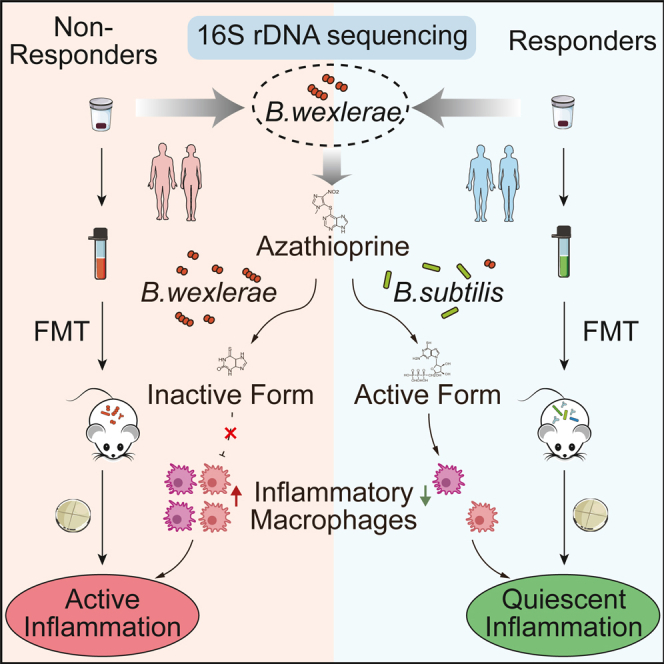
Summary
Azathioprine (AZA) therapy failure, though not the primary cause, contributes to disease relapse and progression in inflammatory bowel disease (IBD). However, the role of gut microbiota in AZA therapy failure remains poorly understood. We found a high prevalence of Blautia wexlerae in patients with IBD with AZA therapy failure, associated with shorter disease flare survival time. Colonization of B. wexlerae increased inflammatory macrophages and compromised AZA’s therapeutic efficacy in mice with intestinal colitis. B. wexlerae colonization reduced 6-mercaptopurine (6-MP) bioavailability by enhancing selenium-dependent xanthine dehydrogenase (sd-XDH) activity. The enzyme sd-XDH converts 6-MP into its inactive metabolite, 6-thioxanthine (6-TX), thereby impairing its ability to inhibit inflammation in mice. Supplementation with Bacillus (B.) subtilis enriched in hypoxanthine phosphoribosyltransferase (HPRT) effectively mitigated B. wexlerae-induced AZA treatment failure in mice with intestinal colitis. These findings emphasize the need for tailored management strategies based on B. wexlerae levels in patients with IBD.
Keywords: inflammatory bowel disease, microbiome, azathioprine therapy failure, biotransformation, 6-mercaptopurine
Graphical abstract
Highlights
-
•
B. wexlerae could predict AZA therapy failure in patients with inflammatory bowel disease
-
•
B. wexlerae weakened AZA’s effect on macrophages in DSS-induced colitis mice
-
•
The sd-XDH gene harbored by B.wexlerae may impact 6-mercaptopurine biotransformation
-
•
B. subtilis reverses B. wexlerae-induced AZA failure in colitis mice
Identification of B. wexlerae by Yan et al. provides potential biomarkers for predicting response to azathioprine (AZA) therapy in inflammatory bowel disease (IBD). B. wexlerae inhibits 6-mercaptopurine biotransformation and attenuates the inhibition of AZA on inflammatory macrophages. Supplementation of HPRT-enriched probiotics shows potential in mitigating the intestinal colitis of mice.
Introduction
Azathioprine (AZA) is one of many therapy options in adult inflammatory bowel disease (IBD) and has been used as the first line of therapy for other medical conditions including autoimmune hepatitis and rheumatological and other disease treatment.1,2 Meanwhile, AZA has its advantages, which are that it is economical and sustainable.3,4 However, the overall remission rates were only 45% in patients with Crohn’s disease (CD) and 58% in patients with ulcerative colitis (UC) after AZA treatment.5 The genotypes of host metabolic enzymes, which include glutathione S-transferase (GST), thiopurine S-methyltransferase (TPMT), and nucleoside diphosphate-linked moiety X-type motif 15 (NUDT15), and the concentration of 6-thioguanine nucleotide (6-TGN) metabolites were routinely examined to evaluate the efficacy and pharmacokinetics of AZA in patients with IBD.6 However, the non-remission rate remains highly in patients with IBD after screening of these biomarkers.
The composition of the gut microbiota could affect different physiological functions of the host, even the uptake and utilization of drugs by the host.7,8 The interaction between microbiota and non-antibiotic drugs is bidirectional, including drug-induced alterations in microbiome composition and microbiota-mediated modulation of the pharmaceutical effect.9,10 The biochemical transformation of the oral drugs to more or less active metabolites were identified and validated by microbiome-encoded enzymes, with consequences for interpersonal variation in drug efficacy and toxicity.11,12,13 In addition, bacteria can bioaccumulate the therapeutic drug intracellularly and modulate the availability and efficacy of it in the gut.14 As an oral immunosuppressant, whether gut microbes affect the metabolism and the curative effect of AZA remains elusive.
This study was performed to reveal the mechanism of AZA therapy failure at the level of intestinal bacteria. We used 16S rDNA sequencing technology to screen out Blautia bacteria, which were associated with ineffective AZA treatment, and verified the bioinformatic data in another independent clinical cohort. The molecular mechanism of Blautia wexlerae (B. wexlerae)-induced AZA therapeutic failure was further explored in in vitro and in vivo models.
Results
Gut microbes were closely associated with therapeutic failure of AZA in patients with IBD
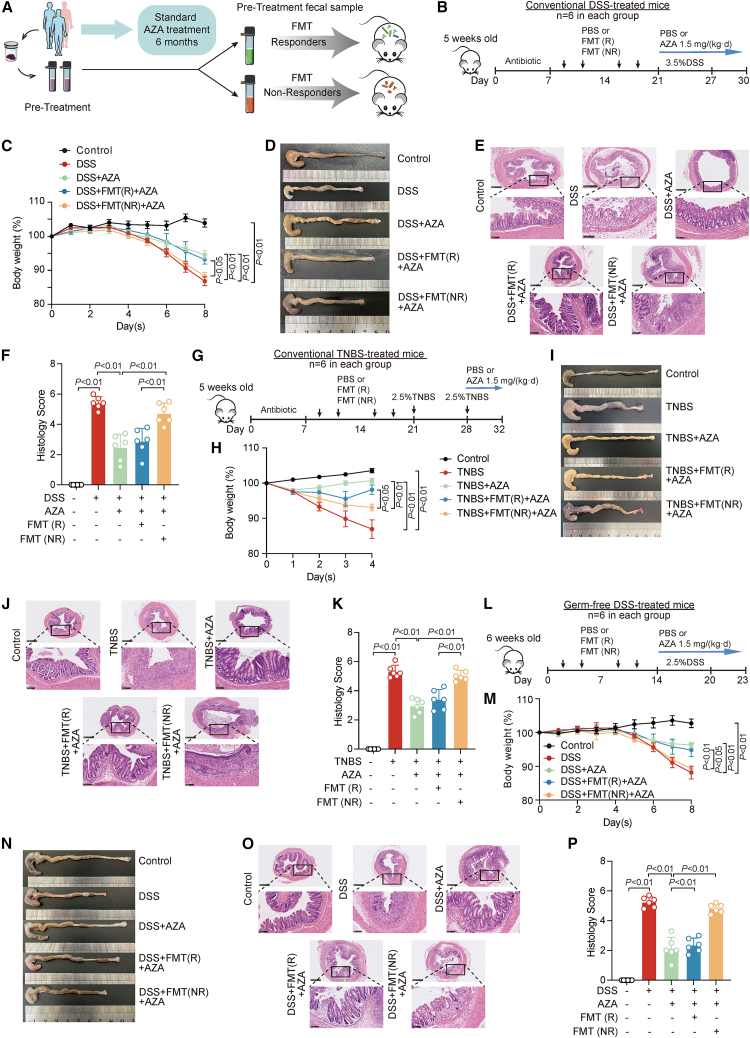
To evaluate the impact of gut microbiota on the efficacy of AZA treatment in patients with IBD, we collected the fecal samples of 49 patients with newly diagnosed IBD, containing 9 patients with UC and 40 patients with CD, before receiving AZA treatment (cohort 1). Patients in remission (R; n = 27) and non-remission (NR; n = 22) were divided based on their endoscopic and imaging evaluations after the initial administration of AZA for 6 months (Figure 1A; Table S1).15 Quantitative PCR analysis of fecal pellets showed that antibiotic-treated mice had a significantly reduced gut bacterial community16,17 (Figure S1A). To address the effect of gut microbiota on AZA treatment in patients with IBD, we performed fecal microbiota transplantation (FMT) experiments with feces from patients in R or NR into C57BL6 mice recipients with dextran sulfate sodium (DSS) treatment (Figure 1B). At sacrifice, AZA treatment failure was observed in DSS-treated mice with transplantation of NR-patient-derived fecal samples as indicated by decreased body weight (Figure 1C), shortened colonic length (Figures 1D and S1B), and increased inflammation pathological score (Figures 1E and 1F) as compared with those mice with transplantation of R-patient-derived fecal samples. These data suggested that the fecal microbiota, which were from NR patients with IBD with AZA treatment, exerted a significant resistant effect on AZA treatment in DSS-induced colitis in mice.
Figure 1.
Gut microbes were closely associated with therapeutic failure of azathioprine (AZA) in patients with inflammatory bowel disease
(A) Flow chart of fecal microbiota transplantation (FMT) experiments with feces from patients in remission (R) or patients in non-remission (NR).
(B) Schematic overview of treatments in conventional DSS-induced colitis model. n = 6 for each group.
(C) The alteration of body weight in conventional DSS-induced colitis mouse. n = 6/group, non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SEM.
(D) Gross morphology of the colon was shown in different treatment groups of conventional DSS-induced colitis mice.
(E) Mouse colonic sections were stained by hematoxylin and eosin in conventional DSS-induced colitis mice. Original magnification, ×5 (top row) and ×20 (bottom row).
(F) The histology scores of colonic sections in conventional DSS-induced colitis. n = 6/group, non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SD.
(G) Schematic overview of treatments in conventional TNBS-induced colitis model. n = 6 for each group.
(H) The alteration of body weight in conventional TNBS-induced colitis mouse. n = 6/group, non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SEM.
(I) Gross morphology of the colon was shown in different treatment group of conventional TNBS-induced colitis mice.
(J) Mouse colonic sections were stained by hematoxylin and eosin in conventional TNBS-induced colitis. Original magnification, ×5 (top row) and ×20 (bottom row).
(K) The histology scores of colonic sections in conventional TNBS-induced colitis. n = 6/group, non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SD.
(L) Schematic overview of treatments in germ-free DSS-induced colitis model. n = 6 for each group.
(M) The alteration of body weight in germ-free DSS-induced colitis mouse. n = 6/group, non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SEM.
(N) Gross morphology of the colon was shown in different treatment group of germ-free DSS-induced colitis mice.
(O) Mouse colonic sections were stained by hematoxylin and eosin in germ-free DSS-induced colitis mice. Original magnification, ×5 (top row) and ×20 (bottom row).
(P) The histology scores of colonic sections in germ-free DSS-induced colitis. n = 6/group, non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SD.
To further evaluate the effect of fecal microbiota-induced AZA treatment failure on colitis, we used another mouse model of 2,4,6-trinitrobenzene sulfonic acid (TNBS)-induced colitis (Figure 1G). Phenotype data showed that transplantation of NR-patient-derived fecal microbiota, but not R-patient-derived fecal microbiota, significantly reduced the therapeutic effect of AZA in the TNBS-induced mice colitis model by decreasing the body weight (Figure 1H) and colon length (Figures 1I and S1C) and exacerbating the inflammatory response (Figures 1J and 1K).
We also investigated the AZA treatment effect on DSS-induced colitis in germ-free mice after transplantation of fecal microbiota derived from different patients with IBD. Transplantation of NR-patient-derived microbiota dramatically weakened the AZA treatment effect in DSS-treated germ-free mice as characterized by body weight loss (Figure 1M), shortened colonic length (Figures 1N and S1D), and increased inflammation score (Figures 1O and 1P). Together, these data demonstrated that the fecal microbiota from NR patients with IBD might participate in AZA treatment failure in different mice with intestinal colitis.
Blautia bacteria could predict the effect of AZA therapy in patients with IBD
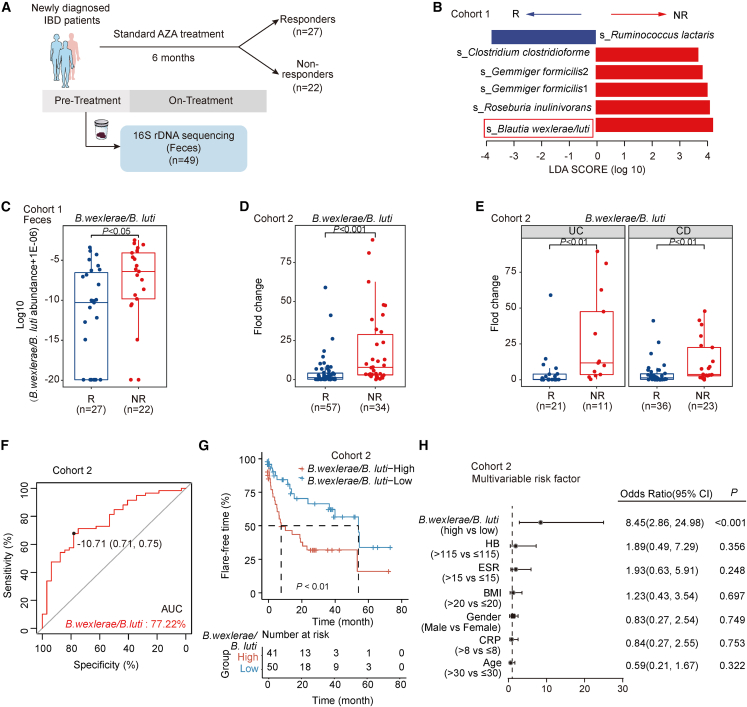
To identify the differentially abundant taxa of microbiota, 16S rDNA sequencing and Linear discriminant analysis Effect Size (LefSe) analysis were then performed between the fecal samples of R and NR patients with IBD in cohort 1 (Figure 2A). The abundances of four bacterial taxa were increased in NR patient groups (linear discriminant analysis [LDA] score > 2, P < 0.05), including B. wexlerae/luti, Roseburia inulinivorans, Gemmiger formicilis, and Clostridum clostridioforme (Figure 2B). Noticeably, the amount of B. wexlerae and B. luti was significantly enriched in the fecal samples (Figures 2C and S2A–S2C) and ileum tissues (Figures S2D–S2F) of the NR patient group compared with that of the R patient group in cohort 1. To validate the association between B. wexlerae and B. luti with the therapeutic efficacy of AZA, we analyzed the intestinal mucosa microbiota and AZA therapeutic efficacy in another independent cohort incorporating 91 patients with newly diagnosed IBD with AZA administration (cohort 2; Table S2). Real-time PCR data showed that the amount of B. wexlerae and B. luti was significantly increased in those patients with IBD (including 11 cases of UC and 23 cases of CD) in NR compared with other patients with IBD in R (Figures 2D, 2E, and S2G–S2J). These data indicated that high levels of B. wexlerae and B. luti were closely associated with the therapeutic failure of AZA in patients with IBD.
Figure 2.
Blautia bacteria could predict the effect of AZA therapy in patients with inflammatory bowel disease (IBD)
(A) Flow chart in cohort 1 of Renji hospital.
(B) LDA effect size analysis in the fecal samples of patients with IBD in NR (n = 22) and in R (n = 27) after performing 16S DNA sequencing.
(C) The amount of B. wexlerae/B. luti species in the feces of patients with IBD in cohort 1. Significance was determined using a non-parametric Wilcoxon rank-sum test (R, n = 27; NR, n = 22).
(D and E) The amount of B. wexlerae/B. luti species was significantly increased in the feces of NR group (D, R, n = 57; NR, n = 34), both in patients with ulcerative colitis (UC; R, n = 21; NR, n = 11) and patients with Crohn’s disease (CD; R, n = 36; NR, n = 23) (E). Non-parametric Wilcoxon rank-sum test.
(F) Classification performance of multivariable logistic regression model using relative abundance of B. wexlerae/B. luti was assessed by area under the receiver operating characteristic (ROC) in cohort 2 (R: n = 57, NR: n = 34, AUC: 77.22%, 95% confidence interval [CI]: 67.32%–87.12%), respectively.
(G) Analysis of the flare-free survival in patients with high (n = 41) versus low (n = 50) B. wexlerae/B. luti abundance in cohort 2.
(H) Multivariable analysis was performed in cohort 2. The bars correspond to 95% CIs.
We next assessed the potential value of predicting AZA efficacy using gut microbiota as biomarkers. We observed that the area under curve (AUC) value of B. wexlerae/B. luti-based prediction was 77.22%. Youden index was used to determine the optimal cut-off point, and −10.71 (−delta CT value) was selected based on the abundance of B. wexlerae/B. luti that provided the best balance between the sensitivity and the specificity to predict AZA treatment failure in patients with IBD (Figure 2F; Table S3). To assess how the variability in the amount of B. wexlerae and B. luti affected disease outcomes, we assessed the clinical flare-free time of patients with IBD with AZA administration categorized as high and low abundance of B. wexlerae/B. luti in cohort 2. A remarkable decrease in flare-free time was observed in patients with IBD with higher levels of B. wexlerae/B. luti compared with those patients with lower levels of B. wexlerae/B. luti (Figure 2G). In addition, univariate (Figure S2K) and multivariable (Figure 2H) regression analyses demonstrated that the amount of B. wexlerae/B. luti was an independent predictor of patients with IBD to predict the clinical outcome of cohort 2. Thus, the data in cohort 2 not only confirmed our observation in cohort 1 but also defined the potential value of the amount of B. wexlerae/B. luti to predict AZA therapy failure in patients with IBD.
B. wexlerae reduced the therapeutic effect of AZA in a DSS-induced acute colitis model
To explore whether B. wexlerae or B. luti is responsible for AZA therapy failure in patients with IBD, we used a DSS-induced acute colitis mice model with AZA treatment for further validation. To exclude the possibility that B. wexlerae and B. luti would aggravate colitis, mice were administrated B. wexlerae and B. luti, respectively, in an acute DSS-treated mice model (Figures S3A–S3C). No significant differences of weight loss (Figure S3D), disease activity index (DAI) score (Figure S3E), colon length (Figure S3F and S3G), pathological score (Figures S3H and S3I), inflammatory factors (Figures S3J–S3N), and immune cells of intestinal mucosa (Figures S3O–S3R) were detected among DSS, (B. wexlerae+DSS), and (B. luti+DSS) treatment mice groups, indicating that B. wexlerae and B. luti have no significant effect on DSS-induced acute colitis in mice.
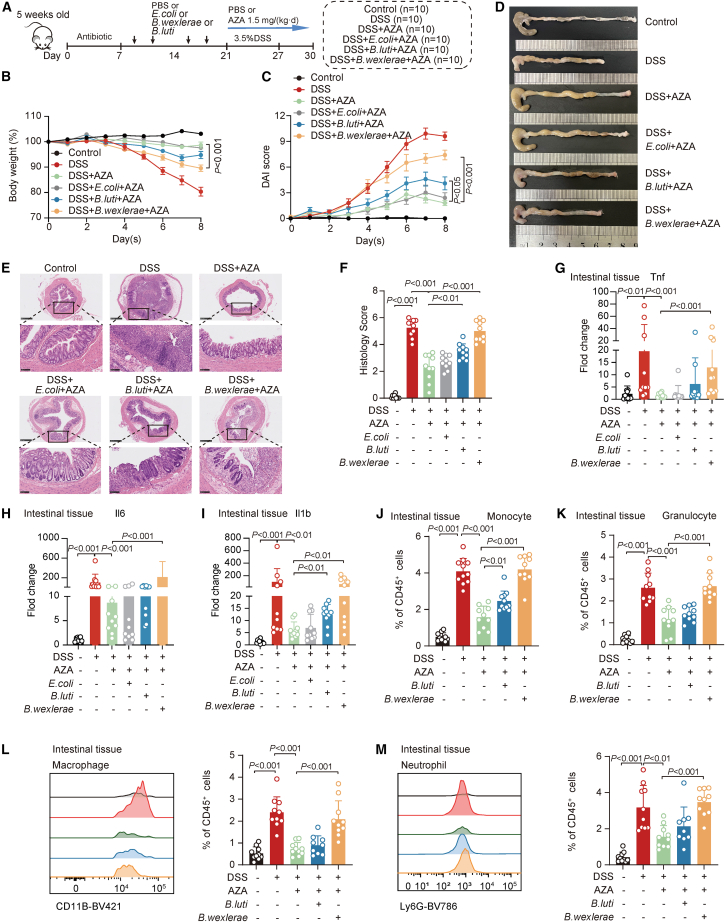
We next investigated the causal relationship between B. wexlerae and B. luti with AZA therapeutic effect on conventional acute DSS-treated mice (Figure 3A). Phenotype data showed that B. wexlerae, but not B. luti, administration significantly reduced the therapeutic effect of AZA characterized by aggravated weight loss (Figure 3B), increased DAI (Figure 3C), shortened colons (Figures 3D and S3S), and increased histology scores (Figures 3E and 3F) in the DSS-induced intestinal colitis mice model. Real-time PCR data showed that the expression levels of intestinal inflammatory factors, including tumor necrosis factor α (TNF-α), interleukin-1β (IL-1β), and IL-6, as well as the markers of monocyte and granulocyte cells, were significantly increased after AZA treatment in mice with B. wexlerae inoculation (Figures 3G–3I and S3T) compared with those in control mice. Flow cytometry validated that the percentages of monocyte and granulocyte cells, but not CD4+T and CD8+T cells, were significantly increased in the intestinal mucosa of the (DSS+B. wexlerae+AZA) treatment mice group (Figures 3J, 3K, and S3U). Further flow cytometry analysis data showed that the proportions of macrophages and neutrophils were also significantly increased in the (DSS+B wexlerae+AZA) treatment mice group compared with the (DSS+AZA) group (Figures 3L and 3M). These data indicated that B. wexlerae might reduce the therapeutic effect of AZA and might increase the amount of macrophage and neutrophil cells in the DSS-induced acute colitis model. Since the B. wexlerae-induced effect of AZA treatment failure is more significant than B. luti in vivo validation, we next focused on B. wexlerae for further validation.
Figure 3.
B. wexlerae reduced the therapeutic effect of AZA in a dextran sulfate sodium-induced acute colitis model
(A) Schematic diagram of the dextran sulfate sodium (DSS)-induced acute colitis model treated with B. wexlerae and B. luti in C57BL/6 mice with AZA treatment including control group (n = 10), DSS group (n = 10), (DSS+AZA) group (n = 10), (DSS+E. coli+AZA) group (n = 10), (DSS+B. luti+AZA) group (n = 10), and (DSS+B. wexlerae+AZA) group (n = 10).
(B and C) The alterations in mouse body weight (B) and DAI evaluation (C). n = 10/group, non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SEM.
(D) Gross morphology of the colon was shown in different treatment mice group.
(E) Mouse colonic sections were stained by hematoxylin and eosin. Original magnification, ×5 (top row) and ×20 (bottom row).
(F) The histology scores of mice in different mice groups. n = 10/group, non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SD.
(G–I) Real-time PCR was performed to detect Tnf (G), Il6 (H), and Il1b (I) in different treatment of mice groups. n = 10/group, non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SD.
(J and K) The flow cytometry analysis of monocyte (J) and granulocyte (K) in different mice groups. n = 10/group, non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SD.
(L and M) The flow cytometry analysis of macrophage (L) and neutrophil (M) in different mice groups. Representative images (left) and statistical proportion (right, n = 10/group, non-parametric Wilcoxon rank-sum test). Data are represented as mean ± SD.
B. wexlerae reduced the therapeutic effect of AZA and elevated the amount IL1b+S100a8+ inflammatory macrophages in a chronic mice colitis model
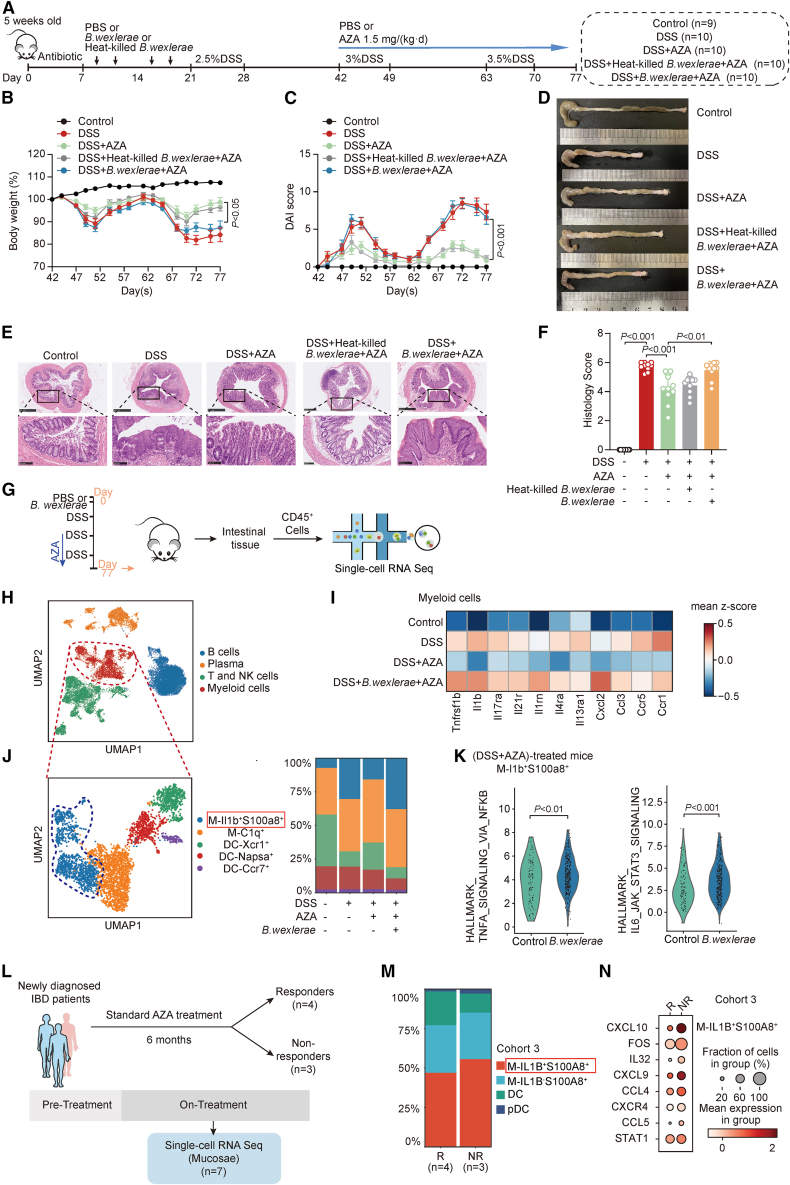
Next, we constructed a conventional chronic DSS-treated mice model, which mimicked the chronic inflammatory progression of patients with IBD more closely. Similarly, the role of B. wexlerae in aggravating colitis was first excluded, and a chronic DSS model was established after the mice were administrated B. wexlerae by gavage as described before (Figures S4A–S4G). B. wexlerae and heat-killed B. wexlerae were then respectively fed to the conventional DSS-induced chronic colitis mice with AZA treatment in different groups (Figure 4A). Phenotype data showed that B. wexlerae, but not heat-killed B. wexlerae, significantly reduced the therapeutic effect of AZA, characterized by aggravated weight loss (Figure 4B), increased DAI (Figure 4C), shortened colons (Figures 4D and S4H), and increased histology scores (Figures 4E and 4F), in the chronic colitis mouse model. These data indicated that B. wexlerae is responsible for AZA therapy invalidation in the DSS-induced chronic colitis mouse model as well.
Figure 4.
B. wexlerae reduced the therapeutic effect of AZA and elevated the amount IL1B+S100A8+ macrophages in chronic mice colitis model
(A) Schematic diagram of the DSS-induced chronic colitis model treated with B. wexlerae and heat-killed B. wexlerae in C57BL/6 mice including control group (n = 9), DSS group (n = 10), (DSS+AZA) group (n = 10), (DSS+heat-killed B. wexlerae+AZA) group (n = 10), and (DSS+B. wexlerae+AZA) group (n = 10).
(B and C) The alterations in mouse body weight (B) and DAI evaluation (C) in the control group (n = 9), DSS group (n = 10), (DSS+AZA) group (n = 10), (DSS+heat-killed B. wexlerae+AZA) group (n = 10), and (DSS+B. wexlerae+AZA) group (n = 10). Non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SEM.
(D) Gross morphology of the colon.
(E) Mouse colonic sections were stained by hematoxylin and eosin. Original magnification, ×5 (top row) and ×20 (bottom row).
(F) The histology scores of mice in different mice groups: control group (n = 9), DSS group (n = 10), (DSS+AZA) group (n = 10), (DSS+heat-killed B. wexlerae+AZA) group (n = 10), and (DSS+B. wexlerae+AZA) group (n = 10). Non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SD.
(G) Experimental design for scRNA-seq analysis of CD45+ immune cells from different groups of mice.
(H) UMAP of major immune cell clusters in mice.
(I) Heatmap showing the expression of proinflammatory genes in myeloid cells of mice.
(J) UMAP plot displaying 3,672 myeloid cells separated into 5 subtypes in mice (left), and bar plots exhibiting the cellular sources for mice myeloid cell subtypes (right). Blocks represent different subtypes and are color coded by their derived types. Block heights are proportional to the number of detected cells.
(K) Violin plots showing the activated pathways in macrophage (M)-Il1b+S100a8+ in (DSS+AZA)-treated mice with high B. wexlerae abundance by gene set-enriched score analysis. Non-parametric Wilcoxon rank-sum test.
(L) Flow chart in cohort 3.
(M) Bar plots exhibiting the myeloid cell subtypes in colon mucosa of patients with IBD. Blocks represent different subtypes and are color coded by their derived types (R, n = 4; NR, n = 3).
(N) Dot plot showing the expression of proinflammatory genes in IL1b+S100A8+ macrophages of patients with IBD with different responses to AZA (R, n = 4; NR, n = 3).
In addition to the gut microbiome, immune system disorder and the imbalance of immune cell proportions are regarded as the main pathogenesis factors in IBD.18,19To discriminate which immune cells participated in B. wexlerae-induced AZA therapy failure in IBD, we collected CD45+ cells from intestinal tissue of the chronic colitis mouse. The transcriptomes of individual cells were obtained by single-cell RNA sequencing (RNA-seq) (Figure 4G). After data pre-processing and quality control, we obtained 16,204 transcriptomes of single cells. These cells were partitioned into four groups using unsupervised graph clustering (Figure 4H), visualized by uniform manifold approximation and projection (UMAP) and labeled according to the marker gene expression (Figure S4I). Considering that the proportion of myeloid cells was the most significantly increased after AZA treatment in acute colitis mice with high B. wexlerae abundance, we firstly verified the functions of myeloid cells in chronic colitis. Higher expression of the proinflammatory genes were observed in all myeloid cell populations of the (DSS+B. wexlerae+AZA) treatment mice groups compared with other groups (Figure 4I). Then, these 3,114 myeloid cells were subgrouped into five subsets (Figures 4J and S4J) to unveil the exact subgroup that promoted intestinal colitis in mice. We found that the amount of Il1b+S100a8+ macrophages, which were reported to be associated with intestinal inflammation,20 was increased in chronic colitis compared with control and that it was rescued after AZA treatment in mice (Figure 4J). Moreover, B. wexlerae treatment significantly increased the amount of Il1b+S100a8+ macrophage cells in the (DSS+AZA)-treatment mice compared with other groups (Figure 4J). Further gene set-enriched score analysis revealed a strong enrichment of inflammation-related pathways in Il1b+S100a8+ macrophage cells of (DSS+AZA)-treatment mice with B. wexlera high abundance compared with those mice with B. wexlerae low abundance (Figure 4K), suggesting that B. wexlerae-induced AZA treatment failure might depend on increasing the proportion of Il1b+S100a8+ inflammatory macrophage cells in the DSS-induced colitis mouse.
Meanwhile, we analyzed another cohort of patients with IBD (n = 7, cohort 3; Table S4) with standard AZA treatment and obtained 28,611 single-cell profiles from their colon biopsies. Patients were divided into responders (n = 4) and non-responders (n = 3) based on 6-month follow-up after the initial AZA administration (Figure 4L). Four clusters of cells were identified according to marker gene expression (Figures S4K–4M). Proportion analysis showed that the percentage of IL1B+S100A8+ macrophage cells was significantly increased in the non-responders group (Figures 4M, S4N, and S4O) compared with the responder group in patients with IBD. Moreover, the expression of the proinflammatory cytokine was higher in Il1B+S100A8+ macrophages of non-responders than in those of responders in patients with IBD (Figure 4N), which is consistent with the data in the chronic colitis mouse model. Further Gene Ontology (GO) analysis data showed that a strong enrichment of immune response pathways was detected in Il1B+S100A8+ macrophage cells of the NR group (Figure S4P). These data suggested that the anti-inflammatory effect of AZA might rely on inhibiting the proportion of IL1B+S100A8+ inflammatory macrophages and that B. wexlerae inoculation significantly impaired the AZA-mediated inhibitory effect on IL1B+S100A8+ macrophages in patients with IBD.
B. wexlerae was closely associated with the biotransformation of 6-mercaptopurine
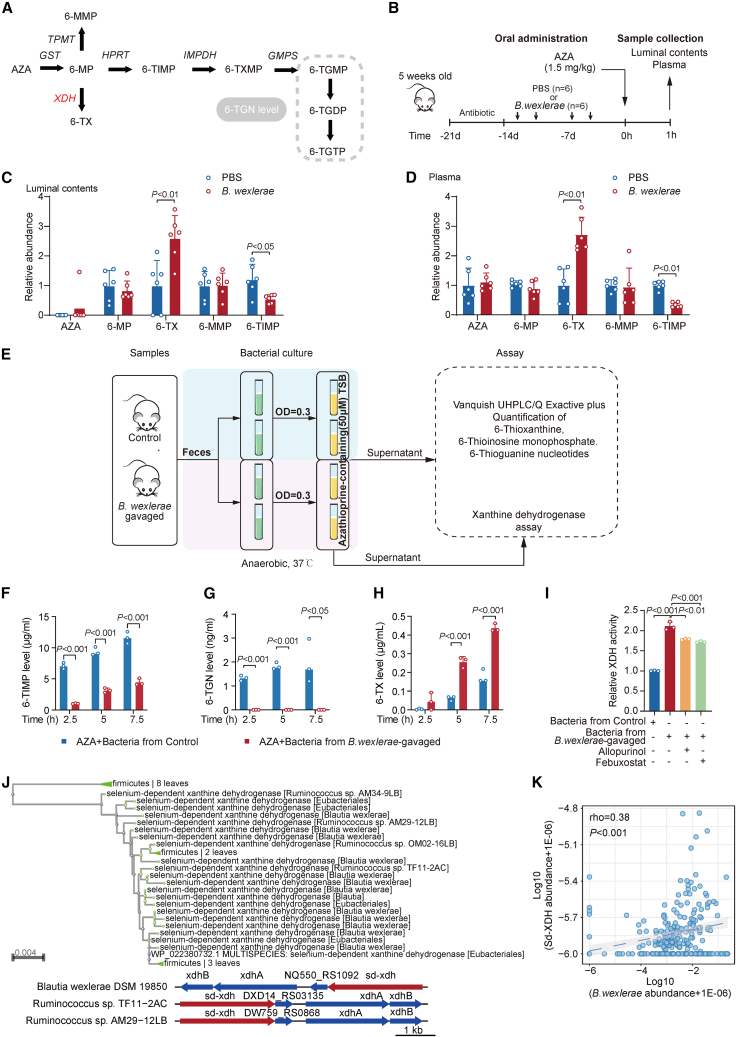
Mass spectrometry analysis and real-time PCR data showed that AZA, the intermediated metabolites of AZA, and B. wexlerae were detectable in the upper, middle, and lower segments of the small intestine in B. wexlerae-enriched mice, respectively (Figures S5A and S5B). Therefore, we considered that B. wexlerae and AZA might meet in the small intestine of mouse. To explore the mechanism of B. wexlerae-induced therapeutic failure of AZA in the intestinal colitis of mouse, we first evaluated whether this bacterium could directly degrade or bioaccumulate AZA. Vanquish ultra-high-performance liquid chromatography (UHPLC)/Q Exactive plus analysis showed that B. wexlerae did not alter the AZA level in the bacterial culture tryptic soy broth (TSB) medium (Figure S5C), indicating that B. wexlerae could not degrade AZA.
Next, we examined whether B. wexlerae affected the drug metabolism of AZA. AZA belongs to a prodrug, which is converted to 6-mercaptopurine (6-MP).21,22 6-MP is then further metabolized by downstream enzymes and is ultimately metabolized to 6-TGNs, consisting of 6-thioguanine-monophosphate (6-TGMP), 6-diphosphate (6-TGDP), and 6-triphosphate (6-TGTP), which are considered to contribute to the immunosuppressive effects in IBD treatment21,22 (Figure 5A). We next evaluated the amount of AZA and its downstream metabolites in mice with or without B. wexlerae treatment. Antibiotic-treated mice were firstly administered B. wexlerae, followed by oral administration of AZA (Figure 5B). Vanquish UHPLC/Q Exactive plus analysis showed that the intermediate metabolite 6-TX (6-thioxanthine), which is the precursor of the non-active form 6-thiouric acid (6-TU), was significantly increased, and another active intermediate, metabolite 6-thioinosine-monophosphate (6-TIMP), which is the precursor of the pharmaceutical active form of 6-TGN (6-TGMP, 6-TGDP, 6-TGTP), is significantly decreased in mice after B. wexlerae inoculation in the luminal contents (Figure 5C) and in the serum samples (Figure 5D). Since xanthine dehydrogenase (XDH) is responsible for 6-MP transforming into 6-TX and hypoxanthine phosphoribosyltransferase (HPRT) is responsible for 6-MP transforming into 6-TIMP in AZA metabolism, we first detected the activity of these two enzymes in mouse intestinal tissue. ELISA data showed that the activity of XDH and HPRT has no significantly alteration in mouse intestinal tissue after B. wexlerae administration compared with control mice (Figure S5D), which suggested that the B. wexlerae-induced therapeutic failure of AZA might depend on decreasing the bioavailability of AZA by the bacteria but not by the host.
Figure 5.
B. wexlerae was closely associated with the biotransformation of AZA
(A) Proposed thiopurine metabolism. AZA, azathioprine; 6-MP, 6-mercaptopurine; 6-TIMP, 6-thioinosine monophosphate; 6-TXMP, 6-thioxanthosine monophosphate; 6-TGMP, 6-thioguanine monophosphate; 6-TGDP, 6-thioguanine diphosphate; 6-TGTP, 6-thioguanine triphosphate; 6-TGN, 6-thioguanine; GST, glutathione S-transferase; HPRT, hypoxanthine phosphoribosyl transferase; IMPDH, inosine monophosphate dehydrogenase; GMPS, guanosine monophosphate synthetase. In therapeutic drug monitoring, a 6-TGN level consists of the sum of 6-TGMP, 6-TGDP, and 6-TGTP levels.
(B) Schematic overview of experimental design for studying the effect of B. wexlerae and B. luti on AZA bioavailability in C57BL/6 mice. n = 6 for each group.
(C and D) Quantitative analysis of AZA and its metabolite levels in luminal contents (C) and plasma (D). n = 6/group, non-parametric Wilcoxon rank-sum test. Data are represented as mean ± SD.
(E) Schematic overview of experimental design for studying the impact of B. wexlerae on AZA in vitro. The green tube contains fecal bacterial suspension (OD = 0.3). The yellow tube contains fecal bacterial suspension (OD = 0.3) with AZA supplement. Control mice (blue block). B. wexlerae-gavaged mice (purple block).
(F–H) Quantitative levels of 6-TIMP (F), 6-TGN (G), and 6-TX (H) in AZA-containing culture medium post-anaerobic incubation with fecal samples. n = 3/group, Student’s t test. Data are represented as mean ± SD.
(I) XDH activity in the medium exposed to fecal samples under anaerobic conditions. n = 3/group, Student’s T test. Data are represented as mean ± SD.
(J) Bioinformatic analysis showing B. wexlere might encode a selenium-dependent xanthine dehydrogenase (sd-XDH) gene.
(K) Correlation analysis of bacterial sd-XDH and B. wexlerae in PRJNA385949.
We next verified this hypothesis in vitro. Fecal samples from B. wexlerae-gavaged and control mice were cultured in AZA-containing TSB medium under anaerobic conditions, following by examination of the intermediate metabolites of AZA in the medium (Figure 5E). We found that the production of 6-TIMP and 6-TGN (Figures 5F and 5G) was significantly impaired in the medium exposed to fecal bacteria from B. wexlerae-gavaged mice compared with that from control mice, while the production of 6-TX was significantly increased in the same treatment mice groups (Figure 5H). These data collectively supported that B. wexlerae-enriched feces promoted the conversion of 6-MP into its non-active form, 6-TX, but not its active form, 6-TGN.
Enzymatic modification of drugs by gut bacteria (biotransformation or metabolization) is a well-known and widely recognized interaction mode.9 Further ELISA data showed that XDH activity, but not HPRT activity, was significantly increased in the medium exposed to fecal bacteria from B. wexlerae-administrated, antibiotic-treated mice than that from control mice under anaerobic culture conditions (Figures 5I and S5E). Since experiments to knockout selenium-dependent (sd)-XDH in B.wexlerae are not feasible currently, we next used the inhibitors of XDH to explore the effect of B.wexlerae-induced AZA therapeutic failure in vitro. Allopurinol or febuxostat (the inhibitors of XDH) treatment significantly reduced the B. wexlerae-induced increase in XDH activity (Figure 5I) and 6-TX levels (Figure S5F) in fecal bacteria from mice. We next analyzed the DNA sequence of B. wexlerae on the NCBI website. The bioinformatic analysis data showed that B. wexlere might encode an sd-XDH (Figures 5J and S5G). Further correlation analysis of the sd-XDH gene abundance in different taxonomic groups across the (74 control and 185 IBD) metagenomes (PRJNA385949) showed that the number of sd-XDH reads per metagenome (rho = 0.38, p < 0.0001) was positively associated with the relative abundance of B. wexlere (Figure 5K). Consistently, these data illustrated that the sd-XDH gene harbored by B. wexlerae might mediate the biotransformation of 6-MP.
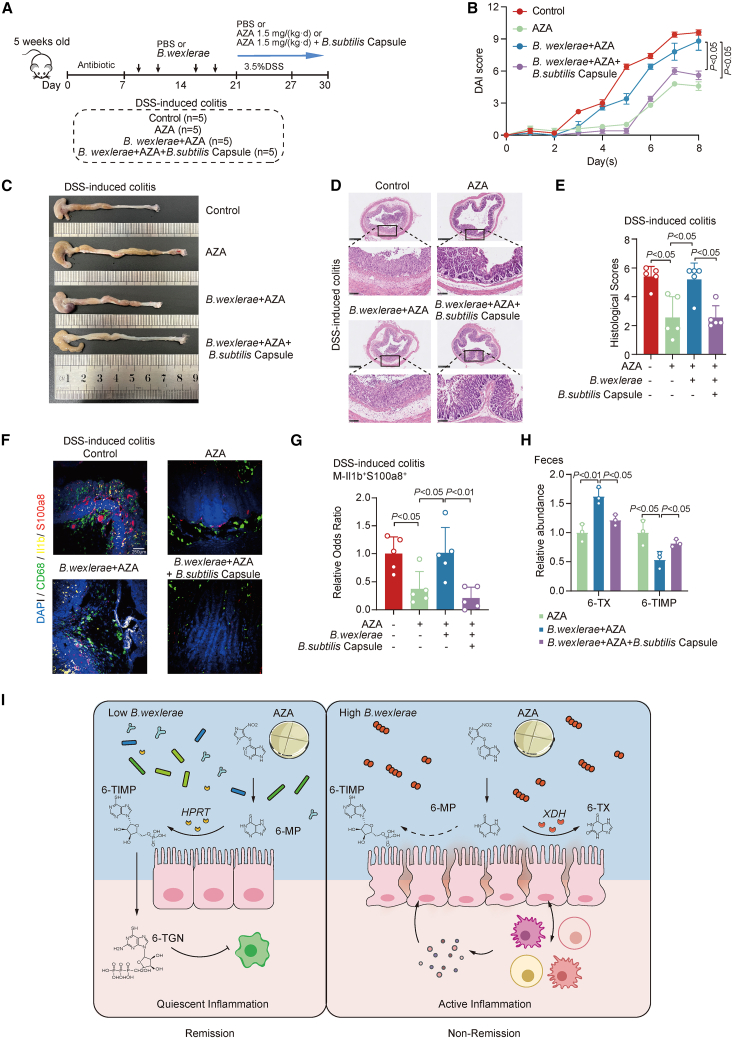
B. subtilis supplementation reversed B. wexlerae-induced AZA therapeutic failure in an intestinal colitis mouse model
The drug interaction between XDH inhibitors and AZA can result in clinically significant adverse events.23,24 Thereby, we next explored whether B. subtilis, which has been reported to encode the HPRT gene before,25 could rescue B. wexlerae-induced AZA treatment failure. Real-time PCR data showed that the B. subtilis capsule, which contained B. subtilis bacteria, could enhance the abundance of B. subtilis in the fecal samples of mice (Figure S6A). Functional validation data showed that B. subtilis treatment significantly rescued B. wexlerae-mediated AZA therapy failure (Figure 6A), characterized by alleviated weight loss (Figure S6B), decreased DAI (Figure 6B), longer colons (Figures 6C and S6C), and decreased histology scores (Figures 6D and 6E), in the intestinal colitis mouse model. In addition, supplementation of B. subtilis significantly reduced the B. wexlerae-induced increase in the percentage of monocytes (Figure S6D) and macrophages (Figure S6E) in colitis mice with AZA treatment. Multi-immunofluorescent staining showed that the signals of Il1b and S100a8 were strongly enriched in macrophages in the intestinal tissues of the B. wexlerae-enriched mice group compared with control mice groups with DSS and AZA treatment (Figures 6F and 6G). B. subtilis treatment dramatically decreased this bacteria-induced upregulation of Il1b and S100a8 in macrophages in vivo (Figures 6F and 6G) and effectively blocked B. wexlerae-induced biotransformation of 6-MP into 6-TX (Figure 6H). These data indicated that B. subtilis treatment effectively reduced B. wexlerae-induced activation of inflammatory macrophage cells in intestinal colitis mice with AZA treatment. These data indicated that supplementation of B. subtilis might effectively and specifically restore B. wexlerae-induced AZA therapy failure in patients with IBD.
Figure 6.
B. subtilis supplementation reversed B. wexlerae-induced AZA therapeutic failure in intestinal colitis mouse model
(A) Schematic diagram of the DSS-induced acute colitis model with different treatments including AZA, B. wexlerae, and B. subtilis capsules. Control group (n = 5), AZA group (n = 5), (B. wexlerae+AZA) group (n = 5), and (B. wexlerae+AZA+B. subtilis capsule) group (n = 5) in DSS-induced colitis model.
(B) The alterations in mouse DAI score. n = 5/group, Student’s t test. Data are represented as mean ± SEM.
(C) Gross morphology of the colon in different treatment groups of DSS-induced colitis mice.
(D) Mouse colonic sections were stained by hematoxylin and eosin. Original magnification, ×5 (top row) and ×20 (bottom row).
(E) The histology scores of colonic sections in C57BL/6 intestinal colitis mice. n = 5/group, Student’s t test. Data are represented as mean ± SD.
(F) Representative images for each marker of Il1b+S100a8+ macrophages were shown in mice colon of different groups.
(G) Relative odds ratios (OR) across samples of inflammatory macrophage markers Il1b and S100a8 co-staining with CD68 macrophage cells versus total cells are shown. n = 5/group, Student’s t test. Data are represented as mean ± SD.
(H) Relative abundance of 6-TX and 6-TIMP in feces of mice. n = 5/group, Student’s t test. Data are represented as mean ± SD.
(I) Schematic diagram of the relationship among B. wexlerae, XDH, and AZA treatment failure in patients with IBD. AZA, azathioprine; 6-MP, 6-mercaptopurine; 6-TIMP, 6-thioinosine-monophosphate; 6-TX, 6-thioxanthine; 6-TGN, 6-thioguanine nucleotide; XDH, xanthine dehydrogenase; HPRT, hypoxanthine phosphoribosyltransferase.
We also explored whether B. wexlere treatment could alter the composition of gut microbiota in DSS-plus-AZA-treated mice. Shotgun metagenomics analysis was performed in fecal samples of DSS-plus-AZA-treated mice with or without B. wexlere inoculation. Principal-component analysis showed that the gut microbiota composition of DSS-plus-AZA-treated mice was significantly different in B. wexlere-inoculated and control mice (Figure S6F). LDA with effect size revealed that the pathogenic bacteria Oscillibacter sp. 57_20 and Oscillibacter sp. KLE 172826 were enriched and that the amount of probiotic bacteria, including Eubacterium sp. 3_1_31 [C] and Lactobacillus animalis,27,28 were reduced in B. wexlere-treated mice compared with control mice (Figure S6G). These data suggested that B. wexlere-induced AZA treatment failure might depend on increasing the amount of pathogenic bacteria in intestinal colitis mice.
Discussion
The gut microorganism is a recognized player in host fitness,29 modulating numerous bioactive molecules, and may affect many drugs’ potency via different mechanisms.9,10,11,12,13,14 We currently bridged the knowledge gap between the gut microbes and AZA efficacy by using different mouse models. Functional assay showed that AZA-mediated immunosuppressive function was weakened after transplantation of NR-patient-derived feces, but not R-patient-derived feces, into antibiotic-treated and germ-free intestinal colitis mice. We further demonstrated that B. wexlerae was involved in AZA therapy failure in patients with IBD via blocking the biotransformation of 6-MP and increasing the amount of Il1b+S100a8+ inflammatory macrophages by using a combination of genomic, bioinformatics, and biological approaches, in vivo models, and clinical studies (Figure 6I).
The gut microbiota has been identified as an essential factor in driving inflammation and the development of mucosal lesions in IBD,30,31 so we first explored whether B. luti and B. wexlerae could promote inflammation in DSS-treated mice. Flow cytometry data showed that both B. luti and B. wexlerae could not aggravate colitis by decreasing the amount of CD8+ T cells in mice under DSS-treatment conditions. The data are consistent that B. luti and B. wexlerae could exert an anti-inflammatory effect in peripheral blood mononuclear cell cultures in vitro32 and that oral administration of B. wexlerae to mice induced metabolic changes and anti-inflammatory effects.33 The mechanism of B. wexlerae and B. luti inducing anti-inflammatory effects or decreasing the amount of CD8+ T cells still needs to be further explored. Furthermore, our findings indicated that while B. wexlerae did not promote DSS-induced colitis in mice, it impaired the efficacy of AZA in treating intestinal inflammation. In addition, Faecalibacterium prausnitzii was also reported to product a 15-kDa protein with anti-inflammatory properties preventing colitis.34 However, the acyl-CoA N-acyltransferases encoded by F. prausnitzii are involved in the metabolism of 5-aminosalicylic acid, diminishing its clinical efficacy in patients with IBD.35 Therefore, the same bacteria may have different effects depending on the environmental conditions.
Gut microbiota has been reported to be involved in the metabolism of a range of drugs, especially orally administered, small-molecule drugs.9,36 In some individual mice, up to 70% of the drug transformation can be ascribed to microbial metabolism,13 further emphasizing that intestinal bacteria may be the second largest reservoir of metabolic enzymes besides the liver. Bacterial levodopa metabolism by tyrosine decarboxylases has been identified, dominantly driven by Enterococcus and Lactobacillus species, which is problematic because dopamine generated in the periphery cannot cross the blood-brain barrier and causes unwanted side effects.37,38 Lysinibacillus sphaericus showed high arylesterase activity, which depicted aspirin degradation and reduced the chemopreventive effects of aspirin in mice.39 Microbiome-derived acarbose kinases from the human gut and oral microbiome encode kinases that selectively phosphorylate acarbose, resulting in its inactivation.40 In this case, our studies demonstrated that B. wexlerae significantly reduced the 6-TIMP level (the active form of AZA metabolism), increased the 6-TX level (the non-active form of AZA metabolism), and further compromised the IBD anti-inflammatory effect of AZA. The B. wexlerae-induced increase in XDH activity and the 6-TX level were significantly decreased by the XDH inhibitors in fecal bacteria of mice. Thus, we supposed that one of the reasons for AZA therapy failure in patients with IBD with a high amount of B. wexlerae might account for increasing this bacteria-derived sd-XDH activity and decreasing the biotransformation efficiency of 6-MP in intestinal lumen. Drug interaction between XDH inhibitors and AZA can result in clinically significant adverse events,23 and combination therapy with low-dose AZA and allopurinol is still under research.24 We hence used another probiotics B. subtilis, which codes HPRT, for in vivo validation. Supplementation of B. subtilis effectively increased the amount of 6-TIMP and successfully rescued B. wexlerae-induced AZA treatment failure in vivo. These findings demonstrated the important role of B. wexlerae in reducing AZA bioavailability and dampening the treatment efficiency of AZA in IBD.
It has been shown that immune cells exposed to specific gut microbes can infiltrate the tumor microenvironment and produce chemotactic factors affecting response to checkpoint inhibitors.10 Since B. wexlerae has been validated to slightly alleviate colitis, we hypothesized that B. wexlerae might affect the composition of intestinal microbiota in colitis mice under AZA treatment. Metagenomic sequencing analysis showed that B. wexlerae treatment resulted in the accumulation of pathogenic bacteria, including Oscillibacter. It is becoming evident that an unbalanced gut microbial structure may trigger or promote inflammation in intestine.41 It is plausible that the enhanced enrichment of pathogenic species by B. wexlerae might contribute to the activation of intestinal inflammation, including IL1B+S100A8+ inflammatory macrophages in IBD with AZA treatment failure. Besides the degrading effect of B. wexlerae on AZA, additional explorations are needed to evaluate the impact of intestinal inflammation activation by B. wexlerae in AZA treatment failure of patients with IBD in the future.
In addition to its biological importance, our data raised an important question: whether regular therapeutic regimens including AZA treatment are suitable for patients with IBD with a high amount of B. wexlerae. Since it is challenging to obtain tissue samples from different parts of the small intestine in clinical settings, this study had a limitation in investigating the distribution of AZA and B. wexlerae in the mucosal tissue of different segments of the human small intestine. Therefore, the impact of B. wexlerae on the effectiveness of AZA in patients with IBD needs to be reevaluated. While B. wexlerae may be considered a beneficial bacterium in other disease models, in our animal model, it was found to inhibit the therapeutic effects of AZA in mouse colitis. Further studies with a larger number of clinical samples are necessary to explore the role of B. wexlerae in inducing treatment failure of AZA in patients with IBD.
Limitations of the study
While this study discovered the impact of gut microbiota on the efficacy of AZA in patients with IBD and identified the key bacterium B. wexlerae, there are several limitations to consider. Firstly, B. wexlerae was screened from the fecal samples of patients with IBD, and although its presence was also found in the terminal ileum tissue, the exact location of B. wexlerae in the upper gastrointestinal tract of patients with IBD and its role in disrupting AZA efficacy cannot be determined due to limitations in sampling the proximal small intestine of patients with IBD. Additionally, although extensive animal and in vitro experiments validated the inhibitory effect of B. wexlerae on AZA biotransformation, this study does not provide sufficient evidence to lead to significant changes in clinical practice, such as predicting the response to AZA in patients with IBD based on microbial composition in different segments of the intestine. In conclusion, the limitations of this study suggest the need for further research in patients with IBD to elucidate the interaction between B. wexlerae and AZA biotransformation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Ms CD45 APC-Cy7 30-F11 | BD Pharmingen | Cat# 557659; RRID:AB_396774 |
| Ms CD3e PerCP-Cy5.5 145-2C11 | BD Pharmingen | Cat# 551163; RRID: AB_394082 |
| Ms CD4 FITC RM4-5 | BD Pharmingen | Cat# 553046; RRID: AB_394582 |
| Ms CD8a PE 53-6.7 | BD Pharmingen | Cat# 553032; RRID: AB_394570 |
| Ms Ly-6C BV605 AL-21 | BD Pharmingen | Cat# 563011; RRID: AB_2737949 |
| ANTI-MO CD11B M1/70 EF450 | Thermo | Cat# 48-0112-82; RRID: AB_1582236 |
| Ms Ly-6G/Ly-6C APC RB6-8C5 | BD Pharmingen | Cat# 553129; RRID: AB_398532 |
| Ms F4/80 APC-R700 T45-2342 | BD Pharmingen | Cat# 565787; RRID: AB_2869711 |
| Brilliant Violet 785 anti-mouse Ly-6G | BioLegend | Cat# 127645; RRID: AB_2566317 |
| Fixable Viability Stain 510 | BD Pharmingen | Cat# 564406; RRID: AB_2869572 |
| CD68 (E3O7V) Rabbit mAb | Cell Signaling Technology | Cat# 97778; RRID: AB_2928056 |
| Anti-IL-1 beta antibody | Abcam | Cat# ab9722; RRID:AB_308765 |
| Anti-S100A8 + S100A9 antibody | Abcam | Cat# ab17050; RRID: AB_443607 |
| Bacterial and virus strains | ||
| Blautia wexlerae | ATCC | 14507 |
| Blautia luti | DSM | 14534 |
| Biological samples | ||
| Human stool specimens | Renji Hospital affiliated to the School of Medicine, Shanghai Jiao Tong University | 2021-skt-004 |
| Chemicals, peptides, and recombinant proteins | ||
| DSS | MP | 216011090 |
| Vancomycin | Meilunbio | MB1260 |
| Ampicillin | Meilunbio | MB1507 |
| Neomycin | Meilunbio | MB1716 |
| Metronidazole | Meilunbio | MB2200 |
| Critical commercial assays | ||
| Mouse CD45 Positive Selection Kit | StemCell | #18945 |
| Xanthine Oxidase (XOD) activity detection Kit | Solarbio | BC1095 |
| QIAamp DNA FFPE Tissue Kit | QIAGEN | 56404 |
| QIAamp PowerFecal Pro DNA Kit | QIAGEN | 51804 |
| Four-color multiplex fluorescent immunohistochemical staining kit | Absin | abs50028 |
| Deposited data | ||
| 16S rDNA sequencing data | This paper | China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (Accession Number: PRJCA015633) |
| scRNA-seq data | This paper | Zenodo (https://doi.org/10.5281/zenodo.7754718, https://zenodo.org/record/7754718#.ZBlHz1lBz9s; https://doi.org/10.5281/zenodo.7754741, https://zenodo.org/record/7754741#.ZBlJ0VlBz9s) |
| Metagenome sequencing raw data of longitudinal stool study of patients with IBD and controls | Hall et al.42 | SRA database: PRJNA385949 |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice | Shanghai Jihui Laboratory Animal Care | N/A |
| Oligonucleotides | ||
| Bacterial universal 16S rDNA gene primers: F: 5′-GGTGAATACGTTCCCGG-3′ R: 5′-TACGGCTACCTTGTTACGACTT-3′ |
This paper | N/A |
| Primers for Blautia wexlerae/Blautia luti: F: 5′- GCATAAGCGCACAGAGCT-3′ R: 5′- CACATCAGACTTGCCACA-3′ |
Touyama et al.43 | N/A |
| Primer for mouse β-actin, Tnf-α, Ifn-γ, Il-1b, Il-17, Il-6. More details see Table S6. | Oancea et al.36 | N/A |
| Primer for Blautia wexlerae, Blautia luti and mouse Cd4, Cd8a, Ptprc, Cd19, Sdc1, Tnfrsf17, Ly6c, Ly6g, Itgax, Ncam1, Itga2. More details see Table S6. | This paper | N/A |
| Primer for pycA. More details see Table S6. | Lee et al.44 | N/A |
| Software and algorithms | ||
| Prism V9.0 | Graph Pad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo | FlowJo LLChttps | https://www.flowjo.com/ |
| R v4.1.2 | R | https://www.r-project.org/ |
| VSEARCH v2.15.0 | Rognes et al.45 | https://github.com/torognes/vsearch |
| Scanpy Package v1.8.1 | Wolf et al.46 | https://github.com/theislab/Scanpy |
| fastp 0.23.2 | Chen et al.47 | https://github.com/OpenGene/fastp |
| curatedMetagenomicData v3.6.2 | Pasolli et al.48 | https://github.com/waldronlab/curatedMetagenomicData |
| mOTUs v2.0.1 | Milanese et al.49 | https://github.com/motu-tool/mOTUs |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jie Hong, PhD (jiehong97@sjtu.edu.cn).
Materials availability
All reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and study participant details
Study design and clinical specimen collection
A total of 227 IBD patients receiving AZA treatment were divided into 4 cohorts. Their basic information was collected before AZA treatment (Tables S1, S3 and S4). In Cohort 1 (n = 49), fecal samples from all the enrolled patients were obtained before standard AZA treatment and 16S rDNA sequencing was conducted on their feces samples for the identification of AZA-treatment-response-related microbe. In Cohort 2 (n = 91), colonoscopy paraffin-embedded mucosa was collected from newly diagnosed patients with IBD before AZA treatment. The 72-month follow-up results in Cohort 2 were acquired for external validation and cutoff value determination. Single-cell RNA sequencing was performed in 7 IBD patients in Cohort 3 in order to illuminate the relationship between immune microenvironment and the response to AZA treatment. The origins of Cohort 1–3 were identified in a summary table (Table S5).
Bacterial strain and culture conditions
Blautia wexlerae (ATCC 14507) and Blautia luti (DSM 14534) were cultured at 37°C under anaerobic conditions (DG 250, Don Whitley Scientific, West Yorkshire, UK) in Tryptic Soy Broth supplemented with Yeast Extract, L-cysteine hydrochloride, Hemin Stock, and Vitamin K1 Stock.
Animal experiments and treatments
Male C57BL/6 mice were purchased from Shanghai Jihui Laboratory Animal Care Co.Ltd. All mice were housed and reared at the animal center of Renji Hospital Affiliated to Shanghai Jiao Tong University School of Medicine in specific pathogen free (SPF) conditions.
To enhance B. wexlerae and B. luti species colonization, 5-week-old mice were administered with a cocktail of vancomycin (0.5 g/L, MB1260, Meilunbio, Dalian, China), ampicillin (1 g/L, MB1507, Meilunbio, Dalian, China), neomycin (1 g/L, MB1716, Meilunbio, Dalian, China), and metronidazole (1 g/L, MB2200, Meilunbio, Dalian, China) 7 days before peroral bacterial inoculations. Afterward, the mice were gavaged with bacterial inoculations twice a week for 2 weeks. B. wexlerae and B. luti colonization in fecal were examined regularly. Heat-killed B. wexlerae was sterilized by high pressure steam (120°C at 30MPa, maintained for 30 min). B. subtilis capsule was administered intragastrically according to the instruction (60 mg/kg or 6 × 106 CFU/kg). Two weeks after inoculation, mice were randomly divided into indicated groups for sets of experiments. The mice were then treated differently according to the purpose.
Additional methods are provided in online supplemental information.
Method details
Clinical assessment
Every participant has received standard therapy including 7-day induction therapy and the subsequent maintenance therapy in accordance with Chinese expert consensus.50 During the induction therapy, patients were treated with 40 mg/day methylprednisolone intravenously. Patients diagnosed with UC or CD followed our inclusion and exclusion criteria were recruited in Renji Hospital affiliated to the School of Medicine, Shanghai Jiao Tong University from 2012 to 2022. The patient cohorts were approved by the ethics committee of Renji Hospital affiliated to the School of Medicine, Shanghai Jiao Tong University, and the protocol was approved by the Ethics Committee of 2021-skt-004.
Inclusion criteria
-
(1)
Participants who have been newly diagnosed with IBD
-
(2)
Participants who have received standard therapy including steroid induction remission and AZA maintenance therapy.
-
(3)
Participants who have given written informed consent and willingly cooperate with the collection of biological samples as well as basic and clinical information.
Exclusion criteria
-
(1)
Participants with cancer history.
-
(2)
Participants with TPMT or NUDT mutation.
-
(3)
Participants who could not tolerate AZA due to side effects.
-
(4)
Participants who have taken antibiotics or probiotics within 3 months before clinical sample collection.
Patients were assessed for outcomes based on histologic findings, clinical and endoscopic examination 6 months after the initial AZA administration.15 Clinical response of UC was defined as a decrease in Mayo score of at least 3 points and at least 30 percent, with an accompanying decrease in the subscore for rectal bleeding of at least 1 point or an absolute subscore for rectal bleeding of 0 or 1.51 Clinical response of CD was defined as a decrease in Crohn’s Disease Activity Index (CDAI) ≥ 100 points.52
Animal experiments and treatments
For Dextran sodium sulfate (DSS)-induced acute colitis model, mice were administrated with 3.5% DSS (molecular weight (MW), 36,000–50,000 Da; MP Biomedicals, LLC, Solon, OH) in drinking water for 6 days and DSS-free water for the next 2 days. Mice were intragastrically gavaged daily with either vehicle control or AZA (1.5 mg/kg, MW, 277.26 Da; MB1758, Meilunbio, Dalian, China) drugs for periods between 21 and 30 days.53,54 The clinical signs of disease activity index (DAI) were scored daily, including of body weight, diarrhea, and bloody stools.55,56
For 2,4,6-trinitrobenzene sulfonic acid (TNBS)-induced acute colitis model, mice were treated by a previous protocol.57 Acetone and olive oil was mixed in a 4:1 volume ratio by rigorous vertexing, and 1 volume of 5% (w/v) TNBS (p2297, Sigma, USA) solution was mixed with these 4 volumes of acetone/olive oil by vertexing rigorously to make TNBS pre-sensitization solution. Mice were immunized with 150 μL of the TNBS presensitization solution on the shaved abdominal skin of mice. After one week, 100 μL of 2.5% (w/v) TNBS (diluted in 50% ethanol) was administered intrarectally through a catheter inserted into the colon 4 cm proximal to the anus. The clinical signs were monitored daily. Colon histology were examined until day 4 after TNBS treatment.
For DSS-induced chronic colitis model, DSS was administered in the drinking water chronically for three cycles (2.5% DSS for 7 days on, followed by 14 days off; 3% DSS for 7 days on, followed by 14 days off; 3.5% DSS for 7 days on, followed by 7 days off). Meanwhile, we intragastrically gavaged mice daily with either vehicle control or AZA (1.5 mg/kg) for periods between 42 and 77 days.36,52 The clinical signs of DAI were scored as described above.
Fecal microbiota transplantation (FMT) was performed according to a reported protocol.58,59 Briefly, feces containing bacteria was mixed and centrifuged at 3200g. We resuspended the pellet containing bacteria in Columbia broth (BD Difco, USA) 25% glycerol pre-incubated in anaerobic atmosphere. Bacterial suspensions were aliquoted and frozen at −80°C in cryo-vials (Cat.430488, Corning, USA). For human FMT administration, frozen aliquots were thawed on ice, centrifuged at 3200g, and resuspended in sterile PBS pre-incubated in anaerobic atmosphere.
For the plasma level of AZA and its downstream metabolites examination model, Mice were intragastrically gavaged daily with either vehicle control or AZA drugs (1.5 mg/kg) and the plasma and cecal contents were collected at 1 h.
The animal experiments were approved by the ethics committee of Renji Hospital affiliated to the School of Medicine, Shanghai Jiao Tong University, and the protocol was approved by the Ethics Committee of 21-E−05.
Histology and immunohistochemical analysis
Mouse colon tissues were fixed in 10% formalin, embedded in paraffin, and cut into 4-μm sections. Hematoxylin and eosin staining was performed according to the standard protocol. Then histological assessment was performed blinded to the mouse genotypes based on inflammatory cell infiltrate and intestinal architecture, as previously described.60
For immunohistochemical staining, endogenous peroxidase of the paraffin-embedded tissue sections was blocked by all-purpose antigen retrieval solution and non-specific binding was blocked by 10% goat serum albumin blocking buffer. The slides were incubated with primary antibodies for 1h, followed by incubation with the secondary antibodies. The color development was performed using a Four-color multiple fluorescent immunohistochemical staining kit (Absin, Shanghai, China) in accordance with the manufacturer’s instructions. Slides were washed in phosphate-buffered saline (PBS), stained with 2-(4- amidinophenyl)-1H-indole-6-carboxamidine (DAPI) and immunofluorescence images were obtained using Olympus Fluorescence Microscopy.
All histological scorings and quantifications were performed in a blinded fashion.
DNA extraction and target microbiota quantification
Genomic DNAs from fecal were extracted using QIAamp PowerFecal DNA Kit (Qiagen, GmbH, Hilden, Germany). Genomic DNAs from formalin-fixed paraffin-embedded tissue were extracted using QIAamp DNA FFPE Tissue Kit (Qiagen, GmbH, Hilden, Germany). Real-time PCR was performed to detect B.wexlerae/B.luti, B.wexlerae, B.luti and B.subtilis species using StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA). Results are represented as 2-ΔΔCt, normalized to 16S rDNA and to NR group.61,62 Each reaction contained 500 ng of DNA and was assayed in triplicate.
Semiquantitative RT-PCR and real-time PCR assays
Total RNA from cells and colon tissues was isolated with Trizol reagent (Invitrogen, Carlsbad, CA, USA). NanoVue spectrophotometer (GE Healthcare) was used to assess the quantity and quality. mRNA was subjected to reverse transcription with 5′All-In-One RT MasterMix kit (TaKaRa). For quantitative analysis of mRNA expression, real-time PCR was performed using StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA). β-actin was used as an internal control for mRNA expression. 2-△△Ct method was used to quantify the relative expression levels. The primers used for the real-time PCR analysis were summarized in Table S6.
Flow cytometric analysis
Single cells were isolated from colon mucosa and cell suspensions were stained. Fixable viability dye and surface markers was stained using fluorescently labeled antibodies for 30 min at 4°C. Flow cytometer data were acquired on FACS Celesta (BD Biosciences, Schwechat, Austria) and analyzed with FlowJo software (FlowJo, LLC, Ashland, OR).
AZA, 6-MP, 6-TX, 6-MMP, 6-TIMP and 6-TGN detection
The bacterial culture supernatant was deposited with 50% methanol and the supernatant was sucked out and tested after centrifugation. The plasma was prepared by methanol and acetonitrile (1:1). After centrifugation, the supernatant was concentrated and centrifuged, and redissolved in 10% methanol to be tested. The levels of AZA, 6-MP, 6-TX, 6-MMP, 6-TIMP and 6-TGN from bacterial culture supernatant, plasma and gut luminal contents were determined by Vanquish ultra-high-performance liquid chromatography-quadrupole (UHPLC/Q) Exactive plus.
Xanthine dehydrogenase and hypoxanthine phosphoribosyltransferase 1 activity assay
Xanthine Dehydrogenase (XDH) and hypoxanthine phosphoribosyltransferase 1 (HPRT1), depicting the metabolism of AZA, in bacterial culture medium was determined by a commercial XDH activity assay kit (SEC608Mu, Cloud-Clone, Wuhan, China) and HPRT1 ELISA kit (SEA717Hu, Cloud-Clone, Wuhan, China). Briefly, the bacterial culture medium was centrifugated (12, 000 r/min) for 2min, and the supernation was detected as described in the kit instruction.
Quantification and statistical analysis
16S rDNA gene sequencing and analysis
Total microbial genomic DNA was extracted from fecal samples using the E.Z.N.A. soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to manufacturer’s instructions. The DNA were then determined by 1.0% agarose gel electrophoresis and a NanoDrop ND-2000 spectrophotometer (Thermo Scientific Inc., USA) for quality and concentration. Prior to further use, the DNA were kept at −80°C. The hypervariable region V3-V4 of the bacterial 16S rDNA gene were amplified with primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R(5′-GGACTACHVGGGTWTCTAAT-3′) by an ABI GeneAmp 9700 PCR thermocycler (ABI, CA, USA). PCR amplification cycling conditions were performed according to manufacturer’s instructions. All samples were amplified in triplicate. The PCR product was extracted from 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to manufacturer’s instructions and quantified using Quantus Fluorometer (Promega, USA). Purified amplicons were pooled in equimolar amounts and paired-end sequenced on an Illumina MiSeq PE300 platform/NovaSeq PE250 platform (Illumina, San Diego,USA) according to the standard protocols by NEO-bio Bio-technology Co.Ltd.
The open-source software VSEARCH v2.15.0 was used for initial bioinformatic analysis of the sequencing data.45 Sequences were trimmed and those with ambiguous bases were discarded. Suspected chimeric sequences were identified using VSEARCH and removed. Phylogenetic identification of each OTU was achieved by aligning sequences to the alignment database rdp_16s_v16_sp (https://www.drive5.com/sintax/rdp_16s_v16_sp.fa.gz). OTU values generated by VSEARCH were further analyzed using R-studio (phyloseq).
Single cell sequencing and analysis
The samples were dissociated to single-cell suspensions and the libraries were generated on a 10x Genomics platform using Chromium Single Cell 3′ library and gel bead kit v3. The single-cell libraries were sequenced on the NovaSeq 6000 System (Illumina) at NEO-bio Bio-technology Co.Ltd with 150 bp paired-end sequencing of reads. Scanpy Package v1.4.4 was used for downstream data analysis. Cells with more than 200 genes, and genes detected in more than 3 cells were retained for further analysis. After quality control, we obtained datasets containing a total of 21,721 cells with 19,252 genes in mice tissues and 32,425 cells with 33,538 genes in IBD patient mucosa. Harmony was used to adjust PCs by sample ID and used to generate the neighborhood graph and embedded using UMAP. In mice samples, clustering was performed using the louvain algorithm with an initial resolution of 0.3. In IBD patient tissues, clustering was performed using the leiden algorithm with an initial resolution of 0.2. For initial clustering, differentially expressed genes were calculated using the Wilcoxon rank-sum test (via ‘tl.rank_genes_groups’ in scanpy).
Shotgun metagenomic sequencing and analysis
The Illumina Nova-seq 6000 paired-end shotgun sequencing was used. Raw data was filtered by fastp with default parameters followed by the quantification of microbial composition based on marker gene based operational taxonomic units (mOTUs, https://github.com/motu-tool/mOTUs).
Statistical analysis
Statistical analysis was conducted using the program R (www.r-project.org). Experiments were performed at least in triplicate and all of the data were showed as mean ± SD or mean ± SEM in this study. Data were examined to determine whether they were normally distributed with the One-Sample Kolmogorov-Smirnov test. If the data were normally distributed, comparisons of measurement data between two groups were performed using independent sample t test and the comparisons among three or more groups were first performed by one-way ANOVA test. If the results showed significant difference, when the data were skewed distribution, comparisons were performed by nonparametric test. To account for the small sample sizes in certain groups (n = 3–5), we were unable to assess normality of the distribution for all samples. As a result, statistical analysis was performed under the assumption that the data were normally distributed.63 Statistical tests were two-tailed and a p value of less than 0.05 was considered statistically significant.
Acknowledgments
The authors thank all members of collaborating laboratories for thoughtful discussions about this research project. This work was supported by grants from the National Natural Science Foundation of China (nos. 82230016, 82272979, 82073115, and 82270550); the Program for Professor of Special Appointment (Eastern Scholar no. 201268) at Shanghai Institutions of Higher Learning; the Shanghai Municipal Education Commission—Gaofeng Clinical Medicine Grant Support (nos. 20152512 and 20161309); the Shanghai Municipal Health Commission Talent Program (no. 2022XD048); and the National Key Research and Development Plan of the Ministry of Science and Technology (2022YFE0125300).
Author contributions
J.H. and Y.Y. conceived the study. Z.W., Z.C., Y.Y., Z.G., and L.N. enrolled patients and managed patient sample collection. Y.Y., Y. Zhou, Z.G., L.N., Y.Z., B.X., and M.H. conducted experiments and analyzed data. H.C. and Y.Y. performed independent statistical analyses and visualization. J.H. and Y.Y. wrote the original draft. J.H., H.C., and J.-Y.F. carried out review and editing. All authors reviewed and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: August 15, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101153.
Contributor Information
Zhe Cui, Email: drcui1568@163.com.
Haoyan Chen, Email: haoyanchen@sjtu.edu.cn.
Jie Hong, Email: jiehong97@sjtu.edu.cn.
Supplemental information
Data and code availability
-
•
Data: 16S rDNA sequencing and scRNA sequencing data generated during this study are available at the China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (https://ngdc.cncb.ac.cn): PRJCA015633, PRJCA018136. The metagenome sequencing raw data of longitudinal stool study of patients with IBD and controls are available at the NCBI Sequence Read Archive (SRA) database: PRJNA385949.
-
•
Code: This study does not report original code.
-
•
General Statement: Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- 1.Mieli-Vergani G., Vergani D., Czaja A.J., Manns M.P., Krawitt E.L., Vierling J.M., Lohse A.W., Montano-Loza A.J. Autoimmune hepatitis. Nat. Rev. Dis. Primers. 2018;4:18017. doi: 10.1038/nrdp.2018.17. [DOI] [PubMed] [Google Scholar]
- 2.Fanouriakis A., Kostopoulou M., Alunno A., Aringer M., Bajema I., Boletis J.N., Cervera R., Doria A., Gordon C., Govoni M., et al. 2019 update of the EULAR recommendations for the management of systemic lupus erythematosus. Ann. Rheum. Dis. 2019;78:736–745. doi: 10.1136/annrheumdis-2019-215089. [DOI] [PubMed] [Google Scholar]
- 3.Ng S.C., Mak J.W.Y., Pal P., Banerjee R. Optimising management strategies of inflammatory bowel disease in resource-limited settings in Asia. Lancet. Gastroenterol. Hepatol. 2020;5:1089–1100. doi: 10.1016/s2468-1253(20)30298-3. [DOI] [PubMed] [Google Scholar]
- 4.Mak L.Y., Ng S.C., Wong I.O.L., Li M.K.K., Lo F.H., Wong M.T.L., Leung C.M., Tsang S.W.C., Chan K.H., Sze S.F., et al. Direct health-care cost utilization in Hong Kong inflammatory bowel disease patients in the initial 2 years following diagnosis. J. Gastroenterol. Hepatol. 2018;33:141–149. doi: 10.1111/jgh.13817. [DOI] [PubMed] [Google Scholar]
- 5.Fraser A.G., Orchard T.R., Jewell D.P. The efficacy of azathioprine for the treatment of inflammatory bowel disease: a 30 year review. Gut. 2002;50:485–489. doi: 10.1136/gut.50.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucafò M., Stocco G., Martelossi S., Favretto D., Franca R., Malusà N., Lora A., Bramuzzo M., Naviglio S., Cecchin E., et al. Azathioprine biotransformation in young patients with inflammatory Bowel disease: contribution of glutathione-S transferase M1 and A1 variants. Genes. 2019;10:277. doi: 10.3390/genes10040277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tremaroli V., Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 9.Lindell A.E., Zimmermann-Kogadeeva M., Patil K.R. Multimodal interactions of drugs, natural compounds and pollutants with the gut microbiota. Nat. Rev. Microbiol. 2022;20:431–443. doi: 10.1038/s41579-022-00681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weersma R.K., Zhernakova A., Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69:1510–1519. doi: 10.1136/gutjnl-2019-320204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570:462–467. doi: 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Javdan B., Lopez J.G., Chankhamjon P., Lee Y.C.J., Hull R., Wu Q., Wang X., Chatterjee S., Donia M.S. Personalized mapping of drug metabolism by the human gut microbiome. Cell. 2020;181:1661–1679.e22. doi: 10.1016/j.cell.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A.L. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science. 2019;363:eaat9931. doi: 10.1126/science.aat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klünemann M., Andrejev S., Blasche S., Mateus A., Phapale P., Devendran S., Vappiani J., Simon B., Scott T.A., Kafkia E., et al. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature. 2021;597:533–538. doi: 10.1038/s41586-021-03891-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peyrin-Biroulet L., Sandborn W., Sands B.E., Reinisch W., Bemelman W., Bryant R.V., D'Haens G., Dotan I., Dubinsky M., Feagan B., et al. Selecting therapeutic targets in inflammatory Bowel disease (STRIDE): determining therapeutic goals for treat-to-target. Am. J. Gastroenterol. 2015;110:1324–1338. doi: 10.1038/ajg.2015.233. [DOI] [PubMed] [Google Scholar]
- 16.Erttmann S.F., Swacha P., Aung K.M., Brindefalk B., Jiang H., Härtlova A., Uhlin B.E., Wai S.N., Gekara N.O. The gut microbiota prime systemic antiviral immunity via the cGAS-STING-IFN-I axis. Immunity. 2022;55:847–861.e10. doi: 10.1016/j.immuni.2022.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Lynn M.A., Tumes D.J., Choo J.M., Sribnaia A., Blake S.J., Leong L.E.X., Young G.P., Marshall H.S., Wesselingh S.L., Rogers G.B., Lynn D.J. Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe. 2018;23:653–660.e5. doi: 10.1016/j.chom.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Chang J.T. Pathophysiology of inflammatory Bowel diseases. N. Engl. J. Med. 2020;383:2652–2664. doi: 10.1056/NEJMra2002697. [DOI] [PubMed] [Google Scholar]
- 19.Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 20.Huang B., Chen Z., Geng L., Wang J., Liang H., Cao Y., Chen H., Huang W., Su M., Wang H., et al. Mucosal profiling of pediatric-onset colitis and IBD reveals common pathogenics and therapeutic pathways. Cell. 2019;179:1160–1176.e24. doi: 10.1016/j.cell.2019.10.027. [DOI] [PubMed] [Google Scholar]
- 21.Derijks L.J.J., Gilissen L.P.L., Hooymans P.M., Hommes D.W. Review article: thiopurines in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2006;24:715–729. doi: 10.1111/j.1365-2036.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W., Modén O., Mannervik B. Differences among allelic variants of human glutathione transferase A2-2 in the activation of azathioprine. Chem. Biol. Interact. 2010;186:110–117. doi: 10.1016/j.cbi.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 23.Logan J.K., Wickramaratne Senarath Yapa S., Harinstein L., Saluja B., Muñoz M., Sahajwalla C., Neuner R., Seymour S. Drug interaction between febuxostat and thiopurine antimetabolites: a review of the FDA adverse event reporting system and medical literature. Pharmacotherapy. 2020;40:125–132. doi: 10.1002/phar.2362. [DOI] [PubMed] [Google Scholar]
- 24.Kiszka-Kanowitz M., Theede K., Thomsen S.B., Bjerrum J.T., Brynskov J., Gottschalck I.B., Akimenko E., Hilsted K.L., Neumann A., Wildt S., et al. Low-dose azathioprine and allopurinol versus azathioprine monotherapy in patients with ulcerative colitis (AAUC): An investigator-initiated, open, multicenter, parallel-arm, randomised controlled trial. EClinicalMedicine. 2022;45:101332. doi: 10.1016/j.eclinm.2022.101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christiansen L.C., Schou S., Nygaard P., Saxild H.H. Xanthine metabolism in Bacillus subtilis: characterization of the xpt-pbuX operon and evidence for purine- and nitrogen-controlled expression of genes involved in xanthine salvage and catabolism. J. Bacteriol. 1997;179:2540–2550. doi: 10.1128/jb.179.8.2540-2550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y., Hwang S.W., Kim S., Lee Y.S., Kim T.Y., Lee S.H., Kim S.J., Yoo H.J., Kim E.N., Kweon M.N. Dietary cellulose prevents gut inflammation by modulating lipid metabolism and gut microbiota. Gut Microb. 2020;11:944–961. doi: 10.1080/19490976.2020.1730149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Guo Y., Chen H., Wei H., Wan C. Potential of Lactobacillus plantarum ZDY2013 and Bifidobacterium bifidum WBIN03 in relieving colitis by gut microbiota, immune, and anti-oxidative stress. Can. J. Microbiol. 2018;64:327–337. doi: 10.1139/cjm-2017-0716. [DOI] [PubMed] [Google Scholar]
- 28.Chemmugil P., Lakshmi P.T.V., Annamalai A. Exploring Morin as an anti-quorum sensing agent (anti-QSA) against resistant strains of Staphylococcus aureus. Microb. Pathog. 2019;127:304–315. doi: 10.1016/j.micpath.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Eberl G. A new vision of immunity: homeostasis of the superorganism. Mucosal Immunol. 2010;3:450–460. doi: 10.1038/mi.2010.20. [DOI] [PubMed] [Google Scholar]
- 30.Ni J., Wu G.D., Albenberg L., Tomov V.T. Gut microbiota and IBD: causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017;14:573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manichanh C., Borruel N., Casellas F., Guarner F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 32.Benitez-Paez A., Gomez Del Pugar E.M., Lopez-Almela I., Moya-Perez A., Codoner-Franch P., Sanz Y. Depletion of Blautia species in the microbiota of obese children relates to intestinal inflammation and metabolic phenotype worsening. mSystems. 2020;5 doi: 10.1128/mSystems.00857-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosomi K., Saito M., Park J., Murakami H., Shibata N., Ando M., Nagatake T., Konishi K., Ohno H., Tanisawa K., et al. Oral administration of Blautia wexlerae ameliorates obesity and type 2 diabetes via metabolic remodeling of the gut microbiota. Nat. Commun. 2022;13:4477. doi: 10.1038/s41467-022-32015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quévrain E., Maubert M.A., Michon C., Chain F., Marquant R., Tailhades J., Miquel S., Carlier L., Bermúdez-Humarán L.G., Pigneur B., et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease. Gut. 2016;65:415–425. doi: 10.1136/gutjnl-2014-307649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehta R.S., Mayers J.R., Zhang Y., Bhosle A., Glasser N.R., Nguyen L.H., Ma W., Bae S., Branck T., Song K., et al. Gut microbial metabolism of 5-ASA diminishes its clinical efficacy in inflammatory bowel disease. Nat. Med. 2023;29:700–709. doi: 10.1038/s41591-023-02217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oancea I., Movva R., Das I., Aguirre de Cárcer D., Schreiber V., Yang Y., Purdon A., Harrington B., Proctor M., Wang R., et al. Colonic microbiota can promote rapid local improvement of murine colitis by thioguanine independently of T lymphocytes and host metabolism. Gut. 2017;66:59–69. doi: 10.1136/gutjnl-2015-310874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maini Rekdal V., Bess E.N., Bisanz J.E., Turnbaugh P.J., Balskus E.P. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science. 2019;364:eaau6323. doi: 10.1126/science.aau6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Kessel S.P., Frye A.K., El-Gendy A.O., Castejon M., Keshavarzian A., van Dijk G., El Aidy S. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson's disease. Nat. Commun. 2019;10:310. doi: 10.1038/s41467-019-08294-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao R., Coker O.O., Wu J., Zhou Y., Zhao L., Nakatsu G., Bian X., Wei H., Chan A.W.H., Sung J.J.Y., et al. Aspirin reduces colorectal tumor development in mice and gut microbes reduce its bioavailability and chemopreventive effects. Gastroenterology. 2020;159:969–983.e4. doi: 10.1053/j.gastro.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Balaich J., Estrella M., Wu G., Jeffrey P.D., Biswas A., Zhao L., Korennykh A., Donia M.S. The human microbiome encodes resistance to the antidiabetic drug acarbose. Nature. 2021;600:110–115. doi: 10.1038/s41586-021-04091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caruso R., Mathes T., Martens E.C., Kamada N., Nusrat A., Inohara N., Núñez G. A specific gene-microbe interaction drives the development of Crohn's disease-like colitis in mice. Sci. Immunol. 2019;4:eaaw4341. doi: 10.1126/sciimmunol.aaw4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall A.B., Yassour M., Sauk J., Garner A., Jiang X., Arthur T., Lagoudas G.K., Vatanen T., Fornelos N., Wilson R., et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9:103. doi: 10.1186/s13073-017-0490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Touyama M., Jin J.S., Kibe R., Hayashi H., Benno Y. Quantification of Blautia wexlerae and Blautia luti in human faeces by real-time PCR using specific primers. Benef. Microbes. 2015;6:583–590. doi: 10.3920/BM2014.0133. [DOI] [PubMed] [Google Scholar]
- 44.Lee G., Heo S., Kim T., Na H.E., Park J., Lee E., Lee J.H., Jeong D.W. Discrimination of Bacillus subtilis from other Bacillus species using specific oligonucleotide primers for the pyruvate carboxylase and shikimate dehydrogenase genes. J. Microbiol. Biotechnol. 2022;32:1011–1016. doi: 10.4014/jmb.2205.05014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasolli E., Schiffer L., Manghi P., Renson A., Obenchain V., Truong D.T., Beghini F., Malik F., Ramos M., Dowd J.B., et al. Accessible, curated metagenomic data through ExperimentHub. Nat. Methods. 2017;14:1023–1024. doi: 10.1038/nmeth.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milanese A., Mende D.R., Paoli L., Salazar G., Ruscheweyh H.J., Cuenca M., Hingamp P., Alves R., Costea P.I., Coelho L.P., et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 2019;10:1014. doi: 10.1038/s41467-019-08844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inflammatory Bowel Disease Group, Chinese Society of Gastroenterology, Chinese Medical Association Chinese consensus on diagnosis and treatment in inflammatory bowel disease (2018, Beijing) J. Dig. Dis. 2021;22:298–317. doi: 10.1111/1751-2980.12994. [DOI] [PubMed] [Google Scholar]
- 51.Rutgeerts P., Sandborn W.J., Feagan B.G., Reinisch W., Olson A., Johanns J., Travers S., Rachmilewitz D., Hanauer S.B., Lichtenstein G.R., et al. Infliximab for induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med. 2005;353:2462–2476. doi: 10.1056/NEJMoa050516. [DOI] [PubMed] [Google Scholar]
- 52.Gomollón F., Dignass A., Annese V., Tilg H., Van Assche G., Lindsay J.O., Peyrin-Biroulet L., Cullen G.J., Daperno M., Kucharzik T., et al. 3rd European evidence-based consensus on the diagnosis and management of Crohn's disease 2016: part 1: diagnosis and medical management. J. Crohns Colitis. 2017;11:3–25. doi: 10.1093/ecco-jcc/jjw168. [DOI] [PubMed] [Google Scholar]
- 53.Cui M., Fang Z., Song M., Zhou T., Wang Y., Liu K. Phragmites rhizoma polysaccharide-based nanocarriers for synergistic treatment of ulcerative colitis. Int. J. Biol. Macromol. 2022;220:22–32. doi: 10.1016/j.ijbiomac.2022.07.245. [DOI] [PubMed] [Google Scholar]
- 54.Gassmann A.E., van Furth R. The effect of azathioprine (Imuran) on the kinetics of monocytes and macrophages during the normal steady state and an acute inflammatory reaction. Blood. 1975;46:51–64. doi: 10.1182/blood.V46.1.51.51. [DOI] [PubMed] [Google Scholar]
- 55.Cooper H.S., Murthy S.N., Shah R.S., Sedergran D.J. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab. Invest. 1993;69:238–249. [PubMed] [Google Scholar]
- 56.Ai L., Ren Y., Zhu M., Lu S., Qian Y., Chen Z., Xu A. Synbindin restrains proinflammatory macrophage activation against microbiota and mucosal inflammation during colitis. Gut. 2021;70:2261–2272. doi: 10.1136/gutjnl-2020-321094. [DOI] [PubMed] [Google Scholar]
- 57.Wirtz S., Neufert C., Weigmann B., Neurath M.F. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2007;2:541–546. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- 58.Pernigoni N., Zagato E., Calcinotto A., Troiani M., Mestre R.P., Calì B., Attanasio G., Troisi J., Minini M., Mosole S., et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science. 2021;374:216–224. doi: 10.1126/science.abf8403. [DOI] [PubMed] [Google Scholar]
- 59.Zhang G., Wang Q., Tao W., Jiang W., Elinav E., Wang Y., Zhu S. Glucosylated nanoparticles for the oral delivery of antibiotics to the proximal small intestine protect mice from gut dysbiosis. Nat. Biomed. Eng. 2022;6:867–881. doi: 10.1038/s41551-022-00903-4. [DOI] [PubMed] [Google Scholar]
- 60.Erben U., Loddenkemper C., Doerfel K., Spieckermann S., Haller D., Heimesaat M.M., Zeitz M., Siegmund B., Kühl A.A. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int. J. Clin. Exp. Pathol. 2014;7:4557–4576. [PMC free article] [PubMed] [Google Scholar]
- 61.Vétizou M., Pitt J.M., Daillère R., Lepage P., Waldschmitt N., Flament C., Rusakiewicz S., Routy B., Roberti M.P., Duong C.P.M., et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Llorente C., Jepsen P., Inamine T., Wang L., Bluemel S., Wang H.J., Loomba R., Bajaj J.S., Schubert M.L., Sikaroodi M., et al. Gastric acid suppression promotes alcoholic liver disease by inducing overgrowth of intestinal Enterococcus. Nat. Commun. 2017;8:837. doi: 10.1038/s41467-017-00796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baris O.R., Ederer S., Neuhaus J.F.G., von Kleist-Retzow J.C., Wunderlich C.M., Pal M., Wunderlich F.T., Peeva V., Zsurka G., Kunz W.S., et al. Mosaic deficiency in mitochondrial oxidative metabolism promotes cardiac arrhythmia during aging. Cell Metab. 2015;21:667–677. doi: 10.1016/j.cmet.2015.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: 16S rDNA sequencing and scRNA sequencing data generated during this study are available at the China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (https://ngdc.cncb.ac.cn): PRJCA015633, PRJCA018136. The metagenome sequencing raw data of longitudinal stool study of patients with IBD and controls are available at the NCBI Sequence Read Archive (SRA) database: PRJNA385949.
-
•
Code: This study does not report original code.
-
•
General Statement: Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.