Summary
Background
Patients with type 1 diabetes (T1D) and type 2 diabetes (T2D) present intestinal disturbances. Recent epidemiological data have showed that, worldwide, over half of newly diagnosed T1D patients were adults. However, the gut microbial alterations in adult-onset T1D are unclear. We aimed to identify the signatures of gut microbiota and metabolites in patients with adult-onset T1D systematically, comparing with T2D patients and healthy controls (HCs).
Methods
This study enrolled 218 subjects from February 2019 to April 2022 (discovery cohort: 36 HCs, 51 patients with adult-onset T1D and 56 patients with T2D; validation cohort: 28 HCs, 27 patients with adult-onset T1D and 20 patients with T2D). Gut microbial profiles of the study subjects were investigated by metagenomic sequencing, and their faecal and serum metabolites were measured with targeted metabolomics. The study was registered on ClinicalTrials.gov (NCT05252728).
Findings
Patients with adult-onset T1D had significant differences in the composition of bacteria and their metabolites, characterized by notable depletion of short-chain fatty acid-producing bacteria, especially Eubacterium rectale. This was associated with a severe loss of phenolic acids and their derivatives, including gallic acid (associated with glucose metabolism) and 3,4-dihydroxyhydrocinnamic acid (linked with glucose metabolism and pancreatic beta cell autoimmunity). A predictive model based on six bacteria and six metabolites simultaneously discriminated adult-onset T1D from T2D and HCs with high accuracy. Interestingly, bacterial-viral or bacterial-fungal trans-kingdom relationships, especially positive correlations between bacteriophages and beneficial bacteria, were significantly reduced in adult-onset T1D compared to HCs.
Interpretation
Adult-onset T1D patients exhibit unique changes in host-microbiota-metabolite interactions. Gut microbiota and metabolite-based algorithms could be used as additional tools for differential diagnosis of different types of diabetes and beyond.
Funding
National Key Research and Development Program of China, the National Natural Science Foundation of China.
Keywords: Adult-onset diabetes, Type 1 diabetes, Type 2 diabetes, Gut microbiota, Metabolites, Diagnosis
Research in context.
Evidence before this study
Over half of newly diagnosed cases of type 1 diabetes (T1D) occur in adults, however, nearly 40% of adult-onset T1D is initially misdiagnosed as type 2 diabetes (T2D), with serious medical consequences. We searched PubMed with the terms “type 1 diabetes”, “metagenome”, “microbiota”, “metabolome” and “metabolites” for original articles and reviews published up to April 13th, 2023. Previous studies have illustrated that gut microbiota and metabolites contribute to the pathogenesis of both childhood-onset T1D and latent autoimmune diabetes in adults (LADA), but none of them mentioned the adult-onset T1D. The efficacy of differential diagnostic methods for different types of diabetes based on gut microbes and metabolites may be underestimated.
Added value of this study
To the best of our knowledge, this is the first study using metagenomics and metabolomics to identify not only the altered gut bacteria but also intestinal viruses and fungi in patients with adult-onset T1D in comparison with patients with T2D and/or healthy controls (HCs). A predictive model based on gut microbiota and metabolic features has been developed using machine learning techniques, demonstrating high efficacy for the diagnosis and classification of diabetes after further validation.
Implications of all the available evidence
Our study highlighted the alteration of gut microbiota and metabolites in adult-onset T1D. The marker panel of bacterial species and faecal metabolites provided a potential diagnostic tool for discriminating adult-onset T1D from T2D and HCs. These predictive models have the potential to be applied in public healthcare settings and community-level populations, thereby promoting precision diagnosis and treatment of diabetes.
Introduction
Type 1 diabetes (T1D) is caused by autoimmune destruction of pancreatic beta cells, leading to severe insulin deficiency and requires life-long insulin therapy.1 It is historically considered as a disease of childhood and adolescence, but recent epidemiological studies showed that approximately half of the new-onset T1D cases occur in adults worldwide.2 The correct diagnosis of childhood-onset T1D is straightforward, however, nearly 40% of adults with T1D are often initially misdiagnosed with type 2 diabetes (T2D),3,4 which is associated with insulin resistance and not absolute insulin deficiency. Misdiagnosing T1D as T2D can lead to poor blood glucose control, increased risk of ketoacidosis, and possibly life-threatening complications.5 Thus, it is critical to identify the molecular basis and explore new diagnostic biomarkers for adult-onset T1D.
It has been reported that there is a dysbiosis of gut microbiota in childhood-onset T1D and adult T2D.6,7 Patients with childhood-onset T1D exhibit compositional alterations in the gut microbiota, which are often seen as a decreased ratio of Firmicutes/Bacteroides and an increased abundance of potential pathobionts, including Bacteroides and Blautia.8,9 Patients with T2D exhibit a loss of butyrate-producing bacteria, especially a decreased abundance of Akkermansia muciniphila,10 and an increased abundance of bacteria (e.g., Prevotella copri and Bacteroides vulgatus) with potential of branched-chain amino acids (BCAAs) biosynthesis which may contribute to increased peripheral BCAAs concentrations and insulin resistance.11,12 However, the change in the gut microbiota in adult-onset T1D has not been elucidated, systemically.
In addition to the direct interaction of gut microbiota with host tissue cells including immune cells, the impact of the gut microbiota can also be mediated by their metabolites that are able to induce host immunological and physipathological responses in the intestine and at distant organs. Microbe-derived metabolites showed obvious alterations in T2D,13 especially BCAAs produced by intestinal bacteria, which are related to the deterioration of insulin sensitivity.11 In patients with latent autoimmune diabetes in adults (LADA), a subtype of T1D, the composition of the gut microbiota and its metabolites were different from those in T2D patients and healthy controls (HCs), especially with a severe deficiency of short-chain fatty acid (SCFA)-producing bacteria in LADA.14 Microbial signatures can help discriminate patients with longstanding T1D from HCs.15 However, thus far, no integrated analysis of multiomics data from the gut microbiome and metabolome has been conducted to identify novel biomarkers differentiating adult-onset T1D patients, T2D patients and HCs.
To bridge the abovementioned knowledge gaps, we applied whole-genome shotgun metagenomic analysis to elucidate the overall landscape of the gut microbiota in patients with different subtypes of diabetes. Furthermore, we identified various metabolites and clarified the relationship between gut microbiota and metabolites, as well as the host phenotypes. Finally, we established a multiomic classifier that discriminates adult-onset T1D patients from T2D patients and HCs.
Methods
Ethics
All procedures were in accordance with the principles of the Declaration of Helsinki and were approved by the Human Ethics Committee of Second Xiangya Hospital of Central South University (No. 2019-Research-30). Written informed consent was obtained from each subject at enrolment. The study was registered at https://clinicaltrials.gov(NCT05252728).
Subject recruitment
A total of 218 study subjects were recruited from February 2019 to April 2022, including a discovery set from Changsha, China (discovery set, n = 143) and an external validation set from Pingjiang, China (validation set, n = 75). In the discovery set, three groups, including HCs (n = 36), adult-onset T1D (n = 51) and T2D (n = 56), were recruited. The validation set enrolled three sex- and age-matched groups, including HCs (n = 28), adult-onset T1D (n = 27) and T2D (n = 20). All HCs enrolled in this study were unrelated to the patients. All patients enrolled in this study fulfilled the 1999 World Health Organization criteria for diabetes.16 The anthropometric and biochemical characteristics of HCs and patients with adult-onset T1D or T2D in the discovery set and validation set are summarized in online Supplementary Table S1. T1D was diagnosed according to the criteria of the American Diabetes Association.17 In addition to the diagnosis of diabetes, the followings were also taken into account in the diagnosis of T1D: symptoms, signs, history, dependence of insulin application, absence of pancreatic beta cell function, and were positive for at least one of islet autoantibodies, including glutamic acid decarboxylase antibody (GADA), insulinoma-associated protein 2 antibody (IA-2A), or zinc transporter-8 antibody (ZnT8A). Adult onset was defined as an onset age ≥20 years. Subjects with the following conditions were excluded: severe gastrointestinal, cardiovascular, cerebrovascular, liver or kidney diseases; other autoimmune diseases; a history of gastrointestinal surgery; tumours; pregnancy; treatments with oral hypoglycaemic agents or immunomodulators; and use of probiotics, prebiotics or antibiotics within one month. All subjects had an in-person interview, in which host metadata, disease status, dietary intake and lifestyle information were collected.
Metagenomic sequencing, taxonomic classification and functional annotation
Faecal samples were directly stored in a −80 °C freezer after sampling. Faecal DNA was extracted following the manufacturer’s protocol.18 DNA was fragmented to construct a paired-end library by using a Covaris M220 sonicator. Samples were sequenced on the Illumina platform according to the standard protocol. Clean raw reads were obtained by removing the raw sequence reads with low quality (paired-end; insert size, 350 bp; read length, 150 bp) and using Burrows-Wheeler alignment (BWA) to remove human-derived reads, and 32.6 million reads per sample on average remained.
The clean reads were assembled into contigs by using MEGAHIT. MetaProdigal was used to predict the open reading frames from each contig. The sequences of predicted genes were clustered using CD-HIT software. High-quality reads were aligned to the nonredundant (NR) gene set using SOAPaligner, and the relative abundance of genes was calculated as described by Qin et al.7 For the taxonomic annotations of bacteria, viruses and fungi at different taxonomic levels, the NR gene set was aligned to the National Centre for Biotechnology Information (NCBI) NR database with an e-value cutoff of 1e-5 using DIAMOND. Genes with a relative abundance of less than 0.01% were removed for the subsequent analysis. The Kyoto Encyclopedia of Genes and Genomes (KEGG) functional annotation of different categories was conducted using DIAMOND with an e-value cutoff of 1e-5.
Metabolomic analysis of faecal and serum samples
Samples were directly stored at −80 °C until analysis. An ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) system (ACQUITY UPLC-Xevo TQ-S, Waters Corp., Milford, MA, USA) was used for targeted metabolomic analysis. Quality control (QC) samples were assembled by mixing equal amounts of each sample and were injected at regular intervals. UPLC-MS/MS was used to acquire raw data files. Peak integration, calibration, and quantitation for each metabolite were performed by using MassLynx software (v4.1, Waters, Milford, MA, USA). Metabolites with a relative SD of >30% across QC samples and present in <80% of samples in any group were removed for further analysis. The missing values were filled with minimum values, and the abundance data were log2-transformed.
Clinical parameter measurements
Demographic and clinical data were collected, including age, sex, body mass index (BMI), and diabetes duration, according to standard measurement methods.19 Venous blood samples were collected in the morning after overnight fasting and 120 minutes after a mixed meal. Fasting blood glucose (FBG), and postprandial blood glucose (PBG) were measured. Haemoglobin A1c (HbA1c) was determined by a Bio-Rad VARIANT II Haemoglobin Testing System (Hercules, CA, USA). Serum levels of fasting C-peptide (FCP) and postprandial C-peptide (PCP) were assessed by the Advia Centaur System (Siemens, Munich, Germany). Homoeostasis model assessment (HOMA) 2 estimates of β-cell function (HOMA2-B), insulin sensitivity (HOMA2-S) and insulin resistance (HOMA2-IR) based on C-peptide concentrations were calculated with the HOMA calculator.20 GADA, IA-2A, and ZnT8A were detected by radioligand assays as previously described.19
Statistics
Differences in clinical parameters were analysed using the chi-square test or Kruskal–Wallis test, and multiple comparisons were corrected with Bonferroni post hoc tests. Due to lack of data on the gut microbiota differences among adult-onset T1D, T2D and HCs, no statistical methods were used to estimate the sample size, but our sample size is similar to those reported in childhood- and adolescent-onset T1D and HCs.21,22 Significant differences in the relative abundances of taxa were identified by linear discriminant analysis (LDA) effect size (LEfSe) analysis, and P values were corrected using the Benjamini and Hochberg false discovery rate (FDR). Taxa with LDA values > 2.0 and P < 0.05 were considered differentially abundant, and taxa with Pfdr < 0.1 were considered significantly changed. Permutational multivariate analysis of variance (PERMANOVA) for microbiota beta diversity comparison and redundancy analysis (RDA) for evaluating the effects of demographical variables on microbiota community variation were conducted using the R package vegan. The multivariate association with linear models (MaAsLin) framework was used to adjust the effects of host factors. Orthogonal partial least squares discriminant analysis (OPLS-DA) algorithms were applied to visualize the comparison in the metabolite profiles. Variable influence on projection (VIP) scores from the OPLS-DA were calculated. Random forest models were built to differentiate different groups (randomForest package in R) based on the microbial features, metabolic features and a combination of the two types of data. All statistical analyses were performed in SPSS version 26.0 and R 4.0.4. Pfdr < 0.1 was considered statistically significant.
Role of the funding source
The funders of the study had no role in study design, data collection, data analyses, data interpretation, or writing of the report. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Demographics and baseline characteristics of subjects
Subjects in these three groups were matched by age and sex. The glucose metabolism parameters of patients with diabetes, including FBG, PBG and HbA1c levels, were significantly higher than those of HCs (P < 0.05), and there was no significant difference between the two diabetic groups. The serum levels of FCP and PCP in patients with adult-onset T1D were significantly lower than those in HCs and patients with T2D (P < 0.05, Supplementary Table S1).
Alterations in the microbial diversity in adult-onset T1D
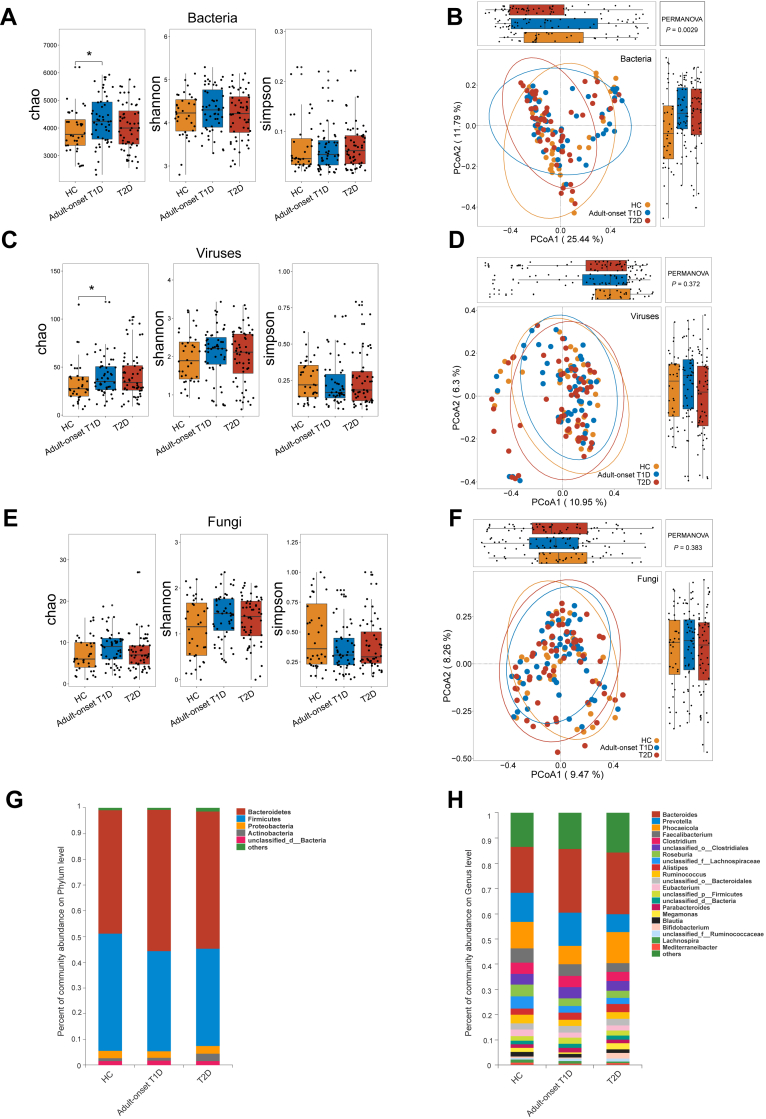
The study design was illustrated in Fig. 1. We analysed the microbial diversity of the three groups. For alpha diversity, significant differences were found in the Chao index of bacteria and viruses between adult-onset T1D patients and HCs, but no significant differences were found in the Shannon and Simpson index (Fig. 2A, C, E). To further identify the overall microbial features of the three groups, beta diversity comparison was performed by PERMANOVA. Principal coordinates analysis (PCoA) based on Bray–Curtis distance showed that the overall bacterial community structure of adult-onset T1D patients was significantly different from that of HCs and T2D patients (PERMANOVA test, adult-onset T1D vs HC: P = 0.020; T2D vs HC: P = 0.013; adult-onset T1D vs T2D: P = 0.030) (Fig. 2B). However, there were no significant differences in the global viral or fungal composition among these three groups (Fig. 2D, F), which is probably due to the limited identification of viral or fungal species.
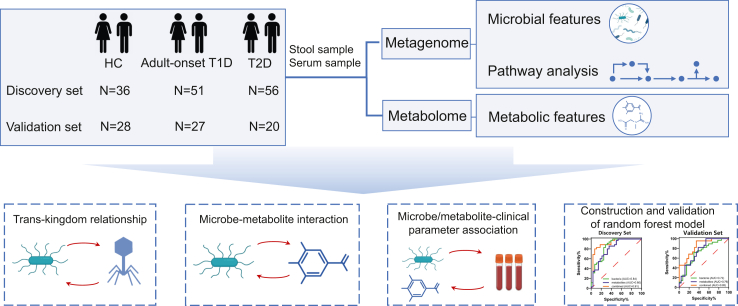
Fig. 1.
Diagram of the study design.
Fig. 2.
Faecal microbiome variations in patients with adult-onset T1D and patients with T2D. Alpha and beta diversity comparison of bacteria (A, B), viruses (C, D) and fungi (E, F) among patients with adult-onset T1D, patients with T2D and HCs. (G, H) Percentage of bacterial community abundance at phylum and genus levels; phyla or genera with a relative abundance <1% in each sample are merged into others. Box plots show median ± quartiles, and the whiskers extend from the hinge to the largest or smallest value no further than 1.5 folds of the inter-quartile range. ∗P < 0.05, Abbreviations: HCs, healthy controls; T1D, type 1 diabetes; T2D, type 2 diabetes; PCoA, principal coordinated analysis; PERMANOVA, permutational multivariate analysis of variance.
Taxonomic changes in microbial composition in adult-onset T1D
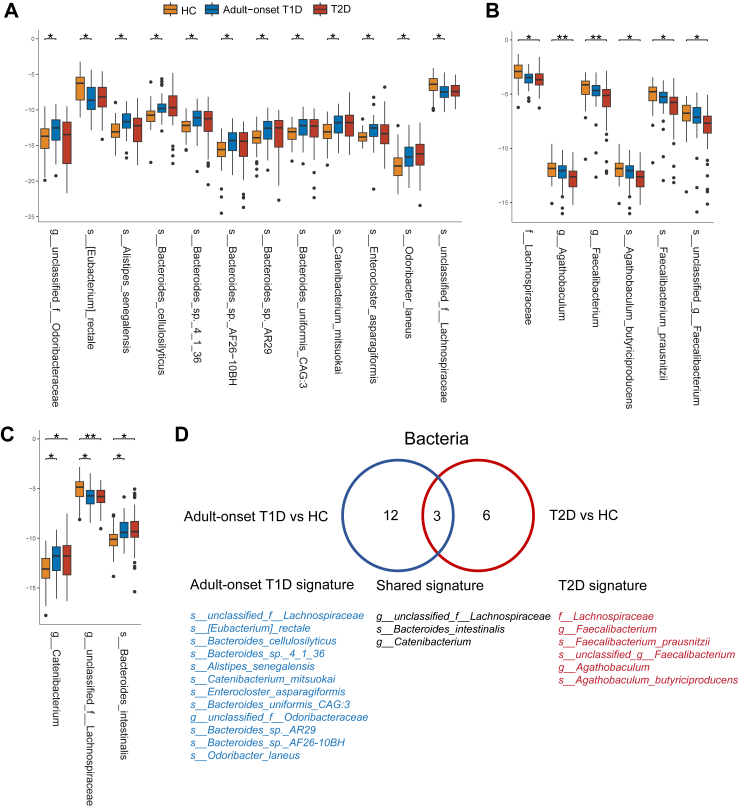
Next, we analysed the microbial composition at different taxonomic levels. The microbial composition at the phylum and genus level were shown in Fig. 2G and H. LEfSe analysis was used to identify the differentially abundant microbial features among HCs, adult-onset T1D patients, and T2D patients. A total of 102 species between adult-onset T1D patients and HCs, 86 species between T2D patients and HCs, and 49 species between adult-onset T1D patients and T2D patients were identified as differentially abundant microbial species (LDA value > 2, P < 0.05) (Supplementary Tables S2 and S3). Given that host factors may influence microbial composition, we applied RDA analysis to identify the potential confounding factors associated with the microbial composition in these groups and then adjusted them by the MaAsLin test. Host factors including age, sex, BMI and duration of diabetes were included in RDA analysis and BMI was identified as a confounding factor (P = 0.038). Regarding the bacterial microbiota, when compared with those in HCs, 15 taxa in adult-onset T1D patients and nine taxa in T2D patients remained significantly differentially altered after multiple testing correction and adjustment for BMI (LDA value > 2, Pfdr < 0.1). Among the 15 taxa that were altered in adult-onset T1D, 12 taxa were exclusively changed in adult-onset T1D patients, and ten of them were enriched in adult-onset T1D, most of which belonged to the genera Bacteroides, Catenibacterium, and Alistipes, while the other two species were depleted in adult-onset T1D patients compared with HCs, including Eubacterium rectale and unclassified Lachnospiraceae (Fig. 3A, D). Among the nine taxa that were altered in T2D, six taxa were exclusively decreased in T2D patients when compared with HCs, and they belonged to the family Ruminococcaceae and family Lachnospiraceae, such as Faecalibacterium prausnitzii, and Agathobaculum butyriciproducens (Fig. 3B, D). Further, three taxa were altered in both adult-onset T1D and T2D patients as shown in Fig. 3C and D. Additionally, we identified functional alterations of gut microbiota in adult-onset T1D. We found that the pathways of “Alanine, aspartate and glutamate metabolism” and “Nucleotide excision repair” were significantly enriched in adult-onset T1D patients when compared with HCs and T2D patients (Supplementary Fig. S1).
Fig. 3.
Gut microbiota signatures in patients with adult-onset T1D and patients with T2D. (A–C) Boxplots show the relative abundance of taxa exclusively altered in patients with adult-onset T1D (A), patients with T2D (B), and both patient groups (C) when compared with HCs. (D) Venn diagram of 21 taxa with altered abundances in patients with adult-onset T1D, patients with T2D or shared in both patient groups. Box plots show median ± quartiles, and the whiskers extend from the hinge to the largest or smallest value no further than 1.5 folds of the inter-quartile range. ∗Pfdr < 0.1, ∗∗Pfdr < 0.05. Abbreviations: HCs, healthy controls; T1D, type 1 diabetes; T2D, type 2 diabetes.
Regarding the viruses, when compared with those in HCs, six taxa were differentially abundant in adult-onset T1D patients, and five taxa were differentially abundant in T2D patients (LDA > 2, P < 0.05, Supplementary Tables S4 and S5). But only three of the taxa which were altered in adult-onset T1D remained significant after multiple testing correction and adjustment for BMI. Regarding the fungi, compared with HCs, six fungal species were found differentially abundant in T2D patients (LDA > 2, P < 0.05, Supplementary Table S6), and two of them remained significantly decreased after multiple testing correction and adjustment for BMI.
Decreased bacterial-viral/fungal associations in adult-onset T1D
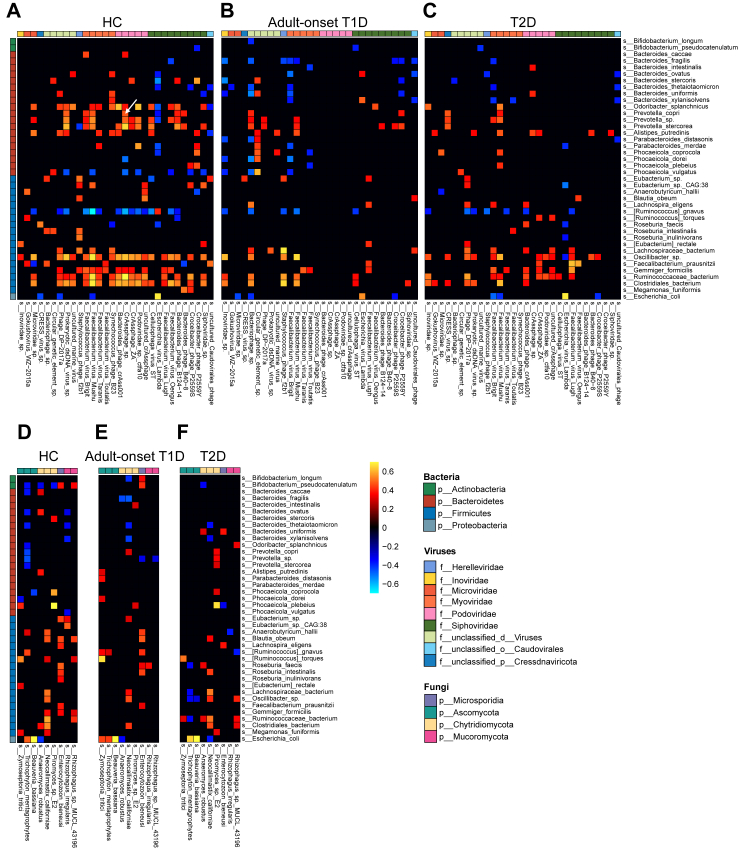
Accumulating evidence suggests that viruses and fungi also play an essential role in maintaining the homoeostasis of gut microecology and contribute to metabolic diseases.23,24 We further explored the microbial relationships among the bacterial, viral, and fungal prevalent species in different study groups. The number of correlations between gut bacterial and viral communities was decreased in adult-onset T1D compared with that in T2D patients and HCs (Fig. 4A–C). Positive bacterial-viral trans-kingdom correlations accounted for 81.5% in HCs and 73.0% in T2D patients but only 56.3% in adult-onset T1D patients (Chi-squared test, P < 0.001 for all). Compared with that in HCs, the strength of correlations between the bacteriome and virome was significantly weaker in the diabetic groups and was the weakest in the adult-onset T1D group (Fig. 4A–C). The significant reductions in bacterial and viral associations in adult-onset T1D occurred mainly between viruses assigned to the family Podoviridae or Siphoviridae and bacteria from the genus Prevotella or Oscillibacter. Interestingly, P. copri, which is the host of crAss-like phages,25 showed a strong positive correlation with CrAssphage sp. in HCs (Fig. 4A). However, the similar correlation disappeared in adult-onset T1D patients (Fig. 4B). Moreover, the strength of correlations between bacterial and fungal communities in the adult-onset T1D and T2D groups was also significantly weaker than that in the HC group (Fig. 4D–F). Taken together, these results indicate that notable changes in the bacterial-viral or bacterial-fungal trans-kingdom relationships present in adult-onset T1D, which may contribute to disease development. The robust positive correlations between phages and their bacterial hosts may be important in maintaining the ecological balance of the gut microbiota.
Fig. 4.
Trans-kingdom correlations in patients with adult-onset T1D, patients with T2D and HCs. The trans-kingdom associations of the top 40 bacterial prevalent species and the top 30 viral prevalent species were calculated with Spearman’s correlation in HCs (A), patients with adult-onset T1D (B), and patients with T2D (C). The trans-kingdom associations of the top 40 bacterial prevalent species and the top nine fungal prevalent species were calculated with Spearman’s correlation in HCs (D), patients with adult-onset T1D (E), and patients with T2D (F). Prevalent species were present in more than 20% of the samples with a relative abundance of ≥0.01%. Only correlations with rho > 0.3 or <−0.3 and P value < 0.05 were selected for visualization. Abbreviations: HCs, healthy controls; T1D, type 1 diabetes; T2D, type 2 diabetes; PCoA, principal coordinated analysis; PERMANOVA, permutational multivariate analysis of variance.
Associations of the microbiota and faecal/serum metabolites
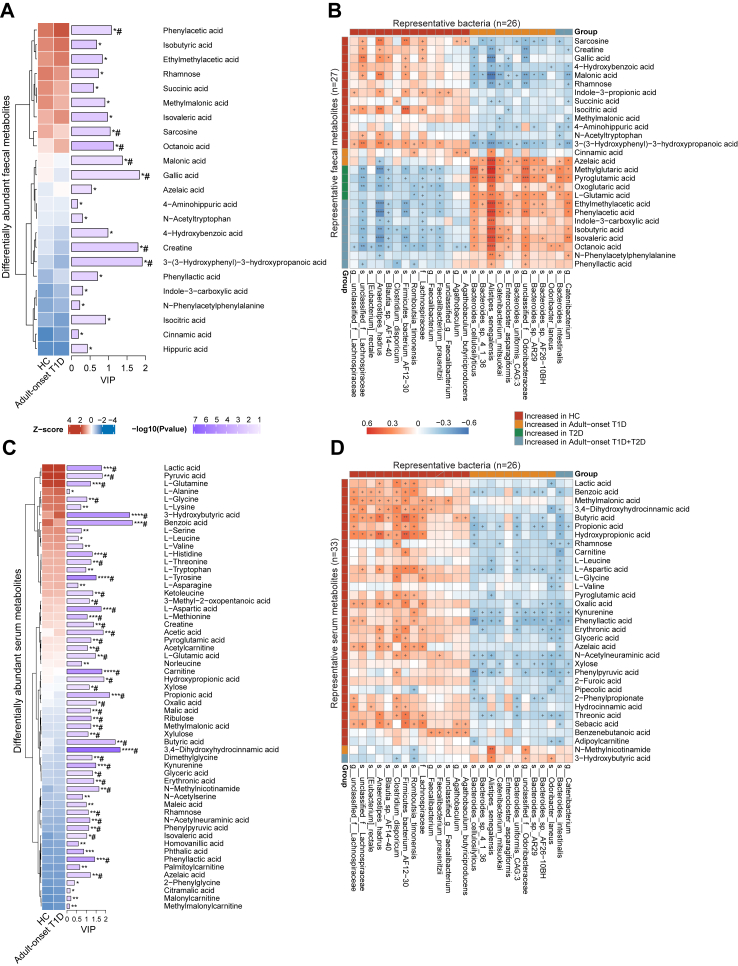
To further elucidate the association between the microbiota and host metabolism, we performed targeted metabolomics using the faecal and serum samples. When compared with the HC group, there were 23 and 25 differentially abundant faecal metabolites, as well as 57 and 71 differentially abundant serum metabolites in adult-onset T1D and T2D, respectively (Pfdr < 0.1, Fig. 5, Supplementary Figs. S2 and S3). Of note, some phenolic acids and their derivatives were markedly decreased in adult-onset T1D patients, especially gallic acid and 3,4-dihydroxyhydrocinnamic acid (Supplementary Tables S7–S10). SCFA-producing bacteria and SCFAs (e.g., butyric acid and propionic acid) also showed significantly decreased abundance in adult-onset T1D patients compared to those of HCs, though the decline of these bacteria and SCFAs were comparable between adult-onset T1D and T2D patients. Next, we performed correlation analysis to investigate the associations between differentially abundant bacteria and metabolites. We found that HC-enriched bacteria had strongly positive correlation with HC-enriched metabolites but were negatively correlated with diabetes-enriched metabolites (Fig. 5B and D). Notably, we found that some less abundant phenolic acids and their derivatives (e.g., gallic acid and 3,4-dihydroxyhydrocinnamic acid) were positively correlated with SCFA-producing bacteria, which were decreased in adult-onset T1D. And most of these SCFA-producing bacteria belonged to family Lachnospiraceae, genus Clostridium, and genus Anaerostipes (Fig. 5D).
Fig. 5.
Differentially abundant faecal/serum metabolites and their associations with gut bacteria. Standardized relative abundance and VIP score of altered faecal (A) and serum (B) metabolites identified by comparison between patients with adult-onset T1D patients and HCs. Altered metabolites were identified by OPLS-DA. (C, D) Associations of representative bacteria and faecal or serum metabolites that were altered in adult-onset T1D patients, T2D patients or both compared with HCs were assessed by Spearman’s correlation analysis. The relative abundance was transformed into Z scores. +P < 0.05, ∗Pfdr < 0.1, ∗∗Pfdr < 0.05, ∗∗∗Pfdr < 0.01, ∗∗∗∗Pfdr < 0.001, #VIP >1. Abbreviations: FC, fold change; OPLS-DA, orthogonal partial least squares discriminant analysis; VIP, variable influence on projection; HCs, healthy controls; T1D, type 1 diabetes; T2D, type 2 diabetes.
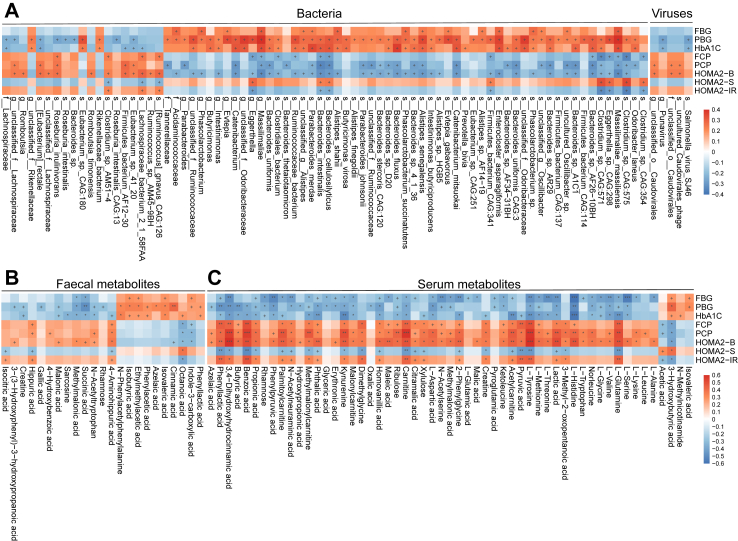
Associations of the altered microbes and metabolites with clinical parameters
To better understand the role of gut microbiota in disease progression, we analysed the associations of clinical parameters and differentially abundant bacteria or metabolites in adult-onset T1D and HC (Fig. 6). We found some adult-onset-T1D-related taxa, including some SCFA-producing bacteria that showed significant correlations with glucose-related parameters and pancreatic beta cell function, which were in accordance with previous study.26 For instance, E. rectale, a butyric acid-producing bacteria, was negatively correlated with PBG and HbA1c (ρ = −0.25, P = 0.029; ρ = −0.26, P = 0.019; Fig. 6A), and was positively correlated with PCP and HOMA2-B (ρ = 0.25, P = 0.024; ρ = 0.26, P = 0.016). Some viruses such as Salmonella virus SJ46, uncultured Caudoviralesphage and unclassified Caudovirales, also showed negative correlations with PBG and HbA1c, or positive correlations with PCP and HOMA2-B (Fig. 6A). In addition, we found some novel links between host phenotypes and faecal metabolites including faecal gallic acid, an adult-onset T1D-depleted phenolic acid, which was negatively correlated with PBG (ρ = −0.31, P = 0.020, Fig. 6B). Regarding serum metabolites, four serum amino acids were decreased in adult-onset T1D, including L-glycine, L-serine, L-threonine, and L-glutamine, which were negatively associated with glucose metabolism-related parameters but positively correlated with pancreatic beta cell function (Fig. 6C). In patients with adult-onset T1D only Bacteroides faecis was negatively correlated with FBG and titres of ZnT8A (ρ = −0.32, P = 0.027; ρ = −0.31, P = 0.029; respectively). Serum rhamnose, which was depleted in adult-onset T1D patients, was positively correlated with PCP and HOMA2-B (ρ = 0.42, P = 0.037; ρ = 0.48, P = 0.016; respectively). Additionally, serum 3,4-dihydroxyhydrocinnamic acid was negatively correlated with titres of ZnT8A (ρ = −0.38, P = 0.047) (Supplementary Fig. S4). These findings suggest potential links between gut microbiota, metabolites and pancreatic beta cell autoimmunity in adult-onset T1D.
Fig. 6.
Interactions of disease-related taxa/metabolites and host clinical parameters in patients with adult-onset T1D and HCs. Associations of differentially abundant taxa (A) or faecal/serum metabolites (B, C) and clinical parameters in patients with adult-onset T1D and HCs. Correlations were calculated with Spearman’s correlation analysis. +P < 0.05, ∗Pfdr < 0.1, ∗∗Pfdr < 0.05, ∗∗∗Pfdr < 0.01. Abbreviations: FBG, fasting blood glucose; PBG, postprandial blood glucose; HbA1c, haemoglobin A1c; FCP, fasting C-peptide; PCP, postprandial C-peptide; HOMA2-B, homoeostasis model assessment 2 estimates of β-cell function; HOMA2-S, homoeostasis model assessment 2 estimates of insulin sensitivity; HOMA2-IR, homoeostasis model assessment 2 estimates of insulin resistance.
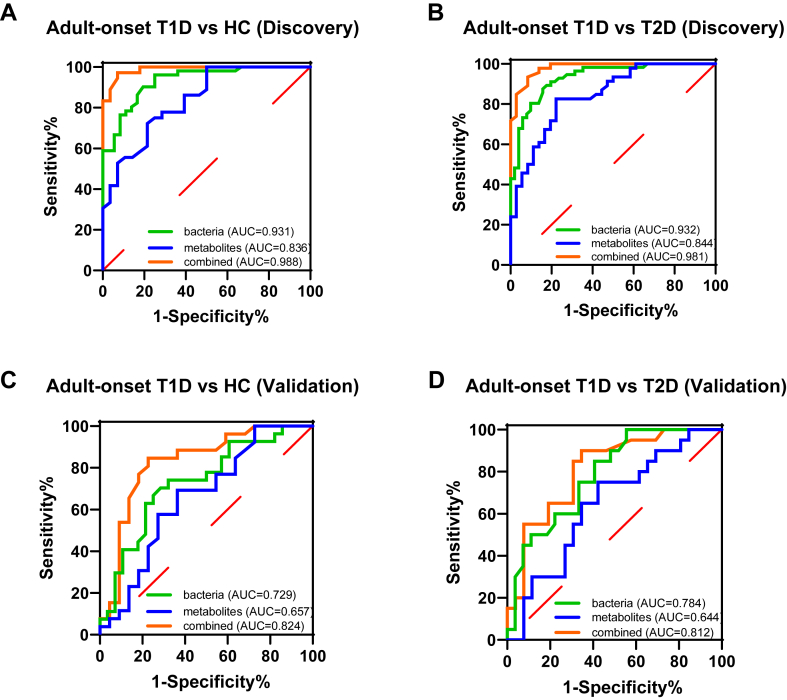
Multiomic classifier discriminating patients with adult-onset T1D from HCs or patients with T2D
We next assessed the potential value of using the gut microbiota and metabolites as biomarkers for the differential diagnosis of diabetes. We built random forest models based on faecal taxonomic or metabolic features that were altered in adult-onset T1D or T2D to discriminate adult-onset T1D patients from HCs and from T2D patients (Supplementary Table S11). A random forest model was employed to select key discriminatory gut microbiota or metabolites constituents. We identified a bacterial signature composed of six bacterial species that could distinguish patients with adult-onset T1D from HCs or patients with T2D (adult-onset T1D vs. HC: area under the curve (AUC) = 0.931 [95% CI 0.881–0.982]; adult-onset T1D vs. T2D: AUC = 0.932 [95% CI 0.887–0.978]). In addition, the diagnostic efficacy of another random forest model based on six identified faecal metabolites showed similar results (adult-onset T1D vs. HC: AUC = 0.836 [95% CI 0.740–0.933]; adult-onset T1D vs. T2D: AUC = 0.844 [95% CI 0.761–0.927]). Notably, using six identified bacterial species and six identified metabolites, the combined model yielded an AUC of 0.988 (95% CI 0.970–1.000) in discriminating between patients with adult-onset T1D and HCs, and the same model reached the AUC of 0.981 (95% CI 0.959–1.000) for discriminating patients with adult-onset T1D from patients with T2D (Fig. 7A and B).
Fig. 7.
Disease classification based on the signatures of gut microbiome and metabolome. Random forest classifiers composed of bacteria, metabolites and their combination were constructed to discriminate patients with adult-onset T1D from HCs (A) and from patients with T2D (B) in the discovery set. Validation of random forest classifiers composed of bacteria, metabolites and their combination in discriminating patients with adult-onset T1D from HCs (C) and from patients with T2D (D). Abbreviations: HCs, healthy controls; T1D, type 1 diabetes; T2D, type 2 diabetes; AUC, area under the curve.
Moreover, we verified the diagnostic performance of the above models in the validation set independently. Accordingly, we found that the combined marker panel of six bacterial species and six faecal metabolites could still effectively differentiate patients with adult-onset T1D from HCs with an AUC of 0.824 (95% CI 0.698–0.951) and differentiate patients with adult-onset T1D from patients with T2D with an AUC of 0.812 (95% CI 0.687–0.936) in the validation set (Fig. 7C and D).
Discussion
In this study, we reported the metagenomic and metabolomic features of adult-onset T1D. To our knowledge, this is the first study using metagenomics and metabolomics to identify not only the altered gut bacteria but also intestinal viruses and fungi in patients with adult-onset T1D in comparison with patients with T2D and/or HCs. The faecal and serum metabolomic profiles were also significantly distinct across the three study groups and were tightly linked with microbiota compositions. Importantly, we found that microbial and metabolic changes in adult-onset T1D were associated with islet autoimmunity and pancreatic beta cell function. Using different models, we independently validated a biomarker panel of gut bacteria and metabolites that can distinguish patients with adult-onset T1D from HCs as well as from patients with T2D with high accuracy. Moreover, we detected different signatures of the gut viral and fungal species in patients with adult-onset T1D and found that the bacterial-viral or bacterial-fungal correlations were significantly reduced in patients with adult-onset T1D in comparison to HCs. Most of the studies have been focusing on intestinal bacteria, little is known about intestinal viral and fungal signatures in diabetes, thus, our findings lay the foundation for better understanding the role of the entire intestinal ecosystem in different types of diabetes and may provide a more precise diagnostic tool for adult-onset T1D.
In the current study, we identified distinctive signatures of microbiota and metabolites in adult-onset T1D. It is striking that the metabolites of patients with adult-onset T1D showed significantly decreased abundance of some phenolic acids and their derivatives (e.g., gallic acid, 3,4-dihydroxyhydrocinnamic acid). Phenolic acids, an important category of polyphenols, exert high antioxidant properties, antidiabetic ability and anti-inflammatory effects on accelerating the differentiation of T cells, increasing the number of Tregs, and repressing the release of inflammatory cytokines from macrophages.27,28 Plant extracts containing polyphenols like gallic acid could reduce insulitis and maintain serum insulin levels in NOD mice.29 Intriguingly, studies have indicated phenolic acids, particularly gallic acid, can increase the levels of SCFAs, which are known to have important beneficial functions locally in the intestine and immuno-modulatory effects systemically.30 In line with those findings, we found an apparent loss of SCFA-producing bacteria accompanied by decreased levels of serum SCFAs in patients with adult-onset T1D, which were linked to pancreatic beta cell function and islet autoimmunity. Animal studies showed that SCFA-rich diets could protect NOD mice from the development of T1D through enhancing gut integrity and increasing the number and function of Tregs.31 It is conceivable to speculate that the severe loss of phenolic acids lead to the decrease in the abundance of SCFAs-producing bacteria and the subsequent reduction of SCFAs, which is a risk factor for the onset of T1D.26,31 Therefore, the supplementation of these phenolic acid compounds could be a promising approach to alleviate the disease severity.
Some recent studies have suggested that enteroviruses and fungi may play essential roles in the pathogenesis of autoimmune diabetes.24,32 Interestingly, we found that viruses from patients with adult-onset T1D showed higher alpha diversity than those from HCs. However, the positive correlations between bacteria and viruses in patients with adult-onset T1D were lower and weaker than those in HCs and in patients with T2D. To a large extent, most of the reduced positive correlations found in patients with adult-onset T1D were between viruses and beneficial bacteria. Interestingly, similar reductions in trans-kingdom correlations were also found in other autoimmune diseases, such as ulcerative colitis.33 Although limited fungal taxa were identified in our study, we did find a mild reduction in bacterial-fungal trans-kingdom relationships in patients with adult-onset T1D compared to those in HCs. Little is known about the role of fungi in autoimmune diseases such as T1D and it is clear that some extensive and in-depth investigations are needed to elucidate their role in the pathogenesis of adult-onset T1D.
Importantly, based on gut microbial signatures and metabolic features of the disease, we built a prediction model of adult-onset T1D, which could accurately identify the disease. We showed that the differentiation power of the model could be further improved with the addition of metabolites, and the use of the “6 + 6” model could simultaneously distinguish patients with adult-onset T1D from patients with T2D and from HCs. Thus, it is possible to differentiate the disease from the perspective of gut microbiota and metabolites. More importantly, the efficacy of the model was sufficiently validated in an external validation cohort. At present, the methods to differentiate adult-onset T1D from T2D are limited to the use of clinical phenotypes such as disease onset pattern, C-peptide levels, BMI and islet autoantibody levels. However, the increasing prevalence of obesity in patients with T1D, due to environment and lifestyle factors, the presence of ketosis-prone in patients with T2D and idiopathic T1D, as well as unavailable facility for autoantibody detection in some clinics make it difficult to accurately classify different types of diabetes. In that sense, our study provides a valuable auxiliary aid to diagnostic precision of diabetes.
Our study has several limitations. First, despite our efforts to address for confounding factors when comparing among the three groups (sex- and age-matched with comparable demographic characteristics, antibiotic exposure and comorbidities), our findings could be influenced by other confounders including described differences such as disease duration, and not well documented dietary intake which was based solely on a questionnaire. Second, there might be missing information on the RNA virome, as our approach could only identify faecal eukaryotic viruses. Additionally, the methods used in this study were not specifically designed to capture viral or fungal changes. Third, our study only evaluated the prediction value of microbial and metabolic features, and a prediction model integrating demographic and clinical variables with gut microbial features and metabolic characteristics is required to achieve higher efficacy and accuracy. Fourth, due to the cross-sectional design, the results cannot establish a causal relationship between the identified gut microbiota and adult-onset T1D. Finally, the sample size of this study was relatively small, and the subjects were restricted to a specific ethnic population and geographic region, which may limit the generalizability of the results. Therefore, the significance of these findings on the new biomarkers for adult-onset T1D remains to be confirmed by larger prospective follow-up studies involving more ethnic populations and geographic regions.
In summary, our study indicates that patients with adult-onset T1D have altered host-microbiota-metabolite interactions, which is likely involved in the pathogenesis of the disease. Moreover, our study provides some insight into gut microbiota and metabolite signatures that could be useful tools in the differential diagnosis of different types of diabetes.
Contributors
Y.X., Z.Z. and A.X. designed the study. J.H. and J.D. recruited subjects, conducted the experiments, performed the statistical analysis, and drafted the manuscript. X.L., L.X., C.L., Y.T., K.G., J.H., S.L., J.Y., W.P., and C.H. participated in the recruitment of subjects and contributed to clinical data acquisition. J.L. and T.Z. contributed to the analysis of the microbiota and metabolites data. L.W., A.X., Z.Z. and Y.X. revised the manuscript. Y.X., Z.Z. and A.X. are the guarantors of this work, had full access to all of the data in the study and were responsible for the decision to submit the manuscript. J.H., J.D., Y.X., Z.Z. and A.X. have assessed and verified the data. All authors contributed to the article and approved the submitted version.
Data sharing statement
Additional data from the analyses presented in this paper are available in the Supplementary material. The raw metagenomic sequence data of this study have been deposited in NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under the accession number PRJNA893406.
Declaration of interests
No potential conflicts of interest relevant to this article were reported.
Acknowledgments
We thank the fundings supported by the National Key Research and Development Program of China (2018YFE0114500), the National Natural Science Foundation of China (82270891, 82200933, 81820108007), the Natural Science Foundation of Hunan Province for Youths (2022JJ40689, 2022JJ40718), and the Natural Science Foundation of Changsha (kq2202404).
Footnotes
Translation: For the language translation of the abstract see Supplementary materials section.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2023.102132.
Contributor Information
Jingyi Hu, Email: hujingyi0066@csu.edu.cn.
Jin Ding, Email: dingjin2020@csu.edu.cn.
Xia Li, Email: lixia2014@vip.163.com.
Jun Li, Email: jun.li@cityu.edu.hk.
Tingting Zheng, Email: tingting.zheng@tum.de.
Lingxiang Xie, Email: xielingxiang@csu.edu.cn.
Chenyu Li, Email: 218211027@csu.edu.cn.
Yingxin Tang, Email: tangyingxin95@csu.edu.cn.
Keyu Guo, Email: 198206037@csu.edu.cn.
Juan Huang, Email: juan.huang@csu.edu.cn.
Shanshan Liu, Email: lss0625@csu.edu.cn.
Jianru Yan, Email: 380102106@qq.com.
Weijun Peng, Email: pengweijun87@csu.edu.cn.
Can Hou, Email: houcan84@csu.edu.cn.
Li Wen, Email: li.wen@yale.edu.
Aimin Xu, Email: amxu@hku.hk.
Zhiguang Zhou, Email: zhouzhiguang@csu.edu.cn.
Yang Xiao, Email: xiaoyang29@csu.edu.cn.
Appendix ASupplementary data
References
- 1.Holt R.I.G., DeVries J.H., Hess-Fischl A., et al. The management of type 1 diabetes in adults. A consensus report by the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD) Diabetes Care. 2021;44(11):2589–2625. doi: 10.2337/dci21-0043. [DOI] [PubMed] [Google Scholar]
- 2.Leslie R.D., Evans-Molina C., Freund-Brown J., et al. Adult-onset type 1 diabetes: current understanding and challenges. Diabetes Care. 2021;44(11):2449–2456. doi: 10.2337/dc21-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas N.J., Lynam A.L., Hill A.V., et al. Type 1 diabetes defined by severe insulin deficiency occurs after 30 years of age and is commonly treated as type 2 diabetes. Diabetologia. 2019;62(7):1167–1172. doi: 10.1007/s00125-019-4863-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muñoz C., Floreen A., Garey C., et al. Misdiagnosis and diabetic ketoacidosis at diagnosis of type 1 diabetes: patient and caregiver perspectives. Clin Diabetes. 2019;37(3):276–281. doi: 10.2337/cd18-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harding J.L., Wander P.L., Zhang X., et al. The incidence of adult-onset type 1 diabetes: a systematic review from 32 countries and regions. Diabetes Care. 2022;45(4):994–1006. doi: 10.2337/dc21-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vatanen T., Franzosa E.A., Schwager R., et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562(7728):589–594. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin J., Li Y., Cai Z., et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 8.Murri M., Leiva I., Gomez-Zumaquero J.M., et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y., Li S.C., Hu J., et al. Gut microbiota profiling in Han Chinese with type 1 diabetes. Diabetes Res Clin Pract. 2018;141:256–263. doi: 10.1016/j.diabres.2018.04.032. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J., Ni Y., Qian L., et al. Decreased abundance of Akkermansia muciniphila leads to the impairment of insulin secretion and glucose homeostasis in lean type 2 diabetes. Adv Sci (Weinh) 2021;8(16) doi: 10.1002/advs.202100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen H.K., Gudmundsdottir V., Nielsen H.B., et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 12.Canfora E.E., Meex R.C.R., Venema K., Blaak E.E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol. 2019;15(5):261–273. doi: 10.1038/s41574-019-0156-z. [DOI] [PubMed] [Google Scholar]
- 13.Thingholm L.B., Rühlemann M.C., Koch M., et al. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe. 2019;26(2):252–264.e210. doi: 10.1016/j.chom.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Y., Zhang C., Shi H., et al. Characteristics of the gut microbiota and metabolism in patients with latent autoimmune diabetes in adults: a case-control study. Diabetes Care. 2021;44(12):2738–2746. doi: 10.2337/dc20-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shilo S., Godneva A., Rachmiel M., et al. The gut microbiome of adults with type 1 diabetes and its association with the host glycemic control. Diabetes Care. 2022;45(3):555–563. doi: 10.2337/dc21-1656. [DOI] [PubMed] [Google Scholar]
- 16.Alberti K.G., Zimmet P.Z. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 17.American Diabetes Association 2. Classification and diagnosis of diabetes. Diabetes Care. 2017;40(Suppl 1):S11–S24. doi: 10.2337/dc17-S005. [DOI] [PubMed] [Google Scholar]
- 18.Yang J., Zheng P., Li Y., et al. Landscapes of bacterial and metabolic signatures and their interaction in major depressive disorders. Sci Adv. 2020;6(49) doi: 10.1126/sciadv.aba8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Xiao Y., Zhong L., et al. Increased neutrophil elastase and proteinase 3 and augmented NETosis are closely associated with β-cell autoimmunity in patients with type 1 diabetes. Diabetes. 2014;63(12):4239–4248. doi: 10.2337/db14-0480. [DOI] [PubMed] [Google Scholar]
- 20.Levy J.C., Matthews D.R., Hermans M.P. Correct homeostasis model assessment (HOMA) evaluation uses the computer program. Diabetes Care. 1998;21(12):2191–2192. doi: 10.2337/diacare.21.12.2191. [DOI] [PubMed] [Google Scholar]
- 21.Leiva-Gea I., Sanchez-Alcoholado L., Martin-Tejedor B., et al. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: a case-control study. Diabetes Care. 2018;41(11):2385–2395. doi: 10.2337/dc18-0253. [DOI] [PubMed] [Google Scholar]
- 22.Alkanani A.K., Hara N., Gottlieb P.A., et al. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes. 2015;64(10):3510–3520. doi: 10.2337/db14-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang K., Niu J., Zuo T., et al. Alterations in the gut virome in obesity and type 2 diabetes mellitus. Gastroenterology. 2021;161(4):1257–1269.e13. doi: 10.1053/j.gastro.2021.06.056. [DOI] [PubMed] [Google Scholar]
- 24.Salamon D., Sroka-Oleksiak A., Gurgul A., et al. Analysis of the gut mycobiome in adult patients with type 1 and type 2 diabetes using next-generation sequencing (NGS) with increased sensitivity-pilot study. Nutrients. 2021;13(4):1066. doi: 10.3390/nu13041066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomofuji Y., Kishikawa T., Maeda Y., et al. Whole gut virome analysis of 476 Japanese revealed a link between phage and autoimmune disease. Ann Rheum Dis. 2022;81(2):278–288. doi: 10.1136/annrheumdis-2021-221267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan X., Wang R., Han B., et al. Functional and metabolic alterations of gut microbiota in children with new-onset type 1 diabetes. Nat Commun. 2022;13(1):6356. doi: 10.1038/s41467-022-33656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang K., Zhang L., Liao P., et al. Impact of gallic acid on gut health: focus on the gut microbiome, immune response, and mechanisms of action. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.580208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yahfoufi N., Alsadi N., Jambi M., Matar C. The immunomodulatory and anti-inflammatory role of polyphenols. Nutrients. 2018;10(11):1618. doi: 10.3390/nu10111618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumacher N.S., Colomeu T.C., de Figueiredo D., et al. Identification and antioxidant activity of the extracts of eugenia uniflora leaves. Characterization of the anti-inflammatory properties of aqueous extract on diabetes expression in an experimental model of spontaneous type 1 diabetes (NOD mice) Antioxidants (Basel, Switzerland) 2015;4(4):662–680. doi: 10.3390/antiox4040662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi X., Xiao C., Wang Y., Tang H. Gallic acid intake induces alterations to systems metabolism in rats. J Proteome Res. 2013;12(2):991–1006. doi: 10.1021/pr301041k. [DOI] [PubMed] [Google Scholar]
- 31.Mariño E., Richards J.L., McLeod K.H., et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017;18(5):552–562. doi: 10.1038/ni.3713. [DOI] [PubMed] [Google Scholar]
- 32.Isaacs S.R., Foskett D.B., Maxwell A.J., et al. Viruses and type 1 diabetes: from enteroviruses to the virome. Microorganisms. 2021;9(7):1519. doi: 10.3390/microorganisms9071519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuo T., Lu X.J., Zhang Y., et al. Gut mucosal virome alterations in ulcerative colitis. Gut. 2019;68(7):1169–1179. doi: 10.1136/gutjnl-2018-318131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.