Abstract
The human carbonic anhydrase IX (CA IX) is a hypoxia-induced transmembrane protein belonging to the α-CA enzyme family. It has a crucial role in pH regulation in hypoxic cells and acts by buffering intracellular acidosis induced by hypoxia. Indeed, it is frequently expressed in cancer cells, where it contributes to tumor progression. CA IX is also able to localize in the nucleus, where it contributes to 47S rRNA precursor genes transcription; however, the mechanisms assisting its nuclear translocation still remain unclear. The aim of our study was to deepen the understanding of the mechanisms involved in CA IX subcellular distribution. To this purpose, we implemented a site-directed mutagenesis approach targeting the C-terminal domain of CA IX and evaluated the subcellular distribution of the wild-type and mutant proteins in the SH-SY5Y cell line. The mutant proteins showed impaired binding ability and altered subcellular distribution in both normoxic and hypoxic conditions. Our data suggest that CA IX nuclear translocation depends on its transit through the secretory and the endocytic pathways.
Keywords: Hypoxia, NLS, Nuclear import, Carbonic anhydrase, Protein glycosylation, Membrane protein
1. Introduction
The human carbonic anhydrase IX (CA IX) is described as a hypoxia-induced transmembrane protein belonging to the α-CA enzyme family. CA IX has a crucial role in pH regulation, especially in hypoxic cells, where it acts in concerted mechanisms with intracellular carbonic anhydrases, by buffering intracellular acidosis induced by hypoxia and contributing to extracellular pH acidification [1]. Tumor cells usually implement this strategy to promote invasiveness and resistance to chemo- and radio-therapy, and to avoid immune surveillance. In fact, CA IX is a major effector of HIF-1α activity in the metabolic adaptation of hypoxic tumors, which results in a worse prognosis for CA IX-expressing tumors [[2], [3], [4], [5], [6]]. In normal tissues, CA IX expression is tightly controlled by oxygen levels, almost undetectable under normoxic conditions and heavily induced by hypoxia. This multidomain protein consists of an extracellular region, a transmembrane segment and an intracytoplasmic tail; the extracellular region comprises an N-terminal proteoglycan-like domain and the catalytic domain [7,8]. As most membrane proteins [9], newly synthesized CA IX is initially translocated to the endoplasmic reticulum (ER) and then imported into the Golgi apparatus; its trafficking results in both N- and O-glycosylation, before being transferred and inserted in the plasma membrane [10].
Thanks to its interaction with the nuclear import/export machinery via its C-terminal region, CA IX is also actively transferred to and from the nuclear compartment [11]. CA IX nuclear localization allows its association to nucleolar chromatin, according to a mechanism regulated by oxygen levels. In fact, CA IX was found on the 47S rDNA gene promoters in normoxic cells, and less represented on these sites in hypoxic cells in association to enriched complexes with exportin-1 (XPO1). 47S rRNA transcript levels are accordingly down-represented in hypoxic cells [12].
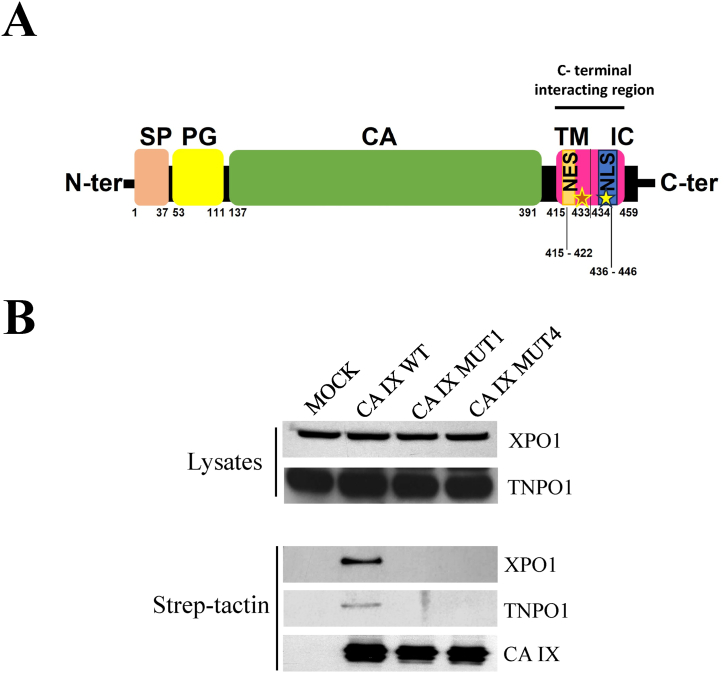
The characterization of the CA IX intracellular interactome revealed a set of CA IX interacting proteins, mainly in hypoxic cells via their HEAT/ARM repeat domains, including importins and exportins [11]. The Fig. 1A shows a schematic representation of the CA IX protein in its canonical transmembrane topology. The putative nuclear localization signal (NLS) of CA IX, RRGHRRTHGG (aa 436–446), resides in the short C-terminal intra-cytosolic domain of the protein; intriguingly, in the canonical membrane-bound topology the putative nuclear export sequence (NES), ILALVFGL (aa 415–422), sits in the transmembrane alpha helix of CA IX. The C-terminal sequence Leu418−Ala459 of CA IX, encompassing both nuclear trafficking sequences was revealed, by means of pull-down experiments with synthetic peptides, as the minimal portion of the protein required for interaction with importins, exportins and CAND1, an additional member of CA IX interactome [11]. Yet, the mechanisms ensuring the nuclear trafficking of CA IX have not been elucidated. Moreover, CA IX has been shown to be endocytosed under certain circumstances [13] and its intracellular tail is involved in localization and extracellular function [14].
Fig. 1.
Co-precipitation analysis of wild-type and mutant CA IX proteins with components of the nucleocytoplasmic transport. A) Schematic representation of CA IX structural features and protein domains in its canonical transmembrane topology. From the N- to the C-terminus: Signal peptide (pink), a.a. 1–37; PG (proteoglycan) domain (yellow), a.a. 53–111; CA (catalytic) domain (green), a.a. 137–391; transmembrane (TM) and intracytosolic (IC) domains (lilac), a.a. 415–459. The asterisks in the C-terminal interacting region, encompassing the putative nuclear export signal (NES) and nuclear localization signal (NLS) denote the mutated sites of MUT1 and MUT 4 proteins used in this study. B) Western blot analysis of HEK-293 lysates expressing mock, CA IX WT, MUT1 (Leu423/Arg-Thr427/Arg), MUT4 (Arg436/Glu-Arg441/Glu), with XPO1 and TNPO1. Co-precipitation analysis of CA IX WT and its mutant proteins with XPO1 and TNPO1 after purification with strep-tactin resin. Uncropped images of the western blots are available as supplementary material (Succoio_supplementary_materials).
Altogether, the available information supports that CA IX protein can traffic to and from the nucleus. This is not surprising, considering that other proteins traditionally described as transmembrane can modify their canonical subcellular localization in response to extracellular stimuli to exert additional biological functions [15,16]. These proteins are transported from the cell surface to the nucleus after endocytosis and participate in multiple biological functions, including transcription regulation, cellular proliferation, tumor progression, DNA repair, and chemo- and radio-resistance [17]. In particular, many cell surface receptor tyrosine kinases (RTKs), such as insulin-like growth factor 1 receptor (IGF-1R), fibroblast growth factor receptor (FGFR), vascular endothelial growth factor receptor (VEGFR), and epidermal growth factor receptor (EGFR), have been shown to localize in the nucleus [17]. Accordingly, EGFR was shown to localize in the nucleus after endocytosis followed by transfer to the Golgi apparatus and to ER, a process known as retrograde trafficking [18]. COPI is the crucial protein complex responsible for the retrograde transport of proteins from cis Golgi to the ER. Brefeldin A (BFA) is an inhibitor of a small GTPase ADP- ribosylation factor (ARF), a component of the COPI multicomplex, whose inactivation causes the disassembly of COPI vesicles, thus suppressing this transport [[19], [20], [21]]. In the case of ErbB-2, a member of the epidermal growth factor receptor family of receptor tyrosine kinases, a mechanism for nuclear entry has been depicted that allows migration from the plasma membrane to the nucleus through endocytosis, importins, and Nup358 [22].
In this study, we undertook an experimental strategy, based on engineered variants of CA IX protein that were previously found unable to bind to the HEAT/ARM protein CAND1 [23], to shed light on CA IX trafficking in mammalian cells. The provided evidence supports the occurrence of a post-endocytic mechanism for CA IX nuclear trafficking in the neuroblastoma cell line SH-SY5Y, used in this study as a simplified neuronal model, for its potential involvement in stroke.
2. Results
2.1. CA IX proteins with mutated C-terminal sequences show altered subcellular distributions
Biochemical analysis previously showed that the C-terminal sequence of CA IX is necessary and sufficient for binding to selected members of its interactome, including CAND1, importin-1 (TNPO1) and XPO1 [11]. These data were further refined by modelling analysis and binding measurements, highlighting the peptide string of CA IX from Leu418 to Ala459 as critically involved in the interactions with CAND1 [23]. In particular, cellular CAND1 was unable to co-immunoprecipitate with CA IX mutant protein (CA IX MUT1) bearing substitutions of the residues Leu423 and Thr427, involved in van der Waals interactions, with arginine. Similarly, the interactions with CAND1 were absent when CA IX residues involving electrostatic interactions, namely Arg436, Arg440, and Arg441, were mutated to alanine or glutamic acid in various combinations [23]. As shown in the co-precipitation analysis displayed in Fig. 1, the CAND1 inability to bind to the mutant CA IX proteins was also shared by TNPO1 and XPO1 proteins. HEK-293 cells were used as reservoir for the high yield in protein production. Based on the biochemical evidence for defective binding properties of the CA IX mutant proteins, we hypothesized that lack of interaction with the natural ligands could generate defects in CA IX cellular trafficking. Thus, the mutant CA IX proteins CA IX-MUT1 (Leu423/Arg-Thr427/Arg) and CA IX-MUT4 (Arg436/Glu-Arg441/Glu) were selected as the representative models for subcellular localization studies in the human neuroblastoma SH-SY5Y cell line.
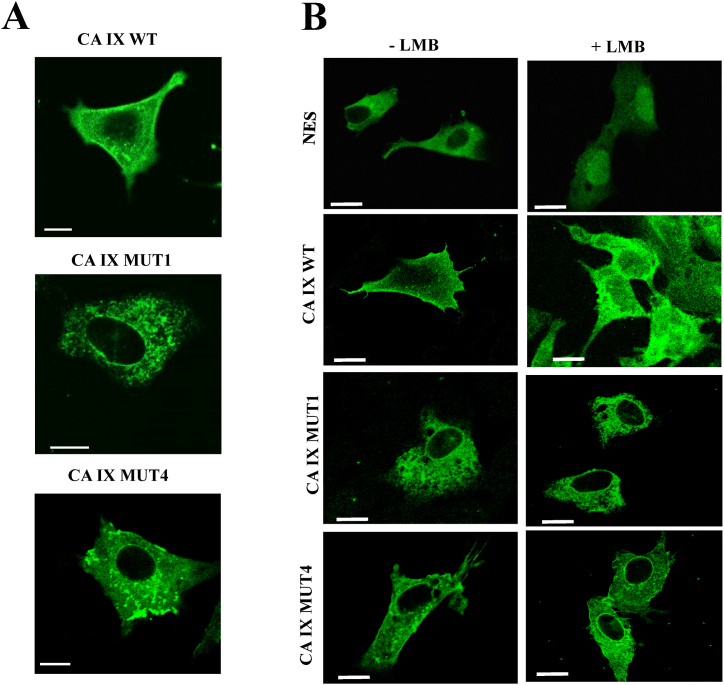
The immunofluorescence analysis of the CA IX proteins exogenously expressed in SH-SY5Y cells (Fig. 2A) confirmed a strong membrane signal for the wild-type protein (CA IX WT), in contrast with the two mutant proteins, where this prominent localization was lost. Namely, the CA IX MUT1 protein showed a very intense intracellular accumulation, involving subcellular structures resembling the endoplasmic reticulum (ER). CA IX MUT4 protein generated intracellular accumulations too, according to a distribution that would seem to highlight vesicles, located mostly in the perinuclear area and below the cytoplasmic membrane. As expected, the CA IX wild-type protein showed a weak nuclear presence, which was partially lost, in the case of CA IX MUT1, and undetectable for the CA IX MUT4 protein; both mutant proteins showed, however, a perinuclear accumulation. Considering these results, the mutant proteins clearly showed altered subcellular distributions, compared to the wild-type CA IX protein; even more interestingly, the subcellular distribution of the two mutant proteins differed deeply from each other.
Fig. 2.
Localization studies of CA IX WT and its mutant forms in SH-SY5Y neuroblastoma cells. A) Immunofluorescence analysis of CA IX WT, CA IX MUT1 and CA IX MUT4 in SH-SY5Y neuroblastoma cell line using monoclonal CA IX specific antibody VII:20 (green). Both mutants show a different intracellular localization compared to the WT protein. B) Immunofluorescence assay of CA IX WT, CA IX MUT1, or CA IX MUT4 was performed using transfected SH-SY5Y cells with or without leptomycin B (LMB) treatment for 5 h at a final concentration of 20 ng/mL, or with 70% methanol as negative control. As positive control, we used a plasmid driving expression of GFP protein with NES sequence. Cells transfected with CA IX WT and exposed to LMB show enrichment of nuclear signal if compared to untreated cells. Cells transfected with CA IX MUT4 showed a weak nuclear enrichment, while in MUT1 the treatment resulted in undetectable nuclear accumulation. Bars = 10 μm.
Membrane proteins, including CA IX, frequently show nuclear localization in cell cultures [24,25]. In order to evaluate the potential trafficking of the mutant proteins through the nuclear compartment, we performed immunofluorescence experiments in SH-SY5Y cells transiently expressing CA IX proteins, through transfection, and after having treated the cell cultures with leptomycin B, an inhibitor of nuclear export. Thus, the nuclear accumulation of proteins as a consequence of leptomycin B treatment would provide an indication of their actual transit through the nuclear compartment. In the Panel B of Fig. 2 a control experiment is shown, in which an EGFP protein fused to a canonical NES was normally excluded from the nucleus, while it accumulated in the nuclear compartment, after treatment with leptomycin B. As expected, wild-type CA IX showed straightforward evidence of nuclear accumulation in the transfected cells exposed to leptomycin B, even if lower than the NES alone. On the contrary, both CA IX MUT1 and CA IX MUT4 mutant proteins showed limited evidence for nuclear representation upon treatment with leptomycin B, indicating that they were actually impaired in their trafficking to the nucleus.
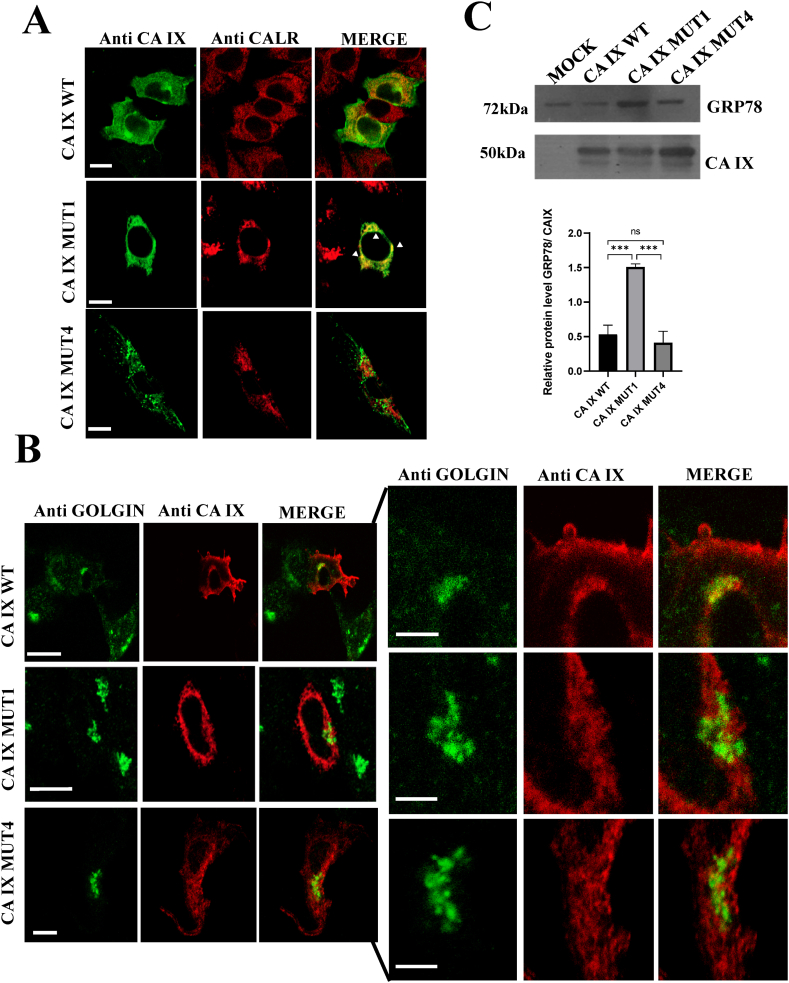
To further investigate the different cellular localizations of the mutant proteins, we performed co-localization studies with well-known markers of intracellular structures, i.e., calreticulin for ER (Fig. 3A) and golgin for Golgi apparatus (Fig. 3B). In the case of wild-type CA IX, the lack of colocalization with the ER marker and the limited dimensions of the CA IX-comprising Golgi apparatus suggested the occurrence of a rapid traffic through these structures in reaching the plasma membrane. The CA IX MUT4 protein did not show significant co-localization, neither with calreticulin nor with golgin. A more pronounced match of the CA IX MUT1 protein was, instead, clear for calreticulin, conversely with no match was observed for golgin, supporting the possibility that CA IX MUT1 accumulated into the ER. In agreement with the latter evidence, cells transfected with the CA IX MUT1 protein showed increased accumulation of GRP78, a marker of ER stress (Fig. 3C).
Fig. 3.
Co-localization studies of CA IX and its mutants with endoplasmic reticulum and Golgi apparatus markers in SH-SY5Y neuroblastoma cells. A) CA IX (green) and calreticulin (red) were visualized by confocal fluorescence microscopy using specific antibodies. The merged images show clear CA IX co-localization with calreticulin only in CA IX MUT1. B) In this panel CA IX is shown in green and golgin in red. The two mutants do not show significant co-localization with golgin while a modest presence is detected for the WT protein. C) Unfolded protein response in the endoplasmic reticulum is shown by GRP78 expression levels in lysates of SH-SY5Y cells transfected with mock, CA IX WT, MUT1 and MUT4 vectors. The Western blot analysis shows a significant increase of GRP78 levels in SH-SY5Y cells overexpressing CA IX MUT1. CA IX protein in the same lysates is used as normalizer. Statistic was performed by unpaired t-test (***p < 0.005). Bars in 3A and 3B = 10 μm (enlargements = 5 μm). Uncropped images of the Western blot analysis are available as supplementary material (Succoio_supplementary_materials).
2.2. Glycosidase shear-sensitivity analyses highlight incomplete maturation of the CA IX MUT1 protein and suggest its accumulation in the ER
The observed findings involving CA XI MUT1 could be associated with an altered maturation of the mutant protein; thus, in order to verify the actual transit of the wild-type and mutant CA IX proteins through the ER and the Golgi apparatus, we performed sensitivity experiments to glycosidase shear. It is in fact well known that CA IX is a substrate for N-glycosylation, which occurs in the ER, at the residue Asn309, and for O-glycosylation on Thr78 [10,26].
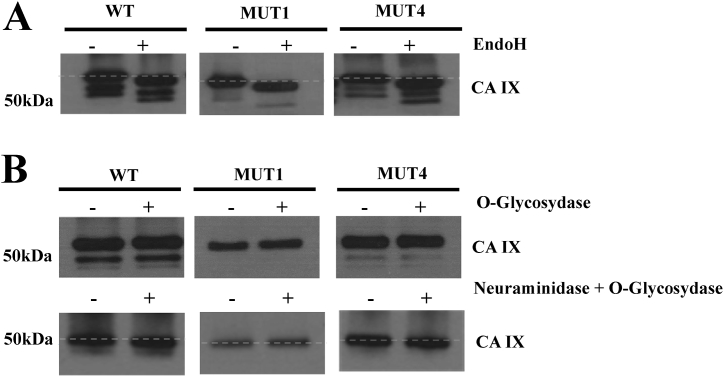
The analysis of N-glycosylation of the three proteins was performed by enzymatic digestion with the enzyme Endoglycosidase H (Endo H). This enzyme cuts between two N-acetylglucosamine residues of a sugar chain bound to asparagine, generating a truncated sugar molecule, consisting of a single N-acetylglucosamine residue bound to Asn. Endo H selectively digests chains with a high content of mannose and simple hybrids (typical to glycoproteins in transit in the ER) but is not able to cut complex oligosaccharides. N-glycosylation analysis was performed on the streptavidin-biotin based immunoprecipitation-enriched proteins CA IX WT, CA IX MUT1 and CA IX MUT4. The results in Fig. 4A show that all the proteins undergo N-glycosylation, since they all showed a shift in the electrophoretic migration as the result of Endo H treatment, in comparison to the corresponding undigested controls.
Fig. 4.
Glycosidase shear-sensitivity analyses of CA IX WT and its mutant forms. A) Western blot analysis of purified proteins CA IX WT, MUT1 and MUT4 digested and non-digested with EndoH. All digested samples present an electrophoretic shift when compared to undigested controls. B) Western blot analysis of purified proteins CA IX WT and mutants digested with O-glycosidase alone and combined with Neuraminidase. Undigested samples are used as negative control. Only CA IX WT and MUT4 show an electrophoretic shift. Uncropped images of the Western blot analyses are available as supplementary material (Succoio_supplementary_materials).
Following enzymatic digestion with O-glycosidase, no differences in migration were observed between the treated or untreated samples (Fig. 4B, upper panel). The lack of digestion of CA IX WT with O-glycosidase led us to think that the enzyme could not work, due to sialic acid residues protecting the potential cleavage sites. For this reason, we made use of neuraminidase, which removes the sialic acid residues. In the bottom panel of Fig. 4B, a slight shift in electrophoretic migration was observed, when compared to undigested samples, for both CA IX WT and CA IX MUT4 proteins, suggesting that they were actually O-glycosylated; no shift was detected, however, in the case of the CA IX MUT1 protein. Taken together, these results show that all the CA IX proteins evaluated transit through the ER and that only the CA IX WT and CA IX MUT4 are capable to pass through the ER and undergo post-translational modifications in the Golgi apparatus; accordingly, we conclude that the CA IX MUT1 protein accumulates within the ER.
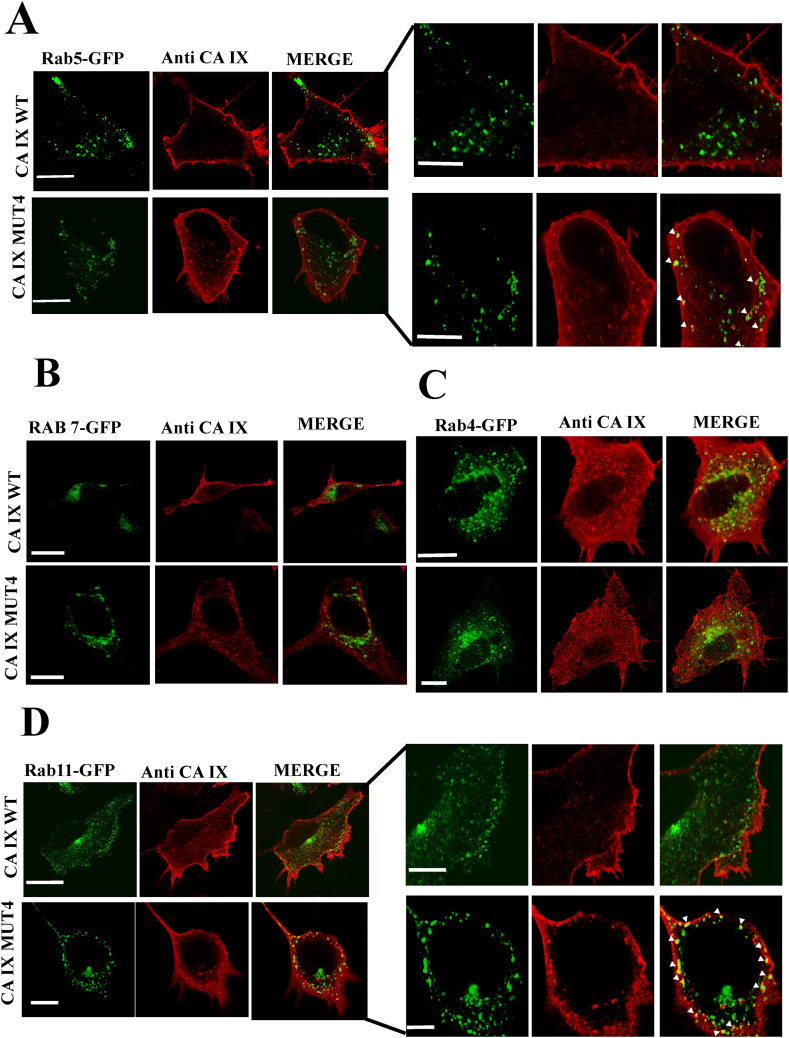
2.3. CA IX MUT4 is enriched in Rab11-containing endosomes
Having established that CA IX WT and MUT4 proteins were able to mature through the ER and the Golgi apparatus, immunofluorescence experiments were performed to verify whether the CA IX MUT4 protein was actually enriched in cytosolic vesicular structures. In this regard, members of the Rab family of small GTPases, fused to EGFP, were used as vesicular markers, while CA IX proteins were detected by immunofluorescence (Fig. 5). Assuming that these vesicles could be part of the endocytic circuit, we used vesicular markers EGFP-Rab5 for early endosomes, EGFP-Rab7 for late endosomes, EGFP-Rab4 for fast recycling endosomes and EGFP-Rab11 for late recycling endosomes.
Fig. 5.
Co-localization analysis of CA IX WT and CA IX MUT4 with Rabs as intracellular and vesicular trafficking markers. Plasmids expressing GFP-conjugated Rabs were co-transfected with CA IX WT and MUT4. CA IX was identified by using specific polyclonal antibodies. A) Fluorescence analysis of Rab5-GFP (green) and CA IX (red) show co-localization spots in proximity of the plasma membrane and the perinuclear area in MUT4 B) Fluorescence analysis of Rab7-GFP (green) and CA IX MUT4 (red) does not show any significant co-localization between the two proteins. C) Immunofluorescence of Rab4-GFP (green) and CA IX (red) does not show any significant difference between CA IX WT and MUT4. D) Rab11-GFP (green) and CA IX (red). The images show an appreciable co-localization signal only in MUT4. Bars in 5A, 5B and 5D = 10 μm, in 5C = 5 μm (enlargements = 5 μm).
Analysis of the fluorescence for EGFP-Rab5 revealed a significant amount of co-localization spots for CA IX MUT4 compared to CA IX WT, mostly due to vesicles set in proximity to the plasma membrane and near the perinuclear area (Fig. 5A). This could mean that CA IX MUT4 reaches the cell membrane and is rapidly internalized in these endosomes. Conversely, the complete lack of co-localization with Rab7 suggests that CA IX MUT4 vesicles are not intended to lysosomal degradation pathway (Fig. 5B).
Further immunofluorescence analysis using Rab4 vesicular marker showed no significant co-localization with both CA IX WT and CA IX MUT4 (Fig. 5C), suggesting that they are not caught in the fast-recycling endosomes toward the cell surface.
Finally, immunofluorescence analysis with Rab11 showed a weak localization with CA IX WT, and a strong localization with CA IX MUT4, especially in the vesicles scattered in the cytoplasm frequently under the cell membrane (Fig. 5D), indicating that after pausing in early endosomes, the CA IX MUT4 protein could get stuck in the slow recycling endosomal trafficking.
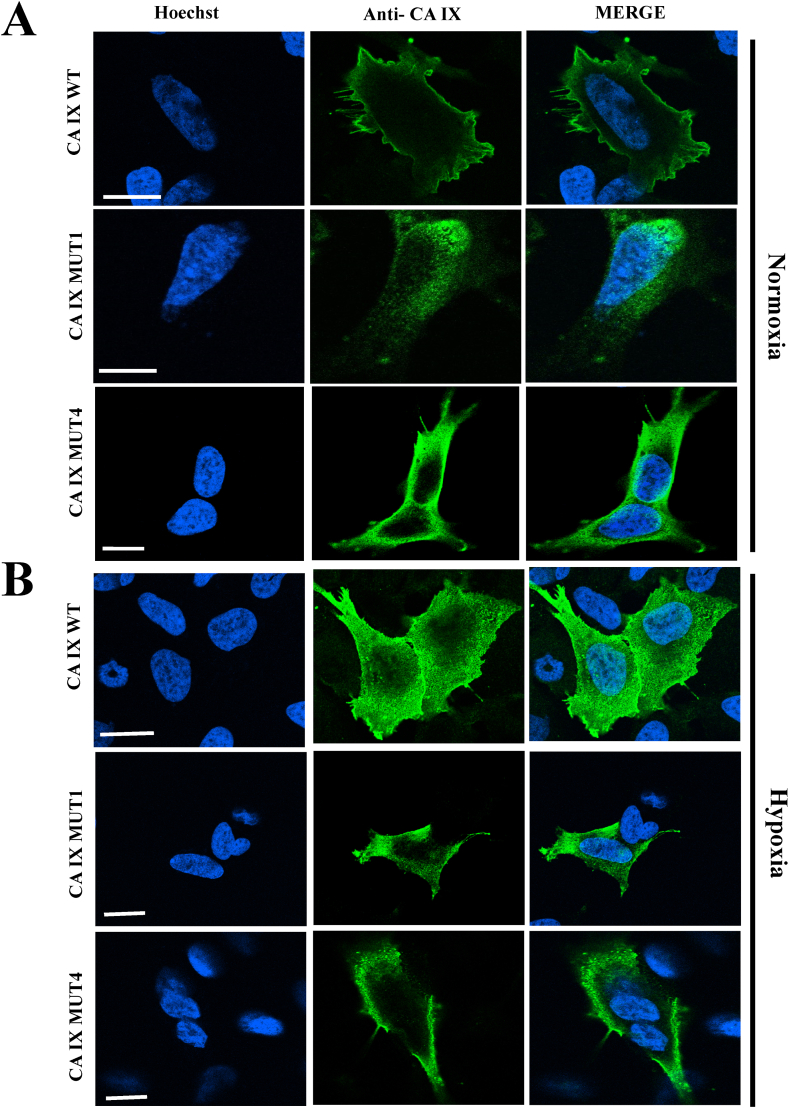
2.4. Analysis of the distribution of CA IX proteins in hypoxic conditions
Although well expressed in normoxic cancer cell cultures, CA IX has relevant functions in hypoxic cells, where it contributes to the acidification of the extracellular space and, by acting in synergy with membrane transporters and intracellular CAs, to the buffering of the intracellular compartments [27,28].
Since CA IX mutants displayed major modifications in cellular distribution in normoxic conditions, an immunofluorescence analysis was performed to investigate the localization of CA IX mutant proteins under normoxic conditions (Fig. 6A) and hypoxic conditions (Fig. 6B). SH-SY5Y cells transfected with CA IX WT and subjected to hypoxia showed, as expected, a stronger membrane signal and an appreciable nuclear staining in comparison to normoxic cells. This pattern was paralleled by the confirmed presence of numerous vesicular structures in the cytoplasm [13]. CA IX MUT1 protein appeared to be still accumulated in the ER, in an even more pronounced manner, after hypoxic treatment. Finally, CA IX MUT4 retained its vesicular appearance, although the vesicles seemed to be larger and denser, especially in the perinuclear region. In both mutants, plasma membrane enrichment and nuclear localization were not discernible.
Fig. 6.
Analysis of CA IX subcellular distribution in normoxic and hypoxic SH-SY5Y cells. A) Normoxic SH-SY5Y. B) Hypoxic SH-SY5Y. The immunofluorescence images show a nuclear enrichment in hypoxia only in CA IX WT, absent in the mutants. MUT1 still has intense staining of the ER widespread in the cytoplasm while MUT 4 retained its intracellular accumulation typical of vesicular structures. Also, the plasma membrane localization is absent in the mutant proteins exposed to hypoxia. Bars = 10 μm.
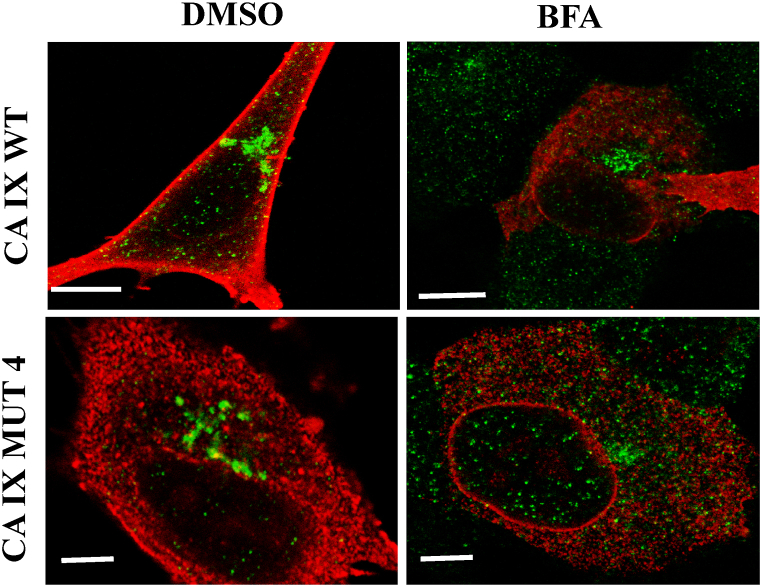
2.5. COPI-mediated retrograde trafficking is involved in CA IX nuclear translocation
To learn more about the potential mechanisms that regulate the membrane flow of CA IX to and from the ER, and to establish whether the altered cellular localization of CA IX MUT 4 was causally related to the disruption of early biosynthetic trafficking, we used brefeldin A, an antibacterial drug that inhibits COPI coat formation, thus blocking retrograde trafficking, for which COPI is essential. Therefore, CA IX WT and MUT4 expressing constructs were individually transfected into SH-SY5Y cells and after 48 h the cells were treated with 1 μg/mL brefeldin A for 6 h. Then, their phenotype was assessed by immunofluorescence analysis using CA IX and Golgin antibodies (Fig. 7).
Fig. 7.
Brefeldin A impairs nuclear translocation of the wild-type CA IX protein. Immunolocalization of golgin (green) with CA IX (red) in SH-SY5Y cells with and without BFA treatment. In CA IX WT, after BFA treatment, cells show a significant increase in CA IX the perinuclear signal and in vesicle quantity, whereas MUT4 shows mostly perinuclear signal. Bars: on left panels = 5 μm, on right panels = 3 μm.
Despite the impaired morphology of the Golgi apparatus in both treated cells, BFA-treated WT cells showed a much more robust phenotypical change, in comparison to DMSO control. Namely, CA IX distribution in treated WT cells resembled that of untreated MUT4 cells, with a significant increase in the perinuclear signal and in vesicle quantity, the hallmarks of MUT4 expressing cells. The treatment in MUT4 exacerbated the vesicular phenotype, with evident perinuclear accumulation. As previously observed, the double immunofluorescence staining of CA IX with the ER marker calreticulin, showed an accentuated perinuclear accumulation both in cells transfected with CA IX MUT4 and treated with brefeldin A, and in control cells. Conversely, CA IX WT assumed a perinuclear localization that resembled CA IX MUT4 subcellular distribution only after the treatment with brefeldin A, compared to the vehicle-treated control group. Then, these experiments suggested that, in transfected cells, CA IX WT traffic may involve the intervention of COPI vesicles that mediate GOLGI-ER retrograde transport, as it happens for some members of the receptor tyrosine kinase family of membrane proteins. An interpretation of these results may suggest that the altered localization of the CA IX mutant could be due to inefficient GOLGI-ER retrograde transport.
3. Discussion
Carbonic anhydrase IX (CA IX) belongs to a wide family of zinc-enzymes differing for cellular localization, tissue distribution, and catalytic properties. CA IX catalyzes the reversible hydration of carbon dioxide into a bicarbonate ion and a proton, it is transcriptionally regulated by HIF-1α, a primary regulator of hypoxic-stress response, and has a crucial role in hypoxia, as it buffers the intracellular compartment, thus ensuring cell survival.
The increase in co-localization and the well-established body of knowledge acquired on CA IX interactions with proteins of the nucleocytoplasmic shuttling and its nuclear localization, prompted us to focus on CA IX nuclear trafficking in a neuroblastoma-derived cellular model, namely the SH-SY5Y cell line. Indeed, several cells express CA IX in the nucleus, where it contributes to transcription modulation of 47S rRNA precursor genes [12]. The identification of the whole machinery of nuclear import/export as the main component of its intracellular interactome and the presence of nuclear localization sequences (NES and NLS) in the C-terminal of the protein, provide a strong rationale for CA IX nuclear localization. With the purpose of elucidating the subcellular trafficking of CA IX, we exploited complementary approaches of molecular modelling, previously implemented by our group [23], and cell biology. These mutants did not simply cause a loss of interaction with endogenous CA IX ligands but also demonstrated very peculiar subcellular localizations, based on the profound modifications (Leu423/Arg-Thr427/Arg for MUT1 and Arg436/Glu-Arg441/Glu for MUT4) introduced in the key regions of the protein.
The modification of the transmembrane alpha helix, containing the NES, characteristic of CA IX MUT1 comes with the most severe phenotype. It is clearly trapped within the ER, which suggests that this region holds the exit signal from the ER. There, MUT1 acquires the N-glycosylation correctly, but it fails to reach the Golgi apparatus, lacking the typical O-glycosylation acquired by membrane proteins through the Golgi transit and, therefore, it is impaired in reaching the membrane. Entangled in the ER, this mutated protein increases the accumulation of the unfolded protein response marker, GRP78. On the contrary, the CA IX MUT4, bearing mutations in the intracytoplasmic tail containing the NLS, is competent for the transit through the secretory pathway and partially reaches the plasma membrane, but it is incompetent for membrane stabilization, showing rapid internalization and characteristic vesicular localization, particularly in endosomes labelled by Rab5 (early) and Rab11 (late recycling). This indicates that the integrity of the intracytoplasmic tail of CA IX is important for the endocytic pathway and for the binding with known and unknown intracellular interactors at the vesicle stage.
Regarding nuclear localization, both mutants showed impairments in trafficking to the nucleus, even in the presence of the nuclear export inhibitor leptomycin B. Although it has been described that hypoxia increases the nuclear mobilization of CA IX [11,12], the hypoxic stimulation caused nuclear translocation of neither of the two mutants, whereas the wild-type CA IX correctly increased its representation in both plasma membrane and in the nucleus. Finally, the blockade of Golgi-ER trafficking inflicted by brefeldin A treatments impaired the nuclear translocation of the wild-type CA IX and caused a subcellular localization resembling the one of MUT4 in normal conditions, where evident perinuclear accumulation was present. Overlapping the data from the intracellular trafficking of the mutants and their lack of interaction with the nucleocytoplasmic machinery suggests that the intracytoplasmic sequence of CA IX contains signals fundamental for its intracellular trafficking and nuclear translocation and that these two processes are closely related, even though the exact mechanism is still unknown.
Increasing evidence suggests that proteins traditionally described as transmembrane, as members of the RTK family, can translocate to the nucleus through retrograde Golgi-ER pathway, indicating that CA IX does not stand alone as a membrane protein with unusual nuclear trafficking and that this localization might follow a similar route. Indeed, a reasonable interpretation of these results is that CA IX requires to transit within the secretory and endocytic pathways in order to localize to the nucleus. This is supported by our model in which mutant CA IX proteins that are stuck into ER (MUT1) and endocytic subcellular compartments (MUT4) were unable to reach the nuclear compartment.
4. Conclusion
In this paper, we have characterized CA IX subcellular trafficking through inactivation of its binding with intracellular ligands by site-directed mutagenesis. The analyzed mutants, MUT1 and MUT4, were shown to be unable to stably reside on the plasma membrane and to reach the nucleus. Their nuclear translocation was not observed even with experiments aimed at stressing this condition, namely the blockade of nuclear export, retrograde trafficking and hypoxia. In addition, the blockade of ER-Golgi trafficking showed impairment in the nuclear mobilization of the wild-type CA IX.
These results allowed us to propose the route to the nucleus of this transmembrane protein, which probably requires the normal transit through the secretory pathway and the following intracellular recycling and binding with intracellular ligands belonging to nucleocytoplasmic shuttling. Although we provided more detailed evidence for CA IX nuclear translocation, further experiments will be required to characterize in greater detail the molecular mechanisms and the proteins involved in this complex trafficking.
5. Materials and methods
5.1. Cell lines and treatments
HEK-293 and SH-SY5Y cell lines were purchased from ATCC. Cells were cultured in standard conditions using DMEM (Sigma) complemented with 15% FBS, 2% glutamine, and 1% penicillin/streptomycin at 37 °C in 5% CO2 humidified atmosphere. Vectors encoding for CA IX WT and mutant proteins have been previously described (Buanne et al., 2013; Buonanno et al., 2017). Transient transfections of these plasmids were performed on cells at 90% confluence using TransFectin Lipid Reagent (Bio-Rad) and cells were incubated for 48 h at 37 °C in humidified air with 5% CO2, cell medium was renewed after 24 h.
Leptomycin B treatment was performed 24 h post-transfection. Cells were incubated for 5 h with leptomycin B (LMB; Sigma) at a final concentration of 20 ng/mL, or with 70% methanol as negative control.
Hypoxia was performed in a hypoxia incubator chamber (STEMCELL Technologies), blowing 95% N2 and 5% CO2 gas for 6 min and repeating the same treatment after 30 min to expel all the O2 from the chamber. Hypoxia lasted for 6 h. Cells incubated for the same time at 37 °C in humidified air with 5% CO2 were used as normoxia control [5].
Brefeldin A (B5936- Sigma-Aldrich) was used in culturing cells for 6 h with a final concentration of 1μg/mL.
5.2. Cell lysis, purification, western blotting
Cells were rinsed with PBS and harvested for extract preparation as described with a modified lysis buffer from Ref. [29], containing 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% Triton X-100, 0.5 mM EDTA, 0.5 mM EGTA, 10% glycerol, 50 mM NaF, 1 mM Na3VO4, 1 mM DTT in the presence of a cocktail of proteases inhibitors (Sigma Aldrich). To obtain purified proteins, each lysate was challenged with 50 μl of Strep-Tactin resin (IBA) and incubated for 2 h at 4 °C. After being washed with 1 M Tris-HCl pH 7.5, 5 M NaCl buffer, purified proteins were eluted with 100 mM Tris-HCl pH 7.5, 150 mM NaCl, 0,1% Triton X-100, 2 mM D-biotin Lysates were clarified by centrifugation at 12.000×g for 20 min at 4 °C and then quantified with BioRad Protein Assay, based on the Bradford method, according to manufacturer's instructions. Proteins were resolved on 10% SDS-PAGE gels and transferred onto a PVDF membrane (Millipore). Target proteins were detected probing the membrane with the following primary antibodies: anti-CA IX M75 (1:300) monoclonal mouse and anti-GRP78 (1:100) mouse monoclonal (Santa Cruz Biotechnology). Then, the peroxidase-conjugated anti-mouse secondary antibody (1:10,000) (Santa Cruz Biotechnology) was used. Immunoreactive bands were detected by ECL (Amersham) and acquired with Chemi Doc Imaging System (BioRad) or with X-ray films (Fuji Film). Statistical comparisons of the Western blot data were performed by unpaired t-test.
5.3. Immunofluorescence analysis
SH-SY5Y cells were plated on glass slides and fixed for 20 min with 3% paraformaldehyde in PBS after 48 h from transfection. After 2 washes with PBS, cells were permeabilized with 0.3% Triton X-100 in PBS for 4 min and then treated with 1% BSA-PBS for 30 min, to reduce nonspecific signals. Cells were then incubated for 2 h with specific primary antibodies in humidified chamber. In the various experiments were used: anti-CA IX VII/20 (1:80) mouse monoclonal; anti-CA IX H-120 (1:100) (Santa Cruz Biotechnology) rabbit polyclonal; anti-Calreticulin (1:120) rabbit polyclonal; anti-Golgin (1:120) mouse monoclonal. To visualize the targeted proteins by fluorescence, after 3 washes with PBS, cells were incubated in humidified chamber for 1 h with secondary antibodies Alexa-488-conjugated donkey anti-mouse and/or Alexa-546-conjugated donkey anti-rabbit (1:200, Jackson Laboratories). Finally, cell nuclei were marked with Hoechst staining (1:3000) for 20 min and after 2 washes with PBS the treated glasses were assembled onto microscope slides with Glycerol/PBS 1:1. Fluorescence was observed at confocal microscope (Zeiss LM510). Each experiment was performed in triplicate and 30 images for each were analyzed with good reproducibility.
The plasmids driving the expression of EGFP-conjugated Rabs, exploited for co-localization studies with CA IX, were kindly provided by Prof. Simona Paladino (Department of Molecular Medicine and Medical Biotechnology, Federico II University of Naples) and Chiara Zurzolo (Pasteur Institute, Paris).
5.4. Glycosidase shear-sensitivity analysis
Glycosidase shear-sensitivity analysis was conducted on CA IX WT, CA IX MUT1, and CA IX MUT 4 purified proteins using New England BioLabs reagents. Protein purification was performed with Strep-Tactin resin (IBA) as described above. Samples (1 μg) were firstly denatured with Glycoprotein Denaturing Buffer (0.5% SDS, 40 mM DTT) at 100 °C for 10 min. For N-glycosidase analysis, GlycoBuffer 3 (50 mM Sodium Acetate, pH 6.0) and EndoH enzyme were added and incubated for 2 h at 37 °C. For O-glycosidase analysis GlycoBuffer 2 (50 mM Sodium Phosphate, pH 7.5), 1% NP40, Neuraminidase and O-Glycosidase were added to denatured samples and incubated at 37 °C for 4 h. Digested samples were visualized on 11% SDS-PAGE followed by western blotting using anti-CA IX M75 as primary antibody and anti-mouse as secondary antibody.
Author contribution statement
Mariangela Succoio: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Sara Amiranda: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Emanuele Sasso: Analyzed and interpreted the data.
Carmen Marciano, Arianna Finizio: Performed the experiments.
Giuseppina De Simone: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Corrado Garbi: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Nicola Zambrano: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
Professor Nicola Zambrano was supported by Ministero dell’Istruzione, dell’Università e della Ricerca {PRIN 2015KRYSJN_003}.
This work was supported by Regione Campania {CEINGE-SATIN, CEINGE-Contributo ordinario}.
Data availability statement
No data was used for the research described in the article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The Authors thank Prof. Simona Paladino for critical discussion. This work was supported by MiUR-PRIN 2015KRYSJN_003 and by Regione Campania, CEINGE SATIN project and Contributo ordinario, to N.Z.
Footnotes
Nicola Zambrano reports a relationship with Heliyon that includes: board membership. Currently serving as Section Editor for the Journal Heliyon.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e18885.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Supuran C.T. Structure and function of carbonic anhydrases. Biochem. J. 2016;473:2023–2032. doi: 10.1042/BCJ20160115. [DOI] [PubMed] [Google Scholar]
- 2.Ilardi G., et al. Histopathological determinants of tumor resistance: a special look to the immunohistochemical expression of carbonic anhydrase IX in human cancers. Curr. Med. Chem. 2014;21:1569–1582. doi: 10.2174/09298673113209990227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wykoff C.C., et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60:7075–7083. [PubMed] [Google Scholar]
- 4.Kaluz S., Kaluzova M., Liao S.Y., Lerman M., Stanbridge E.J. Transcriptional control of the tumor- and hypoxia-marker carbonic anhydrase 9: a one transcription factor (HIF-1) show? Biochim. Biophys. Acta. 2009;1795:162–172. doi: 10.1016/j.bbcan.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miro C., et al. Thyroid hormone enhances angiogenesis and the warburg effect in squamous cell carcinomas. Cancers. 2021;13 doi: 10.3390/cancers13112743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benej M., et al. CA IX stabilizes intracellular pH to maintain metabolic reprogramming and proliferation in hypoxia. Front. Oncol. 2020;10:1462. doi: 10.3389/fonc.2020.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Opavsky R., et al. Human MN/CA9 gene, a novel member of the carbonic anhydrase family: structure and exon to protein domain relationships. Genomics. 1996;33:480–487. doi: 10.1006/geno.1996.0223. [DOI] [PubMed] [Google Scholar]
- 8.Supuran C.T., et al. Inhibition of carbonic anhydrase IX targets primary tumors, metastases, and cancer stem cells: three for the price of one. Med. Res. Rev. 2018;38:1799–1836. doi: 10.1002/med.21497. [DOI] [PubMed] [Google Scholar]
- 9.Rusciano G., Sasso E., Capaccio A., Zambrano N., Sasso A. Publisher correction: revealing membrane alteration in cells overexpressing CA IX and EGFR by surface-enhanced Raman scattering. Sci Rep. 2019;9:9001. doi: 10.1038/s41598-019-45208-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Fiore A., Supuran C.T., Scaloni A., De Simone G. Post-translational modifications in tumor-associated carbonic anhydrases. Amino Acids. 2022;54:543–558. doi: 10.1007/s00726-021-03063-y. [DOI] [PubMed] [Google Scholar]
- 11.Buanne P., et al. Characterization of carbonic anhydrase IX interactome reveals proteins assisting its nuclear localization in hypoxic cells. J. Proteome Res. 2013;12:282–292. doi: 10.1021/pr300565w. [DOI] [PubMed] [Google Scholar]
- 12.Sasso E., et al. Binding of carbonic anhydrase IX to 45S rDNA genes is prevented by exportin-1 in hypoxic cells. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/674920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christianson H.C., et al. Tumor antigen glycosaminoglycan modification regulates antibody-drug conjugate delivery and cytotoxicity. Oncotarget. 2017;8:66960–66974. doi: 10.18632/oncotarget.16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulikova A., et al. Intact intracellular tail is critical for proper functioning of the tumor-associated, hypoxia-regulated carbonic anhydrase IX. FEBS Lett. 2009;583:3563–3568. doi: 10.1016/j.febslet.2009.10.060. [DOI] [PubMed] [Google Scholar]
- 15.Brand T.M., et al. Nuclear EGFR as a molecular target in cancer. Radiother. Oncol. 2013;108:370–377. doi: 10.1016/j.radonc.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 16.Wang Y.N., Hung M.C. Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci. 2012;2:13. doi: 10.1186/2045-3701-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpenter G., Liao H.J. Receptor tyrosine kinases in the nucleus. Cold Spring Harbor Perspect. Biol. 2013;5:a008979. doi: 10.1101/cshperspect.a008979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y.N., et al. COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochem. Biophys. Res. Commun. 2010;399:498–504. doi: 10.1016/j.bbrc.2010.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryant K.L., Baird B., Holowka D. A novel fluorescence-based biosynthetic trafficking method provides pharmacologic evidence that PI4-kinase IIIalpha is important for protein trafficking from the endoplasmic reticulum to the plasma membrane. BMC Cell Biol. 2015;16:5. doi: 10.1186/s12860-015-0049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lippincott-Schwartz J., Yuan L.C., Bonifacino J.S., Klausner R.D. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misumi Y., et al. Novel blockade by brefeldin A of intracellular transport of secretory proteins in cultured rat hepatocytes. J. Biol. Chem. 1986;261:11398–11403. [PubMed] [Google Scholar]
- 22.Giri D.K., et al. Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol. Cell Biol. 2005;25:11005–11018. doi: 10.1128/MCB.25.24.11005-11018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buonanno M., et al. Disclosing the interaction of carbonic anhydrase IX with cullin-associated NEDD8-dissociated protein 1 by molecular modeling and integrated binding measurements. ACS Chem. Biol. 2017;12:1460–1465. doi: 10.1021/acschembio.7b00055. [DOI] [PubMed] [Google Scholar]
- 24.Swinson D.E., et al. Carbonic anhydrase IX expression, a novel surrogate marker of tumor hypoxia, is associated with a poor prognosis in non-small-cell lung cancer. J. Clin. Oncol. 2003;21:473–482. doi: 10.1200/JCO.2003.11.132. [DOI] [PubMed] [Google Scholar]
- 25.Dungwa J.V., Hunt L.P., Ramani P. Carbonic anhydrase IX up-regulation is associated with adverse clinicopathologic and biologic factors in neuroblastomas. Hum. Pathol. 2012;43:1651–1660. doi: 10.1016/j.humpath.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Hilvo M., et al. Biochemical characterization of CA IX, one of the most active carbonic anhydrase isozymes. J. Biol. Chem. 2008;283:27799–27809. doi: 10.1074/jbc.M800938200. [DOI] [PubMed] [Google Scholar]
- 27.Supuran C.T. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008;7:168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- 28.McDonald P.C., Swayampakula M., Dedhar S. Coordinated regulation of metabolic transporters and migration/invasion by carbonic anhydrase IX. Metabolites. 2018;8 doi: 10.3390/metabo8010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbato C., et al. Interaction of Tau with Fe65 links tau to APP. Neurobiol. Dis. 2005;18:399–408. doi: 10.1016/j.nbd.2004.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.