Abstract
Sequence analysis of the murine gammaherpesvirus 68 (γHV68) genome revealed an open reading frame (gene 4) which is homologous to a family of proteins known as the regulators of complement activation (RCA proteins) (H. W. Virgin, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck, J. Virol. 71:5894–5904, 1997). The predicted gene 4 product has homology to other virally encoded RCA homologs, as well as to the complement-regulatory proteins decay-accelerating factor and membrane cofactor protein. Analyses by Northern blotting and rapid amplification of cDNA ends revealed that gene 4 is transcribed as a 5.2-kb bicistronic transcript of the late kinetic class. Three γHV68 RCA protein isoforms (60 to 65 kDa, 50 to 55 kDa, and 40 to 45 kDa) were detected by Western blotting of infected murine NIH 3T12 fibroblast cells. A soluble 40- to 45-kDa isoform was detected in the supernatants of virally infected cells. Flow cytometric analysis revealed that the γHV68 RCA protein was expressed on the surfaces of infected cells. Supernatants from virally infected cells contained an activity that inhibited murine complement activation as measured by inhibition of C3 deposition on activated zymosan particles. Recombinant γHV68 RCA protein, containing the four conserved short consensus repeats, inhibited murine C3 deposition on zymosan via both classical and alternative pathways and inhibited deposition of human C3 on activated zymosan particles. Expression of this inhibitor of complement activation, both at the cell surface and in the fluid phase, may be important for γHV68 pathogenesis via the inhibition of innate and adaptive immunity.
Gammaherpesvirus 68 (γHV68) (also known as murine gammaherpesvirus 68 or MHV-68) is a gammaherpesvirus related to herpesvirus saimiri (HVS), Epstein-Barr virus, and Kaposi’s sarcoma-associated herpesvirus (KSHV) (human herpesvirus 8) (13, 14, 65). γHV68 was first isolated from a bank vole, is a murine pathogen, and infects both inbred and outbred mouse strains (6, 48, 51). γHV68 acutely infects multiple organs in mice (7, 51) and also establishes a latent infection in the spleen and peritoneal cells (60, 69). γHV68 can cause inflammation of the great elastic arteries and has a tropism for vascular smooth muscle cells (68). Although there is evidence that B cells are a reservoir for latent γHV68 (60), B-cell-deficient mice were found to harbor latent virus or viral DNA (59, 63, 66, 67, 69). Recent work from our group has demonstrated γHV68 latency in macrophages and chronic carriage of the γHV68 genome in CD19+ B cells (70). The program of transcription during latency has recently been preliminary characterized (58, 66). Importantly, regions of the γHV68 genome corresponding to known or suspected latency-associated genes of the primate herpesviruses HVS, Epstein-Barr virus, and KSHV are actively transcribed in latent tissues (66). In addition, we have shown that γHV68 (64) shares with KSHV and HVS a functional D-type cyclin homolog (22, 29, 38, 61). These data argue that γHV68 will share molecular mechanisms of pathogenesis with primate gammaherpesviruses.
A further argument for pathogenetic relatedness of γHV68 and the primate gammaherpesviruses KSHV and HVS is the fact that these viruses share an open reading frame (ORF) predicted to encode a protein structurally related to host regulators of complement activation (RCA proteins) (65). The structural hallmark of RCA proteins is the presence of short consensus repeats (SCRs). SCRs are ca. 60-amino-acid motifs with four conserved cysteine residues (disulfide bonded together in the manner, 1-3, 2-4) (39, 53), which are critical for binding C3b and C4b (2, 49). Gene 4 of γHV68, KSHV, and HVS is predicted to encode a protein containing four SCRs homologous to the viral and mammalian RCA proteins (65). The best characterized of these gammaherpesvirus RCA proteins is the HVS complement control protein homolog (CCPH) (4, 5, 18). Northern blot analysis demonstrated two transcripts of 1.5 and 1.7 kb from the CCPH gene (5), and analysis of cDNAs demonstrated two forms, one spliced and one unspliced (5). These two forms encode a membrane-bound form and a secreted form of CCPH (5). The membrane-bound form of CCPH inhibits cell damage mediated by human complement (18). This property is explained by CCPH inhibition of C3 convertase activity and consequent deposition of C3 on the cell surface (18).
Since the HVS homolog of the γHV68 RCA protein has two forms and regulates complement activation, we tested the hypothesis that the γHV68 RCA protein has similar properties. In this paper we show that gene 4 is a late gene, and we demonstrate expression of membrane-bound and soluble isoforms of the γHV68 RCA protein. We found that the γHV68 RCA protein regulates both the classical and alternative pathways of murine complement activation. These findings are consistent with a general strategy for complement evasion shared by some gammaherpesviruses.
MATERIALS AND METHODS
Sequence analysis.
The γHV68 genome sequence and putative ORFs were analyzed on Vector NTI version 4.0 Deluxe (Informax Inc., Gaithersburg, Md.) as previously described (65). The first 275 amino acids of the putative RCA sequence were aligned with other murine and viral RCA sequences by using the DNASTAR MEGALIGN software program (DNASTAR Inc., Madison, Wis.).
Nomenclature for the γHV68 RCA gene and protein.
In the original full-length sequence of γHV68, we designated the region predicted to encode an RCA family member ORF 4 (65). We used the ORF designation, rather than referring to predicted ORFs in the γHV68 genome as genes, since transcriptional data were not available for most regions of the genome. In this paper we show that ORF 4 is both transcribed and translated and that ORFs 6 and M4 are transcribed, thereby meeting the criteria for being genes (gene M4, gene 4, and gene 6). The protein product of gene 4 is homologous to members of the RCA family and, per this paper, regulates complement activation. We therefore refer to the protein product of γHV68 gene 4 as the γHV68 RCA protein. Note that γHV68 ORFs without homology to other gammaherpesvirus ORFs were designated with an M. Thus, gene M4 is the fourth ATG-initiated ORF in the γHV68 genome without clear viral homologs and is adjacent to but distinct from gene 4.
Generation of virus and viral DNA.
γHV68 clone WUMS was cloned by limiting dilution from a stock generously provided by P. Doherty and A. Nash and has been sequenced in full (65). Virus was passaged and grown as previously described (65). γHV68 was grown in NIH 3T12 cells in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum, 100 U of penicillin per ml, 100 mg of streptomycin per ml, and 2 mM l-glutamine. Virus titer was measured by plaque assay (67). Viral DNA was prepared by infecting NIH 3T12 cells at a multiplicity of infection (MOI) of 0.5. Culture supernatants were collected and DNA was isolated as previously described (65).
Northern blot analysis.
Total RNA was isolated from mock-infected and γHV68-infected NIH 3T12 cells infected at an MOI of 5 in the presence or absence of a DNA synthesis inhibitor (phosphonoacetic acid [PAA]) and protein synthesis inhibitors (cycloheximide and anisomycin). Cycloheximide, anisomycin, and PAA were used at final concentrations of 40 μM, 10 μM, and 200 μg/ml, respectively. Total cellular RNA was harvested at 7 and 12 h postinfection by using the single guanidinium thiocyanate-phenol method (50), and 10 μg of total RNA was loaded per lane. Probes to gene M4, gene 4, and gene 6 ORFs were generated by PCR amplification of γHV68 genomic DNA. PCR was carried out in 25-μl reaction mixtures consisting of 1× Taq DNA Polymerase Reaction Buffer (Promega, catalog no. M1882), 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs), 5 U of Taq polymerase enzyme (Promega, catalog no. M186E), 0.2 μM primers, and 1 μg of γHV68 DNA. PCR conditions were as follows: (i) denaturation at 94°C for 5 min; (ii) five cycles of 94°C for 1 min, 55°C for 2 min, and 72°C for 3 min; (iii) 25 cycles of 94°C for 1 min, 60°C for 2 min, and 72°C for 3 min; and (iv) a 4°C hold. PCR primers used for each probe and the corresponding coordinates (65) of the amplified product in the γHV68 genome were as follows: gene M4 (bp 8608 to 8914), 5′-TACGGATCCTCGAGCACCCACCCCTGGAGAAGATGAT-3′ and 5′-TACGGATCGCGGCCGCATTGACTTCTAATCGACCCA-3′; gene 4 (bp 10227 to 10593), 5′-TACGGATCCTCGAGGGCTACAATCTTCTGGGAGAAAGT-3′ and 5′-TACGGATCGCGGCCGCATCGCAATTGACAACAAGTTCTCTA-3′; and gene 6 (bp 11956 to 12307), 5′-TACGGATCCTCGAGACATATCCTGCAGAAAACTGCG-3′ and 5′-TACGGATCGCGGCCGCCAGTCCATGCTGCATATAGTA-3′. For these and all subsequently described primers, nonhomologous bases containing a unique restriction site for cloning purposes are underlined. Probes were labeled by using the Megaprime DNA Labeling System (Amersham, Arlington Heights, Ill.) as per the manufacturer’s instructions. A probe for the rat cyclophilin transcript was used for all Northern blots to control for loading (10).
RACE and S1 nuclease protection analysis.
Poly(A) mRNA was purified from total RNA harvested from mock-infected and γHV68-infected NIH 3T12 cells by using the PolyA Spin mRNA Isolation Kit (New England Biolabs [NEB], catalog no. 1560). Poly(A) mRNA was converted into cDNA and amplified by using a Marathon cDNA Amplification Kit (Clontech, catalog no. K1802-1). Both 5′ and 3′ rapid amplification of cDNA ends (RACE) PCRs were performed as per the kit manufacturer’s protocol. The 5′ RACE primer sequence was as follows: 5′-AATTTACTTTCTCCCAGAAGATTGTAGCCGGGA-3′ (5′ end of primer at γHV68 coordinate bp 10255). The 3′ RACE primer sequence was as follows: 5′-TATCTGAGGCACCCGAGGTTCCCA-3′ (5′ end of primer at γHV68 bp 10720).
S1 nuclease assays were performed as previously described (71, 74) with some modifications. Briefly, analysis of the 5′ transcriptional start site of the RCA mRNA was performed with total RNA isolated from mock- or γHV68-infected NIH 3T12 cells harvested at 24 h postinfection. Four nanograms of 5′-end-labeled S1 oligonucleotide probe was hybridized to 40 μg of total cellular RNA extracted from either mock- or γHV68-infected cells. Hybridization was carried out at 37°C overnight. S1 nuclease (Promega) was then added at concentrations of 100, 300, and 500 U/ml and incubated at 37°C for 30 min, and the protected fragments were separated by electrophoresis on a 10% denaturing polyacrylamide gel. The sequence of the S1 oligonucleotide probe, including the putative gene 4 translational start site, was as follows (5′ end of probe at γHV68 coordinate bp 9911): 5′-CCCCACCAATACTGCGCAGAAAAGTGGAAGTTGGCCATGATTTTGTTGAGAGTAATTTTTATTAACCCT-3′.
Generation of recombinant γHV68 RCA protein and rabbit polyclonal antiserum.
A truncated form of the predicted γHV68 RCA protein was generated by PCR with primers internal to gene 4 such that the expressed protein lacked the amino-terminal and carboxy-terminal hydrophobic domains. PCR was carried out in 25-μl reaction mixtures containing 1× Thermopol Buffer (NEB), 1.5 U of Vent DNA polymerase (NEB, catalog no. 254S), 0.2 mM dNTPs, 40 ng of primers, and 1 μg of γHV68 DNA. PCR conditions were as follows: (i) denaturation at 94°C for 5 min; (ii) five cycles of 94°C for 1 min, 40°C for 1 min, and 72°C for 3 min; (iii) 15 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 3 min; (iv) extension at 72°C for 10 min; and (v) a 4°C hold. The PCR primers used were as follows: 5′-TACGGATCTTCGAAATGTGCCACCATCTTCCTCA-3′ (5′ end of primer at γHV68 coordinate bp 9936) and 5′-TACGGATCCTCGAGCTATAAGGATGACTTTTTGGGCG-3′ (5′ end of primer at γHV68 coordinate bp 10937), which deleted 63 and 99 bp from the 5′ and 3′ ends of the gene 4 ORF, respectively, resulting in a truncated RCA protein containing amino acid residues 22 to 354 of the predicted 388-amino-acid sequence. The primers include unique BstB1 and XhoI restriction enzyme sites which were used to clone it into pET-30a(+) (Novagen), which contains an amino-terminal His6 tag. Escherichia coli BL21 cells (Novagen) were transformed with this construct, grown to an optical density at 600 nm of 0.6 to 0.9, and induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma, catalog no. I-6758) at a final concentration of 0.4 mM. Induction was carried out for 3 to 5 h at 37°C, after which cells were spun down and His-tagged RCA protein was column purified according to the instructions of the manufacturer (Qiagen). His-tagged v-cyclin derived from γHV68 gene 72 protein was similarly purified on an Ni-nitrilotriacetic acid column and eluted with 200 mM imidazole (64). Polyclonal rabbit antiserum (Cocalico, Reamstown, Pa.) was generated to an initial inoculation of 100 μg of recombinant γHV68 RCA protein in complete Freund adjuvant with eight follow-up boosts of 50 μg of incomplete Freund adjuvant 1 month apart. Sera were collected 7 days after the boosts.
Western blot analysis for γHV68 RCA protein expression.
NIH 3T12 cells were infected at an MOI of 5, and total cell lysates were harvested at 24 h postinfection. Cells were either mock infected or infected with γHV68 in the absence or presence of protein or DNA synthesis inhibitors (cycloheximide and PAA, respectively) (see above). A total of 3 × 105 cell equivalents per lane were resuspended in 1× reducing loading buffer, boiled for 10 min, and loaded onto a 10% polyacrylamide gel. The gels were transferred onto Hybond-N membranes (Amersham). Blots were blocked and incubated with a 1:500 dilution of rabbit polyclonal antiserum as previously described (64). The blots were washed three times following incubation with the primary antibody, incubated with horseradish peroxidase-conjugated donkey antirabbit antiserum, and developed with ECL chemiluminescent reagent (Amersham) as previously described (64). Supernatants containing soluble γHV68 RCA protein were prepared as follows. Three T225 flasks containing NIH 3T12 fibroblasts (Costar) were either mock or γHV68 infected at an MOI of 5, and supernatants were harvested 34 h later. Cells were infected in Dulbecco’s modified Eagle medium supplemented with 0.5% fetal calf serum, 100 U of penicillin per ml, 100 mg of streptomycin per ml, and 2 mM l-glutamine. Supernatants were spun at 5,000 rpm for 10 min at 4°C, passed through a 0.2-μm-pore-size filter to remove cell debris, and concentrated 50-fold to a final volume of 2.0 ml by using Centriprep 10 concentrators (Amicon, catalog no. 4304). The supernatants were centrifuged at 100,000 × g for 3.5 h at 4°C in an Optima TLX ultracentrifuge (Beckman). The top 1.0 ml was used as the soluble fraction. The pellet was washed and resuspended in phosphate-buffered saline (PBS). Both the soluble and pellet fractions were aliquoted and frozen in the presence of a protease inhibitor cocktail (Sigma, catalog no. P-8340). Western blot analysis was then performed as described above.
Flow cytometric analysis for detecting surface expression of RCA protein.
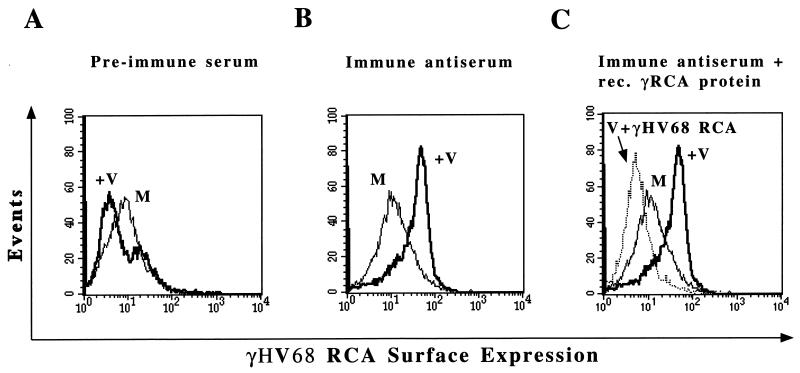
Flow cytometry was performed with a Becton Dickenson FACScan, and 10,000 events were collected per sample. Mock- or γHV68-infected NIH 3T12 cells were scraped from 100-mm-diameter tissue culture dishes, resuspended in complete Dulbecco’s modified Eagle medium, and spun at 230 × g for 5 min. The medium was aspirated, and the cells were resuspended in staining buffer (1% bovine serum albumin [BSA] in PBS). An aliquot of cells were fixed in 2% paraformaldehyde (pH 7.0) and counted, and 106 cells were used per reaction. Cells were blocked for 1 h with 10% fetal calf serum and 5% BSA in PBS and then incubated with rabbit polyclonal antiserum at a 1:750 dilution for 1 h at 4°C. Preimmune serum gave no specific staining (see Fig. 7A). Specific fluorescence-activated cell sorter (FACS) signal from immune antisera was eliminated by the addition of recombinant γHV68 RCA protein (see Fig. 7C) but not by the addition of recombinant v-cyclin (data not shown). Cells were washed three times with staining buffer, incubated with donkey anti-rabbit antibody conjugated to the fluorescein derivative 5-[(4,6-dichlorotriazine-2-yl)amino]-fluorescein (DTAF) at a 1:100 dilution, washed three times, fixed in 2% paraformaldehyde, and analyzed.
FIG. 7.
Surface expression of the γHV68 RCA protein on infected NIH 3T12 cells. (A) Flow cytometric analysis to detect γHV68 RCA protein by using preimmune rabbit polyclonal serum. Mock-infected (M) or γHV68-infected (+V) cells were harvested at 24 h postinfection, and FACS analysis was performed with rabbit polyclonal antiserum followed by donkey anti-rabbit secondary antibody conjugated to DTAF fluorophore. No surface expression was detected when preimmune serum was used. (B) FACS analysis to detect γHV68 RCA protein with immune antiserum. γHV68 RCA protein was detected on virus-infected but not mock-infected cells with immune antiserum. (C) Specificity of FACS signal for γHV68 RCA protein expression. Seventy micrograms of recombinant γHV68 RCA protein was added during incubation of virus-infected cells with immune antiserum prior to FACS analysis (V + γHV68 RCA). The data are representative of two independent experiments.
Assay for regulation of C3 deposition by soluble RCA protein isoform.
Regulation of complement activation was measured by using an assay that detects C3 deposition on activated zymosan A (zymosan), as previously described (19). Briefly, zymosan (Sigma) was suspended in 0.9% NaCl solution and boiled for 30 min. Zymosan particles were then counted on a hemocytometer, spun down at 230 × g for 5 min, and resuspended at 1.3 × 109 particles/ml. Two hundred microliters of concentrated supernatant from mock- or γHV68-infected cells (prepared as described above) was incubated with serum derived from 129Ev/Sv mice as a source of complement and 1 μl of zymosan at 37°C for 30 min. The reaction mixture was centrifuged and washed three times in FACS buffer (1% BSA and 0.1% sodium azide in PBS). Zymosan particles were then incubated with a 1:100 dilution of FITC-conjugated goat anti-mouse C3 antibody (Jackson Immunoresearch) for 30 min on ice, washed three times, and resuspended in FACS buffer. Flow cytometric analysis was performed, and 25,000 events were collected. In other assays, 70 μg of recombinant His-tagged RCA protein was added to medium to assess whether the bacterially expressed RCA protein exhibited complement-regulatory activity. The His-tagged γHV68 v-cyclin (64) was used as a negative control protein purified in the same way as the γHV68 RCA protein. This amount of γHV68 RCA protein was selected based on studies showing that inhibition of complement activation by the γHV68 protein was dose dependent and that 70 μg gave maximal inhibition of complement activation. Protein concentrations were determined by using the Bio-Rad protein assay kit (catalog no. 500-0006) as per the manufacturer’s instructions.
Mouse strains used for serum preparation.
Mice were housed and bred at the Washington University School of Medicine at biosafety level 2 in accordance with all federal government and university policies. Sentinel mice, assessed every 3 to 6 months, were determined to be negative for adventitious mouse pathogens. B-cell-deficient mice, generated by creating a null mutation in the transmembrane exon of the immunoglobulin M (IgM) heavy chain (34), were originally obtained from Jackson Laboratory (C57BL/6J-Igh-6tm1Cgn). C3-deficient and factor B-deficient mice were kindly supplied by Harvey Colten of Northwestern University. Both mice strains are on a mixed 129/J and C57BL/6J background. Heterozygous factor B-deficient mice were crossed, and homozygous progeny were detected by PCR and Southern blotting as previously described (45). PCR primers used for factor B deficiency detection were as follows: 5′-CCGAAGCATTCCTATCCTCC-3′, 5′-CGAATGGGCTGACCGCTTCC-3′, and 5′-GTAGTCTTGTCTGCTTTCTTC-3′. C3-deficient mice were detected by PCR. PCR was carried out in 25 μl of 1× Taq buffer (Promega)–5 U of Taq DNA polymerase (Promega, catalog no. M186B)–0.2 mM dNTPs–150 ng of each of the four primers stated below–2 μg of mouse DNA. PCR conditions were as follows: (i) denaturation at 94°C for 4 min; (ii) 30 cycles of 94°C for 1 min, 60°C for 2 min, and 72°C for 3 min; (iii) 72°C for 3 min; and (iv) a 4°C hold. PCR primers were as follows: 5′-CTTAACTGTCCCACTGCCAAGAAACCGTCCCAGATC-3′, 5′-CTCTGGTCCCTCCCTGTTCCTGCACCAGGGACTGCCCAAAATTTC GCAAC-3′, 5′-ATCGCATCGAGCGAGCACGTACTCGGA-3′, and 5′-AGCTCTTCAGCAATATCACGGGTAGCC-3′. Serum used in the analysis of C3 deposition was prepared fresh each time. The mice were bled, and the blood was allowed to clot at room temperature for 20 min and then centrifuged at 12,000 × g for 5 min. Ten microliters of the serum was then used per reaction as described above.
RESULTS
The γHV68 gene 4 is homologous to other viral and murine members of the RCA gene family.
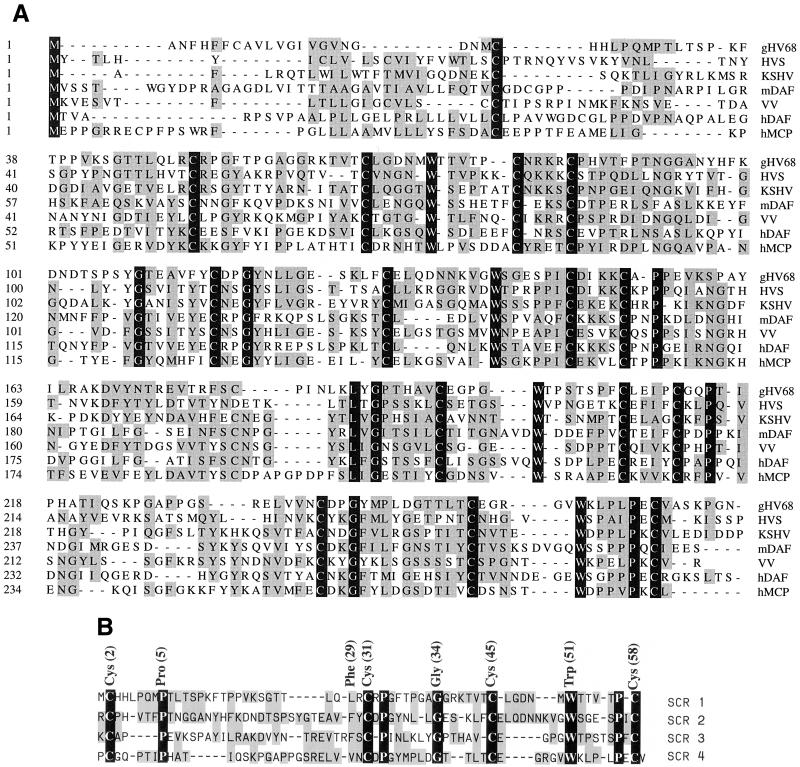
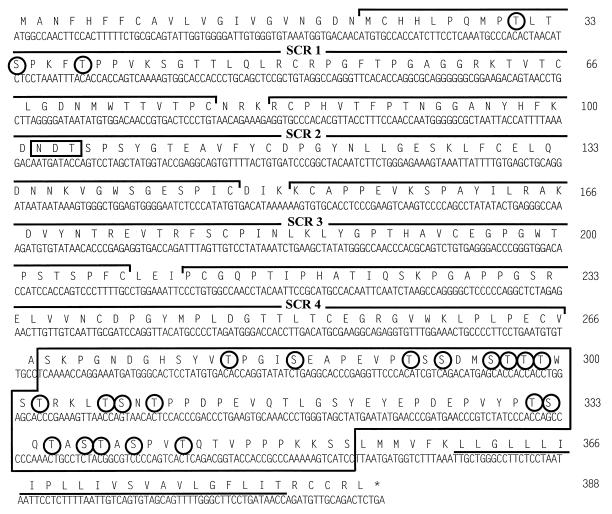
The predicted protein product of γHV68 gene 4 shows significant homology to members of the RCA family of proteins (Fig. 1) (65). The predicted 388-amino-acid γHV68 RCA protein consists of an N-terminal 21-amino-acid putative signal peptide and a 245-amino-acid region containing the four SCRs. This is followed by an 86-amino-acid region which is 32.5% serine and threonine (S/T-rich region) and a 23-amino-acid hydrophobic region (Fig. 2). The overall structure of the γHV68 RCA protein is strikingly homologous to the structure of human decay-accelerating factor (DAF) (39). The γHV68 RCA protein contains four SCRs (Fig. 1 and 2) and shows conservation of the key cysteines, tryptophans, and glycines within each SCR, similar to that seen in the viral and mammalian RCA family members (Fig. 1B). The first 275 amino acids of the predicted γHV68 RCA protein sequence were aligned with RCA family members from HVS, KSHV, and vaccinia virus, as well as with murine DAF and human membrane cofactor protein (MCP) (Fig. 1). The predicted γHV68 RCA protein shows 36 and 33% sequence similarity to the HVS CCPH and human DAF proteins, respectively. Both DAF and MCP are posttranslationally glycosylated. Human MCP contains an extensively O-glycosylated alternatively spliced S/T-rich region (39) which is homologous to the predicted γHV68 RCA protein S/T-rich region (Fig. 2). Analysis of the predicted γHV68 RCA protein was performed with an algorithm for predicting O-glycosylation sites (23a, 23b). Nineteen residues within the S/T-rich region and three residues in SCR1 scored as likely sites for O-linked glycosylation (Fig. 2). In addition to containing an S/T-rich region (9), human DAF has one N-linked complex-type oligosaccharide unit attached at asparagine residue 61 between the first and second SCRs. The predicted γHV68 RCA protein contains one potentially N-glycosylated residue at position 102 within the second SCR (Fig. 2).
FIG. 1.
Alignment of the predicted γHV68 RCA protein sequence with sequences of multiple RCA protein family members. (A) Sequence alignment of the γHV68 putative gene 4 product (gHV68) with RCA homologs from HVS (CCPH), vaccinia virus (VV), and KSHV (gene 4 protein), murine DAF (mDAF), human DAF (hDAF), and human MCP (hMCP). The first 275 amino acids of the gene 4 product, which include the four short consensus repeats, were aligned by using the DNASTAR MEGALIGN program. Amino acids with functional groups similar to those in the consensus sequence are shaded in grey. Identical amino acids are denoted by with white letters in black boxes. (B) Alignment of the four SCRs in the γHV68 RCA protein. Note the conservation of the four cysteines as well as the tryptophans, glycines, and prolines. Approximate positions of conserved amino acids (see Discussion) are signified by the SCR consensus above the sequence alignments of the four SCRs. The shadings are the same as for panel A.
FIG. 2.
Putative γHV68 RCA amino acid sequence. The 388-amino-acid sequence is shown in single-letter code above the DNA sequence. The four SCR regions are shown by the brackets above the amino acid sequence and designated SCR 1 to 4. The S/T-rich region is boxed and is followed by the C-terminal hydrophobic domain (underlined). Circled amino acids are potential sites for O-linked oligosaccharides. The box at amino acid 102 is a potential site for N-linked glycosylation. Since no O-glycosylation consensus sequence exists, the potential sites were determined by neural networks and weight matrix algorithms, based on sites already known to be O-glycosylated (Center for Biological Sequence Analysis).
Northern and 3′ RACE analyses demonstrate that gene 4 is transcribed as a late bicistronic transcript in infected NIH 3T12 murine fibroblasts.
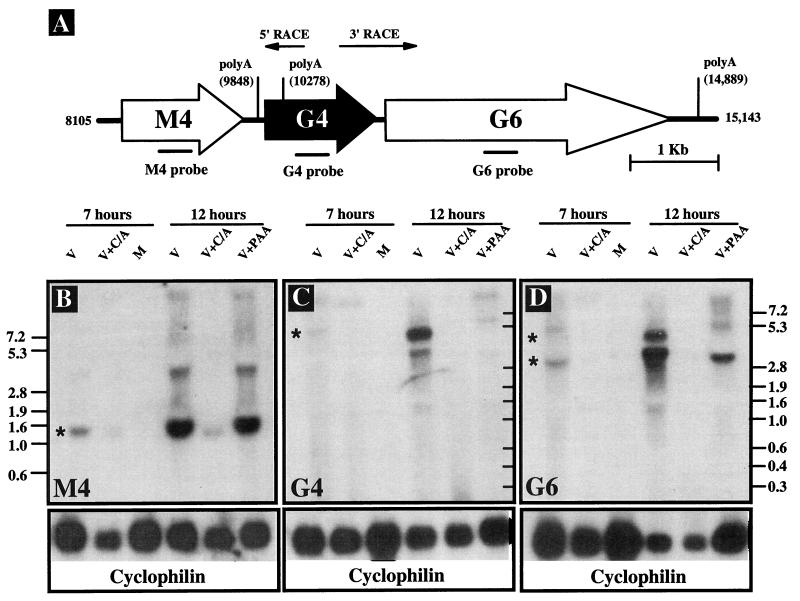
To determine whether γHV68 gene 4 was transcribed during infection, we performed Northern blot analysis on total RNA isolated from mock- or γHV68-infected murine NIH 3T12 fibroblasts. Cells were cultured in the presence or absence of viral DNA or protein synthesis inhibitors, and RNA was harvested at 7 and 12 h postinfection. Blots were probed with glycoprotein B (gB)- and gene 25 (major capsid protein)-specific probes to provide a positive control for PAA treatment. PAA inhibited expression of both gB and the major capsid protein (data not shown). Blots were probed for the cellular cyclophilin gene transcript as a loading control (Fig. 3B to D, lower panels). Using a gene 4-specific probe, we detected a transcript of approximately 5.2 kb (Fig. 3C). A second band running below the 5.2-kb band may represent an additional transcript or may be an artifact caused by the 28S rRNA band at this position. Gene 4 was transcribed as a late gene as shown by its sensitivity to both DNA and protein synthesis inhibitors (Fig. 3C). The presence of different amounts of RNA in the different lanes did not confound assignment of kinetic class (see the cyclophilin results in Fig. 3C). The gene 4 mRNA was significantly larger than the gene 4 ORF, demonstrating that the gene 4 mRNA included sequences either 5′ or 3′ to gene 4.
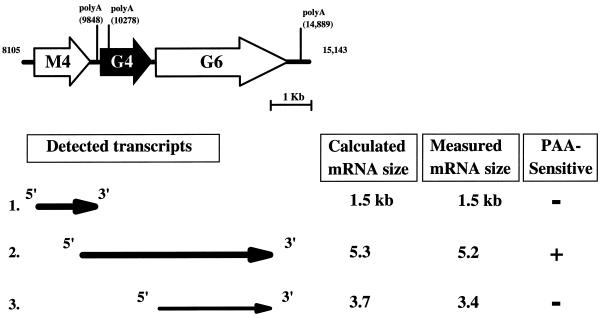
FIG. 3.
Northern blot analysis of gene 4 transcription. (A) Schematic of the genome region surrounding gene 4 (G4), with the known poly(A) sequences on the r strand labeled (AATAAA). Numbers correspond to the genomic sequence coordinates (65). The 5′ and 3′ RACE products are shown as arrows. (B, C, and D) Northern blots were probed with gene M4 (B)-, gene 4 (C)-, and gene 6 (D)-specific probes. Total RNA was harvested at 7 and 12 h postinfection from NIH 3T12 cells either mock (M) or γHV68 (V) infected at an MOI of 5 in the presence or absence of either a viral DNA synthesis inhibitor (PAA) or the protein synthesis inhibitors cycloheximide and anisomycin (C/A). Molecular size markers (in kilobase pairs) are shown to the left and right of Northern blot panels. ∗, transcripts referred to in the text. A probe to rat cyclophilin was used as a loading control. The data are representative of two independent experiments.
γHV68 gene 4 is flanked by two ORFs (Fig. 3A, gene M4 and gene 6). Gene M4 is predicted to encode a protein without clear homologs in the database, while gene 6 is predicted to encode the γHV68 single-stranded DNA binding protein (65). The absence of a polyadenylation sequence immediately downstream of gene 4 and the presence of polyadenylation signals downstream of gene 6 and gene M4 suggested that gene 4 might be part of a bicistronic transcript with gene 6 (Fig. 3A). We tested this hypothesis by using both further Northern analysis and 3′ RACE.
A gene 6 probe hybridized to a 5.2-kb transcript, consistent with gene 4 and gene 6 being part of a bicistronic mRNA (compare Fig. 3C and D). In addition, the gene 6 probe hybridized to a 3.4- to 3.7-kb transcript which was PAA insensitive and thus of the early class. To determine if sequences 5′ to gene 4 were contained in the mRNA, a gene M4-specific probe was used (Fig. 3B). The gene M4 probe hybridized to an abundant early transcript, as shown by its resistance to PAA but not cycloheximide (Fig. 3B). However, the gene M4 probe did not hybridize with the 5.2-kb mRNA detected with gene 4 and gene 6 probes (compare 3B, C, and D). This argues that the gene 4 transcript initiates downstream of gene M4 (see below).
To directly determine whether the gene 4 mRNA extends into the gene 6 ORF, 3′ RACE was performed (Fig. 3A). 3′ RACE generated multiple amplified products. We cloned and sequenced the most abundant product, which was approximately 800 bp in length. This 3′ RACE product extended from 283 bp upstream of the translation stop codon of gene 4 and across the 176 bp between the end of the gene 4 ORF and the beginning of the gene 6 ORF and ended 280 bp downstream of the putative gene 6 translation initiation codon. Since this product contained neither the RACE adapter sequence nor a poly(A) tail, it does not extend to the 3′ end of the gene 4 transcript. This is consistent with the large size of the mRNA, making it difficult to extend through the 3′ end of the mRNA using RACE. Further 3′ RACE analysis with downstream primers would be confounded by the presence of gene 6 transcripts, since primers would be complementary to both the bicistronic transcript containing gene 4 and gene 6 sequences and monocistronic gene 6 transcripts. However, the sequence of this RACE product confirms the prediction from Northern analysis (see above) that gene 4 mRNAs extend into gene 6, indicating that the γHV68 RCA protein (see below) is encoded on a bicistronic mRNA (Fig. 4, transcript 2).
FIG. 4.
Transcripts in the region surrounding gene 4. Calculated mRNA sizes (in kilobase pairs) were based on the actual sequence between potential TATA box sequences and poly(A) recognition sequences (AATAAA) in the γHV68 genome as analyzed with the Vector NTI version 4.0 Deluxe program (Informax Inc.). RNA molecular size markers, in logarithmic scale, were plotted as a function of distance (in centimeters) from the Northern blots, and measured mRNA sizes were extrapolated from those graphs. For gene 4 and gene 6 transcript sizes, the sizes from the independently probed blots were averaged. The data are from a single experiment.
Defining the 5′ end of the gene 4 transcript.
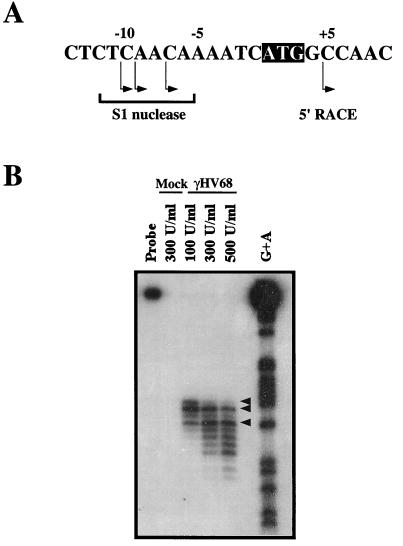
To further analyze the structure of the gene 4 transcript, we performed 5′ RACE and S1 nuclease protection assays. 5′ RACE, using a primer 382 bp 3′ of the first ATG in the gene 4 ORF (corresponding to bp 10223 of the γHV68 genome), gave rise to one predominant product. This product was sequenced, and it extended to a point 4 bp 3′ of the first ATG in the gene 4 ORF (Fig. 5A). While the failure to extend 5′ of the first ATG in the gene 4 ORF could be attributed to incomplete elongation or degradation, it was also possible that the gene 4 transcript initiated within the gene 4 ORF. For example, there is another potential translation start site 22 residues within the gene 4 ORF, located at the start of SCR 1 (Fig. 2). To further map the 5′ end of the gene 4 mRNA, we performed S1 nuclease protection analysis with an oligonucleotide probe complementary to the region of the transcript spanning the first ATG of the gene 4 ORF. Total RNA was isolated from either mock- or γHV68-infected NIH 3T12 cells at 22 h postinfection. The S1 probe did not hybridize to mock-infected RNA, demonstrating specificity of the signal detected from virus-infected cells (Fig. 5B). Analysis of the bands derived from the S1 protection assays showed that the gene 4 mRNA initiates ∼10 nucleotides 5′ of the first ATG in the gene 4 ORF (Fig. 5A). The smaller protected fragments observed upon digestion with high concentrations of S1 nuclease most likely reflect “nibbling” of the protected 3′ end of the probe (Fig. 5). This analysis demonstrates that the first ATG codon of the gene 4 ORF is present in the gene 4 transcript and is likely used to generate the γHV68 RCA protein containing the putative signal sequence (Fig. 2).
FIG. 5.
Identification of the γHV68 gene 4 transcriptional start site by S1 nuclease analysis. (A) Schematic representation of the genomic region surrounding the first ATG in the gene 4 ORF (shown as the black box with white letters). Arrows below the bases denote transcriptional start sites as deduced through S1 nuclease protection assay and 5′ RACE analysis. (B) The S1 oligonucleotide probe was incubated with total RNA isolated from mock- and γHV68-infected NIH 3T12 cells and various concentrations of S1 nuclease enzyme (100 to 500 U/ml). The G+A ladder for the probe is shown at the right. Arrowheads denote the major protected fragments (also indicated in panel A). The data are representative of three independent experiments.
Multiple isoforms of the γHV68 RCA protein in infected fibroblast cell cultures.
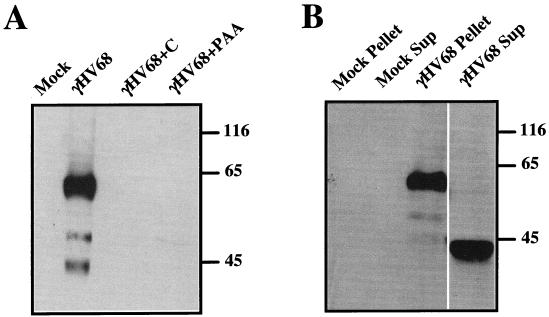
To detect expression of the γHV68 RCA protein, we performed Western blot analyses with rabbit polyclonal antiserum raised against bacterially expressed His-tagged γHV68 RCA fusion protein. The truncated γHV68 RCA protein used as an immunogen lacked the 21 N-terminal amino acids and the 33 C-terminal residues of the predicted γHV68 RCA protein but contained all four of the SCRs and the S/T-rich region. Three prominent bands of approximately 60 to 65, 50 to 55, and 40 to 45 kDa were detected in γHV68-infected, but not mock-infected, NIH 3T12 fibroblast cell lysates using the rabbit polyclonal antiserum (Fig. 6A). The absence of any detectable RCA protein in virally infected cells treated with either cycloheximide or PAA was consistent with Northern blot data demonstrating that the gene 4 transcript is of the late kinetic class (Fig. 6A and 3). No RCA protein was detected in cells infected with UV-inactivated virus or in virus-infected cell lysates probed with preimmune rabbit serum (data not shown). Since other viral RCA proteins have at least two isoforms (see Discussion), one soluble (released or secreted) and the other cell or membrane associated, we determined whether there were soluble forms of the γHV68 RCA protein in supernatants from γHV68-infected cells. A 40- to 45-kDa band was detected in supernatants of virally infected cells after high-speed centrifugation, while a prominent 60- to 65-kDa band was detected in pellets derived from high-speed centrifugation of infected-cell supernatants (Fig. 6B). No protein was detected in mock-infected supernatants or pellets (Fig. 6B) or when preimmune serum was used (data not shown). These data show that there are three predominant forms of the γHV68 RCA protein, one of which is either secreted or released from infected cells.
FIG. 6.
Identification of isoforms of the γHV68 RCA protein by Western blotting. (A) Western blot analysis of NIH 3T12 fibroblasts mock infected or infected with γHV68 in the presence or absence of cycloheximide or PAA. Total cell lysates were harvested at 24 h postinfection and run on sodium dodecyl sulfate–10% polyacrylamide gels. Western analysis was performed with rabbit polyclonal antiserum to bacterially expressed recombinant γHV68 RCA, followed by donkey anti-rabbit secondary antibody conjugated to horseradish peroxidase. (B) Western analysis of proteins present in supernatants or pellets derived from high-speed centrifugation of γHV68-infected or mock-infected 3T12 fibroblasts. Supernatants from cells either mock infected or γHV68 infected were collected at 34 h postinfection, filtered, concentrated 50-fold, spun at 100,000 × g for 3.5 h at 4°C, and loaded onto a 10% polyacrylamide gel. Soluble and pellet fractions from the spin are designated Sup and Pellet, respectively. Molecular mass markers (in kilodaltons) are shown to the right of each gel. In each case, no bands were detected when preimmune serum or UV-inactivated γHV68 was used. The data are representative of those from four independent experiments.
Cell surface expression of the γHV68 RCA protein.
To determine whether the γHV68 RCA protein is expressed on the surfaces of infected cells, we performed FACS analysis on infected and mock-infected cells with polyclonal anti-γHV68 RCA protein antibody. RCA protein was detected on γHV68-infected cell surfaces (Fig. 7). Preimmune serum did not bind significantly to virus-infected cells (Fig. 7A). In contrast, significant binding of polyclonal anti-γHV68 RCA protein antibody was detected on γHV68-infected but not mock-infected cells (Fig. 7B). The specificity of the staining for γHV68 RCA protein was confirmed by adding recombinant γHV68 RCA protein to the FACS staining reaction to compete for specific antibody binding to the infected-cell surface. Addition of 70 μg of recombinant γHV68 RCA protein resulted in a loss of FACS staining of infected cells (Fig. 7C). Addition of 70 μg of recombinant His-tagged γHV68 v-cyclin protein did not alter the FACS signal, providing a negative control (not shown). The recombinant γHV68 RCA protein was prepared under denaturing conditions, making it likely that only a small fraction of the total protein added was in a conformation capable of competing for binding of the immune serum to the native γHV68 RCA protein. No RCA protein expression was detected with UV-inactivated virus (not shown), suggesting that productive viral infection of NIH 3T12 cells was necessary for RCA protein expression on the surfaces of infected cells.
The γHV68 RCA protein inhibits murine and human complement activation.
We next determined whether the γHV68 RCA protein regulates complement activation, using a zymosan-based assay as previously described (19) and mouse serum as a source of complement. Zymosan is a yeast cell wall polysaccharide component known to activate complement. In this assay, activated zymosan is incubated with serum, and the deposition of C3 on the zymosan particle is determined by FACS analysis with anti-C3 antibody (Fig. 8). To provide a negative control, we took advantage of the fact that both the classical and alternative pathways require divalent cations (Ca2+ and Mg2+, respectively) and that addition of a Ca2+ and Mg2+ chelator, such as EDTA at a final concentration of 10 mM, abrogates both pathways.
FIG. 8.
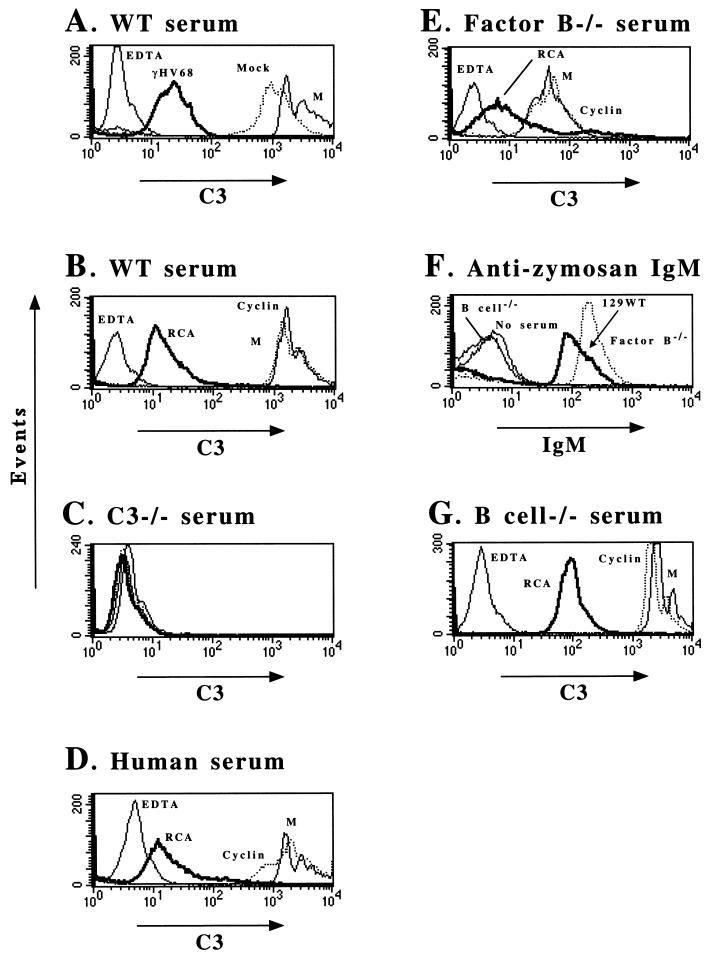
Inhibition of alternative and classical pathway-dependent C3 deposition on zymosan particles by infected cell supernatants or the recombinant γHV68 RCA protein. In all FACS analyses, EDTA addition was used as a negative control. Abbreviations: M, medium; RCA, recombinant γHV68 RCA protein; cyclin, recombinant γHV68 v-cyclin; WT, wild type. The data are representative of those from at least two independent experiments. (A) Inhibition of C3 on zymosan by virus-infected cell supernatant prepared as described in Materials and Methods. γHV68, supernatant from cells infected with γHV68; Mock, supernatant from mock-infected cells. (B) Inhibition of C3 deposition on zymosan by recombinant γHV68 RCA protein but not recombinant v-cyclin. (C) FACS staining for C3 after incubation of zymosan with serum from C3-deficient mice. (D) Inhibition of human complement activation by recombinant γHV68 RCA protein. (E) Analysis of inhibition of the classical pathway of complement activation by the γHV68 RCA protein by using serum lacking factor B as a source of complement. (F) Presence of zymosan-binding IgM in both 129 wild-type and factor B-deficient mice as detected by flow cytometric analysis with an anti-mouse IgM antibody. (G) Analysis of inhibition of the alternative pathway of complement activation by the γHV68 RCA with serum from B-cell-deficient mice as a source of complement.
We first analyzed supernatants harvested from either mock- or γHV68-infected 3T12 cells (Fig. 8A). In the presence of EDTA, no C3 deposition on the zymosan particles was detected (Fig. 8A). In contrast, in the presence of either medium alone or concentrated supernatant from mock-infected cells, significant C3 was deposited on the zymosan particles, as shown by the >100-fold shift in fluorescence intensity (Fig. 8A). Notably, concentrated cell supernatants from γHV68-infected cells inhibited C3 deposition significantly (ca. 50-fold) (Fig. 8A). To determine whether purified γHV68 RCA protein can inhibit complement activation and consequent C3 deposition, we added 70 μg of either purified recombinant γHV68 RCA protein or γHV68 v-cyclin protein to the zymosan assay. γHV68 v-cyclin protein served as a negative control protein purified by using the same protocol used to purify recombinant γHV68 RCA protein. Recombinant γHV68 RCA protein inhibited C3 deposition, as shown by a >100-fold decrease in fluorescence (Fig. 8B), while recombinant γHV68 v-cyclin had no effect (Fig. 8B). To determine whether the zymosan assay was dependent on the presence of C3 in serum, we repeated the assay with serum derived from C3-deficient mice. As expected, no C3 deposition was detected (Fig. 8C). Similar experiments using fresh human serum demonstrated that recombinant γHV68 RCA protein also inhibited deposition of human C3 on activated zymosan (Fig. 8D).
Recombinant γHV68 RCA protein inhibits C3 deposition via both classical and alternative pathways.
To determine whether the soluble γHV68 RCA protein regulated C3 deposition by the classical pathway, the zymosan assay was performed with serum from factor B-deficient mice (45). Factor B is required for the activity of the alternative pathway of complement activation. As expected, C3 deposition on activated zymosan was decreased in factor B-deficient serum compared to serum from normal mice (compare Fig. 8A and E). Recombinant RCA protein inhibited C3 deposition from factor B-deficient serum (Fig. 8E), consistent with the γHV68 RCA protein inhibiting the classical pathway of complement activation. As a negative control, recombinant γHV68 v-cyclin protein had no effect on C3 deposition from factor B-deficient serum (Fig. 8E).
We interpreted C3 deposition in the presence of factor B-deficient serum to reflect activity of the classical pathway of complement activation. However, for the classical pathway to play a role in deposition of complement on zymosan particles, antibody in factor B-deficient serum must bind to zymosan particles. We tested this prediction by incubating zymosan particles with normal serum, factor B-deficient serum, or serum from B-cell-deficient (and therefore antibody-deficient) mice and detected antibody binding to zymosan particles by FACS staining with a secondary antibody specific for mouse IgM (Fig. 8F). This analysis demonstrated that serum derived from both factor B-deficient and normal 129 mice contained zymosan-binding natural antibodies (Fig. 8F). The binding we detected was specific for mouse antibody, since serum from B-cell-deficient mice did not show significant binding to zymosan particles (Fig. 8F, compare B cell−/− and no-serum controls). These data show that C3 deposition on zymosan particles in the presence of factor B-deficient serum can reasonably be attributed to the activity of the classical pathway, and they support the conclusion that the γHV68 RCA protein can inhibit the activity of the classical pathway of complement activation (Fig. 8E).
The lack of antibody that bound zymosan in serum from B-cell-deficient mice allowed us to determine the effect of the γHV68 RCA protein on the alternative pathway independent of the classical pathway (Fig. 8G). Recombinant γHV68 RCA protein inhibited C3 deposition from serum derived from B-cell-deficient mice (Fig. 8G, compare RCA to cyclin). Together, these data show that the γHV68 RCA protein inhibited complement activation in both human and mouse sera and that both the mouse alternative and classical pathways of complement activation were targets for the inhibitory activity of this protein.
DISCUSSION
Murine γHV68 gene 4 is a bone fide gene encoding a complement-regulatory protein of the RCA protein family. Gene 4 is expressed as a bicistronic late mRNA, and there are multiple isoforms of the γHV68 RCA protein, including a membrane-bound form and a soluble form. The γHV68 RCA protein inhibits the activation of murine complement via both the classical and alternative pathways. Thus, the γHV68 RCA protein joins the HVS CCPH as a gammaherpesvirus RCA protein that inhibits complement activation (3, 4, 18).
Transcriptional programs used by gammaherpesviruses to express RCA proteins.
Transcript analysis revealed that gene 4 is transcribed as a 5.2-kb bicistronic mRNA, which also contains sequences from the downstream gene 6 (Fig. 4, transcript 2), as predicted by the lack of a poly(A) signal downstream of gene 4 (65). Northern blot analysis with a gene 6-specific probe also revealed the presence of a smaller, 3.7-kb transcript which was PAA resistant and thus of the early kinetic class (Fig. 4, transcript 3). This early mRNA is most likely the transcript that encodes the single-stranded DNA binding protein. Transcription of the HVS CCPH gene is quite different, despite the homology of the encoded proteins. HVS gene 4 encodes both a 1.7-kb transcript and a 1.5-kb transcript, the shorter of which arises due to alternative splicing of the gene, giving rise to a secreted form as well as a membrane-bound form of the protein (3). One important difference between the γHV68 and HVS genomes is the presence of an ORF (ORF 5) in HVS between gene 4 and gene 6 (4). This ORF is lacking in γHV68 (65), bringing gene 4 and gene 6 into close proximity. Interestingly, although the transcriptions of the γHV68 gene and the HVS gene differ significantly, in both cases multiple isoforms of the RCA protein are expressed, one of which is soluble and one of which is membrane bound. This suggests that both forms have important functions and raises the possibility that the KSHV gene 4 also encodes at least two isoforms of the RCA protein.
Isoforms of the γHV68 RCA protein.
Western blot analysis revealed multiple isoforms of γHV68 RCA antibody-reactive proteins in infected NIH 3T12 fibroblasts. Although the calculated molecular mass of the full-length, 388-amino-acid γHV68 RCA protein is ca. 43 kDa, Western blot analysis revealed one isoform of 60 to 65 kDa. This is consistent with the structure of murine DAF, which is initially synthesized as a precursor of 46 kDa, but gives rise to a membrane-associated form of 70 to 80 kDa (39). The increase in size of the mature DAF protein has been shown to be due to the presence of N- and O-linked glycosylation, corresponding to an addition of 4 and 26 kDa respectively (9, 42). The S/T-rich region in human DAF is extensively O glycosylated, has been shown to be critical for stable surface expression of DAF, and also acts as a nonspecific spacer to project the SCRs above the cell surface (39). Human MCP also contains an extensively O-glycosylated S/T-rich region (39). The presence of high-molecular-weight forms of the γHV68 RCA protein, combined with the presence of an S/T-rich region in the γHV68 RCA protein sequence that contains many potential sites for O-linked glycosylation, strongly suggests that the processing of the γHV68 RCA protein involves glycosylation. Thus, it seems likely that the multiple species of the γHV68 RCA protein detected on Western blots of cell lysates (Fig. 6A) reflect, at least in part, different glycosylated forms of the protein. The 50- to 55-kDa band was detected only in infected cells, raising the possibility that it is a precursor of the 60- to 65-kDa protein. Further analyses of precursor-product relationships are needed to clarify the relationships between the observed γHV68 RCA protein species.
How the soluble and membrane-bound isoforms of the γHV68 RCA protein are generated is not clear. The two forms of the HVS CCPH are generated by alternative splicing in the 3′ portion of the gene 4 transcript (3). It is possible that there are alternatively spliced forms of the γHV68 gene 4 transcript that were not differentiated on Northern blots due to the large size (5.2 kb) of the γHV68 gene 4 mRNA. Further analysis of the structure of the γHV68 gene 4 mRNA in the region corresponding to the 3′ end of the γHV68 RCA protein ORF will be required to address this possibility. An alternative possibility is that the γHV68 RCA soluble form is generated by proteolysis of the membrane-bound form. A precedent for release of a soluble viral RCA protein from a membrane-bound form is provided by the poxvirus B5R (or ps/hr) protein, which contains four SCRs as well as transmembrane and cytoplasmic domains (15, 62). One product of the B5R gene is a 42-kDa protein found in the extracellular enveloped form of poxviruses but not in the intracellular mature form (15, 25, 44, 62). A 38-kDa soluble form is detected in the supernatant of infected cells (43, 44). This soluble form is likely generated by cleavage between SCR 4 and the membrane, and the efficiency of this cleavage is regulated by SCRs 3 and 4 (43, 44). It is possible, therefore, that the soluble form of the γHV68 RCA protein is generated by cleavage of the membrane-bound form.
Mechanism of complement regulation by the γHV68 RCA protein.
The γHV68 RCA protein inhibited complement activation as shown by the capacity of the recombinant γHV68 RCA protein to inhibit C3 deposition from either mouse or human serum on zymosan particles. Supernatant from infected cells, but not mock-infected cells, was also able to inhibit murine complement activation. Whether the cell surface-associated γHV68 RCA protein, as detected by flow cytometric analysis (Fig. 7), regulates C3 deposition is still unknown and is currently being addressed. The structure of the γHV68 RCA protein is consistent with its role as a complement regulator. As a general rule, SCRs in RCA proteins exhibit a consensus amino acid sequence with the following approximate positions: Cys2, Pro5, Tyr/Phe29, Cys31, Gly34, Cys45, Trp51, Ala/Pro56, and Cys58 (26, 39, 52). Alignment of the four SCRs present in the γHV68 RCA protein showed conservation of all cysteines in each SCR, as well as most of the other SCR consensus residues (Fig. 1B). The conservation of SCR structure, together with data demonstrating that the γHV68 RCA protein inhibits both classical and alternative pathways, argues that the γHV68 RCA protein will inhibit complement activation via the same mechanisms used by other host and viral RCA proteins.
The fact that the γHV68 RCA protein inhibits activation of both mouse and human complement suggests that a highly conserved component of the complement system is targeted by this protein. Since both the classical and alternative pathways of complement activation are inhibited by the γHV68 RCA protein, the protein likely targets a component common to the two pathways. Activity against both pathways is a common property of viral regulators of complement activation. For example, the vaccinia virus RCA protein VCP inhibits both alternative and classical pathways of complement activation (27, 35, 36, 46, 55), as does the herpes simplex virus (HSV) gC protein (20, 21, 24, 41, 47). This likely indicates that both the classical and alternative pathways of complement activation have important effects on virus infection, although this has not been directly tested in vivo.
Since the γHV68 RCA protein decreased C3 deposition on zymosan particles, our data are consistent with the γHV68 RCA protein acting at the level of C3 or before C3 in the complement cascade. Together with the close structural homology with other RCA proteins, which in general target C3 convertases, the data are most consistent with the γHV68 RCA protein inhibiting the function of both the classical and alternative pathway C3 convertases. There are two main mechanisms by which RCA molecules regulate activation of C3 (1, 39). First, degradation of the activated fragment of C3, C3b, is mediated by factor I. Factor I is a serine protease that cleaves C3b efficiently, but only in the presence of protein cofactors. Factor I also cleaves C4b, the activated fragment of C4, by a similar mechanism. This C4b inactivation is also an important mechanism by which the RCA proteins regulate the activation of the classical pathway of complement. Second, certain complement-regulatory proteins prevent the formation, or accelerate the dissociation, of the classical and alternative C3 convertase enzyme complex by a process known as decay acceleration. Many host proteins exhibit either decay-accelerating or cofactor activity. A notable exception is the mouse Crry protein, which exhibits both activities (19, 33). In addition, the vaccinia virus VCP protein, which like the γHV68 RCA protein has four SCRs (27, 35, 36), also acts by both decay-accelerating and cofactor activities (35, 46). The precise mechanism by which VCP acts is better detailed than mechanisms for other poxvirus complement regulators, with VCP acting by different mechanisms than either factor H or complement receptor 1 (55). While our data are consistent with the γHV68 RCA protein acting at the level of C3 convertases, the precise mechanism (decay acceleration activity, cofactor activity, or both) remains to be determined.
Function of the γHV68 RCA protein.
The most attractive hypothesis is that the γHV68 RCA protein regulates complement activation by virions or virus-infected cells and thus prevents complement from controlling γHV68 infection. HSV type 1 and type 2 glycoproteins gC-1 and gC-2 bind C3b and inhibit complement-mediated neutralization of virus (20, 21, 24, 41, 47). Similar to HSV gC, VCP inhibits complement-enhanced antibody-dependent neutralization of vaccinia virus (27). Both the HVS CCPH and HSV gC protect cells from lysis by antibody plus complement (18, 24). Thus, expression of the γHV68 RCA protein could alter γHV68 pathogenesis by protecting virions and/or virus-infected cells from complement-mediated damage.
While regulation of complement is an attractive hypothesis for the role of the γHV68 RCA protein, studies of other viral RCA proteins or complement-regulatory proteins provide precedents for γHV68 RCA protein having other functions. For example, the poxvirus B5R protein contains four SCRs and is essential for virion morphogenesis (12, 16, 25, 28, 43, 44, 62, 72). The SCR-containing portion of B5R is not essential for virion morphogenesis (25, 32, 44). Interestingly, the B5R protein is also involved in polymerization of actin on virions in infected cells (44). In addition to the complement-regulatory functions of HSV gC, gC may have additional roles in HSV infection via interactions with molecules such as heparan sulfate (37) or attachment to polarized epithelial cells (41, 56), although the role in binding has not been confirmed for all strains of HSV (23, 41).
Studies of host RCA proteins provide further precedents for complement-independent functions of RCA family members. MCP has alternatively spliced forms with different cytoplasmic domains (39, 40), suggesting a signaling function. MCP interacts with intracellular kinases in macrophages (73), and signaling via MCP can down-regulate the immunomodulatory protein interleukin-12 (31). Similarly, DAF is glycosylphosphatidylinositol linked, and DAF (11, 30) and other glycosylphosphatidylinositol-linked proteins can generate intracellular signals when cross-linked (reviewed in references 8 and 54). These studies raise the possibility that the γHV68 RCA protein functions via induction of intracellular signals. It has furthermore been argued that DAF and/or MCP plays a role in regulation of cell sensitivity to killing by NK cells (17, 57). Since NK cells are important in herpesvirus resistance, it is possible that the γHV68 RCA protein regulates NK cell activity in addition to complement.
The specific role that the γHV68 RCA protein plays in pathogenesis and whether this role is partly or completely dependent on regulation of complement remain to be determined. Interesting in this regard is a recent study using HSV gC mutant viruses which argues for both C3-dependent and C3-independent effects of gC on HSV pathogenesis (41). We expect that these issues can be addressed by generating γHV68 RCA protein mutants and analyzing their pathogenesis in normal mice and mice deficient in C3.
ACKNOWLEDGMENTS
H.W.V. was supported by grant RPG-97-134-01-MBC from the American Cancer Society and NIH grants RO1 CA74730, RO1 HL60090, and RO1AI39616. S.H.S. was supported by NIH grants RO1 CA43143, RO1 CA52004, RO1 CA58524, and RO1 CA74730. H.M. was supported by a Veteran’s Administration Merit Award, an Arthritis Foundation Arthritis Investigator Award, and NIH grant RO1 40576.
We thank Robin Lorenz for generously supplying reagents. We also thank John Atkinson and the members of the laboratories of H.W.V., S.H.S., David Leib, and Lynda Morrison for continued commentary on this project.
REFERENCES
- 1.Abbas A K, Lichtman A H, Pober J S. Cellular and molecular immunology. Philadelphia, Pa: W. B. Saunders; 1991. pp. 294–316. [Google Scholar]
- 2.Adams E M, Brown M C, Nunge M, Krych M, Atkinson J P. Contribution of the repeating domains of membrane cofactor protein (CD46) of the complement system to ligand binding and cofactor activity. J Immunol. 1991;147:3005–3011. [PubMed] [Google Scholar]
- 3.Albrecht J-C, Fleckenstein B. New member of the multigene family of complement control proteins in herpesvirus saimiri. J Virol. 1992;66:3937–3940. doi: 10.1128/jvi.66.6.3937-3940.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albrecht J C, Fleckenstein B. New member of the multigene family of complement control proteins in herpesvirus saimiri. J Virol. 1992;66:3937–3940. doi: 10.1128/jvi.66.6.3937-3940.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaskovic D, Stancekova M, Svobodova J, Mistrikova J. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 1980;24:468–468. [PubMed] [Google Scholar]
- 7.Blaskovic D, Stanekova D, Rajcani J. Experimental pathogenesis of murine herpesvirus in newborn mice. Acta Virol. 1984;28:225–231. [PubMed] [Google Scholar]
- 8.Brown D. The tyrosine kinase connection: how GPI-anchored proteins activate T cells. Curr Opin Immunol. 1993;5:349–354. doi: 10.1016/0952-7915(93)90052-t. [DOI] [PubMed] [Google Scholar]
- 9.Coyne K E, Hall S E, Thompson S, Arce M A, Kinoshita T, Fujita T, Anstee D J, Rosse W, Lublin D M. Mapping of epitopes, glycosylation sites, and complement regulatory domains in human decay accelerating factor. J Immunol. 1992;149:2906–2913. [PubMed] [Google Scholar]
- 10.Danielson P E, Forss-Petter S, Brow M A, Calavetta L, Douglass J, Milner R J, Sutcliffe J G. p1B15: a cDNA clone of the rat mRNA encoding cyclophilin. DNA. 1988;7:261–267. doi: 10.1089/dna.1988.7.261. [DOI] [PubMed] [Google Scholar]
- 11.Davis L S, Patel S S, Atkinson J P, Lipsky P E. Decay-accelerating factor functions as a signal transducing molecule for human T cells. J Immunol. 1988;141:2246–2252. [PubMed] [Google Scholar]
- 12.del Mar L, Herrera E, Blasco R, Isaacs S N. Functional analysis of vaccinia virus B5R protein: role of the cytoplasmic tail. Virology. 1998;252:450–457. doi: 10.1006/viro.1998.9483. [DOI] [PubMed] [Google Scholar]
- 13.Efstathiou S, Ho Y M, Hall S, Styles C J, Scott S D, Gompels U A. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J Gen Virol. 1990;71:1365–1372. doi: 10.1099/0022-1317-71-6-1365. [DOI] [PubMed] [Google Scholar]
- 14.Efstathiou S, Ho Y M, Minson A C. Cloning and molecular characterization of the murine herpesvirus 68 genome. J Gen Virol. 1990;71:1355–1364. doi: 10.1099/0022-1317-71-6-1355. [DOI] [PubMed] [Google Scholar]
- 15.Engelstad M, Howard S T, Smith G L. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- 16.Engelstad M, Smith G L. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 17.Finberg R W, White W, Nicholson-Weller A. Decay-accelerating factor expression on either effector or target cells inhibits cytotoxicity by human natural killer cells. J Immunol. 1992;149:2055–2060. [PubMed] [Google Scholar]
- 18.Fodor W L, Rollins S A, Bianco-Caron S, Rother R P, Guilmette E R, Burton W V, Albrecht J-C, Fleckenstein B, Squinto S P. The complement control protein homolog of herpesvirus saimiri regulates serum complement by inhibiting C3 convertase activity. J Virol. 1995;69:3889–3892. doi: 10.1128/jvi.69.6.3889-3892.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foley S, Li B, Dehoff M, Molina H, Holers V M. Mouse Crry/p65 is a regulator of the alternative pathway of complement activation. Eur J Immunol. 1993;23:1381–1384. doi: 10.1002/eji.1830230630. [DOI] [PubMed] [Google Scholar]
- 20.Friedman H M, Wang L, Fishman N O, Lambris J D, Eisenberg R J, Cohen G H, Lubinski J. Immune evasion properties of herpes simplex virus type 1 glycoprotein gC. J Virol. 1996;70:4253–4260. doi: 10.1128/jvi.70.7.4253-4260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fries L F, Friedman H M, Cohen G H, Eisenberg R J, Hammer C H, Frank M M. Glycoprotein C of herpes simplex virus 1 is an inhibitor of the complement cascade. J Immunol. 1986;137:1636–1641. [PubMed] [Google Scholar]
- 22.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi’s sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths A, Renfrey S, Minson T. Glycoprotein C-deficient mutants of two strains of herpes simplex virus type 1 exhibit unaltered adsorption characteristics on polarized or non-polarized cells. J Gen Virol. 1998;79:807–812. doi: 10.1099/0022-1317-79-4-807. [DOI] [PubMed] [Google Scholar]
- 23a.Hansen, J. 4 August 1998 revision date. [Online.] Center for Biological Sequence Analysis, The Technical University of Denmark, Langby, Denmark. http://NetOglyc@cbs.dtu.dk. [28 June 1999, last date accessed.]
- 23b.Hansen J E, Lund O, Tolstrup N, Gooley A A, Williams K L, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconjugate J. 1998;15:115–130. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- 24.Harris S L, Frank I, Yee A, Cohen G H, Eisenberg R J, Friedman H M. Glycoprotein C of herpes simplex virus type 1 prevents complement-mediated cell lysis and virus neutralization. J Infect Dis. 1990;162:331–337. doi: 10.1093/infdis/162.2.331. [DOI] [PubMed] [Google Scholar]
- 25.Herrera E, del Mar L M, Blasco R, Isaacs S N. Functional analysis of vaccinia virus B5R protein: essential role in virus envelopment is independent of a large portion of the extracellular domain. J Virol. 1998;72:294–302. doi: 10.1128/jvi.72.1.294-302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hourcade D, Holers V M, Atkinson J P. The regulators of complement activation (RCA) gene cluster. Adv Immunol. 1989;45:381–416. doi: 10.1016/s0065-2776(08)60697-5. [DOI] [PubMed] [Google Scholar]
- 27.Isaacs S N, Kotwal G J, Moss B. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc Natl Acad Sci USA. 1992;89:628–632. doi: 10.1073/pnas.89.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isaacs S N, Wolffe E J, Payne L G, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung J U, Stager M, Desrosiers R C. Virus-encoded cyclin. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kammer G M, Walter E I, Medof M E. Association of cytoskeletal re-organization with capping of the complement decay-accelerating factor on T lymphocytes. J Immunol. 1988;141:2924–2928. [PubMed] [Google Scholar]
- 31.Karp C L, Wysocka M, Wahl L M, Ahearn J M, Cuomo P J, Sherry B, Trinchieri G, Griffin D E. Mechanism of suppression of cell-mediated immunity by measles virus. Science. 1996;273:228–231. doi: 10.1126/science.273.5272.228. . (Erratum, 275:1053, 1997.) [DOI] [PubMed] [Google Scholar]
- 32.Katz E, Wolffe E J, Moss B. The cytoplasmic and transmembrane domains of the vaccinia virus B5R protein target a chimeric human immunodeficiency virus type 1 glycoprotein to the outer envelope of nascent vaccinia virions. J Virol. 1997;71:3178–3187. doi: 10.1128/jvi.71.4.3178-3187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y U, Kinoshita T, Molina H, Hourcade D, Seya T, Wagner L M, Holers V M. Mouse complement regulatory protein Crry/p65 uses the specific mechanisms of both human decay-accelerating factor and membrane cofactor protein. J Exp Med. 1995;181:151–159. doi: 10.1084/jem.181.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 35.Kotwal G J, Isaacs S N, McKenzie R, Frank M M, Moss B. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science. 1990;250:827–830. doi: 10.1126/science.2237434. [DOI] [PubMed] [Google Scholar]
- 36.Kotwal G J, Moss B. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature. 1988;335:176–178. doi: 10.1038/335176a0. [DOI] [PubMed] [Google Scholar]
- 37.Laquerre S, Argnani R, Anderson D B, Zucchini S, Manservigi R, Glorioso J C. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J Virol. 1998;72:6119–6130. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M, Lee H, Yoon D-W, Albrecht J-C, Fleckenstein B, Neipel F, Jung J U. Kaposi’s sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liszewski M K, Farries T C, Lublin D M, Rooney I, Atkinson J P. Control of the complement system. Adv Immunol. 1996;61:201–283. doi: 10.1016/s0065-2776(08)60868-8. [DOI] [PubMed] [Google Scholar]
- 40.Liszewski M K, Tedja I, Atkinson J P. Membrane cofactor protein (CD46) of complement. Processing differences related to alternatively spliced cytoplasmic domains. J Biol Chem. 1994;269:10776–10779. [PubMed] [Google Scholar]
- 41.Lubinski J M, Wang L, Soulika A M, Burger R, Wetsel R A, Colten H, Cohen G H, Eisenberg R J, Lambris J D, Friedman H M. Herpes simplex virus type 1 glycoprotein gC mediates immune evasion in vivo. J Virol. 1998;72:8257–8263. doi: 10.1128/jvi.72.10.8257-8263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lublin D M, Krsek-Staples J, Pangburn M K, Atkinson J P. Biosynthesis and glycosylation of the human complement regulatory protein decay-accelerating factor. J Immunol. 1986;137:1629–1635. [PubMed] [Google Scholar]
- 43.Martinez-Pomares L, Stern R J, Moyer R W. The ps/hr gene (B5R open reading frame homolog) of rabbitpox virus controls pock color, is a component of extracellular enveloped virus, and is secreted into the medium. J Virol. 1993;67:5450–5462. doi: 10.1128/jvi.67.9.5450-5462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathew E, Sanderson C M, Hollinshead M, Smith G L. The extracellular domain of vaccinia virus protein B5R affects plaque phenotype, extracellular enveloped virus release, and intracellular actin tail formation. J Virol. 1998;72:2429–2438. doi: 10.1128/jvi.72.3.2429-2438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumoto M, Fukuda W, Circolo A, Goellner J, Strauss-Schoenberger J, Wang X, Fujita S, Hidvegi T, Chaplin D D, Colten H R. Abrogation of the alternative complement pathway by targeted deletion of murine factor B. Proc Natl Acad Sci USA. 1997;94:8720–8725. doi: 10.1073/pnas.94.16.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenzie R, Kotwal G J, Moss B, Hammer C H, Frank M M. Regulation of complement activity by vaccinia virus complement-control protein. J Infect Dis. 1992;166:1245–1250. doi: 10.1093/infdis/166.6.1245. [DOI] [PubMed] [Google Scholar]
- 47.McNearney T A, Odell C, Holers V M, Spear P G, Atkinson J P. Herpes simplex virus glycoproteins gC-1 and gC-2 bind to the third component of complement and provide protection against complement-mediated neutralization of viral infectivity. J Exp Med. 1987;166:1525–1535. doi: 10.1084/jem.166.5.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mistrikova J, Blaskovic D. Ecology of the murine alphaherpesvirus and its isolation from lungs of rodents in cell culture. Acta Virol. 1985;29:312–317. [PubMed] [Google Scholar]
- 49.Molina H, Perkins S J, Guthridge J, Gorka J, Kinoshita T, Holers V M. Characterization of a complement receptor 2 (CR2, CD21) ligand binding site for C3. An initial model of ligand interaction with two linked short consensus repeat modules. J Immunol. 1995;154:5426–5435. [PubMed] [Google Scholar]
- 50.Puglielli M T, Woisetschlaeger M, Speck S H. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J Virol. 1996;70:5758–5768. doi: 10.1128/jvi.70.9.5758-5768.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajcani J, Blaskovic D, Svobodova J, Ciampor F, Huckova D, Stanekova D. Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol. 1985;29:51–60. [PubMed] [Google Scholar]
- 52.Reid K B, Bentley D R, Campbell R D, Chung L P, Sim R B, Kristensen T, Tak B F. Complement system proteins which interact with C3b or C4b. Immunol Today. 1986;7:230–234. doi: 10.1016/0167-5699(86)90110-6. [DOI] [PubMed] [Google Scholar]
- 53.Reid K B, Day A J. Structure-function relationships of the complement components. Immunol Today. 1989;10:177–180. doi: 10.1016/0167-5699(89)90317-4. . (Erratum, 10:217.) [DOI] [PubMed] [Google Scholar]
- 54.Robinson P J. Signal transduction via GPI-anchored membrane proteins. Adv Exp Med Biol. 1997;419:365–370. doi: 10.1007/978-1-4419-8632-0_48. [DOI] [PubMed] [Google Scholar]
- 55.Sahu A, Isaacs S N, Soulika A M, Lambris J D. Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b1 inactivates the alternative complement pathway. J Immunol. 1998;160:5596–5604. [PubMed] [Google Scholar]
- 56.Sears A E, McGwire B S, Roizman B. Infection of polarized MDCK cells with herpes simplex virus 1: two asymmetrically distributed cell receptors interact with different viral proteins. Proc Natl Acad Sci USA. 1991;88:5087–5091. doi: 10.1073/pnas.88.12.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seya T, Kojima A, Hara T, Hazeki K, Sugita Y, Akedo H. Enhancement of lymphocyte-mediated K562 cytotoxicity by antibodies against complement membrane cofactor protein (CD46) and decay-accelerating factor (CD55) Immunobiology. 1991;183:115–124. doi: 10.1016/S0171-2985(11)80191-9. [DOI] [PubMed] [Google Scholar]
- 58.Simas J P, Swann D, Bowden R, Efstathiou S. Analysis of murine gammaherpesvirus-68 transcription during lytic and latent infection. J Gen Virol. 1999;80:75–82. doi: 10.1099/0022-1317-80-1-75. [DOI] [PubMed] [Google Scholar]
- 59.Stewart J P, Usherwood E J, Ross A, Dyson H, Nash T. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J Exp Med. 1998;187:1941–1951. doi: 10.1084/jem.187.12.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sunil-Chandra N P, Efstathiou S, Nash A A. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J Gen Virol. 1992;73:3275–3279. doi: 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- 61.Swanton C, Mann D J, Fleckenstein B, Neipel F, Peters G, Jones N. Herpes viral cyclin/Cdk6 complexes evade inhibition by CDK inhibitor proteins. Nature. 1997;390:184–187. doi: 10.1038/36606. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi-Nishimaki F, Funahashi S, Miki K, Hashizume S, Sugimoto M. Regulation of plaque size and host range by a vaccinia virus gene related to complement system proteins. Virology. 1991;181:158–164. doi: 10.1016/0042-6822(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 63.Usherwood E J, Stewart J P, Robertson K, Allen D J, Nash A A. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J Gen Virol. 1996;77:2819–2825. doi: 10.1099/0022-1317-77-11-2819. [DOI] [PubMed] [Google Scholar]
- 64.Van Dyk L F, Hess J L, Katz J D, Jacoby M, Speck S H, Virgin H W. The murine gammaherpesvirus 68 v-cyclin is an oncogene that promotes cell cycle progression in primary lymphocytes. J Virol. 1999;73:5110–5122. doi: 10.1128/jvi.73.6.5110-5122.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Virgin H W, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Virgin H W, Presti R M, Li X-Y, Liu C, Speck S H. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J Virol. 1999;73:2321–2332. doi: 10.1128/jvi.73.3.2321-2332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weck K E, Barkon M L, Yoo L I, Speck S H, Virgin H W. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J Virol. 1996;70:6775–6780. doi: 10.1128/jvi.70.10.6775-6780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weck K E, Dal Canto A J, Gould J D, O’Guin A K, Roth K A, Saffitz J E, Speck S H, Virgin H W. Murine gammaherpesvirus 68 causes large vessel arteritis in mice lacking interferon-gamma responsiveness: a new model for virus induced vascular disease. Nat Med. 1997;3:1346–1353. doi: 10.1038/nm1297-1346. [DOI] [PubMed] [Google Scholar]
- 69.Weck K E, Kim S S, Speck S H. B cells regulate murine gammaherpesvirus 68 latency. J Virol. 1999;73:4651–4661. doi: 10.1128/jvi.73.6.4651-4661.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weck K E, Kim S S, Virgin IV H W, Speck S H. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J Virol. 1999;73:3273–3283. doi: 10.1128/jvi.73.4.3273-3283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woisetschlaeger M, Strominger J L, Speck S H. Mutually exclusive use of viral promoters in Epstein-Barr virus latently infected lymphocytes. Proc Natl Acad Sci USA. 1989;86:6498–6502. doi: 10.1073/pnas.86.17.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. . (Erratum, 67:5709–5711.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wong T C, Yant S, Harder B J, Korte-Sarfaty J, Hirano A. The cytoplasmic domains of complement regulatory protein CD46 interact with multiple kinases in macrophages. J Leukoc Biol. 1997;62:892–900. doi: 10.1002/jlb.62.6.892. [DOI] [PubMed] [Google Scholar]
- 74.Yoo L I, Mooney M, Puglielli M T, Speck S H. B-cell lines immortalized with an Epstein-Barr virus mutant lacking the Cp EBNA2 enhancer are biased toward utilization of the oriP-proximal EBNA gene promoter Wp1. J Virol. 1997;71:9134–9142. doi: 10.1128/jvi.71.12.9134-9142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]