Abstract
Aerobic nitrification is a key process in the global nitrogen cycle mediated by microorganisms. While nitrification has primarily been studied in near-neutral environments, this process occurs at a wide range of pH values, spanning ecosystems from acidic soils to soda lakes. Aerobic nitrification primarily occurs through the activities of ammonia-oxidising bacteria and archaea, nitrite-oxidising bacteria, and complete ammonia-oxidising (comammox) bacteria adapted to these environments. Here, we review the literature and identify knowledge gaps on the metabolic diversity, ecological distribution, and physiological adaptations of nitrifying microorganisms in acidic and alkaline environments. We emphasise that nitrifying microorganisms depend on a suite of physiological adaptations to maintain pH homeostasis, acquire energy and carbon sources, detoxify reactive nitrogen species, and generate a membrane potential at pH extremes. We also recognize the broader implications of their activities primarily in acidic environments, with a focus on agricultural productivity and nitrous oxide emissions, as well as promising applications in treating municipal wastewater.
Keywords: archaea, metabolism, nitrification
Introduction
During aerobic chemolithoautotrophic nitrification, microorganisms convert ammonia (NH3) to nitrite (NO2−) and then nitrate (NO3−) [1]. The first step is mediated by ammonia-oxidising archaea (AOA) and ammonia-oxidising bacteria (AOB), while the second step is mediated by diverse lineages of nitrite-oxidising bacteria (NOB) [2]. It has also recently been discovered that some Nitrospira species, known as comammox bacteria, mediate complete oxidation of ammonia to nitrate [3,4]. Collectively, these microorganisms support key steps in the biogeochemical cycling of nitrogen on the earth and play important ecological roles in diverse terrestrial and aquatic ecosystems, as well as engineered ecosystems such as wastewater and drinking water treatment plants.
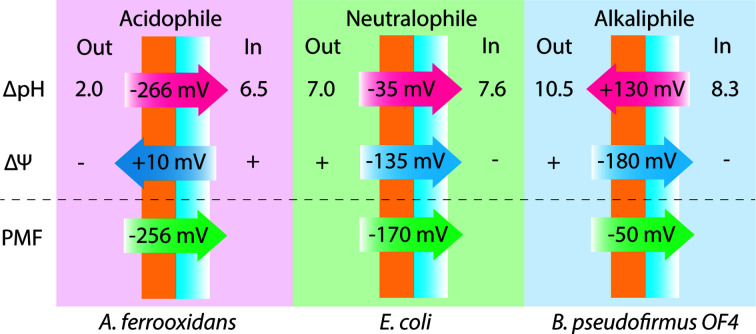
The maintenance of a circumneutral intracellular pH, termed ‘pH homeostasis’, is crucial in any living cell [5–7]. This is because most proteins have distinct ranges of pH within which they can function, while pH also affects the structure of nucleic acid and many other biological molecules [7]. pH also plays a central role in cellular bioenergetics since the establishment of proton-motive force (PMF) depends on both the electrical gradient (membrane potential, ΔΨ) and pH gradient (ΔpH, i.e. pHin − pHout), as per the equation PMF = ΔΨ − 59ΔpH at 25°C [5]. It is therefore vital for microorganisms to sense pH in their milieu and maintain intracellular pH homeostasis [5,8]. Most microorganisms have pH optima between 5 and 9, while a small but significant proportion of microorganisms grow optimally at pH values below 5 or greater than 9 [9]. At low pH, PMF is primarily made up of ΔpH, while ΔΨ detracts from PMF formed by ΔpH. At high pH, alkaliphilic and alkalitolerant microorganisms maintain a higher ΔΨ component of the PMF than neutralophiles or acidophiles because the ΔpH component is reversed and detracts from the PMF [5] (Figure 1).
Figure 1. Differences in proton motive force composition under acidic, neutral and alkaline conditions.
ΔpH contributes to proton motive force (PMF) generation under acidic and neutral conditions, and detracts from proton motive force under alkaline conditions at 25°C. This is illustrated with examples from the obligate acidophile Acidithiobacillus ferrooxidans, the neutralophile Escherichia coli, and facultative alkaliphile Bacillus pseudofirmus OF4; adapted from Krulwich et al., 2011 [5].
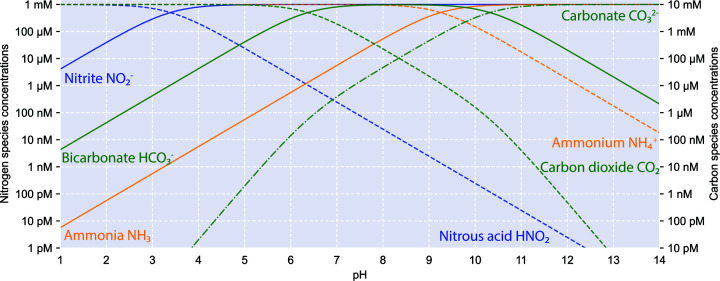
In addition, the shifting equilibrium between acid–base conjugates (determined by acid dissociation constant pKa) of growth substrates at pH extremes imposes nutritional and toxicity stresses on nitrifying microorganisms. Ammonia is the presumed substrate for ammonia monooxygenase [10–14] and it is more abundant than ammonium at pH conditions < 9.25 (pKa NH4+/NH3 = 9.25, at 25°C and zero ionic strength, same conditions used herein). Similarly, nitrous acid (HNO2) dominates over nitrite (NO2−) below pH 3.39 (pKa HNO2/NO2− = 3.39), while bicarbonate (HCO3−) dominates over carbon dioxide (CO2) above pH 6.37 (pKa CO2/HCO3− = 6.37) (Figure 2) [15]. pH drastically influences the speciation of these compounds, as decreasing one pH unit would result in 10 times higher acid/base ratio for each conjugating pair. Thus, the effect of pH on nitrifying microorganisms includes the availability of energy and carbon sources, as well as the toxicity of reactive nitrogen species such as nitrous acid.
Figure 2. Comparisons of concentrations of nitrogen and carbon sources for nitrifying microorganisms from pH 1 to 14.
These comparisons are based on pKa values (NH4+/NH3 = 9.25, HNO2/NO2− = 3.39, CO2/HCO3− = 6.37, HCO3−/CO32− = 10.32), calculated using 1 mM total concentration for ammonia (orange, solid) and ammonium (orange, dashed); nitrite (blue, solid) and nitrous acid (blue, dashed); and 10 mM total concentration for bicarbonate (green, solid), carbon dioxide (green, dashed) and carbonate (green, dash dot).
While the process of nitrification has been studied extensively owing to its biogeochemical and industrial significance, most of our understandings are centered towards neutralophilic nitrifiers or near-neutral environments. An increasing number of culture-dependent and culture-independent studies (summarised in Table 1) have demonstrated that nitrification is active at pH extremes and identified the microorganisms mediating these processes. However, our insights on how nitrifying microorganisms have adapted to acidic and alkaline pH remain rudimentary. This topic has not previously been reviewed, with the exceptions of a consideration of the pH dependency of enzymatic processes regulating N2O production in wastewater treatment systems [16], one study in 2012 that reviewed progress in mechanistic understanding that could explain the dominance of AOA over AOB in certain acidic soils [17], and prior reviews focusing primarily on the distribution and diversity of nitrifiers in acidic soils [18–20]. In the following sections, we describe the process of nitrification, as well as the pH preferences and cellular adaptations of nitrifiers, in diverse acidic and alkaline ecosystems. For a description of the distribution of nitrifiers in acidic and alkaline habitats, we refer to Table 1.
Table 1. Overview of reported nitrification in acidic (≤5.5) and alkaline (≥8.5) environments, including information on the identified nitrifier groups and organisms.
| pH range* | Environments | Nitrifier group | Representative taxa | Technique/type of study** | Citation |
|---|---|---|---|---|---|
| 2.0–7.0 | Acidic tea field | AOB | Ca. Nitrosoglobus terrae | F, I, B, Q, A, O, K, T | [21] |
| 2.2–5.4 | Laboratory reactor | AOB | Nitrosococcus-like | I, B, A, T | [22] |
| 2.5–7.0 | Laboratory reactor | AOB | Ca. Nitrosacidococcus tergens | I, B, A, O, M, K | [23] |
| 2.5–7.0 | Laboratory reactor | AOB | Ca. Nitrosacidococcus urinae | I, B, A, K | [24] |
| 2.9–7.0 | Agricultural soil | AOB | Nitrosococcus-like | F, I, B | [25] |
| 3.0–10.0 | Laboratory incubation | AOB, AOA | Nitrosomonas europaea, Nitrosomonas multiformis, Nitrosospira briensis, and Nitrosopumilus maritimus | I, H | [26]† |
| 3.2–6.6 | Agricultural soil | AOB | Nitrosomonas | F, B, Q | [27] |
| 3.3–4.5 | Permafrost environment | AOA, AOB | NA | F, B, Q, S, O, T | [28] |
| 3.6–4.5 | Heathland soil | AOB, NOB | Nitrosospira spp. AHB1, Nitrobacter spp. NHB1 | F, I, B, M | [29–31,32,33] |
| 3.7–8.8 | Soil | AOA | NA | F, A, T | [34] |
| 3.8–4.3 | Forest soil | AOA | NA | F, B, A, Q | [35] |
| 3.8–5.3 | Agricultural and forest soil | AOA | Nitrosotalea-like | F, I, B, D, O | [36] |
| 3.8–5.7 | Laboratory reactor | AOB | Ca. Nitrosoglobus | I, B, A, T | [37] |
| 3.8–6.2 | Agricultural soil | AOA, AOB | NA | F, I, A, Q, T | [38] |
| 3.8–8.0 | Soil | AOA | NA | R | [17] |
| <4.0–6.0 | Agricultural soil | AOB, NOB, comammox | Nitrosospira, canonical and comammox Nitrospira | F, B, I, A, M | [39] |
| 4.0–4.1 | Forest soil | AOA | NA | I, B, A, Q, T | [40] |
| 4.0–4.3 | Agricultural and restored soil | AOA, AOB | Nitrosomonas, Nitrosospira | F, B, A, Q, S | [41] |
| 4.0–5.5 | Agricultural soil | AOA | Ca. Nitrosotalea devaniterrae | F, B, I, A, M, O, T | [42–45] |
| 4.0–7.5 | Laboratory cultivation | AOB | Nitrosospira sp. NPAV | I, B, Q | [46] |
| 4.0–5.8 | Forest soil | AOA, AOB, NOB | Ca. Nitrosotalea, Nitrosospira, Nitrospira, Nitrobacter, Nitrosococcus, Nitrosospira | F, I, B, A, O, T | [47] |
| 4.1–4.9 | Forest, grassland, and wetland | AOA, NOB | NA | F, O | [48] |
| 4.1–5.2 | Forest soil | AOA, AOB | NA | F, B, A, Q, T | [49] |
| 4.2–4.5 | Agricultural soil | AOA, AOB | NA | F, B, A, Q, T, D | [50] |
| 4.2–7.5 | Glacier foreland soil | AOA | NA | F, A, Q | [51] |
| 4.3 | Laboratory reactor | AOB | Ca. Nitrosoglobus | I, B, A, Q | [52] |
| 4.3–5.2 | Forest soil | NOB | Nitrobacter spp. IOacid | F, I, B, M | [53] |
| 4.7–5.6 | Volcanic soil | AOA, AOB | Nitrososphaera, Ca. Nitrosotalea, Nitrosospira, Nitrosomonas | F, B, A, T | [54] |
| 4.8–6.4 | Peat land | AOA, AOB, NOB | NA | F, A, T | [55] |
| 4.8–6.9 | Agricultural soil | NOB | Nitrospira, Nitrobacter | F, B, A, Q, O, T | [56] |
| 5.0 | Laboratory reactor | AOB | Ca. Nitrosoglobus | I, B, A, K | [57] |
| 5.0–7.0 | Laboratory reactor | AOB | Ca. Nitrosoglobus | I, B, A, K | [58] |
| 5.0–8.0 | Laboratory reactor | AOB | Nitrosomonas europaea | I, B | [13,59] |
| 5.0–9.5 | Soil | AOA | NA | F, A, Q | [60] |
| 5.2–5.9 | Agricultural soil | AOA, AOB | NA | F, I, Q | [61] |
| 5.5–8.5 | Laboratory cultivation | AOA, NOB | Nitrosocosmicus oleophilus, Nitrosotenuis chungbukensis, Nitrosomonas europaea, Nitrobacter winogradskyi | I, S, B, A, T | [62] |
| 5.5–8.0 | Hot spring | NOB | Chloroflexota | F, I, Q, A, O, M | [63] |
| 8.2–8.8 | Glacier soil | NA | NA | F, B, A, T | [64] |
| 8.9 | Laboratory reactor | NA | NA | I, B, M | [65] |
| 9.5–10.2 | Soda lake | NOB | Nitrobacter alkalicus | F, I, B, A, M | [66] |
| 9.7 | Saline-alkaline lake | NA | NA | F, B, S | [67] |
| 9.8 | Soda lake | AOA, AOB | Nitrosomonas | F, B, A, M | [68] |
| 9.0–11.0 | Soda lake | AOB, NOB | Nitrosomonas, Nitrobacter | R | [69] |
| 8.9–10.5 | Saline-alkaline lake | NOB | canonical Nitrospira, Ca. Nitrospira alkalitolerans | F, I, B, A, O, M | [70] |
| 9.6–10.4 | Soda lake | AOA | NA | F, B, A | [71] |
| 9.5–11 | Soda lake | AOB, NOB | Nitrosomonas, Nitrobacter | R | [72] |
| 7.6–11.0 | Saline-alkaline lake | AOA, NOB, and comammox | Nitrosocosmicus, Nitrososphaera, canonical and comammox Nitrospira | F, B, A, S | [73] |
| ∼11.3 | Soda lake | AOB, NOB | Nitrosomonas halophilus, Nitrobacter alkalicus | R | [74] |
Sorted based on lowest or highest reported pH for the detection of nitrification or nitrifiers in acidic or alkaline environments, respectively
**Abbreviations of techniques or study type:
F: Field campaigns
I: Isolation or cultivation
B: Biochemical assays and physiological studies
Q: Auantitative polymerase chain reaction (qPCR)
A: Amplicon-based sequencing analysis using genes such as 16S rRNA gene, amoAB and nitrite oxidoreductase (nxrAB)
O: Omics and meta-omics including genomics, transcriptomics, and proteomics
M: Microscopy including electron microscopy and fluorescence in situ hybridization (FISH)
S: Stable isotopic tracing
T: Statistical analysis
K: Kinetics
D: DNA stable-isotope probing (DNA-SIP)
H: Thermodynamic investigation
R: Review article
This study applies calorimetric and potentiometric titrations to thermodynamically characterise cell surfaces of AOB and AOB from pH 3 to 10, which is beyond the growth pH range of subject microorganisms.
Ecological distribution
To date, there are over 50 studies reporting nitrification activities from a range of acidic and alkaline environments (Table 1). At acidic pH, nitrifiers have been reported to occur in agricultural, forest, coastal, permafrost, and volcanic soils, as well as in various reactor systems treating municipal or mining wastewaters. Nitrification has been less frequently investigated at high pH, but it is known to occur in certain alkaline saltmarshes, saline-alkaline and soda lakes, and bioreactors. The habitats with reported nitrification activities at the lowest pH (2.0) include acidic tea fields and laboratory bioreactors [21–23], while the activity at the highest pH (11.3) detected was in soda lakes [74].
Acidic ecosystems
Some of the highest rates of soil nitrification are found in acidic soils (pH < 5.5) that constitute approximately 30% of the world’s ice-free land [75]. The majority of these soils are naturally acidic, and approximately 5% are used for arable crops [75]. AOA are generally the predominant nitrifiers in such soils, which makes them a crucial subject for investigation [19,75,34]. The best-studied acidophilic AOA belong to the genus Ca. Nitrosotalea [42], which is currently represented by three cultivated strains (Ca. Nitrosotalea devaniterrae, Ca. Nitrosotalea sinensis and Ca. Nitrosotalea okcheonensis) with growth pH restricted to 4 and 5.5 (optima between 4 and 5) [42–45]. This is supported by a regional and global phylogenetic analysis of archaeal amoA gene distribution [34,76], which demonstrate that most lineages of Ca. Nitrosotalea are distributed in acidic environments. In addition, these studies revealed that clades of the Nitrososphaerales (γ and ζ [76]) are also consistently found in acidic soils, revealing patterns of niche speciation driven by low pH in AOA. This view is supported by a study using Bayesian comparative phylogenetics to investigate co-evolutionary relationships between amoA and soil characteristics, which shows that pH is a strong driver for AOA speciation [77]. However, no higher-level AOA clades that are specific to neutral or near-alkaline pH have been identified so far [76].
AOB can outnumber AOA in certain acidic soils and man-made ecosystems containing high levels of ammonium or urea [78,79] (Table 1). Cultivated neutralophilic AOB in the order of Burkholderiales within the Gammaproteobacteria (formerly placed within Betaproteobacteria) have been demonstrated to grow in acidic conditions when using urea as a substrate [29, 30, 46] or when growing in aggregates [31] or biofilms [59]. To date, the most acid-tolerant ammonia oxidizers characterised belong to the two gammaproteobacterial AOB isolates, Ca. Nitrosoglobus terrae TAO100 [21] and Ca. Nitrosacidococcus tergens RJ19 [23]. Both are most closely related to the neutralophilic marine AOB genus Nitrosococcus [23,21]. While both AOB have an optimum growth pH ∼6, they can mediate ammonia oxidation at pH 2–2.5, with Ca. Nitrosacidococcus tergens RJ19 even growing at pH 2.5. Additionally, 16S rRNA gene-based surveys in bioreactors treating wastewater suggest a much higher diversity of acid-tolerant AOB than currently enriched [22,52,24]. Finally, both AOA and AOB are found to be pioneer microorganisms in acidic volcanic soils [54], with their capacity to autotrophically grow on inorganic nitrogen compounds facilitating primary succession of barren environments [80].
A range of NOB have been identified through culture-independent surveys of various acidic systems such as soil and aquatic ecosystems, geothermal sites, and engineered environments. NOB activities and adaptations have been minimally studied under low pH conditions through bioassays and culture-dependent methods. To date, two acidophilic Nitrobacter strains have been isolated: Nitrobacter NHB1 from acidic heath soil [30] with an activity range of pH 5.0–7.5 and Nitrobacter strain IOacid from an acidic forest soil with maximal nitrite oxidation activity at pH 5.5 (range 4.1–7.2) [53]. Furthermore, canonical (only nitrite-oxidizing) and a comammox clade A Nitrospira strain have been enriched from acidic agricultural soils with activity from pH 4 to 6 [81]. To our knowledge, NOB and comammox organisms from acidic environments have not been subject to detailed physiological studies or genomic characterisation.
Alkaline ecosystems
Nitrification also occurs in saline-alkaline lakes and soda lakes ecosystems, as confirmed by ex situ incubations of lake sediments [82,73]. Multiple studies have shown that ammonia oxidisers mediate ammonia oxidation and primary production at high pH in these environments [74,67, 68, 71–73, 83]. Nitrosomonas halophila Ans5, the most alkaliphilic AOB known, was isolated from a Mongolian soda lake in 2001 [84]. This bacterium has an optimal pH range of 8.5–9.5 for growth and can maintain growth up to pH 11.3. In 2008, AOB affiliated with Nitrosomonas and AOA from Nitrososphaerales were suggested to be significant contributors of nitrification in the saline and alkaline Mono Lake, California [68]. Moreover, a recent study implicates both comammox and canonical Nitrospira, as well as several AOA phylotypes, in carrying out rapid nitrification in saline-alkaline lakes at pH up to 11 [73]. Analysis of an enrichment culture of the canonical nitrite oxidiser Ca. Nitrospira alkalitolerans showed it thrives at pH between 8.9 and 10.3, and is tolerant to hyposaline and subsaline conditions [70]. Five strains from Nitrobacter (AN1–AN5) have been isolated from sediments of soda lakes and soda soil at pH 10 with nitrite as the sole electron source; they formed a distinct species cluster denoted Nitrobacter alkalicus [84]. Several excellent reviews have highlighted the central role of nitrifying microorganisms in regulating elemental cycling and enabling primary production in alkaline and saline soda lakes [74,72,69]. However, compared to acidic ecosystems, research on nitrification in alkaline ecosystems is scarce. Given the apparent presence and activity of nitrifiers in alkaline environments, future research on the distribution, physiological speciation and adaptation to elevated pH of these nitrifiers is needed to better understand alkaline nitrification and explore its potential applications.
Physiological adaptations
Ammonia acquisition and oxidation
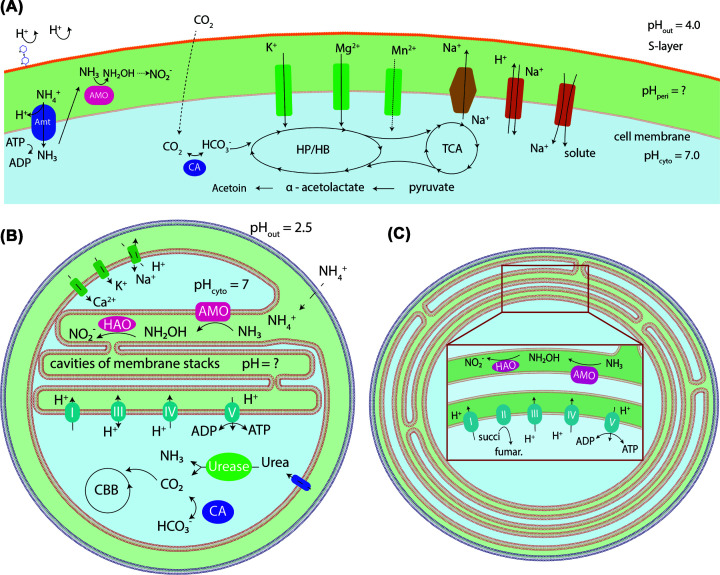
The active sites of ammonia monooxygenase (AMO) and hydroxylamine dehydrogenase (HAO) are thought to be periplasmic in AOA and AOB (Figure 3A–C) [85]. Our current understanding of comammox Nitrospira also favours a periplasmic orientation of AMO and HAO [3,86]. Therefore, the functionalities of these enzymes are likely influenced by environmental pH, as periplasmic pH is likely closer to external pH [5,87]. At acidic pH, ammonia (generally considered to be the substrate of AMO) becomes substantially scarce, posing a major substrate limitation for ammonia oxidisers (Figure 2). Early observations that the AOB Nitrosospira sp. AHB1 and Nitrosospira. sp. NPAV were able to grow on urea alone, but not on ammonium, at pH < 5.5 have led to the hypothesis that intracellular ureolysis enables ammonia acquisition and oxidation to occur independently of extracellular pH [46,29]. However, Ca. Nitrosotalea devaniterrae is not ureolytic [44], and the ability of the AOB Ca. Nitrosacidococcus tergens RJ19 and Ca. Nitrosoglobus terrae TAO100 to grow at low pH in ammonium-fed bioreactors without added urea suggests that ureolysis can play an important, but not essential role, in adaptation to low pH for some ammonia oxidizers. It should be noted that AMO and HAO of known comammox Nitrospira are phylogenetically distinct from those of canonical AOB [3,4,86] and their orientation has yet to be proven. This knowledge is necessary to fully understand the impact of pH on bioenergetics in comammox Nitrospira.
Figure 3. Conceptual diagrams of substrate acquisition and pH homeostasis systems encoded in the genomes of nitrifiers.
(A) AOA Ca. Nitrosotalea devaniterrae based on illustrations in Lehtovira-Morley et al., 2016 [44]. (B) The intracytoplasmic membrane stacks based on illustration and electron micrographs of Ca. Nitrosacidococcus tergens RJ19 in Picone et al., 2020 [23]. The cation transporters and Na+/H+ antiporters provide pH homeostasis capabilities. (C) Proposed allocations of AMO and HAO on the intracytoplasmic membrane stacks according to electron microgram of Nitrosomonas halophila Ans1 [84]. The presence of respiratory complexes is inferred from the model pathway of N. halophila Nm1 at Biocyc.org [88]. Respiratory complexes I to V are indicated by roman numerals in panels (B and C); HP/HB, the hydroxypropionate/hydroxybutyrate cycle; TCA, the tricarboxylic acid cycle; CBB, Calvin–Benson–Bassham cycle; CA, carbonic anhydrase.
The obligately acidophilic AOA Ca. Nitrosotalea devaniterrae is hypothesized to overcome ammonia limitation at picomolar ranges by actively importing NH4+ into the cytoplasm [44]. This is accompanied by deprotonation to NH3 and diffusion of NH3 through the cytoplasmic membrane back into the pseudo-periplasmic space to generate sufficiently high levels of ammonia locally [44] (Figure 3A). Acid-tolerant/acidophilic AOA likely have a competitive advantage in acidic soils, just as they do in the oligotrophic ocean, due to a high ammonia affinity and efficient CO2 fixation pathways [14,89,90]. Indeed, Ca. Nitrosotalea devaniterrae possesses the highest specific affinity for NH3 of all kinetically characterized AOA [14]. Based on simulations, Li and co-authors further proposed that the negatively charged S-layer proteins of AOA may serve as a reservoir for the positively charged NH4+, thus indirectly increasing the NH3 concentration in the pseudo-periplasmic space and around AMO [91,26]. No AmtB-type ammonia transporter could be identified in the genomes of the two AOB species with the highest tolerance to low pH conditions and it has been speculated that these organisms rely on permeases or passive ammonia diffusion across the membrane (Figure 3B) [23].
Unlike AOA, ammonia oxidation in AOB likely occurs primarily around the extensive specialized intracytoplasmic membrane stacks predicted to house enzymes involved in nitrification and respiration to facilitate energy generation (Figure 3B) [23]. The membrane stacks found in many AOB may increase the membrane surface and periplasm volume and provide more space for the ammonia oxidizing enzymes. This would increase the maximal reaction rate at the cellular level and thus the rate of energy generation from NH3. This may help acidophilic AOB to survive under strongly NH3-limited conditions and provide a competitive advantage over other AOB. It is worth noting that the intracellular localization of intracytoplasmic membrane stacks displays a considerable distinction across different phylogenetic lineages of AOB [92]. For instance, in the AOB Nitrosomonas halophila, intracytoplasmic membrane stacks are localized in the immediate periphery of the outer cytoplasmic membrane [84] whereas those in Ca. Nitrosacidococcus tergens RJ19 and Ca. Nitrosoglobus terrae TAO100 are centered towards the cellular center (Figure 3C) [23,21]. Whether there is link between the cellular allocation of intracytoplasmic membrane stacks and adaptation towards acidic conditions remains to be determined.
Nitrite acquisition and oxidation
A low pH also presents challenges for nitrite acquisition in NOB. The chemical equilibrium dictates the dominance of HNO2 over NO2− under acidic conditions (Figure 2), further limiting the availability of nitrite, a transient substrate especially at reduced pH [93,94]. In addition, a high level of HNO2 imposes toxicity, nitrosative stress, and structural damage to the cell [95–98]. Both Nitrobacter and Nitrospira species have been detected in and/or isolated from acidic soils [53,39], where they likely inhabit different niches given their differences in affinity (to substrate NO2−) [99] and sensitivity (towards the toxic NH3 and HNO2) [100]. Moreover, micro-niches in soil might provide different pH micro-environments, allowing coexistence of different NOB. Therefore, kinetic data for the mostly uncultured NOB in soils, and physiological and comparative genomic studies of their mechanisms to combat nitrosative stress, are crucially needed to decipher the respective adaptive traits of different NOB lineages in acidic soils and other habitats. At high pH, the NO2−-HNO2 equilibrium is unlikely to be a limiting factor, because NO2− is the predominant form over HNO2 and therefore HNO2 toxicity plays no significant role at high pH (Figure 2). It is worth noting that the capacity of NOB including Nitrobacter and Nitrospira to use alternative energy sources, including and hydrogen, formate, and acetate, may also enable them to adapt to fluctuations in nitrite availability and fulfill their energetic requirements for intracellular pH homeostasis [101–106]. Indeed, the genome of the NOB Ca. Nitrospira alkalitolerans encodes a high-affinity group 2a [NiFe]-hydrogenase to potentially consume atmospheric H2, suggesting physiological flexibility as demonstrated in the neutralophile Nitrospira moscoviensis [101,105,107].
Intracellular pH homeostasis
Nitrifying microorganisms adapted to acidic environments use several mechanisms to overcome intracellular acidification resulting from inward proton flow due to the high ΔpH. In a recent study, Wang and colleagues demonstrated that AOA adapted to acidic conditions had horizontally acquired V-type ATPases that extrude cytosolic protons; consistently, heterologous expression of this enzyme in Escherichia coli enhanced acid tolerance [36]. Ion transporters and reduced membrane permeability have also been proposed to facilitate pH homeostasis in Ca. Nitrosotalea devaniterrae, including a range of cation transporters predicted for K+, Na+ and divalent cation uptake, a H+/Na+ antiporter, a carbonic anhydrase that may facilitate pH homeostasis in addition to contributing to carbon fixation, and an α-acetolactate decarboxylase suggested to mediate proton scavenging [44]. This archaeon may possess lipid membranes that are less permeable compared with neutralophilic AOA in addition to increased sugar units on the outside of the S-layer that are suggested to prohibit proton entry (Figure 3A) [44]. Several studies have inferred mechanisms for acidic adaptations of AOB based on genomic analysis, though without further validation [23,21]. The AOB Ca. Nitrosacidococcus tergens is proposed to use multiple enzymes for this purpose: carbonic anhydrases to scavenge protons, cation transporters to maintain an inside-positive ΔΨ, and several Na+/H+ transporters that may contribute to pH homeostasis (Figure 3B) [23]. The AOB Ca. Nitrosoglobus terrae TAO100 also encodes cation transporters which may be involved in the creation of an inside positive membrane potential to resist proton flux [21].
At high pH, the energetic challenge is much more severe as ΔpH detracts from PMF and it is therefore crucial to maintain a high ΔΨ with tightly regulated pH and ion homeostasis [5]. Instead of relying solely on PMF for ATP synthesis, the NOB Ca. Nitrospira alkalitolerans can potentially conserve energy through a sodium-motive force (SMF) as inferred by the presence of an N-type ATPase and a sodium-dependent NADH:quinone oxidoreductase in the genome next to the canonical complexes of the electron transport chain, in conjunction with the several H+/Na+ antiporters such as Mrp- and Nha-type antiporters to facilitate pH homeostasis [70].
While complex mechanisms are adopted by different groups of nitrifying microorganisms to regulate intracellular pH at a single cell level, microbes do not live in isolation in nature. Biofilms and aggregates formed by nitrifying communities provide microsites and buffers that may enable nitrification to continue at pH beyond tolerable ranges at the individual level. For example, de Boer et al. reported that aggregated but not individual cells from nitrifying communities in two Dutch acidic heath soils were able to nitrify at pH 4 [31], while Allison and Prosser also observed that biofilm populations of Nitrosomonas europaea oxidized ammonia at lower pH than planktonic populations [59]. Consistently, aggregate cell formation of Ca. Nitrosoglobus terrae TAO100 is induced under strongly acidic conditions [21] and Ca. Nitrospira alkalitolerans cells enriched under high pH densely clustered into consortia [70].
Nitrifier carbon fixation
As chemolithoautotrophs, nitrifying microorganisms build biomass through energy-demanding CO2 fixation pathways. AOA employ a variant of the hydroxypropionate/hydroxybutyrate (HP/HB) cycle. Being the most energy-efficient known aerobic carbon fixation pathway, it facilitates their adaptation to oligotrophic environments [108]. However, the carboxylases involved in this pathway use bicarbonate as a substrate [108], which is limited relative to dissolved CO2 at low pH (Figure 1). Some acidophilic AOA may enhance the rates and efficiency of carbon fixation through carbonic anhydrase to enrich intracellular bicarbonate through the conversion of CO2; consistently, carbonic anhydrases have been identified in two of four Ca. Nitrosotalea genomes [45]. At high pH, importing bicarbonate rather than CO2 is more advantageous due to the higher concentration of bicarbonate (Figure 2).
AOB and Chloroflexota NOB adopt the Calvin–Benson–Bassham (CBB) cycle, which uses CO2 as the substrate [63]. Despite the higher availability of dissolved CO2 at low pH, these nitrifying microorganisms have evolved mechanisms to further facilitate carbon fixation, possibly in part due to the low catalytic and energetic efficiency of the CBB cycle [109]. For instance, the genome of the acid-tolerant AOB Ca. Nitrosacidococcus tergens encodes two types of carbonic anhydrases that may support a ‘buffering’ mechanism by forming bicarbonate from CO2 that has entered the cytoplasm (with proton consumption) and subsequently converting bicarbonate back to CO2 as required by carbon fixation. Ca. Nitrosacidococcus tergens and Ca. Nitrosoglobus terrae TAO100 contain a urease that breaks down urea to generate CO2, in addition to providing NH3 as a source of energy [23,21]. On the other hand, the alkaliphilic Nitrosomonas halophila contains carboxysome-like organelles [84], which may be responsible for concentrating CO2 as suggested for Nitrosomonas eutropha C91 [110,111]. Finally, NOB in the genus Nitrospira employ the reductive tricarboxylic acid cycle for carbon fixation, which utilizes CO2 as the substrate [86,112,113]. The NOB Ca. Nitrospira alkalitolerans is likely able to use the sodium gradient between its saline-alkaline environment and the cytoplasm for bicarbonate uptake with a BicA symporter. Its cytoplasmic carbonic anhydrase may then convert the imported bicarbonate to CO2 in the cytoplasm for carbon fixation [70]. However, genes encoding these enzymes are also present in the genome of the neutralophilic marine species Nitrospira marina [114].
Applications and implications
Agricultural land
Agricultural soils accounted for 38.8% of global N2O emission between 2007 and 2016 [115]; the proportion and the gross emission are expected to intensify continually in the coming decade driven by increased fertilizer application and population growth [116]. Understanding the link between soil pH, productivity and microbial nitrogen cycling in soil is crucial to mitigating N2O emissions and enhancing productivity from agricultural land. Firstly, soil pH and crop productivity are intrinsically linked to nitrifier activities. This is because nitrifiers in agricultural soils compete for ammonium with crops, contribute to soil acidification due to the conversion of ammonia to nitrite in the first step of nitrification, and accelerate nitrogen losses through leaching of the highly soluble end-product nitrate [18]. Secondly, pH is strongly correlated with the emission of N2O from soil [117]. This is because both nitrifiers and denitrifiers are drivers of N2O emission [118,119], and their activities are strongly influenced by soil pH [120–123]. Soil acidification affects N2O emission in opposite ways between nitrifiers and denitrifiers. For instance, low pH impairs the maturation of N2O reductase in the denitrifier Paracoccus denitrificans [124], therefore, N2O emission by denitrifiers is increased when soil pH decreases [125,126], as N2O reductase is the only known enzyme able to reduce N2O to N2 [117]. In contrast, N2O emission by nitrifiers decreases as pH decreases because of niche differentiation; low pH in general favours the dominance of AOA over AOB and AOA produce less N2O [117,118,127]. N2O yields from ammonia oxidation by AOB species (0.1–8%) are generally at least one order of magnitude higher than from AOA species (0.04–0.3%) and comammox bacteria (0.07%) [119], with AOB activity generating approximately double the yield of AOA in soil [127]. Therefore, soils dominated by AOA and comammox Nitrospira are predicted to have lower N2O emission potentials than soils dominated by AOB activity, which are also associated with high inputs of inorganic ammonium fertilizer [118,128].
Various strategies to inhibit nitrification have been developed, which can be used in acidic agricultural soils to reduce N2O emissions and nitrogen loss. The most widely adopted method is the field application of nitrification inhibitors such as dicyandiamide (DCD), nitrapyrin (NP), and dimethylpyrazole phosphate (DMPP). These commercial inhibitors were tested against neutralophilic AOB and developed prior to the discoveries of AOA and comammox, and prior to the isolation of acid-tolerant ammonia oxidisers [19]. Indeed, the efficacy of these inhibitors varies across different groups of nitrifying microorganisms and soils with different pH and physicochemical conditions as reviewed by Li et al. [19] and Beeckman et al. [129]. AOA rather than AOB are often major ammonia oxidisers in agricultural soils [18,130,131] and the improved identification of resident nitrifiers is facilitating the development of targeted approaches to suppress their nitrification activities at various soil pH regimes. For example, a novel potential nitrification inhibitor, quinone imine, has recently been developed and shown to act more effectively than DCD and NP on AOA in acidic soils [132]. The isolation of acid-tolerant and acidophilic strains also offers opportunities for evaluating the efficacy of existing nitrification inhibitors on nitrification under acidic conditions, for understanding their mechanisms of action, as well as for designing and testing novel inhibitory compounds specific to the unique physiology of acidophilic nitrifiers while minimising harmful impacts on non-target organisms and the environment. Besides synthetic nitrification inhibitors, there is a prospect of employing biological nitrification inhibitors (BNIs) released in the rhizosphere of certain plants [131,133,134]. These plant-produced compounds could act as a natural control of nitrifier activities (reviewed in [135]), but it is unclear whether plants produce sufficient amounts of BNI for inhibiting nitrification when growing in acidic/alkaline soils or whether BNI can maintain their function in these soils.
Wastewater treatment at low pH
Acidic nitrification has extensive implications in global wastewater treatment. Biological nitrogen removal from wastewater is based on nitrification and denitrification mediated by autotrophic nitrifiers in oxic tanks and heterotrophic denitrifiers in anoxic tanks, respectively. Compared to a generally low abundance and activity of AOA, AOB usually play a more critical role in converting ammonium to nitrite in these systems [136,137]. The dominant NOB genera in most wastewater treatment plants (WWTPs) are Nitrospira and Nitrotoga [137,138]. Compared with the conventional combination of nitrification and denitrification, the application of either partial nitritation coupled with the anaerobic oxidation of ammonium (anammox) (PN/A) or of the ‘nitrite shunt’ approach can reduce aeration costs by 60% and eliminate the need for organic carbon dosing to heterotrophic denitrifiers [139], thus being more sustainable nitrogen removal techniques. Anammox is a process carried out by Planctomycetes in the Brocadiales order that oxidize ammonium using nitrite as the electron acceptor to produce dinitrogen gas. In contrast, the nitrite shunt approach is the attempt to stop nitrification at nitrite, which is then reduced to dinitrogen gas. Both processes require the selective suppression of NOB while retaining the activities of AOB and of anammox or denitrifiers, respectively [140]. However, this this has been a long-standing technological challenge.
NOB are typically sensitive to acid stress (see above), which has inspired attempts to develop efficient nitrification at low pH using acid-tolerant AOB [37]. Acid-tolerant AOB can provide a stable source of nitrite for anammox bacteria, while suppressing the activity of NOB at low pH [37,141]. This coupling of acidic nitrification with anammox is a highly promising technology that has been demonstrated in both lab-scale [37,57,142,143] and pilot-scale reactors [144]. It can save significant aeration costs, eliminate the need for organic carbon, and allow for the separation and use of digested sludge in bioenergy recovery [141,145]. This technology has the potential to transform sewage treatment from an energy-negative to an energy-neutral service for wastewater treatment plants [139,146,147]. In acidic laboratory reactors treating urine, Ca. Nitrosacidococcus urinae was favored under acidic conditions, while NOB were likely absent [24]. Although aerobic acidic nitritation has the potential to brings tremendous energy sufficiency in wastewater treatment, N2O emissions during this process (likely caused by nitrite detoxification in the microbial consortia) cannot be ignored [123]. Currently, a better understanding of the mechanisms regulating N2O emission is needed for these highly promising systems that aim to operate with low energy consumption and environmental footprint.
Conclusions and outlook
Through a combination of culture-based and culture-independent techniques, we now have an increasing understanding of the ecophysiology, adaptations, and distribution of nitrifying microorganisms in different ecosystems. As pH is intricately linked to substrate availability, bioenergetics and cellular defense against oxidative and nitrosative stress, it is crucial to deepen our understanding of mechanisms allowing nitrifiers to thrive or adapt to extreme pH. It is also critical to translate these ecological and physiological insights into mitigating nitrogen loss in agricultural land and improving energy efficiency in next-generation wastewater treatment processes.
Summary
Nitrification at acidic and alkaline pH has been understudied compared to nitrification at neutral pH conditions.
Ammonia-oxidising bacteria and archaea, nitrite-oxidising bacteria, and comammox bacteria inhabit diverse acidic and alkaline environments.
Physiological adaptations allow them to maintain pH homeostasis, acquire limiting energy and carbon sources, and detoxify toxic compounds at pH extremes.
Nitrification at acidic pH contributes to nitrogen loss and nitrous oxide emissions in agricultural systems, but also has promising applications in acidic wastewater treatment.
Abbreviations
- AOA
ammonia-oxidising archaea
- AOB
ammonia-oxidising bacteria
- BNI
biological nitrification inhibitor
- DCD
dicyandiamide
- DMPP
dimethylpyrazole phosphate
- HP/HB
hydroxypropionate/hydroxybutyrate
- NOB
nitrite-oxidising bacteria
- NP
nitrapyrin
- WWTP
wastewater treatment plant
Contributor Information
Gaofeng Ni, Email: gaofeng.ni@monash.edu.
Chris Greening, Email: chris.greening@monash.edu.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by Early Career Postdoctoral Fellowship [grant number ECPF23-8566329039] at the Faculty of Medicine, Nursing and Health Sciences of Monash University awarded to G.N. A.D. was supported by the Czech Science Foundation (GACR) project 21-17322M. J.G. is supported by Australian Research Council (ARC) Discovery Project [grant number DP230101340]. P.C. and C.G. are supported by an Australian Research Council (ARC) Discovery Project (DP210101595). This work was also supported by National Health & Medical Research Council (NHMRC) [grant number APP1178715 (to C.G.)]. Open access publishing facilitated by Monash University, as part of the Portland Press (Biochemical Society) - Monash University agreement via the Council of Australian University Librarians.
Open Access
Open access for this article was enabled by the participation of Monash University in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with CAUL.
Author Contribution
Conceptualization: C.G., G.N., P.M.L. Supervision: C.G. Writing - original draft: G.N., P.M.L., C.G. Writing - review and editing: G.N., P.M.L., H.D., J.G., A.D., C.G.g, G.W.N., S.H., P.C. Figure preparation: G.N., P.M.L.
References
- 1.Ward B.B. (2013) Nitrification. In Encyclopedia of Ecology Second Edition(Fath B., ed.), pp. 351–358, Elsevier, Oxford: 10.1016/B978-0-12-409548-9.00697-7 [DOI] [Google Scholar]
- 2.Stein L.Y. and Klotz M.G. (2016) The nitrogen cycle. Curr. Biol. 26, R94–R98 10.1016/j.cub.2015.12.021 [DOI] [PubMed] [Google Scholar]
- 3.Daims H., Lebedeva E.V., Pjevac P., Han P., Herbold C., Albertsen M.et al. (2015) Complete nitrification by Nitrospira bacteria. Nature 528, 504–509 10.1038/nature16461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Kessel M.A.H.J., Speth D.R., Albertsen M., Nielsen P.H., Op den Camp H.J.M., Kartal B.et al. (2015) Complete nitrification by a single microorganism. Nature 528, 555–559 10.1038/nature16459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krulwich T.A., Sachs G. and Padan E. (2011) Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9, 330–343 10.1038/nrmicro2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangold S., Jonna V.R. and Dopson M. (2013) Response of Acidithiobacillus caldus toward suboptimal pH conditions. Extremophiles 17, 689–696 10.1007/s00792-013-0553-5 [DOI] [PubMed] [Google Scholar]
- 7.Slonczewski J.L., Fujisawa M., Dopson M. and Krulwich T.A. (2009) Cytoplasmic pH Measurement and Homeostasis in Bacteria and Archaea. In Advances in Microbial Physiologyvol. 55, (Poole R.K., ed.), pp. 1–317, Academic Press; 10.1016/S0065-2911(09)05501-5 [DOI] [PubMed] [Google Scholar]
- 8.Baker-Austin C. and Dopson M. (2007) Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 15, 165–171 10.1016/j.tim.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 9.Madigan M.T., Martinko J.M. and Parker J. (2003) Brock biology of microorganisms, Prentice Hall/Pearson Education [Google Scholar]
- 10.Jones R.D. and Morita R.Y. (1985) Low-temperature growth and whole-cell kinetics of a marine ammonium oxidizer. Marine Ecol. Progress Series 21, 239–243 10.3354/meps021239 [DOI] [Google Scholar]
- 11.Suzuki I., Dular U. and Kwok S.C. (1974) Ammonia or ammonium ion as substrate for oxidation by Nitrosomonas europaea cells and extracts. J. Bacteriol. 120, 556–558 10.1128/jb.120.1.556-558.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunik J.H., Meijer H.J.G. and Tramper J. (1992) Kinetics of Nitrosomonas europaea at extreme substrate, product and salt concentrations. Appl. Microbiol. Biotechnol. 37, 802–807 10.1007/BF00174849 [DOI] [Google Scholar]
- 13.Frijlink M.J., Abee T., Laanbroek H.J., de Boer W. and Konings W.N. (1992) The bioenergetics of ammonia and hydroxylamine oxidation in Nitrosomonas europaea at acid and alkaline pH. Arch. Microbiol. 157, 194–199 10.1007/BF00245290 [DOI] [Google Scholar]
- 14.Jung M.-Y., Sedlacek C.J., Kits K.D., Mueller A.J., Rhee S.-K., Hink L.et al. (2022) Ammonia-oxidizing archaea possess a wide range of cellular ammonia affinities. ISME J. 16, 272–283 10.1038/s41396-021-01064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zumdahl S.S. and Zumdahl S.A. (2005) Chemistry 6th Edition, 6Rev Ed edition, Houghton Mifflin [Google Scholar]
- 16.Blum J.-M., Su Q., Ma Y., Valverde-Pérez B., Domingo-Félez C., Jensen M.M.et al. (2018) The pH dependency of N-converting enzymatic processes, pathways and microbes: effect on net N2O production. Environ. Microbiol. 20, 1623–1640 10.1111/1462-2920.14063 [DOI] [PubMed] [Google Scholar]
- 17.He J.-Z., Hu H.-W. and Zhang L.-M. (2012) Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils. Soil Biol. Biochem. 55, 146–154 10.1016/j.soilbio.2012.06.006 [DOI] [Google Scholar]
- 18.Hu H.-W., Xu Z.-H. and He J.-Z. (2014) Chapter Six - Ammonia-oxidizing archaea play a predominant role in acid soil nitrification. In Advances in Agronomyvol. 125, (Sparks D.L., ed.), pp. 261–302, Academic Press [Google Scholar]
- 19.Li Y., Chapman S.J., Nicol G.W. and Yao H. (2018) Nitrification and nitrifiers in acidic soils. Soil Biol. Biochem. 116, 290–301 10.1016/j.soilbio.2017.10.023 [DOI] [Google Scholar]
- 20.de Boer W. and Kowalchuk G.A. (2001) Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol. Biochem. 33, 853–866 10.1016/S0038-0717(00)00247-9 [DOI] [Google Scholar]
- 21.Hayatsu M., Tago K., Uchiyama I., Toyoda A., Wang Y., Shimomura Y.et al. (2017) An acid-tolerant ammonia-oxidizing γ-proteobacterium from soil. ISME J. 11, 1130–1141 10.1038/ismej.2016.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fumasoli A., Bürgmann H., Weissbrodt D.G., Wells G.F., Beck K., Mohn J.et al. (2017) Growth of Nitrosococcus-related ammonia oxidizing bacteria coincides with extremely low pH values in wastewater with high ammonia content. Environ. Sci. Technol. 51, 6857–6866 10.1021/acs.est.7b00392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picone N., Pol A., Mesman R., van Kessel M.A.H.J., Cremers G., van Gelder A.H.et al. (2021) Ammonia oxidation at pH 2.5 by a new gammaproteobacterial ammonia-oxidizing bacterium. ISME J. 15, 1150–1164 10.1038/s41396-020-00840-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faust V., van Alen T.A., Op den Camp H.J.M., Vlaeminck S.E., Ganigué R., Boon N.et al. (2022) Ammonia oxidation by novel “Candidatus Nitrosacidococcus urinae” is sensitive to process disturbances at low pH and to iron limitation at neutral pH. Water Res. X 17, 100157 10.1016/j.wroa.2022.100157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayatsu M. (1993) The lowest limit of pH for nitrification in tea soil and isolation of an acidophilic ammonia oxidizing bacterium. Soil Sci. Plant Nutr. 39, 219–226 10.1080/00380768.1993.10416993 [DOI] [Google Scholar]
- 26.Gorman-Lewis D., Martens-Habbena W. and Stahl D.A. (2014) Thermodynamic characterization of proton-ionizable functional groups on the cell surfaces of ammonia-oxidizing bacteria and archaea. Geobiology 12, 157–171 10.1111/gbi.12075 [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama K., Kimura H. and Shinozaki H. (2003) Ammonia-oxidizing activity under extremely oligotrophic conditions in strongly acid tea soils. Soil Sci. Plant Nutr. 49, 711–718 10.1080/00380768.2003.10410329 [DOI] [Google Scholar]
- 28.Wang X., Wang S., Yang Y., Tian H., Jetten M.S.M., Song C.et al. (2023) Hot moment of N2O emissions in seasonally frozen peatlands. ISME J. 17, 792–802 10.1038/s41396-023-01389-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Boer W. and Laanbroek H.J. (1989) Ureolytic nitrification at low pH by Nitrosospira spec. Arch. Microbiol. 152, 178–181 10.1007/BF00456098 [DOI] [Google Scholar]
- 30.de Boer W., Duyts H. and Laanbroek H.J. (1989) Urea stimulated autotrophic nitrification in suspensions of fertilized, acid heath soil. Soil Biol. Biochem. 21, 349–354 10.1016/0038-0717(89)90142-9 [DOI] [Google Scholar]
- 31.de Boer W., Gunnewiek P.J.A.K., Veenhuis M., Bock E. and Laanbroek H.J. (1991) Nitrification at Low pH by aggregated chemolithotrophic bacteria. Appl. Environ. Microbiol. 57, 3600–3604 10.1128/aem.57.12.3600-3604.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Boer W., Duyts H. and Laanbroek H.J. (1988) Autotrophic nitrification in a fertilized acid heath soil. Soil Biol. Biochem. 20, 845–850 10.1016/0038-0717(88)90091-0 [DOI] [Google Scholar]
- 33.de Boer W., Klein Gunnewiek P.A. and Laanbroek H.J. (1995) Ammonium-oxidation at low pH by a chemolithotrophic bacterium belonging to the genus Nitrosospira. Soil Biol. Biochem. 27, 127–132 10.1016/0038-0717(94)00157-V [DOI] [Google Scholar]
- 34.Gubry-Rangin C., Hai B., Quince C., Engel M., Thomson B.C., James P.et al. (2011) Niche specialization of terrestrial archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. 108, 21206–21211 10.1073/pnas.1109000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kemnitz D., Kolb S. and Conrad R. (2007) High abundance of Crenarchaeota in a temperate acidic forest soil. FEMS Microbiol. Ecol. 60, 442–448 10.1111/j.1574-6941.2007.00310.x [DOI] [PubMed] [Google Scholar]
- 36.Wang B., Qin W., Ren Y., Zhou X., Jung M.-Y., Han P.et al. (2019) Expansion of Thaumarchaeota habitat range is correlated with horizontal transfer of ATPase operons. ISME J. 13, 3067–3079 10.1038/s41396-019-0493-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z., Zheng M., Meng J., Hu Z., Ni G., Guerrero Calderon A.et al. (2021) Robust nitritation sustained by acid-tolerant ammonia-oxidizing bacteria. Environ. Sci. Technol. 55, 2048–2056 10.1021/acs.est.0c05181 [DOI] [PubMed] [Google Scholar]
- 38.Bossolani J.W., Crusciol C.A.C., Merloti L.F., Moretti L.G., Costa N.R., Tsai S.M.et al. (2020) Long-term lime and gypsum amendment increase nitrogen fixation and decrease nitrification and denitrification gene abundances in the rhizosphere and soil in a tropical no-till intercropping system. Geoderma 375, 114476 10.1016/j.geoderma.2020.114476 [DOI] [Google Scholar]
- 39.Takahashi Y., Fujitani H., Hirono Y., Tago K., Wang Y., Hayatsu M.et al. (2020) Enrichment of comammox and nitrite-oxidizing Nitrospira from acidic soils. Front. Microbiol 11, 1737 10.3389/fmicb.2020.01737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stopnišek N., Gubry-Rangin C., Höfferle Š., Nicol G.W., Mandič-Mulec I. and Prosser J.I. (2010) Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl. Environ. Microbiol. 76, 7626–7634 10.1128/AEM.00595-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying J.-Y., Zhang L.-M. and He J.-Z. (2010) Putative ammonia-oxidizing bacteria and archaea in an acidic red soil with different land utilization patterns. Environ. Microbiol. Rep. 2, 304–312 10.1111/j.1758-2229.2009.00130.x [DOI] [PubMed] [Google Scholar]
- 42.Lehtovirta-Morley L.E., Stoecker K., Vilcinskas A., Prosser J.I. and Nicol G.W. (2011) Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. 108, 15892–15897 10.1073/pnas.1107196108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehtovirta-Morley L.E., Ge C., Ross J., Yao H., Nicol G.W. and Prosser J.I. (2014) Characterisation of terrestrial acidophilic archaeal ammonia oxidisers and their inhibition and stimulation by organic compounds. FEMS Microbiol. Ecol. 89, 542–552 10.1111/1574-6941.12353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehtovirta-Morley L.E., Sayavedra-Soto L.A., Gallois N., Schouten S., Stein L.Y., Prosser J.I.et al. (2016) Identifying potential mechanisms enabling acidophily in the ammonia-oxidizing archaeon “Candidatus Nitrosotalea devanaterra”. Appl. Environ. Microbiol. 82, 2608–2619 10.1128/AEM.04031-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herbold C.W., Lehtovirta-Morley L.E., Jung M.-Y., Jehmlich N., Hausmann B., Han P.et al. (2017) Ammonia-oxidising archaea living at low pH: Insights from comparative genomics. Environ. Microbiol. 19, 4939–4952 10.1111/1462-2920.13971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burton S.A.Q. and Prosser J.I. (2001) Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl. Environ. Microbiol. 67, 2952–2957 10.1128/AEM.67.7.2952-2957.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scarlett K., Denman S., Clark D.R., Forster J., Vanguelova E., Brown N.et al. (2021) Relationships between nitrogen cycling microbial community abundance and composition reveal the indirect effect of soil pH on oak decline. ISME J. 15, 623–635 10.1038/s41396-020-00801-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bay S.K., Dong X., Bradley J.A., Leung P.M., Grinter R., Jirapanjawat T.et al. (2021) Trace gas oxidizers are widespread and active members of soil microbial communities. Nat. Microbiol 6, 246–256 10.1038/s41564-020-00811-w [DOI] [PubMed] [Google Scholar]
- 49.Long X., Chen C., Xu Z., Oren R. and He J.-Z. (2012) Abundance and community structure of ammonia-oxidizing bacteria and archaea in a temperate forest ecosystem under ten-years elevated CO2. Soil Biol. Biochem. 46, 163–171 10.1016/j.soilbio.2011.12.013 [DOI] [Google Scholar]
- 50.Zhang L.-M., Hu H.-W., Shen J.-P. and He J.-Z. (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 6, 1032–1045 10.1038/ismej.2011.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicol G.W., Tscherko D., Embley T.M. and Prosser J.I. (2005) Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ. Microbiol. 7, 337–347 10.1111/j.1462-2920.2005.00698.x [DOI] [PubMed] [Google Scholar]
- 52.Meng J., Hu Z., Wang Z., Hu S., Liu Y., Guo H.et al. (2022) Determining factors for nitrite accumulation in an acidic nitrifying system: influent ammonium concentration, operational pH, and ammonia-oxidizing community. Environ. Sci. Technol. 56, 11578–11588 10.1021/acs.est.1c07522 [DOI] [PubMed] [Google Scholar]
- 53.Hankinson T.R. and Schmidt E.L. (1988) An acidophilic and a neutrophilic Nitrobacter strain isolated from the numerically predominant nitrite-oxidizing population of an acid forest soil. Appl. Environ. Microbiol. 54, 1536–1540 10.1128/aem.54.6.1536-1540.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hernández M., Dumont M.G., Calabi M., Basualto D. and Conrad R. (2014) Ammonia oxidizers are pioneer microorganisms in the colonization of new acidic volcanic soils from South of Chile. Environ. Microbiol. Rep. 6, 70–79 10.1111/1758-2229.12109 [DOI] [PubMed] [Google Scholar]
- 55.Wang H., Bagnoud A., Ponce-Toledo R.I., Kerou M., Weil M., Schleper C.et al. (2021) Linking 16S rRNA gene classification to amoA gene taxonomy reveals environmental distribution of ammonia-oxidizing archaeal clades in peatland soils. MSystems 6, e00546–e00621 10.1128/mSystems.00546-21 [DOI] [PubMed] [Google Scholar]
- 56.Han S., Li X., Luo X., Wen S., Chen W. and Huang Q. (2018) Nitrite-oxidizing bacteria community composition and diversity are influenced by fertilizer regimes, but are independent of the soil aggregate in acidic subtropical red soil. Front. Microbiol. 9, 885 10.3389/fmicb.2018.00885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z., Ni G., Maulani N., Xia J., De Clippeleir H., Hu S.et al. (2021) Stoichiometric and kinetic characterization of an acid-tolerant ammonia oxidizer ‘Candidatus Nitrosoglobus’. Water Res. 196, 117026 10.1016/j.watres.2021.117026 [DOI] [PubMed] [Google Scholar]
- 58.Zhang J., Hu Z., Liu T., Wang Z., Guo J., Yuan Z.et al. (2021) Feasibility of methane bioconversion to methanol by acid-tolerant ammonia-oxidizing bacteria. Water Res. 197, 117077 10.1016/j.watres.2021.117077 [DOI] [PubMed] [Google Scholar]
- 59.Allison S.M. and Prosser J.I. (1993) Ammonia oxidation at low pH by attached populations of nitrifying bacteria. Soil Biol. Biochem. 25, 935–941 10.1016/0038-0717(93)90096-T [DOI] [Google Scholar]
- 60.Ochsenreiter T., Selezi D., Quaiser A., Bonch-Osmolovskaya L. and Schleper C. (2003) Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5, 787–797 10.1046/j.1462-2920.2003.00476.x [DOI] [PubMed] [Google Scholar]
- 61.Shi R.Y., Ni N., Nkoh J.N., Li J.Y., Xu R.K. and Qian W. (2019) Beneficial dual role of biochars in inhibiting soil acidification resulting from nitrification. Chemosphere 234, 43–51 10.1016/j.chemosphere.2019.06.030 [DOI] [PubMed] [Google Scholar]
- 62.Jung M.-Y., Gwak J.-H., Rohe L., Giesemann A., Kim J.-G., Well R.et al. (2019) Indications for enzymatic denitrification to N2O at low pH in an ammonia-oxidizing archaeon. ISME J. 13, 2633–2638 10.1038/s41396-019-0460-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spieck E., Spohn M., Wendt K., Bock E., Shively J., Frank J.et al. (2020) Extremophilic nitrite-oxidizing Chloroflexi from Yellowstone hot springs. ISME J. 14, 364–379 10.1038/s41396-019-0530-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strauss S.L., Garcia-Pichel F. and Day T.A. (2012) Soil microbial carbon and nitrogen transformations at a glacial foreland on Anvers island, Antarctic peninsula. Polar Biol. 35, 1459–1471 10.1007/s00300-012-1184-5 [DOI] [Google Scholar]
- 65.Su H., Zhang D., Antwi P., Xiao L., Deng X., Liu Z.et al. (2021) Exploring potential impact(s) of cerium in mining wastewater on the performance of partial-nitrification process and nitrogen conversion microflora. Ecotoxicol. Environ. Saf. 209, 111796 10.1016/j.ecoenv.2020.111796 [DOI] [PubMed] [Google Scholar]
- 66.Sorokin D.Y., Muyzer G., Brinkhoff T., Gijs Kuenen J. and Jetten M.S.M. (1998) Isolation and characterization of a novel facultatively alkaliphilic Nitrobacter species, N. alkalicus sp. Nov. Arch. Microbiol. 170, 345–352 10.1007/s002030050652 [DOI] [PubMed] [Google Scholar]
- 67.Joye S.B., Connell T.L., Miller L.G., Oremland R.S. and Jellison R.S. (1999) Oxidation of ammonia and methane in an alkaline, saline lake. Limnol. Oceanogr. 44, 178–188 10.4319/lo.1999.44.1.0178 [DOI] [Google Scholar]
- 68.Carini S.A. and Joye S.B. (2008) Nitrification in Mono Lake, California: activity and community composition during contrasting hydrological regimes. Limnol. Oceanogr. 53, 2546–2557 10.4319/lo.2008.53.6.2546 [DOI] [Google Scholar]
- 69.Sorokin D.Y. and Kuenen J.G. (2005) Chemolithotrophic haloalkaliphiles from soda lakes. FEMS Microbiol. Ecol. 52, 287–295 10.1016/j.femsec.2005.02.012 [DOI] [PubMed] [Google Scholar]
- 70.Daebeler A., Kitzinger K., Koch H., Herbold C.W., Steinfeder M., Schwarz J.et al. (2020) Exploring the upper pH limits of nitrite oxidation: diversity, ecophysiology, and adaptive traits of haloalkalitolerant Nitrospira. ISME J. 14, 2967–2979 10.1038/s41396-020-0724-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lanzén A., Simachew A., Gessesse A., Chmolowska D., Jonassen I. and Øvreås L. (2013) Surprising prokaryotic and eukaryotic diversity, community structure and biogeography of Ethiopian soda lakes. PloS ONE 8, e72577 10.1371/journal.pone.0072577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sorokin D.Y., Berben T., Melton E.D., Overmars L., Vavourakis C.D. and Muyzer G. (2014) Microbial diversity and biogeochemical cycling in soda lakes. Extremophiles 18, 791–809 10.1007/s00792-014-0670-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daebeler A., Güell-Bujons Q., Mooshammer M., Zechmeister T., Herbold C.W., Richter A.et al. (2023) Rapid nitrification involving comammox and canonical Nitrospira at extreme pH in saline-alkaline lakes. Environ. Microbiol. 25, 1055–1067 10.1111/1462-2920.16337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sorokin D.Y., Banciu H.L. and Muyzer G. (2015) Functional microbiology of soda lakes. Curr. Opin. Microbiol. 25, 88–96 10.1016/j.mib.2015.05.004 [DOI] [PubMed] [Google Scholar]
- 75.von Uexküll H.R. and Mutert E. (1995) Global extent, development and economic impact of acid soils. Plant Soil 171, 1–15 10.1007/BF00009558 [DOI] [Google Scholar]
- 76.Alves R.J.E., Minh B.Q., Urich T., von Haeseler A. and Schleper C. (2018) Unifying the global phylogeny and environmental distribution of ammonia-oxidising archaea based on amoA genes. Nat. Commun 9, 1517 10.1038/s41467-018-03861-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gubry-Rangin C., Kratsch C., Williams T.A., McHardy A.C., Embley T.M., Prosser J.I.et al. (2015) Coupling of diversification and pH adaptation during the evolution of terrestrial Thaumarchaeota. Proc. Natl. Acad. Sci. 112, 9370–9375 10.1073/pnas.1419329112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wertz S., Leigh A.K.K. and Grayston S.J. (2012) Effects of long-term fertilization of forest soils on potential nitrification and on the abundance and community structure of ammonia oxidizers and nitrite oxidizers. FEMS Microbiol. Ecol. 79, 142–154 10.1111/j.1574-6941.2011.01204.x [DOI] [PubMed] [Google Scholar]
- 79.Petersen D.G., Blazewicz S.J., Firestone M., Herman D.J., Turetsky M. and Waldrop M. (2012) Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environ. Microbiol. 14, 993–1008 10.1111/j.1462-2920.2011.02679.x [DOI] [PubMed] [Google Scholar]
- 80.Ni G., Lappan R., Hernández M., Santini T., Tomkins A.G. and Greening C. (2022) Functional basis of primary succession: Traits of the pioneer microbes. Environ. Microbiol. 25, 171–176 10.1111/1462-2920.16266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takahashi Y., Fujitani H., Hirono Y., Tago K., Wang Y., Hayatsu M.et al. (2020) Enrichment of comammox and nitrite-oxidizing Nitrospira from acidic soils. Front. Microbiol 11, 1737 10.3389/fmicb.2020.01737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sorokin D.Y. (1998) On the possibility of nitrification in extremely alkaline soda biotopes. Microbiology 67, 335–339 [Google Scholar]
- 83.Khmelenina Y.N., Eshinimaev B.Ts., Kalyuzhnaya M.G. and Trotsenko Yu.A. (2000) Potential activity of methane and ammonium oxidation by methanotrophic communities from the soda lakes of southern Transbaikal. Microbiology 69, 460–465 10.1007/BF02756771 [DOI] [PubMed] [Google Scholar]
- 84.Sorokin D.Y., Tourova T., Schmid M.C., Wagner M., Koops H.-P., Kuenen G.J.et al. (2001) Isolation and properties of obligately chemolithoautotrophic and extremely alkali-tolerant ammonia-oxidizing bacteria from Mongolian soda lakes. Arch. Microbiol. 176, 170–177 10.1007/s002030100310 [DOI] [PubMed] [Google Scholar]
- 85.Simon J. and Klotz M.G. (2013) Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim. Biophys. Acta 1827, 114–135 10.1016/j.bbabio.2012.07.005 [DOI] [PubMed] [Google Scholar]
- 86.Palomo A., Pedersen A.G., Fowler S.J., Dechesne A., Sicheritz-Pontén T. and Smets B.F. (2018) Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 12, 1779–1793 10.1038/s41396-018-0083-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilks J.C. and Slonczewski J.L. (2007) pH of the cytoplasm and periplasm of Escherichia coli: Rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 189, 5601–5607 10.1128/JB.00615-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. (2023) Nitrosomonas halophila Nm1 aerobic respiration I (cytochrome c). BiocycOrg https://biocyc.org/GCF_900107165/new-image?object=PWY-3781 (accessed February 9, 2023) [Google Scholar]
- 89.Martens-Habbena W., Berube P.M., Urakawa H., de la Torre J.R. and Stahl D.A. (2009) Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979 10.1038/nature08465 [DOI] [PubMed] [Google Scholar]
- 90.Stahl D.A. and de la Torre J.R. (2012) Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol. 66, 83–101 10.1146/annurev-micro-092611-150128 [DOI] [PubMed] [Google Scholar]
- 91.Li P.-N., Herrmann J., Tolar B.B., Poitevin F., Ramdasi R., Bargar J.R.et al. (2018) Nutrient transport suggests an evolutionary basis for charged archaeal surface layer proteins. ISME J. 12, 2389–2402 10.1038/s41396-018-0191-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koops H.-P., Purkhold U., Pommerening-Röser A., Timmermann G. and Wagner M. (2006) The lithoautotrophic ammonia-oxidizing bacteria. In The Prokaryotes: Volume 5: Proteobacteria: Alpha and Beta Subclasses(Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H. and Stackebrandt E., eds), pp. 778–811, Springer, New York, NY: 10.1007/0-387-30745-1_36 [DOI] [Google Scholar]
- 93.Shen Q.R., Berry W. and Cao Z.H. (2003) Mechanisms of nitrite accumulation occurring in soil nitrification. Chemosphere 50, 747–753 10.1016/S0045-6535(02)00215-1 [DOI] [PubMed] [Google Scholar]
- 94.Burns L.C., Stevens R.J., Smith R.V. and Cooper J.E. (1995) The occurrence and possible sources of nitrite in a grazed, fertilized, grassland soil. Soil Biol. Biochem. 27, 47–59 10.1016/0038-0717(94)00130-S [DOI] [Google Scholar]
- 95.Duan H., Gao S., Li X., Ab Hamid N.H., Jiang G., Zheng M.et al. (2020) Improving wastewater management using free nitrous acid (FNA). Water Res. 171, 115382 10.1016/j.watres.2019.115382 [DOI] [PubMed] [Google Scholar]
- 96.Chislett M., Guo J., Bond P.L., Jones A. and Yuan Z. (2020) Structural changes in cell-wall and cell-membrane organic materials following exposure to free nitrous acid. Environ. Sci. Technol. 54, 10301–10312 10.1021/acs.est.0c01453 [DOI] [PubMed] [Google Scholar]
- 97.Chislett M., Guo J., Bond P.L. and Yuan Z. (2021) Structural changes in model compounds of sludge extracellular polymeric substances caused by exposure to free nitrous acid. Water Res. 188, 116553 10.1016/j.watres.2020.116553 [DOI] [PubMed] [Google Scholar]
- 98.Chislett M., Guo J., Bond P.L., Wang Y., Donose B.C. and Yuan Z. (2022) Reactive nitrogen species from free nitrous acid (FNA) cause cell lysis. Water Res. 217, 118401 10.1016/j.watres.2022.118401 [DOI] [PubMed] [Google Scholar]
- 99.Nowka B., Daims H. and Spieck E. (2015) Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl. Environ. Microbiol. 81, 745–753 10.1128/AEM.02734-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blackburne R., Vadivelu V.M., Yuan Z. and Keller J. (2007) Kinetic characterisation of an enriched Nitrospira culture with comparison to Nitrobacter. Water Res. 41, 3033–3042 10.1016/j.watres.2007.01.043 [DOI] [PubMed] [Google Scholar]
- 101.Leung P.M., Daebeler A., Chiri E., Hanchapola I., Gillett D.L., Schittenhelm R.B.et al. (2022) A nitrite-oxidising bacterium constitutively consumes atmospheric hydrogen. ISME J. 16, 2213–2219 10.1038/s41396-022-01265-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smith A.J. and Hoare D.S. (1968) Acetate assimilation by Nitrobacter agilis in relation to its “obligate autotrophy”. J. Bacteriol. 95, 844–855 10.1128/jb.95.3.844-855.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bock E. (1976) Growth of Nitrobacter in the presence of organic matter. Arch. Microbiol. 108, 305–312 10.1007/BF00454857 [DOI] [PubMed] [Google Scholar]
- 104.Freitag A., Rudert M. and Bock E. (1987) Growth of Nitrobacter by dissimilatoric nitrate reduction. FEMS Microbiol. Lett. 48, 105–109 10.1111/j.1574-6968.1987.tb02524.x [DOI] [Google Scholar]
- 105.Koch H., Galushko A., Albertsen M., Schintlmeister A., Gruber-Dorninger C., Lücker S.et al. (2014) Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science 345, 1052–1054 10.1126/science.1256985 [DOI] [PubMed] [Google Scholar]
- 106.Koch H., Lücker S., Albertsen M., Kitzinger K., Herbold C., Spieck E.et al. (2015) Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl. Acad. Sci. 112, 11371–11376 10.1073/pnas.1506533112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ehrich S., Behrens D., Lebedeva E., Ludwig W. and Bock E. (1995) A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch. Microbiol. 164, 16–23 10.1007/BF02568729 [DOI] [PubMed] [Google Scholar]
- 108.Könneke M., Schubert D.M., Brown P.C., Hügler M., Standfest S., Schwander T.et al. (2014) Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc. Natl. Acad. Sci. 111, 8239–8244 10.1073/pnas.1402028111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Berg I.A. (2011) Ecological Aspects of the Distribution of Different Autotrophic CO2 Fixation Pathways. Appl. Environ. Microbiol. 77, 1925–1936 10.1128/AEM.02473-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stein L.Y., Arp D.J., Berube P.M., Chain P.S.G., Hauser L., Jetten M.S.M.et al. (2007) Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ. Microbiol. 9, 2993–3007 10.1111/j.1462-2920.2007.01409.x [DOI] [PubMed] [Google Scholar]
- 111.Badger M.R. and Bek E.J. (2008) Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 59, 1525–1541 10.1093/jxb/erm297 [DOI] [PubMed] [Google Scholar]
- 112.Daims H., Lücker S. and Wagner M. (2016) A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 24, 699–712 10.1016/j.tim.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lücker S., Wagner M., Maixner F., Pelletier E., Koch H., Vacherie B.et al. (2010) A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. 107, 13479–13484 10.1073/pnas.1003860107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bayer B., Saito M.A., McIlvin M.R., Lücker S., Moran D.M., Lankiewicz T.S.et al. (2021) Metabolic versatility of the nitrite-oxidizing bacterium Nitrospira marina and its proteomic response to oxygen-limited conditions. ISME J. 15, 1025–1039 10.1038/s41396-020-00828-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shukla P.R., Skea J., Calvo Buendia E., Masson-Delmotte V., Pörtner H.O., Roberts D.C.et al. (2019) Climate Change and Land: an IPCC special report on climate change, desertification, land degradation, sustainable land management, food security, and greenhouse gas fluxes in terrestrial ecosystems. IPCC, Available from: https://www.ipcc.ch/srccl/ [Google Scholar]
- 116.Hu H.-W., Chen D. and He J.-Z. (2015) Microbial regulation of terrestrial nitrous oxide formation: understanding the biological pathways for prediction of emission rates. FEMS Microbiol. Rev. 39, 729–749 10.1093/femsre/fuv021 [DOI] [PubMed] [Google Scholar]
- 117.Bakken L.R. and Frostegård Å. (2020) Emerging options for mitigating N2O emissions from food production by manipulating the soil microbiota. Curr. Opin. Environ. Sustain. 47, 89–94 10.1016/j.cosust.2020.08.010 [DOI] [Google Scholar]
- 118.Prosser J.I., Hink L., Gubry-Rangin C. and Nicol G.W. (2020) Nitrous oxide production by ammonia oxidizers: Physiological diversity, niche differentiation and potential mitigation strategies. Global Change Biol. 26, 103–118 10.1111/gcb.14877 [DOI] [PubMed] [Google Scholar]
- 119.Kits K.D., Jung M.-Y., Vierheilig J., Pjevac P., Sedlacek C.J., Liu S.et al. (2019) Low yield and abiotic origin of N2O formed by the complete nitrifier Nitrospira inopinata. Nat. Commun 10, 1836 10.1038/s41467-019-09790-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Frostegård Å., Vick S.H.W., Lim N.Y.N., Bakken L.R. and Shapleigh J.P. (2022) Linking meta-omics to the kinetics of denitrification intermediates reveals pH-dependent causes of N2O emissions and nitrite accumulation in soil. ISME J. 16, 26–37 10.1038/s41396-021-01045-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bakken L.R., Bergaust L., Liu B. and Frostegård Å. (2012) Regulation of denitrification at the cellular level: a clue to the understanding of N2O emissions from soils. Philos. Transact. R. Soc. B: Biol. Sci. 367, 1226–1234 10.1098/rstb.2011.0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bergaust L., Mao Y., Bakken L.R. and Frostegård Å. (2010) Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal pH on nitrogen oxide reductase in Paracoccus denitrificans. Appl. Environ. Microbiol. 76, 6387–6396 10.1128/AEM.00608-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lu X., Wang Z., Duan H., Wu Z., Hu S., Ye L.et al. (2023) Significant production of nitric oxide by aerobic nitrite reduction at acidic pH. Water Res. 230, 119542 10.1016/j.watres.2022.119542 [DOI] [PubMed] [Google Scholar]
- 124.Liu B., Frostegård Å. and Bakken L.R. (2014) Impaired reduction of N2O to N2 in acid soils is due to a posttranscriptional interference with the expression of nosZ. MBio 5, e01383–14, 10.1128/mBio.01383-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Van Den Heuvel R.N., Van Der Biezen E., Jetten M.S.M., Hefting M.M. and Kartal B. (2010) Denitrification at pH 4 by a soil-derived Rhodanobacter-dominated community. Environ. Microbiol. 12, 3264–3271 10.1111/j.1462-2920.2010.02301.x [DOI] [PubMed] [Google Scholar]
- 126.Van Den Heuvel R.N., Bakker S.E., Jetten M.S.M. and Hefting M.M. (2011) Decreased N2O reduction by low soil pH causes high N2O emissions in a riparian ecosystem. Geobiology 9, 294–300 10.1111/j.1472-4669.2011.00276.x [DOI] [PubMed] [Google Scholar]
- 127.Hink L., Gubry-Rangin C., Nicol G.W. and Prosser J.I. (2018) The consequences of niche and physiological differentiation of archaeal and bacterial ammonia oxidisers for nitrous oxide emissions. ISME J. 12, 1084–1093 10.1038/s41396-017-0025-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Verhamme D.T., Prosser J.I. and Nicol G.W. (2011) Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 5, 1067–1071 10.1038/ismej.2010.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Beeckman F., Motte H. and Beeckman T. (2018) Nitrification in agricultural soils: impact, actors and mitigation. Curr. Opin. Biotechnol. 50, 166–173 10.1016/j.copbio.2018.01.014 [DOI] [PubMed] [Google Scholar]
- 130.Gubry-Rangin C., Nicol G.W. and Prosser J.I. (2010) Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol. Ecol. 74, 566–574 10.1111/j.1574-6941.2010.00971.x [DOI] [PubMed] [Google Scholar]
- 131.Coskun D., Britto D.T., Shi W. and Kronzucker H.J. (2017) Nitrogen transformations in modern agriculture and the role of biological nitrification inhibition. Nat. Plants 3, 1–10 10.1038/nplants.2017.74 [DOI] [PubMed] [Google Scholar]
- 132.Papadopoulou E.S., Bachtsevani E., Papazlatani C.V., Rousidou C., Brouziotis A., Lampronikou E.et al. (2022) The Effects of Quinone Imine, a New Potent Nitrification Inhibitor, Dicyandiamide, and Nitrapyrin on Target and Off-Target Soil Microbiota. Microbiol. Spectrum 10, e02403–e02421 10.1128/spectrum.02403-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Subbarao G.V., Sahrawat K.L., Nakahara K., Ishikawa T., Kishii M., Rao I.M.et al. (2012) Chapter six - Biological nitrification inhibition—A novel strategy to regulate nitrification in agricultural systems. In Advances in Agronomyvol. 114, (Sparks D.L., ed.), pp. 249–302, Academic Press [Google Scholar]
- 134.Subbarao G.V., Ito O., Sahrawat K.L., Berry W.L., Nakahara K., Ishikawa T.et al. (2006) Scope and strategies for regulation of nitrification in agricultural systems— Challenges and opportunities. Crit. Rev. Plant Sci. 25, 303–335 10.1080/07352680600794232 [DOI] [Google Scholar]
- 135.Nardi P., Laanbroek H.J., Nicol G.W., Renella G., Cardinale M., Pietramellara G.et al. (2020) Biological nitrification inhibition in the rhizosphere: determining interactions and impact on microbially mediated processes and potential applications. FEMS Microbiol. Rev. 44, 874–908 10.1093/femsre/fuaa037 [DOI] [PubMed] [Google Scholar]
- 136.Wells G.F., Park H.-D., Yeung C.-H., Eggleston B., Francis C.A. and Criddle C.S. (2009) Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ. Microbiol. 11, 2310–2328 10.1111/j.1462-2920.2009.01958.x [DOI] [PubMed] [Google Scholar]
- 137.Wu L., Ning D., Zhang B., Li Y., Zhang P., Shan X.et al. (2019) Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiol 4, 1183–1195 10.1038/s41564-019-0426-5 [DOI] [PubMed] [Google Scholar]
- 138.Saunders A.M., Albertsen M., Vollertsen J. and Nielsen P.H. (2016) The activated sludge ecosystem contains a core community of abundant organisms. ISME J. 10, 11–20 10.1038/ismej.2015.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jetten M.S.M., Horn S.J. and van Loosdrecht M.C.M. (1997) Towards a more sustainable municipal wastewater treatment system. Water Sci. Technol. 35, 171–180 10.2166/wst.1997.0341 [DOI] [Google Scholar]
- 140.Peng Y. and Zhu G. (2006) Biological nitrogen removal with nitrification and denitrification via nitrite pathway. Appl. Microbiol. Biotechnol. 73, 15–26 10.1007/s00253-006-0534-z [DOI] [PubMed] [Google Scholar]
- 141.Wang Z., Zheng M., Duan H., Hu S. and Yuan Z. (2022) Re-configuring mainstream anammox. Chem. Eng. J. 445, 136817 10.1016/j.cej.2022.136817 [DOI] [Google Scholar]
- 142.Wang Z., Ni G., Xia J., Song Y., Hu S., Yuan Z.et al. (2021) Bioleaching of toxic metals from anaerobically digested sludge without external chemical addition. Water Res. 200, 117211 10.1016/j.watres.2021.117211 [DOI] [PubMed] [Google Scholar]
- 143.Wang Z., Zheng M., Duan H., Ni G., Yu W., Liu Y.et al. (2021) Acidic aerobic digestion of anaerobically-digested sludge enabled by a novel ammonia-oxidizing bacterium. Water Res. 194, 116962 10.1016/j.watres.2021.116962 [DOI] [PubMed] [Google Scholar]
- 144.Zheng M., Li H., Duan H., Liu T., Wang Z., Zhao J.et al. (2023) One-year stable pilot-scale operation demonstrates high flexibility of mainstream anammox application. Water Res. X 19, 100166 10.1016/j.wroa.2023.100166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang Z., Liu T., Duan H., Song Y., Lu X., Hu S.et al. (2021) Post-treatment options for anaerobically digested sludge: Current status and future prospect. Water Res. 205, 117665 10.1016/j.watres.2021.117665 [DOI] [PubMed] [Google Scholar]
- 146.Kartal B., Kuenen J.G. and van Loosdrecht M.C.M. (2010) Sewage treatment with anammox. Science 328, 702–703 10.1126/science.1185941 [DOI] [PubMed] [Google Scholar]
- 147.McCarty P.L., Bae J. and Kim J. (2011) Domestic wastewater treatment as a net energy producer-can this be achieved? Environ. Sci. Technol. 45, 7100–7106 10.1021/es2014264 [DOI] [PubMed] [Google Scholar]