ABSTRACT
Aspergillus flavus is a mycotoxigenic fungus that contaminates many important agricultural crops with aflatoxin B1, the most toxic and carcinogenic natural compound. This fungus is also the second leading cause of human invasive aspergillosis, after Aspergillus fumigatus, a disease that is particularly prevalent in immunocompromised individuals. Azole drugs are considered the most effective compounds in controlling Aspergillus infections both in clinical and agricultural settings. Emergence of azole resistance in Aspergillus spp. is typically associated with point mutations in cyp51 orthologs that encode lanosterol 14α-demethylase, a component of the ergosterol biosynthesis pathway that is also the target of azoles. We hypothesized that alternative molecular mechanisms are also responsible for acquisition of azole resistance in filamentous fungi. We found that an aflatoxin-producing A. flavus strain adapted to voriconazole exposure at levels above the MIC through whole or segmental aneuploidy of specific chromosomes. We confirm a complete duplication of chromosome 8 in two sequentially isolated clones and a segmental duplication of chromosome 3 in another clone, emphasizing the potential diversity of aneuploidy-mediated resistance mechanisms. The plasticity of aneuploidy-mediated resistance was evidenced by the ability of voriconazole-resistant clones to revert to their original level of azole susceptibility following repeated transfers on drug-free media. This study provides new insights into mechanisms of azole resistance in a filamentous fungus.
IMPORTANCE Fungal pathogens cause human disease and threaten global food security by contaminating crops with toxins (mycotoxins). Aspergillus flavus is an opportunistic mycotoxigenic fungus that causes invasive and noninvasive aspergillosis, diseases with high rates of mortality in immunocompromised individuals. Additionally, this fungus contaminates most major crops with the notorious carcinogen, aflatoxin. Voriconazole is the drug of choice to treat infections caused by Aspergillus spp. Although azole resistance mechanisms have been well characterized in clinical isolates of Aspergillus fumigatus, the molecular basis of azole resistance in A. flavus remains unclear. Whole-genome sequencing of eight voriconazole-resistant isolates revealed that, among other factors, A. flavus adapts to high concentrations of voriconazole by duplication of specific chromosomes (i.e., aneuploidy). Our discovery of aneuploidy-mediated resistance in a filamentous fungus represents a paradigm shift, as this type of resistance was previously thought to occur only in yeasts. This observation provides the first experimental evidence of aneuploidy-mediated azole resistance in the filamentous fungus A. flavus.
KEYWORDS: Aspergillus flavus, aneuploidy, azoles, drug resistance mechanisms
OBSERVATION
Azoles are among the most widely used antifungal drugs for the management of fungal infections in clinical and agricultural environments. Voriconazole (VRC), a triazole antifungal drug, was approved by the FDA in 2002 as a first-line agent for the treatment of invasive Aspergillus infections due to its efficacy and safety (1). Long-term triazole treatment of patients with aspergillosis can lead to the emergence of drug resistance (2, 3). Additionally, exposure of environmental Aspergillus spp., including A. flavus, to azole fungicides in agriculture can induce cross-resistance to medical triazoles in clinical settings (4–7). Molecular mechanisms of azole resistance have been extensively investigated in A. fumigatus and include alterations in the target protein Cyp51A, upregulation of the target gene cyp51A, and overexpression of efflux pump genes (reviewed in references 8 to 10). Few studies have examined A. flavus clinical isolates for point mutations in azole target genes (cyp51A, cyp51B, and cyp51C) and overexpression of efflux pumps that may explain VRC resistance in this fungus (11–15).
Pathogenic yeasts, such as Cryptococcus neoformans and Candida spp., can acquire resistance to high concentrations of fluconazole, one of the most widely used antifungal azole drugs, through the formation of aneuploidy in response to drug pressure (16–23). This type of acquired resistance is unstable, as aneuploid chromosomes are lost in the absence of drug pressure (20, 24). Aneuploidy has not been described in filamentous fungi as a mechanism of genomic plasticity that mediates azole resistance.
In the present study, we observed that strains of A. flavus quickly developed clones resistant to VRC when exposed to levels above the MIC. These clones lost their resistance upon repeated transfer in drug-free media. Whole-genome sequencing of the resistant clones revealed a duplication of chromosome 8 (Chr8) and a segmental duplication of chromosome 3 (Chr3) in some clones. We hypothesize that aneuploidy is an underlying mechanism of drug resistance that arises rapidly in mycotoxigenic fungi in response to azole stress.
Emergence of resistance to voriconazole in the mycotoxigenic A. flavus.
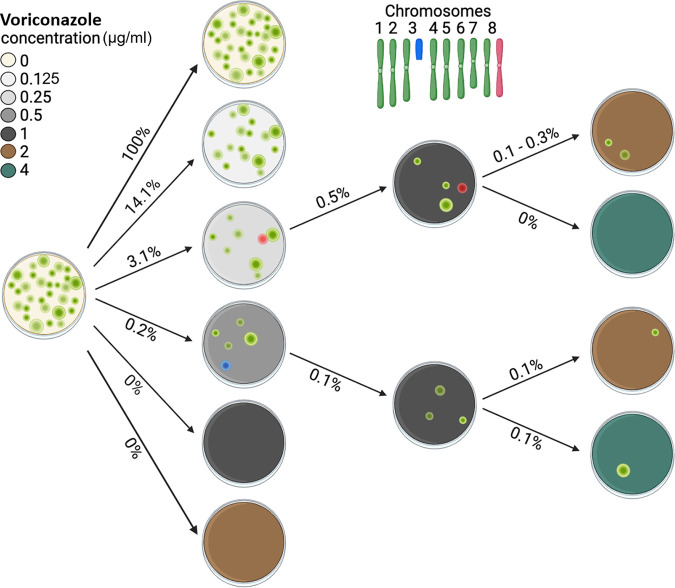
Susceptibility of A. flavus environmental isolates to VRC was measured by Etest strips, and the MIC was confirmed by using the CLSI broth microdilution method (25) (see Table S1 in the supplemental material). After determining that VRC resistance was consistent across a range of isolates (0.125 μg/mL for all tested strains), we chose the highly aflatoxigenic strain of A. flavus (SS1), which was isolated from stored wheat grains, to study the mechanism of resistance. Our preliminary experimentation demonstrated that the VRC resistance levels of A. flavus SS1 wild-type (WT) subpopulations could be increased by exposing them to higher concentrations of the drug in a stepwise manner. We spread 1 × 103 conidia on potato dextrose agar (PDA) plates supplemented with 0.125, 0.25, 0.5, 1, and 2 μg/mL of VRC. After 4 days of incubation on PDA with 0.125 μg/mL VRC at 28°C, approximately 150 CFU were visible, indicating a frequency of resistance of 14%. After 14 days of incubation, 34 colonies grew on PDA plates supplemented with 0.25 μg/mL, and 2 colonies grew on media containing 0.5 μg/mL VRC (Fig. 1). No growth was observed on PDA supplemented with 1 or 2 μg/mL of VRC. However, when the clones grown at 0.25 or 0.5 μg/mL VRC were subcultured on PDA containing 1 μg/mL VRC, resistant colonies emerged at low frequencies (0.1% to 0.5%). Exposure of these subclones to higher VRC concentrations resulted in clones resistant to 2 and 4 μg/mL (Fig. 1).
FIG 1.
Diagram of adaptive resistance in Aspergillus flavus strain SS1 to VRC. Spores (1 × 103) of the A. flavus SS1 strain were inoculated on drug-free PDA plates or plates supplemented with VRC. The resulting colonies showed a high level of variation in VRC resistance at the concentrations of 0.125 to 0.5 μg/mL (percentages indicate the proportion of resistant colonies from each treatment). Serial transfer on media containing increasing concentrations of VRC resulted in the emergence of subclones that were resistant to high concentrations of the drug. Coloration of fungal colonies corresponds to chromosomal states. Chromosomes found in the wild type are colored green. A partial duplication of Chr3 is indicated in blue, and a full duplication of Chr8 is indicated in red. Figure created using BioRender.com.
Aneuploidy confers azole resistance in A. flavus strains.
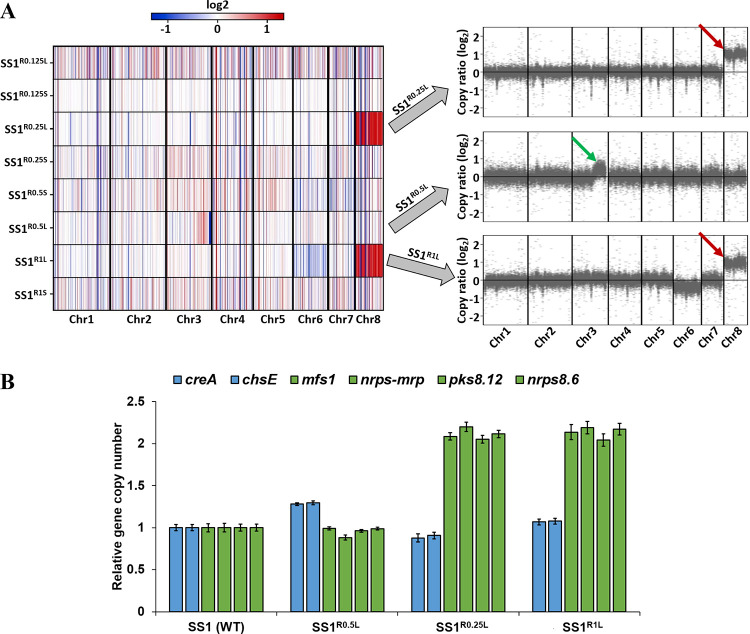
We randomly selected two resistant colonies (R), one large (L) and one small (S), from each concentration of VRC, including 0.125 μg/mL (SS1R0.125), 0.25 μg/mL (SS1R0.25), 0.5 μg/mL (SS1R0.5), and 1 μg/mL (SS1R1, which was derived from SS1R0.25 and SS1R0.5), for whole-genome sequencing (see strain descriptions in Table 1). Genome sequence analysis (see “Materials and Methods” in supplemental material) of these eight resistant clones revealed that three large-colony clones were aneuploid. A clone, resistant to 0.25 μg/mL, and its derived strain, resistant to 1 μg/mL VRC (SS1R0.25L and SS1R1L, respectively), were disomic for Chr8; additionally, a clone that was resistant to 0.5 μg/mL VRC (SS1R0.5L) had a segmental disomy of Chr3 (Fig. 2A). Neither of the clones that were resistant at the MIC level of VRC (0.125 μg/mL) displayed ploidy changes. We further confirmed chromosomal duplications, by quantitative PCR (qPCR) analysis, of copy number variation of four genes located on the left and the right arms of Chr8 (mfs1, nrps-mrp, pks8.12, and nrps8.6) and two more genes located at the partially duplicated segment of Chr3 (creA, chsE). This analysis used the genomic DNA previously prepared for whole-genome sequencing. The copy number of the genes on Chr8 was twice as high as that of genes located on Chr6 (unduplicated control) in the genomes of resistant SS1R0.25L and SS1R1L strains, confirming the duplication of Chr8. The average gene copy number on Chr3 of the SS1R0.5L strain was close to 1.3, confirming partial segmental aneuploidy of Chr3 (Fig. 2B).
TABLE 1.
Strains used in this study
| Strain name | Description |
|---|---|
| SS1 | A. flavus WT isolate |
| SS1R0.125L | Derived from SS1, large colony resistant to 0.125 μg/mL VRC |
| SS1R0.125S | Derived from SS1, small colony resistant to 0.125 μg/mL VRC |
| SS1R0.25L | Derived from SS1, large colony resistant to 0.25 μg/mL VRC |
| SS1R0.25S | Derived from SS1, small colony resistant to 0.25 μg/mL VRC |
| SS1R0.5L | Derived from SS1, large colony resistant to 0.5 μg/mL VRC |
| SS1R0.5S | Derived from SS1, small colony resistant to 0.5 μg/mL VRC |
| SS1R1L | Derived from SS1R0.25L, resistant to 1 μg/mL VRC |
| SS1R1S | Derived from SS1R0.5S, resistant to 1 μg/mL VRC |
| SS1R2-1 | Derived from SS1R1L, resistant to 2 μg/mL VRC |
| SS1R2-2 | Derived from SS1R1L, resistant to 2 μg/mL VRC |
| SS1R2-3 | Derived from SS1R1S, resistant to 2 μg/mL VRC |
| SS1R4 | Derived from SS1R1S, resistant to 4 μg/mL VRC |
| SS1RVT2-1 | Derived from SS1R2-1, transferred six times in drug-free media |
| SS1RVT2-2 | Derived from SS1R2-2, transferred six times in drug-free media |
| SS1RVT2-3 | Derived from SS1R2-3, transferred six times in drug-free media |
| SS1RVT4 | Derived from SS1R4, transferred six times in drug-free media |
FIG 2.
(A) Chromosome duplication events in three (SS1R0.25L, SS1R0.5L, and SS1R1L) of eight VRC-resistant isolates. (A, Left) Heatmap displaying log2-transformed coverage depth from whole-genome sequencing of eight VRC-resistant isolates. Results are normalized to the untreated SS1 wild-type strain. (A, Right) The red and green arrows emphasize the complete duplication of Chr8 in two isolates and partial duplication of Chr3 in one isolate, respectively. (B) Copy numbers of genes on Chr3 and Chr8, determined by qPCR. The copy numbers of two genes on Chr3 (creA, chsE) and four genes on Chr8 (mfs1, nrps-mrp, pks8.12, and nrps8.6) were determined by SYBR green-based qPCR compared to a control gene located on Chr6 (bgt1), using gene-specific primer pairs (see Table S2 in the supplemental material).
Exposure to high concentrations of VRC results in an increase in gene copy number across multiple chromosomes.
We examined gene dosages in resistant clones that emerged on plates containing high concentrations of VRC (pending whole-genome sequencing analysis) by using qPCR. We selected four clones for analysis, each of which was derived from a separate colony growing on media amended with 1 μg/mL VRC. Three of these clones were resistant at 2 μg/mL (denoted SS1R2-1, SS1R2-2, and SS1R2-3), and one was resistant at 4 μg/mL of VRC (denoted SS1R4). We identified variable increases in the copy number of the genes located on different chromosomes (Chr1, Chr2, Chr3, Chr4, Chr5, Chr7, and Chr8) compared to an internal control gene on Chr6 (Fig. S1). Interestingly, serial transfer on drug-free media caused the resistant clones to revert to the parental type. After six serial passages of SS1R2-1, SS1R2-2, SS1R2-3, and SS1R4 on drug-free PDA media, we isolated four clones that reverted (RVT) to their original susceptibility (MIC of 0.125 μg/mL). Gene duplication events were no longer evident in the reverted strains, although one clone (SS1RVT4) retained slightly elevated copy numbers (Fig. S1). We speculate that the duplications still evident in this isolate would resolve with additional passages on drug-free media. These data suggest that transient duplication events of multiple chromosomes occur within individual colonies; nevertheless, we are hesitant to interpret these results, as the status of chromosome copy number must be determined by whole-genome sequencing (as was done for aneuploid clones described above).
Aneuploidy-mediated azole resistance in yeasts, including Candida albicans and C. neoformans, is typically associated with the duplication of chromosomal regions that harbor genes associated with azole resistance (16, 20). The A. flavus ortholog of A. fumigatus transcription factor AtrR, which plays an important role in azole resistance in A. fumigatus (26), resides in a partially duplicated region of Chr3 and is possibly associated with increased VRC resistance in A. flavus. Nevertheless, full duplication of Chr8 in resistant clones does not coincide with any known resistance genes. Furthermore, resistant clones did not have single nucleotide polymorphisms, insertions, or deletions in azole target genes, cyp51A, cyp51B, and cyp51C, or in other genes involved in azole resistance of A. flavus (such as yap1 and efflux pump genes). In conclusion, our observation suggests that aneuploidy evolves readily in A. flavus upon exposure to VRC. Our results provide a foundation for future studies to explore the prevalence and impact of aneuploidy-mediated azole resistance in filamentous fungi.
Data availability.
The genome sequence data are available in the NCBI SRA repository under BioProject accession number PRJNA893458.
Footnotes
Supplemental material is available online only.
Contributor Information
Edward Sionov, Email: edwardsio@volcani.agri.gov.il.
Joshua J. Obar, Geisel School of Medicine at Dartmouth
REFERENCES
- 1.Greer ND. 2003. Voriconazole: the newest triazole antifungal agent. Proc (Bayl Univ Med Cent) 16:241–248. doi: 10.1080/08998280.2003.11927910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meis JF, Chowdhary A, Rhodes JL, Fisher MC, Verweij PE. 2016. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos Trans R Soc B 371:20150460. doi: 10.1098/rstb.2015.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howard SJ, Cerar D, Anderson MJ, Albarrag A, Fisher MC, Pasqualotto AC, Laverdiere M, Arendrup MC, Perlin DS, Denning DW. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg Infect Dis 15:1068–1076. doi: 10.3201/eid1507.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhodes J, Abdolrasouli A, Dunne K, Sewell TR, Zhang Y, Ballard E, Brackin AP, van Rhijn N, Chown H, Tsitsopoulou A, Posso RB, Chotirmall SH, McElvaney NG, Murphy PG, Talento AF, Renwick J, Dyer PS, Szekely A, Bowyer P, Bromley MJ, Johnson EM, Lewis White P, Warris A, Barton RC, Schelenz S, Rogers TR, Armstrong-James D, Fisher MC. 2022. Population genomics confirms acquisition of drug-resistant Aspergillus fumigatus infection by humans from the environment. Nat Microbiol 7:663–674. doi: 10.1038/s41564-022-01091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meireles LM, de Araujo ML, Endringer DC, Fronza M, Scherer R. 2019. Change in the clinical antifungal sensitivity profile of Aspergillus flavus induced by azole and a benzimidazole fungicide exposure. Diagn Microbiol Infect Dis 95:171–178. doi: 10.1016/j.diagmicrobio.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 6.Duong TMN, Nguyen PT, Le TV, Nguyen HLP, Nguyen BNT, Nguyen BPT, Nguyen TA, Chen SC, Barrs VR, Halliday CL, Sorrell TC, Day JN, Beardsley J. 2020. Drug-resistant Aspergillus flavus is highly prevalent in the environment of Vietnam: a new challenge for the management of aspergillosis? J Fungi (Basel) 6:296. doi: 10.3390/jof6040296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermida-Alava K, Brito Devoto T, Sautua F, Gordo M, Scandiani M, Formento N, Luque A, Carmona M, Cuestas ML. 2021. Antifungal susceptibility profile and molecular identification of Cyp51C mutations in clinical and environmental isolates of Aspergillus flavus from Argentina. Mycoses 64:95–101. doi: 10.1111/myc.13193. [DOI] [PubMed] [Google Scholar]
- 8.Rivero-Menendez O, Alastruey-Izquierdo A, Mellado E, Cuenca-Estrella M. 2016. Triazole resistance in Aspergillus spp.: a worldwide problem? J Fungi (Basel) 2:21. doi: 10.3390/jof2030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Cantero A, Lopez-Fernandez L, Guarro J, Capilla J. 2020. Azole resistance mechanisms in Aspergillus: update and recent advances. Int J Antimicrob Agents 55:105807. doi: 10.1016/j.ijantimicag.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Nywening AV, Rybak JM, Rogers PD, Fortwendel JR. 2020. Mechanisms of triazole resistance in Aspergillus fumigatus. Environ Microbiol 22:4934–4952. doi: 10.1111/1462-2920.15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnan-Natesan S, Chandrasekar PH, Alangaden GJ, Manavathu EK. 2008. Molecular characterisation of cyp51A and cyp51B genes coding for P450 14alpha-lanosterol demethylases A (CYP51Ap) and B (CYP51Bp) from voriconazole-resistant laboratory isolates of Aspergillus flavus. Int J Antimicrob Agents 32:519–524. doi: 10.1016/j.ijantimicag.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Liu W, Sun Y, Chen W, Liu W, Wan Z, Bu D, Li R. 2012. The T788G mutation in the cyp51C gene confers voriconazole resistance in Aspergillus flavus causing aspergillosis. Antimicrob Agents Chemother 56:2598–2603. doi: 10.1128/AAC.05477-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul RA, Rudramurthy SM, Meis JF, Mouton JW, Chakrabarti A. 2015. A Novel Y319H substitution in cyp51C associated with azole resistance in Aspergillus flavus. Antimicrob Agents Chemother 59:6615–6619. doi: 10.1128/AAC.00637-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma C, Kumar R, Kumar N, Masih A, Gupta D, Chowdhary A. 2018. Investigation of multiple resistance mechanisms in voriconazole-resistant Aspergillus flavus clinical isolates from a chest hospital surveillance in Delhi, India. Antimicrob Agents Chemother 62:e01928-17. doi: 10.1128/AAC.01928-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi MJ, Won EJ, Joo MY, Park YJ, Kim SH, Shin MG, Shin JH. 2019. Microsatellite typing and resistance mechanism analysis of voriconazole-resistant Aspergillus flavus isolates in South Korean hospitals. Antimicrob Agents Chemother 63:e01610-18. doi: 10.1128/AAC.01610-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selmecki A, Forche A, Berman J. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Yang F, Li D, Zhou M, Wang X, Xu Q, Zhang Y, Yan L, Jiang Y. 2015. Trisomy of chromosome R confers resistance to triazoles in Candida albicans. Med Mycol 53:302–309. doi: 10.1093/mmy/myv002. [DOI] [PubMed] [Google Scholar]
- 18.Anderson MZ, Saha A, Haseeb A, Bennett RJ. 2017. A chromosome 4 trisomy contributes to increased fluconazole resistance in a clinical isolate of Candida albicans. Microbiology (Reading) 163:856–865. doi: 10.1099/mic.0.000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polakova S, Blume C, Zarate JA, Mentel M, Jorck-Ramberg D, Stenderup J, Piskur J. 2009. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci USA 106:2688–2693. doi: 10.1073/pnas.0809793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sionov E, Lee H, Chang YC, Kwon-Chung KJ. 2010. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog 6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone NR, Rhodes J, Fisher MC, Mfinanga S, Kivuyo S, Rugemalila J, Segal ES, Needleman L, Molloy SF, Kwon-Chung J, Harrison TS, Hope W, Berman J, Bicanic T. 2019. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J Clin Invest 129:999–1014. doi: 10.1172/JCI124516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bing J, Hu T, Zheng Q, Munoz JF, Cuomo CA, Huang G. 2020. Experimental evolution identifies adaptive aneuploidy as a mechanism of fluconazole resistance in Candida auris. Antimicrob Agents Chemother 65:e01466-20. doi: 10.1128/AAC.01466-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang F, Gritsenko V, Lu H, Zhen C, Gao L, Berman J, Jiang YY. 2021. Adaptation to fluconazole via aneuploidy enables cross-adaptation to amphotericin B and flucytosine in Cryptococcus neoformans. Microbiol Spectr 9:e00723-21. doi: 10.1128/Spectrum.00723-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F, Todd RT, Selmecki A, Jiang YY, Cao YB, Berman J. 2021. The fitness costs and benefits of trisomy of each Candida albicans chromosome. Genetics 218:iyab056. doi: 10.1093/genetics/iyab056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed, vol 28. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 26.Paul S, Stamnes M, Thomas GH, Liu H, Hagiwara D, Gomi K, Filler SG, Moye-Rowley WS. 2019. AtrR is an essential determinant of azole resistance in Aspergillus fumigatus. mBio 10:e02563-18. doi: 10.1128/mBio.02563-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.04339-22-s0001.pdf, PDF file, 0.5 MB (474.3KB, pdf)
Data Availability Statement
The genome sequence data are available in the NCBI SRA repository under BioProject accession number PRJNA893458.