Abstract
Hcs77 is a putative cell surface sensor for cell integrity signaling in Saccharomyces cerevisiae. Its loss of function results in cell lysis during growth at elevated temperatures (e.g., 39°C) and impaired signaling to the Mpk1 mitogen-activated protein kinase in response to mild heat shock. We isolated the MID2 gene as a dosage suppressor of the cell lysis defect of an hcs77 null mutant. MID2 encodes a putative membrane protein whose function is required for survival of pheromone treatment. Mid2 possesses properties similar to those of Hcs77, including a single transmembrane domain and a long region that is rich in seryl and threonyl residues. We demonstrate that Mid2 is required for cell integrity signaling in response to pheromone. Additionally, we show that Mid2 and Hcs77 serve a redundant but essential function as cell surface sensors for cell integrity signaling during vegetative growth. Both proteins are uniformly distributed through the plasma membrane and are highly O-mannosylated on their extracellular domains. Finally, we identified a yeast homolog of MID2, designated MTL1, which provides a partially redundant function with MID2 for cell integrity signaling during vegetative growth at elevated temperature but not for survival of pheromone treatment. We conclude that Hcs77 is dedicated to signaling cell wall stress during vegetative growth and that Mid2 participates in this signaling, but its primary role is in signaling wall stress during pheromone-induced morphogenesis.
The cell wall of the budding yeast Saccharomyces cerevisiae is required to maintain cell shape and integrity (3, 20). The cell must remodel this rigid structure during vegetative growth and during pheromone-induced morphogenesis. Wall remodeling is monitored and regulated by the cell integrity signaling pathway controlled by the Rho1 GTPase. Two essential functions have been identified for Rho1. First, it serves as an integral regulatory subunit of the 1,3-β-glucan synthase (GS) complex that stimulates GS activity in a GTP-dependent manner (7, 32). A pair of closely related genes, FKS1 and FKS2, encode alternative catalytic subunits of the GS complex (6, 13, 28, 33) that are the presumed targets of Rho1 activity.
A second essential function of Rho1 is to bind and activate protein kinase C (19, 29), which is encoded by PKC1 (26, 39). Loss of PKC1 function, or of any of the components of the mitogen-activated protein (MAP) kinase cascade under its control (25), results in a cell lysis defect that is attributable to a deficiency in cell wall construction (23, 24, 31). The MAP kinase cascade is a linear pathway comprised of a MEK kinase (BCK1 [4, 22]), a pair of redundant MEKs (MKK1/2 [14]), and a MAP kinase (MPK1 [21]). One of the consequences of signaling through the MAP kinase cascade is transcriptional activation of FKS2 (41).
Cell integrity signaling is induced in response to several environmental stimuli. First, signaling is activated persistently in response to growth at elevated temperatures (e.g., 37 to 39°C [18]), consistent with the finding that null mutants in many of the pathway components display cell lysis defects only when cultivated at high temperatures. Second, hypo-osmotic shock induces a rapid but transient activation of signaling (5, 18). Finally, treatment with mating pheromone stimulates signaling at a time that is coincident with the onset of morphogenesis (1, 8). Indeed, mutants defective in cell integrity signaling undergo cell lysis during pheromone-induced morphogenesis (8).
The mechanism by which information regarding the state of the cell wall is transmitted to the intracellular signaling apparatus remains an open question. Recently, a gene encoding a putative cell surface sensor for the activation of cell integrity signaling was described and variously designated HCS77 (9), WSC1 (38), and SLG1 (16). Loss of function of HCS77 results in a cell lysis defect that is less severe than that observed for other components of the cell integrity signaling pathway (9, 16, 38), suggesting that its function may be partially redundant. Indeed, several HCS77 homologs (WSC2, -3, and -4, exist in budding yeast. WSC2 and WSC3 have been reported to contribute to cell integrity signaling to a lesser degree than HCS77 (38). In our hands, mutants in these genes in combination with hcs77 have only a modest effect on the severity of the cell lysis defect. Moreover, residual signaling to the Mpk1 MAP kinase is evident even in an hcs77 wsc2 wsc3 triple mutant (38), indicating that these signaling components serve a partially redundant function with another unidentified sensor. Here we present evidence that the Mid2 protein is a cell surface sensor for cell integrity signaling, which together with Hcs77 provides an essential function during vegetative growth. Additionally, we demonstrate that Mid2 is required for activation of this signaling pathway in response to pheromone-induced morphogenesis.
MATERIALS AND METHODS
Strains, growth conditions, and transformations.
The S. cerevisiae strains used in this study are listed in Table 1. Yeast cultures were grown in YEPD (1% Bacto Yeast Extract, 2% Bacto Peptone, 2% glucose). Synthetic minimal (SD) medium (34) supplemented with the appropriate nutrients was used to select for plasmid maintenance and gene replacement. Yeast transformations were carried out by the lithium acetate method (15). Escherichia coli DH5α was used for the propagation of all plasmids. E. coli cells were cultured in Luria broth medium (1% Bacto Tryptone, 0.5% Bacto Yeast Extract, 1% NaCl) and transformed by standard methods (27).
TABLE 1.
S. cerevisiae strains used
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| 1783 | MATaa | I. Herskowitz |
| 1784 | MATαa | I. Herskowitz |
| 1788 | MATa/MATαa | I. Herskowitz |
| DL1235 | MATα rho1-5b (YOC755) | 19 |
| DL1985 | MATahcs77Δ∷LEU2a | This study |
| DL1987 | MATa/MATα hcs77Δ∷LEU2a | This study |
| DL2040 | MATa hcs77Δ∷LEU2 SUP1a | This study |
| DL2230 | MATa/MATα hcs77Δ∷LEU2 wsc2Δ∷LEU2a | This study |
| DL2256 | MATa/MATα hcs77Δ∷LEU2/HCS77a | This study |
| DL2277 | MATa mid2Δ∷URA3a | This study |
| DL2282 | MATa mid2Δ∷URA3 hcs77Δ∷LEU2a | This study |
| DL2337 | MATa/MATα mid2Δ∷URA3/MID2a | This study |
| DL2355 | MATa mtl1Δ∷HIS4a | This study |
| DL2357 | MATa mid2Δ∷URA3 mtl1Δ∷HIS4a | This study |
| DL2394 | MATa/MATα mid2Δ∷URA3a | This study |
| DL2395 | MATa/MATα mtl1Δ∷HIS4a | This study |
| DL2396 | MATa/MATα mid2Δ∷URA3 mtl1Δ∷HIS4a | This study |
| DL2393 | MATa cdc28-13c (D13au) | 1 |
| DL2435 | MATa cdc28-13 mid2Δ∷URA3c | This study |
EG123 background: leu2-3,112 trp1-1 ura3-52 his4 can1.
YPH500 background: ura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1.
D13au background: ade1 his2 leu2-3,112 trp1-1a ura3ΔNS.
Isolation of MID2 and MTL1.
S. cerevisiae DL1987 (hcs77Δ) was transformed with a plasmid library of genomic yeast DNA (from strain DL2040) in centromeric vector pRS316 (35), constructed as described previously (22). Transformants were grown at room temperature and replicated onto YEPD at 38°C for 2 days. Plasmids were rescued from colonies arising at the nonpermissive temperature. Clone pRM5.2 contained an 8.4-kb insert that included three complete open reading frames (ORFs). In this clone, the MID2 gene was flanked by YLR331C and RPS31. A BglII-BamHI subclone that possesses MID2 and YLR331C was suppression competent. A 3′ truncation of MID2 (by SnaBI) eliminated the suppression activity of this clone.
The MTL1 gene was amplified by PCR from wild-type (1783) genomic DNA with 1.0 kb of 5′ flanking sequence and 290 bp of 3′ flanking sequence and then cloned into the 2μm-based plasmid pRS424 (35). The DNA sequence of the complete insert was determined. DNA sequence analysis was performed by the JHU Biosynthesis and Sequencing Facility, using oligonucleotides synthesized by the Biochemistry Core Facility. PCRs for the purposes of cloning were carried out with Pfu polymerase (Stratagene); Taq polymerase (Perkin-Elmer) was used for analytical purposes (see below). The sequence of MTL1 from strain 1783 contained 84 additional nucleotides compared with the database sequence from S288C. These residues result in an in-frame insertion of 28 amino acid residues (nearly all seryl) at position 265, which is within the Ser/Thr-rich region of this protein (see Fig. 5).
FIG. 5.
Sequence alignment of Mid2 and Mtl1. Identical residues are in bold, similar residues are in shadow type. The underline indicates a 28-residue insertion in the predicted Mtl1 sequence from the EG123 background relative to the S288C sequence available through the yeast genome database.
Genomic deletions of MID2 and MTL1.
For construction of the mid2Δ allele, plasmid pRS315[mid2Δ∷URA3] (provided by H. Iida) was digested with XhoI and SspI, and the resultant 1.6-kb fragment was purified and used to transform yeast strains DL2256 and DL2393. Transformants of DL2256 were sporulated, and tetrad analysis was carried out on YEPD with or without 10% sorbitol. Deletion of MID2 was confirmed by PCR in both strains.
To delete the genomic copy of MTL1, a 900-bp fragment that is immediately 5′ to the MTL1 coding sequence, amplified by PCR from genomic 1783 DNA, was first inserted into pBluescript[HIS4] (gift of Susan Michaelis). Next, a 610-bp fragment that includes 320 bp of coding sequence and 290 bp immediately 3′ to the MTL1 coding sequence was amplified and inserted into the resultant plasmid on the other end of HIS4 in the same orientation as the 5′ sequence. This construct, designated pmtl1Δ∷HIS4, was digested with BssHII to liberate a 6.4-kb fragment, which was used to transform yeast strain DL2337. Deletion of MTL1 was confirmed by PCR.
Pheromone-induced killing.
To test for sensitivity to killing by α-factor, MATa strains were grown in synthetic minimal medium containing limiting calcium (100 μM CaCl2) (12) for 24 h, subcultured in the same medium, and grown to an A600 of 0.5 to 1.0 (5 × 106 to 1 × 107 cells/ml). Cells were then treated with α-factor (8 μg/ml; Sigma), and viability was measured over time by plating onto YEPD.
Construction of MID2 and HCS77 fusions to green fluorescent protein (GFP) and the hemagglutinin (HA) epitope.
Mid2 was tagged at its C terminus with a UV-optimized variant of GFP, GFPuv. In the first of two steps, the GFPuv coding sequence (711 bp, including the stop codon) was amplified by PCR from pGFPuv (Clontech), with three additional guanosyl residues at the 5′ end. This fragment was blunt-end inserted into the SmaI site of pRS315 (35) so as to regenerate a single SmaI site immediately 5′ of the GFPuv sequence. Next, a 1.9-kb fragment including MID2 with 760 bp of 5′ noncoding sequence (amplified from pRM5.2) but without its stop codon was blunt-end inserted into the SmaI site of pRS315[GFPuv]. This construct (pMID2-GFP) expressed GFPuv fused to the C terminus of Mid2 with an additional glycyl residue at the fusion point.
Mid2 and Hcs77 were tagged at their C termini with the 3xHA (three-repeat HA) epitope (40). In the first of two steps, the 3xHA sequence from PKC1-HA (39) was amplified together with 250 bp of 3′ noncoding sequence from the PKC1 transcriptional terminator and inserted into the SmaI site of YEp352 (10) so as to regenerate a single SmaI site immediately 5′ to the epitope sequence. Next, MID2 (same fragment as above) and HCS77 sequences (including 600 bp 5′ to the translational start site) were amplified through the final coding base (excluding the stop codon), and blunt-end inserted into the SmaI site of YEp352[3xHA]. The resultant clones fused the 3xHA epitope, in frame with an additional glycine residue, to the C termini of Mid2 and Hcs77.
Construction of N-glycosylation site mutants.
Hcs77-HA N-glycosylation mutants N4Q and N65Q were constructed by site-directed mutagenesis using the PCR overlap extension method (11). Pairs of complementary mutagenic primers (AAATAATGAGACCGCAAAAAACAAGTCTGC and its reverse complement for the N4Q mutation; CTTTGCCCTTTATCAACATTCAGAATGTTA and its reverse complement for the N65Q mutation) were used in separate PCRs with primers either 5′ or 3′ to the coding sequence to generate overlapping 5′ and 3′ regions of HCS77-HA, using YEp352[HCS77-HA] as the template. The products of those reactions were combined in a third reaction to amplify the full-length mutated alleles, which were then digested with SacI and XbaI and inserted into the cognate sites of YEp352. Construction of the double mutant was done by generating the N65Q mutation in the N4Q mutant.
Fluorescence microscopy of Mid2-GFP.
Haploid strain DL2277 expressing Mid2-GFP from pRS315 was grown in SD medium for 24 h, subcultured in YEPD to an A600 of 0.5 to 1.0, and washed three times with phosphate-buffered saline (137 mM NaCl, 2.68 mM KCl, 10 mM Na2HPO4, 1.47 mM KH2OPO4 [pH 7.4]). The cells were resuspended in Vectashield mounting medium (Vector Laboratories, Inc.) with Hoechst 33342 (10 μg/ml). A Zeiss fluorescence microscope fitted with a fluorescein isothiocyanate filter was used to visualize cells.
Activation of Mpk1-HA by temperature shift and pheromone treatment.
Strains expressing Mpk1-HA were tested for thermal activation (18) or pheromone-induced activation (1) of this protein kinase by immunoprecipitation of the fusion protein from cell extracts with monoclonal antibody 12CA5 (BabCo and Boehringer Mannheim), followed by immunocomplex protein kinase assays. Strains were tested for thermal activation of Mpk1-HA as described previously (18). Strains tested for pheromone-induced activation were grown to a density of ca. 107 cells/ml at room temperature in SD medium. Cells were harvested and resuspended in prewarmed (to 37°C) YEPD containing 100 mM sorbitol. The cultures were incubated at this temperature for 1.5 to 2 h until the cells, which bear a temperature-sensitive cdc28 allele, accumulated at the G1 block as large unbudded cells. At this time (t = 0), samples of each culture were removed for preparation of protein extracts (50 ml), mating pheromone was added to the remaining culture (50 nM α-factor), and the cultures were incubated at 37°C. Samples (50 ml) were removed at the indicated times following pheromone addition for preparation of extracts. Extracts were prepared and protein concentrations were determined as described previously (1). Immunocomplex protein kinase assays were performed as described elsewhere (18).
α-Mannosidase treatment.
Mid2-HA and Hcs77-HA were immunoprecipitated from cell extracts as described previously (18) except that 100 μg of protein was used and the immunoprecipitation buffer contained 1% Triton X-100 instead of Nonidet P-40. Immunoprecipitates were washed twice with the mannosidase assay buffer (100 mM sodium acetate [pH 5.0], 2 mM ZnCl2) and resuspended in 15 μl of buffer. α-Mannosidase (5 μl of 0.1 U/μl) was added to the immunocomplexes and incubated for 3 h at 37°C. Controls were mock treated with assay buffer only under the same conditions. Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on an 8% polyacrylamide gel followed by immunoblotting as described previously (18). Protein concentrations of cell extracts that contained detergent were determined by using the bicinchoninic acid protein assay reagent (Pierce).
Fractionation of Hcs77-HA and Mid2-HA.
Transformants of yeast strain 1788 expressing either Mid2-HA or Hcs77-HA from YEp352 were grown in YEPD to an A600 of 1 to 1.2. Cells were harvested from 400 ml, washed once with 10% sorbitol, and resuspended in 6 ml of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 50 mM KF) with protease inhibitors (20 μg of leupeptin, 20 μg of benzamidine, 10 μg of pepstatin, and 40 μg of aprotinin per ml plus 1 mM phenylmethylsulfonyl fluoride). An equal volume of glass beads (0.3-mm diameter) was added to the suspension, and cells were broken by vigorous vortexing for 5 min at 4°C. The beads and cell debris were removed by centrifugation at 13,000 × g. This crude extract was split, and part was treated with Triton X-100 (1%) prior to centrifugation at 100,000 × g for 1 h at 4°C. The pellet was resuspended in a volume of lysis buffer equal to that of the supernatant, and both were analyzed by SDS-PAGE on an 8% gel.
RESULTS
The HCS77 gene encodes a predicted type 1 membrane protein (9). Its most prominent features include a single transmembrane domain near its C terminus, a long Ser/Thr-rich region (ca. 145 residues), and a potential Cys-zinc finger near its N terminus. Loss of HCS77 function results in a cell lysis defect during growth at 37 to 39°C (9, 16, 38), depending on the genetic background (unpublished results). This defect is accompanied by a deficiency in the ability to activate the Mpk1 MAP kinase in response to mild thermal stress (9). These findings, together with the localization of Hcs77 to the cell periphery (38), support a model in which Hcs77 functions as a sensor for cell integrity signaling (9, 16, 38).
Isolation of MID2 as a dosage suppressor of the hcs77Δ defect.
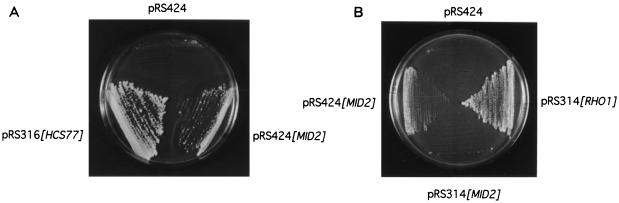
In the course of screening a centromeric library constructed to isolate the gene encoding a mutational suppressor of the hcs77Δ growth defect, we isolated a plasmid that contains the wild-type MID2 gene. An hcs77Δ strain (DL1987) was transformed with a genomic yeast library and screened for suppression of its cell lysis defect at 39°C. We isolated from this screen a single plasmid (pRM5.2) which contains a region of chromosome 12 with three complete ORFs. One of these is MID2 (YLR332W); the others were RPS31 and an uncharacterized ORF, designated YLR331C. Deletion analysis of this plasmid revealed that MID2 (for mating-induced death) was the gene responsible for suppression of hcs77Δ (data not shown). A MID2 isolate from wild-type cells was able to suppress hcs77Δ when cloned into a centromeric vector. Moreover, DNA sequence analysis of the plasmid-borne MID2 revealed no differences from the same region of DNA isolated by PCR from the isogenic wild-type strain, indicating that MID2 acts as a dosage suppressor of the hcs77Δ defect, even when expressed from a low-copy-number plasmid. This suppression was more easily observed in an hcs77Δ wsc2Δ double mutant (Fig. 1A), which has a growth defect slightly more severe than that of the strain with hcs77Δ alone (38).
FIG. 1.
Overexpression of MID2 suppresses the cell lysis defect of an hcs77Δ wsc2Δ mutant and a conditional rho1 mutant. (A) Transformants of yeast strain DL2230 (hcs77Δ wsc2Δ) harboring episomal vector (pRS424), pRS424[MID2], or centromeric plasmid pRS316[HCS77] were streaked onto a YEPD plate and incubated at 38°C for 3 days. (B) Transformants of yeast strain DL1235 (rho1-5) harboring episomal plasmid pRS424 or pRS424[MID2] or centromeric plasmid pRS314[RHO1] or pRS314[MID2] were streaked onto a YEPD plate and incubated at 37°C for 3 days.
Because Hcs77 acts through the cell integrity signaling pathway, we examined the ability of MID2 overexpression to suppress other mutant defects in signaling components thought to be under the control of Hcs77. Figure 1B shows that overexpression of MID2 from a multicopy vector suppresses the growth defect of the rho1-5 mutant (DL1235), which lyses at the restrictive temperature (19). This suppression was allele specific, because MID2 failed to suppress the growth defects of rho1-3 and rho1-4 mutants (not shown), indicating that MID2 overexpression does not bypass the requirement for RHO1. Strains with mutations in other genes that function downstream of RHO1 (e.g., PKC1, BCK1, and MPK1) were not detectably suppressed by MID2 overexpression.
MID2 and HSC77 act in parallel to regulate cell integrity signaling.
Three possibilities exist for the organization of a pathway in which both Hcs77 and Mid2 function. Mid2 might be under the control of Hcs77 such that overexpression of Mid2 drives pathway activation sufficiently to allow survival of thermal stress in the absence of signal from Hcs77. Alternatively, these proteins might act in parallel to provide a partially redundant function in the regulation of cell integrity signaling. Finally, Mid2 might positively regulate the expression of the HCS77-related genes WSC2, WSC3, and WSC4. However, steady-state mRNA levels from these genes were found to be unaffected by either overexpression or deletion of MID2 (data not shown), ruling out this last possibility.
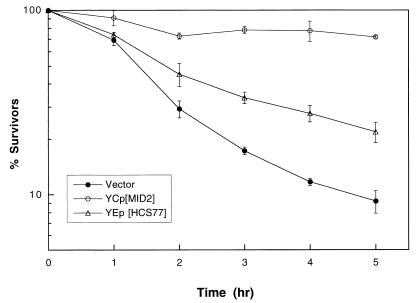
To distinguish between the two remaining possibilities, we first tested for suppression of the mid2Δ defect by HCS77 overexpression. The MID2 gene was isolated initially for its null mutant phenotype, which is failure to survive mating pheromone treatment (30). Treatment of a MATa mid2Δ mutant with α-factor resulted in 90% cell death after 5 h (Fig. 2). Overexpression of either HCS77 (Fig. 2) or RHO1 (not shown) from episomal plasmids partially suppressed this defect. The absence of an epistatic relationship, demonstrated by the reciprocal suppression of hcs77Δ by MID2 and mid2Δ by HCS77, suggests that these genes function in parallel.
FIG. 2.
Overexpression of HCS77 suppresses the pheromone-induced death of a mid2Δ mutant. Yeast strain DL2277 (MATa mid2Δ) transformed with centromeric vector pRS314, pRS314[MID2], or episomal plasmid pRS424[HCS77] was grown in SD with limiting calcium (100 μM CaCl2) at 30°C to an A600 of 0.5 to 1.0. Mating pheromone (α-factor; 8 μg/ml) was added to the cultures, and samples were tested hourly for viability by plating onto YEPD. Plates were scored after 2 days at room temperature.
The possibility that Mid2 and Hcs77 share a redundant function is also supported by structural similarities between these proteins. The Mid2 polypeptide, like Hcs77, is predicted to be a type 1 membrane protein with its transmembrane region near the C terminus. Additionally, it possesses a long Ser/Thr-rich region (ca. 185 residues) that is N terminal to the transmembrane domain. These features, together with the ability of MID2 to suppress hcs77Δ, suggests that these proteins might have overlapping functions, although Hcs77 and Mid2 show no sequence similarity outside these generally related regions.
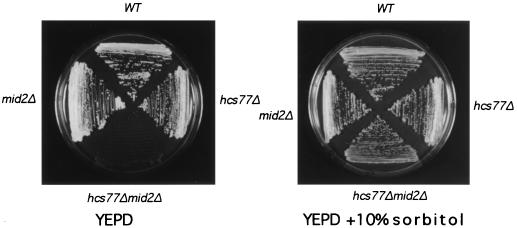
One prediction of a model in which Mid2 and Hcs77 share a function is that the combined defect of mutations in both genes should be more severe than either defect alone. Therefore, we tested the mid2Δ and hcs77Δ mutations for additivity of growth defects. Either mutation alone did not result in a growth defect at temperatures as high as 37°C (not shown). In contrast, the double mutant was inviable even at room temperature (Fig. 3). This growth defect was suppressed by the addition of 10% sorbitol to the growth medium for osmotic support at temperatures as high as 34°C. Removal of osmotic support resulted in immediate cell lysis, as judged microscopically by a high frequency of nonrefractile ghosts. The behavior of the mid2Δ hcs77Δ mutant is characteristic of other mutants in the cell integrity signaling pathway.
FIG. 3.
A mid2Δ hcs77Δ mutant displays a severe cell lysis defect. Haploid yeast strains 1783 (wild type [WT]), DL2277 (mid2Δ), DL1985 (hcs77Δ), and DL2282 (mid2Δ hcs77Δ) were streaked onto YEPD and YEPD supplemented with 10% sorbitol for osmotic support and then incubated at room temperature for 2 days.
Mid2 and Hcs77 are plasma membrane-associated proteins.
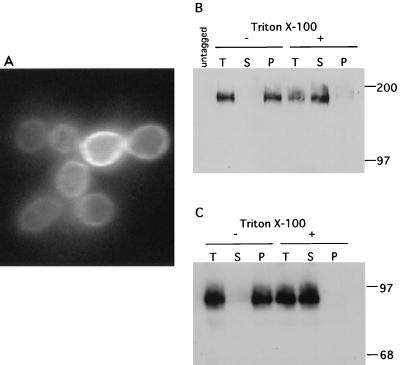
Another prediction of a model in which Mid2 and Hcs77 function as sensors for cell integrity signaling is that they should localize to the plasma membrane. Hcs77 has been shown by fluorescence microscopy to be distributed uniformly around the cell periphery (38). To examine the intracellular localization of Mid2, we first fused GFP to the C terminus of Mid2. This fusion protein was fully functional, as judged by its ability to complement the pheromone sensitivity of the mid2Δ mutant when expressed from a centromeric plasmid (not shown). Fluorescence microscopy of cells expressing the Mid2-GFP fusion from the same plasmid revealed that this protein localizes uniformly around the cell periphery (Fig. 4A). By contrast, GFP alone, when expressed in yeast, is cytoplasmic (38). No fluorescent signal was detected from control cells (not shown). Treatment of cells with mating pheromone for up to 6 h did not appear to alter the localization of Mid2-GFP (not shown).
FIG. 4.
Mid2 and Hcs77 localize to the plasma membrane. (A) Mid2-GFP, expressed in yeast strain DL2277 from a centromeric plasmid (pRS315[MID2-GFP]), was visualized with a fluorescein isothiocyanate filter. (B and C). Mid2-HA (B) and Hcs77-HA (C) were expressed from an episomal vector (YEp352) in yeast strain 1788. Crude extracts were split and subjected to a 100,000 × g centrifugation with or without 1% Triton X-100. Samples containing protein from the total extract (T; 46 μg of the Mid2-HA extract and 25 μg of the Hcs77-HA extract), supernatant (S), and pellet (P) were subjected to SDS-PAGE followed by immunoblot analysis. The control (untagged) lane in panel B contains total extract from cells of strain 1788 without an epitope-tagged protein. Sizes are indicated in kilodaltons.
To examine the intracellular localization of Mid2 and Hcs77 biochemically, we fused the 3xHA epitope to their C termini. These fusions were fully functional, as judged by their ability to complement the defects associated with their deletion mutations when expressed from centromeric plasmids (not shown). Mid2-HA and Hcs77-HA were overexpressed from multicopy plasmids for the purpose of immunodetection. Both proteins migrated on SDS-PAGE as diffuse bands of much greater apparent molecular mass than those predicted by their amino acid sequences, suggestive of modification (see below). Figure 4B and C show that both Mid2-HA and Hcs77-HA fractionated completely with the pellet from a 100,000 × g centrifugation in the absence of detergent. In contrast, both proteins were solubilized in the presence of the nonionic detergent Triton X-100 (1%), indicating that they are associated with a membrane. Taken together with the peripheral localization observed for Hcs77-GFP and Mid2-GFP, these results indicate that both proteins are associated with the plasma membrane. This finding, combined with the strongly additive growth defect of the mid2Δ hcs77Δ mutant and the reciprocal suppression results described above, indicates that these genes provide an essential but redundant function in the maintenance of cell integrity during vegetative growth.
Mid2 has a homolog, Mtl1.
A search of the yeast genome database revealed that MID2 has a homolog on chromosome 7. This uncharacterized ORF (YGR023) shows 50% amino acid sequence identity with Mid2 (Fig. 5). The gene was therefore designated MTL1 (for MID2-like). We constructed a genomic deletion of MTL1 (see Materials and Methods). An mtl1Δ mutant displayed no growth defect at temperatures up to 39°C, nor was it sensitive to pheromone-induced death (data not shown). However, a mid2Δ mtl1Δ mutant displayed an osmotic-remedial lysis defect when grown at 39°C (Fig. 6A), as evidenced by a high frequency of nonrefractile ghosts in the absence of osmotic support at the restrictive temperature. Overexpression of HCS77 weakly suppressed this growth defect (Fig. 6B), further supporting a model in which these genes share a common function during vegetative growth. Loss of MTL1 did not enhance the sensitivity of a mid2Δ mutant to pheromone-induced killing, nor did it confer pheromone sensitivity to an hcs77Δ mutant (not shown). However, overexpression of MTL1 partially suppressed the pheromone sensitivity of mid2Δ. In contrast to mid2Δ, the mtl1Δ mutation did not exacerbate the cell lysis defect of an hcs77Δ mutant, nor did overexpression of MTL1 suppress hcs77Δ. These results, taken in the aggregate, indicate that MID2 plays a more important role in both vegetative growth and pheromone sensitivity than does MTL1.
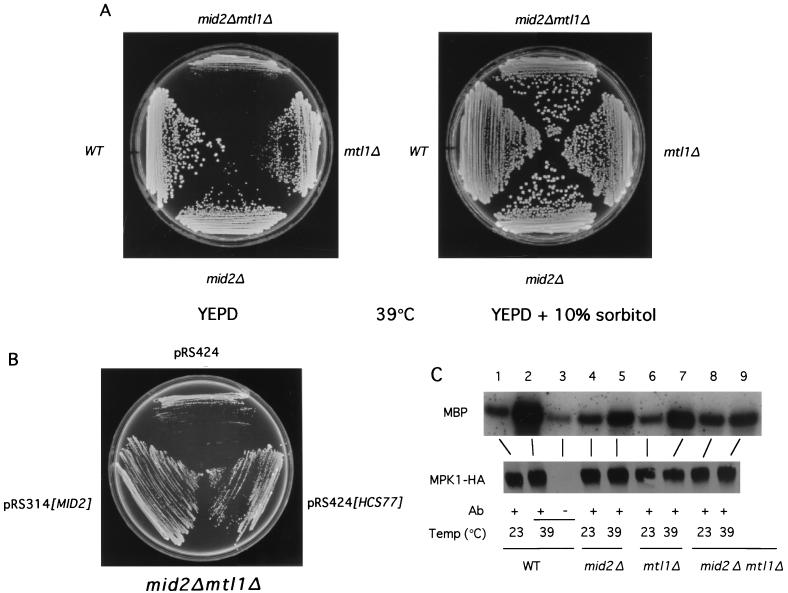
FIG. 6.
A mid2Δ mtl1Δ mutant displays an osmotic-remedial cell lysis defect and is deficient in activating the Mpk1 MAP kinase. (A) Diploid yeast strains 1788 (wild type [WT]), DL2394, DL2395, and DL2396 were streaked onto YEPD or YEPD supplemented with 10% sorbitol and incubated at 39°C for 3 days. (B) Transformants of a mid2Δ mtl1Δ mutant (DL2396) harboring episomal plasmid pRS424, pRS424[HCS77], or centromeric plasmid pRS314[MID2] were streaked onto a YEPD plate and incubated at 39°C for 3 days. (C) The same yeast strains as in panel, A but transformed with YEp351[MPK1-HA], were cultured to mid-log phase at 23°C and shifted to 39°C for 1 h. Mpk1-HA was immunoprecipitated from extracts with monoclonal antibody (Ab) 12CA5, and the immunoprecipitates were subjected to protein kinase assays using MBP as the substrate. The lower panel displays an immunoblot of the Mpk1-HA immunoprecipitates.
Mild thermal stress is a condition known to activate the Mpk1 MAP kinase in wild-type cells (18). To examine the role of Mid2 and Mtl1 in cell integrity signaling during vegetative growth, we compared the activities of Mpk1 in wild-type and mutant strains in response to shift from growth at room temperature to 39°C for 1 h. Mpk1 was strongly activated by this treatment in wild-type cells (Fig. 6C; compare lanes 1 and 2). Temperature-induced activation of this protein kinase in an assay with myelin basic protein (MBP) as the substrate was somewhat diminished in both a mid2Δ mutant and an mtl1Δ mutant (compare lanes 4 and 5 and lanes 6 and 7, respectively). Mpk1 activation was severely impaired in the mid2Δ mtl1Δ mutant but not completely eliminated (compare lanes 8 and 9), consistent with the weak cell lysis defect of the double mutant. It should be noted that the double mutant survived this treatment for the duration of the experiment (data not shown). An hcs77Δ mutant, which is also markedly impaired for Mpk1 activation, also retains some ability to stimulate this kinase. Because an hcs77Δ mid2Δ mutant is inviable in the absence of osmotic support, and high osmolarity prevents activation of Mpk1 in response to thermal stress (18), we were unable to examine Mpk1 activation in this mutant.
MID2 is required for cell integrity signaling in response to mating pheromone-induced morphogenesis.
Cell integrity signaling is activated by pheromone treatment at a time that is coincident with the onset of morphogenesis (8). Moreover, like mid2 mutants, mutants that fail to activate cell integrity signaling are killed by pheromone treatment (8). Therefore, we examined the ability of a mid2Δ mutant to activate cell integrity signaling in response to treatment with mating pheromone. Cells that are synchronized at Start display a strong activation of the Mpk1 MAP kinase in response to α-factor treatment, which peaks approximately 1 h after exposure to the pheromone (1, 8) (Fig. 7). An isogenic mid2Δ mutant was greatly impaired in this activation. It should be noted that the mid2Δ mutant survived pheromone treatment for the duration of this experiment (not shown). Interestingly, the pheromone-induced death of the mid2Δ mutant was not suppressed by osmotic support (1 M sorbitol), in contrast to pheromone-induced death of an mpk1Δ mutant (8). This finding suggests that Mid2 may serve an additional function beyond regulation of cell integrity signaling.
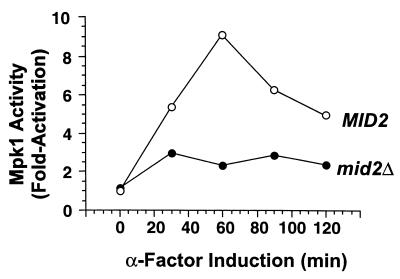
FIG. 7.
Mid2 is required for pheromone-induced activation of Mpk1. Cultures of a cdc28-13 mid2Δ strain (DL2435) and an isogenic MID2 strain (DL2393), both expressing Mpk1-HA from episomal plasmid pNC507 (1), were arrested in G1 at 37°C and treated with mating pheromone (50 nM α-factor; t = 0). Mpk1-HA was immunoprecipitated from extracts of cultures taken at the indicated time points. Immunocomplex protein kinase assays were conducted with MBP as the substrate. Values represent 32P-labeled MBP quantified by phosphorimager at each time point relative to that for the MID2 extract assay at t = 0. Each value is the average of two independent experiments. Immunoblots of Mpk1-HA indicated that the levels of this protein did not change during the course of the experiment (not shown).
Hcs77 and Mid2 are highly glycosylated proteins.
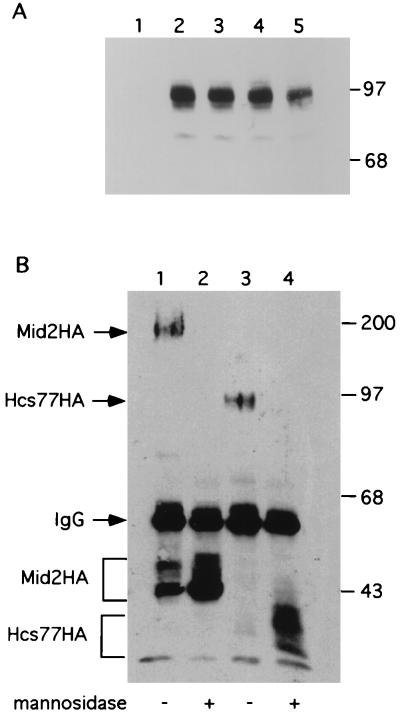
Hsc77-HA is predicted to migrate on SDS-PAGE with a molecular mass of approximately 43 kDa. Instead, it migrated as a diffuse band with apparent molecular mass of 85 to 90 kDa, suggesting that Hcs77 is a highly glycosylated protein (Fig. 4B). This protein possesses two potential N-glycosylation sites (AsnSer/Thr) at positions 4 and 65, as well as many potential O-glycosylation sites in the Ser/Thr-rich region. To examine the glycosylation state of this protein, we first eliminated the potential N-glycosylation sites by mutating the acceptor Asn residues to Gln. Figure 8A shows that neither of the single mutants nor the double mutant had an altered mobility, indicating that Hcs77 is not N-glycosylated. Additionally, these mutants were fully functional for complementation of an hcs77Δ mutant (not shown).
FIG. 8.
Mid2 and Hcs77 are O-mannosylated. (A) Extracts (50 μg of protein) of yeast strain 1788 expressing Hcs77 (lane 1), Hcs77-HA (lane 2), Hcs77N4Q-HA (lane 3), Hcs77N65Q-HA (lane 4), or Hcs77N4Q,N65Q-HA (lane 5) from YEp352 were subjected to SDS-PAGE followed by immunoblot analysis. (B) Mid2-HA (lanes 1 and 2) and Hcs77-HA (lanes 3 and 4) were immunoprecipitated from extracts (100 μg of protein) of yeast strain 1788. Immunoprecipitates were either treated with α-mannosidase at 37°C for 3 h (lanes 2 and 4) or mock treated with buffer only under the same conditions (lanes 1 and 3). Immunoprecipitates were subjected to SDS-PAGE and immunoblot analysis. Sizes are indicated in kilodaltons. IgG, immunoglobulin G.
The Mid2-HA protein behaved similarly to Hcs77-HA, having a predicted molecular mass of approximately 45 kDa, but migrating on SDS-PAGE with a much greater apparent molecular mass (160 to 180 kDa [Fig. 4C]). To examine these proteins for O-glycosylation, we immunoprecipitated them from cell extracts and subjected the immunocomplexes to α-mannosidase treatment. This treatment hydrolyzes the α-linkages between residues of O-linked mannose chains. α-Mannosidase can also hydrolyze mannosyl residues present at the termini of N-linked GlcNAc chains. Figure 8B shows that mannosidase treatment reduced Mid2-HA to a species of apparent molecular mass of approximately 45 kDa and reduced Hcs77-HA to a species of approximately 40 to 42 kDa. The observed values are nearly identical to the predicted masses of the unmodified proteins, confirming that Hcs77 (and strongly suggesting that Mid2) is specifically O-mannosylated.
DISCUSSION
MID2 and HCS77 act in parallel to stimulate cell integrity signaling.
The HCS77 gene is thought to encode a cell surface sensor that detects cell wall weakness during vegetative growth. We isolated MID2 as a low-copy-number dosage suppressor of the growth defect of an hcs77Δ mutant at 39°C. Several additional observations support a model in which Hcs77 and Mid2 are redundant sensors for cell integrity signaling during growth. First, an hcs77Δ mid2Δ double mutant displayed an osmotic remedial cell lysis defect that was much more severe than that of an hcs77Δ mutant, failing to grow at any temperature in the absence of osmotic support. Second, although loss of MID2 function alone does not result in a growth defect (30), deletion of the MTL1 gene, which encodes a Mid2-like protein (see below), in a mid2Δ mutant resulted in an osmotic remedial cell lysis defect at 39°C that was suppressed by overexpression of HCS77. Third, the ability to activate cell integrity signaling at elevated growth temperature, as measured by stimulation of the Mpk1 MAP kinase, was partially impaired by deletion of HCS77, MID2, or MTL1. Fourth, the structural similarity between Hcs77 and Mid2, together with their intracellular colocalization, also suggests redundancy in function. Specifically, they are both uniformly distributed through the plasma membrane, presumably by virtue of their single membrane-spanning regions. Additionally, they were both shown to be highly O-mannosylated through long Ser/Thr-rich regions that are predicted to reside extracellularly.
Overexpression of either RHO1 or PKC1 suppresses the growth defect of hcs77Δ cells (9, 38), suggesting that these components of the cell integrity pathway function downstream of HCS77. Similarly, we found that overexpression of RHO1 also suppresses a mid2Δ mutant. However, overexpression of MID2 was capable of suppressing the growth defect of one rho1 mutant (rho1-5). We interpret this allele-specific suppression to indicate that MID2 is not capable of bypassing the requirement for RHO1 (either by acting downstream of or parallel to this G protein) but instead suppresses the rho1-5 defect by acting upstream of this allele. Therefore, we propose that both Hcs77 and Mid2 contribute to Rho1 activation.
MID2 is required for cell integrity signaling in response to mating pheromone treatment.
The MID2 gene was isolated initially because its loss of function causes cell death in response to treatment with mating pheromone (30). Mutants in MID2 arrest growth in G1 and undergo morphogenesis in response to pheromone but fail to recover from this arrest. Activation of cell integrity signaling is a late response to pheromone treatment that, like MID2, is essential for survival of pheromone-induced morphogenesis (8). In addition to the role that Mid2 plays in signaling cell wall stress during vegetative growth, we demonstrated that it is also critical for cell integrity signaling during pheromone-induced morphogenesis. Interestingly, in contrast to the behavior of an mpk1Δ mutant in response to pheromone (8), the pheromone sensitivity of a mid2Δ mutant is not suppressed by the addition of osmotic support to the medium (to prevent cell lysis), suggesting that Mid2 may have an additional function in the survival of pheromone treatment. Intriguingly, MID2 was also isolated as a dosage suppressor of a strain with a mutation of MPT5 (37), which has been proposed to act through the Cdc28 cyclin-dependent kinase to stimulate passage through G1 in pheromone-arrested cells (2). Therefore, Mid2 may have a cell cycle function that is independent of its role in cell integrity signaling. In contrast to Mid2, Hcs77 appears to be dedicated to signaling cell wall perturbation during vegetative growth and thermal stress, because a null mutant did not display sensitivity to killing by mating pheromone (32a).
Hcs77 and Mid2 as sensors of cell wall stress.
The predicted topography of these plasma membrane proteins, based on the positions of their signal sequences, indicates that they have short C-terminal cytoplasmic domains and longer extracellular domains that are very rich in seryl and threonyl residues. These domains are separated by a single transmembrane sequence. The extracellular domains are highly O-mannosylated. In yeast, this modification consists of several (four or five) mannosyl residues in α-1,2 and 1,3 linkages (36). When many such modifications are present in a stretch of seryl/threonyl residues, as appears to be the case for both Hcs77 and Mid2, they induce the polypeptide to adopt a stiff and extended conformation (17). Therefore, Hcs77 and Mid2 may function as molecular probes that span the periplasmic space to interact directly with the cell wall. If these proteins are glycosylated throughout their Ser/Thr-rich regions, the calculated extents of Mid2 (ca. 470 Å) and Hcs77 (ca. 370 Å) are adequate to span the distance from the plasma membrane to the cell wall (ca. 200 to 300 Å).
Mid2 and Hcs77 are predicted to have overlapping intracellular targets. Paradoxically, however, their predicted intracellular domains are not similar at the level of primary sequence. They may interact with different complexes that share common signaling components, or they may be linked to the cell integrity signaling apparatus through different adaptor molecules.
The Mid2 homolog, Mtl1.
We isolated a gene predicted to encode a polypeptide showing 50% amino acid sequence identity with Mid2. Loss of MTL1 function did not result in any overt defects, either during vegetative growth or in response to mating pheromone, nor did it exacerbate either the pheromone sensitivity of a mid2Δ mutant or the cell lysis defect of an hcs77Δ mutant. However, a mid2Δ mtl1Δ mutant displayed a weak cell lysis defect when cultivated at high temperature and was more impaired for thermal activation of the Mpk1 MAP kinase than was a mid2Δ mutant. Therefore, MTL1 appears to play a minor role in cell integrity signaling during vegetative growth.
ACKNOWLEDGMENTS
We thank H. Iida, J. Gray, and S. Michaelis for strains and plasmids.
This work was supported by grants from the NIH (GM48533 to D.E.L. and GM39852 to B. E. and training grant 5T32CA09110) and the American Cancer Society (Faculty Research Award 446 to D.E.L.) and by Center Grant ES-03819 from the NIEHS.
REFERENCES
- 1.Buehrer B M, Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen T, Kurjan J. Saccharomyces cerevisiae Mpt5p interacts with Sst2p and plays a role in pheromone sensitivity and recovery from pheromone arrest. Mol Cell Biol. 1997;17:3429–3439. doi: 10.1128/mcb.17.6.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cid V J, Duran A, Rey F, Snyder M P, Nombela C, Sanchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costigan C, Gehrung S, Snyder M. A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol. 1992;12:1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davenport K R, Sohaskey M, Kamada Y, Levin D E. A second osmosensing signal transduction pathway in yeast. J Biol Chem. 1995;270:30157–30161. doi: 10.1074/jbc.270.50.30157. [DOI] [PubMed] [Google Scholar]
- 6.Douglas C M, Foor F, Marrinan J A, Morin N, Nielsen J B, Dahl A M, Mazur P, Baginsky W, Li W, El-Sherbeini M, Clemas J A, Mandala S M, Frommer B R, Kurtz M B. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-d-glucan synthase. Proc Natl Acad Sci USA. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drgonova J, Drgon T, Tanaka K, Kollar R, Chen G-C, Ford R A, Chan C S M, Takai Y, Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- 8.Errede B, Cade R M, Yashar B M, Kamada Y, Levin D E, Irie K, Matsomoto K. Dynamics and organization of MAP kinase signal pathways. Mol Reprod Dev. 1995;42:477–485. doi: 10.1002/mrd.1080420416. [DOI] [PubMed] [Google Scholar]
- 9.Gray J V, Ogas J P, Kamada Y, Stone M, Levin D E, Herskowitz I. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 1997;16:4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill J E, Muers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 11.Ho S-N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 12.Iida H, Nakamura H, Ono T, Okumura M S, Anraku Y. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and mating. Mol Cell Biol. 1994;14:8259–8271. doi: 10.1128/mcb.14.12.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue S B, Takewaki N, Takasuka T, Mio T, Adachi M, Fujii Y, Miyamoto C, Arisawa M, Furuichi Y, Watanabe T. Characterization and gene cloning of 1,3-β-d-glucan synthase from Saccharomyces cerevisiae. Eur J Biochem. 1995;231:845–854. doi: 10.1111/j.1432-1033.1995.tb20770.x. [DOI] [PubMed] [Google Scholar]
- 14.Irie K, Takase M, Lee K S, Levin D E, Araki H, Matsumoto K, Oshima Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol Cell Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacoby J J, Nilius S M, Heinisch J J. A screen for upstream components of the yeast protein kinase C signal transduction pathway identifies the product of the SLG1 gene. Mol Gen Genet. 1998;258:148–155. doi: 10.1007/s004380050717. [DOI] [PubMed] [Google Scholar]
- 17.Jentoft N. Why are proteins O-glycosylated. Trends Biochem Sci. 1990;15:291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 18.Kamada Y, Jung U S, Piotrowski J, Levin D E. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 19.Kamada Y, Qadota H, Python C P, Anraku Y, Ohya Y, Levin D E. Activation of yeast protein kinase C by Rho1 GTPase. J Biol Chem. 1996;271:9193–9195. doi: 10.1074/jbc.271.16.9193. [DOI] [PubMed] [Google Scholar]
- 20.Klis F M. Review: cell wall assembly in yeast. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 21.Lee K S, Irie K, Gotoh Y, Watanabe Y, Araki H, Nishida E, Matsumoto K, Levin D E. A yeast mitogen-activated protein kinase homolog (Mpk1) mediates signaling by protein kinase C. Mol Cell Biol. 1993;13:3067–3075. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee K S, Levin D E. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol Cell Biol. 1992;12:172–182. doi: 10.1128/mcb.12.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin D E, Bartlett-Heubusch E. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin D E, Bowers B, Chen C, Kamada Y, Watanabe M. Dissecting the protein kinase C/MAP kinase signalling pathway of Saccharomyces cerevisiae. Cell Mol Biol Res. 1994;40:229–239. [PubMed] [Google Scholar]
- 25.Levin D E, Errede B. The proliferation of MAP kinase signaling pathways in yeast. Curr Opin Cell Biol. 1995;7:197–202. doi: 10.1016/0955-0674(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 26.Levin D E, Fields F O, Kunisawa R, Bishop J M, Thorner J. A candidate protein kinase C gene PKC1, is required for the S. cerevisiae cell cycle. Cell. 1990;62:213–224. doi: 10.1016/0092-8674(90)90360-q. [DOI] [PubMed] [Google Scholar]
- 27.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 28.Mazur P, Morin N, Baginsky W, El-Sherbeini M, Clemas J A, Nielsen J B, Foor F. Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol Cell Biol. 1995;15:5671–5681. doi: 10.1128/mcb.15.10.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono T, Suzuki T, Anraku Y, Iida H. The MID2 gene encodes a putative integral membrane protein with a Ca(2+)-binding domain and shows mating pheromone-stimulated expression in Saccharomyces cerevisiae. Gene. 1994;151:203–208. doi: 10.1016/0378-1119(94)90657-2. [DOI] [PubMed] [Google Scholar]
- 31.Paravicini G, Cooper M, Friedli L, Smith D J, Carpentier J-L, Klig L S, Payton M A. The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol Cell Biol. 1992;12:4896–4905. doi: 10.1128/mcb.12.11.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qadota H, Python C P, Inoue S B, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin D E, Ohya Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 32a.Rajavel, M. Unpublished results.
- 33.Ram A F J, Brekelmans S S C, Oehlen L J W M, Klis F M. Identification of two cell cycle regulated genes affecting the β-1,3-glucan content of cell wall in Saccharomyces cerevisiae. FEBS Lett. 1995;358:165–170. doi: 10.1016/0014-5793(94)01418-z. [DOI] [PubMed] [Google Scholar]
- 34.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 35.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strahl-Bolsinger S, Gentzsch M, Tanner W. Protein O-mannosylation. Biochim Biophys Acta. 1999;1426:297–307. doi: 10.1016/s0304-4165(98)00131-7. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi J, Okada M, Toh-e A, Kikuchi Y. The SMS1 gene encoding a serine-rich transmembrane protein suppresses the temperature sensitivity of the htr1 disruptant of Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1260:94–96. doi: 10.1016/0167-4781(94)00188-9. [DOI] [PubMed] [Google Scholar]
- 38.Verna J, Lodder A, Lee K, Vagts A, Ballester R. A family of genes required for the maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe M, Chen C-Y, Levin D E. Saccharomyces cerevisiae PKC1 encodes a protein kinase C (PKC) homolog with a substrate specificity similar to that of mammalian PKC. J Biol Chem. 1994;269:16829–16836. [PubMed] [Google Scholar]
- 40.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 41.Zhao C, Jung U S, Garrett-Engele P, Roe T Y, Cyert M S, Levin D E. Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol Cell Biol. 1998;18:1013–1022. doi: 10.1128/mcb.18.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]