Abstract
Although melanoma is notorious for its high degree of heterogeneity and plasticity1,2, the origin and magnitude of cell state diversity remains poorly understood. Equally, it is unclear whether growth and metastatic dissemination are supported by overlapping or distinct melanoma subpopulations. By combining mouse genetics, single-cell and spatial transcriptomics, lineage tracing and quantitative modelling, we provide evidence of a hierarchical model of tumour growth that mirrors the cellular and molecular logic underlying embryonic neural crest cell fate specification and differentiation. We show that tumorigenic competence is associated with a spatially localized perivascular niche, a phenotype acquired through a intercellular communication pathway established by endothelial cells. Consistent with a model in which only a fraction of cells are fated to fuel growth, temporal single-cell tracing of a population of melanoma cells harbouring a mesenchymal-like state revealed that these cells do not contribute to primary tumour growth but, instead, constitutes a pool of metastatic-initiating cells that switch cell identity while disseminating to secondary organs. Our data provide a spatially and temporally resolved map of the diversity and trajectories of melanoma cell states and suggest that the ability to support growth and metastasis are limited to distinct pools of cells. The observation that these phenotypic competencies can be dynamically acquired upon exposure to specific niche signals warrant the development of therapeutic strategies that interfere with the cancer cell reprogramming activity of such microenvironmental cues.
Overwhelming evidence indicates that melanoma cells can exhibit either a “proliferative” (or melanocytic) or a mesenchymal-like (or “invasive”) transcriptional cell state1,2. Single-cell RNA-sequencing (scRNA-seq) confirmed the coexistence of these two cell states in clinical samples3-5. However, recent data from in vitro and in vivo models suggested that additional melanoma cell states may exist1,4,6, and that our current view of melanoma IntraTumor Heterogeneity (ITH) is likely incomplete. These observations also raised the possibility that these distinct cell states may not contribute equally to tumour growth and metastatic spreading. An in-depth dissection of melanoma phenotypic heterogeneity in the in vivo relevant context is therefore needed.
Revisiting melanoma cell state diversity
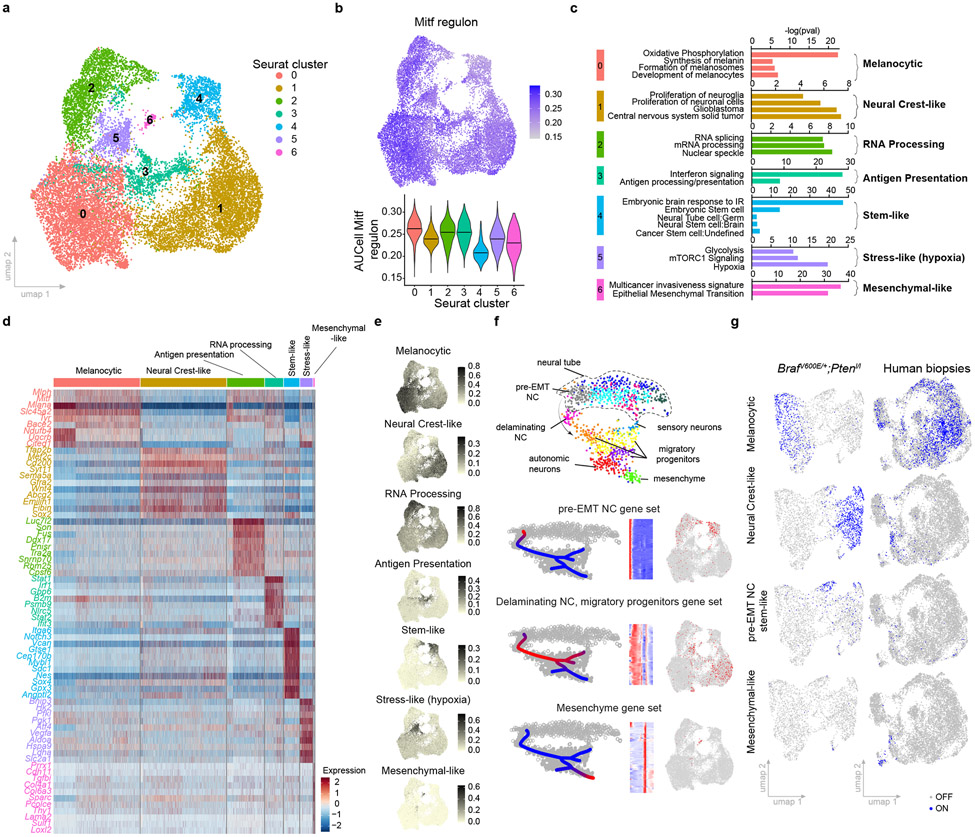
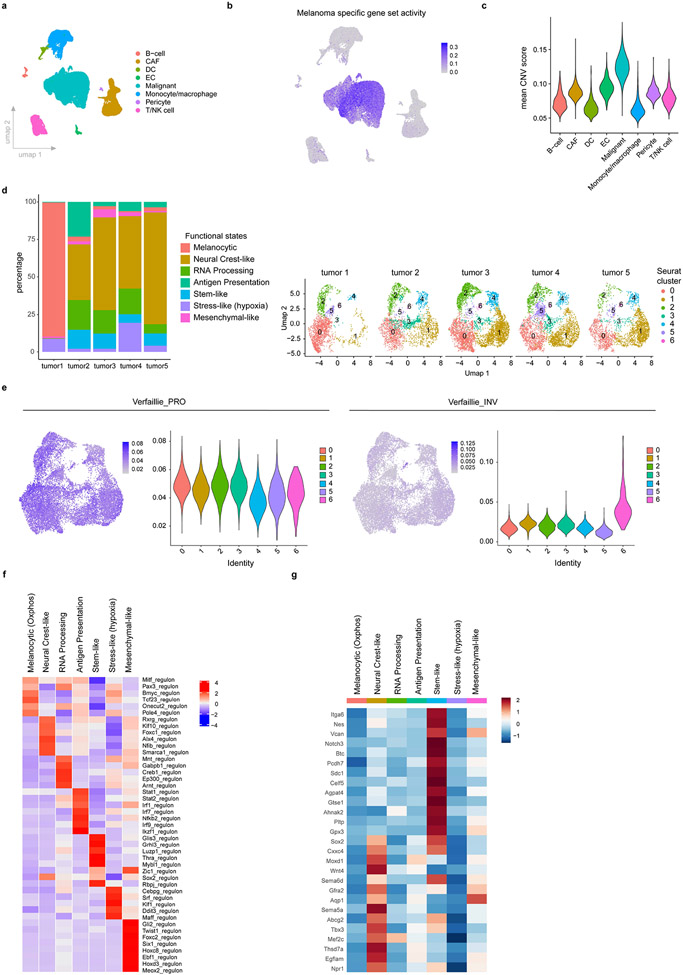
Although mouse models have been instrumental in understanding melanoma biology7, their ability to recapitulate the phenotypic diversity observed in human patients remains unclear. We tested this in a spontaneous mouse model carrying the Tyr::NRASQ61K/° allele8 and lacking p16INK4a (or Ink4a)9. These mice develop cutaneous monoclonal melanoma lesions, and subsequently lymph node, liver and lung metastases. To increase the versatility of this model, we derived syngeneic lines in C57BL/6 mice, and assessed the transcriptomic landscape of such melanoma lesions by scRNA-seq. Using published cell type-specific gene sets, we identified T-cell, B-cell, cancer-associated fibroblast (CAF), dendritic cell (DC), monocyte/macrophage, endothelial cell (EC), pericyte and malignant populations10-13 (Extended Data Fig. 1a), thus highlighting the presence of a complex tumour microenvironment (TME). To refine malignant cell typing, we selected cells expressing a melanoma-specific gene signature and exhibiting high copy number variation (CNV), which was inferred using Honeybadger14 (Extended Data Fig. 1b,c). About 35% of cells were annotated as malignant. Unsupervised Louvain-based clustering and Uniform Manifold Approximation and Projection (UMAP) identified 7 distinct melanoma cell states, which were all detected in each lesion (n=5) analysed (Fig. 1a and Extended Data Fig. 1d).
Fig. 1: Single-cell transcriptomic landscape of NRAS-driven melanoma.
a, Uniform Manifold Approximation and Projection (UMAP) of malignant cells analysed by scRNA-seq and integrated across five different NRASQ61K/°;Ink4a−/− primary lesions. b, Mitf regulon activity in mouse malignant cells projected as UMAP (upper panel) and violin plot (lower panel), respectively. The middle line represents the median. c. Functional enrichment terms of the characteristic Seurat clusters identified in a (Fisher exact test, two-tailed p value). d. Heatmap illustrating discriminative marker genes (n=9) for each functionally enriched state. e. AUCell score of top functionally enriched marker genes per state projected on malignant UMAP (see Supplementary Table 1). f, Mapping of gene expression program activities during murine neural crest development18 onto transcriptional states of NRASQ61K/°Ink4a−/− melanoma. g, Activity (binary AUCell score) of selected mouse malignant gene signatures from NRASQ61K/°;Ink4a−/− melanoma projected onto scRNAseq datasets from BRAFV600E;Ptenl/l mouse melanoma lesions (left panel) and human clinical Metastatic Melanoma samples22 (right panel).
MITF is a key regulator of the differentiation status of melanoma1,15,16. It induces genes implicated in melanogenesis, the primary differentiation-associated function of melanocytes, and low MITF leads to dedifferentiation. A wide range of Mitf levels and transcriptional activities were detected across all malignant cells (Fig. 1b). Distinct transcriptional programs have been associated with differential levels of MITF activity, namely the melanocytic/proliferative (MITF high, PRO) and mesenchymal-like/invasive (MITF low; INV). Melanoma cells expressing these signatures were readily detectable in the NRASQ61K/°;Ink4a−/− lesions (Extended Data Fig. 1e). Notably, cells from cluster 4 exhibited the lowest MITF activity, indicating that these cells are the most de-differentiated of all malignant cells (Fig. 1b).
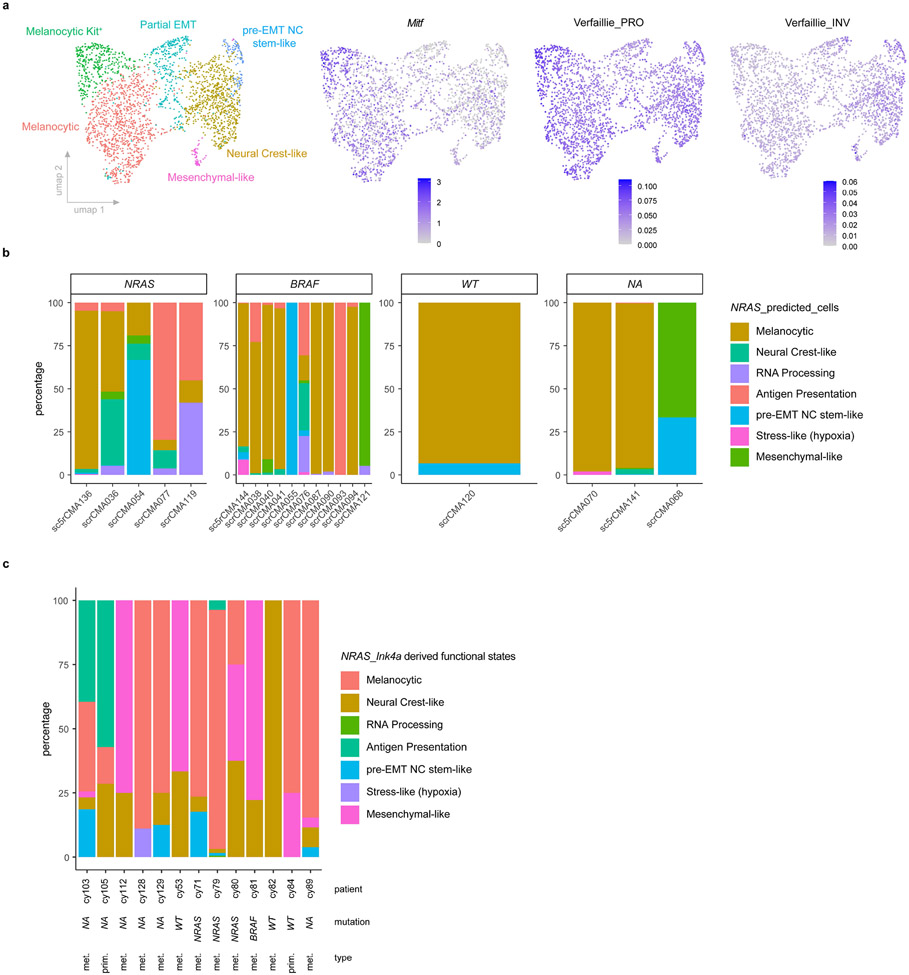
Differential gene expression analysis resulted in characteristic gene lists, and SCENIC17 identified specific regulons, for each cell state (Extended Data Fig. 1f and Supplementary Table 1). Gene set enrichment tools were used to functionally annotate each cluster (Fig.1c and Supplementary Table 2). A gene expression signature for each of these clusters was then established (Fig. 1d and Supplementary Table 3), its activity measured by AUCell17 and projected on the UMAP space (Fig. 1e). This analysis highlighted a transcriptional heterogeneity that goes beyond the melanocytic (cluster 0) and mesenchymal-like (cluster 6) subpopulations. Cells from cluster 4 and 1 shared markers of de-differentiated neural crest stem-like cells (NCSCs), such as Gfra2, Aqp1 or Mef2c (Extended Data Fig. 1g). Although largely overlapping, these two neural crest-like cell populations presented distinct features. Cells from cluster 4, in addition to expressing pluripotency markers such as Sox2 or Klf4 (Supplementary Table 1), uniquely expressed a distinct set of markers, including Notch3, Nestin (or Nes) or Vcan (Extended Data Fig. 1g). Remarkably, the transcriptome of these cells was reminiscent of that found in a pre-EMT (premigratory) neural crest stem cell population described recently18. These premigratory cells are the first neural crest-committed cells to emerge embryonically and constitute a pool of self-renewing neural crest stem cells19 (Fig. 1f). The second population to emerge during development, the delaminating neural crest population, mapped to cells from cluster 0 and 6, corresponding to the NCSC state described previously4,20. As neural crest progenitors migrate, they acquire features of more mature derivatives, with one population moving towards a mesenchymal fate. These cells activate a transcriptional program that was similar to the melanoma mesenchymal-like cell signature (cluster 6). Thus, melanoma cells can hijack the molecular logic underlying the neural crest cell fate specification program and dedifferentiate (all the way back) to a pre-EMT NC stem-like state. By doing so, these cells may acquire stem cell-like phenotypic properties.
Given the high prevalence of BRAF mutations in human melanoma, we also profiled lesions arising in the commonly used Tyr::CreERT2;BRafV600E/+;Ptenl/l mouse model21. scRNA-seq revealed that these lesions comprise cells with contrasting Mitf levels (Extended Data Fig. 2a). Importantly, melanoma cells exhibiting the melanocytic, mesenchymal-like, neural crest-like and pre-EMT NC stem-like signatures could also be identified in these lesions (Fig. 1g). We measured the mouse NRAS malignant cell state signatures in a large scRNA-seq data set from treatment-naïve human metastatic melanoma biopsies22 (Extended Data Fig. 2b). As expected, the melanocytic and mesenchymal-like cell states were identified in multiple patients. We also interrogated a previously reported scRNA-seq dataset10,11, which contains few primary lesions (Extended Data Fig. 2c). Most malignant transcriptional cell states, defined by the clustering analysis of the mouse lesions, were identified in both primary and metastatic human melanoma lesions from both cohorts, and thus irrespective of their oncogenic driver mutations (Fig. 1g and Extended Data Fig. 2c).
A cellular hierarchy in melanoma
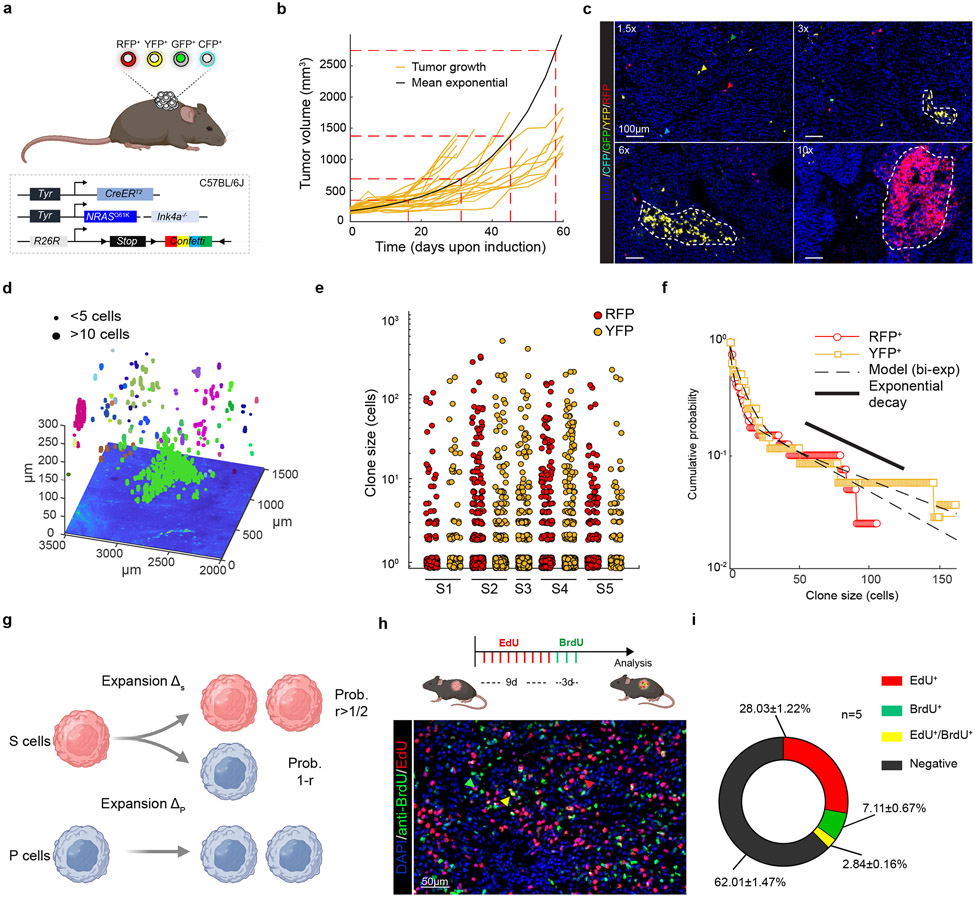
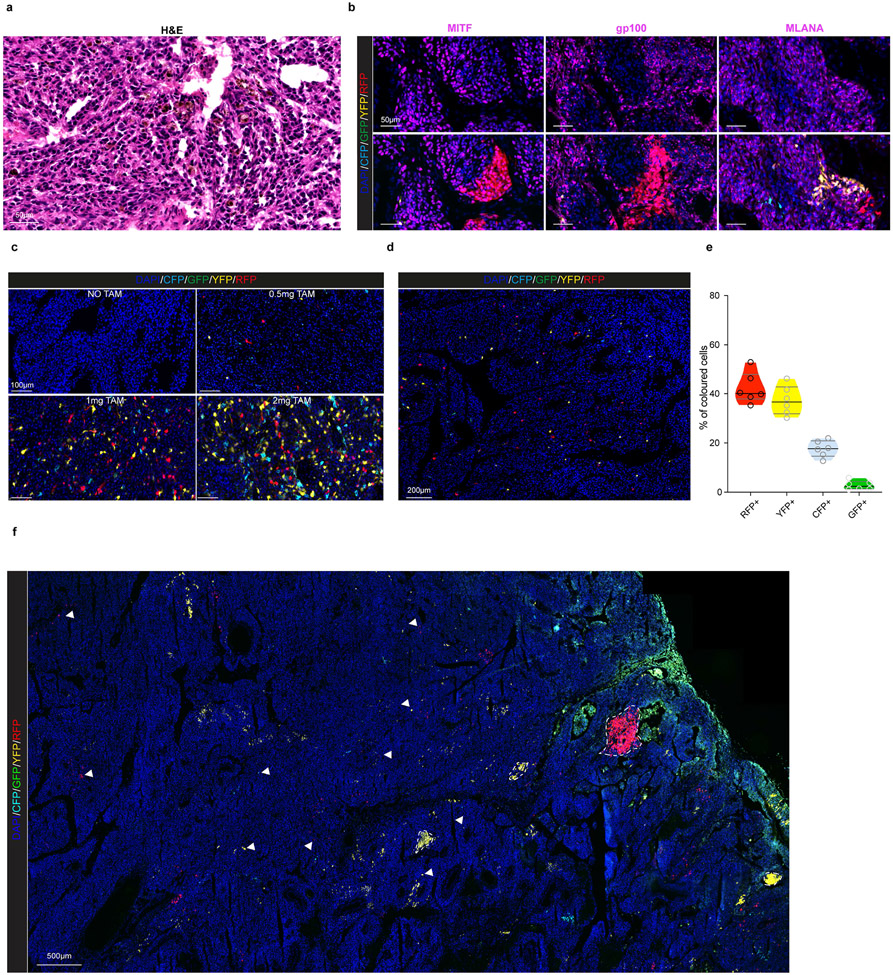
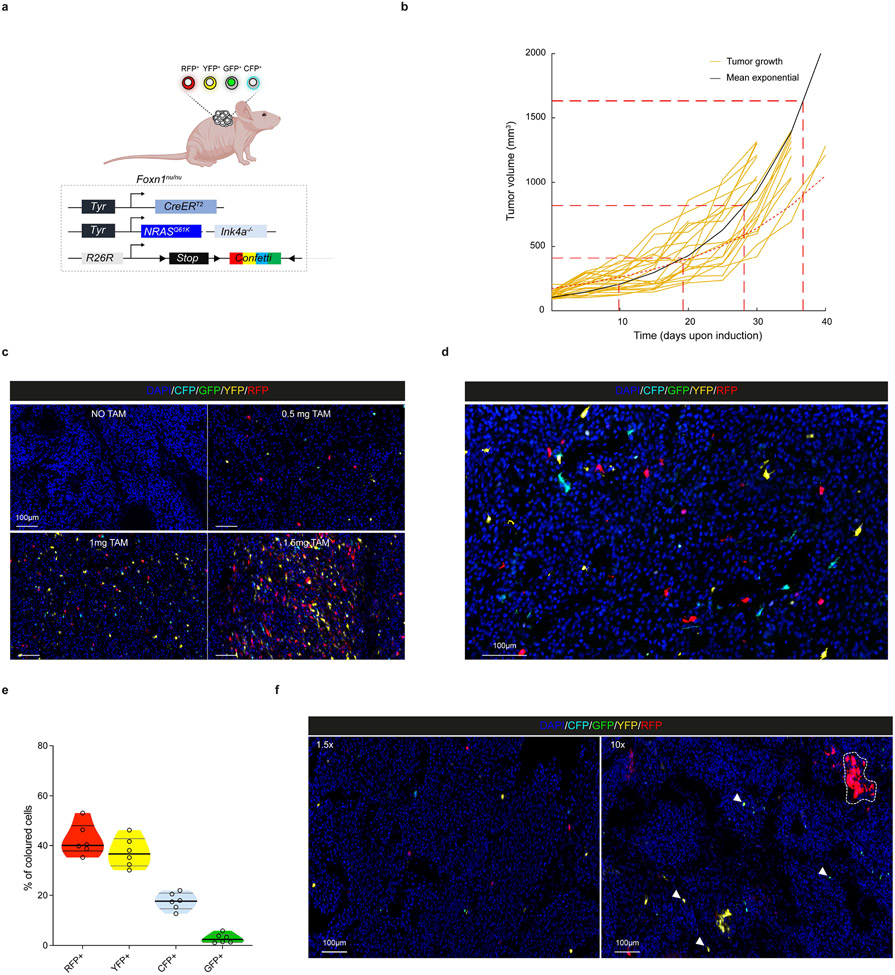
Given their transcriptomic diversity, we questioned whether the growth of these primary melanomas might be hierarchically organized. We turned to an inducible genetic lineage tracing approach based on the multicolour Rosa26-LSL-Confetti23 allele, which allows the stochastic expression of up to 10 different colour combinations, and the tamoxifen (TAM)-inducible CreERT2 allele under the control of the melanocytic promoter Tyr::CreERT2 7. The Tyr::NRASQ61K/°;Ink4a−/−;Tyr::CreERT2/°;Rosa26-LSL-Confetti+/+ compound mice (Fig. 2a) developed primary melanoma lesions at around 8±2 months of age. Tumor pieces of primary lesions were transplanted into cohorts of C57/bl6 mice and lesions grew steadily with a slow, exponential-like, growth characteristic with tumour-to-tumour variability in the expansion rate (Fig. 2b). These lesions were histologically indistinguishable from their tumour-of-origin and expressed heterogeneous melanocytic patterns (Extended Data Fig. 3a, b). Titrating down the dose of TAM, we were able to reach clonal density (Extended Data Fig. 3c), while maintaining the expected colour mosaicism24 (Extended Data Fig. 3d, e).
Figure 2: Multicolour lineage tracing reveals that melanoma growth is hierarchically organized.
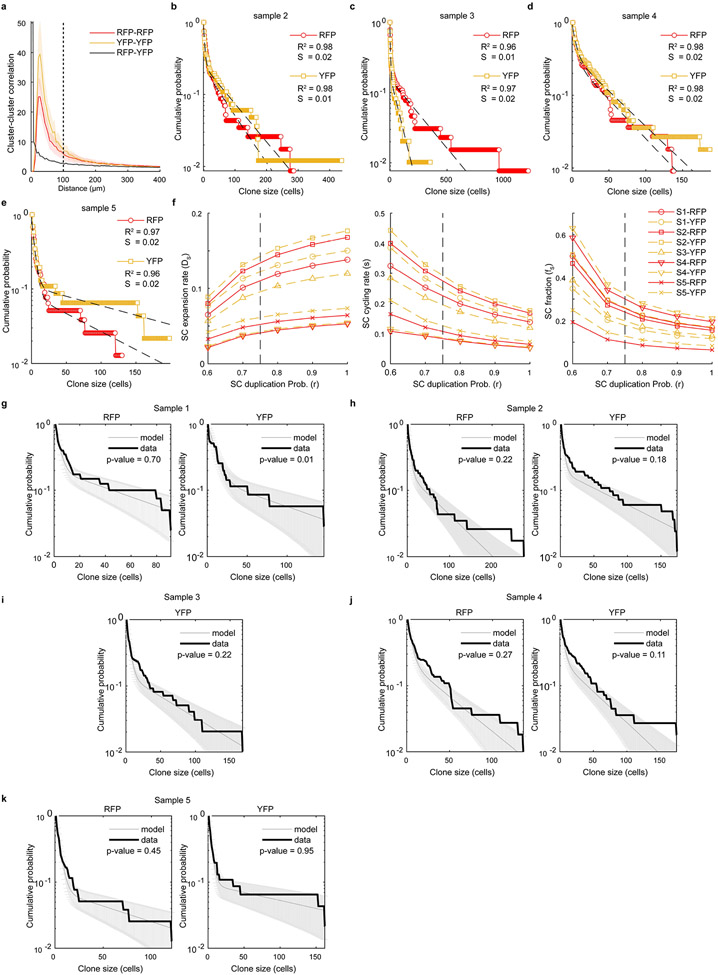
a, Schematic representation of mouse model. b, Tumour growth kinetics (mm3) of individual tumours (yellow), and corrected mean obtained from extrapolating the missing time-points using the model prediction (black curve). c, Confocal images of melanomas exhibiting Confetti expression at different relative growths upon TAM administration (0.5 mg). Representative images from at least 8 mice per group. d, Representative reconstruction of 3D clones. Independent clones appear in different colours. A single z-slice of the corresponding dataset is shown at the bottom of the plot. e. Dot plot showing the distribution of clone sizes, by sample and by channel obtained from 5 cleared tumours (see Supplementary Table 5). f. Cumulative distribution, showing the probability of observing a clone larger than a given size. The corresponding bi-exponential fits (dashed line, R2=0.96, standard error of the fit S=0.03) and exponential decay of the tail (black line) are shown. g, Schematics of the minimal hierarchical model. The parameters ΔS and ΔP correspond to the expansion rates of the S and P populations, respectively. Although the data could accommodate a “line” of fit parameters, fixing the relative transition rate of stem cells into the progenitor cell compartment, r=0.75, the model shows an excellent fit to the data with ΔS~0.09 and ΔP~0.04 and a stem fraction fS~0.21 or 21% (see Supplementary Note). h, Upper panel, experimental procedure for dual pulse labelling assay using EdU and BrdU. Lower panel, image of a double BrdU (green) and EdU (red) immunostaining. Representative image from n=5 tumors. i, Pie-chart illustrating the percentage of single and double labelled cells. Quantification was assessed from 5 different mice and 4 technical replicates per mouse. Schematic in a, g and h made with Biorender.com.
To gain insight into the overall growth pattern, we explored the distribution of marked cells from 2D sections. We found that a small fraction of clones grew large, spanning tens of cells or more, while the majority of clones remained very small, containing few cells (Fig. 2c and Extended Data Fig. 3f).
We next used a more rigorous density-based method of assignment to serially reconstruct and score total clone sizes at single-cell resolution by 3D confocal imaging (Fig. 2d and Extended Data Fig. 4a). Once again, we found significant heterogeneity in clone size across all tumour samples (Fig. 2e and Supplementary Table 4). Further evidence of “bimodal” behaviour was visible in the cumulative distribution of clone sizes (Fig. 2f, Extended Data Fig. 4b-e and Supplementary Table 5, Supplementary Note). Such behaviour was consistent with a proliferative hierarchy in which a stem cell-like population, S, expands stochastically through cell duplication, while giving rise to a second progenitor cell-like state, P, which in turn expands through cell duplication (Fig. 2g and Supplementary Note).
Statistical analysis confirmed that the minimal hierarchical model could capture quantitatively the wide range of clone size data across all tumour samples with stem and progenitor cell expansion rates that were sample-specific but broadly in the around ΔS~0.09 and ΔP~0.04 per day, respectively (Fig. 2g, Extended Data Fig. 4f-k, Supplementary Table 5 and Supplementary Note). Moreover, the analysis indicated that, following a 10-fold expansion of tumour size, around 10-32% of surviving clones are stem cell-derived with the remainder arising from progenitors (or non-cycling cells). Considering possible P cell-derived clone loss, such a figure was broadly consistent with the estimated abundance (6±3%) of pre-EMT NC stem-like cells based on scRNA-seq data, raising the possibility that this population may function as melanoma stem cells.
We also examined the proliferation kinetics of tumour growth using consecutive pulses of cell labelling with 5-Ethynyl-2’-deoxyUridine (EdU) and BromodeoxyUridine (BrdU) (Fig. 2h). The EdU+ and BrdU+ cell fractions scaled in approximate proportion to the incorporation time, consistent with the majority of the tumour being comprised of proliferative cells dividing at a similar rate. Moreover, with the fraction of double-positive cells being approximately equal to the product of the individual fractions, these results supported the prediction of the model that tumour growth is maintained by cells dividing stochastically at a similar slow average rate (Fig. 2i). Finally, with S-phase estimated at around 6-8 hours, these results pointed to an average cell division rate of approximately once per 30 days, a figure broadly consistent with the tracing analysis.
We performed similar lineage tracing experiments following the transplantation of tumours into immunocompromised mice (Extended Data Fig. 5a-e). Qualitative analysis of the 2D cluster size distribution compared to that observed in the immune-proficient background (Extended Data Fig. 5f). Notably, however, tumours grew more rapidly on this background (Extended Data Fig. 5b), indicating that the adaptive immune system does not affect the mode of tumour growth per se, but instead restrains the overall growth by keeping in check the effective proliferation rate of each malignant compartment.
These results support a model in which primary melanoma is organized in a cellular hierarchy, comprising at least two distinct populations, a stem- and progenitor cell-like population, that exhibit different growth kinetics and fate behaviour. Based on the scRNA-seq analysis, we hypothesized that the S population might overlap with the pre-EMT NC stem-like population. The spatial organization and dispersion of clones was suggestive of a niche-like organization in which stem cell competence is associated with a spatially localized environment.
Endothelial cells promote melanoma growth
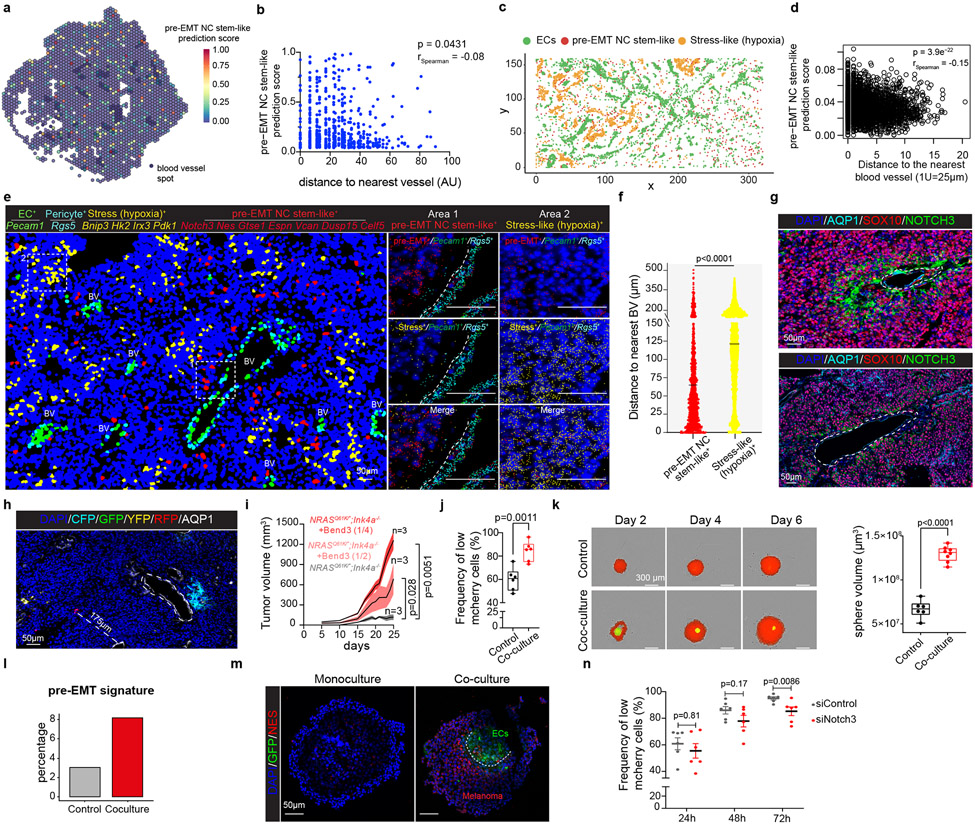
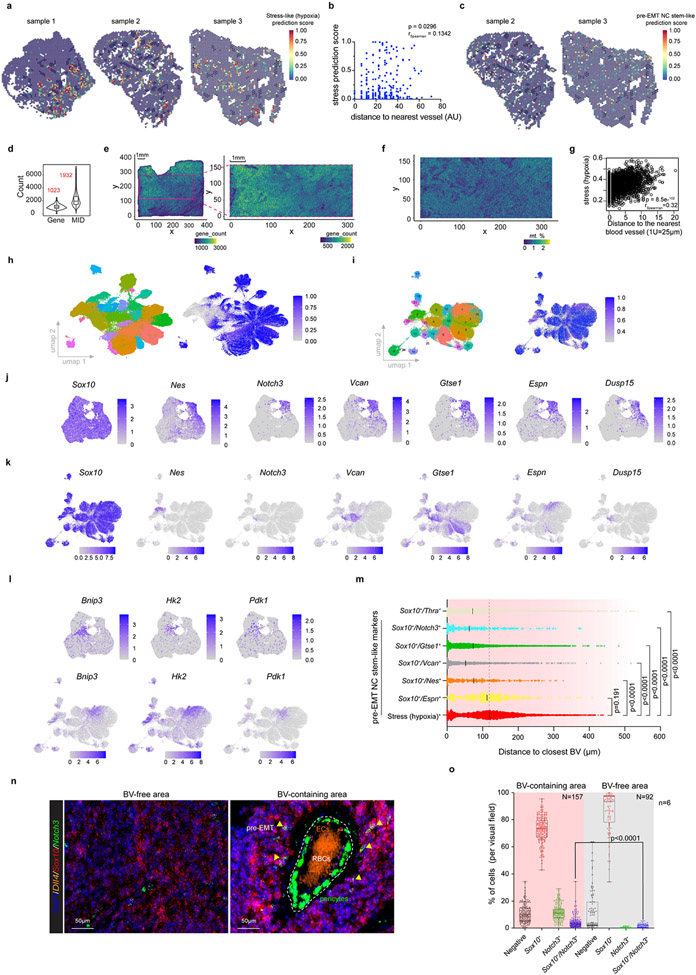
To search for evidence of a niche-like organization that could support a pre-EMT NC stem-like population we leveraged spatially resolved unsupervised transcriptomics (Fig. 3a). The spatial transcriptomics data was integrated with the scRNA-seq data using Seurat-v3 anchor- based integration25. A pathologist (J. v.d.O.) annotated various histopathological features on each section. This analysis revealed an expected positive correlation between the Stress-hypoxia prediction score and the distance to the nearest blood vessels (BVs; Extended Data Fig. 6a, b). In contrast, an inverse correlation was found between the distance to the nearest BV and the pre-EMT NC stem-like prediction score (Fig. 3b and Extended Data Fig. 6c).
Figure 3: A perivascular niche promotes melanoma growth.
a, Quantification of pre-EMT NC stem-like activity onto spatial transcriptomics. Dark blue, BVs annotation. b, Inverse correlation (Spearman) between the pre-EMT NC stem-like cell prediction score and distance to nearest BV. c, Spatial scatter plot shows bins of EC, pre-EMT NC stem-like and hypoxia (binary AUCell score). d, Anti-correlation (Spearman) of pre-EMT NC stem-like activity (AUCell score) and distance to closest BV. e, Molecular cartography for the indicated genes. Left, pseudocolored cells based on the type/state identity (n=33 ROIS). Insets, areas enriched either for pre-EMT NC stem-like (Area 1) or stress-hypoxia cells (Area 2). f, Median distance to nearest BV (n=33 ROIs; p<0.0001). g, Immunostaining for indicated proteins. Upper, a capillary; lower, a dilated tumour BV (n=3 tumors). h, Confocal image of Confetti+ tumours (n=6), immunostained for AQP1. i, DsRed-labeled melanoma cells injected into immunocompromised mice alone or with GFP-labeled Bend3 ECs (ratio 1/2; p= 0.0281 and ratio 1/4; p=0.0051). Tumour volume (mean±SEM) in function of time/days (n=3). j, Percentage of mCherry high versus low watermelon+ cells cultured alone or with Bend3 (n=6, p=0.0011). k, Left, melanospheres (DsRed-labeled) grown alone or with (GFP-labeled) Bend3 ECs. Right, melanosphere volume (n=6 monoculture, n=8 co-culture; p<0.0001). l, Melanoma cells cultured alone (Control) or with Bend3 cells and percentage of malignant cells harbouring the pre-EMT NC stem-like state (binary AUCell score) determined by scRNA-seq. m, Representative images of melanospheres (n=2) grown alone or with GFP-labeled Bend3. The presence of NES+ cells was evaluated. n, Doxycycline induced watermelon+ cells transfected with siControl or siNotch3 supplemented with Bend3 cells. b and d, Fisher exact test, two-tailed p value. f, i and n, two-tailed Mann-Whitney Test. In j and k, boxes extend from the 25th to 75th percentile, middle line represents the median. Whiskers represent min to max values (unpaired two-tailed Student’s t test with Welch’s correction).
To substantiate this finding, we applied SpaTial Enhanced REsolution Omics-sequencing (Stereo-seq), a method that allows transcriptomic profiling with high resolution and sensitivity26. We segmented the data into 50 x 50 DNB bins to retrieve transcriptomic information for 51.2k bins of 25 μm diameter (Extended Data Fig. 6d-f). Measuring a transcriptional endothelial signature (binary AUCell cut-off) revealed the location of the tumor vasculature (Fig. 3c, and Supplementary Table 6), whose external boundaries largely mirrored those manually-annotated by our pathologist. We assigned cell identities to each spatial module, and thus confirmed the significant negative correlation between the pre-EMT NC stem-like state and distance to the nearest BV (Fig. 3d), and the positive correlation between the stress-hypoxia cell state and BVs (Extended Data Fig. 6g).
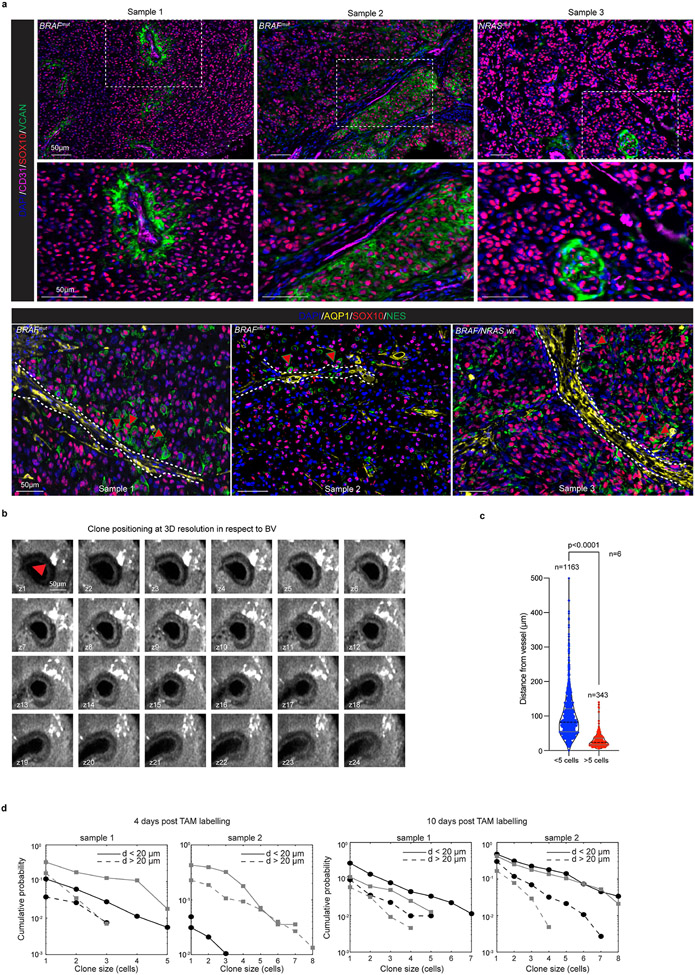
To validate these locations at true single-cell resolution, we employed Molecular Cartography™. In each section, we probed the expression of genes chosen to distinguish cellular compartments of the TME and various melanoma cell states (Supplementary Table 7). A computational framework that automatically computes cell segmentation and annotation was developed (Extended Data Fig. 6h). Approximatively 109,000 malignant cells were identified, among which about 900 cells expressed multiple pre-EMT NC stem-like markers, including Notch3, Nes, Vcan, Dusp15, Celf5 (cluster 23, Extended Data Fig. 6i-k). 90% of cells from cluster 23 co-expressed Sox10 and all three pre-EMT NC stem-like markers Notch3, Vcan and Nes. A similar approach was used to annotate the stress-hypoxia cell population (cluster 1, Extended Data Fig. 6l). Both melanoma cell populations were then pseudo-coloured (Fig. 3e), and their distance to the nearest BV quantified (Fig. 3f). The medium distance to nearest BV for pre-EMT NC stem-like cells was significantly shorter than for those annotated as stress/hypoxia. Note that similar conclusions could be drawn when analysing the localization of cells positive for Sox10 and only one of the pre-EMT NC stem-like markers, such as Notch3, Nes or Vcan for example (Extended Data Fig. 6m). An enrichment of cells positive for both Sox10 and Notch3 in BV-containing areas was further confirmed by RNAscope (Extended Data Fig. 6n,o) and by immunohistochemistry (Fig. 3g). We next performed IHC analysis using thick (>2mm Breslow) primary nodular human melanoma biopsies (n= 13). Cells positive for both SOX10 and VCAN could be identified in 12 out of 13 lesions and their proportion ranged from rare (less than 1%, in 7/13 lesions) to more frequent (between 1 and 5%, in 5/13 lesions). Importantly, these cells occurred in clusters and a fraction localized in proximity to CD31+ BVs (Extended Data Fig. 7a). Additional staining confirmed the presence of SOX10+/NES+ double positive cells in the vicinity of the tumour vasculature (AQP1+, 7/8 lesions) and thus irrespective of the NRAS or BRAF status (Extended Data Fig. 7a).
These findings support a model in which pre-EMT NC stem-like tumour cells localize preferentially at perivascular regions where they renew, producing more S cells that expand to colonize further the vasculature, or detach, giving rise to P-type cells that move away from the niche (Extended Data Fig. 7b). Consistently, we found that the productive Confetti-positive clones were typically either in contact with or in close vicinity to the tumour vasculature (Fig. 3h and Extended Data Fig. 7c,d).
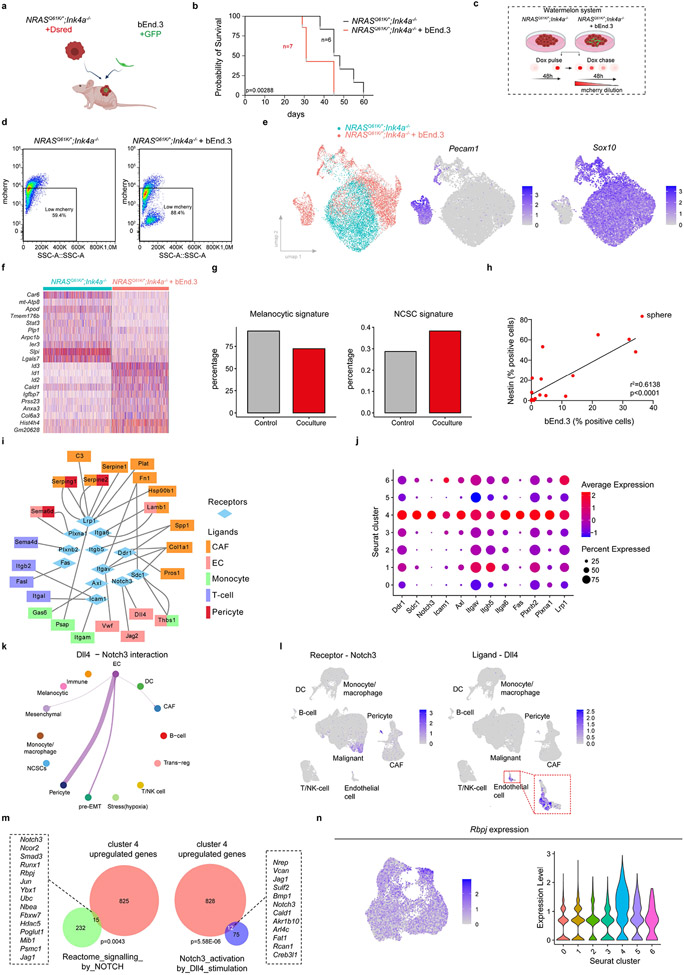
ECs were shown to promote the propagation of brain tumours27. To test whether ECs also support the in vivo growth of melanoma, we co-injected DsRed+ NRASQ61K/°/Ink4a−/− mouse melanoma cells either alone or with GFP+ ECs (Bend3) in immunocompromised mice (Extended Data Fig. 8a). Co-injection of ECs enhanced xenograft growth in a cell-dose-dependent manner (Fig. 3i), and ultimately decreased overall survival (Extended Data Fig. 8b). Bend3 ECs also stimulated the 2D and 3D (melanospheres) in vitro growth of melanoma cells (Fig. 3j,k Extended Data Fig. 8c, d, Methods and Supplementary Fig. 1).
Following the co-culturing of melanoma and Bend3 cells, we analysed the cellular composition by scRNA-seq (Extended Data Fig. 8e). While the parental NRASQ61K/°Ink4a−/− cell line harboured >90% of cells in the melanocytic cell state, the presence of Bend3 caused emergence of cells harbouring the de-differentiated neural crest-like and pre-EMT NC stem-like signatures (Fig. 3l and Extended Data Fig. 8f,g). Moreover, Nes+ cells emerged in melanospheres grown in the presence of Bend3/GFP ECs (Fig. 3m and Extended Data Fig. 8h). These data indicated that ECs contribute to a niche environment that promotes de-differentiation of melanoma cells and, thereby, stimulate melanoma growth.
To dissect the molecular mechanisms underlying the growth-promoting activity of ECs, we applied Nichenet28 and CellChat29, two different computational approaches that predict cell-to-cell communications. We identified cell-type-enriched ligands and searched for matching receptors expressed selectively in the pre-EMT NC stem-like cells. A putative interaction was identified between the Dll4-expressing ECs and NOTCH3 receptor specifically expressed in pre-EMT NC stem-like cells (Extended Data Fig. 8i-l). Notably, significant overlaps were detected when comparing genes upregulated in the pre-EMT NC stem-like melanoma cells with those from the Reactome-based NOTCH signalling signature (GSEA ID: M10189)30 and genes induced in synovial fibroblasts following DLL4 stimulation31 (Extended Data Fig. 8m), indicating that the DLL4-NOTCH3 interaction leads to activation of NOTCH signalling. Consistently, expression of the co-factor Rbpj of the active intracellular domain of NOTCH (ICD) was enriched in the pre-EMT NC stem-like cells (Extended Data Fig. 8n). To test whether the growth-stimulating activity of ECs was dependent on activation of NOTCH signalling, we silenced Notch3 in melanoma cells. Strikingly, the growth of Notch3 knock-down (KD) melanoma cells was no longer stimulated by the presence of Bend3 ECs (Fig. 3n). Together these data support a model in which ECs stimulate melanoma growth by promoting emergence and/or maintenance of the pre-EMT NC stem-like state.
PRRX1 marks cells that fuel metastasis
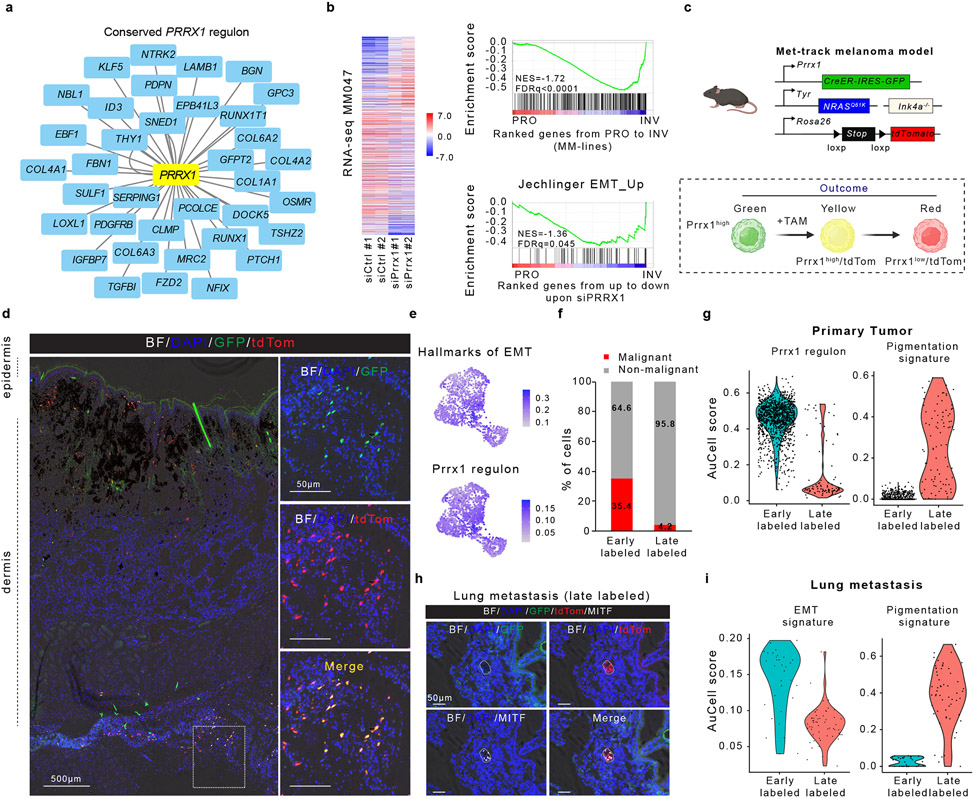
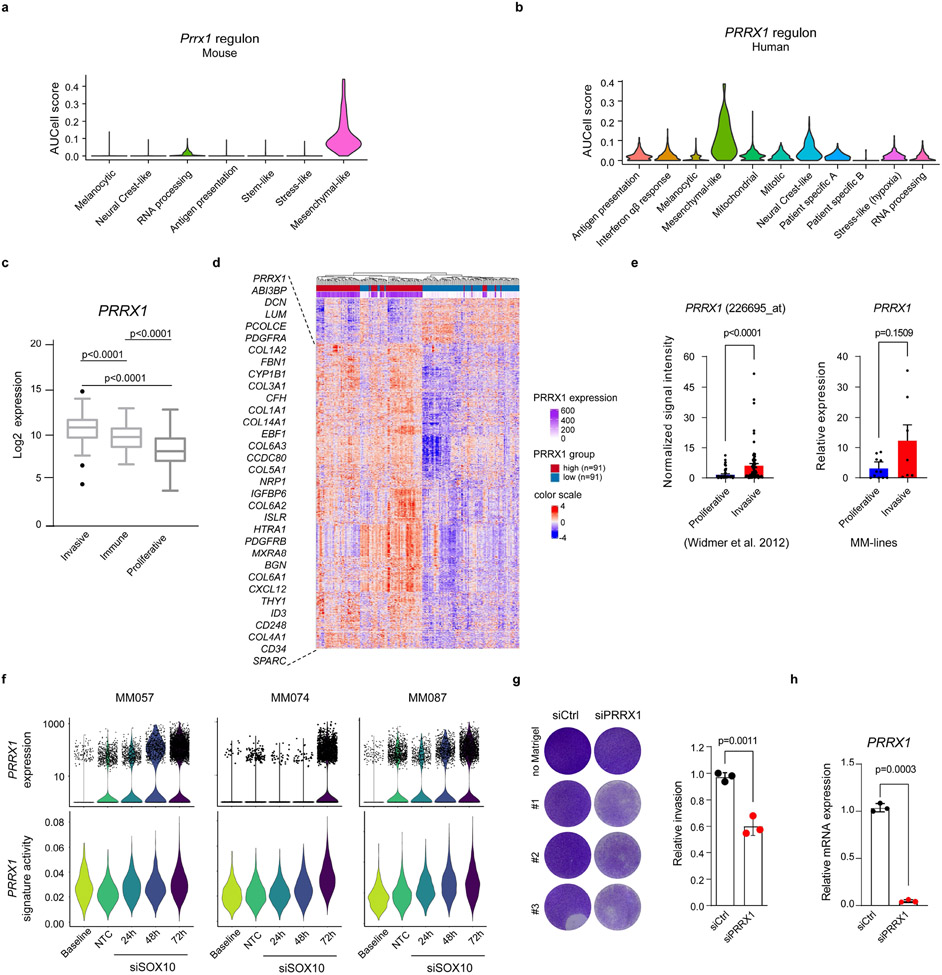
Unsupervised analysis predicted PRRX1, known to promote Epithelial-to-Mesenchymal-Transition (EMT) in epithelial cancers32,33, as a putative driver of both mouse and human mesenchymal-like cells (Fig. 4a, Extended Data Fig. 9a, b). PRRX1 was also highly expressed and/or transcriptionally active in freshly isolated clinical biopsies, TCGA_SKCM human clinical samples, melanoma cell lines and short-term cultures (MM lines) harbouring the invasive/undifferentiated melanoma signature (Extended Data Fig. 9c-e)34-36. A set of common mouse and human PRRX1-target genes was identified, which includes FBN1 and TGFBI (Fig. 4a). Consistently, PRRX1 expression and activity was progressively increased in melanocytic melanoma lines (MM057, MM074, MM087) following SOX10 KD, which leads to a progressive reprogramming into mesenchymal-like cells6 (Extended Data Fig. 9f). Moreover, silencing PRRX1 in mesenchymal-like melanoma cultures caused a dramatic decrease of the mesenchymal and EMT signatures (Fig. 4b), which was accompanied by a decreased ability to invade in short-term in vitro migration assays (Extended Data Fig. 9g,h). These data indicated that PRRX1 may not only mark the melanoma mesenchymal-like cell population, but also contribute to the acquisition and/or maintenance of the mesenchymal-like phenotype.
Figure 4: Temporal single-cell tracing identifies a population of melanoma cells that fuels metastasis but not primary tumour growth.
a, Conserved (mouse and human) PRRX1 regulon (see Methods). b, Left panel, heatmap of differentially expressed genes upon silencing of PRRX1 in MM047 cells (bulk RNAseq, n=2 biological replicates). Right panel, GSEA shows downregulation of mesenchymal-like signature upon siPRRX1 (MM-lines and Jechlinger_EMT_UP gene sets). c, Schematic representation of the Met-Track mouse model and possible lineage tracing outcomes following Tamoxifen (TAM) exposure. Schematic made with Biorender.com. d, Confocal images of a primary tumour 4 weeks post-TAM. Insets showing the majority of GFP/tdTomato-double positive cells in the deepest dermal part of the lesion. BF, Bright Field (n=4 independent mice). e UMAP shows hallmarks of EMT and PRRX1 regulon activity (AUCell score) in tdTomato+ cells 2 days (early labelled) post-TAM. f, Ratio malignant versus non-malignant (as determined by scRNA-seq) of FACS-sorted tdTomato+ fraction from primary tumours 2 days (early labelled) and 30-days (late labelled) post-TAM. g, Expression of PRRX1 regulon and pigmentation signatures (determined by scRNA-seq) of FACS-sorted tdTomato+ fraction 2 days (early labelled) and 30-days (late labelled) upon last TAM administration. h, Confocal images of a lung micro-metastasis 4 weeks post-TAM, immunostained for MITF (white). Red signal is from tdTomato expression; GFP (green) signal. BF, Bright Field (n=3 independent mice). i, Expression of PRRX1 regulon and pigmentation gene signatures (as determined by scRNA-seq) of FACS-sorted tdTomato+ fraction from lung metastatic lesions 2 days (early labelled) and 30-days (late labelled) post-TAM.
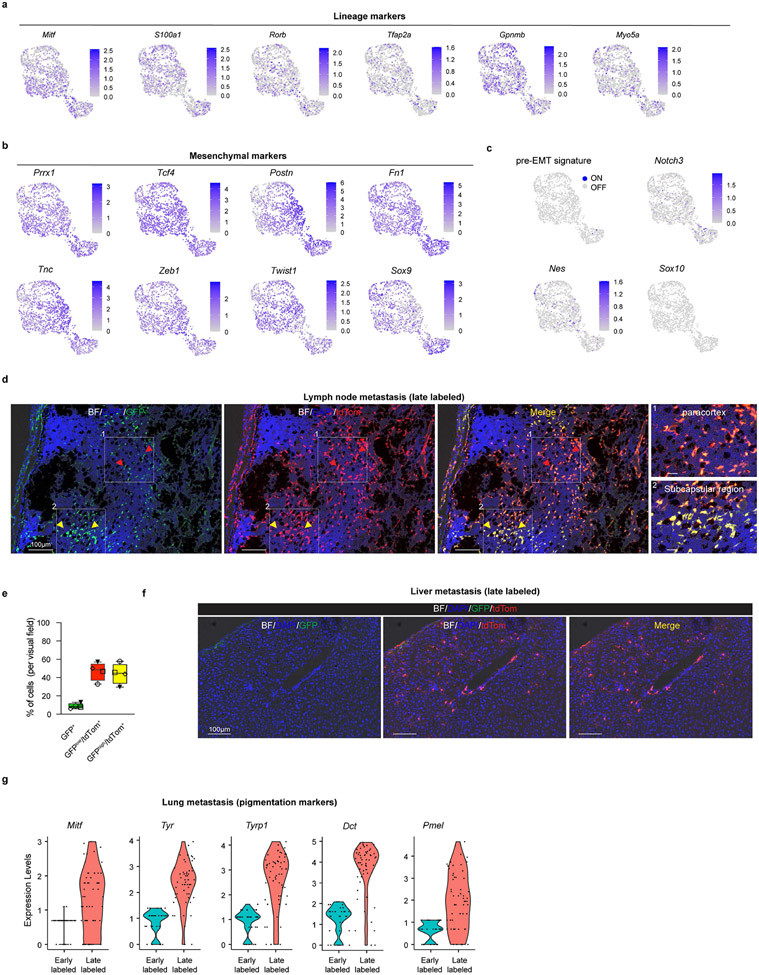
Taking advantage of the Prrx1-CreER-IRES-GFP37, we next devised a strategy to fate map Prrx1-positive cells in vivo. We generated Tyr::NRASQ61K;Ink4a−/−;Prrx1-CreER-IRES-GFP;ROSA26-LSL-tdTomato (referred to as “Met-Track”) mice (Fig. 4c). As expected, these mice developed primary melanomas and a small fraction of GFP+ cells could be detected in the deepest part (e.g. dermis or subcutaneous tissues) of these lesions (data not shown). Several rounds of TAM administrations ensured that the GFP+ cells turned ON tdTomato expression (Fig. 4c,d). While all Prrx1-progenies are expected to remain tdTomato+, a fraction of these cells may lose GFP expression owing to their transcriptional reprogramming into Prrx1-negative cells. We FACS-sorted tdTomato+ cells shortly after TAM exposure and analysed cell composition by scRNA-seq (Methods and Supplementary Fig. 2). All malignant tdTomato+ cells, which maintained low levels of expression of lineage marker genes (Extended Data Fig. 10a), expressed high levels of Prrx1 and other mesenchymal-like markers (Extended Data Fig. 10b). Consistently, high Prrx1 regulon activity as well as elevated expression levels of an EMT gene signature (GSEA-Molecular Signature Database: M5930) were detected in these cells (Fig. 4e). Critically, none of these cells (co-)expressed pre-EMT NC stem-like cell markers (Extended Data Fig. 10c). These data confirmed that this model is well-suited for the specific fate mapping of the Prrx1+ mesenchymal-like cell population.
To study the fate of these cells in primary tumours, we TAM-treated tumour-bearing mice and harvested the melanoma lesions 30-days post-treatment. TdTomato+ cells were isolated by FACS and cell composition was analyzed by scRNA-seq (Methods and Supplementary Fig. 2). Whereas 8% of the entire tumour population expressed tdTomato in tumours harvested shortly after TAM exposure (early labelled), this number dropped to about 2% 30-days post-TAM (late labelled). Moreover, whereas about 35% of the tdTomato+ cells were annotated as malignant at the early time point, this number dropped to about 4% at the late time point (Fig. 4f). These data indicated that the Prrx1-population does not contribute to the expansion of these primary tumours. Interestingly, while none of the tdTomato+ malignant cells expressed the melanocytic program at the early time point, a small number of cells had activated this program at the late time point, indicating that a fraction of the Prrx1-mesenchymal-like cells (or their progeny) can switch phenotype in primary tumours (Fig. 4g). Strikingly, a high fraction of disseminated tumour cells in lymph nodes (late labelled) were found to be tdTomato+ (Extended Data Fig. 10d,e). While a fraction of cells expressed high levels of both GFP and tdTomato in the subcapsular sinuses of lymph nodes, a large fraction of tdTomato+ cells that had invaded/expanded in the cortical region expressed lower levels of GFP. Similarly, whereas tdTomato+ cells could be identified in liver and lungs (late labelled) the majority of these cells exhibited low to undetectable GFP expression. The decrease in GFP expression seemed to occur very early during the metastatic cascade. This is, for instance, illustrated by the low to undetectable GFP signal in a lung micro-metastasis or in disseminated tdTomato+ melanoma cells that infiltrated the liver of one of the MetTrack mice (Fig. 4h and Extended Data Fig. 10f).
To further study cell fate dynamics during metastatic dissemination, we isolated early and late-labelled tdTomato+ cells from the lungs of TAM-treated mice, and the cell composition was analysed by scRNA-seq (Methods and Supplementary Fig. 2). Cells from the early-labelled lungs expressed the EMT/mesenchymal-like signature, but undetectable to very low levels of Mitf and the MITF-driven pigmentation signature (Fig. 4i and Extended Data Fig. 10g). In contrast, whereas expression of Mitf and other pigmentation markers could be detected in cells isolated from late-labelled lungs, these cells did not express high levels of the EMT/Mesenchymal-signature. High levels of MITF expression in these cells was confirmed by IHC (Fig. 4h). This longitudinal analysis indicated that Prrx1 marks a population that does not contribute to primary tumour growth, but instead form a pool of metastatic initiating cells (MICs).
Discussion
We show herein that the growth of melanoma is supported by a relatively limited population of tumorigenic cells and provide several lines of evidence indicating that a dedifferentiated melanoma cell population, exhibiting an embryonic transcriptional program activated in cells at the origin of the embryonic neural crest stem cell lineage, may sit at the apex of this cellular hierarchy. Critically, the non-random spatial distribution of these cells, which we find enriched at the tumour vasculature, raised the possibility that their tumour fuelling capacity may be acquired via exposure to specific extrinsic cues, such as signals emanating from the endothelium, rather than driven by an intrinsic cancer stem cell (CSC) program. Consistent with this possibility, we show that ECs stimulate melanoma de-differentiation, and promote the in vitro and in vivo growth of melanoma in a manner that depends on NOTCH3, a receptor expressed specifically in the pre-EMT NC stem-like cell population. These data therefore support a dynamic model of hierarchical growth in which de-differentiation can serve as a source of CSCs. This permutable CSC paradigm may help reconcile evidence supporting the scarcity of melanoma cells with high tumorigenic potential38-40 with (apparently conflicting) reports showing that a large fraction of melanoma cells can exhibit tumorigenic potential in xenotransplantation experiments40,41.
Consistent with a model in which tumour growth is supported by a limited population of tumorigenic cells, lineage tracing of the mesenchymal-like melanoma cell state indicated that these cells do not contribute to the expansion of primary tumours but, instead, constitute a pool of MICs. We identified Prrx1 as a marker of this state and we generated a novel mouse model, termed Met-Track, which allows for the fate mapping of this Prrx1+ MIC population. Temporal single-cell tracing revealed that a large fraction of MICs (and/or their progeny) change cell identity when (or even before) reaching their metastatic sites. This suggests that metastatic outgrowth can only occur following suppression of the Prrx1+ mesenchymal-like transcriptional program. Such a reprogramming event is reminiscent to the mesenchymal-to-epithelial transition (MET) process described in epithelial cancers42. These data are consistent with previous findings showing that PRRX1 promotes EMT while suppressing stemness traits in breast cancer cells33.
In conclusion, our study provides a high-resolution and spatially-resolved map of the mouse melanoma ecosystem, charting the origin of phenotypic diversity and tumour growth. Together with the Met-Track mouse model we report herein, these resources provide a platform to study extrinsic and systemic factors driving primary tumour growth and metastatic dissemination, with the potential to develop methods for early detection as well as therapeutic strategies that intercept the disease before its spreading to vital organs.
Methods
Compliance with ethical regulations
All mouse colonies were maintained in a certified animal facility in accordance with European guidelines. Specifically, animals were housed in a controlled environment with 14/10hour light/dark cycles, standard diet and water ad libitum. All experiments strictly complied with the protocols approved by the University of Leuven Animal Care and Use ethical committee. Mice were euthanized in different time points and tumor sizes or if the tumour was ulcerated independently of its size, if the mouse lost >20% of the initial weight or any other sign of distress (based on the general health status). None of the experiments performed in this study surpassed the size limit of the tumours (volume did not exceed 2cm3). No randomization and no blinding were performed in this study.
Mice
We inter-crossed the following strains: Tyr::CreERT243, Tyr::NRASQ61K/°8, Ink4a−/−44, R26RLSL-tdTomato (Jackson laboratory, #007909), R26RLSL-confetti23, BRafCA/+45; Ptenl/l45, Prrx1::CreER-IRES2-EGFP37 to generate Tyr::CreERT2/+;NRASQ61K/°;Ink4a−/−;ROSA26RLSL-confetti/LSL-confetti, Tyr::CreERT2/+;BRafCA/+;Ptenl/l;ROSA26LSL-tdTomato/LSL-tdTomato and Prrx1::CreER-GFP;Tyr-NRASQ61K/°;Ink4a−/−;ROSA26RLSL-tdTomato/LSL-tdTomato compound mice.
For the generation of the melanoma allograft models, spontaneous skin melanoma lesions from Tyr::CreERT2/+; Tyr-NRASQ61K/°;Ink4a−/−;ROSA26RLSL-confetti/LSL-confetti animals were dissociated into small pieces using forceps and scissors. Some of these tumour pieces were subcutaneously engrafted into C57bl/6 or Foxn1nu/nu mice and expanded for three generations before starting the clonal analyses. Tumour volume was monitored with a caliper and the volume was calculated using the following formula: V= (π/6) x length x width x height.
For lineage tracing experiments, Tamoxifen (Tam; Sigma Aldrich, #T5648) was dissolved in 10% ethanol and 90% Corn Oil for 4 h at 55°C. Aliquots were kept at −20°C. Stock solutions of 5mg/ml and 20mg/ml were prepared and 100μl were administered intraperitoneally (IP). For the clonal analysis on the Confetti background, the dose of tamoxifen was titrated down up to 0.5mg/ml. Mice were injected once their tumours reached an approximate volume of 150mm3 and sacrificed at the indicated time points following injections or just before tumours reached the humane endpoint. The Prrx1::CreER-GFP;Tyr-NRASQ61K/°;Ink4a−/−;ROSA26RLSL-tdTomato/LSL-tdTomato mice were induced by intraperitoneal injection of 2mg of Tamoxifen for 5 consecutive days when primary melanoma lesions had reached an approximate size of 50mm3. Mice were sacrificed either two days or thirty days post-induction. For tumour induction in the Tyr::CreERT2;BRafV600E/+;Ptenl/l tail mouse model, 6 weeks-old mice were topically treated with 130 mM of 4-Hydroxy-Tamoxifen (4-HT) using 1 μL per cm (tail length) and sacrificed 111 days post-treatment. To induce the formation of the back-skin tumours, mice were topically treated on the back with 1 μL of 130 mM 4-HT and sacrificed 65 days post-treatment.
For the proliferation kinetics experiments, mice were first IP injected daily for 9 consecutive days with EdU (25mg/kg in PBS, Invitrogen, E10415) and then daily for 3 consecutive days with BrdU (50mg/kg in PBS, Sigma, #19-160). Mice were sacrificed 24h after the last BrdU injection.
Co-injection of Melanoma and Endothelial cells in mice
Tyr::NRASQ61K/°;Ink4a−/− mouse melanoma cells stably expressing dsRed-encoding lentiviruses and Bend3 endothelial cells (ECs, ATCC, # CRL-2299) stably expressing GFP encoding lentiviruses were mixed in a ratio of 1:2 (105 melanoma:2x105 ECs) and 1:4 (105 melanoma:4x105 ECs) and resuspended in matrigel (5mg/ml; Thermofisher scientific, #356255). Cells were then injected subcutaneously in the back skin of Foxn1nu mice. Mice were sacrificed and tumors were collected 25 days following melanoma initiation. To measure overall survival, tumors were grown until reaching a humane endpoint of approximately 1,5cm3. Tumor volume was monitored with a caliper and the volume was calculated using the following formula: V = (π/6)*length*width*height.
Immunofluorescence of mouse frozen tissue sections
Tissue samples were dissected and fixed for 20min in 4% paraformaldehyde at RT. Samples were washed in PBS and incubated overnight in 30% sucrose in PBS at 4°C. Tissue samples were then embedded in Tissue-Tek® O.C.T. Compound (Sakura® Finetek, #4583) and stored at −80°C. Sections of 10 μm were cut using the Thermo Scientific CryoStar NX70 Cryostat. For immunofluorescence, tissue sections were fixed for 10min in 4% paraformaldehyde on ice. Sections were washed in PBS for 5min at RT, permeabilized in 1% Triton X-100 in PBS for 10min at RT and washed in 0.1% Tween®20 (VWR chemicals, Amresco, #0777) in TBS. Sections were incubated in blocking buffer (1%BSA, 10% donkey serum, 0.1% Tween in TBS) for 1h at RT. Subsequently, primary antibodies were incubated overnight at 4°C in antibody diluent (1%BSA, 0.1% Tween in TBS). Sections were washed three times using 0.1% Tween®20 in TBS for 5min at RT and incubated with corresponding secondary antibodies diluted 1:250 in antibody diluent for 1h at RT. Sections were washed three times for 5min at RT, nuclei were stained with DAPI (4’,6-diamino-2-phenylindole, dihydrochloride, 1:1000 dilution, Thermofisher, #D3571) solution (0.5mg/ml) diluted 1:1000 in PBS for 5min and mounted in ProLong™ Diamond Antifade Mountant (Thermofisher Scientific, #P36961).
For EdU/BrdU staining, samples were first stained with the primary antibody, washed and stained with the secondary antibody, as described above. In order to denature DNA, the sections were incubated in 2N HCl for 30 minutes at 37 °C and neutralized the acid by immersing sections in 0.1M borate buffer two times for 5 minutes. The sections were washed three times using 0.1% Tween®20 in TBS for 5min and incubated overnight with BrdU antibody in blocking buffer. The next day, samples were washed in 0.1% Tween®20 in TBS and incubated with secondary antibody. Samples were washed 3 times in 0.1% Tween®20 in TBS and proceeded for EdU detection. Edu staining was performed using Click-iT™ EdU Cell Proliferation Kit for Imaging, Alexa Fluor™ 647 dye following the manufacturer’s instructions (Thermofisher Scientific, #C10340).
The following primary antibodies were used: anti-GP100 (1:400, Abcam, #ab137078); anti-MITF (rabbit, 1:200, Sigma-Aldrich, #HPA003259, Sigma); anti-AQP1 (1:2000, Millipore, #AB2219); anti-CD31/PECAM-1 (1:200, Dianova, #DIA-310); anti-MLANA (1:200, Sigma-Aldrich, HPA048662), anti-NES (1:100, Abcam, #ab81462) and anti-BrdU (1:100, Abcam, #ab6326). The following secondary antibodies were used: donkey-anti-Rat IgG (H+L), Alexa Fluor 488 (Thermofisher Scientific, #A-21208); donkey-anti-Rabbit IgG (H+L), Alexa Fluor 647 (Thermofisher Scientific, #A31573) and goat-anti-Rat IgG (H+L), Alexa Fluor 647 (Thermofisher Scientific, #A21247).
Histopathology of mouse tissue specimens
Tissue samples were collected and fixed in 4% paraformaldehyde for 24 hr and processed for paraffin embedding (Thermo Scientific Excelsior AS Tissue Processor and HistoStar Embedding Workstation). 5 μm thick sections were obtained from the paraffin-embedded tissues (Thermo Scientific Microm HM355S microtome) and mounted on Superfrost Plus Adhesion slides (Thermo Scientific) and stained with hematoxylin and eosin (H&E, Diapath #C0302 and #C0362) for histopathological examination.
Immunofluorescence of mouse paraffin-embedded tissue specimens
We used the opal-based approach, which relies on individual tyramide signal amplification (TSA)-conjugated fluorophores to detect various targets. After deparaffinization, antigen retrieval was performed in citrate buffer at pH 6. Sections were incubated 2h (room temperature or RT) or overnight (4°C) with the following primary antibodies: anti-AQP1 (1:2000, Millipore, #AB2219), anti-SOX10 (1:100, Thermofisher Scientific, #703439), anti-NOTCH3 (1:100, Abcam, #ab23426). The slides were then washed and incubated for 5 min with successive washes in TBST at room temperature. The slides were next incubated at RT for 10 min with one of the following Alexa Fluor tyramides (PerkinElmer) to detect antibody staining, prepared according to the manufacturer’s instructions: Opal 520, Opal 570, and Opal 690 (dilution 1:50). Stripping of primary and secondary antibodies was performed by placing the slides in a plastic container filled with antigen retrieval (AR) buffer in citrate buffer pH = 6. A microwave was used to bring the liquid at 100 °C (2 min), and the sections were then microwaved for an additional 15 min at 75 °C. Slides were allowed to cool down in the AR buffer for 15 min at RT and were then rinsed with deionized water and 1 × Tris-buffered saline with Tween 20. After three additional washes in deionized water, the slides were counterstained with DAPI for 5 min and mounted with ProLong Gold Antifade Mountant (Thermofisher Scientific, #P36930).
Immunofluorescence on human biopsies
5μm-sections of formalin-fixed, paraffin-embedded (FFPE) primary nodal melanoma lesions were stained with antibodies against SOX10 (SCBT; #sc-365692; 1 μg/ml), VCAN (Sigma-Aldrich; #HPA004726; 0.2 μg/ml) and CD31 (LSBio; #LS-C173974; 1 μg/ml) according to the Multiple Iterative Labeling by Antibody Neodeposition (MILAN) protocol46. Image registration was performed by applying a homomorphic transformation over a set of matched descriptors using a Harris detector. Autofluorescence (AF) subtraction was performed by subtracting the scaled AF channel from the measured signal (MS). The scaling factor was obtained by applying Gaussian mixture models and linear modeling in the MS/AF hypermatrix. For SOX10/NES double positive cells, opal-based immunofluorescence was used in primary human biopsies as described above. Sections were incubated at 2h (RT) or overnight (4°C) with the following primary antibodies: anti-AQP1 (1:2000, Millipore, #AB2219), anti-SOX10 (1:200, R&D Systems, #AF2864), anti-NES (1:100, Abcam, # ab105389).
Tissue clearing
Tissue clearing was performed using a modified protocol described previously47. Tumour pieces from allograft melanoma model of approximately 1mm thickness were fixed overnight in 4% PFA at 4°C. Samples were washed 3x in PBS and cleared using a modified protocol of CUBIC (Clear, Unobstructed Brain Imaging Cocktails and Computational analysis). Briefly, Sca/eCUBIC-1 (Reagent 1A) was prepared as a mixture of 10 wt% urea (Nacalai Tesque Inc., #35904-45, Japan), 5 wt% N,N,N’,N’-tetrakis(2-hydroxypropyl) ethylenediamine (Tokyo Chemical Industry CO., LTD., #T0781), 10 wt% Triton X-100 (Nacalai Tesque Inc., #25987-85) in 5M NaCl. Sca/eCUBIC-2 (Reagent 2) was prepared as a mixture of 50 wt% sucrose (Nacalai Tesque Inc., #30403-55), 25 wt% urea, 10 wt% 2,20,20’-nitrilotriethanol (Wako Pure Chemical Industries Ltd., #145-05605), 0.1% (v/v) Triton X-100 and 0.1% (v/v) Sodium Azide. Tumour pieces were incubated in 1/2-diluted Reagent-1A for 6h at RT with shaking. Then, samples were incubated in Reagent-1A at RT for 2 days on a rocking plate (100 rpm) protected from the light. The samples then were moved at 37°C under continue shaking with fresh media that was replaced every second day. After 8 days the samples were washed with PBS and immersed in 1/2-diluted Reagent-2 for at least 24h at RT and then immerse in Reagent-2 at RT for 2 days. Immunostaining of CUBIC tissue cleared samples to visualise confetti clones in respect to blood vessels (assessed by CD31 positivity) was performed after incubation with Reagent-1A. Samples were incubated with primary antibody (CD31/PECAM-1, 1:200, Dianova, #DIA-310) resuspended blocking buffer (PBS with 0.1% TritonX-100 and 10% normal goat serum) for 3 days at 37°C with rotation. Then cleared tumours were washed using washing buffer (PBS with 0.1% Triton X-100) several times at 37°C with rotation. Secondary antibody (goat-anti-Rat IgG, Alexa Fluor 647, 1/200, Thermofisher Scientific, #A21247) resuspended in blocking buffer was added and samples were incubated for 3 days at 37°C with rotation. Then samples were washed and counterstained with DAPI (1/1000) for 24h at 37°C with rotation. Then they were immersed in Reagent 2 for 2 days. Samples were acquired using Zeiss LSM 880 Airyscan.
Spatially resolved transcriptomics using Visium 10x
For tissue processing and slide preparation, tumours were dissected, washed with 1x PBS and snap-frozen in liquid nitrogen-chilled isopentane. The frozen tumours were then transferred to a cold tissue mold filled with chilled OCT. The cryomold was then immediately placed on dry ice. Tissue blocks were stored at −80°C in a sealed container. For cryosectioning, both the tissue block and the Visium slide were equilibrated inside the cryostat for 30 min at −12 °C before sectioning. Sections were cut at a thickness of 10 μm and immediately placed onto the Visium array slide (Visium Spatial Gene Expression slides, 10x Genomics). Array slides containing sections were stored at −80°C for 24h before use. For fixation, staining, imaging, and construction of cDNA libraries, samples were processed according to the manufacturer’s instructions (Visium Spatial Gene Expression User Guide.Rev, 10x Genomics) and all reagents were from the Visium Spatial Gene Expression Kit (10x Genomics). Briefly, sections were fixed in chilled methanol for 30 min at −20 °C, stained with hematoxylin and eosin. Imaging was performed on a Nikon-Marzhauser Slide Express 2 whole-slide scanner at 20x magnification. After imaging, sections were permeabilized at 37 °C for 20min. Permeabilization time was determined using the Visium Spatial Tissue Optimization Kit (10x Genomics). After permeabilization, the on-slide reverse transcription (RT) reaction was performed at 53 °C for 45 min. Second strand synthesis was subsequently performed on-slide for 15 min at 65 °C. All on-slide reactions were performed in a thermocycler with a metal slide adapter plate. Following second strand synthesis, samples were transferred to tubes for cDNA amplification and cleanup. Cycle number determination for cDNA amplification was done by qPCR according to the manufacturer’s instructions. Library QC was accessed using a TapeStation (Agilent).
Visium libraries were sequenced on the Illumina Novaseq 6000 instrument (parameters: SP flowcell 100 cycles single reads, lane 1 = 220pM + 1% PhiX / lane 2 = 215pM + 1% PhiX). We obtained 998M raw passed filter reads. BCL2 sequencing files were further processed with SpaceRanger (v1.0.0, 10x genomics) to generate spatial gene expression matrices. In parallel, we performed manual grid-alignment using the loupe browser (v4.2.0, 10x genomics). The SpaceRanger output of the 3 melanoma samples was further analyzed using the Seurat spatial vignette (https://satijalab.org/seurat/vignettes.html). In brief, spots were retained when nFeature_Spatial >1000 & percent.mt <5 and expression data was normalized using SCTransform (Seurat v3.2.3)25. The spatial distribution of the different transcriptional melanoma states was inferred using CCA-based label transfer (k.anchor = 10). The resulting prediction scores (0.0-1.0) for the different melanoma states were further used to correlate the distance of every spot to the nearest vessel. To do so, vessels that have been identified by the pathologist based on matching HE-stained sections were manually selected using the Loupe browser (10x genomics). The Euclidean distance between all spots was then calculated and returned for each spot a distance to the nearest vessel-annotated spot (au= arbitrary units). For Spearman correlation, stress (hypoxia) prediction scores per spot had to bigger or equal to 0.008 and each spot had to contain at least n=10 reads for Sox10. This resulted in n=263 spots for all 3 samples. For putative pre-EMT NC stem-like spots, the pre-EMT NC stem-like prediction score had to be bigger or equal to 0.04, spots needed to be Sox10 positive and negative for Rgs5. This resulted in 626 pre-EMT NC stem-like spots that were correlated with “distance to nearest vessel”.
Spatial transcriptomics using Stereo-seq
Stereo-seq chips have DNA nanoball (DNB) bins with 220 nm diameter and a center-to-center distance of 500 nm. For sample preparation and sequencing, tissue blocks in OCT (Sakura, 4583) were cut into sections of 10 μm thickness using cryostat (Leica, CM1950) and adhered to Stereo-seq chip surface (BGI Research) to generate cDNA library as described previously26. Briefly, a 10 μm tissue section was adhered to the surface of Stereo-seq chip, whereas the adjacent sections were used for HE staining or IHC. The chip was placed on a warming plate (Bel-Art, BELAF370151000) at 37°C for 3 min to dry and immersed in pre-cooled methanol at −20°C for 30 min for fixation. The chip was incubated with 100 μl 0.1% pepsin (Sigma, P7000) at 37°C for 12 min and washed with 0.1 × SSC buffer (Thermo Fisher Scientific, AM9770) containing 0.05 U/μL RNase inhibitor. RNA released from the permeabilized tissue was captured by DNA nanoball (DNB) on the chip and reverse transcribed using 100 μl Superscript reverse transcription II mix (10 U/μl reverse transcriptase, 1 mM dNTPs, 1 M betaine solution PCR reagent, 7.5 mM MgCl2, 5 mM DTT, 2 U/μl RNase inhibitor, 2.5 μM Stereo-seq template switch oligo, and 1× First-Strand buffer) at 42°C for 90 min. Tissue on the chip was removed by incubating with tissue removal buffer (10 mM Tris-HCl, 25 mM EDTA, 100 mM NaCl, 0.5% SDS) at 55°C for 10 min. After washing twice with 0.1× SSC buffer, the chip was incubated with 400 μl cDNA release buffer at 55°C for 4 hours. Then the cDNA was purified using AMPure® XP DNA Clean Beads (Vazyme, N411-03) and amplified using KAPA HiFi Hotstart Ready Mix (Roche, KK2602) and cDNA primer. A total of 20 ng of cDNA PCR product was fragmented to generate cDNA sequencing library. The cDNA library was loaded onto a G400 sequencer (MGI) for sequencing (pair-end, read 1 of 50 bp and read 2 of 100 bp).
For cell-type probability calculation, the probability of a single bin50 matching a specific cell-type was measured using the AUCell package (10.18129/B9.bioc.AUCell) with default parameters. 50 endothelial marker genes, 14 SuperMel markers, and top 100 signature genes for pre-EMT NC stem-like and stress-hypoxia were used (see Supplementary Table 6). Bin50 spots within an area of 320 x 160 bin50 units were selected for further analysis (bin50 resolution, 25 μm diameter). Within the targeted area, the median Molecular Identifier (MID) and gene counts of the bin50 spots were 1932 and 1023, respectively. Maximum percentage of mitochondrial genes was 2.8% with an average of 0.84%.
For statistical analysis, correlations between two variables (e.g., AUCell values of two cell-types) were calculated using the R environment build-in function cor.test with default parameters (Spearman's correlation coefficient). Blood vessels were identified using AUCell binary cutoff and expression of 1 or more endothelial cell markers (Aqp1, Gpihbp1, Pecam1, Kdr, and Cdh5). Both pre-EMT NC stem-like and stress (hypoxia) spots had to be Sox10 positive and negative for Kdr and Cdh5. This resulted in 4153 spots that were correlated with “distance to nearest vessel”.
Molecular Cartography™
To prepare tissue sections, tumor samples were resected and fixed for 24 h in PAXgene tissue fix following an incubation for 2 h with PAXgene Tissue Stabilizer. Three sections from two tumor samples were used. 10μm thick sections were placed within the capture areas of cold Resolve Biosciences slides, thawed and treated with Isopropanol for 1 minute followed by 1 minute washes in 95% Ethanol and 70% Ethanol at RT. Samples were used for Molecular Cartography™ (100-plex combinatorial single molecule fluorescence in-situ hybridization) according to the manufacturer’s instructions (protocol 4.0; available for download from Resolve’s website to registered users), starting with the aspiration of ethanol and the addition of buffer BST1 (step 6 and 7 of the tissue priming protocol). Briefly, tissues were primed followed by overnight hybridization of all probes specific for the target genes (see below for probe design details and target list). Samples were washed the next day to remove excess probes and fluorescently tagged in a two-step color development process. Regions of interest were imaged as described below and fluorescent signals removed during decolorization. Color development, imaging and decolorization were repeated for multiple cycles to build a unique combinatorial code for every target gene that was derived from raw images as described below.
For the probe design of the 97 selected genes, we used the Resolve’s proprietary design algorithm. For every targeted gene all full-length protein coding transcript sequence from the ENSEMBL database was used as design target if the isoform had the GENCODE annotation tag ‘basic’48,49. To filter highly repetitive regions, the abundance of k-mers was obtained from the background transcriptome using Jellyfish50. Every target sequence was scanned once for all k-mers, and those regions with rare k-mers were preferred as seeds for full probe design. A probe candidate was generated by extending a seed sequence until a certain target stability was reached. A set of simple rules were applied to discard sequences that previously found to cause problems experimentally. After these fast screens, every selected probe candidate was mapped to the background transcriptome using ThermonucleotideBLAST51 and probes with stable off-target hits were discarded. Specific probes were then scored based on the number of on-target matches (isoforms), which were weighted by their associated APPRIS level52, favoring principal isoforms over others. A bonus was added if the binding-site was inside the protein-coding region. From the pool of accepted probes, the final set was composed by greedily picking the highest scoring probes (Supplementary Table 7).
33 regions of interest (ROIs) of all samples were imaged on a Zeiss Celldiscoverer 7, using the 50x Plan Apochromat water immersion objective with an NA of 1.2 and the 0.5x magnification changer, resulting in a 25x final magnification. Standard CD7 LED excitation light source, filters, and dichroic mirrors were used together with customized emission filters optimized for detecting specific signals. Excitation time per image was 1000 ms for each channel (DAPI was 20 ms). A z-stack was taken at each region with a distance per z-slice according to the Nyquist-Shannon sampling theorem. The custom CD7 CMOS camera (Zeiss Axiocam Mono 712, 3.45 μm pixel size) was used. For each region, a z-stack per fluorescent color (two colors) was imaged per imaging round. A total of 8 imaging rounds were done for each position, resulting in 16 z-stacks per region. The completely automated imaging process per round (including water immersion generation and precise relocation of regions to image in all three dimensions) was realized by a custom python script using the scripting API of the Zeiss ZEN software (Open application development).
For spot segmentation, the algorithms for spot segmentation were written in Java and are based on the ImageJ library functionalities. Only the iterative closest point algorithm is written in C++ based on the libpointmatcher library (https://github.com/ethz-asl/libpointmatcher).
For preprocessing, as a first step all images were corrected for background fluorescence. A target value for the allowed number of maxima was determined based upon the area of the slice in μm2 multiplied by the factor 0.5. This factor was empirically optimized. The brightest maxima per plane were determined, based upon an empirically optimized threshold. The number and location of the respective maxima was stored. This procedure was done for every image slice independently. Maxima that did not have a neighboring maximum in an adjacent slice (called z-group) were excluded. The resulting maxima list was further filtered in an iterative loop by adjusting the allowed thresholds for (Babs-Bback) and (Bperi-Bback) to reach a feature target value (Babs: absolute brightness, Bback: local background, Bperi: background of periphery within 1 pixel). This feature target values were based upon the volume of the 3D-image. Only maxima still in a z-group of at least 2 after filtering were passing the filter step. Each z-group was counted as one hit. The members of the z-groups with the highest absolute brightness were used as features and written to a file. They resemble a 3D-point cloud. Final signal segmentation and decoding: To align the raw data images from different imaging rounds, images had to be corrected. To do so the extracted feature point clouds were used to find the transformation matrices. For this purpose, an iterative closest point cloud algorithm was used to minimize the error between two point-clouds. The point clouds of each round were aligned to the point cloud of round one (reference point cloud). The corresponding point clouds were stored for downstream processes. Based upon the transformation matrices the corresponding images were processed by a rigid transformation using trilinear interpolation. The aligned images were used to create a profile for each pixel consisting of 16 values (16 images from two color channels in 8 imaging rounds). The pixel profiles were filtered for variance from zero normalized by total brightness of all pixels in the profile. Matched pixel profiles with the highest score were assigned as an ID to the pixel. Pixels with neighbors having the same ID were grouped. The pixel groups were filtered by group size, number of direct adjacent pixels in group, number of dimensions with size of two pixels. The local 3D-maxima of the groups were determined as potential final transcript locations. Maxima were filtered by number of maxima in the raw data images where a maximum was expected. Remaining maxima were further evaluated by the fit to the corresponding code. The remaining maxima were written to the results file and considered to resemble transcripts of the corresponding gene. The ratio of signals matching to codes used in the experiment and signals matching to codes not used in the experiment were used as estimation for specificity (false positives). For downstream analyses, we used customized scripts. The gene expression from Resolve Bioscience has been imported in QuPath53. Cells have been segmented using StarDist54 with the model dsb2018_heavy_augment.pb within QuPath and the gene expression has been measured by cell. Cells have been classified based on their gene expression and counted. Euclidean distances between cell types have been measured. To overcome limitations/artefacts based on segmentation, for cell type and cell state classification a number of criteria were set to characterize with high confidence the segmented cells. A customized script included the following parameters for the classification: 1. minimum number of genes, 2. mandatory genes, 3. number of transcripts per gene, 4. number of genes excluded. To classify pre-EMT NC stem-like cell state a segmented cell was set to express the melanocytic marker Sox10 and minimum three additional genes, characteristic for this state: Nes, Notch3, Celf5, Gtse1, Dusp15, Thra, Vcan, Espn. To classify Hypoxic/Stress cell state a segmented cell was set to express Sox10/Mitf and minimum three additional genes, characteristic for this state: Bnip3, Hk2, Irx3, Pdk1. To classify endothelial cells a segmented cell was set to express minimum three genes, characteristic for this type: Pecam1, Dll4, Kdr, Cdh5. To classify pericytes a segmented cell was set to express Rgs5 and minimum two additional genes, characteristic for this type: Acta2, Kcnj8, Col6a2. In addition, to avoid misclassification, cells were set to express low (or be negative) markers from other cell types or cell states. Blood vessels were assigned based on EC presence and distance of different cell states was calculated based on nearest distance to each BV (object). The customized scripts are available upon request. Furthermore, the gene expression matrix containing cells (>170k) of all 33 ROIs (without spatial information) was log-normalized and scaled while retaining only cells with at least 2 features using a customized R-script (available upon request). Variable features were identified using PCA. Louvain clustering (resolution = 0.3) and subsequent dimension reduction (dims = 1:20) using UMAP were used to project all cells into a two-dimensional UMAP space. To identify malignant melanoma cells, we measured melanocytic gene expression signature (Pmel, Mlana, Mitf, Sox10, Rab27a, Car14, Tspan10 and Gsta4) in all cells using AUCell. Malignant cells were subsetted and re-clustered (dims=1:15, resolution=0.8) when they showed a melanocytic AUCell value > 0.25 and absence of endothelial, pericyte markers and immune markers.
Fluorescent in situ hybridization (RNAscope)
Tumor samples were collected and fixed in 4% PFA for 24h at RT. Then samples were dehydrated using ethanol series followed by xylene and embedded in paraffin using standard procedures. Tissue blocks were cut in 5 μm-thick sections collected onto Superfrost™ Plus slides. Multiplex fluorescent in situ hybridization was performed using the RNAscope Multiplex Fluorescent V2 Assay kit (ACDBio 323100), reagents and probes according to manufacturer’s instructions. RNAscope® probes were designed commercially by the manufacturer and are available from Advanced Cell Diagnostics, Inc.. The following probes were used: Dll4 (#31997), Sox10 (#435931), Notch3 (#425171). Probes were then labelled with TSA opal 520 (PerkinElmer FP1487001KT), TSA opal 570 (PerkinElmer FP1488001KT) and TSA opal 690 (PerkinElmer FP1497001KT). Samples were counterstained with DAPI for 5 min and mounted with ProLong Gold Antifade Mountant (Thermofisher Scientific, #P36930).
Microscope image acquisition
For 2D imaging, slides were acquired on the Zeiss Axio Scan.Z1 (20x objective) or in Vectra® Polaris™ Imaging System. Cryosections with spheres were imaged using Nikon C2 confocal Microscope. For 3D imaging, image acquisition was performed using Zeiss LSM 880 Airyscan. All images acquired with the Zeiss Axio Scan.Z1 or Zeiss LSM 880 Airyscan were stitched using the Zeiss ZEN 2 software. Images were analyzed using Zen 2 (blue edition) or QuPath softwares.
2D quantification of clones with respect to their distance to blood vessels
10μm thick sections were quantified by measuring the distance of the clones to the closest blood vessel using Qupath. Vessels were visualized by immunostaining sections with the endothelial markers AQP1 and PECAM1.
Image segmentation and clone reconstruction at 3D resolution
We quantified cell clusters in 3D section, which corresponded to z-stacks of (x,y)-dimensions between 3.1x3.9 and 5.4x8.4 mm2 with a resolution of 0.83 μm/pixel, and a thickness of 0.45-1.0 mm, with a z-resolution between 5-7 μm/pixel. The images were processed using a purpose-built segmentation algorithm (implemented in MATLAB R2020a, The MathWorks, Inc., Natick, Massachusetts, USA). Every image in the z-stack was transformed to grayscale (Matlab function mat2gray), and its background subtracted using a moving average filter with radius of 500 pixels. (ii) We then applied an intensity threshold to the image, retaining only those pixels that belong to the 30th percentile of intensity in that given image (threshold = 0.3). This protocol was applied on both YFP and RFP channels independently.
With the segmented image, we then proceeded to identify cell clusters. For this, we applied a density-based clustering algorithm (DBSCAN) with a strict 10μm distance threshold such that only groups of cells of a common colour tightly packed together would be considered as part of a given cluster. By measuring the spatial correlation between clusters of the same and different colours, we estimated that clusters separated by a distance of 100 μm or less (equivalent to about 6-7 cell diameters) were likely to belong to the same clone. By reclustering the data with this threshold, we were able to reconstruct three-dimensional clones, and extract their volumetric information. The clone volumes were then normalized by the typical single cell volume of about 1800μm3 (or typical diameter of 15 microns) to construct the clone size distribution as a function of the cell number. The form of the clone size distribution and qualitative features of the data were not affected by variations of the normalization volume. This protocol was applied to both YFP and RFP channels independently.
After clonal reconstruction, we identified a high fraction of singlets, i.e., clones containing a single cell. This fraction was higher than expected for a proliferative cell population and could originate either from a true subpopulation of non-dividing tumour cells or alternatively be fragments of larger clones that could not be assigned to their respective clones due to their large distance of separation. To focus our analysis on the ensemble of clones rooted in tumour cells capable of division, single-cell clones were eliminated from the clonal analysis and statistical modelling, as discussed in the Supplementary Note. The total number of clones and number of single cell clones per sample are shown in Supplementary Table 4.
Single cell suspension of murine tumours
Tumor samples and metastatic organs were isolated from the corresponding mouse melanoma mouse models (allograft or GEMMs) were rinsed in cold Dulbecco’s Phospho-Buffered Saline (DPBS). Tissues were minced into small pieces and with sterile scalpels or scissors and incubated 30 minutes in a heather-shaker at 37°C 800 rpm in digestion buffer. Tissue samples from each mouse model were enzymatically digested using Liberase (300μg/ml, Sigma Aldrich, #5401127001) and DNAseI (1 mg/ml) diluted in serum free medium (DMEM) and incubate for 30 minutes at 37°C. For primary back skin tumors in the Tyr::CreERT2;BRafV600E/+;Ptenl/l model, digestion buffer consisted of 0.8 mg/mL DNase I (Sigma-Aldrich, Cat#11284932001) and 2 ng/mL collagenase P (Sigma-Aldrich, Cat#11249002001) diluted in serum free medium (DMEM). Digestion mix was inactivated with DMEM containing 10% FBS. To remove excess of red blood cells, samples when needed were incubated for 5 min in red blood lysis buffer (Thermo Fisher, Cat#A1049201) at room temperature according to the manufacturer’s instructions. Single cells were separated from remaining tissue using 40 μm Falcon Cell Strainers (Corning) and resuspended either in FACs buffer (PBS supplied with 2% fetal bovine serum and 2 mM EDTA) or in 0.04% BSA. Then, cells were sorted using BD FACSAria™ Fusion Flow Cytometer (see below) or counted (either using Neubauer chamber or LUNA™ Automated Cell Counter) to achieve the optimal target cell number (1000 cells/μl). Then, cells were processed for 10x single cell sequencing RNA sequencing according to the manufacturer’s instructions.
Cell sorting
Tdtomato cells from Prrx1::CreER-GFP;Tyr-NRASQ61K/°;Ink4a−/−;ROSA26RLSL-tdTomato/LSL-tdTomato model and Tyr::CreERT2;BRafV600E/+;Pfenl/l tail model were isolated using a 4-laser BD FACSAria™ Fusion Flow Cytometer (BD Biosciences). Cells were sorted at low pressure (12 psi), using a large 130 μm nozzle to reduce potential cell stress, cells were kept at 4°C at all time. Non-induced mice were used as negative controls for the gating strategy. For the Prrx1::CreER-GFP;Tyr-NRASQ61K/°;Ink4a−/−;ROSA26RLSL-tdTomato/LSL-tdTomato model, to exclude a significant fraction of Cancer Associated Fibroblasts (CAFs) that express PRRX1, tumor samples were first incubated with the CAF marker Alpha-Smooth Muscle Actin Monoclonal Antibody (1A4), eFluor 660 (ACTA2, 1:500 dilution, Invitrogen, #50-9760-82) for 30min at 4 °C. Cells were then washed 3 times with PBS and resuspended in FACS sorting buffer. Cells were incubated with DAPI (4’,6-diamino-2-phenylindole, dihydrochloride, 1:1000 dilution, Thermofisher, #D3571) for 5 min at RT to sort for viable cells. Cell suspensions were kept at 4 °C during the sorts. Cells were sorted based on dead-cell exclusion (DAPI negative), morphological parameters (FSC/SSC), singlet-discrimination, exclusion of the respective markers mentioned above for each model, followed by positive selection of tdTomato-expressing cells. These were collected into 1.5ml tubes containing FBS. Sorted tdTomato+ cells were washed two times with PBS, resuspended in 0.04% BSA and counted in LUNA™ Automated Cell Counter to achieve the optimal target cell (1000 cells/μl). Cell suspensions were then processed for 10x scRNA sequencing. Data were analysed using FlowJo for MAC (V10.7.1; Becton, Dickinson & Company; 2021).
Single cell RNA library preparation of murine tumours
After single cell suspension preparation, the cells were suspended in 0.04% BSA–PBS. And loaded onto each channel of the Chromium Single Cell microfluidic chips (V2-chemistry, 10X Genomics) and individually barcoded with a 10X Chromium controller according to the manufacturer’s recommendations (10X Genomics). RNA from the barcoded cells was reverse transcribed, followed by amplification, shearing 5′ adaptor and sample index attachment. The libraries were prepared using the Chromium Single Cell Library Kit v3 (10x genomics, #1000078) quantified using a low coverage Illumina NextSeq 550 run and sequenced on an Illumina NovaSeq 6000 instrument.
Single cell RNA-seq data acquisition and pre-processing
All libraries were sequenced on Illumina NextSeq, HiSeq4000 or NovaSeq6000 until sufficient saturation was reached (73.8% on average). After quality control, the raw sequencing reads were aligned to the mouse reference genome v. mm10-3.0.0, by application of CellRanger (10X genomics) in order to obtain feature-barcode matrices.
CNV inference from scRNA-seq data
Moreover, we inferred copy number variation (CNV) based on scRNA-seq data by using a R package HoneyBader v. 0.114. The input for HoneyBader was the count matrix from the “RNA” assay of the integrated Seurat object of all cells. The reference for normal cells were the immune cells, which were identified also by AUCell using the immune gene set from Jerby-Arnon et al. The mean CNV score was calculated as below:
where, gene, cell.
Single cell RNA-seq data analysis of NRASQ61K/°;Ink4a−/− melanoma samples
Raw count matrices were analyzed using R package Seurat v. 3.1.325. The matrices were filtered by removing cell barcodes with <1000 expressed genes, >7,500 expressed genes and >10% of reads mapping to mitochondrial reads. Next, SCTransformation was applied for normalization. In order to identify doublets, DoubletFinder v. 2.0.255 was applied to NRAS_1 (CMA001) and NRAS_3 (CMA079, 080, 081, 082, 083, 084, 085, 086) .rds objects separately assuming that the doublet rate in each sample was 6.1%. Next, all the Seurat objects were merged, SCTransform was applied regressing out mitochondrial read percentage per cell. Subsequently, data integration was performed using R package Harmony v. 1.056. After data normalization and integration, cell cycle scoring was performed, data were filtered for singlets and again SCTransform was applied regressing out mitochondrial read percentage and cell cycle scores, followed by data integration of this subset as described above. The number of dimensions for Louvain clustering was chosen based on Harmony embeddings clustering, driven by a clear variation in embeddings across the cells. To identify malignant cells in the scRNA-seq dataset, three stringent filtration steps were applied. Firstly, the data was subset based on the AUCell score of a malignant gene set acquired from Tirosh et al.10, >0.11 or mean CNV score >0.13. Subsequently the cells that passed the first filtration step were filtered based on the superMEL gene set >0.16 or mean CNV score >0.13. Finally, to remove all immune cells, we filtered out cells that were expressing Ptprc (Cd45). The malignant subset (16,786 cells) was subjected to SCTransform (regressing out mitochondrial read percentage and the cell cycle scores,) and harmony integration followed by Seurat (Louvain) clustering (dims=1:15, resolution=0.2). Clustering robustness at different resolutions of the Harmony integrated space as well as the individual tumor samples was assessed by Silhouette scores57.
The marker genes of each Seurat cluster were identified using FindAllMarkers function in Seurat (Wilcoxon Rank Sum test) by comparing each cluster with the remaining ones (Supplementary Table 1).
Top 120-150 overexpressed genes (ranked by adj. p-value) per Seurat cluster were analyzed by multiple enrichment tools, such as HypeR, Reactome, KEGG, IPA and EnrichR (https://maayanlab.cloud/Enrichr/) and manual literature search (Supplementary Table 2). Top ranked genes per cluster that overlapped with functionally enriched terms were used to establish functional gene signatures (Supplementary Table 3).
Gene regulatory network analysis
SCENIC17 analysis was run with raw counts from “SCT” assay of malignant cells. SCENIC uses gene regulatory network inference, followed by a refinement step using cis-regulatory information, to generate a set of refined regulons (i.e. transcription factors and their target genes) in the scRNA-seq data. The python implementation, (pySCENIC: https://github.com/aertslab/pySCENIC, version 0.9.19), was run using a Nextflow (version 20.04.1) pipeline (version 0.19.0) (https://github.com/aertslab/SCENICprotocol), which streamlined the main steps of GRN inference and refinement with pySCENIC, as well as the quantification of cellular activity, and visualization. The Nextflow pipeline also performed a standard analysis in parallel, using highly variable genes selected on the basis of expression. Differentially activated TF regulons of each malignant cluster were identified by the Wilcoxon rank sum test against all the cells of the rest of the clusters. We inferred a “human-mouse conserved” PRRX1 regulon by intersecting the human PRRX1 target genes (≥10 of 50 occurrences) with the top120 overexpressed mouse mesenchymal-like genes, which resulted in overlapping n=38 genes.
Mapping of Neural Crest development similarities
The activity of different gene expression sets during neural crest development18 were quantified in our murine scRNAseq data set using AUCell: neural tube (pre-EMT NC, cluster12), delaminating, premigratory neural crest progenitors (cluster2) and mesenchyme (cluster3). Binary AUCell score activities were plotted on the umap plot (cluster12>0.09(on), cluster2>0.04(on), cluster3>0.05(on)).
Prediction of Ligand-Receptor interactions
To predict potential ligand-receptor interactions between TME populations and different melanoma cell states we used the R implementation of NicheNet available at Github (https://github.com/saeyslab/nichenetr). We focused on the role of endothelial cells (sender) and malignant cluster 4 (receiver) in particular and performed the following dataset and database preprocessing steps: i) preselection of only overexpressed genes in endothelial cells compared to other TME populations (LogFC>0.25); ii) ligand-receptor network generation via https://zenodo.org/record/3260758/files/lr_network.rds, conversion to mouse genes and removal of “ppi_prediction_go” and “ppi_prediction” databases while ligands had to be expressed among sender and receptors among receiver cells; iii) signalling network generation by downloading https://zenodo.org/record/3260758/files/signaling_network.rds, conversion to mouse genes, while all genes had to be expressed in receiver cells; iv) gene regulatory network generation by importing the results of the SCENIC analysis while all genes had to be expressed in receiver cells; v) to generate weighted networks we used as input: ligand-receptor, signalling and gene regulatory networks. The weight of every protein interaction pair is proportional to the number of hits among different databases and then is more likely to be a true interaction pair; vi) to generate the ligand-target matrix we used the weighted networks and ligands as input. This matrix was used for the NicheNet analysis pipeline. To validate the DLL4-NOTCH3 interaction predicted by NicheNet we run another cell-cell interaction algorithm, CellChat29. To do so, we used the Seurat object and run CellChat version 1.1.3 applying 10 % truncated mean for average gene expression per cell group and minimum twenty cells required per cell group for cell-cell communication.
Patient biopsies collection and scRNA-seq data analysis
Baseline tumour biopsies from twenty-two patients (23 samples, of which 20 had malignant cells detected in the single cells pool) with locally advanced or metastatic melanoma (stage IIIB – stage IV) from lymph nodes (n = 14), skin (n = 5) and subcutis (n = 4). Most patients (n = 14) were treatment naïve were collected. Written informed consent was obtained from all patients. Biopsies were collected from different metastatic sites: Two patients had received prior systemic treatment in adjuvant setting. One patient had previously received three cycles of neo-adjuvant chemotherapy for a metachronous non-small cell lung carcinoma (SPECIAL; UZ Leuven; #s62275). This study was approved by UZ Leuven Medical Ethical Committee.
Methods for tumour dissociation, library construction and sequencing, scRNA-seq data acquisition and analysis are previously described22. Jerby-Arnon data11 (cell annotation and count matrix) was acquired from GEO (GSE115978) and the signatures were measured as described above. For both datasets we measured the activity of each functional NRAS model derived state in all individual human cells using the functional signatures (Supplementary Table 3). Next, these signature genes were used to generate scores by AUCell17. A binary cutoff (ON/OFF) was applied based on AUCell score distributions. We discarded all cells in which more than one transcriptional program was evaluated as ON in order to only retain cells that can be assigned to a specific state with high confidence.
Single cell RNA-seq data analysis of BrafV600E/+;Ptenl/l tumour samples
Single cell RNA-seq data from BrafV600E/+;Ptenl/l tail and back skin tumour samples (n=2) were analyzed as described above. Cells with more than 500 and less than 6000 genes and less than 10 percent mitochondrial genes expressed were filtered out for further analyses. Braf/Pten model is considered having lower CNV aberrations and indeed we observed that mean CNV score was not sufficient to separate normal from tumor cells. Therefore, we selected malignant cells based on high AUCell SuperMEL score or positivity of tdTomato expression. The SuperMel signature was derived from the differential gene expression analysis of malignant vs. CAF clusters. It represents the top 14 genes upregulated (Copg2, Cd59a, Nceh1, Cdh19, Gjc3, Cers4, Sort1, Plekhb1, Pax3, Sox10, Rapgef4, Kcnn4, Akr1b7, Syngr1) in malignant clusters (adj. pval <1.0E−300) with at least 90% of malignant cells expressing these markers compared to less than 35% for CAFs. Next, we clustered the cells using same method as described for the NRAS model. Marker genes per cluster were called using standard Seurat pipeline and AUCell scores of functionally enriched top 100 marker genes (Supplementary Table 1) from Melanocytic, Neural Crest-like, pre-EMT NC stem-like, and Mesenchymal-like NRAS model states were measured across all cells.
Single cell RNA-seq data analysis from Prrx1::CreER-GFP;Tyr-NRASQ61K/°;Ink4a−/−;ROSA26RLSL-tdTomato/LSL-tdTomato
TdTomato+ sorted (primary and lung, early and late) cells were analysed as described above. Cells with more than 500 and less than 7500 genes and less than 10 percent mitochondrial genes expressed were used for further analyses. After correcting for multiplets using DoubletFinder (7.6 percent doublet rate), singlets were subjected to SCTransform (regressing out mitochondrial read percentage and the cell cycle scores) and harmony integration followed by Seurat (Louvain) clustering.
Early labeled primary tumor cells (Mitf low expressing cells, negative for Krt14&Pecam1&Lum) were subclustered and interrogated for mesenchymal, melanoma lineage marker expression and pre-EMT NC stem-like signature activities. Early and late labeled lung metastatic melanoma cells (Mitf and Mlana enriched Seurat cluster 6 resolution=0.4, dims=1:15) were subclustered and interrogated for pigmentation marker expression.
Single cell RNA-seq data analysis of NRASQ61K/°;Ink4a−/− and Bend3 cells co-culture
scRNA-seq data from NRASQ61K/°;Ink4a−/− cell line (expressing watermelon construct) as control and NRASQ61K/°;Ink4a−/− cells co-cultured with Bend3 cell line was analysed as described above. Cells with more than 2000 and less than 7000 genes and less than 7 percent of mitochondrial genes expressed were filtered out for further analyses. Moreover, for direct comparison of only malignant cells between test and control samples, endothelial cells (positive for Pecam1 expression) were also removed. Single cell data was processed as described above. However, in this analysis we did not perform harmony integration, as we only measured AUCell scores of top 100 marker genes for Melanocytic, Neural Crest-like, preEMT NC and Mesenchymal-like NRAS model states. Differential gene expression between the two samples was performed using standard Seurat pipeline.
Bulk RNA-seq upon genetic inactivation of PRRX1 in human melanoma cells
Total RNA was extracted using the NucleoSpin RNA kit (Macherey Nagel, cat. no. 740955). The RNA integrity was monitored using Bioanalyzer analysis (Agilent, RIN: 9.7–10). About 500 pg of RNA per sample was reverse-transcribed and amplified using a modified SMARTseq2 protocol (Rambow et al. 2018). Prior to generating sequencing libraries using the NexteraXT kit (Illumina, cat. no. FC-131-10), cDNA profiles were monitored using the Bioanalyzer. Sequencing was performed on a Nextseq500 platform (Illumina, SE75bp). Differential analyses between siPRRX1 and siCtrl samples were executed using the DeSeq2 pipeline.
Human melanoma cell lines
The human melanoma cultures were derived from patient biopsies by the Laboratory of Oncology and Experimental Surgery (Prof. Dr. Ghanem Ghanem, Institute Jules Bordet, Brussels, Belgium). All the cell lines were grown in 5% CO2 at 37°C in F10 supplemented with 10% FBS, 0.25% GlutaMAX and Penicilline/Streptomycin antibiotics. Cells have been tested for mycoplasma contamination.
Generation of a Watermelon-positive NRASQ61K/°;Ink4a−/− cell line
Watermelon library58 and backbone are available at Addgene #155257 and #155258. An established mouse NRASQ61K/°;Ink4a−/− cell line was transduced using the watermelon virus in DMEM medium supplemented with 10% FBS in presence of 16 μg/ml polybrene (Sigma-Aldrich). After a 24h incubation with virus, medium was changed for medium without polybrene. 48h after infection, cells were sorted for mNeon expression and expanded in culture for 3-5 passages before being aliquoted to 10 × 106 cells per vial and stored in −80 °C. Cells have been tested for mycoplasma contamination.
Lentiviral production
HEK293 FT cells were transfected with with dVPR and VSVG packaging plasmids using Lipofectamine™ 2000 reagent (ThermoFisher Scientific) according to the manufacturer’s instructions. 24h after transfection, medium was replaced with DMEM medium (ThermoFisher Scientific) supplemented with 20% FBS. Medium containing virus particles was collected 48 h after transfection. Virus particles were filtered through a 0.45 nm syringe filter and concentrated using Vivaspin 20, 50.000 MWCO 20 ml columns (Sigma-Aldrich) in a cold centrifuge at 3,000 g to a final volume of 2 ml (30-60 min). Virus was aliquoted and stored at −80 °C until use. Virus titer was estimated by measuring the fluorescence by flow cytometry.
mCherry dilution FACS experiments
Watermelon cells were thawed and expanded in DMEM medium supplemented with 10% FBS for 2-3 days before the induction with doxycycline (dox) at a concentration of 2 μg/ml (pulse). After 48 h pulse, mCherry expression were monitored using IncuCyte (Sartorius) live imaging system. Cells were then seeded into 10 cm plates either alone or together with Bend3 cell lines at a ratio of melanoma cells to endothelial cells 1:2 in DMEM medium supplemented with 10% FBS, 1% sodium pyruvate and 1% HEPES. 48 h later (chase) cells were trypsinized for 3 min to detach melanoma cells but not endothelial cells and resuspended in FACS buffer (2% FBS, 1mM EDTA, 25 mM, HEPES pH7.0 in phosphate-buffered saline). Cells were then filtered through a 40-μm strainer. mCherry expression in Watermelon cells (mNeon positive cells) was measured by FACS (SONY MA9000 or BD Fortessa X-20) and analyzed in FlowJo.
3D monoculture and co-culture
NRASQ61K/°;Ink4a−/− cell line stably expressing dsRed and Bend3 cell line stably expressing GFP were seeded at a total number of 3000 cells per well into 96-well ultra-low attachment plate (Thermo Scientific™ Nunclon™) at a different ratio in DMEM medium supplemented with 10% FBS, 1% sodium pyruvate and 1% HEPES. The growth of cells was monitored using IncuCyte Zoom (Sartorius) live imaging system for 10 days. For quantification, the size of spheres was measured in Zoom software. 3 measurements were taken for each sphere and the average of three values has been used to calculate the volume.
3D culture staining
3D spheres were collected and pooled together in the 1,5 ml Eppendorf. Spheres were then transferred into disposable base molds 7x7x5 mm to be embedded in Tissue-Tek® O.C.T. Compound (Sakura® Finetek, 4583) and stored at −80 °C. Sections of 10 μm were cut using the Thermo Scientific CryoStar NX70 Cryostat. Sections were stored short term at −20 °C. Staining was performed following the protocol described above for mouse frozen section immunofluorescence.
siRNA-Mediated Transient Genetic Inactivation
Cells were transfected with the indicated specific short interfering RNA (siRNA) molecules using Lipofectamine 2000 Transfection Reagent (Invitrogen) according to manufacturer’s protocol. siRNAs were used at a final concentration of 100 nM for PPRX1 and 80 nM for Notch3 (Dharmacon, #L-047867-01-0005).
For bulk RNA sequencing of MM047 upon siPPRX1, 2*105 cells were plated one day before transfection in a 6-well plate. Cells were collected 48 hours after transfection. RNA was extracted using the RNA NucleoSpin extraction kit (Macherey&Nagel) according to the manufacturer’s instructions. PRRX1 silencing was achieved by pooling at 1:1 ration siRNAs targeting PRRX1A and PRRX1B isoforms purchased from Dharmacon. For PRRX1A the siRNAs were: 5’-AAGAUGUUGUUUACACGAGGG-3’ (HA09071733) and 5’-CCCUCGUGUAAACAACAUCUU-3’ (HA09071734). For PRRX1B the siRNAs were: 5’-GACAGCGUCUCCGUACAGCGC-3’ (HA09071735) and 5’-GCGCUGUACGGAGACGCUGUC-3’ (HA09071736). For Notch3, a pool of 4 siRNAs at 80 nM final concentration was used: 1. 5’-CCACGUGUCUUGACCGAAU-3’, 2. 5’-GAACGUGUGUAGACGGUGU-3’, 3. 5’-GCACUUUGUGUGAGCGAAA-3’, 4. 5’-GGAAUAGGCUUUCGCUGCA-3’. Control samples were transfected with non-targeting siRNA pool (Dharmacon, #D-001810-10-20).
Matrigel invasion assay and quantification
The invasive activity of melanoma cells was determined by matrigel transwell invasion assays using Boyden chambers (0.8 mm BD BioCoat Matrigel Invasion Chambers; from Corning, 354480), according to the manufacturer’s guidelines. Briefly, MM099 cells were starved overnight with FBS and L-glutamine deprived medium. Next, 2*105 cells were plated in each invasion chamber in FBS-deprived medium, while FBS (10%) and L-glutamine enriched medium (2.5%) was used in the wells placed in the lower chamber. Uncoated inserts were used as a control for proliferation. 72 hours after seeding, membranes were stained with crystal violet and subsequently imaged.
Cells were quantified using the ImageJ plugin Colony Area to quantify the intensity weighted area percentage (intensity of staining for each pixel)59. Each sample was then normalized over the control well and statistical significance was assessed using a t-test (unpaired, two-tailed).
RT-qPCR
Cells resuspended in QIAzol using a miRNeasy Kit and processed according to the manufacturer’s instructions (QIAGEN) or in RA1 lysis buffer using the RNA NucleoSpin extraction kit (Macherey&Nagel). RNA was quantified using a NanoDrop 1000 (Thermo Scientific) and 500–2,000 ng was reverse transcribed with a High-Capacity cDNA Reverse Transcription Kit (Life Technologies). qPCRs were performed using Fast SYBR Green Master Mix (Life Technologies) and run on a Roche LightCycler-480-384. Data processing with qbase+ 2.6 software (Biogazelle) relies on normalization with a minimum of 2 reference genes. RT-qPCR primer sequences are the following: for PRRX1, forward 5’-CAGGACAATGACCAGCTGAACTC-3’ and reverse 5’-TGTGTCCGCTCAAAGACACG-3’; for ACTB, forward 5’- CTGGAACGGTGAAGGTGACA-3’ and reverse 5’-AAGGGACTTCCTGTAACAATGCA-3’; for RPL13A, forward 5’-CCTGGAGGAGAAGAGGAAAGAGA-3’ and reverse 5’-TTGAGGACCTCTGTGTATTTGTCAA-3’; for SDHA, 5’- TGGGAACAAGAGGGCATCTG-3’ and reverse 5’- CCACCACTGCATCAAATTCATG-3’.
Data analysis
Pooled data are presented as mean ± SEM unless other type of representation is indicated. Information regarding sample size, error bars and statistical analysis used is described in each figure legend. P values for statistical analysis of two experimental group or for multiple comparisons were calculated using Microsoft Excel 2016 and Graphpad Prism 9.2.0 (https://www.graphpad.com) software.
Extended Data
Extended Data Fig. 1: Cellular composition and discriminative regulons of NRAS-driven mouse melanoma.
a, UMAP illustrating cell type diversity in NRASQ61K/°;Ink4a−/− lesions (43k cells). b, Activity (AUCell score) of a melanoma specific gene set5 in NRASQ61K/°;Ink4a−/− lesions. c, Violin plot depicting mean Copy Number Variation (CNV) scores for the different cell types, including the malignant compartment, identified in NRASQ61K/°;Ink4a−/− lesions. d, Relative proportions of the melanoma transcriptional cell states highlighting a strong representation across all 5 NRASQ61K/°;Ink4a−/− tumors, illustrated as stacked bar (left panel) and as UMAP (right panel). e, Proliferative (PRO or melanocytic) and invasive (INV or mesenchymal-like) gene sets34 activity (AUCell score) in all mouse malignant cells (umap plots, left panel) or per transcriptional state (violin plots, right panel). f, Heatmap depicts changes in regulon activities for seurat clusters described in NRASQ61K/°;Ink4a−/− lesions g, Heatmap depicting average expression of selected Neural Crest Stem Cell (NCSC) and pre-migratory (pre-EMT) Neural Crest markers.
Extended Data Fig. 2: Single-cell transcriptomic landscape in BRAF-mutant mouse and human melanoma lesions.
a, UMAP visualisation of >2.600 malignant cells analysed by scRNA-seq and integrated across 2 different BRAFV600E;Ptenl/l lesions. The Seurat clusters were annotated using signature-based annotation (left panel). Mitf expression and Proliferative (PRO or Melanocytic) and invasive (INV or mesenchymal-like) gene sets35 activity (AUCell score) across all malignant cells (right panels). b, Stacked bar chart illustrating the distribution of malignant cell states (binary cut-offs) extracted from NRASQ61K/°;Ink4a−/− and projected onto scRNAseq data of drug naive human melanoma lesions. Each graph is categorized based on the main genetic driver mutations identified. WT indicates lesions that do not carry BRAF neither NRAS mutations; N/A indicates lesions for which genetic information is lacking. c, Stacked bar chart illustrating the distribution of malignant cell states (binary cut-offs) extracted from NRASQ61K/°;Ink4a−/− lesions and projected onto a publicly available scRNA-seq dataset (malignant treatment-naïve cells) of human melanoma biopsies11. The patient IDs and main genetic driver mutations are indicated. WT indicates lesions that do not carry BRAF neither NRAS mutations; N/A indicates lesions for which genetic information is lacking; met. stands for metastatic and prim. for primary lesions.
Extended Data Fig. 3: Characterization of the Confetti melanoma mouse model.
a, H&E staining of melanoma model Tyr::NRASQ61K/°;Ink4a−/−;Tyr::CreERT2/°;Rosa26-LSL-Confetti+/+. Representative image from n=3 independent tumors. b, Confocal images of labeled melanoma subclones acquired by confocal imaging upon TAM administration and immunostained for melanocytic lineage markers MITF, gp100 and MLANA (magenta) exhibiting heterogeneous expression. Images from n=4 independent tumors. c, Representative confocal images showing Confetti labelling efficiency in melanoma tumours 3 days after the administration of different doses of TAM in order to achieve the optimal dose to perform clonal analyses. Images from n=4 independent experiments. d, Representative confocal images showing Confetti labelling of melanoma cells 5 days after TAM administration (0.5 mg). Images from n=6 independent tumors. e, Violin plot depicting the proportion of labelled melanoma cells expressing each Confetti fluorophore 5 days after TAM administration (0.5 mg). Six melanoma tumours (two sectional areas of each) were analysed. Black line represents median and grey lines the 25th to 75th percentile. f, Representative confocal image at low magnification illustrating the clone size heterogeneity in tumours that have expanded 10-fold from the initial size after TAM administration (0.5 mg). Arrowheads show several single and/or oligoclones that exhibit minor expansion over time. Images from n=8 independent tumors.
Extended Data Fig. 4: Clone size distributions are consistent with the hierarchical model.
a, Cluster-cluster distance correlation indicating a high likelihood that clusters of a given colour are found near clusters of the same colour for distances below 100 μm. This threshold, indicated by a dashed line, was then used in the association of clones. Solid line and shaded area correspond to mean and ±SEM for n=5 biological repeats. b-e, Cumulative distribution (CDF) of clone sizes for YFP+ and RPF+ cells in samples 2-5 (for number of clones per sample see Supplementary Table 5) Sample 1 is presented in Fig. 2f. The dashed black lines correspond to the bi-exponential fits of the data, as predicted by the hierarchical model, for each dataset we show the R-squared (R2) and standard error of the fit (S) of the theoretical CDFs to the data. f, Sensitivity of the stem cell expansion rate, cycling rate and stem cell fraction to different choices of the stem cell symmetric division probability, r. The vertical dotted line indicates the value r=0.75 used here to estimating the model parameters (see Supplementary Table 5). g-k, Comparison of the empirical CDFs of clone sizes for each of the n=5 biological repeats and the corresponding distributions and SD obtained from performing 10.000 stochastic simulations of the two-compartment stem-progenitor cell model using the parameters in Supplementary Table 5. The p-values from two-sample Kolmogorov-Smirnov tests comparing the empirical and numerical distribution of clone sizes are shown. In f and i, the RFP channel of sample 3 is not shown as it was considered to be below the threshold of clonality. For Extended Data Fig.5 see Supplementary Note in Supplementary Information).
Extended Data Fig. 5: Hierarchical melanoma growth is independent of the adaptive immune system.
a, Schematic representation of the generated melanoma mouse model transplanted in immunocompromised mice (Foxn1nu). Schematic made with Biorender.com. b, Tumour growth kinetics (mm3) of individual tumours in immunodeficient mice (yellow), and corrected mean obtained from fitting an exponential growth to each individual sample (black curve). Dashed lines indicate tumour duplication times. Dotted red curve refers to the mean growth kinetics extracted from Fig. 2b. c, Confocal images showing Confetti labelling efficiency in melanoma tumours 3 days after the administration of different doses of TAM in order to achieve the optimal dose to perform clonal analyses. Representative images from n=3 independent tumors. d, Confocal image showing Confetti labelling of melanoma cells 5 days after TAM administration (0.5 mg) in immunodeficient background. Representative image from n=5 independent tumors. e, Violin plot depicting the proportion of labelled melanoma cells expressing each Confetti fluorophore 5 days after TAM administration (0.5 mg). Six melanoma tumours (two sectional area of each) were analysed. Black line represents median and grey lines the 25th to 75th percentile. f, Confocal images of melanoma tumours in immunodeficient background exhibiting Confetti labelling 5 days after initial induction (1.5-fold tumour increase) and at relative growth of 10-fold upon single low dose intraperitoneal Tamoxifen administration (0.5 mg). Representative images from n=3 independent tumors.
Extended Data Fig. 6: Spatially mapping melanoma heterogeneity.
a, Spatially resolved quantification of stress (hypoxia) state (prediction score) by label transfer of scRNA-seq data onto Visium spots (sample 1,2,3). b, Scatter plot shows a positive correlation (Spearman) between the stress (hypoxia) activity score and the distance to the nearest BV (Fisher exact test, two-tailed p value). c, Spatially resolved quantification of pre-EMT NC stem-like state (prediction score) by label transfer of scRNA-seq data onto Visium spots (sample 2,3). d, Violin plots and box plots showed the number of Stereo-seq detected genes (left) and Molecular Identifier (MID) (right) per bin50 (25 μm diameter). The middle horizontal line of the boxplot represents the median. e, Gene count visualization of Stereo-seq data. f, ROI colored by percentage of mitochondrial gene count (bin50). g, Scatter plot shows correlation (Spearman) of stress (hypoxia) activity (AUCell score) and distance to closest blood vessel per bin based on Stereo-seq bin50 data (Fisher exact test, two-tailed p value). h, UMAP of all segmented cells (170k) from molecular cartography (see Fig. 3f), coloured by Seurat cluster and melanoma (melanocytic state) gene signature activity (AUCell score). i, UMAP of malignant subset (109k) from molecular cartography, coloured by Seurat cluster and melanoma (melanocytic state) gene signature activity (AUCell score). j, Expression of pre-EMT NC stem-like cell markers assessed by scRNA-seq. k, Expression of pre-EMT NC stem-like cell markers assessed by Molecular Cartography. l, Expression of stress (hypoxia) cell markers as assessed by scRNA-seq (upper panels) and Molecular Cartography (lower panels). m, Horizontal histogram showing median distance (black line) to the nearest blood Vessel (BV) for all cells annotated as stress-hypoxia or expressing Sox10 and one of the indicated pre-EMT NC stem-like markers. Exact p values are indicated. n, Representative confocal image showing mRNA expression of the indicated genes, as detected by RNAscope, in blood vessel (BV) free and containing areas. Yellow arrowheads indicate the presence of cells positive for both Sox10 (red) and Notch3 (green). Endothelial Cells (Dll4+) are highlighted in orange. RBCs, Red Blood Cells. o, Quantification of m (n=6; p<0.0001). Boxes extend from the 25th to 75th percentile. The middle line represents the median. Whiskers represent min to max values. In panels m and o significance was assessed by an unpaired nonparametric, two-tailed Mann-Whitney Test (approximate p value, p<0.0001).
Extended Data Fig. 7: Mapping the melanoma perivascular niche in human primary melanomas.
a, Confocal image of immunostaining of human primary melanoma lesions for CD31, SOX10, VCAN (upper panel, 3 representative lesions from 5 tumors) or CD31, SOX10, NES (lower panel, 3 representative lesions from 7 tumors). The main driver mutations are indicated. b, Spatial distribution of Confetti clones in respect to vessels. Panel of xy images of a clone (indicated with the red arrowhead) in close proximity to the vessel at indicated z-stack positions (z1-z24). V corresponds to the vessel. c, Violin plot depicting 2D quantification of subclones categorized in two groups based on their size and their spatial distance (μm) from the BV (visualized by AQP1 staining, n=6; p<0.0001). Dashed line represents median and grey lines the 25th to 75th percentile. Significance was by an unpaired nonparametric, two-tailed Mann-Whitney Test. d. Cumulative distribution of clone sizes for clones directly in contact with the vasculature (distance < 20 m, solid lines) and clones detached from it (distance ≥ 20 m, dashed lines) for RFP (black circles) and YFP (gray squares) channels, obtained from short-term clonal labelling of (upper) 4 days and (lower) 10 days post TAM, where the vasculature was segmented after staining with CD31 staining in n=2 biological repeats for each time-point (shown separately).
Extended Data Fig. 8: Melanoma-Endothelial cell heterotypic interaction promotes growth and induction of the pre-EMT NC stem-like phenotype.
a, Schematic representation of Fig. 3i. b, Kaplan-Meier survival curves from log-rank (Mantel-Cox) test of mice described in a (p=0.00288, n is indicated in the graph). c, Schematic representation of in vitro assays using the Watermelon system. d, Representative FACS profiles from Fig. 3j and experiment described in c. Percentages of low mcherry (mNeon-positive) populations. e, NRASQ61K/°;Ink4a−/− cells cultured either alone or in the presence of Bend3 cells for 48h and processed for scRNA-seq. Left, UMAP by sample identity; middle, expression of the endothelial cell marker Pecam1; and right, panmelanoma marker Sox10. f, Heatmap showing the overall transcriptional reprogramming effect observed following exposure of NRASQ61K/°;Ink4a−/− melanoma cells to Bend3 endothelial cells. g, Percentage of melanoma cells positive for the melanocytic and Neural Crest-like signatures (binary AUCell score). h, Melanospheres of NRASQ61K/°;Ink4a−/− cells grown in the presence of GFP-labeled Bend3 ECs for 10 days and the percentage of Nestin-positive cells was correlated with the amount of Bend3/GFP-positive cells. Linear regression test was used for statistical significance (n=19). i, Predicted cell-to-cell interactions based on known ligand-receptor pairs between TME populations (ligands) and the pre-EMT NC stem-like malignant cells (receptors) in scRNA seq data from mouse tumors as inferred by NicheNet. j, Dotplot showing gene expression intensities of selected receptors over the different Seurat clusters. k, Circos plot illustrating Dll4-Notch3 predicted interaction between cell types in scRNA seq data from mouse tumors and inferred by CellChat. The edge width is proportional to the prediction score. Note the predicted interaction of pre-EMT NC stem-like state with ECs. l, UMAP highlighting the specific expression of the ligand Dll4 in ECs and the receptor Notch3 in subpopulation of malignant cells. m, Left panel shows venn diagram highlighting the overlap of the pre-EMT NC stem-like cell state (cluster 4) upregulated genes with NOTCH signalling signature (Reactome). Right panel shows venn diagram highlighting the significant overlap of the pre-EMT NC stem-like cell state (cluster 4) upregulated genes with a Notch3 activation signature31. Hypergeometric distribution test was used for statistical analysis. P values are indicated in each Venn diagram. n, Rbpj expression in the malignant cluster of scRNA data from NRASQ61K/°;Ink4a−/− lesions projected either as UMAP (left panel) or as violin plot (right panel) for each malignant transcriptional state. Schematic in a and c made with Biorender.com.
Extended Data Fig. 9: PRRX1 is a master regulator of the mesenchymal-like transcriptional program.
a, Violin plots depict conserved Prrx1 regulon activity scores (AUCell) across mouse Seurat clusters. b, Violin plots depict conserved PRRX1 regulon activity scores (AUCell) across human Seurat clusters. c, PRRX1 expression in n=375 TCGA_SKCM patients stratified as invasive (n=123), immune-related (n=116) and proliferative (n=136) phenotypes based on their gene expression profile described previously35. Box plots summarize the values per phenotypic group and show the median. Boxes extend from the 25th to 75th percentiles, whiskers and outliers were plotted according to the Tukey method. Significance was assessed by an unpaired nonparametric, two-tailed Mann-Whitney Test (approximate p value, p<0.0001). d, Heatmap generated using the TCGA database illustrating the gene expression profile of patients with PRRX1 high and low expression (20th percentile cut-off). Genes from the conserved PRRX1 regulon are co-expressed together with PRRX1. e, Left panel, PRRX1 expression (226695_at) in invasive (n=71) and proliferative (n=81) melanoma cell lines as previously described36. Error bars represent ±SEM. Significance was assessed by an unpaired nonparametric, two-tailed Mann-Whitney Test (approximate P value, p<0.0001). Right panel, qPCR analysis for PRRX1 in short term melanoma cells lines categorized as proliferative (n=11) and invasive (n=7) based on their gene expression profile). Error bars represent ±SEM. Significance was assessed by unpaired nonparametric two-tailed Student’s t-test. (p = 0.1509). f, PRRX1 expression (upper panel) and signature activity (lower panel) in short term proliferative/melanocytic melanoma cell lines (MM057, MM074 and MM087) transfected with SOX10 siRNA or non-targeting control. Baseline refers to the non-transfected melanoma cell lines. g, Left panel, matrigel-invasion assay upon silencing of PRRX1 in MM099 cells. Right panel, quantification (n=3; p=0.0011). h, Relative mRNA expression of PRRX1 upon siPRRX1 (n=3; p=0.0003). In g and h significance was assessed by unpaired two-tailed Student’s t-test.
Extended Data Fig. 10: Reprogramming of Prrx1+ melanoma cells during metastatic spreading.
a, Expression of melanocytic lineage markers in malignant FACS-sorted tdTomato+ cells isolated from a primary melanoma lesion of Met-track mice, 2 days (early labelled) post-TAM. b, Expression of Prrx1 and well-established melanoma mesenchymal-like markers in cells described in a. c, Expression of pre-EMT NC stem-like cell markers and activity (AUCell score) of the pre-EMT NC stem-like cell signature in cells described in a. d, Confocal image of a lymph node metastasis 4 weeks post-TAM. Cells positive for both GFP and tdTomato reporters (subcapsular region) as well as tdTomato+ cells expressing lower to undetectable levels of GFP. Black regions correspond to the pigmented melanoma cells. BF, Bright Field. Representative images from 5 different tumors. e, Box plots showing the percentage of tdTomato+/GFPhigh versus and tdTomato+/GFPlow cells in lymph nodes (n=4 mice). Boxes extend from the 25th to 75th percentile. The middle line represents the median. Whiskers represent min to max values. f, Confocal image of single tdTomato+ (and GFPlow) cells in the liver of a Met-Track mouse 4 weeks post-TAM. Representative image from 2 different tumors. g, Violin plots of Mitf expression and pigmentation genes in FACS-sorted tdTomato+ fraction isolated from lung metastases 2 days (early labelled) and 30-days (late labelled) post-TAM administration.
Supplementary Material
Acknowledgments
We thank Prof. G. Ghanem, LOCE-Institut J. Bordet, Université Libre de Bruxelles for providing us with the MM lines and Hans Clevers for providing us with the Confetti-reporter mouse strain. We thank Odessa Van Goethem and Veronique Benne for their assistance with the mouse experiments. P.K. received financial support from Marie Curie Individual Fellowship (H2020-MSCA-IF-2018, #841092), FWO (#1210520N) and Stichting tegen Kanker (Foundation against Cancer, #2021-028). J.P. received financial support from Marie Curie Individual Fellowship (H2020-MSCA-IF-2019, #896897). A.N. received postdoctoral from the KU Leuven (PDMT1/21/035). C.P. received PhD research fellowships from FWO (#11M3822N) and of the Boehringer Ingelheim Fonds. J.W. received a postdoctoral research fellowship from Stichting Tegen Kanker (Foundation against Cancer; 2019-100). O.M-B. was supported by 12T1217N project by FWO at the program under the Marie Skłodowska-Curie grant agreement no. 665501. D.P. and N.V.D. received PhD fellowships from the VIB PhD international program and FWO-SB 1S79619N, respectively. F.R. acknowledges support from the Alexander von Humboldt Foundation. We thank VIB Technology Watch members, Michiel Bontinck and Yu-Chun Wang, for facilitating collaborations with Resolve Biosciences and BGI research, respectively. We thank Yaara Oren and the labs of A. Regev and J. Brugge, for providing the watermelon vector. The computational resources and services used in this work were provided by the VSC (Flemish Supercomputer Centre), funded by the Research Foundation - Flanders (FWO) and the Flemish Government – department EWI. B.D.S. acknowledges funding from the Royal Society E.P. Abraham Research Professorship (RP\R1\180165), EPSRC (EP/P034616/1) and Wellcome Trust (219478/Z/19/Z). This work was supported by FWO (G0C530N and G070622N), Stichting Tegen Kanker (FAF-F/2018/1265), Neftkens foundation, Melanoma Research Alliance (MRA, EIA#623591), KULeuven (C1 grant) and the Belgian Excellence of Science (EOS) program to J-C.M.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information Supplementary information
The file contains information on the Supplementary Table legends, gating strategies for in vitro and in vivo experimental settings and Supplementary Theory explaining clonal analysis and modelling of melanoma growth.
Data availability
Raw data of scRNAseq (and output files from CellRanger) and spatial transcriptomics (Visium and Stereo-seq) are deposited to GEO with the accession number GSE207592. Processed count matrices and cell annotations (scRNAseq and Visium) are available at: https://marinelab.sites.vib.be/en. Molecular cartography data are available at Zenodo: https://zenodo.org/record/6856193#.Ytj-fnZBz-g. All other data supporting the findings of this study are available upon reasonable request. Source data are provided with this paper.
Code availability
Scripts and annotation files for the study have been deposited on github at: https://github.com/MarineLab/Karras-et-al. The raw list of clone sizes and scripts to extract the model parameters as well as running the stochastic simulations of the two-compartment model are available at https://github.com/ibordeu/scripts_Karras_et_al_2022_git. A brief description of how to reproduce the analysis workflows and the figures presented in this paper is included.
References
- 1.Rambow F, Marine JC & Goding CR Melanoma plasticity and phenotypic diversity: Therapeutic barriers and opportunities. Genes and Development Preprint at 10.1101/gad.329771.119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arozarena I & Wellbrock C Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nature Reviews Cancer Preprint at 10.1038/s41568-019-0154-4 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Gulati GS et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science (1979) (2020) doi: 10.1126/science.aax0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rambow F et al. Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell vol. 174 843–855.e19 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Tirosh I et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wouters J et al. Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nature Cell Biology 22, 986–998 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Patton EE et al. Melanoma models for the next generation of therapies. Cancer Cell vol. 39 Preprint at 10.1016/j.ccell.2021.01.011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ackermann J et al. Metastasizing melanoma formation caused by expression of activated N-RasQ61K on an INK4a-deficient background. Cancer Research 65, 4005–4011 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Serrano M et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85, 27–37 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Tirosh I et al. Dissecting the 17ulticellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science (1979) 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jerby-Arnon L et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell (2018) doi: 10.1016/j.cell.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rambow F et al. New Functional Signatures for Understanding Melanoma Biology from Tumor Cell Lineage-Specific Analysis. Cell Reports (2015) doi: 10.1016/j.celrep.2015.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sade-Feldman M et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell (2018) doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan J et al. Linking transcriptional and genetic tumor heterogeneity through allele analysis of single-cell RNA-seq data. Genome Research (2018) doi: 10.1101/gr.228080.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goding CR & Arnheiter H Mitf—the first 25 years. Genes and Development vol. 33 983–1007 Preprint at 10.1101/gad.324657.119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoek KS & Goding CR Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell and Melanoma Research vol. 23 746–759 Preprint at 10.1111/j.1755-148X.2010.00757.x (2010). [DOI] [PubMed] [Google Scholar]
- 17.Aibar S et al. SCENIC: Single-cell regulatory network inference and clustering. Nature Methods (2017) doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soldatov R et al. Spatiotemporal structure of cell fate decisions in murine neural crest. Science (1979) (2019) doi: 10.1126/science.aas9536. [DOI] [PubMed] [Google Scholar]
- 19.Kerosuo L & Bronner ME cMyc Regulates the Size of the Premigratory Neural Crest Stem Cell Pool. Cell Reports 17, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsoi J et al. Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell (2018) doi: 10.1016/j.ccell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köhler C et al. Mouse Cutaneous Melanoma Induced by Mutant Braf Arises from Expansion and Dedifferentiation of Mature Pigmented Melanocytes. Cell Stem Cell 21, 679–693.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Pozniak J et al. A TCF4/BRD4-dependent regulatory network confers cross-resistance to targeted and immune checkpoint therapy in melanoma. bioRxiv 2022.08.11.502598 (2022) doi: 10.1101/2022.08.11.502598. [DOI] [Google Scholar]
- 23.Snippert HJ et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell (2010) doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Reeves MQ, Kandyba E, Harris S, Del Rosario R & Balmain A Multicolour lineage tracing reveals clonal dynamics of squamous carcinoma evolution from initiation to metastasis. Nature Cell Biology (2018) doi: 10.1038/s41556-018-0109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen A et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792.e21 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Calabrese C et al. A Perivascular Niche for Brain Tumor Stem Cells. Cancer Cell (2007) doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 28.Browaeys R, Saelens W & Saeys Y NicheNet: modeling intercellular communication by linking ligands to target genes. Nature Methods (2020) doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 29.Jin S et al. Inference and analysis of cell-cell communication using CellChat. Nature Communications 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the … (2005) doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei K et al. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature (2020) doi: 10.1038/s41586-020-2222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takano S et al. Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes and Development 30, 233–247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ocaña OH et al. Metastatic Colonization Requires the Repression of the Epithelial-Mesenchymal Transition Inducer Prrx1. Cancer Cell 22, 709–724 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Hoek KS et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Research (2008) doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- 35.Verfaillie A et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nature Communications (2015) doi: 10.1038/ncomms7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Widmer DS et al. Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell and Melanoma Research 25, (2012). [DOI] [PubMed] [Google Scholar]
- 37.Kawanami A, Matsushita T, Chan YY & Murakami S Mice expressing GFP and CreER in osteochondro progenitor cells in the periosteum. Biochemical and Biophysical Research Communications (2009) doi: 10.1016/j.bbrc.2009.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boiko AD et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature (2010) doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roesch A et al. A Temporarily Distinct Subpopulation of Slow-Cycling Melanoma Cells Is Required for Continuous Tumor Growth. Cell (2010) doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schatton T et al. Identification of cells initiating human melanomas. Nature (2008) doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quintana E et al. Efficient tumour formation by single human melanoma cells. Nature (2008) doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stemmler MP, Eccles RL, Brabletz S & Brabletz T Non-redundant functions of EMT transcription factors. Nature Cell Biology Preprint at 10.1038/s41556-018-0196-y (2019). [DOI] [PubMed] [Google Scholar]
Additional references
- 43.Bosenberg M et al. Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis 44, 262–267 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A & Berns A Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature (2001) doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 45.Dankort D et al. BRAF V600E cooperates with PTEN silencing to elicit metastatic melanoma. Nat Genet 41, 544–552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maria Bosisio F et al. Functional heterogeneity of lymphocytic patterns in primary melanoma dissected through single-cell multiplexing. Elife (2020) doi: 10.7554/eLife.53008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Susaki EA et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell (2014) doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 48.Frankish A et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Research 47, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yates AD et al. Ensembl 2020. Nucleic Acids Research 48, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marçais G & Kingsford C A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gans JD & Wolinsky M Improved assay-dependent searching of nucleic acid sequence databases. Nucleic Acids Research 36, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez JM et al. APPRIS 2017: Principal isoforms for multiple gene sets. Nucleic Acids Research 46, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bankhead P et al. QuPath: Open source software for digital pathology image analysis. Scientific Reports 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt U, Weigert M, Broaddus C & Myers G Cell detection with star-convex polygons. in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) vol. 11071 LNCS; (2018). [Google Scholar]
- 55.McGinnis CS, Murrow LM & Gartner ZJ DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Systems (2019) doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korsunsky I et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nature Methods (2019) doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rousseeuw PJ Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics 20, 53–65 (1987). [Google Scholar]
- 58.Oren Y et al. Cycling cancer persister cells arise from lineages with distinct programs. Nature 596, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guzmán C, Bagga M, Kaur A, Westermarck J & Abankwa D ColonyArea: An ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS ONE 9, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data of scRNAseq (and output files from CellRanger) and spatial transcriptomics (Visium and Stereo-seq) are deposited to GEO with the accession number GSE207592. Processed count matrices and cell annotations (scRNAseq and Visium) are available at: https://marinelab.sites.vib.be/en. Molecular cartography data are available at Zenodo: https://zenodo.org/record/6856193#.Ytj-fnZBz-g. All other data supporting the findings of this study are available upon reasonable request. Source data are provided with this paper.
Scripts and annotation files for the study have been deposited on github at: https://github.com/MarineLab/Karras-et-al. The raw list of clone sizes and scripts to extract the model parameters as well as running the stochastic simulations of the two-compartment model are available at https://github.com/ibordeu/scripts_Karras_et_al_2022_git. A brief description of how to reproduce the analysis workflows and the figures presented in this paper is included.