Abstract
The Saccharomyces cerevisiae RAD27 gene encodes the yeast homologue of the mammalian FEN-1 nuclease, a protein that is thought to be involved in the processing of Okazaki fragments during DNA lagging-strand synthesis. One of the predicted DNA lesions occurring in rad27 strains is the presence of single-stranded DNA of the template strand for lagging-strand synthesis. We examined this prediction by analyzing the terminal DNA structures generated during telomere replication in rad27 strains. The lengths of the telomeric repeat tracts were found to be destabilized in rad27 strains, indicating that naturally occurring direct repeats are subject to tract expansions and contractions in such strains. Furthermore, abnormally high levels of single-stranded DNA of the templating strand for lagging-strand synthesis were observed in rad27 cells. Overexpression of Dna2p in wild-type cells also yielded single-stranded DNA regions on telomeric DNA and caused a cell growth arrest phenotype virtually identical to that seen for rad27 cells grown at the restrictive temperature. Furthermore, overexpression of the yeast exonuclease Exo1p alleviated the growth arrest induced by both conditions, overexpression of Dna2p and incubation of rad27 cells at 37°C. However, the telomere heterogeneity and the appearance of single-stranded DNA are not prevented by the overexpression of Exo1p in these strains, suggesting that this nuclease is not simply redundant with Rad27p. Our data thus provide in vivo evidence for the types of DNA lesions predicted to occur when lagging-strand synthesis is deficient and suggest that Dna2p and Rad27p collaborate in the processing of Okazaki fragments.
Due to the conserved 5′-to-3′ polarity of all DNA polymerases known to date, newly synthesized DNA at the replication fork is assembled in a discontinuous (lagging) and a continuous (leading) strand. All replicative polymerases also require a primer to efficiently start synthesizing DNA. This priming of DNA synthesis by a short RNA molecule presumably only occurs once, at the origin of replication, for leading-strand synthesis, but priming is required throughout the synthesis of the lagging strand. Thus, the generation of a continuous DNA strand for lagging-strand synthesis is dependent on the activities of a primase, DNA polymerases α, δ, and/or ɛ with accessory proteins such as PCNA and RF-C, as well as activities involved in removal of the RNA primer and resealing of the resulting gap (for reviews, see references 3 and 4).
In the yeast Saccharomyces cerevisiae, as in mammalian cells, removal of the RNA primer is thought to be mediated by an endo/exonucleolytic activity called FEN-1 (flap endonuclease or 5′-exonuclease 1 [20, 21]), presumably aided by RNase H1 (18, 24, 36, 48, 50; for reviews, see references 17, 29, and 32). Recent biochemical analyses of the FEN-1 nuclease indicate that the FEN-1 protein interacts with, and its activity is stimulated by, PCNA (31, 56). The yeast homologue of the mammalian FEN-1 enzyme is encoded by the RAD27/RTH1 gene and belongs in the RAD6 epistasis group (37). Yeast cells lacking Rad27p are sensitive to alkylating agents like methyl methanesulfonate but not to ionizing radiation (37). Mutant strains also display high levels of instability of simple repetitive DNA (14, 26, 28, 41) and a temperature-sensitive growth defect (37, 45). Finally, rad27Δ strains incubated at the restrictive temperature exhibit a checkpoint-dependent cell cycle arrest in late S/G2 (49). These phenotypes, as well as the molecular analysis of the mutation spectra occurring in rad27Δ strains, suggest a crucial deficiency in DNA replication and repair (28, 47). Since mutations in RAD27, when combined with mutations in genes belonging to the RAD52 epistasis group, yield inviable cells, it has been proposed that the lesions occurring in rad27Δ cells are predominantly repaired by a double-strand break repair mechanism (47). Recent studies have also shown that an essential yeast replicative helicase, Dna2p, interacts genetically and biochemically with Rad27p (6, 7). Cells harboring temperature sensitivity alleles of DNA2 arrest in G2/M with a 2C DNA content at the restrictive temperature and contain DNA of low molecular weight (13). Taken together, these results have been interpreted to suggest that Rad27p and Dna2p act together in lagging-strand processing (3, 4). Specifically, based also on the fact that the Rad27p nuclease loads onto, and acts much more efficiently on, a displaced-5′-end single strand, it has been proposed that Dna2p displaces the 5′ ends at the RNA-DNA junction of Okazaki fragments, resulting in a 5′ flap structure that could be removed by the endonucleolytic activity of Rad27p (36). However, direct in vivo evidence for such a mode of action is still lacking.
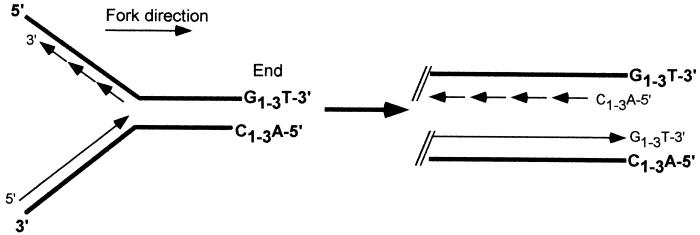
Telomeres, the ends of eukaryotic chromosomes, are essential for chromosome integrity: they protect chromosome ends from degradation and random fusion events (34, 40), and they ensure the complete replication of the chromosome (19, 51, 57). Chromosomes in S. cerevisiae end in 300 bp of short, heterogeneous sequence repeats, commonly abbreviated as C1–3A or TG1–3 (42). These yeast telomeric sequence repeats are similar to those of most other eukaryotes in having clusters of G residues in the strand running 5′ to 3′ from the center toward the end of the DNA molecule (the G-rich strand) (53). Due to the base disparity between the two strands and the polarity of DNA synthesis, the G-rich strand will always be synthesized by leading-strand synthesis and the C-rich strand will always be synthesized by lagging-strand synthesis (Fig. 1). Analyzing the DNA replication intermediates occurring during telomere replication, we have previously shown that chromosome ends in yeast acquire transient ≥30-base single-stranded extensions of the G-rich strand (G tails) (52, 54). Furthermore, these G tails could be detected in cells that were devoid of telomerase, suggesting that the bulk of the G tails are generated in a telomerase-independent fashion (11).
FIG. 1.
Nucleotide composition of templating and newly synthesized strands on telomeric repeats. Due to the conserved polarity of the telomeric repeats on all telomeres, the templating strand for lagging-strand synthesis is always the G-rich strand and the templating strand for leading-strand synthesis is the C-rich strand. Thick lines with nucleotides in boldface, templating strands; thin lines, newly synthesized strands. Note that the identity of the last nucleotide at the end of each strand is unknown; the designation drawn out here is given for simplicity.
In order to understand all the activities required for telomere processing, we are interested in determining how the single-stranded G tails occurring during telomere replication are generated and processed. Since the Rad27p/Dna2p proteins have been proposed to process the 5′ ends of newly generated Okazaki fragments, we investigated the involvement of these proteins in the generation of telomere replication intermediates. If these proteins were involved in lagging-strand synthesis in vivo, we expected aberrant telomere processing and/or a deficiency in synthesizing the new C-rich strands on telomeres in rad27Δ strains. Indeed, terminal single-stranded G-rich strands do occur at abnormally high levels on terminal restriction fragments (TRFs) derived from rad27Δ cells incubated at the restrictive temperature. Conversely, single-stranded DNA of the C-rich strand, which would be indicative of failing leading-strand synthesis, remained undetectable as in wild-type cells. In the same DNA samples, there were no detectable single-stranded Y′ sequences for either strand. The appearance of the single-stranded G tails correlated with an expansion of the heterogeneity of the lengths of the terminal repeat tracts, indicative of abnormal repeat lengthening and shortening in rad27Δ cells. Overexpression of a full-length Dna2p resulted in cell cycle arrest. This arrest in cell growth can be suppressed by cooverexpressing Rad27p in the same cells, supporting the notion of a functional interaction between Rad27p and Dna2p. Most significantly, by analysis of the chromosomal termini derived from cells in which Dna2p was overexpressed, a transient increase in single-stranded G strands was detected. We also show that while overexpression of the yeast exonuclease Exo1p can suppress the temperature sensitivity and mutator phenotypes of rad27Δ cells (47), it does not suppress the appearance of single-stranded DNA in these cells. Our results thus provide in vivo physical evidence for the types of lesions generated by an absence of Rad27p and suggest that Rad27p and Dna2p collaborate in proper Okazaki fragment processing.
MATERIALS AND METHODS
Strains and plasmids.
Yeast strains used were SX46A (MATa RAD27 ade2 his3-532 trp1-289 ura3-52), SX46A rad27Δ::URA3 (MATa rad27Δ::URA3 ade2 his3-532 trp1-289 ura3-52) (37), 7399-3-1 (MATα trp1 leu2 ura3 his3) (obtained by J. Aron and T. Formosa), and MW35a (MATa ade2 ade5 canr 1 cyhr 2 lys5 ura3 trp1 leu2-3,112 his3-Δ200 Gal++).
The 2μm DNA2 plasmid, pTN3, was made by inserting a 5,709-bp EcoRI-PstI fragment containing the DNA2 gene into YEplac195 (16). The pGal-DNA2 plasmid was constructed by using a ScaI-PstI fragment containing the complete DNA2 open reading frame. The ScaI site upstream of DNA2 was converted to a BamHI site, and this fragment was inserted into the Gal-inducible pJS227 vector. pJS227 is YEplac195 (16) containing the Gal1-10 promoter (27). Plasmids pTN3 and pGAL-DNA2 were kindly provided by J. Aron and T. Formosa. pGAL-vector is pGAL-DNA2 from which a 5.9-kb BamHI-PstI fragment was removed and the vector was religated. The 2μm RAD27 plasmid, pR2, was constructed by inserting the 2,500-bp SphI-EcoRI fragment of pMR92393 (37) into EcoRI-SmaI-digested pRS424 (10). The 2μm EXO1 plasmid, pE1, was made by inserting a 2,470-bp NotI-XhoI fragment of pRW200 into NotI-XhoI-digested pRS423 DNA (10). In order to amplify the yeast EXO1 gene from wild-type genomic DNA, we used the following primers for PCR: DF 5′-CTCCTCGAGTCTTTATAGGGCATTATTTGTAC-3′ (the underlined bases are complementary to positions −288 to −265 relative to the EXO1 translational start site, and the 5′ extension contains an XhoI site) and DR 5′-CTCTCTAGATCTTGTCTTGAGGCATTTCG-3′ (the underlined bases are complementary to positions +2585 to +2566 relative to the EXO1 translational start site, and the 5′ extension contains an XbaI site). We used 30 cycles of PCR at 94°C for 1 min, 56°C for 1 min, and 72°C for 3 min, followed by a 15-min 72°C soaking period. The PCR product was digested with XhoI and XbaI and cloned into pRS316 (43) to generate plasmid pRW200.
All plasmids were propagated in bacteria by using standard Escherichia coli strains and growth conditions (39). Yeast cells were transformed by a modification (15) of the lithium acetate method (25) and grown in standard yeast media (38, 58).
DNA isolation and analysis.
For Fig. 2 to 5 and 8, yeast strains were grown in synthetic complete (SC) medium at the permissive temperature for SX46A rad27Δ::URA3 (23°C) to mid-logarithmic phase (optical density at 660 nm [OD660] = 0.6), then shifted to a semipermissive temperature (30°C) or to the restrictive temperature (37°C) for the indicated times. For Fig. 7, the MW35a cells containing pGAL-DNA2 were grown in SC-Ura medium containing glycerol (2%) and lactate (2%) at 30°C to mid-logarithmic phase. Galactose (final concentration, 2%) was then added to induce expression of DNA2, and the cells were incubated for the indicated times. Total genomic DNA from cells was isolated by a modified glass bead procedure (23, 54). Treatment of genomic DNA with E. coli exonuclease I and mung bean nuclease was carried out as described elsewhere (54).
FIG. 2.
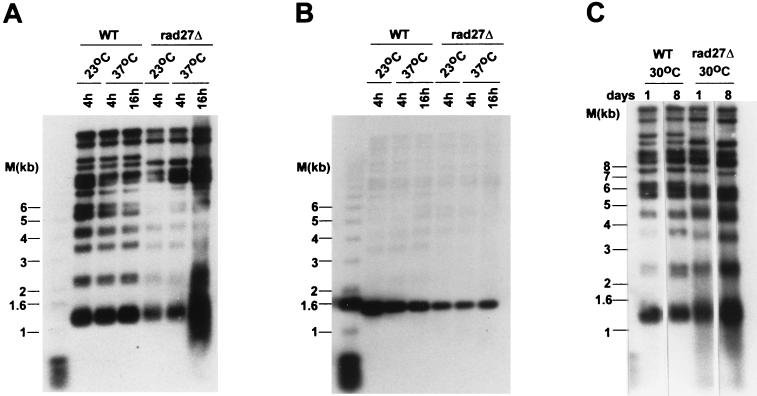
Telomeric repeat instability in rad27Δ strains. (A) Genomic DNA isolated from RAD27 (WT) or rad27Δ cells that were incubated at 23 or 37°C for the indicated times was digested with the restriction endonuclease XhoI and analyzed by Southern blotting. The probe used was randomly labeled telomeric repeat DNA. Y′ TRFs migrate at about 1.3 kb, and some of the other bands correspond to non-Y′ TRFs. M, end-labeled 1-kb ladder DNA serving as a size standard. (B) The same blot as in panel A, rehybridized to a probe specific for CEN4 sequences. (C) The same experiment as in panel A, except that the strains were grown at 30°C. Note that rad27Δ cells do grow at this temperature. Every day, an aliquot of the cultures was diluted into fresh medium and the cells were regrown to stationary phase (days of culturing are indicated on top of the gel). From cell density measurements, it was calculated that the cultures had grown for approximately 100 generations on day 8 (data not shown).
FIG. 5.
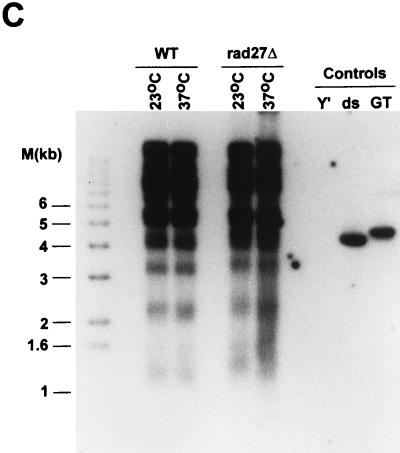
The single strandedness of the telomeric G-rich strand does not extend into neighboring Y′ sequences. DNAs derived from RAD27 (WT) or rad27Δ cells incubated overnight at the indicated temperatures were digested with XhoI and analyzed as in Fig. 3 and 4. (A) The probe consisted of an end-labeled Y′ oligonucleotide, detecting the same strand as the G-rich telomeric repeat strand, and the sequences are about 500 bp from the Y′-telomeric repeat sequences boundary. (B) The same gel as in panel A, rehybridized to the telomeric CA probe. Since the gel was not denatured between probings, both single-stranded controls are visible. (C) The gel shown in panels A and B was then denatured and rehybridized to the telomeric CA oligonucleotide. During the repeated hybridizations and washings, some of the smaller DNA fragments (below 1.2 kb) had diffused out of the gel (note the losses of the DNA size standard bands of 1 kb and less). Thus, the signal for the 1.3-kb Y′ TRFs is somewhat weaker than expected for this gel. Controls and DNA size standards are as in Fig. 3 and 4.
FIG. 8.
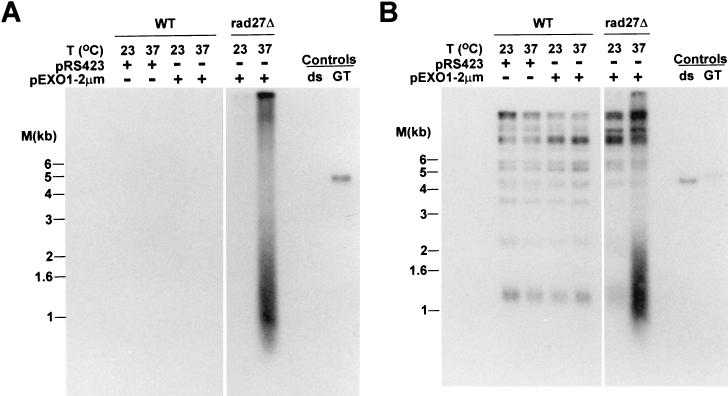
Overexpression of Exo1p does not prevent the formation of single-stranded DNA in rad27Δ cells. RAD27 (WT) or rad27Δ cells were transformed with either pRS423 (empty vector) or pE1 (labeled pExo1-2μm), grown in selective media, and incubated overnight at the indicated temperatures. DNA was isolated, digested with XhoI, and analyzed as in Fig. 3 through 5. (A) Nondenatured gel probed with the CA oligonucleotide. (B) The DNA in the gel shown in panel A was denatured and rehybridized to the same probe. Controls and molecular size standards are as in Fig. 7.
FIG. 7.
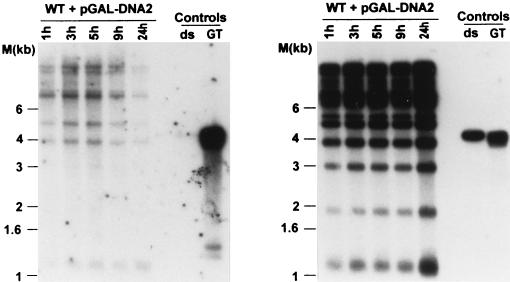
Induction of DNA2 expression leads to a transient increase of single-stranded telomeric G-rich DNA. MW35a cells containing pGAL-DNA2 were pregrown in SC-Ura media containing glycerol and lactate. Expression of DNA2 was then induced by the addition of galactose, and DNA was prepared from the culture at the indicated times after galactose addition. These DNAs were then digested with XhoI and analyzed by nondenaturing in-gel hybridization (left) using an end-labeled CA oligonucleotide as a probe. After denaturation of the DNA, the gel was rehybridized to the same probe as a control (right). Controls and molecular size standards are as in Fig. 2 through 5.
Agarose gel techniques, Southern blot transfer to a nylon membrane, and hybridization conditions were as described previously (54). The nondenaturing in-gel hybridizations were carried out as described in reference 11. DNAs used as probes were a 300-bp fragment containing 280 bp of telomeric repeats derived from pYLPV (54), a 1.4-kb XhoI-XhoI fragment derived from yeast chromosomal CEN4 sequences (55), a 22-mer of the sequence 5′-CCCACCACACACACCCACACCC-3′ (referred to as the CA oligonucleotide [11]), a 22-mer of the sequence 5′-GGGTGTGGGTGTGTGTGGTGGG-3′ (referred to as the GT oligonucleotide), and a 30-mer of the sequence 5′-CCCTCGTGTTATCTGCAGCGAGAACTTCAA-3′ (referred to as the Y′ oligonucleotide). A heat-denatured 0.6-kb KpnI-KpnI fragment of Y′ sequences (33) cloned in the KpnI site of pVZ1 (22) was used as a Y′ control. Single-stranded DNA from pCA75 and pGT75 (54) were obtained by standard procedures using a helper phage (39) to produce CA and GT controls, respectively. The double-stranded control was obtained by the linearization of pMW55 with BamHI. The pMW55 plasmid was made by inserting 55 bp of duplex telomeric repeat DNA into the EcoRV site of pRS303 (43).
RESULTS
Increased heterogeneity of telomeric repeats in rad27Δ cells at the restrictive temperature.
Previous work showed an elevated instability of di- and trinucleotide repeats in yeast strains deficient for Rad27p (14, 26, 28, 41). Telomeres, the ends of the chromosomes, represent one genomic locus that naturally consists of short direct repeats in many eukaryotes. We were therefore interested to know whether telomeric repeat maintenance was affected by an absence of Rad27p. Telomeric repeat length in yeast can be assessed by digesting genomic DNA with XhoI, which generates diagnostic TRFs of approximately 1.3 kb due to the conservation of such a site in the subtelomeric Y′ element that is present on most telomeres (Y′ TRFs) (9). However, about one-third of the telomeres do not harbor a Y′ element, and the TRFs derived from these telomeres (non-Y′ TRFs) will be larger. When the TRFs of RAD27 strains grown at 23°C and shifted to 37°C were analyzed, no apparent change in the TRF lengths could be discerned, as expected (Fig. 2A). However, after rad27Δ strains were shifted to the nonpermissive temperature, the TRFs displayed very heterogeneous sizes: Y′ TRFs ranged from 0.9 to 1.6 kb, and the bands for non-Y′ TRFs were similarly broadened (Fig. 2A). This increase in size heterogeneity did not require extensive outgrowth, as these cells, when incubated at 37°C, arrested as large-budded cells after 2 to 3 generations and did not grow further (data not shown). As a control, the Southern blot shown in Fig. 2A was stripped of the probe and rehybridized to a probe specific for the CEN4 region (see Materials and Methods). This rehybridization yielded the expected distinct band at approximately 1.4 kb for all DNAs, including the DNA derived from rad27Δ cells incubated at the restrictive temperature (Fig. 2B). At 30°C, cells with a deletion of RAD27 grow slowly and display a TRF heterogeneity that is intermediate between those of wild-type and rad27Δ cells incubated at 37°C. However, this intermediate heterogeneity did not change significantly upon further outgrowth of the cells at 30°C for at least 100 generations (Fig. 2C). Thus, in rad27Δ cells incubated at the semipermissive and restrictive temperatures, telomeric repeat length is destabilized, resulting in very heterogeneous TRFs. These results suggest that in rad27Δ cells, the natural yeast telomeric repeats are subject to tract expansions and contractions similar to those reported for artificially inserted short repeated elements in the yeast genome (14, 26, 28, 41).
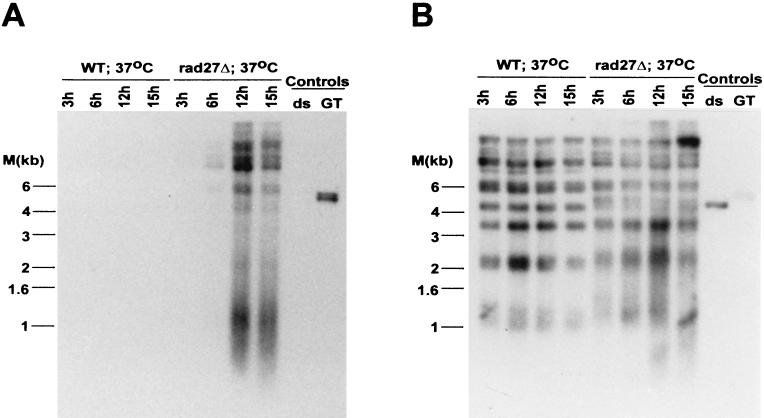
Accumulation of single-stranded DNA of the G-rich telomeric repeats is detected in rad27Δ cells incubated at 37°C.
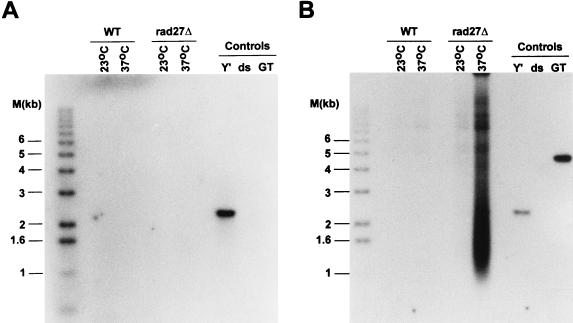
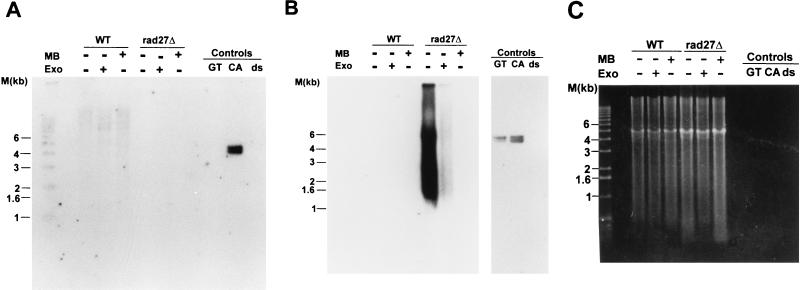
The specific types of mutations observed in rad27Δ cells are most easily rationalized if the Rad27p is involved in processing the 5′ ends of Okazaki fragments (28, 47). The homologous enzyme from mammalian cells will cleave a displaced 5′ strand (flap) in vitro, which is thought to be necessary to allow the ligation of two adjoining Okazaki fragments. The displaced 5′ end of the downstream Okazaki fragment could be generated by strand displacement synthesis from the upstream Okazaki fragment or could be the product of a helicase-mediated strand separation (3, 4). However, the 5′ end of the most distal Okazaki fragment initiated on telomeric repeat DNA cannot be displaced by strand displacement synthesis, as by definition there is no upstream Okazaki fragment that could initiate this process (see Fig. 1). If a helicase displaces the 5′ ends of Okazaki fragments for Rad27p cleavage, the most distal Okazaki fragment could be processed in the same way as the internal ones. In order to establish whether cells lacking Rad27p had a deficiency in telomeric repeat replication, TRFs derived from rad27Δ cells were analyzed for the appearance of single-stranded DNA. In wild-type cells, telomeres acquire single-stranded DNA of the G-rich strand (G tails) late in S phase but have no detectable single-stranded DNA during the rest of the cell cycle (52, 54). Moreover, single-stranded DNA of the C-rich strand always remained undetectable. Thus, when the TRFs derived from asynchronous wild-type cells were analyzed for G tails, only a very faint signal was detected (Fig. 3A). Similarly, rad27Δ cells grown at 23°C and shifted to 37°C for a short time (≤6 h) displayed only minor G tails (Fig. 3A). However, TRFs derived from rad27Δ cells incubated at 37°C for more than 6 h had single-stranded G-rich DNA that was easily detectable in our assay (Fig. 3A). The appearance of this single-stranded DNA correlated in time with the appearance of the repeat heterogeneity (Fig. 2 and 3B) and with the arrest of cell growth (data not shown). The single-stranded DNA detected in Fig. 3A could reflect gaps or could be a terminal G tail, as occurs on telomeres derived from wild-type cells. To distinguish between these possibilities, genomic DNA derived from rad27Δ cells that were incubated at the restrictive temperature was digested with E. coli exonuclease I, a 3′-end-specific single-stranded exonuclease (30), prior to TRF analysis. This treatment abolished the signal for the single-stranded DNA, as did a pretreatment with mung bean nuclease, a single-stranded-specific endonuclease (Fig. 4B). Neither of these treatments caused a general DNA degradation, and approximately the same amount of DNA was loaded in all lanes (Fig. 4C). Moreover, on this same DNA, no single-stranded DNA of the C-rich strand was detectable (Fig. 4A). Finally, a probe specific for Y′ sequences and located about 500 bp proximal to the Y′-to-telomeric repeat transition did not reveal any single-stranded DNA in this region (Fig. 5A). The probe used in Fig. 5A would detect the strand making the 3′ ends of the chromosomes (the G-rich strand on the telomeric repeats), but an analogous probing with a probe from the opposing strand did not detect any single-stranded DNA either (data not shown). Once again, however, the accumulation of single-stranded DNA of the G-rich strand was detected on the DNA derived from rad27Δ cells incubated at 37°C, when this gel was rehybridized to a telomeric C-strand probe (Fig. 5B), and all lanes contained approximately the same amount of DNA (Fig. 5C). Thus, the single-stranded DNA detected on the TRFs derived from rad27Δ cells incubated at the restrictive temperature is specific for the G-rich telomeric repeats and consists of terminal extensions. These results indicate that in rad27Δ cells incubated at the restrictive temperature, excessive telomeric G tails are generated and cannot be processed to normal chromosomal ends.
FIG. 3.
Appearance of single-stranded DNA on telomeres derived from rad27Δ cells. (A) Genomic DNA isolated from RAD27 (WT) or rad27Δ cells incubated at 37°C for the indicated times was digested with XhoI and analyzed by a nondenaturing in-gel hybridization procedure (11) with an end-labeled CA oligonucleotide as a probe. Double-stranded and linearized pMW55 served as a negative control (labeled ds), and single-stranded phagemid DNA containing yeast telomeric repeats of the G-rich strands served as a positive control (labeled GT) (see Materials and Methods). Molecular size standards are as in Fig. 2. (B) The DNA in the gel shown in panel A was denatured, and the same gel was rehybridized to the probe to show all the telomeric repeat-containing fragments.
FIG. 4.
The single-stranded telomeric repeats observed on TRFs derived from rad27Δ cells are specific for the G-rich strand and are terminal extensions. RAD27 (WT) or rad27Δ cells were pregrown at 23°C, then incubated overnight at 37°C, and DNA was isolated from both cultures. This DNA was then digested with mung bean nuclease (lanes labeled + for MB) or E. coli exonuclease I (lanes labeled + for Exo) or was mock treated (no enzyme; indicated with a minus sign). All the DNAs were then digested with XhoI and analyzed by nondenaturing in-gel hybridization as in Fig. 3. For the designation of the control DNAs, see Fig. 3 and Materials and Methods. (A) The gel was hybridized to the end-labeled GT oligonucleotide. (B) The gel shown in panel A was then rehybridized to the end-labeled CA oligonucleotide. Note that due to the fact that the gel was not denatured between the two probings, both the CA and GT single-stranded controls show positive signals after the second probing. (C) A photograph of the ethidium bromide-stained gel prior to the first hybridization is shown to demonstrate approximately equal loading of DNA in all lanes.
Overexpression of Dna2p leads to formation of G tails.
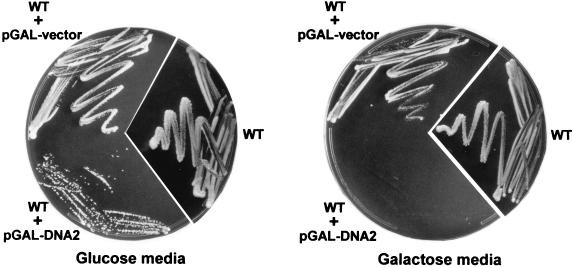
One possible interpretation of the above results could be that in the absence of the Rad27p nuclease, a DNA helicase responsible for the dissociation of the 5′ ends of newly generated Okazaki fragments inappropriately dissociates Okazaki fragments. Given the genetic and biochemical interactions of the Dna2p helicase with Rad27p, it was previously proposed that Dna2p could be the helicase performing this function (3, 4). However, DNA2 has been shown to be an essential gene, and a temperature sensitivity allele of DNA2, dna2-1, is synthetically lethal with a rad27Δ mutation (6). Thus, we were unable to test whether the defects caused by a rad27Δ mutation could be alleviated by mutations in DNA2. Alternatively, one might predict that overexpression of Dna2p in otherwise wild-type cells should yield the same phenotypes as an absence of Rad27p at high temperatures. To test this prediction, the entire coding sequence for Dna2p was inserted downstream of a galactose-inducible promoter and transformed into yeast grown on glucose media. While transformed cells form colonies on plates containing glucose, no growth is discernible on plates containing galactose (Fig. 6), indicating that overexpression of Dna2p yields inviable cells. In addition, a high-copy-number vector harboring the DNA2 gene did not yield any transformants, while control transformations with the empty vector did (Table 1). More significantly, however, cotransformation of the high-copy-number plasmid containing the DNA2 gene with one that contained the RAD27 gene resulted in viable transformants (Table 1). We conclude that overexpression of the Dna2p helicase induces growth arrest and that this arrest can be suppressed by a concomitant overexpression of Rad27p. It was therefore possible that the actual lesions to the chromosomes in cells overexpressing Dna2p were similar to those observed in rad27Δ cells grown at elevated temperatures. To test this hypothesis, DNA was isolated from cells in which DNA2 expression was induced from the pGAL-DNA2 plasmid. This DNA was then analyzed for telomeric end structure (Fig. 7). After 1 h of galactose-induced Dna2p expression, only a slight signal for single-stranded G strands was detectable (Fig. 7). However, after 5 h of induction, signals for single-stranded G strands were readily detectable. In cultures in which Dna2p was induced for longer than 5 h, cells stopped growing, with a concomitant decrease in the signal for single-stranded DNA (Fig. 7). After 24 h of induction, single-stranded DNA was barely detectable, even though somewhat more DNA was loaded in this lane than in the other lanes (Fig. 7). Although we do not know the reasons why the signals decrease upon prolonged Dna2p expression, single-stranded telomeric G-strand DNA is generated at elevated levels when Dna2p is overexpressed for a brief period, and the cells are still able to return to growth on glucose-containing plates. We conclude that induction of Dna2p overexpression results in increased amounts of telomeric G-strand single-stranded DNA, a feature that is also displayed in rad27Δ cells at elevated temperatures. We also examined the chromosomal end structure on DNA derived from cells that contained a temperature sensitivity allele of the DNA2 gene, dna2-1 (5), at the permissive and restrictive temperatures. Contrary to results obtained with DNA derived from the rad27Δ cells, no aberrant single-stranded DNA was detectable at any temperature, but the overall telomeric repeat length appeared slightly increased in these strains (data not shown).
FIG. 6.
Overexpression of DNA2 leads to nongrowing cells. WT, nontransformed MW35a cells grown on SC media; WT+pGAL-vector, MW35a cells containing the empty vector and grown on SC-Ura plates; WT+pGAL-DNA2, MW35a cells transformed with a plasmid that contained the DNA2 gene under the control of a galactose-inducible promoter (see Materials and Methods for construction of the plasmids). The plates contained glucose (left) or galactose (right) as a carbon source.
TABLE 1.
Rescue of Dna2p-induced mortality by Rad27p and Exo1pa
| Plasmid(s) | Formation of coloniesb |
|---|---|
| pRS424 (TRP1) + pRS426 (URA3) | Yes |
| pR2 (RAD27 TRP1) | Yes |
| pR2 (RAD27 TRP1) + pRS426 (URA3) | Yes |
| pTN3 (DNA2 URA3) | No |
| pTN3 (DNA2 URA3) + pRS424 (TRP1) | No |
| pTN3 (DNA2 URA3) + pR2 (RAD27 TRP1) | Yes |
| pE1 (EXO1 HIS3) | Yes |
| pTN3 (DNA2 URA3) + pE1 (EXO1 HIS3) | Yes |
See Materials and Methods for a detailed description of the construction of the plasmids. Yeast strain 7399-3-1 was used for all these transformations.
Yes, >100 colonies/μg of DNA; No, no colonies obtained. Plates always selected for all nutritional markers on plasmids.
Overexpression of Exo1p does not suppress all the phenotypes observed for rad27Δ cells.
The characterization of the EXO1 gene, encoding a 5′-3′ exonuclease, has shown that Exo1p, when overexpressed, can suppress the temperature sensitivity and mutator phenotypes of cells carrying mutations in RAD27 (46). In addition, mutations in EXO1 are lethal when combined with RAD27 mutations (46). Thus, it has been speculated that either EXO1 and RAD27 may encode redundant functions or that the actual lesions occurring in rad27Δ cells need Exo1p for repair (46). Since our physical analysis of the DNA derived from rad27Δ cells uncovered the existence of single-stranded G tails at the telomeres, we investigated whether overexpression of Exo1p would also reverse this effect in rad27Δ cells (Fig. 8). Overexpression of Exo1p from a high-copy-number vector in wild-type cells or deletion of the EXO1 gene had no effect on telomeric end structure or telomere length (Fig. 8 and data not shown). However, while introducing the same plasmid into rad27Δ cells suppressed their temperature sensitivity (46) (data not shown), it did not alleviate the appearance of the terminal single-stranded DNA after rad27Δ cells were shifted to 37°C (Fig. 8A). Furthermore, the length heterogeneity of the TRFs in this strain is very similar to that observed in rad27Δ cells without overexpression of Exo1p (compare the last two lanes in Fig. 8B with those in Fig. 2A). Thus, overexpression of Exo1p does not suppress all the phenotypes observed for rad27Δ cells, suggesting that the EXO1 gene product is not redundant with RAD27 but rather is required for an efficient repair of the lesions induced by the absence of Rad27p.
DISCUSSION
Instability of telomeric repeats in rad27Δ cells.
Previous genetic analyses of yeast cells carrying a deletion of the RAD27 gene and biochemical analyses of the homologous mammalian FEN-1 protein in vitro suggested that Rad27p was involved in the processing of Okazaki fragments during lagging-strand synthesis (see references 3 and 4 for reviews). A particular characteristic of yeast strains harboring a rad27 deletion is the destabilization of micro- and minisatellite sequences (14, 26, 28, 41). This repeat tract destabilization appeared to worsen with increased tract lengths (14, 41). Since the chromosome ends constitute one natural locus in which relatively long tracts of simple, direct repeats occur in many organisms, including yeast, we investigated whether the repeat instability observed in rad27Δ strains also occurred on telomeric repeats. As shown in Fig. 2 and 8, this is indeed the case: telomeric repeat tract lengths display a severe increase in their heterogeneity when rad27Δ cells are grown at elevated temperatures. This increased heterogeneity, indicating tract contractions and expansions, appears to be in contrast to some results that suggested a strong bias for repeat expansions in rad27Δ cells (26, 28). However, this bias was found only for relatively short tracts (≤50 bp) of mono- and dinucleotide repeats, and in larger tracts (≥100 bp) of trinucleotide repeats, such a bias was not observed. In such long tracts, which more closely resemble the telomeric repeat tracts (average length of about 300 bp), there was an approximately equal distribution of repeat contractions and expansions in rad27Δ cells (14, 41). Thus, an absence of Rad27p not only affects the stability of artificially introduced short repeats in the yeast genome or on plasmids but also has a severe effect on the stability of the natural telomeric repeats at chromosome ends. Given that the telomeric repeats are essential for chromosomal stability, it is possible that this tract instability may contribute significantly to the growth arrest observed in rad27Δ strains grown at elevated temperatures.
Single-stranded template DNA of the lagging strand occurs in rad27Δ cells and in cells in which Dna2p is overexpressed.
Several mechanisms have been proposed to explain the generation of repeat tract heterogeneity in rad27Δ cells (17, 26, 28, 47). Most of them rely on the assumption that Rad27p indeed plays an important role in Okazaki fragment processing in vivo, a role that was suggested from the relatively well characterized activities that the mammalian homologue of this enzyme displays in vitro (see reference 32 for a review). Some particularities occurring during the replication of telomeric repeats allowed an examination of this prediction, at least at a qualitative level. First, due to the conserved polarity of the repeats at all telomeres, the G-rich strands will always be synthesized by leading-strand synthesis and the C-rich strands by lagging-strand synthesis (see Fig. 1). Thus, by analyzing and comparing C-rich strand versus G-rich strand synthesis on telomeric repeats, one can gain insights into strand-specific effects of the gene product investigated. Second, after the last primer is synthesized on the most distal location on telomeric repeats, there is no upstream primer that may mask the processing occurring on the 5′ end of the downstream primer. Thus, inappropriate Okazaki fragment processing events that are predicted to yield single-stranded regions on the template strand are detectable on telomeric repeats due to an absence of fill-in synthesis from an upstream Okazaki fragment. We used a nondenaturing in-gel hybridization technique (11) to examine the occurrence of such predicted DNA intermediates on telomeric repeat DNA in strains lacking Rad27p (Fig. 3 through 5). While single-stranded regions on the C-rich strands, which serve as templates for the leading-strand synthesis, were not detectable in any cells (Fig. 4A), a strong increase in single-stranded DNA of the G-rich strand is observed in rad27Δ cells compared to wild-type cells. Control experiments using a strand-specific, single-strand exonuclease demonstrated that this single-stranded DNA consisted of terminal overhangs as opposed to gaps (Fig. 4B). Consistent with this finding, single-stranded DNA of either strand remains undetectable at an internal location that is just about 500 bp proximal to the Y′-to-terminal-repeat junction (Fig. 5A). We interpret these results to suggest that there is no detectable delay between parental-strand separation and new-strand synthesis on leading-strand synthesis either in wild-type or in rad27Δ cells. On the other hand, there appears to be a deficiency in lagging-strand synthesis in rad27Δ cells grown at elevated temperatures, since we observe an accumulation of terminal single-stranded DNA of the G-rich strands in such cells.
At least two possibilities could contribute to the generation of these single-stranded DNA overhangs: either priming itself is inefficient during fork movement, creating at the very ends of the chromosomes single-stranded tails of the templating strand, or primer processing is aberrant in these cells. For instance, priming may actually occur to the very ends of the chromosome, but these newly synthesized Okazaki fragments may be dissociated or degraded in an inappropriate way. Since Dna2p, an essential helicase and/or nuclease (2, 5, 7), interacts genetically and biochemically with Rad27p (6), we examined whether this protein was involved in such a process. Indeed, an increase of single-stranded G-strand DNA can be observed in cells in which Dna2p was overexpressed (Fig. 7). The results also show that overexpression of a full-length Dna2p caused arrest of cell growth (Table 1; Fig. 6), an effect that is not observed when a truncated Dna2 protein lacking the N-terminal 105 amino acids was overexpressed (6). On the other hand, overexpression of an N-terminal portion of Dna2p causes a significant derepression of genes located near a telomere in yeast (44), supporting the notion that the N-terminal part of Dna2p may play a significant regulatory role (2).
Our results are thus consistent with a model in which Rad27p and Dna2p work together to remove the 5′ ribonucleotides from newly generated Okazaki fragments. In the absence of Rad27p, or when Dna2p is overexpressed, there may be excessive primer dissociation or degradation leading to single-stranded DNA of the templating strand. If this model was correct, one might expect that the actual lesions on the DNA in these two experimental settings were similar in nature, triggering growth arrest via similar pathways. Consistent with this idea, rad27Δ cells incubated at 37°C, or cells in which Dna2p was overexpressed, displayed an identical arrest phenotype with predominantly large, dumbbell-shaped cells (data not shown) (37, 45).
The exonuclease Exo1p is not redundant to Rad27p.
Overexpression of Exo1p, a yeast 5′-3′ exonuclease, has been reported to be able to suppress the temperature sensitivity and mutator phenotypes of rad27Δ cells (46). However, overexpression of Exo1p from a high-copy-number vector had no diminishing effect on the appearance of the single-stranded G tails or on the length heterogeneity of the TRFs in rad27Δ cells (Fig. 8). Moreover, there was no detectable increase in such G tails when EXO1 was deleted or overexpressed in wild-type cells (Fig. 8) (12). These results suggest that Exo1p and Rad27p are not simply redundant proteins for Okazaki fragment maturation but rather that the kinds of DNA lesions occurring in the absence of Rad27p require Exo1p for repair (46). Overexpression of Exo1p also suppresses the growth defect of cells in which Dna2p is overexpressed (Table 1). This is consistent with our model that overexpression of Dna2p induces similar lesions as an absence of Rad27p: Exo1p may be required for the repair of the lesions in both situations.
These data thus provide direct in vivo evidence of single-stranded regions that occur on the templating strand when lagging-strand synthesis is impaired. While in our experiments such single-stranded DNA was detectable only on the terminal repeats of the chromosome, we suppose that such single-stranded areas of the template strand also occur in internal locations, as previously proposed (17, 29, 47). Due to continued fill-in synthesis from an upstream primer, these internal lesions may be very transient and therefore not detectable with the techniques used here. Consistent with this idea, newly synthesized DNA isolated from rad27Δ cells contains a large proportion of relatively small DNA molecules, corroborating the evidence that Okazaki fragment maturation is impaired in these cells (35). Chromosome ends in wild-type yeast cells acquire very transient single-stranded G-strand extensions only late in S phase (54). We have previously shown that the generation of at least some of these G tails is independent of an active telomerase activity but that it does require the passage of a replication fork (11, 12). It is thus possible that in normal cells, there is a short delay between the separation of the two parental strands and the time when lagging-strand synthesis reaches the very ends of the template strands. This would create single-stranded G tails which could serve as templates for telomerase-mediated strand elongation. If lagging-strand synthesis is compromised, those G tails could persist and/or become even more extensive due to primer dissociation (see above), leading to the effects reported here.
Thus, our results also emphasize the importance of lagging-strand synthesis in telomeric repeat maintenance. Recently, we reported that chromosome end processing events require the passage of a replication fork (12). Furthermore, deficiencies in the regulation of telomeric repeat lengths in yeast mutants affected in DNA replication have been documented previously (1, 8), but the mechanisms involved remained unclear. Analyzing the specific types of DNA lesions found in those mutants may lead to insights as to how telomeric tract length is regulated and more-detailed insights into the coordination of the conventional replication machinery with telomere maintenance.
ACKNOWLEDGMENTS
We thank T. Formosa, J. Aron, M. Reagan, and E. Friedberg for generous gifts of plasmids and yeast strains. The members of the Wellinger lab are thanked for valuable discussions during this project.
This work was supported by Canadian Medical Research Council grant MT 12616. J.P. received a studentship from the Fonds pour la Formation des Chercheurs et l’Aide à la Recherche (FCAR), and R.J.W. is a Chercheur-Boursier Senior of the Fonds de la Recherche en Santé du Québec (FRSQ).
REFERENCES
- 1.Adams A K, Holm C. Specific DNA replication mutants affect telomere length maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4614–4620. doi: 10.1128/mcb.16.9.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae S-H, Choi E, Lee K-H, Park J S, Lee S-H, Seo Y-S. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J Biol Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 3.Baker T A, Bell S P. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 4.Bambara R A, Murante R S, Henricksen L A. Enzymes and reactions at the eukaryotic DNA replication fork. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 5.Budd M E, Campbell J L. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc Natl Acad Sci USA. 1995;92:7642–7646. doi: 10.1073/pnas.92.17.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budd M E, Campbell J L. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budd M E, Choe W-C, Campbell J L. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J Biol Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 8.Carson M J, Hartwell L. CDC17: an essential gene that prevents telomere elongation in yeast. Cell. 1985;42:249–257. doi: 10.1016/s0092-8674(85)80120-3. [DOI] [PubMed] [Google Scholar]
- 9.Chan C S M, Tye B-K. Organization of DNA sequences and replication origins at yeast telomeres. Cell. 1983;33:563–573. doi: 10.1016/0092-8674(83)90437-3. [DOI] [PubMed] [Google Scholar]
- 10.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 11.Dionne I, Wellinger R J. Cell cycle regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc Natl Acad Sci USA. 1996;93:13902–13907. doi: 10.1073/pnas.93.24.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dionne I, Wellinger R J. Processing of telomeric DNA ends requires the passage of a replication fork. Nucleic Acids Res. 1998;26:5365–5371. doi: 10.1093/nar/26.23.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiorentino D F, Crabtree G R. Characterization of Saccharomyces cerevisiae dna2 mutants suggests a role for the helicase late in S phase. Mol Biol Cell. 1997;8:2519–2537. doi: 10.1091/mbc.8.12.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freudenreich C H, Kantrow S M, Zakian V A. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 15.Gietz R D, Schiestl R H. Transforming yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 16.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 17.Gordenin D A, Kunkel T A, Resnick M A. Repeat expansion—all in a flap. Nat Genet. 1997;16:116–118. doi: 10.1038/ng0697-116. [DOI] [PubMed] [Google Scholar]
- 18.Goulian M, Richards S H, Heard C J, Bigsby B M. Discontinuous DNA synthesis by purified mammalian proteins. J Biol Chem. 1990;265:18461–18471. [PubMed] [Google Scholar]
- 19.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 20.Harrington J J, Lieber M R. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrington J J, Lieber M R. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 1994;8:1344–1355. doi: 10.1101/gad.8.11.1344. [DOI] [PubMed] [Google Scholar]
- 22.Henikoff S, Eghtedarzadeh M K. Conserved arrangement of nested genes at the Drosophila Gart locus. Genetics. 1987;117:711–725. doi: 10.1093/genetics/117.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huberman J A, Spotila L D, Nawotka K A, El-Assouli S M, Davis L R. The in vivo replication origin of the yeast 2μm plasmid. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- 24.Ishimi Y, Claude A, Bullock P, Hurwitz J. Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J Biol Chem. 1988;263:19723–19733. [PubMed] [Google Scholar]
- 25.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 27.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kokoska R J, Stefanovic L, Tran H T, Resnick M A, Gordenin D A, Petes T D. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t) Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel T A, Resnick M A, Gordenin D A. Mutator specificity and disease: looking over the FENce. Cell. 1997;88:155–158. doi: 10.1016/s0092-8674(00)81832-2. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann I R, Nussbaum A L. The deoxyribonucleases of E. coli. V. On the specificity of exonuclease I (phosphodiesterase) J Biol Chem. 1964;239:2628–2634. [PubMed] [Google Scholar]
- 31.Li X, Li J, Harrington J, Lieber M R, Burgers P M J. Lagging strand synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 32.Lieber M R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 33.Louis E J, Haber J E. The subtelomeric Y′ repeat family in Saccharomyces cerevisiae: an experimental system for repeated sequence evolution. Genetics. 1990;124:533–545. doi: 10.1093/genetics/124.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merrill B J, Holm C. The RAD52 recombinational repair pathway is essential in pol30 (PCNA) mutants that accumulate small single-stranded DNA fragments during DNA synthesis. Genetics. 1998;148:611–624. doi: 10.1093/genetics/148.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murante R S, Henrickson L A, Bambara R A. Junction ribonuclease: an activity in Okazaki fragment processing. Proc Natl Acad Sci USA. 1998;95:2244–2249. doi: 10.1073/pnas.95.5.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reagan M S, Pittenger C, Siede W, Friedberg E C. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rose M D, Winston F, Hieter P, editors. Methods in yeast genetics: a laboratory course manual. 1st ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sandell L S, Zakian V A. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 41.Schweitzer J K, Livingston D M. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum Mol Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 42.Shampay J, Szostak J W, Blackburn E H. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- 43.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singer M S, Kahana A, Wolf A J, Meisinger L L, Peterson S E, Goggin C, Mahowald M, Gottschling D E. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sommers C H, Miller E J, Dujon B, Prakash S, Prakash L. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′ to 3′ exonuclease required for lagging strand synthesis in reconstituted systems. J Biol Chem. 1995;270:4193–4196. doi: 10.1074/jbc.270.9.4193. [DOI] [PubMed] [Google Scholar]
- 46.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7478–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 48.Turchi J J, Huang L, Kim Y, Bambara R A. Enzymatic completion of mammalian lagging-strand DNA replication. Proc Natl Acad Sci USA. 1994;91:9803–9807. doi: 10.1073/pnas.91.21.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallen E A, Cross F R. Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol Cell Biol. 1995;15:4291–4302. doi: 10.1128/mcb.15.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 51.Watson J D. Origin of concatemeric DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 52.Wellinger R J, Ethier K, Labrecque P, Zakian V A. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 53.Wellinger R J, Sen D. The DNA structures at the ends of eukaryotic chromosomes. Eur J Cancer. 1997;33:735–749. doi: 10.1016/S0959-8049(97)00067-1. [DOI] [PubMed] [Google Scholar]
- 54.Wellinger R J, Wolf A J, Zakian V A. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 55.Wellinger R J, Zakian V A. Lack of positional requirements for autonomously replicating sequence elements on artificial yeast chromosomes. Proc Natl Acad Sci USA. 1989;86:973–977. doi: 10.1073/pnas.86.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu X, Li J, Li X, Hsieh C-L, Burgers P M J, Lieber M R. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 1996;24:2036–2043. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zakian V A. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu Rev Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]
- 58.Zakian V A, Scott J F. Construction, replication, and chromatin structure of TRP1 RI circle, a multiple-copy synthetic plasmid derived from Saccharomyces cerevisiae chromosomal DNA. Mol Cell Biol. 1982;2:221–232. doi: 10.1128/mcb.2.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]