Abstract
Recently, gene therapy vectors based upon the human immunodeficiency virus type 1 (HIV-1) genome have been developed. Here, we create an HIV-1 vector which is defective for all HIV-1 genes, but which maintains cis-acting elements required for efficient packaging, infection, and expression. In T cells transduced by this vector, vector expression is low but efficiently induced following HIV-1 infection. Remarkably, although the HIV-1 vector does not contain specific anti-HIV-1 therapeutic genes, the presence of the vector alone is sufficient to inhibit the spread of HIV-1 infection. The mechanism of inhibition is likely to be at the level of competition for limiting substrates required for either efficient packaging or reverse transcription, thereby selecting against propagation of wild-type HIV-1. These results provide proof of a concept for potential application of a novel HIV-1 vector in HIV-1 disease.
Gene therapy approaches for the treatment of human immunodeficiency virus type 1 (HIV-1) disease have significant potential as a modality for treatment (5). In theory, cells rendered resistant to the effects of HIV-1 should have a selective advantage in repopulating the host immune system. However, gene therapy applications in general have suffered from inadequate expression and an inability to regulate the expression of therapeutic genes (4, 12, 15, 39, 40). For HIV-1, this is a particularly serious issue, since HIV-1 infection of cells results in high levels of sustained expression of viral genome and gene products. Therefore, for any gene therapy approach to be effective, sufficient levels of expression must be obtained in order to provide protection (32).
A number of groups have developed vectors based upon the HIV-1 genome for various gene therapy applications (2, 6, 10, 19, 20, 25–27, 29, 30, 34, 35, 38). These vectors are capable of infecting nondividing human cells such as macrophages, a target for HIV-1 infection. Many of the known HIV-1 genes are toxic to human cells (8, 16, 17, 23, 24, 37); therefore, the construction of a “safe” vector requires ablation of as many HIV-1 genes and sequences as possible. Thus, the most widely used HIV-1 vector consists of only the HIV-1 sequences required for packaging and infection, and foreign genes are generally expressed through an internal heterologous promoter (19, 20, 25, 26, 28, 29, 34). Since the effectiveness of various promoters is dependent upon the cell type, we reasoned that an effective vector for expression of genes directed against HIV-1 should utilize the HIV-1 long terminal repeats (LTRs) such that the levels and regulation of expression by the vector would be comparable to those of wild-type HIV-1. Therefore, we constructed an HIV-1 vector in which expression is dependent upon the activity of the HIV-1 LTRs. All HIV-1 genes, including tat and rev, required for efficient viral gene expression are ablated; however, cis-acting sequences required for packaging, infection, and expression are retained. Thus expression of this vector is dependent upon coinfection with wild-type HIV-1 to provide trans-acting factors required for efficient infection and expression.
We demonstrate that this vector can transduce human cells and can be efficiently induced when the cells are infected by HIV-1. Remarkably, the presence of this vector in cells efficiently inhibits the spread of HIV-1, presumably by competition for limiting substrates required for replication.
MATERIALS AND METHODS
Construction of vectors.
The nucleotide position numbers used here to describe vector constructions start at the 5′ end of HIV-1 NL4-3 provirus (1). HIV-1-based vectors DAEGFP and DAt1ruEGFP were derived from HIV-1NL4-3-lucenv(−) (31). To make DAEGFP, gag, pol, vif, and vpr genes were removed between the AvrII sites (no. 2012 to 5662). The luciferase gene was replaced with the enhanced green fluorescent protein (EGFP) gene (Clontech) by cloning EGFP between the XhoI and MluI sites of HIV-1NL4-3-lucenv(−). To make DAt1ruEGFP, a frameshift was introduced within the tat gene by insertion of a linker sequence (5′-TTAGGTCTAGACCCGGGCGGCCGATCGATCC-3′) into the Sau1 site (no. 5954). A frameshift was introduced within the rev gene at the BamHI site (no. 8466) by treatment with Klenow fragment. Site-directed mutagenesis was performed to create an NcoI site in the vpu gene (no. 6221 A to C and no. 6225 A to G), which was then treated with Klenow fragment to create a frameshift in the vpu gene. An additional XmaI site was made by site-directed mutagenesis (no. 7624 T to C and no. 7627 A to G).
An HIV-1 vector, pHR′CMVEGFP, was derived from pHR′-CMV-lucif (29) by replacing the luciferase gene with EGFP. A murine retrovirus vector, SRαLEGFP, was derived from SRαLthy (3) by replacing the murine thy1.2 gene with EGFP.
A packaging plasmid for an HIV-1-based vector, pCMVΔR8.2DVPR, was derived from pCMVΔR8.2 (28) by deleting the vpr gene from no. 5625 to 5731 by oligonucleotide-directed mutagenesis.
A packaging plasmid for murine retrovirus vector, pSVψ−env−MLV, was obtained from Dan R. Littman (22).
A vesicular stomatitis virus protein G (VSVG) expression plasmid, pHCMVG, was obtained from Jane C. Burns (7).
Vector production and concentration.
All vector stocks were generated by calcium phosphate-mediated transfection of 293T cells (36). 293T cells were cultured in Dulbecco’s modified Eagle medium with 10% calf serum (CS), 100 U of penicillin per ml, and 100 μg of streptomycin per ml. 293T cells (2 × 107) were plated on 175-cm2 flasks in 25 ml of the medium and transfected the following day with 5 μg of pHCMVG, 12.5 μg of pCMVΔR8.2DVPR, and 12.5 μg of DAt1ruEGFP or HR′CMVEGFP for HIV-1-based vectors. For the murine leukemia virus (MLV)-based vector, 5 μg of pHCMVG, 12.5 μg of pSVψ−env−MLV, and 12.5 μg of SRαLEGFP were used.
At 8 h posttransfection, media were replaced with 35 ml of fresh medium. At 36 and 60 h posttransfection, the medium was harvested, centrifuged at 1,500 rpm for 5 min in a Sorvall RT 6000B (Sorvall, Newtown, Conn.), and filtered through a 0.45-μm-pore-size filter. Further vector concentration was achieved by ultracentrifugation at 50,000 × g for 90 min at 4°C. The pellet was resuspended in Iscove’s modified Dulbecco’s medium with 10% FCS, 100 U of penicillin per ml, and 100 μg of streptomycin per ml overnight at 4°C. The vectors were concentrated 100-fold and kept in liquid nitrogen until use.
Transduction of VSVG-pseudotyped vectors.
CEMx174 cells or SupT1 cells were cultured in Iscove’s modified Dulbecco’s medium with 10% FCS, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Cells (5 × 105) were infected with VSVG-pseudotyped DAt1ruEGFP, HR′CMVEGFP, or SRαLEGFP vector by incubation with 1 ml of 10× concentrated vectors in the presence of Polybrene (8 μg/ml) at 37°C for 2 h. Three days postinfection, cells were analyzed for EGFP expression by flow cytometric analysis, and cells expressing EGFP were sorted by fluorescence-activated cell sorting (FACS) with a FACSstar cell sorter (Becton Dickinson). EGFP-expressing sorted CEMx174 or SupT1 cells (vector-transduced cells) were used for VSVG-pseudotyped HIV-1NL4-3thyenv(−)-vprX (31) or NL-r-HSAS virus (18) infection.
Production and infection of VSVG-pseudotyped HIV-1 reporter virus.
Stocks of VSVG-pseudotyped HIV-1NL4-3thyenv(−)-vprX virus were made by calcium phosphate-mediated transfection with 5 μg of pHCMVG and 10 μg of NLthyΔBglVprX plasmid (33) into 293T cells as described for vector production and concentration. Stocks of VSVG-pseudotyped HIV-1NL4-3thyenv(−)-vprX virus were titrated by infecting HeLa cells (105) with various amounts of the virus and analyzing them for murine Thy1.2 expression by flow cytometry on day 3 postinfection.
Mock-transduced and vector-transduced CEMx174 or SupT1 cells (2 × 105) were infected with VSVG-pseudotyped HIV-1NL4-3thyenv(−)-vprX virus for 2 h at 37°C in the presence of Polybrene (8 μg/ml) at a multiplicity of infection (MOI) of 0.5. After 2 h of infection, cells were centrifuged at 1,500 rpm for 5 min and washed with fresh medium twice and cultured for 3 days.
Monoclonal antibody staining and flow cytometric analysis of murine Thy1.2.
At day 3 postinfection, cells were stained for murine Thy1.2. Cells (2 × 105) were washed with phosphate-buffered saline (PBS) (4°C) and stained with 100 μl of monoclonal antibody to murine Thy1.2 directly conjugated to phycoerythrin (PE) (Caltag, catalog no. MM2004), diluted 20-fold with PBS containing 2% FCS, for 20 min on ice. The cells were then washed with PBS (4°C) and resuspended in 0.5 ml of 1% formaldehyde in PBS. Samples were run on a FACSscan flow cytometer (Becton Dickinson), and data were analyzed with the Cell Quest program (Becton Dickinson). The amount of p24gag in culture supernatants was quantified by enzyme-linked immunosorbent assay (ELISA). (Coulter)
Production and infection of replication-competent HIV-1 reporter virus.
Stocks of NL-r-HSAS virus were made by electroporation of 30 μg of infectious proviral NL-r-HSAS DNA into 107 CEM cells, as previously described (18). Infectious units were determined by limiting dilution on SupT1 and CEMx174 cells, by using a fivefold dilution of virus. Before infection, the NL-r-HSAS virus was treated with RNase-free DNase (20 μg/ml; Worthington) for 30 min at 37°C in the presence of 0.01 M MgCl2.
Mock-transduced and vector-transduced CEMx174 cells or SupT1 cells (7 × 105) were infected with replication-competent NL-r-HSAS virus for 2 h at 37°C in the presence of Polybrene (8 μg/ml) at different MOI (MOI of 0.01, 0.1, and 1 for CEMx174 and 0.01 and 0.1 for SupT1 cell experiments). After 2 h of infection, cells were centrifuged at 1,500 rpm for 5 min, washed with fresh medium twice, and cultured. Every 3 or 4 days, the cultures were divided into fifths, fresh medium was added, and the cells were then cultured. The remaining culture was analyzed for murine heat-stable antigen (HSA) expression by flow cytometry, and the amount of p24 in the supernatant was analyzed by ELISA (Coulter). Cell counts were performed by mixing cells with trypan blue, and live cells were counted.
Monoclonal antibody staining and flow cytometric analysis of murine HSA.
Monoclonal antibodies to murine HSA (Pharmingen, catalog no. 01575A) directly conjugated with PE were diluted 500-fold with PBS containing 2% FCS. Cells (2 × 105) were washed with PBS (4°C) and resuspended in 100 μl of human AB serum (4°C) (Omega Scientific, Inc., Tarzana, Calif.). The cells were mixed with a diluted 100-μl concentration of monoclonal antibodies and incubated for 15 min on ice. The cells were washed with PBS (4°C) and resuspended in 0.5 ml of 1% formaldehyde in PBS. Samples were run on a FACSscan flow cytometer (Becton Dickinson), and data were analyzed with the Cell Quest program (Becton Dickinson).
RNA isolation from virions.
Cell culture supernatants (35 ml) from cultures of vector-transduced CEMx174 cells infected with NL-r-HSAS at an MOI of 0.1 were harvested at day 7 postinfection and subjected to ultracentrifugation through 5 ml of 20% sucrose cushion at 50,000 × g for 90 min at 4°C. Virus pellets were resuspended in 250 μl of TEN (0.1 M NaCl, 10 mM Tris-Cl [pH 8.0], 1 mM EDTA [pH 8.0]). RNAs isolated with Trizol LS reagent (Gibco BRL, Grand Island, N.Y.).
Preparation of RNA standards for EGFP, HSA, and HIV-1 reverse transcription-PCR (RT-PCR).
A BamHI-to-NotI fragment containing 780 bp of EGFP cDNA from plasmid pEGFPN1 (Clontech, Palo Alto, Calif.) was ligated into pGEM-11Zf(−) (Promega Corp., Madison, Wis.). A SacI-to-ApaI fragment (1.5 kb) from the full-length HIV-1 molecular clone pYKJRCSF (21) was ligated into pGEM-11Zf(−) (Promega Corp.). An XbaI-to-EcoRI fragment containing 267 bp of HSA cDNA from the recombinant HIV-1 molecular clone pNL-r-HSAS (18) was ligated into pBluescriptII KS(+) (Stratagene, La Jolla, Calif.). EGFP, HSA, and HIV-1 RNA standards were generated by in vitro transcription with a MEGAscript kit (Ambion Inc., Austin, Tex.). Copy numbers were determined by analysis of the amounts of RNAs by using a spectrophotometer and determining the length of the RNAs on a denaturing formaldehyde agarose gel. Serial half-log dilutions were made in order to generate an EGFP, HSA, and HIV-1 standard curve (range, 30 to 10,000 copies).
Quantitative RT-PCR.
RT-PCR assays for EGFP, HSA, or HIV-1 R/U5 sequences were performed by using a Gene Amp kit (Perkin-Elmer, Branchburg, N.J.). In brief, purified RNAs from virions were amplified by using a recombinant Thermus thermophilus (rTth) DNA polymerase with RT in the presence of 0.75 μM an antisense primer, 200 μM deoxynucleoside triphosphates dNTPs, and 1 mM MgCl2 at 70°C for 15 min, followed by 35 cycles of amplification for EGFP or HSA PCR and 30 cycles of amplification for HIV-1 R/U5 with 0.15 μM 32P-labeled sense primer. The absence of DNA contamination was examined by omitting the RT step in a duplicate sample, and the samples were analyzed in parallel. The nucleotide sequences of the HSA primers used for HSA RT-PCR are as follows: SE3, sense primer, 5′GGCTGGGGTTGCTGCTTCTGG3′; and SE4, antisense primer, 5′CCCCTCTGGTGGTAGCGTTAC3′. The primer pairs used for EGFP and HIV-1 R/U5 RT-PCR were GL5/GL6 and M667/AA55, respectively. The primer sequences are described below.
Quantitative DNA PCR assay.
Cell culture supernatants were harvested from cultures of vector-transduced CEMx174 cells infected with NL-r-HSAS at day 4 (MOI of 1) and day 7 (MOI of 0.1). Supernatants at day 7 (MOI of 0.1) were normalized by the p24 value (84.2 μg/ml) for infection. Supernatants at day 4 (MOI of 1) were used for infection without normalization. Before infection, the supernatants were treated with RNase-free DNase (20 μg/ml; Worthington) for 30 min at 37°C in the presence of 0.01 M MgCl2. Fresh CEMx174 cells (5 × 105) were infected for 2 h with 1 ml of each of the supernatants. After 2 h of infection, cells were centrifuged for 1,500 rpm for 5 min and washed with fresh medium twice and cultured. Cells were harvested 12 h postinfection, and DNA was purified from cells by a urea lysis method (41). The resulting reverse transcriptase products for vectors and NL-r-HSAS were distinguished by quantitative PCR for EGFP and HSA genes, respectively. The amount of DNA used for quantitative PCR was adjusted by the copy number of HIV-1 5′ LTR R/U5 obtained by quantitative HIV-1 5′ LTR R/U5 PCR. PCR amplification was performed as previously described (41). Briefly, to detect DAt1ruEGFP, HR′CMVEGFP, or SRαLEGFP vector DNA sequence or NL-r-HSAS viral DNA sequence, one of the oligonucleotide primers for each pair used was end labeled with 32P, and 25 ng of the 32P-labeled oligonucleotide primers was included in the reaction mixture (usually 5 × 106 to 1 × 107 cpm). The second oligonucleotide primer for each pair was not labeled, and 50 ng was incorporated into each reaction mixture. Each reaction mixture contained a 0.25 mM concentration of each of the four dNTPs, 50 mM NaCl, 25 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 100 μg of bovine serum albumin per ml, and 1.25 U of Taq DNA polymerase (Promega). The reaction mixture was overlaid with 25 μl of mineral oil and then subjected to 1 cycle of denaturation for 5 min at 94°C and then 25 cycles of denaturation for 1 min at 94°C and polymerization for 2 min at 65°C. For HSA PCR, 1 cycle of denaturation for 5 min at 94°C and then 30 cycles of denaturation for 2 min at 94°C and polymerization for 3 min at 65°C were used. The reaction was performed on a Perkin-Elmer thermocycler. Amplified products resulting from the PCR were analyzed by electrophoresis on 6% nondenaturing polyacrylamide gels and visualized by direct autoradiography of the dried gels. Quantitative analysis of the amplified products was performed with a PhosphorImager (Molecular Dynamics), and data were analyzed with the Image QuaNT program (Molecular Dynamics). The nucleotide sequences of the oligonucleotide primers used for detecting DAt1ruEGFP and HR′CMVEGFP were derived from the nucleotide sequence of the EGFP DNA and are as follows: GL5, 5′-ATGGTGAGCAAGGGCGAGGAGC-3′; and GL6, 5′GGGTCAGCTTGCCGTAGGTGGC-3′. The oligonucleotide primers used for detecting HSA sequence were derived from the nucleotide sequence of the NL-r-HSAS DNA and are as follows: CH1, 5′-GAACAAGCCCCAGAAGACC-3′; and CH2, 5′-GTAGGAGCAGTGCCAGAAGC-3′.
The nucleotide sequences of the M667 and AA55 oligonucleotide primers used for detecting HIV-1 R/U5 of LTR DNA sequence were previously described (41).
Quantitation of EGFP during PCR amplification was performed by analyzing standard curves of linearized SRαLEGFP plasmid digested with HindIII. For HSA and HIV-1 R/U5 LTRs linearized NL-r-HSAS plasmid digested with BamHI was used. The DNAs described above were diluted in human peripheral blood mononuclear cell DNA (0.1 μg/μl). The copy numbers of the GFP or NL-r-HSAS and HIV-1 R/U5 LTRs included in the standard curve ranged from 7 to 1,800 and from 3 to 1,000 copies, respectively.
RESULTS
Construction of an inducible HIV-1 vector.
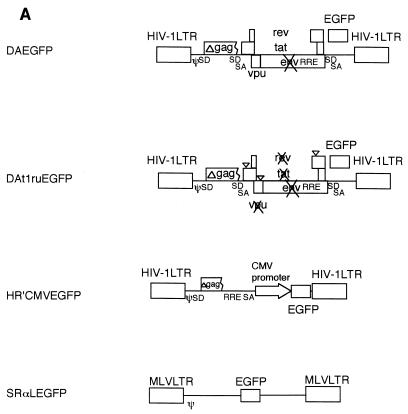
Our strategy to construct an efficiently inducible HIV-1 vector was to first remove essential HIV-1 genes encoding virion products, such as the gag, pol, and env genes, as well as genes not essential for in vitro replication, including vif, vpr, and nef. This construction resulted in a vector, termed DA, bearing an EGFP reporter gene (Fig. 1A) which required gag, pol, and env provided in trans, but which maintained efficient packaging and gene expression. An inducible version (DAt1ru) was then constructed by ablation of tat and rev, followed by ablation of an accessory gene, vpu. It can be rescued into virions by cotransfection with HIV-1 packaging plasmids, which included gag, pol, and env and transactivating genes tat and rev. Infection of SupT1 cells resulted in a population of cells which had low-level expression of EGFP expression by flow cytometry, approximately 2 logs lower than that observed with the parental vector, which was wild type for tat and rev (data not shown), and approximately 1 log lower than that observed with HR′CMV EGFP, a vector which expresses EGFP by using an internal cytomegalovirus (CMV) promoter and SRαLEGFP, a murine retrovirus vector which expresses EGFP.
FIG. 1.
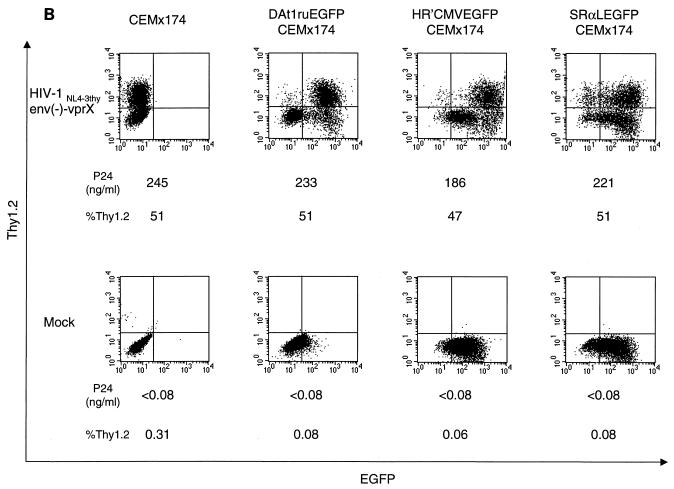
(A) Maps of vectors. An HIV-1 vector, DAEGFP, contains sequences from HIV-1 NL4-3 but lacks the gag, pol, env, vif, vpr, and nef genes, as described in Materials and Methods. The EGFP gene was cloned in place of the nef gene, and is expressed from HIV-1 LTR. The tat and rev genes are intact for gene expression from HIV-1 LTR. An inducible HIV-1 vector, DAt1ruEGFP, was constructed from DAEGFP by ablation of the tat, rev, and vpu genes, as described in Materials and Methods. All genes from HIV-1 were ablated in the DAt1ruEGFP vector. An HIV-1 vector, pHR′CMVEGFP, lacks all HIV-1 genes and expresses EGFP from the CMV internal promoter, as described in Materials and Methods. A murine retrovirus vector, SRαLEGFP, express EGFP from the MLV LTR, as described in Materials and Methods. The LTR, splice donor and acceptor sites (SD and SA, respectively), the packaging signal (ψ), the truncated gag sequence (Δgag), the frameshift mutation (∇), the rev-responsive element (RRE), CMV, and MLV are indicated. (B) Induction of gene expression from HIV-1 vectors by single-round infection with infectious HIV-1 reporter virus. Mock-transduced (CEMx174) and vector-transduced (DAt1ruEGFP CEMx174, HR′CMVEGFP CEMx174, and SRαLEGFP CEMx174) CEMx174 cells (2 × 105) were infected with VSVG-pseudotyped HIV-1NL4-3thyenv(−)-vprX at an MOI of 0.5 (upper panels) or were mock infected (lower panels). At 3 days postinfection, cells (2 × 105) were stained with a monoclonal antibody to murine Thy1.2 conjugated with PE, and 5 × 103 cells were analyzed by flow cytometry for EGFP and Thy1.2 expression. The x axis indicates EGFP fluorescence intensity; the y axis indicates Thy1.2 expression. As indicated, p24 production in cell supernatant was also measured by ELISA at 3 days postinfection. %Thy1.2 indicates Thy1.2-positive populations.
The inducibility of the DAt1ru proviral genome in transduced cells was demonstrated by superinfection of the transduced cells by HIV-1. CEMX 174 cells were subjected to FACS to isolate cells which were expressing low levels of EGFP. This population of cells was then superinfected with HIV-1 bearing the murine thy1.2 reporter gene substituted in place of nef [HIV-1NL4-3 thyenv(−)-vprX] and also defective for env and vpr (33) (Fig. 1B). Due to the deletion in env, this virus requires envelope in trans and therefore permits only a single round of infection. Following infection with HIV-1, each transduced population of cells expressed Thy1.2 on the cell surface and p24 in the supernatant at levels similar to those of mock-transduced cells. In the case of cells transduced with DAt1ru, we observed an approximately 2-log induction of EGFP expression following infection. The majority of the Thy1.2-positive population was also EGFP positive, indicating that superinfection of the transduced cells was responsible for the induction of EGFP expression. As expected, cells infected with the murine retrovirus vector SRαLEGFP did not show any additional induction of EGFP expression. It is noteworthy that the cells transduced with HR′CMVEGFP, although expressing EGFP from an internal CMV promoter, show still greater levels of EGFP expression following infection, possibly reflecting a shift in utilization from the CMV promoter to the HIV LTR of the vector.
Suppression of HIV-1 replication by an inducible HIV-1 vector.
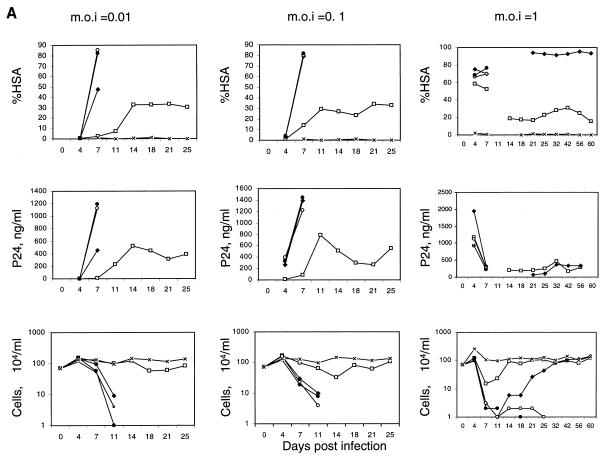
The population of cells transduced with the inducible HIV-1 vector was infected with replication-competent HIV-1 to determine the kinetics of induction in a spreading infection (Fig. 2A). We utilized a replication-competent HIV-1 virus (NL-r-HSAS) bearing the murine HSA gene as a reporter gene substituted in place of the vpr gene (18). CEMx174 cells mock transduced with the vector were rapidly infected, as evidenced by an increasing percentage of HSA-positive cells over time. By day 7, nearly all cells in the culture were HSA positive, and the cultures showed large numbers of syncytia. Death of most of the cells in the culture resulted between 1 and 2 weeks after infection, depending on the initial MOI.
FIG. 2.
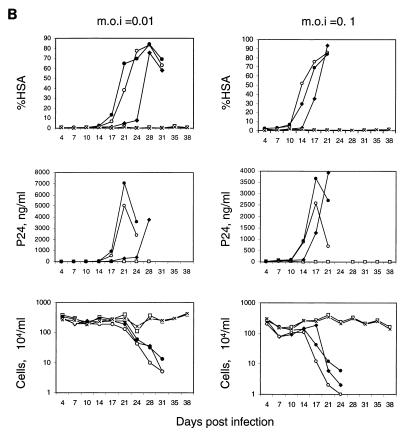
Kinetics of HSA expression, p24 production, and cell count after infection of replication-competent HIV-1 reporter virus. (A) Mock-transduced and vector-transduced CEMx174 cells (DAt1ruEGFP CEMx174, HR′CMVEGFP CEMx174, and SRαLEGFP CEMx174) (7 × 105) were infected with replication-competent NL-r-HSAS at MOI of 0.01, 0.1, and 1. Every 3 or 4 days, the cultures were divided into fifths and recultured in fresh medium. The remaining four-fifths was analyzed for HSA expression by flow cytometry (%HSA), the amount of p24 in the supernatant was measured by ELISA (p24, ng/ml), and the cells were counted (104), as described in Materials and Methods. Due to the low number of cells in cultures at day 11 postinfection at an MOI of 1, analyses of HSA expression and p24 were not performed at that time point. □, DAt1ruEGFP CEMx174 cells infected with NL-r-HSAS; ⧫, HR′CMVEGFP CEMx174 cells infected with NL-r-HSAS; ○, SRαLEGFP CEMx174 cells infected with NL-r-HSAS; ●, CEMx174 cells infected with NL-r-HSAS virus; ×, CEMx174 cells with no NL-r-HSAS infection. (B) Mock-transduced and vector-transduced SupT1 cells (7 × 105) (DAt1ruEGFP SupT1, HR′CMVEGFP SupT1, SRαLEGFP SupT1) were infected with replication-competent NL-r-HSAS at MOI of 0.01 and 0.1. The cultures were analyzed as described for panel A. □, DAt1ruEGFP SupT1 cells infected with NL-r-HSAS; ⧫, HR′CMVEGFP SupT1 cells infected with NL-r-HSAS; ○, SRαLEGFP SupT1 cells infected with NL-r-HSAS; ●, SupT1 cells infected with NL-r-HSAS virus; ×, SupT1 cells with no NL-r-HSAS infection.
Infection of DAt1ru-transduced cells resulted in induction of vector EGFP expression (data not shown). The cells showed an initial level of HSA expression that plateaued and remained at a low level for at least 25 days after infection. The results observed by monitoring HSA expression were paralleled by monitoring p24 production in culture supernatants over time. These cultures showed significantly less syncytia than mock-transduced cells and were significantly protected from cell death. CEMx174 cells transduced with a murine retrovirus vector (SRαLEGFP) and sorted for EGFP expression were infected with NL-r-HSAS and died with kinetics equivalent to that of mock-transduced CEMx174 cells. The constitutively EGFP-expressing HIV-1 vector (HR′CMVEGFP) also showed some inhibition of HIV replication, but was generally nonprotective. At the highest MOI, most cells transduced with this vector died by 11 days after infection, but some cells recovered that expressed HSA but low levels of p24, likely representing selection for resistant survivors containing defective HIV-1. Similar inhibition of HIV-1 replication was observed by utilizing wild-type HIV-1 NL4-3 at an MOI of 0.01. p24 levels comparable to those of NL-r-HSAS were observed, and cells were protected from death; however, more cell death was observed than in infection with NL-r-HSAS, most likely as a result of the presence of the functional vpr gene product, deleted from NL-r-HSAS.
Results similar to those described above were observed with SupT1 cells (Fig. 2B). The kinetics of infection of SupT1 cells is slower than that of CEMx174 cells. Death of most cells in the culture occurred by 3 to 4 weeks for all vectors, except the DAt1ru-inducible vector, which showed suppression of HIV-1 replication and protection from cell death for at least 38 days after infection. We also observed some inhibition of HIV-1 spread in cells transduced with the HR′CMVEGFP vector, but not to the same extent as with DAt1ru, and cells died with kinetics similar to those of mock and murine retrovirus vector-transduced cells. These differences could not be explained by differences in proviral copy numbers in the populations of transduced cells, since both populations had similar levels of provirus (approximately one per cell), as measured by quantitative DNA PCR (data not shown).
Packaging and RT of vector.
The experiments described above utilizing single-step infection with Thy1.2 cells bearing replication-defective HIV-1 (Fig. 1B) indicated that the mechanism of suppression is not a result of competition for tat or rev activity, since cells transduced with the DAt1ru-inducible vector expressed Thy1.2 and p24 with efficiency comparable to that of mock-transduced cells and cells transduced with other vectors. Since inhibition was observed in a spreading infection, but not in the single-step infection, we determined whether HIV-1 produced from DAt1ru-transduced cells was less efficient at establishing infection. Our results show that virus harvested from DAt1ru-transduced cells was less efficient at infecting fresh cells, as monitored by HSA expression 4 and 7 days postinfection (Table 1). One possibility was that the genomic RNA of the vector was packaged and/or reverse transcribed with greater efficiency than that of the replication-competent HIV-1, thereby providing a selective advantage to the vector in a spreading infection.
TABLE 1.
Infection with NL-r-HSAS virus derived from DAt1ruEGFP-transduced cellsa
| Expt | % HSA in CEMx174 cells
|
||||
|---|---|---|---|---|---|
| NL-r-HSAS infected
|
Mock infected, no vector | ||||
| DAt1ruEGFP | HR′CMVEGFP | SRαLEGFP | No vector | ||
| 1 | 2.2 | 22 | 9.7 | 26 | 0.08 |
| 2 | 42 | 74 | 71 | 73 | 0.37 |
Cell culture supernatants were harvested from cultures of vector-transduced CEMx174 cells infected with NL-r-HSAS at day 7 (MOI of 0.1) and at day 4 (MOI of 1) (shown in Fig. 2A). The p24 values of the supernatants from day 7 (MOI of 0.1) were 84.2 ng/ml for NL-r-HSAS-infected DAt1ruEGFP-transduced cells, 1,390 ng/ml for NL-r-HSAS-infected HR′CMVEGFP-transduced cells, 1,220 ng/ml for NL-r-HSAS-infected SRαLEGFP-transduced cells, 1,440 ng/ml for NL-r-HSAS-infected no-vector-transduced cells, and <0.08 ng/ml for mock-infected no-vector-transduced cells. The p24 values for supernatants from day 4 (MOI of 1) were 1,180 ng/ml for NL-r-HSAS-infected DAt1ruEGFP-transduced cells, 1,950 ng/ml for NL-r-HSAS-infected HR′CMVEGFP-transduced cells, 1,120 ng/ml for NL-r-HSAS-infected SRαLEGFP-transduced cells, 920 ng/ml for NL-r-HSAS-infected no-vector-transduced cells, and <0.08 ng/ml for mock-infected no-vector-transduced cells. Supernatants from day 7 (MOI of 0.1) were used for experiment 1. Supernatants from day 4 (MOI of 1) were used for experiment 2. Supernatants from day 7 (MOI of 0.1) were normalized by the p24 value (84.2 μg/ml) for infection. Fresh CEMx174 cells (5 × 105) were infected for 2 h with 1 ml of each supernatant. Four days postinfection (experiment 1) or 7 days postinfection (experiment 2), cells (2 × 105) were stained with a monoclonal antibody to HSA conjugated with PE, and 7 × 103 events were analyzed for the percentage of HSA expression by flow cytometry.
We examined the relative ability of DAt1ru vector and NL-r-HSAS genomic RNAs to be incorporated into virions by measuring EGFP and HSA RNA sequences by quantitative RT-PCR (Table 2). Our results indicate that the DAt1ru RNA genome is packaged at amounts comparable to those of the NL-r-HSAS RNA genome. Therefore, it is unlikely that competition for packaging could account for the suppression of HIV-1 replication observed. In contrast to virions produced from DAt1ru-transduced cells, virions produced from HR′CMVEGFP-transduced cells had less EGFP-containing vector RNA than HSA-containing replication-competent HIV-1 RNA. The reason for this is unclear, but may reflect the presence of the internal CMV promoter transcribing RNA which would not contain packaging signals suitable for incorporation into virions.
TABLE 2.
Quantitative analysis of virion RNAsa
| RNA type | Copy no. in supernatant:
|
||||
|---|---|---|---|---|---|
| NL-r-HSAS infected
|
Mock infected, no vector | ||||
| DAt1ruEGFP | HR′CMVEGFP | SRαLEGFP | No vector | ||
| EGFP | 169 ± 23 | 345 ± 131 | <30 | <30 | <30 |
| HSA | 193 ± 48 | 3,484 ± 1,423 | 6,269 ± 1,811 | 3,650 ± 1,612 | <30 |
RNA was isolated from virions in 35 ml of cell culture supernatants from cultures of vector-transduced (DAt1ruEGFP, HR′CMVEGFP, or SRαLEGFP) or no-vector-transduced CEMx174 cells infected with NL-r-HSAS virus (MOI of 0.1) at day 7 postinfection. RNA was subjected to in vitro RT and PCR amplification for EGFP and HSA sequences, as described in Materials and Methods. The amount of RNA used for RT-PCR was adjusted by the value of p24 in the cell culture supernatants. The copy numbers shown represent the equivalent of 5 pg of p24. The EGFP and HSA RNA copy numbers shown here are the average and standard error of data from six independent RT-PCR assays. The absence of DNA contamination was examined by omitting the RT step in a duplicated sample, analyzed in parallel, and no signals were detected (data not shown). The Kruskal-Wallis test, a nonparametric analysis of variance, was utilized to test for differences of log ratios of EGFP and HSA RNA copy numbers (log10 [EGFP/HSA]) in virions produced from DAt1ruEGFP or HR′CMVEGFP vector-transduced CEMx174 cells infected with NL-r-HSAS virus. This test will give the same answer for a comparison of ratios as it does for a comparison of differences in log counts. The Kruskal-Wallis test does not depend on the assumption of normality as does the two-sample t test. At a 5% significance level (P = 0.037), there is a difference between the two groups.
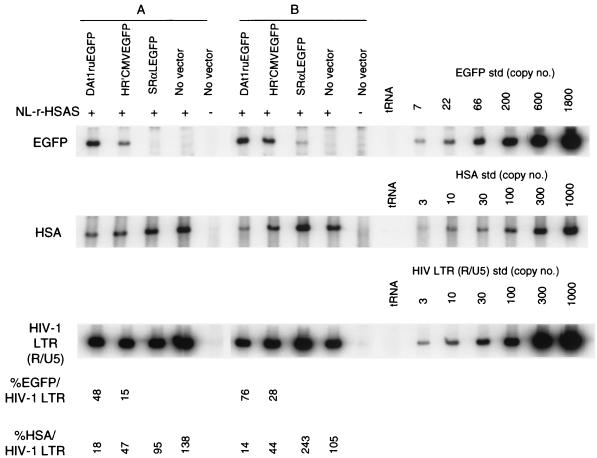
Reverse transcriptase products synthesized 12 h after infection for the DAt1ru vector and NL-r-HSAS were distinguished by quantitative PCR for EGFP- and HSA-containing genomes, respectively (Fig. 3). The DAt1ru vector DNA genome was present at approximately two- to fivefold-greater levels than the NL-r-HSAS virus genome. In contrast, the ratios of HR′CMVEGFP vector DNA to NL-r-HSAS were 0.31 and 0.63 for samples A and B, respectively. These results indicate that the inhibition of HIV-1 infection occurs at least in part at a step of the viral life cycle during RT.
FIG. 3.
Quantitative analysis of de novo-synthesized vector and viral DNA from cells harboring both replication-competent HIV-1 and vectors. Cell culture supernatants were harvested from cultures of vector-transduced CEMx174 cells infected with NL-r-HSAS at day 4 (MOI of 1 [sample A]) and day 7 (MOI of 0.1 [sample B]). The p24 values of each of the supernatants of sample A were as follows: 1,180 ng/ml for the NL-r-HSAS-infected DAt1ruEGFP-transduced cells, 1,950 ng/ml for NL-r-HSAS-infected HR′CMVEGFP-transduced cells, 1,120 ng/ml for NL-r-HSAS-infected SRαLEGFP-transduced cells, 920 ng/ml for NL-r-HSAS-infected no-vector-transduced cells, and <0.08 ng/ml for mock-infected no-vector-transduced cells). The p24 values of each of the supernatants of sample B were as follows: 84.2 ng/ml for NL-r-HSAS-infected DAt1ruEGFP-transduced cells, 1,390 ng/ml for NL-r-HSAS-infected HR′CMVEGFP-transduced cells, 1,220 ng/ml for NL-r-HSAS-infected SRαLEGFP-transduced cells, 1,440 ng/ml for NL-r-HSAS-infected no-vector-transduced cells, and <0.08 ng/ml for mock-infected no-vector-transduced cells). Supernatants of sample B were normalized by the p24 value (84.2 μg/ml) for infection. Supernatants of sample A were used for infection without normalization. Supernatants were treated with DNase before infection, as described in Materials and Methods. Fresh CEMx174 cells (5 × 105) were infected for 2 h with 1 ml of each supernatant. At 12 h postinfection, DNA was purified from cells and subjected to quantitative PCR for EGFP gene, HSA gene, and HIV-1 R/U5 LTR sequences, as described in Materials and Methods. tRNA (0.1 μg/ml) was used as a negative control for PCR. Quantitative EGFP, HSA, and HIV-1 LTR (R/U5) DNA standards (std) were assayed in parallel. The EGFP- and HSA-specific signals were compared with that of the amplified HIV-1 LTR (R/U5) sequence to determine the percentages of EGFP/HIV-1 LTR and HSA/HIV-1 LTR, respectively. The data are representative of two independent PCR analyses.
DISCUSSION
These results represent proof of a concept for modeling a novel gene therapeutic strategy for HIV-1 disease. Similar to other therapeutic strategies, such as the use of protease inhibitors and gene therapeutic strategies, such as those involving ribozymes and dominant-negative HIV-1 proteins, this strategy would not inhibit the initial establishment of HIV-1 infection within a target cell; however, the DAt1ru vector would act to inhibit subsequent rounds of viral infection. Furthermore, the relatively low levels of gene transduction and reconstitution currently achievable may not be as severe limitations as they are for most gene therapeutic applications, since the HIV-1-based vector is packaged and reverse transcribed and would likely be mobilized and passed to other uninfected target cells, thus amplifying the pool of cells which could serve to limit the spread of infectious HIV-1.
We have not yet elucidated the specific step in the viral life cycle which is affected by this vector, although it appears to occur at least in part during RT. In cotransfection studies, other investigators have proposed that competition for tat or rev may be involved (11). In our experiments, we have seen no effect upon establishment of the initial infection and gene expression dependent upon tat and rev. A smaller genome size of the vector resulting in more efficient packaging or RT cannot solely explain the interference observed, since HR′CMVEGFP is smaller than DAt1ru (4.2 versus 6.1 kb), yet does not result in as significant a degree of suppression. Further genetic mapping of viral genetic elements required for suppression would allow us to more optimally design a vector which should have even greater suppressive capabilities. In addition, since this vector is packaged together with the wild-type genome into virions, it would be conceivable that this vector can be further modified to contain genetic elements, such as ribozymes or antisense RNAs, that may act upon wild-type RNA copackaged in the same virion (9, 13, 14). Further understanding of the mechanism of action and optimization of its effects will be critical for consideration of the use of such a vector in clinical applications.
ACKNOWLEDGMENTS
We thank Beth Jamieson, Betty Poon, and Kathie Grovit-Ferbas for reagents, Matthew Leibowitz for technical assistance, and Liz Duarte and Rosie Taweesup for manuscript preparation.
This work was supported by UCLA CFAR grants I AI39975 and AI36555.
ADDENDUM IN PROOF
Recently Bukovsky et al. (A. A. Bukovsky, J.-P. Song, and L. Naldini, J. Virol. 73:7087–7092, 1999) used a vector similar to HR′CMVEGFP that we found to only weakly suppress HIV-1.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akkina R K, Walton R M, Chen M L, Li Q-X, Planelles V, Chen I S Y. High-efficiency gene transfer into CD34+ cells with a human immunodeficiency virus type 1-based retroviral vector pseudotyped with vesicular stomatitis virus envelope glycoprotein G. J Virol. 1996;70:2581–2585. doi: 10.1128/jvi.70.4.2581-2585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An D S, Koyanagi Y, Zhao J-Q, Akkina R, Bristol G, Yamamoto N, Zack J A, Chen I S Y. High-efficiency transduction of human lymphoid progenitor cells and expression in differentiated T cells. J Virol. 1997;71:1397–1404. doi: 10.1128/jvi.71.2.1397-1404.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson W F. Human gene therapy. Nature. 1998;392:25–30. doi: 10.1038/32058. [DOI] [PubMed] [Google Scholar]
- 5.Baltimore D. Intracellular immunization. Nature. 1988;335:395–396. doi: 10.1038/335395a0. [DOI] [PubMed] [Google Scholar]
- 6.Buchschacher G L, Jr, Panganiban A T. Human immunodeficiency virus vectors for inducible expression of foreign genes. J Virol. 1992;66:2731–2739. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J-K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao J, Park I-W, Cooper A, Sodroski J. Molecular determinants of acute single-cell lysis by human immunodeficiency virus type 1. J Virol. 1996;70:1340–1354. doi: 10.1128/jvi.70.3.1340-1354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cara A, Rybak S M, Newton D L, Crowley R, Rottschafer S E, Reitz M S, Jr, Gusella G L. Inhibition of HIV-1 replication by combined expression of gag dominant negative mutant and a human ribonuclease in a tightly controlled HIV-1 inducible vector. Gene Ther. 1998;5:65–75. doi: 10.1038/sj.gt.3300545. [DOI] [PubMed] [Google Scholar]
- 10.Corbeau P, Kraus G, Wong-Staal F. Transduction of human macrophages using a stable HIV-1/HIV-2-derived gene delivery system. Gene Ther. 1998;5:99–104. doi: 10.1038/sj.gt.3300563. [DOI] [PubMed] [Google Scholar]
- 11.Corbeau P, Wong-Staal F. Anti-HIV effects of HIV vectors. Virology. 1998;243:268–274. doi: 10.1006/viro.1998.9089. [DOI] [PubMed] [Google Scholar]
- 12.Crystal R G. Transfer of genes to humans: early lessons and obstacles to success. Science. 1995;270:404–410. doi: 10.1126/science.270.5235.404. [DOI] [PubMed] [Google Scholar]
- 13.Ding S F, Noronha J, Joshi S. Co-packaging of sense and antisense RNAs: a novel strategy for blocking HIV-1 replication. Nucleic Acids Res. 1998;26:3270–3278. doi: 10.1093/nar/26.13.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dropulic B, Hermankova M, Pitha P M. A conditionally replicating HIV-1 vector interferes with wild-type HIV-1 replication and spread. Proc Natl Acad Sci USA. 1996;93:11103–11108. doi: 10.1073/pnas.93.20.11103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedmann T. Human gene therapy—an immature genie, but certainly out of the bottle. Nat Med. 1996;2:144–147. doi: 10.1038/nm0296-144. [DOI] [PubMed] [Google Scholar]
- 16.Gibbs J S, Lackner A A, Lang S M, Simon M A, Sehgal P K, Daniel M D, Desrosiers R C. Progression to AIDS in the absence of a gene for vpr or vpx. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanna Z, Kay D G, Rebai N, Guimond A, Jothy S, Jolicoeur P. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell. 1998;95:163–175. doi: 10.1016/s0092-8674(00)81748-1. [DOI] [PubMed] [Google Scholar]
- 18.Jamieson B D, Zack J A. In vivo pathogenesis of a human immunodeficiency virus type 1 reporter virus. J Virol. 1998;72:6520–6526. doi: 10.1128/jvi.72.8.6520-6526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kafri T, Blomer U, Peterson D A, Gage F H, Verma I M. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- 20.Kim V N, Mitrophanous K, Kingsman S M, Kingsman A J. Minimal requirement for a lentivirus vector based on human immunodeficiency virus type 1. J Virol. 1998;72:811–816. doi: 10.1128/jvi.72.1.811-816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koyanagi Y, Miles S, Mitsuyasu R T, Merril J E, Vinters H V, Chen I S Y. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 22.Landau N R, Littman D R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C J, Friedman D J, Wang C, Metelev V, Pardee A B. Induction of apoptosis in uninfected lymphocytes by HIV-1 tat protein. Science. 1995;268:429–431. doi: 10.1126/science.7716549. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y-Y, Koga Y, Tanaka K, Sasaki M, Kimura G, Nomoto K. Apoptosis induced in CD4+ cells expressing gp160 of human immunodeficiency virus type 1. J Virol. 1994;68:390–399. doi: 10.1128/jvi.68.1.390-399.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyoshi H, Blömer U, Takahashi M, Gage F H, Verma I M. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyoshi H, Takahashi M, Gage F H, Verma I M. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc Natl Acad Sci USA. 1997;94:10319–10323. doi: 10.1073/pnas.94.19.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mochizuki H, Schwartz J P, Tanaka K, Brady R O, Reiser J. High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J Virol. 1998;72:8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naldini L, Blomer U, Gage F H, Trono D, Verma I M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–266. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 30.Parolin C, Dorfman T, Palú G, Göttlinger H, Sodroski J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–3895. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Planelles V, Bachelerie F, Jowett J B M, Haislip A, Xie Y, Banooni P, Masuda T, Chen I S Y. Fate of the human immunodeficiency virus type 1 provirus in infected cells: a role for vpr. J Virol. 1995;69:5883–5889. doi: 10.1128/jvi.69.9.5883-5889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plavec I, Agarwal M, Ho K, Pineda M, Auten J, Baker J, Matsuzaki H, Escaich S, Bonyhadi M, Bohnlein E. High transdominant RevM10 protein levels are required to inhibit HIV-1 replication in cell lines and primary T cells: implication for gene therapy of AIDS. Gene Ther. 1997;4:128–139. doi: 10.1038/sj.gt.3300369. [DOI] [PubMed] [Google Scholar]
- 33.Poon B, Jowett J B M, Stewart S A, Armstrong R W, Rishton G M, Chen I S Y. Human immunodeficiency virus type 1 vpr gene induces phenotypic effects similar to those of the DNA alkylating agent, nitrogen mustard. J Virol. 1997;71:3961–3971. doi: 10.1128/jvi.71.5.3961-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poznansky M, Lever A, Bergeron L, Haseltine W, Sodroski J. Gene transfer into human lymphocytes by a defective human immunodeficiency virus type 1 vector. J Virol. 1991;65:532–536. doi: 10.1128/jvi.65.1.532-536.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiser J, Harmison G, Kluepfel-Stahl S, Brady R O, Karlsson S, Schubert M. Transduction of nondividing cells using pseudotyped defective high-titer HIV type 1 particles. Proc Natl Acad Sci USA. 1996;93:15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 16.32–16.34. [Google Scholar]
- 37.Stewart S A, Poon B, Jowett J B M, Chen I S Y. Human immunodeficiency virus type 1 Vpr induces apoptosis following cell cycle arrest. J Virol. 1997;71:5579–5592. doi: 10.1128/jvi.71.7.5579-5592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutton R E, Wu H T M, Rigg R, Böhnlein E, Brown P O. Human immunodeficiency virus type 1 vectors efficiently transduce human hematopoietic stem cells. J Virol. 1998;72:5781–5788. doi: 10.1128/jvi.72.7.5781-5788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma I M. Gene therapy: hopes, hypes, and hurdles. Mol Med. 1994;1:2–3. [PMC free article] [PubMed] [Google Scholar]
- 40.Verma I M, Somia N. Gene therapy—promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 41.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]