Abstract
The past decade has seen significant advances in our understanding of Hedgehog (HH) signaling pathway in various biological events. HH signaling pathway exerts its biological effects through a complex signaling cascade involved with primary cilium. HH signaling pathway has important functions in embryonic development and tissue homeostasis. It plays a central role in the regulation of the proliferation and differentiation of adult stem cells. Importantly, it has become increasingly clear that HH signaling pathway is associated with increased cancer prevalence, malignant progression, poor prognosis and even increased mortality. Understanding the integrative nature of HH signaling pathway has opened up the potential for new therapeutic targets for cancer. A variety of drugs have been developed, including small molecule inhibitors, natural compounds, and long non-coding RNA (LncRNA), some of which are approved for clinical use. This review outlines recent discoveries of HH signaling in tissue homeostasis and cancer and discusses how these advances are paving the way for the development of new biologically based therapies for cancer. Furthermore, we address status quo and limitations of targeted therapies of HH signaling pathway. Insights from this review will help readers understand the function of HH signaling in homeostasis and cancer, as well as opportunities and challenges of therapeutic targets for cancer.
Subject terms: Cell biology, Cancer
Introduction
HH signaling pathway was originally identified for its role in patterning the Drosophila embryo development.1 When mutated, the larvae of Drosophila were covered with short barbed spines like a hedgehog.2 As an evolutionarily conserved and highly organized signaling, HH signaling regulates numerous processes of vertebrates and invertebrates, such as embryogenesis, cellular development, epithelial-mesenchymal transition (EMT), and varieties of pathological variations.3,4 HH signaling pathway plays a vital role in the regulation of homeostasis in various tissues, such as stem cells in musculoskeletal tissue and digestive system.5 Moreover, Evidence-Based Medicine has shown that abnormal HH signaling is a considerable driver of tumorigenesis and malignancy.6 Understanding the function of HH signaling offers potential therapeutic interventions. Some targeted tumor therapies have entered clinical trials as the mechanism of HH signaling pathway is further explored.7 The current review aims to provide a comprehensive background on HH signaling pathway and highlight the recent advances of its role in homeostasis and tumorigenesis. It will also shed light on the available means and potential challenges of targeted therapies for HH signaling pathway-dependent cancer.
Hedgehog signaling
Introduction of Hedgehog signaling pathway
The initiation of Hedgehog signaling is based on the processing of Hedgehog ligands in secretory cells, where a dual lipid-modified amino-terminal (N-terminal) polypeptide of the autocatalytically cleaved precursor protein is formed.8 Signal reception is achieved by conserved receptors on the cell membrane, including the 12-pass transmembrane protein Patched (Ptch) and the 7-pass transmembrane protein Smoothened (Smo). In the absence of hedgehog ligand, Ptch inhibits the activity of Smo causing pathway inhibition. When the hedgehog ligand binds to Ptch, the inhibition of Smo is lifted, thus continuing to activate downstream signaling.9 In addition to this, the activation of hedgehog signaling requires co-receptors, which can be involved in the inactivation of Ptch.
In mammals, hedgehog signaling activation elicits a transcriptional response in the nucleus of the recipient cell that is mediated by glioma-associated oncogene (Gli) transcription factors.10 Gli proteins include an amino-terminal transcriptional repression domain, a zinc finger DNA binding domain and a carboxy-terminal transcriptional activation domain. When transcription is not activated, Gli is processed by protein hydrolysis into the form of a repressor with a truncated carboxy-terminal activation domain, which inhibits the transcription of the target genes.11 In addition, Suppressor of fused (Sufu), a negative regulator of Gli, is essential for pathway activity in mammals.12
A particular aspect of mammalian hedgehog signaling is the transport of signaling molecules at all levels in the primary cilium.13 The primary cilium consists of ciliary membrane, axoneme and basal body. The presence of intraflagellar transport (Ift) proteins on the axoneme mediates the transport of signaling proteins, and this transport mode can regulate the concentration of the corresponding proteins within the cilium.14 In addition, primary cilia form compartments that are separated from the rest of the cytoplasm of the cell and thus provide a unique intracellular environment for pathway components.15
Components of Hedgehog signaling pathway
In this part, we will briefly introduce the signaling process to understand the mechanism of HH signaling pathway in tissue homeostasis and tumorigenesis. A large body of researches have reported the core components of HH signaling pathway. This cascade includes extracellular Hedgehog ligands; cell surface receptors and co-receptors; Gli family transcription factors (Gli1-Gli3) containing zinc-lipid structures and numerous intracellular and extracellular co-regulators.16
Extracellular Hedgehog ligands
Hedgehog ligands are paracrine signaling factors that can mediate communication between cells up to hundreds of micrometers away.17 Three soluble extracellular Hedgehog ligands have been identified in vertebrates, including Sonic Hedgehog (Shh), Desert Hedgehog (Dhh), and Indian Hedgehog (Ihh), which have distinct distributions in different developmental stages and tissues.18,19
Shh
Shh-triggered signaling is well documented for its critical role during embryonic and postnatal developmental processes. For example, Shh forms a ventral and dorsal concentration gradient in the neural tube, thereby directing neural progenitor cell fate in time and space as well as acting physiologically as a morphogenetic agent.20,21 Moreover, Shh directs the growth of post commissural axons along the longitudinal axis of the spinal cord, acting as a major mediator during central nervous system development.22,23 Collectively, Shh plays a vital role in the developmental regulation of various tissues such as skull, adrenal cortex, hair follicles and sweat glands.24–27
Activation of Shh following synthesis on the endoplasmic reticulum of secretory cells involves lipidation by cholesterol molecules and palmitic acid moieties (Fig. 1).8 The secretion of activated Shh serves a crucial role in regard to its biological function. Shh was first cleaved into a 19 kDa Shh amino-terminal polypeptide (Shh-N) and Shh carboxy-terminal polypeptide (Shh-C). Then, Shh binds cholesterol to the carboxyl terminus of Shh-N through a protein hydrolytic cleavage process, whereas, Shh-C exits the endoplasmic reticulum and enters the proteasome to be degraded, ending its mission.28,29 In general, palmitoylation occurs after cholesterol modification, further increasing Shh activity.30 Palmitoylation of Shh is mediated by HH acyltransferase (Hhat), which transfers palmitic acid to the amino-terminal cysteine residue of Shh-N.31,32 The activated form of Shh is a dual lipidation of cholesterol and palmitic acid by Shh-N (Fig. 2a).20
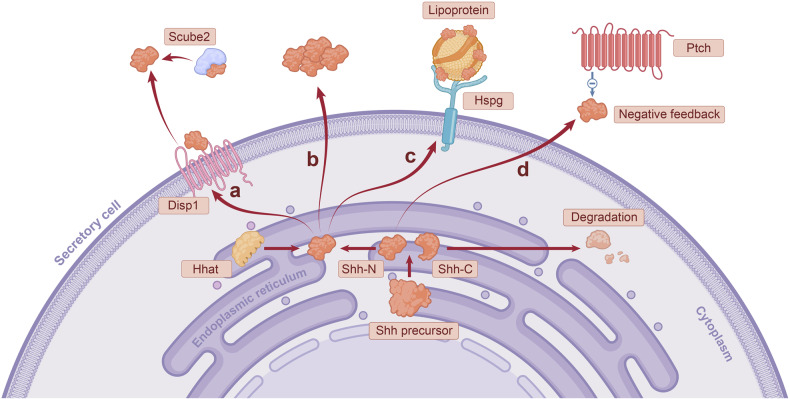
Fig. 1.
Activation and spread of hedgehog ligands in secretory cells. The three vertebrate hedgehog ligands Shh, Dhh and Ihh have similar secretion processes, differing slightly in the tissue distribution of the three. The figure shows the secretion of Shh as an example of the secretion process of hedgehog ligands. Activation of Shh occurs in the endoplasmic reticulum (ER). The Shh precursor is first processed into two parts, Shh-N and Shh-C. The carboxyl terminus of Shh-N is bound to cholesterol by a protein hydrolytic cleavage process, while the amino terminus of Shh-N is bound to palmitic acid mediated by Hhat. In contrast, Shh-C is degraded after being transported out of the ER. There are several mechanisms for the diffusion of Shh-N out of secretory cells after activation: a Shh-N is mainly transported out of secretory cells by the synergistic action of Disp1 and Scube2. b The polymerization of monomer-activated Shh-N into multimolecules facilitates the diffusion of ligands. c Hspg is localized to the secretory cell membrane, which recruits lipoprotein. Shh-N is loaded on the lipoprotein as a "passenger" for long-distance transportation. d Ptch1 on the surface of the receiving cells has a negative feedback regulation on the release of Shh-N
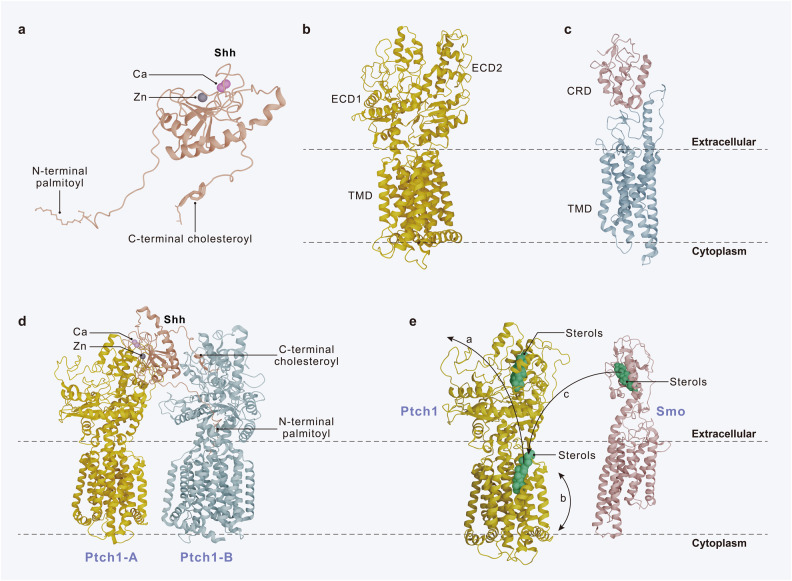
Fig. 2.
Three-dimensional structure of Shh, Ptch1 and Smo. a The structure of the activated Shh. b The structure of Ptch1. c The structure of Smo. d The structure of the 1Shh:2 Ptch1 complex. Ptch1-A binds to Shh at the interface of its calcium and zinc binding sites, which drives Ptch1 degradation. Ptch1-B binds to the N-terminal palmitoyl and C-terminal cholesterol modifications of Shh and anchors to the core of Ptch1 protein, which decreases the protein activity of Ptch1. e Mechanism of Smo inhibition by Ptch1. Ptch1 regulates the binding of sterols to Smo, and hence inhibits the activity of Smo. Three possible mechanisms are shown by the black arrows. a Sterols move from the outer leaflet of the membrane to ECD1 via Ptch1, which depletes the sterols inside the membrane. b Ptchl inhibits Smo activity by decreasing its accessibility to sterols from the inner leaflet of the membrane. c Ptch1 accepts sterols from CRD and transports them to the membrane, thereby inhibiting Smo activity
The mechanism by which the diffusion of activated Shh from secretory cells occurs is more complex (Fig. 1). Dispatched-1 (Disp1), a 12-transmembrane protein, is required for the secretion of activated Shh. Recent studies suggest that Disp1 binds cholesterol molecules of Shh-N and, together with Scube2, a secreted glycoprotein, transports Shh out of the cell.33,34 If the cholesterol and palmitic acid of Shh are broken down, Shh is also degraded.35 In addition, monomer-activated Shh-N can polymerize into multimolecules, a form that naturally facilitates the long-distance transport of ligands.36 Heparan sulfate proteoglycan (Hspg) is localized on the surface of secretory cell lipid membranes, where it interacts with Shh-N and recruits the lipoprotein lipophorin, resulting in the loading of Shh-N on lipoprotein for transport.37–39 In addition to the ability of secretory cells to positively regulate Shh secretion, Ptch1, which receives Shh receptors on the cell surface, is able to participate in the negative feedback regulation of ligands, limiting the range of Shh signal waves.40 In conclusion, Shh is able to assemble into suitable carriers for transport, and other possible mechanisms that help Shh diffusion deserve further investigation.
Dhh and Ihh
Dhh and Ihh act more specifically than Shh and show pronounced tissue specificity. Dhh is a key regulator of gonadal tissue development, especially in ovarian granulosa cells and spermatogenesis.41,42 Expressed by Sertoli cells, Dhh directly mediates the proliferation and differentiation of spermatogonia through the regulation of downstream signaling pathways. Dhh-deficient animals exhibit abnormal testicular mesenchymal cell differentiation and defective peritubular cell morphology, resulting in sterility in male mice.43 The function of Ihh is mostly related to chondrocyte differentiation in the skeleton.44 It regulates the differentiation of growth plate chondrocytes through a negative feedback mechanism of parathyroid hormone-related proteins, and also directly controls chondrocyte proliferation and osteoblast function.45
The activation and secretion of Dhh and Ihh are similar to that of Shh. Dhh and Ihh also undergo autocatalytic cleavage to form Dhh-N and Ihh-N, in which the C-terminal structural domain is removed.46 This N-terminal structural domain is then modified by N-terminal palmitoylation as well as the cholesterol addition of C-terminal amino acids, resulting in the formation of the mature proteins Dhh-N and Ihh-N.47 All three HH ligands share high sequence homology, similar overall fold and are capable of binding to and activating the same signaling pathway receptors. They mainly differ in their N-terminal amino acid sequence; however, their N-terminal sequence similarity may also reach 76–91%.48
Over the years, it has also become increasingly clear that Dhh and Ihh have multiple functions in other tissues. For instance, full-length Dhh plays a critical role in regulating vascular endothelial integrity.49 The potential roles of Dhh and Ihh in varies organs requires further investigation.
Receptors and co-receptors
Ptch
Ptch, the 12th transmembrane HH receptor, has two homologs, Ptch1 and Ptch2.50 The structure of Ptch1 includes the transmembrane structural domain (TMD), which comprises a sterol-sensing structural domain (SSD) and two extracellular structural domains (ECD1 and ECD2) (Fig. 2b).51 Various researchers have suggested that Ptch1 acts as a major receptor for HH ligands. Here, modifications of Shh-N may occur in its interaction with Ptch1.52 In addition, molecular evidence has shown that Ptch1 is able to recognize the calcium-mediated interface of Shh-N. Moreover, the binding mode of Shh and Ptch1 has been reported to be the formation of the 1 Shh:2 Ptch1 complex (Fig. 2d).53 However, in vivo studies have revealed that co-receptors can form a heterotrimeric complex with Ptch1 and HH-N and induce cell proliferation or embryonic development.29 Nonetheless, how HH recognizes and inhibits Ptch1 continues to remain controversial.
While the role of Ptch2 is inadequately understood, it compensates the response to Shh in the absence of Ptch1.54 Ptch2 may be able to bind HH proteins in different cellular environments in order to regulate the subsequent signaling pathway.55
Smo
In-depth structural and functional studies have revealed the role of the two variable ligand binding sites, seven-pass α-helical transmembrane bundle (TMD) and extracellular cysteine-rich domain (CRD) of Smo, a G protein-coupled receptor, in regulating Smo activity (Fig. 2c).56,57 The TMD and extracellular loops form a ligand-binding region.58 CRD at the extracellular amino terminus mediates Smo dimerization. In addition, intracellularly, the carboxy-terminal tail mediates the conformational change of the Smo dimer. In the absence of ligand, the carboxyl terminus is in a closed conformation, obscuring the kinase binding site; in the presence of ligand, the carboxyl terminus ser/thr cluster is highly phosphorylated, forming an open conformation.59–61
In the absence of ligand stimulation, Ptch inhibits Smo to block the cascade reaction. The question of how Ptch inhibits Smo remains relatively mysterious. Most scholars believe that Ptch can modulate the binding of lipid Smo ligands to Smo, which can inhibit Smo activity. These endogenous inhibitory molecules usually refer to sterols or their derivatives (Fig. 2e).20,62,63 The inactive Smo is located in the cytoplasm. However, when HH binds to Ptch, the restrictive effect of Ptch on Smo is lifted to initiate downstream signaling pathways.64,65 Ptch is transported to lysosomes in the cytoplasm for degradation. Smo activation requires two mechanisms: first, Smo needs to be translocated to primary cilia, a microtubule-based organelle (Fig. 3); second, Smo localized to cilia undergoes carboxy-terminal phosphorylation.66,67 A recent study found that cholesterol covalently bound to the D95 residue of the CRD of Smo is required for Smo activation, a process that results from calcium-promoted autocholesterolization of Smo.68,69
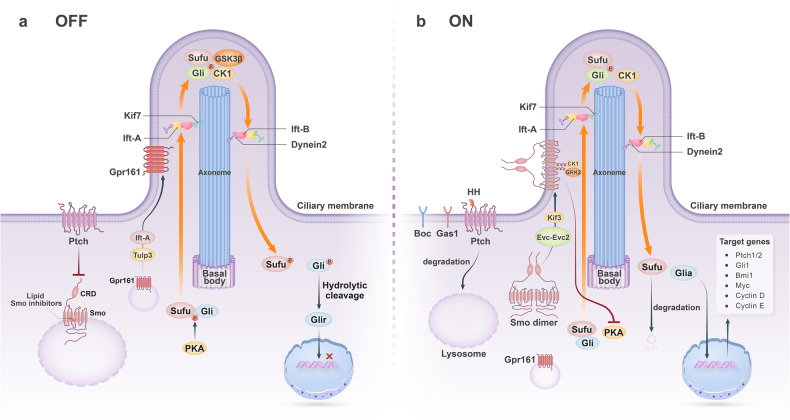
Fig. 3.
Primary cilia play a key role in HH signaling pathway. Primary cilia are composed of ciliary membrane, axoneme and basal body. The proper functioning of HH signaling relies on the Ift mechanism. Ift-B complex protein and Ift-A complex protein together constitute Ift trains that carry motors with HH signaling components sliding on the axoneme. Kinesin mediates transport from the base to the tip of the cilium, while dynein2 mediates transport from the tip to the base of the cilium. a Receptor cells are closed to signal transduction. In the absence of hedgehog ligand, Ptch1 inhibits Smo. Lipid Smo inhibitors bind to the TMD region of Smo, inhibit Smo activity, and constrain Smo to the cytoplasm. Gpr161 localizes to the cilia in the presence of Ift-A and Tulp3. Gpr161 induces PKA activity, and PKA phosphorylates the Sufu/Gli complex. Kinesin Kif7 then transports the complex to the cilia tip. Sufu is further phosphorylated by GSK3β; Gli is further phosphorylated by CK1 and GSK3β. The complex then dissociates and Gli is processed by protein hydrolytic cleavage to Glir, which subsequently enters the nucleus to repress target gene transcription. b Receptor cells are open to signal transduction. In the presence of Hedgehog ligand, the inhibition of Smo by Ptch is released and Ptch is transported to the lysosome for degradation. The co-receptors Boc and Gas1 can interact with Ptch1 and Ptch2 to form a receptor complex respectively, thereby promoting or inhibiting signal transduction. The concentration of intracellular lipid Smo inhibitor ligands decreases and Smo forms an activated dimeric form that is transported by Kinesin Kif3 and Evc-Evc2 proteins to the cilia membrane near the basolateral. The Smo carboxyl terminus is then fully activated by phosphorylation of CK1 and GRK2. After Smo activation, Gpr161 returns to the cytoplasm. Meanwhile, activated Smo inhibits PKA activity and is not sufficient to phosphorylate the Sufu/Gli complex. The unphosphorylated complex is transported to the cilia tip by Kinesin Kif7. The unphosphorylated Sufu is degraded by ubiquitination; Gli is phosphorylated by CK1 to form Glia. Glia enters the nucleus to promote the transcription of related target genes, such as Ptch1/2, which acts as a negative regulator of signal transduction, Gli1, which amplifies signals, Bmi1, which encodes a transcriptional repressor, and Myc, Cyclin D, and Cyclin E, which encode cell cycle regulators
Co-receptors
Brother of Cam-related/downregulated by oncogenes (Boc) and growth arrest-specific 1 (Gas1) have been identified as co-receptors regulating HH signaling pathway activity. The specific heterogeneous complexes formed by their interaction with Ptch1/2 receptors, respectively, can mediate different kinetic Smo depression programs through distinct ligand reception patterns.70,71 Notably, Boc sometimes acts as an inhibitor of HH signaling pathway during craniofacial tissues development.71 In addition, Hspg can act as co-receptors for neuronal cell development through glypican 5 core protein and 2-O-sulfo-iduronic acid structures.72
Gli family transcription factors
Upon excitation of HH signaling pathway, Gli transcription factors are activated thereby regulating target gene expression. The vertebrate homologs of this Drosophila Cubitus interruptus transcription factors include Gli1, Gli2, and Gli3 proteins. Gli1 is exclusively a transcriptional activator, whereas Gli2 and Gli3 have dual activity. Although both contain highly similar zinc finger structural domains that can bind to DNA, Gli3 often acts as a transcriptional repressor because the process of protein hydrolysis is most pronounced at Gli3, while Gli2 acts primarily as a transcriptional activator.73,74 Through protein hydrolysis, they act as bifunctional transcription factors, changing from the full-length transcriptional activator form (Glia) to the carboxy-terminal truncated transcriptional repressor form (Glir).11 GLI-similar (Glis) transcription factors, a subfamily highly similar to Glis, can also undergo nuclear translocation by post-translational modifications in the cytoplasm, and their potential interconnection with Gli1-3 enriches the Gli network.75–77
Gli protein activity can be regulated by protein-protein interactions. Sufu, a negative regulator of Gli1-3, was shown to abduct Gli1-3 in the cytoplasm to prevent it from undergoing nuclear translocation.12,78,79 In cells, Sufu is phosphorylated by Protein kinase A (PKA) to form a stable phosphorylated form of Sufu, which affects its interaction with Gli.80 Sufu and Gli form a complex in the cytoplasm after entering the primary cilia (Fig. 3), resulting in different phosphorylation states of the two proteins and determining Glia or Glir production, depending on the presence or absence of hedgehog ligand stimulation.81 In the absence of ligand stimulation, the Sufu/Gli complex is in a closed conformation; after ligand stimulation, Sufu assumes an open conformation in the complex. The different phosphorylation states of both proteins cause conformational changes that lead to dissociation of the Sufu/Gli complex and entry of Gli into the nucleus to regulate target gene expression.82,83
Hedgehog signaling pathway target genes
HH signaling pathway plays an essential role in cell proliferation and differentiation, tissue development, homeostasis and EMT.3,16 Its complex regulatory network influences the transcription of divergent target genes that vary between tissues and cell types (Table 1). In most cells responsive to HH signaling, the common target genes include Ptch1/2 and Gli1.84 Moreover, increased expression of the Ptch1/2 and Gli1 genes have been shown indicate an activated HH signaling pathway and could provide negative (Ptch1) and positive (Gli1) regulation of HH signaling with feedback loop mechanisms. Furthermore, in tissue-specific stem cells, HH signaling induces Bmi1, which acts as a transcriptional repressor that regulates the proliferative capacity of cells.85 Numerous target genes in this pathway are cell cycle regulators, such as Myc, CyclinD, and CyclinE, as well as development-related regulators, such as Fgf4 and Hhip.86–89 In the developing neural tube, this pathway regulates the Nkx6.1, Olig2, Nkx2.2, and FoxA2 genes, conferring ventral neural fates and has Gli-binding sites in the regulatory regions.90 In addition, HH signaling pathway promotes the expression of certain genes, including Snzi1, Zeb1, Twist2 and Foxc2, which enhance epithelial mesenchymal transition.91 Other target genes include the apoptosis regulator Bcl2, Vegfa, Abcg2, Mycn, Foxm1, Pax6, Pax7, Pax9 and Jag1, as well as members of the Wnt signaling pathway.92–98 In summary, HH signaling pathway can regulate the corresponding target gene transcription in different functional states and cell types.
Table 1.
Hedgehog signaling pathway target genes
| Target gene | Full name of the gene | Function of the gene products |
|---|---|---|
| Ptch184 | Patched 1 | Receptor that initiates signaling pathways, negative feedback regulation |
| Ptch284 | Patched 2 | Receptor that initiates signaling pathways |
| Gli184 | Glioma-associated oncogene 1 | Direct transcriptional activator, positive feedback regulation |
| Bmi185 | Bmi1 proto-oncogene | Transcriptional repressor regulating the proliferative capacity of cells |
| Myc86 | Myc proto-oncogene | Cell cycle progression, apoptosis and cellular transformation |
| CycD and CycE87 | CyclinD and CyclinE | Cell cycle progression |
| Fgf489 | Fibroblast growth factor 4 | Involved in varieties of biological processes including embryonic development, cell growth, morphogenesis, tissue repair and invasion |
| Hhip88 | Hedgehog interacting protein | Involved in varieties of developmental processes including anteroposterior patterns of limbs and regulation of left-right asymmetry in embryonic development |
| Nkx6.190 | NK6 homeobox 1 | Required for the development of beta cells |
| Nkx2.290 | NK2 homeobox 2 | Involved in the morphogenesis of the central nervous system |
| Olig290 | Oligodendrocyte transcription factor 2 | Regulator of ventral neuroectodermal progenitor cell fate |
| FoxA290 | Forkhead box A2 | Transcriptional activators that regulate the specificity and differentiation of dopaminergic neurons |
| Snai191 | Snail family transcriptional repressor 1 | Critical for mesoderm formation in the developing embryo |
| Zeb191 | Zinc finger E-box binding homeobox 1 | Associated with posterior polymorphous corneal dystrophy-3 and late-onset Fuchs endothelial corneal dystrophy |
| Twist291 | Twist family bHLH transcription factor 2 | Inhibitors of osteoblast maturation, maintaining the pre-osteoblast phenotype of cells during osteoblast development |
| Foxc291 | Forkhead box C2 | Involved in the development of mesenchymal tissues |
| Pax6, Pax7 and Pax997 | Paired box 6, 7 and 9 | Involved in the development of neural tissues |
Hedgehog signaling and the primary cilium
Unlike other signaling pathways, the normal maintenance of the vertebrate canonical HH signaling pathway is highly dependent on primary cilia. Primary cilia recruit extracellular signaling components that link the extracellular environment, where signals are complex and diverse, to the intracellular environment.13,99 This section will briefly describe the structural features of primary cilia and the way in which the key components of HH signaling pathway act in primary cilia.
Brief introduction of primary cilia
The primary cilium is a specialized structure located on the cell surface.100 The axoneme of primary cilia includes 9 microtubule doublets in the outer ring, while lacking the central 2 microtubule singlets (known as 9 + 0 axoneme). Primary cilia are anchored to the cell by basal body, which is specialized from the mother centriole and contains a number of proteins for ciliogenesis and assembly.101,102 Between the cilia and basal body is the ciliary transition zone, which regulates protein transport within the cytoplasm and cilia.103
The transport of protein molecules within primary cilia to the cytoplasm is achieved through the Ift mechanism.14 Ift is driven by two molecular motors, involving cytoplasmic dynein2, which mediates transport from tip to base, and the heterotrimeric kinesin-2 complex comprised of Kif3a, Kif3b, and Kap3, which is responsible for transport from base to tip.15,104,105 Mutations in the gene encoding the heavy chain of cytoplasmic dynein2 cause axoneme swelling because Ift-related proteins are trapped in cilia due to disruption of retrograde transport.14 Gli cooperates with Kif3a and Kap3, where the interaction of Kap3 restricts Gli transcriptional activity.106 Ift-B complex protein and Ift-A complex protein together constitute Ift trains that bridge kinesin motors with HH signaling molecules.107 Ift trains slide back and forth along the axoneme and are responsible for cilia structure assembly and balancing. Periodic binding of Ift-A, Ift-B and dynein2 can balance the protein entry and exit of cilia, effectively preventing the accumulation of material.108,109 Huang et al. showed that wimple mutants disrupting Ift172, an Ift-B protein, lacked the normal primary cilia structure, which resulted in the inability of the receiving cells to respond to Shh.110
Changes in the structure and function of primary ciliary protein fractions can have an impact on HH signaling pathway. Defective RNAi of Kinesin Kif14 disrupts the function of distal appendage proteins Sclt1 and Fbf1, which results in a defective basal body that fails to activate HH signaling pathway.111 Disruption of most primary ciliary proteins decreases signaling activity, but some proteins are altered in different ways that enhance pathway activity; thus, changes in ciliary structure have a bidirectional effect on the HH signaling cascade response, with subsequent effects on cell fate.15,112
Hedgehog signaling functions through primary cilia
Smo and primary cilia
The ciliary membrane contains a specific receptor for HH signaling pathway, Ptch, which is essential for receiving the initial signal from the outside.113,114 In the absence of Hedgehog ligand, Ptch induces a closed conformation of Smo by controlling the TMD binding of small lipid Smo ligands to Smo, allowing Smo to remain in the cytoplasm (Fig. 3a).63,115 In the presence of Hedgehog ligand, Ptch derepresses Smo and Smo activates. Activation of Smo involves its localization to the ciliary membrane and carboxy-terminal phosphorylation.59,116–118 First, the concentration of small lipid Smo inhibitors in the cytoplasm decreases and Smo transforms into an open conformation and is transported to the ciliary membrane by the action of the kinesin Kif3.119,120 Besides, after reaching the cilia, the activation of Smo depends on the concentration of Hedgehog ligands. When there is no sufficient ligand to stimulate the signal, the small lipid Smo inhibitor rapidly binds to the TMD again and the Smo changes back to a closed conformation and returns to the cytoplasm.121 When stimulated by sufficient Hedgehog ligands, the Smo carboxyl terminus is fully activated by casein kinase 1 (CK1) and G protein-coupled receptor kinase 2 (GRK2) phosphorylation (Fig. 3b).60,61 It can be seen that Smo acts in a Gαi - coupled Gpcr manner, which is highly sensitive to changes in Hedgehog ligand concentration.
Gli and primary cilia
In the absence of Hedgehog ligands, Gpr161, a G protein-coupled receptor, enters the cilia and localizes to the ciliary membrane via Ift-A and Tulp3, a member protein of the vertebrate tubby-like family.122,123 Then, in the presence of adenylate cyclase 5/6 (Ac5/Ac6), cAMP increases and activates PKA, which phosphorylates both proteins of the Sufu/Gli complex.124,125 Gli is further phosphorylated by CK1 and Glycogen Synthase Kinase-3 (GSK3β) and forms Glir by proteolytic cleavage (Fig. 3a).123,126,127
In the presence of Hedgehog ligands, Kinesin Kif7 dephosphorylates and then aggregates at the tip of primary cilia, in parallel with the return of Gpr161 to the cytoplasm.128,129 Gαi inhibits Ac5/Ac6 and reduces PKA activity after intraciliogenic activation.130–132 In addition, Evc-Evc2 proteins, which localize active Smo at the base of the cilia, also inhibit PKA activity.133 The Sufu/Gli complex, which is not phosphorylated by PKA, is transported to the cilia tip by Kinesin Kif7 and accumulates there.134 Within the cilia, unphosphorylated Sufu is degraded by Scf-mediated ubiquitination; however, Gli is phosphorylated by CK1 to form Glia.135 Then the complex dissociates and Glia enters the nucleus to mediate the transcription of target genes (Fig. 3b).117,121
Because CK1 and GSK3β, which are required for the phosphorylation process of the Sufu/Gli complex, are present in primary cilia, primary cilia are necessary to determine the form of Gli.136 Furthermore, the cilium is a local compartment relative to the cytoplasm, therefore small changes in PKA activity can be detected in the little space, which determines changes in Glia versus Glir.125,137
Hedgehog signaling in homeostasis
Well known for its crucial role in regulating the embryonic and postnatal development of vertebrate, HH signaling pathway has been proven to orchestrate the specific formation and morphogenesis of an organism.5,99,138,139 Meanwhile, HH signaling pathway has been demonstrated to express continually in many adult mammalian tissues and organs, especially in the epithelial and mesenchymal cells, maintaining the homeostasis of adult tissues and organs via regulating a great variety of quiescent stem cell populations as well as the epithelial-mesenchymal interactions.99,140,141 This part of review outlines recent discoveries on the crucial roles of HH signaling pathway in the homeostasis of multiple tissues and organs possessing representative characteristics.
HH signaling pathway maintains the homeostasis in the mesenchyme
HH signaling pathway is one of the major signaling pathways regulating osteogenesis and post-embryonic long bone homeostasis with postnatal expression in the mesenchyme.142,143 Yang et al. found that silencing Ift80 impairs cilia formation and reduces HH signaling pathway expression in bone.144 Ift80 increases the expression of osteoblast markers by regulating HH/Gli pathway, demonstrating that HH signaling pathway plays a remarkable role in maintaining osteoblast differentiation and mineralization. Ihh upregulates mesenchymal stem cells (MSCs) and osteoblast markers. Tissue engineering experiments utilizing Ihh-MSCs-scaffold complex has shown increased bone repair capacity.145 Shh is a potential signaling molecule that regulates osteoblast differentiation. Armstrong et al. have shown that Shh in zebrafish can upregulate the expression of Sp7 in osteoblast lineage cells and increase the proliferation of osteoblasts.146
The calvarial bone has become an emerging model for studying the function of HH signaling pathway in bone homeostasis. Regarded as the primary niches for osteogenesis, craniofacial sutures contain MSCs.140,147,148 Zhao et al. revealed that cells harboring MSCs characteristics were Gli1+. These MSCs play a significant role in regulating calvarial bone formation and injury repair.147 Using diphtheria toxin to disentangle Gli1+ cells led to premature fusion within the cranial sutures, resulting in craniosynostosis, indicating the vital role of HH signaling pathway in controlling the suture homeostasis and calvarial bone patterning and repair.147,149 Moreover, the interplay between BMP and Ihh signaling has been identified in maintaining suture homeostasis via interaction among MSCs, osteoprogenitors, and osteoclasts during calvarial bone formation and repair.150
Skeletal muscle has the ability to repair damage due to the presence of muscle stem cells, namely, myosatellite cells.151 Myosatellite cells normally maintain a quiescent condition and give rise to myogenic cells and reform the myofibers of the muscle in response to injury.152 Study showed that muscle injury could stimulate Shh expression. When Shh signaling was inhibited, the expression of Myf5 and MyoD was impaired, accompanied by reduced number of activated myosatocytes.153 Similarly, Koleva et al. showed that Shh could promote the proliferation of myosatellite cells and myotube fusion.154 Subsequently, Elia et al. found that Shh can promote the differentiation of chicken primary myoblasts, and this effect can be inhibited by cyclopamine, an inhibitor of Shh signaling.155 It has also been reported that Shh promotes the expression of Pax7, Myf5, MyoD, MyoG, and MyHC in mouse myoblasts, and the promotion of these genes by Igf-1 depends on HH signaling pathway.156,157 Further study by Voronova et al. showed that Gli2 formed a protein complex with MyoD and Mef2c to enhance the transcription factor activity of MyoD.158 MyoD regulates the activity of HH signaling pathway, which allows Gli2, Mef2c, and MyoD to form a reciprocally regulated recycling network. Devakanmalai et al. reported that HH signal affects muscle cell differentiation by activating MyoD and Myf5, and then regulating Mef2c. These studies indicate that HH signaling pathway is essential for postnatal muscle homeostasis.159
HH signaling pathway may also participate in smooth muscle regeneration and differentiation. Kramann et al. found that adventitial MSC‐like Gli1+ cells were progenitors of vascular smooth muscle cell and expressed CD34, Sca1, and Pdgfrβ, possessing tri-lineage differentiation capacity towards osteoblasts, adipocytes, and chondrocytes.160
HH signaling pathway maintains the homeostasis in epithelial tissue
During the hair cycle, Shh signaling pathway is essential for maintaining hair follicle stem cell populations and regulating the development of hair follicles and sebaceous glands, meanwhile regulating epithelial-mesenchymal interactions.161,162 Mammalian epidermal metabolism is maintained by the continuous proliferation of epidermal stem cells.163 Adolphe C et al. found that Ihh overexpression did not result in any significant epidermal morphogenetic phenotype, while overexpression of Dhh was indistinguishable from Shh. The phenotypes were resulted from dysregulation of stem cell activity, including hyperplasia of epidermal progenitors and almost complete loss of epidermal tissue renewal capacity, indicating that HH activity is the vital factor to maintain homeostasis of epidermal stem cells.164 Zhou et al. showed that Shh, its receptors Ptch1, Smo and its downstream transcription factor Gli1 were detected in the basal layer of fetal epidermis and in newly sorted human putative epidermal stem cells (HPESCs).165 HPESCs treated with medium containing Shh-N exhibited enhanced cell proliferation. In contrast, cyclopamine inhibits Shh and thus prevents proliferation. Similarly, the mitogenic effect of epidermal growth factor on HPESCs can be eliminated by cyclopamine. In addition, the expression of Bmp-4, a potential downstream effector of Shh signaling, can increase HPESCs proliferation.165 Shh signaling in mammalian skin controls hair follicle epithelial cell growth and morphogenesis by regulating Gli transcription factors.166 Shh expression and the ability of skin cells to respond to Shh signaling pathways are spatially and temporally regulated during the hair cycle.167 Knockout of Shh-dependent Sox9 in the skin results in the appearance of external hair, with severe proliferation defects and difficulty in forming a stem cell microenvironment.168 Additional studies have shown that the demand for substrate formation by β-catenin occurs downstream of Tabby/Downless and upstream of Bmp and Shh.169
Intestinal epithelial homeostasis is regulated by a strictly controlled balance between intestinal stem cell (ISC) proliferation and differentiation.170 Villin-Cre;Ihhflox/flox mutant mice show a significantly reduced number of villi and reduced cell proliferation in the stem cell compartment, suggesting that Ihh is essential for ISC regeneration and differentiation.171 Specifically, deletion of Ihh in intestinal epithelial is accompanied by an increase in epithelial Wnt signaling pathway, while the activation of the Wnt pathway is a common cause of intestinal tumorigenesis.172,173 HH might have a tolerogenic influence as a regulator of inflammation.174 Lees et al. showed the reduced expression of HH signaling pathway in individuals with inflammatory bowel disease and ulcerative colitis.175 Dop et al. conditionally deleted Ihh in adult mice and found an inflammatory response accompanied crypt changes, resulting in influx of macrophages and fibroblasts into the villus core.176 Recent studies suggest that more than one target cell population (e.g., macrophages, dendritic cells) is relevant to HH-modulated inflammation.177,178
HH signaling pathway maintains the homeostasis via regulating epithelial-mesenchymal interaction
Epithelial-mesenchymal interactions coordinated by HH actively maintain homeostasis and regulates repair and regeneration in lung.179 Tien et al. found proliferative expansion of the adjacent lung mesenchyme when specifically deleting Shh in the murine lung.180 Reduced expression of HH signaling pathway is initially detected during the acute phase of epithelial injury as the mesenchyme proliferates in response, whereas being upregulated to the baseline while the homeostasis is restored. However, Liu et al. demonstrated the upregulated HH signaling pathway in lung fibrosis or airway injury, supported by a proliferation in stromal Gli1+ cells in adult mice.181 Moreover, when confronted with an airway injury, Shh overexpression enhances collagen deposition and lung fibrosis.
Both epithelial and mesenchymal compartments of the rodent incisors continue to grow and regenerate throughout the lifespan of animals and requires constant repair. It has been identified that during the growth of the mouse incisor, Shh plays a significant role in maintaining the ability of stem cells to expand the ameloblast lineage in the incisor epithelium, indicating that Shh regulates the homeostasis of mouse incisor enamel, the epithelial compartment of incisors.182 Meanwhile, Shh signaling pathway also plays an important role in the homeostasis and regeneration of dentin, the mesenchymal compartment of incisors. Zhao et al. identified that Gli1+ MSCs were localized around arteries and the accompanying nerves, while the HH ligand that maintains the Gli1+ MSC population was secreted by nerves in the neighboring neurovascular bundle (NVB).183 In addition, Gli1+ cells were also indicated to contribute to incisor injury repair by generating reparative dentin.
Unlike the rodent incisor, the rodent molar does not possess the property of continuous growth, and the majority of Gli1+ cells were distributed within the mesenchyme, mainly in the periodontal ligament near the molar apical. These cells were activated and significantly contributed to the bone remodeling when imposed on the orthodontic forces.184
Hedgehog signaling in cancer
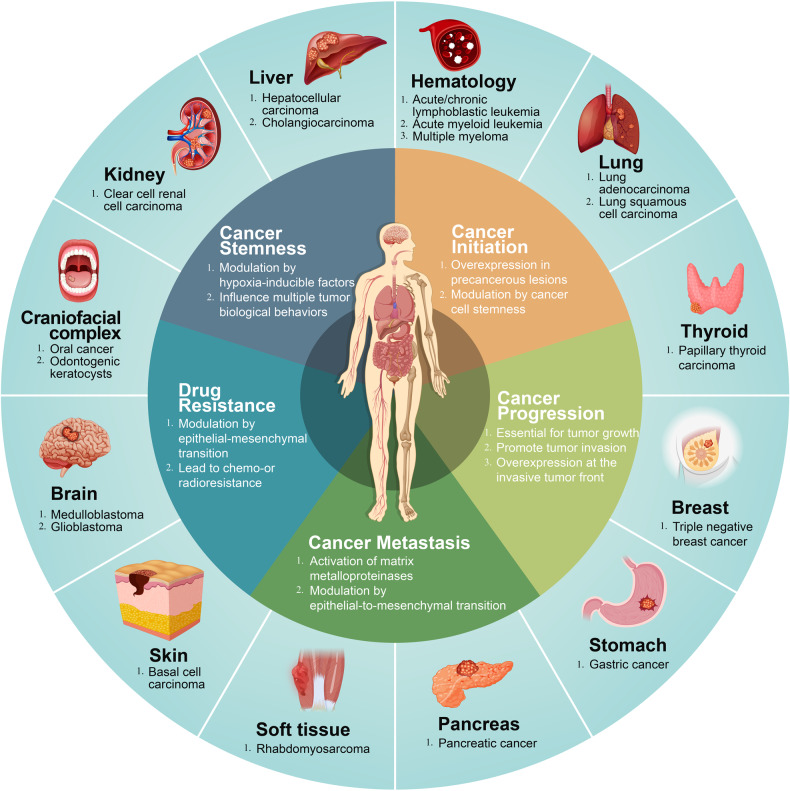
Cancer is one of the major causes of death in the world. Almost ten million cancer deaths were expected in 2020.185 Cancers are highly burdensome diseases, which have negative physical and mental effects on patients, as well as serious economic consequences. HH signaling pathway has been documented to be responsible for tumor initiation and premalignant lesion as well as tumor progression leading to a greater tumor size and more invasive behavior.186–188 Tumor metastasis and the resistance to anti-cancer therapy (radio and chemo-resistance) were the ultimate challenges to fight cancer as a life-threatening disease.189,190 Also, cancer stem cell (CSC) with the self-renewal ability not only has a close relationship with multiple tumor properties mentioned above such as tumor initiation, development, metastasis, and tumor recurrence.191 Therefore, we mainly discussed the decisive role of HH signaling pathway in five aspects including cancer initiation, cancer progression, metastasis, resistance to anti-cancer therapy, and cancer stemness in different organs in this section (Fig. 4).
Fig. 4.
Summary of types of cancer caused by dysregulation of HH signaling pathway. The inner circle present five aspects of tumor biological behavior which are influenced by HH signaling pathway. The outer circle shows the representative cancers discussed in the review. The online resources in the picture were obtained from the website: www.699pic.com and www.vecteezy.com
Cancer cells in conjunction with the surrounding oncogenic stromal components form the tumor. Microvascular components, fibroblasts, endothelial cells and other noncellular elements in the tumor stroma determine the multiple behaviors of tumors, such as tumor initiation, progression, and resistance to anticancer therapy.192 The interplay of HH signaling pathway between cancer cells and the stroma component may be overall described using the following models. Mutations of HH signaling pathway components, such as Ptch and Smo, drive ligand-independent HH signaling pathway activation, which can lead to systemic disease and manifest with a variety of symptoms, such as Gorlin syndrome. Cancer cells have also been shown to secrete HH ligands in an autocrine manner in cancer progression.193,194 In addition, ligand-dependent activation of the HH pathway in tumors have been shown to adopt a paracrine or a reverse-paracrine manner.195,196
A plethora of studies have supported the presence of a dynamic interaction via mutual transmission of HH signaling molecules between tumor cells and stromal cells. Coexistence of such HH ligand interactions in different types of tumors has also been shown to be conclusive. The association of HH signaling pathways with different cancers, including changes in related molecules, secretory mechanisms, and tumor characteristics were summarized (Table 2).
Table 2.
Changes of HH signaling pathway in cancer
| Cancer | Mechanism | HH signaling pathway-related molecules | Biological behavior influenced by HH signaling pathway |
|---|---|---|---|
| Craniofacial Complex | |||
| Oral squamous cell carcinoma198,199 | ligand-dependent | Ptch1 (+), Shh (+), Smo (+), Gli1 (+) | Initiation (+), Progression (+), Metastasis (+) |
| Odontogenic karatocytes211 | ligand-independent | Ptch1 (+), Smo (+) | Progression (+) |
| Brain | |||
| Medulloblastoma226,230,232 | ligand-independent | Ptch1 (+), Shh (-) | Stemness (+), Progression (+), Resistance (+) |
| Glioma237,510 | ligand-dependent | Shh (+), Gli2 (-) | Progression (+), Metastasis (+) |
| Renal | |||
| Renal cell carcinoma249,254,255,511 | ligand-dependent | Shh (+), Smo (+), Gli1 (+), Gli2 (+) | Stemness (+), Progression (+), Resistance (+) |
| Prostate | |||
| Prostate cancer259,263,512 | ligand-dependent | Shh (+), Smo (+) | Stemness (+), Progression (+), Resistance (+) |
| Liver | |||
| Hepatocellular carcinoma266,267,271,274 | ligand-dependent | Ptch1 (+), Smo (+), Gli1 (+) | Progression (+), Metastasis (+), Resistance (+) |
| Cholangiocarcinoma276,282,513 | ligand-dependent | Gli1 (+), Gli2 (+) | Stemness (+), Progression (+), Metastasis (+), Drug resistance (+) |
| Colon | |||
| Colorectal cancer284,287 | ligand-dependent | Ihh (-), Shh (+), | Progression (+), Metastasis (+), Resistance (+) |
| Pancreas | |||
| Pancreatic cancer289–291,294 | ligand-dependent | Shh (+), Smo (+), Gli1 (+) | Initiation (+), Progression (+), Metastasis (+) |
| Lung | |||
| Lung cancer299,304,305,514 | ligand-dependent | Shh (+), Gli (+) | Progression (+), Stemness (+), Metastasis (+), Resistance (+) |
| Blood | |||
| Hematological malignancy308,311,515 | ligand-dependent | Smo (+), Dhh (+), Gli1 (+) | Radiation resistance (+), Stemness (+) |
| Thyroid | |||
| Thyroid cancers320,324,516 | ligand-dependent | Shh (+), Ptch (+), Smo (+), Gli (+) | Stemness (+), Progression (+), Resistance (+) |
| Skin | |||
| Basel cell carcinoma327,329,335 | ligand-independent | Shh (+), Ptch1 (+), Gli1 (+) | Progression (+), Resistance (+) |
| Breast | |||
| Triple negative breast cancer341,342,345,346 | ligand-dependent | Smo (+), Gli1 (+), Shh (+) | Progression (+), Metastasis (+), Resistance (+), Stemness (+) |
| Muscle | |||
| Rhabdomyosarcoma351,357 | ligand-dependent | Gli1(+), Ptch1 (+) | Progression (+), Stemness (+), Drug resistance (+) |
| Stomach | |||
| Gastric cancer364–367 | ligand-independent | Shh (+), Gli1(+) | Initiation (+), Progression (+), Drug resistance (+), Stemness (+) |
Cancers in craniofacial complex
Squamous cell carcinomas account for more than 90% of oral malignant tumors and often begin with precancerous lesions.197 Shh expressed in oral epithelial dysplasia and carcinoma in situ.198 Also, Shh, Gli2, Smo, and Ptch are highly expressed in precancerous lesions of oral mucosa.199 Higher expression levels of Ptch1, Smo, and Gli1 were found in oral squamous cell carcinoma (OSCC) cases when compared to nonneoplastic oral mucosa. Recent study demonstrated that Gli1 was present only in the nuclei of cancer-associated fibroblasts in OSCC, indicating that HH signaling is active in the progression of OSCC.200 Furthermore, endophytic-type parenchyma of OSCC possessed a stronger expression of Shh and Ptch1 than exophytic-type parenchyma of OSCC.201
HH signaling molecules are considered to be prognostic indicators in OSCC as overexpression of related proteins can be indicative to tumor size, metastatic potential, tumor recurrence and even to shorter overall survival.202,203 HH signaling regulates cell proliferation in OSCC.204 Several studies have investigated the molecular mechanisms on the invasion behavior of human OSCC. Ptch1 and Gli expression were found in the microvascular cells in the invasive front of OSCC.205 Takabatake et al. reported that Ptch and CD31 double-positive blood vessels in the OSCC stroma might affect tumor angiogenesis in OSCC.201 Also, Shh overexpression is correlated with cancer stem cell markers CD133 and Sox2 in OSCC specimens.202 Shh might induce OSCC progression and invasion through prolonged half-life of activated leukocyte cell adhesion molecule (ALCAM) which is a transmembrane glycoprotein mediating cell adhesion and multiple other function of cancer cell.206 An increased MMP-9 and downregulated epithelial cadherin (E-cadherin) expression caused by Shh signaling in OSCC might be responsible for the invasiveness and metastatic potential.207
Odontogenic keratocysts (OKCs) accounts for 3.3–17.4% of all jaw cysts, of which neoplastic lesions often involve mandibles with a high clinical risk of recurrence.208,209 Gorlin syndrome (GS) is a multisystem genetic disease caused by gain of function mutation in HH signaling pathway and manifests as multiple OKCs and basal cell carcinomas (BCCs).209,210 The most common gene mutation for GS is Ptch1 while Ptch2, Smo and Sufu are also rare causative genes for GS.211 Although multiple OKCs is a major symptom of GS, even patients with sporadic OKCs were suspected with an underestimated Ptch1 mutation rate.212 Stojanov et al. demonstrated Ptch1 inactivating mutations accounts for 93% of sporadic OKCs.213 Yu et al. indicated that mutations in the intracellular loop of Ptch1 might regulate Cyclin B1 in NBCCS-associated OKCs in a manner of noncanonical HH signaling pathway.214 Ptch1 mutation leads to ligand-independent activation of Smo and subsequently upregulates HH signaling pathway target genes transcription.
Many studies concluded that the aberrant HH signaling pathway is active in the development of OKCs.215,216 Grachtchouk et al. uncovered that elevated expression of HH target genes is detected in lower cell layers of cyst wall of human OKCs.217 Besides, OKCs were found to express lower levels of Sufu gene and higher level of Smo, Ptch1, Cyclin D1 and Bcl2.218 Syndromic OKCs were found to have a higher expression of HH signaling pathway protein than the sporadic OKCs.219 Higher Smo expression was considered to be a risk factor of recurrence of OKCs.220
Brain tumor
Brain tumor and other neoplasm in nervous system account for 1.6% new cases worldwide in 2020 and lead to more than 251,329 deaths.185 Two types of malignant primary brain tumors including medulloblastoma (MB) and glioma were in close relationship with HH signaling pathway.221 MB is a common aggressive malignant tumor in brain during childhood.222 There are four subtypes of MB based on research on their molecular mechanism and clinical characteristics: Wnt-MB, Shh-MB, group 3 MB and group 4 MB.223 The novel feature of Shh-MB is constitutive activation of the Shh signaling pathway.
HH signaling stimulates the proliferation of cerebellar granule neuron precursors (CGNPs) during cerebellar development, which lead to MB formation.224 Researches have indicated that germline and somatic mutation of HH signaling pathway related components leads to MB and such mutation varies in different ages.225 However, Ptch1 represents the most common oncogenic mutations in Shh-MB.226 Nearly half of patients with Shh-MB harbored Ptch1 (45%) alterations while Smo (14%) and Sufu (8%) alterations were also detected.227 Mouse model with conditional deletion of Ptch1 in CGNPs caused MB formation, suggesting that CGNPs play a central role in the origin of Shh-MB.228,229 Astrocytes in the cerebellum perform important functions that support granule cell proliferation and migration in the physiological state while MB-associated astrocytes can secrete Shh ligand that helps maintain proliferation tumor cell in a Ptch1-independent manner.230
HH signaling pathway has effect on multiple targets including Mycn, Snail1, Cyclin D1, Sox2, Sox9 and further influence cancer behavior as tumor growth and tumor cell proliferation during MB tumorigenesis.231 Quiescent cells with a therapy-resistant characteristic can serve as a reservoir for relapse. Sox2, a neural stem cell marker, is recognized as a poor prognosis marker in human Shh-MB. In a mouse model of Shh-MB, cells expressing Sox2 accounts for a proportion of less than 5% of the total tumor cells but might serve as a reservoir for relapse.232
Glioblastoma multiforme (GBM) is a common malignant brain tumor and account for 23% of all gliomas.233 Given to characteristics of high invasiveness and drug resistance, GBM is a neoplasm with a dismal prognosis and an effective practical therapeutic approach remains under investigation.234 Researchers have founded that HH signaling pathway is activated in human glioma cell lines, but not in cultured human astrocytes.235 HH/Gli1 pathway affects the growth of glioma and glioma stem cells are thought to be generated by the population of Gli positive neural stem cells. Researchers reported that Gli1 and Gli2 expression is associated with grades III and IV gliomas with less survival.236 The overall survival of patients with glioblastoma is decreased by upregulation of Shh based on data from the Cancer Genome Atlas glioblastoma.237
Cell migration and proliferation in human GBM cell lines are associated with expression of connexin 43, an integral membrane protein within gap junctions, upon HH signaling pathway modulation.238 Chang et al. enhances HH signaling pathway in GBM cells by recombinant human Shh N-terminal peptide, which increases the production of MMPs to promote cell migration and invasion through the PI3K/AKT pathway.239 Overexpression of fms related tyrosine kinase 1 (FLT1) in GBM cells was related to the invasion and migration of tumor cells through HH signaling pathway.240 Although, Shahi et al. reported a fainter expression of HH signaling pathway component in neuroblastoma (NB) than MB or glioblastoma.241 Studies also reported that HH signaling pathway determines multiple biological behavior of neuroblastoma,242–244 while several researchers proposed theories of a tumor-suppressive functions of HH signaling pathway in NB.243,245
Renal cancer
Renal cell carcinoma (RCC) is the most common renal tumor, accounting for 80–85% of all renal cancers, as clear cell renal cell carcinoma (CRCC) is responsible for the vast majority of RCC.246,247 Dormoy et al. reported HH signaling pathway components are expressed in human CRCC tumor samples, which participates in cell proliferation of CRCC cell lines and related tumor growth. Such reactivation of HH signaling pathway leads to regulation downstream gene transcription of Cyclin D1, PAX2, VEGF, and TGF-β.248 The mRNA levels of Shh, Smo and Gli1 were higher in CRCC tissue compared with control kidney tissue in different degrees.249 Gli3, Ptch1, Dhh, and Shh were highly expressed in in higher grade tumor as Dhh expression can be recognized as an independent predictor of CRCC survival.250 Multiple studies show antitumor potential in human RCC by targeting HH signaling pathway.251,252
HH signaling pathway influence aggressiveness of RCC. Recombinant Shh protein enhanced cell proliferation and suppressed the expression of E-cadherin in RCC cell lines, suggesting the essential role of EMT modulating by HH signaling pathway in RCC.253 HH signaling pathway may interact with hypoxia-inducible factor 2α modulating the radiosensitivity of RCC.254 Furukawa et al. suggested that Gli2 may be related to the underlying mechanism of drug resistance associated with tyrosine kinase inhibitor (TKI) inhibitors sunitinib in CRCC.255
Prostate cancer
Prostate cancer (PCa) is the second most common cancer in male accounting for 14.1% new cases worldwide.185 Paracrine signaling of HH signaling pathway is crucial to prostate development.234 PCa originates from prostatic epithelia and epithelial-mesenchymal interaction plays an important role in PCa progression and metastasis.256,257 Wilkinson et al. has shown the co-localization of Smo with primary cilia in prostatic fibroblasts and confirmed the activated HH signaling pathway in prostatic tumor microenvironment.258 Also, Shh expression level was correlated with cell proliferation in PCa. HH signaling pathway blockade by cycloplamine, a selective inhibitor of Smo, significantly reduced the proliferation of PCa cell lines.259 Enhanced expression of HH signaling pathway components was found in the cancer tissue than in the normal prostatic epithelial tissue, which was correlated with higher Gleason score and worse prognosis.260,261 In animal models, overexpression of Hedgehog protein persistently in mutant mice accelerated the progression of prostatic intraepithelial neoplasia that led to PCa. Hyperplastic basal cells might be the true cellular origin of primary PCa. HH signaling pathway plays important roles in transforming normal prostate basal/stem cells into PCa stem cells.262 In addition, inhibition of Shh signaling pathway showed potential for prevention of drug resistance such as zoledronic acid.263
Liver tumor
HH signaling pathway is considered to play an essential role in liver organogenesis and hepatic tissue repair.264,265 Importantly, HH signaling pathway is influential to multiple biological behaviors of liver tumor including hepatocellular carcinoma (HCC) and Cholangiocarcinoma (CCC).
The most common malignant liver tumor is HCC.185 Ptch1 was found overexpressed in HCC tissue compared with the surrounding non-neoplastic liver tissue. Moreover, increased expression of Smo and Gli1 was directly correlated to a large tumor size of HCC.266,267 Ptch1 and Gli1 can be potential biomarkers for the recurrence of HCC and cumulative survival of HCC patients.268 MMPs promote tumor metastasis by influencing extracellular matrix remodeling. Shh signaling pathway induces MMP-2 and MMP-9 production to promote invasiveness through FAK/AKT signaling in HCC.269 HCC’s invasive behavior was attenuated by treatment with a Smo inhibitor which partially suppressed the expression of MMPs and Gli1/2.270 In addition, target genes of HH signaling pathway such as EMT transcription factors were found overexpressed in poorly differentiated hepatoma cells in HCC. Activation of HH signaling pathway and related EMT factors might be responsible for the invasiveness and chemoresistance in poorly differentiated hepatoma cells.271,272
A specific Gli inhibitor GANT-61 significantly suppressed HH signaling to reverse sorafenib resistance in CD44-positive HCC, suggesting HH signaling pathway can influence HCC drug resistance.273 Further experiments by Zhou et al. confirmed that Gli1/2 binds to the promoter of transporter associated with antigen processing 1 (TAP1) gene, indicating that TAP1 is one of target genes of HH signaling pathway in HCC cell lines. RNAi targeting Gli or TAP1 can alleviating drug resistance.274
CCC is a malignant tumor derived from biliary epithelial cells. According to different anatomical positions, it can be classified into intrahepatic, perihilar, and distal CCC, among which perihilar CCC represents about 50% of all cases.275 Riedlinger et al. revealed a significant activation of HH signaling pathway in CCC samples and cell proliferation of CCC cells was significantly reduced by inhibiting HH signaling pathway.276 The potential effectiveness was also confirmed in a CCC xenograft model that treatment with a combined therapy with BMS-833923 and gemcitabine inhibited tumor growth. Furthermore, HH signaling pathway components such as Gli1 and Gli2, are reliable prognostic factors for CCC.277,278
Hypoxia influences multiple tumor biological behavior. Hypoxia inducible factor-1 (HIF-1) promotes cancer stemness and invasive behavior of CCC by modulating Shh, Smo and Gli1.279,280 HH signaling pathway protein might promote the interaction between cancer cell and stromal cell and eventually further promotes cancer progression. Coculture of CCC cells with hepatic stellate cell line Lx-2 increased cancer cell migration and invasion.281 CCC cells in conventional and hypoxic conditions have been observed to promote tumor-associated macrophage polarization and TGF-β1 secretion via paracrine Shh ligands, thereby promoting CCC cell growth, metastasis and endoplasmic reticulum homeostasis via TGF-β1.282
Colorectal cancers
Among all malignant tumors, colorectal cancer (CRC) ranks third in the world.185 HH signaling pathway in the gastrointestinal tract relies mainly on paracrine secretion for completion. In mouse models, the overexpression of Shh ligands provided advantages in growth to tumor cells by activating HH signaling pathway in the surrounding stroma.196 Further studies have shown that Shh ligands are upregulated in CRC in order to activate the stromal HH signaling pathway, while stromal downstream genes, such as Gli1 and Hhip expression, are decreased.283 This could explain the negative results and Shh ligand overexpression that were evident in clinical trials involving vismodegib for CRC. Several studies have indicated that the downregulation of Ihh can be observed as an early event in the formation of CRC.284,285 Ihh is mainly expressed in the stromal tissue of the colon, and its high expression level has been described to inhibit the development of CRC.283 In a mutant mouse model, colorectal epithelial cells were shown to secrete Ihh to maintain the intestinal stromal phenotype, which was essential to adenoma development, suggesting that Ihh could influence colorectal malignancies in a variety of manners.286 Hypoxic environments can induce the binding of HIF-1α produced by tumor cells and TGF-β2 secreted by cancer-associated fibroblasts to activate Gli2 expression in CRC cells, thus leading to elevated resistance of CRC cells to chemotherapy.287
Pancreatic cancer
Overall 5-year survival of pancreatic cancer is below 10%.288 Shh is aberrantly expressed in pancreatic intraepithelial neoplasia and HH signaling remains active in cell lines established from primary and metastatic pancreatic adenocarcinomas.289 High expression of Shh and Gli1 was an independent prognostic factor for worse survival of patients with pancreatic ductal adenocarcinoma (PDAC).290 Moreover, pancreatic cancer-associated fibroblasts show overexpression of Smo that lead to activation of HH signaling pathway.291
HH signaling pathway influences multiple tumor cell behavior in pancreatic cancer through regulating Gli1 expression. Gli1 expression is essential for PDAC cell survival by facilitating the migration and invasion of cells by promoting EMT. S100A4 is an EMT indicator protein and might be an essential target gene mediated by Gli1 in pancreatic cancer.292,293 However, Lee et al. proposed that stromal response to HH signaling pathway is protective against PDAC and that HH signaling pathway activated by SAG21k can decelerate tumorigenesis.294 Perineural invasion is an important characteristic of pancreatic cancer with an incidence of 70–100%. Cancer cells invade into peripheral nerves in pancreatic tissue and are considered to be associated with poor tumor prognosis and cancer pain.295,296 Moreover, overexpression of Shh activates HH signaling pathway in pancreatic stellate cells in the tumor stroma which is not only essential for tumor growth but also responsible for the nerve invasion in cancer.297
Lung cancer
Similar to many cancers, HH signaling pathway is also important for the prediction of lung cancer prognosis.298 Inhibition of Shh signaling by cycloplamine induces a significant decrease in the proliferation of Non-Small-Cell-Lung-Cancer (NSCLC) cells mediated by Gli through regulation of cyclin expression. Moreover, increased Shh expression in NSCLC might be related to cancer progression mediated by cancer stroma-associated fibroblast.299
Researchers found that in patients with lung adenocarcinomas tumors and lung squamous cell carcinomas had higher Gli expression, accompanied by significantly lower expression of an EMT marker E-Cadherin. Inhibition of HH signaling pathway in vivo decreased tumor growth and induced E-Cadherin expression.300,301 Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) are widely used as first line therapy for NSCLC. Upregulation of HH signaling pathway resulted in EGFR-TKIs resistance by EMT induction. Blockade of HH signaling increased sensitivity to EGFR-TKIs in NSCLC cells.302 HH signaling pathway drives lung adenocarcinoma (LAC) cells growth under stress conditions such as serum-starvation.303 Moreover, microRNA (miRNA) disorders might be related to cell cisplatin resistance in LAC through targeting Gli2.304 Besides, researchers found that Shh+ NSCLC cells produced full-length Shh protein on the membranes of these cells. Shh+ cells exhibited chemo-resistance and showed proliferation and migration stimulatory effect on Shh- cells in a paracrine manner.305
Hematological malignancy
It has been found that HH components are aberrantly activated in a variety of hematological malignancies including multiple biological behavior. HH signaling pathway was upregulated in CD34+ hematopoietic cells from patients with chronic myelomonocytic leukemia (CML).306,307 HH signaling pathway is increased in BCR-ABL+ progenitor cells in CML through enhanced expression of Smo. Smo is responsible for the downregulation of microRNA-326 in patient with CML.308,309 Inhibiting Gli2 abolishes dormancy in human leukemia stem cells.310
Gli expression can be recognized as a prognostic indicator for acute myeloid leukemia (AML) and elevated Dhh plasma level can be detected in AML patient.311 In a murine model of myelodysplastic syndrome (MDS), HH signaling pathway activation led to leukemic transformation to AML by acquiring self-renewal potential.312 Stromal cells show potential of supporting activity for proliferation of leukemic cells and the expression of human Hhip in AML/MDS-derived stromal cells was markedly lower than stromal cells from healthy individuals.313 HH/Gli-1 also plays a key role in resistance to radiation, and that inhibition by LDE225 precipitates a radiation-resistant cell line overcoming radioresistance.314 HH signaling pathway has important impact on B-cell in chronic lymphoblastic leukemia, acute lymphoblastic leukemia (ALL) and multiple myeloma.315–317 HH signaling pathway is associated with progression of these diseases.318 Targeting HH signaling pathway holds promising therapeutic effects.
Thyroid cancer
A total of 80% of thyroid cancers occur as papillary thyroid carcinomas (PTCs).319 A majority of Thyroid tumor specimens are positive for HH signaling pathway component (Shh, Ptch, Smo, and Gli) while inhibition of HH signaling pathway reduced proliferation of thyroid tumor cell lines.320 Shh enhances cell motility and invasiveness of anaplastic thyroid carcinomas.321 Moreover, Shh signaling pathway is important to PTCs occurrence and progression.322,323 It also regulates CSC self-renewal and radiosensitivity in anaplastic thyroid carcinomas cell lines by Snail expression.324 Ma et al. proposed that the loss of primary cilia has been linked to the malignant transformation in thyrocyte.325
Basal cell carcinoma
As the most common skin malignant tumor, BCC is most likely to occur in the skin of head and face.326 Multiple BCC is a major symptom of Gorlin syndrome. 90% of sporadic BCC cases carry somatic mutations in Ptch, others have gain of-function mutations in Smo, leading to over-activation of the pathway.327,328 Overexpression of Ptch1 and Gli1 were observed in BCC compared with normal epidermal tissue and enhanced Shh immunoexpression was found in the aggressive BCC.329,330 Interestingly, Gli1 was specifically upregulated in BCC, while the other skin malignancies such as squamous cell carcinoma showed no Gli1 expression.331
Intrafollicular epidermal stem cells are reported to induce BCC formation through HH signaling pathway due to their enhanced self-renewing ability.332 The tumor epithelium of human BCC with Smo inhibitor resistance possesses reduced primary cilia. Loss of primary cilia might be related to HH signaling pathway inactivation and upregulated RAS/MAPK pathway in resistant BCCs.333 A lower cilia count in the primary lesion might be correlated with BCC recurrence according to a preliminary study.334 Whitson et al. reported non-canonical hedgehog pathway activation also drives drug resistance of Smo inhibitor in BCC.335
Triple negative breast cancer
There were 2.3 million new cases of female breast cancer (BC) in 2020, surpassing lung cancer as the most common cancer worldwide.185 Among all BC subtypes, 10% are triple negative breast cancer (TNBC). Poor prognosis and the lack of efficient targeted therapy make TNBC treatment most challenging.
Multiple studies have shown that HH signaling pathway plays an essential role in normal mammary gland development.336 HH signaling pathway mediates ductal morphogenesis of the mammary gland during puberty and remains downregulated in normal adult mammary tissue.336,337
HH signaling pathway in relation to the aggressive biological behavior of TNBC, including cancer migration, invasion and angiogenesis, has also been discussed.338 Somatic mutations of HH signaling pathway components have been shown to be relatively rare in BC, where ligand-dependent HH signaling pathway activation played a more fundamental role in the pathogenesis of TNBC.339 Moreover, expression of Shh, Dhh, Ptch1, Gli1 has been observed to be upregulated in BC tumor tissue.340 Smo and Gli1 in TNBC tissue is also significantly increased compared to those of non-triple-negative BC.341 Furthermore, TNBC patients with higher Shh expression have worse overall survival.342
HH signaling pathway promotes TNBC progression in an autocrine manner, while promoting tumor vascularization via regulation of vascular endothelial growth factor receptor 2 (VEGFR2) expression in a paracrine manner.343 Another possible mechanism of TNBC chemoresistance is HH signaling pathway upregulated ATP binding cassette (ABC) transporters and other target genes.338 TNBC cells may also harbor dysfunctional stemness pathways compared to non-triple-negative BC cells.338 TNBC associated fibroblasts can activate HH signaling pathway in a paracrine manner and enhance tumor cell growth.344 In addition, HH-activated CAFs have been shown to form a niche for chemo-resistant TNBC stem cells and target HH-activated CAFs via Smo inhibitors, which benefits patients suffering from TNBC.345 Exposure to chemotherapeutics agents of TNBC cell lines results in the release of Shh and HH signaling pathway activation, leading to an increase in the stemness marker of TNBC stem cells, which may explain the chemoresistance and recurrence of TNBC.346 The above evidence may illustrate one possible way that could relate to the aggressiveness and difficulty in treating TNBC by HH signaling pathway.
Rhabdomyosarcoma
Rhabdomyosarcoma (RMS) is known to be the most common type of soft tissue sarcoma in children whose cells tend to undergo myogenic differentiation.347 Two major subtypes of this condition are embryonal subtype of RMS (ERMS) and alveolar subtype of RMS (ARMS).348
The relationship between RMS and HH signaling pathway was first identified in mice heterozygous for Ptch1 as these mutants exhibited a high incidence of ERMS.349 The consistent activation of the HH pathway has been associated with RMS, though there have been relatively few RMS cases in patients with Gorlin syndrome.211 According to the Children’s Oncology Group, 18% of patients with fusion-negative RMS harbor a mutation in HH signaling pathway.350
Controversy is still prevalent pertaining to the exact mechanism of the constitutive activation of HH signaling pathway in RMS. Overexpression of HH signaling pathway components, such as Gli1 and Ptch1, has been detected in RMS patient samples and human RMS cell lines.351 However, activating mutations of HH signaling pathway reported to date only account for a small subset of patients.347 A particular study proposed a different model of RMS in HH signaling pathway activation, where a wide distribution of Dhh and Ihh ligands were detected by immunohistology in RMS samples, in which Shh was only found to be expressed in a small set of RMS cell lines. These findings demonstrated the potential mechanism of the ligand-dependent activation of HH signaling pathway in RMS.352 Hatley et al. utilized aP2-Cre; SmoM2/+ mutant mice and discovered that the activated HH signaling pathway in the adipose lineage led to ERMS in mice.353 Fu et al. reported that primary cilia play a central role in muscle differentiation. Either ciliogenesis or HH signaling pathway are dysregulated in RMS.354
Targeting HH signaling pathway in RMS has demonstrated therapeutic potential in in vitro studies, and its inhibition contributes to the suppression of cell invasion and self-renewal in RMS cell lines.355,356 Meanwhile, upregulation of Gli1 contributes to drug resistance.357
However, the pathogenesis of RMS as well as the oncogenic role of HH signaling pathway remains unclear, and further studies should be urgently conducted in order to develop effective treatment options for RMS
Gastric cancer
Gastric cancer (GC) deaths ranks fourth among all cancers and has led to more than 760,000 deaths in 2020.185 Although the incidence of GC decreases annually, gastric cancer continues to be fatal, with a sharp shortened survival.358
In stomach, HH signaling pathway is essential for differentiation and maturation of gastric epithelial cells under physiological conditions.359 Shh is highly expressed in gastric parietal cells for gastric acid and gastrin production, illustrating that HH signaling pathway is indispensable for normal digestive function.360
HH signaling pathway links progression from chronic inflammation to cancerous lesions.361 Helicobacter pylori (H pylori) infection is one of the main causes of chronic gastritis and is a significant risk factor for gastric cancer. Multiple studies have reported that chronic H pylori infection can interfere in the balance of HH pathway activity within gastric tissue.360 H pylori may also contribute to gastric atrophy and intestinal metaplasia for increased potential of tumorigenesis.362 Mice with parietal cell-specific deletion of Shh were not observed to develop gastritis when infected by H pylori, and Shh in gastric mucosa was considered to act as a chemoattractant for macrophages.363
Shh has been found to be overexpressed in resected GC samples compared to that of adjacent normal tissue.364 Specifically, Shh expressing in H pylori positive patients with early GC was noted to be significantly higher than H pylori negative controls.365 The increased expression of Shh and Gli1 was also shown to be significantly correlated with tumor staging and tumor aggressiveness, suggesting a worse overall survival for patients suffering from GC.193,366
Shh promotes gastric cancer cell proliferation and the survival of lines, which suggests an autocrine progression of gastric cancer. It also regulates gastric cancer migration and invasion of gastric cancer cell lines through EMT.
Shh-Gli1 signaling was activated in CD44+ gastric cancer stem cells, which was responsible for drug resistance in advanced GC. Moreover, Gli interacts with the promoter of ABCG2 and regulates its expression in gastric cancer stem cells.367 Accordingly, targeting HH signaling pathway may improve GC chemosensitivity.368,369 Koh et al. investigated PDL-1 expression mediated by Gli in gastric cancer organoids, which may relate to immune evasion in GC.370
A brief summary on HH signaling pathway in cancer
HH signaling pathway participates in multiple functions in a variety of tumors. According to immunohistology, early exploration of HH signaling pathway was concentrated on the expression and distribution of the HH pathway components in tumor tissue. One potential reason for controversies in early studies that examined the expression patterns of HH signaling pathway proteins in cancer could be differential tumor samples analyzed.371 In light of the rapid understanding of tumor biology, additional features of HH signaling-associated cancers have been determined. Cancer stem cells and the hedgehog pathway are closely related to cancer relapse and cancer resistance to anticancer therapy. In addition, the mechanism of HH signaling pathway activation in cancer is multifactorial, demonstrating that canonical pathway activation and noncanonical pathway activation may exist in a single form of cancer.372 Similarly, ligand-dependent and ligand-independent activation may promote cancer progression cooperatively. Therefore, further studies should focus on the precise mechanism of HH signaling pathway in cancer for better therapeutic targets.
Targeted therapies of HH signaling in cancers
Introduction of targeted therapies
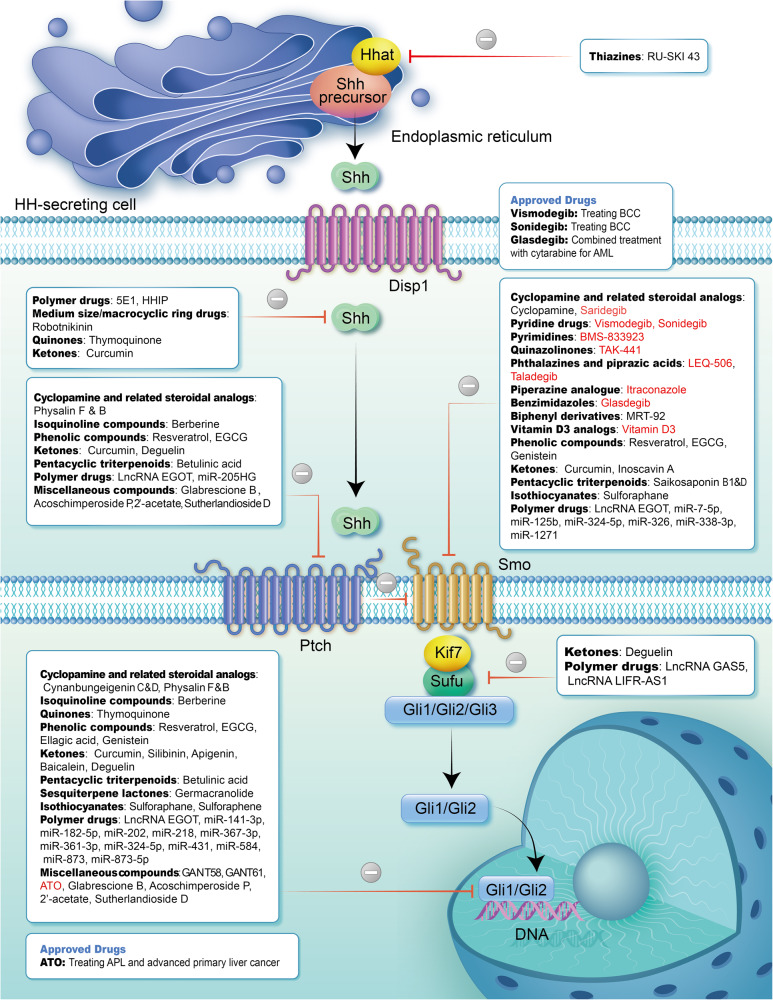
Recent breakthroughs have revealed the carcinogenic function of HH signaling pathway, which makes it an interesting target for cancer treatment. Drugs related to HH signaling pathway target Hhat, Shh, Ptch, Smo, Sufu and Gli1 (Fig. 5). These drugs targeting HH signal transduction can be divided into small molecule inhibitors, natural chemicals, lncRNAs and monoclonal antibodies. Among them, small molecule inhibitors are the most widely studied, usually referring to chemical compounds with molecular weight less than 1 kDa. These drugs have significant advantages: (1) small molecular weight brings more obvious permeability to tissues and cells; (2) there are various forms of drug delivery; (3) oral tolerance and bioavailability are good.
Fig. 5.
Inhibitors targeting HH signaling pathway. In this figure, we mainly show the inhibitors targeting HH signaling pathway, classified according to the properties of the compounds, and the inhibitors in ongoing or completed clinical trials are shown in red. Approved drugs are individually identified, including their indications. BCC basal cell carcinomas, AML acute myeloid leukemia, APL acute promyelocytic leukemia
In 2000, the first drug targeting HH signaling pathway, arsenic trioxide (ATO), was approved for the treatment of acute promyelocytic leukemia (APL).373 ATO not only inhibits HH signaling pathway, but also targets other signaling that involved in the development and progression of APL. The first Food and Drug Administration (FDA) approved drug specifically designed to target HH signaling pathway is vismodegib (GDC-449), which was approved for the treatment of BCC in 2012.374 Since then, sonidegib and glasdegib have been approved for clinical treatment of cancer.375,376 Although vismodegib and sonidegib have achieved satisfactory outcome in the treatment of locally advanced or metastatic BCC, frequent Smo mutations lead to increased drug resistance.377 Therefore, recent studies have revealed that new HH signaling pathway inhibitors and reasonable multi target drugs are applied together to solve this problem, including taladegib, LEQ-506 and TAK-441, which are not sensitive to drug-resistant D473H mutations.378–380 In addition, some studies on Hhat, Shh and Gli1 inhibitors are also expected to inhibit Smo resistant cells.
Hhat inhibitor
Hhat catalyzes the completion of palmitoylation of HH proteins to maintain their stability and normal activity.381 Inhibitors targeting Hhat may therefore block HH protein-mediated pathway activation. Current inhibitor targeting Hhat was RU-SKI 43. Petrova et al. carried out a high throughput screen using a peptide-based assay and identified Hhat specific inhibition by RU-SKI 43 both intracellularly and in vitro.382 They further investigated the potential effect of RU-SKI 43 in cancer. The results suggested that RU-SKI 43 was able to reduce Gli1 activation, Akt and mTOR pathway activity and tumor growth in vitro in pancreatic cancer cell proliferation and in vivo in xenograft models by inhibiting Hhat.383 It should be noted that the inhibition effect of RU-SKI 43 was not mediated through Shh protein.383 Another study showed similar observations that RU-SKI 43 treatment reduced the proliferation of estrogen receptor positive breast cancer cells, showing a dose-dependent effect.384 Overexpression of Hhat protein was able to rescue this inhibition, but the addition of exogenous recombinant Shh protein did not.384 The lack of Hhat inhibitor studies may be due to the need to use radiolabeled fatty acid substrates to measure activity, which has limitations in terms of throughput, cost and safety.385 Recently, Andrei et al. proposed an acylation-coupled lipophilic polarization induction assay. They applied this method to discover the potent Hhat inhibitor IMP-1575.385,386 Future studies will be needed to reveal the role of IMP-1575 in anticancer therapy.
Shh inhibitor
Shh proteins bind to and inhibit Ptch1 receptor, which could unblock Ptch1 for Smo proteins and cause typical HH signaling pathway. Inhibitors targeting Shh proteins may therefore block Shh protein-mediated signaling pathway activation. Inhibitors targeting Shh include 5E1, Hedgehog-interacting protein (Hhip) and Robotnikinin.
5E1
5E1 is a monoclonal antibody targeting Shh and blocks HH signaling pathway by competing with Ptch1 for the Shh binding site.387 Treatment of colorectal cancer cell lines with 5E1 reduced the expression of HH signaling pathway target genes (Gli1, Ptch1, Hip1, Muc5ac) as well as cell cycle protein D1 and mediated the reacquisition of epithelial-like features in cells.388 5E1 was also shown to inhibit the self-renewal capacity and chemoresistance of gastric cancer stem cells, demonstrating a potential role for 5E1 in enhancing chemotherapeutic sensitivity.389 In the non-Smo protein-mediated HH signaling pathway, 5E1 may have a more satisfactory effect. Both cyclopamine and 5E1 could inhibit chronic lymphocytic leukemia (CLL) cells in B-CLL patients.390 Stromal cells could induce activation of the paracrine pathway and activated ERK directly via Ptch1 receptor in CLL cells without Smo.390 In this case, cyclopamine treatment was suboptimal, while 5E1 could completely eliminate ERK phosphorylation.390
Hhip
Hhip is a negative regulator of HH signaling pathway that prevents Ptch1 from binding Shh by binding the cholesterol fraction covalently linked to Shh through the N-terminal.391 Abnormal expression of Hhip has been associated with cancer development. For example, hypermethylation of the Hhip gene promoter has been found in several tumor cell lines, including HCC, pancreatic cancer, MB, and various gastrointestinal tumors.392–395 Down-regulation of Hhip expression using siRNA resulted in a significant increase in colon cancer cell growth and invasion in vitro.395 Therefore, adjustment of Hhip level allows regulation of cancer development. Yu et al. used a lentiviral vector to deliver Hhip. The result showed that gastric cancer cells in the Hhip group had reduced proliferation, migration and invasion compared to the control.396 Interestingly, Hhip overexpression significantly reduced its de novo promoter methylation level in gastric cancer cells.396
Robotnikinin
Robotnikinin is a small molecule that targets Shh-N protein. It has the following features: inhibits HH signaling pathway in a concentration dependent manner; does not show inhibitory activity in the absence of Ptch1 receptors; does not compete with cyclopamine.397 Robotnikinin reduced Gli2 mRNA levels in human keratin forming cells and in primary human synthetic skin tissue.397 There is currently lack of other studies revealing the potential application of robotnikinin in the treatment of cancer.
Ptch activator
Ptch is an inhibitor of HH signaling pathway. Without Shh ligand, Ptch plays a regulatory role by inhibiting the Smo protein.398 Ptch protein silencing or gene deletion is related to the development of many cancers, such as odontogenic keratocyst, BCC and plexiform fibromyxoma.213,399,400 Therefore, activators targeting Ptch can prevent cancer development by inhibiting Smo protein. At present, some natural compounds have been found to increase Ptch1 expression, for example, the combination of epigallocatechin gallery (EGCG) and theaflavin (TF) can reduce the expression of Gli1 and Smo while increase the expression of Ptch1 in mouse liver cancer cells.401
Smo inhibitor
Within HH signaling pathway, Smo protein is often utilized as the drug target because of (1) the ability to transduce downstream signaling pathways in cancers with a loss of function of Ptch1, (2) cancer-causing capacity in the presence of aberrant expression, and (3) the hydrophobic structural transmembrane helical domain capable of binding a variety of small-molecule drugs. The inhibitors targeting Smo mainly include cyclopamine, and cyclopamine derivatives such as vismodegib, sonidegib, etc.
Cyclopamine
Cyclopamine is a steroidal alkaloid metabolite produced by Veratrum californicum, which has been shown to bind and inhibit the activity of Smo proteins.402,403 Cyclopamine may inhibit the development of several cancers by affecting HH signaling pathway. Cyclopamine is able to block HH signaling pathway in vitro, which in turn affects the aggressiveness and motility of human HCC cells.404,405 On the other hand, in an in vivo study, Jeng et al. observed a statistically significant reduction in tumor size in mice with HCC (0.152 ± 0.219 cm2 vs. 0.003 ± 0.009 cm2) following a 2-week cyclopamine injection at a dose of 30 mg/kg/day.406 Cyclopamine has also been shown to inhibit the self-renewal and invasive capacity of cancer stem cells in pancreatic cancer and glioblastoma.407,408 Unfortunately, the poor oral bioavailability and specificity of cyclopamine limit its applications.409 Under this circumstance, semi-synthetic cyclopamine derivatives such as vismodegib, sonidegib and saridegib have been developed, which show better oral bioavailability and perform well in several clinical trials. Of note, cyclopamine may still function as an anti-cancer agent for sensitization to radiotherapy. For instance, Tsai et al. showed that cyclopamine in combination with radiation therapy resulted in a 67% reduction in the average size of in situ tumors compared to radiation therapy alone.410
Vismodegib (GDC-0449)
Vismodegib, a second generation cyclopamine derivative, was approved by the FDA in 2012 as the first Shh inhibitor for BCC treatment.374,411 Vismodegib achieved satisfactory results in clinical trials in patients with locally advanced and metastatic BCC. After 21 months of daily oral administration of 150 mg vismodegib, the objective remission rate was 47.6% for locally advanced BCC and 33.3% for metastatic BCC, with a median remission and progression-free survival of 9.5 months months.412,413 Several phase I and phase II studies have investigated the use of vismodegib in detail, for instance, vismodegib in the treatment of TNBC, multiple BCC, recurrent/refractory MB, and keratinized cystic odontogenic tumors with significant therapeutic benefit.345,414–416 In 2016, Jacobsen et al. evaluated the effect of vismodegib for BCC by meta analysis.417 Data from eight studies with a total of 744 patients showed that: objective response to vismodegib for locally advanced BCC had a weighted average of 64.7% and complete response averaged 31.1%. The objective response for metastatic BCC was 33.6% and complete response averaged 3.9%.417 This meta-analysis showed that vismodegib was identified to have a significant and consistent effect on the median duration of therapy (35.8 weeks) of locally advanced BCC and metastatic BCC. Vismodegib, on the other hand, was not effective in the treatment of metastatic pancreatic cancer, lymphoma, chronic leukemia, and advanced osteosarcoma.418–420 Therefore, more comprehensive clinical trials are needed to validate vismodegib in different populations. Several clinical trials of vismodegib are currently underway as monotherapy as well as in combination therapy for leukemia, meningioma and chondrosarcoma (Table 3). Adverse events associated with vismodegib include muscle cramps, taste disturbances, weight loss, hair loss, and weakness.421 These conditions often subside spontaneously within 4 weeks, but the management of adverse reactions should not be neglected.421
Table 3.
Summary of drugs in clinical trials targeting canonical HH signaling in cancer
| Drugs | Target protein | NCT | N | Status | Phase | Cancer |
|---|---|---|---|---|---|---|
| Vismodegib | ||||||
| Vismodegib | Smo | NCT02436408 | 35 | Completed | Phase IV | BCC |
| NCT01201915 | 74 | Completed | Phase II | BCC | ||
| NCT01267955 | 45 | Active | Phase II | Chondrosarcoma | ||
| NCT01367665 | 1232 | Completed | Phase II | BCC | ||
| NCT01700049 | 28 | Completed | Phase II | BCC | ||
| NCT01815840 | 229 | Completed | Phase II | BCC | ||
| NCT00739661 | 104 | Completed | Phase II | OC | ||
| NCT02366312 | 2 | Completed | Phase II | KCOT | ||
| NCT03052478 | 10 | Completed | Phase II | Stomach neoplasms | ||
| Vismodegib/Chemotherapy/Other | Smo | NCT01878617 | 660 | Active | Phase II | MB |
| Vismodegib/GSK2256098/Capivasertib/Abemaciclib | Smo/ FAK/ AKT / CDK | NCT02523014 | 124 | Recruiting | Phase II | Progressive meningiomas |
| Vismodegib+ Chemotherapy | Smo | NCT01835626 | 24 | Completed | Phase II | Locally advanced BCC; Skin cancer; CM |
| Vismodegib+ Chemotherapy | Smo | NCT02694224 | 40 | Recruiting | Phase II | TNBC |
| Vismodegib+ Pembrolizumab | Smo + PD-1 | NCT02690948 | 16 | Completed | Phase I-II | Metastatic or unresectable BCC |
| Vismodegib+ Ribavirin+ Decitabine | Smo | NCT02073838 | 23 | Completed | Phase II | AML |
| Vismodegib/Other targeted drugs | Smo | NCT02091141 | 670 | Completed | Phase II | Neoplasms |
| Vismodegib/Other targeted drugs | Smo | NCT03158389 | 350 | Recruiting | Phase I-II | Glioblastoma |
| Vismodegib/Other targeted drugs | Smo | NCT03297606 | 720 | Recruiting | Phase II | NHL; MM; Advanced solid tumors |
| Vismodegib+ Bevacizumab | Smo + VEGF | NCT00636610 | 199 | Completed | Phase II | Metastatic CRC |
| Vismodegib+ Gemcitabine Hydrochloride | Smo | NCT01064622 | 118 | Completed | Phase I-II | PC |
| Vismodegib+ Gemcitabine Hydrochloride+ nab-Paclitaxel | Smo | NCT01088815 | 98 | Completed | Phase II | PC |
| Vismodegib+ RO4929097 | Smo + Gamma-Secretase | NCT01154452 | 78 | Completed | Phase I-II | Advanced or metastatic sarcoma |
| Vismodegib+ Gemcitabine Hydrochloride | Smo | NCT01195415 | 25 | Completed | Phase II | PC |
| Sonidegib | ||||||
| Sonidegib | Smo | NCT03534947 | 10 | Recruiting | Phase II | BCC |
| NCT01708174 | 22 | Completed | Phase II | MB | ||
| NCT00961896 | 18 | Completed | Phase II | BCNS | ||
| NCT01125800 | 76 | Completed | Phase I-II | MB | ||
| NCT01350115 | 10 | Completed | Phase II | BCNS | ||
| Sonidegib+ Lenalidomide | Smo + Cereblon | NCT02086552 | 28 | Completed | Phase II | Recurrent/refractory PCM |
| Sonidegib+ Ribociclib | Smo + CDK4/6 | NCT03434262 | 68 | Active | Phase I | Refractory or recurrent MB |
| Sonidegib+ Radiotherapy+Chemotherapy | Smo | NCT04402073 | 205 | Recruiting | Phase II | MB |
| Saridegib | ||||||
| Saridegib | Smo | NCT02762084 | 17 | Completed | Phase II | BCNS |
| NCT02828111 | 36 | Completed | Phase II | BCC | ||
| Saridegib+ Gemcitabine | Smo | NCT01130142 | 122 | Completed | Phase I-II | PC |
| Glasdegib | ||||||
| Glasdegib | Smo | NCT01841333 | 31 | Completed | Phase II | AML |
| NCT01286467 | 23 | Completed | Phase I | Solid tumors | ||
| NCT01842646 | 35 | Completed | Phase II | MDS; CML | ||
| Glasdegib+ Temozolomide Oral Capsule | Smo | NCT03466450 | 75 | Active | Phase I-II | Glioblastoma |
| Glasdegib+ Chemotherapy | Smo | NCT01546038 | 255 | Completed | Phase II | AML |
| Glasdegib+ Other targeted drugs | Smo | NCT03390296 | 50 | Completed | Phase I-II | AML |
| Glasdegib+ Azacitidine | Smo | NCT04842604 | 15 | Completed | Phase III | AML; MDS; CML |
| Taladegib | ||||||
| Taladegib+ Chemotherapy | Smo | NCT02784795 | 94 | Completed | Phase I | Solid tumor; BC; CC; CCC; STS |
| TAK-441 | ||||||
| TAK-441 | Smo | NCT01204073 | 34 | Completed | Phase I | Nonhematologic malignancies |
| Itraconazole | ||||||
| Itraconazole | Smo | NCT01108094 | 29 | Completed | Phase II | BCC |
| NCT01787331 | 21 | Completed | Phase II | Prostate adenocarcinoma | ||
| NCT02354261 | 38 | Completed | Phase II | BCC | ||
| Itraconazole+ AZD9291 | Smo + EGFR | NCT02157883 | 39 | Completed | Phase I | Advanced/inoperable NSCLC |
| Itraconazole+ volasertib | Smo + PLK1 | NCT01772563 | 28 | Completed | Phase I | Neoplasms |
| Itraconazole+ Other drugs | Smo | NCT02770378 | 10 | Completed | Phase I | Glioblastoma |
| Vitamin D3 | ||||||
| Vitamin D3 | Smo | NCT02553447 | 197 | Active | Phase I | CLL; NHL |
| Vitamin D3+ Photodynamic Therapy | Smo | NCT03483441 | 37 | Active | Phase I | BCC; BCNS |
| Vitamin D+ Rituximab | Smo + CD20 | NCT03078855 | 211 | Active | Phase III | NHL |
| ATO | ||||||
| ATO + GO | Gli + CD33 | NCT00274781 | 30 | Completed | Phase II | Advanced MDS |
| ATO + GO + ATRA | Gli + CD33 | NCT01409161 | 151 | Recruiting | Phase II | APML |
BCC Basal cell carcinoma, OC ovarian cancer, BCNS Basal cell nevus syndrome, MB medulloblastoma, KCOT keratocystic odontogenic tumor, CRC colorectal cancer, CM cutaneous malignancy, TNBC triple negative breast cancer, AML acute myeloid leukemia, PCM plasma cell myeloma, NHL non-Hodgkin lymphoma, PC pancreatic cancer, MM multiple myeloma, CML chronic myelomonocytic leukemia, MDS myelodysplastic syndrome, EC esophageal cancer, BC breast cancer, CCC cholangiocarcinoma, CC colon cancer, STS soft tissue sarcoma, NSCLC non-small cell lung cancer, CLL chronic lymphocytic leukemia, APML acute promyelocytic leukemia
Sonidegib (LDE225)
Sonidegib (LDE225) is a Smo antagonist found by high-throughput screening in vitro.422 In vivo and in vitro studies found that sonidegib can effectively reduce epithelial mesenchymal transformation and invasion potential of various types of cancer, such as glioblastoma, PCa and renal cell carcinoma.251,423,424 A phase II clinical trial proved that sonidegib was administered with 200 mg and 800 mg respectively, the objective remission rates of locally advanced BCC were 57.6% and 43.8%, metastatic BCC were 7.7% and 17.4%, and the incidence of adverse events in the 200 mg group was lower.425 Another phase II clinical trial yielded the similar results.426 Based on this, FDA approved sonidegib for the treatment of locally advanced BCC in 2015.375 A meta-analysis of Shh inhibitors for BCC included 22 studies from 2009–2022 and showed overall response rates (ORRs) of 50.1% for sonidegib compared with 68.5% for vismodegib.427 This indicates that the majority of patients receiving sonidegib and vismodegib achieved promising treatment outcomes. In recent years, some clinical studies have focused on applying sonidegib to other cancers, such as MB, ovarian cancer, breast cancer, etc.428–430 Sonidegib showed ideal therapeutic effect and safety in the treatment of these cancers. Sonidegib related adverse events included muscle spasms, taste disorders, nausea, alopecia, and elevated creatine kinase levels.431 These reactions are often mild, but long-term adverse reactions may also lead to the decline of patients’ quality of life and drug withdrawal.431 Compared with vismodegib, patients receiving sonidegib treatment had lower overall adverse events and occurred more slowly.432 At present, several clinical trials of sonidegib as a single therapy and a combination therapy are under way, mainly for the treatment of MB (Table 3).
Saridegib (patidegib, IPI-926)
Saridegib (patidigib, IPI-926) is a semi synthetic derivative of cycloparamide in Smo inhibitor, which has significantly improved drug characteristics, efficacy and good pharmacokinetic characteristics.433 In vivo and in vitro studies show that saridegib can effectively inhibit the proliferation and invasion of chondrosarcoma, serous ovarian cancer, osteosarcoma, acute lymphocytic leukemia and other cancer.434–437 It is worth noting that saridegib also shows a prominent inhibitory effect on the drug-resistant cells obtained by D473H point mutation after the treatment of MB with vismodegib.438 A phase I study used saridegib and FOLFIRINO (5-fluorouracil, leucovorin, irinotecan, oxaliplatin) to treat 15 patients with advanced pancreatic cancer. Four out of five patients treated with saridegib monotherapy observed a continuous decrease in CA19-9 (26.9–97.7%).439 In addition, saridegib and cetuximab also showed anti-tumor activity in the phase I study on the treatment of recurrent/metastatic head and neck squamous cell carcinoma.440 The common adverse reactions of saridegib treatment are fatigue, nausea, muscle spasm, liver dysfunction and alopecia.441 These adverse reactions are similar to other HH signaling pathway inhibitors and may be caused by inhibition of the same pathway.
Glasdegib (PF-04449913)
Glasdegib is a selective small molecule inhibitor that binds to Smo protein.442 A randomized phase II clinical trial showed that compared with cytarabine alone, glasdegib combined with cytarabine can effectively treat AML patients who are not suitable for intensive chemotherapy or have high-risk MDS (median survival, 8.8 vs 4.9 months; 12 month survival, 59.8% vs 38.2%), with better safety and tolerance.443 Based on this study, FDA approved the combined treatment of glasdegib and low-dose cytarabine for newly diagnosed AML patients who are not suitable for intensive induction chemotherapy in 2018.376 In addition, the phase I study of glasdegib in patients with advanced solid tumors shows that it can keep the patient’s condition stable for a long time.444 At present, glasdegib is conducting further clinical trials, including evaluating whether it can effectively treat AML, MDS or CML patients who have not been treated before with or without cytosine arabinoside (NCT04842604) and whether the combination of glasdegib and temozolomide can effectively treat glioblastoma (NCT03466450). Further clinical research will help to reveal the safety and effectiveness of glasdegib in cancer treatment.
Taladegib (LY2940680)
Taladegib binds to the extracellular end of the Smo transmembrane spiral bundle to inhibit the transmission of HH signal transduction.58 The effect of taladegib is not affected by D473 mutation, and it can effectively treat tumor cells resistant to vismodegib.378 Phase I clinical trial proved that taladegib has acceptable safety in the treatment of locally advanced and metastatic BCC patients. The common adverse events are taste disorder, fatigue, nausea and muscle spasm.445 It is worth noting that another phase I study showed that Notch inhibitor crenigacestat combined with taladegib was poorly tolerated in the treatment of patients with advanced or metastatic solid tumors, showing disappointing clinical efficacy.446 The reason why the combination therapy is not ideal may be due to the heterogeneity of the patient population or the history of systemic therapy.446 Further research on taladegib single drug therapy or combination therapy will fully reveal its safety and therapeutic effect.
XL-139 (BMS-833923)
XL-139 (BMS-833923) is a Smo inhibitor, which has been proved to inhibit tumor in many in vitro and in vivo studies. For example, XL-139 can reduce the activity of HH signaling pathway, reduce cell proliferation, while induce apoptosis of esophageal cancer cells in vitro.447 XL-139 can inhibit amyloblastoma cells with SMO-L412F mutation resistant to vismodegib.448 XL-139 combined with gemcitabine can significantly inhibit the growth of CCC in the subcutaneous xenotransplantation model of mice.276 Two clinical studies were designed to explore the effect of XL-139 combined with dasatinib in the treatment of leukemia, but the trial failed to proceed as expected (NCT01218477, NCT01357655). Therefore, further research is needed to reveal the safety and effectiveness of XL-139.
LEQ-506
LEQ-506 is a small molecule inhibitor of Smo, and there are few relevant studies at present. For D473H mutant cells resistant to vismodegib and sonidegib, LEQ-506 was not affected and showed inhibitory effect.379 In addition, in vivo study has proved that LEQ-506 at 1% concentration can effectively inhibit the expression of Gli1 (80–90%) in the skin cells of depilated mice.449
TAK-441
TAK-441 is an oral small molecule Smo inhibitor. Preclinical studies showed that its 50% inhibitory concentration (IC50) on Gli1 transcriptional activity was 4.4 nmol/L.450 Similar to LEQ-506, TAK-441 can be used to treat D473H mutants resistant to vismodegib.380 In vivo studies have proved that TAK-441 can inhibit Gli1 mRNA expression and tumor progression in castration resistant PCa and xenotransplantation of pancreatic tumor in mice.451,452 A phase I clinical trial used TAK-441 to treat 34 patients with solid tumors, including colorectal cancer (26%), BCC (21%) and pancreatic cancer (9%). The results showed that TAK-441 was well tolerated and had preliminary anti-tumor activity.453 The common adverse reactions of TAK-441 during treatment are taste disorder, fatigue, nausea and muscle spasm, mostly mild to moderate.453
Itraconazole
Itraconazole is a drug to treat systemic fungal infection, especially in patients with low immune function and cancer.454 In 2010, it was found that itraconazole could inhibit HH signaling pathway by preventing the accumulation of Smo in primary cilia.455 The unique mechanism of action makes itraconazole a feasible choice for the treatment of other HH signaling pathway inhibitors resistance (such as vismodegib). Many in vivo and in vitro studies have proved that itraconazole can effectively suppress the proliferation of cancer cells, including MB, oral squamous cell carcinoma, gastric cancer, malignant pleural mesothelioma, etc.455–458 Phase II clinical trial showed that after treatment with itraconazole in 19 patients with basal cells, cell proliferation decreased by 45% (P = 0.04), HH signaling pathway activity decreased by 65% (P = 0.03), and tumor area decreased by 24%.459
MRT-92
MRT-92 is a small molecule inhibitor based on acyl guanidine or acyl thiourea scaffold, which can bind to the whole 7-transmembrane domain of Smo, and is insensitive to human D473H.460,461 When inhibiting the proliferation of rat cerebellar granule cells by more than 50%, the required dose of MRT-92 was 0.3 μM and 3 μM for vismodegib.461 Further studies showed that MRT-92 could inhibit the growth of melanoma cells in vitro and xenograft melanoma in mice by inducing DNA damage and G2/M cell cycle arrest.462 Another study identified a new BRD4-SOX2 transcription complex, which is related to the non-standard activation of Gli1 in melanoma.463 The researchers combined the powerful BRD4 degrading agent MZ1 with MRT-92, showing a synergistic anti-proliferation effect on melanoma cells.463 These studies have demonstrated the potential of MRT-92 single drug therapy and multi-drug therapy for melanoma. More studies will help further reveal the safety and efficacy of MRT-92.
PF-5274857
PF-5274857 is an oral Smo antagonist. In vivo studies have shown that it can effectively penetrate the blood-brain barrier and inhibit the activity of Smo protein in the brain of mice with primary MB, thereby improving the survival rate of animals.464 PF-5274857 has potential in the treatment of brain tumors driven by the activated HH signaling pathway or brain metastasis of primary tumors, but there is still a lack of further research.
Gli inhibitor
The nuclear transcription factor Gli is a downstream protein of HH signaling pathway, and inhibitors that target the Gli protein may block its binding to downstream genes and exert an inhibitory effect on HH signaling pathway.465 Although Smo inhibitors have made good progress in the treatment of cancer, the drug-resistant mutations that they cause cannot be ignored.466 Studies have shown that inhibitors targeting Gli proteins are effective against primary and secondary drug resistance induced by Smo inhibitors.467 Currently, inhibitors targeting Gli include GANT compounds, ATO-like compounds, and Hedgehog pathway inhibitors (HPIs).
GANT58 and GANT61
The first inhibitors GANT58 and GANT61, which can inhibit Gli protein production, were identified by representative screening in HEK293 cells transiently expressing Gli1 and Gli-dependent luciferase reporter.468 GANT58 and GANT61 have no effect on other cancer signaling pathways (e.g. TNF signaling/-NF-κB, glucocorticoid and MAPK pathways), and therefore they are highly selective for Gli.468 GANTs can inhibit the proliferation of a variety of cancer cells. For example, GANT58 inhibited spheroid formation and invasion of cervical cancer stem cells undergoing EMT.469 Moreover, it showed more potent cytotoxic effects in the face of cyclopamine-resistant T-ALL cells.470
Compared to GANT58, GANT61 has a significant inhibitory effect on more types of cancer cells. For example, in vitro studies have shown that GANT61 can inhibit the proliferation and invasiveness of breast cancer, head and neck squamous cell carcinoma, HCC, ovarian plasmacytoma, and PCa.471–475 Similar findings were found in in vivo studies, for example, Chang et al. found that the mean volume of cervical cancer in mice after 28 days of GANT61 treatment was significantly reduced compared to the control group (1467.39 ± 403.4 mm3 Vs 460.73 ± 91.01 mm3).476 In addition, GANT61 also inhibited the growth of PCa, MB and rhabdomyosarcoma in vivo.477,478 Currently, an increasing number of studies are using GANT61 as a positive control for identifying Gli-specific drugs. Notably, GANT61 under physiological conditions is unstable and rapidly hydrolyzes to aldehydes (GANT61-A) and diamine derivatives (GANT61-D).479 GANT61-A lacked biological activity on HH signaling while GANT61-D could inhibit Gli transcription.479 This nature limited the applicability of GANT61 and no clinical trials have been conducted so far.
ATO and darinaparsin (ZIO-101)
ATO is an arsenic compound that has been approved by the FDA as a first-line drug for the treatment of APL, with fewer metastases and longer survival after treatment.480 ATO exerts its therapeutic effects by inducing promyelocytic leukemia-retinoic acid receptor multimerization, sumoylation and proteasomal degradation.481 In 2010, ATO was validated as a Gli inhibitor, which inhibited Gli transcription and reduced target gene expression by acting directly on the zinc finger structural domain, thereby preventing Gli accumulation in primary cilia and affecting HH signaling pathway.482,483 ATO, on the other hand, does not affect the interaction between Gli and DNA.484 Yang et al. showed that after 28 months of single ATO treatment, 39 APL patients (86.67%) achieved complete hematological remission, and these patients showed significant downregulation of Gli2 and Smo gene expression.485 In addition, several in vitro studies have shown that ATO can inhibit pancreatic cancer, osteosarcoma, rhabdomyosarcoma, malignant pleural mesothelioma and MB cells by acting on Gli.458,484,486–488 ATO has been used as a first-line agent in the treatment of APL, but its clinical use in solid tumors is limited by a number of factors, including severe side effects, low drug solubility, and rapid renal clearance.489 Pharmacology, drug combinations, and nano-drug delivery systems have all been effective in increasing the therapeutic potential of ATO.490
The toxicity of ATO limits its clinical use in solid tumors, so researchers have developed new arsenic analogs to address this issue. Darinaparsin (S-dimethylarsino-glutathione, ZIO-101) is an organic small molecule arsenic compound, and like other arsenic compounds, the arsenic in darinaparsin can bind glutathione and act on the zinc finger structure of Gli protein.491 Darinaparsin has better antitumor activity and less systemic toxicity than ATO, and intracellular arsenic levels increase faster, higher, and more consistently than ATO.492–494 Furthermore, compared to ATO, darinaparsin showed significantly higher in vitro cytotoxicity and radiosensitizing activity against solid tumor cells under normoxia and hypoxia without affecting normal bone marrow.495 Studies have shown that darinaparsin inhibits the proliferation of cancer cells and the growth of prostate tumors in mice by inhibiting Gli2 transcription.496 Phase I clinical trials of darinaparsin for refractory solid tumors and phase II clinical trials of Hodgkin’s lymphoma have shown satisfied performance.497,498 However, in vivo safety of darinaparsin still requires further study.
HPIs
HPI is a class of small molecules that directly antagonize Gli, independent of PI3K-AKT, PKA, MAPK and other related pathways.499 Each HPI has a unique mechanism of action: (1) HPI-1 inhibits the activation of HH signaling pathway induced by Sufu deletion or Gli overexpression, suppresses Gli protein modification, and affects its function as a transcription factor/cofactor; (2) HPI-2 is less effective against exogenous Gli1 but may interfere with the conversion of full-length Gli2 into a transcriptional activator; (3) HPI-3 inhibits the formation of activated Gli2; (4) HPI-4 acts by disrupting ciliogenesis without affecting Gli2 formation.499 In vitro studies have shown that HPI-1 can effectively inhibit the proliferation of small cell lung cancer, breast cancer and other tumor cells.471,500,501 In addition, Wei et al. showed that HPI-4 treatment of chondrosarcoma cells significantly decreased the expression of ciliary microtubule transport protein Ift88, Gli protein, and Ptch1 protein, and impaired cell proliferation and invasion ability.502 However, the low bioavailability of HPI in vivo due to its high lipophilicity and poor water solubility has limited further studies.503
Other drugs may target HH signaling pathway
In addition to small molecule inhibitors, there are several inhibitors that can target HH signaling pathway, such as natural compounds and LncRNA. Natural compounds are easy to apply, highly available and acceptable therapeutic methods, usually targeting multiple signaling pathways.504 It has been found that many natural compounds can target HH signaling pathway and inhibit the proliferation of cancer cells.504 Among them, vitamin D3, also known as cholecalciferol, is the most studied and it is formed by 7-dehydrocholesterol after dehydrogenation of cholesterol by ultraviolet radiation. The A-ring of vitamin D3 can directly bind to Smo and inhibit HH signal transduction.505 Studies have shown that vitamin D3 can reduce the expression of Gli1 and Ptch and cell proliferation in mouse BCC cell line, and this inhibition is carried out in an independent manner of vitamin D receptor.506 At present, several clinical phase I and phase III trials of vitamin D3 are under way to study the role of vitamin D3 monotherapy or combination therapy in non-Hodgkin’s lymphoma and BCC (NCT03078855, NCT02553447, NCT0343441). Since the function of these natural drugs targeting HH signaling pathway has not been fully revealed, we did not include the discussion of these drugs in the current review. Detailed information on natural medicines can be obtained from Table 4.
Table 4.
Natural compounds and ncRNAs targeting HH signaling in cancers
| Name | Features | HH signaling target | Cancer | Model | Effect |
|---|---|---|---|---|---|
| Novel natural compounds | |||||
| Berberine517 | GI disease treatment | Gli1, Ptch1 | Primary intracranial MB | In vitro and vivo | Tumor growth reduction (37.5%) |
| Cynanbungeigenin C/Cynanbungeigenin D518 | Health care drugs | Gli | MB | In vitro and vivo | Inhibition of cell viability and proliferation |
| Thymoquinone519 | Anti-oxidant, anti-tumor | Shh, Gli1 | PCa | In vitro and vivo | Inhibition of cell viability and proliferation |
| Resveratrol520,521 | Anti-aging, anti-tumor | Gli1 | GC | In vitro | Cancer cell invasion and metastasis inhibition |
| Ptch, Smo | PC | In vitro | Inhibition of cell viability and increase in apoptosis rate | ||
| Curcumin522–525 | Food additives | Gli1 | Malignant glioma | In vitro and vivo | Tumor growth reduction (71.4%) |
| Shh, Smo, Gli1, Gli2 | Lung cancer | In vitro | Inhibition of cell proliferation and increase in apoptosis rate | ||
| Shh, Smo, Gli | PC | In vitro | Inhibition of cell EMT, proliferation and migration | ||
| Shh, Gli1, Ptch1 | MB | In vitro | Inhibition of cell viability and increase in apoptosis | ||
| EGCG401,526–528 | Anti-oxidant, anti-tumor | Gli1, Smo | Tongue and liver cancer | In vivo | Limiting histological carcinogenesis |
| Ptch1, Smo, Gli1 | Liver cancer | In vivo | Limiting histological carcinogenesis | ||
| Smo, Ptch1, Ptch2, Gli | PC | In vitro | Inhibition of cell proliferation and increase in apoptosis | ||
| Gli1, Ptch | Chondrosarcoma | In vitro | Inhibition of cell proliferation and increase in apoptosis | ||
| Ellagic acid529 | Anti-tumor | Gli1, Gli2 | PC | In vivo | Tumor growth and metastasis reduction (41.2%) |
| Saikosaponin B1/Saikosaponin D530 | Anti-tumor | Smo | MB | In vitro and vivo | Inhibition of cell proliferation and tumor growth |
| Genistein531,532 | Estrogen-like activity | Smo, Gli1 | BC | In vivo | Tumor growth reduction (68%) |
| Gli1 | PCa | In vivo | Tumor growth reduction (58.3%) | ||
| Glabrescione B533 | Gli1, Ptch1 | MB | In vivo | Tumor growth reduction (63.6%) | |
| Gli1, Ptch1 | BCC | In vitro | Tumor growth reduction (71.4%) | ||
| Silibinin534 | Hepatoprotective drugs | Gli1, Gli2 | RCC | In vivo | Tumor growth reduction (64.9%) |
| Inoscavin A535 | Smo | CC | In vitro and vivo | Inhibition of cell proliferation and tumor growth | |
| Acoschimperoside P, 2’-acetate536 | Gli1, Ptch1 | PC | In vitro | Inhibition of cancer cell proliferation and migration | |
| Apigenin537 | Anti-cancer, anti-viral | Gli1 | PCa | In vitro | Inhibition of cancer cell proliferation and migration |
| Baicalein537 | Treatment of paralysis after cerebrovascular disease | Gli1 | PCa | In vitro | Inhibition of cancer cell proliferation and migration |
| Betulinic acid538 | Anti-cancer | Gli1, Ptch1 | PC and PCa | In vitro | Inhibition of cancer cell proliferation |
| Deguelin539 | Insect poisoning, anti-cancer | Gli1, Ptch1, Sufu | PC | In vitro | Inhibition of cell migration and increased apoptosis |
| Germacranolide540 | Gli1 | PC | In vitro | Inhibition of cancer cell proliferation | |
| Physalin F & B541 | Anti-inflammatory, immunomodulatory | Gli1, Gli2, Ptch1 | PC | In vitro | Inhibition of cancer cell proliferation |
| Sutherlandioside D542 | Anti-cancer | Gli1, Ptch1 | PCa | In vitro | Inhibition of cancer cell proliferation |
| Sulforaphene543 | Anti-cancer | Gli1 | BC | In vitro | Cell invasion and metastasis inhibition |
| Sulforaphane544 | Anti-oxidant, anti-tumor | Smo, Gli1, Gli2 | PC | In vivo | Tumor weight reduction (45.0%) |
| Vitamin D3252,506 | Promote calcium and phosphorus absorption | Gli2 | RCC | In vivo | Tumor growth reduction (81.4%) |
| Gli1 | BCC | In vitro and vivo | Tumor growth reduction | ||
| NcRNA | |||||
| LncRNA GAS5545 | Sufu | TNBC | In vivo | Increased apoptosis in cancer cells | |
| LncRNA EGOT546 | Gli1, Smo, Ptch1, Hhip | BC | In vitro and vivo | Inhibition of cell viability and migration capacity | |
| LncRNA LIFR-AS1547 | Sufu | BC | In vitro and vivo | Inhibition of cancer cell proliferation and migration | |
| miR-7-5p548,549 | Smo | GC | In vitro | Inhibition of cancer cell proliferation and migration | |
| Gli3 | Bladder cancer | In vitro and vivo | Inhibition of cancer cell proliferation and migration | ||
| miR-125b550 | Smo | MB | In vivo | Inhibition of cancer cell proliferation and migration | |
| miR-141-3p551 | Gli2 | Osteosarcoma | In vitro and vivo | Inhibition of cancer cell proliferation and increased apoptosis in cancer cells | |
| miR-182-5p304 | Gli1, Gli2 | Lung cancer | In vitro | Cell proliferation inhibition and increased sensitivity to cisplatin | |
| miR-202552 | Gli2 | Osteosarcoma | In vitro and vivo | Inhibition of cancer cell proliferation and increased apoptosis in cancer cells | |
| miR-218553,554 | Gli3 | Cervical cancer | In vitro and vivo | Inhibition of cancer cell proliferation and migration | |
| Gli1 | PCa | In vitro | Inhibition of cancer cell proliferation, metastasis and EMT | ||
| miR-205HG555 | Ptch1 | EC | In vitro and vivo | Tumor volume reduction (39.4%) | |
| miR-367-3p556 | Gli1, Gli2 | PCa | In vitro and vivo | Inhibition of cancer cell proliferation and migration | |
| miR-361-3p557 | Gli1, Gli3 | Retinoblastoma | In vivo | Inhibition of cancer cell proliferation and migration | |
| miR-324-3p558 | Gli3 | NC | In vitro and vivo | Inhibition of cancer cell proliferation | |
| miR-324-5p550,559,560 | Smo, Gli1 | MB | In vivo | Inhibition of cancer cell proliferation and migration | |
| Gli1 | Glioma | In vitro | Inhibition of cancer cell survival | ||
| Smo, Gli1 | MM | In vitro and vivo | Inhibition of cancer cell proliferation and increased apoptosis in cancer cells | ||
| miR-326308,550,561,562 | Smo | MB | In vivo | Inhibition of cancer cell proliferation and migration | |
| Smo | Glioblastoma | In vitro and vivo | Inhibition of cancer stem cell self-renewal and stemness | ||
| Smo | Osteosarcoma | In vitro and vivo | Inhibition of cancer cell proliferation and migration | ||
| Smo | CML | In vitro and vivo | Inhibition of cancer cell proliferation and increased apoptosis in cancer cells | ||
| miR-338-3p563,564 | Smo | CRC | In vitro | Inhibition of cancer cell proliferation and migration | |
| Smo | Glioma | In vitro | Inhibition of cancer cell proliferation and migration | ||
| miR-431565 | Gli | Papillary thyroid carcinoma | In vitro and vivo | Inhibition of cancer cell proliferation and migration | |
| miR-506566 | Gli3 | Cervical cancer | In vitro and vivo | Inhibition of cancer cell proliferation and increased apoptosis in cancer cells | |
| miR-584567 | Gli1 | Cervical cancer | In vitro and vivo | Inhibition of cancer cell proliferation and migration | |
| miR-873568 | Gli1 | Cervical cancer | In vitro and vivo | Inhibition of cancer cell proliferation, metastasis and EMT | |
| miR-873-5p569 | Gli1 | GC | In vitro | Inhibition of cancer cell proliferation and migration | |
| miR-1271570 | Smo | MM | In vitro and vivo | Inhibition of cancer cell proliferation and migration | |
BCC Basal cell carcinoma, PCa prostate cancer, MB medulloblastoma, CRC colorectal cancer, TNBC triple negative breast cancer, PC pancreatic cancer, MM multiple myeloma, CML chronic myelomonocytic leukemia, EC esophageal cancer, BC breast cancer, CC colon cancer, GC gastric cancer, RCC renal cell carcinoma, NC nasopharyngeal carcinoma
Non-coding RNA (ncRNA) refers to RNA that does not encode proteins and operates at the RNA level. Recent studies have shown that ncRNA can regulate HH signaling pathway by changing Gli1/2, Smo and Ptch1, such as lncRNA, miRNA and circular RNA (circRNA).507 LncRNA can regulate HH signaling pathway through activation or inhibition, which is related to tumor initiation or progression, chemotherapy resistance, recurrence and other processes.508 The diagnosis or treatment methods based on lncRNA are promising. In Table 4, we summarized ncRNAs that inhibit cancer development through HH signaling pathway. The existing challenge is how to deliver the specificity of oligonucleotide inhibitors to tumor targets. At present, nano based drug delivery systems are being developed to deliver oligonucleotide inhibitors, such as liposomes, exosomes, nanoparticles or viral vectors.509 It is hoped that with the progress of research, more ncRNAs can be approved for clinical treatment.
Challenges of HH-targeted therapies
At present, the selective Smo inhibitors vismodegib and sonidegib have been approved by FDA for clinical treatment for cancers. However, the acquired drug resistance of cancer patients to vismodegib proves the clinical limitation of targeting Smo. Therefore, new inhibitors of HH signaling pathway have been developed to overcome this drug resistance, such as taladegib, LEQ-506 and TAK-441. Their effectiveness and safety need to be further validated in clinical trials. Furthermore, some natural compounds can affect HH signaling transduction through Smo, Gli1, Sufu and related factors. For example, vitamin D3 has entered clinical trials to evaluate its effect on non-Hodgkin’s lymphoma. Yet, these natural compounds have not been proved to be direct targets of HH signaling pathway. This can be further determined by using detailed in vivo and in vitro models. In addition, the bioavailability of many natural products is relatively low, and the concentrations used in in vitro studies cannot be achieved under physiological conditions. Therefore, it is necessary to determine the blood level at which natural products act. NcRNAs can mediate HH signaling pathway by changing Gli1/2, Smo and Ptch1 expression. Nanobased drug delivery systems are developed to deliver oligonucleotide inhibitors, such as liposomes, exosomes, nanoparticles and viral vectors. With the progress of these studies, it will be more conducive to targeted treatment of cancers with ncRNA. Finally, HH signaling transduction pathway also cooperates with other carcinogenic pathways, such as PI3K, MAPK, KRAS/BRAF and TGF-β, etc. It is beneficial to use HH signaling pathway inhibitors as combination or adjuvant therapy in cancer. Existing clinical trials have focused on the combination of HH signaling pathway inhibitors with targeted drugs of other pathways. As such, a future goal should be to define the specific function of drugs targeting HH signaling pathway, including their in vivo effectiveness, safety, side effects, etc. A deeper understanding of the mechanisms of HH-targeted therapies will prompt new treatments and continue to offer potential therapeutic targets for cancer.
Conclusion and future perspective
Early literature primarily focused on defining the function of HH signaling pathway in embryonic development. Recent studies have begun to reveal that the orchestration of HH signaling is critical for healthy cell function. Dysregulation of HH signaling pathway is associated with the development and progression of numerous cancers. Therefore, understanding the integrative nature of HH signaling pathway has opened up the potential for new therapeutic targets for cancer. A variety of drugs targeting HH signaling pathway have been developed, and some of which have been approved as for clinical treatment for cancer. Notably, most research of the inhibitors targeting HH signaling pathway are still in the early stages. The results of several clinical trials are unsatisfactory, owing to the fact that the efficacy and safety of the drugs are not well documented in preclinical studies. Meanwhile, certain drugs targeting HH signaling pathway show limited specificity, which is due to the lack of sufficient studies to verify the mechanism of the specific action on HH signaling pathway. As implied above, with new therapeutic targets being identified at a rapid rate, a more comprehensive understanding of the mechanisms of HH-targeted therapy will solve the existing problems in the application of these drugs and pave the way for a new clinical paradigm.
In summary, our review comprehensively summarizes the function of HH signaling pathway in tissue homeostasis and cancer development. Importantly, we discussed the current application, advantages and challenges of HH signaling pathway targeted therapies, providing valuable insight and important implications for the exciting translational innovations in the future.
Acknowledgements
This work was supported by NSFC grants 82222015, 82171001 and 81800928, Young Elite Scientist Sponsorship Program by CAST No. 2020QNRC001, Research Funding from West China School/Hospital of Stomatology Sichuan University RCDWJS2023-(1).
Author contributions
J.J., Z.W. and J.W. contributed equally to this work. J.J., Z.W., J.W., G.L., H.L. performed literature searching and summary. J.J., Z.W. and J.W. wrote the manuscript; and Y.F. and C.Z. provided valuable discussion and revised the manuscript. All authors have read and approved the article.
Competing interests
The authors declare no competing interests. C.Z., who is a member of the editorial board of Signal Transduction and Targeted Therapy, was not involved in the handling process of this manuscript.
Footnotes
These authors contributed equally: Junjun Jing, Zhuoxuan Wu, Jiahe Wang
Contributor Information
Yi Fan, Email: yifan@scu.edu.cn.
Chenchen Zhou, Email: chenchenzhou5510@scu.edu.cn.
References
- 1.Cavodeassi, F., Creuzet, S. & Etchevers, H. The hedgehog pathway and ocular developmental anomalies. Hum. Genet.138, 917–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingham, P. Drosophila segment polarity mutants and the rediscovery of the hedgehog pathway genes. Curr. Top. Dev. Biol.116, 477–488 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Ingham, P. Hedgehog signaling. Curr. Top. Dev. Biol.149, 1–58 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez, D. & Medici, D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal7, re8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briscoe, J. & Thérond, P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol.14, 416–429 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Jiang, J. Hedgehog signaling mechanism and role in cancer. Semin Cancer Biol.85, 107–122 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katoh, M. Genomic testing, tumor microenvironment and targeted therapy of Hedgehog-related human cancers. Clin. Sci. (Lond.)133, 953–970 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Chen, X. et al. Processing and turnover of the Hedgehog protein in the endoplasmic reticulum. J. Cell Biol.192, 825–838 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lum, L. & Beachy, P. A. The Hedgehog response network: sensors, switches, and routers. Science304, 1755–1759 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Rohatgi, R. & Scott, M. P. Patching the gaps in Hedgehog signalling. Nat. Cell Biol.9, 1005–1009 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Hui, C. & Angers, S. Gli proteins in development and disease. Annu Rev. Cell Dev. Biol.27, 513–537 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Svärd, J. et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell10, 187–197 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Goetz, S. & Anderson, K. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet.11, 331–344 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.May, S. et al. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev. Biol.287, 378–389 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Bangs, F. & Anderson, K. Primary cilia and mammalian hedgehog signaling. Cold Spring Harb. Perspect. Biol.9, a028175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper, J. & Scott, M. Communicating with hedgehogs. Nat. Rev. Mol. Cell Biol.6, 306–317 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Lewis, P. et al. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell105, 599–612 (2001). [DOI] [PubMed] [Google Scholar]
- 18.St-Jacques, B., Hammerschmidt, M. & McMahon, A. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev.13, 2072–2086 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baldinotti, F. et al. Novel familial variant of the desert hedgehog gene: clinical findings in two sisters with 46,XY gonadal dysgenesis or 46,XX karyotype and literature review. Horm. Res. Paediatr.89, 141–149 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Kong, J., Siebold, C. & Rohatgi, R. Biochemical mechanisms of vertebrate hedgehog signaling. Development146, dev166892 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riddle, R., Johnson, R., Laufer, E. & Tabin, C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell75, 1401–1416 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Bourikas, D. et al. Sonic hedgehog guides commissural axons along the longitudinal axis of the spinal cord. Nat. Neurosci.8, 297–304 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Hill, S., Fu, M. & Garcia, A. Sonic hedgehog signaling in astrocytes. Cell Mol. Life Sci.78, 1393–1403 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii, M., Sun, J., Ting, M. & Maxson, R. The development of the calvarial bones and sutures and the pathophysiology of craniosynostosis. Curr. Top. Dev. Biol.115, 131–156 (2015). [DOI] [PubMed] [Google Scholar]
- 25.King, P., Paul, A. & Laufer, E. Shh signaling regulates adrenocortical development and identifies progenitors of steroidogenic lineages. Proc. Natl Acad. Sci. USA106, 21185–21190 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rishikaysh, P. et al. Signaling involved in hair follicle morphogenesis and development. Int J. Mol. Sci.15, 1647–1670 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosoya, A. et al. Sonic hedgehog signaling and tooth development. Int J. Mol. Sci.21, 1587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varjosalo, M. & Taipale, J. Hedgehog signaling. J. Cell Sci.120, 3–6 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Qi, X. & Li, X. Mechanistic insights into the generation and transduction of hedgehog signaling. Trends Biochem. Sci.45, 397–410 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pepinsky, R. et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem.273, 14037–14045 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Buglino, J. & Resh, M. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J. Biol. Chem.283, 22076–22088 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chamoun, Z. et al. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science293, 2080–2084 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Tukachinsky, H. et al. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep.2, 308–320 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehring, K. & Grobe, K. Dispatching plasma membrane cholesterol and Sonic Hedgehog dispatch: two sides of the same coin? Biochem. Soc. Trans.49, 2455–2463 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohlig, S. et al. Sonic hedgehog shedding results in functional activation of the solubilized protein. Dev. Cell20, 764–774 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Zeng, X. et al. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature411, 716–720 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Ramsbottom, S. & Pownall, M. Regulation of Hedgehog signalling inside and outside the cell. J. Dev. Biol.4, 23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takei, Y. et al. Three Drosophila EXT genes shape morphogen gradients through synthesis of heparan sulfate proteoglycans. Development131, 73–82 (2004). [DOI] [PubMed] [Google Scholar]
- 39.The, I., Bellaiche, Y. & Perrimon, N. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol. Cell4, 633–639 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Chen, Y. & Struhl, G. Dual roles for patched in sequestering and transducing Hedgehog. Cell87, 553–563 (1996). [DOI] [PubMed] [Google Scholar]
- 41.La Sala, G. et al. Modulation of Dhh signaling and altered Sertoli cell function in mice lacking the GPR37-prosaposin receptor. FASEB J.29, 2059–2069 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Zhao, C. et al. Desert hedgehog mediates the proliferation of medaka spermatogonia through smoothened signaling. Reproduction163, 209–218 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Clark, A., Garland, K. & Russell, L. Desert hedgehog (Dhh) gene is required in the mouse testis for formation of adult-type Leydig cells and normal development of peritubular cells and seminiferous tubules. Biol. Reprod.63, 1825–1838 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Sugito, H. et al. Ihh signaling regulates mandibular symphysis development and growth. J. Dent. Res.90, 625–631 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Ohba, S. Hedgehog signaling in skeletal development: roles of Indian Hedgehog and the mode of its action. Int. J. Mol. Sci.21, 6665 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King, P. J., Guasti, L. & Laufer, E. Hedgehog signalling in endocrine development and disease. J. Endocrinol.198, 439–450 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Bonn-Breach, R. et al. Structure of Sonic Hedgehog protein in complex with zinc(II) and magnesium(II) reveals ion-coordination plasticity relevant to peptide drug design. Acta Crystallogr D. Struct. Biol.75, 969–979 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pathi, S. et al. Comparative biological responses to human Sonic, Indian, and Desert hedgehog. Mech. Dev.106, 107–117 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Hollier, P. et al. Full-length Dhh and N-terminal Shh act as competitive antagonists to regulate angiogenesis and vascular permeability. Cardiovasc Res.117, 2489–2501 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Carpenter, D. et al. Characterization of two patched receptors for the vertebrate hedgehog protein family. Proc. Natl Acad. Sci. USA95, 13630–13634 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong, X. et al. Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science361, eaas8935 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Ingham, P. et al. Patched represses the Hedgehog signalling pathway by promoting modification of the Smoothened protein. Curr. Biol.10, 1315–1318 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Qi, X., Schmiege, P., Coutavas, E. & Li, X. Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science362, eaas8843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alfaro, A. et al. Ptch2 mediates the Shh response in Ptch1−/− cells. Development141, 3331–3339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rahnama, F., Toftgård, R. & Zaphiropoulos, P. Distinct roles of PTCH2 splice variants in Hedgehog signalling. Biochem. J.378, 325–334 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byrne, E., Luchetti, G., Rohatgi, R. & Siebold, C. Multiple ligand binding sites regulate the Hedgehog signal transducer smoothened in vertebrates. Curr. Opin. Cell Biol.51, 81–88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumari, S., Mitra, A. & Bulusu, G. Structural dynamics of Smoothened (SMO) in the ciliary membrane and its interaction with membrane lipids. Biochim. Biophys. Acta Biomembr.1864, 183946 (2022). [DOI] [PubMed] [Google Scholar]
- 58.Wang, C. et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature497, 338–343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao, Y., Tong, C. & Jiang, J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature450, 252–258 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Chen, Y. et al. Sonic Hedgehog dependent phosphorylation by CK1α and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol.9, e1001083 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jia, J. et al. Hedgehog signalling activity of smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature432, 1045–1050 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Frank-Kamenetsky, M. et al. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J. Biol.1, 10 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharpe, H., Wang, W., Hannoush, R. & de Sauvage, F. Regulation of the oncoprotein smoothened by small molecules. Nat. Chem. Biol.11, 246–255 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Kaushal, J., Batra, S. & Rachagani, S. Hedgehog signaling and its molecular perspective with cholesterol: a comprehensive review. Cell Mol. Life Sci.79, 266 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lacour, J. Carcinogenesis of basal cell carcinomas: genetics and molecular mechanisms. Br. J. Dermatol.Suppl 61, 17–19 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Kim, J. et al. The role of ciliary trafficking in Hedgehog receptor signaling. Sci. Signal8, ra55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen, Y. & Jiang, J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res.23, 186–200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu, A. et al. Cholesterylation of Smoothened is a calcium-accelerated autoreaction involving an intramolecular ester intermediate. Cell Res.32, 288–301 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kong, Z. et al. The cation-π interaction in cysteine-rich domain of smoothened is critical for its cholesterylation and function. Acta Biochim. Biophys. Sin. (Shanghai)54, 1171–1179 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim, Y. et al. Ptch2/Gas1 and Ptch1/Boc differentially regulate Hedgehog signalling in murine primordial germ cell migration. Nat. Commun.11, 1994 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Echevarría-Andino, M. & Allen, B. The hedgehog co-receptor BOC differentially regulates SHH signaling during craniofacial development. Development147, dev189076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Witt, R. et al. Heparan sulfate proteoglycans containing a glypican 5 core and 2-O-sulfo-iduronic acid function as Sonic Hedgehog co-receptors to promote proliferation. J. Biol. Chem.288, 26275–26288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niewiadomski, P. et al. Gli proteins: regulation in development and cancer. Cells8, 147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kasper, M., Regl, G., Frischauf, A. & Aberger, F. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur. J. Cancer42, 437–445 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Lichti-Kaiser, K. et al. Gli-similar proteins: their mechanisms of action, physiological functions, and roles in disease. Vitam. Horm.88, 141–171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma, T. et al. Geniposide alleviates inflammation by suppressing MeCP2 in mice with carbon tetrachloride-induced acute liver injury and LPS-treated THP-1 cells. Int. Immunopharmacol.29, 739–747 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Pearson, R. et al. Krüppel-like transcription factors: a functional family. Int. J. Biochem. Cell Biol.40, 1996–2001 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Liao, H. et al. Protein phosphatase 4 promotes Hedgehog signaling through dephosphorylation of suppressor of fused. Cell Death Dis.11, 686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maloverjan, A. et al. Dual function of UNC-51-like kinase 3 (Ulk3) in the Sonic hedgehog signaling pathway. J. Biol. Chem.285, 30079–30090 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen, Y. et al. Dual phosphorylation of suppressor of fused (Sufu) by PKA and GSK3beta regulates its stability and localization in the primary cilium. J. Biol. Chem.286, 13502–13511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pearse, R., Collier, L., Scott, M. & Tabin, C. Vertebrate homologs of Drosophila suppressor of fused interact with the gli family of transcriptional regulators. Dev. Biol.212, 323–336 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, Y. et al. Structural insight into the mutual recognition and regulation between suppressor of fused and Gli/Ci. Nat. Commun.4, 2608 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou, M. et al. Dose-dependent phosphorylation and activation of Hh pathway transcription factors. Life Sci. Alliance5, e202201570 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bonifas, J. et al. Activation of expression of hedgehog target genes in basal cell carcinomas. J. Invest. Dermatol.116, 739–742 (2001). [DOI] [PubMed] [Google Scholar]
- 85.Yang, D. et al. BMI1 in the heart: novel functions beyond tumorigenesis. EBioMedicine63, 103193 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thompson, E. The many roles of c-Myc in apoptosis. Annu Rev. Physiol.60, 575–600 (1998). [DOI] [PubMed] [Google Scholar]
- 87.Duman-Scheel, M., Weng, L., Xin, S. & Du, W. Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature417, 299–304 (2002). [DOI] [PubMed] [Google Scholar]
- 88.Chuang, P. & McMahon, A. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature397, 617–621 (1999). [DOI] [PubMed] [Google Scholar]
- 89.Bouldin, C. & Harfe, B. Aberrant FGF signaling, independent of ectopic hedgehog signaling, initiates preaxial polydactyly in Dorking chickens. Dev. Biol.334, 133–141 (2009). [DOI] [PubMed] [Google Scholar]
- 90.Sasai, N., Toriyama, M. & Kondo, T. Hedgehog signal and genetic disorders. Front. Genet.10, 1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katoh, Y. & Katoh, M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr. Mol. Med.9, 873–886 (2009). [DOI] [PubMed] [Google Scholar]
- 92.Bigelow, R. L. et al. Transcriptional regulation of bcl-2 mediated by the sonic hedgehog signaling pathway through gli-1. J. Biol. Chem.279, 1197–1205 (2004). [DOI] [PubMed] [Google Scholar]
- 93.Oliver, T. G. et al. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc. Natl Acad. Sci. USA100, 7331–7336 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morrow, D. et al. Sonic Hedgehog induces Notch target gene expression in vascular smooth muscle cells via VEGF-A. Arterioscler Thromb. Vasc. Biol.29, 1112–1118 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Singh, R. R. et al. ABCG2 is a direct transcriptional target of hedgehog signaling and involved in stroma-induced drug tolerance in diffuse large B-cell lymphoma. Oncogene30, 4874–4886 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Teh, M. T. et al. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res.62, 4773–4780 (2002). [PubMed] [Google Scholar]
- 97.Macdonald, R. et al. Midline signalling is required for Pax gene regulation and patterning of the eyes. Development121, 3267–3278 (1995). [DOI] [PubMed] [Google Scholar]
- 98.Katoh, M. & Katoh, M. Notch ligand, JAG1, is evolutionarily conserved target of canonical WNT signaling pathway in progenitor cells. Int. J. Mol. Med.17, 681–685 (2006). [PubMed] [Google Scholar]
- 99.Anvarian, Z. et al. Cellular signalling by primary cilia in development, organ function and disease. Nat. Rev. Nephrol.15, 199–219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Inglis, P., Boroevich, K. & Leroux, M. Piecing together a ciliome. Trends Genet.22, 491–500 (2006). [DOI] [PubMed] [Google Scholar]
- 101.Kim, S. & Dynlacht, B. Assembling a primary cilium. Curr. Opin. Cell Biol.25, 506–511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cui, C. et al. Disruption of Mks1 localization to the mother centriole causes cilia defects and developmental malformations in Meckel-Gruber syndrome. Dis. Model Mech.4, 43–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang, L. et al. Ciliary transition zone proteins coordinate ciliary protein composition and ectosome shedding. Nat. Commun.13, 3997 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Webb, S., Mukhopadhyay, A. & Roberts, A. Intraflagellar transport trains and motors: insights from structure. Semin Cell Dev. Biol.107, 82–90 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ishida, Y., Tasaki, K., Katoh, Y. & Nakayama, K. IFT52Molecular basis underlying the ciliary defects caused by variations found in skeletal ciliopathies. Mol. Biol. Cell33, ar83 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Carpenter, B., Barry, R., Verhey, K. & Allen, B. The heterotrimeric kinesin-2 complex interacts with and regulates GLI protein function. J. Cell Sci.128, 1034–1050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pigino, G. et al. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol.187, 135–148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wingfield, J. et al. In vivo imaging shows continued association of several IFT-A, IFT-B and dynein complexes while IFT trains U-turn at the tip. J. Cell Sci.134, jcs259010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oswald, F., Prevo, B., Acar, S. & Peterman, E. Interplay between ciliary ultrastructure and IFT-train dynamics revealed by single-molecule super-resolution imaging. Cell Rep.25, 224–235 (2018). [DOI] [PubMed] [Google Scholar]
- 110.Huangfu, D. et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature426, 83–87 (2003). [DOI] [PubMed] [Google Scholar]
- 111.Pejskova, P. et al. KIF14 controls ciliogenesis via regulation of Aurora A and is important for Hedgehog signaling. J. Cell Biol.219, e201904107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hildebrandt, F., Benzing, T. & Katsanis, N. Ciliopathies. N. Engl. J. Med.364, 1533–1543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Satir, P. & Christensen, S. Overview of structure and function of mammalian cilia. Annu Rev. Physiol.69, 377–400 (2007). [DOI] [PubMed] [Google Scholar]
- 114.Rohatgi, R., Milenkovic, L. & Scott, M. Patched1 regulates hedgehog signaling at the primary cilium. Science317, 372–376 (2007). [DOI] [PubMed] [Google Scholar]
- 115.Myers, B. et al. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Dev. Cell26, 346–357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rohatgi, R., Milenkovic, L., Corcoran, R. & Scott, M. Hedgehog signal transduction by smoothened: pharmacologic evidence for a 2-step activation process. Proc. Natl Acad. Sci. USA106, 3196–3201 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang, Y., Zhou, Z., Walsh, C. & McMahon, A. Selective translocation of intracellular smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc. Natl Acad. Sci. USA106, 2623–2628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wilson, C., Chen, M. & Chuang, P. Smoothened adopts multiple active and inactive conformations capable of trafficking to the primary cilium. PLoS One4, e5182 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goetz, S., Ocbina, P. & Anderson, K. The primary cilium as a Hedgehog signal transduction machine. Methods Cell Biol.94, 199–222 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hirokawa, N., Tanaka, Y. & Okada, Y. Cilia, KIF3 molecular motor and nodal flow. Curr. Opin. Cell Biol.24, 31–39 (2012). [DOI] [PubMed] [Google Scholar]
- 121.Zhang, J., Liu, Z. & Jia, J. Mechanisms of smoothened regulation in hedgehog signaling. Cells10, 2138 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mukhopadhyay, S. & Rohatgi, R. G-protein-coupled receptors, Hedgehog signaling and primary cilia. Semin Cell Dev. Biol.33, 63–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bachmann, V. et al. Gpr161 anchoring of PKA consolidates GPCR and cAMP signaling. Proc. Natl Acad. Sci. USA113, 7786–7791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cai, E., Zhang, J. & Ge, X. Control of the Hedgehog pathway by compartmentalized PKA in the primary cilium. Sci. China Life Sci.65, 500–514 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Truong, M. et al. Vertebrate cells differentially interpret ciliary and extraciliary cAMP. Cell184, 2911–2926 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pal, K. & Mukhopadhyay, S. Primary cilium and sonic hedgehog signaling during neural tube patterning: role of GPCRs and second messengers. Dev. Neurobiol.75, 337–348 (2015). [DOI] [PubMed] [Google Scholar]
- 127.Happ, J. et al. A PKA inhibitor motif within SMOOTHENED controls Hedgehog signal transduction. Nat. Struct. Mol. Biol.29, 990–999 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu, Y. et al. The PPFIA1-PP2A protein complex promotes trafficking of Kif7 to the ciliary tip and Hedgehog signaling. Sci. Signal7, ra117 (2014). [DOI] [PubMed] [Google Scholar]
- 129.He, M. et al. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol.16, 663–672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Villanueva, H. et al. An essential role for Gα(i2) in smoothened-stimulated epithelial cell proliferation in the mammary gland. Sci. Signal8, ra92 (2015). [DOI] [PubMed] [Google Scholar]
- 131.Ogden, S. et al. G protein Galphai functions immediately downstream of smoothened in Hedgehog signalling. Nature456, 967–970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ogden, S. Genetic evidence for a Smoothened-Gα(i) signaling axis in mammals. Sci. Signal8, fs16 (2015). [DOI] [PubMed] [Google Scholar]
- 133.Yang, C., Chen, W., Chen, Y. & Jiang, J. Smoothened transduces Hedgehog signal by forming a complex with Evc/Evc2. Cell Res.22, 1593–1604 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Endoh-Yamagami, S. et al. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr. Biol.19, 1320–1326 (2009). [DOI] [PubMed] [Google Scholar]
- 135.Raducu, M. et al. SCF (Fbxl17) ubiquitylation of Sufu regulates Hedgehog signaling and medulloblastoma development. EMBO J.35, 1400–1416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fokin Artem, I., Zhapparova Olga, N., Burakov Anton, V. & Nadezhdina Elena, S. Centrosome-derived microtubule radial array, PCM-1 protein, and primary cilia formation. Protoplasma256, 1361–1373 (2019). [DOI] [PubMed] [Google Scholar]
- 137.Li, J. et al. PKA-mediated Gli2 and Gli3 phosphorylation is inhibited by Hedgehog signaling in cilia and reduced in Talpid3 mutant. Dev. Biol.429, 147–157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fernandes-Silva, H., Correia-Pinto, J. & Moura, R. Canonical sonic hedgehog signaling in early lung. Dev. J. Dev. Biol.5, 3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Washington Smoak, I. et al. Sonic hedgehog is required for cardiac outflow tract and neural crest cell development. Dev. Biol.283, 357–372 (2005). [DOI] [PubMed] [Google Scholar]
- 140.Jing, D. et al. The vital role of Gli1 mesenchymal stem cells in tissue development and homeostasis. J. Cell Physiol.236, 6077–6089 (2021). [DOI] [PubMed] [Google Scholar]
- 141.Petrova, R. & Joyner, A. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development141, 3445–3457 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang, J., Andre, P., Ye, L. & Yang, Y. The Hedgehog signalling pathway in bone formation. Int. J. Oral. Sci.7, 73–79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kimura, H., Ng, J. & Curran, T. Transient inhibition of the Hedgehog pathway in young mice causes permanent defects in bone structure. Cancer Cell13, 249–260 (2008). [DOI] [PubMed] [Google Scholar]
- 144.Yang, S. & Wang, C. The intraflagellar transport protein IFT80 is required for cilia formation and osteogenesis. Bone51, 407–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zou, S. et al. Mesenchymal stem cells overexpressing Ihh promote bone repair. J. Orthop. Surg. Res.9, 102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Armstrong, B., Henner, A., Stewart, S. & Stankunas, K. Shh promotes direct interactions between epidermal cells and osteoblast progenitors to shape regenerated zebrafish bone. Development144, 1165–1176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhao, H. et al. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat. Cell Biol.17, 386–396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shi, Y. et al. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun.8, 2043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao, H. & Chai, Y. Stem cells in teeth and craniofacial bones. J. Dent. Res.94, 1495–1501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Guo, Y. et al. BMP-IHH-mediated interplay between mesenchymal stem cells and osteoclasts supports calvarial bone homeostasis and repair. Bone Res.6, 30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lepper, C., Partridge, T. & Fan, C. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development138, 3639–3646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yin, H., Price, F. & Rudnicki, M. Satellite cells and the muscle stem cell niche. Physiol. Rev.93, 23–67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Straface, G. et al. Sonic hedgehog regulates angiogenesis and myogenesis during post-natal skeletal muscle regeneration. J. Cell Mol. Med.13, 2424–2435 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Koleva, M. et al. Pleiotropic effects of sonic hedgehog on muscle satellite cells. Cell Mol. Life Sci.62, 1863–1870 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Elia, D. et al. Sonic hedgehog promotes proliferation and differentiation of adult muscle cells: involvement of MAPK/ERK and PI3K/Akt pathways. Biochim. Biophys. Acta1773, 1438–1446 (2007). [DOI] [PubMed] [Google Scholar]
- 156.Madhala-Levy, D. et al. Cooperation between Shh and IGF-I in promoting myogenic proliferation and differentiation via the MAPK/ERK and PI3K/Akt pathways requires Smo activity. J. Cell Physiol.227, 1455–1464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Anderson, C. et al. Sonic hedgehog acts cell-autonomously on muscle precursor cells to generate limb muscle diversity. Genes Dev.26, 2103–2117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Voronova, A. et al. Hedgehog signaling regulates MyoD expression and activity. J. Biol. Chem.288, 4389–4404 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Devakanmalai, G., Zumrut, H. & Ozbudak, E. Cited3 activates Mef2c to control muscle cell differentiation and survival. Biol. Open2, 505–514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kramann, R. et al. Adventitial MSC-like cells are progenitors of vascular smooth muscle cells and drive vascular calcification in chronic kidney disease. Cell Stem Cell19, 628–642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Boras-Granic, K., Chang, H., Grosschedl, R. & Hamel, P. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev. Biol.295, 219–231 (2006). [DOI] [PubMed] [Google Scholar]
- 162.Sun, X. et al. Coordinated hedgehog signaling induces new hair follicles in adult skin. Elife9, e46756 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Parisi, M. & Lin, H. The role of the hedgehog/patched signaling pathway in epithelial stem cell proliferation: from fly to human. Cell Res.8, 15–21 (1998). [DOI] [PubMed] [Google Scholar]
- 164.Adolphe, C. et al. An in vivo comparative study of sonic, desert and Indian hedgehog reveals that hedgehog pathway activity regulates epidermal stem cell homeostasis. Development131, 5009–5019 (2004). [DOI] [PubMed] [Google Scholar]
- 165.Zhou, J. et al. Role of sonic hedgehog in maintaining a pool of proliferating stem cells in the human fetal epidermis. Hum. Reprod.21, 1698–1704 (2006). [DOI] [PubMed] [Google Scholar]
- 166.Karlsson, L., Bondjers, C. & Betsholtz, C. Roles for PDGF-A and sonic hedgehog in development of mesenchymal components of the hair follicle. Development126, 2611–2621 (1999). [DOI] [PubMed] [Google Scholar]
- 167.Oro, A. & Higgins, K. Hair cycle regulation of Hedgehog signal reception. Dev. Biol.255, 238–248 (2003). [DOI] [PubMed] [Google Scholar]
- 168.Vidal, V. et al. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr. Biol.15, 1340–1351 (2005). [DOI] [PubMed] [Google Scholar]
- 169.Huelsken, J. et al. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell105, 533–545 (2001). [DOI] [PubMed] [Google Scholar]
- 170.Santos, A., Lo, Y., Mah, A. & Kuo, C. The intestinal stem cell niche: homeostasis and adaptations. Trends Cell Biol.28, 1062–1078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kosinski, C. et al. Indian hedgehog regulates intestinal stem cell fate through epithelial-mesenchymal interactions during development. Gastroenterology139, 893–903 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Degirmenci, B. et al. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature558, 449–453 (2018). [DOI] [PubMed] [Google Scholar]
- 173.Coquenlorge, S. et al. GLI2 modulated by SUFU and SPOP induces intestinal stem cell niche signals in development and tumorigenesis. Cell Rep.27, 3006–3018 (2019). [DOI] [PubMed] [Google Scholar]
- 174.Xie, Z. et al. Emerging roles of the Hedgehog signalling pathway in inflammatory bowel disease. Cell Death Discov.7, 314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lees, C. et al. Analysis of germline GLI1 variation implicates hedgehog signalling in the regulation of intestinal inflammatory pathways. PLoS Med.5, e239 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.van Dop, W. et al. Loss of Indian Hedgehog activates multiple aspects of a wound healing response in the mouse intestine. Gastroenterology139, 1665–1676 (2010). 1676.e1661-1610. [DOI] [PubMed] [Google Scholar]
- 177.Westendorp, B. et al. Indian Hedgehog suppresses a stromal cell-driven intestinal immune response. Cell Mol. Gastroenterol. Hepatol.5, 67–82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lee, J. et al. Control of inflammation by stromal Hedgehog pathway activation restrains colitis. Proc. Natl Acad. Sci. USA113, E7545–E7553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang, C., Cassandras, M. & Peng, T. The role of hedgehog signaling in adult lung regeneration and maintenance. J. Dev. Biol.7, 14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Peng, T. et al. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature526, 578–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu, L. et al. Hedgehog signaling in neonatal and adult lung. Am. J. Respir. Cell Mol. Biol.48, 703–710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Seidel, K. et al. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development137, 3753–3761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhao, H. et al. Secretion of shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell14, 160–173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Men, Y. et al. Gli1+ periodontium stem cells are regulated by osteocytes and occlusal force. Dev. Cell54, 639–654.e636 (2020). [DOI] [PubMed] [Google Scholar]
- 185.Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 186.Bach, D., Zhang, W. & Sood, A. Chromosomal instability in tumor initiation and development. Cancer Res.79, 3995–4002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bajaj, J., Diaz, E. & Reya, T. Stem cells in cancer initiation and progression. J. Cell Biol.219, e201911053 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zeng, X. & Ju, D. Hedgehog signaling pathway and autophagy in cancer. Int. J. Mol. Sci.19, 2279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hanna, A. & Shevde, L. Hedgehog signaling: modulation of cancer properies and tumor mircroenvironment. Mol. Cancer15, 24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Carballo, G., Honorato, J., de Lopes, G. & Spohr, T. A highlight on sonic hedgehog pathway. Cell Commun. Signal16, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yang, L. et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target Ther.5, 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Valkenburg, K. C., de Groot, A. E. & Pienta, K. J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol.15, 366–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ertao, Z. et al. Autocrine sonic hedgehog signaling promotes gastric cancer proliferation through induction of phospholipase Cγ1 and the ERK1/2 pathway. J. Exp. Clin. Cancer Res.35, 63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Singh, S. et al. Hedgehog-producing cancer cells respond to and require autocrine Hedgehog activity. Cancer Res.71, 4454–4463 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Skoda, A. M. et al. The role of the Hedgehog signaling pathway in cancer: a comprehensive review. Bosn. J. Basic Med. Sci.18, 8–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yauch, R. L. et al. A paracrine requirement for hedgehog signalling in cancer. Nature455, 406–410 (2008). [DOI] [PubMed] [Google Scholar]
- 197.Neville, B. & Day, T. Oral cancer and precancerous lesions. CA Cancer J. Clin.52, 195–215 (2002). [DOI] [PubMed] [Google Scholar]
- 198.Hasegawa, M. et al. Differential immunohistochemical expression profiles of perlecan-binding growth factors in epithelial dysplasia, carcinoma in situ, and squamous cell carcinoma of the oral mucosa. Pathol. Res. Pract.212, 426–436 (2016). [DOI] [PubMed] [Google Scholar]
- 199.Gonzalez, A. et al. Immunohistochemical evaluation of hedgehog signalling in epithelial/mesenchymal interactions in squamous cell carcinoma transformation: a pilot study. J. Oral. Pathol. Med.45, 173–179 (2016). [DOI] [PubMed] [Google Scholar]
- 200.Guimaraes, V. et al. Hedgehog pathway activation in oral squamous cell carcinoma: cancer-associated fibroblasts exhibit nuclear GLI-1 localization. J. Mol. Histol.51, 675–684 (2020). [DOI] [PubMed] [Google Scholar]
- 201.Takabatake, K. et al. The role of sonic hedgehog signaling in the tumor microenvironment of oral squamous cell carcinoma. Int. J. Mol. Sci.20, 5779 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cierpikowski, P., Lis-Nawara, A. & Bar, J. SHH expression is significantly associated with cancer stem cell markers in oral squamous cell carcinoma. Anticancer Res.41, 5405–5413 (2021). [DOI] [PubMed] [Google Scholar]
- 203.Wang, Y. et al. Expression of hedgehog signaling molecules as a prognostic indicator of oral squamous cell carcinoma. Head Neck34, 1556–1561 (2012). [DOI] [PubMed] [Google Scholar]
- 204.Rodrigues, M. et al. GLI3 knockdown decreases stemness, cell proliferation and invasion in oral squamous cell carcinoma. Int. J. Oncol.53, 2458–2472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kuroda, H. et al. Oral squamous cell carcinoma-derived sonic hedgehog promotes angiogenesis. Anticancer Res.37, 6731–6737 (2017). [DOI] [PubMed] [Google Scholar]
- 206.Chen, G., Yan, M., Li, R. & Chen, W. Sonic hedgehog signalling activation contributes to ALCAM over-expression and poor clinical outcome in patients with oral squamous cell carcinoma. Chin. J. Dent. Res.21, 31–40 (2018). [DOI] [PubMed] [Google Scholar]
- 207.Fan, H. et al. Sonic hedgehog signaling may promote invasion and metastasis of oral squamous cell carcinoma by activating MMP-9 and E-cadherin expression. Med. Oncol.31, 41 (2014). [DOI] [PubMed] [Google Scholar]
- 208.Shear, M. Odontogenic keratocysts: clinical features. Oral. Maxillofac. Surg. Clin. North Am.15, 335–345 (2003). [DOI] [PubMed] [Google Scholar]
- 209.Hashmi, A. et al. Mutiple keratocystic odontogenic tumors (KCOT) in a patient with Gorlin syndrome: a case report with late presentation and absence of skin manifestations. BMC Res. Notes9, 357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gorlin, R. & Goltz, R. Multiple nevoid basal-cell epithelioma, jaw cysts and bifid rib. A syndrome. N. Engl. J. Med.262, 908–912 (1960). [DOI] [PubMed] [Google Scholar]
- 211.Onodera, S., Nakamura, Y. & Azuma, T. Gorlin syndrome: recent advances in genetic testing and molecular and cellular biological research. Int. J. Mol. Sci.21, 7559 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Qu, J. et al. Underestimated PTCH1 mutation rate in sporadic keratocystic odontogenic tumors. Oral. Oncol.51, 40–45 (2015). [DOI] [PubMed] [Google Scholar]
- 213.Stojanov, I. et al. Biallelic PTCH1 inactivation is a dominant genomic change in sporadic keratocystic odontogenic tumors. Am. J. Surg. Pathol.44, 553–560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yu, F. et al. The large intracellular loop of ptch1 mediates the non-canonical Hedgehog pathway through cyclin B1 in nevoid basal cell carcinoma syndrome. Int. J. Mol. Med.34, 507–512 (2014). [DOI] [PubMed] [Google Scholar]
- 215.Barreto, D., Bale, A., De Marco, L. & Gomez, R. Immunolocalization of PTCH protein in odontogenic cysts and tumors. J. Dent. Res.81, 757–760 (2002). [DOI] [PubMed] [Google Scholar]
- 216.Ohki, K. et al. PTC gene mutations and expression of SHH, PTC, SMO, and GLI-1 in odontogenic keratocysts. Int. J. Oral. Maxillofac. Surg.33, 584–592 (2004). [DOI] [PubMed] [Google Scholar]
- 217.Grachtchouk, M. et al. Odontogenic keratocysts arise from quiescent epithelial rests and are associated with deregulated hedgehog signaling in mice and humans. Am. J. Pathol.169, 806–814 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gurgel, C. et al. Transcriptional profiles of SHH pathway genes in keratocystic odontogenic tumor and ameloblastoma. J. Oral. Pathol. Med.43, 619–626 (2014). [DOI] [PubMed] [Google Scholar]
- 219.Hoyos Cadavid, A. et al. Immunohistochemical evaluation of Sonic Hedgehog signaling pathway proteins (Shh, Ptch1, Ptch2, Smo, Gli1, Gli2, and Gli3) in sporadic and syndromic odontogenic keratocysts. Clin. Oral. Investig.23, 153–159 (2019). [DOI] [PubMed] [Google Scholar]
- 220.Yagyuu, T. et al. Recurrence of keratocystic odontogenic tumor: clinicopathological features and immunohistochemical study of the Hedgehog signaling pathway. Pathobiology75, 171–176 (2008). [DOI] [PubMed] [Google Scholar]
- 221.Li, X. et al. The role of Shh signalling pathway in central nervous system development and related diseases. Cell Biochem. Funct.39, 180–189 (2021). [DOI] [PubMed] [Google Scholar]
- 222.Rolland, A. & Aquilina, K. Surgery for recurrent medulloblastoma: a review. Neurochirurgie67, 69–75 (2021). [DOI] [PubMed] [Google Scholar]
- 223.Garcia-Lopez, J., Kumar, R., Smith, K. & Northcott, P. Deconstructing sonic hedgehog medulloblastoma: molecular subtypes, drivers, and beyond. Trends Genet.37, 235–250 (2021). [DOI] [PubMed] [Google Scholar]
- 224.Chen, L. et al. O-GlcNAcylation promotes cerebellum development and medulloblastoma oncogenesis via SHH signaling. Proc. Natl Acad. Sci. USA119, e2202821119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kumar, V. et al. Challenges and recent advances in medulloblastoma therapy. Trends Pharmacol. Sci.38, 1061–1084 (2017). [DOI] [PubMed] [Google Scholar]
- 226.Northcott, P. et al. Medulloblastomics: the end of the beginning. Nat. Rev. Cancer12, 818–834 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kool, M. et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell25, 393–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Yang, Z. et al. Medulloblastoma can be initiated by deletion of patched in lineage-restricted progenitors or stem cells. Cancer Cell14, 135–145 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Selvadurai, H. et al. Medulloblastoma arises from the persistence of a rare and transient Sox2 granule neuron precursor. Cell Rep.31, 107511 (2020). [DOI] [PubMed] [Google Scholar]
- 230.Liu, Y. et al. Astrocytes promote medulloblastoma progression through hedgehog secretion. Cancer Res.77, 6692–6703 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kieran, M. Targeted treatment for sonic hedgehog-dependent medulloblastoma. Neuro Oncol.16, 1037–1047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Vanner, R. et al. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell26, 33–47 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Fine, H. Malignant gliomas: simplifying the complexity. Cancer Discov.9, 1650–1652 (2019). [DOI] [PubMed] [Google Scholar]
- 234.Mittal, S., Pradhan, S. & Srivastava, T. Recent advances in targeted therapy for glioblastoma. Expert Rev. Neurother.15, 935–946 (2015). [DOI] [PubMed] [Google Scholar]
- 235.Cherepanov, S. et al. Effect of hedgehog signaling pathway activation on proliferation of high-grade gliomas. Bull. Exp. Biol. Med.161, 674–678 (2016). [DOI] [PubMed] [Google Scholar]
- 236.Henao-Restrepo, J. et al. Expression of activator proteins of SHH/GLI and PI3K/Akt/mTORC1 signaling pathways in human gliomas is associated with high grade tumors. Exp. Mol. Pathol.122, 104673 (2021). [DOI] [PubMed] [Google Scholar]
- 237.Wang, H. et al. Hedgehog signaling regulates the development and treatment of glioblastoma. Oncol. Lett.24, 294 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Torrisi, F. et al. Connexin 43 and sonic hedgehog pathway interplay in glioblastoma cell proliferation and migration. Biol. (Basel)10, 767 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Chang, L. et al. Activation of sonic hedgehog signaling enhances cell migration and invasion by induction of matrix metalloproteinase-2 and -9 via the phosphoinositide-3 kinase/AKT signaling pathway in glioblastoma. Mol. Med. Rep.12, 6702–6710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jiang, K. et al. Fms related tyrosine kinase 1 (Flt1) functions as an oncogene and regulates glioblastoma cell metastasis by regulating sonic hedgehog signaling. Am. J. Cancer Res.7, 1164–1176 (2017). [PMC free article] [PubMed] [Google Scholar]
- 241.Shahi, M., Lorente, A. & Castresana, J. Hedgehog signalling in medulloblastoma, glioblastoma and neuroblastoma. Oncol. Rep.19, 681–688 (2008). [PubMed] [Google Scholar]
- 242.Chen, S. et al. HIF-1α contributes to proliferation and invasiveness of neuroblastoma cells via SHH signaling. PloS One10, e0121115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Souzaki, R. et al. Hedgehog signaling pathway in neuroblastoma differentiation. J. Pediatr. Surg.45, 2299–2304 (2010). [DOI] [PubMed] [Google Scholar]
- 244.Xu, L. et al. Sonic Hedgehog pathway is essential for neuroblastoma cell proliferation and tumor growth. Mol. Cell Biochem.364, 235–241 (2012). [DOI] [PubMed] [Google Scholar]
- 245.Koeniger, A. et al. Activation of cilia-independent hedgehog/GLI1 signaling as a novel concept for neuroblastoma therapy. Cancers (Basel)13, 1908 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Siegel, R., Miller, K., Fuchs, H. & Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin.71, 7–33 (2021). [DOI] [PubMed] [Google Scholar]
- 247.Signoretti, S., Flaifel, A., Chen, Y. & Reuter, V. Renal cell carcinoma in the era of precision medicine: from molecular pathology to tissue-based biomarkers. J. Clin. Oncol.36, JCO2018792259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Dormoy, V. et al. The sonic hedgehog signaling pathway is reactivated in human renal cell carcinoma and plays orchestral role in tumor growth. Mol. Cancer8, 123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Kotulak-Chrzaszcz, A. et al. Expression of the Sonic Hedgehog pathway components in clear cell renal cell carcinoma. Oncol. Lett.18, 5801–5810 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Jäger, W. et al. DHH is an independent prognosticator of oncologic outcome of clear cell renal cell carcinoma. J. Urol.192, 1842–1848 (2014). [DOI] [PubMed] [Google Scholar]
- 251.D’Amato, C. et al. Inhibition of Hedgehog signalling by NVP-LDE225 (Erismodegib) interferes with growth and invasion of human renal cell carcinoma cells. Br. J. Cancer111, 1168–1179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Dormoy, V. et al. Vitamin D3 triggers antitumor activity through targeting hedgehog signaling in human renal cell carcinoma. Carcinogenesis33, 2084–2093 (2012). [DOI] [PubMed] [Google Scholar]
- 253.Behnsawy, H. et al. Possible role of sonic hedgehog and epithelial-mesenchymal transition in renal cell cancer progression. Korean J. Urol.54, 547–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zhou, J. et al. Reciprocal regulation of hypoxia-inducible factor 2α and GLI1 expression associated with the radioresistance of renal cell carcinoma. Int. J. Radiat. Oncol. Biol. Phys.90, 942–951 (2014). [DOI] [PubMed] [Google Scholar]
- 255.Furukawa, J., Miyake, H. & Fujisawa, M. GLI2 expression levels in radical nephrectomy specimens as a predictor of disease progression in patients with metastatic clear cell renal cell carcinoma following treatment with sunitinib. Mol. Clin. Oncol.5, 186–192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hyuga, T. et al. Hedgehog signaling for urogenital organogenesis and prostate cancer: an implication for the epithelial-mesenchyme interaction (EMI). Int. J. Mol. Sci.21, 58 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Odero-Marah, V., Hawsawi, O., Henderson, V. & Sweeney, J. Epithelial-mesenchymal transition (EMT) and prostate cancer. Adv. Exp. Med. Biol.1095, 101–110 (2018). [DOI] [PubMed] [Google Scholar]
- 258.Wilkinson, S. et al. Hedgehog signaling is active in human prostate cancer stroma and regulates proliferation and differentiation of adjacent epithelium. Prostate73, 1810–1823 (2013). [DOI] [PubMed] [Google Scholar]
- 259.Sanchez, P. et al. Inhibition of prostate cancer proliferation by interference with Sonic Hedgehog-GLI1 signaling. Proc. Natl Acad. Sci. USA101, 12561–12566 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kim, T. et al. Hedgehog signaling protein expression and its association with prognostic parameters in prostate cancer: a retrospective study from the view point of new 2010 anatomic stage/prognostic groups. J. Surg. Oncol.104, 472–479 (2011). [DOI] [PubMed] [Google Scholar]
- 261.Sheng, T. et al. Activation of the hedgehog pathway in advanced prostate cancer. Mol. Cancer3, 29 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Chang, H. et al. Hedgehog overexpression leads to the formation of prostate cancer stem cells with metastatic property irrespective of androgen receptor expression in the mouse model. J. Biomed. Sci.18, 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Acikgoz, E. et al. Sonic hedgehog signaling is associated with resistance to zoledronic acid in CD133high/CD44high prostate cancer stem cells. Mol. Biol. Rep.48, 3567–3578 (2021). [DOI] [PubMed] [Google Scholar]
- 264.Cai, H. et al. Sonic hedgehog signaling pathway mediates development of hepatocellular carcinoma. Tumour Biol.37, 16199–16205 (2016). [DOI] [PubMed] [Google Scholar]
- 265.Machado, M. & Diehl, A. Hedgehog signalling in liver pathophysiology. J. Hepatol.68, 550–562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Dugum, M. et al. Sonic hedgehog signaling in hepatocellular carcinoma: a pilot study. Mol. Clin. Oncol.4, 369–374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Sicklick, J. et al. Dysregulation of the Hedgehog pathway in human hepatocarcinogenesis. Carcinogenesis27, 748–757 (2006). [DOI] [PubMed] [Google Scholar]
- 268.Jeng, K. et al. High expression of patched homolog-1 messenger RNA and glioma-associated oncogene-1 messenger RNA of sonic hedgehog signaling pathway indicates a risk of postresection recurrence of hepatocellular carcinoma. Ann. Surg. Oncol.20, 464–473 (2013). [DOI] [PubMed] [Google Scholar]
- 269.Chen, J. et al. Sonic hedgehog signaling pathway induces cell migration and invasion through focal adhesion kinase/AKT signaling-mediated activation of matrix metalloproteinase (MMP)-2 and MMP-9 in liver cancer. Carcinogenesis34, 10–19 (2013). [DOI] [PubMed] [Google Scholar]
- 270.Fan, Y. et al. Aberrant hedgehog signaling is responsible for the highly invasive behavior of a subpopulation of hepatoma cells. Oncogene35, 116–124 (2016). [DOI] [PubMed] [Google Scholar]
- 271.Chen, X. et al. Epithelial mesenchymal transition and hedgehog signaling activation are associated with chemoresistance and invasion of hepatoma subpopulations. J. Hepatol.55, 838–845 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Yamada, S. et al. Epithelial to mesenchymal transition is associated with shorter disease-free survival in hepatocellular carcinoma. Ann. Surg. Oncol.21, 3882–3890 (2014). [DOI] [PubMed] [Google Scholar]
- 273.Wang, S. et al. Hedgehog signaling promotes sorafenib resistance in hepatocellular carcinoma patient-derived organoids. J. Exp. Clin. Cancer Res.39, 22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Zhou, X. et al. Hedgehog signalling mediates drug resistance through targeting TAP1 in hepatocellular carcinoma. J. Cell Mol. Med.24, 4298–4311 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Razumilava, N. & Gores, G. Cholangiocarcinoma. Lancet383, 2168–2179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Riedlinger, D. et al. Hedgehog pathway as a potential treatment target in human cholangiocarcinoma. J. Hepatobiliary Pancreat. Sci.21, 607–615 (2014). [DOI] [PubMed] [Google Scholar]
- 277.Anichini, G. et al. The Role of the Hedgehog Pathway in Cholangiocarcinoma. Cancers (Basel)13, 4774 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Tang, L. et al. The prognostic significance and therapeutic potential of hedgehog signaling in intrahepatic cholangiocellular carcinoma. Clin. Cancer Res.19, 2014–2024 (2013). [DOI] [PubMed] [Google Scholar]
- 279.Jing, X. et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer18, 157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Bhuria, V. et al. Hypoxia induced Sonic Hedgehog signaling regulates cancer stemness, epithelial-to-mesenchymal transition and invasion in cholangiocarcinoma. Exp. Cell Res.385, 111671 (2019). [DOI] [PubMed] [Google Scholar]
- 281.Kim, Y. et al. Hedgehog signaling between cancer cells and hepatic stellate cells in promoting cholangiocarcinoma. Ann. Surg. Oncol.21, 2684–2698 (2014). [DOI] [PubMed] [Google Scholar]
- 282.Chen, Z. et al. SHH/GLI2-TGF-β1 feedback loop between cancer cells and tumor-associated macrophages maintains epithelial-mesenchymal transition and endoplasmic reticulum homeostasis in cholangiocarcinoma. Pharmacol. Res.187, 106564 (2023). [DOI] [PubMed] [Google Scholar]
- 283.Gerling, M. et al. Stromal Hedgehog signalling is downregulated in colon cancer and its restoration restrains tumour growth. Nat. Commun.7, 12321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Fu, X. et al. Expression of Indian hedgehog is negatively correlated with APC gene mutation in colorectal tumors. Int. J. Clin. Exp. Med.7, 2150–2155 (2014). [PMC free article] [PubMed] [Google Scholar]
- 285.Fu, X. et al. Opposite expression patterns of Sonic hedgehog and Indian hedgehog are associated with aberrant methylation status of their promoters in colorectal cancers. Pathology42, 553–559 (2010). [DOI] [PubMed] [Google Scholar]
- 286.Büller, N. et al. Stromal Indian hedgehog signaling is required for intestinal adenoma formation in mice. Gastroenterology148, 170–180.e176 (2015). [DOI] [PubMed] [Google Scholar]
- 287.Tang, Y. A. et al. Hypoxic tumor microenvironment activates GLI2 via HIF-1α and TGF-β2 to promote chemoresistance in colorectal cancer. Proc. Natl Acad. Sci. USA115, E5990–E5999 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Klein, A. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat. Rev. Gastroenterol. Hepatol.18, 493–502 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Thayer, S. et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature425, 851–856 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Maréchal, R. et al. Sonic hedgehog and Gli1 expression predict outcome in resected pancreatic adenocarcinoma. Clin. Cancer Res.21, 1215–1224 (2015). [DOI] [PubMed] [Google Scholar]
- 291.Walter, K. et al. Overexpression of smoothened activates the sonic hedgehog signaling pathway in pancreatic cancer-associated fibroblasts. Clin. Cancer Res.16, 1781–1789 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Inaguma, S., Kasai, K. & Ikeda, H. GLI1 facilitates the migration and invasion of pancreatic cancer cells through MUC5AC-mediated attenuation of E-cadherin. Oncogene30, 714–723 (2011). [DOI] [PubMed] [Google Scholar]
- 293.Xu, X. et al. Sonic hedgehog-Gli1 signaling pathway regulates the epithelial mesenchymal transition (EMT) by mediating a new target gene, S100A4, in pancreatic cancer cells. PloS One9, e96441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lee, J. et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc. Natl Acad. Sci. USA111, E3091–E3100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Yang, Y. et al. Relationship between autophagy and perineural invasion, clinicopathological features, and prognosis in pancreatic cancer. World J. Gastroenterol.23, 7232–7241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Bapat, A., Hostetter, G., Von Hoff, D. & Han, H. Perineural invasion and associated pain in pancreatic cancer. Nat. Rev. Cancer11, 695–707 (2011). [DOI] [PubMed] [Google Scholar]
- 297.Li, X. et al. Sonic hedgehog paracrine signaling activates stromal cells to promote perineural invasion in pancreatic cancer. Clin. Cancer Res.20, 4326–4338 (2014). [DOI] [PubMed] [Google Scholar]
- 298.Ma, C. et al. Molecular mechanisms involving the sonic hedgehog pathway in lung cancer therapy: recent advances. Front. Oncol.12, 729088 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Bermudez, O. et al. Gli1 mediates lung cancer cell proliferation and Sonic Hedgehog-dependent mesenchymal cell activation. PloS One8, e63226 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Li, H. et al. Gli promotes epithelial-mesenchymal transition in human lung adenocarcinomas. Oncotarget7, 80415–80425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Yue, D. et al. Hedgehog/Gli promotes epithelial-mesenchymal transition in lung squamous cell carcinomas. J. Exp. Clin. Cancer Res.33, 34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Bai, X. et al. Blockade of hedgehog signaling synergistically increases sensitivity to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer cell lines. PloS One11, e0149370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Lin, E. et al. Hedgehog pathway maintains cell survival under stress conditions, and drives drug resistance in lung adenocarcinoma. Oncotarget7, 24179–24193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Seidl, C. et al. MicroRNA-182-5p regulates hedgehog signaling pathway and chemosensitivity of cisplatin-resistant lung adenocarcinoma cells via targeting GLI2. Cancer Lett.469, 266–276 (2020). [DOI] [PubMed] [Google Scholar]
- 305.Giroux Leprieur, E. et al. Membrane-bound full-length Sonic Hedgehog identifies cancer stem cells in human non-small cell lung cancer. Oncotarget8, 103744–103757 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.de Cássia Viu Carrara, R. et al. Expression differences of genes in the PI3K/AKT, WNT/b-catenin, SHH, NOTCH and MAPK signaling pathways in CD34+ hematopoietic cells obtained from chronic phase patients with chronic myeloid leukemia and from healthy controls. Clin. Transl. Oncol.20, 542–549 (2018). [DOI] [PubMed] [Google Scholar]
- 307.Long, B. et al. Activation of the Hedgehog pathway in chronic myelogeneous leukemia patients. J. Exp. Clin. Cancer Res.30, 8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Babashah, S. et al. Targeting of the signal transducer Smo links microRNA-326 to the oncogenic Hedgehog pathway in CD34 + CML stem/progenitor cells. Int. J. Cancer133, 579–589 (2013). [DOI] [PubMed] [Google Scholar]
- 309.Dierks, C. et al. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell14, 238–249 (2008). [DOI] [PubMed] [Google Scholar]
- 310.Sadarangani, A. et al. GLI2 inhibition abrogates human leukemia stem cell dormancy. J. Transl. Med.13, 98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Wellbrock, J. et al. Expression of hedgehog pathway mediator GLI represents a negative prognostic marker in human acute myeloid leukemia and its inhibition exerts antileukemic effects. Clin. Cancer Res.21, 2388–2398 (2015). [DOI] [PubMed] [Google Scholar]
- 312.Lau, B. et al. Hedgehog/GLI1 activation leads to leukemic transformation of myelodysplastic syndrome in vivo and GLI1 inhibition results in antitumor activity. Oncogene38, 687–698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Kobune, M. et al. Stromal cells expressing hedgehog-interacting protein regulate the proliferation of myeloid neoplasms. Blood Cancer J.2, e87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Li, X. et al. Gli-1/PI3K/AKT/NF-kB pathway mediates resistance to radiation and is a target for reversion of responses in refractory acute myeloid leukemia cells. Oncotarget7, 33004–33015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Hegde, G. et al. Hedgehog-induced survival of B-cell chronic lymphocytic leukemia cells in a stromal cell microenvironment: a potential new therapeutic target. Mol. Cancer Res.6, 1928–1936 (2008). [DOI] [PubMed] [Google Scholar]
- 316.Dagklis, A. et al. Hedgehog pathway activation in T-cell acute lymphoblastic leukemia predicts response to SMO and GLI1 inhibitors. Blood128, 2642–2654 (2016). [DOI] [PubMed] [Google Scholar]
- 317.Peacock, C. et al. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc. Natl Acad. Sci. USA104, 4048–4053 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Ghia, E. et al. Activation of hedgehog signaling associates with early disease progression in chronic lymphocytic leukemia. Blood133, 2651–2663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Fagin, J. & Wells, S. Biologic and clinical perspectives on thyroid cancer. N. Engl. J. Med.375, 1054–1067 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Xu, X. et al. Activation of the Sonic Hedgehog pathway in thyroid neoplasms and its potential role in tumor cell proliferation. Endocr. Relat. Cancer19, 167–179 (2012). [DOI] [PubMed] [Google Scholar]
- 321.Williamson, A. et al. The sonic hedgehog signaling pathway stimulates anaplastic thyroid cancer cell motility and invasiveness by activating Akt and c-Met. Oncotarget7, 10472–10485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Bian, X. et al. Expression and clinical significance of Shh/Gli-1 in papillary thyroid carcinoma. Tumour Biol.35, 10523–10528 (2014). [DOI] [PubMed] [Google Scholar]
- 323.Lee, J. et al. GLI1 transcription factor affects tumor aggressiveness in patients with papillary thyroid cancers. Med. (Baltim.)94, e998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Heiden, K. et al. The sonic hedgehog signaling pathway maintains the cancer stem cell self-renewal of anaplastic thyroid cancer by inducing snail expression. J. Clin. Endocrinol. Metab.99, E2178–E2187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Ma, C., Ma, X., Li, Y. & Fu, S. The role of primary cilia in thyroid cancer: from basic research to clinical applications. Front. Endocrinol. (Lausanne)12, 685228 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Rubin, A., Chen, E. & Ratner, D. Basal-cell carcinoma. N. Engl. J. Med.353, 2262–2269 (2005). [DOI] [PubMed] [Google Scholar]
- 327.Xie, J. et al. Activating smoothened mutations in sporadic basal-cell carcinoma. Nature391, 90–92 (1998). [DOI] [PubMed] [Google Scholar]
- 328.Gailani, M. et al. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat. Genet.14, 78–81 (1996). [DOI] [PubMed] [Google Scholar]
- 329.Deng, L. et al. Expression of hedgehog signaling pathway proteins in basal cell carcinoma: clinicopathologic study. Clin. Cosmet. Investig. Dermatol.15, 2353–2361 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Moisejenko-Golubovica, J., Volkov, O., Ivanova, A. & Groma, V. Analysis of the occurrence and distribution of primary and recurrent basal cell carcinoma of head and neck coupled to the assessment of tumor microenvironment and Sonic hedgehog signaling. Rom. J. Morphol. Embryol.61, 821–831 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Kim, H. et al. Expression profile of sonic hedgehog signaling-related molecules in basal cell carcinoma. PloS One14, e0225511 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Sánchez-Danés, A. et al. Defining the clonal dynamics leading to mouse skin tumour initiation. Nature536, 298–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Kuonen, F. et al. Loss of primary cilia drives switching from hedgehog to Ras/MAPK pathway in resistant basal cell carcinoma. J. Invest. Dermatol.139, 1439–1448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Jacquet, A., Dormoy, V., Lorenzato, M. & Durlach, A. Preliminary results on a proposed histopathological assessment of predictive factors for basal cell carcinoma recurrence after primary free margin excision. Ski. Health Dis.2, e88 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Whitson, R. et al. Noncanonical hedgehog pathway activation through SRF-MKL1 promotes drug resistance in basal cell carcinomas. Nat. Med.24, 271–281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Riobo-Del Galdo, N. A., Lara Montero, Á. & Wertheimer, E. V. Role of hedgehog signaling in breast cancer: pathogenesis and therapeutics. Cells8, 375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.McDermott, K. M., Liu, B. Y., Tlsty, T. D. & Pazour, G. J. Primary cilia regulate branching morphogenesis during mammary gland development. Curr. Biol.20, 731–737 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Qayoom, H., Wani, N. A., Alshehri, B. & Mir, M. A. An insight into the cancer stem cell survival pathways involved in chemoresistance in triple-negative breast cancer. Future Oncol.17, 4185–4206 (2021). [DOI] [PubMed] [Google Scholar]
- 339.Bhateja, P., Cherian, M., Majumder, S. & Ramaswamy, B. The hedgehog signaling pathway: a viable target in breast cancer? Cancers (Basel)11, 1126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Riaz, S. K. et al. Involvement of hedgehog pathway in early onset, aggressive molecular subtypes and metastatic potential of breast cancer. Cell Commun. Signal16, 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Tao, Y., Mao, J., Zhang, Q. & Li, L. Overexpression of Hedgehog signaling molecules and its involvement in triple-negative breast cancer. Oncol. Lett.2, 995–1001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Noman, A. S. et al. Overexpression of sonic hedgehog in the triple negative breast cancer: clinicopathological characteristics of high burden breast cancer patients from Bangladesh. Sci. Rep.6, 18830 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Di Mauro, C. et al. Hedgehog signalling pathway orchestrates angiogenesis in triple-negative breast cancers. Br. J. Cancer116, 1425–1435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Reyes-Ramos, A. M. et al. Mesenchymal cells support the oncogenicity and therapeutic response of the hedgehog pathway in triple-negative breast cancer. Cancers (Basel)11, 1522 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Cazet, A. et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat. Commun.9, 2897 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Sims-Mourtada, J. et al. Taxane-induced hedgehog signaling is linked to expansion of breast cancer stem-like populations after chemotherapy. Mol. Carcinog.54, 1480–1493 (2015). [DOI] [PubMed] [Google Scholar]
- 347.Zarzosa, P. et al. Targeting the Hedgehog pathway in rhabdomyosarcoma. Cancers (Basel)15, 727 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Belyea, B. et al. Embryonic signaling pathways and rhabdomyosarcoma: contributions to cancer development and opportunities for therapeutic targeting. Sarcoma2012, 406239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Hahn, H. et al. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat. Med.4, 619–622 (1998). [DOI] [PubMed] [Google Scholar]
- 350.Teot, L. et al. Clinical and mutational spectrum of highly differentiated, paired box 3:forkhead box protein o1 fusion-negative rhabdomyosarcoma: a report from the Children’s Oncology Group. Cancer124, 1973–1981 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Tostar, U. et al. Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J. Pathol.208, 17–25 (2006). [DOI] [PubMed] [Google Scholar]
- 352.Almazán-Moga, A. et al. Ligand-dependent Hedgehog pathway activation in Rhabdomyosarcoma: the oncogenic role of the ligands. Br. J. Cancer117, 1314–1325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Hatley, M. E. et al. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell22, 536–546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Fu, W., Asp, P., Canter, B. & Dynlacht, B. D. Primary cilia control hedgehog signaling during muscle differentiation and are deregulated in rhabdomyosarcoma. Proc. Natl Acad. Sci. USA111, 9151–9156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Ridzewski, R. et al. Hedgehog inhibitors in Rhabdomyosarcoma: a comparison of four compounds and responsiveness of four cell lines. Front. Oncol.5, 130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Satheesha, S. et al. Targeting hedgehog signaling reduces self-renewal in embryonal rhabdomyosarcoma. Oncogene35, 2020–2030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Yoon, J. W. et al. Up-regulation of GLI1 in vincristine-resistant rhabdomyosarcoma and Ewing sarcoma. BMC Cancer20, 511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Patel, T. H. & Cecchini, M. Targeted therapies in advanced gastric cancer. Curr. Treat. Options Oncol.21, 70 (2020). [DOI] [PubMed] [Google Scholar]
- 359.Xu, Y., Song, S., Wang, Z. & Ajani, J. A. The role of hedgehog signaling in gastric cancer: molecular mechanisms, clinical potential, and perspective. Cell Commun. Signal17, 157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Wessler, S., Krisch, L. M., Elmer, D. P. & Aberger, F. From inflammation to gastric cancer - the importance of Hedgehog/GLI signaling in Helicobacter pylori-induced chronic inflammatory and neoplastic diseases. Cell Commun. Signal15, 15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Akyala, A. I. & Peppelenbosch, M. P. Gastric cancer and Hedgehog signaling pathway: emerging new paradigms. Genes Cancer9, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Shiotani, A. et al. Helicobacter pylori-induced atrophic gastritis progressing to gastric cancer exhibits sonic hedgehog loss and aberrant CDX2 expression. Aliment Pharmacol. Ther.24, 71–80 (2006). [DOI] [PubMed] [Google Scholar]
- 363.Schumacher, M. A. et al. Gastric sonic hedgehog acts as a macrophage chemoattractant during the immune response to Helicobacter pylori. Gastroenterology142, 1150–1159.e1156 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Akhtar, K. et al. Role of sonic hedgehog ligand in gastric cancer therapeutics. J. Cancer Res. Ther.18, S267–S272 (2022). [DOI] [PubMed] [Google Scholar]
- 365.Shiotani, A. et al. Evidence that loss of sonic hedgehog is an indicator of Helicobater pylori-induced atrophic gastritis progressing to gastric cancer. Am. J. Gastroenterol.100, 581–587 (2005). [DOI] [PubMed] [Google Scholar]
- 366.Lu, L. et al. Prognostic and clinicopathological value of Gli-1 expression in gastric cancer: a meta-analysis. Oncotarget7, 69087–69096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Yu, B. et al. The role of GLI2-ABCG2 signaling axis for 5Fu resistance in gastric cancer. J. Genet. Genomics44, 375–383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Xu, M. et al. Sonic hedgehog-glioma associated oncogene homolog 1 signaling enhances drug resistance in CD44(+)/Musashi-1(+) gastric cancer stem cells. Cancer Lett.369, 124–133 (2015). [DOI] [PubMed] [Google Scholar]
- 369.Yoon, C. et al. CD44 expression denotes a subpopulation of gastric cancer cells in which Hedgehog signaling promotes chemotherapy resistance. Clin. Cancer Res.20, 3974–3988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 370.Koh, V. et al. Hedgehog transcriptional effector GLI mediates mTOR-Induced PD-L1 expression in gastric cancer organoids. Cancer Lett.518, 59–71 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Wu, C. et al. Hedgehog signaling pathway in colorectal cancer: function, mechanism, and therapy. Onco Targets Ther.10, 3249–3259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Gu, D., Schlotman, K. & Xie, J. Deciphering the role of hedgehog signaling in pancreatic cancer. J. Biomed. Res.30, 353–360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Cohen, M. et al. Drug approval summaries: arsenic trioxide, tamoxifen citrate, anastrazole, paclitaxel, bexarotene. Oncologist6, 4–11 (2001). [DOI] [PubMed] [Google Scholar]
- 374.Axelson, M. et al. U.S. Food and Drug Administration approval: vismodegib for recurrent, locally advanced, or metastatic basal cell carcinoma. Clin. Cancer Res.19, 2289–2293 (2013). [DOI] [PubMed] [Google Scholar]
- 375.Burness, C. Sonidegib: first global approval. Drugs75, 1559–1566 (2015). [DOI] [PubMed] [Google Scholar]
- 376.Hoy, S. Glasdegib: first global approval. Drugs79, 207–213 (2019). [DOI] [PubMed] [Google Scholar]
- 377.Metcalfe, C. & de Sauvage, F. Hedgehog fights back: mechanisms of acquired resistance against smoothened antagonists. Cancer Res.71, 5057–5061 (2011). [DOI] [PubMed] [Google Scholar]
- 378.Wang, C. et al. Structural basis for smoothened receptor modulation and chemoresistance to anticancer drugs. Nat. Commun.5, 4355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Tu, J. et al. Molecular modeling study on resistance of WT/D473H SMO to antagonists LDE-225 and LEQ-506. Pharmacol. Res.129, 491–499 (2018). [DOI] [PubMed] [Google Scholar]
- 380.Ishii, T. et al. Inhibition mechanism exploration of investigational drug TAK-441 as inhibitor against vismodegib-resistant smoothened mutant. Eur. J. Pharmacol.723, 305–313 (2014). [DOI] [PubMed] [Google Scholar]
- 381.Coupland, C. et al. Structure, mechanism, and inhibition of Hedgehog acyltransferase. Mol. Cell81, 5025–5038.e5010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Petrova, E. et al. Inhibitors of hedgehog acyltransferase block sonic hedgehog signaling. Nat. Chem. Biol.9, 247–249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Petrova, E., Matevossian, A. & Resh, M. Hedgehog acyltransferase as a target in pancreatic ductal adenocarcinoma. Oncogene34, 263–268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Matevossian, A. & Resh, M. Hedgehog acyltransferase as a target in estrogen receptor positive, HER2 amplified, and tamoxifen resistant breast cancer cells. Mol. Cancer14, 72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Andrei, S., Tate, E. & Lanyon-Hogg, T. Evaluating hedgehog acyltransferase activity and inhibition using the acylation-coupled lipophilic induction of polarization (Acyl-cLIP) assay. Methods Mol. Biol.2374, 13–26 (2022). [DOI] [PubMed] [Google Scholar]
- 386.Lanyon-Hogg, T. et al. Photochemical probe identification of a small-molecule inhibitor binding site in hedgehog acyltransferase (HHAT)*. Angew. Chem. Int. Ed. Engl.60, 13542–13547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Bosanac, I. et al. The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nat. Struct. Mol. Biol.16, 691–697 (2009). [DOI] [PubMed] [Google Scholar]
- 388.Magistri, P. et al. SMO inhibition modulates cellular plasticity and invasiveness in colorectal cancer. Front. Pharmacol.8, 956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Song, Z. et al. Sonic hedgehog pathway is essential for maintenance of cancer stem-like cells in human gastric cancer. PloS One6, e17687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Decker, S. et al. Trisomy 12 and elevated GLI1 and PTCH1 transcript levels are biomarkers for Hedgehog-inhibitor responsiveness in CLL. Blood119, 997–1007 (2012). [DOI] [PubMed] [Google Scholar]
- 391.Griffiths, S. et al. Hedgehog-interacting protein is a multimodal antagonist of hedgehog signalling. Nat. Commun.12, 7171 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Tada, M. et al. Down-regulation of hedgehog-interacting protein through genetic and epigenetic alterations in human hepatocellular carcinoma. Clin. Cancer Res.14, 3768–3776 (2008). [DOI] [PubMed] [Google Scholar]
- 393.Martin, S. et al. Aberrant methylation of the human hedgehog interacting protein (HHIP) gene in pancreatic neoplasms. Cancer Biol. Ther.4, 728–733 (2005). [DOI] [PubMed] [Google Scholar]
- 394.Shahi, M. et al. Human hedgehog interacting protein expression and promoter methylation in medulloblastoma cell lines and primary tumor samples. J. Neurooncol.103, 287–296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Taniguchi, H. et al. Transcriptional silencing of hedgehog-interacting protein by CpG hypermethylation and chromatic structure in human gastrointestinal cancer. J. Pathol.213, 131–139 (2007). [DOI] [PubMed] [Google Scholar]
- 396.Song, Y. et al. HHIP overexpression suppresses human gastric cancer progression and metastasis by reducing its CpG island methylation. Front. Oncol.10, 1667 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Stanton, B. et al. A small molecule that binds hedgehog and blocks its signaling in human cells. Nat. Chem. Biol.5, 154–156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Taipale, J., Cooper, M., Maiti, T. & Beachy, P. Patched acts catalytically to suppress the activity of smoothened. Nature418, 892–897 (2002). [DOI] [PubMed] [Google Scholar]
- 399.Hasan Ali, O. et al. Genomic profiling of late-onset basal cell carcinomas from two brothers with nevoid basal cell carcinoma syndrome. J. Eur. Acad. Dermatol. Venereol.35, 396–402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Banerjee, S. et al. Loss of the PTCH1 tumor suppressor defines a new subset of plexiform fibromyxoma. J. Transl. Med.17, 246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Sur, S. et al. Tea polyphenols epigallocatechin gallete and theaflavin restrict mouse liver carcinogenesis through modulation of self-renewal Wnt and hedgehog pathways. J. Nutr. Biochem.27, 32–42 (2016). [DOI] [PubMed] [Google Scholar]
- 402.Chen, J., Taipale, J., Cooper, M. & Beachy, P. Inhibition of hedgehog signaling by direct binding of cyclopamine to smoothened. Genes Dev.16, 2743–2748 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Turner, M. et al. Cyclopamine bioactivity by extraction method from veratrum californicum. Bioorg. Med. Chem.24, 3752–3757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Cheng, W. et al. Role of hedgehog signaling pathway in proliferation and invasiveness of hepatocellular carcinoma cells. Int. J. Oncol.34, 829–836 (2009). [DOI] [PubMed] [Google Scholar]
- 405.Chen, X. et al. Expression of sonic hedgehog signaling components in hepatocellular carcinoma and cyclopamine-induced apoptosis through Bcl-2 downregulation in vitro. Arch. Med. Res.41, 315–323 (2010). [DOI] [PubMed] [Google Scholar]
- 406.Jeng, K. et al. Blockade of the sonic hedgehog pathway effectively inhibits the growth of hepatoma in mice: an in vivo study. Oncol. Lett.4, 1158–1162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Miyazaki, Y. et al. Efficient elimination of pancreatic cancer stem cells by hedgehog/GLI inhibitor GANT61 in combination with mTOR inhibition. Mol. Cancer15, 49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Balbous, A. et al. Selective release of a cyclopamine glucuronide prodrug toward stem-like cancer cell inhibition in glioblastoma. Mol. Cancer Ther.13, 2159–2169 (2014). [DOI] [PubMed] [Google Scholar]
- 409.Lin, T. & Matsui, W. Hedgehog pathway as a drug target: smoothened inhibitors in development. Onco Targets Ther.5, 47–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Tsai, C. et al. Sonic hedgehog inhibition as a strategy to augment radiosensitivity of hepatocellular carcinoma. J. Gastroenterol. Hepatol.30, 1317–1324 (2015). [DOI] [PubMed] [Google Scholar]
- 411.Robarge, K. et al. GDC-0449-a potent inhibitor of the hedgehog pathway. Bioorg. Med. Chem. Lett.19, 5576–5581 (2009). [DOI] [PubMed] [Google Scholar]
- 412.Sekulic, A. et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N. Engl. J. Med.366, 2171–2179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Sekulic, A. et al. Long-term safety and efficacy of vismodegib in patients with advanced basal cell carcinoma: final update of the pivotal ERIVANCE BCC study. BMC Cancer17, 332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Dréno, B. et al. Two intermittent vismodegib dosing regimens in patients with multiple basal-cell carcinomas (MIKIE): a randomised, regimen-controlled, double-blind, phase 2 trial. Lancet Oncol.18, 404–412 (2017). [DOI] [PubMed] [Google Scholar]
- 415.Gajjar, A. et al. Phase I study of vismodegib in children with recurrent or refractory medulloblastoma: a pediatric brain tumor consortium study. Clin. Cancer Res.19, 6305–6312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Ally, M. et al. The use of vismodegib to shrink keratocystic odontogenic tumors in patients with basal cell nevus syndrome. JAMA Dermatol.150, 542–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Jacobsen, A. A. et al. Hedgehog pathway inhibitor therapy for locally advanced and metastatic basal cell carcinoma: a systematic review and pooled analysis of interventional studies. JAMA Dermatol.152, 816–824 (2016). [DOI] [PubMed] [Google Scholar]
- 418.Catenacci, D. et al. Randomized phase Ib/II study of gemcitabine plus placebo or vismodegib, a hedgehog pathway inhibitor, in patients with metastatic pancreatic cancer. J. Clin. Oncol.33, 4284–4292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Houot, R. et al. Inhibition of hedgehog signaling for the treatment of lymphoma and CLL: a phase II study from the LYSA. Ann. Oncol.27, 1349–1350 (2016). [DOI] [PubMed] [Google Scholar]
- 420.Italiano, A. et al. GDC-0449 in patients with advanced chondrosarcomas: a French Sarcoma Group/US and French National Cancer Institute single-arm phase II collaborative study. Ann. Oncol.24, 2922–2926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Calzavara Pinton, P. et al. Vismodegib in the treatment of basal cell carcinoma: indications for clinical practice. Future Oncol.11, 1429–1435 (2015). [DOI] [PubMed] [Google Scholar]
- 422.Pan, S. et al. Discovery of NVP-LDE225, a potent and selective smoothened antagonist. ACS Med. Chem. Lett.1, 130–134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Fu, J. et al. NPV-LDE-225 (Erismodegib) inhibits epithelial mesenchymal transition and self-renewal of glioblastoma initiating cells by regulating miR-21, miR-128, and miR-200. Neuro Oncol.15, 691–706 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Nanta, R. et al. NVP-LDE-225 (Erismodegib) inhibits epithelial-mesenchymal transition and human prostate cancer stem cell growth in NOD/SCID IL2Rγ null mice by regulating Bmi-1 and microRNA-128. Oncogenesis2, e42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Dummer, R. et al. The 12-month analysis from basal cell carcinoma outcomes with LDE225 treatment (BOLT): a phase II, randomized, double-blind study of sonidegib in patients with advanced basal cell carcinoma. J. Am. Acad. Dermatol.75, 113–125.e115 (2016). [DOI] [PubMed] [Google Scholar]
- 426.Migden, M. et al. Treatment with two different doses of sonidegib in patients with locally advanced or metastatic basal cell carcinoma (BOLT): a multicentre, randomised, double-blind phase 2 trial. Lancet Oncol.16, 716–728 (2015). [DOI] [PubMed] [Google Scholar]
- 427.Nguyen, A., Xie, P., Litvinov, I. V. & Lefrançois, P. Efficacy and safety of sonic hedgehog inhibitors in basal cell carcinomas: an updated systematic review and meta-analysis (2009–2022). Am. J. Clin. Dermatol.24, 359–374 (2023). [DOI] [PubMed] [Google Scholar]
- 428.Kieran, M. et al. Phase I study of oral sonidegib (LDE225) in pediatric brain and solid tumors and a phase II study in children and adults with relapsed medulloblastoma. Neuro Oncol.19, 1542–1552 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Stathis, A. et al. Phase I trial of the oral smoothened inhibitor sonidegib in combination with paclitaxel in patients with advanced solid tumors. Invest. New Drugs35, 766–772 (2017). [DOI] [PubMed] [Google Scholar]
- 430.Rodon, J. et al. A phase I, multicenter, open-label, first-in-human, dose-escalation study of the oral smoothened inhibitor Sonidegib (LDE225) in patients with advanced solid tumors. Clin. Cancer Res.20, 1900–1909 (2014). [DOI] [PubMed] [Google Scholar]
- 431.Jain, S., Song, R. & Xie, J. Sonidegib: mechanism of action, pharmacology, and clinical utility for advanced basal cell carcinomas. Onco Targets Ther.10, 1645–1653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Gutzmer, R. et al. Key clinical adverse events in patients with advanced basal cell carcinoma treated with sonidegib or vismodegib: a post hoc analysis. Dermatol. Ther. (Heidelb.)11, 1839–1849 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Tremblay, M. et al. Discovery of a potent and orally active hedgehog pathway antagonist (IPI-926). J. Med. Chem.52, 4400–4418 (2009). [DOI] [PubMed] [Google Scholar]
- 434.Campbell, V. et al. Hedgehog pathway inhibition in chondrosarcoma using the smoothened inhibitor IPI-926 directly inhibits sarcoma cell growth. Mol. Cancer Ther.13, 1259–1269 (2014). [DOI] [PubMed] [Google Scholar]
- 435.McCann, C. et al. Inhibition of hedgehog signaling antagonizes serous ovarian cancer growth in a primary xenograft model. PloS One6, e28077 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Lo, W. et al. Involvement and targeted intervention of dysregulated hedgehog signaling in osteosarcoma. Cancer120, 537–547 (2014). [DOI] [PubMed] [Google Scholar]
- 437.Lin, T. et al. Self-renewal of acute lymphocytic leukemia cells is limited by the Hedgehog pathway inhibitors cyclopamine and IPI-926. PloS One5, e15262 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Lee, M. et al. Hedgehog pathway inhibitor saridegib (IPI-926) increases lifespan in a mouse medulloblastoma model. Proc. Natl Acad. Sci. USA109, 7859–7864 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Ko, A. et al. A phase I study of FOLFIRINOX Plus IPI-926, a hedgehog pathway inhibitor, for advanced pancreatic adenocarcinoma. Pancreas45, 370–375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Bowles, D. et al. A pilot study of cetuximab and the hedgehog inhibitor IPI-926 in recurrent/metastatic head and neck squamous cell carcinoma. Oral. Oncol.53, 74–79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Jimeno, A. et al. Phase I study of the hedgehog pathway inhibitor IPI-926 in adult patients with solid tumors. Clin. Cancer Res.19, 2766–2774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Munchhof, M. et al. Discovery of PF-04449913, a potent and orally bioavailable inhibitor of smoothened. ACS Med. Chem. Lett.3, 106–111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Cortes, J. et al. Randomized comparison of low dose cytarabine with or without glasdegib in patients with newly diagnosed acute myeloid leukemia or high-risk myelodysplastic syndrome. Leukemia33, 379–389 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Wagner, A. et al. A phase I study of PF-04449913, an oral hedgehog inhibitor, in patients with advanced solid tumors. Clin. Cancer Res.21, 1044–1051 (2015). [DOI] [PubMed] [Google Scholar]
- 445.Bendell, J. et al. Phase I study of LY2940680, a Smo antagonist, in patients with advanced cancer including treatment-naïve and previously treated basal cell carcinoma. Clin. Cancer Res.24, 2082–2091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Azaro, A. et al. A phase 1b study of the Notch inhibitor crenigacestat (LY3039478) in combination with other anticancer target agents (taladegib, LY3023414, or abemaciclib) in patients with advanced or metastatic solid tumors. Invest. New Drugs39, 1089–1098 (2021). [DOI] [PubMed] [Google Scholar]
- 447.Zaidi, A. et al. Smoothened inhibition leads to decreased proliferation and induces apoptosis in esophageal adenocarcinoma cells. Cancer Invest.31, 480–489 (2013). [DOI] [PubMed] [Google Scholar]
- 448.Nguyen, J. et al. New ameloblastoma cell lines enable preclinical study of targeted therapies. J. Dent. Res.101, 1517–1525 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Lauressergues, E. et al. Pharmacological evaluation of a series of smoothened antagonists in signaling pathways and after topical application in a depilated mouse model. Pharmacol. Res. Perspect.4, e00214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Ohashi, T. et al. Discovery of the investigational drug TAK-441, a pyrrolo[3,2-c]pyridine derivative, as a highly potent and orally active hedgehog signaling inhibitor: modification of the core skeleton for improved solubility. Bioorg. Med. Chem.20, 5507–5517 (2012). [DOI] [PubMed] [Google Scholar]
- 451.Ibuki, N. et al. TAK-441, a novel investigational smoothened antagonist, delays castration-resistant progression in prostate cancer by disrupting paracrine hedgehog signaling. Int. J. Cancer133, 1955–1966 (2013). [DOI] [PubMed] [Google Scholar]
- 452.Kogame, A. et al. Pharmacokinetic and pharmacodynamic modeling of hedgehog inhibitor TAK-441 for the inhibition of Gli1 messenger RNA expression and antitumor efficacy in xenografted tumor model mice. Drug Metab. Dispos.41, 727–734 (2013). [DOI] [PubMed] [Google Scholar]
- 453.Goldman, J. et al. Phase I dose-escalation trial of the oral investigational hedgehog signaling pathway inhibitor TAK-441 in patients with advanced solid tumors. Clin. Cancer Res.21, 1002–1009 (2015). [DOI] [PubMed] [Google Scholar]
- 454.Spanakis, E., Aperis, G. & Mylonakis, E. New agents for the treatment of fungal infections: clinical efficacy and gaps in coverage. Clin. Infect. Dis.43, 1060–1068 (2006). [DOI] [PubMed] [Google Scholar]
- 455.Kim, J. et al. Itraconazole, a commonly used antifungal that inhibits Hedgehog pathway activity and cancer growth. Cancer Cell17, 388–399 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Freitas, R. et al. Inhibition of CAL27 oral squamous carcinoma cell by targeting hedgehog pathway with vismodegib or itraconazole. Front. Oncol.10, 563838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Hu, Q. et al. Itraconazole induces apoptosis and cell cycle arrest via inhibiting hedgehog signaling in gastric cancer cells. J. Exp. Clin. Cancer Res.36, 50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.You, M. et al. Targeting of the hedgehog signal transduction pathway suppresses survival of malignant pleural mesothelioma cells in vitro. J. Thorac. Cardiovasc. Surg.147, 508–516 (2014). [DOI] [PubMed] [Google Scholar]
- 459.Kim, D. et al. Open-label, exploratory phase II trial of oral itraconazole for the treatment of basal cell carcinoma. J. Clin. Oncol.32, 745–751 (2014). [DOI] [PubMed] [Google Scholar]
- 460.Solinas, A. et al. Acylthiourea, acylurea, and acylguanidine derivatives with potent hedgehog inhibiting activity. J. Med. Chem.55, 1559–1571 (2012). [DOI] [PubMed] [Google Scholar]
- 461.Hoch, L. et al. MRT-92 inhibits hedgehog signaling by blocking overlapping binding sites in the transmembrane domain of the smoothened receptor. FASEB J.29, 1817–1829 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Pietrobono, S. et al. Targeted inhibition of Hedgehog-GLI signaling by novel acylguanidine derivatives inhibits melanoma cell growth by inducing replication stress and mitotic catastrophe. Cell Death Dis.9, 142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Pietrobono, S. et al. Targeting non-canonical activation of GLI1 by the SOX2-BRD4 transcriptional complex improves the efficacy of HEDGEHOG pathway inhibition in melanoma. Oncogene40, 3799–3814 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Rohner, A. et al. Effective targeting of hedgehog signaling in a medulloblastoma model with PF-5274857, a potent and selective smoothened antagonist that penetrates the blood-brain barrier. Mol. Cancer Ther.11, 57–65 (2012). [DOI] [PubMed] [Google Scholar]
- 465.Zhang, M., Gao, L., Ye, Y. & Li, X. Advances in glioma-associated oncogene (GLI) inhibitors for cancer therapy. Invest. New Drugs40, 370–388 (2022). [DOI] [PubMed] [Google Scholar]
- 466.Sharpe, H. et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell27, 327–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Atwood, S. et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell27, 342–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Lauth, M., Bergström, A., Shimokawa, T. & Toftgård, R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc. Natl Acad. Sci. USA104, 8455–8460 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Sharma, A. et al. Sonic hedgehog pathway activation regulates cervical cancer stem cell characteristics during epithelial to mesenchymal transition. J. Cell Physiol.234, 15726–15741 (2019). [DOI] [PubMed] [Google Scholar]
- 470.Hou, X. et al. Inhibition of hedgehog signaling by GANT58 induces apoptosis and shows synergistic antitumor activity with AKT inhibitor in acute T cell leukemia cells. Biochimie101, 50–59 (2014). [DOI] [PubMed] [Google Scholar]
- 471.Oladapo, H. et al. Pharmacological targeting of GLI1 inhibits proliferation, tumor emboli formation and in vivo tumor growth of inflammatory breast cancer cells. Cancer Lett.411, 136–149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Zubčić, V. et al. GANT61 and Lithium Chloride inhibit the growth of head and neck cancer cell lines through the regulation of GLI3 processing by GSK3β. Int. J. Mol. Sci.21, 6410 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Harada, K., Ohashi, R., Naito, K. & Kanki, K. Hedgehog signal inhibitor GANT61 inhibits the malignant behavior of undifferentiated hepatocellular carcinoma cells by targeting non-canonical GLI signaling. Int. J. Mol. Sci.21, 3126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Sneha, S. et al. The hedgehog pathway regulates cancer stem cells in serous adenocarcinoma of the ovary. Cell Oncol. (Dordr.)43, 601–616 (2020). [DOI] [PubMed] [Google Scholar]
- 475.Yang, H. et al. Inhibition of Gli1-mediated prostate cancer cell proliferation by inhibiting the mTOR/S6K1 signaling pathway. Oncol. Lett.14, 7970–7976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Chang, Y. et al. The inhibitory effect and safety of GANT61 on HeLa cells in nude mice. Exp. Mol. Pathol.113, 104352 (2020). [DOI] [PubMed] [Google Scholar]
- 477.Chang, J. et al. Downregulation of Rab23 in prostate cancer inhibits tumor growth in vitro and in vivo. Oncol. Res.25, 241–248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Azatyan, A. et al. RITA downregulates Hedgehog-GLI in medulloblastoma and rhabdomyosarcoma via JNK-dependent but p53-independent mechanism. Cancer Lett.442, 341–350 (2019). [DOI] [PubMed] [Google Scholar]
- 479.Calcaterra, A. et al. Chemical, computational and functional insights into the chemical stability of the Hedgehog pathway inhibitor GANT61. J. Enzym. Inhib. Med. Chem.33, 349–358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Cicconi, L. & Lo-Coco, F. Current management of newly diagnosed acute promyelocytic leukemia. Ann. Oncol.27, 1474–1481 (2016). [DOI] [PubMed] [Google Scholar]
- 481.Jeanne, M. et al. PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3. Cancer Cell18, 88–98 (2010). [DOI] [PubMed] [Google Scholar]
- 482.Kim, J. et al. Arsenic antagonizes the hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc. Natl Acad. Sci. USA107, 13432–13437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Zhang, X. et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science328, 240–243 (2010). [DOI] [PubMed] [Google Scholar]
- 484.Beauchamp, E. et al. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J. Clin. Invest.121, 148–160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Yang, D. et al. Arsenic trioxide inhibits the hedgehog pathway which is aberrantly activated in acute promyelocytic leukemia. Acta Haematol.130, 260–267 (2013). [DOI] [PubMed] [Google Scholar]
- 486.Han, J. et al. Arsenic trioxide inhibits viability of pancreatic cancer stem cells in culture and in a xenograft model via binding to SHH-Gli. Onco Targets Ther.6, 1129–1138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Nakamura, S. et al. Arsenic trioxide prevents osteosarcoma growth by inhibition of GLI transcription via DNA damage accumulation. PloS One8, e69466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Boehme, K. et al. Targeting hedgehog signalling by arsenic trioxide reduces cell growth and induces apoptosis in rhabdomyosarcoma. Int. J. Oncol.48, 801–812 (2016). [DOI] [PubMed] [Google Scholar]
- 489.Douer, D. & Tallman, M. Arsenic trioxide: new clinical experience with an old medication in hematologic malignancies. J. Clin. Oncol.23, 2396–2410 (2005). [DOI] [PubMed] [Google Scholar]
- 490.Yu, M. et al. Current advances of nanomedicines delivering arsenic trioxide for enhanced tumor therapy. Pharmaceutics14, 743 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Kramann, R. et al. Pharmacological GLI2 inhibition prevents myofibroblast cell-cycle progression and reduces kidney fibrosis. J. Clin. Invest.125, 2935–2951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Mann, K., Wallner, B., Lossos, I. & Miller, W. J. Darinaparsin: a novel organic arsenical with promising anticancer activity. Expert Opin. Investig. Drugs18, 1727–1734 (2009). [DOI] [PubMed] [Google Scholar]
- 493.Frampton, J. Darinaparsin: first approval. Drugs82, 1603–1609 (2022). [DOI] [PubMed] [Google Scholar]
- 494.Ravi, D. et al. The novel organic arsenical darinaparsin induces MAPK-mediated and SHP1-dependent cell death in T-cell lymphoma and Hodgkin lymphoma cells and human xenograft models. Clin. Cancer Res.20, 6023–6033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Tian, J. et al. Darinaparsin: solid tumor hypoxic cytotoxin and radiosensitizer. Clin. Cancer Res.18, 3366–3376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Bansal, N. et al. Darinaparsin inhibits prostate tumor-initiating cells and Du145 xenografts and is an inhibitor of hedgehog signaling. Mol. Cancer Ther.14, 23–30 (2015). [DOI] [PubMed] [Google Scholar]
- 497.Tsimberidou, A. et al. A phase I clinical trial of darinaparsin in patients with refractory solid tumors. Clin. Cancer Res.15, 4769–4776 (2009). [DOI] [PubMed] [Google Scholar]
- 498.Hosein, P. et al. A multicenter phase II study of darinaparsin in relapsed or refractory Hodgkin’s and non-Hodgkin’s lymphoma. Am. J. Hematol.87, 111–114 (2012). [DOI] [PubMed] [Google Scholar]
- 499.Hyman, J. et al. Small-molecule inhibitors reveal multiple strategies for hedgehog pathway blockade. Proc. Natl Acad. Sci. USA106, 14132–14137 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Kaur, G. et al. Bromodomain and hedgehog pathway targets in small cell lung cancer. Cancer Lett.371, 225–239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Jeng, K. et al. Glioma-associated oncogene homolog inhibitors have the potential of suppressing cancer stem cells of breast cancer. Int. J. Mol. Sci.19, 1375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Xiang, W. et al. Hedgehog pathway inhibitor-4 suppresses malignant properties of chondrosarcoma cells by disturbing tumor ciliogenesis. Oncol. Rep.32, 1622–1630 (2014). [DOI] [PubMed] [Google Scholar]
- 503.Chenna, V. et al. A polymeric nanoparticle encapsulated small-molecule inhibitor of hedgehog signaling (NanoHHI) bypasses secondary mutational resistance to smoothened antagonists. Mol. Cancer Ther.11, 165–173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Bao, C., Kramata, P., Lee, H. & Suh, N. Regulation of hedgehog signaling in cancer by natural and dietary compounds. Mol. Nutr. Food Res.62, 1700621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Deberardinis, A. et al. Structure-activity relationships for vitamin D3-based aromatic a-ring analogues as hedgehog pathway inhibitors. J. Med. Chem.57, 3724–3736 (2014). [DOI] [PubMed] [Google Scholar]
- 506.Tang, J. et al. Vitamin D3 inhibits hedgehog signaling and proliferation in murine Basal cell carcinomas. Cancer Prev. Res. (Philos.)4, 744–751 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Sharma, U. et al. Role of Hedgehog and Hippo signaling pathways in cancer: a special focus on non-coding RNAs. Pharmacol. Res.186, 106523 (2022). [DOI] [PubMed] [Google Scholar]
- 508.Sargazi, M. et al. The crosstalk between long non-coding RNAs and the hedgehog signaling pathway in cancer. Med. Oncol.39, 127 (2022). [DOI] [PubMed] [Google Scholar]
- 509.Chen, Y., Li, Z., Chen, X. & Zhang, S. Long non-coding RNAs: from disease code to drug role. Acta Pharmacol. Sin. B11, 340–354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Takezaki, T. et al. Essential role of the hedgehog signaling pathway in human glioma-initiating cells. Cancer Sci.102, 1306–1312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Zhou, J. et al. Non-canonical GLI1/2 activation by PI3K/AKT signaling in renal cell carcinoma: a novel potential therapeutic target. Cancer Lett.370, 313–323 (2016). [DOI] [PubMed] [Google Scholar]
- 512.Vecchiotti, D. et al. Elevated NF-κB/SHh/GLI1 signature denotes a worse prognosis and represent a novel potential therapeutic target in advanced prostate cancer. Cells11, 2118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Cho, K. et al. Pharmacological inhibition of sonic hedgehog signaling suppresses tumor development in a murine model of intrahepatic cholangiocarcinoma. Int. J. Mol. Sci.22, 13214 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Szczepny, A. et al. The role of canonical and non-canonical hedgehog signaling in tumor progression in a mouse model of small cell lung cancer. Oncogene36, 5544–5550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Zhou, C. et al. GLI1 reduces drug sensitivity by regulating cell cycle through PI3K/AKT/GSK3/CDK pathway in acute myeloid leukemia. Cell Death Dis.12, 231 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Romitti, M. et al. MAPK and SHH pathways modulate type 3 deiodinase expression in papillary thyroid carcinoma. Endocr. Relat. Cancer23, 135–146 (2016). [DOI] [PubMed] [Google Scholar]
- 517.Wang, J. et al. Berberine, a natural compound, suppresses Hedgehog signaling pathway activity and cancer growth. BMC Cancer15, 595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Li, X. et al. Cynanbungeigenin C and D, a pair of novel epimers from Cynanchum bungei, suppress hedgehog pathway-dependent medulloblastoma by blocking signaling at the level of Gli. Cancer Lett.420, 195–207 (2018). [DOI] [PubMed] [Google Scholar]
- 519.Singh, S. et al. Selective targeting of the hedgehog signaling pathway by PBM nanoparticles in docetaxel-resistant prostate cancer. Cells9, 1976 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Gao, Q., Yuan, Y., Gan, H. & Peng, Q. Resveratrol inhibits the hedgehog signaling pathway and epithelial-mesenchymal transition and suppresses gastric cancer invasion and metastasis. Oncol. Lett.9, 2381–2387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Qin, Y. et al. Effect of resveratrol on proliferation and apoptosis of human pancreatic cancer MIA PaCa-2 cells may involve inhibition of the hedgehog signaling pathway. Mol. Med. Rep.10, 2563–2567 (2014). [DOI] [PubMed] [Google Scholar]
- 522.Du, W. et al. Curcumin suppresses malignant glioma cells growth and induces apoptosis by inhibition of SHH/GLI1 signaling pathway in vitro and vivo. CNS Neurosci. Ther.19, 926–936 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Zhu, J. et al. Curcumin suppresses lung cancer stem cells via inhibiting Wnt/β-catenin and sonic hedgehog pathways. Phytother. Res.31, 680–688 (2017). [DOI] [PubMed] [Google Scholar]
- 524.Cao, L. et al. Curcumin inhibits hypoxia-induced epithelial‑mesenchymal transition in pancreatic cancer cells via suppression of the hedgehog signaling pathway. Oncol. Rep.35, 3728–3734 (2016). [DOI] [PubMed] [Google Scholar]
- 525.Elamin, M. et al. Curcumin inhibits the sonic hedgehog signaling pathway and triggers apoptosis in medulloblastoma cells. Mol. Carcinog.49, 302–314 (2010). [DOI] [PubMed] [Google Scholar]
- 526.Sur, S. et al. Tea polyphenols EGCG and TF restrict tongue and liver carcinogenesis simultaneously induced by N-nitrosodiethylamine in mice. Toxicol. Appl. Pharmacol.300, 34–46 (2016). [DOI] [PubMed] [Google Scholar]
- 527.Tang, S. et al. Inhibition of sonic hedgehog pathway and pluripotency maintaining factors regulate human pancreatic cancer stem cell characteristics. Int. J. Cancer131, 30–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Tang, G. et al. (-)-Epigallocatechin-3-gallate induces apoptosis and suppresses proliferation by inhibiting the human Indian Hedgehog pathway in human chondrosarcoma cells. J. Cancer Res. Clin. Oncol.136, 1179–1185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Zhao, M. et al. Ellagic acid inhibits human pancreatic cancer growth in Balb c nude mice. Cancer Lett.337, 210–217 (2013). [DOI] [PubMed] [Google Scholar]
- 530.Luo, J. et al. Saikosaponin B1 and Saikosaponin D inhibit tumor growth in medulloblastoma allograft mice via inhibiting the Hedgehog signaling pathway. J. Nat. Med.76, 584–593 (2022). [DOI] [PubMed] [Google Scholar]
- 531.Fan, P. et al. Genistein decreases the breast cancer stem-like cell population through Hedgehog pathway. Stem Cell Res. Ther.4, 146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Zhang, L. et al. Genistein inhibits the stemness properties of prostate cancer cells through targeting Hedgehog-Gli1 pathway. Cancer Lett.323, 48–57 (2012). [DOI] [PubMed] [Google Scholar]
- 533.Infante, P. et al. Gli1/DNA interaction is a druggable target for Hedgehog-dependent tumors. EMBO J.34, 200–217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Ma, Z. et al. Silibinin induces apoptosis through inhibition of the mTOR-GLI1-BCL2 pathway in renal cell carcinoma. Oncol. Rep.34, 2461–2468 (2015). [DOI] [PubMed] [Google Scholar]
- 535.Qiu, P. et al. Inoscavin A, a pyrone compound isolated from a Sanghuangporus vaninii extract, inhibits colon cancer cell growth and induces cell apoptosis via the hedgehog signaling pathway. Phytomedicine96, 153852 (2022). [DOI] [PubMed] [Google Scholar]
- 536.Rifai, Y. et al. Acoschimperoside P, 2’-acetate: a Hedgehog signaling inhibitory constituent from Vallaris glabra. J. Nat. Med.65, 629–632 (2011). [DOI] [PubMed] [Google Scholar]
- 537.Slusarz, A. et al. Common botanical compounds inhibit the hedgehog signaling pathway in prostate cancer. Cancer Res.70, 3382–3390 (2010). [DOI] [PubMed] [Google Scholar]
- 538.Arai, M. et al. Hedgehog/GLI-mediated transcriptional inhibitors from Zizyphus cambodiana. Bioorg. Med. Chem.16, 9420–9424 (2008). [DOI] [PubMed] [Google Scholar]
- 539.Zheng, W. et al. Deguelin inhibits proliferation and migration of human pancreatic cancer cells in vitro targeting hedgehog pathway. Oncol. Lett.12, 2761–2765 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Lee, H. et al. A sesquiterpene lactone from Siegesbeckia glabrescens suppresses Hedgehog/Gli-mediated transcription in pancreatic cancer cells. Oncol. Lett.12, 2912–2917 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Hosoya, T. et al. Naturally occurring small-molecule inhibitors of hedgehog/GLI-mediated transcription. Chembiochem9, 1082–1092 (2008). [DOI] [PubMed] [Google Scholar]
- 542.Lin, H. et al. Inhibition of Gli/hedgehog signaling in prostate cancer cells by “cancer bush” Sutherlandia frutescens extract. Cell Biol. Int.40, 131–142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Bao, C. et al. Sulforaphene interferes with human breast cancer cell migration and invasion through inhibition of hedgehog signaling. J. Agric. Food Chem.64, 5515–5524 (2016). [DOI] [PubMed] [Google Scholar]
- 544.Li, S. et al. Sulforaphane regulates self-renewal of pancreatic cancer stem cells through the modulation of sonic hedgehog-GLI pathway. Mol. Cell Biochem.373, 217–227 (2013). [DOI] [PubMed] [Google Scholar]
- 545.Zheng, S., Li, M., Miao, K. & Xu, H. lncRNA GAS5-promoted apoptosis in triple-negative breast cancer by targeting miR-378a-5p/SUFU signaling. J. Cell Biochem.121, 2225–2235 (2020). [DOI] [PubMed] [Google Scholar]
- 546.Qiu, S. et al. LncRNA EGOT decreases breast cancer cell viability and migration via inactivation of the Hedgehog pathway. FEBS Open Bio.10, 817–826 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Xu, F., Li, H. & Hu, C. LIFR-AS1 modulates Sufu to inhibit cell proliferation and migration by miR-197-3p in breast cancer. Biosci. Rep.39, BSR20180551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 548.Xin, L. et al. DNA-methylation-mediated silencing of miR-7-5p promotes gastric cancer stem cell invasion via increasing Smo and Hes1. J. Cell Physiol.235, 2643–2654 (2020). [DOI] [PubMed] [Google Scholar]
- 549.Li, J. et al. miR-7-5p acts as a tumor suppressor in bladder cancer by regulating the hedgehog pathway factor Gli3. Biochem. Biophys. Res. Commun.503, 2101–2107 (2018). [DOI] [PubMed] [Google Scholar]
- 550.Ferretti, E. et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J.27, 2616–2627 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Wang, N. et al. miR-141-3p suppresses proliferation and promotes apoptosis by targeting GLI2 in osteosarcoma cells. Oncol. Rep.39, 747–754 (2018). [DOI] [PubMed] [Google Scholar]
- 552.Sun, Z. et al. miR-202 suppresses proliferation and induces apoptosis of osteosarcoma cells by downregulating Gli2. Mol. Cell Biochem.397, 277–283 (2014). [DOI] [PubMed] [Google Scholar]
- 553.Zhang, J. et al. miRNA-218 regulates the proliferation and apoptosis of cervical cancer cells via targeting Gli3. Exp. Ther. Med.16, 2433–2441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Guan, B. et al. MicroRNA-218 inhibits the migration, epithelial-mesenchymal transition and cancer stem cell properties of prostate cancer cells. Oncol. Lett.16, 1821–1826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Song, J. et al. Novel long noncoding RNA miR205HG functions as an esophageal tumor-suppressive hedgehog inhibitor. Cancers (Basel)13, 1707 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Du, W., Li, D., Xie, J. & Tang, P. miR‑367‑3p downregulates Rab23 expression and inhibits hedgehog signaling resulting in the inhibition of the proliferation, migration, and invasion of prostate cancer cells. Oncol. Rep.46, 192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Zhao, D. & Cui, Z. MicroRNA-361-3p regulates retinoblastoma cell proliferation and stemness by targeting hedgehog signaling. Exp. Ther. Med.17, 1154–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Zhang, H. et al. The expression of microRNA-324-3p as a tumor suppressor in nasopharyngeal carcinoma and its clinical significance. Onco Targets Ther.10, 4935–4943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Xu, H. et al. MiR-324-5p inhibits proliferation of glioma by target regulation of GLI1. Eur. Rev. Med. Pharmacol. Sci.18, 828–832 (2014). [PubMed] [Google Scholar]
- 560.Tang, B. et al. MicroRNA-324-5p regulates stemness, pathogenesis and sensitivity to bortezomib in multiple myeloma cells by targeting hedgehog signaling. Int. J. Cancer142, 109–120 (2018). [DOI] [PubMed] [Google Scholar]
- 561.Du, W. et al. Targeting the SMO oncogene by miR-326 inhibits glioma biological behaviors and stemness. Neuro Oncol.17, 243–253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Huang, J., Xu, Y. & Lin, F. The inhibition of microRNA-326 by SP1/HDAC1 contributes to proliferation and metastasis of osteosarcoma through promoting SMO expression. J. Cell Mol. Med.24, 10876–10888 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Xue, Q. et al. MicroRNA-338-3p inhibits colorectal carcinoma cell invasion and migration by targeting smoothened. Jpn. J. Clin. Oncol.44, 13–21 (2014). [DOI] [PubMed] [Google Scholar]
- 564.Xiong, Z. et al. Circular RNA SMO sponges miR-338-3p to promote the growth of glioma by enhancing the expression of SMO. Aging (Albany NY)11, 12345–12360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Liu, Y. et al. Downregulation of MiR-431 expression associated with lymph node metastasis and promotes cell invasion in papillary thyroid carcinoma. Cancer Biomark.22, 727–732 (2018). [DOI] [PubMed] [Google Scholar]
- 566.Wen, S. et al. miR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene34, 717–725 (2015). [DOI] [PubMed] [Google Scholar]
- 567.Wang, T., Feng, J. & Zhang, A. miR-584 inhibits cell proliferation, migration and invasion in vitro and enhances the sensitivity to cisplatin in human cervical cancer by negatively targeting GLI1. Exp. Ther. Med.19, 2059–2066 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Feng, J. & Wang, T. MicroRNA-873 serves a critical role in human cervical cancer proliferation and metastasis via regulating glioma-associated oncogene homolog 1. Exp. Ther. Med.19, 1243–1250 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 569.Cao, D., Yu, T. & Ou, X. MiR-873-5P controls gastric cancer progression by targeting hedgehog-GLI signaling. Pharmazie71, 603–606 (2016). [DOI] [PubMed] [Google Scholar]
- 570.Xu, Z., Huang, C. & Hao, D. MicroRNA-1271 inhibits proliferation and promotes apoptosis of multiple myeloma cells through inhibiting smoothened-mediated hedgehog signaling pathway. Oncol. Rep.37, 1261–1269 (2017). [DOI] [PubMed] [Google Scholar]