Abstract
Using an intragenic complementation screen, we have identified a temperature-sensitive TATA-binding protein (TBP) mutant (K151L,K156Y) that is defective for interaction with certain yeast TBP-associated factors (TAFs) at the restrictive temperature. The K151L,K156Y mutant appears to be functional for RNA polymerase I (Pol I) and Pol III transcription, and it is capable of supporting Gal4-activated and Gcn4-activated transcription by Pol II. However, transcription from certain TATA-containing and TATA-less Pol II promoters is reduced at the restrictive temperature. Immunoprecipitation analysis of extracts prepared after culturing cells at the restrictive temperature for 1 h indicates that the K151L,K156Y derivative is severely compromised in its ability to interact with TAF130, TAF90, TAF68/61, and TAF25 while remaining functional for interaction with TAF60 and TAF30. Thus, a TBP mutant that is compromised in its ability to form TFIID can support the response to Gcn4 but is defective for transcription from specific promoters in vivo.
Transcriptional initiation by eukaryotic RNA polymerase II (Pol II) requires general transcription factors (TFIID, -A, -B, -E, -F, and -H) that nucleate on the TATA promoter element to form the preinitiation complex that is necessary for accurate positioning and initiation by Pol II (for a review, see reference 34). This process begins with the sequence-specific binding of TFIID, a multiprotein complex composed of TATA-binding protein (TBP) and approximately 10 RNA Pol II-specific TBP-associated factors (TAFs) (reviewed in reference 5). TAFs are a set of phylogenetically conserved proteins found in humans, flies, and yeast, ranging in molecular mass from 15 to 250 kDa (for a recent review, see reference 49). Recruitment of TBP, presumably in the TFIID complex, has been proposed to be a rate-limiting step in formation of the preinitiation complex (6, 8, 25, 26, 60).
Two distinct mechanisms have been proposed for assembly of the preinitiation complex, depending on the presence or absence of a canonical TATA element (5). Initiation from a TATA-containing promoter is dependent on strong TBP-DNA contacts for recognition and binding of TFIID to the TATA box. However, for TATA-less promoters, sequence-specific recognition of the core promoter by TBP does not occur, suggesting that TAF-DNA interactions play an important role (5). In vitro, TFIID but not TBP is able to recognize and support transcription from TATA-less promoters (62). Moreover, in Drosophila TFIID, specific TAFs have been implicated in making contacts with DNA in sequences overlapping the transcription start site (4, 5, 52). In addition, mutations that hinder the ability of TBP to recognize a TATA box do not affect transcription from TATA-less promoters even though TFIID is required (29). Consistent with this, depletion of certain individual yeast TAFs in vivo causes a reduction in transcription from promoters lacking consensus TATA boxes (31, 32). Thus, it seems that transcription from TATA-less promoters does not depend on strong TBP-DNA interactions but rather depends on TAF-DNA and TAF-TBP interactions.
In addition to their role in TATA-less transcription, TAFs have been implicated in the response to activators. Transcriptional activators function by increasing recruitment and stabilizing the Pol II machinery at promoters (reviewed in references 40, 48, and 53). In vitro, activators can interact with many components of the transcription machinery, including TBP, TAFs, TFIIA, TFIIB, TFIIF, TFIIH, Pol II, and the SAGA complex (12, 15, 17, 19, 21, 28, 39, 47, 61). Initial biochemical studies suggested that TAFs function as coactivators required for activator-dependent recruitment of TBP, since TFIID but not TBP could support high levels of transcription in the presence of an activator. Further, it was hypothesized that TAFs can serve as direct targets for activators, because individual TAFs can interact directly with activator proteins (for reviews, see references 5 and 53), and partial TFIID complexes reconstituted from a subset of TAFs can mediate activator-dependent transcription (7). However, TAFs are not absolutely required for the response to activators in vitro, because activation can occur in reactions lacking TAFs (24, 27, 37, 58, 59).
In vivo depletion studies indicate that certain individual TAFs are not generally required for response to activators (31, 54). Instead, depletion of certain TAFs results in specific effects on transcription, depending on the particular TAF targeted. In the case of depletion of TAF130, a loss of transcription from genes lacking a canonical TATA element (31), from genes encoding small-subunit ribosomal proteins (RPS genes), and from various cyclin genes (43, 55) was observed. The TAF130 dependence of these genes is mediated by the core promoter, not the initiator or the activator binding sites. In mammalian cells, TAF250, the homolog of yeast TAF130, is important for transcription of selected genes, including those involved in cell cycle control (50, 56). Taken together, these data suggest that TAFs play a critical role in gene-specific as well as TATA-less transcription.
Aside from their role in TFIID, certain TAFs are present in the yeast SAGA and mammalian PCAF histone acetyltransferase complexes (16, 38). In vivo depletion of TAF61/68, a component of yeast SAGA, indicates that this TAF is necessary for normal enzymatic activity but is dispensable for interaction with TBP or activation domains. Furthermore, depletion of TAF17, which is also present in SAGA, has broad effects on transcription in yeast cells (1, 30, 32). Thus, it seems that TAFs can serve a variety of roles in the transcription process, depending on the complexes in which they reside.
Here we present the characterization of a TBP mutant that is defective for TFIID formation in vivo. Our studies differ from previous in vivo studies in that we analyzed transcription after TFIID disruption which removes multiple TAFs, not after individual TAF depletion. This approach permits the removal of TAFs from TFIID but not from SAGA or other TAF-containing complexes. We show that TFIID disruption does not prevent the response to acidic activators, but it causes transcriptional defects at certain cell cycle-dependent and TATA-less promoters. These results support previous findings that the TBP-TAF interactions may not be essential for activated transcription in vivo and may instead play a role in gene-specific transcription.
MATERIALS AND METHODS
Yeast strains and DNA constructs.
The parental strain of Saccharomyces cerevisiae was BYΔ2 (9), which contains a deletion of the chromosomal TBP locus covered by a URA3-marked 2.4-kb EcoRI-BamHI genomic fragment of the TBP gene locus containing the Pol III-defective allele in which the codon for phenylalanine at position 155 is replaced by that for serine (F155S). Briefly, the parental strain, which is temperature sensitive (ts), was transformed with a set of TBP mutant libraries generated by regional codon randomization on a TRP1-marked plasmid (11). Strains that could grow at the restrictive temperature were isolated, the F155S allele was shuffled out by plating to 5-fluoro-orotic acid, and the resulting strains were tested for a ts phenotype. This last step is necessary because wild-type copies of TBP present in the library would also complement the F155S allele by providing Pol III function. Five TBP mutants generated in this screen have been previously described (45, 46). The sixth mutant is described here and has two substitutions: lysine at position 151 is replaced with leucine, and lysine at position 156 is replaced with tyrosine (K151L,K156Y). The K151L and K156Y single mutants were generated by site-directed mutagenesis by PCR. PCR products were generated and cloned by using an engineered BamHI site in the TBP-coding sequence. The plasmid shuffle technique was used to introduce the single-substitution TBP molecules into yeast. The reporter construct used for the lacZ assays is YCp86-Sc3801 (44).
Phenotypic analysis.
The merodiploid strain (containing both the F155S and K151L,K156Y derivatives), along with wild-type TBP- and vector (pRS316)-containing strains, were grown in the appropriate media. Cells (10-fold serial dilutions) were spotted onto dropout plates lacking the appropriate amino acids (uracil and tryptophan) and incubated at the indicated temperatures. For the Pol I assay, the plasmid PNOY103 (36) or vector (pRS316) was transformed into the both K151L,K156Y TBP- and wild-type TBP-containing strains and colony purified on 2% galactose-containing dropout medium lacking uracil and tryptophan. Cells were then spotted onto galactose-containing medium and incubated at the indicated temperatures. Growth assays of both wild-type TBP- and K151L,K156Y TBP-containing strains were performed as described above.
Transcriptional analysis.
In most cases RNA analysis was done by quantitative S1 nuclease analysis with approximately 30 to 75 μg of RNA (20). In the temperature shift experiments, cells were heat shocked for 15 min, returned to 30°C for 1 h, and then shifted to 38°C for 1 h. Total RNA prepared by hot-phenol extraction was quantitated by A260. For 3-aminotriazole (AT) induction done at the permissive temperature, strains were grown overnight in synthetic complete medium in the presence or absence of 15 mM AT. Activation competency under the restrictive conditions was investigated by growing the cells overnight in synthetic complete medium, and the restrictive-condition protocol described above was performed. After the 1-h incubation at 38°C, 15 mM AT was added and the cells were incubated for an additional hour at 38°C. The probes used in the RNA analysis are listed in the figure legends. The RNA amounts in each reaction mixture were normalized to the RNA levels obtained from a probe to the intron of the tryptophan tRNA gene (tRNAW).
Immunoprecipitation and immunoblotting.
Strains were grown to log phase (optical density at 600 nm = 0.1 to 0.2) at 30°C and then subjected to a 1-h heat shock as described above. Cells were harvested before and after the heat shock, and whole-cell extracts were prepared by glass bead lysis in 450 mM Tris-acetate (pH 7.8)–150 mM potassium acetate–60% glycerol, 3 mM EDTA (pH 8.0)–3 mM dithiothreitol–1 mM phenylmethylsulfonyl fluoride (33). Immunoprecipitations were performed as described previously (33), except that 240 μg of extract was used in the immunoprecipitation reaction. Antigen-antibody complexes were recovered by centrifugation and washed four times with 1 ml of buffer A containing 125 mM potassium acetate and 1% Nonidet P-40 (Sigma). Samples were boiled in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer, and proteins were separated on either 7.5 or 12% gels and electroblotted to nitrocellulose. Antibody reactions were performed by standard techniques, and blots were developed by using chemiluminescent detection according to the recommendations of the manufacturer (Pierce). TBP antibodies used in both the immunoprecipitations and Western blots were generated in rabbits by Cocalico Biological (Reamstown, Pa.) with purified recombinant TBP. Yeast TAF antibodies used for immunoblot analysis were kindly provided by Michael Green.
RESULTS
Isolation and characterization of a TBP mutant that complements a Pol III defect.
An intragenic complementation screen was used to isolate ts TBP mutants that are functional for Pol III transcription at the restrictive temperature, thereby eliminating any mutants that are structurally compromised under the restrictive conditions. Complementation depends on the ability of two ts TBP mutants, each of which confers a different functional defect, to support cell viability when they are present in the cell at the same time (10). Six ts mutants were isolated from this screen, five of which have been previously characterized and shown to be activation defective at the permissive temperature (45, 46). Here we describe the final TBP mutant, in which the lysine at position 151 is changed to leucine and the lysine at position 156 is changed to tyrosine (K151L,K156Y).
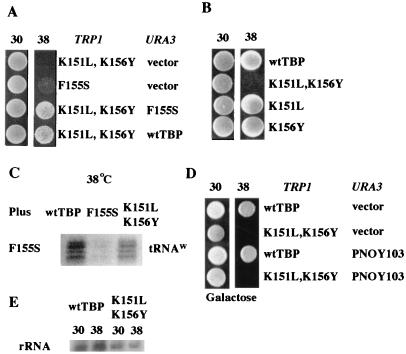
In keeping with the criteria of the screen, the K151L,K156Y derivative is a ts mutant of TBP that is able to complement the growth defect of TBP-F155S, a TBP mutant that is defective for Pol III transcription at the restrictive temperature (Fig. 1A). Substitutions at both lysine 151 and lysine 156 are required to produce the ts phenotype, because the single substitutions support efficient cell growth at the restrictive temperature (Fig. 1B). As expected, the K151L,K156Y derivative can also complement, albeit to lower levels than wild-type TBP, the molecular defect (i.e., the loss in transcription by Pol III) caused by the F155S allele (Fig. 1C).
FIG. 1.
Isolation and characterization of a TBP mutant that complements a TBP mutant with a Pol III defect. (A) Growth conferred by the indicated TBP derivatives and wild-type (wt) TBP at both 30 and 38°C. Cells were grown overnight in liquid medium, and approximately 105 cells were spotted onto the medium lacking the appropriate markers. (B) Growth conferred by TBP derivatives singly substituted at position 151 or 156. (C) S1 nuclease analysis of tRNAW transcription with 30 μg of RNA from the indicated TBP derivative after shifting cultures to 38°C for 1 h. (D) Mutant and wild-type strains containing PNOY103 (see text) were grown overnight in liquid medium (2% galactose), spotted to galactose medium, and incubated at 30 and 38°C. (E) S1 nuclease analysis of an unstable portion of the rRNA transcript with 70 μg of RNA from the indicated strains after incubation at 30 or 38°C for one hour.
To determine if a loss of Pol I transcription was the cause of the ts phenotype conferred by the K151L,K156Y mutant, we utilized a plasmid-based system in which the rRNAs are synthesized from a Pol II promoter (36). Briefly, a construct containing the 35S rRNA gene under the control of a Pol II, galactose-inducible promoter is able to rescue the growth defect (inviability) of a mutant that is lacking the largest subunit of Pol I. If the K151L,K156Y derivative is strictly defective for Pol I transcription, then this Pol II-driven rRNA transcript should rescue the ts phenotype when the cells are cultured in galactose-containing medium. However, the presence of the construct causes no change in viability at the restrictive temperature (Fig. 1D). Transcription of rRNA by Pol I was also examined directly (Fig. 1E). Steady-state levels of an unstable portion of the rRNA transcript were twofold lower under the permissive conditions in the K151L,K156T strain than in the wild type. It should be noted that under these conditions (permissive), the K151L,K156Y strain grows indistinguishably from the wild-type strain. A slight decrease (1.5-fold) in the level of the rRNA transcript was observed in the K151L,K156Y strain after the 1-h temperature shift. Since these changes were relatively minor (especially compared to the more dramatic changes described below), and taken together with the results with the Pol II-driven rRNA construct, this suggests that the ts phenotype is not due strictly to a defect in Pol I transcription.
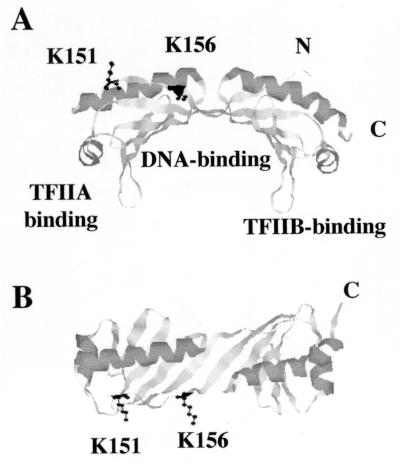
Molecular modeling of lysine 151 and lysine 156 on the TBP-TATA cocrystal structure indicates that both residues are located on the upper surface of TBP and exposed to the solvent (Fig. 2). Consistent with this, gel shift experiments indicate that the K151L,K156Y derivative and wild-type TBP bind the TATA element with comparable affinities (data not shown). In addition, the mutations map to a surface that is distinct from the surfaces that directly interact with TFIIA and TFIIB (13, 35, 51).
FIG. 2.
Molecular modeling of the K151L,K156Y derivative by using the TBP-DNA cocrystal structure (23) (Rasmol 2.6). (A) TBP is shown as a ribbon drawing, with residues K151 and K156 shown in ball-and-stick format. Known binding sites for DNA, TFIIA, and TFIIB and the C and N termini are indicated (13, 35, 51). (B) Top view of TBP (view in panel A rotated 90° forward).
The K151L,K156Y mutant is functional for the response to acidic activators at the permissive temperature.
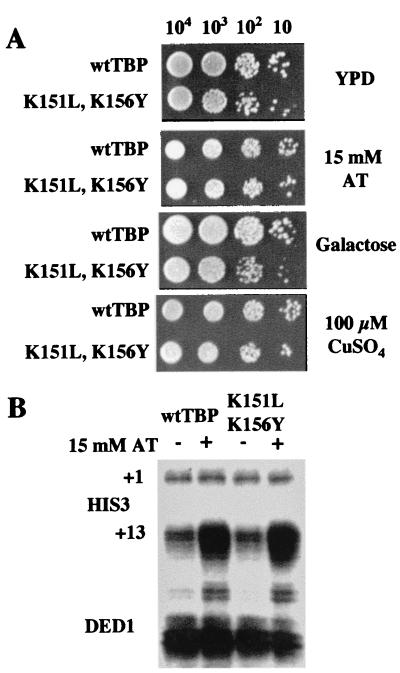
Since the five TBP mutants previously characterized from this complementation screen exhibit activation defects at the permissive temperature (45, 46), we determined whether the K151L,K156Y derivative could respond to acidic activators in vivo. The K151L,K156Y mutant strain grows robustly under conditions that require functional interactions with a number of different acidic activators (Fig. 3A), suggesting that the mutant allele is competent for activated transcription. The response to Gal4, which activates transcription in the presence of galactose, was assayed by using the lacZ reporter YCp86-Sc3801 (44). In the presence of galactose, the wild-type strain produced 430 ± 30 U (mean ± standard deviation) of β-galactosidase activity. Similarly, the K151L,K156Y strain produced 425 ± 20 U of activity. (Activities for culturing in glucose were <1 U for both strains.) Thus, the response to Gal4 conferred by the K151L,K156Y mutant is indistinguishable from the response conferred by wild-type TBP. We also observed that the K151L,K156Y derivative is responsive to the activator Gcn4. Cells grown in the presence of AT, a competitive inhibitor of the HIS3 gene product that results in activation of the HIS3 gene, showed wild-type levels of HIS3 transcription (Fig. 3B). Thus, the K151L,K156Y mutant is functional for activated transcription at the permissive temperature.
FIG. 3.
The K151L,K156Y mutant is functional for activated transcription at the permissive temperature. wt, wild type. (A) Strains were spotted on plates containing the media indicated at the density shown, followed by incubation at 30°C. Yeast extract-peptone-dextrose (YPD) is used as a growth rate indicator on rich media. Growth on 15 mM AT, galactose, and CuSO4 requires functional interactions with the Gcn4, Gal4, and Ace1 transcription factors, respectively. (B) Analysis of Gcn4-dependent activation of HIS3 transcription. Constitutive HIS3 expression initiates equally from both the +1 and +13 start sites, whereas Gcn4-activated transcription is mediated primarily through the +13 initiation site. Strains were grown to log phase in synthetic complete medium in either the presence (+) or absence (−) of 15 mM AT. Total RNA (30 μg) was hybridized with 100-fold excesses of HIS3 and DED1 probes and then subjected to S1 nuclease digestion. The DED1 transcript is not affected by AT and is used as a loading control in this experiment.
The K151L,K156Y mutant is defective for transcription from certain Pol II promoters.
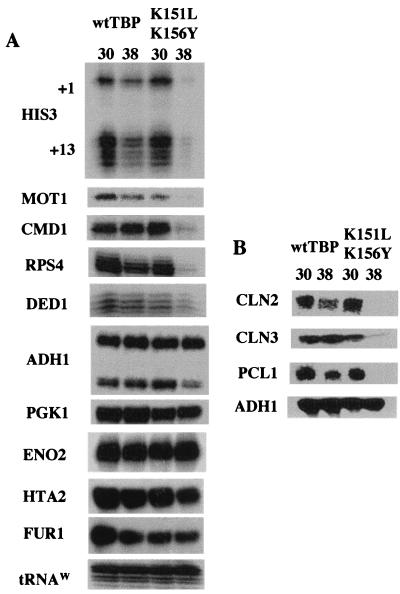
We next compared the transcriptional profiles of wild-type TBP and the K151L,K156Y derivative at the restrictive temperature. After a 1-h incubation at the restrictive temperature, there was a significant reduction in message accumulation of the HIS3 (both the +1 and +13 transcripts), CMD1, MOT1, and RPS4 genes (Fig. 4A). DED1 mRNA levels also decreased (slightly less than twofold). In contrast, levels of the ADH1, PGK1, ENO2, HTA2, and FUR1 messages were not affected at all. It should be noted that the apparent half-life of each of these messages is in the range of 10 to 22 min (18), with the exception of the CMD1 message, which has an apparent half-life of 41 min. Since CMD1 was one of the transcripts that decreased in the mutant strain after the 1-h temperature shift, this longer half-life is evidently not an issue.
FIG. 4.
Transcriptional analysis of the K151L,K156Y strain under restrictive conditions indicates defects for a collection of Pol II-transcribed genes. (A) Mutant and wild-type (wt) TBP-containing strains were grown to log phase and shifted to the restrictive temperature for 1 h. Total RNA isolated before and after the temperature shift was hybridized with a 100-fold excess of the indicated probe and treated with S1 nuclease; tRNAW served as a loading control. The promoter for the +1 transcript from the HIS3 gene is considered TATA-less, while the promoter for the +13 transcript from the HIS3 gene contains a canonical TATA element. The promoters of the other genes tested (MOT1, CMD1, RPS4, ADH1, PGK1, DED1, ENO2, HTA2, and FUR1) have recognizable TATA elements within 250 bp of their translation start sites. The functional relevance of many of these elements is not currently known. (B) Analysis of cyclin genes at the restrictive temperature. Total RNAs from the indicated strains grown at 30°C and for 1 h at 38°C were blotted to nylon membranes and subsequently hybridized with the indicated cyclin probes. ADH1 served as a loading and transfer control.
Can we discern a pattern in the subset of the genes that are very sensitive to the K151L,K156Y substitution under the restrictive conditions? Interestingly, the promoters that are sensitive appear to have transcription rates prior to the temperature shift that are lower than those of promoters that are unaffected. Based on relative RNA levels from our quantitative S1 analyses, the HIS3, MOT1, and CMD1 genes are fairly weakly transcribed. As a frame of reference, the absolute number of HIS3 messages has been calculated to be around seven messages per cell (20). Also, this set of promoters that are sensitive to the K151L,K156Y substitutions includes a TATA-less promoter, in that the HIS3 +1 transcript is decreased in the mutant background under the restrictive conditions. Thus, it may be that weaker promoters have a greater requirement for the factor(s) that is defined by the K151L,K156Y mutant. In contrast, the promoters that are insensitive to the K151L,K156Y mutant exhibit relatively high levels of transcription. For example, ADH1, ENO2, and PGK1 messages are estimated to be present at approximately 50 copies per cell (18). The only exception is the RPS4 gene, which is a highly active gene yet shows a loss of transcription in the K151L,K156L strain under the restrictive condition.
Defects in the transcription of RPS genes and in +1 transcription of the HIS3 gene have been observed after in vivo depletion or inactivation of TAF130 (31, 43). This suggested that the K151L,K156Y mutant may be defective for interaction with TAF130. To determine how closely the molecular phenotype of the K151L,K156Y derivative corresponds to that of depletion of TAF130, we analyzed other genes known to be dependent on TAF130 for transcription.
The K151L,K156Y derivative is defective for transcription of cyclin genes.
Previous TAF depletion experiments demonstrated that certain cell cycle genes require TAF130 for transcription. We analyzed two cell-cycle-regulated genes (CLN2 and PCL1), previously shown to be dependent on TAF130, and a non-cell-cycle-regulated gene (CLN3), shown to be independent of TAF130 inactivation (55). Northern blot analysis revealed that messages for all three genes (CLN2, CLN3, and PCL1) were not detectable at the restrictive temperature in RNA isolated from the K151L,K156Y strain (Fig. 4B). In contrast, ADH1 message amounts remained constant after the temperature shift. Thus, the fact that CLN3 message amounts were affected in the K151L,K156Y strain but not by inactivation of TAF130 indicates that the transcriptional defects are overlapping but not identical. The CLN3, CLN1, and PCL1 messages are each expressed at fairly low levels (predicted message abundance of around one message per cell [reference 18 and citations therein]). Due to the similarities to the TAF130 depletion transcription profile, we next ascertained the ability of the K151L,K156Y TBP mutant to form TFIID.
The K151L,K156Y mutant is defective for TFIID complex formation.
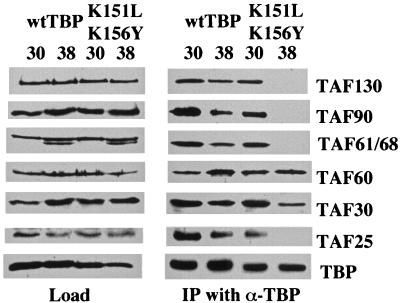
Immunoprecipitation experiments were done to check the integrity of TFIID in the strain harboring the K151L,K156Y mutant. Whole-cell extracts were prepared from mutant and wild-type TBP-containing strains at both the permissive and restrictive temperatures, and antibodies against TBP were used to immunoprecipitate TFIID. Immunoblot analyses of the precipitated complexes with various TAF antibodies indicate that the K151L,K156Y derivative is competent for interaction with all of the TAFs tested at the permissive temperature (Fig. 5). In contrast, after incubation for only 1 h at the restrictive temperature, TFIID isolated from the K151L,K156Y strain lacks detectable amounts of TAF130, TAF90, TAF68/61, and TAF25. It should be noted that although these TAFs are no longer associated with TBP, they remain detectable in the whole-cell extract and hence are likely to be present in SAGA and other TAF complexes. Surprisingly, TAF60 and TAF30 are still associated with TBP even in the absence of the other TAFs. Thus, at the restrictive temperature, the K151L,K156Y TBP mutant exhibits impaired interactions with some TAFs, while interaction with other TAFs remains intact. To our knowledge, this is the first yeast TBP mutant described that is defective for selective TAF interactions and that is capable of forming partial TFIID complexes in vivo. It should be noted that immunoblot analysis with other TAF-directed antibodies (TAF40, TAF17, and TAF170) was attempted, however, these antibodies lacked sufficient titers to detect the corresponding yeast proteins in either the whole-cell extract or the enriched fraction after immunoprecipitation.
FIG. 5.
TFIID is disrupted in the K151L,K156Y strain. Coimmunoprecipitation of TFIID from both mutant (K151L,K156Y) and wild-type (wt) TBP-containing strains is shown. Strains were grown to log phase and shifted to the restrictive temperature for 1 h. Whole-cell extracts were prepared before and after the temperature shift, and TFIID was immunoprecipitated with anti-TBP antibodies (α-TBP) coupled to protein A-Sepharose beads. The immunoprecipitated (IP) complexes were separated with either a 7.5 or 12% acrylamide gel and electroblotted to nitrocellulose. Blots were first probed with anti-TBP antibodies to serve as an internal control. Blots were then stripped and reprobed for the indicated TAF antibodies. The load represents 1/10 of the input in the IP lanes.
The K151L,K156Y mutant supports activated transcription under conditions in which TBP-TAF interactions are compromised.
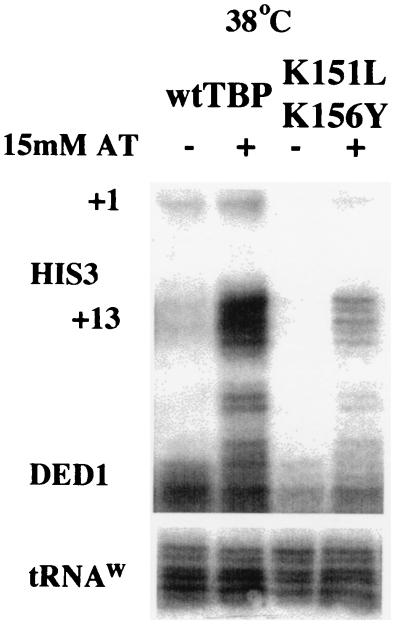
Depletion or inactivation of certain individual TAFs does not affect activated transcription in yeast cells (31, 54). We wished to test whether the K151L,K156Y derivative, which is defective for multiple TAF interactions, is capable of Gcn4-dependent activation of HIS3 transcription at the restrictive temperature. Thus, cells were incubated for 1 h at the restrictive temperature, and then AT was added and the cultures were allowed to incubate for an additional hour. In the absence of AT, HIS3 transcription is not detected at the restrictive temperature (Fig. 4A and 6). However, in the presence of AT, the mutant TBP is able to activate transcription to significant levels (Fig. 6). Thus, although the K151L,K156Y mutant is significantly compromised for interactions with TAF130, TAF90, TAF68/61, and TAF25, this TBP mutant is still competent for the response to the Gcn4 acidic activator in vivo.
FIG. 6.
Maintenance of activated transcription by Gcn4 under the restrictive conditions in the TFIID-disrupted mutant TBP strain. Strains were grown to log phase in synthetic complete medium, and cultures were shifted to the restrictive temperature for 1 h. Cells were then grown for an additional hour in the presence (+) or absence (−) of 15 mM AT, and total RNA was analyzed for HIS3, DED1, and tRNA expression by S1 analysis. wt, wild type.
DISCUSSION
The K151L,K156Y derivative of TBP is defective for TFIID formation.
We have characterized a ts TBP mutant (K151L,K156Y) defective for TFIID formation in vivo. Substitutions at both lysine 151 and 156 are required for the ts mutant phenotype. Unlike other TBP mutants isolated in this screen (45, 46), the K151L,K156Y derivative is responsive to acidic activators at the permissive temperature. Analysis of transcription from Pol II promoters reveals that a subset of TATA-containing genes, as well as several TATA-less genes, are affected at the restrictive temperature. However, the mutant TBP is capable of Gcn4-dependent activation of HIS3 transcription. The transcriptional defects overlap with, but are not identical to, the defects observed for depletion of TAF130 (31, 43, 55). Immunoprecipitation of TBP and its associated factors under the restrictive conditions indicates that although the K151L,K156Y protein is functional for interaction with TAF60 and TAF30, interactions with TAF130, TAF90, TAF68/61, and TAF25 are severely compromised. Because this mutant disrupts only TFIID, leaving SAGA and other potential TAF complexes intact, it provides new information about how TAFs may function in vivo.
Loss of certain TAFs results in promoter-specific defects.
Transcriptional analysis of the K151L,K156Y mutant reveals specific gene defects under conditions of partial TFIID disruption, suggesting that the corresponding promoters are dependent on the TAFs in TFIID for transcription. A subset of these genes (PCL1, CLN2, and the RPS genes) have already been shown to be dependent on TAF130 for transcription (43, 55). Therefore, we would expect them to be affected, as the mutant TBP is defective for interaction with TAF130 at the restrictive temperature. The other promoters affected (CMD1, MOT1, HIS3 +1, and CLN3) exhibit fairly low levels of transcription, providing evidence for a correlation between the requirement for certain TAFs and the transcription of weaker promoters. In addition, the HIS3 +1 promoter exhibits TATA-less features. In higher eukaryotes, TAFs and/or TFIID are critical for in vitro TATA-less transcription (62). Presumably, weak TATA elements need the additional promoter-TAF contacts, as TBP alone is unable to recognize and support transcription from TATA-less promoters. This notion has also been supported by previous in vivo studies, in which depletion of certain individual TAFs causes a decrease in transcription from two genes transcribed from TATA-less promoters (31, 32). We have found a strong positive correlation between the loss of interaction with multiple TAFs and transcription from weaker promoters, in that the K151L,K156Y mutant is defective for the TATA-less promoters tested as well as other weak promoters at the restrictive temperature.
Certain genes are unaffected in the TFIID-disrupted mutant strain.
There are some genes that are not affected in the K151L,K156Y mutant strain under the restrictive conditions (ADH1, PGK, ENO2, HTA2, and FUR1). There are at least two possible mechanisms for transcription from these promoters. The first is that transcription from these genes is independent of the loss of the particular TAFs in the context of TFIID. It may be that the unaffected promoters are dependent on the TAFs that remain associated with the TBP mutant or that TBP is sufficient for expression. The second possible mechanism, based on the premise that TAFs are necessary for transcription, is that unaffected promoters can engage the TAFs in an alternate manner that does not involve TFIID. This model is supported by the observation that TAFs exist in at least one other complex (16, 38).
Interestingly, we have observed one promoter that switches from being defective under TFIID disruption conditions to being functional. At the restrictive temperature, the K151L,K156Y mutant is defective for transcription from the HIS3 promoter, indicating dependence on the missing TAFs. Yet, under conditions of activated transcription by Gcn4, the activity of this promoter is largely independent of the loss of the particular TBP-TAF interactions. It may be that Gcn4 is able to recruit TBP to the promoter, consequently bypassing the dependence on the TAF-TBP interactions observed under the nonactivated conditions. In this scenario, the TAFs in TFIID are not required for activated transcription. Recruitment of TBP by Gcn4 could be direct or could be indirect via interactions with the holoenzyme mediator complex or the SAGA complex. In accord with this model, Gcn4 is able to interact with some of the holoenzyme mediator complex components, as well as with the SAGA complex (12). Moreover, artificial recruitment of members of the holoenzyme mediator complex can activate transcription, indicating that this complex is able to target TBP to a promoter (40). An alternate hypothesis is that the missing TAFs are required for transcription and that by interaction with several redundant targets, the effect of Gcn4 is to stabilize the mutant TFIID complex on the promoter. In a related model, these TAFs are required and necessary, although not as components of the TFIID complex. Thus, the critical TAFs could be supplied to the promoter via an alternate TAF-containing complex. The finding that Gcn4 is unable to interact with purified TFIID (12) suggests that if TAFs are a target of Gcn4, then this activator is likely to interact with the TAFs in the context of the SAGA complex and not in the context of the TFIID complex.
Multiple TFIID complexes versus partial TFIID complexes.
TAF130 is the only yeast TAF that has been shown to contact TBP directly (3, 41), and it is thought to serve as a scaffold for TFIID complex formation. In addition, a ts allele of TAF130 that is rapidly degraded when shifted to the restrictive temperature causes the subsequent degradation of two other yeast TAFs (TAF90 and TAF61/68), while the levels of TAF60 and TAF47 remain unchanged (54). This concomitant degradation suggests that some TAFs are regulated by being part of a TFIID complex, and it is consistent in part with the idea that TAF130 is needed for the overall stability of TFIID. It also suggests the possibility that the TAFs not degraded may exist in a separate TFIID complex not dependent on TAF130 for stability. It should be noted that similar patterns of degradation are seen when other yeast TAFs are depleted (54).
The TBP mutant described in this paper is defective for interaction with TAF130, TAF90, TAF68/61, and TAF25 but is functional for interaction with TAF60 and TAF30. Although many TBP-TAF interactions have been reported for both human and Drosophila TFIIDs, little is known about the TBP-TAF interactions in yeast. Both human TAF70 and Drosophila TAF60/62, the homologs of yeast TAF60, interact with TBP directly (57); thus, it seems likely that TAF60 does contact TBP. More directed experiments, however, are needed to determine if the immunoprecipitated complex represents the remnants of a partially disrupted TFIID complex or a subcomplex that is not affected by the temperature shift. Nevertheless, our studies suggest that these particular TAFs are not dependent on the scaffolding function provided by TAF130 for TFIID formation, perhaps due to direct interactions between TBP and TAF60 and/or TAF30.
Role of TAFs in activated transcription.
Interactions between isolated TAFs and activator proteins occur in vitro, suggesting that TAFs might function as coactivators (53). Furthermore, artificial recruitment of TAFs can bypass the need for an activation domain in vivo (2, 14, 22), and reconstituted TFIID subcomplexes containing only a few TAFs can mediate activated transcription in vitro (42). However, the view that TAFs are required for activated transcription has been challenged by the finding that inactivation (depletion) of single TAFs does not affect the response to many acidic activators in yeast (31, 54). We have expanded these results by demonstrating that a TBP mutant defective for interaction with multiple TAFs remains responsive to the acidic activator Gcn4. The discrepancy between the in vivo and in vitro observations may reflect the redundancy of activator targets in vivo (48).
It appears that in vivo yeast TAFs have developed specialized roles at promoters, perhaps in addition to serving as potential targets for activators. Interestingly, lysines at position 151 and 156 are conserved from yeast to higher eukaryotes. This sequence conservation is likely to reflect functional conservation. This strongly suggests that these residues may also contribute to important TBP-TAF interactions in human TFIID.
ACKNOWLEDGMENTS
We thank Zarmik Moqtaderi for oligonucleotide probes, Michael Green for TAF antibodies, Masayasu Nomura for PNOY103, and Karen Van Orden for critical reading of the manuscript.
This work was supported by research grants from the National Institutes of Health to K.S. (GM30186) and L.A.S. (GM56884).
REFERENCES
- 1.Apone L M, Virbasius C-A, Holstege F C, Wang J, Young R A, Green M R. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol Cell. 1998;2:653–661. doi: 10.1016/s1097-2765(00)80163-x. [DOI] [PubMed] [Google Scholar]
- 2.Apone L M, Virbasius C A, Reese J C, Green M R. Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 3.Bai Y, Perez G M, Beechem J M, Weil P A. Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAFII130. Mol Cell Biol. 1997;17:3081–3093. doi: 10.1128/mcb.17.6.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burk T W, Kadonaga J T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee S, Struhl K. Connecting a promoter-bound protein to the TATA-binding protein overrides the need for a transcriptional activation region. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen J-L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 8.Colgan J, Manley J L. TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 1992;6:304–315. doi: 10.1101/gad.6.2.304. [DOI] [PubMed] [Google Scholar]
- 9.Cormack B P, Strubin M, Ponticelli A S, Struhl K. Functional differences between yeast and human TFIID are localized to the highly conserved region. Cell. 1991;65:341–348. doi: 10.1016/0092-8674(91)90167-w. [DOI] [PubMed] [Google Scholar]
- 10.Cormack B P, Strubin M, Stargell L A, Struhl K. Conserved and nonconserved functions of yeast and human TATA-binding proteins. Genes Dev. 1994;8:1335–1343. doi: 10.1101/gad.8.11.1335. [DOI] [PubMed] [Google Scholar]
- 11.Cormack B P, Struhl K. Regional codon randomization: defining a TATA-binding protein surface required for RNA polymerase III transcription. Science. 1993;262:244–248. doi: 10.1126/science.8211143. [DOI] [PubMed] [Google Scholar]
- 12.Drysdale C M, Jackson B M, McVeigh R, Klebanow E R, Bai Y, Kokubo T, Swanson M, Nakatani Y, Weil P A, Hinnebusch A G. The Gcn-4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol Cell Biol. 1998;18:1711–1724. doi: 10.1128/mcb.18.3.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiger J H, Hahn S, Lee S, Sigler P B. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Couto E, Klages N, Strubin M. Synergistic and promoter-selective activation of transcription by recruitment of transcription factors TFIID and TFIIB. Proc Natl Acad Sci USA. 1997;94:8036–8041. doi: 10.1073/pnas.94.15.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 16.Grant P A, Schieltz D, Pray-Grant M G, Steger D, Reese J C, Yates III J R, Workman J L. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 17.Hoey T, Weinzierl R O J, Gill G, Chen J-L, Dynlacht B D, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 18.Holstege F C P, Jennings E G, Wyrick C J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 19.Ingles C J, Shales M, Cress W D, Triezenberg S J, Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 20.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joliot V, Demma M, Prywes R. Interaction with RAP74 subunit of TFIIF is required for transcriptional activation by serum response factor. Nature. 1995;373:632–635. doi: 10.1038/373632a0. [DOI] [PubMed] [Google Scholar]
- 22.Keaveney M, Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 23.Kim J L, Nikolov D B, Burley S K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 25.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 26.Klein C, Struhl K. Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science. 1994;266:280–282. doi: 10.1126/science.7939664. [DOI] [PubMed] [Google Scholar]
- 27.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y-S, Ha I, Maldonado E, Reinberg D, Green M R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 29.Martinez E, Zhou Q A, Letoile N D, Oelgeschlager T, Berk A J, Roeder R G. Core promoter-specific function of a mutant transcription factor TFIID defective in TATA-box binding. Proc Natl Acad Sci USA. 1995;92:11864–11868. doi: 10.1073/pnas.92.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel B, Komarnitsky P, Buratowski S. Histone-like TAFs are essential for transcription in vivo. Mol Cell. 1998;2:663–672. doi: 10.1016/s1097-2765(00)80164-1. [DOI] [PubMed] [Google Scholar]
- 31.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 32.Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- 33.Moqtaderi Z, Yale J D, Struhl K, Buratowski S. Yeast homologues of higher eukaryotic TFIID subunits. Proc Natl Acad Sci USA. 1996;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikolov D B, Burley S K. RNA polymerase II transcription initiation: a structural view. Proc Natl Acad Sci USA. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 36.Nogi Y, Yano R, Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oelgeschlager T, Tao Y, Kang Y K, Roeder R G. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 38.Ogryzko V V, Kotani T, Zhang X, Schiltz R L, Howard T, Yang X-J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 39.Ozer J, Moore P A, Bolden A H, Lee A, Rosen C A, Lieberman P M. Molecular cloning of the small (g) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- 40.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 41.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Yeast TAFIIs in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 42.Sauer F, Wasserman D A, Rubin G M, Tjian R. TAFIIs mediate activation of transcription in the Drosophila embryo. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 43.Shen W-C, Green M R. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 44.Singer V L, Wobbe C R, Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990;4:636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- 45.Stargell L A, Struhl K. The TBP-TFIIA interaction in the response to acidic activators in vivo. Science. 1995;269:75–78. doi: 10.1126/science.7604282. [DOI] [PubMed] [Google Scholar]
- 46.Stargell L A, Struhl K. A new class of activation-defective TATA-binding protein mutants: evidence for two steps of transcriptional activation in vivo. Mol Cell Biol. 1996;16:4456–4464. doi: 10.1128/mcb.16.8.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stringer K F, Ingles C J, Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 48.Struhl K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 49.Struhl K, Moqtaderi Z. The TAFs in the HAT. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki-Yagawa Y, Guermah M, Roeder R G. The ts13 mutation in the TAF(II)250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan S, Hunziker Y, Sargent D F, Richmond T J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 52.Verrijzer C P, Chen J-L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 53.Verrijzer C P, Tijan R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 54.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;382:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 55.Walker S S, Shen W-C, Reese J C, Apone L M, Green M R. Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 56.Wang E H, Tjian R. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 57.Weinzierl R O J, Ruppert S, Dynlacht B D, Tanese N, Tjian R. Cloning and expression of Drosophila TAFII60 and human TAFII70 reveal conserved interactions with other subunits of TFIID. EMBO J. 1993;12:5303–5309. doi: 10.1002/j.1460-2075.1993.tb06226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Workman J L, Taylor I C A, Kingston R E. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell. 1991;64:533–544. doi: 10.1016/0092-8674(91)90237-s. [DOI] [PubMed] [Google Scholar]
- 59.Wu S-Y, Kershnar E, Chiang C-M. TAFII-independent activation mediated by human TBP in the presence of the positive cofactor PC4. EMBO J. 1998;17:4478–4490. doi: 10.1093/emboj/17.15.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao H, Friesen J D, Lis J T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, Greenblatt J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Q, Lieberman P M, Boyer T G, Berk A J. Holo TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]