Abstract
BACKGROUND:
Vascular smooth muscle cells are key players involved in atherosclerosis, the underlying cause of coronary artery disease. They can play either beneficial or detrimental roles in lesion pathogenesis, depending on the nature of their phenotypic changes. An in-depth characterization of their gene regulatory networks can help better understand how their dysfunction may impact disease progression.
METHODS:
We conducted a gene expression network preservation analysis in aortic smooth muscle cells isolated from 151 multiethnic heart transplant donors cultured under quiescent or proliferative conditions.
RESULTS:
We identified 86 groups of coexpressed genes (modules) across the 2 conditions and focused on the 18 modules that are least preserved between the phenotypic conditions. Three of these modules were significantly enriched for genes belonging to proliferation, migration, cell adhesion, and cell differentiation pathways, characteristic of phenotypically modulated proliferative vascular smooth muscle cells. The majority of the modules, however, were enriched for metabolic pathways consisting of both nitrogen-related and glycolysis-related processes. Therefore, we explored correlations between nitrogen metabolism-related genes and coronary artery disease–associated genes and found significant correlations, suggesting the involvement of the nitrogen metabolism pathway in coronary artery disease pathogenesis. We also created gene regulatory networks enriched for genes in glycolysis and predicted key regulatory genes driving glycolysis dysregulation.
CONCLUSIONS:
Our work suggests that dysregulation of vascular smooth muscle cell metabolism participates in phenotypic transitioning, which may contribute to disease progression, and suggests that AMT (aminomethyltransferase) and MPI (mannose phosphate isomerase) may play an important role in regulating nitrogen and glycolysis-related metabolism in smooth muscle cells.
Keywords: atherosclerosis, coronary artery disease, glycolysis, risk factors, United States
Coronary artery disease (CAD) is the leading cause of death in the United States.1 Although mortality due to CAD has decreased by ≈50% in the United States since the 1980s, the remaining disease burden still has a large socioeconomic impact on our society. Though this decrease may be attributed to therapies that modify CAD risk factors, such as lipid-lowering and anti-hypertensive drugs, these therapies do not target the vessel wall where the disease develops. Vascular smooth muscle cells (VSMCs) make up the medial layer of the vessel wall and have been shown to impact every step of atherosclerosis, the underlying cause of CAD.2
VSMCs show remarkable plasticity in response to vascular injury. Quiescent VSMCs can shift to a highly proliferative and migratory phenotype that promotes VSMCs to migrate into the intimal layer of the vessel wall. VSMCs in the intimal layer then produce extracellular matrix components and promote fibrous cap stability, therefore, protecting against plaque rupture. As VSMCs undergo phenotypic switching, they have been shown to lose the expression of traditional VSMC marker genes and dedifferentiate into both atheroprotective (fibroblast-like) and atherogenic (macrophage-like) cell types.2,3 Macrophage-like VSMCs become proinflammatory, releasing cytokines, enhancing the migration of phagocytic cells, and accelerating the rate of cell necrosis and plaque growth. Despite intensive research, the mechanisms driving the structural and functional phenotypic transformations of VSMCs are not fully understood. Uncovering how gene expression in VSMCs is being reprogrammed could lead to the discovery of new treatments.
Systems biology approaches have been used to describe the components of the cardiovascular system.4,5 These approaches postulate that networks of genes, rather than linear pathways, define complex physiological and pathological processes.6,7 Therefore, we used gene coexpression networks to identify modules that represent highly correlated transcript profiles in both quiescent and proliferative VSMCs to study the properties of the network modules. We then used network-based preservation statistics to quantify within-module topology that are preserved between the 2 conditions. By focusing on the modules with the lowest preservation, we were able to identify networks of genes, and therefore, complex physiological and pathological processes, specific to each phenotypic condition. A deeper investigation of these modules highlighted reprogrammed gene expression profiles that occur during, and potentially drive, VSMC phenotypic transformation. Because loci associated with CAD through genome-wide association studies have been reported to regulate VSMC plasticity,8 we then highlighted genes present in CAD genome-wide association studies loci to further demonstrate that the dysregulated pathways may be contributing to phenotypic plasticity and disease progression.
METHODS
A detailed description of the methods and the experimental procedures are provided in the Supplemental Material. The RNA-sequencing data are available at GEO with the accession number GSE193817. Data published in this article can be queried at http://civeleklab.cphg.virginia.edu. Personalized scripts used in this article can be found at https://github.com/civeleklab/Network-Preservation-VSMC. University of Virginia Institutional Review Board determined that the study is exempt from human subject research regulations since the data were collected from cells derived from cadavers.
RESULTS
Gene Expression Modules in Human VSMCs
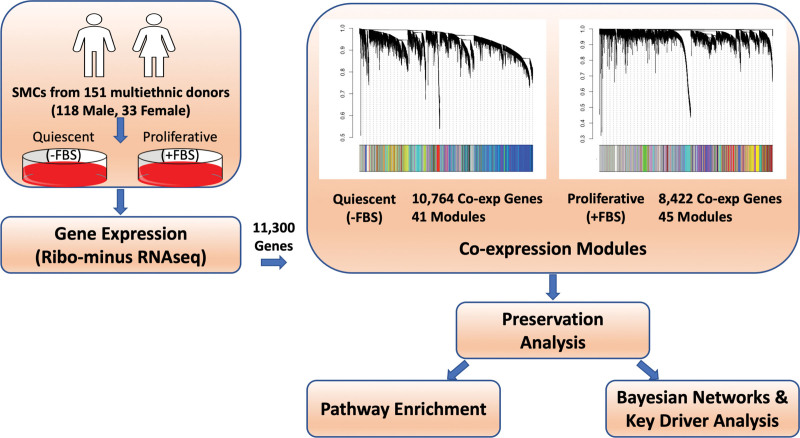
We constructed gene coexpression networks from RNA-sequencing data of aortic smooth muscle cells (SMCs) isolated from 151 heart transplant donors from distinct genetic ancestries cultured in quiescent and proliferative conditions (see Methods). The overall analysis workflow adopted in this work is summarized in Figure 1. After preprocessing and sample outlier detection, 151 samples with gene expression data for 11 330 genes were inputted into the weighted gene coexpression network analysis9 to create gene coexpression modules for both VSMC phenotypic conditions. We performed module detection using iterative weighted gene coexpression network analysis.10 To identify modules of coexpressed genes, we searched for genes with similar patterns of connection strengths to other genes or high topological overlap. A soft-threshold power of 3 and 6 were used for quiescent and proliferative conditions, respectively, to ensure resulting coexpression networks are closer to a scale-free network frequently observed in large-scale biological networks11–13 (Figure S1). An equal soft thresholding power was not used for both conditions as the network connections, and thus the power law distribution that leads to scale-free topology, are unique to each dataset.14 Setting a soft-threshold power of 6 for the quiescent condition results in high reproducibility of coexpression networks, but several modules previously capturing potential biological interactions lose connectivity (Table S1). The quiescent condition resulted in 10 764 coexpressed genes segmented into 41 modules (Q1 - Q41), and the proliferative condition resulted in 8422 coexpressed genes segmented into 45 modules (P1 - P45). The modules ranged in size from 34 to 2134 genes. The contingency table in Figure S2 reports the number of genes that fall into quiescent (rows) and proliferative (columns) modules. This table also shows that some modules possess high gene overlap (preserved) across conditions while others appear to be phenotype-specific (unpreserved). Because coexpression modules can capture genes operating within similar biological pathways and functions, deciphering the modules that are context-specific could lead to understanding genes and pathways operating in phenotype-specific context.
Figure 1.
Schematic representation of the overall study design. Smooth muscle cells (SMCs) from the ascending aortas of 151 multiethnic donors were cultured with and without FBS to mimic the quiescent and proliferative phenotypes of vascular SMCs. Gene expression was measured with RNA-sequencing (RNA-seq). There were 11 300 genes expressed in at least 80% of the samples across both culture conditions. Coexpression modules were created using the expression levels of the 11 300 genes. Network preservation analysis was performed to rank modules based on preservation. To interrogate the modules, pathway enrichment analysis was performed and Bayesian networks were created. Key driver analysis was performed to identify key regulating genes.
Preservation Analysis of VSMC Modules in Distinct Phenotypic States
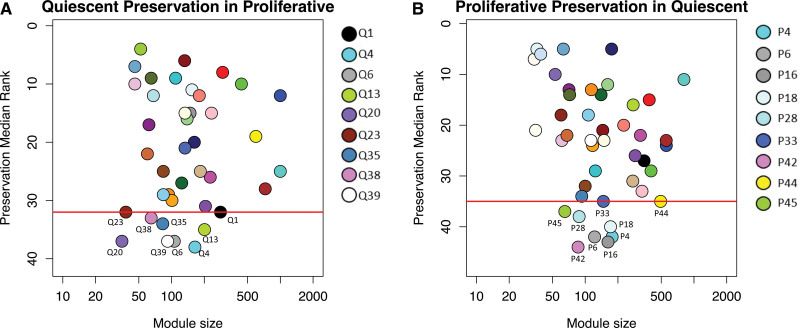
We assessed whether the 41 modules identified in quiescent VSMCs were preserved in the 45 modules identified in proliferative VSMCs. We utilized statistics that do not depend on a gene’s particular module assignment, but rather network properties such as density and connectivity which rely on connection strengths and topology among all genes.15 Using a composite statistic of preservation from the weighted gene coexpression network analysis R package, medianRank, we ranked the preservation of each module across phenotypes. A low medianRank score represented a highly preserved module expected to capture biological pathways that are fundamental to VSMC function. A high medianRank represented a less preserved module expected to identify genes and biological pathways that become rewired and are most likely to be enriched for phenotype-specific functions. Therefore, we identified the modules scoring in the bottom 20th percentile of preservation. This cutoff denoted the 9 least preserved modules in both the quiescent and proliferative conditions, represented by the modules beneath the red line (Figure 2A and 2B). Together these 18 modules contain 2379 unique genes with topological connectivity representing phenotype-specific interactions. Of the 2379 genes, <10% were shared across the conditions (Figure S3). Pathway analysis of the 18 modules revealed pathways representative of differential conditions that were not identified with differential gene expression16 or gene set enrichment analyses17 at both the 0.05 and 0.25 false discovery rate cutoffs. Gene set enrichment analyses and differential gene expression analysis primarily captured pathways that were up or down regulated, such as cell cycle checkpoints, DNA replication, and RNA processing. Pathway analysis of modules scoring in the top 20th percentile of preservation showed enrichment for similar cellular functions (Table S2). This indicates that preservation statistics were able to differentiate between up and downregulated pathways with rewired or dysfunctional biological functions between the quiescent and proliferative conditions.
Figure 2.
Composite preservation statistics for modules between quiescent and proliferative conditions. The composite statistic, medianRank (y axis), as a function of the module size. Each point represents a module, labeled by color and a secondary numeric label. Low numbers on the y axis indicate a high preservation. The red line denotes the bottom 20th percentile of preservation scores. Modules at or below the red line represent the least preserved modules. A, medianRank scores for the preservation of quiescent modules identified in quiescent vascular smooth muscle cells (VSMCs) in proliferative modules identified in proliferative VSMCs. B, medianRank scores for the preservation of proliferative modules in quiescent modules. There are 9 modules at or below the red line cutoff in each condition.
Enrichment Analysis of Unpreserved Modules
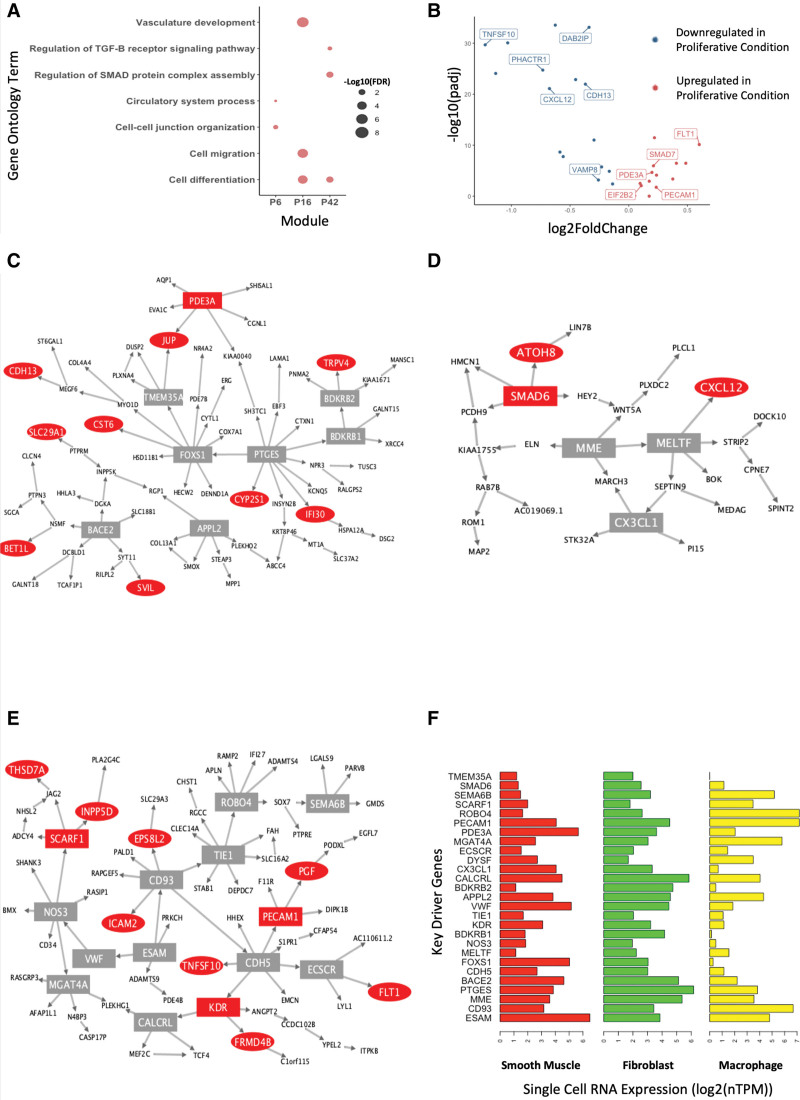
Multiple unpreserved modules captured previously described biological functions that occur during VSMC phenotypic transition from a quiescent to a proliferative state. Gene Ontology enrichment18,19 revealed modules in the proliferative condition to be overrepresented with genes belonging to cell-cell junction organization (P6), vasculature development and migration (P16), and regulation of the Suppressor of Mothers Against Decapentaplegic pathway (P42), all representative of the proliferative phenotype20–22 (Figure 3A). Furthermore, these 3 modules contain 38 CAD candidate genes and 11 CAD prioritized genes (see Methods). Thirty CAD genes are differentially expressed between quiescent and proliferative conditions, including all 11 prioritized genes (Figure 3B). Upregulation of PHACTR1 and CDH13 in the proliferative state is consistent with the direction of their association with CAD risk alleles23,24 (Table S3). Furthermore, the directionality of gene expression of the remaining 8/9 prioritized genes (excluding VAMP8) are consistent with CAD pathogenesis.25–30 For example, upregulation of FLT1 increases cell migration through modulating actin reorganization and downregulation of CXCL12 promotes destabilization of atherosclerotic lesions.31,32 Differential expression of these CAD-associated genes suggests their involvement in the phenotypic plasticity of VSMCs.
Figure 3.
Characterization of modules representative of phenotypic conversion of vascular smooth muscle cells (VSMCs) from a quiescent to a proliferative state. A, Gene Ontology enrichment of proliferative VSMC-related pathways in the P6, P16, and P42 modules. Each point is scaled according to -log10(false discovery rate [FDR]) values. B, Volcano plot displaying differential expression of the 38 coronary artery disease (CAD) candidate genes present in the P6, P16, and P42 modules. The 11 CAD prioritized genes are labeled. Blue points represent genes downregulated and red points represent genes upregulated in proliferative VSMCs compared to quiescent VSMCs. C through E, Bayesian networks created from genes in the (C) P6, (D) P42, and (E) P16 modules. Red nodes represent CAD candidate genes and square nodes represent genes with a key driver score greater or equal to 1. F, Horizontal bar plots of single-cell RNA-sequencing expression data from Protein Atlas of key driver genes in smooth muscle cells, fibroblasts, and macrophages.
We created Bayesian networks (BNs) using the Reconstructing Integrative Molecular BNs algorithm33 from genes within coexpression modules to refine regulatory interactions to predict how CAD genes are being regulated or regulating gene expression (Figure 3C through 3E). We next performed key driver (KD) analysis34 to identify the genes with high regulatory potential. We highlighted genes with a KD score >1 to capture a wide range of genes with regulatory potential based on network topology. These KD genes are expected to have a greater effect in regulating downstream gene expression and the function of biological pathways. In Figure 3C through 3E, genes in CAD genome-wide association studies loci are denoted by red nodes, and genes with a KD score greater than one are denoted by a rectangular node. Gene Ontology Term enrichment analysis of all KD genes across the three modules revealed enrichment for cell differentiation (false discovery rate, 0.031), macrophage activation (false discovery rate, 0.025), and response to TGF (transforming growth factor)-β stimulus (false discovery rate, 0.023), suggesting a role in regulating VSMC dedifferentiation into macrophage-like35,36 and fibroblast-like37 SMCs. We examined RNA single-cell expression profiles of KDs in disease-associated cell types from the Human Protein Atlas38 and found that all KD genes are expressed in dedifferentiated cell types of fibroblasts or macrophages. Furthermore, many of the KDs are also traditional endothelial genes (eg, PECAM1, TIE1, CDH5). Despite still being expressed in VSMCs, the presence of endothelial genes could also represent the dedifferentiation into an SMC-derived intermediate state representative of stem, endothelial, and monocyte cells that eventually transitions into macrophage-like and fibroblast-like phenotypes.39 Together, the pathway analysis and expression profiles suggest that these modules are representative of VSMCs that have dedifferentiated from a quiescent state into a phenotype representative of atherosclerotic behavior (Figure 3F). These analyses show strong evidence that using preservation statistics can capture biologically accurate activity occurring in VSMCs during the transition from a quiescent to a proliferative state.
Metabolic Pathway Enrichment in Unpreserved Modules
Over half of the unpreserved modules representative of phenotype switching were enriched for metabolic pathways. Of these 10 modules, 9 showed enrichment for nitrogen-specific metabolism (Figure 4A). The network preservation analysis strongly supports a novel role of nitrogen metabolism in regulating VSMC plasticity but is further supported by the differential expression of a subset of the canonical cellular nitrogen metabolism pathway genes (Figure S4). Changes in nitrogen metabolism have been demonstrated in endothelial cells to promote endothelial cell phenotypic transition from a quiescent to a proliferative state.40 It is well documented that changes in nitrogen content, specifically nitric oxide, also regulate VSMC proliferation, migration, and calcification,41,42 but it is unclear if changes in nitrogen processes are a byproduct or causal of phenotypic switching in VSMCs. One hundred fourteen of the CAD candidate genes were members of the nitrogen metabolism enriched modules. To further test the potential role of nitrogen metabolism in contributing to phenotypic plasticity, we calculated correlations between expression levels of 8 genes in the Nitrogen Metabolism Kyoto Encyclopedia of Genes and Genomes pathway43,44 expressed in quiescent and proliferative VSMCs and 114 genes in the CAD candidate gene set present in our 9 nitrogen metabolism enriched modules (Figure 4B; Figure S5). Seventy-five percent of the Nitrogen Metabolism Kyoto Encyclopedia of Genes and Genomes pathway genes were significantly correlated (Bonferroni corrected P<5×10-5, Pearson r>|0.3|) with CAD candidate genes in both the quiescent and proliferative conditions. The strongest correlations for both phenotypes were with AMT, which encodes the aminomethyltransferase enzyme. Eleven CAD candidate genes in the quiescent condition and 12 CAD candidate genes in the proliferative condition were moderate to highly correlated with AMT (Pearson r values between |0.5 and 0.75|).45,46 AMT gene expression has previously been shown to be associated with CAD risk.47 AMT was also differentially expressed between quiescent and proliferative conditions, suggesting that gene expression levels of CAD genome-wide association studies genes functioning in nitrogen metabolic enriched modules were reprogrammed due to changes in AMT expression levels, or vice versa. These data suggest that nitrogen metabolism plays a role in the progression of CAD, potentially through regulating VSMC plasticity.
Figure 4.
Presence of nitrogen metabolic processes in the least preserved modules. A, Gene Ontology enrichment of metabolically related pathways in 10 of the least preserved modules. Each point is scaled according to −log10(false discovery rate [FDR]) values. Blue points represent modules from the quiescent condition and red points represent modules from the proliferative condition. B, Heatmap of Pearson correlations (r) between 55 coronary artery disease (CAD) candidate genes in the P4, P18, P28, and P45 modules and 8 genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) Nitrogen Metabolism pathway. Asterisk marks denote genes in the KEGG Nitrogen Metabolism pathway with at least one correlation (r) ≥|0.3| at a Bonferroni corrected P≤5×10-5. C, Expression levels of AMT in quiescent and proliferative conditions. AMT is downregulated in proliferative conditions (P<9.3×10-28).
In addition to modules enriched for nitrogen metabolic processes, another module was enriched for NADH (reduced nicotinamide adenine dinucleotide) regeneration and canonical glycolysis pathways (Q23; Figure 4A). This analysis was also supported by the differential expression of a subset of the canonical glycolysis pathway genes (Figure S6). Glycolysis plays an important role in the proliferation of VSMCs.48,49 Consequently, proliferative VSMCs demonstrate an increase in glycolytic flux so it is unsurprising that we identified a context-specific function of glycolysis.50 Exploring the glycolytic alterations of VSMCs, however, may provide new insights into the genes involved in the quiescent and proliferative functions of glycolysis. To address the potential rewiring of glycolysis, we compared network topology between the unpreserved Module Q23 in the quiescent condition, and a proliferative module, Module P17, that is also enriched for NADH regeneration and canonical glycolysis. Again, using Reconstructing Integrative Molecular BNs, we created BNs for each coexpression module to predict genetic regulatory function and identified KDs to isolate potential genes responsible for driving glycolytic rewiring (Figure 5A and 5B). There were 11 shared genes between the 2 BNs, 7 unique to the quiescent condition, and 11 unique to the proliferative condition. The BNs shared 2 CAD candidate genes, ENO2 and SPAG4, with the addition of RAB20 in the proliferative network. RAB20 expression was downregulated in proliferative VSMCs (P<0.001). Downregulation of RAB20 gene expression has been shown to promote glycolysis and contribute to enhanced cell proliferation and motility.51 KD analysis identified a novel KD gene in the proliferative BN, MPI, which encodes for mannose phosphate isomerase. Gene Ontology Term analysis of the genes present in each BN showed that all enriched pathways present in the quiescent condition were preserved in the proliferative condition. However, in the proliferative condition, there were more genes in each shared pathway and the addition of new pathways. Three of the new Gene Ontology Term pathways in the proliferative BN were represented by the presence of the KD, MPI, suggesting that mannose metabolism could be driving glycolysis rewiring during VSMC transition (Figure 5C through 5E). To validate KD predictions within our networks, we regenerated BNs for Modules Q23 and P17 using bnlearn,52,53 an R package for Bayesian network learning and inference (Figure S7). key driver analysis again identified FAM162A and MPI as KDs in the Q23 and P17 networks and replicated the results of genes downstream of MPI, including ENO2 and RAB20.
Figure 5.
Rewiring of glycolysis metabolic pathway in vascular smooth muscle cell (VSMC) phenotypic transition. Bayesian networks (BNs) of genes in the (A) Q23 module and (B) P17 module. Red nodes represent coronary artery disease (CAD) candidate genes and yellow diamonds represent the highest-scoring key driver gene. Bold node outlines in (B) represent genes unique to the P17 BNs. C through E, Hypergraph representations of enriched Gene Ontology (GO) terms (false discovery rate [FDR] ≤0.05) based on genes present in the (C) Q23 and (E) P17 BNs. Each node represents a GO term (D). Nodes are scaled according to the number of genes functioning in the GO Term. Edges represent the gene or genes present in the enriched GO term (node).
DISCUSSION
VSMCs are a major cell type present at all stages of an atherosclerotic plaque. Lineage-tracing studies have highlighted that VSMCs can adopt alternative phenotypes that positively and negatively contribute to disease progression.35,54,55 VSMCs dedifferentiating away from a quiescent state due to vascular injury have the potential to stabilize the fibrous cap (fibroblast-like) as well as contribute to the advancement of the necrotic core (macrophage-like).56 Investigating the genetic architecture of quiescent VSMCs compared with proliferative VSMCs could identify biological pathways being rewired during phenotypic transformation, leading to mechanistic predictions driving VSMC plasticity in atherosclerosis, the underlying cause of CAD.
Quantification of VSMC phenotypes in cell culture relevant to atherosclerosis has inevitable limitations. For example, the culture conditions lack the key interactions with other cell types and environmental conditions in the vessel wall and atherosclerotic plaque. In addition, atherosclerosis takes decades to develop; therefore, it cannot be adequately replicated in vitro. Despite these challenges, cultured human coronary artery SMCs have been successfully used in previous studies to investigate genetic determinants of CAD.57,58 Due to the difficult nature of capturing vascular wall phenotypes in cellular detail in the arteries of humans, our approach provides a reasonable proxy for in vivo characteristics of VSMCs. We cultured VSMCs isolated from an ancestrally diverse population of 151 heart transplant donors in 2 conditions that are believed to represent the quiescent and proliferative state of the cells. Differential gene expression analysis confirmed the gene expression profiles are consistent with the phenotypic state of the cells.59 To our knowledge, this work is the first to propose a comprehensive approach exploring gene coexpression networks observed in proliferative VSMCs that are not preserved in quiescent VSMCs, and vice versa. Because coexpression networks are representative of functionally related genes, a network preservation approach was able to capture dysregulated pathways whose gene-gene interactions were rewired as a result of phenotypic transition of VSMCs. Our findings, however, must be considered under the current limitations of employing system genetics and network analyses. There are many decisions made during the model-building phase that can affect the results and conclusions. This strategy may not fully detect the dysregulated pathways underlying VSMC phenotypic transitioning.
Preservation analysis identified the least preserved gene coexpression modules between quiescent and proliferative VSMCs. Three of these modules were significantly enriched for biological pathways representative of VSMCs in atherosclerotic lesions, such as proliferation, migration, cell differentiation,3 cell-cell junction assembly,20 and Suppressor of Mothers Against Decapentaplegic regulation.22 Capturing physiologically relevant in vivo biology assured that our in vitro experimental design was able to capture aspects of VSMC plasticity. Furthermore, over half of the unpreserved modules were enriched for metabolic function. Emerging evidence has shown that the metabolism of VSMCs is correlated with the phenotype switching and the progression of atherosclerosis, among other vascular diseases.60 Unpreserved modules enriched for metabolic functions were present in our quiescent and proliferative conditions representing genetic rewiring of metabolic pathways contributing to a phenotype-specific role of metabolism. Thus, our results support the claims that metabolism of VSMCs are correlated with phenotype switching.
Although nitrogen and glycolytic metabolism in VSMCs are not fully understood, previous reports have identified their potential role in VSMCs and atherosclerosis.60 We are the first to hypothesize mannose metabolism as a possible mechanism contributing to proliferative VSMCs. Mannose is not a significant energy source in humans but it is required for protein glycosylation.61 Mannose treatment was shown to attenuate weight gain, improve glucose and lipid homeostasis, and reduce gene expression of inflammatory markers in adipocytes of high-fat diet mice.62 In addition, plasma levels of mannose have recently been shown to be a biomarker of CAD and a more vulnerable plaque phenotype.63 It is not clear whether mannose is related to CAD because it of its role in regulating insulin resistance or because of an intrinsic biological property.64 As VSMCs respond to vascular injury and transition to a more proliferative, disease-like phenotype, mannose metabolism may be mediating changes in metabolism due to imbalances in energy uptake, thus contributing to disease development through a discrete biological mechanism.
This study demonstrates the power of network preservation statistics to identify differences between 2 biological states. We provide new evidence supporting the role of nitrogen metabolism as a potential regulator of VSMC plasticity. Further studies need to be conducted to discern whether dysregulated metabolism in VSMCs is a byproduct or a driving mechanism of phenotypic plasticity. Specifically, considering the role of AMT (aminomethyltransferase) in regulating nitrogen metabolism and MPI (mannose phosphate isomerase) in regulating mannose and glycolysis metabolism in VSMCs.
ARTICLE INFORMATION
Sources of Funding
This work was supported by National Institutes of Health Grants T32 HL007284, F31 HL165772 to R.N. Perry and R01 HL166428 to Dr Civelek, an American Heart Association Postdoctoral Fellowship 18POST33990046 to Dr Aherrahrou, Transformational Project Award 19TPA34910021 to Dr Civelek, and International Network of Excellence Award (22CVD04) from Foundation Leducq to Dr Civelek.
Disclosures
None.
Supplemental Material
Supplemental Methods
Tables S1–S3
Figures S1–S7
Supplementary Material
Nonstandard Abbreviations and Acronyms
- BN
- Bayesian network
- CAD
- coronary artery disease
- KD
- key driver
- SMC
- smooth muscle cell
- VSMC
- vascular smooth muscle cell
For Sources of Funding and Disclosures, see page 379.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCGEN.122.003781.
Contributor Information
R. Noah Perry, Email: rnp4zh@virginia.edu.
Diana Albarracin, Email: da5rb@virginia.edu.
Redouane Aherrahrou, Email: ra2qy@virginia.edu.
REFERENCES
- 1.Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, et al. ; American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics-2018 update: a report from the American Heart Association. Circulation. 2018;137:e67–e492. doi: 10.1161/CIR.0000000000000558 [DOI] [PubMed] [Google Scholar]
- 2.Basatemur GL, Jørgensen HF, Clarke MCH, Bennett MR, Mallat Z. Vascular smooth muscle cells in atherosclerosis. Nat Rev Cardiol. 2019;16:727–744. doi: 10.1038/s41569-019-0227-9 [DOI] [PubMed] [Google Scholar]
- 3.Bennett MR, Sinha S, Owens GK. Vascular smooth muscle cells in atherosclerosis. Circ Res. 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lusis AJ, Weiss JN. Cardiovascular networks. Circulation. 2010;121:157–170. doi: 10.1161/CIRCULATIONAHA.108.847699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Codoni V, Blum Y, Civelek M, Proust C, Franzén O, Björkegren JLM, Le Goff W, Cambien F, Lusis AJ, Trégouët D-A. Preservation analysis of macrophage gene coexpression between human and mouse identifies PARK2 as a genetically controlled master regulator of oxidative phosphorylation in humans. G3 (Bethesda). 2016;6:3361–3371. doi: 10.1534/g3.116.033894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Dam S, Craig T, de Magalhães JP. GeneFriends: a human RNA-seq-based gene and transcript co-expression database. Nucleic Acids Res. 2015;43:D1124–D1132. doi: 10.1093/nar/gku1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter AE, Sabatini DM. Systematic genome-wide screens of gene function. Nat Rev Genet. 2004;5:11–22. doi: 10.1038/nrg1248 [DOI] [PubMed] [Google Scholar]
- 8.Aherrahrou R, Lue D, Perry RN, Aberra YT, Khan MD, Soh JY, Örd T, Singha P, Yang Q, Gilani H, et al. Genetic regulation of SMC gene expression and splicing predict causal CAD genes. Circ Res. 2023;132:323–338. doi: 10.1161/CIRCRESAHA.122.321586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenfest-Allen E, Cartailler J-P, Magnuson MA, Stoeckert CJ. iterativeWGCNA: iterative refinement to improve module detection from WGCNA co-expression networks. bioRxiv. 2017;234062. doi:10.1101/234062 [Google Scholar]
- 11.Barabási A-L. Scale-free networks: a decade and beyond. Science. 2009;325:412–413. doi: 10.1126/science.1173299 [DOI] [PubMed] [Google Scholar]
- 12.Albert R, Barabasi AL. Topology of evolving networks: local events and universality. Phys Rev Lett. 2000;85:5234–5237. doi: 10.1103/PhysRevLett.85.5234 [DOI] [PubMed] [Google Scholar]
- 13.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509 [DOI] [PubMed] [Google Scholar]
- 14.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
- 15.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carbon S, Douglass E, Good BM, Unni DR, Harris NL, Mungall CJ, Basu S, Chisholm RL, Dodson RJ, Hartline E, et al. ; The Gene Ontology Consortium. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sazonova OV, Lee KL, Isenberg BC, Rich CB, Nugent MA, Wong JY. Cell-cell interactions mediate the response of vascular smooth muscle cells to substrate stiffness. Biophys J. 2011;101:622–630. doi: 10.1016/j.bpj.2011.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doran AC, Meller N, McNamara CA. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:812–819. doi: 10.1161/ATVBAHA.107.159327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Low EL, Baker AH, Bradshaw AC. TGFβ, smooth muscle cells and coronary artery disease: a review. Cell Signal. 2019;53:90–101. doi: 10.1016/j.cellsig.2018.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakareko K, Rydzewska-Rosołowska A, Zbroch E, Hryszko T. TRAIL and cardiovascular disease—a risk factor or risk marker: a systematic review. J Clin Med. 2021;10:1252. doi: 10.3390/jcm10061252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Zhou HJ, Ji W, Min W. AIP1-mediated stress signaling in atherosclerosis and arteriosclerosis. Curr Atheroscler Rep. 2015;17:503. doi: 10.1007/s11883-015-0503-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasikara C, Schilperoort M, Gerlach B, Xue C, Wang X, Zheng Z, Kuriakose G, Dorweiler B, Zhang H, Fredman G, et al. Deficiency of macrophage PHACTR1 impairs efferocytosis and promotes atherosclerotic plaque necrosis. J Clin Invest. 2021;131:e145275. doi: 10.1172/JCI145275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toma I, McCaffrey TA. Transforming growth factor-β and atherosclerosis: interwoven atherogenic and atheroprotective aspects. Cell Tissue Res. 2012;347:155–175. doi: 10.1007/s00441-011-1189-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ercu M, Markó L, Schächterle C, Tsvetkov D, Cui Y, Maghsodi S, Bartolomaeus TUP, Maass PG, Zühlke K, Gregersen N, et al. Phosphodiesterase 3A and arterial hypertension. Circulation. 2020;142:133–149. doi: 10.1161/CIRCULATIONAHA.119.043061 [DOI] [PubMed] [Google Scholar]
- 30.Harry BL, Sanders JM, Feaver RE, Lansey M, Deem TL, Zarbock A, Bruce AC, Pryor AW, Gelfand BD, Blackman BR, et al. Endothelial cell PECAM-1 promotes atherosclerotic lesions in areas of disturbed flow in ApoE-deficient mice. Arterioscler Thromb Vasc Biol. 2008;28:2003–2008. doi: 10.1161/ATVBAHA.108.164707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanno S, Oda N, Abe M, Terai Y, Ito M, Shitara K, Tabayashi K, Shibuya M, Sato Y. Roles of two VEGF receptors, Flt-1 and KDR, in the signal transduction of VEGF effects in human vascular endothelial cells. Oncogene. 2000;19:2138–2146. doi: 10.1038/sj.onc.1203533 [DOI] [PubMed] [Google Scholar]
- 32.Akhtar S, Gremse F, Kiessling F, Weber C, Schober A. CXCL12 promotes the stabilization of atherosclerotic lesions mediated by smooth muscle progenitor cells in Apoe-deficient mice. Arterioscler Thromb Vasc Biol. 2013;33:679–686. doi: 10.1161/ATVBAHA.112.301162 [DOI] [PubMed] [Google Scholar]
- 33.Zhu J, Zhang B, Smith EN, Drees B, Brem RB, Kruglyak L, Bumgarner RE, Schadt EE. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40:854–861. doi: 10.1038/ng.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B, Zhu J. Identification of key causal regulators in gene networks. Proceedings of the World Congress on Engineering. 2013;Vol. 2. [Google Scholar]
- 35.Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M, Feil R. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ Res. 2014;115:662–667. doi: 10.1161/CIRCRESAHA.115.304634 [DOI] [PubMed] [Google Scholar]
- 36.Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AAC, Greene ES, Straub AC, et al. KLF4 dependent phenotypic modulation of SMCs plays a key role in atherosclerotic plaque pathogenesis. Nat Med. 2015;21:628–637. doi: 10.1038/nm.3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK, et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019;25:1280–1289. doi: 10.1038/s41591-019-0512-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karlsson M, Zhang C, Méar L, Zhong W, Digre A, Katona B, Sjöstedt E, Butler L, Odeberg J, Dusart P, et al. A single–cell type transcriptomics map of human tissues. Sci Adv. 2021;7:eabh2169. doi: 10.1126/sciadv.abh2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan H, Xue C, Auerbach BJ, Fan J, Bashore AC, Cui J, Yang DY, Trignano SB, Liu W, Shi J, et al. Single-cell genomics reveals a novel cell state during smooth muscle cell phenotypic switching and potential therapeutic targets for atherosclerosis in mouse and human. Circulation. 2020;142:2060–2075. doi: 10.1161/CIRCULATIONAHA.120.048378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eelen G, de Zeeuw P, Simons M, Carmeliet P. Endothelial cell metabolism in normal and diseased vasculature. Circ Res. 2015;116:1231–1244. doi: 10.1161/CIRCRESAHA.116.302855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dubey RK, Jackson EK, Lüscher TF. Nitric oxide inhibits angiotensin II-induced migration of rat aortic smooth muscle cell. Role of cyclic-nucleotides and angiotensin1 receptors. J Clin Invest. 1995;96:141–149. doi: 10.1172/JCI118014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanno Y, Into T, Lowenstein CJ, Matsushita K. Nitric oxide regulates vascular calcification by interfering with TGF-signalling. Cardiovasc Res. 2008;77:221–230. doi: 10.1093/cvr/cvm049 [DOI] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou J, Ye X, Feng W, Zhang Q, Han Y, Liu Y, Li Y, Wei Y. Distance correlation application to gene co-expression network analysis. BMC Bioinf. 2022;23:81. doi: 10.1186/s12859-022-04609-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sotos AEC, Vanhoof S, Noortgate WVD, Onghena P. The transitivity misconception of pearson’s correlation coefficient. STATISTICS EDUCATION RESEARCH JOURNAL. 2009;8:33–55. doi: 10.52041/serj.v8i2.394 [Google Scholar]
- 47.LeBlanc M, Zuber V, Andreassen BK, Witoelar A, Zeng L, Bettella F, Wang Y, McEvoy LK, Thompson WK, Schork AJ, et al. ; CARDIoGRAM Consortium. Identifying novel gene variants in coronary artery disease and shared genes with several cardiovascular risk factors. Circ Res. 2016;118:83–94. doi: 10.1161/CIRCRESAHA.115.306629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall JL, Chatham JC, Eldar-Finkelman H, Gibbons GH. Upregulation of glucose metabolism during intimal lesion formation is coupled to the inhibition of vascular smooth muscle cell apoptosis. Role of GSK3beta. Diabetes. 2001;50:1171–1179. doi: 10.2337/diabetes.50.5.1171 [DOI] [PubMed] [Google Scholar]
- 49.Vesely ED, Heilig CW, Brosius FC. GLUT1-induced cFLIP expression promotes proliferation and prevents apoptosis in vascular smooth muscle cells. Am J Physiol Cell Physiol. 2009;297:C759–C765. doi: 10.1152/ajpcell.00213.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salabei JK, Hill BG. Mitochondrial fission induced by platelet-derived growth factor regulates vascular smooth muscle cell bioenergetics and cell proliferation. Redox Biol. 2013;1:542–551. doi: 10.1016/j.redox.2013.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu BHM, Tey SK, Mao X, Ma APY, Yeung CLS, Wong SWK, Ng TH, Xu Y, Yao Y, Fung EYM, et al. TPI1-reduced extracellular vesicles mediated by Rab20 downregulation promotes aerobic glycolysis to drive hepatocarcinogenesis. J Extracell Vesicles. 2021;10:e12135. doi: 10.1002/jev2.12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scutari M. Learning Bayesian networks with the bnlearn R package. J Stat Softw. 2010;35:1–22. doi: 10.18637/jss.v035.i0321603108 [Google Scholar]
- 53.Nagarajan R, Scutari M, Lèbre S. Bayesian Networks in R: With Applications in Systems Biology. Springer; 2013. [Google Scholar]
- 54.Hoofnagle MH, Thomas JA, Wamhoff BR, Owens GK. Origin of neointimal smooth muscle: we’ve come full circle. Arterioscler Thromb Vasc Biol. 2006;26:2579–2581. doi: 10.1161/01.ATV.0000249623.79871.bc [DOI] [PubMed] [Google Scholar]
- 55.Bentzon JF, Majesky MW. Lineage tracking of origin and fate of smooth muscle cells in atherosclerosis. Cardiovasc Res. 2018;114:492–500. doi: 10.1093/cvr/cvx251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sorokin V, Vickneson K, Kofidis T, Woo CC, Lin XY, Foo R, Shanahan CM. Role of vascular smooth muscle cell plasticity and interactions in vessel wall inflammation. Front Immunol. 2020;11:599415. doi: 10.3389/fimmu.2020.599415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu B, Pjanic M, Wang T, Nguyen T, Gloudemans M, Rao A, Castano VG, Nurnberg S, Rader DJ, Elwyn S, et al. Genetic regulatory mechanisms of smooth muscle cells map to coronary artery disease risk loci. Am J Hum Genet. 2018;103:377–388. doi: 10.1016/j.ajhg.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller CL, Pjanic M, Wang T, Nguyen T, Cohain A, Lee JD, Perisic L, Hedin U, Kundu RK, Majmudar D, et al. Integrative functional genomics identifies regulatory mechanisms at coronary artery disease loci. Nat Commun. 2016;7:12092. doi: 10.1038/ncomms12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aherrahrou R, Guo L, Nagraj VP, Aguhob A, Hinkle J, Chen L, Soh JY, Lue D, Alencar GF, Boltjes A, et al. Genetic regulation of atherosclerosis-relevant phenotypes in human vascular smooth muscle cells. Circ Res. 2020;127:1552–1565. doi: 10.1161/CIRCRESAHA.120.317415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi J, Yang Y, Cheng A, Xu G, He F. Metabolism of vascular smooth muscle cells in vascular diseases. Am J Physiol Heart Circ Physiol. 2020;319:H613–H631. doi: 10.1152/ajpheart.00220.2020 [DOI] [PubMed] [Google Scholar]
- 61.Stanley P, Taniguchi N, Aebi M. N-Glycans. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH, eds. Essentials of Glycobiology. 3rd ed. Cold Spring Harbor Laboratory Press; 2015. [PubMed] [Google Scholar]
- 62.Sharma V, Smolin J, Nayak J, Ayala JE, Scott DA, Peterson SN, Freeze HH. Mannose alters gut microbiome, prevents diet-induced obesity, and improves host metabolism. Cell Rep. 2018;24:3087–3098. doi: 10.1016/j.celrep.2018.08.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferrannini E, Marx N, Andreini D, Campi B, Saba A, Gorini M, Ferrannini G, Milzi A, Magnoni M, Maseri A, et al. ; CAPIRE investigators. Mannose as a biomarker of coronary artery disease: angiographic evidence and clinical significance. Int J Cardiol. 2022;346:86–92. doi: 10.1016/j.ijcard.2021.11.038 [DOI] [PubMed] [Google Scholar]
- 64.Mardinoglu A, Stančáková A, Lotta LA, Kuusisto J, Boren J, Blüher M, Wareham NJ, Ferrannini E, Groop PH, Laakso M, et al. Plasma mannose levels are associated with incident type 2 diabetes and cardiovascular disease. Cell Metab. 2017;26:281–283. doi: 10.1016/j.cmet.2017.07.006 [DOI] [PubMed] [Google Scholar]
- 65.Zimmermann O, Zwaka TP, Marx N, Torzewski M, Bucher A, Guilliard P, Hannekum A, Hombach V, Torzewski J. Serum starvation and growth factor receptor expression in vascular smooth muscle cells. J Vasc Res. 2006;43:157–165. doi: 10.1159/000090945 [DOI] [PubMed] [Google Scholar]
- 66.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200 [Google Scholar]
- 67.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leung A, Stapleton K, Natarajan R. Functional long non-coding RNAs in vascular smooth muscle cells. Curr Top Microbiol Immunol. 2016;394:127–141. doi: 10.1007/82_2015_441 [DOI] [PubMed] [Google Scholar]
- 69.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller JA, Cai C, Langfelder P, Geschwind DH, Kurian SM, Salomon DR, Horvath S. Strategies for aggregating gene expression data: the collapseRows R function. BMC Bioinf. 2011;12:322. doi: 10.1186/1471-2105-12-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uffelmann E, Huang QQ, Munung NS, de Vries J, Okada Y, Martin AR, Martin HC, Lappalainen T, Posthuma D. Genome-wide association studies. Nat Rev Methods Primers. 2021;1:1–21. doi: 10.1038/s43586-021-00056-9 [Google Scholar]
- 72.Koyama S, Ito K, Terao C, Akiyama M, Horikoshi M, Momozawa Y, Matsunaga H, Ieki H, Ozaki K, Onouchi Y, et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat Genet. 2020;52:1169–1177. doi: 10.1038/s41588-020-0705-3 [DOI] [PubMed] [Google Scholar]
- 73.Pers TH, Karjalainen JM, Chan Y, Westra H-J, Wood AR, Yang J, Lui JC, Vedantam S, Gustafsson S, Esko T, et al. ; Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. Morgan Kaufmann Publishers; 1988. [Google Scholar]
- 75.Zhu J, Lum PY, Lamb J, GuhaThakurta D, Edwards SW, Thieringer R, Berger JP, Wu MS, Thompson J, Sachs AB, et al. An integrative genomics approach to the reconstruction of gene networks in segregating populations. Cytogenet Genome Res. 2004;105:363–374. doi: 10.1159/000078209 [DOI] [PubMed] [Google Scholar]
- 76.Zhu J, Wiener MC, Zhang C, Fridman A, Minch E, Lum PY, Sachs JR, Schadt EE. Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS Comput Biol. 2007;3:e69. doi: 10.1371/journal.pcbi.0030069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu J, Sova P, Xu Q, Dombek KM, Xu EY, Vu H, Tu Z, Brem RB, Bumgarner RE, Schadt EE. Stitching together multiple data dimensions reveals interacting metabolomic and transcriptomic networks that modulate cell regulation. PLoS Biol. 2012;10:e1001301. doi: 10.1371/journal.pbio.1001301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cohain A, Divaraniya AA, Zhu K, Scarpa JR, Kasarskis A, Zhu J, Chang R, Dudley JT, Schadt EE. Exploring the reproducibility of probabilistic causal molecular network models. Pac Symp Biocomput. 2017;22:120–131. doi: 10.1142/9789813207813_0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Castelo R, Siebes A. Priors on network structures. Biasing the search for Bayesian networks. Int J Approx Reason. 2000;24:39–57. doi: 10.1016/s0888-613x(99)00041-9 [Google Scholar]
- 80.Schwarz G. Estimating the dimension of a model. The Annals of Statistics. 1978;6:461–464. doi: 10.1214/aos/1176344136 [Google Scholar]
- 81.Zhang L, Rodrigues LO, Narain NR, Akmaev VR. bAIcis: a novel Bayesian network structural learning algorithm and its comprehensive performance evaluation against open-source software. J Comput Biol. 2020;27:698–708. doi: 10.1089/cmb.2019.0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]