Abstract
Rfv3 is a host resistance gene that operates through an unknown mechanism to control the development of the virus-neutralizing antibody response required for recovery from infection with Friend retrovirus. The Rfv3 gene was previously mapped to an approximately 20-centimorgan (cM) region of chromosome 15. More refined mapping was not possible, due to a lack of microsatellite markers and leakiness in the Rfv3 phenotype, which prevented definitive phenotyping of individual recombinant mice. In the present study, we overcame these difficulties by taking advantage of seven new microsatellite markers in the Rfv3 region and by using progeny tests to accurately determine the Rfv3 phenotype of recombinant mice. Detailed linkage analysis of relevant crossovers narrowed the location of Rfv3 to a 0.83-cM region. Mapping of closely linked genes in an interspecific backcross panel allowed us to exclude two previous candidate genes, Ly6 and Wnt7b. These studies also showed for the first time that the Hsf1 gene maps to the Rfv3-linked cluster of genes including Il2rb, Il3rb, and Pdgfb. This localization of Rfv3 to a region of less than 1 cM now makes it feasible to attempt the cloning of Rfv3 by physical methods.
Infection with Friend virus complex (FV) induces rapid polyclonal erythroid cell proliferation and splenomegaly in genetically susceptible adult mice (5, 10). Mice of strains which mount rapid humoral and cell-mediated FV-specific immune responses spontaneously recover from FV-induced splenomegaly without progressing to erythroleukemia. Such recovery from FV disease is dependent on a number of host genes, including several genes of the major histocompatibility complex (MHC), which influence critical CD4+- and CD8+-T-cell responses (5, 13). However, unlike some other viral systems in which either a cellular or humoral immune response alone is sufficient to resolve infection (1, 19, 20), spontaneous recovery from FV requires both FV-specific T-cell responses and virus-neutralizing antibody responses (6, 12, 21).
The non-MHC gene Rfv3 influences the ability of mice to mount FV-neutralizing antibody responses following infection (6). C57BL/10 and C57BL/6 mice have the genotype Rfv3r/Rfv3r, and BALB/c, A.BY, and A/WySn mice have the genotype Rfv3s/Rfv3s (6, 7, 9). At about 2 weeks postinfection, mice carrying at least one dominant Rfv3 resistance allele (Rfv3r), such as (C57BL/10 × A.BY)F1 mice, begin to make FV-neutralizing antibodies, and they usually clear FV plasma viremia by 30 days postinfection (DPI). In contrast, mice with two sensitive alleles (Rfv3s/Rfv3s) fail to make FV-neutralizing antibodies, remain viremic, and eventually succumb to FV-induced erythroleukemic splenomegaly (6, 9). Interestingly, Rfv3s/Rfv3s mice have normal antibody responses to other antigens, suggesting that these mice are not generally immunosuppressed (18). The mechanism whereby Rfv3 controls the FV-specific humoral response remains unknown. The gene has been mapped to a 20-centimorgan (cM) region of mouse chromosome 15, ruling out linkage to genes such as MHC genes, immunoglobulin genes, or T-cell receptor genes (14). In this study, we used progeny testing and microsatellite linkage analysis with seven new markers to define the location of the Rfv3 gene to a region of less than 1 cM. These experiments determined that the previous candidate genes, Ly6 and Wnt7, mapped to regions adjacent to, rather than within, the Rfv3 gene region.
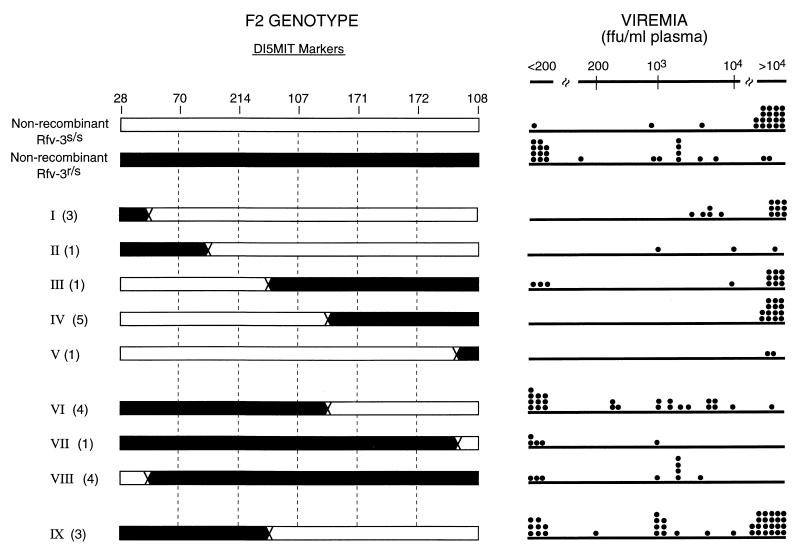
To map the Rfv3 gene, heterozygous (B10.A × A/Wy)F1 mice (Rfv3r/Rfv3s) were intercrossed to produce F2 offspring. Tail tip DNA samples from 181 F2 mice were analyzed by using PCR amplification of simple sequence length polymorphisms (microsatellites) (15). Initially, two markers, D15Mit28 and D15Mit108, which flank the 20-cM region containing the Rfv3 gene (14), were used to identify 45 recombinant F2 mice. DNA from these recombinants with microsatellite markers lying between the flanking markers was further tested, and nine recombination locations (groups I through IX) were identified (Fig. 1). Because the Rfv3 phenotype shows some leakiness even in genetically identical mice (Fig. 1) (14), we could not rely on the accuracy of phenotyping of individual recombinant F2 mice. Rather, the recombinant F2 mice were backcrossed to A/Wy parental mice, and the resulting progeny were genotyped and tested. A total of 23 of the 45 recombinant F2 mice were backcrossed, and all progeny were genotyped prior to phenotypic analysis. For determination of the Rfv3 phenotype, recombinant progeny and a number of nonrecombinant littermate control mice were infected with FV and tested for plasma viremia at 30 DPI. Viremia has been shown to inversely correlate with FV-neutralizing antibody production (9, 22) and is a convenient assay for determining the Rfv3 phenotype. Mice exhibiting less than 200 focus-forming units (FFU)/ml of plasma were scored as nonviremic, whereas mice with more than 104 FFU/ml of plasma were considered highly viremic. Leakiness in the Rfv3 phenotype was manifested by intermediate viremia levels, which were observed at a low incidence in both Rfv3r/Rfv3s and Rfv3s/Rfv3s controls (Fig. 1).
FIG. 1.
Genotypes of recombinant (B10.A × A/Wy)F2 mice and corresponding Rfv3 phenotypes for (B10.A × A/Wy)F2 × A/Wy backcross progeny. (Left) F2 mice were typed for D15Mit markers by PCR. Thirty cycles of PCR were performed with 100 ng of tail DNA template, 1× PCR buffer (Promega), 0.2 μM deoxynucleoside triphosphate, 1 mM MgCl2, 1 μM flanking primers, and 0.05 U of Taq polymerase (Promega). Arabic numbers at the top refer to the markers used in this study. The nine crossover locations detected on chromosome 15 are shown. The markers are evenly spaced for convenience, and crossover is arbitrarily shown halfway between each marker. Black regions denote DNA originating from the B10.A (Rfv3r/Rfv3r) parent; white regions denote DNA originating from the A/Wy (Rfv3s/Rfv3s) parent. Numbers in parentheses indicate the numbers of recombinant F2 mice showing crossover in the same region which were progeny tested. (Right) Plasma viremia data were analyzed at 30 DPI for individual recombinant (B10.A × A/Wy)F2 × A/Wy backcross progeny from each crossover group. Viremia data for nonrecombinant (Rfv3r/Rfv3s or Rfv3s/Rfv3s) littermates from each backcross are grouped at the top. Viremia levels between 200 and 104 FFU/ml are plotted on a log10 scale; values of <200 and values of >104 are grouped. Each dot represents the FV viremia titer for one mouse as detected by focal immunoassay on Mus dunni cells (17). The detection limit of the assay was 200 FFU/ml.
To map Rfv3, we looked for a correlation between the genotypes and phenotypes in the recombinant backcross progeny. High levels of viremia were seen in 35 of 46 (76%) recombinant progeny from groups I, II, III, IV, and V. When the genotypes of these mice were compared, it was observed that the mice had the Rfv3s/Rfv3s genotype at marker D15Mit214 (Fig. 1). In contrast, the low-viremia groups VI, VII, and VIII had the Rfv3r/Rfv3s genotype at this marker (Fig. 1). These results indicated that the Rfv3 gene was located near D15Mit214 in a region between markers D15Mit70 and D15Mit107.
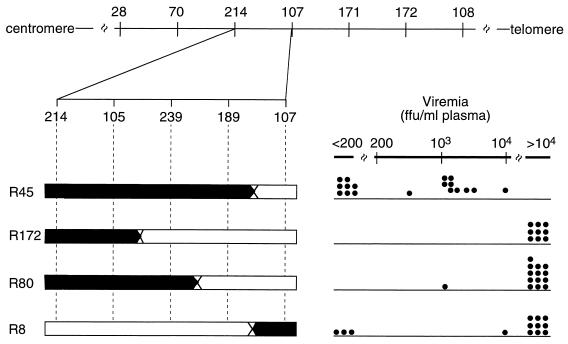
Group IX, which consisted of progeny from three F2 recombinants, R45, R172, and R80, exhibited both high and low levels of viremia (Fig. 1 and 2). This variability prompted further typing of the mice of this group in order to detect possible differences in recombination positions. Typing with three markers located between D15Mit214 and D15Mit107 revealed unique crossover positions in these three recombinants (Fig. 2). Interestingly, 17 of 18 progeny of R45 were either nonviremic or showed only intermediate viremia (<104 FFU/ml of plasma), whereas 20 of 21 progeny of recombinants R172 and R80 were highly viremic (Fig. 2). These data were consistent with a location for Rfv3 that was distal to D15Mit239 and proximal to D15Mit107 (Fig. 2). Progeny testing of another recombinant mouse, R8 (group III), with a recombination in the same region supported this position for Rfv3 (Fig. 2).
FIG. 2.
Fine mapping of the recombination between D15Mit214 and D15Mit107 in recombinant F2 mice (R45, R172, R80, and R8) and Rfv3 phenotype (viremia) data for recombinant backcross progeny. Genotyping data for the intervening markers D15Mit105, D15Mit239, and D15Mit189 are shown. The parental contribution of the recombinant chromosomes is shown as in Fig. 1. Viremia data are the same as those shown in Fig. 1 for the backcross progeny of group IX (R45, R172, and R80) and group III (R8).
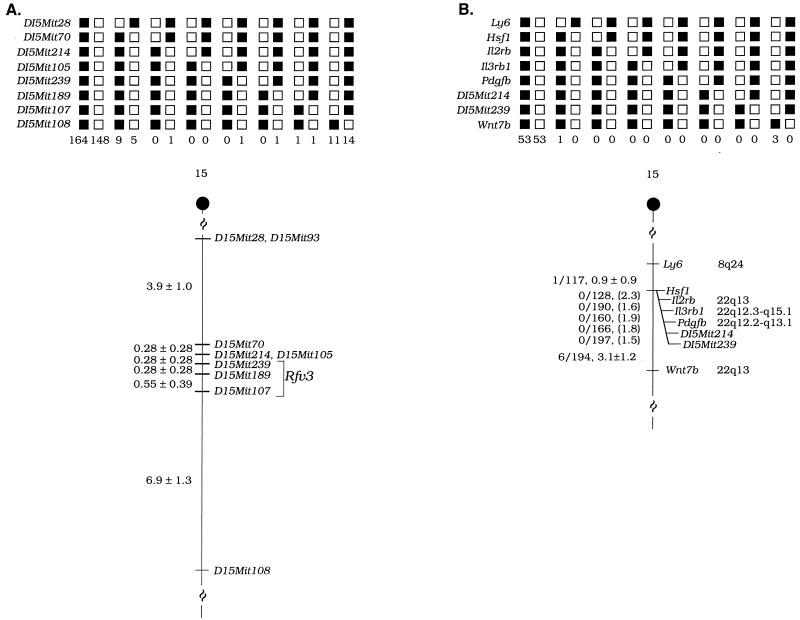
The genotyping of recombinant F2 mice has allowed us to determine the genetic distances between the D15Mit markers used in this study. Our previous results indicated that Rfv3 was located in a 5- to 20-cM region between D15Mit108 and D15Mit93. Here, we genotyped F2 mice with the microsatellite markers D15Mit108 and D15Mit28, which is very closely linked to D15Mit93. We now estimate the distance between these markers to be 12.2 cM (Fig. 3A). Furthermore, our mapping of Rfv3 between D15Mit239 and D15Mit107 localizes the Rfv3 position to a 0.83-cM region of chromosome 15 (Fig. 3A).
FIG. 3.
(A) Genetic distances and predicted position of Rfv3 on mouse chromosome 15 based on (B10.A × A/Wy)F2 recombinant mice. The chromosomes inherited from each F1 parent are shown as columns at the top. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. Shaded boxes represent the presence of a B10.A allele, and white boxes represent the presence of an A/Wy allele. In the partial chromosome 15 linkage map at the bottom, the genetic distances (means ± standard errors) between the D15Mit markers shown were calculated as the percentages of (B10.A × A/Wy)F2 recombinant animals of the total of 181 F2 animals which were typed. Since each F1 parent could contribute a recombinant chromosome, the distances were based on 362 total chromosomes 15. Three mice (six chromosomes) were determined to be recombinant in chromosome 15 but were not typeable at all the loci shown, and so they were omitted from the figure. The Rfv3 gene is predicted to be between D15Mit239 and D15Mit107, based on viremia data for animals in groups IX and III (Fig. 2). (B) D15Mit214 and D15Mit239, markers closely linked to Rfv3, map in the central region of mouse chromosome 15. The markers were placed on mouse chromosome 15 by analysis of the progeny of the interspecific backcross (C57BL/6J × M. spretus)F1 × C57BL/6J (8). Only animals that were recombinant in the Ly6-to-Wnt7b interval of mouse chromosome 15 were typed by PCR under the conditions described in the legend to Fig. 1. The segregation patterns of the markers and flanking genes in 110 backcross animals that were typed for all loci are shown at the top. Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J × M. spretus)F1 parent. The shaded boxes represent the presence of a C57BL/6J allele, and the white boxes represent the presence of an M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 15 linkage map showing the location of D15Mit214 and D15Mit239 in relation to linked genes is shown at the bottom. For individual pairs of loci, more than 110 animals were typed. The number of recombinant animals over the total number of animals typed is shown as a ratio; the recombination frequency, expressed as the mean genetic distance in centimorgans (± standard error), is shown for each pair of loci to the left of the chromosome. Standard errors were determined as described previously (2). When no recombination was detected between the loci, the upper 95% confidence limit of the recombination distance is given in parentheses. The positions of the loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from the Genome Data Base, a computerized database of human linkage information maintained by the William H. Welch Medical Library of The Johns Hopkins University (Baltimore, Md.).
To identify other genes in the interval of the Rfv3 locus, we mapped two closely linked markers, D15Mit214 and D15Mit239, in an interspecific backcross panel derived from the matings of (C57BL/6J × Mus spretus)F1 × C57BL/6J mice (8). This mapping panel has been typed for over 2,700 loci, most of which are genes that are well distributed among all the autosomes as well as the X chromosome. By this analysis, Rfv3 was separable from Ly6 and Wnt7b (Fig. 3B), two genes which cosegregated with Rfv3 in previous experiments (14). However, Rfv3 colocalized with a cluster of genes including the immune-system-related genes Il2rb (interleukin 2 [IL-2] receptor beta), Il3rb1 (IL-3 receptor beta 1), and Pdgfb (platelet-derived growth factor beta) in the central region of mouse chromosome 15 (3, 4, 11). This cluster of loci is 0.9 cM distal to Ly6 and 3.1 cM proximal to Wnt7b (Fig. 3B).
One previously unmapped gene, Hsf1 (heat shock factor 1), was also found to map to the same gene region as Rfv3. The position of Hsf1 was determined by Southern blot analysis with a cDNA probe (16). Major fragments of 11.5 and 2.8 kb were detected in TaqI-digested C57BL/6J DNA, and major fragments of 5.2 and 3.1 kb were detected in TaqI-digested M. spretus DNA. The presence or absence of the M. spretus fragments which cosegregated was monitored in [(C57BL/6 × M. spretus) × C57BL/6]B1 backcross mice. Recombination distances were calculated with Map Manager, version 2.6.5, and the gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
Ideally, it would be desirable to type the critical recombinant mice for crossovers between Rfv3 and the candidate genes Il2rb, Il3rb1, Pdgfb, and Hsf1. However, to our knowledge, no polymorphisms that distinguish the alleles of these genes in the C57BL/10 and A/Wy strains of mice have been reported. Furthermore, no crossovers between any of these genes have been detected in several crosses including Mus musculus and M. spretus, where distinguishing between alleles is more feasible. Because of the obvious importance of IL-2 receptor-mediated signal transduction in many immune responses, we made a preliminary attempt to detect allele-specific polymorphisms in Il2rb mRNAs but found no allelic differences between C57BL/10 and A/Wy mice. Also, we found no significant differences in the levels of expression of Il2rb when spleen RNA was examined by RNase protection assays at multiple time points during the first 3 weeks following infection with FV (data not shown).
The present study has narrowed the location of Rfv3 by a factor of over 20 and has both excluded two previous candidate genes and included Hsf1 as a new candidate. The central region of mouse chromosome 15 has homology with human chromosomes 8q and 22q (Fig. 3B), and further mapping of these regions of the human chromosomes may uncover additional candidate genes. While the analysis of candidates is appealing, the Rfv3 gene region could contain numerous unidentified genes, any of which could be Rfv3. The real advantage of the current fine mapping is that identification of Rfv3 by physical means is now feasible. Bacterial artificial chromosomes containing overlapping sections of the Rfv3 region can be produced and used to breed transgenic mice. The future identification of the Rfv3 gene will aid our understanding of the control of the FV host immune response and may ultimately contribute more generally to understanding human retroviral immunity.
Acknowledgments
The first two authors contributed equally to this study.
We thank Thomas Malek for the generous gift of the Il2rb cDNA clone and Verity Letts for use of her genetic maps. We also thank Clint Kenley, Deborah Householder, and Mary Barnstead for excellent technical assistance and John Portis and Don Lodmell for critical reviews of the manuscript.
This research was supported, in part, by the National Cancer Institute, DHHS, under contract with ABL.
REFERENCES
- 1.Bahmanyar M, Fayaz A, Nour-Salehi S, Mohammadi M, Koprowski H. Successful protection of humans exposed to rabies infection. Postexposure treatment with the new human diploid cell rabies vaccine and antirabies serum. JAMA. 1976;236:2751–2754. [PubMed] [Google Scholar]
- 2.Bancroft H. Introduction to biostatistics. New Orleans, La: Hoeber-Harper; 1962. [Google Scholar]
- 3.Brannan C I, Gilbert D J, Ceci J D, Matsuda Y, Chapman V M, Mercer J A, Eisen H, Johnston L A, Copeland N G, Jenkins N A. An interspecific linkage map of mouse chromosome 15 positioned with respect to the centromere. Genomics. 1992;13:1075–1081. doi: 10.1016/0888-7543(92)90021-j. [DOI] [PubMed] [Google Scholar]
- 4.Campbell H D, Webb G C, Kono T, Taniguchi T, Ford J H, Young I G. Assignment of the interleukin-2 receptor beta chain gene (Il-2rb) to band E on mouse chromosome 15. Genomics. 1992;12:179–180. doi: 10.1016/0888-7543(92)90428-u. [DOI] [PubMed] [Google Scholar]
- 5.Chesebro B, Miyazawa M, Britt W J. Host genetic control of spontaneous and induced immunity to Friend murine retrovirus infection. Annu Rev Immunol. 1990;8:477–499. doi: 10.1146/annurev.iy.08.040190.002401. [DOI] [PubMed] [Google Scholar]
- 6.Chesebro B, Wehrly K. Identification of a non-H-2 gene (Rfv-3) influencing recovery from viremia and leukemia induced by Friend virus complex. Proc Natl Acad Sci USA. 1979;76:425–429. doi: 10.1073/pnas.76.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesebro B, Wehrly K, Doig D, Nishio J. Antibody-induced modulation of Friend virus cell surface antigens decreases virus production by persistent erythroleukemia cells: influence of the Rfv-3 gene. Proc Natl Acad Sci USA. 1979;76:5784–5788. doi: 10.1073/pnas.76.11.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copeland N G, Jenkins N A. Development and applications of a molecular genetic linkage map of the mouse genome. Trends Genet. 1991;7:113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- 9.Doig D, Chesebro B. Anti-Friend virus antibody is associated with recovery from viremia and loss of viral leukemia cell surface antigens in leukemic mice. Identification of Rfv-3 as a gene locus influencing antibody production. J Exp Med. 1979;150:10–19. doi: 10.1084/jem.150.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friend C. Cell-free transmission in adult Swiss mice of a disease having the character of a leukemia. J Exp Med. 1957;105:307–318. doi: 10.1084/jem.105.4.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorman D M, Itoh N, Jenkins N A, Gilbert D J, Copeland N G, Miyajima A. Chromosomal localization and organization of the murine genes encoding the beta subunits (AIC2A and AIC2B) of the interleukin 3, granulocyte/macrophage colony-stimulating factor, and interleukin 5 receptors. J Biol Chem. 1992;267:15842–15848. [PubMed] [Google Scholar]
- 12.Hasenkrug K J, Brooks D M, Chesebro B. Passive immunotherapy for retroviral disease: influence of major histocompatibility complex type and T-cell responsiveness. Proc Natl Acad Sci USA. 1995;92:10492–10495. doi: 10.1073/pnas.92.23.10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasenkrug K J, Chesebro B. Immunity to retroviral infection: the Friend virus model. Proc Natl Acad Sci USA. 1997;94:7811–7816. doi: 10.1073/pnas.94.15.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasenkrug K J, Valenzuela A, Letts V A, Nishio J, Chesebro B, Frankel W N. Chromosome mapping of Rfv3, a host resistance gene to Friend murine retrovirus. J Virol. 1995;69:2617–2620. doi: 10.1128/jvi.69.4.2617-2620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hearne C M, Ghosh S, Todd J A. Microsatellites for linkage analysis of genetic traits. Trends Genet. 1992;8:288–294. doi: 10.1016/0168-9525(92)90256-4. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins N A, Copeland N G, Taylor B A, Lee B K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lander M R, Chattopadhyay S K. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell forus-forming viruses. J Virol. 1984;52:695–698. doi: 10.1128/jvi.52.2.695-698.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison R P, Nishio J, Chesebro B. Influence of the murine MHC (H-2) on Friend leukemia virus-induced immunosuppression. J Exp Med. 1986;163:301–314. doi: 10.1084/jem.163.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oldstone M B. The role of cytotoxic T lymphocytes in infectious disease: history, criteria, and state of the art. Curr Top Microbiol Immunol. 1994;189:1–8. doi: 10.1007/978-3-642-78530-6_1. [DOI] [PubMed] [Google Scholar]
- 20.Perry L L, Lodmell D L. Role of CD4+ and CD8+ T cells in murine resistance to street rabies virus. J Virol. 1991;65:3429–3434. doi: 10.1128/jvi.65.7.3429-3434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson M N, Spangrude G J, Hasenkrug K, Perry L, Nishio J, Wehrly K, Chesebro B. Role and specificity of T-cell subsets in spontaneous recovery from Friend virus-induced leukemia in mice. J Virol. 1992;66:3271–3277. doi: 10.1128/jvi.66.6.3271-3277.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Super H J, Brooks D, Hasenkrug K, Chesebro B. Requirement for CD4+ T cells in the Friend murine retrovirus neutralizing antibody response: evidence for functional T cells in genetic low-recovery mice. J Virol. 1998;72:9400–9403. doi: 10.1128/jvi.72.11.9400-9403.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]