonatal mice on the day of birth (during a sex-differentiating hormone surge) and hormonally-quiescent juveniles.
Conclusions:
Our study provides novel insights into the influence of age, biological sex, and cell type on regulatory variant function, and provides a framework for in vivo parallel assays to functionally define interactions between organismal variables like sex and regulatory variation. Moreover, we experimentally demonstrate that a portion the sex differences seen in MDD occurrence may be a product of sex-differentiated effects at associated regulatory variants.
Keywords: Major depressive disorder, sex differences, reporter assay, hippocampus, transcription factors, noncoding variation
One-Sentence Summary:
Massively parallel assays in vivo identified extensive functional and sex-interacting common variants in depression risk loci.
Introduction
Major depressive disorder (MDD), a profoundly disruptive and sometimes lethal disorder, affects women 2–3 times more frequently than men globally (1). Sex differences are pervasive in MDD, from symptom profiles (2) and effective drug classes (3) to brain-wide gene expression (4, 5). Genome-wide association studies (GWASes) have associated dozens of linkage regions of numerous single-nucleotide polymorphisms (SNPs) with MDD, demonstrating its heritability (6–8). Sex-by-genotype (SxG) analyses of GWASes have revealed that MDD risk loci are the same for men and women, yet these loci explain up to 4-fold greater MDD heritability in females (9, 10). Accordingly, sex-stratified MDD GWAS in UK Biobank identified significantly greater female than male overlap of risk variants with those influencing hippocampal gene expression (11). These findings suggest that sex and perhaps brain region interact with a common pool of SNPs in conferring MDD risk, motivating direct functional genomics testing of these hypotheses.
Disease-associated SNPs are seldom found in protein-coding space, complicating prediction of their molecular consequences. Instead, these SNPs are found in probable transcriptional regulatory elements (REs) predicted from measures such as chromatin marks, accessibility, and conformation (12–14). MDD-associated loci and putative target genes are enriched for such measures in specific brain regions and cell types, including the hippocampus (7, 15) and excitatory neurons (16–22), suggesting sites of action for these REs. Notably, the hippocampus has been a long-standing structure of interest in MDD given its loss of volume in patients (23). Finally, sex influences the biology of the hippocampus throughout the lifespan (24, 25), including its gene expression (4) and volume (26) in MDD.
Identifying functional SNPs from MDD-associated regions is the first step toward understanding how risk genotypes perturb brain physiology, ultimately via improved inference of dysregulated target genes and shared, cross-locus regulatory programs. However, studies connecting MDD-associated SNPs to altered gene regulation, even those considering sex effects (27), have been limited to indirect functional indicators (e.g., chromatin state), or are confounded by linkage disequilibrium (e.g., expression quantitative trait loci (eQTLs)). Recently, Massively Parallel Reporter Assays (MPRAs)—methods for functionally measuring activity of thousands of REs and variants simultaneously—have enabled large-scale in vitro identification of functional regulatory variants. MPRAs, like traditional reporter assays, place REs upstream of an optical reporter (e.g., luciferase), but instead quantify many REs’ activity simultaneously through RNA-sequencing of unique, RE-identifying “barcode” sequences in the reporter’s 3’ untranslated region. MPRAs have identified trait- and disease-associated SNPs affecting REs in disease-relevant cell types in vitro (28–31). However, the complexities of brain cell types and the sex hormonal milieu cannot readily be emulated ex vivo.
To overcome existing limitations in functional regulatory SNP (rSNP) identification, we delivered an MPRA library of MDD-associated variants (32) into the adult mouse hippocampus in vivo. Building on prior brain MPRAs of REs and a variant (33, 34), our approach greatly extends in vivo MPRA methods to identify rSNPs, their sex interactions, and cell type-specific effects. Hippocampal MPRA was combined with translating ribosome affinity purification (TRAP) to simultaneously identify MDD rSNPs in excitatory neurons and the broader hippocampus. Further, we test the hypothesis that rSNPs are subject to sex-by-genotype (SxG) interactions using mice of both sexes. Finally, to characterize the potential role of circulating hormones in sex-differentiated rSNP activity, and to functionally replicate predicted fetal brain RE enrichments at MDD SNPs (19, 35–38), we likewise delivered the library to the mouse brain in utero. Thus, we additionally identify rSNPs neonatally, coinciding with a testosterone surge and critical period for establishing sex-specific brain circuitry (39), and observe loss of SxG effects in juveniles, when hormonal influences are quiescent. rSNP-enriched transcription factor (TF) motif disruptions identified cross-locus ‘predicted altered regulation’ TFs (parTFs), which—along with their shared regulators—consistently implicated sex hormone receptors in MDD rSNP regulatory networks. Our work illustrates that MPRAs can be leveraged in vivo to directly identify not only functional variants, but their context-dependence on age, sex, and cell type. Moreover, we experimentally demonstrate that sex interacts with MDD-associated functional variants, providing a potential genetic mechanism to explain observations of shared risk loci between sexes (9, 10) despite differing degrees of effect on MDD incidence.
Methods
MPRA library design
The MPRA library covered 40 GWAS loci (~1,000 SNPs) in LD R2>0.1 with MDD-associated tag variants. SNPs were prioritized by their overlap with human brain and neural cell type eQTLs, histone marks, enhancer RNA overlap, and chromatin contacts ((32); Data Availability; Supplementary Methods). 966 SNPs were from 35 GWAS loci for MDD (6, 7, 38, 40–45) or related traits (46–49). SNP-centered 126bp genomic sequences for each allele were designed with 10 unique 10bp barcodes for internal replication (Fig. S1). These were inserted into an AAV plasmid, then cloned to contain an hsp68 minimal promoter (50) driving dsRed along with the “woodchuck” hepatitis 3’UTR element to improve recovery of reporter RNAs (51), and packaged into AAV9.
AAV9 delivery and TRAP
The AAV9 library was delivered bilaterally into the hippocampus of Vglut1-TRAP mice ages P60-P80 (n=6 per sex), followed by hippocampal dissection and TRAP (Fig. 1A; Suppplementary Methods) 28 days later to identify rSNPs and their shared regulatory features (Fig. 1B). Immunofluorescence (IF) of hippocampi from additional mice confirmed robust dsRed expression 28 days after injection (Fig. 1C). qPCR confirmed that TRAP RNA was depleted of other cell type markers, the main expectation for TRAP from an abundant cell type (52) (Fig. 1D; Supplementary Results). After quality control, 5 samples per tissue fraction and sex were retained for MPRA analysis covering ~1,000 SNPs (Supplementary Methods).
Figure 1. Experimental design, analysis plan, and quality control: adult mouse hippocampus and its excitatory neurons.
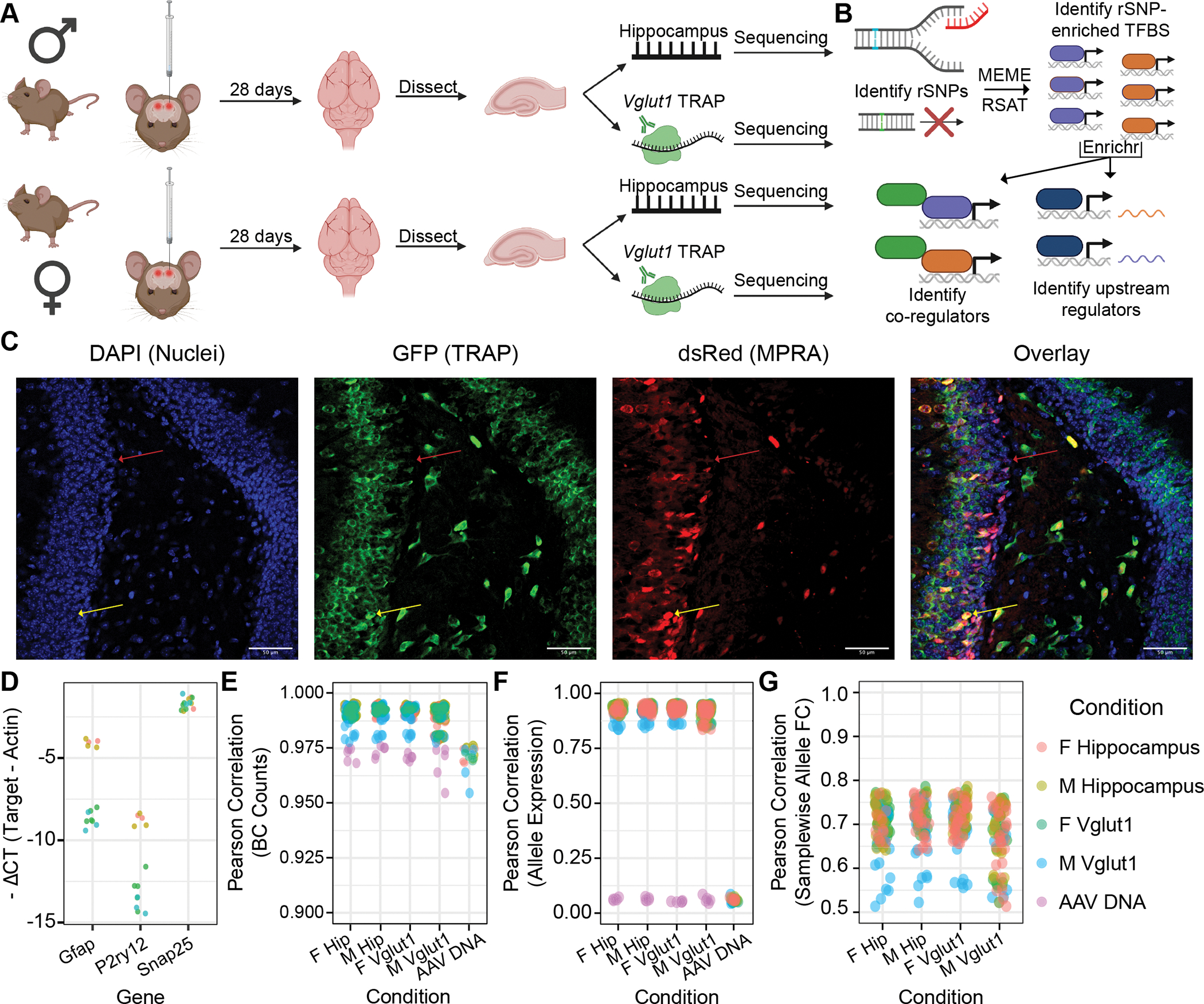
(A) Adult male and female C57BL/6J mice received bilateral stereotactic injections into the hippocampus delivering the AAV9-packaged MPRA. TRAP yielded two RNA fractions per sample: “input” (total hippocampal) and TRAP (Vglut1+). (B) Analyses identified regulatory SNPs (rSNPs), transcription factor (TF) binding sites (TFBSes) enriched at rSNPs, shared protein interactors among these TFs, and shared regulators of these TFs’ expression. (C) IF of Vglut1-TRAP mouse hippocampus 28 days after MPRA-AAV9 delivery, illustrating strong TRAP (GFP) co-expression with dsRed reporter, confirming RNA from the latter is present in the cell type of interest. (D) qPCR confirmed depletion of glial genes (Gfap, P2ry12) and modest enrichment of excitatory neuron marker Gria1 in Vglut1+ RNA. (E) Barcode count correlations between replicates. Each point represents the cross-correlation between one sample of the type on the x-axis and one of the color-coded type. (F) Correlation of mean barcode expression between replicates. (G) Correlation of samplewise allelic differences in expression. PCC: Pearson correlation coefficient (or Pearson’s r).
MPRA data analysis
We assessed SNPs for allelic effects in each individual sex and RNA fraction, and separately for sex-allele interactions in joint-sex datasets, using linear mixed models (LMMs) with barcode as a random effect on expression (Supplementary Methods; Fig. S2–S4). Empirical p values (pemp) for model coefficient(s) were calculated using the appropriate allele or interaction model with 50,000 simulated ‘allelic’ comparisons of random subsets of minimal promoter-coupled barcodes (32, 53) to account for technical and barcode-mediated noise. This approach is not confounded by sex differences, as promoter activity was not sex-differential within age/tissue groups (all t-tests of barcode expression p>0.1; Figs. S5–S6). Except where noted, significance was called at FDR-corrected pemp<0.2 (plotted alongside default LMM p-derived FDRs for each experiment in Fig. S7), a stringency comparable to a recent study of sex-interacting eQTLs (27).
Motif enrichment and parTF analysis with Enrichr
We assessed motif disruptions using motifbreakR (54) and RSAT var-tools (55) (Supplementary Methods) for rSNP sets with allelic (single-sex) or SxG pemp <0.05. Enrichment was tested against assayed SNPs without allelic effects (pemp > 0.05); SxG rSNP sets were also tested against rSNPs with only main (allele) effects. Motifs perturbed by ≥4 rSNPs (for P10, ≥5) with enrichment FDR<0.05 were filtered to cognate TFs expressed in Genotype-Tissue Expression atlas (v8) hippocampus (adult assays) or midfetal human brain (P0) (56) from the corresponding sex.
Results
rSNPs, sex interactions, and their regulatory landscapes: adult hippocampus and excitatory neurons
Given molecular and structural associations of MDD with the hippocampus, we sought to identify mouse hippocampal rSNPs from broad, MDD-associated linkage disequilibrium (LD) regions. First, a feasibility experiment using TRAP to add cell type-specific information was performed with a mini-MPRA library of neuron- and glia-specific REs, delivered as AAV9 to Snap25-TRAP mice, validating reproducibility and specificity (Supplementary Results, Fig S8–S9). We then delivered the AAV9 MDD MPRA library stereotaxically to hippocampus in Slc17a7 (Vglut1) TRAP mice. 28 days later, input (hippocampus) and TRAP RNA (Vglut1+) from each sample was collected and sequenced (Table S1, Supplementary Methods). Sequencing was replicable at levels of barcode counts (pairwise Pearson’s r 0.95–0.99) (Fig. 1E), RE-level expression (r 0.85–0.96) (Fig. 1F), and mean allelic fold changes (log2FCs) between conditions (r 0.90–0.94) (Fig. 1G), confirming we were able to reliably measure SNP-mediated regulatory effects and differences from a defined cell type in vivo.
rSNPs in total hippocampus and hippocampal Vglut1+ neurons
We first assessed ~1,000 SNPs for allelic effects in each individual sex and RNA fraction. In total hippocampus, we identified 36 (male) and 31 (female) rSNPs, 34 and 31 of which were from MDD loci, respectively. While male and female total hippocampi had similar numbers of rSNPs, we observed a striking sex difference in the number of rSNPs in Vglut1+ cells: 58 (female) vs. 7 (male, all likewise female rSNPs; Fig. S10), indicating that a higher proportion of MDD SNPs have discernible allelic effects in excitatory neurons of females. Female Vglut1+ and total hippocampal SNP effects were consistent in magnitude and direction with an additional (n=3) TRAP cohort (Pearson’s r 0.61–0.77) (Supplementary Results, Fig. S11, Data S1). Notable rSNPs from ≥1 condition included rs2563323 and rs250427, putative brain and hippocampal (57) eQTLs for SRA1, a noncoding RNA which activates nuclear receptors without ligand (58). Also notable were rs301806—an MDD GWAS index SNP (41)—and rs301807 (Fig. 2A–B), both of which likely regulate the nearby gene RERE (19, 59). Allele betas (log2FC) and significance at 10 different thresholds are provided (Data S2).
Figure 2. Adult hippocampus rSNPs and complex context-dependent, polygenic architecture of the RSRC1 locus.
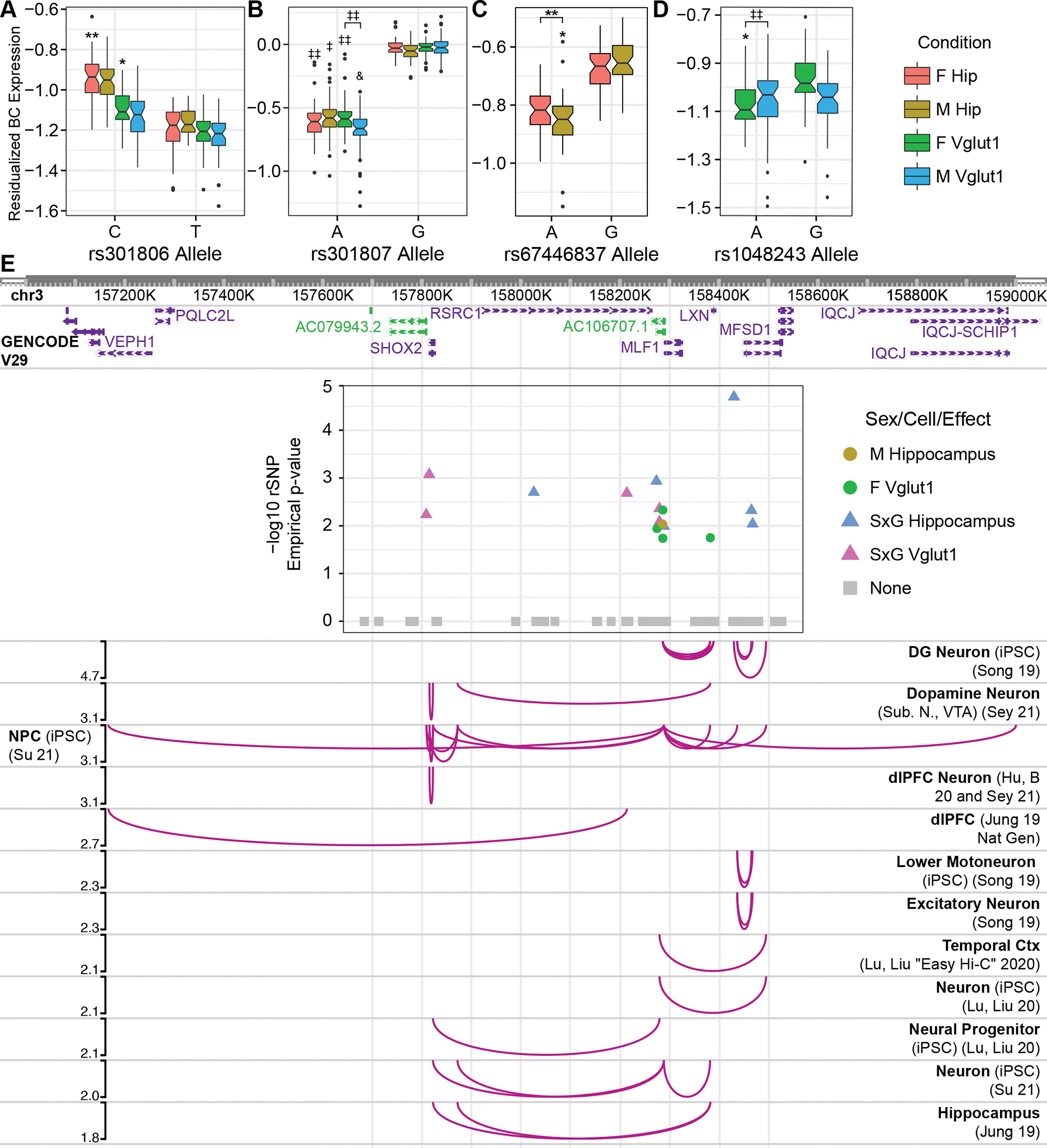
Boxplots show single-barcode (BC) expression levels adjusted for random effects across analyzed replicates. Center bars: median; boxes: 25–75% quantile; whiskers: observations spanning box edges to ± 1.5*interquantile range (IQR); single points: observed values outside whisker range. Notched regions span ± 1.58 * IQR / sqrt (n measurements), approximating the 95% confidence interval for comparing median BC expression (84). rSNPs corresponding to RERE locus (38) tag variants (A) rs301806 and (B) rs301807 are shown. (C, D) Sex-interacting SNPs from the RSRC1 locus (40) included (C) rs67446837 and (D) rs1048243. (E) rSNPs identified in the RSRC1 locus and their regulatory target genes in human tissues ascertained by Hi-C. A plot of rSNP effects, colored by most significant condition, is embedded with its x-axis in human genomic (hg19) coordinates; chromatin contacts between the SNPs and distal gene promoters are illustrated below (60–64). Curve heights correspond to −log10pemp for the plotted rSNP. iPSC: Induced pluripotent stem cell; DG: dentate gyrus; Ctx: cortex. *: pemp-derived FDR < 0.25; **: <0.2; ‡ < 0.15; ‡‡ < 0.1; & < 0.05.
Sex-interacting rSNPs in hippocampus
Given the role of sex in MDD risk and the observed differences in rSNP activity in the adult hippocampus, we next investigated whether sex interacts with MDD risk genotypes, identifying 41 sex-allele interaction rSNPs in each tissue. We confirmed that sex differences in replicability did not drive Vglut1+ results by repeating single-sex analyses without the largest outlier per sex. This identified 30 and 142 rSNPs for male and female Vglut1+, respectively; SxG analysis without the male outlier yielded 23 SxG rSNPs (Data S3).
While only 1 SxG rSNP specifically was shared between total hippocampus and Vglut1+, 16/18 Vglut1+ SxG loci contained hippocampal SxG rSNPs. The tag locus rs1193510 (40), for example, contained 11 sex-interacting rSNPs, including several unique to hippocampus (Fig. 2C) or Vglut1+ (Fig. 2D). This locus is rich in neural chromatin contacts (60–65), implicating several rSNP target genes. Interestingly, SxG effects in hippocampus and Vglut1+ segregated into distinct portions of this LD region (Fig. 2E, see legend). Also notable was a near-significant SxG effect in Vglut1+ for rs2400075 (0.20<FDR<0.25), recently reported significant in SxG GWASes of several body traits (66).
Transcriptional-regulatory systems across hippocampal rSNPs and SxG rSNPs
To test for common transcriptional-regulatory mechanisms underlying hippocampal rSNP effects, enrichment analysis was performed to identify TF binding motifs perturbed more frequently by rSNPs than expected by chance (Data S4A) (32). To ensure adequate depth for enrichment analysis, a relaxed significance threshold of pemp < 0.05 was used, identifying dozens of parTFs for both male and female hippocampus (Fig. 3A; Supplementary Results). To gain network-level insights into these cross-variant effectors, we examined known interactors and upstream regulators of the parTFs using Enrichr (Data S4B–C; Supplementary Results). Among Vglut1+ rSNPs, many parTFs were shared (e.g., DLX1, POU3F1/2/3; Fig. 3B). POU3F2 has been previously shown to be a highly centralized, cross-disorder hub gene in postmortem brain co-expression analysis by PsychENCODE (67). Additionally, parTFs from male glutamatergic neurons were enriched in four known protein-protein interaction networks: SMAD3 and SMAD4 (5/16 parTFs each), consistent with neuronal enrichment by TRAP, and more surprisingly, sex hormone receptors: estrogen receptor α (ESR1; 4/16) and AR (3/16).
Figure 3. Shared regulatory architecture of rSNPs by sex, cell type, and sex-interacting SNP type.
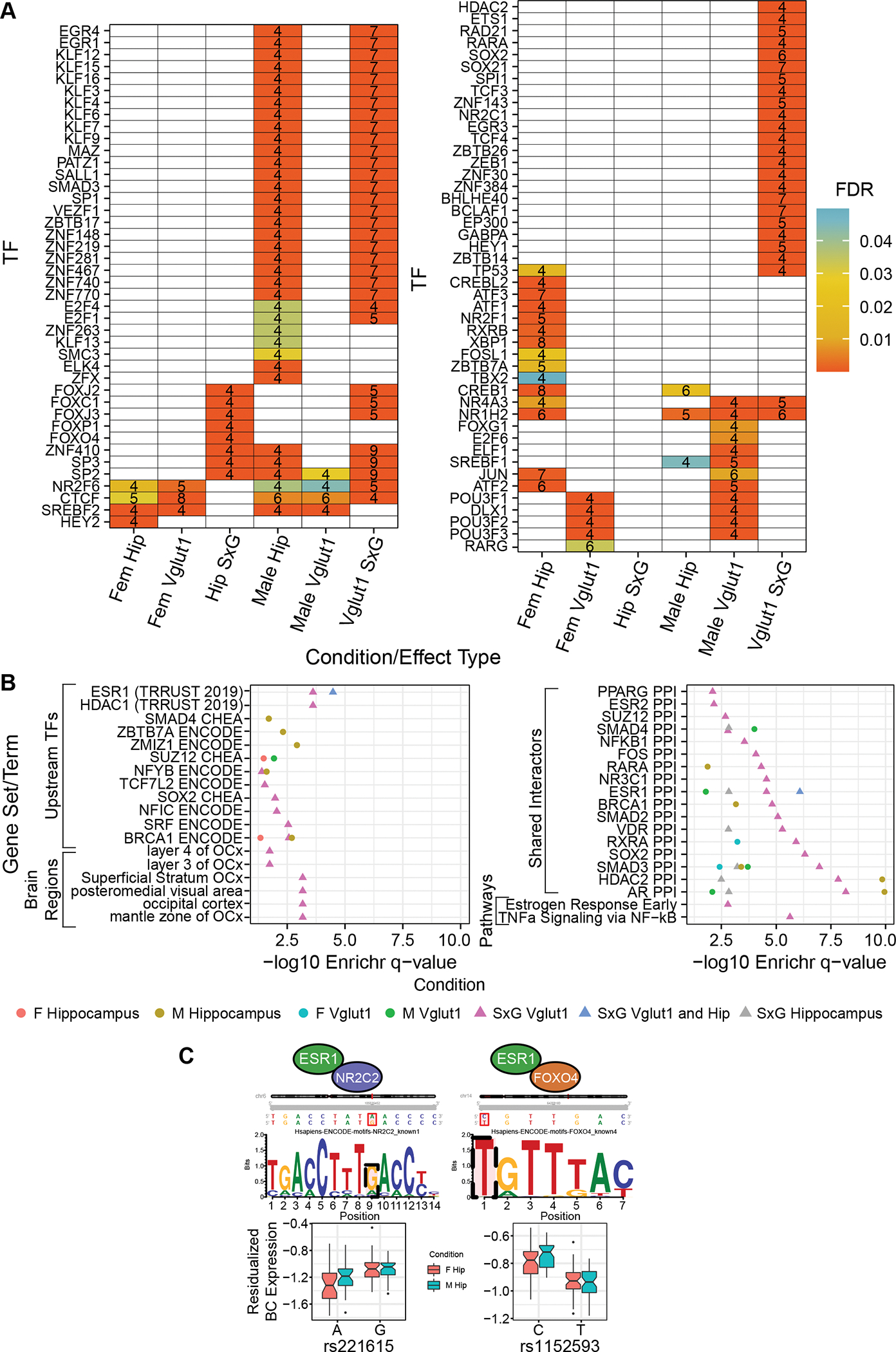
(A) TFs with binding motif disruptions by ≥ 4 nominally significant (empirical p < 0.05) rSNPs or sex-interacting rSNPs, enriched relative to nonfunctional SNPs. Number of rSNPs associated to a given binding site are shown. (B) Terms from Enrichr (85) analysis, identifying shared upstream regulators (TFs controlling expression of several of the TFs in panel A), brain regions enriched for expression of the rSNP-enriched TFs, protein interactors enriched among rSNP-enriched TFs, and MSigDB pathway term enrichment for rSNP-enriched TF sets. Bolded enrichments are discussed in the text.
Using a similar approach to that above, we investigated parTFs enriched at sex-allele interaction rSNPs (pemp<0.05) (Fig. 3A). We identified only 8 parTFs enriched among total hippocampal SxG rSNPs (ZNF410 and five FOX TFs with shared motifs). However, we identified 57 parTFs at glutamatergic SxG rSNPs, including six of those from total hippocampus, suggesting coherent regulatory dynamics spanning MDD loci in this cell type. SxG parTFs included several KLF members, nuclear receptors (NR4A3, NR2C1, NR1H2), and the retinoid receptor RARA.
Enrichr analysis of the Vglut1+ SxG parTFs revealed striking enrichments, including for parTF upstream regulation by CREB1 (15/57), PPIs with AR (11/57), ESR1 (11/57), and HDAC2 (12/57) (Fig. 3B), and the MsigDB term “estrogen response early” (5/57). (The small hippocampal SxG parTF set yielded no notable Enrichr results.) These Vglut1+ SxG regulators implicate sex hormones and histone acetylation in both establishing sex-divergent MDD risk from upstream, e.g., via ESR1 (5/57), and actuating it downstream via rSNP-enriched parTFs and their protein interactors (AR, ESR1, HDAC2, ZBTB7A). Both sex hormones (68) and histone (de-)acetylases (69) have been highlighted in reviews of sex differences in MDD.
Identification of rSNPs in developing whole mouse brain
Given the role of hormones in sexual differentiation of the brain (39), we investigated whether MDD risk variants were subject to sex-differential regulation during vs. after the physiologic perinatal testosterone surge. This surge—exerting acute activational and long-lasting organizational effects (39)—is underway at P0, while sex hormones are effectively absent by P10. Together, these two timepoints allowed assessment of activational hormone effects on MDD SNPs. The AAV9 library was thus delivered intracerebroventricularly to embryonic day 15 (E15) mice before collection at postnatal day 0 (P0) or P10 (Supplementary Methods).
IF verified that dsRed expression was detectable at P0 and P10 following in utero delivery despite the short incubation time (Fig. 4A–D; Fig. S12–S13), consistent with prior reports (70). We subsequently collected whole brains (except cerebellum) for RNA isolation and MPRA sequencing. DNA from n=4 brain samples was sequenced to verify consistent distribution of delivered MPRA barcodes between replicates (r 0.86–0.94) and compared to input virus (r 0.88–0.91) (Fig. 4E). n=15 P0 (6 female/9 male) and n=13 P10 (6 female/7 male) RNA samples were retained for analysis (Supplementary Methods). Replicates from each condition had well-correlated barcode counts (PCC 0.86–0.98) and RE expression values (PCC 0.58–0.96) (Fig. 4F–H).
Figure 4. Validating the in utero MPRA delivery method, and identification of rSNPs and sex-interacting rSNPs in the developing brain.
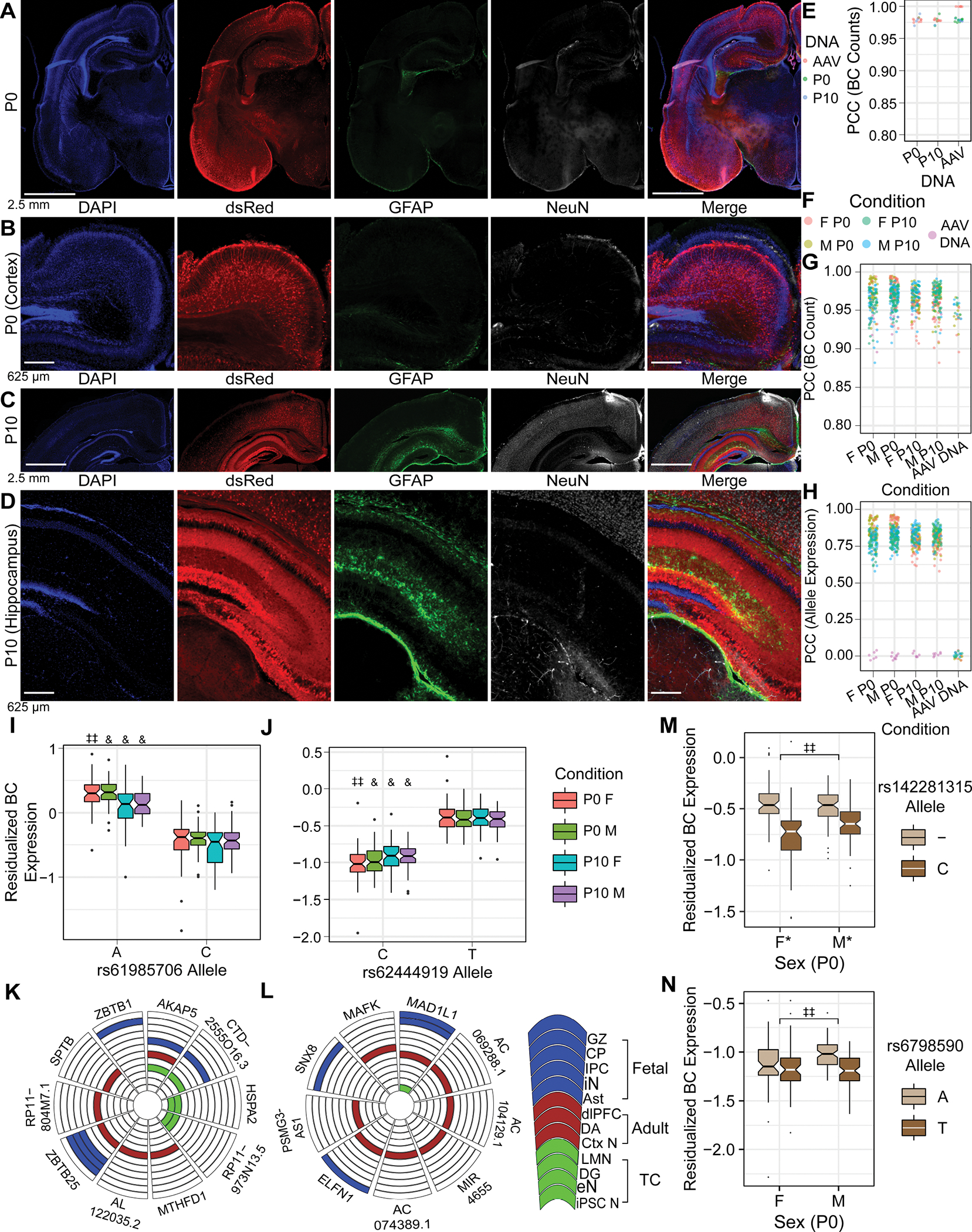
(A, B) IF of P0 brain after E15 MPRA-AAV delivery. (C, D) IF of P10 brain after MPRA-AAV delivery. (E) Comparability of barcode counts in recovered brain DNA and original AAV. (F) Color legend for panels G-H. (G) BC count correlation between samples. (H) Sequence expression correlation between samples. (I, J) rSNPs (I) rs61985706 and (J) rs62444919 showed effects consistent across sexes and ages. (K, L) Putative target genes of the respective rSNPs from Hi-C in human fetal, adult, and cultured neural tissues. (M) Example P0 SxG SNP with comparatively small sex difference in allele effect size. (N) Example P0 SxG SNP with magnitude of sex difference in allelic effect comparable to smaller (female) allelic effect itself. GZ: germinal zone; CP: cortical plate (86); IPC: intermediate progenitor cell; iN: inhibitory neuron (19); Ast: astrocyte (61); dlPFC: dorsolateral prefrontal cortex; DA: dopamine neurons of substantia nigra and ventral tegmental area (65); Ctx N: cortical neuron (60); LMN: lower motor neuron; eN: excitatory neuron (61); iPSC N: iPSC-derived neuron (62). *: pemp-derived FDR < 0.25; **: <0.2; ‡ < 0.15; ‡‡ < 0.1; & < 0.05.
At P0, we identified 5 rSNPs in females and 12 rSNPs in males, respectively, four of which were shared between sexes with concordant effect direction. By contrast, there were 105 female and 72 male rSNPs at P10, with 42 rSNPs identified in both sexes (all concordant). Three rSNPs shared significance and effect direction across all four age-sex conditions (two illustrated, Fig. 4I, J). These rSNPs also have rich chromatin contact evidence supporting gene regulatory roles in fetal, adult, and cultured human neural cell types (Fig. 4K and 4L, respectively), consistent with their detection as rSNPs in whole brain tissue and highlighting the robustness of in utero MPRA delivery for neurodevelopmental functional-regulatory assays.
To test our hypothesis of sex hormone-dependent variant effects, SxG models were performed for P0 and P10. At P0, we identified 31 SxG rSNPs (e.g., Fig. 4M–N). eQTLs from human whole brain (56) during the midfetal testosterone surge (71) significantly overlapped the union of SxG, P0 female, and P0 male rSNPs (33/43 rSNPs; OR=1.19, p < 0.011, Fisher’s exact test). By contrast, we identified no SxG interactions at P10 (pemp FDR<0.25). This finding held when repeating the SxG analysis with the least variable 5 samples per sex (despite similar n and replicability to P0) (Data S5). These findings further support the role of sex hormones in shaping rSNP effects suggested by our adult parTF/Enrichr analyses.
Postnatal whole-brain transcriptional-regulatory systems shared across postnatal whole brain rSNPs
67 parTFs were identified across the four age-sex conditions (Fig 5A, Supplementary Results) and analyzed with Enrichr (Fig 5B; Supplementary Results). Notably, male P10 parTFs were enriched for interactors of ZBTB7A and RARG; retinoids have been implicated in local sex hormone synthesis in the brain at this age (75). Moreover, 6 P0 parTFs are sex-differentially expressed in human midfetal brain (72): E2F2, FOXJ3, NR2C2, REST, SP3, and ZBTB17. P0 SxG parTFs were enriched for PPI targets of ESR1 (as were P0 female parTFs), while only SxG parTFs were enriched for PPI targets of AR and ESR2 (Fig. 5B, Data S2D–E), confirming activational hormone roles in MDD SNP function.
Figure 5. Regulatory architecture of rSNPs at P0 and P10, permutation analysis evaluating the number of detected SxG rSNPs, and the context-dependent landscape of MDD loci.
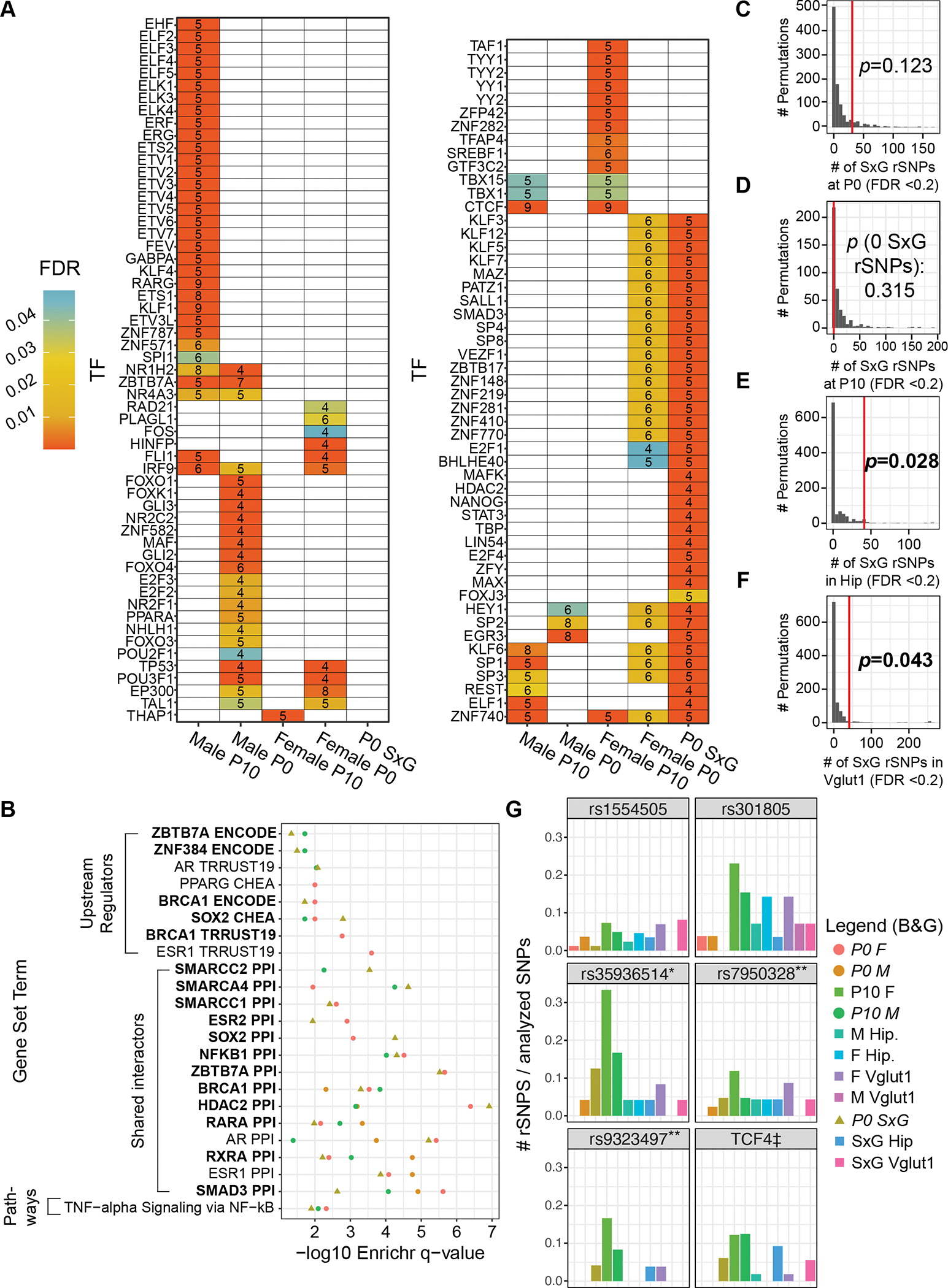
(A) TFs with binding motif disruptions by ≥5 nominally significant (pemp < 0.05) rSNPs or SxG rSNPs, enriched relative to nonfunctional SNPs. (B) Enrichr analysis findings for rSNP-enriched TFs in P0 and P10. Color key is shared with panel G. (C-F) Distribution of significant (FDR < 0.2) SxG rSNPs in 1, 000 permutation analyses per condition (400 for P10); red lines: number of SxG rSNPs identified experimentally; overlaid p-values indicate the probability of observing as many SxG rSNPs by chance. Only conditions italicized in legend are shown in this panel. (G) Six exemplary MDD loci (tag SNPs), with bars representing the percentage of analyzed variants that were rSNPs or SxG rSNPs for each condition at pemp-derived FDR<0.2. Color key shared with panel B; columns are in the same order as the legend. *: all-female MDD GWAS locus (43); **: near-genome-wide significant loci for first-episode MDD in males over age 50 (42); ‡: collapsed results from LD partners of five tag SNPs, comprising two GWAS significant tags and several weaker association peaks covering a span of ~5Mb around the gene TCF4 (6).
Landscape of functional variation differs broadly across age, sex, and brain region/cell type
Overrepresentation of SxG interactions at MDD loci in hippocampus and its Vglut1+ neurons
If increased prevalence of MDD in females is in part caused by interactions between risk genetics and sex, then the number of SxG interactions observed should exceed chance expectations. We therefore randomly scrambled sex labels and repeated our analyses to define a null distribution for the number of SxG rSNPs at a pemp FDR of 0.2. Chance expectations of SxG rSNP count were not exceeded at P0 or P10 (Fig. 5C–D), while they were significantly exceeded in adult total hippocampus (Fig. 5E) and Vglut1+ (Fig. 5F).
Risk loci are characterized by multiple functional variants
Overall, we found the majority of loci to contained multiple functional, context-dependent SNPs, in contrast to the concept of a singular “causal variant” driving a given GWAS association. Indeed, there was a modest, significant correlation between effect sizes of ≥50 directly genotyped MDD GWAS (7) variants and MPRA allele effects in hippocampus and Vglut1+, supporting the notion that functional variants beyond beyond GWAS tags and their near neighbors influence disease risk (73) (Fig S14).
Altogether, our analyses identified 280 rSNPs from 31 LD regions (28 depression-associated), with up to 13 rSNPs in a single locus within a single condition (Fig. 5G). Only 64 rSNPs were functional in more than one sex/age/cell type combination, with sex having stark effects on SNP functionality. 113 rSNPs were identified only in one sex study-wide (excluding interactions), and 109 SNPs had SxG effects (Supplementary Results). In other words, only 21% of rSNPs showed sex-invariant activity in vivo. Finally, our prior MPRA of this library in mouse neuroblastoma cells (28) identified a significant proportion of the rSNPs discovered here (Supplementary Results); however, analogous motif perturbation analyses in those assays did not implicate sex hormone receptors, highlighting the ability of in vivo MPRA to discern regulatory variants subject to effects not easily modeled in vitro.
Discussion
The present study illustrates SxG interactions across MDD-associated variants, providing a possible explanation for the greater impact of risk loci on female disease. Functional differences between sexes at MDD-associated SNPs are marked in the hippocampus, its excitatory neurons, and the whole brain during and after its sexual differentiation. These discoveries expand observed clinical and population-genetic sex differences in MDD to direct identification of sex-interacting variants.
The application of statistical methods controlling for several aspects of experiment- and design-specific technical noise (namely, empirical p-values and random-effect modeling of barcodes) allowed us to obtain results supported by a wide variety of orthogonal datasets, including reporter assays and human brain epigenomic datasets. For example, MDD-associated SNP rs1467013 was functional in luciferase reporter assays (74); indeed, we find this to be an rSNP in hippocampal Vglut1+ neurons of both sexes. A separate in vitro MPRA identified rs301807, but not rs301806, as an rSNP (32), while both were identified as functional here. This highlights the importance of the in vivo context for obtaining relevant insights about functional variation within GWAS loci—in this case, revealing two rSNPs in close (~2kb) proximity that likely influence expression of the same target (RERE).
Human datasets further support the translatability of our mouse approach in identifying regulatory SNPs and sex interactions. Four rSNPs identified here were recently identified as chromatin accessibility QTLs in human midfetal neural progenitors and/or neurons (Supplementary Results) (75), consistent with the allele-differential regulatory activity we observed. Intriguingly, an early attempt to identify sex-interacting GWAS loci for MDD (76) found suggestive significance (GWAS p < 1×10−5) for a tag SNP rs1345818, near TMEM161B and MEF2C, since been sex-agnostically associated to MDD (6). The adult MPRAs identified three SxG variants in LD with rs1345818, confirming that this risk locus indeed has sex-dependent regulatory activity (Supplementary Results). In terms of population genetics, it has recently been demonstrated that heritable variant SxG interactions display “amplification”, acting via the same loci with sex-modulated magnitudes (but not directions) of effect (77). Consistent with thisseveral rSNPs had effects shy of significance in one sex but not the other; for example, of the 58 rSNPs with FDR<0.2 in female Vglut1+, 32 were also near-significant (0.2 < FDR < 0.25) in male Vglut1+, all with shared direction of effect.
Downstream analyses supported both rSNP findings themselves and suggest novel candidate TFs/regulatory networks underlying sex interactions at MDD loci. Our SxG-enriched parTF sets were rich in sex hormone receptors and interactors, indicating a role for sex hormone receptors in co-regulation of MDD risk variants. Our hippocampal analyses revealed male-specific roles for AR: male rSNPs were enriched for binding sequences of ZMIZ1, an AR co-activator, while an AR co-repressor, ZBTB7A (78), was identified as a shared upstream regulator of these parTFs. Notably, ZBTB7A also regulates human non-coding RNA LINC00473 (79), which was recently shown to have sex-differentiated effects on depressive mouse behaviors when overexpressed in cortex (80). Similarly, AR itself has been shown to play a direct and sex-differential role in modulating hippocampal CA1 efferents, impacting depressive-like behaviors (81). Identification of P0—but not P10—SxG rSNPs, their parTFs, and enriched interactors (ESR1, ESR2, AR) verified that our approach recovers known transcriptional regulators where/when expected, and further confirmed the key role of circulating sex hormones in MDD variant function implicated by the adult MPRAs. Steroid receptors have similar binding motifs; as such, these results cannot define which hormone(s)/receptor(s) matter (and when). Nonetheless, the findings are broadly consistent with the idea that sex hormone receptors may underlie an SxG mechanism for increased MDD risk in females.
These experiments have certain limitations, especially in terms of integrated interpretation. P0/P10 and adult findings could not be compared directly (e.g., sex-by-age interactions), as adult experiments were limited to the hippocampus while developmental assays were brain-wide. Unfortunately, hippocampal morphogenesis is postnatal, precluding a hippocampus-specific P0 assay. Experimental differences also confound interpretation of distinct rSNPs at P0/P10 vs. adult, which could result from age- or region-specific factors. Future approaches could incorporate pre-and post-pubertal onset (i.e., in defined regions with a physiologic hormone-negative and hormone-positive condition). Another limitation is that, while benchmark studies indicate comparability, episomal AAVs may not capture all regulatory information of a genome-integrated assay. Lentivirus is much lower titer than AAV and spreads little when delivered intracranially, and base editing methods would be constrained by mouse-human orthology and in vivo efficiency/throughput. While these approaches are tractable in cell lines, such lines cannot be used to generate fully mature, differentiated cells like those studied here (organoids can only transcriptionally approximate prenatal/infant brain (82)), nor would they reproduce the hormonal milieu, precluding an in vitro study or replication of sex effects. Finally, results might be influenced by use of either a different minimal promoter or longer fragments if nearby elements interact with the rSNPs. However, none of the limitations above would be expected to create spurious sex effects, indicating the SxG interactions that were central to supporting our primary hypothesis are likely to be robust. It would, however, be interesting to know if such SxG effects were specifically enriched in MDD loci, or represent a general mechanism leading to sex biases in other psychiatric diseases. Lastly, we note that human gender is a complex variable that model organisms cannot currently emulate, placing possible gender identity effects on SxG interactions beyond the scope of approaches such as ours.
Nonetheless, the presented in vivo MPRAs indicate that critical biological and environmental factors involved in brain gene regulation and regulatory variation can be studied using a high-fidelity model of development, cell types, and biological signals. Our approach provides a framework for direct, functional study of psychiatric risk genetics and interactions with such contexts—along with potential future applications to environmental factors—all of which are imperfectly modeled in vitro. Future uses of this approach could include direct identification of variants subject to other important psychiatric gene-by-environment interactions, such as early life adversity or chronic stress.
Our results provide a novel, experimental demonstration of sex effects on noncoding MDD risk variants, especially during life stages with circulating sex hormones. Consistent with a study showing top female (but not male) MDD risk SNPs were enriched in hippocampal eQTLs (11), we find that the hippocampus and its excitatory neurons are foci of significant sex interactions for MDD variants. Given the broad sharing of risk loci across psychiatric disorders (37, 38, 83), our findings may well implicate similar molecular processes behind sex differences in other disorders; more directly, we identify and highlight SxG interactions in the brain as a potential mechanistic underpinning of sex differences in MDD incidence.
Supplementary Material
Acknowledgments:
Scott Lee for assistance with immunofluorescence of pilot hippocampal injections; Michael Vasek, Ph.D. and Lexi Harris for assistance with hippocampal dissections; Sarah Koester for assistance with RNA purification; Ernesto Gonzalez of the Washington University in St. Louis (WU) Hope Center Animal Surgery Core for performing hippocampal AAV9 delivery; EZH Switzerland Viral Vector Facility (VVF) facility for Sanger sequencing and AAV9 packaging of the MPRA library; McDonnell Genome Institute (sequencing), funded by National Institutes of Health grant UL1TR002345; WU Center for Genome Sciences and Systems Biology (sequencing); Barak Cohen, Ph.D., Tony Fisher, Ph.D., Tomás Lagunas, Jr., Ph.D., and Stephen Plassmeyer for insightful discussions and methodologic guidance; WU Center for Cellular Imaging (Axioscan microscope); Biorender (hippocampus experiment schematic); WU Epigenome Browser (RSRC1 locus visualization); and Michael White, Ph.D. and Rachel Rahn, Ph.D. for reviewing the manuscript. A preprint of this work appeared on Biorxiv (https://doi.org/10.1101/2021.11.01.466849) before undergoing review. JDD has received royalties related to the TRAP method in the past.
Funding:
National Institutes of Mental Health (NIMH) grant 5F30MH116654 (BM)
NIMH grant 1R01MH116999 (JDD)
Simons Foundation grant 571009 (JDD)
Footnotes
Competing interests: Authors declare that they have no competing interests.
Data and materials availability:
The annotation of library SNPs performed at the time of design are available at https://bitbucket.org/jdlabteam/n2a_atra_mdd_mpra_paper/src/master/Library%20Design%20Epigenomic%20Annotations/. Code is available at https://bitbucket.org/jdlabteam/paper-resources-mdd-in-vivo-mpras/src. Raw sequencing files and tabulated barcode counts per sample are available through GEO accession GSE186348 with reviewer token evabumkctpsltut. Outside datasets used for annotation are detailed in Supplementary Materials.
References
- 1.Salk RH, Hyde JS, Abramson LY (2017): Gender Differences in Depression in Representative National Samples: Meta-Analyses of Diagnoses and Symptoms. Psychol Bull 143: 783–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcus SM, Young EA, Kerber KB, Kornstein S, Farabaugh AH, Mitchell J, et al. (2005): Gender differences in depression: Findings from the STAR*D study. J Affect Disorders 87: 141–150. [DOI] [PubMed] [Google Scholar]
- 3.LeGates TA, Kvarta MD, Thompson SM (2018): Sex differences in antidepressant efficacy. Neuropsychopharmacol Official Publ Am Coll Neuropsychopharmacol 44: 140–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labonté B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, et al. (2017): Sex-specific transcriptional signatures in human depression. Nat Med 23: 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagy C, Maitra M, Tanti A, Suderman M, Théroux J-F, Davoli MA, et al. (2020): Single-nucleus transcriptomics of the prefrontal cortex in major depressive disorder implicates oligodendrocyte precursor cells and excitatory neurons. Nat Neurosci 1–11. [DOI] [PubMed] [Google Scholar]
- 6.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. (2018): Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, et al. (2019): Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 22: 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levey DF, Stein MB, Wendt FR, Pathak GA, Zhou H, Aslan M, et al. (2021): Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat Neurosci 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blokland GAM, Grove J, Chen C-Y, Cotsapas C, Tobet S, Handa R, et al. (2021): Sex-Dependent Shared and Non-Shared Genetic Architecture, Across Mood and Psychotic Disorders. Biol Psychiat. 10.1016/j.biopsych.2021.02.972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trzaskowski M, Mehta D, Peyrot WJ, Hawkes D, Davies D, Howard DM, et al. (2019): Quantifying between-cohort and between-sex genetic heterogeneity in major depressive disorder. Am J Medical Genetics Part B Neuropsychiatric Genetics 180: 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silveira PP, Pokhvisneva I, Howard DM, Meaney MJ (2022): A Sex-Specific Genome-Wide Association Study of Depression Phenotypes in UK Biobank. Medrxiv 2022.03.30.22273201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. (2012): Systematic localization of common disease-associated variation in regulatory DNA. Sci New York N Y 337: 1190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M (2012): Linking disease associations with regulatory information in the human genome. Genome Res 22: 1748–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D (2019): Benefits and limitations of genome-wide association studies. Nat Rev Genet 20: 467–484. [DOI] [PubMed] [Google Scholar]
- 15.Fullard JF, Hauberg ME, Bendl J, Egervari G, Cirnaru M-D, Reach SM, et al. (2018): An atlas of chromatin accessibility in the adult human brain. Genome Res 28: 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryois J, Skene NG, Hansen TF, Kogelman LJA, Watson HJ, Liu Z, et al. (2020): Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease. Nat Genet 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauberg ME, Creus-Muncunill J, Bendl J, Kozlenkov A, Zeng B, Corwin C, et al. (2020): Common schizophrenia risk variants are enriched in open chromatin regions of human glutamatergic neurons. Nat Commun 11: 5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong P, Hoffman GE, Apontes P, Bendl J, Rahman S, Fernando MB, et al. (2021): Transcribed enhancers in the human brain identify novel disease risk mechanisms. Biorxiv 2021.05.14.443421. [Google Scholar]
- 19.Song M, Pebworth M-P, Yang X, Abnousi A, Fan C, Wen J, et al. (2020): Cell-type-specific 3D epigenomes in the developing human cortex. Nature 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polioudakis D, Torre-Ubieta L de la, Langerman J, Elkins AG, Shi X, Stein JL, et al. (2019): A Single-Cell Transcriptomic Atlas of Human Neocortical Development during Mid-gestation. Neuron 103: 785–801.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K, Hocker JD, Miller M, Hou X, Chiou J, Poirion OB, et al. (2021): A single-cell atlas of chromatin accessibility in the human genome. Cell. 10.1016/j.cell.2021.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jagadeesh KA, Dey KK, Montoro DT, Mohan R, Gazal S, Engreitz JM, et al. (2022): Identifying disease-critical cell types and cellular processes by integrating single-cell RNA-sequencing and human genetics. Nat Genet 54: 1479–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmaal L, Veltman DJ, Erp TGM van, Sämann PG, Frodl T, Jahanshad N, et al. (2015): Subcortical brain alterations in major depressive disorder: findings from the ENIGMA Major Depressive Disorder working group. Mol Psychiatr 21: 806–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang JM, Tonelli L, Regenold WT, McCarthy MM (2010): Effects of neonatal flutamide treatment on hippocampal neurogenesis and synaptogenesis correlate with depression-like behaviors in preadolescent male rats. Neuroscience 169: 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isgor C, Sengelaub DR (2003): Effects of neonatal gonadal steroids on adult CA3 pyramidal neuron dendritic morphology and spatial memory in rats. J Neurobiol 55: 179–190. [DOI] [PubMed] [Google Scholar]
- 26.Kohli MA, Lucae S, Saemann PG, Schmidt MV, Demirkan A, Hek K, et al. (2011): The Neuronal Transporter Gene SLC6A15 Confers Risk to Major Depression. Neuron 70: 252–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliva M, Muñoz-Aguirre M, Kim-Hellmuth S, Wucher V, Gewirtz ADH, Cotter DJ, et al. (2020): The impact of sex on gene expression across human tissues. Science 369: eaba3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tewhey R, Kotliar D, Park DS, Liu B, Winnicki S, Reilly SK, et al. (2016): Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell 165: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X, Chen X, Forney C, Donmez O, Miller D, Parameswaran S, et al. (2021): Global discovery of lupus genetic risk variant allelic enhancer activity. Nat Commun 12: 1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi J, Zhang T, Vu A, Ablain J, Makowski MM, Colli LM, et al. (2020): Massively parallel reporter assays of melanoma risk variants identify MX2 as a gene promoting melanoma. Nat Commun 11: 2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doan RN, Bae B-I, Cubelos B, Chang C, Hossain AA, Al-Saad S, et al. (2016): Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 167: 341–354.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulvey B, Dougherty JD (2021): Transcriptional-regulatory convergence across functional MDD risk variants identified by massively parallel reporter assays. Transl Psychiat 11: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen SQ, Kim-Han JS, Cheng L, Xu D, Gokcumen O, Hughes AEO, et al. (2019): A candidate causal variant underlying both higher intelligence and increased risk of bipolar disorder. Biorxiv 580258. [Google Scholar]
- 34.Lambert JT, Su-Feher L, Cichewicz K, Warren TL, Zdilar I, Wang Y, et al. (2021): Parallel functional testing identifies enhancers active in early postnatal mouse brain. Biorxiv 2021.01.15.426772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Werling DM, Pochareddy S, Choi J, An J-Y, Sheppard B, Peng M, et al. (2020): Whole-Genome and RNA Sequencing Reveal Variation and Transcriptomic Coordination in the Developing Human Prefrontal Cortex. Cell Reports 31: 107489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kouakou MR, Cameron D, Hannon E, Dempster EL, Mill J, Hill MJ, Bray NJ (2021): Sites of active gene regulation in the prenatal frontal cortex and their role in neuropsychiatric disorders. Am J Medical Genetics Part B Neuropsychiatric Genetics. 10.1002/ajmg.b.32877 [DOI] [PubMed] [Google Scholar]
- 37.Schork AJ, Won H, Appadurai V, Nudel R, Gandal M, Delaneau O, et al. (2019): A genome-wide association study of shared risk across psychiatric disorders implicates gene regulation during fetal neurodevelopment. Nat Neurosci 22: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cross-Disorder Group of the Psychiatric Genomics Consortium, Lee PH, Anttila V, Won H, Feng Y-CA, Rosenthal J, et al. (2019): Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 179: 1469–1482.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnold AP, Breedlove SM (1985): Organizational and activational effects of sex steroids on brain and behavior: A reanalysis. Horm Behav 19: 469–498. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Luo Z, Gu C, Hall LS, McIntosh AM, Zeng Y, et al. (2018): Common variants on 6q16.2, 12q24.31 and 16p13.3 are associated with major depressive disorder. Neuropsychopharmacol 43: 2146–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, et al. (2016): Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 48: 1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Power RA, Tansey KE, Buttenschøn HN, Cohen-Woods S, Bigdeli T, Hall LS, et al. (2017): Genome-wide Association for Major Depression Through Age at Onset Stratification: Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Biol Psychiat 81: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai N, Bigdeli TB, Kretzschmar W, Li Y, Liang J, Song L, et al. (2015): Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523: 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren H, Fabbri C, Uher R, Rietschel M, Mors O, Henigsberg N, et al. (2018): Genes associated with anhedonia: a new analysis in a large clinical trial (GENDEP). Transl Psychiat 8: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. (2019): Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 51: 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith DJ, Escott-Price V, Davies G, Bailey MES, Colodro-Conde L, Ward J, et al. (2016): Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol Psychiatr 21: 1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luciano M, Hagenaars SP, Davies G, Hill WD, Clarke T-K, Shirali M, et al. (2017): Association analysis in over 329, 000 individuals identifies 116 independent variants influencing neuroticism. Nat Genet 50: 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meier SM, Trontti K, Purves KL, Als TD, Grove J, Laine M, et al. (2019): Genetic Variants Associated With Anxiety and Stress-Related Disorders. Jama Psychiat 76: 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward J, Tunbridge EM, Sandor C, Lyall LM, Ferguson A, Strawbridge RJ, et al. (2019): The genomic basis of mood instability: identification of 46 loci in 363, 705 UK Biobank participants, genetic correlation with psychiatric disorders, and association with gene expression and function. Mol Psychiatr 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White MA, Myers CA, Corbo JC, Cohen BA (2013): Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc National Acad Sci 110: 11952–11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inoue F, Kreimer A, Ashuach T, Ahituv N, Yosef N (2019): Identification and Massively Parallel Characterization of Regulatory Elements Driving Neural Induction. Cell Stem Cell. 10.1016/j.stem.2019.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dougherty JD, Schmidt EF, Nakajima M, Heintz N (2010): Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res 38: 4218–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Storey JD, Tibshirani R (2003): Statistical significance for genomewide studies. Proc National Acad Sci 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coetzee SG, Coetzee GA, Hazelett DJ (2015): motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinform Oxf Engl 31: 3847–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santana-Garcia W, Rocha-Acevedo M, Ramirez-Navarro L, Mbouamboua Y, Thieffry D, Thomas-Chollier M, et al. (2019): RSAT variation-tools: An accessible and flexible framework to predict the impact of regulatory variants on transcription factor binding. Comput Struct Biotechnology J 17: 1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Brien HE, Hannon E, Hill MJ, Toste CC, Robertson MJ, Morgan JE, et al. (2018): Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol 19: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaffe AE, Hoeppner DJ, Saito T, Blanpain L, Ukaigwe J, Burke EE, et al. (2020): Profiling gene expression in the human dentate gyrus granule cell layer reveals insights into schizophrenia and its genetic risk. Nat Neurosci 23: 510–519. [DOI] [PubMed] [Google Scholar]
- 58.Coleman KM, Lam V, Jaber BM, Lanz RB, Smith CL (2004): SRA coactivation of estrogen receptor-alpha is phosphorylation-independent, and enhances 4-hydroxytamoxifen agonist activity. Biochem Bioph Res Co 323: 332–8. [DOI] [PubMed] [Google Scholar]
- 59.Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. (2018): Comprehensive functional genomic resource and integrative model for the human brain. Science 362: eaat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu B, Won H, Mah W, Park RB, Kassim B, Spiess K, et al. (2021): Neuronal and glial 3D chromatin architecture informs the cellular etiology of brain disorders. Nat Commun 12: 3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song M, Yang X, Ren X, Maliskova L, Li B, Jones IR, et al. (2019): Mapping cis-regulatory chromatin contacts in neural cells links neuropsychiatric disorder risk variants to target genes. Nat Genet 51: 1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su C, Argenziano M, Lu S, Pippin JA, Pahl MC, Leonard ME, et al. (2021): 3D promoter architecture re-organization during iPSC-derived neuronal cell differentiation implicates target genes for neurodevelopmental disorders. Prog Neurobiol 201: 102000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jung I, Schmitt A, Diao Y, Lee AJ, Liu T, Yang D, et al. (2019): A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat Genet 51: 1442–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu L, Liu X, Huang W-K, Giusti-Rodríguez P, Cui J, Zhang S, et al. (2020): Robust Hi-C Maps of Enhancer-Promoter Interactions Reveal the Function of Non-coding Genome in Neural Development and Diseases. Mol Cell. 10.1016/j.molcel.2020.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sey NYA, Hu B, Iskhakova M, Lee S, Sun H, Shokrian N, et al. (2022): Chromatin architecture in addiction circuitry identifies risk genes and potential biological mechanisms underlying cigarette smoking and alcohol use traits. Mol Psychiatr 1–10. [DOI] [PubMed] [Google Scholar]
- 66.Bernabeu E, Canela-Xandri O, Rawlik K, Talenti A, Prendergast J, Tenesa A (2021): Sex differences in genetic architecture in the UK Biobank. Nat Genet 53: 1283–1289. [DOI] [PubMed] [Google Scholar]
- 67.Chen C, Meng Q, Xia Y, Ding C, Wang L, Dai R, et al. (2018): The transcription factor POU3F2 regulates a gene coexpression network in brain tissue from patients with psychiatric disorders. Sci Transl Med 10: eaat8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams ES, Mazei-Robison M, Robison AJ (2021): Sex Differences in Major Depressive Disorder (MDD) and Preclinical Animal Models for the Study of Depression. Csh Perspect Biol a039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawatake-Kuno A, Murai T, Uchida S (2021): The Molecular Basis of Depression: Implications of Sex-Related Differences in Epigenetic Regulation. Front Mol Neurosci 14: 708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chakrabarty P, Rosario A, Cruz P, Siemienski Z, Ceballos-Diaz C, Crosby K, et al. (2013): Capsid Serotype and Timing of Injection Determines AAV Transduction in the Neonatal Mice Brain ((Qiu J, editor)). Plos One 8: e67680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abramovich DR (1974): HUMAN SEXUAL DIFFERENTIATION—IN UTERO INFLUENCES. Bjog Int J Obstetrics Gynaecol 81: 448–453. [DOI] [PubMed] [Google Scholar]
- 72.O’Brien HE, Hannon E, Jeffries AR, Davies W, Hill MJ, Anney RJ, et al. (2019): Sex differences in gene expression in the human fetal brain. Biorxiv 483636. [Google Scholar]
- 73.Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sal·lari R, et al. (2014): Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res 24: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li S, Li Y, Li X, Liu J, Huo Y, Wang J, et al. (2020): Regulatory mechanisms of major depressive disorder risk variants. Mol Psychiatr 1–20. [DOI] [PubMed] [Google Scholar]
- 75.Liang D, Elwell AL, Aygün N, Krupa O, Wolter JM, Kyere FA, et al. (2021): Cell-type-specific effects of genetic variation on chromatin accessibility during human neuronal differentiation. Nat Neurosci 24: 941–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aragam N, Wang K-S, Pan Y (2011): Genome-wide association analysis of gender differences in major depressive disorder in the Netherlands NESDA and NTR population-based samples. J Affect Disorders 133: 516–521. [DOI] [PubMed] [Google Scholar]
- 77.Zhu C, Ming MJ, Cole JM, Kirkpatrick M, Harpak A (2022): Amplification is the Primary Mode of Gene-by-Sex Interaction in Complex Human Traits. Biorxiv 2022.05.06.490973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui J, Yang Y, Zhang C, Hu P, Kan W, Bai X, et al. (2011): FBI-1 functions as a novel AR co-repressor in prostate cancer cells. Cell Mol Life Sci 68: 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang L, Wang Y, Li X, Xia X, Li N, He R, et al. (2017): ZBTB7A Enhances Osteosarcoma Chemoresistance by Transcriptionally Repressing lncRNA LINC00473-IL24 Activity. Neoplasia New York N Y 19: 908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Issler O, Zee YY van der, Ramakrishnan A, Wang J, Tan C, Loh Y-HE, et al. (2020): Sex-Specific Role for the Long Non-coding RNA LINC00473 in Depression. Neuron. 10.1016/j.neuron.2020.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams ES, Manning CE, Eagle AL, Swift-Gallant A, Duque-Wilckens N, Chinnusamy S, et al. (2019): Androgen-Dependent Excitability of Mouse Ventral Hippocampal Afferents to Nucleus Accumbens Underlies Sex-Specific Susceptibility to Stress. Biol Psychiat 87: 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bhaduri A, Andrews MG, Leon WM, Jung D, Shin D, Allen D, et al. (2020): Cell stress in cortical organoids impairs molecular subtype specification. Nature 578: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Consortium C-DG of the PG (2013): Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381: 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McGill R, Tukey JW, Larsen WA (1978): Variations of Box Plots. Am Statistician 32: 12. [Google Scholar]
- 85.Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, et al. (2021): Gene Set Knowledge Discovery with Enrichr. Curr Protoc 1: e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Won H, Torre-Ubieta L de la, Stein JL, Parikshak NN, Huang J, Opland CK, et al. (2016): Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538: 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The annotation of library SNPs performed at the time of design are available at https://bitbucket.org/jdlabteam/n2a_atra_mdd_mpra_paper/src/master/Library%20Design%20Epigenomic%20Annotations/. Code is available at https://bitbucket.org/jdlabteam/paper-resources-mdd-in-vivo-mpras/src. Raw sequencing files and tabulated barcode counts per sample are available through GEO accession GSE186348 with reviewer token evabumkctpsltut. Outside datasets used for annotation are detailed in Supplementary Materials.


