Abstract
The emergence and re-emergence of antibiotic-resistant bacteria is a potential threat to treating infectious diseases. This study employed a nanometer-scale green synthesis using an extract of Solanum incanum leaves to obtain nanoparticles (NPs) and nanocomposites (NCs) possessing antibacterial properties. The FESEM-EDS elemental mapping analysis proved the novelty of the green synthesis approach in synthesizing a copper-doped ZnO NCs with good dopant distribution. The crystallinity and ZnO bandgap were adjusted by extrinsic copper doping in the ZnO lattice. The optical property adjustments from 3.04 to 2.97 eV for indirect Kubelka–Munk functions were confirmed from DRS-UV-vis analysis. The dopant inclusion in the host lattice was also confirmed by the angle shift on the XRD pattern analysis relative to single ZnO. In addition to doping, the XRD pattern analysis also showed the development of CuO crystals. The lattice fringe values from HRTEM analysis confirmed the existence of both CuO and ZnO crystals with local heterojunctions. Doping and heterojunctions have crucial values in charge transfer and visible light harvesting behaviour, as proved by the PL analysis. The synergistic effects of the doped NCs showed greater antibacterial activity against both Gram-positive and Gram-negative bacteria as a result of more ROS generation through the bacteria–cell–catalyst interaction and release of metal ions. The antioxidant potential of the doped NCs was found to be higher than that of single NPs, using the 2,2-diphenyl-1-picrylhydrazyl free radical scavenging assay and is expected to impart protective effects to the host cells by scavenging destructive free radicals. Thus, the overall analysis leads to the conclusion that the potentiality of synthesized materials has a future outlook for biological applications, especially in the development of antimicrobials to combat antibiotic-resistant bacteria and microbes.
Green synthesis approach-based synthesis of nanoscale-sized doped nanocomposites for enhanced antibacterial activity.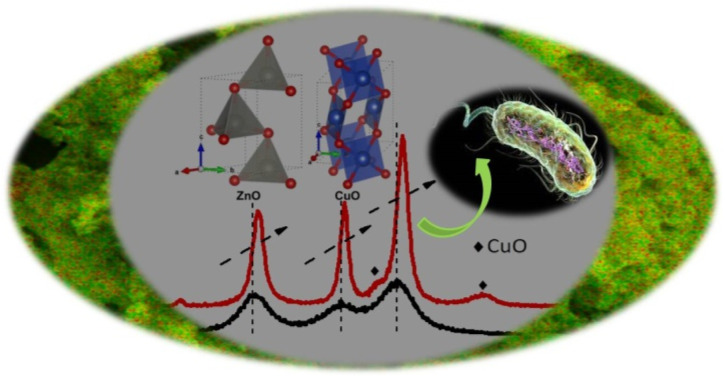
Introduction
The rapid spread of bacterial infections and the massive occurrence of antibiotic-resistant bacteria are increasing the worldwide mortality rate, estimated to reach up to 10 million deaths every year by 2050 with a treatment cost of 100 trillion USD.1,2 In 2014, the World Health Organization (WHO) recognized antimicrobial resistance (AMR) as a major threat to global health in response to the massive increase in populations of multidrug-resistant strains worldwide.3,4 Various factors, including antibiotic misuse and abuse, agricultural antibiotic use, and accessible migration routes, have been reported to drive the spread of antimicrobial resistance.4 Multidrug resistance (MDR) and AMR give rise to infections such as hospital-acquired, ulcerative skin, lungs, urinary tract (UTI), catheters, ear, eye, and Gingivitis, leading to increased medical costs and marked morbidity and mortality. Clinically relevant bacteria such as Escherichia coli, Klebsiella pneumoniae, Enterobacter spp., Proteus spp., Staphylococcus aureus, Pseudomonas aeruginosa, Acinetobacter spp., and methicillin-resistant S. aureus (MRSA), among others, have been implicated in infections, AMR and MDR, respectively.5,6 AMR and MDR patterns in Gram-negative and Gram-positive bacteria give rise to infections that are either difficult to treat or impossible to cure with conventional antimicrobials.5 Thus, finding solutions to combat this widespread challenge associated with bacteria and other microbes becomes an urgent and nonstop issue. Zinc oxide (ZnO) is one of the current choices as a novel antibacterial material that can generate reactive oxygen species (ROS). It is one of the wide-bandgap (3.34 eV) semiconductor materials with an exciton band energy of 60 meV. ZnO has wide applications due to its unique properties, although with some drawbacks, such as being incapable of harvesting the majority of the solar spectrum (the visible range) and having photon-induced e−/h+ recombination properties.7 Transition metal or metal ion inclusion, or creating a defect or shallow state within the ZnO bandgap, has been extensively studied and found to be one of the crucial techniques used to advance the drawbacks of ZnO and increase its biomedical application.8
Doping of copper in the ZnO lattice creates an accepter level near the ZnO conduction band at a 0.17 eV energy level and near the conduction band at about 0.45 eV, which consequently induces green photoluminescence emission.9 The ionic radii of copper ions (0.073 nm) are comparable with those of Zn ions (0.073 nm), which create significant optoelectrical and biological property changes in ZnO after inclusion.10 Besides, oxides of copper, a narrow bandgap material, have extraordinarily good properties for biocidal applications.11 Copper doping causes the creation of independent copper oxide crystals with an increase in copper concentrations. To indicate, the copper-doped ZnO study by Morales-Mendoza and his groups12 and Mahmoud and his groups13 revealed the development of a copper-independent peak above six wt% and four wt% copper levels, respectively. Of course, the formation of CuO–ZnO heterojunction (p–n heterojunction (type II, staggered gap type)) can diminish the e−/h+ recombination properties, consequently enhance the charge separation and visible light harvesting properties, and increase stability and reusability as well.14,15 The synergistic effects of CuO–ZnO heterojunction on antibacterial activity have also been reported.10
There are several nanoscale material synthesis technologies for both chemical and physical approaches. However, costs, hazardous chemical usage, and toxic byproducts are some of the many disadvantages associated with these approaches. Thus, the use of the biosynthesis approach, specifically using plant metabolites such as stabilizing, reducing, and gelating agents, received attention due to its easy availability, low cost, and biocompatibility.16,17Solanum incanum L. leaves, the source of several groups of phytochemicals and multiple metabolites, is one of the medicinal plants used as a reducing and capping agent.18 There are recent reports regarding the green synthesis of ZnO nanoparticles (NPs) and copper-doped ZnO nanocomposites (NCs) for different applications. For example, Doan Thi et al. synthesized ZnO NPs in the presence of orange fruit peel extract as a biological reducing agent.19 Besides, Khan et al.,20 Karthik et al.,8 Bekru et al.,14 and Truong et al.21 synthesised copper-doped ZnO nanocomposites using plant extracts of Clerodendrum infortunatum, Synadium grantii, Erbascum sinaiticum benth, and Piper chaudocanum L., respectively. To the best of our knowledge, S. incanum L. leaf extract has never been used for the green synthesis of copper-doped ZnO NCs. This work also gives insight into the chemistry of doping and heterojunction in detail, which are crucial parameters for the enhancement of the properties of the materials in several applications, such as antibacterials.
Thus, this study aims to synthesize the ZnO-, CuO-NPs, and copper-doped ZnO NCs using a green approach in the presence of S. incanum L. leaves extract for the development of antimicrobial agents. The FESEM-EDS analysis showed the presence of a good dopant distribution with high purity. The nanoscale size, crystallinity, and heterojunction formation between CuO and ZnO were confirmed from the TEM-HRTEM-SAED analyses. The XRD pattern analysis also confirmed the greater crystalline nature and high-angle shift due to doping. The DRS-UV-vis and PL analyses confirmed the presence of visible optical property improvements (redshift) for doped materials compared to single ZnO. In addition, the formation of an intimate local contact between CuO and ZnO was confirmed from the XRD and HRTEM analyses, which is crucial in harvesting the visible light spectrum.15 The doping and heterojunction strategies also improve material charge transfer properties without recombination. The continuous charge transfer process (electron–hole relaxation) results in a reaction with oxygen and water that produces a high oxidizing agent, ROS, which is beneficial for several applications such as catalysis and antibacterial activity. This greater ROS generation and novel antibacterial activity for the doped composite than single ZnO were also confirmed in this study. Besides, we believe that this study gives crucial clues regarding the chemistry of doping in general.
Results and discussion
Characterizations
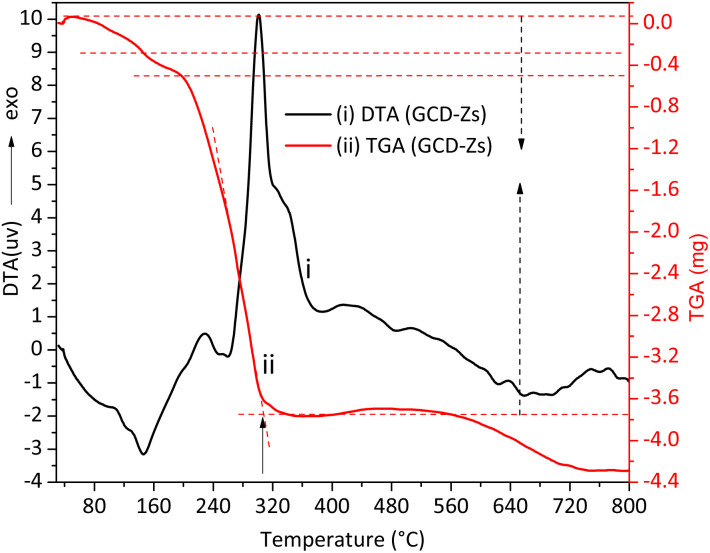
The surface energy of nanoscale-sized materials is much greater than that of their bulk counterparts, which assists in their aggregation with one another.22 Besides, the coalescence or aggregation of NPs is greatly assisted by temperature.23 Thus, the maximum calcination temperature applied for the removal of plant organic matter after its role as a capping, reducing, and gelating agent should be optimized. Fig. 1 shows the TGA-DTA thermal stability analysis result of the green synthesized copper-doped zinc oxide nanocomposites (GCD-Zs) precursor-plant complex organic matter. The amounts taken for the analysis were 7.530 and 10.001 mg for ZnO and GCD-Zs, respectively. The total weight loss for ZnO and GCD-Zs was −3.001 and −4.029 mg, respectively.
Fig. 1. The thermal gravimetric analysis and differential thermal (TGA-DTA) analysis for the GCD-Zs precursor-plant complex before calcination. The total plant decomposition occurred at about 315 °C.
The first two weight losses on the TGA curve within the temperature range of 50–300 °C are due to surface or interstitial absorbed water molecule removal, or both. However, within this 50–300 °C temperature range, the decomposition of some organic substances may also occur.19 The respective DTA stability loss endothermic peaks due to water removal and organic substance decomposition were detected at 150 and 250 °C, respectively. The third greatest mass loss on the TGA curve detected in the temperature range of 250–420 °C (see the respective broad and intense exothermic DTA peak) is due to the total precursor and left-over organic matter decomposition to give pure metal oxide crystal. In the thermal stability analysis result of the zinc precursor-plant complex, the first two weight losses within the temperature range of 50–130 °C are due to water molecule removal and decomposition of some organic substances. The stability peaks were detected at 64 and 117 °C, respectively. The third mass loss within the 190–420 °C range is due to the total decomposition of precursor and left-over organic matter to give pure metal oxide crystal (Fig. S1†). The GCD-Zs precursor-plant complex has greater stability than the zinc precursor-plant complex, which indicates that the copper precursor is acting as a catalyst for the degradation of the plant extract and is also increasing material crystal structure perfection.24 Thus, 450 °C was selected for the next consecutive characterization and application experiments. The selected temperature is greater than the TGA optimal temperature of the zinc precursor-plant complex (403 °C) and the GCD-Zs precursor-plant complex (315 °C) for the purpose of decomposition of the undercapped leftover organic matter.
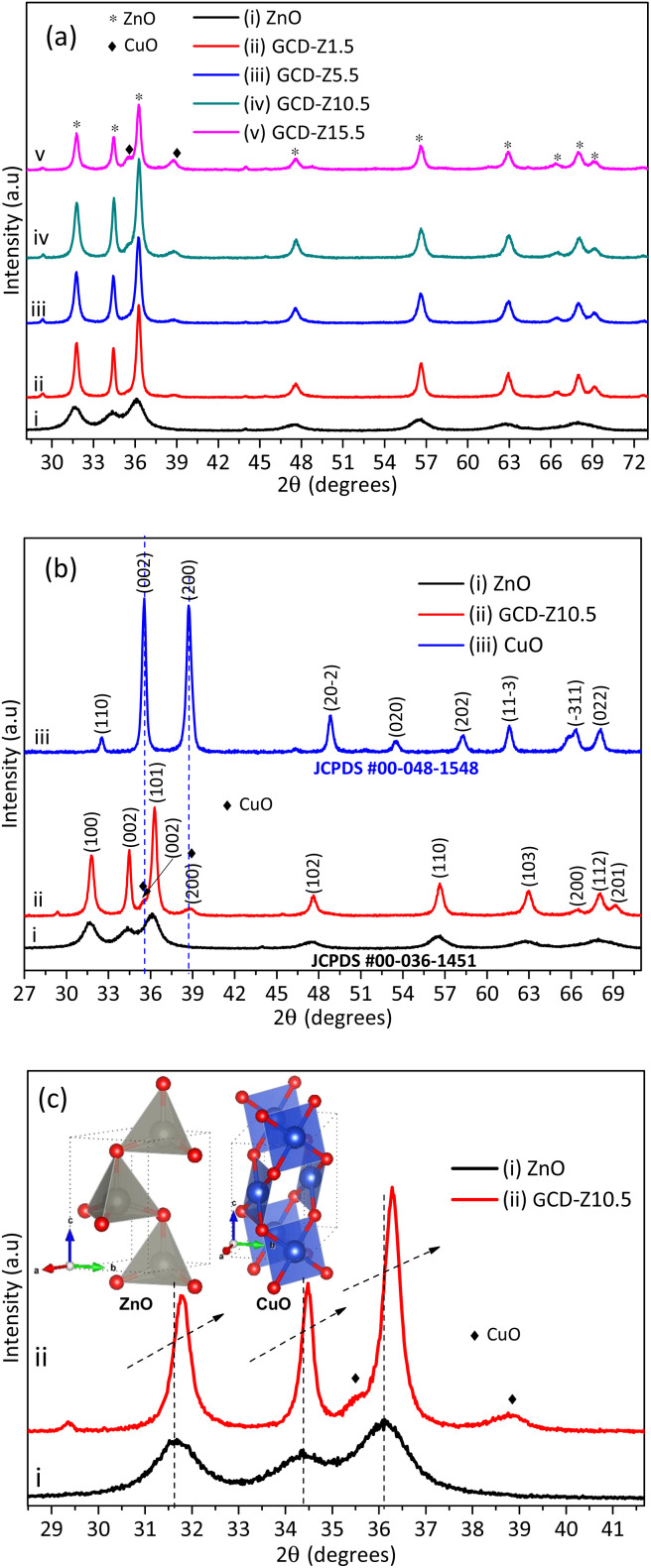
The extract-precursor concentration ratio, temperature, and pH optimization experiments showed that a 2 : 1 precursor plant extract ratio, a temperature of 75 °C, and a pH of 9.8 were obtained as optimal conditions, as shown in Table S1 and Fig. S1.† These conditions were used for the subsequent synthesis, characterization, and application tests. The crystallinity of green synthesized powdered ZnO NPs and GCD-Zs was characterized by the XRD technique, as shown in Fig. 2. Fig. 2a shows the XRD patterns of ZnO and GCD-Zs with different copper dopant percentages of 1.5–15.5%. The crystallinity of GCD-Zs was found to be greater than that of ZnO, and the crystallinity increased with an increase in the dopant concentration. The increase in crystallinity of the material improves antibacterial activity as compared to its relatively amorphous equivalents.25 The additional peak detected at about 35.5 and 38.7° was due to the occurrence of copper oxide. The intensity of the CuO-independent peak also increases as the percentage of dopant increases. It was also observed that as the dopant concentration increased, the average crystallite size also showed a slight reduction. The average crystallite sizes were calculated according to Scherrer's equation, (kλ)/(β cos θ). Where the value of the constant k is taken as 0.9, assuming that the particle has a spherical shape and the X-ray radiation wavelength θ has a value of 0.15406 nm.
Fig. 2. XRD pattern of ZnO and CuO NPs and GCD-Zs nanocomposites: (a) XRD patterns of ZnO and different percentages of GCD-Zs (1.5–15.5%), in which the crystallinity increases with dopant%; (b) XRD patterns of ZnO and CuO NPs and GCD-Z10.5; here, the CuO phase was detected at 35.5 and 38.7° on the GCD-Z10.5; (c) magnified view of the XRD pattern for ZnO NPs and GCD-Z10.5; there is a higher angle shift on the XRD pattern of GCD-Z10.5 than single ZnO, confirming the copper inclusion in the ZnO lattice. The inset in (c) is the polyhedral structure models of ZnO and CuO crystals developed by the software 3D Visualisation for Electronic Structural Analysis (3D-VESTA) from the crystallographic information file (CIF) data.
The information for full width at half maximum of the peak β and Bragg X-ray diffraction angle θ was taken from the XRD pattern analysis data.26 According to Scherrer's calculation, GCD-Zs10.5 has a smaller size (22 nm) and a greater surface area compared to the other composites. However, the average crystallite size for ZnO (7 nm) was found to be much smaller than that of GCD-Zs.
Fig. 2b compares the XRD patterns of single CuO and ZnO NPs with GCD-Z10.5. The peaks at 2θ values of 31.8, 34.5, 36.3, 47.6, 56.7, 62.9, 66.5, 68.0, and 69.2° that have the corresponding crystal planes of (100), (002), (101), (102), (110) (103), (200), (112), and (201) match the hexagonal ZnO geometry (JCPDS 00-036-1451).27 Single CuO has about nine major peaks at 2θ values with its corresponding crystal planes of 32.5 (110), 35.5 (002), 38.7 (200), 48.8 (20−2), 53.5 (020), 58.2 (202), 61.6 (11−3), 66.3 (−311), and 68.1 (022)28 (JCPDS, File no. 00-048-1548). As shown in the XRD pattern of the GCD-Z10.5 (Fig. 2b, inset label ii), the copper peaks detected at 35.5 and 38.7° exactly matched the single monoclinic CuO.15 In all the green synthesized nanoaprticles (GNPs) and green synthesized nanocomposites (GNCs), no impurity peaks other than ZnO and CuO were detected within the detection limit of the XRD instrument, which confirms the purity of the synthesis method. Fig. 2c shows the magnified view of single ZnO and GCD-Z10.5, respectively. The GCD-Z10.5 peak showed a high angle shift of 0.26°, indicating the inclusion of copper in the ZnO lattice. In this study, the copper solubility level in the ZnO lattice was found to be below 5.5%, and above this level, a copper-independent peak was detected on the GCD-Zs XRD pattern. A similar copper-independent peak detection after a 4% copper dopant amount and a higher angle shift starting from 2% copper doping were also reported in the Theyvaraju et al. study.29 Even if the angle shift was detected due to slight radii differences between copper and zinc ions, however, no structural distortion was observed due to the exchange of copper atoms with zinc atoms. The lower solubility level of copper (only up to 5%) was also reported in another study.12 The polyhedral structure models of ZnO and CuO crystals, which have the space groups P63mc (#186-1) and C2/c (#15-1), respectively, were created by the software 3D Visualisation for Electronic Structural Analysis (3D-VESTA) using data from the crystallographic information file (CIF).
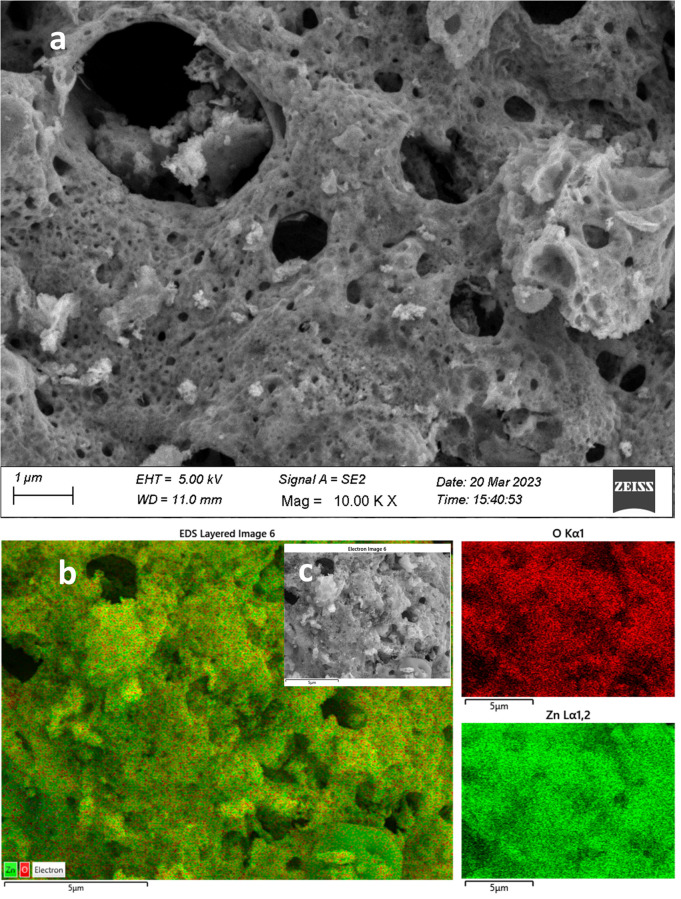
Field emission scanning electron microscopy (FESEM) as an image analysis technique in the presence of energy dispersive X-ray spectroscopy (EDS) as a compositional analysis technique was used for the morphological and elemental composition analysis (Fig. 3 and 4). ZnO GNPs have porous or amorphous morphology or both, which is created due to the evolution of gaseous byproducts as a result of combustion (Fig. 3a). The combustion reaction occurred due to the heat treatment of the complexes formed between the nitrate precursor and plant extract (phytochemicals and multiple metabolite groups). Once the ignition temperature of the complexes is reached, the material starts to combust to form a porous product.17
Fig. 3. FESEM-EDS morphological, crystallinity, elemental composition, and elemental mapping analysis for ZnO NPs: (a) FESEM image of ZnO; (b) EDS layered image taken from the electron image of (c) at a specific point. The red and green images on the right represent the oxygen and zinc mapping results, respectively.
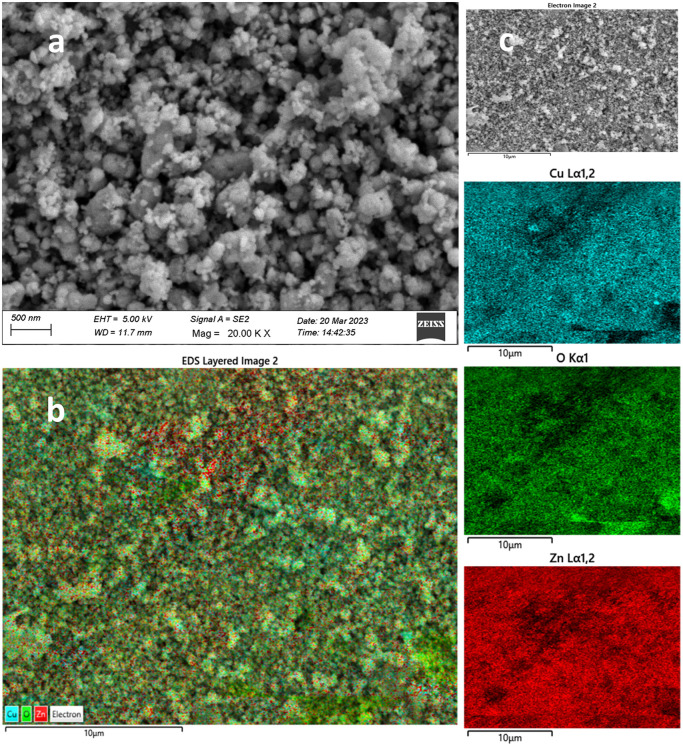
Fig. 4. FESEM-EDS morphological, crystallinity, elemental composition, and elemental mapping analysis for GCD-Z10.5: (a) FESEM image of GCD-Z10.5, (b) EDS layered image taken from the electron image of (c) at a specific point. The aqua, green, and red images on the right represent the copper, oxygen, and zinc elemental mapping results, respectively.
FESEM image analysis clearly confirmed the higher crystalline nature of the GCD-Zs compared to ZnO (Fig. 4a). The total change of ZnO's porous morphological nature to a spherical crystalline morphology in GCD-Z10.5 is probably due to copper inclusion. This crystalline morphology for GCD-Zs and the amorphous nature of ZnO are also consistent with the XRD result interpretation. The crystalline nature of single CuO compared to ZnO synthesized independently following the same procedures was confirmed on the low-magnification SEM image, as shown in Fig. S3a and b,† respectively. The accurate composition and well-doped distribution on the surface of the host material boost crucial host properties such as optoelectricity, which is crucial for antibacterial, catalysis, and several other applications.30 The compositional analysis for ZnO and GCD-Zs was determined by the EDX analysis, as shown in Fig. S4 and S5,† respectively. The EDX spectra show only zinc and oxygen for ZnO and zinc, copper, and oxygen for GCD-Z10.5, without any other impurities. The elements are observed at 1.00, 0.95, and 0.40 keV as major peaks for zinc, copper, and oxygen, respectively. The obtained amounts are fitting and in accordance with the added amount during synthesis (see Fig. S4 and S5† inset tables). In addition, the elemental distribution analyzed by EDS mapping also showed the occurrence of a good dopant distribution. The EDS layered mapping images (Fig. 3b and 4b) for both ZnO and GCD-Zs were analyzed from the electron images taken at the specific surface (Fig. 3c and 4c). The mapping images for zinc and oxygen are given in red and green for ZnO, respectively. The mapping images for GCD-Zs, presented in red, green, and aqua colours, are for zinc, copper, and oxygen, respectively.
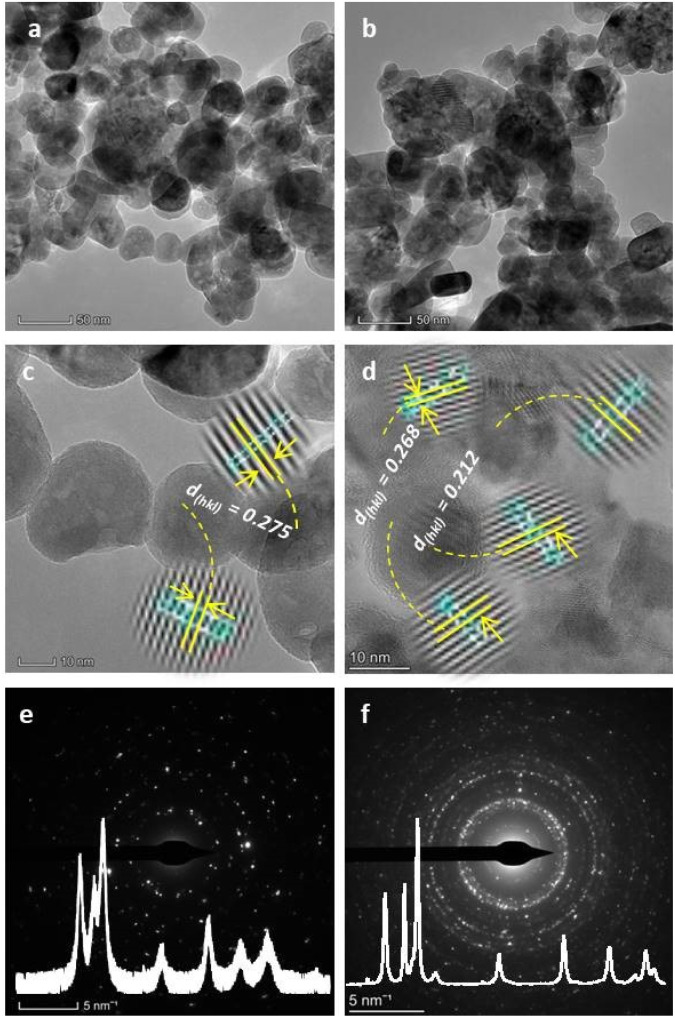
The particle size, morphology, and crystallinity of nanoscale-sized ZnO and GCD-Z10.5 materials were further analyzed using transmission electron microscopy, high-resolution transmission electron microscopy, and selected area electron diffraction (TEM-HRTEM-SAED) analytical techniques, as shown in Fig. 5. The TEM images of ZnO seem to have a less agglomerated, non-perfect spherical-type morphology (Fig. 5a), while a relatively greater agglomerated mixture of spherical and cylindrical shapes is seen for the GCD-Z10.5 (Fig. 5b). The particle sizes for ZnO and GCD-Z10.5 were in the range of 5–20 nm and 20–40 nm, respectively, which is consistent with the XRD pattern analysis. The d-spacing value for the ZnO crystal is 0.275 nm (Fig. 5c, inset image), which corresponds to the ZnO (002) crystallographic plane. The HRTEM image for GCD-Z10.5 confirmed the presence of 0.268 and 0.212 nm lattice fringe values (Fig. 5d, inset image), which are also matching with the (002) and (200) crystal planes of ZnO and CuO, respectively.31,32 There is a slight ZnO crystallographic plane lattice fringe value difference between single ZnO and GCD-Z10.5 that is probably due to the effect of copper inclusion in the ZnO lattice as a dopant, leading to slight distortion.33,34 The HRTEM image also confirmed the more crystalline nature of the GCD-Z10.5 crystal than that of ZnO, which is also clearly observed as shown in the SAED ring analysis (Fig. 5e and f, respectively).
Fig. 5. TEM/HRTEM/SAED morphological, particle size, and crystallinity analysis for ZnO and GCD-Z10.5: (a and b) TEM images; (c and d) HRETM images; and (e and f) SAED images of ZnO and GCD-Z10.5, respectively. The insets in (e) and (f) are the respective XRD patterns of ZnO and GCD-Z10.5.
The XRD pattern analysis results of ZnO and GCD-Z10.5 are also depicted in the respective ZnO and GCD-Z10.5 SAED rings as insets for comparison.
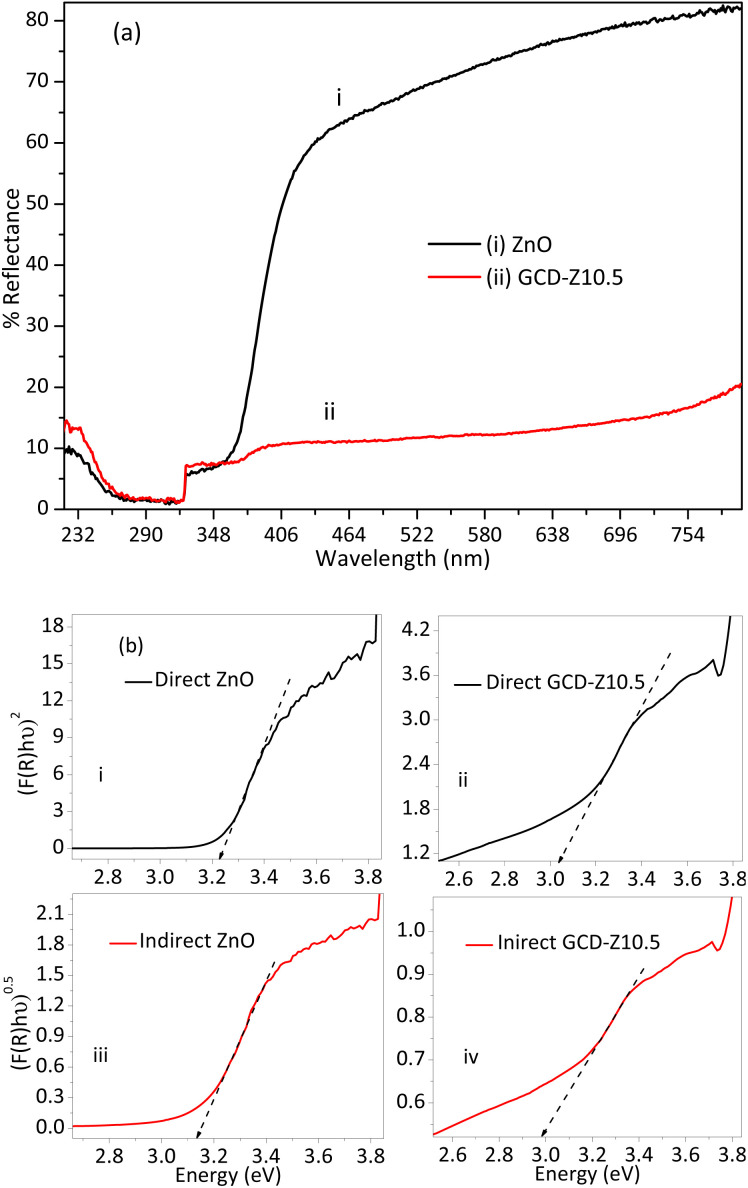
Doping and heterojunction are crucial strategies used for tuning the properties of host materials, which are also beneficial for several applications such as antibacterial activity, sensors, and catalysis. The optical property of ZnO was tuned by copper doping and forming a CuO–ZnO heterojunction, as analyzed by DRS-UV-vis spectra analysis and shown in Fig. 6a and b. The percentage reflection vs. wavelength plots of the DRS-UV-vis analysis showed greater light absorption properties for GCD-Z10.5 than ZnO (Fig. 6a). The reflection observed within the wavelength range of 370–410 nm may be attributed to the conduction band-to-valence band electron–hole recombination characteristics of ZnO.35 The greater absorption properties of the NCs are a result of the shallow level d–d transition created within the ZnO energy gap after copper inclusion. The shallow level creation is further confirmed by the Kubelka–Munk (K–M) function plots,36 which were shown by interpolation of the linear plots towards the x-axis. The direct electron transition occurred when photon-induced electron–hole recombination occurred without relaxation.
Fig. 6. Optical properties study using the DRS-UV-vis spectroscopic technique and Kubelka–Munk function analysis: (a) % reflectance versus wavelength plot of ZnO and GCD-Z10.5 NCs, with greater light harvesting behaviour for the GCD-Z10.5 NCs. (b) Kubelka–Munk function plots: (inset labels i and ii) direct and (inset labels iii and iv) indirect Kubelka–Munk plots for ZnO and GCD-Z10.5 NCs, respectively. Redshift for both direct and indirect GCD-Z10.5 plots confirms the inclusion of copper in the ZnO lattice of GCD-Z10.5.
Conversely, in the indirect electron transition, electrons have a relaxation time before recombination. The crystal momentum, k factors, for a direct transition is zero, unlike the indirect transition, which has a non-zero value.37 Electron relaxation and transition from the host conduction band to the shallow level or CuO crystal have great importance for ROS generation, which in turn is important for bactericidal activities.38 The direct bandgap values for ZnO and GCD-Z10.5 are 3.23 and 3.04 eV, respectively (Fig. 6b, inset labels i and ii). The indirect bandgap values for ZnO and GCD-Z10.5 are 3.13 and 2.97 eV, respectively (Fig. 6b, inset labels iii and iv). The redshift on both the direct and indirect bandgaps for GCD-Z10.5, compared to ZnO, confirms the inclusion of copper in the ZnO lattice and visible light absorption enhancement.39
The optical properties of CuO and ZnO, and GCD-Z10.5 materials were further confirmed using photoluminescence (PL) spectroscopy, as shown in Fig. S6.† ZnO has two broad peaks at 367 and 465 nm and one intense band at about 403 nm, respectively. The peak detected at 403 nm is a characteristic near-band edge (NBE) transition in ZnO that occurred due to free exciton electron–hole recombination.40 The green emission that occurred at about 465 nm is probably due to intrinsic defects (oxygen vacancies).41 CuO also has two short and broad emission peaks at 369 and 473 nm and one relatively intense and broad peak at 409 nm, respectively. These CuO emission peaks occurred as a result of the ultraviolet, green, and violet emissions, respectively. The GCD-Z10.5 also showed one ultraviolet and two visible bands at 370, 406, and 470 nm, respectively. The two broad peaks on the GCD-Z10.5 NCs showed an emission intensity rise and a slight shift due to the combined effects of copper and zinc. The green emission is attributed to the electron transition from the dopant shallow level to the host valence band after relaxation.40 All the emissions detected on the copper oxide spectrum were also detected on GCD-Z10.5, which indicate the presence of copper crystals as a local contact (heterojunction) with ZnO.42 This has importance in improving the charge transfer properties and consequently enhancing the generation of ROS species through electron and hole reactions with oxygen and water, respectively.
The functional groups and purity of the GNPs and GNCs were analyzed by Fourier-transform infrared (FTIR) spectroscopy. Fig. S7a† shows the FTIR spectra of ZnO before and after calcination, and CuO and GCD-Z10.5 NCs after calcination. The distinctive signal detected at the approximate band positions of 3440 and 3245 cm−1 on the uncalcined ZnO is attributed to the plant extract alkyne C–H stretching, carboxylic acid, and phenol O–H stretching from the hemicellulose, cellulose, and lignin groups.18,43 The two sharp but short bands detected at about 2360 and 2330 cm−1 are attributed to carbon dioxide absorption. The other new peak at 1360 cm−1 for uncalcined ZnO is the result of a symmetric or asymmetric stretching vibration of C O.44 The peaks at 890 and 625 cm−1 are probably due to the bending vibration of other organic residues left over.19 However, the broad peak detected at 3370 and 1640 cm−1 on the calcined ZnO are due to O–H stretching and bending vibration of the surface/interstitial adsorbed water molecules, respectively.29 The effect of copper doping resulted in a peak intensity reduction compared to a single ZnO peak, with the occurrence of copper bands on the GCD-Z10.5 NCs spectrum. Our concern is that the fingerprint region (400–500 cm−1) shows the presence of metal–oxygen bonds (Cu-, Zn-)O. Fig. S7b† shows the magnified view of the lower wavenumber FTIR spectrum, in which 450 and 430 cm−1 for Zn–O bending vibration; 440 and 415 cm−1 for the GCD-Z10.5; and 440 and 405 cm−1 for CuO, respectively. These bending vibration peaks for GCD-Z10.5 resemble those of the copper spectrum with shifts after copper inclusion compared to ZnO, which is caused by copper inclusion.29
Antibacterial activity
The antibacterial activity of all GNPs and GNCs was examined using the disc diffusion method and different bacterial strains, including two Gram-positive (S. aureus and S. pyogenes) and two Gram-negative (E. coli and P. aeruginosa) species, at varying concentrations of GNPs/GNCs (2.5, 5, and 10 mg mL−1). Ciprofloxacin (10 μg mL−1) and DMSO were used as a positive control (PC) and negative control (NC), respectively. The results of antibacterial tests conducted on all GNPs and GNCs are presented in Table 1. In general, all the GNPs and GNCs showed antibacterial activity against both the Gram-positive and Gram-negative bacterial strains tested with dose-dependency in the following order: ZnO < CuO < GCD-Z10.5. The measurable zone of inhibition (MZI) values for ZnO varied from 6.1 ± 0.33 mm to 13.3 ± 0.58 mm (PC: 23.3 ± 0.58–39.3 ± 0.88), CuO 6.1 ± 0.33 mm–12.7 ± 0.33 mm (PC: 22.1 ± 0.33–26.9 ± 0.33) and GCD-Z10.5 6.4 ± 0.33 mm–16.7 ± 0.33 mm (PC: 17.7 ± 0.33–28.6 ± 0.57), respectively for the different strains. The control normalized percent inhibition for ZnO was found to be 34.5–43.3%, CuO 23.7–49.5%, and GCD-Z10.5 24.5–79.5%, respectively (Table 1). The ZnO caused an observable inhibitory effect against the tested pathogenic bacterial strains, which may be due to the interaction between the negatively charged membrane of the bacteria and the positively charged ions of ZnO.45 Based on XRD analysis, ZnO synthesized is very small (7 nm), allowing it to penetrate bacteria's membranes and damage their intracellular structures. As reported,46 these GNPs can disrupt biological processes and damage DNA. This result is also supported by the PL and DRS results (Fig. S6† and 6), which show high surface defects indicating roughness, which can generate ROS and damage bacteria cells.47 It was also observed that CuO showed significant toxicity to the bacterial strains. According to a previous report, CuO has the potential to penetrate the cells of microorganisms as well as accumulate in the cell.48,49 Cu has a high affinity to fix oxygen, and generating ROS causes intracellular damage.48 Doping of ZnO and CuO has been shown to impart a synergistic effect and improve antibacterial property.47 We also observed a significant enhancement of antibacterial activity by GNCs compared to ZnO and CuO GNPs. At a concentration of 10 mg mL−1, the percentage inhibition for ZnO-GNP was found to be 34.5–43.3%, CuO-GNP was 36.7–49.5%, and GCD-Z10.5 showed the highest inhibition in the range of 54.8–79.5%, respectively (Fig. S8† and Table 1).
The antibacterial activities of all GNPs and GNCs materials determined by disc diffusion assay.
| []a (mg mL−1) | E. coli | % RIb | P. aeruginosa | %RI | S. aureus | %RI | S. pyogenes | % RI |
|---|---|---|---|---|---|---|---|---|
| Measurable zone of inhibition (MZI) of ZnO (mm) | ||||||||
| 10 | 10 ± 0.58 | 40 | 8.1 ± 0.33 | 34.5 | 12 ± 0.58 | 43.3 | 13.3 ± 0.58 | 34.7 |
| 5 | 7.9 ± 0.58 | 31.5 | 7.2 ± 0.33 | 30.8 | 11.1 ± 0.33 | 40.1 | 9.2 ± 0.58 | 33.5 |
| 2.5 | 7.2 ± 0.88 | 28.6 | 6.1 ± 0.33 | 26.2 | 9.5 ± 0.58 | 34.6 | 6.9 ± 0.33 | 27.5 |
| NCc | 6 ± 0.00 | 24.0 | 6 ± 0.00 | 25.7 | 6 ± 0.00 | 21.6 | 6 ± 0.00 | 15.3 |
| PCd | 25 ± 1.00 | 100 | 23.3 ± 0.58 | 100 | 27.7 ± 1.20 | 100 | 39.3 ± 0.88 | 100 |
| Measurable zone of inhibition (MZI) of CuO (mm) | ||||||||
| 10 | 11.7 ± 0.33 | 43.5 | 8.1 ± 0.33 | 36.7 | 12.7 ± 0.33 | 49.5 | 12.3 ± 0.33 | 47.5 |
| 5 | 9.3 ± 0.33 | 34.5 | 7.5 ± 0.33 | 34.0 | 8.1 ± 0.88 | 31.2 | 8.2 ± 0.58 | 31.5 |
| 2.5 | 6.9 ± 0.33 | 25.8 | 6.1 ± 0.33 | 27.7 | 7.1 ± 0.58 | 27.6 | 6.2 ± 0.33 | 23.7 |
| NC | 6 ± 0.00 | 23.3 | 6.0 ± 0.00 | 27.1 | 6.0 ± 0.00 | 23.3 | 6.0 ± 0.00 | 23.3 |
| PC | 26.9 ± 0.33 | 100 | 22.1 ± 0.33 | 100 | 25.7 ± 0.33 | 100 | 25.8 ± 0.33 | 100 |
| Measurable zone of inhibition (MZI) of GCD-Z10.5 (mm) | ||||||||
| 10 | 15.7 ± 0.33 | 54.8 | 13.3 ± 0.33 | 75.1 | 16.3 ± 0.58 | 79.5 | 16.7 ± 0.33 | 75.2 |
| 5 | 12.2 ± 0.58 | 42.5 | 7.5 ± 0.58 | 42.1 | 11.2 ± 0.33 | 54.7 | 14.0 ± 0.88 | 63.1 |
| 2.5 | 7.0 ± 0.88 | 24.5 | 6.4 ± 0.33 | 36.2 | 6.7 ± 0.58 | 32.6 | 9.9 ± 0.58 | 44.6 |
| NC | 6.00 ± 0.00 | 20.9 | 6.00 ± 0.00 | 33.9 | 6.0 ± 0.00 | 29.3 | 6.0 ± 0.00 | 27.0 |
| PC | 28.6 ± 0.57 | 100 | 17.7 ± 0.33 | 100 | 20.5 ± 0.58 | 100 | 22.2 ± 1.20 | 100 |
Concentration.
Relative percentage inhibition.
Negative control.
Positive control.
Among the bacterial strains tested, all GNPs and GNCs showed higher activity against Gram-positive bacteria (S. aureus and S. pyogenes) compared to Gram-negative bacteria (E. coli and P. aeruginosa). This may be attributed to the difference in cell wall composition. The Gram-positive bacterial cell is composed of teichoic acids that partially impart a negative charge to the cell surface and facilitate the passage of metal ions, thereby plausibly rupture the cells, resulting in higher antibacterial activity of GNPs and GNCs.50 When tested at the highest concentration (10 mg mL−1), S. aureus and S. pyogenes showed 79.5% and 75.2% inhibition, respectively (higher susceptibility). Gram-negative bacteria, however, have a complex cell wall composition. It consists of a second membrane that restricts molecules' penetration and confers resistance to antibacterial agents.44
Among the two Gram-negative strains tested, P. aeruginosa was found to be less susceptible to ZnO and CuO GNPs than E. coli. P. aeruginosa has been categorized as a multidrug-resistant strain, which may explain this resistance against the GNPs and GNCs.51 It is interesting to note that both strains showed higher susceptibility (54.8 and 75.1% inhibition) against GCD-Z10.5 at the concentration of 10 mg mL−1, up from 36.7% (CuO, P. aeruginosa) and 40% (ZnO, E. Coli), respectively, at the same concentration (Table 1). The increased potency of GCD-Z10.5 compared to single GNPs may be plausibly due to the synergistic effect of ZnO and CuO GNPs.
MIC and MBC analysis
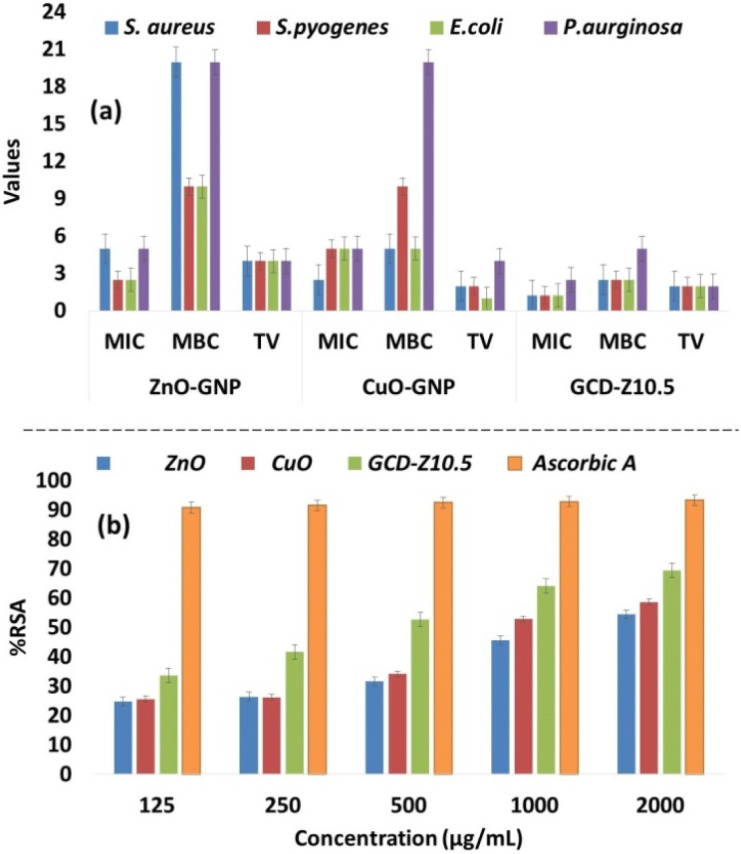
The determination of the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) is necessary for evaluating GNPs and GNCs for their cytotoxicity.52 The MIC is the lowest concentration of a substance needed to prevent the visible growth of a bacterium, while the MBC is the lowest concentration required to kill the bacterium. Thus, the smallest concentration value indicates the material's effectiveness at low concentrations. A tolerance test provides information about the potency of the test materials; higher values indicate bacteriostatic action, while lower values indicate bactericidal action. Fig. 7a and Tables S2–S4† show the MIC, MBC, and tolerance tests of various GNPs and GNCs against different bacterial strains. The MIC of the synthesized GNPs and GNCs was found to vary from 1.25–5 mg mL−1, the MBC from 5–20 mg mL−1, and the tolerance value (TV) from 1–4, respectively. The minimal concentration of NPs suspension that releases efficient metal ions was detected, suggesting that the bacterial growth inhibition is primarily due to the interaction of GNPs and GNCs with the membrane of the microorganism's.53
Fig. 7. Graphical representation of antibacterial MIC, MBC, and TV for GNPs and GNCs against S. aureus, E. coli, S. pyogenes, and P. aeruginosa bacteria. The values on the y-axis are in mg mL−1 for MIC and MBC and numericals for TV, respectively. The lowest MIC values for GCD-Z10.5 indicate its effectiveness in bacterial growth prevention at low concentrations. In general, here also, the doped GCD-Z10.5 showed greater activity, which is due to the synergistic effects. (b) The antioxidant activity test was determined using a DPPH free radical scavenging assay.
The results obtained in this study showed a good correlation with the percentage inhibition of the antibacterial assay in the following order: ZnO < CuO < GCD-Z10.5. In particular, P. aeruginosa was more resistant compared to others and generated higher MIC/MBC/TV. ZnO and CuO showed equal potency in terms of MIC/MBC/TV against P. aeruginosa, however, it is interesting to note that GCD-Z10.5 showed higher potency in reducing the TV of P. aeruginosa, which indicates higher bactericidal activity. The results obtained here are comparable and in good agreement with the previous reports.55 This suggests the synergistic effects of CuO and ZnO on antibacterial activity.
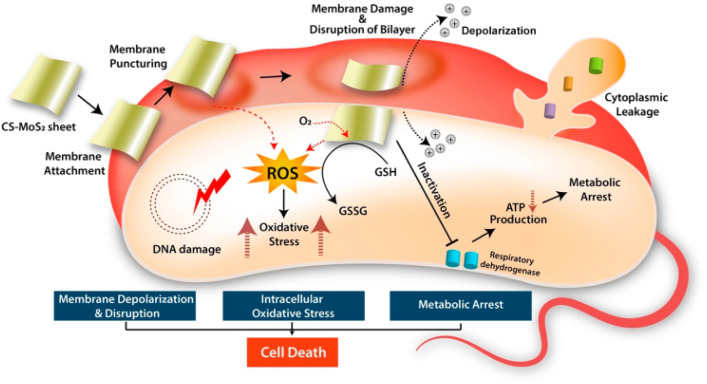
Even if the antibacterial activity of NPs is based on a different mechanism, it is reported to be mainly due to the release of antimicrobial ions, interactions of NPs with microorganisms, and the production of ROS due to light radiation.38 According to this study, the minimal effects of metal ions were confirmed from the MIC and MBC analyses, and the experiments also did not use light as a radion source. Thus, the mechanism for bacterial deactivation is the interaction of bacterial cells with GNPs or GNCs, although with some release of ions. It has been reported that oxidative stress is induced by the generation of ROS as a result of electrostatic or van der Waals interactions or both between NPs and bacterial cells.56 Similar ROS generation through chitosan-MoS2 nanosheets with bacterial cell interaction, ROS generation, and then oxidative stress was also reported, as shown in Scheme 1.54 Upon internalization inside the cells, ROS can target organelles, DNA synthesis, proteins, and metabolic pathways. These events may cumulatively lead to cytotoxicity and cell death, thereby reducing the GNPs- and GNCs-treated bacterial populations. DNA is an integral component of cells, and genomic stability is vital to maintaining the integrity of cells as it regulates most of the cellular processes, including protein synthesis and metabolic pathways. GNPs and GNCs can generate intermediate ROS in the presence of oxygen and water. These ROS may potentially cause oxidative damage to DNA (genotoxicity), impair DNA-regulated processes, and eventually lead to cell death. Further studies using the NPs and NCs of this study are required to elucidate this mechanism.57
Scheme 1. The antibacterial mechanism by which bacterial cell activity is inhibited by the electrostatic and/or van der Waals interaction of NPs and NCs with the bacterial cells, which leads to ROS generation and oxidative stress creation. Reprinted with permission from ref. 54 Copyright 2019. American Chemical Society.
Antioxidation potential
The antioxidant activity was determined by a DPPH free radical scavenging assay that works on the principle of reduction of DPPH by hydrogen donors to 2,2-diphenyl-1-picrylhydrazyl (DPPH).58 The GNPs and GNCs used in this study were found to show an effective range of radical scavenging at the different concentrations examined (0.125–2 μg mL−1) (Fig. 7b and Table S5†). In general, the activities of the samples ranged from 24.8–54.4% RSA for ZnO, 25.6–58.6% RSA for CuO, and 33.6–69.4 for GCD–Z10.5% RSA, and were found to be dose-dependent. Ascorbic acid was used as a standard reference with the same concentrations, and the generated values were in the range of 90.8–93.3% RSA. The antioxidation potential was observed in the following order: ZnO < CuO < GCD-Z10.5, which may be due to an increased surface-to-volume ratio and small particle size as observed by XRD and TEM analysis (Fig. 1 and 4).47 The higher antioxidation potential is beneficial in scavenging free radicals involved in several diseases and metabolic pathways and imparts neuroprotective, immunoprotective, and hepatoprotective effects.59
Conclusions
The S. incanum L. extract-based green approach was successfully employed to synthesize Cu-doped ZnO (GCD-Zs) and intimately contact CuO–ZnO NCs. The XRD pattern and TEM analysis of the synthesized materials showed nanometer-scale size (in the range of 10–50 nm). The presence of a redshift for the doped GCD-Zs NCs over single ZnO (from 3.23 to 3.13 on the direct and from 3.04 to 2.97 eV on the indirect Kubelka–Munk function) was confirmed. The PL analysis showed the presence of electron–hole recombination hindrance in the GCD-Zs composites over ZnO, which is the result of shallow-level copper inclusion and heterojunction. The HRTEM analysis confirmed the nearby contact within the CuO–ZnO composite, with d-spacing values of 0.212 and 0.268 nm, which match the (200) and (002) crystal planes of CuO and ZnO, respectively. The overall doping and heterojunction effects proved to improve the crystallinity of GCD-Zs and ROS generation and consequently enhanced the antibacterial activity and lowered the MIC/MBC/TV of the GCD-Zs NCs over single ZnO in both Gram-positive and negative bacteria by the synergy effect. Furthermore, the significant decrease of the tolerance test values to the lowest by the doped mixture on the multidrug-resistant strain, P. Aeruginosa, was notable. We believe that this green approach to the synthesis of NPs and NCs may contribute to and add possible solutions in the pursuit of antimicrobials for antimicrobial resistant, multidrug-resistant bacteria, and possibly other microbes.
Experimental
The usual procedures for the preparation of plant extract, phytochemical analysis, and UV-vis-based green synthesis parameter optimization (extract and precursor concentration ratio, temperature, and pH) on the host zinc precursor were used.60 During the above optimizations, a 2 : 1 precursor plant extract ratio, a temperature of 75 °C, and a pH of 9.8 were selected by observing the peak intensity of absorbance that integrated with the concentration of synthesizing NPs and were used for the next consecutive synthesis, characterization, and application tests. To optimize the GCD-Z synthesis, we have varied the concentration (wt%) of copper acetate (0, 1.5, 5.5, 10.5, and 15.5 wt%) in a 2 : 1 volume ratio of precursor to plant extract mixture. For this purpose, 40 mL of copper acetate and zinc nitrate mixture and 20 mL of plant extract were mixed for each copper content. The plant extract was added to the precursors and stirred for 20 min at 25 °C, and then the pH of the mixtures was adjusted to 9.8 by drop-wise addition of 1 M sodium hydroxide solution.10 The stirring was continued for 2 hours after the pH was adjusted until the white precipitation was observed visually. The resulting colloidal solution was aged at room temperature for 24 hours for more precipitation, followed by centrifugation. The product thus obtained was washed well with ethanol and distilled water, transferred to a Petri dish, and dried overnight in an electric oven at 45 °C. The resulting powder was subjected to calcination in the muffle furnace for 2 hours at 400 °C. To obey the single-precursor nanocrystal doping strategy, the host and dopant salt precursors were mixed once.61 Here, the precursors were dispersed in the pre-extracted plant extract, which plays a significant role in achieving crucial properties such as cations/colloidal particle dispersion and nanoparticle prevention from aggregation.19 Of course, the doping process follows LaMer's model, which means (i) precursor decomposition, increase in the concentration of constituent ions, and reduction of ions into atoms so as to form a supersaturated solution; (ii) nucleation either homogenously or heterogeneously via random aggregation of atoms; and (iii) finally growth to nanoparticles starts via diffusion of atoms towards the pre-formed nuclei.62–64 During heating, the precursor-plant extract complexes combust with an increase in temperature when they reach the point of complex ignition temperature.65 Within a few seconds of starting the combustion, porous materials are created, yielding stable metal oxides, or metals.66
Experiments on biological activities were performed on four conventional antibiotic-resistant bacterial strains: Gram-negative: Escherichia coli (E. coli, ATCC 25922) and Pseudomonas aeruginosa (P. aeruginosa, ATCC 27853); Gram-positive: Staphylococcus aureus (S. aureus, ATCC 25923) and Streptococcus pyogenes (S. pyogenes, ATCC 13311). First, all the stock-cultured bacteria were activated on Mueller–Hinton (MH) agar plates. After activation, the cultures were subjected to inocula development by inoculating a loop full of cells from a single colony into MH broth and incubated for 24 h at 37 °C. Then the active pure bacterial cultures and sub-cultures were maintained according to the 0.5 McFarland standards. This process was repeated every time before starting a new experiment. The antibacterial properties of the biosynthesized GNPs and GNCs were investigated using the standard Kirby–Bauer disc diffusion method.67 The fresh overnight cultures of each strain were swabbed uniformly over the sterilized and cooled Mueller–Hinton agar medium (MHA) Petridishes. Then, 6 mm diameter sterile discs impregnated with different green synthesized NP/NC solutions (10, 5, and 2.5 mg mL−1) were placed onto these plates and incubated for 24 h at 37 °C. The discs impregnated with Ciprofloxacin solution (10 μg mL−1) and DMSO were used as positive and negative controls, respectively. After incubation, the antimicrobial activity was determined by measuring the diameter of the zone of inhibition. The antibacterial activity was calculated in terms of percentage inhibition according to: % inhibition = diameter of sample/diameter of control × 100.
The MIC of green synthesized NPs and NCs was investigated by broth assay and measuring the absorbance of bacterial growth. For this purpose, six double dilutions of the NP/NC suspension were prepared, varying from 0.625 mg mL−1 to 20 mg mL−1, respectively. The 10 mL of Muller–Hinton broth was inoculated with 0.1 mL of bacterial culture of 0.5 McFarland standard and 0.5 mL of various concentrations of prepared GNPs/GNCs suspensions in each test tube and incubated at 37 °C for 24 h. The absorbance of the mixture was observed before and after the incubation for each test tube by taking a sufficient amount into a cuvette and measuring the OD at 610 nm (Table S3†).68 The last concentration that appears clear visually and shows approximately the same or less absorbance value than the earlier concentration after incubation is taken as the MIC. The MBC is the lowest antibacterial concentration that prevents the growth of an organism after subculture on growth media. To determine MBC, the test tubes that contain MIC and higher concentrations were considered. A 0.1 mL mixture of GNPs/GNCs, bacteria, and broth was taken from each MIC sample and spread on the corresponding MHA agar plates. The appearance of colonies on the agar plate that indicate the viability of bacteria was observed after incubation at 37 °C for 24 h. The lowest concentration that showed no formation of bacterial colonies on the agar plate was considered as MBC. Tolerance test: the values of MBC were divided by the values of MIC to obtain Tolerance test values.
The antioxidant activity test was performed using the DPPH free radical scavenging assay. A 0.1 mM DPPH solution was prepared in methanol by dissolving 0.04 g DPPH in 100 mL methanol. The solution was kept in the dark for 30 minutes to complete the reaction. The powdered GNPs and GNCs were diluted in the five vials at 2000, 1000, 500, 250, and 125 μg mL−1 in methanol. From these concentrations, 1 mL of each was mixed with 3 mL of 0.1 mM DPPH. Ascorbic acid was prepared with the same concentration of samples and used as a positive control in the experiment. The absorbances were measured using a UV-vis spectrophotometer at 517 nm after 30 min of incubation in a dark place.58 The radical scavenging activity was calculated according to: % of radical scavenging activity = (Abs of DPPH (control) − absorbance of samples)/absorbance of DPPH (control) × 100.
Author contributions
Writing – original draft and laboratory work by Adam Mengistu. The writing – review & editing and advising were done by Mohammed Naimuddin and Buzuayehu Abebe. All authors read the final manuscript in detail and approved it.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The authors acknowledge the aid of the Departments of Applied Biology, Chemistry, Institute of Pharmaceutical Science, and Materials Engineering in completing this work, as well as Adama Science and Technology University for providing funds (ASTU/AS-R/052/2022).
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04488b
Notes and references
- Maruthapandi M. Saravanan A. Das P. Natan M. Jacobi G. Banin E. Luong J. H. T. Gedanken A. ACS Appl. Polym. Mater. 2020;2:5878–5888. [Google Scholar]
- Hassan I. A. Sathasivam S. Nair S. P. Carmalt C. J. ACS Omega. 2017;2:4556–4562. doi: 10.1021/acsomega.7b00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. E. Patel N. G. Levy M. A. Storeygard A. Balk D. Gittleman J. L. Daszak P. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J. Kristiansson E. Larsson D. G. J. FEMS Microbiol. Rev. 2018;42:68–80. doi: 10.1093/femsre/fux053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulani M. S. Kamble E. E. Kumkar S. N. Tawre M. S. Pardesi K. R. Front. Microbiol. 2019;10:539. doi: 10.3389/fmicb.2019.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso J. O. Lamaro-Cardoso J. Neves L. S. Borges L. F. A. Pires C. H. Lamaro L. Guerreiro T. C. Ferreira E. M. A. André M. C. P. APMIS. 2019;127:717–726. doi: 10.1111/apm.12989. [DOI] [PubMed] [Google Scholar]
- Saravanan R. Karthikeyan S. Gupta V. K. Sekaran G. Narayanan V. Stephen A. Mater. Sci. Eng. Carbon. 2013;33:91–98. doi: 10.1016/j.msec.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Karthik K. V. Raghu A. V. Reddy K. R. Ravishankar R. Sangeeta M. Shetti N. P. Reddy C. V. Chemosphere. 2022;287:132081. doi: 10.1016/j.chemosphere.2021.132081. [DOI] [PubMed] [Google Scholar]
- Agarwal D. C. Singh U. B. Gupta S. Singhal R. Kulriya P. K. Singh F. Tripathi A. Singh J. Joshi U. S. Avasthi D. K. Sci. Rep. 2019;9:6675. doi: 10.1038/s41598-019-43184-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid A. Ahmad P. Alharthi A. I. Muhammad S. Khandaker M. U. Faruque M. R. I. Din I. U. Alotaibi M. A. Khan A. PLoS One. 2021;16:e0251082. doi: 10.1371/journal.pone.0251082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N. Kumar N. ACS Biomater. Sci. Eng. 2019;5:1170–1188. doi: 10.1021/acsbiomaterials.8b01092. [DOI] [PubMed] [Google Scholar]
- Morales-Mendoza J. E. Paraguay-Delgado F. Mater. Lett. 2021;291:129494. [Google Scholar]
- Mahmoud A. Echabaane M. Omri K. Boudon J. Saviot L. Millot N. Ben Chaabane R. B. R. Molecules. 2021;26:929. doi: 10.3390/molecules26040929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekru A. G. Tufa L. T. Zelekew O. A. Goddati M. Lee J. Sabir F. K. ACS Omega. 2022;7:30908–30919. doi: 10.1021/acsomega.2c02687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas A. Florica C. Preda N. Besleaga C. Kuncser A. Enculescu I. Sci. Rep. 2022;12:6834. doi: 10.1038/s41598-022-10879-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momeni S. S. Nasrollahzadeh M. Rustaiyan A. J. Colloid Interface Sci. 2016;472:173–179. doi: 10.1016/j.jcis.2016.03.042. [DOI] [PubMed] [Google Scholar]
- Manikandan A. Sridhar R. Arul Antony S. Ramakrishna S. J. Mol. Struct. 2014;1076:188–200. [Google Scholar]
- Lashin I. Fouda A. Gobouri A. A. Azab E. Mohammedsaleh Z. M. Makharita R. R. Biomolecules. 2021;11:341. doi: 10.3390/biom11030341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan Thi T. U. Nguyen T. T. Thi Y. D. Ta Thi K. H. Phan B. T. Pham K. N. RSC Adv. 2020;10:23899–23907. doi: 10.1039/d0ra04926c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A. Noreen F. Kanwal S. Iqbal A. Hussain G. Mater. Sci. Eng. Carbon. 2018;82:46–59. doi: 10.1016/j.msec.2017.08.071. [DOI] [PubMed] [Google Scholar]
- Truong T. T. Le T. H. Pham T. D. Colloid Polym. Sci. 2022;300:1343–1354. [Google Scholar]
- Nanda K. K. Maisels A. Kruis F. E. Fissan H. Stappert S. Phys. Rev. Lett. 2003;91:106102. doi: 10.1103/PhysRevLett.91.106102. [DOI] [PubMed] [Google Scholar]
- Zhang G. Shen X. Yang Y. J. Phys. Chem. C. 2011;115:7145–7152. [Google Scholar]
- Ren M. Frimmel F. H. Abbt-Braun G. J. Mol. Catal. A: Chem. 2015;400:42–48. [Google Scholar]
- Perelshtein I. Lipovsky A. Perkas N. Gedanken A. Moschini E. Mantecca P. Nano Res. 2015;8:695–707. [Google Scholar]
- Sharma D. Jha R. Ceram. Int. 2017;43:8488–8496. [Google Scholar]
- Paraguay-Delgado F. Hermida-Montero L. A. Morales-Mendoza J. E. Durán-Barradas Z. Mtz-Enriquez A. I. Pariona N. RSC Adv. 2022;12:9898–9908. doi: 10.1039/d2ra00863g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D. Wang L. Yu W. Xie H. Sci. Rep. 2018;8:5282. doi: 10.1038/s41598-018-23174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theyvaraju D. Muthukumaran S. Phys. E. 2015;74:93–100. [Google Scholar]
- Abebe B. Murthy H. C. A. RSC Adv. 2022;12:5816–5833. doi: 10.1039/d1ra09152b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abebe B. Kefale B. Leku D. T. RSC Adv. 2023;13:4523–4529. doi: 10.1039/d2ra07845g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. C. Kim A. Y. Kim J. Y. Park S. Park K. H. Song H. Chem. Commun. 2012;48:8484. doi: 10.1039/c2cc31654d. [DOI] [PubMed] [Google Scholar]
- Majeed Khan M. A. Siwach R. Kumar S. Ahmed M. Ahmed J. J. Mater. Sci.: Mater. Electron. 2020;31:6360–6371. [Google Scholar]
- Kazmi J. Ooi P. C. Raza S. R. A. Goh B. T. Karim S. S. A. Samat M. H. Lee M. K. Mohd M. F. Wee R. Taib M. F. M. Mohamed M. A. J. Alloys Compd. 2021;872:159741. [Google Scholar]
- Liu J. Li J. Wei F. Zhao X. Su Y. Han X. ACS Sustainable Chem. Eng. 2019;7:11258–11266. [Google Scholar]
- Abebe B. Zereffa E. A. Murthy H. C. A. ACS Omega. 2021;6:954–964. doi: 10.1021/acsomega.0c05597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K. J. Mater. Sci.: Mater. Electron. 2020;31:10478–10488. [Google Scholar]
- Abebe B. Zereffa E. A. Tadesse A. Murthy H. C. A. Nanoscale Res. Lett. 2020;15:190. doi: 10.1186/s11671-020-03418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thabit H. A. Kabir N. A. Ismail A. K. Alraddadi S. Bafaqeer A. Saleh M. A. Nanomaterials. 2022;12:3068. doi: 10.3390/nano12173068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G. P. Aman A. K. Singh R. K. Roy M. K. Optik. 2020;203:163966. [Google Scholar]
- Ma Y. Choi T.-W. Cheung S. H. Cheng Y. Xu X. Xie Y.-M. Li H.-W. Li M. Luo H. Zhang W. So S. K. Chen S. Tsang S.-W. Nanoscale. 2019;11:8736–8743. doi: 10.1039/c9nr02020a. [DOI] [PubMed] [Google Scholar]
- Parui S. S. Kumar N. Tiwari P. Tiwari N. Chauhan R. N. Mater. Today: Proc. 2021;41:233–236. [Google Scholar]
- AL-Shehri H. S. Alanazi H. S. Shaykhayn A. M. ALharbi L. S. Alnafaei W. S. Alorabi A. Q. Alkorbi A. S. Alharthi F. A. Sustainability. 2022;14:2644. [Google Scholar]
- Ali H. M. Ibrahim S. M. Abo Zeid E. F. Al-Hossainy A. F. El-Aal M. A. RSC Adv. 2022;12:16496–16509. doi: 10.1039/d2ra02515a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-López E. Gomes D. Esteruelas G. Bonilla L. Lopez-Machado A. L. Galindo R. Cano A. Espina M. Ettcheto M. Camins A. Silva A. M. Durazzo A. Santini A. Garcia M. L. Souto E. B. Nanomaterials. 2020;10:292. doi: 10.3390/nano10020292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y. He Y. Irwin P. L. Jin T. Shi X. Appl. Environ. Microbiol. 2011;77:2325–2331. doi: 10.1128/AEM.02149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta J. Bahadur D. ACS Omega. 2018;3:2956–2965. doi: 10.1021/acsomega.7b01953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas A. M. Aboelmagd A. Kishk S. M. Nasrallah H. H. Boyd W. C. Kalil H. Orabi A. S. Molecules. 2022;27:7540. doi: 10.3390/molecules27217540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemi J. O. Onwudiwe D. C. Oyedeji A. O. Molecules. 2022;27:3206. doi: 10.3390/molecules27103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnwal A. Kumar G. Pant G. Hossain K. Ahmad A. Alshammari M. B. ACS Omega. 2023;8:13492–13508. doi: 10.1021/acsomega.3c00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marner M. Kolberg L. Horst J. Böhringer N. Hübner J. Kresna I. D. M. Liu Y. Mettal U. Wang L. Meyer-Bühn M. Mihajlovic S. Kappler M. Schäberle T. F. von Both U. Microbiol. Spectrum. 2023;11:e04437. doi: 10.1128/spectrum.04437-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi K. S. ur Rahman A. Tajuddin Husen A. Nanoscale Res. Lett. 2018;13:141. doi: 10.1186/s11671-018-2532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo L. A. Zapata P. A. Vejar N. D. Azócar M. I. Gulppi M. A. Zhou X. Thompson G. E. Rabagliati F. M. Páez M. A. Mater. Sci. Eng. Carbon. 2014;40:24–31. doi: 10.1016/j.msec.2014.03.037. [DOI] [PubMed] [Google Scholar]
- Roy S. Mondal A. Yadav V. Sarkar A. Banerjee R. Sanpui P. Jaiswal A. ACS Appl. Bio Mater. 2019;2:2738–2755. doi: 10.1021/acsabm.9b00124. [DOI] [PubMed] [Google Scholar]
- Govindasamy G. A. Mydin R. B. S. M. N. Sreekantan S. Harun N. H. Sci. Rep. 2021;11:99. doi: 10.1038/s41598-020-79547-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applerot G. Lellouche J. Lipovsky A. Nitzan Y. Lubart R. Gedanken A. Banin E. Small. 2012;8:3326–3337. doi: 10.1002/smll.201200772. [DOI] [PubMed] [Google Scholar]
- Diez-Pascual A. M. Polymers. 2019;11:1790. [Google Scholar]
- Kalia A. Kaur M. Shami A. Jawandha S. K. Alghuthaymi M. A. Thakur A. Abd-Elsalam K. A. Biomolecules. 2021;11:224. doi: 10.3390/biom11020224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romay C. Gonzalez R. Ledon N. Remirez D. Rimbau V. Curr. Protein Pept. Sci. 2003;4:207–216. doi: 10.2174/1389203033487216. [DOI] [PubMed] [Google Scholar]
- Mat Yusuf S. N. A. Che Mood C. N. A. Ahmad N. H. Sandai D. Lee C. K. Lim V. R. Soc. Open Sci. 2020;7:200065. doi: 10.1098/rsos.200065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonsanti R. Milliron D. J. Chem. Mater. 2013;25:1305–1317. [Google Scholar]
- Polte J. CrystEngComm. 2015;17:6809–6830. [Google Scholar]
- Dunne P. W. Munn A. S. Starkey C. L. Huddle T. A. Lester E. H. Philos. Trans. R. Soc., A. 2015;373:20150015. doi: 10.1098/rsta.2015.0015. [DOI] [PubMed] [Google Scholar]
- Qin D. He Z. Li P. Zhang S. Front. Chem. 2022;10:1–11. doi: 10.3389/fchem.2022.834503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. Meng F. Li X. Wen J. Z. Li Z. Catal. Sci. Technol. 2016;6:7800–7811. [Google Scholar]
- Varma A. Mukasyan A. S. Rogachev A. S. Manukyan K. V. Chem. Rev. 2016;116:14493–14586. doi: 10.1021/acs.chemrev.6b00279. [DOI] [PubMed] [Google Scholar]
- Muthuvel A. Jothibas M. Manoharan C. Nanotechnol. Environ. Eng. 2020;5:14. [Google Scholar]
- Akbar N. Aslam Z. Siddiqui R. Shah M. R. Khan N. A. AMB Express. 2021;11:104. doi: 10.1186/s13568-021-01261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.