Abstract
The effect of three independent variables (i.e., tyrosine, sucrose, and incubation time) on melanin production by Aureobasidium pullulans AKW was unraveled by two distinctive approaches: response surface methodology (i.e. Box Behnken design (BBD)) and artificial neural network (ANN) in this study for the first time ever using a simple medium. Regarding BBD, sucrose and incubation intervals did impose a significant influence on the output (melanin levels), however, tyrosine did not. The validation process exhibited a high consistency of BBD and ANN paradigms with the experimental melanin production. Concerning ANN, the predicted values of melanin were highly comparable to the experimental values, with minor errors competing with BBD. Highly comparable experimental values of melanin were achieved upon using BBD (9.295 ± 0.556 g/L) and ANN (10.192 ± 0.782 g/L). ANN accurately predicted melanin production and showed more improvement in melanin production by about 9.7% higher than BBD. The purified melanin structure was verified by scanning electron microscopy (SEM), energy-dispersive X-ray spectroscopy (EDX), X-ray diffraction pattern (XRD), and thermogravimetric analysis (TGA). The results verified the hierarchical architecture of the particles as small compasses by SEM analysis, inter-layer spacing in the XRD analysis, maximal atomic % for carbon, and oxygen atoms in the EDX analysis, and the great thermal stability in the TGA analysis of the purified melanin. Interestingly, the current novel endophytic strain was tyrosine-independent, and the uniquely applied ANN paradigm was more efficient in modeling the melanin production with appreciate amount on a simple medium in a relatively short time (168 h), suggesting additional optimization studies for further maximization of melanin production.
Subject terms: Biotechnology, Microbiology
Introduction
The curiosity towards natural pigments, especially those from microorganisms, has attracted consumers, since the widespread of synthetic dyes in cosmetics, food processing, textile, and pharmaceuticals became limited, owing to their carcinogenicity, hyper allergenicity, and toxigenicity1,2. The value of microbial pigments includes their high stability, yield, and easy production, as well as their low cost and ease of microbial cultivation3. Microbial melanin production is a talented field of investigation, due to the growth of the industry demanding natural pigments as safe products, effortlessly degradable and eco-friendly1.
Melanin is a dark-brown pigment that is formed by the oxidative polymerization of phenolic compounds, such as glutaminyl-3,4-dihydroxybenzene, catechol, 3,4-dihydroxynaphthalene, or 3,4-dihydroxy-phenylalanine, during the metabolic pathways of fungi, another route is the 3,4-dihydroxyphenylalanine pathway, where tyrosine is catalyzed by tyrosinase and laccase to form dopaquinone, which is oxidized and auto polymerized to melanin3,4. Additionally, melanin production depends upon intra and/or extracellular tyrosinase enzymes, by which several studies attained the kinetic process of activation, inactivation, inducer, and inhibitors of tyrosinase enzyme during growth of Penicillium chrysogenium, Trichderma reesei and Trichoderma harzianum2,5.
Among microbial melanin producers, fungi have a superiority in melanin construction with availability as a potent source, because they are proficient in producing a high yield of the substance in the low-cost culture medium, which crafting the bioprocess to economically practicable on the industrial scale3,4. Several ascomycetous fungi were reported to produce melanin e.g., Aureobasidium pullulans, Neophaeotheca triangularis, Trimmatostroma salinum, and Hortaea werneckii6,7. In comparison to bacteria, these fungi have potential advantages due to the predominancy of two key enzymes, (tyrosinase, and laccase) that play a major role in melanin production8.
Another virgin area of microorganisms had to be investigated and explored for melanin production i.e., the endophytic microbes, which are more efficient in the production of unique biomolecules in appreciated amounts. Endophytes refer to microbial communities that live and grow within plant tissues without causing any harm to the host9.
Melanin has several feasible applications in the medicinal field e.g., antimicrobial, antiviral, antioxidant, antitumor, and anti-inflammatory, as well as, in cosmetics as a protective agent for the skin and eye3,10, and environmental bioremediation of polluted sites by heavy metals11. Owing to these melanin benefits in a variety of applications, there is a growing request for melanin production in a low cost-medium. The bioproduction process could be substantially reduced using a new optimization concept12.
Response surface methodology (RSM) is a multivariate statistical design applied for evaluating the effects of factors, constructing models, and predicting optimal conditions with the minimum number of runs9,13,14. RSM is a statistical technique that models the correlation between multiple independent variables and the response of bioprocess. It is commonly applied to optimize bioprocess performance by identifying the optimal settings for a set of variables that influence the response. RSM is an efficient way for the design and analysis of experiments to optimize bioprocess performance. This, in turn, allows the determination of a strong spot where the parameters can achieve the highest yield and the lowest possible operation parameters, and error9,13.
Central composite designs (CCD), and BBD are both RSM that can be used to fit a full quadratic model. BBD was selected because it does not include axial points that are located outside the cube of the design space to estimate the curvature of the response surface; thus, BBD has fewer design points and saves time and money, compared to that of CCD. These points are not necessary if the region of interest is well-behaved, as it is the case in our study here. For the same reason, BBD is safe to run, because there are no extreme runs, and the factors performed well at their tested limits. Because of the rotatable nature of BBD, it is more robust to noise, meaning that not sensitive to the order of the experimental runs, i.e., does not need to run the design in a specific order15–17.
BBD involves varying the input variables (factors) within certain predetermined ranges and levels, allowing for efficient use of resources and reduction in experimental runs18. The design can also be useful for studying the interactions between variables and identifying the optimal combination of variables that maximizes the response variable19. The review of literature contains a plethora of reports highlighting the potential of RSM, in particular, the BBD, in the optimization of numerous bioproducts like enzymes (α-amylase15, β-xylanase20, pectic oligosaccharides production21, biodegradation of antibiotics in the environment17, and monascus pigment production16, etc.
No previous work was carried out on the optimization of melanin production using artificial intelligence. Artificial neural network (ANN) serves as a core element and one of the basic tools utilized in machine learning. ANN could be considered an advanced extension of RSM and could terrifically, substitute the polynomial regression-based modeling approach of RSM22–24. Like the human brain, an ANN is capable of sophistically analyzing and processing data by efficiently constructing computational models with fully connected nodes within the hidden layer(s). This type of modeling facilitates the learning of data patterns and, as a result, enables accurate decision-making based on historical data. When constructing an ANN model, the network architecture is first selected, followed by the creation of hidden layer(s) with sufficient neurons. The network then undergoes a learning and training process until it grasps the data pattern. Once completed, the resulting ANN model is validated and verified before it is approved as a predictive model. The paradigm of ANN is based on identifying various patterns in the data and detecting any differences to determine the pattern that achieves the desired outcome. This process is regulated through intelligent backpropagation, which generates the desired output model to achieve the objective (target). This procedure is more accurate and can effectively replace other modeling methods22,25.
In this study we aimed to develop a simple medium that contains the minimal requirements for melanin production (i.e., tyrosine, and sucrose, in addition to the incubation period). The reason behind the selection of tyrosine is that the biosynthesis of melanin depends upon tyrosine as the precursor for melanin production3. Many sources of carbon could be metabolized by fungi, but no documented data indicated the role of sucrose or carbon sources in melanin synthesis by fungi, but they can be involved in various metabolic processes by most fungi for energy and growth26. Herein sucrose was selected as a readily available and cheap carbon source. Fermentation time is crucial for melanin production as it determines the duration and extent of melanin synthesis.
However, although the commercial production of melanin from microbial origin has seen significant advancements27, there is still room for improvement. Therefore, our study aimed to optimize melanin production using a combination of BBD and ANN methodologies. To the best of our knowledge, this is the first study to utilize ANN procedure to enhance melanin production from the endophytic fungus, A. pullulans, while utilizing an economical medium source.
Materials and methods
Fungus and inoculum preparation
Aureobasidium pullulans strain AKW was isolated and identified in our previous study28. The stock culture was preserved on potato dextrose agar plates after being incubated at 30 °C for 5 days under dark conditions, and sub-cultured periodically. For the preparation of the standard inoculum, A. pullulans AKW was grown in the same fermentation medium under shaking (100 rpm, 30 °C, 72 h) to obtain a cell count of approximately 3 × 107 cells/mL.
Core melanin production medium
For the melanin production process, a simple fermentation medium i.e., potato sucrose broth was used. The medium was prepared using potato infusion as a base for the fermentation medium. To prepare potato infusion, 200 g of sliced, and unpeeled potatoes were boiled in one-liter distilled water for 30 min, then filtered to remove debris of potato residuals. Different concentrations of tyrosine (as an inducer of melanin production) and sucrose (as a carbon source) were added to the potato infusion (Table 1). The pH of the medium was adjusted to 6.0. Portions of 100 mL medium were transferred into canonical flasks and autoclaved at 121 °C for 15 min. The medium was inoculated by fungal inoculum in a proportion of 5%. The inoculated culture medium was incubated at 30 °C for different periods with a rotation speed of 200 rpm.
Table 1.
Three-factors matrix used for melanin production using A. pullulans AKW, showing the actual and the predicted values of each point of Box-Behnken and ANN models.
| Run | Independent variable | Melanin (g/L) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Actual ± SD | Box-Behnken | ANN | |||||||
| No | Pattern | Tyrosine (%, w/v) | Sucrose (%, w/v) | Time (h) | Predicted | Error | Predicted | Error | |
| 1* | − − 0** | 0.1 | 3 | 168 | 5.871 ± 0.490 | 5.695 | 0.176 | 5.857 | 0.014 |
| 2* | − + 0 | 0.1 | 7 | 168 | 6.954 ± 0.426 | 6.983 | − 0.029 | 6.984 | − 0.030 |
| 3* | + − 0 | 0.5 | 3 | 168 | 5.529 ± 0.152 | 5.501 | 0.029 | 5.494 | 0.035 |
| 4 | + + 0 | 0.5 | 7 | 168 | 6.783 ± 0.227 | 6.959 | − 0.176 | 6.959 | − 0.176 |
| 5 | 0 − − | 0.3 | 3 | 120 | 8.436 ± 0.325 | 8.505 | − 0.069 | 8.375 | 0.061 |
| 6* | 0 − + | 0.3 | 3 | 216 | 3. 743 ± 0.255 | 3.878 | − 0.135 | 3.749 | − 0.006 |
| 7* | 0 + − | 0.3 | 7 | 120 | 8.341 ± 0.321 | 8.206 | 0.135 | 8.221 | 0.120 |
| 8 | 0 + + | 0.3 | 7 | 216 | 6.992 ± 0.673 | 6.923 | 0.069 | 7.074 | − 0.082 |
| 9* | − 0 − | 0.1 | 5 | 120 | 7.904 ± 0.734 | 8.011 | − 0.107 | 7.967 | − 0.063 |
| 10* | + 0 − | 0.5 | 5 | 120 | 8.075 ± 0.523 | 8.035 | 0.040 | 8.167 | − 0.092 |
| 11 | − 0 + | 0.1 | 5 | 216 | 5.149 ± 0.645 | 5.189 | − 0.040 | 5.134 | 0.015 |
| 12 | + 0 + | 0.5 | 5 | 216 | 5.054 ± 0.747 | 4.947 | 0.107 | 4.955 | 0.099 |
| 13* | 000 | 0.3 | 5 | 168 | 9.804 ± 0.850 | 10.184 | − 0.380 | 10.177 | − 0.373 |
| 14* | 000 | 0.3 | 5 | 168 | 10.811 ± 0.539 | 10.184 | 0.627 | 10.177 | 0.634 |
| 15* | 000 | 0.3 | 5 | 168 | 9.973 ± 0.412 | 10.184 | − 0.247 | 10.177 | − 0.240 |
*Ten runs were used for the training process, whereas the other 5 runs were used for the validation process.
**The pattern represents the level composition of each variable within each of the tested runs, where + is the high level, − is the low level, and 0 is the middle concentration.
Separation and purification of melanin
The protocol of El-Gamal et al.29 and Müjdeci30 were used for the separation and purification of melanin with minor modifications. Briefly, melanin was separated from the microbial cell pellets by centrifugation for 15 min at 3000×g. The resulting cell-free filtrate containing melanin was precipitated by reducing the pH down to 2.0 by HCl (6 M) and kept for 4 h under cooling. The precipitate was centrifuged at 7000×g for 15 min. The precipitated melanin was washed with distilled water. The process was repeated four times to obtain purified melanin. The purified pigment was stored (at − 20 °C) after lyophilization.
Design of Box-Behnken matrix
This investigation aimed to study the best combination of the most important factors affecting melanin production on a simple medium. The study hypotheses that tyrosine (the precursor for melanin production), sucrose (the most common carbon source), and incubation time are significant factors. The three independent variables were optimized for maximization of melanin production using the BBD of RSM31. Based on the matrix of BBD, the independent factors were tested at three levels (low, middle, or high). Accordingly, 15 runs were generated that contain three center points (middle levels). Table 1 displays the experimental design matrix, which includes the various combinations of the three variables. Each combination was repeated three times. Upon performing the experimental trials as previously stated, melanin production was measured as the target response to the three-factors matrix. Then, the collected data were statistically analyzed to find out the relationship between the three independent variables and melanin production to predict the optimum level of each factor, the next Eq. (1) was applied:
| 1 |
where Y is the forecasted melanin production, β0 is the model constant, βi is the linear coefficient, βij is the cross-product coefficient, βii is the quadratic coefficient, and Xi, and Xj are the independent variables.
The equation was used to find out the connection between independent variables and melanin production, as well as the prediction of the optimum concentration of each factor. The analysis of variance (ANOVA) and coefficient of determination (R2) were performed to determine the significance of variables and ensure the goodness of the model. Moreover, the BBD precision was visually assessed by plotting the predicted values against the actual values and examining the distribution of the data points.
Modeling melanin production with ANN
The obtained data from the BBD matrix was utilized for developing a predictive model with machine learning. The ANN model was created by training a fully connected ANN with two hidden layers, in which all nodes were equipped with a hyperbolic tangent sigmoid; exp(-x2) activation function. The established forecast model was a fully connected algorithm of multilayer perceptron.
Accordingly, the data were split into (i) the training set that was used to reduce the error and establish weights at each neuron of the ANN, (ii) the validation set that was used to tune hyperparameters and monitor model performance during training to halt the ANN training and select the best model, and finally (iii) the testing set that was used to evaluate the final trained model, as an external dataset that was solely used to test the ANN robustness and serve as the final valuation of prediction capacities; this external dataset was not used during the model development process32.
The ANN topology was investigated utilizing several hidden layers and nodes. The ANN topology was designated as 3-h-1. Three neurons of the tested factors (i.e., tyrosine, sucrose, and incubation time), represent the input layer. The output layer is composed of one neuron (fungal melanin production). Additional hidden layers were created between the two input and output layers, in which a range of neurons from 3 to 10 was tested.
ANN was trained using trial-and-error at various specific parameters (learning rate, learning method, and the ideal neurons number in the hidden layer). For each trial phase, a 10,000 tour was used. Then, the optimum architectural structure that fits the best model was identified. The trial-and-error process was employed untill the lowest error was for the root average squared error (RASE) and average absolute error (AAE). The coefficient of variation (R2) between the predicted and actual values was also used to evaluate the training process.
The precision of the trained ANN was determined by comparing its predicted outputs to the actual values of melanin production. If the predicted values were close to or equivalent the actual values, then the ANN was deemed to have high precision in its predictions. This evaluation was usually done by calculating a statistical measure of accuracy, RASE, AAE, and R2. Additionally, the ANN's precision was visually assessed by plotting the predicted values against the actual values and examining the distribution of the data points24.
Verification
The optimum level of each of the tested factors that maximize melanin production was theoretically estimated using the fitted model of both BBD and ANN, then the expected melanin production was estimated. A verification trial was performed, applying the optimum settings in the laboratory in triplicates and compared with the predicted melanin values to test the accuracy of both models.
Software and statistical analysis
Data of melanin production were presented as the mean ± standard deviation. The design of the BBD matrix and the statistical analysis were performed using the JMP® Pro 17 software (JMP®, SAS Institute Inc., Cary, NC). The software was also utilized for conducting machine learning procedures, constructing the ANN topology, and implementing the training, validation, and testing procedures.
Characterization methods
The particles’ surface morphology of the purified melanin pigment was inspected by scanning electron microscope (SEM)33. The gold-coated melanin that had been previously lyophilized was examined using SEM-type FEI Czech (accelerating voltage at 25 kV), which consists of an electron optical column, vacuum system, and electronics. The electromagnetic lenses focused the electrons into a fine spot onto melanin. The electron gun, tungsten filament at the top of the column produced the electron beam, which is focused into a fine spot less than 4 nm in diameter on the melanin specimen. This beam is scanned in rectangular rashers. The compositional pattern was determined by energy-dispersive X-ray (EDX) spectroscopy33, which is connected to SEM.
Thermogravimetric analysis (TGA) was run on Perkin Elmer 400033. The X-Ray diffraction pattern analysis (XRD) was run at Pan Analytical Philips according to Khouqeer et al.34. The Bragg equation (Eq. 2) was applied to calculate the inter-layer spacing d. The average grain (crystalline) size of melanin was estimated by the Dedye-Schrerrer equation (Eq. 3). The percentage of the melanin crystallinity was correspondingly calculated as stated in (Eq. 4), considering glass substrate as a background.
| 2 |
where, θ ≡ diffraction angle; m ≡ diffraction order; λ ≡ X-ray wavelength at the considered first-order diffraction (m = 1).
| 3 |
where, λ denotes the wavelength (1.54) and FWHM ≡ full width at half maximum of a diffraction peak.
| 4 |
Results
A simple medium of the yeast-like fungus, A. pullulans AKW, was sequentially optimized for melanin production via BBD and ANN paradigms. The resulting melanin was characterized based on SEM, XRD, EDX, and TGA properties.
Modeling melanin production by BBD
Data from Table 1 displays the pattern of a 15-run design matrix of the different combinations of the three variables together with the experimental melanin production. Data obtained revealed a substantial variation in melanin production with the best levels combination at the center point (middle level), being 5.69 ± 0.39 (run number 14).
The statistical weight of the tested variables
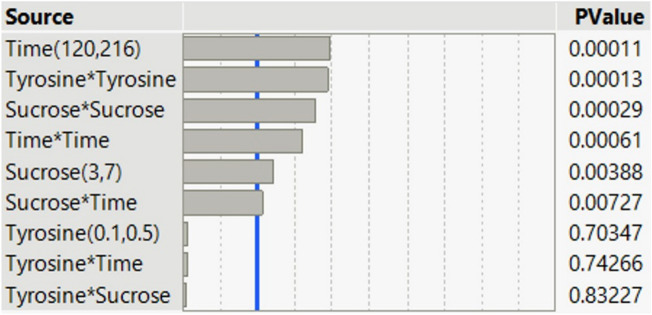
The relative single, interaction and quadratic importance of the tested factors were quantified using the P-value for comparison. The statistical weight (Fig. 1) of the tested variables was used to detect which predictor (variable) is the most significant. The most important variable has the lowest P-value. Likewise, the single effect of incubation time and sucrose was significant, whereas tyrosine level did not show any significant influence on melanin production. Regarding the interaction effect, the interaction between tyrosine with the other two factors (incubation time and sucrose level) showed a non-significant effect. Eventually, the three variables showed a significant quadratic effect.
Figure 1.
The relative single, interaction, and quadratic importance of the tested factors on melanin production by A. pullulans AKW.
ANOVA and regression analysis
The data of BBD were analyzed by ANOVA (Table 2). The overall model is significant (F-value = 42.4805) with a very low P-value (0.0003), on the other side, the lack of fit error is not statistically significant (P-value = 0.9206). Other model evaluation statistics were additionally estimated i.e., the R2, adjusted-R2, and pridected-R2 values, which were found to be extremely high, being 0.9871, 0.9639, and 0.9382, respectively.
Table 2.
ANOVA analysis of Box-Behnken design for melanin production by A. pullulans AKW.
| Source | Degree of freedom | Sum of Squares | F-ratio | Prob > F |
|---|---|---|---|---|
| Overall model | 9 | 56.139 | 42.481 | 0.001 |
| Error | ||||
| Lack of fit | 3 | 0.136 | 0.151 | 0.9206 |
| Pure error | 2 | 0.599 | ||
| Total error | 5 | 0.734 | ||
| Single | ||||
| Tyrosine (0.1, 0.5%, w/v) | 1 | 0.024 | 0.163 | 0.7035 |
| Sucrose (3, 7%, w/v) | 1 | 3.769 | 25.667 | 0.0039 |
| Time (120, 216 h) | 1 | 17.458 | 118.895 | 0.0001 |
| Interaction | ||||
| Tyrosine*Sucrose | 1 | 0.007 | 0.050 | 0.8323 |
| Tyrosine*Time | 1 | 0.018 | 0.121 | 0.7427 |
| Sucrose*Time | 1 | 2.796 | 19.039 | 0.0073 |
| Quadratic | ||||
| Tyrosine*Tyrosine | 1 | 16.534 | 112.602 | 0.0001 |
| Sucrose*Sucrose | 1 | 11.746 | 79.996 | 0.0003 |
| Time*Time | 1 | 8.557 | 58.278 | 0.0006 |
| Corrected total | 14 | 56.873528 | ||
| Model evaluation statistics | ||||
| Determination coefficient (R2) | 0.9871 | |||
| Adjusted-R2 | 0.9639 | |||
| Pridected-R2 | 0.9382 | |||
The single, interaction, and quadratic terms were also investigated by ANOVA. Apart from tyrosine, sucrose and incubation time were significant. Concerning the interaction among variables, the interaction between sucrose level and incubation time has a significant effect (P-value = 0.0073). Otherwise, all the quadratic terms showed a significant effect (P-value ≤ 0.05) on melanin production mediated by A. pullulans AKW.
Consequently, the regression coefficients of the various terms of the model were computed and obeyed to a polynomial function of the second order. The predicted melanin production by A. pullulans AKW was estimated by the next equation:
where Y is the predicted melanin production, X1 is tyrosine (%, w/v), X2; is sucrose (%, w/v), and X3 is the incubation time (h).
As could be noticed, some model terms (X1, X3, X1X3, and all quadratic terms) showed negative coefficient values, whereas the others had positive coefficient values. However, such an equation was used for the prediction of melanin production values, which were found to be very close to the experimental ones and therefore had lower errors. Based on the above regression model, the values predicted by BBD were estimated and compared with actual values. Both estimated and actual values at each data point exhibited high agreement (Table 2).
Modeling melanin production by ANN
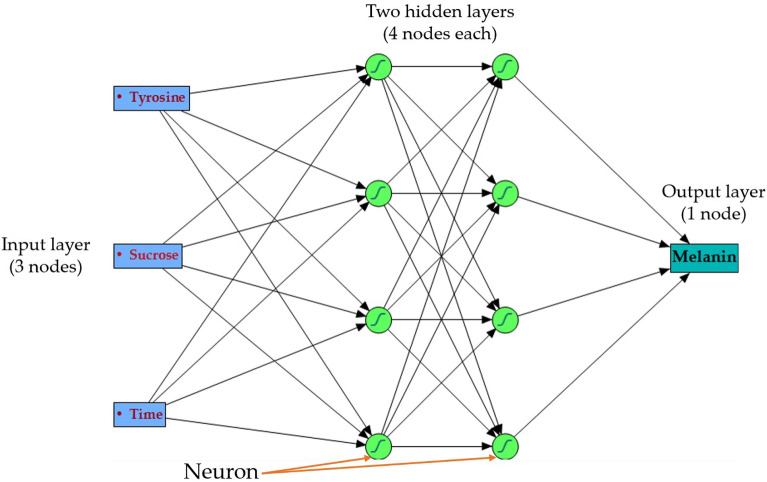
A multilayer feed-forward ANN design was employed in a neural network platform that is fully connected to construct a predictive ANN model. During the trial-and-error procedure, the holdback propagation ratio was used at 0.3333, which divided the data of BBD into two sets, the first set having 10 runs for training to reduce, prediction error and create neural weights. The second set is composed of 5 runs for validation to stop ANN training and choose the finest model. After several trials (each of 10,000 tours) with various ANN arrangements. The penalty of ANN topology was best achieved at a squared learning rate of 0.2, with two hidden layers each with 4 nodes. The activation function in all nodes of both hidden layers of the network was hyperbolic tangent sigmoid (NTanH). As a result, a four-layer ANN topology structure (Fig. 2), denoted as 3–4–4–1, was determined to be the optimal architecture. The input layer consists of three neurons (independent factors), which are tyrosine, sucrose, and incubation time. The neuron-output layer represents the response factor of melanin production. The two hidden layers demonstrated superior performance when employing the activation function of NTanH(4)NTanH2(4) in the hidden neurons.
Figure 2.
The ANN layout of melanin production by A. pullulans AKW. The ANN contains an input layer (three neurons), two hidden layers (4 neurons, each), and the output layer (one neuron).
To test the generality of the model, trials were made to minimize errors during the machine-learning process. The ANN was trained until the R2 value was maximized, achieving a score of 0.9857 for training, and 0.9935 for validation.
The trained network's ability to accurately predict outputs was evaluated. The predicted values for each point of the BBD data were computed using the ANN (Table 1). The values predicted by ANN exhibited high consistency with the investigational values and displayed lower error values than those of the BBD model.
Assessment of BBD and ANN models
Residual analysis, and linear relationships
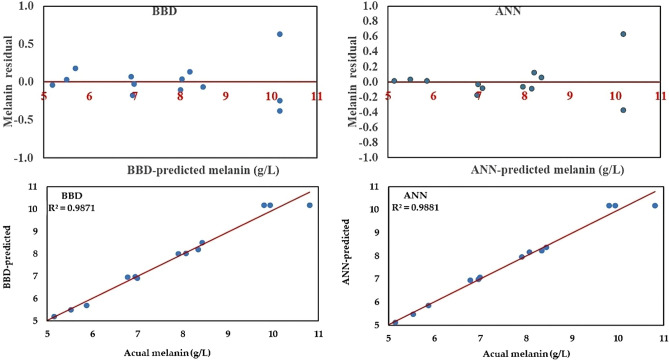
Residual analysis was performed for further assessment of both models to predict melanin production by A. pullulans AKW. The plotted residuals versus predicted values by both models (Fig. 3) revealed an equal distribution of the residual points that were scattered closely, and equally around the 0-axis without exhibiting linearity.
Figure 3.
Plots of the BBD and ANN, showing the predicted values of melanin production versus residual, and actual data.
The predicted model points against the actual experimental values of both models (Fig. 3) were also investigated. The points of both models lie near the line of perfect prediction, implying that the model can approach the actual experimental data faithfully. However, the linear regression showed that the ANN model's predictions are significantly closer to the prediction line compared to the BBD model.
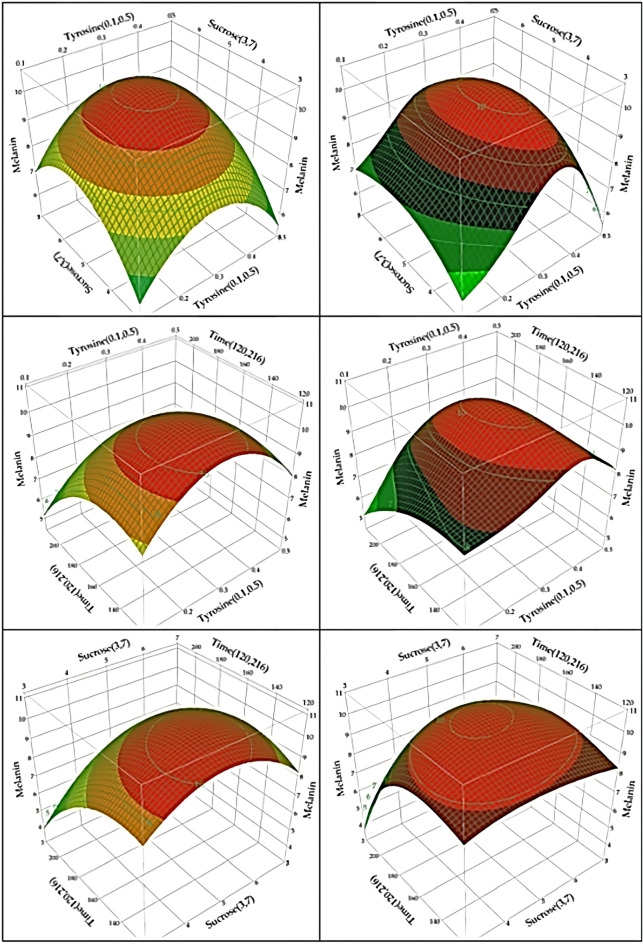
Three-dimensional (3D) plots
To know the mutual, and interactive effects between every pair of variables, the 3D plots of both BBD and ANN were constructed (Fig. 4). The 3D plots depicted the relationships between the variables, demonstrating an increase in melanin production as the concentration of the three variables (i.e., tyrosine, sucrose, and incubation time) approached the optimum level. However, once the optimum level was exceeded, a decline in melanin production was observed. It can be noticed that higher levels of the three variables have a negative effect on the response.
Figure 4.
Three-dimensional response surface plots of melanin production by A. pullulans AKW, represented as a pairwise combination of the tested factors, keeping the third-factor constant at its center (middle) level, based on the models of BBD (represented by the right column) and ANN (represented by the left column).
Compared with BBD, ANN demonstrates an elliptical curve of the response surface plot between each pair of the tested factors. The 3D plots of BBD showed various patterns, and no specific prototype could be drawn for every pair of the tested factors, where the elliptical curve appeared only on the interaction between tyrosine and sucrose levels. In general, the 3D plot indicates that the melanin production process reached its maximum value around the central points of the experimental design.
Comparison of the models
Both the BBD and ANN models exhibited strong predictive ability and lower residuals, indicating that they can accurately fit actual investigational data and make highly reliable predictions. To evaluate the precision of these models in predicting melanin production, various statistical parameters were assessed during both the training and validation processes, as well as for the overall models (Table 3). Based on the performance statistics, the ANN model was more confident than the BBD model, where the R2 value of the overall ANN model was higher than these of the BBD model. On the other hand, RASE and AAE values recorded lower values for the ANN model than for the BBD model.
Table 3.
Measuring the performance statistics, prediction, and the actual response of BBD and ANN models for melanin production by A. pullulans AKW.
| Model | R2 | RASE | AAE | Frequency | |
|---|---|---|---|---|---|
| Training | BBD | 0.9845 | 0.2609 | 0.1905 | 10 |
| ANN | 0.9857 | 0.2506 | 0.1607 | 10 | |
| Validation | BBD | 0.9933 | 0.1034 | 0.0921 | 5 |
| ANN | 0.9935 | 0.1016 | 0.0868 | 5 | |
| Overall | BBD | 0.9871 | 0.2212 | 0.1577 | 15 |
| ANN | 0.9881 | 0.2128 | 0.1360 | 15 | |
| Variable | Predicted value | ||||
|---|---|---|---|---|---|
| BBD | ANN | ||||
| Predicted optimum conditions | |||||
| Tyrosine (%) | 0.299 | 0.329 | |||
| Sucrose (%) | 5.168 | 4.870 | |||
| Incubation time (h) | 145.825 | 167.994 | |||
| Validation of response | |||||
| Predicted melanin production (g/L) | 10.554 | 10.286 | |||
| Experimental melanin production (g/L) | 9.295 ± 0.556 | 10.192 ± 0.782 | |||
RASE root average squared error, AAE average absolute error.
Model verification
A verification experiment was conducted based on the predicted values of each factor by both models, to assess the modeling process used in the optimization study of melanin production. The predicted optimal conditions were calculated separately by BBD and ANN models (Table 3). Then the calculated values of the three tested factors were experimentally validated. Under the theoretical optimum conditions for the three variables, the response of melanin production was calculated to be 10.555 and 10.286 g/L for BBD and ANN, respectively. The validation process revealed high consistency with the experimental melanin production, being 9.295 ± 0.556 g/L (BBD) and 10.192 ± 0.782 g/L (ANN). It is worth mentioning that both models showed high accuracy, but ANN surpassed the BBD model in predicting melanin production, achieving a 9.7% increase in melanin production over BBD.
Characterization of melanin
Following the preceding conditions, a mass production of melanin was conducted and subsequently isolated for the investigation of the chemical structure. The obtained melanin exhibits a black, granular appearance (Fig. 5). Generally, melanin is a heteropolymer consisting of diverse subunits, resulting in a complex, amorphous, and insoluble molecular structure. However, the specific composition of melanin varies depending on its source. Therefore, traditional methods face challenges when attempting to examine its constitution. Consequently, researchers employ several methods to explore melanin structure.
Figure 5.

A close-up vision of melanin particles biosynthesized by A. pullulans AKW.
Scanning electron microscopy investigation
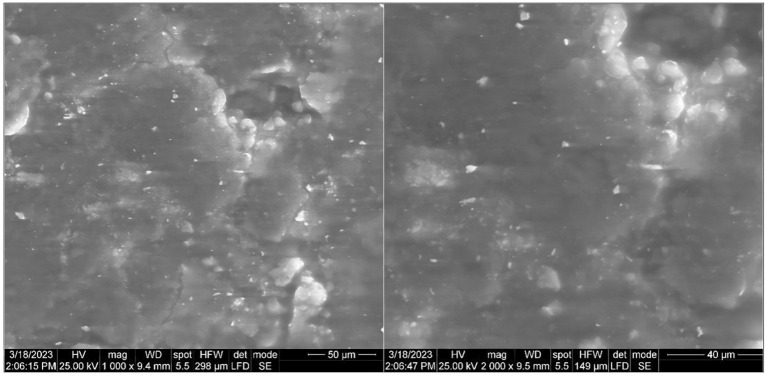
SEM analysis was applied to investigate the morphological features and architecture of melanin. The analysis provided a topography with high-magnification images of the surface of the melanin particles (Fig. 6). The hierarchical architecture of melanin particles produced by A. pullulans AKW was investigated by SEM. Generally, the particles of melanin polymer were shown in small compasses and aggregated.
Figure 6.
SEM micrographs of melanin pigment particles produced by A. pullulans AKW at the different indicated magnifications.
X-ray diffraction pattern analysis
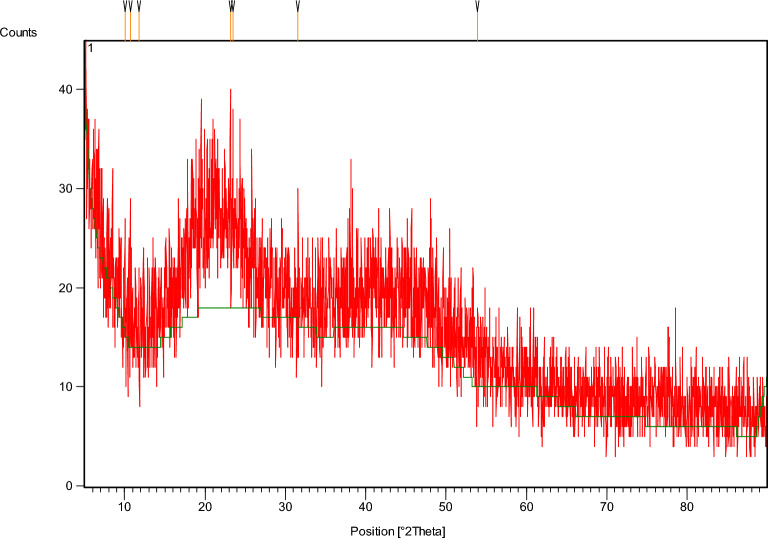
The prepared melanin pigment was analyzed by XRD analysis to investigate its material type, phase, crystallographic properties, chemical composition, and physical features. The analysis depends on the constructive interference between the crystalline melanin and the monochromatic X-rays. The chart of XRD analysis is shown in Fig. 7, and the data is presented in Table 4.
Figure 7.
The XRD analysis of melanin.
Table 4.
The XRD analysis data of melanin.
| Pos. (°2Theta) | Height (cts) | FWHM (°2Theta) | d-spacing (Å) | Rel. int. (%) | Tip width (°2Theta) |
|---|---|---|---|---|---|
| 10.0341 | 11.50 | 0.0787 | 8.81557 | 52.27 | 0.0945 |
| 10.6947 | 14.69 | 0.0590 | 8.27246 | 66.76 | 0.0708 |
| 11.7830 | 7.72 | 0.0984 | 7.51074 | 35.09 | 0.1181 |
| 23.1501 | 22.00 | 0.0787 | 3.84216 | 100.00 | 0.0945 |
| 23.4513 | 11.89 | 0.1968 | 3.79350 | 54.02 | 0.2362 |
| 31.5697 | 13.60 | 0.0394 | 2.83406 | 61.82 | 0.0472 |
| 53.8896 | 7.41 | 0.0960 | 1.69994 | 33.66 | 0.1152 |
Energy-dispersive X-ray spectroscopy
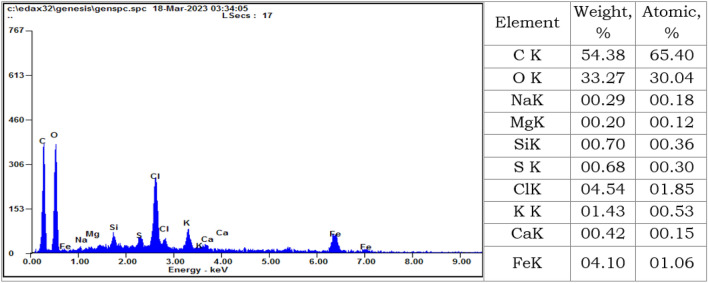
The EDX analysis as an analytical technique was implemented for melanin pigment as shown in Fig. 8. The EDX analysis was applied to estimate the elemental analysis and chemical composition of the purified melanin pigment.
Figure 8.
The EDX analysis of the purified melanin pigment.
Thermogravimetric analysis
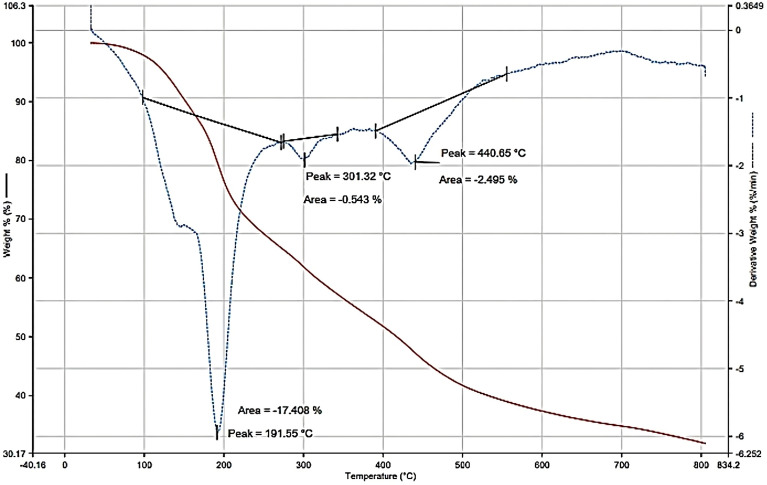
The TGA was applied to investigate the thermal properties of the purified melanin pigment. Thus, the results as shown in Fig. 9 demonstrated the initial loss in the mass with a peak at 191.55 °C with indication of no residual adsorbed water.
Figure 9.
Thermogravimetric analysis of melanin.
Discussion
The microbial fermentation process for optimizing melanin production is an auspicious procedure since microbial melanin is eco-friendly, biocompatible, and economical. Additionally, melanin polymer has a broad spectrum of applications, such as bioactive molecule (antioxidant, sunscreen, and antimicrobial agent), and could also be used in functional biomaterials as a sensitizer in dye-sensitized solar cells, and frame polymer for metal nanoparticles, as well as in bioremediation of metal-pollutant sites and extracellular electronic materials in microbial fuel cells3,35. Thus, the current study has been organized to maximize melanin production by the endophytic; A. pullulans AKW, using BBD, while ANN paradigm was applied for the first time.
Concerning the BBD paradigm, the center (middle) points of the tested factors achieved the maximum melanin production, being 10.811 ± 0.539 g/L (run 14). These results indicate that the selected level of each factor was accurate enough. As evidenced by the high F-values and low P-values in ANOVA, most of the model terms demonstrated a significant effect and lack of significant model error in comparison to the pure error, confirming the accuracy of the obtained data and the experimental design. Another, when the model has a high F-value and a non-significant lack of fit, it is indicative of adequate fitness. Contrary to tyrosine, incubation time and sucrose level attain a significant trend.
Commonly, the P-value is employed to detect the significance of the coefficients of various model terms. The significance level at the alpha ≤ 0.05 threshold signifies the incidence of a strong association between the tested variables and the response (melanin production)25.
The F-test hypothesizes no variations (null hypothesis, H0) between the model terms on the response. Instead, rejected H0 indicates the model terms are varied from each other (alternative hypothesis, H1). A large P-value (> 0.05) provides strong evidence to accept H0.13,36. Based on ANOVA, the H0 was rejected meaning the significance of the tested factors and reliability of the model in predicting melanin production by A. pullulans AKW within the investigated space.
Furthermore, for the model to be reliable, R2, adjusted R2, and pridected-R2 must be ≥ 0.75 and relatively near to each other36. The high R2 (0.9871), adjusted-R2 (0.9639), and pridected-R2 (0.9382) imply a good regression model, further, the relationship between the three tested variables and the melanin production is tight. A much lower predicted R2 than R2 is a warning sign that the model is overfitting the data, this is not applied to our case, where they are very close to each other25.
Based on the estimated coefficients the optimum level of the three variables, and the melanin response were predicted, which was very close to the actual melanin production, suggesting additional evidence for the accuracy of the model, and the design space, as well. The negative coefficient value indicates an antagonistic relationship between melanin production and the variable concentration in the tested design space, and vice versa. However, several studies reported BBD as an operative in signifying the factors reliable for melanin maximization, for instance, the optimization of the medium composition enhanced the production of melanin by Aspergillus fumigatus12.
The optimal ANN topology structure was denoted as 3–4–4–1, consisting of three input neurons, one output neuron, and two hidden layers of 4 neurons each, employing the activation function of NTanH(4)NTanH2(4).
Although artificial intelligence has become increasingly prevalent in recent scientific research, there has been no prior work on the modulation of microbial melanin production using ANN. This study supports this type of modeling.
ANN paradigm is elastic, able to predict well, and capable to learn any nonlinear function. Therefore, ANN can generate an efficient model from any type of data. The ANN platform utilizes a machine learning algorithm together with a flexible function for the input variable(s) for building a fully connected multilayer perceptron. This architecture uses indirect intermediate layers to predict the target melanin production by A. pullulans AKW. This modeling approach is more suitable when it is not necessary to elucidate the correlation between the response, and the inputs22–24.
Comparing the predicted vis residual values of both BBD and ANN models showed an even distribution along the 0-axis sides. A tight cluster of points was noted around the diagonal line. This distribution pattern confirms the high precision and adequacy of both models. Moreover, the residuals and the linear regression analysis of the ANN declared that both predicted and actual points were located very close to the line of ideal prediction than those of the BBD model, designating a well-trained ANN model, with better generality and higher precision.
The oval-shaped curve of the 3D plot for both BBD, and ANN indicates a strong and clear interaction between every pair of the tested factors, suggesting a careful choice of the factors, and the model fits the design well9.
However, contrarily to ANN, the prototype of ANN was not identical to BBD because ANN can discover complex nonlinear relationships between the inputs (tyrosine, sucrose, and incubation period) and the melanin production. ANN can identify concealed connections between input and output variables, even when they don't seem to have an explicit correlation. This is due to the two intermediate layers in ANN that manage unique correlation between inputs and output, rather than finding a direct route like observed in BBD. Thus, ANN is an excellent forecaster when the relationships between the output and input are not needed or identifiable22–25.
The performance of both models was evaluated by measuring RASE and AAE. ANN showed lower values for both errors’ measurements. Although the variation between the statistics of each model was small, the ANN model has a better generalization capability than BBD in melanin production. Experimental validation of the theoretical response of melanin production that was calculated by both models indicates the validity of both models.
ANN can predict with high accuracy because it can handle the system's nonlinearity better than other models that need only one-step calculation to approach the target22,23. The comparison of models from ANN and BBD requires some consideration of the differences between these two approaches. ANN is a type of machine learning model that mimics the behavior of the human brain, while BBD is a statistical approach used to optimize response variables based on experimental designs. Therefore, to compare models from ANN and BBD, it is required to clearly define the prediction accuracy, model interpretability, computational complexity, or ease of implementation. ANN models are often considered black-box models, while BBD models are typically more interpretable. Moreover, the modeling technique of ANN involves the building of network architecture, instituting the hidden layers and node numbers in each layer, training, and lastly, justification and authenticating the obtained model. Thus, the ANN paradigm is putatively more accurate because it encompasses all data points of experiments22–25. Furthermore, the robustness and generalization performance of ANN, especially on the new data, is better than the BBD model. Indeed, the ANN-robustness to noise or outliers surpassed BBD22,23. Therefore, the decision on the optimum conditions for melanin production was made based on the ANN model.
However, ANN has some limitations, such as taking longer computational time, for training and prediction, due to many iterations, and not showing the importance and influence of each factor in the model, which makes it hard to remove or simplify any factors from the model22,32.
Commonly, the regulation of melanin production depends on certain enzymes, i.e. tyrosinase with essentiality of sulfhydryl and lysyl residues in the enzyme active site5. Other nutritional conditions, e.g. glucose, peptone, and yeast extract are widely utilized in the nutritional medium. Moreover, numerous recent studies aimed to exploit agricultural residues for melanin productivity, such as corn steep liquor, fruit waste extract, and wheat bran, to minimize the cost, with a high yielding of melanin37. Additionally, there are several studies attaining optimization of fungal melanin, for example, Ribera et al.38 optimized melanin synthesis by Armillaria borealis with a yield of 11.58 g/L in a medium containing glucose, yeast extract, and tyrosine, over 97 days. Another study optimized melanin synthesis by the fungus, Aspergillus fumigatus (0.1 g/L) in a medium containing dextrose and peptone without tyrosine after 10 days12. The optimized melanin has been reported by Auricularia auricula (2.97 g/L), in a medium containing lactose, yeast extract, and tyrosine after 8 days39. Whereas, Gliocephalotrichum simplex produced melanin at 6.6 g/L after 6 days in a medium containing peptone and yeast extract40. However, in Table 5, the productivity of organisms in previous studies was calculated as g/day and compared with the current melanin yield by A. pullulans AKW was stated.
Table 5.
Productivity of melanin by microorganisms including A. pullulans AKW relative to other previous studies.
| Organism | Yield (g/L) | Incubation period (h) | Productivity (g/day) | References |
|---|---|---|---|---|
| Fungi | ||||
| Aureobasidium pullulans AKW | 10.19 | 168 | 1.46 | Current study |
| Gliocephalotrichum simplex | 6.60 | 144 | 1.10 | Jalmi et al.40 |
| Aspergillus fumigatus | 0.10 | 240 | 0.01 | Raman et al.12 |
| Auricularia auricula | 2.97 | 192 | 0.37 | Sun et al.39 |
| Armillaria borealis | 11.58 | 2328 | 0.12 | Ribera et al.38 |
| Armillaria cepistipes | 27.98 | 3864 | 0.17 | |
| Daldinia concentrica | 1.78 | 1752 | 0.02 | |
| Bacteria | ||||
| Zymomonas mobilis | 3.20 | 120 | 0.64 | Chávez-Béjar et al.41 |
| Nocardiopsis alba MSA10 | 3.40 | 168 | 0.49 | Kiran et al.42 |
| Bacillus safensis | 6.90 | 24 | 6.90 | Tarangini and Mishra43 |
| Pseudomonas stutzeri | 6.70 | 10 | 16.08 | Choi8 |
| Actinomycetes | ||||
| Streptomyces kathirae SC-1 | 13.70 | 120 | 2.74 | Guo et al.44 |
| Streptomyces sp. ZL-24 | 4.20 | 120 | 0.84 | Wang et al.45 |
| Streptomyces puniceus RHPR9 | 0.39 | 168 | 0.06 | Polapally et al.46 |
In comparison to these previous studies, based on the ANN paradigm, our strain (A. pullulans AKW) produced an appreciated amount of melanin (10.192 g/L) after a shorter incubation period (168 h) on a simple potato infusion medium, containing tyrosine (0.329%), and sucrose (4.870%). The biotechnological potential of the current strain in melanin production may be attributed to its constitutive production by the fungus that emerged as an endophytic fungus.
The endophytic microbiome, also known as endosymbiont, refers to a wide range of bacteria and/or fungi that live and develop inside or between cells of all types of plants without causing any harm to the host plant during at least some part of their life cycle. Endophytes are a newly emerged area of interest owing to their unique features9.
As revealed by BBD, the current A. pullulans AKW is tyrosine independent for melanin production. Following our results, Pseudomonas stutzeri produced melanin (6.7 g/L) within 10 h of incubation without tyrosine supplementation8. Contrarily, Surwase et al.47 found that L-tyrosine at 1.872 g/L enhanced melanin production by Brevundimonas sp SGJ, being 6.811 g/L. Generally, the metabolic pathways of melanin are depending upon the tyrosinase enzyme, which is a copper-dependent biocatalyst involved in ortho-specific hydroxylation and subsequent oxidation of monophenols like tyrosine8, in addition to the laccase enzyme which catalyzes the oxidation of a broad range of substrates like tyrosine including dihydroxy phenols and quinones48. Additionally, the mechanism of melanin could be conducted throughout different pathways, depending upon the random polymerization of a few building blocks e.g., L-tyrosine metabolites of indole-5,6-quinone, 5,6-dihydroxyindole carboxylic acid, 5,6-dihydroxyquinone carboxylic acid, dopamine, homogentisate, and some phenolic precursors49.
In general, two pathways are suggested for the production of microbial melanin, the first is the DOPA pathway which is catalyzed by tyrosinase and laccase, in which tyrosine is converted to L-DOPA, then to dopaquinone. The latter is extremely active and spontaneously oxidized and auto-polymerized to create melanin3. In the second DHN-pathway, malonyl-CoA is generated endogenously, then catalyzed by polyketide synthases, the sequential decarboxylative condensation of five malonyl-CoA molecules generates 1,3,6,8-tetrahydroxynaphthalene, which is subjected to cycles of reduction and dehydration reaction, generating 1,8-dihydroxynaphthalene (DHN), that polymerized finally to DHN melanin3. Another hypothesis for melanin synthesis in microorganisms suggested the formation of hydroxylated aromatic molecules, during the catabolic metabolism, e.g., such as homogentisic acid that accumulates due to enzymatic imbalance or interruption, this process generates other various sorts of melanin50. On this base, our data suggest that the current melanin of A. pullulans AKW is biosynthesized independently from tyrosine.
It is already known that the structure of melanin is complicated, in which it has various forms depending upon the polymerization pathway, building blocks, and enzymes, melanin is classified into several groups, including eumelanin, pyomelanin, pheomelanin, neuromelanin and allomelanin8. Therefore, the current melanin characteristics were elucidated.
The SEM investigation of melanin particle architecture confirmed the particles in small compasses in a definite crystal shape. Herein, the extracted pigmented melanin particles with average size lie between 150 and 160 nm. Commonly, the graphic particles of melanin showed to have a definite crystal shape, besides the particles shown in small compasses.
The XRD spectrum of the produced melanin sample is characterized by a broad peak, which is frequently perceived in amorphous and disordered materials balanced at about 23.1501. The Bragg reflection peak 2θ values were found for seven diffraction peaks at 10.0341, 10.6947, 11.7830, 23.1501, 23.4513, 31.5697, and 53.8896 consistent with tip width (°2Theta) 0.0945, 0.0708, 0.1181, 0.0945, 0.2362, 0.0472, and 0.1152, respectively. This peak is recorded owing to X-ray diffraction from parallel planer layers.
The Bragg equation was applied to calculate the inter-layer spacing d. The calculated values of d were found at 8.81557, 8.27246, 7.51074, 3.84216, 3.79350, 2.83406, and 1.69994 for the melanin sample. These findings are matched with the literature value of the inter-layer spacing in the stacked sheets model of melanin. The quality of the purified melanin was indicated by the proximity of the values of the grain size.
Moreover, the percentage of the crystallinity of melanin was calculated considering a glass substrate as a background. The melanin sample recorded the relative intensity percentages at 52.27, 66.76, 35.09, 100.0, 54.02, 61.82, and 33.66% for the seven diffraction peaks, respectively. These findings, regarding the lack of diffraction patterns related to the sample crystallinity, are in agreement with the results reported by Tarangini and Mishra43, and Xin et al.51. The crystallinity lack referred to the reliable physical features of melanin52.
The EDX analysis is particularly dependent on the interaction of X-ray excitation with the analyzed sample. The results indicated the amplest elements with the superlative compositions for carbon and oxygen atoms with weight of 54.38, and 33.27%, and atomic percent at of 65.40, and 30.04%, respectively. Our findings are in perfect conformity in all aspects with the results reported by Tarangini and Mishra53. The data also specified minor elements in the analysis with a total atomic % of 4.55%, this verified the purity of the produced melanin. The quantified minor elements were inferred as sodium/K (0.18%), magnesium/K (0.12%), silicon/K (0.36%), sulfur/K (0.30%), chlorine/K (1.85%), potassium/K (0.53%), calcium/K (0.15%), and iron/K (1.06%). The variation in the composition of melanin is supported by the factor of the change in the composition of the medium. Ribera et al.38 have supposed that infrequent elements such as sodium, potassium, and sulfur are interrelated to the growth medium composition. In addition, a considerable preliminary indication for the sample purity is the increased percentage of carbon besides the attendance of residual peptone in the raw melanin.
Likewise, the mass loss at 191.55 °C in the TGA analysis referred to the decomposition of peptone beside other residual growth media. Based on the literature reports54, the aliphatic chains or constituents decomposed below 400 °C, but the aromatic constituents started to decompose above 400 °C. regarding our melanin sample, the first decomposition occurred with the weight loss rate induced by 33% at 191.55 °C. The results in all characteristics indicated respectable thermal stability of melanin with a rate of weight loss of less than 20% unit 800 °C. These results regarding the intensive resistance of melanin to thermal degradation are in harmony with the previous literature55. It was worth mentioning that the raw melanin decomposed with loss of weight at 325 °C recording the highest loss in mass, while melanin was decomposed at 350 °C, these findings were formerly reported by Ribera et al.38 and approved with our results. Accordingly, the second loss in mass was recorded at 301.32 °C with a loss in mass lower than 80%, and the third loss in mass was recorded at 440.65 °C without significant loss in mass. Consequently, the residual-mass quantity for the solid sample of melanin did not show substantial variance (78%).
Conclusion
To sum up, the optimization process of melanin was conducted by BBD and ANN models. l-tyrosine, sucrose, and incubation interval were the independent variables encountered in the BBD matrix. Both models were efficient in the determination of the best conditions for maximum melanin production. However, the ANN paradigm showed a superiority in the optimization process, yielding 10.192 ± 0.782 g/L. ANN accurately predicted melanin production and showed more improvement in melanin production by about 9.7% higher than BBD. Interestingly, L-tyrosine has no significant effect during the optimization process of melanin. The structure features of the produced melanin were established by SEM, XRD, EDX, and TGA analysis. SEM analysis depicted the aggregation of the melanin particles in small compasses. The XRD analysis specified the inter-layer spacing in the stacked sheets, and the crystallinity lack is referred to as a reliable physical feature. The EDX analysis identified the maximal atomic % for carbon and oxygen atoms at 65.40, and 30.04%, respectively. The peptone decomposition in the TGA was recorded with the weight loss rate induced by 33% at 191.55 °C. Importantly, our results are thus the first to optimize melanin production by artificial intelligence paradigm on a simple medium using the new endophytic A pullulans AKW. The current work suggests developing new methods for the production of melanin by A. pullulans AKW on a higher scale, thus allowing its involvement in different products. Furthermore, a research paper could be performed to focus on the current state of the genetics behind microbial melanin production, as well as the potential for genetic engineering to improve microbial melanin production (Supplementary Information S1 and S2).
Supplementary Information
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSP2023R114), King Saud University, Riyadh, Saudi Arabia.
Author contributions
Conceptualization, W.I.A.S., A.G., F.A.-O., F.M., and K.E.; data curation, M.E.-H., F.M., and K.E.; formal analysis, A.G., M.E.-H., N.E., F.M., and K.E.; investigation, A.G., F.A.-O., N.E., and K.E.; methodology, W.I.A.S., A.G., N.E., and K.E.; project administration, F.A.-O.; resources, F.A.-O.; software, W.I.A.S.; supervision, W.I.A.S., M.E.-H., N.E., and K.E.; validation, F.A.-O., M.E.-H., N.E., and F.M.; visualization, W.I.A.S., A.G. and F.M.; writing—review and editing, W.I.A.S., A.G., M.E.-H., and F.M. All authors have read and agreed to the published version of the manuscript.
Data availability
All data generated or analyzed during this study are included in this published article and the supplementary files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
WesamEldin I. A. Saber, Email: wesameldin.saber@arc.sci.eg, Email: wiasaber@gmail.com
Fatimah O. Al-Otibi, Email: falotibi@ksu.edu.sa
Noha M. Eldadamony, Email: nohamohamadt@gmail.com
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-40549-z.
References
- 1.Shindy A. Problems and solutions in colors, dyes and pigments chemistry: A review. Chem. Int. 2017;3:97–105. [Google Scholar]
- 2.El-Shora HM, Khateb AM, Darwish DB, El-Sharkawy RM. Thiolation of myco-synthesized Fe3O4-NPs: A novel promising tool for Penicillium expansium laccase immobilization to decolorize textile dyes and as an application for anticancer agent. J. Fungi. 2022;8:71. doi: 10.3390/jof8010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Naggar NE-A, Saber WI. Natural melanin: current trends, and future approaches, with especial reference to microbial source. Polymers. 2022;14:1339. doi: 10.3390/polym14071339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venil CK, Lakshmanaperumalsamy P. An insightful overview on microbial pigment, prodigiosin. Electron. J. Biol. 2009;5:49–61. [Google Scholar]
- 5.El-Shora HM, El-Sharkawy RM. Tyrosinase from Penicillium chrysogenum: Characterization and application in phenol removal from aqueous solution. J. Gen. Appl. Microbiol. 2020;66:323–329. doi: 10.2323/jgam.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Chung D, Kim H, Choi HS. Fungi in salterns. J. Microbiol. 2019;57:717–724. doi: 10.1007/s12275-019-9195-3. [DOI] [PubMed] [Google Scholar]
- 7.Nazareth, S. W. The world of halophilic fungi. (2014).
- 8.Choi, K.-Y. Bioprocess of microbial melanin production and isolation. Front. Bioeng. Biotechnol. 1127 (2021). [DOI] [PMC free article] [PubMed]
- 9.Al-Askar, A. A. et al. A novel endophytic Trichoderma longibrachiatum WKA55 with biologically active metabolites for promoting germination and reducing mycotoxinogenic fungi of peanut. Front. Microbiol. 769 (2022). [DOI] [PMC free article] [PubMed]
- 10.Kumar CG, Mongolla P, Pombala S, Kamle A, Joseph J. Physicochemical characterization and antioxidant activity of melanin from a novel strain of Aspergillus bridgeri ICTF-201. Lett. Appl. Microbiol. 2011;53:350–358. doi: 10.1111/j.1472-765X.2011.03116.x. [DOI] [PubMed] [Google Scholar]
- 11.Mahmoud YA-G. Uptake of radionuclides by some fungi. Mycobiology. 2004;32:110–114. [Google Scholar]
- 12.Raman NM, Shah PH, Mohan M, Ramasamy S. Improved production of melanin from Aspergillus fumigatus AFGRD105 by optimization of media factors. AMB Express. 2015;5:1–9. doi: 10.1186/s13568-015-0161-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saber WI, El-Naggar NE, El-Hersh MS, El-Khateeb AY. An innovative synergism between Aspergillus oryzae and Azotobacter chroococcum for bioconversion of cellulosic biomass into organic acids under restricted nutritional conditions using multi-response surface optimization. Biotechnology. 2015;14:47–57. [Google Scholar]
- 14.El-Sharkawy RM, Swelim MA, Hamdy GB. Aspergillus tamarii mediated green synthesis of magnetic chitosan beads for sustainable remediation of wastewater contaminants. Sci. Rep. 2022;12:9742. doi: 10.1038/s41598-022-13534-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matrawy, A. A., Khalil, A. I. & Embaby, A. M. Bioconversion of bread waste by marine psychrotolerant Glutamicibacter soli strain AM6 to a value-added product: Cold-adapted, salt-tolerant, and detergent-stable α-amylase (CA-AM21). Biomass Convers. Biorefinery, 1–18 (2022).
- 16.Embaby AM, Hussein MN, Hussein A. Monascus orange and red pigments production by Monascus purpureus ATCC16436 through co-solid state fermentation of corn cob and glycerol: An eco-friendly environmental low cost approach. PLoS ONE. 2018;13:e0207755. doi: 10.1371/journal.pone.0207755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anan A, Ghanem KM, Embaby AM, Hussein A, El-Naggar MY. Statistically optimized ceftriaxone sodium biotransformation through Achromobacter xylosoxidans strain Cef6: An unusual insight for bioremediation. J. Basic Microbiol. 2018;58:120–130. doi: 10.1002/jobm.201700497. [DOI] [PubMed] [Google Scholar]
- 18.El-Metwally M, El-Hersh M, Saber W. A dual microbial culture for improving C/N ratio and multi-response optimization of rice straw fermentation for bioethanol production. Res. J. Microbiol. 2017;12:210–217. [Google Scholar]
- 19.Al-Askar A, Saber W, Ghoneem K, Rashad Y. Oxalic acid as the main molecule produced by Trichoderma asperellum MG323528 fermented on corn stover based medium. Biotechnology. 2018;17:95–103. [Google Scholar]
- 20.Matrawy AA, Khalil AI, Marey HS, Embaby AM. Use of wheat straw for value-added product xylanase by Penicillium chrysogenum strain A3 DSM105774. J. Fungi. 2021;7:696. doi: 10.3390/jof7090696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Embaby, A. M., Melika, R. R., Hussein, A., El-Kamel, A. H. & Marey, H. S. A Novel non-cumbersome approach towards biosynthesis of pectic-oligosaccharides by non-aflatoxigenic Aspergillus sp. Section Flavi Strain EGY1 DSM 101520 through citrus pectin fermentation. Plos one11, e0167981 (2016). [DOI] [PMC free article] [PubMed]
- 22.Saber WI, et al. Rotatable central composite design versus artificial neural network for modeling biosorption of Cr6+ by the immobilized Pseudomonas alcaliphila NEWG-2. Sci. Rep. 2021;11:1–15. doi: 10.1038/s41598-021-81348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ram Talib, N. S., Halmi, M. I. E., Abd Ghani, S. S., Zaidan, U. H. & Shukor, M. Y. A. Artificial neural networks (ANNs) and response surface methodology (RSM) approach for modelling the optimization of chromium (VI) reduction by newly isolated Acinetobacter radioresistens strain NS-MIE from agricultural soil. BioMed. Res. Int.2019 (2019). [DOI] [PMC free article] [PubMed]
- 24.Moussa Z, Darwish DB, Alrdahe SS, Saber WI. Innovative artificial-intelligence-based approach for the biodegradation of feather keratin by Bacillus paramycoides, and cytotoxicity of the resulting amino acids. Front. Microbiol. 2021;12:731262. doi: 10.3389/fmicb.2021.731262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elsayed, A. et al. Optimization of heavy metals biosorption via artificial neural network: A case study of cobalt (II) sorption by pseudomonas alcaliphila NEWG-2. Front. Microbiol.13 (2022). [DOI] [PMC free article] [PubMed]
- 26.Suwannarach N, Kumla J, Watanabe B, Matsui K, Lumyong S. Characterization of melanin and optimal conditions for pigment production by an endophytic fungus, Spissiomyces endophytica SDBR-CMU319. PLoS ONE. 2019;14:e0222187. doi: 10.1371/journal.pone.0222187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran-Ly AN, Reyes C, Schwarze FW, Ribera J. Microbial production of melanin and its various applications. World J. Microbiol. Biotechnol. 2020;36:1–9. doi: 10.1007/s11274-020-02941-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saber WI, Al-Askar AA, Ghoneem KM. Exclusive biosynthesis of pullulan using Taguchi’s approach and decision tree learning algorithm by a novel endophytic Aureobasidium pullulans strain. Polymers. 2023;15:1419. doi: 10.3390/polym15061419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-Gamal M, El-Bialy H, Elsayed M, Khalifa M. Isolation and characterization of melanized yeast form of Aureobasidium pullulans and physiological studies on the melanization process. J. Nucl. Sci. Technol. 2017;5:57–72. [Google Scholar]
- 30.Müjdeci GN. Experimental modeling and optimization of melanin production by Aureobasidium pullulans NBRC 100716 in carrot peel extract. Environ. Prog. Sustain. Energy. 2022;41:e13919. [Google Scholar]
- 31.Box GE, Behnken DW. Some new three level designs for the study of quantitative variables. Technometrics. 1960;2:455–475. [Google Scholar]
- 32.Elsayed, M. S., Eldadamony, N. M., Alrdahe, S. S. & Saber, W. I. Definitive screening design and artificial neural network for modeling a rapid biodegradation of date palm fronds by a new Trichoderma sp. PWN6 into citric acid. Molecules26, 5048 (2021). [DOI] [PMC free article] [PubMed]
- 33.Rudrappa, M. et al. Bioproduction, purification and physicochemical characterization of melanin from Streptomyces sp. strain MR28. Microbiol. Res.263, 127130 (2022). [DOI] [PubMed]
- 34.Khouqeer G, et al. Preparation and characterization of natural melanin and its nanocomposite formed by copper doping. Nano Select. 2022;3:1598–1608. [Google Scholar]
- 35.Lee, H. S. et al. Melanin biopolymer synthesis using a new melanogenic strain of Flavobacterium kingsejongi and a recombinant strain of Escherichia coli expressing 4-hydroxyphenylpyruvate dioxygenase from F. kingsejongi. Microb. Cell Fact.21, 75 (2022). [DOI] [PMC free article] [PubMed]
- 36.El-Hersh MS, Saber W, El-Fadaly HA, Mahmoud MK. Lysine and glutamic acids as the end products of multi-response of optimized fermented medium by Mucor mucedo KP736529. Pak. J. Biol. Sci.: PJBS. 2016;19:279–288. doi: 10.3923/pjbs.2016.279.288. [DOI] [PubMed] [Google Scholar]
- 37.Zou Y, Tian M. Fermentative production of melanin by Auricularia auricula. J. Food Process. Preserv. 2017;41:e12909. [Google Scholar]
- 38.Ribera J, et al. Scalable biosynthesis of melanin by the basidiomycete Armillaria cepistipes. J. Agric. Food Chem. 2018;67:132–139. doi: 10.1021/acs.jafc.8b05071. [DOI] [PubMed] [Google Scholar]
- 39.Sun S, Zhang X, Chen W, Zhang L, Zhu H. Production of natural edible melanin by Auricularia auricula and its physicochemical properties. Food Chem. 2016;196:486–492. doi: 10.1016/j.foodchem.2015.09.069. [DOI] [PubMed] [Google Scholar]
- 40.Jalmi P, Bodke P, Wahidullah S, Raghukumar S. The fungus Gliocephalotrichum simplex as a source of abundant, extracellular melanin for biotechnological applications. World J. Microbiol. Biotechnol. 2012;28:505–512. doi: 10.1007/s11274-011-0841-0. [DOI] [PubMed] [Google Scholar]
- 41.Chávez-Béjar MI, et al. Metabolic engineering of Escherichia coli to optimize melanin synthesis from glucose. Microb. Cell Fact. 2013;12:1–12. doi: 10.1186/1475-2859-12-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiran GS, et al. Melanin-templated rapid synthesis of silver nanostructures. J. Nanobiotechnol. 2014;12:1–13. doi: 10.1186/1477-3155-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarangini K, Mishra S. Production of melanin by soil microbial isolate on fruit waste extract: Two step optimization of key parameters. Biotechnol. Rep. 2014;4:139–146. doi: 10.1016/j.btre.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo J, et al. High-level production of melanin by a novel isolate of Streptomyces kathirae. FEMS Microbiol. Lett. 2014;357:85–91. doi: 10.1111/1574-6968.12497. [DOI] [PubMed] [Google Scholar]
- 45.Wang, L., Li, Y. & Li, Y. Metal ions driven production, characterization and bioactivity of extracellular melanin from Streptomyces sp. ZL-24. Int. J. Biol. Macromol.123, 521–530 (2019). [DOI] [PubMed]
- 46.Polapally R, et al. Melanin pigment of Streptomyces puniceus RHPR9 exhibits antibacterial, antioxidant and anticancer activities. PLoS ONE. 2022;17:e0266676. doi: 10.1371/journal.pone.0266676. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Surwase, S. N., Jadhav, S. B., Phugare, S. S. & Jadhav, J. P. Optimization of melanin production by Brevundimonas sp. SGJ using response surface methodology. 3 Biotech3, 187–194 (2013). [DOI] [PMC free article] [PubMed]
- 48.Nagai M, et al. Important role of fungal intracellular laccase for melanin synthesis: Purification and characterization of an intracellular laccase from Lentinula edodes fruit bodies. Microbiology. 2003;149:2455–2462. doi: 10.1099/mic.0.26414-0. [DOI] [PubMed] [Google Scholar]
- 49.Li S, Yang L, Li J, Chen T, Ye M. Structure, molecular modification, and anti-radiation activity of melanin from Lachnum YM156 on ultraviolet B-induced injury in mice. Appl. Biochem. Biotechnol. 2019;188:555–567. doi: 10.1007/s12010-018-2898-9. [DOI] [PubMed] [Google Scholar]
- 50.Eisenman HC, Casadevall A. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 2012;93:931–940. doi: 10.1007/s00253-011-3777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xin, C. et al. Preparation of melanin from Catharsius molossus L. and preliminary study on its chemical structure. J. Biosci. Bioeng.119, 446–454 (2015). [DOI] [PubMed]
- 52.Thathachari Y, Blois M. Physical studies on melanins: II X-ray diffraction. Biophys. J. 1969;9:77–89. doi: 10.1016/S0006-3495(69)86370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tarangini, K. & Mishra, S. Production, characterization and analysis of melanin from isolated marine Pseudomonas sp. using vegetable waste. Res. J. Eng. Sci.2278, 9472 (2013).
- 54.Abthagir PS, Saraswathi R. Charge transport and thermal properties of polyindole, polycarbazole and their derivatives. Thermochim. Acta. 2004;424:25–35. [Google Scholar]
- 55.Pralea I-E, et al. From extraction to advanced analytical methods: The challenges of melanin analysis. Int. J. Mol. Sci. 2019;20:3943. doi: 10.3390/ijms20163943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and the supplementary files.