Summary
Background
When sepsis is detected, organ damage may have progressed to irreversible stages, leading to poor prognosis. The use of machine learning for predicting sepsis early has shown promise, however international validations are missing.
Methods
This was a retrospective, observational, multi-centre cohort study. We developed and externally validated a deep learning system for the prediction of sepsis in the intensive care unit (ICU). Our analysis represents the first international, multi-centre in-ICU cohort study for sepsis prediction using deep learning to our knowledge. Our dataset contains 136,478 unique ICU admissions, representing a refined and harmonised subset of four large ICU databases comprising data collected from ICUs in the US, the Netherlands, and Switzerland between 2001 and 2016. Using the international consensus definition Sepsis-3, we derived hourly-resolved sepsis annotations, amounting to 25,694 (18.8%) patient stays with sepsis. We compared our approach to clinical baselines as well as machine learning baselines and performed an extensive internal and external statistical validation within and across databases, reporting area under the receiver-operating-characteristic curve (AUC).
Findings
Averaged over sites, our model was able to predict sepsis with an AUC of 0.846 (95% confidence interval [CI], 0.841–0.852) on a held-out validation cohort internal to each site, and an AUC of 0.761 (95% CI, 0.746–0.770) when validating externally across sites. Given access to a small fine-tuning set (10% per site), the transfer to target sites was improved to an AUC of 0.807 (95% CI, 0.801–0.813). Our model raised 1.4 false alerts per true alert and detected 80% of the septic patients 3.7 h (95% CI, 3.0–4.3) prior to the onset of sepsis, opening a vital window for intervention.
Interpretation
By monitoring clinical and laboratory measurements in a retrospective simulation of a real-time prediction scenario, a deep learning system for the detection of sepsis generalised to previously unseen ICU cohorts, internationally.
Funding
This study was funded by the Personalized Health and Related Technologies (PHRT) strategic focus area of the ETH domain.
Keywords: Sepsis, Machine learning, Deep learning, Intensive care, ICU, Early prediction, Early warning
Research in context.
Evidence before this study
We searched PubMed with no language restrictions for research articles published up to July 22, 2022. We used the terms “sepsis” and “machine learning” and “external validation”.
We found no international, multi-centre study that externally validated prediction models for the early detection of sepsis using machine learning. While the early diagnosis and timely management of sepsis could improve prognosis, there is evidence that currently-deployed proprietary models (a) lead to alarm fatigue, and (b) exhibit poor discrimination for predicting sepsis onset when subjected to external validation.
Added value of this study
We report the first multi-national, multi-centre study for the prediction of sepsis in the intensive care unit using machine learning. Our cohort features 136,478 ICU admissions corresponding to 708 patient years of closely monitored patients in ICUs from the US, the Netherlands, and Switzerland. We developed a deep learning system to detect sepsis onset and performed an extensive internal and external validation to assess model transferability across sites and even continents. Our model raised 1.4 false alerts per true alert and detected 80% of the septic patients 3.7 h (95% CI, 3.0–4.3) prior to the onset of sepsis, providing a time window for intervention.
Implications of all the available evidence
To our knowledge, this study represents the first successful attempt to validate accurate sepsis prediction with deep learning across hospitals in countries and continents different from the training sites. By creating the largest public and harmonised international ICU dataset, this work facilitates further statistical validations of the early prediction of sepsis and other clinical complications.
Introduction
Sepsis remains a major public health issue associated with high mortality, morbidity, and related health costs.1, 2, 3 From sepsis onset, each hour of delay before an effective antimicrobial therapy is initiated increases mortality.4, 5, 6 However, identifying bacterial species in the blood can take up to 48 h after blood sampling.7 Meanwhile, an abundance of clinical and laboratory data is being routinely collected, the richest set of which is accumulated in the intensive care unit (ICU). While it has become harder for intensivists to manually process the increasing quantities of patient information,8 machine learning (ML) systems have been developed to leverage these data to raise early warnings of imminent complications.9,10
Currently, there is no clinical gold standard for the early identification of sepsis. Furthermore, when compared to other endpoints such as mortality or length of stay, sepsis is a hard-to-define outcome, which has resulted in the development of a diverse set of strategies to define sepsis onset. These range from consensus definitions (Sepsis-2,11 Sepsis-312) to more ad-hoc approaches combining international classification of disease (ICD) billing codes with clinical and laboratory signs of infection and inflammation.13 The use of different definitions in the literature complicates the task of comparing quantitative results concerning the early predictability of sepsis. There is a general lack of systematically annotated, multi-centre data and international external validations of predictive models for sepsis.13,14 In fact, a widely adopted proprietary sepsis prediction model was recently found to perform surprisingly poorly when externally validated.15
The goal of this study therefore was to address these challenges by unifying ICU data from multiple sources to build an open-access platform for developing and externally validating sepsis prediction approaches. After harmonising, cleaning, and filtering these data, we implemented sepsis annotations based on Sepsis-312 and developed sepsis early warning systems using state-of-the-art machine learning (ML) algorithms. We further devised an evaluation strategy that accounts for the inherent trade-off between accurate and early alarms for sepsis while keeping false alarms (and therefore alarm fatigue) at bay. Finally, our unique disposition, with harmonised ICU data from four international data sources, enabled us to perform an extensive external validation to assess transferability of models between hospitals, countries, and even continents.
Methods
Study design
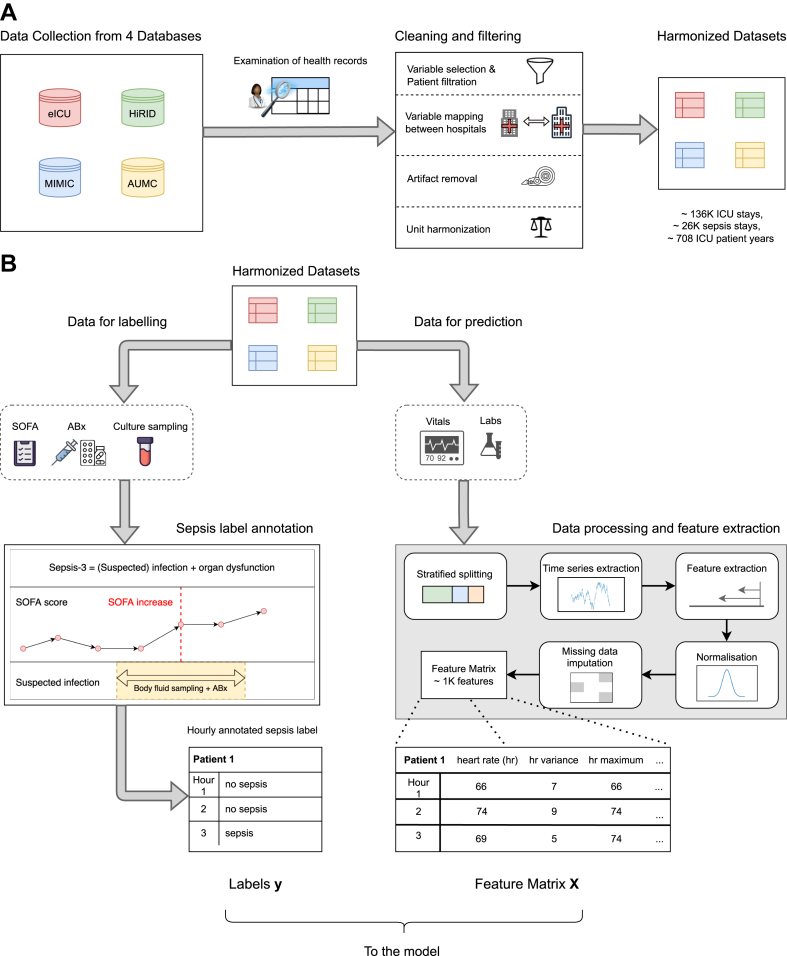
This was a retrospective, observational, multi-centre cohort study. The study involved the creation of a harmonised multi-centre annotated ICU cohort, as well as the development, internal validation, and external testing of a sepsis early warning system. The study cohort was constructed using four large electronic health record databases representing clinical and laboratory ICU data that was routinely collected between 2001 and 2016 in three nations and two continents: HiRID9 from Switzerland, AUMC16 from the Netherlands, as well as MIMIC-III,17 and eICU18 from the US. In all datasets, the Sepsis-3 definition was implemented.12 Fig. 1 gives an overview of data processing pipeline. More details regarding the cohorts as well as a list of inclusion and exclusion criteria are provided in the Supplementary Appendix (Supplementary Section S1a–S1c).
Fig. 1.
Overview of the preprocessing pipeline. Data from four ICU EHR databases are collected, cleaned and harmonised (Panel A). In Panel B, we illustrate how data are extracted for sepsis label annotation (left) as well as feature extraction (right) resulting in labels and features that are used for training the machine learning model.
Our system was designed to monitor a comprehensive set of vital, laboratory, and static patient variables that were i) potentially relevant for sepsis, while ii) consistently measured and iii) not reliant on waiting for clinicians to treat suspected sepsis, which can lead to spurious modelling outcomes. To account for the last point, we excluded therapeutic variables such as antibiotics, intravenous fluids or vasopressors from the set of input data used for making predictions. To ensure interoperability, we resampled all datasets to an hourly resolution, reporting the median value per hour and patient for each variable. Table 1 lists all input variables and indicates their availability per dataset. Unit synchronisation and filtering of values outside of clinically valid ranges (determined by an experienced ICU clinician) were applied (Supplementary Table S5). Furthermore, we manually inspected the distributions of biomarkers to assert that they were visually similar across all datasets, confirming that the units of measurement were properly harmonised. Statistical non-discernability of the biomarkers between the datasets cannot be expected due to slight shifts and variations in the underlying data distributions. It would not even be desirable for a credible simulation of model deployments in different countries. We plotted the density of all biomarkers, stratified by dataset. An example of such a plot, for some vital and laboratory parameters, is given in Supplementary Fig. S2A.
Table 1.
Demographic and patient characteristics of our multi-center ICU cohort.
| Variable | MIMIC-III | eICU | HiRID | AUMC |
|---|---|---|---|---|
| Cohort size (n) | 36,591 | 56,765 | 27,278 | 15,844 |
| Sepsis-3 prevalence (n (%)) | 9541 (26) | 4708 (8) | 10,170 (37) | 1275 (8) |
| Age, years (Median (IQR)) | 65 (52–77) | 65 (53–76) | 65 (55–75) | 65 (55–75) |
| In-hospital mortality (n (%)) | 2829 (8) | 3962 (7) | 1399 (5) | 745 (5) |
| ICU LOS, days (Median (IQR)) | 1.99 (1.15–3.63) | 1.71 (0.95–3.01) | 0.97 (0.8–1.95) | 0.97 (0.81–1.82) |
| Hospital LOS, days (Median (IQR)) | 6.43 (3.82–11.14) | 5.53 (2.99–9.89) | – | – |
| Sex, female (n (%)) | 15,944 (44) | 25,740 (45) | 9977 (37) | 5350 (35) |
| Sex, male (n (%)) | 20,647 (56) | 31,011 (55) | 17,301 (63) | 10,089 (65) |
| Ventilated patients (n (%)) | 16,499 (45) | 24,534 (43) | 14,021 (51) | 10,469 (66) |
| Patients on vasopressors (n (%)) | 9669 (26) | 6769 (12) | 7721 (28) | 7980 (50) |
| Patients on antibiotics (n (%)) | 21,598 (59) | 21,847 (38) | 17,152 (63) | 11,165 (70) |
| Patients with suspected infection (n (%)) | 16,349 (45) | 9739 (17) | 15,160 (56) | 1639 (10) |
| Initial SOFA (Median (IQR)) | 3 (1–4) | 3 (1–5) | 5 (3–8) | 6 (3–7) |
| SOFA components (Median (IQR)) | ||||
| Respiratory | 1 (0–2) | 1 (0–2) | 3 (2–4) | 2 (1–3) |
| Coagulation | 0 (0–1) | 0 (0–1) | 0 (0–1) | 0 (0–1) |
| Hepatic | 0 (0–1) | 0 (0–0) | 0 (0–1) | 0 (0–0) |
| Cardiovascular | 1 (1–1) | 1 (0–1) | 1 (1–4) | 2 (1–4) |
| CNS | 0 (0–1) | 0 (0–2) | 0 (0–1) | 0 (0–1) |
| Renal | 0 (0–1) | 0 (0–1) | 0 (0–0) | 0 (0–1) |
| Admission type (n (%)) | ||||
| Surgical | 13,836 (38) | 9865 (19) | – | 11,905 (80) |
| Medical | 22,346 (61) | 41,674 (79) | – | 2172 (15) |
| Other | 408 (1) | 1346 (3) | – | 786 (5) |
CNS, central nervous system; LOS, length of stay.
Outcome and prediction problem
We considered the onset of sepsis as determined by the Sepsis-3 definition12 as the primary outcome in this study. To fulfill this definition, a co-occurrence of suspected infection and a SOFA score increase of two or more points are required. A detailed account of the label implementation, including the treatment of missing values of SOFA components, is provided in the Supplementary Appendix (Supplementary Section S1b). Our model was designed to continuously monitor 59 vital and laboratory parameters in hourly intervals together with 4 static variables in order to raise an alarm when sepsis is about to occur (the list of variables is provided in Supplementary Table S1). Our intended use group are ICU patients within the first seven days of their ICU stay (Supplementary Section S1c, Supplementary Fig. S12). We incentivised our model to recognise if sepsis will start within the next 6 h (Supplementary Fig. S12). The employed Sepsis-3 definition subsumes the SOFA score, which captures additional treatment information (e.g., vasopressors) that the models were not intended to rely upon. Still, since the clinical baseline scores encode valuable domain knowledge, our model was provided partial scores by only including laboratory and vital parameters that belong to our list of readily measured, non-therapeutic 63 input variables (Supplementary Section S1f).
Prediction methods
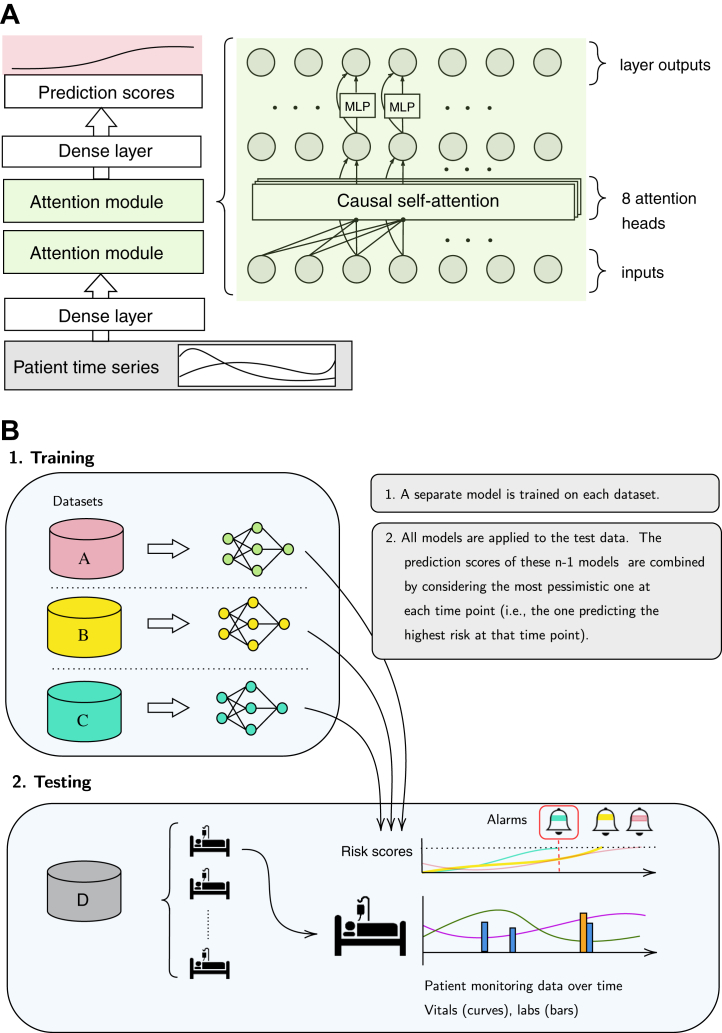
To improve upon clinical baselines, we devised a deep learning-based early warning system, specifically a deep self-attention model (attn).19 For comparison, we further investigated a range of state-of-the-art machine learning approaches. These included further deep learning approaches, i.e., machine learning algorithms that use deep neural networks, as well as classical machine learning approaches based on statistical learning concepts. For additional deep learning models, we considered recurrent neural networks employing Gated Recurrent Units (gru).20 Both these methods are intrinsically capable of leveraging sequential data. Next, we included LightGBM (lgbm)21 and a LASSO-regularised Logistic regression (lr).22 In order to make temporal dynamics governing the data accessible to these two methods, they were supplied with a total of 1269 features that incorporated statistical moments and temporal trends as extracted from the 63 input variables. Further details about the construction and standardisation of these features are provided in Supplementary Section S1f. As for clinical baselines, we investigated how well sepsis could be predicted with a range of early-warning scores, including NEWS,23 MEWS,24 SIRS,25 SOFA,26 and qSOFA.12 Further details regarding missing data handling (Supplementary Section S1f) and model development (Supplementary Section S1e) are provided in the Supplementary Appendix.
Statistical analysis
We first trained our deep learning system and all baselines on the development split of each individual database, resulting in a model for each prediction method and dataset. For an internal validation, we evaluate the performance on the held-out test split of the same database that the model was trained on, respectively. For the external validations, for a given testing dataset and prediction method we apply the models that were fitted individually on the remaining datasets to the testing dataset and take the maximal prediction score at each point in time (i.e., the earliest alarm among these models), and refer to this setting as pooled prediction (see Fig. 2). As an ablation, we also report the performance for training and testing across pairs of datasets, i.e., pair-wise prediction. To assess performance characteristics, we calculated the area under the receiver-operating-characteristic (ROC) curve (AUC). Next, for a fixed 80% sensitivity threshold, we reported the positive predictive value (PPV) as well as the median number of hours the alarm preceded sepsis onset (median earliness). All measures were computed on the patient level. All results are presented with 95% confidence intervals (CI) when appropriate (Supplementary Section S1i). Details about significance tests are provided in Supplementary Section S1i. We devised an evaluation strategy in which repeated alarms are not permissible to prevent alarm fatigue (Supplementary Section S1i). While this made the task to recognise sepsis cases more challenging, it also guaranteed that at most a single false alarm could be raised in a control stay. To ensure comparability between internal and external evaluations, identical test splits are used in both settings. In order to make performance metrics comparable across datasets, upon testing time we harmonised the prevalence of sepsis cases to the across-dataset average of 18.8% via repeated subsampling upon testing time (Supplementary Section S1h). To further assess the transferability of our model across sites, we simulated a fine-tuning scenario where a small portion (10%) of the target cohort is made available for fine-tuning a pre-trained model before testing on the held-out split of the target cohort (Fig. 3). Doing this across 4 cohorts is more extensive than previous fine-tuning experiments where typically a fixed development and fine-tuning cohort is used.27,28
Fig. 2.
Illustration of the deep learning model and the pooling strategy. In Panel A, the deep self-attention model (attn) is shown. The input stream of data is fed through an initial dense layer. This is followed by two attention modules, each comprising a causal self-attention layer and a Multilayer perceptron (MLP). A final dense layer maps to a sequence of prediction scores. In Panel B, the pooling strategy is illustrated. We combined information from n-1 (training) datasets to predict on the n-th (test) dataset. For n = 4, we developed n-1 models, each optimised on a different training dataset. Second, we applied all these models to the test dataset, resulting in n-1 prediction scores (i.e., a predicted probability for sepsis) for each hour of the patients in the test dataset. We aggregated these n-1 predictions into a single risk score by taking the maximal value at each point in time. Hence, we raise the most pessimistic alarm as soon as the first model would raise an alarm (dark green bell with red frame). This strategy was referred to as pooled predictions.
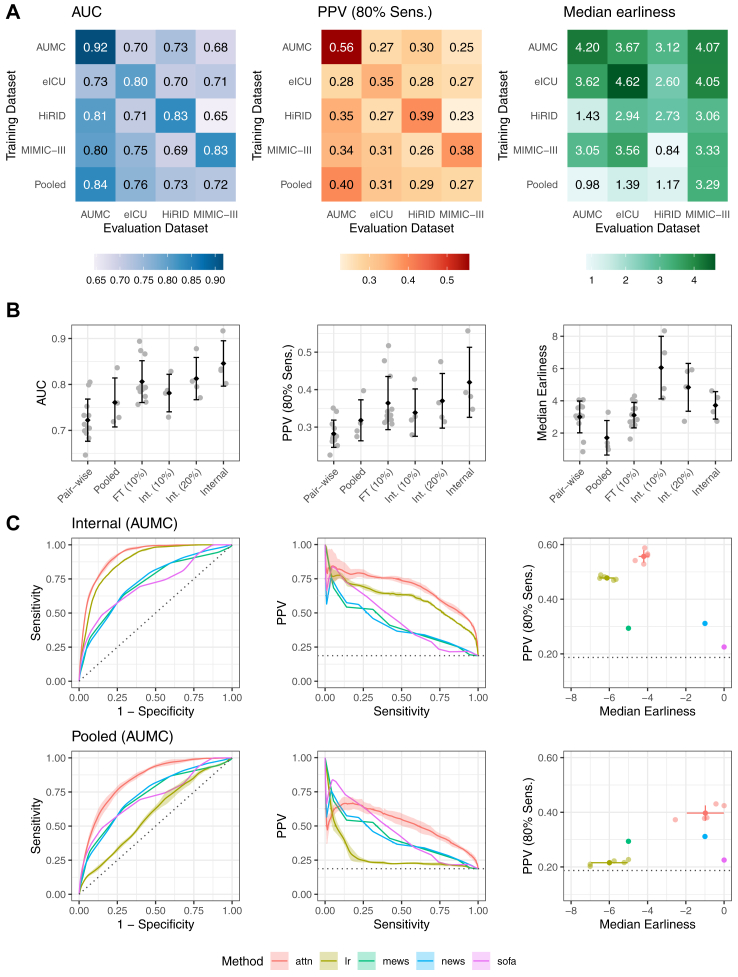
Fig. 3.
Performance of the deep learning system for the prediction of sepsis. In Panel A, heatmaps of the performance of our deep learning system are shown for AUC as well as PPV and median Earliness, whereas the latter two metrics are displayed at 80% Sensitivity. For a given heatmap, the rows indicate the training dataset, the columns refer to the testing dataset. In the last row, the externally pooled predictions are shown. Averaged across datasets, we observe an internally validated AUC of 0.846 (95% CI, 0.841–0.852), PPV of 42.0% (95% CI, 40.5–44.1), and median lead time to sepsis onset of 3.7 (95% CI, 3.0–4.3) hours. In the bottom row of the heatmaps, we observed an average externally validated AUC of 0.761 (95% CI, 0.746–0.770), PPV of 31.8% (95% CI, 30.3–34.0), and median lead time to sepsis onset of 1.71 (95% CI, 0.75–2.69) hours. The pooling approach improved the generalisability to new datasets by outperforming or being on par with predictions derived from the best (a priori unknown) training dataset in terms of AUC and PPV at 80% Sensitivity. Panel B illustrates how the metrics behave with different model transfer strategies: AUC performance of the internal validaton is increasingly approached when using pooling of models (pooled), and more so when instead fine-tuning a model to a small fine-tuning set of 10% of the testing site (FT (10%)), reflecting the realistic scenario that only small sample has been collected in a novel target hospital. By comparison, Int. (10%) and Int. (20%) displays the internal validation performance when training on only 10% or 20% of the training site, respectively. The error bars indicate the standard deviation of the metrics as calculated over the four datasets. A black diamond indicates the mean over the datasets. In Panel C, more detailed performance curves of the internal validation (top row) and external validation via pooled predictions (bottom row) performance are shown for an example dataset (AUMC). Our deep learning approach (attn) is visualised together with a subset of all included baselines, including clinical baselines (SOFA, MEWS, NEWS), and LASSO-regularised logistic regression (lr). Error bands indicate standard deviation over 5 repetitions of train-validation splitting. All baselines are shown in Supplementary Figs. S4–S7.
Next, to explain our model’s predictions, we calculated Shapley values,29 which provide a measure of the contribution and importance of individual variables to the overall prediction (Supplementary Section S1g). In an auxiliary analysis (Supplementary Section S2a), we investigated whether the more effortful approach, to pool the actual datasets for training, would be superior to combining only predictors (in a federated way). Finally, we detail an ablation analysis of our model and assess model calibration in Supplementary Sections S1i and S2b. Analyses were performed with R software, version 4.1, and Python software, version 3.7.4.
Ethics approval
Ethics approval to conduct a machine learning-based study on the early prediction of sepsis was obtained from the “Ethikkommission Nordwest-und Zentralschweiz EKNZ” (BASEC ID 2019-01088). We obtained deidentified data from the critical care research databases AUMC, MIMIC-III, eICU, and HiRID to conduct a retrospective secondary analysis.
Role of funding source
The funders of this study had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. All authors had access to the data, and jointly decided to publish the study findings.
Results
After cleaning, filtering and processing, our fully interoperable cohort comprised a total of 136,478 unique ICU stays (amounting to 708 ICU admission years) of which 25,694 (18.8%) developed sepsis. A summary of the cohort statistics is provided in Table 1 (more detailed in Supplementary Table S4).
First, we consider the internal validation, where all models were trained on all datasets separately in order to assess performance on a held-out test split of the training dataset, respectively. Our deep learning model (attn) achieved an average test AUC of 0.846 (95% CI, 0.841–0.852) when internally validating on the four core datasets that were harmonised to the list of 63 variables (Supplementary Table S1). At 80% sensitivity and a harmonised sepsis prevalence of 18.8% (for further details see Supplementary Section S1h), our model detected septic patients with 42.0% (95% CI, 40.5–44.1) PPV and a median lead time to sepsis onset of 3.7 (95% CI, 3.0–4.3) hours in advance. This corresponds to raising 1.4 false alerts (95% CI, 1.3–1.5) per true alert. The results for our deep attention model (attn) are shown in Fig. 3, whereas the full set of comparison methods and datasets is displayed in Supplementary Figs. S4–S7.
Next, we consider our external validation, where models previously trained on one database were applied to independent test databases. The pair-wise predictions (transfer from one database to another one using the harmonised variable set), are displayed for our deep attention model in Fig. 3A. Fig. 3A depicts a heatmap of AUC values, with rows corresponding to the training database and columns corresponding to the testing database. The pooled predictions are displayed in the last row of the heatmap in Fig. 3A. Using this pooling strategy in our external validation (Supplementary Figs. S4–S7), we achieve an average AUC of 0.761 (95% CI, 0.746–0.770). When fixing the prediction threshold at 80% sensitivity, on average this resulted in a PPV of 31.8% (95% CI, 30.3–34.0) with a lead time to sepsis onset of 1.71 (95% CI, 0.75–2.69) hours (Supplementary Figs. S4–S7).
Overall, Fig. 3A shows that applying the pooling strategy for a given testing database achieves better or equivalent performance as compared to the best-performing model that was trained on a single database that could only be determined post hoc (P = 0.087 for HiRID and P < 0.0001 for the remaining core datasets). As shown in Fig. 3B, when additionally given access to a small fine-tuning set of the target testing site (10% of the target site [FT (10%)]), we observe an AUC of 0.807 (95% CI, 0.801–0.813). Overall, Fig. 3B serves as an ablation demonstrating that the predictive performance gradually increases over the increasingly involved transfer strategies (pair-wise predictions, pooled predictions, and fine-tuned predictions) approaching the internal validation performance.
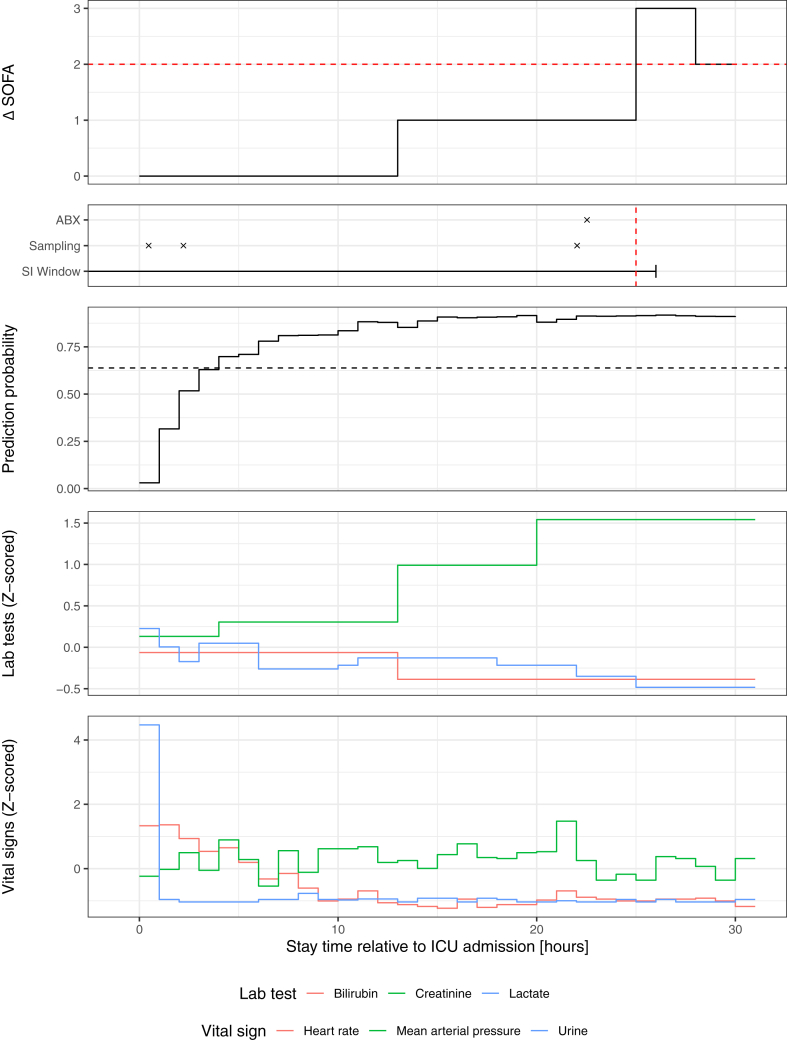
In our ablation analysis (Supplementary Fig. S10), we found that the higher performance in AUMC can be attributed to the predominantly surgical cohort, however we could not generalise this finding to an external dataset. In the auxiliary analysis (Supplementary Section S2a, Supplementary Fig. S11), where we retrained our deep learning model by pooling all datasets except for the respective testing dataset, we found no improvement over our pooling strategy (P = 0.99) which combines predictors without the need for a) costly retraining on larger datasets and b) sharing patient data across sites. Furthermore, a temporal analysis of the alarms over the course of the ICU stay suggests that it may be useful to consider time-dependent alarm thresholds (Supplementary Section S2c). In Fig. 4, our deep learning system is illustrated for an example patient in an unseen testing site.
Fig. 4.
Illustration of the deep learning system for recognising sepsis. Our deep learning system is illustrated for one sample patient (of an unseen testing database) together with a subset of vital and laboratory parameters that were used for prediction. In the top two rows, the sepsis label is shown decomposed into its components, the suspected infection (SI) window (consisting of antibiotics [ABX] administration coinciding with body fluid sampling), and an acute increase in SOFA (ΔSOFA) of two or more points. The third row illustrates the hourly predictions as probability of sepsis. The last two rows show laboratory and vital parameters (Z-scored units for joint visualisation). Red dotted lines indicate the point at which the SOFA criterion is fulfilled. A decision threshold based on 80% sensitivity is indicated by the black horizontal dashed line. The displayed model was trained on eICU and here applied to a patient of the AUMC dataset.
Variable importance
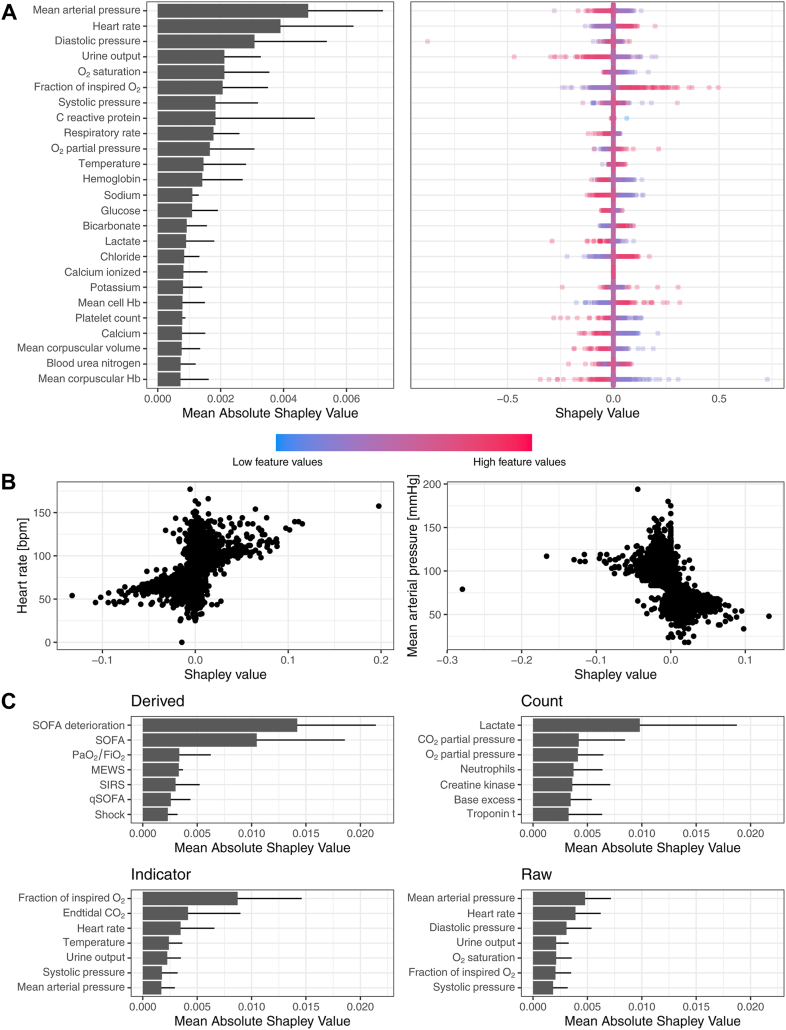
Explanations of the deep learning system’s predictions are provided in a Shapley analysis in Fig. 5. In Fig. 5A, variable importances are shown in terms of the mean absolute Shapley value averaged over all datasets, displayed for the top 20 raw variables. Mean arterial pressure (MAP), followed by heart rate exhibit the largest overall contributions to predictions of our model, which suggests that across datasets, the model has learned to attend to variables relevant to the assessment of hemodynamic instability. On a more detailed level, the right panel of Fig. 5A depicts distributions of Shapley values for a single dataset (eICU), revealing the effect increased (or decreased) values of individual measurements have on the prediction score. High values in MAP resulted in a lower prediction score, while high heart rate values led to a higher prediction score, thus encouraging a positive prediction, i.e., an alarm for sepsis. In Fig. 5B, individual MAP measurements were scattered against their Shapley value. We observe that low values in this variable (below 60 mmHg) are associated with high Shapley values, meaning they are associated with positive predictions of the model. This is in line with the definition of (septic) shock, which associates low MAP values with adverse outcomes.30 In Fig. 5C, mean absolute Shapley values are shown for individual feature groups, highlighting that depending on the variable, different feature representations are most informative. E.g., for MAP the raw measurement value is informative, whereas for Lactate the number of measurements is more informative due to sampling information. When comparing the Shapley distributions across datasets (Supplementary Fig. S8), we observe that depending on the dataset, the top-ranking effects are more (e.g., eICU) or less (e.g., AUMC) aligned with clinical assumptions about sepsis. Please refer to Supplementary Fig. S8 for more visualisations on other datasets and Supplementary Fig. S9 for a depiction of all feature types. In our ablation analysis (Supplementary Fig. S10), we found that lab tests carry relevant sampling information whereas this was less the case for vital signs.
Fig. 5.
Shapley analysis for variable importance and explanation of predictions. Panel A shows the mean absolute Shapley (SHAP) values averaged over all datasets (error bars indicate standard deviation over datasets). The top 20 variables are displayed. Large values indicate large contributions to the model’s prediction of sepsis. In the subpanel on the right, Shapley value distributions are exemplified for the eICU dataset. Positive Shapley values are indicative of positive predictions of the system and vice versa. In Panel B, Shapley values of individual heart rate and mean arterial pressure (MAP) measurements are shown. Panel C shows the Shapley values of the individual feature groups averaged across all datasets, whereas the available components of SOFA (labs and vitals), the count of lactate measurements, indication of oxygenation and ventilation as well as raw MAP values were most informative.
Discussion
In this study, we constructed the first international multi-centre ICU dataset with hourly sepsis labels to date, using data from four databases from three countries. For this, we undertook the effort to harmonise the largest and most widely used13 ICU databases that to date are publicly accessible. Using this data, an early warning system based on deep learning was developed and subsequently validated, both internally and externally. Upon internal validation, that is when applying the model to a hold-out set coming from the same hospital centres as the training data, we observed excellent model performance that can be rendered clinically useful (1.4 false alarms raised per each true alarm at 80% sensitivity). When externally validated, that is, applied to a cohort from a different hospital (not included in the training data), we still observed good performance, indicating that the model leverages a signal that can generalise to new hospital centres in different countries and even continents. On top of that, we found that fine-tuning a pre-trained model on only a small fraction of the target site boosts performance and facilitates model transfer across sites especially when the collected data is initially scarce at a new testing site.
Sepsis is one of the most challenging conditions in ICU and the leading cause of mortality in critically ill patients.31 Therefore, the possible benefits of an automated early warning system able to predict sepsis are manifold. Multiple studies attempted to address this problem using machine learning tools, but many of them either lack external validation, or are based on restricted-access datasets, limiting the ability of external validation for researchers in the field.13,14 Moreover, one of the most widely implemented proprietary tools for sepsis prediction in the US was recently found to perform poorly upon external validation,15 once again emphasising the fundamental importance of our study.32 Existing multi-centric studies carrying out external validation are limited to hospitals in the US33,34 and do not validate across country borders where shifts in policies, measurement devices, provider infrastructures, and patient cohorts are to be expected to make successful model transferability considerably more challenging. By carrying out a large external validation, and by developing an open-access, international dataset with sepsis labels, our work aims to complement the current literature in precisely this way, and it also allows other clinicians to externally validate their prediction models. Previous cohort studies treated sepsis prediction as a retrospective problem (time windows before sepsis onset were compared to time windows in control patients) such that high AUC values may be achieved without the guarantee that this translates to a real-time monitoring scenario.13,14 In contrast, in our study real-time predictions were simulated by making predictions in hourly intervals, rendering our performance assessment closer to a bed-side monitoring scenario. Finally, to raise an early alarm is typically more challenging than the task of a regular diagnostic test, which is why can expect lower AUCs and PPVs compared to (later) diagnostic tests, which may result in alarm fatigue (due low PPV).9 Here, we explicitly addressed alarm fatigue by devising a system that has an upper bound of at most one single false alarm for an entire ICU stay. For comparison, Shashikumar et al.33 report 0.04 false alarms per patient hour, which for a stay of 100 h would on average amount to 4 false alarms.
A major implication of our study is that a sepsis prediction model can generalise internationally to new hospital sites, which opens the door for prospective evaluations of such tools that were extensively validated beforehand. Next, we found that the combination of models that were trained on different databases has a beneficial effect on the external validity, implying that the integration of heterogeneous cohorts originating from different hospitals leads to early warning systems that can generalise to new settings (hospitals and countries). Interestingly, we found that this can be achieved without the need for sharing (and anonymising) sensitive data and without the need for costly retraining of models on large unions of datasets, but only by means of sharing and combining trained models across centres. This finding is promising and well-aligned with recent studies that employ federated learning to leverage multi-centric data in a differentially private manner.35,36
The availability of the harmonised and annotated dataset used in this study allows for other researchers and clinicians to evaluate their prediction models on external hospital sites, which could be a valuable consideration when making decisions about implementing early warning systems in new hospital centres. Finally, while in our external validations we observed a moderate reduction in PPV, we found that alarm earliness suffers when applying a model to a previously-unseen data distribution, which can be addressed by fine-tuning already on a small sample of the target data distribution. When considering deployment of an early warning system in a new hospital site, an on-site fine-tuning and recalibration of pretrained models will be necessary, in particular to account for a new (and possibly unknown) prevalence of sepsis in the target hospital.
Our study has several strengths. The first is the size and the heterogeneity of the cohort, coming from multiple countries and hospital centres with a varying proportion of medical and surgical admissions (MIMIC-III predominantly medical, AUMC predominantly surgical). The second strength of the study is the depth of the external validation performed, in which a model trained on one database was validated externally on all other databases, giving a high degree of external validity to the study findings. The third strength is the nature of the prediction problem we investigated. We simulated a real-time prediction scenario, in which a model obtains new data every hour and is able to raise an alarm at any given time-point. Such a setting is more closely aligned with a possible clinical implementation of an early warning system as opposed to the majority of existing sepsis prediction studies using ML.13
We also acknowledge some limitations to our study. This was a retrospective, observational study. Even though we simulated a real-time prediction scenario, a prospective international evaluation is necessary in order to assess the clinical utility of bed-side sepsis predictions. Despite the large resulting sample size, many patients and even sites (in the case of the eICU dataset) had to be excluded from all analyses due to their insufficient data quality. Such exclusions may introduce selection bias, which could affect the model performance for certain subgroups of patients in future applications. Finally, another limitation was the difference in reporting of body fluid sampling information across databases. Due to this, on two databases we had to use an alternative definition of suspected infection, which relied on multiple administrations of antibiotics. However, on databases where this was possible, we successfully validated this definition against the original definition, showing that the two definitions have a good overlap.
In a large international cohort of more than 136,000 patient ICU admissions, we successfully developed and externally validated a deep learning system that recognised sepsis patients in previously unseen hospitals, using information on vital signs and laboratory measurements. We hope that the harmonised dataset resulting from our study and the performed analyses will help pave the way for international clinical validation studies to deploy sepsis prediction models that were externally statistically validated.
Contributors
M.M., N.B., D.P., and K.B. conceived the study. N.M., P.B., and K.B. supervised the study. M.M., N.B., D.P., M.H., B.R., P.B., and K.B. designed the experiments. N.B. and D.P. performed the cleaning, harmonization, and label annotation. M.M. and M.H. implemented the filtering and feature extraction. M.H. and M.M. implemented the deep learning models. M.M., N.B., and D.P. implemented the non-deep ML models. N.B. implemented the clinical baselines. M.M. designed the patient-focused evaluation. M.M. and B.R. implemented patient-focused evaluation plots. B.R. and M.M. implemented and designed the performance plots. M.H. and B.R. implemented the Shapley value calculation. B.R. designed, implemented, and performed the Shapley value analysis. M.H. and M.M. analysed the calibration of the models. M.M. ran the internal and external validation experiments for all methods. M.M. ran the hyperparameter search of the deep learning models and LightGBM. B.R. ran the hyperparameter search of Logistic regression. N.B. investigated different feature sets. M.M. implemented and ran the max pooling strategy. M.M. designed the pipeline overview figure. D.P. and B.R. designed the data harmonisation figure. N.B. designed the risk score illustration and the study flow chart. D.P. devised the dataset table. P.B. and K.B. advised on algorithmic modelling, statistical interpretation and evaluation. N.B. and M.M. verified the underlying data. All authors had full access to all the data. All authors contributed to the interpretation of the findings and to the writing of the manuscript.
Data sharing statement
All the raw data used in this manuscript is publicly available for accredited researchers. MIMIC-III, eICU, and HiRID are available via Physionet.37 Data from the AUMC database is available via the website of Amsterdam Medical Data Science. The data loading was performed using the “ricu” R-package.38 All code used for extracting, cleaning, filtering and modelling will be made available immediately upon publication, ensuring end-to-end reproducibility of all results presented.
Declaration of interests
Karsten Borgwardt has a patent application submitted for a biomarker for Long COVID, with no thematic link to the current manuscript. The authors declare no competing interests.
Acknowledgements
This study was supported by the grant #2017-110 of the Strategic Focal Area “Personalized Health and Related Technologies (PHRT)” of the ETH Domain for the SPHN/PHRT Driver Project “Personalized Swiss Sepsis Study”. The study was also funded by the Alfried Krupp Prize of the Alfried Krupp von Bohlen und Halbach-Stiftung (K.B.). We thank https://diagrams.net/ for their open-access tool for creating figures of unrestricted usage.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2023.102124.
Appendix A. Supplementary data
References
- 1.Dellinger R.P., Levy M.M., Rhodes A., et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med. 2013;41:580–637. doi: 10.1097/CCM.0b013e31827e83af. [DOI] [PubMed] [Google Scholar]
- 2.Hotchkiss R.S., Moldawer L.L., Opal S.M., Reinhart K., Turnbull I.R., Vincent J.-L. Sepsis and septic shock. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaukonen K.-M., Bailey M., Suzuki S., Pilcher D., Bellomo R. Mortality related to severe sepsis and septic shock among critically ill patients in Australia and New Zealand, 2000-2012. JAMA. 2014;311:1308–1316. doi: 10.1001/jama.2014.2637. [DOI] [PubMed] [Google Scholar]
- 4.Ferrer R., Martin-Loeches I., Phillips G., et al. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: results from a guideline-based performance improvement program. Crit Care Med. 2014;42:1749–1755. doi: 10.1097/CCM.0000000000000330. [DOI] [PubMed] [Google Scholar]
- 5.Pruinelli L., Westra B.L., Yadav P., et al. Delay within the 3-hour surviving sepsis campaign guideline on mortality for patients with severe sepsis and septic shock∗. Crit Care Med. 2018;46:500–505. doi: 10.1097/CCM.0000000000002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seymour C.W., Gesten F., Prescott H.C., et al. Time to treatment and mortality during mandated emergency care for sepsis. N Engl J Med. 2017;376:2235–2244. doi: 10.1056/NEJMoa1703058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osthoff M., Gürtler N., Bassetti S., et al. Impact of MALDI-TOF-MS-based identification directly from positive blood cultures on patient management: a controlled clinical trial. Clin Microbiol Infect. 2017;23:78–85. doi: 10.1016/j.cmi.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Pickering B.W., Gajic O., Ahmed A., Herasevich V., Keegan M.T. Data utilization for medical decision making at the time of patient admission to ICU. Crit Care Med. 2013;41:1502–1510. doi: 10.1097/CCM.0b013e318287f0c0. [DOI] [PubMed] [Google Scholar]
- 9.Hyland S.L., Faltys M., Hüser M., et al. Early prediction of circulatory failure in the intensive care unit using machine learning. Nat Med. 2020;26:364–373. doi: 10.1038/s41591-020-0789-4. [DOI] [PubMed] [Google Scholar]
- 10.Tomašev N., Glorot X., Rae J.W., et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019;572:116–119. doi: 10.1038/s41586-019-1390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy M.M., Fink M.P., Marshall J.C., et al. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med. 2003;31:1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- 12.Singer M., Deutschman C.S., Seymour C.W., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moor M., Rieck B., Horn M., Jutzeler C.R., Borgwardt K. Early prediction of sepsis in the ICU using machine learning: a systematic review. Front Med. 2021;8 doi: 10.3389/fmed.2021.607952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleuren L.M., Klausch T.L.T., Zwager C.L., et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med. 2020;46:383–400. doi: 10.1007/s00134-019-05872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong A., Otles E., Donnelly J.P., et al. External validation of a widely implemented proprietary sepsis prediction model in hospitalized patients. JAMA Intern Med. 2021;181(8):1065–1070. doi: 10.1001/jamainternmed.2021.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thoral P.J., Peppink J.M., Driessen R.H., et al. Sharing ICU patient data responsibly under the Society of Critical Care Medicine/European Society of Intensive Care Medicine Joint Data Science Collaboration: the Amsterdam University Medical Centers Database (AmsterdamUMCdb) example. Crit Care Med. 2021;49:e563–e577. doi: 10.1097/CCM.0000000000004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson A.E.W., Pollard T.J., Shen L., et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3 doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollard T.J., Johnson A.E.W., Raffa J.D., Celi L.A., Mark R.G., Badawi O. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci Data. 2018;5 doi: 10.1038/sdata.2018.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaswani A., Shazeer N., Parmar N., et al. In: Advances in neural information processing systems. Guyon I., Luxburg U.V., Bengio S., et al., editors. Vol. 30. Curran Associates, Inc.; 2017. Attention is all you need; pp. 5998–6008. [Google Scholar]
- 20.Cho K., van Merrienboer B., Bahdanau D., Bengio Y. 2014. On the properties of neural machine translation: encoder-decoder approaches. arXiv [cs.CL]http://arxiv.org/abs/1409.1259 [Google Scholar]
- 21.Ke G., Meng Q., Finley T., et al. In: Advances in neural information processing systems. Guyon I., Luxburg U.V., Bengio S., et al., editors. Curran Associates, Inc.; 2017. LightGBM: a highly efficient gradient boosting decision tree. [Google Scholar]
- 22.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Ser B Stat Methodol. 1996;58:267–288. [Google Scholar]
- 23.Jones M. NEWSDIG: the national early warning score development and implementation group. Clin Med. 2012;12:501–503. doi: 10.7861/clinmedicine.12-6-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subbe C.P., Kruger M., Rutherford P., Gemmel L. Validation of a modified early warning score in medical admissions. QJM. 2001;94:521–526. doi: 10.1093/qjmed/94.10.521. [DOI] [PubMed] [Google Scholar]
- 25.Bone R.C., Balk R.A., Cerra F.B., et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 26.Vincent J.L., Moreno R., Takala J., et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the working group on sepsis-related problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22:707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 27.Wardi G., Carlile M., Holder A., Shashikumar S., Hayden S.R., Nemati S. Predicting progression to septic shock in the emergency department using an externally generalizable machine-learning algorithm. Ann Emerg Med. 2021;77:395–406. doi: 10.1016/j.annemergmed.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin Y., Cho K.-J., Lee Y., et al. Multicenter validation of a deep-learning-based pediatric early-warning system for prediction of deterioration events. Acute Crit Care. 2022;37:654–666. doi: 10.4266/acc.2022.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundberg S.M., Lee S.-I. Proceedings of the 31st international conference on neural information processing systems. 2017. A unified approach to interpreting model predictions; pp. 4768–4777. [Google Scholar]
- 30.Leone M., Asfar P., Radermacher P., Vincent J.-L., Martin C. Optimizing mean arterial pressure in septic shock: a critical reappraisal of the literature. Crit Care. 2015;19:101. doi: 10.1186/s13054-015-0794-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genga K.R., Russell J.A. Update of sepsis in the intensive care unit. J Innate Immun. 2017;9:441–455. doi: 10.1159/000477419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Habib A.R., Lin A.L., Grant R.W. The epic sepsis model falls short—the importance of external validation. JAMA Intern Med. 2021;181:1040–1041. doi: 10.1001/jamainternmed.2021.3333. [DOI] [PubMed] [Google Scholar]
- 33.Shashikumar S.P., Wardi G., Malhotra A., Nemati S. Artificial intelligence sepsis prediction algorithm learns to say “I don’t know”. NPJ Digit Med. 2021;4:134. doi: 10.1038/s41746-021-00504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams R., Henry K.E., Sridharan A., et al. Prospective, multi-site study of patient outcomes after implementation of the TREWS machine learning-based early warning system for sepsis. Nat Med. 2022;28(7):1455–1460. doi: 10.1038/s41591-022-01894-0. [DOI] [PubMed] [Google Scholar]
- 35.Dayan I., Roth H.R., Zhong A., et al. Federated learning for predicting clinical outcomes in patients with COVID-19. Nat Med. 2021;27:1735–1743. doi: 10.1038/s41591-021-01506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rieke N., Hancox J., Li W., et al. The future of digital health with federated learning. NPJ Digit Med. 2020;3:119. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldberger A.L., Amaral L.A., Glass L., et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000;101:E215–E220. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 38.Bennett N., Plečko D., Ukor I.-F., Meinshausen N., Bühlmann P. ricu: R’s interface to intensive care data. GigaScience. 2023;12:p.giad041. doi: 10.1093/gigascience/giad041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.