Integrating HTS with molecular profiling is a powerful tool for expanding precision medicine to support drug treatment recommendations and broaden the therapeutic options available to high-risk pediatric cancers.
Abstract
For one-third of patients with pediatric cancer enrolled in precision medicine programs, molecular profiling does not result in a therapeutic recommendation. To identify potential strategies for treating these high-risk pediatric patients, we performed in vitro screening of 125 patient-derived samples against a library of 126 anticancer drugs. Tumor cell expansion did not influence drug responses, and 82% of the screens on expanded tumor cells were completed while the patients were still under clinical care. High-throughput drug screening (HTS) confirmed known associations between activating genomic alterations in NTRK, BRAF, and ALK and responses to matching targeted drugs. The in vitro results were further validated in patient-derived xenograft models in vivo and were consistent with clinical responses in treated patients. In addition, effective combinations could be predicted by correlating sensitivity profiles between drugs. Furthermore, molecular integration with HTS identified biomarkers of sensitivity to WEE1 and MEK inhibition. Incorporating HTS into precision medicine programs is a powerful tool to accelerate the improved identification of effective biomarker-driven therapeutic strategies for treating high-risk pediatric cancers.
Significance:
Integrating HTS with molecular profiling is a powerful tool for expanding precision medicine to support drug treatment recommendations and broaden the therapeutic options available to high-risk pediatric cancers.
Graphical Abstract
Introduction
Currently, most precision medicine programs aim to molecularly profile patients’ tumors to identify the drivers of their disease and make treatment recommendations based on these findings. Tumor molecular profiling has identified actionable driver events in 34% to 87% of pediatric cancers (1–11). Zero Childhood Cancer (ZERO), Australia's national childhood cancer precision medicine program, identified at least one genetic driver in 94% of the enrolled patients, while 71% of the patients had a potentially druggable target (12). Although promising, this still means that genomic analysis alone does not result in a targeted treatment recommendation for one-third of the patients. Crucially, the number of drivers for which therapeutic intervention has resulted in significant clinical responses are limited. For example, CDK4/6 inhibitors such as palbociclib or ribociclib to target biallelic loss of CDKN2A/B in high-risk pediatric cancers have shown minimal clinical efficacy (13, 14). Tumor heterogeneity and the acquisition of resistance mutations after treatment further complicate the translation of tumor molecular characteristics into effective personalized treatments (15, 16). A better understanding of the relationships between the complex genomes and transcriptomes of cancer and responses to targeted agents would help address these limitations.
Patient-derived in vitro tumor models that accurately recapitulate the tumor characteristics have been successfully developed for adult (17–19) and pediatric (20–23) cancers. These models can support clinical treatment decision making and identify associations between the molecular characteristics of a tumor and its drug sensitivity profile. ZERO undertakes in vitro high-throughput drug screening (HTS) and in vivo drug efficacy testing in patient-derived xenograft (PDX) models as an additional strategy for personalized treatment identification, in conjunction with comprehensive molecular profiling. Recently, we published our pilot trial of in vitro HTS on patient-derived samples and showed the clinical benefit (24).
In this study, we confirm known associations between tumor drivers and their inhibitors, but also highlight the need to improve our understanding of molecular drug targets in pediatric cancer. Thus, we also show that integrating the drug data with molecular profiling can lead to the identification of novel predictive biomarkers. Our in vitro drug testing pipeline confirms that drug efficacy profiles are retained through in vitro or in vivo expansion of tumor cells and responses are consistent with in vivo PDX models and clinical responses in treated patients. In addition, we describe how we use in vitro HTS on patient samples to identify potentially effective combinations for individual patients.
Materials and Methods
Patient specimen procurement
The PRecISion Medicine (PRISM) clinical trial (NCT03336931) was conducted as part of the Australian ZERO Childhood Cancer program from September 2017 and was approved by the Hunter New England Human Research Ethics Committee of the Hunter New England Local Health District (reference no. 17/02/015/4.06). Patients ≤21 years old with suspected or confirmed diagnosis of a very rare or high-risk malignancy and patients >21 years old with a suspected or confirmed diagnosis of high-risk pediatric-type cancer could be enrolled in the study, as described previously (12). Written informed consent was obtained for each participant prior to inclusion in the study. Fresh tumor tissues from biopsy or surgical resection were processed for molecular profiling, as described previously (12), and the establishment of patient-derived models for preclinical drug testing (See Supplementary Materials and Methods).
Tissue dissociation and establishment of patient-derived cultures
A Human Dissociation Kit (Miltenyi Biotec) was used to enzymatically digest all extracranial primary tumor specimens or PDXs into single-cell suspensions. Central nervous system (CNS) tumors were dissociated as described previously (25). Tumor-type specific culture media and supplements were used for maintaining and expanding PDX or patient-derived cultures (Supplementary Fig. S1A). Media were obtained from Life Technologies, growth factors from Jomar Life Research, and other supplements from Invitrogen, unless stated otherwise. Brain tumor cells were grown in 50% Dulbecco's Modified Eagle Medium/Nutrient Mixture F12 (DMEM/F12) plus 50% Neurobasal-A Medium supplemented with 2 mmol/L GlutaMAX-I, 1 mmol/L Pyruvate, 100 μmol/L non-essential amino acids (NEAA), 10 μmol/L 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 1× antibiotic/antimycotic solution (AAS), 20 ng/mL basic fibroblast growth factor (bFGF), and 20 ng/mL epidermal growth factor (EGF) with or without 2 μg/mL Heparin, 10 ng/mL platelet-derived growth factor (PDGF)-AA, and 10 ng/mL PDGF-BB. Neuroblastoma cells were grown in Iscove's Modified Dulbecco's Medium (IMDM) supplemented with 20% (v/v) fetal bovine serum (FBS; Life Technologies), 1× insulin-transferrin-selenium (ITS; Life Technologies), and 10 μg/mL gentamicin (Life Technologies), while dissociated cells from other solid tumor types were grown in Minimum Essential Medium (MEM) α supplemented with 10% (v/v) FBS, 1× ITS, 1× AAS, and 10 nmol/L Y-27632 (Selleck Chemicals). Cells from hematologic malignancies were cultured in Roswell Park Memorial Institute (RPMI) 1640 Medium supplemented with 20% (v/v) FBS and penicillin–streptomycin (lymphoma), Mononuclear Cell Medium [acute myeloid leukemia (AML); PromoCell], StemSpan Serum-Free Expansion Medium (SFEM) II supplemented with 1× StemSpan CD34+ Expansion Supplement and 1× AAS (AML), AIM V Serum-Free Medium on a layer of human telomerase reverse transcriptase-immortalized mesenchymal stromal cells [MSC-hTERT cells; acute lymphoblastic leukemia (ALL)], or AIM V Serum-Free Medium supplemented with 10% GM-CSF (other hematologic malignancies; Stem Cell Technologies). Cells were maintained at 37°C under 5% CO2. See Supplementary Fig. S1A for the oxygen conditions used for the different tumor types. CNS and neuroblastoma tumor cells were expanded in conventional cell culture flasks. All other types of extracranial solid tumor cells were initially grown in 96-well ultra-low attachment (ULA) U-bottom plates for 14 to 21 days to prevent the growth of nontumor cells. Cells were then collected and disaggregated using Accutase (Sigma-Aldrich) before propagating in culture flasks adherently. Cell viability and proliferation were evaluated by trypan blue cell count throughout the expansion. No in vitro expansion was conducted for lymphoma and leukemia cells. MSC-hTERT cells were maintained in RPMI1640 media supplemented with 10% FBS and were plated in AIMV media overnight prior to seeding of leukemia cells.
In vitro HTS
Freshly dissociated or expanded tumor cells were seeded in 384- or 1,536-well plates as single-cell suspensions using the Multidrop Combi Reagent Dispenser (Thermo Fisher Scientific). ALL cells were seeded on a layer of human telomerase reverse transcriptase-immortalized bone marrow mesenchymal stem (MSC-hTERT) cells (seeded 24 hours before). After 3 hours, one day or three days incubation, cells were exposed to a drug library containing up to 126 drugs (MedChem Express; Selleck Chemicals) approved or in (pre)clinical development for pediatric cancer. Depending on material availability samples were exposed to the full or a smaller version of the drug library (i.e., 56, 70, 111, or 112 drugs). Drugs were tested in duplicate and added in final concentrations of 0.5 nmol/L–5 μmol/L (10-fold serial dilutions), using the Hamilton STAR liquid handling robot or the Echo 550 acoustic liquid handler (Labcyte). Thonzonium bromide and DMSO were included as positive and negative control, respectively. Three days after drug addition, metabolic activity was measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). Drug effects for ALL samples were measured using the CyQUANT Direct Cell Proliferation Assay (Thermo Fisher Scientific) and the images were captured using the Operetta (PerkinElmer). Data analysis was performed using the ActivityBase (IDBS) software platform. Raw values for each data point were converted into viabilities using the following formula: ([readout value drug – average readout positive controls]/[average readout negative controls – average readout positive controls] × 100%). Resulting cell viabilities were used to generate dose–response curves using a four-parameter logistic function and to calculate the area under the dose–response curve (AUC) and half-maximal inhibitory concentrations (IC50) values. See Supplementary Materials and Methods for a detailed description of the authentication and validation of each sample for HTS.
In vivo efficacy studies
All animal studies were approved by the Animal Care and Ethics Committee of UNSW Sydney, approval numbers 17/101B, 19/82B and 20/82B. PDX tumor tissues were subcutaneously xenotransplanted in recipient NSG mice. Tumor growth was monitored twice weekly by digital caliper measurements, and when tumors reached a volume of approximately 100 mm3, mice were randomized to vehicle control or indicated single drugs or drug combinations. Mice were treated for up to four weeks unless stated otherwise. Best responses were calculated using the following formula: [(lowest measured tumor volume during treatment/tumor volume at the start of treatment) − 1] × 100%. Event-free survival (EFS) was calculated as described in ref. 26. Xenografts from the vehicle control groups were used for authentication by STR profiling and validation by SNP profiling and cytopathology examination. For the in vivo efficacy study in the HGG PDX model (zcc116), tumor cells were injected intracranially, and on day 82 postinjection, mice were randomized to vehicle control or indicated single drugs or drug combination. Mice were treated for up to four weeks, and EFS was defined by 20% weight loss and/or lethargy. See Supplementary Materials and Methods for the dosing schedules and routes of administration for the in vivo tested drugs and drug combinations.
Whole genome and RNA sequencing data
Of the 125 authenticated and validated samples that underwent successful HTS, 98 had matching whole genome sequencing (WGS) + RNA sequencing (RNA-seq), 13 WGS only and 6 RNA-seq only. No matching molecular profiling was available for the remaining samples. To be considered a match the original sample needed to have come from the patient's same biomaterial. WGS and RNA-seq of tumor biopsies have been performed as described in detail previously (12). RNA-seq expression data used in the current study was reported in transcripts per million (TPM), and WGS data were utilized to study germline and somatic single-nucleotide variants and copy number variations and to establish tumor ploidy. Normalized copy numbers for a gene were calculated using the ratio of the actual copy number and the corresponding tumor ploidy.
Gene expression correlation analysis with drug efficacy
Correlations with gene expression were analyzed for the top 50 drugs with the most varying AUC values for all nonhematologic samples within our cohort with matching RNA-seq data (N = 90) based on the highest coefficient variations (CV). CV analysis was also performed on the gene expression data in the 90-sample cohort, of which, the top 75% most variable genes (N = 19,750) were then filtered to only select genes for which TPM > 1 in ≥5 samples (N = 14,865). Pearson correlation analysis was performed between the log2-transformed TPM values of these genes and AUC Z scores for each of the 50 drugs.
Gene set enrichment analysis
We performed gene set enrichment analysis (GSEA; software version v4.2.1; ref. 27) using the Pearson correlation ranking metric comparing gene expression against drug AUC Z score with 1000 permutations for both adavosertib and trametinib. For adavosertib, gene ontology (GO), hallmark and curated gene sets enriched in adavosertib sensitive solid tumor samples containing 15–1,500 genes with a normalized enrichment score (NES) ≤ 2, list of >10%, and FDR q value < 0.01 were used for further investigation and visualization in Cytoscope (software version 3.8.2; ref. 28). For trametinib, GO biological processes (GO:BP) gene sets containing 15–1,500 genes with an NES <0 were used for further analysis.
In vitro culture of SHEP-21N
MYCN dependency of adavosertib in neuroblastoma cells was studied using the SHEP-21N cell line (RRID:CVCL_9812), which expresses MYCN under a doxycycline repressible promoter (29). SHEP-21N cells were kindly provided by M. Schwab. Cells were tested for Mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza; negative result on 31/03/2023) and kept in culture for no longer than one month after thawing. SHEP-21N cells were cultured in RPMI medium 1640 (Gibco, #22400–089), supplemented with 10% FBS (Gibco, #10100147) and 1× of Antibiotic Antimycotic Solution (Sigma-Aldrich, #A5955). MYCN expression was suppressed by 1 μg/mL doxycycline (Sigma-Aldrich #D3447) at least 72 hours prior to the experiments. Cell densities for the experiments were optimized separately for the MYCN-expressing SHEP-21N cells and SHEP-21N cells in which MYCN was suppressed in the presence of doxycycline (data not shown).
MYCN-dependent effects of adavosertib on neuroblastoma cell viability
Untreated SHEP-21N cells (750 cells/well) and SHEP-21N cells under doxycycline pressure for ≥3 days (1,000 cells/well) were seeded in 384-well microplates (Greiner Bio-One, #781080) and incubated overnight. Next, cells were exposed to 14.6 nmol/L to 15 μmol/L adavosertib in 2-fold serial dilutions using the Tecan D300e Digital Dispenser. DMSO and thonzonium bromide (20 μmol/L) were included as negative and positive control, respectively. Readout was conducted on days 1 and 3 after treatment initiation using the CellTiter-Glo 2.0 Luminescent Cell Viability Assay (Promega) and the EnSpire Alpha multimode plate reader (PerkinElmer) for luminescence detection. Cell viabilities were calculated as described for in vitro HTS and dose–response curves were generated using GraphPad Prism (version 9.3.1). All experiments were conducted in triplicate in three independent experiments. Each data point represents the mean ± standard deviation (SD). The DepMap GDSC dataset (https://depmap.org/portal/; ref. 30) was utilized to explore the efficacy of adavosertib (i.e., MK-1775) in an independent cohort. A one-sided Wilcoxon test was performed to statistically compare the AUC (31) difference between MYCN amplified (N = 6) versus MYCN wild-type (N = 14) classical neuroblastoma cell lines.
In vitro flow cytometry
Untreated SHEP-21N cells (75,000 cells/well) and SHEP-21N cells under doxycycline pressure for ≥3 days (100,000 cells/well) were seeded in 6-well plates and incubated overnight. Cells were subsequently treated with 1.88 μmol/L adavosertib or DMSO (control). Twenty-four hours after treatment, cells were trypsinized and fixed with 70% ethanol. Fixed cells were permeabilized with 0.5% Triton X-100 in 0.1% BSA/PBS and incubated at room temperature for 30 minutes. Next, the samples were stained with 25 μg/mL propidium iodide (Sigma-Aldrich, #P4864) in the presence of 2 μg/mL DNase-free RNase (Roche, #11119915001) in 0.1% BSA/PBS. Cell-cycle analysis was performed using the BD FACSCanto A Flow Cytometer (BD Biosciences). Twenty-five thousand viable single cells were gated, and the results were analyzed by FlowJo 10.8.1 software (BD Biosciences).
In vitro Western blotting
The following anti-human primary monoclonal antibodies were used: mouse MYCN (B8.4.B) (1:1,000, Santa Cruz Biotechnology: sc-53993), mouse β-actin (AC-74; 1:10,000, Sigma: A2228), rabbit phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) XP (1:1,000, Cell Signaling Technology: #4370), rabbit p44/42 MAPK (Erk1/2) (137F5) (1:1,000, Cell Signaling Technology: #4695), rabbit phospho-Akt (Ser473) (193H12) (1:1,000, Cell Signaling Technology: #4058), rabbit Akt (1:1,000, Cell Signaling Technology: #9272), rabbit phospho-S6 ribosomal protein (Ser235/236) (1:1,000, Cell Signaling Technology: #2211), mouse S6 ribosomal protein (54D2) (1:1,000, Cell Signaling Technology: #2317), and rabbit β-actin (D6A8) (1:2,000, Cell Signaling Technology: #8457). The following secondary antibodies were used: HRP-linked sheep anti-mouse IgG (1:10,000, GE Healthcare: NA931V), HRP-linked goat anti-rabbit IgG (7074p2) (1:2,000, Cell Signaling Technology: #7074), and HRP-linked horse anti-mouse IgG (1:2,000, Cell Signaling Technology: #7076). See Supplementary Materials and Methods for a detailed protocol.
Statistical analysis
Statistical analyses were performed by either the Pearson correlation coefficient test, the Wilcoxon rank sum test for unpaired data followed by the Benjamini–Hochberg (BH) procedure for multiple tests correction if needed, or the Kruskal–Wallis test followed by a post hoc Dunn test for multiple groups comparison. Statistical differences between in vivo event-free survival rates were calculated using the Gehan–Breslow–Wilcoxon test. Statistical analysis was either performed in R (version 4.0.3) or GraphPad Prism (version 9.1.0). A P value below 0.05 was considered significant.
Data availability
The datasets used in this study that were previously published are available from the European Genome-phenome Archive under accession number EGAS00001004572 (https://ega-archive.org/studies/EGAS00001004572) and EGAS00001007029 (https://ega-archive.org/studies/EGAS00001007029). All other raw data generated in this study are available upon request from the corresponding author.
Results
Different tumor sample sources were equally evaluable and informative following in vitro HTS
ZERO performs molecular profiling (WGS, RNA-seq, and methylation array) on high-risk childhood cancers (12). Our TARGET pilot study showed that additional in vitro HTS and in vivo PDX drug testing of the patient's own tumor cells can also be performed on patients with available fresh tumor tissue (24). We expanded this platform nationally through ZERO's national PRISM study. Here, we present the results of patients that were enrolled in the PRISM study between September 2017 and September 2022. In vitro HTS was attempted for 169 tumor samples from 166 patients. A library containing 126 drugs was utilized as they were either approved by the Food and Drug Administration and/or Therapeutic and Goods Administration or in late preclinical or early experimental trial stages of development for pediatric cancer patients (Supplementary Table S1). Tumor samples for HTS were generated by dissociation of tumor biopsies into viable single cells, in vitro expansion of dissociated tumor cells, in vivo expansion of tumor pieces, or in vivo expansion followed by in vitro expansion of PDX-dissociated tumor cells (Fig. 1A). Authentication was performed by STR profiling to ensure sample identity (screened cells are from the original patient). SNP array, morphologic assessment of hematoxylin and eosin– and IHC-stained sections, flow cytometry (FC), or, for screens on freshly dissociated tumor cells only, WGS was used to validate tumor content and whether the screened cells matched the original tumor (Fig. 1A and B; Supplementary Fig. S1B–S1E). HTS was successfully undertaken for 125 of the 169 tumor samples covering a wide spectrum of pediatric cancers: brain tumors, hematologic malignancies, neuroblastoma, sarcoma, and other solid tumors (Fig. 1C and D; Supplementary Table S2). Three patients had two samples screened that were taken at different time points before and/or after treatment initiation: a diagnosis and relapse sample for one hematologic malignancy and two relapse samples for one sarcoma and one brain tumor. The remaining 44 samples failed HTS (Fig. 1D). In 24 of these cases, they failed due to the outgrowth of non-neoplastic cells during expansion of the dissociated tumor cells or the lack of tumor cells in the original sample. Two samples failed authentication and the remaining 18 samples failed because of either poor-quality dose–response curves caused by, for example, low luminescence values due to insufficient cell numbers or variable viability of tumor cells in multi-well plates independent of drug treatment (13/18), bacterial/fungal contamination (3/18), or technical failure (2/18).
Figure 1.
Drug response profiles are independent of tumor cell expansion. A, Workflow for in vitro HTS on tumor cells derived from patients enrolled in the ZERO childhood cancer precision medicine program. HTS is performed on freshly dissociated tumor cells or tumor cells expanded by in vitro culturing and/or in vivo growth. Samples are authenticated by STR profiling and validated by at least one of the following methods: SNP array, IHC, flow cytometry (FC) or, for screens on freshly dissociated tumor cells only, WGS. B, Hematoxylin and eosin (H&E) staining of representative patient tumor samples (left) and matching 3D primary cultures (middle), and IHC of tumor markers in the 3D primary cultures (right). Tumor markers used: CD99 for Ewing sarcoma sample zcc38, inhibin for adrenocortical carcinoma (ACC) sample zcc292, and WT1 for Wilms tumor (WT) sample zcc384. Bottom images show Ki67 staining results for matching tumor biopsy and 3D primary culture of WT zcc384. C, Cancer types and subtypes with successful in vitro HTS (N = 125). D, Number of patient-derived samples for main types of pediatric cancer that underwent in vitro HTS. Percentages indicate samples for which HTS, authentication by STR profiling and validation by SNP array profiling, IHC, flow cytometry, or WGS was successful. E, Successful in vitro HTS on freshly dissociated and expanded samples highlighted by cancer types. F, Number of samples screened by different sized libraries and the associated methodology used to generate the samples for HTS (i.e., fresh dissociation of tumor cells or in vitro and/or in vivo expansion of tumor cells). G, Heatmap of the targeted drug response profiles across the 125-sample cohort. Drug response profiles were established by calculating the median AUC Z scores for targeted drugs grouped according to MOA. Median AUC Z scores are represented by a color scale from blue (resistant) to red (sensitive). MOAs and tumor samples are ordered by unsupervised hierarchical clustering. Top annotations indicate tumor type and type of sample. Tumor type key: brain tumors (BT), hematologic malignancies (HM), neuroblastoma (NB), sarcoma tumors (SAR), solid other (SO). Cancer subtype key: diffuse midline glioma (DMG), high-grade glioma (HGG), medulloblastoma (MB), B-precursor acute lymphoblastic leukemia (Pre-B-ALL), acute myeloid leukemia (AML), T-lymphoblastic leukemia (T-LBL), non-MYCN-amplified (NonMYCNA), MYCN-amplified (MYCNA), alpha thalassemia/mental retardation syndrome X-linked (ATRX), Ewing sarcoma (ES), rhabdomyosarcoma (RMS), osteosarcoma (OS), Wilms tumor (WT), adrenocortical carcinoma (ACC), malignant rhabdoid tumor (MRT).
HTS on freshly dissociated tumor cells was done for 40 of 125 tumor samples, representing all tumor groups (Fig. 1E). In 24 of these cases cell numbers were sufficient to screen the full 126-compound library (Fig. 1F). Of the remaining 85 samples, 83 were expanded in vitro or in vivo only, with median expansion times of 112 and 92 days, respectively (Supplementary Fig. S1F). Notably, 70 (82.4%) of the expanded tumor samples completed successful HTS while the patient was still alive. This suggests that HTS, even when it requires in vitro or in vivo expansion, can still support clinical treatment decision making for individual patients.
To assess whether the expansion of tumor cells influenced the nature of observed drug responses, we performed unsupervised clustering of the tumor samples based on their drug sensitivity profiles. Drug sensitivity profiles were determined by establishing the area under the dose–response curve (AUC) and calculating the AUC Z score for each compound for each model. The drugs were then grouped according to mechanism of action (MOA), and median AUC Z scores were calculated. For both targeted agents (Fig. 1G) and chemotherapeutics (Supplementary Fig. S2A), there was no clear clustering of drug responses with either of the four strategies used in generating the samples for HTS. Importantly, a strong correlation was observed between drug responses of freshly dissociated or in vivo expanded cells from the same patient (Supplementary Fig. S2B). Taken together, these data show that drug sensitivity is independent of the type of tumor sample used for HTS.
Drug–drug sensitivity correlations identify potential effective combination therapies
We next sought to determine whether drugs in the same class have similar efficacy profiles, and to identify potential interactions between different drug classes. Using a correlation analysis of AUC Z scores, we identified 340 drug pairs from a possible 7,875 combinations that had similar efficacy profiles across the cohort (r ≥ 0.6; Fig. 2; Supplementary Table S3). In 114 of the 340 drug pairs involved agents in the same class and most of these cases shared the same target(s), such as microtubule stabilizers in mitosis, DNA topoisomerase I and II inhibitors in DNA replication, mTORC1 inhibitors in PI3K–AKT–mTOR signaling and FGFR inhibitors in receptor tyrosine kinase signaling.
Figure 2.
Correlations between drug response profiles. Pearson correlation coefficient heatmap of the relationship between drug AUC Z scores across the 125-sample cohort. Pearson r values are represented by a color scale from red (positive linear relationship) to blue (negative linear relationship). Chemotherapeutics (gray; top) and targeted drugs (black; bottom) are ordered by unsupervised hierarchal cluster analysis using Euclidian distance. Drugs are annotated by their MOA.
Exceptions included PARP inhibitors, as exemplified by the moderate AUC Z score correlations between veliparib and the other PARP inhibitors olaparib (r = 0.45) and talazoparib (r = 0.43; Fig. 3A). Veliparib, olaparib, and talazoparib are potent inhibitors of PARP1/2 catalytic activities, but have variable DNA entrapment potency, veliparib being the weakest and talazoparib the strongest (32, 33). Accordingly, we observed that veliparib was the least and talazoparib the most potent PARP inhibitor with the highest and lowest AUC values in our cohort, respectively (Fig. 3B). Importantly, a very strong correlation was observed between talazoparib AUC Z scores in vitro and effects on event-free survival in vivo (Fig. 3C). Another exception was the only moderate correlation between the efficacy profile of selumetinib and the efficacy profiles of the other MEK inhibitors trametinib and cobimetinib (r = 0.56 for both), while a very strong AUC Z score correlation was observed between trametinib and cobimetinib (r = 0.88; Supplementary Fig. S2C).
Figure 3.
In vitro PARP inhibitor responses correlate with in vivo responses and correlated drug response profiles identify effective combinations for individual samples. A, AUC Z score correlations across the cohort between PARP inhibitors: olaparib versus veliparib (left), talazoparib versus veliparib (middle), and talazoparib versus olaparib (right). B, Comparison of veliparib, olaparib, and talazoparib AUC values shows increased efficacy with increasing PARP DNA entrapment potency. C, Correlation between in vitro talazoparib AUC Z scores and in vivo responses in matching PDX models as established by calculating T/C values for each model using the following formula: [medium EFS time after talazoparib treatment (=T)]/[medium EFS time when untreated (=C)]. D, Correlation between the AUC Z scores of AURKA inhibitor alisertib and irinotecan metabolite SN-38. Samples sensitive to both alisertib and SN-38 that are used for in vivo combination testing in D are depicted in orange. E,In vivo effects of vehicle control (black), irinotecan plus temozolomide (green), monotherapy alisertib (blue), and alisertib in combination with irinotecan plus temozolomide (red) in PDX models for MRT (left), Wilms tumor (middle), and neuroectodermal tumor (NET; right). Top graphs show percentage change in tumor model for each PDX and bottom graphs show EFS. MRT, malignant rhabdoid tumor; WT, Wilms tumor; NET, neuroectodermal tumor. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. *, versus vehicle control; *, versus alisertib; *, versus irinotecan (IRN) plus temozolomide (TMZ).
Unexpected strong correlations across the 125-sample cohort were also observed between the efficacy profiles of some chemotherapeutic drugs and targeted agents. For example, the chemotherapeutic SN-38, the active metabolite of DNA topoisomerase I inhibitor irinotecan, correlated with PARP inhibitor talazoparib (r = 0.84), AURKA inhibitor alisertib (r = 0.79), and PLK1 inhibitor volasertib (r = 0.79; Supplementary Table S3). We used the correlation between SN-38 and alisertib to assess whether sensitivity to both drugs provided a rationale for combination treatment. A malignant rhabdoid tumor (MRT), a Wilms tumor, and neuroectodermal tumor (NET) sample demonstrated in vitro sensitivity to SN-38 and alisertib (Fig. 3D). Therefore, we performed in vivo drug testing on the matched PDXs using either alisertib as a single agent, irinotecan plus temozolomide or alisertib plus irinotecan and temozolomide, because of the clinical experience with alisertib in combination with irinotecan plus temozolomide in pediatric cancer treatment (34, 35). The combination therapy resulted in an improvement in overall survival in all three models (Fig. 3E; Supplementary Fig. S2D). Importantly, combining alisertib with irinotecan plus temozolomide did not result in increased toxicity, as shown by the limited changes in body weight for both alisertib monotherapy and the triple combination (Supplementary Fig. S2E). Results are in line with our previous observation that a Wilms tumor patient with in vitro sensitivity to both talazoparib and SN-38 responded to clinical combination treatment with talazoparib and irinotecan (36) and suggest that correlating drug sensitivity profiles identifies potential individualized drug combination strategies.
High-throughput drug testing more accurately reflects in vivo responses than molecular markers
The most potent targeted agents with the lowest median AUC and log2[IC50] values for the 125-sample cohort were: proteasome inhibitors bortezomib, carfilzomib and ixazomib; CDK1/2/5/9 inhibitor dinaciclib; pan-HDAC inhibitor panobinostat; and PI3K-AKT-mTOR signaling inhibitors gedatolisib, TAK228, and vistusertib (Fig. 4A; Supplementary Fig. S3A). We studied tumor-type specific sensitivity or resistance to drugs by calculating the difference between average AUC (Fig. 4B) and log2[IC50] (Supplementary Fig. S3B) Z scores of each drug for samples of the same tumor type compared to the remaining tumor samples. In keeping with their prioritization for clinical trials (NCT03194932, NCT03236857, NCT04161885), venetoclax was significantly more effective against hematologic malignancies compared with other types of pediatric cancer (Fig. 4B; Supplementary Fig. S3B; Supplementary Table S4). Interestingly, sarcomas were generally sensitive to ceritinib and crizotinib (Fig. 4B; Supplementary Fig. S3B; Supplementary Table S4). Because no increased sensitivity to the selective ALK inhibitor alectinib was observed, the increased efficacy of ceritinib and crizotinib against sarcomas is most likely the result of blocking targets other than ALK (i.e., IGF-1R and/or InsR for ceritinib and c-Met and/or ROS1 for crizotinib).
Figure 4.
In vitro HTS on patient-derived tumor samples confirms known associations between drug sensitivity and driver aberrations. A, AUC distribution across all samples for targeted drugs. Drugs are ordered from most (left) to least (right) effective based on lowest median AUC, followed by lowest quartile AUC values. Colors indicate the mechanism of action of each drug. B, Volcano plots of tumor type-specific sensitivity or resistance to drugs based on the AUC Z scores for each drug, using the following formula: [average AUC Z score for tumor type of interest] − [average AUC Z score for the remaining cohort]. A difference in average AUC Z score ≤ 0.5 and adjusted P < 0.01 is considered relatively resistant (blue) and a difference in average AUC Z score > 0.5 and adjusted P < 0.01 is considered relatively sensitive (red). Only significant targeted drugs are named in the figure. Circle size represents the number of samples within the given tumor type on which the indicated drug has been tested. C, AUC values for TRK inhibitor larotrectinib sulfate, BRAF inhibitors dabrafenib and vemurafenib, and ALK inhibitors ceritinib, crizotinib, and alectinib with samples harboring an NTRK fusion, BRAF V600E mutation, or ALK aberration (i.e., ALK F1245I mutation or EML4-ALK fusion) highlighted in red, respectively. Arrows indicate the BRAF V600E–mutated HGG and ALK F1245I–mutated NB sample used for in vivo efficacy testing in D and E, respectively. D,In vivo effects of vehicle control (saline; black), dabrafenib monotherapy (green), trametinib monotherapy (blue), and dabrafenib plus trametinib combination therapy (red) on EFS in the matching HGG PDX model of the BRAF V600E–mutated sample with in vitro sensitivity to BRAF inhibition. E,In vivo effects of vehicle control (black) and ALK inhibitors ceritinib (green), crizotinib (blue), alectinib (orange), and lorlatinib (red) in the matching NB PDX model of the ALK F1245I–mutated NB sample with in vitro sensitivity to ALK-targeting inhibitors. Left graph shows percentage change in tumor model for each PDX and right graph EFS. F, AUC values for MEK inhibitors trametinib (left), cobimetinib (middle), and selumetinib (right) in samples with (colored) and without (gray) driver aberrations in RAS–MAPK signaling. Shapes of the symbols indicate tumor type. Tumor type key: brain tumors (BT), hematologic malignancies (HM), neuroblastoma (NB), sarcoma tumors (SAR), solid other (SO). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. *, versus vehicle control.
Next, we examined correlations between pediatric cancer drivers and drug sensitivity. As expected, larotrectinib (NTRK inhibitor) was only effective in our sole high-grade glioma (HGG) sample that harbored a KANK1-NTRK2 fusion with AUC and IC50 values of 348 and 0.16 μmol/L, respectively, compared with AUC values >418 and IC50 values >0.84 μmol/L for all other models tested (Fig. 4C; Supplementary Fig. S3C and S3D). Similarly, the BRAF inhibitors dabrafenib and vemurafenib were most potent against the BRAF V600E-mutant HGG and melanoma samples, and the ALK F1245I-mutant neuroblastoma sample was among the samples most sensitive to ALK-targeting inhibitors ceritinib and crizotinib (Fig. 4C; Supplementary Fig. S3C and S3D). A HGG sample that harbored an EML4-ALK fusion was also tested and did not respond to ALK inhibition. This was in line with the previously described observation that patients harboring shorter EML4-ALK variants without parts of the tandem atypical β-propeller (TAPE) domain of EML4 might be less sensitive to ALK inhibition (37). Biomarker-driven positive responses to BRAF and ALK inhibition in vitro were in accordance with positive in vivo PDX responses (Fig. 4D and E) and clinical responses including the HGG patient harboring the NTRK fusion (Supplementary Fig. S3E–S3G). Finally, models responded significantly better to trametinib and cobimetinib in the presence of alterations in the RAS–MAPK pathway, including aberrations in oncogenes ALK, BRAF, KRAS, or NRAS and/or tumor suppressor genes NF1 or PTPN11 (Fig. 4F; Supplementary Fig. S3H). In agreement with the moderate correlation with trametinib and cobimetinib efficacy profiles (Supplementary Fig. S2C), the difference in potency for samples with versus without RAS–MAPK pathway alterations was less pronounced for selumetinib without significant difference based on log2[IC50] values (Fig. 4F; Supplementary Fig. S3H).
Driver variants in PI3K–AKT–mTOR and cell-cycle signaling are among the most frequently observed actionable events in pediatric cancers, often leading to the recommendation of mTOR or CDK4/6 inhibitors as potential therapies (10, 12). Our data showed that inhibitors of PI3K–AKT–mTOR signaling had varying AUC and log2[IC50] Z scores across the 125-sample cohort, independent of the presence of bona fide aberrations in PI3K–AKT–mTOR signaling genes (PIK3CA, PIK3R1, PTEN, or TSC2; Fig. 5A; Supplementary Fig. S4A and S4B). To validate this observation, the efficacy of the clinically used mTORC1 inhibitor temsirolimus was tested in vivo in matching PDX models for an insensitive osteosarcoma sample harboring a TSC2 mutation and a sensitive Ewing sarcoma sample harboring a PIK3CA mutation. Although no significant effects on best responses were observed in both PDX models (Supplementary Fig. S4C), temsirolimus treatment significantly improved EFS in the Ewing sarcoma and not the osteosarcoma model (Fig. 5B). In vitro and in vivo efficacies were consistent with the clinical observations in matching patients who received temsirolimus plus temozolomide and irinotecan backbone therapy where the TSC2-mutated osteosarcoma patient had progressive disease and the PIK3CA-mutated Ewing sarcoma patient had a partial response (Fig. 5C). Similar to PI3K–AKT–mTOR pathway inhibition, sensitivity to the CDK4/6 inhibitors ribociclib, palbociclib, and abemaciclib could not be predicted by the presence or absence of CDKN2A/B deletions, CDK4 amplification, or RB1 loss (Fig. 5D; Supplementary Fig. S4D and S4E). Together, these results illustrate that tumor mutational and copy number profiles are not always sufficient to predict all targeted drug responses, and that tumor-agnostic approaches such as HTS can identify previously unrecognized therapeutic susceptibilities.
Figure 5.
Sensitivity to mTORC1 and CDK4/6 inhibitors is not predicted by clinically applied predictive biomarkers. A, Dot plots of the AUC Z scores of PI3K-AKT-mTOR inhibitors for samples with (colored) and without (gray) bona fide genomic alterations in PI3K–AKT–mTOR signaling. Arrows indicate the sarcoma samples harboring a TSC2 mutation (green arrow; average sensitivity to temsirolimus) and a PIK3CA mutation (purple arrow; high sensitivity to temsirolimus) used for in vivo and clinical validation in B and C, respectively. B,In vivo effects of temsirolimus on EFS in matching sarcoma PDX models for the two sarcoma samples indicated in A (N = 4 mice/treatment arm). C, Observed responses for temsirolimus addition to backbone chemotherapy (irinotecan plus temozolomide) in matching sarcoma patients. Top images, 18F-FDG PET/CT images demonstrating progressive disease in the anterior chest wall of the patient with a TSC2-mutated osteosarcoma. Bottom images, computed tomography (CT) scans of the chest demonstrating partial regression of pleural metastases in the patient with a PIK3CA-mutated Ewing sarcoma. D, Dot plots of CDK4/6 inhibitors for samples with (colored) and without (gray) bona fide genomic alterations in cell-cycle regulation. Dot sizes in A and D indicate log2[IC50] Z score values. *, P < 0.05.
Sensitivity to WEE1 inhibition is correlated with genes involved in G2–M checkpoint and DNA replication
WEE1 inhibitor adavosertib showed a large coefficient of variation (CV) using AUC Z scores across all nonhematologic samples in the cohort, indicating that there is sufficient diversity of responders and non-responders to potentially identify novel predictive biomarkers. WEE1 is a key regulator of the G2–M transition and stalled replication fork stabilization during S phase (38) and is a promising target for personalized cancer treatment (39).
Adavosertib was tested against 63 nonhematologic tumor samples with available matching RNA-seq data. Using Pearson correlation coefficients between adavosertib AUC Z scores and log-transformed gene expression values, we performed GSEA to study the biological processes linked to adavosertib sensitivity. The most highly enriched gene ontology biological processes (GO:BP) and hallmark gene sets were G2–M checkpoint, (DNA-templated) DNA replication, organelle fission, DNA metabolic process, and MYC targets version 1 (V1) and version 2 (V2; Fig. 6A and B; Supplementary Fig. S5A; Supplementary Table S5). Genes correlating with adavosertib sensitivity were also enriched for targets of the MuvB (DREAM) multiprotein complex (Fig. 6A), a known master regulator of cell cycle (40, 41).
Figure 6.
Integrative analysis identifies novel biomarkers predictive of adavosertib efficacy. A, GSEA enrichment plots for top enriched hallmark, GO biological processes (GO:BP), and curated gene sets for the positively correlating genes with adavosertib efficacy. Gene sets enriched among the genes for which log2[TPM] values negatively correlated with adavosertib AUC Z scores (high expression correlates with increased efficacy) with FDR q value < 0.01, list > 10%, and NES ≤ −2 were selected as most relevant. Genes with |r| ≥ 0.3 are highlighted in the barcode regions. B, Clustering of the MSigDB GO:BP gene sets enriched in the adavosertib-sensitive nonhematologic tumor samples with FDR q value < 0.01, list > 10%, and NES ≤ −2. Nodes are grouped and colored based on same or similar functional indication and their size indicates the NES. The two largest clusters are shown. C, Twenty-five top correlating genes with adavosertib efficacy based on Pearson correlation coefficient between adavosertib AUC Z scores and gene log2[TPM] values across the 63-sample tumor cohort. Negatively (high expression correlates with sensitivity) and positively (high expression correlates with resistance) correlating genes with P < 0.01 are indicated in red and blue, respectively. D, Principal component (PC) analysis of the adavosertib response based on AUC Z score with red being relatively sensitive and blue relatively resistant. Shapes of the symbols indicate tumor type. E, Twenty top GO, hallmark, and KEGG gene sets identified from the 25-gene set shown in C correlating with adavosertib efficacy (FDR q value < 0.01). Blue, GO biological process; black, GO cellular component; green, hallmark. F, Adavosertib dose–response curves for SHEP-21N neuroblastoma cells with MYCN on [green; −doxycycline (Dox)] versus MYCN off (red; +Dox). Effects on cell viability were established after 24-hour treatment in four independent experiments with three technical replicates in each experiment. Dots indicate the average cell viabilities ± SD and lines represent the fitted dose-response curves using nonlinear regression. Western blot on top shows MYCN repression upon doxycycline treatment after 0, 24, and 96 hours. G, Adavosertib AUC (left) and log2[IC50] (right) values in SHEP-21N neuroblastoma cells with MYCN off (−Dox) versus MYCN on (+Dox) after 24-hour treatment. Horizontal lines indicate median values. Tumor type key: brain tumors (BT), neuroblastoma (NB), sarcoma tumors (SAR), solid other (SO). *, P < 0.05.
Top 25 genes with the strongest association with adavosertib efficacy included 24 negatively (r ≤ −0.48) and 1 positively (r = 0.49) correlated gene(s) predictive of sensitivity and resistance, respectively (Fig. 6C; Supplementary Fig. S5B; Supplementary Table S6). Clustering analysis showed no bias for specific tumor types (Fig. 6D). Sixteen of the 24 genes predictive of adavosertib sensitivity are involved in processes related to mitotic cell-cycle regulation and chromosome organization (Fig. 6E). Further exploration of the biological interconnections to identify functional interactions of the 25-gene set using Reactome revealed TP53, PLK1, CDK1, EP300, and CDC42 as key linker nodes between WEE1 and the 25-gene set (Supplementary Fig. S6A). We looked for germline and somatic single nucleotide variants (SNV), small insertions and deletions (InDels), and copy number (CN) status in the 25-gene set, linker node genes and WEE1. SNVs and InDels identified in our gene lists were independently curated to assign pathogenicity status and those predicted to be benign or likely benign were filtered out. TP53 was the only gene with variants considered to be pathogenic, which were present in 19 of the 59 samples with matching WGS data but was not associated with adavosertib response (Supplementary Fig. S6B). There was no strong correlation between adavosertib AUC Z scores and genomic alterations of the 25-gene set, linker node genes and WEE1.
Interestingly, an adavosertib-sensitive neuroblastoma sample with MYCN amplification did not conform to the expression profile of the 25-gene set (Supplementary Fig. S6B). When we compared neuroblastoma samples with and without MYCN amplification or activating MYCN mutations, those with MYCN aberrations had significantly lower AUC and log2[IC50] values (P = 0.016 and 0.024, respectively) when treated with adavosertib (Supplementary Fig. S6C). This observation was validated using the publicly available Genomics of Drug Sensitivity in Cancer project database (Supplementary Fig. S6D). MYCN dependency was further explored using the human neuroblastoma cell line SHEP-21N, which expresses high levels of MYCN under a doxycycline repressible promoter (29). MYCN repression in SHEP-21N cells diminished adavosertib effects on cell viability (Fig. 6F), resulting in significantly higher AUC and log2[IC50] values (Fig. 6G). Adavosertib caused a shift from the G1 phase to the G2–M phase checkpoint of the cell cycle (Supplementary Fig. S6E). In line with the higher proliferation rate observed in SHEP-21N with MYCN on versus MYCN off (Supplementary Fig. S6F), adavosertib-induced G2–M phase cell-cycle arrest was most pronounced in SHEP-21N cells expressing high levels of MYCN. Supported by the observed enrichment of hallmarks MYC targets versions 1 and 2 in adavosertib-sensitive samples, this suggests the potential efficacy of adavosertib in tumors with MYCN amplification or activating mutations. Taken together, the activation of MYCN in neuroblastoma is a potential biomarker of WEE1 inhibitor efficacy.
MEK inhibitor efficacy is associated with alterations in PI3K signaling
Our data showed that trametinib had efficacy in HTS of WT, HGG, and diffuse midline glioma (DMG), which do not harbor bona fide driver mutations or CN alterations in RAS–MAPK signaling (Supplementary Fig. S7; Fig 4F). We took advantage of the HTS of CNS tumors in which no tumor content was present as negative controls to compare the AUC and log2[IC50] values of trametinib in HGG and DMG samples. Significantly higher trametinib efficacy was observed in tumors compared to controls (Fig. 7A). To understand the unexpected trametinib responses, we looked for CN and SNV alterations in a more extensive network of RAS–MAPK signaling genes. Only weak correlations between trametinib AUC Z scores and normalized copy numbers of NTRK1 (r = −0.36), MET (r = −0.24), ALK (r = −0.22), and EGFR (r = −0.21) were identified. In line with this, GSEA did not identify enrichment for RAS–MAPK signaling in sensitive samples (Fig. 7B; Supplementary Table S7). We also studied links with alterations in PI3K–AKT–mTOR signaling because of the cross-talk between RAS–MAPK and PI3K–AKT–mTOR signaling (42). Three of the five most sensitive glioma samples harbor a heterozygous PIK3R1 mutation, of which, two samples have an additional gain in AKT3 and/or activating mutation in PIK3CA (Supplementary Fig. S7). A fourth glioma sample harboring a biallelic PIK3R1 mutation was less sensitive. Further investigation showed increased activity of AKT but not ribosomal protein S6K (a negative feedback regulator) in glioma cells with versus without PIK3R1, PIK3CA, and/or AKT3 genomic alterations (Fig. 7C), thus confirming PI3K–AKT–mTOR pathway activation. Although PIK3R1 mutations were distinct from nSH2 domain mutations associated with neomorphic activated RAS–MAPK signaling and increased sensitivity to MEK inhibition (Fig. 7D; ref. 43), higher phosphorylated ERK levels were observed in glioma cells harboring genomic PI3K pathway alterations compared with the earlier discussed BRAF V600E–harboring HGG sample (Fig. 7C). ERK phosphorylation was inhibited upon trametinib treatment. No enrichment of gene sets related to active PI3K signaling was observed in trametinib-sensitive solid tumor samples (Fig. 7B; Supplementary Table S7). These results suggest that sensitivity of brain tumor samples to MEK inhibition is potentially linked to genomic alterations in PI3K signaling but require further functional validation.
Figure 7.
PIK3R1 mutations predict sensitivity in pediatric HGG and DMG samples to MEK inhibition. A, Trametinib AUC (left) and log2[IC50] (right) values for noncancerous brain samples versus pediatric HGG and DMG samples without bona fide alterations in RAS–MAPK signaling. Horizontal lines represent median values. B, GSEA of MSigDB GO:BP gene sets on the genes positively correlated with trametinib sensitivity (NES < 0). Dot size indicates NES and dots above the dotted line (FDR q value > 0.01) are not significant. Biological processes associated with RAS–MAPK and PI3K signaling are highlighted in red and orange, respectively. C, Activation status of RAS–MAPK and PI3K signaling pathways in trametinib-sensitive DMG samples zcc372 and zcc135 harboring a PIK3R1 mutation. DMG sample zcc135 harbors an additional activating mutation in PIK3CA and both DMG samples zcc372 and zcc135 harbor gain of AKT3. RAS–MAPK and PI3K pathway activation was established after 40-minute treatment with DMSO (= baseline levels) or 50 nmol/L trametinib by Western blot analysis of phosphorylated and total levels of ERK1/2 and AKT and S6K, respectively. Trametinib-sensitive DMG sample zcc116 harboring a BRAF V600E mutation and trametinib-insensitive DMG sample zcc92 without bona fide alterations in RAS–MAPK or PI3K signaling have been included as controls. β-Actin was used as loading control. The color of the crosses indicates the sensitivity of the sample to trametinib. Red, sensitive; blue, insensitive. D, Lollipop diagram of the PIK3R1 mutations in brain tumors in our cohort (indicated in black) that are associated with trametinib sensitivity. nSH2 domain mutations associated with neomorphic-activated RAS–MAPK signaling and increased sensitivity to MEK inhibition are indicated in gray. Genome coordinates are in hg19.
Discussion
This study demonstrates the potential contribution in vitro HTS embedded in a personalized medicine program can make to clinical treatment decision making. We confirmed that drug response profiles are independent of the methodology used to obtain material for screening (freshly dissociated cells versus in vitro or in vivo expanded cells). Moreover, in vitro responses could be achieved within timeframes that permitted data being made available to treating clinicians. In well-established indications, such as NTRK fusions and BRAF mutations, the in vitro responses were entirely consistent with responses in matching PDX models and patients. Excitingly, by applying our novel methodology that uses drug responses as a starting point, we provide new biological insights into drug responses, which, when validated, can be utilized for prediction of favorable clinical responses, or identifying non-responders.
We previously showed that in vitro and in vivo drug testing is an effective strategy to identify treatments for pediatric cancer patients independently from molecular profiles (24). A key challenge is doing this in a clinically relevant timeframe. The most time-efficient method is using freshly dissociated tumor cells to perform HTS, relate this to the contemporaneous molecular profiling, and inform treating clinicians using both sets of data. However, in many cases, in vitro or in vivo expansion prior to HTS was required due to limited tumor material. Although patient-derived cells and PDXs have been shown to recapitulate the molecular and phenotypic features of tumors (17, 44, 45), the concern is this may introduce variables that may influence drug responses and limit the relevance of the drug screen data. Our finding that drug responses were consistent across the different methodologies used is important as this means that HTS can be performed on more tumor samples. In addition, this is relevant as 82% of the screens on expanded tumor cells were completed while the patient was still alive, and HTS results could be used to guide clinical decision making.
Although we have previously shown that HTS can identify potential therapeutic responses not identified using genomic and transcriptomic analyses alone (24), our experience with ZERO has shown that clinicians are hesitant to recommend treatment based solely on HTS data without molecular features supporting the drug response. Our results demonstrate that HTS on patient samples validate known gene–drug interactions, such as larotrectinib sensitivity associated with a KANK1–NTRK2 fusion (46), or dabrafenib and vemurafenib sensitivity in pediatric tumors that harbor BRAF V600E mutations (47, 48). The corollary of this is that in vitro drug screening provides an unprecedented opportunity to explore the biology of drug responses, particularly when biomarkers are not well established or potentially misleading. For example, cell-cycle aberrations such as loss of CDKN2A/B and PI3K–AKT–mTOR signaling pathway aberrations are the most common alterations in pediatric cancer genomes (10, 12). However, inhibitors such as palbociclib and ribociclib and mTORC1 inhibitors such as everolimus and sirolimus are yet to have significant clinical impact in pediatric cancer (13, 14, 49, 50). We observed significant responses to CDK4/6 and PI3K–AKT–mTOR pathway inhibitors in a subset of samples without variants in cell cycle or PI3K–AKT–mTOR signaling genes. Importantly, in vitro sensitivity or resistance of tumor samples with aberrations in PI3K–AKT–mTOR signaling genes to mTORC1 inhibition was consistent with matched in vivo and clinical responses. Further data from in vitro drug testing programs, with follow-up of clinical responses to HTS treatment recommendations will be required to determine how well in vitro screens match clinical utility. In the shorter term, inclusion of in vitro drug assays to support targeting a genomic feature provides powerful evidence to support drug treatment recommendations.
We have used the HTS response data to highlight several associations between genomic features in high-risk pediatric cancer and drug responses, which are not currently clinically applied, but which could potentially improve patient selection. For example, we identified MYCN activation as a potential predictive biomarker for adavosertib efficacy in neuroblastoma. This is in line with the previously reported observation that neuroblastoma cells homozygous for MYCN were more sensitive to adavosertib compared to neuroblastoma cells heterozygous for the oncogene (51). The 25-gene signature of sensitivity to adavosertib (Fig. 6C) is independent of MYCN activation, but requires much more extensive validation before it could be employed to select patients for this treatment. Also, sensitivity to MEK1/2 inhibitor trametinib in a subset of HGG and DMG samples is potentially associated with PIK3R1 mutations and/or AKT3 CN gain, consistent with the previous observation that a N564D mutation in PIK3R1 confers in vitro sensitivity to trametinib in a patient-derived diffuse intrinsic pontine glioma model (52). With the availability of MEK inhibitors with improved blood–brain barrier (BBB) permeability, such as pimasertib, the latter finding opens new avenues for the development of biomarker-driven trials involving MEK inhibitors to improve outcomes for challenging tumor types such as HGG and DMG. Each of these examples provide evidence that utilizing HTS alongside genomic and transcriptomic analysis can lead to better predictive biomarkers, which are validated ultimately by clinical follow-up.
Single-agent therapy is rarely efficacious in achieving adequate responses due to genetic intratumor heterogeneity or acquired resistance (53). A critical challenge for HTS programs is to identify combination therapies that can be matched to individual tumors or tumor genomic profiles. The availability of limited tumor material for preclinical drug testing and the need to obtain results in a clinically relevant timeframe makes it all but impossible to test all drug combinations. We devised a potential methodology to tackle this challenge by using correlations between the efficacy profiles of single agents to identify drugs with different MOAs but similar activity across our cohort. Although we validated a successful combination of SN-38 and alisertib through this approach, further studies are required to test the hypothesis that sensitivity to two single agents can be used to identify effective drug combinations for individual patients. This kind of innovative approach can provide a rationale for selection of preclinical combination testing.
In conclusion, we have shown the value of incorporating HTS into precision medicine programs to broaden therapeutic options available to high-risk pediatric cancers. Importantly, this is feasible while patients are currently undergoing clinical management. Expanding from our TARGET pilot study (24) to the PRISM clinical trial, we showed the sustainability of such an approach, which can be a valuable asset to the health care system. Furthermore, our findings explore and integrate the interplay between complementary drug screening and molecular profiling data, which provides a higher level of clinical confidence in treatment recommendations. Therefore, by expanding cohorts through global pediatric precision medicine programs, this will develop a rich source of data that will provide additional power and confidence to better identify biomarkers of drug response and more effective combination therapies can be identified and trialed.
Supplementary Material
Supplementary Materials and Methods
Supplementary Tables S1-S7
Acknowledgments
The authors thank Tour de Cure for supporting CCI Tumour Bank personnel. They thank the Australian Federal Government Department of Health, the New South Wales State Government, and the Australian Cancer Research Foundation for funding to establish infrastructure to support the Zero Childhood Cancer personalized medicine program. The authors thank the Cancer Therapeutics Cooperative Research Centre, for supporting the development of a personalized medicine program; The Cure Brain Cancer Foundation, Robert Connor Dawes Foundation and Cancer Institute NSW for the brain tumor work; Neuroblastoma Australia for the neuroblastoma work; The Steven Walter Children's Cancer Foundation and The Hyundai Help 4 Kids Foundation for supporting G.M. Marshall and P.G. Ekert; The Samuel Nissen Charitable Foundation for supporting P.G. Ekert. The authors would like to acknowledge Luminesce Alliance – Innovation for Children's Health for its contribution and support (C. Mayoh, M. Wong, and M.J. Cowley). Luminesce Alliance – Innovation for Children's Health, is a not-for-profit cooperative joint venture between the Sydney Children's Hospitals Network, the Children's Medical Research Institute, and the Children's Cancer Institute. It has been established with the support of the NSW Government to coordinate and integrate pediatric research. Luminesce Alliance is also affiliated with the University of Sydney and the University of New South Wales Sydney; The Medical Research Future Fund – Australian Brain Cancer Mission, the Minderoo Foundation's Collaborate Against Cancer Initiative and funds raised through the Zero Childhood Cancer Capacity Campaign, a joint initiative of the Children's Cancer Institute and the Sydney Children's Hospital Foundation, supported the national clinical trial and associated clinical and research personnel. The authors thank the National Health and Medical Research Council of Australia (fellowships APP1059804 and APP1157871 to R.B. Lock and APP2017898 to D.S. Ziegler).
The authors thank patients and parents for participating in this study. The authors thank the many clinicians and health professionals for their time acquiring consenting patients and for collection and coordination of samples and associated clinical data at Sydney Children's Hospital, Randwick; The Children's Hospital at Westmead; the John Hunter Hospital; the Queensland Children's Hospital; the Royal Children's Hospital Melbourne; the Monash Children's Hospital; the Adelaide Women's & Children's Hospital; and the Perth Children's Hospital. The authors also thank the Sydney Children's Tumour Bank Network [Children's Cancer Institute (CCI) Tumour Bank and/or Children's Hospital at Westmead Tumour Bank] for providing samples and related clinical information for this study. They thank the staff of the Personalized Medicine Theme of the Children's Cancer Institute for their dedicated work on Zero Childhood Cancer. The authors acknowledge Compounds Australia (www.compoundsaustralia.com) for their provision of specialized compound management and logistics research services to the project. Zero Childhood Cancer is a joint initiative led by the Children's Cancer Institute and Sydney Children's Hospital, Randwick.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
C. Mayoh reports grants from Medical Research Future Funds and Luminesce Alliance during the conduct of the study. D.S. Ziegler reports grants from NHMRC and Cancer Institute NSW during the conduct of the study, personal fees from Bayer, Amgen, Day One, Novartis, Alexion, and FivepHusion, grants and personal fees from Accendatech, and personal fees from AstraZeneca outside the submitted work. J.I. Fletcher reports other support from Walter and Eliza Hall Institute outside the submitted work. R.B. Lock reports grants from National Health and Medical Research Council of Australia during the conduct of the study. M. Dolman reports grants from Tour de Cure, The Australian Federal Government Department of Health, The New South Wales State Government, The Australian Cancer Research Foundation, The Kids Cancer Alliance, Cancer Therapeutics Cooperative Research Centre, The Cure Brain Cancer Foundation, The Robert Connor Dawes Foundation, Cancer Institute NSW, Neuroblastoma Australia, The Steven Walter Children's Cancer Foundation, The Hyundai Help 4 Kids Foundation, The Samuel Nissen Charitable Foundation, Luminesce Alliance & Innovation for Children's Health, The Medical Research Future Fund, The Australian Brain Cancer Mission, The Minderoo Foundation's Collaborate Against Cancer Initiative, and The National Health and Medical Research Council of Australia during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
C. Mayoh: Conceptualization, data curation, software, formal analysis, supervision, investigation, methodology, writing–original draft. J. Mao: Software, formal analysis, investigation, methodology, writing–original draft. J. Xie: Formal analysis, methodology. G. Tax: Investigation. S.O. Chow: Investigation, methodology. R. Cadiz: Investigation. K. Pazaky: Investigation. P. Barahona: Formal analysis. P. Ajuyah: Formal analysis. P. Trebilcock: Investigation. A. Malquori: Investigation. K. Gunther: Investigation. A. Avila: Investigation. D.Y. Yun: Investigation. S. Alfred: Investigation. A. Gopalakrishnan: Investigation. A. Kamili: Investigation, methodology. M. Wong: Software, methodology. M.J. Cowley: Software, methodology. S. Jessop: Resources. L.M.S. Lau: Conceptualization, resources, data curation. T.N. Trahair: Conceptualization, resources. D.S. Ziegler: Conceptualization, resources, funding acquisition. J.I. Fletcher: Resources, supervision. A.J. Gifford: Formal analysis, investigation. M. Tsoli: Resources, supervision. G.M. Marshall: Conceptualization, resources, funding acquisition. M. Haber: Conceptualization, funding acquisition. V. Tyrrell: Funding acquisition, project administration. T.W. Failes: Methodology. G.M. Arndt: Supervision, methodology. R.B. Lock: Resources, supervision, methodology. P.G. Ekert: Conceptualization, data curation, formal analysis, supervision, writing–original draft. M.E.M. Dolman: Conceptualization, data curation, formal analysis, supervision, investigation, methodology, writing–original draft.
References
- 1. Harris MH, DuBois SG, Glade Bender JL, Kim A, Crompton BD, Parker E, et al. Multicenter feasibility study of tumor molecular profiling to inform therapeutic decisions in advanced pediatric solid tumors: the individualized cancer therapy (iCat) study. JAMA Oncol 2016;2:608–15. [DOI] [PubMed] [Google Scholar]
- 2. Harttrampf AC, Lacroix L, Deloger M, Deschamps F, Puget S, Auger N, et al. Molecular screening for cancer treatment optimization (MOSCATO-01) in pediatric patients: a single-institutional prospective molecular stratification trial. Clin Cancer Res 2017;23:6101–12. [DOI] [PubMed] [Google Scholar]
- 3. Khater F, Vairy S, Langlois S, Dumoucel S, Sontag T, St-Onge P, et al. Molecular profiling of hard-to-treat childhood and adolescent cancers. JAMA Netw Open 2019;2:e192906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marks LJ, Oberg JA, Pendrick D, Sireci AN, Glasser C, Coval C, et al. Precision medicine in children and young adults with hematologic malignancies and blood disorders: the columbia university experience. Front Pediatr 2017;5:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mody RJ, Wu YM, Lonigro RJ, Cao X, Roychowdhury S, Vats P, et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA 2015;314:913–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parsons DW, Roy A, Yang Y, Wang T, Scollon S, Bergstrom K, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol 2016;2:616–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pincez T, Clement N, Lapouble E, Pierron G, Kamal M, Bieche I, et al. Feasibility and clinical integration of molecular profiling for target identification in pediatric solid tumors. Pediatr Blood Cancer 2017;64. [DOI] [PubMed] [Google Scholar]
- 8. Worst BC, van Tilburg CM, Balasubramanian GP, Fiesel P, Witt R, Freitag A, et al. Next-generation personalised medicine for high-risk paediatric cancer patients - the INFORM pilot study. Eur J Cancer 2016;65:91–101. [DOI] [PubMed] [Google Scholar]
- 9. Chang W, Brohl AS, Patidar R, Sindiri S, Shern JF, Wei JS, et al. MultiDimensional ClinOmics for Precision therapy of children and adolescent young adults with relapsed and refractory cancer: a report from the center for cancer research. Clin Cancer Res 2016;22:3810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Tilburg CM, Pfaff E, Pajtler KW, Langenberg KPS, Fiesel P, Jones BC, et al. The pediatric precision oncology INFORM registry: clinical outcome and benefit for patients with very high-evidence targets. Cancer Discov 2021;11:2764–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berlanga P, Pierron G, Lacroix L, Chicard M, Adam de Beaumais T, Marchais A, et al. The european MAPPYACTS trial: precision medicine program in pediatric and adolescent patients with recurrent malignancies. Cancer Discov 2022;12:1266–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wong M, Mayoh C, Lau LMS, Khuong-Quang DA, Pinese M, Kumar A, et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat Med 2020;26:1742–53. [DOI] [PubMed] [Google Scholar]
- 13. Geoerger B, Bourdeaut F, DuBois SG, Fischer M, Geller JI, Gottardo NG, et al. A phase I study of the CDK4/6 inhibitor ribociclib (LEE011) in pediatric patients with malignant rhabdoid tumors, neuroblastoma, and other solid tumors. Clin Cancer Res 2017;23:2433–41. [DOI] [PubMed] [Google Scholar]
- 14. Van Mater D, Gururangan S, Becher O, Campagne O, Leary S, Phillips JJ, et al. A phase I trial of the CDK 4/6 inhibitor palbociclib in pediatric patients with progressive brain tumors: a pediatric brain tumor consortium study (PBTC-042). Pediatr Blood Cancer 2021;68:e28879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81–94. [DOI] [PubMed] [Google Scholar]
- 16. Xiong S, Feng Y, Cheng L. Cellular reprogramming as a therapeutic target in cancer. Trends Cell Biol 2019;29:623–34. [DOI] [PubMed] [Google Scholar]
- 17. Lee JK, Liu Z, Sa JK, Shin S, Wang J, Bordyuh M, et al. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat Genet 2018;50:1399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018;359:920–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015;161:933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calandrini C, Schutgens F, Oka R, Margaritis T, Candelli T, Mathijsen L, et al. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat Commun 2020;11:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Braekeveldt N, Wigerup C, Gisselsson D, Mohlin S, Merselius M, Beckman S, et al. Neuroblastoma patient-derived orthotopic xenografts retain metastatic patterns and geno- and phenotypes of patient tumours. Int J Cancer 2015;136:E252–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bate-Eya LT, Ebus ME, Koster J, den Hartog IJ, Zwijnenburg DA, Schild L, et al. Newly-derived neuroblastoma cell lines propagated in serum-free media recapitulate the genotype and phenotype of primary neuroblastoma tumours. Eur J Cancer 2014;50:628–37. [DOI] [PubMed] [Google Scholar]
- 23. Stewart E, Federico SM, Chen X, Shelat AA, Bradley C, Gordon B, et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 2017;549:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lau LMS, Mayoh C, Xie J, Barahona P, MacKenzie KL, Wong M, et al. In vitro and in vivo drug screens of tumor cells identify novel therapies for high-risk child cancer. EMBO Mol Med 2022;14:e14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin GL, Monje M. A protocol for rapid post-mortem cell culture of diffuse intrinsic pontine glioma (DIPG). J Vis Exp 2017:55360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Houghton PJ, Morton CL, Tucker C, Payne D, Favours E, Cole C, et al. The pediatric preclinical testing program: description of models and early testing results. Pediatr Blood Cancer 2007;49:928–40. [DOI] [PubMed] [Google Scholar]
- 27. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lutz W, Stohr M, Schurmann J, Wenzel A, Lohr A, Schwab M. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene 1996;13:803–12. [PubMed] [Google Scholar]
- 30. Picco G, Chen ED, Alonso LG, Behan FM, Goncalves E, Bignell G, et al. Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat Commun 2019;10:2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER III, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Slade D. PARP and PARG inhibitors in cancer treatment. Genes Dev 2020;34:360–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pilie PG, Gay CM, Byers LA, O'Connor MJ, Yap TA. PARP inhibitors: extending benefit beyond BRCA-mutant cancers. Clin Cancer Res 2019;25:3759–71. [DOI] [PubMed] [Google Scholar]
- 34. DuBois SG, Marachelian A, Fox E, Kudgus RA, Reid JM, Groshen S, et al. Phase I study of the aurora a kinase inhibitor alisertib in combination with irinotecan and temozolomide for patients with relapsed or refractory neuroblastoma: A NANT (New Approaches to Neuroblastoma Therapy) trial. J Clin Oncol 2016;34:1368–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. DuBois SG, Mosse YP, Fox E, Kudgus RA, Reid JM, McGovern R, et al. Phase II trial of alisertib in combination with irinotecan and temozolomide for patients with relapsed or refractory neuroblastoma. Clin Cancer Res 2018;24:6142–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xie J, Kumar A, Dolman MEM, Mayoh C, Khuong-Quang DA, Cadiz R, et al. The important role of routine cytopathology in pediatric precision oncology. Cancer Cytopathol 2021;129:805–18. [DOI] [PubMed] [Google Scholar]
- 37. Sabir SR, Yeoh S, Jackson G, Bayliss R. EML4-ALK variants: biological and molecular properties, and the implications for patients. Cancers 2017;9:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elbaek CR, Petrosius V, Benada J, Erichsen L, Damgaard RB, Sorensen CS. WEE1 kinase protects the stability of stalled DNA replication forks by limiting CDK2 activity. Cell Rep 2022;38:110261. [DOI] [PubMed] [Google Scholar]
- 39. Ghelli Luserna di Rora A, Cerchione C, Martinelli G, Simonetti G. A WEE1 family business: regulation of mitosis, cancer progression, and therapeutic target. J Hematol Oncol 2020;13:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ 2018;25:114–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer 2013;13:585–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci 2011;36:320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cheung LW, Yu S, Zhang D, Li J, Ng PK, Panupinthu N, et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer Cell 2014;26:479–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov 2017;7:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun H, Cao S, Mashl RJ, Mo CK, Zaccaria S, Wendl MC, et al. Comprehensive characterization of 536 patient-derived xenograft models prioritizes candidates for targeted treatment. Nat Commun 2021;12:5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hong DS, DuBois SG, Kummar S, Farago AF, Albert CM, Rohrberg KS, et al. Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol 2020;21:531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hargrave DR, Bouffet E, Tabori U, Broniscer A, Cohen KJ, Hansford JR, et al. Efficacy and safety of dabrafenib in pediatric patients with BRAF V600 mutation-positive relapsed or refractory low-grade glioma: results from a phase I/IIa study. Clin Cancer Res 2019;25:7303–11. [DOI] [PubMed] [Google Scholar]
- 48. Nicolaides T, Nazemi KJ, Crawford J, Kilburn L, Minturn J, Gajjar A, et al. Phase I study of vemurafenib in children with recurrent or progressive BRAF(V600E) mutant brain tumors: pacific pediatric neuro-oncology consortium study (PNOC-002). Oncotarget 2020;11:1942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fouladi M, Laningham F, Wu J, O'Shaughnessy MA, Molina K, Broniscer A, et al. Phase I study of everolimus in pediatric patients with refractory solid tumors. J Clin Oncol 2007;25:4806–12. [DOI] [PubMed] [Google Scholar]
- 50. Geoerger B, Kieran MW, Grupp S, Perek D, Clancy J, Krygowski M, et al. Phase II trial of temsirolimus in children with high-grade glioma, neuroblastoma and rhabdomyosarcoma. Eur J Cancer 2012;48:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Russell MR, Levin K, Rader J, Belcastro L, Li Y, Martinez D, et al. Combination therapy targeting the Chk1 and Wee1 kinases shows therapeutic efficacy in neuroblastoma. Cancer Res 2013;73:776–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Izquierdo E, Carvalho DM, Mackay A, Temelso S, Boult JKR, Pericoli G, et al. DIPG harbors alterations targetable by MEK inhibitors, with acquired resistance mechanisms overcome by combinatorial inhibition. Cancer Discov 2022;12:712–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhong L, Li Y, Xiong L, Wang W, Wu M, Yuan T, et al. Small molecules in targeted cancer therapy: advances, challenges, and future perspectives. Signal Transduct Target Ther 2021;6:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Supplementary Tables S1-S7
Data Availability Statement
The datasets used in this study that were previously published are available from the European Genome-phenome Archive under accession number EGAS00001004572 (https://ega-archive.org/studies/EGAS00001004572) and EGAS00001007029 (https://ega-archive.org/studies/EGAS00001007029). All other raw data generated in this study are available upon request from the corresponding author.



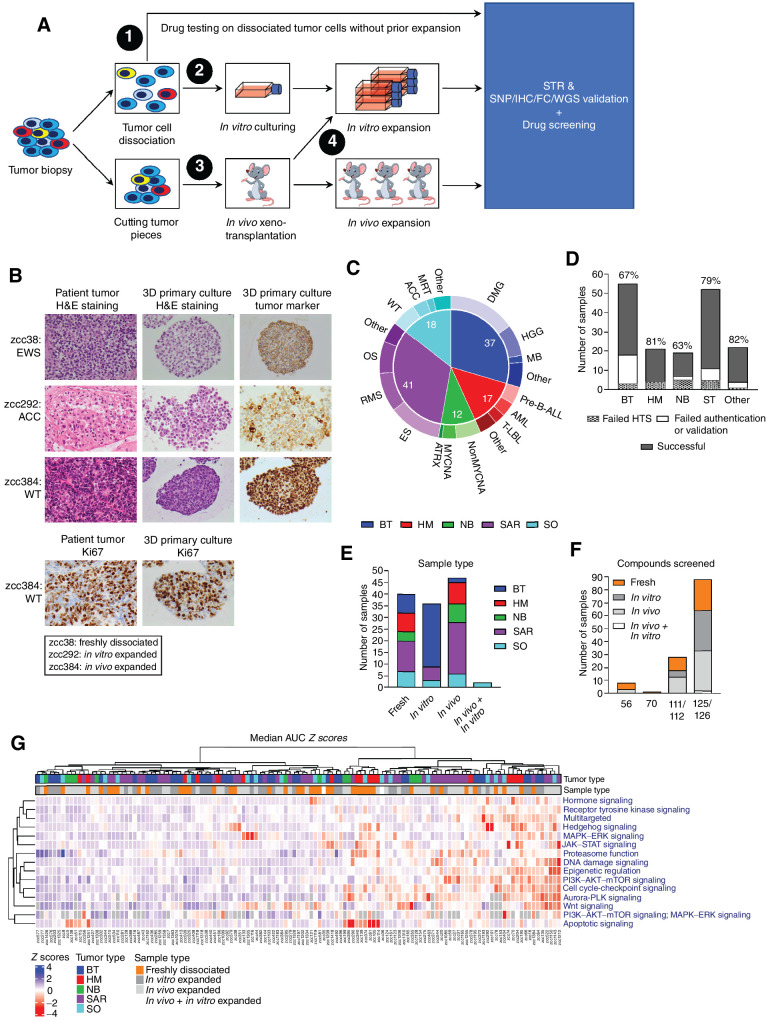
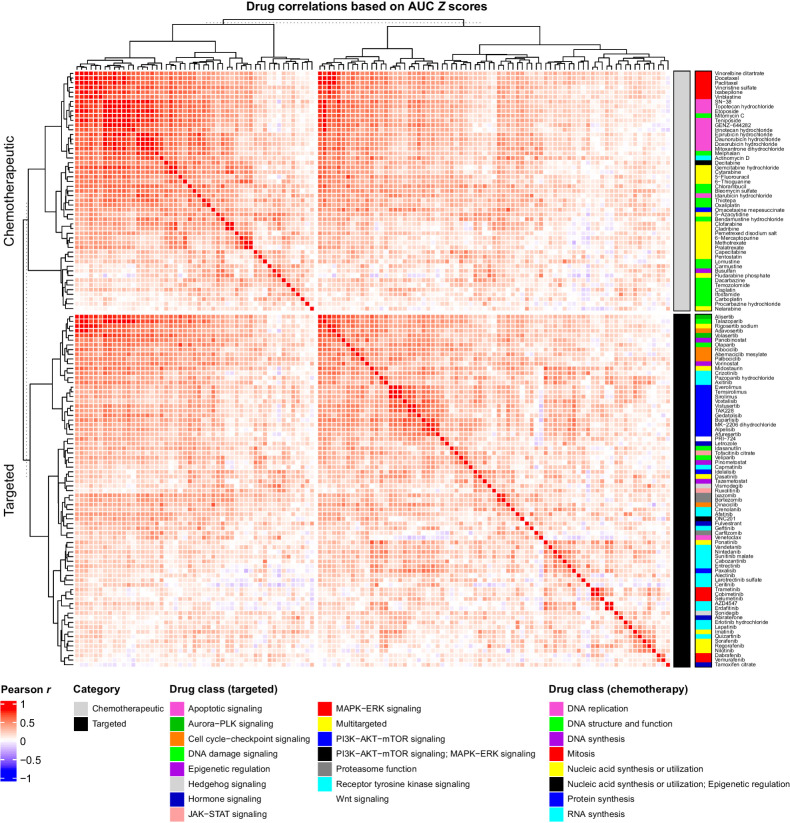
![Figure 3. In vitro PARP inhibitor responses correlate with in vivo responses and correlated drug response profiles identify effective combinations for individual samples. A, AUC Z score correlations across the cohort between PARP inhibitors: olaparib versus veliparib (left), talazoparib versus veliparib (middle), and talazoparib versus olaparib (right). B, Comparison of veliparib, olaparib, and talazoparib AUC values shows increased efficacy with increasing PARP DNA entrapment potency. C, Correlation between in vitro talazoparib AUC Z scores and in vivo responses in matching PDX models as established by calculating T/C values for each model using the following formula: [medium EFS time after talazoparib treatment (=T)]/[medium EFS time when untreated (=C)]. D, Correlation between the AUC Z scores of AURKA inhibitor alisertib and irinotecan metabolite SN-38. Samples sensitive to both alisertib and SN-38 that are used for in vivo combination testing in D are depicted in orange. E, In vivo effects of vehicle control (black), irinotecan plus temozolomide (green), monotherapy alisertib (blue), and alisertib in combination with irinotecan plus temozolomide (red) in PDX models for MRT (left), Wilms tumor (middle), and neuroectodermal tumor (NET; right). Top graphs show percentage change in tumor model for each PDX and bottom graphs show EFS. MRT, malignant rhabdoid tumor; WT, Wilms tumor; NET, neuroectodermal tumor. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. *, versus vehicle control; *, versus alisertib; *, versus irinotecan (IRN) plus temozolomide (TMZ).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/0cc5/10425737/8eacc4672414/2716fig3.jpg)
![Figure 4. In vitro HTS on patient-derived tumor samples confirms known associations between drug sensitivity and driver aberrations. A, AUC distribution across all samples for targeted drugs. Drugs are ordered from most (left) to least (right) effective based on lowest median AUC, followed by lowest quartile AUC values. Colors indicate the mechanism of action of each drug. B, Volcano plots of tumor type-specific sensitivity or resistance to drugs based on the AUC Z scores for each drug, using the following formula: [average AUC Z score for tumor type of interest] − [average AUC Z score for the remaining cohort]. A difference in average AUC Z score ≤ 0.5 and adjusted P < 0.01 is considered relatively resistant (blue) and a difference in average AUC Z score > 0.5 and adjusted P < 0.01 is considered relatively sensitive (red). Only significant targeted drugs are named in the figure. Circle size represents the number of samples within the given tumor type on which the indicated drug has been tested. C, AUC values for TRK inhibitor larotrectinib sulfate, BRAF inhibitors dabrafenib and vemurafenib, and ALK inhibitors ceritinib, crizotinib, and alectinib with samples harboring an NTRK fusion, BRAF V600E mutation, or ALK aberration (i.e., ALK F1245I mutation or EML4-ALK fusion) highlighted in red, respectively. Arrows indicate the BRAF V600E–mutated HGG and ALK F1245I–mutated NB sample used for in vivo efficacy testing in D and E, respectively. D, In vivo effects of vehicle control (saline; black), dabrafenib monotherapy (green), trametinib monotherapy (blue), and dabrafenib plus trametinib combination therapy (red) on EFS in the matching HGG PDX model of the BRAF V600E–mutated sample with in vitro sensitivity to BRAF inhibition. E, In vivo effects of vehicle control (black) and ALK inhibitors ceritinib (green), crizotinib (blue), alectinib (orange), and lorlatinib (red) in the matching NB PDX model of the ALK F1245I–mutated NB sample with in vitro sensitivity to ALK-targeting inhibitors. Left graph shows percentage change in tumor model for each PDX and right graph EFS. F, AUC values for MEK inhibitors trametinib (left), cobimetinib (middle), and selumetinib (right) in samples with (colored) and without (gray) driver aberrations in RAS–MAPK signaling. Shapes of the symbols indicate tumor type. Tumor type key: brain tumors (BT), hematologic malignancies (HM), neuroblastoma (NB), sarcoma tumors (SAR), solid other (SO). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. *, versus vehicle control.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/0cc5/10425737/655e0013fc35/2716fig4.jpg)
![Figure 5. Sensitivity to mTORC1 and CDK4/6 inhibitors is not predicted by clinically applied predictive biomarkers. A, Dot plots of the AUC Z scores of PI3K-AKT-mTOR inhibitors for samples with (colored) and without (gray) bona fide genomic alterations in PI3K–AKT–mTOR signaling. Arrows indicate the sarcoma samples harboring a TSC2 mutation (green arrow; average sensitivity to temsirolimus) and a PIK3CA mutation (purple arrow; high sensitivity to temsirolimus) used for in vivo and clinical validation in B and C, respectively. B, In vivo effects of temsirolimus on EFS in matching sarcoma PDX models for the two sarcoma samples indicated in A (N = 4 mice/treatment arm). C, Observed responses for temsirolimus addition to backbone chemotherapy (irinotecan plus temozolomide) in matching sarcoma patients. Top images, 18F-FDG PET/CT images demonstrating progressive disease in the anterior chest wall of the patient with a TSC2-mutated osteosarcoma. Bottom images, computed tomography (CT) scans of the chest demonstrating partial regression of pleural metastases in the patient with a PIK3CA-mutated Ewing sarcoma. D, Dot plots of CDK4/6 inhibitors for samples with (colored) and without (gray) bona fide genomic alterations in cell-cycle regulation. Dot sizes in A and D indicate log2[IC50] Z score values. *, P < 0.05.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/0cc5/10425737/051d9781bfdf/2716fig5.jpg)
![Figure 6. Integrative analysis identifies novel biomarkers predictive of adavosertib efficacy. A, GSEA enrichment plots for top enriched hallmark, GO biological processes (GO:BP), and curated gene sets for the positively correlating genes with adavosertib efficacy. Gene sets enriched among the genes for which log2[TPM] values negatively correlated with adavosertib AUC Z scores (high expression correlates with increased efficacy) with FDR q value < 0.01, list > 10%, and NES ≤ −2 were selected as most relevant. Genes with |r| ≥ 0.3 are highlighted in the barcode regions. B, Clustering of the MSigDB GO:BP gene sets enriched in the adavosertib-sensitive nonhematologic tumor samples with FDR q value < 0.01, list > 10%, and NES ≤ −2. Nodes are grouped and colored based on same or similar functional indication and their size indicates the NES. The two largest clusters are shown. C, Twenty-five top correlating genes with adavosertib efficacy based on Pearson correlation coefficient between adavosertib AUC Z scores and gene log2[TPM] values across the 63-sample tumor cohort. Negatively (high expression correlates with sensitivity) and positively (high expression correlates with resistance) correlating genes with P < 0.01 are indicated in red and blue, respectively. D, Principal component (PC) analysis of the adavosertib response based on AUC Z score with red being relatively sensitive and blue relatively resistant. Shapes of the symbols indicate tumor type. E, Twenty top GO, hallmark, and KEGG gene sets identified from the 25-gene set shown in C correlating with adavosertib efficacy (FDR q value < 0.01). Blue, GO biological process; black, GO cellular component; green, hallmark. F, Adavosertib dose–response curves for SHEP-21N neuroblastoma cells with MYCN on [green; −doxycycline (Dox)] versus MYCN off (red; +Dox). Effects on cell viability were established after 24-hour treatment in four independent experiments with three technical replicates in each experiment. Dots indicate the average cell viabilities ± SD and lines represent the fitted dose-response curves using nonlinear regression. Western blot on top shows MYCN repression upon doxycycline treatment after 0, 24, and 96 hours. G, Adavosertib AUC (left) and log2[IC50] (right) values in SHEP-21N neuroblastoma cells with MYCN off (−Dox) versus MYCN on (+Dox) after 24-hour treatment. Horizontal lines indicate median values. Tumor type key: brain tumors (BT), neuroblastoma (NB), sarcoma tumors (SAR), solid other (SO). *, P < 0.05.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/0cc5/10425737/3e3ccac990af/2716fig6.jpg)
![Figure 7. PIK3R1 mutations predict sensitivity in pediatric HGG and DMG samples to MEK inhibition. A, Trametinib AUC (left) and log2[IC50] (right) values for noncancerous brain samples versus pediatric HGG and DMG samples without bona fide alterations in RAS–MAPK signaling. Horizontal lines represent median values. B, GSEA of MSigDB GO:BP gene sets on the genes positively correlated with trametinib sensitivity (NES < 0). Dot size indicates NES and dots above the dotted line (FDR q value > 0.01) are not significant. Biological processes associated with RAS–MAPK and PI3K signaling are highlighted in red and orange, respectively. C, Activation status of RAS–MAPK and PI3K signaling pathways in trametinib-sensitive DMG samples zcc372 and zcc135 harboring a PIK3R1 mutation. DMG sample zcc135 harbors an additional activating mutation in PIK3CA and both DMG samples zcc372 and zcc135 harbor gain of AKT3. RAS–MAPK and PI3K pathway activation was established after 40-minute treatment with DMSO (= baseline levels) or 50 nmol/L trametinib by Western blot analysis of phosphorylated and total levels of ERK1/2 and AKT and S6K, respectively. Trametinib-sensitive DMG sample zcc116 harboring a BRAF V600E mutation and trametinib-insensitive DMG sample zcc92 without bona fide alterations in RAS–MAPK or PI3K signaling have been included as controls. β-Actin was used as loading control. The color of the crosses indicates the sensitivity of the sample to trametinib. Red, sensitive; blue, insensitive. D, Lollipop diagram of the PIK3R1 mutations in brain tumors in our cohort (indicated in black) that are associated with trametinib sensitivity. nSH2 domain mutations associated with neomorphic-activated RAS–MAPK signaling and increased sensitivity to MEK inhibition are indicated in gray. Genome coordinates are in hg19.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/0cc5/10425737/cb03d024340a/2716fig7.jpg)