Abstract
For decades, marine plankton have been investigated for their capacity to modulate biogeochemical cycles and provide fishery resources. Between the sunlit (epipelagic) layer and the deep dark waters, lies a vast and heterogeneous part of the ocean: the mesopelagic zone. How plankton composition is shaped by environment has been well-explored in the epipelagic but much less in the mesopelagic ocean. Here, we conducted comparative analyses of trans-kingdom community assemblages thriving in the mesopelagic oxygen minimum zone (OMZ), mesopelagic oxic, and their epipelagic counterparts. We identified nine distinct types of intermediate water masses that correlate with variation in mesopelagic community composition. Furthermore, oxygen, NO3− and particle flux together appeared as the main drivers governing these communities. Novel taxonomic signatures emerged from OMZ while a global co-occurrence network analysis showed that about 70% of the abundance of mesopelagic plankton groups is organized into three community modules. One module gathers prokaryotes, pico-eukaryotes and Nucleo-Cytoplasmic Large DNA Viruses (NCLDV) from oxic regions, and the two other modules are enriched in OMZ prokaryotes and OMZ pico-eukaryotes, respectively. We hypothesize that OMZ conditions led to a diversification of ecological niches, and thus communities, due to selective pressure from limited resources. Our study further clarifies the interplay between environmental factors in the mesopelagic oxic and OMZ, and the compositional features of communities.
Subject terms: Microbial ecology, Community ecology, Community ecology
Introduction
Below the ocean’s sunlit layer lies the mesopelagic zone that occupies around 20% of the global ocean volume [1]. The mesopelagic zone is biologically defined as starting where photosynthesis no longer occurs (<1% irradiance; around 200 m depth), down to its lower boundary where there is no detectable sunlight (around 1000 m depth) [2]. This twilight ecosystem cannot rely on photoautotrophy, but sustains its energetic requirements by the combination of heterotrophic, chemoautotrophic, and chemo-mixotrophic metabolisms, together with physicochemical processes. Among the latter, the fraction of upper ocean productivity that escapes epipelagic recycling and sinks by gravity or is delivered by the daily vertical migration of zooplankton constitutes an essential energy source in deep waters and is a vector for attached organisms [3].
Considerable attention has been devoted to the mesopelagic layer in recent years, given its recognized potential for exploitation for bioresources and fisheries [4], potentially becoming an important source of goods for the global bioeconomy [5]. So far, efforts have been made to increase the knowledge of mesopelagic macrofauna by studying the abundance and diversity of nekton. Concerning the mesopelagic community’s microscopic fraction, previous reports have shown a stratification of planktonic communities by the water column. In this regard, the mesopelagic zone displays a distinct assemblage of dsDNA viruses [6], Nucleo-Cytoplasmic Large DNA viruses [7], prokaryotes [8–10], and eukaryotes [11]. These studies have highlighted a global organization different from that of the surface. For example, the mesopelagic plankton diversity does not show a latitudinal diversity gradient trend from pole-to-pole, peaking at lower latitudes [12], and also displays a higher heterogeneity compared to epipelagic waters [13]. Conversely, the mesopelagic microbiome seems to make crucial links in the food web between phototrophic primary production from the sunlit layer and dark ocean specialized consumers [10, 14, 15].
Among the studies conducted in mesopelagic zones, particular efforts have been made to explore regions characterized by extreme conditions, such as oxygen minimum zones (OMZs). These zones are formed by relatively old, slowly upwelling waters, often lying below highly productive surface zones [16], and are currently increasing in volume in the ocean [17]. OMZ prokaryotic communities are well documented and taxa such as Nitrospira, Marinimicrobia, and anammox bacteria from the phylum Planctomycetes have been reported as typical taxonomic features for OMZ regions studied so far [18–23]. In contrast, knowledge about viruses and eukaryotic diversity in OMZs is still rudimentary. A prevalence of specific eukaryotic taxa such as Ciliophora, Dinoflagellata, MALV, and Acantharia has been reported, together with a higher metabolic activity of these taxa [24–26]. Viruses may have a key role in OMZ ecosystem feedback by modulating the local community (host-virus relationship) [27–29].
The last decades have seen a significant increase in large-scale oceanic surveys [30–32]. Despite the advances reported in previous studies [10, 33, 34], most mesopelagic community studies have been limited to geographically or ecologically fragmented regions, or to specific taxonomic groups, mainly because of the inherent difficulties of accessing this zone on a global scale [35]. Moreover, the combination of biotic and abiotic factors influencing community structure [36, 37], has been poorly explored in the mesopelagic zone. Extending this knowledge by including more comprehensive and homogeneous datasets from assorted geographical and oceanographic systems should lead to a global understanding of this layer. Understanding plankton community structure and dynamics is fundamental to anticipate the impact of global warming and acidification in these regions.
Here, we present a trans-kingdom omics-based comparative study of epipelagic, oxic- and OMZ-mesopelagic communities. To this end, we compiled Tara Oceans survey taxonomic DNA barcodes data from four oceanographic basins using standardized sampling protocols. In particular, we focused on mesopelagic environmental drivers, ecology, and taxa associations networks in both oxic and OMZ.
Materials and methods
Sample collection and pre-processing
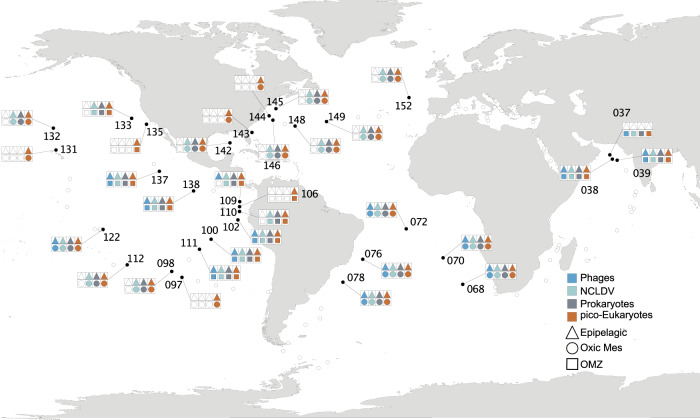
The environmental and biological data were obtained during the Tara Oceans expedition (2009–2012) in 32 oceanographic stations located in the Indian Ocean (IO - 037, 038, 039), Pacific Ocean (PO - 097, 098, 100, 102, 106, 109, 110, 111, 112, 122, 131, 132, 133, 135, 137, 138), South Atlantic Ocean (SAO - 068, 070, 072, 076, 078) and North Atlantic Ocean (NAO - 142, 143, 144, 145, 146, 148, 149, 152) comprising tropical and subtropical regions (Fig. 1). Physico-chemical environmental data were obtained along a vertical profile at each station. Temperature, salinity, and oxygen were measured using a CTD-rosette system with a coupled dissolved oxygen sensor. Chlorophyll-a concentrations were measured using high-performance liquid chromatography. Nutrient concentrations were determined using segmented flow analysis. All these metadata are available at PANGAEA [38–43] (10.1594/PANGAEA.875582).
Fig. 1. Geographical locations of Tara Oceans epipelagic and mesopelagic sampling sites included in this study.
Symbol colors represent organism groups evaluated in the present study. Shape formats represent eco-regions, Epipelagic, Oxic MES = oxicmesopelagic, OMZ = oxygen minimum zone mesopelagic.
The vertical distribution of marine particles was investigated with an Underwater Vision Profiler (UVP [44, 45],) mounted on the CTD-Rosette. The UVP acquires images in a coherent water volume (1L) delimited by a light sheet issued from red light-emitting diodes. Automatic identification of objects was made using Ecotaxa [46], based on a learning set of visually identified, manually classified objects and associated features. Images were classified to distinguish mesozooplankton from non-living objects and artifacts (e.g., detrital particles, fibers, and out-of-focus objects).
Water vertical profiles of temperature and salinity generated from the CTD were used to identify the water masses by plotting a temperature x salinity (T/S) diagram using the Ocean Data View V 5.0 (ODV) software package [47].
Three different water layers were sampled: surface (SRF, 3–7 m), deep chlorophyll maximum (DCM - depth identified according to the peak of chlorophyll-a fluorescence obtained in situ), and mesopelagic (ranging from 200–1000 m) [48]. The planktonic community was sampled by partitioning the pumped seawater by filtering each sampled depth with different filter sizes, the on-board sampling methodology is detailed by Pesant et al. [42].
Among the sampled mesopelagic zones, 13 of them were identified as deficient in oxygen and classified as oxygen minimum zone (OMZ, stations IO - 037, 038, 039 / PO - 100, 102, 106, 109, 110, 111, 133, 135, 137, 138). The OMZ were categorized as suboxic: <10 μM O2/kg seawater and anoxic: (<0.003 μM/kg seawater or undetectable with most sensitive techniques, e.g., STOX sensors) [Units of O2 concentration: 1 mL.L−1 = 1.43 mg. L−1 or 1 mL.L−1 = 44.64 μM] [25].
Our dataset comprises different organismal size-fractions from viruses (two dsDNA-virus bacteriophage families Podoviridae and Myoviridae - hereafter named as phages and Nucleo-Cytoplasmic Large DNA viruses - hereafter named as NCLDV) to pico-eukaryotes.
Phage libraries were constructed from seawater samples filtered at 0.22 μm, concentrated using iron chloride flocculation, and treated with deoxyribonuclease (DNase). NCLDV polB and prokaryotic 16S rRNA gene sequences were extracted from plankton metagenomes sequenced from 0.22–1.6 or 0.22–3 μm filters, and the pico-eukaryote dataset was obtained by V9-18S rRNA gene marker amplification from 0.8–3 or 0.8–5 μm filters. Details of sample preparation and sequencing procedures are fully described in Alberti et al. [49].
Phage read counts was accessed through the search for the marker genes gp23 (Myoviridae) and polA (Podoviridae) in the protein collection GOV2.0 derived from metagenomic sequencing described in Gregory et al. [6]. The NCLDV read counts profile was obtained from the polB marker gene gathered from the OM-RGC.v2 catalog [9] as described in Endo et al. [7]. The Prokaryotic read counts was assessed from a metagenomic dataset called 16S mitag, as described in Sunagawa et al. [8], which does not rely on PCR amplification [50]. Sequences matching “Eukaryota”, “chloroplasts”, and “mitochondria” were removed from the final Prokaryotic OTU table. Clustering and annotation of pico-eukaryote V9-18S rRNA gene PCR-amplicons are described in de Vargas et al. [51], and functional annotation of taxonomically assigned V9-18S rRNA gene metabarcodes was improved afterwards; in this case, we conserved in the final data only sequences assigned to the “Eukaryota” domain. More details concerning the acquisition and pre-processing of the sequence data used in the present study were compiled from earlier publications and are provided in Supplementary Methods. Throughout the manuscript, we use the classical term “OTU” to denote taxonomic units, although we did not employ the clustering technique historically associated with this acronym. More comments about “OTU” usage are available in Supplementary Methods.
To simplify the biological information and obtain a concise epipelagic dataset (EPI), we merged redundant SRF and DCM OTUs by summing their read counts for each taxonomic group and preserved non-redundant OTUs. Putative biases produced by this merging procedure were tested and discarded as shown in the Supplementary Methods; they did not affect the conclusions.
Afterward, to deal with the compositional nature of the data, OTU count matrices (EPI and MES) were transformed using robust CLR (centered log ratio) after adding the value of one as a pseudo count. Robust CLR transformation considers only values greater than 0 to calculate the geometric mean and avoid biases due to sparse data [52].
Epipelagic and mesopelagic community and environmental differences
We applied an NMDS analysis based on the Bray–Curtis dissimilarity matrix on CLR transformed data. The ‘metaNMDS’ function from the vegan R package [53] was applied to confirm community differences between epipelagic and mesopelagic layers. Homogeneity of the sampled environmental parameters was checked using the ‘betadisper’ function (homogeneity of multivariate dispersions in the vegan package). The analysis was conducted using the Euclidean distance matrix of the environmental variables with the depths (epipelagic, mesopelagic) as group factor. A permutation test statistically confirmed the results.
Ecological inferences and statistics
Ecological patterns were inferred using environmental variables to constrain the variation observed in biological data (CLR transformed) for planktonic samples using Canonical Correspondence Analysis (CCA) in the vegan R package. A set of physico-chemical variables for the discrete depths were selected for the ecological inferences, such as nitrate (NO3−), oxygen, temperature, salinity, density, and particles using particle flux UVP data. In order to avoid collinearity among factors, the selected variables were checked for variance inflation factor using the vif.cca function and tested for significance by ANOVA-like tests performed by ‘anova’ implemented in vegan with 999 permutations. The significance of the effect of each variable was tested individually using all others parameters as covariables (independently from their order in the model) by applying the option ‘margin’ to the ‘anova’ function in vegan.
Permutational multivariate analysis of variance (PERMANOVA) was performed with the function ‘adonis’ in vegan to determine the relationship between mesopelagic community composition and predefined water masses based on 999 permutations.
Classification of organisms eco-region
In order to detect organisms specific to epipelagic (EPI), oxic mesopelagic (Oxic MES), and OMZ eco-regions, only sampling sites containing both epipelagic and mesopelagic information were considered: in total, 25 stations for NCLDV, prokaryotes, and pico-eukaryotes, and 13 for phages. We ran a Kruskal–Wallis test (‘kruskal.test’ from stats R package [54]) to detect differential OTU abundances between eco-regions, followed by a Benjamini & Hochberg correction to avoid false discovery rate (FDR - p.value.bh). Organisms with a p-value < 0.05, indicating a difference within groups, were subject to a post-hoc Dunn test (‘dunn.test’ from dunn.test R package [55]) to identify preferential eco-regions for each OTU. From these results, OTUs non-significant Kruskal–Wallis tests (p.value.bh >0.05) were assigned to the “ubiquitous” group. In contrast, those with significant p-values.bh were classified as EPI, Oxic MES, or OMZ if only the corresponding eco-region was elected according to the Dunn test. Organisms with no significant differences between Oxic MES and OMZ were assigned to Core MES.
Co-occurrence network inference
For investigation of ecological associations between organisms across eco-regions, a co-occurrence network was inferred. In this analysis, phage samples were not included due to the lower number of stations sampled. Therefore, samples for NCLDV, prokaryotes and pico-eukaryotes from stations 038, 039, 068, 070, 072, 076, 078, 098, 100, 102, 109, 110, 111, 112, 122, 132, 133, 137, 138, 142, 145, 146, 148, 149 and 152 were retained. OTUs with a relative abundance lower than10−4 and counting fewer than 5 observations were discarded. Network inferences were performed on CLR transformed data using FlashWeave version 0.18 implemented in Julia version 1.2 [56], using the sensitive and heterogeneous mode. FlashWeave assumes features to be multivariate Gaussian distributed in CLR-transformed space.
We analyzed this global co-occurrence network by delineating communities (or modules) using the Clauset-Newman-Moore algorithm [57]. These modules are subsets of OTUs, obtained by maximizing the co-occurrences within modules and minimizing connections between them. Next, we investigated modules enriched in OTUs from specific eco-regions using Fisher’s exact test using the “fisher.test” function from the stats R package, followed by the Benjamini & Hochberg correction to control the FDR due to multiple testing.
Results and discussion
Leveraging the resources produced by the Tara Oceans project, we deciphered differences between epipelagic and mesopelagic beta-diversity stratification, with a particular emphasis on the role of environmental variables such as temperature, oxygen, salinity, NO3−, chlorophyll-a, and particle flux (see Methods). Here, we combined the diversity information obtained for surface and DCM samples to consider a single epipelagic group. We observed a clear distinction between the mesopelagic and epipelagic communities without loss of signal, as shown in previous studies [6–11], supporting our subsequent analyses reported here (Supplementary Fig. 1, Supplementary Methods Figs. 1 and 2). Consequently, we first investigated differences among physicochemical characteristics of the mesopelagic and epipelagic sampling sites. We observed a high dissimilarity gradient among sites for both layers (Supplementary Fig. 2a, b). Mesopelagic samples were heterogeneously distributed, with most of the points placed distant from the group centroid (located in the center of the cloud of points identified for each group) (Supplementary Fig. 2a). In contrast, epipelagic points displayed a large variance due to the samples positioned apart from the main cluster (Supplementary Fig. 2a). These results underlie the heterogeneity of environmental conditions encountered in both sampled layers, and this environmental variation may be an important factor that can directly influence community composition.
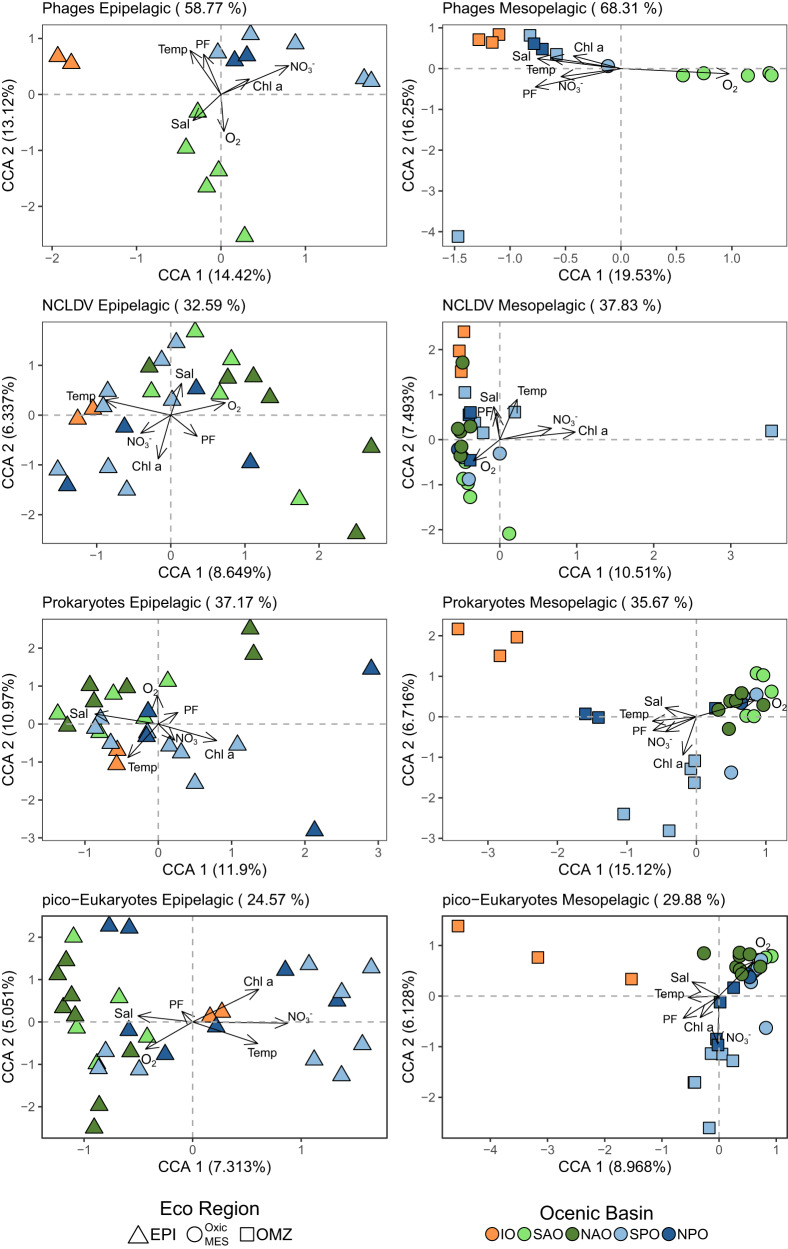
Next, to quantify how much of the differences in the assemblages’ variance can be explained by environmental conditions, we employed canonical correspondence analysis (CCA) using the environmental variables measured at discrete depths as constraint variables. The results showed that abiotic factors explained 40.5% on average of community variance for both layers (Fig. 2). The phage assemblage was the exception, for which about 58% of the epipelagic variation and 68% of the mesopelagic variation could be explained by the variables investigated (Fig. 2). Our analysis also demonstrated a clustering according to the different oceanic basins studied for all the assemblages, confirmed by a PERMANOVA analysis (Fig. 2, Supplementary Table S1). A basin-scale biogeographical structure was already shown for virus, bacteria and protist in the epipelagic layer [58, 59]. Here, we showed that this structuration appears even more pronounced in the mesopelagic samples at global scale.
Fig. 2. Ordination plot of canonical correspondence analysis (CCA) from epipelagic (left) and mesopelagic (right) communities based on OTU composition.
Percentages in parentheses are the amount of variation constrained - in titles represent the total in each analysis, and in the axis represent the correspondent value for each dimension. Arrows represent environmental quantitative explanatory variables with arrowheads indicating their direction of increase. Shapes represent sampling sites. Shape formats represent eco-regions, epi epipelagic, Oxic MES oxic mesopelagic, OMZ oxygen minimum zone mesopelagic. IO Indian Ocean, NAO North Atlantic Ocean, NPO North Pacific Ocean, SAO South Atlantic Ocean, SPO South Pacific Ocean.
Furthermore, we assessed the variance of communities regarding each environmental parameter as explanatory variables individually. In contrast with epipelagic communities mainly structured by temperature, as observed elsewhere [6, 8, 11, 12, 60, 61], temperature was only a significant variable structuring the viruses (phage and NCLDV) in the mesopelagic layer. However, oxygen, NO3− and particle flux appeared as common environmental drivers governing at least three out of four mesopelagic assemblages (Table 1, complete analysis in Supplementary Table S2).
Table 1.
ANOVA p-value of the variance explained by environmental variables for global model including all tested environmental variables (Global) and for each explanatory variable individually with all the others used as covariables (independently from their order in the model).
| Assemblages | Depth | Global | Temp °C | Salinity | O2 [µmol/Kg] | NO3− [µmol/L] | Chl-a [µmol/m3] | Particle flux |
|---|---|---|---|---|---|---|---|---|
| Phages | Epi | 0.001 | 0.048 | 0.132 | 0.126 | 0.001 | 0.308 | 0.013 |
| Phages | MES | 0.001 | 0.006 | 0.151 | 0.001 | 0.230 | 0.306 | 0.001 |
| NCDLV | Epi | 0.001 | 0.001 | 0.033 | 0.036 | 0.083 | 0.019 | 0.197 |
| NCDLV | MES | 0.002 | 0.043 | 0.186 | 0.005 | 0.031 | 0.001 | 0.048 |
| Prokaryotes | Epi | 0.035 | 0.062 | 0.088 | 0.303 | 0.152 | 0.106 | 0.606 |
| Prokaryotes | MES | 0.006 | 0.410 | 0.590 | 0.004 | 0.039 | 0.575 | 0.418 |
| pico-Eukaryotes | Epi | 0.025 | 0.108 | 0.576 | 0.373 | 0.065 | 0.173 | 0.807 |
| pico-Eukaryotes | MES | 0.001 | 0.168 | 0.304 | 0.033 | 0.001 | 0.016 | 0.009 |
Previous studies have identified oxygen as one of the main drivers of the eukaryotic community structure in OMZ regions [25, 26, 62]. These studies mainly compared community composition along the oxygen gradient within the water column depth, from the surface downwards. However, depth stratification of plankton communities is evident even in regions with high oxygen concentrations, so distinct parameters co-varying with depth must be taken into account in addition to the oxygen gradients [63].
In addition to the physicochemical parameters, our results show that particle flux (derived from UVP measurements) was a significant variable structuring phage, NCLDV and pico-eukaryote mesopelagic assemblages (Table 1, complete analysis in Supplementary Table S2). These observations support previous reports about the high correlation of this environmental factor with phages, finding possible relevance for the carbon pump’s functioning in epipelagic layers [14]. This observation may also reflect the association with virus (phages and NCLDV) inputs from overlaying water layers via sinking particles [64, 65]. Furthermore, Bettarel et al. [66] suggested that marine aggregates can act as virus-factories, where these entities use the adsorbed bacteria to replicate and therefore be massively exported through the water column (one-way motion). The authors also showed that adsorbed bacteria can easily detach from aggregates (two-way motion), which can explain the lack of correlation between prokaryotes and particle flux observed here. On the other hand, pico-eukaryotes were also driven by particle flux. Durkin et al. [67], demonstrated that about 25% of epipelagic diversity can be detected on marine sinking particles and that the particle associated diversity is linked to the size and type of particle (fecal pellet loose or dense, aggregates and detritus).
In situ physico-chemical measurements have revealed the dynamics and fluctuating nature of the ocean, even over a short time scale [68]. The heterogeneity in mesopelagic layers given by deep currents, the impact of surface production, and the low mixing levels may favor a diversification in the mesopelagic community living in different water masses, leading to species adaptation-acclimation. The Tara Oceans expedition route included samples from common or distinct water masses defined by temperature/salinity profiles - T/S, comprising regionally connected or unconnected stations. We identified nine different water masses in the mesopelagic sampled locations (Fig. 3). We could confirm significant differences among mesopelagic communities sampled in these different water masses based on the PERMANOVA test (Table 2). This result indicates that the oceanic patchiness created by distinct water masses significantly shape community beta-diversity in the mesopelagic layer, which would imply it to be a critical component for mesopelagic community variation for all the assemblages studied (phages, NCLDV, prokaryotes, and pico-eukaryotes). Thus, we hypothesize that this result may be explained by two non-exclusive causes related to water masses: (i) past common origin among water masses that have drifted or (ii) constant connectivity by ocean circulation between sampled sites belonging to the same water mass.
Fig. 3. Temperature and salinity plot indicating water mass designation for all mesopelagic samples.

Formats represent the different oceanic basins (■ - North Atlantic Ocean, ● - South Atlantic Ocean, ▲- Pacific Ocean, ★- Indian Ocean). Colors indicate the oxygen concentration at the sampling depth. LSW Labrador Sea Water, AAIW Antarctic Intermediate Water, tNPIW transitional North Pacific Intermediate Water, SAMW Subantarctic Mode Water, SPSTMW South Pacific Subtropical Mode Water, modAAIW modified Antarctic Intermediate Water, PGW Persian Gulf Water mass, RSW Red Sea Water mass, NASTMW North Atlantic Subtropical Mode Water.
Table 2.
Proportion of the assemblages variation explained by water masses using the Permutation multivariate analysis of variance (PERMANOVA)
| Assemblage | DF | Sum of squares | Mean squares | F | R2 | Pr |
|---|---|---|---|---|---|---|
| Phages | 4 | 1513.1 | 378.27 | 1.36 | 0.37 | 0.1 |
| NCDLV | 8 | 3136.6 | 392.08 | 1.72 | 0.45 | 0.01 |
| Prokaryotes | 8 | 13584 | 1698.02 | 2.37 | 0.53 | 0.001 |
| pico-Eukaryotes | 8 | 71998 | 8999.8 | 1.54 | 0.35 | 0.001 |
We addressed another lingering question, resolving planktonic community signatures of Oxic MES and OMZ regions from those observed in epipelagic layer. For this, we classified OTUs into three eco-regions: 1) EPI, 2) Oxic MES, and 3) OMZ. OTUs were classified as Core MES when commonly present in Oxic MES and OMZ samples. Taxa that were either equally abundant in all three eco-regions or not statistically confirmed to a single eco-region were classified as ubiquitous (Supplementary Fig. 3, Supplementary Tables S3–S7). Using this approach, we could identify ubiquitous taxa that are likely to thrive in a wide range of environmental conditions, or that may be detected in mesopelagic samples due to the simple vertical movement of sinking particles. This classification should help avoid putative biases inherent to the metabarcoding methodology.
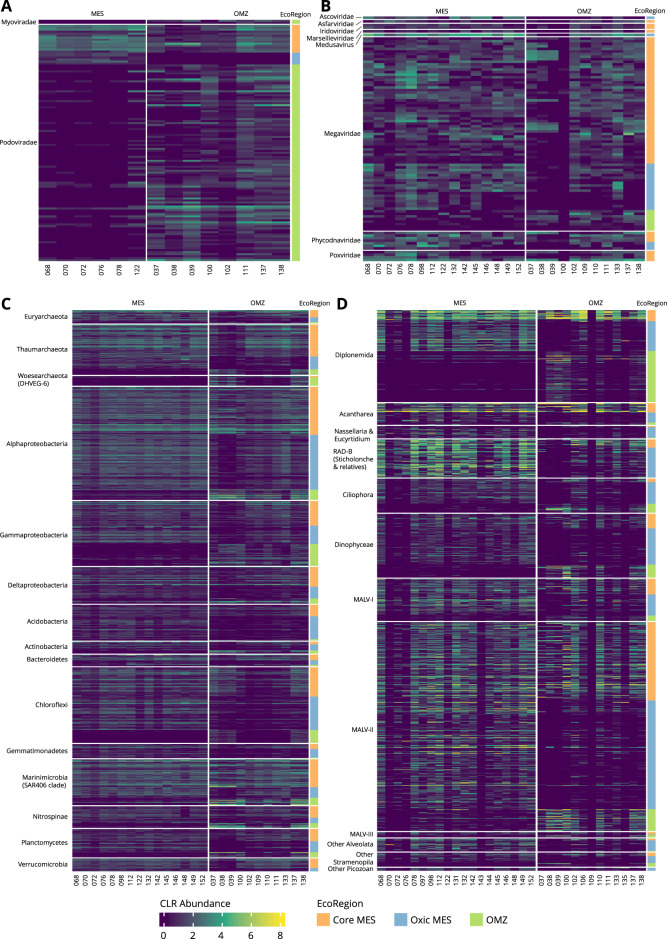
More specifically, we were able to identify Oxic MES and OMZ signatures mainly at the infra-taxonomic level (OTU-species) for all biotic groups investigated (Fig. 4, Supplementary Figs. 3–7, Supplementary Tables S3–S7). This reflected the wide ecological niche occupied by the different species at a higher taxonomic level (i.e. family). At the species level, we observed large taxonomic plasticity of OTUs that occurred equally in both Oxic MES and OMZ samples, called Core MES. However, most OTUs are not yet classified at the infra-taxonomic level (Supplementary Tables S3–S7). This observation reflects the knowledge gap about the biodiversity and functional plasticity of species thriving in this ecosystem.
Fig. 4. Heatmaps occurrence of OTUs assigned to mesopelagic eco-regions.
A Phages, B NCLDV, C Prokaryotes and D pico-Eukaryotes.
The great majority of phage taxa (93.51%) occurred at similar abundance in all eco-regions (ubiquitous). Surprisingly, we did not identify any OTUs assigned specifically to an EPI eco-region, meaning that almost all taxa observed in the epipelagic layer were also present in the mesopelagic layer (Supplementary Figs. 3, 4). This observation supports the seed-bank hypothesis raised by Brum et al. [60], and the correlation to the sinking particles observed here. On the other hand, we detected taxa specific to the mesopelagic layer, mostly related to the OMZ eco-region (Fig. 4a, Supplementary Figs. 3–4). This mesopelagic specificity agrees with the sharp increase in marine phage micro-diversity following depth, as previously shown by Gregory et al. [6]. Our results emphasize that one cause for phage stratification in the water column might be the adaptation to the mesopelagic environment. Two hypotheses arise here, 1) the environment acts as a strong driver, directly selecting phages independently of their hosts, and 2) there is higher phage-host specificity in the mesopelagic layer, promoting phage selection. Following the first hypothesis, we can posit that the environment can directly impact phage assemblage composition. The direct contact with the environment of free phage entities (released from their hosts) may reduce infectivity, degrade, or remove virus particles, and adversely affect adsorption to the host [69]. This direct environmental effect over marine phages was reported for different ionic gradients [70], daylight conditions, and temperature [71]. However, the enrichment of prokaryotic OTUs specific to mesopelagic regions (Fig. 4, Supplementary Figs. 3, 6), especially in OMZs, does not exclude the phage-host indirect selection relationship.
We found 136 mesopelagic-specific NCLDV OTUs out of 5538 in both oxic and OMZ eco-regions. Even though it represents a small number, these OTUs were highly abundant (Fig. 4b, Supplementary Figs. 3, 5). Most of the mesopelagic-specific NCLDV OTUs corresponded to the Core MES group (OMZ = 18 OTUs, Oxic MES = 31 OTUs, Core MES = 87 OTUs- Supplementary Table S3). NCLDV can encode genes such as transporters for ammonium, magnesium, and phosphate that are important in marine oligotrophic areas [72]. This characteristic can improve the host’s fitness in the short-term and ultimately favor NCLDV fecundity and endurance. This property is named NCLDV-mediated host reprogramming [72]. Our results therefore indicate that these entities are less diverse in mesopelagic waters and may successfully infect a wide range of hosts adapted to different oxygen concentrations.
Among the planktonic microorganisms, prokaryotes have been, so far, the most investigated group in OMZ regions, especially in the Pacific Ocean [20, 21]. We could better distinguish the prokaryotic mesopelagic signatures between Oxic MES and OMZ, confirming the influence of oxygen reported here and in previous studies [20–22] (Fig. 4c, Supplementary Figs. 3, 6). We observed similar occurrences and abundances for the OMZ signature taxa in the Indian Ocean stations (IO - 037, 038, 039) and in stations PO - 100, 137, and 138 from the Pacific Ocean (Fig. 4c). These Pacific stations are located in the open ocean (PO - 137 and 138 located in the Equatorial upwelling zone and station PO - 100 in the South Pacific Subtropical Gyre). They present a strong upwelling signature, disclosing an intense decrease in oxygen concentration almost reaching shallow waters. Likewise, the sampling stations in the Indian Ocean are located in well-stratified waters, markedly characterized by the abrupt decrease of oxygen concentration below the thermohaline at 100–120 m depth, especially for stations 038 and 039. At the mesopelagic layer of the Indian Ocean stations, the oxygen concentration ranges from 0.83 to 3 μmol/kg, characterizing functionally anoxic waters since aerobic metabolisms cannot be sustained at this oxygen level [73]. The other OMZ stations in the Pacific Ocean (PO - 102, 109, 110, 111) are located in coastal areas. Although they are also under the influence of upwellings, with low oxygen content, the oxygen level does not correspond to anoxic conditions, so they are classified as suboxic waters. This microoxic condition of this environment is sufficient to completely alter the microbial metabolism delineating the community composition in these sites. In addition, differences in the formation of offshore and coastal upwelling, for instance, or the influence of river runoffs, transporting anthropogenic nutrient enrichment from the continent to coastal areas [73], could be crucial in supporting the differences we observed in OMZ communities.
The same clear enrichment in both OMZ anoxic and suboxic samples was observed for the pico-eukaryotic groups Diplonemida, MALV-II, and Dinophyceae, suggesting these OTUs as the true OMZ eukaryotic signatures (Fig. 4d). Some OTUs of these groups exhibited similar occurrences in the anoxic Indian and Pacific Oceans but not in suboxic samples from the Pacific Ocean. However, we observed a lower number of pico-eukaryotic taxa in the OMZ eco-region, the prevailing OTUs being specific to Oxic MES locations in most cases.
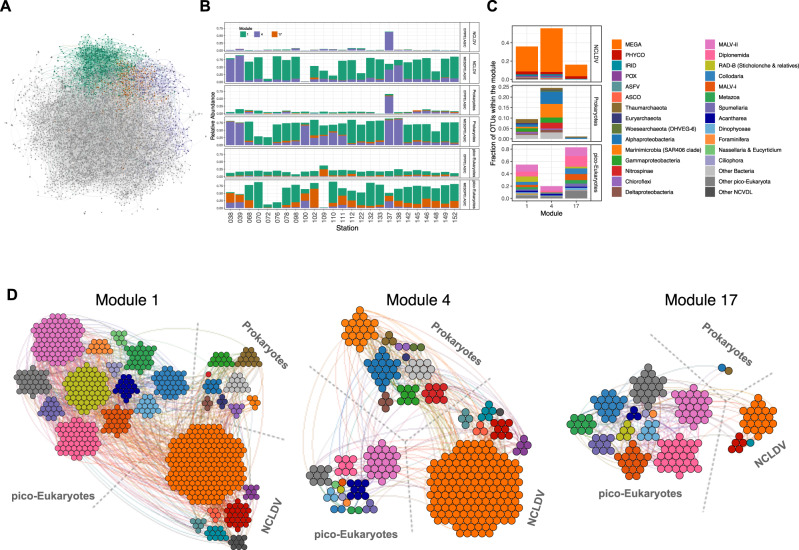
Another step to better understand mesopelagic community dynamics is to dissect the ecological relationships among species that thrive in this layer. Co-occurrence networks can indicate how the environment may structure the community acting as a filter for resident species [74]. They can also give us glimpses of organisms’ potential ecological interactions based on species connectivity [74, 75]. Combining the NCLDV, prokaryote, and pico-eukaryote data, we inferred a network containing 6154 nodes and 12,935 edges (Fig. 5a, Table 3). Due to the lower number of stations sampled for phages, we excluded this group from the analysis. We found mainly positive relationships (94%), suggesting a predominance of putative biotic interactions (e.g. competition, symbiosis) rather than taxa avoidance or exclusion. This dominance of positive relations was also reported for epipelagic plankton communities [36, 76]. The global network had a modularity value greater than 0.4 (Table 3), indicating that the network has a modular structure [73]. Applying a community detection algorithm [57] on this global graph, we were able to delineate 36 distinct modules (or subnetworks), presumably corresponding to ecological communities. Three of them were mainly composed of OTUs significantly enriched in mesopelagic OTUs (Oxic MES enriched module 1, p-value.bh = 1.29e−153 and OMZ enriched modules 4, p-value.bh = 1.90e−51, and 17, p-value.bh = 2.1e−13; Fig. 5). Together, these three modules covered almost the total richness found in the mesopelagic zone (Fig. 5b), and presented similar values for the average degree, clustering coefficient, and average path length (Table 3). These parameters indicate a network complexity [74], hinting at distinct ecological niches within the mesopelagic layer.
Fig. 5. Co-occurrence network in epipelagic and mesopelagic communities.
A Global network, with connected modules for OMZ (purple and orange) and Oxic MES (green) highlighted. B Relative taxa abundance in each module in each station and depth. C Relative number of OTUs classified in taxonomic groups. D Network representation of modules.
Table 3.
Network topological features derived from global analysis including NCDLV, prokaryotes and pico-Eukaryotes samples in epipelagic and mesopelagic depths.
| Parameter | Global | Mod 1 | Mod 4 | Mod 17 |
|---|---|---|---|---|
| Nodes | 6154 | 731 | 323 | 175 |
| Positive edges | 12193 | 1236 | 480 | 223 |
| Negative edges | 742 | 70 | 49 | 9 |
| Av. Degree | 4.20 | 3.57 | 3.28 | 2.65 |
| Clustering | 0.03 | 0.03 | 0.09 | 0.05 |
| Density | 0.00 | 0.00 | 0.01 | 0.02 |
| Average path length | 7.28 | 6.01 | 6.27 | 6.30 |
| Betweenness | 0.01 | 0.05 | 0.10 | 0.22 |
| Degree Centralization | 0.00 | 0.01 | 0.02 | 0.04 |
| Modularity | 0.47 | 0.60 | 0.67 | 0.66 |
More precisely, the OMZ modules were composed of a few connected nodes (323 and 175 nodes for modules 4 and 17, respectively - Table 3), potentially indicating two distinct OMZ community niches. The Oxic MES module 1 counted more nodes composing the network associations (731 nodes), and all modules presented a variation in taxonomic composition and proportions. OMZ module 4 (OMZ-4) contained mainly prokaryotic (23%) and NCLDV (55%) OTUs (Fig. 5c, d). Among the prokaryotes, we detected taxa previously determined as representing OMZ signatures (Nitrospinae, Marinimicrobia SAR 406, and Planctomycetes). As this module contains both prokaryotes and NCLDV OTUs, it suggests the existence of a confounding factor not captured in the dataset (such as large eukaryotes), as NCLDV are known to be specific to eukaryotes. Nevertheless, this correlation has been observed independently in another study [12]. On the other hand, module 17 (OMZ-17) is mainly composed of pico-eukaryotes OTUs (83%) (notably MALV-II (14%) and Diplonemida (17%) previously indicated as OMZ signatures) and NCLDV (16%) as expected due to virus-host relationships. Module 1 is taxonomically more diverse but consisted mainly of NCLDV and pico-eukaryotes. These groups accounted for 36 and 55%, respectively, of OTUs in this module (Fig. 5c, d). NCLDV contributed to 598 associations (edges) in mesopelagic module 1, of which 177 occurred between NCLDV and pico-eukaryotes. NCLDV from the Mimiviridae family are the most numerous taxa in all three mesopelagic modules. Mimiviridae is a very abundant family in the ocean, present in various size ranges from piconanoplankton (0.8–5 μm) up to mesoplankton (180–2000 μm) [9, 75]. This observation supports our finding that NCLDV are a prosperous group in mesopelagic waters, undertaking different strategies to endure in such environmental conditions. In all three modules, we observed the presence of Foraminifera, of which some species can use nitrate over oxygen as an electron acceptor, favoring their survival in OMZ regions [77].
Our converging results suggest that the mesopelagic zone can be characterized by at least three well-defined ecological niches (Oxic MES, OMZ-4 and OMZ-17), with established conditions and resources (abiotic and biotic) that allow the survival of a specific communities in these environments. Observed differences between OMZ and Oxic MES networks suggest a potential loss of connections and interactions among mesopelagic community members, directly affecting ecosystem stability due to habitat change.
Conclusions
In this study, we explored mesopelagic pico-plankton ecological structuring and concluded that this component of oceanic plankton is heterogeneous with respect to environmental conditions. We could pinpoint the relevance of oxygen for all assemblages and the relation of particle flux with phages, NCLDV and pico-eukaryotes. These results reinforce the need to better understand the mesopelagic ecosystem in order to improve our comprehension of carbon export through the biological carbon pump in the twilight zone. Also, we show that intermediated water masses defined by their T/S profiles can explain the differences in the observed mesopelagic pico-plankton structure, pointing to the role of a set of environmental parameters for community composition.
By establishing eco-regions (Epipelagic, Oxic MES, and OMZ), we were able to discriminate specific mesopelagic signatures OTUs across all Life’s domains. While we recovered known markers for Oxic MES and OMZ regions at high taxonomic levels, we also found that most of these OTU signatures are observed at low taxonomic levels, which sometimes cannot be resolved using existing databases. Combining these OTU profiles within co-occurrence networks, we proposed three niches with biotic and abiotic conditions that appear to characterize mesopelagic ecosystems.
Limited access to data is usually the bottleneck for knowledge of mesopelagic dynamics. Our study benefits from a larger number of organism samples and distinct oceanic provinces. This allowed us to integrate these data and thus obtain an expanded vision of mesopelagic community structure and dynamics. Our results emphasize the need for a better understanding of mesopelagic life, particularly by improving our knowledge of oxic and oxygen-low mesopelagic-dwelling communities. This effort is especially necessary as climate change can be expected to expand marine OMZs in the future.
Supplementary Information
Acknowledgements
This study is part of the “Ocean Plankton, Climate and Development” project conducted by the Tara Ocean Foundation with the support of the French Facility for the Global Environment (FFEM). JR, MB, AAM, MCB, JJPK and YDS received financial support from FFEM to execute the project. MCB also received financial support from Coordination for the Improvement of Higher Education Personnel of Brazil (CAPES 99999.000487/2016-03). MB and SC received financial support from the H2020 European Commission project AtlantECO (award number 862923). Tara Oceans (which includes both the Tara Oceans and Tara Oceans Polar Circle expeditions) would not exist without the leadership of the Tara Ocean Foundation and the continuous support of 23 institutes (https://fondationtaraocean.org). We further thank the commitment of the following sponsors: CNRS (in particular Groupement de Recherche GDR3280 and the Research Federation for the study of Global Ocean Systems Ecology and Evolution, FR2022/Tara Oceans-GOSEE), European Molecular Biology Laboratory (EMBL), Genoscope/CEA, The French Ministry of Research, and the French Government ‘Investissements d’Avenir’ programs OCEANOMICS (ANR- 11-BTBR-0008), FRANCE GENOMIQUE (ANR-10-INBS-09-08), MEMO LIFE (ANR-10-LABX-54), CLIMACLOCK (ANR-20-CE20-0024) and PSL* Research University (ANR-11-IDEX-0001-02). This study benefits from the European Union’s Horizon 2020 Blue Growth research and innovation programme under grant agreement number 862923 (project AtlantECO). We also thank the support and commitment of agnès b. and Etienne Bourgois, the Prince Albert II de Monaco Foundation, the Veolia Foundation, Region Bretagne, Lorient Agglomeration, Serge Ferrari, World Courier. The global sampling effort was enabled by countless scientists and crew who sampled aboard the schooner Tara from 2009–2013, and we thank MERCATOR-CORIOLIS and ACRIST for providing daily satellite data during the expeditions. We are also grateful to the countries who graciously granted sampling permission. We want to thank to the reviewers and Editor for all the critics and suggestions that certainly improved the quality of the manuscript. We also acknowledge Noan Le Bescot (Ternog Design) for assistance in preparing figures. The authors declare that all data reported herein are fully and freely available from the date of publication, with no restrictions, and that all of the analyses, publications, and ownership of data are free from legal entanglement or restriction by the various nations whose waters the Tara Oceans expeditions sampled in. This article is contribution number 145 of Tara Oceans.
Author contributions
JR, MB and OJ wrote the manuscript with help of AAM, MCB, JJPK, DI, MG, DV, PF, CB, HO, PB, DE, LS, SSunagawa, EK, SC, PW. JR, MB, AAM, MCB, JJPK, YDS, LS, SC and OJ designed the study. CD, MP, SS, JP, SP processed plankton sampling. EK, SK and JP coordinated and managed sampling logistics. KL, JP and PW coordinated the sequencing. SP, LS, and FL processed environmental parameters. FL, YDS and MCB managed and processed imagery data. FK, DI, FL, LS and MG analyzed physical oceanography data. NH and CdV processed and coordinated eukaryote taxonomic markers OTU acquisition. ACG, DV, AAZ, HO, HE, and MBS processed and coordinated viral taxonomic marker acquisitions. AAM, PB and SSunagawa processed and coordinated bacterial taxonomic marker acquisitions. JR, MB, AAM, MCB, JJPK, YDS, PF, HE, LS, SC, OJ and HO performed ecological and global analysis. Tara Oceans coordinators conceptualized and organized sampling efforts for the Tara Oceans expedition. All authors have read and approved the manuscript.
Data availability
All data analyzed during this study are included in this published article in the Supplementary Information file.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Janaina Rigonato, Marko Budinich.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Janaina Rigonato, Email: jrigonat@genoscope.cns.fr.
Olivier Jaillon, Email: ojaillon@genoscope.cns.fr.
Tara Oceans coordinators:
Silvia G. Acinas, Emmanuel Boss, Guy Cochrane, Colomban de Vargas, Gabriel Gorsky, Nigel Grimsley, Lionel Guidi, Pascal Hingamp, Lee Karp-Boss, Fabrice Not, Jeroen Raes, Christian Sardet, and Sabrina Speich
Supplementary information
The online version contains supplementary material available at 10.1038/s43705-023-00279-9.
References
- 1.Proud R, Cox MJ, Brierley AS. Biogeography of the global ocean’s mesopelagic zone. Curr Biol. 2017;27:113–9. doi: 10.1016/j.cub.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Robinson C, Steinberg DK, Anderson TR, Arístegui J, Carlson CA, Frost JR, et al. Mesopelagic zone ecology and biogeochemistry – a synthesis. Deep Sea Res Part II. 2010;57:1504–18. [Google Scholar]
- 3.Mestre M, Ruiz-González C, Logares R, Duarte CM, Gasol JM, Sala MM. Sinking particles promote vertical connectivity in the ocean microbiome. Proc Natl Acad Sci. 2018;115:E6799–807.. doi: 10.1073/pnas.1802470115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.St. John MA, Borja A, Chust G, Heath M, Grigorov I, Mariani P, et al. A dark hole in our understanding of marine ecosystems and their services: perspectives from the mesopelagic community. Front Marine Sci. 2016;3:31.
- 5.Glover AG, Wiklund H, Chen C, Dahlgren TG. Managing a sustainable deep-sea ‘blue economy’ requires knowledge of what actually lives there. eLife. 2018;7:e41319. doi: 10.7554/eLife.41319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory AC, Zayed AA, Conceição-Neto N, Temperton B, Bolduc B, Alberti A, et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell. 2019;177:1109–23.e14. doi: 10.1016/j.cell.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endo H, Blanc-Mathieu R, Li Y, Salazar G, Henry N, Labadie K, et al. Biogeography of marine giant viruses reveals their interplay with eukaryotes and ecological functions. Nat Ecol Evol. 2020;4:1639–49. [DOI] [PubMed]
- 8.Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, et al. Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 9.Salazar G, Paoli L, Alberti A, Huerta-Cepas J, Ruscheweyh H-J, Cuenca M, et al. Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell. 2019;179:1068–83.e21. doi: 10.1016/j.cell.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giovannoni SJ, Vergin KL. Seasonality in ocean microbial communities. Science. 2012;335:671–6. doi: 10.1126/science.1198078. [DOI] [PubMed] [Google Scholar]
- 11.Giner CR, Pernice MC, Balagué V, Duarte CM, Gasol JM, Logares R, et al. Marked changes in diversity and relative activity of picoeukaryotes with depth in the world ocean. ISME J. 2020;14:437–49. doi: 10.1038/s41396-019-0506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibarbalz FM, Henry N, Brandão MC, Martini S, Busseni G, Byrne H, et al. Global trends in marine plankton diversity across kingdoms of life. Cell. 2019;179:1084–97.e21. doi: 10.1016/j.cell.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baltar F, Arístegui J, Gasol JM, Herndl GJ. Microbial functioning and community structure variability in the mesopelagic and epipelagic waters of the subtropical Northeast Atlantic Ocean. Appl Environ Microbiol. 2012;78:3309–16. doi: 10.1128/AEM.07962-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guidi L, Chaffron S, Bittner L, Eveillard D, Larhlimi A, Roux S, et al. Plankton networks driving carbon export in the oligotrophic ocean. Nature. 2016;532:465–70. doi: 10.1038/nature16942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landry Z, Swan BK, Herndl GJ, Stepanauskas R, Giovannoni SJ. SAR202 genomes from the dark ocean predict pathways for the oxidation of recalcitrant dissolved organic matter. mBio. 2017;8:10–1128. [DOI] [PMC free article] [PubMed]
- 16.Capone DG, Hutchins DA. Microbial biogeochemistry of coastal upwelling regimes in a changing ocean. Nat Geosci. 2013;6:711–7. [Google Scholar]
- 17.Breitburg D, Levin LA, Oschlies A, Grégoire M, Chavez FP, Conley DJ, et al. Declining oxygen in the global ocean and coastal waters. Science. 2018;359:eaam7240. doi: 10.1126/science.aam7240. [DOI] [PubMed] [Google Scholar]
- 18.Stevens H, Ulloa O. Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbiol. 2008;10:1244–59. doi: 10.1111/j.1462-2920.2007.01539.x. [DOI] [PubMed] [Google Scholar]
- 19.Stewart FJ, Ulloa O, DeLong EF. Microbial metatranscriptomics in a permanent marine oxygen minimum zone: OMZ community gene expression. Environ Microbiol. 2012;14:23–40. doi: 10.1111/j.1462-2920.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- 20.Ulloa O, Wright JJ, Belmar L, Hallam SJ. Pelagic oxygen minimum zone microbial communities. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. pp. 113–22.
- 21.Ulloa O, Pantoja S. The oxygen minimum zone of the eastern South Pacific. Deep Sea Res Part II. 2009;56:987–91. [Google Scholar]
- 22.Wright JJ, Konwar KM, Hallam SJ. Microbial ecology of expanding oxygen minimum zones. Nat Rev Microbiol. 2012;10:381–94. doi: 10.1038/nrmicro2778. [DOI] [PubMed] [Google Scholar]
- 23.Divya B. Bacterial community profiling of the Arabian Sea oxygen minimum zone sediments using cultivation independent approach. EIMBO. 2017;1:22–28.
- 24.Duret MT, Pachiadaki MG, Stewart FJ, Sarode N, Christaki U, Monchy S, et al. Size-fractionated diversity of eukaryotic microbial communities in the Eastern Tropical North Pacific oxygen minimum zone. FEMS Microbiol. Ecol. 2015;91:fiv037. [DOI] [PubMed]
- 25.Orsi W, Song YC, Hallam S, Edgcomb V. Effect of oxygen minimum zone formation on communities of marine protists. ISME J. 2012;6:1586–601. doi: 10.1038/ismej.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parris DJ, Ganesh S, Edgcomb VP, DeLong EF, Stewart FJ. Microbial eukaryote diversity in the marine oxygen minimum zone off northern Chile. Front Microbiol. 2014;5:543. [DOI] [PMC free article] [PubMed]
- 27.Vik D, Gazitúa MC, Sun CL, Zayed AA, Aldunate M, Mulholland MR, et al. Genome-resolved viral ecology in a marine oxygen minimum zone. Environ Microbiol. 2021;23:2858–74. doi: 10.1111/1462-2920.15313. [DOI] [PubMed] [Google Scholar]
- 28.Gazitúa MC, Vik DR, Roux S, Gregory AC, Bolduc B, Widner B, et al. Potential virus-mediated nitrogen cycling in oxygen-depleted oceanic waters. ISME J. 2021;15:981–98. doi: 10.1038/s41396-020-00825-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chow C, Winget DM, White RA, Hallam SJ, Suttle CA. Combining genomic sequencing methods to explore viral diversity and reveal potential virus-host interactions. Front Microbiol. 2015;6:265. [DOI] [PMC free article] [PubMed]
- 30.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II global ocean sampling expedition: northwest atlantic through eastern tropical pacific. PLOS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karsenti E, Acinas SG, Bork P, Bowler C, De Vargas C, Raes J, et al. A holistic approach to marine eco-systems biology. PLoS Biol. 2011;9:e1001177. doi: 10.1371/journal.pbio.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pernice MC, Forn I, Gomes A, Lara E, Alonso-Sáez L, Arrieta JM, et al. Global abundance of planktonic heterotrophic protists in the deep ocean. ISME J. 2015;9:782–92. doi: 10.1038/ismej.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris RM, Rappé MS, Urbach E, Connon SA, Giovannoni SJ. Prevalence of the Chloroflexi -Related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl Environ Microbiol. 2004;70:2836–42. doi: 10.1128/AEM.70.5.2836-2842.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehrshad M, Rodriguez-Valera F, Amoozegar MA, López-García P, Ghai R. The enigmatic SAR202 cluster up close: shedding light on a globally distributed dark ocean lineage involved in sulfur cycling. ISME J. 2018;12:655–68. doi: 10.1038/s41396-017-0009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hidalgo M, Browman HI. Developing the knowledge base needed to sustainably manage mesopelagic resources. ICES J Marine Sci. 2019;76:609–15. [Google Scholar]
- 36.Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F, et al. Determinants of community structure in the global plankton interactome. Science. 2015;348:1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 37.Louca S, Hawley AK, Katsev S, Torres-Beltran M, Bhatia MP, Kheirandish S, et al. Integrating biogeochemistry with multiomic sequence information in a model oxygen minimum zone. Proc Natl Acad Sci. 2016;113:E5925–33.. doi: 10.1073/pnas.1602897113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sunagawa S, Acinas SG, Bork P, Bowler C, Acinas SG, Babin M, et al. Tara Oceans: towards global ocean ecosystems biology. Nat Rev Microbiol. 2020;18:428–45. doi: 10.1038/s41579-020-0364-5. [DOI] [PubMed] [Google Scholar]
- 39.Guidi L, Picheral M, Pesant S, Tara Oceans Consortium, Coordinators; Tara Oceans Expedition, Participants. Environmental context of all samples from the Tara Oceans Expedition (2009–2013), about sensor data in the targeted environmental feature. PANGAEA - Data Publisher for Earth & Environmental Science; 2017. 10.1594/PANGAEA.875576.
- 40.Guidi L, Ras J, Claustre H, Pesant S, Tara Oceans Consortium, Coordinators; Tara Oceans Expedition, Participants. Environmental context of all samples from the Tara Oceans Expedition (2009-2013), about pigment concentrations (HPLC) in the targeted environmental feature. PANGAEA - Data Publisher for Earth & Environmental Science; 2017. 10.1594/PANGAEA.875569.
- 41.Guidi L, Morin P, Coppola L, Tremblay J-É, Pesant S, Tara Oceans Consortium, Coordinators; Tara Oceans Expedition, Participants. Environmental context of all samples from the Tara Oceans Expedition (2009–2013), about nutrients in the targeted environmental feature. PANGAEA - Data Publisher for Earth & Environmental Science; 2017. 10.1594/PANGAEA.875575.
- 42.Pesant S, Tara Oceans Consortium, Coordinators; Tara Oceans Expedition, Participants. Methodology used on board to prepare samples from the Tara Oceans Expedition (2009–2013). PANGAEA - Data Publisher for Earth & Environmental Science; 2017. 10.1594/PANGAEA.875580.
- 43.Speich S, Chaffron S, Ardyna M, Pesant S, Tara Oceans Consortium, Coordinators; Tara Oceans Expedition, Participants. Environmental context of all samples from the Tara Oceans Expedition (2009–2013), about the water column features at the sampling location. PANGAEA - Data Publisher for Earth & Environmental Science; 2017. 10.1594/PANGAEA.875579.
- 44.Picheral M, Guidi L, Stemmann L, Karl DM, Iddaoud G, Gorsky G. The underwater vision profiler 5: an advanced instrument for high spatial resolution studies of particle size spectra and zooplankton. Limnol Oceanogr Methods. 2010;8:462–73. [Google Scholar]
- 45.Picheral M, Searson S, Taillandier V, Bricaud A, Boss E, Ras J, et al. Tara Oceans Consortium, Coordinators; Tara Oceans Expedition, Participants. Vertical profiles of environmental parameters measured on discrete water samples collected with Niskin bottles during the Tara Oceans expedition 2009–2013. PANGAEA - Data Publisher for Earth & Environmental Science; 2014. 10.1594/PANGAEA.836319.
- 46.Picheral M, Colin S & Irisson JO. EcoTaxa, a tool for the taxonomic classification of images. 2017. https://ecotaxa.obs-vlfr.fr.
- 47.Schlitzer R. Interactive analysis and visualization of geoscience data with Ocean Data View. Comput Geosci. 2002;28:1211–8. [Google Scholar]
- 48.Pesant S, Not F, Picheral M, Kandels-Lewis S, Le Bescot N, Gorsky G, et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci Data. 2015;2:150023. doi: 10.1038/sdata.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alberti A, Poulain J, Engelen S, Labadie K, Romac S, Ferrera I, et al. Viral to metazoan marine plankton nucleotide sequences from the Tara Oceans expedition. Sci Data. 2017;4:170093. doi: 10.1038/sdata.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Logares R, Sunagawa S, Salazar G, Cornejo-Castillo FM, Ferrera I, Sarmento H, et al. Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environ Microbiol. 2014;16:2659–71. doi: 10.1111/1462-2920.12250. [DOI] [PubMed] [Google Scholar]
- 51.de Vargas C, Audic S, Henry N, Decelle J, Mahe F, Logares R, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 52.Martino C, Morton JT, Marotz CA, Thompson LR, Tripathi A, Knight R, et al. A novel sparse compositional technique reveals microbial perturbations. mSystems. 2019;4:e00016–19. doi: 10.1128/mSystems.00016-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oksanen J. Vegan: ecological diversity. R project. 2018;368:1–11.
- 54.R Core Team. R a language and environment for statistical computing: reference index. Vienna: R Foundation for Statistical Computing; 2010.
- 55.Dinno A. dunn. test: Dunn’s test of multiple comparisons using rank sums. R package version, 2017;1:1.
- 56.Tackmann J, Matias Rodrigues JF, von Mering C. Rapid inference of direct interactions in large-scale ecological networks from heterogeneous microbial sequencing data. Cell Syst. 2019;9:286–96.e8. doi: 10.1016/j.cels.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 57.Clauset A, Newman MEJ, Moore C. Finding community structure in very large networks. Phys Rev E. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 58.Frémont P, Gehlen M, Vrac M, Leconte J, Delmont TO, Wincker P, et al. Restructuring of plankton genomic biogeography in the surface ocean under climate change. Nat Clim Chang. 2022;12:393–401. [Google Scholar]
- 59.Richter DJ, Watteaux R, Vannier T, Leconte J, Frémont P, Reygondeau G, et al. Genomic evidence for global ocean plankton biogeography shaped by large-scale current systems. eLife. 2022;11:e78129. doi: 10.7554/eLife.78129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brum JR, Ignacio-Espinoza JC, Roux S, Doulcier G, Acinas SG, Alberti A, et al. Patterns and ecological drivers of ocean viral communities. Science. 2015;348:1261498. doi: 10.1126/science.1261498. [DOI] [PubMed] [Google Scholar]
- 61.Ghiglione J-F, Galand PE, Pommier T, Pedrós-Alió C, Maas EW, Bakker K, et al. Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc Natl Acad Sci. 2012;109:17633–8. doi: 10.1073/pnas.1208160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De la Iglesia R, Echenique-Subiabre I, Rodríguez-Marconi S, Espinoza JP, von Dassow P, Ulloa O, et al. Distinct oxygen environments shape picoeukaryote assemblages thriving oxygen minimum zone waters off central Chile. J Plankton Res. 2020;42:514–29. [Google Scholar]
- 63.Schnetzer A, Moorthi SD, Countway PD, Gast RJ, Gilg IC, Caron DA. Depth matters: microbial eukaryote diversity and community structure in the eastern North Pacific revealed through environmental gene libraries. Deep Sea Res Part I. 2011;58:16–26. [Google Scholar]
- 64.Parada V, Sintes E, van Aken HM, Weinbauer MG, Herndl GJ. Viral abundance, decay, and diversity in the meso- and bathypelagic waters of the North Atlantic. Appl Environ Microbiol. 2007;73:4429–38. doi: 10.1128/AEM.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaneko H, Blanc-Mathieu R, Endo H, Chaffron S, Delmont TO, Gaia M, et al. Eukaryotic virus composition can predict the efficiency of carbon export in the global ocean. iScience. 2021;24:102002. doi: 10.1016/j.isci.2020.102002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bettarel Y, Motegi C, Weinbauer MG, Mari X. Colonization and release processes of viruses and prokaryotes on artificial marine macroaggregates. FEMS Microbiol Lett. 2016;363:fnv216. [DOI] [PubMed]
- 67.Durkin CA, Cetinić I, Estapa M, Ljubešić Z, Mucko M, Neeley A, et al. Tracing the path of carbon export in the ocean though DNA sequencing of individual sinking particles. ISME J. 2022;16:1896–906. doi: 10.1038/s41396-022-01239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bates AE, Helmuth B, Burrows MT, Duncan MI, Garrabou J, Guy-Haim T, et al. Biologists ignore ocean weather at their peril. Nature. 2018;560:299–301. doi: 10.1038/d41586-018-05869-5. [DOI] [PubMed] [Google Scholar]
- 69.Mojica KDA, Brussaard CPD. Factors affecting virus dynamics and microbial host–virus interactions in marine environments. FEMS Microbiol Ecol. 2014;89:495–515. doi: 10.1111/1574-6941.12343. [DOI] [PubMed] [Google Scholar]
- 70.Kukkaro P, Bamford DH. Virus–host interactions in environments with a wide range of ionic strengths. Environ Microbiol Rep. 2009;1:71–7. doi: 10.1111/j.1758-2229.2008.00007.x. [DOI] [PubMed] [Google Scholar]
- 71.Bettarel Y, Bouvier T, Bouvy M. Viral persistence in water as evaluated from a tropical/temperate cross-incubation. J Plankton Res. 2009;31:909–16. [Google Scholar]
- 72.Schulz F, Roux S, Paez-Espino D, Jungbluth S, Walsh DA, Denef VJ, et al. Giant virus diversity and host interactions through global metagenomics. Nature. 2020;578:432–6. doi: 10.1038/s41586-020-1957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thamdrup B, Dalsgaard T, Revsbech NP. Widespread functional anoxia in the oxygen minimum zone of the Eastern South Pacific. Deep Sea Res Part I. 2012;65:36–45. [Google Scholar]
- 74.Berry D, Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol. 2014;5:219. [DOI] [PMC free article] [PubMed]
- 75.Zhu W, Qin C, Ma H, Xi S, Zuo T, Pan W, et al. Response of protist community dynamics and co-occurrence patterns to the construction of artificial reefs: a case study in Daya Bay, China. Sci Total Environ. 2020;742:140575. doi: 10.1016/j.scitotenv.2020.140575. [DOI] [PubMed] [Google Scholar]
- 76.Chaffron S, Delage E, Budinich M, Vintache D, Henry N, Nef C, et al. Environmental vulnerability of the global ocean epipelagic plankton community interactome. Sci Adv. 2021;7:eabg1921. doi: 10.1126/sciadv.abg1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glock N, Roy A-S, Romero D, Wein T, Weissenbach J, Revsbech NP, et al. Metabolic preference of nitrate over oxygen as an electron acceptor in foraminifera from the Peruvian oxygen minimum zone. PNAS. 2019;116:2860–5. doi: 10.1073/pnas.1813887116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data analyzed during this study are included in this published article in the Supplementary Information file.