Abstract
The U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins (snRNPs) form essential components of spliceosomes, the machinery that removes introns from pre-mRNAs in eukaryotic cells. A critical initial step in the complex process of snRNP biogenesis is the assembly of a group of common core proteins (Sm proteins) on spliceosomal snRNA. In this study we show by multiple independent methods that the protein pICln associates with Sm proteins in vivo and in vitro. The binding of pICln to Sm proteins interferes with Sm protein assembly on spliceosomal snRNAs and inhibits import of snRNAs into the nucleus. Furthermore, pICln prevents the interaction of Sm proteins with the survival of motor neurons (SMN) protein, an interaction that has been shown to be critical for snRNP biogenesis. These findings lead us to propose a model in which pICln participates in the regulation of snRNP biogenesis, at least in part by interfering with Sm protein interaction with SMN protein.
The function of pICln has been debated since it was cloned by screening a Madin-Darby canine kidney (MDCK) cell library for the induction of a novel chloride current in Xenopus laevis oocytes. Although we initially proposed that pICln is an integral chloride channel component, pICln lacks predicted hydrophobic transmembrane domains and structural homology to known channel proteins (31). Furthermore, pICln was not detected in the plasma membrane; it is localized predominantly in the cytoplasm and nucleus, while a small fraction is associated with the cytoskeleton (1, 15, 26). Thus, the structure and localization of pICln suggest that it is not itself a chloride channel. Nevertheless, a consistent conclusion from previous studies of pICln is that its overexpression induces the appearance of a chloride conductance (1, 2, 28, 31). The pathway connecting pICln overexpression to activation of the chloride current is not known. It is possible that activation of this chloride current is a side effect of exogenous protein expression in oocytes rather than an effect related to the physiologic function of pICln (1, 2). In order to improve our understanding of the physiologic role of pICln, we have identified several proteins that associate with pICln. In this study, we demonstrate that pICln binds several proteins which are components of spliceosomal small nuclear ribonucleoproteins (snRNPs).
The small nuclear ribonucleoproteins U1, U2, U4, and U5 are essential components of the pre-mRNA splicing machinery (13). Each snRNP is made up of U RNA (U1, U2, U4, or U5), a set of core snRNP proteins (Sm proteins) which are common to all snRNPs, and a set of snRNP-specific proteins. There are at least eight Sm proteins, named B′ (29 kDa), B (28 kDa), D1 (16 kDa), D2 (16.5 kDa), D3 (18 kDa), E (12 kDa), F (11 kDa), and G (9 kDa) (21). While the snRNP-specific proteins have snRNP-specific functions in the splicing reaction, Sm proteins are only known to be involved in the biogenesis of snRNPs (reviewed in reference 23). U1, U2, U4, and U5 snRNAs are transcribed in the nucleus by RNA polymerase II and acquire a monomethyl cap. The RNAs are then exported to the cytoplasm, where they bind the Sm proteins to form the Sm core (3, 36). Subsequently, the monomethyl cap is hypermethylated to a trimethyl cap (22), and the assembled U snRNP is actively imported into the nucleus. Nuclear import depends on the presence of at least two import signals, the Sm core and the trimethyl cap (4, 9). SnRNP nuclear import requires importin β and snurportin1, a nuclear import receptor which binds to both trimethyl cap structures and importin β (12, 29).
Sm proteins form a complex with the protein SMN (survival of motor neurons [20]), the protein which is altered in the autosomal recessive disease spinal muscular atrophy (16). SMN is tightly associated with the protein SIP1 (SMN-interacting protein 1 [20]). In Xenopus oocytes, antibodies directed against SIP1 blocked assembly of Sm proteins with U RNA and inhibited snRNP import into the nucleus. Antibodies specific for SMN enhanced the assembly of Sm proteins on U RNA and had no influence on snRNP nuclear import (6). These results were interpreted as evidence that SMN and SIP1 have a role in U snRNP assembly and are consistent with a model in which the SMN-SIP1 complex interacts with Sm proteins and thereby mediates their assembly onto U RNA.
In the process of identifying proteins which bind to pICln, we found that pICln interacts with several Sm proteins. We show that Sm protein binding to pICln inhibits their assembly on U RNA and interferes with snRNP biogenesis. Furthermore, we show that pICln inhibits SMN binding to Sm proteins.
MATERIALS AND METHODS
Constructs and yeast two-hybrid analysis.
pT7-U RNA plasmids (9) for in vitro transcription of U RNAs were kindly provided by I. Mattaj. U1Δ was generated by using PCR to delete bases 127 to 164 of Xenopus U1, which contains the Sm binding site (24). Clones used for coupled in vitro transcription and translation of SmB′, SmD2, SmD3, and SmF were derived from full-length human expressed sequence tagged (EST) clones (GenBank accession no. aa084844, aa280959, aa452419, and n71826, respectively). SmD1, SmE, and SmG coding regions were amplified from a human fetal brain cDNA library (Stratagene) by PCR. SmE-H6, SmF-H6, and SmG-H6 were expressed from PCR products which included a T7 promoter, the entire coding sequence, and a C-terminal hexahistidine (H6) sequence.
Plasmids expressing glutathione-S-transferase (GST) fused to full-length rat pICln (GST-ICln), Xenopus pICln (GST-xICln), or deletion mutants of human pICln were generated by subcloning the appropriate PCR products into pGEX-2T (Pharmacia). The deletion mutant hICln[1-34] was generated by using a naturally occurring restriction site, resulting in the addition of two amino acids onto the C terminus. The plasmids expressing SMN fused to GST or to thioredoxin (Trx) were constructed by using PCR to subclone the SMN coding region from an EST clone (accession no. n73104) into pGEX6P-2 (Pharmacia) or pET32a (Novagen), respectively.
The plasmid expressing LexA-pICln was constructed by subcloning the pICln coding region into pBTM116KN (11). A library expressing the Gal4 activation domain fused to cDNA from human brain (Clontech) was screened (2 × 106 colonies) in the yeast strain L40 (11). All EST clones and constructs made by PCR were verified by DNA sequencing.
Recombinant protein purification.
GST fusion proteins were expressed in Escherichia coli BL21, purified over glutathione-Sepharose according to the manufacturer’s protocols, and dialyzed against intracellular buffer (IB; 5 mM HEPES, 100 mM KCl [pH 7.4]). Trx fusion proteins were expressed in E. coli BL21(DE3)pLys and purified over an immobilized nickel resin (Qiagen) according to the manufacturer’s protocol.
Purification of pICln-bound proteins.
pICln-bound proteins (IBPs) were purified in one step by affinity chromatography over immobilized GST-ICln fusion protein. Purified GST or GST-ICln was covalently immobilized on ω-aminobutyl agarose (Sigma) by using sulfo-SMPB (Pierce) as described previously (30) at 2 mg of protein per ml of resin. Cytosolic MDCK proteins were isolated from 1.5 × 108 cells as described previously (15) and precleaned by batch rotation with immobilized GST resin for 3 h. Supernatant was supplemented with NaCl to 0.4 M and rotated overnight with 200 μl of GST-ICln agarose. After vigorous washing with isolation buffer containing 0.4 M NaCl, bound proteins were eluted with sodium dodecyl sulfate (SDS) loading buffer, separated by SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride film (Millipore), and visualized by Coomassie staining. Excised protein bands were trypsinized, peptides were separated by high-pressure liquid chromatography, and two to three peptides from each protein band were microsequenced.
Immunochemistry.
Generation and purification of rabbit polyclonal anti-pICln antibody and monoclonal anti-pICln antibody were described previously (15). Y12 anti-Sm antibody and anti-TMG antibody were purchased commercially (Y12 from Labvision; anti-TMG from Calbiochem). 2B1 anti-SMN monoclonal antibody (5) was a gift from G. Dreyfuss. Immunoprecipitation from MDCK cell cytosolic and nuclear extracts was performed as described previously (15). When RNA from immunoprecipitates of cytosolic and nuclear extracts was analyzed, the immunoprecipitates were washed extensively with a buffer containing 20 mM Tris (pH 7.6), 350 mM NaCl, and 1% Triton X-100. RNA from immunoprecipitates was recovered by proteinase K digestion, phenol extraction, chloroform extraction, and ethanol precipitation. The RNA was analyzed on a 10% polyacrylamide gel containing 7 M urea, electroblotted onto nylon, and detected with a radiolabeled U1-specific probe. For immunoprecipitation of Sm-bound RNA from oocytes, oocytes were homogenized in phosphate-buffered saline (PBS) by pipetting, and soluble proteins were recovered by centrifugation. RNA was recovered and analyzed as described above.
Cell culture, [35S]methionine labeling, and cell lysis.
MDCK cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). Proteins were labeled by growing cells in DMEM minus methionine supplemented with 10% dialyzed FCS and containing 50 μCi of [35S]methionine (Amersham)/ml. Cell extracts were prepared as described previously (15).
In vitro translation and binding assays.
Coupled in vitro transcription and translation were performed by using the TNT system (Promega) and [35S]methionine. SmE, SmF, and SmG were expressed as C-terminal hexahistidine fusions so that their correct synthesis could be demonstrated by purification on a metal affinity resin (data not shown). GST binding assays were performed by incubating 5 μg of purified GST fusion protein with 10 μl of in vitro-translated protein in binding buffer (25 mM Tris, 150 mM NaCl, 1 mM MgCl2, 1% Triton X-100 [pH 7.5]) containing 20 μl of glutathione-Sepharose beads (Pharmacia) for 1 h at room temperature. The beads were then washed five times with binding buffer, once with 25 mM Tris (pH 7.5), and then boiled in SDS sample buffer. The bound proteins were analyzed by electrophoresis on an SDS–15% polyacrylamide gel. The lower chamber buffer contained standard running buffer supplemented with 0.1 M sodium acetate. For binding assays involving both SMN and pICln fusion proteins, the thioredoxin fusion protein was preincubated with in vitro-translated Sm proteins for 30 min. Glutathione-Sepharose beads coated with 1 μg of GST fusion protein were then added, and incubation was continued for 30 more min. The samples were then washed and analyzed as described above.
In vitro transcription of U RNA and injection of oocytes.
pT7 U RNA plasmids were linearized at restriction enzyme sites designed to generate U RNAs of an appropriate length. In vitro transcription was performed by incubating 40 U of T7 RNA polymerase with 1 μg of linearized plasmid in 1× transcription buffer (Promega) containing 10 mM dithiothreitol (DTT); a 500 μM concentration (each) of ATP, CTP, and UTP; 50 μM GTP; 30 μCi of [32P]GTP (Amersham); 1 mM m7GpppG (Pharmacia); and 20 U of RNasin (Promega). After 2 h at 37°C, the reaction was terminated by incubating with DNase for 15 min, followed by phenol extraction, chloroform extraction, G-50 spin column chromatography (Pharmacia), and ethanol precipitation. The pellet was resuspended at 106 cpm/μl. Stage 5 to 6 Xenopus oocytes were injected (Drummond microinjector) in the cytoplasm with 50 nl of either IB, GST (4 mg/ml), or GST fusion proteins (4 to 5 mg/ml) in the animal pole. Four hours later, 20 nl of radiolabeled U RNA was injected into the vegetal pole. After 16 h at 18°C, the oocytes were fixed by boiling for 3 min. The nucleus was manually dissected from the cytoplasm. RNA from each fraction was purified by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation and analyzed by electrophoresis through an 8% polyacrylamide gel containing 7 M urea.
For bandshift analysis, five injected oocytes per sample were homogenized by pipetting in 50 μl of ice-cold buffer (25 mM Tris, 50 mM NaCl, 250 μg of yeast tRNA/ml). Ten microliters of the soluble fraction was incubated with 1 μl of 100 mM DTT, 40 U of RNasin (Promega), and 2 μl of either buffer, purified anti-Sm monoclonal antibody (2 μg), or purified anti-pICln monoclonal antibody (21 μg). After 15 min at room temperature, an 8-μl aliquot was mixed with 1 μl of loading buffer (30% glycerol, 0.25% bromphenol blue, 0.25% xylene cyanol) and run on a 6% polyacrylamide gel buffered with 0.5× Tris-borate-EDTA (TBE). RNA was recovered from a 4-μl aliquot of the bandshift reaction mixture as described above and analyzed on an 8% polyacrylamide gel containing 7 M urea.
RESULTS
pICln binds Sm proteins.
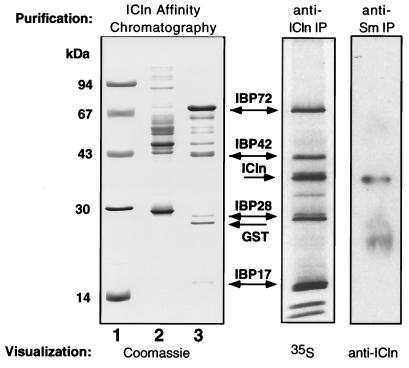
pICln was immunoprecipitated from cytosolic extracts of metabolically labeled MDCK cells by using a polyclonal antibody generated to pICln. Several proteins consistently coimmunoprecipitated with pICln (IBPs), with electrophoretic mobilities of the major proteins corresponding to molecular masses of 72, 42, 28, and 17 kDa (Fig. 1, middle panel). We have previously reported the purification and preliminary characterization of IBP72 (14). This novel protein has significant homology to yeast Skb1 protein, which binds a yeast homolog of p21Cdc42/Rac-activated protein kinase and is implicated in the regulation of cell morphology (8).
FIG. 1.
pICln associates with several cytosolic proteins in vivo, including Sm proteins. (Left panel) MDCK cell cytosolic proteins purified by binding to immobilized GST (lane 2) or GST-pICln (lane 3). Lane 1, molecular size standards. To better visualize nonspecifically bound proteins, lane 2 was loaded with five times more sample than lane 3. (Middle panel) Proteins coimmunoprecipitated with endogenous pICln from the cytosolic fraction of [35S]methionine-labeled MDCK cells. (Right panel) MDCK cell cytosolic extract was immunoprecipitated with a monoclonal anti-Sm antibody and immunoblotted with a polyclonal anti-pICln antibody.
IBP42, IBP28, and IBP17 were purified from MDCK cell cytosolic extracts by affinity chromatography on immobilized GST-ICln fusion protein. The IBPs were significantly enriched in eluates from the pICln affinity resin (Fig. 1, left panel). The interaction was specific, since these proteins did not bind to immobilized GST, and their electrophoretic mobilities matched those of the proteins which coimmunoprecipitated with pICln. The purified IBP42, IBP28, and IBP17 proteins were digested with trypsin, and the resulting peptides were microsequenced. Using peptide sequences obtained from IBP42, we cloned the corresponding cDNA. IBP42 is a novel protein which contains several WD40 motifs (26) and is most similar to the β-subunit of photoreceptor G-protein transducin. The significance of this protein interaction is currently under study.
Several peptide sequences identified IBP28 and IBP17 as SmB/B′ and SmD3, respectively. SmB and SmB′ are products of alternative splicing of the same precursor RNA and differ only by 11 carboxy-terminal residues (34). The peptide sequences from IBP28 correspond to the region common to SmB and SmB′. Both SmB and SmB′ may bind pICln, given that a second protein with a molecular weight slightly higher than that of IBP28 coimmunoprecipitated with pICln (Fig. 1).
To confirm the interaction of pICln with Sm proteins, we used several independent approaches. First, Sm proteins were immunoprecipitated from MDCK cell cytosolic extract by using the anti-Sm monoclonal antibody Y12 (19), and pICln was detected in the precipitate by immunoblotting with an anti-pICln monoclonal antibody (Fig. 1, right panel). In control experiments, pICln was not detected in immunoprecipitates generated with an unrelated monoclonal antibody (data not shown). Second, we used a LexA-pICln fusion protein as bait to screen a human brain library in the yeast two-hybrid system. From this screen, we isolated five clones that contained full-length SmD3 cDNA. Clones encoding the other Sm proteins were not isolated, although not all positives were examined due to a high degree of background activation by the LexA-pICln fusion protein itself. Third, we tested the ability of recombinant, purified pICln to associate with in vitro-translated Sm proteins. When in vitro-translated Sm proteins were incubated with GST-ICln immobilized on glutathione-Sepharose beads, SmB′, SmD1, and SmD3 bound to GST-ICln but not to GST alone (Fig. 2). The finding that in vitro-translated SmD1 binds to GST-ICln is compatible with our coimmunoprecipitation experiments, since SmD1, SmD2, and SmD3 have similar electrophoretic mobilities (18), and there is clearly more than one protein with an electrophoretic mobility corresponding to 17 kDa which coimmunoprecipitates with pICln and which binds to pICln affinity resin (Fig. 1). There was no detectable binding of SmF and SmG to GST-ICln. SmD2 and SmE bound to GST-ICln, although the ratio of specific to nonspecific binding for these two proteins was lower than for SmB′, SmD1, and SmD3. We did not investigate the possible interaction of SmD2 and SmE with pICln further, because these interactions appeared to be the weakest, and we lacked independent data to substantiate these interactions.
FIG. 2.
pICln interacts with Sm proteins in vitro. (a) (Left panel) 35S-labeled Sm protein in vitro translation products input into binding assays. (Right panel) GST-ICln fusion proteins used in binding assays, separated on an SDS–10% polyacrylamide gel and stained with Coomassie blue. GST-hICln deletion mutants are labeled with the numbers of the pICln amino acids present in the fusion protein. (b) GST or GST-ICln immobilized on glutathione-Sepharose beads was incubated with in vitro-translated 35S-labeled Sm. After extensive washing, bound proteins were analyzed by polyacrylamide gel electrophoresis and visualized by using a phosphorimager. In vitro translation of SmD3 consistently resulted in two products, both of which bound to GST-ICln. (b) Deletion analysis of pICln-Sm interaction. In vitro-translated SmB′, SmD1, or SmD3 was incubated with deletion mutants of pICln fused to GST and immobilized on glutathione-Sepharose beads. Input levels of in vitro-translated proteins are shown in panel a, and binding to GST or GST-ICln from the same experiment is shown in panel b. 35S-labeled proteins were visualized by using a phosphorimager.
To outline the region of pICln that interacts with Sm proteins, we generated proteins consisting of various deletions of pICln fused to GST and analyzed the ability of these proteins to bind in vitro-translated SmB′, SmD1, and SmD3 (Fig. 2c). hICln[1-199] binds to SmB′, SmD1, and SmD3 as efficiently as the wild type. However, hICln[1-36] and hICln[175-237] do not bind Sm proteins, indicating that the extreme N-terminal and C-terminal ends of the pICln are not necessary for Sm protein binding. We have previously shown that the C-terminal portion of pICln is necessary and sufficient for pICln interaction with IBP72 (14). The failure of the truncated pICln fusion proteins to binding Sm proteins also provides additional negative controls for binding specificity in the in vitro-binding assay.
pICln is not a component of snRNPs.
We next asked whether pICln interacts with Sm proteins assembled in snRNPs in addition to free Sm proteins. To determine if pICln is a component of snRNPs, we used two independent methods. First, we immunoprecipitated RNAs containing a trimethylguanine (TMG) from MDCK cell nuclear and cytoplasmic extracts by using a TMG antibody. Since U1, U2, U4, and U5 RNAs contained in snRNPs possess a TMG cap, while free U RNAs possess a monomethylguanine cap, TMG immunoprecipitates contain predominantly snRNPs. When the TMG immunoprecipitates were immunoblotted with a pICln-specific antibody, we failed to detect pICln (Fig. 3a), suggesting that it is not a component of snRNPs. In contrast, a pICln antibody immunoprecipitated pICln from the same extracts, indicating that pICln was detectable by this assay. Second, we completely immunoprecipitated pICln from MDCK cell nuclear and cytoplasmic extracts and probed for the presence of U1 RNA. U1 RNA was not coimmunoprecipitated by the pICln-specific antibody above the background amount precipitated by an unrelated antibody (Fig. 3b). In contrast, U1 RNA was immunoprecipitated by anti-TMG and coimmunoprecipitated by anti-Sm antibodies. Since U1 RNA is an integral component of U1 snRNPs, this result further demonstrates that pICln is not a part of snRNPs.
FIG. 3.
pICln is not a component of snRNPs. (a) SnRNPs were immunoprecipitated from MDCK cell cytosolic (C) or nuclear (N) extracts with TMG-specific antibody or pICln-specific antibody. The immunoprecipitate, the supernatant (Sup), and the untreated extract were analyzed by immunoblotting with a monoclonal pICln-specific antibody. (b) MDCK cell cytosolic (C) and nuclear (N) extracts were immunoprecipitated with antibodies specific for pICln, TMG, Sm, and an unrelated antigen. Ctrl Ab, control antibody. RNA from the immunoprecipitates and from untreated extract was deproteinized, electrophoresed on a denaturing polyacrylamide gel, and transferred to a nylon membrane. U1 RNA was detected by hybridization to a U1-specific probe. U1 content in the pICln immunoprecipitate was negligible and indistinguishable from the control immunoprecipitate, in contrast to prominent U1 signals in TMG and Sm immunoprecipitates.
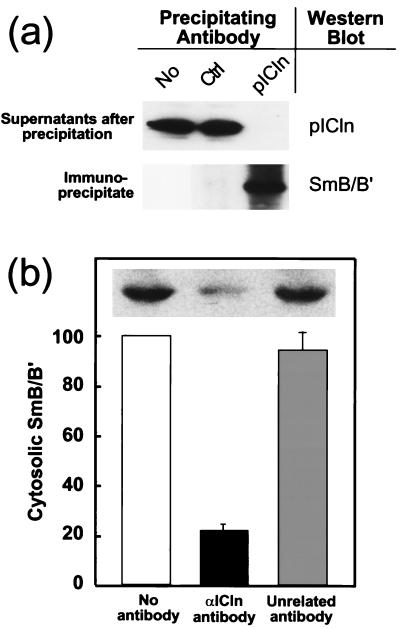
If pICln binding to non-snRNP Sm proteins is functionally important, then it would be reasonable to expect that a significant fraction of cytosolic Sm proteins is bound to pICln. To measure the fraction of cytoplasmic Sm proteins bound to pICln, we immunodepleted cytosolic extracts of pICln with an anti-pICln-specific antibody and measured SmB/B′ content of the depleted extract by immunoblotting with an Sm-specific antibody (Fig. 4). Greater-than-95% pICln depletion was verified by immunoblotting with a monoclonal anti-pICln antibody (Fig. 4a). We found that immunodepletion of pICln reduced the amount of SmB/B′ by 80% (Fig. 4b), indicating that the majority of cytoplasmic SmB/B′ was bound to pICln. These immunodepletion experiments were performed under stringent conditions (1% Triton X-100 and 350 mM NaCl) to ensure minimal nonspecific binding SmB/B′ to the immunoprecipitates. Under more-physiological conditions (150 mM NaCl and no detergent), more than 95% of SmB/B′ was depleted by anti-pICln antibody (data not shown). We were unable to perform a similar experiment with SmD1 or SmD3, since our preparation of Y12 antibody poorly recognized these proteins in cytosolic extract. However, SmD3 has been shown to form stable RNA-free complexes with SmB/B′ (7, 33) and therefore it is likely that the majority of SmD3 also coimmunoprecipitates with pICln.
FIG. 4.
The majority of cytosolic SmB/B′ is bound to pICln. (a) pICln was immunoprecipitated from MDCK cell cytosolic extract with a purified polyclonal anti-pICln antibody. More than 95% of pICln was immunoprecipitated, as demonstrated by immunostaining the supernatant with an anti-pICln antibody (top panel). Immunostaining the pICln immunoprecipitate with anti-Sm protein antibody shows that SmB/B′ coimmunoprecipitated with pICln (bottom panel). An unrelated antibody (Ctrl) did not immunoprecipitate pICln or SmB/B′. (b) Immunodepletion of pICln results in depletion of SmB/B′. Cytosolic extract from MDCK cells was immunodepleted of pICln as shown in panel a. SmB/B′ content in the extract prior to (No antibody) and after immunoprecipitation with pICln or unrelated antibody was assessed by Western blotting with Y12 antibody (insert) and quantitated by densitometry in four independent experiments. The amount of SmB/B′ in pICln-depleted samples was normalized to the amount in control samples in each experiment. Treatment of sample with an unrelated antibody and protein A beads did not change SmB/B′ content in the extract. Bars indicate standard errors of the means.
pICln inhibits Sm protein assembly on U RNA.
Our finding that pICln binds to Sm proteins only when they are not assembled in snRNPs raised the possibility that pICln may affect Sm protein assembly on U RNA. Much of our understanding of snRNP biogenesis has come from experiments in the X. laevis oocyte model. Oocytes differ from most cell types in that the vast majority of Sm and SMN proteins are localized in the cytoplasm (6, 36). The oocyte model has been used successfully to study snRNP biogenesis in vivo, including Sm assembly on U RNA, U RNA cap hypermethylation, and snRNP nuclear import (3, 24, 36). We elected to use this model to study the effect of pICln on snRNP biogenesis. For these experiments, we used pICln cloned from an ovarian Xenopus cDNA library (15). pICln is expressed in Xenopus oocytes (15) and is associated with Sm proteins, as was demonstrated by coimmunoprecipitation experiments (data not shown).
To determine if pICln interaction with Sm proteins influences Sm core domain formation on U RNA, we injected oocytes with Xenopus pICln fused to GST (GST-xICln). The oocytes were then injected with 32P-labeled, in vitro-transcribed U1, U2, U4, U5, and U6 snRNAs. Subsequently, the oocytes were homogenized, and U RNA bound by Sm proteins was immunoprecipitated by using an anti-Sm antibody and analyzed by denaturing polyacrylamide gel electrophoresis (Fig. 5a). In control oocytes injected with buffer or GST, more than 80% of injected U1, U2, U4, and U5 RNA coimmunoprecipated with Sm proteins. The immunoprecipitation was specific, because U6 RNA, which does not bind Sm proteins (10), is not significantly precipitated by the anti-Sm antibody (Fig. 5a). Injection of GST-xICln significantly inhibited binding of U1, U2, U4, and U5 RNAs to Sm proteins (P < 0.05; Fig. 5a and b), with U5 association with Sm protein most affected (85% inhibition). The degrees of inhibition were different for each U RNA (P < 0.01; Fig. 5b), except for those for U2 and U4.
FIG. 5.
pICln inhibits Sm binding to U RNA. (a) Oocytes were injected with buffer, GST, or GST-xICln, followed by a mixture of 32P-labeled U1, U2, U4, U5, and U6 RNA. The oocytes were then homogenized, and Sm-associated U RNA was immunoprecipitated with the anti-Sm antibody Y12. The immunoprecipitate and supernatant were deproteinized and analyzed on a denaturing polyacrylamide gel. Each sample represents a pool of four oocytes. Ctrl indicates the mixture of U RNAs prior to injection. The different amounts of each U RNA reflect different amounts of U RNA produced from each template in in vitro transcription reactions. (b) Quantitation of the results of panel a. The amount of U RNA present in the immunoprecipitate and supernatant was quantitated by measuring radioactivity in gels on a phosphorimager. The percentage of U RNA found in the precipitate of pICln-injected oocytes was plotted relative to the percentage of U RNA in the precipitate in the buffer-injected control oocytes. •, significant inhibition due to GST-xICln injection compared to buffer or GST controls (P < 0.03). Each column represents the average (error bar, standard error of the mean) for five (GST) or six (buffer, GST-xICln) samples. (c) Bandshift analysis of Sm binding to U RNA. Oocytes were injected with GST (G) or GST-xICln (I) and subsequently injected with either 32P-labeled U1 (1), U5 (5), or U1Δ (Δ), a U1 mutant that lacks the Sm binding site. The oocytes were homogenized, and either buffer, anti-Sm antibody, or anti-pICln antibody was added. An aliquot was analyzed by nondenaturing polyacrylamide gel electrophoresis (top panel). RNA was recovered from another aliquot of the same bandshift reaction mixture and analyzed by denaturing gel electrophoresis (bottom panel). The square bracket indicates U RNA with reduced mobility due to protein binding. The arrow indicates complexes with very low mobility due to antibody binding. The positions of free U1, U5, and U1Δ probes are shown.
Although pICln inhibition of U RNA immunoprecipitation by anti-Sm antibody strongly suggests that pICln association with Sm proteins prevents Sm protein binding to U RNA, an alternative explanation is that pICln binding to Sm proteins prevents them from being recognized by the anti-Sm antibody. To exclude this possibility and confirm the inhibitory effect of pICln by an independent method, we used the mobility shift assay to assess Sm protein binding to U RNA. Oocytes were injected with GST-xICln, followed by injection with 32P-labeled, in vitro-transcribed U1. Extracts from these oocytes were then analyzed on a nondenaturing polyacrylamide gel. In control oocytes injected with GST, several discrete complexes were present (square bracket, Fig. 5c). The complex contained Sm proteins specifically bound to U1 RNA, since the mobility of the complex was reduced by incubation with anti-Sm antibody (arrow, Fig. 5c). Furthermore, this complex does not form on U1Δ, a U1 deletion mutant that lacks the Sm binding site (lane 1). Although a small amount of U1Δ is bound in a lower mobility complex, this complex does not involve Sm proteins because the mobility of the complex is not further reduced by anti-Sm antibody (lane 4). When oocytes were injected with GST-xICln, the amount of U1 RNA incorporated into the Sm-U1 RNA complex was decreased (lane 2 versus lane 3), as was the amount which could be supershifted by the addition of anti-Sm antibody (lane 5 versus lane 6). Similar results were found by using a U5 RNA probe. U5 RNA forms protein complexes with reduced mobility (lane 9); these complexes involve Sm protein since they can be supershifted by the addition of anti-Sm antibody (lane 12), and pICln interferes with the U5-Sm protein interaction (lane 9 versus lane 10). The inhibitory effect of pICln was not due to degradation of the labeled U RNA probe, since the same amount of intact U RNA probe was recovered from the GST and GST-xICln bandshift reactions (Fig. 5c, bottom panel). Thus, in agreement with the previous experiment, pICln inhibited Sm protein binding to U RNA. Finally, the anti-pICln antibody had no effect on the electrophoretic mobility of the Sm-containing complex (Fig. 5c, lanes 7, 8, and 11), consistent with our finding that pICln was not associated with snRNPs (Fig. 3).
pICln inhibits U RNA nuclear transport in Xenopus oocytes.
Since snRNP biogenesis requires Sm core domain formation on U RNA and pICln inhibits this assembly, we predicted that pICln should influence snRNP biogenesis in vivo. To test this hypothesis, we studied the nuclear import of U RNA, the final step of snRNP biogenesis. We injected oocytes in the cytoplasm with GST-xICln and then with 32P-labeled, in vitro-transcribed U snRNAs. We measured U RNA nuclear import by quantitating labeled U RNA recovered separately from nuclear and cytoplasmic fractions. In control oocytes injected with buffer or GST, 25 to 30% of the injected RNA was imported into the nucleus (Fig. 6a). In contrast, following injection of GST-xICln, nuclear import of U RNA was significantly inhibited (Fig. 6a). The effect of pICln varies for the different U RNAs (Fig. 6b), with transport of U1 and U5 most inhibited. U2 and U4 nuclear import was not substantially inhibited by pICln. Thus, the degree to which pICln inhibited U RNA interaction with Sm proteins (Fig. 5b) correlated with the ability of pICln to inhibit nuclear import (Fig. 6b), suggesting that inhibition of nuclear import by pICln is due at least in part to inhibition of Sm core domain assembly.
FIG. 6.
pICln inhibits nuclear import of U RNA. (a) Oocytes were injected with buffer, GST, or GST-xICln and then with 32P-labeled, in vitro-transcribed U1, U2, U4, and U5 RNAs. RNA was recovered from nuclear (N) and cytoplasmic (C) fractions and analyzed by denaturing polyacrylamide gel electrophoresis. Each sample represents a pool of five oocytes. The lane labeled control contains the mixture of RNAs prior to injection. U RNAs imported into the nucleus were slightly smaller than the injected U RNAs due to 3′ end trimming (27). The standard is a labeled RNA that was added in equal amounts to the nuclear and cytoplasmic fractions following dissection to ensure equal recovery during processing of the samples. (b) Quantitation of the effect of GST-xICln on individual U RNAs. The amount of each U RNA in the nuclear and cytoplasmic fractions of GST or GST-xICln injected oocytes was determined by analyzing gels, such as shown in panel a, on a phosphorimager. Results are displayed as the percentage of nuclear import of each U RNA in GST-xICln injected oocytes relative to GST injected oocytes. Each column represents the average (error bar, standard deviation) of two independent experiments.
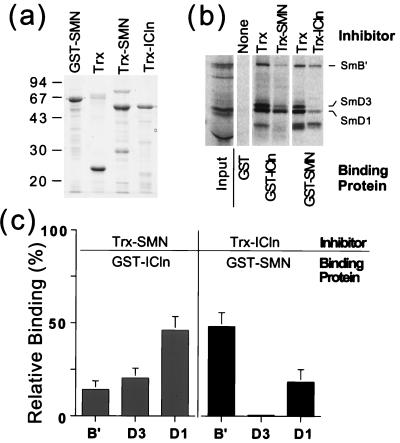
pICln inhibits SMN binding to Sm proteins.
We have shown that pICln binds to Sm proteins and inhibits their assembly on U RNA. The protein SMN also binds Sm proteins and was suggested to mediate their assembly on U RNA (5, 20). SMN and pICln do not coexist in a single complex with Sm proteins, since an anti-SMN antibody failed to detect SMN in pICln immunoprecipitates (data not shown). We hypothesized that a complex containing all three proteins was not detected because pICln and SMN binding to Sm protein was mutually exclusive. To test this hypothesis, we asked whether pICln binding to Sm proteins could prevent these proteins from interacting with SMN. For this experiment, we measured in vitro binding of in vitro-translated Sm proteins to SMN fused to GST (GST-SMN) in the presence of pICln competitor (a thioredoxin-ICln fusion protein, Trx-ICln). SmB′, SmD1, and SmD3 binding to SMN was significantly decreased by Trx-ICln compared to an equal amount of Trx alone (Fig. 7). This inhibitory effect of pICln was most pronounced for SmD1 and SmD3, which reduced binding to SMN by 82 and 99%, respectively. We also performed the reverse experiment, in which we measured the binding of in vitro-translated Sm proteins to pICln (GST-ICln) in the presence of SMN competitor (Trx-SMN). SMN inhibited the binding of SmB′, SmD1, and SmD3 by pICln. Since in both experiments we used nearly saturating amounts of the inhibitory proteins but the inhibitory effect was not complete and not identical for pICln and SMN, the inhibitory effect may not reflect simple competition for Sm binding. These results, together with the absence of SMN in pICln immunoprecipitates, lead us to conclude that pICln interferes with SMN binding to Sm proteins in vivo and in vitro.
FIG. 7.
pICln interferes with SMN binding to Sm proteins. (a) Fusion proteins used, separated on an SDS–10% polyacrylamide gel and stained with Coomassie blue. (b) In vitro-transcribed and -translated, 35S-labeled SmB′, SmD1, and SmD3 were preincubated with 12 μg of either Trx, Trx-ICln, or Trx-SMN. The mixture was then incubated with 1 μg of either GST, GST-SMN, or GST-ICln bound to glutatione beads, and the beads were vigorously washed. Bound Sm proteins were analyzed by SDS-polyacrylamide gel electrophoresis. (c) Quantitation of the results in panel b. A phosphorimager was used to measure the amount of each Sm protein bound in the presence of Trx or inhibitor protein (Trx-SMN or Trx-ICln). Relative binding was calculated as the percent of Sm protein bound in the presence of inhibitor compared to the amount bound in the presence of Trx (i.e., binding in the presence of Trx was taken as 100%). Each column represents the average (error bar, standard deviation) of five to six independent measurements.
DISCUSSION
We have demonstrated that pICln binds to Sm proteins by several independent methods. First, microsequencing of proteins purified by affinity to pICln identified two pICln-binding proteins as SmB/B′ and SmD3. Second, immunoprecipitation of Sm proteins coimmunoprecipitated pICln. Third, a bait containing pICln extracted SmD3 from a cDNA library in a yeast two-hybrid screen. Fourth, purified GST-ICln bound to in vitro-translated SmB′, SmD1, and SmD3. Finally, proteins which coimmunoprecipitated with pICln from MDCK cell cytoplasmic extracts had electrophoretic mobilities appropriate for SmB/B′ and SmD1 and SmD3. Although these experiments do not define which Sm proteins directly bind to pICln, they clearly demonstrate that Sm proteins and pICln form a complex. Furthermore, this interaction is likely to be functionally important, since immunodepletion experiments showed that in MDCK cells the majority of cytoplasmic Sm protein (or at least SmB/B′) is bound to pICln.
Binding of Sm proteins to pICln inhibited their assembly on U RNA to form the Sm core domain, as determined by immunoprecipitation and mobility shift assays. By inhibiting Sm core domain assembly on U RNA, high concentrations of pICln interfered with snRNP nuclear import in Xenopus oocytes. At the maximal GST-xICln concentration tested (5.5 μM), there was a 2.5-fold inhibition of nuclear import. Although this concentration of pICln was 25-fold higher than the normal oocyte pICln concentration (0.2 μM [15]), this was expected given the high concentration of Sm proteins in the oocyte cytoplasm. In previous studies, 1.5 to 15 μM U1 RNA was required to completely bind the endogenous cytoplasmic pool of free Sm proteins (4). Using this as a minimal estimate of the oocyte cytoplasmic Sm protein concentration, the concentration of pICln necessary for effective inhibition of nuclear import was reasonable if inhibition required stoichiometric binding of pICln to Sm proteins.
Although lack of U RNA, rather than the presence of pICln, is likely the primary factor which regulates snRNP assembly in oocytes (36), the nuclear import assay demonstrates clearly a functional consequence of pICln binding to Sm proteins. In contrast to oocytes, in the majority of mammalian cells the cytoplasmic Sm concentration is low and pICln is relatively abundant, and the majority of cytoplasmic Sm is bound to pICln. Under these conditions, pICln may play an important role in the regulation of snRNP assembly.
Much has been learned recently about the assembly pathway of the Sm core domain. The Sm proteins have been found in three stable RNA-free complexes (D1-D2, E-F-G, and D3-B/B′) (5, 7, 17, 33). The D1-D2 and E-F-G Sm protein complexes can together form a stable complex with U RNA. This subcore particle then binds the D3-B/B′ complex to form the complete core domain (33). Our finding that pICln binding to complexes containing SmB/B′, D1, and D3 interfered with the ability of Sm proteins to associate with U RNA is consistent with this model.
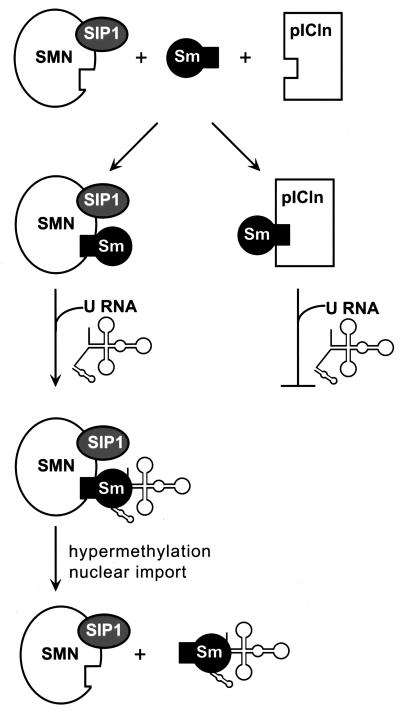
We have also found that pICln binding to Sm proteins inhibited their association with SMN protein. This protein forms a complex with the protein SIP1, and the SMN-SIP1 complex binds to SmB/B′, D1-3, and E (20). Antibodies directed against SIP1 blocked assembly of the Sm core domain on U RNA, while anti-SMN antibodies enhanced this process. These results implicate the SMN-SIP1 complex in Sm core domain assembly and suggest that by binding Sm proteins, the complex may facilitate their binding to U RNA (5). In combination with our results, these data lead us to propose a model in which pICln inhibits Sm protein assembly on U RNA, at least in part by inhibiting Sm protein interaction with SMN (Fig. 8). Consistent with this model, U1 and U5 RNAs efficiently coimmunoprecipitated with SIP1 or SMN and were most inhibited by pICln, while U2 and U4 RNAs were weakly coimmunoprecipitated with SIP1 or SMN (5) and were relatively insensitive to pICln (Fig. 5 and 6). Although this model is compatible with the existing data, alternative models are possible. For instance, we have depicted SMN as an integral component of the assembly pathway, whereas it may actually be a regulatory element. SMN and pICln both influence snRNP biogenesis at the level of Sm core domain assembly, but a role for these proteins in regulating other steps in the biogenesis pathway has not been excluded.
FIG. 8.
Model of pICln and SMN regulation of Sm protein assembly on U RNA. pICln is depicted as inhibiting Sm core domain assembly and U RNA nuclear import by preventing Sm interaction with SMN. Hypermethylation and nuclear import of U RNA occur after Sm core domain assembly, although the timing of these events relative to SMN dissociation is not known.
In addition to promoting snRNP biogenesis, SMN may also be essential for proper spliceosome function, since a dominant-negative SMN mutant inhibits pre-mRNA splicing in vitro and causes dramatic nuclear snRNP reorganization in vivo (32). The inhibitory effect of the dominant-negative mutant on in vitro splicing was observed only when the SMN mutant was preincubated with splicing extract prior to addition of pre-mRNA (32). One interpretation of these results is that splicing results in snRNP rearrangement to an inactive form and that SMN is necessary for the regeneration of functional snRNPs. Since pICln is also present in the nucleus, pICln could potentially regulate snRNP recycling by modulating the interaction of SMN with snRNPs. In keeping with the participation of pICln in a critical cellular function, loss of pICln in mice results in embryonic lethality between 3.5 and 7.5 days postcoitus, and embryonic stem cells lacking both pICln alleles cannot be generated in tissue culture (35).
In summary, we have described a novel interaction between pICln and several Sm proteins, and we have demonstrated that this interaction inhibits association of Sm proteins with SMN, Sm protein binding to U RNA, and snRNP biogenesis. We have presented a model that accounts for these observations. Experiments are in progress to test this model and to determine the mechanism by which loss of pICln leads to embryonic lethality.
ACKNOWLEDGMENTS
W.T.P. and G.B.K. contributed equally to this work.
We thank John Leszyk for his peptide microsequencing expertise and Kevin Wickman for helpful discussions. I. Mattaj kindly provided T7-U RNA in vitro transcription plasmids. G. Dreyfuss generously provided us with anti-SMN antibody.
REFERENCES
- 1.Buyse G, Voets T, Tytgat J, Degreef C, Droogmans G, Nilius B, Eggermont J. Expression of human pICln and CIC-6 in Xenopus oocytes induces an identical endogenous chloride conductance. J Biol Chem. 1997;272:3615–3621. doi: 10.1074/jbc.272.6.3615. [DOI] [PubMed] [Google Scholar]
- 2.Clapham D E. The list of potential volume-sensitive chloride channels continues to swell (and shrink) J Gen Physiol. 1998;111:623–624. doi: 10.1085/jgp.111.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeRobertis E M. Nucleocytoplasmic segregation of proteins and RNAs. Cell. 1983;32:1021–1025. doi: 10.1016/0092-8674(83)90285-4. [DOI] [PubMed] [Google Scholar]
- 4.Fischer U, Sumpter V, Sekine M, Satoh T, Luhrmann R. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 1993;12:573–583. doi: 10.1002/j.1460-2075.1993.tb05689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 6.Fisher D E, Conner G E, Reeves W H, Wisniewolski R, Blobel G. Small nuclear ribonucleoprotein particle assembly in vivo: demonstration of a 6S RNA-free core precursor and posttranslational modification. Cell. 1985;42:751–758. doi: 10.1016/0092-8674(85)90271-5. [DOI] [PubMed] [Google Scholar]
- 7.Fury M G, Zhang W, Christodoulopoulos I, Zieve G W. Multiple protein: protein interactions between the snRNP common core proteins. Exp Cell Res. 1997;237:63–69. doi: 10.1006/excr.1997.3750. [DOI] [PubMed] [Google Scholar]
- 8.Gilbreth M, Yang P R, Wang D, Frost J, Polverino A, Cobb M H, Marcus S. The highly conserved skb1 gene encodes a protein that interacts with Shk1, a fission yeast Ste20/PAK homolog. Proc Natl Acad Sci USA. 1996;93:13802–13807. doi: 10.1073/pnas.93.24.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamm J, Darzynkiewicz S, Tahara M, Mattaj I W. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartitie nuclear targeting signal. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- 10.Hamm J, Mattaj I W. An abundant U6 snRNP found in germ cells and embryos of Xenopus laevis. EMBO J. 1989;8:4179–4187. doi: 10.1002/j.1460-2075.1989.tb08603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M, Luhrmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–4126. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 14.Krapivinsky G, Pu W, Wickman K, Krapivinsky L, Clapham D E. pICln binds to a mammalian homolog of a yeast protein involved in regulation of cell morphology. J Biol Chem. 1998;273:10811–10814. doi: 10.1074/jbc.273.18.10811. [DOI] [PubMed] [Google Scholar]
- 15.Krapivinsky G B, Ackerman M J, Gordon E A, Krapivinsky L, Clapham D E. Molecular characterization of a swelling-induced chloride conductance regulatory protein, pICln. Cell. 1994;76:439–448. doi: 10.1016/0092-8674(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 16.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;89:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 17.Lehmeier T, Raker V, Hermann H, Luhrmann R. cDNA cloning of the Sm proteins D2 and D3 from human small nuclear ribonucleoproteins: evidence for a direct D1-D2 interaction. Proc Natl Acad Sci USA. 1994;91:12317–12321. doi: 10.1073/pnas.91.25.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmeier T, Foulaki K, Luhrmann R. Evidence for three distinct D proteins, which react differentially with anti-Sm autoantibodies, in the cores of the major snRNPs U1, U2, U4/U6 and U5. Nucleic Acids Res. 1990;18:6475–6484. doi: 10.1093/nar/18.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lerner E A, Lerner M R, Hardin J A, Janeway C A, Steitz J A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 21.Lührmann R, Kastner B, Bach M. Structure of snRNPs and their role in pre-mRNA splicing. Biochim Biophys Acta. 1990;1087:265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 22.Mattaj I. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 23.Mattaj I W. U snRNP assembly and transport. In: Birnstiel M L, editor. Structure and function of major and minor small nuclear ribonucleoprotein particles. Heidelberg, Germany: Springer-Verlag; 1988. pp. 100–114. [Google Scholar]
- 24.Mattaj I W, De Robertis E M. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985;40:111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- 25.Musch M W, Davis-Amaral E M, Vandenburgh H H, Goldstein L. Hypotonicity stimulates translocation of ICln in neonatal rat cardiac myocytes. Pflugers Arch. 1998;436:415–422. doi: 10.1007/s004240050651. [DOI] [PubMed] [Google Scholar]
- 26.Neer E J, Schmidt C J, Nambudripad R, Smith T F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 27.Neuman De Vegvar H E, Dahlberg J E. Nucleocytoplasmic transport and processing of small nuclear RNA precursors. Mol Cell Biol. 1990;10:3365–3375. doi: 10.1128/mcb.10.7.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okada H, Ishii K, Nunoki K, Taira N. Cloning of a swelling-induced chloride current related protein from rabbit heart. Biochim Biophys Acta. 1995;1234:145–148. doi: 10.1016/0005-2736(95)00015-u. [DOI] [PubMed] [Google Scholar]
- 29.Palacios I, Hetzer M, Adam S A, Mattaj I W. Nuclear import of U snRNPs requires importin β. EMBO J. 1997;16:6783–6792. doi: 10.1093/emboj/16.22.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang I-H, Sternweis P C. Isolation of the α subunits of GTP-binding regulatory proteins by affinity chromatography with immobilized βγ subunits. Biochemistry. 1989;86:7814–7818. doi: 10.1073/pnas.86.20.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulmichl M, Li Y, Wickman K, Peralta E, Clapham D. New mammalian chloride channel identified by expression cloning. Nature. 1992;356:238–241. doi: 10.1038/356238a0. [DOI] [PubMed] [Google Scholar]
- 32.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 33.Raker V A, Plessel G, Luhrmann R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 1996;15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- 34.Van Dam A, Winkel I, Zijlstra-Baalbergen J, Smeenk R, Cuypers H T. Cloned human snRNP proteins B and B′ differ only in the carboxy-terminal part. EMBO J. 1989;8:3853–3860. doi: 10.1002/j.1460-2075.1989.tb08563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickman, K., W. T. Pu, and D. E. Clapham. Unpublished data.
- 36.Zeller R, Nyffenegger T, DeRobertis E M. Nucleocytoplasmic distribution of snRNPs and stockpiled snRNA-binding proteins during oogenesis and early development in Xenopus laevis. Cell. 1983;32:425–434. doi: 10.1016/0092-8674(83)90462-2. [DOI] [PubMed] [Google Scholar]