Abstract
BACKGROUND & AIMS:
Pancreatic cancer has the highest prevalence of cancer-associated cachexia among all cancers. ZIP4 promotes pancreatic cancer progression by regulating oncogenic miR-373, and perturbation of circular RNAs (circRNAs) is associated with cancer aggressiveness. This study aimed to identify circRNAs involved in ZIP4/miR-373–driven cancer growth and cachexia and decipher the underlying mechanism.
METHODS:
Differentially expressed circRNAs and potential targets of microRNA were identified through in silico analysis. The RNA interactions were determined by means of biotinylated microRNA pulldown, RNA immunoprecipitation, and luciferase reporter assays. The function of circRNA in ZIP4–miR-373 signaling axis were examined in human pancreatic cancer cells, 3-dimensional spheroids and organoids, mouse models, and clinical specimens. Mouse skeletal muscles were analyzed by means of histology.
RESULTS:
We identified circANAPC7 as a sponge for miR-373, which inhibited tumor growth and muscle wasting in vitro and in vivo. Mechanistic studies showed that PHLPP2 is a downstream target of ZIP4/miR-373. CircANAPC7 functions through PHLPP2-mediated dephosphorylation of AKT, thus suppressing cancer cell proliferation by down-regulating cyclin D1 and inhibiting muscle wasting via decreasing the secretion of transforming growth factor–β through STAT5. We further demonstrated that PHLPP2 induced dephosphorylation of CREB, a zinc-dependent transcription factor activated by ZIP4, thereby forming a CREB–miR-373–PHLPP2 feed-forward loop to regulate tumor progression and cancer cachexia.
CONCLUSION:
This study identified circANAPC7 as a novel tumor suppressor, which functions through the CREB–miR-373–PHLPP2 axis, leading to AKT dephosphorylation, and cyclin D1 and transforming growth factor–β down-regulation to suppress tumor growth and muscle wasting in pancreatic cancer.
Keywords: CircRNA, MicroRNA, Proliferation, Cachexia
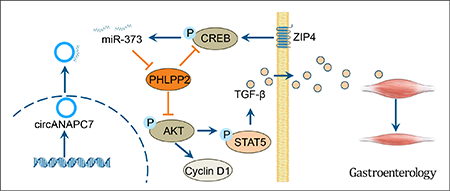
Graphical Abstract
Pancreatic cancer is predicted to become the second leading cause of cancer-associated death in the United States by 2030, with a 5-year survival rate of no more than 10%.1 Surgery plus chemotherapy provides the best chance of cure for patients with pancreatic cancer. Unfortunately, >80% of patients are diagnosed at advanced stages and are not eligible for surgery.2 In addition, approximately 70% of patients with pancreatic cancer develop cancer cachexia, a systematic metabolic dysfunction characterized by body weight loss with ongoing muscle wasting (with or without adipose loss), which cannot be reversed by conventional nutritional support.3,4 Muscle wasting is the major hallmark of cancer cachexia caused by substantially increased muscle protein breakdown.5 Cancer cachexia reduces quality of life, decreases tolerance to chemotherapy, and ultimately leads to tumor progression in patients with pancreatic cancer.3 However, there is no effective treatment for cancer-associated cachexia and the underlying mechanism remains mostly unclear. Therefore, there is an urgent need to further explore the biology and molecular mechanism of pancreatic cancer progression and develop efficient therapeutic targets.
Zinc is an essential metal ion and nutrient that plays important physiological and pathologic roles.6 Abnormal zinc homeostasis is associated with increased cell death and resistance to chemotherapy in pancreatic cancer.7 Therefore, illustrating the mechanism of aberrant expression of zinc transporter and its regulation in pancreatic cancer may lead to more effective therapeutics.8 We recently found that ZIP4 plays critical roles in cell proliferation, metastasis, drug resistance, and cancer-associated cachexia in pancreatic cancer.9–14 Therefore, it is crucial to further explore the signaling network on ZIP4-induced cancer progression and cachexia to develop novel therapeutic strategies for pancreatic cancer.
Circular RNA (circRNA) is a closed-loop RNA with a junction between the 3′ and 5′ ends from reverse splicing of precursor messenger RNA (mRNA).15 MicroRNA (miRNA) is a class of small noncoding RNA involved in multiple biological processes by inhibiting the translation of mRNA.16 The miRNA sponge is an RNA transcript with complementary binding sites to this specific miRNA, thus inhibiting the function of the miRNA.17 CircRNAs sponge miRNAs more effectively, as they are more stable and have more binding sites than linear RNAs.18 CircRNAs are involved in multiple functions of pancreatic cancer, including tumor growth, differentiation, apoptosis, invasion, metastasis, lymph angiogenesis, and chemotherapy resistance.19–22 However, the mechanism of circRNA in pancreatic cancer remains largely elusive, especially in the context of ZIP4-mediated tumor progression. Previously, we showed that the expression of an oncogenic miRNA, miR-373, is regulated by ZIP4 and is essential for ZIP4-induced pancreatic cancer progression. However, whether circRNAs could act as miR-373 sponges, and their impact on ZIP4 signaling, is unknown.
In the present study, we found that circRNA ANAPC7 suppressed pancreatic cancer growth and ameliorated cachexia by sponging miR-373. We identified PHLPP2, a protein phosphatase involved in the regulation of AKT signaling, as a novel miR-373 target gene in pancreatic cancer. Furthermore, we found that PHLPP2 decreased CREB phosphorylation, a zinc-dependent transcription factor regulated by ZIP4, which promotes miR-373 expression by transcriptional regulation. We identified an uncharacterized CREB–miR-373–PHLPP2 feed-forward loop of the ZIP4-mediated signaling axis in pancreatic cancer. Moreover, we found circANAPC7 ameliorated cachexia by reversing ZIP4-induced muscle wasting through down-regulating transforming growth factor (TGF)-β expression and secretion via STAT5. Our findings suggest novel insights on circANAPC7 in ZIP4-mediated pancreatic cancer progression and cachexia, thus providing a potential therapeutic strategy for cancer treatment and clinical management of cancer cachexia.
Materials and Methods
Cell Lines and Clinical Specimens
Human pancreatic cancer cell lines AsPC-1, MIA PaCa-2, BxPC-3, Panc-1, and CFPAC-1 were purchased from American Type Culture Collection. The human pancreatic duct epithelial (HPDE) cell line was a gift from Dr Ming-Sound Tsao (Ontario Cancer Institute).23 These cells were cultured as described previously.9 MIA-V/ZIP4, MIA-ZIP4-anti-C/anti-373, AsPC-shV/shZIP4, AsPC-shZIP4-PreC/Pre373 stable cell lines were constructed in our laboratory previously.10 MIA-V/ZIP4, MIA-ZIP4-anti-C/anti-373 cells were cultured in complete media supplemented with 0.5 μg/mL puromycin (#A1113803; Gibco). AsPC-shV/shZIP4, AsPC-shZIP4-PreC/Pre373 cells were cultured in complete media supplemented with 1 μg/mL puromycin. Murine C2C12 myoblasts (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum. All cells were cultured at 37°C under 5% CO2. Banked de-identified human pancreatic cancer tissue specimens were obtained from the University of Oklahoma Health Sciences Center according to an approved Institutional Review Boards human protocol. Written consent was obtained from all subjects.
3-Dimensional Spheroid and Organoid Culture
The 3-dimensional (3D) spheroid and organoid cultures were performed as described previously.13,14 Briefly, pancreatic cancer cells were resuspended in medium containing 0.24% methylcellulose (#M0512; Sigma-Aldrich). Cells were seeded on the inner lid of a 10-cm Petri dish with 20 μL per drop. The lid was put on the Petri dish containing 10 mL phosphate buffer solution. The suspended droplets were incubated for 5–7 days in 5% CO2 at 37°C to form 3D cell spheroids. Matrigel-based 3D embedded and suspended organoids were generated based on a similar method, except that the cell suspension was mixed with Matrigel (#354234; Corning). The spheroids were embedded with HistoGel and paraffin, then sectioned for H&E staining.
Biotinylated MicroRNA Pulldown Assay
The miRNA pulldown assay was performed as described by Lal et al.24 Briefly, the 3′ end biotinylated miRCURY LNA Premium miRNA Mimic or control RNA (#39178/3BIO; Qiagen) were transfected into cells at a final concentration of 30 nM for 24 hours. The cells were washed with phosphate-buffered saline and incubated in lysis buffer. The biotin-coupled RNA complex was pulled down by incubating the cell lysates with Dynabeads MyOne Streptavidin T1 (#65601; Invitrogen). The beads were blocked with RNase-free bovine serum albumin (#AM2616; Invitrogen) and yeast transfer RNA (#15401029, Invitrogen) on a rotator at 4°C for 2 hours to prevent nonspecific binding of RNA and protein complexes. The blocked beads were incubated with cell lysates at 4°C overnight, washed 3 times with ice-cold lysis buffer. The RNA complexes on the beads were eluted and extracted by TRIzol reagent (#15596026; Invitrogen). The abundance of circANAPC7 and miR-373 of RNA complexes was evaluated by reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) analysis.
RNA Immunoprecipitation
miRNA mimics were transfected into cells for 24 hours after transfection, and AGO2 antibody (#SAB4200085; MilliporeSigma) or negative control IgG antibody (#10500C; Invitrogen) was used to perform AGO2 immunoprecipitation. Cells were lysed in cOmplete Lysis-M (#04719956001; Roche) supplemented with protease inhibitor (#04693116001; Roche) and RNase inhibitor (#AM2694; Invitrogen) for 10 minutes. The antibodies were incubated with Dynabeads magnetic beads of a Dynabeads Protein G Immunoprecipitation Kit (#10007D; Invitrogen) at room temperature for 30 minutes. The lysate was mixed with antibody-beads complexes and incubated under rotation overnight at 4°C. After treating with proteinase K (#03115836001; Roche), RNA complexes were extracted using TRIzol reagent. The abundance of RNAs was evaluated by RT-qPCR analysis.
Luciferase Reporter Assay
Luciferase reporter assay was performed using the Dual-Luciferase Reporter Assay System (#E1960; Promega) following the manufacturer’s protocol. Briefly, cells were seeded in a 24-well plate and co-transfected with the plasmid of 500 ng and miRNA mimics of 10 pmol per well using Lipofectamine 3000 Reagent (#L3000015; Invitrogen). At 48 hours after transfection, the cells were lysed and firefly luciferase activity and Renilla luciferase activity were measured using a Microplate Reader (Synergy H1; BioTek).
RNA Location Assay
The nuclear and cytoplasmic RNAs were extracted using the Nuclear and Cytoplasmic Extraction Kit (PARIS, AM1921; Invitrogen). One picogram of DNA spike-in molecules was added to each sample for RT-qPCR normalization. DNA spike-in was produced from the multiple cloning sites in pcDNA3.1(+) CircRNA Mini Vector (#60648; Addgene).
Transcription Block Assay
Transcription was blocked by adding 2 μg/mL actinomycin D (A1410; Sigma-Aldrich) or dimethyl sulfoxide control to the cell culture medium and cultured for 0, 4, 8, 12, or 24 hours. Then the cells were harvested and total RNA was extracted. The stability of circANAPC7 and ANAPC7 mRNA was analyzed by means of RT-qPCR.
Ribonuclease R Treatment
The total RNA was incubated with or without 2 U/μg ribonuclease R (#RNR07250; Epicentre Technologies) for 15 minutes at 37°C and purified using the RNeasy MinElute Cleanup Kit (#74204; Qiagen), then analyzed by RT-qPCR.
Orthotopic Xenograft Mouse Model
MIA-ZIP4-EV/circANAPC7, AsPC-shZIP4-Pre373-EV/circANAPC7 stable cell lines were used to establish the pancreatic cancer orthotopic xenograft mouse model as described previously.10 Briefly, 3 × 106 cells in 50 μL Dulbecco’s modified Eagle medium or RPMI medium were injected into the tail of the pancreas of 5-week athymic nude mice. The mice study was performed under an animal protocol approved by the Animal Welfare Committee at University of Oklahoma Health Sciences Center. Four weeks post injection, mice were euthanized and tissues were collected.
Statistical Analysis
Statistical analysis was performed with Prism 8 (GraphPad Software). Comparisons of 2 groups were conducted using a 2-tailed Student t test. One-way analysis of variance was used for multiple conditions compared for 1 variable. Two-way analysis of variance was performed for multiple sets of multivariate comparisons. A P value < .05 was considered statistically significant.
Results
CircANAPC7 Acts as a miR-373 Sponge in Pancreatic Cancer Cells
To identify the potential circRNA that acts as a miR-373 sponge, we analyzed differentially expressed circRNAs (DECs) from the circRNA microarray dataset of pancreatic cancer and screened circRNAs with miR-373 binding sites from circBank online database.25,26 We identified 223 up-regulated DECs and 214 down-regulated DECs (Supplementary Table 1) in pancreatic cancer, and the top 100 DECs are shown in Figure 1A. Among the 214 down-regulated DECs, we focused on 5 circRNAs containing miR-373 binding sites for further investigation (Supplementary Figure 1A). To validate which circRNA interacts with miR-373, we performed miRNA pulldown assay and found only hsa_circ_0005785 (also known as circANAPC7) was enriched in the miR-373–captured fraction compared with the negative control, indicating miR-373 could interact with circANAPC7 in pancreatic cancer cells (Figure 1B, Supplementary Figure 1B). Next, we confirmed the circular structure of circANAPC7, derived from exon 3 to exon 9 of a protein-coding gene ANAPC7, in pancreatic cancer cell lines by Sanger sequencing and PCR analysis (Figure 1C and Supplementary Figure 1C). We found that circANAPC7 was down-regulated in pancreatic cancer cells compared with HPDE cells (Supplementary Figure 1D). Consistent with the circRNA microarray results, circANAPC7 level was significantly lower in human pancreatic cancer tissues than tumor-adjacent tissues (Supplementary Figure 1E). The analysis of nuclear and cytoplasmic circANAPC7 revealed that circANAPC7 is preferentially localized to the cytoplasm (Figure 1D). To analyze the stability of circANAPC7 and ANAPC7 mRNAs, we treated pancreatic cancer cells with actinomycin D, an inhibitor of transcription, and found that circANAPC7 transcript is more stable than linear ANAPC7 mRNA transcript (Supplementary Figure 1F). Furthermore, we found circANAPC7 was resistant to ribonuclease R exonuclease digestion, which further supported the circular structure of this RNA transcript (Supplementary Figure 1G). We next determined whether circANAPC7 serves as a binding platform for miR-373 and AGO2, the essential component in the process of miRNA repression of specific target RNA. We performed RNA immunoprecipitation of AGO2 in pancreatic cancer cells transfected with miR-373 mimics and found that endogenous circANAPC7 was enriched in the AGO2 group (Figure 1E). We further constructed luciferase reporter vectors by inserting circANAPC7 wild-type or mutant fragment right behind the reporter gene (Figure 1F) and found a significantly decreased luciferase activity in the circANAPC7 wild-type group compared with the circANAPC7 mutant group in miR-373 overexpressed cells (Figure 1G). These results suggest that circANAPC7 binds to miR-373 and may serve as a miR-373 sponge to suppress its function in pancreatic cancer.
Figure 1.
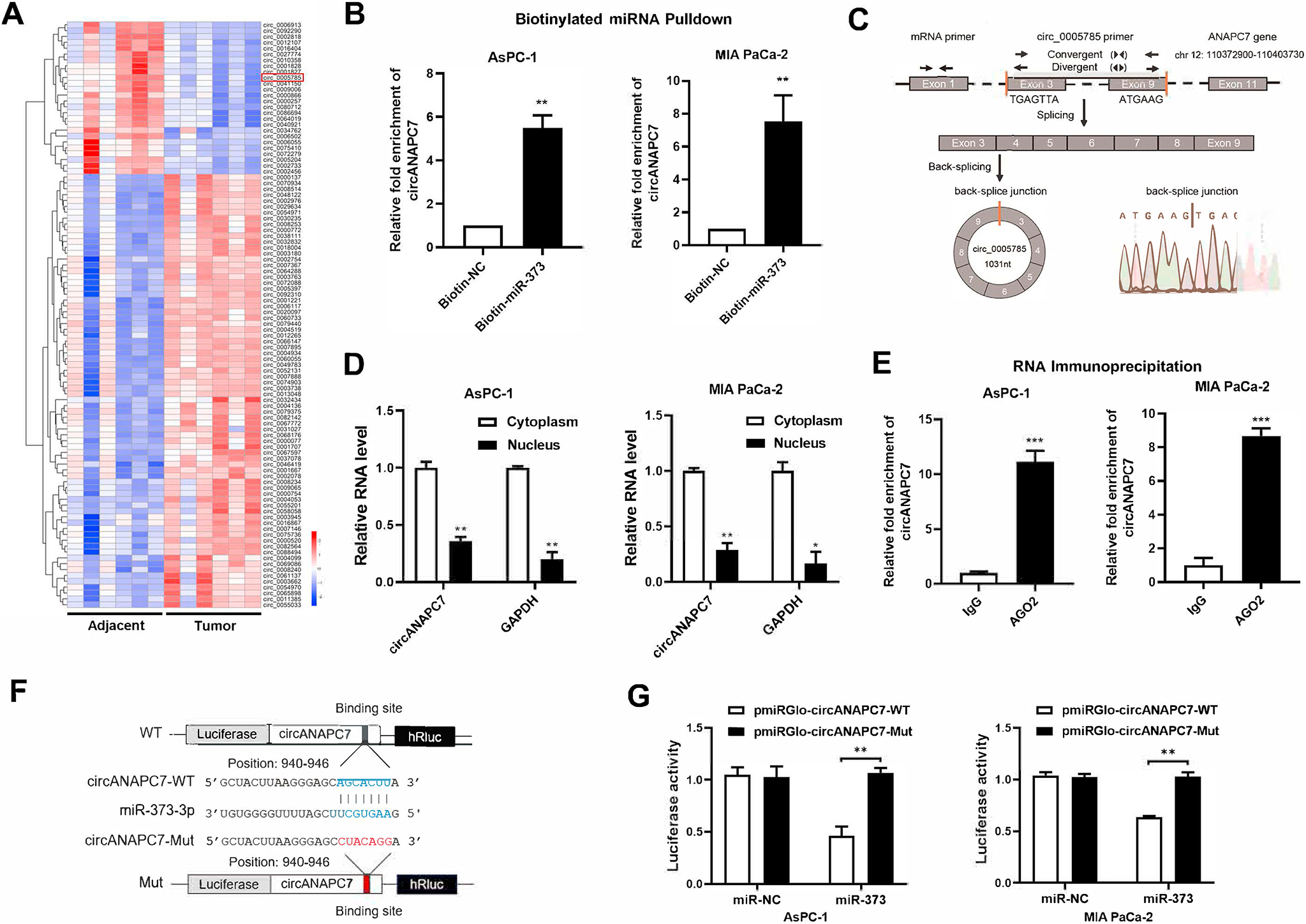
CircANAPC7 interacts with miR-373 in pancreatic cancer cells. (A) Heatmap of the top 100 differentially expressed circRNAs between 6 pairs of pancreatic cancer and adjacent tissues (GSE69362). (B) CircANAPC7 levels detected by RT-qPCR analysis of miRNA pulldown products. (C) CircANAPC7 structure and the validation strategy. (D) CircANAPC7 and GAPDH mRNA level in pancreatic cancer cells. (E) RNA immunoprecipitation assay was performed with cell lysate of AsPC-1 and MIA PaCa-2 cells transfected with miR-373 mimics using either anti-AGO2 or IgG as the immunoprecipitating antibody. (F) Schematic diagram of luciferase reporter vectors with circANAPC7 wild-type (WT) (blue) or mutated (Mut) (red) putative miR-373 binding sites. (G) Luciferase activity of circANAPC7 reporter vectors (WT and Mut) co-transfected with negative control (NC) or miR-373 mimics in AsPC-1 and MIA PaCa-2 cells. Firefly luciferase activity was normalized to Renilla luciferase activity. Error bars represent SDs. *P < .05; **P < .01; ***P < .001.
CircANAPC7 Inhibits Pancreatic Cancer Cell Proliferation in 2-Dimensional and 3-Dimensional Models
To further investigate the function of circANAPC7, we overexpressed circANAPC7 in AsPC-1 and MIA PaCa-2 cells (Supplementary Figure 2A). Overexpression of circANAPC7 significantly suppressed pancreatic cancer cell proliferation and colony formation (Figure 2A and B). The 5-ethynyl-2′-deoxyuridine incorporation assay revealed that circANAPC7 overexpression impaired DNA synthesis in human pancreatic cancer cells (Figure 2C). We also observed a similar inhibitory effect of circANAPC7 on cell growth in 3D spheroid and organoid models. CircANAPC7 showed no significant impact on the aggregation of spheroids, however, circANAPC7 overexpression inhibited spheroid growth compared with the control group (Supplementary Figure 2B–D and Figure 2D). Consistent with 2D culture and 3D spheroids, circANAPC7 overexpression suppressed the growth of organoids in both Matrigel-suspended and Matrigel-embedded culture conditions (Figure 2E, Supplementary Figure 2E). These results demonstrated that circANAPC7 inhibits pancreatic cancer cell growth as a tumor suppressor.
Figure 2.
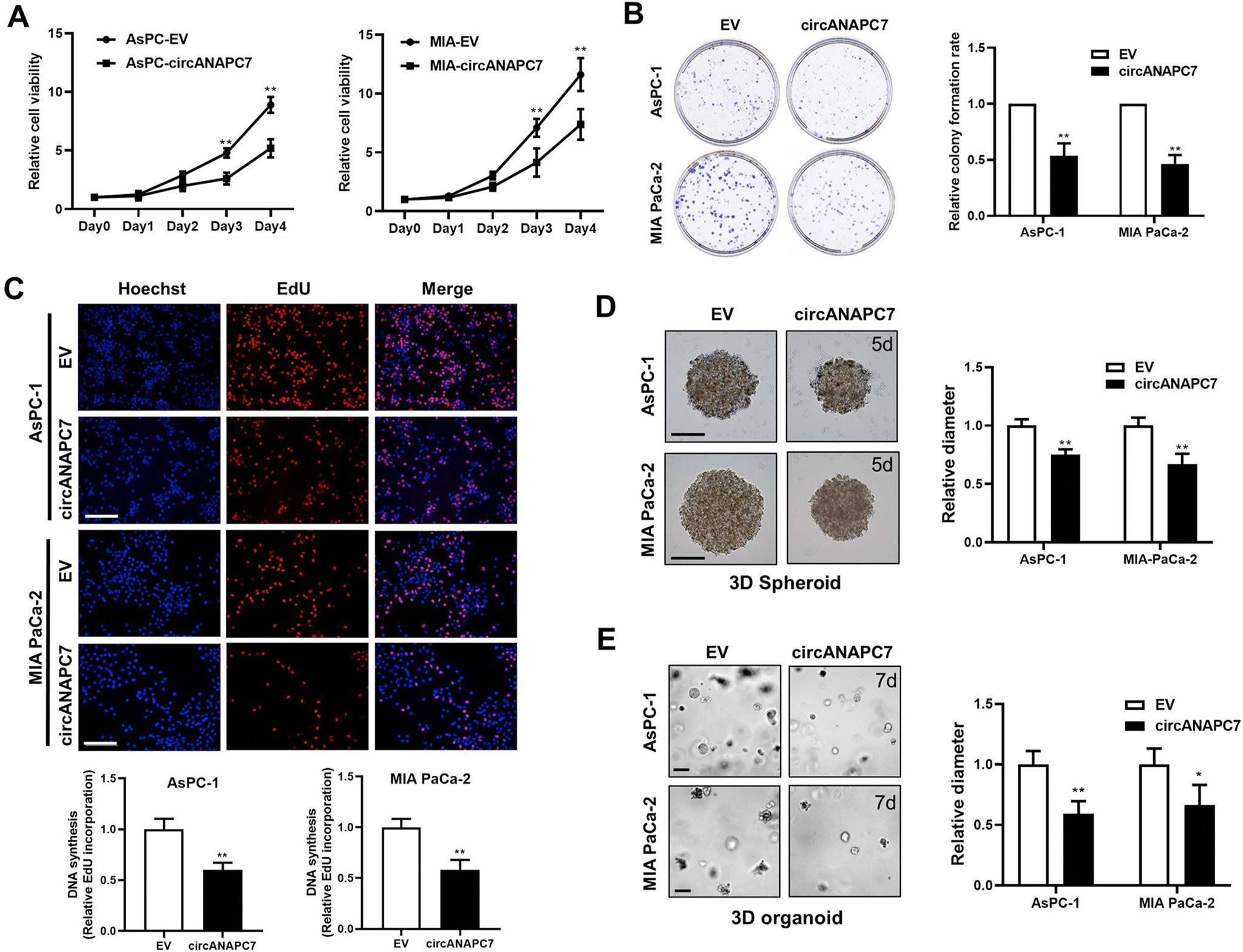
CircANAPC7 inhibits pancreatic cancer cell growth in 2D and 3D cultures. (A) Cell viability of AsPC-EV/circANAPC7, MIA-EV/circANAPC7 cells were assessed using the MTT reagent at the indicated days. (B) Colony-formation assay (left panel). The analysis of relative colony-formation rate (right panel). (C) DNA synthesis assessed using a 5-ethynyl-2′-deoxyuridine (EdU) assay in indicated cells (top panel). Scale bar: 100 μm. Quantitative data of EdU assay (bottom panel). (D) Representative images of methylcellulose-based 3D cancer cell spheroids established from AsPC-EV/circANAPC7, MIA-EV/circANAPC7 cells cultured for 5 days. Scale bar: 200 μm. (E) Representative images of Matrigel-suspended 3D organoids established from AsPC-EV/circANAPC7, MIA-EV/circANAPC7 cells cultured for 7 days. Scale bar: 50 μm. *P < .05; **P < .01.
PHLPP2 Is a Downstream Target of miR-373
To identify the downstream targets responsible for mediating the oncogenic function of miR-373, we analyzed the human tumor suppressor genes with miR-373 binding sites and found 18 genes occurred simultaneously within the 3 categories (Figure 3A and Supplementary Figure 3A). Among the 18 potential downstream targets of miR-373, we selected 3 candidates (SASH1, PHLPP2, and PAFAH1B1) for further validation because they are negatively correlated with ZIP4 in human pancreatic cancer tissues (Supplementary Figure 3B–D). Then we evaluated whether circANAPC7 regulates miR-373 target gene expression through sponging miR-373. The RT-qPCR analysis demonstrated that overexpression of circANAPC7 up-regulated the mRNA level of PHLPP2 but not SASH1 or PAFAH1B1 (Figure 3B and C). CircANAPC7 overexpression also increased the protein level of PHLPP2 in pancreatic cancer cells (Figure 3D, Supplementary Figure 3E). miR-373 blocking up-regulated PHLPP2, while miR-373 overexpression down-regulated PHLPP2 (Figure 3E and Supplementary Figure 3F–H). These results indicated that circANAPC7 serves as a miR-373 sponge to regulate PHLPP2 expression in pancreatic cancer cells. To examine whether miR-373 binds to the 3′UTR of PHLPP2, we performed a biotin-labeled miR-373 pulldown assay and found an 8-fold enrichment of the 3′UTR of PHLPP2 (Figure 3F). We also used the luciferase reporter system to detect whether miR-373 regulated the expression of PHLPP2 by binding to its 3′UTR. We cloned the PHLPP2–3′UTR with wild-type or mutant miR-373–binding site right behind the luciferase reporter vector for the luciferase reporter assay (Figure 3G) and observed that miR-373 expression could reduce luciferase activity of the wild-type PHLPP2–3′UTR reporter vector, but not the mutant reporter vector (Figure 3H). Collectively, these data suggested that PHLPP2 is a downstream target of miR-373, and circANAPC7 increased the PHLPP2 level by sponging miR-373.
Figure 3.
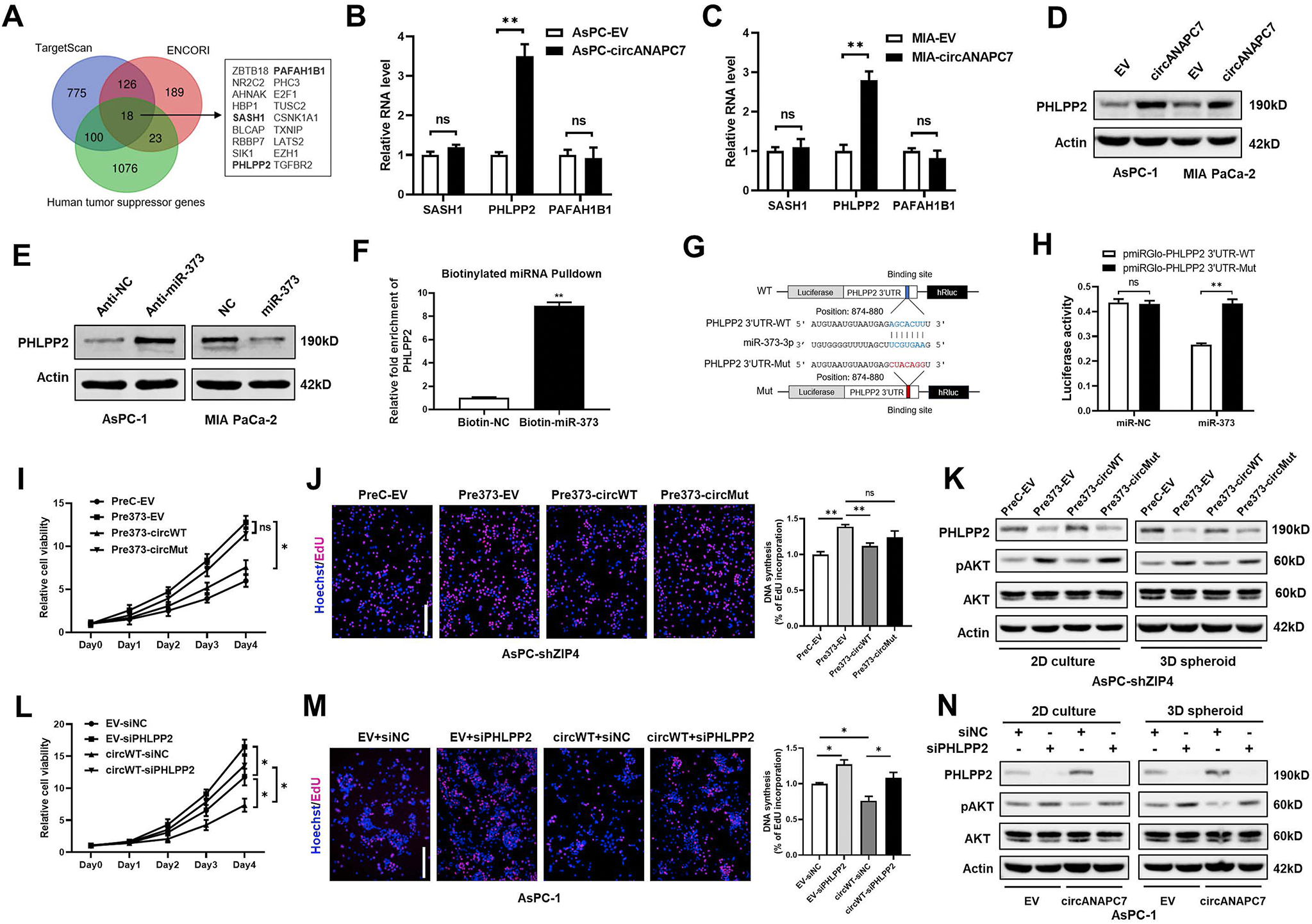
CircANAPC7 inhibits cell proliferation by serving as a miR-373 sponge to increase PHLPP2 level in pancreatic cancer. (A) Schematic diagram showing the identification of tumor suppressor genes that contain predicted binding sites of miR-373 in indicated databases. (B) mRNA levels of SASH1, PHLPP2, and PAFAH1B1 in AsPC-EV/circANAPC7 cells. (C) mRNA levels of SASH1, PHLPP2, and PAFAH1B1 in MIA-EV/circANAPC7 cells. (D) PHLPP2 protein levels in AsPC-EV/circANAPC7, MIA-EV/circANAPC7 cells. (E) PHLPP2 protein levels in AsPC-1 cells transfected with anti-NC/miR-373 oligonucleotides (left panel). PHLPP2 protein levels in MIA PaCa-2 cells transfected with miR-NC/miR-373 mimics (right panel). (F) RT-qPCR analysis of RNA pulldown products detecting PHLPP2 mRNA level in the streptavidin captured fractions from the MIA PaCa-2 cell lysates after transfection with biotinylated miR-373 or NC probes. (G) Schematic diagram of PHLPP2–3′UTR reporter vectors. (H) Luciferase activity of PHLPP2–3′UTR reporters (WT and Mut) co-transfected with NC or miR-373 mimics in MIA PaCa-2 cells. (I) Cell viability was assessed by MTT. (J) DNA synthesis was assessed by 5-ethynyl-2′-deoxyuridine (EdU) assay. (K) Protein levels of PHLPP2, pAKT, and total AKT. (L) Cell viability was assessed by MTT. (M) DNA synthesis was assessed by EdU assay. (N) Protein level of PHLPP2, pAKT, and total AKT in AsPC-EV/circANAPC7 stable cells transfected with small interfering RNAs of PHLPP2 or control under 2D culture and 3D spheroid culture. circWT, circANAPC7-WT; circMut, circANAPC7-Mut; NC, negative control; ns, not significant; *P < .05; **P < .01.
We further examined whether circANAPC7 suppresses pancreatic cancer cell proliferation by up-regulating PHLPP2. Cell viability assays showed that miR-373–induced cell proliferation was partially reversed by wild-type, but not mutant, circANAPC7 (Figure 3I). In addition, 5-ethynyl-2′-deoxyuridine incorporation assays also revealed that the wild-type, but not mutant, circANAPC7 could reverse miR-373–mediated increase of DNA synthesis (Figure 3J). PHLPP2 was recently identified as an important regulator of AKT, functioning as an AKT-Ser473 phosphatase and resulting in AKT inactivation.27 We also observed phosphorylated AKT level was increased after PHLPP2 silencing, but decreased by PHLPP2 overexpression in pancreatic cancer cells (Supplementary Figure 4A–F). The overexpression of miR-373 significantly decreased the PHLPP2 level and increased phosphorylated AKT, and these effects were reversed by wild-type circANAPC7 overexpression under both 2D culture and 3D spheroid culture conditions (Figure 3K). Furthermore, PHLPP2 silencing attenuated the inhibition of cancer cell growth by circANAPC7 (Figure 3L and M). CircANAPC7–mediated inhibition of AKT phosphorylation was also attenuated by PHLPP2 silencing (Figure 3N). These results demonstrated that circANAPC7 suppressed pancreatic cancer cell proliferation through miR-373/PHLPP2.
ZIP4 Inhibits PHLPP2 Expression by Activating miR-373
The expression of miR-373 is regulated by ZIP4 through a zinc-dependent transcription factor CREB, as we demonstrated previously.10 The survival analysis of miR-373 target gene PHLPP2 in The Cancer Genome Atlas database showed that PHLPP2 is a prognostic marker in pancreatic cancer. Patients with a lower level of PHLPP2 had a worse prognosis in pancreatic cancer (5-year survival, low vs high: 15% vs 34%, P = .0002, Figure 4A). Thus, we further investigated whether ZIP4 regulates expression of PHLPP2. We observed that ZIP4 knockout increased both mRNA and protein levels of PHLPP2, while ZIP4 overexpression decreased PHLPP2 levels (Supplementary Figure 5A–C and Figure 4B). Furthermore, the protein level of PHLPP2 was reduced in pancreatic cancer stable cells with ZIP4 knockdown and miR-373 overexpression, while PHLPP2 level increased in miR-373 blocked pancreatic cancer cells after ZIP4 overexpression (Figure 4C and D and Supplementary Figure 5D and E). Accordingly, the PHLPP2 level was decreased by miR-373 mimics and increased by anti–miR-373 oligonucleotides transfection with or without ZIP4 present in MIA-ZIP4 and AsPC-koZIP4 cells (Figure 4E and F and Supplementary Figure 5F and G). Next, we confirmed the regulatory role of the ZIP4–miR-373–PHLLP2 signaling axis in pancreatic tumor growth using an orthotopic xenograft mouse model. Immunohistochemistry (IHC) staining showed that tumor tissues of the AsPC-shZIP4-Pre373 group had higher levels of Ki67 but lower levels of PHLPP2 compared with the AsPC-shZIP4-PreC group (Figure 4G). Compared with the MIA-ZIP4-anti-C group, tumor tissues of the MIA-ZIP4-anti-373 group had a lower level of Ki67 and a higher level of PHLPP2 (Figure 4H). To further analyze the downstream of the ZIP4-miR-373-PHLLP2 signaling axis, we performed Kyoto Encyclopedia of Genes and Genomes pathway analysis on differentially expressed genes (|fold change| ≥ 2; P < .05) in The Cancer Genome Atlas pancreatic cancer tissues with ZIP4 high vs ZIP4 low expression. These data suggested that AKT is an important downstream target of this ZIP4 signaling axis in pancreatic cancer as well (Supplementary Figure 5H). Consistently, the level of phosphorylated AKT, a downstream target of PHLPP2, was reduced in the ZIP4 knockout pancreatic cancer cells (Figure 4I, Supplementary Figure 5I). Expression of PHLPP2 was also confirmed to be negatively correlated with ZIP4 in human pancreatic cancer tissues by means of IHC staining (Figure 4J). These results demonstrated that ZIP4 regulates expression of PHLPP2 through miR-373 in pancreatic cancer.
Figure 4.
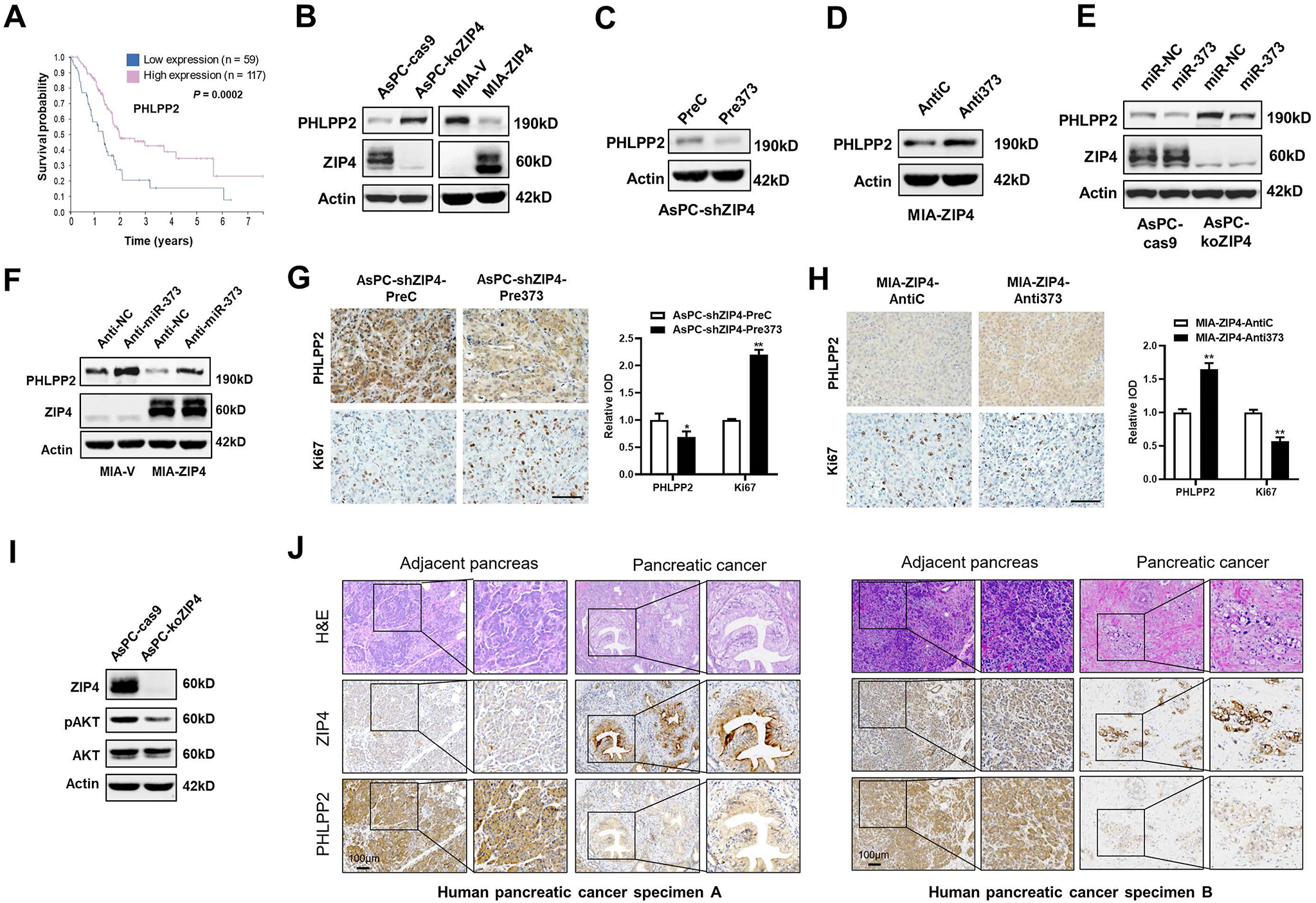
ZIP4 inhibits PHLPP2 expression by increasing the miR-373 level in pancreatic cancer. (A) Kaplan-Meier curve for overall survival of patients (n = 176) with pancreatic cancer with low vs high expression of PHLPP2 from The Cancer Genome Atlas database. (B) Protein levels of PHLPP2 and ZIP4 in AsPC-cas9/koZIP4, MIA-V/ZIP4 cells. (C) Protein levels of PHLPP2 in AsPC-shZIP4-PreC/Pre373 cells. (D) Protein levels of PHLPP2 in MIA-ZIP4-anti-C/anti-373 cells. (E) Protein levels of PHLPP2 in AsPC-cas9/koZIP4 cells transfected with either miR-NC or miR-373. (F) Protein levels of PHLPP2 and ZIP4 in MIA-V/ZIP4 cells transfected with either anti-NC or anti-373. (G, H) PHLPP2 and Ki67 expression in orthotopic models implanted with AsPC-shZIP4-PreC/Pre373 cells and MIA-ZIP4-anti-C/anti-373 cells. Scale bar: 50 μm. (I) Protein levels of ZIP4, pAKT, and total AKT in AsPC-cas9/koZIP4 cells. (J) Representative H&E and IHC staining images of ZIP4 and PHLPP2 in human pancreatic cancer and cancer-adjacent pancreas tissues. *P < .05; **P < .01.
CircANAPC7 Is Involved in ZIP4-Mediated CREB–miR-373–PHLPP2 Feed-Forward Loop and Inhibits Cell Proliferation in Pancreatic Cancer
To identify the role of circANAPC7 in the ZIP4-mediated malignant phenotype in pancreatic cancer, we performed rescue experiments. Cell viability and colony-formation assays showed that the increased cell proliferation and colony formation mediated by ZIP4 was reversed by wild-type circANAPC7 overexpression, but not the mutant circANAPC7 (Figure 5A and B). DNA synthesis assay revealed that wild-type circANAPC7 overexpression, but not the mutant circANAPC7, reversed ZIP4-mediated cell proliferation (Supplementary Figure 6A). Subsequently, we used MIA-ZIP4 and AsPC-shZIP4-Pre373 cells with stable overexpression of circANAPC7 to investigate the impact of circANAPC7 in vivo. CircANAPC7 overexpression in both AsPC-shZIP4-Pre373 and MIA-ZIP4 cells significantly inhibited tumor growth in an orthotopic xenograft mouse model (Figure 5C and D). Because cyclin D1 plays a central role in regulating cell proliferation, and our previous study showed that cyclin D1 is an important downstream target of ZIP4 in pancreatic cancer,28 therefore, we investigated whether circANAPC7 regulates cell proliferation through cyclin D1 in vitro and in vivo. CircANAPC7 decreased mRNA and protein levels of cyclin D1 in pancreatic cancer cells and mouse xenografts (Supplementary Figure 6B and Figure 5E and F). ZIP4 knockdown down-regulated cyclin D1, while ZIP4 overexpression up-regulated cyclin D1 (Supplementary Figure 6C). Moreover, miR-373 blocking could also down-regulate cyclin D1 in vitro (Supplementary Figure 6D). These results indicate that circANAPC7 is involved in ZIP4/miR-373–mediated cell proliferation through cyclin D1.
Figure 5.
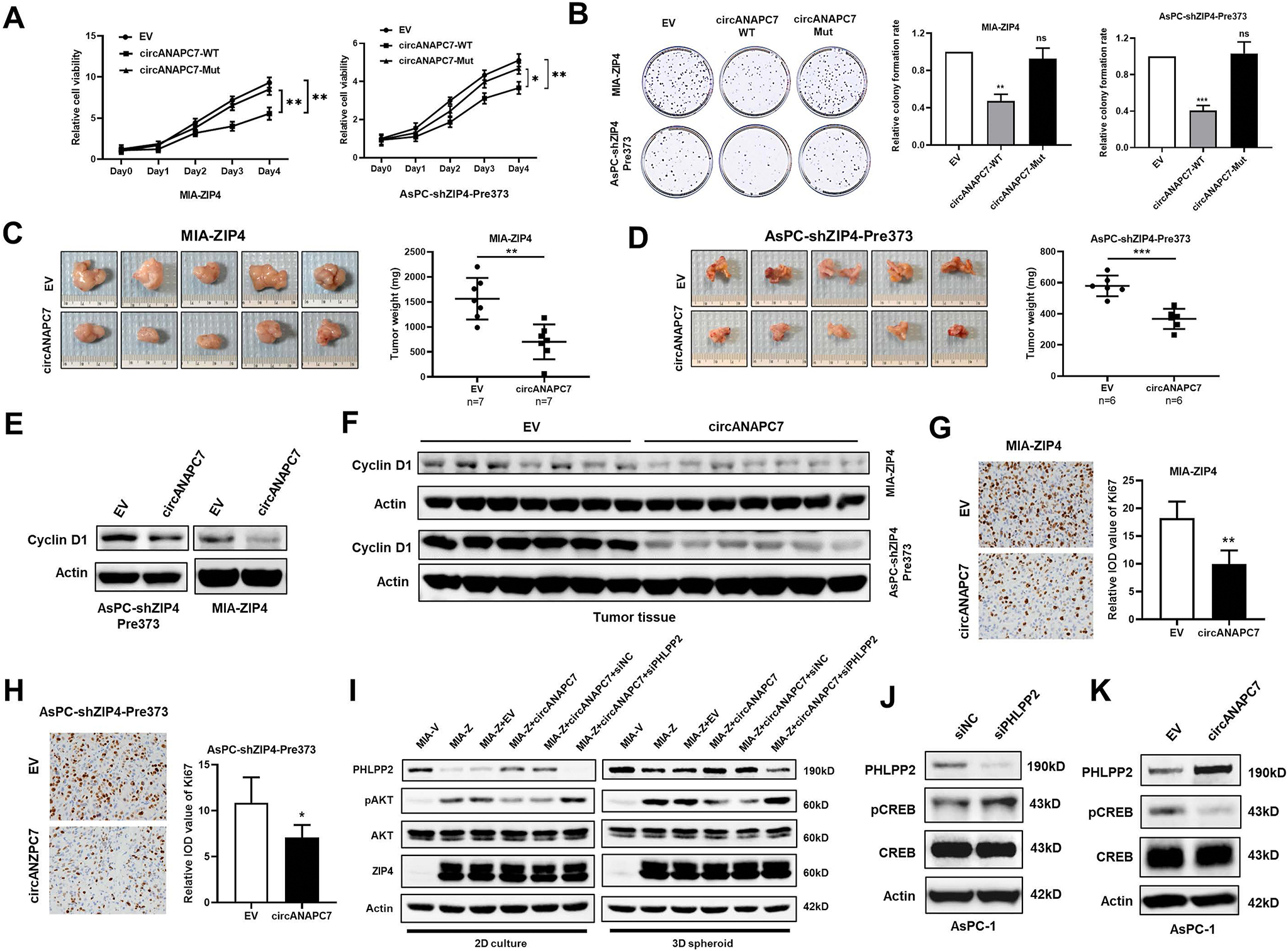
CircANAPC7 suppresses tumor growth in vivo by regulating CREB–miR-373–PHLPP2 feed-forward loop. (A) Cell viability of indicated cells was examined by means of MTT assay. (B) Colony-formation assay. (C) Representative tumor images and tumor weight plot of MIA-ZIP4-EV/circANAPC7 orthotopic models. (D) Representative tumor images and tumor weight plot of AsPC-shZIP4-Pre373-EV/circANAPC7 orthotopic models. (E) Cyclin D1 level was assessed in cell lysates. (F) Cyclin D1 level in orthotopic xenografts. (G, H) Ki67 IHC staining analysis of orthotopic xenografts. (I) Protein levels of PHLPP2, pAKT, and total AKT under 2D and 3D spheroid culture. (J, K) PHLPP2, pCREB, and total CREB expression was assessed by means of Western blot. ns, not significant; *P < .05; **P < .01; ***P < .001.
The IHC staining revealed significantly reduced Ki67 level in the circANAPC7 overexpression xenografts (Figure 5G and H). Furthermore, ZIP4-mediated elevation of phosphorylated AKT level can be reversed by circANAPC7 and PHLPP2 in both 2D and 3D spheroid culture conditions (Figure 5I and Supplementary Figure 6E and F). Because ZIP4 activates miR-373 through transcriptional regulation by increasing phosphorylated CREB and that PHLPP2 is a phosphatase, we examined whether PHLPP2 regulated CREB phosphorylation. We found PHLPP2 silencing increased CREB phosphorylation (Figure 5J and Supplementary Figure 6G). Moreover, circANAPC7 overexpression decreased the phosphorylated CREB level in pancreatic cancer cells (Figure 5K and Supplementary Figure 6H). We also validated that ZIP4 is positively correlated with pCREB and negatively correlated with PHLPP2 in human pancreatic cancer tissues by means of IHC staining (Supplementary Figure 7A). These results demonstrated the presence of a feed-forward loop in the ZIP4–CREB–miR-373–PHLPP2 signaling axis, and circANAPC7 inhibits pancreatic cancer cell proliferation through the regulation of this signaling axis.
CircANAPC7 Inhibits ZIP4/miR-373–Mediated Muscle Wasting Through Down- Regulation of Transforming Growth Factor–β
In the cachectic xenograft mouse model, we noticed the body weight of mice implanted with AsPC-shZIP4-Pre373-circANAPC7 cells is significantly higher than the AsPC-shZIP4-Pre373-EV group (Figure 6A and B). We also examined the body weight of mice injected with MIA PaCa-2 cells, a noncachectic pancreatic cancer cell line, and found that circANAPC7 overexpression cells did not affect murine body weight (Supplementary Figure 7B). Cancer-associated body weight loss is mostly due to muscle wasting, we further evaluated muscle wasting in mice implanted with AsPC-shZIP4-Pre373-EV/circANAPC7 cells. At 4 weeks after tumor implantation, the circANAPC7 group showed stronger grip strength compared with the EV group (Figure 6C). In addition, skeletal muscle weight was increased considerably in the AsPC-shZIP4-Pre373-circANAPC7 group compared with the AsPC-shZIP4-Pre373-EV group (Figure 6D and E). Morphometric analysis of the cross-sectional area of tibialis anterior muscle showed the AsPC-shZIP4-Pre373-circANAPC7 group had a larger cross-sectional area than those in the control group (Figure 6F). The increased myofibrillar protein, myosin heavy chain (MyHC), and decreased muscle-specific E3 ubiquitin ligases, Atrogin-1 were also observed in the mice muscle tissues of the AsPC-shZIP4-Pre373-circANAPC7 group (Figure 6G). In addition, Kaplan-Meier survival curves showed that circANAPC7 significantly increased median survival of the mice with pancreatic cancer xenografts compared with the control mice, suggesting the role of circANAPC7 in pancreatic cancer growth and cachexia (Supplementary Figure 7C). To determine the impact of circANAPC7 on ZIP4-mediated muscle wasting in vitro, we used conditioned media of AsPC-shZIP4-Pre373-EV/circANAPC7 cells to treat C2C12 myotubes and examined the level of muscle atrophy markers and MyHC, the motor protein of muscle thick filaments. The conditioned media from AsPC-shZIP4-Pre373-circANAPC7 cells down-regulated Atrogin-1, MuRF1, and UBR2, but increased MyHC levels (Figure 6H and I). Conversely, we found conditioned media from miR-373 overexpressed cells up-regulated muscle atrophy markers but decreased MyHC level (Supplementary Figure 7D and E).
Figure 6.
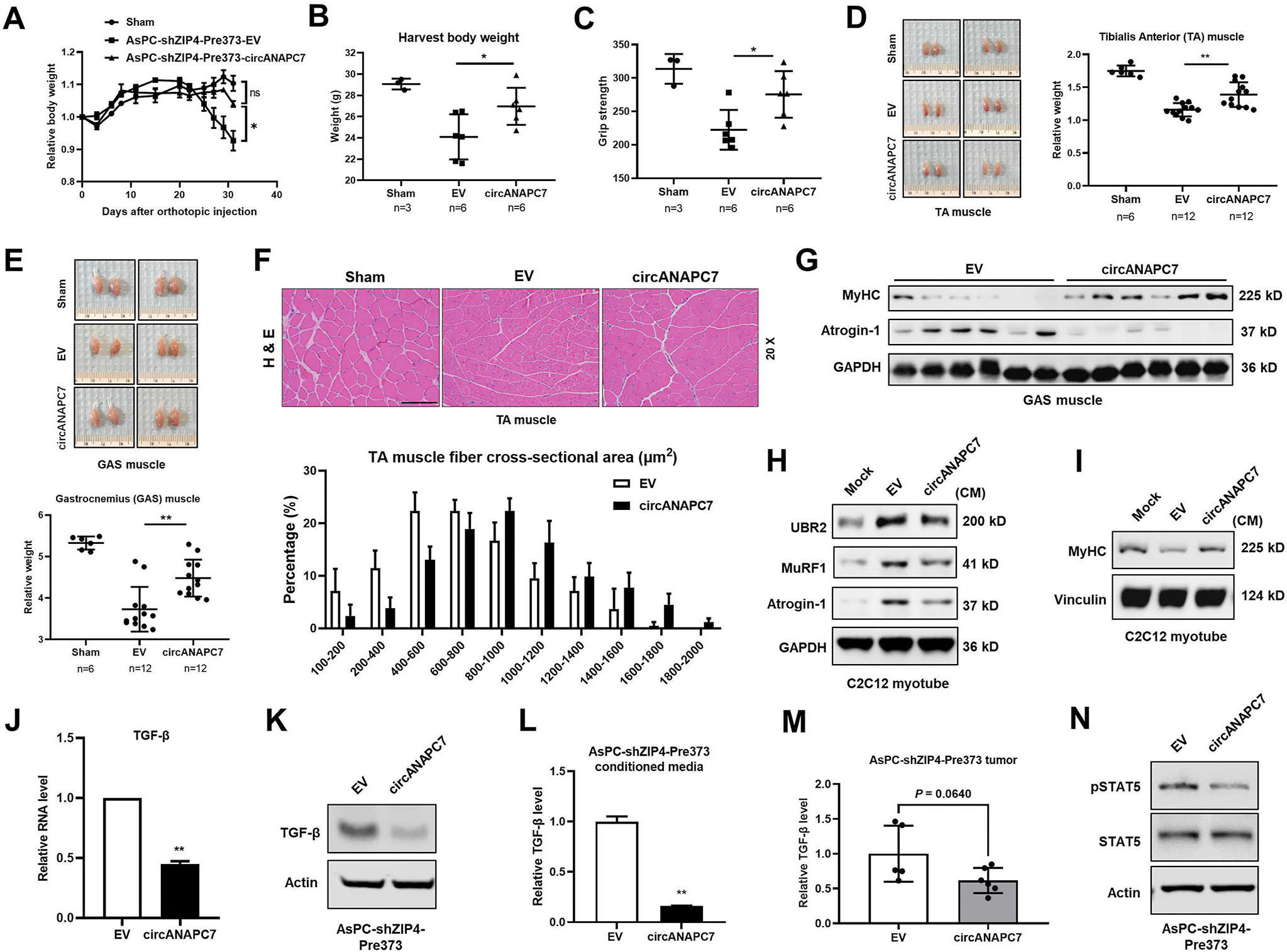
CircANAPC7 inhibits ZIP4/miR-373–mediated muscle wasting in vitro and in vivo. (A) Relative body weight curve of mice injected with AsPC-shZIP4-Pre373-EV/circANAPC7 cells. (B) Mice body weight. (C) Grip strength plot of mice. (D) Representative images of tibialis anterior (TA) muscle and TA muscle weight relative to body weight. (E) Representative images of gastrocnemius (GAS) muscle and GAS muscle weight relative to body weight. (F) Representative images of H&E-stained TA muscle sections and quantitative cross-sectional area. (G) MyHC and Atrogin-1 protein levels were detected in GAS muscle tissues of mice. (H) UBR2, MuRF1, and atrogin-1 levels in C2C12 myotubes cultured in conditioned media for 8 hours. (I) MyHC levels in C2C12 myotubes cultured in conditioned media for 72 hours. (J, K) mRNA and protein levels of TGF-β in AsPC-shZIP4-Pre373-EV/circANAPC7 cells. (L, M) TGF-β levels in cell-conditioned media and tumor tissues were detected by enzyme-linked immunosorbent assay. (N) STAT5 and pSTAT5 levels were assessed by Western blot.
We then examined potential soluble cachectic factors in the context of circANAPC7 mediated anti-cachexia. The multifunctional cytokine TGF-β could activate several procachexia signaling pathways in breast and colon cancer cachexia models and in patients as described previously.29
We found TGF-β level was significantly decreased in AsPC-shZIP4-Pre373-circANAPC7 group both in the cell lysate and the conditioned media (Figure 6J–L). We also observed a reduced level of TGF-β in tumor tissues of mice injected with AsPC-shZIP4-Pre373-circANAPC7 cells, but not MIA-ZIP4-circANAPC7 cells compared with the control group (Figure 6M and Supplementary Figure 7F). Because STAT5 is a transcriptional regulator mediating AKT induced TGF-β expression and secretion,30,31 we examined the phosphorylation of STAT5 in pancreatic cancer cells and found AsPC-shZIP4-Pre373-circANAPC7 cells showed reduced STAT5 phosphorylation (Figure 6N), which confirmed that STAT5 is the mediator down-regulating TGF-β in AsPC-shZIP4-Pre373-circANAPC7 cells. Taken together, these results suggest circANAPC7 inhibited ZIP4/miR-373 mediated muscle wasting in vitro and in vivo, at least partially through STAT5/TGF-β signaling in pancreatic cancer (Figure 7).
Figure 7.
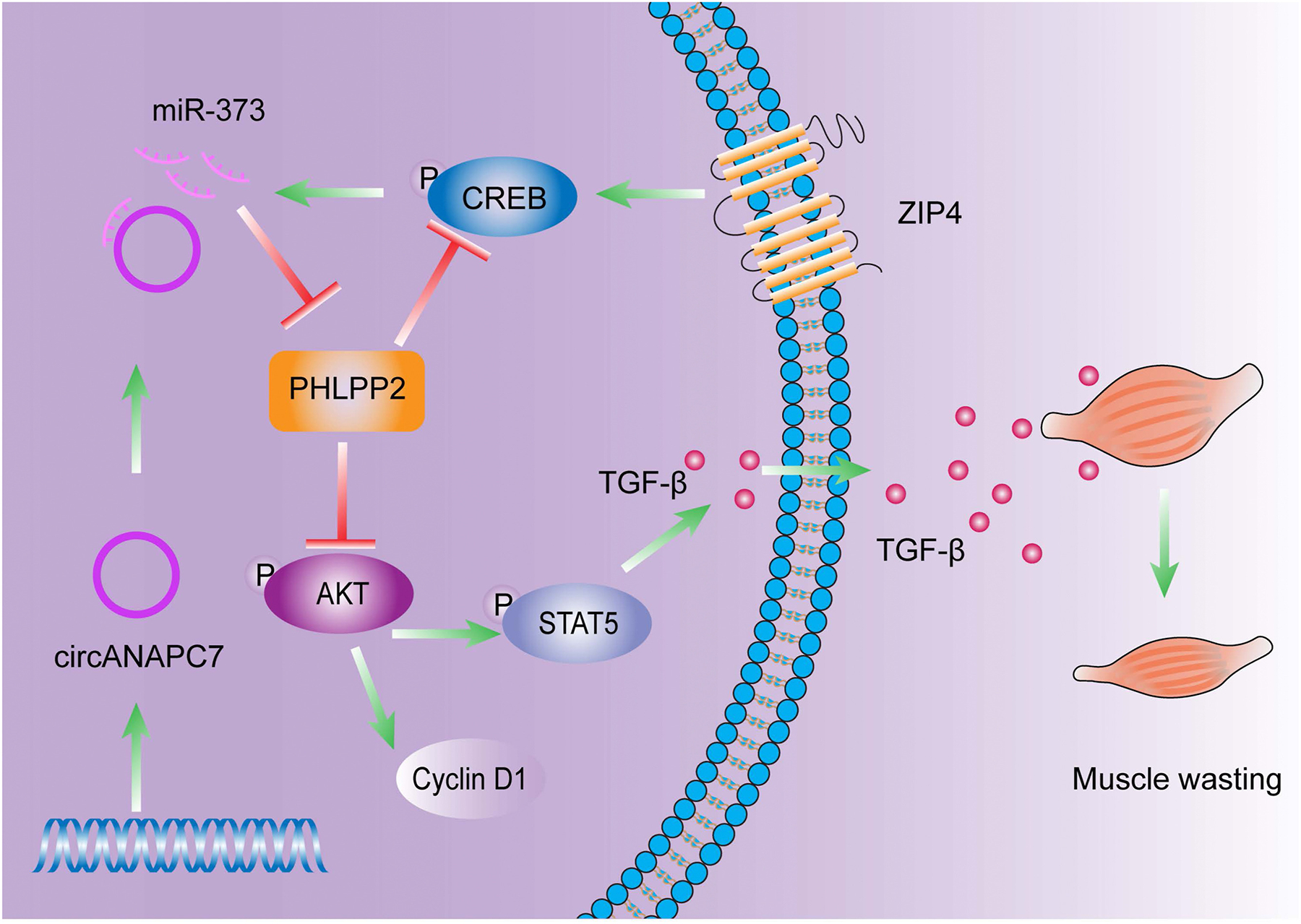
Schematic diagram of circANAPC7 suppressed tumor growth and muscle wasting through PHLPP2–AKT–TGF-β signaling axis in pancreatic cancer. This study identified circANAPC7 as a novel tumor suppressor, which functions through the CREB–miR-373–PHLPP2 axis, leading to AKT dephosphorylation and cyclin D1 and TGF-β down-regulation to suppress tumor progression in pancreatic cancer.
Discussion
ZIP4 plays important roles in cell proliferation, metastasis, drug resistance, and cancer-associated cachexia in pancreatic cancer.9–14,32 These findings prompted us to further investigate the signaling network involved in ZIP4-mediated pancreatic cancer progression. In this study, we found that circANAPC7 inhibited ZIP4-mediated cell proliferation through miR-373–PHLPP2–AKT signaling axis in 2D cultured cancer cell lines, 3D spheroid/organoid model, and an orthotopic xenograft mouse model. In addition, we demonstrated that circANAPC7 inhibited ZIP4-mediated cancer cachexia by inhibiting TGF-β secretion in cancer cells. These findings suggested the involvement of circANAPC7 in ZIP4-mediated pancreatic cancer progression and cachexia, thereby providing a novel therapeutic strategy for pancreatic cancer treatment.
miR-373 is a human embryonic stem cell–specific miRNA and has diverse functions in human cancer.33 Previously, we found that ZIP4 up-regulated miR-373 by activating CREB, and that miR-373 was responsible for ZIP4-promoted cell proliferation in pancreatic cancer.10 In this study, we identified PHLPP2 as a novel target of miR-373. We found circANAPC7 serves as a miR-373 sponge, resulting in the increased expression of PHLPP2 and decreased AKT phosphorylation thereby inhibiting pancreatic cancer cell proliferation. Moreover, we found the conditioned media from miR-373 overexpressed cells increased the protein level of muscle atrophy markers in C2C12 myotubes, suggesting miR-373 also plays an important role in cancer cachexia.
Previous studies showed that circRNAs play critical roles in cancer growth, metastasis, stemness, and resistance to therapy.34,35 Guo et al20 reported that circBFAR promotes tumor growth and metastasis in pancreatic cancer by up-regulating MET through competitively binding to miR-34b. Chen et al19 demonstrated that circPTN promotes stemness in glioma by increasing the level of stemness markers (Nestin, CD133, SOX9, and SOX2). Zhang et al21 reported that circUHRF1 was associated with resistance to anti-PD1 therapy in hepatocellular carcinoma through inhibiting natural killer cell function by increasing TIM-3 level. However, the function of circRNAs in pancreatic cancer muscle wasting and cachexia is largely unknown. Recently, a microarray analysis suggested that circANAPC7 was down-regulated in pancreatic cancer tissues compared with adjacent benign tissues.26 Here, we systematically evaluated circANAPC7 function in pancreatic cancer progression and cachexia and found that circANAPC7 suppressed pancreatic cancer cell proliferation in 2D and 3D spheroid/organoid conditions. Furthermore, circANAPC7 suppressed cell growth and cancer cachexia in pancreatic cancer cell lines and mouse models. CircRNAs have been reported to play critical roles in cancer progression; however, little is known about circRNA in muscle wasting and cancer cachexia. We found circANAPC7 reduced the levels of muscle degradation–associated ligases, thus reversed the muscle wasting in cachectic mice. Intriguingly, we also identified circANAPC7 acts as a sponge for miR-373, which serves as the downstream of CREB and upstream of PHLPP2. We further showed that PHLPP2 could dephosphorylate CREB, forming a CREB–miR-373–PHLPP2 feed-forward loop to inhibit pancreatic cancer cell progression.
PHLPP2 is a member of the PHLPP phosphatase family and is known to inhibit cell growth by inhibiting proliferation and promoting apoptosis.27,32,36,37 Oncogenic kinases AKT, PKC, S6K, and pro-apoptotic kinase Mst1 are functional targets of the PHLPP family.38 AKT was found to be a downstream target of ZIP4-mediated EMT transition in human nasopharyngeal carcinoma cells, but the underlying mechanism is not clear.39 In this study, we revealed the ZIP4-dependent regulation of AKT is mediated by PHLPP2. CircANAPC7 inhibits pancreatic cancer cell proliferation by reducing the AKT phosphorylation through regulating PHLPP2 expression. Moreover, we found PHLPP2 could regulate CREB phosphorylation, forming a CREB–miR-373–PHLPP2 feed-forward loop. These results suggest circANAPC7 regulated PHLPP2/AKT is critical for ZIP4/CREB/miR-373 mediated pancreatic cancer cell proliferation and cachexia.
Cancer cachexia usually presents with >5% of body weight loss within 6 months and develops from pre-cachexia to cachexia to refractory cachexia.4,40 Muscle wasting is a signature event of cancer cachexia, which severely affects patients’ mortality because of weakness and fatigue.41 Increased myofibrillar degradation was observed in skeletal muscle wasting caused by activation of the protein-degradation pathway, including the ubiquitin-proteasome pathway and autophagy-lysosome pathway.42 The inflammatory factor was considered as a key mediator triggering the activation of the protein-degradation pathway in muscle.29,43 Currently, very few studies focus on the role of circRNA in cancer cachexia, especially muscle wasting. Zhang et al44 reported that exosomal circular RNA ciRS-133 derived from gastric tumor promotes browning of white adipose tissue by targeting the miR-133/PRDM16 pathway. Recently, Ding et al45 demonstrated the role of circPTK2 in promoting lipolysis and reducing adipogenesis. To our knowledge, this study is the first to report the impact of circRNA on skeletal muscle wasting and cachexia of pancreatic cancer. We found the overexpression of circANAPC7 in pancreatic cancer cells relieved muscle wasting in mice, as indicated by increased muscle mass, decreased muscle atrophy proteins (atrogin-1, MuRF1, and UBR2) level, and increased myofibrillar protein (MyHC). Emerging evidence has shown that TGF-β plays critical roles in cancer-associated cachexia, including muscle wasting and adipose loss.29,43 We found that circANAPC7 up-regulates PHLPP2, leading to dephosphorylation of AKT and CREB. TGF-β blockade reduced the metabolic changes of pancreatic cancer cachexia and improved overall survival.46 Consistent with previous study,30 our results showed that circANAPC7 decreased TGF-β by dephosphorylating STAT5. These results suggest that circANAPC7 could effectively ameliorate muscle wasting of pancreatic cancer via PHLPP2–AKT–STAT5–TGF-β signaling axis.
In conclusion, our current study demonstrated that circANAPC7 inhibits ZIP4-mediated cell proliferation by serving as a miR-373 sponge to increase PHLPP2 levels, which in turn results in decreased AKT phosphorylation and cyclin D1 expression. CircANAPC7 also inhibits ZIP4-mediated cancer cachexia by decreasing TGF-β expression and secretion through regulating the AKT/STAT5 signaling. Our results may provide a novel strategy for pancreatic cancer treatment by ameliorating cancer cachexia.
Supplementary Material
WHAT YOU NEED TO KNOW.
BACKGROUND AND CONTEXT
ZIP4 promotes pancreatic cancer progression by regulating miR-373. Perturbation of circular RNAs (circRNAs) is associated with cancer aggressiveness. We aimed to identify circRNAs involved in ZIP4/miR-373–driven cancer cachexia and decipher the underlying mechanism.
NEW FINDINGS
CircANAPC7 is a tumor suppressor that functions through PHLPP2-mediated dephosphorylation of AKT, resulting in down-regulation of cyclin D1 and transforming growth factor–β. PHLPP2 also induced dephosphorylation of CREB, a zinc-dependent transcription factor activated by ZIP4, thus forming a CREB–miR-373–PHLPP2 feed-forward loop to regulate cancer progression and cachexia.
LIMITATIONS
The role of other miR-373–related circRNAs were not included in this study.
IMPACT
This study identifies CircANAPC7 as a novel tumor suppressor and provides a potential therapeutic strategy to suppress tumor growth and muscle wasting in pancreatic cancer.
Funding
This work was supported in part by the William and Ella Owens Medical Research Foundation (Min Li), United States; Department of Medicine at University of Oklahoma Health Sciences Center, United States.
Abbreviations used in this paper:
- circRNA
circular RNA
- 2D
2-dimensional
- 3D
3-dimensional
- DEC
differentially expressed circular RNA
- HPDE
human pancreatic duct epithelial
- IHC
immunohistochemistry
- mRNA
messenger RNA
- MyHC
myosin heavy chain
- RT-qPCR
reverse transcription quantitative real-time polymerase chain reaction
- TGF
transforming growth factor
Footnotes
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://doi.org/10.1053/j.gastro.2022.02.017.
CRediT Authorship Contributions
Xiuhui Shi, MD, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Project administration: Lead; Writing – original draft: Lead; Writing – review & editing: Lead).
Jingxuan Yang, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Methodology: Lead; Project administration: Equal; Supervision: Equal; Validation: Lead; Visualization: Lead; Writing – original draft: Equal; Writing – review & editing: Equal).
Mingyang Liu, MD, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Lead; Investigation: Equal; Validation: Lead; Visualization: Equal; Writing – review & editing: Equal).
Yuqing Zhang, PhD (Conceptualization: Lead; Data curation: Equal; Formal analysis: Equal; Methodology: Lead; Supervision: Equal; Visualization: Equal; Writing – review & editing: Lead).
Zhijun Zhou, MD (Conceptualization: Equal; Data curation: Supporting; Formal analysis: Equal; Investigation: Supporting; Methodology: Equal; Project administration: Equal; Software: Equal; Visualization: Lead; Writing – review & editing: Lead).
Wenyi Luo, MD, PhD (Formal analysis: Equal; Investigation: Supporting; Methodology: Equal; Resources: Equal; Visualization: Supporting; Writing – review & editing: Supporting).
Kar-Ming A. Fung, MD, PhD (Formal analysis: Supporting; Methodology: Equal; Resources: Equal; Visualization: Supporting; Writing – review & editing: Supporting).
Chao Xu, PhD (Formal analysis: Supporting; Methodology: Supporting; Resources: Supporting; Software: Equal; Visualization: Supporting; Writing – review & editing: Equal).
Michael S. Bronze, MD (Resources: Equal; Supervision: Equal; Writing – review & editing: Equal).
Courtney W. Houchen, MD (Resources: Equal; Supervision: Equal; Writing – review & editing: Equal).
Min Li, PhD (Conceptualization: Lead; Funding acquisition: Lead; Resources: Lead; Supervision: Lead; Writing – review & editing: Lead).
Conflicts of interest
The authors disclose no conflicts.
References
- 1.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 2.Mizrahi JD, Surana R, Valle JW, et al. Pancreatic cancer. Lancet 2020;395:2008–2020. [DOI] [PubMed] [Google Scholar]
- 3.Baracos VE, Martin L, Korc M, et al. Cancer-associated cachexia. Nat Rev Dis Primers 2018;4:17105. [DOI] [PubMed] [Google Scholar]
- 4.Fearon K, Strasser F, Anker SD, et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol 2011;12:489–495. [DOI] [PubMed] [Google Scholar]
- 5.Petruzzelli M, Wagner EF. Mechanisms of metabolic dysfunction in cancer-associated cachexia. Genes Dev 2016;30:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bafaro E, Liu Y, Xu Y, et al. The emerging role of zinc transporters in cellular homeostasis and cancer. Signal Transduct Target Ther 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Z, Yu Z, Chen ZB, et al. Zinc chelator TPEN induces pancreatic cancer cell death through causing oxidative stress and inhibiting cell autophagy. J Cell Physiol 2019; 234:20648–20661. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Zhang Y, Cui X, et al. Gene profile identifies zinc transporters differentially expressed in normal human organs and human pancreatic cancer. Curr Mol Med 2013;13:401–409. [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Zhang Y, Liu Z, et al. Aberrant expression of zinc transporter ZIP4 (SLC39A4) significantly contributes to human pancreatic cancer pathogenesis and progression. Proc Natl Acad Sci U S A 2007;104:18636–18641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Yang J, Cui X, et al. A novel epigenetic CREB-miR-373 axis mediates ZIP4-induced pancreatic cancer growth. EMBO Mol Med 2013;5:1322–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M, Yang J, Zhang Y, et al. ZIP4 promotes pancreatic cancer progression by repressing ZO-1 and claudin-1 through a ZEB1-dependent transcriptional mechanism. Clin Cancer Res 2018;24:3186–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu M, Zhang Y, Yang J, et al. ZIP4 increases expression of transcription factor ZEB1 to promote integrin alpha3beta1 signaling and inhibit expression of the gemcitabine transporter ENT1 in pancreatic cancer cells. Gastroenterology 2020;158:679–692.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M, Zhang Y, Yang J, et al. Zinc-dependent regulation of ZEB1 and YAP1 coactivation promotes epithelialmesenchymal transition plasticity and metastasis in pancreatic cancer. Gastroenterology 2021; 160:1771–1783.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Zhang Z, Zhang Y, et al. ZIP4 promotes muscle wasting and cachexia in mice with orthotopic pancreatic tumors by stimulating RAB27B-regulated release of extracellular vesicles from cancer cells. Gastroenterology 2019;156:722–734.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675–691. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Lei C, He Q, et al. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol Cancer 2018;17:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong YX, Du YJ, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 2018;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Chen T, Zhu Y, et al. circPTN sponges miR-145–5p/miR-330–5p to promote proliferation and stemness in glioma. J Exp Clin Cancer Res 2019;38:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo X, Zhou Q, Su D, et al. Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis. Mol Cancer 2020;19:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang PF, Gao C, Huang XY, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer 2020; 19:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao F, Huang M, Meng F, et al. Circular RNA signature predicts gemcitabine resistance of pancreatic ductal adenocarcinoma. Front Pharmacol 2018;9:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong OY, Mou LJ, Luk C, et al. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am J Pathol 2000; 157:1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal A, Thomas MP, Altschuler G, et al. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet 2011;7:e1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu M, Wang Q, Shen J, et al. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol 2019;16:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Hao X, Wang H, et al. Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cell Physiol Biochem 2016; 40:1334–1344. [DOI] [PubMed] [Google Scholar]
- 27.Brognard J, Sierecki E, Gao T, et al. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell 2007;25:917–931. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Bharadwaj U, Logsdon CD, et al. ZIP4 regulates pancreatic cancer cell growth by activating IL-6/STAT3 pathway through zinc finger transcription factor CREB. Clin Cancer Res 2010;16:1423–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G, Biswas AK, Ma WC, et al. Metastatic cancers promote cachexia through ZIP14 upregulation in skeletal muscle. Nat Med 2018;24:770–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang S, Wang J, Brand DD, et al. Role of TNF-TNF receptor 2 signal in regulatory T cells and its therapeutic implications. Front Immunol 2018;9:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones N, Vincent EE, Cronin JG, et al. Akt and STAT5 mediate naive human CD4+ T-cell early metabolic response to TCR stimulation. Nat Commun 2019; 10:2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YQ, Chen CY, Yao QZ, et al. ZIP4 upregulates the expression of neuropilin-1, vascular endothelial growth factor, and matrix metalloproteases in pancreatic cancer cell lines and xenografts. Cancer Biol Ther 2010; 9:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006; 124:1169–1181. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792–806. [DOI] [PubMed] [Google Scholar]
- 35.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016;17:205–211. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Eckert MA, Harada BT, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol 2018;20:1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nitsche C, Edderkaoui M, Moore RM, et al. The phosphatase PHLPP1 regulates Akt2, promotes pancreatic cancer cell death, and inhibits tumor formation. Gastroenterology 2012;142:377–387.e1–e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan Y, Krecke KN, Bapat AS, et al. Phosphatase PHLPP2 regulates the cellular response to metabolic stress through AMPK. Cell Death Dis 2021;12:904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng Q, Liu YM, Liu J, et al. Inhibition of ZIP4 reverses epithelial-to-mesenchymal transition and enhances the radiosensitivity in human nasopharyngeal carcinoma cells. Cell Death Dis 2019;10:588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fearon K, Arends J, Baracos V. Understanding the mechanisms and treatment options in cancer cachexia. Nat Rev Clin Oncol 2013;10:90–99. [DOI] [PubMed] [Google Scholar]
- 41.Poulia KA, Sarantis P, Antoniadou D, et al. Pancreatic cancer and cachexia-metabolic mechanisms and novel insights. Nutrients 2020;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, Sin KWT, Ding H, et al. p38beta MAPK mediates ULK1-dependent induction of autophagy in skeletal muscle of tumor-bearing mice. Cell Stress 2018;2:311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guttridge DC. A TGF-beta pathway associated with cancer cachexia. Nat Med 2015;21:1248–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Zhu L, Bai M, et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int J Cancer 2019;144:2501–2515. [DOI] [PubMed] [Google Scholar]
- 45.Ding Z, Sun D, Han J, et al. Novel noncoding RNA CircPTK2 regulates lipolysis and adipogenesis in cachexia. Mol Metab 2021;53:101310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greco SH, Tomkotter L, Vahle AK, et al. TGF-beta blockade reduces mortality and metabolic changes in a validated murine model of pancreatic cancer cachexia. PLoS One 2015;10:e0132786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


