Summary
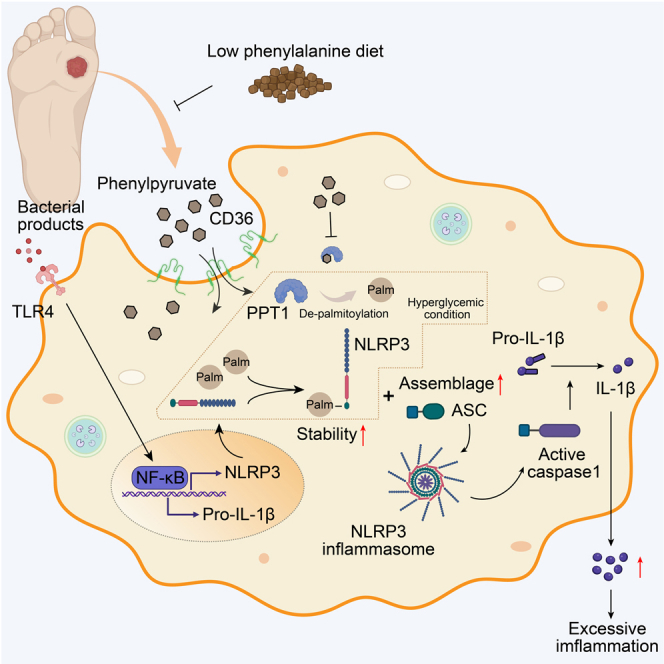
Moderate inflammation is essential for standard wound healing. In pathological conditions, such as diabetes, protracted and refractory wounds are associated with excessive inflammation, manifested by persistent proinflammatory macrophage states. However, the mechanisms are still unclear. Herein, we perform a metabolomic profile and find a significant phenylpyruvate accumulation in diabetic foot ulcers. Increased phenylpyruvate impairs wound healing and augments inflammatory responses, whereas reducing phenylpyruvate via dietary phenylalanine restriction relieves uncontrolled inflammation and benefits diabetic wounds. Mechanistically, phenylpyruvate is ingested into macrophages in a scavenger receptor CD36-dependent manner, binds to PPT1, and inhibits depalmitoylase activity, thus increasing palmitoylation of the NLRP3 protein. Increased NLRP3 palmitoylation is found to enhance NLRP3 protein stability, decrease lysosome degradation, and promote NLRP3 inflammasome activation and the release of inflammatory factors, such as interleukin (IL)-1β, finally triggering the proinflammatory macrophage phenotype. Our study suggests a potential strategy of targeting phenylpyruvate to prevent excessive inflammation in diabetic wounds.
Graphical abstract
Highlights
-
•
Phenylpyruvate mediates inflammatory macrophage infiltration in diabetic wounds
-
•
Dietary phenylalanine restriction is a strategy to reduce phenylpyruvate accumulation
-
•
Phenylpyruvate enters macrophages via CD36 and promotes M1 macrophage polarization
-
•
Phenylpyruvate binds to PPT1 protein and increases NLRP3 palmitoylation level
The broad roles of immunomodulatory metabolites in excessive inflammation in diabetic wounds are not completely understood. Lv et al. identify that phenylpyruvate-mediated NLRP3 palmitoylation results in pathologic inflammation in diabetic wounds. This discovery provides a potential strategy of targeting phenylpyruvate for diabetic wound healing.
Introduction
Diabetic foot ulcers (DFUs) are one of the most common and challenging chronic complications of diabetes and occur over a prolonged disease and refractory treatment cycle.1,2 Over the past few decades, DFUs have been a serious global healthcare issue and remain the primary cause of nontraumatic lower limb loss. In addition, DFUs are closely associated with an increased risk of disability and mortality in diabetes mellitus, and the 5-year survival rate of DFU patients is poorer than that of the most common cancers.3 One exciting area of ongoing research about DFUs concentrates on the abnormal polarization of macrophages, which plays a critical role in mediating chronic inflammation in diabetic wounds.4,5 Specifically, an impaired transition from proinflammatory phenotypes into pro-healing phenotypes leads to the aggregation of inflammatory mediators and hinders diabetic wound healing. Therefore, it is of great scientific importance to explore the pathogenic factors of chronic inflammation in diabetic wounds and develop better therapeutic methods for individualized prevention and treatment.
Nucleotide-binding oligomerization domain-like receptor (NLR) family pyrin domain-containing 3 (NLRP3) interacts with adaptor protein apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) and caspase-1 to assemble an intracellular protein complex known as the NLRP3 inflammasome. It acts as an innate immune trigger initiating inflammatory responses in the presence of sterile danger signals or certain microbial products.6,7 When the inflammasome is activated, bioactive interleukin (IL)-1β and IL-18 are generated under the cleavage of caspase-1, and then released into the microenvironment to enhance the adaptive immune response. Chronic activation of the NLRP3 inflammasome underlines the pathogenesis of several autoinflammatory diseases, including rheumatoid arthritis8,9 and gout.10,11 New evidence suggests that elevated NLRP3 levels in people with diabetes contribute to chronic inflammation and oxidative stress, thus impairing wound healing.12,13 Even so, further research is needed for fully understanding the regulation of NLRP3 in diabetic wounds and determining the best approaches to treat this condition.
Cumulative studies have revealed that substantial metabolic perturbations exist in diabetes mellitus and its complications, which may be aroused by the onset of hyperglycemia or occur independently.14,15 Changes in metabolome are supposed to reflect the molecular characteristics most mimicking to individual phenotypes because they integrate information from exogenous exposures as well as genome, transcriptome, and proteome. Metabolic reprogramming is closely associated with adaptive immunity and inflammation regulation.16,17,18 For example, α-ketoglutarate (αKG) generated from glutaminolysis orchestrates macrophage activation via Jmjd3-dependent metabolic and epigenetic reprogramming of M2 genes.19 Itaconate and its derivatives impair alternative activation of macrophages by directly modifying cysteine residues on JAK1 and inhibiting its phosphorylation.20 However, how metabolic perturbations in DFUs regulate uncontrolled local adaptive inflammatory responses has not been elucidated.
Herein, we mapped metabolic profiles for both DFU wounds and nondiabetic wounds (NDWs) and identified a remarkable perturbation in phenylalanine metabolism, especially an obvious accumulation of phenylpyruvate (PPA) in DFU wounds, which was positively correlated with NLRP3 levels. Phenylpyruvate was found to increase NLRP3 expression levels, sustain the polarization of proinflammatory macrophages, and impair wound healing in vivo. Administration of a restricted phenylalanine (Phe) diet abrogated the increase of phenylpyruvate, improved the inflammatory response, and accelerated diabetic wound healing. Mechanistically, phenylpyruvate is taken into macrophages via the scavenger receptor CD36 and then binds to and inhibits the enzyme activity of PPT1 protein, resulting in increased palmitoylation of the NLRP3 protein, enhanced protein stability, and decreased lysosome degradation. It also promoted the assembly of the NLRP3 inflammasome and increased the release of inflammatory factors, thus maintaining the continuous polarization of the proinflammatory phenotype of macrophages. Our study suggests phenylpyruvate modulation as a potential therapeutic strategy for chronic inflammation in diabetic wound healing.
Results
Global comparative metabolomic analysis between DFU wounds and nondiabetic wounds
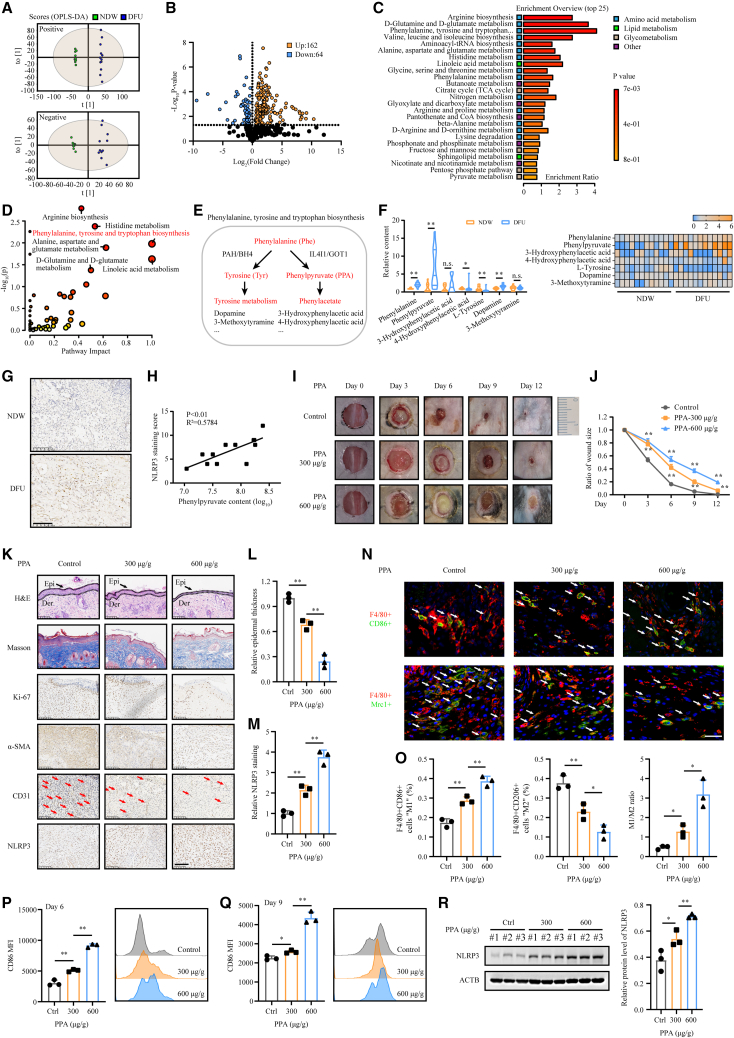
To explore important metabolic patterns and distinguish DFU wounds and nondiabetic wounds, UHPLC-HRMS-based metabolomics analysis was performed in both the DFU group and NDW group (n = 12). The orthogonal projections to latent structures discriminant analysis (OPLS-DA) model effectively distinguished between groups in both positive and negative ion models (Figure 1A). Compared to the NDW group, the DFU group was found to have many altered metabolites. Among the 226 metabolites identified following manual filtering (Table S1), 162 metabolites were found to be increased in DFUs and the rest were decreased (Figure 1B). According to the metabolic enrichment analysis, the differentially expressed metabolites were mainly involved in amino acid metabolism, including arginine biosynthesis, D-glutamine and D-glutamate metabolism, and phenylalanine, tyrosine, and tryptophan biosynthesis (Figure 1C). In the metabolic pathway analysis, considering the p value and pathway impact value, the phenylalanine, tyrosine, and tryptophan biosynthesis pathways were chosen for further investigation (Figure 1D).
Figure 1.
Metabolome profile identifies accumulation of phenylpyruvate in DFUs, which is found to impair wound healing and prolong inflammation
(A) Score plot presenting the metabolic separation for pair splitting between the NDW group and the DFU group through OPLS-DA (n = 12).
(B) A volcano plot shows the differential metabolites in the comparison of DFUs to NDWs.
(C) Overview of the top 25 terms in the metabolic enrichment analysis of the differential metabolites.
(D) Pathway enrichment analysis of differential metabolites between DFUs and NDWs. The ordinate represents the p value of pathway enrichment analysis (negative logarithm). The abscissa represents the influencing factor of pathway topological analysis. The larger the circle is, the higher the influence factor. A deeper red color indicates a larger −log(p) value, indicating more enrichment.
(E) Schematic representation of phenylalanine, tyrosine, and tryptophan biosynthesis.
(F) Histogram and heatmap generated by relative fold change of differential metabolites in the phenylalanine, tyrosine, and tryptophan biosynthesis pathways.
(G) Representative IHC staining images of NLRP3 in the DFU group and NDW group.
(H) Correlation analysis between phenylpyruvate levels and NLRP3 expression in DFUs (n = 12).
(I) Representative images of cutaneous wounds of age-matched C57BL/6JGpt mice on days 0, 3, 6, 9, and 12 after wound model generation by surgical excision. n = 12 for each group.
(J) Ratio of wound sizes were quantified by using ImageJ software and were calculated by the percentages of wound closure compared to day 0 wound size.
(K) Cutaneous wound sections were subjected to H&E and Masson’s trichrome staining, and IHC staining using Ki-67, α-SMA, CD31, and NLRP3 antibodies was performed. Samples were collected 6 days after wound model generation. n = 3 mice for sampling at the indicated time points. Scale bar, 100 μm.
(L) Histological staining analysis showing relative epidermal thickness (n = 3).
(M) Statistical analysis of NLRP3 staining in cutaneous wounds (n = 3).
(N and O) Representative immunofluorescence images and analysis showing macrophage infiltration in cutaneous wounds on day 6. n = 3 mice in each group. Scale bar, 20 μm.
(P and Q) Flow cytometry analysis of macrophage infiltration in cutaneous wounds on days 6 and 9. The representative histogram shows the levels of F4/80+CD86+ cells in cutaneous wounds from these groups. Statistical analysis indicates the mean fluorescence intensity (MFI) of CD86 (n = 3).
(R) Western blot image and analysis showing NLRP3 protein expression levels in cutaneous wounds from these groups (n = 3). Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01; n.s., not significant.
Phenylalanine is a necessary amino acid for human nutrition. When phenylalanine is consumed, it normally enters the tyrosine metabolism pool by hydroxylation. When the hydroxylation of phenylalanine is dysfunctional, the accumulation of phenylalanine and its harmful metabolic products, such as phenylpyruvate, leads to a series of pathologies, such as the well-known phenylketonuria (PKU) (Figure 1E). In our metabolomic profile analysis, we discovered that the phenylalanine level in DFU wounds was slightly increased, accompanied by a significant elevation of phenylpyruvate levels (Figure 1F). Metabolite correlation analysis found a strong positive correlation between phenylpyruvate and phenylalanine, but a negative correlation between phenylpyruvate and tyrosine, indicating that the classical catabolism of phenylalanine was dysregulated and the significantly increased phenylpyruvate was derived from phenylalanine (Figure S1A). Persistent NLRP3 inflammasome activity was found to contribute to chronic inflammation in diabetic wounds,12 and we confirmed through immunohistochemistry (IHC) staining that the NLRP3 expression level was higher in DFU wounds than in NDWs (Figure 1G). Surprisingly, we observed a strong positive correlation between NLRP3 levels and phenylpyruvate levels in the DFU group (Figure 1H) but not for other significantly altered metabolites in the phenylalanine metabolic pathway (Figures S1B–S1E). Furthermore, targeted metabolite analysis in mouse wound models also revealed a robust increase in phenylpyruvate levels in the wound edge of diabetic mice compared to those in the wound edge of nondiabetic mice (Figures S1F and S1G). However, according to previous investigations, no significant alteration in phenylpyruvate levels was found in the serum of individuals with diabetic wounds.21,22 Overall, our global metabolomic profile analysis revealed a significant accumulation of phenylpyruvate in DFU wounds, which positively correlated with the levels of the inflammatory protein NLRP3.
Phenylpyruvate interferes with macrophage polarization proportion and impedes wound healing
To further elucidate the effect of phenylpyruvate on wound healing dynamics, different concentrations of phenylpyruvate were injected into the wound edges of the mice using microsyringes. Delayed wound healing was observed in the treated mice in a phenylpyruvate dose-dependent manner (Figures 1I and 1J). Histological examination revealed thinner epidermises in the phenylpyruvate-treated groups than in the control group (Figures 1K and 1L). In addition, we observed a decreased collagen volume fraction and reduced α-smooth muscle actin (α-SMA) accumulation after injection of phenylpyruvate (Figures S1H and S1J). A lower proportion of Ki-67-positive cells and less angiogenesis were also found in the phenylpyruvate-treated groups (Figures S1I and S1K). These results demonstrated that phenylpyruvate could greatly suppress wound healing. Subsequently, immunofluorescence staining was performed to explore macrophage infiltration in the wound tissues. Phenylpyruvate injection increased the number of F4/80+CD86+ cells, which represented the proinflammatory M1 phenotype, and reduced the number of F4/80+CD206+ cells, which represented the pro-healing M2 phenotype, resulting in an elevated ratio of M1/M2 macrophages (Figures 1N and 1O). As expected, flow cytometry assays showed that phenylpyruvate treatment promoted the persistent polarization of proinflammatory macrophages on days 6 and 9 (Figures 1P and 1Q). In addition, we found that phenylpyruvate injection significantly increased the NLRP3 expression levels in the wound (Figures 1M and 1R) and induced elevated levels of inflammatory factors, including IL-1β and tumor necrosis factor alpha (TNFα) (Figures S1L and S1M). In conclusion, phenylpyruvate can trigger an imbalance in macrophage polarization, resulting in increased inflammation and impaired wound healing.
Dietary phenylalanine restriction improves persistent inflammation and benefits diabetic wound healing
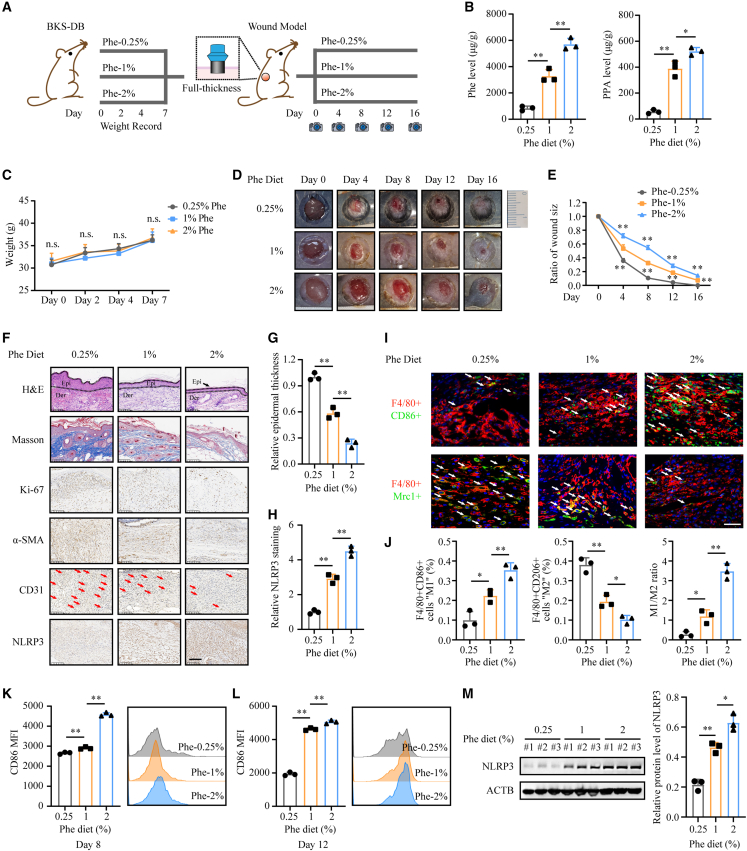
Based on the identification of phenylpyruvate accumulation in diabetic wounds, we further explored whether it can be used as a therapeutic target to improve the healing of diabetic wounds. Since phenylpyruvate is a metabolite of phenylalanine, dietary phenylalanine restriction has been the main treatment of patients with PKU and achieves a satisfactory outcome23; therefore, we speculated that dietary phenylalanine restriction may be helpful in reducing the accumulation of phenylpyruvate in diabetic wounds. We designed a diet that differed only in the phenylalanine ratio (Table S2). According to a previous study, phenylalanine deficiency can cause weight loss in mice, while supplementation with 0.2%–0.25% phenylalanine can eliminate this effect.24 Prior to wound modeling in BKS-DB mice, the mice were fed a diet with different phenylalanine concentrations for 1 week, and body weight, blood glucose level, and food intake were recorded. This feeding pattern continued throughout the experiment (Figure 2A). We demonstrated by metabolic mass spectrometry analysis that a phenylalanine-restricted diet reduced the accumulation of phenylpyruvate in diabetic wounds (Figure 2B). Moreover, body weight, blood glucose level, and food intake were not influenced by this feeding diet (Figures 2C, S2A, and S2B). No significant histological changes were observed in the heart, kidney, liver, lung, muscle, pancreas, or spleen (Figure S2I). We found that dietary phenylalanine restriction clearly accelerated diabetic wound healing (Figures 2D and 2E). Histological examination revealed thicker epidermises, an increased collagen volume fraction, more α-SMA accumulation, a higher proportion of Ki-67-positive cells, and more angiogenesis in the phenylalanine-restricted diet group (Figures 2F, 2G, and S2C–S2F). Subsequently, immunofluorescence staining identified that phenylalanine limitation reduced the number of proinflammatory macrophages and increased the number of pro-healing macrophages on day 8, with a smaller ratio of M1/M2 macrophages (Figures 2I and 2J). Flow cytometry assays demonstrated that higher phenylalanine intake levels led to more polarization of proinflammatory macrophages on days 6 and 9, but the phenylalanine restriction diet did not (Figures 2K and 2L). In addition, dietary phenylalanine restriction reduced NLRP3 expression levels to a great extent (Figures 2H and 2M) and decreased the levels of IL-1β and TNFα (Figures S2G and S2H). Overall, dietary phenylalanine limitation can reduce phenylpyruvate accumulation in diabetic wounds, reduce excessive inflammation, and promote healing, with little effect on the general condition of the body.
Figure 2.
Dietary phenylalanine restriction can reduce the accumulation of phenylpyruvate and promote diabetic wound healing
(A) Schematic representation of diabetic wound model mice fed different phenylalanine diets. n = 12 for each group.
(B) Detection of the indicated metabolites in cutaneous wounds from different groups by metabolic mass spectrometry (n = 3).
(C) Recorded weights of diabetic mice fed a phenylalanine restriction diet for 1 week before wound model generation (n = 12).
(D) Representative images of cutaneous wounds of diabetic mice on days 0, 4, 8, 12, and 16 after wound model generation by surgical excision.
(E) Percentage of wound sizes were quantified by using ImageJ software and were calculated by the percentages of wound closure compared to day 0 wound size.
(F) Cutaneous wound sections from different groups were subjected to H&E and Masson’s trichrome staining and IHC staining with Ki-67, α-SMA, CD31, and NLRP3 antibodies. Samples were collected at 8 days after wound model generation. n = 3 mice for sampling at the indicated time points. Scale bar, 100 μm.
(G) Histological staining analysis showing relative epidermal thickness (n = 3).
(H) Statistical analysis of NLRP3 staining in cutaneous wounds from different groups (n = 3).
(I and J) Representative immunofluorescence images and statistical analysis showing macrophage infiltration in cutaneous wounds on day 8. n = 3 mice in each group. Scale bar, 20 μm.
(K and L) Flow cytometry analysis of macrophage infiltration in cutaneous wounds from different groups on days 8 and 12. The representative histogram shows the levels of F4/80+CD86+ cells in cutaneous wounds from these groups. Statistical analysis indicates the MFI of CD86 (n = 3).
(M) Western blot images and analysis showing NLRP3 protein expression levels in cutaneous wounds from these groups (n = 3). Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01; n.s., not significant.
Phenylpyruvate can be taken up by macrophages and promote a proinflammatory macrophage phenotype
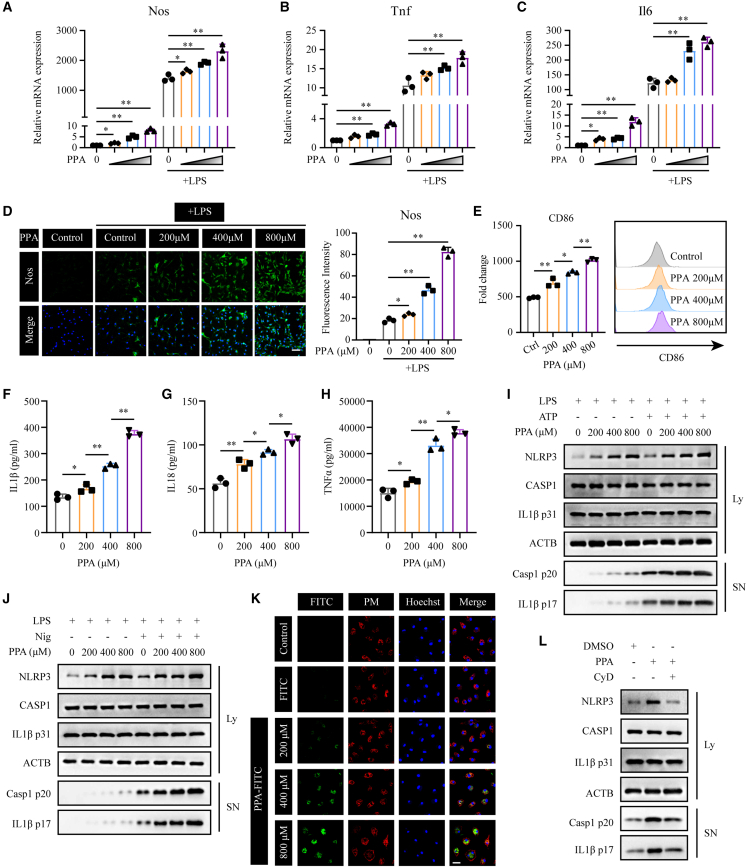
Based on the observations in vivo, phenylpyruvate regulates macrophage polarization. In diabetic wounds, early macrophages are differentiated from infiltrating bone marrow-derived monocytes. To investigate how phenylpyruvate regulates macrophage activation, we first determined the effect of phenylpyruvate treatment on diabetic mouse bone marrow-derived macrophages (BMDMs) treated with LPS or IL-4 in high-glucose (30 mM) culture medium25 (Figure S3A). Phenylpyruvate treatment increased the LPS-induced expression of the M1 signature genes Nos, Tnf, and Il6 in BMDMs in a dose-dependent manner (Figures 3A–3C). In contrast, phenylpyruvate treatment did not significantly alter the IL-4-induced M2-signature genes, including Arg1, Chil3, and Retnla (Figures S3B–S3D). Immunocytochemistry analysis showed that phenylpyruvate treatment upregulated Nos expression in LPS-treated BMDMs (Figure 3D) but there was no change in Mrc1 expression in IL-4-treated BMDMs (Figure S3E). Additionally, there was an obviously elevation of the M1 marker CD86 protein on the surface of BMDMs (Figure 3E), with no alternation in the M2 surface marker CD163 (Figure S3F). We also found that treatment of LPS + ATP-stimulated macrophages with phenylpyruvate increased the secretion of IL-1β, IL-18, and TNFα into the supernatant (Figures 3F–3H). We next stimulated LPS-primed BMDMs with ATP or nigericin (Nig) and observed that phenylpyruvate treatment increased the protein level of NLRP3 in a dose-dependent manner, as well as increasing the levels of cleaved caspase-1 and mature IL-1β in the culture medium (Figures 3I and 3J). However, RT-qPCR analyses revealed that the RNA expression levels of NLRP3 remained unaffected by phenylpyruvate treatment (Figure S3G). No significant difference was observed in the upstream events of NLRP3 inflammasome activation in BMDMs treated with phenylpyruvate (Figure S3H).
Figure 3.
Phenylpyruvate is taken up by BMDMs and triggers a proinflammatory macrophage phenotype
(A–C) BMDMs were pretreated with dimethyl sulfoxide (DMSO, control) or increasing concentrations of phenylpyruvate for 4 h and then stimulated with media or LPS (100 ng/mL) for 24 h. The black triangle indicates increasing phenylpyruvate concentrations starting from the left: 200, 400, and 800 μM. The RT-qPCR results showing relative mRNA expression of Nos, Tnf, and Il6 normalized to Actb (n = 3).
(D) Immunostaining and statistical analysis of Nos in BMDMs after pretreatment with phenylpyruvate and stimulation with LPS (n = 3). Scale bar, 150 μm.
(E) BMDMs were treated as in (A). Flow cytometry data for the M1 surface marker CD86, showing MFI (left) and representative histograms of CD86 (right) (n = 3).
(F–H) BMDMs were pretreated with increasing phenylpyruvate concentrations for 4 h and then stimulated with LPS (100 ng/mL) for 24 h and ATP (3 mM) for 45 min. Inflammatory factors IL-1β, IL-18, and TNFα in the supernatant were measured using ELISA assay (n = 3).
(I) Immunoblots of supernatants (SN) and cell extracts (lysate) of BMDMs treated as in (F) (n = 3).
(J) Immunoblots of supernatants (SN) and cell extracts (lysate) of BMDMs, which were pretreated with increasing phenylpyruvate concentrations for 4 h and then stimulated with LPS (100 ng/mL) for 24 h and nigericin (10 μM) for 45 min (n = 3).
(K) Immunostaining showing the uptake of phenylpyruvate into BMDMs. The FITC antibody indicated phenylpyruvate, and the lipid raft antibody indicated plasma membrane. Scale bar, 50 μm.
(L) BMDMs were pretreated with phenylpyruvate (400 μM) with or without cytochalasin D (10 μM) and then stimulated with LPS (100 ng/mL) for 24 h and ATP (3 mM) for 45 min. Immunoblots of the supernatants (SN) and cell extracts (lysate) (n = 3). Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01.
To clarify whether phenylpyruvate acts as an intercellular or intracellular signaling agent, we investigated whether phenylpyruvate could enter macrophages. We first synthesized phenylpyruvate-coupled fluorescein isothiocyanate (FITC) complex by chemical synthesis. We found that the majority of phenylpyruvate entered macrophages and distributed uniformly in the cytoplasm, and a small part of phenylpyruvate remained in the cell membrane (Figure 3K). Blocking phagocytosis in BMDMs with cytochalasin D (CyD) markedly reduced the uptake of phenylpyruvate (Figure S3I). In addition, blocking phagocytosis prevented phenylpyruvate from increasing NLRP3 protein levels and the secretion of cleaved caspase-1 and mature IL-1β (Figure 3L). In conclusion, phenylpyruvate can enter macrophages and promote the proinflammatory phenotype.
Since phenylpyruvate is derived from phenylalanine, we spontaneously wonder whether phenylalanine could also promote macrophage inflammatory phenotypes as phenylpyruvate does. We treated BMDMs with phenylalanine at the same concentration as phenylpyruvate but found no differential effect of phenylalanine on the M1 signature genes (Figures S3J–S3L). Phenylalanine also had no effect on NLRP3 protein levels (Figure S3M). These prove that the changes are specific to phenylpyruvate.
PPT1 is a bona fide target of phenylpyruvate
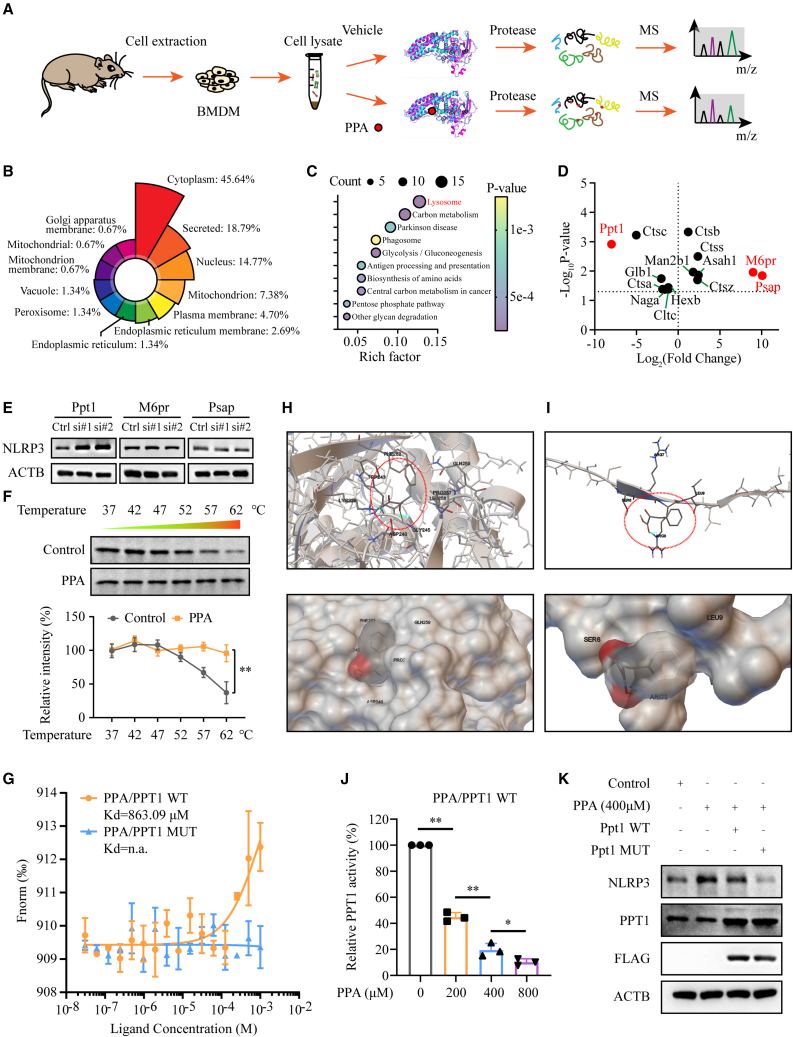
To directly identify that molecular mechanism by which phenylpyruvate regulates macrophage phenotype, we employed a chemical proteomics screening following an LiP-small-molecule mapping (LiP-SMap) workflow26 using BMDM cell lysate to explore cellular proteins that potentially bind to phenylpyruvate (Figure 4A). Significant alternations in the abundance of half tryptic peptides (fold change ≤0.5 or ≥2, p value or p value chitest <0.05) were screened out for structural alternation induced by phenylpyruvate binding. In total, 193 proteins fulfilled these criteria (Table S3). Further bioinformatics analysis indicated that most of these proteins were localized in the cytoplasm (Figure 4B), which was consistent with our previous tracer results. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of these candidate proteins demonstrated that the lysosome pathway was the most enriched term (Figure 4C). Previous studies have revealed that lysosome damage resulting in the release of its contents can activate the NLRP3 inflammasome.27 We then explored whether phenylpyruvate could damage lysosomes to activate the NLRP3 inflammasome. No significant changes in staining with LysoTracker, a reagent allowing the identification of the lysosomal compartment, and LysoSensor, a reagent indicating the pH in lysosomes, were found following exposure to phenylpyruvate (Figures S4A and S4B), suggesting that phenylpyruvate had little effect on lysosomes. We then examined the three most prominent proteins in this pathway, namely Ppt1, M6pr, and Psap (Figure 4D). We knocked down the corresponding targets using small interfering RNA (siRNA) (Figures S4C and S4D) and found that NLRP3 protein levels were significantly increased only after Ppt1 knockdown, while knockdown of the other proteins did not result in NLRP3 expression changes (Figure 4E). In addition, the mRNA level of NLRP3 did not change after Ppt1 knockdown (Figure S4E). These data suggested that the PPT1 protein might be a potential target that mediates the effect of phenylpyruvate on macrophage polarization.
Figure 4.
Identification of potential phenylpyruvate targets in macrophages using LiP-SMap
(A) Flow chart depicting the LiP-SMap assay. The LiP-SMap assay is a chemical proteomics screening method for investigating potential protein-metabolite interactions.
(B) The rose chart reflects the proportion of differentially expressed polypeptide-dependent proteins mapped in each subcellular location.
(C) The bubble plot shows the KEGG pathway enrichment analysis. The abscissa represents the enrichment value, and the ordinate represents the enriched KEGG pathways. The larger the circle is, the more the differentially expressed polypeptide-dependent proteins in the mapping pathway are enriched. The color of the circle indicates the p value.
(D) A spot chart showing the differential proteins mapped in the lysosome pathway. Fold changes greater than 2 or less than 0.5 both indicate potential interactions between proteins and metabolites.
(E) Immunoblot analysis of NLRP3 expression levels in BMDMs with knockdown of Ppt1, M6pr, and Psap (n = 3).
(F) Cellular thermal shift assay and statistical analysis of PPT1 in BMDMs treated with or without phenylpyruvate (n = 3).
(G) Microscale thermophoresis assay of phenylpyruvate binding to the PPT1 WT or MUT protein.
(H) Representative images of autodocking for phenylpyruvate to the principal chain structure of the PPT1 protein.
(I) Representative images of autodocking for phenylpyruvate to the signal domain of the PPT1 protein.
(J) Enzymatic activity measurement of PPT1 incubated with increasing phenylpyruvate concentrations (n = 3).
(K) BMDMs were treated with phenylpyruvate and then transfected with the Ppt1 WT plasmid or the MUT plasmid for 24 h. Immunoblot analysis showing NLRP3 expression in BMDMs (n = 3). Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01; n.a., not applicable.
To validate the binding of phenylpyruvate to PPT1, a thermal shift assay was carried out and showed that phenylpyruvate treatment significantly changed PPT1 protein stability after heating (Figure 4F). We then performed a microscale thermophoresis (MST) assay with PPT1 protein and a dilution series of phenylpyruvate. As expected, PPT1 protein was readily bound by phenylpyruvate, and the dissociation constant (KD) of phenylpyruvate/PPT1 was calculated as 863.09 μM in MST analysis, which was indicative of an interaction between phenylpyruvate and PPT1 (Figure 4G).
PPT1 consists of a signal domain and the principal chain structure, with three essential sites, namely S115, D233, and H289, for its palmitoyl-coenzyme A (CoA) hydrolase activity (Figure S4F). To assess the exact binding site of phenylpyruvate on PPT1, we performed molecular docking using Autodock to detect possible binding modes on the two domains of PPT1. The predicted binding mode of phenylpyruvate to the principal chain structure revealed a suitable shape match; residues Lys229 and Gly245 of PPT1 participated in forming hydrogen bonds with phenylpyruvate (Figure 4H). The predicted binding mode to the signal domain involved hydrogen bonds between phenylpyruvate and residue Arg8 (Figure 4I). Nevertheless, it was obvious that phenylpyruvate fit more favorably to the former binding pocket, as the docking energy was significantly lower in the former mode (−5 kcal/M) than in the latter mode (−3.39 kcal/M).
Since the binding mode of residues Lys229 and Gly245 is adjacent to the catalytic site D233, we wondered whether phenylpyruvate could influence PPT1 activity. As expected, the specific activity of PPT1 was strongly decreased after treatment with phenylpyruvate (Figure 4J). Then, we generated a Ppt1 mutant (MUT) with phenylpyruvate-binding site mutations (Figure S4G), and no binding of phenylpyruvate with the Ppt1 mutant protein was detected (Figure 4G). Moreover, we found that phenylpyruvate did not affect the activity of the Ppt1 mutant (Figure S4H). In addition, Ppt1 MUT plasmid transfection reversed the increase of NLRP3 protein levels induced by phenylpyruvate exposure, while Ppt1 wild-type (WT) transfection did not (Figure 4K). We also identified that phenylpyruvate treatment had no impact on the mRNA and protein levels of PPT1 (Figures S4I and S4J). In conclusion, phenylpyruvate binds and inhibits PPT1 protein activity, which mediates phenylpyruvate-induced upregulation of NLRP3 expression.
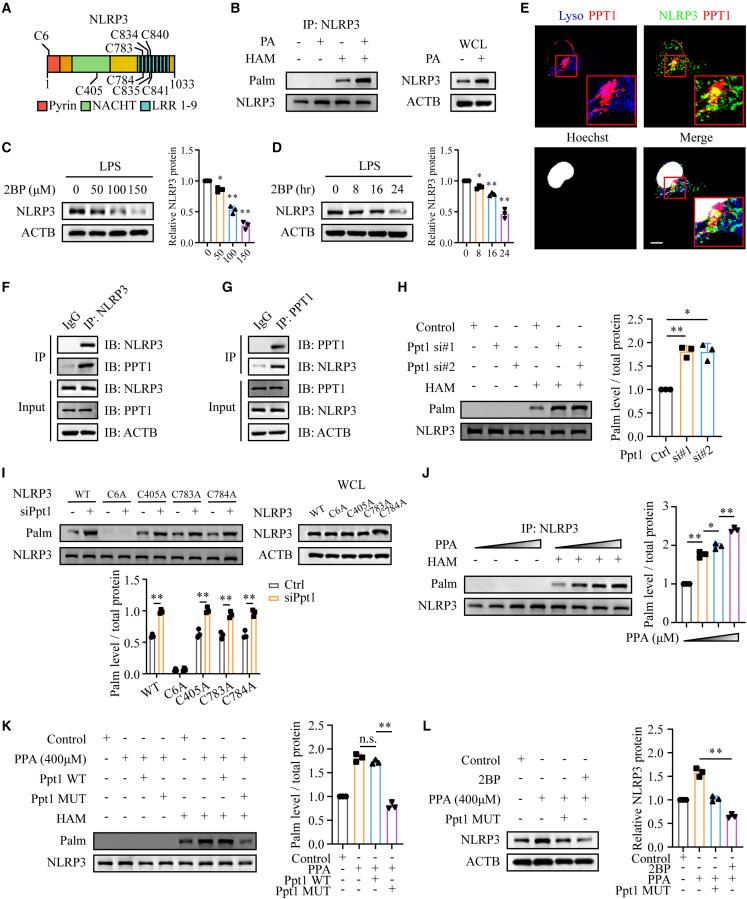
Phenylpyruvate regulates NLRP3 palmitoylation by binding to the PPT1 protein
PPT1, as a thioesterase, can remove palmitic acid from S-acylated proteins, which is essential for dynamic palmitoylation.28 Since phenylpyruvate was proved to inhibit PPT1 activity, we wonder whether phenylpyruvate could regulate NLRP3 palmitoylation. To explore the role of palmitoylation on NLRP3, we first predicted the palmitoylation site(s) on NLRP3 through the online software CSS-Palm (http://csspalm.biocuckoo.org/). The results indicated several palmitoylation sites on NLRP3 that were highly conserved in different species (Figures 5A and S5A). Using an acyl-biotin exchange (ABE) assay in BMDMs (Figure S5B), we verified that NLRP3 was palmitoylated (Figure 5B). Since S-palmitoylation can control protein stability and trafficking, we then explored whether palmitoylation affects NLRP3 protein level by treating cells with a general S-palmitoylation inhibitor, 2-bromopalmitate (2BP). The protein levels of NLRP3 were reduced by 2BP in both a dose-dependent and time-dependent manner (Figures 5C and 5D).
Figure 5.
Phenylpyruvate upregulated NLRP3 palmitoylation by binding to the PPT1 protein
(A) Schematic view of NLRP3 (full length) and the location of the S-palmitoylation sites.
(B) ABE assay and immunoblot analysis were used to evaluate NLRP3 palmitoylation in BMDMs treated with or without palmitic acid (n = 3).
(C) Immunoblot and statistical analysis showing NLRP3 expression in BMDMs treated with 2BP at the indicated concentrations (0, 50, 100, and 150 μM) for 24 h (n = 3).
(D) Immunoblot and statistical analysis showing NLRP3 expression in BMDMs treated with 2BP (100 μM) for 0, 8, 16, and 24 h (n = 3).
(E) Immunostaining of the location of LAMP2 (indicating lysosomes), PPT1, and NLRP3 in macrophages. The nuclei were stained with Hoechst dye. Scale bar, 10 μm.
(F and G) Immunoprecipitation (IP) and immunoblot analysis were used to detect the endogenous NLRP3 and PPT1 association in macrophages (n = 3).
(H) ABE assay and immunoblot analysis were used to evaluate NLRP3 palmitoylation in BMDMs with Ppt1 knockdown (n = 3).
(I) Mouse embryo fibroblasts (MEFs) were transfected with WT NLRP3 or the indicated NLRP3 mutants for 24 h with or without knockdown of Ppt1. ABE assay and immunoblot analysis showing palmitoylation levels of the indicated NLRP3 mutants (n = 3).
(J) ABE assay and immunoblot analysis were used to determine NLRP3 palmitoylation levels in BMDMs treated with increasing phenylpyruvate concentrations (n = 3).
(K) Determination of NLRP3 palmitoylation levels in BMDMs treated with 400 μM phenylpyruvate and then transfected with the Ppt1 WT plasmid or the MUT plasmid for 24 h (n = 3).
(L) BMDMs were treated with 400 μM phenylpyruvate with or without 100 μM 2BP and then transfected with or without the Ppt1 MUT plasmid. ABE assay and immunoblot analysis determining NLRP3 palmitoylation levels in the indicated cells (n = 3). Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01; n.s., not significant.
To further verify our hypothesis, we then focused on whether PPT1 could regulate the palmitoylation of NLRP3. PPT1 was first described to localize in the lysosomal lumen, but recent studies have shown its biology function with extralysosomal localization.29,30,31 Our immunofluorescence staining showed that there was co-localization between PPT1 and NLRP3, both in lysosomal lumen and extralysosomal sites (Figure 5E). Additionally, an immunoprecipitation assay demonstrated an interaction between PPT1 and NLRP3 (Figures 5F and 5G). We also performed an ABE assay and revealed that PPT1 deficiency increased the palmitoylation of NLRP3 (Figure 5H). Based on palmitoylation site prediction analysis, we determined cysteine residues 6, 405, 783, 784, 834, 835, 840, and 841 of NLRP3 as potential palmitoylation sites, which are conserved in other species. Consistent with the largest prediction score of cysteine residues 6, we found that mutation of C6A in NLRP3 mostly abrogated its palmitoylation, and PPT1 deficiency could no longer increase its palmitoylation, demonstrating C6 as the major palmitoylation site in NLRP3 (Figures 5I and S5C). Subsequently, we found that phenylpyruvate treatment can increase the palmitoylation of NLRP3 (Figure 5J). In addition, Ppt1 MUT plasmid transfection reversed the increased palmitoylation of the NLRP3 protein induced by phenylpyruvate, while Ppt1 WT transfection did not (Figure 5K). We also found that, in the presence of 2BP, which blocks palmitoylation, treatment with phenylpyruvate could not increase the expression level of NLRP3 (Figure 5L). Overall, phenylpyruvate can increase the palmitoylation of NLRP3 by binding and inhibiting PPT1 activity.
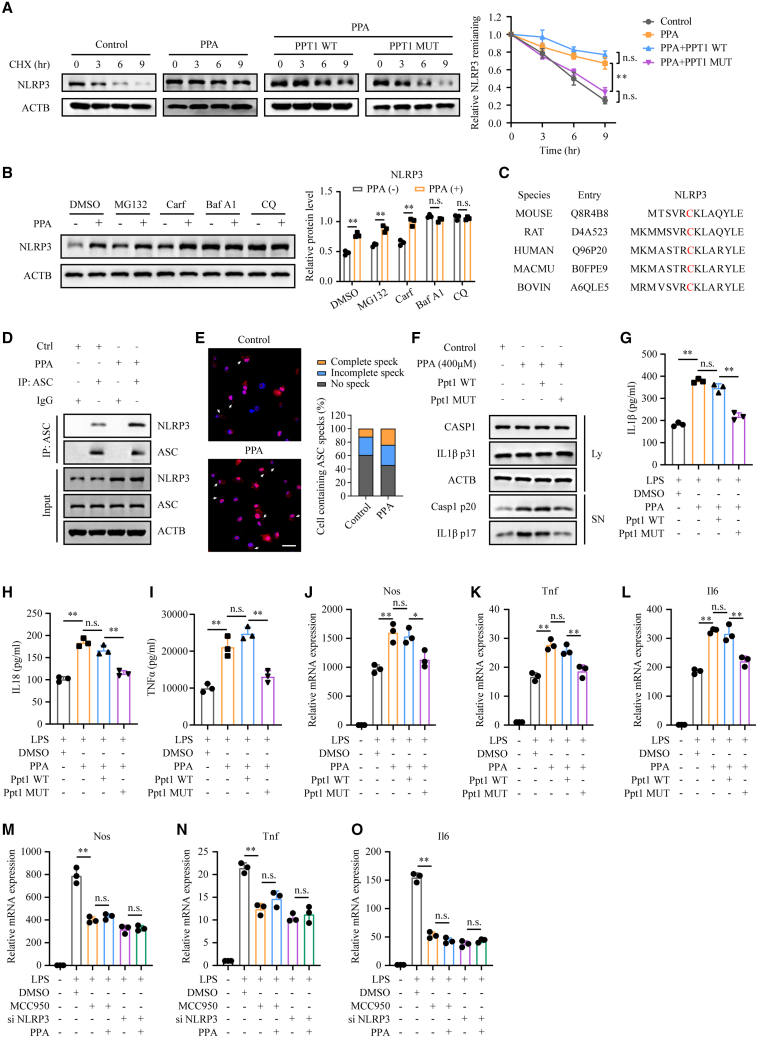
Phenylpyruvate stabilized the NLRP3 protein and promoted inflammasome activation to trigger a proinflammatory macrophage phenotype
According to previous studies, S-palmitoylation is essential for protein stability of immunity effectors, such as NOD2 and programmed death-ligand 1 (PD-L1).32,33 Therefore, we wondered whether the increase in NLRP3 palmitoylation induced by phenylpyruvate could affect its protein stability. We observed that phenylpyruvate treatment markedly enhanced the protein stability of NLRP3, while transfection of the Ppt1 MUT plasmid, but not the WT plasmid, attenuated its stability (Figure 6A). Previous studies have shown that the NLRP3 protein is mainly degraded through the proteasome and lysosome pathways. To determine which degradation pathway of NLRP3 is dominantly regulated by S-palmitoylation, we investigated whether the degradation of NLRP3 under phenylpyruvate treatment could be abrogated by inhibitors of the proteasome or the autophagy degradation pathway. We found that the degradation of NLRP3 by phenylpyruvate treatment could be blocked by autophagy and the autolysosome inhibitors bafilomycin A1 (Baf A1) and chloroquine (CQ) but not the proteasome inhibitors MG132 and carfilzomib (Carf), which indicated that phenylpyruvate treatment prevents NLRP3 from autophagic degradation (Figure 6B).
Figure 6.
Phenylpyruvate increased NLRP3 protein stability and promoted inflammasome activation
(A) Immunoblot assay and statistical analysis showing the stability of NLRP3 protein in the indicated cells. BMDMs were treated with CHX (100 μg/mL) for 0, 3, 6, and 9 h and then collected for immunoblot analysis (n = 3).
(B) Immunoblot analysis showing NLRP3 protein levels in BMDMs treated with proteasome inhibitors MG132 (10 μM), carfilzomib (100 nM), or autophagy and autolysosome inhibitors Baf A1 (0.2 μM) or CQ (50 μM) for 8 h (n = 3).
(C) Alignment of NLRP3 pyrin domain-containing palmitoylation sites among different species.
(D) BMDMs were treated with 400 μM phenylpyruvate for 4 h and LPS (100 ng/mL) for 24 h and then stimulated with ATP (3 mM) for 45 min. Cell lysates were collected to determine the interaction between NLRP3 and ASC by IP and immunoblot analysis (n = 3).
(E) Arrows showing ASC immunofluorescence in BMDMs treated with phenylpyruvate (400 μM) for 4 h and LPS (100 ng/mL) for 24 h, followed by stimulation with ATP (3 mM) for 45 min. Scale bar, 50 μm.
(F) Immunoblot analysis of the supernatant and cell extracts in the indicated BMDMs, which were treated with phenylpyruvate for 4 h and LPS for 24 h, with transfection of Ppt1 WT or MUT plasmid, followed by stimulation with ATP for 45 min (n = 3).
(G–I) BMDMs were treated with phenylpyruvate (400 μM) for 4 h and LPS for 24 h, with transfection of Ppt1 WT or MUT plasmid, followed by stimulation with ATP for 45 min. Levels of the inflammatory factors IL-1β, IL-18, and TNFα in the supernatant were measured using ELISA (n = 3).
(J–L) BMDMs were treated with phenylpyruvate (400 μM) for 4 h and LPS for 24 h and transfected with the Ppt1 WT or MUT plasmid. The RT-qPCR results showing relative mRNA expression of Nos, Tnf, and Il6 normalized to Actb (n = 3).
(M–O) RT-qPCR analysis of relative mRNA expression of Nos, Tnf, and Il6 when giving phenylpyruvate (400 μM) to either NLRP3 inhibitor MCC950-treated (1 μM) or NLRP3-knockdown BMDMs (n = 3). Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01; n.s., not significant.
Based on the above findings, the palmitoylation site, cysteine residue 6, of NLRP3 is located in its pyrin domain, which is the binding region for the adaptor protein ASC. This cysteine residue is highly conserved among different species (Figure 6C). Given that S-palmitoylation also has a regulatory effect on protein-protein interaction,34,35 surprisingly, we observed that phenylpyruvate treatment augmented the interaction between NLRP3 and ASC (Figure 6D). Besides, the formation of ASC specks was significantly increased after phenylpyruvate treatment, demonstrating that phenylpyruvate availed NLRP3 inflammasome activation in LPS-primed BMDM after ATP treatment (Figure 6E). Western blot assay results showed that transfection of the Ppt1 MUT plasmid, but not the WT plasmid, reversed the increased release of cleaved caspase-1 and mature IL-1β in the supernatant induced by phenylpyruvate treatment (Figure 6F). We also identified that only transfection of the Ppt1 MUT attenuated the secretion of IL-1β, IL-18, and TNFα into the culture medium induced by phenylpyruvate treatment (Figures 6G–6I). Additionally, the M1 signature genes Nos, Tnf, and Il6, which were upregulated by phenylpyruvate treatment, were downregulated upon transfection with the Ppt1 MUT plasmid but not the WT plasmid (Figures 6J–6L), demonstrating that phenylpyruvate treatment promoted the proinflammatory phenotype in macrophages in a manner dependent on its association with PPT1. Meanwhile, when inhibiting NLRP3 in BMDMs using MCC950 (an NLRP3 inhibitor) or siRNA, we found that phenylpyruvate treatment could no longer upregulate the M1 signature genes (Figure 6M-O), elucidating that phenylpyruvate promotes macrophage inflammatory phenotype through the NLRP3 inflammasome. In conclusion, phenylpyruvate contributes to a proinflammatory phenotype by strengthening the protein stability of NLRP3 and promoting inflammasome activation.
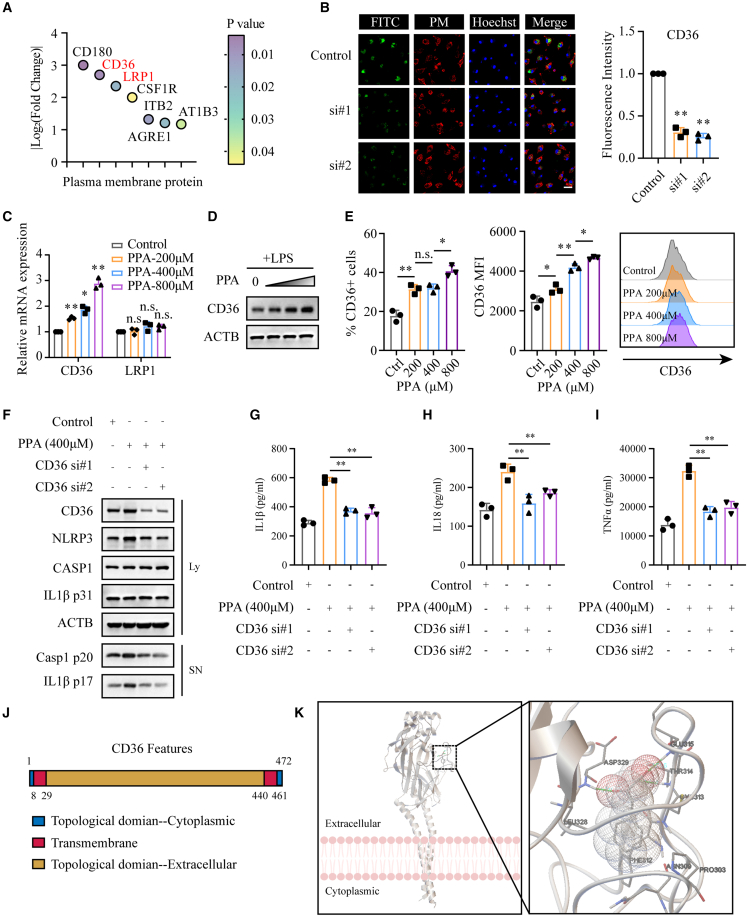
Phenylpyruvate enters macrophages in a CD36-dependent manner
As revealed in Figure 4B, we noticed the potential binding proteins that were localized in the plasma membrane, which led us to explore the key factors involved in phenylpyruvate uptake in BMDMs. As shown in Figure 7A, two scavenger receptors, CD36 and LRP1, were identified as potential phenylpyruvate-binding proteins. We knocked down the two scavenger receptors (Figures S7A and S7B) and found that only knockdown of CD36 markedly decreased phenylpyruvate uptake in BMDMs (Figures 7B and S7C). The RT-qPCR results indicated that phenylpyruvate treatment significantly increased the CD36 mRNA expression level but not the expression level of LRP1 (Figure 7C). Moreover, western blot analysis and flow cytometry assay results suggested that phenylpyruvate treatment was associated with upregulated expression of the CD36 receptor on the surface of BMDMs but not LRP1 expression (Figures 7D, 7E, and S7D). Additionally, after downregulating CD36 expression via siRNA, phenylpyruvate treatment did not increase NLRP3 expression levels or activate inflammasomes (Figure 7F). We also found that reducing CD36 expression reversed the phenylpyruvate-induced increase in IL-1β, IL-18, and TNFα secretion (Figures 7G–7I). The CD36 protein possesses a large extracellular region containing ligand-binding sites, two transmembrane domains, and a single short cytoplasmic tail at each terminal (Figure 7J). Molecular docking analysis demonstrated that the binding sites of phenylpyruvate with the CD36 extracellular region were located in the residues Thr314, Glu315, and Asp329, with minimum binding energy (−6.28 kcal/M) (Figure 7K). In general, CD36 plays a key role in phenylpyruvate uptake in macrophages and its proinflammatory effects.
Figure 7.
CD36 mediates the uptake of phenylpyruvate in macrophages
(A) The bubble plot shows the plasma membrane proteins identified in the LiP-SMap assay. The color of the circle indicates the p value.
(B) Immunostaining and statistical analysis showing the uptake of phenylpyruvate in BMDMs with or without knockdown of CD36. The FITC antibody indicated phenylpyruvate, and the lipid raft antibody indicated plasma membrane (n = 3). Scale bar, 50 μm.
(C) RT-qPCR results showing the relative mRNA expression of CD36 and LRP1 in BMDMs treated with increasing phenylpyruvate concentrations (n = 3).
(D) Immunoblot analysis of total CD36 protein expression in BMDMs treated with increasing phenylpyruvate concentrations. The black triangle indicates increasing phenylpyruvate concentrations starting from the left: 200, 400, and 800 μM (n = 3).
(E) Statistical analysis and representative histograms from flow cytometry data showing CD36 protein expression on the surface of BMDMs treated with increasing phenylpyruvate concentrations (n = 3).
(F) Immunoblot analysis of the supernatant and cell extracts in the indicated BMDMs, with or without knockdown of CD36, and then treated with phenylpyruvate for 4 h and LPS for 24 h, followed by stimulation with ATP for 45 min (n = 3).
(G–I) ELISA measuring the levels of the inflammatory factors IL-1β, IL-18, and TNFα in the supernatant of BMDMs, with or without knockdown of CD36, and then treated with phenylpyruvate for 4 h and LPS for 24 h, followed by stimulation with ATP for 45 min (n = 3).
(J) Schematic view of CD36 (full length).
(K) Representative images of autodocking for phenylpyruvate to the extracellular section of CD36 protein. Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01; n.s., not significant.
Discussion
Delayed healing and pathologic inflammation are the main characteristics of diabetic wounds, and there are limited available options for diabetic wound treatment.36 Macrophage plasticity plays a critical role in normal tissue repair after injury. However, under the stimulation of various intrinsic and exogenous factors, there exists an impaired Mφ phenotype transformation in diabetic wound, leading to the persistent activation of the proinflammatory M1 phenotype.4 Drugs targeting M1/M2 macrophage in DFUs have shown great translational value in a recent large randomized clinical trial.37 Therefore, it is promising to explore the key pathogenic factors regulating macrophage conversion in diabetic wound. Herein, we performed a metabolic profile analysis in DFUs and NDWs and identify a marked accumulation of phenylpyruvate in DFUs compared to NDWs. Following up on previous diabetes-related metabolomics analyses, we found that patients with diabetes had higher levels of phenylalanine in both serum and urine, while no difference in phenylpyruvate was found, further suggesting that increased phenylpyruvate was a local feature of diabetic wounds.21,22,38,39,40,41 Importantly, phenylpyruvate was found to be positively correlated with the inflammatory protein NLRP3, which mediated inflammasome is a powerful booster of local inflammatory storms.42 Phenylalanine is primarily metabolized to tyrosine by phenylalanine hydroxylase (PAH). However, in patients with PKU, phenylalanine cannot be converted to tyrosine and is instead converted to phenylpyruvate, which causes to mental retardation and brain damage.43 In addition, phenylpyruvate has been found to inhibit glycolysis in hepatocytes.44 In our study, we found that injection of phenylpyruvate locally into the wound in vivo impaired wound healing and caused intense inflammatory responses, demonstrating that the local accumulation of phenylpyruvate in diabetic wounds may be a key pathological factor in persistent inflammation.
Phenylalanine is an essential amino acid and cannot be synthesized in the human body, meaning that the phenylalanine concentration in the body is dependent on dietary intake. In the past 60 years, a phenylalanine-restricted diet has been shown to be effective for the treatment of PKU.23 We then thought that a phenylalanine-restricted diet may alleviate the burden of phenylalanine catabolism, reduce phenylpyruvate accumulation, and increase diabetic wound healing. We designed a phenylalanine-restricted diet based on a recent study.24 Since the phenylalanine concentration in a general mouse diet is close to 1%, we tested the feasibility of our hypothesis by using diets containing 0.25% and 2% phenylalanine. We found that a phenylalanine-restricted diet effectively reduced phenylpyruvate levels in diabetic wound. Specifically, a phenylalanine-restricted diet effectively advanced the transition of M1/M2 Mφ, reduced the inflammatory response, and accelerated wound healing. This finding is invigorating, suggesting that, in addition to strict sugar intake control, phenylalanine diet management may be a potential treatment strategy for diabetic wounds. Future clinical studies are necessary to determine how to restrict natural protein intake and supplement protein substitutes to achieve therapeutic goals while reducing the side effects in the body.
Mechanistically, in the presence of exogenous pathogens such as LPS, phenylpyruvate treatment enhanced the polarization of macrophages toward the proinflammatory phenotype but has no significant effect on the pro-healing macrophage phenotype induced by IL-4, which is supported by our observations of persistent inflammation induced by phenylpyruvate treatment in the wounds. As for the reduction in the proportion of pro-healing M2 macrophages in the wounds caused by phenylpyruvate treatment, it is possible that the phenotypic transition is hindered by the continuous polarization of proinflammatory macrophages. We also found that phenylpyruvate treatment increased NLRP3 protein levels and enhanced inflammasome activation but did not affect NLRP3 transcription. It is not surprising that specific metabolites regulate macrophage phenotypes.19,20,45 LiP-SMap is a chemical proteomics screening method that integrates the advantages of high-throughput quantitative proteomics technology and high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify authentic metabolite-targeted proteins.46,47 Using this means, we identified PPT1 as a target of phenylpyruvate, supported biophysically by MST assay results. PPT1 is a thioesterase that removes palmitic acid from S-acylated proteins and PPT1 exhibits broad substrate specificity.48 There are three essential sites, namely S115, D233, and H289, for PPT1 palmitoyl-CoA hydrolase activity. Our molecular dynamics simulations suggested that phenylpyruvate favorably binds to PPT1 residues Lys229 and Gly245. Given the spatial conformational proximity of the binding sites and the active site, phenylpyruvate treatment was found to markedly alleviate PPT1 palmitoyl-CoA hydrolase activity. Importantly, phenylpyruvate treatment had no effect on PPT1 activity after mutation of the binding sites, and the mutant PPT1 abrogated the phenylpyruvate-mediated upregulation of NLRP3 expression. Overall, these results demonstrate that alternation of PPT1 activity may explain the effect of phenylpyruvate on macrophage polarization.
S-palmitoylation is a reversible cotranslational and posttranslational modification of proteins in which palmitates covalently bind to cysteine residues mostly via thioester linkages.49 Palmitoylation modification is determined by the homeostasis of palmitoyltransferases and depalmitoylases. It plays an essential role in human physiological and pathological processes, including protein membrane anchoring, trafficking, degradation, and interaction.50,51,52 We found that NLRP3 was palmitoylated in macrophages, and the major palmitoylation site in NLRP3 is cysteine residue 6. In addition, the palmitoylation of NLRP3 was upregulated after PPT1 knockdown, and there was an interaction between PPT1 and NLRP3, demonstrating that PPT1 is a depalmitoylase of the NLRP3 protein. Furthermore, we observed that phenylpyruvate treatment increased NLRP3 palmitoylation, and the binding site mutant of PPT1 reversed the increased palmitoylation induced by phenylpyruvate treatment, demonstrating that phenylpyruvate treatment increases NLRP3 palmitoylation by binding and inhibiting PPT1. Surprisingly, we observed that the phenylpyruvate-mediated increase of NLRP3 palmitoylation enhanced NLRP3 protein stability and increased the interaction between NLRP3 and ASC during inflammasome activation. These results revealed that phenylpyruvate increases NLRP3 abundance and promotes inflammasome activation by interacting with PPT1 and then increasing NLRP3 palmitoylation, ultimately contributing to the formation of a proinflammatory macrophage phenotype. We notice that a recent study has earlier reported palmitoylation on NLRP3 and showed that zDHHC12-mediated palmitoylation promotes NLRP3 degradation through HSC70-mediated CMA pathway.53 However, we did not observe an enhanced interaction between NLRP3 and HSC70 induced by phenylpyruvate-mediated increased palmitoylation (Figure S6). Additionally, the NLRP3 palmitoylation site we observed in this study was different from the identified site in their study. However, we do not consider these reports contradictory, and the discrepancy between them may reflect the disruption of intracellular homeostasis between palmitoyltransferase levels and depalmitoylase levels in the high-glucose environment, thus leading to different roles of palmitoylation in NLRP3. Therefore, the regulation and biological function of palmitoylation on NLRP3 in different environments are worthy of further exploration.
CD36, also known as the scavenger receptor B2, is widely expressed in various immune and nonimmune cells. It functions not only as a long-chain free fatty acid transporter but also as a signaling receptor that responds to PAMPs and DAMPs.54,55,56 CD36 can regulate immune cell differentiation and activation by integrating cell metabolic and signaling pathways.57 In our study, we identified CD36 as a mediator of phenylpyruvate uptake into macrophages. Phenylpyruvate treatment upregulated the cell surface expression of CD36, and knockdown of CD36 alleviated the phenylpyruvate-induced proinflammatory macrophage phenotype, giving us a better understanding of how phenylpyruvate works. However, the cellular instability associated with CD36 knockdown limits the possibility of using it as a target to ameliorate the phenylpyruvate effect in clinical applications. In future studies, the design of specific molecular compounds blocking the binding of phenylpyruvate to CD36 using a computational phenylpyruvate-CD36 model may be a suitable therapeutic approach.
Multiple elevated proinflammatory cytokines caused by activated NLRP3 inflammasome in diabetic wounds, including IL-1β and TNFα, induce local hyperglycemia, accumulation of advanced glycation end products (AGEs), and reactive oxygen species (ROS).12,58 High levels of ROS in a hyperglycemic environment can induce premature senescence in endothelial progenitor cells (EPCs), leading to inadequate repair and angiogenesis.59 NLRP3 inflammasome activity driven by exogenous irritants induces excessive infiltration of innate immune cells into the diabetic wound and damages the proliferation and migration of keratinocytes, thus severely impairing wound healing.60,61 These studies suggest that the inflammatory protein NLRP3 is an effective therapeutic target for diabetic wounds. Based on our results, targeting phenylpyruvate to control NLRP3-mediated inflammation is a potential translational strategy for diabetic wounds.
Limitations of the study
This study uncovered the role of phenylpyruvate as an essential proinflammatory pathogenic factor in diabetic wound. Given that no increase in phenylpyruvate levels was observed in the previous serum metabolome analysis of patients with diabetes mellitus and the early wound macrophages are reported to mainly differentiate from infiltrating bone marrow-derived monocytes in diabetic wound, we still need to further elucidate which pathological process leads to the accumulation of phenylpyruvate in the wound. In addition, we reveal the different roles of NLRP3 palmitoylation in a high-glucose environment, demonstrating the complexity of the so-called immune metabolic reprogramming. In future studies, it is worthwhile to evaluate modifications of intracellular palmitoylation modification homeostasis in a high-glucose environment.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-NLRP3 (for WB, IP) | Cell Signaling Technology | Cat# 15101; RRID: AB_2722591 |
| Rabbit anti-NLRP3 (for IHC) | Affinity Biosciences | Cat# DF7438; RRID: AB_2839376 |
| Mouse anti-NLRP3 (for IF) | AdipoGen Life Sciences | Cat# AG-20B-0014-C100; RRID: AB_2885199 |
| Rabbit anti-ASC | Cell Signaling Technology | Cat# 67824; RRID: AB_2799736 |
| Rabbit anti-Cleaved-IL-1β | Cell Signaling Technology | Cat# 63124; RRID: AB_2799639 |
| Rabbit anti-IL-1β | Cell Signaling Technology | Cat# 31202; RRID: AB_2799001 |
| Rabbit anti-Cleaved Caspase-1 | Cell Signaling Technology | Cat# 89332; RRID: AB_2923067 |
| Rabbit anti-Caspase-1 | Cell Signaling Technology | Cat# 24232; RRID: AB_2890194 |
| Rabbit anti-β-Actin | Cell Signaling Technology | Cat# 4970; RRID: AB_2223172 |
| Rabbit anti-GAPDH | Cell Signaling Technology | Cat# 2118; RRID: AB_561053 |
| Rabbit anti-p-p65 | Abcam | Cat# ab76302; RRID: AB_1524028 |
| Rabbit anti-p65 | Abcam | Cat# ab32536; RRID: AB_776751 |
| Rabbit anti-p-ERK | Abcam | Cat# ab201015; RRID: AB_2934088 |
| Rabbit anti-ERK | Abcam | Cat# ab184699; RRID: AB_2802136 |
| Rabbit anti-p-AKT | Cell Signaling Technology | Cat# 4060; RRID: AB_2315049 |
| Mouse anti-AKT | Proteintech | Cat# 60203-2-Ig; RRID: AB_10912803 |
| Rabbit anti-p-p38 | Abcam | Cat# ab195049; RRID: AB_2576214 |
| Rabbit anti-p38 | Abcam | Cat# ab170099 |
| Rabbit anti-PPT1 | Proteintech | Cat# 29653-1-AP; RRID: AB_2923599 |
| Rabbit anti-FLAG | Abcam | Cat# ab205606; RRID: AB_2916341 |
| Rabbit anti-M6PR | Abcam | Cat# ab124767; RRID: AB_10974087 |
| Rabbit anti-PSAP | Abcam | Cat# ab300469 |
| Rabbit anti-HSC70 | Proteintech | Cat# 10654-1-AP; RRID: AB_2120153 |
| Rabbit anti-CD36 | Abcam | Cat# ab252923 |
| Rabbit anti-LRP1 | Abcam | Cat# ab92544; RRID: AB_2234877 |
| Rabbit anti-Ki-67 | Abcam | Cat# ab15580; RRID: AB_443209 |
| Rabbit anti-α-SMA | Proteintech | Cat# 14395-1-AP; RRID: AB_2223009 |
| Rabbit anti-CD31 | Proteintech | Cat# 11265-1-AP; RRID: AB_2299349 |
| Rabbit anti-Mrc1 | Servicebio | Cat# GB113497 |
| Rabbit anti-F4/80 | Servicebio | Cat# GB11027; RRID: AB_2814687 |
| Rabbit anti-Nos | Cell Signaling Technology | Cat# 13120; RRID: AB_2687529 |
| Rabbit anti-CD206 | Cell Signaling Technology | Cat# 24595; RRID: AB_2892682 |
| Cholera Toxin Subunit B | Thermo Fisher Scientific | Cat# C22843 |
| Rat anti-mouse LAMP2 | Proteintech | Cat# 65052-1-Ig; RRID: AB_2881468 |
| LysoTracker red | Beyotime | Cat# C1046 |
| LysoSensor™ Green DND-189 | Yeasen | Cat# 40767ES50 |
| APC anti-mouse CD86 | Biolegend | Cat# 105012; RRID: AB_493342 |
| PE anti-mouse CD163 | Biolegend | Cat# 156703; RRID: AB_2860724 |
| Brilliant Violet 421™ anti-mouse F4/80 | Biolegend | Cat# 123137; RRID: AB_2563102 |
| APC anti-mouse/human CD11b | Biolegend | Cat# 101212; RRID: AB_312795 |
| APC anti-mouse CD36 | Biolegend | Cat# 102611; RRID: AB_571994 |
| TruStain FcX™ (anti-mouse CD16/32) | Biolegend | Cat# 101319; RRID: AB_1574973 |
| ABflo™ 555-conjugated Goat Anti-Mouse IgG (H + L) | Abclonal | Cat# AS057; RRID: AB_2768321 |
| Alexa Fluor 647-conjugated Goat Anti-Rabbit IgG (H + L) | Abclonal | Cat# AS060; RRID: AB_2768330 |
| Fluorescein (FITC)–conjugated Goat Anti-Rat IgG(H + L) | Proteintech | Cat# SA00003-11; RRID: AB_2890989 |
| Biological samples | ||
| Human nondiabetic wound tissue samples | the First Affiliated Hospital of Sun Yat-sen University | N/A |
| Human diabetic wound tissue samples | the First Affiliated Hospital of Sun Yat-sen University | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| LPS | Sigma Aldrich | Cat# L2880 |
| Recombinant Murine IL-4 | Peprotech | Cat# 214-14 |
| Phenylpyruvate | Sigma Aldrich | Cat# 286958 |
| Phenylalanine | MedChemExpress | Cat# HY-N0215 |
| Lipofectamine 2000 Transfection Reagent | Invitrogen | Cat# 11668019 |
| Lipofectamine RNAi Max Transfection Reagent | Invitrogen | Cat# 13778030 |
| Penicillin‒streptomycin | Sigma Aldrich | Cat# V900929 |
| Fetal bovine serum | ThermoFisher Scientific | Cat# 10099-141 |
| MCC950 | Selleck | Cat# S8930 |
| ATP | Selleck | Cat# S5260 |
| Recombinant M-CSF | PeproTech | Cat# 315-02 |
| Red cell lysis buffer | Beyotime | Cat# C3702 |
| Palmitic acid | MedChemExpress | Cat# HY-N0830 |
| DMEM | ThermoFisher Scientific | Cat# C11995500BT |
| DL-dithiothreitol | MedChemExpress | Cat# HY-15917 |
| 4-methylumbelliferyl-6-thio-βd-gluco-pyranoside | Medbio | Cat# MED20989 |
| β-glucosidase | Sigma Aldrich | Cat# G4511 |
| Critical commercial assays | ||
| IP-ABE Palmitoylation Kit | AIMS | Cat# AM101314 |
| HiScript 1st Strand cDNA Synthesis Kit | Vazyme, China | Cat# R111 |
| Real-time Fluorescent Quantitative PCR Kit | Vazyme, China | Cat# Q321 |
| ECL detection reagent | EpiZyme | Cat# SQ202 |
| BCA assay kit | Thermo Fisher Scientific | Cat# 23225 |
| IL-1β ELISA KIT | MEIMIAN | Cat# MM-0040M2 |
| IL-18 ELISA KIT | MEIMIAN | Cat# MM-0169M2 |
| TNF-α ELISA KIT | MEIMIAN | Cat# MM-0132M2 |
| Deposited data | ||
| LC-MS raw data files | https://www.ebi.ac.uk/metabolights/ | MTBLS7853 |
| See Table S3 for LiP-SMap determination of PPA-bound proteins | This paper | N/A |
| Experimental models: Cell lines | ||
| Murine BMDM | This paper | N/A |
| Mouse embryonic fibroblasts | This paper | N/A |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice | GemPharmatech | N/A |
| BKS-DB mice | GemPharmatech | N/A |
| Oligonucleotides | ||
| Primers for qRT-PCR, see Table S4 | Ruibiotech | N/A |
| siRNA, see Table S5 | GenePharma | N/A |
| Recombinant DNA | ||
| pIRES2-Flag-Ppt1 | Umine Biotechnology | N/A |
| pIRES2-Flag-Ppt1-MUT | Umine Biotechnology | N/A |
| Software and algorithms | ||
| FlowJo software Version 10.8.1 | BD Biosciences | https://www.flowjo.com/ |
| ImageJ | National Institutes of Health (NIH) | https://imagej.nih.gov/ij/ |
| GraphPad Prism 8 | GraphPad Software | https://www.graphpad.com/scientificsoftware/prism/ |
| Autodock | Autodock Software | https://autodock.scripps.edu/ |
| Adobe Illustrator | Adobe | https://www.adobe.com/ |
| Other | ||
| Phenylalanine diets, see Table S2 | This paper | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Bing Tang (tangbing@mail.sysu.edu.cn) upon reasonable request.
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human skin samples collection
All patients were enrolled and followed at the department of Burn and Plastic Surgery, the First Affiliated Hospital of Sun Yat-sen University. This study was approved by the Research Medical Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University in accordance with the Declaration of Helsinki (Reference number: [2023]040). Informed consent was obtained from all subjects. Biopsies from diabetic foot ulcers (n = 12) and non-diabetic wound (n = 12) were collected. The inclusion criteria for DFUs were (1) diabetes mellitus >1 year; (2) a foot ulcer larger than 0.5 cm2; (3) neuropathy; (4) wound duration >1 month; (5) hemoglobin A1c: ≤13.0%; and (6) no surgery. Exclusion criteria for DFUs were (1) active cellulitis; (2) gangrene; (3) osteomyelitis; (4) vascular insufficiency; and (5) any experimental drugs applied to the wound for 4 weeks preceding the biopsy. Wounds were obtained using a sterile 3-mm punch biopsy. Nondiabetic wound samples were collected as previously described.62 A sterile 3-mm punch biopsy was used to establish a uniform, full thickness biopsy in the posterior axillary area of the arm of the nondiabetic participants. Three days later, the wounds were again biopsied and specimens were collected. Skin samples were collected and frozen at −80°C until sample preparation.
In vivo wound healing
Animal experiments were approved by the Animal Research Committee of First Affiliated Hospital of Sun Yat-sen University and were conducted in accordance with established guidelines for the Use and Care of Laboratory Animals. Eight-week-old C57BL/6J female mice and BKS-DB female mice (GemPharmatech, Nanjing, China) were used to establish an in vivo wound model. All mice were maintained in specific pathogen-free conditions. All mice were separated randomly into three groups, with 12 mice in each group. Mice were anesthetized, the dorsal hair was removed, and the area was rinsed with sterile water. Two full-thickness back wounds were made using 10-mm punch biopsy with or without wound splinting. Wound treatment was performed as previously described, and the wounds were photographed at the indicated time points. In the phenylpyruvate dosing experiment, local injection was conducted at four points along the wound edge,63 and the injection of saline was used as the control group. In the dietary phenylalanine restriction experiment, the weight, blood glucose level, and food intake of mice were recorded. Mice that died from other causes, such as infection and fighting, were excluded from the study. The wound area was analyzed using ImageJ software and normalized as a percentage of the initial area. The researchers were blinded during the experiment and data analysis.
Murine BMDM isolation and cell culture
Eight-week-old BKS-DB female mice were used to obtain BMDMs, which were flushed by DMEM through the femur and tibia with a 25-gauge needle. The cell suspension was filtered using 70 μm cell filters and centrifuged at 1,000 rpm for 5 min. The cell pellet was subsequently resuspended in red cell lysis buffer (Beyotime, China) for 3 min. After being centrifuged and resuspended, cells were cultured in DMEM containing 1% L-glutamine, 30 mM glucose, 10% fetal bovine serum (FBS, ThermoFisher Scientific), 1% penicillin‒streptomycin (Sigma‒Aldrich) and 20 ng/mL macrophage colony stimulating factor (PeproTech). On day 3, half of the medium was replaced, and on day 5, all medium was replaced with fresh medium. On day 7, BMDMs were used for related experiments. Mouse embryonic fibroblasts (MEFs) were cultured in DMEM containing 30 mM glucose, 10% FBS and 1% penicillin‒streptomycin. All cells were cultured at 37°C in a 5% CO2 incubator.
Method details
Metabolomic profile
Metabolite extractions
Biopsy samples were homogenized into powder in liquid nitrogen and vortexed with cold extraction solvent. For absolute quantification of the metabolites in targeted metabolomics, stock solutions of stable-isotope internal standards were also added to the extraction solvent simultaneously. After vortexing, the mixture was incubated on ice for 20 min and then centrifuged at 14,000 × g for 20 min at 4°C. The supernatant was then collected and flowed through 96-well protein precipitation plates, and the elution was collected and dried in a vacuum centrifuge at 4°C. For LC‒MS analysis, the samples were redissolved in 100 μL acetonitrile/water solvent and transferred to LC vials.
LC-MS analysis
Biopsy extracts were analyzed using a quadrupole time-of-flight mass spectrometer coupled to hydrophilic interaction chromatography via electrospray ionization. LC separation was performed on an ACQUITY UPLC BEH Amide column using a gradient of solvent A (25 mM ammonium acetate and 25 mM ammonium hydroxide in water) and solvent B (acetonitrile). The mass spectrometer was operated in both positive ion and negative ionizations modes. In MS acquisition, the instrument was set to acquire over the m/z range 60–1000 Da, and the accumulation time for the time-of-flight (TOF) MS scan was set at 0.20 s/spectra. The product ion scan was acquired using information-dependent acquisition (IDA) with high sensitivity mode selected.
Data analysis
The raw data were converted to MzXML files by ProteoWizard MSConvert before importing into XCMS software. In the extracted ion features, only the variables with more than 50% of the nonzero measurement values in at least one group were retained. Compound identification of metabolites was performed with an in-house database established based on available authentic standards. After normalization to the total peak intensity, the processed data were then subjected to multivariate data analysis. The variable importance in the projection (VIP) value in the OPLS-DA model was determined to indicate the contribution to classification. Significance was calculated using an unpaired Student’s t test. Metabolites with VIP values > 1 and p < 0.05 were considered as statistically significant.
Coimmunoprecipitation and immunoblotting assay
In the coimmunoprecipitation assay, whole cell extracts were prepared after treatment and then incubated overnight with Protein A/G beads (Pierce), which were pretreated with the indicated antibodies or rabbit IgG for 3 h at 4°C with gentle shaking. The beads were washed five times on ice and boiled in sample buffer for SDS‒PAGE. Proteins were separated and transferred to PVDF membranes (Merck Millipore) at 350 mA and 4°C for an hour. After blocking, the membranes were incubated with appropriate primary antibodies overnight at 4°C and then with HRP-conjugated secondary antibodies at room temperature for an hour. ECL detection reagent (EpiZyme) was used for protein detection in a MiniChrome Chemiluminescence imager (SAGECREATION, Beijing).
RNA extraction and RT-qPCR
Total RNA was extracted by TRIzol reagent (Invitrogen), and reverse-transcribed cDNA was generated with the HiScript 1st Strand cDNA Synthesis Kit (Vazyme, R111) according to the manufacturer’s instructions. RT‒qPCR assays were conducted with the Real-time Fluorescent Quantitative PCR Kit (Vazyme, Q321) using an ABI7900HT Fast Real-Time PCR system (Applied Biosystems, CA, USA). Actb was used as an endogenous control. The comparative Ct (2−▵▵CT) method was used to calculate the relative mRNA expression. The primers used in our study are listed in Table S4.
Measurement of cytokine levels
For mouse wound skin tissues, 15 mg samples with cold lysis buffer were homogenized with a tissue homogenizer and centrifuged at 10000 × g for 10 min at 4°C. For BMDM-derived conditioned medium, samples were collected and centrifuged to remove the precipitate. IL1β, IL18 and TNFα levels were measured in the tissue and supernatants using the indicated Mouse Quantikine ELISA Kit (MEIMIAN) according to the manufacturer’s protocol.
Flow cytometry assay
Before surface staining with antibodies, Fc-γ receptors were blocked by incubating samples with anti-mouse CD16/32 antibodies (Biolegend) for 10 min. Cells were then stained with appropriate fluorochrome-conjugated antibodies for surface proteins diluted in FACS buffer for 20 min on ice. Cells were washed and analyzed using a FACS Canto II Cell analyser (BD Biosciences). Acquired data were analyzed with FlowJo 10 software.
Immunohistochemistry and immunofluorescence
Mouse skin tissue slides were deparaffinized and rehydrated, and antigen retrieval was performed. Skin tissues were blocked with 6% goat serum with 0.1% Triton X-100 and 3% H2O2 in PBS and then incubated with appropriate antibodies overnight at 4°C. IHC staining proceeded with horseradish peroxidase (HRP) conjugates using 3,3′-diaminobenzidine (DAB) detection. The evaluation of IHC staining was performed by two independent pathologists. Images of the sections were captured under a microscope (EVOS FL Auto Cell Imaging System, USA).
For immunofluorescence staining, BMDMs were incubated on 35 mm confocal dishes and stimulated by the indicated reagents. Cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min, and then permeabilized in 0.3% Triton X-100 for 10 min at room temperature. After washing with PBS three times, the cells were blocked with 6% fetal goat serum (Solarbio) for 1 h and incubated with the indicated primary antibodies and subsequently the fluorescently labelled secondary antibody. Confocal images were visualized by a confocal microscope (Zeiss LSM880 with Airyscan). The images were analyzed using Zen 2.6 or ImageJ software.
Identification of proteins interacting with phenylpyruvate
Chemical proteomics using LiP-SMap screening was performed as previously described.26 Briefly, cell lysates from three independent replicates were aliquoted in equivalent volumes containing 100 mg of proteome sample and incubated for 10 min at 25°C with vehicle or phenylpyruvate (final concentration: 0.33 nM per μg of total protein). Proteinase K from Tritirachium album was added simultaneously to all the proteome-phenylpyruvate samples at a proteinase K to substrate mass ratio of 1:100 and incubated at 25°C for 5 min. Digestion reactions were stopped by heating samples for 5 min at 98°C in a thermocycler followed by the addition of sodium deoxycholate to a final concentration of 2%.
Protein fragments from the proteinase K-limited proteolysis step were reduced with 10 mM DTT for 30 min at 37°C and then alkylated by incubation with 40 mM iodoacetamide for 45 min at 25°C in the dark. The samples were digested with trypsin at an enzyme-substrate ratio of 1:50 overnight at 37°C under agitation at 800 rpm. The digestion was stopped and the DOC was precipitated by adding a 50% TFA solution at a final concentration of 2%. The pH of the samples was adjusted to less than 3 with formic acid. Acidified peptide mixtures were loaded onto Sep-Pak C18 cartridges, desalted, and eluted with 70% acetonitrile–0.1% formic acid. The samples were dried in a vacuum centrifuge, solubilized in 0.1% formic acid, and immediately analyzed by mass spectrometry.
siRNA transfection
siRNA oligonucleotides were synthesized by GenePharma Co., Ltd. (Suzhou, China). siRNA (50 nM) was mixed with 5 μL Lipofectamine RNAi Max Transfection Reagent (Invitrogen), and 100 μL Opti-MEM (Thermo Fisher Scientific) was subsequently added and incubated for 20 min. The prepared mixture was added to the 6-well plate lined with cells. The indicated target sequences are listed in Table S5.
Transfection of plasmid
The expression vector pIRES2-Flag-Ppt1 encodes full-length mouse PPT1 protein, and the corresponding vector pIRES2-Flag-Ppt1-MUT encodes phenylpyruvate-binding site-deficient PPT1 protein containing K229N and G245A (Umine Biotechnology Co., Ltd., Guangzhou, China). Transient transfections of the indicated plasmid DNA were conducted with Lipofectamine 2000 (Invitrogen) at 80–90% confluence cells in serum-free Opti-MEM according to the manufacturer’s instructions.
Thermal shift assay
A thermal shift assay was conducted as previously described.64 First, BMDMs were treated with phenylpyruvate or dimethyl sulfoxide (DMSO). Second, the intact cell suspensions were harvested and heated at each temperature point from 37°C to 62°C for 3 min. Finally, protein extraction was performed, and protein expression was assessed by immunoblotting analysis.
Molecular docking
The interactions between phenylpyruvate and PPT1 and between phenylpyruvate and CD36 were calculated using a computational docking method. Crystal structures of PPT1 and CD36 were obtained from UniProt (https://www.uniprot.org/). Induced-fit docking of phenylpyruvate to each potential binding site on PPT1 or CD36 was conducted using Autodock (https://autodock.scripps.edu/). From docking data, the best-fit docking configuration, binding energy values, potential conformations, type of interaction, and bond distances were predicted. The lowest-energy binding configuration in the docking data was selected as the representative binding configuration for further analysis.
MST assay
PPT1 Protein acts as a Target and is labeled according to the instruction manual of the Monolith protein Labeling Kit RED-NHS 2nd Generation Kit. After labelling was completed, a preliminary experiment was conducted to evaluate the labelling effect. After the preliminary experiment, binding affinity detection was performed if there was no adsorption and aggregation of proteins, and the signal-to-noise ratio was up to standard. 10 μL of gradient-diluted unlabeled molecules (phenylpyruvate) and 10 μL of fixed concentration labelled molecules (PPT1 protein) were incubated for 5 min. The mixed samples were loaded into a glass capillary tube and analyzed by the MST-NT.115 instrument.
PPT1 activity quantification assay
The quantification assay of PPT1 activity was performed as previously described.65 Specifically, palmitate linked to 4-methylumbelliferyl-6-thio-βd-gluco-pyranoside (MU-6S-Palm-βGlc, MedBio) was selected as a substrate. PPT1 cleaved the thioester bond and then released the intermediate 4-methylumbelliferyl-6thio-β-d-gluco-pyranoside and palmitate. The intermediate was further hydrolyzed by exogenous almond β-glucosidase (Sigma) to 4-methylumbelliferone, the fluorescence of which was measured to quantify PPT1 cleavage of palmitate groups.
Acyl-biotin exchange (ABE) assay
The ABE assay was performed using the IP-ABE Palmitoylation Kit (AIMS, China) according to the manufacturer’s instructions. Briefly, the procedure includes blocking, reduction, labelling, elution and detection. Cells were harvested and suspended in lysis buffer followed by incubation with anti-NLRP3 beads overnight at 4°C. N-ethylmaleimide (NEM) was used to block the unmodified cysteines for 30 min. Then, the beads were washed and incubated with hydroxylamine (HAM) for 1 h at room temperature. Each group was divided into two parts, one including the HAM step (+HAM) and one omitting the HAM cleavage step (-HAM). After washing, the beads were treated with thiol-reactive biotin molecules for 1 h at room temperature. The immunoprecipitated samples were analyzed by immunoblotting assay.
Quantification and statistical analysis
Statistical analyses and mapping were carried out using GraphPad Prism 8.0 or SPSS 25.0 software. Each experiment was repeated three times independently. The images presented in the manuscript are representative plots of 3 independent experiments. All data are presented as the mean ± standard deviation. The sample sizes among groups were determined according to the previous literature in the same research area for adequate power to detect a pre-specified effect. Student’s t test was used to compare two groups with a single variable, whereas one-way ANOVA with Tukey’s multiple comparisons test was used to compare multiple groups. For experimental set-ups with a second variable, two-way ANOVA with Tukey’s multiple comparisons test was performed. Correlations were assessed with linear regression analysis. p < 0.05 was considered significant.
Acknowledgments
We are grateful for all of the patients who participated in our study. We thank Shanghai Applied Protein Technology Co., Ltd. for the assistance with the metabolomic analysis; Shanghai Biotree Biotech Co., Ltd. for the LiP-SMap analysis; and Xi’an Qiyue Biotechnology Co., Ltd. for the generation of the FITC-phenylpyruvate compound. This study was supported by grants from the National Natural Science Foundation of China (nos. 82272273, 82172949, 82072181, 81972569, 81972623), the Guangdong Basic and Applied Basic Research Foundation (nos. 2022A1515010083, 2022A1515012508), and the Sun Yat-sen University Clinical Research 5010 Program (no. 2018003).
Author contributions
Conceptualization, B.T. and W.D.; methodology, Z.H., M.C., D.L., and R.Y.; validation, D.L., X.C., and L.Z.; formal analysis, D.L., Y.D., Z.X., and Y.R.; investigation, D.L., L.Z., H.X., and Z.W.; resources, D.L., X.C., H.Y., and M.C.; writing – original draft, D.L. and X.C.; writing – review & editing, C.K., B.T., W.D., and L.Z.; visualization, D.L., Z.W., and Y.R.; supervision, L.Z., H.X., and M.C.; funding acquisition, B.T.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: July 21, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101129.
Contributor Information
Zhicheng Hu, Email: huzhch5@mail.sysu.edu.cn.
Wuguo Deng, Email: dengwg@sysucc.org.cn.
Bing Tang, Email: tangbing@mail.sysu.edu.cn.
Supplemental information
Data and code availability
-
•
The LC-MS data were deposited in the MetaboLights repository (www.ebi.ac.uk/metabolights/) under the accession number MTBLS7853. Data generated and/or analyzed in this study, excluding identifying personal information, are available from the lead contact, Bing Tang (tangbing@mail.sysu.edu.cn) with reasonable request to protect research participant privacy.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- 1.Doğruel H., Aydemir M., Balci M.K. Management of diabetic foot ulcers and the challenging points: An endocrine view. World J. Diabetes. 2022;13:27–36. doi: 10.4239/wjd.v13.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grennan D. Diabetic Foot Ulcers. JAMA. 2019;321:114. doi: 10.1001/jama.2018.18323. [DOI] [PubMed] [Google Scholar]
- 3.Jeffcoate W.J., Vileikyte L., Boyko E.J., Armstrong D.G., Boulton A.J.M. Current Challenges and Opportunities in the Prevention and Management of Diabetic Foot Ulcers. Diabetes Care. 2018;41:645–652. doi: 10.2337/dc17-1836. [DOI] [PubMed] [Google Scholar]
- 4.Louiselle A.E., Niemiec S.M., Zgheib C., Liechty K.W. Macrophage polarization and diabetic wound healing. Transl. Res. 2021;236:109–116. doi: 10.1016/j.trsl.2021.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Wolf S.J., Melvin W.J., Gallagher K. Macrophage-mediated inflammation in diabetic wound repair. Semin. Cell Dev. Biol. 2021;119:111–118. doi: 10.1016/j.semcdb.2021.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma B.R., Kanneganti T.D. NLRP3 inflammasome in cancer and metabolic diseases. Nat. Immunol. 2021;22:550–559. doi: 10.1038/s41590-021-00886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paik S., Kim J.K., Silwal P., Sasakawa C., Jo E.K. An update on the regulatory mechanisms of NLRP3 inflammasome activation. Cell. Mol. Immunol. 2021;18:1141–1160. doi: 10.1038/s41423-021-00670-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spel L., Martinon F. Inflammasomes contributing to inflammation in arthritis. Immunol. Rev. 2020;294:48–62. doi: 10.1111/imr.12839. [DOI] [PubMed] [Google Scholar]
- 9.Jäger E., Murthy S., Schmidt C., Hahn M., Strobel S., Peters A., Stäubert C., Sungur P., Venus T., Geisler M., et al. Calcium-sensing receptor-mediated NLRP3 inflammasome response to calciprotein particles drives inflammation in rheumatoid arthritis. Nat. Commun. 2020;11:4243. doi: 10.1038/s41467-020-17749-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renaudin F., Orliaguet L., Castelli F., Fenaille F., Prignon A., Alzaid F., Combes C., Delvaux A., Adimy Y., Cohen-Solal M., et al. Gout and pseudo-gout-related crystals promote GLUT1-mediated glycolysis that governs NLRP3 and interleukin-1beta activation on macrophages. Ann. Rheum. Dis. 2020;79:1506–1514. doi: 10.1136/annrheumdis-2020-217342. [DOI] [PubMed] [Google Scholar]
- 11.Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 12.Ding Y., Ding X., Zhang H., Li S., Yang P., Tan Q. Relevance of NLRP3 Inflammasome-Related Pathways in the Pathology of Diabetic Wound Healing and Possible Therapeutic Targets. Oxid. Med. Cell. Longev. 2022;2022 doi: 10.1155/2022/9687925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirza R.E., Fang M.M., Weinheimer-Haus E.M., Ennis W.J., Koh T.J. Sustained inflammasome activity in macrophages impairs wound healing in type 2 diabetic humans and mice. Diabetes. 2014;63:1103–1114. doi: 10.2337/db13-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira P.R., Carrageta D.F., Oliveira P.F., Rodrigues A., Alves M.G., Monteiro M.P. Metabolomics as a tool for the early diagnosis and prognosis of diabetic kidney disease. Med. Res. Rev. 2022;42:1518–1544. doi: 10.1002/med.21883. [DOI] [PubMed] [Google Scholar]
- 15.Pillon N.J., Loos R.J.F., Marshall S.M., Zierath J.R. Metabolic consequences of obesity and type 2 diabetes: Balancing genes and environment for personalized care. Cell. 2021;184:1530–1544. doi: 10.1016/j.cell.2021.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou W.C., Rampanelli E., Li X., Ting J.P.Y. Impact of intracellular innate immune receptors on immunometabolism. Cell. Mol. Immunol. 2022;19:337–351. doi: 10.1038/s41423-021-00780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.SantaCruz-Calvo S., Bharath L., Pugh G., SantaCruz-Calvo L., Lenin R.R., Lutshumba J., Liu R., Bachstetter A.D., Zhu B., Nikolajczyk B.S. Adaptive immune cells shape obesity-associated type 2 diabetes mellitus and less prominent comorbidities. Nat. Rev. Endocrinol. 2022;18:23–42. doi: 10.1038/s41574-021-00575-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearce E.L. Metabolism as a driver of immunity. Nat. Rev. Immunol. 2021;21:618–619. doi: 10.1038/s41577-021-00601-3. [DOI] [PubMed] [Google Scholar]
- 19.Liu P.S., Wang H., Li X., Chao T., Teav T., Christen S., Di Conza G., Cheng W.C., Chou C.H., Vavakova M., et al. alpha-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 2017;18:985–994. doi: 10.1038/ni.3796. [DOI] [PubMed] [Google Scholar]
- 20.Runtsch M.C., Angiari S., Hooftman A., Wadhwa R., Zhang Y., Zheng Y., Spina J.S., Ruzek M.C., Argiriadi M.A., McGettrick A.F., et al. Itaconate and itaconate derivatives target JAK1 to suppress alternative activation of macrophages. Cell Metab. 2022;34:487–501.e8. doi: 10.1016/j.cmet.2022.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Hung S.Y., Tsai J.S., Yeh J.T., Chen K.H., Lin C.N., Yang H.M., Lin C.W., Chen H.Y., Huang C.H., Huang Y.Y. Amino acids and wound healing in people with limb-threatening diabetic foot ulcers. J. Diabetes Complications. 2019;33 doi: 10.1016/j.jdiacomp.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Floegel A., Stefan N., Yu Z., Mühlenbruch K., Drogan D., Joost H.G., Fritsche A., Häring H.U., Hrabě de Angelis M., Peters A., et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62:639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Spronsen F.J., Blau N., Harding C., Burlina A., Longo N., Bosch A.M. Phenylketonuria. Nat. Rev. Dis. Primers. 2021;7:36. doi: 10.1038/s41572-021-00267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Czibik G., Mezdari Z., Murat Altintas D., Bréhat J., Pini M., d'Humières T., Delmont T., Radu C., Breau M., Liang H., et al. Dysregulated Phenylalanine Catabolism Plays a Key Role in the Trajectory of Cardiac Aging. Circulation. 2021;144:559–574. doi: 10.1161/CIRCULATIONAHA.121.054204. [DOI] [PubMed] [Google Scholar]
- 25.Liang D., Lin W.J., Ren M., Qiu J., Yang C., Wang X., Li N., Zeng T., Sun K., You L., et al. m(6)A reader YTHDC1 modulates autophagy by targeting SQSTM1 in diabetic skin. Autophagy. 2022;18:1318–1337. doi: 10.1080/15548627.2021.1974175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piazza I., Kochanowski K., Cappelletti V., Fuhrer T., Noor E., Sauer U., Picotti P. A Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication. Cell. 2018;172:358–372.e23. doi: 10.1016/j.cell.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Katsnelson M.A., Lozada-Soto K.M., Russo H.M., Miller B.A., Dubyak G.R. NLRP3 inflammasome signaling is activated by low-level lysosome disruption but inhibited by extensive lysosome disruption: roles for K+ efflux and Ca2+ influx. Am. J. Physiol. Cell Physiol. 2016;311:C83–C100. doi: 10.1152/ajpcell.00298.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin H. Protein cysteine palmitoylation in immunity and inflammation. FEBS J. 2021;288:7043–7059. doi: 10.1111/febs.15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahtiainen L., Van Diggelen O.P., Jalanko A., Kopra O. Palmitoyl protein thioesterase 1 is targeted to the axons in neurons. J. Comp. Neurol. 2003;455:368–377. doi: 10.1002/cne.10492. [DOI] [PubMed] [Google Scholar]
- 30.Lyly A., Marjavaara S.K., Kyttälä A., Uusi-Rauva K., Luiro K., Kopra O., Martinez L.O., Tanhuanpää K., Kalkkinen N., Suomalainen A., et al. Deficiency of the INCL protein Ppt1 results in changes in ectopic F1-ATP synthase and altered cholesterol metabolism. Hum. Mol. Genet. 2008;17:1406–1417. doi: 10.1093/hmg/ddn028. [DOI] [PubMed] [Google Scholar]
- 31.Bagh M.B., Peng S., Chandra G., Zhang Z., Singh S.P., Pattabiraman N., Liu A., Mukherjee A.B. Misrouting of v-ATPase subunit V0a1 dysregulates lysosomal acidification in a neurodegenerative lysosomal storage disease model. Nat. Commun. 2017;8 doi: 10.1038/ncomms14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou L., He X., Wang L., Wei P., Cai Z., Zhang S., Jin S., Zeng H., Cui J. Palmitoylation restricts SQSTM1/p62-mediated autophagic degradation of NOD2 to modulate inflammation. Cell Death Differ. 2022;29:1541–1551. doi: 10.1038/s41418-022-00942-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y., Hsu J.M., Sun L., Chan L.C., Li C.W., Hsu J.L., Wei Y., Xia W., Hou J., Qiu Y., et al. Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res. 2019;29:83–86. doi: 10.1038/s41422-018-0124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y.C., Lee S.E., Kim S.K., Jang H.D., Hwang I., Jin S., Hong E.B., Jang K.S., Kim H.S. Toll-like receptor mediated inflammation requires FASN-dependent MYD88 palmitoylation. Nat. Chem. Biol. 2019;15:907–916. doi: 10.1038/s41589-019-0344-0. [DOI] [PubMed] [Google Scholar]
- 35.Delandre C., Penabaz T.R., Passarelli A.L., Chapes S.K., Clem R.J. Mutation of juxtamembrane cysteines in the tetraspanin CD81 affects palmitoylation and alters interaction with other proteins at the cell surface. Exp. Cell Res. 2009;315:1953–1963. doi: 10.1016/j.yexcr.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon D., Edmonds M. Managing Diabetic Foot Ulcers: Pharmacotherapy for Wound Healing. Drugs. 2021;81:29–56. doi: 10.1007/s40265-020-01415-8. [DOI] [PubMed] [Google Scholar]
- 37.Huang Y.Y., Lin C.W., Cheng N.C., Cazzell S.M., Chen H.H., Huang K.F., Tung K.Y., Huang H.L., Lin P.Y., Perng C.K., et al. Effect of a Novel Macrophage-Regulating Drug on Wound Healing in Patients With Diabetic Foot Ulcers: A Randomized Clinical Trial. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.22607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y., Gao H.Y., Fan Z.Y., He Y., Yan Y.X. Metabolomics Signatures in Type 2 Diabetes: A Systematic Review and Integrative Analysis. J. Clin. Endocrinol. Metab. 2020;105 doi: 10.1210/clinem/dgz240. [DOI] [PubMed] [Google Scholar]
- 39.Merino J., Leong A., Liu C.T., Porneala B., Walford G.A., von Grotthuss M., Wang T.J., Flannick J., Dupuis J., Levy D., et al. Metabolomics insights into early type 2 diabetes pathogenesis and detection in individuals with normal fasting glucose. Diabetologia. 2018;61:1315–1324. doi: 10.1007/s00125-018-4599-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galderisi A., Pirillo P., Moret V., Stocchero M., Gucciardi A., Perilongo G., Moretti C., Monciotti C., Giordano G., Baraldi E. Metabolomics reveals new metabolic perturbations in children with type 1 diabetes. Pediatr. Diabetes. 2018;19:59–67. doi: 10.1111/pedi.12524. [DOI] [PubMed] [Google Scholar]
- 41.Drogan D., Dunn W.B., Lin W., Buijsse B., Schulze M.B., Langenberg C., Brown M., Floegel A., Dietrich S., Rolandsson O., et al. Untargeted metabolic profiling identifies altered serum metabolites of type 2 diabetes mellitus in a prospective, nested case control study. Clin. Chem. 2015;61:487–497. doi: 10.1373/clinchem.2014.228965. [DOI] [PubMed] [Google Scholar]
- 42.Kong P., Cui Z.Y., Huang X.F., Zhang D.D., Guo R.J., Han M. Inflammation and atherosclerosis: signaling pathways and therapeutic intervention. Signal Transduct. Target. Ther. 2022;7:131. doi: 10.1038/s41392-022-00955-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorland L., Poll-The B.T., Duran M., Smeitink J.A., Berger R. Phenylpyruvate, fetal damage, and maternal phenylketonuria syndrome. Lancet. 1993;341:1351–1352. doi: 10.1016/0140-6736(93)90866-f. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki R., Sato Y., Fukaya M., Suzuki D., Yoshizawa F., Sato Y. Energy metabolism profile of the effects of amino acid treatment on hepatocytes: Phenylalanine and phenylpyruvate inhibit glycolysis of hepatocytes. Nutrition. 2021;82 doi: 10.1016/j.nut.2020.111042. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J., Muri J., Fitzgerald G., Gorski T., Gianni-Barrera R., Masschelein E., D'Hulst G., Gilardoni P., Turiel G., Fan Z., et al. Endothelial Lactate Controls Muscle Regeneration from Ischemia by Inducing M2-like Macrophage Polarization. Cell Metab. 2020;31:1136–1153.e7. doi: 10.1016/j.cmet.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L., Jiang L., Yu L., Li Q., Tian X., He J., Zeng L., Yang Y., Wang C., Wei Y., et al. Inhibition of UBA6 by inosine augments tumour immunogenicity and responses. Nat. Commun. 2022;13:5413. doi: 10.1038/s41467-022-33116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S., Liu X., Peng C., Tan C., Sun H., Liu H., Zhang Y., Wu P., Cui C., Liu C., et al. The phytochemical hyperforin triggers thermogenesis in adipose tissue via a Dlat-AMPK signaling axis to curb obesity. Cell Metab. 2021;33:565–580.e7. doi: 10.1016/j.cmet.2021.02.007. [DOI] [PubMed] [Google Scholar]
- 48.Bellizzi J.J., 3rd, Widom J., Kemp C., Lu J.Y., Das A.K., Hofmann S.L., Clardy J. The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis. Proc. Natl. Acad. Sci. USA. 2000;97:4573–4578. doi: 10.1073/pnas.080508097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y., Qin Z., Sun W., Chu F., Zhou F. Function of Protein S-Palmitoylation in Immunity and Immune-Related Diseases. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.661202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu Y., Zheng Y., Coyaud É., Zhang C., Selvabaskaran A., Yu Y., Xu Z., Weng X., Chen J.S., Meng Y., et al. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science. 2019;366:460–467. doi: 10.1126/science.aau6391. [DOI] [PubMed] [Google Scholar]
- 51.Zhou B., Hao Q., Liang Y., Kong E. Protein palmitoylation in cancer: molecular functions and therapeutic potential. Mol. Oncol. 2023;17:3–26. doi: 10.1002/1878-0261.13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong G., Adak S., Spyropoulos G., Zhang Q., Feng C., Yin L., Speck S.L., Shyr Z., Morikawa S., Kitamura R.A., et al. Palmitoylation couples insulin hypersecretion with beta cell failure in diabetes. Cell Metab. 2023;35:332–344.e7. doi: 10.1016/j.cmet.2022.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L., Cai J., Zhao X., Ma L., Zeng P., Zhou L., Liu Y., Yang S., Cai Z., Zhang S., et al. Palmitoylation prevents sustained inflammation by limiting NLRP3 inflammasome activation through chaperone-mediated autophagy. Mol. Cell. 2023;83:281–297.e10. doi: 10.1016/j.molcel.2022.12.002. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y., Zhang J., Cui W., Silverstein R.L. CD36, a signaling receptor and fatty acid transporter that regulates immune cell metabolism and fate. J. Exp. Med. 2022;219 doi: 10.1084/jem.20211314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shu H., Peng Y., Hang W., Nie J., Zhou N., Wang D.W. The role of CD36 in cardiovascular disease. Cardiovasc. Res. 2022;118:115–129. doi: 10.1093/cvr/cvaa319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J., Li Y. CD36 tango in cancer: signaling pathways and functions. Theranostics. 2019;9:4893–4908. doi: 10.7150/thno.36037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y., Yang M., Huang W., Chen W., Zhao Y., Schulte M.L., Volberding P., Gerbec Z., Zimmermann M.T., Zeighami A., et al. Mitochondrial Metabolic Reprogramming by CD36 Signaling Drives Macrophage Inflammatory Responses. Circ. Res. 2019;125:1087–1102. doi: 10.1161/CIRCRESAHA.119.315833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geng K., Ma X., Jiang Z., Huang W., Gao C., Pu Y., Luo L., Xu Y., Xu Y. Innate Immunity in Diabetic Wound Healing: Focus on the Mastermind Hidden in Chronic Inflammatory. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.653940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li X., Xie X., Lian W., Shi R., Han S., Zhang H., Lu L., Li M. Exosomes from adipose-derived stem cells overexpressing Nrf2 accelerate cutaneous wound healing by promoting vascularization in a diabetic foot ulcer rat model. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee M.K.S., Sreejit G., Nagareddy P.R., Murphy A.J. Attack of the NETs! NETosis primes IL-1beta-mediated inflammation in diabetic foot ulcers. Clin. Sci. 2020;134:1399–1401. doi: 10.1042/CS20200240. [DOI] [PubMed] [Google Scholar]
- 61.Feldmeyer L., Keller M., Niklaus G., Hohl D., Werner S., Beer H.D. The inflammasome mediates UVB-induced activation and secretion of interleukin-1beta by keratinocytes. Curr. Biol. 2007;17:1140–1145. doi: 10.1016/j.cub.2007.05.074. [DOI] [PubMed] [Google Scholar]
- 62.Iglesias-Bartolome R., Uchiyama A., Molinolo A.A., Abusleme L., Brooks S.R., Callejas-Valera J.L., Edwards D., Doci C., Asselin-Labat M.L., Onaitis M.W., et al. Transcriptional signature primes human oral mucosa for rapid wound healing. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aap8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Audu C.O., Melvin W.J., Joshi A.D., Wolf S.J., Moon J.Y., Davis F.M., Barrett E.C., Mangum K.D., Deng H., Xing X., et al. Macrophage-specific inhibition of the histone demethylase JMJD3 decreases STING and pathologic inflammation in diabetic wound repair. Cell. Mol. Immunol. 2022;19:1251–1262. doi: 10.1038/s41423-022-00919-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peng L., Jiang J., Zhou L., Nice E.C., Huang C. A cellular thermal shift assay for detecting amino acid sites involved in drug target engagement. STAR Protoc. 2022;3 doi: 10.1016/j.xpro.2022.101423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Diggelen O.P., Keulemans J.L., Winchester B., Hofman I.L., Vanhanen S.L., Santavuori P., Voznyi Y.V. A rapid fluorogenic palmitoyl-protein thioesterase assay: pre- and postnatal diagnosis of INCL. Mol. Genet. Metab. 1999;66:240–244. doi: 10.1006/mgme.1999.2809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The LC-MS data were deposited in the MetaboLights repository (www.ebi.ac.uk/metabolights/) under the accession number MTBLS7853. Data generated and/or analyzed in this study, excluding identifying personal information, are available from the lead contact, Bing Tang (tangbing@mail.sysu.edu.cn) with reasonable request to protect research participant privacy.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.