Abstract
Acetylcholine plays an essential role in fundamental aspects of cognition. Studies that have mapped the activity and functional connectivity of cholinergic neurons have shown that the axons of basal forebrain cholinergic neurons innervate the pallium with far more topographical and functional organization than was historically appreciated. Together with the results of studies using new probes that allow release of acetylcholine to be detected with high spatial and temporal resolution, these findings have implicated cholinergic networks in ‘binding’ diverse behaviours that contribute to cognition. Here, we review recent findings on the developmental origins, connectivity and function of cholinergic neurons, and explore the participation of cholinergic signalling in the encoding of cognition-related behaviours.
Introduction
Acetylcholine (ACh) contributes to numerous physiological functions. Since its discovery more than 100 years ago1,2, ACh has been recognized as a key neurotransmitter in the mammalian peripheral nervous system (PNS) and central nervous system (CNS). In the periphery, ACh is the primary mediator of nerve muscle transmission3,4. In the CNS, ACh plays a more complex role, directly depolarizing or hyperpolarizing neurons and modulating the actions of a wide array of neurotransmitters to drive or inhibit synaptic activity5,6. The widespread distribution of ACh and the diversity of its receptors in the brain has led to the proposal that cholinergic signalling is highly dynamic and, by critically contributing to network coordination, mediates behavioural outcomes7,8.
In the mammalian CNS, cholinergic neurons can be divided into three major classes: motor neurons, interneurons and projection neurons. Cholinergic motor neurons are dispersed throughout the hindbrain and spinal cord and are essential for motor control9,10. Cholinergic interneurons are spread across the striatal complex and some areas of the cortex and hippocampus, depending on the species examined11–17. Cholinergic projection neurons are organized into two major pools, one located within the brainstem and a second that is broadly distributed across the base of the forebrain18–20. Cholinergic neurons in the brain participate in essential functions (including the control of respiration, sleep, attention, mood and memory)21–29 and act via two major classes of receptors: G protein-coupled muscarinic ACh receptors (mAChRs) and ionotropic nicotinic ACh receptors (nAChRs)30–35.
In the adult mammalian brain, cholinergic projection neurons of the basal forebrain (usually known as basal forebrain cholinergic neurons (BFCNs)) span the entire rostrocaudal extent of the telencephalon19,36 but loosely cluster in several broad subregions (defined on the basis of anatomical landmarks), including the medial septum, the vertical and horizontal subdivisions of the diagonal band, the ventral pallidum, the substantia innominata and nucleus basalis19,36. However, increasing evidence now points to a functional organization that extends beyond these anatomical boundaries and suggests that this, together with their subregional heterogeneity, enables them to coordinate diverse actions across the brain.
In this Review, we focus on recent advances (derived primarily from rodent studies) in our understanding of the heterogeneous development, functional organization and signalling of BFCNs. In recognition of the fundamental importance of other cholinergic neuronal populations that are not discussed here, we refer the reader to several excellent reviews that focus on striatal cholinergic interneurons and brainstem cholinergic projection neurons17,37–42. Here, we review features of basal forebrain cholinergic circuits and signalling of BFCNs that we believe contribute to their diverse functions. We summarize the factors that lead to the developmental specification of distinct populations of BFCNs (Table 1 and Fig. 1) and describe what is known about their unique anatomical organization, their specific projection patterns and their predominant inputs and outputs (Fig. 2). Finally, we review methodological advances (Box 1) that have improved our understanding of the timescales of ACh release and its role in cognitive behaviour (Fig. 3). We believe that understanding these themes, and the heterogeneity within BFCN populations across species (Box 2), will be important in understanding the role of ACh in normal cognition and in determining the factors that contribute to BFCN vulnerability in disease (Box 3).
Table 1 |.
Effects of selected genes on cholinergic neuron fate
| Gene | Gene expression in specific progenitor domainsa | Manipulation | Effects of manipulation | Major BFCN population affected | Ref. |
|---|---|---|---|---|---|
| Nkx2.1 | pMGE1 and pMGE5, pPOA1 and pPOA2, and pSe4, pSe5 and pSe6 | Loss-of-function mutant | No TRKA-expressing neurons in the brain | All cholinergic neurons | 63 |
| cKO in the septum (using Zic4–Cre mice) | Large loss of BFCNs in MS and DB and loss of septohippocampal cholinergic innervation Moderate losses in other BFCN regions Preserved innervation of the neocortex and amygdala | MS and DB | 61 | ||
| Isl1 | SVZ and MZ of pMGE, pLGE, pSe and pPOA | cKO in cholinergic neurons (using Six3–Cre mice) | Large loss of BFCNs in nBM, VP and SI Cortical cholinergic innervation lost Hippocampal cholinergic innervation largely intact | VP, SI and nBM | 59 |
| Lhx8 | pMGE4 and pMGE5, SVZ and MZ of entire pMGE and pSe | Loss-of-function mutant | Large loss of NGFR-expressing BFCNs in nBM Cholinergic markers preserved in small neurons of the VP and SI and in large neurons of the MS and DB Septohippocampal cholinergic innervation to hippocampal CA regions preserved but innervation of DG lost | nBM, MS and DB | 65 |
| KO and dominant negative mutationb | Massive loss of all CHAT-expressing and NGFR-expressing neurons | All BFCNs | 62 | ||
| Overexpression | GABAergic–cholinergic bipotential progenitors sequestered towards a cholinergic fate and cholinergic differentiation later in development promoted | All bipotential cholinergic neurons (scattered across rostrocaudal axis) | 58 | ||
| Tbr1 | Ventral pallium (bordering LGE) | Loss-of-function mutant | Loss of BFCNs in nBM, SI and hDB | hDB, SI and nBM | 47 |
| O1ig2 | pMGE1 and pMGE2 and pPOH1 | KO | ~40% reduction of CHAT-expressing cells (largely in nBM and SI) | VP, SI and nBM | 60 |
| Ldb1 | pMGE, pSe and pLGE | cKO in Nfcx2.7-lineage cells | Major loss of CHAT-expressing cells in MS and SI Some large cells preserved in vDB and nBM | MS and SI | 64 |
BFCN, basal forebrain cholinergic neuron; CHAT, choline acetyltransferase; cKO, conditional knockout; DB, diagonal band complex; DG, dentate gyrus; hDB, horizontal subdivision of the diagonal band; KO, knockout; LGE, lateral ganglionic eminence; MS, medial septum; MZ, marginal zone; nBM, nucleus basalis of Meynert; pLGE, progenitor domain from the lateral ganglionic eminence; pMGE, progenitor domain from the medial ganglionic eminence; pPOA, progenitor domain from the anterior preoptic area; pPOH, progenitor domain from the preoptic hypothalamic border region; pSe, progenitor domain from the septum; SI, substantia innominata; SVZ, subventricular zone; vDB, vertical subdivision of the diagonal band; VP, ventral pallidum.
Information on gene expression derived from data in ref.48.
HX6 function was also potentially blocked.
Fig. 1 |. A model for BFCN fate specification.
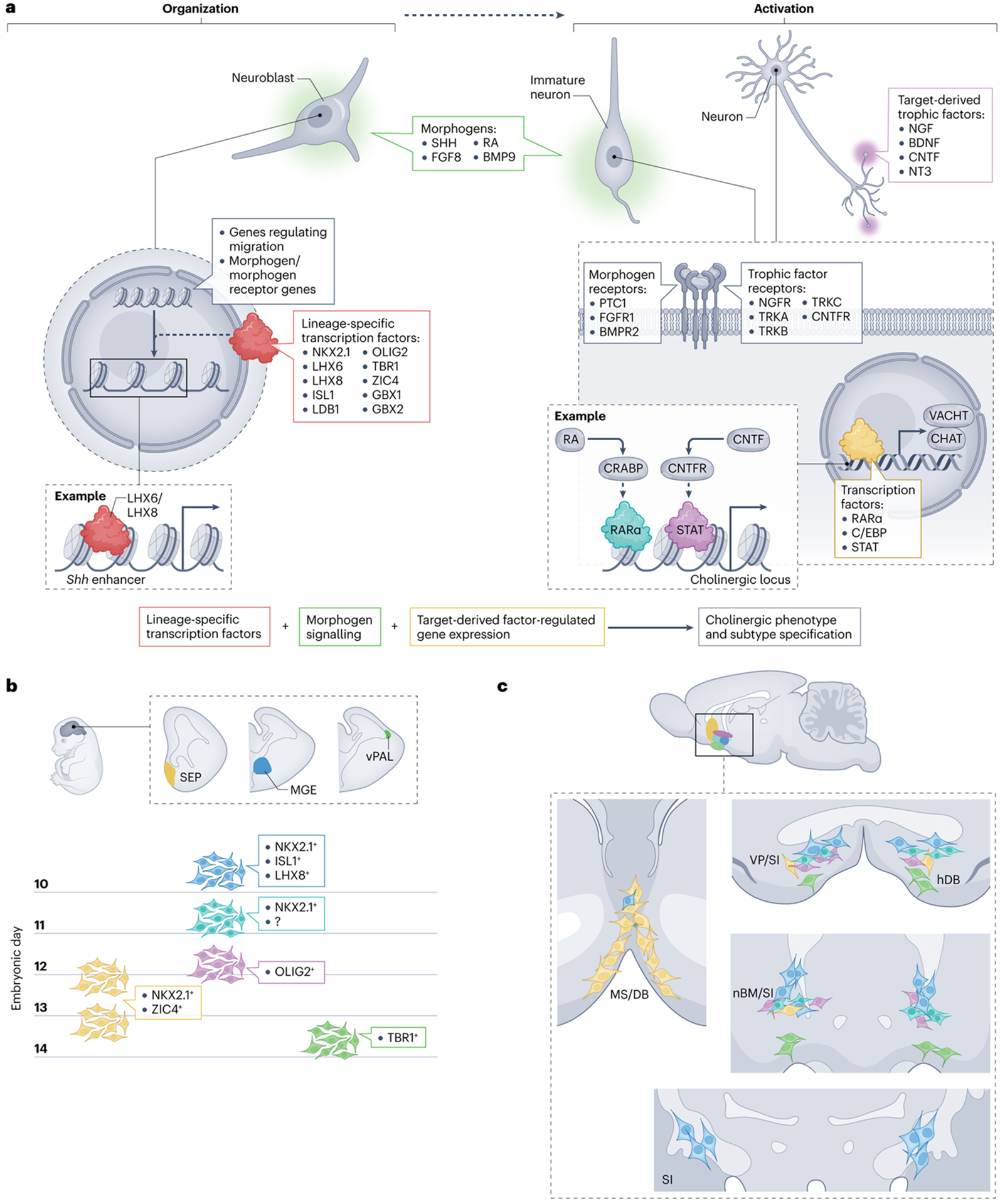
A proposed model for cholinergic neuron fate commitment based on evidence from genetic studies (Table 1), common developmental mechanisms among cholinergic cell populations and trends in developmental regulation43–111. a, The model proposes that lineage-specific transcription factors ‘organize’ the genome to regulate the expression of genes involved in the migration (radial or tangential) of immature neurons and/or neuroblasts and the expression of receptors for morphogens and trophic factors. Exposure to morphogens present in the basal forebrain or (later in development) to trophic factors present in the terminal fields ‘activates’ the genome. For example, the cholinergic gene locus has been shown to contain cis-regulatory elements that respond to retinoic acid (RA) signalling and a ciliary neurotrophic factor (CNTF) response element, STAT, that responds to CNTF signalling, meaning that treatment with either RA or CNTF increases choline acetyltransferase (CHAT) and vesicular acetylcholine transporter (VACHT) expression. b, The different germinal zones in the embryonic brain that give rise to basal forebrain cholinergic neurons (BFCNs) and our current understanding of the timing of the birth of the neurons in each of these zones and some of the contributing regulators. These regulators shown were selected on the basis of their expression in the progenitor zones and may represent master regulators. We propose that the earliest-born BFCNs (shown in blue) are generated from progenitors in the medial ganglionic eminence (MGE) and rely on the transcription factors NKX2.1, LHX8 and ISL1 for their specification, whereas subsequent generations of neurons born in this domain (shown in turquoise and purple) do not rely strongly on LHX8. The latest population to be derived from this zone is proposed to require OLIG2; however, it is also possible that this population is derived from a neighbouring germinal zone such as the anterior entopeduncular area or preoptic area. Most late-born BFCNs (shown in yellow) are generated in the septum (SEP) from NKX2.1-expressing and ZIC4-expressing progenitors and rely on NKX2.1 expression for their specification. However, one late-born population that expresses TBR1 (shown in green) migrates into the basal forebrain from the ventral pallium (vPAL). c, The proposed final location of the BFCNs derived from the progenitor populations proposed in part b. We propose that early-born MGE-derived progenitors produce the large cholinergic neurons that reside in the substantia innominata (SI), nucleus basalis of Meynert (nBM) and ventral pallidum (VP) (blue), whereas subsequently generated progenitors might produce the smaller cholinergic neurons in these regions (turquoise). The OLIG2-expressing lineage might generate the more medially located cholinergic neurons in these regions (purple). Late-born septal progenitors differentiate into large neurons that continue to populate the medial septum (MS) and diagonal band (DB) and migrate tangentially to populate the basal forebrain and striatum (yellow). Finally, the TBR1-expressing population (green) that arrives from the vPAL represents the most ventrally located neurons within the horizontal diagonal band (hDB), SI, nBM and VP. CNTFR, ciliary neurotrophic factor receptor; NGF, nerve growth factor; NT3, neurotrophin 3; SHH, sonic hedgehog protein.
Fig. 2 |. Spatial localization and projection patterns of BFCNs.
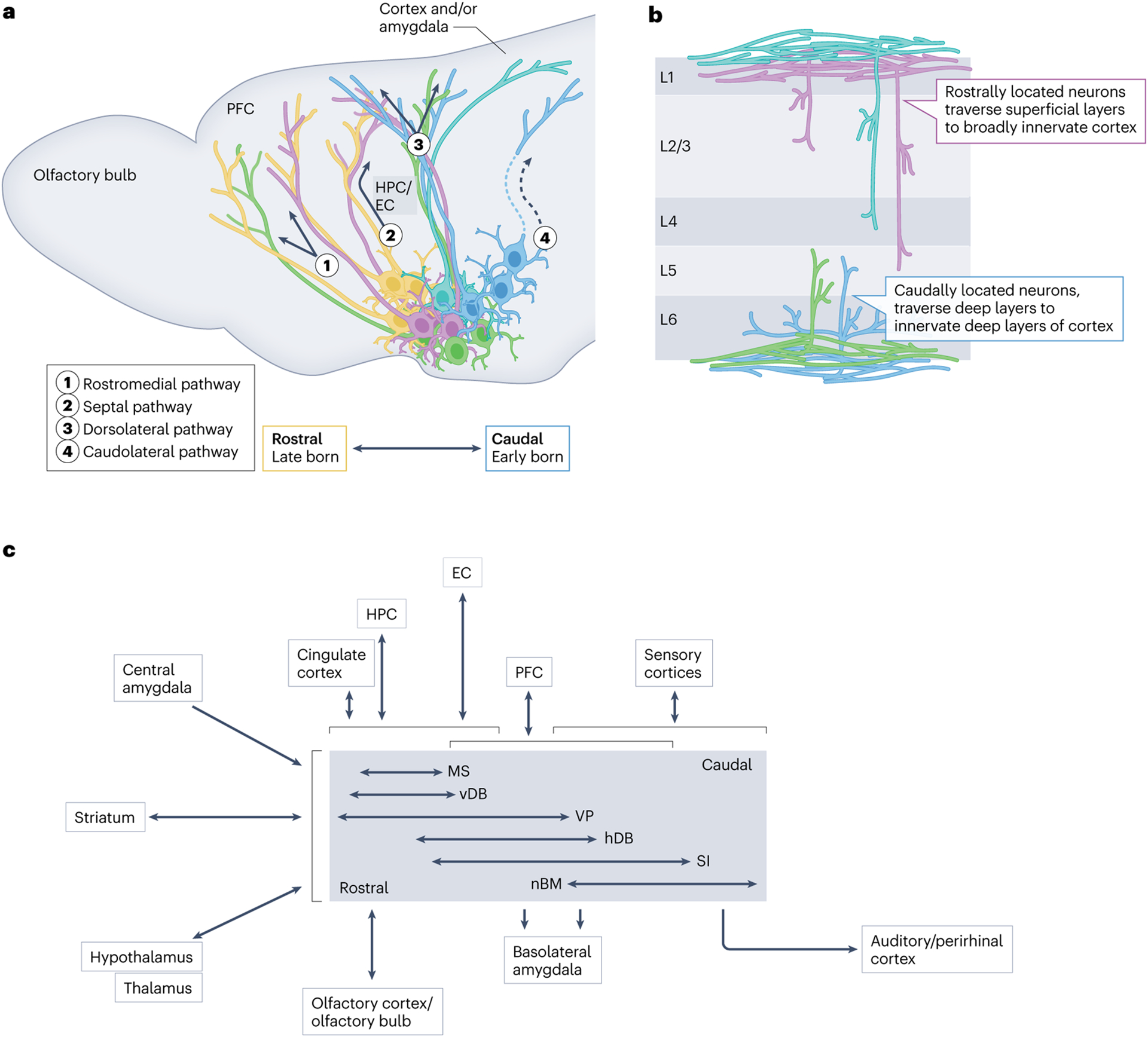
a, Overlapping pools of cholinergic neurons are located along the rostrocaudal extent of the basal forebrain in a manner that corresponds to their birthdate and site of origin162,216 (Fig. 1b,c). The axons of the basal forebrain cholinergic neurons (BFCNs) that make up these different rostrocaudal pools take four distinct projection paths to innervate their targets: the rostromedial pathway, the septal pathway, the dorsolateral pathway and the caudolateral pathway. In the schematic, arrows depict the relative trajectories of each of these fibre paths. Solid projections indicate fibres that traverse the sagittal plane, whereas those depicted with dashed lines are tracts that break from the sagittal plane and move laterally46,158. Developmentally diverse populations and their expected projection paths are shown in colours that correspond to their relative birth order (Fig. 1b,c). The major targets of each of the projections are noted. b, BFCNs innervate cortical layers distinctly on the basis of their rostrocaudal location in the basal forebrain46,158. Rostrally located, cortically projecting cholinergic neurons innervate both superficial and deeper layers of the cortex (purple and turquoise projection populations), whereas caudally located, cortically projecting cholinergic neurons primarily innervate deep layers of the cortex46,158 (blue and green). In both cases, fibre bundles traverse superficial or deep layers of the cortex as they find their targets46,158. c, An input and output wiring diagram for BFCN populations, derived from studies on the connectivity of cholinergic neurons34,64,144–176. The centre grey box organizes BFCNs beside horizontal arrows denoting their varying rostral (left) to caudal (right) extent. Regions shown to the left of the central BFCN box broadly innervate all BFCNs. Regions above and below the central BFCN box connect to specific BFCN population(s) denoted by the vertical arrows, highlighting approximate regional specificity in wiring. Brackets denote broad (rostrocaudal) overlap in connectivity across BFCN populations. The double-headed arrows denote reciprocal projections to a region, and the single-headed arrows denote unidirectional projections. EC, entorhinal cortex; hDB, horizontal subdivision of the diagonal band; HPC, hippocampus; L, layer; MS, medial septum; nBM, nucleus basalis of Meynert; PFC, prefrontal cortex, SI, substantia innominata; vDB, vertical diagonal band; VP, ventral pallidum.
Box 1. New tools for studying the cholinergic system.
Transgenic mouse models and viral tracing strategies have greatly contributed to our understanding of the cholinergic system, but they have limitations. New tools and improvements to existing methods now allow us to examine acetylcholine (ACh) release in vivo with much greater spatial and temporal resolution148,195,217 and to explore the gene-expression profiles that make cholinergic neurons unique.
ACh biosensors
A major advance in the ability to measure ACh release in vivo has been the development of genetically encoded ACh biosensors. G protein-coupled receptor activation-based (GRAB) ACh sensors couple a circularly permutated green fluorescent protein (GFP) with the M3 muscarinic receptor195,196. These sensors detect ACh release in vitro with kinetics identical to endogenous ACh receptors, a minimal signal-to-noise ratio and a length constant of 9–15 μm — all without coupling to downstream G protein-coupled receptor-mediated signalling pathways195. One of these sensors, GRABACh3.0, was used in recent studies to detect ACh release in the basolateral amygdala during a cue-induced reward learning task156 or a cued fear conditioning task157 (Fig. 3c) and in the hippocampus during exposure to stress15.
The development of an intensity-based ACh-sensing fluorescent reporter (iAChSnFR) that uses circularly permutated GFP and periplasmic binding proteins148 (similarly to previously developed sensors for glutamate218, GABA219 and glucose220) was recently reported. One proposed advantage of iAChSnFR over GRABACh3.0 is that it allows the effects of cholinergic drugs on ACh activity to be monitored, because it is not based on a mutated M3 receptor.
ACh biosensors allow the direct detection of ACh release in vivo on a millisecond timescale (rate-limited by the imaging technique rather than the sensors per se)148,195. These biosensors have the flexibility to signal ACh release via different wavelengths of fluorescence, allowing them to be used in conjunction with other fluorescent biomarkers (such as calcium sensors, other neurotransmitter sensors and voltage sensors). The newest ACh biosensors can probe the effects of ACh release on circuit activity, co-transmission of ACh with other neurotransmitters148,156,195 and, when combined with high-resolution multiphoton imaging, the detailed spatiotemporal dynamics of ACh release (Fig. 3b) in head-fixed mice or even awake, behaving mice195,221. In addition, ACh biosensors can be used to investigate behaviours associated with either transient or fast, phasic release of ACh217.
The development of new tools with greater spatiotemporal resolution always calls into question the interpretation of previous studies that had more-limited resolution. Continued, directed use of ACh biosensors in behaviourally relevant contexts with high-resolution imaging may thus lead to a reinterpretation of previous results. Finally, it is important to note that these methods are new and may have unknown limitations with regard to the interpretation of the findings. Sharing of collected datasets and in-depth discussion of region-specific and behaviour-specific use of these sensors will be essential to iron out these details.
Genomic technologies
The introduction and enhanced accessibility of single-cell genomics techniques have made it possible to assess changes in the transcriptomic signatures of individual neural precursors (and neurons) across developmental stages. Technologies such as RNAscope can provide quantitative fluorescent in situ hybridization (multiplexing signals from more than 48 distinct probes), allowing the integration of marker discovery by sequencing with the annotation of the anatomical location across time222,223. Additionally, the increased rate of development of genomic techniques has increased their detection sensitivity. Therefore, the protocols for most genomic technologies have been optimized for neuronal cells, with the capability to scale down to single-cell resolution76,224.
The mechanisms by which the chromatin landscape is regulated in developing cholinergic neurons and how that interacts with different transcription factors and the signalling triggered by morphogen exposure await detailed explanation. The development of techniques such as CUT&RUN (cleavage under targets and release using nuclease) sequencing may replace traditional crosslinked chromatin immunoprecipitation assays by providing an increased signal-to-noise ratio225. Furthermore, the introduction of ATAC-seq (assay for transposase-accessible chromatin using sequencing) for profiling chromatin accessibility at single-cell resolution226 and chromosome conformation capture techniques227 allows improved characterization of 3D chromatin structure and its relation to transcriptional regulation. These techniques together will be key to dissecting the genetic diversity of basal forebrain cholinergic neurons.
Fig. 3 |. Improvements in measurement of in vivo ACh dynamics.
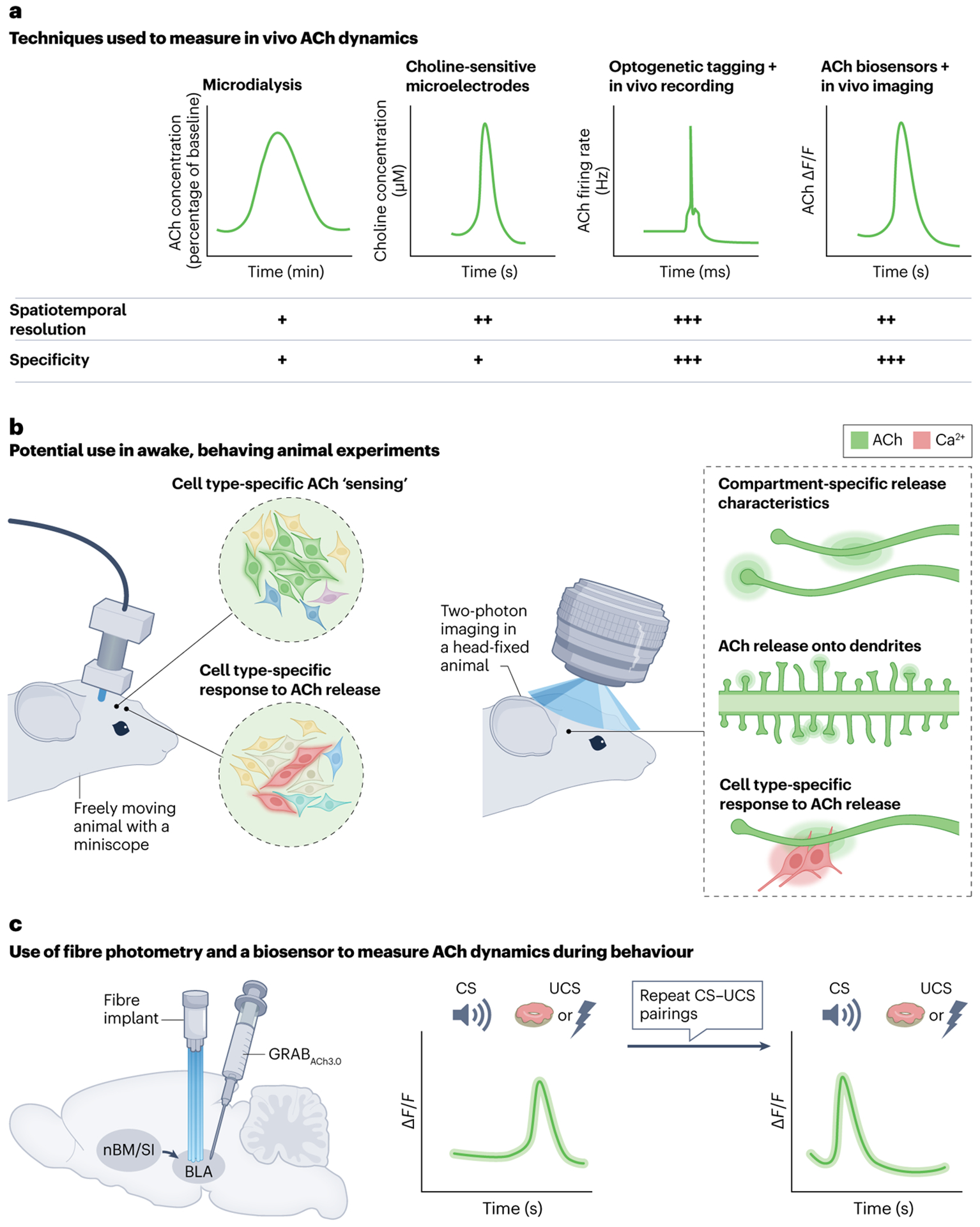
a, Different techniques used to measure acetylcholine (ACh) levels or dynamics in vivo are listed and are accompanied by schematic representations of typical measurements and a comparison of the spatiotemporal resolution and specificity of these measurements. Microdialysis can be used to measure extracellular ACh concentrations184,185. Choline-sensitive microelectrodes indirectly measure ACh levels using electrochemistry182,183. Optogenetic tagging uses Cre-dependent opsins to identify cholinergic neurons, and in vivo recordings from those neurons monitor their activity198. Finally, ACh biosensors directly measure ACh release using fibre photometry156,157,195. More recently, ACh biosensors have been used in conjunction with widefield imaging to visualize ACh dynamics across the cortex during behaviour7. For spatiotemporal resolutions, a single plus sign represents slow (minutes) measurements over a wide area (more than 250 μm), two plus signs represents measurements over seconds in a smaller area (~10–100 μm) and three plus signs represents fast (milliseconds) single-unit recordings. For specificity, a single plus sign indicates an indirect measure of ACh and three plus signs represents a direct measure of ACh. b, Schematics highlighting possible directions of future study using ACh biosensors and high-resolution or dynamic imaging techniques, which will allow us to better understand behaviourally relevant ACh dynamics. The left image depicts possible experiments in freely moving animals with a miniscope (a miniature microscope that can be head-fixed to a moving animal) that allows cell type-specific ‘sensing’ of ACh release as in fibre photometry, with the added potential for single-cell resolution. ACh measurements can be made in specific cell types (represented by green glowing neurons) (top). Alternatively, cell type-specific calcium dynamics can be measured with a calcium indicator (red) in response to ACh release (bottom). The right image shows possible experiments in head-fixed animals using two-photon imaging. This method offers the potential for deeper imaging at higher resolution to detect compartment-specific ACh dynamics. This includes measurement of ACh release by cholinergic axons (top) or ‘sensing’ of ACh release by dendrites (represented by green glowing circles, middle image). In addition, cell type-specific responses to compartment-specific ACh release can be measured. c, An example of the application of fibre photometry to measure ACh dynamics in a target brain region156,157. An ACh biosensor (GRABACh3.0) is injected into the basolateral amygdala (BLA). A cannula is implanted in the BLA to measure in vivo time-locked changes in ACh release. The traces to the right are schematized representations of the findings of two recent studies which demonstrated initial increases in ACh release in the BLA in response to an unconditioned stimulus (UCS) (including both positive valence156 and negative valence157 stimuli) but not the conditioned stimulus (CS) (left). After repeated CS–UCS presentations, however, ACh release in the BLA shifted from the UCS to the CS156,157. nBM, nucleus basalis of Meynert; SI, substantia innominata.
Box 2. Primate BFCNs.
This Review focuses largely on advances and findings from rodent research, due to the methodological diversity of the studies and their large number. However, primate studies have also contributed to our understanding of basal forebrain cholinergic neurons (BFCNs). Indeed, some of the earliest reports on BFCNs described the highly conserved organization of magnocellular cholinergic neurons throughout the basal forebrain of primates18,228, their conserved expression of NGFR and/or TRKA229–231 (with the exception of a specific basolateral amygdala-projecting population)228–230,232 and their broad innervation of the pallium18,228,231. Nevertheless, there are key differences across species that are important to consider when one is translating findings. These include a greater percentage of cholinergic neurons in the nucleus basalis, substantia innominata and medial septum228 and greater anatomical diversity within the nucleus basalis and substantia innominata233 of primates compared with other mammalian species. There are also differences in innervation density across cortical layers228,234 and in receptor expression and localization235.
Recent advances in the technologies used for transcriptomics and in vivo imaging will greatly aid our understanding of BFCNs in primates. For example, efforts to generate transgenic enhancer mouse lines to label specific neuronal populations have identified hs192 as an enhancer that is specifically active in TRKA-expressing cells that also express Gbx1, Gbx2, Isl1, Zic1 and Zic4 (ref.86). This may prove to be an important tool to identify and selectively manipulate BFCNs in primate species. Advances in in vivo imaging modalities have also improved our ability to selectively and repeatedly image the cholinergic system in primates, including humans. Early studies relied heavily on positron emission tomography ligands that targeted acetylcholine receptors236–239. Although informative, these ligands label cholinoceptive cells, not cholinergic neurons. The development of multiple positron emission tomography ligands that target vesicular acetylcholine transporter (VACHT) now allows direct assessment of the integrity of the cholinergic neuron and/or its terminal in vivo and over time240–243. Through combinatorial use of magnetic resonance imaging techniques, we can also reliably localize and delineate small BFCN somatic regions244,245. Using these techniques, we are able to answer questions about an ageing primate cholinergic system in vivo246–248. We will be able to investigate how the system changes upon onset of disease, and whether any of these changes display sexual dimorphism. Combining these metrics with behavioural assays in primates will be critical for translational interpretation across species. Use of a road map such as that provided by the cross-species investigations in the BRAIN Initiative Cell Census Network collection will surely improve our understanding of the components that are conserved, as well as those that are diverse, in BFCNs of primates and rodents.
Box 3. BFCNs and cognitive decline.
It is well established that loss of basal forebrain cholinergic neuron (BFCNs) and deterioration of cholinergic terminal fields are hallmarks of cognitive impairment249–254. Many studies have identified interactions between BFCNs (or acetylcholine receptors) and factors that may make them vulnerable to neurodegeneration, including amyloid precursor protein (APP) processing255,256, amyloid plaque accumulation257–259, tau hyperphosphorylation259–261 and the brain’s immune response262–264. What remains unclear is whether an insult to cholinergic neurons drives pathology or vice versa.
Recent positron emission tomography and magnetic resonance imaging studies have found that changes to the integrity of cholinergic terminal field synapses occur early in cognitive impairment and are a better predictor of cognitive impairment than amyloid plaque load or even brain glucose metabolism246–248. Careful examination of these changes also revealed that some projection fields are affected far sooner and to a much greater extent than others251,252. Given the tremendous heterogeneity across BFCNs in developmental origin, connectivity and function, it is possible that some subpopulations are more susceptible to pathological ageing than others.
BFCNs send extensive, highly branched projections to many cortical and subcortical regions which are metabolically demanding to maintain161. Some of the longest of these project to temporal lobe structures which are known to be vulnerable to ageing265,266. In humans, these projections are even longer than they are in smaller mammals; however, there is only a modest corresponding increase in soma size and the overall number of projections, which probably increases their energetic load267–269. It is possible that ageing contributes to a loss of cholinergic projections, which alters cholinergic tone and leads to a cascade of pathology. It is important to investigate why this occurs earlier in some regions than in others, and in some subsets of cholinergic neurons before others. Understanding the factors that contribute to the differential resilience and vulnerability of cholinergic circuits in pathological ageing260 will therefore be a critical next step towards the development and deployment of next-generation therapeutics.
Development of cholinergic neurons
During development, neuron types are specified through an interplay between different patterns of expression of homeodomain transcription factors and the signalling induced by gradients of extracellular morphogens (Fig. 1a). This process is best documented in the developing spinal cord, where different progenitor domains give rise to distinct types of neurons and glia, including cholinergic motor neurons43. BFCNs are derived from progenitors in the ventricular zone of the ventral telencephalon (also known as the subpallium), which comprises various subregions: the ganglionic eminences (with medial, caudal and lateral subdivisions known as the medial ganglionic eminence (MGE), caudal ganglionic eminence (CGE) and lateral ganglionic eminence (LGE), respectively), the anterior entopeduncular area, the preoptic area and the septum44,45. BFCNs are born predominantly in the MGE, anterior entopeduncular area, preoptic area and septum46. In addition, the ventral pallium — located between the LGE and the lateral pallium — gives rise to cholinergic neurons that will eventually reside within the subpallium47. Genetic mapping of transcription factor expression has illustrated a spatial delineation of different progenitor domains within each of these zones48 (Table 1).
Cholinergic identity is typically defined by the expression of choline acetyltransferase (CHAT; the synthetic enzyme involved in the production of ACh) and vesicular ACh transporter (VACHT; encoded by the Slc18a3 gene and responsible for packaging ACh into vesicles). The Chat and Slc18a3 genes are clustered in all animals and constitute the cholinergic gene locus49. This locus is regulated by many transcriptional regulators, including the LIM homeodomain (LHX) family of transcription factors, RE1-silencing transcription factor (REST) and the signalling induced by some extracellular cues50–57. Thus, during specification, these factors can drive or suppress the cholinergic gene locus to promote or impede cholinergic cell fate.
Specification of cholinergic neurons
The consequences of deleting some of the genes that mark specific progenitor domains in the developing telencephalon for BFCNs47,58–65 are summarized in Table 1. The related studies indicate that no single transcription factor is essential for generating all cholinergic neurons (or even all cholinergic neurons in a single basal forebrain cholinergic subregion), implying there are multiple mechanisms by which progenitors from diverse lineages commit to a cholinergic fate. A general model of cholinergic neuronal specification suggests that a combination of developmentally regulated transcription factors, interacting with the intracellular signalling pathways triggered by a cocktail of morphogens, directs the choice of multipotent progenitors towards the cholinergic fate. This type of interaction has been well documented in the generation of cholinergic motor neurons in the spinal cord, where OLIG2-expressing progenitors give rise to motor neurons, interneurons or oligodendrocytes, depending on the level of sonic hedgehog protein (SHH; a morphogen) signalling in the progenitor domain and the developmental age at which the progenitors are born66–69. On the basis of this, we propose a model for the specification of BFCNs in which the chromatin landscape of neuronal progenitors and/or immature neurons is first organized by lineage-specific transcription factors (Fig. 1a) to potentially allow the expression of cholinergic markers. Next, this gene expression programme is activated via signalling induced by morphogens and trophic factors, resulting in a stable cholinergic phenotype. According to this model, diversity within BFCNs arises from different combinations of transcription factors and signalling driven by different target-derived and/or local extracellular cues.
The data on cholinergic fate specification show that birth order and the location of birth dictate how the neurons populate regions of the basal forebrain46. Distinct transcriptional regulators (Table 1 and Fig. 1b) are associated with cholinergic neurogenesis at different rostrocaudal loci (each of which overlaps broadly with adjacent regions), generating a developmentally heterogenous basal forebrain (Figs. 1b,c,2a). Most cholinergic neurons are born between embryonic day 11 (E11) and E16 in mice: the early-born cholinergic neurons are caudally located (in the nucleus basalis and/or substantia innominata), whereas later-born neurons are rostrally located (in the medial septum and/or vertical subdivision of the diagonal band)70. Neurons in the caudal cholinergic regions and in posterior aspects of the ventral pallidum rely on transcriptional regulation by a hexameric complex formed by the transcription factors LHX8 (also known as LHX7 and L3) and ISL1 to achieve cholinergic fate71. These caudal cholinergic neuron clusters undergo radial migration from the ventricular zone to their final location, a process that is also regulated by LHX family members72. Meanwhile, progenitors expressing both ZIC4 and NKX2.1 generate cholinergic neurons in the vertical subdivision of the diagonal band, medial septum and anterior horizontal subdivision of the diagonal band61 from a locally generated pool of progenitors within the developing septum (without the need for extensive radial migration)61,73. A subset of cholinergic neurons within the ventral pallidum, anterior substantia innominata and nucleus basalis rely on OLIG2 for their specification60 and may arise from a pool of progenitors that appear transiently at around E12.5 (ref.60) and/or from a TBR1-expressing, NKX2.1-negative population that migrates to the substantia innominata and nucleus basalis from the ventral pallium47. After their initial birth and migration, exposure of the neurons to local trophic factors and/or to retrograde signals in their terminal fields may further stabilize cholinergic fate.
Transcriptional factors mediating initial organization.
A key protein that may drive the initial organization step of cholinergic specification across the basal forebrain is the homeobox transcription factor NKX2.1. Loss of NKX2.1 function results in a complete loss of TRKA-expressing neurons (including virtually all cholinergic neurons) in the basal forebrain and the striatum63. However, as the loss of NKX2.1 function also converts the MGE into an LGE-like structure63, the loss of cholinergic neurons derived from this region could be a consequence of both a loss of NKX2.1 and a loss of a variety of other transcriptional regulators in the MGE that regulate cholinergic fate (Table 1).
A subset of cholinergic neurons derive from bipotential progenitors that can take on either a GABAergic fate or a cholinergic fate, depending on expression of specific LHX family transcription factors58,74. Expression of LHX8 directs these progenitors towards a cholinergic fate71 (Fig. 1). A ubiquitous role for LHX8 in cholinergic specification is doubtful though, because of the lack of sensitivity of some cholinergic neurons to disruptions in LHX8 function62,65.
As noted earlier herein, LHX8 interacts with ISL1 to regulate genes encoding cholinergic markers71. Loss of the function of either transcription factor in mice primarily affects large, cortically projecting cholinergic neurons within the nucleus basalis and substantia innominata, along with a subset of cholinergic neurons within the medial septum and diagonal band complex that project to the dentate gyrus59,65. Lhx8-knockout mice have no cholinergic innervation of the hippocampus and cortex and are missing neuronal expression of NGFR (normally expressed by nearly all cortical- and hippocampal-projecting cholinergic neurons)62,75. It is important to note that these results were seen in an Lhx8-knockout mouse that also expresses a dominant negative form of LHX8 to prevent compensation by endogenous LHX6. Recent single-cell RNA sequencing analyses show that a subset of cholinergic neurons in the basal ganglia co-express Lhx6 and Lhx8 (ref.76). Thus, it is possible that NGFR-expressing BFCNs rely on both LHX6 and LHX8 to directly regulate their migration and cholinergic fate77,78. Finally, it has been discovered that different subregions of the MGE express different relative levels of LHX8 and LHX6. For example, most BFCNs derive from progenitors that arise at the junction of the developing septum and the rostomedioventral MGE, an Lhx8-expressing region in which Lhx6 is not expressed79. However, the directly overlying section of the MGE expresses both Lhx6 and Lhx8. Thus, BFCNs generated from distinct subregions of the MGE may rely on different LHX family transcription factors for specification, further diversifying BFCN subtypes48.
LDB1 (a co-transcriptional regulator that interacts with LHX6 and LHX8) is highly expressed in BFCNs64. Loss of Ldb1 results in loss of BFCNs throughout the basal forebrain, with the large cholinergic neurons of the medial septum (those that are spared by Lhx8 loss-of-function mutations) most notably affected74. Interestingly, the large cholinergic neurons of the nucleus basalis and substantia innominata that rely heavily on LHX8 function were spared in Ldb1-knockout mice. LHX, LIM-only (LMO) and LDB proteins interact competitively to form protein complexes with different stabilities, gene targeting abilities and transcriptional activation abilities80. In the developing spinal cord, the competition between LDB1 and ISL1 for binding to LHX3 regulates the specification of cholinergic motor neurons81. Thus, it is likely that similar competitive interactions between LDB1 and LHX or ISL transcription factors may determine subregion-specific BFCN fate80,81.
The transcription factor genes Gbx1 and Gbx2 are differentially expressed in cholinergic neuron populations82. In the developing basal ganglia of mice, the expression of Gbx1 versus Gbx2 dictates the type of migration that cholinergic neurons undergo (radial migration or tangential migration)83. Most TRKA-expressing BFCNs express Gbx1; however, a subset of laterally situated ventral pallidum and substantia innominata cholinergic neurons, along with striatal cholinergic interneurons, selectively express Gbx2 (refs.84–86). Although the phenotype resulting from Gbx1 deletion has not yet been tested in BFCNs, Gbx1 deletion in the spinal cord results in loss of cholinergic motor neurons87, further highlighting its importance in cholinergic neuron fate.
Olig2 is expressed by progenitors and neurons in the ventricular zone of the MGE and anterior entopeduncular area, a small fraction of which co-express Lhx8 (ref.60). As these neurons migrate away from the ventricular zone, Olig2 expression decreases and Lhx8 expression increases60. The Olig2 lineage in the ventral telencephalon of mice is born around E12.5 and probably includes cholinergic neurons fated to populate the posterior ventral pallidum, substantia innominata and anterior nucleus basalis60. By E18.5, nearly half of the Olig2 lineage-derived neurons in these regions also express Lhx8 (ref.60). Olig2-knockout mice have normal embryonic patterns of Nkx2.1 (also known as Nkx2-1) and Lhx8 expression, but show an approximately 40% reduction in the number of cholinergic neurons in the ventral pallidum and the substantia innominata60. Further investigation is needed to understand the mechanisms that commit this population to a cholinergic fate.
As noted earlier herein, although most BFCNs are generated in the embryonic subpallium, the Tbr1-expressing population that does not express Nkx2.1 (ref.47) migrates into the basal forebrain at around E14.5 from the ventral pallium. These neurons are attracted by FGF8, secreted from the anterior neural ridge, and predominantly populate ventral aspects of the horizontal subdivision of the diagonal band and substantia innominata, as well as contributing some cells to the nucleus basalis47. This population is probably the cholinergic neurons that are spared in Lhx8-knockout mice and/or Isl1-knockout mice. These neurons are, however, noticeably absent from Nkx2.1-knockout mice, despite not being derived from the MGE63, possibly as a result of the loss of the morphogens that attract them into the ventral forebrain. It is important to mention that no transcription factor discussed here is uniquely expressed in BFCNs. These factors are involved in specifying various neuronal subtypes across a variety of brain regions. Instead, we suggest that it is the coordinated interaction between morphogen signalling and specific complements of transcription factors that specifies unique cholinergic identities.
Activation: beyond transcription factors.
BFCNs develop near sources of morphogens, including SHH (secreted from the floorplate), FGF8 (secreted from the anterior neural ridge), bone morphogenetic proteins (secreted from the roof plate) and retinoic acid (secreted from the anterior surface ectoderm) and express morphogen receptors for each of these factors57,88–90. Several studies have demonstrated that addition of these factors to neuronal or stem cell cultures promotes cholinergic differentiation79,91–93. Additionally, the transcriptional regulators associated with cholinergic fate specification (see earlier) regulate expression of both morphogens and morphogen receptors77,87.
Trophic factors, such as nerve growth factor (NGF), act synergistically with morphogens to regulate cholinergic marker expression55. SHH and other morphogens also regulate the expression of neurotrophin receptors88,94–97. BFCNs express all the neurotrophin receptors: NGFR, TRKA, TRKB, TRKC and ciliary neurotrophic factor (CNTF) receptor (CNTFR)98,99. Neurons in their target fields express the corresponding ligands, with subsets of cortical interneurons supplying NGF and various classes of excitatory neurons supplying BDNF, CNTF and neurotrophin 3 (NT3)76,100,101. It is possible that BFCNs innervating distinct targets transmit retrograde signals that are dependent on different combinations of neurotrophin and morphogen exposure to drive cholinergic differentiation57,102. Most cholinergic markers are detected postnatally, consistent with a requirement for retrograde signalling from terminal fields to stabilize their fate and phenotype. In spinal motor neurons, it is known that distinct combinations of LHX family transcription factors direct projections to distinct muscle groups and that target-specific cues subsequently support differentiation into distinct subtypes103. We propose a similar sequence of events for BFCNs, in which target-derived trophic factors support differentiation of diverse BFCN populations.
Finally, it is important to mention that although the VACHT coding sequence is located within intron 1 of the Chat gene, the expression of each of these genes shows distinct developmental regulation104. Although transcribed from a common promotor, the genes have additional individual start sites with differential capabilities to respond to signalling factors that105,106 can either promote or diversify expression from the cholinergic gene locus107. Furthermore, although both CHAT and VACHT are critical determinants of cholinergic identity, cholinergic fate is decided long before their expression is detectable. Whether and (if so) how CHAT and VACHT contribute to BFCN developmental diversity remains to be determined.
Developmental influences on diversity
It is not yet known what specific combinations of factors mediate the developmental diversity of BFCNs. Recent single-cell or single-nucleus RNA sequencing and ATAC-seq (assay for transposase-accessible chromatin using sequencing) analyses of the ganglionic eminences show that there are multiple types of progenitors within the Nkx2.1-expressing and Lhx8-expressing lineages108–110. Similarly, single-cell transcriptomics analyses from subpallial enhancer-driven transgenic mouse lines suggest that cholinergic neurons might arise from various sets of precursors86,111. However, other data are consistent with cholinergic neurons evolving from a uniform progenitor state, with postmitotic divergence in expression of transcriptional regulators13. The availability of new techniques (and further examination of these sequencing datasets) will allow us to ask how and when the cholinergic gene locus (as well as other markers of BFCNs, such as the high-affinity choline transporter CHT, TRKA and NGFR) become accessible and which transcriptional regulators access the locus at different times in development.
The physiology of cholinergic neurons
Critical to understanding how a neuron functions is an understanding of the molecular and cellular processes that underlie its activity. Here we briefly discuss the molecular features that further diversify BFCN signalling, the time course over which ACh signalling occurs and additional features that define the actions of ACh in the brain.
Co-transmission and co-release
Many neurons can synthesize, store and release more than one transmitter112,113; cholinergic neurons are no exception. Outside the basal forebrain, cholinergic neurons demonstrate the capacity for both co-release and co-transmission112,114. For example, cholinergic neurons in the medial habenula115,116 and striatal cholinergic interneurons co-release ACh and glutamate11,12,117,118, whereas starburst amacrine cells in the retina and cortical cholinergic interneurons both co-transmit ACh and GABA14,16,119–124.
BFCNs are likely also capable of co-release and/or co-transmission, although evidence for this is limited. Markers of BFCNs (CHAT and/or VACHT) colocalize with vesicular transporters for glutamate (VGLUT1, VGLUT2 and VGLUT3)125–127, GABA (VGAT)128,129 and, to a lesser degree, monoamines (VMAT2)130 and zinc (ZNT3)131. The co-expression of different combinations of neurotransmitter markers in BFCNs is also evident from immunohistochemistry125–127, in situ hybridization126 and more recently (and in a broader dataset) immunogold electron microscopy131 and single-cell transcriptomics studies130,132. Whether this co-expression consistently corresponds to co-transmission or co-release of these neurotransmitters is less clear. Nevertheless, the diverse patterns of neurotransmitter co-expression throughout the basal forebrain further highlight the heterogeneity of BFCNs and may be useful in distinguishing between different projection neuron populations126,127,133,134.
A subset of BFCNs that project to the cortex co-express VACHT and VGAT128,129. Deletion of the VGAT gene specifically within these cholinergic neurons does not affect stimulus-evoked excitatory postsynaptic currents, but abolishes the ability of their terminals to elicit inhibitory postsynaptic currents in the neurons to which they project129. Whether the release of ACh and GABA occurs from the same or distinct presynaptic vesicles and/or by shared mechanisms in these neurons remains unclear.
Another subset of BFCNs that project to the hippocampus demonstrate co-transmission of ACh and GABA134. At the terminals of these neurons, ACh and GABA are stored in distinct presynaptic vesicles, and the release of each is dependent on different calcium channels (and probably different stimuli)134. Likewise, cholinergic projections from the basal forebrain to the entorhinal cortex133 and the olfactory bulb126 exhibit co-transmission of ACh and GABA. However, whether co-release of ACh with other neurotransmitters occurs remains to be determined.
The functional significance of co-transmission of different neurotransmitters by different populations of BFCNs is unknown. One possible functional advantage of co-transmitting populations is better control over net excitation and/or inhibition in target brain regions. For example, co-transmission of ACh and GABA from septal cholinergic fibres decreases oscillatory patterns of activity in the hippocampus134. In contrast, co-transmission of ACh and glutamate might potentially enhance excitation, strengthening the effects of efferent cholinergic projections. Further studies are required to determine the functional role of cholinergic co-transmission and its relevance in behaviour. A significant advance towards answering these questions may be provided by the combinatorial use of in vivo fluorescent biosensors to measure the release of distinct neurotransmitters simultaneously during behavioural assays (Box 1).
Timescales of cholinergic transmission
Current technology has yet to allow visualization of ACh release sites and ACh receptor-studded membranes in the CNS. However, an extensive literature documents the various cholinergic receptor subtypes, their mechanisms of action and how they transduce signals from BFCNs30,31,35,135,136. ACh acts via two classes of receptor: nAChRs and mAChRs30,31,137. nAChRs are pentameric, non-selective cation channels, comprising a combination of subunits (α2–α10 and β2–β4) ordered around a single pore. Several combinations of subunits exist, but the most common subtypes in the brain are α7* (where the asterisk denotes receptors that contain the specific subunit) and α4β2* (ref.138). mAChRs signal via activation of G proteins that affect the kinetics and opening or closing of cation channels. Within this class, some receptor subtypes are Gq-coupled, whereas others are Gi/o-coupled. In the CNS, these ACh receptors are targeted to multiple subcellular compartments including axons, dendrites and somas139,140.
As a neuromodulator, ACh utilizes its diverse receptor subtypes to either increase or decrease synaptic release probability. This signalling depends on several factors, including the receptor subtypes present on the target neuron, the downstream signalling cascades with which they interact, the cellular (subcompartment, presynaptic or postsynaptic) localization of those receptors, the cell types containing those receptors, the target regions in the brain, and the network activity and consequent brain state5,141–143. With a wide array of cholinergic receptors and broad connectivity (see later), it is clear that the cholinergic system has the capacity to coordinate diverse functions across the brain. Given the extensive heterogeneity of nAChR-expressing and/or mAChR-expressing targets, as well as the time course, duration and impact of ACh release, this neurotransmitter elicits a diverse range of actions throughout the brain.
Cholinergic signalling occurs at both point-to-point synapses144 and en passant release sites along cholinergic axons141,145–147, although the morphology of these synapses and release sites has been difficult to define. The development of genetically encoded ACh receptors (Box 1) allows ACh release to be imaged with greatly increased spatial and temporal resolution. It is now clear that ACh signalling occurs across a broad range of spatiotemporal timescales that depend on the stimulus and the time course of receptor activation and consequent downstream signalling. Indeed, this is supported by several recent studies that used fluorescent cholinergic biosensors to demonstrate both tonic signalling and phasic signalling by cholinergic neurons144,148,149. Other studies have identified at least two distinct types of cholinergic neurons within the basal forebrain that differ in both their firing pattern and their connectivity150–152. Additional improvements in technologies for imaging ACh release and ACh receptor engagement are needed to understand the functional implications of these different timescales and how they support cholinergic neurons to coordinate cellular and network activity.
Cholinergic neuron connectivity
BFCNs form a broad network of projections across many neocortical and allocortical domains. These highly organized projections allow ACh to play a central role in many behaviours142,153–157 (see later). One of the major challenges to understanding how the basal forebrain cholinergic system works (and its deterioration in disease) (Box 3) has been the anatomical complexity of the basal forebrain. The basal forebrain is a large structure comprising a variety of cell types, within which cholinergic neurons are a minority. These cholinergic neurons are organized into distinct subclusters that can be segregated by birthdate, are specified by different factors following birth and are organized according to their projection targets and function46,158,159 (Figs. 1,2).
Outputs of BFCNs
Cholinergic projection neurons of the basal forebrain are organized into overlapping ‘pools’ of neurons that share common sets of projection targets159 (Fig. 2a). Each projection target has a primary source of cholinergic input; however, cholinergic neurons with somas immediately adjacent, anterior and/or posterior to that primary source region provide residual input. Thus, conventional, anatomically defined regions project to multiple, sometimes overlapping, target regions that are linked by a common function. One excellent example of this is the prefrontal cortex (PFC), which receives cholinergic input from much of the rostrocaudal extent of the basal forebrain to its many functional subregions, rather than having one source region of input158,160.
BFCNs innervate their targets with long, highly branched, elaborate axonal arbors, with individual axons in the mouse being up to 30 cm long (and in humans up to 100 m long)161. These projection neurons innervate their target fields by traversing one of four distinct pathways46,158. In the rostromedial pathway, axons project medially, rostral to the septum and genu of the corpus callosum and then dorsally towards the olfactory bulb or PFC, entering the superficial layers of these regions. Axons of the septal pathway project medially to enter the fornix and then project caudally towards the hippocampus or enter the corpus callosum to travel dorsally through deep layers of the cortex before terminating in superficial cortical layers. In the dorsolateral pathway, axons project laterally and then dorsally: some of these axons join the stria terminalis, whereas others traverse the internal capsule, to broadly innervate the cortex. Finally, in the caudolateral pathway, fibres traverse the substantia innominata laterally and then turn dorsally towards the cortex (Fig. 2a).
Birthdate largely predicts the path that the axon of a cholinergic neuron takes to reach its target46. Early-born (E10 to E11) cholinergic neurons project via the caudolateral pathway and innervate deep cortical layers46, whereas later-born (E11 to E12) cholinergic neurons take the rostrolateral and septal pathways to reach their targets46. The latest-born (E12 to E13 or later) cholinergic neurons take the septal or rostromedial pathway to reach their targets46. Generally, rostral clusters of cholinergic neurons take rostral pathways and caudal clusters of cholinergic neurons take caudal pathways46. This projection distribution matches the ‘back to front’ birth order of cholinergic neurons, such that cells take the closest projection path to innervate their final targets.
The cholinergic neurons that ultimately innervate the cortex follow a rostromedial, caudolateral projection pattern: that is, rostrally located cholinergic neurons project to medial neocortical regions, whereas caudally located cholinergic neurons project to lateral neocortical regions158,162–164. Cortically projecting cholinergic neurons also display laminar selectivity46 (Fig. 2b). Early-born, caudally located cholinergic neurons innervate deep layers, whereas later-born, rostrally located cholinergic neurons are more variable in their distribution46 (Fig. 2b).
In sensory cortices, cholinergic projections do not appear to be organized on the basis of primary sensory modality. For example, in the visual cortex cholinergic projections are not retinotopically organized165. Similarly, the findings reported in a recent preprint indicate that cholinergic projections are not tonotopically organized in the auditory cortex166.
BFCNs also send extensive projections to both the amygdala and the hippocampal formation. Cholinergic neurons extending through the ventral pallidum, substantia innominata and nucleus basalis send dense projections to the basolateral amygdala (BLA) as well as some input to the centromedial amygdala155,167,168. The lateral and centrolateral nuclei of the amygdala lack basal forebrain cholinergic input. Rostrally located cholinergic neurons of the medial septum and diagonal band complex project to the hippocampal formation, where they broadly innervate all CA subfields (with the densest innervation observed in CA3)169,170. Cholinergic neurons of the medial septum and diagonal band complex also project to anatomically and functionally connected regions adjacent to the hippocampus, such as the entorhinal cortices and the subiculum36,170–172.
Inputs to BFCNs
Identification of the regions and cell types that control the activity of BFCNs has lagged behind that of its projections. However, studies of two types have begun to address this in rodents: rabies virus-based mapping studies, which provide information on anatomical inputs to subsets of BFCNs, and region-specific studies, which offer information about the functional organization of the system.
Rabies virus-based mapping has revealed that BFCNs receive input from the striatum (both dorsal and ventral components, as well as the nucleus accumbens), the hypothalamus (including the lateral hypothalamus and the subthalamic nucleus) and the amygdala (primarily the central nucleus)159,173,174. Additional input regions identified include the hippocampal formation, subregions of the frontal cortex, thalamic subdivisions and regions within the basal forebrain itself159,173,174. One study targeted not only inputs to cholinergic neurons of the basal forebrain, but also inputs to three additional basal forebrain cell types: glutamatergic neurons, parvalbumin-expressing GABAergic neurons and somatostatin-expressing GABAergic neurons173. Interestingly, that study found that the pattern of anatomical input deriving from the striatum, hypothalamus and amygdala was also conserved across these basal forebrain cell types173.
A recent study159 mapped the afferents of cholinergic neurons that specifically project to the motor cortex, the orbitofrontal cortex and/or medial PFC (mPFC) and the amygdala159, showing that cholinergic input–output partners are functionally organized. Specifically, amygdala-projecting cholinergic neurons receive input from subregions of the amygdala (such as the central amygdala)159, whereas motor cortex-projecting and orbitofrontal cortex-projecting cholinergic neurons receive input directly from other neocortical areas159 and mPFC-projecting cholinergic neurons receive input directly from allocortical regions (with sparse input from the neocortex)159. In addition, each of these regions receives some input from dorsal and ventral regions of the striatal complex. Importantly, the locations of the cholinergic neurons that coordinate these distinct input–output streams are intermingled, rather than clustered by input or output partner, further supporting the notion that cholinergic neurons are organized in overlapping, functional pools across the rostrocaudal axis159.
Similar virus-based mapping studies have shown that sensory cortex-projecting and accessory sensory cortex-projecting cholinergic neurons receive different inputs that are consistent with the functional demands of their terminal fields. That is, caudally located cholinergic neurons that project to the primary auditory cortex receive input mainly from the globus pallidus, striatum and thalamus175. In contrast, accessory auditory cortex-projecting cholinergic neurons receive input from the same major regions (globus pallidus, striatum and thalamus), but also from other sensory cortices, the hypothalamus, the substantia nigra and the central amygdala175. Since accessory sensory cortices integrate diverse types of sensory information, it is likely that accessory auditory cortex-projecting cholinergic neurons need to receive more diverse input than the primary auditory cortex-projecting cholinergic neurons. This is consistent with cholinergic neuron connectivity being carefully modelled around the functional needs of the target region.
Studies have shown that medially located BFCNs of the horizontal subdivision of the diagonal band and substantia innominata that are functionally engaged in olfactory discrimination receive strong input from all major olfactory processing regions151,174. By contrast, rostral cholinergic neurons of the medial septum and diagonal band complex receive input primarily from the hippocampus, whereas caudally located cholinergic neurons of the nucleus basalis and posterior substantia innominata received input primarily from the central amygdala151. Both of these populations are not significantly engaged during olfactory discrimination tasks. This is again consistent with the notion that cholinergic connectivity is functionally organized.
Additional considerations
One striking observation that can be made when evaluating data on the inputs and outputs of cholinergic neurons is their extensive projection profile161 and the high level of interconnectivity of BFCNs with neurons in their projection fields151,159,174. This organization may make these neurons ideally situated to a role in monitoring ongoing brain activity, thereby coordinating complex functions.
Retrograde-tracing studies in rodents have found that individual cholinergic neurons typically send collateral projections to regions that are physically distant but functionally related (such as projections to the primary somatosensory cortex and the primary motor cortex that represent common parts of the body)8. Sixty per cent of PFC-projecting cholinergic neurons innervate more than one PFC subregion, and around 20% innervate at least three subregions160. Thus, the level of organization within the basal forebrain cholinergic system extends beyond the location of the soma or projection paths to the individual branches of a single cell (in an already highly branched axonal arbor)159,160, and individual axon branches may even be functionally distinguishable units. This multilevel organization situates the cholinergic system in an ideal position to coordinate cognitive processes in functionally connected regions.
Entorhinal, cingulate and orbital cortices receive cholinergic input from rostral cholinergic neurons, whereas other cortical areas are primarily innervated by more caudal BFCN regions176, consistent with BFCN cortical projections being parsed between neocortical and allocortical domains. In addition, distant clusters of cholinergic neurons innervate target fields that are anatomically adjacent (such as the entorhinal and perirhinal cortices), whereas adjacent cholinergic neurons send projections to distant non-overlapping target fields8,162. Thus, soma location has only a limited relationship with target area, further underscoring the pattern of overlapping ‘bands’ of cholinergic neurons innervating target regions.
It is not clear to what extent the anatomically defined inputs described above are functional177. Rodent studies indicate that, in addition to the strong reciprocal connections made by BFCNs, the predominant anatomical input to these neurons is the striatum159,173,174. Most outputs of the striatum (and the central amygdala, another major anatomical input to BFCNs) are inhibitory159,174,178–180, raising the possibility that most BFCNs are under strong inhibitory control. How BFCNs integrate inputs from areas with which they are reciprocally connected with these inhibitory inputs in behaviourally relevant contexts remains to be determined. In addition, it is becoming increasingly apparent that different pools of cholinergic neurons with common projection targets play unique roles in promoting behavioural outcomes157,181. Going forward, it is therefore critical that we learn how anatomical localization and connectivity (Fig. 2c) of BFCNs map onto functional topography.
ACh and behaviour
With the development of novel in vivo imaging tools for measuring ACh release (Box 1 and Fig. 3a,b) and the array of technologies available for selective activation of cholinergic neurons, our appreciation of the functional heterogeneity of BFCNs is expanding.
Attention and cue detection
Studies using choline-sensitive microelectrodes182,183, microdialysis184,185, pharmacological tools186,187 and transgenic mice187 have established that BFCN projections to the cortex play an important role in top-down attention and cue detection142,154. BFCNs also contribute to the processing of afferent sensory information. In primary sensory cortices, an increase in basal forebrain-derived ACh levels desynchronizes cortical activity6,188,189, leading to decorrelation of noise and a corresponding increase in the signal-to-noise ratio6. For example, activation of cholinergic axons in the visual cortex using optogenetics desynchronized neuronal firing and increased the detection of visual stimuli189,190, whereas inhibition of cholinergic projections to the same area synchronized neuronal firing and decreased detection of visual stimuli189,190.
Similar studies underscore the role of BFCN signalling to the cortex in driving attention towards relevant stimuli. In the mPFC, transient increases in cholinergic signalling enhance cue detection142,153,182,191. ACh release in the mPFC leads to an increased top-down influence on neuronal activity in sensory cortices6. Furthermore, as in sensory cortices, an increase in ACh levels in the mPFC can engage brain networks to drive attention towards salient sensory information. Thus, in sensory cortices, ACh efflux enhances the detection of relevant sensory information188–190, whereas coordinated BFCN signalling in the sensory cortices and the mPFC can enhance cue detection and promote behavioural outcomes.
Valence and emotional memory
BFCNs play an important role in encoding valence192–194. With use of the GRABACh3.0 sensor (Box 1), it was shown that mild foot shocks elicit robust increases in ACh release in the BLA157,195,196. ACh release in the hippocampus has also been observed after exposure to stress paradigms5,15,197, whereas exposure to an aversive air puff directly activates cholinergic neurons in the nucleus basalis and the horizontal subdivision of the diagonal band198. BFCNs also play a prominent role in encoding positive valence. One study found that optogenetic stimulation of cholinergic terminals in the BLA increased the time spent in the stimulation-paired chamber, suggesting that ACh release in the BLA can be directly rewarding155. In addition, some cholinergic neurons in the nucleus basalis and the horizontal subdivision of the diagonal band respond to water reward198, and this response is scaled to the unexpectedness of the reinforcement, consistent with greater BFCN activity corresponding to more salient stimuli198. Similarly, a preprint describing work using another ACh biosensor (iAChSnFR) (Box 1), reports reliable increases in ACh release in the hippocampus time-locked to reward (sucrose) delivery148. Furthermore, another recent study used GRABACh3.0 to examine ACh release in the BLA during an operant conditioning task156, in which mice were trained to perform a nose poke in response to an auditory cue to receive an appetitive reward. Initially, ACh release was observed in response to reward-related tasks; however, as learning improved, an increase in ACh release was observed only in response to the tone and no longer during reward-related events156. Thus, dynamic changes in ACh release are associated with cue–reward contingency learning.
In addition to being activated in response to positive valence and negative valence stimuli, BFCNs contribute to the encoding of emotionally salient memories. This has been studied using contextual and cued fear conditioning, and has been reviewed extensively6,8,25,199–202. Recent experiments have considered which cholinergic projections from the basal forebrain are important for the acquisition, recall and extinction of these fear memories8,199,200 and have identified the mPFC, the dorsal and ventral hippocampus, and the amygdala as key targets8,199,200. Of these, perhaps the best characterized are the cholinergic projections to the BLA. A study reported in a recent preprint found that BLA-projecting nucleus basalis and substantia innominata cholinergic neurons are activated during both training and recall in a cue-conditioned fear task157. With use of GRABACh3.0 and fibre photometry in the BLA, an increase in ACh release was observed during tone–shock pairing (training) and subsequent tone-alone (recall) sessions. During training, mice initially showed an increase in ACh release in response to the negative valence stimulus (shock) but not the tone. However, after three tone–shock pairings, the increase in ACh release shifted to the tone157 and could be elicited with only tone presentation during the recall session. Thus, dynamic changes in ACh release are associated with both positive valence and negative valence learning (Fig. 3c). Future work should determine the mechanism underlying the experience-dependent shift in the timing of ACh release in the BLA during learning.
At first glance, it may seem paradoxical that BFCNs are activated and release ACh in response to both positive valence and negative valence stimuli. A simple explanation is that BFCNs may encode emotionally salient stimuli, regardless of valence. In support of this hypothesis, in vivo recordings in the basal forebrain have revealed that some burst-firing cholinergic neurons located in the rostral part of the basal forebrain fire in response to both aversive and appetitive stimuli150,198. Another possibility is that, although all BFCNs are activated in response to salient stimuli, their co-expression with other neuronal markers regulates their response to positive valence versus negative valence stimuli. For instance, co-release or co-transmission of ACh with GABA or glutamate may differentially signal negative valence versus positive valence, and this is an interesting area for future research. An alternative possibility is that distinct subsets of BFCNs respond to positive and negative stimuli. In this case, distinct BLA-projecting subpopulations would uniquely encode reward versus punishment. Given the tremendous functional heterogeneity of BFCNs, this plausible (but unproven) idea also warrants further investigation. Finally, it is important to consider cell type diversity and projection distribution within the regions targeted by BFCNs203–205. For example, the inputs from BFCNs activated in response to negative and positive stimuli may lead to distinct behavioural outputs depending on the functional organization of cholinergic terminal fields in the BLA or which synaptic targets respond to this ACh release. Mapping cholinergic inputs and outputs and mapping the recruitment of subpopulations of cholinergic neurons in behaviourally relevant contexts thus remains an important area for future research.
Cholinergic signalling in other circuits may further contribute to the encoding of valence and may influence subsequent behavioural outcomes. For example, one study has demonstrated coordinated phasic release of ACh in the PFC and hippocampus during reward delivery in a spatial memory task206. This is consistent with the notion that coordinated ACh release across brain regions is important for normal behavioural output7. Further investigations using in vivo recording, biosensors and high-resolution imaging in behaving animals will be critical if we are to understand the differential roles of BFCNs in specific behavioural responses.
BFCN network activity
In addition to examining behaviourally relevant BFCN activity at the single-cell and/or population level, a surge of recent work has examined the contributions of cholinergic transmission at the network level. Cholinergic neurons promote memory formation by sustaining theta oscillations, thereby decreasing aberrant network activity (increasing the signal-to-noise ratio) in the hippocampus207–209. Selective ablation of medial septum and diagonal band complex cholinergic neurons in rodents significantly reduces theta power209, whereas a lesion that encompasses the whole of these regions can completely abolish theta oscillations210. High levels of hippocampal ACh correspond to increased levels of hippocampal theta oscillations, whereas low levels of ACh correspond to non-theta states206. Indeed, ACh input controls changes in the oscillatory states of the hippocampus. For example, activation of cholinergic projections to the hippocampus211,212 (or of mAChRs)213 suppresses sharp-wave ripples, the specific, synchronous activity patterns that support many cognitive functions and occur in the absence of theta waves214. Recent studies using new techniques to monitor ACh release (Box 1) found that optogenetic stimulation of hippocampus-projecting cholinergic neurons increases ACh signalling and theta power and correspondingly decreases the incidence of sharp-wave ripples143.
Stimulation of cholinergic neurons also results in different behavioural outcomes, depending on brain state. Stimulation of cholinergic neurons during an ‘offline’ period of dorsal hippocampal activity (that is, during sharp-wave ripples) impaired performance in a behavioural task143. When cholinergic neurons were stimulated during on ‘online’ (or learning) period (that is, during theta activity), performance was not affected. Conversely, inhibition of cholinergic neurons during the learning phase affected memory performance, whereas inhibition of cholinergic neurons during the ‘offline’ period did not affect performance143. Similar network effects have been reported in the amygdala: optogenetic stimulation of cholinergic projections in the BLA during the learning phase of a cued fear conditioning task strengthened the emotionally salient memories215, whereas inhibition of cholinergic neurons during the learning phase impaired learning and accelerated extinction learning157,215. These examples highlight the importance of timing and brain state on BFCN signalling and the effects of the modulation of network activity on specific aspects of behaviour.
Role of BFCNs in coordinating activity
We propose that the heterogeneity of BFCNs allows them to participate in and mediate a wide array of behaviours by coordinating/binding activity across the brain. As described earlier herein, this diversity of function probably arises from multiple factors, including developmental origin and the unique target-specific cues that support terminal differentiation, as well as the diverse anatomical connectivity and physiology of these neurons.
To achieve this coordination in activity, we suggest that subpopulations of BFCNs likely form functional connections with other BFCNs to regulate their activity. These circuits are modulated by experience and are supported by the capacity of cholinergic neurons to form long-range collaterals to monitor activity in distant regions as well as their ability to signal over broad timescales. These functionally connected regions are coordinated by common pools of cholinergic neurons which prioritize critical stimuli to promote behavioural outcomes. In this manner, cholinergic neurons are able to ‘bind’ diverse behaviours through their connectivity and signalling. Unique molecular signatures, likely heterogeneous subpopulations, that are specifically tuned towards target regions and their functional demands coordinate this process. The next step towards an understanding of these circuits is to identify these unique gene-expression profiles to allow selective targeting of BFCN subpopulations. This will allow tagging and monitoring during behavioural assays. Given the combined power of multisite in vivo recording, ACh sensors and optogenetic tagging approaches during behaviour, we should soon be able to assess the kinetics of ACh activity and determine precisely how ACh participates in network activity and behavioural output (Fig. 3b).
Conclusions and gaps for future study
In this Review, we have outlined factors that are important to the heterogeneity of BFCN circuits. We have presented emerging themes in our understanding of the development and specification of cholinergic neuron subtypes, the factors that diversify cholinergic signalling, the general rules by which these neurons contact their targets and the functional importance of these neurons in attention, learning and memory.
There are many avenues left to explore in understanding the diversity and complexity of cholinergic signalling in health and disease. Mercifully, advances in the methods available are finally beginning to catch up with the field’s long-standing questions. Using advanced transcriptomic and epigenomic profiling (including spatial transcriptomics methods), we can investigate the diversity of cholinergic neuron subtypes and their relationship with anatomical location across species (Boxes 1,2). Studies identifying the input–output connectivity of cholinergic neurons coupled with their functional importance will be critical to our understanding of how the cholinergic system contributes to cognition. Equally important, advances in experimental imaging modalities (Fig. 3), improved ligands to detect ACh terminal field integrity in humans and other primates (Box 2) and compartment-specific measurement of cholinergic dynamics during behaviourally relevant tasks (Fig. 3) are critical. These innovations will allow us to take the next steps towards understanding the role of ACh in cognition and, eventually, its vulnerability in disease (Box 3).
Acknowledgements
All authors are supported by funds from the NINDS Intramural Research Program.
Glossary
- Chromatin landscape
An umbrella term encompassing chromatin accessibility, long-range contacts and looping, higher-order chromatin structure and DNA/histone modifications.
- Collateral projections
Branching neuronal projections from a single neuron targeting multiple brain regions.
- Co-release
The release of two (and possibly more) neurotransmitters from the same presynaptic vesicle using the same mechanism.
- Co-transmission
The release of multiple neurotransmitters from different presynaptic vesicles via distinct mechanisms but leading to an aggregate effect on synaptic transmission.
- En passant release
Neurotransmitter release along axonal projections rather than from an axon terminal.
- Fluorescent biosensors
A class of tools used to visualize and measure changes in biological processes using sensitive fluorescence readout in response to a conformational change.
- Homeodomain transcription factors
Transcription factors with a conserved helix–turn–helix motif, called a ‘homeodomain’, involved in regulating gene expression early in development.
- Morphogens
Secreted signalling molecules that pattern cell and tissue fates during early development.
- Neuromodulator
A messenger (including peptides and certain neurotransmitters) that can change the state of a neuron or group of neurons to alter their excitability.
- Optogenetics
A technique that uses light to manipulate neuronal activity.
- Point-to-point synapses
Synapses at which individual sites of presynaptic release are closely apposed to postsynaptic terminals.
- Phasic signalling
Fast, transient signalling in response to the onset of a stimulus.
- Radial migration
Migration radially from the ventricular zone towards the directly overlying surface of the brain.
- Retrograde signals
Signalling initiated in axon terminals influencing signalling/gene expression in the soma/nucleus.
- Single-cell RNA sequencing
Sequencing technology that allows transcriptomic profiling of individual cells/nuclei.
- Specification
The process involved in the specification of cell identity.
- Synaptic release probability
The likelihood that a neurotransmitter will be released from a presynaptic vesicle.
- Tangential migration
Migration parallel to the ventricular zone.
- Terminal fields
A collection of axon terminals used to describe innervation of an anatomically defined region.
- Theta oscillations
Synchronous neuronal activity between 4 Hz and 8 Hz that corresponds to a behavioural state of alertness and attention in the brain.
- Tonic signalling
Slow and graded signalling.
- Trophic factors
Peptides (often secreted) that promote growth and survival of cells.
- Top-down attention
Intentional and goal-driven attention, as opposed to ‘bottom-up’ attention in response to sensory stimuli.
- Valence
The hedonic value (positive or negative) of a stimulus.
- Virus-based mapping
The use of viral vectors (anterograde and/or retrograde) to delineate anatomical connections between different brain regions.
Footnotes
Competing interests
The authors declare no competing interests.
Related link
BRAIN Initiative Cell Census Network collection: https://www.nature.com/collections/cicghheddj
References
- 1.Dale HH The action of certain esters and ethers of choline, and their relation to muscarine. J. Pharmacol. Exp. Ther 6, 147–190 (1914). [Google Scholar]
- 2.Ewins AJ Acetylcholine, a new active principle of ergot. Biochem. J 8, 44 (1914). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katz B & Miledi R The measurement of synaptic delay, and the time course of acetylcholine release at the neuromuscular junction. Proc. R. Soc. Lond. B 161, 483–495 (1965). [DOI] [PubMed] [Google Scholar]
- 4.Peper K, Bradley RJ & Dreyer F The acetylcholine receptor at the neuromuscular junction. Physiol. Rev 62, 1271–1340 (1982). [DOI] [PubMed] [Google Scholar]
- 5.Picciotto MR, Higley MJ & Mineur YS Acetylcholine as a neuromodulator: cholinergic signaling shapes nervous system function and behavior. Neuron 76, 116–129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballinger EC, Ananth M, Talmage DA & Role LW Basal forebrain cholinergic circuits and signaling in cognition and cognitive decline. Neuron 91, 1199–1218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohani S et al. Spatiotemporally heterogeneous coordination of cholinergic and neocortical activity. Nat. Neurosci 25, 1706–1713 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses in vivo wide-field imaging of cortical activity and ACh release to show distinct activity profiles associated with specific behavioural states.
- 8.Záborszky L et al. Specific basal forebrain-cortical cholinergic circuits coordinate cognitive operations. J. Neurosci 38, 9446–9458 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stifani N Motor neurons and the generation of spinal motor neuron diversity. Front. Cell Neurosci 8, 293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alkaslasi MR et al. Single nucleus RNA-sequencing defines unexpected diversity of cholinergic neuron types in the adult mouse spinal cord. Nat. Commun 12, 2471 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higley MJ et al. Cholinergic interneurons mediate fast VGluT3-dependent glutamatergic transmission in the striatum. PLoS ONE 6, e19155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kljakic O, Janickova H, Prado VF & Prado MAM Cholinergic/glutamatergic co-transmission in striatal cholinergic interneurons: new mechanisms regulating striatal computation. J. Neurochem 142 (Suppl. 2), 90–102 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Mayer C et al. Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obermayer J et al. Prefrontal cortical ChAT-VIP interneurons provide local excitation by cholinergic synaptic transmission and control attention. Nat. Commun 10, 5280 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mineur YS et al. Hippocampal acetylcholine modulates stress-related behaviors independent of specific cholinergic inputs. Mol. Psychiatry 27, 1829–1838 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granger AJ et al. Cortical ChAT+ neurons co-transmit acetylcholine and GABA in a target- and brain-region-specific manner. eLife 9, e57749 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudai A, Yayon N, Soreq H & London M Cortical VIP+/ChAT+ interneurons: from genetics to function. J. Neurochem 158, 1320–1333 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Mesulam MM, Mufson EJ, Levey AI & Wainer BH Cholinergic innervation of cortex by the basal forebrain: cytochemistry and cortical connections of the septal area, diagonal band nuclei, nucleus basalis (substantia innominata), and hypothalamus in the rhesus monkey. J. Comp. Neurol 214, 170–197 (1983). [DOI] [PubMed] [Google Scholar]
- 19.Woolf NJ Cholinergic systems in mammalian brain and spinal cord. Prog. Neurobiol 37, 475–524 (1991). [DOI] [PubMed] [Google Scholar]
- 20.Eckenstein F & Baughman RW Two types of cholinergic innervation in cortex, one co-localized with vasoactive intestinal polypeptide. Nature 309, 153–155 (1984). [DOI] [PubMed] [Google Scholar]
- 21.Geula C et al. Basal forebrain cholinergic system in the dementias: vulnerability, resilience, and resistance. J. Neurochem 158, 1394–1411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertrand D & Wallace TL A review of the cholinergic system and therapeutic approaches to treat brain disorders. Curr. Top. Behav. Neurosci 45, 1–28 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Ferreira-Vieira TH, Guimaraes IM, Silva FR & Ribeiro FM Alzheimer’s disease: targeting the cholinergic system. Curr. Neuropharmacol 14, 101–115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solari N & Hangya B Cholinergic modulation of spatial learning, memory and navigation. Eur. J. Neurosci 48, 2199–2230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knox D The role of basal forebrain cholinergic neurons in fear and extinction memory. Neurobiol. Learn. Mem 133, 39–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar Yadav R & Mallick BN Dopaminergic- and cholinergic-inputs from substantia nigra and pedunculo-pontine tegmentum, respectively, converge in amygdala to modulate rapid eye movement sleep in rats. Neuropharmacology 193, 108607 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Vanini G & Torterolo P Sleep-wake neurobiology. Adv. Exp. Med. Biol 1297, 65–82 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Jones BE Arousal and sleep circuits. Neuropsychopharmacology 45, 6–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shao X & Feldman J Central cholinergic regulation of respiration: nicotinic receptors. Acta Pharmacol. Sin 30, 761–770 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dani JA & Bertrand D Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annu. Rev. Pharmacol. Toxicol 47, 699–729 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Thiele A Muscarinic signaling in the brain. Annu. Rev. Neurosci 36, 271–294 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Nathanson NM Regulation of muscarinic acetylcholine receptor expression and function. Prog. Brain Res 109, 165–168 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Gaimarri A et al. Regulation of neuronal nicotinic receptor traffic and expression. Brain Res. Rev 55, 134–143 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Wess J Molecular basis of muscarinic acetylcholine receptor function. Trends Pharmacol. Sci 14, 308–313 (1993). [DOI] [PubMed] [Google Scholar]
- 35.Zoli M, Pucci S, Vilella A & Gotti C Neuronal and extraneuronal nicotinic acetylcholine receptors. Curr. Neuropharmacol 16, 338–349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaborszky L, van den Pol A & Gyengesi E in The Mouse Nervous System (eds Watson C, Paxinos G & Puelles L) 684–718 (Academic, 2012). [Google Scholar]
- 37.Huerta-Ocampo I, Dautan D, Gut NK, Khan B & Mena-Segovia J Whole-brain mapping of monosynaptic inputs to midbrain cholinergic neurons. Sci. Rep 11, 9055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poppi LA, Ho-Nguyen KT, Shi A, Daut CT & Tischfield MA Recurrent implication of striatal cholinergic interneurons in a range of neurodevelopmental, neurodegenerative, and neuropsychiatric disorders. Cells 10, 907 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mena-Segovia J & Bolam JP Rethinking the pedunculopontine nucleus: from cellular organization to function. Neuron 94, 7–18 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Gut NK & Mena-Segovia J Midbrain cholinergic neurons signal negative feedback to promote behavioral flexibility. Trends Neurosci. 45, 502–503 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abudukeyoumu N, Hernandez-Flores T, Garcia-Munoz M & Arbuthnott GW Cholinergic modulation of striatal microcircuits. Eur. J. Neurosci 49, 604–622 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nosaka D & Wickens JR Striatal cholinergic signaling in time and space. Molecules 27, 1202 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanabe Y & Jessell TM Diversity and pattern in the developing spinal cord. Science 274, 1115–1123 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Marin O & Rubenstein JL in Mouse Development (eds Rossant J & Tam PPL) 75–106 (Elsevier, 2002). [Google Scholar]
- 45.Marín O & Rubenstein JL A long, remarkable journey: tangential migration in the telencephalon. Nat. Rev. Neurosci 2, 780–790 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Allaway KC et al. Cellular birthdate predicts laminar and regional cholinergic projection topography in the forebrain. eLife 9, e63249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the most comprehensive analysis of the relationship between developmental origin and circuit integration of BFCNs to date.
- 47.Pombero A et al. Pallial origin of basal forebrain cholinergic neurons in the nucleus basalis of Meynert and horizontal limb of the diagonal band nucleus. Development 138, 4315–4326 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Flames N et al. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J. Neurosci 27, 9682–9695 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work examines expression of several transcription factors using in situ hybridization to define subdomains of the embryonic subpallium, thereby generating a spatial combinatorial code.
- 49.Eiden LE The cholinergic gene locus. J. Neurochem 70, 2227–2240 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Brock M, Nickel A-C, Madziar B, Blusztajn JK & Berse B Differential regulation of the high affinity choline transporter and the cholinergic locus by cAMP signaling pathways. Brain Res. 1145, 1–10 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimojo M, Wu D & Hersh LB The cholinergic gene locus is coordinately regulated by protein kinase A II in PC12 cells. J. Neurochem 71, 1118–1126 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Shimojo M & Hersh LB Regulation of the cholinergic gene locus by the repressor element-1 silencing transcription factor/neuron restrictive silencer factor (REST/NRSF). Life Sci. 74, 2213–2225 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Dorn R, Loy B, Dechant G & Apostolova G Neurogenomics of the sympathetic neurotransmitter switch indicates that different mechanisms steer cholinergic differentiation in rat and chicken models. Dataset Pap. Sci 2013, 520930 (2013). [Google Scholar]
- 54.Berse B & Blusztajn JK Modulation of cholinergic locus expression by glucocorticoids and retinoic acid is cell-type specific. FEBS Lett. 410, 175–179 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Tanaka H, Zhao Y, Wu D & Hersh LB The use of DNase I hypersensitivity site mapping to identify regulatory regions of the human cholinergic gene locus. J. Neurochem 70, 1799–1808 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Kania A Concocting cholinergy. PLoS Genet. 10, e1004313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopez-Coviella I et al. Bone morphogenetic protein 9 induces the transcriptome of basal forebrain cholinergic neurons. Proc. Natl Acad. Sci. USA 102, 6984–6989 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bachy I & Rétaux S GABAergic specification in the basal forebrain is controlled by the LIM-hd factor Lhx7. Dev. Biol 291, 218–226 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Elshatory Y & Gan L The LIM-homeobox gene Islet-1 is required for the development of restricted forebrain cholinergic neurons. J. Neurosci 28, 3291–3297 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Furusho M et al. Involvement of the Olig2 transcription factor in cholinergic neuron development of the basal forebrain. Dev. Biol 293, 348–357 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Magno L et al. NKX2–1 is required in the embryonic septum for cholinergic system development, learning, and memory. Cell Rep. 20, 1572–1584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work identifies progenitors co-expressing Nkx2.1 and Zic4 as the major source of septal cholinergic neurons and examines the role of Nkx2.1 in their function.
- 62.Mori T et al. The LIM homeobox gene, L3/Lhx8, is necessary for proper development of basal forebrain cholinergic neurons. Eur. J. Neurosci 19, 3129–3141 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Sussel L, Marin O, Kimura S & Rubenstein J Loss of Nkx2. 1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development 126, 3359–3370 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Zhao Y et al. Ldb1 is essential for development of Nkx2.1 lineage derived GABAergic and cholinergic neurons in the telencephalon. Dev. Biol 385, 94–106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Y et al. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc. Natl Acad. Sci. USA 100, 9005–9010 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun T, Pringle NP, Hardy AP, Richardson WD & Smith HK Pax6 influences the time and site of origin of glial precursors in the ventral neural tube. Mol. Cell Neurosci 12, 228–239 (1998). [DOI] [PubMed] [Google Scholar]
- 67.Ravanelli AM & Appel B Motor neurons and oligodendrocytes arise from distinct cell lineages by progenitor recruitment. Genes Dev. 29, 2504–2515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scott K, O’Rourke R, Winkler CC, Kearns CA & Appel B Temporal single-cell transcriptomes of zebrafish spinal cord pMN progenitors reveal distinct neuronal and glial progenitor populations. Dev. Biol 479, 37–50 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xing L et al. Expression of myelin transcription factor 1 and lamin B receptor mediate neural progenitor fate transition in the zebrafish spinal cord pMN domain. J. Biol. Chem 298, 102452 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Semba K & Fibiger HC Time of origin of cholinergic neurons in the rat basal forebrain. J. Comp. Neurol 269, 87–95 (1988). [DOI] [PubMed] [Google Scholar]
- 71.Cho H-H et al. Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet. 10, e1004280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alifragis P, Liapi A & Parnavelas JG Lhx6 regulates the migration of cortical interneurons from the ventral telencephalon but does not specify their GABA phenotype. J. Neurosci 24, 5643–5648 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Magno L et al. Fate mapping reveals mixed embryonic origin and unique developmental codes of mouse forebrain septal neurons. Commun. Biol 5, 1137 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fragkouli A, van Wijk NV, Lopes R, Kessaris N & Pachnis V LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development 136, 3841–3851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koh S & Loy R Localization and development of nerve growth factor-sensitive rat basal forebrain neurons and their afferent projections to hippocampus and neocortex. J. Neurosci 9, 2999–3018 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saunders A et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Flandin P et al. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron 70, 939–950 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou C et al. Lhx6 and Lhx8: cell fate regulators and beyond. FASEB J. 29, 4083–4091 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoch RV, Clarke JA & Rubenstein JL Fgf signaling controls the telencephalic distribution of Fgf-expressing progenitors generated in the rostral patterning center. Neural Dev. 10, 8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matthews JM et al. Competition between LIM-binding domains. Biochem. Soc. Trans 36, 1393–1397 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Lee S et al. A regulatory network to segregate the identity of neuronal subtypes. Dev. Cell 14, 877–889 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Asbreuk CH et al. The homeobox genes Lhx7 and Gbx1 are expressed in the basal forebrain cholinergic system. Neuroscience 109, 287–298 (2002). [DOI] [PubMed] [Google Scholar]
- 83.Waters ST, Wilson CP & Lewandoski M Cloning and embryonic expression analysis of the mouse Gbx1 gene. Gene Expr. Patterns 3, 313–317 (2003). [DOI] [PubMed] [Google Scholar]
- 84.Chen L, Chatterjee M & Li JY The mouse homeobox gene Gbx2 is required for the development of cholinergic interneurons in the striatum. J. Neurosci 30, 14824–14834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanchez-Ortiz E et al. TrkA gene ablation in basal forebrain results in dysfunction of the cholinergic circuitry. J. Neurosci 32, 4065–4079 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Su-Feher L et al. Single cell enhancer activity distinguishes GABAergic and cholinergic lineages in embryonic mouse basal ganglia. Proc. Natl Acad. Sci. USA 119, e2108760119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work uses subpallial enhancer lines combined with single-cell transcriptomics to identify postmitotic cells of the cholinergic lineage and reveals transcriptomes of striatal cholinergic interneurons as distinct from BFCNs, with hints of more heterogeneity within the BFCNs.
- 87.Buckley DM, Burroughs-Garcia J, Lewandoski M & Waters ST Characterization of the Gbx1−/− mouse mutant: a requirement for Gbx1 in normal locomotion and sensorimotor circuit development. PLoS ONE 8, e56214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reilly JO, Karavanova ID, Williams KP, Mahanthappa NK & Allendoerfer KL Cooperative effects of sonic hedgehog and NGF on basal forebrain cholinergic neurons. Mol. Cell Neurosci 19, 88–96 (2002). [DOI] [PubMed] [Google Scholar]
- 89.Fagan AM et al. Endogenous FGF-2 is important for cholinergic sprouting in the denervated hippocampus. J. Neurosci 17, 2499–2511 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Halilagic A et al. Retinoids control anterior and dorsal properties in the developing forebrain. Dev. Biol 303, 362–375 (2007). [DOI] [PubMed] [Google Scholar]
- 91.Garel S & Rubenstein JL Intermediate targets in formation of topographic projections: inputs from the thalamocortical system. Trends Neurosci. 27, 533–539 (2004). [DOI] [PubMed] [Google Scholar]
- 92.Kang YH et al. Comparative analysis of three different protocols for cholinergic neuron differentiation in vitro using mesenchymal stem cells from human dental pulp. Anim. Cell Syst 23, 275–287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Storm EE et al. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development 133, 1831–1844 (2006). [DOI] [PubMed] [Google Scholar]
- 94.Boskovic Z et al. Regulation of cholinergic basal forebrain development, connectivity, and function by neurotrophin receptors. Neuronal Signal. 3, NS20180066 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lindholm D Models to study the role of neurotrophic factors in neurodegeneration. J. Neural Transm. Suppl 49, 33–42 (1997). [DOI] [PubMed] [Google Scholar]
- 96.Nilbratt M, Porras O, Marutle A, Hovatta O & Nordberg A Neurotrophic factors promote cholinergic differentiation in human embryonic stem cell-derived neurons. J. Cell Mol. Med 14, 1476–1484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schnitzler AC et al. BMP9 (bone morphogenetic protein 9) induces NGF as an autocrine/paracrine cholinergic trophic factor in developing basal forebrain neurons. J. Neurosci 30, 8221–8228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ginsberg SD, Che S, Wuu J, Counts SE & Mufson EJ Down regulation of trk but not p75NTR gene expression in single cholinergic basal forebrain neurons mark the progression of Alzheimer’s disease. J. Neurochem 97, 475–487 (2006). [DOI] [PubMed] [Google Scholar]
- 99.Hashimoto Y, Abiru Y, Nishio C & Hatanaka H Synergistic effects of brain-derived neurotrophic factor and ciliary neurotrophic factor on cultured basal forebrain cholinergic neurons from postnatal 2-week-old rats. Dev. Brain Res 115, 25–32 (1999). [DOI] [PubMed] [Google Scholar]
- 100.Biane J, Conner JM & Tuszynski MH Nerve growth factor is primarily produced by GABAergic neurons of the adult rat cortex. Front. Cell Neurosci 8, 220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.La Manno G et al. Molecular architecture of the developing mouse brain. Nature 596, 92–96 (2021). [DOI] [PubMed] [Google Scholar]
- 102.Nonomura T, Nishio C, Lindsay RM & Hatanaka H Cultured basal forebrain cholinergic neurons from postnatal rats show both overlapping and non-overlapping responses to the neurotrophins. Brain Res. 683, 129–139 (1995). [DOI] [PubMed] [Google Scholar]
- 103.di Sanguinetto SADT, Dasen JS & Arber S Transcriptional mechanisms controlling motor neuron diversity and connectivity. Curr. Opin. Neurobiol 18, 36–43 (2008). [DOI] [PubMed] [Google Scholar]
- 104.Hollera T, Berse B, Cermak JM, Diebler M-F & Krzysztof Blusztajn J Differences in the developmental expression of the vesicular acetylcholine transporter and choline acetyltransferase in the rat brain. Neurosci. Lett 212, 107–110 (1996). [DOI] [PubMed] [Google Scholar]
- 105.Mallet J et al. The cholinergic locus: ChAT and VAChT genes. J. Physiol 92, 145–147 (1998). [DOI] [PubMed] [Google Scholar]
- 106.Berse B & Blusztajn JK Coordinated up-regulation of choline acetyltransferase and vesicular acetylcholine transporter gene expression by the retinoic acid receptor α, cAMP, and leukemia inhibitory factor/ciliary neurotrophic factor signaling pathways in a murine septal cell line. J. Biol. Chem 270, 22101–22104 (1995). [DOI] [PubMed] [Google Scholar]
- 107.Berse B, Lopez-Coviella I & Blusztajn JK Activation of TrkA by nerve growth factor upregulates expression of the cholinergic gene locus but attenuates the response to ciliary neurotrophic growth factor. Biochem. J 342, 301–308 (1999). [PMC free article] [PubMed] [Google Scholar]
- 108.Lee DR et al. Transcriptional heterogeneity of ventricular zone cells in the ganglionic eminences of the mouse forebrain. eLife 11, e71854 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mi D et al. Early emergence of cortical interneuron diversity in the mouse embryo. Science 360, 81–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rhodes CT et al. An epigenome atlas of neural progenitors within the embryonic mouse forebrain. Nat. Commun 13, 4196 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Silberberg SN et al. Subpallial enhancer transgenic lines: a data and tool resource to study transcriptional regulation of GABAergic cell fate. Neuron 92, 59–74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tritsch NX, Granger AJ & Sabatini BL Mechanisms and functions of GABA co-release. Nat. Rev. Neurosci 17, 139–145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nusbaum MP, Blitz DM & Marder E Functional consequences of neuropeptide and small-molecule co-transmission. Nat. Rev. Neurosci 18, 389–403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vaaga CE, Borisovska M & Westbrook GL Dual-transmitter neurons: functional implications of co-release and co-transmission. Curr. Opin. Neurobiol 29, 25–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Frahm S et al. An essential role of acetylcholine-glutamate synergy at habenular synapses in nicotine dependence. eLife 4, e11396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ren J et al. Habenula “cholinergic” neurons co-release glutamate and acetylcholine and activate postsynaptic neurons via distinct transmission modes. Neuron 69, 445–452 (2011). [DOI] [PubMed] [Google Scholar]
- 117.Amilhon B et al. VGLUT3 (vesicular glutamate transporter type 3) contribution to the regulation of serotonergic transmission and anxiety. J. Neurosci 30, 2198–2210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gras C et al. A third vesicular glutamate transporter expressed by cholinergic and serotoninergic neurons. J. Neurosci 22, 5442–5451 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kosaka T, Tauchi M & Dahl JL Cholinergic neurons containing GABA-like and/or glutamic acid decarboxylase-like immunoreactivities in various brain regions of the rat. Exp. Brain Res 70, 605–617 (1988). [DOI] [PubMed] [Google Scholar]
- 120.Fisher RS & Levine MS Transmitter cosynthesis by corticopetal basal forebrain neurons. Brain Res. 491, 163–168 (1989). [DOI] [PubMed] [Google Scholar]
- 121.O’Malley DM, Sandell JH & Masland RH Co-release of acetylcholine and GABA by the starburst amacrine cells. J. Neurosci 12, 1394–1408 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.O’Malley DM & Masland RH Co-release of acetylcholine and gamma-aminobutyric acid by a retinal neuron. Proc. Natl Acad. Sci. USA 86, 3414–3418 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Santos PF, Carvalho AL, Carvalho AP & Duarte CB Differential acetylcholine and GABA release from cultured chick retina cells. Eur. J. Neurosci 10, 2723–2730 (1998). [DOI] [PubMed] [Google Scholar]
- 124.Sethuramanujam S et al. A central role for mixed acetylcholine/GABA transmission in direction coding in the retina. Neuron 90, 1243–1256 (2016). [DOI] [PubMed] [Google Scholar]
- 125.Hur EE, Edwards RH, Rommer E & Zaborszky L Vesicular glutamate transporter 1 and vesicular glutamate transporter 2 synapses on cholinergic neurons in the sublenticular gray of the rat basal forebrain: a double-label electron microscopic study. Neuroscience 164, 1721–1731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Case DT et al. Layer- and cell type-selective co-transmission by a basal forebrain cholinergic projection to the olfactory bulb. Nat. Commun 8, 652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nickerson Poulin A, Guerci A, El Mestikawy S & Semba K Vesicular glutamate transporter 3 immunoreactivity is present in cholinergic basal forebrain neurons projecting to the basolateral amygdala in rat. J. Comp. Neurol 498, 690–711 (2006). [DOI] [PubMed] [Google Scholar]
- 128.Granger AJ, Mulder N, Saunders A & Sabatini BL Cotransmission of acetylcholine and GABA. Neuropharmacology 100, 40–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saunders A, Granger AJ & Sabatini BL Corelease of acetylcholine and GABA from cholinergic forebrain neurons. eLife 4, e06412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article demonstrates co-transmission of ACh and GABA by forebrain cholinergic neurons.
- 130.Brunet Avalos C & Sprecher SG Single-cell transcriptomic reveals dual and multi-transmitter use in neurons across metazoans. Front. Mol. Neurosci 14, 623148 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Upmanyu N et al. Colocalization of different neurotransmitter transporters on synaptic vesicles is sparse except for VGLUT1 and ZnT3. Neuron 110, 1483–1497.e7 (2022). [DOI] [PubMed] [Google Scholar]
- 132.Ximerakis M et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci 22, 1696–1708 (2019). [DOI] [PubMed] [Google Scholar]
- 133.Desikan S, Koser DE, Neitz A & Monyer H Target selectivity of septal cholinergic neurons in the medial and lateral entorhinal cortex. Proc. Natl Acad. Sci. USA 115, E2644–E2652 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Takács VT et al. Co-transmission of acetylcholine and GABA regulates hippocampal states. Nat. Commun 9, 2848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Colangelo C, Shichkova P, Keller D, Markram H & Ramaswamy S Cellular, synaptic and network effects of acetylcholine in the neocortex. Front. Neural Circuits 13, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Changeux JP, Corringer PJ & Maskos U The nicotinic acetylcholine receptor: from molecular biology to cognition. Neuropharmacology 96, 135–136 (2015). [DOI] [PubMed] [Google Scholar]
- 137.Gotti C, Zoli M & Clementi F Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol. Sci 27, 482–491 (2006). [DOI] [PubMed] [Google Scholar]
- 138.McKay BE, Placzek AN & Dani JA Regulation of synaptic transmission and plasticity by neuronal nicotinic acetylcholine receptors. Biochem. Pharmacol 74, 1120–1133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jones CK, Byun N & Bubser M Muscarinic and nicotinic acetylcholine receptor agonists and allosteric modulators for the treatment of schizophrenia. Neuropsychopharmacology 37, 16–42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wess J, Eglen RM & Gautam D Muscarinic acetylcholine receptors: mutant mice provide new insights for drug development. Nat. Rev. Drug Discov 6, 721–733 (2007). [DOI] [PubMed] [Google Scholar]
- 141.Disney AA & Higley MJ Diverse spatiotemporal scales of cholinergic signaling in the neocortex. J. Neurosci 40, 720–725 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sarter M & Lustig C Cholinergic double duty: cue detection and attentional control. Curr. Opin. Psychol 29, 102–107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang Y et al. Cholinergic suppression of hippocampal sharp-wave ripples impairs working memory. Proc. Natl Acad. Sci. USA 118, e2016432118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows that behavioural outcome in response to ACh signalling is critically dependent on brain state.
- 144.Kramer PF et al. Synaptic-like axo-axonal transmission from striatal cholinergic interneurons onto dopaminergic fibers. Neuron 110, 2949–2960.e4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]; In this technical tour de force, the authors use patch clamp recording from dopaminergic axons in the striatum to demonstrate axonal/presynaptic ACh signalling regulated dopamine release independently of soma-derived action potentials.
- 145.Muller JF, Mascagni F, Zaric V & McDonald AJ Muscarinic cholinergic receptor M1 in the rat basolateral amygdala: ultrastructural localization and synaptic relationships to cholinergic axons. J. Comp. Neurol 521, 1743–1759 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Turrini P et al. Cholinergic nerve terminals establish classical synapses in the rat cerebral cortex: synaptic pattern and age-related atrophy. Neuroscience 105, 277–285 (2001). [DOI] [PubMed] [Google Scholar]
- 147.Umbriaco D, Watkins KC, Descarries L, Cozzari C & Hartman BK Ultrastructural and morphometric features of the acetylcholine innervation in adult rat parietal cortex: an electron microscopic study in serial sections. J. Comp. Neurol 348, 351–373 (1994). [DOI] [PubMed] [Google Scholar]
- 148.Borden PM et al. A fast genetically encoded fluorescent sensor for faithful in vivo acetylcholine detection in mice, fish, worms and flies. Preprint at bioRxiv 10.1101/2020.02.07.939504 (2020). [DOI] [Google Scholar]
- 149.Liu C et al. An action potential initiation mechanism in distal axons for the control of dopamine release. Science 375, 1378–1385 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Laszlovszky T et al. Distinct synchronization, cortical coupling and behavioral function of two basal forebrain cholinergic neuron types. Nat. Neurosci 23, 992–1003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; Using in vivo electrophysiology, this study demonstrates that distinct populations of BFCNs that differ in their firing modes are associated with unique behavioural profiles.
- 151.Zheng Y et al. Different subgroups of cholinergic neurons in the basal forebrain are distinctly innervated by the olfactory regions and activated differentially in olfactory memory retrieval. Front. Neural Circuits 12, 99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li X et al. Molecularly defined and functionally distinct cholinergic subnetworks. Neuron 110, 3774–3788.e7 (2022). [DOI] [PubMed] [Google Scholar]
- 153.Gritton HJ et al. Cortical cholinergic signaling controls the detection of cues. Proc. Natl Acad. Sci. USA 113, E1089–E1097 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Thiele A & Bellgrove MA Neuromodulation of attention. Neuron 97, 769–785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Aitta-Aho T et al. Basal forebrain and brainstem cholinergic neurons differentially impact amygdala circuits and learning-related behavior. Curr. Biol 28, 2557–2569.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 156.Crouse RB et al. Acetylcholine is released in the basolateral amygdala in response to predictors of reward and enhances the learning of cue-reward contingency. eLife 9, e57335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses an ACh biosensor and shows that dynamic changes in ACh release in the BLA are associated with cue–reward contingency learning.
- 157.Rajebhosale P et al. Basal forebrain cholinergic neurons are part of the threat memory engram. Preprint at bioRxiv 10.1101/2021.05.02.442364 (2021). [DOI] [Google Scholar]; This study demonstrates that BFCNs are engaged during training and again during recall of the cued fear conditioning task, and are also necessary for proper defensive behaviours.
- 158.Bloem B et al. Topographic mapping between basal forebrain cholinergic neurons and the medial prefrontal cortex in mice. J. Neurosci 34, 16234–16246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gielow MR & Zaborszky L The input-output relationship of the cholinergic basal forebrain. Cell Rep. 18, 1817–1830 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; Using output-specific monosynaptic tracing, this study reveals that the inputs to and outputs from cholinergic neurons are functionally organized.
- 160.Chandler D & Waterhouse B Evidence for broad versus segregated projections from cholinergic and noradrenergic nuclei to functionally and anatomically discrete subregions of prefrontal cortex. Front. Behav. Neurosci 6, 20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wu H, Williams J & Nathans J Complete morphologies of basal forebrain cholinergic neurons in the mouse. eLife 3, e02444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zaborszky L et al. Neurons in the basal forebrain project to the cortex in a complex topographic organization that reflects corticocortical connectivity patterns: an experimental study based on retrograde tracing and 3D reconstruction. Cereb. Cortex 25, 118–137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Saper CB Organization of cerebral cortical afferent systems in the rat. II. Magnocellular basal nucleus. J. Comp. Neurol 222, 313–342 (1984). [DOI] [PubMed] [Google Scholar]
- 164.Saper CB Diffuse cortical projection systems: anatomical organization and role in cortical function. Compr. Physiol 10.1002/cphy.cp010506 (2011). [DOI] [Google Scholar]
- 165.Huppé-Gourgues F, Jegouic K & Vaucher E Topographic organization of cholinergic innervation from the basal forebrain to the visual cortex in the rat. Front. Neural Circuits 12, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhu F, Elnozahy SE, Lawlor J & Kuchibhotla KV A parallel channel of state-dependent sensory signaling from the cholinergic basal forebrain to the auditory cortex. Preprint at bioRxiv 10.1101/2022.05.05.490613 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Alheid GF Extended amygdala and basal forebrain. Ann. NY Acad. Sci 985, 185–205 (2003). [DOI] [PubMed] [Google Scholar]
- 168.Root DH, Melendez RI, Zaborszky L & Napier TC The ventral pallidum: subregion-specific functional anatomy and roles in motivated behaviors. Prog. Neurobiol 130, 29–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Teles-Grilo Ruivo LM & Mellor JR Cholinergic modulation of hippocampal network function. Front. Synaptic Neurosci 5, 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Slomianka L & Geneser FA Distribution of acetylcholinesterase in the hippocampal region of the mouse: II. Subiculum and hippocampus. J. Comp. Neurol 312, 525–536 (1991). [DOI] [PubMed] [Google Scholar]
- 171.Kondo H & Zaborszky L Topographic organization of the basal forebrain projections to the perirhinal, postrhinal, and entorhinal cortex in rats. J. Comp. Neurol 524, 2503–2515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Slomianka L & Geneser FA Distribution of acetylcholinesterase in the hippocampal region of the mouse: I. Entorhinal area, parasubiculum, retrosplenial area, and presubiculum. J. Comp. Neurol 303, 339–354 (1991). [DOI] [PubMed] [Google Scholar]
- 173.Do JP et al. Cell type-specific long-range connections of basal forebrain circuit. eLife 5, e13214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study maps monosynaptic inputs to four distinct cell populations within the basal forebrain and identifies common patterns of input to basal forebrain cell types.
- 174.Hu R, Jin S, He X, Xu F & Hu J Whole-brain monosynaptic afferent inputs to basal forebrain cholinergic system. Front. Neuroanat 10, 98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chavez C & Zaborszky L Basal forebrain cholinergic–auditory cortical network: primary versus nonprimary auditory cortical areas. Cereb. Cortex 27, 2335–2347 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zaborszky L et al. in The Rat Nervous System 4th edn (ed. Paxinos G) 491–507 (Academic, 2015). [Google Scholar]
- 177.Shu SY et al. A new neural pathway from the ventral striatum to the nucleus basalis of Meynert with functional implication to learning and memory. Mol. Neurobiol 56, 7222–7233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yager LM, Garcia AF, Wunsch AM & Ferguson SM The ins and outs of the striatum: role in drug addiction. Neuroscience 301, 529–541 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Swanson LW & Petrovich GD What is the amygdala? Trends Neurosci. 21, 323–331 (1998). [DOI] [PubMed] [Google Scholar]
- 180.Kreitzer AC & Berke JD Investigating striatal function through cell-type-specific manipulations. Neuroscience 198, 19–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Robert B et al. A functional topography within the cholinergic basal forebrain for encoding sensory cues and behavioral reinforcement outcomes. eLife 10, e69514 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Parikh V, Kozak R, Martinez V & Sarter M Prefrontal acetylcholine release controls cue detection on multiple timescales. Neuron 56, 141–154 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Howe WM et al. Acetylcholine release in prefrontal cortex promotes gamma oscillations and theta-gamma coupling during cue detection. J. Neurosci 37, 3215–3230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dalley JW et al. Distinct changes in cortical acetylcholine and noradrenaline efflux during contingent and noncontingent performance of a visual attentional task. J. Neurosci 21, 4908–4914 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Himmelheber AM, Sarter M & Bruno JP Increases in cortical acetylcholine release during sustained attention performance in rats. Brain Res. Cogn. Brain Res 9, 313–325 (2000). [DOI] [PubMed] [Google Scholar]
- 186.Levin ED & Simon BB Nicotinic acetylcholine involvement in cognitive function in animals. Psychopharmacology 138, 217–230 (1998). [DOI] [PubMed] [Google Scholar]
- 187.Klinkenberg I, Sambeth A & Blokland A Acetylcholine and attention. Behav. Brain Res 221, 430–442 (2011). [DOI] [PubMed] [Google Scholar]
- 188.Meir I, Katz Y & Lampl I Membrane potential correlates of network decorrelation and improved SNR by cholinergic activation in the somatosensory cortex. J. Neurosci 38, 10692–10708 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pinto L et al. Fast modulation of visual perception by basal forebrain cholinergic neurons. Nat. Neurosci 16, 1857–1863 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Minces V, Pinto L, Dan Y & Chiba AA Cholinergic shaping of neural correlations. Proc. Natl Acad. Sci. USA 114, 5725–5730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sarter M, Parikh V & Howe WM Phasic acetylcholine release and the volume transmission hypothesis: time to move on. Nat. Rev. Neurosci 10, 383–390 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mineur YS & Picciotto MR The role of acetylcholine in negative encoding bias: too much of a good thing? Eur. J. Neurosci 53, 114–125 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dulawa SC & Janowsky DS Cholinergic regulation of mood: from basic and clinical studies to emerging therapeutics. Mol. Psychiatry 24, 694–709 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tye KM Neural circuit motifs in valence processing. Neuron 100, 436–452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jing M et al. An optimized acetylcholine sensor for monitoring in vivo cholinergic activity. Nat. Methods 17, 1139–1146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jing M et al. A genetically encoded fluorescent acetylcholine indicator for in vitro and in vivo studies. Nat. Biotechnol 36, 726–737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Jing et al. (2020), this article describes the design and utility of genetically encoded fluorescent sensors that allow high spatiotemporal monitoring of ACh release in vitro and in vivo, providing a major advance in our understanding of cholinergic transmission relative to circuit function.
- 197.Mineur YS et al. Cholinergic signaling in the hippocampus regulates social stress resilience and anxiety- and depression-like behavior. Proc. Natl Acad. Sci. USA 110, 3573–3578 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hangya B, Ranade SP, Lorenc M & Kepecs A Central cholinergic neurons are rapidly recruited by reinforcement feedback. Cell 162, 1155–1168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses optogenetic tagging to identify cholinergic neurons in the basal forebrain and uses in vivo recordings to demonstrate that BFCNs respond to both negative valence and positive valence stimuli.
- 199.Wilson MA & Fadel JR Cholinergic regulation of fear learning and extinction. J. Neurosci. Res 95, 836–852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kellis DM, Kaigler KF, Witherspoon E, Fadel JR & Wilson MA Cholinergic neurotransmission in the basolateral amygdala during cued fear extinction. Neurobiol. Stress 13, 100279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Unal CT, Pare D & Zaborszky L Impact of basal forebrain cholinergic inputs on basolateral amygdala neurons. J. Neurosci 35, 853–863 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Robinson L, Platt B & Riedel G Involvement of the cholinergic system in conditioning and perceptual memory. Behav. Brain Res 221, 443–465 (2011). [DOI] [PubMed] [Google Scholar]
- 203.Huang L et al. Organizational principles of amygdalar input-output neuronal circuits. Mol. Psychiatry 26, 7118–7129 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hájos N Interneuron types and their circuits in the basolateral amygdala. Front. Neural Circuits 15, 687257 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.O’Leary TP et al. Extensive and spatially variable within-cell-type heterogeneity across the basolateral amygdala. eLife 9, e59003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Teles-Grilo Ruivo LM et al. Coordinated acetylcholine release in prefrontal cortex and hippocampus is associated with arousal and reward on distinct timescales. Cell Rep. 18, 905–917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Buzsáki G Theta oscillations in the hippocampus. Neuron 33, 325–340 (2002). [DOI] [PubMed] [Google Scholar]
- 208.Hasselmo ME The role of acetylcholine in learning and memory. Curr. Opin. Neurobiol 16, 710–715 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lee MG, Chrobak JJ, Sik A, Wiley RG & Buzsáki G Hippocampal theta activity following selective lesion of the septal cholinergic system. Neuroscience 62, 1033–1047 (1994). [DOI] [PubMed] [Google Scholar]
- 210.Lawson VH & Bland BH The role of the septohippocampal pathway in the regulation of hippocampal field activity and behavior: analysis by the intraseptal microinfusion of carbachol, atropine, and procaine. Exp. Neurol 120, 132–144 (1993). [DOI] [PubMed] [Google Scholar]
- 211.Ma X et al. The firing of theta state-related septal cholinergic neurons disrupt hippocampal ripple oscillations via muscarinic receptors. J. Neurosci 40, 3591–3603 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Vandecasteele M et al. Optogenetic activation of septal cholinergic neurons suppresses sharp wave ripples and enhances theta oscillations in the hippocampus. Proc. Natl Acad. Sci. USA 111, 13535–13540 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Norimoto H, Mizunuma M, Ishikawa D, Matsuki N & Ikegaya Y Muscarinic receptor activation disrupts hippocampal sharp wave-ripples. Brain Res. 1461, 1–9 (2012). [DOI] [PubMed] [Google Scholar]
- 214.Joo HR & Frank LM The hippocampal sharp wave–ripple in memory retrieval for immediate use and consolidation. Nat. Rev. Neurosci 19, 744–757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jiang L et al. Cholinergic signaling controls conditioned fear behaviors and enhances plasticity of cortical-amygdala circuits. Neuron 90, 1057–1070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Schmitz TW & Zaborszky L Spatial topography of the basal forebrain cholinergic projections: organization and vulnerability to degeneration. Handb. Clin. Neurol 179, 159–173 (2021). [DOI] [PubMed] [Google Scholar]
- 217.Zhu PK et al. Nanoscopic visualization of restricted nonvolume cholinergic and monoaminergic transmission with genetically encoded sensors. Nano Lett. 20, 4073–4083 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Marvin JS et al. Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nat. Methods 15, 936–939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Marvin JS et al. A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat. Methods 16, 763–770 (2019). [DOI] [PubMed] [Google Scholar]
- 220.Díaz-García CM et al. Quantitative in vivo imaging of neuronal glucose concentrations with a genetically encoded fluorescence lifetime sensor. J. Neurosci. Res 97, 946–960 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zong W et al. Large-scale two-photon calcium imaging in freely moving mice. Cell 185, 1240–1256.e30 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mead TJ & Apte SS Expression analysis by RNAscope™ in situ hybridization. Methods Mol. Biol 2043, 173–178 (2020). [DOI] [PubMed] [Google Scholar]
- 223.Dikshit A, Zong H, Anderson C, Zhang B & Ma X-J in In Situ Hybridization Protocols (eds Nielsen BS & Jones J) 301–312 (Springer, 2020). [Google Scholar]
- 224.Booeshaghi AS et al. Isoform cell-type specificity in the mouse primary motor cortex. Nature 598, 195–199 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Meers MP, Bryson TD, Henikoff JG & Henikoff S Improved CUT&RUN chromatin profiling tools. eLife 8, e46314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Preissl S et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat. Neurosci 21, 432–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Liu H et al. DNA methylation atlas of the mouse brain at single-cell resolution. Nature 598, 120–128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mesulam MM Cholinergic circuitry of the human nucleus basalis and its fate in Alzheimer’s disease. J. Comp. Neurol 521, 4124–4144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mufson EJ, Ginsberg SD, Ikonomovic MD & DeKosky ST Human cholinergic basal forebrain: chemoanatomy and neurologic dysfunction. J. Chem. Neuroanat 26, 233–242 (2003). [DOI] [PubMed] [Google Scholar]
- 230.Boissière F, Lehéricy S, Strada O, Agid Y & Hirsch EC Neurotrophin receptors and selective loss of cholinergic neurons in Alzheimer disease. Mol. Chem. Neuropathol 28, 219–223 (1996). [DOI] [PubMed] [Google Scholar]
- 231.Boskovic Z et al. The role of p75NTR in cholinergic basal forebrain structure and function. J. Neurosci 34, 13033–13038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Mufson EJ, Bothwell M, Hersh LB & Kordower JH Nerve growth factor receptor immunoreactive profiles in the normal, aged human basal forebrain: colocalization with cholinergic neurons. J. Comp. Neurol 285, 196–217 (1989). [DOI] [PubMed] [Google Scholar]
- 233.Liu AK, Chang RC, Pearce RK & Gentleman SM Nucleus basalis of Meynert revisited: anatomy, history and differential involvement in Alzheimer’s and Parkinson’s disease. Acta Neuropathol. 129, 527–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Mesulam MM Behavioral neuroanatomy of cholinergic innervation in the primate cerebral cortex. EXS 57, 1–11 (1989). [DOI] [PubMed] [Google Scholar]
- 235.Coppola JJ & Disney AA Is there a canonical cortical circuit for the cholinergic system? Anatomical differences across common model systems. Front. Neural Circuits 12, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kimes AS et al. Quantification of nicotinic acetylcholine receptors in the human brain with PET: bolus plus infusion administration of 2-[18F]F-A85380. Neuroimage 39, 717–727 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wong DF et al. Brain PET imaging of α7-nAChR with [18F]ASEM: reproducibility, occupancy, receptor density, and changes in schizophrenia. Int. J. Neuropsychopharmacol 21, 656–667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Horti AG, Kuwabara H, Holt DP, Dannals RF & Wong DF Recent PET radioligands with optimal brain kinetics for imaging nicotinic acetylcholine receptors. J. Label. Comp. Radiopharm 56, 159–166 (2013). [DOI] [PubMed] [Google Scholar]
- 239.Tiepolt S et al. PET imaging of cholinergic neurotransmission in neurodegenerative disorders. J. Nucl. Med 63, 33S–44S (2022). [DOI] [PubMed] [Google Scholar]
- 240.Jin H et al. Kinetic modeling of [18F]VAT, a novel radioligand for positron emission tomography imaging vesicular acetylcholine transporter in non-human primate brain. J. Neurochem 144, 791–804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Petrou M et al. In vivo imaging of human cholinergic nerve terminals with (–)-5-18F-fluoroethoxybenzovesamicol: biodistribution, dosimetry, and tracer kinetic analyses. J. Nucl. Med 55, 396–404 (2014). [DOI] [PubMed] [Google Scholar]
- 242.Hillmer AT et al. PET imaging of α7 nicotinic acetylcholine receptors: a comparative study of [18F]ASEM and [18F]DBT-10 in nonhuman primates, and further evaluation of [18F]ASEM in humans. Eur. J. Nucl. Med. Mol. Imaging 44, 1042–1050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lao PJ et al. [18F]Nifene test-retest reproducibility in first-in-human imaging of α4β2* nicotinic acetylcholine receptors. Synapse 10.1002/syn.21981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yuan R, Biswal BB & Zaborszky L Functional subdivisions of magnocellular cell groups in human basal forebrain: test-retest resting-state study at ultra-high field, and meta-analysis. Cereb. Cortex 29, 2844–2858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Fritz HJ et al. The corticotopic organization of the human basal forebrain as revealed by regionally selective functional connectivity profiles. Hum. Brain Mapp 40, 868–878 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Aghourian M et al. Quantification of brain cholinergic denervation in Alzheimer’s disease using PET imaging with [18F]-FEOBV. Mol. Psychiatry 22, 1531–1538 (2017). [DOI] [PubMed] [Google Scholar]
- 247.Schmitz TW, Mur M, Aghourian M, Bedard MA & Spreng RN Longitudinal Alzheimer’s degeneration reflects the spatial topography of cholinergic basal forebrain projections. Cell Rep. 24, 38–46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Xia Y et al. Reduced cortical cholinergic innervation measured using [18F]-FEOBV PET imaging correlates with cognitive decline in mild cognitive impairment. Neuroimage Clin. 34, 102992 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Bartus RT, Dean RL 3rd, Beer B & Lippa AS The cholinergic hypothesis of geriatric memory dysfunction. Science 217, 408–414 (1982). [DOI] [PubMed] [Google Scholar]
- 250.Drachman DA & Leavitt J Human memory and the cholinergic system. A relationship to aging? Arch. Neurol 30, 113–121 (1974). [DOI] [PubMed] [Google Scholar]
- 251.Geula C & Mesulam M-M Systematic regional variations in the loss of cortical cholinergic fibers in Alzheimer’s disease. Cereb. Cortex 6, 165–177 (1996). [DOI] [PubMed] [Google Scholar]
- 252.Geula C & Mesulam MM Cortical cholinergic fibers in aging and Alzheimer’s disease: a morphometric study. Neuroscience 33, 469–481 (1989). [DOI] [PubMed] [Google Scholar]
- 253.Whitehouse PJ et al. Alzheimer’s disease and senile dementia: loss of neurons in the basal forebrain. Science 215, 1237–1239 (1982). [DOI] [PubMed] [Google Scholar]
- 254.Newhouse PA et al. Intravenous nicotine in Alzheimer’s disease: a pilot study. Psychopharmacology 95, 171–175 (1988). [DOI] [PubMed] [Google Scholar]
- 255.Ovsepian SV, O’Leary VB & Zaborszky L Cholinergic mechanisms in the cerebral cortex: beyond synaptic transmission. Neuroscientist 22, 238–251 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Fisher A Cholinergic modulation of amyloid precursor protein processing with emphasis on M1 muscarinic receptor: perspectives and challenges in treatment of Alzheimer’s disease. J. Neurochem 120 (Suppl. 1), 22–33 (2012). [DOI] [PubMed] [Google Scholar]
- 257.Gil-Bea FJ et al. Cholinergic denervation exacerbates amyloid pathology and induces hippocampal atrophy in Tg2576 mice. Neurobiol. Dis 48, 439–446 (2012). [DOI] [PubMed] [Google Scholar]
- 258.Wang YJ et al. p75NTR regulates Aβ deposition by increasing Aβ production but inhibiting Aβ aggregation with its extracellular domain. J. Neurosci 31, 2292–2304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Majdi A et al. Amyloid-β, tau, and the cholinergic system in Alzheimer’s disease: seeking direction in a tangle of clues. Rev. Neurosci 31, 391–413 (2020). [DOI] [PubMed] [Google Scholar]
- 260.Cantero JL et al. Atrophy of basal forebrain initiates with tau pathology in individuals at risk for Alzheimer’s disease. Cereb. Cortex 30, 2083–2098 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wysocka A, Palasz E, Steczkowska M & Niewiadomska G Dangerous liaisons: tau interaction with muscarinic receptors. Curr. Alzheimer Res 17, 224–237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Gamage R et al. Cholinergic modulation of glial function during aging and chronic neuroinflammation. Front. Cell Neurosci 14, 577912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Piovesana R, Salazar Intriago MS, Dini L & Tata AM Cholinergic modulation of neuroinflammation: focus on α7 nicotinic receptor. Int. J. Mol. Sci 22, 4912 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Nazmi A et al. Cholinergic signalling in the forebrain controls microglial phenotype and responses to systemic inflammation. Preprint at bioRxiv 10.1101/2021.01.18.427123 (2021). [DOI] [Google Scholar]
- 265.Leng K et al. Molecular characterization of selectively vulnerable neurons in Alzheimer’s disease. Nat. Neurosci 24, 276–287 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Igarashi KM Entorhinal cortex dysfunction in Alzheimer’s disease. Trends Neurosci. 46, 124–136 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Ovsepian SV, O’Leary VB, Hoschl C & Zaborszky L Integrated phylogeny of the human brain and pathobiology of Alzheimer’s disease: a unifying hypothesis. Neurosci. Lett 755, 135895 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Li S & Sheng ZH Energy matters: presynaptic metabolism and the maintenance of synaptic transmission. Nat. Rev. Neurosci 23, 4–22 (2022). [DOI] [PubMed] [Google Scholar]
- 269.Cavanagh JB The problems of neurons with long axons. Lancet 1, 1284–1287 (1984). [DOI] [PubMed] [Google Scholar]


