Abstract
The 1.6-megabase deletion at chromosome 3q29 (3q29Del) is the strongest identified genetic risk factor for schizophrenia, but the effects of this variant on neurodevelopment are not well understood. We interrogated the developing neural transcriptome in two experimental model systems with complementary advantages: isogenic human cortical organoids and isocortex from the 3q29Del mouse model. We profiled transcriptomes from isogenic cortical organoids that were aged for 2 and 12 months, as well as perinatal mouse isocortex, all at single-cell resolution. Systematic pathway analysis implicated dysregulation of mitochondrial function and energy metabolism. These molecular signatures were supported by analysis of oxidative phosphorylation protein complex expression in mouse brain and assays of mitochondrial function in engineered cell lines, which revealed a lack of metabolic flexibility and a contribution of the 3q29 gene PAK2. Together, these data indicate that metabolic disruption is associated with 3q29Del and is conserved across species.
Single-cell transcriptomic profiling reveals effects of the schizophrenia-associated 3q29 deletion on mitochondrial function.
INTRODUCTION
Rare variants have now been identified that confer extraordinarily high risk for schizophrenia (SCZ). Functional study of these variants may yield insights into the molecular and cellular impairments that ultimately give rise to psychosis. By restricting investigation to a single variant, etiologic heterogeneity is vastly reduced, which may lead to better discrimination of causal mechanisms. To date, the strongest identified single genetic risk factor for SCZ is the 3q29 deletion (3q29Del), a copy number variant (CNV) that encompasses 22 protein-coding genes and is located near the telomeric end of human chromosome 3 (1, 2). Hemizygous loss of this set of genes is associated with at least a 40-fold increase in risk for SCZ (3, 4); this deletion also increases risk for additional neurodevelopmental and psychiatric conditions, including intellectual disability, autism spectrum disorder, and attention-deficit/hyperactivity disorder [Online Mendelian Inheritance in Man (OMIM) #609425] (5).
Exciting developments in molecular neuroscience have led to powerful new tools for the investigation of neurobiology of mental health disorders. In this study, we leverage two state-of-the-art experimental model systems, which together amplify the rigor of our approach. The starting substrate for these experiments are two CRISPR-engineered experimental systems: newly generated isogenic human induced pluripotent stem cells (iPSCs), where we have precisely introduced the 3q29Del using CRIPSR-Cas9 and the 3q29 mouse model (B6.Del16+/Bdh1-Tfrc), which bears complete synteny to the human 3q29 interval and displays neurodevelopmental and somatic correlates of human syndromic phenotypes (6–8). These experimental systems offer complementary advantages; cortical organoids are the current gold standard model of early human cortical development in vitro, whereas syntenic 3q29Del mice provide a source of brain tissue from a physiological context. We hypothesized that a transcriptomic analysis of differentially expressed genes (DEGs) in developing cortical tissue would provide relatively unbiased insights into underlying mechanisms of cellular dysfunction. We reasoned that the set of transcriptomic changes observed in both model systems are likely attributable to 3q29Del and may underlie core phenotypes.
To investigate the biological effects of 3q29Del, we performed single-cell mRNA sequencing (scRNA-seq) in isogenic human cortical organoids at both early (2-month) and late (12-month) developmental time points and in perinatal [postnatal day 7 (P7)] mouse isocortex. We devised a strategy to systematically identify the most salient transcriptomic effects, both globally and in specific neural cell types, to identify molecular phenotypes for functional analysis. This strategy led us to a dysregulated transcriptome linked to mitochondria, which displayed both early and prolonged changes that were subsequently supported by orthogonal analyses of protein expression and functional assays in engineered human embryonic kidney (HEK) cells and multiple cohorts of human iPSC-derived neural progenitor cells (NPCs) including lines from 3q29Del study participants. In the context of emerging reports of mitochondrial phenotypes associated with other risk alleles such as 22q11.2 deletion (22q11.2Del) (9, 10), these results point to mitochondria as a possible site of convergent biology downstream of discrete neurodevelopmental variants.
RESULTS
Generating isogenic 3q29Del iPSC lines
To isolate the effects of 3q29Del from variable human genetic backgrounds, we introduced the full 1.6 Mb deletion into an iPSC line derived from a neurotypical female individual by adapting a method previously used to generate isogenic iPSC lines carrying other neurodevelopmental CNVs (11). Similar to most recurrent CNVs, 3q29Del is flanked by low copy repeats (LCRs) or segmental duplications (SDs), which are multikilobase stretches of highly homologous sequence (12) that are likely involved in the formation of structural variants such as CNVs (13). We targeted this homologous sequence with a single guide RNA (gRNA) that is predicted to cut at one site within each 3q29 SD (14) and isolated three clonal lines carrying the 3q29Del (fig. S1). A neurotypical male iPSC line (one clone) was also engineered to carry the 3q29Del using the methods described above. All clones retained normal iPSC morphology and karyotype, and genome-wide optical mapping analyses revealed no off-target structural variants.
Single-cell transcriptomics in developing mouse and human cortical tissue
Four deletion clones and four clones of the parent cell lines (three female and one male per group) were differentiated to dorsal forebrain cortical organoids by established methods (fig. S1F) (15). Single-cell transcriptomes were produced from multiple organoids from two clonal female lines per genotype at 2 and 12 months into in vitro differentiation to capture a broad diversity of developing and mature cell types (figs. S2 and S3). A total of 54,255 cells were included in the human cortical organoid analysis (54% control; fig. S4). A mean of 2805 genes were detected in each cell.
The 3q29Del mouse has been previously reported by two independent groups to express neuropsychiatric phenotypes including alterations in startle responses and social interactions (6, 8). P7 was chosen for tissue dissociation and single-cell sequencing to capture an array of mature and developing cell types (figs. S5 and S6). A total of 71,066 cortical cells were isolated from four male mice per genotype and included in the mouse scRNA-seq analysis (52.9% control; fig. S7). The mean number of genes detected in each cell was 2920.
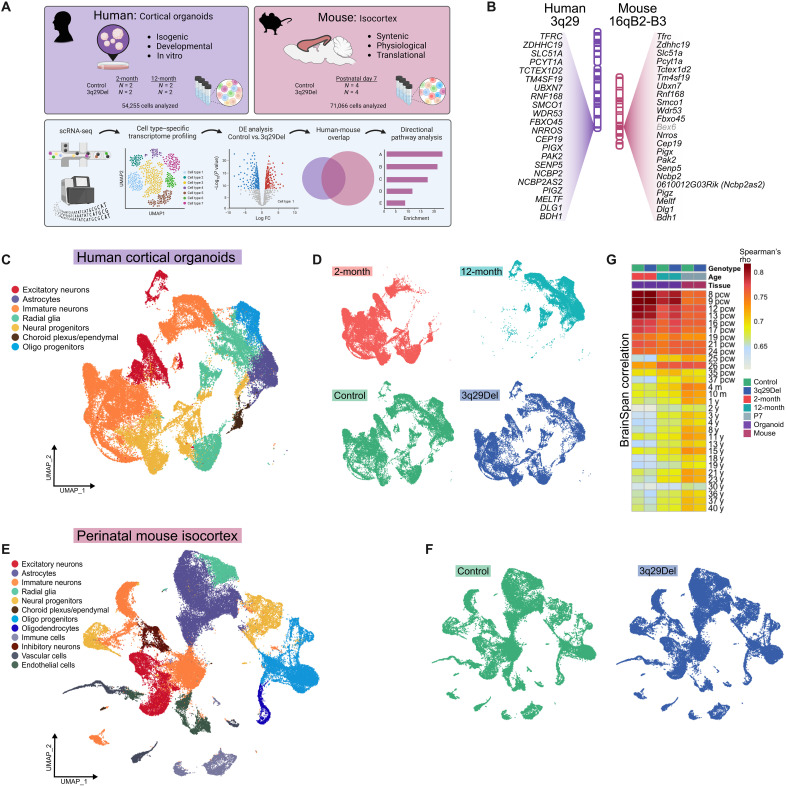
Figure 1 (C and E) shows the major cell types with distinct expression profiles that were isolated in each sequencing experiment. As expected, 2- and 12-month human cortical organoids contained many of the cell types that were also found in the perinatal mouse isocortex including excitatory neurons, astrocytes, immature neurons, radial glia/neural stem cells (NSCs), neural progenitors, choroid plexus/ependymal cells, and oligodendrocyte progenitor cells (OPCs) (fig. S8). In addition to these cells, we also identified immune cells, inhibitory neurons, vascular cells, and endothelial cells in the mouse experiment (fig. S9). Notably, in both experiments, we did not observe large-scale changes in cell clustering by genotype (Fig. 1, D and F) but did observe a stark division in human cell clustering by time point (Fig. 1D). Most human clusters were composed almost entirely of cells from a single time point, and only one cluster (cl. 2, annotated as migrating neuroblasts) was nearly evenly split by time point, consistent with developmentally regulated shifts in cell type composition (fig. S4). Predictably, astrocytes, OPCs, and upper-layer excitatory neurons were not yet present in 2-month organoids (15) but were found in 12-month organoids.
Fig. 1. Cross-species single-cell sequencing.
(A) A scRNA-seq experiment was performed in isogenic human iPSC-derived cortical organoids at two time points and in P7 mouse isocortex. An overview of the strategy to collect and filter differential gene expression data from both model systems is illustrated. (B) The human 3q29Del locus is nearly perfectly syntenic with a region of mouse chromosome 16, with the same gene order inverted. Corresponding loci are illustrated in the same orientation to facilitate clearer cross-species comparison. Bex6 (in gray) is the only gene present in the mouse, but not in the human locus. (C and E) Uniform manifold approximation and projection (UMAP) for dimensionality reduction, colored by the main cell types identified in human (C) and mouse (E) experiments. Human and mouse cells showed no obvious difference in gross distribution by genotype, (D and F) but human cells were clearly divided in their transcriptomic clustering patterns by time point (D) (top). The average expression profile of each sample was correlated (Spearman) to BrainSpan gene expression data, profiling the human brain transcriptome in postmortem specimens across the life span (16) (G). pcw, postconception weeks (prenatal); m, months (postnatal); y, years (postnatal).
To better understand the window of cortical development that our experimental models best reproduce, we compared the average expression profile of each sample to postmortem human brain transcriptomes from the BrainSpan database (Fig. 1G) (16). Spearman correlations revealed that both 2- and 12-month organoids best matched very early phases of human brain development [8 to 9 postconception weeks (pcw); Spearman’s r > 0.80 for 2 months, r > 0.78 for 12 months) and that, across 2-month organoids, 12-month organoids and P7 mouse cortical cells, strong correlations were maintained through the second trimester of human gestation (up to 24 pcw; r > 0.74 for 2 months, r > 0.73 for 12 months, r > 0.73 for mouse cortex). From 25 pcw onwards, the average concordance between human brain tissue and 2-month organoids began a greater decline (r > 0.60) compared to that of 12-month organoids (r > 0.63), which overall remained lower than that of mouse cortex (r > 0.66). As a control experiment, we compared the gene expression profiles of the human homologs of control and 3q29Del mouse liver (7) to the same BrainSpan data and, as expected, found that all correlations were markedly poorer than with any human or mouse cortical sample (r < 0.56; fig. S7). Together, these data suggest that the gene expression profiles of 2- to 12-month human cortical organoids and P7 mouse cortex best model the first two trimesters of human gestation.
We performed differential expression analysis in each cluster of both experiments by genotype. In both mouse and human cells, all 3q29Del transcripts were observed to be decreased to approximately match the copy number in nearly every cell type (figs. S10 to S12). The only mouse-specific gene located in the syntenic locus (Bex6) was either not detected or not differentially expressed in any cluster. Across all clusters and two time points, there were 5244 unique DEGs in human organoids and 3482 DEGs across all mouse clusters. To test for a similar global impact of the 3q29Del, all unique DEGs were compiled for human and mouse experiments. Notably, we found that more than half of the 3253 strictly matched human homologs of mouse DEGs were also found to be DEGs in the human dataset (fold enrichment = 1.53, hypergeometric P = 1.64 × 10−162; fig. S13A).
We explored whether the total number of DEGs in a given cluster was determined by cluster size (i.e., number of cells assigned to a cluster) or the number of 3q29 locus DEGs found in that cluster (fig. S13B). In both mouse and human datasets, the number of 3q29 DEGs within the cluster (but not cluster size) was found to be a significant predictor of the total number of DEGs (negative binomial regression; P < 0.0001), suggesting that haploinsufficiency of genes in the 3q29 locus is a significant driver of total differential gene expression.
To further test the degree of similarity in differential gene expression across mouse and human experiments, we determined the average log fold change of DEGs in 10 comparable clusters and calculated Pearson correlations (fig. S13, C to L). From proliferating neural progenitors to deep-layer excitatory neurons, we found significant positive correlations (moderate to strong) in mouse and human 3q29Del gene expression changes in all comparisons except in astrocytes (fig. S13H), which may reflect differences in maturation state between 12-month cortical organoid cells and postnatal mouse brain.
Effects of 3q29Del on expression of mitochondrial and metabolic genes
We developed two systematic approaches to understand the most salient effects of 3q29Del on the developing cortical transcriptome in mouse and human models. First, we sought to determine the pathways that were most frequently enriched across mouse and human clusters regardless of cell type. To identify these frequently implicated pathways, DEGs from each cluster were split by direction of change (up-regulated versus down-regulated), and pathway analysis was performed as described in Materials and Methods. All significantly enriched Gene Ontology:Biological Process (GO:BP) pathways were compiled and filtered by Revigo (17) to identify umbrella terms that were frequently dysregulated across multiple clusters.
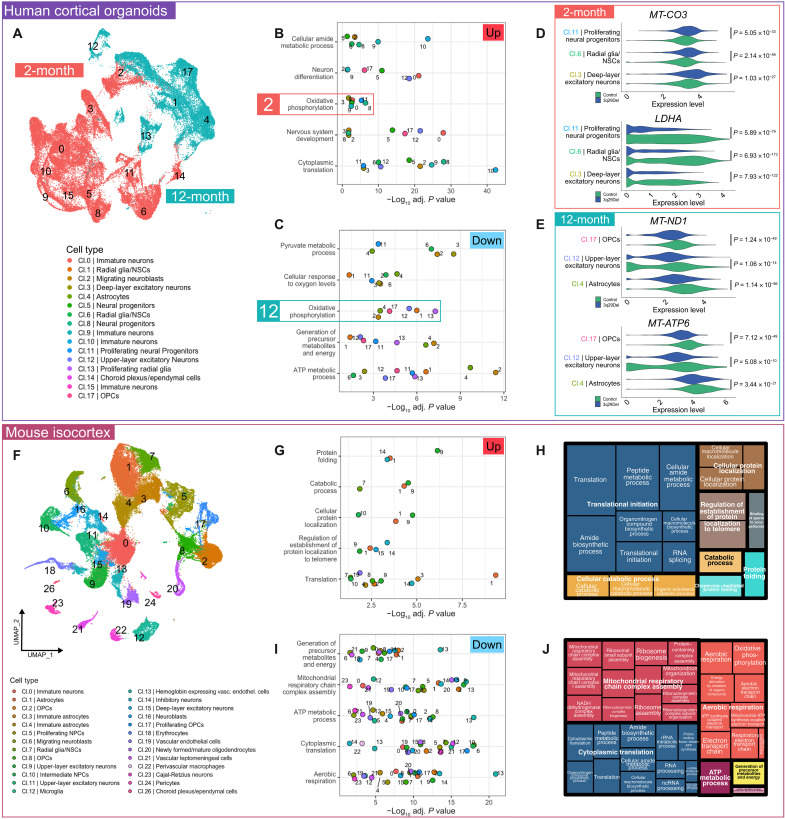
We found that oxidative phosphorylation (OXPHOS) was both down-regulated and up-regulated across multiple clusters (Fig. 2B). A closer examination revealed that all down-regulated OXPHOS clusters were found in 12-month organoids and all up-regulated OXPHOS clusters were found in 2-month organoids. Moreover, the glycolysis-related pyruvate metabolic process was found to be down-regulated in several of the 2-month clusters (cl. 3 and cl. 11) that also up-regulated OXPHOS. Glycolytic process specifically was found to be among the top 10 down-regulated pathways in 2-month clusters of 3q29Del deep-layer excitatory neurons (cl. 3), radial glia/NSCs (cl. 6), and proliferating neural progenitors (cl. 11; fig. S14). This observation inspired the hypothesis that 3q29Del cells may exhibit altered metabolic maturation. A critical stage of neuronal differentiation is the switch from the heavily glycolytic progenitor state to mitochondrial aerobic respiration in mature neurons, which involves down-regulation of several key genes including LDHA (18), which encodes the enzyme lactate dehydrogenase A. Cluster-level analysis revealed a notable decrease in the expression of LDHA in 3q29Del early-born deep-layer excitatory neurons (cl. 3) and radial glia/neural progenitors (cl. 6 and cl. 11), which also showed increased mitochondrial gene expression (Fig. 2D). This result indicates a possible alteration in neuronal metabolic transition. Notably, all 12-month clusters displayed down-regulated OXPHOS (Fig. 2, C and E), which may indicate a long-term effect of 3q29Del on cellular aerobic respiration across multiple neural cell types.
Fig. 2. Transcriptomic evidence of metabolic changes in 3q29Del.
The umbrella pathways most frequently found to be differentially expressed based on up-regulated (B) and down-regulated (C) genes in cortical organoids (A). OXPHOS was enriched among both increased and decreased genes, but all clusters contributing to up-regulated OXPHOS were from 2-month organoids and all clusters contributing to down-regulated OXPHOS were from 12-month organoids. (D) Example violin plots visualizing log-normalized expression data of genes dysregulated in 2-month organoid clusters: MT-CO3 (increased in 3q29Del) encodes the respiratory chain complex IV subunit COX3, and LDHA (decreased in 3q29Del) is a key enzyme in glycolysis. (E) Example violin plots visualizing log-normalized expression data of genes dysregulated in 12-month organoid clusters: MT-ND1 (decreased in 3q29Del) encodes a component of respiratory chain complex I, and MT-ATP6 (decreased in 3q29Del) encodes a component of the ATP synthase complex. The most frequently up-regulated (G) and down-regulated (I) umbrella pathways in mouse isocortex (F) are shown. Treemaps derived from Revigo analysis (H and J) display the hierarchical organization of specific GO:BP terms identified in pathway analysis. Similar colors denote semantic similarity. The size of each rectangle is proportional to the number of clusters exhibiting overrepresentation of a given GO:BP term. (All P values are adjusted for multiple comparisons).
Differential pathway analysis of the mouse cortical data further supported the notion of a long-term decrease in mitochondria-related gene expression. Of the top five most frequently down-regulated umbrella terms, four were related to mitochondrial function and energy metabolism (Fig. 2, I and J). Most mouse clusters were found to have down-regulated genes enriched for at least one of aerobic respiration, adenosine triphosphate (ATP) metabolic process, mitochondrial respiratory chain complex assembly, and generation of precursor metabolites and energy (fig. S15).
Cross-species analysis in astrocytes and neurons
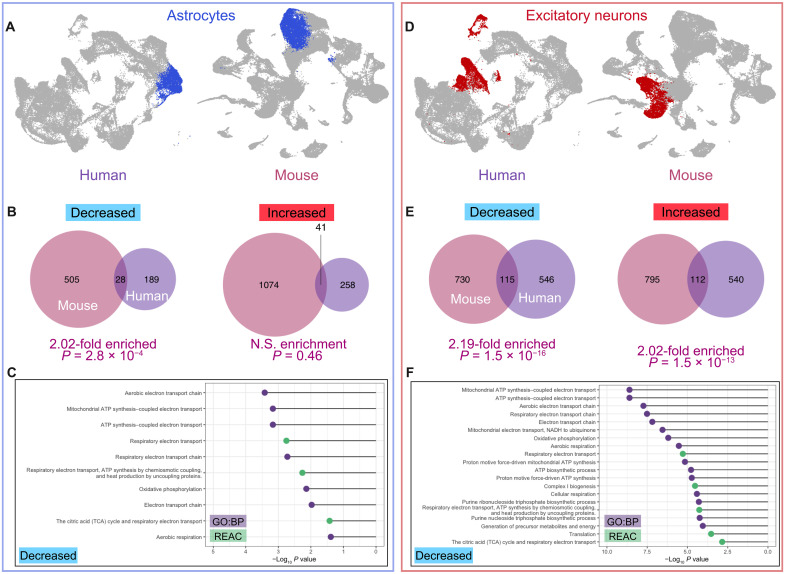
A second, parallel analysis strategy that we used was to stringently filter exact-match DEGs in homologous human and mouse clusters by direction of change. First, we identified the human homologs of mouse DEGs in astrocytes [Mus musculus (mm) cl. 1] and compared them to human DEGs in organoid cl. 4 (Fig. 3A). We found a 2.02-fold enrichment of commonly down-regulated genes (hypergeometric P = 2.8 × 10−4) but no significant overlap among up-regulated DEGs (Fig. 3B). Pathway analysis revealed that the 28 commonly down-regulated genes were heavily enriched for terms related to the electron transport chain and OXPHOS (Fig. 3C).
Fig. 3. Common patterns of differential gene expression in two major mouse and human cell types.
Astrocytes were identified in human cortical organoids (12-month) and mouse isocortex. Corresponding clusters are color coded in blue in UMAP visualizations (A). The human homologs of mouse DEGs identified by MAST analysis were compared to organoid DEGs based on direction of change, and a significant overlap was observed between the down-regulated DEGs of mouse and organoid astrocyte clusters (B). Pathway analysis of overlapping DEGs showed that all significantly enriched GO:BP and Reactome (REAC) terms were related to mitochondrial function and metabolism (C). Upper- and deep-layer excitatory neuron DEGs were pooled, and unique organoid DEGs were compared to the human homologs of mouse DEGs based on direction of change. Corresponding clusters are color coded in red in UMAP visualizations (D). There was a significant overlap between the DEGs of mouse and organoid excitatory neuron clusters for both up-regulated and down-regulated genes (E). Decreased genes were heavily enriched for GO:BP and REAC terms related to mitochondrial function and cellular respiration (F). N.S., not significant.
We performed a parallel analysis in excitatory neurons pooling the unique DEGs of organoid cl. 3 (deep layer) and cl. 12 (upper layer) and compared this list to the human homologs of mouse DEGs from excitatory neuron cl. 9, 11, and 15 (Fig. 3D). In this case, we found approximately twice the overlap that would be expected by chance among both increased and decreased genes (P = 1.5 × 10−16). Nearly all significantly enriched GO:BP and Reactome (REAC) terms among down-regulated genes are related to cellular energy metabolism and mitochondrial function (Fig. 3F; top 20 enriched pathways by P value are shown). Statistically significant intersections were also observed across human and mouse OPCs, radial glia, and migrating neuroblasts (fig. S16).
Altered respiratory chain components in the mouse brain
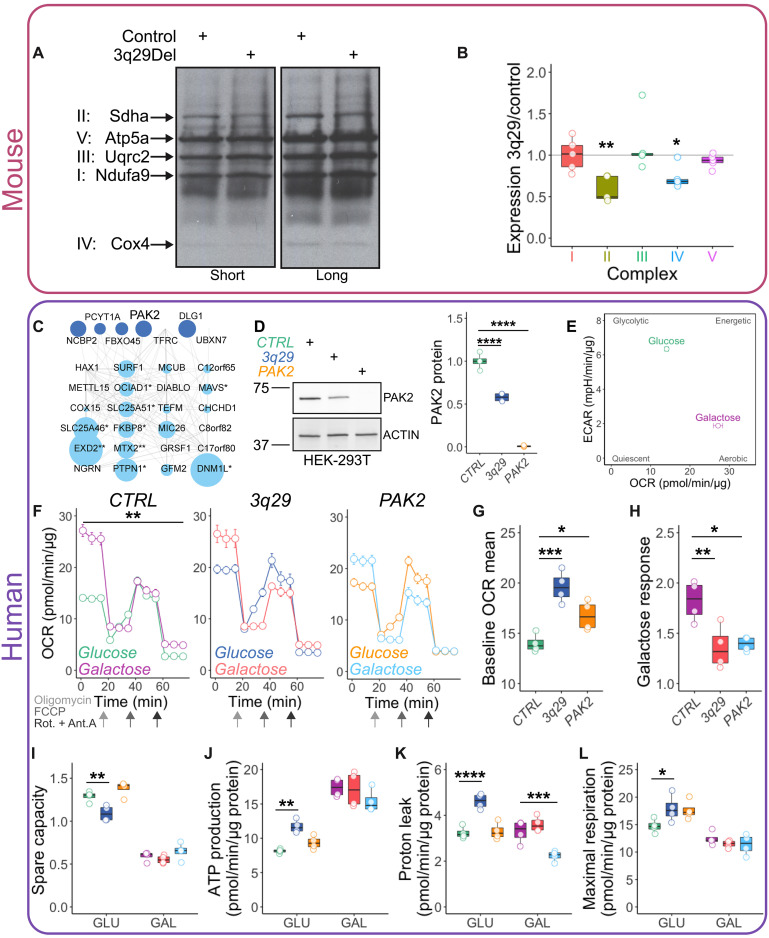
The single-cell transcriptomic data from human 12-month organoids and mouse isocortex strongly indicated a long-term effect of the 3q29Del on mitochondrial function. More specifically, a top pathway dysregulated across 17 mouse clusters was mitochondrial respiratory chain complex assembly. To test the hypothesis that the 3q29Del compromises the integrity of the mitochondrial respiratory chain at the protein level, we probed by Western blot for OXPHOS complex components in Percoll-isolated mitochondrial fractions from adult male and female mouse brains in control and 3q29Del backgrounds. Across five independent experiments with two mice per genotype pooled in each replicate, we observed selective decreases in components of complexes II and IV (one-sample Wilcoxon signed-rank test, P < 0.05; Fig. 4, A and B), indicating a shift in the stoichiometry of respiratory chain complexes.
Fig. 4. Mitochondrial phenotypes in 3q29 mice and engineered cell lines.
Mitochondrial fractions from adult mouse brain lysates were found to have selective decreases in components of OXPHOS complexes II and IV [(A) quantified in (B); N = 5]. At least seven 3q29-encoded proteins interact with mitochondria-localized proteins (C) [from Antonicka et al. (23)]. Symbol size reflects topological coefficients. HEK cell lines were engineered to carry either the heterozygous 3q29Del or completely lack PAK2 as shown by Western blot (D) [one-way analysis of variance (ANOVA), F(2,9) = 237.7, CTRL versus 3q29 or PAK2 ****P < 0.0001]. Control HEK-293T cells (CTRL) transition from a glycolytic to more aerobic cellular respiration state in galactose medium (E). Oxygen consumption rate (OCR) is significantly increased by 48-hour galactose medium challenge in CTRL cells (F) [two-way repeated measures (RM) ANOVA, main effect of medium F(1,6) = 23.99, **P = 0.0027] but not in 3q29 [two-way RM ANOVA, F(1,6) = 0.08808, P = 0.7766] or PAK2 cells [two-way RM ANOVA, F(1,6) = 0.6221, P = 0.4603]. Both 3q29 and PAK2 cells displayed increased baseline OCR (G) [one-way ANOVA, effect of genotype F(2,9) = 17.24, P = 0.0008; CTRL versus 3q29 ***P = 0.0004, CTRL versus PAK2 *P = 0.0332] and decreased response to galactose (H) [one-way ANOVA, effect of genotype F(2,9) = 8.838, P = 0.0075; CTRL versus 3q29 **P = 0.0074, CTRL versus PAK2 *P = 0.0138]. In glucose medium, 3q29 cells showed reduced spare capacity (I) [two-way ANOVA effect of genotype, F(2,18) = 13.04, P = 0.0003; CTRL versus 3q29 **P = 0.0047) and increased ATP production (J) [two-way ANOVA effect of genotype, F(2,18) = 4.309, P = 0.0296; CTRL versus 3q29 **P = 0.0079). Proton leak (K) was found to be increased in 3q29 cells in glucose [two-way ANOVA, main effect of genotype F(2,18) = 31.16, P < 0.0001; CTRL versus 3q29 ****P < 0.0001] and decreased in PAK2 cells in galactose (CTRL versus PAK2 ***P = 0.0007). Maximal respiration was significantly elevated in 3q29 cells in glucose (L) [two-way ANOVA interaction of genotype and medium, F(2,18) = 4.219, P = 0.0314; CTRL versus 3q29 *P = 0.0364) but was unchanged from CTRL under galactose conditions.
Mitochondrial function in 3q29Del engineered cell lines
Transcriptomic data from human 2-month 3q29Del organoids suggested an alteration in the timing and/or efficiency of the canonical glycolysis to OXPHOS shift in developing neural cells. We sought to mimic this shift by challenging engineered HEK-293T cell lines with galactose medium, which forces cells to use OXPHOS for energy production (19–21). Forty-eight-hour galactose treatment markedly shifted control HEK cells (CTRL) from glycolytic to aerobic metabolism (Fig. 4E). For these experiments, we used similar CRISPR-Cas9 methods to engineer a complete version of the hemizygous 3q29Del into HEK-293T cells (TFRC-BDH1). In a separate HEK line, we ablated expression of the 3q29 gene PAK2 (Fig. 4D and fig. S17), which was found to be among the most highly expressed genes in the 3q29Del interval in nearly all mouse and human cell types and was recently shown to be involved in cellular energy metabolism (22). We found that P21 (RAC1) Activated Kinase 2 (PAK2) is one of the seven 3q29-encoded proteins identified in a proximity labeling protein-protein interaction mitochondria map (23).
After a 48-hour galactose challenge, we measured mitochondrial function in the Seahorse mitochondrial stress test, which isolates contributions of the respiratory chain complexes to oxygen consumption in cultured cells through sequential addition of complex-specific inhibitor molecules. Galactose medium significantly increased oxygen consumption rate (OCR) in CTRL cells (Fig. 4F) but not in 3q29 or PAK2 cells, consistent with the disrupted stoichiometry of OXPHOS protein complexes identified in 3q29 mouse brain (Fig. 4). Both 3q29 and PAK2 cells displayed increased baseline OCR (Fig. 4G) and decreased response to galactose (Fig. 4H). Not all 3q29 phenotypes were recapitulated by PAK2 knockout. For example, 3q29 cells showed reduced spare capacity (Fig. 4I) and increased ATP production dependent on respiration (Fig. 4J) in glucose medium, which were unaffected by loss of PAK2. Proton leak (Fig. 4K) was found to be increased in 3q29 cells in glucose and decreased in PAK2 cells in galactose. Maximal respiration (Fig. 4L) was significantly elevated in 3q29 cells in glucose but was unchanged from CTRL under galactose conditions. Thus, PAK2 is likely contributing to, but not solely sufficient for, the metabolic phenotypes of 3q29Del.
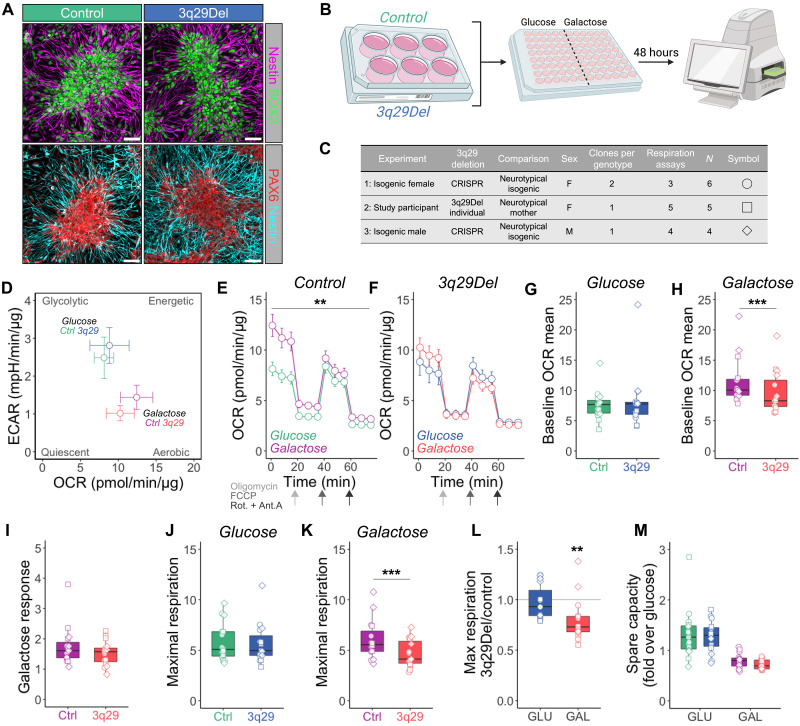
Disrupted metabolic flexibility in 3q29Del neural progenitors
To directly test the hypothesis that the 3q29Del disrupts the metabolic transition from the glycolytic state of NPCs to aerobic respiration in differentiated neurons, we challenged control and 3q29Del NPCs with galactose medium for 48 hours before Seahorse mitochondrial stress assays. These assays were performed in eight clones of six independent cell lines differentiated from both isogenic cell lines and lines derived from 3q29Del study participants, over 12 respiration assays (Fig. 5C and Table 1). As we observed in HEK cell lines, galactose medium shifted NPCs toward a more aerobic metabolic profile (Fig. 5D), and while control NPCs significantly increased OCR in response to galactose challenge, 3q29Del cells did not (Fig. 5E). Unlike HEK cells, we found no differences in OCR under glucose conditions but found that 3q29Del NPCs had a lower baseline OCR mean (Fig. 5H) and decreased maximal respiration (Fig. 5K) in galactose medium. These data demonstrate that similar to HEK-293T cells, 3q29Del NPCs are impaired in the transition from glycolysis to oxidative metabolism induced by galactose challenge. These changes were observed in patient-derived lines and in our laboratory-generated isogenic lines. Together, these experiments reveal that the 3q29Del produces a convergent mitochondrial phenotype at the level of transcriptome, protein expression, and function.
Fig. 5. Lack of metabolic flexibility in 3q29Del NPCs.
(A) Control and 3q29Del NPCs exhibited normal morphology and stained positive for the neurofilament protein Nestin, multipotency marker SOX2, and NPC marker PAX6 (quantified in fig. S15). Scale bars, 50 μm. (B) Illustration of experimental design. NPCs were challenged for 48 hours in neural medium containing glucose (GLU) or galactose (GAL). (C) Table of cell lines used in this experiment. Data from three separate cohorts were combined in plots (D) to (M). N = 15 from six independent NPC lines for all experiments. (D) Energy map indicates that galactose treatment pushes cells from more glycolytic to more aerobic metabolic profile. (E) Control NPCs significantly increase OCR in galactose medium [two-way RM ANOVA main effect of medium, F(1,28) = 9.295, **P = 0.0050]. (F) 3q29Del NPCs exhibited no significant change in OCR in galactose medium [two-way RM ANOVA, medium effect F(1,28) = 0.01219, P = 0.9129]. (G) No significant difference in baseline OCR mean in glucose medium was observed (two-tailed ratio paired t test, P = 0.7015), but 3q29 NPCs displayed significantly lower baseline OCR mean in galactose medium (H) (two-tailed ratio paired t test, ***P = 0.0009). (I) Galactose response (i.e., basal OCR fold change over glucose) was unchanged in 3q29Del NPCs (two-tailed ratio paired t test, P = 0.0935). (J) Maximal respiration was unchanged in glucose medium (two-tailed ratio paired t test, P = 0.5028) but was significantly decreased in 3q29Del NPCs in galactose medium (K) (two-tailed ratio paired t test, ***P = 0.0007). Similarly, (L) the maximal respiration ratio of 3q29Del:control NPCs was unchanged in glucose medium (GLU) but was significantly reduced under galactose conditions (GAL; one sample two-tailed t test, **P = 0.0026). There was no significant change in spare capacity in 3q29Del NPCs (M) [two-way ANOVA genotype effect F(1,56) = 0.5930, P = 0.4445].
DISCUSSION
As the strongest known genetic risk factor for SCZ, 3q29Del is a high-priority target for mechanistic investigation. This study used a rigorous cross-species strategy to identify transcriptomic phenotypes in human and mouse 3q29Del neural tissue. By leveraging the complementary advantages of two highly relevant experimental model systems, we have isolated the effects of the 3q29Del in the developing cortex. We further designed an analysis strategy to systematically filter widespread DEG findings down to the most likely processes and pathways for in-depth functional analysis. This approach led to neural mitochondria as a site of consistent gene dysregulation. Subsequent testing revealed changes in mitochondrial protein expression and function at the cellular level, consistently observed in multiple cell types including study participant–derived cell lines. These data strongly implicate the mitochondrion as an organelle affected by 3q29Del. These results also highlight the strength of our cross-species approach, which rapidly and efficiently led to productive avenues for functional study.
In human cortical organoids, we found that OXPHOS was among the most frequently implicated pathways across cell type clusters (Fig. 2). However, the direction of change was dependent on developmental time point: It was up-regulated in 2-month cells and decreased in 12-month cells. Moreover, the key glycolysis gene LDHA was found to be strongly decreased in multiple 2-month clusters that were overexpressing OXPHOS genes. These findings suggest that the glycolysis to OXPHOS transition, which is critical for neuronal differentiation and maturation (18), may be disrupted in 3q29Del cells. We tested this prediction in an independent cellular model system, 3q29-engineered HEK cells, and found further evidence for a lack of metabolic flexibility; 3q29Del cells showed almost no spare capacity under baseline conditions and, when challenged with a galactose-based medium to force aerobic respiration via OXPHOS, 3q29Del cells had a notably blunted response (Fig. 4). In addition, ablation of the 3q29-encoded gene PAK2 recapitulated two of these effects, decreased response to galactose medium and increased baseline aerobic respiration, while leaving spare capacity intact. These results suggest that PAK2 is likely one of multiple 3q29 locus-encoded genes that contribute to metabolic phenotypes. We further tested the hypothesis of an altered metabolic transition in 3q29Del cells by challenging neural progenitor cultures from two independent isogenic pairs (male and female) and a pair of study participant–derived cell lines in a similar galactose medium paradigm. We found that 3q29Del neural progenitors did not significantly increase oxygen consumption in galactose medium and had reduced maximal respiration compared to CTRL cells. These data further indicate that the 3q29Del imparts a vulnerability to metabolic challenges. Further studies will be required to dissect the contributions of 3q29Del locus genes to this phenotype.
A major strength of the design of the current study is the utilization of human and mouse models as well as two time points in vitro. In both 12-month human cortical organoids and perinatal mouse cortical tissue, we found a widespread decrease in expression of genes related to mitochondrial energy production. In particular, down-regulated gene lists from 17 mouse clusters were enriched for mitochondrial respiratory chain complex assembly (Fig. 2). In support of this transcriptomic prediction, we found evidence for a shift in the stoichiometry of respiratory chain complex proteins in mitochondrial fractions from mouse brain with specific decreases in protein components of complexes II and IV (Fig. 4). This result indicates that the transcriptomic changes that we observed in perinatal mouse cortical tissue and 12-month in vitro human cortical organoids may translate into long-term, persistent effects at the protein level. Future work may be aimed in determining whether there is a mechanistic connection between the cellular energy metabolism phenotypes that we have described and the consistent finding that mouse and human individuals with 3q29Del are significantly smaller than expected (5, 6, 8).
Mitochondria have been previously implicated in the pathophysiology of neurodevelopmental CNV disorders and idiopathic SCZ (24–27). A CNV disorder with perhaps the most similar phenotypic profile to 3q29Del in human carriers, 22q11.2Del, harbors at least eight genes that encode mitochondria-linked proteins, several of which are also enriched at synapses (28): MRPL40, SLC25A1, PRODH, TXNRD2, AIFM3, COMT, RTL10, and SNAP29 (29). Several mitochondrial phenotypes have now been reported in 22q11.2Del models. Similar to our findings, the activity of OXPHOS complexes I and IV was found to be decreased in human 22q11.2Del iPSC-derived neurons, which resulted in reduced ATP production (9, 10). This suggests a convergent biology of mitochondrial dysfunction between 3q29Del and 22q11 deletion. This phenotype was attributed to haploinsufficiency of the 22q11.2Del locus gene MRPL40, which is a component of mitochondrial ribosome. Loss of one copy of Mrpl40 in mice is sufficient to produce short-term neuroplasticity phenotypes (30), potentially linking mitochondrial phenotypes to more well-established synaptic defects in SCZ models. A separate study that used a cross-species strategy to prioritize 22q11.2Del-associated effects in mouse brain and human patient fibroblasts identified the 22q11.2 gene encoding the mitochondrial citrate transporter SLC25A1 as a key component of a dysregulated mitochondrial protein hub (31). Further studies indicated an interaction between SLC25A1 and MRPL40 at the protein level (32). In addition, a large transcriptomic study of 22q11.2Del cortical organoids also found enrichment of DEGs related to mitochondrial function (33).
Other neurodevelopmental CNVs have been associated with mitochondrial phenotypes as well. The Williams syndrome (deletion) and SCZ-associated (duplication) locus 7q11.23 contains the gene DNAJC30, which encodes a protein that interacts with the ATP synthase complex (34). Complete loss of DNAJC30 was found to disrupt sociability in mice and severely impair mitochondrial function in mouse neurons (34). Human fibroblasts from individuals with Williams syndrome (i.e., hemizygous for DNAJC30) were also found to have impaired mitochondrial function and reduced ATP production (34). In addition, a recent study of reciprocal CNVs at the neurodevelopmental disorder–associated locus 16p11.2 also found a strong signal for enrichment of DEGs related to energy metabolism and mitochondrial function in both mouse brain and cultured human neural cell lines (35), and loss of the 16p11.2 locus–encoded gene TAOK2 was found to disrupt mitochondrial morphology and function in mouse neurons (36). Another study of convergent biology in mouse models of the neurodevelopmental CNVs 1q21.1, 15q13.3, and 22q11.2 found dysregulation of a transcriptomic module related to neuronal energetics (37). Last, recent data indicate that complete loss of one of the top single-gene risk factors for SCZ, SETD1A (38), impairs basal glycolysis and respiratory capacity in human neurons (39). Together, these data indicate that our findings in 3q29Del mouse and human cortical tissue fit with reports of other mitochondrial phenotypes associated with neurodevelopmental variants and suggest that neural mitochondria may be a key site of biological convergence downstream of these high-risk alleles.
Unlike 22q11.2, which encodes several proteins that function within mitochondria, the mechanistic link to 3q29 genes is not known. Our data implicate the highly expressed kinase PAK2 in the metabolic phenotypes associated with 3q29Del, although likely in conjunction with additional driver genes, as predicted by our earlier network-based inferences on 3q29 neuropathology emerging upon loss of multiple functionally connected genes in the interval (40). Mitochondria are involved in many cellular pathways and processes in addition to energy production including apoptosis signaling. Thus, our findings of transcriptomic and functional phenotypes at 3q29Del mitochondria fit with a previous report of increased susceptibility to apoptosis in Drosophila models (41). Further studies will be required to determine whether mitochondrial phenotypes are a primary consequence of 3q29Del and the specific driver genes for these effects.
Given the hierarchical structure of scRNA-seq data, treating individual cells as independent sampling units can yield false positives in differential expression results because of the underestimation of true SEs. To improve the reproducibility and validity of our findings, we focused our investigation on disrupted gene expression signals and corresponding signaling pathways with independent statistical support from two separate model systems. We note that, while this approach increases our confidence in capturing true associations, additional signals relevant to disease mechanism may be hidden among unshared findings between model systems. We also note that noncoding genes make up a small fraction of the detected transcripts in our present library construction approach; hence, the extent to which noncoding genes of the 3q29Del interval contribute to pathogenesis is unknown; more work is needed to decode the involvement of the noncoding elements of this region in future studies.
Bridging the gap between genetic risk and biological mechanisms is a major challenge for psychiatry. In this study, we sought to use systematic methods to identify the most salient, conserved transcriptomic effects of the SCZ-associated 3q29Del in disease-relevant tissues as an important step toward determining cellular and molecular phenotypes of this important variant. These findings should motivate further work to determine the mechanisms of these 3q29Del sequelae and their relevance to various clinical phenotypes.
MATERIALS AND METHODS
Cell culture and genome engineering
Whole blood samples of 5 to 10 ml were collected in EDTA Vacutainer test tubes and processed for the isolation of erythroid progenitor cells (EPCs) using the Erythroid Progenitor Kit (STEMCELL Technologies). EPCs were reprogrammed using Sendai particles (CytoTune-iPS 2.0 reprogramming kit, Invitrogen) and plated onto Matrigel-coated six-well plates (Corning). Cultures were transitioned from erythroid expansion media to ReproTeSR (STEMCELL Technologies) and then fed daily with ReproTeSR until clones were isolated. iPSCs were maintained on Matrigel-coated tissue culture plates with mTeSR Plus (STEMCELL Technologies).
Cell lines were characterized for stem cell markers by reverse transcription polymerase chain reaction (RT-PCR) and immunocytochemistry after at least 10 passages in culture. Total RNA was isolated from each cell line with the RNeasy Plus Kit (QIAGEN) according to the manufacturer’s protocol. mRNA was reverse transcribed into cDNA using the High Capacity cDNA Synthesis Kit (Applied Biosystems). Expression of pluripotency genes OCT4, SOX2, REX1, and NANOG was determined by RT-PCR. Sendai virus inactivity was confirmed using Sendai genome–specific primers.
Isogenic 3q29Del iPSC and HEK cell lines were generated using the SCORE method (11). To identify LCR target sequences, the reference sequence (hg38) between TNK2 to TFRC (centromeric) and BDH1 to RUBCN (telomeric) was downloaded and aligned in the National Center for Biotechnology Information (NCBI) BLAST. A ~20 kb segment was found to be 97% identical and was searched for gRNA sequences using CHOPCHOP (https://chopchop.cbu.uib.no) (14). Three single gRNA sequences (IDT) that were predicted to each cut at a single site in both LCRs were identified and cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0, which was a gift from F. Zhang (Addgene plasmid #62988; http://n2t.net/addgene:62988; RRID: Addgene_62988) (42).
Single gRNA plasmids were transfected into a neurotypical control iPSC line (IRB#CR002-IRB00088012), maintained in mTeSR or mTeSR+ (STEMCELL Technologies, Vancouver) on Matrigel (Corning)–coated plates using a reverse transfection method and Mirus TransIT-LT1 reagent (Mirus Bio, Madison, WI), and transfected cells were transiently selected for puromycin resistance. Genome cleavage efficiency for each gRNA was calculated using the GeneArt Genomic Cleavage Detection Kit (Thermo Fisher Scientific), and gRNA_2 (5′-CAGTCTTGGCTACATGACAA-3′, directed to −strand; hg38 chr3:195,996,820 to chr3:197,634,397) was found to be most efficient with cleaved bands at the predicted sizes. Cells transfected with gRNA_2 were dissociated and cloned out by limiting dilution in mTeSR supplemented with 10% CloneR (STEMCELL Technologies). Putative clonal colonies were manually transferred to Matrigel-coated 24-well plates for expansion and screened for change in the copy number of the 3q29Del locus gene PAK2 (Hs03456434_cn) using TaqMan Copy Number Assays (Thermo Fisher Scientific). Three (of 100) clones showed an apparent loss of one copy of PAK2 and were subsequently screened for loss of the 3q29 genes TFRC (Hs03499383_cn), DLG1 (Hs04250494_cn), and BDH1 (Hs03458594_cn) and for no change in copy number to external (nondeleted) 3q29 genes TNK2 (Hs03499383_cn) and RUBCN (Hs03499806_cn) all referenced to RNASEP (Thermo Fisher Scientific, #4401631). All cell lines retained normal karyotypes (WiCell, Madison, WI) and were free of mycoplasma contamination (LookOut, Sigma-Aldrich).
To generate 3q29Del HEK-293T cell lines, HEK cells (RRID:CVCL_0063) were transfected with either empty px459 or px459 + gRNA_1 (5′-ttagatgtatgccccagacg-3′; directed to the +strand) and screened and verified with TaqMan copy number assays as described above. PAK2 was deleted from a control HEK-293T line as detailed above (PAK2 gRNA 5′-TTTCGTATGATCCGGTCGCG-3′; directed to −strand). Clones were screened by Western blot (rabbit monoclonal PAK2 from Abcam; ab76293; RRID AB_1524149; 1:5000 dilution) and confirmed by Sanger sequencing PCR-amplified gDNA. HEK cell lines were also negative for mycoplasma contamination.
Genome-wide optical mapping
A total of 1.5 × 106 iPSCs were pelleted, washed with Dulbecco’s phosphate-buffered saline (PBS), and frozen at −80°C following aspiration of all visible supernatant. A total of 750 ng of DNA was labeled, stained, and homogenized using the DNA Labeling Kit-DLS (Bionano, 80005). Stained DNA was loaded onto the Saphyr chip G1.2, and the chip was scanned to image the labeled DNA using the Saphyr System. Structural variants were called relative to the reference genome (hg38) using Bionano Solve. Structural variants were compared to the parent (unedited) cell line using the Bionano Solve Variant Annotation Pipeline.
Cortical organoid differentiation
Engineered isogenic 3q29Del iPSC lines and the unedited parent line, along with two additional clonal lines from the same donor, were expanded in mTeSR or mTeSR+ on Matrigel-coated plates. On DIV (day in vitro) 0, colonies were gently released from plates in Dispase (0.35 mg/ml) according to an established protocol (15). Floating colonies were resuspended in mTeSR supplemented with 10 μM Y-27632 (Reprocell, Beltsville, MD) in ultralow attachment 10 cm dishes (Corning). After 48 hours, spheroids were transitioned to neural induction medium [20% knockout serum replacement, 1% nonessential amino acids, penicillin/streptomycin (Pen/Strep) (100 U/ml), 0.5% GlutaMAX, and 0.1 mM 2-mercaptoethanol in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12) with Hepes], supplemented with 5 μM dorsomorphin and 10 μM SB-431542 (added fresh) with daily medium changes through DIV 6. On DIV 7, neural induction medium was replaced with neural medium [Neurobasal-A with 2% B-27 without vitamin A, 1% GlutaMAX, and Pen/Strep (100 U/ml)] supplemented with fresh epidermal growth factor (EGF) (20 ng/ml; R&D Systems) and fibroblast growth factor (FGF) (20 ng/ml; R&D Systems) for daily medium changes through day 16. From day 17 to 25, organoids were fed neural medium with EGF and FGF every 2 days. From day 26 to 42, neural medium was supplemented with brain-derived neurotrophic factor (20 ng/ml; R&D Systems) and Neurotrophin-3 (NT-3) (20 ng/ml; R&D Systems) every 2 days. From day 43 onwards, organoids were fed Neural Medium without supplements twice weekly.
Mouse genotyping and maintenance
All animal experiments were performed under guidelines approved by the Emory University Institutional Animal Care and Use Committee. Mice were genotyped as described previously (6) and noted as either control (wild-type, C57BL/6 N Charles River Laboratories) or 3q29Del (B6.Del16+/Bdh1-Tfrc, MGI:6241487). Male 3q29Del mice and control littermates were included in the scRNA-seq study. Both male and female mice were included in mitochondrial fractionation experiments.
Tissue dissociation and sorting
Single-cell suspensions from cortical organoids (DIV 50 = “2-month,” N = 2 control, N = 2 3q29Del; DIV 360 = “12-month,” N = 2 control, N = 2 3q29Del) and P7 mouse cortices (N = 4 control, N = 4 3q29Del) were produced by a papain dissociation method based on a published protocol (43). Organoids were dissociated in three batches that were each balanced for genotype and “age.” Mouse samples were also dissociated in three batches each balanced by genotype. In both sets, the experimenter was blinded to genotype. Tissue was coarsely chopped with a sterile scalpel and digested for 1 hour at 34°C in a pH-equilibrated papain solution (Worthington, Lakewood, NJ) with constant CO2 flow over the enzyme solution. Digested tissue was gently spun out of papain, through ovomucoid solutions, and sequentially triturated with P1000 and P200 pipet tips. Live cells were counted by manual and automated methods (Countess II, Thermo Fisher Scientific) and, in organoid, samples were isolated from cellular debris by fluorescence-activated cell sorting on a FACSAria II instrument (calcein AM–high, Ethidium Homodimer-1 low).
Single-cell library prep and RNA-seq
Single-cell suspensions were loaded into the 10x Genomics Controller chip for the Chromium Next GEM Single Cell 3′ kit workflow as instructed by the manufacturer with a goal capture of 10,000 cells per sample. The resulting 10x libraries were sequenced using Illumina chemistry. Mouse samples and libraries were prepared and sequenced at a separate time from human samples.
scRNA-seq data processing and analysis
To quantify gene expression at a single-cell resolution, the standard Cell Ranger (10x Genomics) and Seurat (44) data processing pipelines were followed for demultiplexing base call files into FASTQ files, alignment of scRNA-seq reads to species-specific reference transcriptomes with STAR (mouse: mm10; human: GRCh38), cellular barcode and unique molecular identifier (UMI) counting, and gene- and cell-level quality control (QC). To filter out low-quality cells, empty droplets and multiplets, genes expressed in <10 cells, cells with >30% reads mapping to the mitochondrial genome, and cells with unique feature (gene) counts >7000 were removed on the basis of manual inspection of the distributions of each QC metric individually and jointly. Outlier cells with low unique feature counts were further removed via sample-specific thresholding of corresponding distributions (<250 for mice; <700 for organoids). Thresholds were set as permissive as possible to avoid filtering out viable cell populations, consistent with current best-practice recommendations (45).
The sctransform function in Seurat was used for normalization and variance stabilization of raw UMI counts based on regularized negative binomial regression models of the count by cellular sequencing depth relationship for each gene, while controlling for mitochondrial mapping percentage as a confounding source of variation (46). Resulting Pearson’s residuals were used to identify the most variable features in each dataset (n = 3000 by default), followed by dimensionality reduction by principal component analysis and uniform manifold approximation and projection, shared nearest neighbor graph construction on the basis of the Euclidean distance between cells in principal component space, and unbiased clustering of cells with Louvain modularity optimization. Optimal clustering solutions for each dataset was determined by building cluster trees and evaluating the SC3 stability index for every cluster iteratively at 10 different clustering resolutions with the clustree function in R (47). The effect of cell cycle variation on clustering was examined by calculating and regressing out cell cycle phase scores in a second iteration of sctransform, based on the expression of canonical G2-M and S phase markers (48). Consistent with the developmental context of the interrogated datasets, cell cycle differences were found to covary with cell type and retained in final analyses as biologically relevant sources of heterogeneity. Cluster compositions were checked to confirm comparable distributions of experimental batch, replicate ID, and genotype metadata. Cluster annotations for cell type were determined on the basis of the expression of known cell type and cortical layer markers curated from the literature (49–53). Clusters exhibiting cell type ambiguity were further subclustered to refine annotations or dropped from downstream analysis in case of inconclusive results (human cl. 7 and cl. 16; mouse cl. 25 and cl. 27).
Seahorse mitochondrial stress assay
HEK-293T cells that had been engineered to carry the 3q29Del (3q29Del), PAK2 knockout (PAK2), and mock-edited CTRL cells were plated on poly-d-lysine–coated 96-well Seahorse assay plates (XF96, Agilent) in DMEM (Gibco, A144300) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, and either 10 mM d-(+)-glucose (“Glu”; 7.5 × 103 cells per well) or 10 mM galactose (“Gal”; 15 × 103 cells per well). After 48 hours, cells were washed twice in XF DMEM Assay Medium (Agilent) with either glucose or galactose (10 mM) supplemented with 1 mM pyruvate and 2 mM glutamine.
NPCs were plated at 5 × 104 cells per well in poly-l-ornithine (Sigma-Aldrich, P4957; 15 μg/ml)– and laminin (Sigma-Aldrich, 23017-015; 5 μg/μl)–coated 96-well Seahorse assay plates in STEMdiff Neural Progenitor Medium (STEMCELL Technologies, 05834). After 24 hours, all media were aspirated and exchanged for neural medium (Neurobasal minus glucose, glutamine, pyruvate; Thermo Fisher Scientific, A2477501) supplemented with B-27 minus insulin (Thermo Fisher Scientific, A1895601), 2 mM l-glutamine, and 1 mM sodium pyruvate with either 17.5 mM glucose or galactose for 48 hours.
Mitochondrial stress test compounds were loaded into injection ports as indicated by the manufacturer to achieve the following final concentrations for HEK cells: 1 μM oligomycin, 0.25 μM carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), 0.5 μM rotenone, and 0.5 μM antimycin A (all sourced from Sigma-Aldrich). NPC assays were performed with 2 μM oligomycin, 0.5 μM FCCP, 1 μM rotenone, and 1 μm antimycin A. Cells equilibrated at 37°C with ambient CO2 for approximately 1 hour before assay initiation. At the end of the experiment, cells were washed twice in PBS + Ca2+ + Mg2+ and lysed at 4°C for 30 min in 0.5% Triton X-100 protein buffer (150 mM NaCl, 10 mM Hepes, 0.1 mM MgCl2, 1 mM EGTA, and 1× HALT protease + phosphatase inhibitor). Protein concentrations in each well were determined by BCA (Pierce) to normalize OCR data. Data were analyzed in Wave (Agilent). Assay wells that did not show responses to drug injections were excluded from analysis.
Mouse brain mitochondrial isolation
A protocol for mitochondrial isolation was adapted from prior work (54). Two whole brains per genotype were dissected from adult mice (2 to 6 months) and pooled in 2.5 ml of ice-cold medium I [0.32 M sucrose, 5 mM Hepes (pH 7.5), 0.1 mM EDTA, and cOmplete protease inhibitor] and homogenized with 16 strokes at approximately 800 rpm in a Teflon glass homogenizer (0.125 mm clearance) with rest on ice midway through. Crude homogenate was cleared by centrifugation at 1000g for 10 min, and the supernatant was further centrifuged at 12,000g for 20 min. All centrifugations were carried out at 4°C.
Isoosmotic Percoll [nine parts Percoll to one part 2.5 M sucrose (v/v)] gradients were prepared in medium II [0.25 M sucrose, 5 mM Hepes (pH 7.2), and 0.1 mM EDTA]. The second pellet was carefully resuspended in an appropriate volume of 8.5% Percoll to produce a 7.5% Percoll solution and then was gently homogenized by twisting the Teflon pestle through the solution. The 7.5% Percoll solution containing the resuspended tissue fraction was carefully layered on top of a gradient containing 16 and 10% Percoll. Gradients were centrifuged for 20 min at 15,000g, and mitochondrial fractions were extracted from the bottom of the tube and solubilized in 0.5% Triton X-100 protein buffer (150 mM NaCl, 10 mM Hepes, 0.1 mM MgCl2, 1 mM EGTA, and 1× cOmplete). Protein concentrations were determined by BCA (Pierce) and normalized. Twenty micrograms of protein was loaded to each lane of Criterion gels for SDS–polyacrylamide gel electrophoresis. Gels were transferred onto polyvinylidene difluoride membranes by standard protocols and blocked in 5% milk. OXPHOS complex component proteins were probed for with an OXPHOS antibody cocktail (1:250; Abcam, ab110412). Protein levels were determined by band densitometry and quantified by normalizing to the most stable complex component (V).
Statistical analysis
Differential gene expression testing for genotype was performed on log-normalized expression values (scale.factor = 10,000) of each cluster separately with a two-part generalized linear model that parameterizes the bimodal expression distribution and stochastic dropout characteristic of scRNA-seq data, using the MAST algorithm, while controlling for cellular detection rate (55). A threshold of 0.1 was implemented as the minimum cutoff for average log fold change (logfc.threshold) and detection rates (min.pct) of each gene in either genotype to increase the stringency of differential expression analysis. Multiple hypothesis testing correction was applied conservatively using the Bonferroni method to reduce the likelihood of type 1 errors, based on the total number of genes in the dataset. To facilitate comparative transcriptomics, human homologs (including multiple paralogs) were identified for all DEGs in the mouse dataset via the NCBI’s HomoloGene database (ncbi.nlm.nih.gov/homologene/). Data processing and analysis pipelines were harmonized across the mouse and organoid datasets, yielding parallel computational approaches for cross-species comparison of differential expression signals. The BrainSpan Developmental Transcriptome dataset used for developmental stage estimations was obtained by bulk RNA-seq of postmortem human brain specimens collected from donors with no known history of neurological or psychiatric disorders, as described previously (16, 56). This large-scale resource is accessible via the Allen Brain Atlas data portal (www.brainspan.org/static/download/; file name: “RNA-Seq Gencode v10 summarized to genes”); dbGaP accession number: phs000755.v2.p1. All statistical analyses of scRNA-seq data were performed in R (v.4.0.3).
To interpret differential gene expression results, pathways likely affected by the 3q29Del were determined on the basis of statistically overrepresented gene sets with known functions using g:Profiler (57). DEGs (Bonferroni adj. P < 0.05) for each cluster were identified as described above and input with an experiment-specific background gene set (genes with min.pct > 0.1 in any cluster). GO:BP and REAC databases were searched with 10 < term size < 2000. Significantly enriched pathways below a threshold of g:SCS < 0.05 (58) were compiled and filtered in Revigo (17) to reduce redundancy and to determine umbrella terms. Western blot analysis of mouse brain OXPHOS complex components (Fig. 4B) used a one-sample Wilcoxon signed-rank test (two-tailed), *P < 0.05 and **P < 0.01; N = 5.
Seahorse mitochondrial stress test analysis (Fig. 4F) was performed by two-way repeated measures (RM) analysis of variance (ANOVA), main effect of medium F(1,6) = 23.99, **P = 0.0027; (Fig. 4G) one-way ANOVA, effect of genotype F(2,9) = 17.24, P = 0.0008; CTRL versus 3q29 ***P = 0.0005, CTRL versus *PAK2 P = 0.0332; (Fig. 4H) one-way ANOVA, effect of genotype F(2,9) = 8.838, P = 0.0075; CTRL versus 3q29 **P = 0.0074, CTRL versus PAK2 *P = 0.0138; (Fig. 4I) two-way ANOVA effect of genotype, F(2,18) = 13.04, P = 0.0003; CTRL versus 3q29 **P = 0.0047; (Fig. 4J) two-way ANOVA effect of genotype, F(2,18) = 4.309, P = 0.0296; CTRL versus 3q29 **P = 0.0079; (Fig. 4K) two-way ANOVA, main effect of genotype F(2,18) = 31.16, P < 0.0001; CTRL versus 3q29 ****P < 0.0001, galactose CTRL versus PAK2 ***P = 0.0007; (Fig. 4L) two-way ANOVA interaction of genotype and medium, F(2,18) = 4.219, P = 0.0314; CTRL versus 3q29 *P = 0.0364; N = 4 for all HEK Seahorse experiments; (Fig. 5E) two-way RM ANOVA main effect of medium, F(1,28) = 9.295, **P = 0.0050; (Fig. 5F) two-way RM ANOVA, medium effect F(1,28) = 0.01219, P = 0.9129; (Fig. 5G) two-tailed ratio paired t test, P = 0.7015; (Fig. 5H) two-tailed ratio paired t test, ***P = 0.0009; (Fig. 5I) two-tailed ratio paired t test, P = 0.0935; (Fig. 5J) two-tailed ratio paired t test, P = 0.5028; (Fig. 5K) two-tailed ratio paired t test, ***P = 0.0007; (Fig. 5L) one-sample two-tailed t test, **P = 0.0026; (Fig. 5M) two-way ANOVA genotype effect, F(1,56)=0.5930, P = 0.4445; N = 15 for all NPC Seahorse experiments according to Table 1.
Table 1. Respiration assay cell lines.
| Parent cell line | Clone | Genotype | Group | Respiration assays |
|---|---|---|---|---|
| 1003-0031 | c27 | Control | Control | 3 |
| 1003-0031 | c33 | Control | Control | 3 |
| 1003-0031 | 3G2b | 3q29Del | Isogenic | 3 |
| 1003-0031 | 11D7 | 3q29Del | Isogenic | 3 |
| 4258-2096 | c13 | Control | Control | 5 |
| 4258-1031 | c11 | 3q29Del | Study participant | 5 |
| 4258-2046 | c17 | Control | Control | 4 |
| 4258-2046 | 1C7 | 3q29Del | Isogenic | 4 |
Acknowledgments
Funding: Research reported here was supported in part by Imagine, Innovate, and Impact (I3) from the Emory School of Medicine; a gift from Woodruff Fund Inc.; and through the Georgia CTSA NIH award (UL1-TR002378). This study was also supported in part by the Emory Integrated Genomics Core (EIGC), Emory Integrated Computational Core (EICC), Emory Integrated Cellular Imaging, Emory Flow Cytometry Core, and the Emory Stem Cell Core, which are subsidized by the Emory University School of Medicine and are part of the Emory Integrated Core Facilities. Figure illustrations generated with Biorender.com. This work was supported by National Institutes of Health grant F32MH124273 (to R.H.P.), a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation (to R.H.P.), the University Research Committee of Emory University (to R.H.P.), National Institutes of Health grant R56MH116994 (to S.T.W. and J.G.M.), National Institutes of Health grant R01MH110701 (to G.J.B. and J.G.M.), and National Institutes of Health grant R01MH118534 (to J.G.M.).
Author contributions: Conceptualization: R.H.P., E.S., S.T.W., S.A.S., G.J.B., and J.G.M. Methodology: R.H.P., E.S., E.W., A.T.K., T.J.M., M.E.M.-G., P.C., Z.T.M., N.R., B.J.V., D.S., B.L.F., K.T., Z.W., V.F., S.A.S., G.J.B., and J.G.M. Investigation: R.H.P., E.W., T.J.M., S.K., M.I.R., and V.F. Visualization: R.H.P., E.S., E.W., and V.F. Supervision: R.H.P., G.J.B., and J.G.M. Writing—original draft: R.H.P., E.S., and J.G.M. Writing—review and editing: R.H.P., E.S., E.W., T.J.M., N.R., V.F., S.A.S., G.J.B., and J.G.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data, code, and materials used in the experiments and analyses are available to interested researchers at doi.org/10.5061/dryad.b2rbnzsmm. Cell lines are available for research use upon a material transfer agreement. Requests for cell lines should be submitted to R.H.P., G.J.B., and J.G.M. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S18
Legends for tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.J. G. Mulle, M. J. Gambello, R. Sanchez Russo, M. M. Murphy, T. L. Burrell, C. Klaiman, S. White, C. A. Saulnier, E. F. Walker, J. F. Cubells, S. Shultz, L. Li, 3q29 recurrent deletion, in GeneReviews [Internet], M. P. Adam, G. M. Mirzaa, R. A. Pagon, S. E. Wallace, L. J. H. Bean, K. W. Gripp, A. Amemiya, Eds. (University of Washington, 2016). [Google Scholar]
- 2.Mulle J. G., Dodd A. F., McGrath J. A., Wolyniec P. S., Mitchell A. A., Shetty A. C., Sobreira N. L., Valle D., Rudd M. K., Satten G., Cutler D. J., Pulver A. E., Warren S. T., Microdeletions of 3q29 confer high risk for schizophrenia. Am. J. Hum. Genet. 87, 229–236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mulle J. G., The 3q29 deletion confers >40-fold increase in risk for schizophrenia. Mol. Psychiatry 20, 1028–1029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall C. R., Howrigan D. P., Merico D., Thiruvahindrapuram B., Wu W., Greer D. S., Antaki D., Shetty A., Holmans P. A., Pinto D., Gujral M., Brandler W. M., Malhotra D., Wang Z., Fajarado K. V. F., Maile M. S., Ripke S., Agartz I., Albus M., Alexander M., Amin F., Atkins J., Bacanu S. A., Belliveau R. A. Jr., Bergen S. E., Bertalan M., Bevilacqua E., Bigdeli T. B., Black D. W., Bruggeman R., Buccola N. G., Buckner R. L., Bulik-Sullivan B., Byerley W., Cahn W., Cai G., Cairns M. J., Campion D., Cantor R. M., Carr V. J., Carrera N., Catts S. V., Chambert K. D., Cheng W., Cloninger C. R., Cohen D., Cormican P., Craddock N., Crespo-Facorro B., Crowley J. J., Curtis D., Davidson M., Davis K. L., Degenhardt F., Del Favero J., DeLisi L. E., Dikeos D., Dinan T., Djurovic S., Donohoe G., Drapeau E., Duan J., Dudbridge F., Eichhammer P., Eriksson J., Escott-Price V., Essioux L., Fanous A. H., Farh K. H., Farrell M. S., Frank J., Franke L., Freedman R., Freimer N. B., Friedman J. I., Forstner A. J., Fromer M., Genovese G., Georgieva L., Gershon E. S., Giegling I., Giusti-Rodriguez P., Godard S., Goldstein J. I., Gratten J., de Haan L., Hamshere M. L., Hansen M., Hansen T., Haroutunian V., Hartmann A. M., Henskens F. A., Herms S., Hirschhorn J. N., Hoffmann P., Hofman A., Huang H., Ikeda M., Joa I., Kahler A. K., Kahn R. S., Kalaydjieva L., Karjalainen J., Kavanagh D., Keller M. C., Kelly B. J., Kennedy J. L., Kim Y., Knowles J. A., Konte B., Laurent C., Lee P., Lee S. H., Legge S. E., Lerer B., Levy D. L., Liang K. Y., Lieberman J., Lonnqvist J., Loughland C. M., Magnusson P. K. E., Maher B. S., Maier W., Mallet J., Mattheisen M., Mattingsdal M., McCarley R. W., McDonald C., McIntosh A. M., Meier S., Meijer C. J., Melle I., Mesholam-Gately R. I., Metspalu A., Michie P. T., Milani L., Milanova V., Mokrab Y., Morris D. W., Muller-Myhsok B., Murphy K. C., Murray R. M., Myin-Germeys I., Nenadic I., Nertney D. A., Nestadt G., Nicodemus K. K., Nisenbaum L., Nordin A., O'Callaghan E., O'Dushlaine C., Oh S. Y., Olincy A., Olsen L., O'Neill F. A., Van Os J., Pantelis C., Papadimitriou G. N., Parkhomenko E., Pato M. T., Paunio T.; Psychosis Endophenotypes International Consortium, Perkins D. O., Pers T. H., Pietilainen O., Pimm J., Pocklington A. J., Powell J., Price A., Pulver A. E., Purcell S. M., Quested D., Rasmussen H. B., Reichenberg A., Reimers M. A., Richards A. L., Roffman J. L., Roussos P., Ruderfer D. M., Salomaa V., Sanders A. R., Savitz A., Schall U., Schulze T. G., Schwab S. G., Scolnick E. M., Scott R. J., Seidman L. J., Shi J., Silverman J. M., Smoller J. W., Soderman E., Spencer C. C. A., Stahl E. A., Strengman E., Strohmaier J., Stroup T. S., Suvisaari J., Svrakic D. M., Szatkiewicz J. P., Thirumalai S., Tooney P. A., Veijola J., Visscher P. M., Waddington J., Walsh D., Webb B. T., Weiser M., Wildenauer D. B., Williams N. M., Williams S., Witt S. H., Wolen A. R., Wormley B. K., Wray N. R., Wu J. Q., Zai C. C., Adolfsson R., Andreassen O. A., Blackwood D. H. R., Bramon E., Buxbaum J. D., Cichon S., Collier D. A., Corvin A., Daly M. J., Darvasi A., Domenici E., Esko T., Gejman P. V., Gill M., Gurling H., Hultman C. M., Iwata N., Jablensky A. V., Jonsson E. G., Kendler K. S., Kirov G., Knight J., Levinson D. F., Li Q. S., McCarroll S. A., McQuillin A., Moran J. L., Mowry B. J., Nothen M. M., Ophoff R. A., Owen M. J., Palotie A., Pato C. N., Petryshen T. L., Posthuma D., Rietschel M., Riley B. P., Rujescu D., Sklar P., Clair D. S., Walters J. T. R., Werge T., Sullivan P. F., O'Donovan M. C., Scherer S. W., Neale B. M., Sebat J.; Cnv and Schizophrenia Working Groups of the Psychiatric Genomics Consortium , Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 49, 27–35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russo R. S., Gambello M. J., Murphy M. M., Aberizk K., Black E., Burrell T. L., Carlock G., Cubells J. F., Epstein M. T., Espana R., Goines K., Guest R. M., Klaiman C., Koh S., Leslie E. J., Li L., Novacek D. M., Saulnier C. A., Sefik E., Shultz S., Walker E., White S. P.; Emory 3q29 Project, Mulle J. G., Deep phenotyping in 3q29 deletion syndrome: Recommendations for clinical care. Genet. Med. 23, 872–880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutkowski T. P., Purcell R. H., Pollak R. M., Grewenow S. M., Gafford G. M., Malone T., Khan U. A., Schroeder J. P., Epstein M. P., Bassell G. J., Warren S. T., Weinshenker D., Caspary T., Mulle J. G., Behavioral changes and growth deficits in a CRISPR engineered mouse model of the schizophrenia-associated 3q29 deletion. Mol. Psychiatry 26, 772–783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollak R. M., Purcell R. H., Rutkowski T. P., Malone T., Pachura K. J., Bassell G. J., Epstein M. P., Dawson P. A., Smith M. R., Jones D. P., Zwick M. E., Warren S. T., Caspary T., Weinshenker D., Mulle J. G., Metabolic effects of the schizophrenia-associated 3q29 deletion. Transl. Psychiatry 12, 66 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baba M., Yokoyama K., Seiriki K., Naka Y., Matsumura K., Kondo M., Yamamoto K., Hayashida M., Kasai A., Ago Y., Nagayasu K., Hayata-Takano A., Takahashi A., Yamaguchi S., Mori D., Ozaki N., Yamamoto T., Takuma K., Hashimoto R., Hashimoto H., Nakazawa T., Psychiatric-disorder-related behavioral phenotypes and cortical hyperactivity in a mouse model of 3q29 deletion syndrome. Neuropsychopharmacology 44, 2125–2135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J., Ryan S. K., Deboer E., Cook K., Fitzgerald S., Lachman H. M., Wallace D. C., Goldberg E. M., Anderson S. A., Mitochondrial deficits in human iPSC-derived neurons from patients with 22q11.2 deletion syndrome and schizophrenia. Psychiatry 9, 302 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J., Tran O. T., Crowley T. B., Moore T. M., Zackai E. H., Emanuel B. S., McDonald-McGinn D. M., Gur R. E., Wallace D. C., Anderson S. A., Association of mitochondrial biogenesis with variable penetrance of schizophrenia. JAMA Psychiatry 78, 911–921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tai D. J. C., Ragavendran A., Manavalan P., Stortchevoi A., Seabra C. M., Erdin S., Collins R. L., Blumenthal I., Chen X., Shen Y., Sahin M., Zhang C., Lee C., Gusella J. F., Talkowski M. E., Engineering microdeletions and microduplications by targeting segmental duplications with CRISPR. Nat. Neurosci. 19, 517–522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey J. A., Gu Z., Clark R. A., Reinert K., Samonte R. V., Schwartz S., Adams M. D., Myers E. W., Li P. W., Eichler E. E., Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Stankiewicz P., Lupski J. R., Genome architecture, rearrangements and genomic disorders. Trends Genet. 18, 74–82 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Labun K., Montague T. G., Krause M., Cleuren Y. N. T., Tjeldnes H., Valen E., CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sloan S. A., Andersen J., Pasca A. M., Birey F., Pasca S. P., Generation and assembly of human brain region-specific three-dimensional cultures. Nat. Protoc. 13, 2062–2085 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang H. J., Kawasawa Y. I., Cheng F., Zhu Y., Xu X., Li M., Sousa A. M., Pletikos M., Meyer K. A., Sedmak G., Guennel T., Shin Y., Johnson M. B., Krsnik Z., Mayer S., Fertuzinhos S., Umlauf S., Lisgo S. N., Vortmeyer A., Weinberger D. R., Mane S., Hyde T. M., Huttner A., Reimers M., Kleinman J. E., Sestan N., Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Supek F., Bosnjak M., Skunca N., Smuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L., Ma L., Hamm M., Gage F. H., Hunter T., Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. eLife 5, e13374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson B. H., Petrova-Benedict R., Buncic J. R., Wallace D. C., Nonviability of cells with oxidative defects in galactose medium: A screening test for affected patient fibroblasts. Biochem. Med. Metab. Biol. 48, 122–126 (1992). [DOI] [PubMed] [Google Scholar]
- 20.Gohil V. M., Sheth S. A., Nilsson R., Wojtovich A. P., Lee J. H., Perocchi F., Chen W., Clish C. B., Ayata C., Brookes P. S., Mootha V. K., Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat. Biotechnol. 28, 249–255 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S. J., Capaldi R. A., Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 64, 985–993 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Koranova T., Dvoracek L., Grebenova D., Roselova P., Obr A., Kuzelova K., PAK1 and PAK2 in cell metabolism regulation. J. Cell. Biochem. 123, 375–389 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Antonicka H., Lin Z. Y., Janer A., Aaltonen M. J., Weraarpachai W., Gingras A. C., Shoubridge E. A., A high-density human mitochondrial proximity interaction network. Cell Metab. 32, 479–497.e9 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Ni P., Noh H., Park G. H., Shao Z., Guan Y., Park J. M., Yu S., Park J. S., Coyle J. T., Weinberger D. R., Straub R. E., Cohen B. M., McPhie D. L., Yin C., Huang W., Kim H. Y., Chung S., iPSC-derived homogeneous populations of developing schizophrenia cortical interneurons have compromised mitochondrial function. Mol. Psychiatry 25, 2873–2888 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajasekaran A., Venkatasubramanian G., Berk M., Debnath M., Mitochondrial dysfunction in schizophrenia: Pathways, mechanisms and implications. Neurosci. Biobehav. Rev. 48, 10–21 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Prabakaran S., Swatton J. E., Ryan M. M., Huffaker S. J., Huang J. T., Griffin J. L., Wayland M., Freeman T., Dudbridge F., Lilley K. S., Karp N. A., Hester S., Tkachev D., Mimmack M. L., Yolken R. H., Webster M. J., Torrey E. F., Bahn S., Mitochondrial dysfunction in schizophrenia: Evidence for compromised brain metabolism and oxidative stress. Mol. Psychiatry 9, 684–697 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Ben-Shachar D., Mitochondrial dysfunction in schizophrenia: A possible linkage to dopamine. J. Neurochem. 83, 1241–1251 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Maynard T. M., Meechan D. W., Dudevoir M. L., Gopalakrishna D., Peters A. Z., Heindel C. C., Sugimoto T. J., Wu Y., Lieberman J. A., Lamantia A. S., Mitochondrial localization and function of a subset of 22q11 deletion syndrome candidate genes. Mol. Cell. Neurosci. 39, 439–451 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T. J., Goodman R. P., Grabarek Z., Haas M. E., Hung W. H. W., Joshi P. R., Jourdain A. A., Kim S. H., Kotrys A. V., Lam S. S., McCoy J. G., Meisel J. D., Miranda M., Panda A., Patgiri A., Rogers R., Sadre S., Shah H., Skinner O. S., To T.-L., Walker M. A., Wang H., Ward P. S., Wengrod J., Yuan C. C., Calvo S. E., Mootha V. K., MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devaraju P., Yu J., Eddins D., Mellado-Lagarde M. M., Earls L. R., Westmoreland J. J., Quarato G., Green D. R., Zakharenko S. S., Haploinsufficiency of the 22q11.2 microdeletion gene Mrpl40 disrupts short-term synaptic plasticity and working memory through dysregulation of mitochondrial calcium. Mol. Psychiatry 22, 1313–1326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gokhale A., Hartwig C., Freeman A. A. H., Bassell J. L., Zlatic S. A., Sapp Savas C., Vadlamudi T., Abudulai F., Pham T. T., Crocker A., Werner E., Wen Z., Repetto G. M., Gogos J. A., Claypool S. M., Forsyth J. K., Bearden C. E., Glausier J., Lewis D. A., Seyfried N. T., Kwong J. Q., Faundez V., Systems analysis of the 22q11.2 microdeletion syndrome converges on a mitochondrial interactome necessary for synapse function and behavior. J. Neurosci. 39, 3561–3581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gokhale A., Lee C. E., Zlatic S. A., Freeman A. A. H., Shearing N., Hartwig C., Ogunbona O., Bassell J. L., Wynne M. E., Werner E., Xu C., Wen Z., Duong D., Seyfried N. T., Bearden C. E., Olah V. J., Rowan M. J. M., Glausier J. R., Lewis D. A., Faundez V., Mitochondrial proteostasis requires genes encoded in a neurodevelopmental syndrome locus. J. Neurosci. 41, 6596–6616 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan T. A., Revah O., Gordon A., Yoon S. J., Krawisz A. K., Goold C., Sun Y., Kim C. H., Tian Y., Li M. Y., Schaepe J. M., Ikeda K., Amin N. D., Sakai N., Yazawa M., Kushan L., Nishino S., Porteus M. H., Rapoport J. L., Bernstein J. A., O'Hara R., Bearden C. E., Hallmayer J. F., Huguenard J. R., Geschwind D. H., Dolmetsch R. E., Pasca S. P., Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nat. Med. 26, 1888–1898 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tebbenkamp A. T. N., Varela L., Choi J., Paredes M. I., Giani A. M., Song J. E., Sestan-Pesa M., Franjic D., Sousa A. M. M., Liu Z. W., Li M., Bichsel C., Koch M., Szigeti-Buck K., Liu F., Li Z., Kawasawa Y. I., Paspalas C. D., Mineur Y. S., Prontera P., Merla G., Picciotto M. R., Arnsten A. F. T., Horvath T. L., Sestan N., The 7q11.23 protein DNAJC30 interacts with ATP synthase and links mitochondria to brain development. Cell 175, 1088–1104.e23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tai D. J. C., Razaz P., Erdin S., Gao D., Wang J., Nuttle X., de Esch C. E., Collins R. L., Currall B. B., O'Keefe K., Burt N. D., Yadav R., Wang L., Mohajeri K., Aneichyk T., Ragavendran A., Stortchevoi A., Morini E., Ma W., Lucente D., Hastie A., Kelleher R. J., Perlis R. H., Talkowski M. E., Gusella J. F., Tissue- and cell-type-specific molecular and functional signatures of 16p11.2 reciprocal genomic disorder across mouse brain and human neuronal models. Am. J. Hum. Genet. 109, 1789–1813 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murtaza N., Cheng A. A., Brown C. O., Meka D. P., Hong S., Uy J. A., El-Hajjar J., Pipko N., Unda B. K., Schwanke B., Xing S., Thiruvahindrapuram B., Engchuan W., Trost B., Deneault E., Calderon de Anda F., Doble B. W., Ellis J., Anagnostou E., Bader G. D., Scherer S. W., Lu Y., Singh K. K., Neuron-specific protein network mapping of autism risk genes identifies shared biological mechanisms and disease-relevant pathologies. Cell Rep. 41, 111678 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Gordon A., Forsingdal A., Klewe I. V., Nielsen J., Didriksen M., Werge T., Geschwind D. H., Transcriptomic networks implicate neuronal energetic abnormalities in three mouse models harboring autism and schizophrenia-associated mutations. Mol. Psychiatry 26, 1520–1534 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Singh T., Poterba T., Curtis D., Akil H., Al Eissa M., Barchas J. D., Bass N., Bigdeli T. B., Breen G., Bromet E. J., Buckley P. F., Bunney W. E., Bybjerg-Grauholm J., Byerley W. F., Chapman S. B., Chen W. J., Churchhouse C., Craddock N., Cusick C. M., DeLisi L., Dodge S., Escamilla M. A., Eskelinen S., Fanous A. H., Faraone S. V., Fiorentino A., Francioli L., Gabriel S. B., Gage D., Taliun S. A. G., Ganna A., Genovese G., Glahn D. C., Grove J., Hall M. H., Hamalainen E., Heyne H. O., Holi M., Hougaard D. M., Howrigan D. P., Huang H., Hwu H. G., Kahn R. S., Kang H. M., Karczewski K. J., Kirov G., Knowles J. A., Lee F. S., Lehrer D. S., Lescai F., Malaspina D., Marder S. R., McCarroll S. A., McIntosh A. M., Medeiros H., Milani L., Morley C. P., Morris D. W., Mortensen P. B., Myers R. M., Nordentoft M., O'Brien N. L., Olivares A. M., Ongur D., Ouwehand W. H., Palmer D. S., Paunio T., Quested D., Rapaport M. H., Rees E., Rollins B., Satterstrom F. K., Schatzberg A., Scolnick E., Scott L. J., Sharp S. I., Sklar P., Smoller J. W., Sobell J. L., Solomonson M., Stahl E. A., Stevens C. R., Suvisaari J., Tiao G., Watson S. J., Watts N. A., Blackwood D. H., Borglum A. D., Cohen B. M., Corvin A. P., Esko T., Freimer N. B., Glatt S. J., Hultman C. M., McQuillin A., Palotie A., Pato C. N., Pato M. T., Pulver A. E., Clair D. S., Tsuang M. T., Vawter M. P., Walters J. T., Werge T. M., Ophoff R. A., Sullivan P. F., Owen M. J., Boehnke M., O'Donovan M. C., Neale B. M., Daly M. J., Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 604, 509–516 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chong Z. S., Khong Z. J., Tay S. H., Ng S. Y., Metabolic contributions to neuronal deficits caused by genomic disruption of schizophrenia risk gene SETD1A. Schizophrenia (Heidelb) 8, 115 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sefik E., Purcell R. H.; Emory 3q29 Project, Walker E. F., Bassell G. J., Mulle J. G., Convergent and distributed effects of the 3q29 deletion on the human neural transcriptome. Trans. Psychiatry 11, 357 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh M. D., Jensen M., Lasser M., Huber E., Yusuff T., Pizzo L., Lifschutz B., Desai I., Kubina A., Yennawar S., Kim S., Iyer J., Rincon-Limas D. E., Lowery L. A., Girirajan S., NCBP2 modulates neurodevelopmental defects of the 3q29 deletion in Drosophila and Xenopus laevis models. PLOS Genet. 16, e1008590 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foo L. C., Purification of rat and mouse astrocytes by immunopanning. Cold Spring Harb. Protoc. 2013, 421–432 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luecken M. D., Theis F. J., Current best practices in single-cell RNA-seq analysis: A tutorial. Mol. Syst. Biol. 15, e8746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hafemeister C., Satija R., Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zappia L., Oshlack A., Clustering trees: A visualization for evaluating clusterings at multiple resolutions. Gigascience 7, giy083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tirosh I., Izar B., Prakadan S. M., Wadsworth M. H. 2nd, Treacy D., Trombetta J. J., Rotem A., Rodman C., Lian C., Murphy G., Fallahi-Sichani M., Dutton-Regester K., Lin J. R., Cohen O., Shah P., Lu D., Genshaft A. S., Hughes T. K., Ziegler C. G., Kazer S. W., Gaillard A., Kolb K. E., Villani A. C., Johannessen C. M., Andreev A. Y., Van Allen E. M., Bertagnolli M., Sorger P. K., Sullivan R. J., Flaherty K. T., Frederick D. T., Jane-Valbuena J., Yoon C. H., Rozenblatt-Rosen O., Shalek A. K., Regev A., Garraway L. A., Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenberg A. B., Roco C. M., Muscat R. A., Kuchina A., Sample P., Yao Z., Graybuck L. T., Peeler D. J., Mukherjee S., Chen W., Pun S. H., Sellers D. L., Tasic B., Seelig G., Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y., Chen K., Sloan S. A., Bennett M. L., Scholze A. R., O'Keeffe S., Phatnani H. P., Guarnieri P., Caneda C., Ruderisch N., Deng S., Liddelow S. A., Zhang C., Daneman R., Maniatis T., Barres B. A., Wu J. Q., An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loo L., Simon J. M., Xing L., McCoy E. S., Niehaus J. K., Guo J., Anton E. S., Zylka M. J., Single-cell transcriptomic analysis of mouse neocortical development. Nat. Commun. 10, 134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y., Sloan S. A., Clarke L. E., Caneda C., Plaza C. A., Blumenthal P. D., Vogel H., Steinberg G. K., Edwards M. S., Li G., Duncan J. A. 3rd, Cheshier S. H., Shuer L. M., Chang E. F., Grant G. A., Gephart M. G., Barres B. A., Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ximerakis M., Lipnick S. L., Innes B. T., Simmons S. K., Adiconis X., Dionne D., Mayweather B. A., Nguyen L., Niziolek Z., Ozek C., Butty V. L., Isserlin R., Buchanan S. M., Levine S. S., Regev A., Bader G. D., Levin J. Z., Rubin L. L., Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 22, 1696–1708 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Nagy A., Delgado-Escueta A. V., Rapid preparation of synaptosomes from mammalian brain using nontoxic isoosmotic gradient material (Percoll). J. Neurochem. 43, 1114–1123 (1984). [DOI] [PubMed] [Google Scholar]
- 55.Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A. K., Slichter C. K., Miller H. W., McElrath M. J., Prlic M., Linsley P. S., Gottardo R., MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.BrainSpan, “Technical white paper: Transcriptome profiling by RNA sequencing and exon microarray (v.5)” (BrainSpan Atlas of the Developing Human Brain, 2013).
- 57.Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J., g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reimand J., Kull M., Peterson H., Hansen J., Vilo J., g:Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193–W200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S18
Legends for tables S1 and S2
Tables S1 and S2