Abstract
In eukaryotes, RNA polymerase II transcribes messenger RNAs and several small nuclear RNAs. Like RNA polymerases I and III, polymerase II cannot act alone. Instead, general initiation factors [transcription factor (TF) IIB, TFIID, TFIIE, TFIIF, and TFIIH] assemble on promoter DNA with polymerase II, creating a large multiprotein–DNA complex that supports accurate initiation. Another group of accessory factors, transcriptional activators and coactivators, regulate the rate of RNA synthesis from each gene in response to various developmental and environmental signals. Our current knowledge of this complex macromolecular machinery is reviewed in detail, with particular emphasis on insights gained from structural studies of transcription factors.
Eukaryotic RNA polymerase II (pol II) is a 12-subunit DNA-dependent RNA polymerase that is responsible for transcribing nuclear genes encoding messenger RNAs and several small nuclear RNAs (1). Despite its obvious structural complexity, this multisubunit enzyme requires two groups of auxiliary proteins to solve two critical biochemical problems. First, pol II cannot recognize its target promoters directly. Second, pol II must be able to modulate production of the RNA transcripts of individual genes in response to developmental and environmental signals.
Promoter Anatomy and the Preinitiation Complex (PIC)
Class II nuclear gene promoters contain combinations of DNA sequences, which include core or basal promoter elements, promoter proximal elements, and distal enhancer elements. Transcription initiation by pol II is precisely regulated by transcription factors (proteins) that interact with these three classes of DNA targets and also with each other (reviewed in refs. 2–4). The best characterized core promoter elements, which can function independently or synergistically, are the TATA element (located 25 bp upstream of the transcription start site with consensus sequence TATAa/tAa/t), and a pyrimidine-rich initiator element (located at the start site). The core promoter constitutes the principal DNA target for pol II, and accurate initiation of transcription depends on assembling pol II and transcription factors (TFs) IID, IIB, IIF, IIE, and IIH into a PIC (Table 1; Fig. 1). It is believed that these transcription factors are required to position pol II on most class II nuclear gene promoters, and they are usually referred to as general initiation factors (reviewed in ref. 2). Thus, the PIC is functionally equivalent to the much simpler Escherichia coli holoenzyme, which is composed of the core RNA polymerase subunits and a σ-factor (reviewed in ref. 6). Promoter proximal elements occur anywhere between 50 and 200 bp upstream of the start site and transcriptional activators binding to these sequences regulate transcription. Finally, distal enhancer elements, which can be found far from the transcription initiation site in either direction and orientation, constitute another group of DNA targets for factors modulating pol II activity.
Table 1.
General class II transcription initiation factors from human cells
| Factor | Subunits, kDa | (no.) | Function | |
|---|---|---|---|---|
| TFIID | /TBP | 38 | (1) | Binds to TATA, promotes TFIIB binding |
| \TAFs* | 15–250 | (12) | Regulatory functions (+ and −) | |
| TFIIB | 35 | (1) | Promotes TFIIF–pol II binding | |
| TFIIF | 30, 74 | (2) | Targets pol II to promoter | |
| RNA pol II | 10–220 | (12) | Catalytic function | |
| TFIIE | 34, 57 | (2) | Stimulates TFIIH kinase and ATPase activities | |
| TFIIH | 35–89 | (9) | Helicase, ATPase, CTD kinase activities | |
| All class II GTFs | > 2 MDa | (>42) |
Function and subunit composition of the human class II general initiation factors. The factor denoted with an asterisk is not absolutely required for in vitro basal or core promoter-dependent pol II transcription initiation. TBP, TATA box-binding protein; TAF, TBP-associated factor; GTF, general transcription factor.
Figure 1.
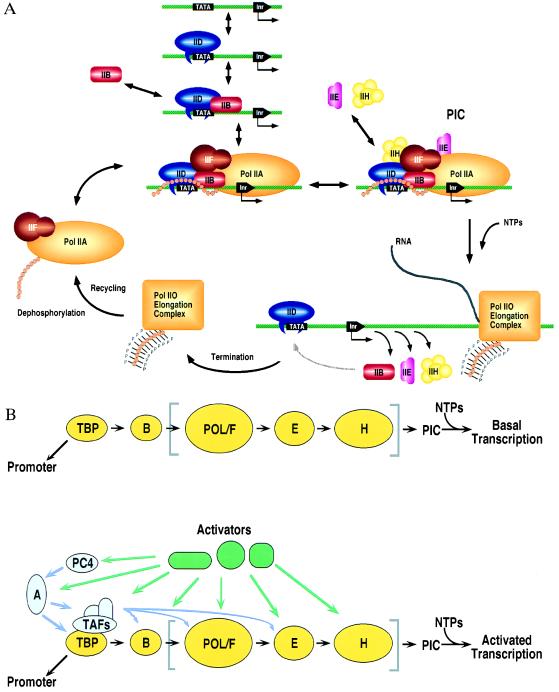
(A) PIC assembly begins with TFIID recognizing the TATA element, followed by coordinated accretion of TFIIB, the nonphosphorylated form of pol II (pol IIA) plus TFIIF, TFIIE, and TFIIH. Before elongation pol II is phosphorylated (pol IIO). Following termination, a phosphatase recycles pol II to its nonphosphorylated form, allowing the enzyme to reinitiate transcription in vitro. TBP (and TFIID) binding to the TATA box is an intrinsically slow step, yielding a long-lived protein–DNA complex. Efficient reinitiation of transcription can be achieved if recycled pol II reenters the preinitiation complex before TFIID dissociates from the core promoter. (Adapted from ref. 5.) (B) Schematic representation of functional interactions that modulate basal (Upper) and activator-dependent transcription (Lower). The basal factors TBP, TFIIB, TFIIF, TFIIE, and TFIIH and pol II are denoted by yellow symbols, with the general initiation factor contents of a “pol II holoenzyme” enclosed by square brackets. TAFII and non-TAFII coactivators (purple) and transcriptional activators (green) are shown interacting with their targets in the PIC. (Figure courtesy of R. G. Roeder and S. Stevens, The Rockefeller University.)
Transcription Factor IID
In the most general case, messenger RNA production begins with TFIID recognizing and binding tightly to the TATA element (Fig. 1). TFIID’s critical role has made it the focus of considerable biochemical and genetic study since its discovery in human cells in 1980 (7). Our current census of cloned TFIID subunits includes more than a dozen distinct polypeptides, ranging in mass from 15 to 250 kDa (reviewed in ref. 8). The majority of these TFIID subunits display significant conservation among human, Drosophila, and yeast, implying a common ancestral TFIID, and gene disruption studies of four yeast TFIID subunits revealed that they are essential for viability (9, 10).
DNA binding by human TFIID was first demonstrated with the adenovirus major late promoter (AdMLP) (11). DNase I footprinting studies of the AdMLP and selected human gene promoters revealed sequence-specific interactions between human TFIID and the TATA element, which are primarily mediated by the TBP subunit of TFIID (see below). In contrast, protection both upstream and downstream of the TATA element is largely sequence independent, displays a nucleosome-like pattern of DNase I hypersensitivity, varies radically between promoters, and can be induced by some activators (reviewed in ref. 8). It is remarkable that TATA box binding by either TFIID or TBP precludes packaging of the core promoter with the nonlinker histone proteins (H2A, H2B, H3, and H4). Conversely, core promoter packaging by histone octamers into nucleosomes prevents TFIID or TBP binding to the TATA element, effectively repressing transcription (reviewed in ref. 12). In vivo, chromatin-mediated transcriptional repression is overcome by various ATP-dependent macromolecular machines (e.g., the SWI/SNF complex) that remodel chromatin in the vicinity of the core promoter (reviewed in ref. 13).
TATA Box-Binding Protein
Publication of the sequence of yeast TBP in 1989 was followed rapidly by the sequences of homologous genes from various eukaryotes and an archaebacterium (amino acid identities within the phylogenetically conserved 180 residue portion range between 38% and 100%, reviewed in ref. 14). Recombinant TBP alone can bind both general and regulatory factors and direct PIC assembly in vitro and basal transcription (reviewed in ref. 2). Basal or core promoter-dependent transcription is a relatively inefficient in vitro reaction that has served as an important tool for characterizing pol II’s minimal requirement for TBP, TFIIB, TFIIF, TFIIE, and TFIIH and the core region of the promoter immediately upstream of the transcription start site. In addition to defining the minimal factors required for accurate pol II initiation, the basal transcription system has been used to establish the order of PIC assembly depicted in Fig. 1A.
Fig. 1B contrasts basal and activated transcription (reviewed in ref. 8). Activated transcription in vivo requires the entire promoter, which includes the core region plus promoter proximal and distal enhancer regions. Transcriptional activators regulate the efficiency of pol II initiation by recognizing their target DNA sequences within promoter proximal or distal enhancer elements and interacting with the PIC (possibly via intermediaries known as coactivators). Activated transcription requires TBP and the remaining subunits of TFIID (the TAFIIs), the other general initiation factors TFIIB, TFIIF, TFIIE, and TFIIH, plus transcriptional activators and coactivators.
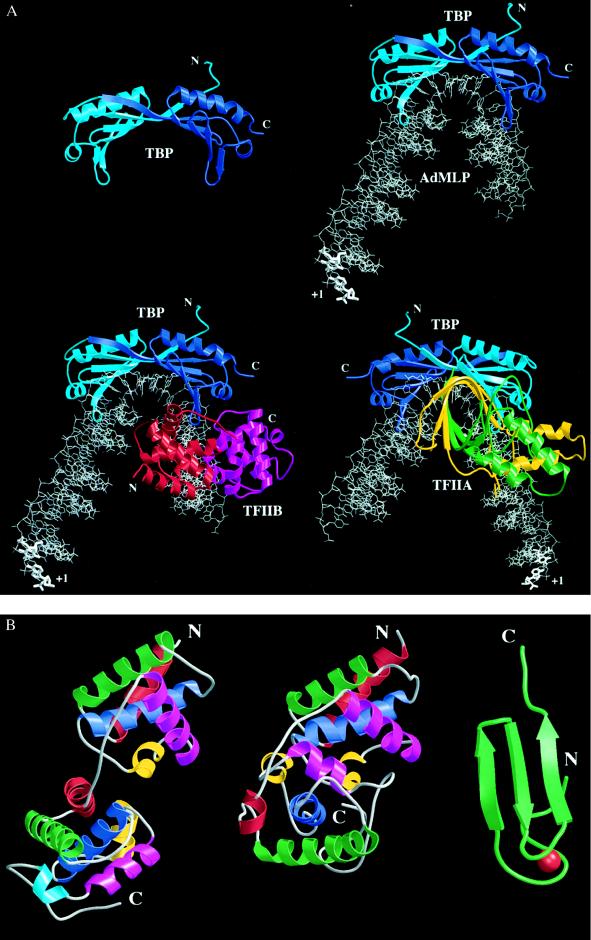
The three-dimensional structure of the conserved portion of TBP is strikingly similar to a saddle (14–16) (Fig. 2A), which correlates perfectly with TBP’s biochemical function as a protein that sits on DNA creating a stable platform for binding other transcription factors. DNA binding is supported by the concave underside of the saddle, while the convex upper surface or seat of the saddle binds various components of the transcription machinery (reviewed in ref. 14). TBP consists of two quasi-identical domains (Fig. 2A), corresponding to the two direct repeats found in the conserved portion of TBP (15). TBP’s ancestor may, therefore, have functioned as a dimer, with gene duplication and fusion giving rise to a monomeric, quasisymmetric TBP.
Figure 2.
(A) Three-dimensional structures of TBP (14–16) (Upper Left), TBP complexed with the TATA element (17–19) (Upper Right), C terminal or core TFIIB (cTFIIB)–TBP–TATA element ternary complex (20) (Lower Left), and TFIIA–TBP–TATA element ternary complex (21, 22) (Lower Right). The proteins are depicted as ribbon drawings, with their N and C termini labeled when visible. The DNA is shown as a stick figure, with hypothetical, linear, B-form extensions at both ends. The transcription start site of the AdMLP is labeled with +1. TBP, and the TBP–DNA and cTFIIB–TBP–DNA complexes are shown from the same vantage point downstream of the transcription start site. The TFIIA–TBP–DNA complex is viewed from upstream of the TATA element, looking toward the transcription start site. Molecules are color coded as follows: red, cTFIIB first repeat; magenta, cTFIIB second repeat; light blue, TBP N terminus and first repeat; dark blue, TBP second repeat; green, TFIIA small subunit; yellow, TFIIA large subunit; and gray, DNA. When TBP recognizes the minor groove of the TATA element, the DNA is kinked and unwound to present the minor groove edges of the bases to the underside of the molecular saddle. On cTFIIB or TFIIA binding to the TBP-DNA complex there is essentially no change in the structure of the binary complex. (B) Structural details of TFIIB. The relative orientation of the cTFIIB’s two domains in the free and bound form is completely different. The bound and free cTFIIBs are drawn with their first domains aligned. The N and C termini of the protein fragments used in the structural studies are labeled, and the α-helices of each cTFIIB domain are colored in order red, green, blue, yellow, and magenta. A helix present only in the second domain of cTFIIB in the ternary complex is colored light blue. Structure of cTFIIB in the cTFIIB–TBP–TATA element ternary complex (20) (Left). Structure of free cTFIIB (23) (Center). Structure of the N-terminal, Zn2+ binding region of TFIIB (24) (Right). The Zn atom is colored in red. The 60 residues between the C terminus of the Zn2+ binding domain and the N terminus of cTFIIB are flexible and have not been visualized in high-resolution structural studies.
Structures of plant (17, 25), yeast (18), and human (19) TBPs complexed with various TATA elements have also been determined (Fig. 2A). These three cocrystal structures are very similar and demonstrate a common induced-fit mechanism (26) of protein–DNA recognition (G. Patikoglou, J. L. Kim, and S.K.B., unpublished data). DNA binding is mediated by the curved, antiparallel β-sheet, which provides a large concave surface for minor groove and backbone contacts with the 8-bp TATA element. The 5′ end of standard B-form DNA enters the underside of the molecular saddle, where TBP produces an abrupt transition to an unprecedented, partially unwound form of the right-handed double helix induced by insertion of two phenylalanine residues into the first T:A base step. Thereafter, the widened minor groove face of the unwound, smoothly bent DNA is approximated to the underside of the molecular saddle, permitting direct interactions between protein side chains and the minor groove edges of the central 6 bp. A second large kink is induced by insertion of two phenylalanine residues into the base step between the last two base pairs of the TATA element, and there is a corresponding abrupt return to B-form DNA. Despite this massive distortion, Watson–Crick base pairing is preserved throughout, and there appears to be no helical strain induced in the DNA because partial unwinding has been compensated for by right-handed supercoiling of the double helix.
DNA packaging into nucleosomes involves wrapping a double helix around the histone octamer. Sequence-dependent nucleosome positioning correlates with bending A+T-rich sequences toward the minor groove (28, 29), and packaging of TATA elements into nucleosomes probably results in minor groove compression precluding TBP binding. Conversely, a preformed PIC remains transcriptionally active after nucleosome assembly (30), and recombinant yeast TBP alone prevents nucleosome-mediated repression of transcription (31). Thus, the cocrystal structures of the TBP–DNA complexes may provide a simple mechanical explanation for the mutual exclusion of DNA packaging and transcription. It is widely believed that transcriptional activators bound to promoter proximal and/or distal enhancer elements target nucleosome-remodeling factors to the core promoters of genes slated for expression. Once chromatin has been remodeled, TFIID would be able to recognize the TATA element and begin PIC assembly (reviewed in ref. 32).
DNA deformation by TBP may also be important for coordinating and/or stabilizing PIC assembly and activator–PIC interactions. PIC assembly around a bend could produce a more compact multiprotein–DNA complex. Moreover, DNA bending by TBP could aid in the looping of DNA to bring remotely bound transcriptional activators closer to the core promoter for interactions with components of the PIC.
Complementary biophysical methods have been used to study interactions between TBP and DNA. Site-selection experiments with Acanthamoeba TBP showed a marked preference for a site very similar to those studied crystallographically (33). DNA bending by TBP in solution was confirmed using circular permutation assays (34). TBP binding was also shown to be enhanced by prebending of DNA toward the major groove, and inhibited by prebending toward the minor groove (35). TBP–DNA association kinetics have been studied by various techniques (36–38), which gave results consistent with formation of an initial collision complex followed by a slow isomerization step with a second-order rate constant of about 106 M−1·s−1. Once formed, the TBP–TATA box complex is very stable and the measured half-life of the yeast TBP–AdMLP complex in aqueous solution is approximately 2 h (36). Finally, a novel chemical modification study has demonstrated that core promoter distortion transiently extends beyond the 3′ end of the TATA element during TBP binding (39).
Transcription Factor IIB
TFIIB is the next general initiation factor to enter the PIC. The resulting TFIIB–TFIID–DNA platform is in turn recognized by a complex of pol II and TFIIF, followed by TFIIE and TFIIH (Fig. 1). In vitro studies with a negatively supercoiled immunoglobulin gene promoter demonstrated that accurate transcription initiation can be reconstituted with TBP, TFIIB, and pol II, suggesting that together TBP and TFIIB position pol II (40). Presumably, the energy provided by negative supercoiling contributes to promoter melting at the transcription start site, which is normally facilitated by the ATP-dependent DNA helicase subunit of TFIIH (see below). Mutations in TFIIB alter pol II start sites in yeast, as do mutations in the large subunit of pol II, providing compelling evidence for its function as a precise spacer/bridge between TFIID and pol II on the core promoter that determines the transcription start site (reviewed in ref. 20).
The second step of PIC assembly has also proved amenable to x-ray crystallographic study. The structure of a TFIIB–TBP–TATA element ternary complex was reported in 1995 (20) (Fig. 2A). C terminal or core TFIIB (cTFIIB) is a two domain α-helical protein that is a structural homolog of the cell cycle protein cyclin A (41, 42) (Fig. 2B). Despite this remarkable structural similarity, there is no evidence that TFIIB regulates the activity of any cyclin-dependent kinase. Moreover, the presence of a cyclin/cyclin-dependent kinase pair within TFIIH would seem to make the prospect of TFIIB having cyclin-like behavior unlikely.
The cTFIIB–TBP–DNA ternary complex is formed by cTFIIB clamping the acidic C-terminal stirrup of TBP in its basic cleft, and interacting with the phosphoribose backbone upstream and downstream of the center of the TATA element. The first domain of cTFIIB forms the downstream surface of the cTFIIB–TBP–DNA ternary complex, where together with the N-terminal domain of TFIIB (24) (illustrated in Fig. 2B) it could readily act as a bridge between TBP and pol II to fix the transcription start site. The remaining solvent-accessible surfaces of TBP and the TFIIB are extensive, providing ample recognition sites for binding of TAFIIs, other class II initiation factors, and transcriptional activators and coactivators. The structure of the TBP–TATA element complex itself is essentially unchanged by ternary complex formation. cTFIIB recognizes the preassembled TBP–DNA complex, including the path of the phosphoribose backbone created by the unprecedented DNA deformation induced by binding of TBP. In addition to stabilizing the TBP–DNA complex, TFIIB binding may contribute to the polarity of TATA element recognition. If TBP were to bind to the quasisymmetric TATA box in the wrong orientation (i.e., if the N-terminal half of the molecular saddle were to interact with the 5′ end of the TATA element), the basic/hydrophobic surface of the N-terminal stirrup would make unfavorable electrostatic interactions with the basic cleft of TFIIB.
The solution NMR structure of cTFIIB alone has also been determined (Fig. 2B) (23). Although each domain in the NMR structure is very similar to its counterpart in the x-ray structure, the two structures demonstrate a different spatial arrangement of the two domains. These data suggest that the oligopeptide linker between the two domains is flexible, and that TFIIB undergoes a conformational change on recognizing the preformed TBP–DNA complex. Thus, TFIIB, like TBP, recognizes its target via induced fit (G. Patikoglou, J. L. Kim, and S.K.B., unpublished data).
Transcription Factors IIE, -IIF, and -IIH
After formation of the TFIIB–TFIID–DNA complex, three other general initiation factors and pol II complete the growing PIC. TFIIF is a heterodimer of subunits with masses of 30 and 74 kDa (reviewed in ref. 43). Among the general initiation factors, TFIIF is unique in its ability to form a very stable complex with pol II, referred to as pol/F (Fig. 1). Although there is no high-resolution structural information available for TFIIF, the results of site-directed mutagenesis and protein–DNA crosslinking studies provide some information about its location within the PIC. Alanine-scanning mutagenesis of human TBP revealed a single residue essential for TFIIF binding, which is located on the convex upper surface of the molecular saddle on its downstream face (44). Photocrosslinking studies identified crosslinks between both TFIIF subunits and positions −5, −15, and −19 (45). Together, these data localize TFIIF within the PIC to the region of the core promoter between the 3′ end of the TATA box (position −24) and the transcription start site (Fig. 2A). TFIIE is an α2β2 heterotetramer of subunits with masses of 34 and 56 kDa (reviewed in ref. 46). Photocrosslinking studies identified crosslinks between the 34-kDa TFIIE subunit and positions −2 and −14 (45), which localize TFIIE to the same portion of the core promoter as TFIIF. TFIIH is a large multiprotein assembly, consisting of nine subunits that range in mass from 39 to 89 kDa (reviewed in ref. 47). Unlike the other general initiation factors, TFIIH supports various catalytic activities, including DNA-dependent ATPase, ATP-dependent DNA helicase, and a serine/threonine kinase that is capable of phosphorylating the C-terminal domain of the large subunit of pol II and is regulated by the cyclin H subunit. At least two of the TFIIH subunits (ERCC2 and ERCC3) are also components of the DNA excision repair machinery, which suggests that the TFIIH multiprotein complex may also participate in DNA repair (reviewed in ref. 48).
The PIC assembly steps detailed above were established in vitro using the minimal transcription system depicted in Fig. 1B Upper. They are not necessarily the only means by which a functional PIC can be assembled. Recently, a number of large multiprotein complexes containing pol II and most of the general initiation factors (other than TFIID and TFIIB), plus the SRB complex and other proteins have been purified from nuclear extract (reviewed in ref. 49). Such complexes are commonly referred to as “pol II holoenzymes,” which is not strictly correct because they cannot function alone. These exciting discoveries suggest that in vivo the PIC could be assembled in only a few steps (e.g., TFIID plus DNA, followed by addition of TFIIB and then the “pol II holoenzyme,” as depicted in Fig. 1B).
Cycling of RNA Pol II Transcription Initiation
Once PIC assembly is complete, and in the presence of nucleoside triphosphates, strand separation at the transcription start site occurs to give an open complex, the C-terminal domain of the large subunit of pol II is phosphorylated (presumably by the kinase subunit of TFIIH), and pol II initiates transcription and is released from the promoter. During elongation in vitro, TFIID can remain bound to the core promoter supporting reinitiation of transcription by pol II and the other general initiation factors (Fig. 1A; reviewed in ref. 5). Because core promoter binding by the TBP subunit of TFIID is an intrinsically slow step, the transcription cycle illustrated in Fig. 1A may represent the mechanism of pol II initiation in vivo. Both the need for chromatin remodeling, which requires ATP, and the slow isomerization step during TBP-induced DNA deformation would be amortized over multiple initiation events if TFIID remained stably associated with the core promoter between successive rounds of transcription. This scenario is particularly attractive in the context of an abbreviated PIC assembly mechanism involving the “pol II holoenzyme.”
Regulation of RNA Pol II Transcription Initiation
Regulation of transcription from a class II nuclear gene in response to developmental or environmental signals is achieved by controlling assembly of the PIC or the catalytic efficiency of pol II during initiation, elongation, or termination. When transcriptional activators interact with TAFIIs, increased recruitment and/or stabilization of TFIID on the promoter is observed (reviewed in ref. 8). The results of studies with hybrid proteins consisting of TBP fused with heterologous DNA-binding domains suggest that TFIID recruitment to the promoter can be a rate limiting step (50–52), which is overcome by activator–TAFII interactions. In vivo footprinting of the promoter proximal regions of some liver-specific genes have demonstrated that many transcriptional activators appear to be bound simultaneously (53), which is consistent with the view that two or more activators can exert synergistic effects on transcription through concerted interactions with multiple components of the PIC. Tjian and coworkers (54, 55) have recently provided direct support for this hypothesis by demonstrating that synergy between two different activators (Bicoid and Hunchback) bound to the same promoter results, at least in part, from specific interactions with two distinct Drosophila TAFIIs that enhance TFIID recruitment.
In their simplest form, protein–protein interactions that regulate pol II activity involve components of the preinitiation complex (TBP, TAFIIs, TFIIB, pol II, TFIIF, TFIIE, and TFIIH) and transcriptional activators (bound either to promoter proximal or distal enhancer elements). Our current picture of activator–TFIID interactions suggests that the TAFIIs can be regarded as a large multiprotein complex that sits atop TBP and integrates signals from many activators and non-TAFII coactivators. The remaining general initiation factors and pol II represent distinct targets within the PIC for interactions with transcriptional activators. Indeed, it seems likely that every component of the PIC is the target of at least one transcriptional activator during transcription from one or more of the estimated 100,000 class II nuclear gene promoters. Indirect interactions between the PIC and transcriptional activators mediated by non-TAFII coactivators have also been observed (Fig. 1B). Coactivators, such as human PC4, human OCA-B, and the yeast SRB complex, can serve as adaptors between activators and basal factors (reviewed in ref. 8).
Transcription Factor IIA
TFIIA was first described as a general initiation factor (7) and was originally thought to be essential for transcription from many if not all class II nuclear gene promoters. Following extensive mechanistic characterization and cloning of the genes encoding the subunits of TFIIA, however, it is now clear that TFIIA is best defined as a coactivator that supports regulation of pol II transcription (reviewed in ref. 3). Early in PIC assembly, TFIIA can associate with and stabilize the TFIID–DNA or the TFIIB–TFIID–DNA complexes, allowing them to ward off the deleterious effects of inhibitory negative cofactors and enhance the stimulatory effects of transcriptional activators (reviewed in ref. 56).
Recently, the structure of a TFIIA–TBP–TATA element ternary complex has been determined by x-ray crystallography (21, 22) (Fig. 2A). Yeast TFIIA consists of two α/β subunits of 14 and 32 kDa, which form an intimate heterodimer via a 12-stranded β-barrel structure. The ternary complex is formed by TFIIA recognizing the N-terminal stirrup of TBP and interacting with the phosphoribose backbone upstream of the TATA element on the opposite face of the double helix from cTFIIB (Fig. 2A). As in the cTFIIB–TBP–DNA complex, TFIIA recognizes the preformed TBP–DNA complex, explaining TFIID–DNA complex stabilization by TFIIA.
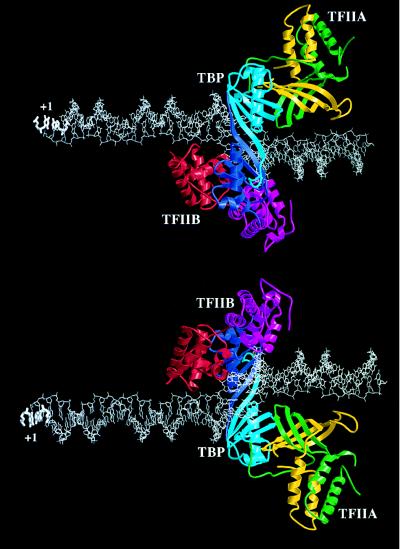
When the structures of the cTFIIB–TBP–DNA and TFIIA–TBP–DNA complexes are combined to create a model of the TFIIA–TFIIB–TBP–DNA quaternary complex (Fig. 3), the mechanism by which TFIIB and TFIIA act synergistically in stabilizing the TFIID–DNA complex can be rationalized. Instead of interacting with one another directly, the basic surfaces of TFIIB and TFIIA make contacts with the negatively charged phosphoribose backbone on opposite faces of the double helix immediately upstream of the TATA element. The model of the TFIIA–TFIIB–TBP–DNA complex also provides critical insights into the role TFIIA as a coactivator, or bridge between transcriptional activators and the PIC. Both subunits of TFIIA form the upstream surface of the TFIIA–TBP–DNA ternary complex, where they are available for interactions with transcriptional activators bound to promoter proximal or distal enhancer elements. It is, therefore, not surprising that the residues on the surface of TBP that are involved in contacts with TFIIA are essential for activated transcription in vivo (56, 57).
Figure 3.
Model of the TFIIA–TFIIB–TBP–DNA complex based on the structures of the cTFIIB–TBP–TATA element (20), and the TFIIA–TBP–TATA element (21, 22) complexes (see Fig. 2A). The transcription start site is labeled with +1. The color coding scheme is the same as in Fig. 2A. (Upper) Viewed along TBP’s axis of approximate intramolecular symmetry from above the saddle. (Lower) Viewed from below the molecular saddle.
Conclusions and Perspectives
It has been more that a quarter of a century since the complexity of eukaryotic transcription was first revealed by Roeder’s discovery of the three RNA polymerases (58). Since then, technically difficult biochemical work and elegant genetic studies have identified and functionally characterized many of the components that together facilitate and regulate pol II production of messenger RNA. Three-dimensional structures of TBP and its complex with the core promoter, cTFIIB, TFIIBn, and TFIIB–TBP–DNA and TFIIA–TBP–DNA ternary complexes have revealed novel protein–DNA interactions, and a detailed mechanistic appreciation of how these polypeptides support transcription initiation. Structural biologists are now tackling even larger transcription factor assemblies, and there is every reason to believe that we will soon see structures of TFIIE, TFIIF, TFIID, TFIIH, and RNA pol II. Transcription factor biologists are currently directing their efforts toward the problem of understanding how transcription initiation is controlled at the level of an individual gene. There is considerable evidence that the PIC and transcriptional activators and coactivators can assemble on a promoter into a stereospecific nucleoprotein complex or “transcriptosome” that supports transcriptional activation (reviewed in ref. 59).
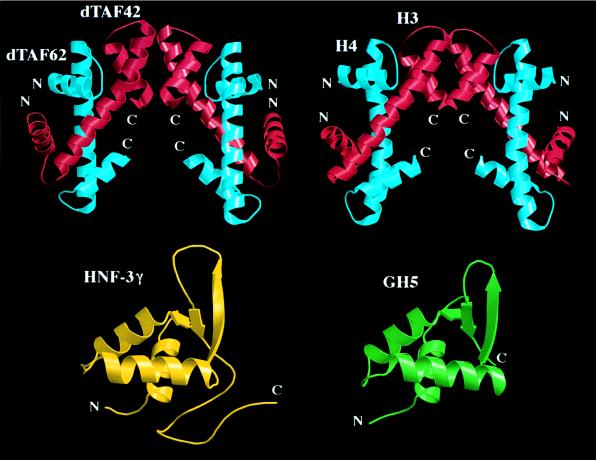
The other important challenge that must be addressed is the need to understand the complicated interplay between DNA packaging and transcription. Unexpectedly, recent crystallographic studies have documented direct structural connections between transcription factors and histone proteins (Fig. 4), suggesting that the macromolecular machines responsible for DNA packaging and transcription are, at some level, evolutionarily related. The structural relationships illustrated in Fig. 4 also raise intriguing questions concerning the mechanisms by which histone-like transcription factors work (reviewed in ref. 64). TFIID may contain a TAFII substructure that resembles the histone octamer and mediates some of TFIID’s nonspecific interactions with DNA (11). Direct evidence of DNA wrapping around TFIID has been obtained by Roeder and coworkers (65), who demonstrated TAFII–DNA crosslinks immediately upstream of the TATA element and downstream of the TATA element extending into the 5′ untranslated region of the gene, and TFIID-induced DNA supercoiling of a closed circular plasmid. In contrast, TBP binding to the same plasmid does not alter the linking number, because DNA supercoiling by TBP is compensated for by partial unwinding of the double helix (reviewed in ref. 25). Finally, the structural similarity of hepatocyte nuclear factor (HNF)-3γ and histone H5 may be functionally significant. HNF-3 binding to two adjacent, high-affinity sites in the mouse serum albumin gene enhancer (66) has been shown to induce phasing of the arrangement of nucleosomes within the enhancer (27).
Figure 4.
Structural similarities between transcription factors and histone proteins. (Upper) Heterotetrameric assembly of the N-terminal portions of two Drosophila TAFIIs (dTAFII42/dTAFII62)2 (60), and the corresponding view of the histone H3/H4 heterotetramer derived from the structure of the histone octamer (61) (the additional N-terminal helix of H3 visualized in this study has been omitted for clarity). (Lower) The DNA binding domain of hepatocyte nuclear factor-3γ (62), and the corresponding view of the globular domain of the linker histone H5 (GH5) (63).
Acknowledgments
We thank Drs. G. Arents, S. Bagby, J. Geiger, M. Ikura, J. L. Kim, E. N. Moudrianakis, P. B. Sigler, M. Summers, and X. Xie for help with figure preparation. This work was supported by the Howard Hughes Medical Institute (S.K.B.) and a Rockefeller University Graduate Fellowship (D.B.N.).
Footnotes
Abbreviations: pol II, polymerase II; PIC, preinitiation complex; AdMLP, adenovirus major late promoter; cTFIIB, C terminal or core TFIIB.
References
- 1.Sentenac A. CRC Crit Rev Biochem. 1985;18:31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- 2.Roeder R G. Trends Biochem Sci. 1991;16:402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- 3.Zawel L, Reinberg D. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]
- 4.Hori R, Carey M. Curr Opin Genet Dev. 1994;4:236–244. doi: 10.1016/s0959-437x(05)80050-4. [DOI] [PubMed] [Google Scholar]
- 5.Zawel L, Kumar K, Reinberg D. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 6.Helmann J, Chamberlin M. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 7.Matsui T, Segall J, Weil P, Roeder R. J Biol Chem. 1980;255:11992–11996. [PubMed] [Google Scholar]
- 8.Burley S K, Roeder R G. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 9.Reese J, Apone L, Walker S, Griffin L, Green M. Nature (London) 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 10.Poon D, Bai Y, Campbell A, Bjorklund S, Kim Y-J, Zhou S, Kornberg R, Weil P. Proc Natl Acad Sci USA. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawadogo M, Roeder R G. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 12.Owen-Hughes T, Workman J. Crit Rev Eukaryotic Gene Expression. 1994;4:403–441. [PubMed] [Google Scholar]
- 13.Peterson C L, Tamkun J W. Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 14.Nikolov D B, Burley S K. Nat Struct Biol. 1994;1:621–637. doi: 10.1038/nsb0994-621. [DOI] [PubMed] [Google Scholar]
- 15.Nikolov D B, Hu S-H, Lin J, Gasch A, Hoffmann A, Horikoshi M, Chua N-H, Roeder R G, Burley S K. Nature (London) 1992;360:40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- 16.Chasman D, Flaherty K, Sharp P, Kornberg R. Proc Natl Acad Sci USA. 1993;90:8174–8178. doi: 10.1073/pnas.90.17.8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J L, Nikolov D B, Burley S K. Nature (London) 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y, Geiger J H, Hahn S, Sigler P B. Nature (London) 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 19.Nikolov D B, Chen H, Halay E D, Hoffmann A, Roeder R G, Burley S K. Proc Natl Acad Sci USA. 1996;93:4956–4961. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikolov D B, Chen H, Halay E, Usheva A, Hisatake K, Lee D, Roeder R G, Burley S K. Nature (London) 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 21.Tan S, Hunziker Y, Sargent D F, Richmond T J. Nature (London) 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 22.Geiger J H, Hahn S, Lee S, Sigler P B. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 23.Bagby S, Kim S, Maldonado E, Tong K, Reinberg D, Ikura M. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 24.Zhu W L, Zeng Q D, Colangelo C M, Lewis L M, Summers M F, Scott R A. Nat Struct Biol. 1995;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]
- 25.Kim J L, Burley S K. Nat Struct Biol. 1994;1:638–653. doi: 10.1038/nsb0994-638. [DOI] [PubMed] [Google Scholar]
- 26.Koshland D E., Jr Proc Natl Acad Sci USA. 1958;44:98–114. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McPherson C, Shin E-Y, Friedman D, Zaret K. Cell. 1993;75:387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- 28.Drew H, Travers A. J Mol Biol. 1985;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- 29.Satchwell S, Drew H, Travers A. J Mol Biol. 1986;191:659–679. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 30.Workman J L, Roeder R G. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 31.Meisterernst M, Horikoshi M, Roeder R G. Proc Natl Acad Sci USA. 1990;87:9153–9157. doi: 10.1073/pnas.87.23.9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolffe A. Curr Opin Genet Dev. 1994;4:245–254. doi: 10.1016/s0959-437x(05)80051-6. [DOI] [PubMed] [Google Scholar]
- 33.Wong J, Bateman E. Nucleic Acids Res. 1994;22:1890–1896. doi: 10.1093/nar/22.10.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starr D, Hoopes B, Hawley D. J Mol Biol. 1995;250:434–446. doi: 10.1006/jmbi.1995.0388. [DOI] [PubMed] [Google Scholar]
- 35.Parvin J, McCormick R, Sharp P, Fisher D. Nature (London) 1995;273:724–727. doi: 10.1038/373724a0. [DOI] [PubMed] [Google Scholar]
- 36.Hoopes B, LeBlanc J, Hawley D. J Biol Chem. 1992;267:11539–11546. [PubMed] [Google Scholar]
- 37.Perez-Howard G, Weil P, Beechem J. Biochemistry. 1995;34:8005–8017. doi: 10.1021/bi00025a006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parkhurst K, Brenowitz M, Parkhurst L. Biochemistry. 1996;35:7459–7465. doi: 10.1021/bi9530301. [DOI] [PubMed] [Google Scholar]
- 39.Sun D, Hurley L. Chem Biol. 1995;2:457–469. doi: 10.1016/1074-5521(95)90263-5. [DOI] [PubMed] [Google Scholar]
- 40.Parvin J, Sharp P. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 41.Jeffrey P, Russo A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N. Nature (London) 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 42.Brown N, Noble M, Endicott J, Garman E, Wakatsuki S, Mitchell E, Rasmussen B, Hunt T, Johnson L. Structure (London) 1995;3:1235–1247. doi: 10.1016/s0969-2126(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 43.Tan S, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1995;92:6042–6046. doi: 10.1073/pnas.92.13.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang H, Sun X, Reinberg D, Ebright R H. Proc Natl Acad Sci USA. 1996;93:1119–1124. doi: 10.1073/pnas.93.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 46.Okhuma Y, Roeder R G. Nature (London) 1994;368:160–163. doi: 10.1038/368160a0. [DOI] [PubMed] [Google Scholar]
- 47.Drapkin R, Reinberg D. Trends Biochem Sci. 1994;19:504–508. doi: 10.1016/0968-0004(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 48.Svejstrup J Q, Wang Z, Feaver W J, Wu X, Bushnell D A, Donahue T F, Frieberg E C, Kornberg R D. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- 49.Koleske A, Young R. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 50.Chatterjee S, Struhl K. Nature (London) 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 51.Klages N, Strubin M. Nature (London) 1995;374:822–824. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 52.Xiao H, Friesen J, Lis J. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rigaud G, Roux J, Pictet R, Grange T. Cell. 1991;67:977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- 54.Sauer F, Hansen S, Tjian R. Science. 1995A;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 55.Sauer F, Hansen S, Tjian R. Science. 1995B;270:1825–1828. doi: 10.1126/science.270.5243.1825. [DOI] [PubMed] [Google Scholar]
- 56.Bryant G O, Martel L, Burley S K, Berk A J. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 57.Stargell L A, Struhl K. Science. 1995;269:75–78. doi: 10.1126/science.7604282. [DOI] [PubMed] [Google Scholar]
- 58.Roeder R G, Rutter W J. Nature (London) 1969;224:234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- 59.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 60.Xie X, Kokubo T, Cohen S L, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Nature (London) 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 61.Arents G, Burlingame R W, Wang B-C, Love W E, Moudrianakis E N. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clark K L, Halay E D, Lai E, Burley S K. Nature (London) 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 63.Ramakrishnan V, Finch J, Graziano V, Sweet R. Nature (London) 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 64.Burley, S. K., Xie, X., Clark, K. L. & Shu, F. (1997) Curr. Opin. Struct. Biol. 7, in press. [DOI] [PubMed]
- 65.Oelgeschlager T, Chiang C-M, Roeder R G. Nature (London) 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 66.Liu J-K, DiPersio C M, Zaret K S. Mol Cell Biol. 1991;11:773–784. doi: 10.1128/mcb.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]