Abstract
The precise duplication of eukaryotic genetic material takes place once and only once per cell cycle and is dependent on the completion of the previous mitosis. Two evolutionarily conserved kinases, the cyclin B (Clb)/cyclin-dependent kinase (Cdk/Cdc28p) and Cdc7p along with its interacting factor Dbf4p, are required late in G1 to initiate DNA replication. We have determined that the levels of Dbf4p are cell cycle regulated. Dbf4p levels increase as cells begin S phase and remain high through late mitosis, after which they decline dramatically as cells begin the next cell cycle. We report that Dbf4p levels are sensitive to mutations in key components of the anaphase-promoting complex (APC). In addition, Dbf4p is modified in response to DNA damage, and this modification is dependent upon the DNA damage response pathway. We had previously shown that Dbf4p interacts with the M phase polo-like kinase Cdc5p, a key regulator of the APC late in mitosis. These results further link the actions of the initiator protein, Dbf4p, to the completion of mitosis and suggest possible roles for Dbf4p during progression through mitosis.
Eukaryotic DNA replication is initiated during the synthesis (S) phase of the mitotic cell cycle at multiple chromosomal sites designated origins. Segregation of newly replicated chromosomes occurs during mitosis (M phase) and is separated from S phase by two gaps, G1 and G2. The multiple origins on a chromosome initiate replication at various times from early to late in S phase (16, 17, 19). Initiation from individual origins happens at most once per S phase, and reinitiation is prevented until the cell has finished M phase (36). The mechanism by which the cell regulates the timing of initiation of DNA replication and prevents reinitiation until the completion of mitosis has not been defined.
In budding yeast there are at least two complexes present at origins (13). A postreplication complex, which consists of at least the origin recognition complex, is detected after initiation during S phase and is present until late in M (13). This complex is modified late in M, giving rise to the prereplication complex. The prereplication complex is present from late M to S phase and is most probably formed through the association of Cdc6p and the MCM family of six proteins with origins (3, 8, 40). Cdc6p becomes associated with chromatin and origins late in M and dissociates late in G1 or early in S (40). The MCM proteins associate with origin sequences late in mitosis or early in G1, and their release from origins is coincident with replication initiation (3, 40). Precisely how the cell cycle machinery is linked to replication through regulation of the transitions between these two complexes late in M and again late in G1 is not known.
Two evolutionarily conserved kinases, the cyclin B (Clb)/cyclin-dependent kinase (Cdk/Cdc28p) (13) and Cdc7p along with its interacting factor Dbf4p, are required late in G1 to initiate DNA replication (5, 22, 25, 31). Cdc7p kinase activity peaks late in the G1 phase of the cell cycle (25), and Cdc7p is required for origin firing during S phase (4, 10). Based on a series of one-hybrid studies, Dbf4p is thought to be targeted to origins (12, 21). We reported the identification in Saccharomyces cerevisiae of a recessive loss-of-function mutation in MCM5/CDC46, cdc46-bob1, which bypasses the requirement for Dbf4p-Cdc7p (20). A recent report from Tanaka et al. shows that dbf4-1 cells fail to release MCM proteins from origins under restrictive growth conditions (40). These results suggest a role for Dbf4p-Cdc7p as a modifier of MCM function at origins, late in G1 or early in S. Consistent with this model, Mcm2p is a target of Dbf4p-Cdc7p during initiation of DNA replication (32).
Our previous studies indicate that Cdc5p interacts with Dbf4p (21). Cdc5p is a member of the polo-like kinase family. It accumulates in the nuclei of M cells (7, 35) and is thought to play roles both in anaphase-promoting complex (APC) regulation late in M (6, 35) and in adaptation to the DNA damage checkpoint in metaphase-arrested cells (41). The interaction with Cdc5p suggested that Dbf4p might have an M phase function in addition to its S phase roles (21). In this study we have further developed links between Dbf4p and mitosis. Specifically, we show that the levels of Dbf4p fluctuate during the cell cycle and that Dbf4p accumulates in the nuclei of late G1, S, and M phase cells. The levels of Dbf4p decline dramatically as cells exit mitosis, and they are sensitive to mutations in key APC components. In addition, we show that Dbf4p is modified in cdc13-1 cells grown at the restrictive temperature and that this modification is dependent on key components of the DNA damage checkpoint pathway. We speculate that Dbf4p may provide a link between mitosis and S phase through its interactions with the M and S phase regulators, Cdc5p and Cdc7p, respectively.
MATERIALS AND METHODS
Plasmids and strains.
Yeast strains used in this study are listed in Table 1. Plasmid DNA was transformed into yeast by the lithium acetate method as described previously (24). Yeast strains without plasmids were grown in yeast extract-peptone-dextrose. YEP medium contained 1% yeast extract and 2% Bacto Peptone. Yeast strains bearing plasmids were grown in selective synthetic medium (SC) with 2% sugar (galactose or raffinose as indicated). Strains with plasmids to be induced with galactose were first grown in synthetic medium with 2% raffinose to an A600 of 0.2 to 0.4. Wild-type strains were grown at 30°C unless noted otherwise. pCH920 carries the DBF4 open reading frame ligated into the BamHI-XhoI site of pCH765 (pRS423-GAL1-HA). The GAL-HA-dbf4Δ70 plasmid (pCH955) was created by deleting DBF4 sequences from position +1 to +210 by PCR.
TABLE 1.
Strains
| Strain | Genotype (source) |
|---|---|
| W303-1A | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 LYS2 (R. Rothstein) |
| W303-1B | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 LYS2 (R. Rothstein) |
| YCH192 | W303-1A bar1::URA3 |
| YCH201 | W303-1A DBF4-proA-HIS3-URA3 bar1::URA3 |
| H14C1A1 | MATa cdc14-1 his7 ura1 (L. Hartwell) |
| YCH189 | W303-1A cdc14-1 (H5C1A1 backcrossed two times to W303) |
| YCH219 | W303-1A cdc14-1 DBF4-proA-HIS3-URA3 (ProA-tagged DBF4 in YCH189) |
| K1983 | MATa cdc4-1 ade2 ura3 his3 (backcrossed four times to W303) (K. Nasmyth) |
| YCH215 | MATa cdc4-1 ade2 ura3 his3 DBF4-proA-HIS3-URA3 (ProA-tagged DBF4 in K1983) |
| K2034-CH-α | MATα cdc13-1 ade2 ura3 his3 (backcrossed five times to W303) (K. Nasmyth backcrossed three times, and then we backcrossed it an additional three times to W303) |
| YCH216 | MATα cdc13-1 ade2 ura3 his3 DBF4-proA-HIS3-URA3 (ProA-tagged DBF4 in K2034) |
| K2531 | MATa cdc23-1 ade2 leu2 ura3 his3 TRP1 (backcrossed three times to W303) (K. Nasmyth) |
| YCH217 | MATa cdc23-1 ade2 leu2 ura3 his3 TRP1 DBF4-proA-HIS3-URA3 (ProA-tagged DBF4 in K2531) |
| YCH208 | W303-1A cdc23-1 bar1::URA3 (segregant from YCH192 × K2531) |
| H5C1A1 | MATa cdc5-1 his7 ura1 (B. Gavin and L. Hartwell) |
| YCH187 | W303-1A cdc5-1 (H5C1A1 backcrossed five times to W303) |
| YCH218 | W303-1A cdc5-1 DBF4-proA-HIS3-URA3 (ProA-tagged DBF4 in YCH187) |
| YCH288 | W303-1A DBF4-proA-HIS3-URA3 (ProA-tagged DBF4 in W303-1A) |
| YOC1523 | W303-1A bar1 PDS1-HA::URA3 mec1-1 (O. Cohen-Fix and D. Koshland) |
| YOC1524 | W303-1A bar1 PDS1-HA::URA3 rad53-21 (O. Cohen-Fix and D. Koshland) |
| YCH324 | W303-1A cdc13-1 DBF4-proA-HIS3-URA3 mec1-1 (segregant of YCH216 and YOC1523) |
| YCH325 | W303-1A cdc13-1 DBF4-proA-HIS3-URA3 rad53-21 (segregant of YCH216 and YOC1524) |
| H16C1A1 | MATa cdc16-1 his7 ura1 (B. Gavin and L. Hartwell) |
| YCH407 | W303-1B cdc16-1 (H16C1A1 backcrossed five times to W303) |
| YCH406 | W303-1A cdc16-1 DBF4-proA-HIS3-URA3 bar1::URA3 (segregant from YCH201 × YCH407) |
| YCH404 | W303-1A cdc16-1 bar1::URA3 (segregant from YCH201 × YCH407) |
ProA tagging of Dbf4p.
The chromosomal copy of the DBF4 gene was tagged by a C-terminal, in-frame integration of a DNA fragment encoding the immunoglobulin G (IgG) binding domains of protein A (42). The protein A (ProA) gene and adjacent HIS3 and URA3 markers were amplified by PCR with pProA-HIS3-URA3 (a gift from Mike Rout and John Aitchison) (1). The following primers were used for the PCR: DBF4 sense primer, 5′-GAA AAC GAT TTA AAT TTT GAG GCT ATT GAC TCG TTA ATT GAA AAT CTC AGA TTT CAA ATA GGT GAA GCT CAA AAA CTT AAT-3′; DBF4 antisense primer, 5′-CAA TAA CAT TGC CGT TGA TAG CTT TTT TGT TCC GAG CAG CAA TGG AAA CGG AAC AAT ACT TTT ACT TAT AAT ACA GTT TTT TAG-3′. The 5′ region of the sense primer encodes the carboxy-terminal 20 amino acids of Dbf4p (up to but not including the stop codon) and continues in frame to encode the 7 amino acids of ProA beginning with the glycine at amino acid residue 24 (42). The 5′ region of the antisense primer corresponds to nucleotides 2220 to 2161 of the untranslated region of DBF4 (1 is the A of the initiation codon) and continues with 24 nucleotides, 1050 to 1027, of the reverse complement of the URA3 gene. The PCR products were transformed into yeast, and HIS+ URA+ transformants were screened by Western analysis for expression of Dbf4-ProA.
Immunofluorescence.
Indirect immunofluorescence was carried out exactly as described previously (45). For localization of the epitope-tagged Dbf4p (Dbf4-ProA), wild-type strains were grown to early log phase and prepared for immunofluorescence microscopy. Cells from the various temperature-sensitive strains used in this study were grown to early log phase at 24°C and then transferred to 37°C for 3 to 4 h, at which time greater than 90% of the population exhibited the arrest morphology. α-factor and nocodazole arrests were conducted with cells in early log phase as described previously (18) and as described by Jacobs et al. (26), respectively. The DNA content of these samples was determined by fluorescence-activated cell sorter (FACS) analysis (see Fig. 2d). When Dbf4-ProA was visualized, cells were fixed for 10 min. The fixed cells were incubated first with affinity-purified rabbit anti-mouse IgG (Cappel catalog no. 55480) and then with donkey anti-rabbit IgG (Chemicon catalog no. AP182F) fluorescein-conjugated secondary antibody. Photographs were taken with a 100× objective on a Olympus microscope with Kodak T-MAX400 film processed at 400 ASA.
FIG. 2.
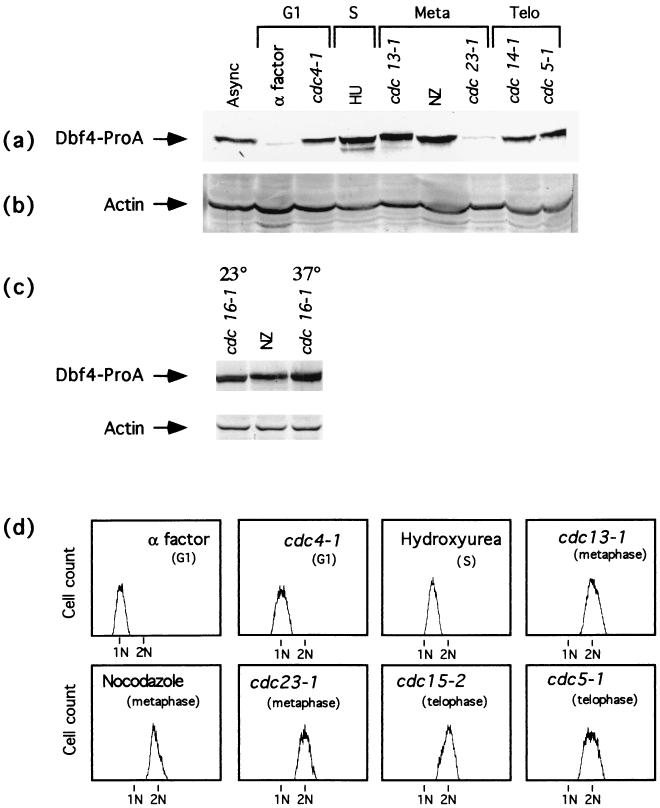
Dbf4p is present in late G1 to late M phase synchronized cells. Wild-type and cdc mutant cells expressing an integrated ProA tagged copy of Dbf4p were grown asynchronously or arrested in different stages of the cell cycle either by using chemicals or by growth of the cdc mutant at the restrictive temperature. Late G1, α-factor (YCH201) and cdc4-1 (YCH215); S, hydroxyurea (HU) (YCH201); metaphase, cdc13-1 (YCH216), nocodazole (NZ) (YCH201), and cdc23-1 (YCH217); telophase, cdc14-1 (YCH219) and cdc5-1 (YCH218). Extracts were derived from these cells, and the levels of Dbf4-ProA (a) and actin (b) were determined by immunoblotting with anti-IgG and antiactin antibodies, respectively. (c) Immunoblot analysis of Dbf4-ProA in cdc16-1 (YCH406) cells at permissive (23°C) and restrictive (37°C) temperatures. (d) FACS analysis of the DNA content at the different arrest points.
Immunoprecipitation.
Cells were pelleted, washed once in water, and lysed or frozen in liquid nitrogen. Pellets were resuspended in 0.3 ml of lysis buffer containing 5% glycerol, 20 mM Tris-HCl (pH 8.0), 1 mM EDTA, 10 mM MgCl2, 0.3 M AmSO4, 1 mM dithiothreitol, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, 5 mg of leupeptin per ml, 2 mM pepstatin A, 50 mM NaF, 10 mM sodium pyrophosphate, and 0.5 mM Na-VO4. The cells were lysed by adding 0.5 ml of acid-washed glass beads and vortexing in pulses until 90% lysis was achieved, and then 0.35 ml of immunoprecipitation buffer (1 M LiCl, 2% Triton X-100, 10% glycerol, 0.5 mM Na-VO4, and protease inhibitors as described for lysis buffer) was added and vortexed for 1 min. The lysate was spun for 10 min at 3,000 rpm (Heraeus-Picofuge) and the supernatant was aliquoted and frozen in liquid nitrogen. Protein concentrations were determined with the Bio-Rad protein assay. Four hundred milligrams of lysate was incubated with 0.1 ml of IgG-Sepharose (Pharmacia) at 4°C for 1.5 h. Immunoprecipitates were pelleted by centrifugation, washed two times with immunoprecipitation buffer and then two times with CIP buffer (50 mM HEPES, 10 mM MgCl2, and 5 mM MnCl2), and divided into two samples. One hundred units of calf intestinal phosphatase (CIP) was added to one of the samples but not to the other. To this suspension an equal volume of sodium dodecyl sulfate (SDS) gel-loading buffer was added, and the mixture was incubated for 5 min at 100°C and subjected to electrophoresis on an SDS–7% polyacrylamide gel. Proteins were electrophoretically transferred to nitrocellulose membranes. Blots were incubated with primary antibody (rabbit anti-mouse IgG [Cappel catalog no. 55480]) for 1 h at room temperature in 10 mM Tris-Cl (pH 7.5)–150 mM NaCl–0.05% Tween 20 (TBST) with 2% nonfat milk. The immunoblots were washed with TBST and then incubated for 1 h with secondary antibody (alkaline phosphatase-conjugated anti-rabbit IgG) in TBST. The immunoblots were washed again in TBST and developed via color visualization with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-1-phosphate (Promega).
Other methods.
Hydroxyurea was obtained from Sigma and used at a final concentration of 200 mM. α-factor was obtained from Sigma and for bar1 mutant and wild-type cells was used at final concentrations of 0.2 μM and 5 mg/ml, respectively. Nocodazole was obtained from Aldrich and was added to medium from a 20-mg/ml stock solution in dimethyl sulfoxide. It was used at a final concentration of 20 μg of nocadazole per ml and 1% dimethyl sulfoxide, as described by Jacobs et al. (26). The DNA content of cells was measured on a Becton Dickinson (San Jose, Calif.) FACSsan as described by Epstein and Cross (15).
RESULTS
Dbf4p is cell cycle regulated.
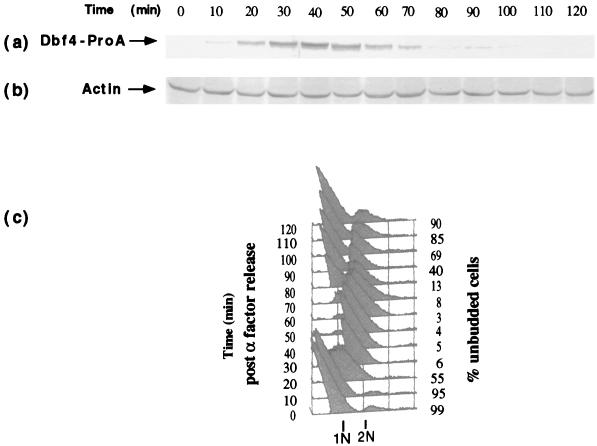
The DBF4 message is constitutively present during the cell cycle, peaking in G1/S (5). To determine the pattern of Dbf4p localization and its regulation during the cell cycle, the chromosomal copy of DBF4 was replaced with sequence encoding an ProA epitope-tagged version (the five IgG binding domains of ProA fused in frame at the C terminus) (42). The Dbf4-ProA-tagged strain exhibited no growth defects, and therefore the Dbf4-ProA fusion performed all the essential functions of Dbf4p. We initially examined the fluctuations of Dbf4p levels in a population of G1 cells synchronously released from an α-factor-induced G1 arrest state. Progression through a single cell cycle was monitored by FACS analysis to determine DNA content and by budding index (Fig. 1c). Immunoblot analysis was also performed with extracts derived from these synchronized cells, and Dbf4-ProA was detected as a 120-kDa band. As shown in Fig. 1a, Dbf4-ProA was not detected in extracts derived from cells arrested in G1 with α-factor. Dbf4-ProA began to accumulate as cells were released from the block, completed G1, and entered S phase. Dbf4p levels remained high as cells progressed through mitosis and declined dramatically as the cells completed mitosis and entered G1 of the next cell cycle.
FIG. 1.
Dbf4p levels fluctuate during the cell cycle. DBF4-proA (YCH288) cells were synchronized in G1 by addition of α-factor and released into fresh medium lacking α-factor at 25°C. Samples for FACS analysis and extract preparation were taken at the times shown. (a and b) Dbf4-ProA (a) and actin (b) were detected by immunoblotting with anti-IgG and antiactin antibodies, respectively. (c) FACS analysis of DNA content. The percentage of unbudded cells is shown on the right of the FACS analysis profile.
Dbf4p accumulates in the nuclei of G1-, S-, and M-phase-arrested cells.
To more fully determine the subcellular pattern of Dbf4p localization and accumulation during the cell cycle, DBF4-proA was expressed in a panel of cdc mutant strains, and immunoblot and immunofluorescence analyses were performed with the temperature-arrested cells. The arrest state of these cells was determined by FACS analysis (Fig. 2d). As shown in Fig. 1a and 2a, Dbf4-ProA was not detected in extracts derived from cells arrested in late G1 with α-factor. However, Dbf4-ProA was detected in cdc4-1 late-G1-arrested cells (Fig. 2a). Dbf4-ProA was also detected by immunoblot analysis in extracts derived from cells synchronized in S phase (with hydroxyurea), in metaphase (cdc13-1 cells grown at the restrictive temperature and cells treated with nocadazole), and late in M (cdc14-1 and cdc5-1 cells grown at the restrictive temperature) (Fig. 2a). The levels of Dbf4-ProA in arrested cdc23-1 cells were significantly lower than the levels of Dbf4p in nocadazole-arrested cells. As Cdc23p is a component of the APC, we analyzed the Dbf4p levels in two other mutant strains that are defective for other components of the APC. We examined Dbf4p levels in cdc16-1 (Fig. 2a) and cdc27-2 (data not shown) cells grown at the permissive and restrictive temperatures. By immunoblotting, the levels of Dbf4p in arrested cdc16-1 and cdc27-2 cells were comparable to, if not higher than, the level of Dbf4p present in nocadazole-arrested cells.
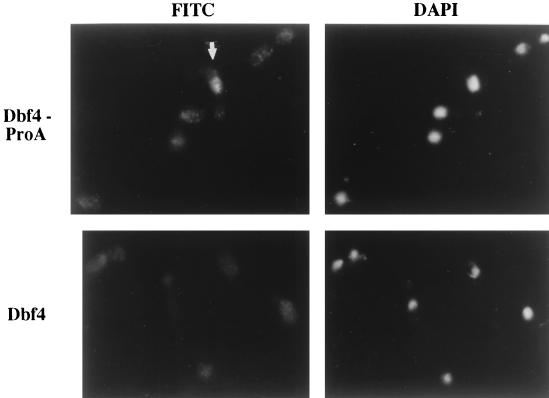
Indirect immunofluorescence showed that Dbf4p-ProA was localized predominantly in the nucleus, and an asynchronous cell population was detected in only a subset of the cells (Fig. 3, top panels). The signal was strongest in budded cells containing a single nucleus (possible S, G2, and early M phase cells) and was never observed in either single cells (G1 cells) or large budded cells containing two nuclei (cells in late mitosis or undergoing cytokinesis) (Fig. 3 and data not shown). The background signal in the untagged parental strain (W303) is shown in Fig. 3, bottom panels.
FIG. 3.
Detection of Dbf4p by immunofluorescence in mitotic cells. Cells derived from asynchronous cultures were fixed with methanol-formaldehyde and processed for indirect immunofluorescence. (Top) Left panel, population of asynchronous Dbf4-ProA cells (YCH201) stained with anti-IgG to detect Dbf4-ProA. FITC, fluorescein isothiocyanate. Right panel, DAPI (4′,6-diamidino-2-phenylindole) staining to detect DNA. An arrow points to a large budded single cell with a single nucleus. (Bottom) Left panel, population of asynchronous Dbf4p (untagged) cells (W303-1A) stained with anti-IgG. Right panel, DAPI staining to detect DNA. All images were taken with a 100× objective and printed at the same magnification.
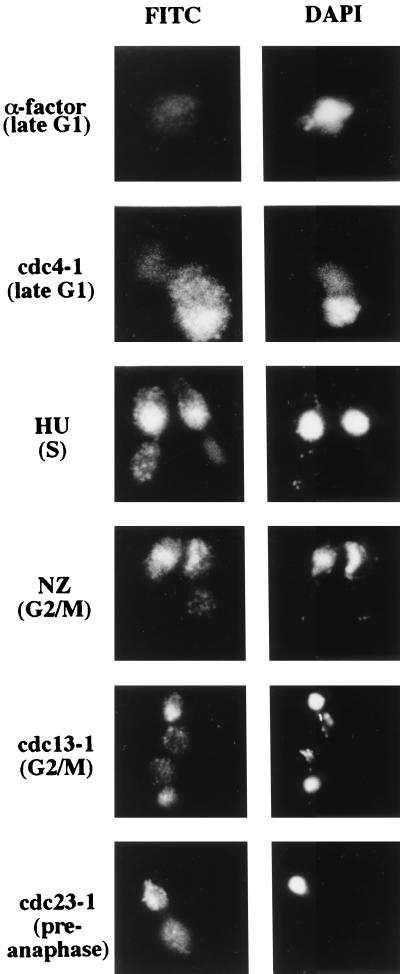
Indirect immunofluorescence analysis was also performed with synchronized cells. Dbf4-ProA was observed only in the nucleus, and it was not present in cells synchronized in G1 with α-factor. It was, however, detected in cdc4-1 cells grown at the restrictive temperature, as shown in Fig. 4. A Dbf4-ProA signal was also present in cells synchronized in S phase (cells arrested with hydroxyurea) and in cells synchronized early in M, during metaphase (cdc13-1, nocodazole-treated, and cdc23-1) cells (Fig. 4). The localization results were in general agreement with the immunoblot analysis, with the exception of the cdc23-1 cells (YCH217). Whereas the immunoblotting detected a low level of Dbf4-ProA in arrested cdc23-1 cells, a strong Dbf4-ProA signal was detected by immunofluorescence in the arrested cdc23-1 cells. The reason for this lack of correlation is being further investigated. We speculate that the immunofluorescence strategy may be more sensitive and have a lower threshold for detection of Dbf4p than the immunoblot.
FIG. 4.
Dbf4p is present in the nuclei of late G1- to metaphase-arrested cells. Wild-type and cdc mutant cells expressing an integrated ProA-tagged copy of Dbf4p were arrested in different stages of the cell cycle either by using chemicals or by growth of the cdc mutant at the restrictive temperature. Immunofluorescence was performed on these cells as described in Materials and Methods. Two views of each field are shown, Dbf4-ProA staining (fluorescein isothiocyanate [FITC]) and staining of DNA (DAPI). All photographs were taken with a 100× objective and printed at the same magnification. cdc4-1 mutants form abnormal buds (44). The same cultures were used for immunoblot analysis (Fig. 2). HU, hydroxyurea; NZ, nocodazole.
APC mediation of Dbf4p degradation in G1.
Progression through and completion of M requires the ubiquitin-dependent proteolysis of specific substrates (29). For example, the metaphase-to-anaphase transition requires degradation of Pds1p in S. cerevisiae and of cut2 in Schizosaccharomyces pombe (9, 18). Completion of M is dependent on the degradation of mitotic cyclins (23, 29, 37, 39, 43). Stage-specific degradation of these cyclins is dependent on a 20S particle designated the APC or cyclosome, a ubiquitin protein ligase (30, 38). In addition, the Dbf4p-interacting kinase Cdc5p is a substrate for the APC late in M (6, 21, 35). The cell cycle-regulated pattern of Dbf4p protein levels suggested that the APC/cyclosome might also target Dbf4p for degradation. If APC function is required for the degradation of Dbf4p, we predicted that the stability of Dbf4p would be enhanced in cells compromised for APC function compared to wild-type cells.
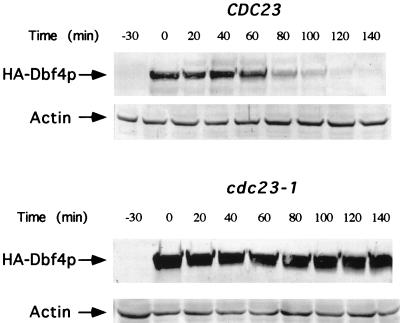
To examine whether the APC is involved, the stability of Dbf4p in cells arrested in G1, a phase of the cell cycle in which the APC is active in Pds1p, Clb2p, and Ase1p degradation (2, 9, 28, 46), was examined. CDC23 and cdc23-1 cells containing plasmids expressing hemagglutinin (HA)-Dbf4p under control of the GAL1 promoter were grown in glucose at 23°C, arrested in G1 with α-factor, and then shifted to 37°C to inactivate the cdc23-1 gene product (7, 9). At this point HA-Dbf4p expression was induced by the addition of galactose, followed after 30 min by the addition of glucose to turn off its expression. The restrictive temperature (37°C) was maintained while these steps were performed. As shown in the immunoblot, HA-Dbf4p was not detected after 80 min in the wild-type cells (Fig. 5, top). In contrast, the majority of HA-Dbf4p was still present after 2 h in the cdc23-1 cells (Fig. 5, bottom). These results provide evidence that Dbf4p proteolysis is APC mediated. It is interesting to note the possible distinction between the immunoblot levels of Dbf4p in arrested cdc23-1 versus cdc16-1 cells in Fig. 2 (cells which were not α-factor arrested before the temperature shift). This may suggest that for APC-mediated degradation of Dbf4p outside G1, the function of Cdc16p may be more important than that of Cdc23p.
FIG. 5.
Degradation of Dbf4p is dependent on Cdc23p function. Wild-type (YCH192) and cdc23-1 (YCH208) cells were transformed with a 2μm-based plasmid carrying GAL1-HA-DBF4 (pCH920). For both strains an overnight culture was grown selectively at 23°C in SC lacking histidine. This culture was inoculated into YEP plus raffinose, grown to early log phase at 23°C, and arrested in G1 with α-factor. When >90% of the cells, as determined by microscopy, exhibited the arrest state, the temperature of the culture was shifted to 37°C, the restrictive temperature for cdc23-1 cells. FACS analysis of these blocked samples indicated that >90% of the cells had a 1N content of DNA and were in G1 (data not shown). After 30 min at the restrictive temperature, galactose was added at time −30 to induce expression of HA-DBF4 from the GAL promoter. Thirty minutes later, at time zero, glucose was added to turn off expression of HA-DBF4 from the GAL promoter while maintaining the G1 arrest at the restrictive temperature. FACS analysis of these blocked samples also indicated that >90% of the cells had a 1N content of DNA and were in G1 (data not shown). Samples of equal density were taken at the indicated time points and assayed by immunoblotting for HA-Dbf4p and actin. Actin detection was used as an internal loading control.
The results in Fig. 5 suggested that Dbf4p may have a half-life of as long as 1 h. Alternatively, the persistence of Dbf4p in the cdc23-1 cells seen in Fig. 5 could have been due to a long half-life for the DBF4 RNA message. To distinguish between these possibilities, we repeated the experiment with the following modification: following a 15-min galactose induction of HA-Dbf4p, glucose (2%) and cycloheximide (10 μg/ml) were added to repress transcription and translation, respectively (Fig. 6B). The majority of the Dbf4p disappeared within 10 min of the addition of glucose and cycloheximide. In contrast, under the same conditions, we determined that the stability of Dbf4p in α-factor-arrested cells was greatly enhanced in arrested cdc16-1 mutant cells (Fig. 6D). This indicates a Dbf4p half-life of approximately 5 min and further suggests that Dbf4p degradation is APC dependent. Such a short half-life for Dbf4p correlates well with the measured stabilities of other known APC substrates, including Ase1p and Cdc5p, which also have 5-min half lives (2, 6, 9, 28, 46).
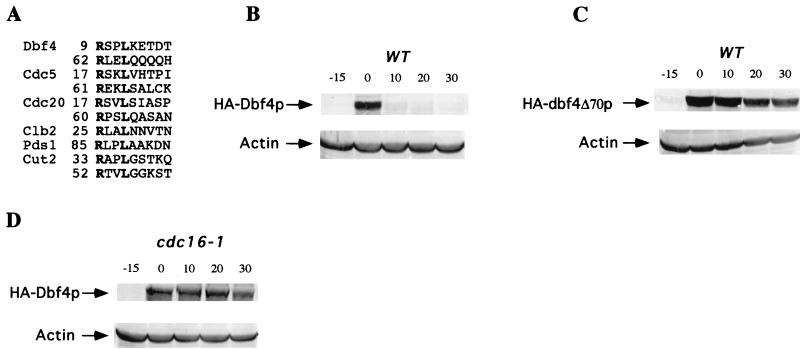
FIG. 6.
Degradation of Dbf4p depends on sequences containing destruction boxes. (A) Localization of putative destruction boxes in Dbf4p and alignment with destruction boxes of other APC substrates. (B and C) Degradation of Dbf4p in G1 depends on the N-terminal 70 amino acids. Wild-type (WT) cells expressing either HA-tagged Dbf4p (GAL-HA-DBF4, pCH920) or a version lacking the N-terminal 70 amino acids (GAL-HA-dbf4ΔN70, pCH955) under the control of the GAL promoter were arrested in G1 with α-factor. The Dbf4p proteins were detected after induction and subsequent repression of the GAL promoter as described for Fig. 5 with the following modification: following a 15-min galactose induction of HA-Dbf4p, glucose (2%) and cycloheximide (10 μg/ml) were added to repress transcription and translation, respectively. (D) Degradation of Dbf4p in G1 depends on the APC gene component, CDC16. cdc16-1 cells (YCH404) expressing HA-tagged Dbf4p (GAL-HA-DBF4, pCH920) were arrested in G1 with α-factor and then shifted to 37°C for 15 min prior to induction. Dbf4p was detected after induction and subsequent transcriptional and translational repression as described for panels B and C.
All known substrates of the APC contain a destruction box motif characterized by conserved arginine and leucine residues in positions 1 and 4 of a 9- to 10-amino-acid sequence. This destruction box is required for APC-mediated proteolysis and was first identified in vertebrate Clb cyclins (29). The N-terminal region of Dbf4p contains two putative destruction boxes (Fig. 6A). To test their role in Dbf4p degradation, the region containing these motifs was deleted, resulting in a deletion of the N-terminal 70 amino acid residues of Dbf4p (dbf4ΔN70). When expressed in wild-type cells as for Fig. 5, the Dbf4ΔN70 protein was greatly stabilized compared to Dbf4p (Fig. 6C). Thus, the region harboring the destruction box motifs is necessary for degradation of Dbf4p. These results provide further support for the proposal that Dbf4p degradation is dependent on the APC.
Modification of Dbf4p in cdc13-1 cells is dependent on Mec1p.
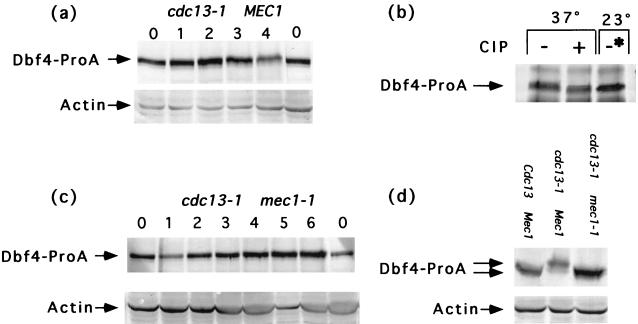
During the immunoblot analysis of Dbf4p levels in cell cycle-arrested cells (Fig. 2), we observed that the electrophoretic mobility of Dbf4p was modified in cdc13-1 cells. The migration of Dbf4p in arrested cdc13-1 cells was notably slow. In contrast, the shifted form of Dbf4p was not observed in cycling cells (Fig. 1) or in cells grown in either hydroxyurea or nocodazole, which arrest in S phase and metaphase, respectively (Fig. 2). The appearance of the modified form of Dbf4p in cdc13-1 cells was further monitored in a time course following a shift of the cdc13-1 culture to the restrictive temperature (Fig. 7a). We determined that the modified form of Dbf4p was sensitive to phosphatase treatment (Fig. 7b), and therefore the shift in arrested cdc13-1 cells was due to phosphorylation of Dbf4p.
FIG. 7.
Dbf4p is modified in cdc13-1 cells in a MEC1-dependent manner. (a) DBF4-proA cdc13-1 (YCH216) cells were incubated at 37°C, the nonpermissive temperature, for 4 h. Samples for extract preparation were taken at 1-h intervals. (b) DBF4-proA cdc13-1 (YCH216) cells were grown at 37°C for 4 h. Dbf4-ProA was immunoprecipitated from extract by using IgG-Sepharose, washed into CIP buffer, and divided into two samples. One hundred units of CIP was added to one of the samples (+) but not to the other (−). Dbf4-ProA was also immunoprecipitated from extracts derived from DBF4-proA cdc13-1 (YCH216) cells grown at the permissive temperature. This sample was not exposed to CIP (−∗). All three tubes were placed at 37°C for 30 min. The samples were loaded onto an SDS–7% polyacrylamide gel, and immunoblot analysis was performed with anti-IgG to detect Dbf4-ProA. (c) DBF4-proA cdc13-1 mec1-1 (YCH317) cells were incubated at 37°C for 6 h. Samples for extract preparation were taken at 1-h-intervals. (d) DBF4-proA CDC13 MEC1 (YCH201) cells were incubated at 37°C. DBF4-proA cdc13-1 MEC1 (YCH216) and DBF4-proA cdc13-1 mec1-1 (YCH324) cells were incubated at 37°C for 4 and 6 h, respectively. Samples for extract preparation were taken at these times. Dbf4-ProA and actin were detected by immunoblotting with anti-IgG and antiactin antibodies, respectively.
The cdc13-1 mutant cells are known to activate the DNA damage checkpoint when they are grown under restrictive conditions (14). Therefore, we next asked whether the MEC1 gene was necessary for the modification. The MEC1 gene product is required for the DNA damage checkpoint pathway (14). We monitored the shift of Dbf4p in the cdc13-1 mec1-1 checkpoint-defective strain by immunoblotting following 6 h of growth at the restrictive temperature (Fig. 7b). In this strain, the shifted form of Dbf4p was not observed (Fig. 7c and d). The shift was also not observed in the cdc13-1 rad53-21 checkpoint-defective strain (data not shown). Thus, accumulation of the shifted phosphorylated form of Dbf4p is dependent upon Mec1p and Rad53p function.
DISCUSSION
Here we report that the levels of the initiator factor Dbf4p are cell cycle regulated and decline as cells complete mitosis. Based on immunoblot and immunofluorescence analyses, Dbf4p accumulates in the nuclei of late G1, S, and M phase cells, and its levels decline as cells complete M and begin the next cell cycle. It is well established that progression through and completion of M requires the ubiquitin-dependent proteolysis of specific substrates (29). Stage-specific degradation of these factors is dependent on a 20S particle designated the APC or cyclosome, a ubiquitin protein ligase (30, 38). We show that the level of Dbf4p is sensitive to mutations in genes for key APC components, CDC16 and CDC23, in a manner similar to that for other APC substrates, including Pds1p, Ase1p, Clb cyclins, and the polo-like kinase Cdc5p (6, 7, 9, 28, 35, 46). The pattern of Dbf4p disappearance from cells that are completing mitosis and the sensitivity of Dbf4p degradation during G1 to mutations in components of the APC suggest that Dbf4p is also targeted for degradation by the APC. In addition, the destruction of Dbf4p is dependent on prototypic APC destruction box sequences present in its N-terminal region. We also show that Dbf4p is modified in arrested cdc13-1 cells which activate the DNA damage response pathway and that this modification requires the key DNA damage checkpoint genes MEC1 and RAD53 (14).
Previous studies of Dbf4p function have characterized its interaction with Cdc7p (12, 21, 25). The Cdc7p protein kinase is required for firing of origins during S phase (4, 10). Cdc7p kinase activity peaks during late G1 and is positively regulated by Dbf4p (25). Moreover, Dbf4p is thought to be targeted to origins, based on a one-hybrid assay (12, 21). These results have led to a proposal that the Cdc7p-Dbf4p complex is recruited to origin complexes where it modifies the prereplication complex, leading to initiation (12, 13, 21). These results are consistent with an S phase role for Dbf4p. The results from this paper, combined with our earlier report documenting a Dbf4p interaction with the M phase kinase Cdc5p (21), suggest that Dbf4p may play roles outside S phase.
Dbf4p may mark origins that have initiated DNA replication.
As discussed above, Dbf4p is required just prior to initiation and likely interacts with the prereplication complex (27). The formation of the prereplication complex late in M requires the recruitment of Cdc6p and MCM proteins (13). Interestingly, Dbf4p function can be bypassed by mcm5-bob1, which contains a point mutation in MCM5 (20). In addition, Mcm2p is a target of regulation by Dbf4-Cdc7p during initiation of replication (32). These results suggest that Dbf4p and MCM functions are linked. Dbf4p could play a role in the recruitment of MCM proteins to origins and/or in their removal from origins coincident with initiation of replication. We propose that the degradation of Dbf4p late in M may also be part of the process which mediates origin remodeling late in M. In other words, the destruction of Dbf4p late in M phase and its coincident removal from postreplication complexes may facilitate the recruitment of MCM proteins to origins. If this model is correct, then Dbf4p would be present at origins from just prior to initiation until late in M, when origins transition from the post- to the preinitiation complex. It is clear from the evidence in this report that Dbf4p is present during this interval of the cell cycle (from just prior to initiation to late in M). If Dbf4p is associated with origins during this entire interval, it could act as a marker for origins that have undergone replication during the same cell cycle.
Is Dbf4p a regulator of Cdc5p kinase activity during M?
Dbf4p interacts with Cdc5p, a member of the polo-like kinase family (21). The kinase activity of Cdc5p is cell cycle regulated and peaks during M phase (6, 7). Cdc5p plays a key role in the regulation of the APC late in M (6, 35). Therefore, Dbf4p’s main role during mitosis might be as a regulator of Cdc5p kinase activity. What support is there from this work that Cdc5p and Dbf4p act in concert during mitosis? First, we have determined that Dbf4p is modified in arrested cdc13-1 cells in a MEC1- and RAD53-dependent manner. Cdc5p is also required for adaptation to the DNA damage response. Wild-type yeast cells exposed to DNA damage activate the DNA damage checkpoint, resulting in a cell cycle arrest (14). These cells eventually reenter the cell cycle after an extended metaphase arrest through a process designated adaptation (41). Restrictive growth of cdc13-1 CDC5 cells activates the DNA damage checkpoint and arrests these cells for an extended period in metaphase. In contrast, cdc13-1 cdc5-ad cells remain arrested in G2/M following growth at the restrictive temperature (41). Cdc5p, like Dbf4p, is modified in arrested cdc13-1 cells in a MEC1- and RAD53-dependent manner (7). It will be interesting to determine if Dbf4p plays any role in checkpoint adaptation and if so whether the checkpoint-dependent modifications of Dbf4p and/or Cdc5p are related to any roles they might play in this adaptation.
Second, strikingly, Dbf4p is targeted for destruction late in M by the APC, in a manner similar to that for Cdc5p. The other known substrates of the APC play key roles during mitosis, and their removal late in the cell cycle allows the cell to reset the cell cycle stage (29). A number of APC substrates, including Cdc5p, Cdc20p, and Clb/Cdk, also play key roles in regulating APC function (6, 29, 35, 44). Taken together, these results hint at a role for Dbf4p in regulating APC activity late in M, perhaps through its interactions with Cdc5p. However, other results suggest that Dbf4p does not play a role in APC regulation. Overexpression of Clb2p in mutant cells with APC defects is lethal (6), whereas overexpression of Clb2p does not adversely affect the growth of a dbf4-1 mutant (data not shown). In addition, the APC activity level in dbf4-1 cells grown at the permissive temperature is identical to that in wild-type cells (6a). Finally, overexpression of APC regulators CDC5 and HCT1 in yeast increases APC activity, leading to a decrease in Clb2p levels (6, 44). In contrast, overexpression of DBF4 has no affect on APC activity or Clb2 levels in cells (6a). These results suggest that Dbf4p’s interactions with Cdc5p are not involved in regulating the APC.
In conclusion, the cell cycle regulation of Dbf4p and its interactions with the G1/S and M phase kinases Cdc7p and Cdc5p, respectively, suggest that Dbf4p is a link between the factors which control M and those which control initiation of DNA replication. In order to clarify these interactions, our future studies will be directed at understanding both the dynamics of Dbf4p at origins and the role of Dbf4p-Cdc5p during mitosis.
ACKNOWLEDGMENTS
We thank L. Hartwell, K. Nasmyth, J. Cooper, and M. Rout for strains and plasmids, and we thank S. Wente for comments on the manuscript.
This work was supported by grant GM5678801 from NIH to G.F.J.H.
REFERENCES
- 1.Aitchison J D, Rout M P, Marelli M, Blobel G, Wozniak R W. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis resists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–66. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 4.Bousset K, Diffley J. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman J W, Johnston L H. The yeast gene, DBF4, essential for entry into S phase is cell cycle regulated. Exp Cell Res. 1989;180:419–428. doi: 10.1016/0014-4827(89)90068-2. [DOI] [PubMed] [Google Scholar]
- 6.Charles J, Jaspersen S, Tinker-Kulberg R, Hwang L, Szidon A, Morgan D O. The polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- 6a.Charles, J., C. F. J. Hardy, and D. Morgan. Unpublished data.
- 7.Cheng L, Hunke L, Hardy C F J. Cell cycle regulation of the Saccharomyces cerevisiae polo-like kinase Cdc5p. Mol Cell Biol. 1998;18:7360–7370. doi: 10.1128/mcb.18.12.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffley J F X. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 9.Cohen-Fix O, Peters J M, Kirschner M W, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 10.Donaldson A, Fangman W, Brewer B. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donovan S, Harwood J, Drury L S, Diffley J F X. Cdc6p dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowell S J, Romanowski P, Diffley J F X. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- 13.Dutta A, Bell S P. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- 14.Elledge S. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 15.Epstein C B, Cross F R. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- 16.Fangman W L, Brewer B. A question of time; replication origins of eukaryotic chromosomes. Cell. 1992;71:363–366. doi: 10.1016/0092-8674(92)90505-7. [DOI] [PubMed] [Google Scholar]
- 17.Fangman W L, Brewer B J. Activation of replication origins within yeast chromosomes. Annu Rev Cell Biol. 1991;7:375. doi: 10.1146/annurev.cb.07.110191.002111. [DOI] [PubMed] [Google Scholar]
- 18.Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yangida M. Cut2 proteolysis is required for sister-chromatid exchange separation in fission yeast. Nature. 1996;381:438–41. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 19.Hand R. Eukaryotic DNA: organization of the genome for replication. Cell. 1978;15:317–325. doi: 10.1016/0092-8674(78)90001-6. [DOI] [PubMed] [Google Scholar]
- 20.Hardy C F J, Dryga O, Seematter S, Pahl P M B, Sclafani R A. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardy C F J, Pautz A. A novel role for Cdc5p in DNA replication. Mol Cell Biol. 1996;16:6775–6782. doi: 10.1128/mcb.16.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hereford L M, Hartwell L M. Sequential gene function in the initiation of Saccharomyces cerevisiae DNA synthesis. J Mol Biol. 1974;84:445–461. doi: 10.1016/0022-2836(74)90451-3. [DOI] [PubMed] [Google Scholar]
- 23.Holloway S L, Glotzer M, King R W, Murray A W. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;73:1393–402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- 24.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson A L, Pahl P M B, Harrison K, Rosamond J, Sclafani R A. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs C W, Adams A E M, Szaniszlo P J, Pringle J R. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston L H, Thomas A P. A further two mutants defective in initiation of the S phase in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1982;186:445–448. doi: 10.1007/BF00729467. [DOI] [PubMed] [Google Scholar]
- 28.Juang Y L, Huang J, Peters J M, Mclaughlin M E, Tai C Y, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- 29.King R W, Deshaies R J, Peters J M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 30.King R W, Peter J M, Tugendreich S, Rolfe M, Heiter P, Kirschner M W. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 31.Kitada K, Johnston L H, Sugino T, Sugino A. Temperature-sensitive cdc7 mutations of Saccharomyces cerevisiae are suppressed by the DBF4 gene, which is required for the G1/S cell cycle transition. Genetics. 1992;131:501–567. doi: 10.1093/genetics/131.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei M, Kawasaki Y, Young M R, Kihara M, Sugino A, Tye B K. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–74. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pringle J R, Hartwell L H. The life cycle of yeast. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 97–142. [Google Scholar]
- 35.Shirayama M, Zachariae W, Coisk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20/fizzzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–49. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 37.Stueland C S, Lew D J, Cismowski M J, Reed S I. Full activation of p34CDC28 histone H1 kinase activity is unable to promote entry into mitosis in checkpoint-arrested cells of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3744–3755. doi: 10.1128/mcb.13.6.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca F C, Ruderman J V, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 41.Toczyski D P, Galgoczy D J, Harwell L H. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- 42.Uhlen M, Guss B, Nillson B, Gatenback S, Philipson L, Lindberg M. Compete sequence of the staphylococcal gene encoding protein A: a gene evolved through multiple duplications. J Biol Chem. 1984;259:13628–13638. [PubMed] [Google Scholar]
- 43.Van der Velden H M, Lohka M J. Mitotic arrest caused by the amino terminus of Xenopus cyclin B2. Mol Cell Biol. 1993;13:1480–1488. doi: 10.1128/mcb.13.3.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 45.Wente S R, Rout M P, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zachariae W, Nasmyth K. TPR proteins required for anaphase progression mediate ubiquitination of mitotic B-type cyclins in yeast. Mol Biol Cell. 1996;7:791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]